Summary
Aging is characterized by progressive decline in tissue function and represents the greatest risk factor for many diseases. Nevertheless, many fundamental mechanisms driving human aging remain poorly understood. Aging studies using model organisms are often limited in their applicability to humans. Mechanistic studies of human aging rely on relatively simple cell culture models that fail to replicate mature tissue function, making them poor surrogates for aged tissues. These culture systems generally lack well-controlled cellular microenvironments that capture the changes in tissue mechanics and microstructure that occur during aging. Biomaterial platforms presenting dynamic, physiologically relevant mechanical, structural, and biochemical cues can capture the complex changes in the cellular microenvironment in a well-defined manner, accelerating the process of cellular aging in model laboratory systems. By enabling selective tuning of relevant microenvironmental parameters, these biomaterials systems may enable identification of new therapeutic approaches to slow or reverse the detrimental effects of aging.
Subject areas: Bioengineering, Biomaterials, Health sciences
Graphical abstract

Bioengineering; Biomaterials; Health sciences
Background
Aging is characterized by progressive decline in function of cells and tissues, and is a significant risk factor for a poor prognosis in many diseases.1 Manifestations of this functional decline include impaired mobility, decreased cognitive function, diminished immune response, and dysfunctional tissue regeneration, among many others. Understanding the mechanisms by which aging gives rise to this loss in tissue function is necessary to develop approaches to counteract the effects of aging and increase the length of life with good health or “healthspan.” Such therapeutic approaches will prove transformational as the proportion of the world’s population living to advanced age continues to increase.2 Extending the healthspan for aged individuals not only will improve the quality of life for a large proportion of the population but it will also reduce the burden on the healthcare system in terms of cost and human resources that are currently required to care for the aged. Despite the correlation between advanced age and poor clinical outcomes, many of the underlying causes of reduced tissue function in aging remain poorly understood.
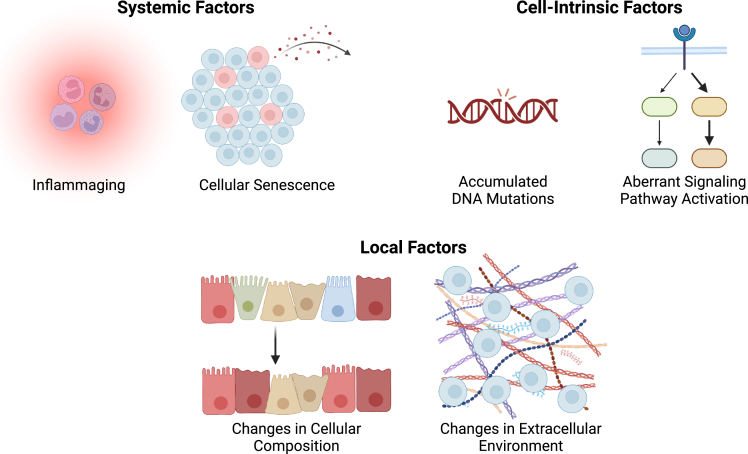
Aging is a multifactorial process that results in significant heterogeneity, even in model organisms that are otherwise genetically identical.1 The potential mechanistic drivers of aging largely arise spontaneously over time, resulting in diverse aged phenotypes despite identical genetic makeup (Figure 1). Some of the causes of tissue dysfunction are systemic in nature, occurring throughout the organism. For instance, a chronic overactivated inflammatory state, known as “inflammaging,” has been observed in both humans and model organisms.3,4 This aberrant immune response is thought to be a cause of cardiovascular disease in aging and may play a role in numerous other conditions ranging from increased incidence of cancer to age-associated muscle wasting, known as sarcopenia.3 Inflammaging is closely related to another contributor of systemic dysregulation in aging: cellular senescence. Senescent cells are characterized by growth arrest, resistance to apoptosis, and altered gene expression compared to healthy cells of the same type.5,6 Senescence arises during aging for a variety of reasons, including cells reaching their proliferative limit due to telomere shortening from successive rounds of DNA replication or the activation of oncogenes due to accumulation of DNA mutations that trigger an anti-proliferative response to prevent cancer growth.5 Senescent cells secrete a collection of factors known as senescence-associated secretory phenotypes (SASPs) that can have a detrimental effect on tissue function systemically, similar to inflammaging.5,6
Figure 1.
Aging-related changes that influence cellular function can be grouped into three categories
These categories include: (1) systemic factors, such as inflammaging and secretion of SASPs by senescent cells, (2) cell-intrinsic factors, such as accumulated DNA mutations from replication or oxidative damage and aberrant activation of cellular signaling pathways, and (3) local factors, such as changes in the cellular composition of tissues or alterations in the extracellular environment.
Other causes of aging-related dysfunction are more specific to a given tissue or organ. Intrinsic changes to individual cells are well known to cause tissue dysfunction.7 For instance, accumulated genetic mutations can give rise to undesirable cellular behavior, ranging from disrupted tissue function to malignant cancers.8 Intrinsic changes can also include aberrant signaling pathway activation, such as the overactivation of the p38 MAP kinase pathway that impairs the regenerative function of aged skeletal muscle stem cells.9,10 Aged tissues additionally often exhibit altered cellular composition compared to healthy, young tissues. Perhaps, the most famous example is the myeloid bias that occurs during hematopoiesis, wherein aged organisms produce more myeloid cells, such as red blood cells, macrophages, and neutrophils, at the expense of lymphoid cells, such as B and T cells.11 Beyond cellular changes, aging is associated with significant changes to the non-cellular microenvironment, such as the extracellular matrix (ECM).12,13 The composition and microstructure of the ECM are altered by changes in the type of ECM deposited (e.g., in tissue fibrosis), altered patterns of proteolysis (e.g., due to SASP-associated proteases), and changes in glycosylation patterns (e.g., from advanced glycation end products, or AGEs).14 These alterations in the ECM in turn impact cellular mechanotransduction, potentially resulting in aberrant cellular behavior.14 In addition to changes in the ECM, protein misfolding and aggregation contribute to microenvironmental changes with age, particularly in the context of neurodegenerative diseases.15
Despite the inherent heterogeneity of whole organism aging, various animal models are the current gold standard for investigating the cellular and molecular mechanisms underlying aging.16 The model organisms chosen for aging studies are most often those with relatively short lifespans, ranging from ∼20 days in worms to 2–3 years in mice.16 Each model organism has unique limitations in its applicability to longer-lived organisms such as humans.1,16 Studies of human aging have been limited to observation of human subjects or simplistic cell culture models.16 The advent of human cellular reprogramming has opened the door to improve in vitro models of relevant human cell types to study “aging in a dish.”17 However, these models fail to adequately capture most aging-related effects on biological systems. Simple 2D cell culture on plastic dishes is a poor representation of the in vivo microenvironment, and 3D organotypic cultures are often arrested at an immature phenotype not reminiscent of adult, let alone aged, tissue.18 A major limitation of these systems is the lack of a dynamic, physiologically relevant mechanical, chemical, and structural microenvironment that mimics the progression of aging.
The majority of mechanistic studies of aging have focused on identifying biochemical changes in tissues and whole organisms that can be targeted to arrest or reverse the progression of aging. Some of the most famous examples are experiments using heterochronic parabiosis, in which the blood supplies of young and aged animals are joined, to identify “Fountain of Youth” factors in young animals that can restore the function of aged tissues.19 These approaches have had limited success, likely due in large part to the fact that the physical microenvironment in which aged cells reside is markedly different from the young cellular microenvironment.14 Young biochemical factors may not be sufficient to rejuvenate the physical features of aged tissues. Recent studies have highlighted the importance of biophysical properties on restoring function to aged cells, as both aged skeletal muscle stem cells and aged oligodendrocyte progenitor cells require culture on relatively compliant substrates with elastic moduli comparable to the measured stiffness of young tissue, as opposed to more rigid aged tissue, to restore their stem cell function.9,20
This Perspective highlights the opportunities presented by recent innovations in dynamic biomaterials platforms to recapitulate key features of human aging in model systems. While the cumulative changes to the cellular microenvironment that constitutes the aging phenotype take years to decades to accumulate, engineered systems can accelerate this aging process, allowing observation of altered cellular behaviors over days to weeks. By reducing experimental lengths to laboratory-relevant timescales, these engineered systems may enable identification of previously unrecognized therapeutic targets to combat the detrimental effects of aging and increase healthspan.
