Abstract
The previously characterized monoclonal antibodies (MAbs) A1, A69, B1, and A20 are directed against assembled or nonassembled adeno-associated virus type 2 (AAV-2) capsid proteins (A. Wistuba, A. Kern, S. Weger, D. Grimm, and J. A. Kleinschmidt, J. Virol. 71:1341–1352, 1997). Here we describe the linear epitopes of A1, A69, and B1 which reside in VP1, VP2, and VP3, respectively, using gene fragment phage display library, peptide scan, and peptide competition experiments. In addition, MAbs A20, C24-B, C37-B, and D3 directed against conformational epitopes on AAV-2 capsids were characterized. Epitope sequences on the capsid surface were identified by enzyme-linked immunoabsorbent assay using AAV-2 mutants and AAV serotypes, peptide scan, and peptide competition experiments. A20 neutralizes infection following receptor attachment by binding an epitope formed during AAV-2 capsid assembly. The newly isolated antibodies C24-B and C37-B inhibit AAV-2 binding to cells, probably by recognizing a loop region involved in binding of AAV-2 to the cellular receptor. In contrast, binding of D3 to a loop near the predicted threefold spike does not neutralize AAV-2 infection. The identified antigenic regions on the AAV-2 capsid surface are discussed with respect to their possible roles in different steps of the viral life cycle.
Adeno-associated viruses (AAVs) are small, icosahedral viruses of the Parvoviridae family with a capsid of 20 to 25 nm in diameter. The capsid harbors a linear, single-stranded DNA genome of 4.7 kb which contains two open reading frames flanked by inverted terminal repeats. The left and right open reading frames encode four nonstructural proteins (Rep78, Rep68, Rep52, and Rep40) and three structural proteins (VP1, VP2, and VP3), respectively (for a review, see reference 5). The three overlapping capsid proteins differ only in their N-terminal sequences and have molecular masses of 90 kDa (VP1), 72 kDa (VP2), and 60 kDa (VP3). VP1, VP2, and VP3 assemble in the nucleus (52, 53) into mature virions in a 1:1:20 stoichiometry (33). Capsid assembly can occur independently of VP1 (36), but VP1 is essential for formation of infectious AAV type 2 (AAV-2) particles (17, 42, 50). VP2 cotransports VP3 into the nucleus before capsid assembly (18, 36). VP3 alone also forms capsids but only when targeted to the nucleus (18). Encapsidation of the AAV-2 genome likely occurs in the nucleoplasm in areas where capsids, Rep proteins, and DNA colocalize (52). Detailed analysis of the protein-protein interactions of Rep and VP proteins favors a model by which Rep-tagged DNA initiates packaging by interaction with capsid proteins (11). Several of the above-mentioned studies of the AAV-2 capsid assembly process were aided by using monoclonal antibodies (MAbs) directed against the capsid proteins.
AAV-2 infects a broad range of cells by binding to its primary receptor, heparan sulfate proteoglycan (47). Two types of coreceptors, αvβ5 integrin and fibroblast growth factor receptor 1, have been implicated in the subsequent internalization process (27, 46). However, conflicting results have raised doubts about the general role of these coreceptors in the AAV-2 infection process (28, 29). Analysis of insertion mutants of AAV-2 capsids suggests that the heparin binding site resides within the VP3 portion of the capsid proteins (30). After binding, AAV-2 enters the cell by a dynamin-dependent endosomal pathway (4, 10). Acidification of endosomes leads to the release of AAV-2 particles after which they move rapidly through the cytosol toward the nucleus and accumulate at a perinuclear site followed by slow entry of capsids into the nucleus (4). However, none of the sequences on the capsid surface involved in attachment or subsequent entry steps have been determined.
The study of the basic biology of AAV-2 has been accelerated due to the increased use of AAV-derived viral vectors in gene therapy applications. The advantages of AAV-2 vectors are based on the nonpathogenic nature of the wild-type (wt) virus, the ability to infect dividing and nondividing cells, and the establishment of long-term expression of heterologous genes by recombinant AAV (for reviews, see references 12, 20, 24, and 37). One avenue for improvement of these vector systems is targeting of recombinant particles to nonpermissive cells. Initial attempts using either genetic or immunologic modifications of the capsid look promising (3, 13, 55). However, the use of well-characterized antibodies binding to and possibly preventing the native tropism of AAV-2 capsids is a critical parameter in the immunologic approach. Another important consideration of using this vector system is the high prevalence of anti-AAV-2 antibodies in humans. Many of these antibodies are neutralizing (6, 8, 22). Several mechanisms of neutralization have been described (for a review, see reference 41): (i) interference with receptor attachment (19, 25), (ii) inhibition of uncoating (23), (iii) induction of structural changes in the capsid (51), and (iv) interparticle cross-linking (aggregation). In an effort to study the effect of neutralizing antibodies on readministration, Moskalenko et al. (22) characterized immunogenic epitopes on the AAV-2 capsid surface recognized by polyclonal antibodies in human serum. The identification of neutralizing epitopes could aid the development of less immunogenic vectors. In related autonomous parvoviruses, highly accessible regions of the capsid form antigenic sites which are generally found (i) on the shoulder of the threefold spike, (ii) between the twofold depression and the threefold spike, and (iii) near the center of the threefold spike (7). These capsid structures are in part formed by the G-H loop which is a general region of antigenicity (7).
In this report, we use the characteristics of previously described MAbs (A1, A69, B1, and A20) and newly generated MAbs (C24-B, C37-B, and D3) to study early steps of the viral life cycle. The antibodies C24-B and C37-B inhibit AAV-2 binding to cells, while A20 neutralizes AAV-2 infection at a postbinding step. The antibody D3 has no neutralizing ability. These data in combination with the determination of linear or conformational epitopes of six of these antibodies using gene fragment phage display and/or peptide scanning and peptide competition experiments led to the proposal of antigenic regions on the AAV-2 capsid involved in receptor binding and neutralization.
MATERIALS AND METHODS
Cell culture and virus stocks.
HeLa, 293, 293T, and GMK cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum and penicillin-streptomycin. The AAV-1 stock was generated on 293 cells, the AAV-4 stock was generated on GMK cells and AAV-2, -3, and -5 stocks were generated on HeLa cells. Cells were coinfected with AAV-1, -3, or -5 and adenovirus type 2 (Ad2) at a multiplicity of infection (MOI) of 1 and incubated for 5 days. AAV-2 stocks were prepared by coinfection of AAV-2 (MOI = 1) and Ad5 (MOI = 2) for 2 days. The AAV-4 stock used for propagation on GMK cells already contained the simian helper virus SV15. All AAV serotypes were CsCl purified using standard protocols (43) and dialyzed against phosphate-buffered saline (PBS). AAV-2 insertion mutants and empty AAV-2 particles were generated as previously described (in references 13 and 44, respectively). Recombinant AAV-2 particles containing the green fluorescent protein (GFP) reporter gene (rAAV-2-GFP) were prepared by transfection of plasmids pDG and pTRUF3 on 293T cells as previously described (15).
Generation of MAbs and purification of antibodies and Fab fragments.
The generation of MAbs A1, A69, B1, and A20 has been described previously (53). The same procedure was used to generate MAbs D3, C24-B, and C37-B with the following modifications. For D3, CsCl-purified AAV-2 particles were used for subcutaneous immunization (50 μg) and two booster injections (25 μg) and the resulting hybridoma supernatants were screened by an AAV-2 capsid ELISA. C24-B and C37-B were generated to isolate MAbs, which inhibit AAV-2 binding to cells. Therefore, a subcutaneous immunization and booster injection were performed with 200 μg of each synthetic peptide. The peptides were selected on the basis of sequence differences between the AAV-2 and AAV-3 capsid genes, which do not compete for the same receptor (21). The peptides had the following amino acid sequences: AAV2-1, GPPPPKPAERHKDDS (amino acids [aa] 28 to 42 in VP1); AAV2-2, SRTNTPSGTTTQSRLQFSQAGASDIRDQS (aa 446 to 474 in VP1); AAV2-3, QSGVLIFGKQGSEKTNVDIEK (aa 536 to 556 in VP1); and AAV2-4, SVSTNLQRGNRQAATADVNTQ (aa 578 to 598 in VP1). Two additional booster injections followed, each with 30 μg of purified empty AAV-2 particles to account for possible nonlinear epitopes. The resulting hybridoma supernatants were screened in a nonradioactive binding assay (see below).
Each MAb was purified from hybridoma supernatant by affinity chromatography on protein G Sepharose columns (Amersham Pharmacia, Braunschweig, Germany) according to the manufacturer's protocol. Fab fragments were generated and isolated using the ImmunoPure Fab purification kit (Pierce, Rockford, Ill.).
Neutralization assay and AAV-2 binding assay.
To determine the neutralizing capability of individual antibodies, rAAV-2-GFP particles (MOI = 10 transducing units) were incubated with the respective hybridoma supernatant at a final dilution of 1:10 on HeLa cells. GFP was assayed 20 to 22 h postinfection by fluorescence microscopy. In the presence of nonneutralizing antibodies, up to 0.5% of cells were GFP positive in the absence of Ad.
