Abstract
Background & Aims
Loss-of-function variants in the PTPN2 gene are associated with increased risk of inflammatory bowel disease. We recently showed that Ptpn2 is critical for intestinal epithelial cell (IEC) barrier maintenance, IEC–macrophage communication, and modulation of the gut microbiome in mice, restricting expansion of a small intestinal pathobiont associated with inflammatory bowel disease. Here, we aimed to identify how Ptpn2 loss affects ileal IEC subtypes and their function in vivo.
Methods
Constitutive Ptpn2 wild-type, heterozygous, and knockout (KO) mice, as well as mice with inducible deletion of Ptpn2 in IECs, were used in the study. Investigation was performed using imaging techniques, flow cytometry, enteroid culture, and analysis of gene and protein levels of IEC markers.
Results
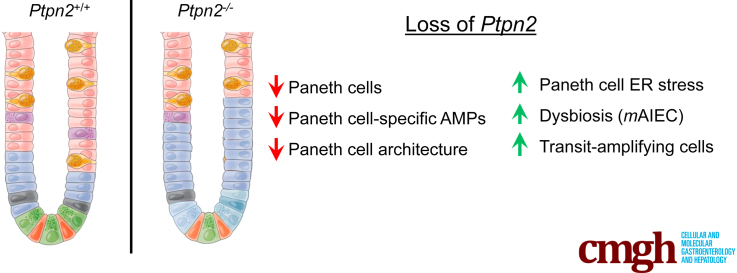
Partial transcriptome analysis showed that expression of Paneth cell–associated antimicrobial peptides Lyz1, Pla2g2a, and Defa6 was down-regulated markedly in Ptpn2-KO mice compared with wild-type and heterozygous. In parallel, Paneth cell numbers were reduced, their endoplasmic reticulum architecture was disrupted, and the endoplasmic reticulum stress protein, C/EBP-homologous protein (CHOP), was increased in Ptpn2-KO mice. Despite reduced Paneth cell number, flow cytometry showed increased expression of the Paneth cell–stimulatory cytokines interleukin 22 and interferon γ+ in CD4+ T cells isolated from Ptpn2-KO ileum. Key findings in constitutive Ptpn2-KO mice were confirmed in epithelium-specific Ptpn2ΔIEC mice, which also showed impaired lysozyme protein levels in Paneth cells compared with Ptpn2fl/fl control mice.
Conclusions
Constitutive Ptpn2 deficiency affects Paneth cell viability and compromises Paneth cell–specific antimicrobial peptide production. The observed effects may contribute to the increased susceptibility to intestinal infection and dysbiosis in these mice.
Keywords: Antimicrobial Peptides, TCPTP, Lysozyme, Intestinal Epithelial Cells, Inflammatory Bowel Disease, Microbiome
Graphical abstract
Summary.
PTPN2 has been associated with inflammatory bowel diseases. Constitutive deletion of this gene in mice depletes ileal Paneth cells, whereas specific deletion in intestinal epithelial cells impairs production of the antimicrobial peptide lysozyme.
The inflammatory bowel diseases (IBDs), Crohn’s disease (CD) and ulcerative colitis (UC), are characterized by recurrent episodes of chronic inflammation of the gastrointestinal tract. Both etiologies show increased levels of intestinal dysplasia, placing patients at an increased risk of developing intestinal cancer.1,2 Current therapies include administration of broad anti-inflammatory and immunosuppressive drugs, but the efficacy of these approaches decreases after long-term administration.3, 4, 5 In addition, IBD onset is multifactorial, which involves environmental factors, alterations in the microbiome, and genetic susceptibility. Individually and collectively, these factors can initiate or worsen underlying defects in the intestinal epithelial barrier function. Increased intestinal permeability is a hallmark of IBD pathogenesis that contributes to intestinal fluid loss, while also permitting translocation of microbes and microbial products from the luminal space into the intestinal mucosa, leading to immune cell activation and cytokine production.6,7
Genome-wide association studies have identified 240 gene loci associated with IBD to date.8 Many of these genes are associated with modulation of immune responses, maintenance of the epithelial barrier, autophagy, interactions between host cells and bacteria, and production of antimicrobial factors, thereby emphasizing the complexity and heterogeneity of IBD pathophysiology.9,10 Of interest, IBD-associated single-nucleotide polymorphisms were identified in the PTPN2 gene locus,11 which encodes T-cell protein tyrosine phosphatase (TCPTP), a protein responsible for inactivating several substrates, including proteins of the Janus activated kinase family (JAK) and signal transducer and activator of transcription (STAT) proteins.12,13 A number of studies have reported on functional roles of TCPTP in mouse models and the physiological consequences of Ptpn2 deletion.14 With respect to intestinal function, we and others have shown in vitro and in vivo that Ptpn2 loss increases intestinal barrier permeability,13,15 changes the composition of tight junction proteins,16 modulates IEC–macrophage communication,17 and alters the intestinal microbiota, which in turn allows the expansion of pathobionts associated with IBD.18 In contrast, naïve constitutive Ptpn2-heterozygous (HET) mice appear normal with no marked physiological or morphologic alterations, but display increased susceptibility to colitis and systemic inflammation when challenged with dextran sulfate sodium.19 These studies show the pivotal role Ptpn2 exerts on intestinal homeostasis and modulation of the microbiome, although the mechanisms by which Ptpn2 influences the intestinal flora are yet to be elucidated.
The intestinal barrier comprises 3 layers of protection: the mucus layer, intestinal epithelial cell (IEC) lining, and immune cells in the lamina propria. A healthy IEC layer functions as a selectively permeable barrier at the host–microbiome interface, and is essential for generating and maintaining ion-solute concentration gradients, absorption of nutrients, sampling bacterial antigens, restricting entry of pathogens and toxins, modulating the microbial intestinal flora by secreting antimicrobial components, and mediating communication with immune cells.20 These functions are accomplished by a set of specialized and highly differentiated IEC subtypes that are located strategically along the crypt–villus axis of the small intestinal mucosa. Among them, Paneth cells normally are present only in the small intestines, dwelling at the crypt base intermingled with intestinal stem cells, and are critically involved in innate immune responses. Paneth cells secrete several antimicrobial peptides (AMPs), such as α-defensins (cryptdins in mice), lysozyme, and phospholipase A2 group IIA. In addition, Paneth cells are an important source of stem cell niche factors, such as proto-oncogene wingless-type 3 (WNT3), Notch ligands, and epidermal growth factor (EGF), which are required for intestinal stem cell function.21 Moreover, dysfunction of Paneth cells may play a role in intestinal inflammation.22 For example, a subset of CD patients display reduced expression of AMPs and defective Paneth cells, suggesting that loss of critical Paneth cell functions alters the host–bacterial interaction in ways that increase the risk of chronic inflammation.23, 24, 25
IEC subtypes are functionally altered in IBD, although such impairments are dependent on IBD manifestation (CD vs UC), genetic susceptibility, and site of disease activity.23 In CD ileitis, Paneth cell number and function are affected whereas reduced numbers of goblet cells and a defective mucus layer have been reported in UC.24, 25, 26, 27 Furthermore, studies examining IBD patient biopsy specimens and transgenic mice show that genetic variants or deletion of IBD-associated genes can functionally impair IEC subtypes, thereby increasing susceptibility to intestinal infections and promoting dysbiosis.28, 29, 30, 31
Here, we show that constitutive Ptpn2 deficiency alters the expression of ileal IEC markers in mice. Notably, the number of Paneth cells was reduced dramatically in Ptpn2-knockout (KO) mice, negatively affecting the production of AMPs that directly modulate the intestinal microbiota. These findings indicate a new mechanism by which Ptpn2 loss of function might increase susceptibility to infection and contribute to intestinal dysbiosis and disease onset.
Results
Whole-Body Ptpn2-Deficient Mice Display Epithelial Structural Changes and Unique Gene Expression in Isolated Ileal IECs
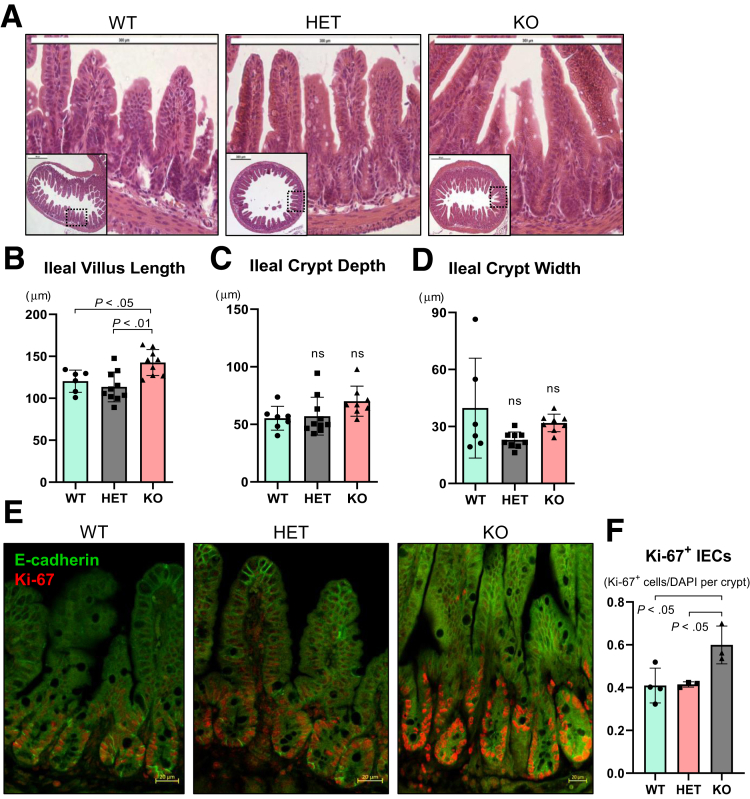
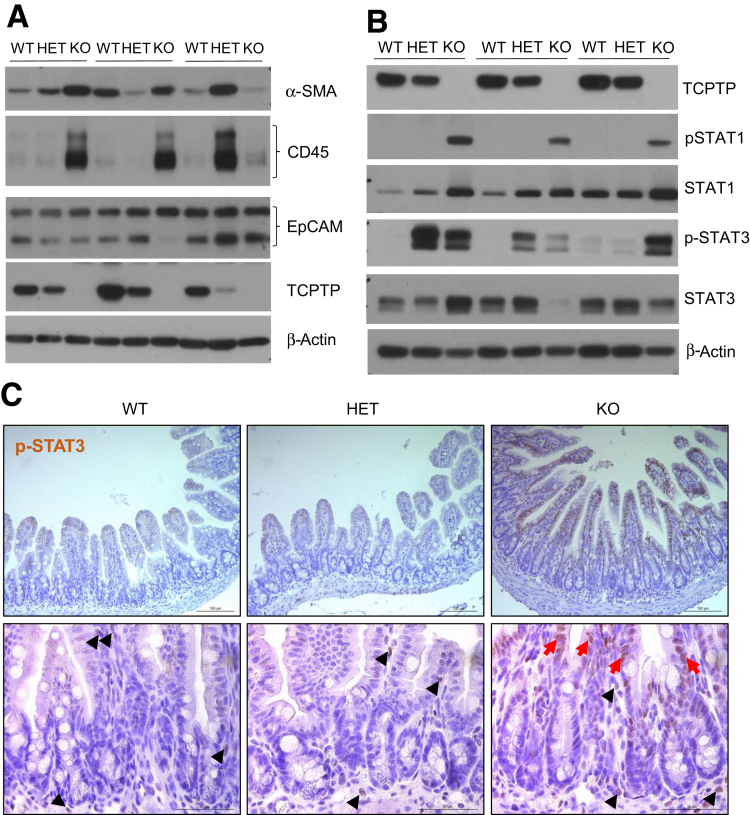
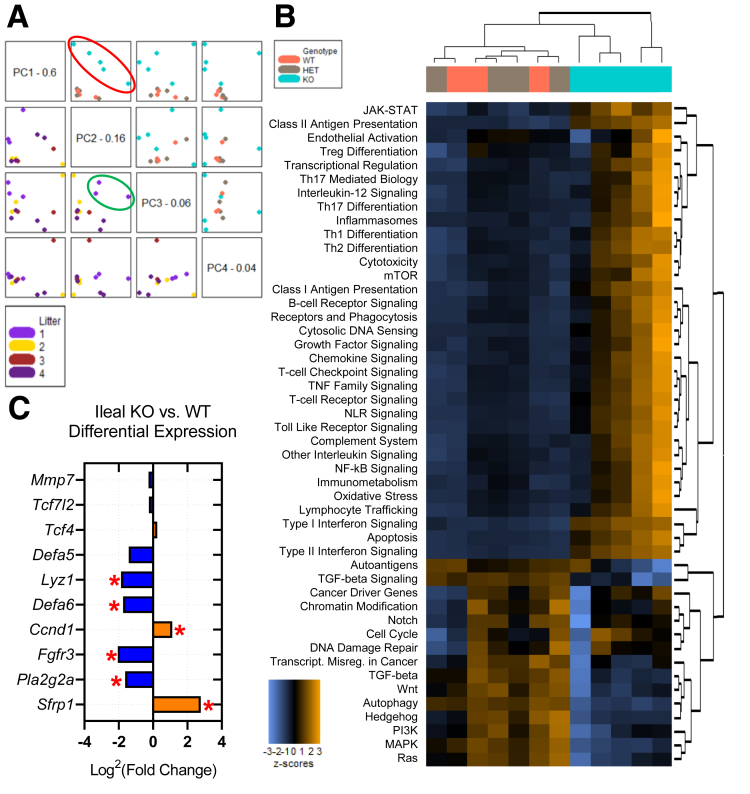
Our group recently showed that whole-body Ptpn2-KO mice display increased crypt depth in the cecum and proximal colon, although no other gross alteration was present in the intestinal epithelium.32 Here, we show that ileal villus length was increased in Ptpn2-KO mice when compared with wild-type (WT) and HET (Figure 1A and B). Ileal crypt depth and width were not affected (Figure 1C and D). Increased villus length in the Ptpn2-KO mice was accompanied by increased numbers of proliferating cells detected by Ki-67 immunofluorescence (Figure 1E and F). Next, to investigate whether constitutive Ptpn2 deletion also affects IEC subtypes and their function, we isolated ileal IECs from Ptpn2-WT, -HET, and -KO mice. Purity of IEC samples was assessed by abundance of epithelial cell adhesion molecule (EpCAM), mesenchymal α-smooth muscle actin (αSMA), and immune cell (CD45) protein markers, confirming that samples are IEC-enriched (Figure 2A). Loss of the Ptpn2 gene product, TCPTP, and increased phosphorylation of downstream substrates, STAT1 and STAT3, was confirmed by Western blot of enriched ileal IECs and immunohistochemistry (Figure 2B and C). Partial transcriptomic analysis was performed using Nanostring profiling technology (nCounter® SPRINT Profiler, NanoString Technologies, Seattle, WA). Results of 2 separate panels with a predefined set of targets, AutoImmune Profiling and PanCancer Pathways, were combined, with the addition of 60 customized targets comprising IEC markers, function and differentiation factors, host–bacteria interaction, autophagy, immune response, and iron transport, totaling more than 1500 targets (Supplementary Table 1). Principal component analysis showed unique gene expression patterns in Ptpn2-KO IECs that clustered separately from IECs of WT or HET mice (Figure 3A, red ellipse), indicating that genotype corresponds to 76% of the gene expression variance. Furthermore, the same analysis showed that at least one set of littermates clustered together when data points were compared by this parameter, indicating that co-housing contributes to gene expression variance (Figure 3A, green ellipse). When adjusted for the littermate effect, 97 genes were dysregulated markedly (false discovery rate, <0.1) of 1398 targets with detectable levels (Supplementary Table 1). A heatmap of pathway scores summarizes the overall effect of constitutive Ptpn2 deletion on cellular pathways in each sample (Figure 3B). Figure 3C lists all detected genes associated with Paneth cell function and differentiation factors. Notably, the Paneth cell–associated AMP genes Defa6, Lyz1, and Pla2g2a, were down-regulated (blue) in KO IECs relative to WT, suggesting that the function of Paneth cells could be impaired by constitutive Ptpn2 deficiency.