Mechanical and structural changes in the aged microenvironment
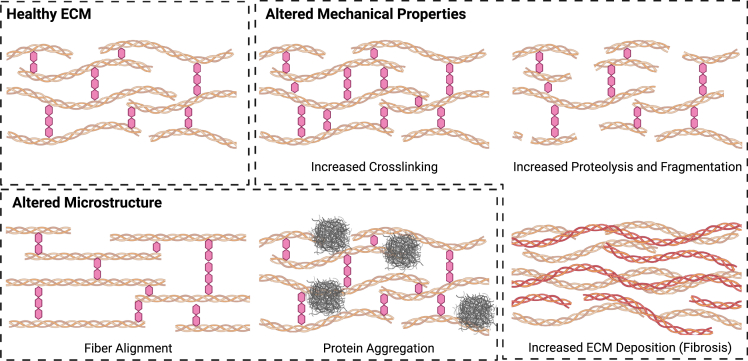
The extracellular microenvironment is a structurally and compositionally diverse collection of macromolecules, predominantly proteins and polysaccharides, that provides a complicated combination of biophysical and biochemical cues that act in concert to regulate cell fate.21 The ECM is comprised of fibrillar (e.g., collagens I and III), networked (e.g., collagen IV and laminin), and intrinsically disordered (e.g., elastin) proteins and glycosaminoglycans (e.g., hyaluronic acid).21 In addition to providing structural and mechanical stability, the ECM serves as a scaffold that regulates biochemical signaling arising from morphogen and cytokine binding.21 In aging, the mechanics, microstructure, and composition of the ECM become dysregulated (Figure 2),12 thereby impacting a myriad of biological processes that may give rise to tissue dysfunction. Because of the interconnected nature of microenvironmental cues, well-controlled systems in which the contributions of individual parameters to cell fate can be assessed will be required to deconvolve the web of signaling changes that result in detrimental aging phenotypes.
Figure 2.
The extracellular environment exhibits multiple aging-related changes that impact both the mechanical and structural properties of the microenvironment
While these simplified schematics use collagen triple helices and glycans to represent ECM polymers and crosslinks, respectively, other ECM proteins and crosslinking chemistries may be implicated, depending on the tissue of interest.
One of the major changes in the aged ECM is altered stiffness. As cells are well known to respond to changes in their mechanical microenvironment,22 aging-associated changes in ECM stiffness may play a causative role in cellular dysfunction. Changes in ECM stiffness with aging are often heterogeneous, even within a given tissue, but can generally be attributed to three different causes: (1) increased crosslinking, (2) increased proteolysis, and (3) excessive deposition of new ECM material (fibrosis).14 A major cause of increased crosslinking is due to reactions between ECM proteins like collagen and elastin with AGEs.23 AGEs arise from chemical reactions of sugars and tend to accumulate by reacting with long-lived ECM proteins. Crosslinks formed between these proteins lead to increased ECM stiffness. A second cause of altered tissue elasticity in aging is enzymatic dysregulation, which plays a two-pronged role in altering ECM stiffness. In some cases, crosslinking enzymes, such as lysyl oxidase, are upregulated, resulting in increased crosslinking and stiffness.14 In other cases, upregulation of ECM-degrading enzymes such as matrix metalloproteinases (MMPs; or equivalently, downregulation of their inhibitors, TIMPs) leads to increased fragmentation of the ECM.14 Both of these changes have been observed in skin aging.13 Fibrosis is the third major cause of altered tissue stiffness with aging. Fibrosis is a scarring response arising from improper tissue repair and resulting in the aberrant and excess deposition of new ECM material. The prevalence of fibrosis in various tissues increases with aging, especially in the lungs and cardiovascular system.24 These mechanisms for increasing tissue stiffness result in increases in measured elastic moduli on the order of 2- to 5-fold, depending on the tissue of interest.25,26,27,28
In addition to time-independent (elastic) mechanical properties such as stiffness, the time-dependent (viscoelastic) properties of the ECM are increasingly being recognized as important determinants of cell fate.29 While changes in tissue viscoelasticity with aging remain largely unexplored, several studies have demonstrated that the viscoelasticity of the brain is altered with aging.30,31,32 However, such studies have not reached a consensus for how viscoelasticity changes in the aging brain, likely due to the differences in techniques used to measure viscoelastic properties (e.g., oscillatory rheology on dissected brain slices versus in situ techniques like magnetic resonance elastography (MRE)). Direct mechanical measurements using indentation or bulk rheology report an increase in both storage and loss moduli as a function of increasing age,30,32 while MRE studies have converged on a decrease in the elastic component with increasing age and diverged on the relative viscous component.31,33 Some MRE studies considering the damping ratio as a metric for viscous vs. elastic behavior report an increase in the damping ratio, and thus increased viscous nature, as a function of aging.33,34 Such studies have suggested a link between increased damping ratio in aged brain tissue and decreased cognitive function in human subjects.33 Therefore, identifying mechanistic links between altered brain viscoelasticity and neuronal function may provide new insights into aging-associated cognitive decline.
Beyond changes in bulk mechanical properties like stiffness and viscoelasticity, aging-related changes in the extracellular microenvironment also impact the microstructural properties of tissues. As mentioned above, increased proteolysis with aging leads to fragmentation of collagen fibrils, for example, in aged skin.13 This not only locally decreases tissue stiffness but also results in shorter overall fiber length. Despite this increased fragmentation, the alignment of collagen fibers in aged skin is increased relative to young skin,35 altering long-distance force transduction through the tissue and impacting directional processes such as cell migration. These changes in turn may alter immune cell recruitment and enhance cancer metastasis.35 Similar alignment of collagen fibrils is seen in aged tendon,36 which may impair both bulk tissue mechanics (e.g., lower strain until failure) and have similar deleterious effects on immune cell migration as described above. Non-ECM proteins also substantially contribute to microstructural changes in aging. Increased incidence of protein misfolding and aggregation are correlated with aging. This is particularly evident in multiple neurodegenerative diseases that are characterized by the appearance of insoluble extracellular plaques of aggregated proteins.15 Understanding how the biophysical changes caused by these aggregates impact cellular function has gained increased importance as new clinical trials targeting the aggregates themselves have not yet resulted in dramatic improvement in patients.37
If mechanical and microstructural changes are found to be causative for aging-related tissue dysfunction, treatments that reverse the microenvironmental changes may not be sufficient to restore a youthful cellular phenotype. Recent studies have demonstrated that cells can possess a “mechanical memory” wherein exposure to a particular mechanical microenvironment results in persistent alteration of cellular behavior, even after the cell has been moved to a different microenvironment.38 This memory phenomenon was first described in the context of persistent changes in regulatory molecules, such as transcriptional regulatory proteins and micro RNAs, upon exposure of cells to an excessively stiff environment.39,40 More recent approaches have sought to link the apparent memory to changes in chromatin organization.41,42 Because this memory may persist even after the microenvironment is restored to a healthy state, therapies targeting microenvironmental-related dysfunction in aging may also need to “erase” the cellular mechanical memory. Engineered biomaterials platforms will also play a substantial role in elucidating the underlying mechanisms of the memory phenomenon.
Leveraging dynamic biomaterials systems to recapitulate the aged microenvironment
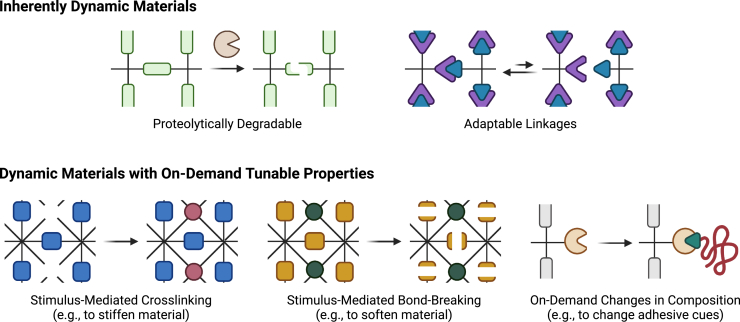
Aging is an inherently time-dependent progress. Microenvironmental changes that contribute to aging phenotypes accumulate over timescales ranging from weeks to decades. Thus, in order to adequately capture aging phenotypes in model systems, biomaterials systems with time-dependent properties are required. There are opportunities to apply two different types of dynamic materials to the study of aging biology: (1) inherently dynamic materials that can alter their properties in response to cell-initiated or other naturally occurring stimuli and (2) dynamic materials with on-demand tuning of properties in response to user-directed stimuli (Figure 3).
Figure 3.
Biomaterials with dynamic properties can be used to recapitulate the temporal nature of aging in engineered systems
These dynamic properties can be either inherent to the material, enabling a direct response to cellular behavior, or user-controlled, enabling on-demand tuning of mechanics and biochemical composition to mimic the aging process.
