Abstract
Vaccinia virus encodes two glutaredoxins, O2L and G4L, both of which exhibit thioltransferase and dehydroascorbate reductase activities in vitro. Although O2L was previously found to be dispensable for virus replication, we now show that G4L is necessary for virion morphogenesis. RNase protection and Western blotting assays indicated that G4L was expressed at late times after infection and was incorporated into mature virus particles. Attempts to isolate a mutant virus with a deleted G4L gene were unsuccessful, suggesting that the protein was required for virus replication. This interpretation was confirmed by the construction and characterization of a conditional lethal recombinant virus with an inducible copy of the G4L gene replacing the original one. Expression of G4L was proportional to the concentration of inducer, and the amount of glutaredoxin could be varied from barely detectable to greater than normal amounts of protein. Immunogold labeling revealed that the induced G4L protein was associated with immature and mature virions and adjacent cytoplasmic depots. In the absence of inducer, the production of infectious virus was severely inhibited, though viral late protein synthesis appeared unaffected except for decreased maturation-dependent proteolytic processing of certain core components. Electron microscopy of cells infected under nonpermissive conditions revealed an accumulation of crescent membranes on the periphery of electron-dense globular masses but few mature particles. We concluded that the two glutaredoxin homologs encoded by vaccinia virus have different functions and that G4L has a role in virion morphogenesis, perhaps by acting as a redox protein.
Vaccinia virus, the prototypic member of the Orthopoxvirus genus in the family Poxviridae, has a double-stranded DNA genome of approximately 185 kbp with nearly 200 open reading frames (ORFs) that are likely to encode proteins (15). Clues regarding the functions of some of these genes have been obtained from the identification of cellular homologs. For example, O2L and G4L resemble glutaredoxins in sequence and were therefore predicted to have redox capabilities. This idea was confirmed, as both viral proteins exhibited thioltransferase and dehydroascorbate reductase activities in vitro (3, 12). In addition, the O2L glutaredoxin was shown to serve as a cofactor in the reduction of ribonucleotides to the corresponding deoxyribonucleotides by the viral ribonucleotide reductase with glutathione serving as the hydrogen donor (16). The finding that O2L was nonessential for vaccinia virus replication (16) was consistent with similar findings for other vaccinia virus-encoded enzymes with primary roles in DNA precursor biosynthesis. Nevertheless, the late expression of O2L is atypical for such enzymes and leaves open other biological roles. Still less is known about the glutaredoxin encoded by the G4L gene, which is no more closely related to O2L than to other members of the glutaredoxin family. Unlike O2L, G4L is conserved in poxviruses from other genera including Molluscum contagiosum virus (18), Shope rabbit fibroma virus (20), Myxoma virus (7), Fowlpoxvirus (2), and an entomopoxvirus (1). As the majority of highly conserved genes are essential for virus replication, we suspected that G4L might have a different role than that of O2L despite their similar in vitro activities. In addition to serving as a cofactor for ribonucleotide reductase, cellular glutaredoxins have numerous functions related to the maintenance of proteins at the correct redox state including the regulation of transcription, oxidative stress pathways, and the metabolism of sulfur. Here we show that G4L is essential for the morphogenesis of vaccinia virions.
MATERIALS AND METHODS
Cells and viruses.
Cells and recombinant vaccinia viruses were propagated as previously described (10) using mycophenolic acid, xanthine, and hypoxanthine to select for xanthine-guanine phosphoribosyltransferase (gpt); Geneticin to select for neomycin resistance; and 50 μM isopropylthiogalactoside (IPTG) for inducer-dependent strains. Virus was purified by centrifugation through a 36% sucrose cushion or by sucrose gradient centrifugation (11).
Antibodies.
Antiserum was raised to the C-terminal 13 amino acids of the predicted G4L sequence preceded by a cysteine (CEKATYGVWPPVTE). A purified murine monoclonal antibody (MAb) to the influenza virus hemagglutinin (HA) epitope tag (HA.11) was obtained from BABco/Covance Research Products Inc., Richmond, Calif.
RNase protection assays.
BS-C-1 cells were infected with vaccinia virus strain WR at a multiplicity of 5. Total cellular RNA was purified from infected cells using the RNAqueous kit as described by the manufacturer (Ambion Inc., Austin, Tex.). A 610-nucleotide region encompassing G4L and G5R sequences was amplified by PCR using the primers 5′-GACACTAAGCTTCACCAGCGATTTATCGGTTTTGG-3′ and 5′-GACACTGGATCCTAGCATTTTTCTGTCCTTGGTTA-3′ with the HindIII and BamHI restriction enzyme sites underlined. The resulting PCR product was digested with BamHI and HindIII and inserted into pGem-4z (Promega) adjacent to the T7 RNA polymerase promoter to produce pGem-4z-G4L. Labeled riboprobes were made by linearizing pGem-4z-G4L with BamHI and transcribing the DNA in vitro with T7 RNA polymerase (Promega) and [α-32P]UTP (3,000 Ci/mmol, 10 mCi/ml). Total cell RNA was hybridized overnight at 42°C with excess labeled complementary RNA probe in 40 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (pH 6.4)–400 mM NaCl–1 mM EDTA–80% deionized formamide. Single-stranded RNA was digested with RNase A (4 μg/ml) and RNase T1 (10 U/ml) in 10 mM Tris HCl (pH 7.5)–5 mM EDTA–0.3 M NaCl for 1 h at room temperature. Protected fragments were analyzed by electrophoresis in a 6% polyacrylamide–8 M urea gel and visualized by autoradiography.
Transfer vector construction.
A copy of the vaccinia virus G4L gene, with an influenza virus HA epitope tag at the N terminus, was amplified by PCR from vaccinia virus strain WR DNA which had been purified using the Qiagen blood kit. The HA tag was engineered using overlapping oligonucleotide primers in sequential PCRs. The first PCR amplification used the primers 5′-GATGTTCCAGACTATGCTATGAAGAACGTACTGATT-3′ (endogenous G4L translation initiation codon indicated in boldface and HA epitope tag sequence indicated in italics) and 5′-CTCAGTTACTGGAGGCCAGACACCGTAAGTAGCTTT-3′. The second amplification used the primers 5′-TGAGCCATGGCATATCCATATGATGTTCCAGACTAT-3′ (NcoI site underlined, translation initiation codon from the NcoI site in boldface, and epitope tag sequence in italics) and 5′-GCGGGATCCCTACTCAGTTACTGGAGGCCAGAC-3′ (BamHI restriction site underlined). Because 29 nucleotides at the C terminus of the G4L ORF overlapped with G2R, silent mutations were made in the third nucleotide in each codon of overlapping G4L sequence to prevent the occurrence of homologous recombination. The G4L gene PCR product was digested with NcoI and BamHI and inserted into pVOTE.1 (19) to generate pVOTE.1G4L-HA.
