Summary
We report genome-wide DNA data for 73 individuals from five archaeological sites across the Bronze and Iron Ages Southern Levant. These individuals, who share the “Canaanite” material culture, can be modeled as descending from two sources: (1) earlier local Neolithic populations, and (2) populations related to the Chalcolithic Zagros or the Bronze Age Caucasus. The non-local contribution increased over time, as evinced by three outliers who can be modeled as descendants of recent migrants. We show evidence that different “Canaanite” groups genetically resemble each other more than other populations. We find that Levant-related modern populations typically have substantial ancestry coming from populations related to the Chalcolithic Zagros and the Bronze Age Southern Levant. These groups also harbor ancestry from sources we cannot fully model with the available data, highlighting the critical role of post-Bronze Age migrations into the region over the past 3000 years.
eTOC blurb
Genome-wide data from Bronze Age individuals across nine sites in the Southern Levant show strong genetic resemblance including a component from populations related to Chalcolithic Zagros and Early Bronze Age Caucasus introduced by gene flow lasting at least until the late Bronze Age and impacting modern Levantine population architecture.
Graphical Abstract
Introduction
The Bronze Age (ca. 3500–1150 BCE) was a formative period in the Southern Levant, a region that includes present-day Israel, Jordan, Lebanon, the Palestinian Authority, and southwest Syria. This era, which ended in a large-scale civilization collapse across this region (Cline, 2014), shaped later periods both demographically and culturally. The following Iron Age (ca. 1150–586 BCE) saw the rise of territorial kingdoms such as biblical Israel, Judah, Ammon, Moab and Aram Damascus, as well as the Phoenician city-states. In much of the Late Bronze Age, the region was ruled by imperial Egypt, while in later phases of the Iron Age it was controlled by the Mesopotamian-centered empires of Assyria and Babylonia. Archaeological and historical research has documented major changes during the Bronze and Iron Ages, such as the cultural influence of the northern (Caucasian) populations related to the Kura-Araxes tradition during the Early Bronze Age (Greenberg and Goren, 2009) and impacts from the “Sea Peoples” (such as Philistines) from the west in the beginning of the Iron Age (Yasur-Landau, 2010).
The inhabitants of the Southern Levant in the Bronze Age are commonly described as “Canaanites”, that is, residents of the Land of Canaan. The term appears in several 2nd millennium BCE sources (e.g., Amarna, Alalakh and Ugarit tablets) and in biblical texts dating to the 8th-7th centuries BCE and later (Bienkowski, 1999; Lemche, 1991; Na’aman, 1994a). In the latter, the Canaanites are referred to as the pre-Israelite inhabitants of the land (Na’aman, 1994a). Canaan of the 2nd millennium BCE was organized in a system of city-states (Goren et al., 2004), with elites ruling from urban hubs over rural (and in some places pastoral) countryside. The material culture of these city-states was relatively uniform (Mazar, 1992), but whether this uniformity extends to their genetic ancestry is unknown. Whereas genetic ancestry and material culture are unlikely to ever match perfectly, past ancient DNA analyses show that they might sometimes be strongly associated. In other cases, a direct correspondence between genetics and culture cannot be established. We discuss several examples in the Discussion.
Previous ancient DNA studies published genome-scale data for 13 individuals from four Bronze Age sites in the Southern Levant: three individuals from ‘Ain Ghazal in present-day Jordan, dated to ~2300 BCE (Intermediate Bronze Age) (Lazaridis et al., 2016); five from Sidon in present-day Lebanon, dated to ~1750 BCE (Middle Bronze Age) (Haber et al., 2017); two from Tel Shadud in present-day Israel, dated to ~1250 BCE (Late Bronze Age) (van den Brink et al., 2017); and three from Ashkelon in present-day Israel, dated to ~1650–1200 BCE (Middle and Late Bronze Age) (Feldman et al., 2019). The ancestry of these individuals could be modeled as a mixture of earlier local groups and groups related to the Chalcolithic people of the Zagros Mountains, located in present-day Iran and designated in previous studies as Iran_ChL (Haber et al., 2017; Lazaridis et al., 2016). The Bronze Age Sidon group could be modeled as a major (93±2%) ancestral source for present-day groups in the region (Haber et al., 2017). A study of Chalcolithic individuals from Peqi’in cave in the Galilee (present-day Israel) showed that the ancestry of this earlier group included an additional component related to earlier Anatolian farmers, which was excluded as a substantial source for later Bronze Age groups from the Southern Levant, with the exception of the coastal groups from Sidon and Ashkelon (Feldman et al., 2019; Harney et al., 2018). These observations point to a degree of population turnover in the Chalcolithic-Bronze Age transition, consistent with archaeological evidence for a disruption between local Chalcolithic and Early Bronze cultures (de Miroschedji, 2014).
Here, we set out to address three issues. First, we sought to determine the extent of genetic homogeneity among the sites associated with Canaanite material culture. Second, we analyzed the data to gain insights into the timing, extent and origin of gene flow that brought Zagros and Caucasus-related ancestry to the Bronze Age Southern Levant. Third, we assessed the extent to which additional gene flow events have impacted the region since that time.
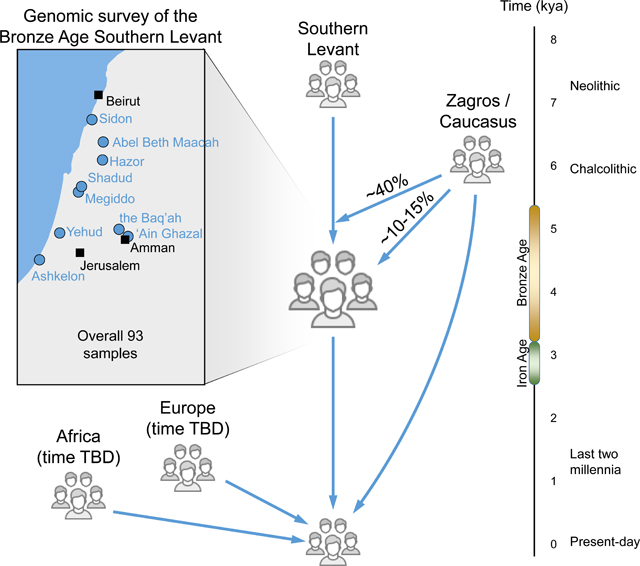
To address these questions, we generated genome-wide ancient DNA data for 71 Bronze Age and two Iron Age individuals, spanning roughly 1,500 years, from the Intermediate Bronze Age to the Early Iron Age. Combined with previously published data on the Bronze and Iron Ages in the Southern Levant, we assembled a dataset of 93 individuals from nine sites across present-day Israel, Jordan and Lebanon, all demonstrating Canaanite material culture. We show that the sampled individuals from the different sites are usually genetically similar, albeit with subtle but in some cases significant differences, especially in residents of the coastal regions of Sidon and Ashkelon. Almost all individuals can be modeled as a mixture of local earlier Neolithic populations and populations from the northeastern part of the Near East. However, the mixture proportions change over time, revealing the demographic dynamics of the Southern Levant during the Bronze Age. Finally, we show that the genomes of present-day groups geographically and historically linked to the Bronze Age Levant, including the great majority of present-day Jewish groups and Levantine Arabic-speaking groups, are consistent with having 50% or more of their ancestry from people related to groups who lived in the Bronze Age Levant and the Chalcolithic Zagros. These present-day groups also show ancestries that cannot be modeled by the available ancient DNA data, highlighting the importance of additional major genetic impacts on the region since the Bronze Age.
Results
Data Set
We extracted DNA from the bones of 73 individuals from five archaeological sites in the Southern Levant (Table S1, STAR Methods, Figure 1A):
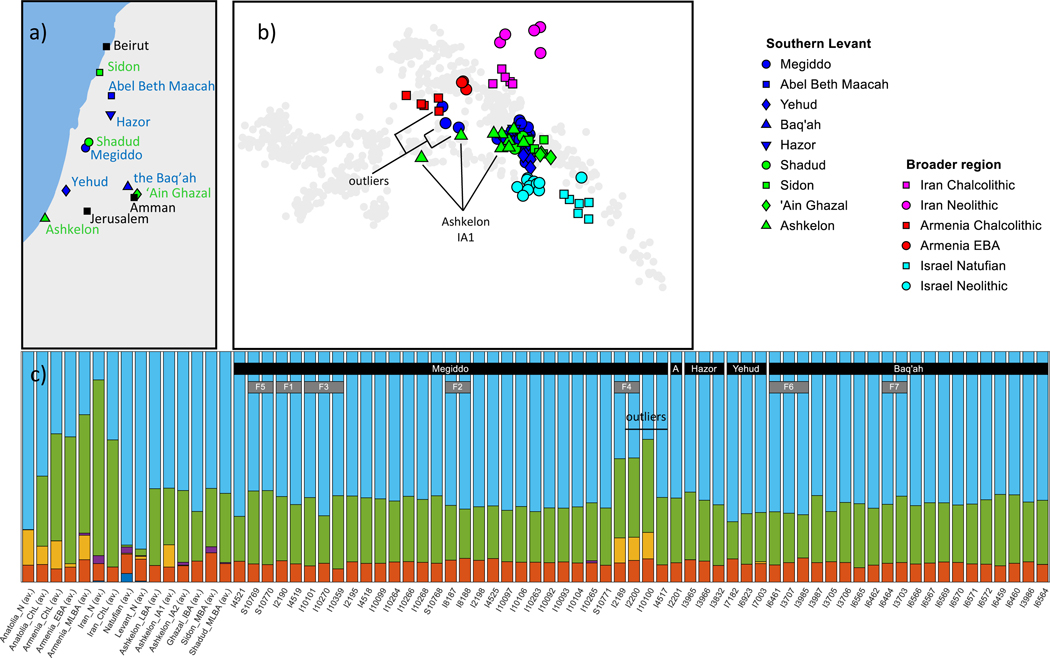
Figure 1. Bronze and Iron Age individuals analyzed in this study.
(A) Location of archaeological sites. (Blue) Sites with individuals first reported in this paper. (Green) Sites with individuals reported in previous studies. (B) PCA plot, showing present-day Eurasian individuals in grey (taken from Lazaridis et al., 2014) and ancient individuals in color. Only individuals with at least 30,000 autosomal SNPs were plotted. All Bronze and Iron Age individuals cluster (blue and green marks), except for the three denoted as “outliers.” (C) ADMIXTURE plot with , showing Bronze and Iron Age individuals, as well as other selected populations. Only individuals with at least 30,000 autosomal SNPs were plotted. The seven families are marked by F1-F7. “A” stands for “Abel.” See also Figure S1, Table S1.
Thirty-five individuals from Tel Megiddo (northern Israel), most of whom date to the Middle-to-Late Bronze Age, except for one dating to the Intermediate Bronze Age and one dating to the Early Iron Age.
Twenty-one individuals from the Baq‛ah in central Jordan (northeast of Amman), mostly from the Late Bronze Age.
Thirteen individuals from Yehud (central Israel), dating to the Middle Bronze Age.
Three individuals from Tel Hazor (northern Israel) dating to the Middle-to-Late Bronze Age.
One individual from Tel Abel Beth Maacah (northern Israel), dating to the Iron Age.
For all analyzed samples but one, DNA was extracted from petrous bones. The DNA was converted to double-indexed half UDG-treated libraries that we enriched for about 1.2 million single nucleotide polymorphism (SNPs) before sequencing (see STAR Methods). The median number of autosomal SNPs covered was 288,863 (range 4,883–945,269). In addition to genetic data, we measured values of strontium isotopes for 12 individuals (and for eight additional individuals that did not produce DNA, STAR Methods, Methods S1A), and generated accelerator mass spectrometry radiocarbon dates for 20 individuals (Table S1). We combined our newly generated data with published data for 13 Bronze Age Southern Levant individuals from ‘Ain-Ghazal, Sidon, Tel-Shadud, and Ashkelon (van den Brink et al., 2017; Feldman et al., 2019; Haber et al., 2017; Lazaridis et al., 2016), and seven Iron Age Southern Levant Individuals from Ashkelon (Feldman et al., 2019).
