Abstract
SSU72 is an essential gene encoding a phylogenetically conserved protein of unknown function that interacts with the general transcription factor TFIIB. A recessive ssu72-1 allele was identified as a synthetic enhancer of a TFIIB (sua7-1) defect, resulting in a heat-sensitive (Ts−) phenotype and a dramatic downstream shift in transcription start site selection. Here we describe a new allele, ssu72-2, that confers a Ts− phenotype in a SUA7 wild-type background. In an effort to further define Ssu72, we isolated suppressors of the ssu72-2 mutation. One suppressor is allelic to RPB2, the gene encoding the second-largest subunit of RNA polymerase II (RNAP II). Sequence analysis of the rpb2-100 suppressor defined a cysteine replacement of the phylogenetically invariant arginine residue at position 512 (R512C), located within homology block D of Rpb2. The ssu72-2 and rpb2-100 mutations adversely affected noninduced gene expression, with no apparent effects on activated transcription in vivo. Although isolated as a suppressor of the ssu72-2 Ts− defect, rpb2-100 enhanced the transcriptional defects associated with ssu72-2. The Ssu72 protein interacts directly with purified RNAP II in a coimmunoprecipitation assay, suggesting that the genetic interactions between ssu72-2 and rpb2-100 are a consequence of physical interactions. These results define Ssu72 as a highly conserved factor that physically and functionally interacts with the RNAP II core machinery during transcription initiation.
Eukaryotic RNA polymerase II (RNAP II) is a multisubunit enzyme that is responsible for transcription of all protein-encoding genes. Saccharomyces cerevisiae RNAP II is a 12-subunit complex encoded by the genes RPB1 to RPB12 (reviewed in references 2, 53, and 55). The two largest subunits, Rpb1 and Rpb2, are homologous to the β′ and β subunits of bacterial RNA polymerase (RNAP), respectively (23, 49). The yeast counterpart of the bacterial α subunit appears to be shared between the Rpb3 and Rpb11 subunits (26, 57). Accordingly, Rpb1, Rpb2, Rpb3, and Rpb11 comprise the functional equivalent of the bacterial α2ββ′ core RNAP. The functions of the remaining RNAP II subunits are less clear, although five subunits, Rpb5, Rpb6, Rpb8, Rpb10, and Rpb12, are shared among all three forms of RNAP and therefore function in transcription of all genes. With the exception of Rpb4 and Rpb9, all subunits are essential for cell viability. Rpb4 forms a subcomplex with Rpb7 that is required for promoter-specific initiation but is dispensable for elongation (13).
RNAP II is unable to initiate promoter-specific transcription on its own. Promoter recognition requires the general transcription factors (GTFs), which include the TATA binding protein (TBP), TFIIB, TFIIE, TFIIF, and TFIIH (reviewed in references 17 and 33). TBP nucleates assembly of a transcription preinitiation complex by binding the TATA element and inducing a sharp bend in the DNA template. TFIIB binds the TATA-TBP complex and is responsible for defining the polarity of transcription by binding asymmetrically to the BRE element, located immediately upstream of the TATA box (28, 50). RNAP II, in association with TFIIF, binds the TATA-TBP-TFIIB ternary complex, followed by TFIIE and TFIIH. TFIIH is a multisubunit complex with catalytic activities responsible for phosphorylation of the carboxy-terminal repeat domain of the Rpb1 subunit of RNAP II, promoter melting and promoter clearance (25).
The GTFs were discovered based on their requirement for accurate initiation in vitro (29, 52) and are highly conserved among eukaryotic organisms (reviewed in references 17, 30, 33, and 38). Furthermore, the TBP and TFIIB requirements for transcription predate the divergence of Eukarya and Archaea (reviewed in reference 5). Nonetheless, the RNAP II requirement for GTFs is not universal but is instead dictated by promoter sequence and architecture (16, 35, 51). Indeed, genome-wide expression analysis revealed that most components of the RNAP II transcriptional machinery are dispensable for expression of subsets of genes (21). Conversely, since only a limited number of promoters have been analyzed in vitro, it is conceivable that additional GTFs might be identified based on their requirement for accurate initiation from specific promoters. Other factors might be identified based on their ability to stimulate activator-independent transcription. For example, the yeast Tsp1 (Sub1) protein was identified as a positive effector of in vitro transcription in the absence of a sequence-specific activator (20). Tsp1 interacts physically and genetically with TFIIB and is homologous to the human transcriptional cofactor PC4 (20, 27). Additional basal factors are likely to be identified, either by their requirement for promoter-specific initiation, by their ability to stimulate activator-independent transcription, or by genetic interactions with GTFs or RNAP II subunits.
The yeast gene encoding TFIIB (SUA7) was identified as an effector of transcriptional accuracy (36). Replacements at either of two phylogenetically invariant residues, glutamate-62 (E62) or arginine-78 (R78), caused a marked start site shift downstream of the normal site at the CYC1 and ADH1 promoters (37). Nearly identical effects on transcriptional accuracy are conferred by the sua8 alleles of RPB1, suggesting a functional interaction between TFIIB and Rpb1 during start site selection (6). In an effort to identify other factors that affect initiation, we isolated suppressors of the sua7-1 (E62K) mutation. The ssu71 and ssu73 suppressors encode altered forms of the largest subunit of TFIIF (Tfg1) and the Rpb9 subunit of RNAPII, respectively (46, 47). In contrast, ssu72 was identified as an enhancer of the TFIIB E62K defect: the ssu72-1 allele confers a heat-sensitive (Ts−) growth defect and a dramatic downstream start site shift, with both effects being dependent upon the sua7-1 allele (48).
The SSU72 gene is essential for cell viability and encodes a novel protein of undefined function (48). To further characterize Ssu72, we generated a new ssu72 allele that confers a tight Ts− phenotype, independent of a TFIIB defect. We have taken advantage of the ssu72 Ts− phenotype to isolate a new rpb2 mutation encoding an altered from of the Rpb2 subunit of RNAP II. The results presented here demonstrate that Ssu72 is a transcription factor that interacts with the core RNAP II machinery both in vivo and in vitro.
MATERIALS AND METHODS
Yeast strains.
The yeast strains used in this study were derived from LRB535 (MATa his3Δ200 leu2-3,112 ura3-52). YZS84 is isogenic to LRB535, except for the ssu72-2 allele, which was generated by site-directed mutagenesis and subsequent allele replacement, as described below. YDP19 (ssu72-2 rpb2-100 shs2-1) is a spontaneous Ts+ revertant of YZS84; YDP135 (ssu72-2 rpb2-100) is a segregant derived from a cross of YDP19 with YDP87 (ssu72-2 RPB2 SHS2). The plasmid-shuffle strain YZS89A (ssu72::LEU2 [SSU72-URA3]) was described previously (48).
Growth media, genetic methods, and phenotypes.
All growth media were prepared according to standard recipes (43). −Ino medium is synthetic complete medium lacking inositol; +Ino control medium contains 55 μM inositol. Standard yeast genetic methods were used for making crosses, selecting diploids, inducing sporulation, and dissecting tetrads (44). The following designations are used for phenotypes: Ts−, temperature sensitivity, defined by impaired growth on YPD medium at 37°C; Cs−, cold sensitivity, defined by impaired growth on YPD medium at 16°C; and Ino−, impaired growth on −Ino medium at 30°C relative to growth on +Ino control medium. Plasmids carrying the URA3 marker were counterselected on synthetic medium containing 5-fluoro-orotic acid (8).
Construction of the ssu72-2 mutant.
The ssu72-2 allele was constructed by site-directed mutagenesis as described previously (48) using synthetic oligonucleotide oZS-200 (5′-GAAGATTTGATGAATGCAGGTGGGAAATTAAAC-3′). The underlined GC bases of the GCA triplet encode alanine at amino acid position 129. The EcoRI-KpnI DNA fragment encompassing the ssu72-2 open reading frame (ORF) was cloned into the corresponding sites of plasmid pRS314 (45), creating the ssu72-2 TRP1 CEN plasmid pM721. The phenotype of ssu72-2 was initially determined by plasmid shuffle using host strain YZS89A.
