Abstract
Landmark genome-wide association studies (GWAS) identified that mutations in autophagy genes correlated with inflammatory bowel disease (IBD), a heterogenous disease characterised by prolonged inflammation of the gastrointestinal tract, that can reduce a person's quality of life. Autophagy, the delivery of intracellular components to the lysosome for degradation, is a critical cellular housekeeping process that removes damaged proteins and turns over organelles, recycling their amino acids and other constituents to supply cells with energy and necessary building blocks. This occurs under both basal and challenging conditions such as nutrient deprivation. An understanding of the relationship between autophagy, intestinal health and IBD aetiology has improved over time, with autophagy having a verified role in the intestinal epithelium and immune cells. Here, we discuss research that has led to an understanding that autophagy genes, including ATG16L, ATG5, ATG7, IRGM, and Class III PI3K complex members, contribute to innate immune defence in intestinal epithelial cells (IECs) via selective autophagy of bacteria (xenophagy), how autophagy contributes to the regulation of the intestinal barrier via cell junctional proteins, and the critical role of autophagy genes in intestinal epithelial secretory subpopulations, namely Paneth and goblet cells. We also discuss how intestinal stem cells can utilise autophagy. Importantly, mouse studies have provided evidence that autophagy deregulation has serious physiological consequences including IEC death and intestinal inflammation. Thus, autophagy is now established as a key regulator of intestinal homeostasis. Further research into how its cytoprotective mechanisms can prevent intestinal inflammation may provide insights into the effective management of IBD.
Keywords: autophagy, Crohns disease, disease, gastrointestinal physiology, homeostasis, molecular basis of health and disease
Introduction
Inflammatory bowel disease (IBD) is considered a modern disease, having only emerged in the last 150 years [1] and being of higher prevalence in socio-economically advantaged populations [2]. Studies show that prenatal or perinatal exposure to antibiotics, use of oral contraceptives, or a Westernised diet of highly processed foods and high sugar content, can correlate with IBD [3–6]. These are factors that may explain how IBD disproportionately affects individuals in urbanised regions, although whether these are causative of IBD is yet to be proven unequivocally [3–6]. Approximately 6.8 million cases of IBD were reported across the globe in 2017 [2]. There are two clinically distinct classes of IBD — ulcerative colitis (UC) and Crohn's disease (CD) — although 5–15% of cases are classed as ‘indeterminate colitis’ [7]. The current paradigm for IBD aetiology is that individuals with genetic predispositions are exposed to triggering substances, for instance, bacteria or viruses, resulting in unresolved chronic inflammatory conditions driven by deregulated immune activity. Treatment options for IBD thus include an arsenal of both broad-acting and targeted immunosuppressive or immune-modulatory drugs that act to reduce inflammation. Still, the precise mechanisms of IBD aetiology remain unclear, and complete remission rates are generally reported to be below 50% [8–10].
In 2007, genome-wide association studies (GWAS) uncovered the core autophagy gene ATG16L to be a CD susceptibility locus [11–14]. Similarly, polymorphisms in other autophagy genes and autophagy-associated genes have been shown to be IBD-associated, including in MTMR3 and GPR65 [15,16], and also in ULK1, IRGM, NDP52 and PTPN2 for CD in particular [14,17–20]. This discovery of a link between ATG16L and CD catalysed research into the role of autophagy in the intestine and IBD. Here, we provide an updated review of the concepts that have emerged from this research, specifically the mechanisms and functions of autophagy proteins in intestinal epithelial cells (IECs) and intestinal stem cells (ISCs) and how these contribute to intestinal homeostasis and pathophysiology. We refer readers to other excellent reviews on how autophagy in immune cells contributes to IBD [21,22].
Molecular features of autophagy
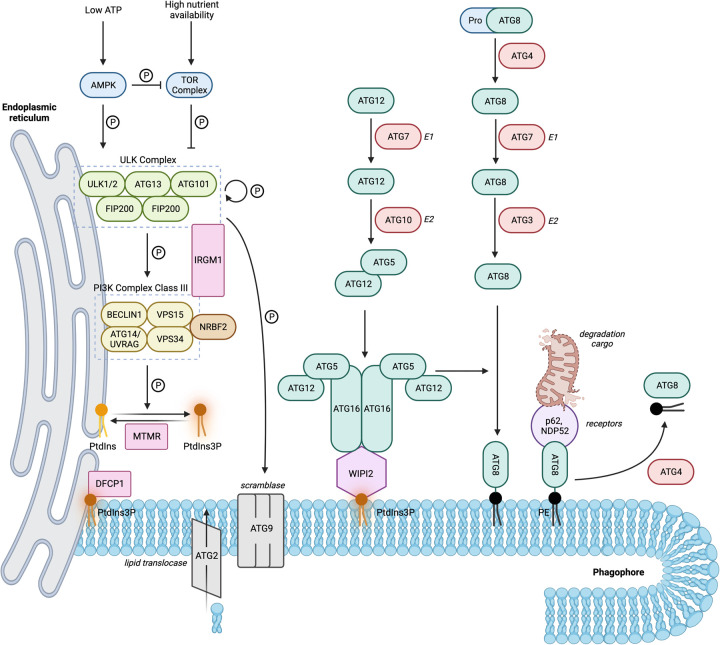
Autophagy is defined as the lysosomal degradation of intracellular entities. This pathway can selectively degrade a diverse range of cargo, for example, protein aggregates or entire organelles such as mitochondria. Alternatively, non-specific targets can be degraded in bulk [23]. Three types of autophagy have been described based on the route of cargo delivery to the lysosome. Whilst chaperone-mediated autophagy (CMA) and microautophagy involve direct translocation through, or invagination of, the lysosomal membrane, respectively, macroautophagy (hereafter referred to as autophagy) is arguably more complex as it involves the formation of a de novo double membrane that encloses cargo. Cargo-enclosed vesicles can then mature and fuse with the lysosome. The overall pathway is summarised in Figure 1. The full names of all proteins are provided in the Abbreviations section.
Figure 1. Schematic of the autophagy molecular network showing the major complexes involved in autophagosome initiation (ULK and PI3K Class III) and the two ubiquitin-like conjugation systems (ATG12–ATG5–ATG16L, and ATG8 lipidation) required for autophagosome expansion and capture of cargo.
Autophagy initiation
Low energy or nutrient status of cells may be detected by AMPK to initiate autophagy via phosphorylation at distinct sites on ULK1. Conversely, the phosphorylation of ULK1 by TOR at a different site can restrict autophagy under high nutrient conditions [24]. Initiation of autophagy drives the serine/threonine kinase activity of ULK1/2, which is stabilised as a multimeric complex consisting of ULK1/2, FIP200, ATG101, and ATG13 to phosphorylate itself, ATG9 and members of the Class III PI3K complex [25–33]. The Class III PI3K complex, consisting of the lipid kinase VPS34, and subunits BECLIN1, VPS15 and either ATG14 or UVRAG, is then activated to convert phosphatidylinositol (PtdIns) to PtdIns3P, an important lipid constituent of autophagosome double membranes that recruits downstream effectors such as the tether WIPI2, and DFCP1 [34–41]. ATG2 has recently been uncovered to be a lipid translocase [42–44], whilst ATG9 is a transmembrane protein located on various vesicular compartments and described to have scramblase activity [45]. Together, ATG2 and ATG9 traffic lipids to supply the growing autophagosome membrane [42–44].
Autophagosome expansion
Cargo sequestration and membrane expansion are supported by two ubiquitin-like conjugation systems that are localised to the autophagosome nucleation site by the ULK complex [46–49] and WIPI2 [40,50–53]. Firstly, the E1-like enzyme ATG7 activates and transfers ATG12 to the E2-like protein ATG10. This catalyses the conjugation of ATG12 to ATG5. ATG16L then interacts with ATG5 and self-associates to enable the formation of a multimer consisting of two ATG12–ATG5:ATG16L dimers [54]. Secondly, ATG4 cleaves pro-ATG8 proteins (LC3 or GABARAP) which enables them to be activated by the E1-like enzyme ATG7, passed onto the E2-like ATG3, before finally being conjugated to phosphatidylethanolamine (PE) by the E3 activity of the ATG12–ATG5:ATG16L multimer [54]. Lipidated ATG8s are incorporated into the growing phagophore membrane and serve to bind cargo with the assistance of cargo receptors and adaptors such as p62 and NDP52 [55,56].
Autophagosome maturation and fusion
Steps following the sealing of the autophagosome are considered part of the maturation process. This can include the removal of some (but not all) ATG proteins from the phagophore surface [57], the transport of autophagosomes to, and fusion with, late endosomes and/or lysosomes, and the acquisition of amphisome (autophagosome fused with endosome) and autophagolysosome acidity and degradative hydrolase activity [58]. The molecular machinery employed in the transport and fusion processes is shared with the endosomal trafficking network and includes SNAREs, tethers, adaptor proteins, and RAB proteins. Here, SNARES on the autophagosome (for example, STX17 and YTK6) form complexes with those on the late endosome or lysosome (for example, VAMP8 and STX7), typically using the SNARE SNAP29 as an intermediary [58]. Tethers (including the HOPs complex, PLEKHM1 and EPG5) are SNARE chaperones that interact with autophagosome membrane components (for instance PtdIns, GABARABs, WIPI proteins), to promote SNARE complexes formation, but can also be multifunctional, particularly as RAB7 modulators. RAB7, a late endosome and lysosomal marker which interacts with LC3, PtdIns, NRBF (a subsidiary of PI3K complex 1), and others, appears essential to autophagosome maturation through its GTP hydrolysis function mediated by a range of effectors [59,60], and is additionally involved with autophagosome positioning due to its interactions with the transport adaptor protein FYCO1 [61].
Other players
Several other proteins that are not directly involved in the generation of autophagosome membranes or the capture of cargo but can regulate some of these ‘core’ proteins to enhance or inhibit autophagy have also been investigated. Myotubularin phosphatases (MTMRs) are lipid phosphatases that can dephosphorylate PtdIns3P (and other lipids), to either promote autophagosome formation or reduce autophagic flux [62]. IRGM is an interferon (IFN)-inducible protein with pro-autophagic activity through interactions with both ULK1 and BECLIN1 to promote the assembly of autophagy initiation complexes [63]. Interactions between BECLIN1 and other proteins, such as pro-survival BCL-2 family members or AMBRA1, inhibit or promote autophagy, respectively [64]. The PI3K complex also binds additional auxiliary proteins such as NRBF2, forming subcomplexes that can be pro-autophagic or autophagy-inhibiting [64]. LRRK2, which harbours both GTPase and RAB-targeting kinase activity, is associated with both autophagy and endosomal trafficking, with context-dependent repression or activation of autophagy [65,66]. A G protein-coupled receptor involved in cAMP and Rho signalling pathways, and proton sensing, GPR65, was shown to be important for controlling lysosomal pH and the degradative capacity of this organelle [16]. Finally, the tyrosine phosphatase PTPN2 is required for efficient autophagosome formation but the mechanisms are still poorly understood [67].