For removal of the endogenous G4L gene, a 652-bp region corresponding to sequence following the G4L ORF (left flank) was generated by PCR from WR DNA, using the oligonucleotide primers 5′-ATAAGAATGCGGCCGCCTAAAATCCGAAATAGAAAAAGCTACC-3′ (NotI site underlined) and 5′-GCTTAAAGATCTGGATCCGAATCGTTGGAGATAGTGTCTTCC-3′ (BglII site underlined). The PCR product was digested with NotI and BglII and inserted into the pZippy-neo/gus plasmid (gift of T. Shors) downstream of the gus gene to generate pZippy-neo/gus-G4L(LF). A second PCR product of 544 bp corresponding to the sequence prior to the G4L ORF (right flank) was made using the oligonucleotide primers 5′-TATCCCAAGCTTTGTGAGTTTATCGATTTTTAATTGC-3′ (HindIII site underlined) and 5′-CGTCGTGTCGACCTTTAAAAAAAATGATAAGATATCAACATGGAG-3′ (SalI site underlined). The PCR product was digested with HindIII and SalI and inserted into pZippy-neo/gus-G4L(LF) upstream of the neomycin resistance gene to produce pZippy-G4L(RF)-neo/gus-G4L(LF).
Generation of recombinant vaccinia viruses.
vG4Li was constructed in two steps. BS-C-1 cells were infected with vT7lacOI at a multiplicity of 5 for 1 h at 37°C. The cells were then washed twice with Opti-MEM (Life Technologies) and transfected with 2 μg of pVOTE.1G4L-HA using Lipofectamine (Life Technologies). After 5 h, the transfection mixture was removed and replaced with complete Eagle minimal essential medium (EMEM) containing 5% fetal calf serum (FCS). The cells were harvested at 48 h after infection, frozen and thawed three times, and sonicated for 30 s. Recombinant vaccinia viruses were plaque purified three times in BS-C-1 cells in the presence of mycophenolic acid, xanthine, and hypoxanthine. The presence of the G4L ORF in the HA locus was confirmed by PCR and agarose gel electrophoresis. The virus vG4L/G4Li contained two G4L ORFs, the endogenous ORF and an inducible copy in the A56R (HA) locus.
vG4Li was constructed by infecting BS-C-1 cells with vG4L/G4Li at a multiplicity of 5 and then transfecting the cells with 2 μg of pZippy-G4L(RF)-neo/gus-G4L(LF) as described above. Recombinant vaccinia viruses were selected in the presence of 0.64 mg of Geneticin per ml, 4 mM HEPES (pH 7.4; Life Technologies), and 50 μM IPTG, and plaques were stained using X-Gluc (5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid) (0.2 mg/ml; Clontech Laboratories, Palo Alto, Calif.) (8). After six rounds of plaque purification, recombinant viruses were assessed for the presence of the neo/gus cassette by PCR and agarose gel electrophoresis.
Plaque assay and one-step virus growth.
BS-C-1 cell monolayers in six-well tissue culture plates were infected with 10-fold serial dilutions of virus. After 1 h, the inocula were removed, and the monolayers were washed twice with medium and then incubated for 2 days at 37°C with complete EMEM containing 2.5% FCS and 50 μM IPTG. The monolayers were stained with crystal violet, and the plaques were counted.
BS-C-1 cells were inoculated with 10 PFU of virus per cell for 1 h at 37°C. The inoculum was then removed, and the cells were washed twice with complete EMEM containing 2.5% FCS. The cells were then incubated in complete EMEM containing 2.5% FCS with or without 50 μM IPTG. At various times postinfection, the cells were harvested, frozen and thawed three times, sonicated, and stored at −80°C. Subsequently, virus titers were determined by plaque assay in the presence of 50 μM IPTG. The zero time point was obtained in the same way except that removal of the inoculum and washing were done after 5 min.
Complementation assays.
Plasmids containing the G4L gene controlled by its natural promoter or a synthetic early-late vaccinia virus promoter were constructed. The G4L gene with its natural promoter was amplified by PCR from vaccinia virus strain WR DNA with the primers 5′-GGAATTCCCAGCAGTAACGATTTTAAGTTTTTGATACCCATAAATGAAGAACGTACTGATT ATTTTCGG-3′ (EcoRI site underlined and translation initiation codon in boldface) and 5′-ACAGCGGGATCCTTATTCGGTAACAGGTGGCCAAAC-3′ (BamHI site underlined). The G4L ORF with a synthetic early-late promoter was amplified by PCR with the primers 5′-GGAATTCTAAATTGAAATTTTATT TTTTTTTTTTGGAATATAAATGAAGAACGTACTGATTATTTTCGG-3′ (EcoRI site underlined and translation initiation site in boldface) and the second primer (described above), which contained a BamHI restriction enzyme site. The PCR products were digested with EcoRI and BamHI and inserted into pUC-19 to produce the clones pUC-G4L native and pUC-G4L E/L.
BS-C-1 cells were infected with vG4Li at a multiplicity of 5 in the presence or absence of 50 μM IPTG for 1 h. The cells were washed with Opti-MEM (Life Technologies) and transfected with 2 μg of pUC-G4L native, pUC-G4L E/L, or pUC-19 using Lipofectamine (Life Technologies) in the presence or absence of 50 μM IPTG. After 5 h, the medium was removed and replaced with complete EMEM containing 2.5% FCS with or without IPTG. At 24 h after infection, the cells were harvested, frozen and thawed three times, and sonicated for 30 s and the virus titers were determined by plaque assay.
Western blotting.
Cells were lysed in 0.06 M Tris HCl (pH 6.8)–3% sodium dodecyl sulfate (SDS)–10% (vol/vol) glycerol–0.002% bromophenol blue unless otherwise specified and analyzed by SDS-polyacrylamide gel electrophoresis (SDS-PAGE; Owl Scientific) in the presence or absence of 5% β-mercaptoethanol. Proteins were electrophoretically transferred to a polyvinylidene difluoride membrane (Millipore), and the membrane was blocked overnight in 2.5% nonfat dried milk in TBS-T (100 mM Tris HCl [pH 7.5], 150 mM NaCl, and 0.1% [vol/vol] Tween 20). The membranes were incubated with either the G4L peptide antiserum or an anti-HA MAb at a 1:1,000 dilution for 1 h. The blots were washed in TBS-T and incubated with the secondary antibody, either anti-rabbit or anti-mouse immunoglobulin G conjugated to horseradish peroxidase (Amersham). Proteins were detected by chemiluminescence (West-Pico; Pierce).
