Abstract
A clear relationship exists between histone acetylation and transcriptional output, the balance of which is conferred by opposing histone acetyltransferases (HATs) and histone deacetylases (HDACs). To explore the role of HDAC activity in determining the transcriptional competency of chromatin, we have exploited the biological features of Tetrahymena as a model. Each vegetative cell contains two nuclei: a somatic, transcriptionally active macronucleus containing hyperacetylated chromatin and a transcriptionally silent, germ line micronucleus containing hypoacetylated histones. Using a PCR-based strategy, a deacetylase gene (named THD1) encoding a homolog of the yeast HDAC Rpd3p was cloned. Thd1p deacetylates all four core histones in vitro. It resides exclusively in the macronucleus during vegetative growth and is asymmetrically distributed to developing new macronuclei early in their differentiation during the sexual pathway. Together, these data are most consistent with a potential role for Thd1p in transcriptional regulation and suggest that histone deacetylation may be important for the differentiation of micronuclei into macronuclei during development.
In eukaryotes, DNA is assembled with histones to form nucleosomes, the fundamental repeating subunit of chromatin (27). Further compaction of DNA into dynamic higher-order structures is necessary for organization of complex genomes within nuclei, and can influence DNA-templated processes such as transcription and replication (21, 56). Nucleosomal histones possess a globular core domain and projecting amino-terminal “tails” known to contain a diverse array of posttranslational modifications including acetylation, phosphorylation, and methylation (15, 55). A rapidly expanding body of evidence suggests that these covalent histone modifications, acting singly or in combination, play important roles in many DNA processes, although their exact function remains to be determined (46).
A strong correlation exists between histone acetylation and the transcriptional output of resident genes packaged in a chromatin template (10, 21, 47). Transcriptionally inert chromatin is most often hypoacetylated, relative to transcriptionally active or competent regions, which are hyperacetylated. Regulation of the steady-state levels of histone acetylation is a dynamic process under the control of two competing enzymatic activities: histone acetyltransferases (HATs), which catalyze the transfer of acetyl moieties from acetyl coenzyme A to the conserved lysines in the histone tails, and histone deacetylases (HDACs), which catalyze the removal of acetyl moieties from these residues. Recently, the discovery that many HATs and HDACs had previously been identified as transcriptional regulators provided a clear mechanistic link between chromatin and gene expression (9, 10, 47).
In addition to the transcription process, histone acetylation is associated with the assembly of histones onto DNA. Newly synthesized histones H3 and H4 are both deposited into replicating chromatin in acetylated forms (39). On nascent histone H4, a diacetylation pattern involving lysine 5 and lysine 12 (K5 and K12) is conserved among a wide group of eukaryotes (1, 13, 44, 45) that probably includes yeast (25, 36). Once assembled into chromatin, newly synthesized histones are deacetylated, an event thought to be important for chromatin maturation (4, 43). Thus far, an HDAC responsible for the removal of deposition acteyl groups in vivo is not known, and the relationship, if any, between deposition- and transcription-related acetylation and deacetylation is unclear.
HDACs have been identified from a number of organisms and fall into one of three general families based on sequence homology to yeast Rpd3p, Hda1p, or the maize nucleolar phosphoprotein Hd2p (31). Rpd3p and Hda1p are catalytic subunits of distinct yeast HDAC complexes (40). Rpd3p and its homologs are capable of deacetylating lysines used in deposition-related acetylation (26, 41), but their activities are also involved in targeted gene repression, as are some Hda1p-type activities (37, 54).
To begin to explore the role of HDACs in deposition- versus transcription-related acetylation, we took advantage of the biological features of the ciliated protozoan Tetrahymena thermophila. During vegetative growth, each cell has a transcriptionally active macronucleus that contains highly acetylated chromatin and a micronucleus that is transcriptionally inert and contains unacetylated chromatin except during periods of active replication and chromatin assembly (1, 12, 51). It has been suggested that differences in chromatin structure and function between these two nuclei may be related to their global acetylation state, as determined by the histone-modifying activities present in each (12).
In this report, we describe the identification and cloning of the first ciliate HDAC gene, a gene (called THD1) encoding a Rpd3p-type HDAC. Although Thd1p is able to deacetylate newly synthesized histone H4 in vitro, it localizes specifically to the macronucleus, indicating that it is not the activity responsible for maintaining unacetylated chromatin in micronuclei by removing deposition-related acetylation. The macronuclear localization of Thd1p during vegetative growth and its selective recruitment to developing new macronuclei during the sexual pathway at a time when these nuclei become transcriptionally active raise the intriguing possibility that Thd1p plays an important role in the establishment of transcriptionally competent chromatin during nuclear differentiation.
MATERIALS AND METHODS
Strains and plasmids.
Genetically marked strains of T. thermophila, CU427 (Mpr/Mpr[6-mp-s]VI) and CU428 (Chx/Chx-[cy-s]VII), provided by Peter Bruns (Cornell University, Ithaca, N.Y.) were used in all experiments. The DNA sequences reported were derived from strain CU428. For green fluorescent protein (GFP) localization, constructs were made in the GFP vector pVGF-1 (M.-C. Yao and C.-H. Yao, unpublished data), consisting of the micronuclear rDNA vector pD5H8 (19) containing the GFP coding sequence controlled by flanking sequences of the Tetrahymena rpL29 gene, and unique cloning sites at the 3′ end. For construction of GFP-THD1, the entire THD1 open reading frame (ORF), with flanking restriction sites, was obtained by PCR oligonucleotides THD1-F (5′-AAACCTCGAGATGAAGGGATTAGATTTGTATCC-3′) and THD1-B (5′-GATGGGCCCTCACTCGATATCCATTTTTTTATC-3′). The resulting PCR product was restriction digested with ApaI and XhoI and ligated to XhoI-ApaI-digested pVGF-1 with T4 DNA ligase (New England BioLabs). The resulting plasmid was named pVGF-T.
Gene cloning and sequencing of THD1.
Degenerate oligonucleotide primers were designed against highly conserved amino acid sequences within the HDAC family (see Fig. 1A). Their sequences are as follows: Rpd3-5′, 5′-CAYCCYATGAARCCYCAYAGA-3′; and Rpd3-3′, 5′-RTAWCCWCCWCCWCC-3′ (degenerate bases in oligonucleotide primers: R, A or G; Y, C or T; and W, A or T). Total cDNA from vegetatively growing cells, produced by the method of Barnard et al. (7), was used to amplify a product, which was cloned and sequenced. An internal, gene-specific sequence was used to design a third primer, Rpd3-B (5′-CTGTCGTTTTATAATGTGG-3′), that was used in conjunction with an oligo(dT) primer to amplify a cDNA copy of the 3′ end of the gene. The 5′ end of the gene was obtained using rapid amplification of cDNA ends (cRACE), as previously described (34). For this DNA, the 3′ cDNA was synthesized by PCR using a sequence-specific oligonucleotide primer, RACE-P (5′-GCTATATAAACGATTGTGTT-3′). The product was circularized by end ligation with RNA ligase (New England BioLabs) and amplified by PCR using the oligonucleotide primers RACE-1 (5′-GTTGATATTGCACTTAACTGG-3′) and RACE-2 (5′-GGTTTGCATCATGCTAAGC-3′). All PCR products were cloned and sequenced as described previously (11).
