Summary
New Omicron subvariants continue to emerge throughout the world. In particular, the XBB subvariant, which is a recombinant virus between BA.2.10.1.1 and BA.2.75.3.1.1.1, as well as the BA.2.3.20 and BR.2 subvariants that contain mutations distinct from BA.2 and BA.2.75, are currently increasing in proportion of variants sequenced. Here we show that antibodies induced by 3-dose mRNA booster vaccination as well as BA.1- and BA.4/5-wave infection effectively neutralize BA.2, BR.2, and BA.2.3.20 but have significantly reduced efficiency against XBB. In addition, the BA.2.3.20 subvariant exhibits enhanced infectivity in the lung-derived CaLu-3 cells and in 293T-ACE2 cells. Overall, our results demonstrate that the XBB subvariant is highly neutralization resistant, which highlights the need for continued monitoring of the immune escape and tissue tropism of emerging Omicron subvariants.
Keywords: SARS-CoV-2, XBB, BR.2, BA.2.3.20, neutralizing antibody, infectivity, immune evasion, mRNA vaccine
Graphical abstract
Highlights
-
•
3-dose mRNA vaccination-induced antibodies barely neutralize XBB
-
•
BA.1 and BA.4/5 infection-induced antibodies weakly neutralize XBB
-
•
BR.2 and BA.2.3.20 have similar neutralization to the parental BA.2
-
•
BA.2.3.20 exhibits enhanced infectivity in the lung-derived CaLu-3 cells
Faraone et al. investigate the infectivity and neutralization of Omicron subvariants BR.2, BA.2.3.20, and XBB. They show remarkable neutralization escape by XBB, but not BR.2 and BA.2.3.20, for 3-dose mRNA vaccination or natural BA.1 or BA.4/5 infection. BA.2.3.20 also exhibits enhanced infectivity in the lung-derived CaLu-3 cells.
Introduction
Since the emergence of the Omicron variant of severe acute respiratory syndrome virus 2 (SARS-CoV-2) in late 2021, several Omicron subvariants with varying degrees of immune evasion have emerged. The Omicron subvariant BA.2 has become the ancestral strain of several diverse lineages of subvariants including BA.4, BA.5 (the spike [S] proteins of BA.4 and BA.5 are identical and will be referred to as BA.4/5 hereafter), and BA.2.75. BA.4/5 exhibited an especially strong immune evasion phenotype and became the main circulating variant in the fall of 2022 in the United States.1
More recently, a series of subvariants derived from BA.4/5 and BA.2.75 have emerged, and some have grown steadily in case proportions across the United States.2 BA.4.6, BF.7, BQ.1, and BQ.1.1 (BA.4/5 derived) and BA.2.75.2 (BA.2.75 derived) have all exhibited marked immune evasion, especially BQ.1 and BQ.1.1.3 Previously, this phenotype has been attributed to some substitution mutations in the spike protein, particularly N460K in BQ.1 and BQ.1.1 and F486S in BA.2.75.2.3 The strength of immune evasion of vaccine-induced as well as BA.1- and BA.4/5-infection-induced immunity has allowed BQ.1 and BQ.1.1 to become the currently dominating variants in the United States.2
New Omicron subvariants continue to emerge and are closely monitored throughout the world. Of particular note, the XBB subvariant, which is a recombinant virus between BA.2.10.1.1 and BA.2.75.3.1.1.1, as well as the BA.2.3.20 and BR.2 subvariants that contain mutations distinct from BA.2 and BA.2.75, are currently increasing in proportion of variants sequenced.4 XBB emerged in Southeast Asia in September of 2022 and quickly raised concerns in other parts of world due to an increased reinfection risk.5 BA.2.3.20 has largely remained confined to Southeast Asia since its emergence in November of 2022, with some cases reported in the United States and United Kingdom, while BR.2 has become slightly more prevalent, especially in Australia, since its emergence in September 2022.6 Here we examine the sensitivity of these newly emerging subvariants to neutralization by vaccine- and infection-induced sera samples and compare them to the ancestral D614G, BA.2, and BA.2.75 subvariants.
