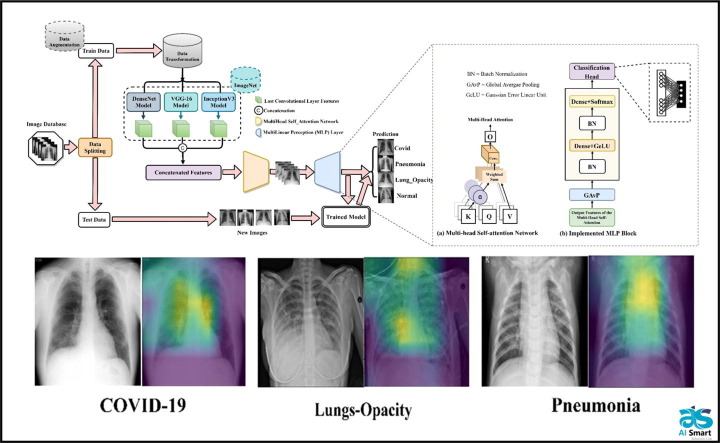
Graphical abstract
Keywords: COVID-19 prediction, Hybrid ensemble deep feature extraction, Multi-head self-attention network, Visual explainable saliency maps, Explainable Artificial Intelligence (XAI)
Abstract
COVID-19 is a contagious disease that affects the human respiratory system. Infected individuals may develop serious illnesses, and complications may result in death. Using medical images to detect COVID-19 from essentially identical thoracic anomalies is challenging because it is time-consuming, laborious, and prone to human error. This study proposes an end-to-end deep-learning framework based on deep feature concatenation and a Multi-head Self-attention network. Feature concatenation involves fine-tuning the pre-trained backbone models of DenseNet, VGG-16, and InceptionV3, which are trained on a large-scale ImageNet, whereas a Multi-head Self-attention network is adopted for performance gain. End-to-end training and evaluation procedures are conducted using the COVID-19_Radiography_Dataset for binary and multi-classification scenarios. The proposed model achieved overall accuracies (96.33% and 98.67%) and F1_scores (92.68% and 98.67%) for multi and binary classification scenarios, respectively. In addition, this study highlights the difference in accuracy (98.0% vs. 96.33%) and F_1 score (97.34% vs. 95.10%) when compared with feature concatenation against the highest individual model performance. Furthermore, a virtual representation of the saliency maps of the employed attention mechanism focusing on the abnormal regions is presented using explainable artificial intelligence (XAI) technology. The proposed framework provided better COVID-19 prediction results outperforming other recent deep learning models using the same dataset.
1. Introduction
COVID-19 is a deadly and contagious disease that affects the human respiratory system it was declared as a global pandemic on March 11, 2020 by the Word Health Organization (WHO) (Sethy et al., 2020, Al-antari et al., 2021, Al-antari et al., 2021) with its origin traced to Wuhan province China. It was identified as a coronavirus with more than 70% similarity with the SARS-CoV and much more than 95% similarity to bat Coronavirus that was identified on January 7, 2020. COVID-19 symptoms range from mild to severe and result in multi-organ damage related to respiratory illnesses, such as Middle East respiratory syndrome (MERS) and severe acute respiratory syndrome (SARS). Symptoms range from cold to fever, breathing issues, and respiratory syndromes (Danet 2021). So far, health practitioners have used two distinctive ways to diagnose COVID-19 patients: (i) diagnostic tests such as reverse transcription polymerase chain reaction (RT-PCR) and (ii) antigen testing (Al-antari et al., 2021, Al-antari et al., 2021). Since the antigen test results are mostly false-positive, RT-PCR is generally accepted as the most suitable way to diagnose the disease. However, RT-PCR requires extensive laboratory work and an experienced medical practitioner to obtain and analyze the results; as a result, the test is expensive and time-consuming, thus raising concerns for researchers to address the issue. Artificial Intelligence (AI) and medical imaging have proven to be more effective, especially in the assessment of critical body parts, such as the lungs, heart, and brain (Jacobi et al., 2020, Doo et al., 2021, Ukwuoma et al., 2022a, Ukwuoma et al., 2022b, Ukwuoma et al., 2022c, Ukwuoma et al., 2022d); thus, researchers have focused on the development of AI-based methods to facilitate cheaper, faster, and home-based COVID-19 detection to help mitigate the spread of the disease. However, quicker and more efficient AI tools are required to detect COVID-19 using limited medical imaging datasets. Radiological images, such as computed tomography (CT) (Zheng et al., 2018) and chest radiography (X-ray) (Zheng et al., 2019) are promising for the diagnosis of lung disease. Extensive research has been conducted to highlight that COVID-19 is analyzed using CT (Zheng et al., 2018). However, chest X-rays are preferred over CT due to the less radiation exposure, low cost (Zheng et al., 2019), and extensive availability (Zheng et al., 2020). Therefore, in this study, we have used chest X-Ray to diagnose COVID-19.
The use of deep learning in vision tasks, especially image detection (Abubakar, et al., 2022, Ukwuoma et al., 2022a, Ukwuoma et al., 2022b, Ukwuoma et al., 2022c, Ukwuoma et al., 2022d, Cai et al., 2023) and classification (Ukwuoma et al., 2022a, Ukwuoma et al., 2022b, Ukwuoma et al., 2022c, Ukwuoma et al., 2022d, Ukwuoma et al., 2022a, Ukwuoma et al., 2022b, Ukwuoma et al., 2022c, Ukwuoma et al., 2022d), has received tremendous attention from researchers Moreover, it is a widely adopted technique for image classification problems owing to the outstanding results. In medical imaging (Gupta et al., 2018, Ukwuoma et al., 2022a, Ukwuoma et al., 2022b, Ukwuoma et al., 2022c, Ukwuoma et al., 2022d), deep learning has demonstrated robust and efficient results in computer-aided diagnosis (Ortiz et al., 2016). Deep learning models are used as feature extractors to enhance classification accuracy. Diabetic retinopathy, skin lesions, X-ray bone suppression, and detection of tumor regions in the lungs are examples of the contribution of deep learning in medical sector (Chen et al., 2019, Lakshmanaprabu et al., 2019). For the diagnosis of COVID-19, researchers have proposed several architectures that use deep learning techniques. Some researchers developed architectures from scratch or modified the convolutional neural network (CNN) models, such as COVIDiagnosis-Net (Ucar et al., 2020), CoroNet (Khan et al., 2020), and Xception with depthwise separable convolution (Chollet 2017) while others used transfer learning; that is, using the pre-trained model as feature weights for the task of COVID-19, such as CovidGAN (Waheed et al., 2020), transfer learning with CNN (Apostolopoulos et al., 2020), Deep CNN (Liu et al., 2015), and MobileNet (Howard et al., 2017).
Several strategies have been developed to automate COVID-19 detection; however, these strategies still face challenges. In addition, several issues have been identified that result in poor performance. These issues can be traced to the extraction of input image features by the proposed model. Input feature extraction is an essential step in any AI-based deep learning model. A small and imbalanced dataset results in poor deep feature generation and yields a poor diagnostic performance. To mitigate these issues, researchers have attempted to resolve the use of ensemble models. Ensemble models are obtained by concatenating two or three models, whether pre-trained or from scratch, for better feature extraction from the input image and result generalization. Previous studies have attempted to solve such challenges using different scenarios of ensemble models, such as CNN + HOG + VGG19 (Ahsan et al., 2021), VGG16 + VGG19 (Khan et al., 2022a, Khan et al., 2022b), Xception architecture and ResNet50V2 (Rahimzadeh et al., 2020), and ResNet + SVM (Sethy et al., 2020). Other studies have used the SGDM + SqueezeNet architecture for rapid and accurate COVID-19 prediction (Chowdhury et al., 2020). Despite the promising results of these ensemble models compared to single models, there is still a need to improve the outcome of the ensemble model. Attention mechanism is a current state-of-the-art approach to visual tasks. It is the process of evaluating the relevance of input features to an assigned task and using this relative weight to assist in accomplishing the target and identifying significant aspects from other features of the input images. Ensemble modeling techniques employ convolutional layers to extract visual information from input images. These convolutional layers either extract undesirable elements with necessary components or ignore vital features (Yang 2020). However, because the extracted characteristics affect the outcome and prediction, ignoring these factors may result in inaccurate image evaluation. Consequently, the properties derived from the convolutional layers of the ensemble models are sometimes insufficient for categorizing COVID-19. Therefore, we have described an attention-based deep learning system for detecting COVID-19 using global and local characteristics.
This study proposes an end-to-end deep learning framework based on deep feature concatenation and a Multi-head Self-attention network for COVID-19 detection in chest X-ray images. Because the proposed model involves feature concatenation as the network backbone, we explored six pre-trained deep learning models (i.e., InceptionV3, EfficientNetB7, VGG16, Xception, InceptionResNetV2, and DensetNet201 networks) based on a transfer learning approach on chest X-ray images to select the models for the proposed network. Among the explored and pre-trained deep learning models, the results of DenseNet201, InceptionV3, and VGG-16 architectures were more promising; therefore, we used them in a concatenating approach as our model backbone. In the implementation of the Multi-head Self-attention network, this study followed the implementation techniques of a vision transformer. In implementing vision transformers, the input images are first patch-split before passing them to position embedding. The learnable position-embedding vectors and patches are joined before being fed into the transformer encoder. The input and output must be the same as those of the transformer encoder. Multi-head Self-attention and Multi-linear Perceptron (MLP) block computations occur within the encoder network. The output of the encoder is then fed to the classification layer to output the final classification result. For the MLP implementation, the output features of Multi-head Self-attention are first sent to GlobalAveragePooling1D before the normalization layer. Subsequently, a dense layer (GeLu activation) is considered before the last normalization layer and dense layer (Softmax activation). Our contributions are summarized as follows:
-
•
This study proposes an end-to-end deep-learning framework based on the deep feature concatenation and a Multi-head Self-attention network for rapid and accurate COVID-19 prediction using chest X-ray images.
-
•
The backbone network is built based on a fusion concatenation strategy to generate excellent and stronger deep features. The multi-head Self-attention network is built based on positional embedding and patching mechanisms to mitigate the CNN max-pooling layers’ issues of discarding important information about the precise location; that is, the composition and position of the components contained in an image, whereas MLP is used for model prediction.
-
•
A comprehensive experimental study was conducted to evaluate the capability of the proposed framework with various dataset categorizations: binary and multi-class prediction scenarios with four and three different respiratory disease classes, including a Grad-CAM strategy, to visually show the explicable saliency maps and emphasize the capability of the proposed AI framework to accurately predict different respiratory diseases simultaneously.
The remainder of this paper is organized as follows; Section 2 highlights related works. Section 3 describes the materials and methods in detail, and Section 4 summarizes the experimental setup, results, and comparisons. Finally, the conclusions and future work are presented in Section 5.
