Abstract
The first component known to recognize and discriminate among potential 5′ splice sites (5′SSs) in pre-mRNA is the U1 snRNP. However, the relative levels of U1 snRNP binding to alternative 5′SSs do not necessarily determine the splicing outcome. Strikingly, SF2/ASF, one of the essential SR protein-splicing factors, causes a dose-dependent shift in splicing to a downstream (intron-proximal) site, and yet it increases U1 snRNP binding at upstream and downstream sites simultaneously. We show here that hnRNP A1, which shifts splicing towards an upstream 5′SS, causes reduced U1 snRNP binding at both sites. Nonetheless, the importance of U1 snRNP binding is shown by proportionality between the level of U1 snRNP binding to the downstream site and its use in splicing. With purified components, hnRNP A1 reduces U1 snRNP binding to 5′SSs by binding cooperatively and indiscriminately to the pre-mRNA. Mutations in hnRNP A1 and SF2/ASF show that the opposite effects of the proteins on 5′SS choice are correlated with their effects on U1 snRNP binding. Cross-linking experiments show that SF2/ASF and hnRNP A1 compete to bind pre-mRNA, and we conclude that this competition is the basis of their functional antagonism; SF2/ASF enhances U1 snRNP binding at all 5′SSs, the rise in simultaneous occupancy causing a shift in splicing towards the downstream site, whereas hnRNP A1 interferes with U1 snRNP binding such that 5′SS occupancy is lower and the affinities of U1 snRNP for the individual sites determine the site of splicing.
Alternative splicing of pre-mRNA is responsible for the production of multiple mRNA and protein products from individual genes. In many cases, different protein isoforms have unique functions, and their production is tightly regulated at the splicing level. Although a common form of alternative splicing involves the omission or skipping of specific exons during splicing, there are many examples of alternative splicing in which two or more alternative 5′ splice sites (5′SSs) compete for joining to a single 3′ splice site. In such cases, both sites may be used ubiquitously, or their use may be stringently regulated. Similarly, the deliberate introduction of a duplicate 5′SS may result in use of either site or both sites, depending on the precise context, including the sequence of the sites, their relative 5′-3′ order, separation, adjacent sequences, or secondary structures (9, 28, 31, 33, 46, 57, 58, 66, 73). Some of these influences reflect the activity of trans-acting factors.
One trans-acting factor is the U1 snRNP, the RNA component of which forms base pairs across the 5′SSs. The strength of base pairing correlates with the choice of 5′SSs in vivo, at least in some circumstances (39, 77, 95), suggesting that a low probability of binding by U1 snRNP and different affinities for various 5′SSs can dictate 5′SS preferences. This idea is consistent with the observation in Saccharomyces cerevisiae that interactions by components of the U1 snRNP with the cap-binding complex or adjacent pre-mRNA sequences can influence 5′SS selection (34, 71). However, this simple explanation is contradicted in mammals by two sets of results: selective splicing with duplicated sites and a poor correlation of splicing preferences with U1 snRNP binding in vitro (24, 58, 65, 66, 92). Ribonuclease H protection assays have shown that two alternative consensus 5′SSs are occupied simultaneously on each molecule of pre-mRNA by U1-dependent complexes at the initial stages of a splicing reaction, even though only one site (the downstream or intron-proximal site) is used for splicing (32). Thus occupation of one site does not affect binding to another. This finding suggests a model in which U1 snRNP particles bind independently to all alternative 5′ splice sites, but the splicing outcome depends on the numbers of sites occupied on each molecule; when the level of binding is low, then whichever site is occupied at a critical point will be used, and the ratio of use depends primarily on the affinity or probability of occupancy at each site (which will depend on the sequence of the site and also other factors, such as the flanking sequences, cap proximity, etc.); at the opposite extreme, when multiple sites are occupied simultaneously by U1 snRNP particles, the downstream site will be used (32, 68). This model offers an explanation for some of the strong polarity effects seen in 5′SS selection, without invoking any inherent polarity in 5′SS complex formation.
After U1 snRNP binding, a 5′SS is incorporated into a commitment complex (47, 60, 61, 78, 93). It is not known whether it is at this stage that the downstream site is selected when U1 snRNP particles are bound to several alternative sites on a molecule of pre-mRNA. After formation of the spliceosome, U1 snRNA is displaced by U6 snRNA (64). Surprisingly, the extent of U6 base pairing to close alternative 5′SSs can determine preferences if there is a sequence nearby to which U1 snRNA can form base pairs (39). Similarly, there are circumstances in which base pairing of U5 snRNA can affect alternative 5′SS preferences (27, 67). However, in neither case is it clear whether the effect is of any significance for the selection of competent alternative sites.
Other factors appear to be able to affect the choice of sites on the basis of the position of the sites rather than their sequences. The first proteins shown to affect splice site selection were the SR proteins (36, 37, 43, 91). These proteins have one or two RNA recognition motif (RRM)-type RNA-binding domains and a characteristic C-terminal domain rich in SR or RS dipeptides (7). They generally stimulate splicing of either constitutive or optional exons (reviewed in references 15, 52, 86), and at least some of their functions are mediated by recognition of specific sequences in the exons (25, 45, 49–51, 59, 72, 74, 75, 82, 84, 85). The SR proteins affect the choice of alternative 5′ splice sites as well, and generally they stimulate use of the downstream alternative site both in vitro (14, 22, 32, 37, 43, 54, 91, 96) and in vivo (16, 17, 62, 76, 87, 88, 94). Only p54, which is the most divergent family member, has never been reported to cause a shift in splicing to the downstream 5′SS but only to the upstream one (94).
The effect of most SR proteins on the selection of alternative 5′SSs raises two important questions: do they, as with the inclusion of exons, require specific recognition of sequences in the pre-mRNA, and do they act via U1 snRNP redistribution on the alternative sites? Although only a few substrates have been tested, some results indicate that the effects of SRp20 (16, 76), SRp40, and SRp55 (91, 92) vary with the substrate. Interestingly, exon enhancer sequences have been identified that do affect the choice of alternative 5′SSs in caldesmon and adenovirus E1a pre-mRNA (11, 31). Based on the model for selection of 5′SSs by U1 snRNP, described above, we predicted that SF2/ASF shifted splicing to the downstream site because it tightened U1 snRNP binding indiscriminately, increasing the proportion of pre-mRNA in which the alternative sites were bound simultaneously. This prediction was confirmed by showing that the protein did increase the number of pre-mRNA molecules in which two alternative 5′SSs were occupied simultaneously in nuclear extract (32). Further support for this model came from experiments showing that SF2/ASF directly promoted U1 snRNP binding to a single 5′SS to form a ternary complex (41, 42). This activity requires the C-terminal RS domain of SF2/ASF (41, 42). However, the domain is not required for alternative splicing activity (14, 16, 87, 96). The implication is that the alternative splicing activity is not mediated by effects on U1 snRNP binding. However, no other mechanisms have been proposed, and it is important to reexamine the possibility that the result depended on the native gel electrophoresis assay used.
The other major class of proteins shown to affect 5′SS selection comprises the hnRNP A/B proteins. hnRNP A1, the best characterized member of the group, has activities including nucleocytoplasmic shuttling (69), a possible role in mRNA export (40, 81), and accelerating RNA annealing (21, 63, 70). The proteins have two RRM-type RNA-binding domains and a C-terminal region rich in arginine and glycine. Like other hnRNP proteins, they bind nascent RNA in vivo (30) and are components of the nonspecific H complex that assembles on exogenous RNA in nuclear extracts (6). The intrinsic specificity with which hnRNP A1 binds RNA is controversial (1, 2, 12); purified hnRNP A1 binds RNA cooperatively (21).
In pre-mRNA splicing, the hnRNP A/B proteins generally promote the use of the 5′-most of two alternative 5′SSs and exon skipping both in vitro and in vivo, counteracting SF2/ASF (17, 53–55, 79, 82, 90). Whether these actions depend in general on the recognition of specific sequences is an open question. In certain cases, it has been shown that hnRNP A1 recognizes specific sites within an exon (19, 29) or in flanking intron sequences (10, 23), as a prelude to exon skipping. However, although the specific sites enhanced the effect of exogenous hnRNP A1 on the use of alternative 5′SSs derived from the hnRNP A1 pre-mRNA itself, they were not essential (10). An extensive analysis of the effect of mutations in hnRNP A1 has shown that the strengths of sequence-specific binding and annealing activities are correlated with the ability of mutants to affect alternative splicing of a β-globin-derived substrate but that the alternative splicing activity is much more sensitive to the mutations, i.e., that these activities of hnRNP A1 are insufficient for modulation of 5′SS selection (55, 56).
In terms of the model based on U1 snRNP binding described above, the effects of hnRNP A1 on 5′SS selection suggest that it might reduce binding of U1 snRNP and thus the likelihood of simultaneous occupancy of alternative 5′SSs. As a result, the use of specific sites would depend on the individual probabilities that they are occupied, i.e., on their affinity for U1 snRNP, rather than their position. Consequently, we have tested the prediction that hnRNP A1 reduces U1 snRNP binding, and we have determined the mechanisms of this effect. We conclude that the effects of both SF2/ASF and hnRNP A1 are consistent with our model and involve modulations of U1 snRNP binding to alternative 5′SSs, shifting the balance between simultaneous and single occupancy of the alternative 5′SSs on each molecule of pre-mRNA.
MATERIALS AND METHODS
Proteins and RNA.
hnRNP A1, SF2/ASF, and their mutants were all prepared as recombinant proteins from Escherichia coli as previously described (14, 26, 44, 54) and dialyzed against buffer D. The proteins had no extraneous tags, except for oligohistidine on the RRM1/RS and RRM2/RS mutant forms of SF2/ASF. U1 snRNP was generously supplied by B. Kastner, C. L. Will, and R. Luhrmann (4). HeLa nuclear extracts were supplied by 4C (Mons, Belgium). C175G and AdML WW transcripts have been described (32, 68); CE1a transcripts were derived from pS10 (23); human β-globin sequences comprising exon 1, intron 1 (IVS-1), and part of exon 2 were transcribed from plasmid 3′D-205 (73). Mutants of AdML WW containing potential target sites for hnRNP A1 were prepared by incorporating the sequence TAGGGCAGGC, from the K-SAM exon of the fibroblast growth factor receptor 2 gene (29), between nucleotides 40 and 41 from the transcription start site (producing Ad 40), between nucleotides 132 and 133 (producing Ad 132), and at both positions (Ad 40/132). Ad 40/132 contains an additional difference at position 125, which is T rather than C.
Splicing and psoralen cross-linking.
AdML transcripts were prepared with m7G caps as previously described (68). The final concentrations of components in the splicing reaction mixtures were 0.4 mM ATP, 17 mM phosphocreatine, 2.7 mM MgCl2, 1.8% (wt/vol) low-molecular-weight polyvinyl alcohol (PVA), 33% nuclear extract, 2.5% (vol/vol) RNasin (Promega), additional 17 mM HEPES-KOH (pH 7.5), 0.005% Tween 20, 33% (vol/vol) buffer D or protein supplements, and AMT-psoralen (HRI) at 0.016 μg/μl. The protein concentrations were as noted in the figure legends. Mixtures were preincubated at 30°C for an hour to permit phosphorylation of SF2/ASF before pre-mRNA was added (38). Cross-linking and quantification were performed as previously described (68), using a PhosphorImager (Molecular Dynamics) and a Cyclone (Canberra Packard) for measurements.
Ribonuclease H cleavage assays.
For kinetic assays, incubations were done routinely in volumes of 15 μl, comprising U1 snRNP, hnRNP A1, or mutant proteins, at concentrations given in the figure legends, C175G pre-mRNA, approximately 66% (vol/vol) buffer D [with 0.1 M KCl or 0.08 M potassium glutamate (8)], 3.2 mM MgCl2, 20 mM phosphocreatine, 1.7 mM ATP, 1.7% (wt/vol) PVA, and 2.7% (vol/vol) Rnasin. In some experiments, 0.05% Nonidet P-40 (NP40) was included (80). These were standard in vitro-binding conditions. Cleavage was initiated typically by addition to all reaction mixtures simultaneously of 5 to 8 μl containing 300 pmol of 14-mer oligodeoxyribonucleotide, 0.05% NP40, and approximately 1 U of RNase H (Pharmacia) in buffer D. Samples of 1 to 1.5 μl were withdrawn simultaneously from the reactions and mixed with 50 μl of proteinase K digestion mixture on ice before incubation, precipitation with ethanol, and gel electrophoresis. Quantification of the extent of cleavage was done with a PhosphorImager, correcting the signals for the numbers of labeled nucleotides in the RNA. The simulations and fitting of reaction kinetics were done with the program KfitSim.
Solid-phase assays.
