Abstract
Regeneration is the regrowth of damaged tissues or organs, a vital process in response to damages from primitive organisms to higher mammals. Planarian possesses active whole‐body regenerative capability owing to its vast reservoir of adult stem cells, neoblasts, providing an ideal model to delineate the underlying mechanisms for regeneration. RNA N 6‐methyladenosine (m6A) modification participates in many biological processes, including stem cell self‐renewal and differentiation, in particular the regeneration of haematopoietic stem cells and axons. However, how m6A controls regeneration at the whole‐organism level remains largely unknown. Here, we demonstrate that the depletion of m6A methyltransferase regulatory subunit wtap abolishes planarian regeneration, potentially through regulating genes related to cell–cell communication and cell cycle. Single‐cell RNA‐seq (scRNA‐seq) analysis unveils that the wtap knockdown induces a unique type of neural progenitor‐like cells (NP‐like cells), characterized by specific expression of the cell–cell communication ligand grn. Intriguingly, the depletion of m6A‐modified transcripts grn, cdk9 or cdk7 partially rescues the defective regeneration of planarian caused by wtap knockdown. Overall, our study reveals an indispensable role of m6A modification in regulating whole‐organism regeneration.
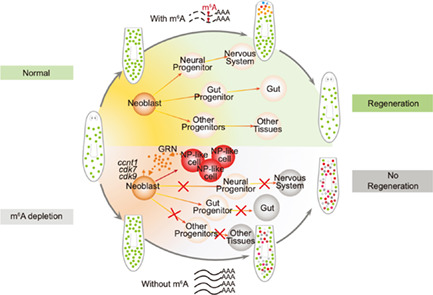
As depicted in our model, during planarian homeostasis, the expression level of grn is held at a moderate level to inhibit overgrowth, as equivalent to grn expression level before amputation at 0 hpa. Upon injury, m6A modification selectively targets several important gene transcripts, including grn for degradation, manifested as its reduced expression level after 6 hpa, as well as transcripts of cell cycle‐related genes including ccnt1, cdk7 and cdk9. m6A‐mediated downregulation of these genes, especially the grn, may halt the initial proliferation of the stem cell pool at the first stage of regeneration, which in turn promotes their differentiation process, including the differentiation of neoblasts to the progenitor cells and eventually to different tissue types. After wtap knockdown, grn transcript accumulates to form a unique cell cluster resulting in increased GRN secretion, which may lead to cell cycle dysregulation and the loss of neoblast differentiation and the failure of planarian regeneration.
1. INTRODUCTION
Regeneration is a process of replacing or restoring damaged or missing cells and tissues to full function, which widely exists in the animal kingdoms. 1 For example, hydra can regenerate from tiny body fragments to entire organisms, 1 and zebrafish can regenerate large portions of the heart. 2 For humans, some tissues also have regenerative potential throughout life, including the liver and muscle. 3
Planarians are being considered a popular model system to study regeneration at the whole‐organism level. 4 , 5 The discovery of the planarian stem cell (neoblast) marker piwi (smedwi‐1), 6 the development of neoblast isolation 7 and RNA interference (RNAi) knockdown 8 methods have opened up opportunities to unravel the mechanisms of planarian regeneration. A number of pathways and regulatory factors have been identified to be essential for planarian regeneration, such as Wnt 9 , 10 , 11 , 12 , 13 , 14 and EGFR signalling. 15 Especially along with the advances of third‐generation DNA 16 and large‐scale single‐cell RNA sequencing technologies, 17 , 18 accumulating evidence suggests that the pluripotent neoblasts are cellular sources for regeneration. 19 , 20 In addition, some studies have revealed that epigenetic modifications also exert functions in planarian regeneration. For instance, the COMPASS family MLL3/4 histone methyltransferases are essential for the differentiation and regeneration of the planarian. 21 , 22 , 23 In addition, the CREB‐binding protein (CBP) and p300 family of histone acetyltransferases homologues Smed‐CBP2 and Smed‐CBP3 displayed distinct roles in stem cell maintenance and functions. 24 , 25
N 6‐methyladenosine (m6A), the most abundant dynamic internal mRNA modification, has been shown to be an epi‐transcriptomic marker playing diverse regulatory roles under physiological and/or pathological conditions. 26 Studies in various model systems have revealed that m6A regulates stem cell self‐renewal and differentiation. 27 , 28 , 29 , 30 Depletion of either m6A methyltransferases or its demethylases dramatically affects gene expression profiles. 31 , 32 , 33 , 34 High‐throughput sequencing technologies enable the detection of the m6A location within the transcriptome. 31 , 35 Subsequently, m6A has been identified in various RNA species and has been implicated in stem cell biology, developmental and cancer biology. 26 For example, two separate studies showed that m6A regulates the transition of embryonic stem cells from a pluripotent state to a differentiated state. In this case, m6A selectively marks transcripts that code for key transcription factors involved in differentiation. 27 , 36 This demonstrates that m6A is a molecular switch that regulates stem cell differentiation, a fundamental mechanism in development and stem cell biology. Moreover, recent work showed that m6A regulates haematopoietic stem cell regeneration 37 and axon regeneration in the mouse nervous system. 38 While those previous studies have provided important insights into the underlying molecular mechanism of m6A‐mediated regulation of regeneration at the cellular level, whether and how m6A is implicated in regeneration in tissues and entire organisms is unknown.
Here, we employed planarian Schmidtea mediterranea as the model to investigate the role of m6A in stem cell function and whole‐body regeneration. The results illustrated that depletion of the m6A methyltransferase regulatory subunit wtap abolished planarian regeneration, which is mainly mediated by wtap‐dependent m6A genes functioning in cell–cell communication and cell cycle through m6A‐seq analysis. Further, single‐cell RNA‐seq (scRNA‐seq) demonstrated that a unique type of neural progenitor‐like cells (NP‐like cells) specifically expressing a cell–cell communication ligand granulin (grn) was induced upon wtap depletion. Intriguingly, depletion of wtap with any one of GRN, cyclin‐dependent kinase 9 (CDK9) and CDK7, a signalling axis modified by m6A, partially rescues the defective regeneration of wtap knockdown planarian. Collectively, we reveal an indispensable role of m6A for whole‐organism regeneration. These findings improve our current understanding of the critical molecular events controlling regeneration and have the potential to benefit future cell‐ or tissue‐based replacement therapies.
2. RESULTS
2.1. m6A methyltransferase complex upregulated during planarian regeneration
During planarian regeneration, gene expression profiles of all regeneration time points undergo continuous changes that can be represented by three distinct timepoints including 6 h post‐amputation (hpa), 3 days post‐amputation (dpa), and 7 dpa. 20 These timepoints display a unique composition of piwi‐1 (smedwi‐1) positive population and smedwi‐1 negative population, 20 resembling the combination of both processes of tissue regeneration from proliferating neoblasts (epimorphosis) and remodelling of the existing tissues (morphallaxis). 39 In order to gain an overview of the gene expression dynamics from our own perspective during planarian regeneration, we performed RNA‐seq at five different time points after amputation (0 hpa, 6 hpa, 3 dpa, 7 dpa, 11 dpa; 0 hpa means sample collected immediately after amputation; Figure 1A). To investigate the gene expression changes of the planarian during regeneration, we grouped all the expressed genes into five clusters according to their distinct expression pattern, with each cluster containing groups of genes upregulated specifically at a certain timepoint after amputation (Figure 1B). Since m6A modification is catalysed by three key components of the methyltransferases, including mettl3, mettl14 and indispensable regulatory component wtap, 40 , 41 we found that all these three genes displayed upregulated expression at 3, 7 and 11 dpa (Figure 1C), and all belong to the clusters 3 and 4. Based on the gene ontology (GO) functional analysis, several genes in cluster 4 were observed to be related to the regulation of system process, signalling and cell communication (Figure 1D, Table S1), including growth factors and transcription factors such as wnt2 and egfr (Figure 1D). Furthermore, GO enrichment analysis revealed that genes from cluster 4 also involve in nervous system development and differentiation (Figure 1E, Table S1). Since the components of m6A enzymes have a similar expression trend as genes in cluster 4, these findings suggest the potentially important roles of m6A in regulating planarian regeneration through the aforementioned pathways.
FIGURE 1.
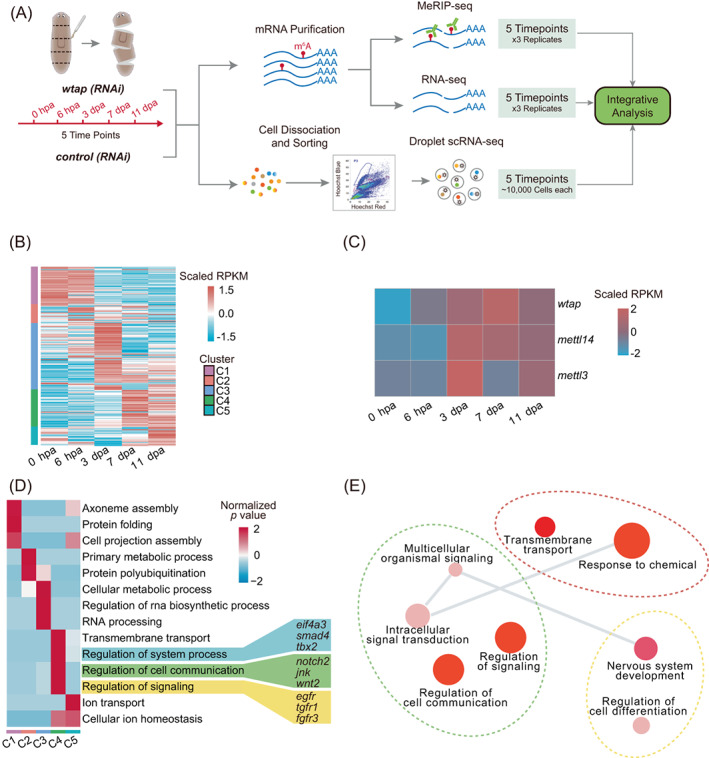
m6A methyltransferase complex upregulated during planarian regeneration. (A) Experimental outline of multi‐omics data including methylated RNA immunoprecipitation sequencing (MeRIP‐seq), bulk RNA‐seq and single‐cell RNA‐seq. (B) Heatmap showing the expression levels of all expressed genes and clustered into five categories based on expression pattern during planarian regeneration (left side of the heatmap). Genes were clustered by MEV with parameter—distance‐metric‐selection = Pearson correlation, number of cluster = 5, maximum iterations = 50 and plot with the parameter ‘pheatmap (matrix, scale =“row”)’ through pheatmap function (R package). (C) Heatmap showing the expression levels of major components of m6A writers during regeneration. (D) Heatmap showing the enriched gene ontology (GO) and important genes of five different categories as shown in (B). The colour bar of heatmap represents z‐score of GO's p‐value. (E) Gene ontology of genes belongs to fourth category (C4) shown in (B). Visualization was performed using Revigo with default parameters (semantic similarity measure = SimRel).
2.2. m6A changes dynamically during planarian regeneration
To determine the regulatory role of m6A in planarian regeneration, we first measured the relative level of m6A in planarian mRNA, which shows a much higher level of over 0.5% relative to another modification on adenosine (N 1‐methyladenosine, m1A) by using UHPLC‐MRM‐MS/MS (ultra‐high‐performance liquid chromatography‐triple quadrupole mass spectrometry coupled with multiple‐reaction monitoring; Figure 2A).
FIGURE 2.
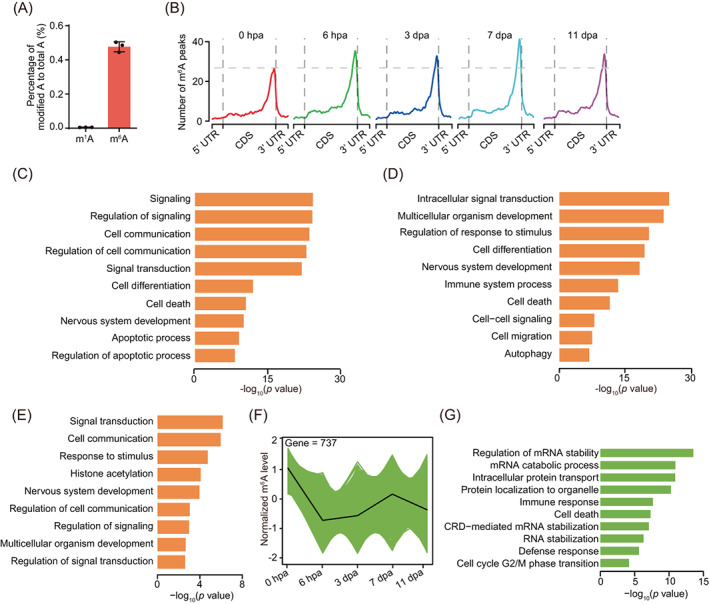
m6A changes dynamically during planarian regeneration. (A) Barplot showing the abundance of m1A and m6A along mRNA. The methylation level of each RNA methylation was quantified via UHPLC‐MRM‐MS/MS. The abundance of each modification was calculated with the percentage of modified A to total A. (B) Metagene profiles of m6A peaks along transcripts with three non‐overlapping segments (5′UTR, CDS and 3′UTR) for five regeneration stages. (C) Barplot showing the significant GO terms for 6385 mRNAs that conserved with m6A modification of five different timepoints shown in Figure S1C. (D) Barplot showing the significant gene ontology (GO) terms for 12,016 mRNAs that with m6A modification of at least one timepoint during regeneration showing in Figure S1C. (E) Barplot showing the significant GO terms for 1532 mRNAs that with m6A modification of four different regeneration timepoints (6 hpa, 3 dpa, 7 dpa and 11 dpa) showing in Figure S1C. (F) Line chart showing one of the trends (fourth category) of mRNAs m6A level during regeneration, which with gradual decreased m6A level from 0 hpa to 11 dpa. mRNAs with different expression pattern were defined by MEV with parameter—distance‐metric‐selection = Pearson correlation, number of cluster = 4, maximum‐iterations = 50. (G) Barplot showing the significant GO terms for mRNAs shown in Figure 2F. See also Figure S1.