2. Related works
This section reviews the state-of-the-art research involving chest X-ray imaging for Deep Learning-based COVID-19 identification. Shi et al. (2020) have described different methods that used AI approaches to assess COVID-19 (Shi et al., 2021). Nayak et al. examined the MobileNetV2, ResNet-50, InceptionV3, ResNet34, VGG16, AlexNet, SqueezeNet, and Inception V3 models for the early identification of COVID-19 infection using chest X-ray images (Nayak et al., 2021). Parameters, such as learning rate, number of epochs, and batch size were considered for optimal model selection. The evaluation results showed that the ResNet34 model, with an accuracy of 98.33%, surpassed all other evaluated models. Soares et al. used CNN-based transfer learning (TL) to diagnose COVID-19 using chest X-ray images (Soares et al., 2020). The images were fed into the Inception-V3 model with no preprocessing and an accuracy of 96% was achieved. Based on a COVID-19 radiography dataset, Das et al. proposed a COVID-19 test model (ResNet-50 and VGG-16) with three classes: COVID-19, normal, and other types of pneumonia (Das et al., 2021). Based on the accuracy, VGG-16 model performed better with an accuracy of 97.67%. In addition, Monshi et al. revealed that employing data augmentation and adjusting the parameters of the CNN model improved the COVID-19 detection performance using chest X-ray images (Monshi et al., 2021). Moreover, this method improves the efficiency of VGG-19 and ResNet-50 models. Furthermore, CovidXrayNet was proposed by the authors which is based on optimization and EfficientNet-B0, thus recording an accuracy of 95.82% when evaluated using two distinct databases. The COVID-19 multi-class detection problem was studied by (Rajpal et al., 2021). The proposed structure was divided into three experiments. First, ResNet-50 with TL was used to obtain 2048 parameters. Second, principal component analysis (PCA) was employed to select 64 features from the 252 chosen features. The characteristics gathered in the previous two sections were integrated and categorized in the third module, thus achieving a classification accuracy of 98%. Kumar et al. proposed SARS-Net for identifying COVID-19 using chest X-ray (Kumar et al., 2022). The accessible COVIDx database comprising chest X-ray data was employed in that investigation. A quantitative study has shown that the proposed approach exhibited higher accuracy of 97.60%.
Transfer learning plays a pivotal role in medical imaging by leveraging pre-trained deep learning models on large datasets to extract meaningful features, enhance model performance, and enable efficient training on limited medical image data, ultimately aiding in more accurate and efficient diagnosis and treatment. Using a resampling technique and five-fold cross-examination approach, Ali et al. implemented an experiment using the ResNet50 on a small database (50 COVID-19 and 50 normal), thus achieving an accuracy of 98% (Alimadadi et al., 2020). CNN were employed by (Saba et al., 2021) to detect COVID-19 instances via TL (Xception, VGG-16, and ResNet models). The collection included 175 COVID-19, 100 pneumonias, and 100 normal chest X-ray images. Their findings demonstrated that all algorithms functioned well, thus exhibiting high accuracy, with the VGG-16 model performing better than the others. However, they proposed further research to improve the methods proposed by them in terms of “evaluating variations architectures, variables, and databases that apply augmentation techniques”. Linda et al. introduced Covid-Net based on TL by constructing a dataset with 8066 (normal), 183 (COVID-19), and 5538 (pneumonia) samples, whereas the test set comprised 100 (normal and pneumonia) and 31 (COVID-19) samples (Wang et al., 2020, Wang et al., 2020). An accuracy of 92% was achieved. It was specifically mentioned that there was no duplication between the exam and training sets of the participants, which is critical while performing such tasks (Farooq et al., 2020) fine-tuned the ResNet-50 model using TL for COVID-19 identification using chest X-ray images. They employed a three-class dataset consisting of COVID-19, bacterial pneumonia, and viral pneumonia data. (Minaee et al., 2020) applied TL to create a deep COVID-19 detector using chest X-ray images. They employed a subset of 5000 chest X-ray images to train four CNNs: Squeeze-Net, DenseNet-121, ResNet-18, and ResNet50. Ucar et al. proposed a Squeeze-Net + Bayesian optimization additive to diagnose COVID-19 (Ucar et al., 2020). Because of the fine-tuned parameters and additional datasets, they exhibited improved performance and increased COVID-19 diagnostic accuracy. Novitasari et al. integrated CNNs and SVM to detect COVID-19 using chest X-ray images (Novitasari et al., 2020).
Data augmentation and the use of attention mechanism plays a crucial role in medical imaging by enhancing the diversity and volume of training data, improving the robustness and generalization of deep learning models, and facilitating more accurate and reliable diagnosis and analysis. Rajaraman et al. demonstrated a method for augmenting COVID-19 chest X-ray images using weakly labeled data (Rajaraman et al., 2020). Khan et al. proposed CoroNet for COVID-19 detection using chest X-ray images (Khan et al., 2020). They employed Xception model pretrained using TL and evaluated using a four-fold cross-validation approach. Classifiers, such as SVMs, decision trees, KNNs, and random forests were used in a feature extraction network (InceptionV3). Pereira et al. classified the chest X-ray images used to detect COVID-19 such that F1_Score of 89% was achieved using a stratified analysis (Pereira et al., 2020). Hussain et al. used chest X-ray images to distinguish COVID-19 from non-COVID-19, bacterial pneumonia, and viral pneumonia (Hussain et al., 2020). A recently published paper by (Ukwuoma et al., 2022a, Ukwuoma et al., 2022b, Ukwuoma et al., 2022c, Ukwuoma et al., 2022d) employed a dual-path CNN, attention mechanism, and second-order pooling for COVID-19 detection from images. The dual path serves as feature extraction (RGB and grey image features), whereas second-order pooling is employed to capture the second-order derivative of the generated features before passing it into the attention model, thus achieving a significant result for both datasets tested. Deep_Pachi (Ukwuoma et al., 2022a, Ukwuoma et al., 2022b, Ukwuoma et al., 2022c, Ukwuoma et al., 2022d) implemented the same idea as proposed by us; however, it was implemented in a different scenario. Deep_Pachi was used for breast cancer multi-classification. Thus, feature concatenation is based on the channel and spatial features. An image enhancement technique was considered in (Ukwuoma et al., 2022a, Ukwuoma et al., 2022b, Ukwuoma et al., 2022c, Ukwuoma et al., 2022d, Ukwuoma et al., 2022a, Ukwuoma et al., 2022b, Ukwuoma et al., 2022c, Ukwuoma et al., 2022d) before passing the model to the proposed deep learning model. The recorded results show that image enhancement improves detection accuracy. Khan et al. developed a new channel-based network to improve CNN models and achieved an accuracy of 96.5 binary classifications and F1_score of 95% (Khan et al., 2022a, Khan et al., 2022b). Regarding the issue of accurate feature extraction for computer vision tasks, research has shown that features of palm print with a two-dimensional discrete cosine transform to constitute a dual-source space resulted in improved performance of recognition models (Leng et al., 2017). In addition, deep learning models are prone to adversarial attacks; hence, security guidance is necessary (Leng et al., 2017). We intend to integrate these ideas into a model, which is a part of future research.
3. Material and methods
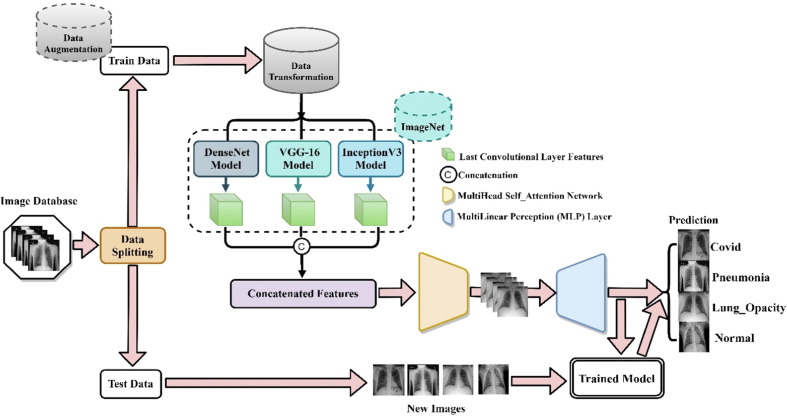
The proposed architecture is shown in Fig. 1 . The benchmark dataset is first split into training (training and validation) and test set, followed by augmentation of the training set because it contains a large number of unequal sample size among the classes. The training sets are further preprocessed using different image-processing strategies, such as rotation, resizing, cropping, and normalization. The processed data are fed into selected pretrained models (DenseNet, VGG-16, and InceptionV3), which are originally trained on a large-scale ImageNet dataset. The features of the pre-trained models are concatenated at their last convolutional layer. The output shape of the last convolutional layer of DenseNet201 is (None, 7, 7, 1920), VGG-16 is (None, 7, 7, 512) while that of InceptionV3 is (None, 5, 5, 2048). Since the output shapes have different sizes, concatenation of outputs of the three models results in an error; hence, we have employed a zero-padding layer to the output, thus resulting in an output shape of (None, 7, 7, 2048. After concatenation, the output feature shape becomes (None, 7, 7, 4480), which is a strong and deep feature for further prediction. Because it has been proven that most CNNs ignore vital features (Yang, 2020) of the input image due to the max-pooling layer, we have employed a Multi-head Self-attention network to precisely distinguish the different symptoms and focus on the regional and global deep features simultaneously. The concatenated features are then passed through the attention block. Finally, we have adopted the multi-layer perceptron (MLP) block to improve false symptom prediction, classify the different respiratory diseases, and prove the final prediction score per class. Finally, the trained model is saved and used to evaluate the test class as shown in Fig. 1.
Fig. 1.
Framework of the proposed deep learning model for COVID-19 prediction from chest X-Ray images.
3.1. Concatenated features framework
While training a deep learning network, the convolutional layer selects the extracted characteristics used to perform the classification procedure. Therefore, it is essential to acquire only the useful characteristics to achieve high classification efficiency (Al-antari et al., 2021, Al-antari et al., 2021, Ukwuoma et al., 2022a, Ukwuoma et al., 2022b, Ukwuoma et al., 2022c, Ukwuoma et al., 2022d).
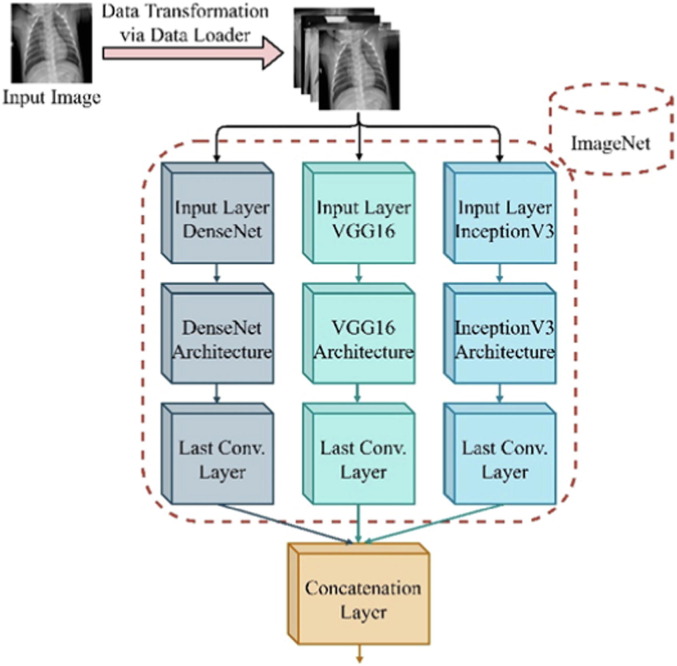
Fig. 2 shows the proposed backbone model, which incorporates the concatenation of multiple deep features extracted from three deep learning models (DenseNet, VGG-16, and InceptionV3) using the concatenation feature strategy. The models are originally trained on ImageNet, which is first introduced in the ImageNet Large Scale Visual Recognition Challenge (ILSVRC) for image classification/detection tasks. The concatenated deep features from the pretrained DenseNet, VGG-16, and InceptionV3 are computed in Equation (1).
| (1) |
where is the number of selected pre-trained models and signifies the extracted features. The extracted features are concatenated and represented in a single vector given in Eq. (2).
| (2) |
Fig. 2.
