Abstract
Nonstructural protein 5B (NS5B) of hepatitis C virus (HCV) possesses an RNA-dependent RNA polymerase activity responsible for viral genome RNA replication. Despite several reports on the characterization of this essential viral enzyme, little is known about the reaction pathway of NS5B-catalyzed nucleotide incorporation due to the lack of a kinetic system offering efficient assembly of a catalytically competent polymerase/template/primer/nucleotide quaternary complex. In this report, specific template/primer requirements for efficient RNA synthesis by HCV NS5B were investigated. For intramolecular copy-back RNA synthesis, NS5B utilizes templates with an unstable stem-loop at the 3′ terminus which exists as a single-stranded molecule in solution. A template with a stable tetraloop at the 3′ terminus failed to support RNA synthesis by HCV NS5B. Based on these observations, a number of single-stranded RNA templates were synthesized and tested along with short RNA primers ranging from two to five nucleotides. It was found that HCV NS5B utilized di- or trinucleotides efficiently to initiate RNA replication. Furthermore, the polymerase, template, and primer assembled initiation-competent complexes at the 3′ terminus of the template RNA where the template and primer base paired within the active site cavity of the polymerase. The minimum length of the template is five nucleotides, consistent with a structural model of the NS5B/RNA complex in which a pentanucleotide single-stranded RNA template occupies a groove located along the fingers subdomain of the polymerase. This observation suggests that the initial docking of RNA on NS5B polymerase requires a single-stranded RNA molecule. A unique β-hairpin loop in the thumb subdomain may play an important role in properly positioning the single-stranded template for initiation of RNA synthesis. Identification of the template/primer requirements will facilitate the mechanistic characterization of HCV NS5B and its inhibitors.
Infection by hepatitis C virus (HCV) is a significant human medical problem. HCV is recognized as the causative agent for most cases of non-A and non-B hepatitis (8), with an estimated prevalence of 170 million cases (i.e., 2 to 3%) globally (36). Four million individuals may be infected in the United States alone. Upon first exposure to HCV only about 10% of infected individuals develop acute clinical hepatitis, while others appear to resolve the infection spontaneously. In the most instances, however, the virus establishes a chronic infection that persists for decades. This usually results in recurrent and progressively worsening liver inflammation, which often leads to more severe disease states such as cirrhosis and hepatocellular carcinoma (31, 32). Currently, there are no broadly effective treatments for the debilitating progression of chronic HCV.
HCV is an enveloped positive-stranded RNA virus belonging to the Flaviviridae family (24). The HCV genome encodes a polyprotein of 3,010 to 3,033 amino acids (30). The nonstructural (NS) proteins and the catalytic machinery for viral replication are derived by proteolytic cleavage of this polyprotein. One of the NS proteins is NS5B, which has been shown to be an RNA-dependent RNA polymerase (RdRp) responsible for HCV genome replication (5, 10, 12, 18, 28, 37). Although the exact mechanism of HCV replication remains unclear, evidence suggests that de novo initiation is likely to play a critical role in HCV replication in vivo (21, 28, 33, 38). Recent studies revealed several unique structural features of HCV NS5B which might help to advance our understanding of viral replication at the molecular and structural level (1, 6, 17).
A comprehensive understanding of the differences between HCV and cellular polymerases will facilitate the design of specific inhibitors of HCV replication. Detailed kinetic information is expected to play an important role in understanding the molecular basis of HCV NS5B-catalyzed nucleotide incorporation and subsequently the mechanistic characterization of the inhibitors. Currently, such information is very limited due to the lack of suitable RNA template and primer pairs, which can assemble properly with the enzyme and permit efficient nucleotide incorporation to be followed by extension of end-labeled primers in vitro. Previous studies (5, 9, 10, 18, 20) provided little information with regard to the proportion of the polymerase-RNA complexes that are competent for catalysis. Furthermore, these reports did not demonstrate the stoichiometric assembly of enzyme and template/primer, which is required for accurate measurement of the elementary steps of the polymerase reaction.
In this report, the template and primer requirements for HCV NS5B-directed RNA replication were investigated. We found that templates with 3′ termini free of secondary structures and short primers 2 or 3 nucleotides (nt) long were preferred for efficient initiation of RNA synthesis as detected by extension of the radiolabeled primers. This finding is consistent with a structural model of NS5B complexed with the template/primer/nucleotide substrates. Future advances in our understanding of the structure as well as the kinetics of NS5B should aid in the development of highly specific and potent inhibitors of viral RdRp as the anti-HCV agents.
MATERIALS AND METHODS
Protein expression and purification.
DNA sequences encoding HCV (BK), bovine viral diarrhea virus (BVDV; NADL), and poliovirus (PV; Mahoney) RdRp proteins were cloned in bacterial expression vectors (pET-22b or pET28a). To improve solubility of HCV and BVDV NS5B proteins, their C-terminal hydrophobic regions, consisting of 21 and 24 amino acids, respectively, were removed (10, 16). Additional sequences (coding for a methionine at the N terminus and a polyhistidine tag, GSHHHHHH, at the C terminus) were engineered to facilitate the cloning, expression, and purification. Protein production was induced in freshly transformed Escherichia coli JM109(DE3) cells (optical density of 0.6) by isopropylthio-β-d-galactoside at a final concentration of 0.2 mM. Soluble cell lysates were batch adsorbed onto a nickel-chelated (Ni-nitrolotriacetic acid) resin. After 10 column volumes of wash at 1 M sodium chloride, the protein was eluted from the column with a buffer containing 0.3 M imidazole. The protein was further purified as described previously (17).
Copy-back RNA replication assay.
Synthetic RNAs were used as the templates in the copy-back RNA replication assay. Standard reactions for HCV NS5B (in a volume of 40 μl) contained 20 mM HEPES (pH 7.3), 7.5 mM dithiothreitol, 50 mM NaCl, 5 mM MgCl2, 0.05% glycerol, 0.2 to 0.5 μM RNA template, 100 μM UTP and CTP, 10 μCi of [α-33P]UTP label, and 300 ng of HCV NS5B. Reaction conditions for BVDV NS5B and PV 3Dpol are as described previously (16, 29). All reactions were performed at 30°C for 30 min and terminated by phenol-chloroform extraction. Labeled product was precipitated with ethanol in the presence of glycogen as carrier. The pellet was dissolved in diethylpyrocarbonate-treated water and resolved in a 15% polyacrylamide–urea–Tris-borate-EDTA (TBE) gel (Novex, San Diego, Calif.) according to the manufacturer's instructions. After electrophoresis, the gel was fixed, vacuum dried, and subjected to autoradiography.
End labeling of RNA template and primer.
Synthetic RNA templates and primers were chemically synthesized (Oligos, Etc., Wilsonville, Oreg.). The RNAs were all gel purified except for those shorter than 7 nt (about 90% pure for the latter RNAs). End labeling of RNA template or primer was performed using T4 polynucleotide kinase and [γ-33P]ATP. Labeling reactions were carried out at 37°C for 30 min in a 50-μl volume containing 50 pmol of RNA, 100 μCi of [γ-33P]ATP, and 10 U of T4 polynucleotide kinase. After labeling, the reaction mixture was extracted with phenol-chloroform and precipitated with ethanol in the presence of glycogen as carrier. The labeled RNA was dissolved in diethylpyrocarbonate-treated water to required concentrations. Unlabeled RNA template or primer with a higher concentration was supplemented to constitute a working stock with an accurate concentration.