Allele replacement of the normal SSU72 chromosomal locus was made by integration-excision (40). The EcoRI-KpnI fragment encompassing ssu72-2 was cloned into the corresponding sites of plasmid pRS306 (45), yielding the ssu72-2-integrating plasmid pM728. pM728 was linearized at the unique ClaI site located with the SSU72 coding region and introduced into strain LRB535. Ura+ transformants were isolated and subsequently counterselected on 5-fluoro-orotic acid medium. Strains retaining the ssu72-2 allele were identified based on the Ts− phenotype associated with this allele (see above) and confirmed by the ability of the Ts− phenotype to be complemented by plasmid-borne SSU72. Southern blot analysis confirmed integration and excision at the SSU72 locus (D. L. Pappas, Jr., unpublished results).
Isolation and sequence analysis of the rpb2-100 allele of RPB2.
The rpb2-100 allele of RPB2 was recovered by gap repair (40). Plasmid RY2119 (RPB2 URA3 CEN) was digested to completion with SnaBI, which removes most of the RPB2 ORF. Vector DNA flanked by RPB2 sequences was isolated by agarose gel electrophoresis, purified, and introduced into strain YDP135 (rpb2-100). Ura+ colonies were selected and subsequently screened for retention of the Cs− and Ino− phenotypes conferred by the rpb2-100 allele of RPB2. Plasmid DNA was recovered, amplified in Escherichia coli, and analyzed by restriction digestion to confirm gap repair by the rpb2-100 locus. This resulting plasmid, pDP73, failed to rescue the Cs− and Ino− phenotypes when introduced into strains YDP19 and YDP135, confirming recovery of rpb2-100. To facilitate sequence analysis, the 1.88-kb KpnI-EcoRI, 1.05-kb EcoRI-BglII, and 2.5-kb PstI-SalI rpb2-100 fragments from pDP73 were cloned into the KpnI-EcoRI sites of pRS316, EcoRI-BamHI sites of pRS316, and PstI-SalI sites of pRS424, respectively. Single-stranded DNA was generated using VCS-M13 helper phage in the presence of kanamycin as described previously (41). The DNA sequence of the entire RPB2 ORF was determined using an ABI Prism automated DNA sequencer. DNA sequence comparisons were performed using the BLAST algorithm accessed via the Saccharomyces Genome Database (http://genome-www.stanford.edu/Saccharomyces/).
RNA analyses.
CYC1 transcript levels were analyzed by S1 nuclease protection. Strains were grown at 30°C in synthetic complete medium containing 2% glucose to mid-log phase (optical density at 600 nm [OD600] = 0.6). Cells were collected, washed, and used to inoculate two 50-ml cultures of synthetic complete medium containing either 2% glucose or 3% glycerol plus 2% ethanol as the sole carbon sources. Cultures were incubated at 30°C for 8 h, and cells were harvested. Total RNA was isolated and S1 nuclease protection assays were performed as described previously (22), using synthetic oligonucleotides 5′-GTGTAGCACCTTTCTTAGCAGAACCGGCCTTGAATTCAGTCGGACGG and 5′-GGAATTTCCAAGATTTAATTGGAGTCGAAAGCTCGCCTTA for CYC1 and tRNAW, respectively.
INO1 transcript levels were analyzed by Northern blotting. Cells were grown at 30°C in synthetic complete medium to mid-log phase, collected by centrifugation, washed, and used to inoculate two 50-ml cultures of synthetic complete medium either lacking or containing inositol (55 μM). Cultures were incubated at 30°C for 8 h, and cells were harvested. Total RNA was extracted and analyzed as described previously (36). The INO1 probe was generated by nick translation of a 1.0-kb INO1 EcoRI-HindIII fragment derived from plasmid pJM310 in the presence of [α-32P]dCTP. The U6 control probe was a nick-translated 450 bp fragment encompassing the SNR6 gene. CYC1 and INO1 transcript levels were quantified using the National Institutes of Health Image program (http://rsb.info.nih.gov/nih-image/) and normalized to the tRNAW and U6 RNA loading control signals, respectively.
β-Galactosidase assays.
Strains harboring CYC1-lacZ (pLG265-UP1 [15]), INO1-lacZ (pJS325 [42]), or PGK1-lacZ (pN1086 [7]) reporter plasmids were grown at 30°C in −Ura medium to mid-log phase (OD600 = 0.6). Cells were harvested and resuspended in −Ura medium containing either (i) 2% glucose or 3% glycerol plus 2% ethanol for CYC1-lacZ strains, (ii) synthetic complete medium either containing or lacking 55 μM inositol for INO1-lacZ strains, or (iii) no addition for PGK1-lacZ strains. Cells were incubated for 8 h, harvested by centrifugation, and resuspended in 500 μl of breaking buffer (100 mM Tris-HCl [pH 8], 1 mM dithiothreitol, 20% glycerol; 2 mM phenylmethylsulfonyl fluoride). Cell extracts were prepared by vortexing with 0.5-mm-diameter glass beads six times in 15-s bursts. β-Galactosidase assays were done as described previously (24). Activities were determined by duplicate assays of three independent transformants and are expressed according to the following formula: (1.7 ml × OD420 units)/(0.0045 × cell extract volume [milliliters] × reaction time [minutes] × protein concentration [milligrams per milliliter]).
In vitro transcription and translation reactions.
Radiolabeled TBP, TFIIB, and Ssu72 were generated using the TNT kit (Promega) in the presence of [35S]methionine according to vendor specifications. Template DNAs were purified using a Mini-Prep kit (Qiagen). Plasmids pT7-IID (9) and pET-SUA7 were used as templates for synthesis of TBP and TFIIB, respectively. SSU72 and ssu72 allelic DNAs were amplified by PCR using oligonucleotides 5′-GGAATTCCATATGCCTAGTCATCGC and 5′-CGCGGATCCTTAGTGATGGTGATGGTGATGTTTGTAATATGAAGGAGCG. This amplifies the SSU72 ORF with sequences coding for a hexahistidine tag at the C terminus and flanking NdeI and BamHI sites (underlined). The PCR products were digested and ligated into the NdeI and BamHI sites in pCITE-4a(+) (Novagen), generating the ssu72 templates pM1540 (SSU72), pM1541 (ssu72-1), pM1542 (ssu72-2), or pM1543 (ssu72-4).
RNAP II immunoprecipitation.
Immunoprecipitation assays were performed as described previously (34), with minor modifications. Highly purified yeast RNAP II (425 ng) and in vitro-transcribed and translated input proteins (10 μl) were incubated in 250 μl of P100 buffer (50 mm Tris-acetate [pH 7.9], 10% glycerol, 100 mM potassium acetate, 14 mM magnesium acetate, 4 mM dithiothreitol, 1 mM EDTA, 50 μg of bovine serum albumin per ml, and protease inhibitors [0.5 μg of pepstatin A per ml, 0.5 μg of leupeptin per ml, 2 μg of chymostatin per ml, 2.5 μg of antipain per ml, 1 mM phenylmethylsulfonyl fluoride, 2 mM benzamidine]) at 4°C for 18 h. Pansorbin (Calbiochem) was harvested by centrifugation and resuspended in blocking buffer (50 mM Tris-acetate [pH 7.9], 100 mM potassium acetate, 25 mg of PVP-40 [Sigma] per ml and 30 mg of bovine serum albumin per ml [pH 7.9]). Following incubation for 20 min at room temperature, Pansorbin was centrifuged and resuspended to its original volume in P100 buffer. Twenty-five microliters of preblocked Pansorbin was added to each reaction mixture. Samples were incubated at 4°C for 30 min and centrifuged at 15,000 rpm for 2 min. Supernatants were transferred to fresh tubes containing 5 μg of anti-RNAP II monoclonal antibody (8WG16; Covance, Inc.) and incubated for 90 min at 4°C. Preblocked Pansorbin (25 μl) was added, and the reaction mixtures were incubated for 60 min at 4°C. The samples were centrifuged at 16,000 × g for 2 min. The pellets were washed three times with 250 μl of P100 buffer containing 0.1% NP-40 and once with 250 μl of P100 buffer. Pellets were resuspended in 40 μl of sample buffer, and 20 μl of each was resolved in a sodium dodecyl sulfate–12% polyacrylamide gel. Following electrophoresis, the gel was treated with En3Hance (Dupont NEN), dried, and exposed to a phosphor screen. Products were visualized using a PhosphorImager and ImageQuant software (Molecular Dynamics).
