Abstract
How molecules interact governs how they move. Single-molecule tracking (SMT) thus provides a unique window into the dynamic interactions of biomolecules within live cells. Using transcription regulation as a case study, we describe how SMT works, what it can tell us about molecular biology, and how it has changed our perspective on the inner workings of the nucleus. We also describe what SMT cannot yet tell us and how new technical advances seek to overcome its limitations. This ongoing progress will be imperative to address outstanding questions about how dynamic molecular machines function in live cells.
Keywords: live cell biochemistry, molecular interactions, proximity assisted activation, single molecule tracking, transcription
Introduction
An increasing number of labs around the world deploy various methods to track single endogenous protein molecules in cells, tissues, and even whole organisms to probe their biochemical behavior in their native environment. Here we will discuss how single-molecule tracking (SMT) is performed, what we can learn from it, and how ongoing technical advances are making SMT more informative. SMT has diverse applications in cell and molecular biology, from studying the translocation of molecular motors along the cytoskeleton to monitoring interactions of membrane receptors. Each biological problem poses its own distinct challenges and requires different methodologies [1–3]. In this review, we will cover two specific applications of SMT: fast tracking of diffusing molecules and slower tracking of bound molecules in the nucleus. These methods, which require specialized analysis tools, enable a new form of ‘live-cell biochemistry’ that can distinguish complexes and measure kinetics. Here, we will focus on applications in the field of transcription regulation and discuss a few selected examples.
Transcription regulation as a case study: what do we want to understand?
Switching a gene on or off involves multitudinous interactions between hundreds of proteins. Protein-encoding genes are transcribed by RNA polymerase II (Pol II), which initiates transcription following assembly of general transcription factors (GTFs) into a pre-initiation complex (PIC) at the gene promoter [4,5]. Transcription initiation by Pol II and release from promoter-proximal pausing are tightly regulated by sequence-specific transcription factors (TFs), which bind short DNA sequences either adjacent to the transcription start site or thousands of base pairs away within enhancers [6]. TFs in turn bind an assortment of proteins and protein complexes called coactivators and corepressors, that activate or repress transcription through diverse mechanisms [7–9]. A critical and outstanding challenge in molecular biology is to understand how this vast network of molecular interactions gives rise to precise patterns of gene expression [10–12]. Making progress toward this goal will require developing new ways to measure protein–DNA and protein–protein interactions (PPIs) of TFs, GTFs, and cofactors in live cells. While SMT has provided a new window into these interactions, its practitioners are still learning to peer through that window and decipher what we see.
How SMT works
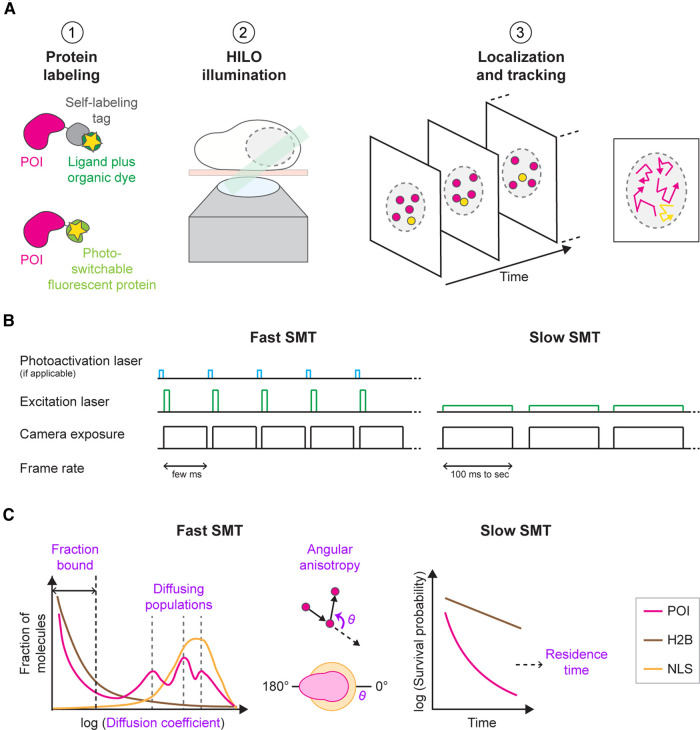
SMT involves localizing and tracking individual fluorescently labeled molecules over time. More than a decade of advances in microscopy [13,14], genome editing [15] self-labeling tags [16], and photoswitchable fluorophores [17–20] have expanded the scope of SMT from in vitro mixtures [21–23] to cell membranes [24,25], to the interior of nuclei in live cells and embryos [26–40]. Several challenges had to be overcome: First, superresolution methods were developed to resolve protein molecules closer together than the diffraction limit of light, using sparse and stochastic labeling with either photoactivatable fluorophores or self-labeling tags [41,42]. Second, detecting the miniscule amount of light emitted from each fluorophore required the development of sensitive electron-multiplying charge-coupled devices (EMCCD) and complementary metal oxide semiconductor (CMOS) cameras [43–46]. Third, optical methods were developed to minimize the background fluorescence and scattered light that would otherwise overwhelm the weak fluorescence from single molecules. The most commonly used approach, highly inclined and laminated optical sheet (HILO) illumination [47], passes a laser beam obliquely through the sample, making it possible to visualize fluorophores in the cell interior. Another option, light-sheet microscopy, illuminates a single plane using a thin sheet of light and detects emission from only that plane [48–50]. A schematic of a typical SMT experiment in the mammalian cell nucleus is shown in Figure 1A.
Figure 1. Schematic of live-cell SMT methodology and measurements.
(A) Typical workflow for SMT of tagged nuclear-localizing TFs. Step 1: The protein of interest (POI, magenta) is tagged either with a self-labeling tag (e.g. HaloTag, gray) and sparsely labeled with a tag-specific ligand (dark green) coupled to a bright and photostable organic dye (yellow star; top) or with a photoswitchable fluorescent protein (light green with yellow star; bottom). Step 2: Cells growing in a monolayer on top of a glass coverslip (beige) are imaged by HILO illumination (green) to visualize single molecules in the nucleus (round dashed line) over time. Step 3: Single molecules (magenta, yellow) are then localized in each frame of a single-molecule movie and tracked over consecutive frames (left), resulting in single-molecule trajectories (right). (B) Illumination schemes for fast (left) and slow SMT (right). Fast SMT employs short camera frame rates of only a few milliseconds (black) combined with short stroboscopic excitation pulses at high laser power at the beginning of each frame (green). If required, short photoactivation pulses (blue) are placed during the camera dead time between frames. Slow SMT uses longer frame rates of hundreds of milliseconds to seconds (black), combined with continuous excitation at low laser power (green), to visualize bound molecules. (C) Data analysis of single-molecule trajectories. Fast SMT (left) provides the population distribution of diffusion coefficients (‘diffusion spectrum') for the POI (magenta). Chromatin-bound histone H2B (brown) and freely diffusing nuclear localization sequence (NLS; orange) serve as controls. This analysis reveals differently diffusing subpopulations. Most TFs exhibit a chromatin-bound population and one or more diffusing subpopulations. Fast SMT also reveals whether the distribution of angles (theta) between successive displacements exhibits anisotropy, i.e. whether it is uniform in all directions (orange) or backward-biased (magenta). Slow SMT (right) can be used to measure residence times of immobile molecules, e.g. of a TF bound to chromatin.