Figure 1.
Intestinal morphometry and proliferation analysis in whole-body constitutive Ptpn2-KO mice. (A) H&E staining of ileal epithelium of Ptpn2-WT, HET, and KO mice. (B) Morphometric analysis of ileal structures show that villi are longer in KO mice in comparison with WT and HET counterparts. Ileal (C) crypt depth and (D) crypt width were similar between groups. Each data point represents the average of a single mouse. Columns show means ± SD. One-way analysis of variance and Tukey post hoc test. WT = 6, HET = 10, KO = 8. (E) Immunofluorescence imaging of ileal sections staining for proliferation marker Ki-67 (red) and IEC marker E-cadherin (green). (F) Quantification of Ki-67+ cells indicates that the number of proliferating IECs is increased in KO mice. WT = 4, HET = 3, KO = 3. Columns show means ± SD. One-way analysis of variance and Tukey post hoc test.
Figure 2.
Assessment of STAT activation in isolated IEC samples from Ptpn2-deficient mice. (A) Western blot of ileal IECs probing for α-smooth muscle actin (α-SMA) to assess the presence of mesenchymal cells; CD45 to assess the presence of leukocytes; and epithelial cell adhesion molecule (EpCAM) to assess an abundance of epithelial cells. (B) Western blot of ileal IECs showing the absence of TCPTP and abundance of TCPTP substrates STAT1 and STAT3 in their total and phosphorylated isoforms. WT = 5, HET = 5, KO = 5. (C) Staining of ileal mucosa by immunohistochemistry for phosphorylated STAT3. Black arrowheads indicate positive staining for phosphorylated STAT3 (p-STAT3) in the mesenchyme, whereas red arrows indicate IECs positive for p-STAT3. WT = 3, HET = 3, KO = 3.
Figure 3.
Partial transcriptome analysis of isolated Ptpn2-KO mouse ileal IECs. Data were consolidated from 2 different Nanostring panels: AutoImmune Profiling and PanCancer Pathways with the addition of 60 custom targets. (A) Principal component (PC) analysis is a statistical method that transforms large data sets with multiple variables into a linear set of principal components retaining most of the information from the large set. Principal component 1 (PC1) captures the highest level of variance, with variance decreasing from PC2 to PC3 to PC4. The resulting image plots each PC vs another twice and colors the points by the selected covariate, helping to identify clusters in the data associated with a covariate. The boxes on the diagonal each contain a PC name and value representing the variance percentage; all plots in the same row will have this PC on their y-axis and all plots in the same column will have this PC on their x-axis. Upper quadrants: Color-coded with the genotype of each sample indicating that gene expression of Ptpn2-KO mice is unique. The red ellipse indicates how data points from KO mice cluster apart from WT and HET mice. Lower quadrants: Color-coded with littermates, indicating that littermates number 1 clustered together as indicated by the green ellipse. Data were normalized by littermates (co-housed mice) as a covariate. (B) Heatmap of pathway scores is a high-level overview of how the cellular pathway scores change across samples. Orange indicates high scores; blue indicates low scores. Scores are displayed in the same scale by Z-transformation. Each column represents an individual sample. (C) KO vs WT differential expression of ileal Paneth cell markers, differentiation factors, and function. WT = 3, HET = 4, KO = 5. ∗P value < .05 and false discovery rate (FDR) <0.1.
Whole-Body Ptpn2 Deletion Depletes Paneth Cells and AMP Production
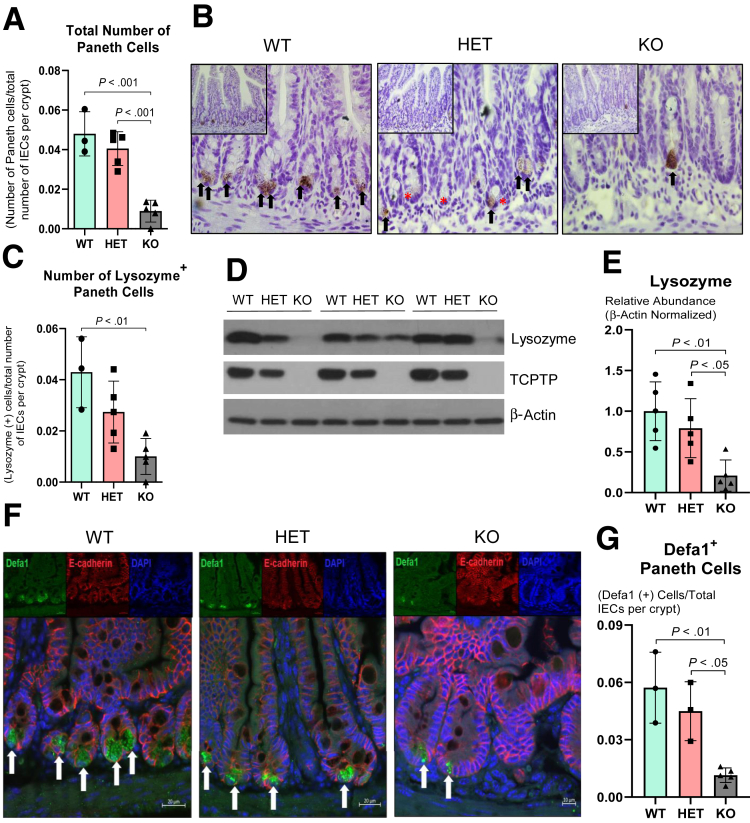
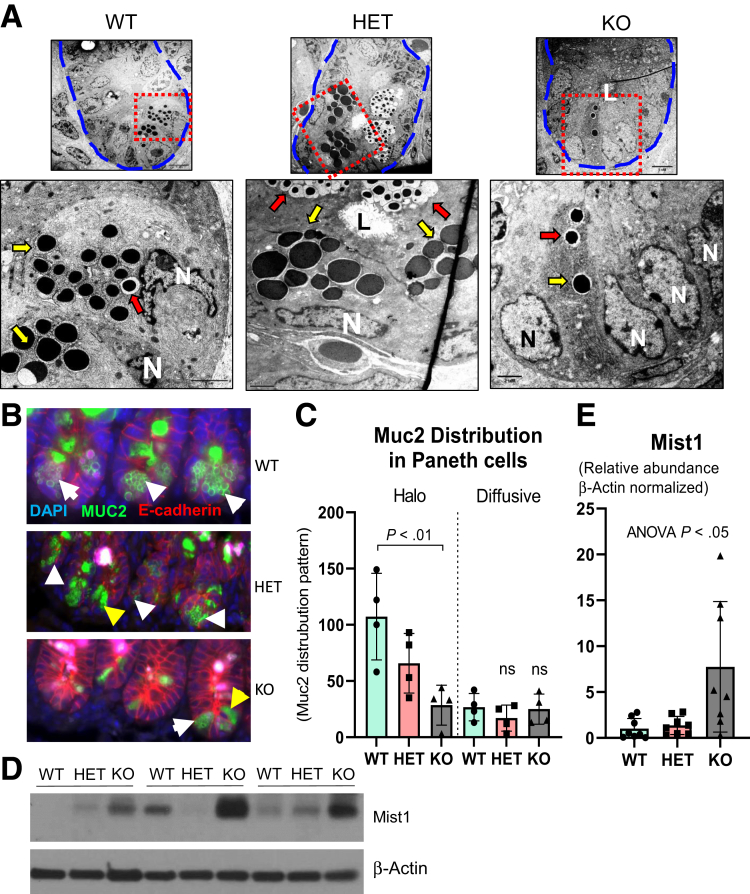
Given the established importance of Paneth cells in shaping host–microbial interactions in the gut, we investigated whether whole-body Ptpn2 deficiency compromises Paneth cell function in mice.25,33 Counting Paneth cells on H&E sections (Figure 1A), discriminated from other IECs by the presence of characteristic large cytosolic granules, showed that their number was decreased in Ptpn2-KO mice (Figure 4A). In parallel, immunohistochemistry staining showed a marked reduction of Paneth cell–associated lysozyme in the ileal mucosa of Ptpn2-KO mice (Figure 4B and C), a finding that was confirmed by Western blot analysis of isolated ileal IECs (Figure 4D and E). Moreover, immunofluorescence for another Paneth cell–specific AMP, defensin alpha 1 (Defa1; cryptdin-1 in mice), showed a dramatic reduction of Defa1-positive Paneth cells in Ptpn2-KO mice in comparison with WT and HET mice (Figure 4F and G). To further confirm that Ptpn2-KO mice lack Paneth cells, we assessed the presence/absence of dense core vesicles, a cytoplasmic structure present in Paneth cells responsible for packing and storing AMPs, by transmission electron microscopy (TEM). Figure 5A confirmed that Ptpn2-KO mice have fewer Paneth cells at the crypt base (delineated by blue dashed lines), with fewer dense core vesicles (yellow arrows). Satoh et al34 reported that large cytoplasmic vacuoles were formed when dense core vesicles released their contents into the luminal space after Paneth cell stimulation. However, we did not observe cytoplasmic vacuole formation in any of the constitutive Ptpn2 mouse genotypes (WT, HET, KO) (Figure 5A). We also observed some dense core vesicles with a surrounding low-density halo, indicated by red arrows, a feature that has been suggested to be associated with increased expression of the mucin-2 (Muc2) protein in Paneth cells (Figure 5A).35,36 Muc2 staining showed a halo shape distribution of this mucin protein in cells found at the crypt bottom of WT mice (Figure 5B). However, although the total number of Muc2+ cells was reduced in Ptpn2-KO mice, the number of cells with a Muc2+ diffuse distribution in the cytosol was unchanged between genotypes (Figure 5C). Next, Western blot analysis indicated that muscle, intestine and stomach expression 1 (Mist1), a protein shown to be important in the secretory apparatus and maturation of exocrine cells, including Paneth cells,37, 38, 39 was increased in KO mice (Figure 5D and E). Although it is unclear why Mist1 was induced in Ptpn2-KO mice, it could suggest an attempt to rescue the normal secretory function of Paneth cells in these mice. Supporting this idea, expression of transcription factor 4 (Tcf4) and Tcf7l2, regulators of α-defensin expression,40,41 and Mmp7, a Paneth cell–specific matrix metalloproteinase-7 (MMP7) responsible for cleavage and activation of pro–α-defensins in mice,42 remained unchanged in the Nanostring analysis (Figure 3C and Supplementary Table 1), indicating that some of the Paneth cell molecular markers were retained despite the loss of many phenotypic features of this IEC subtype. Collectively, these data suggest that constitutive Ptpn2 loss results in a dramatic reduction of mature Paneth cells and depletion of Paneth cell–specific AMPs in Ptpn2-KO mice.
Figure 4.
Ptpn2-KO mice display loss of Paneth cells and antimicrobial peptide expression. (A) Counting of Paneth cells by calculating the ratio of the total number of Paneth cells by the total number of IECs per crypt. Columns show means ± SD. One-way analysis of variance and Tukey post hoc test. (B) Ileal Paneth cells were stained for antimicrobial peptide lysozyme through immunohistochemistry. Black arrows indicate Paneth cells positively stained for lysozyme, whereas redasterisks indicates Paneth cells without lysozyme staining (characterized by large cytosolic granules at the crypt bottom). WT = 3, HET = 5, KO = 5. (C) Ratio of the number of Paneth cells positively stained for lysozyme by total number of IECs per crypt. (D) Western blot of ileal IECs probing for lysozyme. (E) Densitometry of lysozyme protein levels in ileal IECs. WT = 5, HET = 5, KO = 5. (F) Immunofluorescence of ileal sections staining for Defa1 (green), a Paneth cell–specific AMP; E-cadherin (red), a marker of IECs; and 4′,6-diamidino-2-phenylindole (DAPI) (blue) a marker of cell nuclei. White arrows indicate Paneth cells positive for Defa1 staining. (G) Counting and ratio of Defa1- (cryptdin-1) positive cells over the total number of IECs per crypt. WT = 3, HET = 3, KO = 5
Figure 5.
Distortion of Paneth cell secretory vesicle in Ptpn2-KO mice. (A) TEM images of the ileal crypt bottom (delineated by dashed blue lines). Higher-magnification images were captured from red dashed lines displaying Paneth cells and their dense core vesicles (indicated by yellow arrows) while many vesicles are surrounded by a translucid halo (red arrows). Note the fewer number of dense core vesicles in the KO mice. WT = 3, HET = 3, KO = 4. (B) Muc2 staining shows halo-shaped distribution of mucin-2 protein (white arrowheads) and diffused (yellow arrowheads) in Paneth cells. (C) Counting of Muc2+ Paneth cells according to cytosolic distribution pattern. Only well-oriented crypts were used for quantification by counting only the crypt bottom IECs (∼10 cells representing the stem cell zone) that were Muc2+; n = 4. (D) Western blot of ileal IECs probing for Mist1 protein. (E) Densitometry of Mist1 protein levels in ileal IECs, a protein shown to be important in the organization of dense core vesicles in secretory cells. WT = 8, HET = 8, KO = 8. Columns display means ± SD. Brown-Forsythe analysis of variance (ANOVA) test and Dunnett T3 post hoc test. DAPI, 4′,6-diamidino-2-phenylindole; L, luminal space; N, nucleus.
