Abstract
Infection of cells by many viruses affects the cell division cycle of the host cell to favor viral replication. We examined the ability of the paramyxovirus simian parainfluenza virus 5 (SV5) to affect cell cycle progression, and we found that SV5 slows the rate of proliferation of HeLa T4 cells. The SV5-infected cells had a delayed transition from G1 to S phase and prolonged progression through S phase, and some of the infected cells were arrested in G2 or M phase. The levels of p53 and p21CIP1 were not increased in SV5-infected cells compared to mock-infected cells, suggesting that the changes in the cell cycle occur through a p53-independent mechanism. However, the phosphorylation of the retinoblastoma protein (pRB) was delayed and prolonged in SV5-infected cells. The changes in the cell cycle were also observed in cells expressing the SV5 V protein but not in the cells expressing the SV5 P protein or the V protein lacking its unique C terminus (VΔC). The unique C terminus of the V protein of SV5 was shown previously to interact with DDB1, which is the 127-kDa subunit of the multifunctional damage-specific DNA-binding protein (DDB) heterodimer. The coexpression of DDB1 with V can partially restore the changes in the cell cycle caused by expression of the V protein.
Infection of cells by many viruses affects the cell division cycle of the host cell to favor viral replication. The DNA tumor viruses promote progression through the cell cycle by the specific interaction of viral and cellular proteins. For example, it is known that the simian virus 40 large T antigen protein, adenovirus E1A protein, and papillomavirus E7 protein interact with the cellular tumor repressor protein, retinoblastoma protein (pRB) to allow pRB to release the transcription factor E2F from a pRB-E2F complex (11, 19, 46, 77). Release of E2F from the pRB-E2F complex permits E2F-mediated transactivation of genes necessary for DNA replication and cell proliferation and promoting the entry of the host cell into S phase (reviewed in reference 49). Other viruses encode proteins which block cell cycle progression: human cytomegalovirus UL69 protein prevents progression from G1 to S phase (15, 23, 45), herpes simplex virus blocks G1 to S phase progression by blocking pRB phosphorylation (20, 70) and the human immunodeficiency virus Vpr protein causes cells to accumulate in G2-M phase by preventing cyclin B/cdc2 activation (26, 60, 61). Less is known about the interaction of RNA viruses with the host cell cycle, but it is known that measles virus, a paramyxovirus, causes G1 arrest in virus-infected T lymphocytes (47).
Simian virus 5 (SV5) is a prototype of the genus Rubulavirus of the virus family Paramyxoviridae which includes Sendai virus, human parainfluenza virus types 1 to 4, mumps virus, Newcastle disease virus, and measles virus. The paramyxoviruses are enveloped viruses with a nonsegmented, single-stranded, negative-sense RNA genome of ∼15,000 nucleotides. The SV5 genome contains 15,246 nucleotide, and it has eight proteins encoded from seven genes: the V and P proteins (50, 55) are both transcribed from the V/P gene by a process termed pseudotemplated transcription (54, 73). The SV5 V mRNA is a faithful transcript of the V/P gene, whereas the P mRNA contains two nontemplated G residues which are added cotranscriptionally by the viral polymerase as it stutters at a specific site, 3′-AAAAUUCU-5′, located just upstream of the G nucleotide insertion site (73, 75) (reviewed in reference 36). The consequence of the addition of two G residues to create the P mRNA is that the translational reading frame is changed relative to the V protein. SV5 V and P proteins share an N-terminal domain of 164 amino acids but have unique C-terminal domains (73). The process of pseudotemplated transcription at a specific site in the V/P gene occurs for almost all members of the subfamily Paramyxovirinae. However, whereas for the rubulaviruses the V mRNA is transcribed directly from the genome RNA, for the respiroviruses and the morbilliviruses the P mRNA is transcribed directly from the genome RNA and the V mRNA contains the additional pseudotemplated G nucleotide(s) (reviewed in reference 29). The sequence of the unique C-terminal domain of the V proteins is highly conserved among the paramyxoviruses. It contains seven cysteine residues, reminiscent of a zinc finger domain (73), and it has been shown that the measles virus, SV5, Newcastle disease virus V proteins bind atomic zinc (43, 52, 72).
The SV5 V protein is incorporated into virions (∼350 molecules per virion), and it is found associated with the nucleocapsid (52). However, for Sendai virus and measles virus the V protein does not appear to be incorporated into virions. The SV5 V protein has been shown to interact with soluble NP (58) and the shared N-terminal domain of V and P has been shown to bind RNA through a basic region (41). Despite extensive effort recovery of SV5 from an infectious cDNA (25) containing a deletion of the V gene has not been possible (B. He, unpublished observations), suggesting an essential role for the V protein in the SV5 life cycle. Recently, it has been shown that the SV5 V protein is involved in inhibition of the interferon pathway by targeting the transcription factor STAT-1, directly or indirectly, for proteosome-mediated degradation (14). Recombinant Sendai virus, measles virus, and rinderpest virus unable to synthesize the V protein [V(−) viruses] have been recovered from cloned DNA, indicating that the V protein is not essential for these viruses for replication in tissue culture (3, 13, 30, 64). However, the V proteins are important for pathogenicity of these viruses in animals (3, 30, 31, 74).
The SV5 V protein has been shown to interact, via its C-terminal zinc binding domain, with a cellular protein (DDB1), the 127-kDa subunit of the damage-specific DNA-binding protein (DDB; also known as the UV-damaged DNA binding protein [UV-DDB], xeroderma pigmentosum group E binding factor [XPE-BF], and the hepatitis B virus X-associated protein 1 [XAP-1]) (42). Purified DDB consists of a weakly associated heterodimer of a 127-kDa subunit (DDB1) and a 48-kDa subunit (DDB2) (33). The DDB complex has been found to interact with the transcription factor E2F1 and, in in vitro assays, DDB was found to overcome the inhibition by pRB of E2F1-mediated transactivation (24). It has been suggested that when damaged DNA is present in a cell, DDB binds to the damaged DNA and no longer interacts with E2F1 (24). As the E2F family of transcription factor proteins have been found to have a very important role in the G1-S phase transition, it has been suggested that the lack of interaction with DDB may be important in slowing down cell growth during the repair process (24). DDB has also been found to interact with CUL-4A (68), a member of the cullin family of proteins. The function of CUL-4A is not known, but CUL-1 and CUL-3 are involved in the ubiquitin-mediated degradation of cyclin E (reviewed in reference 79) and cyclin D and p21CIP (80).
We report here that infection of HeLa T4 cells with SV5 slows their proliferation. SV5-infected cells have a prolonged G1-S phase transition and S phase, and some of the infected cells are arrested in G2-M phase. The changes in the cell cycle were also observed in cells expressing the V protein but were not observed in cells expressing the P protein or a V protein lacking its unique C terminus of V (VΔC). The changes in the cell cycle can be partially restored by coexpression of DDB1. Thus, although the functions of the SV5 V protein are not fully elucidated, the V protein appears to be a multifunctional protein.
MATERIALS AND METHODS
Cells and viruses.
HeLa T4 cells were grown in Dulbecco modified Eagle's medium (DMEM) (Gibco BRL, Gaithersburg, Md.), containing 10% fetal bovine serum (FBS), and penicillin and streptomycin (P/S). MDBK cells were grown as described previously (51). The W3A strain of SV5 was grown in MDBK cells as described previously (55).
Plasmids.
The SV5 V and P cDNA and the VΔC mutant (deletion of residues 168 to 222 of the V protein) were subcloned into pCAGGS-MCS (48) from pGEM2-P/V-203, pGEM2-P/V103 (73), and pGEM-VΔ168-222 (42), respectively, to yield pCAGGS-V, pCAGGS-P, and pCAGGS-VΔC. pCAGGS-NP was constructed by subcloning from pBH269 (25) and was kindly provided by Anthony Schmitt. pCAGGS-HN and pCAGGS-F were subcloned from pGEM-HN and pGEM-F (53). pCAGGS-SH and pCAGGS-M were constructed by subcloning from pBH276 (25) and were kindly provided by Biao He. pCAGGS-DDB1 was constructed by subcloning from pBluescript SK(−) DDB1 (42). The pCAGGS-HA-DDB1 plasmids were constructed by adding an oligonucleotide encoding the hemagglutinin (HA)-epitope tag sequence to the 5′ (HA-DDB1) or the 3′ (DDB1-HA) end of the DDB1 cDNA by PCR using Vent polymerase (New England Biolabs, Beverly, Mass.). Oligonucleotides were synthesized at the Northwestern University Biotechnology facility.
Cells proliferation assays.
Monolayer cultures (∼10% confluent) of HeLa T4 cells were either mock infected or infected with SV5 at a multiplicity of infection (MOI) of 3 PFU/cell. After 1 h the inoculum was removed and replaced with DMEM–0.5% FBS–P/S. At 12-h intervals, the cells were harvested by trypsinization and resuspended in phosphate-buffered saline deficient in calcium and magnesium (PBS−; Gibco BRL). The cells were counted using a hemacytometer.
The proliferation of cells was assessed by determining the decrease in carboxyfluorescein succinimidyl ester (CFSE; Molecular Probes, Eugene, Oreg.) staining essentially as described previously (47). Monolayer cultures (∼10% confluent) of HeLa T4 cells were infected with SV5 or were mock infected as described above. After 1 h the inoculum was removed, and the cells were washed twice in phosphate-buffered saline (PBS+; Gibco BRL). A 5 mM CFSE stock solution, in dimethyl sulfoxide, was made immediately before labeling cells and diluted in PBS+ to a final concentration of 50 μM before addition to cells. Cells were labeled in 50 μM CFSE for 30 min, washed three times with PBS+, and then grown in DMEM–0.5% FBS–P/S. At 24-h intervals, the cells were harvested by trypsinization and fixed in 0.25% paraformaldehyde in PBS−. The cells were then analyzed by flow cytometry on a FACSCalibur flow cytometer (Becton Dickinson Immunocytometry Systems, San Jose, Calif.), and data were analyzed by using the Proliferation Wizard module of the ModFIT LT program, version 2.0 (Verity Software House, Inc., Topsham, Maine).
For expression of individual SV5 cDNAs in the proliferation assays and in all other experiments, monolayer cultures were transfected with pCAGGS plasmids expressing the SV5 genes using Lipofectamine Plus (Gibco BRL), essentially according to the manufacturer's instructions. At 16 to 18 h posttransfection, the cells were washed twice with PBS+ and then labeled with CFSE as described above. The cells were harvested at 24-h intervals by trypsinization and fixed in 0.25% formaldehyde in PBS−. The pCAGGS-P-, pCAGGS-V-, pCAGGS-VΔC-, pCAGGS-NP-, and pCAGGS-M-transfected cells were permeabilized in 1 ml 50% DMEM–50% FBS and 3 ml of 70% ethanol. The pCAGGS-SH-transfected cells were permeabilized in PBS+ with 0.1% saponin (Sigma-Aldrich, St. Louis, Mo.) and incubated with antibodies in the presence of 0.1% saponin. To examine protein expression, the cells were treated with rabbit antisera raised against SH (27) or one of the following mouse monoclonal antibodies (MAb): P-k, which recognizes the shared N-terminal domains of P and V; NP-a; M-f; F1a; or HN5a (59). The secondary antibodies used were goat antisera to mouse antibodies conjugated to phycoerythrin or goat antisera to rabbit antibodies conjugated to phycoerythrin (Molecular Probes). The cells were then analyzed on the FACSCalibur flow cytometer. The transfected cells were gated on a FL2-H (viral protein expression) versus FL1-H (CFSE staining) plot. The generation of the cells was determined using the Proliferation Wizard module of the ModFIT LT program.
Cell cycle analysis.