To obtain m6A landscape during planarian regeneration, we performed transcriptome‐wide m6A mapping on samples of five time points (0 hpa, 6 hpa, 3 dpa, 7 dpa, 11 dpa) of regenerative process (Figure 1A). We carried out methylated RNA immunoprecipitation sequencing (MeRIP‐seq) to investigate the features and distribution dynamics of m6A in mRNA. In total, we identified 9057–13,362 m6A peaks over all five different regeneration time points. We found that most genes have one MeRIP peak (Figure S1A). The total number of m6A peaks at each stage (especially at 6 hpa and 7 dpa) of regeneration is slightly higher than that at 0 hpa (Figure S1B). Consistent with previous observations in mammals, 31 we found that m6A peaks were markedly enriched near the stop codon. Also, the distribution pattern of m6A is similar among different regeneration time points (Figure 2B). While transcripts of 6385 genes contain m6A peaks at all regeneration time points including the one before amputation (0 hpa), 1532 genes show transcript being modified only after amputation (i.e., only in timepoints other than 0 hpa). We can also find m6A‐modified transcripts that are unique to each particular time point during regeneration (Figure S1C, Table S2).
GO enrichment analysis revealed that the 6385 shared m6A‐modified gene transcripts are associated with various essential biological processes, including regulation of signalling, regulation of cell communication and cell differentiation (Figure 2C, Table S1). The m6A‐modified genes during regeneration were related to signal transduction, cell differentiation, and nervous system development (Figure 2D, Table S1), indicating an important role of m6A in these regeneration‐related processes. The overlap of all m6A‐modified transcripts with five different clusters indicates that the cluster 4 genes with upregulation at 7 and 11 dpa show the highest overlap mostly with m6A‐modified gene transcripts (Figure S1D). GO analysis shows that the genes with their transcripts exclusively modified during the regeneration period are related to response to stimulus, cell communication and signal transduction pathways (Figure 2E, Table S1).
By classifying genes based on different patterns of the change of m6A modification level during regeneration (Figures S1E‐J and S2F–G), we found that genes with m6A level change following the same trend of wtap expression change during regeneration were related to cell communication, signal transduction and system development (Figure S1E‐J, Table S1). Conversely, genes with decreased level of m6A during regeneration showed involvement in the regulation of mRNA stability and cell cycle G2/M phase transition‐related pathways (Figure 2F–G, Table S1). These results may imply a potential role of m6A in cell–cell communication and neuron development.
2.3. m6A methyltransferase wtap depletion impairs regeneration in planarians
To further study the functional roles of m6A modification in planarians, we first performed whole‐mount in situ hybridization of m6A methyltransferase regulatory subunit, wtap, and found that wtap specifically colocalizes with the neoblast marker smedwi‐1, especially at the posterior region, suggesting a regulatory role of m6A in neoblast (Figure 3A,B). To study whether m6A modification is indispensable for planarian regeneration, the key regulatory unit, wtap, was knocked down by RNAi, and then the worm was amputated at both anterior and posterior regions (Figure 3C). Protein expression and mRNA levels of wtap were significantly reduced compared to the control, as shown by whole‐mount immunostaining (Figure 3D), western blot (Figure S2A) and quantitative reverse‐transcription PCR (qRT‐PCR) (Figure S2B). Accordingly, the mRNA m6A level was reduced by nearly 80% at 7 dpa (Figure S2C).
FIGURE 3.
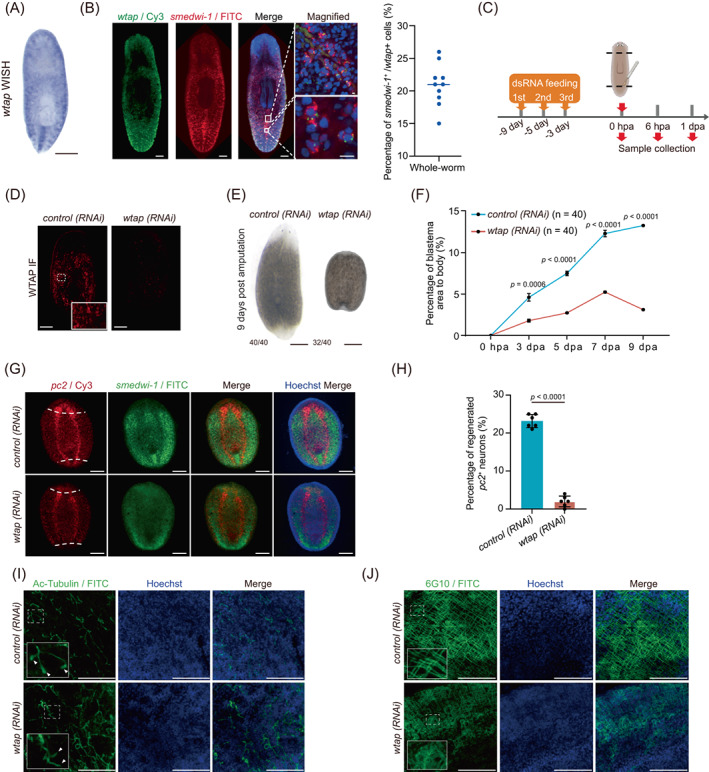
wtap depletion leads to regeneration defects. (A) WISH showing the expression and localization of wtap transcripts in planarians. Scale bar, 300 μm. (B) Whole‐mount fluorescent in situ hybridization showing the expression and localization of wtap and smedwi‐1 transcripts. Scale bar, 100 μm (left panels), 10 μm (right upper row), 10 μm (right lower row). Right panel, scatter plot showing the percentage of smedwi‐1 +/wtap + cells. (C) Schematic diagram showing the knockdown strategy and amputation position. (D) Immunofluorescence showing expression and localization of WTAP protein in control (control) and wtap knockdown (wtap RNA interference [RNAi]) planarians (n ≥ 3). Scale bar, 200 μm. (E) Bright‐field image showing total body sizes for control (control) and wtap knockdown (wtap RNAi) planarians at 9 dpa. Scale bar, 500 μm. Bottom left number, planarians with phenotype of total tested. (F) Percentage of blastema area to total body size in control (control) and wtap knockdown (wtap RNAi) planarians. Error bars represent standard deviation. Data are the mean ± S.D (n ≥ 3 independent experiments). The p values were determined using a two‐sided unpaired Student's t‐test. (G) Whole‐mount fluorescent in situ hybridization showing expressions and localizations of pc2 (red) and smedwi‐1 (green) along whole body of control (control) and wtap knockdown (wtap RNAi) planarians at 7 dpa. Dotted line, amputation plane (n ≥ 3). Scale bar, 200 μm. (H) Quantification analysis of the percentage of regenerated pc2 positive neuron in control (control) and wtap knockdown (wtap RNAi) planarians at 7 dpa. Error bars represent standard deviation. Data are the mean ± S.D. (n ≥ 3 independent experiments). The p values were determined using a two‐sided unpaired Student's t‐test. (I) Immunofluorescence showing the distribution of Ac‐Tubulin protein in control (control) and wtap knockdown (wtap RNAi) planarians at 7 dpa. Arrow indicates the presence of flame cell in the protonephridia of control (control) planarians, and missing in the wtap knockdown (wtap RNAi) planarians (n ≥ 3). Scale bar, 75 μm. (J) Immunofluorescence shows the distribution of 6G10 protein distribution in control (control) and wtap knockdown (wtap RNAi) planarians at 7 dpa. (n ≥ 3) Scale bar, 75 μm. See also Figure S2.
Most importantly, wtap‐deficient planarians failed to fully regenerate the amputated tissue parts compared to the controls, which regrow head and tail tissues at 9 dpa (Figure 3E). Moreover, newly regenerated tissue in the wtap‐deficient planarian area was significantly smaller at 3 dpa than that of the control planarians (Figure S2D). The blastema formed at the wound region of wtap‐deficient planarians was also smaller and failed to grow the missing anterior and posterior tissues in comparison with the control planarians (Figure 3F). Especially at 7 dpa, when control planarians had regrown photoreceptors, the wtap knockdown planarians appeared to be still unable to give rise to any new tissues from their healed wounds (Figure S2D). We noticed that smedwi‐1 RNA expression did not change obviously in wtap knockdown planarians (Figure 3G). Since smedwi‐1 RNA is one of the canonical markers of stem cells in planarians, wtap‐mediated m6A depletion may affect neoblast function. At the same time, the signatures of the level of regeneration are characterized by the regeneration of the nervous system, containing a pair of ventral nerve cords and the brain. 42 However, neither part was regrown in wtap knockdown planarians, shown by whole‐mount fluorescence in situ hybridization (FISH) staining of pc2 (Figures 3G,H and S2E), a neuropeptide processor expressed throughout the planarian central nervous system (CNS). Apart from the failure to regenerate new tissues, the existing tissues were also abnormal upon wtap knockdown, such as the missing ciliated ‘flame cells’ of protonephridia in the excretory system (Figure 3I) and disorganized muscle fibres (Figure 3J), which were indicated by the immunofluorescence staining of Ac‐tubulin and 6G10, respectively.
Impaired regeneration and abnormal tissue morphology may occur due to the abnormality of the proliferation of the neoblast and progenitor cells, or apoptosis of terminally differentiated cells. Thus, we investigated if proliferation and apoptosis are impaired in wtap knockdown planarians. We first performed BrdU pulse‐chase labelling assay and revealed BrdU‐labelled cells planarians were more enriched near the amputation site in wtap knockdown than that in control planarians, indicating the cell proliferation near the amputation site is more pronounced under wtap knockdown relative to control (Figure S2F,G). Then, we performed TUNEL assay to analyse apoptosis. The results showed a remarkable increase in cells undergoing apoptosis starting from 3 dpa in wtap‐deficient in comparison to control planarians (Figure S2H,I). These results indicate that m6A is essential for planarian regeneration, and its depletion leads to abnormal cell proliferation accompanied by increased apoptosis.
2.4. WTAP‐mediated m6A controls cell cycle‐ and cell–cell communication‐related factors essential for regeneration
To investigate the underlying molecular mechanism for the defective planarian regeneration mediated by a decreased m6A level upon wtap knock‐down, we performed m6A MeRIP‐seq in control and wtap knockdown planarians (Figure 1A). Even though the distribution of m6A along the CDS only slightly differs in wtap knockdown compared to controls, m6A peaks near stop codons become visibly reduced in wtap knockdown compared to the control worms at each time point during regeneration (Figure 4A). We found that among the genes having upregulation trends in cluster 4 (Figure 1B), many genes showed a disordered expression pattern after wtap knockdown (Figure 4B). Further, GO functional analysis indicated that these genes are related to the regulation of signalling, cell communication and signal transduction‐associated pathways (Figure 4B, Table S1). We then analysed the differentially expressed genes in each time point between control and wtap knockdown planarians. During the regeneration process, the number of both upregulated and downregulated genes went up starting from 0 hpa with the highest number of genes changed at late stages, suggesting that wtap deficiency ultimately leads to regeneration failure through affecting a series of genes in multiple stages of regeneration (Figure S3A). This indicates a sustaining, persisting and specific effect of wtap deficiency during the regeneration process. Gene set enrichment analyses revealed a marked upregulation of the cell cycle checkpoint signature and downregulation of cell–cell signalling in wtap deficient samples compared to control during regeneration (Figures 4C and S3B).
FIGURE 4.
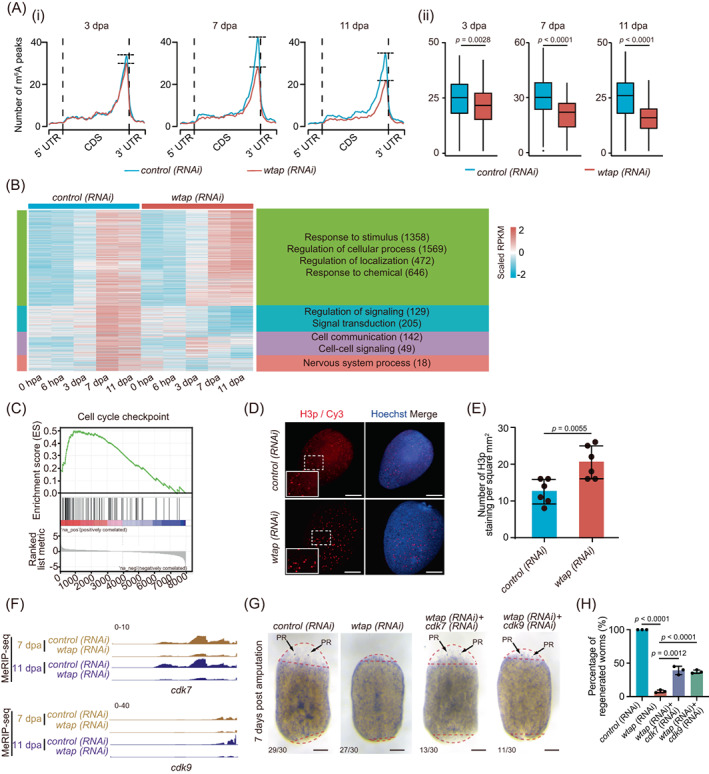
WTAP‐mediated m6A controls cell cycle‐ and cell–cell communication‐related factors. (A) (i) Metagene profiles of m6A peaks along transcripts with three non‐overlapping segments (5′‐UTR, CDS and 3′‐UTR) for control (blue) and wtap knockdown (red) planarians of different regeneration timepoints (3, 7, and 11 dpa). (ii) Boxplot showing the difference in the number of m6A peaks that located near stop codon of transcripts of control (blue) and wtap knockdown (red) planarians at 3, 7 and 11 dpa. The p values were determined using Wilcoxon‐test. (B) For genes from cluster 4 (C4) in Figure 1B, the expression level during regeneration were displayed for both control and wtap knockdown planarians of five timepoints. Then those genes were separated into four new sub‐groups based on expression features and the enriched GO terms were shown for each sub‐group. Genes with different expression pattern were defined by MEV with parameter–distance‐metric‐selection = Pearson‐correlation–number‐of‐cluster = 4–maximum‐iterations = 50. (C) GSEA plots evaluating the changes in cell cycle checkpoint pathway upon wtap depletion. Normalized p value <0.01. (D) Immunofluorescence showing the distribution of H3P protein in control (control) and wtap knockdown (wtap RNA interference [RNAi]) planarians at 5 dpa. Scale bar, 200 μm. Enlarged field was selected to best represent the statistical mean of staining signals. (E) Statistical analysis for H3p immunostaining. Error bars represent standard deviation. Data are the mean ± S.D. (n ≥ 3 independent experiments). The p values were determined using a two‐sided unpaired Student's t‐test. (F) Integrative genomics viewer tracks displaying the distributions of MeRIP‐seq data that normalized by RNA‐seq data along cdk7 and cdk9 in both control (top) and wtap knockdown (bottom) planarians at 7 dpa and 11 dpa. (G) Bright‐field images showing the phenotypes of control, wtap, cdk7 + wtap and cdk9 + wtap knockdown planarians at 7 dpa. Scale bar, 200 μm. Bottom left number, number of planarians with phenotype versus total number tested. (H) Barplot showing percentage of regenerated worms to total sample size in control, wtap, cdk7 + wtap and cdk9 + wtap knockdown planarians at 7 dpa. Error bars represent standard deviation. Data are the mean ± S.D. (n ≥ 3 independent experiments). The p values were determined using a two‐sided unpaired Student's t‐test. See also Figure S3.