Inhibition of AAV-2 binding to HeLa cells by antibodies was determined in a nonradioactive binding assay. HeLa cells (104 cells/well) were grown in 96-well plates overnight, and unspecific binding sites were blocked by incubation with 0.2% casein in PBS for 90 min at 37°C. Hybridoma supernatant (final dilution, 1:10) was preincubated with 109 AAV-2 particles for 30 min on ice in HEPES-buffered saline containing 1% bovine serum albumin before a 90-min incubation with HeLa cells at 4°C was performed. Cells were washed with PBS and fixed in methanol for 15 min at −20°C, and bound AAV-2 particles were detected by an A20 enzyme-linked immunosorbent assay (ELISA) (14).
Western blot analysis, immunofluorescence microscopy, and immunoprecipitations.
For Western blot analysis, extracts of HeLa cells infected with AAV-2 and Ad5 were separated on a sodium dodecyl sulfate–15% polyacrylamide gel (48) and electrophoretically transferred onto a nitrocellulose membrane (16). Incubation with hybridoma supernatant was performed, and proteins were visualized with an alkaline phosphatase-coupled secondary antibody, following established protocols (16). Immunofluorescence analysis of hybridoma supernatants on AAV-2-Ad5- or Ad5-infected HeLa cells was performed as previously described (52). To analyze the ability of several MAbs to inhibit AAV-2 binding to cells, purified empty AAV-2 capsids were labeled with FITC (fluorescein isothiocyanate) (Sigma-Aldrich Chemie, Deisenhofen, Germany) using standard protocols (16). FITC-labeled capsids (109 particles) were preincubated with 0.1 μg of antibody in PBS containing 1% bovine serum albumin for 25 min at room temperature. To prevent internalization of capsids, labeled capsids were incubated with HeLa cells grown on coverslips in the presence or absence of competing antibody for 30 min at 4°C. Cells were fixed and visualized as described previously (52). Immunoprecipitation experiments were performed as described previously (52).
ELISA.
Binding of MAbs to AAV serotypes and AAV-2 insertion mutants was quantified in an ELISA. Titers of determined CsCl-purified particles of AAV serotypes and AAV-2 insertion mutants were determined by counting of negatively stained particles in electron micrographs (14). Particles were diluted in PBS, and equal amounts (8 × 107 particles/well) were used to coat the wells of microtiter plates (Polysorb; Nunc Nalgene International, Roskilde, Denmark) overnight at 4°C. After blocking, hybridoma supernatants diluted in PBS containing 0.05% Tween 20 were incubated for 1 h at 37°C, followed by another hour of incubation with a biotinylated anti-mouse secondary antibody (Amersham Pharmacia). Detection and quantification were performed as described previously (14).
Epitope mapping.
A VP1-VP2-VP3 gene fragment phage display library was constructed (40) with the modifications described previously (26). The library contained 3 × 105 primary transformants. Phages were amplified and used for biopanning with the different antibodies as described previously (26) with some modifications. For the first round of panning, 10 μg of biotinylated A1, A69, or B1 was coated and incubated overnight with 7 × 108 transforming units. After a second round of amplification, 1.5 μg of biotinylated antibody and all phages recovered from the first round of panning (1011 transforming units) were used for biopanning in solution. Immunopositive clones were further characterized by sequencing using the T7 sequencing system (Amersham Pharmacia).
For fine mapping of epitopes by peptide scanning, overlapping 10- or 15-mer peptides offset by 2 aa were synthesized on activated membranes using the SPOT system (Genosys, Cambridge, England) according to the manufacturer's instructions. For some experiments, amino acids were systematically replaced by glycine (glycine walk). Membranes were probed with specific antibodies as described previously (26). Signal intensities of immunopositive peptides of an individual antibody were determined using the National Institutes of Health image program for Macintosh.
The linear epitopes of MAbs A1, A69, and B1 were confirmed in peptide competition experiments using Western blot analyses of AAV-2-Ad5-infected HeLa cell extracts (see above). An optimal dilution of hybridoma supernatant as determined by a titration curve was incubated in the presence or absence of synthetic peptides (1 mg/ml) on membranes. Proteins were visualized by an alkaline phosphatase-coupled anti-mouse secondary antibody (Dianova, Hamburg, Germany) using standard protocols (16).
The involvement of short peptide sequences in the conformational epitope of MAbs A20, C37-B, and D3 as determined by peptide scanning was confirmed by a competition assay using an AAV-2 capsid ELISA. Wells on microtiter plates were coated with CsCl-purified AAV-2 particles and handled as described above. The optimal concentration of purified A20 (100 ng/well), C37-B (20 ng/well), or D3 (7 μg/well) as determined by a titration curve was preincubated with 400 μg of either a single peptide or a mixture of peptides per well for 30 min at room temperature before increasing the volume to 100 μl/well with PBS. This mixture was incubated in wells coated with AAV-2 for 1 h at 37°C. The amount of MAb able to bind to AAV-2 particles was detected by an ELISA as described above.
The following peptides were synthesized by the DKFZ peptide synthesis support facility for use in peptide competition experiments: A1 peptide (aa 123 to 131), KRVLEPLGLV; A69 peptide (aa 171 to 182), LNFGQTGDADSV; B1 peptide (aa 726 to 733), IGTRYLTRNL; C37-1 peptide (aa 461 to 470), QFSQAGASDI; C37-2 peptide (aa 492 to 503), SADNNNSEYSWT; C37-2M peptide, SADNGGSEYSWT; C37-3 peptide (aa 601 to 610), LPGMVWQDRD; A20-1 peptide (aa 272 to 281), HYFGYSTPWG; A20-2 peptide (aa 369 to 378), VFMVPQYGYL; A20-3 peptide (aa 533 to 542), FFPQSGVLIF; A20-4 peptide (aa 566 to 575), RTTNPVATEQ; D3-1 peptide (aa 204 to 216), ATGSGAPMADNNE; D3-2 peptide (aa 234 to 240), WMGDRVI; D3-3 peptide (aa 259 to 269), QISSQSGASND; D3-4 peptide (aa 283 to 292), DFNRFHCHFS; D3-5 peptide (aa 323 to 332), VTQNDGTTT; D3-6 peptide (aa 354 to 363), VLGSAHQGCL; D3-7 peptide (aa 474 to 483), SRNWLPGPCY; D3-8 peptide (aa 705 to 714), NKSVNVDFTV; E42 peptide (aa 315 to 324), LFNIQVKEVT; and E43 peptide (aa 333 to 342), IANNLTSTVQ. Peptides were dissolved in PBS (20 mg/ml) or were first dissolved in methanol and then diluted with PBS to a final concentration of 10 mg/ml, depending on the solubility of the peptide, and used for peptide competition experiments. Peptides were chosen for each antibody based on serotype reactivity (see Results) and numbered consecutively.
The capsid protein structure of canine parvovirus (CPV) was taken from Xie et al. (54). Based on the alignment of Chapman and Rossmann (7), the determined epitopes and important antigenic amino acids of parvoviruses were visualized using Rasmol (version 2.6) (38).
RESULTS
Epitope mapping of MAbs A1, A69, and B1.
MAbs A1, A69, and B1 were isolated and characterized previously (52). They specifically detect VP1 (A1), VP1 and VP2 (A69), or VP1, VP2, and VP3 (B1) by Western blotting (Table 1), suggesting they recognize linear epitopes specific to VP1 or common to VP1 and VP2 or to VP1, VP2, and VP3, respectively.
TABLE 1.
Characteristics of MAbs recognizing linear epitopes
| Assay or parameter | Characteristic of MAb
|
||
|---|---|---|---|
| A1 | A69 | B1 | |
| Specificity in Western blot analysesa | VP1 | VP1, VP2 | VP1, VP2, VP3 |
| Specificity in immunoprecipitationsa | VP1, complexes with VP1 | VP1, VP2, complexes containing VP1 or VP2 | VP1, VP2, VP3 |
| IgG subclassb | IgG2a | IgG1 | IgG1 |
| Recognized epitope sequencec | KRVLEPLGL | LNFGQTGDADSV | IGTRYLTR |
Data from Wistuba et al. (52).
Determined using the Amersham MAb isotyping kit.
Determined by gene fragment phage display library and peptide scanning and verified by peptide competition as described in Materials and Methods.