Backbone ensemble network of the proposed framework based on DenseNet, VGG-16, and InceptionV3.
3.2. The proposed framework
is passed through Conv2D, Kernal_size = 1, padding = “same” and Activation = “ReLU.” The output of the convolution layer is zero-padded with Zeropadding2D (padding = {(0, 5), (0, 5)}. Thus, the deep feature entering the Multi-head Self-attention is expressed in Eq. (3).
| (3) |
Fig. 1 shows that the extracted features are implemented by integrating two networks, namely the Multi-head Self-attention network and MLP layer. This hybrid paradigm supports the proposed model in achieving promising prediction performance. Each layer in the proposed model is constructed with a skip connection and normalization layer to generate the output given by Eq. (4).
| (4) |
where denotes the output of layer (indicating the extracted features) in Eq. (3) and input of layer , depicts the norm layer, and is the functionality of multi-head attention , which is also depicted as MLP . is used to extract the input dependency among tokens, which is implemented using scaled dot-product attention (to identify the data pertinent to the target order from the source order).
Assuming that is the span of the source and target order and is the hidden dimension, the target order query is formulated as whereas the source order is demonstrated by key as and value as . The scaled dot-product attention (Q, K, and V) output is deduced in Eq. (5).
| (5) |
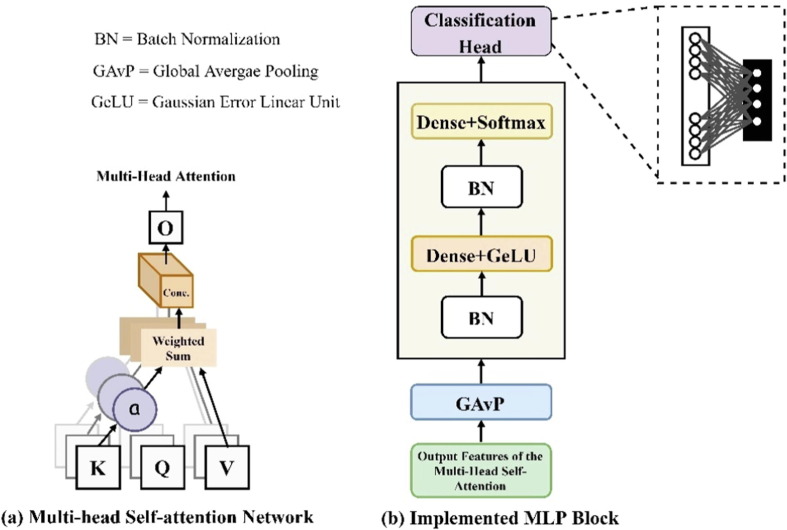
where the row-wise regression function of Softmax is represented by . Each Q, K, and V attend to only one position in each row (i.e., each target token has one position in a row) because the Softmax output is one-dimensional (larger than other individual row dimensions). For multiple position settings, is employed via multiple Q, K, and V in parallel as seen in Fig. 3 (a). This can be mathematically represented as given below in Eq. (6).
| (6) |
where , = number of attention heads, and represents the learnable weight parameters. The MLP layer setting is defined in Eq. (7) where the non-linear function = and = learnable features.
| (7) |
Fig. 3.
Building blocks of the proposed model.
The GeLU activation function is used in the first layer after the GAvP whereas the second block is built using the Softmax activation function after the batch norm as shown in Fig. 3(b). From the AI model perspective, GeLU stands for Gaussian Error Linear Unit, GAvP stands for global average pooling, and BN is batch normalization. Therefore, the output of the previous layers is more realistic, scales the activations of the input layer, enables subsequent levels of the model to adapt more autonomously, and serves as a model regularizer to overcome overfitting. The proposed experiment consists of two optimizers: Adam and stochastic gradient descent (SGD). The two loss functions are: categorically cross-entropy and categorical smooth loss (Cengil et al., 2022). We typically run across the following issues when employing deep learning algorithms for classification: overconfidence and overfitting. Overfitting has been extensively investigated and can be prevented through early termination, dropouts, and weight regulation. However, we do not have many resources to combat this overconfidence. Both issues are addressed using a regularization method known as label smoothing. Categorical smooth loss represents the addition of the label-smoothing function to cross-entropy loss function as expressed in Eq. (8).
| (8) |
The attention layer setup computes outputs as given in Eq. (9), whereas the MLP layer setup computes outputs as stated in Eq. (10).
| (9) |
| (10) |
From Eq. (9), the attention layer sets values are equal to , thus capturing the dependency among the tokens within the same sequence, which is referred to as Self-attention.
3.3. Dataset
The chest X-ray medical dataset used in this study is the COVID-19_Radiography_Dataset (Chowdhury et al., 2020). For research purpose, this dataset is open source and consists of four classes: COVID-19, pneumonia, lung opacity, and normal. Fig. 4 depicts the examples of chest X-ray images with a pictorial view for each class. Table 1 lists the randomly balanced partitions of the dataset distribution for each class using the training, validation, and test sets. This dataset contains 3,616 COVID-19 samples, 1,345 viral pneumonia samples, 6,012 lung opacity samples (i.e., non-COVID lung infection), and 10,192 normal samples. All chest X-ray images are saved in portable network graphics “png” file format with a spatial resolution of pixels. An augmentation strategy is used to enlarge the pneumonia samples to match the target balanced size of 3,000 chest X-ray images per class. The data transformation includes a rotation range set to 1, horizontal flip set to true, and zoom range set to 0.2. For the other classes, a balanced sample size is randomly extracted from the original dataset. A total of 3,000 training samples are chosen to match the scarce samples of the COVID-19 chest X-ray images.
Fig. 4.
Pictorial representation of the chest X-Ray image samples of COVID-19, pneumonia, lung opacity, and normal.
Table 1.
Dataset arrangement used to build and evaluate the proposed approach.
| Partition | COVID-19 | Pneumonia | Lung Opacity | Normal | Total | Total |
|---|---|---|---|---|---|---|
| Training | 3000 | 3000 | 3000 | 3000 | 12,000 | 14,400 |
| Validation | 300 | 300 | 300 | 300 | 1,200 | |
| Testing | 300 | 300 | 300 | 300 | 1,200 |
3.4. Experimental setup
-
•
Loss Function: A loss function is crucial in training a deep learning model because it estimates the forecasting efficiency of the model using a specified set of variables. Loss is the computed outcome, which is the difference between the model's estimate and measured ground truth. This study uses a predetermined loss called “categorical cross-entropy loss” and modified loss called “categorical smooth loss”.
-
•
Optimizer: The basic aim of training a neural network is to minimize the error in the network estimation, which is accomplished by adjusting the weights. The optimizer minimizes the loss function by updating the model parameters according to the loss function outcome, thus resulting in a global minimum with the smallest and most precise outcome. In this study, we have used two optimizers: Adaptive moment estimation (Adam) and stochastic gradient descent (SGD).
-
•
Learning Rate: The learning rate is a variable that controls the model's response with respect to the weight change over the yielded error. As a result, learning rate selection is very challenging because, neither too low nor too high is good for the model. We used learning rates of 1 × 10−4 and 1 × 10−3 in this study. Other training hyper-parameters used are summarized in Table 2 .
Table 2.
Experimental hyper-parameters.
| Model parameters | Loss functions | Categorical cross-entropy loss and Categorical smooth loss |
| Optimizers | Adam and SGD | |
| Learning rates | 0.0001 and 0.001 | |
| Input size | 224 × 224 | |
| Epoch | 100 | |
| Batch size | 8 | |
| Multi-head attention model Parameters | Patch size | (2, 2) |
| Window size | Window Size//2 | |
| Number of heads | 8 | |
| Number of MLP | 256 | |
| Embed_dim | 64 | |
| Drop rate | 0.01 | |
| Learning rate selection regulating parameters | Reduce learning rate | 0.2 |
| Verbose | 1 | |
| Epsilon | 0.001 | |
| Es-Callback (Patience) | 10 | |
| Clip value | 0.2 | |
| Patience | 10 |
3.5. Evaluation metrics
In this study, we have used the widely accepted classification task evaluation metrics, which are mathematically expressed as given in Eqs. (9), (10), (11), (12). TP = True positive, TN = True negative, FP = False positive, FN = False negative, N = Negative and P = Positive. In addition, the receiver operating characteristic (ROC) curve with its area under the curve (AUC) and precision-recall (PR) curves were used to assess and evaluate the AI models.
| (11) |
| (12) |
| (13) |
| (14) |
| (15) |
3.6. Execution environment
All the experiments in this study were conducted using a personal computer (PC) with an Intel(R) Core (TM) i9-10850 K CPU @ 3.60 GHz, 64.0 GB RAM, and NVIDIA GEFORCE RTX-3080 Ti 10 GB graphical processing unit. Open-source libraries Keras and TensorFlow (Abadi et al., 2016) are used for implementation.
4. Experimental results
This section presents the prediction results of all the experiments conducted in this study. First, an optimization parameter study is conducted using various pre-trained AI models to select the best combination for the ensemble strategy. Multiple optimizers, learning rates, and loss functions are also investigated. Second, the experimental results of the ensemble AI model are compared with those of the proposed AI framework. Finally, the experimental results for binary and multi-class prediction scenarios are presented and summarized.
4.1. Backbone model selection
The backbone model is selected based on six pretrained models: DenseNet201, EfficientNetB7, InceptionV3, InceptResNetV2, VGG-16, and Xception to select the most prominent combination of models to build the proposed backbone network. The results for all the models are evaluated using Adam and SGD optimizers. The experiment was repeated twice for learning rates of 0.001 and 0.0001 with the same training settings and model structures. Table 3 presents the evaluation performance of all the models. It is clear that DenseNet201 achieves the best evaluation results because it exhibits its superiority by using different optimizers and different learning rates. However, InceptionResNetv2 records the second-best evaluation results followed by InceptionV3.
Table 3.