HeLa T4 cells were synchronized at the G1-S phase boundary essentially as described previously (71). The cells were initially blocked in DMEM–10% FBS–P/S with 2 mM thymidine (Sigma-Aldrich) for 12 to 14 h, the cells were released from the first block by incubation with DMEM–10% FBS–P/S for 8 h, and then the cells were blocked by incubation with DMEM–10% FBS–P/S with 0.4 mM mimosine (Sigma-Aldrich) for 12 to 14 h. After synchronization the cells were either infected with SV5, mock infected, or transfected with expression plasmids. For the SV5 infections, HeLa T4 cells were infected with SV5 at an MOI of 3 or 10 PFU/cell. After 1 h the inoculum was removed and replaced with DMEM–0.5% FBS–P/S.
The pCAGGS plasmids were transfected into synchronized HeLa T4 cells. For transfections with single plasmids, 2 μg of pCAGGS, pCAGGS-P, pCAGGS-V, or pCAGGS-VΔC was used. For the coexpression experiments, 2 μg of pCAGGS-V, 3 μg of pCAGGS-HA-DDB1, or 3 μg of pCAGGS-DDB1-HA was used, and 5 μg of DNA total was present in each transfection with the balance amount made up using pCAGGS vector. After 6 h, the transfection mixes were replaced with DMEM–0.5% FBS–P/S.
At 3- or 4-h intervals, the mock- or SV5-infected or transfected cells were harvested by trypsinization and fixed in 0.25% paraformaldehyde for 1 h at 4°C. The cells were then permeabilized in 1 ml of 50% DMEM–50% FBS and 3 ml of 70% ethanol at 4°C overnight or longer as described above, and the cells washed with PBS−. A portion of the cells was then treated for cell cycle analysis by DNA content or treated for measuring the expression of cyclins E, A, or B.
For cell cycle analysis by DNA content, cells were treated essentially as described elsewhere (Source Book, section 2.20.1; Becton Dickinson). To determine if the cells were infected or transfected, the primary antibodies used included the following MAb: P-k or 12CA5 (78). The secondary antibody used was goat antisera to mouse antibodies conjugated to fluorescein isothiocyanate (FITC) (Organon Teknika Corp., Charlotte, N.C.). The cells were treated with 0.5 mg of RNase A (Sigma-Aldrich) and 50 μg of propidium iodide (Sigma-Aldrich) per ml for at least 30 min at 4°C. The cells were then analyzed on the FACSCalibur flow cytometer. The data were analyzed using the Synch Wizard module of the ModFit LT program. Since single cells in G2-M are smaller than an aggregate of two G0-G1 cells or two cells fused together, the single cells were selected on an FL2-Width (FL2-W; size of cells) versus FL2-Area (FL2-A; DNA content) plot, and the infected or transfected cells were gated on a FL2-A (DNA content) versus FL1-H plot (P-k or 12CA5 staining). The percentage of cells in each phase of the cell cycle was then determined by modeling with the Synch Wizard module algorithm.
For analysis of expression of cell cyclins, a portion of the cells, which were synchronized, infected or transfected, and harvested as described above, were stained for cyclin expression. To determine if the cells (SV5 infected or transfected) expressed the V protein, cells were stained with MAb P-k. To determine if the cells were expressing the cyclin proteins, the following primary antibodies were used: rabbit polyclonal antisera to cyclin A (H-432; Santa Cruz Biotechnology, Inc., Santa Cruz, CA) or mouse MAb to cyclin B1 (14541C) or mouse MAb (14591C) to cyclin E (PharMingen, San Diego, Calif.). The bound MAb P-k was then detected with goat anti-mouse immunoglobulin G2a (IgG2a) conjugated to R-phycoerythrin (Southern Biotechnologies Associates, Inc., Birmingham, Ala.). The other secondary antibodies were goat anti-rabbit antiserum conjugated to FITC (Jackson ImmunoResearch Laboratories, Inc., West Grove, Pa.) which recognizes the cyclin A antisera or goat antimouse IgG1 conjugated to FITC (Southern Biotechnologies Associates, Inc.) which recognizes the cyclin B1 and E antibodies. The percentage of cells expressing P, V, or VΔC and the cyclins was determined by flow cytometry on the FACSCalibur. The data were analyzed with the Cell Quest program (Becton Dickinson) where quadrant markers were set with mock-infected or vector-transfected cells which were treated with MAb P-k and irrelevant rabbit antisera or irrelevant mouse MAb of the same isotype.
Immunoblotting.
HeLa T4 cells were synchronized as described above and then mock-infected or SV5-infected (MOI of ∼10 PFU/cell). At each time point two 6-cm-diameter plates of cells were harvested and treated as described above for DNA content analysis. At each time point, one plate of cells was lysed in protein lysis buffer (2% sodium dodecyl sulfate SDS; 62.5 mM Tris-HCl, pH 6.8; 2% dithiothreitol [DTT]), briefly sonicated, and an aliquot of cell lysate was analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) on a 15% gel for the p53 and p21 blots or a 9.25% acrylamide-DATD cross-linked gel (70) for the pRB blots. The proteins were transferred to Immobilon-P (Millipore Corp., Bedford, MA) using a Transblot SD apparatus (Bio-Rad, Hercules Calif.). Immunoblotting was performed essentially as previously described (6, 51). The primary antibodies used were a mixture of MAb to p53 (DO-1) and p21 (MAb 187) (Santa Cruz Biotechnology, Inc.) or p21 (65951A; PharMingen). For pRB immunoblotting, the primary antibody used was a mouse MAb (14001A; PharMingen). The secondary antibody used was a goat-anti mouse antisera conjugated to alkaline phosphatase (Amersham Pharmacia, Piscataway, N.J.) or goat anti-mouse antisera conjugated to horseradish peroxidase (Promega Biotech, Madison, Wis.). The immobilized protein was detected with either by using the ECF kit (Amersham Pharmacia) according to the manufacturer's instructions and chemifluorescence detected using a Storm Phosphorimager (Molecular Dynamics, Sunnyvale, Calif.) or by using the SuperSignal Plus kit (Pierce, Rockford, Ill.).
Quantification of V expression in SV5-infected cells and pCAGGS-V-transfected cells.
HeLa T4 cells were either mock infected, SV5 infected at an MOI of 3 PFU/cell, or transfected with pCAGGS-V. At 24 h after infection or transfection, the cells were trypsinized, fixed, and permeabilized as described for the cell cycle analysis. The cells were stained with an MAb (V MAb 11) unique to the C-terminal domain of V (52), followed by goat anti-mouse antisera conjugated to FITC. The cells were then analyzed by single-wavelength (color) flow cytometry.
Apoptosis assays.
Asynchronous populations of HeLa T4 cells were transfected with 2 μg each of pCAGGS, pCAGGS-P, pCAGGS-V, and pCAGGS-VΔC with or without 3 μg of pCAGGS-HA-DDB1. Additional pCAGGS was added if necessary to ensure that 5 μg of DNA was present in each transfection. At 1 or 2 days after transfection, the cells were harvested by trypsinization, fixed in paraformaldehyde, and permeabilized as described above for the cell cycle analysis. The cells were stained with MAb P-k for the P-, V-, and VΔC-expressing cells and with MAb 12CA5 for the HA-DDB-expressing cells. The cells were then treated with 0.5 mg of RNase A (Sigma-Aldrich) and 50 μg of propidium iodide per ml for at least 30 min at 4°C and analyzed by flow cytometry. The single cells were selected on FL2-W versus FL2-A plots as described above, but the aggregated cells and debris which have a signal for FL2-A but no signal for FL2-W were not selected. The percentage of cells with sub-G0-G1 DNA content was determined using the Cell Quest software (Becton Dickinson).
RESULTS
HeLa T4 cells infected with SV5 proliferate more slowly than mock-infected cells.
To determine if SV5 infection affects the growth rate of HeLa T4 cells, the number of viable mock- or SV5-infected cells was determined at 12-h intervals (Fig. 1A). By 36 and 48 h postinfection (p.i.) there were fewer cells present on the SV5-infected plates than on the mock-infected plates (Fig. 1A), suggesting that infection of cells with SV5 slows their rate of multiplication.
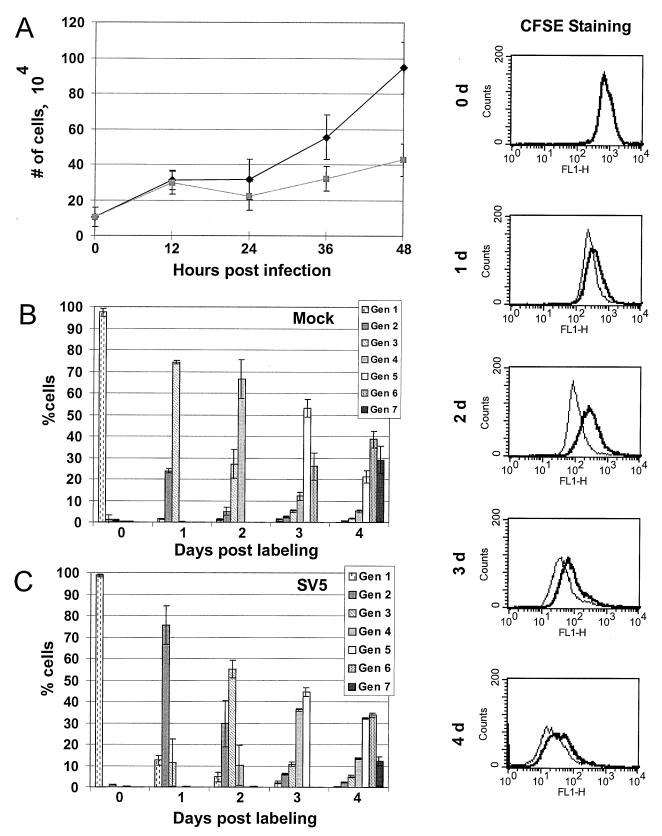
FIG. 1.
Cell growth and proliferation in SV5-infected and mock-infected cells. (A) Cell growth assay. Equal numbers of HeLa T4 cells were either mock infected (⧫) or SV5 infected ( ) and then harvested at 12-h intervals, and the cells were counted. Each time point represents the average of three separate plates of cells. (B and C) Proliferation assay. HeLa T4 cells were either mock infected or SV5 infected and then labeled with 50 μM CFSE. At various times after labeling, cell were harvested by trypsinization, fixed in paraformaldehyde, and analyzed by flow cytometry. Primary data are shown in side panels: thick line, SV5-infected cells; thin line, mock-infected cells. (B and C) Computed data for mock-infected cells (B) and SV5-infected cells (C). With each successive generation of cells the amount of label in each cell decreases. Cell generations were modeled using the Proliferation Wizard algorithm of the ModFIT LT program. Averaged data from three plates of cells for each time point are presented.
To examine further the growth of SV5-infected HeLa T4 cells, proliferation assays were performed in which living cells were labeled with the fluorescent dye CFSE. As the cells divide, each successive generation of cells contains a decreasing amount of CFSE. Mock- or SV5-infected cells were analyzed by flow cytometry, and the percentage of cells in each generation was determined using an algorithm (see Materials and Methods). The primary data for mock-infected cells and SV5-infected cells are shown in the righthand-side panels (Fig. 1) and the computed data are in Fig. 1B and C, respectively. The mock-infected cells showed a progressive dilution of the dye, and the algorithm modeled ∼75% of the cells in the third generation after 1 day and 65% of the cells in the fourth generation on day 2. In contrast, although the SV5-infected cells did not remain blocked in any generation, indicating that SV5 does not cause a pronounced growth arrest in the cells, SV5 infection caused a decrease in the proliferation rate of the HeLa T4 cells (Fig. 1C). After 1 day ∼75% of the SV5-infected cells were in the second generation, and on day 2 ∼55% of the SV5-infected cells were still in the third generation. The SV5-infected cells continued to progress through the generations more slowly than the mock-infected cells for up to 4 days after labeling (Fig. 1C), before the cells became confluent and the cells stopped proliferating.