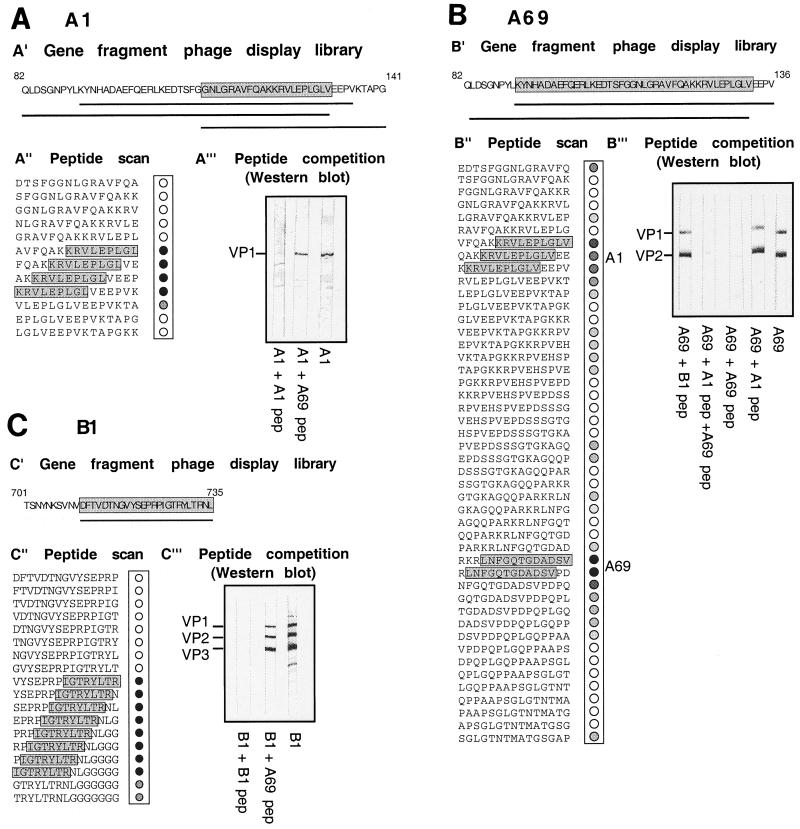
In order to determine the A1 epitope, a VP1-VP2-VP3 gene fragment phage display library was screened with MAb A1 and sequence analysis of immunopositive clones after the first round of biopanning revealed three different types of inserts containing an overlapping sequence of 21 aa (aa 111 to 132) (Fig. 1A′, gray box) (Please note that all of the following amino acid positions are with respect to the VP1 sequence.). To further narrow down the A1 epitope, a peptide scan was performed. Overlapping synthetic 15-mer peptides spanning the 21-aa sequence were immunostained with MAb A1. Nine amino acids (KRVLEPLGL) were common to all immunoreactive spots corresponding to the minimal epitope recognized by MAb A1 (Fig. 1A", gray box). These results were confirmed by using synthetic peptides in Western blot competition experiments (Fig. 1A′"). A1 binding to VP1 was inhibited upon addition of peptide KRVLEPLGLV (A1 peptide) in immunoblots, while a control peptide (A69 peptide) had no effect. Hence, A1 recognizes the amino acid sequence KRVLEPLGL in the N-terminal VP domain specific to VP1.
FIG. 1.
Identification of linear epitopes of MAbs A1, A69, and B1. The linear epitopes of MAbs A1 (A), A69 (B), and B1 (C) were determined in a similar fashion. In each case, the gene fragment phage display library (′) identified immunopositive clones and sequence analysis of these clones identified different inserts (black bars) after one round of panning with specific amino acid overlaps (gray box). To further fine map the epitope, a peptide scan (") was carried out. Overlapping 15-mer peptides were synthesized on a membrane and incubated with the respective antibody followed by peroxidase-coupled secondary antibodies. Immunoreaction of peptides is shown on the right next to the sequence, and amino acids common to all are marked by a gray box. Note also the weaker immunoreaction with other peptides in the case of A69. To confirm these epitopes, peptide competition in Western blot analysis (′") were performed. Extracts of AAV-2-Ad5-infected HeLa cells were separated on sodium dodecyl sulfate-polyacrylamide gels and blotted. In each case, hybridoma supernatant was incubated on membranes alone and in the presence of 1 mg of the peptide (pep) corresponding to the epitope or a control peptide per ml as follows: MAb A1 with A1 peptide (KRLVLEPLGLV) and the control peptide (A69 peptide [LNFGQTGDADSV]) (A′"); MAb A69 with A1 peptide (KRLVLEPLGLV) or A69 peptide (LNFGQTGDADSV) alone, a combination of both peptides, or a control peptide (B1 peptide [IGTRYLTRNL]) (B′"); and MAb B1 with B1 peptide (IGTRYLTRNL) or a control peptide (A69 peptide [LNFGQTGDADSV]) (C′"). The antibody reaction was visualized by an alkaline phosphatase-coupled secondary antibody.
The A69 epitope was delineated accordingly. Two different inserts containing a 41-aa overlap located in VP1 (aa 91 to 132) were identified after the first round of panning (Fig. 1B′, gray box). This was surprising because MAb A69 recognizes VP1 and VP2, implying an epitope location in the sequence common to VP1 and VP2 (aa 138 to 202) but not present in VP3. To clarify this discrepancy, a peptide scan of aa 91 to 210 was probed with MAb A69 (partially shown in Fig. 1B"). The antibody reacted strongest with spots corresponding to aa 169 to 184 located in VP2 (A69 peptide). In addition, three reactive spots albeit with reduced intensity were obtained corresponding to aa 123 to 136 located in the VP1-specific region (A1 peptide). Moreover, several additional peptides weakly reacted with A69, suggesting that the antibody is promiscuous. The inability to enrich phages containing VP2 amino acid sequence inserts might be due to a toxic effect exerted by these amino acids. Nevertheless, peptide competition experiments with both peptides clearly demonstrated that the VP2-specific peptide (A69 peptide) alone was able to abolish binding of MAb A69 to VP1 and VP2, while the VP1-specific peptide (A1 peptide) had no effect (Fig. 1B′"). On the basis of this result, we concluded that the primary epitope of A69 is LNFGQTGDADSV (aa 171 to 182) located in VP2.
To determine the B1 epitope, biopanning of the phage display library identified immunopositive clones containing the same 25-aa insert (aa 711 to 735) (Fig. 1C′, gray box). Peptide scans reduced this epitope sequence to IGTRYLTR, located in the C terminus of the VP protein (aa 726 to 733) (Fig. 1C", gray box). This epitope was validated in Western blot competition experiments where the synthesized peptide B1 (IGTRYLTRNL) prevented binding of B1 hybridoma supernatant, while the A69 peptide had no effect (Fig. 1C′"). An epitope sequence common to all three VP proteins is consistent with recognition of VP1, VP2, and VP3 by B1 in Western blots. In conclusion, B1 recognizes eight C-terminal amino acids of the VP protein. This very C terminus is likely buried in the capsid, since B1 reacts only very weakly with assembled capsids (52).
Characterization of MAbs D3, A20, C24-B, and C37-B.
Conformational epitopes are defined as continuous or discontinuous sequences, which are not detected by a given antibody when the antigen is denatured. MAbs A20, C24-B, C37-B, and D3 fulfilled this criteria. However, they specifically recognized AAV-2 capsids in immunofluorescence analysis of AAV-2-Ad5-infected HeLa cells (Table 2). A20 recognizes only assembled AAV-2 particles in immunoprecipitations (52), whereas C24-B, C37-B, and D3 also precipitated nonassembled capsid proteins, albeit to differing degrees. All four MAbs belong to the same immunoglobulin G (IgG) subclass (IgG1) and bound empty and full AAV-2 particles in an ELISA. In a neutralization assay, incubation of rAAV-2-GFP with A20, C24-B, or C37-B on HeLa cells prevented transgene expression, while antibody D3 did not influence the GFP fluorescence pattern (Table 2).
TABLE 2.
Characteristics of MAbs recognizing conformational epitopes
| Assay or parameter | Characteristic of MAb
|
|||
|---|---|---|---|---|
| D3 | A20 | C24-B | C37-B | |
| Neutralization of AAV-2 infectiona | − | + | + | + |
| Inhibition of binding of AAV-2 to HeLa cellsb | − | − | + | + |
| Western blotc | − | − | − | − |
| Immunofluorescenced on: | ||||
| AAV-2-Ad5-infected HeLa cells | + | + | + | + |
| Ad5-infected HeLa cells | − | − | − | − |
| Specificity in immunoprecipitationse | VP1, -2, and -3; capsids | Capsids | Capsids; VP1, -2, and -3 (weakly) | Capsids; VP1, -2, and -3 (weakly) |
| Capsid ELISAf | + | + | + | + |
| AAV-2 ELISAg | + | + | + | + |
| IgG subclassh | IgG1 | IgG1 | IgG1 | IgG1 |
| Recognized epitope sequencesi | SRNWLPGPCY | HYFGYSTPWG, VFMVPQYGYL, RTTNPVATEQ | ND | SADNNNSEYSWT, LPGMVWQDRD |
Neutralization of AAV-2 GFP infection was performed as described in Materials and Methods. +, no GFP fluorescence seen after 20 to 22 h.
Binding assays were performed as described in Materials and Methods. +, inhibition of binding.
Western blot of AAV-2-Ad5-infected HeLa cell extracts were performed as described in Materials and Methods. −, no detectable signal.
Immunofluorescence microscopy was performed as described in Materials and Methods. +, red fluorescence staining of cells.
Experiments were performed by the method of Wistubal et al. (52).
ELISA performed on plates coated with purified empty AAV-2 capsids. +, signal greater than an optical density of 0.2.
ELISA performed on plates coated with CsCl-purified AAV-2. +, signal greater than an optical density of 0.2.
Determined using the Amersham monoclonal antibody isotyping kit.
Conformational epitope sequences were determined as described in Results, and sequences representing major sites in the epitopes are indicated by underlining. ND, not determined.