Ptpn2-KO Mice Show Contrasting Activation of Apoptosis Between Immune Cells and Epithelium
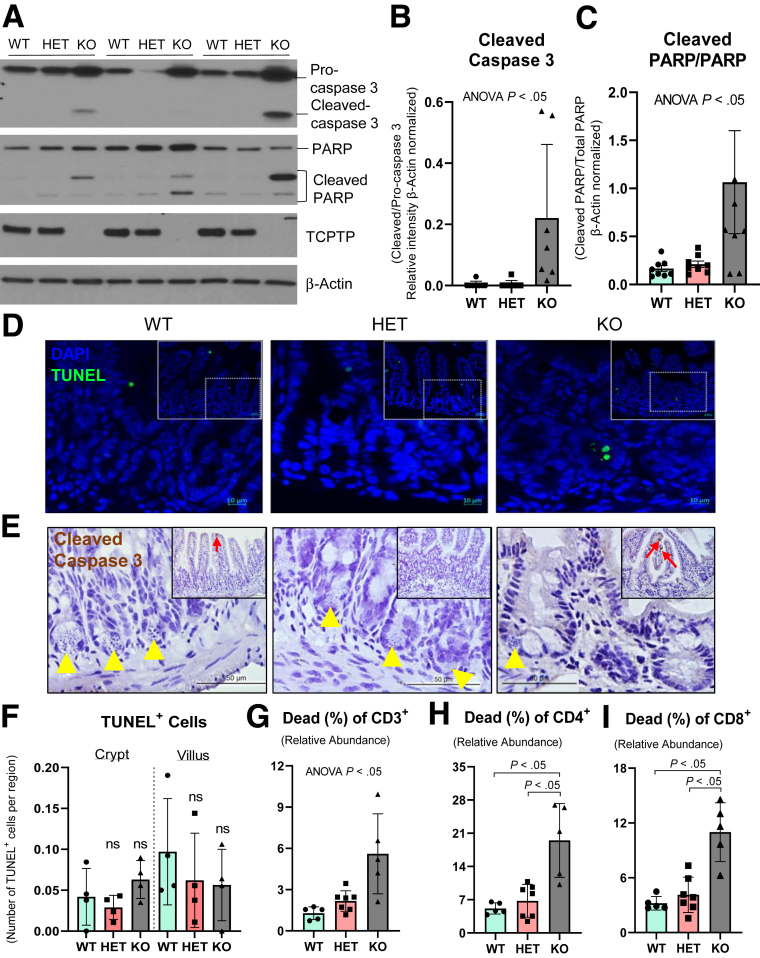
Given the dramatic defects in Paneth cell phenotype in mice that lack whole-body Ptpn2 activity, we sought to investigate whether the absence of Paneth cells in Ptpn2-KO mice was owing to apoptotic cell death. Western blot analysis of IECs showed that levels of both total and activated (cleaved) isoforms of the pro-apoptotic marker caspase-3 were increased in KO mice compared with WT and HET (Figure 6A and B). Moreover, cleaved poly-(adenosine diphosphate-ribose) polymerase (PARP), a critical protein during DNA repair, was increased in IECs from KO mice, further suggesting activation of apoptosis (Figure 6A and C). However, there was no difference in the number of terminal deoxynucleotidyl transferase–mediated deoxyuridine triphosphate nick-end labeling (TUNEL)+ cells, a measure of DNA fragmentation in the late stage of cell apoptosis, in IECs between mouse genotypes (Figure 6D and F). In addition, staining for cleaved caspase 3 showed no indication of increased presence of this apoptosis marker in cells at the ileal crypt base of Ptpn2-deficient mice (Figure 6E). Because the few TUNEL+ cells did not appear to be IECs, we characterized immune cell viability in ileal tissues of these mice. Flow cytometric analysis identified increased proportions of dead CD3+ immune cells and this was associated with an increased number of dead CD4+ and CD8+ T lymphocytes (Figure 6G and I). Collectively, these data identify a quantifiable increase in lymphocyte death in Ptpn2-KO mouse ileum with no indication of caspase-dependent apoptosis occurring in epithelial cells.
Figure 6.
Measurement of epithelial apoptosis markers and immune cell viability in ileum from Ptpn2-KO mice. (A) Western blot of ileal IECs from constitutive Ptpn2-KO mice probing for apoptosis markers caspase 3 and PARP. (B) Densitometry and ratio of caspase 3 isoforms. Columns show means ± SD of relative intensity. Brown-Forsythe analysis of variance (ANOVA) test and Dunnett T3 post hoc test. (C) Densitometry and ratio of PARP isoforms. Brown-Forsythe ANOVA test and Dunnett T3 post hoc test. (D) TUNEL staining of ileal sections showing few apoptotic cells (green) and cell nuclei (blue). N = 4 per group. (E) Immunohistochemistry for cleaved caspase 3 in ileal sections. Yellow arrowheads indicate Paneth cells, red arrows indicate cleaved caspase 3+ IECs; n = 4. (F) Quantification of TUNEL+ IECs in the crypt and villus compartment. Flow cytometry of ileal mucosa cells stained for immune cell types and apoptotic marker showing increased abundance of apoptotic (G) lymphocytes (CD3+), (H) T helper cells (CD4+), and (I) cytotoxic T cells (CD8+). WT = 8, HET = 8, KO = 8. Columns show means ± SD. Brown-Forsythe ANOVA test and Dunnett T3 post hoc test. DAPI, 4′,6-diamidino-2-phenylindole.
Constitutive Ptpn2 Deficiency Increases Abundance of Ileal Immune Cells and Production of Paneth Cell Stimulatory Cytokines
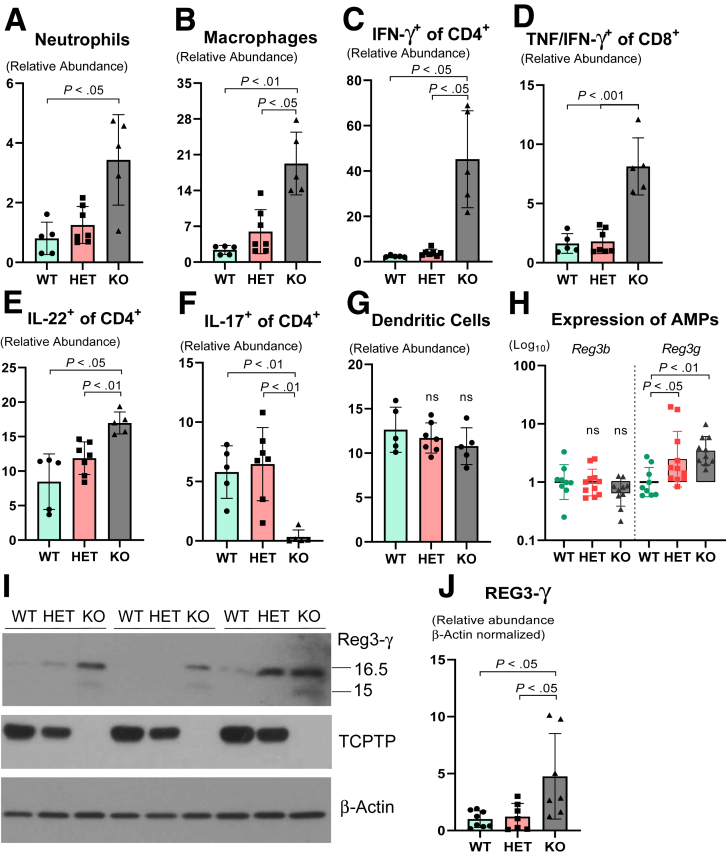
Because whole-body Ptpn2-KO mice failed to express several Paneth cell–associated AMPs, and were unable to form cytosolic granules, we investigated whether the ileal mucosa lacks stimulatory factors required for Paneth cell maturation, expression, and secretion of AMPs by evaluating immune cell infiltration and expression of immune cell–derived cytokines. Flow cytometry analysis showed increased abundance of neutrophils, macrophages, and tumor necrosis factor (TNF)-α/interferon (IFN)-γ expressing cytotoxic (CD8+) and T-helper (CD4+) cells in the ileal mucosa of Ptpn2-KO mice in comparison with WT and HET mice (Figure 7A–D). Moreover, the abundance of CD4+ T cells positive for IFN-γ and interleukin (IL)22 also was increased (Figure 7C–E). These data indicate that the ileal mucosa contains abundant immune cells that express stimulatory cytokines that can promote Paneth cell function and stimulate secretion of AMPs. Furthermore, gene expression of cytokine receptors, Il22ra1, Il10rb, and Ifngr1, in IECs was unchanged (Supplementary Table 1), suggesting that IECs were not functionally uncoupled from cytokine stimulation, at least at the gene expression level. Conversely, abundance of CD4+ T cells primed to express IL17 was reduced dramatically in Ptpn2-KO mice in comparison with WT and HET (Figure 7F), whereas no change was detected in the abundance of ileal dendritic cells, a specialized antigen-presenting cell that orchestrates innate and adaptive immune responses43 (Figure 7G). Because proinflammatory cytokines should exert a global response on the ileal mucosa rather than targeting an individual IEC subtype, we analyzed the expression of 2 AMPs that are not Paneth cell–specific, Reg3b and Reg3g, by reverse-transcription quantitative polymerase chain reaction (RT-qPCR). Expression of Reg3b was unchanged, whereas the expression of Reg3g was induced in both Ptpn2-HET and KO mice compared with WT (Figure 7H). Western blots of ileal IECs confirmed that the regenerating islet-derived protein 3-gamma (REG3-γ) protein level was increased in Ptpn2-KO mice compared with WT and HET mice (Figure 7I and J), suggesting that the deficit in the expression of Paneth cell–specific AMPs is a selective effect on these cells.
Figure 7.
Quantification of immune cell subtypes and non–Paneth cell–specific antimicrobial peptides in the ileal mucosa of Ptpn2-KO mice. Flow cytometric quantification of (A) relative abundance of neutrophils, (B) relative abundance of macrophages, (C) relative abundance of T-helper cells (CD4+) positive for IFN-γ, (D) relative abundance of cytotoxic cells (CD8+) positive for TNF and IFN-γ, (E) relative abundance of T-helper cells (CD4+) positive for IL22, (F) relative abundance of T helper cells (CD4+) positive for IL17, and (G) relative abundance of dendritic cells (CD11c+). WT = 5, HET = 7, KO = 5. All columns show average ± SD. Brown-Forsythe analysis of variance and Dunnett T3 post hoc test. (H) RT-qPCR gene expression of Paneth cell nonspecific antimicrobial peptides: Reg3b and Reg3g. WT = 9, HET = 11, KO = 10. Columns show geometric mean ± geometric SD in a log10 scale. One-way analysis of variance and Tukey post hoc test. (I) Western blot of ileal IECs probing for Reg3-γ. (J) Densitometry of Reg3-γ protein levels in ileal IECs. Only the upper band (16.5 kilodaltons) was measured, as has been shown that cleavage of Reg3-γ by trypsin occurs only after both proteins are released into the luminal space generating the shorter (active) isoforms of approximately 15 kilodaltons. WT = 8, HET = 7, KO = 7. Columns show means ± SD. One-way analysis of variance and Tukey post hoc test.
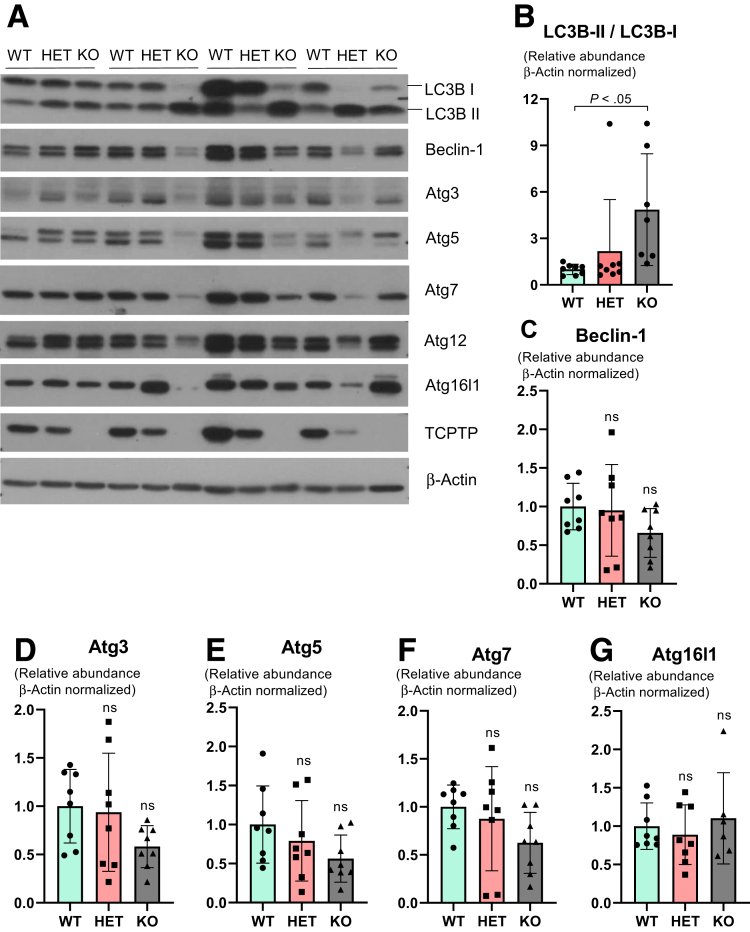
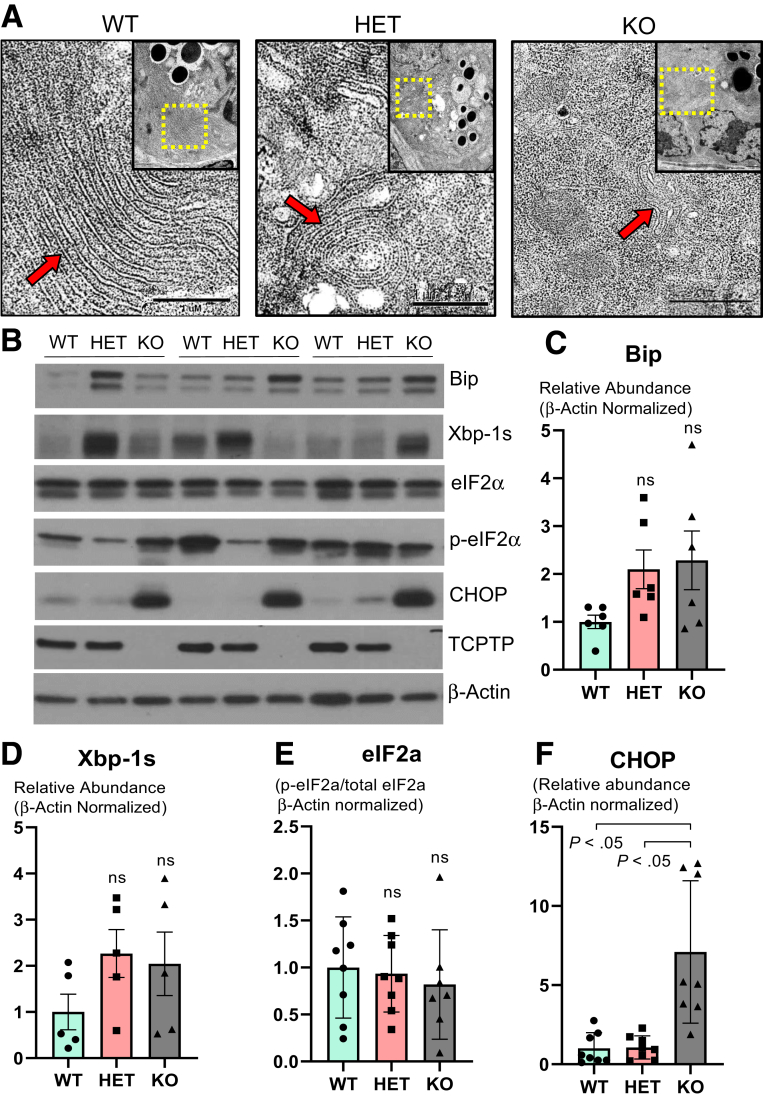
Constitutive Ptpn2 Deletion Disrupts Endoplasmic Reticulum Architecture and Increases Levels of Endoplasmic Reticulum Stress Without Affecting Abundance of Autophagy Proteins
Paneth cell secretion of lysozyme is orchestrated via the secretory autophagy pathway during bacterial infection of the intestine, and when autophagy was disrupted in Atg16l1-deficient mice, lysozyme secretion was hindered.44 Therefore, we tested whether the lysozyme deficit observed in constitutive Ptpn2-deficient mice was associated with impaired autophagy activation and/or autophagosome formation. Levels of the autophagy-related proteins Beclin-1, autophagy-related protein 3 (ATG3), ATG5, ATG7, and ATG12, and the product of another IBD-associated gene, ATG16L1, were unchanged in whole-body Ptpn2-KO mice (Figure 8). Surprisingly, we observed increased levels of autophagosome formation as evidenced by an increased ratio of microtubule-associated protein 1A/1B light chain 3B (LC3B) isoforms, the lipidated LC3B-II over LC3B-I, in Ptpn2-KO mice (Figure 8B). Although unexpected, our data suggest that the impairment in lysozyme production is independent of autophagy. To identify if the endoplasmic reticulum (ER)-driven protein secretory pathway was disrupted,45,46 we investigated whether Paneth cells in whole-body Ptpn2-deficient mice were undergoing ER stress. TEM images showed that the ER architecture in Paneth cells was disrupted dramatically in Ptpn2-KO mice, with ribosomes floating in the cytosol having dissociated from ER cisternae (Figure 9A), which is a typical feature of cells undergoing ER stress. Protein levels of the ER stress–associated pathways, binding-immunoglobulin protein (Bip), an endoplasmic reticulum-associated chaperone, and the unfolded protein response marker, X-box-binding protein 1 (Xbp-1s), were variable in Ptpn2-KO and HET mice, whereas no change in phosphorylated eukaryotic translation initiation factor 2A (eIF2-α) was observed (Figure 9B–E). Conversely, levels of C/EBP-homologous protein (CHOP), a downstream ER stress marker and inducer of apoptosis, were increased in IECs from constitutive Ptpn2-KO mice in comparison with WT and HET mice (Figure 9B and F). In summary, Ptpn2 loss in vivo provoked increased ER stress with compromised ER architecture in Paneth cells. Moreover, the increased levels of CHOP protein in IECs suggest suppression of the unfolded protein response by increasing protein synthesis, which could lead to cell death through oxidative stress and adenosine triphosphate depletion.47,48
Figure 8.