Results
Infectivity of XBB, BR.2, and BA.2.3.20 in CaLu-3 and HEK293T-ACE2 cells
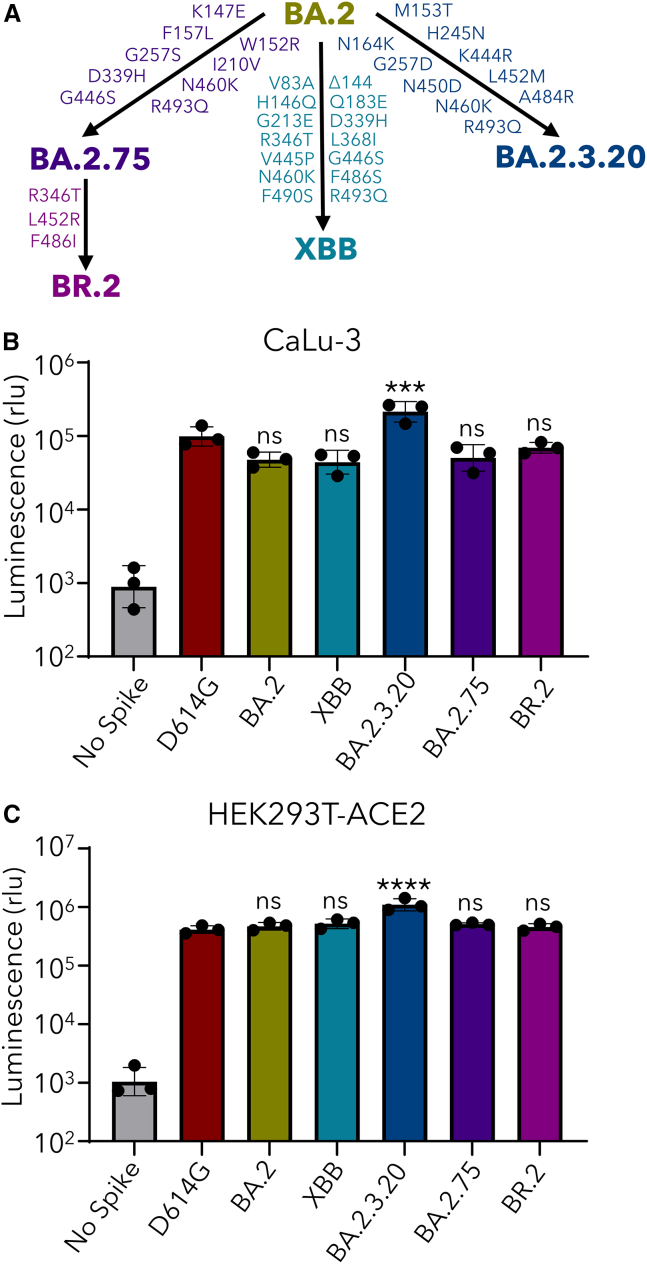
The relationship of XBB, BR.2, and BA.2.3.20 with parental BA.2 and a close relative BA.2.75 is depicted in Figure 1A, with key amino acid mutations shown. We first examined the infectivity of lentivirus pseudotyped with each of the subvariant S proteins in lung-derived CaLu-3 epithelial cells and HEK293T cells overexpressing ACE2 (HEK293T-ACE2). In CaLu-3 cells, XBB and BR.2 exhibited diminished infectivity compared with D614G, as having been shown previously for the prototype Omicron and subvariants1,3,7,8 (Figure 1B). However, the BA.2.3.20 subvariant exhibited enhanced infectivity, with 4.6 times (p < 0.0001) and 2.2 times (p < 0.001) as high infectivity as BA.2 and D614G, respectively (Figure 1B). The titers of these subvariants in HEK293T-ACE2 cells were comparable, again except BA.2.3.20, which had 2.7 times (p < 0.0001) higher titer than D614G (Figure 1C).
Figure 1.
Infectivity of Omicron XBB, BA.2.3.20, and BR.2 subvariants in 293T-ACE2 and Calu-3 cells
(A) Schematic depictions of variants investigated, including mutations that characterize each subvariant.
(B and C) Infectivity of lentiviruses pseudotyped with S from each of the indicated SARS-CoV-2 subvariants in (B) lung-derived CaLu-3 epithelial cells and (C) HEK293T-ACE2 cells. Bars in (B) and (C) represent geometric means ± standard deviation. Dots represent 3 biological replicates. Significance relative to D614G was determined by a one-way repeated measures ANOVA with Bonferroni’s multiple testing correction (n = 3). p values are displayed as ns p > 0.05, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
XBB exhibits an almost complete escape of neutralizing antibodies in sera from 3-dose-vaccinated individuals
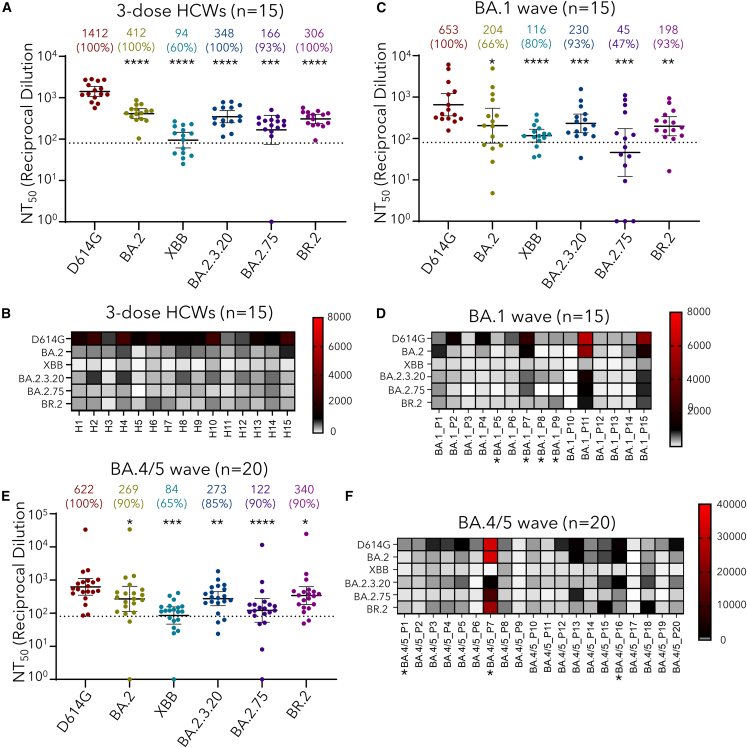
Utilizing our previously reported pseudotyped lentivirus neutralization assay,9 we examined the neutralization resistance of these SARS-CoV-2 Omicron subvariants to sera from The Ohio State University Wexner Medical Center health care workers (HCWs) vaccinated with 3 doses of mRNA vaccine (n = 15) (Table 1). The cohort included 15 HCWs that received homologous mRNA vaccine and booster doses. Sera were collected between 2 and 13 weeks after receiving a third dose of either the monovalent Moderna mRNA-1273 (n = 3) or the Pfizer BioNTech BNT162b2 (n = 12) vaccines. HCWs ranged from 26 to 61 years of age (median 33) and included 10 male and 5 female individuals. Most notably, the XBB subvariant exhibited the strongest neutralization resistance, with mean neutralizing antibody (nAb) titers 15.0 times (p < 0.0001) as low as D614G (Figures 2A and 2B). Critically, XBB exhibited significantly more immune evasion than its parent variant BA.2, with nAb titers 4.4 times lower than BA.2 (p < 0.0001). Neutralization escape by XBB was comparable to BA.2.75, with nAb titers 1.8 times lower than BA.2.75 (p > 0.05).
Table 1.
Demographic and sample collection information of HCWs and BA.1- or BA.4/5-wave patients
| Vaccinated HCWs (n = 15) | BA.1-wave hospitalized patients (n = 15) |
BA.4/5-wave first responders/household contacts (n = 20) | |