Evaluation performance of the backbone AI model selection with different optimizers and learning rates. The evaluation results are derived as an average over all classes: COVID-19, pneumonia, lung opacity, and normal.
| AI Models | Optimizer | ACC | SEN | SPE | PRE | F1_Score | AUC |
|---|---|---|---|---|---|---|---|
| DenseNet201 | Adam, Learning Rate: | 0.96 | 0.92 | 0.97 | 0.92 | 0.92 | 0.95 |
| EfficientNetB7 | 0.87 | 0.75 | 0.92 | 0.83 | 0.74 | 0.83 | |
| InceptionV3 | 0.93 | 0.86 | 0.95 | 0.87 | 0.86 | 0.91 | |
| InceptResNetV2 | 0.95 | 0.89 | 0.96 | 0.91 | 0.89 | 0.93 | |
| VGG-16 | 0.91 | 0.82 | 0.94 | 0.87 | 0.82 | 0.88 | |
| Xception | 0.90 | 0.80 | 0.93 | 0.83 | 0.80 | 0.87 | |
| DenseNet201 | Adam, Learning Rate: | 0.92 | 0.85 | 0.95 | 0.86 | 0.85 | 0.90 |
| EfficientNetB7 | 0.86 | 0.71 | 0.90 | 0.78 | 0.72 | 0.81 | |
| InceptionV3 | 0.89 | 0.78 | 0.93 | 0.81 | 0.78 | 0.85 | |
| InceptResNetV2 | 0.93 | 0.86 | 0.95 | 0.87 | 0.86 | 0.90 | |
| VGG-16 | 0.90 | 0.79 | 0.93 | 0.82 | 0.80 | 0.86 | |
| Xception | 0.89 | 0.77 | 0.92 | 0.80 | 0.78 | 0.85 | |
| DenseNet201 | SGD, Learning Rate: | 0.90 | 0.79 | 0.93 | 0.83 | 0.80 | 0.86 |
| EfficientNetB7 | 0.84 | 0.67 | 0.89 | 0.81 | 0.67 | 0.78 | |
| InceptionV3 | 0.87 | 0.74 | 0.91 | 0.76 | 0.75 | 0.83 | |
| InceptResNetV2 | 0.87 | 0.73 | 0.91 | 0.79 | 0.74 | 0.82 | |
| VGG-16 | 0.86 | 0.72 | 0.91 | 0.76 | 0.73 | 0.81 | |
| Xception | 0.89 | 0.79 | 0.93 | 0.82 | 0.79 | 0.86 | |
| DenseNet201 | SGD, Learning Rate: | 0.95 | 0.90 | 0.97 | 0.91 | 0.90 | 0.93 |
| EfficientNetB7 | 0.86 | 0.73 | 0.91 | 0.80 | 0.73 | 0.82 | |
| InceptionV3 | 0.89 | 0.78 | 0.93 | 0.80 | 0.78 | 0.85 | |
| InceptResNetV2 | 0.91 | 0.82 | 0.94 | 0.84 | 0.82 | 0.88 | |
| VGG-16 | 0.88 | 0.75 | 0.92 | 0.80 | 0.76 | 0.83 | |
| Xception | 0.88 | 0.76 | 0.92 | 0.81 | 0.77 | 0.84 |
4.2. Backbone concatenated features against the proposed model
A comparison of the evaluation results of the backbone model and proposed framework is summarized in Table 4 . This experiment was conducted using two different optimizers (Adam and SGD), two learning rates (0.001 and 0.0001), and two loss functions (categorical smooth loss and categorical cross-entropy). The results obtained using the learning rate of are better than those using in both implemented loss functions and optimizers. Comparing the results of the employed optimizers, the Adam optimizer supersedes the SGD optimizer with the same learning rate and loss function. For a learning rate of 0.001 and categorical smooth loss, the Adam optimizer exhibits a lower performance than the SGD optimizer. For categorical cross-entropy, the Adam optimizer performs better than the SGD optimizer. In all other settings, a learning rate of and categorical cross-entropy are preferred for the Adam optimizer, whereas a learning rate of and categorical cross-entropy loss are preferred for the SGD optimizer.
Table 4.
Comparison of evaluation results of the backbone model against the proposed model.
| AI Model | Optimizer | Learning Rate/Loss Function | ACC | SEN | SPE | PRE | F1_Score | AUC |
|---|---|---|---|---|---|---|---|---|
| Backbone | Adam | /Categorical smooth loss | 0.95 | 0.91 | 0.97 | 0.91 | 0.91 | 0.94 |
| Proposed | 0.96 | 0.92 | 0.97 | 0.93 | 0.92 | 0.95 | ||
| Backbone | Categorical smooth loss | 0.94 | 0.88 | 0.96 | 0.89 | 0.88 | 0.92 | |
| Proposed | 0.97 | 0.93 | 0.98 | 0.94 | 0.93 | 0.96 | ||
| Backbone | /Categorical cross-entropy | 0.96 | 0.93 | 0.98 | 0.93 | 0.93 | 0.95 | |
| Proposed | 0.98 | 0.96 | 0.97 | 0.96 | 0.96 | 0.97 | ||
| Backbone | Categorical cross-entropy | 0.84 | 0.67 | 0.89 | 0.78 | 0.65 | 0.78 | |
| Proposed | 0.98 | 0.95 | 0.99 | 0.96 | 0.95 | 0.97 | ||
| Backbone | SGD | /Categorical smooth loss | 0.94 | 0.88 | 0.96 | 0.89 | 0.88 | 0.92 |
| Proposed | 0.94 | 0.88 | 0.96 | 0.89 | 0.88 | 0.92 | ||
| Backbone | Categorical smooth loss | 0.94 | 0.88 | 0.96 | 0.90 | 0.88 | 0.92 | |
| Proposed | 0.97 | 0.94 | 0.98 | 0.94 | 0.94 | 0.96 | ||
| Backbone | /Categorical cross-entropy | 0.94 | 0.89 | 0.96 | 0.90 | 0.89 | 0.92 | |
| Proposed | 0.95 | 0.89 | 0.96 | 0.91 | 0.90 | 0.93 | ||
| Backbone | Categorical cross-entropy | 0.70 | 0.40 | 0.80 | 0.58 | 0.32 | 0.60 | |
| Proposed | 0.95 | 0.89 | 0.96 | 0.92 | 0.89 | 0.93 |
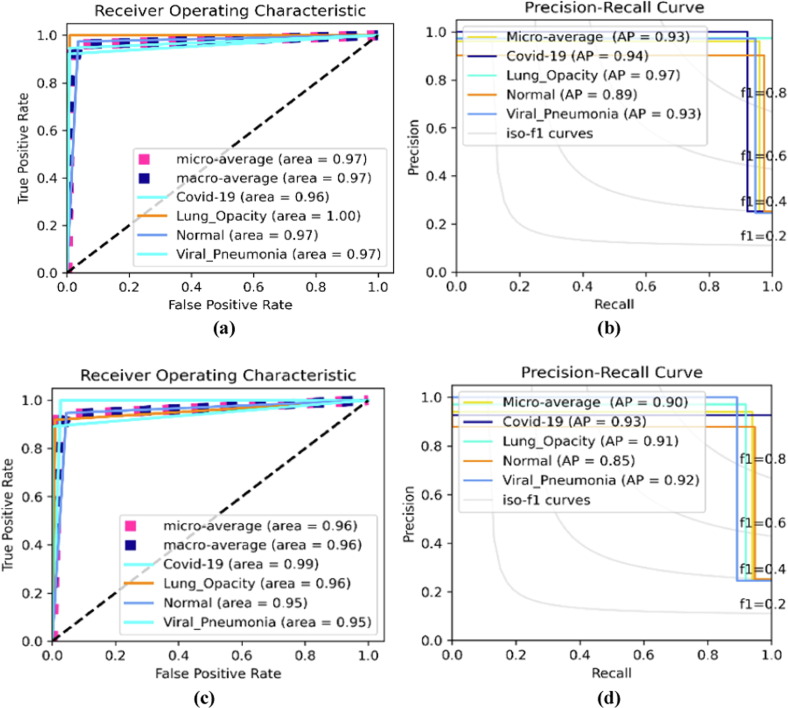
These results were also validated using the ROC and PR evaluation curves as shown in Table 5 and Table 6 using the Adam and SGD optimizers, respectively. ROC and PR curves are used to estimate the exact prediction rate of different respiratory diseases, namely COVID-19, lung opacity, and pneumonia against the normal class. However, the exact prediction rate of the models is highly influenced by the hyper-parameters because, the Adam optimizer settings performed better than the SGD settings except at learning rate and categorical smooth loss where the SGD performance superseded the Adams performance. In general, the Adam optimizer with other settings performed significantly better than the SGD optimizer. The prediction of COVID-19 received better ROC and AP AUCs for most of the optimized settings.
Table 5.
Comparison of the evaluation performances of ROC and PR curves for backbone and proposed models using Adam optimizer.
| Learning Rate/Loss Function | Macro-Average | Micro-Average | COVID-19 | Lung Opacity | Normal | Pneumonia | |
|---|---|---|---|---|---|---|---|
| (1) ROC Evaluation Curve | |||||||
| Backbone | /Categorical smooth loss | 0.94 | 0.94 | 0.95 | 0.97 | 0.91 | 0.93 |
| Proposed | 0.95 | 0.95 | 0.99 | 0.98 | 0.93 | 0.89 | |
| Backbone | /Categorical smooth loss | 0.92 | 0.92 | 0.95 | 0.90 | 0.93 | 0.91 |
| Proposed | 0.96 | 0.96 | 0.96 | 0.98 | 0.96 | 0.93 | |
| Backbone | /Categorical cross-entropy | 0.95 | 0.95 | 0.96 | 0.99 | 0.94 | 0.91 |
| Proposed | 0.97 | 0.97 | 0.96 | 0.99 | 0.97 | 0.97 | |
| Backbone | /Categorical cross-entropy | 0.78 | 0.78 | 0.68 | 0.89 | 0.83 | 0.73 |
| Proposed | 0.94 | 0.94 | 0.90 | 0.98 | 0.95 | 0.94 | |
| (2) Precision-Recall (PR) Evaluation Curve | |||||||
| Backbone | /Categorical smooth loss | 0.85 | 0.92 | 0.84 | 0.77 | 0.87 | |
| Proposed | 0.87 | 0.95 | 0.93 | 0.77 | 0.84 | ||
| Backbone | /Categorical smooth loss | 0.80 | 0.83 | 0.81 | 0.75 | 0.86 | |
| Proposed | 0.89 | 0.92 | 0.95 | 0.83 | 0.87 | ||
| Backbone | /Categorical cross-entropy | 0.88 | 0.92 | 0.95 | 0.81 | 0.85 | |
| Proposed | 0.93 | 0.94 | 0.97 | 0.89 | 0. 93 | ||
| Backbone | /Categorical cross-entropy | 0.54 | 0.50 | 0.64 | 0.52 | 0.59 | |
| Proposed | 0.90 | 0.97 | 0.95 | 0.80 | 0.87 | ||
Table 6.