Inherently dynamic materials
One aspect of the native ECM’s inherent dynamism arises from its cell responsiveness. The protein and polysaccharide components of the ECM are susceptible to degradation and remodeling by cell-secreted enzymes, such as MMPs. In engineered biomaterials, proteolysis and remodeling of the material has been shown to be required for cell spreading and migration,43 maintenance of cell-cell contacts,44 and force transduction in 3D.45 Biomaterials derived from natural proteins generally retain their proteolytic remodelability, making such systems the most straightforward method for integrating cell responsiveness into engineered culture systems. A significant limitation of fully natural systems, including collagen, fibrin, and reconstituted basement membrane mixtures like Matrigel, to studying the impact of changing ECM mechanics during aging is the narrow window of accessible mechanical properties and a general inability to decouple mechanics from the total content of protein and thus of cell adhesive ligands.46 Chemically modified, naturally occurring protein materials, such as methacrylated gelatin47 or methacrylated tropoelastin,48 can overcome this limitation by permitting the crosslink density to be tuned independent of protein content, while still permitting cell-mediated degradation.
The use of fully synthetic materials is often preferred over natural materials for mechanobiology studies, as such materials are inherently non-cell-interactive, so all adhesive and degradation interactions are directly programmed into the material. The most common way for introducing proteolytic degradability into fully synthetic biomaterials is by introducing crosslinkers comprised of short peptide sequences that can be cleaved by specific proteases. Such peptides can be readily incorporated into step-growth hydrogel networks formed using cytocompatible crosslinking chemistries, such as thiol-vinyl sulfone reactions,43 strain-promoted azide-alkyne cycloaddition,49 or photocatalyzed thiol-ene reactions.50 By altering the peptide sequence used, the specificity for particular enzymes and the kinetics of bond cleavage can be controlled.51,52
An intermediate class of materials combining many of the benefits of naturally occurring proteins and fully synthetic materials is recombinantly expressed engineered proteins.53 Such proteins are often based on naturally occurring amino acid sequences, such as from elastin, silk, and resilin, with added domains containing cell adhesive and proteolytically degradable sequences.53 These materials allow for native biochemical functionality like naturally derived proteins while providing precise control over network architecture, similar to synthetic systems like multi-arm poly(ethylene glycol), by genetically encoding specific crosslinking sites into the protein sequence. This level of control enables both the degradation kinetics and the maximal extent of network remodeling to be tuned simultaneously.54
Materials with precise control over proteolytic remodelability will be required to study the effects of altered proteolysis occurring with aging. Increased degradation and turnover of the ECM with aging is impacted by several of the general mechanisms implicated in aging. For instance, cell-intrinsic changes can result in upregulation of ECM-degrading proteins. Activation of the AP-1 family of transcription factors was shown to upregulate expression of MMP-1, which led to fragmentation of collagen fibrils, reduced cell spreading, and increased AP-1 activity in a detrimental positive feedback loop.55 MMPs are also a classic component of SASPs,6 so an increased proportion of senescent cells in aged tissues will contribute to degradation and remodeling of the aged ECM. Finally, persistent inflammation and tissue damage can recruit immune cells that degrade the ECM.
A second aspect of the dynamic nature of the native ECM arises from its viscoelastic character. The supramolecular assemblies that comprise the ECM consist in part of physical crosslinks that can be broken and reformed by applied force. As a result, native tissues exhibit a spectrum of stress relaxation behavior in response to applied forces.29 As the crosslinks are rearranged to dissipate stresses, the material can be physically remodeled by cells. Such physical remodeling of engineered systems has been shown to regulate stem cell differentiation,56 neuronal protrusion and cell-cell contact,44,57 and growth of organoids.58 The crosslinking kinetics of polymeric networks have recently been shown to regulate the self-assembly of collagen interpenetrating networks,59 suggesting that changes in ECM viscoelasticity and remodeling with aging may further contribute to the progression of dysfunctional ECM phenotypes by impacting the organization of newly deposited ECM molecules.
The viscoelastic character of the ECM can be recapitulated in engineered systems by incorporating adaptable crosslinks into polymer networks.60 These adaptable crosslinks can arise from physical interactions or covalent bonds between adjacent polymer chains. One of the most common examples of physically crosslinked adaptable materials is calcium-crosslinked alginate gels. The viscoelastic stress relaxation in these materials can be tuned by varying the molecular weight of the alginate chains or adding poly(ethylene glycol) spacers while keeping the overall crosslink density constant.56 Other physically crosslinked materials make use of molecular recognition between peptide or protein domains that can dock and undock with different affinities to enable tunable stress relaxation.61 Host-guest chemistry employs a similar molecular docking mechanism whereby a hydrophobic “guest” molecule inserts itself into a cavity within a “host” molecule.62,63 Similar to protein-based molecular recognition, the binding affinities of the host-guest pair dictate the viscoelastic properties and stability of the hydrogel networks.
Adaptable covalent bonds provide an alternative approach to regulate hydrogel viscoelasticity.64 The primary requirement for covalent systems to maintain adaptability under physiologically relevant conditions is for the two separate reactive partners and their resulting adduct to exist in dynamic equilibrium, with forward and reverse reaction rates over biologically relevant timescales (seconds to hours).64 The most widely used adaptable covalent crosslinks are hydrazone linkages formed by the reaction of organic hydrazines and aldehydes.64 These systems exhibit stress relaxation over minutes to hours, and the relaxation rate can be tuned by varying the chemical structure of the aldehydes, with aromatic aldehydes relaxing more slowly than aliphatic aldehydes.65 These materials have been used with a variety of cell culture systems, including neural stem cells,57 mesenchymal stromal cells,66 and intestinal organoids.67 More recently, boronic esters have emerged as another class of adaptable covalent crosslinks with tunable stress relaxation.68,69
On-demand tunable dynamic systems
The inherently dynamic materials described above will predominantly find applications in mimicking fixed time points in the aging process (e.g., young vs. middle-aged vs. geriatric). These materials are preprogrammed with specific time-evolving properties that can be tailored to particular tissues at particular ages. However, these systems cannot capture the long timescale dynamics of aging in a single sample. To sufficiently accelerate the decades-long aging process to occur over laboratory timescales (days to weeks), materials with on-demand tuning of key properties are required. Such systems can enable researchers to trigger substantial changes in the properties of the extracellular matrix over short timescales and enable real-time evaluation of cellular responses to these properties.
As described above, one of the hallmarks of aging in many tissues is increased tissue stiffness. In some instances, this is caused by aberrant and excess deposition of extracellular matrix during fibrosis. In other cases, increased crosslinking of existing ECM proteins by AGEs or increased expression of crosslinking enzymes like lysyl oxidase leads to increased stiffness. To capture these progressive changes in ECM stiffness, hydrogel biomaterials with user-controlled stiffening can be used.
One of the most common strategies for introducing user-directed stimuli responsiveness into biomaterials platforms is the incorporation of light-sensitive chemistries. Light is generally a cytocompatible stimulus that can be added to most biological experimental workflows without substantial changes needing to be made.70 To cover the broad range of tissue stiffness that is observed in aged tissue, spanning up to 5-fold compared to young tissues,25,26,27,28 the addition of new crosslinks to the polymeric network is likely required. Approaches that use light to tune material stiffness by changing molecular conformation to alter crosslink spacing are generally unable to span such a broad range of stiffness.71,72,73,74 To increase material stiffness multi-fold, one of the most successful approaches is to introduce secondary crosslinks into an existing polymeric network by photoinitiated radical chain growth polymerization. Methacrylate groups in the hydrogel network are polymerized using UV-sensitive photoinitiators, resulting in the formation of a secondary polymer network that reinforces and stiffens the existing material.75,76 One potential drawback of this approach is the use of free radical polymerization, which may cause damage to sensitive cell types and provide a confounding variable during the accelerated aging process. More recent studies have added covalent crosslinks to existing networks by light-triggered [4 + 2] or [2 + 2] cycloaddition reactions that do not rely on radical-based chemistries.77,78 An alternative approach is to use ionically crosslinked systems with triggered ion release to cause stiffening. By co-encapsulating calcium ions and near-infrared absorbing gold nanorods within lipid nanoparticles suspended within an alginate network, exposure to near-IR light can trigger release of the calcium ions, which add crosslinks to and stiffen the alginate network.79 While extracellular calcium release can provide a confounding variable due to crosstalk with calcium-sensitive cell signaling pathways, the use of near-IR light has several advantages. It is lower energy and thus less likely to be cytotoxic than UV light, and near-IR light is less scattered by cells and tissues than UV and visible light, making near-IR light an attractive stimulus for use in the 3D organotypic culture systems described below.80
In addition to light, other cytocompatible stimuli, such as enzymes or magnetic fields, have been used to change the stiffness of biomaterial cell culture platforms on demand. The enzyme-mediated approaches are similar to the light-mediated approaches described above, in that stiffening is accomplished by addition of crosslinks to a hydrogel network. To limit the potential off-target reactivity of the enzyme-mediated approach with cellular biochemical processes, the enzymes used should not be natively present in the cells of interest and should have low degrees of reactivity with cell-produced proteins. The bacterial enzyme, sortase A, meets these criteria and has been previously used to catalyze transpeptidation reactions to stiffen hydrogels in the presence of live cells.81 One drawback of this approach is that the timescale of the stiffening reaction is limited by the diffusion of the enzyme into the hydrogel network; although for aging studies designed to mimic years or decades of accumulated matrix changes, the slightly longer time taken by the enzymatic approach compared to the light-mediated approach still substantially accelerates the process.