SV5-infected HeLa T4 cells have a prolonged cell cycle.
To analyze the point in the cell cycle where the delay to cell proliferation occurred, cell cycle analysis of mock- and SV5-infected HeLa T4 cells was performed. Cells were synchronized at the G1-S phase boundary with thymidine and mimosine blocks. Following synchronization, the cells were released from the G1-S phase block, and the cells were either mock or SV5 infected for 1 h. At various times the cells were harvested and analyzed for their position in the cell cycle by measuring DNA content using propidium iodide staining. To distinguish between SV5-infected cells and uninfected cells, cells were stained with MAb P-k, which recognizes the common N-terminal domain of the P and V proteins. The cells were analyzed by flow cytometry detecting two wavelengths (two color), and the portion of the cells in each phase of the cell cycle was calculated by an algorithm (see Materials and Methods). Since the expression of the P or V proteins was not detected until 12 h p.i., initially all of the cells were selected for modeling on FL2-A (DNA content) versus FL1-H (P-k staining) plots, but after 12 h only the P-k-stained cells were selected.
It was found that the mock-infected and SV5-infected cells grew similarly for the first 18 h after release from the synchronization block, suggesting that the effect of SV5 on the cell cycle requires synthesis of viral proteins (primary data are shown in Fig. 2 side panels and the computed data are shown in Fig. 2A, B, and C). After 18 h, the SV5-infected cells were observed to proceed more slowly through the cell cycle than the mock-infected cells. SV5-infected cells exited G0-G1 more slowly and peaked in S phase 3 h later than mock-infected cells (Fig. 2A and B), indicating that the progression from G1 to S phase was slower in SV5-infected cells than in mock-infected cells. The number of cells in S phase decreased more slowly in SV5-infected cells than in mock-infected cells, suggesting that S phase was prolonged (Fig. 2B). Furthermore, about 20% of the SV5-infected cells remained in G2-M phase compared to less than 5% of the mock-infected cells (Fig. 2C), suggesting that the progression through G2 or M phase may be blocked in some of the SV5-infected cells. To examine the generality of the cell cycle changes observed in SV5-infected HeLa T4 cells, we also analyzed the cell cycle progression in SV5-infected MDBK cells, and very similar data were obtained (data not shown).
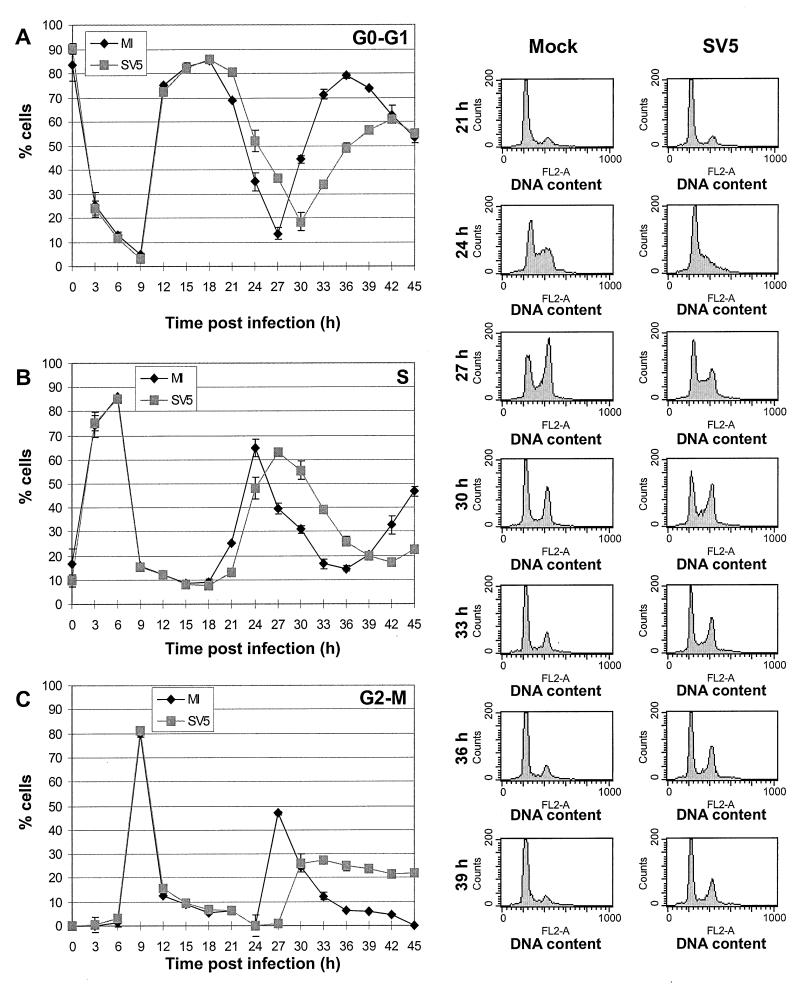
FIG. 2.
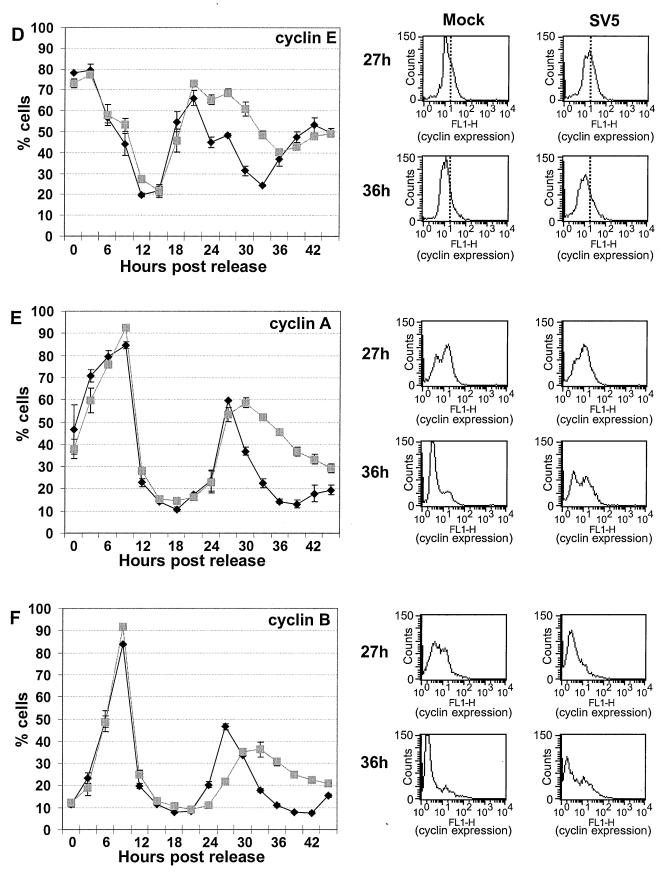
Cell cycle analysis of mock-infected or SV5-infected HeLa T4 cells by DNA content and by cyclin expression levels. HeLa T4 cells were synchronized at the G1-S-phase boundary by a 2 mM thymidine block, released, and then synchronized at the G1-S-phase boundary by a 0.4 mM mimosine block. Cells were released from the mimosine block and then either mock infected or SV5 infected at an MOI of ∼3 PFU/cell. After 1 h the inoculum was replaced by DMEM–0.5% FBS–P/S. Cells were harvested by trypsinization, fixed, and permeabilized. The cells were treated with MAb P-k, which recognizes the shared N-terminal domain of P and V, to stain cells which were virus infected. The cells were treated with propidium iodide for DNA content analysis and analyzed by two-color flow cytometry. The side panels show preliminary data for 21 to 39 h. The percentage of cells in each phase of the cell cycle was then computed by using the Synch Wizard algorithm of the ModFIT program, and the cells in G0-G1 phase (A), S phase (B), and G2-M phase (C) are shown in the line graphs. Each time point represents data averaged from three plates of cells. Symbols: ⧫, mock-infected cells; , SV5-infected cells. (D, E, and F) Cyclin expression levels. Aliquots of the cells form the DNA content experiment were analyzed for expression of cyclin E (D), cyclin A (E), and cyclin B (F) using antibodies to the specific cyclin proteins. To analyze only cells infected with SV5, cells were stained with MAb P-k. The cells were analyzed by two-color flow cytometry, and the percentage of cells positive for expression of cyclins and P or V proteins was determined. Each time point represents the average from three plates of cells. Symbols: ⧫, mock-infected cells; , SV5-infected cells. The four panels at the right of each line graph show the raw histogram data of cyclin expression from mock-infected (left) or SV5-infected (right) cells at 27 or 36 h after release from the mimosine block. The level of cyclin expression (FL1-H) is graphed along the x axis, and the number of cells is graphed on the y axis. For the cyclin E primary data, vertical lines are drawn to show the difference between the levels of cyclin E in mock- and SV5-infected cells.
To investigate further changes in the cell cycle, the expression of the cyclin proteins was examined. An aliquot of the same cells that were synchronized and infected with SV5 as described above was stained with MAb P-k to select for SV5-infected cells and then stained with antibodies to cyclin E (Fig. 2D), cyclin A (Fig. 2E), or cyclin B (Fig. 2F). The cells were then analyzed by two-color flow cytometry. The side panels show the primary flow cytometry histograms for mock- and SV5-infected cells at 27 and 36 h p.i. with cyclin expression shown on the FL1-H axis. For the primary data for cyclin E, vertical lines are drawn to show the differences between the levels of cyclin E in mock- and SV5-infected cells.
Cyclin E, which interacts with the cyclin-dependent protein kinase 2 (CDK2) to promote entry into S phase (17, 21; reviewed in references 10 and 66), was expressed to similar levels and in an approximately equal percentage of mock- or SV5-infected cells through the first 21 h after release from the synchronization block (Fig. 2D). After 24 h, a greater number of SV5-infected cells than mock-infected cells continued to express cyclin E (Fig. 2D). The similar rates of accumulation of cyclin E in mock- and SV5-infected cells 15 h after the release of the synchronization block suggest that the slower G1-to-S-phase transition observed in SV5-infected cells (Fig. 2B) is not due to a slower or a decreased accumulation of cyclin E. Cyclin E levels normally decrease through S phase (reviewed in reference 10), so the prolonged expression of cyclin E in SV5-infected cells corresponds with the prolonged S phase as determined by DNA content analysis.
The fraction of cells expressing cyclin A, which interacts with CDK2 to promote passage through S phase (17, 21; reviewed in references 10 and 66), was similar for up to 27 h after release of the synchronization block (Fig. 2E). These data indicate that the prolonged G1-S-phase transition and prolonged S phase of SV5-infected cells, as observed by propidium iodide staining, is not due to lack of cyclin A expression. After 27 h the amount of SV5-infected cells that expressed cyclin A remained elevated through 30 h and then decreased at a rate slower than observed in mock-infected cells (Fig. 2E). Cyclin A normally accumulates during S phase, reaches a peak at the end of G2, and is degraded during prometaphase of mitosis (17, 21, 56, 57; reviewed in references 10 and 66); thus, the continued expression of cyclin A in the SV5-infected cells suggests that the cells did not pass prometaphase of mitosis. These data provide further evidence that some of the SV5-infected cells remained in G2-M, confirming the DNA content analysis.
The expression of cyclin B, which interacts with CDK1 to form the maturation promoting factor (reviewed in references 10 and 35), was also examined. The percentage of cells that expressed cyclin B over the first 21 h post-release of synchronization block was similar in mock- and SV5-infected cells. However, after 24 h the expression of cyclin B was delayed and then prolonged compared to mock-infected cells (Fig. 2F). Cyclin B levels usually begin to accumulate during S phase and reach maximum levels as the cell enters mitosis (17, 57; reviewed in reference 10); therefore, the delay in cyclin B expression observed in SV5-infected cells further suggests that progression through S phase is perturbed in SV5-infected cells. Since degradation of cyclin B during anaphase is necessary for the cell to complete telophase and re-enter G1 phase (reviewed in reference 34), the slower decrease of cyclin B in SV5-infected cells also indicates that progression through mitosis is affected, confirming the DNA content analysis.
