Significance
Bacterial division is an essential cellular process that involves the formation of a septum made of peptidoglycan. The septum is initially shared between daughters and must be processed to complete division. Septal splitting has long been known to be mediated by enzymes called amidases that are controlled by an activator protein and the ABC-transporter-like complex called FtsEX. However, the mechanism of amidase regulation by this system has remained unclear. Here, we report the structure of FtsEX in complex with an amidase and amidase activator, revealing how ATP binding to the complex promotes amidase activation and providing structural information that may help target the activation mechanism for the development of cell lysis inducing antibiotics.
Keywords: cell division, peptidoglycan, antimicrobial, cryo-EM
Abstract
The peptidoglycan (PG) cell wall produced by the bacterial division machinery is initially shared between the daughters and must be split to promote cell separation and complete division. In gram-negative bacteria, enzymes that cleave PG called amidases play major roles in the separation process. To prevent spurious cell wall cleavage that can lead to cell lysis, amidases like AmiB are autoinhibited by a regulatory helix. Autoinhibition is relieved at the division site by the activator EnvC, which is in turn regulated by the ATP-binding cassette (ABC) transporter-like complex called FtsEX. EnvC is also known to be autoinhibited by a regulatory helix (RH), but how its activity is modulated by FtsEX and the mechanism by which it activates the amidases have remained unclear. Here, we investigated this regulation by determining the structure of Pseudomonas aeruginosa FtsEX alone with or without bound ATP, in complex with EnvC, and in a FtsEX–EnvC–AmiB supercomplex. In combination with biochemical studies, the structures reveal that ATP binding is likely to activate FtsEX-EnvC and promote its association with AmiB. Furthermore, the AmiB activation mechanism is shown to involve a RH rearrangement. In the activated state of the complex, the inhibitory helix of EnvC is released, freeing it to associate with the RH of AmiB, which liberates its active site for PG cleavage. These regulatory helices are found in many EnvC proteins and amidases throughout gram-negative bacteria, suggesting that the activation mechanism is broadly conserved and a potential target for lysis-inducing antibiotics that misregulate the complex.
Nearly all prokaryotic cells are surrounded by a layer of peptidoglycan (PG) that protects them from osmotic lysis. PG is a polymer of N-acetylglucosamine and N-acetylmuramic acid repeating units that is cross-linked by stem peptides attached to N-acetylmuramic acid, forming a cell-shaped sacculus that encases the cytoplasmic membrane (1). During the cell cycle, PG undergoes extensive remodeling to expand and divide the cell envelope (2, 3). In many bacteria, PG synthesis enzymes are organized into one of two supramolecular complexes called the Rod complex (or elongasome) and the divisome, which are responsible for synthesizing PG during elongation and division, respectively. Divisome assembly begins with the formation of the Z-ring formed by dynamic polymers of FtsZ, which then recruits dozens of cell division proteins to the midcell. During Z-ring formation, early divisome proteins like FtsA and ZipA anchor FtsZ to the cytoplasmic membrane at the future division site. Z-ring formation is then followed by the recruitment of a number of division proteins including FtsEX, FtsQLB, FtsWI, and FtsN. Once the divisome is matured, it is activated to initiate cell constriction and the synthesis of the PG that will eventually fortify the daughter cell poles. This so-called septal PG (sPG) formed by the divisome is initially shared by the daughters and must be split to complete division (4–7).
Splitting of the sPG is mediated by the three periplasmic N-acetylmuramyl L-alanine amidases called AmiA, AmiB, and AmiC in Escherichia coli (3, 8, 9). To avoid PG damage, these amidases are autoinhibited by their regulatory helix (RH) (10). Their activity is restricted to the septum via activation by divisome components with dLytM domains (defective LytM, aka peptidase_M23). Once activated, the amidases remove the stem peptides from the glycan strands, which will break cross-links in the matrix connecting the daughter cells. Additionally, the peptide-free (denuded) glycans generated promote the recruitment of division proteins bearing the sporulation-related repeat (SPOR) domains to the division site such as FtsN as part of a positive feedback loop that stimulates sPG synthesis (11, 12).
The best understood dLytM factor is EnvC, which stimulates the activity of AmiA and AmiB. Notably, EnvC is also autoinhibited by its restraining arm (13), which prevents it from interacting with the amidases. The autoinhibition is thought to be released by a widely conserved ATP-binding cassette (ABC)-transporter-like complex called FtsEX (14–16). The periplasmic loops of the membrane component FtsX interact with EnvC and recruit it to the divisome. The ATPase activity of the cytoplasmic component FtsE is required for the activation, but the mechanism of activation and what triggers ATP hydrolysis remain unclear. Besides activating the amidases, FtsEX also interacts with FtsZ and FtsA to promote constriction (17–22). Similar FtsEX systems were identified in Gram-positive bacteria (23, 24) and actinobacteria (25, 26). In these organisms, FtsX interacts directly with their cognate PG hydrolases. Partial structures of FtsEX and their interacting partners are available, including FtsE (27) and the large periplasmic loop of FtsX with (13, 26, 28) or without (29) its interacting partners. While these structures are valuable, the other periplasmic loop of FtsX and the transmembrane domains are missing. Solving the structure of FtsEX with its full complement of associated factors is therefore necessary for elucidating the activation mechanism.
Here, we report the structure of the Pseudomonas aeruginosa FtsEX complex in the presence or absence of ATP, in complex with EnvC, and in complex with EnvC and AmiB. In addition, we characterized the reconstituted FtsEX complex biochemically and demonstrated that EnvC binding to FtsEX stimulates ATP hydrolysis. ATP binding by the FtsEX complex triggers conformational changes in EnvC and promotes AmiB binding. This relieves the autoinhibition through a series of RH rearrangements, subsequently activating the PG hydrolytic activity of the supercomplex.
Results
Biochemical Reconstitution and Structural Studies of FtsEX from P. aeruginosa.
FtsEX belongs to the type VII family of ABC transporters (14), which includes members like MacB (PDB code: 5LJ7) (30) and LolCDE (PDB code: 7MDX) (31, 32). Attempts to reconstitute the FtsEX complex of E. coli were unsuccessful due to low yield. To overcome this issue, we cloned the FtsEX homologs from several bacteria and tested their solubility. Among them, FtsEX from P. aeruginosa was stable, forming a complex with a 1:1 molar ratio of FtsE:FtsX. The purified complex was also readily reconstituted in peptidiscs (SI Appendix, Fig. S1A). Notably, this form of the FtsEX complex had very little ATPase activity, suggesting that it is autoinhibited and may require activation (see below) (SI Appendix, Fig. S1B). We also purified two ATPase-defective complexes, one with FtsED162N that cannot bind ATP and one with FtsEE163Q that can bind but not hydrolyze ATP (SI Appendix, Fig. S1B) (19).