Comparison evaluation performances of ROC and PR curves for backbone and proposed models using SGD optimizer.
| Learning Rate/Loss Function | Macro-Average | Micro-Average | COVID-19 | Lung Opacity | Normal | Pneumonia | |
|---|---|---|---|---|---|---|---|
| (1) ROC Evaluation Curve | |||||||
| Backbone | /Categorical smooth loss | 0.92 | 0.92 | 0.87 | 0.96 | 0.91 | 0.92 |
| Proposed | 0.92 | 0.92 | 0.86 | 0.96 | 0.93 | 0.93 | |
| Backbone | /Categorical smooth loss | 0.92 | 0.92 | 0.96 | 0.97 | 0.89 | 0.86 |
| Proposed | 0.96 | 0.96 | 0.99 | 0.96 | 0.95 | 0.95 | |
| Backbone | /Categorical | ||||||
| cross-entropy | 0.92 | 0.92 | 0.90 | 0.92 | 0.86 | 0.92 | |
| Proposed | 0.93 | 0.93 | 0.92 | 0.97 | 0.93 | 0.89 | |
| Backbone | /Categorical cross-entropy | 0.60 | 0.60 | 0.63 | 0.53 | 0.47 | 0.77 |
| Proposed | 0.93 | 0.93 | 0.88 | 0.96 | 0.94 | 0.93 | |
| (2) Precision-Recall (AP) Evaluation Curve | |||||||
| Backbone | /Categorical smooth loss | 0.79 | 0.77 | 0.88 | 0.70 | 0.88 | |
| Proposed | 0.80 | 0.75 | 0.82 | 0.77 | 0.90 | ||
| Backbone | /Categorical smooth loss | 0.80 | 0.92 | 0.88 | 0.67 | 0.80 | |
| Proposed | 0.90 | 0.93 | 0.91 | 0.85 | 0.92 | ||
| Backbone | /Categorical cross-entropy | 0.81 | 0.81 | 0.86 | 0.74 | 0.88 | |
| Proposed | 0.82 | 0.83 | 0.93 | 0.74 | 0.84 | ||
| Backbone | /Categorical cross-entropy | 0.31 | 0.31 | 0.29 | 0.25 | 0.65 | |
| Proposed | 0.82 | 0.82 | 0.86 | 0.76 | 0.90 | ||
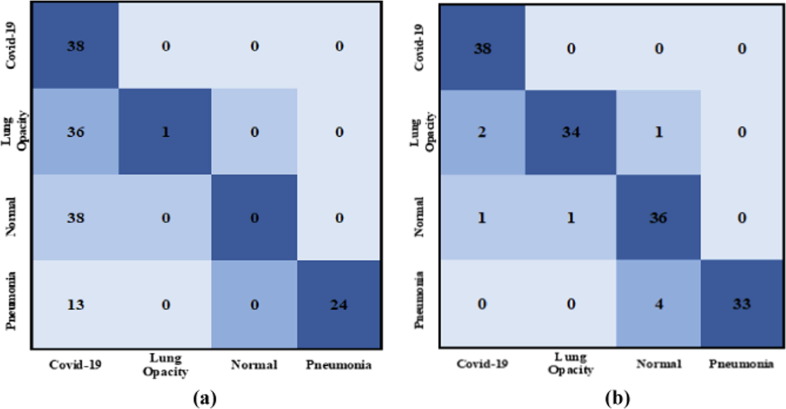
Fig. 5 depicts the graphical ROC and AP evaluation curves for this study as presented in Table 5, Table 6. To analyze the proposed AI model further, Hit Rate matrices are used as shown in Fig. 6 . “Hit Rate” is derived by the entire order number division when the changes are made (subtracting the targets from mistakes). “Miss Rate” is expressed as . COVID-19 has a hit rate of 38 for both the Adam and SGD Optimizers. The experiment with the Adam optimizer, learning rate, and categorical cross-entropy loss is preferred because the normal class hit rate is 38 against the SGD optimizer, learning rate, and categorical smooth loss function.
Fig. 5.
Optimized multi-class scenario for ROC and PR evaluation curves (a) and (b) depict the curves using learning rate, categorical cross-entropy loss, and Adam optimizer. (c) and (d) depict the curves using , categorical smooth loss, and SGD optimizer.
Fig. 6.
Examples of the HIT Rate (Batch size of 4) diagram of some chest X-ray images from the testing sets. (a) The Adam optimizer is used with learning rate of and categorical cross-entropy loss function. (b) The SGD optimizer is used with learning rate of and categorical smooth loss.
4.3. Binary vs. multi-class prediction evaluation results
To further evaluate the robustness of the proposed model for a more confined class, the employed dataset was decomposed into binary and multi-class scenarios. For the binary scenario, we investigated the capability of the proposed AI model for normal and COVID-19 classes. For the multi-class scenarios, the dataset was decomposed into three major classes: COVID-19, pneumonia, and normal.
4.3.1. Binary prediction Scenario: Normal vs. COVID-19
Table 7 presents the evaluation prediction performance of the binary scenario with respect to different optimization conditions. Table 8 presents the binary classification results of the ROC and PR curves using the Adam and SGD optimizers. This clearly shows that the prediction evaluation result for COVID-19 class performed better than the other classes using the proposed framework. This indicated that the proposed model is sufficiently robust, reliable, and feasible for predicting COVID-19 when compared with normal cases.
Table 7.
Comparison of the evaluation results of the binary scenario using the backbone model against the proposed approach.
| AI Model | Optimizer | Learning Rate/Loss Function | ACC | SEN | SPE | PRE | F1_Score | AUC |
|---|---|---|---|---|---|---|---|---|
| Backbone | Adam | categorical cross-entropy | 0.81 | 0.82 | 0.82 | 0.86 | 0.81 | 0.82 |
| Proposed | 0.89 | 0.89 | 0.89 | 0.91 | 0.89 | 0.89 | ||
| Backbone | SGD | categorical cross-entropy | 0.92 | 0.92 | 0.92 | 0.93 | 0.92 | 0.92 |
| Proposed | 0.99 | 0.99 | 0.99 | 0.98 | 0.99 | 0.99 |
Table 8.
Performance of ROC and PR curves for the binary scenario depicting the backbone against the proposed model using the Adam and SDG optimizers.
| Learning Rate/Loss Function | Macro-Average | Micro-Average | COVID-19 | Normal | Macro-Average | Micro-Average | COVID-19 | Normal | |
|---|---|---|---|---|---|---|---|---|---|
| (1) ROC Evaluation Curve | |||||||||
| Adam Optimizer | SGD Optimizer | ||||||||
| Backbone | /Categorical cross-entropy | 0.81 | 0.82 | 0.82 | 0.82 | 0.92 | 0.92 | 0.92 | 0.92 |
| Proposed | 0.89 | 0.89 | 0.89 | 0.90 | 0.99 | 0.99 | 0.99 | 0.99 | |
| (2) Precision-Recall (AP) Evaluation Curve | |||||||||
| Backbone | /Categorical cross-entropy | 0.75 | 0.82 | 0.73 | 0.87 | 0.90 | 0.85 | ||
| Proposed | 0.85 | 0.90 | 0.82 | 0.98 | 0.99 | 0.97 | |||
4.3.2. Multi-class prediction Scenario: Normal vs. COVID-19 vs. Pneumonia
Table 9 presents the comparison of the results of the backbone model with those of the proposed model. The results are recorded for multi-class category performance; however, the performance was influenced by the reduced number of training sets because, the deep learning model requires larger training samples for better performance. In this study, the classification results of multi-class scenarios are derived using Adam and SGD optimizers, and learning rates, and categorical cross-entropy functions. The ROC and PR evaluation curves of the proposed and backbone models are derived using both the Adam and SGD optimizers as shown in Table 10 . It is clear that the COVID-19 class can be identified easily compared to other respiratory diseases, such as pneumonia, or even normal classes. This implies that the proposed model is also acceptable for practical applications in multi-class scenarios.
Table 9.
Comparison of the evaluation results of the multi-class scenario using the backbone model against the proposed approach.
| AI Model | Optimizer | Learning Rate/Loss Function | ACC | SEN | SPE | PRE | F1_Score | AUC |
|---|---|---|---|---|---|---|---|---|
| Backbone | Adam | /Categorical cross-entropy | 0.93 | 0.89 | 0.95 | 0.91 | 0.89 | 0.92 |
| Proposed | 0.96 | 0.93 | 0.98 | 0.93 | 0.93 | 0.95 | ||
| Backbone | Categorical cross-entropy | 0.88 | 0.82 | 0.91 | 0.85 | 0.82 | 0.87 | |
| Proposed | 0.95 | 0.93 | 0.96 | 0.93 | 0.92 | 0.94 | ||
| Backbone | SGD | /Categorical cross-entropy | 0.92 | 0.88 | 0.94 | 0.90 | 0.88 | 0.90 |
| Proposed | 0.95 | 0.92 | 0.96 | 0.93 | 0.92 | 0.94 | ||
| Backbone | Categorical cross-entropy | 0.79 | 0.69 | 0.84 | 0.80 | 0.69 | 0.77 | |
| Proposed | 0.93 | 0.89 | 0.95 | 0.91 | 0.90 | 0.90 |
Table 10.
Evaluation Performance of ROC and PR Curves for multi-class scenario depicting the backbone model when compared with the proposed model using the Adam and SDG optimizers.
| Learning Rate/Loss Function | Micro -Average | COVID-19 | Normal | Pneumonia | Micro -Average | COVID-19 | Normal | Pneumonia | |
|---|---|---|---|---|---|---|---|---|---|
| (1) ROC Evaluation Curve | |||||||||
| Adam Optimizer | SGD Optimizer | ||||||||
| Backbone | /Categorical cross-entropy | 0.92 | 0.97 | 0.93 | 0.86 | 0.94 | 0.93 | 0.95 | 0.95 |
| Proposed | 0.94 | 0.98 | 0.94 | 0.90 | 0.95 | 0.95 | 0.95 | 0.93 | |
| Backbone | /Categorical cross-entropy | 0.87 | 0.85 | 0.90 | 0.85 | 0.77 | 0.81 | 0.77 | 0.73 |
| Proposed | 0.95 | 0.95 | 0.96 | 0.94 | 0.92 | 0.89 | 0.92 | 0.95 | |
| (2) Precision-Recall (AP) Evaluation Curve | |||||||||
| Backbone | /Categorical cross-entropy | 0.83 | 0.94 | 0.77 | 0.82 | 0.87 | 0.89 | 0.82 | 0.93 |
| Proposed | 0.93 | 0.95 | 0.89 | 0.92 | 0.89 | 0.92 | 0.84 | 0.91 | |
| Backbone | /Categorical cross-entropy | 0.73 | 0.71 | 0.72 | 0.80 | 0.58 | 0.70 | 0.51 | 0.64 |
| Proposed | 0.93 | 0.95 | 0.93 | 0.90 | 0.83 | 0.83 | 0.77 | 0.93 | |
4.4. Discussions
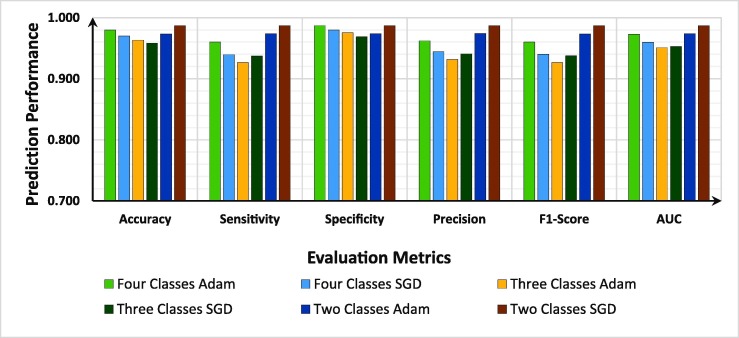
The capability of the proposed model was investigated by conducting a comprehensive evaluation under different scenarios: four (COVID-19 vs. Pneumonia vs. Lung Opacity vs. Normal), three (COVID-19 vs. Pneumonia vs. Normal), and two (COVID-19 vs. Normal). Each category was implemented using the Adam and SGD optimizers. We investigated the effect of the employed loss functions for Adam optimizer versus SGD optimizer and a learning rate of over . The experimental results indicated that the Adam optimizer performed better than SGD when trained with categorical cross-entropy loss and learning rate whereas the SGD optimizer performed significantly better when trained with categorical cross-entropy loss and learning rate. The results indicate that the proposed model exhibits better performance using the categorical cross-entropy loss; thus, it is neither overfitting nor overconfidence. As stated earlier, various studies have suggested that label smoothing prevents the model from overfitting; however, in our case, the use of batch normalization at the MLP block solved the overfitting issues; thus, the proposed model is robust in predicting the outcome. Fig. 7 shows the comparison of the evaluation results of this study in terms of precision, F1_score, sensitivity, accuracy, area under the ROC curve, and specificity.