Detergent extraction and phase separation.
Purified virus or BS-C-1 cells infected with virus at a multiplicity of 5 for 24 h were incubated in 50 mM Tris HCl (pH 7.5)–1% (vol/vol) Nonidet P-40 (NP-40), with or without 50 mM dithiothreitol (DTT), for 1 h at 37°C. The insoluble material was pelleted by centrifugation at 20,000 × g for 30 min at 4°C, and the supernatants were analyzed by Western blotting.
Analysis of [35S]methionine-labeled polypeptides by SDS-PAGE.
BS-C-1 cells were infected with vG4Li or vG4L/G4Li at a multiplicity of 5 for 1 h at 37°C. The inoculum was removed, and the infected cells were incubated in complete medium containing 2.5% FCS with or without 50 μM IPTG. At 8.75 h postinfection, the cells were incubated in methionine-free medium for 15 min and then labeled with 50 μCi of [35S]methionine/ml for 1 h. The cells were then harvested in 0.06 M Tris HCl (pH 6.8)–3% SDS–10% (vol/vol) glycerol–0.002% bromophenol blue. The labeled cells were chased in complete medium containing unlabeled methionine for 14 h prior to lysis.
Electron microscopy.
Infected monolayers were fixed with 2% glutaraldehyde, embedded in Epon resin, and viewed on a Philips CM100 electron microscope as previously described (21). For immunoelectron microscopy, infected cells were fixed with increasing concentrations of paraformaldehyde from 2 to 8% and prepared for cryosectioning (6). Ultrathin frozen sections were cut on a Leica/Reichert Ultracyromicrotome FCS. Thawed sections were incubated with an anti-HA MAb at a 1:300 dilution followed by rabbit anti-mouse immunoglobulin (1:500) which was detected with 10-nm colloidal gold conjugated to protein A (1:65) (17, 21).
Immunofluorescence and confocal microscopy.
Infected cells were fixed in 3% paraformaldehyde in phosphate-buffered saline (PBS) and then washed with 0.02 M glycine–0.1 M phosphate buffer. Cells were permeabilized with 0.05% saponin in PBS and blocked with 0.1% bovine serum albumin or 1% preimmune rabbit serum in PBS. G4L protein was labeled using the G4L peptide antiserum and detected with a rhodamine-conjugated anti-rabbit immunoglobulin (Dako Corporation, Carpinteria, Calif.). DNA was detected with Hoechst stain (5 μg/ml). The endoplasmic reticulum was labeled using an anti-protein disulfide isomerase (PDI) mouse MAb (Stressgen, Victoria, British Columbia, Canada) and detected using an Oregon green-conjugated anti-mouse immunoglobulin (Molecular Probes, Eugene, Oreg.). Samples were mounted in PBS, sealed with clear nail varnish, and visualized on a Leica DM IRBE confocal microscope.
RESULTS
Transcription of the G4L gene.
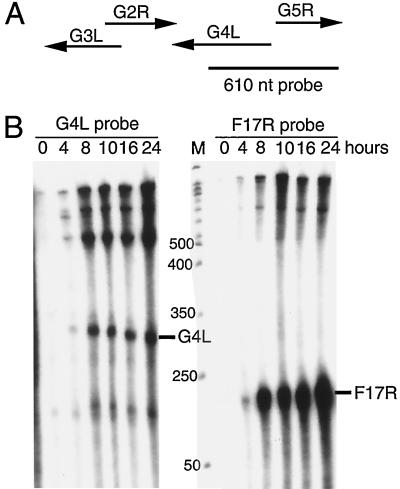
RNA analyses were performed to determine the time of transcription of the G4L gene and locate the transcription start site for subsequent genetic engineering. Inspection of the G4L ORF revealed the characteristic TAAATG late promoter transcription initiator element overlapping the translation initiation codon. To determine the RNA start site, we made a 610-nucleotide riboprobe that was complementary to part of the G4L ORF and extended 305 nucleotides upstream (Fig. 1A). If transcription initiated within the TAAATG, then the downstream 305-nucleotide segment of the probe should be protected by G4L mRNA. Transcripts initiated at other sites should protect smaller or larger segments of the probe. We also expected the probe to be fully protected by RNAs that initiated from the upstream G7L gene. Control experiments indicated that the probe was unprotected when hybridized with RNAs extracted from cells at time zero (Fig. 1B). Faint bands detected when the RNA was isolated at 4 h after infection became more intense with RNA extracted at later times. The prominent bands of approximately 300 and 600 nucleotides corresponded to a G4L transcript that initiated at the TAAAT and a transcript that protected the entire probe, respectively. The steady-state concentrations of the G4L transcript did not greatly increase after 8 h, consistent with the rapid turnover of late RNAs. For comparison, a second probe that was complementary to the well-characterized late transcript of the F17R gene (4) was hybridized to the same RNA preparations. An expected 126-nucleotide protected fragment was detected at 4 h and increased at later times (Fig. 1B), paralleling the results obtained with the G4L probe. Thus, the viral glutaredoxin was regulated as a typical late gene and RNA synthesis was initiated at the predicted site.
FIG. 1.
Transcriptional analysis. (A) Schematic diagrams of the G4L and adjacent ORFs with arrows indicating direction of transcription. The size and position of the uniformly 32P-labeled complementary G4L riboprobe relative to the ORFs are shown. nt, nucleotide. (B) RNase protection assays of G4L and F17R transcripts. Total RNA, extracted from 0 to 24 h after vaccinia virus infection of BS-C-1 cells, was hybridized with the 32P-labeled riboprobes specific for G4L or F17R sequences. Following RNase digestion, the protected probe fragments were analyzed by PAGE and autoradiography. The marker track (M) is a 50-nucleotide end-labeled DNA ladder (sizes in nucleotides on the left). The predicted protected fragment for each gene is indicated on the right.
Synthesis of the G4L protein.
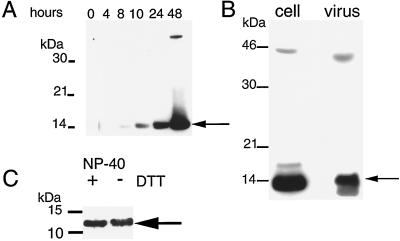
We raised antiserum to the C-terminal 13 amino acids of the predicted G4L ORF in order to facilitate the characterization of the protein. When total-cell lysates from infected cells were analyzed by Western blotting, a band of approximately 14 kDa was detected as a faint band at 8 h postinfection and accumulated over a 48-h period (Fig. 2A). An additional faint band of approximately 45 kDa was noted but not identified. No difference in the mobility of G4L protein was discerned when analyzed under nonreducing conditions using the anti-G4L peptide antiserum (data not shown), indicating that the protein did not form disulfide-bonded oligomers.