Intestinal physiology
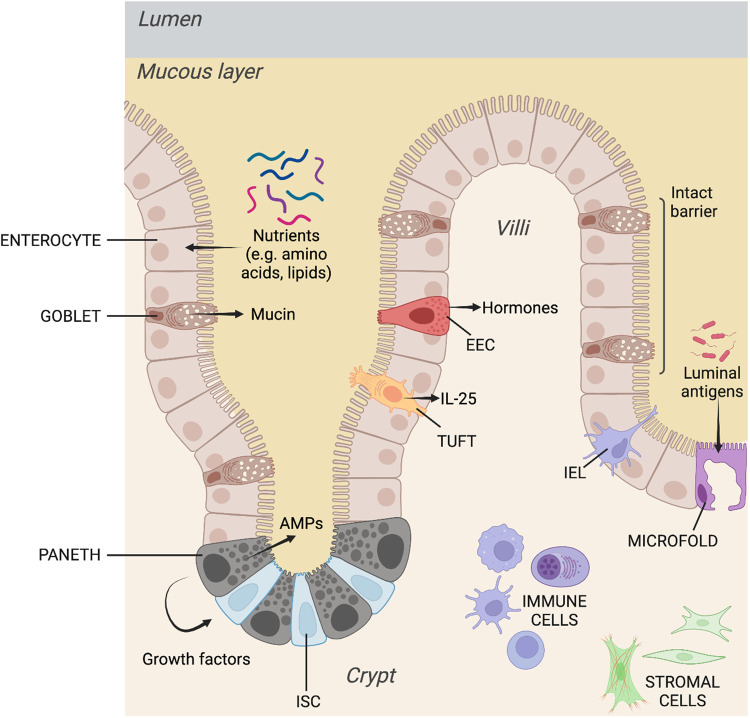
The small intestinal mucosa is organised into repeating crypt-villi structural units where a monolayer of polarised intestinal epithelial cells (IECs) reside over the stroma (Figure 2). Intestinal stem cells (ISCs) in the crypt divide and differentiate to give rise to the absorptive and secretory IEC lineages which perform different functional roles. Enterocytes are the most abundant IEC subtype and carry out digestive and nutrient-absorptive functions. Microfold (M) cells reside over intestinal lymphoid patches and sample the intestinal lumen, thus regulating immunotolerance to luminal antigens.
Figure 2. Anatomy and physiology of the small intestinal mucosa showing the major cell types and their relative locations within the repeating crypt-villi structures.
There are four secretory IEC subpopulations. Paneth cells reside in the crypt interspersed between ISCs and secrete antimicrobial peptides (AMPs) such as lysozyme, as well as growth factors to support the stem cell niche. Goblet cells are critical for generating mucins, which form the principal component of the protective physicochemical mucous barrier between the lumen and IECs. Enteroendocrine cells (EECs) secrete hormones in response to luminal conditions, contributing to gut mobility, nutrient absorption, satiety, and other effects. Finally, tuft cells have a chemosensory role and can secrete IL-25 to initiate type 2 innate immune responses, ultimately resulting in the expulsion of invasive helminths and protozoan parasites from the intestinal lumen [68].
Intestinal barrier permeability is also a key factor determining intestinal homeostasis and affects the development, pathogenicity and severity of IBD [69]. Maintenance of this barrier is controlled largely by intercellular junctional complexes (and their associated transmembrane proteins). This consists of adherens junctions (catenins, cadherins), tight junctions (ZO-1, claudin-2, occludins, Junctional Adhesion Molecules), and desmosomes (desmoglein, desmocollins, etc.). Junctional complexes regulate intercellular contact between adjacent IECs, sealing the intercellular space to protect the host from potentially harmful luminal agents, whilst being selectively permeable to water, nutrients, and electrolytes [70,71].
Structural support for IECs is provided by the underlying stromal cells that can also secrete factors to regulate IECs in a paracrine fashion. The relationship between IECs and the stroma is also bi-directional, where IECs can also, in turn, regulate stromal activity. Immune cells are also present in the mucosa as intraepithelial lymphocytes (IELs) residing between adjacent IECs, as well as the presence of both the myeloid and lymphoid immune compartments in the stroma. These three branches, IECs, stroma and immune cell-mediated effects, together with the microbiome, form a system that enables the intestine to perform its digestive and absorptive functions whilst maintaining intestinal barrier integrity and immunity against the pathogenic or noxious agents they might be exposed to in the lumen.
While the symptoms of UC and CD are similar (bloody stool, weight loss, abdominal pain), the diseases are characterised by several different features. UC is localised to the colon and rectum, and inflammation is restrained to the mucosal layer, resulting in an ulcerated appearance of the intestinal wall with complications that can include toxic megacolon. The UC inflammatory cytokine signature consists of those involved in the immune pathways for Th2 (IL-5, IL-6, IL-13, IL-33 and TNF), Th9 (IL-9, IL-33), and Th17 (IL-1β, IL-6, IL-17, IL-23, (tumour necrosis factor) TNF) response [72]. In contrast, CD can affect any part of the gastrointestinal tract from the mouth to the rectum in a non-contiguous manner. Inflammation is transmural and can result in the intestinal wall having a ‘cobblestone' appearance with fistulas as likely complications. CD is considered to be characterised to have a Th1 (IL-6, IL-12, IL-18, INFγ, TNF) as well as a Th17 response [72]. Microbiota appears to have less diversity for both UC and CD patients compared with healthy subjects, although dysbiosis is reportedly greater in CD than UC [73–75]. Different genetic risk variants and likely further heterogeneity even within these two IBD entities [76] underscore aetiological differences that may drive the development of these different features. Interestingly, from GWAS alone, autophagy gene polymorphisms appear to be more strongly linked to CD.
Xenophagy in IECs is an innate defence mechanism against pathogens and pathobionts
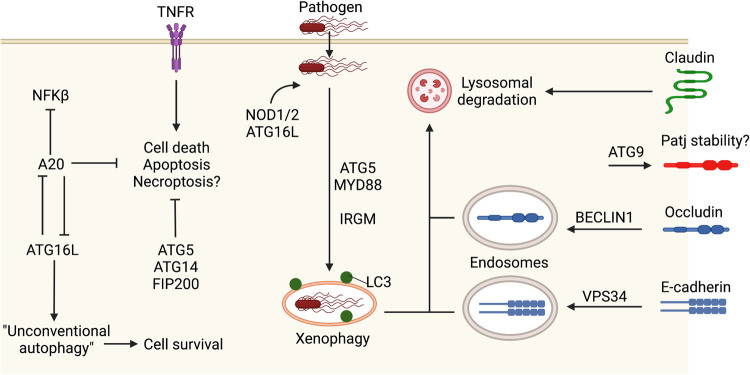
Perhaps the most straightforward mechanism by which autophagy protects the intestinal epithelium is through xenophagy in IECs, or the autophagic degradation of bacteria that have invaded IECs. This can be visualised by the colocalisation of LC3 puncta with bacteria, which can become diminished when autophagy proteins are compromised. Expression of mutant ATG16L harbouring the CD-associated risk polymorphism, ATG16LT300A, in the Caco2 colorectal carcinoma line shows that these cells have impaired capture of Salmonella typhimurium in LC3-positive autophagosomes [77], a bacteria that causes gastroenteritis and is associated with an increased risk of CD or UC [78]. Loss of Atg5 in the intestinal epithelium was also associated with increased S. typhimurium burden in IECs and the extraintestinal dissemination of this pathogenic bacteria [79]. Similarly, the expression of IRGM (which also harbours CD risk variants) is associated with autophagic flux and attenuating the replication of the CD-associated adherent-invasive Escherichia coli (AIEC) in cell lines [80]. The capture of bacteria appears to utilise the intracellular sensors NOD1 and NOD2, which recruit ATG16L to the plasma membrane at the site of bacterial entry (Figure 3). Cells homozygous for a CD-associated NOD2 frameshift mutation fail to recruit ATG16L to the plasma membrane and show impaired bacterial capture in autophagosomes [81]. The major signalling adaptor molecule of the toll-like-receptors (TLRs) family, MyD88, also appears essential in IECs for the xenophagy response to S. typhimurium (Figure 3). In mice deficient for MyD88, a challenge with S. typhimurium does not induce the formation of LC3 puncta [79]. Thus, carrying CD risk variants of autophagy genes that impair clearance of IBD-associated pathogens appears to be an underlying contributor to Crohn's development.
Figure 3. Autophagy genes and cellular processes such as intestinal epithelial barrier function and protection from cell death associated with xenophagy in response to pathogens.
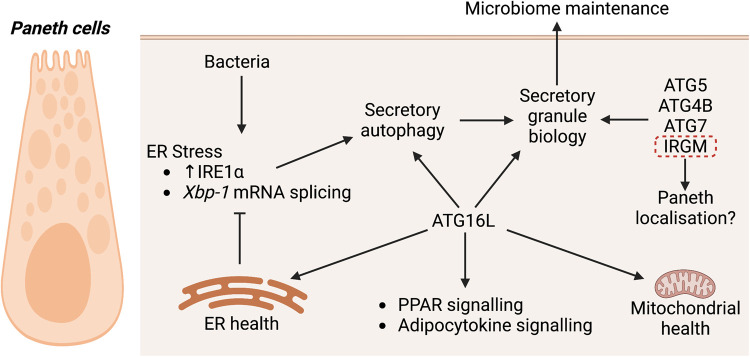
ATG16L and the conjugation machinery maintain Paneth cells
As briefly described above, ATG16L forms part of the multimeric ATG12–ATG5:ATG16L complex that acts as an E3-like enzyme for ATG8 lipidation (Figure 1). The function of the ATG16L protein has been a major focus of studies on intestinal autophagy since its identification as a core autophagy candidate in GWAS studies for IBD. In addition to its role in xenophagy, evidence from multiple studies demonstrates that there is a clear role for ATG16L in maintaining Paneth cell secretory granules, which contain AMPs and immunomodulating proteins, and Paneth cell homeostasis (Figure 4). In mice with an IEC-specific knock-out of Atg16L, Paneth cell numbers were reduced [82], with diminution of both their morphology and size of secretory granules [83], and an accumulation of the endoplasmic reticulum (ER) stress sensor IRE1α [83,84]. Mice with a knock-in of hypomorphic Atg16L showed Paneth cells characterised by degenerating mitochondria and transcriptional up-regulation of the peroxisome proliferator-activated receptor (PPAR) and adipocytokine signalling pathways [85]. Generation of mice bearing a knock-in of the genetic equivalent to the human CD susceptibility allele ATG16LT300A showed that Paneth cells also had reduced amounts of lysozyme and that these mice had an altered microbiota composition [86,87].