Fig. 7.
Comparison of the evaluation results of the proposed model to optimize the training parameters via different dataset categories.
The proposed model uses patches and positional embedding, which enables it to concentrate on all the affected regions in a single patch while considering the possibilities for restoration. The concatenated features that serve as the backbone use a patch-and-attention strategy to obtain global features that assist and improve the prediction performance of the model. We also observed that using the SGD optimizer, the proposed model performed better with the label smoothing loss function technique. This is because, using easy targets, which are a cumulative total of the specific targets and homogeneous distribution across labels, may significantly increase the adaptability and learning pace of a multi-class deep learning model. This label smoothing rendered the network overly optimistic. However, because the proposed model's performance using label smoothing strategy works better with learning rate compared to , we need to first investigate the model hyper-parameter settings (such as learning rate and optimizer), which will be in accordance with the label smoothing optimal performance. The learning rate of the model has a greater influence on the model performance for particular loss functions and optimizers. Compared with other loss functions, the optimal models trained using categorical cross-entropy had the fewest trainable parameters. Based on our findings, the proposed model achieved the best performance by employing a categorical cross-entropy loss, Adam optimizer, and learning rate. By utilizing the appropriate parameters for the proposed approach, excellent classification results were achieved. This study highlights the significance of feature extraction and hyper-parameter adjustment for deep learning models before adding more sophisticated deep layers to the network structure.
We have summarized our discussion by presenting the optimized setting performance of the proposed model on the two optimizers for both binary and multi-class (four and three classes) scenarios as shown in Fig. 7. Our research could be useful for the rapid formation of approachable AI frameworks for swift, accurate, and economical identification of COVID-19 infections.
4.4.1. Ablation study of the proposed model
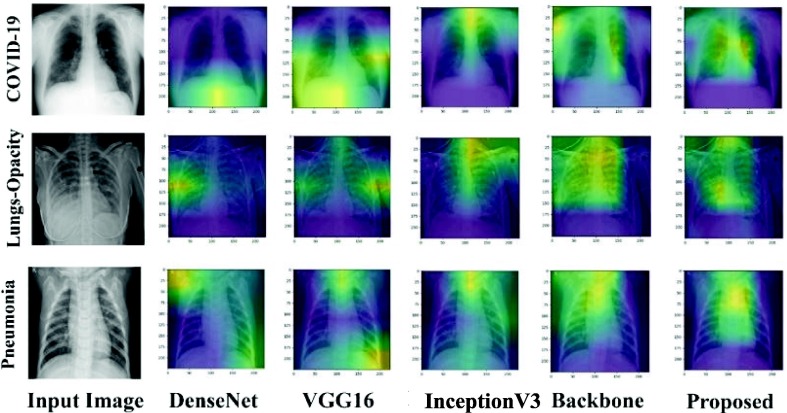
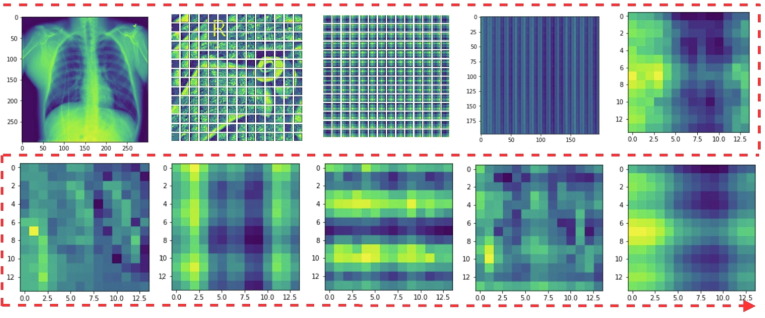
The visually explicable results in terms of heat maps or saliency maps could provide a better interpretation of the disease locations that are focused by the deep learning model to provide the final prediction decision. Such results are important for providing logical reasons for decisions taken using the black model. In addition, the visual results can make models readable and understandable in terms of delivering more explicable clinical results to the end user. In this study, we used Grad-CAM to generate saliency maps for the input chest X-ray images. It uses the gradients from the target idea and feeds them into the final convolution layer of deep learning models. Fig. 8 depicts some of the heat maps generated for each deep learning model in comparison with the proposed model. It is clearly shown in Fig. 8 that the proposed model achieved the best visually explicable results in terms of identifying the proper location of respiratory diseases. It shows reliable and feasible visual saliency maps compared to individual deep learning models (i.e., DenseNet, VGG16, or InceptionV3) or even the backbone model (concatenated features). Fig. 9 summarizes the internal working structure of the proposed model attention mechanism, which assists the visual semantic features used for the model's predictions.
Fig. 8.
Visual explainable saliency maps of the proposed model against other individual ones. These depicted heat maps are generated using the Grad-CAM technique for the abnormal images: COVID-19, Lung-Opacity, and Pneumonia.
Fig. 9.
Visual feature information generated based on the internal structure of the proposed model starting from the input image, patch splitting of the input image, addition of positional embedding to the patches, and different visual features of the employed attention mechanism.
4.4.2. Comparison of evaluation results with the state-of-the-art deep learning models
This section presents a comparison of the results obtained in this study with those by other researchers using the same dataset. To determine the efficacy of the proposed model, it is reasonable to compare it with other models that use the same dataset. We compared the results based on the given task (i.e., class-class classification, three-class classification, and binary classification). For multi-classification involving four classes (COVID-19 vs. pneumonia vs. lung opacity vs. normal), we computed the F1_Score, accuracy, sensitivity, and precision evaluation metrics as shown in Table 11 . The results obtained are then compared with those reported in other studies. Wang et al. introduced the use of COVIDNet network (Wang et al., 2020, Wang et al., 2020) whereas (Khan et al., 2020) proposed the CoroNet for the same task. Based on their reported result, the COVIDNet architecture had a better result in terms of accuracy, precision, and F1_Score (90.78 vs 89.60%, 91.10 vs. 90.0%, and 90.81 vs. 89.80%) while in terms of sensitivity, the CoroNet result was better (96.40 vs. 90.56%). Additionally, both models explored the use of pre-trained models similar to our research on enhanced feature extraction (initially trained on the ImageNet dataset). In accordance with our earlier claims that CNN layers overlook some vital features of the input images (Yang, 2020, Mondal, 2022, Lee et al., 2019, Shi et al., 2021) employed an attention mechanism, which is also in line with our proposed model, to improve the features extracted from the convolutional layers. Shi et al. termed this approach as the teacher-student attention (Shi et al., 2021). The results exhibited an accuracy of 91.38%, which is higher than previously implemented approaches. This approach referred as a multiscale attention-guided network. The reported result was higher than that reported by (Shi et al., 2021) for all evaluation metrics, with an accuracy of 92.35%.
Table 11.
Comparison of evaluation performance of the proposed model against the state-of-the-art models that used the same dataset. This comparison is conducted using the multi-class prediction scenario with four respiratory class diseases: COVID-19 vs. Pneumonia vs. Lung Opacity vs. Normal.
| Reference | AI Architecture | ACC | PRE | SEN | F1_Score |
|---|---|---|---|---|---|
| (Khan et al., 2020) | CoroNet | 0.89 | 0.90 | 0.96 | 0.90 |
| (Wang et al., 2020, Wang et al., 2020) | COVIDNet | 0.90 | 0.91 | 0.91 | 0.91 |
| (Lee et al., 2019), | Multiscale Attention Guided Network | 0.92 | 0.93 | 0.92 | 0.92 |
| (Shi et al., 2021) | Teacher Student Attention | 0.91 | 0.92 | 0.91 | 0.91 |
| (Mondal 2022) | Local Global Attention Network | 0.96 | 0.96 | 0.96 | 0.96 |
| (Khan et al., 2022a, Khan et al., 2022b) (Strategy 1) | EfficientNetB1 | 0.92 | 0.92 | 0.95 | 0.93 |
| NasNetMobile | 0.89 | 0.89 | 0.92 | 0.91 | |
| MobileNetV2 | 0.90 | 0.92 | 0.92 | 0.92 | |
| (Khan et al., 2022a, Khan et al., 2022b) (Strategy 2) | EfficientNetB1 | 0.96 | 0.97 | 0.97 | 0.98 |
| NasNetMobile | 0.95 | 0.95 | 0.95 | 0.95 | |
| MobileNetV2 | 0.94 | 0.94 | 0.95 | 0.95 | |
| Ours (2023) | The Proposed XAI Model | 0.98 | 0.96 | 0.96 | 0.96 |
Mondal et al. introduced a local–global attention network that performed better than the previously implemented models (accuracy = 95.87%, sensitivity = 95.99%, precision = 95.56%, and F1_score = 95.74% (Mondal 2022). Khan et al. tackled the issue of multi-classification of COVID-19 disease with other lung diseases using two strategies (Khan et al., 2020). However, both strategies involved the use of TL from pretrained models, such as EfficiientNetB1, NasNetMobile, and MobileNetV2.) The results of strategy two were higher compared to those of strategy one with the EfficientNetB1 having the best performance in terms of F1_score, accuracy, sensitivity, and precision (97.50%, 96.13%, 96.50%, and 97.25%). However, the results of second strategy by (Khan et al., 2022a, Khan et al., 2022b) using the were slightly higher than the results obtained in this study, except for accuracy where we had a better result with 98% against 96%, which means that the proposed classifier rarely identifies a COVID-19 negative sample as a positive sample.
To compare models that used the same dataset as ours in a three-class scenario (i.e., COVID-19 vs. pneumonia vs. normal), we deployed the experimental models presented by (Pham 2020), which are Inception-V3 and MobileNetV2. The MobileNetV2 architecture yielded the best classification performance. However, the Inception-V3 architecture’s sensitivity performance was superior to that of MobileNetV2 with 0.91 vs. 0.89. Popular pre-trained models were used to perform the COVID-19 classification task in a modified version. Luz et al. employed EfficientNet-B0 (Luz et al., 2021), Wu et al. employed the ResNet-V2 model (Wu et al., 2020), Perumal et al. employed the VGG-16 architecture (Perumal et al., 2021), Khan et al. employed the Xception model (Khan et al., 2020), and Oh et al. employed the DenseNet-121 architecture (Oh et al., 2020). Among the architectures employed, Xception architecture recorded the highest accuracy of 0.90, precision of 0.92, and an F1_score of 0.90. However, its sensitivity was poor (0.87). EfficientNet-Bo yielded the best sensitivity of 0.89. Attention techniques have received considerable interest in computer vision, especially in the medical field. Shome et al. (Shome et al., 2021) used the COVID-transformer network to attain accuracy, precision, sensitivity, and F1_score of 0.92, 0.93, 0.89, and 0.91, respectively.
