Abstract
Catalase (CAT) is an important antioxidant enzyme that breaks down H2O2 into water and oxygen. Inhibitor-modulating CAT activity in cancer cells is emerging as a potential anticancer strategy. However, the discovery of CAT inhibitors towards the heme active center located at the bottom of long and narrow channel has made little progress. Therefore, targeting new binding site is of great importance for the development of efficient CAT inhibitors. Here, the first NADPH-binding site inhibitor of CAT, BT-Br, was designed and synthesized successfully. The cocrystal structure of BT-Br-bound CAT complex was determined with a resolution of 2.2 Å (PDB ID:8HID), which showed clearly that BT-Br bound at the NADPH-binding site. Furthermore, BT-Br was demonstrated to induce ferroptosis in castration-resistant prostate cancer (CRPC) DU145 cells and eventually reduce CRPC tumors in vivo effectively. The work indicates that CAT has potential as a novel target for CRPC therapy based on ferroptosis inducing.
Keywords: Catalase inhibitors, NADPH-Binding site, Ferroptosis, Castration-resistant prostate cancer
Graphical abstract
Highlights
-
•
BT-Br binds at the NADPH-binding site of CAT.
-
•
BT-Br acts as an efficient CAT inhibitor in CRPC DU145 cells.
-
•
BT-Br rises intracellular Fe2+ levels via autophagy-mediated degradation of FTH1.
-
•
BT-Br induces ferroptosis in DU145 cells and reduces CRPC tumors effectively.
1. Introduction
Human catalase (CAT) is a well-known crucial enzyme in protecting cells against the toxic effects of hydrogen peroxide (H2O2). Deficient or inactive CAT will increase the production of reactive oxygen species (ROS) within cells [1,2]. Since cancer cells are particularly sensitive to oxidative stress, targeting the redox status of cancer cells by regulating CAT expression as well as CAT activity, is emerging as a potential anticancer strategy or a novel approach to potentiate chemotherapy [3]. Human erythrocyte CAT has been reported to have four identical subunits, each containing a heme active center and a cofactor NADPH molecule [1,4]. Thereinto, the heme prosthetic group locates at the bottom of a long and narrow channel, with a hydrophobic constriction (2–3 Å wide) immediately above it, which is significant for the molecular ruler recognition of H2O2. For NADPH this is different, it binds at a cleft between the helical domain and the β-barrel on the surface of CAT. Although the function of the bound NADPH is not fully understood, the present studies suggest that NADPH both prevents and reverses the accumulation of inactive intermediate compound II during the catalytic process, protecting the enzyme against oxidation by H2O2 [1,5].
In the proposed catalytic mechanism of CAT [1], the amino-acid residues next to the heme active center, His 75 and Asn 148, are very important for the catalytic activity via fixing the substrate H2O2. In this context, direct interaction with these residues, for instance, the widely recognized specific CAT inhibitor 3-amino-1,2,4-triazole (3-AT), or competition with H2O2 for heme-iron binding, such as the non-specific inhibitor cyanide, can inhibit the activity of CAT. However, due to the long and narrow channel to the active center, it is very difficult for most molecules to access the active center. Therefore, the CAT inhibitors discovery towards the heme active center has made little progress during last decades, and identifying new binding site is critical for the discovery of novel and efficient CAT inhibitors. In our study, we designed and synthesized a benzaldehyde thiosemicarbazone derivative BT-Br with high inhibitory activity against CAT. In depth analysis of the cocrystal structure of CAT-BT-Br complex showed clearly that BT-Br located in the NADPH-binding site of CAT, significantly different from 3-AT or cyanide.
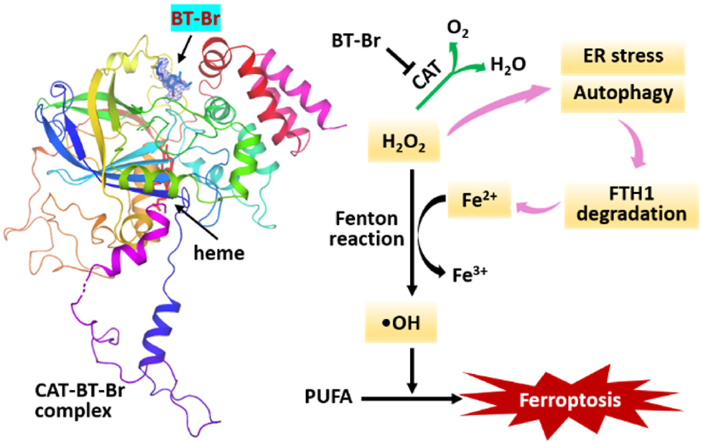
Ferroptosis is a new programmed cell death process driven by cellular metabolism and iron-dependent peroxidation of polyunsaturated fatty acids (PUFAs) [6,7]. According to the previous conclusion, both lipid peroxides (LPO, also called lipid ROS) and iron metabolism are two critical processes involved in the mechanism of ferroptosis [8,9]. As new types of cancer treatment, the strategies of ferroptosis-inducing have been intensively investigated by researchers, mainly including inhibiting the activity of glutathione peroxidase 4 (GPx4) [10,11], accelerating intracellular Fenton reaction [[11], [12], [13]], and increasing the exogenous delivery of LPO [14].
Considering that elevated ROS levels caused by CAT inhibition may induce over production of LPO, leading to ferroptosis of cancer cells, we further investigated the application of BT-Br in castration-resistant prostate cancer (CRPC) therapy. The results demonstrated that BT-Br was able to induce endoplasmic reticulum (ER) stress and subsequent autophagy in CRPC DU145 cells, causing the degradation of ferritin heavy peptide 1 (FTH1) thus enhancement of labile iron pool (LIP) levels. Consequently, the dual effects promoted the Fenton reaction and produced highly toxic product hydroxyl radicals (•OH), hence induced ferroptosis in DU145 cells, and eventually reduced CRPC tumors in vivo effectively. To our knowledge, BT-Br is the first small-molecule CAT inhibitor that binds to NADPH-binding site. Our work also reveals that CAT inhibition is potential to be a novel strategy to induce ferroptosis in CRPC cells via dual regulation of ROS levels and iron ions.
2. Materials and methods
2.1. Materials
Bafilomycin A1 (BafA1), Chloroquine and Deferiprone (DFO) were purchased from MedChemExpress. Propidium iodide (PI) was purchased from Sigma Aldrich, USA. Fetal bovine serum (FBS), Trypsin, penicillin/streptomycin, Phosphate buffered saline (PBS), Minimum Essential Medium (MEM), and Dulbecco's modified Eagle's medium (DMEM) were purchased from Gibco, USA. All the tested compounds were dissolved in DMSO as stock solution, and diluted to the expected experimental concentrations containing 5‰ (v/v) DMSO. In each experiment, vehicle control (5‰ (v/v) DMSO) was used as the reference group unless otherwise specified.
2.2. General instruments
1H NMR and 13C NMR was measured by a Mercury-Plus 400 MHz and 600 MHz. Flow cytometry was performed on a BECKMAN CytoFlex Flow Cytometer. Fluorescence images were then acquired with an inverted fluorescence microscope (Olympus IX71, Japan).
2.3. Syntheses of BT-br
-
i)
The intermediate 3-bromo-4-(diethylamino) benzaldehyde (compound 1) was prepared according to the literature [15]. To a mixture of 4-(diethylamino) benzaldehyde and a catalytic amount of NH4OAc in MeCN, NBS was added and the mixture was stirred at room temperature. After completion of the reaction as indicated by TLC, the mixture was concentrated in vacco and extracted with EtOAc-H2O (1:1) (3 × 5 ml). The organic portion was separated from the extract, dried and concentrated. The residue was subjected to column chromatography (silica gel, hexane-EtOAc, 10:1) to obtain pure 1.
-
ii)
A dried 50 mL round-bottom flask was charged with 1 (500 mg, 1.95 mmol), ethanol (10 mL) and hydrazinecarbothioamide (177.7 mg, 1.95 mmol). Then, the reaction mixture was reflux overnight. After completion of the reaction as monitored by TLC analysis, the mixture was cooled to 25 °C and filtered. The filter cake was dried to afford the desired product BT-Br as light yellow solid (385.2 mg, 60%). 1H NMR (600 MHz, DMSO‑d6, δ): 11.39 (s, 1H), 8.20–8.14 (m, 2H), 8.10 (s, 1H), 7.95 (s, 1H), 7.64–7.60 (m, 1H), 7.19–7.13 (m, 1H), 3.10 (q, J = 7.2 Hz, 4H), 0.97 (t, J = 7.2 Hz, 6H); 13C NMR (101 MHz, DMSO‑d6, δ): 178.25, 150.39, 141.13, 131.89, 130.66, 128.02, 124.02, 121.53, 46.39, 12.54.
2.4. Plasmid construction and protein expression and purification
cDNA of Human CAT was purchased from Yingrun Biotechnologies Inc (Changsha, China). For protein expression, the cDNA (NCBI Gene ID: 847) was introduced a NdeI restriction site and a BamHI restriction site, the gene was constructed into a vector pET15b with an N_terminal His tag, the amino acid sequence (UniProtKB P04040). The recombinant plasmids were expressed in BL21(DE3) Escherichia coli cells. The cells were grown at 37 °C in Luria-Bertani broth supplemented with 100 μg mL−1 of ampicillin. Temperature was adjusted to 20 °C when OD600 reached 0.6, and IPTG was added to a final concentration of 0.1 mM to induce the protein expression. After another 12 h, cells were harvested by centrifugation (5000 rpm, 10 min). The pellets were resuspended in buffer (20 mm HEPES, 150 mm NaCl, pH 7.0) and sonicated. The cell-free supernatant was obtained by centrifugation at 13 500 rpm for 45 min. Human CAT was purified in three chromatographic steps. Firstly, the supernatant was loaded onto a Ni2+ metal chelating column (QIAGEN) which was equilibrated with cell resuspended buffer. Human CAT was eluted with 20 mm HEPES pH 7.0, 150 mm NaCl, and 250 mm imidazole buffer. Secondly, the enzyme was loaded onto a Source 15Q column (GE Healthcare) and eluted with 20 mm HEPES pH 7.0 mixed with NaCl linear gradient buffer. Finally, the protein was loaded onto a Superdex 200 column (GE Healthcare) and eluted with 20 mm HEPES pH 7.0, 100 mm NaCl. The purity of protein was checked by SDS/PAGE after each step. Enzyme was stored at −80 °C, and the concentration was determined by BCA Protein Assay Kit (Biosharp).