SV5 infection does not induce either accumulation of p53 or expression of p21.
Stress to cells, including viral infection, has been shown to cause activation of p53 transcription and stabilization of p53, which increases the amount of p53 in the cell. In turn, increased levels of p53 causes transactivation of p21CIP1 which can bind to cyclin and cyclin-dependent kinase complexes and inactivate them, leading to cell cycle arrest (16, 22; reviewed in reference 39). p53 has also been implicated in G2-M-phase arrest (2, 5; reviewed in reference 39). For HeLa cells the caveat has to be added that they contain sequences from human papillomavirus type 18 (HPV18), including sequences encoding the p53 interacting protein, E6 (65), but HeLa cells do have cell cycle regulation. Although the E6 protein was not detected in HeLa cells (65), it is known that the E6 protein of HPV18 causes ubiquitin-mediated degradation of p53 (62). To determine if SV5 infection affected the levels of p53 and p21CIP1 in the HeLa T4 variant of HeLa cells, cells were synchronized and infected with SV5, and p53 and p21CIP1 levels determined by immunoblotting. As shown in Fig. 3, p53 was detected, and the p53 levels were found to be similar between the mock- and SV5-infected cells (Fig. 3A), but p21CIP1 expression was not detected (data not shown). These results suggest that the changes in the cell cycle in SV5-infected cells are not due to increased accumulation of p53 and p53-induced expression of p21CIP1.
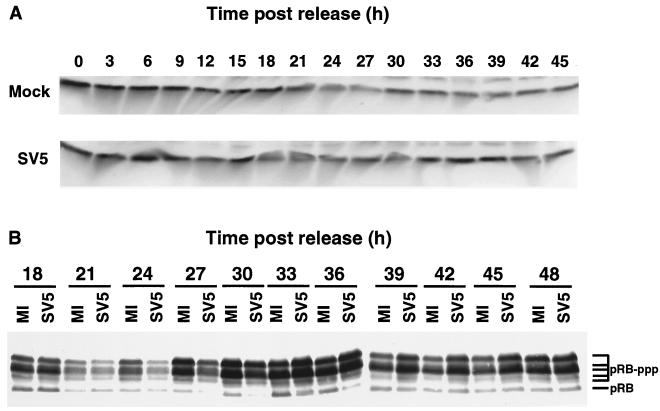
FIG. 3.
Analysis of p53 and pRB expression in synchronized mock-infected or SV5-infected HeLa T4 cells. Cells were synchronized as described in the legend to Fig. 2, released from the block, and infected at an MOI of ∼10 PFU/cell. The cells were harvested in protein lysis buffer containing DTT and sonicated briefly, and the polypeptides were analyzed by SDS-PAGE followed by immunoblotting. (A) The blots were probed with an MAb specific for p53. (B) The polypeptides were separated on a 9.25% acrylamide–DATD cross-linked gel, and the polypeptides were immunoblotted using an MAb specific for pRB. MI, mock-infected cells; SV5, SV5-infected cells; pRB, hypophosphorylated pRB; pRB-ppp, phosphorylated forms of pRB.
SV5 infection and pRB phosphorylation.
pRB is a cell cycle regulatory protein important for progression though the G1-to-S transition. pRB mediates its action by binding to the E2F transcription factor family of proteins. The hypophosphorylated form of pRB is present during the G0 and G1 phases and binds to E2F. pRB is phosphorylated at multiple sites at the G1-S-phase boundary and through S and G2 phases by the cyclin E-CDK2 and cyclin A-CDK2 complexes with the consequence that E2F is released (4, 12, 17, 28, 38, 40; reviewed in references 18 and 66). The HPV18 E7 protein, which binds to pRB (19), has been detected in HeLa cells (65), but the presence of phosphorylated pRB during S phase and G2-M in HeLa cells was found to be similar to primary T cells and to cell lines which have not been transformed by viruses (4, 12).
To determine if pRB phosphorylation is affected in SV5-infected HeLa T4 cells, cells were synchronized, infected with SV5, and lysed at various times p.i., and the polypeptides were analyzed by immunoblotting. As shown in Fig. 3B, less of the phosphorylated forms of pRB was present in the SV5-infected cells at 24, 27, and 30 h p.i. compared to the mock-infected cells, and increasing amounts of pRB were present as the cell enters S phase as seen previously (12). Furthermore, the amount of the slowest-migrating form of pRB began to decrease by 33 h in the mock-infected cells, and there was less of the phosphorylated forms in the mock-infected cells at 39, 42, and 45 h p.i. than in the SV5-infected cells (Fig. 3B). As an indicator of cell cycle progression and of cyclin-CDK activity, the delay in accumulation of the phosphorylated forms of pRB and the prolonged presence of phosphorylated pRB in SV5-infected cells suggests that there is a delay in the G1-S-phase transition and that cells are not returning to G0-G1 as quickly as did the mock-infected cells. Although cyclin E and cyclin A begin to accumulate similarly in mock- and SV5-infected cells, the cyclin-CDK complexes begin to phosphorylate pRB later. The delay in pRB phosphorylation can prevent release of the members of the E2F transcription factor family and delay E2F-mediated transactivation of genes necessary for S phase. However, the direct relationship of the phosphorylation changes observed to the changes in the cell cycle is confounded by the fact that HeLa cells contain DNA sequences that could express HPV18 E7 protein (65). Thus, it would be anticipated that the binding of E7 to pRB may interfere with the normal function of pRB.
Determination of the SV5 gene product that slows cell proliferation.
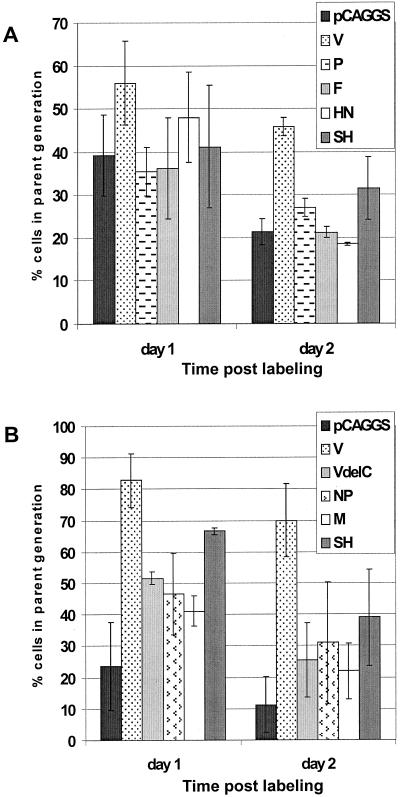
To determine which of the SV5-encoded proteins slows cell proliferation, the SV5-specific cDNAs were expressed transiently in HeLa T4 cells using the pCAGGS expression plasmid. At 16 h posttransfection, cells were labeled with CFSE as described above. The expression of P, F, HN, SH, NP, and M did not significantly (Student two-tailed t test; P > 0.05) alter the proliferation of the HeLa T4 cells compared to the vector-transfected cells (Fig. 4). However, more of the V-expressing cells remained in the parent generation after 1 and 2 days compared to vector-transfected cells, indicating that many (45 to 70%) of the V-expressing cells were not proliferating (Student two-tailed t test; P < 0.0005). Since the P and V proteins share 164 N-terminal amino acids but have unique C-terminal domains, these results suggested that the unique C-terminal domain of V is involved in the arrest of cell proliferation. To determine if the V C-terminal domain is involved in cell cycle regulation, a plasmid expressing VΔC (which lacks the cysteine-rich domain of V) was constructed. The VΔC-expressing cells were then tested for cell proliferation and found to proliferate similarly to vector-transfected cells (Fig. 4), further indicating that the unique C-terminal domain of V is involved in the arrest of cell proliferation.
FIG. 4.
Proliferation assays of HeLa T4 cells transiently expressing individual viral proteins. Cells were transfected with pCAGGS plasmids expressing the individual SV5-specific cDNAs. At 16 to 18 h post transfection, the cells were labeled with CFSE. At various times after labeling, the cells were harvested by trypsinization and fixed in formaldehyde. All of the cells except those expressing HN or F were permeabilized and then stained with antisera recognizing the viral proteins. The cells were then analyzed by flow cytometry. The loss of CFSE signal in cells expressing the viral proteins were then modeled using the Proliferation Wizard algorithm of the ModFIT LT program. The fraction of cells containing the original amount of CFSE staining (the parent generation) after 1 or 2 days is shown. Averaged data are from three plates for each time point. Panels A and B represent data from two different experiments.
HeLa T4 cells expressing the V protein, but not the P or VΔC proteins, have a prolonged cell cycle.
The pCAGGS expression vector exhibits a broad spectrum of expression levels of the foreign gene. Therefore, to compare levels of expression of the V protein in transfected cells to SV5-infected cells, the cells were stained with an MAb specific for the unique C terminus of V (V-MAb 11) (52), and the cells were analyzed by flow cytometry (Fig. 5). The higher-expressing population of V-transfected cells (see below) contained an amount of V protein greater than or equal to the amount of V protein contained in SV5-infected cells. This higher-expressing population of V-transfected cells was selected for cell cycle an analysis.
FIG. 5.
Expression levels of V protein in SV5-infected or pCAGGS-V transfected HeLa T4 cells. Cells were infected with SV5 at an MOI of 3 PFU/cell or transfected with 2 μg of pCAGGS-V. After 24 h the cells were harvested by trypsinization, fixed, permeabilized, and stained with an MAb specific for the unique C terminus of V (V MAb 11) (52). Expression levels were analyzed by flow cytometry. Dotted line, mock-infected cells; narrow line, SV5-infected cells; bold line, pCAGGS-V-transfected cells.