|---|---|---|---|
| Age in years at sample collection [median (range)] | 33 (26–61) | 57 (28–77) | 44 (27–58) |
| Gender [n (% of total)] | |||
| Male | 10 (66.7%) | 12 (80.0%) | 4 (20.0%) |
| Female | 5 (33.3%) | 3 (20.0%) | 15 (75.0%) |
| Sample collection window | Oct 2021–Feb 2022 | Jan 2022–Feb 2022 | Mar 2022–Sept 2022 |
| Type of vaccine [n (% of total)] | |||
| Unvaccinated | N/A | 6 (40.0%) | 17 (85.0%) |
| 3-dose Moderna | 3 (20.0%) | N/A | 2 (10.0%) |
| 3-dose Pfizer | 12 (80.0%) | 4 (26.7%) | 1 (5.0%) |
| 2-dose Moderna | N/A | 3 (20.0%) | N/A |
| 2-dose Pfizer | N/A | 2 (13.3%) | N/A |
| Sample collection timing [median (range)] | |||
| Days post third dose for recipients of 3 doses | 40 (21–86) | 166.5 (158–183) | 158 (64–183) |
| Days post diagnosis | N/A | 4 (1–7) | DNC |
| Prior SARS-CoV-2 infection confirmed by PCR [n (% of total)] | 2 (13.3%) | DNC | 2 (10.0%) |
Summary information for the HCW sera samples collected after 3 doses of mRNA vaccine is shown. In addition, summary information of the hospitalized BA.1-wave patients and first responder and household contacts testing positive for COVID-19 by PCR during the BA.4/5 wave is also provided. N/A means “not applicable” and DNC means “data not collected.”
Figure 2.
Neutralization resistance of XBB, BA.2.3.20, and BR.2 Omicron subvariants
(A–F) Neutralizing antibody titers against lentivirus pseudotyped with S from the indicated Omicron subvariants as well as ancestral D614G were determined (A and B) for sera from health care workers (HCWs) (n = 15) who received a single homologous monovalent Moderna mRNA-1273 (n = 3) or Pfizer/BioNTech BNT162b2 (n = 12) mRNA booster vaccination, (C and D) for sera from BA.1-wave hospitalized COVID-19 patients (n = 15), and (E and F) for sera from BA.4/5-wave SARS-CoV-2-infected Columbus, Ohio, first responders and household contacts (n = 20). Bars represent geometric means with 95% confidence intervals. Geometric mean NT50 values are displayed for each subvariant, with percentages of samples above the threshold of detection indicated. Dashed lines indicate the threshold of detection. Heatmaps in (B), (D), and (F) depict neutralization resistance of emerging XBB, BA.2.3.20, and BR.2 Omicron subvariants; asterisks (∗) in (D) and (F) denote individuals in the BA.1-wave and BA.4/5-wave cohorts that had received 3 doses of mRNA vaccine (Pfizer or Moderna formulations). p values are displayed as ns p > 0.05, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
In comparison, BR.2 and BA.2.3.20 demonstrated neutralization resistance comparable to BA.2, with mean nAb titers 1.3 times (p > 0.05) and 1.2 times (p > 0.05) as low as BA.2, respectively (Figures 2A and 2B). Neutralization capacity of both BR.2 and BA.2.3.20 was slightly higher compared with BA.2.75, with nAb titers 1.8 times (p > 0.05) and 2.1 times (p > 0.05) higher than BA.2.75, respectively. Overall, compared with more than 90% of HCWs exhibiting neutralizing antibody titers above the limit of detection (i.e., 1:80) for D614G and other Omicron subvariants, only 60% of HCWs had a detectable nAb titer (Figure 2A), which was also reflected in the heatmap plotting showing much lower titers for all HCWs (Figure 2B).
