Abstract
We previously showed that the yeast three-hybrid system provides a genetic assay of both RNA and protein components for avian retroviral RNA encapsidation. In the current study, we used this assay to precisely define cis-acting determinants involved in avian leukosis sarcoma virus packaging RNA binding to Gag protein. In vivo screening of Rous sarcoma virus mutants was performed with randomly mutated minimal packaging sequences (MΨ) made using PCR amplification after cotransformation with GagΔPR protein into yeast cells. Colonies with low β-galactosidase activity were analyzed to locate mutations in MΨ sequences affecting binding to Gag proteins. This genetic assay delineated secondary structural elements that are important for efficient RNA binding, including a single-stranded small bulge containing the initiation codon for uORF3, as well as adjacent stem structures. This implies a possible tertiary structure favoring the high-affinity binding sites for Gag. In most cases, results from the three-hybrid assay were well correlated with those from the viral RNA packaging assays. The results from random mutagenesis using the rapid three-hybrid binding assay are consistent with those from site-directed mutagenesis using in vivo packaging assays.
Retroviruses specifically incorporate two copies of genomic RNA into viral particles, despite the fact that cells contain less than 1% viral RNA in the cytoplasm. The precise encapsidation process requires specific mechanisms by which cis-acting sequences in the viral RNA (termed Ψ) are selectively recognized and specifically bound to trans-acting domains in the viral protein and are packaged into virions. In the absence of the Pol and Env proteins, the Gag protein is sufficient to encapsidate RNA (22, 32). The avian retroviral Gag protein is synthesized as a polyprotein, consisting of matrix (MA), p2, p10, capsid (CA), nucleocapsid (NC), and protease (PR), which is cleaved shortly after, or concomitant with, viral budding (21, 33). Previous studies using deletion analyses and chimeric proteins, which contain the NC domain of one retrovirus substituted for the cognate region of another retroviral Gag, have shown that the NC domain confers the specificity of RNA encapsidation, in which selection of RNA occurs through the Gag precursor protein (1, 11, 12, 18, 26, 34, 35, 41).
In the Moloney murine leukemia virus and human immunodeficiency virus type 1 retroviruses, packaging signals for encapsidation are located in the 5′ untranslated region of the genome. Targeted mutational analyses using these packaging signals showed that the primary sequence is not being recognized as a signal for packaging; rather, the secondary structures, including stem-loops, are the determinants for packaging (4, 10). A 160-nucleotide (nt) minimal packaging sequence of avian leukosis sarcoma virus (ALSV MΨ) was identified in the 5′ end of the genome between the primer binding site and the Gag start codon (7). Unlike other retroviruses, ALSVs contain three short open reading frames (ORFs) upstream of the Gag ORF, and the third one, uORF3, resides within MΨ. Since many mutations showing a decrease in uORF3 translation efficiency also reduced the RNA packaging efficiency, it has been suggested that there is a functional coupling between translation regulation and RNA packaging (14, 15, 28). On the other hand, Sonstegard et al. (37) did not find a direct correlation but rather showed that the secondary structure of uORF3 RNA is important for efficient packaging and is critical to maintaining the balance between translation and packaging, which is mediated by the level of the Gag protein.
We previously used the yeast three-hybrid RNA binding assay (see Fig. 1A) to identify domains in the Gag protein that are involved in specific encapsidation (26). We found that the interactions of the MΨ RNA with Gag are of high affinity and specificity. Using a number of MΨ and Gag mutants, we showed a direct correlation between the yeast three-hybrid binding assay and in vivo packaging assays.
FIG. 1.
(A) Schematic diagram of a yeast three-hybrid system. The specific binding of MS2-RSV MΨ RNA to the protease-deleted Gag polyprotein, GagΔPR, would reconstitute the activity of the transcriptional activator and lead to the expression of a reporter gene, lacZ. DB, DB of the LexA protein; AD, AD of the Gal4 protein; UAS, binding site for the transcriptional activator upstream of the reporter gene. (B) Strategy for in vivo screening of RSV mutants with randomly mutated MΨ sequences using the yeast three-hybrid system. Pools of randomly mutagenized MΨ sequences using PCR were digested with XmaI and SphI and ligated with the XmaI-SphI fragment of the pIIIA/ms2-1 RNA hybrid expression plasmid. The ligation mixture was used to cotransform yeast cells along with the AD-GagΔPR hybrid protein expression plasmid and was plated on the plates lacking Ura and Leu to select both plasmids. The β-Gal activity of each transformant colony was qualitatively measured after colonies were transferred onto filters. Colonies with low β-Gal activities were screened to detect MΨ inserts by using PCR. The positive clones were further characterized to locate mutations in MΨ sequences.
In the current study, we used the yeast binding assay to more precisely define cis-acting determinants for RNA packaging. In vivo random screening of mutants defective in Gag binding indicates that there are secondary structural elements that are important for Gag interactions and points to a possible tertiary structure.
MATERIALS AND METHODS
RSV MΨ random mutagenesis.
PCR-based random mutagenesis of Rous sarcoma virus (RSV) MΨ sequences was based on manganese-induced misincorporation of nucleotides by AmpliTaq DNA polymerase (Perkin-Elmer, Norwalk, Conn.) (9, 19). This procedure also uses an increased concentration of magnesium to stabilize noncomplementary pairs, a higher concentration of DNA polymerase, and an increased number of PCR cycles to decrease the fidelity of PCR amplification (23, 27). In order to introduce only one or two base substitutions in the 160-nt target DNA sequences, we designed two PCR protocols employing different combinations of the above-described modifications. The first protocol uses a 100-μl volume of mutagenesis buffer containing 1× PCR Buffer II (10 mM Tris-HCl [pH 8.3], 50 mM KCl [Perkin-Elmer]), 7 mM MgCl2, 200 μM of each dNTP, 10 pmol of each primer, and 5 U of AmpliTaq DNA polymerase. The second mutagenesis PCR protocol uses 0.5 mM MnCl2 in reaction buffer containing 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 200 μM of each dNTP, 10 pmol of each primer, and 5 U of AmpliTaq DNA polymerase. The wild-type (wt) templates were also prepared using a PCR buffer containing 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 200 μM of each dNTP, 10 pmol of each primer, and 1.25 U of AmpliTaq DNA polymerase. All samples were subjected to heating at 94°C for 4 min followed by 40 cycles of PCR (30 s at 94°C, 1 min at 45°C, 1 min at 72°C) and finished at 72°C for 10 min. The sequences from wt-PCR products were verified by direct sequencing after removing unincorporated nucleotides and primers through Microcon filters (Amicon, Inc., Beverly, Mass.).