Nucleotide incorporation assay using end-labeled primers.
The standard nucleotide incorporation reaction in a volume of 20 μl contained 50 mM HEPES (pH 7.3), 10 mM β-mercaptoethanol, 50 mM NaCl, 5 mM MgCl2, 5 μM template RNA, 10 μM end-labeled primer, 5 μM polymerase protein, and 100 μM nucleoside triphosphate (NTP) substrate as indicated. The reaction mixture was incubated at 30°C for 30 min, followed by phenol-chloroform extraction and ethanol precipitation in the presence of glycogen carrier. The product was dissolved in urea-TBE sample buffer (Novex) and separated in a 25% acrylamide–1.7% N,N′-methylenebisacrylamide–6 M urea–1× TBE gel. The gel was electrophoresed at 25 to 50 W until the bromophenol blue dye reached the bottom. The gel was exposed to X-ray film, and the RNA bands were quantified by a PhosphorImager.
RESULTS
Several enzymatic assays for HCV NS5B based on the use of viral RNA or homopolymeric RNA as templates have been described (5, 9, 10, 12, 18, 20, 28). These assays measured the cumulative incorporation of nucleotides and the average steady-state catalytic activity of the polymerase. For the most part, however, these assays did not allow characterization of a single polymerization reaction, i.e., a single nucleotidyl transfer reaction. More importantly, these studies were unable to determine the proportion of enzyme and RNA substrate that were engaged in productive binding to form complexes competent for catalysis. To address these issues, we focused on developing short and well-defined synthetic RNA templates in an effort to identify the specific requirements for efficient assembly of various catalytic components (enzyme, template/primer, and nucleotide) involved in HCV NS5B-directed RNA replication.
Copy-back RNA synthesis using a template with a stable tetraloop near the 3′ terminus.
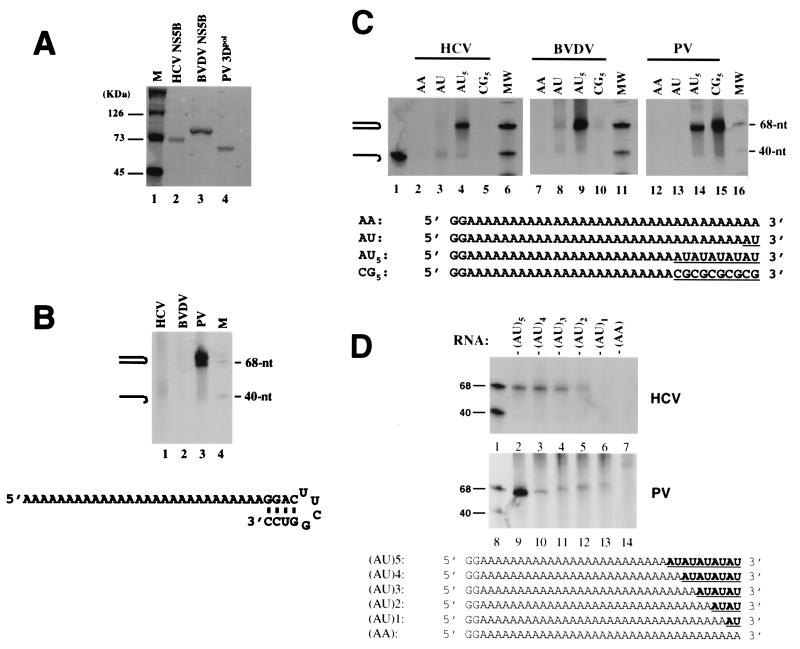
Based on previous reports, viral RdRps, including HCV NS5B, were capable of using RNA templates that folded back intramolecularly at the 3′ terminus to produce a near-dimer-size hairpin product. To test whether HCV NS5B can use small synthetic RNA of defined sequence as the template for copy-back RNA synthesis, a 40-nt RNA (5′-A28GGACUUCGGUCC-3′) which forms a stable tetraloop (3, 25) near the 3′ terminus (base paired as shown in Fig. 1B) was synthesized. This tetraloop has a calculated melting temperature of about 71°C (3). For comparison, RdRps from (3Dpol) and BVDV (NS5B) were also produced and tested in parallel with HCV NS5B (Fig. 1A, lanes 2 to 4). As shown in Fig. 1B, only PV 3Dpol was able to use the tetraloop efficiently as a copy-back substrate and produced a near-dimer-size hairpin product (lane 3). In contrast, little activity was detected for HCV and BVDV NS5B (Fig. 1B, lanes 1 and 2, respectively). The lack of product formation by HCV and BVDV NS5B was not due to insufficient amounts of enzyme used in the reactions because the RdRp activity for each polymerase was normalized by using a standard scintillation proximity assay (data not shown) (10). These results suggest that flavivirus RdRps might require different features at the 3′ terminus of the template for copy-back RNA replication.
FIG. 1.
Template preference for copy-back RNA replication directed by HCV NS5B. (A) Expression and purification of the polymerase proteins of HCV, BVDV, and PV. Approximately 1 μg of each purified protein was analyzed on a sodium dodecyl sulfate-polyacrylamide gel and visualized by Coomassie blue staining. (B) Copy-back RNA synthesis using an RNA with a stable tetraloop at the 3′ terminus. Lane 1, HCV NS5B; lane 2, BVDV NS5B; lane 3, PV 3Dpol; lane 4, size markers. Positions of the template and product are indicated. (C) Correlation between stem-loop stability and copy-back activity. Four synthetic RNAs (AA, AU, AU5, and CG5) with different 3′-terminal sequences were tested for copy-back RNA replication by HCV (lanes 2 to 5), BVDV (lanes 7 to 10), and PV RdRp (lanes 12 to 15). Lane 1, end-labeled template-length RNA marker; lanes 6, 11, and 16, size markers. (D) Correlation between the stem-loop size and copy-back activity. Lanes 1 and 8, end-labeled template-length RNA marker; lanes 2 to 7, RNA synthesis from HCV NS5B; lanes 9 to 14, RNA synthesis from PV 3Dpol.
HCV NS5B utilized RNA with an unstable stem-loop at the 3′ terminus.