Evaluation of autophagy markers in the constitutive Ptpn2-deficient mice. (A) Western blot of ileal IECs probing for autophagy markers LC3B-I and LC3B-II, Beclin-1, autophagy-related proteins ATG3, ATG5 (antibody detects total levels of free ATG5 and conjugated with ATG12), ATG7, ATG12, and ATG16L1. Densitometry of (B) LC3B-I and LC3B-II depicted as a ratio between the 2 isoforms; (C) Beclin-1; (D) ATG3; (E) total ATG5; (F) ATG7; and (G) ATG16L1 protein levels. WT = 8, HET = 8, KO = 8. Columns show means ± SD. One-way analysis of variance and Tukey post hoc test.
Figure 9.
Whole-body PTPN2 loss in mice increases markers of endoplasmic reticulum stress. (A) TEM images of Paneth cell endoplasmic reticulum architecture. The upper right quadrant outlined in yellow shows the Paneth cell cytosol. Red arrows indicate localization of the ER cisternae. WT = 3, HET = 3, KO = 4. (B) Western blot of ileal IECs probing for early ER stress marker Bip, unfolded protein response marker Xbp-1s, eukaryotic initiation factor (eIF) 2A isoforms eIF2-α and p-eIF2α, and downstream ER stress marker C/EBP-homologous protein (CHOP) in constitutive Ptpn2-deficient mice. Densitometry of (C) Bip, (D) Xbp-1s, (E) eIF2-α ratio, and (F) CHOP protein levels. WT = 8, HET = 8, KO = 8. Columns show means ± SD. One-way analysis of variance and Tukey post hoc test.
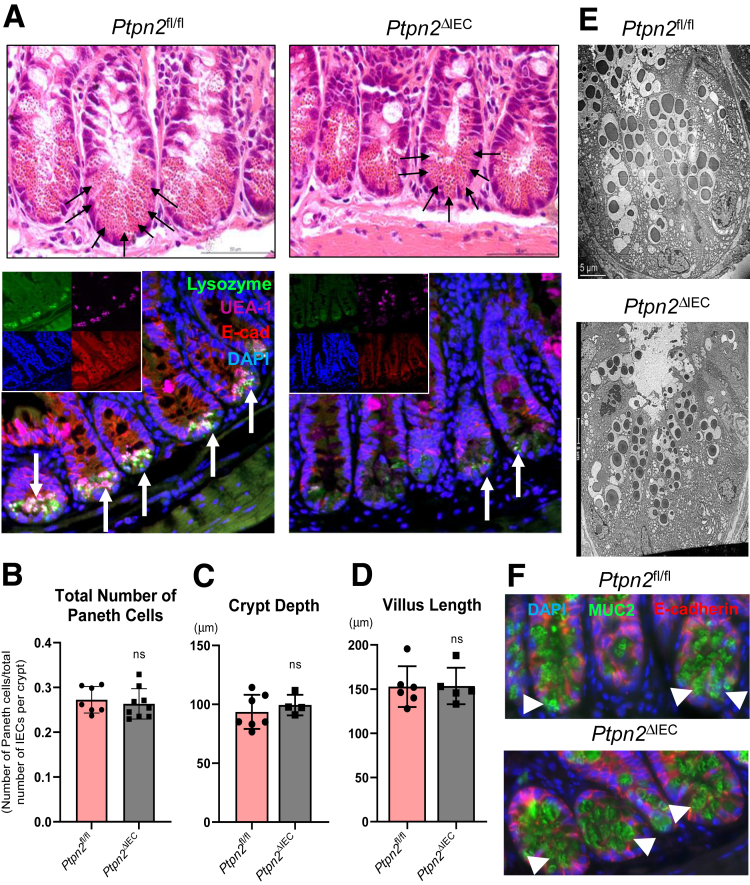
IEC-Specific PTPN2 Deletion Impairs Lysozyme Protein Levels Without Affecting Abundance of Paneth Cells
To determine whether the impact of whole-body Ptpn2 loss on Paneth cells is a direct consequence of Ptpn2 loss in epithelial cells, or is mediated indirectly through effects of Ptpn2 loss in other cells, we investigated Paneth cell phenotypes in a tamoxifen-inducible Villin-Cre transgenic Ptpn2 deletion mouse line (Ptpn2ΔIEC). This mouse model does not display the pronounced systemic inflammation shown by whole-body Ptpn2-KO mice.32 Intestinal tissues from Ptpn2fl/fl and Ptpn2ΔIEC were harvested >30 days after recombinase induction by tamoxifen administration to minimize any potential residual effects from estrogen-receptor activation.32 H&E staining of ileal sections showed that Paneth cells were present at the crypt base, displaying abundant cytosolic granules in both Ptpn2ΔIEC mice and Ptpn2fl/fl controls (Figure 10A). The overall number of Paneth cells was not reduced by IEC-specific Ptpn2 deletion (Figure 10B). Morphometric analysis of intestinal parameters, such as crypt depth and villus length, were similar between both groups (Figure 10C and D). However, when ileal sections were stained for lysozyme and Ulex europaeus agglutinin-1, a marker of lectins and normally used to stain Paneth cell granules, both markers were reduced dramatically at the crypt base in Ptpn2ΔIEC mice compared with controls (Figure 10A). Of note, TEM images of the crypt bottom showed normal Paneth cell architecture and morphology of the intracellular dense core vesicles in both groups (Figure 10E), while Muc2 staining showed no alteration of this mucin protein between groups (Figure 10F).
Figure 10.
Intestinal epithelial-specific Ptpn2 deletion in mice reduces antimicrobial lysozyme expression without affecting Paneth cell number or architecture. (A) H&E staining of ileal sections showing abundant Paneth cells and cytosolic granules at the base of the crypt of Ptpn2fl/fl and Ptpn2ΔIEC mice (black arrows). Staining of ileal sections from Ptpn2fl/fl and Ptpn2ΔIEC mice with Paneth cell marker lysozyme (green); Ulex europaeus agglutinin-1 (UEA-1), a marker of Paneth cell dense core vesicles (magenta); cell nuclei (blue); and marker of intestinal epithelial cell E-cadherin (red). Ptpn2fl/fl = 7, Ptpn2ΔIEC = 9. (B) Counting of total number of Paneth cells present in the ileum of Ptpn2fl/fl and Ptpn2ΔIEC mice displayed as a ratio of Paneth cells over the total number of IECs per crypt. Morphometric analysis of intestinal structures in the ileum of inducible tissue-specific Ptpn2-deletion showing (C) crypt depth, and (D) villus length. Each data point indicates the average value per mouse. Ptpn2fl/fl = 7, Ptpn2ΔIEC = 5. Columns show means ± SD. Two-tailed unpaired t test. (E) TEM images of the ileum crypt base showing an abundant number of Paneth cells and their respective cytosolic granules; n = 4. (F) Muc2 immunofluorescence of ileal crypt base showing abundant mucin-2 protein localization in the cytosol of Paneth cells (white arrowheads) in both genotypes; n = 4. DAPI, 4′,6-diamidino-2-phenylindole; E-cad, E-cadherin.
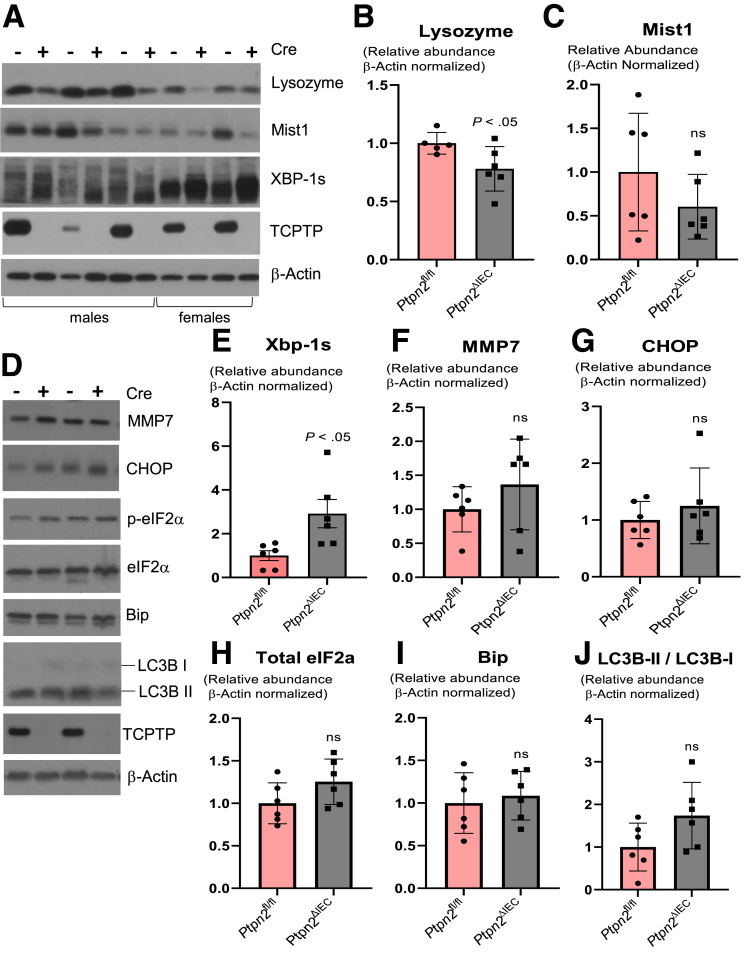
Next, by Western blot of IECs we detected that levels of lysozyme protein in Ptpn2ΔIEC mice were reduced (Figure 11A and B), whereas Mist1 protein levels were unchanged between groups (Figure 11A and C). We observed a substantial difference in the abundance of Xbp-1s protein between males and females (higher) in Ptpn2ΔIEC and Ptpn2fl/fl mice (Figure 11A). When normalized by sex, levels of Xbp-1s were significantly higher in Ptpn2ΔIEC compared with Ptpn2fl/fl mice (Figure 11E), indicating activation of the unfolded protein response in IECs of Ptpn2ΔIEC mice. Conversely, protein levels of the Paneth cell marker matrix metalloproteinase-7, and the abundance of ER stress proteins Bip, C/EBP-homologous protein CHOP, eukaryotic translation initiation factor 2A and p-eIF2α, and the autophagy activation marker LC3B-II/LC3B-I ratio, were unchanged between groups (Figure 11D–J). Together, these data show that epithelial Ptpn2 is critical for Paneth cell function with respect to lysozyme protein levels, however, loss of epithelial Ptpn2 is not sufficient to cause Paneth cell depletion.
Figure 11.
Analysis of ER stress and autophagy markers in intestinal epithelial-specific Ptpn2 mouse IECs. (A) Western blot of IECs from Ptpn2fl/fl and Ptpn2ΔIEC mice probed for lysozyme-1, Mist1, and unfolded protein response marker Xbp-1s. (B) Densitometry of lysozyme protein levels in IECs comparing Ptpn2fl/fl and Ptpn2ΔIEC. (C) Densitometry of Mist1 protein levels in IECs comparing Ptpn2fl/fl and Ptpn2ΔIEC. (D) Western blot and densitometry of IECs from Ptpn2fl/fl and Ptpn2ΔIEC mice probing for (E) Xbp-1s; (F) Mmp7, a Paneth cell marker; (G) downstream ER stress inducer C/EBP-homologous protein; (H) relative ratio of eukaryotic translation initiation factor 2A isoforms p-eIF2α and total eIF2α; (I) ER stress marker Bip protein; and (J) relative ratio of autophagy activation marker LC3B-II/LC3B-I. Columns show means ± SD. Two-tailed unpaired t test.