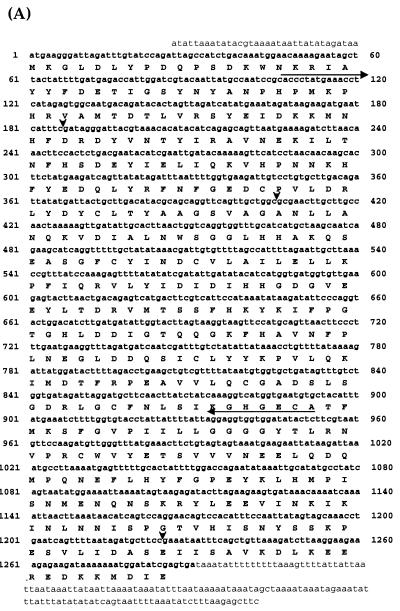
FIG. 1.
Identification and cloning of a ciliate RPD3 homolog. (A) Cloned THD1 cDNA and translated amino acid sequence beginning at the first AUG. Horizontal arrows indicate highly conserved HDAC sequences used to design degenerate oligonucleotide primers used in initial PCRs. Vertical arrowheads mark the positions of introns. (B) Protein sequence alignment of Thd1p, Rpd3p, and Hda1. All three proteins were aligned using ClustalW version 1.8 and printed using BoxShade version 3.21. Identical residues are shaded in black, and conserved residues are shaded in gray. The carboxyl-terminal peptide sequence used to generate Thd1p antiserum is underlined.
Northern blot analyses.
Total RNA from 5 × 106 cells was isolated by Trizol extraction as specified by the manufacturer (GIBCO-BRL). RNA (30 μg) was electrophoresed on a 2.2 M formaldehyde–1% agarose gel, blotted onto Magnagraph nylon membranes (MSI, Inc.), and hybridized with the indicated probes at 42°C in hybridization buffer containing 50% formamide.
Generation, characterization, and affinity purification of polyclonal antibodies against Thd1p.
A peptide corresponding to the amino terminus of Thd1p (see Fig. 1B) was synthesized with an additional amino-terminal cysteine for coupling to the carrier protein, keyhole limpet hemocyanin, as described by Lerner et al. (28). The coupling reaction was allowed to proceed at room temperature for several hours; the progress of the reaction was monitored by reverse-phase high-pressure liquid chromatography. Approximately 500 μg of conjugate was used in total for four separate injections into one rabbit. Preimmune serum was obtained from the same rabbit before immunization. In immunoblotting experiments, crude serum was used at a 1:1,000 dilution. Antibodies against Thd1p were purified by affinity chromatography using immunizing peptide coupled to SulphoLink (Pierce) as specified by the manufacturer.
Immunoblot analysis.
Total nuclei were collected as described previously (20) or further fractionated and isolated over a sucrose gradient by the method described by Allis and Dennison (3). The purity of different nuclei populations was assessed by fluorescence-activated cell sorter analysis of Dounce-homogenized nuclei suspended in nucleus buffer (0.25 M sucrose, 1 mM MgCl2 [pH 7.5]) and stained with 50 μg of propidium iodide per ml. Cytoplasmic fractions were prepared as previously described (38). The resulting nuclear or cytoplasmic fractions were resuspended in sodium dodecyl sulfate (SDS) gel loading buffer, boiled, and subjected by protein separation by SDS-polyacrylamide gel electrophoresis (PAGE) on an 8% polyacrylamide gel, which was then transferred to nitrocellulose and probed with a 1:50 dilution of affinity-purified antibody solution (∼20 μg of anti-Thd1p antibodies), or a 1:1,000 dilution of crude antiserum or preimmune serum as indicated. Immunoreactivity was detected by using alkaline phosphatase-conjugated secondary antibodies or by using horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G (Amersham; 1:5,000 dilution) for enhanced chemiluminescence detection as indicated.
Immunoprecipitation, sucrose density fractionation, and deacetylase assays.
The substrate for all deacetylase assays was Tetrahymena histones that were acetylated in vivo in the presence of [3H]acetate. Following extraction, all four core histones possessed multiple [3H]acetyllysine residues (determined by acid-urea PAGE [53] and fluorography [data not shown]). Core histones were isolated from the nuclei as previously described (53). Tetrahymena histones were labeled in vivo by adding sodium [3H]acetate (12.5 μCi/ml, 15.2 Ci/mmol) to a culture of growing cells at 3 × 105 cells/ml as previously described (50) or by adding [3H]lysine (2 μCi/ml, 50Ci/mmol) to a 5-h conjugating cell population as previously described (1).
For immunoprecipitations, total nuclei from 108 cells were isolated and extracted by DNase I digestion as described previously (35). For each immunoprecipitation, 1/10 of the resulting supernatant (107 cell equivalent) was incubated overnight at 4°C with 10 μg of affinity-purified Thd1p antibodies (preincubated for 2 h at room temperature with or without the immunizing peptide). For a control, 10 μl of the corresponding preimmune serum was used. A 20-μl volume of a protein A-Sepharose slurry (Pierce) was added and allowed to incubate for 1 h at 4°C. Immune complexes were pelleted by centrifugation and washed three times with 1 ml of Tris-buffered saline (TBS) (pH 7.5) containing 1% NP-40. Beads were resuspended in 200 μl of deacetylase buffer (15 mM Tris-HCl [pH 8.0], 15 mM NaCl, 1 mM EDTA), and a deacetylase assay was performed as described previously (49), except that for the substrate, 2 × 104 dpm of Tetrahymena 3H-acetylated histones was used per assay. For nuclear fractionation experiments, total macronuclei and micronuclei were collected, extracted by DNase I digestion, and fractionated over a sucrose density gradient, all as described by Ohba et al. (35). A 20-μl volume of each fraction tested was added to 180 μl of deacetylase buffer containing the 2 × 104 dpm of acetylated histone substrate and assayed as described previously (49). For visualization of [3H]acetyllysine on individual histones, histones separated through SDS-PAGE or acid-urea PAGE gels were stained with Coomassie brilliant blue R 250 and fluorographed.
Nucleotide sequence accession number.
The THD1 sequence has been submitted to GenBank and assigned accession no. AF276431.
RESULTS
Cloning of a RPD3 homolog from Tetrahymena.
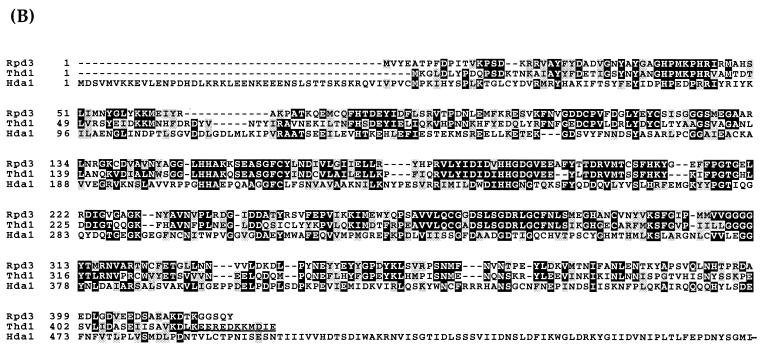
Most organisms express multiple HDACs in both the Rpd3p and Hda1p families, but little is known about the individual deacetylase enzyme(s) of Tetrahymena. HDAC activity can be detected in both the macronuclei and micronuclei of vegetatively growing cells (50; E. Wiley, unpublished data). Initial efforts to elucidate the complexity of deacetylase activities in these cells revealed two peaks of activity in total (combined macronuclear and micronuclear) nuclear extracts (see Fig. 3A).
FIG. 3.
Thd1p is in one of at least two HDAC complexes. (A) Fractions from a sucrose gradient sedimentation of combined macro- and micronuclear extracts (numbered in brackets) were assayed for HDAC activity using in vivo-labeled histones as the substrate and measuring the counts released. The fractionation profile of standard proteins with known molecular masses, run on a parallel gradient, is shown in parentheses. (B) Immunoblot analysis of the above fractions. Individual fractions (brackets) (25 μl) were resolved by SDS-PAGE and analyzed by immunoblotting with affinity-purified Thd1p antibodies.