We then performed immunofluorescence analysis using phosphorylated histone 3 (H3p), a typical marker for M phase, and found a significant increase in the percentage of M phase cells upon wtap knockdown, and no significant difference after 5 dpa in control planarians (Figure 4D, E). One of the key regulators of G2/M phase, Cyclin B1, was shown to have an elevated level of whole‐mount FISH staining during regeneration (Figure S3C). Therefore, combining the results that unaltered proliferation near the wound site but increased apoptosis upon wtap knockdown, we propose that wtap deficiency might drive the entry of the cells into the cell cycle resulting in M phase stalling and eventually cell death. We further observed that the mRNA expression of cyclin‐dependent kinase 7 (cdk7) and cdk9, both important cell cycle‐related factors, 43 , 44 , 45 , 46 , 47 was upregulated upon wtap knockdown, while the level of m6A modification decreased (Figure 4F). In addition, genes related to cell–cell communication, such as notum and kcnd2, were downregulated and their m6A level also decreased after wtap knockdown (Figure S3E,F). Furthermore, double knockdown of cdk7 or cdk9 with wtap partially rescues the wtap knockdown phenotype (Figure 4G) from 9% regenerated worms in wtap knockdown group up to 39% and 37% in cdk7/wtap and cdk9/wtap double knockdown groups, respectively (Figure 4H), while neither of cdk7 nor cdk9 single knockdown affects regeneration (Figure S3G). This indicates that wtap knockdown phenotype is mediated by cdk7 or cdk9. Taken together, the results imply that WTAP controls planarian regeneration potentially through m6A‐modified genes functioning in cell–cell communication and cell cycle.
2.5. scRNA‐seq unveils impact of WTAP on specific cell types essential for regeneration
To decipher the detailed mechanism of m6A in regulating planarian regeneration, we conducted single‐cell transcriptome sequencing of planarian Schmidtea mediterranea from five‐time points (0 hpa, 6 hpa, 3 dpa, 7 dpa, 11 dpa) in control and wtap knockdown samples. In total, 86,758 cells were successfully detected from all sequencing datasets of all samples. These cells were pooled together to generate a whole cell‐type specific atlas. To identify individual cell types, we first performed unsupervised clustering using highly variable genes, and obtained 116 clusters by the Uniform Manifold Approximation and Projection (UMAP) method. We then elucidated the cell type identity of each cluster by identifying marker genes and compared them to markers in previous studies 18 , 20 (Figure S4A). Overall, seven cell type groups such as neoblast, neuron, muscle, gut, secretory, epidermal and parenchymal cells were identified in both control and wtap knockdown samples (Figure 5A, Table S3).
FIGURE 5.
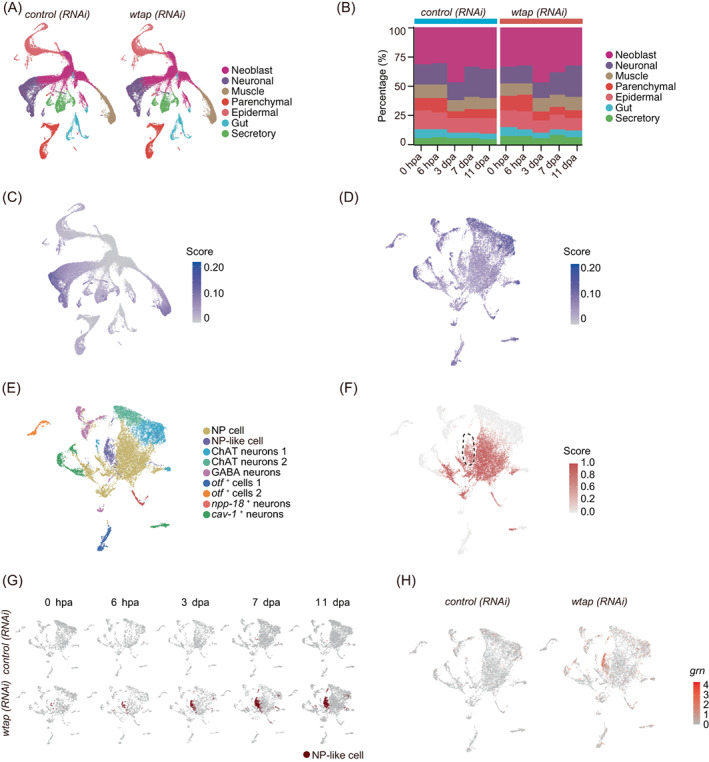
Single‐cell atlas unveils cell‐type specific regulation of WTAP essential for regeneration. (A) Based on single‐cell RNA‐seq data, Uniform Manifold Approximation and Projection (UMAP) detect major cell types in both control (left) and wtap knockdown (right) planarians of 5 timepoints during regeneration (0 hpa, 6 hpa, 3 dpa, 7 dpa and 11 dpa). Each point depicts a single cell, coloured according to cell types. (B) The percentage of each cell type in control (left) and wtap knockdown (right) planarians during regeneration. (C) Expression score of transcripts with m6A modifications are plotted onto the UMAP map. Expression score is calculated by using the AddModuleScore function from Seurat, and the transcripts used to score cells are from cluster 4 (C4) of Figure 1B and required with m6A modification. (D) Expression score of transcripts with m6A modifications are plotted onto the UMAP map in neuronal cells. Expression score is calculated by using the AddModuleScore function from Seurat, and the transcripts used to score cells are from cluster 4 (C4) of Figure 1B and required with m6A modification. (E) UMAP plot demonstrates nine sub‐clusters of neuronal cells in control and wtap knockdown planarians. Each point depicts a single cell, coloured according to cell types. (F) The NP cell prediction score of neuronal cells. The score is calculated by using the TransferData function from Seurat and the score represents the degree of similarity to neuronal progenitor cells. (G) UMAP plot showing a novel cell type, NP‐like cells, mainly occur in wtap knockdown planarians but rarely in controls. Each point depicts a single cell. (H) Expression level of grn in neuronal cells. Control samples on the left, wtap knockdown samples on the right. See also Figure S4.
In order to explore the regulation of m6A in cell lineage formation during planarian regeneration, we compared the cell–cell composition within control and wtap knockdown planarians. We found that the proportion of neural cells changes during regeneration. At 7 dpa, the proportion of regenerated neuronal cells in control planarians is about 25.03%, while it was 18.65% in wtap knockdown planarians, namely compared with control planarians was about 25.50% (6.38%/25.03%) decreased of neuronal cells proportion upon wtap knockdown. Notably, about one‐fourth of neuronal cells could not repopulate normally after wtap knockdown planarians (Figure 5B, Table S4). Except for neuronal cells, the distribution of the other cell types between control and wtap knockdown samples was similar. In order to explore the cellular functional changes after wtap knockdown, we compared the control and wtap‐deficient samples of each cell type and found that epidermal and neoblast contain the most differentially expressed genes (DEGs) in total (Table S4). GO analysis revealed that during the regeneration, after wtap knockdown, the upregulated genes in epidermal were enriched in the nucleic acid metabolic process and cell cycle‐related functions (Figure S4B, Table S7), and related to cell cycle regulation functions in neoblast (Figure S4C). In addition, the downregulated genes are distributed in epidermal, neoblast and neuronal cells from 3 to 11 dpa. Moreover, the downregulated genes in epidermal cells are related to the establishment of tissue polarity, translation, and ATP biosynthetic process (Figure S4D). The functions of downregulated genes in the neoblast are associated with RNA splicing, DNA replication initiation, protein folding, and so forth (Figure S4E). Neuronal cells are associated with protein folding, neurotransmitter secretion and signal release from synapse functions (Figure S4F). These data suggest that wtap knockdown‐induced regeneration retardation might through affecting the cell cycle and signal transduction pathways of neoblast and neuronal cells.
2.6. WTAP‐mediated m6A depletion increases GRN levels, which potentially control cell–cell communication essential for planarian regeneration
The results of single‐cell data showed that the proportion of neural system showed dominant changes after wtap knockdown, and the m6A modified genes with the same expression trend as the known m6A methyltransferases were enriched in the neuronal cells (Figure 5C,D). In addition, planarians could not regenerate nerve cord structure after wtap knockdown (Figure 3G,H). In order to explore the underlying mechanisms for abnormal regeneration of the planarian nervous system, we further classified neuronal cells according to the reported markers and also transferred the reported cluster labels onto our data by Seurat. 18 We clustered neuronal cells into nine subtypes, neural progenitor cells (NPCs), NP‐like cells, ChAT neurons 1, ChAT neurons 2, GABA neurons, otf + cells 1, otf + cells 2, npp‐18 + neurons, cav‐1 + neurons (Figure 5E). Among them, we identified a unique NP‐like cell cluster during regeneration after wtap knockdown (Figure 5E,F), which similar to neural progenitor cells, gradually increases during regeneration but is rarely detected in the control planarians (Figure 5G). We further identified grn (SMED30003962), ubiqp (SMED30009873) and dph1 (SMED30026267) as specific m6A‐modified markers of NP‐like cells (Figures 5H and S5A–D, Table S5).
To further investigate the functions of NP‐like cells in planarian regeneration, we analysed the effect of wtap knockdown on the expression of NP‐like cell markers. First, the IGV and enrichment analyses showed that the levels of m6A modification on grn and ubiqp transcripts were significantly decreased upon wtap knockdown at both 7 and 11 dpa (Figures 6A and S5C). Consistently, WTAP‐mediated m6A depletion leads to a drastic increase in grn and ubiqp expression in wtap‐deficient planarians (Figure S5B). Furthermore, through both whole‐mount fluorescent in situ hybridization and immunofluorescence assays, we observed an increase in grn mRNA and protein levels (Figure 6B–D), as well as increased colocalization of two markers of NP‐like cells, grn and dph1 (Figure 6B,C). In addition, the expression of dph1 has been restored by the double knockdown of wtap and grn (Figure S6A). It has been reported that GRN is a secretory protein important for neuronal functions 48 , 49 , 50 and can exert function over a distance. 48 , 51 Furthermore, GRN can stimulate cell division by promote G1 phase to M phase directly. 52 We found that the double knockdown of wtap and grn remarkably rescued the regeneration failure of planarians induced by wtap knockdown compared to the control (Figures 6B–F and S5E), even though grn knockdown did not affect regeneration (Figure S5F). Moreover, NP‐like cell cluster disappeared upon the double knockdown of wtap with grn, cdk7 or cdk9 as demonstrated by FISH staining of its markers (Figure 6B–D). In addition, co‐staining of BrdU and H3p revealed restored cell proliferation pattern and cell cycle state in the double knockdown worms of wtap with grn, cdk7 or cdk9 (Figure S6B–D). Notably, the whole mount FISH staining of pc2 also showed restored regeneration of CNS in double knockdown worms (Figure 6G,H). Our single‐cell sequencing data showed that Cyclin T1 (ccnt1), cdk7 and cdk9 have relative higher transcript abundance in the neoblast than any other cell types in the wtap knockdown planarians (Figure S7A), and NP‐like cells communicate with other cell types potentially through grn targeting on its receptor sort1 based on cell–cell communication analysis using CellphoneDB (Figure S7B, C). These results suggest that GRN excreted by NP‐like cells might potentially act on neoblast. At the same time, for other cell–cell communication‐related gene transcripts containing m6A modification, such as mtnr1b, their ligands are also modified by m6A and upregulated upon wtap knockdown and seem to participate in planarian regeneration (Figure S7D, E). These data suggest that WTAP‐mediated m6A controls GRN expression levels, which further influences the regeneration of planarians.
FIGURE 6.