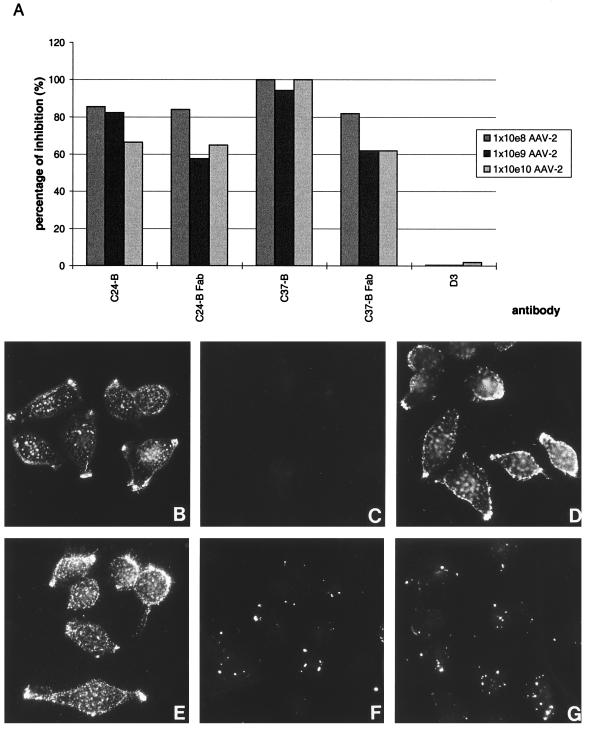
To determine if neutralization occurred at the step of receptor attachment, a nonradioactive binding assay was developed. AAV-2 particles able to bind to cells in the presence of hybridoma supernatant were detected by biotinylated A20. D3 was unable to inhibit binding of AAV-2 to cells, while C24-B and C37-B showed strong inhibition of binding (Fig. 2A). The inhibition of C24-B was somewhat weaker likely due to a lower affinity of this antibody. Fab fragments of C24-B and C37-B also inhibited binding, albeit with a lower efficiency, suggesting that inhibition of cell binding and neutralization were not a result of virus aggregation. This binding assay was not suitable for analysis of A20, as biotinylated A20 was used for detection of bound particles, resulting in a competition for A20 binding sites. To clarify the influence of A20 on receptor binding, the different antibodies were tested in a fluorescence assay using FITC-labeled empty AAV-2 capsids. Control experiments revealed that labeling of capsids did not interfere with antibody binding, since each MAb was able to recognize FITC-labeled capsids in an ELISA (data not shown). Cells not incubated with FITC-labeled capsids were used as a negative control (Fig. 2C). Labeled capsids bound to HeLa cells in the absence of competing antibody (Fig. 2B) showed the same fluorescence pattern as capsids preincubated with antibodies D3 (Fig. 2D) and A20 (Fig. 2E), demonstrating that D3 and A20 did not inhibit AAV-2-cell binding. In contrast, C24-B (Fig. 2F) and C37-B (Fig. 2G) showed strongly reduced fluorescence intensities, verifying earlier observations that these two antibodies inhibit AAV-2 attachment to cells.
FIG. 2.
Ability of MAbs A20, C24-B, C37-B, and D3 to inhibit binding of AAV-2 particles to cells. (A) Nonradioactive binding assay. Purified antibodies and Fab fragments were incubated with increasing amounts of AAV-2 particles on HeLa cells. Bound AAV-2 particles were detected by an A20 ELISA. The absorbance values were used to determine the percentage of inhibition. This graph is a representative of three independent experiments. 1 × 10e8, 1 × 108. (B to G) Immunofluorescence with FITC-labeled AAV-2 capsids. Binding of labeled capsids to HeLa cells (B) and HeLa cells without capsids (C) were used as controls. FITC-labeled capsids were preincubated with purified antibody D3 (D), A20 (E), C24-B (F), or C37-B (G) before addition to cells at 4°C. Photographs were taken with a ×63 lens.
MAbs C24-B, C37-B, D3, and A20 show different reaction patterns with AAV serotypes and AAV-2 insertion mutants.
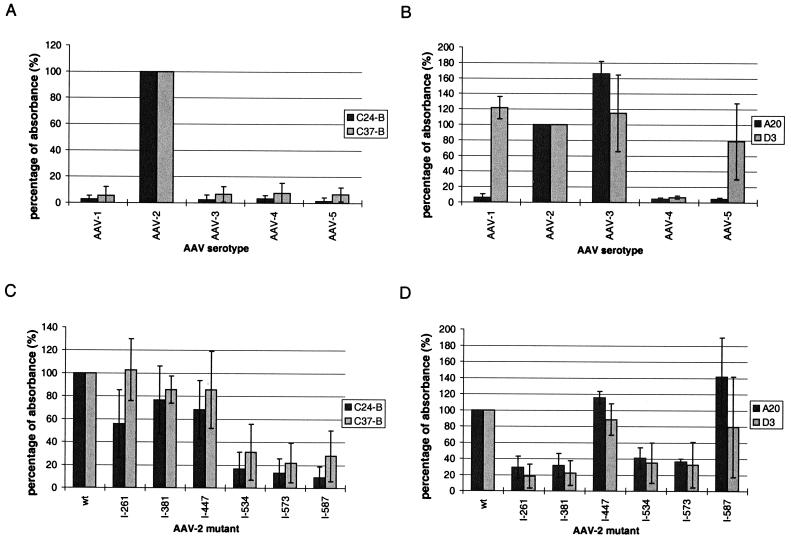
To determine domains involved in the epitopes of these antibodies, the binding affinity toward CsCl-purified AAV serotypes or AAV-2 insertion mutants was evaluated in an ELISA (Fig. 3). MAbs C24-B and C37-B specifically recognized AAV-2 but not AAV-1, -3, -4, or -5 (Fig. 3A). A20 specifically reacted with AAV-2 and AAV-3, while D3 reacted with all serotypes except AAV-4 (Fig. 3B). Analysis of the antibodies with AAV-2 capsid mutants harboring a 14-aa peptide insertion at distinct sites (13; also see Fig. 6A) demonstrated that binding of C37-B and C24-B to the capsid was prevented by insertions at amino acid positions 534, 573, and 587 (Fig. 3C). An insertion at position 261 had no effect on C37-B binding but partially reduced binding of C24-B to the capsid, indicating a slight difference in the epitope recognized by these two antibodies. Taken together, the reactivities of C24-B and C37-B with AAV serotypes and insertion mutants point toward an epitope sequence in the C-terminal half of VP3 that is distinctive for AAV-2. Binding of the A20 antibody was not affected by insertions at positions 447 and 587, while the insertion mutants I-261, I-381, I-534, and I-573 were not recognized as efficiently as the wt AAV-2 capsid (Fig. 3D). This was taken as an indication that sequences in VP3 with homology between AAV-2 and AAV-3 located in the N-terminal half (aa 258 to 275 and 372 to 393) as well as a region(s) in the C-terminal half of VP3 (aa 442 to 601) may be involved in A20 epitope formation (for sequence comparisons, see reference 2 for example). D3 recognized the same AAV-2 mutants, I-447 and I-587, as A20 (Fig. 3D). Mutants I-261, I-381, I-534, and I-573 are located in regions of sequence homology between AAV-2 and -3 but not AAV-5. As the D3 epitope is predicted to include an area of VP with amino acid sequence homology among AAV-1, -2, -3, and -5 and heterology to AAV-4, it is possible that insertions changed the epitope availability on the AAV-2 capsid surface.
FIG. 3.
Binding of MAbs to AAV serotypes and AAV-2 capsid insertion mutants. Specific binding of MAbs to different AAV serotypes and AAV-2 insertion mutants was assayed in an ELISA. Plates were coated with either CsCl-purified AAV-1 through -5 or AAV-2 insertion mutants (Girod et al. 13) at equal concentrations as determined by negative staining of particles. Hybridoma supernatant of either C24-B, C37-B (A and C), A20 or D3 (B and D) was incubated with particles, visualized by a biotinylated secondary antibody followed by streptavidin-peroxidase, and quantified by measuring the absorbance at 450 nm. Binding of the antibody to AAV-2 capsids was set at 100% (y axis). Each experiment was repeated independently at least three times, and the mean values and standard deviations (indicated by the error bars) are shown.
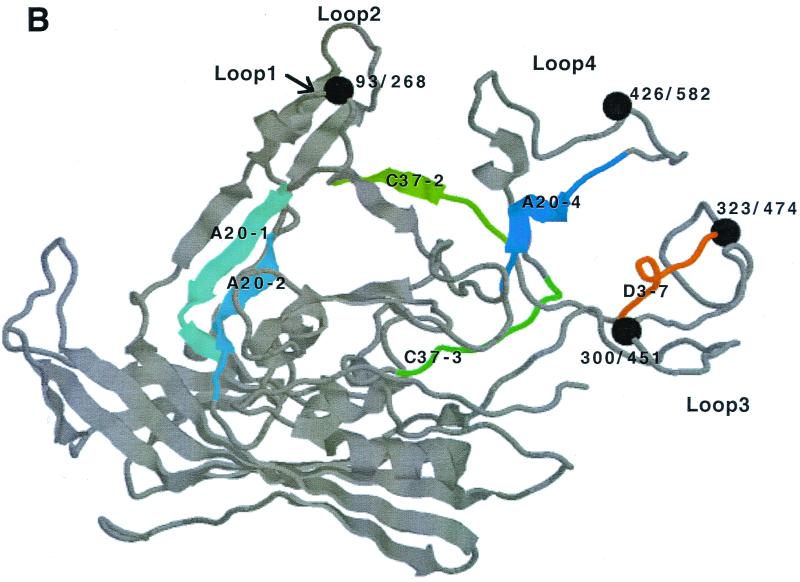
FIG. 6.