We projected the autosomal genetic data onto the plane spanned by the first two principal components of 777 present-day West Eurasian individuals genotyped for roughly 600,000 SNPs on the Affymetrix Human Origins SNP array (Lazaridis et al., 2014). We restricted the plot to 68 individuals represented by at least 30,000 autosomal SNPs (Figure 1B), a coverage threshold where the ability to infer ancestry was robust to sampling noise (Methods S1B). All Bronze and Iron Age Levant individuals (blue and green shapes) form a tight cluster, except for three outliers from Megiddo, and previously identified outliers from the Ashkelon population known as IA1 (Iron Age I; Feldman et al., 2019). We also ran ADMIXTURE on a set of 1,663 present-day and ancient individuals (see STAR Methods, Figure S1). The ADMIXTURE results are qualitatively consistent with the PCA, suggesting that all individuals but the outliers from Megiddo and the Ashkelon IA1 population have similar ancestry (Figure 1C).
We used the method described in (Olalde et al., 2019) to identify 17 individuals as being first, second, or third degree relatives of other individuals in the dataset. They fall within seven families: five in Tel Megiddo and two in the Baq‛ah. In most families, we used only the member with the highest SNP coverage in subsequent analyses (Table S1). Two of the three Megiddo outliers are a brother and a sister (Family 4, I2189 and I2200), leaving in the final dataset two individuals marked as outliers. After removing low-coverage individuals and closely related family members, 62 individuals were left for further analysis (Table S1).
High degree of genetic affinities between multiple sites
We divided the 26 high-coverage individuals from Tel Megiddo into the following groups, based on geographic location, archaeological period, and genetic clustering in PCA (Table S1): Intermediate Bronze Age (Megiddo_IBA, a single individual), Middle-to-Late Bronze Age (Megiddo_MLBA, 22 individuals), Iron Age (Megiddo_IA, a single individual), as well as the two outliers, Megiddo_I2200 and Megiddo_I10100, which were each treated as a separate group. We compared these groups and the other populations in our dataset to previously published data from other sites in the broader region and from earlier periods, including the Early Bronze Age Caucasus (Armenia_EBA), the Middle-to-Late Bronze Age Caucasus (Armenia_MLBA), the Chalcolithic Zagros Mountains (Iran_ChL), the Chalcolithic Caucasus (Armenia_ChL), the Neolithic of the Southern Levant (Levant_N), the Neolithic of the Zagros Mountains (Iran_N), and the Neolithic of Anatolia (Anatolia_N) (Lazaridis et al., 2016).
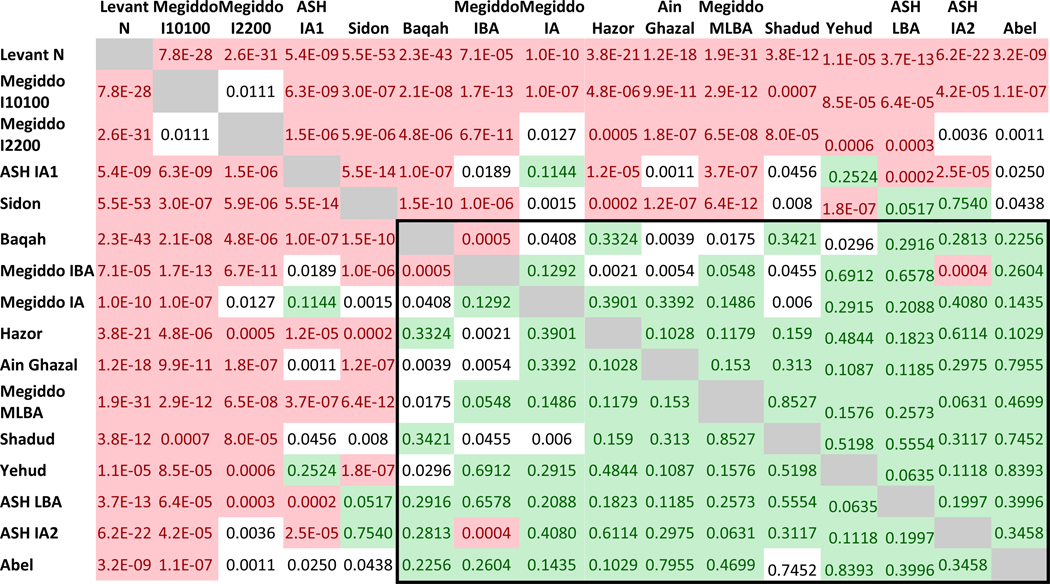
To test for variation in ancestry proportions among the Levant Bronze and Iron Age groups we used qpWave. qpWave tests whether each possible pair of groups (Testi, Testj) is consistent with descending from a common ancestral population – that is, consistent with being a clade – since separation from the ancestors of a set of outgroup populations. qpWave works by computing symmetry test statistics of the form f4(Testi, Testj; Outgroupk, Outgroupl), which have an expected value of zero if (Testi, Testj) form a clade with respect to the outgroups. qpWave then generates a single p-value corrected for the empirically measured correlation among the statistics (Reich et al., 2012). Using a distantly related set of outgroups, we found that with the exception of the outliers from Megiddo, Ashkelon IA1 and Sidon, all Bronze and Iron Age Levant groups are consistent with being pairwise clades with respect to the outgroups (Figure 2).
Figure 2. P-values of qpWave for each pair of populations.
Values greater than 0.05 are shaded in light green, and values lower than 0.001 are shaded in light red. The rectangle shows all Levant populations excluding Sidon, the outliers from Megiddo and Ashkelon IA1. Computations were based on the o9a outgroup set (o9 + Anatolia_N).
We discuss each of qpWave’s findings of significant population substructure in turn. The Megiddo outliers not only fail to form a clade with the other populations, but also with each other. Ashkelon IA1 has previously been reported to harbor European ancestry and so our finding that it is genetically differentiated from contemporary groups is unsurprising (Feldman et al., 2019). The significant differentiation of the Sidon individuals in qpWave—despite the fact that they roughly cluster with the other Southern Levant Bronze Age groups in PCA and ADMIXTURE—is notable especially as we find that they are consistent with forming a clade with the two groups from coastal Ashkelon that do not have European-related admixture (the Bronze Age and later Iron Age groups ASH_LBA and ASH_IA2). Speculatively, this observation could be related to the fact that both Sidon and Ashkelon were port towns with connections to other Mediterranean coastal groups outside the Southern Levant, which could have introduced ancestry components that are absent from inland Levantine Bronze Age groups, although it is difficult to test this hypothesis in the absence of high resolution ancient DNA sampling from the eastern Mediterranean rim. The genetic distinctiveness of the Sidon individuals is also compatible with previous findings that Chalcolithic Levantine individuals from Peqi’in Cave are consistent with contributing some ancestry to the Sidon individuals, but not to the ‘Ain Ghazal ones (Harney et al., 2018). We considered the possibility that the significantly different genetic patterns we detect in the Sidon individuals could reflect their different experimental treatment compared to the other individuals in this study (shotgun sequencing of non-UDG-treated libraries compared to enrichment of UDG-treated libraries). To test this, we repeated the analyses using only transversion SNPs, which are less prone to characteristic ancient DNA errors, but found no indication of systematic bias (Wang et al., 2015). However, we did find evidence of substructure within the Sidon individuals, with some but not all consistent with forming a clade with inland Southern Levant populations, a finding that could reflect substantial cosmopolitan nature of this coastal site (Methods S1C, see Discussion).
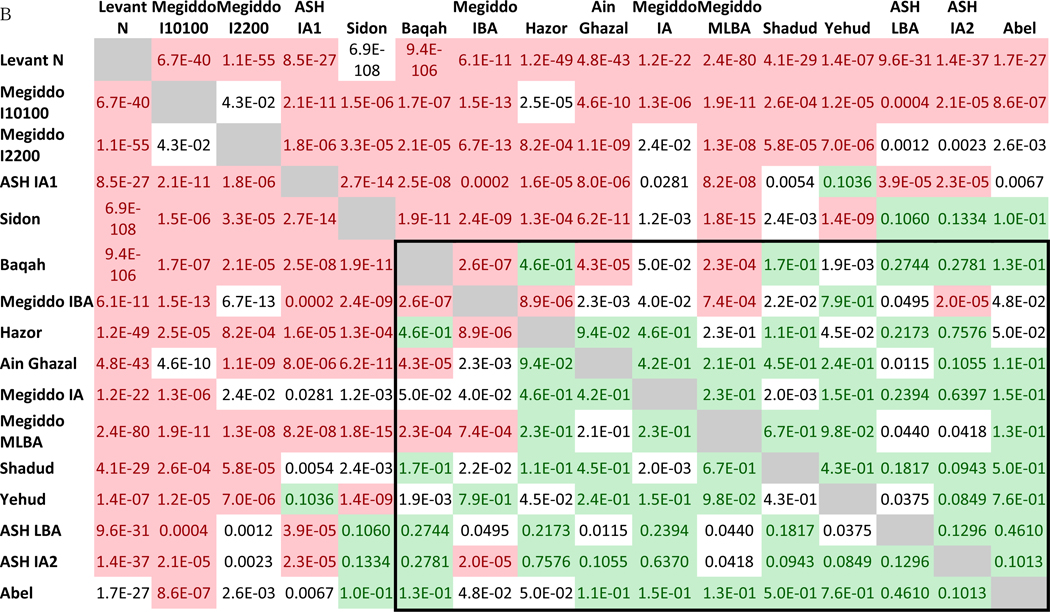
To reveal subtler population structure, we repeated the qpWave analysis adding outgroups that are genetically closer to the test groups, such as Armenia_MLBA and Natufian (Figure 3). With this more powerful set of outgroups, Baqah and Megiddo_IBA also provide evidence of not being pairwise clades with the remaining groups. Thus, beyond the broad observation of genetic affinities between sites, we also observe subtle ancestry heterogeneity across the region during the Bronze Age (see Discussion).
Figure 3. P-values of qpWave for each pair of populations.
Values greater than 0.05 are shaded in light green, and values lower than 0.001 are shaded in light red. The rectangle shows all Levant populations excluding Sidon, the outliers from Megiddo and Ashkelon IA1. Computations were based on the extended outgroup set (o9 + Anatolia_N + Armenia_MLBA + Caucasus Hunter Gatherers (CHG) + Natufians).
Gene flow into the Southern Levant during the Bronze Age
Two previous studies of Bronze Age individuals from ‘Ain Ghazal and Sidon modeled them as derived from a mixture of earlier local groups (Levant_N) and groups related to peoples of the Chalcolithic Zagros mountains (Iran_ChL) (Haber et al., 2017; Lazaridis et al., 2016). These groups were estimated to harbor around 56%±3% and 48%±4% Neolithic Levant-related ancestry for ‘Ain Ghazal (Lazaridis et al., 2016) and Sidon (Haber et al., 2017), respectively. We used qpAdm to estimate that Bronze and Iron Age Ashkelon (ASH_LBA and ASH_IA2) carry 54%±5% and 42%±5% Neolithic Levant-related ancestry, respectively. Next, we used qpAdm to test the same model for the data reported here, and found that most MLBA groups fit the model, with point estimates of 48–57% Levant_N ancestry (except for a point estimate of 66% Levant_N ancestry in Yehud). These ancestry proportions are statistically indistinguishable (Bonferroni-corrected z-test), which corroborates the fact that they are consistent with forming pairwise clades in qpWave (Table S2, Methods S1D). The only group that failed to fit this model was Baqah (P = 0.0003), even when using a wide range of outgroup populations (Table S2). This may be a result of ancestry heterogeneity across the Baqah individuals (see below).
To obtain insight into the Zagros-related ancestry component, we focused on two questions: what is the likely origin of this ancestry component, and what is its likely timing. Whereas people of the Chalcolithic Zagros are so far the best proxy population for this ancestry component, there is no archaeological evidence for cultural spread directly from the Zagros into the Southern Levant during the Bronze Age. In contrast, there is archaeological support for connections between Bronze Age Southern Levant groups and the Caucasus (Greenberg and Goren, 2009), a term we use to represent both present-day Caucasus, as well as neighboring regions such as eastern Anatolia (see Discussion). With regard to the timing of these events, archaeology points to cultural affinities between the Kura-Araxes (Caucasus) and Khirbet Kerak (Southern Levant) archaeological cultures in the first half of the 3rd millennium BCE (Greenberg and Goren, 2009), and textual/linguistic evidence documents a number of non-Semitic, Hurrian (from the northeast of the ancient Near East) personal names in the 2nd millennium BCE, for example in the Amarna archive of the 14th century BCE (Na’aman, 1994b). We therefore reasoned that the Chalcolithic Zagros component might have arrived into the Southern Levant through the Caucasus (and even more proximately the northeastern areas of the ancient Near East, although we have no ancient DNA sampling from this region). This movement might not have been limited to a short pulse, and instead could have involved multiple waves throughout the Bronze Age.