To identify HDACs that are expressed during vegetative growth, PCR was performed on cDNA isolated from growing cells by using degenerate oligonucleotide primers corresponding to evolutionarily conserved HDAC sequences. One set of primers corresponding to the sequences most highly conserved within the Hda1p family failed to amplify detectable amounts of cDNA. Another primer set, corresponding to sequences highly conserved within the Rpd3p family, amplified a single cDNA fragment (Fig. 1A). The entire cDNA, as well as upstream and downstream sequences, was cloned using additional PCR-based techniques (see Materials and Methods for details).
The genomic copy was identified and cloned by similar techniques and was named THD1 (for “Tetrahymena histone deacetylase 1”). Comparison of the genomic and cDNA sequences indicated that THD1 contained three introns (Fig. 1A). Southern blot analysis of genomic DNA using THD1 cDNA as a probe indicated that THD1 is present in single copy in the genome (data not shown). The predicted translation of the largest ORF of the THD1 cDNA, beginning at the first AUG codon, is shown in Fig. 1A. This ORF is believed to be correct for the following reasons: (i) the DNA immediately upstream of this AUG is highly AT rich, a characteristic of most untranslated regions of Tetrahymena genes; (ii) the size of the predicted translated protein is similar to the size of homologs from other organisms (for example, see Fig. 1B); and (iii) the predicted molecular weight of the encoded protein is in agreement with that observed by immunoblotting of Tetrahymena proteins resolved by SDS-PAGE (Fig. 2). The THD1 gene encodes a 429-amino-acid polypeptide with 49% identity (66% similarity) to yeast Rpd3p and 24% identity (48% similarity) to yeast Hda1p. From this comparison, represented in Fig. 1B, Thd1p was considered to be a member of the Rpd3p family of HDACs.
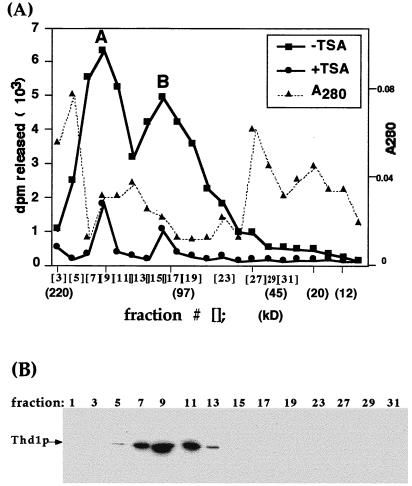
FIG. 2.
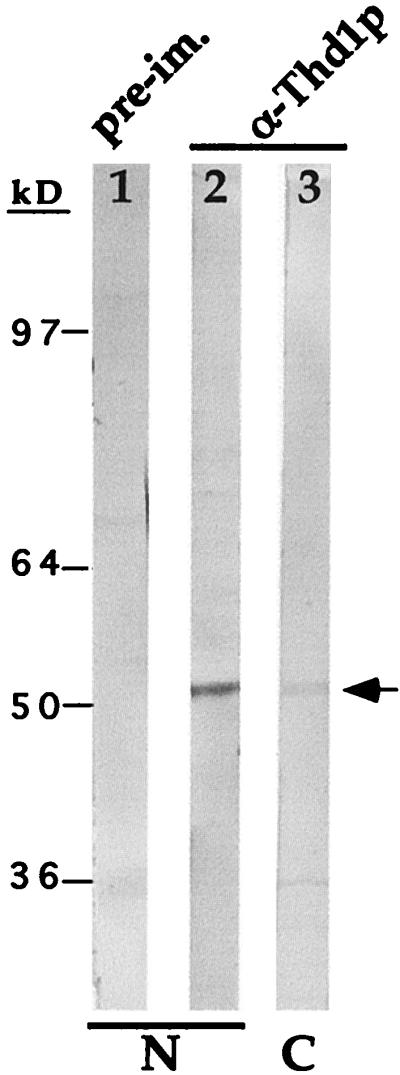
Thd1p antiserum identifies a single ∼52-kDa polypeptide in immunoblot analysis. Total nuclear proteins (macro- and micronuclear) were resolved by SDS-PAGE and transferred to nitrocellulose, and the membrane was cut into parallel strips for immunodetection with either antiserum raised against the COOH-terminal Thd1p peptide (lane 2) or preimmune serum (lane 1) and with alkaline phosphatase-conjugated secondary antibodies. A separate lane was loaded with the cytoplasmic fraction from the same cells and probed with the anti-Thd1p antiserum (α-Thd1p) (lane 3).
Thd1p antiserum identifies a 52-kDa polypeptide present in one deacetylase activity peak from total nuclear extracts.
Like other known HDACs the extreme COOH terminus of Thd1p is divergent from that of other deacetylases, making it feasible to generate antiserum specific for Thd1p. Polyclonal antiserum was raised against a COOH-terminal synthetic peptide (amino acids 414 to 429 [Fig. 1B]) and purified by affinity chromatography. In immunoblot analyses, both the crude and purified antiserum bound to a single polypeptide band present in total nuclear extracts of vegetative cells. This polypeptide, which migrated as a ∼52-kDa molecule in SDS-PAGE, was not detected by preimmune serum (Fig. 2, compare lanes 1 and 2). A smaller amount of the same polypeptide was detected in a cytoplasmic extract from the same cell equivalent (lane 3). Thus, as with Rpd3p homologs in human and mouse cells, Thd1p is concentrated in nuclei. This interpretation was later confirmed by in vivo localization of GFP-tagged Thd1p (see Fig. 6).
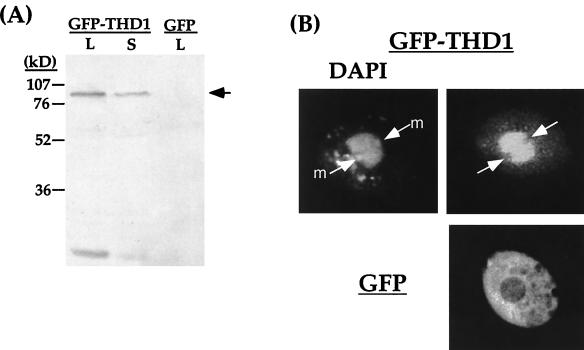
FIG. 6.
GFP-tagged Thd1p is concentrated in macronuclei and is undetectable in micronuclei in vivo. (A) Immunoblot of transformant populations expressing GFP-THD1 (L, mid-log phase; S, near-stationary phase) or GFP alone (mid-log phase) probed with Thd1p antibodies. The arrow indicates the position of GFP-Thd1p. The band detected at the bottom of the gel is probably a degradation product of GFP-Thd1p. (B) Fluorescence micrographs of transformants expressing GFP-THD1 or GFP alone. Arrows point to micronuclei. The presence of two micronuclei in these cells is discussed in the text (see Discussion). The cytoplasmic punctate DAPI staining is due to the presence of DAPI in the vacuoles of living cells.