SMT experiments fall into two broad categories: ‘Slow' SMT observes molecules bound to slow-moving scaffolds, such as chromatin [33,51–53]. A relatively long exposure time — hundreds of milliseconds to seconds — blurs out fast-moving molecules, leaving only bound molecules visible as discrete spots. Slow SMT can measure how long molecules bind chromatin, i.e. their residence time, and how they move while bound [54,55]. In contrast, ‘fast’ SMT detects both fast- and slow-moving molecules and can measure their diffusion coefficient, chromatin-bound fraction, and angular anisotropy [40,56–58]. Even with brief exposure times of 10 ms or less, fast-moving molecules appear blurred, but this motion blur can be reduced by pulsing the excitation laser like a strobe light for only 1–2 ms per frame [27]. Notably, trajectories are much shorter in fast SMT than in slow SMT due to faster photobleaching at the higher laser power used for fast SMT. (Figure 1B,C)
The first step in SMT data analysis is to localize and track single molecules, which can be done using various algorithms [59–61]. In slow SMT, a residence time distribution is obtained by classifying ‘bound' molecules that move less than a certain distance and tabulating how many frames they persist. Interpreting this distribution is complicated by the fact that fluorophores may disappear not only due to dissociation but also due to photobleaching, defocalization, or tracking errors [62]. This can be corrected for by comparison to a control protein, such as histone H2B, that remains bound to chromatin on the timescale of the experiment [36], or by pausing illumination for various time intervals between frames [39,63,64]. While residence time distributions are conventionally fit to single- or multi-exponential models to determine dissociation rate constants for discrete states [36,39,64], others have proposed that a power law distribution provides a better model of dissociation from binding sites with a range of affinities [55]. An alternative method infers distributions of dissociation rates using a numerical inverse Laplace transform [65].
For fast SMT, fitting the mean squared displacement (MSD) as a function of time provides a simple way to estimate diffusion coefficients. However, this approach suffers from systematic bias when applied to three-dimensional samples, because faster-moving molecules are more rapidly lost to defocalization. This bias can be corrected mathematically [36,39,56]. Spot-On, an analysis algorithm from our group, fits displacement histograms to infer the diffusion coefficient, localization error, and relative abundance of two or three distinct populations of molecules, such as those that are freely diffusing or chromatin bound [56]. Bayesian analysis approaches have also been developed that can infer the number of discrete diffusive states, along with their diffusion coefficients and transition rates [66,67]. An approach recently developed in our lab relies on Bayesian inference of posterior probabilities over a large two-dimensional array of states with different values of diffusion coefficient and localization error. Discrete populations are distinguishable as peaks in the resulting probability distribution [68–70].
TF dynamics: what has SMT revealed about transcription?
SMT has enabled a new form of live-cell biochemistry, illuminating previously unseen behaviors of transcription regulators. For instance, it is tempting when gazing at chromatin immunoprecipitation sequencing (ChIP-seq) peaks to imagine that proteins form stable complexes on chromatin. However, SMT has shown that this is generally not the case. Residence time measurements by slow SMT have revealed that different transcription regulators interact with chromatin over a wide range of timescales, from milliseconds to minutes [33,37,39,55,64,71]. For example, residence times of GTFs in yeast and mammalian cells ranged from under 10 s to slightly over a minute [72–74]. These results, and others, have been collected in a database (www.mir-lab.com/dynamics-database). It should be noted that the accuracy of residence time measurements depends sensitively on correction for photobleaching and defocalization of fluorophores, and it will be important to validate the consistency of these measurements across multiple labs.
Fast SMT measurements can reveal how the chromatin bound fraction of a protein changes in response to genetic or chemical perturbations, providing a powerful way to dissect the biochemistry of protein–DNA interactions in live cells. In the case of Type I nuclear receptors, fast SMT revealed an increase in chromatin binding upon ligand treatment [52,63,69,75,76]. Combining SMT with chemical genetics in yeast revealed the in vivo order of assembly of GTFs and Pol II into the PIC [74].
Fast SMT has also allowed us to directly visualize how TFs locate their binding sites. The classic ‘facilitated search' model of von Hippel and Berg proposes that DNA-binding proteins find specific sites by alternating between three-dimensional diffusion and one-dimensional sliding along DNA [77]. While single-molecule studies in vitro have directly observed 1D sliding by TFs, this would be difficult to resolve in live-cell SMT experiments [27,78,79]. However, SMT has revealed rapid transitions between mobile and immobile states of TFs, and analysis of TF mutants suggests that most immobile molecules associate nonspecifically with chromatin [33,39,80,81], consistent with a rapidly alternating 1D and 3D search.
Another intriguing feature of target search has emerged from analyzing angles between successive displacements. The motion of the transcriptional activator c-Myc is well described by regular Brownian motion, in which these displacement angles have a uniform (isotropic) distribution [40]. In contrast, fast SMT of the pause-release factor P-TEFb and DNA-binding protein CTCF revealed a highly anisotropic, backward-biased angular distribution [40,57]. Anisotropic diffusion of CTCF depends on a putative RNA binding domain, suggesting that interactions with RNA transiently trap CTCF in small zones. Like 1D sliding along DNA, anisotropic diffusion could in theory increase the rate at which a protein binds target sites by reducing the effective dimensionality of the space that it explores. Modeling of CTCF binding and diffusion, for instance, suggested that anisotropic movement could increase its on-rate for specific sites by ∼2.5-fold [57]. However, the mechanisms underlying anisotropic motion and its functional consequences remain to be further explored.
SMT also provides a way to detect weak interactions between proteins that are otherwise difficult to study, such as those mediated by intrinsically disordered regions (IDRs). Particularly enriched in TF activation domains, IDRs mediate weak, flexible, and multivalent interactions between many types of proteins [82–84]. IDRs can profoundly influence protein behavior. A recent study from our lab revealed that it is differences in the IDR, not the DNA-binding domain, that dictate the chromatin-bound fraction of different paralogs of the HIFα transcription factor [70]. Weak, multivalent interactions can also drive liquid–liquid phase separation, which is often invoked to explain formation of nuclear sub-compartments that regulate transcription. SMT can help discern if this occurs in specific cases. SMT experiments have shown that various proteins diffuse more slowly inside nucleoli than outside, as predicted for a distinct, higher-viscosity liquid phase [68,85]. In contrast, enrichment of Pol II in herpesvirus replication compartments (RCs) involves elevated binding to DNA with the same diffusion coefficient inside and outside RCs [86]. Conversely, depletion of Pol II molecules from inactive X chromosomes involved reduced chromatin binding, with no change in diffusion coefficient, arguing against physical exclusion from a distinct condensed phase [87]. RCs, inactive X chromosomes, and nucleoli have the experimental advantage that they are relatively large. Given that the localization error of moving molecules is typically tens of nanometers, new methods will be required to discern the material properties of smaller, sub-diffraction-limited protein assemblies, such as those that form at enhancers.
Pushing the envelope: learning more from SMT
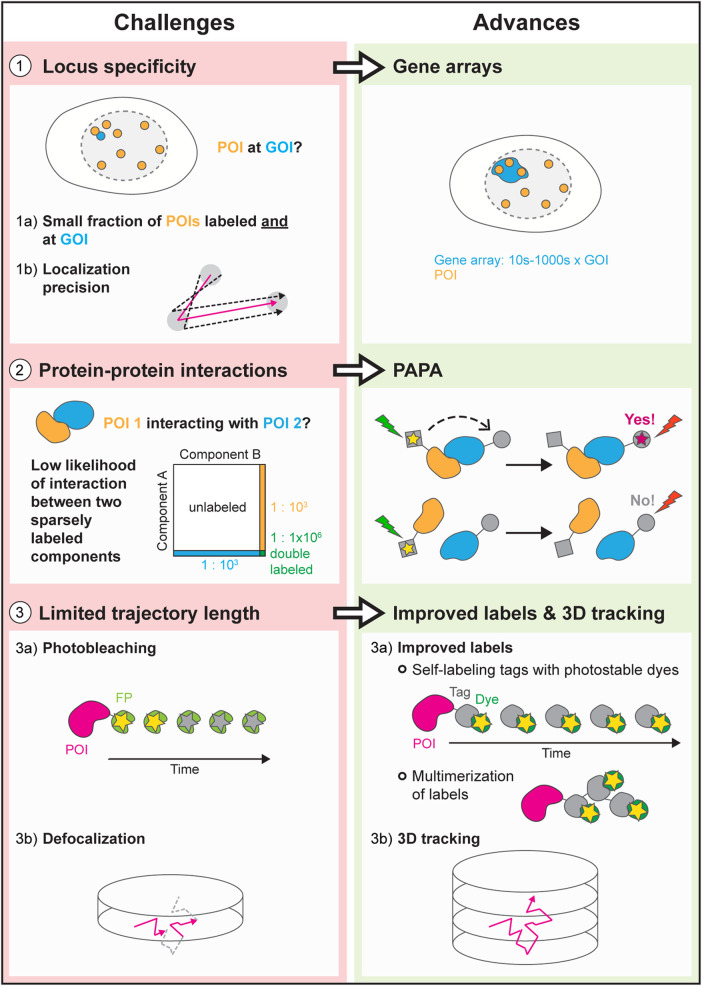
Although SMT experiments are data-rich, there is much more that we would like to discern. We can determine what fraction of a given transcription regulator binds chromatin and measure its residence time, but we don't know where on the genome it binds. We can resolve populations of TFs with different diffusion coefficients, but this does not reveal what those complexes are biochemically. We would like to monitor transitions between protein complexes, yet photobleaching and camera frame rates limit how fast and how long we can track individual molecules. Molecules move three-dimensionally in the nucleus, but we typically track them in two dimensions. Fortunately, technical advances and new approaches provide opportunities to overcome many of these limitations (Figure 2).