Constitutive Ptpn2 Deletion Does Not Affect Expression of Small Intestinal Epithelial Stem Cell Markers
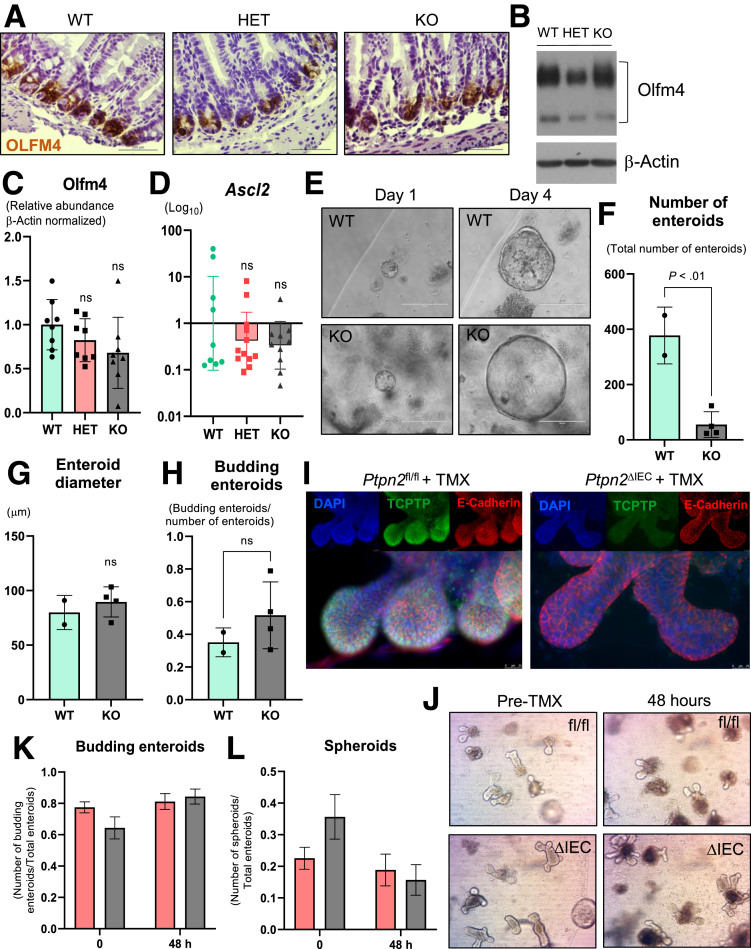
Paneth cells play a critical role in supporting the small intestinal stem cell niche.49 Hence, we wanted to determine whether the abundance of intestinal stem cells was also compromised in whole-body Ptpn2-KO mice. Immunohistochemistry staining for the stem cell marker Olfactomedin-4 (Olfm4) showed abundant numbers of stem cells confined at the crypt base in all groups (Figure 12A), while Western blot analysis of Olfm4 in isolated IECs did not reveal alterations in the protein levels of this intestinal stem cell marker (Figure 12B and C). Expression of Ascl2, a gene that controls stem cell renewal in the crypts, was unchanged (Figure 12D). In addition, gene expression of Lgr5, a marker of intestinal stem cells, Ephb3, a marker of the intestinal stem cell compartment, and Bmi1, a marker of quiescent stem cells, was unaltered (Supplementary Table 1). This indicates that the expression of the stem cell compartment markers was not affected, even though the increased number of proliferative cells suggest higher activity of stem cells, while markers of quiescent intestinal stem cells also remained unchanged. We next investigated effects of Ptpn2 loss on IEC differentiation markers. Expression of fibroblast growth factor receptor 3, Fgfr3, which is involved in the commitment of secretory vs absorptive lineages and is critical for Paneth cell development, was down-regulated in whole-body Ptpn2-KO mice (Figure 3C).50,51 Conversely, Sfrp1, which encodes secreted frizzled related protein 1, and Ccnd1, which encodes cyclin-D1, genes with direct roles in cell cycle and differentiation of the intestinal epithelium, were up-regulated markedly in whole-body Ptpn2-KO mice (Figure 3C). To test whether depletion of Paneth cells negatively impacts intestinal stem cell function, we cultured enteroids derived from the small intestine of Ptpn2-WT and KO mice. Although the number of viable enteroids from Ptpn2-KO mice was reduced dramatically, we did not observe differences in the diameter or number of budding enteroids, suggesting that Paneth cell depletion associated with whole-body Ptpn2 loss negatively impacts the yield of viable enteroids, but not the subsequent development of successfully formed enteroids (Figure 12E–H). Of note, ex vivo deletion of Ptpn2 by tamoxifen treatment of enteroids from inducible tissue-specific Ptpn2ΔIEC mice showed no reduction in enteroid viability or budding (Figure 12I and J). This suggests that epithelial Ptpn2 loss does not compromise IEC stem cell function. Altogether, our data suggest an imbalance in the expression of differentiation factors associated with the Paneth cell lineage, whereas expression of intestinal stem cell markers does not seem to be affected.
Figure 12.
Effect of Ptpn2loss on mouse intestinal stem cell markers and enteroid growth. (A) Staining of intestinal stem cell marker Olfm4 in ileal tissue of whole-body Ptpn2-deficient mice (n = 3). (B) Western blot of ileal IECs probing for Olfm4. Per the antibody manufacturer, Olfm4 in the small intestine typically displays 2 bands that belong to Olfm4 protein, with molecular weight ranging from 85 to 90 and 70 kilodaltons. (C) Densitometry of both bands from Olfm4 protein level in ileal IECs. Columns show means ± SD. WT = 8, HET = 8, KO = 8. One-way analysis of variance and Tukey post hoc test. (D) RT-qPCR analysis of the Ascl2 gene, which is responsible for intestinal stem cell renewal in the crypts. WT = 9, HET = 11, KO = 10. Columns show geometric means ± geometric SD. One-way analysis of variance and Tukey post hoc test. (E) Enteroid cultures derived from small intestinal crypts of Ptpn2-WT and Ptpn2-KO mice showing enteroid size at days 1 and 4 after tissue harvest: WT = 2; KO = 4. (F) Counting of total number of viable enteroids on day 4. (G) Enteroid diameter on day 4. (H) Total number of budding enteroids on day 4 normalized by the number of total enteroids. Columns show means ± SD. Two-tailed unpaired t test. (I) Ptpn2 deletion was confirmed by TCPTP staining (green), 4′,6-diamidino-2-phenylindole (DAPI) (blue), and E-cadherin (red). (J) Bright-field microscopy of Ptpn2fl/fl and Ptpn2ΔIEC-derived enteroids induced with tamoxifen for 48 hours. (K) Number of budding enteroids before and 48 hours after tamoxifen induction. (L) Number of spheroids before and 48 hours after tamoxifen induction. TMX, tamoxifen.
Discussion
PTPN2 modulates the intestinal microbiome and restricts the expansion of a mouse-specific adherent-invasive E coli (AIEC), which has >90% genetic similarity to the human IBD-associated AIEC, LF82.18,52 PTPN2 also is critically involved in the maintenance of intestinal barrier function through epithelial–macrophage crosstalk.15,17 Here, we identified a possible mechanism to explain how loss of PTPN2 contributes to bacterial dysbiosis. We report that ileal IECs showed dramatic down-regulation of critical genes associated with Paneth cell function, whereas analysis of morphologic features and molecular markers confirmed that these cells were nearly ablated in the ileum of constitutive Ptpn2-KO mice. Given the essential role of Paneth cells in the first line of enteric defense by modulating the gut microbiome through a large spectrum of AMPs secreted into the luminal space, the loss of AMP production by Paneth cells is likely a critical factor in the observed changes in microbiota composition.
Paneth cells respond to intestinal microbes by discharging AMP-filled granules into the luminal space.53 Human Paneth cells abundantly express 2 α-defensin genes, DEFA5 and DEFA6, as well as lysozyme, secretory phospholipase A2, and regenerating islet-derived protein 3-α (REG3A).54 DEFA5 has antibiotic activity against Gram+ and Gram- bacteria, including Staphylococcus aureus and Salmonella typhimurium, as shown in transgenic mice expressing human DEFA5.55,56 DEFA6 lacks direct antimicrobial activity, forming nanonets that trap bacteria instead.57 Murine Paneth cells, on the other hand, express at least 19 different α-defensin isoforms.58,59 The expression of these α-defensins in mice does not seem to be dependent on bacterial stimuli, but rather is regulated by transcription factors linked to IEC differentiation, including Tcf4 and Tcf7l2, which are under control of the Wnt/β-catenin pathway.41 In our study, we observed that these critical transcription factors in the development of Paneth cells were not affected in constitutive Ptpn2-KO mice, suggesting that the deficit in the expression of AMPs could be connected inherently to loss of Paneth cells in the ileal mucosa. Moreover, Paneth cells are the primary source of intestinal C-type lysozyme, encoded by the Lyz1 gene, which hydrolyzes the bacterial wall component peptidoglycan, common to both Gram+ and Gram- bacteria, resulting in the release of muramyl dipeptide (MDP), an important agonist of nucleotide-binding oligomerization domain-containing protein 1 (Nod1) and Nod2.60 Unlike α-defensins, lysozyme levels are regulated by commensal bacteria stimulation of Nod2, as shown by germ-free and Nod2-KO mice, in which lysozyme is rerouted from secretion to protein degradation.30 Although Lyz1-/- mice displayed expansion of the mucolytic bacteria Ruminococcus gnavus, a CD-associated pathobiont, they had diminished mucosal sensing and responses to MDP, accompanied by a reduced basal inflammatory response, suggesting the importance of lysozyme activity in tuning of the immune system.61 Consequently, the impact caused by a lack of mature Paneth cells could go beyond the impairment in the production of AMP, also affecting lysozyme-mediated priming of the enteric immune response to bacterial products.
Although Paneth cell–specific AMPs (Lyz, Defa6, and Pla2g2a) were reduced, expression of AMP produced by other IECs were either induced (Reg3g) or unchanged (Reg3b), suggesting that the deficit in the expression of AMPs is Paneth cell–specific. With respect to the different effects on Reg3g and Reg3b expression, this could be owing to a lack of IL17 stimulation, which is required for Reg3b induction. Paneth cell stimulation by IL22 is essential for its maturation and regulation of microbiota-dependent Il17 immune responses. IL17 and IL22 tend to act synergistically in Paneth cells, albeit by distinct cellular mechanisms.62,63 Despite abundant IL22-expressing CD4+ cells in the ileal mucosa, this was not sufficient to promote Paneth cell maturation or expression of Paneth cell–specific AMPs.
Studies with IBD patient and transgenic mice have shown that genetic variants, or deletion of IBD-associated genes, such as ATG16L1, NOD2, and LRRK2, are detrimental to Paneth cell morphology and function, leading to dysbiosis.28, 29, 30, 31 In addition, deletion of the transcription factor X-box binding protein-1 (Xbp1) in IECs, a marker of unfolded protein response, leads to Paneth cell impairment and spontaneous enteritis.64 However, mice with constitutive homozygotic Ptpn2 deficiency develop severe systemic inflammation, dying between 3 and 5 weeks of age.65 Our group recently showed that constitutive Ptpn2-KO mice have increased proinflammatory cytokines (IFN-γ, TNF-α, IL6) in the serum and in large intestinal mucosa.32 Here, we showed that immune cells primed to secrete stimulatory cytokines (IFN-γ, TNF-α, and IL22) were abundant in the ileal mucosa of Ptpn2-KO mice, indicating that Paneth cells seem to be extremely sensitive to inflammatory conditions. For example, IFN-γ induces potent extrusion of secretory granules and Paneth cell nucleus expulsion, leading to apoptosis, if stimulation by IFN-γ is sustained.66,67 We also identified that Ptpn2 deletion in IECs alone is sufficient to impair lysozyme production without affecting overall Paneth cell number, in addition to promoting activation of the unfolded protein response. Thus, the ablation of Paneth cells in constitutive Ptpn2-KO mice may reflect the increased inflammatory status, and likely a contribution of nonepithelial cells (ie, immune cells) in these mice that is not seen in Ptpn2ΔIEC mice.
In our study, gene expression of Tcf4 and Tcf7l2, transcription factors that control expression of α-defensins, and Mmp7, another Paneth cell–associated gene critical in cleaving and activating AMPs, was unaltered in ileal IECs. This could indicate an impairment in Paneth cell maturation. Moreover, levels of Mist1 protein were increased in Ptpn2-KO mice compared with WT and HET counterparts. Mist1 belongs to a basic helix-loop-helix family of transcription factors that bind DNA in several developmentally regulated genes,68 playing a critical role in the regulation of differentiation and maturation of the secretory machinery of exocrine cells.69 Studies using Mist1-KO mice showed that Paneth cells develop an immature phenotype with abnormal dense core vesicles and secretory apparatus without affecting the expression of lysozyme.37 Because Mist1 seems to be critical in maturation of Paneth cells and in the organization of their secretory granules, this might suggest that the increased levels of Mist1 could be an attempt to rescue the normal secretory function of Paneth cells in whole-body Ptpn2-deficient mice.
Loss-of-function PTPN2 variants impair autophagosome formation in human colonic carcinoma T84 epithelial cells and the human monocytic cell line THP-1 in response to the bacterial wall component MDP, or the inflammatory cytokines IFN-γ and TNF in vitro.70,71 However, we did not observe changes in the levels of autophagy proteins, or in autophagosome formation in isolated IECs from Ptpn2-KO mice, indicating that the lysozyme deficit in Paneth cells is not caused by autophagy impairment. We have shown that AIECs replicate and survive in macrophages lacking Ptpn2 activity, however, autophagosome formation was rescued after stimulation with rapamycin, showing that the defective autophagy present in Ptpn2-deficient macrophages in vivo can be bypassed by stimulatory factors.72 One possible explanation for the different effects on autophagy in whole-body vs macrophage-specific loss of Ptpn2 is that the increased ER stress could be the stimulus to activate autophagy in whole-body Ptpn2-KO mice because there is extensive crosstalk between these 2 cellular processes.73 Constitutive Ptpn2-deletion resulted in disruption of the ER architecture in Paneth cells, indicating ER stress that could compromise correct protein processing and folding. Of note, we did not observe increased phosphorylation of eIF2-α, which normally is increased during ER stress. IFN-γ induces potent phosphorylation of eIF2-α at serine 51 in exocrine cells.74 However, in agreement with our findings, PTPN2 knockdown in HT-29 IECs showed no change in eIF2-α phosphorylation after tunicamycin-induced ER stress.75 Moreover, we observed a dramatic increase in the levels of CHOP, which plays an essential role in inducing cell-cycle arrest and apoptosis during ER stress.48,76 CHOP promotes cell death by growth arrrest and DNA damage-induceble protein 34 (GADD34)-mediated dephosphorylation of eIF2-α,48 thus stimulating protein synthesis under ER stress, increasing oxidative stress, and adenosine triphosphate depletion, which collectively culminates in apoptosis.47 In addition, Bettaieb et al77 reported that Ptpn2 knockdown in MIN6 cells mitigated ER stress–induced phosphorylation of eIF2-α. Consequently, it is possible that Ptpn2 deficiency could promote cell death by sustaining ER stress instead of inhibiting it. This provides an interesting mechanistic hypothesis that will require further investigation to address whether Ptpn2-deficient Paneth cells undergo ER stress–mediated cell death.