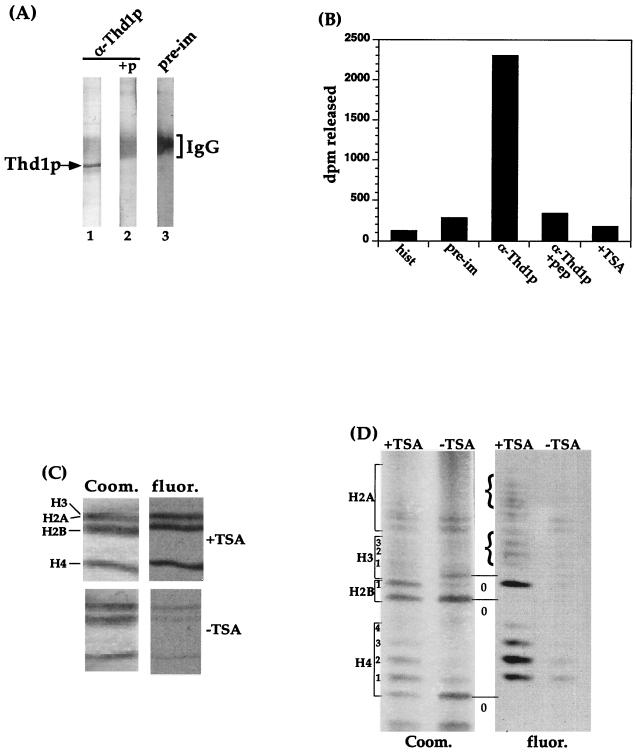
The overall complexity of Tetrahymena HDAC activity was elucidated by assaying sucrose density gradient fractions of total nuclear (micro- and macronuclear) extracts for HDAC activity using in vivo-acetylated histones as substrate. Two clear peaks of deacetylase activity were evident (Fig. 3A). By comparison with size standards fractionated on a parallel gradient, the largest peak of activity (peak A) eluted with a size corresponding to ∼150 to 160 kDa. The smaller peak (peak B) eluted with a mass of ∼100 to 110 kDa. Both activity peaks were sensitive to the noncompetitive HDAC inhibitor trichostatin A (TSA), which is a potent and specific inhibitor of many mammalian and yeast histone deacetylases in vitro (42, 57, 58). The presence of 50 nm TSA in the assay mixture reduced the activities of peaks A and B by 72 and 80%, respectively. Taken together, these data provide evidence for multiple deacetylases in vegetative Tetrahymena nuclei.
To test whether Thd1p was a component of either or both deacetylase complexes, the same density gradient fractions were subjected to immunoblot analysis with anti-Thd1p antibodies. As shown in Fig. 3B, Thd1p was detected only in the fractions comprising activity peak A, indicating that the majority of Thd1p molecules exist in a complex of ∼160 kDa. Moreover, the deacetylase activity in each fraction correlated closely with the relative amount of Thd1p detected, suggesting that Thd1p is responsible for the activity of peak A. These results also indicate that there is at least one other, as yet unknown, HDAC that is responsible for the deacetylase activity peak B.
Thd1p deacetylates each of the four core histones.
To confirm that Thd1p possessed HDAC activity, Thd1p was immunoprecipitated from nuclear extracts by using affinity-purified Thd1p antibodies (Fig. 4A, lane 1). Immunoprecipitation of Thd1p was significantly reduced by preincubating antibodies with a 10-fold molar excess of the Thd1p COOH-terminal peptide (p414–429) used to generate antiserum (lane 2, +P). The resulting immunoprecipitates were then incubated with in vivo-acetylated histone substrate, and the amount of released [3H]acetate was quantified. The anti-Thd1p immunoprecipitate released ∼14-fold more [3H]acetate than did preimmune serum (Fig. 4A, lane 3; Fig. 4B, compare the second and third bars). This activity was inhibited by the presence of 50 nM TSA in the assay reactions mixtures (Fig. 4B, right-hand bar), demonstrating that release of [3H]acetate is due to deacetylase activity. Furthermore, preincubation of anti-Thd1p antibodies with the Thd1p COOH-terminal peptide prevented both the immunoprecipitation Thd1p (Fig. 4A, lane 2) and the release of [3H]acetate from histones (Fig. 4B, fourth bar). Together, these results indicated that the deacetylase activity immunoprecipitated with anti-Thd1p antibodies (Fig. 4B, third bar) was specific to Thd1p.
FIG. 4.
Thd1p deacetylates each of the four core histones. (A) Anti-Thd1p immunoprecipitates Thd1p. Immunoprecipitation was carried out on extract from total nuclei with affinity-purified Thd1p antibodies (α-Thd1p) (lane 1), or the Thd1p antibodies preincubated with Thd1p peptide (+p) (amino acids 414 to 429) (lane 2), or with preimmune serum (pre-im) (lane 3). An equal fraction of each immunoprecipitate was analyzed by SDS-PAGE and immunoblotting with Thd1p antibodies. “IgG” marks the position of signal due to the secondary antibody reaction with immunoglobulin G. (B) An equal fraction of each immunoprecipitate was used for detection of HDAC activity as described in the legend to Fig. 3A. Bars represent the counts (in disintegrations per minute) released and extracted following incubation. The immunoprecipitates tested were preimmune serum (pre-im), anti-Thd1p antibodies (α-Thd1p), the same but with immunoprecipitate incubated with TSA prior to and during the reaction (+TSA), anti-Thd1p antibodies incubated with Thd1p COOH-peptide prior to immunoprecipitation (α-Thd1p +pep), and histones incubated alone without immunoprecipitate (hist). (C) Following incubation with immunoprecipitated Thd1p, with or without TSA in the reaction mixture, reaction products were resolved by SDS-PAGE and subjected to fluorography; Coom., Coomassie blue-stained gel; fluor., fluorographed gel. (D) Following incubation with immunoprecipitated Thd1p, with or without TSA, reaction products were analyzed as in panel C, except that acid-urea PAGE was used to resolve the histones. Numbers indicate the positions of acetylated isoforms.
To test whether Thd1p had selective specificity for any of the core histones, substrates from the reactions with immunoprecipitated Thd1p (Fig. 4B, third and fifth bars) were resolved by SDS-PAGE. [3H]acetate remaining on the resolved histones was detected by fluorography. As shown in Fig. 4C, incubation with anti-Thd1p immunoprecipitate clearly removed [3H]acetate from H2B and H4 compared with control incubations in the presence of TSA. The protein band representing both H3 and H2A also lost acetyl modifications; however, the comigration of Tetrahymena H3 and H2A in SDS-PAGE led us to resolve these histones by acid-urea PAGE followed by fluorography. This analysis demonstrated that both histones H3 and H2A lost acetyl modifications compared with the controls containing TSA (Fig. 4D). Thus, Thd1p is capable of deacetylating all four core histones in vitro with no obvious preference for any single histone under these assay conditions.
Thd1p is specific to transcriptionally active macronuclei.
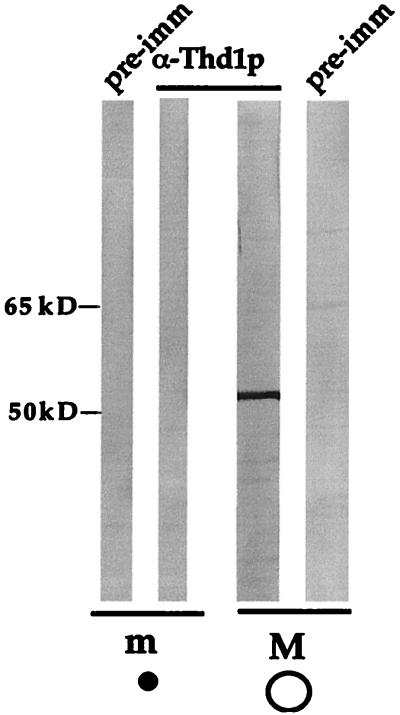
Like Rpd3p homologs in other organisms, Thd1p appeared to be concentrated in the nucleus (Fig. 2). We next assessed whether there was specificity in nuclear localization (i.e., selective partitioning to either micro- or macronuclei). In immunoblot analyses of total protein from micro- and macronuclei, which were separated by unit-gravity sedimentation, anti-Thd1p antibodies bound strongly to a single polypeptide present in extracts from macronuclei but failed to bind any polypeptide from micronuclei (Fig. 5). Therefore, Thd1p appeared to be present only in transcriptionally active macronuclei.