A library of RSV MΨ mutant sequences of each reaction was digested with XmaI and SphI, the restriction enzyme sites encoded by the primers, and ligated with the XmaI-SphI fragment of the pIIIA/ms2-1 plasmid (Fig. 1B), which is a derivative of the pIII/ms2-1 hybrid RNA expression plasmid (36) containing the two unique restriction sites for directional cloning. The ligated DNAs were cotransformed into the yeast strain L40-coat (36) along with the pACTII-GagΔPR hybrid protein expression plasmid (26).
Yeast transformation.
The yeast strain L40-coat (36), which stably expresses the LexA-MS2 coat protein fusion gene in the genome along with the TRP1 marker, was used to obtain double transformants expressing the RNA hybrid plasmid and the activation domain (AD) hybrid protein plasmid. The genotype of this strain is MATa ura3-52 leu2-3,112 his3D200 trp1D1 ade2 LYS2::(lexA-op)-HIS3 ura3::(lexA-op)-lacZ lexA-MS2 coat (TRP1). Transformation was performed by using the frozen-EZ yeast transformation II kit (Zymo Research, Orange, Calif.). Plates lacking uracil and leucine were used to select transformants carrying both the RNA hybrid plasmid and the protein hybrid plasmid.
Yeast β-Gal activity assay.
The liquid β-galactosidase (β-Gal) assay was performed to quantitatively measure the enzyme activity using chlorophenol 5-bromo-4-chloro-3-indolyl β-d-galactopyranoside (X-Gal) as a substrate (8). Enzymatic activity represents the average of results for three to four transformants, and assays were repeated three to four times. The filter β-Gal assay was used to measure β-Gal activity of yeast transformants after directly transferring colonies to filters. Cells were permeabilized by three cycles of freeze-and-thaw treatment of filters in a pool of liquid nitrogen, and the filters were placed on another filter that was presoaked in Z buffer–X-Gal solution (110 mM Na2HPO4, 46 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, 330 μg of X-gal/ml, β-mercaptoethanol). Filters were incubated at 30°C and monitored for the appearance of blue β-Gal-positive colonies.
Subcloning MΨ mutations into the heterologous packaging vector.
The 160 nt of MΨ sequences were PCR amplified from the vector pIIIA/MS2-MΨ as a template in which different MΨ mutations were located and then inserted into the unique MluI site of pASY161, a pCMVneo derivative (3), for the heterologous packaging assay.
Cell cultures and transfection.
The quail packaging cell line Q2bn-4D (38) was grown in GM+D+CK (Ham's F10 medium containing 10% tryptose phosphate broth [Difco], 5% calf serum, 1% heat-inactivated chicken serum, and 1% dimethyl sulfoxide). The modified calcium phosphate method (13) was used for DNA transfections on cells seeded in Dulbecco modified Eagle medium supplemented with 10% calf serum. Stably transfected mass cultures of G418-resistant cells at 0.15 mg/ml were obtained after 2 weeks of selection.
Heterologous packaging assay.
Q2bn-4D cells transfected with MΨ mutant packaging vectors were labeled with 250 μCi of [35S]methionine (EXPRESS 35S protein labeling mix; >1,000 Ci/mmol; NEN Research Products) in 2 ml of serum-free Dulbecco modified Eagle medium without methionine and cysteine (DME−met−cys) for 5 h and chased with DME−met−cys medium supplemented with 10% fetal bovine serum for 18 to 24 h. The supernatants were collected, and virus-like particles were harvested by first removing cell debris by low-speed centrifugation and then concentrating viruses by high-speed centrifugation through a 20% sucrose cushion. The viral pellet was then resuspended in isotonic buffer as described previously (3). Half of the concentrated virions were set aside for RNase protection assays (RPA), and the remaining half were immunoprecipitated. The labeled cells were washed twice with cold isotonic buffer, scraped from the plates, and then directly lysed with the lysis buffer (Direct protect kit; Ambion, Inc., Austin, Texas).
For quantitation of virion levels of Gag protein, radioimmunoprecipitation assays (RIPA) were performed. Concentrated [35S]methionine-labeled viral particles were incubated for 2.5 h at room temperature in 1.0 ml of Ab buffer (20 mM Tris-HCl [pH 7.4], 50 mM NaCl, 0.5% NP-40, 0.5% deoxycholic acid (DOC), 0.5% sodium dodecyl sulfate (SDS), 0.5% aprotinin, 1 mM EDTA [pH 8.0]) with 5 μl of polyclonal rabbit anti-RSV PRB antibody and 30 μl of protein A-Sepharose beads (Pharmacia LKB Biotechnology, Inc.). The antigen-antibody complexes were washed twice in RIPA buffer (10 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1% NP-40, 1% DOC, 0.1% SDS, 0.5% aprotinin), once in high-salt buffer (10 mM Tris-HCl [pH 7.4], 2 M NaCl, 1% NP-40, 0.5% DOC), and then once more with RIPA buffer. The bound proteins were eluted in SDS sample buffer and loaded onto SDS–12.5% polyacrylamide gels. The dried gels were scanned directly by the Molecular Dynamics PhosphorImager after overnight exposure. Radioactive bands were quantitated by using the ImageQuant software.