TUNEL staining did not detect substantial differences in the number of apoptotic cells between genotypes. Although unexpected, our data align with previous findings showing that Ptpn2-KO mice displayed no increase in intestinal permeability to rhodamine B isothiocyanate-70 kilodaltons of dextran (RD70), a molecular probe for the unrestricted permeability route associated with epithelial loss or damage.17,32 Despite the lack of functional evidence of epithelial loss as reported previously, or immunostaining for TUNEL (Figure 6D and F) or cleaved caspase 3 (Figure 6E), we did observe an apparent increase in cleavage of caspase-3 and PARP by Western blot (Figure 6A–C). Given the more robust confirmation of immune cell death as detected by flow cytometry (Figure 6G–I), we suspect that the increased caspase 3 and PARP cleavage in Western blot samples is owing to immune cell contamination of IEC isolates, which is indicated by the presence of the CD45 immune cell marker (Figure 6A). Collectively, our data indicate a more rapid turnover of ileal IECs without causing nonspecific defects in epithelial barrier permeability.
IEC lineage tracing studies, and mouse models of dextran sulfate sodium colitis, have identified that Paneth cells possess some level of cell plasticity, and can reacquire intestinal stem cell characteristics to repopulate the intestinal stem cell compartment depleted of leucine-rich repeat-containing G-protein coupled receptor 5 (Lgr5)+ stem cells.78,79 Thus, we hypothesized that depletion of Paneth cells could be owing to their ability to dedifferentiate and repopulate the stem cell zone, shifting to a stem-like cell state. Consistent with our Nanostring data set showing that gene expression of intestinal stem cell markers, Lgr5 and Olfm4, were not altered significantly, protein levels of Olfm4 were unchanged while its localization was restricted to the bottom of the crypt. In addition, gene expression of the stem cell compartment marker (Ephb3) and of intestinal stem cell renewal activity (Ascl2), were not altered significantly. Moreover, gene expression of a quiescent intestinal stem cell marker, Bmi1, was unchanged, suggesting that quiescent cells remained dormant.80 Therefore, it seems unlikely that depletion of Paneth cells in Ptpn2-deficient mice is the result of a phenotypic shift to rescue/repopulate the stem cell zone.
In conclusion, the present study shows that Ptpn2 is a regulator of ileal intestinal epithelial cell homeostasis and a critical mediator of Paneth cell viability and antimicrobial protein expression in mice. Collectively, our data suggest that the ileal mucosa of Ptpn2-deficient mice undergoes rapid turnover, likely affecting the maturation of Paneth cells. Conversely, the remnant Paneth cells undergo ER stress that potentially could lead to cell death. Thus, our study identifies that loss of Ptpn2 activity causes Paneth cell dysfunction, which may explain the intestinal dysbiosis and pathobiont expansion observed in Ptpn2-deficient mice and IBD patients carrying PTPN2 loss-of-function variants.
Materials and Methods
Animals
All animal experiments were performed according to, and approved by, the Institutional Animal Care and Use Committee at the University of California, Riverside, under protocol A20190032E. Mice colonies were held in a specific–pathogen free facility with free access to food and water.
BALB/c mice with constitutive Ptpn2 deficiency were provided by Michel Tremblay (McGill University). Tissues were harvested when mice reached 21 days old because whole-body homozygotic Ptpn2-deficient mice die between 4 and 5 weeks of age as a result of systemic inflammation.65
Inducible tissue-specific Ptpn2-KO mice were generated as previously described.32,81 Briefly, a mouse line in which critical Ptpn2 exons were flanked by LoxP sites (Ptpn2tm1a[EUCOMM]Wtsi), with a transgenic mouse line expressing the Cre recombinase enzyme under the villin-1 promoter (Tg[Vil-cre/ERT2]23Syr). Recombinase activity was induced by tamoxifen administration intraperitoneally (50 mg/kg body weight) for 5 consecutive days.
Imaging
Distal ileal segments were excised and fixed in 4% paraformaldehyde overnight at 4ºC. Tissues then were rinsed with phosphate-buffered saline (PBS) and dehydrated with increasing concentrations of ethanol washes using Shandon Excelsior ES Tissue Processor (Thermo Fisher Scientific, Kalamazoo, MI). Paraffin embedding was performed using Histoplast LP (Richard-Allan Scientific, Kalamazoo, MI) in a Tissue-Tek station set (Miles Scientific, Naperville, IL). Paraffin blocks were sectioned in a rotary microtome (RM2235; Leica, Nussloch, Germany) 5-μm wide and placed on a charged slide (Fisher Scientific, Pittsburg, PA). Slides were deparaffinized, rehydrated, and stained with H&E. Similarly, goblet cells were visualized using the periodic acid–Schiff kit (procedure no. 395; Sigma) per the manufacturer’s guidelines. Slides were visualized and images were acquired with a Leica microscope (model DM5500B) coupled with a DFC450C camera (Leica).
Heat-induced antigen retrieval was performed for 20 minutes at approximately 96ºC, with appropriate buffer depending on the primary antibody/epitope. In short, endogenous peroxide was quenched with 3% hydrogen peroxide in PBS for 10 minutes. Nonspecific antigens were blocked with blocking buffer (2% normal donkey serum, 1% bovine serum albumin, 0.1% Triton-X (Thermo Fisher Scientific Cat No. BP151), 0.05% Tween-20 (Thermo Fisher Scientific Cat No. BP337), and 0.05% sodium azide (Research Products Internation Cat No. S24080-250) in PBS) for 30 minutes at room temperature. Primary antibodies were incubated for 1 hour at room temperature in PBS with 5% normal donkey serum. Detection was performed using the biotin–streptavidin detection system. For immunohistochemistry, horseradish peroxidase signal was developed by 3,3′-diaminobenzidine tetra hydrochloride incubation according to the manufacturer’s protocol, and incubation time was optimized for each protein target (#8059; Cell Signaling Technology, Beverly, MA). Sections were counterstained with hematoxylin. Slides were mounted with Permount (Electron Microscopy Sciences Cat No. 17986-01) and visualized on a Leica microscope (model DM5500B with a DFC450C camera). For immunofluorescence, Prolong Gold with 4′,6-diamidino-2-phenylindole was applied to the slides according to the manufacturer’s guidelines (Invitrogen Cat. no. P36936). Antibody concentration, manufacturer, and antigen retrieval method are listed in Table 1. Confocal images were acquired on an inverted Zeiss 880 microscope. Cryptdin-1 (Defa1) monoclonal antibody was generated and kindly provided by Tokiyoshi Ayabe and assayed as previously described.66
Table 1.
Antibodies and Reagents for Immunohistochemistry and Immunofluorescence
| Antibody | Company | Cat. no. | Antigen retrieval buffer | Primary concentration |
|---|---|---|---|---|
| Lysozyme | Abcam | ab108508 | Na-citrate pH 6 | 1:3000 |
| Ki-67 | Abcam | ab16667 | Na-citrate pH 6 | 1:400 |
| UEA1-649 | Vector Labs | DL-1068 | Na-citrate pH 6 | 1:500 |
| Tunel | Millipore | S7110 | Proteinase K | Data sheet manufacturer |
| E-cadherin | R&D Systems | AF748 | Na-Citrate pH 6 | 1:200 |
| Phosphorylated STAT3 | Cell Signaling Technology | 9145 | Tris-EDTA pH 9 | 1:50 |
| Muc2 | GeneTex | GTX100664 | Na-Citrate pH 6 | 1:800 |
| Cleaved caspase 3 | Cell Signaling Technology | 9664 | Na-Citrate pH 6 | 1:800 |
| Olfm4 | Cell Signaling Technology | 39141 | Na-Citrate pH 6 | 1:400 |
UEA-1, Ulex europaeus agglutinin-1.
Intestinal cross-sections were used for morphometric measurements. Only well-oriented intestinal structures (ie, crypt and villi) with bottom-to-top axis visibility were used in the analysis. The straight-line tool from FIJI software was calibrated using the scale bar from the image being measured.82 A straight line was drawn to measure crypt depth, villus length, and crypt width. All data points from each parameter were averaged per mouse. A minimum of 4 measurements per parameter was set as a cut-off value to provide a more accurate representation of the mouse. For cell counting, the total number of IECs per crypt was counted using the Cell Counter tool on FIJI software. Paneth cells were counted at the crypt base as cells with cytosolic granules and they were differentiated by the presence of lysozyme staining. The ratio of Paneth cells/total IECs per crypt then was calculated and averaged to represent a single mouse.
Ileal sections were excised, cut open longitudinally, and fixed in 1.25% glutaraldehyde and 4% paraformaldehyde in 0.1 mol/L cacodylate buffer for 5–6 hours at room temperature. Samples then were processed at the Facility for Advanced Microscopy & Microanalysis at the University of California, Riverside using an established protocol.35 Samples embedded and plasticized in Spurr’s resin were sectioned into 80-nm, gold-colored sections, with a surface area of approximately 0.25 mm2. Images were acquired using a Tecnai T12 transmission electron microscope (FEI, Hillsboro, OR). Imaging was performed on Paneth cells localized at the bottom of the intestinal crypts focusing at the dense core vesicles and the endoplasmic reticulum. Linear adjustments (brightness and contrast) were performed in original images in its entirety using FIJI software to enhance the visibility of cell organelles.
Intestinal Epithelial Cell Isolation
Distal ileal segments were excised, cut open longitudinally, and immersed in 500 μL Cell Recovery Solution (#354253; Corning) for 2 hours on ice, and then shaken with forceps to release the intestinal crypts/villi into the solution. Samples were centrifuged at 1.5 × g for 10 minutes at 4ºC. The supernatant was aspirated and IECs were washed with PBS (×2). The solution with IECs then was split: 70% of the IEC suspension was centrifuged and cells were lysed using RIPA buffer (150 mmol/L NaCl, 5 mmol/L EDTA, 50 mmol/L Tris, 1% NP-40, 0.5% sodium-deoxycholate, and 0.1% sodium dodecyl sulfate) supplemented with protease inhibitor cocktail (Roche, Mannheim, Germany), 2 mmol/L sodium fluoride, 1 mmol/L phenylmethylsulfonyl fluoride, 2 mmol/L sodium orthovanadate, and phosphatase inhibitor cocktails 2 and 3 (P5726 and P0044, respectively; Millipore Sigma, Israel) and stored at -80ºC; the remaining 30% was centrifuged, 200 μL of RNAlater solution (#AM7020; Invitrogen, Lithuania) was added to the pelleted cells according to the manufacturer’s guidelines, and stored in -80ºC. The purity of IEC samples was assessed by evaluating the abundance of non-IEC markers α-smooth muscle actin and CD45 (Figure 2).
Western Blot
Frozen IEC samples in RIPA buffer were thawed on ice and lysed using a sonicator (model Q125; QSONICA, Newtown, CT) with the following settings: 30% amplitude for a total time of 40 seconds with 10-second intervals. Lysates were centrifuged at 13,000 rpm for 10 minutes at 4ºC and the supernatant was transferred to new tubes. The total amount of protein was estimated by a Pierce BCA assay (cat no. 23225; Thermo Scientific, Rockford, IL). Equal amounts of protein were loaded on polyacrylamide gels, and after separation by gel electrophoresis was transferred onto polyvinylidene difluoride membranes. Nonspecific epitopes were blocked with 5% milk in Tris-buffered saline with 0.1% Tween-20 added for 1 hour at room temperature. Membranes were incubated overnight with primary antibody at 4ºC, washed (×3) with Tris-buffered saline with 0.1% Tween-20, and incubated with horseradish-peroxidase–conjugated secondary antibody antiprimary species for 1 hour at room temperature. Antibody information is listed in Table 2. Immunoreactive proteins were detected with x-ray films (Labscientific, Inc, Highlands, NJ) using the SuperSignal West Pico PLUS chemiluminescence detection kit (cat. no. 34580; Thermo Fisher Scientific).
Table 2.
Antibodies and Reagents for Western Blot
| Antibody | Company | Cat. no. |
|---|---|---|
| Lysozyme | Abcam | ab108508 |
| β-actin | SIGMA | A 5316 |
| TCPTP | Cell Signaling Technology | 58935 |
| Caspase 3 | Cell Signaling Technology | 14220 |
| Cleaved caspase 3 | Cell Signaling Technology | 9664 |
| C/EBP-homologous protein (CHOP) | Cell Signaling Technology | 2895 |
| PARP | Cell Signaling Technology | 9542 |
| Bip | Cell Signaling Technology | 3117 |
| Xbp-1s | Cell Signaling Technology | 40435 |
| α-Smooth muscle actin | SIGMA | A 2547 |
| CD45 | Cell Signaling Technology | 72787 |
| Epithelial cell adhesion molecule (EpCAM) | Cell Signaling Technology | 42515 |
| Mist1 | Santa Cruz | sc-80984 |
| Reg3-γ | Abcam | ab198216 |
| eIF2-α | Cell Signaling Technology | 5324 |
| Phosphorylated eIF2-α | Cell Signaling Technology | 3398 |
| Olfm4 | Cell Signaling Technology | 39141 |
| STAT1 | Cell Signaling Technology | 14994 |
| Phosphorylated STAT1 | Cell Signaling Technology | 9167 |
| STAT3 | Cell Signaling Technology | 9139 |
| Phosphorylated STAT3 | Cell Signaling Technology | 9145 |
| Beclin-1 | Cell Signaling Technology | 3495 |
| LC3B | Cell Signaling Technology | 2775 |
| Atg3 | Cell Signaling Technology | 3415 |
| Atg5 | Cell Signaling Technology | 12994 |
| Atg7 | Cell Signaling Technology | 8558 |
| Atg12 | Cell Signaling Technology | 4180 |
| Atg16l1 | Cell Signaling Technology | 8089 |
| Mmp7 | Abcam | ab232737 |
RNA Extraction, Complementary DNA Synthesis, and RT-qPCR
Total RNA was isolated using the RNeasy Mini Kit (cat. no. 74106; Qiagen, Venlo, The Netherlands), according to the manufacturer's instructions. The RNA concentration was quantified by measuring the absorbance at 260 nm and 280 nm using a NanoDrop 2000 spectrophotometer (Thermo Scientific, Wilmington, DE). Complementary DNA synthesis was performed using the qScript cDNA SuperMix (cat. no. 95048; Quantabio, Beverly, MA) following the manufacturer’s instructions. Real-time qPCR was performed using iQ SYBR Green Supermix (Bio-Rad, Hercules, CA) on a C1000 Thermal cycler (Bio-Rad) equipped with a CFX96 Real-Time PCR system using Bio-Rad CFX Manager 3.1 software following the manufacturer’s protocol. Each PCR target was assayed in triplicate. Tbp was used as a reference gene. All primers were designed using NCBI Primer-BLAST, and the specificity and optimal annealing temperature were tested empirically. RT-qPCR contained an initial enzyme activation step (3 minutes, 95°C) followed by 45 cycles consisting of denaturing (95°C, 10 seconds), annealing (53°C–60°C, 10 seconds), and extension (72°C, 10 seconds) steps. The primers are listed in Table 3. Results were analyzed by the ΔΔCT method and graphs display geometric means ± SD of the geometric mean in a log10 scale.
Table 3.