FIG. 5.
Thd1p is present in macronuclei but not in micronuclei of vegetative cells. Highly purified populations of macro- and micronuclei were isolated by unit-gravity sedimentation. Equivalent amounts of nuclear extract (2 × 106 macronuclei and 2 × 107 micronuclei) were resolved by SDS-PAGE and transferred to nitrocellulose. Each lane was split in two and separately probed with affinity-purified Thd1p antibodies, or with pre-immune serum. m, micronuclei (small filled circle); M, macronuclei (large open circle). The positions of size standards are indicated on the left.
However, the macronucleus-specific localization pattern observed in immunoblot analyses would also be obtained if Thd1p were selectively degraded in micronuclei during nuclear isolation. We therefore determined whether the same localization pattern was observed in vivo, using GFP-tagged Thd1p. For these experiments, GFP was fused to the entire THD1 ORF and the integrity of the GFP-THD1 fusion junction was confirmed by sequence analysis (data not shown). This fusion construct was contained within an rDNA-based vector. Upon transformation with this vector, the rDNA and flanking constructs are processed, amplified, and maintained as rDNA minichromosomes (19).
Immunoblot analysis of transformants with anti-Thd1p antibodies (Fig. 6A) confirmed that the entire Thd1 protein was synthesized in transformants, since the Thd1p antiserum, raised against the COOH terminus, detected a polypeptide of the approximate predicted molecular weight of the full-length fusion protein. Moreover, this analysis confirmed the expression of the GFP-Thd1p fusion protein at high levels during logarithmic vegetative growth. As expected, a similar polypeptide was not detected in cells expressing GFP alone. Also as expected, a reduction of GFP-Thd1p expression in transformants nearing the stationary phase was observed due to reduced rpL29 promoter function in stationary-phase cells.
In transformants expressing GFP-Thd1p, fluorescence was brightest in the macronucleus and undetectable in the micronuclei. In contrast, control transformants expressing GFP alone showed the brightest fluorescence in the cytoplasm (Fig. 6B). These results lend further support to the conclusion that Thd1p is present only in macronuclei, not in micronuclei, during vegetative growth. This localization pattern suggests that Thd1p functions specifically within the context of a transcriptionally competent nucleus.
Thd1p expression is developmentally regulated.
In Tetrahymena, the micronucleus, acting as the germ line nucleus, undergoes a developmental pathway that ultimately produces both new macronuclei and new micronuclei during the sexual process termed conjugation (reference 32 and references therein). Throughout conjugation, micronuclei undergo replication and chromatin assembly at defined intervals during mitotic and meiotic (micronuclear) divisions, and endoreplications during other intervals are required by the developing new macronucleus (2, 17). To further examine whether Thd1p is linked to deposition- versus transcription-related acetylation, Northern blot analyses of total RNA, taken at hourly intervals through conjugation, were performed.
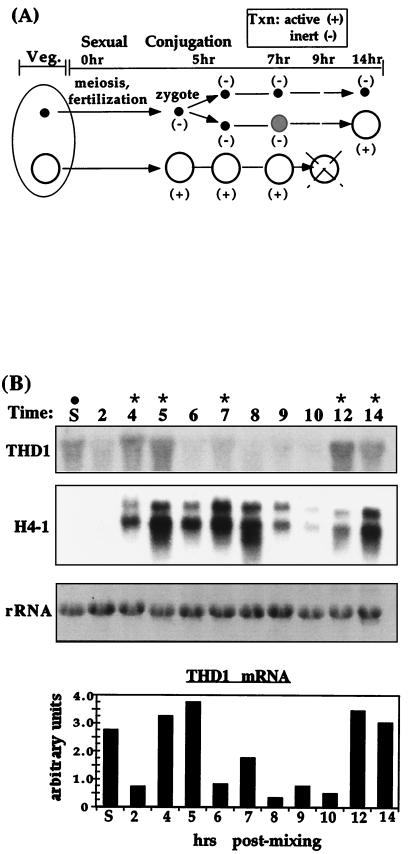
Our analyses clearly revealed that expression of Thd1p mRNA does not correlate only with the development of the new macronucleus, which occurs between 7 and 14 h of conjugation (Fig. 7A) (see below). Thd1p mRNA levels fluctuated throughout the time course examined, with peaks observed at 4 to 5, 7, and 12 to 14 h. Quantitation of THD1 mRNA (Fig. 7B, bottom) revealed a ca. five-fold increase from 4- to 5-h conjugants, a ca. twofold increase in 7-h conjugants, and a ca. sixfold increase in 12- to 14-h conjugants relative to the preceding time point shown. Interestingly, these intervals have each previously been shown to correspond closely to periods of active DNA replication and chromatin assembly (2, 17), as supported by the presence of coincident peaks in histone H4 transcript levels detected in the same RNA samples (Fig. 7B). These data demonstrate that Thd1p expression is regulated throughout development and raise the intriguing possibility that it may play an additional role in deacetylating newly synthesized histones during DNA replication. In support, Thd1p levels are elevated ca. twofold in replicating micronuclei from 5-h conjugating cells compared to the surrounding time points (Fig. 8).
FIG. 7.
Thd1p expression and nuclear localization are developmentally regulated. (A) Schematic representation of the timing of nuclear development and transcriptional activity through sexual conjugation (see reference 52 and the text for details). Micronuclei are denoted by small filled circles; developing new macronuclei (anlagen) are denoted by gray-shaded circles; and macronuclei and parental or old macronuclei (marked by a cross) are denoted by large open circles. (B) Northern hybridization analysis of total RNA from different time points throughout conjugation. Actual times postmixing are indicated by the numbers at the top. Samples were resolved by formaldehyde-agarose gel electrophoresis, transferred to a nylon membrane, and hybridized with cDNA probes of the THD1, histone H4-I, and rDNA genes. Asterisks indicate the intervals of DNA synthesis previously described (2, 17). The two bands detected with the H4-I probe are transcripts from both H4-I and H4-II genes, as previously noted (6). S, starved (premixed) cells. Note that THD1 transcript levels in S cells are reproducibly higher than in 2-h conjugants, but the difference varies (∼1.5- to ∼4-fold) from preparation to preparation. Also, macronuclear Thd1 protein levels are slightly higher but variable. Quantitation of the THD1 signal corrected for the amount of sample loaded (rDNA) was determined by PhosphorImager analysis for each sample; values (in arbitrary units) are represented by the bar graph.
FIG. 8.
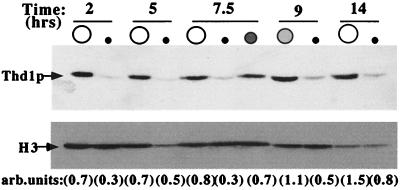
Immunodetection of Thd1p in synchronous populations of developing nuclei. Nuclei isolated by unit-gravity sedimentation were subjected to SDS-PAGE, transferred to a nitrocellulose membrane, and immunoblotted with Thd1p antibodies and anti-histone H3 antiserum as indicated. As a control, the same immunoblot was probed with antiserum against p55, a macronucleus- and anlagen-specific GCN5-type HAT, to rule out cross-contamination of micronuclear samples with macronuclei (data not shown). Fluorescence-activated cell sorter analyses (see Materials and Methods) also revealed that all micronuclear and anlagen samples were >95% pure (not shown). Numbers in parentheses indicate the relative amounts of anti-Thd1p signal corrected for anti-histone H3 signal.
Thd1p is targeted to developing new macronuclei.