2.5. Crystallography
Human CAT (8 mg/mL) was incubated with 0.2 mM BT-Br for 1 h on ice before crystallization. The crystal of CAT-BT-Br complex was grown via hanging drop diffusion method under 18 °C. During crystallization, the CAT-BT-Br complex (1.5 μL) was mixed with an equal volume of the reservoir solution [0.1 M MES, pH 6.1, 8% (w/v) PEG20000]. After 3–4 months of crystallization, the crystal was rapidly frozen in liquid nitrogen for data collection. Diffraction data were collected at beamline 18U in Shanghai Synchrotron Radiation Facility (SSRF).
The raw data was processed with XDS [16], and the complex structure was solved by molecular replacement using Phaser [17] with human CAT (PDB accession code: 1DGH). Standard refinement was performed with COOT [18] and PHENIX [19]. The data processing and refinement statistics were summarized in Table S1. The structure related figures were prepared with PyMOL (http://www.pymol.org).
2.6. Computational alanine scanning mutagenesis
Computational alanine scanning mutagenesis was conducted by a combination of molecular dynamics simulation and molecular mechanics/Poisson-Boltzmann surface area (MM-PBSA). The starting structure for the wild type CAT protein in complex with BT-Br was obtained from the crystal structure deposited in the RCSB Protein Data Bank (PDB ID: 8HID). Molecular dynamics simulation for CAT complex was performed using AMBER 16 program. The CAT protein was parameterized with the AMBER ff14 S B force field, and the BT-Br compound was parameterized with the general AMBER force field (gaff). A 5 ns standard production simulation was performed to obtain the equilibrium trajectory. The interfacial residues in CAT protein were identified by a distance criteria, which are within 5 Å of the BT-Br complex. Based on the equilibrium simulation trajectory, we conducted the computational alanine scanning using the MM-PBSA.py tool provided within the AMBER 16 program. Each of the interfacial residues was mutated to alanine residue, and then the changes of binding free energies (ΔΔG = ΔGALA - ΔGwild-type) caused by the alanine substitutions were calculated by MM-PBSA method.
2.7. MST (MicroScale thermophoresis) assay
The purified Human CAT was in HEPES buffer (20 mM HEPES pH 7.0, 100 mM NaCl), and then labeled according to the protocol of Protein labelling kit RED-NHS (Nanotemper, cat. no. L001). All the tested stock solutions (1 mM) were prepared by dissolving BT-1 or BT-Br into DMSO, and then were diluted into the HEPES buffer (20 mM HEPES pH 7.0, 100 mM NaCl) for the MST binding assays. The MST experiment was performed using Monolith NT.115 instrument (NanoTemper Technologies). Labeled proteins (10 μM) were mixed with the indicated concentrations of compounds (BT-1 or BT-Br) in reaction buffer containing 20 mM HEPES pH 7.0, 100 mM NaCl. The Measurements were performed at room temperature and MST power of 40%. The MST data were analyzed by Nanotemper analysis software (v.1.5.41), then the Kd was determined.
2.8. Cell culture and RNA interference
DU145 cells were obtained from Cell Bank of Type Culture Collection of Chinese Academy of Sciences (Shanghai, China), RWPE-1 and MCF-7 cells were obtained from China Center for Type Culture Collection (CCTCC). RWPE-1 cells were cultured in Defined Keratinocyte-SFM (K-SFM) medium supplemented with 1% penicillin/streptomycin, as well as 2‰ Defined Keratinocyte-SFM Growth Supplement (Gibco). DU145 cells were cultured in MEM medium supplemented with 1% penicillin/streptomycin, and 10% FBS. MCF-7 cells were cultured in DMEM medium supplemented with 1% penicillin/streptomycin, and 10% FBS. Cells were cultured at 37 °C in a humidified incubator with an atmosphere of 5% CO2 and 95% air (normoxia) or 1% oxygen, and 5% carbon dioxide. Transient transfections of siRNAs were conducted with the RNAiMAX lipid transfection reagent (Invitrogen, USA), according to the manufacturer's instructions. The siRNA we designed was chemically synthesized by Genepharma (Shanghai, China), and the sequence of the siRNA is GCTACAACGTCAAATTCGA.
2.9. Western blot
DU145 cell lines were seeded (3.5 × 105 cells/well) on 6-well transparent plates (Corning) for 24 h, and then added the BT-Br, after incubation for another 48 h, cells were washed with cold-ice PBS for 2 times and 60 μL ice-cold lysis buffer containing 1% protease and phosphatase inhibitors (Roche). Cells were scraped off after 30 min on ice and centrifuged for 12 000 rpm for 15 min at 4 °C to obtain the protein lysate. The protein extract was denatured at 100 °C bath and analyzed on 10%–15% SDS-PAGE gels. The gels were blotted onto PVDF membrane (Thermo Scientific; 88 585) and blocked with 5% BSA Buffer (5% Albumin Bovine V from Bovine serumin TBST) for 2 h at room temperature. The primary antibodies used for Western blotting were: FTH1 (Bioswamp, PAB30698), GPx4 (Bioswamp, PAB31461), CAT (Bioswamp, PAB30818), CHOP (Proteintech Group, 66 741), eIF2α (Cell Signaling Technology, 9722), phospho-eIF2α (Cell Signaling Technology, 9721) and β-actin (Bioss, bsm-33036). Then, the membranes were infrared with the secondary antibodies. After washing with TBST for three times, blots were scanned on a HRP imaging system.
2.10. Measurement of LIP
Trypsinized cells were washed twice with 0.5 ml of PBS and incubated with 0.05 μM calceinacetoxymethyl ester (AnaSpec, Fremont, CA USA) for 15 min at 37 °C. Cells were then washed twice with 0.5 ml PBS and incubated with or without deferiprone (100 μM) for 1 h at 37 °C. Cells were analyzed using flow cytometer. Calcein was excited at 488 nm, and the fluorescence was measured at 525 nm. The difference in the mean cellular fluorescence with or without deferiprone incubation reflects the quantity of LIP.
2.11. Measurement of ROS and lipid ROS
Total ROS measurement: Cells were treated as indicated, and then 5 μM 2′,7′-dichlorodihydrofluorescein diacetate (H2DCFDA, Life Technologies, Cat# D-399) was added and incubated for 15 min. Excess H2DCFDA was removed by washing the cells twice with PBS. Labeled cells were trypsinized and resuspended in PBS. Oxidation of H2DCFDA to the highly fluorescent 2′,7′-dichlorofluorescein (DCF) is proportional to ROS generation and was analyzed using a flow cytometer (Fortessa, BD Biosciences). A minimum of 10 000 cells was analyzed per condition.
Lipid ROS measurement: Cells were treated as indicated, and then 5 μM C11-BODIPY (Invitrogen, USA) was added and incubated for 15 min. Excess C11-BODIPY was removed by washing the cells twice with PBS. Labeled cells were trypsinized and resuspended in PBS. Oxidation of the polyunsaturated butadienyl portion of C11-BODIPY resulted in a shift of the fluorescence emission peak from ∼590 nm to ∼510 nm proportional to lipid ROS generation and was analyzed using a flow cytometer.
2.12. Assays of CAT activity
Cells were seeded in a 6-well plate at a density of 2 × 105 cells/well in completed MEM (DU145) culture medium. After washing cells with PBS, the corresponding medium mixed with different concentrations of BT-Br was incubated with cells for 48 h. CAT activity was determined using the Catalase Assay Kit (Beyotime Institute of Biotechnology, S0051). The activity was measured by the production of N-(4-antipyryl)-3-chloro-5-sulfonatep-benzoquinonemonoimine, which would be recorded at 520 nm. The protein contents of all samples were measured using BCA Protein Assay Kit (Thermo Fisher; 23 225). Three parallel experiments were performed in each experiment, and the relative activity was normalized using the control group.
2.13. GSH assay
The relative GSH concentration in cell lysates was assessed using a kit from Solarbio (Cat#BC1175) according to the manufacturer's instructions. The measurement of GSH used a kinetic assay in which catalytic amounts (nmoles) of GSH caused a continuous reduction of 5,50 -dithiobis (2-nitrobenzoic acid) to 5-thio-2- nitrobenzoic acid and the GSSG formed was recycled by glutathione reductase and NADPH. The reaction rate was proportional to the concentration of glutathione up to 2 mmol/L. The yellow product (5-thio-2-nitrobenzoic acid) was measured by spectrophotometrically at 412 nm.
2.14. -qPCR
After extracting total RNA using High Pure RNA Isolation Kit (Roche; 11828665001), reverse transcription was performed from 1 μg of total RNA using Transcriptor First Strand cDNA Synthesis Kit (Roche; 04897030001). Finally, quantitative PCR was performed in a Light Cycler 96 (Roche) with Fast Start Essential DNA Green Master (Roche; 06402712001). All reactions were performed as triplicate, and then the quantity of mRNA was calculated by 2-△△Ct method. The following primers were used.
| GAPDH-F ACCATCTTCCAGGAGCGAGATC |
| GAPDH-R TGATGACCCTTTTGGCTCCCC |
| CAT-F ATACCTGTGAACTGTCCCTACCG |
| CAT-R TAGAATGCCCGCACCTGAGTA |
| FTH1-F AGTTTCTCAGCATGTTCCCTC |
| FTH1-R TCTACGCCTCCTACGTTTACC |
| GPx4 –F GAGGCAAGACCGAAGTAAACTAC |
| GPx4 –R CCGAACTGGTTACACGGGAA |
2.15. Transmission Electron Microscope
DU-145 cells were seeded in 6-well plate for 24 h, and then treated with BT-Br (5 μM) for 4 h and 12 h, respectively. The cells were trypsinized, washed with PBS and fixed with glutaric dialdehyde. The samples were stained with Osmium tetroxide before imaging. All the images were obtained by the Transmission Electron Microscope (FEI Tecnai Spirit, USA).