Figure 4. Autophagy genes and associated processes implicated in the maintenance of Paneth cells in response to bacterial infection.
Similar phenotypes have been reported for genetic knock-out models of other autophagy conjugation proteins. In mice with IEC-specific Atg5 loss, Paneth cells also had a diminished morphology, were reduced in numbers, and showed dissipating granules [85,88]. These mice were also associated with an altered microbiome [89]. Reduced Paneth granule size was also observed in IEC-specific Atg7 knock-out mice [83,90], and reduced lysozyme staining was observed in the Paneth cells of whole-body Atg4B-deficient mice [91]. Each of these strains of mice with modified autophagy conjugation genes, namely Atg16L, Atg5, Atg7 and Atg4B, showed worsened prognostic outcomes to chemical, physical or bacterial intestinal insults that resulted in the development of IBD-like pathologies. This included lower survival and poorer histological scores of the intestinal tract compared with control mice in response to the intestinal barrier-disrupting agent dextran sodium sulfate (DSS, which gives rise to a UC-like phenotype), the development of spontaneous age-dependent CD-like transmural ileitis, chronic colitis (indeterminate if UC or CD) in response to the opportunistic bacteria Helicobacter hepaticus, increased mortality from enteritis induced by Toxoplasma gondii, and poorer clinical disease scores against murine norovirus which results in an indeterminate, possibly transmural form of enterocolitis [79,82–93]. Notably, as the colon does not have any villi, Paneth cells are generally not found in the colon, although there are analogous Paneth-like cells [94]. This may provide an explanation for why autophagy genes do not exclusively protect the colon, but are protective of the larger gastrointestinal tract.
Combined, the data indicates that on a basal level, ATG16L in Paneth cells protects against ER stress, mitochondrial degeneration, regulates PPAR and adipocytokine signalling, and is important in the biogenesis of the secretory granules in these cells. However, the mechanistic pathways between ATG16L, other conjugation proteins, and these functional outcomes are still not well understood. In IECs, it has been shown that the WD40 domain of ATG16L interacts with the anti-inflammatory protein A20 [95]. This was proposed to be a mutually reciprocal relationship, where ATG16L promoted the lysosomal degradation of A20, and A20 promoted the ubiquitin-mediated degradation of ATG16L. Loss of both these proteins in the mouse intestinal epithelium resulted in spontaneous enterocolitis, marked by increased IL-1β and TNF and thickening of the jejunal wall, features suggestive of CD pathology. These were attributed to up-regulated NFκβ signalling and cell death (which is suppressed by A20) and a lack of ATG16L1 WD40 domain-mediated ‘unconventional' autophagy (Figure 3) [95].
The enhanced levels of intestinal pro-inflammatory cytokines as just described above is a common occurrence under challenging conditions where ATG16L or conjugation protein integrity are additionally compromised, even if these genetic modifications target the IEC compartment rather than immune cells. For instance, increased TNFα and IL-1β levels are observed following LPS-stimulated NFκβ signalling in IEC-specific ATG7-deficient mice [96]. Conjugation protein-deficient intestinal cells appear particularly further sensitised to death triggered by TNF. Intestinal organoids derived from IEC-specific ATG16-deficient mice showed increased necroptosis [82] or apoptosis [92] when treated with TNFα. Similarly, TNF-treated ATG5-deficient intestinal organoids had decreased viability [88]. Thus, conjugation proteins enable IEC survival under duress from inflammatory cytokines and a pathogenic or chemically insulting environment (Figure 3). This cell death likely contributes to, and/or further exacerbates, intestinal inflammatory symptoms.
During bacterial infection which induces ER stress, ATG16L is leveraged to help lysozyme secretion from Paneth cells in another unconventional autophagy process termed ‘secretory' autophagy (Figure 4) [97]. Loss of both Atg16L1 and the ER stress/unfolded protein response (UPR)-mediator Xbp-1 (also a risk locus for both UC and CD [98]) in the intestinal epithelium results in spontaneous severe transmural enteritis [83]. In a DSS-induced colitis model, stimulation of colonocytes with TNFα and NOD ligands promotes IKKα to phosphorylate ATG16L1 (Ser278), stabilising ATG16L1 against degradation, which is associated with protection against IREα-mediated ER stress and activation of caspase-12, which causes a loss of cytoprotective IL-18 [99]. Interestingly, the CD-associated ATG16LT300A mutated protein is more susceptible to caspase-3 or caspase-7 mediated degradation resulting in diminished autophagy [86,100].
Although limited data currently exists, studies on IRGM suggest that non-conjugation autophagy proteins work differently from conjugation proteins in Paneth cells. Mice with a systemic deficiency of Irgm1 have Paneth cells with a different morphology to Atg16L-deficient mice. These Paneth cells can be ectopically located further up the villi and appear swollen, with a smaller granule core and a halo that is electron lucent when viewed with transmission electron microscopy [101]. Mice have down-regulated expression of the Paneth antimicrobial genes Lys (lysozyme) and Defa20 and are susceptible to both DSS-induced colitis and ileitis [101]. The role of IRGM1 has only more recently been elucidated and promotes ULK1 and BECLIN1 association to enhance autophagosome nucleation [63]. IRGM1 can also interact with NOD2 [63], and is reported to interact with the inflammasome NLRP3 and ASL proteins to prevent their oligomerisation, as well as with p62 to mediate p62-dependent selective autophagy of NLRP3 and ASL, thus suppressing IL-1β maturation, pyroptosis and protection against caspase-1 activity in a DSS-colitis model [102]. IRGM is associated with mitophagy [101] and has affinity for the mitochondrial lipid cardiolipin that promotes mitochondrial fission through autophagy [103]. The full pathway between these IRGM interactions and Paneth granules and other biology is yet to be elucidated.
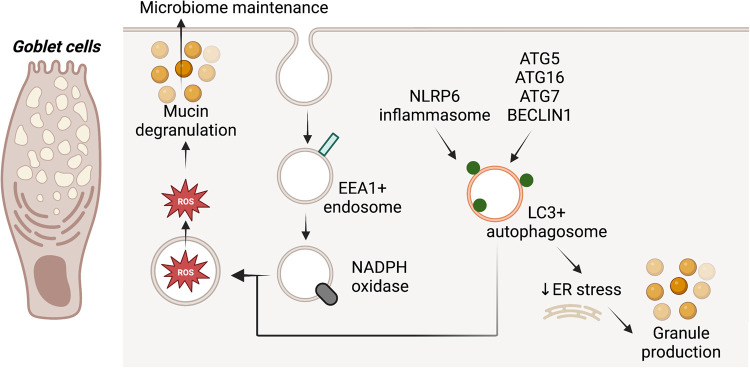
Autophagy maintains goblet cells and promotes their degranulation
Goblet cells are a second secretory cell type that depend on the autophagy machinery for their function. Atg16LT300A mice show colonic and ileal goblet cells with increased mucin area and decreased mucin secretion [86,87]. Similarly, enlarged mucin granules were observed in colonic goblet cells of mice with an intestinal-specific deficiency of Atg5 [104]. In colon epithelial conditional Atg7 knock-out mice, decreased mucin expression and secretion were also observed, together with transcriptional decreases in several antimicrobial and antiparasitic peptides and microbiome changes [105]. Concordantly, Becn1F121A knock-in mice, which express a mutant BECLIN1 with decreased BCL-2 binding that enhances its availability for autophagic flux, have a thicker colonic mucosal layer and reduced ER stress levels [106]. Conversely, Bcl2AAA knock-in mice, which express a mutant BCL-2 that constitutively binds BECLIN1 and reduces its availability for autophagic flux, have a thinner mucosal layer in the colon [106]. As mentioned above, Atg16LT300A mice and IEC-specific Atg5 knock-out mice are sensitised to bacterial or DSS-induced forms of intestinal inflammation, and this is also reflected in Bcl2AAA mice which have an altered microbiome and are sensitive to DSS or AIEC colitis [106].
Common to both Paneth and goblet cells is the protection offered by autophagy against ER-stress that likely contributes to the proper generation of secretory granules, given the importance of the ER in protein production (Figure 5). Mucin-2 (MUC2) production was associated with high constitutive levels of autophagy in colonoids or goblet cell lines, and as such, autophagy was proposed to promote cell survival during production-associated metabolic stress [107]. In addition to granule generation, degranulation may also utilise autophagy proteins. Reactive oxygen species (ROS)-dependent degranulation in goblet cells was reported to require the convergence of LC3-, NADPH- and EEA1-containing compartments (Figure 5) [104]. Secretion also appears to depend on the NLRP signalling pathway-mediated induction of autophagy or formation of LC3-puncta (Figure 5) [108]. It is unclear if similar degranulation mechanisms are also employed by Paneth cells. It is also uncertain if small intestinal goblet cells are moderated in the same manner as colonic goblet cells.
Figure 5. Autophagy genes are associated with goblet cell granule production and degranulation.