To identify the specific template requirement for copy-back RNA synthesis by HCV NS5B, four synthetic RNAs with different 3′-terminal sequences were tested (Fig. 1C). Two of these templates, AA and AU, were unable to form stem-loops, whereas the other two had the ability to form either a weak stem-loop (AU5) or a relatively stable stem-loop (CG5, with a calculated melting temperature of approximately 53°C) at the 3′ terminus. The results shown in Fig. 1C demonstrated that template AA failed to yield any products by all three viral polymerases (lanes 2, 7, and 12), indicating a lack of terminal transferase activity in these polymerases. De novo RNA synthesis from the AA template was not observed since UTP was not a preferred initiating nucleotide (21, 38). The weak RNA synthesis detected for BVDV (lane 8) and HCV NS5B (lane 3) with the template AU probably resulted from the remote base pairing between the terminal uridylate and an internal adenylate or from inefficient elongation. In the case of template AU5, all three viral RdRps exhibited copy-back activity and produced the near-dimer-size hairpin products (lanes 4, 9, and 14), indicating that the alternating A-U pairs at the 3′ terminus were able to support copy-back RNA synthesis. Replacement of the alternating A-U pairs with C-G pairs significantly reduced the RdRp activity for both HCV and BVDV NS5B (lanes 5 versus 4 and 10 versus 9). This observation suggests that a more stable copy-back primer at the 3′ terminus (formed by the stronger base pairing between the C and G bases) was detrimental to the copy-back RNA synthesis directed by flavivirus RdRps. In contrast, the more stable copy-back primer in template CG5 enhanced the activity of PV 3Dpol (Fig. 1C; lane 15 versus 14), consistent with the observation (Fig. 1B) that PV 3Dpol preferred a more stable stem-loop as the copy-back primer. The size of the 3′ stem-loop was next varied by shortening the number of alternating A-U pairs from five to one while maintaining a constant 40-mer RNA template (Fig. 1D). HCV NS5B required a minimum of three A-U pairs for RNA synthesis (Fig. 1D, lanes 2 to 4), while PV 3Dpol preferred five A-U pairs for optimal RdRp activity (Fig. 1D, lane 9). Furthermore, HCV NS5B produced RNA hairpin products of similar size, while PV 3Dpol generated RNA hairpins of increasing lengths from templates with A-U stem-loops of decreasing size (Fig. 1D, compare lanes 9 to 13). This suggests that HCV NS5B can accommodate a smaller and constant size of stem-loop formed by three A-U pairs, while PV 3Dpol prefers to accommodate a larger and more stable stem-loop. The above results indicate that HCV NS5B polymerase requires a template which has a very weak or no stem-loop structure at the 3′ terminus in solution but has the ability to fold back intramolecularly upon binding to the polymerase for copy-back RNA synthesis.
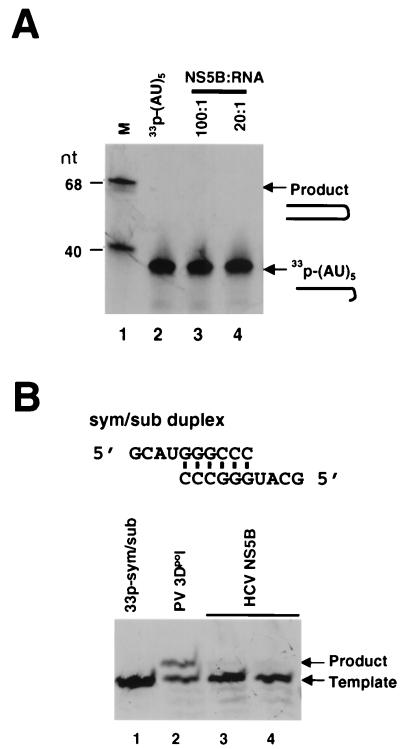
The recently published crystal structures of HCV NS5B demonstrated that this enzyme adopts a unique overall structure with a fully encircled active site cavity, rather than the more open U-shaped structure like that of human immunodeficiency virus type 1 (HIV-1) reverse transcriptase (RT) and other polymerases (1, 6, 17). In addition, a unique β-hairpin structure (formed within amino acids 441 to 456) in the thumb subdomain of HCV NS5B is positioned near the polymerase active site (6, 17). This β-hairpin, which is absent in both PV 3Dpol and HIV-1 RT, may sterically prevent the NS5B apoenzyme from accommodating efficiently double-stranded RNA (dsRNA) duplexes or RNA with a stable secondary structure at the 3′ terminus (17). This notion was further supported by the observation that a dsRNA molecule known as sym/sub, which can be utilized efficiently by PV 3Dpol (4), failed to support nucleotide incorporation by HCV NS5B (Fig. 2B). These results, taken collectively, suggest that a preannealed duplex RNA or any RNA with a stable stem-loop at the 3′ end will not be preferred by HCV NS5B for initiation of RNA synthesis, probably due to the low efficiency of the apoenzyme to accommodate dsRNA molecules. This β hairpin makes few contact with the body of the protein and thus has some intrinsic flexibility (17). Occasionally, at a very low frequency, the β hairpin may undergo a conformational change and move away from the active site, allowing the HCV polymerase to accommodate a dsRNA or a 3′ stem-loop and resulting in the observed dsRNA-mediated or copy-back RNA synthesis (5, 18, 35).
FIG. 2.
Determination of template utilization by HCV NS5B. (A) For AU5 copy-back RNA, each reaction contained 0.2 μM end-labeled template, 100 μM UTP and CTP, and HCV NS5B protein as indicated. Lane 2, no enzyme; lane 3, with 4 μM HCV NS5B; lane 4, with 20 μM HCV NS5B. Lane 1 contained size markers. Reaction products were separated on a 15% polyacrylamide-urea-TBE gel (Novex). (B) For preannealed sym/sub RNA, each reaction contained 0.5 μM end-labeled sym/sub RNA, 100 μM ATP, and 5 μM HCV or PV polymerase protein. Lane 1, no enzyme; lane 2, with PV 3Dpol; lanes 3 and 4, with HCV NS5B. Reaction products were resolved on a 23 to 25% polyacrylamide-urea-TBE gel.
Template utilization by HCV NS5B.
Efficient assembly of polymerase-RNA template/primer complexes that permit nucleotide incorporation is an essential step prior to the determination of kinetics of HCV NS5B-catalyzed RNA synthesis. The experiments described earlier (Fig. 1) were performed in the presence of unlabeled template RNA and radiolabeled NTP substrate ([α-33P]UTP). Thus, the polymerase activity was measured based on the incorporation of radiolabeled UMP into the nascent RNA product. However, this type of assay did not reveal the proportion of RNA substrates bound productively by the enzyme to form catalytically competent complexes. To address this issue, radiolabeled RNA templates (in the case of copy-back RNA synthesis) or primers (in the case of bimolecular primer-dependent RNA synthesis) were used so that the proportion of the radiolabeled RNA substrates that were extended due to incorporation of unlabeled nucleotides could be easily determined. As shown in Fig. 2A, when the end-labeled template AU5 and unlabeled NTPs were tested, the amount of the template RNA which was extended by HCV NS5B was too low to be detected, even when up to 100 molar excess HCV NS5B was used (Fig. 2A, lanes 3 and 4). As a control, the sym/sub RNA, which is an efficient template/primer pair for PV 3Dpol (4), was also tested. PV 3Dpol efficiently utilized this duplex RNA and catalyzed template-dependent single-nucleotide incorporation (Fig. 2B, lane 2). However, little primer extension was observed in reactions containing HCV NS5B (Fig. 2B, lanes 3 and 4). These results revealed that only a negligible amount of the template and primer was assembled correctly in the active site of HCV NS5B (i.e., forming the enzyme-substrate complexes competent for catalysis). As a result, the products generated in these reactions were detectable only by incorporation of radiolabeled nucleotides (Fig. 1). This low efficiency in proper assembly of the template/primer in the active site of HCV NS5B prevented quantitative measurement of single-nucleotide incorporation, which is required for kinetic analysis of NS5B-catalyzed polymerization reaction.
Initiation of RNA synthesis using a dinucleotide primer.