FIG. 2.
Synthesis of the G4L protein and association with purified virions. (A) SDS-PAGE and Western blotting analysis of G4L synthesis. BS-C-1 cells were infected with vaccinia virus at a multiplicity of 5. At the indicated times after infection, cells were lysed with SDS and analyzed by Western blotting using antiserum directed to the C-terminal 13 amino acids of the G4L ORF. Proteins were detected by chemiluminescence. The G4L protein (arrow) and marker proteins are indicated on the right and left, respectively. (B) Association of the G4L protein with purified virions. Sucrose gradient-purified vaccinia virus (107 PFU) and lysates of cells infected for 24 h were analyzed by Western blotting as described for panel A. (C) Detergent extraction of G4L protein from purified vaccinia virus. Approximately 107 PFU of sucrose gradient-purified WR virus was incubated with buffer containing NP-40 with or without DTT. Insoluble material was removed by centrifugation, and the supernatants were analyzed as described for panel A. Only the lower part of the gel is shown. The G4L protein bands are indicated by arrows in all panels.
Association of the G4L protein with virions.
Many late proteins are incorporated into virus particles, although for most it is not known whether this occurs by specific or by nonspecific mechanisms. The G4L protein was associated with sucrose gradient-purified virus particles (Fig. 2B), and electron microscopic evidence confirming this will be shown later. G4L protein was released upon incubation of the virions with NP-40 detergent with or without DTT (Fig. 2C), but some remained in the core pellet (data not shown). Assessment of the hydrophobicity of the G4L protein by phase partitioning with the detergent Triton X-114 (5) did not support or rule out the possibility that G4L associates with membranes as G4L partitioned to both the detergent-rich and the aqueous phases (data not shown).
Construction of a recombinant vaccinia virus with an inducible G4L gene.
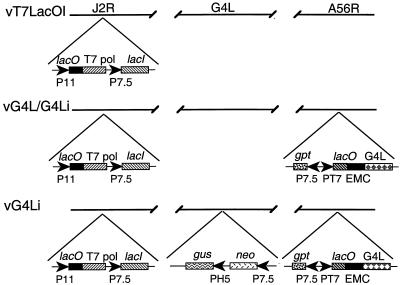
Genetic studies were needed to determine the role of the glutaredoxin. Our attempts to isolate a recombinant virus in which the G4L ORF was replaced by antibiotic selection and color markers failed, suggesting that it was essential for virus growth in BS-C-1 cells. As an alternative, we decided to make a recombinant virus with an inducible G4L gene with the expectation that it would have a conditional lethal, inducer-dependent phenotype (23). A modified version of this system that employs a regulated bacteriophage T7 RNA polymerase (19) was used. The first step in this construction was to form vG4L/G4Li by the insertion of the G4L ORF preceded by a T7 promoter, an Escherichia coli lac operator, and encephalomyocarditis virus leader into the HA locus (A56R) of vT7lacOI. vT7lacOI already contains an inducible copy of the T7 RNA polymerase gene and the E. coli lac repressor in the thymidine kinase locus (J2R) (Fig. 3). The coding sequence of the G4L ORF was modified to contain a 9-amino-acid influenza virus HA epitope tag at the N terminus. The second step was to delete the original G4L gene from vG4L/G4Li in the presence of IPTG. The resulting recombinant virus, called vG4Li, has a single inducible G4L gene (Fig. 3). Antibiotic selection was used to isolate both vG4L/G4Li and vG4Li, and the genotypes of the plaque-purified recombinant viruses were confirmed by PCR (data not shown).
FIG. 3.
Schematic diagram of recombinant vaccinia viruses. The genomes of recombinant vaccinia viruses vT7lacOI, vG4L/G4Li, and vG4Li are shown with the J2R (thymidine kinase), G4L, and A56R (HA) loci depicted. Insertions into these loci are indicated below. Abbreviations: P11, vaccinia virus late promoter; P7.5, vaccinia virus early-late promoter; PH5, vaccinia virus early-late promoter; PT7, bacteriophage T7 promoter; T7 pol, T7 RNA polymerase; lacO, E. coli lac operator; lacI, E. coli lac repressor gene; EMC, encephalomyocarditis virus cap-independent translation enhancer element; neo, neomycin resistance gene; gus, color selection marker; gpt, mycophenolic acid resistance gene.
Inducible expression of G4L.
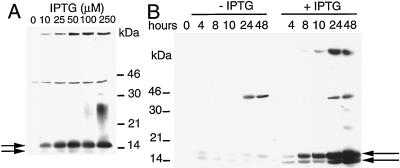
BS-C-1 cells were infected with vG4Li in the presence of a range of IPTG concentrations. In the absence of IPTG, G4L protein was barely detected on a Western blot as a faint band that did not become more intense with time (Fig. 4). The amount of G4L protein synthesized increased proportionally with IPTG concentration and at 25 to 50 μM IPTG was comparable to that of wild-type virus (Fig. 4A and data not shown). In the presence of 50 μM IPTG, the time course of G4L protein synthesis was similar to that of wild-type virus (Fig. 4B). The induced G4L protein appeared as a doublet of 15- and 14-kDa bands. The larger and more abundant band represented the epitope-tagged form, whereas the lower one evidently resulted from ribosome scanning past the first ATG and translation initiation at the retained original ATG codon.
FIG. 4.
Inducer-dependent expression of G4L. The figure shows the effect of IPTG concentration. BS-C-1 cells were infected with 5 PFU of vG4Li per cell and incubated with 0 to 250 μM IPTG and harvested at 24 h (A) or incubated with 0 or 50 μM IPTG and harvested at the times indicated (B). The cells were lysed with SDS and analyzed by Western blotting as described in the legend to Fig. 2. The positions of marker proteins are indicated at the sides. The upper and lower arrows point to epitope-tagged and nontagged G4L proteins, respectively.
Cytoplasmic localization of the G4L protein.