A third approach utilizes magnetic nanoparticles embedded within the hydrogels to change the stiffness of the material in response to an applied magnetic field. When a magnetic field is applied, the nanoparticles align along the field lines and limit motion of the polymer chains comprising the hydrogel, thereby stiffening the gel network.82 More recent optimizations have extended this approach to synthetic fibrous materials that are reminiscent of the native ECM.83 Advantages of magnetically tunable systems include mild magnetic stimuli and inherent reversibility of the stiffness increase by removing the magnetic field. One limitation is that the inclusion of the magnetic nanoparticles often renders the hydrogels opaque, so imaging real-time changes in cells in response to changes in mechanics can be challenging.
As mentioned previously, cells have been shown to exhibit a mechanical memory, wherein previously experiencing a stiff microenvironment can continue to impact cellular behavior even after the microenvironment is rendered soft. A key technology enabling these studies is hydrogels that can soften, rather than stiffen, on demand. The first studies demonstrating mechanical memory used hydrogels with photocleavable crosslinking groups that would break apart when exposed to UV light.40,84 More recently, this general strategy was expanded to new photocleavable groups that respond to more cytocompatible visible light.85 While this initial class of materials only enabled softening, other approaches have combined the free radical-mediated stiffening reactions described previously with the photocleavable crosslinking groups to prepare materials that can be softened and then subsequently stiffened on demand.86 Such materials with reversible stiffening will provide greater insight into the extent mechanical memory is reversible, potentially identifying targets to help aged cells “forget” that they were in a stiffened tissue environment. The [2 + 2] photocycloaddition chemistry referenced previously results in stiffening by triggering cycloaddition reactions with 400–500 nm light that can be reversed by exposure to 340 nm UV light.78 Other strategies to prepare reversibly stiffening and softening materials leverage photoswitchable moieties, including both small molecules72 and proteins,73,74 although the range of stiffness tuning accessible in these materials is limited and does not span the multi-fold changes in stiffness observed in many aged tissues.
The emerging interest in the role of viscoelastic properties on cell fate has heralded the development of new biomaterials systems that exhibit on-demand tuning of viscoelasticity, and not only stiffness. For instance, the stress relaxation rates of hydrogels crosslinked by thioesters were able to be controlled by the amount of excess thiol present, enabling force dissipation in the material through thioester exchange.87 By consuming the excess thiols in the hydrogel networks through phototriggered thiol-ene reactions, thioester exchange could be blocked, rendering the materials unable to relax applied stresses.87 Another strategy employs soluble small-molecule competitors that can be added to hydrogels to simultaneously and reversibly change stiffness and stress relaxation in a cytocompatible manner by varying crosslink density.88 Future approaches will likely build on this work by accessing a broader range of stress relaxation timescales without changing stiffness for on-demand tuning. For instance, DNA-based crosslinkers have begun to show promise as an alternative chemical approach to regulating viscoelasticity on demand.89 One outstanding challenge is the timescale over which the stress relaxation rate of the hydrogel networks is varied. Aging-related changes in ECM mechanics occur gradually, rather than as an abrupt transition. Thus, it will be important to develop materials systems in which the triggering stimulus (e.g., light or small molecules) can be titrated to provide more gradual changes in stress relaxation rates.
The predominant method by which cells sense the mechanical properties of the ECM is through engagement between cell surface receptors, such as integrin proteins, and cell adhesive peptide motifs on ECM proteins. Because the composition of the ECM changes during aging, the identity and concentration of cell adhesive ligands presented to cells also changes.14 For example, in aged skeletal muscle, the laminin-rich basal lamina has been shown to exhibit increased levels of collagen IV90,91 and decreased levels of collagen VI92 and fibronectin.93 Thus, materials that mimic changes in the ECM of skeletal muscle with aging should be designed to allow tuning the balance of collagen- vs. fibronectin-engaging integrin ligands, independent of changes in overall mechanical properties.
Stimuli-responsive chemistries similar to those described above used to change the stiffness of engineered ECM materials can also be used to regulate the presentation of adhesive cues on demand. For instance, photocleavable groups can be used to “cage” adhesive ligands, making them initially inaccessible to cell surface receptors.94 Upon light exposure and photocleavage, the ligands are uncaged and available for cell adhesion.94 In addition to directly uncaging the adhesive cues, photocaging approaches have been used to selectively make available reactive sites within the material to which newly added adhesive cues can chemically couple.95,96 Sortase-mediated transpeptidation has also been applied to alter the presentation of cell adhesive cues within hydrogel materials on demand.97 In contrast to the light-mediated approaches, the sortase reaction is reversible, so the number and type of adhesive sequences present can be changed multiple times using the same stimulus. Another reversible approach to tuning ligand presentation on demand makes use of host-guest supramolecular assembly whereby addition of guests with different binding affinities can trigger exchange of tethered adhesive ligands.98 Such reversible systems may be more amenable to mimicking the changes in multiple ECM components seen during aging.
The microstructure of the extracellular environment also changes extensively during aging. While cell-compatible techniques to manipulate ECM microstructure on demand are not broadly studied, such systems will be key to understanding how cells alter their behavior in response to aberrant structural cues in aging. As discussed previously, the organization of fibrillar ECM components is often altered in aged tissues, for instance, becoming more aligned in aged skin and tendon. Cytocompatible magnetic stimuli have recently been used to generate composite hydrogel biomaterials with aligned fibers.99 Combining physically adaptable interfiber crosslinking chemistries with these magnetic approaches may allow for reorganization of the fibrous network structure on demand. Beyond changes in ECM architecture, aberrant extracellular protein aggregation significantly alters the microstructure of the tissue environment in several organs, notably the brain. Prior efforts have largely focused on engineered cell lines or patient-derived stem cells that produce these protein aggregates in culture to study their effects,100,101 but little has been done in terms of biomaterial engineering to generate cell-independent mechanisms of protein aggregation to study the biophysical effects of these aggregates on cells. As intrinsically disordered protein sequences that can undergo this aggregation phenomenon have been extensively studied,102 there is an opportunity for protein-engineered materials to be employed to generate materials with spatiotemporal control over protein aggregation behavior.
Integrating dynamic biomaterials with organotypic cultures
While the focus of this Perspective has been how materials systems can be engineered to capture mechanical, structural, and biochemical changes that occur in aging, it is important to remember that these microenvironmental changes do not occur in isolation for a single cell type. Interactions between multiple cell types and physiologically relevant spatial configurations of these cell types will undoubtedly play an important role in capturing the complexities of aging in engineered systems (Figure 4). The advent of organotypic 3D cultures, or organoids, has made it possible to recapitulate many key cell-cell interactions in vitro.103 By definition, organoids must capture the cellular composition and organization that gives rise to key tissue functions.
Figure 4.
Organotypic cultures capture much of the in vivo complexity of tissues, including heterogeneous cell populations and native-like cellular organization
By combing organotypic culture models with dynamic biomaterials, progressive changes in the tissue microenvironment can be recapitulated, potentially enabling accelerated aging of tissue-like structures to gain mechanistic insight into the determinants of aging.