The regulation of ISCs by autophagy
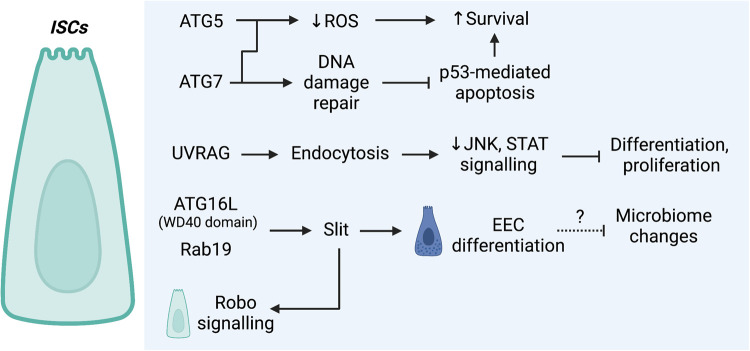
Autophagy appears to be utilised by ISCs to moderate ROS and DNA damage-associated stress to ensure ISC survival (Figure 6). In mice with an intestinal-specific loss of Atg5, numbers of ISCs are reduced and surviving ISCs show increased ROS levels [93]. In mice with an ISC-specific loss of Atg7, a similar lack of ROS clearance was observed as well as inefficient DNA damage repair that was associated with p53-mediated apoptosis of ISCs [109]. Interestingly, an autophagy-independent role in ISC differentiation into EECs via signalling has been reported for ATG16L (Figure 6). It was found in drosophila that the WD40 domain of Atg16L is required to bind Rab19, and these proteins together maintain the production of the ligand Slit, which can activate Robo receptor-mediated signalling in ISCs. Loss of this signalling pathway resulted in a decline in the number of mature EECs and was associated with a spontaneous intestinal inflammatory signature in these flies [110]. Another autophagy-independent mechanism has also been reported for UVRAG in ISCs. A loss of drosophila UVRAG in ISCs results in an accumulation of endocytosed ligands and sustained activation of JNK and STAT signalling [111]. Animals suffer from impaired differentiation and uncontrolled proliferation associated with gut dysfunction and reduced lifespan [111]. As a colorectal cancer-associated tumour suppressor, this suggests a mechanism involving the deregulation of endocytic trafficking functions rather than autophagy functions of UVRAG as a mechanism of colorectal carcinoma development in humans.
Figure 6. In ISCs, autophagy genes are associated with the regulation of ROS and DNA damage-associated stress, and through non-autophagy mechanisms that have been associated with differentiation.
Autophagy genes regulate the intestinal barrier
It is emerging that autophagy has a crucial role in the regulation of the intestinal epithelial barrier, where a loss of autophagy gene expression can lead to alterations in the expression or distribution of intestinal junctional proteins [112–117]. Induction of autophagy via starvation reduces gut epithelial barrier permeability due to the increased lysosomal degradation of claudin-2 in Caco-2 cells (Figure 3) [112]. Similarly, ablation of Atg9 in drosophila results in the aberrant formation of the midgut, accompanied by dramatically enlarged enterocytes as well as barrier dysfunction and reduced fly lifespan. This was attributed to the loss of PALS1-associated tight junction protein (Patj), which is an Atg9-interacting protein (Figure 3) [113].
It is increasingly apparent that many autophagy proteins are multifunctional and that this has physiological consequences. More specifically, members of the Class III PI3K complexes have endosomal trafficking functions that can help maintain intestinal homeostasis through regulation of the intestinal barrier. Loss of Vps34 in zebrafish resulted in disrupted barrier integrity due to defective E-cadherin trafficking to the cell surface (Figure 3), giving rise to epithelial injury, a malformed intestinal adhesion belt, inflammation, and premature death [115]. Additionally, BECLIN1 also plays an autophagy-independent role by associating with the tight junction protein, occludin, and regulating its constitutive endocytosis from the membrane (Figure 3), which subsequently modulates gut barrier permeability [116,117]. As described above, UVRAG in drosophila ISCs is also required for endosomal trafficking of cell surface receptors and ligands and leads to gut dysfunction and shortened lifespan when disrupted, though the impacts on barrier permeability were not directly investigated [111]. It is interesting that whilst ATG14 is a member of the Class III PI3K Complex 1 not typically associated with endosomal trafficking, there is some evidence it can play a role in this process [118]. Accordingly, ATG14 (and FIP200) intestinally deficient mice develop a spontaneous intestinal atrophy phenotype six weeks after birth due to severe loss of villi from an increase in intestinal epithelial apoptosis triggered by TNF [119]. This is unlike the knock-out of conjugation proteins in the intestinal epithelium, which have a milder, stimulus-induced intestinal inflammatory phenotype. This suggests that autophagy genes may have additional functions essential for the regulation of intestinal development and basal homeostasis.
Targeting autophagy to treat IBD
Beyond surgery, approved treatment options for IBD generally act to suppress the inflammatory response associated with UC and CD. These include 5-aminosalicylic acid, corticosteroids and immunomodulators such as methotrexate, thiopurines and calcineurin inhibitors that act on a range of targets to suppress immune cell activation, migration and proliferation via transcriptional and other mechanisms that reduce the production of pro-inflammatory cytokines [120,121]. Such treatments are efficacious in UC and/or CD, however, most are associated with side effects that are often serious and patients can become refractory to treatment. More recently, more selective biologic therapies, including monoclonal antibodies targeting inflammatory cytokines TNF, interleukin IL-12 and IL-23 or leukocyte homing integrin α4β7, and small molecule therapies (e.g. Janus kinase inhibitors, sphingosine-1-phosphate receptor modulators) have been shown to be relatively effective in the management of IBD, however, a significant proportion of patients lose response to these advanced therapies over time [120].
Interestingly, some of the aforementioned immunosuppressive drugs can also modulate (either inhibiting or activating) autophagy, and this ‘secondary' activity may account for at least part of their mechanism of action [122]. Activities such as exercise and caloric restriction that can induce autophagy have shown benefits in IBD models and patients in some studies, however, it is unclear whether autophagy induction itself has any role in these outcomes [123,124]. Moreover, despite the clear involvement of deregulated autophagy in IBD, no treatments that directly target autophagy pathway components have yet to be approved for UC or CD. An early study investigated hydroxychloroquine, an inhibitor of lysosomal degradation, post-surgery in a small number of patients but, perhaps not surprisingly, was ineffective [125] given the more recent knowledge regarding the impact of defective autophagy in IBD. The obvious rationale that induction of autophagy may be therapeutically useful in treating IBD led to the testing of rapamycin (Sirolimus), an inhibitor of mTOR, in CD patients, and was shown to be effective in some cases [126,127]. However, it is unclear if these benefits are due to the known immunosuppressive effects of rapamycin or its induction of autophagy, though a recent study in an IL-10 knock-out model of CD suggested it could decrease intestinal and colonic permeability through its pro-autophagy activity [128]. Otherwise, very few inducers of autophagy have been developed that are highly specific for the components of the autophagy machinery. One exception is the Tat-Beclin peptide derived from the BECLIN1 protein, although its mechanism is via interaction with autophagy regulator Golgi-associated plant pathogenesis related protein (GAPR-1) rather than with BECLIN1 itself [129]. Nevertheless, this peptide induces autophagy in vivo and is effective in disease models where autophagy is impaired (e.g. neurodegenerative disease) [129]. Compounds such as BH3-mimetics which target the BCL-2 proteins that negatively regulate BECLIN1, could also be a therapeutic option, although they may potentially activate apoptotic pathways [130]. Interestingly, a recent report suggested that BH3-mimetics can be developed that selectively target the BCL-2:BECLIN1 interaction over interactions with pro-apoptotic proteins [131], whilst another study showed cell-type selective induction of autophagy (albeit autophagic death) versus apoptosis in response to BH3-mimetics [132]. However, neither BH3-mimetics nor Tat-Beclin is yet to be investigated in IBD models. One ever-present consideration, however, is whether such an approach will work in the context of mutations in autophagy genes such as ATG16L where it may not be possible to (re-)activate the pathway. Such issues may eventually be negated through the use of gene editing approaches which are starting to be investigated in models of IBD [133,134], however, there are many significant hurdles and associated risks that would need to be surmounted before these techniques could be applied in patients.
Concluding remarks
IBD is a multifactorial disease in which deregulated autophagy has been established as one of the possible factors contributing to disease aetiology. The physiological roles of autophagy in the intestine are diverse. It mediates IEC xenophagy, maintains the intestinal barrier (attributed at least in part to emerging roles of key core proteins in endosomal trafficking), and promotes cell survival. In Paneth and goblet cells, the presence of autophagy is required for proper secretory granule biogenesis and impacts granule release. Autophagy proteins are important for the regulation of ROS levels and even have unconventional roles in cell signalling that regulate proliferation and differentiation. Importantly, although not discussed in detail in this manuscript, autophagy regulates myeloid and lymphoid intestinal immune populations to regulate a plethora of functions including cytokine secretion, bacterial clearance, antigen processing, and cell survival, where a loss of autophagy protein integrity is also associated with susceptibility to DSS-induced colitis in mice. Little has been published on how autophagy proteins function in non-immune cells of the intestinal stroma, however recently, it was shown that ATG5 or FIP200 was essential in PDGFRα-positive mesenchymal stem cells in order to generate Wnt ligands necessary for maintaining the stem cell niche [135]. Loss of these proteins in these cells resulted in intestinal epithelial apoptosis, villi blunting and the rapid and fatal deterioration of mice [135], highlighting the critical importance of these proteins in this compartment. This is an area where further investigation is warranted. Similarly, research into the role of autophagy in tuft cells and M cells currently has not been reported. Given the known links between autophagy and secretion, and autophagy and antigen processing, it is likely that autophagy will play a role in the biology of these two cell types.
Despite its multifactorial pathophysiology, dual therapy with current treatments for IBD is relatively novel [136]. Although targeting deregulated autophagy, such as through anti-TNF, is considered effective at inducing and maintaining clinical remission, agents that directly promote autophagy have yet to be clinically tested as monotherapy or in combination with existing therapies in IBD. A greater understanding of the molecular pathways of autophagy in intestinal physiology may provide profound insight into innovative drug development and precision-based strategies in IBD.
Perspectives
Inflammatory bowel disease (IBD) is a complex disease that impacts millions of people globally. Promising treatments do exist, but patients frequently relapse and there is a poor understanding of the specific mechanisms in disease aetiology.
Mutations in autophagy genes are associated with IBD and deregulated autophagy is strongly associated with the loss of intestinal homeostasis due to the role of autophagy proteins in IECs, immune cells and stromal cells.
Multifunctional autophagy proteins have an emerging role in IEC health through their endosomal trafficking roles but have had little investigation so far. Elucidating how autophagy proteins contribute to the maintenance of intestinal cell subpopulations and intestinal physiology offers insights into how we can potentially treat IBD patients, particularly those harbouring autophagy gene mutations.
Acknowledgements
Figures in this article have been created with Biorender.com.