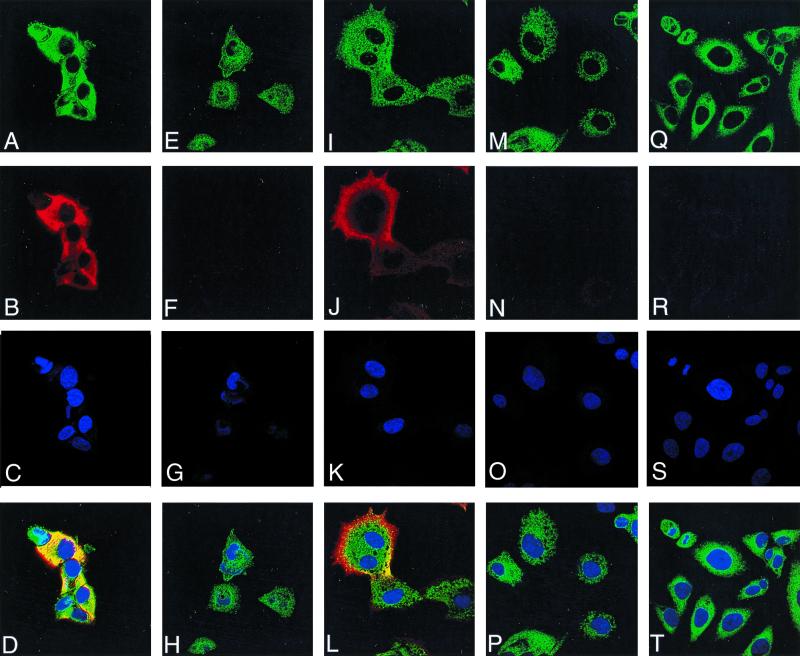
Laser scanning confocal microscopy was used to determine the intracellular distribution of the G4L protein. Using the G4L peptide antiserum, there was no significant background in uninfected cells (Fig. 5R) or cells infected with vG4Li under nonpermissive conditions at either 10 (Fig. 5F) or 16 (Fig. 5N) h postinfection. In cells infected with vG4Li in the presence of inducer for 10 h, G4L staining was distributed throughout the cytoplasm in a reticular pattern (Fig. 5B) but did not specifically colocalize with the virus factories, which appear as irregular juxtanuclear bodies (Fig. 5C). There was, however, some overlap of G4L with PDI, a protein that localizes in the endoplasmic reticulum (Fig. 5A). At 16 h postinfection in the presence of inducer, G4L was localized to the periphery of the cell (Fig. 5J) and did not colocalize with the endoplasmic reticulum (Fig. 5I).
FIG. 5.
Detection of G4L by immunofluorescence. HeLa cells were uninfected (Q to T) or infected with vG4Li in the presence of inducer for 10 h (A to D) or 16 h (I to L) or in the absence of inducer for 10 h (E to H) or 16 h (M to P). After 10 or 16 h, the cells were fixed; permeabilized; stained with G4L peptide antiserum and an antirabbit rhodamine conjugate (B, F, J, N, and R), an anti-PDI MAb and an anti-mouse Oregon green conjugate (A, E, I, M, and Q), or Hoechst stain (blue) (C, G, K, O, and S); and viewed by confocal microscopy. Merged images of cells stained with anti-PDI, anti-G4L, and Hoechst stain are also shown (D, H, L, P, and T).
Effect of IPTG on virus replication.
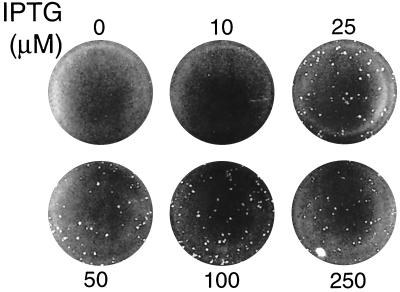
To determine the relationship between G4L expression and plaque formation, BS-C-1 cells were infected with vG4Li in the presence of 0 to 250 μM IPTG. In the absence of IPTG, no plaques were visible, and at 10 μM IPTG, a few pinpoint-sized plaques were present (Fig. 6). At 25 to 50 μM IPTG, the plaques were nearly the size of those formed by the parental vT7lacOI or the intermediate virus vG4L/G4Li and did not increase in size with higher IPTG concentrations (Fig. 6 and data not shown).
FIG. 6.
IPTG dependence of plaque formation. Cell monolayers were inoculated with vG4Li and overlaid with medium containing 0 to 250 μM IPTG. After incubation at 37°C for 48 h, the plates were stained with crystal violet.
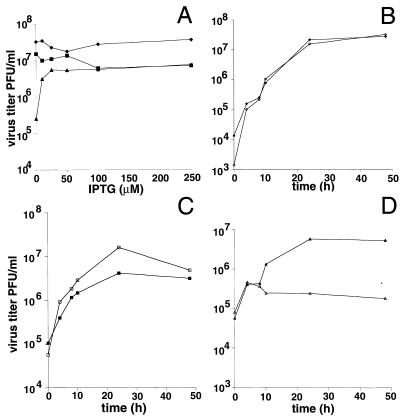
Small plaque size can be due to a defect in virus replication or spread. To assess the effect of G4L expression on virus replication, 24-h virus yields were determined after infection of BS-C-1 cells with vaccinia virus strain WR, vG4L/G4Li, or vG4Li in the presence of 0 to 250 μM IPTG. A sharp increase in the yield of vG4Li occurred between 0 and 25 μM IPTG with smaller increases at higher concentrations (Fig. 7A). In contrast, neither WR nor vG4L/G4Li showed an increase in titer with IPTG (Fig. 7A). The lower maximal titers obtained for vG4Li and vG4L/G4Li than for WR reflect the lower yield obtained with the parental vT7lacOI virus. One-step growth experiments resulted in a level curve out to 48 h for vG4Li in the absence of IPTG (Fig. 7D) and kinetics similar to those of WR and vG4L/G4Li in the presence of IPTG (Fig. 7B and C). The apparent rise between 0 and 4 h is an indicator of virus adsorption from the inoculum, not replication.
FIG. 7.
Effect of IPTG on virus yields. (A) Effect of IPTG concentration on 24-h yields. BS-C-1 cells were infected with WR (⧫), vG4L/G4Li (■), or vG4Li (▴) at a multiplicity of 5 and incubated with medium containing 0 to 250 μM IPTG for 24 h. Virus yields were determined by plaque assay in the presence of 50 μM IPTG for all viruses. (B to D) Time course of virus production in the presence (filled symbols) or absence (unfilled symbols) of 50 μM IPTG. The virus was WR (B), vG4L/G4Li (C), or vG4Li (D). Virus titers were determined by plaque assay as described for panel A.
trans complementation of vG4Li.
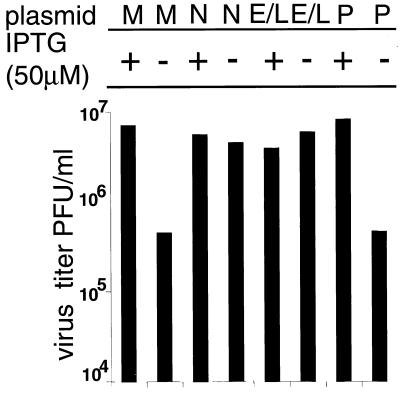
We considered it desirable to demonstrate by an independent method that the defect of vG4Li was solely due to the lack of expression of G4L and that the inducer did not rescue virus replication by some other means. This was accomplished by transfecting plasmids containing the G4L ORF either under its natural promoter or under a synthetic early-late promoter into cells infected with vG4Li in the absence of IPTG. Both of these plasmids increased virus replication more than 10-fold compared to that for the vector alone, and the virus yields approached that obtained by addition of IPTG (Fig. 8).