2.16. Animals
Uncastrated Balb/c Nude mice were obtained from Shanghai SLAC Laboratory Animals and Shanghai Sippr-BK Laboratory Animal (China), respectively. Five-to eight-week-old animals were used for the studies and were allowed unrestricted access to food and water. The animal experiments were approved by the Association for Assessment and Accreditation of Laboratory Animal Care International (AAALAC International), and the Care and Use of Laboratory Animals of the Chinese Animal Welfare Committee.
2.17. Xenograft tumor growth
DU145 cells (2.8*106) were mixed with Matrigel (BD Biosciences) injected into the forelimb armpit of nude mice (BALB/c nu, 5–8 weeks old) to generate xenografts. The mice were grouped randomly (n = 5 or 8 per group) and treatment was initiated when the mean tumor volume was approximately 125 mm3. The mice were treated intraperitoneally with 0.9% saline solution containing 0.5% 2-phenylethanol and 1.5% Tween 80 (vehicle), 10 or 20 mg/kg of BT-Br daily for 14 days (the drug was stopped twice, on the second and the third day, respectively, due to the poor state and decreased activity of mice), or 6 mg/kg of CDDP weekly for two weeks. Tumor volume was recorded by the measurement of two perpendicular diameters of the tumors using formula V = length (mm)3 width (mm)3/2, and the body weight of the animal was measured every 3 days. The volumes of tumors treated with the vehicle control and the compounds were compared and p values were determined by a two-tailed Student's t-test. At the completion of the study, the mice were killed and necropsied, and tumor tissue was removed for further studies.
2.18. Statistical analysis
Data analyses and graphing were accomplished using PyMOL software (Version 1.2, Schrödinger, LLC) and GraphPad Prism Software (GraphPad Software Inc.). Animal group sizes were chosen on the basis of extensive previous experience with the animal models used. No data points were excluded from the analysis in any experiment. Details of each statistical test used are included in the figure legends.
3. Results
3.1. Identification of the inhibitory activity and binding site of BT-Br on CAT
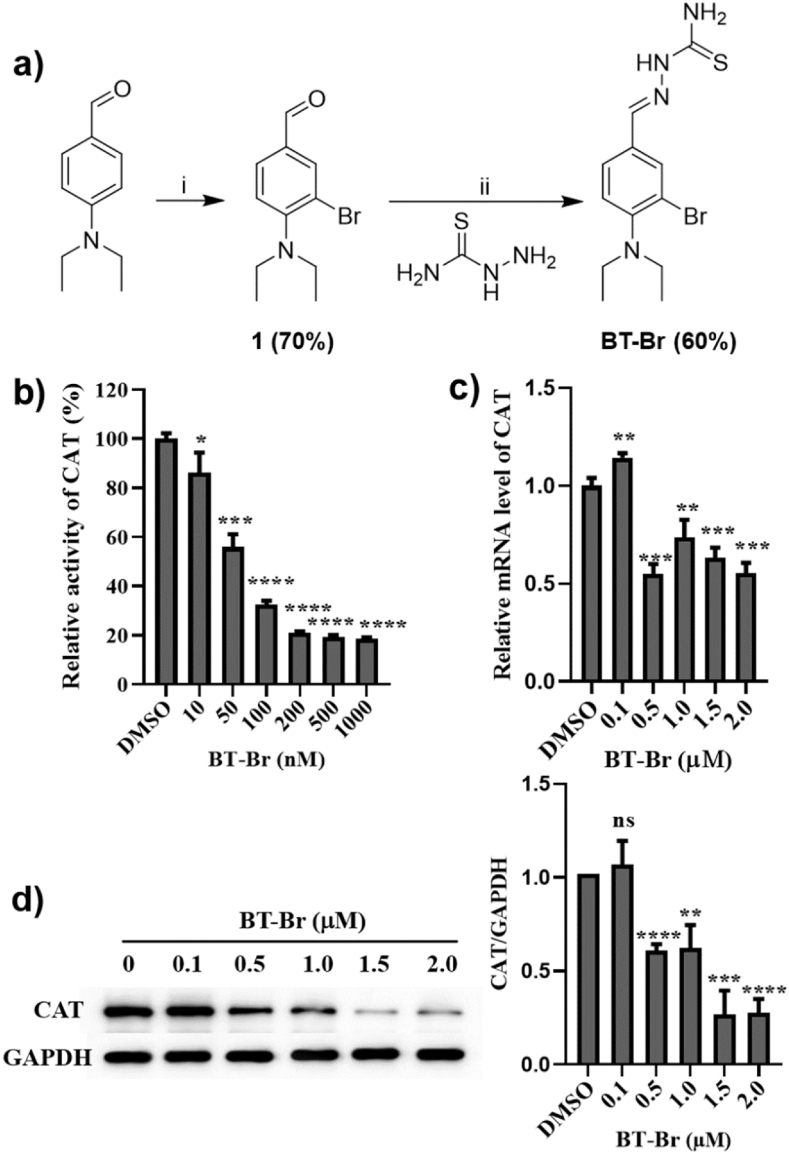
Previously, we reported a CAT inhibitor BT-1 with the non-cyclic structure of 3-AT [20], which displayed dual pro-oxidant effect in cancer cells. During the structural optimization of BT-1, BT-Br with the same skeleton structure was synthesized by a two-step protocol (Fig. 1a, structural characterization data was put in Figs. S1–S3) [15,21]. Then, MST experiment was used to compare the affinity between BT-Br and BT-1 binding to CAT (Fig. S4), and the Kd values were 31.44 ± 9.50 μM and 104.29 ± 47.13 μM, respectively. Furthermore, the inhibitory activity of BT-Br against CAT was determined in PBS buffer, and the IC50 value of BT-Br on CAT was 59.05 ± 5.32 nM (Fig. 1b), which was much better than that of BT-1 (IC50 = 12.55 ± 1.22 μM) [20]. Here, another compound phlorizin was taken as a negative control (Fig. S5), which has been suggested to bind to the NADPH-binding site of CAT with high affinity by docking simulations, but without inhibitory activity on CAT [22].
Fig. 1.
BT-Br acts as an efficient CAT inhibitor in DU145 cells. (a) The synthesis and structural formula of BT-Br. Reagents and conditions are as following: i) NBS, NH4OAc, MeCN, r.t.; ii) EtOH, reflux, overnight. The yields of intermediate and final product are shown in brackets. (b) The inhibition effects of different concentrations of BT-Br on CAT activity in PBS buffer, IC50 = 59.05 ± 5.32 nM. (c) The relative mRNA levels of CAT upon different concentrations of BT-Br in DU145 cells are measured by RT-qPCR. (d) Western blot and the corresponding quantization of CAT/GAPDH ratios in DU145 cells in the presence of different concentrations of BT-Br (0, 0.1, 0.5, 1.0, 1.5, 2.0 μM). The Image J software is used to quantify the grayscale of bands. Error bars represent means ± SD (n = 3); Fig. 1b–d: two-tailed unpaired t-test, *p < 0.05, **p < 0.01, ***p < 0.001, or ****p < 0.0001, ns, not significant.
We analyzed CAT expression in normal and cancerous prostate cells according to the patient data analysis from The Human Proteome Atlas. As shown in Figs. S6a and a summary of normalized single cell RNA (nTPM) displays that CAT mRNA expression level in prostatic glandular cells based on scRNA-seq data from normal tissues is 86.3 nTPM. Furthermore, Fig. S6b shows that the average CAT mRNA expression level of seven prostate cancer cells is 45.6 nTPM. Thereinto, CAT mRNA expression levels are relatively lower in DU145 (23.7 nTPM), NCI–H660 and VCap cells compared with that in LNCaP-clone-FGC, PC-3, 22rv1 and MDA-PCa-2b cells (Fig. S6c), which suggests that prostate cancer cell lines DU145, NCI–H660 and VCap may be more sensitive to CAT inhibitors.
After investigating the cell toxicity of BT-Br in DU145 cells (Fig. S7), we evaluated CAT inhibition activity of BT-Br within DU145 cells. The results showed that both the relative mRNA levels (Fig. 1c) and the protein content of CAT in DU145 cells (Fig. 1d) reduced obviously when treated by different concentrations of BT-Br from 0 to 2.0 μM, which indicated that BT-Br acted as an efficient CAT inhibitor in DU145 cells.
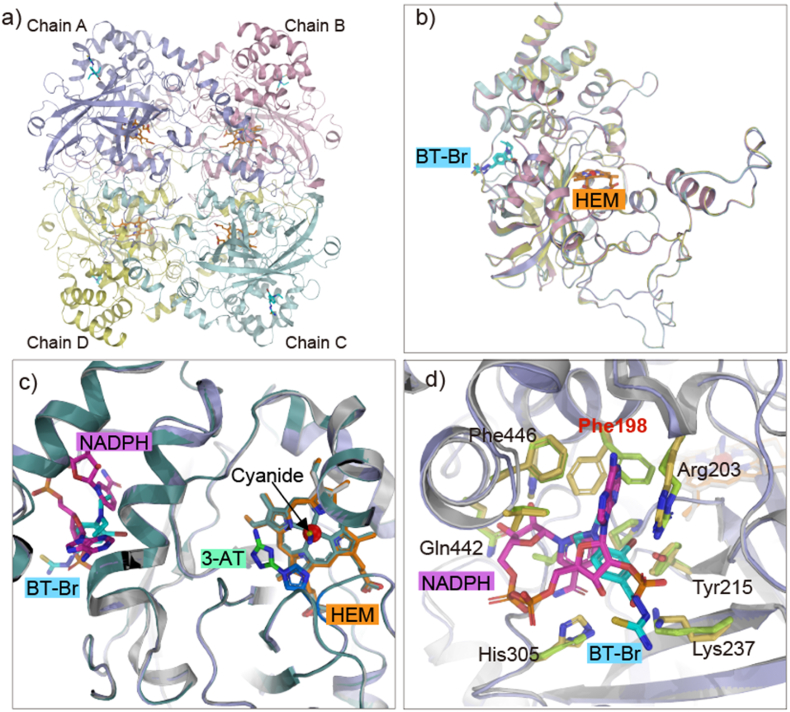
To better understand the binding of BT-Br with CAT, we got pure CAT through plasmid construction and protein expression and purification (Fig. S8), then incubated with BT-Br and obtained the cocrystallization. Finally, the complex structure was solved at a resolution of 2.2 Å (PDB ID: 8HID, Table S1). As shown in Fig. 2, Fig. 3, BT-Br is well defined in the electron density, the binding ratio of CAT to BT-Br is 1:4 (Fig. 2a), exactly the same as Heme or NADPH [1,4]. And the superposition of the four chains presents a high degree of coincidence among the four subunits of CAT-BT-Br complex (Fig. 2b). After comparing with the crystal structure of other CAT-inhibitor complex, such as CAT-3-AT (PDB ID:1DGH) and CAT-cyanide (PDB ID:1DGG) [1], we identified that BT-Br located in the NADPH-binding site of CAT (Fig. 2c). Moreover, we performed a comparison of the surrounding residues bound with NADPH (PDB ID: 1DGH) vs BT-Br based on the two complex crystal structures (Fig. 2d), and found that the conformation of Phe198 changed obviously. Making further superposition of the B chain (without NADPH), A chain (with NADPH) of CAT-3-AT complex (PDB ID:1DGH) and CAT-BT-Br complex crystal structures (Fig. S9), the result displays that the binding of NADPH instead of BT-Br changes the conformation of Phe198 of CAT.