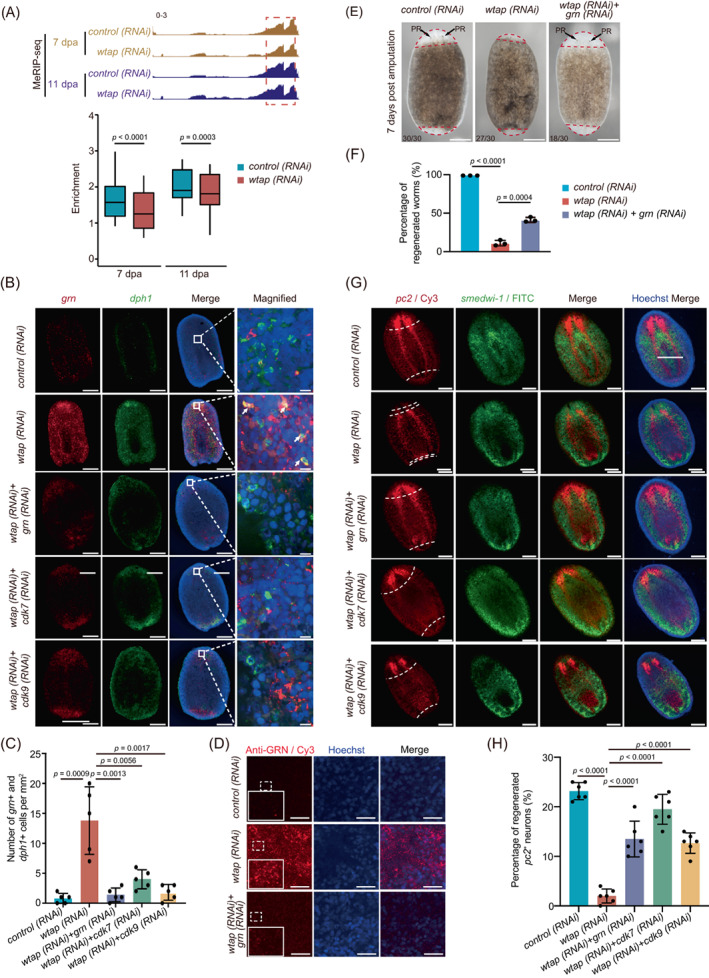
WTAP‐mediated m6A depletion increases GRN levels, and influences cell–cell communication essential for planarian regeneration. (A) Integrative genomics viewer tracks displaying the distributions of methylated RNA immunoprecipitation sequencing (MeRIP‐seq) data that normalized by RNA‐seq data along grn in both control and wtap knockdown planarians at 7 and 11 dpa (top). Barplot showing the enrichment of m6A modification on grn. The p values were determined using Wilcoxon‐test. (B) Whole‐mount fluorescent in situ hybridization showing expressions of grn (red) and dph1 (green) in control, wtap knockdown (wtap RNA interference [RNAi]), double knockdown of wtap and grn (wtap + grn RNAi), double knockdown of wtap and cdk7 (wtap + cdk7 RNAi), double knockdown of wtap and cdk9 (wtap + cdk9 RNAi) planarians at 7 dpa. Hoechst indicates DNA staining. Scale bar, 200 μm (left panels), 10 μm (magnified panels). (C) Barplot showing the number of grn +/dph1 + cells of control (control), wtap knockdown (wtap RNAi), double knockdown of wtap and grn (wtap + grn RNAi), double knockdown of wtap and cdk7 (wtap + cdk7 RNAi), double knockdown of wtap and cdk9 (wtap + cdk9 RNAi) planarians in (B). Data were analysed by the two‐tailed unpaired Student's t‐test (bottom). (D) Immunofluorescence showing the expression and localization of GRN protein in control (control), wtap knockdown (wtap RNAi), double knockdown of wtap and grn (wtap + grn RNAi) planarians at 7 dpa. Hoechst indicates DNA staining. Scale bar, 100 μm. (E) Bright‐field images showing the phenotypes of control (control), wtap knockdown (wtap RNAi), double knockdown of wtap and grn (wtap + grn RNAi) planarians at 7 dpa. Scale bar, 300 μm. Bottom left number, number of planarians with phenotype versus total number tested. (F) Barplot showing percentage of regenerated worms in control (control), wtap knockdown (wtap RNAi), double knockdown of wtap and grn (wtap + grn RNAi) planarians at 7 dpa. Error bars represent standard deviation. Data are the mean ± SD (n ≥ 3 independent experiments). The p values were determined using a two‐sided unpaired Student's t‐test. (G) Whole‐mount fluorescent in situ hybridization showing expressions and localizations of pc2 (red) and smedwi‐1 (green) of control (control), wtap knockdown (wtap RNAi), double knockdown of wtap and grn (wtap + grn RNAi), double knockdown of wtap and cdk7 (wtap + cdk7 RNAi) and double knockdown of wtap and cdk9 (wtap + cdk9 RNAi) planarians at 7 dpa. Dotted line, amputation plane (n ≥ 3). Scale bar, 200 μm. (H) Quantification of percentage of pc2 positive regenerated neurons in each group of (G). Error bars represent standard deviation. Data are the mean ± SD (n ≥ 3 independent experiments). The p values were determined using a two‐sided unpaired Student's t‐test. See also Figures S5–S7.
3. DISCUSSION
Regeneration is a dynamic and tightly controlled process involving a complex yet well‐orchestrated gene regulation network. However, the potential significance of RNA m6A modification in regeneration is not well understood. Recently, one study reported that kiaa1429 knockdown resulted in failed regeneration and decreased m6A level as well as accordingly changed gene expression pattern. 53 In our study, we discovered that abundant RNA m6A modification exists in the planarian Schmidtea mediterranea, with a single high peak near the stop codon region and enrichment at CDS and 3′‐UTR region of mRNA, which are similar to the features in most model organisms. Interestingly, this m6A peak increases along the regeneration process, and functional analysis further demonstrated that the m6A‐modified genes are related to pathways of cell–cell communications, nervous system development, mRNA stability and cell cycle G2/M phase transition, suggesting important regulatory roles of m6A modification in planarian regeneration. Moreover, m6A depletion by wtap knockdown led to a complete loss of the regenerative ability of planarians attributing to the induction of cell cycle related genes cdk7 and cdk9 as well as the emergence of NP‐Like cell cluster with aberrant expression of cell–cell communication‐related secretory gene grn, identified by m6A sequencing analysis and rescue assays. Therefore, the signal axis of wtap‐m6A‐regulated grn/cdk9/cdk7 determines the planarian regenerative potential through modulating cell–cell communication‐related gene expression.
RNA m6A modification has been reported to regulate stem cell differentiation and tissue regeneration. 27 , 29 , 36 , 37 , 38 , 54 It is required for axon regeneration by promoting protein translation of regeneration‐associated genes in the rodent model. 38 Meanwhile, deletion of m6A reader YTH domain‐containing family protein 2 (YTHDF2) in mouse haematopoietic stem cells (HSC) was shown to facilitate HSC regeneration by enhancing the stability of mRNAs related to both Wnt signal and survival. 37 In this study, using planarian Schmidtea mediterranea model system, we found the m6A peaks increase at CDS and 3′‐UTR junction during the regeneration progression. Moreover, knockdown of m6A regulatory unit, wtap, resulted in a significant reduction in mRNA m6A level and abolished the regenerative capability of planarian to regrow missing head and tail. These findings suggest that wtap‐mediated m6A is imperative for the whole‐organism regeneration of planarian.
Regeneration involves cell proliferation and the consequent generation of new tissues, which requires tight control of the cell cycle. 55 In our study, we found that cell cycle‐related genes such as cdk7, cdk9 and ccnt1 are mainly expressed in neoblasts and upregulated upon wtap knockdown. Both CDK7 and CDK9 are members of the cyclin‐dependent protein kinase family with effector CDK activity to phosphorylate Pol II and other targets within the transcriptional machinery, and also serve as a CDK‐activating kinase for at least one other essential CDK involved in transcription. 56 , 57 , 58 Both CDK7 and CDK9 have been shown to be required for the cell cycle regulation, such as the blockage of all cell divisions in C. elegans by Cdk7 loss 43 and G1 cell cycle arrest of D.melanogaster cells by RNAi‐mediated Cdk9 silencing. 47 Moreover granulin (grn)/progranulin has been shown to stimulate cell division, 52 and grn inhibition decreases gata1 expression and further the differentiation of erythroid in zebrafish. 59 In support, we found an induced expression of grn, cdk7 and cdk9 post wtap knockdown accompanied by impaired regeneration, suggesting that m6A modification are critical for planarian regeneration through destabilizing the transcripts of these three genes.
Importantly, accumulative studies revealed that regeneration is a complicated process involving cell–cell communication in various tissue types. 60 The flow of long‐range patterning information in regenerative morphogenesis is crucial to control stem cell behaviour in vivo. 61 For example, muscle regeneration in response to injury, is a non‐specific inflammatory response to trauma, involving interaction between muscle and the immune system. 60 Specifically, growth factors, such as insulin‐like growth factor I (IGF1), released by M1 macrophage, stimulate the growth of muscle progenitor cells at the terminal differentiation stage of regeneration. Notably, because of the limitation of GO terms annotated in planarian, we aligned the planarian transcriptome with the uniport database through blastx program, and selected the best comparison result. According to the comparison results and the related GO annotations, the GO annotations of planarian were enriched. And based on the GO analysis, we found that the cell communication pathway increased upon wtap knockdown. Finally, we identified a specific NP‐like cell cluster secreting growth factor GRN after wtap knockdown, suggesting that GRN might selectively target the neoblast to inhibit their differentiation into neural progenitor cells and block the eventual formation of individual organs.
Among all major cell types identified in planarian by scRNA‐seq, 17 , 18 , 20 the most important cell type among them is the neoblasts, which can provide the cellular source for robust planarian regeneration. This totipotent stem cell can differentiate into all other cell types after injury or stimulation. Soon after amputation, neoblast starts to proliferate and migrate to wound site to form regenerative blastemal tissue, which becomes the foundation for re‐growing the missing tissue. 5 However, both blastema formation and remodelling of pre‐existing tissue (morphallaxis) are required for the planarian regeneration. 55 These intricate processes require correct cell–cell communications and precise coordination between stem cells and pre‐existing tissue to ensure the regeneration is finished on the right track. 5 In our study, we identified a neural progenitor‐like cell cluster accumulated during the regeneration upon wtap knockdown, and further experiments demonstrated that the aberrant high expression of m6A modified gene, grn, secreted from this cell cluster disrupts the regeneration process. It is likely that such an abnormal cell cluster is supposed to be a group of neural progenitor cells at a special state during normal neural regeneration, which at the same time is also critical for subsequent events of regeneration of other tissue types to proceed. Therefore, we speculate that this special state of neural progenitor cell cluster, as we amplified its cell number by wtap knockdown, triggers a critical checkpoint regulated during the entire regeneration process that involves cell–cell communication mediated by GRN to ensure the correct path for regeneration, and wtap knockdown induced aberrant grn expression will stop the regeneration at this checkpoint.
As depicted in our model (Figure S7F), during planarian homeostasis, the expression level of grn is held at a moderate level to inhibit overgrowth, as equivalent to grn expression level before amputation at 0 hpa (Figure S5B). Upon injury, m6A modification selectively targets several important gene transcripts, including grn for degradation, manifested as its reduced expression level after 6 hpa (Figure S5B), as well as transcripts of cell cycle‐related genes including ccnt1, cdk7 and cdk9. m6A‐mediated downregulation of these genes, especially the grn, may halt initial proliferation of stem cell pool at the first stage of regeneration, which in turn promotes their differentiation process, including the differentiation of neoblasts to the progenitor cells and eventually to different tissue types. After wtap knockdown, grn transcript accumulates to form a unique cell cluster resulting in increased GRN secretion, which may lead to cell cycle dysregulation and the loss of neoblast differentiation and the failure of planarian regeneration.
Nevertheless, the detailed mechanisms of cell–cell communication need further investigations and experimental proof. For example, it is uncertain how the expression levels of grn, cdk7 or cdk9 regulate the cell cycle and cell‐to‐cell communication, which eventually lead to regeneration failure. Also, it is worth noting that the wtap knockdown phenotype can produce not only regeneration‐specific phenotype, but also lysis phenotype under homeostatic conditions (data not shown). Therefore, it would be interesting to dissect further which wtap‐mediated m6A genes are regeneration specific. Other model systems which can harness overexpression systems may be able to answer these questions in depth.
Collectively, our study uncovered an essential role of WTAP‐mediated m6A modification in the regeneration of an entire organism. We further discovered that WTAP‐mediated m6A modification is particularly important for the neural progenitor cell population. So far, this process has not been well understood. Our study may have uncovered a novel mechanism in regulating planarian regeneration that utilizes GRN to monitor and permit the regeneration to happen correctly. Therefore, our study highlights the potential importance of cell–cell communication during the planarian regeneration and provides important insights for future studies in regenerative medicine.
4. MATERIALS AND METHODS
4.1. Planarian culture
Animals Clonal asexual (CIW4) and sexual strains of Schmidtea mediterranea were maintained in Montjuïch salts as previously described (1.6 mM NaCl, 1.0 mM CaCl2, 1.0 mM MgSO4, 0.1 mM MgCl2, 0.1 mM KCl and 1.2 mM NaHCO3 prepared in autoclaved Milli‐Q water). 62 Animals were fed weekly with homogenized pig liver. All animals, 3–6 mm in length, were starved 1 week before any experiments.
4.2. Replication, size estimation and randomization
For every assay, at least three independent replicates with a minimum of three animals per experiment were performed. For RNAi phenotype characterization, numbers of animals used are indicated in each panel. No sample size estimation was performed. Animals for all experiments were randomly selected. All animals were included in statistical analyses, and no exclusions were done. Images were randomized before quantifications.