We determined the structure of different forms of FtsEX using cryoelectron microscopy (cryo-EM). After three-dimensional (3D) reconstruction, the resolution of the ATP-free wild-type (WT) FtsEX and ATP-bound form of FtsEE163QX mutant was 4.2 Å and 4.0 Å, respectively (SI Appendix, Figs. S2–S4). The FtsEX complex is composed of two FtsE monomers and two FtsX monomers. Each FtsX chain has four transmembrane helices (TM1-4), a large periplasmic loop we refer to as the periplasmic domain (PLD), and a small periplasmic loop (Fig. 1 A–C). TM4 is fully embedded in the membrane, while the other three TM helices are extended into the periplasmic space. The dimerization interface of FtsX comprises mostly TM1 and TM2 (Fig. 1 D and G). Interaction between FtsE and FtsX is mediated by a coupling helix (CH; residues 235 to 247) between TM2 and TM3 and a C-terminal tail (C tail) of FtsX (Fig. 1H). The N-terminal region of FtsX (residue 35 to 54), named the elbow helix, is amphipathic and is parallel to the cell membrane (Fig. 1 C and H). FtsE has a canonical nucleotide-binding domain fold (Fig. 1I). Electron density corresponding to the PLD of FtsX was only visible in the presence of ATP (SI Appendix, Fig. S4), suggesting that its conformation is stabilized by ATP-binding. The PLD (residues 93 to 189) connects TM1 with TM2. It consists of a 2βαβ motif called the Porter subdomain and an X lobe (residues 129 to 157) (Fig. 1 D and E). Notably, the two PLDs in the dimer form a groove accessible for binding to partner proteins.
Fig. 1.
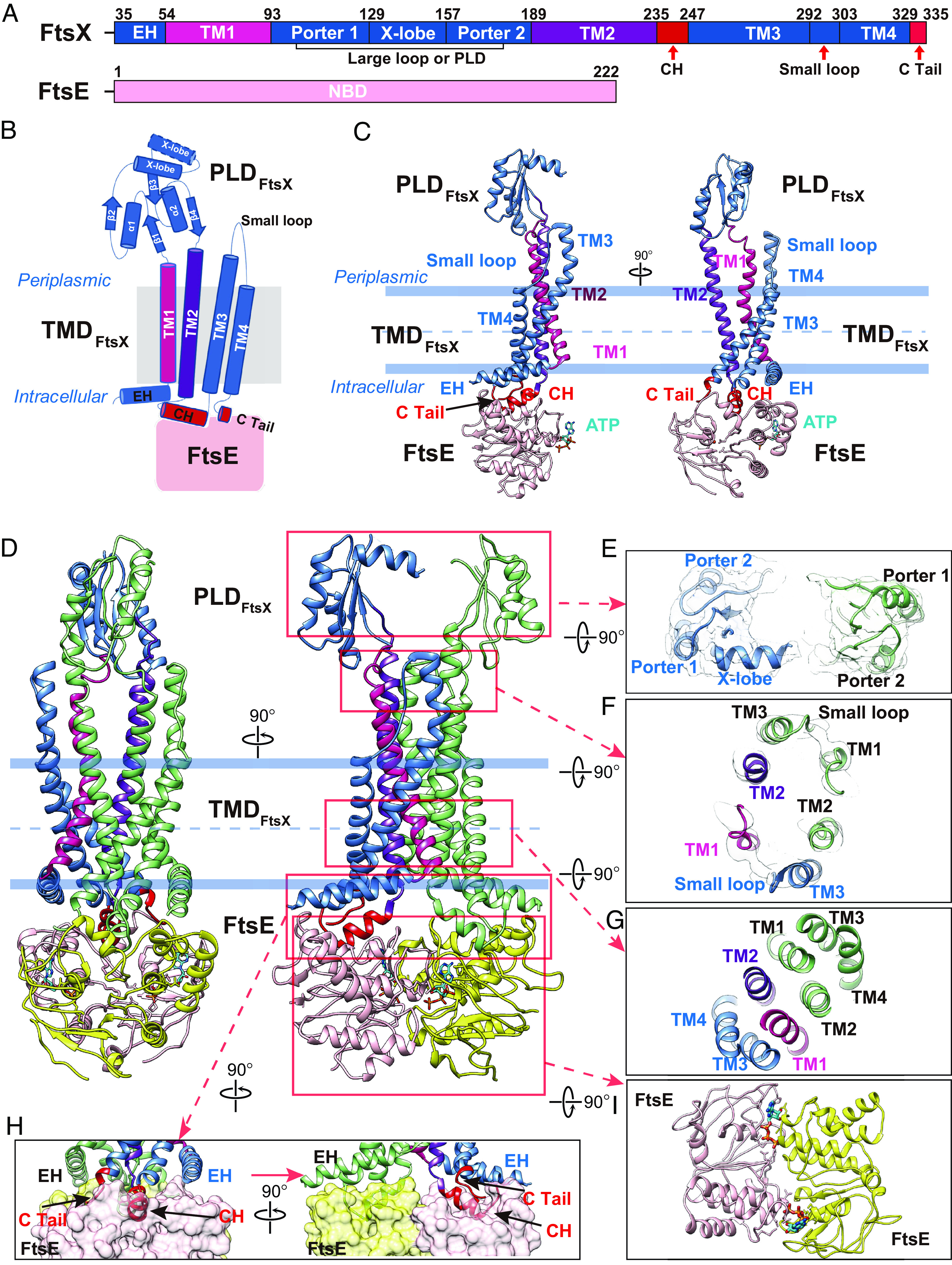
Overall structure of FtsEE163QX complex. (A) Domain arrangements of FtsX and FtsE. (B) Topology diagram of FtsX. α helices are shown as cylinder, β sheets are shown as arrows. (C) Protomer structure of FtsEX in two orientations. Color scheme: FtsE in pink, TM 1 in magenta, TM 2 in purple, CH and C-terminal tail are colored in red, other parts of FtsX are colored in blue, ATP is colored in cyan. (D) Overall structure of FtsEE163QX dimer. (E) Top-Down view of PLD. In the presence of ATP, PLD domain form a groove for EnvC binding. (F) Top-Down view of interacting part of TMDs with PLD. (G) Dimerization of FtsX is mainly mediated by TM1 and TM2. (H) Interaction between FtsE and FtsX is mediated by CH and C-terminal tail. CH is located between TM2 and TM3. (I) Top-Down view of FtsE dimer bound with ATP. Color scheme for panels D–H: FtsE is colored in yellow and pink, FtsX is colored in light green and blue.
Structure of FtsEX Complexed with EnvC.
EnvC has been shown to directly interact with the PLD of FtsX (16). We demonstrated that the purified FtsEX complex can also interact with EnvC in a pull-down assay (SI Appendix, Fig. S5A). Adding Adenosine Triphosphate (ATP) to the mix had no noticeable effect on the interaction. However, the complex appeared to be more stable if FtsEX is trapped in the ATP-bound state (FtsEE163QX-ATP), as evidenced by the result that most of the FtsEX under this condition remained intact without aggregation (SI Appendix, Fig. S5B).