Abbreviations
- AIEC
adherent-invasive Escherichia coli
- AMBRA1
activating molecule in BECN1-regulated autophagy protein 1
- AMPK
AMP-activated protein kinase
- AMPs
antimicrobial peptides
- ATG
autophagy-related protein
- BCL-2
B cell lymphoma 2
- CD
Crohn's disease
- CMA
chaperone-mediated autophagy
- DFCP-1
double FYVE-containing protein-1
- DSS
dextran sodium sulfate
- EEA1
early endosome antigen 1
- EEC
enteroendocrine cell
- EPG5
ectopic P-granules 5 autophagy tethering factor
- ER
endoplasmic reticulum
- FIP200
focal adhesion kinase family interacting protein of 200 kDa
- GABARAP
gamma-aminobutyric acid type A receptor-associated protein
- GAPR-1
Golgi-associated plant pathogenesis related protein
- GPR65
G protein-coupled receptor 65
- GWAS
genome-wide association studies
- HOPS
homotypic fusion and protein sorting
- IBD
inflammatory bowel disease
- IEC
intestinal epithelial cell
- IEL
intra-epithelial lymphocyte
- IFN
interferin
- IRE1α
inositol-requiring enzyme 1α
- IRGM
immunity-related GTPase family M protein
- ISC
intestinal stem cell
- LC3
microtubule-associated protein 1A/1B-light chain 3
- LPS
lipopolysaccharide
- LRRK
leucine-rich repeat serine/threonine-protein kinase 1
- MTMR
myotubularin phosphatase
- mTOR
mammalian target of rapamycin
- MUC2 mucin 2
oligomeric mucus/gel-forming
- NADPH
nicotinamide adenine dinucleotide phosphate
- NDP52
nuclear domain 10 protein 52
- NFκB
nuclear factor kappa B
- NLRP
NOD-, LRR- and pyrin domain-containing protein
- NOD
nucleotide-binding oligomerisation domain-containing protein
- NRBF2
nuclear receptor binding factor 2
- p62
sequestosome-1
- PDGFRα
platelet-derived growth factor receptor α
- PE
phosphatidylethanolamine
- PI3K
phosphoinositide 3-kinase
- PLEKHM1
pleckstrin homology domain containing protein family member 1
- PPAR
peroxisome proliferator-activated receptor
- PtdIns
phosphatidylinositol
- PTPN2
tyrosine-protein phosphatase non-receptor type 2
- ROS
reactive oxygen species
- SNAP29
synaptosome-associated protein 29
- SNARE
soluble N-ethylmaleimide-sensitive factor attachment protein receptor
- STX
syntaxin
- TNF
tumour necrosis factor
- TOR
target of rapamycin
- UC
ulcerative colitis
- ULK
UNC-51-like kinase
- UVRAG
UV radiation resistance-associated gene product
- VAMP
vesicle-associated membrane protein
- VPS
vacuolar protein sorting-associated gene product
- WIPI
WD-repeat protein interacting with phosphoinositides
- XBP-1
X-box-binding protein-1
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
W.D.F. and E.F.L. are supported by the Australian Research Council (ARC) (Discovery Project Grant DP190102612). E.F.L. is a fellowship recipient from the Victorian Cancer Agency (Mid-Career Fellowship MCRF19045).
Open Access
Open access for this article was enabled by the participation of La Trobe University in an all-inclusive Read & Publish agreement with Portland Press and the Biochemical Society under a transformative agreement with CAUL.
Author Contributions
S.T., J.J., W.D.F. and E.F.L. conceptualised the idea for the review, as well as wrote and reviewed the manuscript. All authors have read and agreed to the published version of the manuscript.
References
- 1.Hodson, R. (2016) Inflammatory bowel disease. Nature 540, S97 10.1038/540S97a [DOI] [PubMed] [Google Scholar]
- 2.Alatab, S., Sepanlou, S.G., Ikuta, K., Vahedi, H., Bisignano, C., Safiri, S.et al. (2020) The global, regional, and national burden of inflammatory bowel disease in 195 countries and territories, 1990-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet Gastroenterol. Hepatol. 5, 17–30 10.1016/S2468-1253(19)30333-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Narula, N., Wong, E.C.L., Dehghan, M., Mente, A., Rangarajan, S., Lanas, F.et al. (2021) Association of ultra-processed food intake with risk of inflammatory bowel disease: prospective cohort study. BMJ 374, n1554 10.1136/bmj.n1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Agrawal, M., Sabino, J., Frias-Gomes, C., Hillenbrand, C.M., Soudant, C., Axelrad, J.E.et al. (2021) Early life exposures and the risk of inflammatory bowel disease: systematic review and meta-analyses. EClinicalMedicine 36, 100884 10.1016/j.eclinm.2021.100884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singh, N. and Bernstein, C.N. (2022) Environmental risk factors for inflammatory bowel disease. United European Gastroenterol. J. 10, 1047–1053 10.1002/ueg2.12319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levine, A., Sigall Boneh, R. and Wine, E. (2018) Evolving role of diet in the pathogenesis and treatment of inflammatory bowel diseases. Gut 67, 1726–1738 10.1136/gutjnl-2017-315866 [DOI] [PubMed] [Google Scholar]
- 7.Eisenstein, M. (2016) Biology: a slow-motion epidemic. Nature 540, S98–S99 10.1038/540S98a [DOI] [PubMed] [Google Scholar]
- 8.Bryant, R.V., Costello, S.P., Schoeman, S., Sathananthan, D., Knight, E., Lau, S.Y.et al. (2018) Limited uptake of ulcerative colitis “treat-to-target” recommendations in real-world practice. J. Gastroenterol. Hepatol. 33, 599–607 10.1111/jgh.13923 [DOI] [PubMed] [Google Scholar]
- 9.Volk, N. and Siegel, C.A. (2018) Defining failure of medical therapy for inflammatory bowel disease. Inflamm. Bowel Dis. 25, 74–77 10.1093/ibd/izy238 [DOI] [PubMed] [Google Scholar]
- 10.Lamb, C.A., Kennedy, N.A., Raine, T., Hendy, P.A., Smith, P.J., Limdi, J.K.et al. (2019) British society of gastroenterology consensus guidelines on the management of inflammatory bowel disease in adults. Gut 68, s1–s106 10.1136/gutjnl-2019-318484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hampe, J., Franke, A., Rosenstiel, P., Till, A., Teuber, M., Huse, K.et al. (2007) A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat. Genet. 39, 207–211 10.1038/ng1954 [DOI] [PubMed] [Google Scholar]
- 12.Prescott, N.J., Fisher, S.A., Franke, A., Hampe, J., Onnie, C.M., Soars, D.et al. (2007) A nonsynonymous SNP in ATG16L1 predisposes to ileal Crohn's disease and is independent of CARD15 and IBD5. Gastroenterology 132, 1665–1671 10.1053/j.gastro.2007.03.034 [DOI] [PubMed] [Google Scholar]
- 13.Rioux, J.D., Xavier, R.J., Taylor, K.D., Silverberg, M.S., Goyette, P., Huett, A.et al. (2007) Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat. Genet. 39, 596–604 10.1038/ng2032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burton, P.R., Clayton, D.G., Cardon, L.R., Craddock, N., Deloukas, P., Duncanson, A.et al. (2007) Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447, 661–678 10.1038/nature05911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lahiri, A., Hedl, M. and Abraham, C. (2015) MTMR3 risk allele enhances innate receptor-induced signaling and cytokines by decreasing autophagy and increasing caspase-1 activation. Proc. Natl Acad. Sci. U.S.A. 112, 10461–10466 10.1073/pnas.1501752112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lassen, K.G., McKenzie, C.I., Mari, M., Murano, T., Begun, J., Baxt, L.A.et al. (2016) Genetic coding variant in GPR65 alters lysosomal pH and links lysosomal dysfunction with colitis risk. Immunity 44, 1392–1405 10.1016/j.immuni.2016.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parkes, M., Barrett, J.C., Prescott, N.J., Tremelling, M., Anderson, C.A., Fisher, S.A.et al. (2007) Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn's disease susceptibility. Nat. Genet. 39, 830–832 10.1038/ng2061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morgan, A.R., Lam, W.J., Han, D.Y., Fraser, A.G. and Ferguson, L.R. (2012) Association analysis of ULK1 with Crohn's disease in a New Zealand population. Gastroenterol. Res. Pract. 2012, 715309 10.1155/2012/715309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henckaerts, L., Cleynen, I., Brinar, M., John, J.M., Van Steen, K., Rutgeerts, P.et al. (2011) Genetic variation in the autophagy gene ULK1 and risk of Crohn's disease. Inflamm. Bowel Dis. 17, 1392–1397 10.1002/ibd.21486 [DOI] [PubMed] [Google Scholar]
- 20.Ellinghaus, D., Zhang, H., Zeissig, S., Lipinski, S., Till, A., Jiang, T.et al. (2013) Association between variants of PRDM1 and NDP52 and Crohn's disease, based on exome sequencing and functional studies. Gastroenterology 145, 339–347 10.1053/j.gastro.2013.04.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haq, S., Grondin, J., Banskota, S. and Khan, W.I. (2019) Autophagy: roles in intestinal mucosal homeostasis and inflammation. J. Biomed. Sci. 26, 19 10.1186/s12929-019-0512-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Larabi, A., Barnich, N. and Nguyen, H.T.T. (2020) New insights into the interplay between autophagy, gut microbiota and inflammatory responses in IBD. Autophagy 16, 38–51 10.1080/15548627.2019.1635384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Galluzzi, L., Baehrecke, E.H., Ballabio, A., Boya, P., Bravo-San Pedro, J.M., Cecconi, F.et al. (2017) Molecular definitions of autophagy and related processes. EMBO J. 36, 1811–1836 10.15252/embj.201796697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim, J., Kundu, M., Viollet, B. and Guan, K.-L. (2011) AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 13, 132–141 10.1038/ncb2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ganley, I.G., Lam du, H., Wang, J., Ding, X., Chen, S. and Jiang, X. (2009) ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J. Biol. Chem. 284, 12297–12305 10.1074/jbc.M900573200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hosokawa, N., Hara, T., Kaizuka, T., Kishi, C., Takamura, A., Miura, Y.et al. (2009) Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol. Biol. Cell 20, 1981–1991 10.1091/mbc.e08-12-1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jung, C.H., Jun, C.B., Ro, S.H., Kim, Y.M., Otto, N.M., Cao, J.et al. (2009) ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol. Biol. Cell 20, 1992–2003 10.1091/mbc.e08-12-1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Egan, D.F., Chun, M.G., Vamos, M., Zou, H., Rong, J., Miller, C.J.et al. (2015) Small molecule inhibition of the autophagy kinase ULK1 and identification of ULK1 substrates. Mol. Cell 59, 285–297 10.1016/j.molcel.2015.