PCR Primers (Mus musculus)
| Gene | Primer sequence | GC, % | Length | Melting Temperature (C) | Amplicon | Reference Sequence |
|---|---|---|---|---|---|---|
| Ascl2 | F: TCTTGGGGCTTAAGGGCTGA R: GTCAAGGTGTGCTTCCATGC |
55 55 |
20 20 |
59 | 192 | NM_008554.3 |
| Reg3b | F: GAATATACCCTCCGCACGCA R: TCTTTTGGCAGGCCAGTTCT |
55 50 |
20 20 |
60 | 119 | NM_011036.1 |
| Reg3g | F: TGCAAGGTGAAGTTGCCAAG R: GGTTCATAGCCCAGTGTCGG |
50 60 |
20 20 |
60 | 274 | NM_011260.2 |
| Tbp (HK) | F: CCTTGTACCCTTCACCAATGAC R: ACAGCCAAGATTCACGGTAGA |
50 47 |
22 21 |
61 | 119 | NM_013684.3 |
F, forward; R, reverse.
Nanostring Analysis
Total RNA was extracted as described earlier. Estimation of total RNA was determined by a Qubit 2.0 Fluorometer (Thermo Fisher Scientific) and sample purity was determined by a NanoDrop 2000 spectrophotometer by measuring the 230/260 and 260/280 ratios. Samples were diluted to 10 ng/μL with RNAse-free water. A hybridization reaction was performed following the manufacturer’s guidelines using 50 ng total RNA from each sample. In short, gene expression was explored by using the PanCaner Pathways Panel and the AutoImmune Profiling Panel, totaling more than 1500 targets. Samples were hybridized with a reporter and capture probe for 18 hours at 65ºC. The reaction was ramped down to 4ºC, 12 μL RNAse-free water was added to each sample and immediately pipetted into a cartridge and processed by the nCounter SPRINT Profiler (NanoString Technologies). Data sets were normalized by nSolver Analysis Software 4.0 (NanoString Technologies) for each panel separately, and then combined using the advanced analysis tools. The α-value was set at .05 and the false-discovery rate was calculated by the Benjamini–Yekutieli method, setting a cut-off value at 10% (Q = 0.1) for multiple-comparisons testing.
Flow Cytometry
For flow cytometry of immune cells, lamina propria immune cells were isolated as described.83 For analysis of myeloid immune cells, the cells were washed in PBS, incubated with FcR blocking antibody (Miltenyi Biotec, Bergisch Gladbach, Germany) for 10 minutes, and stained with anti-CD45–Pacific Blue, anti-CD3–BV650, anti-NK1.1–BV650, anti-B220–BV650, anti-CD11b–BV605, anti-CD11c–PECy7, anti-Ly6C–PerCPCy5.5, anti-F4/80–APC, anti-CD64–PE, and anti–MHC-II–AF700 (all from BioLegend, San Diego, CA) for 15–30 minutes. ZOMBI-NIR live dead stain (BioLegend) was used for discrimination between live and dead cells. For cytokine staining, the cells were incubated with ionomycin and PMA in the presence of Brefeldin A for 3.5 hours before surface staining with anti-CD25–AlexaFluor700, anti-CD3–PerCPCy5.5, anti-CD4–BV510, and anti-CD8–BV570 for 15 minutes. Cells then were fixed with the FoxP3 staining kit (eBioscience) according to the manufacturer’s instructions, stained with anti-FoxP3–Pacific Blue, anti–IFN-γ–PECy7, anti-IL17–APC, anti-TNFα–BV650, and anti-IL22–PE for 30 minutes, washed in PermWash buffer (eBioscience), samples were acquired on an LSRII cytometer (BD, Franklin Lakes, NJ), and analyzed using FlowJo (Tree Star, Inc, Ashland, OR). Gating strategy for T and myeloid cells was performed as described previously.32
Small Intestinal Crypt Isolation and Enteroid Culture
Small intestines were excised from 21-day-old Ptpn2-WT and KO mice, washed with cold Dulbecco's PBS (#8662; Sigma), and cut open longitudinally into 2-cm fragments. The tissues were placed in chelation buffer (10 mmol/L EDTA in DPBS) and incubated on a rocker for 45 minutes at 4ºC. Intestines then were moved to a tube with cold DPBS and shaken vigorously for 3 minutes to release crypts. The supernatant then was filtered through a 70-μm cell strainer and centrifuged at 300 × g for 5 minutes. Pellets were resuspended in phenol red–free Matrigel (#356230; Corning), seeded in small 50-μL domes in 24-well plates, and incubated at 37ºC for 20 minutes to allow Matrigel to polymerize. Enteroid expansion complete media composed of advanced Dulbecco’s modified Eagle medium/F12 (#12634-028; Invitrogen) with 100 U/mL Penicillin-Streptomycin (#15140-122; Invitrogen), 2 mmol/L GlutaMAX (35050-079; Invitrogen), and 10 mmol/L HEPES (#15630-56; Invitrogen) supplemented with R-Spondin-1 conditioned media (#3710-001-01; R&D Systems), Noggin conditioned media (#6997-NG; R&D Systems), 1× B-27 (#17504-044; Invitrogen), 1.25 mmol/L N-acetylcysteine (#9165; Sigma), 125 μg/mL primocin (#ant-pm-1; InvivoGen), 0.5 μg/mL epidermal growth factor (#236-EG-01M; R&D Systems), Wnt3a conditioned media (ATCC #CRL-2647), 1× gastrin (#64149; AnaSpec), 50 μmol/L A83 (#2939; Tocris), and 10 μmol/L SB202190 (#1264; Tocris). After 1 week, enteroids were dissociated mechanically by pipetting and seeded in 50-μL domes in 24-well plates and 10-μL in 4-well chambers for staining (Nunc LabTek Chamber Slide #177399; Thermo Scientific). Genetic recombination was induced by 1 μmol/L 4-hydroxytamoxifen (#T176; Millipore) added to the media for 48 hours.
Statistics
Data sets were tested for 2 parameters: normality (Shapiro–Wilk test) and variance homogeneity (Brown–Forsythe F test) using GraphPad Prism 9.0.0 software by Dotmatics. Statistical and post hoc tests were chosen according to these parameters and are indicated on the respective Figure legends. In general, the 2-tailed Student t test was applied when comparing 2 groups and ordinary 1-way analysis of variance was applied for comparison of 3 experimental groups. The critical significance level was set at α = .05. Data are expressed as means ± SD for n independent observations per group unless stated otherwise. Outliers were identified using robust regression and outlier removal (ROUT) method, with Q set at 1% and excluded when appropriate.
All authors had access to the study data and reviewed and approved the final manuscript.
Acknowledgments
The authors thank Dr David Carter for his assistance with confocal imaging. The authors thank Dr Krassimir Bozhilov and Matthew Dickson from the University of California Riverside’s Facility for Advanced Microscopy and Microanalysis for their collaboration with TEM image acquisition. The authors thank Dr Tokiyoshi Ayabe from Hokkaido University for kindly providing the Cryptdin-1 monoclonal antibody. The authors thank Dr Meli’sa Crawford (University of California, Riverside) for insightful discussions.
CRediT Authorship Contributions
Vinicius Canale, PhD (Data curation: Lead; Formal analysis: Lead; Investigation: Lead; Methodology: Lead; Software: Lead; Validation: Lead; Writing – original draft: Lead)
Marianne R Spalinger, PhD (Formal analysis: Supporting; Investigation: Supporting; Methodology: Supporting; Validation: Supporting; Writing – review & editing: Supporting)
Rocio Alvarez, BS (Formal analysis: Supporting; Investigation: Supporting; Methodology: Supporting; Project administration: Supporting; Writing – review & editing: Supporting)
Anica Sayoc-Becerra, PhD (Formal analysis: Supporting; Investigation: Supporting; Methodology: Supporting; Validation: Supporting; Writing – review & editing: Supporting)
Alina N Santos, BS (Investigation: Supporting; Methodology: Supporting; Project administration: Supporting; Writing – review & editing: Supporting)
Hillmin Lei, BS, MS (Investigation: Supporting; Methodology: Supporting; Writing – review & editing: Supporting)
Sharon Jahng (Investigation: Supporting; Methodology: Supporting; Writing – review & editing: Supporting)
Timothy Chu (Investigation: Supporting; Methodology: Supporting; Writing – review & editing: Supporting)
Ali Shawki, PhD (Investigation: Supporting; Methodology: Supporting; Writing – review & editing: Supporting)
Elaine Hanson, BS (Resources: Supporting; Writing – review & editing: Supporting)
Lars Eckmann, MD (Resources: Supporting; Writing – review & editing: Supporting)
Andre J Oullette, PhD (Conceptualization: Supporting; Methodology: Supporting; Resources: Supporting; Writing – review & editing: Supporting)
Declan F. McCole, Ph.D. (Conceptualization: Lead; Formal analysis: Supporting; Funding acquisition: Lead; Project administration: Lead; Resources: Lead; Supervision: Lead; Writing – review & editing: Lead)
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding This work was supported by the Crohn’s and Colitis Foundation Senior Research Award (D.F.M.); National Institutes of Health grants 2R01-DK091281, 1R01AI153314-01, R21AI152017, 1R01DK130373, and R01AI165490 (D.F.M.), and P30 DK120515 (L.E.); Schweizer Nationalfonds zur Förderung der wissenschaftlichen Forschung P300PB_177932 (M.R.S.); American Gastroenterological Association Inflammatory Bowel Disease Research Award (D.F.M.); Science Without Borders Program from CAPES #99999.013456/2013-00, and a training program to advance interdisciplinary stem cell research, education, and workforce diversity (TRANSCEND) fellowship from the California Institute of Regenerative Medicine EDUC4-12752 (V.C.).
Data Availability Statement Partial Transcriptome dataset is available in the NCBI database GEO accession number: GSE181914.
Supplementary Material
References
- 1.Axelrad J.E., Olén O., Sachs M.C., et al. Inflammatory bowel disease and risk of small bowel cancer: a binational population-based cohort study from Denmark and Sweden. Gut. 2021;70:297–308. doi: 10.1136/gutjnl-2020-320945. [DOI] [PubMed] [Google Scholar]
- 2.Biancone L., Armuzzi A., Scribano M.L., et al. Cancer risk in inflammatory bowel disease: a 6-year prospective multicenter nested case-control IG-IBD study. Inflamm Bowel Dis. 2020;26:450–459. doi: 10.1093/ibd/izz155. [DOI] [PubMed] [Google Scholar]
- 3.Murthy S.K., Begum J., Benchimol E.I., et al. Introduction of anti-TNF therapy has not yielded expected declines in hospitalisation and intestinal resection rates in inflammatory bowel diseases: a population-based interrupted time series study. Gut. 2020;69:274–282. doi: 10.1136/gutjnl-2019-318440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roblin X., Williet N., Boschetti G., et al. Addition of azathioprine to the switch of anti-TNF in patients with IBD in clinical relapse with undetectable anti-TNF trough levels and antidrug antibodies: a prospective randomised trial. Gut. 2020;69:1206–1212. doi: 10.1136/gutjnl-2019-319758. [DOI] [PubMed] [Google Scholar]
- 5.Kaser A., Zeissig S., Blumberg R.S. Inflammatory bowel disease. Annu Rev Immunol. 2010;28:573–621. doi: 10.1146/annurev-immunol-030409-101225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.König J., Wells J., Cani P.D., et al. Human intestinal barrier function in health and disease. Clin Transl Gastroenterol. 2016;7:e196. doi: 10.1038/ctg.2016.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Souza H.S.P., Fiocchi C. Immunopathogenesis of IBD: current state of the art. Nat Rev Gastroenterol Hepatol. 2016;13:13–27. doi: 10.1038/nrgastro.2015.186. [DOI] [PubMed] [Google Scholar]
- 8.De Lange K.M., Moutsianas L., Lee J.C., et al. Genome-wide association study implicates immune activation of multiple integrin genes in inflammatory bowel disease. Nat Genet. 2017;49:256–261. doi: 10.1038/ng.3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lees C.W., Barrett J.C., Parkes M., Satsangi J. New IBD genetics: common pathways with other diseases. Gut. 2011;60:1739–1753. doi: 10.1136/gut.2009.199679. [DOI] [PubMed] [Google Scholar]
- 10.Khor B., Gardet A., Xavier R.J. Genetics and pathogenesis of inflammatory bowel disease. Nature. 2011;474:307–317. doi: 10.1038/nature10209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glas J., Wagner J., Seiderer J., et al. PTPN2 gene variants are associated with susceptibility to both Crohn’s disease and ulcerative colitis supporting a common genetic disease background. PLoS One. 2012;7 doi: 10.1371/journal.pone.0033682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simoncic P.D., Lee-Loy A., Barber D.L., et al. The T cell protein tyrosine phosphatase is a negative regulator of Janus family kinases 1 and 3. Curr Biol. 2002;12:446–453. doi: 10.1016/s0960-9822(02)00697-8. [DOI] [PubMed] [Google Scholar]
- 13.Scharl M., Paul G., Weber A., et al. Protection of epithelial barrier function by the Crohn’s disease associated gene protein tyrosine phosphatase N2. Gastroenterology. 2009;137:2030–2040.e5. doi: 10.1053/j.gastro.2009.07.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doody K.M., Bourdeau A., Tremblay M.L. T-cell protein tyrosine phosphatase is a key regulator in immune cell signaling: lessons from the knockout mouse model and implications in human disease. Immunol Rev. 2009;228:325–341. doi: 10.1111/j.1600-065X.2008.00743.x. [DOI] [PubMed] [Google Scholar]
- 15.Sayoc-Becerra A., Krishnan M., Fan S., et al. The JAK-inhibitor tofacitinib rescues human intestinal epithelial cells and colonoids from cytokine-induced barrier dysfunction. Inflamm Bowel Dis. 2020;26:407–422. doi: 10.1093/ibd/izz266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krishnan M., McCole D.F. T cell protein tyrosine phosphatase prevents STAT1 induction of claudin-2 expression in intestinal epithelial cells. Ann N Y Acad Sci. 2017;1405:116–130. doi: 10.1111/nyas.13439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spalinger M.R., Sayoc-Becerra A., Santos A.N., et al. PTPN2 regulates interactions between macrophages and intestinal epithelial cells to promote intestinal barrier function. Gastroenterology. 2020;159:1763–1777.e14. doi: 10.1053/j.gastro.2020.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shawki A., Ramirez R., Spalinger M.R., et al. The autoimmune susceptibility gene, PTPN2, restricts expansion of a novel mouse adherent-invasive E. coli. Gut Microbes. 2020;11:1547–1566. doi: 10.1080/19490976.2020.1775538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hassan S.W., Doody K.M., Hardy S., et al. Increased susceptibility to dextran sulfate sodium induced colitis in the T cell protein tyrosine phosphatase heterozygous mouse. PLoS One. 2010;5 doi: 10.1371/journal.pone.0008868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okumura R., Takeda K. Roles of intestinal epithelial cells in the maintenance of gut homeostasis. Exp Mol Med. 2017;49:338. doi: 10.1038/emm.2017.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sato T., Van Es J.H., Snippert H.J., et al. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature. 2011;469:415–418. doi: 10.1038/nature09637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wehkamp J., Stange E.F. Paneth’s disease. J Crohns Colitis. 2010;4:523–531. doi: 10.1016/j.crohns.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 23.Antoni L., Nuding S., Wehkamp J., Stange E.F. Intestinal barrier in inflammatory bowel disease. World J Gastroenterol. 2014;20:1165–1179. doi: 10.3748/wjg.v20.i5.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stappenbeck T.S., McGovern D.P.B. Paneth cell alterations in the development and phenotype of Crohn’s disease. Gastroenterology. 2017;152:322–326. doi: 10.1053/j.gastro.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Riba A., Olier M., Lacroix-Lamandé S., et al. Paneth cell defects induce microbiota dysbiosis in mice and promote visceral hypersensitivity. Gastroenterology. 2017;153:1594–1606.e2. doi: 10.1053/j.gastro.2017.08.044. [DOI] [PubMed] [Google Scholar]