RPA were performed on viral and whole-cell lysates. To make the antisense neo probe to detect the heterologous RNA, pASY185 (5) was linearized with NcoI and in vitro transcribed with T7 RNA polymerase to produce a probe which protects 249 nt. To make the antisense glyceraldehyde-3-phosphate dehydrogenase (gapdh) probe, pGEM-GAPDH (17) was linearized with HindIII and in vitro transcribed with T7 RNA polymerase to produce a probe which protects 169 nt of gapdh. The probes were gel purified on a 6% polyacrylamide gel. After hybridization of RNAs with probes and RNase treatment, protected RNAs were separated on a 6% polyacrylamide gel. The gel was scanned directly by a Molecular Dynamics PhosphorImager after overnight exposure. RNA bands were quantitated, in machine units, by using ImageQuant software.
Calculation of packaging efficiency.
Packaging efficiencies for the heterologous RNAs were determined by calculating the amount of neo RNA in virions (as measured by RPA) normalized to the level of neo RNA in the cells, relative to a constitutive cellular message, gapdh (as measured by RPA of whole cell lysates). This calculated level of RNA was then normalized to the number of virions (as measured by RIPA).
RESULTS AND DISCUSSION
To define cis-acting determinants for RSV MΨ RNA binding to Gag protein involved in packaging, we randomly mutated MΨ RNA sequences using PCR methods to obtain a low frequency of mutation (as described in Materials and Methods). We then introduced mutated plasmids into yeast cells along with the Gag protein and used the yeast three-hybrid system to select mutants defective in binding to Gag. Like the two-hybrid approach for protein-protein interactions, the yeast three-hybrid system is based on the transcriptional activation of separable domains of regulatory proteins, such as GAL4 and LexA, and is composed of three hybrid molecules (36) (Fig. 1A). One hybrid molecule consists of the DNA binding domain (DB) of LexA fused to a known RNA binding domain, the MS2 coat protein in this case. The other protein hybrid contains the activation domain (AD) of GAL4 hybridized with the protein of interest, the RSV Gag polyprotein. The third hybrid is an RNA-RNA hybrid. The N-terminal MS2 RNA, which binds specifically to the MS2 coat protein, is fused to the target RNA, RSV MΨ RNA. If specific interactions between RSV MΨ RNA and the Gag polyprotein occur, they drive the transcriptional activation of the reporter gene, lacZ. We previously found that use of the GagΔPR protein led to a higher level of sensitivity than wt Gag in the three-hybrid assay, and this construct could be used for screening of defined mutants in both RNA and protein components. GagΔPR assays yielded results which correlated well with the in vivo packaging assay results (26). Thus, in the present studies we also used GagΔPR.
After transforming of yeast cells with a library of RSV MΨ mutant sequences along with the GagΔPR protein was done, selected colonies were incubated at 30°C for 3 days and were directly transferred to filters and assayed for β-Gal activity. In an initial screen, of 115 transformant colonies screened, 89 colonies were as blue as wt cells carrying the MΨ plasmid. We also obtained 13 pale blue, 2 paler blue, and 11 white colonies. All pale blue and paler blue colonies, as well as five blue and two white colonies as controls, were PCR amplified in order to screen MΨ inserts in the RNA hybrid expression plasmid (Fig. 1B). All blue colonies and 10 out of the 15 pale blue and paler blue colonies contained MΨ inserts; the remaining 5 pale blue colonies as well as all of the white colonies did not contain MΨ inserts. All clones that had MΨ inserts, including blue colonies, were sequenced to locate the mutations in MΨ RNA.
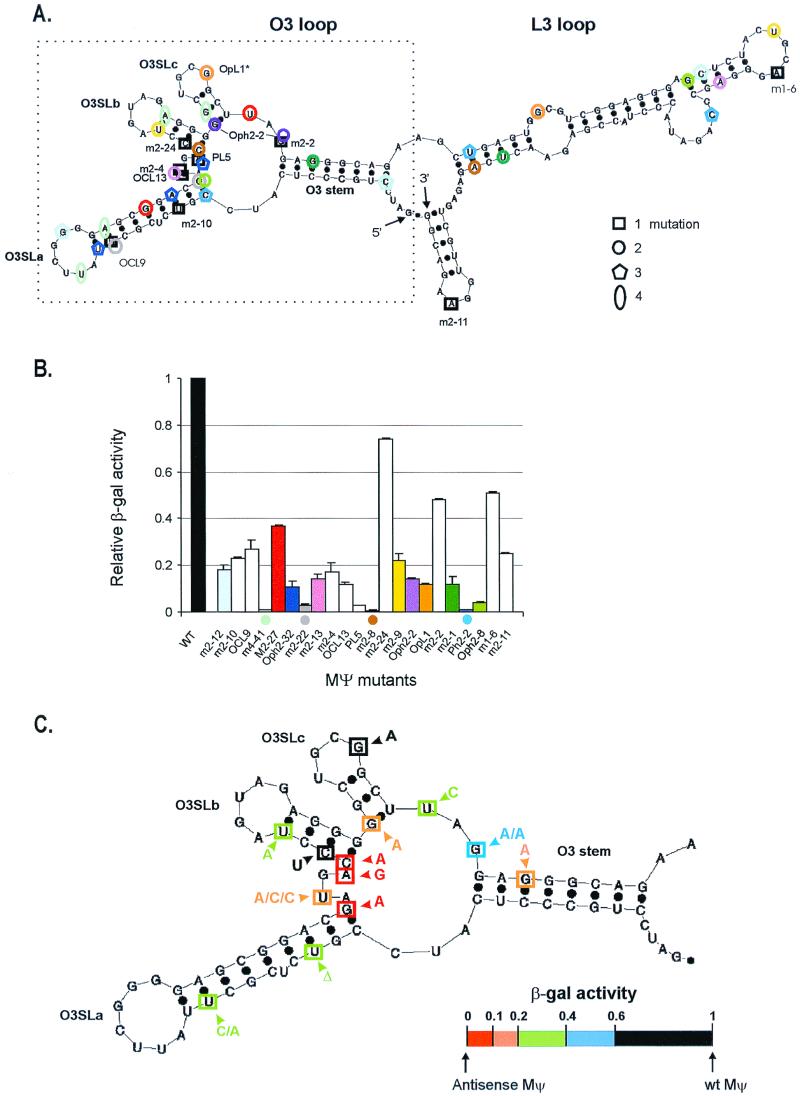