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mercer, T.J., Ohashi, Y., Boeing, S., Jefferies, H.B.J., De Tito, S., Flynn, H.et al. (2021) Phosphoproteomic identification of ULK substrates reveals VPS15-dependent ULK/VPS34 interplay in the regulation of autophagy. EMBO J. 40, e105985 10.15252/embj.2020105985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wold, M.S., Lim, J., Lachance, V., Deng, Z. and Yue, Z. (2016) ULK1-mediated phosphorylation of ATG14 promotes autophagy and is impaired in Huntington's disease models. Mol. Neurodegener. 11, 76 10.1186/s13024-016-0141-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Russell, R.C., Tian, Y., Yuan, H., Park, H.W., Chang, Y.Y., Kim, J.et al. (2013) ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat. Cell Biol. 15, 741–750 10.1038/ncb2757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park, J.M., Jung, C.H., Seo, M., Otto, N.M., Grunwald, D., Kim, K.H.et al. (2016) The ULK1 complex mediates MTORC1 signaling to the autophagy initiation machinery via binding and phosphorylating ATG14. Autophagy 12, 547–564 10.1080/15548627.2016.1140293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou, C., Ma, K., Gao, R., Mu, C., Chen, L., Liu, Q.et al. (2017) Regulation of mATG9 trafficking by Src- and ULK1-mediated phosphorylation in basal and starvation-induced autophagy. Cell Res. 27, 184–201 10.1038/cr.2016.146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Itakura, E., Kishi, C., Inoue, K. and Mizushima, N. (2008) Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol. Biol. Cell 19, 5360–5372 10.1091/mbc.e08-01-0080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matsunaga, K., Saitoh, T., Tabata, K., Omori, H., Satoh, T., Kurotori, N.et al. (2009) Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat. Cell Biol. 11, 385–396 10.1038/ncb1846 [DOI] [PubMed] [Google Scholar]
- 36.Sun, Q., Fan, W., Chen, K., Ding, X., Chen, S. and Zhong, Q. (2008) Identification of Barkor as a mammalian autophagy-specific factor for Beclin 1 and class III phosphatidylinositol 3-kinase. Proc. Natl Acad. Sci. U.S.A. 105, 19211–19216 10.1073/pnas.0810452105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhong, Y., Wang, Q.J., Li, X., Yan, Y., Backer, J.M., Chait, B.T.et al. (2009) Distinct regulation of autophagic activity by Atg14L and rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat. Cell Biol. 11, 468–476 10.1038/ncb1854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fan, W., Nassiri, A. and Zhong, Q. (2011) Autophagosome targeting and membrane curvature sensing by Barkor/Atg14(L). Proc. Natl Acad. Sci. U.S.A. 108, 7769–7774 10.1073/pnas.1016472108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liang, C., Feng, P., Ku, B., Dotan, I., Canaani, D., Oh, B.H.et al. (2006) Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat. Cell Biol. 8, 688–699 10.1038/ncb1426 [DOI] [PubMed] [Google Scholar]
- 40.Dooley, H.C., Razi, M., Polson, H.E., Girardin, S.E., Wilson, M.I. and Tooze, S.A. (2014) WIPI2 links LC3 conjugation with PI3P, autophagosome formation, and pathogen clearance by recruiting Atg12-5-16L1. Mol. Cell 55, 238–252 10.1016/j.molcel.2014.05.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nähse, V., Raiborg, C., Tan, K.W., Mørk, S., Torgersen, M.L., Wenzel, E.M.et al. (2022) ATPase activity of DFCP1 controls selective autophagy. bioRxiv 10.1101/2022.02.24.481614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maeda, S., Otomo, C. and Otomo, T. (2019) The autophagic membrane tether ATG2A transfers lipids between membranes. eLife 8, e45777 10.7554/eLife.45777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Osawa, T., Kotani, T., Kawaoka, T., Hirata, E., Suzuki, K., Nakatogawa, H.et al. (2019) Atg2 mediates direct lipid transfer between membranes for autophagosome formation. Nat. Struct. Mol. Biol. 26, 281–288 10.1038/s41594-019-0203-4 [DOI] [PubMed] [Google Scholar]
- 44.Valverde, D.P., Yu, S., Boggavarapu, V., Kumar, N., Lees, J.A., Walz, T.et al. (2019) ATG2 transports lipids to promote autophagosome biogenesis. J. Cell Biol. 218, 1787–1798 10.1083/jcb.201811139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maeda, S., Yamamoto, H., Kinch, L.N., Garza, C.M., Takahashi, S., Otomo, C.et al. (2020) Structure, lipid scrambling activity and role in autophagosome formation of ATG9A. Nat. Struct. Mol. Biol. 27, 1194–1201 10.1038/s41594-020-00520-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nishimura, T., Kaizuka, T., Cadwell, K., Sahani, M.H., Saitoh, T., Akira, S.et al. (2013) FIP200 regulates targeting of Atg16L1 to the isolation membrane. EMBO Rep. 14, 284–291 10.1038/embor.2013.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gammoh, N., Florey, O., Overholtzer, M. and Jiang, X. (2013) Interaction between FIP200 and ATG16L1 distinguishes ULK1 complex-dependent and -independent autophagy. Nat. Struct. Mol. Biol. 20, 144–149 10.1038/nsmb.2475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Turco, E., Witt, M., Abert, C., Bock-Bierbaum, T., Su, M.Y., Trapannone, R.et al. (2019) FIP200 claw domain binding to p62 promotes autophagosome formation at ubiquitin condensates. Mol. Cell 74, 330–346.e311 10.1016/j.molcel.2019.01.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shi, X., Chang, C., Yokom, A.L., Jensen, L.E. and Hurley, J.H. (2020) The autophagy adaptor NDP52 and the FIP200 coiled-coil allosterically activate ULK1 complex membrane recruitment. eLife 9, e59099 10.7554/eLife.59099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Polson, H.E., de Lartigue, J., Rigden, D.J., Reedijk, M., Urbé, S., Clague, M.J.et al. (2010) Mammalian Atg18 (WIPI2) localizes to omegasome-anchored phagophores and positively regulates LC3 lipidation. Autophagy 6, 506–522 10.4161/auto.6.4.11863 [DOI] [PubMed] [Google Scholar]
- 51.Fracchiolla, D., Chang, C., Hurley, J.H. and Martens, S. (2020) A PI3K-WIPI2 positive feedback loop allosterically activates LC3 lipidation in autophagy. J. Cell Biol. 219, e201912098 10.1083/jcb.201912098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Strong, L.M., Chang, C., Riley, J.F., Boecker, C.A., Flower, T.G., Buffalo, C.Z.et al. (2021) Structural basis for membrane recruitment of ATG16L1 by WIPI2 in autophagy. eLife 10, e70372 10.7554/eLife.70372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fujita, N., Itoh, T., Omori, H., Fukuda, M., Noda, T. and Yoshimori, T. (2008) The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol. Biol. Cell 19, 2092–2100 10.1091/mbc.e07-12-1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mizushima, N. (2020) The ATG conjugation systems in autophagy. Curr. Opin. Cell Biol. 63, 1–10 10.1016/j.ceb.2019.12.001 [DOI] [PubMed] [Google Scholar]
- 55.Kirkin, V., Lamark, T., Sou, Y.S., Bjørkøy, G., Nunn, J.L., Bruun, J.A.et al. (2009) A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol. Cell 33, 505–516 10.1016/j.molcel.2009.01.020 [DOI] [PubMed] [Google Scholar]
- 56.Gatica, D., Lahiri, V. and Klionsky, D.J. (2018) Cargo recognition and degradation by selective autophagy. Nat. Cell Biol. 20, 233–242 10.1038/s41556-018-0037-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reggiori, F. and Ungermann, C. (2017) Autophagosome maturation and fusion. J. Mol. Biol. 429, 486–496 10.1016/j.jmb.2017.01.002 [DOI] [PubMed] [Google Scholar]
- 58.Zhao, Y.G., Codogno, P. and Zhang, H. (2021) Machinery, regulation and pathophysiological implications of autophagosome maturation. Nat. Rev. Mol. Cell Biol. 22, 733–750 10.1038/s41580-021-00392-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jaber, N., Mohd-Naim, N., Wang, Z., DeLeon, J.L., Kim, S., Zhong, H.et al. (2016) Vps34 regulates Rab7 and late endocytic trafficking through recruitment of the GTPase-activating protein Armus. J. Cell Sci. 129, 4424–4435 10.1242/jcs.192260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cai, C.Z., Yang, C., Zhuang, X.X., Yuan, N.N., Wu, M.Y., Tan, J.Q.et al. (2021) NRBF2 is a RAB7 effector required for autophagosome maturation and mediates the association of APP-CTFs with active form of RAB7 for degradation. Autophagy 17, 1112–1130 10.1080/15548627.2020.1760623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pankiv, S., Alemu, E.A., Brech, A., Bruun, J.A., Lamark, T., Overvatn, A.et al. (2010) FYCO1 is a Rab7 effector that binds to LC3 and PI3P to mediate microtubule plus end-directed vesicle transport. J. Cell Biol. 188, 253–269 10.1083/jcb.200907015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Licheva, M., Raman, B., Kraft, C. and Reggiori, F. (2022) Phosphoregulation of the autophagy machinery by kinases and phosphatases. Autophagy 18, 104–123 10.1080/15548627.2021.1909407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chauhan, S., Mandell Michael, A. and Deretic, V. (2015) IRGM governs the core autophagy machinery to conduct antimicrobial defense. Mol. Cell 58, 507–521 10.1016/j.molcel.2015.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tran, S., Fairlie, W.D. and Lee, E.F. (2021) BECLIN1: protein structure, function and regulation. Cells 10, 1522 10.3390/cells10061522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Madureira, M., Connor-Robson, N. and Wade-Martins, R. (2020) LRRK2: autophagy and lysosomal activity. Front. Neurosci. 14, 498 10.3389/fnins.2020.00498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Boecker, C.A., Goldsmith, J., Dou, D., Cajka, G.G. and Holzbaur, E.L.F. (2021) Increased LRRK2 kinase activity alters neuronal autophagy by disrupting the axonal transport of autophagosomes. Curr. Biol. 31, 2140–2154.e2146 10.1016/j.cub.2021.02.