Figure 2. Challenges and advances in live-cell SMT.
Current challenges in live-cell SMT, with a focus on transcription regulation, and selected recent advances in overcoming each problem. POI, protein of interest. GOI, gene of interest. PAPA, proximity-assisted photoactivation.
Locus specificity-where are the TFs binding?
To understand nuclear processes like transcription, it is crucial to measure the binding of proteins to the specific genomic loci where these processes occur. While various tools exist for locus labeling — lacO and tetO arrays, ParB-parS (also referred to as ‘ANCHOR’), and dCas9 [88–96], monitoring locus-specific binding of proteins remains challenging. One problem is statistical: SMT typically entails labeling only a small fraction of molecules, and only a small fraction of those bind a given locus, yielding small sample sizes. The second problem is that localization precision is limited, especially for moving molecules. This means that a molecule that colocalizes with a labeled locus might actually be bound to a nearby segment of chromatin within the localization error.
Gene array imaging
One solution is to image arrays of multiple gene copies. This increases the number of localizations detectable at the specific target while spatially separating those localizations from molecules bound to the rest of the genome. Locus-specific binding of single proteins has been visualized on Drosophila polytene chromosomes, naturally occurring gene arrays consisting of bundles of aligned, endo-replicated sister chromatids [97]. Naturally occurring arrays in yeast have also been used to monitor TF binding to specific target genes [53]. Slow SMT of glucocorticoid receptors (GR) in mammalian cells revealed a longer residence time and lower mobility within an array of GR-responsive genes [32,51]. Slow SMT was used to measure IDR-mediated interactions between proteins in solution and proteins bound to a large array of lacO sites [98]. A drawback of gene arrays in mammalian cells is that they are synthetic, and arrayed genes could in principle behave differently from naturally occurring single-copy genes.
Protein–protein interactions: resolving complexes
In some cases, PPIs can be inferred from SMT data by analyzing the effect of chemical or genetic perturbations. However, these perturbations may have unanticipated effects on the conformational and binding equilibria of proteins, possibly resulting in inaccurate interpretation of results. It would therefore be advantageous to detect PPIs directly.
Two-color SMT
For some proteins, such as cell surface receptors imaged by total internal reflection fluorescence (TIRF) [99–101], PPIs can be inferred by labeling the interacting partners with different fluorophores and observing whether they colocalize and move in tandem. However, this approach only works if the proteins of interest (POIs) are so dilute that single molecules are detectable even when all molecules are labeled. As discussed above, SMT more often relies on sparse labeling, making it unlikely that both interacting partners within a single complex are labeled. Moreover, it is difficult to quantify precisely what fraction of each protein is labeled. Thus, detecting complexes by two-color SMT is neither efficient nor quantitative in most cases. A similar problem limits the use of single-molecule Förster resonance energy transfer (smFRET) as a PPI sensor in live cells [69].
Bimolecular fluorescence complementation (BiFC)
PPIs can also be detected by fusing the two interacting partners to two halves of a split fluorescent protein or HaloTag, which are reconstituted into a complete tag when the fusion proteins come together [102,103]. While this approach has been applied to single-molecule imaging [104–106], a major disadvantage is that it irreversibly locks together the two binding partners [102,104,107], making it incapable of accurately measuring dynamic interactions.
Proximity-assisted photoactivation (PAPA)
We recently reported a new method to detect PPIs by SMT [69]. While imaging a multimeric protein labeled with two different Janelia Fluor (JF) dyes, we noticed that exciting JF549 causes the reappearance of JFX650 molecules that had apparently been lost to photobleaching. Because this phenomenon requires that the two dyes be close together, we dubbed it ‘proximity-assisted photoactivation' (PAPA).
Many fluorescent dyes can undergo chemical reactions when excited, which render them non-fluorescent or ‘dark'. Dark fluorophores can reactivate either spontaneously or through direct reactivation (DR) by near-UV light. Additionally, excitation of one cyanine (Cy) dye (e.g. Cy3) can reactivate a second nearby Cy dye (e.g. Cy5) [108]. Cy dye photoswitching enabled early stochastic optical reconstruction microscopy (STORM) imaging [13], yet its application as a proximity sensor was limited by its shorter distance range than FRET (<2 nm). Additionally, Cy dyes are cell impermeable, and their photoswitching requires an oxygen scavenging system and high thiol concentrations, precluding applications to live-cell imaging.
Although the mechanism of PAPA between JF dyes is unclear, it offers distinct advantages as a proximity sensor. Unlike Cy dye photoswitching, it occurs in live cells with membrane-permeable dyes, and it is more permissive of large inter-dye separations than either Cy dye photoswitching or FRET [69]. Proof-of-concept experiments showed that PAPA can detect PPIs, both in ensemble and single-molecule fluorescence imaging, and that it can be combined with SMT to characterize specific protein complexes [69]. While much work remains to understand PAPA and to refine PAPA-SMT, this approach provides a valuable foot in the door for investigating interactions of single protein molecules in live cells.
Limited trajectory length: monitoring transitions
Tracking a single protein molecule throughout its lifetime, from synthesis to degradation, would reveal how fast it enters and exits the nucleus, how its diffusion changes over time due to association with binding partners, and how often it binds and dissociates from chromatin. Unfortunately, tracking a single molecule for an extended period of time is difficult, particularly in fast SMT, where trajectories of diffusing molecules last no more than a few camera frames [56]. While analysis methods have been devised to glean useful information from short trajectories, collecting longer trajectories would help to resolve diffusion coefficients, residence times, and transition rates between different complexes or conformations.
Two technical limitations stand in the way: First, fluorophores photobleach rapidly at the laser powers required for SMT. Approaches to overcome photobleaching include (1) using more stable fluorophores, (2) labeling each protein with multiple fluorophores, and (3) using a structured illumination approach called MINFLUX. Second, while molecules move three-dimensionally in the cell, SMT typically images a single two-dimensional focal plane, and molecules that exit this plane can no longer be tracked [39,56].
Improved labels
Fluorescent proteins (FPs) transformed microscopy by making it possible to label target proteins using simple molecular cloning. Synthetic organic fluorophores, although much brighter and more photostable than FPs, have historically required chemical conjugation to target proteins, mostly restricting their use to in vitro experiments. The development of genetically encoded, self-labeling tags like Halo and SNAP, together with cell-permeable fluorophores like JF dyes, has combined the best of both worlds, making it possible to label target proteins with bright, photostable synthetic fluorophores in live cells [109–114]. These tools continue to be improved [115,116].
Multimerization of labels
Labeling a protein with multiple fluorophores increases the total fluorescence signal and allows it to be tracked longer before it is lost to photobleaching. A simple option is to fuse multiple tandem copies of HaloTag to the target protein [117]. The SunTag and MoonTag comprise 24 tandem copies of short epitopes bound by different single-chain antibodies fused to a FP [118,119]. ArrayG consists of arrays of nanobodies that bind wild-type green fluorescent protein (GFP) and enhance its brightness ∼26-fold [120]. Exchange of photobleached GFP with newly synthesized GFP enables long-term slow tracking of single proteins. A disadvantage of these large tags, especially for fast tracking, is that they can greatly slow diffusion of the target protein and impact its function [118].