RESULTS
The Ssu72 protein is phylogenetically conserved.
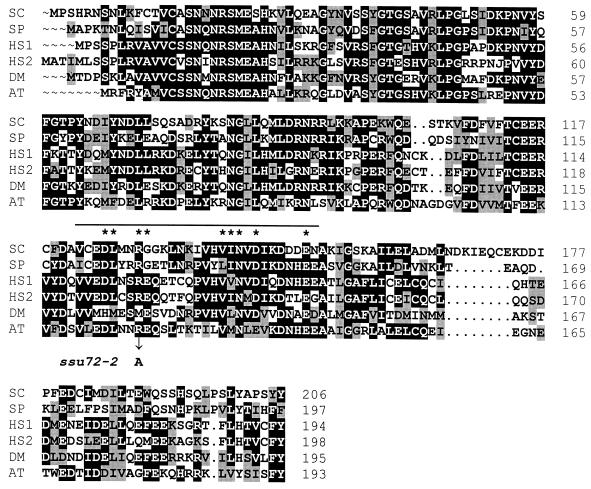
Ssu72 was reported to be a novel protein with no apparent homologs in the databases (48). Genome sequencing projects have since identified homologs of Ssu72 in Schizosaccharomyces pombe, human, Drosophila melanogaster, Arabidopsis thaliana, and Caenorhabditis elegans cells. Indeed, two distinct homologs of Ssu72 have been found in human cells. The yeast, human, fly, and plant proteins are all approximately 200 residues in length and exhibit 46 to 54% sequence identity (BLAST values of <e−39) to S. cerevisiae Ssu72 (Fig. 1). The C. elegans protein is much larger, comprising 1,357 residues, although the region of sequence similarity is confined to the N terminus, showing 43% identity to S. cerevisiae Ssu72 (data not shown). No proteins with structural similarity to Ssu72 were found among the bacterial or archaeal protein sequence databases, suggesting that Ssu72 function is common to eukaryotes but is not conserved in either eubacteria or archaea.
FIG. 1.
Sequence similarities among eukaryotic Ssu72 proteins. The Ssu72 proteins from S. cerevisiae (SC), S. pombe (SP), human (HS1 and HS2), fruit fly (DM), and A. thaliana (AT) were identified using the BLAST algorithm (1) and aligned using the University of Wisconsin Genetics Computer Group programs PILEUP and BOXSHADE. Identical and similar amino acid residues are highlighted in black and gray, respectively. The overbar encompassing residues 122 to 150 of S. cerevisiae Ssu72 indicates the region of sequence similarity to ATP-dependent RNA helicases noted previously (48). The asterisks indicate residues within this region that are phylogenetically invariant among the helicases. The ssu72-2 allele encodes an alanine replacement of the invariant arginine at position 129 (R129A).
Construction of the ssu72-2 mutant.
The original ssu72 allele (ssu72-1) enhances the sua7-1 defect by conferring a Ts− growth defect and by dramatically shifting transcription initiation downstream of the normal site at the ADH1 gene (48). These are pronounced effects but are dependent upon the sua7-1 mutation. Indeed, the ssu72-1 mutation confers no apparent phenotype in a SUA7 wild-type background.
As part of our efforts to characterize SSU72, we sought an ssu72 mutant that exhibits phenotypes independent of sua7-1. The Ssu72 amino acid sequence includes a region, encompassing residues 122 to 150, that is highly conserved among ATP-dependent RNA helicases (Fig. 1). Although other sequences common to this class of helicases, including the DEAD box and HRIGRXXR motif, are not found in Ssu72, all of the invariant residues within the conserved region are present in Ssu72 (Fig. 1). Since SSU72 is essential for cell viability, we reasoned that replacement of a conserved amino acid within this region might confer a growth phenotype. Accordingly, we constructed an allele, ssu72-2, that encodes an alanine replacement of the invariant arginine at position 129 (R129A). Using a plasmid-shuffle assay, we found that the ssu72-2 mutant was viable but failed to grow at 37°C. The plasmid-borne ssu72-2 allele was then used to replace the chromosomal copy of SSU72, yielding strain YZS84. Consistent with the phenotypes associated with the plasmid-borne ssu72-2 mutation, YZS84 was viable but distinctly Ts− (Fig. 2).
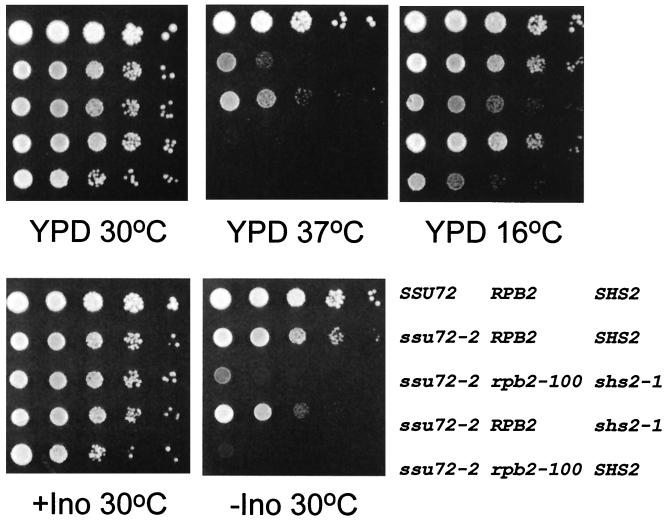
FIG. 2.
Growth defects associated with ssu72, rpb2-100, and shs2 mutations. Relevant genotypes are indicated for the wild-type strain LRB535 (SSU72 SHS1 SHS2), primary mutant YZS84 (ssu72-2 RPB2 SHS2), revertant YDP19 (ssu72-2 rpb2-100 shs2-1), a YDP19 derivative carrying a plasmid-borne copy of RPB2 (ssu72-2 RPB2 shs2-1), and the YDP135 segregant (ssu72-2 rpb2-100 SHS2). The complete genotypes of these strains and the growth media are defined in Materials and Methods.
Genetic suppression of ssu72-2.
To identify factors that interact with Ssu72, we isolated suppressors of the ssu72-2 Ts− defect. Forty spontaneous Ts+ revertants of strain YZS84 were isolated at 37°C. When scored for pleiotropic phenotypes, four revertants failed to grow in the absence of inositol yet grew normally on synthetic complete medium. This phenotype is denoted Ino− and is often associated with defects in components of the general transcriptional machinery (18). Two of the Ino− revertants were also cold sensitive (Cs−), growing poorly on YPD medium at 16°C. One of these strains, YDP19, exhibited distinct Ts+ Cs− Ino− phenotypes (Fig. 2) and was chosen for further characterization.
Strain YDP19 (Ts+ Cs− Ino−) was backcrossed with strain YDP87 (Ts− Cs+ Ino+), and the resulting diploid strain was phenotypically Ts− Cs+ and Ino+, indicating that all YDP19 phenotypes are the result of a recessive mutation(s). When this strain was sporulated and dissected (11 four-spore tetrads), the Ts+:Ts− phenotypes segregated 0:4, 1:3, or 2:2, whereas the Cs+:Cs− and Ino+:Ino− phenotypes segregated 2:2. Furthermore, all Ts+ segregants were Cs− and Ino−. These data indicate that suppression of the ssu72-2 Ts− phenotype is the result of mutations in two unlinked genes and that mutation in one of these genes confers the Cs− and Ino− phenotypes. We tentatively designated these two suppressor genes shs1-1 (rpb2-100 [see below]) and shs2-1. The shs1-1 allele is responsible for the Cs− and Ino− phenotypes, whereas shs2-1 confers no pronounced phenotype in the absence of shs1-1.