Dramatically reduced neutralization of XBB by sera from BA.1 and BA.4/5 convalescents
We next examined the neutralization resistance of these Omicron subvariants to sera from BA.1-wave hospitalized COVID-19 patients (n = 15) as well as BA.4/5-wave Columbus, Ohio, first responders and household contacts testing positive for COVID-19 (n = 20) (Table 1). BA.1-wave COVID-19 patient samples were collected 1–7 days after hospitalization during a BA.1 subvariant-dominant period in Columbus, Ohio (end of January 2022 through February 2022). Patients ranged from 28 to 77 years of age (median 57) and included 12 male and 3 female individuals. Among these, 6 were unvaccinated, 5 received 2 doses of either Pfizer BioNTech BNT162b2 (n = 2) or Moderna mRNA-1273 (n = 3), and 4 patients had received 3 doses of Pfizer BioNTech BNT162b2. BA.4/5-wave samples were collected from first responders and household contacts that tested positive for COVID-19 during the BA.4/5 wave in Columbus, Ohio (late July 2022 through late September 2022). For all patients, nasal swab samples were sequenced to confirm the specific variant that mediated infection, with 4 patients having been infected by BA.4 and 7 patients having been infected with BA.5. The variant could not be determined in the remaining 9 patients, but the dates of sample collection aligned with a BA.4- and BA.5-dominant period in Columbus, Ohio (late July 2022 through late September 2022). Patients ranged from 27 to 58 years of age (median 44) and included 4 male and 15 female individuals. The age and gender of one individual are unknown. This cohort included 17 unvaccinated individuals and 3 individuals who had received 3 doses of either the Pfizer BioNTech BNT162b2 (n = 1) or Moderna mRNA-1272 (n = 2) vaccines.
The neutralization titers of BA.1 and BA.4/5 infection-induced antibodies were generally lower than that of mRNA vaccination, consistent with our previous report.3 Similar to the 3-dose booster mRNA vaccination, XBB exhibited a dramatically increased neutralization resistance to infection-induced sera, with mean nAb titers 5.6 times (p < 0.0001) as low as D614G for the BA.1-wave patients (Figures 2C and 2D) and 7.4 times (p < 0.001) as low as D614G for the BA.4/5-wave COVID-19-positive first responders and household contacts (Figures 2E and 2F), respectively. In particular, only 65% of BA.4/5-infected patients had a nAb titer above the limit of detection (Figure 2F). Of note, XBB exhibited approximately comparable escape from BA.1-induced neutralizing antibodies compared with BA.2, with nAb titers 1.8 times (p > 0.05) lower than BA.2 (Figures 2C and 2D). For the BA.4/5 convalescent samples, XBB exhibited significantly more escape than BA.2, with nAb titers 3.2 times (p < 0.05) lower than BA.2 (Figures 2E and 2F). nAb titers against XBB were comparable with BA.2.75 in both sets of convalescent samples (p > 0.05) (Figures 2C–2F), although BA.2.75 showed a relatively lower nAb titer in BA.1-infected patients compared with BA.4/5 due to the fact that more than 50% of BA.1 patients had no detectable level of nAb (Figure 2C–2F). nAb titers against BR.2 and BA.2.3.20 did not exhibit significant differences compared with BA.2 for both the BA.1 and BA.4/5 convalescent samples, though BR.2 did exhibit significantly higher nAb titers than BA.2.75 in the BA.4/5 samples (p < 0.005) (Figures 2C–2F). Although 4 in 15 of BA.1-wave and 3 in 20 of BA.4/5-wave patients were boosted with mRNA vaccination, which did show generally higher titers (Figures 2D and 2F; denoted by ∗), we were unable to distinguish the effect of booster vaccination on neutralizing antibody titers due to limited sample sizes.