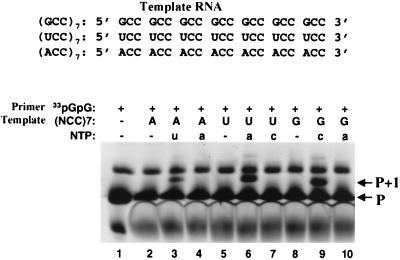
Previous results revealed that a stable stem-loop at the 3′ terminus of the RNA template or a preannealed RNA duplex prevented productive binding or proper entry of the RNA into the polymerase active site. The inability of HCV NS5B to utilize these substrates efficiently supports the observation that the unique β hairpin interferes with the proper binding of dsRNA molecules to the NS5B apoenzyme in the absence of any conformational changes (6, 17). We predicted therefore that the correct docking of RNA to HCV NS5B required a single-stranded 3′ terminus of the template RNA. To test this hypothesis, we designed and evaluated a series of templates and short primers. These short RNA primers will not form stable duplexes with the template RNA in solution and will therefore not interfere with the template docking to the active site of NS5B. Thus, these short primers are conceptually different from the traditional polymerase primers which are preannealed to the templates and form duplex RNAs prior to binding to the polymerases. Rather, they behave more like the initiating nucleotide for de novo synthesis and access the active site independently to prime the RNA synthesis against the bound single-stranded template RNA. Three 21-nt RNAs consisting of trinucleotide repeats [(ACC)7, (UCC)7, or (GCC)7] were tested for the ability to direct RNA synthesis using the end-labeled diguanylate (33pGpG) (Fig. 3). These templates were designed to (i) avoid formation of stable secondary structure, (ii) maximize the base-pairing capability with the pGpG dinucleotide, and (iii) allow detection of single-nucleotide incorporation [UMP, AMP, and CMP incorporation for (ACC)7, (UCC)7, and (GCC)7, respectively]. Each reaction contained either no NTP (lanes 2, 5, and 8) or the indicated NTP substrate (lanes 3, 4, 6, 7, 9, and 10). Single-nucleotide incorporation was monitored by the migration shift of the radiolabeled primers (from P to P+1). In the case of (ACC)7, single-nucleotide incorporation was observed only when UTP was used as the nucleotide substrate (Fig. 3, lane 3; compare with lane 4). Similar results were observed for template (UCC)7 or (GCC)7, in which only the correct nucleotide, ATP or CTP, resulted in extension of the pGpG dinucleotide (lanes 6 and 9). The upper band present in all lanes was a contaminant of the pGpG preparation. These results demonstrated that HCV NS5B was able to utilize a significant portion of the RNA template/primer and catalyze the template-dependent nucleotide incorporation. Under the reaction conditions used approximately 20 to 30% of the labeled dinucleotides were utilized by the polymerase, a level comparable to the extent of sym/sub RNA utilization by PV 3Dpol under similar reaction conditions (Fig. 2B, lane 2).
FIG. 3.
Nucleotide incorporation using radiolabeled diguanylate (33pGpG) as the primer. Three RNA templates, (ACC)7, (UCC)7, and (GCC)7, were tested for nucleotide incorporation using 33pGpG as the primer. Each reaction contained 5 μM RNA, 5 μM 33pGpG, 100 μM NTP as indicated, and 3 μM HCV NS5B. Lane 1, 33pGpG primer alone; lanes 2 to 4, with (ACC)7 RNA as the template plus either no NTP (lane 2), 100 μM UTP (lane 3), or 100 μM ATP (lane 4); lanes 5 to 7, with (UCC)7 RNA as the template plus either no NTP (lane 5), 100 μM ATP (lane 6), or 100 μM CTP (lane 7); lanes 8 to 10, with (GCC)7 RNA as the template plus either no NTP (lane 8), 100 μM CTP (lane 9), or 100 μM ATP (lane 10). The samples were resolved on a 25% polyacrylamide-urea-TBE gel.
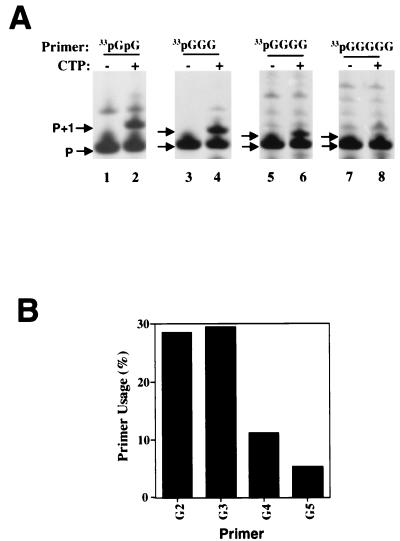
Optimal length of the initiating nucleotides.
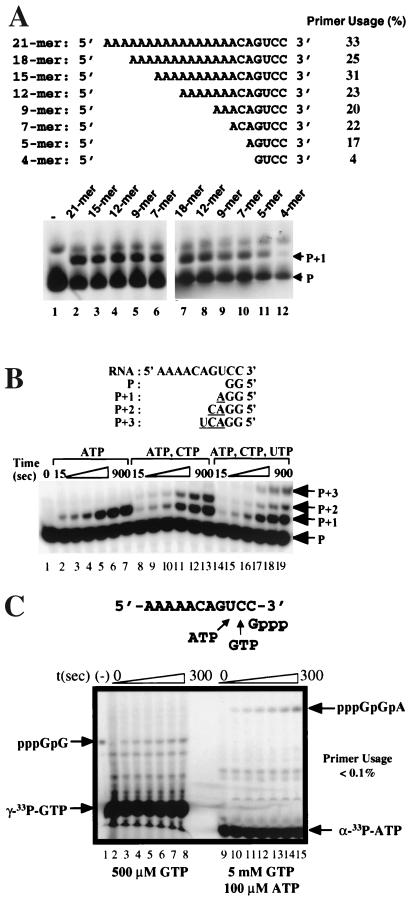
To determine the optimal length of the guanylate primer that allows efficient nucleotide incorporation, we synthesized four template and primer pairs [(GCC)7/33pGpG, (GCCC)6/33pGpGpG, (GCCCC)5/33pGGGG, and (GCCCCC)5/33pGGGGG)] in which the length of the primer, as well as the complementary sequence in the template RNA, varied from 2 to 5 nt. Di- and triguanylate primers supported nucleotide incorporation with the highest efficiency (Fig. 4A, lanes 2 and 4; Fig. 4B). Further increases in primer length to 4 and 5 nt were detrimental to the activity (Fig. 4A, lanes 6 and 8; Fig. 4B). We thus chose to use the diguanylate primer to further characterize this system.
FIG. 4.
Efficiency of guanylate primers of different length in priming nucleotide incorporation. (A) Gel-based analysis. Lanes 1, 3, 5, and 7, no nucleotide substrate; lanes 2, 4, 6, and 8, with 100 μM of CTP. (B) Efficiency of single-nucleotide (CMP) incorporation quantified with a PhosphorImager. The percentage of product (P+1) formation was calculated from the amount of P+1 divided by the total amount of labeled primers including P and P+1. (C) RNA synthesis from a single initiating nucleotide.
The primer base pairs with the 3′ terminus of the template.
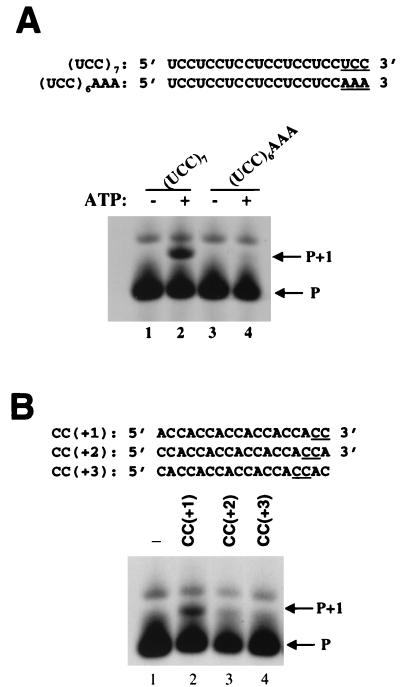
The RNA templates tested earlier (Fig. 3) could base pair with the diguanylate primer at either the 3′ terminus or an internal position. To determine whether the polymerase initiated RNA synthesis from the 3′ terminus or from an internal location, we designed a new RNA template in which the terminal sequence of template (UCC)7 was modified from UCC to AAA (Fig. 5A). This modification rendered the RNA incapable of base pairing with pGpG at the terminal bases. When this RNA was tested, little nucleotide incorporation was detected (Fig. 5A, compare lanes 4 and 2), indicating that the primer base pairs with the 3′ terminus of the template RNA to initiate RNA synthesis.