Fig. 2.
Structure comparison of Human CAT bound with different ligands. a) Overall structure of Human CAT complexed with BT-Br (colored in cyan, PDB ID: 8HID). b) Superposition of the four chains in CAT (RMSD values of 0.17 Å to 0.21 Å for alignment with different chains). c) The comparison of binding modes of BT-Br (colored in cyan), NADPH (colored in magenta, PDB ID:1DGH), 3-AT (colored in green, PDB ID:1DGH) and Cyanide (colored in yellow, PDB ID:1DGG) with CAT. The RMSD value of superposition is 0.19 Å between BT-Br bound CAT and 3-AT bound CAT, or 0.20 Å between BT-Br bound CAT and Cyanide bound CAT. d) Conformational change of one key residue upon BT-Br (colored in cyan) or NADPH (colored in magenta) binding. Key binding residues are shown as sticks. RMSD value of the superposition is 0.19 Å. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
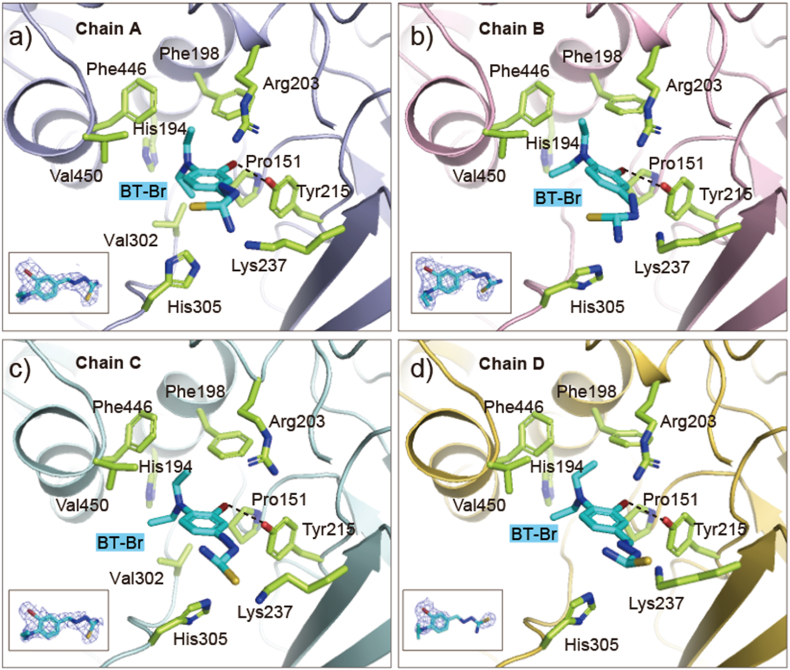
Fig. 3.
The binding of BT-Br with Human CAT. (a–d) The interaction between BT-Br (colored in cyan) and CAT in four chains. Key binding residues are shown as sticks (colored in light green). The 2Fo-Fc omit maps of BT-Br (contoured at 1.0σ) in each chain are also shown. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
In-depth structure analysis (Fig. 3) further illustrates that there is a medium-strength halogen bonding between the Br atom of BT-Br and the OH group of Tyr215, with distance of 2.9 Å. Besides, the hydrophobic interactions between the diethylamino group of BT-Br and the hydrophobic cavity composed by Phe446, Val450 and Pro 151 also contribute to the stability of the complex. Moreover, we have conducted a computational alanine scanning mutagenesis for the interfacial residues of CAT complex with BT-Br to quantify their effects on the binding affinity. As shown in Fig. S10, a total of 15 interfacial residues within a distance cutoff of 5.0 Å were tested. The alanine substitutions of Phe198, Tyr215, Phe446 and Val450 showed unfavorable interaction with BT-Br, which indicated their positive contributions to the binding. To our knowledge, this is the first small-molecule CAT inhibitor that binds to NADPH-binding site, and the analysis mentioned above suggests that the inhibitory activity of BT-Br may be implicated with the absent of NADPH due to the occupation by BT-Br in CAT.
3.2. -Br induces ferroptosis in DU145 cells
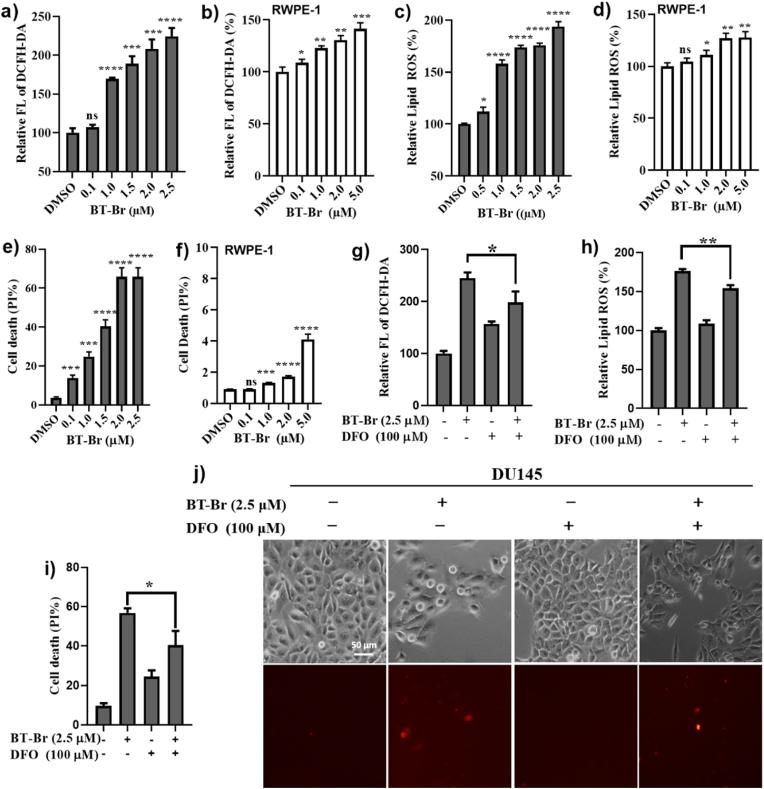
We studied the effects of BT-Br on intracellular ROS levels, lipid ROS and cell death in both DU145 cells and normal prostate cells RWPE-1. Here, the levels of endogenous ROS were evaluated through the fluorescence intensity of the dye 2′,7′-dichlorofluorescein diacetate (DCFH-DA), which was reported extremely sensitive to the changes of cellular redox state and thus used to follow changes in ROS over time [23,24]. Since H2O2 is the major cellular ROS, more intense fluorescence indicates the higher level of H2O2, and thus more potent inhibition on CAT within DU145 cells. When incubated DU145 or RWPE-1 cells with different concentrations of BT-Br from 0 to 2.5 μM for 48 h, the intracellular ROS levels (Fig. 4a), lipid ROS levels (Fig. 4c) as well as cell death of DU145 (Fig. 4e) enhanced gradually in a dose-dependent manner, indicating the effects of ferroptosis induced by BT-Br in DU145 cells. In contrast, the ferroptosis indicators induced by BT-Br, including ROS levels, lipid ROS levels and cell death, were not obvious on RWPE-1 cells (Fig. 4b, d, f).
Fig. 4.
BT-Br induces ferroptosis in DU145 cells. (a–f) Evaluation of the effects of BT-Br on intracellular ROS levels (a, b), the lipid ROS levels (c, d) and cell death (e, f) in DU145 or RWPE-1 cells. Here, the levels of ROS are evaluated through the fluorescent intensity of 2′,7′-dichlorofluorescein diacetate (DCFH-DA). And cell deaths were quantified by propidium iodide (PI) staining coupled with flow cytometry, after incubating cells with different concentrations of BT-Br for 48 h. (g–i) Evaluation of the effects of DFO on the ROS levels (g), lipid ROS levels (h) as well as cell death (i) induced by BT-Br in DU145 cells. (j) Images of DU145 cells treated with BT-Br or BT-Br/DFO respectively, quantified using PI via inverted microscope. The upper panels show the representative cell morphological changes in each group of cells (scale bar = 50 μm). The lower panels show the corresponding cell death in the upper panels. Cell death was analyzed by PI staining. Error bars represent means ± SD (n = 3); Fig. 4a–i: two-tailed unpaired t-test. *p < 0.05, **p < 0.01, ***p < 0.001, or ****p < 0.0001, ns, not significant.
Furthermore, the iron chelator deferoxamine (DFO), a ferroptosis inhibitor in cells [25], was used to confirm the effect of ferroptosis induced by BT-Br. After treated by 2.5 μM BT-Br in DU145 cells, we observed that the ROS levels, lipid ROS levels as well as cell death decreased evidently in the presence of 100 μM DFO (Fig. 4g–i). Besides, the images also showed an increase in the proportion of viable cells when treated with the combination of DFO and BT-Br, in contrast with the use of BT-Br alone (Fig. 4j).