Discussion
Overall, we have demonstrated a remarkable immune evasion capability of the emerging Omicron subvariant XBB, but not BR.2 and BA.2.3.20, by testing sera samples from 3-dose mRNA vaccinees and BA.1- and BA.4/5-wave infected individuals. An almost complete immune evasion phenotype of XBB has recently been corroborated by several groups,10,11,12 further highlighting the importance of continued surveillance of emerging SARS-CoV-2 variants. While infection-induced immunities can be effective against some earlier Omicron subvariants such as BA.2, they are unlikely to protect against newly emerged Omicron subvariants including XBB and BA.2.75. Hence, development of updated COVID-19 vaccines and/or application of the currently FDA-approved bivalent mRNA vaccines may help mitigate this problem.
Of additional note is the increase in viral infectivity for BA.2.3.20 in Calu-3 cells. Since Omicron’s emergence, low infectivity in this human lung-derived epithelial cell line has been a characteristic of Omicron subvariants, including the parental variants of XBB, i.e., BA.2 and BA.2.75.8 This drop in infectivity has been associated with Omicron’s decreased pathogenicity13,14 and a shift in tissue tropism toward the upper respiratory tract.15 Our results show that further examination of in vivo tissue tropism and pathogenicity of some newly emerged Omicron subvariants including the XBB and BA.2.3.20 subvariants is critical. As a whole, this study serves to emphasize the importance of continued efforts in monitoring and studying emerging variants of SARS-CoV-2.
Limitations of the study
One limitation of this work is the relatively small sample size; however, previous studies using similar cohorts have generated reliable data and conclusions that have been repeatedly confirmed. Another limitation is the use of pseudotyped virus rather than infectious SARS-CoV-2; however, this assay has been previously validated using authentic SARS-CoV-29 as have many other groups utilizing pseudotyped lentivirus-based neutralization assays. We do recognize the variability in time between booster/infection and sample collection times among different participants, and this was largely due to their specific clinical arrangements. In addition, because of the variable or limited numbers of vaccinees in the BA.1- and BA.4/5-wave infection cohorts, we did not perform subgroup analyses to separate the samples of vaccinees from the total infected populations. Nevertheless, our results demonstrate the extraordinary neutralization resistance of the emerging recombinant XBB Omicron subvariant of SARS-CoV-2 to vaccine- and infection-induced sera as well as the enhanced infectivity of the emerging BA.2.3.20 subvariant. This strong immune evasion phenotype aligns with previous reports.16 Continued monitoring and testing of newly emerging SARS-CoV-2 variants, especially recombinants, is critical to inform public health responses and elucidate SARS-CoV-2 evolution.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological samples | ||
| 3-dose HCWs Sera | 1,17 | N/A |
| Omicron BA.1-wave Infected Patient Sera | 1,17 | N/A |
| Omicron BA.4/5-wave Infected First Responders and Household Contacts Sera | 3 | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Transporter 5 Transfection Reagent | Polysciences | Cat# 26008-5 |
| Dulbecco’s Modified Eagles Medium (DMEM) | Sigma-Aldrich | Cat#: 11965-092 |
| Fetal Bovine Serum (FBS) | Thermo Fisher Scientific | Cat#: F1051 |
| 0.05% Trypsin +0.53 mM EDTA | Corning | Cat# 25-052-CI |
| Penicillin-Streptomycin | HyClone | Cat#: SV30010 |
| QIAprep Spin Miniprep Kit | QIAGEN | Cat# 27106 |
| Coelenterazine | GoldBio | Cat#: CZ2.5, CAS: 55779-48-1 |
| Deposited data | ||
| NT50 Values and De-identified patient data | SeroNet Coordinating Center, NCI, NIH | N/A∗ |
| Experimental models: Cell lines | ||
| HEK293T | ATCC | Cat#: CRL-11268, RRID: CVCL_1926 |
| HEK293T-ACE2 | BEI Resources | Cat#: NR-52511, RRID: CVCL_A7UK |
| Calu-3 | ATCC | RRID: CVCL_0609 |
| Recombinant DNA | ||
| pNL4-3-inGluc | David Derse, NIH18 | N/A |
| pcDNA3.1-SARS-CoV-2-Flag-S-Flag_D614G | GenScript Biotech19 | N/A |
| pcDNA3.1-SARS-CoV-2-Flag-S-Flag_BA.2 | GenScript Biotech17 | N/A |
| pcDNA3.1-SARS-CoV-2-Flag-S-Flag_BA.2.75 | GenScript Biotech8 | N/A |
| pcDNA3.1-SARS-CoV-2-Flag-S-Flag_XBB | This paper | N/A |
| pcDNA3.1-SARS-CoV-2-Flag-S-Flag_BA.2.3.20 | This paper | N/A |
| pcDNA3.1-SARS-CoV-2-Flag-S-Flag_BR.2 | This paper | N/A |
| Software and algorithms | ||
| GraphPad Prism | Version 9.0.0 | GraphPad |
| Other | ||
| Cytation 5 Imaging Reader | BioTek | N/A |
Accession numbers have not been provided to the lab yet from the NIH SeroNet program because data must first be published before deposition. De-identified information is available upon request made to the corresponding author and will be deposited with the NIH SeroNet Coordinating Center.