From the initial screen and another screen, 227 clones were analyzed and 22 individual mutations were identified. The locations of mutations are indicated on the secondary structure diagram of MΨ in Fig. 2A. This secondary structure was obtained by computer modeling and verified by both phylogenetic analysis and in vivo heterologous packaging assays (6). The secondary structure is predicted to contain two major stem-loops (O3 and L3 loops), and the O3 loop can be divided into three smaller stem-loops: O3SLa, O3SLb, and O3SLc. Of the 22 mutations examined, the majority were single or double mutants (10 single mutations and 8 double mutations). We also obtained three triple mutations (pentagons) and one quadruple mutation (oval). The mutation frequency (number of mutations/number of sequence nucleotides multiplied by the number of total clones analyzed) was 1.1 × 10−3. More mutations mapped to the 5′ half of the MΨ (O3 loop) sequence than to the 3′ half (L3 loop), and some individual mutations were isolated multiple times. To quantitate the effect of each MΨ mutant on binding to the GagΔPR protein, we measured the β-Gal activities, using a liquid β-Gal assay. The β-Gal activity of each mutant relative to that of wt MΨ RNA is shown in Table 1 and plotted in Fig. 2B. Fourteen mutants showed large reductions in β-Gal activity (at least fivefold) with respect to the wild type, while five mutants still had a substantial amount of β-Gal activity and the other three mutants bound to the GagΔPR protein only twofold less efficiently than wt MΨ RNA.
FIG. 2.
Summary of MΨ random mutants. (A) Location of mutations in the putative secondary structure of MΨ RNA. The MΨ RNA is predicted to have two major stem-loops (the O3 and L3 loops), and from the O3 loop are extended three smaller stem-loops, noted as O3SLa, O3SLb, and O3SLc. The number of mutations is indicated by different shapes: □, single mutation; ○, double mutation; ⌂, triple mutation; 0, quadruple mutation. Each color represents a different mutant. Single mutations are marked with their respective names. The 5′ 82-nt segment of the MΨ sequence (designated μΨ [6]) is shown in the box with a dotted line. (B) The β-Gal activity of each mutant relative to that of wt MΨ RNA. Single mutations are shown in white, and the color of each multiple mutant shown in the column graph is matched with that in panel A. For lower β-Gal activity one, the dot with matching color shown in panel A is redrawn at the bottom of the corresponding bar graph. (C) Recapitulation of mutations in the μΨ sequence. Each box represents a different mutation. The new sequence is indicated. Some mutations at the same specific nucleotides were isolated more than once, and these were indicated by more than one letter at the arrowhead. The β-Gal activity of each mutant relative to that of wt MΨ RNA, from three independent assays, is indicated with a different color. The colors in panel C do not mean the same things as in panels A and B. All mutations which led to a binding efficiency of 0.2 or less are indicated by red or orange. No detectable β-Gal activity above background was obtained with antisense MΨ RNA.
TABLE 1.
β-Gal activity of RSV MΨ mutants in the yeast three-hybrid assay
| MΨ constructa | β-gal activityb (U) | β-Gal activity relative to that of wt |
|---|---|---|
| wt | 4,720 ± 70 | 1 |
| m2-12 | 870 ± 90 | 0.18 |
| m2-10 | 1,090 ± 120 | 0.23 |
| OCL9 | 1,270 ± 50 | 0.27 |
| m4-41 | 44 ± 7 | 0.01 |
| m2-27 | 1,750 ± 140 | 0.37 |
| Oph2-32 | 530 ± 110 | 0.11 |
| m2-22 | 140 ± 10 | 0.03 |
| m2-13 | 680 ± 110 | 0.14 |
| m2-4 | 790 ± 180 | 0.17 |
| OCL13 | 560 ± 30 | 0.12 |
| PL5 | 150 ± 5 | 0.03 |
| m2-8 | 33 ± 5 | 0.007 |
| m2-24 | 3,490 ± 260 | 0.74 |
| m2-9 | 870 ± 140 | 0.22 |
| Oph2-2 | 680 ± 40 | 0.14 |
| OpL1 | 550 ± 20 | 0.12 |
| OpL1c | 5,240 ± 510 | 1.11 |
| m2-2 | 2,270 ± 300 | 0.48 |
| m2-1 | 590 ± 140 | 0.12 |
| Ph2-2 | 60 ± 8 | 0.01 |
| Oph2-8 | 200 ± 20 | 0.04 |
| m1-6 | 2,410 ± 130 | 0.51 |
| m2-11 | 1,180 ± 220 | 0.25 |
Mutants listed from left to right in Fig. 2B.
The enzymatic activity represents the mean of three to four assays from three to four independent transformants with the standard deviation indicated. The β-Gal activity of cells cotransformed with the RNA hybrid plasmid lacking MΨ sequences and the GagΔPR protein hybrid plasmid is about 15 U.
Site-directed single mutation in OpL1 (G→A in O3SLc).