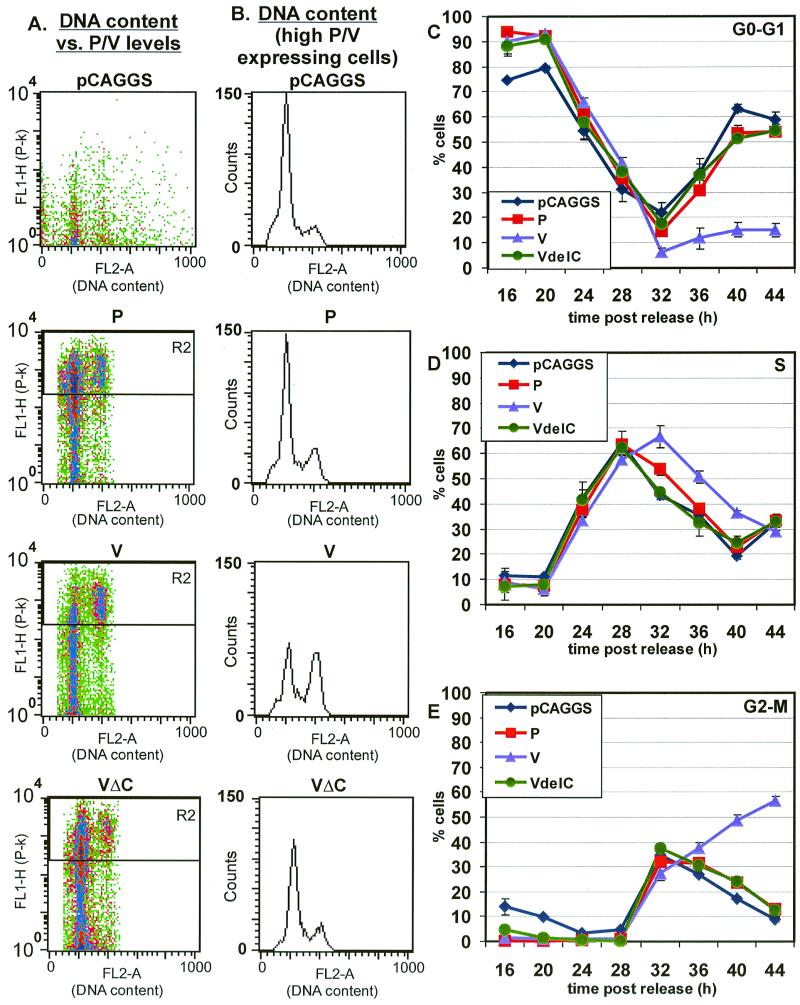
To determine whether transiently expressed P, V, or VΔC proteins have an effect on the cell cycle, HeLa T4 cells were synchronized at the G1-S-phase boundary with thymidine and mimosine blocks and then transfected with the plasmids expressing the P, V, or VΔC proteins. An aliquot of the cells was treated with MAb P-k to identify transfected cells and stained with propidium iodide to enable determination of DNA content and the cells analyzed by flow cytometry. The transfected cells were examined for DNA content (FL2-A) versus P-k staining (FL1-H), and examples of the raw data at 44 h are shown in Fig. 6A. For the cells expressing V protein, there appeared to be two populations of cells: low-level V-protein-expressing cells and high-level V-protein-expressing cells (Fig. 6A). At 44 h post-release from the synchronization block, the population expressing lower amounts of V protein was determined to be in the G0-G1 stage of the cell cycle, whereas the high-expressing V protein population of cells was determined to be in the G2-M stage of the cell cycle. The population of cells expressing the low level of V protein showed a cell cycle progression similar to that of mock-transfected cells (data not shown). Thus, the populations of cells expressing the high levels of P, V, or VΔC were selected for analysis, and the raw histograms for DNA content (FL2-A) of these cells 44 h after release of the synchronization block are shown in Fig. 6B. The average of the computed data of cells in each phase of the cell cycle is shown in Fig. 6C, D, and E.
FIG. 6.
Cell cycle analysis of HeLa T4 cells expressing P, V, or
VΔC proteins. HeLa T4 cells were synchronized as described in the
legend to Fig. 2. After release from the G1-S-phase block,
cells were transfected with 2 μg of the following plasmids: pCAGGS,
pCAGGS-P, pCAGGS-V, or pCAGGS-VΔC. The cells were treated and
analyzed as described in the legend for Fig. 3. The population of cells
expressing high levels of V protein were selected for analysis. (A) The
panels show representative flow cytometry data at the 44-h time
point. The density plots graph DNA content (FL2-A) versus the level of
P or V protein expression (FL1-H). The density plots show the selection
of the high-expressing population of transfected cells (gate R2). (B)
The DNA content (FL2-A) histograms of the high-expressing population of
cells at 44 h post-release of synchronization block are shown. (C,
D, and E) The percentage of cells in G0-G1
phase (C), S phase (D), and G2-M phase (E) are shown. Each
time point is the average of three plates of cells. (F, G, and H) Cell
cycle analysis and cyclin expression for HeLa T4 cells expressing P, V,
or VΔC. Cells were treated as described in the legend for Fig. 2.
Each time point represents the average of three plates of cells.
Symbols: ⧫, control vector-transfected cells;
, P-expressing
cells; ▴, V-expressing cells;
 ,
VΔC-transfected cells.
,
VΔC-transfected cells.
The cell cycle progression for control, P, V, and VΔC-expressing cells was similar for the first 24 h after release of the synchronization block, but at that time expression of proteins from the transfected plasmids was at very low levels (data not shown). After 24 h, the cells expressing the P protein (or the VΔC protein) progressed through the cell cycle at a rate similar to that of the control vector-transfected cells (Fig. 6C, D, and E). In contrast, for the cells expressing the V protein the peak of cells in S phase occurred at 32 h, which is 4 h later than the control cells, suggesting there was a delay in the G1-S-phase transition (Fig. 6D). The number of cells in S phase decreases more slowly in V-expressing cells than in control vector-transfected cells (Fig. 6D). Furthermore, half of the V-protein-expressing cells remained in G2-M after 36 h after release of block. The changes in the cell cycle in the V-protein-expressing cells were similar to those seen in SV5-infected cells. The amount of V protein expressed in the transfected cells was greater than or equal to the amount expressed in SV5-infected cells (Fig. 5), and the cells expressing high levels of V protein had a more pronounced G2-M arrest than that found in SV5-infected cells. Thus, these data indicate that the levels of the V protein in the cell affect the extent of cell cycle changes observed.
To characterize further the changes in the cell cycle of transfected cells, a portion of the cells, which were synchronized and transfected as described above, was stained with antibodies to cyclin E (Fig. 6F), cyclin A (Fig. 6G), or cyclin B (Fig. 6H) and P/V protein-specific MAb P-k, and the cells were then analyzed by flow cytometry. The cells expressing high levels of P, V, or VΔC were selected for analysis.
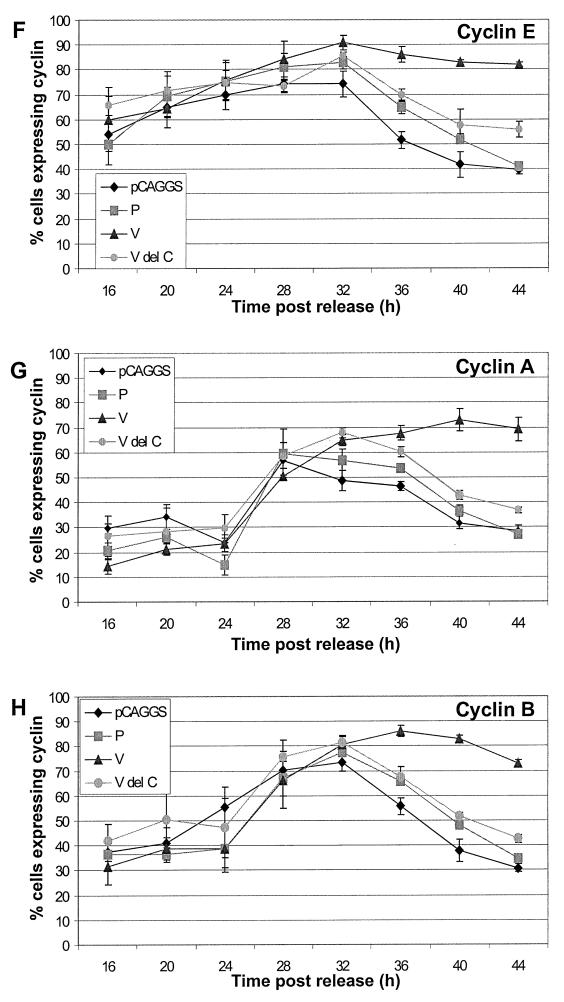
The control vector-transfected cells and P-protein- and VΔC-protein-expressing cells showed similar expression patterns for cyclin E (Fig. 6F). The V-expressing cells showed a cyclin E protein expression pattern similar to that of control cells up to 28 h post-release of the synchronization block. However, after 32 h the level of cyclin E did not decline as observed in control cells (Fig. 6F). The continued expression of cyclin E in V-protein-expressing cells corresponds with the prolonged S phase observed by DNA content analysis.
Analysis of the cyclin A expression levels in control cells and cells expressing the P, V, and VΔC proteins showed very similar levels of cyclin A for the first 28 to 32 h after release of the synchronization block (Fig. 6G). However, after 28 to 32 h the cyclin A level began to decline in control and P- and VΔC-expressing cells. In contrast, the cyclin A level did not decline in V-protein-expressing cells (Fig. 6G). The altered cyclin A expression pattern in V-protein-expressing cells parallels the cyclin A expression pattern observed in SV5-infected cells. Furthermore, the continued expression of cyclin A in V-protein-expressing cells is consistent with the G2-M arrest observed by the DNA content analysis in V-protein-expressing cells.
Analysis of the cyclin B levels in control and in P-, V-, and VΔC-protein-expressing cells (Fig. 6H) paralleled the data obtained for cyclin A expression levels. The expression of cyclin B in control and in P- and VΔC-expressing cells began to decrease after 32 h post-release of the synchronization block, but the cyclin B level in V-protein-expressing cells expressing cyclin B continued to increase until 36 h and then decreased more slowly than control cells (Fig. 6H). The delayed peak of expression and the prolonged expression of cyclin B in the V-protein-expressing cells suggest that the V-protein-expressing cells progress through S phase more slowly than control cells and that the cells were not progressing through mitosis when cyclin B would normally be degraded.
Taken together, the changes in the cell cycle as determined by DNA content analysis and cyclin expression in the V-protein-expressing cells were, for the most part, similar to those observed in SV5-infected cells. Thus, these data suggest that the SV5 V protein is involved in prolonging the cell cycle. Furthermore, since the expression of P and VΔC did not affect the cell cycle, the data indicate that the unique C-terminal domain of the SV5 V protein is important for prolonging the cell cycle.
The effect of V and DDB1 coexpression on the cell cycle.
The SV5 V protein was found previously to interact with DDB1 through its unique C-terminal domain (42). To facilitate determining if the V and DDB1 protein interaction is involved in cell cycle progression, the DDB1 protein was tagged at either its N or its C terminus with an epitope tag (HA) to yield the HA-DDB1 and DDB1-HA proteins, respectively. Alternative tags were used to reduce the possibility that the tag interfered with DDB1 functions. When both tagged DDB1 proteins were expressed in HeLa cells, using pCAGGS, both proteins showed primarily a diffuse cytoplasmic distribution (data not shown). The primarily cytosolic expression of DDB1 confirms recent data indicating that DDB1 is found primarily in the cytoplasm (67, 76), despite the fact that the protein was initially purified from nuclear extracts of HeLa cells (9). The coexpression of V and DDB1-HA or HA-DDB1 did not affect the localization of DDB1 (data not shown).
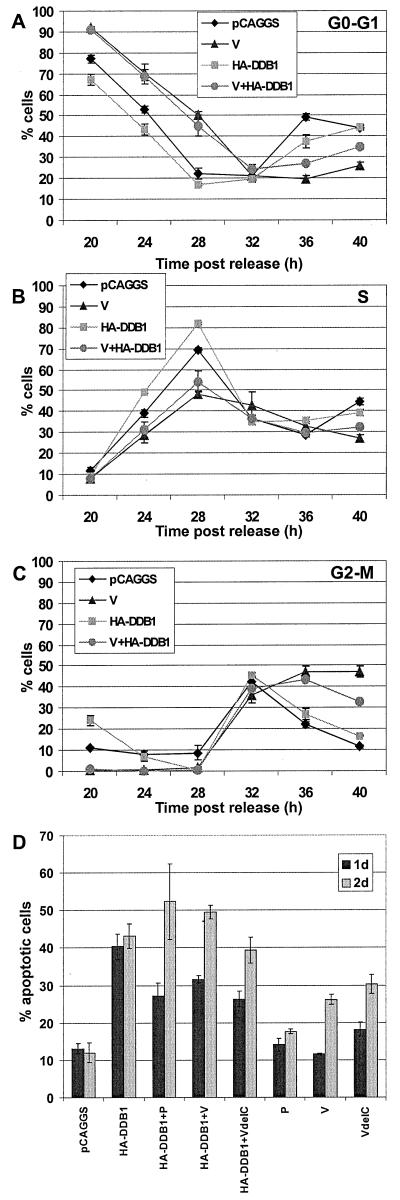
If the V protein interaction with DDB1 reduces the effective concentration of DDB1 available to interact with E2F, then overexpression of HA-DDB1 protein might block the V protein effect on the cell cycle. To test this hypothesis, HeLa T4 cells were synchronized at the G1-S-phase border and then transfected with control plasmid or with plasmids expressing V, HA-DDB1, or both plasmids to express the V and HA-DDB1 proteins (Fig. 7A, B, and C). A range of HA-DDB1 plasmid DNA from 0.5 to 3 μg of DNA was used. The cells were stained with MAb types specific for V and for the HA epitope tag. The V- or V–HA-DDB1-expressing cells were gated for the population of cells expressing a high level of V protein. Interpretation of the DDB-1 and V coexpression data is confounded by the fact that overexpression of DDB1 is deleterious to cells. A total of 45 to 50% of cells transfected with HA-DDB1 were in the sub-G0-G1 population, indicating that overexpression of HA-DDB1 probably causes apoptosis (Fig. 7D). Coexpression of P, V, or VΔC had much smaller effects on the apoptotic population by 2 days posttransfection (Fig. 7D).