3.3. -Br regulates iron ions via autophagy-mediated degradation of FTH1
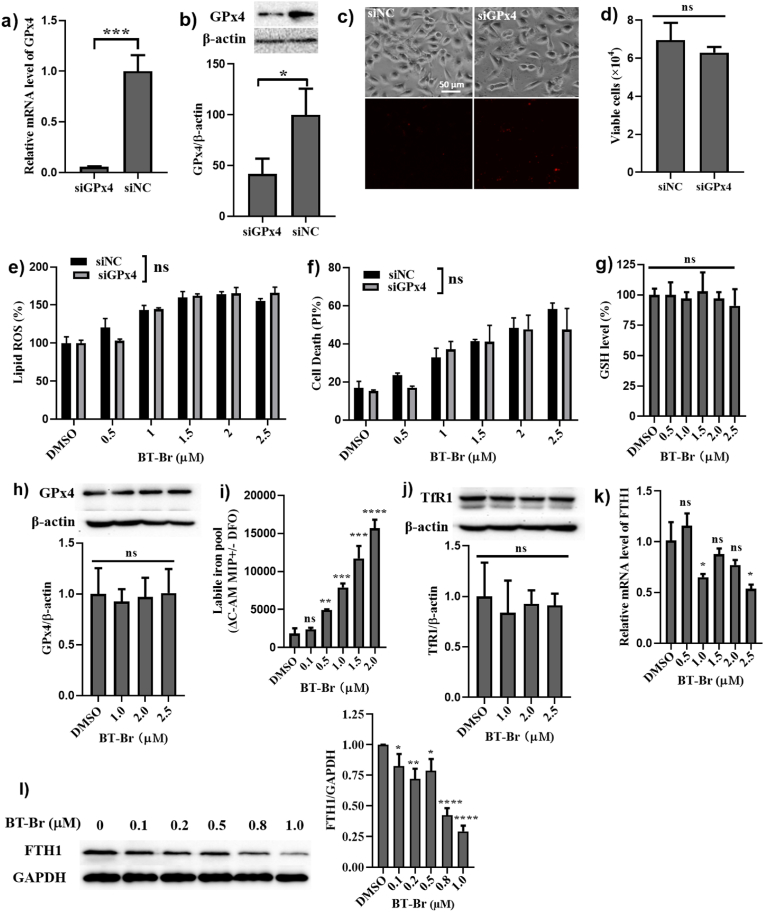
So far, four cellular protective systems have been reported to resist ferroptosis by neutralizing LPO: the cyst(e)ine/glutathione (GSH)/GPX4 axis [26,27], the ferroptosis suppressor protein 1 (FSP1)/coenzyme Q10 (CoQ10, ubiquinol) axis [28,29], the dihydroorotate dehydrogenase (DHODH)/CoQ10 axis [30] and the GTP cyclohydrolase 1 (GCH1)/tetrahydrobiopterin (BH4)/dihydrofolate reductase (DHFR) axis [31]. Thereinto, the initial reported and most classic approach to trigger ferroptosis is inactivation of GPx4 through direct GPx4 inhibitor (such as small molecule RSL3), or indirect inhibition by depletion of its substrate GSH (such as Erastin) [6,32]. To further ascertain whether BT-Br suppresses the activity of GPx4, GPx4 knockdown was first carried out in DU145 cells through siRNA technology. As expected, the data of qPCR and Western blot verified that both the mRNA and protein levels of GPx4 decreased remarkably in siGPx4 DU145 cells, compared with that in siNC DU145 cells (Fig. 5a and b). However, the results of images and detection of viable cells showed that GPx4 knockdown had very litter effect on the proportion of living cells (Fig. 5c and d). Moreover, there were almost no visible differences in both lipid ROS and cell death between siGPx4 and siNC DU145 cells, when treated by the same concentration of BT-Br (Fig. 5e and f). In addition, when incubated DU145 cells with BT-Br for 48 h, the detection results of cellular GSH/GPx4 confirmed that BT-Br neither caused GSH depletion nor suppressed the activity of GPx4 (Fig. 5g and h).
Fig. 5.
BT-Br regulates iron ions via degradation of FTH1 in DU145 cells. (a–d) Evaluation of the effects of GPx4 knockdown on cytotoxicity in DU145 cells through siRNA technology. (a) The detection of GPx4 mRNA levels in siGPx4 and siNC DU145 cells. (b) Western blot and the corresponding quantization of GPx4/β-actin ratios in siGPx4 and siNC DU145 cells. (c) Images of siNC and siGPx4 DU145 cells, quantified using PI via inverted microscope. (d) Detection the viable cells in siNC and siGPx4 DU145 cells. (e, f) Evaluation of the effects of BT-Br on lipid ROS level (e) and cell death (f) in siNC and siGPx4 DU145 cell lines. (g) The relative GSH levels upon different concentrations of BT-Br in DU145 cells are measured by spectrophotometrically at 412 nm. (h) Western blot and the corresponding quantization of GPx4/β-actin ratios in DU145 cells in the presence of different concentrations of BT-Br (0, 1.0, 2.0, 2.5 μM). (i) Detection LIP levels of DU145 cells upon treatment with increasing concentrations of BT-Br from 0 to 2.0 μM. (j) Western blot and the corresponding quantization of TfR1/β-actin ratios in DU145 cells in the presence of different concentrations of BT-Br (0, 1.0, 2.0, 2.5 μM). (k) The relative mRNA levels of FTH1 upon different concentrations of BT-Br in DU145 cells are measured by RT-qPCR. (l) Western blot and the corresponding quantization of FTH1/GAPDH ratios in DU145 cells in the presence of different concentrations of BT-Br (0, 0.1, 0.2, 0.5, 0.8, 1.0 μM). The Image J software is used to quantify the grayscale of WB bands. Error bars represent means ± SD (n = 3); Fig. 5a,b,d,e,f,i,k,l: two-tailed unpaired t-test. Fig. 5g,h,j: one-way ANOVA followed by Dunnett post hoc. *p < 0.05, **p < 0.01, ***p < 0.001, or ****p < 0.0001, ns, not significant.
The ferroptosis-inducing effect of BT-Br on DU145 cells could be attributed to accelerating intracellular Fenton reaction. Thus, we detected the levels of cellular LIP in DU145 cells treated by BT-Br, which is defined as a pool of chelatable and redox-active iron that comprises the entirety of every heterogenous sub-pools of iron [20,33]. The results showed that the levels of LIP rose dramatically after treatment with different concentrations of BT-Br from 0 to 2.0 μM (Fig. 5i). According to the literatures [34,35], the increase of iron ions was probably originated from the degradation of cellular iron-containing protein, especially the transferrin receptor 1 (TfR1) or FTH1. Herein, we detected the influences of BT-Br on the expression of TfR1 and FTH1 within DU145 cells, and the results of Western blot analysis illustrated that the content of TfR1 almost maintained upon addition of BT-Br up to 2.5 μM (Fig. 5j), while both the mRNA and protein levels of FTH1 in DU145 cells decreased obviously when treated by different concentrations of BT-Br (Fig. 5k and l). By contrast, BT-Br had little influence on the expression of FTH1 in RWPE-1 cells (Fig. S11).
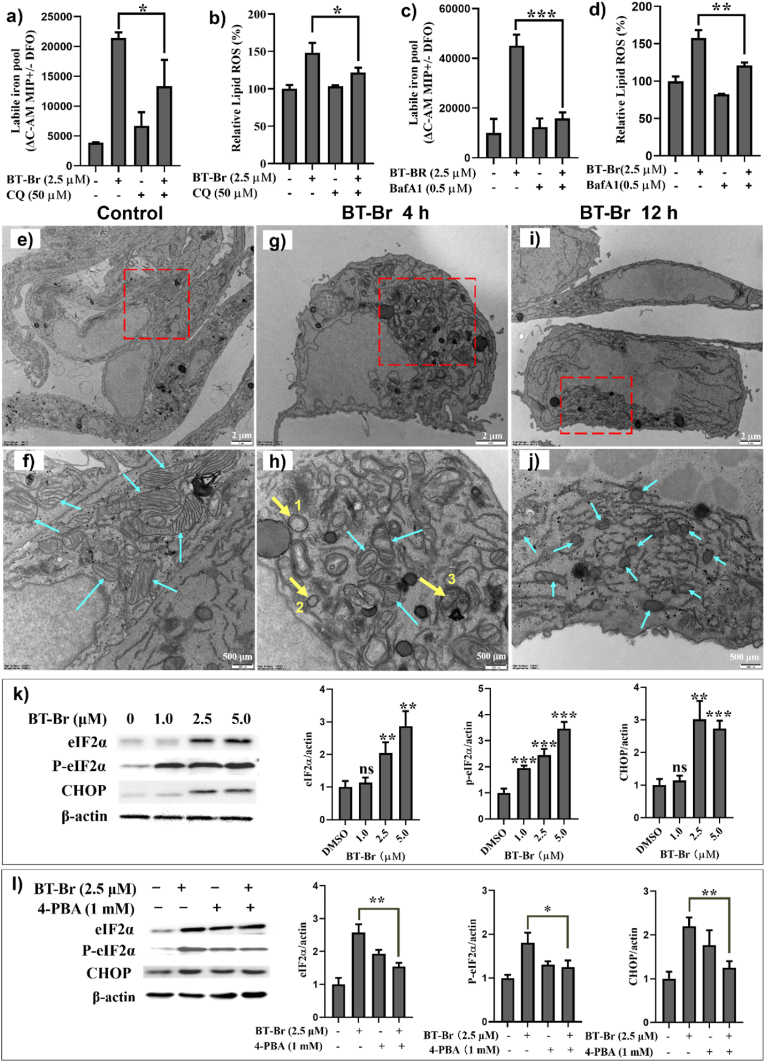
According to the former research, the degradation of FTH1 could be attributed to a cell self-digestive, lysosomal degradation pathway autophagy, which promotes ferrroptosis and can be activated when cells encounter stresses such as nutrient starvation, hypoxia, as well as a wide range of anticancer therapies [[36], [37], [38]]. In order to ascertain whether BT-Br induced autophagy in DU145 cells, the levels of LIP as well as lipid ROS were detected by combined using of BT-Br and chloroquine (CQ) or bafilomycin A1 (BafA1), which were reported to suppress autophagy as lysosomal inhibitors [37,39]. And the results displayed that the addition of CQ (Fig. 6a and b) or BafA1 (Fig. 6c and d) blocked the increase of LIP as well as lipid ROS, suggested the autophagy-dependent effects induced by BT-Br in DU145 cells.