For immobilization of U1 snRNP in microtiter plates, 10-μl aliquots containing 1 μg of U1 snRNP in the standard in vitro-binding buffer were incubated at 4°C in each well of a high-protein-binding vinyl microtiter plate for 2 to 3 h. The wells were blocked for several hours with 2 mg of acetylated bovine serum albumin (BSA) (Sigma) per ml in buffer D–0.05% NP40, washed with the same buffer, and incubated at 30°C for 30 min in standard in vitro-binding buffer with labeled pre-mRNA, SF2/ASF, or hnRNP A1 at concentrations given in the figure legends and 2 mg of acetylated BSA per ml. The unbound RNA was removed by washing with buffer D–0.05% NP40 (and BSA in some experiments). For the experiment summarized in Fig. 2B (see below), one set of wells was washed three times in 1 min, a second set was washed five times in 5 min, and a third set was washed seven times in 10 min. The residual RNA was measured by scintillation counting. For RNase H cleavage of the 5′ end of U1 snRNA (see Fig. 7A below), the unbound U1 snRNP was removed after blocking, and then 10-μl portions of standard in vitro-binding buffer were added to each well, containing 43 pmol of specific or arbitrary oligonucleotide, 0.6 U of RNase H, and 2 mg of acetylated BSA per ml. After 30 min at 30°C, wells were washed three times, and the RNA and recombinant-protein components were added as above.
FIG. 2.
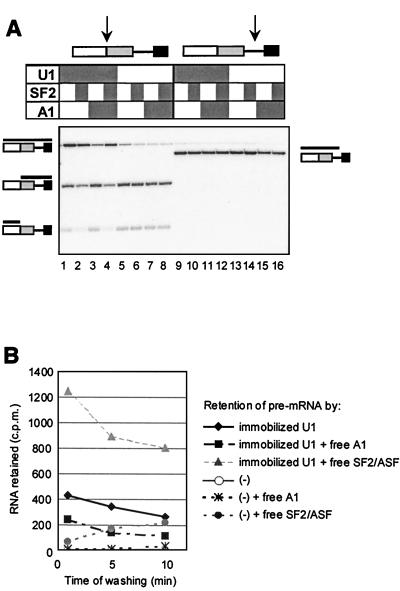
hnRNP A1 reduces binding of U1 snRNP to 5′SSs. (A) Effects of hnRNP A1 and SF2/ASF on U1 snRNP-dependent protection of a consensus 5′SS against RNase H cleavage. C175G pre-mRNA was incubated with RNase H and purified U1 snRNP (0.035 μM), recombinant hnRNP A1 (0.59 μM), or recombinant SF2/ASF (0.24 μM). The components in each reaction are shown by shaded boxes above each lane. Portions of each mixture were incubated for 15 min with one of two cleavage oligonucleotides (sites of cleavage shown by arrows). Reaction mixtures 1 through 8 were incubated with an oligonucleotide complementary to a consensus 5′SS, which is protected by U1 snRNP binding; reaction mixtures 9 through 16 were incubated with an oligonucleotide complementary to an unprotected site. The diagrams at the sides show the substrate, with black bars indicating the structure of the RNA fragment in the corresponding position of the gel. (B) Effects of hnRNP A1 and SF2/ASF on binding of RNA to and dissociation from immobilized U1 snRNP. The labeled RNA retained (cpm) is plotted against the approximate time of washing. Purified U1 snRNP was immobilized in microtiter plate wells (ca. 1 μg per well) and incubated in splicing buffer with 32P-labeled C175G RNA and either hnRNP A1 or SF2/ASF at 0.5 μM. Parallel wells were treated identically but without U1 snRNP [curves labeled (−)].
FIG. 7.
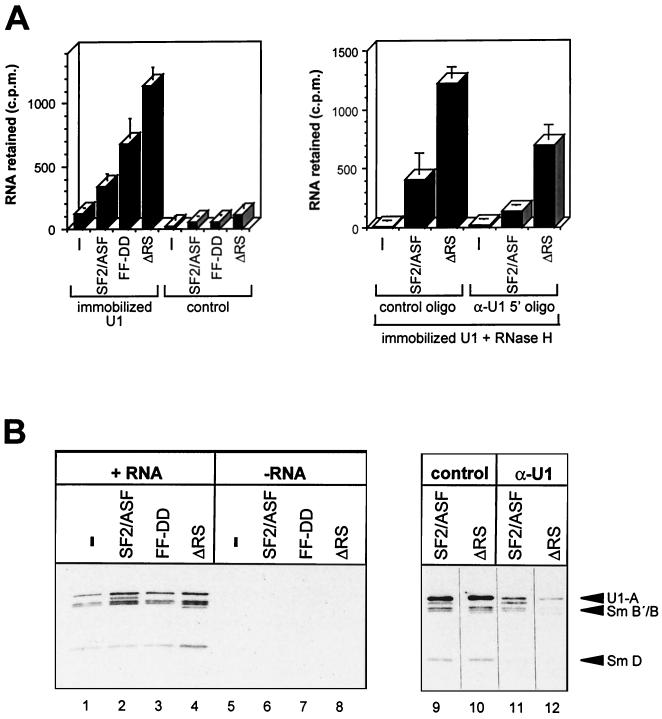
Absence of the RS domain of SF2/ASF does not compromise the enhancement of U1 snRNP binding to 5′SSs. (A) Effects of SF2/ASF mutant proteins on binding of human β-globin IVS-1 pre-mRNA to immobilized U1 snRNP. (Left panel) U1 snRNP was immobilized in microtiter plate wells and, after blocking and washing, incubated with 32P-labeled RNA in splicing buffer in the presence of SF2/ASF proteins at 0.5 μM; reactions were done in triplicate, and the mean value for labeled RNA retained was plotted, with the standard deviations shown; control wells were treated identically, but U1 snRNP was omitted during immobilization. (Right panel) Involvement of the 5′ end of U1 snRNA in the enhancement of RNA binding by SF2/ASF was determined; reactions were done in triplicate, as above, but before addition of the labeled RNA, the wells were treated with RNase H and either an oligonucleotide complementary to the 5′ end of U1 snRNA (α-U1 5′ oligo) or an arbitrary control oligonucleotide. (B) Binding of U1 snRNP to immobilized RNA was examined. (Left panel) U1 snRNP (0.04 μM) and SF2/ASF proteins (0.5 μM) were incubated with streptavidin-coated beads that had or had not been previously bound by biotinylated C175G RNA; bound U1 snRNPs were eluted and detected after SDS-PAGE by Western blotting with anti-Sm antibodies. (Right panel) U1 snRNP was incubated with α-U1 5′ or control oligonucleotides and RNase H before addition to the binding mixtures; U1 snRNP was detected by both anti-Sm and anti-U1-A antibodies; the U1 snRNP proteins detected are labeled; the light additional bands are produced by cross-reactivity with the SF2/ASF proteins.
For recovery of U1 snRNP on immobilized RNA, labeled biotinylated C175G RNA was incubated with M-280 streptavidin Dynabeads (Dynal) in buffer D–0.05% NP40, with 4 mg of acetylated BSA per ml and RNasin. Three-microliter portions of the beads were distributed among wells of a silanized microtiter plate; 15 μl of standard binding buffer containing U1 snRNP (0.2 μg), 2 mg of BSA per ml, and SF2/ASF at 0.5 μM was added to each well. In some cases, U1 snRNA was cleaved by RNase H in the buffer before the addition of SF2/ASF and transfer to the beads. Bound U1 snRNP was detected by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), Western blotting, incubation with anti-Sm and anti-U1 A antibodies, and enhanced chemiluminescence.
U1 snRNP was immobilized on nitrocellulose pieces, 6 mm by 2 mm, by incubation of 0.5 μg of U1 snRNP in 5 μl of standard in vitro-binding buffer (lacking ATP and phosphocreatine) overnight at 4°C. Blocking was done with 5% milk (wt/vol) in buffer D–0.05% NP40–1.3% (vol/vol) RNasin at 4°C for 3 h. After washing, the pieces were incubated with 20 μl of the same standard binding buffer containing the four transcripts and recombinant proteins. After washing, the bound RNA was eluted in 80% formamide–2% SDS at 80°C and loaded on a 5 to 20% discontinuous polyacrylamide denaturing gel containing 0.1% SDS. The electrode buffer (Tris-borate-EDTA) also contained 0.1% SDS.
Protein cross-linking.
The binding reactions in Fig. 6B and C (see below) were done in 5 μl of standard in vitro-binding buffer (without ATP or phosphocreatine). Each sample was irradiated for 30 s with a broad-wavelength UV source (SpotCure, UVP). After RNase treatment, the samples were run on SDS-PAGE and transferred electrophoretically onto nitrocellulose for phosphorimage analysis.
FIG. 6.
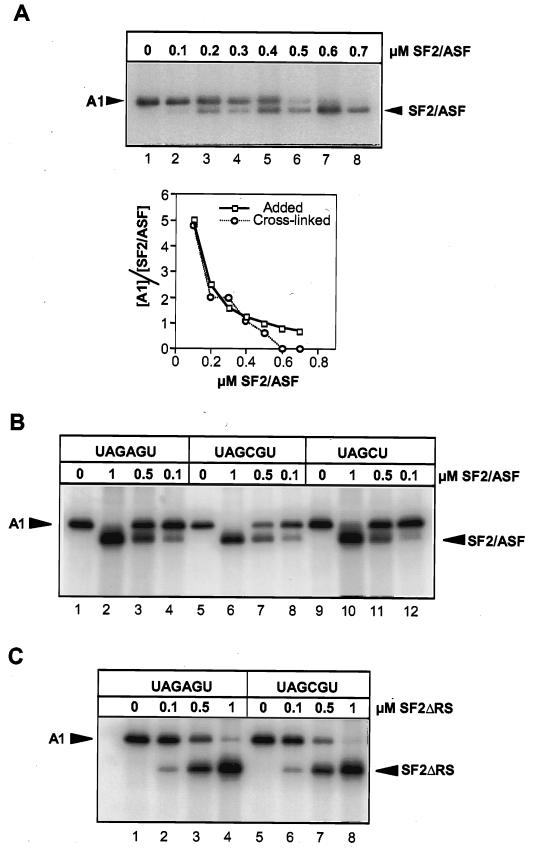
SF2/ASF and hnRNP A1 compete for binding to pre-mRNA. (A) Cross-linking at equilibrium of proteins mixed in various proportions to labeled C175G pre-mRNA. Each 10-μl reaction contained hnRNP A1 at 0.5 μM and SF2/ASF as indicated. After UV irradiation and RNase digestion, samples were analyzed by SDS-PAGE. The right panel shows the ratio of the signals from the cross-linked proteins (y axis) at the different SF2/ASF concentrations, compared with the molar ratios of the proteins added to the RNA. (B) Lack of effect of a high-affinity site for hnRNP A1 on competition for binding. Cross-linking was done with labeled CE1a RNA, containing either a high-affinity site (UAGAGU) or one of two mutant sites (UAGCGU and UAGCU). Reactions were in 5 μl with hnRNP A1 at 0.5 μM. (C) Reactions were done as in panel B but with mutant SF2ΔRS.
Other methods.
Incubation mixtures for nitrocellulose-binding assays were set up with various volumes rather than masses of macromolecules, and the active proportion of RNA was estimated. The analysis of binding to partially hydrolyzed, end-labeled RNA was done as previously described (5), with the difference that the complexes were recovered on nitrocellulose filters, and the RNA was eluted for analysis by gel electrophoresis.
RESULTS
SF2/ASF and hnRNP A1 alter 5′SS preferences, but neither causes selective redistribution of U1 snRNP binding to 5′SS.
To determine whether the effect of hnRNP A1 on alternative 5′SS selection is associated with a general reduction in U1 snRNP binding, splicing reactions were done with an AdML substrate (AdML WW) that has two copies of the wild-type 5′SS (68). Splicing of this substrate in nuclear extract responded as expected to both SF2/ASF and hnRNP A1, with approximately threefold changes in the ratio of use of the upstream and downstream sites (Fig. 1A, lanes 4, 8, and 12, and B).
FIG. 1.