FIG. 7.
Cell cycle analysis on coexpression of V and
HA-DDB1 proteins. Cells were synchronized as described in the legend
for Fig. 2. After the release from the mimosine block, the cells were
transfected with pCAGGS, 2 μg of pCAGGS-V, 3 μg of pCAGGS-HA-DDB1,
or 2 μg of pCAGGS-V plus 3 μg of pCAGGS-HA-DDB1. Additional pCAGGS
DNA was added to the transfections if necessary to ensure that 5 μg
of DNA was present in each transfection. The cells were harvested,
fixed, and permeabilized. The cells were stained with MAb P-k for the
V-expressing cells or with MAb 12CA5, which recognizes the HA-epitope
tag of the cells expressing HA-DDB1. The cells were stained with
propidium iodide and analyzed by flow cytometry as described above. The
high-expressing population of the V-expressing cells was selected for
analysis. The percentages of cells in the G0-G1
(A), S phase (B), and G2-M phase (C) are shown. Each time
point is the average of three plates of cells. Symbols: ⧫,
vector-only-transfected cells; ▴, V-expressing cells;
,
HA-DDB1-expressing cells;
 , V and
HA-DDB1-expressing cells. (D) DDB1 expression causes cells to
accumulate in a sub-G0-G1 state. Asynchronous
populations of HeLa T4 cells were transfected with 2 μg each of
pCAGGS, pCAGGS-P, pCAGGS-V, or pCAGGS-VΔC with or without 3 μg of
HA-DDB1. Additional pCAGGS was added if necessary to ensure that 5 μg
of DNA was present in each transfection. At 1 or 2 days after
transfection, cells were harvested by trypsinization, fixed in
paraformaldehyde, and permeabilized. The cells were stained with MAb
P-k for the P-, V-, and VΔC-expressing cells and with MAb 12CA5 for
the HA-DDB-expressing cells. The cells were then stained with propidium
iodide and analyzed by flow cytometry. The percentage of cells in the
sub-G0-G1 population is shown. Each time point
is the average of three plates of cells.
, V and
HA-DDB1-expressing cells. (D) DDB1 expression causes cells to
accumulate in a sub-G0-G1 state. Asynchronous
populations of HeLa T4 cells were transfected with 2 μg each of
pCAGGS, pCAGGS-P, pCAGGS-V, or pCAGGS-VΔC with or without 3 μg of
HA-DDB1. Additional pCAGGS was added if necessary to ensure that 5 μg
of DNA was present in each transfection. At 1 or 2 days after
transfection, cells were harvested by trypsinization, fixed in
paraformaldehyde, and permeabilized. The cells were stained with MAb
P-k for the P-, V-, and VΔC-expressing cells and with MAb 12CA5 for
the HA-DDB-expressing cells. The cells were then stained with propidium
iodide and analyzed by flow cytometry. The percentage of cells in the
sub-G0-G1 population is shown. Each time point
is the average of three plates of cells.
For the viable cells, DNA content analysis indicated that somewhat more of the HA-DDB1-expressing cells (3 μg of DNA) than the control-vector-transfected cells entered S phase by 24 h post-release of the synchronization block (Fig. 7B), but both entered G2-M by 32 h (Fig. 7C) and reentered G0-G1 by 36 h (Fig. 7A). These data suggest that overexpression of HA-DDB1 causes a small increase in the rate of progression from G1 to S phase. The cells expressing the V protein showed a delayed G1-to-S phase progression (Fig. 7A and B) and a G2-M arrest as found above (Fig. 7C). The delay in the G1-to-S phase transition was similar in cells coexpressing V and DDB1 and cells expressing V alone. However, by 40 h ∼30% of the V- and HA-DDB1-coexpressing cells were arrested in G2-M, compared to ∼45% of the V-expressing cells (Fig. 7C), but the DNA content of these cells remained elevated compared to the DNA content of control-plasmid-transfected cells. Increasing the amount of the HA-DDB1 plasmid from 0.5 to 3 μg of DNA cotransfected with 2 μg of V plasmid DNA overcame the effect of expressing V alone and resulted in decreasing amounts of cells arrested in G2-M phase (e.g., see Fig. 7C). Coexpression of HA-DDB1 and P (or VΔC) protein did not affect cell cycle progression (data not shown). Thus, the data suggest that the coexpression of HA-DDB1 with V protein partially restores progression through the cell cycle. Similar data was obtained when the V protein was coexpressed with DDB1 tagged with the HA epitope at its C terminus (DDB1-HA) (data not shown). Triple transfections using plasmids expressing V, DDB1, and FLAG-epitope-tagged-DDB2 were also performed but the changes in the cell cycle were similar to those found for V and DDB1 coexpression (data not shown). Thus, although the effect of DDB1 overexpression on the cell is more complex than a direct effect on G2-S-phase transition, it is possible that DDB-1 could ameliorate the V protein effects on the cell cycle but these are largely masked by DDB1 overexpression-induced apoptosis.
DISCUSSION
Many viruses have been shown previously to affect progression of the cell cycle of host cells to favor viral replication (reviewed in reference 49). Here we have shown that HeLa-T4 cells, infected with the paramyxovirus SV5, proliferate more slowly than mock-infected cells. DNA content analysis of synchronized cells infected with SV5 and measurement of the accumulation of cyclins A, B, and E indicated that, for the first 18 h p.i., SV5 infection had little effect on the cell cycle. However, after 18 h p.i., a time when viral RNA and protein synthesis reaches a plateau (8), DNA content analysis indicated all phases of the cell cycle were prolonged. The G1-S-phase transition was delayed compared to that found in mock-infected cells, and the duration of S phase was prolonged. In uninfected cells, cyclin E expression usually peaks around the G1-S-phase boundary and its accumulation decreases through S phase, and cyclin A and B accumulate through S phase before their expression decreases during mitosis (reviewed in reference 10). In SV5-infected HeLa-T4 cells, at between 18 and 48 h p.i., cyclin E and A levels reached a peak similar to mock-infected cells but declined more slowly. Thus, the data indicate that the prolonged G1-S-phase transition and prolonged S phase occurring at 18 to 48 h p.i. was not caused by a lack of cyclin E and A expression. Taken together, the DNA content analysis and measurement of cyclin levels indicate that SV5 infection causes a delay in both the beginning of S phase and a delay in the progression through S phase.
A fraction of the SV5-infected cells appeared to be arrested in G2 or M phase based on DNA content analysis. In uninfected cells, mitosis is initiated by the cyclin B-CDK1 (CDC2) complex; thus, a delay in expression of cyclin B would be expected to delay the onset of mitosis (reviewed in reference 34). Indeed, cyclin B expression observed in SV5-infected cells peaked later and declined more slowly. In uninfected cells, cyclin A is degraded during prometaphase, and cyclin B is degraded during anaphase (17, 21, 56, 57; reviewed in references 10 and 34). Direct analysis of the stability of cyclin A in pulse-labeling and chase experiments suggested that cyclin A degradation was decreased in SV5-infected cells compared to control cells (data not shown). The prolonged expression of cyclin A and B in SV5-infected cells suggests that some of the cells do not progress through mitosis and are arrested in G2 phase.
Activation of p53 increases the amount of p53 in the cell by stabilizing the protein from degradation. Activation of p53 has also been shown to cause transcriptional transactivation of expression of p21CIP1. This results in p21CIP1 binding to cyclin and cyclin-dependent kinase complexes, resulting in their inactivation which in turn causes cell cycle arrest (reviewed in reference 39). Therefore, we examined whether the delay in the cell cycle observed in SV5-infected cells were related to altered p53 and p21CIP1 accumulation. The levels of p53 and p21CIP1 expression were not increased in SV5-infected cells compared to mock-infected cells, suggesting that the delay in entry into S phase and progression through S phase occurs through a p53-independent mechanism. pRB phosphorylation is important for progression through the G1-S transition. In SV5-infected cells the phosphorylation of pRB was delayed and prolonged. These data suggest that although cyclin E and cyclin A begin to accumulate, the activity of the cyclin-CDK complexes is delayed even though p21CIP was not detected.
To analyze the viral protein responsible for the effects on the cell cycle, viral cDNAs were expressed transiently in HeLa-T4 cells. In cells expressing high levels of V protein (but not control transfection or P-transfected cells) delays through the stages of the cell cycle occurred that were very similar to those observed in SV5-infected cells, except that the G2-M arrest was more pronounced. Expression of VΔC which lacks the V protein unique C-terminal cysteine-rich domain exhibited a cell cycle progression comparable to control-transfected or P-transfected cells. Thus, the data indicate the alteration to the cell cycle is mediated through the V-protein cysteine-rich C-terminal domain. The amount of V protein expressed transiently in the high-expressing population of transfected cells was higher than that found in SV5-infected cells. The extent of G2-M arrest in V-transfected cells was also higher than in SV5-infected cells. Thus, these data suggest that the level of V protein expression affects the extent of the G2-M arrest. Therefore, it is possible that in SV5-infected cells the expression level of V is regulated such that it is only sufficient to prolong the cell cycle but not to cause G2-M arrest.
SV5 V protein interacts with the DDB1 subunit of the weakly associated DDB heterodimer complex, through the unique C-terminal domain of V (42). The V proteins of mumps virus, human parainfluenza virus type 2, and measles virus also interact with DDB1 (42). DDB1 was initially characterized as a protein that bound to damaged DNA and was proposed to be a part of the nucleotide excision repair pathway (9). However, DDB was found subsequently to be nonessential for nucleotide excision repair in vitro (1, 32). DDB may be a multifunctional protein involved in multiple transcription-related events since it has been shown to interact with the hepatitis B virus X protein (7, 37), and recently the DDB complex has been found to interact with the transcription factor E2F1 (24). In in vitro assays DDB was found to overcome the inhibition by pRB of E2F1-mediated transactivation (24). It has been suggested that when damaged DNA is present in a cell, DDB binds to damaged DNA and no longer associates with E2F, and thus E2F remains associated with pRB and is unable to activate transcription (24). As the E2F family of proteins have been found to be essential for the G1-S-phase transition (reviewed in reference 69), it has been suggested that the lack of interaction of E2F with DDB may be important in slowing cell growth during the DNA repair process (24). The DDB complex has also been found to interact with CUL-4A, a member of the cullin family of proteins (68). The function of CUL-4A is not known but other members of the cullin family, CUL-1 and CUL-3, are involved in the ubiquitin-mediated degradation of cell cycle proteins such as cyclin D, cyclin E, and p21CIP (reviewed in references 79 and 80).