To test whether the origin of the gene flow was from the Caucasus, rather than directly from the Zagros region, we ran qpAdm, replacing Iran_ChL with Early Bronze Age Caucasus (Armenia_EBA). We found that the Caucasus model received similar support to that of the Zagros model (Table S2, Methods S1E). Next, we modeled Armenia_EBA as a mixture of an earlier Caucasus population (Chalcolithic Armenia, Armenia_ChL) and Iran_ChL, and found that indeed Armenia_EBA is compatible with this model (Table S2). Taken together, we conclude that our data is also compatible with a model in which Zagros-related ancestry in the Southern Levant arrived through the Caucasus, either directly or via intermediates.
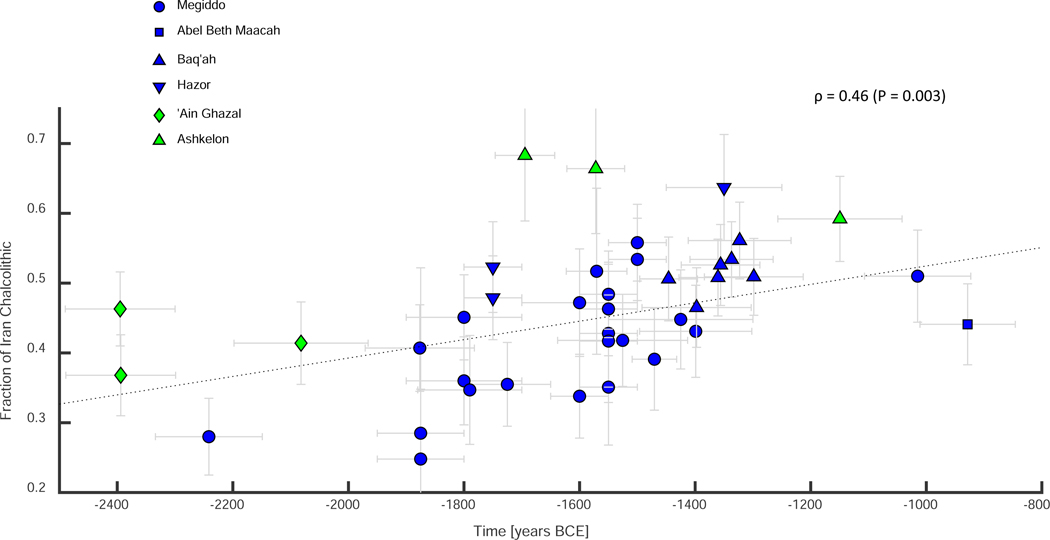
To study the timing of the admixture of Zagros-related ancestry in the Southern Levant, we leveraged the large time span of individuals in our dataset, extending across roughly 1,500 years, from the Intermediate Bronze Age to the Early Iron Age. Using qpAdm-based ancestry estimates for each of the individuals, we found that almost all are compatible with being an admixture of groups related to the Neolithic Levant and Chalcolithic Zagros. One exception to this is an individual in Megiddo_MLBA that is weakly compatible with the model. Another exception is three individuals in the Baq’ah (Table S2), which suggests that the difficulty in modelling individuals from this site as a mixture of Neolithic Levant and Chalcolithic Zagros might reflect ancestry heterogeneity (Figure 3). These results do not change qualitatively when we used a larger set of outgroup populations (Table S2). We observed that the oldest individuals in our collection, from the Intermediate Bronze Age, already carried significant Zagros-related ancestry, suggesting that gene flow into the region started before ca. 2400 BCE. This is consistent with the hypothesis that people of Kura-Araxes archaeological complex of the 3rd millennium BCE might have affected the Southern Levant not only culturally, but also through some degree of movement of people. Our data also imply an increase in the proportion of Zagros-related ancestry following the Intermediate Bronze Age, as reflected in a significantly positive slope in a linear regression of the Chalcolithic Zagros-related ancestry over the calendar year (, Jackknife), amounting to an increase of ≈14% per thousand years (Figure 4, Figure S2A). However, we caution that the number of individuals and their time span are insufficient to determine whether the increase in the Zagros-related ancestry happened continuously during the Middle and Late Bronze Age, or whether there were multiple distinct migration events.
Figure 4. Temporal changes in the genetic makeup of individuals in the Bronze and Iron Age Levant.
Fraction of Chalcolithic Iran-related component in each individual as computed by qpAdm, modeling each individual as a mixture of Neolithic Levant and Chalcolithic Iran, and using the o9a outgroup set (o9 + Anatolia_N). Vertical error bars denote one standard error in each direction. Horizontal error bars denote estimated time ranges. Dashed line describes the linear regression. Only individuals whose time range does not exceed 250 years are plotted and used in the regression. Note that the two well-dated Ash_LBA individuals happen to harbor the highest Iran_ChL component. See also Figures S2 and S3, Tables S2 and S3.
The two outliers from Megiddo (three including the sibling pair) provide additional evidence for the timing and origin of gene flow into the region. The three were found in close proximity to each other at Level K-10, which is radiocarbon dated to 1581–1545 BCE (domestic occupation) and 1578–1421 BCE (burials; both ) (Martin et al., 2020; Toffolo et al., 2014), while the bone of one of the three (I10100) was directly dated (1688–1535 BCE, ). The reason these individuals are distinct from the rest is that their Caucasus- or Zagros-related genetic component is much higher, reflecting ongoing gene flow into the region from the northeast (Table S2, Figure S2B). The Neolithic Levant component is ≈22–27% in I2200, and ≈9–26% in I10100. These individuals are unlikely to be first generation migrants, as strontium isotope analysis on the two outlier siblings (I2189 and I2200, Methods S1A) suggests that they were raised locally. This implies that the Megiddo outliers may be descendants of people who arrived in recent generations. Direct support for this hypothesis comes from the fact that in sensitive qpAdm modeling (including closely related sets of outgroups), the only working northeast source population for these two individuals is the contemporaneous Armenia_MLBA, while the earlier Iran_ChL and Armenia_EBA do not fit (Table S2). The addition of Iran_ChL to the set of outgroups does not change this result or cause model failure. Finally, no other Levantine group shows a similar admixture pattern (Table S2). This shows that some level of gene flow into the Levant took place during the later phases of the Bronze Age, and suggests that the source of this gene flow was the Caucasus.
Taken together, our analyses show that gene flow into the Levant from people related to those in the Caucasus or Zagros was already occurring by the Intermediate Bronze Age, and that it lingered, episodically or continuously, at least in inland sites, during the Middle-to-Late Bronze Age.
Further change in Levantine populations since the Bronze Age
To develop a sense of population changes in the Levant since the Bronze Age, we attempted to model groups that have a tradition of descent from ancient people in the region (Jews) as well as Levantine Arabic-speakers as mixtures of various ancient source populations. qpAdm assumes no admixture between groups related to the outgroups and the source populations, but almost all present-day Levantine and Mediterranean populations have significant sub-Saharan African-related admixture that the ancient groups did not. This eliminates many key outgroups for qpAdm and reduces the utility of the method in this context. In particular, we were not able apply qpAdm to get a single working model for the majority of present-day West Eurasian populations. As an alternative, we developed a methodology we call LINADMIX, which relies on the output of ADMIXTURE (Alexander et al., 2009) and uses constrained least squares to estimate the contribution of given source populations to a target population (see STAR Methods). As a complementary approach, we developed a tool we call pseudo-haplotype ChromoPainter (PHCP), which is an adaptation of the haplotype-based method ChromoPainter (Lawson et al., 2012) to ancient genomes (see STAR Methods, Methods S1F). We first established that these methods provide meaningful estimates of ancestry in the context of this study by using them to re-compute the ancestry proportions that we were able to model with qpAdm. Both LINADMIX and PHCP (Table S3, Figure S3, Methods S1F) produce qualitatively similar estimates as qpAdm (Table S2). To further establish the methods, we performed simulations that were designed to test the methods’ abilities to infer ancestry proportions in present-day populations in a setup similar to the current study (Methods S1H). For this, we generated present-day populations as a mixture of two closely related ancient populations with and without a third, more distant, population. Both methods estimated the ancestry proportion of the distant source population with errors of up to 4% and the proportions of the closely related source populations with errors of up to 10%. Thus, although ADMIXTURE, the basis of LINADMIX, is known to have certain pitfalls as a tool for quantifying ancestry proportions (Lawson et al., 2018), in the case of individuals with ancestry sources similar to those we have analyzed here, our results suggest that both LINADMIX and PHCP are highly informative.
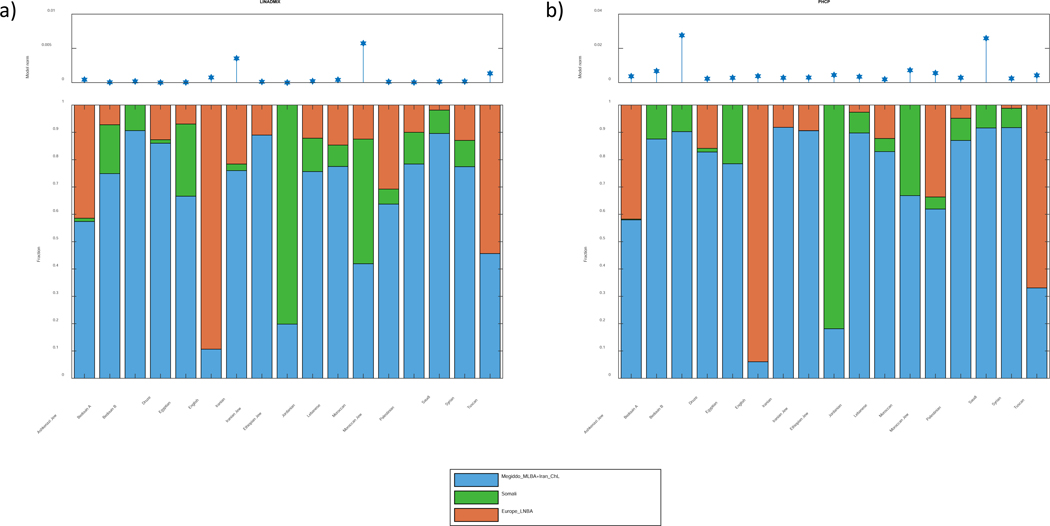
For the LINADMIX analysis of present-day populations we used a background dataset of 1,663 present-day and ancient individuals from 239 populations genotyped using SNP arrays, and focused our analysis on 14 Jewish and Levantine present-day populations, along with modern English, Tuscan and Moroccans that were used as controls. We used LINADMIX to model each of the 17 present-day populations as an admixture of four sources: (1) Megiddo_MLBA (the largest group) as a representative of the Middle to Late Bronze Age component; (2) Iran_ChL as a representative of the Zagros and the Caucasus; (3) Present-day Somalis as representatives of an Eastern African source (in the absence of genetic data on ancient populations from the region); and (4) Europe_LNBA as a representative of ancient Europeans from the Late Neolithic and Bronze Age (Methods S1I, Table S4, Figure S4). We also applied PHCP to these 17 present-day populations (Methods S1G, Table S4, Figure S4). Comparison of PHCP and LINADMIX shows that they agree well with respect to the Somali and Europe_LNBA component, and hence also for the combined contribution of Iran_ChL and Megiddo_MLBA (Methods S1G, Figure S4). However, they deviate regarding the respective contributions of Iran_ChL and Megiddo_MLBA (Figure S4), likely because of the fact that the Megiddo_MLBA and Iran_ChL are already very similar populations (Table S3). To only consider results that are robust and shared by LINADMIX and PHCP, we have combined Megiddo_MLBA and Iran_ChL to a single source population representing the Middle East for our main results (Figure 5). We further verified these conclusions, as well as the robustness of the estimations, using a different representative for the Bronze Age Levantine groups as a source (Tables S4–S5, Methods S1J), and using perturbations to the ADMIXTURE parameters (Table S4, Methods S1K). Combined, these results suggest that modern populations related to the Levant are consistent with having a substantial ancestry component from the Bronze Age Southern Levant and the Chalcolithic Zagros. Nonetheless, other potential ancestry sources are possible, and more ancient samples may enable a refined picture (Table S4).