Identification of the shs1-1 suppressor.
We exploited the Cs− and Ino− phenotypes of shs1-1 to clone the wild-type gene from a YCp50 genomic library (39). From approximately 90,000 Ura+ transformants of YDP135 (ssu72-2 shs1-1), 25 Cs+ strains were isolated, 4 of which were restored to Ino+. Plasmid DNAs were isolated from each of these four transformants and were found to include overlapping DNA inserts within the YCp50 vector. One plasmid, pDP99, was reintroduced into YDP135, and all of the scored transformants were Cs+ and Ino+, indicating that complementation of the Cs− and Ino− phenotypes was a consequence of plasmid DNA rather than strain reversion.
Restriction analysis of plasmid pDP99 defined an insert of approximately 8.0 kb in the YCp50 vector. Partial DNA sequence analysis identified a segment of the yeast genome from chromosome XV encompassing RPB2, the gene encoding the second-largest subunit of RNAP II. To further define the relationship between shs1-1 and RPB2, plasmid RY2119, which includes the entire RPB2 coding region but neither of the flanking ORFs of the original clone, was introduced into YDP135. The resulting transformants were phenotypically Ts−, Cs+, and Ino+, confirming that RPB2, rather than a flanking gene, complements the suppressor mutation. To confirm that the suppressor is allelic to RPB2, the URA3 gene was integrated by homologous recombination adjacent to the RPB2 locus of strain YDP87. The resulting strain (ssu72-2 ura3 RPB2-URA3) was crossed with YDP19 (ssu72-2 ura3 shs1-1 shs2-1), and a diploid strain was sporulated and dissected. A total of 44 segregants were obtained and scored. All Ura+ progeny were phenotypically Cs+ and Ino+, and all Ura− progeny were Cs− and Ino−. Thus, the suppressor segregates opposite to RPB2, thereby confirming that shs1-1 is allelic to RPB2. Accordingly, we have renamed the shs1-1 suppressor rbp2-100.
The rpb2-100 mutation.
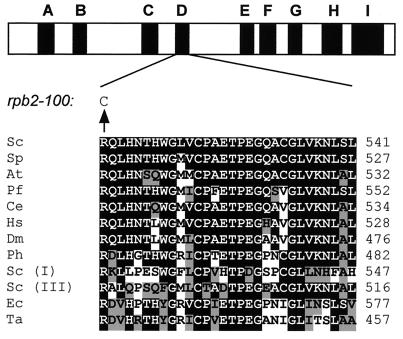
The rpb2-100 allele was cloned by gap repair, and the entire ORF was sequenced using a collection of primers that span the RPB2 ORF. A single-base-pair substitution at position 1534 (C1534T) was identified, encoding replacement of arginine-512 by cysteine (R512C). The Rpb2 subunit of RNAP II is structurally conserved among eukaryotic, archaeal, and bacterial RNAPs, including eukaryotic RNAPs I, II, and III. Sequence comparisons identified nine blocks, designated A to I, which are highly conserved among all RNAPs (49). R512 is the first amino acid within homology block D and is phylogenetically invariant (Fig. 3). The results presented here indicate that R512, despite its phylogenetic invariance, is not essential for cell viability. Indeed an rpb2-100 allele in an otherwise normal (SSU72+) genetic background displays only weak growth impairment at 30°C but is severely Cs− and Ino− (data not shown).
FIG. 3.
The rpb2-100 mutation. The rpb2-100 allele of RPB2 encodes an R512C replacement at a phylogenetically invariant position within homology block D of Rpb2. The alignment of homology block D (30 residues) is shown for S. cerevisiae (Sc), S. pombe (Sp), A. thaliana (At), Plasmodium falciparum (Pf), C. elegans (Ce), Homo sapiens (Hs), Drosophila melanogaster (Dm), Pyrococcus horikoshii (Ph), the second-largest subunits (Rpa135 and Rpc128) of RNAP I and RNAP III of S. cerevisiae [Sc (I) and Sc (III), respectively], and the β subunits of E. coli (Ec) and T. aquaticus (Ta) RNAPs. Amino acids are indicated by the single-residue code; the numbers at the right indicate the position of the C-terminal residue of block D.
The ssu72-2 and rpb2-100 alleles affect noninduced transcription in vivo.
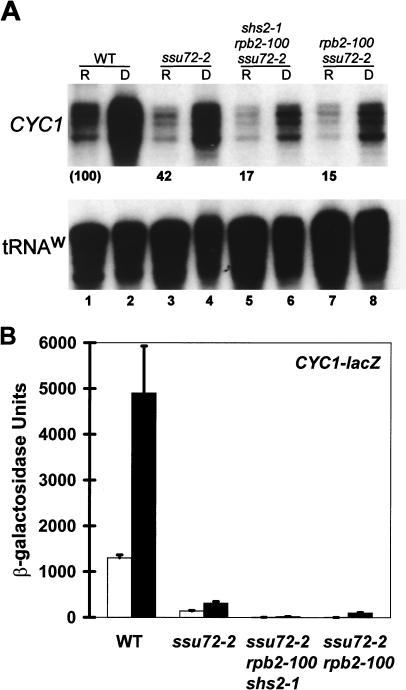
The effects of ssu72-2 and rpb2-100 mutations on transcription from specific promoters were determined in vivo using the wild-type strain (LRB535), an ssu72-2 primary mutant (YZS84), an ssu72-2 rpb2-100 shs2-1 suppressor strain (YDP19), and an ssu72-2 rpb2-100 segregant (YDP135). Effects on CYC1 expression are presented in Fig. 4A. Under repressing conditions (2% glucose), the CYC1 transcript level was reduced in the ssu72-2 mutant to 42% of normal (compare lanes 1 and 3). Expression was further diminished in the ssu72-2 rpb2-100 shs2-1 revertant to 17% of normal (lane 5), an effect that could be attributed to rpb2-100 rather than shs2-1, since the same level of expression was observed in the ssu72-2 rpb2-100 mutant (lane 7). These effects occur at the level of noninduced transcription, since all strains retained the ability to respond to derepressing conditions.
FIG. 4.
Effects of ssu72 and shs mutations on CYC1 expression. (A) The top panel indicates CYC1 transcript levels, determined by S1 nuclease protection, for strains grown under repressing (2% glucose) (lanes R) and derepressing (3% glycerol and 2% ethanol) (lanes D) conditions. Strains are LRB535 (wild type [WT]), YZS84 (ssu72-2), YPD19 (ssu72-2 rpb2-100 shs2-1), and YDP135 (ssu72-2 rpb2-100). Transcript levels under repressing conditions are presented as percentages, normalized to the level for the wild-type strain. The bottom panel is a loading control and indicates tRNAW levels. (B) β-Galactosidase activities expressed from the CYC1-lacZ plasmid pLG265-UP1. Activity units under repressing and derepressing conditions, respectively, are 1,300 ± 66 and 4,900 ± 1030 (WT), 140 ± 13 and 310 ± 40 (YZS84), 4.8 ± 1.2 and 19 ± 3.5 (YDP19), and 3.3 ± 0.54 and 97 ± 14 (YDP135). The indicated units of β-galactosidase activity represent the means for three independent transformants assayed in duplicate; standard deviations are indicated by error bars.
The effects on CYC1 transcription were confirmed using a CYC1-lacZ reporter. In the wild-type strain 1,300 U of activity was measured under repressing conditions, and this activity was enhanced 3.5-fold under derepressing conditions (Fig. 4B). Noninduced expression was diminished 9-fold in the ssu72-2 primary mutant (140 U) and was reduced an additional 30-fold in the ssu72-2 rpb2-100 shs2-1 suppressor strain (4.8 U). The shs2-1 allele did not contribute to this effect, since activity was equally low in an ssu72-2 rpb2-100 strain (3.3 U). Each mutant responded to derepressing conditions, although the absolute activities were diminished in parallel with effects on basal activity. These results are consistent with the RNA analyses (Fig. 4A), confirming that ssu72-2 and rpb2-100 diminish noninduced transcription from the CYC1 promoter.