(A) Summary of identified epitopes. Identified epitopes of different MAbs are indicated along the amino acid sequence of the VP protein. The start sites of VP1, VP2, and VP3 are indicated by arrows. The linear epitopes of MAbs A1, A69, and B1 are drawn as open boxes. The peptides contributing to the conformational epitopes of MAbs A20, C37-B, and D3 are indicated by the boxes and different shades of gray. The different sites forming the epitopes of A20, C37-B, and D3 are numbered according to the designation used in peptide competition experiments (Fig. 4). Based on ELISA competition experiments, sites contributing to the epitope to a major extent are indicated by bold lettering, while important amino acids are underlined. Sites of insertion for the AAV-2 capsid mutants as described by Girod et al. (13) are indicated in italics. (B) Extrapolation of epitope location. Based on a sequence alignment of several parvoviruses (7), the identified epitope sequences were located in the capsid protein of CPV using Rasmol (38). Spheres show key amino acids of antigenicity of feline and canine parvoviruses; numbers indicate amino acids positions in CPV (VP2) (before the slash) and AAV-2 (VP1) (after the slash) (7). The sites forming the epitopes of A20, C37-B, and D3 are numbered according to the designation used in peptide competition experiments (see Fig. 4). Epitopes are indicated by different colors as follows: A20 in blue, C37-B in green, and D3 in orange.
C37-B, A20, and D3 epitope mapping by peptide scanning and peptide competition.
Similar to the identification of linear epitopes, gene fragment phage display libraries were first screened with antibodies C37-B and A20 but without success. This implies that the epitope sequences are separated by more than 100 aa, as only sequences of up to 100 aa can be presented on the phage surface in this system. Since conformational epitopes can also be identified by peptide scanning (31, 32, 35, 39), this method was employed for mapping of the A20, C24-B, C37-B, and D3 epitopes. For each antibody, specific regions of the VP3 protein sequence based on their reactivity with specific serotypes were synthesized as overlapping 10-mer peptides and probed with A20, C24-B, C37-B, or D3. Immunopositive peptides were synthesized and verified in peptide competition experiments in an AAV-2 capsid ELISA (Fig. 4). Each MAb was preincubated with a large excess of peptide(s) (400 μg) before incubation with AAV-2 capsids in an ELISA to account for the very low affinities from individual short peptides representing parts of a discontinuous epitope. The amount of nonspecific competition was assessed by unrelated control peptides and testing for cross-inhibition of the different MAbs.
FIG. 4.
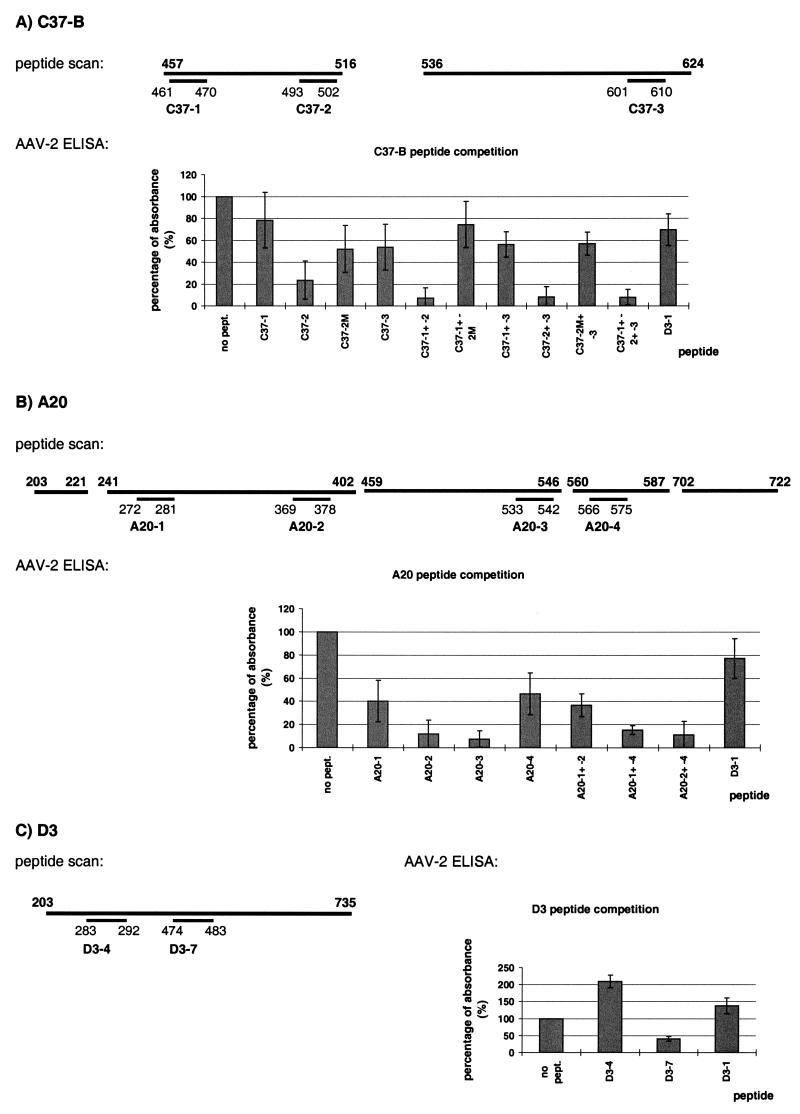
Epitope mapping of MAbs C37-B, A20, and D3. For the peptide scans, overlapping 10-mer peptides were synthesized on a membrane and incubated with C37-B (A), A20 (B), or D3 (C) followed by a peroxidase-coupled secondary antibody. The different domains of the AAV-2-VP amino acid sequence covered by the peptide scan are indicated by a thick black line with the representative amino acid numbers above the line. Immunoreactive peptides are represented by an additional black line with their amino acid positions and peptide names as used in peptide competition experiments (indicated below the line). For AAV-2 ELISA, the identified peptides were verified in peptide competition experiments using an ELISA of CsCl-purified AAV-2 particles. Peptides were preincubated with C37-B (A), A20 (B), or D3 (C) before addition to a well. The amount of MAb binding to AAV-2 particles was measured at 450 nm after incubation with a biotinylated secondary antibody followed by streptavidin peroxidase and addition of substrate. The competing peptide is shown along the x axis. The antibody reaction in the absence of any peptide (no pept.) was set at 100% (y axis), and peptide D3-1 was used as a control peptide. For each experiment, the reactions were performed in triplicate and each competition was repeated independently at least three times, and the mean values and standard deviations (indicated by the error bars) are shown. The peptide sequences are outlined in Materials and Methods.
C37-B.
Peptide scans of selected regions (aa 457 to 516 and aa 536 to 624) were probed with C37-B, and three immunoreactive regions of differing signal intensities were identified (Fig. 4A). Two peptides were located in the vicinity of insertion mutants I-534 (peptide C37-2) and I-587 (peptide C37-3) which showed reduced reactivity with the antibody. When these peptides were used in competition ELISA experiments, peptide C37-2 strongly competed MAb binding to the capsid, alone or in combination with C37-1, C37-3, or both (Fig. 4A). (Please note that the total peptide concentration remained constant in all peptide competition experiments.) Peptide C37-3 alone competed little, while peptide C37-1, which reacted weakly in peptide scan analysis, did not compete for antigen binding in this ELISA. It is therefore unlikely that the amino acid sequence corresponding to peptide C37-1 is involved in epitope formation. In addition, peptide scanning of mutant peptides was done to identify amino acids particularly important for interactions with the antibody. Individual amino acids in peptide C37-2 (SADNNNSEYSWT) were sequentially replaced by glycine. Replacement of two asparagines (positions 4 and 5) resulted in a reduced reactivity of this peptide with the antibody (data not shown). Thus, a mutant peptide, called peptide C37-2M (see Materials and Methods), was synthesized, and in peptide competition experiments, peptide C37-2M partially reversed the competition (Fig. 4A). This indicated that the second and third asparagine residues are amino acids important for interaction of C37-B with the AAV-2 capsid. When peptides C37-2 and C37-3 were tested for their specificity toward the C37-B antibody, they interacted specifically with C37-B but not with D3, A20 or C24-B (Fig. 5 and data not shown). Therefore, based on the ELISA analysis, the sequence SADNNNSEYSWT (aa 493 to 502) (C37-2) contributes to the C37-B epitope to a major extent, while the sequence LPGMVWQDRD (aa 601 to 610) (C37-3) is involved to a lesser extent (Fig. 4A).
FIG. 5.