During conjugation, one transcriptionally inert product of a single postzygotic mitotic division of the micronucleus ultimately differentiates into a transcriptionally active new macronucleus while the other, genetically equivalent, daughter remains transcriptionally inert and condensed. At this time, the former “old” macronucleus is no longer needed, and it is degraded by an apoptosis-like mechanism (Fig. 7A) (see reference 16 for details and additional references). The discovery that Thd1p is confined to the macronucleus during vegetative growth (Fig. 5 and 6) prompted us to determine the point in the development of new macronuclei (between 7 and 14 h [Fig. 7A]) when Thd1p levels dramatically increase.
Synchronous populations of developing nuclei recovered at different time points were collected and analyzed for the presence of Thd1p by immunoblotting with Thd1p antibodies following SDS-PAGE (Fig. 8). Unlike vegetative cells, a small and somewhat variable amount of Thd1p was detected in micronuclei at each time point of conjugation examined. At 5 h postmixing, when ∼80% of the micronuclei were undergoing division (determined by cytological staining with 4′,6-diamidino-2-phenylindole [DAPI] [see Materials and Methods]), a modest peak in micronuclear Thd1p staining was observed (detected relative to staining by general H3 antiserum used as an internal control for the amount of chromatin loaded [see bar graph]). Whether this modest peak reflects a role in removal of deposition-related acetylation sites from nascent micronuclear histones remains unknown and is an intriguing possibility.
After fertilization, two postzygotic divisions lead to the establishment of anteriorly positioned micronuclei that immediately swell and begin to differentiate into new macronuclei, becoming transcriptionally competent. In these nuclei (referred to as anlagen and shaded gray at 7.5 and 9 h [Fig. 8]), the amount of Thd1p is ca. twofold higher than in the micronuclei, which arose from the same nuclear divisions (5-h micronuclei). At the subsequent time points analyzed (9 and 14 h), the levels of Thd1p remained asymmetrically distributed between developing micro- and macronuclei. Taken together, these analyses provide evidence for developmental regulation of Thd1p in the nuclear populations. The increase in the level of Thd1 in developing new macronuclei, compared to micronuclei, early in their differentiation (7.5 h) suggests that it may play a role in the establishment of transcriptionally active chromatin in these nuclei.
DISCUSSION
In this study we describe the cloning and characterization of the first HDAC from a ciliated protozoan. The enzyme from T. thermophila, called Thd1p, is a member of the Rpd3p family. As is the case for other Rpd3p homologs, Thd1p can deacetylate all four core histones (18, 22, 26). Its localization to the macronucleus, but not the micronucleus during vegetative growth, suggests that this activity is not responsible for maintaining micronuclear chromatin in an unacetylated, condensed state. This result is consistent with earlier findings that the extremely low levels of histone acetylation observed in micronuclei are probably due in part to the absence of nuclear HAT-mediated histone acetylation (50). Our data are most consistent with a role of Thd1p functioning within the context of a transcriptionally active nucleus (see below). This interpretation agrees well with the results of studies demonstrating that Rpd3p-type enzymes are components of transcription regulatory complexes, whose activity is necessary for targeted gene repression (5, 23).
Similar to mammalian cells overexpressing the Rpd3p homolog HDAC1, which exhibit a reduced growth rate due to a severe delay during the G2 and M phases of the cell cycle (8), Tetrahymena cells producing high levels of GFP-Thd1p grew slowly and were quite large (data not shown). Moreover, many of these cells contained two micronuclei instead of one during vegetative growth (Fig. 6B), a phenotype that occurs when a macronucleus is delayed in entering or progressing through the M phase (in wild-type cells, macronuclear division immediately follows micronuclear division). In addition, GFP fluorescence in these transformants was detected for only ∼30 to 80 cell generations and was accompanied by a marked decrease in GFP-Thd1p production (data not shown). None of these observations were made for transformants expressing GFP alone, raising the interesting possibility that overproduction of Thd1p is deleterious to vegetative cells, causing several phenotypic aberrations by mechanisms that are not understood.
It is likely that only one Rpd3p-type enzyme exists in Tetrahymena, since PCR with degenerate oligonucleotides encoding amino acid sequences highly conserved within the Rpd3p family of HDACs detected the expression of only THD1. Supporting this idea, a THD1 cDNA probe failed to hybridize with other Tetrahymena genomic DNA fragments under low-stringency Southern hybridization conditions that allowed the hybridization of THD1 to yeast RPD3 (Wiley, unpublished). In addition, unlike RPD3 in yeast, THD1 appears to be an essential gene (Wiley, unpublished), indicating that it probably has no redundant functions with other Tetrahymena HDACs.
Thd1p is clearly not the only Tetrahymena HDAC, however. Fractionation experiments indicated that at least one other deacetylase activity exists in vegetative cells. This activity might be from an Hda1-type enzyme, although PCR with oligonucleotides corresponding to highly conserved Hda1 peptide sequences failed to identify such a cDNA (data not shown). Our results indicate that Thd1p expression is regulated in a dynamic fashion throughout the sequential events of sexual conjugation and nuclear differentiation. In contrast to the situation in vegetative cells, Thd1p may play a role in histone metabolism in micronuclei during early periods of the sexual pathway. Low levels of Thd1p are detected in micronuclei early in conjugation (Fig. 8), a time when micronuclei are transcriptionally active for a brief period during meiotic prophase (33, 48). From this, Thd1p activity may be important for aspects of meiosis, a possibility consistent with results of recent studies in yeast documenting a need for HDAC activity during meiosis (24).
The amino-terminal “tail” of newly synthesized histone H4 is diacetylated on lysines 5 and 12 (Ac K5/K12) in a wide range of organisms (analogous to positions 4 and 11 on Tetrahymena H4 [1, 13, 44, 45]). These “deposition-related” acetyl groups are then removed by an unknown deacetylase activity coincident with maturation of the nascent histones into chromatin (see the introduction). The ability of Rpd3p homologs to deacetylate Ac K5/K12 on H4 has been previously described (30, 41), although, as yet, none have been linked to replicating chromatin. In this study, THD1 transcript levels fluctuated in a developmentally regulated manner, where peaks coincided with intervals of DNA replication, and a modest increase in micronuclear Thd1p was detected in 4- to 5-h conjugants where micronuclei are rapidly dividing. In addition, newly synthesized, diacetylated (Ac K4/K11) H4 histones isolated from this time point are partially deacetylated by Thd1p in vitro to produce many monoacetylated molecules (data not shown), and previous experiments showed that newly synthesized H4 is deacetylated stepwise, through a clear monoacetylated intermediate in vivo (1). Together, these observations raise the possibility that Thd1p activity may play a role in the deacetylation of newly synthesized histones, although clear evidence is lacking. The suggestion that Thd1p plays a role in deacetylating nascent H4 in germ line nuclei, however, seems at odds with our failure to detect Thd1p in vegetative micronuclei. Although this apparent discrepancy remains unresolved, we note that acetylated H4 is not detected in situ in vegetative micronuclei during S phase (see the insert to Fig. 3b of reference 29), even though it is easily detected by the same assay in micronuclei from 5-h mating cells. Future investigations utilizing cells reduced in Thd1p expression will more directly address possible relationships between Thd1p activity and histone deposition in both vegetative and conjugating cells.