Organoids have been generated for most mature tissue types, although more work is required to make engineered biomaterials systems compatible with organoid growth. The most extensively studied organoids in the biomaterials context are intestinal in origin.104,105,106 These initial studies serve as a powerful proof of principle that dynamic material properties play causal roles in regulating multicellular behavior. For instance, materials that are inherently remodelable through hydrolysis or viscoelastic force dissipation are required to enable self-organization and proper differentiation of intestinal organoids.58,105 Furthermore, materials with user-directed spatiotemporal control over network degradation enabled detailed studies of intestinal crypt development in organoids.107
Organotypic cultures are uniquely positioned to study aging-related phenomena, as their multiple constituent cell types are better able to capture the complexity of cell-cell interactions that go awry during aging. As discussed previously, immune dysfunction is a key hallmark of aging that may play a causative role in aging-related phenotypes. For instance, an improper immune response is thought to detrimentally impact normal tissue regeneration during aging, and a recent study demonstrated in a skeletal muscle microtissue model that immune cells were essential for proper regeneration,108 recapitulating in vivo findings. A 3D organotypic culture system has also been used to model neuroinflammation in Alzheimer's disease.109
Dynamic biomaterials systems will not only be useful for studying the effects of matrix properties on individual cell types but also may enable carefully controlled studies in organotypic systems that capture the complicated effects of aging on multiple cell types. As seen in the case of intestinal organoids,58,105 inherently dynamic materials will likely be required to enable self-organization of cells into tissue-like structures. The three-dimensional and multicellular nature of these culture systems will also necessitate the judicious choice of materials to enable reliable readouts of cellular phenotypes. Single-cell RNA sequencing has become the gold standard for quantitatively studying transcriptional changes in heterogeneous cell populations, so any materials system used must enable facile recovery of single cells or nuclei. Live cell imaging with fluorescent reporters will also be crucial for tracking cell fate changes as the cultures are “aged,” making compatibility with fluorescence microscopy a key design criterion for new dynamic biomaterials.
Finally, while a major promise of employing well-defined biomaterial systems to study aging is decreased sample-to-sample heterogeneity that can obscure causal relationships, heterogeneous phenotypes are inherent to the aging process. Thus, therapeutic strategies targeting molecular pathways identified using engineered systems may not be one-size-fits-all solutions. It will therefore be important to design these engineered materials to account for not only a variety of cell types that would be represented in organotypic cultures but also to be scalable to allow for testing and validation using patient-derived samples from diverse genetic backgrounds and overall health status.
Conclusion
Aging is a multifactorial process that results in both systemic and local changes that lead to diminished cell and tissue function. This inherent complexity, along with limitations of model organisms, has made determining the underlying mechanisms of human aging a daunting challenge. The use of engineered platforms to study aging using human cells is an attractive approach to surmount these challenges, as such systems can enable selective, independent tuning of multiple parameters to allow causal relationships to be assigned. Biomaterials with dynamic properties are uniquely positioned to recapitulate the inherently time-dependent nature of aging. Materials that respond to both cell-mediated and user-directed stimuli will likely be required to enable native-like cell-matrix interactions and to allow the aging process to be accelerated to occur over an observable timescale in the lab. Ultimately, these biomaterials will likely need to be optimized for use with organotypic culture models that additionally capture the heterogeneous cell populations and the cellular organization in living tissues to achieve a sufficiently representative model of human aging. If the proper balance between engineering control and biological complexity can be obtained, in vitro models of aging may identify new therapeutic approaches to mitigate the effects of aging and increase healthspan.
Acknowledgments
C.M.M. is supported by funding from the U.S. National Institutes of Health (NIH: R00 AG071738) and a seed grant from the Center for Engineering Mechanobiology (CEMB), a U.S. National Science Foundation-funded Science and Technology Center (NSF: CMMI 1548571). Figures were generated using BioRender.
Author contributions
Writing – original draft, C.M.M.; writing – review & editing, C.M.M.; funding acquisition, C.M.M.
Declaration of interests
The author declares no competing interests.
References
- 1.Mitchell S.J., Scheibye-Knudsen M., Longo D.L., de Cabo R. Animal models of aging research: implications for human aging and age-related diseases. Annu. Rev. Anim. Biosci. 2015;3:283–303. doi: 10.1146/annurev-animal-022114-110829. [DOI] [PubMed] [Google Scholar]
- 2.Christensen K., Doblhammer G., Rau R., Vaupel J.W. Ageing populations: the challenges ahead. Lancet. 2009;374:1196–1208. doi: 10.1016/S0140-6736(09)61460-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferrucci L., Fabbri E. Inflammageing: chronic inflammation in ageing, cardiovascular disease, and frailty. Nat. Rev. Cardiol. 2018;15:505–522. doi: 10.1038/s41569-018-0064-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Franceschi C., Garagnani P., Parini P., Giuliani C., Santoro A. Inflammaging: a new immune–metabolic viewpoint for age-related diseases. Nat. Rev. Endocrinol. 2018;14:576–590. doi: 10.1038/s41574-018-0059-4. [DOI] [PubMed] [Google Scholar]
- 5.Campisi J., d’Adda di Fagagna F. Cellular senescence: when bad things happen to good cells. Nat. Rev. Mol. Cell Biol. 2007;8:729–740. doi: 10.1038/nrm2233. [DOI] [PubMed] [Google Scholar]
- 6.Sharpless N.E., Sherr C.J. Forging a signature of in vivo senescence. Nat. Rev. Cancer. 2015;15:397–408. doi: 10.1038/nrc3960. [DOI] [PubMed] [Google Scholar]
- 7.Liu L., Rando T.A. In: Handbook of the Biology of Aging. Seventh Edition. Masoro E.J., Austad S.N., editors. Academic Press; 2011. Chapter 6 - aging of stem cells: intrinsic changes and environmental influences; pp. 141–161. (Handbooks of Aging). [DOI] [Google Scholar]
- 8.Rossi D.J., Jamieson C.H.M., Weissman I.L. Stems cells and the pathways to aging and cancer. Cell. 2008;132:681–696. doi: 10.1016/j.cell.2008.01.036. [DOI] [PubMed] [Google Scholar]
- 9.Cosgrove B.D., Gilbert P.M., Porpiglia E., Mourkioti F., Lee S.P., Corbel S.Y., Llewellyn M.E., Delp S.L., Blau H.M. Rejuvenation of the muscle stem cell population restores strength to injured aged muscles. Nat. Med. 2014;20:255–264. doi: 10.1038/nm.3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bernet J.D., Doles J.D., Hall J.K., Kelly Tanaka K., Carter T.A., Olwin B.B. p38 MAPK signaling underlies a cell-autonomous loss of stem cell self-renewal in skeletal muscle of aged mice. Nat. Med. 2014;20:265–271. doi: 10.1038/nm.3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pang W.W., Price E.A., Sahoo D., Beerman I., Maloney W.J., Rossi D.J., Schrier S.L., Weissman I.L. Human bone marrow hematopoietic stem cells are increased in frequency and myeloid-biased with age. Proc. Natl. Acad. Sci. USA. 2011;108:20012–20017. doi: 10.1073/pnas.1116110108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Birch H.L. Extracellular matrix and ageing. Subcell. Biochem. 2018;90:169–190. doi: 10.1007/978-981-13-2835-0_7. [DOI] [PubMed] [Google Scholar]
- 13.McCabe M.C., Hill R.C., Calderone K., Cui Y., Yan Y., Quan T., Fisher G.J., Hansen K.C. Alterations in extracellular matrix composition during aging and photoaging of the skin. Matrix Biol. 2020;8:100041. doi: 10.1016/j.mbplus.2020.100041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Phillip J.M., Aifuwa I., Walston J., Wirtz D. The mechanobiology of aging. Annu. Rev. Biomed. Eng. 2015;17:113–141. doi: 10.1146/annurev-bioeng-071114-040829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ross C.A., Poirier M.A. Protein aggregation and neurodegenerative disease. Nat. Med. 2004;10:S10–S17. doi: 10.1038/nm1066. [DOI] [PubMed] [Google Scholar]