Resource availability
Lead contact
Further information and requests for reagents and resources can be requested from the lead contact, Dr. Shan-Lu Liu (liu.6244@osu.edu).
Material availability
Plasmids generated for this study can be made available upon request from the lead contact.
Experimental model and subject details
Samples and patient information
Informed consent was obtained for each of the study groups described. Vaccinated HCW samples were collected under approved IRB protocols (2020H0228, 2020H0527, and 2017H0292). The cohort included 15 HCWs that received homologous vaccine and booster doses. Sera were collected between 2 and 13 weeks after receiving a third dose of either the monovalent Moderna mRNA-1273 (n = 3) or the Pfizer BioNTech BNT162b2 (n = 12) vaccines. HCWs ranged from 26 to 61 years of age (median 33) and included 10 male and 5 female individuals.
BA.1-wave COVID-19 patient samples were collected from individuals hospitalized in Columbus, OH under an approved IRB protocol (2020H0527). Samples were collected when the BA.1 subvariant was dominant in Columbus, OH (end of January 2022 through February 2022) 1–7 days after hospitalization with COVID-19. Patients ranged from 28 to 77 years of age (median 57) and included 12 male and 3 female individuals. Among these, 6 were unvaccinated, 5 received 2 doses of either Pfizer BioNTech BNT162b2 (n = 2) or Moderna mRNA-1273 (n = 3), and 4 patients had received 3 doses of Pfizer BioNTech BNT162b2.
BA.4/5-wave samples were collected from first responders and household contacts in Columbus, OH who had tested positive for SARS-CoV-2 infection (n = 20) under approved IRB protocols (2020H0527, 2020H0531, and 2020H0240). For all patients, nasal swab samples were sequenced to confirm the specific variant that mediated infection, with 4 patients having been infected by BA.4 and 7 patients having been infected with BA.5. The variant could not be determined in the remaining 9 patients, but the dates of sample collection align with when BA.4 and BA.5 were dominant in Columbus, OH (late July 2022 through late September 2022). Patients ranged from 27 to 58 years of age (median 44) and included 4 male and 15 female individuals. The age and gender of one individual are unknown. This cohort included 17 unvaccinated individuals and 3 individuals who had received 3 doses of either the Pfizer BioNTech BNT162b2 (n = 1) or Moderna mRNA-1272 (n = 2) vaccines.
Cell lines and maintenance
Human embryonic kidney cell line HEK293T (ATCC CRL-11268, RRID: CVCL_1926) and HEK293T stably expressing human ACE2 (HEK293T-ACE2) (BEI NR-52511, RRID: CVCL_A7UK) were maintained in DMEM (Gibco, 11965–092) with 10% FBS (Sigma, F1051) and 0.5% penicillin-streptomycin (HyClone, SV30010). Human adenocarcinoma lung epithelial cell line CaLu-3 (RRID:CVCL_0609) were maintained in EMEM (ATCC, 30–2003) with 10% FBS and 0.5% penicillin-streptomycin. All cell lines were passaged by washing in Dulbecco’s phosphate buffer saline (Sigma, D5652–10X1L) and detached with 0.05% Trypsin+0.53 mM EDTA (Corning, 25-052-CI). Cells were maintained at 37°C and 5.0% CO2 in 10cm cell culture dishes.
Method details
Plasmids
Our pseudotyped lentiviral vectors were produced as previous described.9,18,20 The vector is based in the HIV-1 pNL4-3 backbone and includes a deletion of the Env gene. Additionally, it carries a Gaussia luciferase reporter gene that is expressed and secreted in target cells. SARS-CoV-2 spike constructs were synthesized and cloned into the pcDNA3.1 plasmid backbone through restriction enzyme cloning (Kpn I and BamH I) by GenScript Biotech (Piscataway, NJ) in the case of XBB and BA.2.3.20. BR.2 was generated through PCR mutagenesis and confirmed by sequencing. All spike constructs bear N- and C-terminal FLAG tags.
Pseudotyped lentivirus production and infectivity
Pseudotyped lentiviral vectors were produced as previously reported.9 HEK293T cells were transfected in a 2:1 ratio with the pNL4-3-inGluc vector and the spike constructs using polyethyleneimine transfection (Transporter 5 Transfection Reagent, Polysciences) to generate pseudotyped lentiviral particles. Media from the transfected cells was collected 48 and 72 h post-transfection. Relative infectivity was measured in HEK293T-ACE2 and CaLu-3 cells. Readout of Gaussia luciferase activity at 72 h (HEK293T-ACE2) and 120 h (CaLu-3) post-infection was used to determine relative infectivity. To measure Gaussia luciferase activity, equal volumes of infected cell media and Gaussia luciferase substrate (0.1 M Tris pH 7.4, 0.3 M sodium ascorbate, 10 μM coelenterazine) were combined and luminescence signal recorded immediately by a BioTek Cytation plate reader.