Furthermore, in line with our proposed approach, (Chowdhury et al., 2021) introduced an ensemble-based architecture using EfficientNet and a CNN termed ECOVNet. The EfficientNet model serves as the feature extractor because it is trained on a large ImageNet Dataset. Following several model frames, the derived attributes were moved to a specially designed fine-tuned layer. The projections of the model frames were aggregated using two ensemble techniques to boost classification performance. (Chowdhury et al., 2021) recorded the same performance results as our proposed model, except for accuracy, where we obtained a better result. Chakraborty et al. (Chakraborty et al., 2022) suggested the use of ResNet18 via TL based on the idea that ResNet18 uses the idea of skip connections in the deep layers to address the issue of disappearing gradients, thus recording accuracy, sensitivity, and F1_score of 96, 94, and 93%, respectively. Perumal et al. combined InceptionNet and CNN models to search for fast and robust predictions (Perumal et al., 2022). This approach recorded a 94% of all the evaluation metrics. According to (Ho et al., 2022), merging shallow handmade features, texture-based features, and feature levels from models that have already been trained is effective and performs better than methods that simply employ these features separately. Thus, the proposed model exhibits better performance with F1_score, accuracy, sensitivity, and precision of 0.95, 0.96, 0.95, and 0.95, respectively. Table 12 summarizes this comparison using recently published findings.
Table 12.
Multi-class prediction task comparison of three respiratory class diseases: COVID-19 vs. Pneumonia vs. Normal.
| Reference | AI Architecture | ACC | PRE | SEN | F1_Score |
|---|---|---|---|---|---|
| (Pham 2020) | Inception-V3 | 0.90 | 0.89 | 0.91 | 0.89 |
| MobileNet-V2 | 0.90 | 0.90 | 0.89 | 0.90 | |
| (Wu et al., 2020) | ResNet-V2 | 0.88 | 0.87 | 0.86 | 0.86 |
| (Khan et al., 2020) | Xception | 0.90 | 0.92 | 0.87 | 0.90 |
| (Oh et al., 2020) | DenseNet-121 | 0.88 | 0.90 | 0.85 | 0.87 |
| (Luz et al., 2021), | EfficientNet-B0 | 0.89 | 0.88 | 0.89 | 0.88 |
| (Perumal et al., 2021) | VGG-16 | 0.87 | 0.87 | 0.85 | 0.86 |
| (Shome et al., 2021) | COVID-Transformer | 0.92 | 0.93 | 0.89 | 0.91 |
| (Chowdhury et al., 2021) | ECOVNet (pre-trained EfficientNet) | 0.95 | 0.95 | 0.95 | 0.95 |
| (Chakraborty et al., 2022) | ResNet18 + TransferLearning | 0.96 | – | 0.94 | 0.93 |
| (Perumal et al., 2022) | INASNET (Inception Nasnet) | 0.94 | 0.94 | 0.94 | 0.94 |
| (Ho et al., 2022) | Feature-based Ensemble | 0.94 | 0.95 | 0.94 | 0.94 |
| Ours (2023) | The Proposed XAI Model | 0.96 | 0.95 | 0.95 | 0.95 |
Considering a more compact classification task (Binary Classification), (Sethy et al., 2020) employed ResNet50 + SVM classifiers and achieved accuracy and F1_score of 95.33% and 95.34%, respectively as shown in Table 13 . Ismael et al. used an SVM classifier and several pretrained models as feature extractors (leveraging the pretrained weights from ImageNet) (Ismael et al., 2021). Among the several implemented models, the ResnET50 Feature extractor + SVM classifier exhibited the highest accuracy of 94.7%. Likewise, Hemdan et al. employed several deep learning models (Hemdan et al., 2020). Among them, the performances of VGG19 and DenseNet201 were a highlight with F1_score, recall, precision and accuracy of 91.5%, 90%, 90%, and 92%, respectively. Sahinbas et al. are of the opinion that the VGG16 architecture is not an efficient detector of COVID-19 using chest X-Ray images based on their experimental result (Sahinbas et al., 2021).
Table 13.
Binary prediction task scenario with two respiratory classes: COVID-19 vs. Normal.
| Reference | AI Architecture | ACC | PRE | SEN | F1_Score |
|---|---|---|---|---|---|
| (Sethy et al., 2020) | ResNet50 + SVM | 0.95 | 0.95 | – | – |
| (Hemdan et al., 2020) | VGG-19 | 0.90 | 0.90 | 0.92 | 0.90 |
| DenseNet201 | 0.90 | 0.90 | 0.92 | 0.92 | |
| (Kumar et al., 2021) | DeQueezeNet | 0.95 | – | 0.90 | 0.97 |
| (Jaiswal et al., 2020) | COVIDPEN | 0.96 | 0.94 | 0.92 | 0.96 |
| (Ismael et al., 2021) | ResNet50 + SVM | 0.95 | 0.95 | – | 0.91 |
| (Das et al., 2021) | DenseNet201 + Resnet50V2 + Inceptionv3 | 0.96 | – | – | 0.96 |
| (Sahinbas et al., 2021) | VGG16 | 0.80 | 0.80 | 0.80 | 0.80 |
| (Ji et al., 2021) | VGG19 + Xception + ResNet152 + InceptionResNetV2 | 0.96 | 0.96 | 0.96 | 0.96 |
| (Khan et al., 2022a, Khan et al., 2022b) | New Channel Boosted CNN | 0.96 | – | – | 0.95 |
| Ours (2023) | Proposed Model | 0.99 | 0.99 | 0.99 | 0.99 |
Das et al. achieved 91.62% accuracy using a cascade network comprising DenseNet201, Inception v3, and ResNet50V2 (Das et al., 2021). Kumar et al. suggested DeQueezeNet (integration of DenseNet121 and SqueezeNet features), which achieved 94.52% precision and 90.48% accuracy (Kumar et al., 2021). Jaiswal et al. introduced a strategy-based TL using the EfficientNet framework with an accuracy of 96% and lower F1_score, precision, and recall performance (Jaiswal et al., 2020). Ensemble network is used by (Ji et al., 2021) in two scenarios, in which the model achieved a percentage classification accuracy of over 96%. A recently published study by (Khan et al., 2022a, Khan et al., 2022b) used a new channel-addition strategy to boost the performance of CNN Models. Based on their reported findings, the proposed model yielded an accuracy of 96% and F1_score of 95%. Thus, based on our reported results, the proposed model can discriminate COVID-19 positive instances more accurately than the state-of-the-art models in all class instances.
5. Conclusion, limitations, and future works
COVID-19 identification is a vital phase in the epidemic diagnosis, and numerous computer-aided analytical methodologies have recently been applied for faster and more accurate analyses. Effective extraction of chest X-ray characteristics contributes to the accurate diagnosis of COVID-19, thus allowing for early detection and therapy. In this study, we presented an end-to-end deep learning framework with excellent capability to generate and extract strong deep features via a backbone feature concatenation strategy. Meanwhile, the attention mechanism employed was a multi-head Self-attention network with an MLP block that was adapted specifically for better prediction performance of COVID-19 identification. First, the original chest X-ray images were processed using pretrained models (DenseNet, VGG-16, and InceptionV3) trained on a large-scale ImageNet dataset that considered generic attributes. Next, the model features were concatenated at the last convolutional layer. The attention network was then used to analyze the regional characteristics before the generated features were processed through a novel MLP block for improved, more accurate, and generalized prediction. A comprehensive experiment was conducted using dual optimizers, learning rates, and loss functions to verify the robustness of the model and develop an optimum setup for the proposed model. We trained the model in an end-to-end manner using the COVID-19 radiography dataset and evaluated it using several evaluation metrics. The proposed model performed well in the experiments, thus yielding sensitivity, specificity, precision, accuracy, F1_score, and ROC score of 96.01%, 98.67%, 96.20%, 98.00%, 96.03%, and 97.34%, respectively. Furthermore, the proposed model's binary and multi-class (three classes) classification performance as well as that of the six pre-trained models were investigated in this study. In accordance with our findings, the proposed model performed better than all the other suggested networks, providing superior classification performance outcomes when compared to other recent models that used the same dataset. The attention visualization approach validated the multi-head Self-attention model's capacity to learn correct characteristics rather than features of the imaging procedure itself or other pointless data because the choice was made based on the supplied generic and regional aspects of the chest X-ray images.
The proposed model can be used to classify additional medical imaging modalities, such as magnetic resonance imaging, ultrasound, and CT for the prediction of skin cancer, breast cancer, oral cancer, and pneumonia. It is noteworthy that the experiment was evaluated using only chest X-ray images. Regarding the issue of accurate feature extraction for computer vision tasks, research has shown that features of palm print with a two-dimensional discrete cosine transform to constitute a dual-source space resulted in improved performance of recognition models. In addition, deep learning models are prone to adversarial attacks; hence, security guidance is necessary. We intend to integrate these ideas into a model, which is a part of future research.
Code Availability/Availability of Data
The dataset used in this study is publicly accessible at https://www.kaggle.com/datasets/tawsifurrahman/covid19-radiography-database. The TensorFlow/Keras code will be made available upon request from the corresponding author.
Ethical Approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (No. RS-2022-00166402).
Footnotes
Peer review under responsibility of King Saud University.
References
- Abadi, M., et al., 2016. Tensorflow: Large-scale machine learning on heterogeneous distributed systems. arXiv preprint arXiv:1603.04467.
- Abubakar, H.S., et al., 2022. 3D-Based facial emotion recognition using depthwise separable convolution. In: 2022 14th International Conference on Machine Learning and Computing (ICMLC).
- Al-antari M.A., et al. Fast deep learning computer-aided diagnosis of COVID-19 based on digital chest x-ray images. Appl. Intell. 2021;51(5):2890–2907. doi: 10.1007/s10489-020-02076-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-antari, M. A., et al., 2021. A rapid deep learning computer-aided diagnosis to simultaneously detect and classify the novel COVID-19 pandemic. In: 2020 IEEE-EMBS Conference on Biomedical Engineering and Sciences (IECBES), IEEE.
- Alimadadi A., et al. American Physiological Society; Bethesda MD: 2020. Artificial Intelligence and Machine Learning to Fight COVID-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostolopoulos I.D., et al. Covid-19: automatic detection from x-ray images utilizing transfer learning with convolutional neural networks. Phys. Eng. Sci. Med. 2020;43:635–640. doi: 10.1007/s13246-020-00865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai D., et al. Anti-occlusion multi-object surveillance based on improved deep learning approach and multi-feature enhancement for unmanned smart grid safety. Energy Rep. 2023;9:594–603. [Google Scholar]
- Cengil E., et al. The effect of deep feature concatenation in the classification problem: An approach on COVID-19 disease detection. Int. J. Imaging Syst. Technol. 2022;32(1):26–40. doi: 10.1002/ima.22659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty S., et al. An efficient deep learning model to detect COVID-19 using chest X-ray images. Int. J. Environ. Res. Public Health. 2022;19(4):2013. doi: 10.3390/ijerph19042013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., et al. Bone suppression of chest radiographs with cascaded convolutional networks in wavelet domain. IEEE Access. 2019;7:8346–8357. [Google Scholar]
- Chollet, F., 2017. Xception: Deep learning with depthwise separable convolutions. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition.