- 16.Brunet A. Old and new models for the study of human ageing. Nat. Rev. Mol. Cell Biol. 2020;21:491–493. doi: 10.1038/s41580-020-0266-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mertens J., Reid D., Lau S., Kim Y., Gage F.H. Aging in a dish: iPSC-derived and directly induced neurons for studying brain aging and age-related neurodegenerative diseases. Annu. Rev. Genet. 2018;52:271–293. doi: 10.1146/annurev-genet-120417-031534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Madl C.M., Heilshorn S.C., Blau H.M. Bioengineering strategies to accelerate stem cell therapeutics. Nature. 2018;557:335–342. doi: 10.1038/s41586-018-0089-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Conboy M.J., Conboy I.M., Rando T.A. Heterochronic parabiosis: historical perspective and methodological considerations for studies of aging and longevity. Aging Cell. 2013;12:525–530. doi: 10.1111/acel.12065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Segel M., Neumann B., Hill M.F.E., Weber I.P., Viscomi C., Zhao C., Young A., Agley C.C., Thompson A.J., Gonzalez G.A., et al. Niche stiffness underlies the ageing of central nervous system progenitor cells. Nature. 2019;573:130–134. doi: 10.1038/s41586-019-1484-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frantz C., Stewart K.M., Weaver V.M. The extracellular matrix at a glance. J. Cell Sci. 2010;123:4195–4200. doi: 10.1242/jcs.023820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Discher D.E., Mooney D.J., Zandstra P.W. Growth factors, matrices, and forces combine and control stem cells. Science. 2009;324:1673–1677. doi: 10.1126/science.1171643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chaudhuri J., Bains Y., Guha S., Kahn A., Hall D., Bose N., Gugliucci A., Kapahi P. The role of advanced glycation end products in aging and metabolic diseases: bridging association and causality. Cell Metabol. 2018;28:337–352. doi: 10.1016/j.cmet.2018.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murtha L.A., Morten M., Schuliga M.J., Mabotuwana N.S., Hardy S.A., Waters D.W., Burgess J.K., Ngo D.T., Sverdlov A.L., Knight D.A., Boyle A.J. The role of pathological aging in cardiac and pulmonary fibrosis. Aging Dis. 2019;10:419–428. doi: 10.14336/AD.2018.0601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oishi Y., Miyoshi H., Mizuguchi Y., Iuchi A., Nagase N., Oki T. Aortic stiffness is strikingly increased with age ≥50 years in clinically normal individuals and preclinical patients with cardiovascular risk factors: assessment by the new technique of 2D strain echocardiography. J. Cardiol. 2011;57:354–359. doi: 10.1016/j.jjcc.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 26.Sicard D., Haak A.J., Choi K.M., Craig A.R., Fredenburgh L.E., Tschumperlin D.J. Aging and anatomical variations in lung tissue stiffness. Am. J. Physiol. Lung Cell Mol. Physiol. 2018;314:L946–L955. doi: 10.1152/ajplung.00415.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lacraz G., Rouleau A.-J., Couture V., Söllrald T., Drouin G., Veillette N., Grandbois M., Grenier G. Increased stiffness in aged skeletal muscle impairs muscle progenitor cell proliferative activity. PLoS One. 2015;10:e0136217. doi: 10.1371/journal.pone.0136217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koester J., Miroshnikova Y.A., Ghatak S., Chacón-Martínez C.A., Morgner J., Li X., Atanassov I., Altmüller J., Birk D.E., Koch M., et al. Niche stiffening compromises hair follicle stem cell potential during ageing by reducing bivalent promoter accessibility. Nat. Cell Biol. 2021;23:771–781. doi: 10.1038/s41556-021-00705-x. [DOI] [PubMed] [Google Scholar]
- 29.Chaudhuri O., Cooper-White J., Janmey P.A., Mooney D.J., Shenoy V.B. Effects of extracellular matrix viscoelasticity on cellular behaviour. Nature. 2020;584:535–546. doi: 10.1038/s41586-020-2612-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gefen A., Gefen N., Zhu Q., Raghupathi R., Margulies S.S. Age-dependent changes in material properties of the brain and braincase of the rat. J. Neurotrauma. 2003;20:1163–1177. doi: 10.1089/089771503770802853. [DOI] [PubMed] [Google Scholar]
- 31.Sack I., Beierbach B., Wuerfel J., Klatt D., Hamhaber U., Papazoglou S., Martus P., Braun J. The impact of aging and gender on brain viscoelasticity. Neuroimage. 2009;46:652–657. doi: 10.1016/j.neuroimage.2009.02.040. [DOI] [PubMed] [Google Scholar]
- 32.Xue B., Wen X., Kuwar R., Sun D., Zhang N. Age-dependent viscoelastic characterization of rat brain cortex. Brain Multiphys. 2022;3:100056. doi: 10.1016/j.brain.2022.100056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hiscox L.V., Schwarb H., McGarry M.D.J., Johnson C.L. Aging brain mechanics: progress and promise of magnetic resonance elastography. Neuroimage. 2021;232:117889. doi: 10.1016/j.neuroimage.2021.117889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hiscox L.V., Johnson C.L., McGarry M.D.J., Perrins M., Littlejohn A., van Beek E.J.R., Roberts N., Starr J.M. High-resolution magnetic resonance elastography reveals differences in subcortical gray matter viscoelasticity between young and healthy older adults. Neurobiol. Aging. 2018;65:158–167. doi: 10.1016/j.neurobiolaging.2018.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaur A., Ecker B.L., Douglass S.M., Kugel C.H., Webster M.R., Almeida F.V., Somasundaram R., Hayden J., Ban E., Ahmadzadeh H., et al. Remodeling of the collagen matrix in aging skin promotes melanoma metastasis and affects immune cell motility. Cancer Discov. 2019;9:64–81. doi: 10.1158/2159-8290.CD-18-0193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van Gulick L., Saby C., Morjani H., Beljebbar A. Age-related changes in molecular organization of type I collagen in tendon as probed by polarized SHG and Raman microspectroscopy. Sci. Rep. 2019;9:7280. doi: 10.1038/s41598-019-43636-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Panza F., Lozupone M., Logroscino G., Imbimbo B.P. A critical appraisal of amyloid-β-targeting therapies for Alzheimer disease. Nat. Rev. Neurol. 2019;15:73–88. doi: 10.1038/s41582-018-0116-6. [DOI] [PubMed] [Google Scholar]
- 38.Lele T.P., Brock A., Peyton S.R. Emerging concepts and tools in cell mechanomemory. Ann. Biomed. Eng. 2020;48:2103–2112. doi: 10.1007/s10439-019-02412-z. [DOI] [PubMed] [Google Scholar]
- 39.Li C.X., Talele N.P., Boo S., Koehler A., Knee-Walden E., Balestrini J.L., Speight P., Kapus A., Hinz B. MicroRNA-21 preserves the fibrotic mechanical memory of mesenchymal stem cells. Nat. Mater. 2017;16:379–389. doi: 10.1038/nmat4780. [DOI] [PubMed] [Google Scholar]
- 40.Yang C., Tibbitt M.W., Basta L., Anseth K.S. Mechanical memory and dosing influence stem cell fate. Nat. Mater. 2014;13:645–652. doi: 10.1038/nmat3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Killaars A.R., Grim J.C., Walker C.J., Hushka E.A., Brown T.E., Anseth K.S. Extended exposure to stiff microenvironments leads to persistent chromatin remodeling in human mesenchymal stem cells. Adv. Sci. 2019;6:1801483. doi: 10.1002/advs.201801483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Price C.C., Mathur J., Boerckel J.D., Pathak A., Shenoy V.B. Dynamic self-reinforcement of gene expression determines acquisition of cellular mechanical memory. Biophys. J. 2021;120:5074–5089. doi: 10.1016/j.bpj.2021.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lutolf M.P., Raeber G.P., Zisch A.H., Tirelli N., Hubbell J.A. Cell-responsive synthetic hydrogels. Adv. Mater. 2003;15:888–892. [Google Scholar]