MINFLUX
MINFLUX localizes one fluorophore at a time by continuously re-centering a ‘doughnut' beam to find the point of minimum excitation [121]. Fewer photons are required for precise localization than in widefield imaging, reducing the rate of photobleaching and enabling much longer tracking. While so far mostly applied to fixed samples, it can also be used to track moving molecules for several seconds at sampling rates of 100s of kHz [121,122]. Though more technically complex than widefield imaging and currently able to track only one molecule at a time, MINFLUX will enable applications that require long, high-resolution trajectories.
Three-dimensional tracking
Loss of trajectories due to defocalization could be prevented by imaging and tracking in 3D. While light-sheet microscopy can be used for 3D slow SMT [37], its volumetric imaging rate is typically too slow to track 3D diffusion of molecules. One solution is a form of multifocus microscopy (MFM) that uses specialized diffractive optics to image several z-planes simultaneously on a camera [33,112,123]. While MFM decreases the signal per molecule by splitting light over multiple focal planes, more efficient diffraction grating designs and brighter labels are making this less of a problem [124,125].
Glossary
Angular anisotropy: Deviation of the angles of successive single-molecule displacements from the uniform distribution characteristic of regular Brownian motion.
Brownian motion: An idealized random walk in which the subsequent position of an object is drawn from a Gaussian distribution centered on its current position.
Defocalization: Movement of objects out of the focal plane.
Diffusion coefficient: A measure of how fast a molecule diffuses. More precisely, the constant of proportionality between its MSD and 2nt, where t is time and n is the number of dimensions. E.g. in two dimensions, <r2> = 4Dt.
Direct reactivation (DR): Reactivation of a fluorophore from a dark state in response to absorption of UV or near-UV light.
Highly inclined and laminated optical sheet (HILO): A type of illumination similar to TIRF in which a laser beam is allowed to propagate obliquely through a sample, illuminating the region closest to the coverslip. Unlike TIRF, the beam propagates through the sample rather than undergoing total internal reflection, permitting excitation of fluorophores slightly farther from the coverslip (e.g. in the cell nucleus).
Intrinsically disordered regions (IDRs): protein sequences that do not adopt a fixed three-dimensional fold. IDRs have been dubbed the ‘dark matter' of the proteome—extremely common, yet difficult to characterize using the standard tools of biochemistry and structural biology.
Localization error: Uncertainty in the spatial localization of a molecule, arising from a finite number of emitted photons, molecular motion, camera noise, etc.
Mean-squared displacement (MSD): the mean of the squared displacement from the starting position of a random walk. For regular Brownian motion, MSD increases linearly with time.
MINFLUX: A method in which fluorophores are localized and tracked by actively re-centering a ‘doughnut' beam on the fluorophore. This is accomplished by fast electronic feedback, which locates the beam position that gives a minimal flux of emitted photons.
Photobleaching: destruction of fluorophores by chemical reactions that occur in the excited state.
Proximity-assisted photoactivation (PAPA): Reactivation of one fluorophore (e.g. JFX650) from a dark state upon excitation of a second nearby fluorophore (e.g. JF549).
Residence time: How long a molecule remains bound to its target (e.g. chromatin).
- Single-molecule tracking (SMT): a method that monitors the motions of individual fluorescently labeled molecules under a microscope. SMT is a type of single-particle tracking (SPT), which includes the tracking of objects larger than a single molecule, although in practice the terms SMT and SPT are often used interchangeably.
- o Slow SMT: Tracking molecules at a relatively slow frame rate (hundreds of milliseconds to seconds) to monitor how long molecules remain bound to a slow-moving scaffold (e.g. chromatin) and how they move while bound.
- o Fast SMT: Tracking molecules with a relatively fast frame rate (typically ≤ 10 ms) to measure their diffusion. Short 1–2 ms stroboscopic excitation pulses can also be used to reduce motion blur.
Total internal reflection fluorescence (TIRF): A type of illumination in which light is reflected off the interface between the coverslip and the sample at an angle greater than the critical angle. The resulting total internal reflection produces an exponentially decaying evanescent field that excites fluorophores within a few hundred nanometers of the interface.
Perspectives
Single-molecule tracking (SMT) probes interactions of biomolecules in live cells.
Key observables of SMT include diffusion rate and anisotropy, chromatin-bound fraction and residence time, subcellular localization, and proximity to other labeled molecules. SMT can distinguish subpopulations of a biomolecule based on differences in these parameters.
New technologies are enhancing our ability to track molecules for extended periods of time and study specific protein–protein and protein–nucleic acid interactions.
Acknowledgements
We would like to thank John J. Ferrie for insightful comments on this manuscript and apologize to our colleagues whose work we could not cite due to limited space.
Abbreviations
- BiFC
Bimolecular fluorescence complementation
- ChIP-seq
chromatin immunoprecipitation sequencing
- CMOS
complementary metal oxide semiconductor
- DR
direct reactivation
- EMCCD
electron-multiplying charge-coupled device
- FP
fluorescent protein
- GFP
green fluorescent protein
- GTFs
general transcription factors
- HILO
highly inclined and laminated optical sheet
- IDRs
intrinsically disordered regions
- JF
Janelia Fluor
- MFM
multifocus microscopy
- MSD
mean squared displacement
- PAPA
proximity-assisted photoactivation
- PIC
pre-initiation complex
- POI
protein of interest
- Pol II
RNA polymerase II
- PPIs
protein–protein interactions
- RC
replication compartment
- smFRET
single-molecule Förster resonance energy transfer
- SMT
single-molecule tracking
- STORM
stochastic optical reconstruction microscopy
- TFs
transcription factors
- TIRF
total internal reflection fluorescence
Competing Interests
X.D. and R.T. are cofounders of Eikon Therapeutics. Inc.
Funding
This work was supported by National Institutes of Health grants U54-CA231641-01659 (to X.D.), the Howard Hughes Medical Institute (to R.T.). N.W. acknowledges funding from the German Research Foundation (DFG) via a Walter Benjamin Fellowship.
Open Access
Open access for this article was enabled through a transformative open access agreement between Portland Press and the University of California.
Author Contributions
L.D. and T.G.W.G. wrote the manuscript with input from N.W., R.T., and X.D. N.W. prepared the figures.