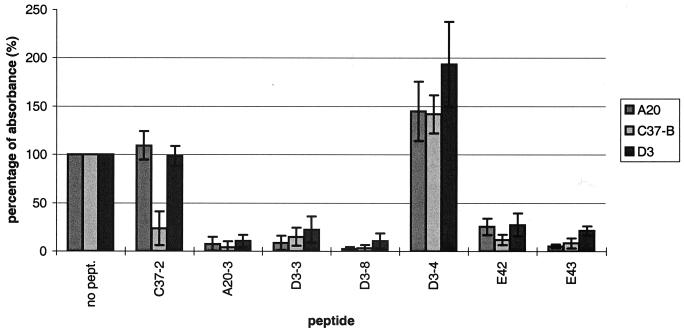
Specificity of selected peptides. Selected examples of peptides that were tested for their specificity toward individual antibodies in peptide competition experiments using an ELISA of CsCl-purified AAV-2 particles are shown. Synthesized peptides were preincubated with C37-B (A), A20 (B), or D3 (C) before addition to a well. The amount of MAb binding to AAV-2 particles was measured at 450 nm after incubation with a biotinylated secondary antibody followed by streptavidin peroxidase and addition of substrate. The competing peptide is shown along the x axis. The antibody reaction in the absence of any peptide (no pept.) was set at 100% (y axis). For each experiment, the reactions were performed in triplicate, each competition was repeated independently at least three times, and the mean values and standard deviations (indicated by the error bars) are shown. The peptide sequences are given in Materials and Methods.
C24-B.
We were unable to detect immunopositive peptides by peptide scanning for C24-B. However, peptides identified for C37-B (peptides C37-1, C37-2, and C37-3) did not prevent C24-B binding to AAV-2 in the ELISA (data not shown). We therefore concluded that C24-B and C37-B recognize distinct epitopes despite their otherwise similar characteristics.
A20.
Five different regions (aa 203 to 221, 241 to 402, 459 to 546, 560 to 587, and 702 to 722) were analyzed by peptide scanning and probed with A20, identifying four different immunoreactive peptides (Fig. 4B). Peptides A20-1 and A20-2 were located close to the point of insertion for mutants I-261 and I-381, while peptides A20-3 and A20-4 directly spanned the point of insertion of mutants I-534 and I-573, respectively. In contrast to C37-B, all four peptides identified by peptide scanning individually showed some competition in the ELISA, with peptides A20-2 and A20-3 competing A20 binding to AAV-2 the most (Fig. 4B). Whereas a combination of peptide A20-4 with peptide A20-2 further increased inhibition of the antibody, peptides A20-1 and A20-2 together did not result in increased competition. When each peptide was tested for its specificity toward A20, peptide A20-3 showed nonspecific inhibition of D3 and C37-B binding to AAV-2 capsids in the ELISA (Fig. 5 and data not shown). Although peptide A20-3 overlaps mutant I-534, the possibility that inhibition of binding of the antibody to capsids might occur by unknown mechanisms, mechanisms other than competition for epitope binding, cannot be excluded. Taken together, the ELISA data indicated that the sequence VFMVPQYGYL (aa 369 to 378, corresponding to peptide A20-2) constitutes the major part of the A20 epitope, while the other two regions corresponding to peptides A20-1 and A20-4 contribute to the epitope to a minor extent (Fig. 4B).
D3.
A peptide scan of the entire VP3 sequence (aa 203 to 735) was probed with D3 and two immunoreactive peptides (DFNRFHCHFS) (aa 283 to 292, corresponding to peptide D3-4) and SRNWLPGPCY (aa 474 to 483, corresponding to peptide D3-7) were identified (Fig. 4C). Both peptides are located far from the points of insertion of the analyzed AAV-2 mutants. When these peptides were used in peptide competition experiments in an AAV-2 ELISA, only peptide D3-7, which matched the identified AAV serotype reactivity pattern, was able to partially compete the interaction of D3 with the capsid. In an effort to identify further amino acid sequences contributing to the conformational D3 epitope, peptides D3-1 (aa 204 to 216), D3-2 (aa 234 to 240), D3-3 (aa 259 to 269), D3-5 (aa 323 to 332), D3-6 (aa 354 to 363), and D3-8 (aa 705 to 714) were synthesized from regions of the AAV-2 sequence that showed homology to AAV-1, -3, and -5 but not AAV-4. When tested, peptides D3-3 and D3-8 competed D3 binding to AAV-2 capsids in an ELISA. However, subsequent control experiments showed that peptides D3-3 and D3-8 nonspecifically competed the binding of all tested antibodies, while peptide D3-7 was specific for its competition of D3 (Fig. 5 and data not shown). Peptide D3-4 did not interfere with antibody binding to the capsid, instead, this peptide appeared to enhance the binding of each antibody with AAV-2 capsids (Fig. 5). We were unable to identify epitope sequences in addition to D3-7, although previous biochemical analysis indicated that D3 recognized a conformational epitope. We therefore concluded that sequence SRNWLPGPCY is part of the D3 epitope. In addition, there may be other sequences that could not be identified using the methods applied in this report.
DISCUSSION
Here, we report the epitope mapping of MAbs directed against nonassembled and assembled AAV-2 capsid proteins. Based on their characteristics and identified epitope sequences, regions that are probably buried in the capsid after assembly and domains involved in binding of AAV-2 to the cellular receptor or essential postbinding infection steps were identified.
MAbs A1, A69, and B1 recognize linear epitopes in VP1, VP1 or VP2, or VP1, VP2, and VP3, respectively (Fig. 1 and 6A). While these antibodies efficiently precipitate monomeric and oligomeric capsid proteins, they recognize capsids with a rather low efficiency compared to that of A20. This indicates that the availability of the respective epitope on the capsid surface changes (i.e., becomes partially masked) during capsid assembly. Thus, these antibodies might provide useful tools for studying AAV-2 assembly and disassembly during the infection process.
Characterization of MAbs D3, A20, C24-B, and C37-B identified the latter three as neutralizing antibodies. Here, we show that C24-B and C37-B interfere with virus attachment to cells. This is not due to interparticle cross-linking, as Fab fragments of these antibodies are still able to prevent virus binding to cells (Fig. 2A). Inhibition of binding might not be complete, since some fluorescent material remained attached to cells after incubation of labeled capsids with C24-B or C37-B (Fig. 2E and F). The antibody A20 neutralizes infection following receptor attachment, because binding of fluorescently labeled capsids to HeLa cells was not prevented by this antibody. However, the precise mode of neutralization remains unknown.
The sequence alignment of several parvoviruses (7) and the crystal structure of CPV (54) were used to extrapolate the locations of identified antigenic sites in the capsid protein of AAV-2 (Fig. 6B). Our data indicated that the A20 epitope is formed through recognition of three different binding sites distributed across the linear VP sequence. This is not unusual as conformational epitopes can be made up of two to five different binding sites (9). Site 1 (HYFGYSTPWG) (A20-1), site 2 (VFMVPQYGYL) (A20-2), and site 3 (RTTNPVATEQ) (A20-4) are located near loops 1, 2, and 4, respectively (Fig. 6B). However, depending on the alignment of CPV and AAV-2 capsid proteins, the epitopes will move closer to the tips of loops 1, 2, and 4. In CPV, residues from all three loops form the threefold spike, a major region of antigenicity. One antigenic site shows a strong conformational dependence due to the spatial vicinity of amino acids from loop 1 and loop 2 of one VP with amino acids in loop 4 of another VP in the assembled capsid (45). The same might be true for AAV-2. Therefore, the exclusive recognition of assembled capsids by the A20 antibody could be caused by a spatial juxtaposition of identified sites on separate capsid proteins only after completion of capsid assembly.
C37-B recognizes two sites which are exposed on the surface of nonassembled and assembled capsid proteins (Fig. 6B; see Table 2 for immunoprecipitation data). Both sites are located in the G-H loop, a general region of antigenicity in autonomous parvoviruses (7). By binding to the major antigenic site (SADNNNSEYEWT) (C37-2) and the minor site (LPGMVWQDRD) (C37-3), this antibody is directly or indirectly (i.e., by steric hindrance) able to prevent virus-cell binding. Girod et al. (13) identified a site in the viral capsid where the native tropism of AAV-2 was extended to AAV-2-resistant cells by insertion of an RGD motif (mutant I-587), indicating that this loop is able to promote interaction of the virus with a cellular receptor. C37-B was unable to bind to this mutant, and the antibody binding sites mapped to either side of this insertion (aa 493 to 502 and aa 601 to 610). This is also consistent with the canyon hypothesis (34), which would support binding of C37-B to an elevated surface structure(s) causing indirect (steric) inhibition of AAV-2 binding to cells. In agreement with this is also the recent identification of an AAV-2 capsid mutant unable to bind heparin (30) which contains an insertion at a site which is located between the two epitope recognition sites identified for C37-B. Similarly, insertions at aa 534 and 578 resulted in a steric separation of partial epitope sequences leading to the loss of antibody binding to the capsid.
The C24-B epitope could not be mapped by the present approach. A different approach for mapping of conformational epitopes, for example, the flock house virus antigen-presenting system (49) or cryoelectron microscopy in conjunction with three-dimensional image reconstruction techniques (1), could be used to define this epitope. However, because of the ability of C24-B to inhibit binding of AAV-2 to cells like C37-B and the similar reactivity with AAV serotypes and AAV-2 insertion mutants, we expect the C24-B epitope to map to the same general region of the capsid proteins.