Figure 5. Estimated fractions contributed by different ancient populations to present-day groups.
Seventeen present-day populations were modeled as an admixture of groups related to four source populations. The upper panel shows the norm of residuals of the models, while the lower panel shows the relative contribution of each of the source populations to the present-day target population listed on the x-axis. (A) LINADMIX. (B) PHCP. See also Figure S4, Tables S4 and S5.
The results show that since the Bronze Age an additional East African-related component was added to the region (on average ~10.6%, excluding Ethiopian Jews that harbor ~80% East African component), as well as a European-related component (on average ~8.7%, excluding Ashkenazi Jews who harbor a ~41% European-related component). The East African-related component is highest in Ethiopian Jews and North Africans (Moroccans and Egyptians). It exists in all Arabic-speaking populations (apart from the Druze). The European-related component is highest in the European control populations (English and Tuscan), as well as in Ashkenazi and Moroccan Jews, both having a history in Europe (Atzmon et al., 2010; Carmi et al., 2014; Schroeter, 2008). This component is present, although in smaller amount, in all other populations except for Bedouin B and Ethiopian Jews. As expected, the English and Tuscan populations have a very low Middle Eastern-related component. Whereas LINADMIX and PHCP have high uncertainty in estimating the relative contributions of Megiddo_MLBA and Iran_ChL, the results and simulations nevertheless suggest that additional Zagros-related ancestry has penetrated the region since the Bronze Age (Methods S1I). Except for the populations with the highest Zagros-related component, PHCP estimates lower magnitudes of this component (Figure S4A), and therefore detection by PHCP of a Zagros-related ancestry is likely an indication for the presence of this component. Indeed, examining the results of LINADMIX and PHCP on all four source populations (Figure S4), we observe a relatively large Zagros-related component in many Arabic-speaking groups, suggesting that gene flow from populations related to those of the Zagros and Caucasus (although not necessarily from these specific regions) continued even after the Iron Age (Methods S1I).
Taken together, the patterns of the present-day populations reflect demographic processes that occurred after the Bronze Age, and are plausibly related to processes known from the historical literature (Methods S1I). These include an Eastern African-related component that is present in Arabic-speaking groups but is lower in non-Ethiopian Jewish groups, as well as Zagros-related contribution to Levantine populations, which is highest in the northernmost population examined, suggesting a contribution of populations related to the Zagros even after the Bronze and Iron Ages.
Discussion
Our results provide a comprehensive genetic picture of the primary inhabitants of the Southern Levant during the 2nd millennium BCE, known in the historical record and based on shared material culture as “Canaanites.” We carried out a detailed analysis aimed at answering three basic questions: how genetically homogeneous were these people, what were their plausible origins with respect to earlier peoples, and how much change in ancestry has there been in the region since the Bronze Age.
Earlier genetic analyses modeled the genomes of Middle to Late Bronze Age people of the Southern Levant as having almost equal shares of earlier local populations (Levant_N) and populations that are related to the Chalcolithic Zagros (Feldman et al., 2019; Haber et al., 2017; Lazaridis et al., 2016), suggesting a movement from the northeast into the Southern Levant. Here, we provide more details on this process, taking into account evidence from both archaeology and our temporally and geographically diverse genetic data. As there is little archaeological evidence of a direct cultural connection between the Southern Levant and the Zagros region in this period, the Caucasus is a more likely source for this ancestry. We used our data to compare these two scenarios, and concluded that the genetic data are compatible with both.
The Megiddo outliers, which we inferred to be relative newcomers to the region, are particularly important in demonstrating that the gene flow continued throughout the Bronze Age, and that at least some of the gene flow likely came from the Caucasus rather than the Zagros. These two individuals have the highest proportions of Zagros-/Caucasian-related ancestry in our dataset. Analysis of these outliers gave significantly stronger evidence of a Caucasus source compared to a Zagros one, although this conclusion may be revised once ancient DNA data from the Middle-to-Late Bronze Age in the Zagros region become available. The two Megiddo individuals with the next lowest Neolithic Levant component (I10769 and I10770, brothers) were found near the monumental tomb that was likely related to the palace at Megiddo, raising the possibility that they might be associated with the ruling caste. Indeed, a ruler of Taanach (a town located immediately to the south of Megiddo) mentioned in a 15th century BCE cuneiform tablet found at the site, and the rulers of Megiddo and Taanach mentioned in the 14th century BCE Amarna letters (found in Egypt) carry Hurrian names (a language spoken in the northeast of the ancient Near East, possibly including the Caucasus) (Na’aman, 1994b). This provides some evidence—albeit so far only suggestive—that at least some of the ruling groups in these (and other) cities may have originated from the northeast of the ancient Near East.
The Caucasus is represented in this study by ancient groups from the present-day country of Armenia, but the region known to have had cultural ties with the Southern Levant is much broader. Evidence of cultural impacts on the Southern Levant is mainly focused on the Kura-Araxes culture during the Early Bronze Age (archaeology) and on the Hurrians during the Middle-to-Late Bronze Age (linguistic testimony). These two complexes were spread over the Caucasus, Eastern Anatolia and neighboring regions. The Armenian sites we analyzed are the best representatives to date of these cultures. The Early Bronze Age individuals from Armenia (Armenia_EBA) come from an Early Bronze Age Kura-Araxes burial ground, and the later Middle-to-Late Bronze Age individuals (Armenia_MLBA) come from the Aragatsotn Province in northwestern Armenia. It is important to note that the Neolithic and Chalcolithic Anatolian individuals analyzed in this study come from the northwestern part of Anatolia, which is not part of the Caucasus. The Chalcolithic Zagros individuals come from the Kangavar Valley in Iran, which is located on the border of the Kura-Araxes influence.
The term “Canaanites” is loosely defined, referring to a collection of groups (which in the Bronze Age were organized in a city-state system), and thus in principle could lack genetic coherence. The individuals examined here cover a wide geographic span – coming from nine sites in present-day Lebanon, Israel and Jordan. Our analyses revealed that, with the exception of Sidon (and to a smaller extent the individuals of the Baq‛ah), they are homogeneous in the sense of being closer to each other than to other contemporary and neighboring populations. This suggests that the archaeological and historical category of “Canaanites” correlates with shared ancestry (Eisenmann et al., 2018). This resembles the pattern observed in the Aegean basin during the 2nd millennium BCE, where the cultural categories of “Minoan” and “Mycenaean” show evidence of genetic homogeneity across multiple sites albeit with potentially subtle ancestry differences within these groupings (Lazaridis et al., 2017). Another example is the “Yamnaya” pastoralists of late 3rd and early 2nd millennium BCE in the western Eurasian Steppe (Allentoft et al., 2015; Haak et al., 2015). This contrasts with the pattern seen in other places, such as for the Bell Beaker cultural complex of the 2nd millennium BCE (Olalde et al., 2018), where people sharing similar cultural practices had widely varying ancestry. In any case, the detection of such associations — as we do here — cannot by itself prove that group identities in the past were related to genetics.
From the groups we have examined, the only one that is somewhat diverged from the rest is Sidon. We provide evidence against the possibility that this observation is a batch effect (Methods S1C). Rather, we suggest that the relative remoteness of Sidon stems from the fact that this population is genetically heterogeneous, with different individuals showing resemblance to different Southern Levantine groups (Methods S1C). During the 2nd millennium BCE, Sidon was a major port city, and was connected in trading relations with the eastern Mediterranean basin, which could have led to a significant genetic inflow, making its population more heterogeneous than that of inland cities. This may also be the reason that the site that most resembles Sidon is Ashkelon, which is another coastal site. The only inland population that resembles Sidon is Abel Beth Maacah, perhaps because of its geographic proximity (Figure 1A, Figure 2). Apart from Sidon, Baqah also shows some minor deviations from the rest when taking a richer set of outgroup populations (Figure 3). The Baq‛ah is located on the fringe of the Syrian desert, hence this population might be admixed with more eastern groups, which are not yet genetically sampled. This might be reflected by the fact that the individuals of the Baq‛ah also show some degree of variability in their ancestry patterns (Table S2).
Whereas this study focuses on the Bronze Age, it also reports two new samples from the Iron Age – one from Megiddo and the other from Abel Beth Maacah. These two individuals show ancestry patterns that are very similar to those observed in the Middle and Late Bronze Age individuals (Figure 4), suggesting that the destruction at the end of the Bronze Age in the region did not necessarily lead to genetic discontinuity in each and every sites. Notably, both Abel Beth Maacah and Megiddo are inland cities, and their genetic continuity throughout the transition from the Bronze Age to the Iron Age might not be representative of other sites in the region. For example, one of the two Iron Age populations in the Philistine coastal city of Ashkelon (ASH_IA1) showed evidence of mobility of populations related to southern Europe around the Bronze Age to Iron Age transition (Feldman et al., 2019).
Estimating the ancestry proportions in present-day Middle-Eastern populations with substantial sub-Saharan African admixture (as well as multiple sources of admixture from different parts of the Mediterranean), is difficult. We addressed the problem by developing two statistical techniques, and then testing the robustness of our inference based on a comparison between these methods, simulations and perturbations of the input (see STAR Methods, Methods S1F–K). We examined 14 present-day populations that are historically or geographically linked to the Southern Levant, and tested the contributions of East Africa, Europe and the Middle East (combining Southern Levant Bronze Age populations and Zagros-related Chalcolithic ones) to their ancestry. We found that both Arabic-speaking and Jewish populations are compatible with having more than 50% Middle Eastern-related ancestry. This does not mean that any these present-day groups bear direct ancestry from people who lived in the Middle to Late Bronze Age Levant or in Chalcolithic Zagros; rather, it indicates that they have ancestries from populations whose ancient proxy can be related to the Middle East. The Zagros-/Caucasian-related ancestry flow into the region apparently continued after the Bronze Age. We also see an Eastern African-related ancestry entering the region after the Bronze Age, with an approximate south-to-north gradient. In addition, we observe a European-related ancestry with the opposite gradient (north-to-south). Given the difficulties in separating the ancestry components arriving from the Southern Levant and the Zagros, an important direction for future work will be to reconstruct in high resolution the ancestry trajectories of each present-day group, and to understand how people from the Southern Levant Bronze Age mixed with other people in later periods in the context of processes known from the rich archaeological and historical records of the last three millennia.
STAR Methods
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for materials should be directed to and will be fulfilled by the Lead Contact, Liran Carmel (liran.carmel@huji.ac.il).