FIG. 8.
trans complementation of vG4Li. BS-C-1 cells were infected with vG4Li at a multiplicity of 5 in the presence (+) or absence (−) of 50 μM IPTG and then mock transfected (M), transfected with pUC-19 (P), or transfected with plasmids containing the G4L gene under its natural promoter (N) or a synthetic early-late viral promoter (E/L). After 24 h, the cells were harvested and the virus titers were determined by plaque assay in the presence of 50 μM IPTG.
Synthesis and processing of viral late proteins under nonpermissive conditions.
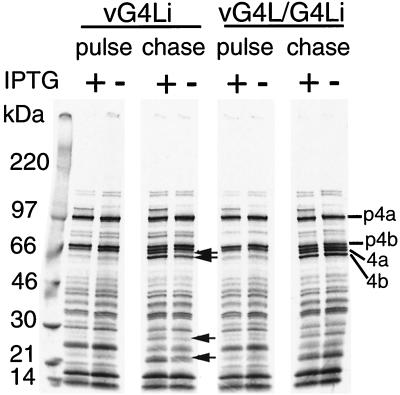
Vaccinia virus early, intermediate, and late genes are expressed in an obligatory sequence, and a defect at any stage precludes transition to the next one. Viral early and intermediate-stage proteins are difficult to resolve by pulse-labeling with radioactive amino acids because of their low expression and the continued labeling of cell proteins. By 6 to 8 h after infection, however, cell protein synthesis has been inhibited, and from this time on, the highly expressed viral late proteins are the only bands detected by autoradiography. Identical patterns of viral proteins were resolved when cells infected for 8.75 h with vG4Li or vG4L/G4Li in the presence or absence of IPTG were pulse-labeled with [35S]methionine and analyzed by SDS-PAGE (Fig. 9). Thus, G4L expression was not required for viral protein synthesis. When the cells were chased with unlabeled amino acids, however, the maturational processing of certain core proteins was partially inhibited under nonpermissive conditions (Fig. 9), suggesting an assembly block.
FIG. 9.
Synthesis and processing of viral late proteins. BS-C-1 cells were infected at a multiplicity of 5 in the presence (+) or absence (−) of 50 μM IPTG. At 8.75 h after infection, the cells were labeled with [35S]methionine for 1 h. One set of cells (pulse) were harvested, and another set were incubated with excess unlabeled methionine for an additional 14 h (chase). The proteins were analyzed by SDS-PAGE and autoradiography. Arrows indicate bands that increase during the chase in the presence of IPTG. The positions of the major core precursor proteins (p4a and p4b) and their mature processed forms (4a and 4b) are indicated on the right.
Effect of G4L repression on vaccinia virus morphogenesis.
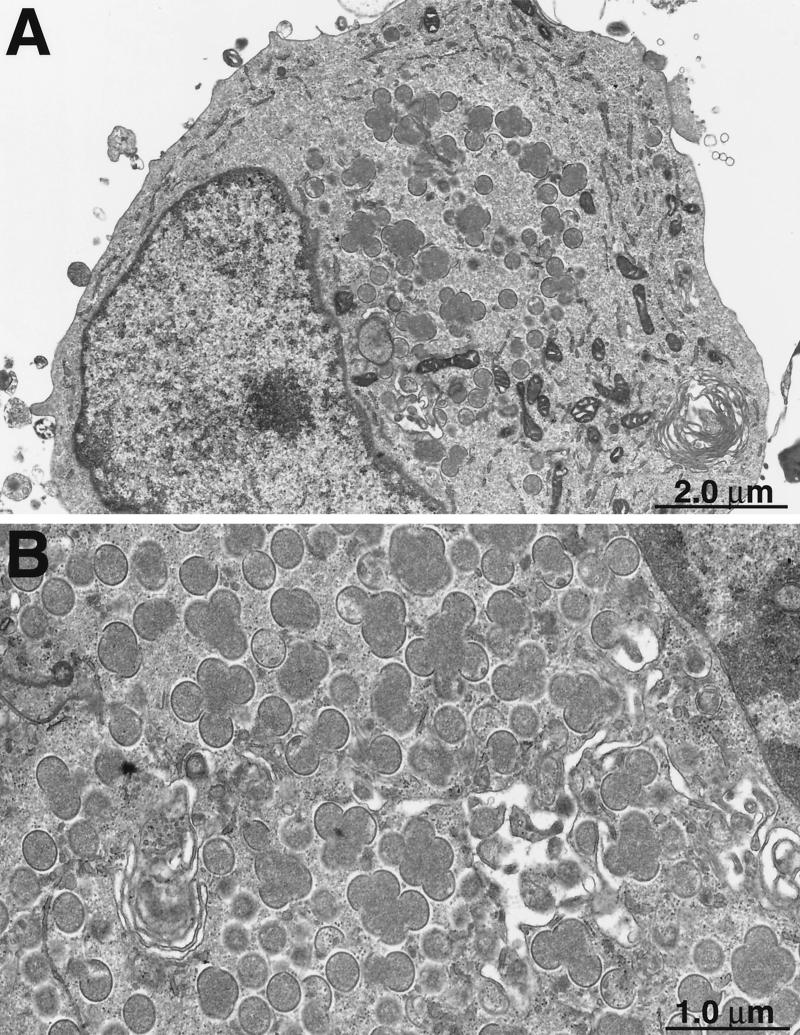
Ultrathin sections of cells infected with vG4Li in the presence and absence of IPTG were examined by electron microscopy to determine the role of G4L in virus morphogenesis. At 36 h after infection in the presence of IPTG, mature virions were seen in more than 70% of the cell sections examined, whereas circular immature forms or crescents were present in less than 30%. In contrast, only 9% of the sections of cells infected with vG4Li in the absence of IPTG for 36 h contained mature forms, and nearly 80% contained immature forms consisting mostly of crescents as well as immature virions. Most typical was the accumulation of large globules of electron-dense material surrounded by crescents in the absence of IPTG (Fig. 10). This phenotype is characteristic of a block in virus maturation.
FIG. 10.
Interruption of virus morphogenesis. BS-C-1 cells were infected with vG4Li in the absence of IPTG. After 36 h, cells were fixed and embedded in Epon. Ultrathin sections were prepared for transmission electron microscopy. Crescent membranes adjacent to electron-dense globules are shown at low (A) and high (B) magnifications.