References
- 1.Kusumi, A., Tsunoyama, T.A., Hirosawa, K.M., Kasai, R.S. and Fujiwara, T.K. (2014) Tracking single molecules at work in living cells. Nat. Chem. Biol. 10, 524–532 10.1038/nchembio.1558 [DOI] [PubMed] [Google Scholar]
- 2.Li, N., Zhao, R., Sun, Y., Ye, Z., He, K. and Fang, X. (2017) Single-molecule imaging and tracking of molecular dynamics in living cells. Natl Sci. Rev. 4, 739–760 10.1093/nsr/nww055 [DOI] [Google Scholar]
- 3.Elf, J. and Barkefors, I. (2019) Single-molecule kinetics in living cells. Annu. Rev. Biochem. 88, 635–659 10.1146/annurev-biochem-013118-110801 [DOI] [PubMed] [Google Scholar]
- 4.Thomas, M.C. and Chiang, C.M. (2006) The general transcription machinery and general cofactors. Crit. Rev. Biochem. Mol. Biol. 41, 105–178 10.1080/10409230600648736 [DOI] [PubMed] [Google Scholar]
- 5.Sainsbury, S., Bernecky, C. and Cramer, P. (2015) Structural basis of transcription initiation by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 16, 129–143 10.1038/nrm3952 [DOI] [PubMed] [Google Scholar]
- 6.Lambert, S.A., Jolma, A., Campitelli, L.F., Das, P.K., Yin, Y., Albu, M.et al. (2018) The human transcription factors. Cell 172, 650–665 10.1016/j.cell.2018.01.029 [DOI] [PubMed] [Google Scholar]
- 7.Näär, A., Lemon, B. and Tjian, R. (2001) Transcriptional coactivator complexes. Cell 70, 475–501 10.1146/annurev.biochem.70.1.475 [DOI] [PubMed] [Google Scholar]
- 8.Zabidi, M.A. and Stark, A. (2016) Regulatory enhancer – core-promoter communication via transcription factors and cofactors. Trends Genet. 32, 801–814 10.1016/j.tig.2016.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reiter, F., Wienerroither, S. and Stark, A. (2017) Combinatorial function of transcription factors and cofactors. Curr. Opin. Genet. Dev. 43, 73–81 10.1016/j.gde.2016.12.007 [DOI] [PubMed] [Google Scholar]
- 10.Levine, M., Cattoglio, C. and Tjian, R. (2014) Looping back to leap forward: transcription enters a new era. Cell 157, 13–25 10.1016/j.cell.2014.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dean, A., Larson, D.R. and Sartorelli, V. (2021) Enhancers, gene regulation, and genome organization. Genes Dev. 35, 427–433 10.1101/gad.348372.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karr, J.P., Ferrie, J.J., Tjian, R. and Darzacq, X. (2022) The transcription factor activity gradient (TAG) model: contemplating a contact-independent mechanism for enhancer–promoter communication. Genes Dev. 36, 7–16 10.1101/GAD.349160.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rust, M.J., Bates, M. and Zhuang, X. (2006) Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Methods 3, 793–796 10.1038/nmeth929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Betzig, E., Patterson, G.H., Sougrat, R., Lindwasser, O.W., Olenych, S., Bonifacino, J.S.et al. (2006) Imaging intracellular fluorescent proteins at nanometer resolution. Science 313, 1642–1645 10.1126/science.1127344 [DOI] [PubMed] [Google Scholar]
- 15.Doudna, J.A. and Charpentier, E. (2014) The new frontier of genome engineering with CRISPR-Cas9. Science 346, 1258096 10.1126/science.1258096 [DOI] [PubMed] [Google Scholar]
- 16.Liss, V., Barlag, B., Nietschke, M. and Hensel, M. (2016) Self-labelling enzymes as universal tags for fluorescence microscopy, super-resolution microscopy and electron microscopy. Sci. Rep. 5, 17740 10.1038/srep17740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Patterson, G.H. and Lippincott-Schwartz, J. (2002) A photoactivatable GFP for selective photolabeling of proteins and cells. Science 297, 1873–1877 10.1126/science.1074952 [DOI] [PubMed] [Google Scholar]
- 18.Lukyanov, K., Chudakov, D., Lukyanov, S. and Verkhusha, V. (2005) Photoactivatable fluorescent proteins. Nat. Rev. Mol. Cell Biol. 6, 885–890 10.1038/nrm1741 [DOI] [PubMed] [Google Scholar]
- 19.Grimm, J.B., Klein, T., Kopek, B.G., Shtengel, G., Hess, H.F., Sauer, M.et al. (2015) Synthesis of a far-red photoactivatable silicon-containing rhodamine for super-resolution microscopy. Angew. Chem. Int. Ed. Engl. 55, 1723–1727 10.1002/anie.201509649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frei, M.S., Hoess, P., Lampe, M., Nijmeijer, B., Kueblbeck, M., Ellenberg, J.et al. (2019) Photoactivation of silicon rhodamines via a light-induced protonation. Nat. Commun. 10, 4580 10.1038/s41467-019-12480-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ghosh, R.N. and Webb, W.W. (1994) Automated detection and tracking of individual and clustered cell surface low density lipoprotein receptor molecules. Biophys. J. 66, 1301–1318 10.1016/S0006-3495(94)80939-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Funatsu, T., Harada, Y., Tokunaga, M., Saito, K. and Yanagida, T. (1995) Imaging of single fluorescent molecules and individual ATP turnovers by single myosin molecules in aqueous solution. Nature 374, 555–559 10.1038/374555a0 [DOI] [PubMed] [Google Scholar]
- 23.Yildiz, A., Forkey, J.N., McKinney, S.A., Ha, T., Goldman, Y.E. and Selvin, P.R. (2003) Myosin V walks hand-over-hand: single fluorophore imaging with 1.5-nm localization. Science 300, 2061–2065 10.1126/science.1084398 [DOI] [PubMed] [Google Scholar]
- 24.Barak, L.S. and Webb, W.W. (1981) Fluorescent low density lipoprotein for observation of dynamics of individual receptor complexes on cultured human fibroblasts. J. Cell Biol. 90, 595–604 10.1083/jcb.90.3.595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sako, Y., Minoghchi, S. and Yanagida, T. (2000) Single-molecule imaging of EGFR signalling on the surface of living cells. Nat. Cell Biol. 2, 168–172 10.1038/35004044 [DOI] [PubMed] [Google Scholar]
- 26.Yang, W., Gelles, J. and Musser, S.M. (2004) Imaging of single-molecule translocation through nuclear pore complexes. Proc. Natl Acad Sci. U.S.A. 101, 12887–12892 10.1073/pnas.0403675101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Elf, J., Li, G.W. and Xie, S.X. (2007) Probing transcription factor dynamics at the single-molecule level in a living cell. Science 316, 1191–1194 10.1126/science.1141967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sugo, N., Morimatsu, M., Arai, Y., Kousoku, Y., Ohkuni, A., Nomura, T.et al. (2015) Single-molecule imaging reveals dynamics of CREB transcription factor bound to its target sequence. Sci. Rep. 5, 10662 10.1038/srep10662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Loffreda, A., Jacchetti, E., Antunes, S., Rainone, P., Daniele, T., Morisaki, T.et al. (2017) Live-cell p53 single-molecule binding is modulated by C-terminal acetylation and correlates with transcriptional activity. Nat. Commun. 8, 313 10.1038/s41467-017-00398-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Callegari, A., Sieben, C., Benke, A., Suter, D.M., Fierz, B., Mazza, D.et al. (2019) Single-molecule dynamics and genome-wide transcriptomics reveal that NF-kB (p65)-DNA binding times can be decoupled from transcriptional activation. PLoS Genet. 15, e1007891 10.1371/journal.pgen.1007891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Popp, A.P., Hettich, J. and Gebhardt, J.C.M. (2021) Altering transcription factor binding reveals comprehensive transcriptional kinetics of a basic gene. Nucleic Acids Res. 49, 6249–6266 10.1093/nar/gkab443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garcia, D.A., Johnson, T.A., Presman, D.M., Fettweis, G., Wagh, K., Rinaldi, L.et al. (2021) An intrinsically disordered region-mediated confinement state contributes to the dynamics and function of transcription factors. Mol. Cell 81, 1484–1498.e6 10.1016/j.molcel.2021.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen, J., Zhang, Z., Li, L., Chen, B.C., Revyakin, A., Hajj, B.et al. (2014) Single-molecule dynamics of enhanceosome assembly in embryonic stem cells. Cell 156, 1274–1285 10.1016/j.cell.2014.01.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Teves, S.S., An, L., Hansen, A.S., Xie, L., Darzacq, X. and Tjian, R. (2016) A dynamic mode of mitotic bookmarking by transcription factors. eLife 5, e22280 10.7554/eLife.22280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mir, M., Reimer, A., Haines, J.E., Li, X.Y., Stadler, M., Garcia, H.et al. (2017) Dense bicoid hubs accentuate binding along the morphogen gradient. Genes Dev. 31, 1784–1794 10.1101/gad.305078.