All data generated in this study is available, see Key Resource Table.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited Data | ||
| Raw and analyzed data from 73 newly reported ancient humans (see Table S1 for details) | This paper | ENA: PRJEB37057 |
| Chemicals, Peptides, Recombinant Proteins | ||
| Pfu Turbo Cx Hotstart DNA Polymerase | Agilent Technologies | 600412 |
| Herculase II Fusion DNA Polymerase | Agilent Technologies | 600679 |
| 2x HI-RPM hybridization buffer | Agilent Technologies | 5190-0403 |
| 0.5 M EDTA pH 8.0 | BioExpress | E177 |
| Sera-Mag™ Magnetic Speed-beads™ Carboxylate-Modified (1mm, 3EDAC/PA5) | GE LifeScience | 65152105050250 |
| USER enzyme | New England Biolabs | M5505 |
| UGI | New England Biolabs | M0281 |
| Bst DNA Polymerase2.0, large frag. | New England Biolabs | M0537 |
| PE buffer concentrate | QIAGEN | 19065 |
| Proteinase K | Sigma Aldrich | P6556 |
| Guanidine hydrochloride | Sigma Aldrich | G3272 |
| 3M Sodium Acetate (pH 5.2) | Sigma Aldrich | S7899 |
| Water | Sigma Aldrich | W4502 |
| Tween-20 | Sigma Aldrich | P9416 |
| Isopropanol | Sigma Aldrich | 650447 |
| Ethanol | Sigma Aldrich | E7023 |
| 5M NaCl | Sigma Aldrich | S5150 |
| 1M NaOH | Sigma Aldrich | 71463 |
| 20% SDS | Sigma Aldrich | 5030 |
| PEG-8000 | Sigma Aldrich | 89510 |
| 1 M Tris-HCl pH 8.0 | Sigma Aldrich | AM9856 |
| dNTP Mix | Thermo Fisher Scientific | R1121 |
| ATP | Thermo Fisher Scientific | R0441 |
| 10x Buffer Tango | Thermo Fisher Scientific | BY5 |
| T4 Polynucleotide Kinase | Thermo Fisher Scientific | EK0032 |
| T4 DNA Polymerase | Thermo Fisher Scientific | EP0062 |
| T4 DNA Ligase | Thermo Fisher Scientific | EL0011 |
| Maxima SYBR Green kit | Thermo Fisher Scientific | K0251 |
| 50x Denhardt’s solution | Thermo Fisher Scientific | 750018 |
| SSC Buffer (20x) | Thermo Fisher Scientific | AM9770 |
| GeneAmp 10x PCR Gold Buffer | Thermo Fisher Scientific | 4379874 |
| Dynabeads MyOne Streptavidin T1 | Thermo Fisher Scientific | 65602 |
| Salmon sperm DNA | Thermo Fisher Scientific | 15632-011 |
| Human Cot-I DNA | Thermo Fisher Scientific | 15279011 |
| DyNAmo HS SYBR Green qPCR Kit | Thermo Fisher Scientific | F410L |
| Methanol, certified ACS | VWR | EM-MX0485-3 |
| Acetone, certified ACS | VWR | BDH1101-4LP |
| Dichloromethane, certified ACS | VWR | EMD-DX0835-3 |
| Hydrochloric acid, 6N, 0.5N & 0.01N | VWR | EMD-HX0603-3 |
| Critical Commercial Assays | ||
| High Pure Extender from Viral Nucleic Acid Large Volume Kit | Roche | 5114403001 |
| MinElute PCR Purification Kit | QIAGEN | 28006 |
| NextSeq® 500/550 High Output Kit v2 (150 cycles) | Illumina | FC-404-2002 |
| Hydrogen peroxide 30% Suprapur ® | Merck | 107298 |
| Hydrochloric acid, 1N | Biolab-chemicals.com | 084105 |
| Nitric Acid 3.5N, 0.05N | Biolab-chemicals.com | 147005 |
| Software and Algorithms | ||
| Samtools | Li, 2011; Li et al., 2009 | http://samtools.sourceforge.net/ |
| BWA | Li and Durbin, 2010 | http://bio-bwa.sourceforge.net/ |
| ADMIXTOOLS | Patterson et al., 2012 | https://github.com/DReichLab/AdmixTools |
| SeqPrep | https://github.com/jstjohn/SeqPrep | https://github.com/jstjohn/SeqPrep |
| smartpca | Patterson et al., 2006 | https://www.hsph.harvard.edu/alkes-price/software/ |
| ADMIXTURE | Alexander et al., 2009 | http://dalexander.github.io/admixture/download.html |
| PMDtools | Skoglund et al., 2014 | https://github.com/pontussk/PMDtools |
| Haplogrep2 | Weissensteiner et al., 2016 | http://haplogrep.uibk.ac.at/index.html |
| ContamMix | Fu et al., 2013 | https://github.com/DReichLab/ADNA-Tools |
| ANGSD | Korneliussen et al., 2014 | https://github.com/ANGSD/angsd |
| mapDamage2.0 | Jónsson et al., 2013 | https://ginolhac.github.io/mapDamage/ |
| PHCP | This paper | https://github.com/ShamamW/PHCP |
| LINADMIX | This paper | liran.carmel@huji.ac.il |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
The 73 Bronze and Iron Age individuals newly reported in this work come from five archaeological sites in Israel and Jordan. Below, we provide details on these sites, and the individuals they harbored and that we analyzed.
Tel Megiddo
Tel Megiddo has been excavated by four expeditions, starting in the early 20th century. The site was inhabited from Neolithic times to the Persian period. Remains of over 30 settlements have been uncovered. Megiddo is a key site for the study of the Bronze and Iron Ages in the Levant and beyond. This is due to the combination of tight control over stratigraphy, ceramic typology and radiocarbon dating, and vast exposures of remains, including significant monuments. Megiddo was the hub of a Canaanite city-state in the Bronze and Iron I Ages and an administrative center of the biblical kingdom of Israel in the Iron II. It is mentioned in Egyptian and Assyrian texts (possibly also in one Hittite text) and in various associations in the Hebrew Bible. This is the location of Armageddon (a Greek corruption of the Hebrew har-Megiddo = the mound of Megiddo) of the Book of Revelations in the New Testament.
The samples discussed here, 35 altogether (Table S1), cover a long period of time, from the Intermediate Bronze Age (ca. 2500–2000 BCE) to the late Iron I (ca. 1000 BCE). With the exception of sample I4517 (late Iron I), all were retrieved from intramural burials scattered over the mound. The majority of the samples date to the Middle Bronze III-Late Bronze I (ca. 1650–1400 BCE). The Megiddo samples come from four excavation fields:
Area J: the cult compound of Megiddo, in the eastern sector of the mound;
Area K: domestic buildings, burials and fortifications in the southeastern sector of the mound;
Area H in the north: buildings (and burials) immediately to the west of the Middle Bronze III-Late Bronze palaces of Megiddo;
Area M: in the center of the mound, remains of a public Late Bronze building (palace?) and stone-built Late Middle Bronze/Early Late Bronze tombs.
Samples originated from the following layers (the letter designates the area and the number represents the stratum):
Level J-8 dating to the Middle Bronze I in the early second millennium BCE.
Level J-11/12 dating to the Middle Bronze II.
Level K-12, Middle Bronze II, domestic architecture, burials under a house near the brick-built fortification wall.
Level K-11, Middle Bronze III, same type of context as Level K-12.
Level K-10, Middle Bronze III-Late Bronze I, same type of context as Level K-12. Habitation terminated at the end of the Middle Bronze III, while burials continue in the Late Bronze I.
Level H-16, Middle Bronze III; the sample (I10771) comes from a monumental stone-built tomb, possibly associated with the palace to its east.
Level H-15, Late Bronze I. The sampled burial, with two individuals (I10769 and I10770), was found above the monumental tomb of Level H-16; those who dug it were probably aware of this association.
Level M-7, Late Bronze I; elaborate stone-built tombs nearby.
Level K-4, late Iron I, large courtyard house destroyed in big fire. The individual (I4517) seems to have perished during the devastation of the city.
Two of the outliers, the brother and sister (I2189, I2220), were buried next to each other, while the third outlier, I10100, was buried in the same grave—a stone-lined cist—as the sister, but in an earlier deposit in that grave.
Tel Abel Beth Maacah
Tel Abel Beth Maacah (Tell Abil el-Qameh) is a large site located in northern Israel, commanding several roads leading to the Lebanese inland Beka҅, the Phoenician coast and Damascus. The town of Abel is mentioned in several Egyptian 2nd millennium BCE sources and also three times in the bible, twice in relation to Aramean and Assyrian conquests in the 9th and 8th centuries BCE, as well as in the story about a rebellion against King David in the 10th century BCE.
Surveys and six seasons of excavation since 2012 have shown that the mound was occupied from the Early Bronze II-III (ca. 3000–2500 BCE) until modern times, with apparent hiatuses in the Middle Bronze I (ca. 1950–1750 BCE), Iron IIB-C (ca. 800–586 BCE) and parts of the Middle Ages. In the Middle Bronze II (ca. 1750–1650 BCE), the site was fortified and co-existed with other larger fortified sites nearby, including Dan and Hazor. During the Late Bronze Age, the town was apparently located mainly in the lower part of the mound, reusing the Middle Bronze fortifications, and probably was part of the kingdom of nearby Hazor. With the destruction of Hazor in the 13th century BCE, Abel Beth Maacah grew in size and occupation complexity, demonstrating one of the most intense sequences known in Iron Age I (12th mid-10th centuries BCE). At this time, the site included a large public complex with administrative, storage, industrial and cultic functions. A destruction at the end of the Iron I is radiocarbon-dated to the 10th century BCE. Iron IIA remains were built directly above this destruction, demonstrating substantial architecture, including a large citadel at the top of the mound in the north, radiocarbon-dated to the 9th century BCE.
The sample described here was extracted from a complete skeleton of an adult male excavated in Area O on the western slope of the lower mound. The grave was cut into a room belonging to a courtyard building dated to the Middle Bronze II. Nearby, in topsoil, a seal dated to Iron IIA (ca. 950–800 BCE) was found. Other pits cut into the building contained pottery that can be dated to this period, along with pottery from Iron I.
Tel Hazor
Hazor is the largest Bronze and Iron Age site in Israel, covering some 200 acres. The mound is composed of an upper mound (acropolis) adjoining a huge lower mound (lower city) to its north. Occupation began in the upper mound during the Early Bronze Age II (early third millennium BCE), while the lower city was founded in the Middle Bronze Age II (approximately the 18th century BCE). Both continued to be settled until a later phase of the Late Bronze Age (13th century), when the upper and lower cities were violently destroyed or abandoned. Following this destruction, only the upper part of the mound was resettled and fortified, becoming a major city in the 10th to 8th centuries BCE, as part of the Israelite kingdom.
Canaanite Hazor is mentioned on several occasions in ancient Near Eastern texts, the earliest being the Egyptian Execration texts of the 19th century BCE. Hazor is the only Canaanite site mentioned in the archive discovered in Mari in Syria (18th century BCE according to the middle chronology, or the 17th century BCE according to the ultra-low chronology). The Mari documents clearly demonstrate the importance, wealth and far-reaching commercial ties of Hazor. In the Amarna archive (14th century BCE) there are several references to Hazor, as well as in records of the military campaigns conducted by Egyptian pharaohs, during the 15th-14th centuries BCE.
Of the three individuals included in this study, one was found on the acropolis (I3965), in a cist grave with 2–3 individuals buried with a rich assemblage of goods. The tomb is related to Stratum pre-XVII, dated to the Middle Bronze I-II transition (ca. 1750 BCE). The two other samples were derived from burials in Area M, on the northern slope of the acropolis. Both contained young individuals or children. One (I3966) was buried beside the monumental Middle Bronze staircase; this burial too dates to the Middle Bronze I-II transition. The other was buried into a drainage channel (I3832) associated with the administrative palace of the Late Bronze Age and is dated to the Late Bronze II (ca. 14th century BCE).
Yehud
Tel Yehud (Tell el-Yehudia) is situated on the northeastern side of the Ono valley in the eastern part of the central coastal plain of Israel, ca. 12 km east of the Mediterranean Sea. Rescue excavations were carried out in 2008, followed by an excavation season in 2009 in Areas A and B, in the location of an underground parking lot. Archaeological findings at these areas include a deep shaft filled with refuse dated to the Chalcolithic Period, a cemetery from the Intermediate Bronze Age, a Late Roman-Byzantine pottery workshop, and Early Islamic cist graves.
The human remains analyzed in this study are of 13 individuals from the Middle Bronze Age I.
Baq҅ah
Human skeletal remains were excavated from the Baq҅ah Valley, Jordan, approximately 20 kilometers NW from Amman between 1977 and 1981 (McGovern, 1986). The skeletal materials spanned from the Late Bronze to the Iron Age (Caves A2, B3, and A4). Only materials from the Cave B3 context are included in these analyses. Skeletal materials from Cave A4 (Iron Age IA) were shipped to the Smithsonian Institution and the Penn Museum received the materials from A2 and B3 in 1982 from Jordan (University of Pennsylvania Museum Archival correspondence).
The tomb at Cave B3 (Late Bronze Age II) is one of a series of caves, and the only cave excavated. It is located on the lower slopes of Jebel al-Qeşīr. Human remains in B3 occur in 2 units: the lower and upper burial remains. The skeletal remains are disassociated and fragmentary; some human skeletal material is also burned/charred, as a result of local depositional history (alkaline soil, percolation of subsoil groundwater). The B3 collection (Finnegan et al., 1986) includes a minimum number estimate of 64 individuals.
Sampling took place at the University of Pennsylvania Museum in March 2015. All samples were derived from Cave B3 petrous temporals. 11 males and 10 female temporal bones were sampled. The age range estimate is from late adolescent to mature adult.