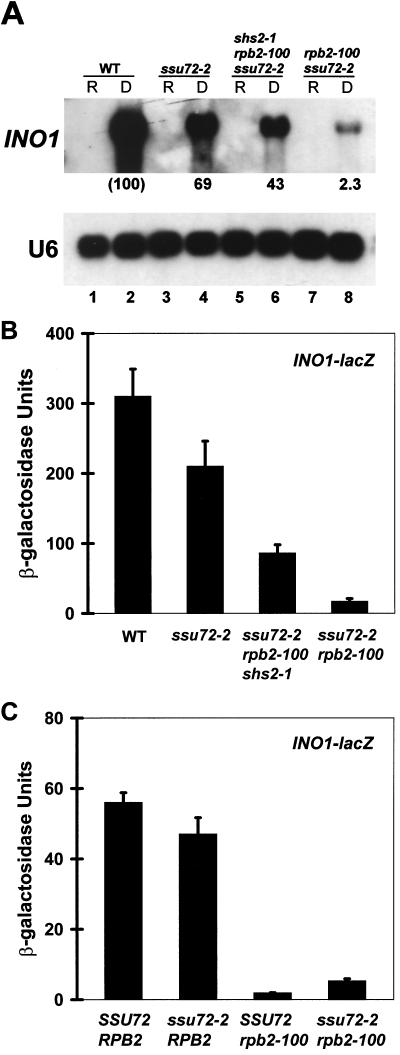
INO1 expression was determined by Northern blot analysis (Fig. 5A). Strong induction of INO1 mRNA was observed in the wild-type strain (compare lanes 1 and 2). Induced levels were diminished to approximately 70% of normal in the ssu72-2 mutant (lane 4) and were further reduced to 43% of normal in the ssu72-2 rpb2-100 shs2-1 suppressor strain (lane 6). Interestingly, rpb2-100 had a more dramatic effect in the absence of shs2-1, nearly eliminating INO1 expression (lane 8). These results are consistent with the effects of ssu72-2 and rpb2-100 on INO1-lacZ reporter gene expression under repressing conditions (Fig. 5B). Although the Northern analysis does not distinguish between effects on noninduced versus activated expression, the measurable levels of β-galactosidase activity from the INO1-lacZ reporter under repressing conditions demonstrate that the effects of ssu72-2 and rpb2-100 on INO1 expression can be accounted for by effects on noninduced transcription.
FIG. 5.
Effects of ssu72 and shs mutations on INO1 expression. (A) The top panel indicates INO1 transcript levels, determined by Northern blot analysis, for strains grown under repressing (+Ino medium) (lanes R) and derepressing (−Ino medium) (lanes D) conditions. Strains are identical to those in Fig. 4. The bottom panel is a loading control showing U6 RNA levels. (B) β-Galactosidase activities expressed from the INO1-lacZ plasmid pJS325 under repressing conditions. Activity units are 310 ± 39 (wild type [WT]), 210 ± 36 (YZS84), 86 ± 12 (YDP19), and 17 ± 4.1 (YDP135). Error bars indicate standard deviations. (C) Same as panel B, except the host strain in each case is YDP135 (ssu72-2 rpb2-100) carrying the INO1-lacZ reporter and plasmid-borne copies of wild-type SSU72, RPB2, or vectors alone, as indicated by the genotypes. Activity units are 56 ± 2.8 (WT), 47 ± 4.7 (ssu72-2), 1.9 ± 0.07 (rpb2-100), and 5.3 ± 0.63 (ssu72-2 rpb2-100).
The INO1-lacZ reporter assays were repeated using strain YDP135 that had been complemented with plasmid-borne copies of SSU72 and/or RPB2, allowing the effects of ssu72-2 and rpb2-100 to be assessed in an isogenic background independent of shs2-1. Consistent with the previous results, ssu72-2 alone had only a minor effect on INO1-lacZ expression, whereas the effects of the rpb2-100 single and ssu72-2 rpb2-100 double mutations were more dramatic, yielding less than 10% of normal activity (Fig. 5C). Thus, ssu72-2 exerts adverse effects on gene expression that are markedly enhanced by the rpb2-100 allele, and these effects are manifest at the level of noninduced transcription.
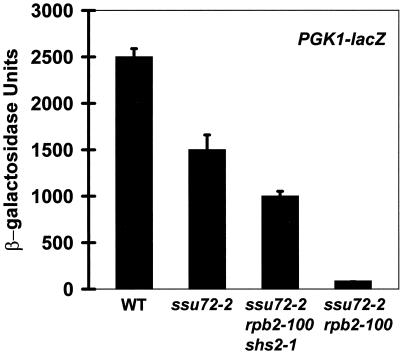
The effects of ssu72-2 and rpb2-100 were also assessed using the PGK1 promoter, which is expressed at a high constitutive level. A PGK1-lacZ reporter plasmid was introduced into each strain, and β-galactosidase activities were measured (Fig. 6). Expression from the PGK1 promoter was diminished to 60% of normal in the ssu72-2 mutant and to 40% of normal in the ssu72-2 rpb2-100 shs2-1 triple mutant. Expression was markedly diminished in the ssu72-2 rpb2-100 double mutant, to 3% of normal. Although these experiments do not distinguish noninduced from activated expression, the dramatic effect of the double mutant indicates that ssu72-2 and rpb2-100 can exert synergistic effects on gene expression. The more adverse effect of rpb2-100 in the SHS2+ background is consistent with its effect on INO1 expression (Fig. 5). These results demonstrate that shs2-1, which is required in addition to rpb2-100 for suppression of the ssu72-2 Ts− phenotype (Fig. 2), also suppresses the adverse effects of rpb2-100 on noninduced transcription.
FIG. 6.
β-Galactosidase activities expressed from a PGK1-lacZ plasmid. Strains are identical to those in Fig. 4 and in Fig. 5A and B, except that each was transformed with the PGK1-lacZ reporter plasmid pN1086. Activity units are 2,500 ± 88 (wild type [WT]), 1,500 ± 160 (YZS84), 1,000 ± 52 (YDP19), and 86 ± 3.6 (YDP135).
Physical interaction between Ssu72 and RNAP II.
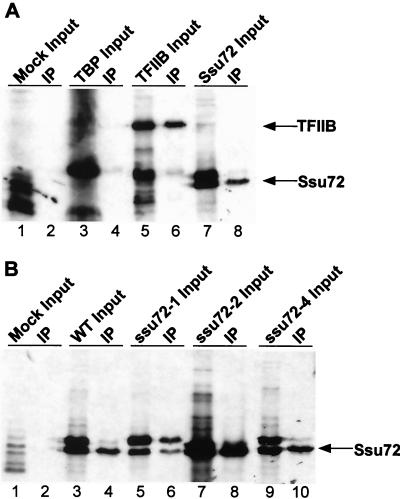
The genetic interactions among ssu72, sua7, and rpb2 suggest that Ssu72 interacts directly with the core transcriptional machinery. Indeed, Ssu72 directly binds TFIIB, consistent with allele-specific interactions between ssu72 and sua7 (54). Physical interaction between Ssu72 and RNAP II was investigated by a coimmunoprecipitation assay using purified yeast RNAP II and a monoclonal antibody directed against the Rpb1 subunit of RNAP II. Input proteins were 35S-labeled Ssu72, with labeled TBP and TFIIB as controls. RNAP II is known to bind TFIIB, but not TBP, whose interaction with RNAP II requires TFIIB as a bridging factor. The Ssu72 input included two forms of the protein, corresponding to full-length Ssu72 and N-terminally truncated Ssu72 resulting from initiation at AUG codon 23. The results demonstrate that RNAP II binds stably to TFIIB and Ssu72 but not to the TBP control (Fig. 7A). Thus, the genetic interaction between ssu72-2 and rpb2-100 is likely to reflect a physical interaction between Ssu72 and RNAP II.