To solve the structure of EnvC-bound FtsEX, purified WT FtsEX was reconstituted in peptidiscs before incubation with purified EnvC. The resulting complex was then purified by size exclusion chromatography (SEC) (SI Appendix, Fig. S5 C and D) and visualized by cryo-EM (SI Appendix, Fig. S6). From the two-dimensional (2D) classifications, the coiled-coil domain of EnvC was found to form a rod with a span of ~140 Å (Fig. 2A). After 3D reconstruction, the overall resolution of the complex is 3.8 Å (SI Appendix, Fig. S6). Electron microscopy (EM) density of almost all sidechains of FtsEX and the coiled-coil domain of EnvC were well resolved in this complex (SI Appendix, Figs. S7–S9). As expected, the complex is composed of 2 FtsE, 2 FtsX, and 1 EnvC subunits (13). Although the resolution of the dLytM domain and the RH is low, they were found near the top of the EnvC coiled–coiled domain (Fig. 2B). This domain arrangement is similar to the reported FtsX PLD–EnvC complex from E. coli (13). In our EnvC–FtsEX structure, the interface between EnvC and FtsX was well resolved, which allowed us to visualize how EnvC interacts with FtsX (Fig. 2C). The first interface is between the coiled-coil region of EnvC (residues 103 to 117, residues 135 to 152) and the X lobe of FtsX (Fig. 2D). This interface consists of extensive hydrophobic contacts between residues F99, L137, L143, L150, and L155 of the X lobe and residues I108, Y114, Y140, Y141, Y143, and I144 of EnvC. The second interface, which was not seen previously, is located at the hinge region of the EnvC coiled-coil domain, TM1-3, and the small periplasmic loop of FtsX (Fig. 2E). This interface is also hydrophobic in nature and comprises Y121, L122, L125, L126, and Q128 of EnvC and W90, W193, and Y292 of FtsX. In addition, a salt bridge (EnvC/K123-FtsX/D190) is found at the second interface, which likely stabilizes the complex through electrostatic interactions.
Fig. 2.
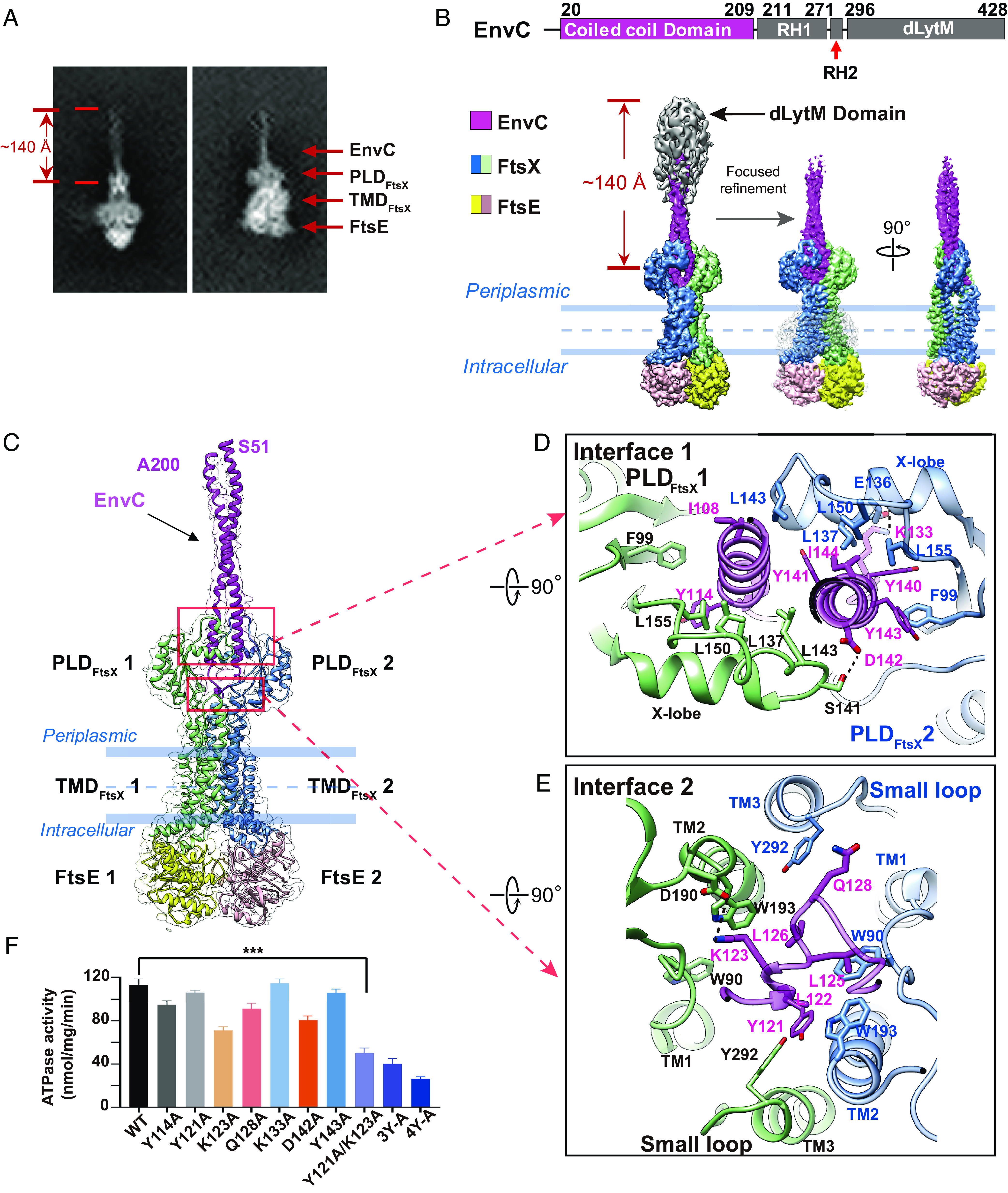
Overall structure of FtsEX in complex with EnvC. (A) 2D averages of EM particles of FtsEX in complex with EnvC. (B) Cryo-EM density map of FtsEX-EnvC in the absence of ATP. EnvC contains four domains: coiled-coil domain, RH1, RH2, dLytM domain. Domains with lower resolution are colored in gray. Domain arrangement of EnvC is shown on top. Bottom Left:density map obtained with larger box dimension (700 Å). Bottom Middle and Right: front- and side-views of the cryo-EM density map obtained with smaller box dimension (270 Å). (C) Overall structure of FtsEX–EnvC. FtsE is colored in yellow and pink, FtsX is colored in light green and blue, and bound EnvC is colored in magenta. (D) Detailed interactions between FtsX and EnvC at interface 1 (Top-Down view). (E) Detailed interactions between FtsX and EnvC at interface 2 (Top-Down view). (F) ATPase activity of FtsEX complexed with various EnvC mutants, where interacting residues have been mutated. “3Y-A” refers to the mutant of Y140A/Y141A/Y143A, and “4Y-A” refers to the mutant of Y114A/Y140A/Y141A/Y143A. The triple asterisks indicate a P-value of less than 0.001.