- 26.Van Der Post S., Jabbar K.S., Birchenough G., et al. Structural weakening of the colonic mucus barrier is an early event in ulcerative colitis pathogenesis. Gut. 2019;68:2142–2151. doi: 10.1136/gutjnl-2018-317571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gersemann M., Becker S., Kübler I., et al. Differences in goblet cell differentiation between Crohn’s disease and ulcerative colitis. Differentiation. 2009;77:84–94. doi: 10.1016/j.diff.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 28.Vandussen K.L., Liu T.C., Li D., et al. Genetic variants synthesize to produce Paneth cell phenotypes that define subtypes of Crohn’s disease. Gastroenterology. 2014;146:200–209. doi: 10.1053/j.gastro.2013.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adolph T.E., Tomczak M.F., Niederreiter L., et al. Paneth cells as a site of origin for intestinal inflammation. Nature. 2013;503:272–276. doi: 10.1038/nature12599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Q., Pan Y., Yan R., et al. Commensal bacteria direct selective cargo sorting to promote symbiosis. Nat Immunol. 2015;16:918–926. doi: 10.1038/ni.3233. [DOI] [PubMed] [Google Scholar]
- 31.Rocha J.D.B., Schlossmacher M.G., Philpott D.J. LRRK2 and Nod2 promote lysozyme sorting in Paneth cells. Nat Immunol. 2015;16:898–900. doi: 10.1038/ni.3255. [DOI] [PubMed] [Google Scholar]
- 32.Marchelletta R.R., Krishnan M., Spalinger M.R., et al. T cell protein tyrosine phosphatase protects intestinal barrier function by restricting epithelial tight junction remodeling. J Clin Invest. 2021;131 doi: 10.1172/JCI138230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu T.-C., Gurram B., Baldridge M.T., et al. Paneth cell defects in Crohn’s disease patients promote dysbiosis. JCI Insight. 2016;1 doi: 10.1172/jci.insight.86907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Satoh Y., Ishikawa K., Oomori Y., et al. Bethanechol and a G-protein activator, NaF/AlCl3, induce secretory response in Paneth cells of mouse intestine. Cell Tissue Res. 1992;269:213–220. doi: 10.1007/BF00319611. [DOI] [PubMed] [Google Scholar]
- 35.Satoh Y., Yamano M., Matsuda M., Ono K. Ultrastructure of Paneth cells in the intestine of various mammals. J Electron Microsc Tech. 1990;16:69–80. doi: 10.1002/jemt.1060160109. [DOI] [PubMed] [Google Scholar]
- 36.Stahl M., Tremblay S., Montero M., et al. The Muc2 mucin coats murine Paneth cell granules and facilitates their content release and dispersion. Am J Physiol Liver Physiol. 2018;315:G195–G205. doi: 10.1152/ajpgi.00264.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dekaney C.M., King S., Sheahan B., Cortes J.E. Mist1 expression is required for Paneth cell maturation. Clin Mol Gastroenterol Hepatol. 2019;8:549–560. doi: 10.1016/j.jcmgh.2019.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pin C.L., Rukstalis M.J., Johnson C., Konieczny S.F. The bHLH transcription factor Mist1 is required to maintain exocrine pancreas cell organization and acinar cell identity. J Cell Biol. 2001;155:519–530. doi: 10.1083/jcb.200105060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnson C.L., Kowalik A.S., Rajakumar N., Pin C.L. Mist1 is necessary for the establishment of granule organization in serous exocrine cells of the gastrointestinal tract. Mech Dev. 2004;121:261–272. doi: 10.1016/j.mod.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 40.Koslowski M.J., Kübler I., Chamaillard M., et al. Genetic variants of Wnt transcription factor TCF-4 (TCF7L2) putative promoter region are associated with small intestinal Crohn’s disease. PLoS One. 2009;4:e4496. doi: 10.1371/journal.pone.0004496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Es J.H., Jay P., Gregorieff A., et al. Wnt signalling induces maturation of Paneth cells in intestinal crypts. Nat Cell Biol. 2005;7:381–386. doi: 10.1038/ncb1240. [DOI] [PubMed] [Google Scholar]
- 42.Weeks C.S., Tanabe H., Cummings J.E., et al. Matrix metalloproteinase-7 activation of mouse Paneth cell pro-α-defensins: SER43 ↓ ILE44 proteolysis enables membrane-disruptive activity. J Biol Chem. 2006;281:28932–28942. doi: 10.1074/jbc.M602041200. [DOI] [PubMed] [Google Scholar]
- 43.Artis D. Epithelial-cell recognition of commensal bacteria and maintenance of immune homeostasis in the gut. Nat Rev Immunol. 2008;8:411–420. doi: 10.1038/nri2316. [DOI] [PubMed] [Google Scholar]
- 44.Bel S., Pendse M., Wang Y., et al. Paneth cells secrete lysozyme via secretory autophagy during bacterial infection of the intestine. Science. 2017;357:1047–1052. doi: 10.1126/science.aal4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Holly M.K., Smith J.G. Paneth cells during viral infection and pathogenesis. Viruses. 2018;10:225. doi: 10.3390/v10050225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yokoi Y., Nakamura K., Yoneda T., et al. Paneth cell granule dynamics on secretory responses to bacterial stimuli in enteroids. Sci Rep. 2019;9:2710. doi: 10.1038/s41598-019-39610-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Han J., Back S.H., Hur J., et al. ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nat Cell Biol. 2013;15:481–490. doi: 10.1038/ncb2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marciniak S.J., Yun C.Y., Oyadomari S., et al. CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes Dev. 2004;18:3066–3077. doi: 10.1101/gad.1250704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cray P., Sheahan B.J., Dekaney C.M. Secretory sorcery: Paneth cell control of intestinal repair and homeostasis. Cell Mol Gastroenterol Hepatol. 2021;12:1239–1250. doi: 10.1016/j.jcmgh.2021.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brodrick B., Vidrich A., Porter E., et al. Fibroblast growth factor receptor-3 (FGFR-3) regulates expression of Paneth cell lineage-specific genes in intestinal epithelial cells through both TCF4/β-catenin-dependent and -independent signaling pathways. J Biol Chem. 2011;286:18515–18525. doi: 10.1074/jbc.M111.229252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vidrich A., Buzan J.M., Brodrick B., et al. Fibroblast growth factor receptor-3 regulates Paneth cell lineage allocation and accrual of epithelial stem cells during murine intestinal development. Am J Physiol Gastrointest Liver Physiol. 2009;297:G168–G178. doi: 10.1152/ajpgi.90589.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Darfeuille-Michaud A., Boudeau J., Bulois P., et al. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn’s disease. Gastroenterology. 2004;127:412–421. doi: 10.1053/j.gastro.2004.04.061. [DOI] [PubMed] [Google Scholar]
- 53.Ayabe T., Satchell D.P., Wilson C.L., et al. Secretion of microbicidal α-defensins by intestinal Paneth cells in response to bacteria. Nat Immunol. 2000;1:113–118. doi: 10.1038/77783. [DOI] [PubMed] [Google Scholar]
- 54.Salzman N.H., Underwood M.A., Bevins C.L. Paneth cells, defensins, and the commensal microbiota: a hypothesis on intimate interplay at the intestinal mucosa. Semin Immunol. 2007;19:70–83. doi: 10.1016/j.smim.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 55.Ericksen B., Wu Z., Lu W., Lehrer R.I. Antibacterial activity and specificity of the six human α-defensins. Antimicrob Agents Chemother. 2005;49:269–275. doi: 10.1128/AAC.49.1.269-275.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Salzman N.H., Ghosh D., Huttner K.M., et al. Protection against enteric salmonellosis in transgenic mice expressing a human intestinal defensin. Nature. 2003;422:522–526. doi: 10.1038/nature01520. [DOI] [PubMed] [Google Scholar]
- 57.Chu H., Pazgier M., Jung G., et al. Human α-defensin 6 promotes mucosal innate immunity through self-assembled peptide nanonets. Science. 2012;337:477–481. doi: 10.1126/science.1218831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ouellette A.J., Darmoul D., Tran D., et al. Peptide localization and gene structure of cryptdin 4, a differentially expressed mouse Paneth cell α-defensin. Infect Immun. 1999;67:6643–6651. doi: 10.1128/iai.67.12.6643-6651.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wehkamp J., Chu H., Shen B., et al. Paneth cell antimicrobial peptides: topographical distribution and quantification in human gastrointestinal tissues. FEBS Lett. 2006;580:5344–5350. doi: 10.1016/j.febslet.2006.08.083. [DOI] [PubMed] [Google Scholar]
- 60.Balasubramanian I., Gao N. From sensing to shaping microbiota: insights into the role of NOD2 in intestinal homeostasis and progression of Crohn’s disease. Am J Physiol Gastrointest Liver Physiol. 2017;313:G7–G13. doi: 10.1152/ajpgi.00330.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yu S., Balasubramanian I., Laubitz D., et al. Paneth cell-derived lysozyme defines the composition of mucolytic microbiota and the inflammatory tone of the intestine. Immunity. 2020;53:398–416.e8. doi: 10.1016/j.immuni.2020.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liang S.C., Tan X.Y., Luxenberg D.P., et al. Interleukin (IL)-22 and IL-17 are coexpressed by Th17 cells and cooperatively enhance expression of antimicrobial peptides. J Exp Med. 2006;203:2271–2279. doi: 10.1084/jem.20061308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Eyerich S., Eyerich K., Cavani A., Schmidt-Weber C. IL-17 and IL-22: siblings, not twins. Trends Immunol. 2010;31:354–361. doi: 10.1016/j.it.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 64.Kaser A., Lee A.-H., Franke A., et al. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell. 2008;134:743–756. doi: 10.1016/j.cell.2008.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.You-Ten K.E., Muise E.S., Itié A., et al. Impaired bone marrow microenvironment and immune function in T cell protein tyrosine phosphatase–deficient mice. J Exp Med. 1997;186:683–693. doi: 10.1084/jem.186.5.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Eriguchi Y., Nakamura K., Yokoi Y., et al. Essential role of IFN-γ in T cell-associated intestinal inflammation. JCI Insight. 2018;3 doi: 10.1172/jci.insight.121886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Farin H.F., Karthaus W.R., Kujala P., et al. Paneth cell extrusion and release of antimicrobial products is directly controlled by immune cell-derived IFN-γ. J Exp Med. 2014;211:1393–1405. doi: 10.1084/jem.20130753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lemercier C., To R.Q., Swanson B.J., et al. Mist1: a novel basic helix-loop-helix transcription factor exhibits a developmentally regulated expression pattern. Dev Biol. 1997;182:101–113. doi: 10.1006/dbio.1996.8454. [DOI] [PubMed] [Google Scholar]
- 69.Acosta-Alvear D., Zhou Y., Blais A., et al. XBP1 controls diverse cell type- and condition-specific transcriptional regulatory networks. Mol Cell. 2007;27:53–66. doi: 10.1016/j.molcel.2007.06.011. [DOI] [PubMed] [Google Scholar]
- 70.Scharl M., Rogler G. The role for protein tyrosine phosphatase nonreceptor type 2 in regulating autophagosome formation. Ann N Y Acad Sci. 2012;1257:93–102. doi: 10.1111/j.1749-6632.2012.06578.x. [DOI] [PubMed] [Google Scholar]
- 71.Scharl M., Wojtal K.A., Becker H.M., et al. Protein tyrosine phosphatase nonreceptor type 2 regulates autophagosome formation in human intestinal cells. Inflamm Bowel Dis. 2012;18:1287–1302. doi: 10.1002/ibd.21891. [DOI] [PubMed] [Google Scholar]
- 72.Spalinger M.R., Shawki A., Chatterjee P., et al. Autoimmune susceptibility gene PTPN2 is required for clearance of adherent-invasive Escherichia coli by integrating bacterial uptake and lysosomal defence. Gut. 2022;71:88–99. doi: 10.1136/gutjnl-2020-323636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rashid H.O., Yadav R.K., Kim H.R., Chae H.J. ER stress: autophagy induction, inhibition and selection. Autophagy. 2015;11:1956–1977. doi: 10.1080/15548627.2015.1091141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gil J., Esteban M. Induction of apoptosis by the dsRNA-dependent protein kinase (PKR): mechanism of action. Apoptosis. 2000;5:107–114. doi: 10.1023/a:1009664109241. [DOI] [PubMed] [Google Scholar]
- 75.Kasper S.H., Spalinger M.R., Raselli T., Scharl M. A cell type-specific role of protein tyrosine phosphatase non-receptor type 2 in regulating ER stress signalling. Digestion. 2015;91:248–256. doi: 10.1159/000375459. [DOI] [PubMed] [Google Scholar]
- 76.Zinszner H., Kuroda M., Wang X.Z., et al. CHOP is implicated in programmed cell death in response to impaired function of the endoplasmic reticulum. Genes Dev. 1998;12:982–995. doi: 10.1101/gad.12.7.982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bettaieb A., Liu S., Xi Y., et al. Differential regulation of endoplasmic reticulum stress by protein tyrosine phosphatase 1B and T cell protein tyrosine phosphatase. J Biol Chem. 2011;286:9225–9235. doi: 10.1074/jbc.M110.186148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yu S., Tong K., Zhao Y., et al. Paneth cell multipotency induced by Notch activation following injury. Cell Stem Cell. 2018;23:46–59.e5. doi: 10.1016/j.stem.2018.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schmitt M., Schewe M., Sacchetti A., et al. Paneth cells respond to inflammation and contribute to tissue regeneration by acquiring stem-like features through SCF/c-Kit signaling. Cell Rep. 2018;24:2312–2328.e7. doi: 10.1016/j.celrep.2018.07.085. [DOI] [PubMed] [Google Scholar]
- 80.Yan K., Chia L., Li X. The intestinal stem cell markers Bmi1 and Lgr5 identify two functionally distinct populations. Proc Natl Acad Sci U S A. 2012;109:466–471. doi: 10.1073/pnas.1118857109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.El Marjou F., Janssen K.P., Chang B.H.J., et al. Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis. 2004;39:186–193. doi: 10.1002/gene.20042. [DOI] [PubMed] [Google Scholar]
- 82.Schindelin J., Arganda-Carreras I., Frise E., et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Spalinger M.R., Manzini R., Hering L., et al. PTPN2 regulates inflammasome activation and controls onset of intestinal inflammation and colon cancer. Cell Rep. 2018;22:1835–1848. doi: 10.1016/j.celrep.2018.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.