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hansen, A.S., Pustova, I., Cattoglio, C., Tjian, R. and Darzacq, X. (2017) CTCF and cohesin regulate chromatin loop stability with distinct dynamics. eLife 6, e25776 10.7554/eLife.25776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mir, M., Stadler, M.R., Ortiz, S.A., Hannon, C.E., Harrison, M.M., Darzacq, X.et al. (2018) Dynamic multifactor hubs interact transiently with sites of active transcription in drosophila embryos. eLife 7, e40497 10.7554/eLife.40497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reisser, M., Palmer, A., Popp, A.P., Jahn, C., Weidinger, G. and Gebhardt, J.C.M. (2018) Single-molecule imaging correlates decreasing nuclear volume with increasing TF-chromatin associations during zebrafish development. Nat. Commun. 9, 5218 10.1038/s41467-018-07731-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mazza, D., Abernathy, A., Golob, N., Morisaki, T. and McNally, J.G. (2012) A benchmark for chromatin binding measurements in live cells. Nucleic Acids Res. 40, e119 10.1093/nar/gks701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Izeddin, I., Récamier, V., Bosanac, L., Cissé, I.I., Boudarene, L., Dugast-Darzacq, C.et al. (2014) Single-molecule tracking in live cells reveals distinct target-search strategies of transcription factors in the nucleus. eLife 3, e02230 10.7554/eLife.02230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Patterson, G., Davidson, M., Manley, S. and Lippincott-Schwartz, J. (2010) Superresolution imaging using single-molecule localization. Annu. Rev. Phys. Chem. 61, 345–367 10.1146/annurev.physchem.012809.103444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang, S., Moffitt, J.R., Dempsey, G.T., Xie, X.S. and Zhuang, X. (2014) Characterization and development of photoactivatable fluorescent proteins for single-molecule-based superresolution imaging. Proc. Natl Acad Sci. U.S.A. 111, 8452–8457 10.1073/pnas.1406593111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Long, F., Zeng, S. and Huang, Z.L. (2012) Localization-based super-resolution microscopy with an sCMOS camera Part II: experimental methodology for comparing sCMOS with EMCCD cameras. Opt. Express 20, 17741–17759 10.1364/OE.20.017741 [DOI] [PubMed] [Google Scholar]
- 44.Wang, Y., Zhao, L., Hu, Z., Wang, Y., Zhao, Z., Li, L.et al. (2017) Quantitative performance evaluation of a back-illuminated sCMOS camera with 95% QE for super-resolution localization microscopy. Cytom. Part A 91, 1175–1183 10.1002/cyto.a.23282 [DOI] [PubMed] [Google Scholar]
- 45.Mandracchia, B., Hua, X., Guo, C., Son, J., Urner, T. and Jia, S. (2020) Fast and accurate sCMOS noise correction for fluorescence microscopy. Nat. Commun. 11, 94 10.1038/s41467-019-13841-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Diekmann, R., Deschamps, J., Li, Y., Deguchi, T., Tschanz, A., Kahnwald, M.et al. (2022) Photon-free (s)CMOS camera characterization for artifact reduction in high- and super-resolution microscopy. Nat. Commun. 13, 3362 10.1038/s41467-022-30907-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tokunaga, M., Imamoto, N. and Kumiko, S.S. (2008) Highly inclined thin illumination enables clear single-molecule imaging in cells. Nat. Methods 5, 159–161 10.1038/nmeth1171 [DOI] [PubMed] [Google Scholar]
- 48.Chen, B.C., Legant, W.R., Wang, K., Shao, L., Milkie, D.E., Davidson, W.et al. (2014) Lattice light sheet microscopy: imaging molecules to embryos at high spatiotemporal resolution. Science 346, 1257998 10.1126/science.1257998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sapoznik, E., Chang, B.J., Huh, J., Ju, R.J., Azarova E, V., Pohlkamp, T.et al. (2020) A versatile oblique plane microscope for large-scale and high-resolution imaging of subcellular dynamics. eLife 9, e57681 10.7554/eLife.57681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Galland, R., Grenci, G., Aravind, A., Viasnoff, V., Studer, V. and Sibarita, J.B. (2015) 3D high-and super-resolution imaging using single-objective SPIM. Nat. Methods 12, 641–644 10.1038/nmeth.3402 [DOI] [PubMed] [Google Scholar]
- 51.Morisaki, T., Müller, W.G., Golob, N., Mazza, D. and McNally, J.G. (2014) Single-molecule analysis of transcription factor binding at transcription sites in live cells. Nat. Commun. 5, 4456 10.1038/ncomms5456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Paakinaho, V., Presman, D.M., Ball, D.A., Johnson, T.A., Schiltz, R.L., Levitt, P.et al. (2017) Single-molecule analysis of steroid receptor and cofactor action in living cells. Nat. Commun. 8, 15896 10.1038/ncomms15896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mehta, G.D., Ball, D.A., Eriksson, P.R., Chereji R, V., Clark, D.J., McNally, J.G.et al. (2018) Single-molecule analysis reveals linked cycles of RSC chromatin remodeling and Ace1p transcription factor binding in yeast. Mol. Cell 72, 875–887.e9 10.1016/j.molcel.2018.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lerner, J., Gomez-Garcia, P.A., McCarthy, R.L., Liu, Z., Lakadamyali, M. and Zaret, K.S. (2020) Two-parameter mobility assessments discriminate diverse regulatory factor behaviors in chromatin. Mol. Cell 79, 677–688.e6 10.1016/j.molcel.2020.05.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Garcia, D.A., Fettweis, G., Presman, D.M., Paakinaho, V., Jarzynski, C., Upadhyaya, A.et al. (2021) Power-law behavior of transcription factor dynamics at the single-molecule level implies a continuum affinity model. Nucleic Acids Res. 49, 6605–6620 10.1093/nar/gkab072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hansen, A.S., Woringer, M., Grimm, J.B., Lavis, L.D., Tjian, R. and Darzacq, X. (2018) Robust model-based analysis of single-particle tracking experiments with spot-on. eLife 7, e33125 10.7554/eLife.33125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hansen, A.S., Amitai, A., Cattoglio, C., Tjian, R. and Darzacq, X. (2020) Guided nuclear exploration increases CTCF target search efficiency. Nat. Chem. Biol. 16, 257–266 10.1038/s41589-019-0422-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Woringer, M. and Darzacq, X. (2018) Protein motion in the nucleus: from anomalous diffusion to weak interactions. Biochem. Soc. Trans. 46, 945–956 10.1042/BST20170310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sergé, A., Bertaux, N., Rigneault, H. and Marguet, D. (2008) Dynamic multiple-target tracing to probe spatiotemporal cartography of cell membranes. Nat. Methods 5, 687–694 10.1038/nmeth.1233 [DOI] [PubMed] [Google Scholar]
- 60.Chenouard, N., Smal, I., De Chaumont, F., Maška, M., Sbalzarini, I.F., Gong, Y.et al. (2014) Objective comparison of particle tracking methods. Nat. Methods 11, 281–289 10.1038/nmeth.2808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Heckert, A. (2022) Quot: a simple single molecule tracking pipeline with a graphic user interface for quality control [Internet]. GitHub. 2022. https://github.com/alecheckert/quot
- 62.Kuhn, T., Hettich, J., Davtyan, R. and Gebhardt, J.C.M. (2021) Single molecule tracking and analysis framework including theory-predicted parameter settings. Sci. Rep. 11, 9465 10.1038/s41598-021-88802-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gebhardt, J.C.M., Suter, D.M., Roy, R., Zhao, Z.W., Chapman, A.R., Basu, S.et al. (2013) Single molecule imaging of transcription factor binding to DNA in live mammalian cells. Science 10, 421–426 10.1038/nmeth.2411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hipp, L., Beer, J., Kuchler, O., Reisser, M., Sinske, D., Michaelis, J.et al. (2019) Single-molecule imaging of the transcription factor SRF reveals prolonged chromatin-binding kinetics upon cell stimulation. Proc. Natl Acad Sci. U.S.A. 116, 880–889 10.1073/pnas.1812734116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Reisser, M., Hettich, J., Kuhn, T., Popp, A.P., Große-Berkenbusch, A. and Gebhardt, J.C.M. (2020) Inferring quantity and qualities of superimposed reaction rates from single molecule survival time distributions. Sci. Rep. 10, 1758 10.1038/s41598-020-58634-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen, Z., Geffroy, L. and Biteen, J.S. (2021) NOBIAS: analyzing anomalous diffusion in single-molecule tracks with nonparametric Bayesian inference. Front. Bioinform. 1, 742073 10.3389/fbinf.2021.742073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Persson, F., Lindén, M., Unoson, C. and Elf, J. (2013) Extracting intracellular diffusive states and transition rates from single-molecule tracking data. Nat. Methods 10, 265–269 10.1038/nmeth.2367 [DOI] [PubMed] [Google Scholar]