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Spalinger, M.R., McCole, D.F., Rogler, G. and Scharl, M. (2016) Protein tyrosine phosphatase non-receptor type 2 and inflammatory bowel disease. World J. Gastroenterol. 22, 1034–1044 10.3748/wjg.v22.i3.1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hendel, S.K., Kellermann, L., Hausmann, A., Bindslev, N., Jensen, K.B. and Nielsen, O.H. (2022) Tuft cells and their role in intestinal diseases. Front. Immunol. 13, 822867 10.3389/fimmu.2022.822867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pastorelli, L., De Salvo, C., Mercado, J., Vecchi, M. and Pizarro, T. (2013) Central role of the gut epithelial barrier in the pathogenesis of chronic intestinal inflammation: lessons learned from animal models and human genetics. Front. Immunol. 4, 280 10.3389/fimmu.2013.00280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stolfi, C., Maresca, C., Monteleone, G. and Laudisi, F. (2022) Implication of intestinal barrier dysfunction in gut dysbiosis and diseases. Biomedicines 10, 289. 10.3390/biomedicines10020289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schneeberger, E.E. and Lynch, R.D. (1992) Structure, function, and regulation of cellular tight junctions. Am. J. Physiol. 262, L647–L661 10.1152/ajplung.1992.262.6.L647 [DOI] [PubMed] [Google Scholar]
- 72.Neurath, M.F. (2019) Targeting immune cell circuits and trafficking in inflammatory bowel disease. Nat. Immunol. 20, 970–979 10.1038/s41590-019-0415-0 [DOI] [PubMed] [Google Scholar]
- 73.Zhu, S., Han, M., Liu, S., Fan, L., Shi, H. and Li, P. (2022) Composition and diverse differences of intestinal microbiota in ulcerative colitis patients. Front. Cell Infect. Microbiol. 12, 953962 10.3389/fcimb.2022.953962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pascal, V., Pozuelo, M., Borruel, N., Casellas, F., Campos, D., Santiago, A.et al. (2017) A microbial signature for Crohn's disease. Gut 66, 813–822 10.1136/gutjnl-2016-313235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Serrano-Gómez, G., Mayorga, L., Oyarzun, I., Roca, J., Borruel, N., Casellas, F.et al. (2021) Dysbiosis and relapse-related microbiome in inflammatory bowel disease: a shotgun metagenomic approach. Comput. Struct. Biotechnol. J. 19, 6481–6489 10.1016/j.csbj.2021.11.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chang, J.T. (2020) Pathophysiology of inflammatory bowel diseases. N. Engl. J. Med. 383, 2652–2664 10.1056/NEJMra2002697 [DOI] [PubMed] [Google Scholar]
- 77.Kuballa, P., Huett, A., Rioux, J.D., Daly, M.J. and Xavier, R.J. (2008) Impaired autophagy of an intracellular pathogen induced by a Crohn's disease associated ATG16L1 variant. PLoS ONE 3, e3391 10.1371/journal.pone.0003391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schultz, B.M., Paduro, C.A., Salazar, G.A., Salazar-Echegarai, F.J., Sebastián, V.P., Riedel, C.A.et al. (2017) A potential role of Salmonella infection in the onset of inflammatory bowel diseases. Front. Immunol. 8, 191 10.3389/fimmu.2017.00191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Benjamin, J.L., Sumpter, Jr, R., Levine, B. and Hooper, L.V. (2013) Intestinal epithelial autophagy is essential for host defense against invasive bacteria. Cell Host Microbe 13, 723–734 10.1016/j.chom.2013.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Brest, P., Lapaquette, P., Souidi, M., Lebrigand, K., Cesaro, A., Vouret-Craviari, V.et al. (2011) A synonymous variant in IRGM alters a binding site for miR-196 and causes deregulation of IRGM-dependent xenophagy in Crohn's disease. Nat. Genet. 43, 242–245 10.1038/ng.762 [DOI] [PubMed] [Google Scholar]
- 81.Travassos, L.H., Carneiro, L.A.M., Ramjeet, M., Hussey, S., Kim, Y.-G., Magalhães, J.G.et al. (2010) Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat. Immunol. 11, 55–62 10.1038/ni.1823 [DOI] [PubMed] [Google Scholar]
- 82.Matsuzawa-Ishimoto, Y., Shono, Y., Gomez, L.E., Hubbard-Lucey, V.M., Cammer, M., Neil, J.et al. (2017) Autophagy protein ATG16L1 prevents necroptosis in the intestinal epithelium. J. Exp. Med. 214, 3687–3705 10.1084/jem.20170558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Adolph, T.E., Tomczak, M.F., Niederreiter, L., Ko, H.J., Böck, J., Martinez-Naves, E.et al. (2013) Paneth cells as a site of origin for intestinal inflammation. Nature 503, 272–276 10.1038/nature12599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tschurtschenthaler, M., Adolph, T.E., Ashcroft, J.W., Niederreiter, L., Bharti, R., Saveljeva, S.et al. (2017) Defective ATG16L1-mediated removal of IRE1α drives Crohn's disease-like ileitis. J. Exp. Med. 214, 401–422 10.1084/jem.20160791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cadwell, K., Liu, J.Y., Brown, S.L., Miyoshi, H., Loh, J., Lennerz, J.K.et al. (2008) A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature 456, 259–263 10.1038/nature07416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lassen, K.G., Kuballa, P., Conway, K.L., Patel, K.K., Becker, C.E., Peloquin, J.M.et al. (2014) Atg16l1 T300A variant decreases selective autophagy resulting in altered cytokine signaling and decreased antibacterial defense. Proc. Natl Acad. Sci. U.S.A. 111, 7741–7746 10.1073/pnas.1407001111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu, H., Gao, P., Jia, B., Lu, N., Zhu, B. and Zhang, F. (2021) IBD-associated Atg16L1T300A polymorphism regulates commensal microbiota of the intestine. Front. Immunol. 12, 772189 10.3389/fimmu.2021.772189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Burger, E., Araujo, A., López-Yglesias, A., Rajala, M.W., Geng, L., Levine, B.et al. (2018) Loss of paneth cell autophagy causes acute susceptibility to Toxoplasma gondii-mediated inflammation. Cell Host Microbe 23, 177–190.e174 10.1016/j.chom.2018.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yang, L., Liu, C., Zhao, W., He, C., Ding, J., Dai, R.et al. (2018) Impaired autophagy in intestinal epithelial cells alters gut microbiota and host immune responses. Appl. Environ. Microbiol. 84, e00880-18 10.1128/aem.00880-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wittkopf, N., Günther, C., Martini, E., Waldner, M., Amann, K.U., Neurath, M.F.et al. (2012) Lack of intestinal epithelial atg7 affects paneth cell granule formation but does not compromise immune homeostasis in the gut. Clin. Dev. Immunol. 2012, 278059 10.1155/2012/278059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cabrera, S., Fernández, A.F., Mariño, G., Aguirre, A., Suárez, M.F., Español, Y.et al. (2013) ATG4B/autophagin-1 regulates intestinal homeostasis and protects mice from experimental colitis. Autophagy 9, 1188–1200 10.4161/auto.24797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pott, J., Kabat, A.M. and Maloy, K.J. (2018) Intestinal epithelial cell autophagy is required to protect against TNF-induced apoptosis during chronic colitis in mice. Cell Host Microbe 23, 191–202.e194 10.1016/j.chom.2017.12.017 [DOI] [PubMed] [Google Scholar]
- 93.Asano, J., Sato, T., Ichinose, S., Kajita, M., Onai, N., Shimizu, S.et al. (2017) Intrinsic autophagy is required for the maintenance of intestinal stem cells and for irradiation-induced intestinal regeneration. Cell Rep. 20, 1050–1060 10.1016/j.celrep.2017.07.019 [DOI] [PubMed] [Google Scholar]
- 94.Schmitt, M., Schewe, M., Sacchetti, A., Feijtel, D., van de Geer, W.S., Teeuwssen, M.et al. (2018) Paneth cells respond to inflammation and contribute to tissue regeneration by acquiring stem-like features through SCF/c-Kit signaling. Cell Rep. 24, 2312–2328.e2317 10.1016/j.celrep.2018.07.085 [DOI] [PubMed] [Google Scholar]
- 95.Slowicka, K., Serramito-Gómez, I., Boada-Romero, E., Martens, A., Sze, M., Petta, I.et al. (2019) Physical and functional interaction between A20 and ATG16L1-WD40 domain in the control of intestinal homeostasis. Nat. Commun. 10, 1834 10.1038/s41467-019-09667-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Fujishima, Y., Nishiumi, S., Masuda, A., Inoue, J., Nguyen, N.M., Irino, Y.et al. (2011) Autophagy in the intestinal epithelium reduces endotoxin-induced inflammatory responses by inhibiting NF-κB activation. Arch. Biochem. Biophys. 506, 223–235 10.1016/j.abb.2010.12.009 [DOI] [PubMed] [Google Scholar]
- 97.Bel, S., Pendse, M., Wang, Y., Li, Y., Ruhn, K.A., Hassell, B.et al. (2017) Paneth cells secrete lysozyme via secretory autophagy during bacterial infection of the intestine. Science 357, 1047–1052 10.1126/science.aal4677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kaser, A., Lee, A.H., Franke, A., Glickman, J.N., Zeissig, S., Tilg, H.et al. (2008) XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell 134, 743–756 10.1016/j.cell.2008.07.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Diamanti, M.A., Gupta, J., Bennecke, M., De Oliveira, T., Ramakrishnan, M., Braczynski, A.K.et al. (2017) IKKα controls ATG16L1 degradation to prevent ER stress during inflammation. J. Exp. Med. 214, 423–437 10.1084/jem.20161867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Murthy, A., Li, Y., Peng, I., Reichelt, M., Katakam, A.K., Noubade, R.et al. (2014) A Crohn's disease variant in Atg16l1 enhances its degradation by caspase 3. Nature 506, 456–462 10.1038/nature13044 [DOI] [PubMed] [Google Scholar]