METHOD DETAILS
Data Generation
We prepared powder from skeletal remains in dedicated clean rooms. Of the samples that produced working data, all but one came from petrous bones, which are known to yield up to one hundred times more DNA than other skeletal elements (Gamba et al., 2014; Pinhasi et al., 2015, 2019). We extracted DNA using a method designed to retain short and highly degraded fragments (Dabney et al., 2013; Korlević et al., 2015; Skoglund et al., 2014), and built the DNA into individual barcoded double-stranded libraries in the presence of the enzyme Uracil–DNA–glycosylase (UDG) to reduce the rate of cytosine-to-thymine errors typical of ancient DNA (Rohland et al., 2014). We enriched the libraries for sequences overlapping the mitochondrial genome as well as about 1.2 million single nucleotide polymorphisms (SNPs), and sequenced on an Illumina NextSeq500 instrument using 2×76 base pair reads and 2×7 base pair indices. We assigned reads to samples based on the match rate to expected barcodes and indices. We merged read pairs that overlapped by at least 15 nucleotides (allowing up to one mismatch), and represented each nucleotide by the read giving higher quality data. We mapped merged sequences to either the mitochondrial reference genome rsrs (Behar et al., 2012; Weissensteiner et al., 2016) or to the human genome reference sequence hg19 using bwa (v.0.6.1) (Li and Durbin, 2010), and removed sequences that were duplicates as assessed by having the same barcode, as well as the same start and stop position when mapped to the reference genome. We determined the genotype of each SNP based on a randomly selected single read (tools used include (Fu et al., 2013; Jónsson et al., 2013; Korneliussen et al., 2014; Li, 2011; Li et al., 2009)).
Chemical and isotopic measurements
Tooth enamel was carefully separated from dentin, following the protocol described in Beherec et al., 2016. Bones and enamel samples were then cleaned using the methodology of Patterson and co-workers (Ericson et al., 1979; Patterson et al., 1991), which involved several steps of sample pre-cleaning and the use of clean laboratory techniques and reagents. Following the mechanical and chemical cleaning process, the samples were dissolved by concentrated, distilled HNO3 (25–100 mg sample in 1 ml HNO3). After sample dissolution, Sr concentrations were determined with an ICP-MS (Agilent 7500cx), following the protocol outlined in Beherec et al., 2016. Then, all samples were subjected to Strontium isotope analysis in order to trace migration of the individuals (Beherec et al., 2016; Bentley, 2006; Hartman and Richards, 2014; Horstwood et al., 2008; Perry, 2002; Perry et al., 2008, 2011). For this, the samples went through ion exchange columns according to the protocol outlined in Erel et al., 2006. Then, the samples were analyzed by a MC-ICP-MS (Thermo, NEPTUNE Plus) as described in Zipori et al., 2015. Replicate measurements of 87Sr/86Sr of SRM-987 standard over the course of this study yielded 87Sr/86Sr = 0.710273 ± 10 (), n = 29.
QUANTIFICATION AND STATISTICAL ANALYSES
PCA
For PCA plots we used SMARTPCA (Patterson et al., 2006).
ADMIXTURE
We ran ADMIXTURE (Alexander et al., 2009) on a set of 1,663 individuals from Europe, Western Asia, and Africa. For analyses solely involving ancient populations, we pruned SNPs that are in linkage disequilibrium using the indep-pairwise flag in PLINK (Purcell et al., 2007), with parameters 200, 25 and 0.5, as in Lazaridis et al., 2014 (Lazaridis et al., 2014). This gave a set of 357,334 SNPs, denoted here ‘ancient’. For analyses involving modern populations, we used the stricter pruning recommended in the ADMIXTURE manual, using the parameters 50, 10 and 0.1, and only using SNPs that are missing in fewer than 10% of the individuals. This resulted in a list of 50,165 SNPs denoted here as the ‘modern’ panel. To find an optimal we followed the default procedure of running 5-fold cross validation. For the ancient SNPs we tested values ranging from 4 to 11, and for the modern SNPs we tested values ranging from 4 to 14. We selected for both analyses (Figure S1A).
f4-statistics
We computed f-statistics using the package ADMIXTOOLS (Patterson et al., 2012). To test for homogeneity we ran f4-statistics and qpWave using the allsnps:YES parameter. To estimate the ancestry proportion for a test population given a set of source populations and a set of outgroups, we used the qpAdm methodology (Lazaridis et al., 2016) in ADMIXTOOLS with the allsnps:YES parameter.
LINADMIX
The ADMIXTURE algorithm (Alexander et al., 2009) takes as input a set of genotypes and assumes that each is a mixture of ancestral populations. For each genotype , it estimates the fraction contributed by ancestral population , denoted . As a result, ADMIXTURE may be viewed as representing a genotype as a vector of length ,
Similar to Leslie et al. (Kozlov et al., 2015; Leslie et al., 2015), we sought to utilize this representation of genotypes in order to find how a target genotype might be related to a list of n source genotypes . We do it by relating to a linear model, whereby we seek to model the target as a linear combination of the source populations,
Where the coefficients are called the mixing coefficients, and we required that they be nonnegative. Formally, the mixing coefficients are the solutions of the constrained non-negative least squares problem
subject to
where and is the matrix .
In many cases the target and source genotypes are populations rather than individuals. In these cases we take the vectors as the average across all individuals that belong to the ’th population. More formally, if population is made of the individuals , then
In order to find the standard error of the mixing coefficients we used parametric bootstrap. We sampled -vectors and computed the mixing coefficients for the sampled vectors. Given that is an individual that belongs to population , his/her bootstrap -vectors, , are computed by the following steps:
Estimation of the covariance matrix of the population, , representing variation between individuals in the population. For populations with a single individual we use the covariance matrix of another close population.
Estimation of the covariance matrix of the individual, representing variation in the vector estimated by ADMIXTURE due to the finite size of the genome. This matrix is approximated by the inverse of the empirical Fisher information matrix, , where is the empirical Fisher information matrix and is the ADMIXTURE output for the frequencies of the minor alleles in each theoretical ancestral population (see below for details).
Drawing a vector from .
Finally, we used the results of all bootstrap samples to calculate the standard error of the mixing coefficients.
The covariance matrix defined in step (2) above is computed as follows. In the ADMIXTURE model there is a large set of individuals , a large set of markers , and a much smaller set of theoretical ancestral populations. The genotype of individual at marker is , the number of occurrences of the minor allele at . The proportion of the genome of individual that comes from ancestral population is , which satisfies . The frequency of the minor allele at marker in ancestral population is . The ADMIXTURE output comprises the matrices and .
The joint log-likelihood of the parameters given the data is (up to constant terms)
Because there are only free parameters for each individual .
The empirical Fisher information matrix is calculated from the second-order partial derivatives with respect to the parameters. The first-order derivative with respect to is:
The derivative of this with respect to is 0 when . Otherwise, it is equal to
We calculated the empirical Fisher information matrix for single individuals. Therefore the empirical Fisher information for individual is given by
From large sample theory the vectors are asymptotically consistent estimators, are independent of each other, and are normally distributed with the covariance matrix estimated by .
In practice, we included in the bootstrap only ADMIXTURE ancestry components greater than 1%. Also, negative simulated bootstrap values were changed to zero and the sampled vectors were normalized to sum to one.
PHCP
We used a modified version of ChromoPainter (Lawson et al., 2012) to “paint” an ancient genome as an imperfect mosaic of modern “donor” haplotypes. ChromoPainter is based on the hidden Markov model (HMM) of Li and Stephens (Li and Stephens, 2003), where each SNP in the target sequence is “copied” from one haplotype (the hidden state) of the donors. Transitions between hidden states are due to ancestral recombination events, and the imperfection of the copying process is due to mutations/genotyping errors.
Our modified version includes changes in the hidden states of the HMM and hence also in the transition and emission probabilities. The modifications were needed because ChromoPainter requires phased data, whereas ancient DNA is typically sequenced/genotyped to very low coverage, and thus cannot be phased using standard tools. Additionally, in many ancient genomes (such as the genomes analyzed in this study), at most one allele is reported at each SNP, based on a randomly selected read. This results in a “pseudo-haploid” sequence that represents both haplotypes. To account for this, the hidden states in our modified HMM represent pairs of haplotypes instead of a single haplotype.
Formally, let denote the donor haplotypes, and let denote the ancient DNA sequence. We assume that each haplotype has sites. Let denote the vector of hidden states, where is an ordered pair of indices of haplotypes (, ) from which copies at site . Each pair of haplotypes has an equal a priori probability, , to be chosen as donors. The probability of a recombination event (in each haplotype) between sites and , is where is the genetic distance in Morgan between sites and , and is a parameter related to the effective population size. Hence, the transition probability between states and is:
The emission probability is the probability to observe a certain allele, A or B, at site of the ancient genome. It depends on the genotypes of the two haplotypes representing the hidden state and on a mutation rate parameter . This probability is calculated conditionally on which of the two copied haplotypes was sequenced (each with probability 0.5). Given the donor haplotype that was sequenced, we observe the same allele as in the donor haplotype with probability , and the other allele with probability . Thus,
Note that , the mutation rate, incorporates the effects of both mutations and sequencing errors. If there was no reported allele at site , we omitted this site from the analyzed sequence.
As in the original ChromoPainter, the forward-backward algorithm was used to compute the posterior probability of each SNP of to copy from each pair of donor haplotypes. The running time can be made linear in the number of states by adapting the “shortcut” offered by Li and Stephens (Li and Stephens, 2003). The expected total length of genetic material (in Morgan) copied from each possible pair of donor haplotypes was calculated based on Equation (S4) of Lawson et al. (Lawson et al., 2012). The expected length of genetic material copied from haplotype was calculated as the sum of the lengths of genetic material copied from all pairs or . The expected proportion of genetic material copied from a diploid individual was calculated as the expected lengths of genetic material copied from the two haplotypes of the individual, divided by the genome length. Finally, the average proportion of genetic material copied from a donor population was calculated as the average proportion over all individuals belonging to the popualtion.
Our initial application of the modified ChromoPainter showed that differences between the proportion of genetic material copied from each donor population were small, implying that the “copying profiles” cannot be directly interpreted as ancestry proportions (Methods S1F). To reduce the noise and obtain more interpretable results, we used the regression technique based on Leslie et al. (Leslie et al., 2015), as above for LINADMIX. In this approach, the copying profile of a target population (i.e., the average copying profile over all individuals in the population) is modeled as a linear combination of the copying profiles of a set of source populations. Formally, let be a set of distinct donor populations and let denote the source populations. Let denote the copying vector (of length ) of source population – the average proportion of the genome that individuals from population copy from each of the donor populations, as inferred by our modified ChromoPainter. The linear model is
such that for all , and (as for LINADMIX). is the copying vector of the target population and are the copying vectors of the source populations. We interpret as the average proportion of the ancestry of the target population coming from source population . We formulated the equations as a quadratic programming problem, using the quadprog package in R. For studying the ancestry of ancient and modern Middle Eastern populations, we chose the donor populations as a set of populations from Europe, Middle East, West Asia, and East Africa. We inferred the ChromoPainter parameters and with ChromoPainter’s EM algorithm on modern populations.
For practical details on refining the PHCP parameters, and on methods for selection of SNPs and donor populations, see Methods S1F.
DATA AND CODE AVAILABILITY
Genomic data first reported here is available at http://carmelab.huji.ac.il/data.html.
Raw and analyzed data from the 73 newly reported ancient samples were deposited at ENA: PRJEB37057.
Codes used for LINADMIX is available upon request from Liran Carmel (liran.carmel@huji.ac.il).
Code for PHCP is available from https://github.com/ShamamW/PHCP
Supplementary Material
Figure S1. Cross-validation errors in ADMIXTURE as a function of K. Related to Figure 1C. (A) Using 1,663 individuals. (Blue) ADMIXTURE was run on 357,334 SNPs according to the ancient samples protocol. (Orange) ADMIXTURE was run on 50,165 SNPs according to the present-day samples protocol. (B) Using 3,515 individuals. Only the present-day samples protocol was used.
Figure S2. Fraction of Chalcolithic Iran-related component in each individual as computed by qpAdm. Related to Figure 4. Modeling each individual as a mixture of Neolithic Levant and Chalcolithic Iran, and using either (A) the o9aensw outgroup set (o9 + Anatolia_N + EHG + Natufian + Switzerland_HG + WHG) or (B) the o9a outgroup set (o9 + Anatolia_N). Vertical error bars denote one standard error. Horizontal error bars denote estimated time ranges. Dashed line describes the linear regression. Only individuals whose time range does not exceed 250 years are plotted and used in the regression.