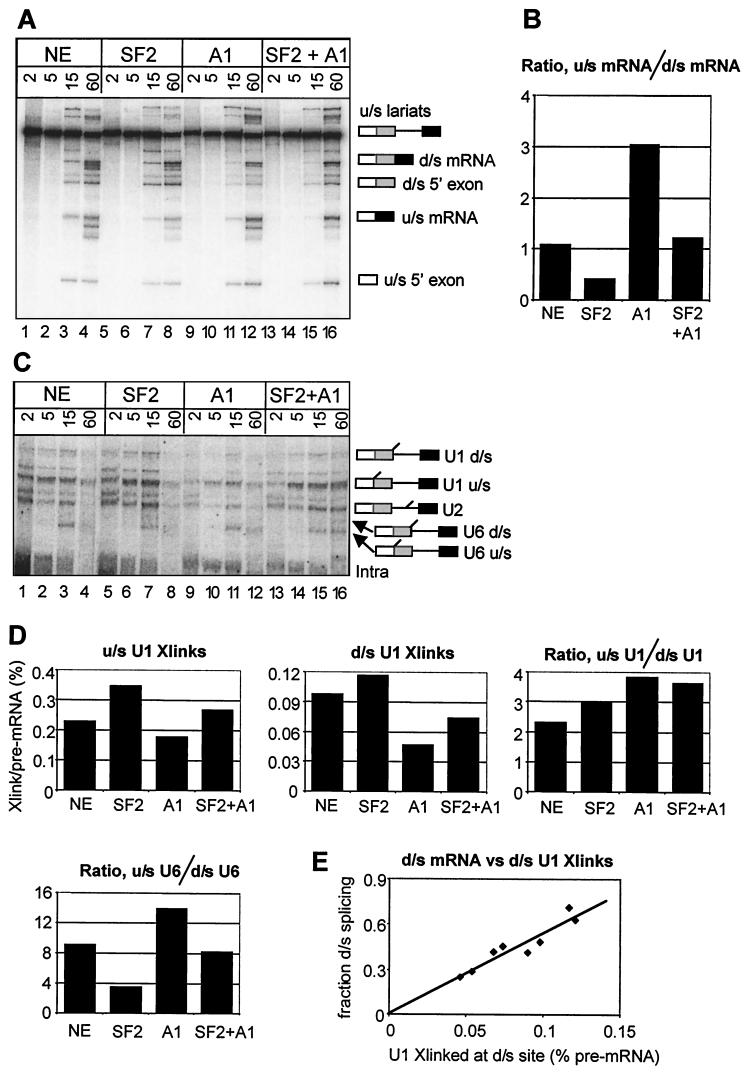
Effects of hnRNP A1 and SF2/ASF on selection of alternative 5′SSs and U1 snRNP binding. (A) Splicing in vitro of AdML WW in nuclear extract (NE) supplemented with SF2/ASF (SF2 [1.2 μM]) or hnRNP A1 (A1 [5.5 μM]) or both (SF2+A1). The splicing reaction mixtures contained AMT-psoralen and were incubated for the times shown (in minutes) above each lane before irradiation and electrophoresis of a portion of each reaction on an 8% denaturing polyacrylamide gel. (B) Ratios of splicing efficiency at the upstream (u/s) and downstream (d/s) 5′SSs. The signals from u/s and d/s mRNA in panel A at 60 min were measured, corrected for label incorporation, and expressed as the ratios of the two isoforms of mRNA produced in each reaction. (C) Analysis of U snRNA cross-links formed in the reactions. Portions of the reactions in panel A were analyzed by electrophoresis on a 5% denaturing polyacrylamide gel. (D) Abundance of the cross-linked adducts. The intensities of the cross-linked U1 snRNA bands at 15 min in panel C are shown as percentages of the pre-mRNA intensities and as a ratio for the two sites; the ratio of the U6 cross-links is plotted also. (E) Correlation between U1 snRNP binding and splicing at the downstream (d/s) site. The graph shows on the ordinate the fraction of splicing to the d/s site at 60 min (d/s mRNA/[u/s mRNA + d/s mRNA]) versus the values for d/s U1 cross-links at 15 min (as in the bar chart). Four points are derived from the experiment shown in panels A and C; four others are derived from an independent experiment (not shown).
The interactions of snRNA with the pre-mRNA substrate in these splicing-reaction mixtures were detected by psoralen-mediated UV cross-linking (Fig. 1C). The bands corresponding to U1 and U6 snRNA cross-links to the alternative 5′SSs have been assigned (68). Their levels in the 15-min reactions (Fig. 1C, lanes 3, 7, 11, and 15) were measured and are shown in Fig. 1D as percentages of the level of the pre-mRNA. Comparisons between the levels of cross-linking at the upstream and downstream sites do not reveal the relative levels of snRNA binding, because there appears to be an intrinsic difference in cross-linking efficiency between the two sites, but changes produced at each site or changes in the relative levels of cross-linking at the sites in response to exogenous protein are informative.
Figure 1D shows that the levels of U1 snRNA cross-linking at both sites were reduced by hnRNP A1. SF2/ASF addition increased cross-linking, as expected (32, 42). Addition of both proteins produced little change. The ratios of U1 snRNA cross-linking at the two sites did not change in accord with splicing preferences (cf. Fig. 1B and D). In contrast, the results for U6 snRNA suggested that its interactions are selectively redistributed according to the pattern of splicing. Our model for the role of U1 snRNP in splice site selection predicted that the proportion of splicing from the downstream site would be proportional to its occupancy by U1 snRNP, whereas splicing from the upstream site would depend on the combined probability that it is occupied and that the downstream one is not. To test the former prediction, we plotted the proportion of splicing from the downstream site at 60 min against the level of U1 cross-linking there at 15 min, combining data from the experiment in Fig. 1A through D with data from a separate but identical experiment (Fig. 1E). The result suggests very strongly that use of the downstream site depends directly on the level of U1 snRNP occupancy (R2 = 0.88). This correlation is in striking contrast to the data for the upstream site; its use decreased when cross-linking at both sites was increased by SF2/ASF and increased when hnRNP A1 caused a reduction in cross-linking.
hnRNP A1 directly reduces U1 snRNP binding to 5′SS.
To test whether the effects of hnRNP A1 are mediated directly, we used an RNase H cleavage assay to measure U1 snRNP binding to consensus 5′ splice sites. The substrate for these assays was C175G, a β-globin pre-mRNA derivative with a consensus 5′SS 175 nucleotides upstream of a natural 5′SS (32). Binding of U1 snRNP at the consensus 5′SS protects the site against cleavage directed by an oligonucleotide complementary to that site. The proportion of uncut (protected) RNA in a reaction was measured. The consensus 5′SS was protected by purified U1 snRNP (Fig. 2A, lane 1 versus lane 5), unlike a control site (lane 9 versus lane 13). The presence of hnRNP A1 caused a marked reduction in protection (cf. lane 3 with lane 1; 62% of the U1-dependent signal was lost), consistent with a loss of U1 snRNP binding or an enhanced rate of dissociation. SF2/ASF reversed this effect (cf. lane 4 with lane 1), suggesting that this simplified system might contain a sufficient number of components to explain the known antagonistic effects of the two proteins. Time-course analyses of RNase H cleavage confirmed that the effect of hnRNP A1 on U1 snRNP-dependent protection could not be caused by enhanced RNase H activity (data not shown; see below).
The effects of hnRNP A1 and SF2/ASF on interactions between U1 snRNP and pre-mRNA were confirmed by a different assay. U1 snRNP was immobilized in microtiter plate wells and incubated with 32P-labeled C175G pre-mRNA in the presence or absence of either protein. The radioactivity retained was measured after washing with buffer for various times (Fig. 2B). hnRNP A1 approximately halved the amount of pre-mRNA retained. Strikingly, SF2/ASF substantially increased the retention of pre-mRNA, confirming the results of other assays (41, 42).
We conclude that hnRNP A1 reduces the binding of U1 snRNP to 5′SSs. The effect is recapitulated by purified components and is therefore likely to be direct.
Mutations in hnRNP A1 that blocked alternative splicing activity also blocked its effects on U1 binding.
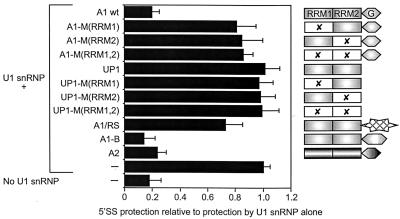
The preceding results showed that hnRNP A1 can reduce the occupancy of 5′SSs by U1 snRNP. It was not clear whether this effect caused the change in splicing patterns, although it occurred when splicing was affected and was predicted by our model. To test whether this property is sufficient for hnRNP A1 to affect alternative 5′SS selection, mutant proteins that lack alternative splicing activity were assayed to determine whether any had retained the ability to reduce U1 snRNP binding. The same mutants have been used to show that the activities of hnRNP A1 in general RNA binding and annealing are not sufficient and may or may not be necessary for alternative splicing function (55). The results of multiple RNase H cleavage assays with purified U1 snRNP are summarized in Fig. 3. All of the hnRNP A1 mutants lost much of their ability to reduce U1 snRNP binding; only one (A1/RS, a chimera of the two RNA-binding domains of hnRNP A1 with the RS domain of SF2/ASF) had any effect, but the reduction was very small compared with that caused by wild-type hnRNP A1. In addition, two other recombinant hnRNP A/B proteins were tested: hnRNP A1B, an alternatively spliced isoform of hnRNP A1, and hnRNP A2, which is encoded by a separate gene and is 70% identical to hnRNP A1. These proteins have effects on 5′SS selection like that of hnRNP A1, although the effect of hnRNP A1B is severalfold weaker (55). Like hnRNP A1, both proteins allowed nearly complete cleavage (cf. absence of U1 snRNP), but separate time course experiments showed that hnRNP A1B and hnRNP B1 (an isoform of hnRNP A2) affected U1 snRNP binding to a lesser extent than did hnRNP A1 and A2 (data not shown). Thus, the results with both the mutant forms of hnRNP A1 and the related proteins were consistent with the hypothesis that the inhibition of U1 snRNP binding may be necessary and sufficient for the effects of hnRNP A1 on alternative splicing.
FIG. 3.
hnRNP A1 mutants that are unable to affect alternative splicing do not block U1 snRNP binding. C175G pre-mRNA was subjected to RNase H digestion at the consensus 5′SS for a fixed time after incubation in the presence of U1 snRNP (0.04 μM) and either hnRNP A1, mutant proteins, or other hnRNP A/B proteins (1.0 μM). The proportion of uncut (protected) RNA in each case is expressed relative to the high proportion protected by U1 snRNP in the absence of hnRNP A1. The values shown are the means of 12 determinations in each case, and the error bars represent the standard deviation. The proteins tested are represented to the right of the chart. The two RRMs (boxed) and the Gly-rich C-terminal domain (hexagon) are shown. X indicates mutations in the conserved RNP-1 submotif of an RRM; A1/RS contains the C-terminal RS domain of SF2/ASF (hatched star) instead of the Gly-rich domain; A1-B is an alternatively spliced variant of A1 with an insertion within the C-terminal domain.
hnRNP A1 blocked U1 snRNP association with the pre-mRNA.
We considered three mechanisms for the effects of hnRNP A1 on U1 snRNP interaction with 5′SSs: (i) the protein might bind to the RNA and prevent U1 snRNP binding, (ii) it might bind U1 snRNP and prevent it from binding, or (iii) it might interact with U1 snRNP-RNA complexes and stimulate U1 snRNP dissociation.
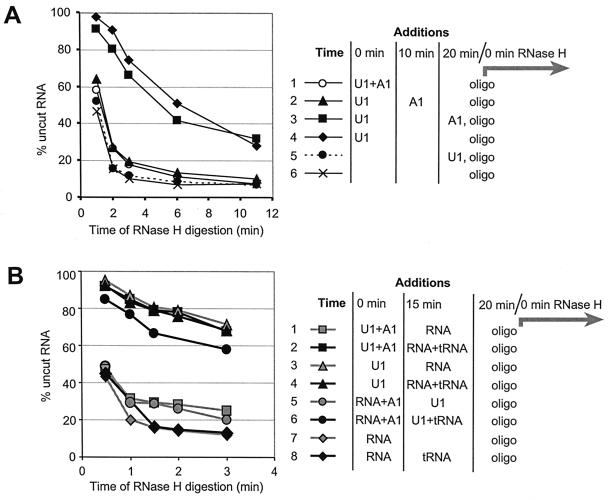
We distinguished between mechanisms in which hnRNP A1 might prevent U1 snRNP binding (i and ii) and the mechanism in which it might stimulate dissociation (iii) by measuring the rate of U1 snRNP dissociation via time courses of digestion at a 5′SS by RNase H. The reaction can be modeled as two first-order steps (slow U1 snRNP dissociation and fast oligonucleotide annealing/cleavage), provided that the steps are irreversible and that the two steps differ sufficiently in rate. Irreversibility was tested as shown in Fig. 4A by the simultaneous addition of U1 snRNP and oligonucleotide (Fig. 4A, curve 5). This procedure produced a cleavage curve almost identical to that produced in the absence of U1 snRNP (curve 6). Thus, U1 snRNP could not bind during cleavage, and its dissociation would be irreversible under these conditions. Furthermore, when cleavage was initiated after U1 snRNP was incubated with the RNA for 20 min (curve 4), the rate of cleavage was much slower. The rate constant for U1 snRNP dissociation in a number of experiments was calculated to be approximately 0.1 min−1, and for RNase H cleavage it was typically around 1.2 min−1. The rate constant for U1 snRNP dissociation was confirmed by a separate method, in which U1 snRNP was incubated with C175G RNA in the presence of psoralen; fresh binding was then blocked by the addition of an oligonucleotide complementary to the 5′ end of U1, and dissociation was followed by irradiation of aliquots at intervals with 1-min pulses of UV light. A similar value was obtained (data not shown). We concluded that the two steps are irreversible and differ significantly in rate, satisfying the conditions for kinetic analysis.