FIG. 7.
Physical interaction between Ssu72 and RNAP II. (A) Equal amounts of highly purified yeast RNAP II were incubated with 35S-labeled mock, TBP, TFIIB, and Ssu72 input proteins. Samples were immunoprecipitated (IP) using a monoclonal antibody directed against the C-terminal repeat domain of Rpb1. Following centrifugation, samples were washed three times with buffer containing 100 mM potassium acetate plus 0.1% NP-40 and once with buffer containing 100 mM potassium acetate and then resuspended in sample buffer. Input proteins (5% of the total; lanes 1, 3, 5, and 7, respectively) and the precipitates (50% of the total; lanes 2, 4, 6, and 8, respectively) were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and analyzed by autoradiography. WT, wild type. (B) Same as panel A, except that different forms of Ssu72 were used as input proteins. The ssu72-1 allele encodes a duplication of amino acid residues 9 to 18, ssu72-2 encodes the R129A replacement, and ssu72-4 encodes an inviable C15S replacement. The upper and lower bands of Ssu72 input proteins correspond, respectively, to the full-length protein and a truncated protein whose N terminus corresponds to methionine at position 23. In the case of Ssu72-1, RNAP II interacts with both the full-length and truncated forms of the protein. Control reactions lacking RNAP II failed to coimmunoprecipitate Ssu72 (data not shown).
The N terminus of Ssu72 is essential for Ssu72 function and cell viability, yet RNAP II binds the smaller form of Ssu72, lacking the N-terminal 22 residues. This was further investigated by asking whether specific mutations in the Ssu72 N terminus affect RNAP II binding. The ssu72-1 allele, which enhances the sua7-1 mutation, encodes a 10-amino-acid duplication in the N terminus of Ssu72 (residues 9 to 18), and ssu72-4 encodes a serine replacement of cysteine 15 that abolishes cell viability (48). The interaction between Ssu72 and RNAP II appeared to be unaffected by the ssu72-2 (R129A) and ssu72-4 (C15S) mutations (Fig. 7B, lanes 5 to 8). Interestingly, however, the 10-amino-acid duplication resulted in preferential binding of RNAP II to the longer form of Ssu72 (Fig. 7B, lanes 1 to 4). These results confirm that Ssu72 binds directly to RNAP II and imply that the Ssu72 N terminus affects this interaction.
DISCUSSION
The results in this paper define genetic and physical interactions between Ssu72 and RNAP II and support a role for Ssu72 in basal (noninduced) transcription by RNAP II. Ssu72 was initially identified based on a genetic interaction with TFIIB. The ssu72-1 allele confers a synthetic Ts− growth defect that is dependent upon the sua7-1 mutation (48). The interaction of ssu72-1 with sua7 is specific, resulting in synthetic phenotypes only in combination with sua7 alleles that affect transcription start site selection (54). Ssu72 is also genetically linked to Sub1 (Tsp1), a homolog of the human general transcriptional cofactor PC4, in that a sub1 deletion also enhances sua7 mutations and does so in an allele-specific manner that is identical to the specificity of the ssu72-1 sua7 interactions (54). Consistent with the genetic data, TFIIB directly interacts with the Ssu72 and Sub1 proteins (27, 54). Here we demonstrate that Ssu72 binds stably to RNAP II and that the ssu72-2 and rpb2-100 alleles affect basal transcription. Taken together, these data demonstrate functional interactions of Ssu72 with TFIIB, Sub1, and Rpb2 and provide strong support for Ssu72 as an RNAP II transcription factor.
We do not know the specific function of Ssu72. Our efforts to characterize Ssu72 have been hampered because sequence tags added to the N terminus appear to expose a cryptic proteolytic site just within the Ssu72 N terminus, thereby removing the tag, whereas sequences added to the C terminus render Ssu72 nonfunctional in vivo, perhaps due to the conserved, tandem aromatic residues at the C terminus (Fig. 1). Interestingly, there are no apparent Ssu72 homologs in bacterial or archaeal genomes, implying that Ssu72 function is specific to eukaryotes. In yeast, depletion of Ssu72 causes growth arrest and diminishes total poly(A) RNA levels by about 20 to 40% but is without effect on RNAP I and RNAP III transcription (Pappas, unpublished results). These results suggest that Ssu72 is critical for transcription of many, but not all, RNAP II genes yet plays no role in RNAP I or RNAP III transcription.
All evidence points to a role for Ssu72 in core promoter function. The original ssu72-1 allele, which encodes a 10-amino-acid duplication near the N terminus, affects transcription start site selection (48). This effect is dramatic, but it is dependent upon a sua7 allele (TFIIB) that also affects initiation. Here we demonstrate that the ssu72-2 allele adversely affects noninduced transcription. The rpb2-100 suppressor of ssu72-2 also diminishes noninduced transcription, and this effect can be synergistic with ssu72-2. The magnitude of the ssu72-2 effect on transcription appears to be promoter specific: whereas CYC1 transcription is diminished nearly 10-fold, less than 2-fold effects are observed for INO1 and PGK1 expression. The promoter elements responsible for these effects are unknown but can readily be addressed by whole-genome microarray analysis using wild-type and ssu72-2 strains.
It is noteworthy that rpb2-100 was isolated as a suppressor of the ssu72-2 Ts− growth defect, yet rpb2-100 and ssu72-2 exert similar effects on transcription. This anomaly can be explained by the results from genetic analysis of the YDP19 revertant, revealing that mutations in two genes, rpb2-100 and shs2-1, are required for suppression of the ssu72-2 Ts− phenotype. Indeed, the effect of either allele in the absence of the other is to enhance the ssu72-2 phenotype, essentially eliminating growth at 37°C (Fig. 2). This result not only accounts for the similar effects of ssu72-2 and rpb2-100 on transcription but also is consistent with their synergistic effects on transcription from the PGK1 promoter (Fig. 6). Although isolated as a suppressor, the rpb2-100 allele is an enhancer of ssu72-2 in the absence of shs2-1.
The rpb2-100 allele encodes a single amino acid replacement, R512C, located at the first position within homology block D of Rpb2 (Fig. 3). The viability of the R512C mutant indicates that R512, despite its phylogenetic invariance, is not functionally invariant. Not only does the rpb2-100 allele (in combination with shs2-1) suppress the ssu72-2 Ts− phenotype, but the ssu72-2 rpb2-100 shs2-1 strain (YDP19) exhibits only marginally impaired growth on rich (YPD) medium at 30°C. The viability of rpb2-100 mutants cannot be attributed to a suppressive effect of ssu72-2 on rpb2-100, because an rpb2-100 mutant is also viable in an SSU72+ background, with a growth rate comparable to that of YDP19 (Pappas, unpublished results). Furthermore, the rpb2-100 SSU72+ strain exhibits the same severe Cs− and Ino− phenotypes as YDP19, thereby confirming that these phenotypes are due to the R512C replacement.
Other rpb2 alleles have been isolated in genetic selections for mutations that affect core promoter function (3, 4, 14, 19). The spt alleles of RPB2 were identified as suppressors of δ-element insertions at the HIS4 and LYS2 promoters (19). These alleles encode amino acid replacements in the region between homology blocks B and C of Rpb2 and underscore a role for the Rpb2 subunit in core promoter recognition. The sit1 and sit2 alleles of RPB1 and RPB2, respectively, were isolated as enhancers of HIS4 transcription in the absence of the trans activators Gcn4, Bas1, and Bas2 (4). These mutations are comparable to rpb2-100 in that they also affect noninduced transcription, albeit with opposite effects. Furthermore, the sit1 and sit2 mutants, like the rpb2-100 mutant, are inositol auxotrophs. The sit1 alleles encode Rpb1 amino acid replacements in either region D or region F, both of which are within or near the active center of RNAP (3). In the Thermus aquaticus RNAP structure, the sit1 region F replacements are adjacent to R428, the counterpart of R512 in Rpb2. The opposite effects of the sit1 and rpb2-100 mutations on transcription might be accounted for by their proximity to the secondary channel, where the distinction between the stimulatory or inhibitory effects might be accounted for by structural changes that affect substrate access to the active site and, consequently, the efficiency of promoter clearance.