D3 is a nonneutralizing antibody that binds to a conformational epitope including amino acids SRNWLPGPCY which are highly exposed on the surface of VP3 (Fig. 6B). Based on a sequence alignment with CPV (7), this sequence is found near the spike region protruding from the threefold axis, a region of antigenicity in CPV. The nonneutralizing D3 epitope corresponds to epitope 61-62 identified by Moskalenko et al. (22) which is part of a peptide pool having no effect on the AAV-2 neutralizing activity of human sera.
The observation of a crosswise and probably nonspecific competition of the antibody reaction in an ELISA by synthetic peptides raises questions about the validity of epitope mapping using solely peptide competition experiments. In a recent study, neutralizing epitopes of AAV-2 were identified based on peptide competition experiments only and the identified pool of neutralizing peptides was able to prevent neutralization of an AAV-2 infection by A20 (22). The peptides proposed to compete the conformational epitope of A20 do not include any of the epitope sequences mapped for A20 in this study. When 10-mer peptides overlapping the competing peptides described by Moskalenko et al. (22) were tested at the described concentration in our assay, no inhibition of A20 binding to capsids was observed. When the peptide concentration was increased to match the concentration used in our study, peptides E42 (LFNIQVKEVT, aa 315 to 324) and E43 (IANNLTSTVQ, aa 333 to 342) nonspecifically inhibited binding of A20, C37-B, and D3 to AAV-2 capsids (Fig. 5). This raises questions about the involvement of these sequences in the A20 epitope. In contrast, our study relies not only on peptide competition experiments but also on data from a comparison of different serotypes and AAV-2 mutants and peptide scans to support the conclusions. Other peptides, A20-3, D3-3, and D3-8 (which overlaps peptide 90 identified by Moskalenko et al. [22]), also nonspecifically inhibited all of the tested antibodies (Fig. 5), underlining the importance of testing peptides for nonspecific cross-competition of different antibodies.
In conclusion, determination of linear epitopes allowed us to identify sequences accessible on nonassembled capsid proteins but not accessible in the assembled capsid. In addition, determination of conformational epitopes of antibodies directed against AAV-2 capsid proteins led to the proposal of sites on the viral surface involved in neutralization and receptor binding. This study extends the known amino acid sequences which constitute antigenic regions on the AAV-2 capsid. It will be interesting to compare our findings to the three-dimensional structure of AAV-2 when it becomes available. The use of these antibodies for studies of the basic biology of AAV-2 (e.g., viral disassembly or elucidation of residues involved in AAV-2 binding to cells) and in the development of immunologically targeted AAV vectors will further underline their biological relevance.
ACKNOWLEDGMENTS
We thank U. Bantel-Schaal for the generous gift of GMK cells and AAV-1, -3, -4, and -5 and Ad2 virus stocks and B. Hub for excellent electron microscopy work. In addition, we are indebted to W. von der Lieth for the graphical epitope presentation. Furthermore, we thank E. Bautz for helpful discussions.
C.E.W. is supported by a DKFZ fellowship.
REFERENCES
- 1.Adrian M, Dubochet J, Lepault J, McDowall A W. Cryo-electron microscopy of viruses. Nature. 1984;308:32–36. doi: 10.1038/308032a0. [DOI] [PubMed] [Google Scholar]
- 2.Bantel-Schaal U, Delius H, Schmidt R, zur Hausen H. Human adeno-associated virus type 5 is only distantly related to other known primate helper-dependent parvoviruses. J Virol. 1999;73:939–947. doi: 10.1128/jvi.73.2.939-947.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartlett J S, Kleinschmidt J, Boucher R C, Samulski R J. Targeted adeno-associated virus vector transduction of nonpermissive cells mediated by a bispecific F(ab′γ)2. Nat Biotechnol. 1999;17:181–186. doi: 10.1038/6185. [DOI] [PubMed] [Google Scholar]
- 4.Bartlett J S, Wilcher R, Samulski R J. Infectious entry pathway of adeno-associated virus and adeno-associated virus vectors. J Virol. 2000;74:2777–2785. doi: 10.1128/jvi.74.6.2777-2785.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berns K I. Parvovirus replication. Microbiol Rev. 1990;54:316–329. doi: 10.1128/mr.54.3.316-329.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blacklow N R, Hoggan M D, Rowe W P. Serologic evidence for human infection with adenovirus-associated viruses. J Natl Cancer Inst. 1968;40:319–327. [PubMed] [Google Scholar]
- 7.Chapman M S, Rossmann M G. Structure, sequence, and function correlations among parvoviruses. Virology. 1993;194:491–508. doi: 10.1006/viro.1993.1288. [DOI] [PubMed] [Google Scholar]
- 8.Chirmule N, Propert K, Magosin S, Qian Y, Qian R, Wilson J. Immune responses to adenovirus and adeno-associated virus in humans. Gene Ther. 1999;6:1574–1583. doi: 10.1038/sj.gt.3300994. [DOI] [PubMed] [Google Scholar]
- 9.Davies D R, Padlan E A, Sheriff S. Antibody-antigen complexes. Annu Rev Biochem. 1990;59:439–473. doi: 10.1146/annurev.bi.59.070190.002255. [DOI] [PubMed] [Google Scholar]
- 10.Duan D, Li Q, Kao A W, Yue Y, Pessin J E, Engelhardt J F. Dynamin is required for recombinant adeno-associated virus type 2 infection. J Virol. 1999;73:10371–10376. doi: 10.1128/jvi.73.12.10371-10376.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dubielzig R, King J A, Weger S, Kern A, Kleinschmidt J A. Adeno-associated virus type 2 protein interactions: formation of pre-encapsidation complexes. J Virol. 1999;73:8989–8998. doi: 10.1128/jvi.73.11.8989-8998.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flotte T R, Carter B J. Adeno-associated virus vectors for gene therapy. Gene Ther. 1995;2:357–362. [PubMed] [Google Scholar]
- 13.Girod A, Ried M, Wobus C, Lahm H, Leike K, Kleinschmidt J, Deleage G, Hallek M. Genetic capsid modifications allow efficient re-targeting of adeno-associated virus type 2. Nat Med. 1999;5:1052–1056. doi: 10.1038/12491. [DOI] [PubMed] [Google Scholar]
- 14.Grimm D, Kern A, Pawlita M, Ferrari F, Samulski R, Kleinschmidt J. Titration of AAV-2 particles via a novel capsid ELISA: packaging of genomes can limit production of recombinant AAV-2. Gene Ther. 1999;6:1322–1330. doi: 10.1038/sj.gt.3300946. [DOI] [PubMed] [Google Scholar]
- 15.Grimm D, Kern A, Rittner K, Kleinschmidt J A. Novel tools for production and purification of recombinant adenoassociated virus vectors. Hum Gene Ther. 1998;9:2745–2760. doi: 10.1089/hum.1998.9.18-2745. [DOI] [PubMed] [Google Scholar]
- 16.Harlow E, Lane D. Antibodies. A laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 17.Hermonat P L, Labow M A, Wright R, Berns K I, Muzyczka N. Genetics of adeno-associated virus: isolation and preliminary characterization of adeno-associated virus type 2 mutants. J Virol. 1984;51:329–339. doi: 10.1128/jvi.51.2.329-339.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoque M, Ishizu K, Matsumoto A, Han S I, Arisaka F, Takayama M, Suzuki K, Kato K, Kanda T, Watanabe H, Handa H. Nuclear transport of the major capsid protein is essential for adeno-associated virus capsid formation. J Virol. 1999;73:7912–7915. doi: 10.1128/jvi.73.9.7912-7915.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iorio R M, Syddall R J, Sheehan J P, Bratt M A, Glickman R L, Riel A M. Neutralization map of the hemagglutinin-neuraminidase glycoprotein of Newcastle disease virus: domains recognized by monoclonal antibodies that prevent receptor recognition. J Virol. 1991;65:4999–5006. doi: 10.1128/jvi.65.9.4999-5006.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kotin R M. Prospects for the use of adeno-associated virus as a vector for human gene therapy. Hum Gene Ther. 1994;5:793–801. doi: 10.1089/hum.1994.5.7-793. [DOI] [PubMed] [Google Scholar]
- 21.Mizukami H, Young N S, Brown K E. Adeno-associated virus type 2 binds to a 150-kilodalton cell membrane glycoprotein. Virology. 1996;217:124–130. doi: 10.1006/viro.1996.0099. [DOI] [PubMed] [Google Scholar]