- 44.Madl C.M., LeSavage B.L., Dewi R.E., Dinh C.B., Stowers R.S., Khariton M., Lampe K.J., Nguyen D., Chaudhuri O., Enejder A., Heilshorn S.C. Maintenance of neural progenitor cell stemness in 3D hydrogels requires matrix remodelling. Nat. Mater. 2017;16:1233–1242. doi: 10.1038/nmat5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Khetan S., Guvendiren M., Legant W.R., Cohen D.M., Chen C.S., Burdick J.A. Degradation-mediated cellular traction directs stem cell fate in covalently crosslinked three-dimensional hydrogels. Nat. Mater. 2013;12:458–465. doi: 10.1038/nmat3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tibbitt M.W., Anseth K.S. Hydrogels as extracellular matrix mimics for 3D cell culture. Biotechnol. Bioeng. 2009;103:655–663. doi: 10.1002/bit.22361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nichol J.W., Koshy S.T., Bae H., Hwang C.M., Yamanlar S., Khademhosseini A. Cell-laden microengineered gelatin methacrylate hydrogels. Biomaterials. 2010;31:5536–5544. doi: 10.1016/j.biomaterials.2010.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Annabi N., Mithieux S.M., Zorlutuna P., Camci-Unal G., Weiss A.S., Khademhosseini A. Engineered cell-laden human protein-based elastomer. Biomaterials. 2013;34:5496–5505. doi: 10.1016/j.biomaterials.2013.03.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.DeForest C.A., Polizzotti B.D., Anseth K.S. Sequential click reactions for synthesizing and patterning three-dimensional cell microenvironments. Nat. Mater. 2009;8:659–664. doi: 10.1038/nmat2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aimetti A.A., Machen A.J., Anseth K.S. Poly(ethylene glycol) hydrogels formed by thiol-ene photopolymerization for enzyme-responsive protein delivery. Biomaterials. 2009;30:6048–6054. doi: 10.1016/j.biomaterials.2009.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lutolf M.P., Lauer-Fields J.L., Schmoekel H.G., Metters A.T., Weber F.E., Fields G.B., Hubbell J.A. Synthetic matrix metalloproteinase-sensitive hydrogels for the conduction of tissue regeneration: engineering cell-invasion characteristics. Proc. Natl. Acad. Sci. USA. 2003;100:5413–5418. doi: 10.1073/pnas.0737381100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Straley K.S., Heilshorn S.C. Dynamic, 3D-pattern formation within enzyme-responsive hydrogels. Adv. Mater. 2009;21:4148–4152. [Google Scholar]
- 53.DiMarco R.L., Heilshorn S.C. Multifunctional materials through modular protein engineering. Adv. Mater. 2012;24:3923–3940. doi: 10.1002/adma.201200051. [DOI] [PubMed] [Google Scholar]
- 54.Madl C.M., Katz L.M., Heilshorn S.C. Tuning bulk hydrogel degradation by simultaneous control of proteolytic cleavage kinetics and hydrogel network architecture. ACS Macro Lett. 2018;7:1302–1307. doi: 10.1021/acsmacrolett.8b00664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Qin Z., Voorhees J.J., Fisher G.J., Quan T. Age-associated reduction of cellular spreading/mechanical force up-regulates matrix metalloproteinase-1 expression and collagen fibril fragmentation via c-Jun/AP-1 in human dermal fibroblasts. Aging Cell. 2014;13:1028–1037. doi: 10.1111/acel.12265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chaudhuri O., Gu L., Klumpers D., Darnell M., Bencherif S.A., Weaver J.C., Huebsch N., Lee H.P., Lippens E., Duda G.N., Mooney D.J. Hydrogels with tunable stress relaxation regulate stem cell fate and activity. Nat. Mater. 2016;15:326–334. doi: 10.1038/nmat4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McKinnon D.D., Domaille D.W., Brown T.E., Kyburz K.A., Kiyotake E., Cha J.N., Anseth K.S. Measuring cellular forces using bis-aliphatic hydrazone crosslinked stress-relaxing hydrogels. Soft Matter. 2014;10:9230–9236. doi: 10.1039/C4SM01365D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chrisnandy A., Blondel D., Rezakhani S., Broguiere N., Lutolf M.P. Synthetic dynamic hydrogels promote degradation-independent in vitro organogenesis. Nat. Mater. 2022;21:479–487. doi: 10.1038/s41563-021-01136-7. [DOI] [PubMed] [Google Scholar]
- 59.Vining K.H., Stafford A., Mooney D.J. Sequential modes of crosslinking tune viscoelasticity of cell-instructive hydrogels. Biomaterials. 2019;188:187–197. doi: 10.1016/j.biomaterials.2018.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang H., Heilshorn S.C. Adaptable hydrogel networks with reversible linkages for tissue engineering. Adv. Mater. 2015;27:3717–3736. doi: 10.1002/adma.201501558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wong Po Foo C.T.S., Lee J.S., Mulyasasmita W., Parisi-Amon A., Heilshorn S.C. Two-component protein-engineered physical hydrogels for cell encapsulation. Proc. Natl. Acad. Sci. USA. 2009;106:22067–22072. doi: 10.1073/pnas.0904851106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rodell C.B., Kaminski A.L., Burdick J.A. Rational design of network properties in guest–host assembled and shear-thinning hyaluronic acid hydrogels. Biomacromolecules. 2013;14:4125–4134. doi: 10.1021/bm401280z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Park K.M., Yang J.-A., Jung H., Yeom J., Park J.S., Park K.-H., Hoffman A.S., Hahn S.K., Kim K. In situ supramolecular assembly and modular modification of hyaluronic acid hydrogels for 3D cellular engineering. ACS Nano. 2012;6:2960–2968. doi: 10.1021/nn204123p. [DOI] [PubMed] [Google Scholar]
- 64.Rizwan M., Baker A.E.G., Shoichet M.S. Designing hydrogels for 3D cell culture using dynamic covalent crosslinking. Adv. Healthc. Mater. 2021;10:2100234. doi: 10.1002/adhm.202100234. [DOI] [PubMed] [Google Scholar]
- 65.McKinnon D.D., Domaille D.W., Cha J.N., Anseth K.S. Biophysically defined and cytocompatible covalently adaptable networks as viscoelastic 3D cell culture systems. Adv. Mater. 2014;26:865–872. doi: 10.1002/adma.201303680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang H., Zhu D., Paul A., Cai L., Enejder A., Yang F., Heilshorn S.C. Covalently adaptable elastin-like protein–hyaluronic acid (ELP–HA) hybrid hydrogels with secondary thermoresponsive crosslinking for injectable stem cell delivery. Adv. Funct. Mater. 2017;27:1605609. doi: 10.1002/adfm.201605609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hunt D.R., Klett K.C., Mascharak S., Wang H., Gong D., Lou J., Li X., Cai P.C., Suhar R.A., Co J.Y., et al. Engineered matrices enable the culture of human patient-derived intestinal organoids. Adv. Sci. 2021;8:2004705. doi: 10.1002/advs.202004705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tang S., Ma H., Tu H.-C., Wang H.-R., Lin P.-C., Anseth K.S. Adaptable fast relaxing boronate-based hydrogels for probing cell–matrix interactions. Adv. Sci. 2018;5:1800638. doi: 10.1002/advs.201800638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Marozas I.A., Anseth K.S., Cooper-White J.J. Adaptable boronate ester hydrogels with tunable viscoelastic spectra to probe timescale dependent mechanotransduction. Biomaterials. 2019;223:119430. doi: 10.1016/j.biomaterials.2019.119430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li L., Scheiger J.M., Levkin P.A. Design and applications of photoresponsive hydrogels. Adv. Mater. 2019;31:1807333. doi: 10.1002/adma.201807333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rosales A.M., Rodell C.B., Chen M.H., Morrow M.G., Anseth K.S., Burdick J.A. Reversible control of network properties in azobenzene-containing hyaluronic acid-based hydrogels. Bioconjugate Chem. 2018;29:905–913. doi: 10.1021/acs.bioconjchem.7b00802. [DOI] [PubMed] [Google Scholar]
- 72.Rosales A.M., Mabry K.M., Nehls E.M., Anseth K.S. Photoresponsive elastic properties of azobenzene-containing poly(ethylene-glycol)-based hydrogels. Biomacromolecules. 2015;16:798–806. doi: 10.1021/bm501710e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu L., Shadish J.A., Arakawa C.K., Shi K., Davis J., DeForest C.A. Cyclic stiffness modulation of cell-laden protein–polymer hydrogels in response to user-specified stimuli including light. Adv. Biosyst. 2018;2:1800240. doi: 10.1002/adbi.201800240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hörner M., Raute K., Hummel B., Madl J., Creusen G., Thomas O.S., Christen E.H., Hotz N., Gübeli R.J., Engesser R., et al. Phytochrome-based extracellular matrix with reversibly tunable mechanical properties. Adv. Mater. 2019;31:1806727. doi: 10.1002/adma.201806727. [DOI] [PubMed] [Google Scholar]
- 75.Guvendiren M., Burdick J.A. Stiffening hydrogels to probe short- and long-term cellular responses to dynamic mechanics. Nat. Commun. 2012;3:792. doi: 10.1038/ncomms1792. [DOI] [PubMed] [Google Scholar]
- 76.Ondeck M.G., Engler A.J. Mechanical characterization of a dynamic and tunable methacrylated hyaluronic acid hydrogel. J. Biomech. Eng. 2016;138:021003–021006. doi: 10.1115/1.4032429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Günay K.A., Ceccato T.L., Silver J.S., Bannister K.L., Bednarski O.J., Leinwand L.A., Anseth K.S. PEG–Anthracene hydrogels as an on-demand stiffening matrix to study mechanobiology. Angew. Chem. Int. Ed. Engl. 2019;58:9912–9916. doi: 10.1002/anie.201901989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Truong V.X., Li F., Ercole F., Forsythe J.S. Wavelength-selective coupling and decoupling of polymer chains via reversible [2 + 2] photocycloaddition of styrylpyrene for construction of cytocompatible photodynamic hydrogels. ACS Macro Lett. 2018;7:464–469. doi: 10.1021/acsmacrolett.8b00099. [DOI] [PubMed] [Google Scholar]