4.3. RNA interference
Double‐stranded RNA (dsRNA) was synthesized by in vitro transcription as previously described. 63 In summary, whole worm cDNA was generated and subcloned into a vector. In vitro transcription was performed using T7 polymerase (Promega, P2075). Planarians were fed by dsRNA mixed with pig liver paste three times every 4 days and were amputated into three fragments pre‐ and post‐pharyngeal 24 h after the last feeding. For double knockdown of two genes (double RNAi), dsRNAs of two genes were mixed in liver paste each at the same final concentration as single gene knockdown assay, and delivered three times at a 4‐day interval. The C. elegans unc‐22 gene was used as a negative RNAi control for single knockdown experiments, and was mixed with wtap dsRNA as control for double knockdown. The knockdown efficiency of double RNAi or single RNAi was verified by RT‐qPCR. Primers used for the dsRNA synthesis are listed in Table S6. ImageJ (V1.5) were used to measure the pixel area of regenerated blastema divided by that of the whole tissue fraction, and the same size of the amputated tissue fractions were chosen for quantification.
4.4. BrdU labelling and whole‐mount immunofluorescent staining
Animals were fed with 20 mg/mL BrdU (Sigma, 19160) as described before. 6 Specimens were harvested after labelling for 6 h and fixed in 4% formaldehyde (FA). The rat anti‐BrdU (Abcam, ab6326) antibody was used for detection. Immunofluorescence staining was performed as described before. 63 Different treatments were used for different antibodies. Following antibodies were used: Phospho‐histone H3 (Ser10; H3P; Abcam, ab32107). Muscle fibres were stained with the 6G10 antibody (1:1000, DSHB, 2C7; RRID:BDSC_9315). GRN protein was stained with anti‐GRN (1:1000, Proteintech, 10053‐1‐AP). Planarians were killed in 5% N‐acetyl cysteine (NAC) for 5 min at room temperature, and incubated in reduction buffer at 37°C for 8 min. After bleaching with 6% H2O2 overnight, planarians were washed with PBSTx (containing 0.3% Triton X‐100) over 6 h. The excretory system was stained with anti‐acetylated‐tubulin (1:1000, Sigma, T6793; RRID:AB_477585). Planarians were treated with 5% NAC for 5 min and 4% fixation for 20 min at room temperature followed by bleaching overnight and treated with proteinase K (1 μg/mL, Roche, 03115844001). After 2 h of blocking with 1% BSA, planarians were incubated with primary antibody overnight, then washed with PBSTx (containing 0.3% TritonX‐100) for more than 6 h. Blocking with 1% BSA for 1 h was performed before the fluorescent secondary antibody incubation overnight. After washing with PBSTx (containing 0.3% TritonX‐100) for more than 6 h, the slide was mounted with 80% glycerol containing Hoechst 33342 (10 μg/mL, Invitrogen, H3570). Quantification of fluorescent signals was analysed by ImageJ software (V1.5) as previously described. 64
4.5. TUNEL assay
The TUNEL experiment was performed with ApopTag TUNEL Kit (Millipore, S7165) to determine apoptotic cell numbers in planarians following the manufacturer's instructions. Five per cent NAC (diluted in PBS) was used to remove the mucus coat of planarians. Planarians were fixed with 4% formaldehyde (with PBST containing 0.3% TritonX‐100) for 30 min at room temperature. The samples were then bleached in 6% H2O2 (diluted in PBST) in direct light overnight. The animals were incubated with TdT reaction mixture for 4 h at 37°C. The antibody was incubated for 4 h at room temperature. Animals were washed every half an hour for 4 h in PBST to reduce nonspecific labelling. Images were obtained with Leica SP8 confocal microscope. ImageJ was used for quantifications.
4.6. Whole‐mount in situ hybridization
DIG‐labelled riboprobes were synthesized using an in vitro transcription kit (Roche, 11277073910), according to the manufacturer's instructions. Animals were treated with 5% NAC for 5 min to clear epidermal mucus, and fixed in 4% formaldehyde in PBST for 30 min at room temperature. Then animals were dehydrated and stored in 100% methanol at −20°C for at least 1 h. The animals were bleached in 6% H2O2 under bright light overnight. Bleached samples were rehydrated through a graded series of methanol, and were incubated in probe mix at 56°C for 16 h. After the animals were washed and blocked, they were further incubated in anti‐Digoxigenin‐POD, Fab fragments (Roche, 11207733910; RRID:AB_514500) or anti‐Digoxigenin‐AP, Fab fragments (Roche, 11093274910; RRID:AB_514497) overnight at 4°C for 16 h, followed by extensive washing. For Colorimetric whole‐mount in situ hybridization, BCIP/NBT was used as substrates. For fluorescent development, a tyramide signal amplification system was used. 65 For double‐colour FISH, POD inactivation was performed between signal developments in 100 mM NaN3 for 60 min. The primers used are listed in Table S6. Quantification of fluorescent signals was analysed by ImageJ software (V1.5) as previously described. 64
4.7. Total RNA extraction
Total RNA was extracted from planarians with 1 mL TRIzol® reagent (Invitrogen, 15596018) through homogenizing on a tissue disruptor. mRNAs were purified from total RNAs using Dynabeads® mRNA Purification Kit (Ambion, 61006) and subjected to TURBO™ DNase (Invitrogen) treatment at 37°C for 30 min and ethanol precipitation. After centrifugation and extensive washing with 75% ethanol, the mRNA was dissolved and quantified using Qubit 3.0 (Thermo Fisher).
4.8. Western blot
Planarians were dissolved and homogenized in RIPA buffer (Cell Signalling Technology, 9806s) on a tissue disruptor. After incubation on ice for 10 min, the samples were centrifuged at 13,000 rpm for 30 min. Supernatant was collected and the Bradford method was used to measure protein concentrations. Western blot was performed as previously reported 66 using the following antibodies: anti‐WTAP antibody (1:500, Proteintech, 10200‐1‐AP; RRID:AB_2216349), anti‐ß‐actin antibody (1:1000, Cell Signalling Technology, 4967s; RRID:AB_330288). The density of each band was quantified with ImageJ (version 1.8.0).
4.9. Quantitative reverse‐transcription PCR
To confirm knockdown efficiency and assess the relative RNA expression level, we conducted quantitative reverse‐transcription PCR (RT‐qPCR). All RNA templates were digested with TURBO™ DNase. cDNA synthesis was performed using the RevertAid™ First Strand cDNA Synthesis Kit (Invitrogen, 18064014). Takara SYBR Premix Ex Taq (Takara) was used according to the manufacturer's instructions and quantified by a CFX96 Real‐Time PCR System (Bio‐Rad). β‐actin was used as an internal control. Values of p were calculated using the two‐tailed unpaired Student's t‐test, ns, no significance. The primers used for qPCR are listed in Table S6.
4.10. UHPLC‐MRM‐MS/MS analysis
UHPLC‐MRM‐MS/MS analysis was performed as previously reported. 67 In brief, 200 ng mRNAs were purified from planarian total RNA at indicated time points. The mRNAs samples were digested overnight with 0.1 U Nuclease P1 (Sigma‐Aldrich, N8630) and 1.0 U calf intestinal phosphatase (NEB, M0290) in a 0 μL reaction volume at 37°C. The mixture was subjected to UHPLC‐MRM‐MS/MS analysis to detect m6A after filtering the samples through ultra‐filtration tubes (MW cutoff: 3 kDa, Pall, Port Washington, New York). The Agilent 1290 UHPLC system coupled with the 6495 triple quadrupole mass spectrometer (Agilent Technologies) was used for the analysis. UHPLC separation of mono‐nucleosides was performed with a Zorbax Eclipse Plus C18 column (100 mm × 2.1 mm I.D., 1.8 μm particle size, Agilent Technologies). The mass spectrometer was operated in a positive ion mode. Three replicates were analysed for each sample, with an injection volume of 5 μL. The amount of m6A and adenosine (A) was determined by using a calibration curve as a standard. Nitrogen was used for nebulization and desolvation at 40 psi with a flow‐rate of 9 L/min, a source temperature of 300°C, capillary voltage of 3500 V and high purity nitrogen of 99.999%. The ribonucleoside standards for m6A and A were purchased from TCI, China.
4.11. RNA‐seq and MeRIP‐seq
mRNAs were purified from total RNAs using the Dynabeads® mRNA purification kit (Ambion, 61006). cDNA libraries were made according to the TruSeq RNA Sample Prep Kit (Illumina, FC‐122‐1001) protocol. All samples were sequenced by Illumina Nova‐seq with paired‐end 150 bp read length. For m6A MeRIP‐seq, the procedure is based on a published protocol. 68 Briefly, purified mRNA was fragmented to a size of about 200 nt using the fragmentation reagent (Life Technologies, AM8740). Thirty microlitres of protein A magnetic beads (Thermo Fisher Scientific, 10002D) were washed twice in 1 mL IP buffer (150 mM NaCl, 10 mM Tris–HCl [pH 7.5], 0.1% NP‐40 in nuclease‐free H2O), resuspended in 500 μL IP buffer mixed with 5 μg anti‐m6A antibody (Millipore, ABE572) and incubated at 4°C with gentle rotation for at least 6 h. After two washes in IPP buffer (10 mM Tris–HCl [pH 7.4], 150 mM NaCl, 0.1% NP‐40 in DEPC‐treated), antibody‐bead mixture was resuspended in 500 μL of the IPP reaction mixture containing 500 ng fragmented mRNA, 100 μL of 5× IP buffer, and 5 μL of RNasin Plus RNase Inhibitor (Promega, N2611) and incubated for 2 h at 4°C. The beads were then washed twice each with 1 mL IPP buffer, low‐salt IPP buffer (50 mM NaCl, 10 mM Tris–HCl [pH 7.5], 0.1% NP‐40 in nuclease‐free H2O) and high‐salt buffer (500 mM NaCl, 10 mM Tris–HCl [pH 7.5], 0.1% NP‐40 in nuclease‐free H2O) for 10 min at 4°C. The beads were then eluted with 300 μL 0.5 mg/mL m6A in IPP buffer (with RNasin) with gentle rotation at RT for 1 h. The m6A‐modified RNAs were eluted using 200 μL of RLT buffer supplied in RNeasy Mini Kit (QIAGEN, 74106) for 2 min at room temperature. Supernatant was collected to a new tube while beads were pulled on magnetic rack. Four hundred microlitres of 100% ethanol was added to the supernatant. The mixture was applied to a RNeasy spin column and centrifuged at 13,000 rpm at 4°C for 30 s. The spin column was washed with 500 μL of RPE buffer, then 500 μL of 80% ethanol, and centrifuged at top speed for 5 min at 4°C to dry the column. m6A‐modified RNAs were eluted with 10 μL nuclease‐free H2O. For a second round of IP, eluted RNA was re‐incubated with new protein A magnetic beads prepared with new anti‐m6A antibody, followed by washes, elution and purification as above. Purified RNAs were used to construct library using the KAPA Standard RNA‐Seq Kit according to the manufacturer's instruction (KAPA, KR1139). Libraries were PCR amplified for 8–12 cycles and size‐selected on the 8% TBE gel. Sequencing was carried out on the Illumina Nova 6000 platform according to the manufacturer's instructions.
4.12. Single‐cell RNA‐seq library construction
For single‐cell RNA‐seq from 10× Chromium platform, Hoechst‐stained and PI‐negative cells (200,000 cells) from wild‐type animals, and cells from wtap knockdown animals were collected on ice using Influx sorter. Approximately 9000 counted cells were loaded per channel. The libraries were made using the Chromium platform and Chromium Single Cell 3′ v3 chemistry. Sequencing libraries were loaded on an Illumina nova 6000 flowcell with two 150 bp paired‐end kits.
4.13. RNA‐seq data analysis
Adaptor sequences were trimmed off for all raw reads using the Cutadapt software (version 1.2.1). 69 Reads that were less than 18 nt in length or contained an ambiguous nucleotide were discarded by Trimmomatic (version 0.30). 70 The remaining reads were aligned to the Schmidtea mediterranea transcriptome smed_20140614 using Bowtie2 (v2.2.9) 71 default parameters. Counts were calculated for the sum of reads of each transcript. R package ‘DEseq2’ 72 was used to identify differentially expressed transcripts. Genes with different expression patterns (Figure 1B and Figure 4B) were defined by the K‐means algorithm in MEV software. In detail, genes were clustered by MEV with parameter—distance‐metric‐selection = Pearson correlation, number of cluster = N, maximum iterations = 50 and the gene expression level of each time point was compared to that of the previous one. 73
4.14. MeRIP‐seq data analysis
For MeRIP‐seq, m6A‐enriched peaks in each m6A immunoprecipitation sample were identified by MACS2 peak‐calling software (version 2.0.10) 74 with the corresponding input sample serving as control. MACS2 was run with default options except for ‘–nomodel’ to turn off fragment size estimation. A stringent cut‐off threshold for a p‐value of 0.001 was used to obtain high‐confidence peaks. In each stage, peaks with over 50% overlap in three biological replicates were used in the downstream analysis. The overlapping peaks were re‐annotated by the highest peak within the region of all three peaks. The m6A level with different patterns was defined by the K‐means algorithm in MEV software (version 4.4). In detail, genes were clustered by MEV with parameter–distance‐metric‐selection = Pearson correlation, maximum‐iterations = 50 and m6A level of each timepoint was compared to that of the previous one. 73
4.15. Single‐cell RNA‐seq data processing
Reads were processed using the Cell Ranger 3.0.0 pipeline 75 with default and recommended parameters. FASTQ files generated from Illumina sequencing were aligned to the Schmidtea mediterranea transcriptome smed_20140614 using the STAR algorithm. 76 Next, Gene‐Barcode matrices were generated for each individual sample by counting unique molecular identifiers and filtering non‐cell associated barcodes. Only genes that can be translated into proteins were retained. We generated a gene‐barcode matrix containing barcoded cells and gene expression counts. This output was then imported into the Seurat (v3.1.2) 77 R toolkit for quality control and downstream analysis of single‐cell RNA‐seq data. All functions were run with default parameters, unless specified otherwise. Low‐quality cells (<500 genes/cell, >6000 genes/cell, <15 cells/ gene and >20% mitochondrial genes) were excluded. In order to exclude multiple captures, which is a major concern in microdroplet‐based experiments, DoubletFinder (version 2.0.2) 78 was employed to remove top N cells with the highest pANN score for each library separately, where N represents the doublet rates. Then all the datasets were merged using the ‘merge’ function in Seurat.