Figure S3. LINADMIX and PHCP results on ancient populations. Related to Figure 4 and to STAR Methods, LINADMIX, PHCP. (A) Comparison between qpAdm and LINADMIX. Bars show the fraction of Neolithic Levant in different populations. Error bars show one standard deviation. (B) LINADMIX of individual samples. Bars show the fraction of Neolithic Levant in different individuals, when the other source population is either Iran_ChL or Armenia_EBA. (C) Fraction of Levant_N in different ancient populations (when the other source population is Iran_ChL) as computed by LINADMIX and PHCP.
Figure S4. LINADMIX and PHCP models on modern populations. Related to Figure 5. (A) The contribution of each of the source populations to the examined present-day populations, using LINADMIX and PHCP. (B) LINADMIX and PHCP relative contribution of each of the source populations to the present-day target population listed on the x-axis.
Table S1. Overview of the 73 individuals newly reported in this study. Related to Figure 1. (we use commas to indicate by-library results in the case of four individuals where data are derived from multiple libraries.) (A) General information. (B) Archaeological information on the Tel Megiddo individuals. (C) Radiocarbon dating information. (D) Family relationships among individuals. In grey are individuals that were selected to represent the family in subsequent analyses
Table S2. P-values, mixing coefficients and standard errors as computed by qpAdm. Related to Figure 4. Testing different target populations as a mixture of Neolithic Levant (Levant N) and either Chalcolithic Iran (Iran_ChL) or Eearly Bronze Age Armenia (Armenia_EBA) using the outgroup sets of o9 and o9a (A) or o9nw, o9ensw and o9aensw (B). (C-M) testing different individuals from different populations as a mixture of Neolithic Levant (Levant N) and either Chalcolithic Iran (Iran_ChL) or Eearly Bronze Age Armenia (Armenia_EBA). The set of outgroups includes either o9a or o9aensw. (N) Modeling early Bronze Age Armenia (Armenia_EBA) as a mixture of Chalcolithic Armenia (Armenia_ChL) and Chalcolithic Iran (Iran_ChL). (O and P) Modeling the outliers from Megiddo I10100 (O) and I2200 (P) as different two-population admixtures. (Q) Testing different target populations as a mixture of Neolithic Levant (Levant N) and Middle-to-Late Bronze Age Armenia (Armenia_MLBA).
Table S3. Model norms, mixing coefficients and standard errors as computed by LINADMIX and PHCP. Related to Figure 4 and to STAR Methods, LINADMIX, PHCP. (A) Testing different target populations as a mixture of Neolithic Levant (Levant N) and either Chalcolithic Iran (Iran_ChL), Eearly Bronze Age Armenia (Armenia_EBA), Neolithic Iran (Iran_N), or Chalcolithic Armenia (Armenia_ChL). (B) LINADMIX results testing different target individuals as a mixture of Neolithic Levant (Levant N) and either Chalcolithic Iran (Iran_ChL) or Eearly Bronze Age Armenia (Armenia_EBA). (C) LINADMIX results of modeling the outliers from Megiddo as different two-population admixtures.
Table S4. Model norms, mixing coefficients and standard errors a testing different target present-day populations. Related to Figure 5. (A) LINADMIX and PHCP results when the target population are modeled as a mixture of Megiddo_MLBA, Iran_ChL, Somali and Europe_LNBA. (B) LINADMIX results when the target populations are Megiddo_Medium, Iran_ChL, Somali and Europe_LNBA. (C) LINADMIX results on modern populations with some ADMIXTURE perturbations. (D) Model norms of LINADMIX, testing different target present-day populations as a mixture of Iran_ChL, Somali, Europe_LNBA and a fourth Anatolian component as indicated in the table.
Table S5. P-values, mixing coefficients and standard errors testing different subpopulations from Megiddo as a mixture of Neolithic Levant (Levant N) and either Chalcolithic Iran (Iran_ChL) or Early Bronze Age Armenia (Armenia_EBA). Related to Figure 5. (A) computed by qpAdm. (B) computed by LINADMIX.
Highlights.
Analysis of genome-wide data for nine sites from the Bronze Age Southern Levant.
Contemporaneous samples from multiple sites are genetically similar.
Migration from the Zagros/Caucasus to the Levant between 2500–1000 BCE.
People related to these individuals contributed to all present-day Levantine populations.
Acknowledgements
We thank Reuven Amitai, Menachem Ben-Sasson, Tom Booth, Pontus Skoglund, Philipp Stockhammer, Jason Ur and several anonymous reviewers for critical comments. This study was funded by the Israel Science Foundation (ISF grant 1009/17 to L.C. and B.Y.; ISF grant 407/17 to S.C.) D.R. is an Investigator of the Howard Hughes Medical Institute and his ancient DNA laboratory work was supported by National Science Foundation HOMINID grant BCS-1032255, by National Institutes of Health grant GM100233, by an Allen Discovery Center grant, and by grant 61220 from the John Templeton Foundation. Work at Megiddo is supported by the Dan David Foundation, the Shmunis Family Foundation, Mark Weismann and Vivian and Norman Belmonte.
Footnotes
Declaration of Interests
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alexander DH, Novembre J, and Lange K. (2009). Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19, 1655–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allentoft ME, Sikora M, Sjögren K-G, Rasmussen S, Rasmussen M, Stenderup J, Damgaard PB, Schroeder H, Ahlström T, Vinner L, et al. (2015). Population genomics of Bronze Age Eurasia. Nature 522, 167–172. [DOI] [PubMed] [Google Scholar]
- Atzmon G, Hao L, Pe’er I, Velez C, Pearlman A, Palamara PF, Morrow B, Friedman E, Oddoux C, Burns E, et al. (2010). Abraham’s Children in the Genome Era: Major Jewish Diaspora Populations Comprise Distinct Genetic Clusters with Shared Middle Eastern Ancestry. Am. J. Hum. Genet 86, 850–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behar DM, van Oven M, Rosset S, Metspalu M, Loogväli E-L, Silva NM, Kivisild T, Torroni A, and Villems R. (2012). A "Copernican" reassessment of the human mitochondrial DNA tree from its root. Am. J. Hum. Genet 90, 675–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beherec MA, Levy TE, Tirosh O, Najjar M, Knabb KA, and Erel Y. (2016). Iron Age Nomads and their relation to copper smelting in Faynan (Jordan): Trace metal and Pb and Sr isotopic measurements from the Wadi Fidan 40 cemetery. J. Archaeol. Sci 65, 70–83. [Google Scholar]
- Bentley RA (2006). Strontium Isotopes from the Earth to the Archaeological Skeleton: A Review. J. Archaeol. Method Theory 13. [Google Scholar]
- Bienkowski P. (1999). Jonathan N. Tubb. Canaanites (Peoples of the Past). 160 pages, 18 colour, 106 black-and-white illustrations. 1998 London: British Museum Press; 0–7141-2089–8 hardback £20. Antiquity 73, 708–709. [Google Scholar]
- van den Brink ECM, Beeri R, Kirzner D, Bron E, Cohen-Weinberger A, Kamaisky E, Gonen T, Gershuny L, Nagar Y, Ben-Tor D, et al. (2017). A Late Bronze Age II clay coffin from Tel Shaddud in the Central Jezreel Valley, Israel: context and historical implications. Levant 49, 105–135. [Google Scholar]
- Carmi S, Hui KY, Kochav E, Liu X, Xue J, Grady F, Guha S, Upadhyay K, Ben-Avraham D, Mukherjee S, et al. (2014). Sequencing an Ashkenazi reference panel supports population-targeted personal genomics and illuminates Jewish and European origins. Nat. Commun 5, 4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline EH (2014). 1177 B.C. : the Year Civilization Collapsed (Princeton: Princeton University Press; ). [Google Scholar]
- Dabney J, Meyer M, and Paabo S. (2013). Ancient DNA Damage. Cold Spring Harb. Perspect. Biol 5, a012567–a012567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenmann S, Bánffy E, van Dommelen P, Hofmann KP, Maran J, Lazaridis I, Mittnik A, McCormick M, Krause J, Reich D, et al. (2018). Reconciling material cultures in archaeology with genetic data: The nomenclature of clusters emerging from archaeogenomic analysis. Sci. Rep 8, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ericson JE, Shirahata H, and Patterson CC (1979). Skeletal Concentrations of Lead in Ancient Peruvians. N. Engl. J. Med 300, 946–951. [DOI] [PubMed] [Google Scholar]
- Feldman M, Master DM, Bianco RA, Burri M, Stockhammer PW, Mittnik A, Aja AJ, Jeong C, and Krause J. (2019). Ancient DNA sheds light on the genetic origins of early Iron Age Philistines. Sci. Adv 5, eaax0061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan M, Husted JJ, Rolston SL, and Saul MB (1986). The human skeletal remains. In The Late Bronze and Early Iron Ages of Central Transjordan: The Baqàh Valley Project, 1977–1981, McGovern PE, ed. (Philadelphia, PA: University Museum Monograph 65. The University Museum Press; ), pp. 295–314. [Google Scholar]
- Fu Q, Meyer M, Gao X, Stenzel U, Burbano H. a, Kelso, J., and Pääbo, S. (2013). DNA analysis of an early modern human from Tianyuan Cave, China. Proc. Natl. Acad. Sci. U.S.A 110, 2223–2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamba C, Jones ER, Teasdale MD, McLaughlin RL, Gonzalez-Fortes G, Mattiangeli V, Domboróczki L, Kővári I, Pap I, Anders A, et al. (2014). Genome flux and stasis in a five millennium transect of European prehistory. Nat. Commun 5, 5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goren Y, Finkelstein I, Naʼaman N, Artzy M, and Makhon le-arkheʼologyah ʻa. sh. Nadler Sonyah u-Marḳo. (2004). Inscribed in clay : provenance study of the Amarna tablets and other ancient Near Eastern texts (Emery and Claire Yass Publications in Archaeology; ). [Google Scholar]
- Greenberg R, and Goren Y. (2009). Transcaucasian Migrants and the Khirbet Kerak Culture in the Third Millennium BCE (Tel-Aviv). [Google Scholar]
- Haak W, Lazaridis I, Patterson N, Rohland N, Mallick S, Llamas B, Brandt G, Nordenfelt S, Harney E, Stewardson K, et al. (2015). Massive migration from the steppe was a source for Indo-European languages in Europe. Nature 522, 207–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber M, Doumet-Serhal C, Scheib C, Xue Y, Danecek P, Mezzavilla M, Youhanna S, Martiniano R, Prado-Martinez J, Szpak M, et al. (2017). Continuity and Admixture in the Last Five Millennia of Levantine History from Ancient Canaanite and Present-Day Lebanese Genome Sequences. Am. J. Hum. Genet 101, 274–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harney É, May H, Shalem D, Rohland N, Mallick S, Lazaridis I, Sarig R, Stewardson K, Nordenfelt S, Patterson N, et al. (2018). Ancient DNA from Chalcolithic Israel reveals the role of population mixture in cultural transformation. Nat. Commun 9, 3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman G, and Richards M. (2014). Mapping and defining sources of variability in bioavailable strontium isotope ratios in the Eastern Mediterranean. Geochim. Cosmochim. Acta 126, 250–264. [Google Scholar]
- Horstwood MSA, Evans JA, and Montgomery J. (2008). Determination of Sr isotopes in calcium phosphates using laser ablation inductively coupled plasma mass spectrometry and their application to archaeological tooth enamel. Geochim. Cosmochim. Acta 72, 5659–5674. [Google Scholar]
- Jónsson H, Ginolhac A, Schubert M, Johnson PLF, and Orlando L. (2013). mapDamage2.0: fast approximate Bayesian estimates of ancient DNA damage parameters. Bioinformatics 29, 1682–1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korlević P, Gerber T, Gansauge M-T, Hajdinjak M, Nagel S, Aximu-Petri A, and Meyer M. (2015). Reducing microbial and human contamination in DNA extractions from ancient bones and teeth. Biotechniques 59, 87–93. [DOI] [PubMed] [Google Scholar]