In the case of the Ψ sequences containing more than one mutation, it was important to determine the contribution of each mutation to the binding phenotype. Our laboratory has recently found that the 3′ half of MΨ can be deleted without affecting the efficiency of packaging of a heterologous RNA, defining an 82-nt packaging signal called μΨ (6) (Fig. 2A). We compared μΨ and MΨ β-Gal activities in the three-hybrid assay and found that they were the same (data not shown). Computer modeling of μΨ suggested that the predicted secondary structure is identical to the 5′ half of MΨ. We could therefore eliminate the contribution of mutations in the 3′ half of MΨ (L3) to the binding phenotype. Two single mutations in the L3 region (m2-11 and m1-6) gave significant amounts of β-Gal activities (Fig. 2A and B). The locations of the mutations that are present in μΨ are shown in Fig. 2C, which summarizes the effects of the individual nucleotide changes. Since multiple defects are expected with the triple and quadruple mutants, we excluded these from the summary. In the case of the double mutants, we marked only the mutational change predicted to contribute the greatest effect. Two criteria were used to determine which mutations are predicted to be the major contributors. As stated above, we eliminated the contribution of mutations in the 3′ half of MΨ. In five of the eight double mutants, one mutation was in the 5′ half. The other three double mutants had both mutations in the μΨ sequence. However, in the case of two mutants that showed severe defects in binding (7- and 30-fold reduction of β-Gal activity from that of the wt, respectively), one mutational change was the same as a single mutant which showed a substantial amount of β-Gal activity. For example, the Oph2-2 mutant (Fig. 2) contained one mutation identical to that found in m2-2, which has high β-Gal activity and efficient packaging (Fig. 3). Thus, we could map the important mutation to the other nucleotide in the O3SLc stem. Another double mutant, m2-27 (Fig. 2A), had one mutation in the O3SLa stem and the other at the single-stranded region between O3SLc and the O3 stem. Banks and Linial (6) mutated 2 nt in the middle of the O3SLa stem (a G in O3SLa was at the same nucleotide as one of these two), and this directed mutant packaged heterologous RNA as efficiently as wt MΨ RNA. Thus, in this double mutant, the important mutational change is most likely the U in the single-stranded region between the O3SLc and the O3 stems.
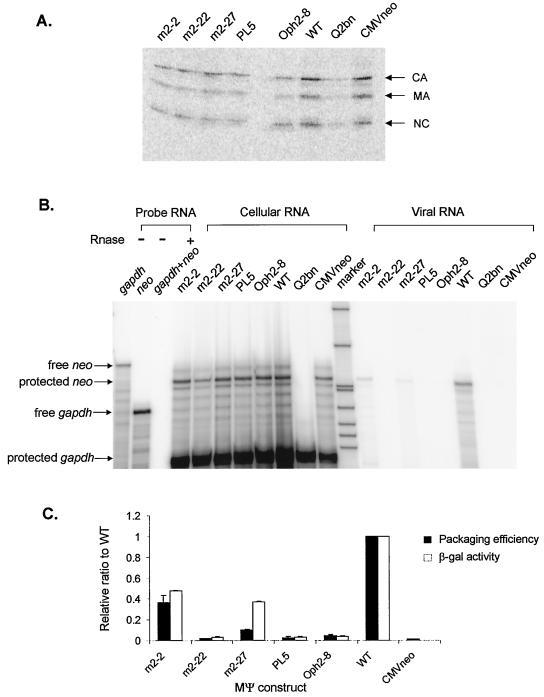
FIG. 3.
(A) RIPA to determine the number of viral particles released to the medium. Q2bn, untransfected cells; CMVneo, Q2bn-4D transfected cells with the parental plasmid pASY161, a pCMVneo derivative (3). (B) RPA to measure the amount of neo RNA in virions and the amounts of neo RNA and cellular gapdh RNA in transfected cells. (C) Comparison of the β-Gal activity measured in the yeast three-hybrid assay and the packaging efficiency determined in vivo. The packaging efficiency was calculated as follows: the amount of neo RNA in virions (as measured by RPA [B]) was normalized to the amount of cellular neo RNA, relative to the level of cellular gapdh RNA (as measured by RPA of whole-cell lysates [B]). This calculated neo RNA was then normalized to the number of virions (as measured by RIPA [A]). Both β-Gal activities and packaging efficiencies were normalized to those for wt MΨ. Each experiment was done three or four times, and the bars show the standard deviations.
As a complementary experiment, we measured in vivo packaging efficiencies of a set of MΨ mutants to correlate results from the yeast three-hybrid binding assay with those of an in vivo packaging assay. The yeast MΨ mutants of various levels of β-Gal activities (in the range of 0.5- to 0.03-fold of wt MΨ RNA activities) were selected. MΨ confers efficient packaging when tethered to heterologous RNAs (6). To measure the ability of MΨ RNAs to confer packaging in the heterologous assay, we amplified mutant MΨ RNAs using PCR and placed them 3′ of the neo gene in the plasmid pCMVneo (3). The constructs were then transfected into the Q2bn-4D packaging cell line, and stably transfected mass cultures were obtained after selection with G418. Quantitative RIPA and RPA were performed with viral particles collected from culture supernatants and transfected cells. Supernatants from Q2bn-4D packaging cells transfected with the five mutants yielded somewhat lower amounts of pelletable capsid protein in the cell supernatants than transfected cells with CMVneo wt MΨ RNA or the parental vector without MΨ sequences (0.4- to 0.6-fold) (Fig. 3A). Although all transfected cells showed equivalent amounts of cellular neo RNAs, relative to a constitutive cellular message, gapdh, these mutants had different effects on packaging of neo RNA into particles (Fig. 3B). The packaging efficiency was calculated as described in Materials and Methods and compared with β-Gal activity of each mutant (Fig. 3C). In five out of six tested, results from the β-Gal assay were consistent with those from the in vivo packaging assay. However, the mutant m2-27, which exhibited only a 2.7-fold reduction in β-Gal activity relative to that of wt MΨ, had a packaging efficiency about 10-fold less than that of wt MΨ. This mutant may have a defect in an event downstream of RNA binding. It is likely that cytoplasmic or membrane cellular proteins act as chaperones for both particle assembly and encapsidation downstream of RNA binding to the Gag protein. Possibly, m2-27 delineates an RNA region which is not absolutely required for Gag binding but is required for binding of such a cellular factor. Previously, using a set of mutants both in MΨ RNA and in the Gag protein, we found a good correlation between β-Gal activity and in vivo packaging efficiency. We found that a level of β-Gal activity that was ≤20% of that of the wt led to a large reduction in RNA packaging (26). Thus, all mutations which led to a binding efficiency of 0.2 or less are indicated in Fig. 2C.