Fig. 6.
BT-Br induces ER stress and subsequent autophagy in DU145 cells. (a–d) Evaluation of the effects of the autophagy inhibitor CQ or BafA1 on BT-Br-induced LIP (a, c) and relative lipid ROS (b, d). (e–j) TEM images of DU145 cells treated with vehicle (1% DMSO) or 5 μM BT-Br (incubated for 4 h and 12 h, respectively). Red rectangles represent the region enlarged, the cyan arrows indicate mitochondria, and the yellow arrows indicate autophagosomes. (k) The expression of ER stress-related proteins eIF2a, P-eIF2a and CHOP in DU145 cells treated with different concentrations of BT-Br (0, 1.0, 2.5, 5.0 μM). DU145 cells were incubated with BT-Br for 24 h. The bar graphs were the corresponding quantization of protein/β-actin ratios to the Western blot images. (l) The expression of eIF2a, P-eIF2a, CHOP (l) in DU145 cells treated with 2.5 μM BT-Br with or without 1 mM 4-PBA. Error bars represent means ± SD (n = 3). Fig. 6a–d,k,l: Two-tailed unpaired t-test. *p < 0.05, **p < 0.01, ***p < 0.001, ns, not significant. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Furthermore, the alternations in morphology of DU145 cells and the formation of autophagosomes upon BT-Br treatment were detected by transmission electron microscopy (TEM) [12,40]. In contrast with the control groups (Fig. 6e and f), the images of cells treated with 5.0 μM BT-Br showed typical ultrastructural characteristics of ferroptosis, including shrunken mitochondria (marked in cyan arrows) and increased density of bilayer membrane [41]. And these characteristics became more obvious when the incubation time extended from 4 h (Fig. 6g and h) to 12 h (Fig. 6i and j). Moreover, apparent autophagosomes were obtained in TEM images after incubation DU145 cells with BT-Br for 4 h (Fig. 6h, marked with yellow arrows and numbers 1, 2, 3), indicating the occurrence of autophagy during the process of ferroptosis induced by BT-Br within DU145 cells.
ER stress is defined as the accumulation of unfolded or misfolded proteins in ER, and caused by conditions such as heat or agents. Since ER stress has been reported to induce an autophagic response in cells [42], we further detected two ER stress-related proteins, eukaryotic initiation factor 2a (eIF2a) and C/EBP homologous protein (CHOP) by Western blot [43], to ascertain whether BT-Br induced ER stress in DU145 cell. As shown in Fig. 6k, after exposure to 0, 1.0, 2.5 and 5.0 μM of BT-Br for 24 h, the expressions of eIF2a, phosphorylation of eIF2a (P-eIF2a) and CHOP were up-regulated in a dose-dependent manner, which were typical signs of ER stress. In addition, DU145 cells were further treated by combined using of BT-Br and 4-phenylbutyric acid (4-PBA), an inhibitor of ER stress [44]. Compared with the control group, the up-regulation of eIF2a, P-eIF2a and CHOP induced by 2.5 μM BT-Br were markedly decreased in the presence of 1 mM 4-PBA (Fig. 6l), confirming that BT-Br induced ER stress in DU145 cells. Therefore, the degradation of FTH1 in DU145 cells could be attributed to the ER stress and subsequent autophagy induced by BT-Br.
3.4. -Br reduces the tumor growth of CRPC in nude mice
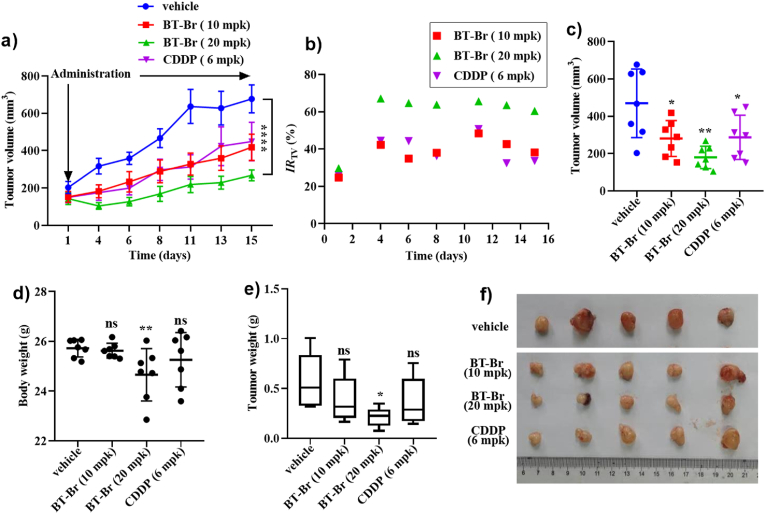
As BT-Br was able to induce ferroptosis in DU145 cells, we further evaluated its anticancer effects on CRPC in vivo. Firstly, DU145 cells were inoculated to 5 ∼ 8-week-old male Balb/c Nude mice and formed transplanted tumors of CRPC. Then BT-Br was administered by way of intraperitoneal injection at daily doses of 10 or 20 mg/kg (mpk), taking 70% PEG400 as vehicle control group and cis-diamminedichloro-platinum (II) (CDDP, weekly administration at the dose of 6 mpk) as the positive control group, respectively. The tumor volumes and body weight of mice were routinely monitored and recorded for 15 days (n = 5 per group). At the end of the experiment, all the mice were killed to get their transplanted tumors to weigh and calculate the inhibitory rate.
Compared with the vehicle control group, the tumor volume of both the two BT-Br groups increased more slowly (Fig. 7a), and the inhibition rate of tumor volume (IRTV) on the 15th day were about 38% for low-doses of BT-Br (10 mpk), up to 60% for high-doses of BT-Br (20 mpk), and 33% for CDDP, respectively (Fig. 7b), indicating better inhibition capacity of BT-Br than CDDP at the given doses. Besides, among the seven monitored data, about half of the tumor volume in vehicle group were over 600 mm3, while it was around 200 mm3 after BT-Br treatment (Fig. 7c). Moreover, almost no significant body weight changed at the end of BT-Br treatment (Fig. 7d), implying that BT-Br possessed low potential systemic toxicity. At the endpoint, the analysis of tumor weight displayed that the inhibition rates of tumor weight (IRTw) were about 24% for low-doses of BT-Br, 58% for high-doses of BT-Br, and 28% for CDDP, respectively (Fig. 7e). It is noteworthy that the significant difference in tumor weight could only be observed between high-doses of BT-Br and vehicle group (Fig. 7e and f). Therefore, we concluded that BT-Br was able to inhibit the growth of CRPC tumors formed by injection of DU145 cells.
Fig. 7.
BT-Br reduces DU145 prostate cancer tumors effectively in vivo. (A) Tumor growth inhibition by BT-Br intraperitoneal injection at 70% PEG400 (Vehicle), 10 and 20 mpk (daily), or CDDP at 6 mpk (weekly). (B) The inhibition rate of tumor volume (IRTV) analysis of BT-Br and CDDP corresponding to the data of a. IRTV (%) = 100 × (CTV-TTV)/CTV. CTV: the tumor volume of vehicle group; TTV: the tumor volume of treatment group. Tumor volumes (C) and body weight (D) analyses, at 1, 4, 6, 8, 11, 13, 15 days post-injection in male Balb/c Nude mice injected with DU145 cells, in Vehicle, BT-Br and CDDP groups, respectively. The analysis of tumor endpoint weight (E) and the tumor endpoint photo (F) at 15 days post-injection in mice upon treatment with 70% PEG400, BT- Br and CDDP, respectively. Error bars represent means ± SEM (n = 5 per group); Fig. 7a: two-way ANOVA followed by Bonferroni post hoc test; Fig. 7c,d,e: two-tailed unpaired t-test. *p < 0.05, **p < 0.01, or ****p < 0.0001, ns, not significant.
4. Discussion
Although CAT is the oldest known and first discovered antioxidant enzyme, the development of efficient CAT inhibitors has made little progress. Except for 3-AT and cyanide [1], few cocrystal structures of CAT-inhibitor complex have been reported so far. In our opinion, targeting new binding site instead of the heme active center of CAT is of great importance for the discovery of novel CAT inhibitors. Here, BT-Br was demonstrated to be the first NADPH-binding site inhibitor with high inhibitory activity against CAT, and the determination of the cocrystal structures showed that BT-Br didn't change the conformations of the surrounding residues around the NADPH-binding site of CAT. In this context, we inferred that the binding of BT-Br may affect the function of NADPH thus inhibits the activity of CAT, although the function of NADPH is not fully understood.
BT-Br was further investigated for CRPC therapy. Here, the results of GPx4 knockdown experiments indicated that the cellular protective systems of LPO neutraliztion were GPx4-independent in DU145 cells. Besides, similar results could be obtained when we carried the same experiments in breast cancer cell line MCF-7 (Fig. S12), which were consistent with the findings reported in the literatures [[28], [29], [30],45,46]. In our study, BT-Br was demonstrated to induce ferroptosis in both DU145 cells and MCF-7 cells (Fig. S13). On one hand, compared with the normal prostate cells RWPE-1, the expressions of both CAT and ferroportin (FPN) are lower in DU145 cells [20], making it more sensitive to CAT inhibitors, and easier for intracellular iron ions to accumulate. On the other hand, the expression of acyl-CoA synthetase long chain family member 4 (ACSL4), a lipid metabolism enzyme identified as a pro-ferroptotic gene, is particularly increased in CRPC relative to hormone naive prostate cancer [47], which enhances tumor growth and proliferation of DU145 cells but also promotes the sensitivity to ferroptosis [[48], [49], [50]]. Therefore, the ferroptosis-inducing strategies possess a distinct advantage in selectivity in CRPC therapy.