FIG. 4.
HnRNP A1 does not facilitate U1 snRNP dissociation from 5′SSs or form stable complexes with U1 snRNP or pre-mRNA. Time courses for oligonucleotide-directed RNase H cleavage at the consensus 5′SS in C175G are shown, with the proportion of uncut RNA plotted against the time of RNase H digestion. Components were incubated for some or all of a 20-min period, as shown at the right of the time courses, before RNase H cleavage was initiated by addition of the oligonucleotide complementary to the 5′SS. (A) Effect of hnRNP A1 on U1 snRNP dissociation rates. U1 snRNP was present at 0.04 μM, and hnRNP A1 was present at 1 μM. HnRNP A1 was added with the U1 snRNP (curve 1), after 10 min (curve 2) or just before the cleavage oligonucleotide was added (curve 3). U1 snRNP was added with the oligonucleotide in one sample (curve 5). In this case, RNase H cleavage was unaffected by the addition, indicating that the dissociation of U1 snRNP was irreversible once the oligonucleotides were added. (B) Addition of unlabeled RNA to test whether hnRNP A1 forms stable complexes with pre-mRNA or U1 snRNP. The components were added for the times shown in the diagram. “RNA” is pre-mRNA; “tRNA” is yeast tRNA. The final concentrations, before addition of the oligonucleotides, were as follows: U1 snRNP, 0.04 μM; hnRNP A1, 0.5 μM; yeast RNA, 20 ng/μl.
To test whether hnRNP A1 affected the rate of dissociation, U1 snRNP was allowed to bind to RNA for 20 min, and hnRNP A1 was added at various times during this period before initiation of RNase H cleavage of the 5′SSs by addition of the oligonucleotide. We observed that hnRNP A1 had little effect on protection by U1 snRNP if added just before the cleavage oligonucleotide (Fig. 4A, curve 3), although it almost eliminated protection if added before the oligonucleotide (Fig. 4A, curves 1 and 2). We conclude that hnRNP A1 has no significant effect on the rate of dissociation of U1 snRNP but blocks its binding. In other experiments, hnRNP A1 was shown to have a small effect if added 2 min before the oligonucleotide (data not shown), the magnitude of this effect being approximately as expected if it is assumed that hnRNP A1 blocks rebinding by the proportion of U1 snRNP that dissociates during the 2-min period. We conclude that hnRNP A1 affects the binding rather than the dissociation of U1 snRNP, and it is likely that this effect involves either hnRNP A1 binding to the RNA or to U1 snRNP but not specifically to the complex of the two.
hnRNP A1 did not block U1 snRNP binding by forming stable complexes.
Chase experiments were done to test whether hnRNP A1 formed stable complexes with either the pre-mRNA (RNA) or U1 snRNP. Pure hnRNP A1 was preincubated with either of these components, and then the missing component was added with yeast tRNA for a short time before RNase H cleavage was initiated. The yeast tRNA had no effect on U1 snRNP binding (Fig. 4B, curve 4; cf. curve 3) but blocked the effect of hnRNP A1 (Fig. 4B, curve 2 versus curve 1). If hnRNP A1 formed stable complexes with either component, before the free hnRNP A1 was sequestered by the tRNA, then the hnRNP A1 effect would persist. When hnRNP A1 was preincubated with U1 snRNP, the carrier tRNA eliminated the hnRNP A1 activity (curve 2), and we inferred that the effect of hnRNP A1 is not mediated via stable complexes with U1 snRNP. A small effect was seen in some experiments when hnRNP A1 was preincubated with C175G RNA (Fig. 4B, curve 6), but it was not consistently observed. The reason for this variability may be that the interactions of hnRNP A1 with RNA are very labile (UV laser cross-linking showed that the bulk of hnRNP A1 dissociates from RNA in seconds [I. C. Eperon and O. Makarova, unpublished data).
hnRNP A1 competed with U1 snRNP for binding to pre-mRNA.
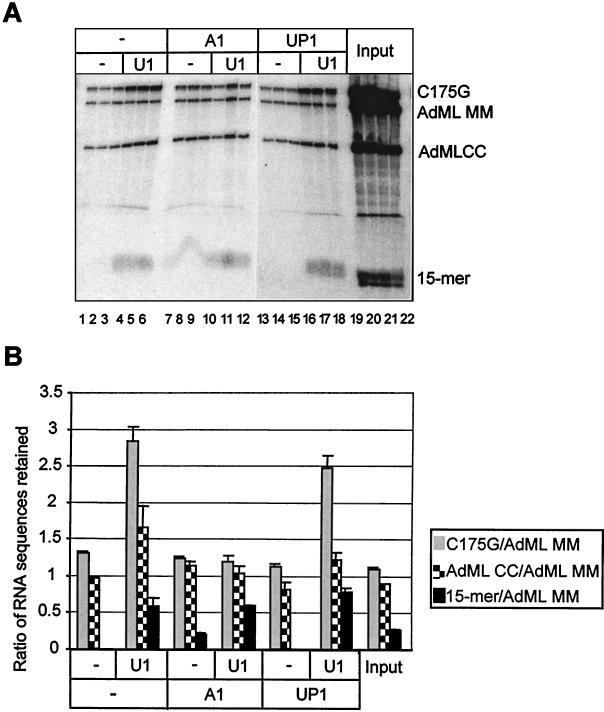
Definitive evidence in favor of mechanism i, direct competition between hnRNP A1 and U1 snRNP for binding to the pre-mRNA, could be achieved by demonstrating that the effect of hnRNP A1 depends on the length of the RNA. If hnRNP A1 formed an inactive complex with the snRNP, the length of the target RNA would be irrelevant; if hnRNP A1 acted by binding to the RNA, then, because binding to RNA is highly cooperative (21), it ought to be much less effective at interfering with U1 snRNP binding to 5′SSs on short RNA sequences. This prediction was tested using as substrates C175G and a short RNA of 15 nucleotides containing a consensus 5′SS. Preliminary experiments with both psoralen cross-linking and RNase H assays showed that U1 binding to the shorter RNA was scarcely affected by hnRNP A1. However, to eliminate variables it was necessary to test both substrates in the same reaction and analyze the outcomes simultaneously. Several different RNA sequences were incubated simultaneously with U1 snRNP immobilized on nitrocellulose in the presence or absence of hnRNP A1. The bound RNA was eluted and analyzed by electrophoresis on a discontinuous denaturing gel. One of the RNA sequences contained no high-affinity binding sites for U1 snRNP (AdML MM), and thus the recovery of the other sequences could be expressed relative to it. Results are shown in Fig. 5A and quantified in Fig. 5B. U1 snRNP immobilized on nitrocellulose retained preferentially the three RNA sequences containing 5′SSs (Fig. 5A, lanes 4 through 6 [cf. lanes 1 through 3]). The inclusion of hnRNP A1 blocked the U1-dependent retention of the two longer RNA sequences, C175G and AdML CC, but the binding of the short RNA was unaffected (lanes 10 through 12 [cf. lanes 4 through 6]). The UP1 fragment of hnRNP A1 was used as a control for nonspecific effects of RNA-binding proteins. UP1 has the RNA-binding domains of hnRNP A1 but no C-terminal domain, and it was shown earlier to have no effect on U1 snRNP binding to 5′SS (Fig. 3). It had a smaller effect than hnRNP A1 in this assay (Fig. 5A, lanes 16 through 18 versus 10 through 12). Thus, hnRNP A1 acts more effectively to block U1 snRNP binding to 5′SSs in long RNA substrates; this observation excludes mechanism ii, in which hnRNP A1 would bind directly to U1 snRNP and sequester it, and it also shows that hnRNP A1 does not act primarily by recognition of and binding to 5′SSs. Instead, we infer that hnRNP A1 blocks U1 snRNP binding as a consequence of binding to RNA either indiscriminately or at specific sites away from the 5′SSs.
FIG. 5.
HnRNP A1 does not reduce binding of short 5′SS RNA to immobilized U1 snRNP. Pieces of nitrocellulose were incubated with or without 0.5 μg of U1 snRNP. After blocking and washing, the filters were incubated in splicing buffer with four RNA sequences and either hnRNP A1, UP1 (both at 0.5 μM), or no protein. Reactions were done in triplicate. After washing, the RNA was eluted. (A) Analysis of the RNA on biphasic denaturing gels of 5 and 20% polyacrylamide. AdML MM and AdML CC were derived from AdML WW by mutation of the 5′SSs to respectively prevent recognition or produce consensus sequences (68). (B) Quantification of the results in panel A. The band intensities were expressed relative to the intensity of the AdML MM RNA in each lane, and the mean was determined for each set of triplicates. The standard deviations are shown.
If hnRNP A1 were to require specific sequences other than 5′SSs for its effect on U1 snRNP binding, the most probable reason would be that sites for which it had high affinity nucleated cooperative binding. We tested this by measuring hnRNP A1 binding to an RNA substrate known as CE1a, a portion of the hnRNP A1 pre-mRNA that contains a site to which hnRNP A1 binds specifically in nuclear extracts (23). CE1a-based substrates were end labeled, subjected to partial hydrolysis, and incubated with hnRNP A1. The bound RNA molecules were recovered by nitrocellulose filtration and analyzed by gel electrophoresis. Molecules of all lengths were retained, including those that lacked the high-affinity site (data not shown). We conclude that a specific site is not necessary for binding. Thus, cooperative, nonspecific binding of hnRNP A1 to the RNA is the most likely mechanism by which it interferes with the binding of U1 snRNP at 5′SS.
Molecular mechanisms for the antagonistic effects of SF2/ASF and hnRNP A1 on alternative 5′SS selection.
SF2/ASF enhances U1 snRNP binding, and its effects are antagonized by hnRNP A1. The enhancement may require initial interactions with the pre-mRNA and then with U1 snRNP (41, 42). Since hnRNP A1 does not interact with SF2/ASF (20) or U1 snRNP (I. C. Eperon, unpublished observations), it is probable that its antagonistic effects arise from its binding to pre-mRNA. The mechanism of action proposed in the previous section for hnRNP A1 suggests two simple possibilities: SF2/ASF might facilitate the binding of U1 snRNP by binding to the RNA at a small number of specific sites, without serious disruption of the hnRNP A1 complex, or it might bind more generally, displacing hnRNP A1 and perhaps opening up any interfering secondary or protein-mediated structures in the RNA. These alternatives were tested with purified proteins.
The binding parameters of the individual proteins to C175G were studied using RNase H protection, nitrocellulose binding, and UV cross-linking. Incubation with SF2/ASF protected pre-mRNA against RNase H at all sites tested (data not shown), consistent with data from RNase T1 protection (41). Nonspecific inhibition of enzyme activity was ruled out by controls in which the oligonucleotide was preincubated with the RNA and the enzyme was preincubated with SF2/ASF. Binding affinities were measured by UV cross-linking. The fractional saturation was determined and used to prepare a Hill plot. The plot was nonlinear for SF2/ASF, with half saturation at approximately 6 × 10−8 M protein (data not shown). This result suggests that SF2/ASF binds to a range of sites with different affinities. For hnRNP A1, the Hill plot was linear, with n > 1, consistent with the expectation of positive cooperativity, but half saturation was reached at about the same concentration as with SF2/ASF. Chase assays (addition of unlabeled RNA to equilibrium binding mixtures, followed by UV cross-linking at various times [data not shown]) showed that most of the hnRNP A1 dissociated within 10 s, and dissociation was complete by 30 s. SF2/ASF dissociated more slowly, and about 30% of the complex could not be exchanged.
If SF2/ASF binds to only a limited set of specific sites, while hnRNP A1 binds promiscuously as described above, then both proteins in a mixture of the two should bind RNA over a broad range of relative concentrations. This prediction was tested by UV cross-linking of C175G RNA (Fig. 6A). Increasing concentrations of SF2/ASF displaced hnRNP A1, showing that the two proteins bound competitively at all cross-linkable sites.
To test whether a binding site with high affinity for hnRNP A1 alters the outcome, the experiment was repeated with CE1a sequences (23). Two variants containing mutations known to affect the specific hnRNP A1-binding site were tested in parallel. The results showed that the site had little or no effect; 0.5 μM hnRNP A1 predominated compared with 0.1 μM SF2/ASF, but 1 μM SF2/ASF replaced hnRNP A1 almost completely (Fig. 6B). To exclude artifactual aggregation of the RNA with the unphosphorylated RS domain, the experiment was repeated with SF2/ASF lacking the RS domain. However, the result was the same (Fig. 6C). Thus, over a rather narrow concentration range, SF2/ASF completely replaced hnRNP A1. We conclude that there are two aspects to the mechanism of action of SF2/ASF; binding to the pre-mRNA both antagonizes hnRNP A1 binding and facilitates U1 snRNP binding.
The RS domain of SF2/ASF was not required for the enhancement of U1 snRNP binding to 5′SSs.