4.16. Identification of cell types and subtypes by nonlinear dimensional reduction
The Seurat package implemented in R was applied to identify major cell types. 77 Highly variable genes were generated and used to perform PCA. Significant principle components were determined using JackStraw 77 analysis and finally focusing on PCs 1–20. We grouped cell types based on their expression profile and matched them to known markers. To identify the subtype of neuronal cells, we used the reported markers of neuronal cells and also using ‘FindTransferAnchors’ and ‘TransferData’ function 77 which provided by Seurat to predict the similarity between the subcluster and the reported cell types that identified by Plass. 18 The gene name equivalencies beyond SMED to the transcriptome that used by Plass were transferred using the correspondence table which was downloaded from https://planosphere.stowers.org/chado/analysis?name=&program=&sourcename.
4.17. GO analysis
To enrich the GO annotation of planarian, we aligned the planarian transcriptome with the uniport database through blastx programme, and selected the best comparison result. According to the comparison results and the related GO annotation, the GO annotations of planarians were enriched. Then, GO analysis was performed using topGO, 79 with all genes as background.
4.18. Cell–cell communication analysis
In order to explore cell–cell communication networks via ligand–receptor interactions, we initially identified the homologous genes of planarian and human genes by blastx (version 2.7.1). 80 Then, we retained the genes with high homology with humans. For the gene corresponding to two annotations, we retained the result with the highest e‐value. After all, CellPhoneDB (v2.1.7) 81 with default parameters (cellphonedb method statistical_analysis–threshold 0.01) was used to identify the interaction among cells and the related receptor–ligand pairs.
4.19. Quantification and statistical analysis
All statistical analyses of qPCR and imaging were performed at least three biological replicates. Student's two‐tailed unpaired t‐test was used for statistical comparisons and data were shown as mean ± SD. Values of p were used for significance analysis; ns, no significance. All statistical analyses were performed using GraphPad Prism (version 7.0).
AUTHOR CONTRIBUTIONS
Yun‐Gui Yang conceived this project. Yun‐Gui Yang and Ying Yang supervised the study and analysed data. Guanshen Cui, Xin‐Yang Ge, Ge‐Ge Song, Xing Wang and Xiu‐Zhi Wang performed the experiments. Jia‐Yi Zhou and Bao‐Fa Sun performed bioinformatics analysis. Rui Zhang and Hai‐Lin Wang performed the UHPLC‐MRM‐MS/MS analysis. Yun‐Gui Yang, Ying Yang, Da‐Li Han, Wei‐Qi Zhang, Guanshen Cui, Jia‐Yi Zhou, Xin‐Yang Ge, Ge‐Ge Song, An Zeng, Yong‐Liang Zhao and Magdalena J. Koziol discussed and integrated the data. Qing Jing provided the planarians. Yun‐Gui Yang, Ying Yang, Guanshen Cui, Jia‐Yi Zhou, Xin‐Yang Ge, Ge‐Ge Song and Magdalena J. Koziol wrote the manuscript. All co‐authors provided feedback on the final manuscript.
FUNDING INFORMATION
This work was supported by grants from the Strategic Priority Research Program of the Chinese Academy of Sciences, China (XDA16010501, XDA16010108), CAS Project for Young Scientists in Basic Research (YSBR‐073), the National Natural Science Foundation of China (91940304, 32121001, 31770872, 32070828, 31922017), the National Key R&D Program of China (2018YFA0801200), the Youth Innovation Promotion Association of CAS (Y2022040), the Beijing Nova Program (Z201100006820104, 20220484210), and Shanghai Municipal Science and Technology Major Project (2017SHZDZX01). This result is supported by the CAMS Innovation Fund for Medical Sciences (CIFMS) (2019‐I2M‐5‐015).
CONFLICT OF INTEREST STATEMENT
The authors declare no competing interests.
Supporting information
Figure S1. Changes of m6A modified genes during regeneration, related to Figure 2. (A) Histogram showing the percentage of mRNAs with different m6A peak numbers. The x‐axis represents the number of m6A peak on one mRNA and y‐axis represents the percentage. (B) Histogram showing the number of genes with m6A modification in different periods. (C) Venn plot showing the overlap of m6A‐modified mRNAs in five regeneration timepoints. (D) Venn plot showing the overlap of mRNAs from different clusters that shown in Figure 1B (yellow pool) and total m6A‐modified mRNAs at all timepoints (green pool). (E) Line chart showing one of the trends (first category) of mRNAs m6A level during regeneration, which with increased m6A level from 0 hpa to 7 dpa and decreased m6A level from 7 to 11 dpa. mRNAs with different expression pattern were defined by MEV with parameter––distance‐metric‐selection = Pearson‐correlation––number‐of‐cluster = 4––maximum‐iterations = 50. (F) Barplot showing the significant GO terms for genes shown in (E). (G) Line chart showing one of the trends (second category) of mRNAs m6A level during regeneration, which with increased m6A level from 0 hpa to 3 dpa and then decreased m6A level from 3 to 11 dpa. mRNAs with different expression pattern were defined by MEV with parameter––distance‐metric‐selection = Pearson‐correlation––number‐of‐cluster = 4––maximum‐iterations = 50. (H) Barplot showing the significant GO terms for genes shown in (G). (I) Line chart showing one of the trends (third category) of mRNAs m6A level during regeneration, with increased m6A level from 0 to 6 hpa and then decreased m6A level from 6 to 11 dpa. mRNAs with different expression pattern were defined by MEV with parameter––distance‐metric‐selection = Pearson‐correlation––number‐of‐cluster = 4––maximum‐iterations = 50. (J) Barplot showing the significant GO terms for genes shown in (I).
Figure S2. Detection of wtap knockdown efficiency and m6A methylation levels, related to Figure 3. (A) Western blot showing the knockdown efficiency of WTAP. Actin is used as a loading control. (B) Quantitative qPCR showing the relative expression of wtap in control and wtap knockdown planarians at 7 dpa. Error bars represent standard deviation. Data are the mean ± SD (n ≥ 3 independent experiments). The p values were determined using a two‐sided unpaired Student's t‐test. (C) UHPLC‐MRM‐MS/MS showing m6A level of mRNA extracted from control and WTAP knockdown (wtap RNAi) planarians at 7 dpa. Error bars represent standard deviation. Data are the mean ± SD (n ≥ 3 independent experiments). The p values were determined using a two‐sided unpaired Student's t‐test. (D) Bright‐field images showing WTAP knockdown (wtap RNAi) phenotype of head, trunk, tail tissue fragment at 1, 3, 5, 7 and 9 dpa. Scale bar, 500 μm. (E) Whole‐mount fluorescent in‐situ hybridization showing the expression and distribution of pc2 and smedwi‐1 transcript of trunk tissue fragment at 7 dpa. Arrow, amputated position. Scale bar, 500 μm. (F) Whole‐mount fluorescent BrdU immunostaining of control and wtap knockdown (wtap RNAi) planarians at 5 dpa. Dotted line, amputation plane. Scale bar, 300 μm. (G) Quantification of BrdU+ nuclei/mm2 at 7 dpa in (F). Error bars represent standard deviation. Data are the mean ± SD (n ≥ 3 independent experiments). The p values were determined using a two‐sided unpaired Student's t‐test. (H) Whole‐mount TUNEL assay showing the late apoptotic cells of control and wtap knockdown (wtap RNAi) planarians at 9 dpa. Scale bar, 200 μm. (I) Quantification of TUNEL+ nuclei/mm2 at 4 hpa, 1 dpa, 3 dpa, 5 dpa, 7 dpa and 9 dpa. Error bars represent standard deviation. Data are the mean ± SD (n ≥ 3 independent experiments). The p values were determined using a two‐sided unpaired Student's t‐test.
Figure S3. WTAP‐mediated m6A controls cell cycle‐ and cell–cell communication‐related factors, related to Figure 4. (A) Barplot showing the total number of up‐ and down‐regulated transcripts (wtap knockdown vs. control planarians) that with m6A modification at 0 hpa, 6 hpa, 3 dpa, 7 dpa and 11 dpa. (B) GSEA plots evaluating the changes in multicellular organismal signalling, compared wtap knockdown (wtap RNAi) with control samples at 3 dpa. Normalized p value = 0.05. (C) Whole‐mount fluorescent in situ hybridization showing the expression and distribution of ccnb1 and smedwi‐1 transcript of trunk tissue fragment at 5 dpa. Scale bar, 200 μm. (D) Barplot showing the enrichment of m6A modification on cdk7 and cdk9. The p values were determined using Wilcoxon‐test. (E) Integrative genomics viewer (IGV) tracks displaying the distribution of MeRIP‐seq data that normalized by RNA‐seq data along notum mRNA in both control and wtap knockdown (wtap RNAi) samples. (F) IGV tracks displaying the distribution of MeRIP‐seq data that normalized by RNA‐seq data along kcnd2 mRNA in both control and wtap knockdown (wtap RNAi) samples. (G) The phenotype of control (control RNAi), cdk7 knockdown (cdk7 RNAi) or cdk9 knockdown (cdk9 RNAi) planarians at 7 dpa. Bottom left number, planarians with phenotype of total tested. Scale bar, 500 μm.
Figure S4. Functional analysis in different cell types upon wtap knockdown during regeneration, related to Figure 5. (A) Violin plots showing expression levels of canonical markers for each cell types. (B) Barplot showing enriched gene ontology terms for up‐regulated genes upon wtap knockdown at 3 dpa in epidermal cells. (C) Barplot showing enriched gene ontology terms for up‐regulated genes upon wtap knockdown at 3 dpa in neoblast cells. (D) Heatmap showing enriched gene ontology terms for downregulated genes upon wtap knockdown in epidermal cells at different regeneration stages. The colour bar of heatmap represents z‐score of GO's p value. (E) Heatmap showing enriched gene ontology terms for downregulated genes upon wtap knockdown in neoblast cells at different regeneration stages. The colour bar of heatmap represents z‐score of GO's p value. (F) Heatmap showing enriched gene ontology terms for downregulated genes upon wtap knockdown in neuronal cells at different regeneration stages. The colour bar of heatmap represents z‐score of GO's p value.
Figure S5. Identification of NP‐like cell markers, related to Figure 6. (A) Expression level of ubiqp in control (control) and wtap knockdown (wtap RNAi) planarians during regeneration. (B) Lineplot showing the dynamic expression level of grn and ubiqp expression levels in both control and wtap knockdown planarians during the five regeneration stages. The expression level is quantified from bulk RNA‐seq. Error bars represent standard deviation. Data are the mean ± SD. The p values were determined using a two‐sided unpaired Student's t‐test. (C) Integrative genomics viewer (IGV) tracks displaying the distributions of MeRIP‐seq data that normalized by RNA‐seq data along ubiqp in both control (top) and wtap knockdown (bottom) planarians at 7 and 11 dpa. Barplot showing the enrichment of m6A modification on ubiqp. The p values were determined using Wilcoxon‐test. (D) IGV tracks displaying the distributions of MeRIP‐seq data that normalized by RNA‐seq data along dph1 in both control (top) and wtap knockdown (bottom) planarians at 7 and 11 dpa. Barplot showing the enrichment of m6A modification on ubiqp. The p values were determined using Wilcoxon‐test. (E) qPCR showing the relative expression of wtap and grn in control (RNAi), wtap knockdown (RNAi) and double knockdown of wtap and grn at 7 dpa. Error bars represent standard deviation. Data are the mean ± SD (n ≥ 3 independent experiments). The p values were determined using a two‐sided unpaired Student's t‐test. (F) Phenotype of control and grn knockdown (grn RNAi) planarian at 7 dpa. Bottom left number, planarians with phenotype of total tested. Scale bar, 500 μm.
Figure S6. wtap regulates the regeneration of planarian through cdk7, cdk9 and grn. Related to Figure 6. (A) qPCR showing the relative expression of wtap, grn, cdk7, cdk9, dph1 and ubiqp in control (RNAi), wtap knockdown (RNAi), double knockdown of wtap and grn (wtap + grn RNAi) planarians at 7 dpa. Error bars represent standard deviation. Data are the mean ± S.D. (n ≥ 3 independent experiments). The p values were determined using a two‐sided unpaired Student's t‐test. (B) Whole‐mount fluorescent BrdU and H3p immunostaining of control, wtap knockdown (wtap RNAi), double knockdown of wtap and grn (wtap + grn RNAi), double knockdown of wtap and cdk7 (wtap + cdk7 RNAi) and double knockdown of wtap and cdk9 (wtap + cdk9 RNAi) planarians at 5 dpa. Dotted line, amputation plane. Scale bar, 300 μm. (C) Quantification of BrdU+ nuclei/mm2 of (B) at 5 dpa. Error bars represent standard deviation. Data are the mean ± S.D. (n ≥ 3 independent experiments). The p values were determined using a two‐sided unpaired Student's t‐test. (D) Quantification of H3p+ nuclei/mm2 of (B) at 7 dpa. Error bars represent standard deviation. Data are the mean ± SD (n ≥ 3 independent experiments). The p values were determined using a two‐sided unpaired Student's t‐test.