- 101.Liu, B., Gulati, A.S., Cantillana, V., Henry, S.C., Schmidt, E.A., Daniell, X.et al. (2013) Irgm1-deficient mice exhibit Paneth cell abnormalities and increased susceptibility to acute intestinal inflammation. Am. J. Physiol. Gastrointest. Liver Physiol. 305, G573–G584 10.1152/ajpgi.00071.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mehto, S., Jena, K.K., Nath, P., Chauhan, S., Kolapalli, S.P., Das, S.K.et al. (2019) The Crohn's disease risk factor IRGM limits NLRP3 inflammasome activation by impeding its assembly and by mediating its selective autophagy. Mol. Cell 73, 429–445.e427 10.1016/j.molcel.2018.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Singh, S.B., Ornatowski, W., Vergne, I., Naylor, J., Delgado, M., Roberts, E.et al. (2010) Human IRGM regulates autophagy and cell-autonomous immunity functions through mitochondria. Nat. Cell Biol. 12, 1154–1165 10.1038/ncb2119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Patel, K.K., Miyoshi, H., Beatty, W.L., Head, R.D., Malvin, N.P., Cadwell, K.et al. (2013) Autophagy proteins control goblet cell function by potentiating reactive oxygen species production. EMBO J. 32, 3130–3144 10.1038/emboj.2013.233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tsuboi, K., Nishitani, M., Takakura, A., Imai, Y., Komatsu, M. and Kawashima, H. (2015) Autophagy protects against colitis by the maintenance of normal gut microflora and secretion of mucus. J. Biol. Chem. 290, 20511–20526 10.1074/jbc.M114.632257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Naama, M., Telpaz, S., Awad, A., Ben-Simon, S., Harshuk-Shabso, S., Modilevsky, S.et al. (2023) Autophagy controls mucus secretion from intestinal goblet cells by alleviating ER stress. Cell Host Microbe 31, 433–446.e4 10.1016/j.chom.2023.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tiwari, S., Begum, S., Moreau, F., Gorman, H. and Chadee, K. (2021) Autophagy is required during high MUC2 mucin biosynthesis in colonic goblet cells to contend metabolic stress. Am. J. Physiol. Gastrointest. Liver Physiol. 321, G489–G499 10.1152/ajpgi.00221.2021 [DOI] [PubMed] [Google Scholar]
- 108.Wlodarska, M., Thaiss Christoph, A., Nowarski, R., Henao-Mejia, J., Zhang, J.-P., Brown Eric, M.et al. (2014) NLRP6 inflammasome orchestrates the colonic host-microbial interface by regulating goblet cell mucus secretion. Cell 156, 1045–1059 10.1016/j.cell.2014.01.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Trentesaux, C., Fraudeau, M., Pitasi, C.L., Lemarchand, J., Jacques, S., Duche, A.et al. (2020) Essential role for autophagy protein ATG7 in the maintenance of intestinal stem cell integrity. Proc. Natl Acad. Sci. U.S.A. 117, 11136–11146 10.1073/pnas.1917174117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Nagy, P., Szatmári, Z., Sándor, G.O., Lippai, M., Hegedűs, K. and Juhász, G. (2017) Drosophila Atg16 promotes enteroendocrine cell differentiation via regulation of intestinal slit/Robo signaling. Development 144, 3990–4001 10.1242/dev.147033 [DOI] [PubMed] [Google Scholar]
- 111.Nagy, P., Kovács, L., Sándor, G.O. and Juhász, G. (2016) Stem-cell-specific endocytic degradation defects lead to intestinal dysplasia in Drosophila. Dis. Model. Mech. 9, 501–512 10.1242/dmm.023416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Nighot, P.K., Hu, C.A. and Ma, T.Y. (2015) Autophagy enhances intestinal epithelial tight junction barrier function by targeting claudin-2 protein degradation. J. Biol. Chem. 290, 7234–7246 10.1074/jbc.M114.597492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wen, J.K., Wang, Y.T., Chan, C.C., Hsieh, C.W., Liao, H.M., Hung, C.C.et al. (2017) Atg9 antagonizes TOR signaling to regulate intestinal cell growth and epithelial homeostasis in Drosophila. eLife 6, e29338 10.7554/eLife.29338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Liu, H., Lou, J., Liu, Y., Liu, Z., Xie, J., Sun, J.et al. (2022) Intestinal epithelial cell autophagy deficiency suppresses inflammation-associated colon tumorigenesis. Mol. Ther. Nucleic Acids 28, 35–46 10.1016/j.omtn.2022.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhao, S., Xia, J., Wu, X., Zhang, L., Wang, P., Wang, H.et al. (2018) Deficiency in class III PI3-kinase confers postnatal lethality with IBD-like features in zebrafish. Nat. Commun. 9, 2639 10.1038/s41467-018-05105-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wong, M., Ganapathy, A.S., Suchanec, E., Laidler, L., Ma, T. and Nighot, P. (2019) Intestinal epithelial tight junction barrier regulation by autophagy-related protein ATG6/beclin 1. Am. J. Physiol. Cell Physiol. 316, C753–C765 10.1152/ajpcell.00246.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Saha, K., Ganapathy, A.S., Wang, A., Morris, N.M., Suchanec, E., Ding, W.et al. (2023) Autophagy Reduces the Degradation and Promotes Membrane Localization of Occludin to Enhance the Intestinal Epithelial Tight Junction Barrier against Paracellular Macromolecule Flux. J Crohns Colitis. 17, 433–449 10.1093/ecco-jcc/jjac148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kim, H.J., Zhong, Q., Sheng, Z.H., Yoshimori, T., Liang, C. and Jung, J.U. (2012) Beclin-1-interacting autophagy protein Atg14L targets the SNARE-associated protein Snapin to coordinate endocytic trafficking. J. Cell Sci. 125, 4740–4750 10.1242/jcs.100339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Jung, H., Leal-Ekman, J.S., Lu, Q. and Stappenbeck, T.S. (2019) Atg14 protects the intestinal epithelium from TNF-triggered villus atrophy. Autophagy 15, 1990–2001 10.1080/15548627.2019.1596495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Cai, Z., Wang, S. and Li, J. (2021) Treatment of inflammatory bowel disease: a comprehensive review. Front. Med. (Lausanne) 8, 765474 10.3389/fmed.2021.765474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Retnakumar, S.V. and Muller, S. (2019) Pharmacological autophagy regulators as therapeutic agents for inflammatory bowel diseases. Trends Mol. Med. 25, 516–537 10.1016/j.molmed.2019.03.002 [DOI] [PubMed] [Google Scholar]
- 122.Nys, K., Agostinis, P. and Vermeire, S. (2013) Autophagy: a new target or an old strategy for the treatment of Crohn's disease? Nat. Rev. Gastroenterol. Hepatol. 10, 395–401 10.1038/nrgastro.2013.66 [DOI] [PubMed] [Google Scholar]
- 123.Rangan, P., Choi, I., Wei, M., Navarrete, G., Guen, E., Brandhorst, S.et al. (2019) Fasting-mimicking diet modulates microbiota and promotes intestinal regeneration to reduce inflammatory bowel disease pathology. Cell Rep. 26, 2704–2719.e2706 10.1016/j.celrep.2019.02.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Baker, K.A., Miller, T.D., Marino, F.E. and Hartmann, T.E. (2022) The exercise-induced inflammatory response in inflammatory bowel disease: a systematic review and meta-analysis. PLoS ONE 17, e0262534 10.1371/journal.pone.0262534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Louis, E. and Belaïche, J. (1995) Hydroxychloroquine (Plaquenil) for recurrence prevention of Crohn's disease after curative surgery. Gastroenterol. Clin. Biol. 19, 233–234 PMID: [PubMed] [Google Scholar]
- 126.Mutalib, M., Borrelli, O., Blackstock, S., Kiparissi, F., Elawad, M., Shah, N.et al. (2014) The use of sirolimus (rapamycin) in the management of refractory inflammatory bowel disease in children. J. Crohns Colitis 8, 1730–1734 10.1016/j.crohns.2014.08.014 [DOI] [PubMed] [Google Scholar]
- 127.Zhong, M., Cui, B., Xiang, J., Wu, X., Wen, Q., Li, Q.et al. (2020) Rapamycin is effective for upper but not for lower gastrointestinal Crohn's disease-related stricture: a pilot study. Front. Pharmacol. 11, 617535 10.3389/fphar.2020.617535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zhao, J., Wang, H., Yang, H., Zhou, Y. and Tang, L. (2020) Autophagy induction by rapamycin ameliorates experimental colitis and improves intestinal epithelial barrier function in IL-10 knockout mice. Int. Immunopharmacol. 81, 105977 10.1016/j.intimp.2019.105977 [DOI] [PubMed] [Google Scholar]
- 129.Shoji-Kawata, S., Sumpter, R., Leveno, M., Campbell, G.R., Zou, Z., Kinch, L.et al. (2013) Identification of a candidate therapeutic autophagy-inducing peptide. Nature 494, 201–206 10.1038/nature11866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Fairlie, W.D., Tran, S. and Lee, E.F. (2020) Crosstalk between apoptosis and autophagy signaling pathways. Int. Rev. Cell Mol. Biol. 352, 115–158 10.1016/bs.ircmb.2020.01.003 [DOI] [PubMed] [Google Scholar]
- 131.Dong, X., Liang, Q., Pan, Y.Z., Wang, X., Kuo, Y.C., Chiang, W.C.et al. (2022) Novel Bcl-2 inhibitors selectively disrupt the autophagy-specific Bcl-2-Beclin 1 protein-protein interaction. ACS Med. Chem. Lett. 13, 1510–1516 10.1021/acsmedchemlett.2c00309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Calis, S., Dogan, B., Durdagi, S., Celebi, A., Yapicier, O., Kilic, T.et al. (2022) A novel BH3 mimetic Bcl-2 inhibitor promotes autophagic cell death and reduces in vivo Glioblastoma tumor growth. Cell Death Discov. 8, 433 10.1038/s41420-022-01225-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wang, R., Graham, S., Sun, N., McCarthy, D., Peng, R., Erickson, J.et al. (2020) CRISPR/Cas9-targeting of CD40 in hematopoietic stem cells limits immune activation mediated by anti-CD40. PLoS ONE 15, e0228221 10.1371/journal.pone.0228221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Yan, X., Pan, Q., Xin, H., Chen, Y. and Ping, Y. (2021) Genome-editing prodrug: targeted delivery and conditional stabilization of CRISPR-Cas9 for precision therapy of inflammatory disease. Sci. Adv. 7, eabj0624 10.1126/sciadv.abj0624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Yang, Y., Gomez, M., Marsh, T., Poillet-Perez, L., Sawant, A., Chen, L.et al. (2022) Autophagy in PDGFRα+ mesenchymal cells is essential for intestinal stem cell survival. Proc. Natl Acad. Sci. U.S.A. 119, e2202016119 10.1073/pnas.2202016119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Stalgis, C., Deepak, P., Mehandru, S. and Colombel, J.F. (2021) Rational combination therapy to overcome the plateau of drug efficacy in inflammatory bowel disease. Gastroenterology 161, 394–399 10.1053/j.gastro.2021.04.068 [DOI] [PubMed] [Google Scholar]