The inhibition of BT-Br on CAT in DU145 cells caused the overproduction of H2O2 and the rise of ROS levels, which induced ER stress and subsequent autophagy within DU145 cells, resulting in the degradation of FTH1 and accumulation of iron ions. In this condition, the elevated H2O2 and Fe2+ accelerated the Fenton reaction and then induced ferroptosis in CRPC cells, which eventually reduced CRPC tumors in vivo effectively. Herein, our work reveals that CAT inhibition has potential to be a novel strategy to induce CRPC ferroptosis through dual regulation of ROS levels and iron ions.
Except for the antitumor applications, the effective ferroptosis inducer BT-Br could be used to promote intracellular bacterial suppression when added into macrophages, as the spatiotemporal response to ferroptosis stress is reported as an efficient pathway for macrophage defense against bacterial invasion [51]. On other hand, since heat stress induces an iron-dependent, oxidative cell death process in plants, with biochemical and morphological similarities to ferroptosis [52], BT-Br might be used as a kind of herbicide, inducing ferroptosis stress and regulating plant cell death like the response to heat stress.
Data and materials availability
All data needed to evaluate the conclusions in the paper are present in the main text and/or the Supplementary Materials.
Author contributions
Prof. Guang Fu Yang and Dr. Dan Zhang conceived and designed the study. Dr. Wei Huang and Ming Shu Wang carried out the synthesis. Ya Cao, Yuan Chen, Meng Xu, Prof. Jing Tong, Wen Lin Zhang and Yu Xuan Qin conducted the in vitro assays and in vivo experiments. Dr. Hong Yan Lin, Chi Zhao, Long Can Mei and Jin Dong performed crystallization along with data analysis and in silico molecular docking studies. Dr. Dan Zhang and Prof. Guang Fu Yang wrote the manuscript. All authors reviewed, provided substantive input, and approved the final manuscript. Ya Cao, Yuan Chen and Ming Shu Wang contribute equally.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was financially supported by the National Key Research and Development Program of China (2017YFA0505203), the National Natural Science Foundation of China (Nos. 21877045). The authors also acknowledged the support from 111 Program (B17019) and Chemical Biology Center, College of Chemistry, Central China Normal University.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2023.102751.
Contributor Information
Dan Zhang, Email: danzhang@mail.ccnu.edu.cn.
Guang Fu Yang, Email: gfyang@mail.ccnu.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
Data availability
Data will be made available on request.
References
- 1.Putnam C.D., Arvai A.S., Bourne Y., Tainer J.A. Active and inhibited human catalase structures: ligand and NADPH binding and catalytic mechanism. J. Mol. Biol. 2000;296:295–309. doi: 10.1006/jmbi.1999.3458. [DOI] [PubMed] [Google Scholar]
- 2.Galasso M., Gambino S., Romanelli M.G., Donadelli M., Scupoli M.T. Browsing the oldest antioxidant enzyme: catalase and its multiple regulation in cancer. Free Radical Biol. Med. 2021;172:264–272. doi: 10.1016/j.freeradbiomed.2021.06.010. [DOI] [PubMed] [Google Scholar]
- 3.Glorieuxa C., Calderon P.B. Catalase, a remarkable enzyme: targeting the oldest antioxidant enzyme to find a new cancer treatment approach. Biol. Chem. 2017;398:1095–1108. doi: 10.1515/hsz-2017-0131. [DOI] [PubMed] [Google Scholar]
- 4.Fan H., Wang B., Zhang Y., Zhu Y., Song B., Xu H., Zhai Y., Qiao M., Sun F. A cryo-electron microscopy support film formed by 2D crystals of hydrophobin HFBI. Nat. Commun. 2021;12:7257. doi: 10.1038/s41467-021-27596-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kirkman H.N., Galiano S., Gaetani G.F. The function of cata-lase-bound NADPH. J. Biol. Chem. 1987;262:660–666. doi: 10.1016/0006-291x(89)90811-5. [DOI] [PubMed] [Google Scholar]
- 6.Dixon S.J., Lemberg K.M., Lamprecht M.R., Skouta R., Zaitsev E.M., Gleason C.E., Patel D.N., Bauer A.J., Cantley A.M., Yang W.S., Morrison B., Stockwell B.R. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149:1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stockwell B.R., Friedmann Angeli J.P., Bayir H., Bush A.I., Conrad M., Dixon S.J., Fulda S., Gascon S., Hatzios S.K., Kagan V.E., Noel K., Jiang X., Linkermann A., Murphy M.E., Overholtzer M., Oyagi A., Pagnussat G.C., Park J., Ran Q., Rosenfeld C.S., Salnikow K., Tang D., Torti F.M., Torti S.V., Toyokuni S., Woerpel K.A., Zhang D.D. Ferroptosis: a regulated cell death nexus linking metabolism, redox biology, and disease. Cell. 2017;171:273–285. doi: 10.1016/j.cell.2017.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu B., Chen X.B., Ying M.D., He Q.J., Cao J., Yang B. The role of ferroptosis in cancer development and treatment response. Front. Pharmacol. 2018;8:992. doi: 10.3389/fphar.2017.00992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galluzzi L., Vitale I., Aaronson S., et al. Molecular mechanisms of cell death: recommendations of the nomenclature committee on cell death. Cell Death Differ. 2018;25:486–541. doi: 10.1038/s41418-017-0012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang W.S., Kim K.J., Gaschler M.M., Patel M., Shchepinov M.S., Stockwell B.R. Peroxidation of polyunsaturated fatty acids by lipoxygenases drives ferroptosis. Proc. Natl. Acad. Sci. U. S. A. 2016;113:4966–4975. doi: 10.1073/pnas.1603244113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yin L., Liu P., Jin Y., Ning Z., Yang Y., Gao H. Ferroptosis-related small-molecule compounds in cancer therapy: strategies and applications. Eur. J. Med. Chem. 2022;244 doi: 10.1038/s41467-021-27596-8. [DOI] [PubMed] [Google Scholar]
- 12.Wang W.J., Ling Y.Y., Zhong Y.M., Li Z.Y., Tan C.P., Mao Z.W. Ferroptosis-enhanced cancer immunity by a ferrocene-appended iridium(III) diphosphine complex. Angew. Chem., Int. Ed. 2022;134 doi: 10.1002/ange.202115247. [DOI] [PubMed] [Google Scholar]
- 13.Dong L., Wang C., Zhen W., Jia X., An S., Xu Z., Zhang W., Jiang X. Biodegradable iron-coordinated hollow polydo-pamine nanospheres for dihydroartemisinin delivery and se-lectively enhanced therapy in tumor cells. J. Mater. Chem. B. 2019;7:6172–6180. doi: 10.1039/c9tb01397k. [DOI] [PubMed] [Google Scholar]
- 14.Zhou Z., Song J., Tian R., Yang Z., Yu G., Lin L., Zhang G., Fan W., Zhang F., Niu G., Nie L., Chen X. Activatable singlet oxygen gen-eration from lipid hydroperoxide nanoparticles for cancer therapy. Angew. Chem., Int. Ed. 2017;56:6492–6496. doi: 10.1002/anie.201701181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Das B., Venkateswarku K., Majhi A., Siddaiah V., Reddy K.R. A facile nuclear bromination of phenols and aznilines using NBS in the presence of ammonium acetate as a catalyst. J. Mol. Catal. Chem. 2007;267:30–31. doi: 10.1016/j.molcata.2006.11.002. [DOI] [Google Scholar]
- 16.Kabsch W., XDS Acta Crystallogr. 2010;66:125–132. doi: 10.1107/s0108767317096726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/s2053273317086934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. 2004;60:2126–2132. doi: 10.18520/cs/v114/i11/2362-2368. [DOI] [PubMed] [Google Scholar]
- 19.Adams P.D., Grosse-Kunstleve R.W., Hung L.W., Ioerger T.R., McCoy A.J., Moriarty N.W., Read R.J., Sacchettini J.C., Sauter N.K., Terwilliger T.C. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 20.Chen Y.Y., Liang H.Q., Liu M.T., Huang W., Yi P.F., Yuan S.Q., Mei L.C., Hu Y.H., Cao Y.Y., Hao G.F., Zhang D., Yang G.F. Catalase inhibitors with dual pro‐oxidant effect as new therapeutic agents in castration‐resistant prostate. Adv. Ther. 2021;4:1–13. doi: 10.1002/adtp.202000164. [DOI] [Google Scholar]
- 21.Govindaraj V., Ungati H., Jakka S.R., Bose S., Mugesh G. Directing traffic: halogen-bond-mediated membrane transport. Chem. Eur J. 2019;25:11180–11192. doi: 10.1002/chem.201902243. [DOI] [PubMed] [Google Scholar]
- 22.Kitlar T., Döring F., H Kinne R.K., Deutscher J., F Diedrich D., Frank R., Wallmeier H. Interaction of phlorizin, a potent inhibitor of the Na+/D-glucose cotransporter, with the NADPH-binding site of mammalian catalases. Protein Sci. 1994;3:696–700. doi: 10.1002/pro.5560030417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eruslanov E., Kusmartsev S. In: Armstrong D., editor. vol. 594. Humana Press; Totowa, NJ: 2010. Identification of ROS using oxidized DCFDA and flow-cytometry; pp. 57–72. (Advanced Protocols in Oxidative Stress II. Methods Mol Biol). [DOI] [PubMed] [Google Scholar]