If SF2/ASF affects the polarity of 5′SS selection by strengthening U1 snRNP binding to both sites, then mutants of SF2/ASF that affect alternative splicing should still affect U1 snRNP binding. Gel mobility assays indicated that this prediction may not hold for proteins lacking the RS domain (41, 42). Because binding assays based on gel electrophoresis may be affected by the net charge of the protein, this issue was reexamined with different assays. Purified U1 snRNP was immobilized in microtiter plate wells and incubated with 32P-labeled human β-globin pre-mRNA, comprising the first intron and flanking sequences. This substrate was used because the level of U1 snRNP binding to the weak 5′SS was low, which increased the proportional response to SF2/ASF. The results (Fig. 7A) showed that the levels of RNA binding to the U1 snRNP were enhanced considerably by the presence of SF2/ASF and even more so by mutant derivatives that are active in alternative splicing: FF-DD, in which two phenylalanine groups in the RNP-1 motif in the first RNA-binding domain were changed to aspartate, and ΔRS, in which the C-terminal RS domain was deleted (14). It was possible that this effect was caused by RNA binding to the protein which, in turn, had bound to U1 snRNP, and that it did not result from the enhancement of U1 snRNA base-pairing interactions with the β-globin RNA. To test this possibility, immobilized U1 snRNP was treated with oligonucleotides and RNase H before incubation with the labeled RNA. As shown in Fig. 7B, the level of RNA bound was halved when the oligonucleotide was complementary to the 5′ end of U1 snRNA. It is probable that at least half of the stimulation in binding seen in the presence of SF2/ASF and the mutant proteins was produced by an enhancement of U1-5′SS interactions. A further test was done by binding biotinylated C175G RNA to streptavidin-coated beads, followed by incubation with U1 snRNP, recovery of the bound material, and detection of the U1 snRNP polypeptides by Western blotting. The results show that binding of U1 snRNP was enhanced by SF2/ASF and mutant proteins (Fig. 7B). Most of the binding was eliminated if the U1 snRNP was treated beforehand with RNase H and an oligonucleotide complementary to the U1 snRNA but not if it was treated with a control oligonucleotide. Additional controls were done with two further mutants of SF2/ASF (RRM1/RS and RRM2/RS) in which the first or second RNA-binding domains of SF2/ASF had been deleted (14, 16). The mutants bind RNA weakly but lack alternative splicing activity in vitro (14). As predicted, RNase H cleavage assays showed that neither protein affected U1 snRNP binding, even when present at concentrations up to 15 μM (results not shown). We conclude that there is a good correlation between the abilities of SF2/ASF mutants to promote alternative splicing and their abilities to enhance U1 snRNP binding.
hnRNP A1 high-affinity sites flanking a splice site redirected splicing without a corresponding change in U1 snRNP binding.
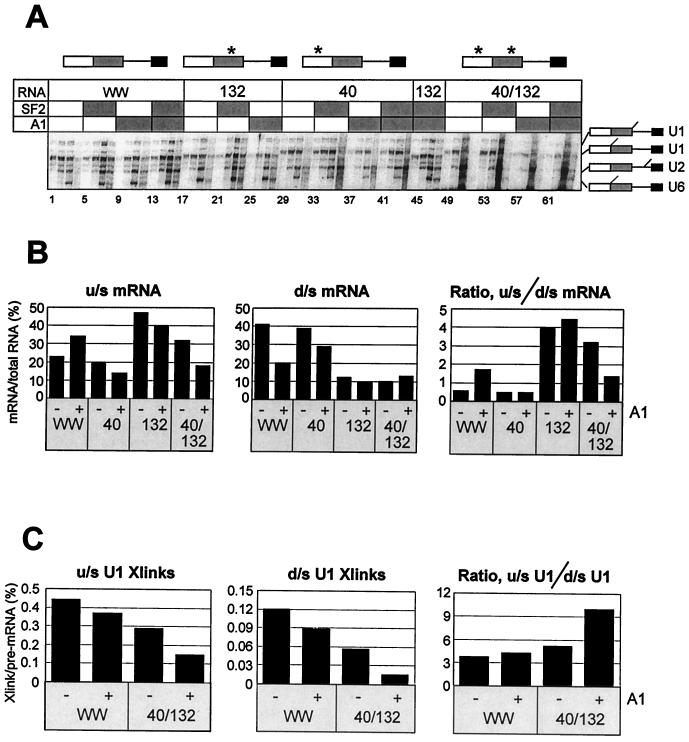
The binding studies with partially hydrolyzed CE1a RNA had shown that high-affinity sites were not required for efficient binding of hnRNP A1. Likewise, the same site did not reduce the displacement of hnRNP A1 by SF2/ASF (Fig. 6B). This was surprising in view of the effects of such sites on splicing (10, 19, 23, 29). To test whether, in nuclear extracts, such sites would enhance recruitment of hnRNP A1 and magnify the effects of hnRNP A1 on 5′SS selection, we prepared derivatives of AdML WW pre-mRNA in which the 10-nucleotide exon repressor and hnRNP A1 target from the K-SAM exon of fibroblast growth factor receptor 2 (29) were inserted into either or both of two positions: between the cap and the upstream 5′SS (Ad 40) and/or between the two 5′SSs (Ad 132). After UV irradiation of psoralen-containing splicing reactions, splicing and cross-linking in each reaction were measured as in Fig. 1. The results of UV irradiation after incubation in splicing conditions for various times are shown in Fig. 8A, and the effects of exogenous hnRNP A1 on splicing preferences are shown in Fig. 8B.
FIG. 8.
Effects of high-affinity binding sites for hnRNP A1 on splicing and U1 snRNP binding. (A) Derivatives of AdML WW containing 10-nucleotide hnRNP A1 target sites from the fibroblast growth factor receptor 2 gene at position 40 or 132 or both positions were incubated in splicing reaction mixtures supplemented where shown (shaded) with SF2/ASF (1.2 μM) or hnRNP A1 (5.5 μM) or both. The reactions contained AMT-psoralen and were irradiated after incubation for approximately 2, 5, 15, or 60 min (left to right in each block of four lanes). Samples were analyzed by electrophoresis on 5 and 8% denaturing polyacrylamide gels. The portions of the 5% gel that resolved the cross-linked adducts are shown. The asterisks above the diagrams of the pre-mRNA show the sites of insertion of the target sites. (B) Effects of hnRNP A1 on splicing efficiencies at the upstream (u/s) and downstream (d/s) 5′SS. The signals from u/s and d/s mRNA in the 60-min reactions (lanes 4, 12, 20, 28, etc., in panel A) were measured and corrected for label incorporation, and they are shown both separately, as percentages of the total RNA (pre-mRNA and splicing intermediates and products) in the reaction and as the ratios of the two. (C) Abundance of the cross-linked adducts of AdML WW and Ad 40/132. The intensities of the cross-linked U1 snRNA bands at 5 min in panel A (lanes 2, 10, 50, and 58) are shown as percentages of the pre-mRNA intensities and as a ratio for the two sites.
The insertion of the hnRNP A1 target sequence in Ad 40 had little effect on splicing, with a reproducible but small decline in the use of the upstream 5′SS (Fig. 8B, u/s mRNA). However, exogenous hnRNP A1 did not increase use of this site, as in the original AdML WW, but reduced it, so the added protein produced little or no shift in splicing. The insertion in Ad 132, placing the hnRNP A1 target between the two 5′SSs, produced a striking increase in use of the upstream 5′SS, but there was once again a slight reduction in use of both sites when hnRNP A1 was added and thus only a small upstream shift was seen. When the target sequence was present at both sites (Ad 40/132), there was a basal increase in use of the upstream 5′SS. However, the addition of hnRNP A1 reproducibly produced a marked drop in use of the upstream site and a small increase in use of the downstream 5′SS, causing an overall shift towards the downstream site. With Ad 40 and Ad 132, exogenous SF2/ASF produced the expected decrease in use of the upstream site and an increase in use of the downstream site (data not shown); with Ad 40/132, there was little change in upstream-site use and a small increase in use of the downstream site, resulting in a downstream shift in the ratio that was less marked than that produced by hnRNP A1.
The levels of cross-linking to U1 snRNA were quantified in these reactions after incubation for 5 min. In all of the mutants, the addition of SF2/ASF produced an increase in the levels of cross-linking at both sites, whereas hnRNP A1 reduced the levels. The quantification of levels with mutant Ad 40/132 was difficult because the cross-linked bands were unusually diffuse in the presence of exogenous hnRNP A1 (Fig. 8A, lanes 57 through 64). Nonetheless, a comparison of the levels of cross-linking to AdML WW and Ad 40/132 pre-mRNA (Fig. 8C) shows that the opposite effects of hnRNP A1 on splicing of these two substrates did not result from an unusual fall in U1 snRNP binding to the upstream 5′SS or an increase in binding to the downstream 5′SS of the mutant. We conclude that the presence of a potential high-affinity-binding site for hnRNP A1 reduced the efficiency of use of even the upstream site in the presence of exogenous hnRNP A1 and that, with two such sites flanking the upstream 5′SS, the shift away from that site did not result primarily from a reduction in U1 snRNP binding but from sequestration of the site from other components.
DISCUSSION
The roles of U1 snRNP in selection of alternative 5′SSs.
We have proposed that spliceosome assembly generally incorporates the U1 snRNP-5′SS complex furthest downstream; when U1 snRNP-binding levels are low, then the affinity of binding will determine the likelihood that a site is occupied and used; when binding levels are higher and more than one site is occupied, then the relative positions of the sites determine the outcome (32). Proteins that affect splicing patterns might do so, according to this scheme, without any intrinsic specificity or polarity by affecting the balance between these two conditions. We have shown previously that U1 snRNP-dependent complexes can form simultaneously on two alternative consensus 5′SSs on each molecule of pre-mRNA and that the levels of these complexes were increased by exogenous SF2/ASF, which shifts splicing to the downstream site (32). However, the RNase H cleavage assay used for those experiments does not detect initial (U2-independent) complexes at nonconsensus sites, and it is only an indirect assay of U1 snRNP interactions. Thus, to test whether the model was informative for modulation of splicing by hnRNP A1, we used psoralen-mediated cross-linking in nuclear extracts instead.
The results of the cross-linking assays in Fig. 1C and D were consistent with our model. At both alternative 5′SSs, SF2/ASF increased U1 snRNA binding as expected (32, 42) and hnRNP A1 reduced binding as predicted. The ratio of U1 snRNA binding to the two sites did not change pro rata with the ratio of splicing. This result may be taken either as being consistent with our model or as indicating that U1 snRNP binding is irrelevant for selection; it is not consistent with the simple view that U1 snRNP binds a single site on pre-mRNA and that this site is used for splicing. There are two further ways in which the results in Fig. 1 are informative. First, reduced levels of U1 snRNP binding and a switch to selection based on affinity rather than position would be expected to produce a shift in the ratio of U1 snRNA binding towards the site that is used for splicing. The shift in U1 snRNA cross-linking in the presence of hnRNP A1 towards the upstream site is consistent with this expectation. More striking support for our model comes from the remarkable correlation in Fig. 1E between the fraction of splicing at the downstream site and the level of U1 snRNA cross-linking there. Correspondingly, there is a more-or-less inverse correlation between U1 snRNA cross-linking to the upstream site and its use for splicing. This result clearly supports our proposal that, if the downstream site is occupied by U1 snRNP, it will be used for splicing, whereas the upstream site will be used only when the downstream one is vacant.
The link between 5′SS position and use has interesting implications. In general, an upstream alternative site must have an intrinsically greater affinity for U1 snRNP if it is to be used significantly. For example, if two sites were used equally and the efficiency of removal of an intron were close to 100%, then the probability of U1 snRNP binding at the downstream site would be 50%, but at the upstream site it would be 100%. This affinity depends on factors such as the sequence of the 5′SS, absence of impeding secondary structure in the pre-mRNA, proximity of enhancers, etc. The presence of the 5′ cap or upstream intron complexes may affect the affinity of binding to the upstream site also (48), although other results suggest that the cap interaction has indiscriminate effects (68). The nature of the components that interact with the U1-5′SS complex furthest downstream is still unknown. Our results with U6 snRNA cross-linking (Fig. 1D) appear to show that it cross-links to one site, i.e., after the site has been selected for splicing, but the cross-links to the downstream site are very faint, and the quantification is not reliable. Nonetheless, it seems likely that selection involves the early complexes that link the 5′SS and branch site (3).