Association of G4L protein with immature and mature virions.
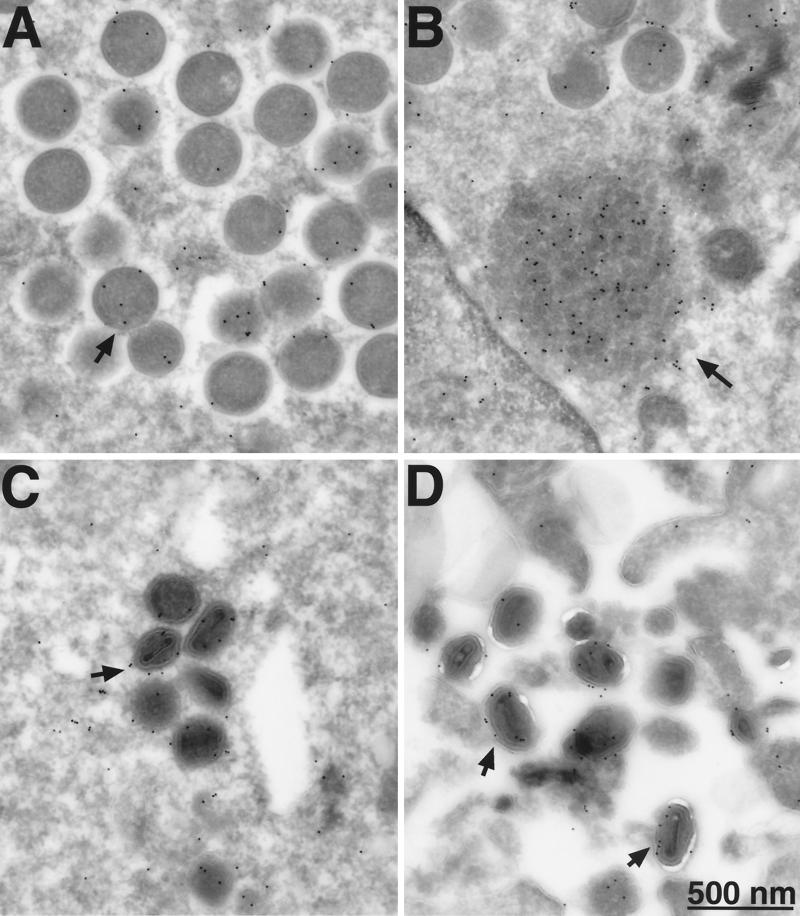
Our earlier biochemical experiments indicated that the G4L protein was associated with purified vaccinia virions. The finding that the G4L protein is required for morphogenesis led us to investigate the stage at which association occurred. For this purpose, we infected cells with vG4Li in the presence or absence of IPTG and visualized the G4L protein by incubating cryosections with the HA epitope antibody followed by protein A conjugated to gold grains. When IPTG was present, gold grains were distributed throughout the cytoplasm but were concentrated on both immature (Fig. 11A) and mature (Fig. 11C and D) virus particles. In addition, there was a high concentration of grains on amorphous structures located near immature virions (Fig. 11B). The specificity of the labeling was demonstrated by the absence of significant background when IPTG was omitted or on cells infected with WR virus (data not shown).
FIG. 11.
Electron microscopy of immunogold-labeled G4L protein. BS-C-1 cells were infected with vG4Li in the presence of 50 μM IPTG. Frozen sections were stained with a MAb to the HA epitope tag and protein A-conjugated gold grains and examined by electron microscopy. Fields show predominantly immature virions (A), depot containing G4L (B), intracellular mature virions (C), and extracellular virions (D). Arrows point to representative gold grains.
DISCUSSION
Vaccinia virus contains two distantly related glutaredoxins encoded by the O2L and G4L ORFs (3, 12). The O2L ORF was previously shown to be dispensable for virus replication (12), a result that was consistent with its suggested role in nucleotide precursor biosynthesis and its absence from poxviruses of other genera. The situation with G4L seemed quite different, however, as it is conserved in all poxviruses examined thus far. Here we have shown that G4L is expressed late in infection, is associated with immature and mature virus particles, and is required for vaccinia virus morphogenesis.
The G4L ORF is preceded by a typical late promoter consensus sequence including a putative overlapping transcription and translation initiation site. Transcriptional analysis demonstrated late expression of the G4L ORF and an RNA start site that was at or near the TAAATG motif. Western blotting also confirmed that G4L was expressed at a late stage of infection. This timing makes it less likely that the principal role of the glutaredoxin is in nucleotide precursor synthesis, although it remains possible that virion-associated G4L protein is released into the cytoplasm to function in this way.
We attempted to delete the G4L gene in order to determine its role. Our inability to isolate such a mutant supported the likelihood that the gene was essential. We therefore made a mutant in which the original G4L gene was replaced by one that was under the stringent control of the E. coli lac repressor. In the absence of inducer, virus replication was severely inhibited. Rescue could be achieved either by addition of IPTG or by transfection of a plasmid containing a G4L gene under the control of the natural G4L promoter or a synthetic early-late viral promoter. Although viral protein synthesis was not significantly affected in the absence of inducer, the proteolytic processing of some core proteins was reduced. Previous studies demonstrated that the processing of these proteins is inhibited when virus assembly is blocked at or before the immature virion stage (13, 22). Such a block was confirmed by electron microscopic studies when cells were infected with the G4L inducer-dependent virus in the absence of IPTG. Under these nonpermissive conditions, numerous, large electron-dense masses surrounded by crescent-shaped membranes accumulated in the cytoplasm. The large numbers of crescents indicated that the G4L protein is not required for the initial stages of membrane assembly. Although the formation of mature virus particles was severely inhibited, it was not totally blocked. This very low virus production could be due to traces of G4L protein produced in the absence of inducer, perhaps due to leakiness or low amounts of IPTG introduced with the virus inoculum. Alternatively, very inefficient maturation might occur in the absence of the G4L protein. Since a similar block in morphogenesis did not occur when O2L was deleted, G4L must have a specific role in the maturation process. Biochemical analysis indicated that G4L protein was present in purified virus particles and could be released with a nonionic detergent. Furthermore, an association with immature and mature virions was demonstrated by immunoelectron microscopy. The G4L protein, however, lacks a typical transmembrane domain, and the absence of a signal peptide was suggested by construction of a stable N-terminal epitope-tagged form of the protein. Moreover, no membrane association was demonstrated by in vitro translation in the presence of canine pancreatic microsomes (C. L. White, unpublished data). Whether the G4L protein is specifically targeted to viral particles or is merely present in the “viroplasm” that is engulfed during the early stages of assembly cannot be determined at this time.