Virus neutralization assay
Neutralization assays using pseudotyped lentiviral vectors were performed as previously described.9 All serum samples were serially diluted 4-fold (final dilutions 1:80, 1:320, 1:1280, 1:5120, 1:20480, and no serum control) and equal amounts of SARS-CoV-2 pseudotyped lentivirus was added. The diluted sera and vector mix was incubated for 1 h at 37°C and then used to infect HEK293T-ACE2 cells. Gaussia luciferase activity was measured 48 and 72 h post infection as described in the previous section. 50% neutralization titers (NT50) were determined by least-squares-fit, non-linear regression in GraphPad Prism 9 (San Diego, CA).
Quantification and statistical analysis
All statistical analysis was performed using GraphPad Prism 9 and are described in the figure legends. NT50 values were determined by least-squares fit non-linear regression in GraphPad Prism 9. Throughout, statistical significance was determined using log10 transformed NT50 values to better approximate normality. Error bars in (Figures 1B and 1C) represent geometric means ± geometric standard deviation. Dots represent three biological replicates. Significance relative to D614G was determined by a one-way repeated measures ANOVA with Bonferroni’s multiple testing correction (n = 3). Bars in (Figure 2) represent geometric means with 95% confidence intervals.
Acknowledgments
We thank the NIH AIDS Reagent Program and BEI Resources for providing important reagents for this work. We also thank the Clinical Research Center/Center for Clinical Research Management of The Ohio State University Wexner Medical Center and The Ohio State University College of Medicine in Columbus, Ohio, specifically Francesca Madiai, Dina McGowan, Breona Edwards, Evan Long, and Trina Wemlinger, for logistics, collection, and processing of samples. In addition, we thank Sarah Karow, Madison So, Preston So, Daniela Farkas, and Finny Johns in the clinical trials team of The Ohio State University for sample collection and other supports. This work was supported by a fund provided by an anonymous private donor to OSU. S.-L.L., F.S., D.J., G.L., A.P., R.J.G., L.J.S., and E.M.O. were supported by the National Cancer Institute of the NIH under award no. U54CA260582. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. J.P.E. was supported by Glenn Barber Fellowship from the Ohio State University College of Veterinary Medicine. R.J.G. was additionally supported by the Robert J. Anthony Fund for Cardiovascular Research and the JB Cardiovascular Research Foundation, and L.J.S. was partially supported by NIH R01 HD095881.
Author contributions
S.-L.L. conceived and directed the project. R.J.G. led the clinical study/experimental design and implementation. J.N.F. and P.Q. performed most of the experiments. J.P.E. assisted in experiments. J.N.F., P.Q., and J.P.E. performed data processing and analyses. C.C., M.A., P.S., S.F., D.J., and G.L. provided clinical samples and related information. J.P.E., J.N.F., P.Q., and S.-L.L. wrote the paper. Y.-M.Z., L.J.S., E.M.O., and R.J.G. provided insightful discussion and revision of the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: April 25, 2023
Data and code availability
This paper does not report original code. NT50 values and de-identified patient information will be shared by the lead contact upon request. Any other additional data can be provided for reanalysis if requested from the lead contact.
References
- 1.Qu P., Faraone J., Evans J.P., Zou X., Zheng Y.-M., Carlin C., Bednash J.S., Lozanski G., Mallampalli R.K., Saif L.J., et al. Neutralization of the SARS-CoV-2 omicron BA.4/5 and BA.2.12.1 subvariants. N. Engl. J. Med. 2022;386:2526–2528. doi: 10.1056/NEJMc2206725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention Variant Proportions. COVID Data Tracker. 2022. https://covid.cdc.gov/covid-data-tracker/#variant-proportions
- 3.Qu P., Evans J.P., Faraone J., Zheng Y.M., Carlin C., Anghelina M., Stevens P., Fernandez S., Jones D., Lozanski G., et al. Enhanced neutralization resistance of SARS-CoV-2 omicron subvariants BQ.1, BQ.1.1, BA.4.6, BF.7 and BA.2.75.2. Cell Host Microbe. 2022;31:9–17.e3. doi: 10.1016/j.chom.2022.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen C., Nadeau S., Yared M., Voinov P., Xie N., Roemer C., Stadler T. CoV-Spectrum: analysis of globally shared SARS-CoV-2 data to identify and characterize new variants. Bioinformatics. 2022;38:1735–1737. doi: 10.1093/bioinformatics/btab856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.TAG-VE statement on Omicron sublineages BQ.1 and XBB . World Health Organization (WHO); 2022. [Google Scholar]