Levels of Thd1p increase in developing new macronuclei, but this increase is not observed in new micronuclei at 7.5 h of conjugation (Fig. 8), the time when the two types of nuclei first become morphologically and functionally distinct (32). Thus, an asymmetric distribution of Thd1p is established early in micro- and macronuclear differentiation. Previous investigations demonstrated that developing new macronuclei initiate transcription detectable by [3H]uridine autoradiography and that bulk levels of acetylated histone increase dramatically in anlagen shortly after this time (12, 29, 52). One intriguing possibility is that Thd1p is involved in creating transcription-related acetylation patterns in new macronuclei, thus establishing transcriptionally competent chromatin in developing new macronuclei. Regulation of steady-state histone acetylation levels appears to be extremely complex, with a wide range of levels maintained by different acetate turnover rates. Histones that are rapidly acetylated and deacetylated are preferentially associated with transcriptionally active or competent chromatin (14, 50). We suspect that the role of Thd1p may be closely tied to the rapid deacetylation of this class of histone as part of the transcription process.
ACKNOWLEDGMENTS
We gratefully acknowledge James Smothers and Craig Mizzen for their helpful discussions and technical advice on the cloning and biochemical characterization of Thd1p, and we thank Douglas Chalker for his critical reading of the manuscript. We give special thanks to David Goldfarb for the time and accommodations he devoted to this research.
This research was supported by grants from the National Institutes of Health to C.D.A. (GM40922), to M.C.Y. (GM26210), and to E.A.W. (GM18785-04).
REFERENCES
- 1.Allis C D, Chicoine L G, Richman R, Schulman I G. Deposition-related histone acetylation in micronuclei of conjugating Tetrahymena. Proc Natl Acad Sci USA. 1985;82:8048–8052. doi: 10.1073/pnas.82.23.8048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allis C D, Colavito-Shepanski M, Gorovsky M A. Scheduled and unscheduled DNA synthesis during development in conjugating Tetrahymena. Dev Biol. 1987;124:469–480. doi: 10.1016/0012-1606(87)90500-8. [DOI] [PubMed] [Google Scholar]
- 3.Allis C D, Dennison D K. Identification and purification of young macronuclear anlagen from conjugating cells of Tetrahymena thermophila. Dev Biol. 1982;93:519–533. doi: 10.1016/0012-1606(82)90139-7. [DOI] [PubMed] [Google Scholar]
- 4.Annunziato A T, Seale R L. Histone deacetylation is required for the maturation of newly replicated chromatin. J Biol Chem. 1983;258:12675–12684. [PubMed] [Google Scholar]
- 5.Ayer D. Histone deacetylases: transcriptional repression with SINers and NuRDs. Trends Cell Biol. 1999;9:193–198. doi: 10.1016/s0962-8924(99)01536-6. [DOI] [PubMed] [Google Scholar]
- 6.Bannon G A, Calzone F J, Bowen J K, Allis C D, Gorovsky M A. Multiple, independently regulated, polyadenylated messages for histone H3 and H4 in Tetrahymena. Nucleic Acids Res. 1983;25:3903–3917. doi: 10.1093/nar/11.12.3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barnard R, Southard J N, Talamantes F. Two-step PCR amplification of multiple specific products from cDNA using one specific primer and oligo dT. BioTechniques. 1994;16:251–252. [PubMed] [Google Scholar]
- 8.Bartl S, Taplick J, Lagger G, Khier H, Kuchler K, Seiser C. Identification of mouse histone deacetylase 1 as a growth factor-inducible gene. Mol Cell Biol. 1997;17:5033–5043. doi: 10.1128/mcb.17.9.5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Belotserkovskaya R, Sterner D E, Deng M, Sayre M H, Lieberman P M, Berger S L. Inhibition of TATA-binding protein function by SAGA subunits Spt3 and Spt8 at Gcn4-activated promoters. Mol Cell Biol. 2000;20:634–647. doi: 10.1128/mcb.20.2.634-647.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown C E, Lechner I, Howe I, Workman J L. The many HATs of transcription coactivators. Trends Biochem Sci. 2000;25:15–19. doi: 10.1016/s0968-0004(99)01516-9. [DOI] [PubMed] [Google Scholar]
- 11.Brownell J E, Zhou J, Ranalli T, Kobayashi R, Edmondson D G, Roth S Y, Allis C D. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 12.Chicoine L G, Allis C D. Regulation of histone acetylation during macronuclear differentiation in Tetrahymena: evidence for control at the level of acetylation and deacetylation. Dev Biol. 1986;116:477–485. doi: 10.1016/0012-1606(86)90148-x. [DOI] [PubMed] [Google Scholar]
- 13.Chicoine L G, Schulman I G, Richman R, Cook R G, Allis C D. Nonrandom utilization of acetylation sites in histones isolated from Tetrahymena. J Biol Chem. 1986;261:1071–1076. [PubMed] [Google Scholar]
- 14.Covault J, Chalkley R. The identification of distinct populations of acetylated histone. J Biol Chem. 1980;255:9110–9116. [PubMed] [Google Scholar]
- 15.Davie J R. Histone modifications, chromatin structure, and the nuclear matrix. J Cell Biochem. 1996;62:149–157. doi: 10.1002/(sici)1097-4644(199608)62:2<149::aid-jcb2>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 16.Davis M C, Ward J G, Herrick G, Allis C D. Programmed nuclear death: apoptotic degradation of specific nuclei in conjugating Tetrahymena. Dev Biol. 1992;154:419–432. doi: 10.1016/0012-1606(92)90080-z. [DOI] [PubMed] [Google Scholar]
- 17.Doerder F P, DeBault L E. Cytofluorimetric analysis of nuclear DNA during meiosis, fertilization and macronuclear development in the ciliate Tetrahymena pyriformis, syngen 1. J Cell Sci. 1975;17:471–493. doi: 10.1242/jcs.17.3.471. [DOI] [PubMed] [Google Scholar]
- 18.Emiliani S, Fischle W, Van Lint C, Al-Abed Y, Verdin E. Characterization of a human RPD3 ortholog, HDAC3. Proc Natl Acad Sci USA. 1997;95:2795–2800. doi: 10.1073/pnas.95.6.2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Godiska R, Yao M-C. A programmed site-specific DNA rearrangement in Tetrahymena thermophila requires flanking polyurine tracts. Cell. 1990;61:1237–1246. doi: 10.1016/0092-8674(90)90688-b. [DOI] [PubMed] [Google Scholar]
- 20.Gorovsky M A, Yao M-C, Keevert J B, Pleger G L. Isolation of micro- and macronuclei of Tetrahymena pyriformis. Methods Cell Biol. 1975;9:311–327. doi: 10.1016/s0091-679x(08)60080-1. [DOI] [PubMed] [Google Scholar]
- 21.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature (London) 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 22.Hassig C, Tong J, Fleischer T, Owa T, Grable P, Ayer D, Schreiber S. A role for histone deacetylase activity in HDAC1-mediated transcriptional repression. Proc Natl Acad Sci USA. 1998;95:3519–3524. doi: 10.1073/pnas.95.7.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hassig C A, Schreiber S L. Nuclear histone acetylases and deacetylases and transcriptional regulation: HATs off to HDACs. Curr Opin Chem Biol. 1997;1:300–308. doi: 10.1016/s1367-5931(97)80066-x. [DOI] [PubMed] [Google Scholar]
- 24.Kim Y B, Honda A, Yoshida M, Horinouchi S. phd1+, a histone deacetylase gene of Schizosaccharomyces pombe, is required for the meiotic cell cycle and resistance to trichostatin A. FEBS Lett. 1998;436:193–196. doi: 10.1016/s0014-5793(98)01124-7. [DOI] [PubMed] [Google Scholar]
- 25.Kleff S, Andrulis E D, Anderson C W, Sternglanz R. Identification of a gene encoding a yeast histone H4 acetyltransferase. J Biol Chem. 1995;270:24674–24677. doi: 10.1074/jbc.270.42.24674. [DOI] [PubMed] [Google Scholar]