There have been relatively few other attempts to determine the mechanisms by which alternative 5′SSs are selected and by which SR proteins or other alternative splicing factors might act. In an S100 extract (in which there was no U1 snRNP binding in the absence of added SR protein), SRp40 was observed to promote splicing to an upstream site in simian virus 40 pre-mRNA and to enhance U1 snRNA cross-linking there (92). This appears to be a specific effect enhancing the affinity at this site. Of more importance to our argument was the finding that SRp30b enhanced binding at both sites but spliced mainly from the downstream site. This finding is consistent with the model. Psoralen cross-linking was used to investigate U1 snRNP binding to a normal and a cryptic 5′SS in an adenovirus pre-mRNA (83). Addition of SR protein increased cross-linking at both sites and enhanced use of the downstream site (to 97%), in agreement with our model. However, in extracts depleted of the U1-A protein, parallel treatments increased U1 snRNA cross-linking at both sites, but use of the downstream site increased only slightly, from 2 to 15%. This result has been interpreted as being inconsistent with our model, in that the response to SR addition was incomplete even though U1 snRNP binding rose (86). However, we note that the 7-fold increase in the level of U1 snRNP binding to the downstream site was matched by a 7.5-fold increase in splicing there. The much reduced switch compared with the native extract might be attributed to the much lower proportion of pre-mRNA bound by U1 snRNA in the depleted extract, although no figures for the proportions of pre-mRNA cross-linked were given (83).
If our model for the mechanisms of splice site modulation by SF2/ASF and hnRNP A1 is correct, then there ought to be a strong connection between the activities of mutant or related proteins in alternative splicing and their effects on U1 snRNP binding. We have tested this with a comprehensive panel of hnRNP A1 mutations and several other hnRNP A/B proteins. Overall, the correspondence is very good. For SF2/ASF, previous work had shown that removal of the RS domain (ΔRS) left its alternative splicing activity intact or even increased it (14, 96) but abolished the protein's activity in constitutive splicing and eliminated its ability to form a U1 snRNP-containing complex on native gels (41, 42). This last result contradicts both our postulated mechanism for the action of SF2/ASF and our results in Fig. 7, but the assays used were different. The native gel electrophoresis assay did not detect a complex between pre-mRNA and SF2/ASF, even though nuclease cleavage experiments showed that guanines throughout the RNA were protected, and no complex of RNA with U1 snRNP was detected in the absence of SF2/ASF. Thus, one possible explanation of the results might be that the His-tagged ΔRS mutant enhances binding but that the complex is too labile to be detected by the electrophoresis assay. The full SF2/ASF protein might have conferred sufficient stability inappropriately by RNA-bridged interactions between the unphosphorylated SR domain and the U1 70K protein (89), a possibility that can be excluded in the case of our assays because the RS domain mutants were active. Indeed, it appears from our results that an unphosphorylated RS domain of SF2/ASF may interfere with the protein's activity. A clearer understanding of the role of the RS domain is unlikely to emerge until the structures and stoichiometries of the complexes on the pre-mRNA are characterized.
Mechanisms of action of hnRNP A1 and SF2/ASF.
We demonstrated in Fig. 2 to 4 that purified hnRNP A1 can reduce U1 snRNP binding to 5′SS and that it does so by preventing binding. The results in Fig. 5 and other data (not shown) indicate that long pre-mRNA substrates are more susceptible, which tends to argue against the possibility that hnRNP A1 blocks by forming stable binary complexes with free U1 snRNP. Although far-Western blots did suggest that hnRNP A1 could interact with some U1 snRNP proteins, such interactions were not detected by kinetic assays (Fig. 4B) or by a variety of other techniques. U2 and U4 (but not U1) snRNPs have been found to coimmunoprecipitate with hnRNP A1 in vitro (13), but the interactions involved were not identified. Thus, we assume that the primary cause of the reduction in U1 snRNP binding is competition for binding to the RNA.
The mechanism is not so clear for SF2/ASF, which might enhance U1 snRNP binding either by forming a binary complex first with the U1 snRNP or by binding to the RNA and recruiting U1 snRNP. The relatively high stability of the SF2/ASF-RNA complex that we measured by UV cross-linking and the ability of SF2/ASF to replace hnRNP A1 suggest that SF2/ASF binds independently, possibly forming higher-order interactions. This direct analysis of binding rates and the stability of the complex is consistent with inferences from assays of splicing after preincubation (18, 35) but not with inferences from the formation of gel-resistant U1 complexes (41). How the complex accommodates U1 snRNP is unclear, particularly given that the purine-rich sites preferred by the purified protein can include the actual 5′SS (97). The properties of SF2/ASF may be affected by phosphorylation in vivo or after incubation in nuclear extracts (38), but the effectiveness of the ΔRS mutant in enhancing U1 snRNP binding to 5′SS and in competing with hnRNP A1 for binding to pre-mRNA demonstrates that these fundamental properties of SF2/ASF are not artifacts of an unphosphorylated RS domain.
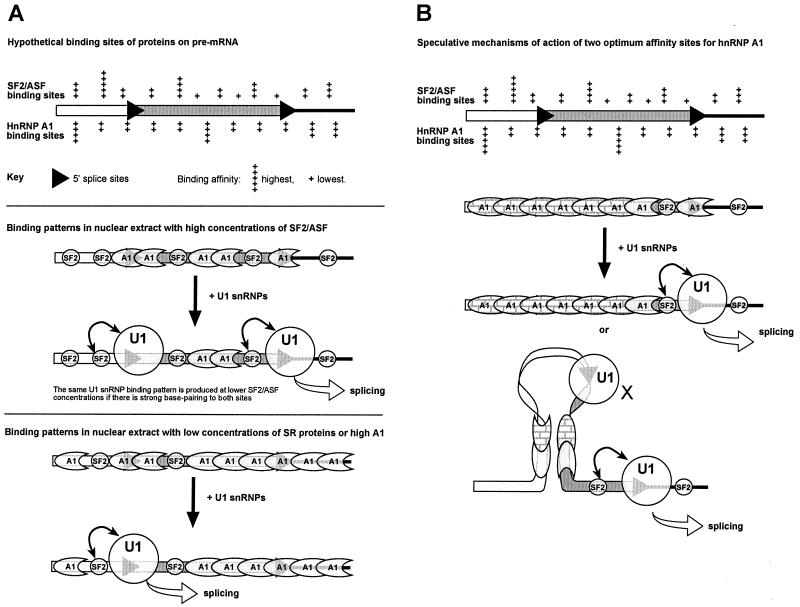
The binding properties of the purified proteins seem at first to be incompatible with the observations of preferred sites of action in extracts or in vivo. Our results suggest that purified hnRNP A1 binds extensively in the absence of SELEX consensus sequences; SF2/ASF competes for binding to most or all of the pre-mRNA. Our current understanding suggests a model such as that shown in Fig. 9A. The diagram shows that the specificity is not stringent; the range of affinities of each protein for various sequences is quite narrow, and thus the proteins compete for binding to virtually all sequences on the pre-mRNA. In a nuclear extract or in vivo, there will be far more competing proteins, and the proportion of the pre-mRNA occupied by any given protein is likely to be smaller. Nonetheless, changes in the relative abundance of the proteins are likely to have significant consequences for the range of sites bound by each protein. The diagram illustrates the effect of SF2/ASF and hnRNP A1 concentrations upon U1 snRNP binding, based on the model described and validated above. It is not yet clear whether preferred SF2/ASF sites do need to be close to the 5′SS, as suggested, but the diagram shows a site of high affinity near the upstream 5′SS for the reasons explained above.
FIG. 9.
Models for the molecular mechanisms by which hnRNP A1 and SR proteins affect 5′SS selection. (A) The top panel shows hypothetical binding sites of various affinities for two proteins on a pre-mRNA containing two alternative 5′SSs. The distributions take account of the fact that the proteins have marked sequence preferences and yet appear to be able to occupy all available sites on the RNA at concentrations used in alternative splicing assays in vitro. hnRNP A1 is shown to bind in a more regular fashion because of its cooperativity. The affinity of interaction at each site is indicated by a range from + to +++++. The middle panel depicts a possible binding pattern in nuclear extract at a high concentration of SF2/ASF. The high-, middle-, and low-affinity sites for SF2/ASF are occupied by SF2/ASF protein (small circles), although hnRNP A1 (among other proteins) competes for binding (crescent shape) and excludes SF2/ASF from the latter's lowest-affinity sites. Thus, both splice sites are close to a sequence bound by SF2/ASF, and U1 snRNP binding is enhanced by interactions (double-headed arrow) at both sites. Double occupancy leads to use of the downstream site. Double occupancy also results even at normal concentrations of SF2/ASF if the 5′SSs have the consensus sequence, because U1 snRNP binding outcompetes hnRNP A1. The lower panel illustrates the effect of elevated hnRNP A1 concentrations. Only at the highest-affinity sites can SF2/ASF or other SR proteins bind in preference to hnRNP A1. U1 snRNP binding is reduced by competition with hnRNP A1, and so double occupancy of the 5′SSs is unlikely. Thus, splicing reflects the probability of occupancy of the individual sites. In this case, the upstream site is favored because it is close to a high-affinity site for SF2/ASF, permitting some U1 snRNP binding. The equilibrium between hnRNP A1, SF2/ASF, and U1 snRNP at the downstream site is dynamic, and, on molecules in which U1 snRNP bound initially at the upstream 5′SS, it would be expected that U1 snRNP would also bind eventually to the downstream site. By this time, the upstream site might be committed to splicing. This may account for the observation of a slower rate of nonproductive and cap-independent U1 snRNP binding to unused sites (68). (B) The introduction of two high-affinity binding sites (+++++) for hnRNP A1 resulted in an inversion of the normal effects of hnRNP A1 on alternative splicing. Although the shift to the downstream 5′SS was akin to the effect of SF2/ASF, the reductions in U1 snRNP cross-linking at both 5′SSs and the results with the individual high-affinity sites showed that the mechanism is quite different. We propose that the two sites flanking the upstream 5′ splice site allow it to be inactivated when hnRNP A1 is elevated, either because they mark out a domain for unusually stable cooperative hnRNP A1 binding (patterned hnRNP A1 proteins) or because two bound sites form a stable interacting complex that mitigates against splicing. U1 snRNP occupancy at the downstream site would be low, because of competition with hnRNP A1, but this site would be used eventually. U1 snRNP may bind also to the upstream site, but the sequestered site is unable to interact with other components. It is not known whether the cooperative binding of hnRNP A1 is propagated in both directions along the RNA or is unidirectional.
Figure 9A does not show any high-affinity sites for hnRNP A1. Preliminary results with the hydrolysis assay showed that the SELEX consensus target for hnRNP A1 in CE1a was not required for binding. However, molecules containing the site were bound slightly more efficiently, particularly with higher ratios of protein to RNA and in the absence of competitor RNA. Although further experiments are required, a possible interpretation is that a single occupied high-affinity site has little effect on binding, because the overall probability of a molecule of RNA being bound anywhere by protein is scarcely affected, until a second molecule of hnRNP A1, bound elsewhere on the same RNA molecule, interacts with it, and a conformational change produces a stable complex. Two potential high-affinity sites would be more potent, on this basis, because the chance of simultaneous occupancy and collision would be higher. The results of our splicing assays with high-affinity sites (Fig. 8B) can be interpreted in this light. Single K-SAM repressor sites seem to reduce the use of the nearest downstream 5′SS, possibly by competition with adjacent SR protein-binding sites. In the case of Ad 40, this effect might weaken the ability of the upstream site to recruit U1 snRNPs in the face of an increased concentration of hnRNP A1. The effects of the double insertion suggest that the upstream 5′SS was sequestered between the two sites in a proportion of the pre-mRNA when the concentration of hnRNP A1 was raised. This effect is consistent with studies on hnRNP A1 pre-mRNA (10), and two possible mechanisms, one based on that work, are shown in Fig. 9B. The observation that hnRNP A1 actually has less effect on U1 snRNP binding to the sequestered site than the site used for splicing (Fig. 8C), while tentative, inclines us to favor the lower model and is consistent with results on U1 snRNP binding reported for hnRNP A1 pre-mRNA (10). We have not yet established that these sites are bound in extracts by hnRNP A1.
In summary, the present experiments offer substantial support for our model of the roles of U1 snRNP in 5′SS selection, and this model provides a good general framework for explaining the effects of hnRNP A1 and SF2/ASF on 5′SS selection. Competition between these proteins for binding to the pre-mRNA is likely to determine the outcome of U1 snRNP binding and thus splicing. Studies of the distribution and dynamics of binding by these and other proteins under splicing conditions should be a high priority for future studies.
ACKNOWLEDGMENTS
We are very grateful to Reinhard Lührmann, Berthold Kastner, and Cindy Will for the generous gifts of purified U1 snRNP; Gideon Dreyfuss, David Williams, Silvano Riva, and Iain Mattaj for antibodies; Benoit Chabot for clones containing CE1a sequences; Clive Bagshaw for advice on the use of the program KfitSim; and Evgeny Makarov for advice and criticism.
This research was supported by the Medical Research Council (United Kingdom) and the Wellcome Trust (to I.C.E.) and in part by National Cancer Institute grant CA13106 (to A.R.K.). The Wellcome Trust supported the purchase of a PhosphorImager and a Cyclone imager.