FIG. 5.
The primer base pairs with the 3′ terminus of the template RNA. (A) (UCC)6AAA, identical to (UCC)7 except for the last three bases, was tested for directing nucleotide incorporation using 10 μM 33pGpG as the primer. Reactions contained 5 μM (UCC)7 alone (lane 1), (UCC)7 plus 100 μM of ATP (lane 2), (UCC)6AAA alone (lane 3), or (UCC)6AAA plus 100 μM ATP. (B) CC(+1), CC(+2), and CC(+3) RNA were tested for directing nucleotide incorporation. Lane 1, 33pGpG primer alone; lane 2, 33pGpG plus CC(+1) and 100 μM of UTP; lane 3, 33pGpG plus CC(+2) and 100 μM of UTP; lane 4, 33pGpG plus CC(+3) and 100 μM of UTP.
The sequence of the template RNA was further modified such that one or two additional nucleotides were added to the 3′-terminal dicytidylates. As a result, the template bases CC were no longer terminally located. Accordingly, one or two residues were removed from the 5′ terminus to retain the same template length (Fig. 5B). When these newly modified RNA templates, CC(+2) and CC(+3), were tested, a significant reduction in activity was observed (Fig. 5B, compare lanes 3 and 4 with lane 2), confirming that the dinucleotide base pairs with the template only at the 3′ terminus during assembly of catalytically competent complexes with the enzyme.
Minimal template length requirement for RNA synthesis.
As demonstrated earlier, the initiating dinucleotide base paired with the 3′ terminus of the template RNA for nucleotide incorporation (Fig. 5). What is not clear is the minimal length of the template required for this interaction. To address this issue experimentally, we designed a series of synthetic RNA templates consisting of two cytidylates at the 3′ terminus (for base pairing with the 33pGpG dinucleotide), a heterogeneous sequence (CAGU) in the middle, and a stretch of adenylates of various lengths at the 5′ end (Fig. 6A). These templates ranged from 4 to 21 nt in length. Efficient nucleotide incorporation was observed with the template as short as 5 nt (Fig. 6A, lanes 2 to 11). However, when the template was shortened to 4 nt, a significant reduction in nucleotide incorporation was observed (Fig. 6A, lane 12). These results demonstrated that the minimal length of the template RNA to retain sufficient template activity was approximately 5 nt. Shorter templates might lack sufficient interaction with the enzyme to support initiation of RNA synthesis. This is consistent with the model of enzyme-RNA interaction in which at least 5 nt are in direct contact with polymerase (6).
FIG. 6.
(A) Minimum length requirement for the template RNA. A number of RNA templates of decreasing length (from 21 to 4 nt) were tested for the ability to direct single-nucleotide incorporation using 33pGpG as the primer. All reactions contained 100 μM ATP. Lane 1, no RNA; lanes 2 to 12, template RNA as indicated. Efficiencies of primer utilization, quantified with a PhosphorImager, are listed at the right. (B) Multiple cycles of nucleotide incorporation. A 10-mer RNA was used for multiple rounds of nucleotide incorporation with 33pGpG as the primer (P). Reaction contained either 100 μM ATP alone (lanes 2 to 7), 100 μM each ATP and CTP (lanes 8 to 13), or 100 μM each ATP, CTP, and GTP (lanes 14 to 19). The reactions were quenched at different time points from 15 to 900 s. (C) RNA synthesis from a single initiating nucleotide. The template used is 5′-AAAAACAGUCC-3′. Two reactions were performed: one with 5 μM HCV NS5B, 5 μM template RNA, and 500 μM (10 μCi) [γ-33P]GTP (lanes 2 to 8); the other with 5 μM HCV NS5B, 5 μM template RNA, 5 mM GTP, and 100 μM (10 μCi) [α-33P]ATP (lanes 9 to 15). The reactions were quenched at various time points from 0 to 300 s. Lane 1 contains γ-33P-labeled pppGpG as a size marker. The labeled nucleotides were spiked into large pools of unlabeled nucleotides to ensure constant final concentrations.
We next tested whether this type of RNA template/dinucleotide pair could support multiple cycles of nucleotide incorporation in a time-dependent manner. In the presence of appropriate NTP substrates, the labeled dinucleotide was extended to P+1 (lanes 2 to 7), P+2 (lanes 8 to 13), and P+3 (lanes 14 to 19). Interestingly, significant accumulation of P+1 products was observed in all cases, suggesting that dissociation of the NS5B-RNA complexes may occur after incorporation of the first nucleotide. In contrast, the formation of P+3 products paralleled that of P+2 products (lanes 17 to 19), indicating that the incorporation of the third nucleotide from P+2 to P+3 may be faster, possibly due to a more stable NS5B-RNA complex.
We further determined the efficiency of RNA synthesis from a single initiating nucleotide, GTP. Two RNA products synthesized from the template RNA (5′-AAAAACGUCC-3′) were quantified: pppGpG in the presence of 500 μM [γ-33P]GTP or pppGpGpA in the presence of 5 mM GTP and 100 μM [α-33P]ATP (Fig. 6C). In both cases, the primer usage was below 0.1%, more than 100-fold less efficient than the dinucleotide-initiated RNA synthesis (20 to 30% primer usage [Fig. 6B, lanes 5 to 7; Fig. 4]). This suggests that the first nucleotidyl transfer to the initiating nucleotide is rate limiting, while the dinucleotide, an elongative product of the initiating nucleotide, is much more efficient in priming RNA synthesis. Similar observations were reported for T7 RNA polymerase-catalyzed de novo RNA transcription (13, 26).
Use of dinucleotides other than pGpG to initiate RNA synthesis.
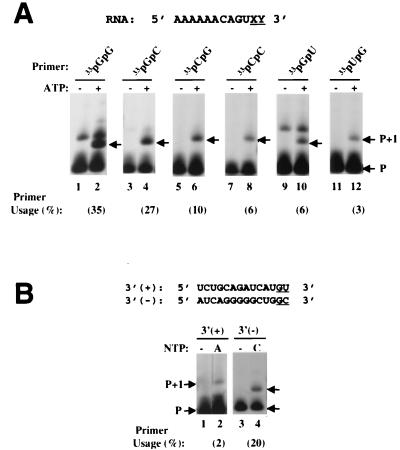
The above experiments were performed by using 33pGpG to initiate synthesis. To determine whether this could apply to other dinucleotides, a series of 12-nt templates with variations at the 3′-terminal positions (5′-AAAAAACAGUXY-3′) were tested along with the matching dinucleotides (pGpG, pGpC, pCpG, pCpC, pGpU, or pUpG). As shown in Fig. 7A, all of the dinucleotides were capable of priming nucleotide (AMP) incorporation (lanes 2, 4, 6, 8, 10, and 12). The relative order of nucleotide incorporation efficiency was pGpG ∼ pGpC > pCpG > pCpC ∼ pGpU > pUpG (Fig. 7A). HCV NS5B utilized dinucleotides with a guanylate at the 5′ terminus more readily than other bases. Whether this has any correlation with the previous reports that HCV NS5B prefers GTP as the initiation nucleotide during de novo initiation of RNA synthesis remains to be addressed (21, 38).