- 26.Kolle D, Brosch G, Lechner T, Pipal A, Helliger W, Taplick J, Loidl P. Different types of maize histone deacetylases are distinguished by a highly complex substrate and site specificity. Biochemistry. 1999;38:6769–6773. doi: 10.1021/bi982702v. [DOI] [PubMed] [Google Scholar]
- 27.Kornberg R D, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryotic chromosome. Cell. 1999;98:285–294. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- 28.Lerner R A, Green N, Alexander H, Liu F T, Sutcliffe J G, Shinnick T M. Chemically synthesized peptides predicted from the nucleotide sequence of the hepatitis B virus genome elicit antibodies reactive with the native envelope protein of Dane particles. Proc Natl Acad Sci USA. 1981;78:3403–3407. doi: 10.1073/pnas.78.6.3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin R, Leone J W, Cook R G, Allis C D. Antibodies specific to acetylated histones document the existence of deposition- and transcription-related histone acetylation in Tetrahymena. J Cell Biol. 1989;108:1577–1588. doi: 10.1083/jcb.108.5.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loidl P. Histone acetylation: facts and questions. Chromosoma. 1994;103:441–449. doi: 10.1007/BF00337382. [DOI] [PubMed] [Google Scholar]
- 31.Lusser A, B G, Loidl A, Haas H, Loidl P. Identification of maize histone deacetylase HD2 as an acidic nucleolar phosphoprotein. Science. 1997;277:88–91. doi: 10.1126/science.277.5322.88. [DOI] [PubMed] [Google Scholar]
- 32.Martindale D W, Allis C D, Bruns P J. Conjugation in Tetrahymena thermophila. A temporal analysis of cytological stages. Exp Cell Res. 1982;140:227–236. doi: 10.1016/0014-4827(82)90172-0. [DOI] [PubMed] [Google Scholar]
- 33.Martindale D W, Allis C D, Bruns P J. RNA and protein synthesis during meiotic prophase in Tetrahymena thermophila. J Protozool. 1985;32:644–649. doi: 10.1111/j.1550-7408.1985.tb03094.x. [DOI] [PubMed] [Google Scholar]
- 34.Maruyama I N, Rakow T L, Maruyama H I. cRACE: a simple method for identification of the 5′ end of mRNAs. Nucleic Acids Res. 1995;23:3796–3797. doi: 10.1093/nar/23.18.3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ohba R, Steger D J, Brownell J E, Mizzen C A, Cook R G, Cote J, Workman J L, Allis C D. A novel H2A/H4 nucleosomal histone acetyltransferase in Tetrahymena thermophila. Mol Cell Biol. 1999;19:2061–2068. doi: 10.1128/mcb.19.3.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parthun M R, Widom J, Gottschling D E. The major cytoplasmic histone acetyltransferase in yeast: links to chromatin replication and histone metabolism. Cell. 1996;87:85–94. doi: 10.1016/s0092-8674(00)81325-2. [DOI] [PubMed] [Google Scholar]
- 37.Pazin M J, Kadonaga J T. What's up and down with histone deacetylation and transcription? Cell. 1997;89:325–328. doi: 10.1016/s0092-8674(00)80211-1. [DOI] [PubMed] [Google Scholar]
- 38.Richman T, Chicoine L G, Collini M P, Cook R G, Allis C D. Micronuclei and the cytoplasm of growing Tetrahymena contain a histone acetylase activity which is highly specific for free histone H4. J Cell Biol. 1988;106:1017–1026. doi: 10.1083/jcb.106.4.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roth S Y, Allis C D. Histone acetylation and chromatin assembly: a single escort, multiple dances? Cell. 1996;87:5–8. doi: 10.1016/s0092-8674(00)81316-1. [DOI] [PubMed] [Google Scholar]
- 40.Rundlett S E, Carmen A A, Kobayashi R, Bavykin S, Turner B M, Grunstein M. HDA1 and RPD3 are members of distinct yeast histone deacetylase complexes that regulate silencing and transcription. Proc Natl Acad Sci USA. 1996;93:14503–14508. doi: 10.1073/pnas.93.25.14503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rundlett S E, Carmen A A, Suka N, Turner B M, Grunstein M. Transcriptional repression by UME6 involves deacetylation of lysine 5 of histone H4 by RPD3. Nature. 1998;392:831–835. doi: 10.1038/33952. [DOI] [PubMed] [Google Scholar]
- 42.Sanchez Del Pino M, Lopez-Rodas G, Sendra R, Tordera V. Properties of the yeast nuclear histone deacetylase. Biochem J. 1994;303:723–729. doi: 10.1042/bj3030723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shimamura A, Worcel A. The assembly of regularly spaced nucleosomes in the Xenopus oocyte S-150 extract is accompanied by deacetylation of histone H4. J Biol Chem. 1989;264:14524–14530. [PubMed] [Google Scholar]
- 44.Sobel R E, Cook R G, Allis C D. Non-random acetylation of histone H4 by a cytoplasmic histone acetyltransferase as determined by novel methodology. J Biol Chem. 1994;269:18576–18582. [PubMed] [Google Scholar]
- 45.Sobel R E, Cook R G, Perry C A, Annunziato A T, Allis C D. Conservation of deposition-related acetylation sites in newly synthesized histones H3 and H4. Proc Natl Acad Sci USA. 1995;92:1237–1241. doi: 10.1073/pnas.92.4.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Strahl B D, Allis C D. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 47.Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 48.Sugai T, Hiwatashi K. Cytologic and autoradiographic studies of the micronucleus at meiotic prophase in Tetrahymena pyriformis. J Protozool. 1974;21:542–548. doi: 10.1111/j.1550-7408.1974.tb03695.x. [DOI] [PubMed] [Google Scholar]
- 49.Taunton J, Hassig C A, Schreiber S L. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- 50.Vavra K J, Allis C D, Gorovsky M A. Regulation of histone acetylation in Tetrahymena macro- and micronuclei. J Biol Chem. 1982;257:2591–2598. [PubMed] [Google Scholar]
- 51.Vavra K J, Pederson D S, Gorovsky M A. Nuclease sensitivity of chromatin containing active genes: kinetic analyses utilizing continuous elution of digestion products from an ultrafiltration cell. Nucleic Acids Res. 1981;9:5825–5843. doi: 10.1093/nar/9.21.5825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wenkert D, Allis C D. Timing of the appearance of macronuclear-specific histone variant hv1 and gene expression in developing new macronuclei of Tetrahymena thermophila. J Cell Biol. 1984;98:2107–2117. doi: 10.1083/jcb.98.6.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wiley E A, Mizzen C A, Allis C D. Isolation and characterization of in vivo modified histones and an activity gel assay for identification of histone acetyltransferases. Methods Cell Biol. 2000;62:379–394. doi: 10.1016/s0091-679x(08)61544-7. [DOI] [PubMed] [Google Scholar]
- 54.Wolffe A P. Transcriptional control. Sinful repression. Nature. 1997;387:16–17. doi: 10.1038/387016a0. [DOI] [PubMed] [Google Scholar]
- 55.Wolffe A P, Hayes J J. Chromatin disruption and modification. Nucleic Acids Res. 1999;27:711–720. doi: 10.1093/nar/27.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Workman J L, Kingston R E. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu Rev Biochem. 1998;67:545–579. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- 57.Yoshida M, Horinouchi S, Beppu T. Trichostatin A and trapoxin: novel chemical probes for the role of histone acetylation in chromatin structure and function. Bioessays. 1995;17:423–430. doi: 10.1002/bies.950170510. [DOI] [PubMed] [Google Scholar]
- 58.Yoshida M, Kijima M, Akita M, Beppu T. Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J Biol Chem. 1990;265:17174–17179. [PubMed] [Google Scholar]