- Korneliussen TS, Albrechtsen A, and Nielsen R. (2014). ANGSD: Analysis of Next Generation Sequencing Data. BMC Bioinformatics 15, 356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov K, Chebotarev D, Hassan M, Triska M, Triska P, Flegontov P, and Tatarinova TV (2015). Differential Evolution approach to detect recent admixture. BMC Genomics 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson DJ, Hellenthal G, Myers S, and Falush D. (2012). Inference of population structure using dense haplotype data. PLoS Genet. 8, e1002453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson DJ, van Dorp L, and Falush D. (2018). A tutorial on how not to over-interpret STRUCTURE and ADMIXTURE bar plots. Nat. Commun 9, 3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaridis I, Patterson N, Mittnik A, Renaud G, Mallick S, Sudmant PH, Schraiber JG, Castellano S, Kirsanow K, Economou C, et al. (2014). Ancient human genomes suggest three ancestral populations for present-day Europeans. Nature 513, 409–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaridis I, Nadel D, Rollefson G, Merrett DCC, Rohland N, Mallick S, Fernandes D, Novak M, Gamarra B, Sirak K, et al. (2016). Genomic insights into the origin of farming in the ancient Near East. Nature 536, 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaridis I, Mittnik A, Patterson N, Mallick S, Rohland N, Pfrengle S, Furtwängler A, Peltzer A, Posth C, Vasilakis A, et al. (2017). Genetic origins of the Minoans and Mycenaeans. Nature 548, 214–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemche NP (1991). The Canaanites and their land : the tradition of the Canaanites (Sheffield: JSOT Press; ). [Google Scholar]
- Leslie S, Winney B, Hellenthal G, Davison D, Boumertit A, Day T, Hutnik K, Royrvik EC, Cunliffe B, Lawson DJ, et al. (2015). The fine-scale genetic structure of the British population. Nature 519, 309–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. (2011). A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27, 2987–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, and Durbin R. (2010). Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, and Stephens M. (2003). Modeling linkage disequilibrium and identifying recombination hotspots using single-nucleotide polymorphism data. Genetics 165, 2213–2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, and Durbin R. (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin MAS, Finkelstein I, and Piasetzky E. (2020). Radiocarbon dating the Late Bronze Age: Cultural and Historical Considerations on Megiddo and Beyond. Bull. Am. Sch. Orient. Res in press. [Google Scholar]
- Mazar A. (1992). Archaeology of the land of the Bible. [Google Scholar]
- McGovern P. (1986). The late bronze and early iron ages of central Transjordan, the Baqʻah Valley project, 1977–1981 (Philadelphia, PA: University Museum, University of Pennsylvania; ). [Google Scholar]
- de Miroschedji P. (2014). The Southern Levant (Cisjordan) during the Early Bronze Age. In The Oxford Handbook of the Archaeology of the Levant, c. 8000–332 BCE, Steiner ML, and Killebrew AE, eds. (Oxford University Press; ), p. Ch. 22. [Google Scholar]
- Na’aman N. (1994a). The Canaanites and Their Land. In Ugarit-Forschungen 26, pp. 397–418. [Google Scholar]
- Na’aman N. (1994b). The Hurrians and the End of the Middle Bronze Age in Palestine. Levant 26, 175–187. [Google Scholar]
- Olalde I, Brace S, Allentoft ME, Armit I, Kristiansen K, Booth T, Rohland N, Mallick S, Szécsényi-Nagy A, Mittnik A, et al. (2018). The Beaker phenomenon and the genomic transformation of northwest Europe. Nature 555, 190–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olalde I, Mallick S, Patterson N, Rohland N, Villalba-Mouco V, Silva M, Dulias K, Edwards CJ, Gandini F, Pala M, et al. (2019). The genomic history of the Iberian Peninsula over the past 8000 years. Science (80-. ). 363, 1230–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson C, Ericson J, Manea-Krichten M, and Shirahata H. (1991). Natural skeletal levels of lead in Homo sapiens sapiens uncontaminated by technological lead. Sci. Total Environ 107, 205–236. [DOI] [PubMed] [Google Scholar]
- Patterson N, Price AL, and Reich D. (2006). Population structure and eigenanalysis. PLoS Genet. 2, 2074–2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson N, Moorjani P, Luo Y, Mallick S, Rohland N, Zhan Y, Genschoreck T, Webster T, and Reich D. (2012). Ancient admixture in human history. Genetics 192, 1065–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry MA (2002). Life and Death in Nabataea: The North Ridge Tombs and Nabataean Burial Practices. Near East. Archaeol 65, 265. [Google Scholar]
- Perry MA, Coleman D, and Delhopital N. (2008). Mobility and exile at 2nd century A.D. khirbet edh-dharih: Strontium isotope analysis of human migration in western Jordan. Geoarchaeology 23, 528–549. [Google Scholar]
- Perry MA, Coleman DS, Dettman DL, Grattan JP, and Halim al-Shiyab A. (2011). Condemned to metallum? The origin and role of 4th–6th century A.D. Phaeno mining campresidents using multiple chemical techniques. J. Archaeol. Sci 38, 558–569. [Google Scholar]
- Pinhasi R, Fernandes D, Sirak K, Novak M, Connell S, Alpaslan-Roodenberg S, Gerritsen F, Moiseyev V, Gromov A, Raczky P, et al. (2015). Optimal ancient DNA yields from the inner ear part of the human petrous bone. PLoS One 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinhasi R, Fernandes DM, Sirak K, and Cheronet O. (2019). Isolating the human cochlea to generate bone powder for ancient DNA analysis. Nat. Protoc 14, 1194–1205. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, et al. (2007). PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet 81, 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich D, Patterson N, Campbell D, Tandon A, Mazieres S, Ray N, Parra MV, Rojas W, Duque C, Mesa N, et al. (2012). Reconstructing Native American population history. Nature 488, 370–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohland N, Harney E, Mallick S, Nordenfelt S, and Reich D. (2014). Partial uracil-DNA-glycosylase treatment for screening of ancient DNA. Philos. Trans. R. Soc. B Biol. Sci 370, 20130624–20130624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeter DJ (2008). The Shifting Boundaries of Moroccan Jewish Identities. In Jewish Social Studies, (Conference on Jewish Relations), pp. 145–164. [Google Scholar]
- Skoglund P, Northoff BH, Shunkov MV, Derevianko AP, Pääbo S, Krause J, and Jakobsson M. (2014). Separating endogenous ancient DNA from modern day contamination in a Siberian Neandertal. Proc. Natl. Acad. Sci. U. S. A 111, 2229–2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toffolo MB, Arie E, Martin MAS, Boaretto E, and Finkelstein I. (2014). Absolute Chronology of Megiddo, Israel, in the Late Bronze and Iron Ages: High-Resolution Radiocarbon Dating. Radiocarbon 56, 221–244. [Google Scholar]
- Wang J, Raskin L, Samuels DC, Shyr Y, and Guo Y. (2015). Genome measures used for quality control are dependent on gene function and ancestry. Bioinformatics 31, 318–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissensteiner H, Pacher D, Kloss-Brandstätter A, Forer L, Specht G, Bandelt H-J, Kronenberg F, Salas A, and Schönherr S. (2016). HaploGrep 2: mitochondrial haplogroup classification in the era of high-throughput sequencing. Nucleic Acids Res. 44, W58–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasur-Landau A. (2010). The Philistines and Aegean Migration at the End of the Late Bronze Age (Cambridge University Press; ). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Cross-validation errors in ADMIXTURE as a function of K. Related to Figure 1C. (A) Using 1,663 individuals. (Blue) ADMIXTURE was run on 357,334 SNPs according to the ancient samples protocol. (Orange) ADMIXTURE was run on 50,165 SNPs according to the present-day samples protocol. (B) Using 3,515 individuals. Only the present-day samples protocol was used.
Figure S2. Fraction of Chalcolithic Iran-related component in each individual as computed by qpAdm. Related to Figure 4. Modeling each individual as a mixture of Neolithic Levant and Chalcolithic Iran, and using either (A) the o9aensw outgroup set (o9 + Anatolia_N + EHG + Natufian + Switzerland_HG + WHG) or (B) the o9a outgroup set (o9 + Anatolia_N). Vertical error bars denote one standard error. Horizontal error bars denote estimated time ranges. Dashed line describes the linear regression. Only individuals whose time range does not exceed 250 years are plotted and used in the regression.
Figure S3. LINADMIX and PHCP results on ancient populations. Related to Figure 4 and to STAR Methods, LINADMIX, PHCP. (A) Comparison between qpAdm and LINADMIX. Bars show the fraction of Neolithic Levant in different populations. Error bars show one standard deviation. (B) LINADMIX of individual samples. Bars show the fraction of Neolithic Levant in different individuals, when the other source population is either Iran_ChL or Armenia_EBA. (C) Fraction of Levant_N in different ancient populations (when the other source population is Iran_ChL) as computed by LINADMIX and PHCP.
Figure S4. LINADMIX and PHCP models on modern populations. Related to Figure 5. (A) The contribution of each of the source populations to the examined present-day populations, using LINADMIX and PHCP. (B) LINADMIX and PHCP relative contribution of each of the source populations to the present-day target population listed on the x-axis.
Table S1. Overview of the 73 individuals newly reported in this study. Related to Figure 1. (we use commas to indicate by-library results in the case of four individuals where data are derived from multiple libraries.) (A) General information. (B) Archaeological information on the Tel Megiddo individuals. (C) Radiocarbon dating information. (D) Family relationships among individuals. In grey are individuals that were selected to represent the family in subsequent analyses
Table S2. P-values, mixing coefficients and standard errors as computed by qpAdm. Related to Figure 4. Testing different target populations as a mixture of Neolithic Levant (Levant N) and either Chalcolithic Iran (Iran_ChL) or Eearly Bronze Age Armenia (Armenia_EBA) using the outgroup sets of o9 and o9a (A) or o9nw, o9ensw and o9aensw (B). (C-M) testing different individuals from different populations as a mixture of Neolithic Levant (Levant N) and either Chalcolithic Iran (Iran_ChL) or Eearly Bronze Age Armenia (Armenia_EBA). The set of outgroups includes either o9a or o9aensw. (N) Modeling early Bronze Age Armenia (Armenia_EBA) as a mixture of Chalcolithic Armenia (Armenia_ChL) and Chalcolithic Iran (Iran_ChL). (O and P) Modeling the outliers from Megiddo I10100 (O) and I2200 (P) as different two-population admixtures. (Q) Testing different target populations as a mixture of Neolithic Levant (Levant N) and Middle-to-Late Bronze Age Armenia (Armenia_MLBA).
Table S3. Model norms, mixing coefficients and standard errors as computed by LINADMIX and PHCP. Related to Figure 4 and to STAR Methods, LINADMIX, PHCP. (A) Testing different target populations as a mixture of Neolithic Levant (Levant N) and either Chalcolithic Iran (Iran_ChL), Eearly Bronze Age Armenia (Armenia_EBA), Neolithic Iran (Iran_N), or Chalcolithic Armenia (Armenia_ChL). (B) LINADMIX results testing different target individuals as a mixture of Neolithic Levant (Levant N) and either Chalcolithic Iran (Iran_ChL) or Eearly Bronze Age Armenia (Armenia_EBA). (C) LINADMIX results of modeling the outliers from Megiddo as different two-population admixtures.
Table S4. Model norms, mixing coefficients and standard errors a testing different target present-day populations. Related to Figure 5. (A) LINADMIX and PHCP results when the target population are modeled as a mixture of Megiddo_MLBA, Iran_ChL, Somali and Europe_LNBA. (B) LINADMIX results when the target populations are Megiddo_Medium, Iran_ChL, Somali and Europe_LNBA. (C) LINADMIX results on modern populations with some ADMIXTURE perturbations. (D) Model norms of LINADMIX, testing different target present-day populations as a mixture of Iran_ChL, Somali, Europe_LNBA and a fourth Anatolian component as indicated in the table.
Table S5. P-values, mixing coefficients and standard errors testing different subpopulations from Megiddo as a mixture of Neolithic Levant (Levant N) and either Chalcolithic Iran (Iran_ChL) or Early Bronze Age Armenia (Armenia_EBA). Related to Figure 5. (A) computed by qpAdm. (B) computed by LINADMIX.
Data Availability Statement
Genomic data first reported here is available at http://carmelab.huji.ac.il/data.html.
Raw and analyzed data from the 73 newly reported ancient samples were deposited at ENA: PRJEB37057.
Codes used for LINADMIX is available upon request from Liran Carmel (liran.carmel@huji.ac.il).
Code for PHCP is available from https://github.com/ShamamW/PHCP