- Chowdhury M.E., et al. Can AI help in screening viral and COVID-19 pneumonia? IEEE Access. 2020;8:132665–132676. [Google Scholar]
- Chowdhury N.K., et al. ECOVNet: a highly effective ensemble based deep learning model for detecting COVID-19. PeerJ Comput. Sci. 2021;7:e551. doi: 10.7717/peerj-cs.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danet A.D. Psychological impact of COVID-19 pandemic in Western frontline healthcare professionals. A systematic review. Medicina Clínica (English Edition) 2021;156(9):449–458. doi: 10.1016/j.medcle.2020.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A.K., et al. Automatic COVID-19 detection from X-ray images using ensemble learning with convolutional neural network. Pattern Anal. Appl. 2021;24:1111–1124. doi: 10.1007/s10044-021-00984-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doo E.Y., et al. Influence of anxiety and resilience on depression among hospital nurses: a comparison of nurses working with confirmed and suspected patients in the COVID-19 and non-COVID-19 units. J. Clin. Nurs. 2021;30(13–14):1990–2000. doi: 10.1111/jocn.15752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farooq, et al., 2020. COVID-ResNet: A Deep Learning Framework for Screening of COVID19 from Radiographs. arXiv:2003.14395 [eess.IV] 2003(14395).
- Gupta, P., et al., 2018. Using deep learning to enhance head and neck cancer diagnosis and classification. In: 2018 IEEE International Conference on System, Computation, Automation and Networking (icscan), IEEE.
- Hemdan, E. E.-D., et al., 2020. Covidx-net: A framework of deep learning classifiers to diagnose covid-19 in x-ray images. arXiv preprint arXiv:2003.11055.
- Ho T.K.K., et al. Feature-level ensemble approach for COVID-19 detection using chest X-ray images. PLoS One. 2022;17(7):e0268430. doi: 10.1371/journal.pone.0268430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard, A. G., et al., 2017. Mobilenets: Efficient convolutional neural networks for mobile vision applications. arXiv preprint arXiv:1704.04861.
- Hussain L., et al. Machine-learning classification of texture features of portable chest X-ray accurately classifies COVID-19 lung infection. Biomed. Eng. Online. 2020;19:1–18. doi: 10.1186/s12938-020-00831-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismael A.M., et al. Deep learning approaches for COVID-19 detection based on chest X-ray images. Expert Syst. Appl. 2021;164 doi: 10.1016/j.eswa.2020.114054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobi A., et al. Portable chest X-ray in coronavirus disease-19 (COVID-19): A pictorial review. Clin. Imaging. 2020;64:35–42. doi: 10.1016/j.clinimag.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal, A. K., et al., 2020. Covidpen: A novel covid-19 detection model using chest x-rays and ct scans. Medrxiv: 2020.2007. 2008.20149161.
- Ji D., et al. Research on classification of COVID-19 chest X-ray image modal feature fusion based on deep learning. J. Healthcare Eng. 2021;2021 doi: 10.1155/2021/6799202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A.I., et al. CoroNet: A deep neural network for detection and diagnosis of COVID-19 from chest x-ray images. Comput. Methods Programs Biomed. 2020;196 doi: 10.1016/j.cmpb.2020.105581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan E., et al. Chest X-ray classification for the detection of COVID-19 using deep learning techniques. Sensors. 2022;22(3):1211. doi: 10.3390/s22031211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S.H., et al. COVID-19 detection in chest X-ray images using a new channel boosted CNN. Diagnostics. 2022;12(2):267. doi: 10.3390/diagnostics12020267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., et al. Deep Transfer Learning-based COVID-19 prediction using Chest X-rays. J. Health Manag. 2021;23(4):730–746. [Google Scholar]
- Kumar A., et al. SARS-Net: COVID-19 detection from chest x-rays by combining graph convolutional network and convolutional neural network. Pattern Recogn. 2022;122 doi: 10.1016/j.patcog.2021.108255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakshmanaprabu S., et al. Optimal deep learning model for classification of lung cancer on CT images. Futur. Gener. Comput. Syst. 2019;92:374–382. [Google Scholar]
- Lee H., et al. Interpretability of Machine Intelligence in Medical Image Computing and Multimodal Learning for Clinical Decision Support. Springer:; 2019. Generation of multimodal justification using visual word constraint model for explainable computer-aided diagnosis; pp. 21–29. [Google Scholar]
- Leng L., et al. Dual-source discrimination power analysis for multi-instance contactless palmprint recognition. Multimed. Tools Appl. 2017;76:333–354. [Google Scholar]
- Liu, S., et al., 2015. Very deep convolutional neural network based image classification using small training sample size. In: 2015 3rd IAPR Asian conference on pattern recognition (ACPR), IEEE.
- Luz E., et al. Towards an effective and efficient deep learning model for COVID-19 patterns detection in X-ray images. Res. Biomed. Eng. 2021:1–14. [Google Scholar]
- Minaee, et al. Deep-COVID: Predicting COVID-19 from chest X-ray images using deep transfer learning. Med. Image Anal. 2020;65 doi: 10.1016/j.media.2020.101794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondal A.K. COVID-19 prognosis using limited chest X-ray images. Appl. Soft Comput. 2022;122 doi: 10.1016/j.asoc.2022.108867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monshi M.M.A., et al. CovidXrayNet: Optimizing data augmentation and CNN hyperparameters for improved COVID-19 detection from CXR. Comput. Biol. Med. 2021;133 doi: 10.1016/j.compbiomed.2021.104375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak S.R., et al. Application of deep learning techniques for detection of COVID-19 cases using chest X-ray images: A comprehensive study. Biomed. Signal Process. Control. 2021;64 doi: 10.1016/j.bspc.2020.102365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novitasari D.C.R., et al. Detection of COVID-19 chest X-ray using support vector machine and convolutional neural network. Commun. Math. Biol. Neurosci. 2020;2020 Article ID 42. [Google Scholar]
- Oh Y., et al. Deep learning COVID-19 features on CXR using limited training data sets. IEEE Trans. Med. Imaging. 2020;39(8):2688–2700. doi: 10.1109/TMI.2020.2993291. [DOI] [PubMed] [Google Scholar]
- Ortiz A., et al. Ensembles of deep learning architectures for the early diagnosis of the Alzheimer’s disease. Int. J. Neural Syst. 2016;26(07):1650025. doi: 10.1142/S0129065716500258. [DOI] [PubMed] [Google Scholar]
- Pereira R.M., et al. COVID-19 identification in chest X-ray images on flat and hierarchical classification scenarios. Comput. Methods Programs Biomed. 2020;194 doi: 10.1016/j.cmpb.2020.105532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perumal V., et al. Detection of COVID-19 using CXR and CT images using Transfer Learning and Haralick features. Appl. Intell. 2021;51:341–358. doi: 10.1007/s10489-020-01831-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perumal M., et al. INASNET: Automatic identification of coronavirus disease (COVID-19) based on chest X-ray using deep neural network. ISA Trans. 2022;124:82–89. doi: 10.1016/j.isatra.2022.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham T.D. A comprehensive study on classification of COVID-19 on computed tomography with pretrained convolutional neural networks. Sci. Rep. 2020;10(1):1–8. doi: 10.1038/s41598-020-74164-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahimzadeh M., et al. A modified deep convolutional neural network for detecting COVID-19 and pneumonia from chest X-ray images based on the concatenation of Xception and ResNet50V2. Inf. Med. Unlocked. 2020;19 doi: 10.1016/j.imu.2020.100360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajaraman S., et al. Weakly labeled data augmentation for deep learning: a study on COVID-19 detection in chest X-rays. Diagnostics. 2020;10(6):358. doi: 10.3390/diagnostics10060358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajpal S., et al. Using handpicked features in conjunction with ResNet-50 for improved detection of COVID-19 from chest X-ray images. Chaos Solitons Fractals. 2021;145 doi: 10.1016/j.chaos.2021.110749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saba L., et al. Six artificial intelligence paradigms for tissue characterisation and classification of non-COVID-19 pneumonia against COVID-19 pneumonia in computed tomography lungs. Int. J. Comput. Assist. Radiol. Surg. 2021;16:423–434. doi: 10.1007/s11548-021-02317-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahinbas K., et al. Data science for COVID-19. Elsevier; 2021. Transfer learning-based convolutional neural network for COVID-19 detection with X-ray images; pp. 451–466. [Google Scholar]
- Sethy, P. K., et al., 2020. Detection of coronavirus disease (covid-19) based on deep features.
- Shi W., et al. COVID-19 automatic diagnosis with radiographic imaging: explainable attention transfer deep neural networks. IEEE J. Biomed. Health Inform. 2021;25(7):2376–2387. doi: 10.1109/JBHI.2021.3074893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shome D., et al. Covid-transformer: Interpretable covid-19 detection using vision transformer for healthcare. Int. J. Environ. Res. Public Health. 2021;18(21):11086. doi: 10.3390/ijerph182111086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares, L. P., et al., 2020. Automatic detection of covid-19 cases on x-ray images using convolutional neural networks. arXiv preprint arXiv:2007.05494.
- Ucar F., et al. COVIDiagnosis-Net: Deep Bayes-SqueezeNet based diagnosis of the coronavirus disease 2019 (COVID-19) from X-ray images. Med. Hypotheses. 2020;140 doi: 10.1016/j.mehy.2020.109761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ukwuoma C.C., et al. Multi-classification of breast cancer lesions in histopathological images using DEEP_Pachi: multiple self-attention head. Diagnostics. 2022;12(5):1152. doi: 10.3390/diagnostics12051152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ukwuoma C.C., et al. Dual_Pachi: attention-based dual path framework with intermediate second order-pooling for Covid-19 detection from chest X-ray images. Comput. Biol. Med. 2022;151 doi: 10.1016/j.compbiomed.2022.106324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ukwuoma C.C., et al. LCSB-inception: reliable and effective light-chroma separated branches for Covid-19 detection from chest X-ray images. Comput. Biol. Med. 2022;150 doi: 10.1016/j.compbiomed.2022.106195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ukwuoma C.C., et al. A hybrid explainable ensemble transformer encoder for pneumonia identification from chest X-ray images. J. Adv. Res. 2022 doi: 10.1016/j.jare.2022.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waheed A., et al. Covidgan: data augmentation using auxiliary classifier gan for improved covid-19 detection. IEEE Access. 2020;8:91916–91923. doi: 10.1109/ACCESS.2020.2994762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., et al. COVID-Net: a tailored deep convolutional neural network design for detection of COVID-19 cases from chest X-ray images. Sci Rep. 2020;10:19549. doi: 10.1038/s41598-020-76550-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L., et al., 2020. COVID-Net: A tailored deep convolutional neural network design for detection of COVID-19 cases from chest radiography images. arXiv preprint arXiv:2003.09871. [DOI] [PMC free article] [PubMed]
- Wu, X., et al., 2020. Recognizing COVID-19 positive: through CT images. In: 2020 Chinese Automation Congress (CAC), IEEE.
- Yang, X., 2020. An overview of the attention mechanisms in computer vision. Journal of Physics: Conference Series, IOP Publishing.
- Zheng Q., et al. Improvement of generalization ability of deep CNN via implicit regularization in two-stage training process. IEEE Access. 2018;6:15844–15869. [Google Scholar]
- Zheng Q., et al. Layer-wise learning based stochastic gradient descent method for the optimization of deep convolutional neural network. J. Intell. Fuzzy Syst. 2019;37(4):5641–5654. [Google Scholar]
- Zheng Q., et al. PAC-Bayesian framework based drop-path method for 2D discriminative convolutional network pruning. Multidim. Syst. Sign. Process. 2020;31(3):793–827. [Google Scholar]