The interface residues of EnvC with FtsX were mutagenized and their effect on the ATPase activity of FtsEX was determined (Fig. 2F). Consistent with our structural studies, a double mutant of Y121A/K123A at interface two, as well as a triple-mutant of Y140A/Y141A/Y144A and a quadruple-mutant of Y114A/Y140A/Y141A/Y144A at interface one, largely abolished ATPase activation. We hypothesized that the decrease in ATPase activity is due to a weakened interaction between FtsEX and the EnvC mutants. To test this possibility, we performed a pull-down assay (SI Appendix, Fig. S10A). Our results showed that the FtsEX complex bound well to EnvC mutants that stimulated ATPase activity normally, whereas the binding was weak for EnvC mutants defective in ATPase stimulation. The SEC profiles of these mutants were similar to WT EnvC, suggesting that these mutants were not misfolded (SI Appendix, Fig. S10B). Thus, the weakened interactions between FtsEX and the EnvC mutant accounted for the decrease in the ATPase activity. Together, these results suggest the coiled-coil domain of EnvC acts as a hydrophobic wedge that is inserted into the slot formed by the FtsX dimer. Comparison with the previous structure of the FtsX PLD–EnvC complex (PDB code: 6TPI) (SI Appendix, Fig. S11) (13), indicates that i) the bound EnvC in the complex is likely in the autoinhibited state and ii) that in the presence of the complete EnvC-binding site (with both the PLD and the small periplasmic loop of FtsX) the coiled-coil domain of EnvC adopts a tighter conformation than in the FtsX PLD–EnvC complex lacking the small loop.
Biochemical Characterization of the AmiB-EnvC-FtsEX Supercomplex.
We examined whether EnvC binding would affect the ATPase activity of FtsEX. Strikingly, EnvC binding increased ATPase activity by 25-fold. Adding AmiB did not further stimulate the ATPase activity (Fig. 3A). Next, we determined the Michaelis–Menten constants (Km) and the turnover rates (kcat) of FtsEX ATPase activity (Fig. 3B). At 37 °C, the Vmax is ~121 nmol/mg/min and the Km is ~0.23 mM in the presence of EnvC; In contrast, when EnvC is absent, the ATPase activity is almost negligible. These data demonstrate the ATPase activity of FtsEX is activated by EnvC binding.
Fig. 3.
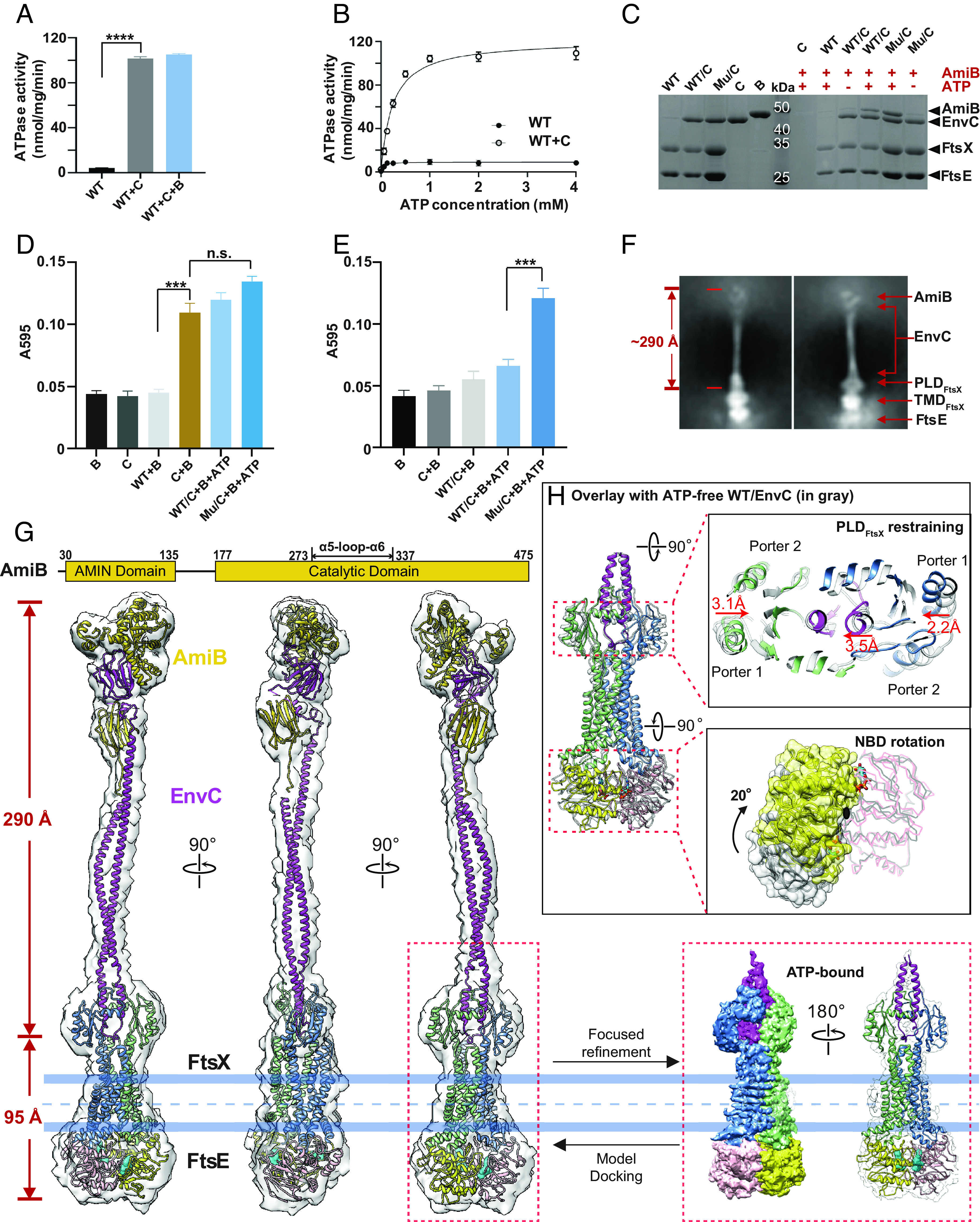
Biochemical and structural study of AmiB activation. (A) ATPase activity of FtsEX is activated by EnvC. The addition of AmiB has no effect on the ATPase activity of FtsEX/EnvC. “WT” refers to the WT FtsEX, “B” refers to AmiB, and “C” refers to EnvC. Four asterisks signify a P-value of less than 0.0001. (B) ATPase activity of FtsEX and FtsEX/EnvC in peptidiscs. The activity of FtsEX alone is too low to detect, but is included here as a reference to show the highly stimulated activity of FtsEX in the presence of EnvC. “Mu” refers to the FtsEE163QX mutant. (C) Pull-down study of AmiB binding to EnvC-coupled FtsEX or its ATPase mutant (E163Q), in the presence or absence of nucleotide. Only in the presence of ATP, a significant amount of AmiB is recruited, and the FtsEE163QX mutant trapped at the ATP-binding state showed clear binding of AmiB, confirming the role of ATP binding in complex formation with AmiB. (D) PG hydrolysis activity of AmiB under different conditions. AmiB in complex with FtsEX/EnvC shows obvious activity, including the FtsEE163QX mutant trapped at the ATP-binding state; however, AmiB in the presence of EnvC alone also exhibits PG hydrolysis at a similar level. Triple asterisks indicate a P-value of less than 0.001; n.s. means a P-value of larger than 0.05. (E) PG hydrolysis activity of AmiB with cross-linked samples. AimB coupled to the FtsEE163QX mutant trapped at the ATP-binding state still showed obvious activity, but the activity of AimB with EnvC alone is almost undetectable. (F) 2D averages of ATP-bound FtsEE163QX/EnvC/AmiB particles, showing a surprisingly extended EM density from the periplasmic region with a span of approximately 29 nm, directly capturing the activated state of EnvC and suggesting the binding of AmiB. (G) Front-, side-, and back-views of the cryo-EM density map of the FtsEE163QX/EnvC/AmiB supercomplex in the presence of ATP, with the structure docked shown in ribbon representation. AmiB domain arrangement is shown on top. With focused refinement, the resolution of FtsEX and part of EnvC is increased to 4.5 Å with side-chains clearly resolved from the FtsX and bound EnvC region at the PLD domain. Color scheme: FtsX in light green and blue, FtsE in yellow and pink, EnvC in magenta, AmiB in gold, ATP in cyan. (H) Overlay with the structure of ATP-free FtsEX/EnvC. The ATP-free structure is colored in gray. ATP binding induces a 20° rotation of one FtsE and restrains the PLD domain by approximately 3.1 and 2.2 Å from two sides, resulting in a pinch of one of the helices from the coiled-coil domain (CCD) domain of EnvC by 3.5 Å. The black dot at the bottom right panel denotes a twofold symmetry axis. These conformational changes trigger the following conformation changes of EnvC and AmiB, as discussed in Fig. 4.