Since V and DDB1 interact and because of the possible role of DDB1 in E2F1-mediated transactivation and DDB complex interaction with a cullin protein, the effect of transient coexpression of V and DDB1 was examined. The overexpression of DDB1 was found to restore partially normal progression through the cell cycle, especially by decreasing the fraction of cells which remained in G2-M, suggesting that the interaction between V and DDB1 may play a role in the cell cycle changes. It is also possible that the V protein prevents DDB1 from interacting with E2F1 and thus decreasing transactivation of S-phase-specific genes by E2F1; the slower increase in S-phase-specific gene transcription would affect the G1-S-phase transition and prolong the S phase. There are several possible reasons for the partial effect on cell cycle progression. (i) There may be varying levels of V and DDB1 expression in each cell due to the wide range of expression of proteins from the pCAGGS expression system. Although several concentrations of DDB1 cDNA were used, it is possible that not enough DDB was synthesized to interact with all of the V protein present in the cell. (ii) After overexpression of DDB1, >40% of the transfected cells were in the sub-G0-G1 population, indicating that these cells were apoptotic. (iii) It is also possible that V interacts directly with other cellular proteins, yet to be identified, that are involved in cell cycle regulation. To facilitate future analysis, it would be useful to derive mouse embryonic stem cells with DDB1 deleted to analyze cell cycle progression in uninfected and SV5-infected cells. It would also be useful to obtain a viable SV5 containing a deletion of V or VΔC, but despite repeated attempts this has not been successful (He, unpublished observations), suggesting an essential role for V in the SV5 life-cycle.
A rapidly dividing cell would not provide a very favorable environment for SV5 assembly since the peak of virus release occurs 18 to 24 h p.i., a time frame similar to that of the cell cycle for many cell types. In mitotic cells, the Golgi apparatus fragments during mitosis causing an inhibition of vesicle fusion and hence glycoprotein transport is blocked (reviewed in reference 44). Fragmentation of the Golgi using ilimaquinone, a metabolite that has been used as a model of Golgi fragmentation, causes the predicted block in transport of the F and HN proteins (data not shown). Furthermore, transport of the SV5 glycoproteins to the plasma membrane is required for virus assembly to occur (63). Therefore, the observed prolongation of the cell cycle in SV5-infected cells that is mediated by the V protein cysteine-rich domain, is consistent with the hypothesis that the prolonged cell cycle serves to promote transport of the glycoproteins to the cell surface and thus permits viral assembly and budding at the cell surface.
ACKNOWLEDGMENTS
We are very grateful to Pradip Raychaudhuri (University of Illinois Medical School, Chicago), Stuart Linn (University of California, Berkeley), Laimonis Laimins (Northwestern University Medical School, Chicago, Ill.), Joseph Nevins (HHMI, Duke University, Durham, N.C.), and Daniel Linzer (Northwestern University, Evanston, Ill.) for helpful discussions.
This work was supported in part by Research Grant AI-23173 from the National Institute of Allergy and Infectious Diseases. G.Y.L. was supported by National Institutes of Health Medical Scientist Training Program Grant T32 GM-08152. R.A.L. is an Investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Aboussekhra A, Biggerstaff M, Shivji M K K, Vilpo J A, Moncollin V, Podust V N, Protic M, Hubscher U, Egly J-M, Wood R D. Mammalian DNA nucleotide excision repair reconstituted with purified protein components. Cell. 1995;80:859–868. doi: 10.1016/0092-8674(95)90289-9. [DOI] [PubMed] [Google Scholar]
- 2.Agarwal M L, Agarwal A, Taylor W R, Stark G R. p53 controls both the G2/M and the G1cell cycle checkpoints and mediates reversible growth arrest in human fibroblasts. Proc Natl Acad Sci USA. 1995;92:8493–8497. doi: 10.1073/pnas.92.18.8493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baron M D, Barrett T. Rinderpest viruses lacking the C and V proteins show specific defects in growth and transcription of viral RNAs. J Virol. 2000;74:2603–2611. doi: 10.1128/jvi.74.6.2603-2611.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buchkovich K, Duffy L A, Harlow E. The retinoblastoma protein is phosphorylated during specific phases of the cell cycle. Cell. 1989;58:1097–1105. doi: 10.1016/0092-8674(89)90508-4. [DOI] [PubMed] [Google Scholar]
- 5.Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown J P, Sedivy J M, Kinzler K W, Vogelstein B. Requirement for p53 and p21 to sustain G2arrest after DNA damage. Science. 1998;282:1497–1501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- 6.Burnette W N. Western blotting: electrophoretic transfer of proteins from sodium dodecyl sulfate-polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal Biochem. 1981;112:195–203. doi: 10.1016/0003-2697(81)90281-5. [DOI] [PubMed] [Google Scholar]
- 7.Butel J S, Lee T-H, Slagle B L. Is the DNA repair system involved in hepatitis-B-virus-mediated hepatocellular carcinogenesis? Trends Microbiol. 1996;4:119–124. doi: 10.1016/0966-842X(96)81529-0. [DOI] [PubMed] [Google Scholar]
- 8.Choppin P W, Compans R W. Reproduction of paramyxoviruses. In: Fraenkel-Conrat H, Wagner R R, editors. Comprehensive virology. Vol. 4. New York, N.Y: Plenum Press; 1975. pp. 95–178. [Google Scholar]
- 9.Chu G, Chang E. Xeroderma pigmentosum group E cells lack a nuclear factor that binds to damaged DNA. Science. 1988;242:564–567. doi: 10.1126/science.3175673. [DOI] [PubMed] [Google Scholar]
- 10.Darzynkiewicz Z, Gong J, Juan G, Ardelt B, Traganos F. Cytometry of cyclin proteins. Cytometry. 1996;25:1–13. doi: 10.1002/(SICI)1097-0320(19960901)25:1<1::AID-CYTO1>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 11.DeCaprio J A, Ludlow J W, Figge J, Shew J Y, Huang C M, Lee W H, Marsilio E, Paucha E, Livingston D M. SV40 large tumor antigen forms a specific complex with the product of the retinoblastoma susceptibility gene. Cell. 1988;54:275–283. doi: 10.1016/0092-8674(88)90559-4. [DOI] [PubMed] [Google Scholar]
- 12.DeCaprio J A, Ludlow J W, Lynch D, Furukawa Y, Griffin J, Piwnica-Worms H, Huang C M, Livingston D M. The product of the retinoblastoma susceptibility gene has properties of a cell cycle regulatory element. Cell. 1989;58:1085–1095. doi: 10.1016/0092-8674(89)90507-2. [DOI] [PubMed] [Google Scholar]
- 13.Delenda C, Hausmann S, Garcin D, Kolakofsky D. Normal cellular replication of Sendai virus without the trans-frame, nonstructural V protein. Virology. 1997;228:55–62. doi: 10.1006/viro.1996.8354. [DOI] [PubMed] [Google Scholar]
- 14.Didcock L, Young D F, Goodbourn S, Randall R E. The V protein of simian virus 5 inhibits interferon signalling by targeting STAT1 for proteasome-mediated degradation. J Virol. 1999;73:9928–9933. doi: 10.1128/jvi.73.12.9928-9933.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dittmer D, Mocarski E S. Human cytomegalovirus infection inhibits G1/S transition. J Virol. 1997;71:1629–1634. doi: 10.1128/jvi.71.2.1629-1634.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dulic V, Kaufmann W K, Wilson S J, Tlsty T D, Lees E, Harper J W, Elledge S J, Reed S I. p53-dependent inhibition of cyclin-dependent kinase activities in human fibroblasts during radiation-induced G1arrest. Cell. 1994;76:1013–1023. doi: 10.1016/0092-8674(94)90379-4. [DOI] [PubMed] [Google Scholar]
- 17.Dulic V, Lees E, Reed S I. Association of human cyclin E with a periodic G1-S phase protein kinase. Science. 1992;257:1958–1961. doi: 10.1126/science.1329201. [DOI] [PubMed] [Google Scholar]
- 18.Dyson N. The regulation of E2F by pRB-family proteins. Genes Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- 19.Dyson N, Howley P M, Munger K, Harlow E. The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science. 1989;243:934–937. doi: 10.1126/science.2537532. [DOI] [PubMed] [Google Scholar]
- 20.Ehmann G L, McLean T I, Bachenheimer S L. Herpes simplex virus type 1 infection imposes a G1/S block in asynchronously growing cells and prevents G1entry in quiescent cells. Virology. 2000;267:335–349. doi: 10.1006/viro.1999.0147. [DOI] [PubMed] [Google Scholar]
- 21.Girard F, Strausfeld U, Fernandez A, Lamb N J. Cyclin A is required for the onset of DNA replication in mammalian fibroblasts. Cell. 1991;67:1169–1179. doi: 10.1016/0092-8674(91)90293-8. [DOI] [PubMed] [Google Scholar]
- 22.Harper J W, Adami G R, Wei N, Keyomarsi K, Elledge S J. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 23.Hayashi M L, Blankenship C, Shenk T. Human cytomegalovirus UL69 protein is required for efficient accumulation of infected cells in the G1phase of the cell cycle. Proc Natl Acad Sci USA. 2000;97:2692–2696. doi: 10.1073/pnas.050587597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hayes S, Shiyanov P, Chen X, Raychaudhuri P. DDB, a putative DNA repair protein, can function as a transcriptional partner of E2F1. Mol Cell Biol. 1998;18:240–249. doi: 10.1128/mcb.18.1.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He B, Paterson R G, Ward C D, Lamb R A. Recovery of infectious SV5 from cloned DNA and expression of a foreign gene. Virology. 1997;237:249–260. doi: 10.1006/viro.1997.8801. [DOI] [PubMed] [Google Scholar]
- 26.He J, Choe S, Walker R, Di Marzio P, Morgan D O, Landau N R. Human immunodeficiency virus type 1 protein R (Vpr) blocks cells in the G2 phase of the cell cycle by inhibiting p34cdc2activity. J Virol. 1995;69:6705–6711. doi: 10.1128/jvi.69.11.6705-6711.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hiebert S W, Paterson R G, Lamb R A. Identification and predicted sequence of a previously unrecognized small hydrophobic protein, SH, of the paramyxovirus simian virus 5. J Virol. 1985;55:744–751. doi: 10.1128/jvi.55.3.744-751.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hinds P W, Mittnacht S, Dulic V, Arnold A, Reed S I, Weinberg R A. Regulation of retinoblastoma protein functions by ectopic expression of human cyclins. Cell. 1992;70:993–1006. doi: 10.1016/0092-8674(92)90249-c. [DOI] [PubMed] [Google Scholar]
- 29.Jacques J P, Kolakofsky D. Pseudo-templated transcription in prokaryotic and eukaryotic organisms. Genes Dev. 1991;5:707–713. doi: 10.1101/gad.5.5.707. [DOI] [PubMed] [Google Scholar]
- 30.Kato A, Kiyotani K, Sakai Y, Yoshida T, Nagai Y. The paramyxovirus, Sendai virus, V protein encodes a luxury function required for viral pathogenesis. EMBO J. 1997;16:578–587. doi: 10.1093/emboj/16.3.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kato A, Kiyotani K, Sakai Y, Yoshida T, Shioda T, Nagai Y. Importance of the cysteine-rich carboxyl-terminal half of V protein for Sendai virus pathogenesis. J Virol. 1997;71:7266–7272. doi: 10.1128/jvi.71.10.7266-7272.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kazantsev A, Mu D, Nichols A F, Zhao X, Linn S, Sancar A. Functional complementation of xeroderma pigmentosum complementation group E by replication protein A in an in vitrosystem. Proc Natl Acad Sci USA. 1996;93:5014–5018. doi: 10.1073/pnas.93.10.5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keeney S, Chang G J, Linn S. Characterization of a human DNA damage binding protein implicated in xeroderma pigmentosum E. J Biol Chem. 1993;268:21293–21300. [PubMed] [Google Scholar]