REFERENCES
- 1.Abdul-Manan N, O'Malley S M, Williams K R. Origins of binding specificity of the A1 heterogeneous nuclear ribonucleoprotein. Biochemistry. 1996;35:3545–3554. doi: 10.1021/bi952298p. [DOI] [PubMed] [Google Scholar]
- 2.Abdul-Manan N, Williams K R. hnRNP A1 binds promiscuously to oligoribonucleotides: utilization of random and homo-oligonucleotides to discriminate sequence from base-specific binding. Nucleic Acids Res. 1996;24:4063–4070. doi: 10.1093/nar/24.20.4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abovich N, Rosbash M. Cross-intron bridging interactions in the yeast commitment complex are conserved in mammals. Cell. 1997;89:403–412. doi: 10.1016/s0092-8674(00)80221-4. [DOI] [PubMed] [Google Scholar]
- 4.Bach M, Bringmann P, Luhrmann R. Purification of small nuclear ribonucleoprotein particles with antibodies against modified nucleosides of small nuclear RNAs. Methods Enzymol. 1990;181:232–257. doi: 10.1016/0076-6879(90)81125-e. [DOI] [PubMed] [Google Scholar]
- 5.Beck J, Nassal M. A sensitive procedure for mapping the boundaries of RNA elements binding in vitro translated proteins defines a minimal hepatitis B virus encapsidation signal. Nucleic Acids Res. 1996;24:4364–4366. doi: 10.1093/nar/24.21.4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bennett M, Piñol-Roma S, Staknis D, Dreyfuss G, Reed R. Differential binding of heterogeneous nuclear ribonucleoproteins to mRNA precursors prior to spliceosome assembly in vitro. Mol Cell Biol. 1992;12:3165–3175. doi: 10.1128/mcb.12.7.3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Birney E, Kumar S, Krainer A R. Analysis of the RNA-recognition motif and RS and RGG domains: conservation in metazoan pre-mRNA splicing factors. Nucleic Acids Res. 1993;21:5803–5816. doi: 10.1093/nar/21.25.5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Black D L. Activation of c-src neuron-specific splicing by an unusual RNA element in vivo and in vitro. Cell. 1992;69:795–807. doi: 10.1016/0092-8674(92)90291-j. [DOI] [PubMed] [Google Scholar]
- 9.Blanchette M, Chabot B. A highly stable duplex structure sequesters the 5′ splice site region of hnRNP A1 alternative exon 7B. RNA. 1997;3:405–419. [PMC free article] [PubMed] [Google Scholar]
- 10.Blanchette M, Chabot B. Modulation of exon skipping by high-affinity hnRNP A1-binding sites and by intron elements that repress splice site utilization. EMBO J. 1999;18:1939–1952. doi: 10.1093/emboj/18.7.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bourgeois C F, Popielarz M, Hildwein G, Stevenin J. Identification of a bidirectional splicing enhancer: differential involvement of SR proteins in 5′ or 3′ splice site activation. Mol Cell Biol. 1999;19:7347–7356. doi: 10.1128/mcb.19.11.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burd C G, Dreyfuss G. RNA binding specificity of hnRNP A1: significance of hnRNP A1 high-affinity binding sites in pre-mRNA splicing. EMBO J. 1994;13:1197–1204. doi: 10.1002/j.1460-2075.1994.tb06369.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buvoli M, Cobianchi F, Riva S. Interaction of hnRNP A1 with snRNPs and pre-mRNAs: evidence for a possible role of A1 RNA annealing activity in the first steps of spliceosome assembly. Nucleic Acids Res. 1992;20:5017–5025. doi: 10.1093/nar/20.19.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caceres J F, Krainer A R. Functional analysis of pre-mRNA splicing factor SF2/ASF structural domains. EMBO J. 1993;12:4715–4726. doi: 10.1002/j.1460-2075.1993.tb06160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cáceres J F, Krainer A R. Mammalian pre-mRNA splicing factors. In: Krainer A R, editor. Eukaryotic mRNA processing. Oxford, England: IRL Press; 1997. pp. 174–203. [Google Scholar]
- 16.Caceres J F, Misteli T, Screaton G R, Spector D L, Krainer A R. Role of the modular domains of SR proteins in subnuclear localization and alternative splicing specificity. J Cell Biol. 1997;138:225–238. doi: 10.1083/jcb.138.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caceres J F, Stamm S, Helfman D M, Krainer A R. Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science. 1994;265:1706–1709. doi: 10.1126/science.8085156. [DOI] [PubMed] [Google Scholar]
- 18.Cao W H, Jamison S F, Garcia-Blanco M A. Both phosphorylation and dephosphorylation of ASF/SF2 are required for pre-mRNA splicing in vitro. RNA. 1997;3:1456–1467. [PMC free article] [PubMed] [Google Scholar]
- 19.Caputi M, Mayeda A, Krainer A R, Zahler A M. hnRNP A/B proteins are required for inhibition of HIV-1 pre-mRNA splicing. EMBO J. 1999;18:4060–4067. doi: 10.1093/emboj/18.14.4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cartegni L, Maconi M, Morandi E, Cobianchi F, Riva S, Biamonti G. hnRNP A1 selectively interacts through its Gly-rich domain with different RNA-binding proteins. J Mol Biol. 1996;259:337–348. doi: 10.1006/jmbi.1996.0324. [DOI] [PubMed] [Google Scholar]
- 21.Casas-Finet J R, Smith J D, Jr, Kumar A, Kim J G, Wilson S H, Karpel R L. Mammalian heterogeneous ribonucleoprotein A1 and its constituent domains: nucleic acid interaction, structural stability and self-association. J Mol Biol. 1993;229:873–889. doi: 10.1006/jmbi.1993.1093. [DOI] [PubMed] [Google Scholar]
- 22.Cavaloc Y, Popielarz M, Fuchs J P, Gattoni R, Stevenin J. Characterization and cloning of the human splicing factor 9G8: a novel 35 kDa factor of the serine/arginine protein family. EMBO J. 1994;13:2639–2649. doi: 10.1002/j.1460-2075.1994.tb06554.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chabot B, Blanchette M, Lapierre I, La Branche H. An intron element modulating 5′ splice site selection in the hnRNP A1 pre-mRNA interacts with hnRNP A1. Mol Cell Biol. 1997;17:1776–1786. doi: 10.1128/mcb.17.4.1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chabot B, Steitz J A. Recognition of mutant and cryptic 5′ splice sites by the U1 small nuclear ribonucleoprotein in vitro. Mol Cell Biol. 1987;7:698–707. doi: 10.1128/mcb.7.2.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chiara M D, Gozani O, Bennett M, Champion-Arnaud P, Palandjian L, Reed R. Identification of proteins that interact with exon sequences, splice sites, and the branchpoint sequence during each stage of spliceosome assembly. Mol Cell Biol. 1996;16:3317–3326. doi: 10.1128/mcb.16.7.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cobianchi F, Karpel R L, Williams K R, Notario V, Wilson S H. Mammalian heterogeneous nuclear ribonucleoprotein complex protein A1: large-scale overproduction in Escherichia coli and cooperative binding to single-stranded nucleic acids. J Biol Chem. 1988;263:1063–1071. [PubMed] [Google Scholar]
- 27.Cortes J J, Sontheimer E J, Selwert S D, Steitz J A. Mutations in the conserved loop of human U5 snRNA generate use of novel cryptic 5′ splice sites in vivo. EMBO J. 1993;12:5181–5189. doi: 10.1002/j.1460-2075.1993.tb06213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cunningham S A, Else A J, Potter B, Eperon I C. Influences of separation and adjacent sequences on the use of alternative 5′ splice sites. J Mol Biol. 1991;217:265–281. doi: 10.1016/0022-2836(91)90541-d. [DOI] [PubMed] [Google Scholar]
- 29.Del Gatto-Konczak F, Olive M, Gesnel M C, Breathnach R. hnRNP A1 recruited to an exon in vivo can function as an exon splicing silencer. Mol Cell Biol. 1999;19:251–260. doi: 10.1128/mcb.19.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dreyfuss G, Matunis M J, Pinol-Roma S, Burd C G. hnRNP proteins and the biogenesis of mRNA. Annu Rev Biochem. 1993;62:289–321. doi: 10.1146/annurev.bi.62.070193.001445. [DOI] [PubMed] [Google Scholar]
- 31.Elrick L L, Humphrey M B, Cooper T A, Berget S M. A short sequence within two purine-rich enhancers determines 5′ splice site specificity. Mol Cell Biol. 1998;18:343–352. doi: 10.1128/mcb.18.1.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eperon I C, Ireland D C, Smith R A, Mayeda A, Krainer A R. Pathways for selection of 5′ splice sites by U1 snRNPs and SF2/ASF. EMBO J. 1993;12:3607–3617. doi: 10.1002/j.1460-2075.1993.tb06034.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eperon L P, Graham I R, Griffiths A D, Eperon I C. Effects of RNA secondary structure on alternative splicing of pre-mRNA: is folding limited to a region behind the transcribing RNA polymerase? Cell. 1988;54:393–401. doi: 10.1016/0092-8674(88)90202-4. [DOI] [PubMed] [Google Scholar]
- 34.Fortes P, Bilbao-Cortes D, Fornerod M, Rigaut G, Raymond W, Seraphin B, Mattaj I W. Luc7p, a novel yeast U1 snRNP protein with a role in 5′ splice site recognition. Genes Dev. 1999;13:2425–2438. doi: 10.1101/gad.13.18.2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fu X D. Specific commitment of different pre-mRNAs to splicing by single SR proteins. Nature. 1993;365:82–85. doi: 10.1038/365082a0. [DOI] [PubMed] [Google Scholar]
- 36.Fu X D, Mayeda A, Maniatis T, Krainer A R. General splicing factors SF2 and SC35 have equivalent activities in vitro, and both affect alternative 5′ and 3′ splice site selection. Proc Natl Acad Sci USA. 1992;89:11224–11228. doi: 10.1073/pnas.89.23.11224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ge H, Manley J L. A protein factor, ASF, controls cell-specific alternative splicing of SV40 early pre-mRNA in vitro. Cell. 1990;62:25–34. doi: 10.1016/0092-8674(90)90236-8. [DOI] [PubMed] [Google Scholar]
- 38.Hanamura A, Caceres J F, Mayeda A, Franza B R, Krainer A R. Regulated tissue-specific expression of antagonistic pre-mRNA splicing factors. RNA. 1998;4:430–444. [PMC free article] [PubMed] [Google Scholar]
- 39.Hwang D Y, Cohen J B. U1-snRNA promotes the selection of nearby 5′ splice sites by U6 snRNA in mammalian cells. Genes Dev. 1996;10:338–350. doi: 10.1101/gad.10.3.338. [DOI] [PubMed] [Google Scholar]
- 40.Izaurralde E, Jarmolowski A, Beisel C, Mattaj I W, Dreyfuss G, Fischer U. A role for the M9 transport signal of hnRNP A1 in mRNA nuclear export. J Cell Biol. 1997;137:27–35. doi: 10.1083/jcb.137.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jamison S F, Pasman Z, Wang J, Will C, Luhrmann R, Manley J L. U1 snRNP-ASF/SF2 interaction and 5′ splice site recognition—characterization of required elements. Nucleic Acids Res. 1995;23:3260–3267. doi: 10.1093/nar/23.16.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kohtz J D, Jamison S F, Will C L, Zuo P, Luhrmann R, Garcia-Blanco M A, Manley J L. Protein-protein interactions and 5′-splice-site recognition in mammalian mRNA precursors. Nature. 1994;368:119–124. doi: 10.1038/368119a0. [DOI] [PubMed] [Google Scholar]
- 43.Krainer A R, Conway G C, Kozak D. The essential pre-mRNA splicing factor SF2 influences 5′ splice site selection by activating proximal sites. Cell. 1990;62:35–42. doi: 10.1016/0092-8674(90)90237-9. [DOI] [PubMed] [Google Scholar]
- 44.Krainer A R, Mayeda A, Kozak D, Binns G. Functional expression of cloned human splicing factor SF2: homology to RNA-binding proteins, U1 70K, and Drosophila splicing regulators. Cell. 1991;66:383–394. doi: 10.1016/0092-8674(91)90627-b. [DOI] [PubMed] [Google Scholar]
- 45.Lavigueur A, La B H, Kornblihtt A R, Chabot B. A splicing enhancer in the human fibronectin alternate ED1 exon interacts with SR proteins and stimulates U2 snRNP binding. Genes Dev. 1993;7:2405–2417. doi: 10.1101/gad.7.12a.2405. [DOI] [PubMed] [Google Scholar]
- 46.Lear A L, Eperon L P, Wheatley I M, Eperon I C. Hierarchy for 5′ splice site preference determined in vivo. J Mol Biol. 1990;211:103–115. doi: 10.1016/0022-2836(90)90014-D. [DOI] [PubMed] [Google Scholar]