O3 stem.
Previous site-directed mutagenesis of three nucleotides in the O3 stem had shown that the base pairing in the O3 stem is required for efficient RNA encapsidation (6, 24). In our random screening using the yeast-three hybrid system, only one single mutant mapped to this region, and it caused a great reduction in binding to GagΔPR (less than 20% of the binding efficiency of the wt). This reflects the important role of the secondary structure of the O3 stem for specific RNA encapsidation.
O3SLa stem-loop.
The two mutants which had changes in the stem structure of O3SLa (including one which deleted a nucleotide) had only a modest effect on β-Gal activity (about a threefold reduction). Our previous work (26) showed that mutation of the four G's in the loop had a less than twofold effect on binding and little effect on packaging. These results are also consistent with results of Banks and Linial (6) using site-directed mutants in this region. The O3SLa region is less conserved among ALSV isolates than other portions of MΨ, and another stable secondary structure has been described for this region (20). Taken together, it is unlikely that this region plays a critical role in RNA binding and packaging.
AUG bulge.
The region upstream of Gag contains three AUGs followed by small potential ORFs. There is no evidence that any of the peptides encoded by these ORFs are actually made in infected cells. The three ORFs have been studied for their effects on the translation of the downstream Gag gene and on genome packaging (14, 15, 28, 29, 37). In some studies, many, but not all, mutations decreased the translation efficiency of uORF3 and also decreased the RNA packaging efficiency, suggesting that the regulation of translation appears to be functionally linked to the packaging (14, 15, 28). But other investigators found no direct functional coupling between translation and packaging. Instead, their data suggested that the secondary structure around the uORF3 is important for packaging (37). In our screen, four mutations were isolated around the uORF3 initiation codon, and all of these drastically decreased the RNA binding efficiency. Three of the mutations disrupted the initiation codon for uORF3, while the change in one mutant (PL5) (Fig. 2A and B) resulted in a better Kozak consensus sequence (AXXAUGA→AXXAUGG, where X is unknown) (25), predicted to increase translation of uORF3. However, this mutational change (PL5) completely abolished RNA binding and packaging (Fig. 2 and 3). The phenotype of this mutant is not consistent with a role for control of uORF3 translation in RNA packaging efficiency but rather suggests that the secondary structure of this region surrounding the initiation codon for uORF3 is important for efficient packaging.
O3SLb and O3SLc stems.
One O3SLb mutant (m2-24) (Fig. 2A) that is located close to the bottom of the stem, in which a predicted G-C base pair is changed to a G-U base pair, behaved like wt MΨ in the three-hybrid assay. In phylogenetic comparisons of different ALSV strains, G-U base pairing is more predominant than G-C base pairing in the O3 stem structure, which is suggested to be one of the most important secondary structures for efficient RNA encapsidation (6, 20). In one study (16), randomized mutations in the Ψ region which allowed viral replication were selected. These authors found that in the O3 stem, only sequences which preserved base pairing permitted replication (and therefore packaging). Both A-U and G-U base pairs allowed viral replication. However, one mutation we isolated in the O3SLc stem (Oph2-2) (Fig. 2A and B) had a change from a G-U base pair to an A-U base pair and decreased β-Gal activity to less than 20% of that of the wt. A G-U base pair is thermodynamically as stable as an A-U pair by making two hydrogen bonds, but it causes a shift from the canonical Watson-Crick pair in the geometry of an RNA helix (2). Thus, a G-U wobble base pair conformation might disrupt the RNA tertiary structure formation, such as is shown at the splice site helix from group I self-splicing introns (39). The other possible explanation for the phenotype of Oph2-2 could be the need for specific recognition of the G residue for Gag protein interactions. However, the latter is less likely, since a previously identified double mutant, in which a G-U base pair is changed to a C-G base pair, was packaging competent (6). Taken together, these results suggest that the stem structures of O3SLb and O3SLc, and particularly the bottom part of the stems, are involved in efficient packaging, consistent with the previous mutational analysis using in vivo heterologous packaging assays. It was previously shown that a substitution mutation at the five G nucleotides spanning the O3SLb and O3SLc stems almost completely abolished packaging (26), and two changes out of five nucleotides in each stem decreased the packaging efficiency, which was restored by compensatory mutations restoring base pairing (6).
Single-stranded regions.
In this study, we isolated very few mutations in the single-stranded regions. The three mutations located in the single-stranded region between O3SLc and the O3 stem still retained significant amounts of β-Gal activity, indicating that this single-stranded region is not critical for protein binding. Other than one mutation in the O3SLc loop, all such mutations which decreased binding to Gag were in the AUG bulge. The effect we found with the mutation changing UGCG in the O3SLc loop to UGCA (OpL1) (Fig. 2A and B) is not consistent with results of Banks and Linial (6) in which a more drastic mutation in this region did not affect packaging. In order to determine whether the combination of the two mutations in the OpL1 mutant has a subtle and unpredictable effect on folding or stability of the Ψ structure, we created a mutant containing only the single change in the O3SLc loop (OpL1*) (Fig. 2A). We found that OpL1* has as high a level of β-Gal activity as wt MΨ RNA in the three-hybrid binding assay (1.11-fold of that of the wt in Table 1).