A crystal structure of yeast RNAP II was recently solved at a resolution of 3 Å (10). Unfortunately, R512 is located within an approximately 13-residue gap that precludes interpretation of the R512C replacement in the context of the RNAP II three-dimensional structure. However, a crystallographic structure of RNAP from E. coli has been determined at a 12-Å resolution (11, 12), and, more recently, that of RNAP from T. aquaticus has been determined at a 3.3-Å resolution (56). R512 is highly conserved, corresponding to R548 and R428 of the β subunits of RNAPs from E. coli and T. aquaticus, respectively (Fig. 3). The sequence conservation among all forms of RNAP and the nearly superimposable structures of E. coli and T. aquaticus RNAPs (32) allow for the R512C replacement to be interpreted in the context of the T. aquaticus crystal structure. Moreover, a comprehensive description of site-specific interactions between catalytically competent RNAP and promoter open DNA was recently defined by protein-DNA photo-cross-linking (31). Accordingly, R428 forms part of the binding site for double-stranded DNA downstream of position +1 and is positioned to make direct contact with the phosphate 5′ to position +6 of the nontemplate strand. The position of this residue suggests possible roles for R428 in stabilizing RNAP-DNA interaction, in facilitating clamping of RNAP on duplex DNA, in facilitating RNAP translocation, and/or in defining the downstream boundary of the transcription bubble. Furthermore, the catabolite gene activator protein has no effect on cross-linking between RNAP and the core promoter region (31). These results readily account for the effects of rpb2-100 on noninduced transcription and underscore our premise that Ssu72 is a core RNAP II transcription factor.
ACKNOWLEDGMENTS
We are especially grateful to Zu-Wen Sun for constructing the ssu72-2 mutant and to Sung-Joon Kim and Danny Reinberg for their generous gift of purified yeast RNAP II. We are also grateful to Richard Ebright, Danny Reinberg and David Gross for valuable discussions and comments on the manuscript and to Steve Buratowski, Bryan Cullen, Leonard Guarente, Lucy Robinson, Hans-Joachim Schüller, and Nancy Woychik for strains and plasmids.
This work was supported by NIH grant GM39484.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Archambault J, Friesen J D. Genetics of eukaryotic RNA polymerases I, II, and III. Microbiol Rev. 1993;57:703–724. doi: 10.1128/mr.57.3.703-724.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Archambault J, Jansma D B, Kawasoe J H, Arndt K T, Greenblatt J, Friesen J D. Stimulation of transcription by mutations affecting conserved regions of RNA polymerase II. J Bacteriol. 1998;180:2590–2598. doi: 10.1128/jb.180.10.2590-2598.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arndt K T, Styles C A, Fink G R. A suppressor of a HIS4 transcriptional defect encodes a protein with homology to the catalytic subunit of protein phosphatases. Cell. 1989;56:527–537. doi: 10.1016/0092-8674(89)90576-x. [DOI] [PubMed] [Google Scholar]
- 5.Bell S D, Jackson S P. Transcription in Archaea. Cold Spring Harb Symp Quant Biol. 1998;63:41–51. doi: 10.1101/sqb.1998.63.41. [DOI] [PubMed] [Google Scholar]
- 6.Berroteran R W, Ware D E, Hampsey M. The sua8 suppressors of Saccharomyces cerevisiae encode replacements of conserved residues within the largest subunit of RNA polymerase II and affect transcription start site selection similarly to sua7 (TFIIB) mutations. Mol Cell Biol. 1994;14:226–37. doi: 10.1128/mcb.14.1.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blair W S, Cullen B R. A yeast TATA-binding protein mutant that selectively enhances gene expression from weak RNA polymerase II promoters. Mol Cell Biol. 1997;17:2888–2896. doi: 10.1128/mcb.17.5.2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boeke J D, Lacroute F, Fink G R. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197:345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- 9.Buratowski S, Sopta M, Greenblatt J, Sharp P A. RNA polymerase II-associated proteins are required for a DNA conformation change in the transcription initiation complex. Proc Natl Acad Sci USA. 1991;88:7509–7513. doi: 10.1073/pnas.88.17.7509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cramer P, Bushnell D A, Fu J, Gnatt A L, Maier-Davis B, Thompson N E, Burgess R R, Edwards A M, David P R, Kornberg R D. Architecture of RNA polymerase II and implications for the transcription mechanism. Science. 2000;288:640–649. doi: 10.1126/science.288.5466.640. [DOI] [PubMed] [Google Scholar]
- 11.Darst S A, Polyakov A, Richter C, Zhang G. Insights into Escherichia coli RNA polymerase structure from a combination of x-ray and electron crystallography. J Struct Biol. 1998;124:115–122. doi: 10.1006/jsbi.1998.4057. [DOI] [PubMed] [Google Scholar]
- 12.Darst S A, Polyakov A, Richter C, Zhang G. Structural studies of Escherichia coli RNA polymerase. Cold Spring Harb Symp Quant Biol. 1998;63:269–276. doi: 10.1101/sqb.1998.63.269. [DOI] [PubMed] [Google Scholar]
- 13.Edwards A M, Kane C M, Young R A, Kornberg R D. Two dissociable subunits of yeast RNA polymerase II stimulate the initiation of transcription at a promoter in vitro. J Biol Chem. 1991;266:71–75. [PubMed] [Google Scholar]
- 14.Fan H Y, Cheng K K, Klein H L. Mutations in the RNA polymerase II transcription machinery suppress the hyperrecombination mutant hpr1 Delta of Saccharomyces cerevisiae. Genetics. 1996;142:749–759. doi: 10.1093/genetics/142.3.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Forsburg S L, Guarente L. Identification and characterization of HAP4: a third component of the CCAAT-bound HAP2/HAP3 heteromer. Genes Dev. 1989;3:1166–1178. doi: 10.1101/gad.3.8.1166. [DOI] [PubMed] [Google Scholar]
- 16.Goodrich J A, Tjian R. Transcription factors IIE and IIH and ATP hydrolysis direct promoter clearance by RNA polymerase II. Cell. 1994;77:145–156. doi: 10.1016/0092-8674(94)90242-9. [DOI] [PubMed] [Google Scholar]
- 17.Hampsey M. Molecular genetics of the RNA polymerase II general transcriptional machinery. Microbiol Mol Biol Rev. 1998;62:465–503. doi: 10.1128/mmbr.62.2.465-503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hampsey M. A review of phenotypes in Saccharomyces cerevisiae. Yeast. 1997;13:1099–1133. doi: 10.1002/(SICI)1097-0061(19970930)13:12<1099::AID-YEA177>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 19.Hekmatpanah D S, Young R A. Mutations in a conserved region of RNA polymerase II influence the accuracy of mRNA start site selection. Mol Cell Biol. 1991;11:5781–5791. doi: 10.1128/mcb.11.11.5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henry N L, Bushnell D A, Kornberg R D. A yeast transcriptional stimulatory protein similar to human PC4. J Biol Chem. 1996;271:21842–21847. doi: 10.1074/jbc.271.36.21842. [DOI] [PubMed] [Google Scholar]
- 21.Holstege F C, Jennings E G, Wyrick J J, Lee T I, Hengartner C J, Green M R, Golub T R, Lander E S, Young R A. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 22.Iyer V, Struhl K. Absolute mRNA levels and transcriptional initiation rates in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1996;93:5208–5212. doi: 10.1073/pnas.93.11.5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jokerst R S, Weeks J R, Zehring W A, Greenleaf A L. Analysis of the gene encoding the largest subunit of RNA polymerase II in Drosophila. Mol Gen Genet. 1989;215:266–275. doi: 10.1007/BF00339727. [DOI] [PubMed] [Google Scholar]