How the G4L protein participates in virus morphogenesis is an interesting question for the future. All of the poxvirus G4L orthologs retain the characteristic CXXC motif of glutaredoxins, and the protein has been shown previously to have thiol reductase activity in vitro (12), suggesting a redox role. Although disulfide bonds are usually formed in the lumen of the endoplasmic reticulum, some structural proteins of vaccinia virus are apparently disulfide bonded within the cytoplasm (9, 14). It is intriguing to consider that the G4L glutaredoxin may participate in this novel aspect of vaccinia virus morphogenesis.
ACKNOWLEDGMENTS
We thank Norman Cooper for cells; Elizabeth Wolffe, Brian Ward, and Owen Schwartz for advice and help with immunofluorescence and confocal microscopy; and Teri Shors for pZippy neo-gus.
REFERENCES
- 1.Afonso C L, Tulman E R, Lu Z, Oma E, Kutish G F, Rock D L. The genome of Melanoplus sanguinipes entomopoxvirus. J Virol. 1999;73:533–552. doi: 10.1128/jvi.73.1.533-552.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Afonso C L, Tulman E R, Lu Z, Zsak L, Kutish G F, Rock D L. The genome of fowlpox virus. J Virol. 2000;74:3815–3831. doi: 10.1128/jvi.74.8.3815-3831.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahn B Y, Moss B. Glutaredoxin homolog encoded by vaccinia virus is a virion-associated enzyme with thioltransferase and dehydroascorbate reductase activities. Proc Natl Acad Sci USA. 1992;89:7060–7064. doi: 10.1073/pnas.89.15.7060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baldick C J, Jr, Moss B. Characterization and temporal regulation of mRNAs encoded by vaccinia virus intermediate-stage genes. J Virol. 1993;67:3515–3527. doi: 10.1128/jvi.67.6.3515-3527.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bordier C. Phase separation of integral membrane proteins in Triton X-114 solution. J Biol Chem. 1981;256:1604–1607. [PubMed] [Google Scholar]
- 6.Bosshart H, Humphrey J, Deignan E, Davidson J, Drazba J, Yuan L, Oorschot V, Peters P J, Bonifacino J S. The cytoplasmic domain mediates localization of furin to the trans-Golgi network en route to the endosomal/lysosomal system. J Cell Biol. 1994;126:1157–1172. doi: 10.1083/jcb.126.5.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cameron C, Hota-Mitchell S, Chen L, Barrett J, Cao J X, Macaulay C, Willer D, Evans D, McFadden G. The complete DNA sequence of myxoma virus. Virology. 1999;264:298–318. doi: 10.1006/viro.1999.0001. [DOI] [PubMed] [Google Scholar]
- 8.Carroll M W, Moss B. E. coli beta-glucuronidase (GUS) as a marker for recombinant vaccinia viruses. BioTechniques. 1995;19:352–356. [PubMed] [Google Scholar]
- 9.Dallo S, Rodriguez J F, Esteban M. A 14K envelope protein of vaccinia virus with an important role in virus-host cell interactions is altered during virus persistence and determines the plaque size phenotype of the virus. Virology. 1987;159:423–432. doi: 10.1016/0042-6822(87)90481-8. [DOI] [PubMed] [Google Scholar]
- 10.Earl P L, Cooper N, Moss B. Generation of recombinant vaccinia viruses. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 2. New York, N.Y: Greene Publishing Associates and Wiley International Science; 1991. pp. 16.17.1–16.17.16. [Google Scholar]
- 11.Earl P L, Cooper N, Moss B. Preparation of cell cultures and vaccinia virus stocks. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 2. New York, N.Y: Greene Publishing Associates and Wiley International Science; 1991. pp. 16.16.1–16.16.7. [Google Scholar]
- 12.Gvakharia B O, Koonin E K, Mathews C K. Vaccinia virus G4L gene encodes a second glutaredoxin. Virology. 1996;226:408–411. doi: 10.1006/viro.1996.0669. [DOI] [PubMed] [Google Scholar]
- 13.Katz E, Moss B. Formation of a vaccinia virus structural polypeptide from a higher molecular weight precursor: inhibition by rifampicin. Proc Natl Acad Sci USA. 1970;6:677–684. doi: 10.1073/pnas.66.3.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Locker J K, Griffiths G. An unconventional role for cytoplasmic disulfide bonds in vaccinia virus proteins. J Cell Biol. 1999;144:267–279. doi: 10.1083/jcb.144.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moss B. Poxviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2637–2672. [Google Scholar]
- 16.Rajagopal I, Ahn B Y, Moss B, Mathews C K. Roles of vaccinia virus ribonucleotide reductase and glutaredoxin in DNA precursor biosynthesis. J Biol Chem. 1995;270:27415–27418. doi: 10.1074/jbc.270.46.27415. [DOI] [PubMed] [Google Scholar]
- 17.Roos N, Cyrklaff M, Cudmore S, Blasco R, Krijnse-Locker J, Griffiths G. A novel immunogold cryoelectron microscopic approach to investigate the structure of the intracellular and extracellular forms of vaccinia virus. EMBO J. 1996;15:2343–2355. [PMC free article] [PubMed] [Google Scholar]
- 18.Senkevich T G, Bugert J J, Sisler J R, Koonin E V, Darai G, Moss B. Genome sequence of a human tumorigenic poxvirus: prediction of specific host response-evasion genes. Science. 1996;273:813–816. doi: 10.1126/science.273.5276.813. [DOI] [PubMed] [Google Scholar]
- 19.Ward G A, Stover C K, Moss B, Fuerst T R. Stringent chemical and thermal regulation of recombinant gene expression by vaccinia virus vectors in mammalian cells. Proc Natl Acad Sci USA. 1995;92:6773–6777. doi: 10.1073/pnas.92.15.6773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Willer D O, McFadden G, Evans D H. The complete genome sequence of Shope (rabbit) fibroma virus. Virology. 1999;264:319–343. doi: 10.1006/viro.1999.0002. [DOI] [PubMed] [Google Scholar]
- 21.Wolffe E J, Moore D M, Peters P J, Moss B. Vaccinia virus A17L open reading frame encodes an essential component of nascent viral membranes that is required to initiate morphogenesis. J Virol. 1996;70:2797–2808. doi: 10.1128/jvi.70.5.2797-2808.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y, Moss B. Immature viral envelope formation is interrupted at the same stage by lac operator-mediated repression of the vaccinia virus D13L gene and by the drug rifampicin. Virology. 1992;187:643–653. doi: 10.1016/0042-6822(92)90467-4. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Y, Moss B. Inducer-dependent conditional-lethal mutant animal viruses. Proc Natl Acad Sci USA. 1991;88:1511–1515. doi: 10.1073/pnas.88.4.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]