Next, we tested whether AmiB binding to the FtsEX–EnvC complex is ATP-dependent. The His6-tagged FtsEX–EnvC complex was immobilized on Ni-NTA resin before untagged AmiB was added in the presence or absence of 2 mM ATP. Intriguingly, ATP promoted the interaction between AmiB and the FtsEX–EnvC complex whether or not the complex was trapped in the ATP-bound state (i.e., EnvC-FtsEE163QX-ATP)(Fig. 3C). Thus, AmiB is likely recruited to the FtsEX–EnvC complex when ATP is bound but not hydrolyzed. To investigate the role of ATP binding and hydrolysis in the recruitment and dissociation of AmiB, we conducted a pull-down assay using the FtsEX/EnvC complex, AmiB, 2 mM of ATP or nonhydrolyzable analogs of ATP (SI Appendix, Fig. S12). ATP-vanadate and Adenosine Diphosphate (ADP) were used to mimic the hydrolyzed state, while AMPPNP and ATPγS were added to trap the FtsEX complex in the ATP-bound prehydrolysis state. Additionally, we included the FtsEE163QX/EnvC mutant for comparison. Our results indicate that the FtsEX complex trapped in the prehydrolysis state had the highest levels of AmiB binding, whereas the posthydrolysis state showed the least binding. These findings suggest that ATP binding, rather than hydrolysis, plays a key role in the recruitment of AmiB, while ATP hydrolysis likely triggers the dissociation of AmiB from the complex.
To determine whether ATP binding triggers the activation of AmiB in the complex with FtsEX–EnvC, we used a dye-release assay (31) to measure its activity. Although AmiB showed clear PG hydrolysis activity in the complex with both WT FtsEX/EnvC and the E163Q mutant, we observed similar activity in the condition with EnvC alone (Fig. 3D), as previously reported (33). This result is probably due to the highly flexible dLytM domain and RH of EnvC in solution, which can shift the equilibrium towards the formation of an EnvC–AmiB complex without coupling to FtsEX, leading to the activation of AmiB by EnvC alone in the in vitro system. To address this possibility, we added glutaraldehyde cross-linker to the reaction mixture of FtsEX/EnvC or EnvC alone, hoping to trap EnvC in the inactivated state and make it dependent on FtsEX conformational changes for activity. After applying the cross-linked sample to the PG hydrolysis assay, only the FtsEE163QX complex with ATP trapped but not hydrolyzed showed clear PG hydrolytic activity by AmiB (Fig. 3E), while AmiB activity with WT FtsEX/EnvC or EnvC alone was greatly reduced. Therefore, our results suggest that the ATP binding, but not hydrolysis, stabilizes the FtsEX–EnvC–AmiB complex, resulting in the activation of AmiB.
Structure of FtsEX Complexed with EnvC and AmiB.
We next reconstituted (SI Appendix, Fig. S13) and determined the structure of FtsEX–EnvC–AmiB by cryo-EM (SI Appendix, Fig. S14). Here, we employed the FtsEE163Q mutant to trap the ATP-bound state. Strikingly, the density corresponding to the periplasmic region was extended to a ~290 Å rod, 150 Å longer than in the FtsEX–EnvC complex (Fig. 3F and SI Appendix, Fig. S14). This repositioned the folded dLytM domain at the tip of the coiled-coil domain. To understand the conformational change associated with AmiB and ATP binding, we refined the structural model with a smaller box size. The resolution of this refined model improved from 5.8 Å to 4.5 Å (Fig. 3G and SI Appendix, Fig. S14) and contains FtsE, FtsX, and part of EnvC. To generate a model of the supercomplex, we first docked the model of ATP-bound FtsEX with the adjacent coiled-coil domain of EnvC into the 5.8 Å EM map. For AmiB and dLytMEnvC, we were able to construct a working model including most of the domains despite the limited EM density. The model revealed a possible interacting interface between dLytMEnvC and the AmiB catalytic domain. However, the orientation of the N-terminal Amidase N-terminal (AMIN) domain of AmiB (AMINAmiB) could not be definitively determined. In addition, we could not identify the residues that are important for the interaction between EnvC and AmiB in this model. To address these issues, we employed AlphaFold (34) to refine our structural model with a detailed EnvC-AmiB interaction interface (SI Appendix, Fig. S15). The structural model of the activated supercomplex comprises two FtsE, two FtsX, one EnvC, one AmiB, and two ATP molecules.
The binding of ATP induced a rotation of one of the FtsE subunits by approximately 20° relative to another FtsE subunit, resulting in a more symmetrical conformation of the FtsE dimer compared to the ATP-free state (as shown in the bottom right panel in Fig. 3H). This conformational change propagates through the coupling helices and the TMDs of FtsX (SI Appendix, Fig. S16A), leading to an inward movement of the two PLDs, with a shift of 3.1 and 2.2 Å, respectively (Fig. 3H and Movie S1). This pinches the coiled-coil domain of EnvC by ~3.5 Å (Fig. 3H). Several new interactions formed between FtsX and EnvC, including R118 (EnvC)-S97 (FtsX1), R136 (EnvC)-S97 (FtsX2), and a salt bridge formed between R148 (EnvC)-E149 (FtsX2) (SI Appendix, Fig. S16B). Substitutions of these residues with alanine showed minor inhibition of ATPase activity (SI Appendix, Fig. S16C). This structural rearrangement appears to be further transduced to the other end of the coiled-coil domain, resulting in a stretched conformation that unlocks the dLytM domain. We conclude that ATP and AmiB binding leads to a series of conformational changes that tighten the PLD of FtsX, resulting in an elongated EnvC coiled-coil domain. The released dLytM domain then binds AmiB, likely positioning it for PG hydrolysis.