- 24.Kaiser C, Michaelis S, Mitchell A. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- 25.Kim T K, Ebright R H, Reinberg D. Mechanism of ATP-dependent promoter melting by transcription factor IIH. Science. 2000;288:1418–1422. doi: 10.1126/science.288.5470.1418. [DOI] [PubMed] [Google Scholar]
- 26.Kimura M, Ishiguro A, Ishihama A. RNA polymerase II subunits 2, 3, and 11 form a core subassembly with DNA binding activity. J Biol Chem. 1997;272:25851–25855. doi: 10.1074/jbc.272.41.25851. [DOI] [PubMed] [Google Scholar]
- 27.Knaus R, Pollock R, Guarente L. Yeast SUB1 is a suppressor of TFIIB mutations and has homology to the human co-activator PC4. EMBO J. 1996;15:1933–1940. [PMC free article] [PubMed] [Google Scholar]
- 28.Lagrange T, Kapanidis A N, Tang H, Reinberg D, Ebright R H. New core promoter element in RNA polymerase II-dependent transcription: sequence-specific DNA binding by transcription factor IIB. Genes Dev. 1998;12:34–44. doi: 10.1101/gad.12.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsui T, Segall J, Weil P A, Roeder R G. Multiple factors required for accurate initiation of transcription by purified RNA polymerase II. J Biol Chem. 1980;255:11992–11996. [PubMed] [Google Scholar]
- 30.Myer V E, Young R A. RNA polymerase II holoenzymes and subcomplexes. J Biol Chem. 1998;273:27757–27760. doi: 10.1074/jbc.273.43.27757. [DOI] [PubMed] [Google Scholar]
- 31.Naryshkin N, Revyakin A, Kim Y, Mekler V, Ebright R H. Structural organization of the RNA polymerase-promoter open complex. Cell. 2000;101:601–611. doi: 10.1016/s0092-8674(00)80872-7. [DOI] [PubMed] [Google Scholar]
- 32.Opalka N, Mooney R A, Richter C, Severinov K, Landick R, Darst S A. Direct localization of a beta-subunit domain on the three-dimensional structure of Escherichia coli RNA polymerase. Proc Natl Acad Sci USA. 2000;97:617–622. doi: 10.1073/pnas.97.2.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Orphanides G, LaGrange T, Reinberg D. The general initiation factors of RNA polymerase II. Genes Dev. 1996;10:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- 34.Pardee T S, Bangur C S, Ponticelli A S. The N-terminal region of yeast TFIIB contains two adjacent functional domains involved in stable RNA polymerase II binding and transcription start site selection. J Biol Chem. 1998;273:17859–17864. doi: 10.1074/jbc.273.28.17859. [DOI] [PubMed] [Google Scholar]
- 35.Parvin J D, Shykind B M, Meyers R E, Kim J, Sharp P A. Multiple sets of basal factors initiate transcription by RNA polymerase II. J Biol Chem. 1994;269:18414–18421. [PubMed] [Google Scholar]
- 36.Pinto I, Ware D E, Hampsey M. The yeast SUA7 gene encodes a homolog of human transcription factor TFIIB and is required for normal start site selection in vivo. Cell. 1992;68:977–988. doi: 10.1016/0092-8674(92)90040-j. [DOI] [PubMed] [Google Scholar]
- 37.Pinto I, Wu W-H, Na J G, Hampsey M. Characterization of sua7 mutations defines a domain of TFIIB involved in transcription start site selection in yeast. J Biol Chem. 1994;269:30569–30573. [PubMed] [Google Scholar]
- 38.Roeder R G. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem Sci. 1996;21:327–335. [PubMed] [Google Scholar]
- 39.Rose M D, Novick P, Thomas J H, Botstein D, Fink G R. A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene. 1987;60:237–243. doi: 10.1016/0378-1119(87)90232-0. [DOI] [PubMed] [Google Scholar]
- 40.Rothstein R. Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 1991;194:281–301. doi: 10.1016/0076-6879(91)94022-5. [DOI] [PubMed] [Google Scholar]
- 41.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory handbook. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 42.Schwank S, Ebbert R, Rautenstrauss K, Schweizer E, Schuller H J. Yeast transcriptional activator INO2 interacts as an Ino2p/Ino4p basic helix-loop-helix heteromeric complex with the inositol/choline-responsive element necessary for expression of phospholipid biosynthetic genes in Saccharomyces cerevisiae. Nucleic Acids Res. 1995;23:230–237. doi: 10.1093/nar/23.2.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- 44.Sherman F, Hicks J. Micromanipulation and dissection of asci. Methods Enzymol. 1991;194:21–37. doi: 10.1016/0076-6879(91)94005-w. [DOI] [PubMed] [Google Scholar]
- 45.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun Z-W, Hampsey M. Identification of the gene (SSU71/TFG1) encoding the largest subunit of transcription factor TFIIF as a suppressor of a TFIIB mutation in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1995;92:3127–3131. doi: 10.1073/pnas.92.8.3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun Z-W, Tessmer A, Hampsey M. Functional interaction between TFIIB and the Rpb9 (Ssu73) subunit of RNA polymerase II in Saccharomyces cerevisiae. Nucleic Acids Res. 1996;24:2560–2566. doi: 10.1093/nar/24.13.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun Z-W, Hampsey M. Synthetic enhancement of a TFIIB defect by a mutation in SSU72, an essential gene encoding a novel protein that affects transcription start site selection in vivo. Mol Cell Biol. 1996;16:1557–1566. doi: 10.1128/mcb.16.4.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sweetser D, Nonet M, Young R A. Prokaryotic and eukaryotic RNA polymerases have homologous core subunits. Proc Natl Acad Sci USA. 1987;84:1192–1196. doi: 10.1073/pnas.84.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsai F T, Sigler P B. Structural basis of preinitiation complex assembly on human Pol II promoters. EMBO J. 2000;19:25–36. doi: 10.1093/emboj/19.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tyree C M, George C P, Lira-DeVito L M, Wampler S L, Dahmus M E, Zawel L, Kadonaga J T. Identification of a minimal set of proteins that is sufficient for accurate initiation of transcription by RNA polymerase II. Genes Dev. 1993;7:1254–1265. doi: 10.1101/gad.7.7a.1254. [DOI] [PubMed] [Google Scholar]
- 52.Weil P A, Luse D S, Segall J, Roeder R G. Selective and accurate initiation of transcription at the Ad2 major late promoter in a soluble system dependent on purified RNA polymerase II and DNA. Cell. 1979;18:469–484. doi: 10.1016/0092-8674(79)90065-5. [DOI] [PubMed] [Google Scholar]
- 53.Woychik N A. Fractions to functions: RNA polymerase II thirty years later. Cold Spring Harb Symp Quant Biol. 1998;63:311–317. doi: 10.1101/sqb.1998.63.311. [DOI] [PubMed] [Google Scholar]
- 54.Wu W-H, Pinto I, Chen B-S, Hampsey M. Mutational analysis of yeast TFIIB: a functional relationship between Ssu72 and Sub1/Tsp1 defined by allele-specific interactions with TFIIB. Genetics. 1999;153:643–652. doi: 10.1093/genetics/153.2.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Young R A. RNA polymerase II. Annu Rev Biochem. 1991;60:689–715. doi: 10.1146/annurev.bi.60.070191.003353. [DOI] [PubMed] [Google Scholar]
- 56.Zhang G, Campbell E A, Minakhin L, Richter C, Severinov K, Darst S A. Crystal structure of Thermus aquaticus core RNA polymerase at 3.3 A resolution. Cell. 1999;98:811–824. doi: 10.1016/s0092-8674(00)81515-9. [DOI] [PubMed] [Google Scholar]
- 57.Zhang G, Darst S A. Structure of the Escherichia coli RNA polymerase alpha subunit amino-terminal domain. Science. 1998;281:262–266. doi: 10.1126/science.281.5374.262. [DOI] [PubMed] [Google Scholar]