- 34.King R W, Deshaies R J, Peters J M, Kirschner M W. How proteolysis drives the cell cycle. Science. 1996;274:1652–1659. doi: 10.1126/science.274.5293.1652. [DOI] [PubMed] [Google Scholar]
- 35.Kishimoto T, Okumura E. In vivo regulation of the entry into M-phase: initial activation and nuclear translocation of cyclin B/Cdc2. Prog Cell Cycle Res. 1997;3:241–249. doi: 10.1007/978-1-4615-5371-7_19. [DOI] [PubMed] [Google Scholar]
- 36.Lamb R A, Kolakofsky D. Paramyxoviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 1177–1204. [Google Scholar]
- 37.Lee T-H, Elledge S J, Butel J S. Hepatitis B virus X protein interacts with a probable cellular DNA repair protein. J Virol. 1995;69:1107–1114. doi: 10.1128/jvi.69.2.1107-1114.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lees J A, Buchkovich K J, Marshak D R, Anderson C W, Harlow E. The retinoblastoma protein is phosphorylated on multiple sites by human cdc2. EMBO J. 1991;10:4279–4290. doi: 10.1002/j.1460-2075.1991.tb05006.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Levine A J. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 40.Lin B T, Gruenwald S, Morla A O, Lee W H, Wang J Y. Retinoblastoma cancer suppressor gene product is a substrate of the cell cycle regulator cdc2 kinase. EMBO J. 1991;10:857–864. doi: 10.1002/j.1460-2075.1991.tb08018.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin G, Paterson R G, Lamb R A. The RNA binding region of the paramyxovirus SV5 V and P proteins. Virology. 1997;238:460–469. doi: 10.1006/viro.1997.8866. [DOI] [PubMed] [Google Scholar]
- 42.Lin G Y, Paterson R G, Richardson C D, Lamb R A. The V protein of the paramyxovirus SV5 interacts with damage-specific DNA binding protein. Virology. 1998;249:189–200. doi: 10.1006/viro.1998.9317. [DOI] [PubMed] [Google Scholar]
- 43.Liston P, Briedis D J. Measles virus V protein binds zinc. Virology. 1994;198:399–404. doi: 10.1006/viro.1994.1050. [DOI] [PubMed] [Google Scholar]
- 44.Lowe M, Nakamura N, Warren G. Golgi division and membrane traffic. Trends Cell Biol. 1998;8:40–44. doi: 10.1016/s0962-8924(97)01189-6. [DOI] [PubMed] [Google Scholar]
- 45.Lu M, Shenk T. Human cytomegalovirus UL69 protein induces cells to accumulate in G1phase of the cell cycle. J Virol. 1999;73:676–683. doi: 10.1128/jvi.73.1.676-683.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ludlow J W, DeCaprio J A, Huang C M, Lee W H, Paucha E, Livingston D M. SV40 large T antigen binds preferentially to an underphosphorylated member of the retinoblastoma susceptibility gene product family. Cell. 1989;56:57–65. doi: 10.1016/0092-8674(89)90983-5. [DOI] [PubMed] [Google Scholar]
- 47.Naniche D, Reed S I, Oldstone M B. Cell cycle arrest during measles virus infection: a G0-like block leads to suppression of retinoblastoma protein expression. J Virol. 1999;73:1894–1901. doi: 10.1128/jvi.73.3.1894-1901.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants by a novel eukaryotic vector. Gene. 1991;108:193–200. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 49.Op De Beeck A, Caillet-Fauquet P. Viruses and the cell cycle. Prog Cell Cycle Res. 1997;3:1–19. doi: 10.1007/978-1-4615-5371-7_1. [DOI] [PubMed] [Google Scholar]
- 50.Paterson R G, Harris T J R, Lamb R A. Analysis and gene assignment of mRNAs of a paramyxovirus, simian virus 5. Virology. 1984;138:310–323. doi: 10.1016/0042-6822(84)90354-4. [DOI] [PubMed] [Google Scholar]
- 51.Paterson R G, Lamb R A. The molecular biology of influenza viruses and paramyxoviruses. In: Davidson A, Elliott R M, editors. Molecular virology: a practical approach. Oxford, England: IRL Oxford University Press; 1993. pp. 35–73. [Google Scholar]
- 52.Paterson R G, Leser G P, Shaughnessy M A, Lamb R A. The paramyxovirus SV5 V protein binds two atoms of zinc and is a structural component of virions. Virology. 1995;208:121–131. doi: 10.1006/viro.1995.1135. [DOI] [PubMed] [Google Scholar]
- 53.Paterson R G, Russell C J, Lamb R A. Fusion protein of the paramyxovirus SV5: destabilizing and stabilizing mutants of fusion activation. Virology. 2000;270:17–30. doi: 10.1006/viro.2000.0267. [DOI] [PubMed] [Google Scholar]
- 54.Paterson R G, Thomas S M, Lamb R A. Specific nontemplated nucleotide addition to a simian virus 5 mRNA: prediction of a common mechanism by which unrecognized hybrid P-cysteine-rich proteins are encoded by paramyxovirus “P” genes. In: Kolakofsky D, Mahy B W J, editors. Genetics and pathogenicity of negative strand viruses. London, England: Elsevier; 1989. pp. 232–245. [Google Scholar]
- 55.Peluso R W, Lamb R A, Choppin P W. Polypeptide synthesis in simian virus 5-infected cells. J Virol. 1977;23:177–187. doi: 10.1128/jvi.23.1.177-187.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pines J, Hunter T. Human cyclin A is adenovirus E1A-associated protein p60 and behaves differently from cyclin B. Nature. 1990;346:760–763. doi: 10.1038/346760a0. [DOI] [PubMed] [Google Scholar]
- 57.Pines J, Hunter T. Human cyclins A and B1 are differentially located in the cell and undergo cell cycle-dependent nuclear transport. J Cell Biol. 1991;115:1–17. doi: 10.1083/jcb.115.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Randall R E, Bermingham A. NP:P and NP:V interactions of the paramyxovirus simian virus 5 examined using a novel protein:protein capture assay. Virology. 1996;224:121–129. doi: 10.1006/viro.1996.0513. [DOI] [PubMed] [Google Scholar]
- 59.Randall R E, Young D F, Goswami K K A, Russell W C. Isolation and characterization of monoclonal antibodies to simian virus 5 and their use in revealing antigenic differences between human, canine and simian isolates. J Gen Virol. 1987;68:2769–2780. doi: 10.1099/0022-1317-68-11-2769. [DOI] [PubMed] [Google Scholar]
- 60.Re F, Braaten D, Franke E K, Luban J. Human immunodeficiency virus type 1 Vpr arrests the cell cycle in G2 by inhibiting the activation of p34cdc2-cyclin B. J Virol. 1995;69:6859–6864. doi: 10.1128/jvi.69.11.6859-6864.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rogel M E, Wu L I, Emerman M. The human immunodeficiency virus type 1 vprgene prevents cell proliferation during chronic infection. J Virol. 1995;69:882–888. doi: 10.1128/jvi.69.2.882-888.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Scheffner M, Werness B A, Huibregtse J M, Levine A J, Howley P M. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 63.Schmitt A P, He B, Lamb R A. Involvement of the cytoplasmic domain of the hemagglutinin-neuraminidase protein in assembly of the paramyxovirus simian virus 5. J Virol. 1999;73:8703–8712. doi: 10.1128/jvi.73.10.8703-8712.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schneider H, Kaelin K, Billeter M A. Recombinant measles viruses defective for RNA editing and V protein synthesis are viable in cultured cells. Virology. 1997;227:314–322. doi: 10.1006/viro.1996.8339. [DOI] [PubMed] [Google Scholar]
- 65.Seedorf K, Oltersdorf T, Krammer G, Rowekamp W. Identification of early proteins of the human papilloma viruses type 16 (HPV 16) and type 18 (HPV 18) in cervical carcinoma cells. EMBO J. 1987;6:139–144. doi: 10.1002/j.1460-2075.1987.tb04731.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sherr C J. Cancer cell cycles. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 67.Shiyanov P, Hayes S A, Donepudi M, Nichols A F, Linn S, Slagle B L, Raychaudhuri P. The naturally occurring mutants of DDB are impaired in stimulating nuclear import of the p125 subunit and E2F1-activated transcription. Mol Cell Biol. 1999;19:4935–4943. doi: 10.1128/mcb.19.7.4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shiyanov P, Nag A, Raychaudhuri P. Cullin 4A associates with the UV-damaged DNA-binding protein DDB. J Biol Chem. 1999;274:35309–35312. doi: 10.1074/jbc.274.50.35309. [DOI] [PubMed] [Google Scholar]
- 69.Slansky J E, Farnham P J. Introduction to the E2F family: protein structure and gene regulation. Curr Top Microbiol Immunol. 1996;208:1–30. doi: 10.1007/978-3-642-79910-5_1. [DOI] [PubMed] [Google Scholar]
- 70.Song B, Liu J J, Yeh K C, Knipe D M. Herpes simplex virus infection blocks events in the G1phase of the cell cycle. Virology. 2000;267:326–334. doi: 10.1006/viro.1999.0146. [DOI] [PubMed] [Google Scholar]
- 71.Spector D L, Goldman R D, Leinwand L A, editors. Cells: a laboratory manual. Vol. 1. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1998. [Google Scholar]
- 72.Steward M, Samson A C R, Errington W, Emmerson P T. The Newcastle disease virus V protein binds zinc. Arch Virol. 1995;140:1321–1328. doi: 10.1007/BF01322759. [DOI] [PubMed] [Google Scholar]
- 73.Thomas S M, Lamb R A, Paterson R G. Two mRNAs that differ by two nontemplated nucleotides encode the amino coterminal proteins P and V of the paramyxovirus SV5. Cell. 1988;54:891–902. doi: 10.1016/S0092-8674(88)91285-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tober C, Seufert M, Schneider H, Billeter M A, Johnston I C, Niewiesk S, ter Meulen V, Schneider-Schaulies S. Expression of measles virus V protein is associated with pathogenicity and control of viral RNA synthesis. J Virol. 1998;72:8124–8132. doi: 10.1128/jvi.72.10.8124-8132.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vidal S, Curran J, Kolakofsky D. Editing of the Sendai virus P/C mRNA by G insertion occurs during mRNA synthesis via a virus-encoded activity. J Virol. 1990;64:239–246. doi: 10.1128/jvi.64.1.239-246.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Watanabe T, Sukegawa J, Sukegawa I, Tomita S, Iijima K, Oguchi S, Suzuki T, Nairn A C, Greengard P. A 127-kDa protein (UV-DDB) binds to the cytoplasmic domain of the Alzheimer's amyloid precursor protein. J Neurochem. 1999;72:549–556. doi: 10.1046/j.1471-4159.1999.0720549.x. [DOI] [PubMed] [Google Scholar]
- 77.Whyte P, Williamson N M, Harlow E. Cellular targets for transformation by the adenovirus E1A proteins. Cell. 1989;56:67–75. doi: 10.1016/0092-8674(89)90984-7. [DOI] [PubMed] [Google Scholar]
- 78.Wilson I A, Niman H L, Houghten R A, Cherenson A R, Connolly M L, Lerner R A. The structure of an antigenic determinant in a protein. Cell. 1984;37:767–778. doi: 10.1016/0092-8674(84)90412-4. [DOI] [PubMed] [Google Scholar]
- 79.Winston J T, Chu C, Harper J W. Culprits in the degradation of cyclin E apprehended. Genes Dev. 1999;13:2751–2757. doi: 10.1101/gad.13.21.2751. [DOI] [PubMed] [Google Scholar]
- 80.Yu Z K, Gervais J L, Zhang H. Human CUL-1 associates with the SKP1/SKP2 complex and regulates p21CIP1/WAF1and cyclin D proteins. Proc Natl Acad Sci USA. 1998;95:11324–11329. doi: 10.1073/pnas.95.19.11324. [DOI] [PMC free article] [PubMed] [Google Scholar]