- 79.Stowers R.S., Allen S.C., Suggs L.J. Dynamic phototuning of 3D hydrogel stiffness. Proc. Natl. Acad. Sci. USA. 2015;112:1953–1958. doi: 10.1073/pnas.1421897112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rapp T.L., DeForest C.A. Visible light-responsive dynamic biomaterials: going deeper and triggering more. Adv. Healthc. Mater. 2020;9:1901553. doi: 10.1002/adhm.201901553. [DOI] [PubMed] [Google Scholar]
- 81.Arkenberg M.R., Moore D.M., Lin C.-C. Dynamic control of hydrogel crosslinking via sortase-mediated reversible transpeptidation. Acta Biomater. 2019;83:83–95. doi: 10.1016/j.actbio.2018.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Abdeen A.A., Lee J., Bharadwaj N.A., Ewoldt R.H., Kilian K.A. Temporal modulation of stem cell activity using magnetoactive hydrogels. Adv. Healthc. Mater. 2016;5:2536–2544. doi: 10.1002/adhm.201600349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chen W., Zhang Y., Kumari J., Engelkamp H., Kouwer P.H.J. Magnetic stiffening in 3D cell culture matrices. Nano Lett. 2021;21:6740–6747. doi: 10.1021/acs.nanolett.1c00371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kloxin A.M., Kasko A.M., Salinas C.N., Anseth K.S. Photodegradable hydrogels for dynamic tuning of physical and chemical properties. Science. 2009;324:59–63. doi: 10.1126/science.1169494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Truong V.X., Li F., Forsythe J.S. Photolabile hydrogels responsive to broad spectrum visible light for selective cell release. ACS Appl. Mater. Interfaces. 2017;9:32441–32445. doi: 10.1021/acsami.7b11517. [DOI] [PubMed] [Google Scholar]
- 86.Rosales A.M., Vega S.L., DelRio F.W., Burdick J.A., Anseth K.S. Hydrogels with reversible mechanics to probe dynamic cell microenvironments. Angew. Chem., Int. Ed. 2017;56:12132–12136. doi: 10.1002/anie.201705684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Carberry B.J., Rao V.V., Anseth K.S. Phototunable viscoelasticity in hydrogels through thioester exchange. Ann. Biomed. Eng. 2020;48:2053–2063. doi: 10.1007/s10439-020-02460-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lou J., Xia Y. Using competitor molecules to reversibly modulate the mechanical properties of viscoelastic hydrogels. ACS Macro Lett. 2022;11:1312–1316. doi: 10.1021/acsmacrolett.2c00527. [DOI] [PubMed] [Google Scholar]
- 89.Peng Y.-H., Gupta K., Hsiao S.K., Ruland A., Auernhammer G.K., Maitz M.F., Boye S., Gerri C., Honigmann A., Werner C., et al. 2022. Dynamic Matrices with DNA-Encoded Viscoelasticity for Advanced Cell and Organoid Culture. [DOI] [Google Scholar]
- 90.Kovanen V., Suominen H., Risteli J., Risteli L. Type IV collagen and laminin in slow and fast skeletal muscle in rats–effects of age and life-time endurance training. Collagen Relat. Res. 1988;8:145–153. doi: 10.1016/s0174-173x(88)80026-8. [DOI] [PubMed] [Google Scholar]
- 91.Alexakis C., Partridge T., Bou-Gharios G. Implication of the satellite cell in dystrophic muscle fibrosis: a self-perpetuating mechanism of collagen overproduction. Am. J. Physiol. Cell Physiol. 2007;293:C661–C669. doi: 10.1152/ajpcell.00061.2007. [DOI] [PubMed] [Google Scholar]
- 92.Scimè A., Desrosiers J., Trensz F., Palidwor G.A., Caron A.Z., Andrade-Navarro M.A., Grenier G. Transcriptional profiling of skeletal muscle reveals factors that are necessary to maintain satellite cell integrity during ageing. Mech. Ageing Dev. 2010;131:9–20. doi: 10.1016/j.mad.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 93.Lukjanenko L., Jung M.J., Hegde N., Perruisseau-Carrier C., Migliavacca E., Rozo M., Karaz S., Jacot G., Schmidt M., Li L., et al. Loss of fibronectin from the aged stem cell niche affects the regenerative capacity of skeletal muscle in mice. Nat. Med. 2016;22:897–905. doi: 10.1038/nm.4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lee T.T., García J.R., Paez J.I., Singh A., Phelps E.A., Weis S., Shafiq Z., Shekaran A., del Campo A., García A.J. Light-triggered in vivo activation of adhesive peptides regulates cell adhesion, inflammation and vascularization of biomaterials. Nat. Mater. 2015;14:352–360. doi: 10.1038/nmat4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Luo Y., Shoichet M.S. A photolabile hydrogel for guided three-dimensional cell growth and migration. Nat. Mater. 2004;3:249–253. doi: 10.1038/nmat1092. [DOI] [PubMed] [Google Scholar]
- 96.DeForest C.A., Tirrell D.A. A photoreversible protein-patterning approach for guiding stem cell fate in three-dimensional gels. Nat. Mater. 2015;14:523–531. doi: 10.1038/nmat4219. [DOI] [PubMed] [Google Scholar]
- 97.Cambria E., Renggli K., Ahrens C.C., Cook C.D., Kroll C., Krueger A.T., Imperiali B., Griffith L.G. Covalent modification of synthetic hydrogels with bioactive proteins via sortase-mediated ligation. Biomacromolecules. 2015;16:2316–2326. doi: 10.1021/acs.biomac.5b00549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Boekhoven J., Rubert Pérez C.M., Sur S., Worthy A., Stupp S.I. Dynamic display of bioactivity through host–guest chemistry. Angew. Chem. Int. Ed. 2013;52:12077–12080. doi: 10.1002/anie.201306278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hiraki H.L., Matera D.L., Rose M.J., Kent R.N., Todd C.W., Stout M.E., Wank A.E., Schiavone M.C., DePalma S.J., Zarouk A.A., Baker B.M. Magnetic alignment of electrospun fiber segments within a hydrogel composite guides cell spreading and migration phenotype switching. Front. Bioeng. Biotechnol. 2021;9:679165. doi: 10.3389/fbioe.2021.679165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Choi S.H., Kim Y.H., Hebisch M., Sliwinski C., Lee S., D’Avanzo C., Chen H., Hooli B., Asselin C., Muffat J., et al. A three-dimensional human neural cell culture model of Alzheimer’s disease. Nature. 2014;515:274–278. doi: 10.1038/nature13800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Grenier K., Kao J., Diamandis P. Three-dimensional modeling of human neurodegeneration: brain organoids coming of age. Mol. Psychiatr. 2020;25:254–274. doi: 10.1038/s41380-019-0500-7. [DOI] [PubMed] [Google Scholar]
- 102.Ruff K.M., Roberts S., Chilkoti A., Pappu R.V. Advances in understanding stimulus-responsive phase behavior of intrinsically disordered protein polymers. J. Mol. Biol. 2018;430:4619–4635. doi: 10.1016/j.jmb.2018.06.031. [DOI] [PubMed] [Google Scholar]
- 103.Lancaster M.A., Knoblich J.A. Organogenesis in a dish: modeling development and disease using organoid technologies. Science. 2014;345:1247125. doi: 10.1126/science.1247125. [DOI] [PubMed] [Google Scholar]
- 104.DiMarco R.L., Dewi R.E., Bernal G., Kuo C., Heilshorn S.C. Protein-engineered scaffolds for in vitro 3D culture of primary adult intestinal organoids. Biomater. Sci. 2015;3:1376–1385. doi: 10.1039/c5bm00108k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gjorevski N., Sachs N., Manfrin A., Giger S., Bragina M.E., Ordóñez-Morán P., Clevers H., Lutolf M.P. Designer matrices for intestinal stem cell and organoid culture. Nature. 2016;539:560–564. doi: 10.1038/nature20168. [DOI] [PubMed] [Google Scholar]
- 106.Cruz-Acuña R., Quirós M., Farkas A.E., Dedhia P.H., Huang S., Siuda D., García-Hernández V., Miller A.J., Spence J.R., Nusrat A., García A.J. Synthetic hydrogels for human intestinal organoid generation and colonic wound repair. Nat. Cell Biol. 2017;19:1326–1335. doi: 10.1038/ncb3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gjorevski N., Nikolaev M., Brown T.E., Mitrofanova O., Brandenberg N., DelRio F.W., Yavitt F.M., Liberali P., Anseth K.S., Lutolf M.P. Tissue geometry drives deterministic organoid patterning. Science. 2022;375:eaaw9021. doi: 10.1126/science.aaw9021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Juhas M., Abutaleb N., Wang J.T., Ye J., Shaikh Z., Sriworarat C., Qian Y., Bursac N. Incorporation of macrophages into engineered skeletal muscle enables enhanced muscle regeneration. Nat. Biomed. Eng. 2018;2:942–954. doi: 10.1038/s41551-018-0290-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Park J., Wetzel I., Marriott I., Dréau D., D’Avanzo C., Kim D.Y., Tanzi R.E., Cho H. A 3D human triculture system modeling neurodegeneration and neuroinflammation in Alzheimer’s disease. Nat. Neurosci. 2018;21:941–951. doi: 10.1038/s41593-018-0175-4. [DOI] [PMC free article] [PubMed] [Google Scholar]