Discussion
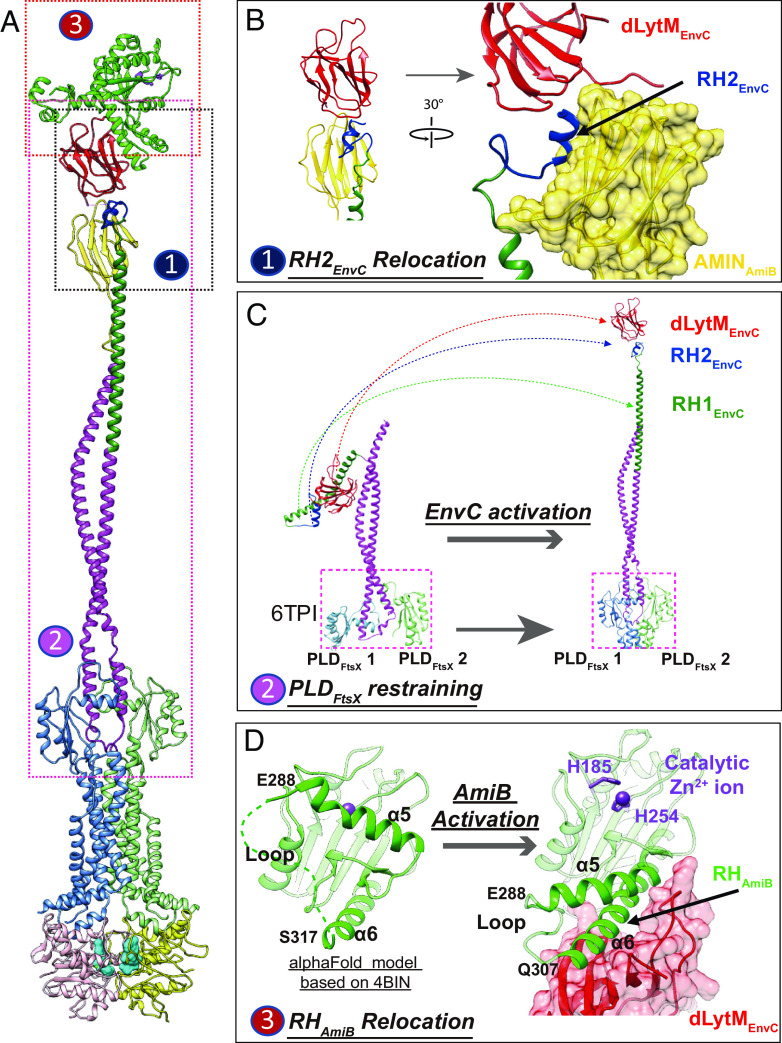
The structures presented here provide important mechanistic insights into the EnvC-mediated activation of AmiB by the membrane complex FtsEX during cell division. We show that the activation cascade involves an extension of the EnvC coiled-coiled domain that releases the dLytM domain and unblocks the AmiB catalytic domain through a cascade of changes involving regulatory helices (Fig. 4). The first relocation involves a RH in EnvC called RH2EnvC (residue 278 to 292). The structure agrees with the prediction by Alphafold that this helix interacts with AMINAmiB (Fig. 4 A and B). The AMINAmiB domain is known to bind PG, and here we suggest it may also regulate its PG hydrolase activity. Compared to the AlphaFold model of AmiB alone, which is superimposable to the crystal structure of a related PG hydrolase AmiC in the inactivated state (PDB code: 4BIN) (35), a large relocation of the AMIN domain likely takes place in the FtsEX–EnvC–AmiB supercomplex via a flexible linker region (residues 135 to 175) between the catalytic and AmiN domains (SI Appendix, Fig. S17A). Interestingly, RH2EnvC is only 8 residues away from the previously identified EnvC RH (RH1EnvC) (13), which apparently locks the dLytM domain of EnvC in the autoinhibited state as a molecular hook. In the activated state, RH1EnvC is released and becomes part of the coiled-coil domain. This change likely promotes AmiB binding. The extended coiled-coil domain is extremely long with a length of ~25 nm (Fig. 4C and Movie S2). The extended state of the coiled-coil domain seems to be stabilized by the interaction between RH2EnvC and the AMIN domain of AmiB.
Fig. 4.
Structure of activated PG supercomplex of FtsEE163QX/EnvC/AmiB. (A) Overall structure of the FtsEE163QX/EnvC/AmiB supercomplex. (B) Predicted interaction between AMINAmiB and RH2EnvC. (C) Activation of EnvC. Left: crystal structure of the autoinhibited EnvC–PLD structure (PDB code: 6TPI); Right: relocation of RH1 (green), RH2 (cyan) and dLytM (red) domain upon activation in the FtsEX–EnvC–AmiB supercomplex (this work). (D) RHAmiB (α6) relocation from catalytic domain liberate the catalytic site of AmiB for PG hydrolysis. Left: The AlphaFold predicted structure of the catalytic domain of AmiB with α5 blocking the catalytic site; Right: α5 relocates with the catalytic site exposed and α6 interacting with dLytM in the activated supercomplex.
The second RH relocation (Fig. 4D) involves the loop region of AmiB (residues 307 to 316), which is disordered in the predicted structure of AmiC in the inactivated state, but upon activation refolds as a helical structure extending the α6 helix (residues 317 to 337). This extended α6 helix interacts with the exposed binding pocket of the dLytM domain of EnvC (SI Appendix, Fig. S17B). The other side of this loop is α5 (residues 273 to 288), the inhibitory helix of AmiB, which is dislodged from the catalytic site via the changes in α6 and the intervening loop. Interestingly, residues of E. coli EnvC previously reported (36) to be required for amidase activation (K321, V324, Y350, V353, Y401, and R405) are conserved in P. aeruginosa (K302, V305, Y331, L328, Y382, and R386). In our model, these residues would interact directly with AmiB (SI Appendix, Fig. S17C). Thus, autoinhibition of AmiB is relieved upon changes to α5 mediated by the binding of α6 of AmiB by the dLytM domain of EnvC.
Our biochemical and structural studies suggest a revised molecular model for FtsEX regulated PG hydrolysis at the division site (Fig. 5). We propose that EnvC complexed with FtsEX in the ATP-free state is in a compact form with the dLytM domain autoinhibited. Structures in the ATP-bound state suggest that ATP binding induces a series of conformational changes starting with FtsE in the cytosol that are propagated through the TMD of FtsX to its PLD (Movie S1). The changes in the PLD in turn promote an extended conformation of EnvC, freeing its dLytM domain from autoinhibition and allowing it to associate with AmiB via its α6 helix (Movie S2). This ultimately opens the amidase active site of AmiB to cleave PG. ATP hydrolysis is then expected to recycle the FtsEX–EnvC complex back to the autoinhibited state, releasing AmiB. The lifetime of the activated state of the FtsEX–EnvC–AmiB complex remains unknown. Whether it can survive multiple rounds of PG cleavage, or one molecule of ATP is hydrolyzed to cut each PG peptide, remains to be determined. Nevertheless, the structures reported here support the idea that the FtsEX–EnvC complex may provide a mechanism to promote a fixed number of PG cleavage events per ATP hydrolyzed for the safe processing of sPG at the division site as suggested previously (16, 24). Additionally, several divisome proteins like FtsZ and FtsA interact with FtsEX and may stabilize a certain conformation. Thus, they may further influence the ATPase activity of the complex and prevent spurious activation of the complex outside of the division site.