FIG. 7.
(A) Use of various dinucleotide primers for RNA synthesis. End-labeled dinucleotides 33pGpG (lanes 1 and 2), 33pGpC (lanes 3 and 4), 33pCpG (lanes 5 and 6), 33pCpC (lanes 7 and 8), 33pGpU (lanes 9 and 10), and 33pUpG (lanes 11 and 12) were paired with appropriate templates in the absence (lanes with odd numbers) or presence (lanes with even numbers) of 100 μM ATP substrate. Efficiencies of primer utilization, quantified with a PhosphorImager, are listed below. (B) Nucleotide incorporation using HCV 3′-terminal sequences as the template. RNA templates 3′(+) and 3′(−) were derived as described in Results. 33pApC and 33pGpC were used as the primers for 3′(+) and 3′(−) RNAs, respectively. Lanes 1 and 3 are reactions in the absence of nucleotide substrate, and lanes 2 and 4 are reactions in the presence of ATP (lane 2) or CTP (lane 4).
Last, we tested whether HCV-derived RNAs could serve as the templates for dinucleotide-initiated RNA synthesis. For this purpose, 3′(+) and 3′(−) RNAs, corresponding to the 3′ termini of HCV positive-strand and negative-strand RNA genomes, were synthesized (Fig. 7B). When these RNAs were tested in the presence of appropriate end-labeled dinucleotide and NTP substrate [33pApC and ATP for 3′(+); 33pGpC and CTP for 3′(−)], single-nucleotide incorporation was detected for both RNA templates, though with different efficiencies. 3′(−) RNA was used about 10-fold more readily than 3′(+) RNA (Fig. 7B, compare lanes 4 and 2). The observation that pGpC initiated RNA synthesis from 3′(−) RNA more efficiently than pApC from 3′(+) RNA was in good agreement with recent results showing that positive-strand RNA is in about 10-fold excess over the negative-strand RNA in the HCV RNA replicon cell lines (19).
DISCUSSION
Several reports have characterized the enzymatic activity of HCV NS5B in various degrees of detail (2, 5, 9, 10, 12, 18, 20, 28). However, further characterization of the reaction pathway of NS5B-catalyzed nucleotide incorporation has been hindered in part due to the lack of template/primer pairs capable of efficiently assembling catalytically competent complexes with the enzyme. In this work, we demonstrate that stable, preannealed dsRNAs are poor substrates for HCV NS5B. Instead, the HCV polymerase utilizes more efficiently short oligonucleotides, 2 or 3 nt in length, to prime nucleotide incorporation which can be followed by extension of radiolabeled RNAs. We further demonstrate that initiation of RNA synthesis preferentially occurs from the 3′ terminus of the template RNA, suggesting that the replicase assembles at the 3′ terminus of viral RNA. Consistent with this possibility was the finding that 3′ termini of both HCV positive-strand and negative-strand RNAs can serve as templates for dinucleotide-initiated RNA synthesis.
Recent structural studies revealed that HCV NS5B has a fully encircled active site with a relatively rigid interdomain structure, resembling the nucleic acid-bound conformation of several other polymerases (1, 6, 17). The encircled overall structure of this enzyme is the result of the extensive interactions between the fingers and thumb subdomains which are likely to be unique to viral RdRps. In addition, an HCV-specific β-hairpin structure located in the thumb subdomain, absent in PV 3Dpol and HIV RT, protrudes toward the active site and may impose a steric barrier to prevent binding to dsRNA molecules (6, 17). A highly conserved RNA binding groove bordered by the fingers subdomain and the interdomain loops provides a positively charged molecular surface to be occupied by the 5′ overhang of the template (6, 17). Upon template/primer binding, NS5B is expected to undergo local conformational changes including those proposed for the β hairpin and the thumb subdomain (6) (Fig. 8). No large-scale domain movements, such as those observed in other polymerases upon nucleic acid binding, are expected. At present, it is not clear how such conformational changes, in particular those required for accommodation of the nascent double-stranded RNA, can be induced.
FIG. 8.
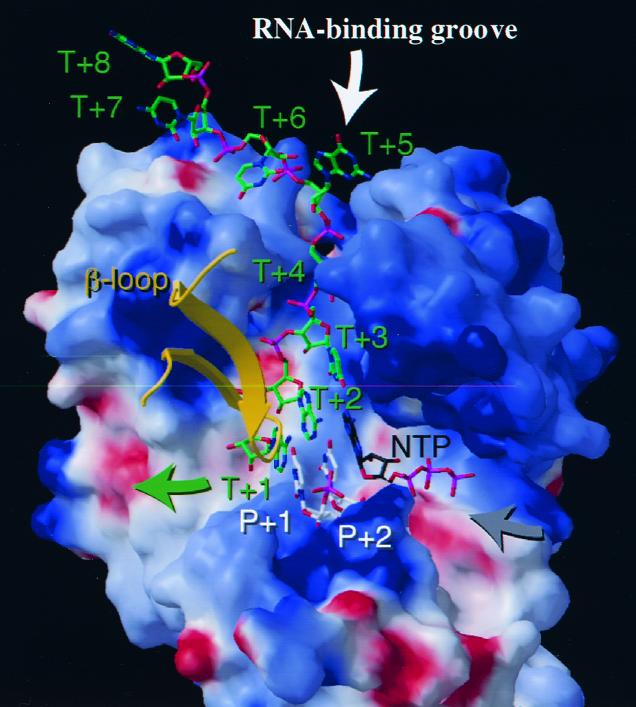
Quaternary complex model for HCV NS5B. The crystal structure containing HIV-1 RT, template/primer DNA pair, and an incoming dNTP (PDB code 1RTD) (11) was used to guide the construction of a model for the analogous complex of HCV NS5B with RNAs and an incoming NTP. This hypothetical model is based on active site superposition between the two polymerases (17). Atoms of RNA are shown and denoted T+1 through T+8 on the template strand and P+1 to P+2 on the primer strand. P+1 and P+2 base pair with T+1 and T+2, respectively. The incoming NTP base pairs with T+3. The HCV NS5B protein is depicted by a molecular surface colored by local electrostatic potential from red to blue as the potential ranges from negative to positive (27). The thumb subdomain is omitted for clarity. The β hairpin containing residues 441 to 456 is shown in yellow. Using NS5B as a fixed frame of reference, the directions of motion of the nucleic acid and the NTP are indicated by arrows. The figure was produced using Molscript and Raster3D (15, 23).