- 68.Heckert, A., Dahal, L., Tijan, R. and Darzacq, X. (2022) Recovering mixtures of fast-diffusing states from short single-particle trajectories. eLife 11, e70169 10.7554/eLife.70169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Graham, T.G.W., Ferrie, J.J., Dailey, G.M., Tjian, R. and Darzacq, X. (2022) Detecting molecular interactions in live-cell single-molecule imaging with proximity-assisted photoactivation (PAPA). eLife 11, e76870 10.7554/eLife.76870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen, Y., Cattoglio, C., Dailey, G.M., Zhu, Q., Tjian, R. and Darzacq, X. (2022) Mechanisms governing target search and binding dynamics of hypoxia-inducible factors. eLife 11, e75064 10.7554/eLife.75064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tatavosian, R., Duc, H.N., Huynh, T.N., Fang, D., Schmitt, B., Shi, X.et al. (2018) Live-cell single-molecule dynamics of PcG proteins imposed by the DIPG H3.3K27M mutation. Nat. Commun. 9, 2080 10.1038/s41467-018-04455-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang, Z., English, B.P., Grimm, J.B., Kazane, S.A., Hu, W., Tsai, A.et al. (2016) Rapid dynamics of general transcription factor TFIIB binding during preinitiation complex assembly revealed by single-molecule analysis. Genes Dev. 30, 2106–2118 10.1101/gad.285395.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Teves, S.S., An, L., Bhargava-Shah, A., Xie, L., Darzacq, X. and Tjian, R. (2018) A stable mode of bookmarking by TBP recruits RNA polymerase II to mitotic chromosomes. eLife 7, e35621 10.7554/eLife.35621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nguyen, V.Q., Ranjan, A., Liu, S., Tang, X., Ling, Y.H., Wisniewski, J.et al. (2021) Spatiotemporal coordination of transcription preinitiation complex assembly in live cells. Mol. Cell 81, 3560–3575.e6 10.1016/j.molcel.2021.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Swinstead, E.E., Miranda, T.B., Paakinaho, V., Baek, S., Goldstein, I., Hawkins, M.et al. (2016) Steroid receptors reprogram FoxA1 occupancy through dynamic chromatin transitions. Cell 165, 593–605 10.1016/j.cell.2016.02.067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wagh, K., Stavreva, D.A., Jensen, R.A.M., Paakinaho, V., Fettweis, G., Schiltz, R.L.et al. (2022) Single-molecule tracking reveals two low-mobility states for chromatin and transcriptional regulators within the nucleus. bioRxiv 1–34 10.1101/2022.07.25.501476 [DOI] [Google Scholar]
- 77.Von Hippel, P.H. and Berg, O.G. (1989) Facilitated target location in biological systems. J. Biol. Chem. 264, 675–678 10.1016/S0021-9258(19)84994-3 [DOI] [PubMed] [Google Scholar]
- 78.Tafvizi, A., Huang, F., Fersht, A.R., Mirny, L.A. and van Oijen, A.M. (2011) A single-molecule characterization of p53 search on DNA. Proc. Natl Acad Sci. U.S.A. 108, 563–568 10.1073/pnas.1016020107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hammar, P., Leroy, P., Mahmutovic, A., Marklund, E., Berg, O. and Elf, J. (2012) The lac repressor displays facilitated diffusion in living cells. Science 336, 1595–1598 10.1126/science.1221648 [DOI] [PubMed] [Google Scholar]
- 80.Normanno, D., Boudarène, L., Dugast-Darzacq, C., Chen, J., Richter, C., Proux, F.et al. (2015) Probing the target search of DNA-binding proteins in mammalian cells using tetR as model searcher. Nat. Commun. 6, 7357 10.1038/ncomms8357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Caccianini, L., Normanno, D., Izeddin, I. and Dahan, M. (2015) Single molecule study of non-specific binding kinetics of LacI in mammalian cells. Faraday Discuss 184, 393–400 10.1039/C5FD00112A [DOI] [PubMed] [Google Scholar]
- 82.Ross, J.L. (2016) The dark matter of biology. Biophys. J. 111, 909–916 10.1016/j.bpj.2016.07.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lu, F. and Lionnet, T. (2021) Transcription factor dynamics. Cold Spring Harb. Perspect. Biol. 13, a040949 10.1101/cshperspect.a040949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ferrie, J.J., Karr, J.P., Tjian, R. and Darzacq, X. (2022) ‘Structure’-function relationships in eukaryotic transcription factors: the role of intrinsically disordered regions in gene regulation. Mol. Cell 82, 3970–3984 10.1016/j.molcel.2022.09.021 [DOI] [PubMed] [Google Scholar]
- 85.Chong, S., Graham, T.G.W., Dugast-Darzacq, C., Dailey, G.M., Darzacq, X. and Tjian, R. (2022) Tuning levels of low-complexity domain interactions to modulate endogenous oncogenic transcription. Mol. Cell 82, 2084–2097.e5 10.1016/j.molcel.2022.04.007 [DOI] [PubMed] [Google Scholar]
- 86.McSwiggen, D.T., Hansen, A.S., Marie-Nelly, H., Teves, S., Heckert, A.B., Dugast-Darzacq, C.et al. (2019) Evidence for DNA-mediated nuclear compartmentalization distinct from phase separation. eLife 8, e47098 10.7554/eLife.47098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Collombet, S., Rall, I., Dugast-Darzacq, C., Heckert, A., Halavatyi, A., Le Saux, A.et al. (2021) RNA polymerase II depletion from the inactive X chromosome territory is not mediated by physical compartmentalization. bioRxiv 1–27 10.1101/2021.03.26.437188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Robinett, C.C., Straight, A., Li, G., Willhelm, C., Sudlow, G., Murray, A.et al. (1996) In vivo localization of DNA sequences and visualization of large-scale chromatin organization using lac operator/repressor recognition. J. Cell Biol. 135, 1685–1700 10.1083/jcb.135.6.1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Saad, H., Gallardo, F., Dalvai, M., Tanguy-le-Gac, N., Lane, D. and Bystricky, K. (2014) DNA dynamics during early double-strand break processing revealed by non-intrusive imaging of living cells. PLoS Genet. 10, e1004187 10.1371/journal.pgen.1004187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tasan, I., Sustackova, G., Zhang, L., Kim, J., Sivaguru, M., HamediRad, M.et al. (2018) CRISPR/Cas9-mediated knock-in of an optimized TetO repeat for live cell imaging of endogenous loci. Nucleic Acids Res. 46, e100 10.1093/nar/gky501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gu, B., Swigut, T., Spencley, A., Bauer, M.R., Chung, M., Meyer, T.et al. (2018) Transcription-coupled changes in nuclear mobility of mammalian cis-regulatory elements. Science 359, 1050–1055 10.1126/science.aao3136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Eykelenboom, J.K., Gierlinski, M., Yue, Z., Hegarat, N., Pollard, H., Fukagawa, T.et al. (2019) Live imaging of marked chromosome regions reveals their dynamic resolution and compaction in mitosis. J. Cell Biol. 218, 1531–1552 10.1083/jcb.201807125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chen, B., Gilbert, L.A., Cimini, B.A., Schnitzbauer, J., Zhang, W., Li, G.W.et al. (2013) Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell 155, 1479–1491 10.1016/j.cell.2013.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ma, H., Tu, L.C., Naseri, A., Huisman, M., Zhang, S., Grunwald, D.et al. (2016) Multiplexed labeling of genomic loci with dCas9 and engineered sgRNAs using CRISPRainbow. Nat. Biotechnol. 34, 528–530 10.1038/nbt.3526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Clow, P.A., Du, M., Jillette, N., Taghbalout, A., Zhu, J.J. and Cheng, A.W. (2022) CRISPR-mediated multiplexed live cell imaging of nonrepetitive genomic loci with one guide RNA per locus. Nat. Commun. 