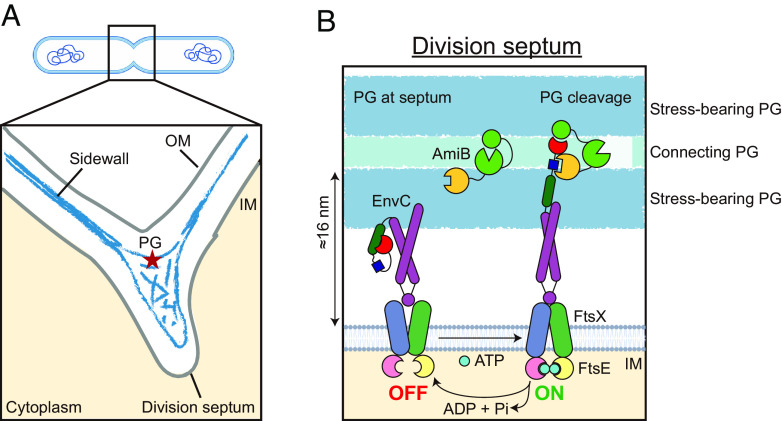
Fig. 5.
A working model of AmiB activation by EnvC and FtsEX. (A) Shown is a schematic of a dividing cell (SI Appendix, Fig. S18). The distance between the inner membrane (IM) and PG is much larger at the division septum compared to the sidewall. (B) FtsEX/EnvC adopts an autoinhibited state in the absence of ATP and AmiB (Left). In the presence of AmiB, ATP binding at the cytosolic side triggers a series of conformational changes that lead to activation of the complex (Right). The activated state is stabilized by the interaction between the AMIN domain of AmiB (yellow circle) with the RH2 of EnvC (dark blue square). ATP hydrolysis recycles the complex back to the autoinhibited state. At the division septum, the increased spacing between the inner membranes of the two daughter cells allows the activated complex to effectively cleave the connecting PG materials located in between the two layers of stress-bearing PG. This cleavage allows for the successful separation of the daughter cells.
During cell division in gram-negative bacteria, the inner membrane is invaginated as an sPG material is synthesized by the divisome (Fig. 5). Recent cryo-EM imaging of the sPG architecture in E. coli (37) revealed two layers of electron density corresponding to the sPG at the leading edge of the membrane invagination. These layers likely form the future stress-bearing layers of the polar PG of the daughter cells. Whether these layers are connected by PG material that is not ordered or dense enough to be visualized is not clear. At the lagging edge of the developing septum where the outer membrane is invaginating, a wedge of PG material was also visualized that appears to be shared between the developing daughter cells. This material along with any material connecting the two future stress-bearing layers of the daughter cell poles must be cleaved to allow continued outer membrane invagination and daughter cell separation. A remarkable feature of the sPG cleavage process is that it must somehow cleave the connecting PG material without compromising the integrity of the future stress-bearing layer. The FtsEX–EnvC–AmiB structure presented here suggests the mechanism by which this delicate surgery may be achieved. In this structure, the amidase active site is placed ~290 Å (29 nm) from the inner membrane by the coiled-coil domain of EnvC, which is beyond the normal distance of the PG layer from the membrane (<20 nm) (SI Appendix, Fig. S18) (37, 38). We propose that the length of the EnvC rod facilitates the delivery of the activated amidase active site such that it can only cleave the connecting layers of the sPG while leaving the future polar (stress-bearing) PG untouched (Fig. 5). Indeed, the thickness of the envelope at the septum is increased to roughly 60 nm (SI Appendix, Fig. S18) such that the amidase active site would be almost perfectly positioned at its center to cleave material between the future stress bearing layers.
FtsEX interacts with a variety of proteins with coiled-coil domains. In gram-positive bacteria and Corynebacterineae, instead of PG hydrolase activators these proteins are usually PG hydrolases, such as PcsB in Streptococcus pneumoniae (24), CwlO in Bacillus subtilis (23, 39), and RipA (and RipC) in Mycobacterium tuberculosis (25, 26, 40). As many of these proteins lack PG hydrolytic activity in vitro, they are likely to be autoinhibited and required activation by FtsEX to release their autoinhibitory helices, similar to that observed for EnvC. The activation may be triggered by PG synthesis by the FtsQLB–FtsWI complex (41) or other division proteins that directly interact with FtsEX (17–22). Although some of these bacteria display a rapid mechanical daughter cell splitting process following enzymatic weakening of the septum as opposed to gradual the gradual sPG processing observed for E. coli and gram-negative bacteria (42, 43), the overall regulatory mechanism of PG hydrolysis by FtsEX is likely to be broadly conserved, making it an attractive target for lysis-inducing antibiotics that misregulate the complex. Indeed, dysregulation of the FtsEX pathway is proposed to be the mechanism of action accounting for the antimicrobial activity of IFN-ɣ induced chemokines CXCL9 and CXCL10 because cells lacking FtsEX and the cognate hydrolase are resistant to these factors (44, 45). Although this mechanism appears to be specific to Bacillus anthracis (46), it is conceivable that further understanding the molecular interactions regulating FtsEX will open the door for antibiotic development.
Materials and Methods
Cloning and Mutagenesis.
Primers and templates used in this study are listed in SI Appendix, Table S2. PCR amplicon containing ftsEX in P. aeruginosa was made using Phusion DNA polymerase. It was then purified and digested with NdeI and HindIII. After gel purification, the product was ligated to pTD68 (33) digested with the same enzymes to produce pCS80 [PT7::his6-ftsEX]. pCS157 (PT7::his6-sumo-envC∆19) was constructed by ligating pTD68 digested with BamHI and HindIII to a PCR amplicon synthesized by EnvC F and EnvC R digested with the same enzymes. pAAY68 (PT7::his6-sumo-amiB∆29) was constructed by Gibson’s assembly using pTD68 digested with NdeI and HindIII and a PCR amplicon generated from primers AmiB_F and AmiB_R. Mutants of ftsE and EnvC were generated using the QuikChange kit (Agilent Technologies) using pCS80 as the template and primers listed in SI Appendix, Table S2. The changes were verified by DNA sequencing.
Protein Expression, Purification of FtsEX Complex and Their Mutants.
FtsEX, FtsED162AX, and FtsEE163QX were expressed in E. coli strain Lemo21(DE3). Cells were grown in lysogeny broth (LB) supplemented with 50 µg/mL ampicillin and 10 µg/mL chloramphenicol at 37 °C until the optical density at 600 nm (OD600) reached 0.8. The expression of the recombinant proteins was induced by adding isopropyl-β-D-thiogalactoside (IPTG) to a final concentration of 0.5 mM at 16 °C for 16 to 18 h. Cells were harvested by centrifugation at 20,000 × g for 15 min at room temperature (RT). Pellets were stored at −80 °C.