In the absence of a liganded structure, a model for the HCV NS5B quarternary complex was created (Fig. 8), based on the complex structure of HIV-1 RT containing the enzyme, template/primer DNA pair, and an incoming dNTP (11). A single-stranded RNA template and dinucleotide primer were modeled into the complex with few structural overlaps. The single-stranded template occupies the RNA binding groove with its 3′ terminus stacked against a tyrosine residue (Y448) at the tip of the β hairpin. The template region in direct contact with the RNA binding groove consists of 5 nt (T+1 to T+5), which is consistent with our observation that the minimal template length is 5 nt (Fig. 6A). We propose that the unique β hairpin in the thumb subdomain may play an important role in positioning the 3′ terminus of the template for proper initiation of RNA synthesis. The side chain of the strictly conserved isoleucine residue (I160) in motif F (17) packed against the T+3 template base and stabilized the base pairing between T+3 and the incoming NTP. Within the space above the active site at the base of the palm subdomain, the dinucleotide primer (P+1 to P+2) forms a short duplex with the terminal template bases (T+1 and T+2). The 5′ phosphate group of the dinucleotide is in close proximity of motif E and a highly conserved arginine (R386) which may play a role in stabilizing the short initiating nucleotide primer. The size of the NS5B active site in this model can accommodate only a short duplex RNA with up to 3 bp, including the one between the incoming nucleotide and the T+3 base. This structural feature is reminiscent of the quaternary complex structure of the bacteriophage T7 RNA polymerase (7) in which a trinucleotide primer base pairs with the single-stranded template DNA and the incoming NTP base pairs with the T+4 template base. Based on the HCV NS5B model, a second nucleotide incorporation (base pairing with the T+4 base) would require that the template translocate towards the β hairpin so that the T+4 ribose will be within the distance to the active site for catalysis to occur. How this template translocation is accomplished requires additional studies. However, this may explain our hypothesis of a rapid dissociation after incorporation of the first nucleotide (Fig. 6B), perhaps a result of steric hindrance imposed by the β hairpin toward the passage of the duplexed template/primer beyond the β-hairpin structure.
Based on the unliganded NS5B structure, the β-hairpin loop is located within a space similar to that of the N-terminal domain of T7 RNA polymerase (6, 7). In the quaternary complex of T7 RNA polymerase, this N-terminal domain functions as a wedge to separate the nascent RNA strand from the DNA template (7). The β hairpin in NS5B may serve a similar function and will be the focus of future studies.
So far, two forms of in vitro RNA synthesis activities have been demonstrated for HCV NS5B. One is from a preannealed primer (2, 5, 9, 10, 12, 18, 20); the other initiates de novo (21, 28, 33, 38). It is generally believed that de novo initiation is the mode of HCV RNA replication in vivo since this mode ensures the faithful replication of the entire viral genome by initiating RNA synthesis from the exact 3′ terminus of the template RNA. However, it is not clear what mechanisms are involved in the initial priming steps of the replication process. We propose the following model for initiation of HCV RNA replication: HCV NS5B binds viral RNA containing a 3′ single-stranded overhang free of secondary structures (Fig. 8). Although this model appears to contradict the prediction that the 3′ terminus of the HCV genome is occluded (14, 34), we believe that the HCV 3′ X region represents a preinitiation structure. With the help of NS3 helicase or other factor(s), the 3′ stem-loop could be melted or unwound to allow initiation as proposed in this study.
Following RNA binding, the initiating nucleotide (ATP for negative-strand synthesis and GTP for positive-strand synthesis) enters the active site through the conserved NTP channel (6) and base pairs with the 3′-terminal template base of the viral RNA. This step has been shown to be sensitive to NTP concentration and is thus rate limiting (21, 38). Subsequently, the polymerase will add one or more nucleotides to the initiating nucleotide to produce short RNA transcripts. These short RNA transcripts may dissociate from the polymerase/template complexes, a process known as abortive initiation/cycling observed in both prokaryotic and eukaryotic RNA polymerases (22). These abortive transcripts, predominantly di- or trinucleotides, can then be used by the enzyme to initiate new rounds of RNA synthesis in a fashion as described in this study. The dinucleotide may significantly accelerate the RNA synthesis by circumventing the first nucleotidyl transfer reaction from the initiating nucleotide which is rate limiting. Similar effects have been reported for T7 RNA polymerase in that the abortive product, a dinucleotide tetraphosphate (pppGpA), is much more efficient in initiating RNA synthesis (13, 26). The biological role of abortive initiation is not clear, although it is proposed that these abortive RNA transcripts may serve as primers for DNA replication (22). An intracellular pool of di- or trinucleotides may exist as a result of abortive cellular RNA transcription. Whether or not these di- or trinucleotides prime HCV RNA replication in vivo can be addressed only in the context of a viral infection. It is conceivable that the initiating dinucleotide is a product of de novo synthesis and allows efficient RNA synthesis to a level suitable for in vitro kinetic analysis.
The single-nucleotide initiation may reflect the first step of replication initiation but lacks the efficiency for in vitro characterization. Identification of the optimal template and primer requirements for HCV NS5B RdRp will facilitate the mechanistic characterization of nucleotide incorporation catalyzed by this enzyme.
ACKNOWLEDGMENTS
We thank Gregory R. Reyes and Bahige M. Baroudy for support and Jacquelyn Wright-Minogue and Anthony Mannarino for technical assistance.
C.E.C. is a recipient of a Howard Temin Award (CA75118) and supported in part by NIAID grant AI47350.
REFERENCES
- 1.Ago H, Adachi T, Yoshida A, Yamamoto M, Habuka N, Yatsunami K, Miyano M. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Structure. 1999;7:1417–1426. doi: 10.1016/s0969-2126(00)80031-3. [DOI] [PubMed] [Google Scholar]
- 2.Al R H, Xie Y, Wang Y, Staercke C D, van Beers E H, Hagedorn C H. Expression of recombinant hepatitis C virus NS5B. Nucleic Acids Symp Ser. 1997;36:197–199. [Google Scholar]
- 3.Antao V P, Lai S Y, Tinoco I. A thermodynamic study of unusually stable RNA and DNA hairpins. Nucleic Acids Res. 1991;19:5901–5905. doi: 10.1093/nar/19.21.5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arnold J J, Cameron C E. Poliovirus RNA-dependent RNA polymerase (3Dpol): assembly of stable, elongation-competent complexes by using a symmetrical primer/template substrate (sym/sub) J Biol Chem. 2000;275:5329–5336. doi: 10.1074/jbc.275.8.5329. [DOI] [PubMed] [Google Scholar]
- 5.Behrens S-E, Tomei L, De Francesco R. Identification and properties of the RNA-dependent RNA polymerase of hepatitis C virus. EMBO J. 1996;15:12–22. [PMC free article] [PubMed] [Google Scholar]