To confirm and expand previous secondary structure analyses using computer modeling and in vivo heterologous packaging assays (6), our present study used a yeast three-hybrid binding assay to examine the binding of mutated Ψ RNAs to Gag. Unlike the DNA double helix, where the major groove is accessible for protein interactions, double-stranded RNA helices form a different geometry (A-form versus B-form of the DNA helix), and so they create different surfaces for protein binding; a narrow, deep major groove and a shallow, wide minor groove (31). Most RNA sites for protein binding are found at the ends of helices, or more likely in helices with loops, bulges, and non-Watson-Crick base pairs, which open up the major groove of the adjacent double helix facilitating the recognition of major groove functional groups (40). Three external O3 loop regions (O3SLa, O3SLb, and O3SLc) were directly mutated in our laboratory and showed only modest defects in packaging efficiency (6, 26). Work described here and previous work from our laboratory (6) points to the single-stranded AUG uORF3 bulge as being a likely protein binding site in a predicted tertiary structure consisting of the O3 stem, the AUG bulge, and the bottoms of the O3SLb and O3SLc stems. However, we cannot completely rule out a role for the loop between the O3 stem and O3SLa as a protein contact point. Some studies have reported that the minor groove of an RNA double helix is recognized for RNA-protein interactions of the alanine tRNA synthetase-tRNA binding (30) or RNA-RNA interactions in the tertiary structure of the tetrahymena ribozyme reactive site (39). We thus predict that the minor groove in one of O3 loop stems could serve as part of the Gag binding site.
In summary, the in vivo random screening using the yeast three-hybrid system highlights several important structures of MΨ RNA for efficient packaging, including a small single-stranded bulge containing the initiation codon for uORF3 and adjacent stem structures, in addition to the O3 stem structure. These finding are fully consistent with those from in vivo heterologous packaging assays that previously defined the MΨ mutations (6).
ACKNOWLEDGMENTS
We are grateful to Annie Alidina and Cynthia May for technical assistance. We also thank Adrian Ferré-D'Amaré and Michael Emerman for critical reviews of the manuscript.
This work was supported by a grant from the National Cancer Institute (CA 18282) to M.L.L. E-G.L. was partially supported by an NIH postdoctoral training grant in viral oncology (T32CA09229).
REFERENCES
- 1.Aldovini A, Young R A. Mutations of RNA and protein sequences involved in human immunodeficiency virus type 1 packaging result in production of noninfectious virus. J Virol. 1990;64:1920–1926. doi: 10.1128/jvi.64.5.1920-1926.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allain F H, Varani G. Structures of the P1 helix from group I self-splicing introns. J Mol Biol. 1995;250:333–353. doi: 10.1006/jmbi.1995.0381. [DOI] [PubMed] [Google Scholar]
- 3.Aronoff R, Linial M L. Specificity of retroviral RNA packaging. J Virol. 1991;65:71–80. doi: 10.1128/jvi.65.1.71-80.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Banks J D, Beemon K L, Linial M L. RNA regulatory elements in the genomes of simple retroviruses. Semin Virol. 1997;8:194–204. [Google Scholar]
- 5.Banks J D, Kealoha B O, Linial M L. An MΨ-containing heterologous RNA, but not env mRNA, is efficiently packaged into avian retroviral particles. J Virol. 1999;73:8926–8933. doi: 10.1128/jvi.73.11.8926-8933.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Banks J D, Linial M L. Secondary structure analysis of a minimal avian leukosis-sarcoma virus packaging signal. J Virol. 2000;74:456–464. doi: 10.1128/jvi.74.1.456-464.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Banks J D, Yeo A, Green K, Cepeda F, Linial M L. A minimal avian retroviral packaging sequence has a complex structure. J Virol. 1998;72:6190–6194. doi: 10.1128/jvi.72.7.6190-6194.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bartel P L, Roecklein J A, SenGupta D, Fields S. A protein linkage map of Escherichia coli bacteriophage T7. Nat Genet. 1996;12:72–77. doi: 10.1038/ng0196-72. [DOI] [PubMed] [Google Scholar]
- 9.Beckman R A, Mildvan A S, Loeb L A. On the fidelity of DNA replication: manganese mutagenesis in vitro. J Biochem. 1983;24:5810–5817. doi: 10.1021/bi00342a019. [DOI] [PubMed] [Google Scholar]
- 10.Berkowitz R, Fisher J, Goff S P. RNA packaging. Curr Top Microbiol Immunol. 1996;214:177–218. doi: 10.1007/978-3-642-80145-7_6. [DOI] [PubMed] [Google Scholar]
- 11.Berkowitz R D, Ohagen A, Hoglund S, Goff S P. Retroviral nucleocapsid domains mediate the specific recognition of genomic viral RNAs by chimeric Gag polyproteins during RNA packaging in vivo. J Virol. 1995;69:6445–6456. doi: 10.1128/jvi.69.10.6445-6456.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bowles N E, Damay P, Spahr P F. Effect of rearrangements and duplications of the cys-his motifs of Rous sarcoma virus nucleocapsid protein. J Virol. 1993;67:623–631. doi: 10.1128/jvi.67.2.623-631.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Donze O, Damay P, Spahr P F. The first and third uORFs in RSV leader RNA are efficiently translated: implications for translational regulation and viral RNA packaging. Nucleic Acids Res. 1995;23:861–868. doi: 10.1093/nar/23.5.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Donze O, Spahr P F. Role of the open reading frames of Rous sarcoma virus leader RNA in translation and genome packaging. EMBO J. 1992;11:3747–3757. doi: 10.1002/j.1460-2075.1992.tb05460.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doria-Rose N A, Vogt V M. In vivo selection of Rous sarcoma virus mutants with randomized sequences in the packaging signal. J Virol. 1998;72:8073–8082. doi: 10.1128/jvi.72.10.8073-8082.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dugaiczyk A, Haron J A, Stone E M, Dennison O E, Rothblum K N, Schwartz R J. Cloning and sequencing of a deoxyribonucleic acid copy of glyceraldehyde-3-phosphate dehydrogenase messenger ribonucleic acid isolated from chicken muscle. Biochemistry. 1983;22:1605–1613. doi: 10.1021/bi00276a013. [DOI] [PubMed] [Google Scholar]