- 22.Moskalenko M, Chen L, van Roey M, Donahue B A, Snyder R O, McArthur J G, Patel S D. Epitope mapping of human anti-adeno-associated virus type 2 neutralizing antibodies: implications for gene therapy and virus structure. J Virol. 2000;74:1761–1766. doi: 10.1128/jvi.74.4.1761-1766.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mosser A G, Leippe D M, Rueckert R R. Neutralization of picornaviruses: support for the pentamer bridging hypothesis. In: Semler B L, Ehrenfeld E, editors. Molecular aspects of picornavirus infection and detection. Washington, D.C.: American Society for Microbiology; 1989. pp. 155–167. [Google Scholar]
- 24.Muzyczka N. Use of adeno-associated virus as a general transduction vector for mammalian cells. Curr Top Microbiol Immunol. 1992;158:97–129. doi: 10.1007/978-3-642-75608-5_5. [DOI] [PubMed] [Google Scholar]
- 25.Nicola A V, Ponce de Leon M, Xu R, Hou W, Whitbeck J C, Krummenacher C, Montgomery R I, Spear P G, Eisenberg R J, Cohen G H. Monoclonal antibodies to distinct sites on herpes simplex virus (HSV) glycoprotein D block HSV binding to HVEM. J Virol. 1998;72:3595–3601. doi: 10.1128/jvi.72.5.3595-3601.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petersen G, Song D, Hugle-Dorr B, Oldenburg I, Bautz E K. Mapping of linear epitopes recognized by monoclonal antibodies with gene-fragment phage display libraries. Mol Gen Genet. 1995;249:425–431. doi: 10.1007/BF00287104. [DOI] [PubMed] [Google Scholar]
- 27.Qing K, Mah C, Hansen J, Zhou S, Dwarki V, Srivastava A. Human fibroblast growth factor receptor 1 is a co-receptor for infection by adeno-associated virus 2. Nat Med. 1999;5:71–77. doi: 10.1038/4758. [DOI] [PubMed] [Google Scholar]
- 28.Qiu J, Mizukami H, Brown K E. Adeno-associated virus 2 co-receptors? Nat Med. 1999;5:467. doi: 10.1038/8328. [DOI] [PubMed] [Google Scholar]
- 29.Qiu J, Brown K E. Integrin αVβ5 is not involved in adeno-associated virus type 2 (AAV2) infection. Virology. 1999;264:436–440. doi: 10.1006/viro.1999.0010. [DOI] [PubMed] [Google Scholar]
- 30.Rabinowitz J E, Xiao W, Samulski R J. Insertional mutagenesis of AAV2 capsid and the production of recombinant virus. Virology. 1999;265:274–285. doi: 10.1006/viro.1999.0045. [DOI] [PubMed] [Google Scholar]
- 31.Reineke U, Sabat R, Kramer A, Stigler R D, Seifert M, Michel T, Volk H D, Schneider-Mergener J. Mapping protein-protein contact sites using cellulose-bound peptide scans. Mol Divers. 1996;1:141–148. doi: 10.1007/BF01544952. [DOI] [PubMed] [Google Scholar]
- 32.Reineke U, Sabat R, Volk H D, Schneider-Mergener J. Mapping of the interleukin-10/interleukin-10 receptor combining site. Protein Sci. 1998;7:951–960. doi: 10.1002/pro.5560070412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rose J A, Maizel J V, Jr, Inman J K, Shatkin A J. Structural proteins of adenovirus-associated viruses. J Virol. 1971;8:766–770. doi: 10.1128/jvi.8.5.766-770.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rossmann M G. The canyon hypothesis. Hiding the host cell receptor attachment site on a viral surface from immune surveillance. J Biol Chem. 1989;264:14587–14590. [PubMed] [Google Scholar]
- 35.Rudiger S, Germeroth L, Schneider-Mergener J, Bukau B. Substrate specificity of the DnaK chaperone determined by screening cellulose-bound peptide libraries. EMBO J. 1997;16:1501–1507. doi: 10.1093/emboj/16.7.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ruffing M, Zentgraf H, Kleinschmidt J A. Assembly of viruslike particles by recombinant structural proteins of adeno-associated virus type 2 in insect cells. J Virol. 1992;66:6922–6930. doi: 10.1128/jvi.66.12.6922-6930.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Samulski R J, Sally M, Muzyczka N. Adeno-associated viral vectors. In: Friedman T, editor. The development of human gene therapy. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1999. pp. 132–172. [Google Scholar]
- 38.Sayle R A, Milner-White E J. RASMOL: biomolecular graphics for all. Trends Biochem Sci. 1995;20:374. doi: 10.1016/s0968-0004(00)89080-5. [DOI] [PubMed] [Google Scholar]
- 39.Schreiber M, Wachsmuth C, Muller H, Odemuyiwa S, Schmitz H, Meyer S, Meyer B, Schneider-Mergener J. The V3-directed immune response in natural human immunodeficiency virus type 1 infection is predominantly directed against a variable, discontinuous epitope presented by the gp120 V3 domain. J Virol. 1997;71:9198–9205. doi: 10.1128/jvi.71.12.9198-9205.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith G P, Scott J K. Libraries of peptides and proteins displayed on filamentous phage. Methods Enzymol. 1993;217:228–257. doi: 10.1016/0076-6879(93)17065-d. [DOI] [PubMed] [Google Scholar]
- 41.Smith T J, Mosser A G. Antibody-mediated neutralization of picornaviruses. In: Chiu W, Burnett M R, Garcea R L, editors. Structural biology of viruses. Oxford, United Kingdom: Oxford University Press; 1997. pp. 134–156. [Google Scholar]
- 42.Smuda J W, Carter B J. Adeno-associated viruses having nonsense mutations in the capsid genes: growth in mammalian cells containing an inducible amber suppressor. Virology. 1991;184:310–318. doi: 10.1016/0042-6822(91)90847-5. [DOI] [PubMed] [Google Scholar]
- 43.Snyder R O, Xiao X, Samulski R J. Production of recombinant adeno-associated viral vectors. In: Dracopoli J H N, Krof B, Moir D, Morton C, Seidman C, Seidman J, Smith D, editors. Current protocols in human genetics. New York, N.Y: John Wiley and Sons Publisher; 1996. pp. 12.1.1–12.24. [Google Scholar]
- 44.Steinbach S, Wistuba A, Bock T, Kleinschmidt J A. Assembly of adeno-associated virus type 2 capsids in vitro. J Gen Virol. 1997;78:1453–1462. doi: 10.1099/0022-1317-78-6-1453. [DOI] [PubMed] [Google Scholar]
- 45.Strassheim M L, Gruenberg A, Veijalainen P, Sgro J Y, Parrish C R. Two dominant neutralizing antigenic determinants of canine parvovirus are found on the threefold spike of the virus capsid. Virology. 1994;198:175–184. doi: 10.1006/viro.1994.1020. [DOI] [PubMed] [Google Scholar]
- 46.Summerford C, Bartlett J S, Samulski R J. αVβ5 integrin: a co-receptor for adeno-associated virus type 2 infection. Nat Med. 1999;5:78–82. doi: 10.1038/4768. [DOI] [PubMed] [Google Scholar]
- 47.Summerford C, Samulski R J. Membrane-associated heparan sulfate proteoglycan is a receptor for adeno-associated virus type 2 virions. J Virol. 1998;72:1438–1445. doi: 10.1128/jvi.72.2.1438-1445.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thomas J O, Kornberg R D. An octamer of histones in chromatin and free in solution. Proc Natl Acad Sci USA. 1975;72:2626–2630. doi: 10.1073/pnas.72.7.2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tisminetzky S G, Scodeller E A, Evangelisti P, Chen Y, Schiappacassi M, Porro F, Bizik F, Zacchi T, Lunazzi G, Miertus S, et al. Immunoreactivity of chimeric proteins carrying the HIV-1 epitope IGP bpGRAF. Correlation between predicted conformation and antigenicity. FEBS Lett. 1994;353:1–4. doi: 10.1016/0014-5793(94)00972-4. [DOI] [PubMed] [Google Scholar]
- 50.Tratschin J D, Miller I L, Carter B J. Genetic analysis of adeno-associated virus: properties of deletion mutants constructed in vitro and evidence for an adeno-associated virus replication function. J Virol. 1984;51:611–619. doi: 10.1128/jvi.51.3.611-619.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wien M W, Filman D J, Stura E A, Guillot S, Delpeyroux F, Crainic R, Hogle J M. Structure of the complex between the Fab fragment of a neutralizing antibody for type 1 poliovirus and its viral epitope. Nat Struct Biol. 1995;2:232–243. doi: 10.1038/nsb0395-232. [DOI] [PubMed] [Google Scholar]
- 52.Wistuba A, Kern A, Weger S, Grimm D, Kleinschmidt J A. Subcellular compartmentalization of adeno-associated virus type 2 assembly. J Virol. 1997;71:1341–1352. doi: 10.1128/jvi.71.2.1341-1352.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wistuba A, Weger S, Kern A, Kleinschmidt J A. Intermediates of adeno-associated virus type 2 assembly: identification of soluble complexes containing Rep and Cap proteins. J Virol. 1995;69:5311–5319. doi: 10.1128/jvi.69.9.5311-5319.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xie Q, Chapman M S. Canine parvovirus capsid structure, analyzed at 2.9 A resolution. J Mol Biol. 1996;264:497–520. doi: 10.1006/jmbi.1996.0657. [DOI] [PubMed] [Google Scholar]
- 55.Yang Q, Mamounas M, Yu G, Kennedy S, Leaker B, Merson J, Wong-Staal F, Yu M, Barber J R. Development of novel cell surface CD34-targeted recombinant adenoassociated virus vectors for gene therapy. Hum Gene Ther. 1998;9:1929–1937. doi: 10.1089/hum.1998.9.13-1929. [DOI] [PubMed] [Google Scholar]