13, 1871 10.1038/s41467-022-29343-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Stanyte, R., Nuebler, J., Blaukopf, C., Hoefler, R., Stocsits, R., Peters, J.M.et al. (2018) Dynamics of sister chromatid resolution during cell cycle progression. J. Cell Biol. 217, 1985–2004 10.1083/jcb.201801157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gomez-Lamarca, M.J., Falo-Sanjuan, J., Stojnic, R., Abdul Rehman, S., Muresan, L., Jones, M.L.et al. (2018) Activation of the notch signaling pathway in vivo elicits changes in CSL nuclear dynamics. Dev. Cell 44, 611–623.e7 10.1016/j.devcel.2018.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chong, S., Dugast-Darzacq, C., Liu, Z., Dong, P., Dailey, G.M., Cattoglio, C.et al. (2018) Imaging dynamic and selective low-complexity domain interactions that control gene transcription. Science 361, eaar2555 10.1126/science.aar2555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wilmes, S., Hafer, M., Vuorio, J., Tucker, J.A., Winkelmann, H., Löchte, S.et al. (2020) Mechanism of homodimeric cytokine receptor activation and dysregulation by oncogenic mutations. Science 367, 643–652 10.1126/science.aaw3242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Asher, W.B., Geggier, P., Holsey, M.D., Gilmore, G.T., Pati, A.K., Meszaros, J.et al. (2021) Single-molecule FRET imaging of GPCR dimers in living cells. Nat. Methods 18, 397–405 10.1038/s41592-021-01081-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sotolongo Bellón, J., Birkholz, O., Richter, C.P., Eull, F., Kenneweg, H., Wilmes, S.et al. (2022) Four-color single-molecule imaging with engineered tags resolves the molecular architecture of signaling complexes in the plasma membrane. Cell Rep. Methods 2, 100165 10.1016/j.crmeth.2022.100165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kerppola, T.K. (2008) Bimolecular fluorescence complementation (BiFC) analysis as a probe of protein interactions in living cells. Annu. Rev. Biophys. 37, 465–487 10.1146/annurev.biophys.37.032807.125842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Makhija, S., Brown, D., Rudlaff, R.M., Doh, J.K., Bourke, S., Wang, Y.et al. (2021) Versatile labeling and detection of endogenous proteins using tag-assisted split enzyme complementation. ACS Chem. Biol. 16, 671–681 10.1021/acschembio.0c00925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nickerson, A., Huang, T., Lin, L.J. and Nan, X. (2014) Photoactivated localization microscopy with bimolecular fluorescence complementation (BiFC-PALM) for nanoscale imaging of protein-protein interactions in cells. PLoS ONE 9, e1005489 10.1371/journal.pone.0100589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mao, S., Ying, Y., Ma, Z., Yang, Y. and Chen, A.K. (2021) A background assessable and correctable bimolecular fluorescence complementation system for nanoscopic single-molecule imaging of intracellular protein-protein interactions. ACS Nano 15, 14338–14346 10.1021/acsnano.1c03242 [DOI] [PubMed] [Google Scholar]
- 106.Shao, S., Zhang, H., Zeng, Y., Li, Y., Sun, C. and Sun, Y. (2021) TagBiFC technique allows long-term single-molecule tracking of protein-protein interactions in living cells. Commun. Biol. 4, 1–14 10.1038/s42003-021-01896-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kodama, Y. and Hu, C.D. (2012) Bimolecular fluorescence complementation (BiFC): a 5-year update and future perspectives. Biotechniques 53, 285–298 10.2144/000113943 [DOI] [PubMed] [Google Scholar]
- 108.Bates, M., Blosser, T.R. and Zhuang, X. (2005) Short-range spectroscopic ruler based on a single-molecule optical switch. Phys. Rev. Lett. 94, 108101 10.1103/PhysRevLett.94.108101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Los G, V., Encell, L.P., Mcdougall, M.G., Hartzell, D.D., Karassina, N., Zimprich, C.et al. (2008) Halotag: a novel protein labeling technology for cell imaging and protein analysis. ACS Chem. Biol. 3, 373–382 10.1021/cb800025k [DOI] [PubMed] [Google Scholar]
- 110.Gautier, A., Juillerat, A., Heinis, C., Corrêa, I.R., Kindermann, M., Beaufils, F.et al. (2008) An engineered protein tag for multiprotein labeling in living cells. Chem. Biol. 15, 128–136 10.1016/j.chembiol.2008.01.007 [DOI] [PubMed] [Google Scholar]
- 111.Grimm, J.B., English, B.P., Chen, J., Slaughter, J.P., Zhang, Z., Revyakin, A.et al. (2015) A general method to improve fluorophores for live-cell and single-molecule microscopy. Nat. Methods 12, 244–250 10.1038/nmeth.3256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Grimm, J.B., English, B.P., Choi, H., Muthusamy, A.K., Mehl, B.P., Dong, P.et al. (2016) Bright photoactivatable fluorophores for single-molecule imaging. Nat. Methods. 13, 985–988 10.1038/nmeth.4034 [DOI] [PubMed] [Google Scholar]
- 113.Lukinavičius, G., Umezawa, K., Olivier, N., Honigmann, A., Yang, G., Plass, T.et al. (2013) A near-infrared fluorophore for live-cell super-resolution microscopy of cellular proteins. Nat. Chem. 5, 132–139 10.1038/nchem.1546 [DOI] [PubMed] [Google Scholar]
- 114.Wang, L., Tran, M., D'Este, E., Roberti, J., Koch, B., Xue, L.et al. (2020) A general strategy to develop cell permeable and fluorogenic probes for multicolour nanoscopy. Nat. Chem. 12, 165–172 10.1038/s41557-019-0371-1 [DOI] [PubMed] [Google Scholar]
- 115.Lavis, L.D. (2017) Teaching old dyes new tricks: biological probes built from fluoresceins and rhodamines. Annu. Rev. Biochem. 86, 825–843 10.1146/annurev-biochem-061516-044839 [DOI] [PubMed] [Google Scholar]
- 116.Kompa, J., Bruins, J., Glogger, M., Wilhelm, J., Frei, M.S., Tarnawski, M.et al. (2023) Exchangeable HaloTag Ligands (xHTLs) for multi-modal super-resolution fluorescence microscopy. J. Am. Chem. Soc. 145, 3075–3083 10.1021/jacs.2c11969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Liu, H., Dong, P., Ioannou, M.S., Li, L., Shea, J., Pasolli, H.A.et al. (2018) Visualizing long-term single-molecule dynamics in vivo by stochastic protein labeling. Proc. Natl Acad. Sci. U.S.A. 115, 343–348 10.1073/pnas.1713895115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Tanenbaum, M.E., Gilbert, L.A., Qi, L.S., Weissman, J.S. and Vale, R.D. (2014) A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell. 159, 635–646 10.1016/j.cell.2014.09.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Boersma, S., Khuperkar, D., Verhagen, B.M.P., Sonneveld, S., Grimm, J.B., Lavis, L.D.et al. (2019) Multi-color single-molecule imaging uncovers extensive heterogeneity in mRNA decoding. Cell 178, 458–472.e19 10.1016/j.cell.2019.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ghosh, R.P., Franklin, J.M., Draper, W.E., Shi, Q., Beltran, B., Spakowitz, A.J.et al. (2019) A fluorogenic array for temporally unlimited single-molecule tracking. Nat. Chem. Biol. 15, 401–409 10.1038/s41589-019-0241-6 [DOI] [PubMed] [Google Scholar]
- 121.Balzarotti, F., Eilers, Y., Gwosch, K.C., Gynnå, A.H., Westphal, V., Stefani, F.D.et al. (2017) Nanometer resolution imaging and tracking of fluorescent molecules with minimal photon fluxes. Science 355, 606–612 10.1126/science.aak9913 [DOI] [PubMed] [Google Scholar]
- 122.Schmidt, R., Weihs, T., Wurm, C.A., Jansen, I., Rehman, J., Sahl, S.J.et al. (2021) MINFLUX nanometer-scale 3D imaging and microsecond-range tracking on a common fluorescence microscope. Nat. Commun. 12, 1478 10.1038/s41467-021-21652-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Abrahamsson, S., Chen, J., Hajj, B., Stallinga, S., Katsov, A.Y., Wisniewski, J.et al. (2013) Fast multicolor 3D imaging using aberration-corrected multifocus microscopy. Nat. Methods 10, 60–63 10.1038/nmeth.2277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hajj, B., Oudjedi, L., Fiche, J.B., Dahan, M. and Nollmann, M. (2017) Highly efficient multicolor multifocus microscopy by optimal design of diffraction binary gratings. Sci. Rep. 7, 5284 10.1038/s41598-017-05531-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hirata-Miyasaki, E., Pettersson, G.M., Bajor, A., Fouke, K., John, D.D., Thibeault, B.et al. (2022) Ultrafast live 3D imaging with 25-plane camera array multifocus microscopy. In Imaging and Applied Optics Congress 2022 (3D, AOA, COSI, ISA, pcAOP), Optica Publishing Group. p. IW1C.2, Washington, DC, USA 10.1364/ISA.2022.IW1C.2 [DOI] [Google Scholar]