- 18.Dupraz P, Spahr P-F. Specificity of Rous sarcoma virus nucleocapsid protein in genomic RNA packaging. J Virol. 1992;66:4662–4670. doi: 10.1128/jvi.66.8.4662-4670.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goodman M F, Keener S, Guidotti S. On the enzymatic basis for mutagenesis by manganese. J Biol Chem. 1983;258:3469–3475. [PubMed] [Google Scholar]
- 20.Hackett P B, Dalton M W, Johnson D P, Petersen R B. Phylogenetic and physical analysis of the 5′ leader RNA sequences of avian retroviruses. Nucleic Acids Res. 1992;19:6929–6934. doi: 10.1093/nar/19.24.6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaplan A H, Manchester M, Swanstrom R. The activity of the protease of human immunodeficiency virus type 1 is initiated at the membrane of infected cells before the release of viral proteins and is required for release to occur with maximal efficiency. J Virol. 1994;68:6782–6786. doi: 10.1128/jvi.68.10.6782-6786.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaye J F, Lever A M L. trans-acting proteins involved in RNA encapsidation and viral assembly in human immunodeficiency virus type 1. J Virol. 1996;70:880–886. doi: 10.1128/jvi.70.2.880-886.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keohavong P, Thilly W G. Fidelity of DNA polymerase in DNA amplification. Proc Natl Acad Sci USA. 1989;86:9253–9257. doi: 10.1073/pnas.86.23.9253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Knight J B, Si Z H, Stoltzfus C M. A base-paired structure in the avian sarcoma virus 5′ leader is required for efficient encapsidation of RNA. J Virol. 1994;68:4493–4502. doi: 10.1128/jvi.68.7.4493-4502.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- 26.Lee E-G, Yeo A, Kraemer B, Wickens M, Linial M L. The Gag domains for avian retroviral RNA encapsidation determined using two independent methods. J Virol. 1999;73:6282–6292. doi: 10.1128/jvi.73.8.6282-6292.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin-Goerke J L, Robbins D J, Burczak J D. PCR-based random mutagenesis using manganese and reduced dNTP concentration. BioTechniques. 1997;23:409–412. doi: 10.2144/97233bm12. [DOI] [PubMed] [Google Scholar]
- 28.Moustakas A, Sonstegard T S, Hackett P B. Alterations of the three short open reading frames in the Rous sarcoma virus leader RNA modulate viral replication and gene expression. J Virol. 1993;67:4337–4349. doi: 10.1128/jvi.67.7.4337-4349.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moustakas A, Sonstegard T S, Hackett P B. Effects of the open reading frames in the Rous sarcoma virus leader RNA on translation. J Virol. 1993;67:4350–4357. doi: 10.1128/jvi.67.7.4350-4357.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Musier-Forsyth K, Schimmel P. Functional contacts of a transfer RNA synthetase with 2′-hydroxyl groups in the RNA minor groove. Nature. 1992;357:513–515. doi: 10.1038/357513a0. [DOI] [PubMed] [Google Scholar]
- 31.Nowakowski J, Tinoco I., Jr RNA structure and stability. Semin Virol. 1997;8:153–165. [Google Scholar]
- 32.Oertle S, Spahr P F. Role of the Gag polyprotein precursor in packaging and maturation of Rous sarcoma virus genomic RNA. J Virol. 1990;64:5757–5763. doi: 10.1128/jvi.64.12.5757-5763.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oroszlan S, Luftig R B. Retroviral proteinases. In: Swanstrom R, Vogt P K, editors. Retroviruses—strategies of replication. Berlin, Germany: Springer-Verlag; 1990. pp. 153–186. [Google Scholar]
- 34.Poon D T, Li G, Aldovini A. Nucleocapsid and matrix protein contributions to selective human immunodeficiency virus type 1 genomic RNA packaging. J Virol. 1998;72:1983–1993. doi: 10.1128/jvi.72.3.1983-1993.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sakalian M, Wills J W, Vogt V M. Efficiency and selectivity of RNA packaging by Rous sarcoma virus Gag deletion mutants. J Virol. 1994;68:5969–5981. doi: 10.1128/jvi.68.9.5969-5981.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.SenGupta D J, Zhang B, Kraemer B, Pochart P, Fields S, Wickens M. A three-hybrid system to detect RNA-protein interactions in vivo. Proc Natl Acad Sci USA. 1996;93:8496–8501. doi: 10.1073/pnas.93.16.8496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sonstegard T S, Hackett P B. Autogenous regulation of RNA translation and packaging by Rous sarcoma virus Pr76gag. J Virol. 1996;70:6642–6652. [PMC free article] [PubMed] [Google Scholar]
- 38.Stoker A W, Bissell M J. Development of avian sarcoma and leukosis virus-based vector-packaging cell lines. J Virol. 1988;62:1008–1015. doi: 10.1128/jvi.62.3.1008-1015.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Strobel S, Cech T R. Minor groove recognition of the conserved G-U pair at the tetrahymena ribozyme reaction site. Science. 1995;267:675–679. doi: 10.1126/science.7839142. [DOI] [PubMed] [Google Scholar]
- 40.Weeks K M, Crothers D M. Major groove accessibility of RNA. Science. 1993;261:1574–1577. doi: 10.1126/science.7690496. [DOI] [PubMed] [Google Scholar]
- 41.Zhang Y Q, Barklis E. Nucleocapsid protein effects on the specificity of retrovirus RNA encapsidation. J Virol. 1995;69:5716–5722. doi: 10.1128/jvi.69.9.5716-5722.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]