- 6.Gangavarapu K., Latif A.A., Mullen J., Alkuzweny M., Hufbauer E., Tsueng G., Haag E., Zeller M., Aceves C.M., Zaiets K., et al. SARS-CoV-2 (hCoV-19) Mutation Reports. outbreak.info. 2023. https://outbreak.info/situation-reports [DOI] [PMC free article] [PubMed]
- 7.Zeng C., Evans J.P., Qu P., Faraone J., Zheng Y.-M., Carlin C., Bednash J.S., Zhou T., Lozanski G., Mallampalli R., et al. Neutralization and stability of SARS-CoV-2 omicron variant. bioRxiv. 2021 doi: 10.1101/2021.12.16.472934. Preprint at. [DOI] [Google Scholar]
- 8.Qu P., Evans J.P., Zheng Y.-M., Carlin C., Saif L.J., Oltz E.M., Xu K., Gumina R.J., Liu S.-L. Evasion of neutralizing antibody responses by the SARS-CoV-2 BA.2.75 variant. Cell Host Microbe. 2022;30:1518–1526.e4. doi: 10.1016/j.chom.2022.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zeng C., Evans J.P., Pearson R., Qu P., Zheng Y.M., Robinson R.T., Hall-Stoodley L., Yount J., Pannu S., Mallampalli R.K., et al. Neutralizing antibody against SARS-CoV-2 spike in COVID-19 patients, health care workers, and convalescent plasma donors. JCI Insight. 2020;5 doi: 10.1172/jci.insight.143213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Uraki R., Ito M., Furusawa Y., Yamayoshi S., Iwatsuki-Horimoto K., Adachi E., Saito M., Koga M., Tsutsumi T., Yamamoto S., et al. Humoral immune evasion of the omicron subvariants BQ.1.1 and XBB. Lancet Infect. Dis. 2023;23:30–32. doi: 10.1016/S1473-3099(22)00816-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Imai M., Ito M., Kiso M., Yamayoshi S., Uraki R., Fukushi S., Watanabe S., Suzuki T., Maeda K., Sakai-Tagawa Y., et al. Efficacy of a fourth dose of covid-19 mRNA vaccine against omicron subvariants BQ.1.1 and XBB. N. Engl. J. Med. 2023:1–3. doi: 10.1056/NEJMc2214302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Q., Iketani S., Li Z., Liu L., Guo Y., Huang Y., Bowen A.D., Liu M., Wang M., Yu J., et al. Alarming antibody evasion properties of rising SARS-CoV-2 BQ and XBB subvariants. Cell. 2023;186:279–286.e8. doi: 10.1016/j.cell.2022.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shuai H., Chan J.F.W., Hu B., Chai Y., Yuen T.T.T., Yin F., Huang X., Yoon C., Hu J.C., Liu H., et al. Attenuated replication and pathogenicity of SARS-CoV-2 B.1.1.529 Omicron. Nature. 2022;603:693–699. doi: 10.1038/s41586-022-04442-5. [DOI] [PubMed] [Google Scholar]
- 14.Suzuki R., Yamasoba D., Kimura I., Wang L., Kishimoto M., Ito J., Morioka Y., Nao N., Nasser H., Uriu K., et al. Attenuated fusogenicity and pathogenicity of SARS-CoV-2 Omicron variant. Nature. 2022;603:700–705. doi: 10.1038/s41586-022-04462-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yuan S., Ye Z.-W., Liang R., Tang K., Zhang A.J., Lu G., Ong C.P., Man Poon V.K., Chan C.C.S., Mok B.W.Y., et al. Pathogenicity, transmissibility, and fitness of SARS-CoV-2 Omicron in Syrian hamsters. Science. 2022;377:428–433. doi: 10.1126/science.abn8939. [DOI] [PubMed] [Google Scholar]
- 16.Cao Y., Jian F., Wang J., Yu Y., Song W., Yisimayi A., Wang J., An R., Chen X., Zhang N., et al. Imprinted SARS-CoV-2 humoral immunity induces convergent Omicron RBD evolution. bioRxiv. 2022 doi: 10.1101/2022.09.15.507787. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Evans J.P., Zeng C., Qu P., Faraone J., Zheng Y.-M., Carlin C., Bednash J.S., Zhou T., Lozanski G., Mallampalli R., et al. Neutralization of SARS-CoV-2 omicron sub-lineages BA.1, BA.1.1, and BA.2. Cell Host Microbe. 2022 doi: 10.1016/j.chom.2022.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mazurov D., Ilinskaya A., Heidecker G., Lloyd P., Derse D. Quantitative comparison of HTLV-1 and HIV-1 cell-to-cell infection with new replication dependent vectors. PLoS Pathog. 2010;6:e1000788. doi: 10.1371/journal.ppat.1000788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zeng C., Evans J.P., Faraone J.N., Qu P., Zheng Y.M., Saif L., Oltz E.M., Lozanski G., Gumina R.J., Liu S.L. Neutralization of SARS-CoV-2 variants of concern harboring Q677H. mBio. 2021;12 doi: 10.1128/mBio.02510-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goerke A.R., Loening A.M., Gambhir S.S., Swartz J.R. Cell-free metabolic engineering promotes high-level production of bioactive Gaussia princeps luciferase. Metab. Eng. 2008;10:187–200. doi: 10.1016/j.ymben.2008.04.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This paper does not report original code. NT50 values and de-identified patient information will be shared by the lead contact upon request. Any other additional data can be provided for reanalysis if requested from the lead contact.