- 6.Bressanelli S, Tomei L, Roussel A, Incitti I, Vitale R L, Mathieu M, De Francesco R, Rey F A. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Proc Natl Acad Sci USA. 1999;96:13034–13039. doi: 10.1073/pnas.96.23.13034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheetham G M, Steitz T A. Structure of a transcribing T7 RNA polymerase initiation complex. Science. 1999;286:2305–2309. doi: 10.1126/science.286.5448.2305. [DOI] [PubMed] [Google Scholar]
- 8.Choo Q-L, Kuo G, Weiner A J, Overby L R, Bradley D W, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359–364. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 9.De Francesco R, Behrens S E, Tomei L, Altamura S, Jiricny J. RNA-dependent RNA polymerase of hepatitis C virus. Methods Enzymol. 1996;275:58–67. doi: 10.1016/s0076-6879(96)75006-1. [DOI] [PubMed] [Google Scholar]
- 10.Ferrari E, Wright-Minogue J, Fang J W S, Baroudy B M, Lau J Y N, Hong Z. Characterization of soluble hepatitis C virus RNA-dependent RNA polymerase expressed in Escherichia coli. J Virol. 1999;73:1649–1654. doi: 10.1128/jvi.73.2.1649-1654.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang H, Chopra R, Verdine G L, Harrison S C. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science. 1998;282:1669–1675. doi: 10.1126/science.282.5394.1669. [DOI] [PubMed] [Google Scholar]
- 12.Ishii K, Tanaka Y, Yap C C, Aizaki H, Matsuura Y, Miyamura T. Expression of hepatitis C virus NS5B protein: characterization of its RNA polymerase activity and RNA binding. Hepatology. 1999;29:1227–1235. doi: 10.1002/hep.510290448. [DOI] [PubMed] [Google Scholar]
- 13.Jia Y, Patel S S. Kinetic mechanism of transcription initiation by bacteriophage T7 RNA polymerase. Biochemistry. 1997;36:4223–4232. doi: 10.1021/bi9630467. [DOI] [PubMed] [Google Scholar]
- 14.Kolykhalov A A, Feinstone S M, Rice C M. Identification of a highly conserved sequence element at the 3′ terminus of hepatitis C virus genome RNA. J Virol. 1996;70:3363–3371. doi: 10.1128/jvi.70.6.3363-3371.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kraulis P J. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structure. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 16.Lai V C H, Kao C C, Ferrari E, Park J, Uss A S, Wright-Minogue J, Hong Z, Lau J Y N. Mutational analysis of bovine viral diarrhea virus RNA-dependent RNA polymerase. J Virol. 1999;73:10129–10136. doi: 10.1128/jvi.73.12.10129-10136.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lesburg C A, Cable M B, Ferrari E, Hong Z, Mannarino A F, Weber P C. Crystal structure of the RNA-dependent RNA polymerase from hepatitis C virus reveals a fully encircled active site. Nat Struct Biol. 1999;6:937–943. doi: 10.1038/13305. [DOI] [PubMed] [Google Scholar]
- 18.Lohmann V, Korner F, Herian U, Bartenschlager R. Biochemical properties of hepatitis C virus NS5B RNA-dependent RNA polymerase and identification of amino acid sequence motifs essential for enzymatic activity. J Virol. 1997;71:8416–8428. doi: 10.1128/jvi.71.11.8416-8428.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lohmann V, Korner F, Koch J-O, Herian U, Theilmann L, Bartenschlager R. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science. 1999;285:110–113. doi: 10.1126/science.285.5424.110. [DOI] [PubMed] [Google Scholar]
- 20.Lohmann V, Roos A, Korner F, Koch J O, Bartenschlager R. Biochemical and kinetic analyses of NS5B RNA-dependent RNA polymerase of the hepatitis C virus. Virology. 1998;249:108–118. doi: 10.1006/viro.1998.9311. [DOI] [PubMed] [Google Scholar]
- 21.Luo G, Hamatake R K, Mathis D M, Racela J, Rigat K L, Lemm J, Colonno R J. De novo initiation of RNA synthesis by the RNA-dependent RNA polymerase (NS5B) of hepatitis C virus. J Virol. 2000;74:851–863. doi: 10.1128/jvi.74.2.851-863.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsumoto J. Evolutionary role of abortive transcript as a primer for DNA replication. J Mol Evol. 1994;39:620–624. doi: 10.1007/BF00160407. [DOI] [PubMed] [Google Scholar]
- 23.Merritt E, Bacon D J. Raster3D: photorealistic molecular graphics. Methods Enzymol. 1997;276:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- 24.Miller R H, Purcell R H. Hepatitis C virus shares amino acid sequence similarity with pestiviruses and flaviruses as well as members of two plant virus supergroups. Proc Natl Acad Sci USA. 1990;87:2057–2061. doi: 10.1073/pnas.87.6.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Molinaro M, Tinoco I. Use of ultra stable UNCG tetraloop hairpins to fold RNA structures: thermodynamic and spectroscopic applications. Nucleic Acids Res. 1995;23:3056–3063. doi: 10.1093/nar/23.15.3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moroney S E, Piccirilli J A. Abortive products as initiating nucleotides during transcription by T7 RNA polymerase. Biochemistry. 1991;30:10343–10349. doi: 10.1021/bi00106a036. [DOI] [PubMed] [Google Scholar]
- 27.Nicholls A, Sharp K A, Honig B. Protein folding and association: insights from interfacial and thermodynamic properties of hydrocarbons. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 28.Oh J W, Ito T, Lai M M. A recombinant hepatitis C virus RNA-dependent RNA polymerase capable of copying the full-length viral RNA. J Virol. 1999;73:7694–7702. doi: 10.1128/jvi.73.9.7694-7702.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Plotch S J, Palant O, Gluzman Y. Purification and properties of poliovirus RNA polymerase expressed in Escherichia coli. J Virol. 1989;63:216–225. doi: 10.1128/jvi.63.1.216-225.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rice C M. Flaviviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Virology. 3rd ed. New York, N.Y: Raven Press; 1996. pp. 931–960. [Google Scholar]
- 31.Saito I, Miyamura T, Ohbayashi A, Harada H, Katayama T, Kikuchi S, Watanabe T Y, Koi S, Onji M, Ohta Y, Choo Q-L, Houghton M, Kuo G. Hepatitis C virus infection is associated with the development of hepatocellular carcinoma. Proc Natl Acad Sci USA. 1990;87:6547–6549. doi: 10.1073/pnas.87.17.6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shimotohno K. Hepatitis C virus as a causative agent of hepatocellular carcinoma. Intervirology. 1995;38:162–169. doi: 10.1159/000150427. [DOI] [PubMed] [Google Scholar]
- 33.Sun X-L, Johnson R B, Hockman M A, Wang Q M. De novo RNA synthesis catalyzed by HCV RNA-dependent RNA polymerase. Biochem Biophys Res Commun. 2000;268:798–803. doi: 10.1006/bbrc.2000.2120. [DOI] [PubMed] [Google Scholar]
- 34.Tanaka T, Kato N, Cho M-J, Sugiyama K, Shimotohno K. Structure of the 3′ terminus of the hepatitis C virus genome. J Virol. 1996;70:3307–3312. doi: 10.1128/jvi.70.5.3307-3312.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tomei L, Vitale R L, Incitti I, Serafini S, Altamura S, Vitelli A, De Francesco R. Biochemical characterization of a hepatitis C virus RNA-dependent RNA polymerase mutant lacking the C-terminal hydrophobic sequence. J Gen Virol. 2000;81:759–767. doi: 10.1099/0022-1317-81-3-759. [DOI] [PubMed] [Google Scholar]
- 36.World Health Organization. Hepatitis C. Seroprevalence of hepatitis C virus (HCV) in a population sample. Wkly Epidemiol Rec. 1996;71:346–349. [PubMed] [Google Scholar]
- 37.Yamashita T, Kaneko S, Shirota Y, Qin W, Nomura T, Kobayashi K, Murakami S. RNA-dependent RNA polymerase activity of the soluble recombinant hepatitis C virus NS5B protein truncated at the C-terminal region. J Biol Chem. 1998;273:15479–15486. doi: 10.1074/jbc.273.25.15479. [DOI] [PubMed] [Google Scholar]
- 38.Zhong W, Uss A S, Ferrari E, Lau J Y, Hong Z. De novo initiation of RNA synthesis by hepatitis C virus nonstructural protein 5B polymerase. J Virol. 2000;74:2017–2022. doi: 10.1128/jvi.74.4.2017-2022.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]