- 24.Halliwell B., Whiteman M. Measuring reactive species and oxidative damage in vivo and in cell culture: how should you do it and what do the results mean? Br. J. Pharmacol. 2004;142:231–255. doi: 10.1038/sj.bjp.0705776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sarma B., Willmes C., Angerer L., Adam C., Becker J.C., Kervarrec T., Schrama D., Houben R. Artesunate affects T antigen expression and survival of virus-positive merkel cell carcinoma. Cancers. 2020;12:919. doi: 10.3390/cancers12040919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang W.S., Sriramaratnam R., Welsch M.E., Shimada K., Skouta R., Viswanathan V.S., Cheah J.H., Clemons P.A., Shamji A.F., Clish C.B., Brown L.M., Girotti A.W., Comish V.W., Schreiber S.L., Stockwell B.R. Regulation of ferroptotic cancer cell death by GPX4. Cell. 2014;156:317–331. doi: 10.1016/j.cell.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Friedmann Angeli J.P., Schneider M., Proneth B., Tyurina Y.Y., Tyurin V.A., Hammond V.J., Herbach N., Aichler M., Walch A., Eggenhofer E., Basavarajappa D., Radmark O., Kobayashi S., Seibt T., Beck H., Neff F., Esposito I., Wanke R., Forster H., Yefremova O., Heinrichmeyer M., Bornkamm G.W., Geissler E.K., Thomas S.B., Stockwell B.R., O'Donnell V.B., Kagan V.E., Schick J.A., Conrad M. Inactivation of the ferroptosis regulator Gpx4 triggers acute renal failure in mice. Nat. Cell Biol. 2014;16:1180–1191. doi: 10.1038/ncb3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bersuker K., Hendricks J.M., Li Z., Magtanong L., Ford B., Tang P.H., Roberts M.A., Tong B., Maimone T.J., Zoncu R., Bassik M.C., Nomura D.K., Dixon S.J., Olzmann J.A. The CoQ oxidoreductase FSP1 acts parallel to GPX4 to inhibit ferroptosis. Nature. 2019;575:688–692. doi: 10.1038/s41586-019-1705-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Doll S., Freitas F.P., Shah R., Aldrovandi M., da Silva M.C., Ingold I., Goya Grocin A., Xavier da Silva T.N., Panzilius E., Scheel C.H., Mourao A., Buday K., Sato M., Wanninger J., Vignane T., Mohana V., Rehberg M., Flatley A., Schepers A., Kurz A., White D., Sauer M., Sattler M., Tate E.W., Schmitz W., Schulze A., O'Donnell V., Proneth B., Popowicz G.M., Pratt D.A., Angeli J.P.F., Conrad M. FSP1 is a glutathione-independent ferroptosis suppressor. Nature. 2019;575:693–698. doi: 10.1038/s41586-019-1707-0. [DOI] [PubMed] [Google Scholar]
- 30.Mao C., Liu X., Zhang Y., Lei G., Yan Y., Lee H., Koppula P., Wu S., Zhuang L., Fang B., Poyurovsky M.V., Olszewski K., Gan B. DHODH-mediated ferroptosis defence is a targetable vulnerability in cancer. Nature. 2021;593:586–590. doi: 10.1038/s41586-021-03539-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kraft V.A.N., Bezjian C.T., Pfeiffer S., Ringelstetter L., Müller C., Zandkarimi F., Merl-Pham J., Bao X., Anastasov N., Kössl J., Brandner S., Daniels J.D., Schmitt-Kopplin P., Hauck S.M., Stockwell B.R., Hadian K., Schick J.A. GTP cyclohydrolase 1/tetrahydrobiopterin counteract ferroptosis through lipid remodeling. ACS Cent. Sci. 2020;6:41–53. doi: 10.1021/acscentsci.9b01063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang W.S., Kim K.J., Gaschler M.M., Patel M., Shchepinov M.S., Stockwell B.R. Peroxidation of polyunsaturated fatty acids by lipoxygenases drives ferroptosis. Proc. Natl. Acad. Sci. U. S. A. 2016;113:4966–4975. doi: 10.1073/pnas.1603244113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kakhlon O., Cabantchik Z.O. The labile iron pool: characterization, measurement, and participation in cellular processes. Free Radical Biol. Med. 2002;33:1037–1046. doi: 10.1016/s0891-5849(02)01006-7. [DOI] [PubMed] [Google Scholar]
- 34.Ma S., Henson E.S., Chen Y., Gibson S.B. Ferroptosis is induced following siramesine and lapatinib treatment of breast cancer cells. Cell Death Dis. 2016;7:230–237. doi: 10.1038/cddis.2016.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cosialls E., El Hage R., Dos Santos L., Gong C., Mehrpour M., Hamai A. Ferroptosis: cancer stem cells rely on iron until "to die for" it. Cells. 2021;10:2981–3003. doi: 10.3390/cells10112981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Theil E.C. Iron, ferritin, and nutrition. Annu. Rev. Nutr. 2004;24:327–343. doi: 10.1146/annurev.nutr.24.012003.132212. [DOI] [PubMed] [Google Scholar]
- 37.Gao M., Monian P., Pan Q., Zhang W., Xiang J., Jiang X. Ferroptosis is an autophagic cell death process. Cell Res. 2016;26:1021–1032. doi: 10.1038/cr.2016.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hou W., Xie Y.C., Song X.X., Sun X.F., Lotze M.T., Zeh H.J., III, Kang R., Tang D.L. Autophagy promotes ferroptosis by degradation of ferritin. Autophagy. 2016;12:1425–1428. doi: 10.1080/15548627.2016.1187366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim H.N., Seo B.R., Kim H., Koh J.Y. Cilostazol restores autophagy flux in bafilomycin A1-treated, cultured cortical astrocytes through lysosomal reacidification: roles of PKA, zinc and metallothionein 3. Sci. Rep. 2020;10:9175. doi: 10.1038/s41598-020-66292-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qiu W., Zhang A.L., Tian Y. Tetrandrine triggers an alternative autophagy in DU145 cells. Oncol. Lett. 2017;13:3734–3738. doi: 10.3892/ol.2017.5897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Doll S., Proneth B., Tyurina Y.Y., Panzilius E., Kobayashi S., Ingold I., Irmler M., Beckers J., Aichler M., Walch A., Prokisch H., Trumbach D., Mao G., Qu F., Bayir H., Fullekrug J., Scheel C.H., Wurst W., Schick J.A., Kagan V.E., Angeli J.P., Conrad M. ACSL4 dictates ferroptosis sensitivity by shaping cellular lipid composition. Nat. Chem. Biol. 2017;13:91–98. doi: 10.1038/nchembio.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yorimitsu T., Nair U., Yang Z.F., Klionsky D.J. Endoplasmic reticulum stress triggers autophagy. J. The function of catalase bound NADPH. 2006;281:30299–30304. doi: 10.1074/jbc.M607007200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang L.L., Guan R.L., Xie L.N., Liao X.X., Xiong K., Rees T.W., Chen Y., Ji L.N., Chao H. An ER‐targeting iridium(III) complex that induces immunogenic cell death in non-small-cell lung cancer. Angew. Chem. Int. Ed. 2021;60:4657–4665. doi: 10.1002/ange.202013987. [DOI] [PubMed] [Google Scholar]
- 44.Yam G.H., Gaplovska-Kysela K., Zuber C., Roth J. Sodium 4-phenylbutyrate acts as a chemical chaperone on misfolded myocilin to rescue cells from endoplasmic reticulum stress and apoptosis. Invest. Ophthalmol. Vis. Sci. 2007;48:1683–1690. doi: 10.1167/iovs.06-0943. [DOI] [PubMed] [Google Scholar]
- 45.Tang D., Chen X., Kang R., Kroemer G. Ferroptosis: molecular mechanisms and health implications. Cell Res. 2021;31:107–125. doi: 10.1038/s41422-020-00441-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chu B., Kon N., Chen D., Li T., Liu T., Jiang L., Song S., Tavana O., Gu W. ALOX12 is required for p53-mediated tumour suppression through a distinct ferroptosis pathway. Nat. Cell Biol. 2019;21:579–591. doi: 10.1038/s41556-019-0305-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu X.Y., Deng F.M., Li Y.R., Daniels G., D X.X., Ren Q.H., Wang J.H., Wang L.H., Yang Y., Zhang V., Zhang D., Ye F., Melamed J., Monaco M.E., Lee P. ACSL4 promotes prostate cancer growth, invasion and hormonal resistance. Oncotarget. 2015;6:44849–44863. doi: 10.18632/oncotarget.6438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li D., Li Y. The interaction between ferroptosis and lipid metabolism in cancer. Signal Transduct. Targeted Ther. 2020;5:108–118. doi: 10.1038/s41392-020-00216-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Doll S., Proneth B., Tyurina Y.Y., Panzilius E., Kobayashi S., Ingold I., Irmler M., Beckers J., Aichler M., Walch A., Prokisch H., Trumbach D., Mao G., Qu F., Bayir H., Fullekrug J., Scheel C.H., Wurst W., Schick J.A., Kagan V.E., Angeli J.P., Conrad M. ACSL4 dictates ferroptosis sensitivity by shaping cellular lipid composition. Nat. Chem. Biol. 2017;13:91–98. doi: 10.1038/nchembio.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kagan V.E., Mao G., Qu F., Angeli J.P., Doll S., Croix C.S., Dar H.H., Liu B., Tyurin V.A., Ritov V.B., Kapralov A.A., Amoscato A.A., Jiang J., Anthonymuthu T., Mohammadyani D., Yang Q., Proneth B., Klein-Seetharaman J., Watkins S., Bahar I., Greenberger J., Mallampalli R.K., Stockwell B.R., Tyurina Y.Y., Conrad M., Bayir H. Oxidized arachidonic and adrenic PEs navigate cells to ferroptosis. Nat. Chem. Biol. 2017;13:81–90. doi: 10.1038/nchembio.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ma R., Fang L., Chen L., Wang X., Jiang J., Gao L. Ferroptotic stress promotes macrophages against intracellular bacteria. Theranostics. 2022;12:2266–2289. doi: 10.7150/thno.66663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Distéfano A.M., Martin M.V., Córdoba J.P., Bellido A.M., D'Ippólito S., Colman S.L., Soto D., Roldán J.A., Bartoli C.G., Zabaleta E.J., Fiol D.F., Stockwell B.R., Dixon S.J., Pagnussat G.C. Heat stress induces ferroptosis-like cell death in plants. J. Cell Biol. 2017;216:463–476. doi: 10.1083/jcb.201605110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.