Figure S7. wtap regulates the regeneration of planarian by cell communication. Related to Figure 6. (A) Expression level of ccnt1, cdk7 and cdk9 in wtap knockdown (wtap RNAi) samples. (B) Network visualization showing neuronal cell communicates with other cell types through secreting GRN in wtap knockdown (wtap RNAi) samples. (C) Expression level of sort1 in all of the single‐cell RNA datasets. (D) Network visualization of ligand‐receptor pair numbers among different cell types. The interaction among cells were calculated by CellphoneDB with default parameters (cellphonedb method statistical_analysis––threshold 0.01). (E) Network visualization of mtnr1b ligand‐receptor pair among different cell types in wtap knockdown (wtap RNAi) samples. (F) Working model for WTAP‐mediated m6A in regulating planarian regeneration.
Table S1. GO analysis of different expression genes.
Table S2. Genes with m6A modification at different time points.
Table S3. Cell classification in single‐cell data.
Table S4. During the regeneration, the cell proportions in control and wtap knockdown samples.
Table S5. The markers of NP‐like cells.
Table S6. Primers used for dsRNA synthesis, ISH and qPCR assays.
Table S7. GO analysis of different expression genes identified in single cell RNA sequencing data.
Cui G, Zhou J‐Y, Ge X‐Y, et al. m6A promotes planarian regeneration. Cell Prolif. 2023;56(5):e13481. doi: 10.1111/cpr.13481
Guanshen Cui, Jia‐Yi Zhou, Xin‐Yang Ge, Bao‐Fa Sun, Ge‐Ge Song and Xing Wang contributed equally to this study.
Contributor Information
Wei‐Qi Zhang, Email: zhangwq@big.ac.cn.
Da‐Li Han, Email: handl@big.ac.cn.
Yun‐Gui Yang, Email: ygyang@big.ac.cn.
Ying Yang, Email: yingyang@big.ac.cn.
DATA AVAILABILITY STATEMENT
The datasets including RNA‐seq, MeRIP‐seq, and single cell sequencing data generated and analyzed during the current study are available in the Genome Sequence Archive under accession number CRA004040 linked to the project PRJCA004712, and also the Gene Expression Omnibus database under accession number GSE171253.
REFERENCES
- 1. Gierer A, Berking S, Bode H, et al. Regeneration of hydra from reaggregated cells. Nat New Biol. 1972;239:98‐101. [DOI] [PubMed] [Google Scholar]
- 2. Poss KD, Wilson LG, Keating MT. Heart regeneration in zebrafish. Science. 2002;298:2188‐2190. [DOI] [PubMed] [Google Scholar]
- 3. Wells JM, Watt FM. Diverse mechanisms for endogenous regeneration and repair in mammalian organs. Nature. 2018;557:322‐328. [DOI] [PubMed] [Google Scholar]
- 4. Alvarado AS, Tsonis PA. Bridging the regeneration gap: genetic insights from diverse animal models. Nat Rev Genet. 2006;7:873‐884. [DOI] [PubMed] [Google Scholar]
- 5. Reddien PW. The cellular and molecular basis for planarian regeneration. Cell. 2018;175:327‐345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Reddien PW, Oviedo NJ, Jennings JR, Jenkin JC, Alvarado AS. SMEDWI‐2 is a PIWI‐like protein that regulates planarian stem cells. Science. 2005;310:1327‐1330. [DOI] [PubMed] [Google Scholar]
- 7. Hayashi T, Asami M, Higuchi S, Shibata N, Agata K. Isolation of planarian X‐ray‐sensitive stem cells by fluorescence‐activated cell sorting. Dev Growth Differ. 2006;48:371‐380. [DOI] [PubMed] [Google Scholar]
- 8. Newmark PA, Reddien PW, Cebrià F, Sánchez Alvarado A. Ingestion of bacterially expressed double‐stranded RNA inhibits gene expression in planarians. Proc Natl Acad Sci USA. 2003;100(Suppl 1):11861‐11865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Petersen CP, Reddien PW. A wound‐induced Wnt expression program controls planarian regeneration polarity. Proc Natl Acad Sci USA. 2009;106:17061‐17066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sikes JM, Newmark PA. Restoration of anterior regeneration in a planarian with limited regenerative ability. Nature. 2013;500:77‐80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Umesono Y, Tasaki J, Nishimura Y, et al. The molecular logic for planarian regeneration along the anterior–posterior axis. Nature. 2013;500:73‐76. [DOI] [PubMed] [Google Scholar]
- 12. Liu SY, Selck C, Friedrich B, et al. Reactivating head regrowth in a regeneration‐deficient planarian species. Nature. 2013;500:81‐84. [DOI] [PubMed] [Google Scholar]
- 13. Petersen CP, Reddien PW. Smed‐betacatenin‐1 is required for anteroposterior blastema polarity in planarian regeneration. Science. 2008;319:327‐330. [DOI] [PubMed] [Google Scholar]
- 14. Gurley KA, Rink JC, Sánchez Alvarado A. Beta‐catenin defines head versus tail identity during planarian regeneration and homeostasis. Science. 2008;319:323‐327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fraguas S, Barberán S, Cebrià F. EGFR signaling regulates cell proliferation, differentiation and morphogenesis during planarian regeneration and homeostasis. Dev Biol. 2011;354:87‐101. [DOI] [PubMed] [Google Scholar]
- 16. Grohme MA, Schloissnig S, Rozanski A, et al. The genome of Schmidtea mediterranea and the evolution of core cellular mechanisms. Nature. 2018;554:56‐61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fincher CT, Wurtzel O, de Hoog T, Kravarik KM, Reddien PW. Cell type transcriptome atlas for the planarian Schmidtea mediterranea . Science. 2018;360:eaaq1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Plass M, Solana J, Wolf FA, et al. Science. 2018;360:eaaq1723. [DOI] [PubMed] [Google Scholar]
- 19. Wagner DE, Wang IE, Reddien PW. Clonogenic neoblasts are pluripotent adult stem cells that underlie planarian regeneration. Science. 2011;332:811‐816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zeng A, Li H, Guo L, et al. Prospectively isolated tetraspanin+ neoblasts are adult pluripotent stem cells underlying planaria regeneration. Cell. 2018;173:1593‐1608.e1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hubert A, Henderson JM, Ross KG, Cowles MW, Torres J, Zayas RM. Epigenetic regulation of planarian stem cells by the SET1/MLL family of histone methyltransferases. Epigenetics. 2013;8:79‐91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Duncan EM, Chitsazan AD, Seidel CW, Sánchez Alvarado A. Set1 and MLL1/2 target distinct sets of functionally different genomic loci in vivo. Cell Rep. 2015;13:2741‐2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mihaylova Y, Abnave P, Kao D, et al. Conservation of epigenetic regulation by the MLL3/4 tumour suppressor in planarian pluripotent stem cells. Nat Commun. 2018;9:1‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Stelman CR, Smith BM, Chandra B, Roberts‐Galbraith RH. CBP/p300 homologs CBP2 and CBP3 play distinct roles in planarian stem cell function. Dev Biol. 2021;473:130‐143. [DOI] [PubMed] [Google Scholar]
- 25. Fraguas S, Cárcel S, Vivancos C, et al. CREB‐binding protein (CBP) gene family regulates planarian survival and stem cell differentiation. Dev Biol. 2021;476:53‐67. [DOI] [PubMed] [Google Scholar]
- 26. Shi H, Wei J, He C. Where, when, and how: context‐dependent functions of rna methylation writers, readers, and erasers. Mol Cell. 2019;74:640‐650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Batista PJ, Molinie B, Wang J, et al. m6A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell. 2014;15:707‐719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang Y, Li Y, Toth JI, Petroski MD, Zhang Z, Zhao JC. N 6‐methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat Cell Biol. 2014;16:191‐198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang C, Chen Y, Sun B, et al. m6A modulates haematopoietic stem and progenitor cell specification. Nature. 2017;549:273‐276. [DOI] [PubMed] [Google Scholar]
- 30. Cui Q, Shi H, Ye P, et al. m6A RNA methylation regulates the self‐renewal and tumorigenesis of glioblastoma stem cells. Cell Rep. 2017;18:2622‐2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dominissini D, Moshitch‐Moshkovitz S, Schwartz S, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A‐seq. Nature. 2012;485:201‐206. [DOI] [PubMed] [Google Scholar]
- 32. Jia G, Fu Y, Zhao X, et al. N 6‐Methyladenosine in nuclear RNA is a major substrate of the obesity‐associated FTO. Nat Chem Biol. 2011;7:885‐887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li HB, Tong J, Zhu S, et al. m6A mRNA methylation controls T cell homeostasis by targeting the IL‐7/STAT5/SOCS pathways. Nature. 2017;548:338‐342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liu J, Yue Y, Han D, et al. A METTL3‐METTL14 complex mediates mammalian nuclear RNA N 6‐adenosine methylation. Nat Chem Biol. 2014;10:93‐95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012;149:1635‐1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Geula S, Moshitch‐Moshkovitz S, Dominissini D, et al. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science. 2015;347:1002‐1006. [DOI] [PubMed] [Google Scholar]
- 37. Wang H, Zuo H, Liu J, et al. Loss of YTHDF2‐mediated m6A‐dependent mRNA clearance facilitates hematopoietic stem cell regeneration. Cell Res. 2018;028:1035‐1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Weng Y‐L, Wang X, An R, et al. Epitranscriptomic m6A regulation of axon regeneration in the adult mammalian nervous system. Neuron. 2018;97:313‐325.e316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Adell T, Cebrià F, Saló E. Gradients in planarian regeneration and homeostasis. Cold Spring Harb Perspect Biol. 2010;2:a000505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ping X‐L, Sun BF, Wang L, et al. Mammalian WTAP is a regulatory subunit of the RNA N 6‐methyladenosine methyltransferase. Cell Res. 2014;24:177‐189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schwartz S, Mumbach MR, Jovanovic M, et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5′ sites. Cell Rep. 2014;8:284‐296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cebrià F, Nakazawa M, Mineta K, Ikeo K, Gojobori T, Agata K. Dissecting planarian central nervous system regeneration by the expression of neural‐specific genes. Dev Growth Differ. 2002;44:135‐146. [DOI] [PubMed] [Google Scholar]
- 43. Wallenfang MR, Seydoux G. Cdk‐7 is required for mRNA transcription and cell cycle progression in Caenorhabditis elegans embryos. Proc Natl Acad Sci. 2002;99:5527‐5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Harper JW, Elledge SJ. The role of Cdk7 in CAK function, a retro‐retrospective. Genes Dev. 1998;12:285‐289. [DOI] [PubMed] [Google Scholar]
- 45. Fisher RP. Secrets of a double agent: CDK7 in cell‐cycle control and transcription. J Cell Sci. 2005;118:5171‐5180. [DOI] [PubMed] [Google Scholar]
- 46. Ganuza M, Sáiz‐Ladera C, Cañamero M, et al. Genetic inactivation of Cdk7 leads to cell cycle arrest and induces premature aging due to adult stem cell exhaustion. EMBO J. 2012;31:2498‐2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bettencourt‐Dias M, Giet R, Sinka R, et al. Genome‐wide survey of protein kinases required for cell cycle progression. Nature. 2004;432:980‐987. [DOI] [PubMed] [Google Scholar]
- 48. Ahmed Z, Sheng H, Xu YF, et al. Accelerated lipofuscinosis and ubiquitination in granulin knockout mice suggest a role for progranulin in successful aging. Am J Pathol. 2010;177:311‐324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Arrant AE, Filiano AJ, Unger DE, Young AH, Roberson ED. Restoring neuronal progranulin reverses deficits in a mouse model of frontotemporal dementia. Brain. 2017;140:1447‐1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pottier C, Zhou X, Perkerson RB III, et al. Potential genetic modifiers of disease risk and age at onset in patients with frontotemporal lobar degeneration and GRN mutations: a genome‐wide association study. Lancet Neurol. 2018;17:548‐558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hoque M, Young TM, Lee CG, Serrero G, Mathews MB, Pe'ery T. The growth factor granulin interacts with cyclin T1 and modulates P‐TEFb‐dependent transcription. Mol Cell Biol. 2003;23:1688‐1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bateman A, Bennett HPJ. The granulin gene family: from cancer to dementia. Bioessays. 2009;31:1245‐1254. [DOI] [PubMed] [Google Scholar]
- 53. Dagan Y, Yesharim Y, Bonneau AR, et al. m6A is required for resolving progenitor identity during planarian stem cell differentiation. EMBO J. 2022;41:e109895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Liu J, Gao M, He J, et al. The RNA m6A reader YTHDC1 silences retrotransposons and guards ES cell identity. Nature. 2021;591:322‐326. [DOI] [PubMed] [Google Scholar]
- 55. Reddien PW, Alvarado AS. Fundamentals of planarian regeneration. Annu Rev Cell Dev Biol. 2004;20:725‐757. [DOI] [PubMed] [Google Scholar]
- 56. Fisher RP. Cdk7: a kinase at the core of transcription and in the crosshairs of cancer drug discovery. Transcription. 2019;10:47‐56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Huang CH, Lujambio A, Zuber J, et al. CDK9‐mediated transcription elongation is required for MYC addiction in hepatocellular carcinoma. Genes Dev. 2014;28:1800‐1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Leucci E, de Falco G, Onnis A, et al. The role of the Cdk9/cyclin T1 complex in T cell differentiation. J Cell Physiol. 2007;212:411‐415. [DOI] [PubMed] [Google Scholar]
- 59. Campbell CA, Fursova O, Cheng X, et al. A zebrafish model of granulin deficiency reveals essential roles in myeloid cell differentiation. Blood Adv. 2021;5:796‐811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Tidball JG. Regulation of muscle growth and regeneration by the immune system. Nat Rev Immunol. 2017;17:165‐178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Oviedo NJ, Morokuma J, Walentek P, et al. Long‐range neural and gap junction protein‐mediated cues control polarity during planarian regeneration. Dev Biol. 2010;339:188‐199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Han X‐S, Wang C, Guo FH, et al. Neoblast‐enriched zinc finger protein FIR1 triggers local proliferation during planarian regeneration. Protein Cell. 2019;10:43‐59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Shibata N, Agata K. RNA interference in planarians: feeding and injection of synthetic dsRNA. Methods Mol Biol. 2018;1774:455‐466. [DOI] [PubMed] [Google Scholar]
- 64. Shihan MH, Novo SG, Le Marchand SJ, Wang Y, Duncan MK. A simple method for quantitating confocal fluorescent images. Biochem Biophys Rep. 2021;25:100916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Forsthoefel DJ, Waters FA, Newmark PA. Generation of cell type‐specific monoclonal antibodies for the planarian and optimization of sample processing for immunolabeling. BMC Dev Biol. 2014;14:1‐22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ziman B, Oviedo NJ. Measuring protein levels in planarians using western blotting. STAR Protocols. 2021;2:100274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Yang X, Yang Y, Sun BF, et al. 5‐methylcytosine promotes mRNA export—NSUN2 as the methyltransferase and ALYREF as an m5C reader. Cell Res. 2017;27:606‐625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zeng Y, Wang S, Gao S, et al. Refined RIP‐seq protocol for epitranscriptome analysis with low input materials. PLoS Biol. 2018;16:e2006092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Martin M. Cutadapt removes adapter sequences from high‐throughput sequencing reads. EMBnet J. 2011;17:10‐12. [Google Scholar]
- 70. Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114‐2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Langmead B, Salzberg SL. Fast gapped‐read alignment with bowtie 2. Nat Methods. 2012;9:357‐359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA‐seq data with DESeq2. Genome Biol. 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Howe EA, Sinha R, Schlauch D, Quackenbush J. RNA‐Seq analysis in MeV. Bioinformatics. 2011;27:3209‐3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Zhang Y, Liu T, Meyer CA, et al. Model‐based analysis of ChIP‐Seq (MACS). Genome Biol. 2008;9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Zheng GX, Terry JM, Belgrader P, et al. Massively parallel digital transcriptional profiling of single cells. Nat Commun. 2017;8:14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA‐seq aligner. Bioinformatics. 2013;29:15‐21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Hao Y, Hao S, Andersen‐Nissen E, et al. Integrated analysis of multimodal single‐cell data. Cell. 2021;184:3573‐3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. McGinnis CS, Murrow LM, Gartner ZJ. DoubletFinder: doublet detection in single‐cell RNA sequencing data using artificial nearest neighbors. Cell Syst. 2019;8:329‐337.e324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Alexa A, Rahnenfuhrer J. topGO: enrichment analysis for gene ontology. R Package Version 2.2010. 2010.