- 47.Legrain P, Seraphin B, Rosbash M. Early commitment of yeast pre-mRNA to the spliceosome pathway. Mol Cell Biol. 1988;8:3755–3760. doi: 10.1128/mcb.8.9.3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lewis J D, Izaurralde E, Jarmolowski A, McGuigan C, Mattaj I W. A nuclear cap-binding complex facilitates association of U1-snRNP with the cap-proximal 5′ splice site. Genes Dev. 1996;10:1683–1698. doi: 10.1101/gad.10.13.1683. [DOI] [PubMed] [Google Scholar]
- 49.Liu H X, Zhang M, Krainer A R. Identification of functional exonic splicing enhancer motifs recognized by individual SR proteins. Genes Dev. 1998;12:1998–2012. doi: 10.1101/gad.12.13.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lynch K W, Maniatis T. Assembly of specific SR protein complexes on distinct regulatory elements of the Drosophila doublesex splicing enhancer. Genes Dev. 1996;10:2089–2101. doi: 10.1101/gad.10.16.2089. [DOI] [PubMed] [Google Scholar]
- 51.Lynch K W, Maniatis T. Synergistic interactions between 2 distinct elements of a regulated splicing enhancer. Genes Dev. 1995;9:284–293. doi: 10.1101/gad.9.3.284. [DOI] [PubMed] [Google Scholar]
- 52.Manley J L, Tacke R. SR proteins and splicing control. Genes Dev. 1996;10:1569–1579. doi: 10.1101/gad.10.13.1569. [DOI] [PubMed] [Google Scholar]
- 53.Mayeda A, Helfman D M, Krainer A R. Modulation of exon skipping and inclusion by heterogeneous nuclear ribonucleoprotein A1 and pre-mRNA splicing factor SF2/ASF. Mol Cell Biol. 1993;13:2993–3001. doi: 10.1128/mcb.13.5.2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mayeda A, Krainer A R. Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell. 1992;68:365–375. doi: 10.1016/0092-8674(92)90477-t. [DOI] [PubMed] [Google Scholar]
- 55.Mayeda A, Munroe S H, Caceres J F, Krainer A R. Function of conserved domains of hnRNP A1 and other hnRNP A/B proteins. EMBO J. 1994;13:5483–5495. doi: 10.1002/j.1460-2075.1994.tb06883.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mayeda A, Munroe S H, Xu R M, Krainer A R. Distinct functions of the closely related tandem RNA-recognition motifs of hnRNP A1. RNA. 1998;4:1111–1123. doi: 10.1017/s135583829898089x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mayeda A, Ohshima Y. Beta-globin transcripts carrying a single intron with three adjacent nucleotides of 5′ exon are efficiently spliced in vitro irrespective of intron position or surrounding exon sequences. Nucleic Acids Res. 1990;18:4671–4676. doi: 10.1093/nar/18.16.4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mayeda A, Ohshima Y. Short donor site sequences inserted within the intron of β-globin pre-mRNA serve for splicing in vitro. Mol Cell Biol. 1988;8:4484–4491. doi: 10.1128/mcb.8.10.4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mayeda A, Screaton G R, Chandler S D, Fu X D, Krainer A R. Substrate specificities of SR proteins in constitutive splicing are determined by their RNA recognition motifs and composite pre-mRNA exonic elements. Mol Cell Biol. 1999;19:1853–1863. doi: 10.1128/mcb.19.3.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Michaud S, Reed R. An ATP-independent complex commits pre-mRNA to the mammalian spliceosome assembly pathway. Genes Dev. 1991;5:2534–2546. doi: 10.1101/gad.5.12b.2534. [DOI] [PubMed] [Google Scholar]
- 61.Michaud S, Reed R. A functional association between the 5′ and 3′ splice sites is established in the earliest prespliceosome complex (E) in mammals. Genes Dev. 1993;7:1008–1020. doi: 10.1101/gad.7.6.1008. [DOI] [PubMed] [Google Scholar]
- 62.Misteli T, Caceres J F, Spector D L. The dynamics of a pre-mRNA splicing factor in living cells. Nature. 1997;387:523–527. doi: 10.1038/387523a0. [DOI] [PubMed] [Google Scholar]
- 63.Munroe S H, Dong X F. Heterogeneous nuclear ribonucleoprotein A1 catalyzes RNA·RNA annealing. Proc Natl Acad Sci USA. 1992;89:895–899. doi: 10.1073/pnas.89.3.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Murray H L, Jarrell K A. Flipping the switch to an active spliceosome. Cell. 1999;96:599–602. doi: 10.1016/s0092-8674(00)80568-1. [DOI] [PubMed] [Google Scholar]
- 65.Nelson K K, Green M R. Mechanism for cryptic splice site activation during pre-mRNA splicing. Proc Natl Acad Sci USA. 1990;87:6253–6257. doi: 10.1073/pnas.87.16.6253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nelson K K, Green M R. Splice site selection and ribonucleoprotein complex assembly during in vitro pre-messenger RNA splicing. Genes Dev. 1988;2:319–329. doi: 10.1101/gad.2.3.319. [DOI] [PubMed] [Google Scholar]
- 67.Newman A, Norman C. Mutations in yeast U5 snRNA alter the specificity of 5′ splice-site cleavage. Cell. 1991;65:115–123. doi: 10.1016/0092-8674(91)90413-s. [DOI] [PubMed] [Google Scholar]
- 68.O'Mullane L, Eperon I C. The pre-mRNA 5′ cap determines whether U6 small nuclear RNA succeeds U1 small nuclear ribonucleoprotein particle at 5′ splice sites. Mol Cell Biol. 1998;18:7510–7520. doi: 10.1128/mcb.18.12.7510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pinol-Roma S, Dreyfuss G. hnRNP proteins: localization and transport between the nucleus and the cytoplasm. Trends Cell Biol. 1993;3:151–155. doi: 10.1016/0962-8924(93)90135-n. [DOI] [PubMed] [Google Scholar]
- 70.Pontius B W, Berg P. Rapid assembly and disassembly of complementary DNA strands through an equilibrium intermediate state mediated by A1 hnRNP protein. J Biol Chem. 1992;267:13815–13818. [PubMed] [Google Scholar]
- 71.Puig O, Gottschalk A, Fabrizio P, Seraphin B. Interaction of the U1 snRNP with nonconserved intronic sequences affects 5′ splice site selection. Genes Dev. 1999;13:569–580. doi: 10.1101/gad.13.5.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ramchatesingh J, Zahler A M, Neugebauer K M, Roth M B, Cooper T A. A subset of SR proteins activates splicing of the cardiac troponin T alternative exon by direct interactions with an exonic enhancer. Mol Cell Biol. 1995;15:4898–4907. doi: 10.1128/mcb.15.9.4898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Reed R, Maniatis T. A role for exon sequences and splice-site proximity in splice-site selection. Cell. 1986;46:681–690. doi: 10.1016/0092-8674(86)90343-0. [DOI] [PubMed] [Google Scholar]
- 74.Schaal T D, Maniatis T. Multiple distinct splicing enhancers in the protein-coding sequences of a constitutively spliced pre-mRNA. Mol Cell Biol. 1999;19:261–273. doi: 10.1128/mcb.19.1.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schaal T D, Maniatis T. Selection and characterization of pre-mRNA splicing enhancers: identification of novel SR protein-specific enhancer sequences. Mol Cell Biol. 1999;19:1705–1719. doi: 10.1128/mcb.19.3.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Screaton G R, Caceres J F, Mayeda A, Bell M V, Plebanski M, Jackson D G, Bell J I, Krainer A R. Identification and characterization of 3 members of the human SR family of pre-mRNA splicing factors. EMBO J. 1995;14:4336–4349. doi: 10.1002/j.1460-2075.1995.tb00108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Seraphin B, Kandels-Lewis S. 3′ splice site recognition in S. cerevisiae does not require base pairing with U1 snRNA. Cell. 1993;73:803–812. doi: 10.1016/0092-8674(93)90258-r. [DOI] [PubMed] [Google Scholar]
- 78.Seraphin B, Rosbash M. Identification of functional U1 snRNA-pre-mRNA complexes committed to spliceosome assembly and splicing. Cell. 1989;59:349–358. doi: 10.1016/0092-8674(89)90296-1. [DOI] [PubMed] [Google Scholar]
- 79.Shen J, Zu K, Cass C L, Beyer A L, Hirsh J. Exon skipping by overexpression of a Drosophila heterogeneous nuclear ribonucleoprotein in vivo. Proc Natl Acad Sci USA. 1995;92:1822–1825. doi: 10.1073/pnas.92.6.1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Stevenson B, DiSpirito A A, Schmidt T M. Reduction of enzyme adsorption to polypropylene surfaces in the presence of a nonionic detergent. BioTechniques. 1994;17:1048–1050. [PubMed] [Google Scholar]
- 81.Stutz F, Rosbash M. Nuclear RNA export. Genes Dev. 1998;12:3303–3319. doi: 10.1101/gad.12.21.3303. [DOI] [PubMed] [Google Scholar]
- 82.Sun Q, Mayeda A, Hampson R K, Krainer A R, Rottman F M. General splicing factor SF2/ASF promotes alternative splicing by binding to an exonic splicing enhancer. Genes Dev. 1993;7:2598–2608. doi: 10.1101/gad.7.12b.2598. [DOI] [PubMed] [Google Scholar]
- 83.Tarn W Y, Steitz J A. Modulation of 5′ splice site choice in pre-messenger RNA by 2 distinct steps. Proc Natl Acad Sci USA. 1995;92:2504–2508. doi: 10.1073/pnas.92.7.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tian M, Maniatis T. A splicing enhancer complex controls alternative splicing of doublesex pre-mRNA. Cell. 1993;74:105–114. doi: 10.1016/0092-8674(93)90298-5. [DOI] [PubMed] [Google Scholar]
- 85.Tian M, Maniatis T. A splicing enhancer exhibits both constitutive and regulated activities. Genes Dev. 1994;8:1703–1712. doi: 10.1101/gad.8.14.1703. [DOI] [PubMed] [Google Scholar]
- 86.Valcarcel J, Green M R. The SR protein family: pleiotropic functions in pre-mRNA splicing. Trends Biochem Sci. 1996;21:296–301. [PubMed] [Google Scholar]
- 87.Wang J, Manley J L. Overexpression of the SR proteins ASF/SF2 and SC35 influences alternative splicing in vivo in diverse ways. RNA. 1995;1:335–346. [PMC free article] [PubMed] [Google Scholar]
- 88.Wang J, Xiao S-H, Manley J L. Genetic analysis of the SR protein ASF/SF2: interchangeability of RS domains and negative control of splicing. Genes Dev. 1998;12:2222–2223. doi: 10.1101/gad.12.14.2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Xiao S H, Manley J L. Phosphorylation of the ASF/SF2 RS domain affects both protein-protein and protein-RNA interactions and is necessary for splicing. Genes Dev. 1997;11:334–344. doi: 10.1101/gad.11.3.334. [DOI] [PubMed] [Google Scholar]
- 90.Yang X, Bani M R, Lu S J, Rowan S, Ben-David Y, Chabot B. The A1 and A1(B) proteins of heterogeneous nuclear ribonucleoparticles modulate 5′ splice site selection in vivo. Proc Natl Acad Sci USA. 1994;91:6924–6928. doi: 10.1073/pnas.91.15.6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zahler A M, Neugebauer K M, Lane W S, Roth M B. Distinct functions of SR proteins in alternative pre-mRNA splicing. Science. 1993;260:219–222. doi: 10.1126/science.8385799. [DOI] [PubMed] [Google Scholar]
- 92.Zahler A M, Roth M B. Distinct functions of SR proteins in recruitment of U1 small nuclear ribonucleoprotein to alternative 5′ splice sites. Proc Natl Acad Sci USA. 1995;92:2642–2646. doi: 10.1073/pnas.92.7.2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhang D, Rosbash M. Identification of eight proteins that cross-link to pre-mRNA in the yeast commitment complex. Genes Dev. 1999;13:581–592. doi: 10.1101/gad.13.5.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhang W J, Wu J Y. Functional properties of p54, a novel SR protein active in constitutive and alternative splicing. Mol Cell Biol. 1996;16:5400–5408. doi: 10.1128/mcb.16.10.5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhuang Y, Weiner A M. A compensatory base change in U1 snRNA suppresses a 5′ splice site mutation. Cell. 1986;46:827–835. doi: 10.1016/0092-8674(86)90064-4. [DOI] [PubMed] [Google Scholar]
- 96.Zuo P, Manley J L. Functional domains of the human splicing factor ASF/SF2. EMBO J. 1993;12:4727–4737. doi: 10.1002/j.1460-2075.1993.tb06161.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zuo P, Manley J L. The human splicing factor ASF/SF2 can specifically recognize pre-mRNA 5′ splice sites. Proc Natl Acad Sci USA. 1994;91:3363–3367. doi: 10.1073/pnas.91.8.3363. [DOI] [PMC free article] [PubMed] [Google Scholar]