- 80. Altschul SF, Madden TL, Schäffer AA, et al. Gapped BLAST and PSI‐BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389‐3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Efremova M, Vento‐Tormo M, Teichmann SA, Vento‐Tormo R. CellPhoneDB: inferring cell–cell communication from combined expression of multi‐subunit ligand–receptor complexes. Nat Protoc. 2020;15:1484‐1506. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Changes of m6A modified genes during regeneration, related to Figure 2. (A) Histogram showing the percentage of mRNAs with different m6A peak numbers. The x‐axis represents the number of m6A peak on one mRNA and y‐axis represents the percentage. (B) Histogram showing the number of genes with m6A modification in different periods. (C) Venn plot showing the overlap of m6A‐modified mRNAs in five regeneration timepoints. (D) Venn plot showing the overlap of mRNAs from different clusters that shown in Figure 1B (yellow pool) and total m6A‐modified mRNAs at all timepoints (green pool). (E) Line chart showing one of the trends (first category) of mRNAs m6A level during regeneration, which with increased m6A level from 0 hpa to 7 dpa and decreased m6A level from 7 to 11 dpa. mRNAs with different expression pattern were defined by MEV with parameter––distance‐metric‐selection = Pearson‐correlation––number‐of‐cluster = 4––maximum‐iterations = 50. (F) Barplot showing the significant GO terms for genes shown in (E). (G) Line chart showing one of the trends (second category) of mRNAs m6A level during regeneration, which with increased m6A level from 0 hpa to 3 dpa and then decreased m6A level from 3 to 11 dpa. mRNAs with different expression pattern were defined by MEV with parameter––distance‐metric‐selection = Pearson‐correlation––number‐of‐cluster = 4––maximum‐iterations = 50. (H) Barplot showing the significant GO terms for genes shown in (G). (I) Line chart showing one of the trends (third category) of mRNAs m6A level during regeneration, with increased m6A level from 0 to 6 hpa and then decreased m6A level from 6 to 11 dpa. mRNAs with different expression pattern were defined by MEV with parameter––distance‐metric‐selection = Pearson‐correlation––number‐of‐cluster = 4––maximum‐iterations = 50. (J) Barplot showing the significant GO terms for genes shown in (I).
Figure S2. Detection of wtap knockdown efficiency and m6A methylation levels, related to Figure 3. (A) Western blot showing the knockdown efficiency of WTAP. Actin is used as a loading control. (B) Quantitative qPCR showing the relative expression of wtap in control and wtap knockdown planarians at 7 dpa. Error bars represent standard deviation. Data are the mean ± SD (n ≥ 3 independent experiments). The p values were determined using a two‐sided unpaired Student's t‐test. (C) UHPLC‐MRM‐MS/MS showing m6A level of mRNA extracted from control and WTAP knockdown (wtap RNAi) planarians at 7 dpa. Error bars represent standard deviation. Data are the mean ± SD (n ≥ 3 independent experiments). The p values were determined using a two‐sided unpaired Student's t‐test. (D) Bright‐field images showing WTAP knockdown (wtap RNAi) phenotype of head, trunk, tail tissue fragment at 1, 3, 5, 7 and 9 dpa. Scale bar, 500 μm. (E) Whole‐mount fluorescent in‐situ hybridization showing the expression and distribution of pc2 and smedwi‐1 transcript of trunk tissue fragment at 7 dpa. Arrow, amputated position. Scale bar, 500 μm. (F) Whole‐mount fluorescent BrdU immunostaining of control and wtap knockdown (wtap RNAi) planarians at 5 dpa. Dotted line, amputation plane. Scale bar, 300 μm. (G) Quantification of BrdU+ nuclei/mm2 at 7 dpa in (F). Error bars represent standard deviation. Data are the mean ± SD (n ≥ 3 independent experiments). The p values were determined using a two‐sided unpaired Student's t‐test. (H) Whole‐mount TUNEL assay showing the late apoptotic cells of control and wtap knockdown (wtap RNAi) planarians at 9 dpa. Scale bar, 200 μm. (I) Quantification of TUNEL+ nuclei/mm2 at 4 hpa, 1 dpa, 3 dpa, 5 dpa, 7 dpa and 9 dpa. Error bars represent standard deviation. Data are the mean ± SD (n ≥ 3 independent experiments). The p values were determined using a two‐sided unpaired Student's t‐test.
Figure S3. WTAP‐mediated m6A controls cell cycle‐ and cell–cell communication‐related factors, related to Figure 4. (A) Barplot showing the total number of up‐ and down‐regulated transcripts (wtap knockdown vs. control planarians) that with m6A modification at 0 hpa, 6 hpa, 3 dpa, 7 dpa and 11 dpa. (B) GSEA plots evaluating the changes in multicellular organismal signalling, compared wtap knockdown (wtap RNAi) with control samples at 3 dpa. Normalized p value = 0.05. (C) Whole‐mount fluorescent in situ hybridization showing the expression and distribution of ccnb1 and smedwi‐1 transcript of trunk tissue fragment at 5 dpa. Scale bar, 200 μm. (D) Barplot showing the enrichment of m6A modification on cdk7 and cdk9. The p values were determined using Wilcoxon‐test. (E) Integrative genomics viewer (IGV) tracks displaying the distribution of MeRIP‐seq data that normalized by RNA‐seq data along notum mRNA in both control and wtap knockdown (wtap RNAi) samples. (F) IGV tracks displaying the distribution of MeRIP‐seq data that normalized by RNA‐seq data along kcnd2 mRNA in both control and wtap knockdown (wtap RNAi) samples. (G) The phenotype of control (control RNAi), cdk7 knockdown (cdk7 RNAi) or cdk9 knockdown (cdk9 RNAi) planarians at 7 dpa. Bottom left number, planarians with phenotype of total tested. Scale bar, 500 μm.
Figure S4. Functional analysis in different cell types upon wtap knockdown during regeneration, related to Figure 5. (A) Violin plots showing expression levels of canonical markers for each cell types. (B) Barplot showing enriched gene ontology terms for up‐regulated genes upon wtap knockdown at 3 dpa in epidermal cells. (C) Barplot showing enriched gene ontology terms for up‐regulated genes upon wtap knockdown at 3 dpa in neoblast cells. (D) Heatmap showing enriched gene ontology terms for downregulated genes upon wtap knockdown in epidermal cells at different regeneration stages. The colour bar of heatmap represents z‐score of GO's p value. (E) Heatmap showing enriched gene ontology terms for downregulated genes upon wtap knockdown in neoblast cells at different regeneration stages. The colour bar of heatmap represents z‐score of GO's p value. (F) Heatmap showing enriched gene ontology terms for downregulated genes upon wtap knockdown in neuronal cells at different regeneration stages. The colour bar of heatmap represents z‐score of GO's p value.
Figure S5. Identification of NP‐like cell markers, related to Figure 6. (A) Expression level of ubiqp in control (control) and wtap knockdown (wtap RNAi) planarians during regeneration. (B) Lineplot showing the dynamic expression level of grn and ubiqp expression levels in both control and wtap knockdown planarians during the five regeneration stages. The expression level is quantified from bulk RNA‐seq. Error bars represent standard deviation. Data are the mean ± SD. The p values were determined using a two‐sided unpaired Student's t‐test. (C) Integrative genomics viewer (IGV) tracks displaying the distributions of MeRIP‐seq data that normalized by RNA‐seq data along ubiqp in both control (top) and wtap knockdown (bottom) planarians at 7 and 11 dpa. Barplot showing the enrichment of m6A modification on ubiqp. The p values were determined using Wilcoxon‐test. (D) IGV tracks displaying the distributions of MeRIP‐seq data that normalized by RNA‐seq data along dph1 in both control (top) and wtap knockdown (bottom) planarians at 7 and 11 dpa. Barplot showing the enrichment of m6A modification on ubiqp. The p values were determined using Wilcoxon‐test. (E) qPCR showing the relative expression of wtap and grn in control (RNAi), wtap knockdown (RNAi) and double knockdown of wtap and grn at 7 dpa. Error bars represent standard deviation. Data are the mean ± SD (n ≥ 3 independent experiments). The p values were determined using a two‐sided unpaired Student's t‐test. (F) Phenotype of control and grn knockdown (grn RNAi) planarian at 7 dpa. Bottom left number, planarians with phenotype of total tested. Scale bar, 500 μm.
Figure S6. wtap regulates the regeneration of planarian through cdk7, cdk9 and grn. Related to Figure 6. (A) qPCR showing the relative expression of wtap, grn, cdk7, cdk9, dph1 and ubiqp in control (RNAi), wtap knockdown (RNAi), double knockdown of wtap and grn (wtap + grn RNAi) planarians at 7 dpa. Error bars represent standard deviation. Data are the mean ± S.D. (n ≥ 3 independent experiments). The p values were determined using a two‐sided unpaired Student's t‐test. (B) Whole‐mount fluorescent BrdU and H3p immunostaining of control, wtap knockdown (wtap RNAi), double knockdown of wtap and grn (wtap + grn RNAi), double knockdown of wtap and cdk7 (wtap + cdk7 RNAi) and double knockdown of wtap and cdk9 (wtap + cdk9 RNAi) planarians at 5 dpa. Dotted line, amputation plane. Scale bar, 300 μm. (C) Quantification of BrdU+ nuclei/mm2 of (B) at 5 dpa. Error bars represent standard deviation. Data are the mean ± S.D. (n ≥ 3 independent experiments). The p values were determined using a two‐sided unpaired Student's t‐test. (D) Quantification of H3p+ nuclei/mm2 of (B) at 7 dpa. Error bars represent standard deviation. Data are the mean ± SD (n ≥ 3 independent experiments). The p values were determined using a two‐sided unpaired Student's t‐test.
Figure S7. wtap regulates the regeneration of planarian by cell communication. Related to Figure 6. (A) Expression level of ccnt1, cdk7 and cdk9 in wtap knockdown (wtap RNAi) samples. (B) Network visualization showing neuronal cell communicates with other cell types through secreting GRN in wtap knockdown (wtap RNAi) samples. (C) Expression level of sort1 in all of the single‐cell RNA datasets. (D) Network visualization of ligand‐receptor pair numbers among different cell types. The interaction among cells were calculated by CellphoneDB with default parameters (cellphonedb method statistical_analysis––threshold 0.01). (E) Network visualization of mtnr1b ligand‐receptor pair among different cell types in wtap knockdown (wtap RNAi) samples. (F) Working model for WTAP‐mediated m6A in regulating planarian regeneration.
Table S1. GO analysis of different expression genes.
Table S2. Genes with m6A modification at different time points.
Table S3. Cell classification in single‐cell data.
Table S4. During the regeneration, the cell proportions in control and wtap knockdown samples.
Table S5. The markers of NP‐like cells.
Table S6. Primers used for dsRNA synthesis, ISH and qPCR assays.
Table S7. GO analysis of different expression genes identified in single cell RNA sequencing data.
Data Availability Statement
The datasets including RNA‐seq, MeRIP‐seq, and single cell sequencing data generated and analyzed during the current study are available in the Genome Sequence Archive under accession number CRA004040 linked to the project PRJCA004712, and also the Gene Expression Omnibus database under accession number GSE171253.