The frozen cell pellets were thawed at 4 °C and resuspended in buffer A (25 mM Tris-HCl pH 7.5, 150 mM NaCl, and 10% glycerol). Cells were lysed using a high-pressure homogenizer (AH-100D, ATS). The mixture was centrifuged at 100,000 × g for 1 h at 4 °C to extract the membrane. The membrane was further solubilized using buffer B containing 25 mM Tris-HCl, 150 mM NaCl, 5% glycerol, 1.3% n-dodecyl b-D-maltoside, pH 7.5 for 2 h at 4 °C, and centrifuged at 100,000 × g for 30 min to remove the insoluble fraction. The solubilized membrane fraction was incubated with the TALON SuperFlow resin (Takara 635502) and washed with buffer B supplemented with 20 mM imidazole, and 2 mM ATP/MgCl2. The detergent was replaced by a peptide (from Peptidisc Biotech) at 1 mg/mL ( to produce peptidiscs using the “on-beads” method described by Carlson et al. (47). FtsEX complex reconstituted in peptidiscs was washed with buffer B with 20 mM imidazole and eluted with buffer B supplemented with 200 mM imidazole. The purified protein was applied to a gel filtration chromatography column (Superose 6 increase 10/300) and eluted with buffer C containing 25 mM Tris-HCl, 150 mM NaCl, pH 7.5. Fractions corresponding to the FtsEX complex were pooled, concentrated to 4 mg/mL, and stored at −80 °C.
Protein Expression, Purification of EnvC, AmiB, and Their Mutants.
EnvC, AmiB, and their variants were expressed in E. coli strain Lemo21(DE3). Cells were grown in LB medium supplemented with 50 µg/mL ampicillin at 37 °C until the OD600 reached 0.8. IPTG was added to a final concentration of 0.5 mM and the culture was incubated at 16 °C for 16 to 18 h to induce protein expression. Cells were harvested using centrifugation at 20,000 × g for 15 min at RT and the pellets were stored at −80 °C. The frozen cell pellets were thawed at 4 °C in buffer A (25 mM Tris-HCl pH 7.5, 150 mM NaCl, and 10% glycerol) and lysed using a high-pressure homogenizer (AH-100D, ATS). The mixture was centrifuged at 27,000 × g for 1 h at 4 °C, then the supernatant was incubated with the TALON resin and washed with buffer A supplemented with 20 mM imidazole. After washing, His-tagged small Ubiquitin-related modifier (SUMO) protease Ulp1 was added to the resin-buffer mixture (0.1 mg Ulp1 per 1 L culture) to cleave the SUMO tag at RT for 2 h. Cleaved samples were collected, concentrated, and applied to the Superose 6 Increase 10/300 column equilibrated in buffer B. Fractions containing the target protein were identified by Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis (SDS-PAGE), pooled, concentrated to 3 mg/mL, and stored at −80 °C.
Protein Interaction Assay.
To investigate the interactions between FtsEX and EnvC, we added 15 μg FtsEX or its ATPase-defective variant FtsEE163QX to preequilibrated TALON resin at RT for 15 min. Unbound FtsEX was removed by washing the resin with 100 μL of buffer A with 20 mM imidazole. Next, 6 µg EnvC or its mutants (Y114A, Y121A, K123A, Q128A, K133A, D142A, Y143A, 121A/K123A, 3Y-A, and 4Y-A) were added, followed by washing with buffer A supplemented with 20 mM imidazole. Bound proteins and potential interacting partners were eluted with 20 μL of buffer A with 300 mM imidazole and analyzed by SDS-PAGE. For this assay, 2 mM ATP/Mg2+ was added throughout the process.
To investigate the interactions between FtsEX/EnvC and AmiB, 20 μg FtsEX/EnvC or its ATPase-defective variants FtsEE163QX/EnvC were added to TALON resin in the presence or absence of 2 mM ATP/MgCl2 at RT for 15 min. Unbound FtsEX/EnvC or FtsEE163QX/EnvC was removed by washing the resin with 100 μL of buffer A with 20 mM imidazole. Next, 20 μg AmiB was added to the resin and incubated for 2 min before washing five times with 200 μL of buffer A with 20 mM imidazole in the presence or absence of 2 mM ATP/MgCl2. Bound protein was eluted by 20 μL of buffer A with 300 mM imidazole and analyzed by SDS-PAGE.
To test the effect of ATP analogues (ATPγS/MgCl2 or AMPPNP/MgCl2 or ATP/Vanadate/MgCl2 or ADP/MgCl2) on the interactions between FtsEX/EnvC or FtsEE163QX/EnvC and AmiB, we performed a similar assay in the presence or absence of 2 mM ATP or its analogues. Briefly, ATP or its analogues were added to the FtsEX/EnvC-resin mixture during their incubation. After removing free FtsEX, 20 μg AmiB was added to the resin and incubated for 2 min before washing five times with 200 μL of buffer A with 20 mM imidazole. Bound protein was eluted by 20 μL of buffer A with 300 mM imidazole and analyzed by SDS-PAGE.
Reconstitution of FtsEX Complexes.
To reconstitute the EnvC–FtsEX complex, excess EnvC (threefold amount of FtsEX) was added to the resin-bound FtsEE163QX-peptidiscs during purification. The mixture was incubated at RT for 15 min. Unbound EnvC was removed by washing with buffer A with 20 mM imidazole. The FtsEX–EnvC complex was eluted with buffer A containing 200 mM imidazole. The eluted protein was concentrated and further purified by gel filtration (Superose 6 increase 10/300) in buffer B. Fractions with the FtsEX–EnvC complex were confirmed by SDS-PAGE, pooled, concentrated to 3 mg/mL, and stored at −80 °C.
To reconstitute the AmiB–EnvC–FtsEX supercomplex, AmiB was added into the FtsEX–EnvC complex in peptidiscs at a ratio of 3:1. After incubation for 15 min, the supercomplex was purified by gel filtration. Fractions with the FtsEX–EnvC–AmiB complex were pooled and concentrated to 3 mg/mL, and stored at −80 °C.
Protein Chemical Cross-Linking.
Glutaraldehyde was diluted immediately in ddH2O before use. Proteins were buffer exchanged to 25 mM 4-(2-Hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), 150 mM NaCl using a desalting column (Thermofisher). Samples were diluted to 0.1 mg/mL and were chemically cross-linked with 0.025% glutaraldehyde for 10 min on ice. The cross-linking reaction was quenched by the addition of 20 mM glycine (pH 7.4). Cross-linked samples were concentrated prior to the experiment.
ATP Hydrolysis Assay.
The ATPase activity is measured using the malachite green phosphate assay (48). Briefly, 2 μg FtsEX or its variants were added into a 20-μL reaction buffer containing 25 mM Tris-HCl pH 7.5, 150 mM NaCl, and 2 mM ATP/MgCl2. To determine the Michaelis–Menten constants, different amounts of ATP/MgCl2 (0/0.0625/0.125/0.25/0.5/1/2/4 mM) were added to initiate the reaction. After incubating at 37 °C for 10 min, 80 μL Malachite green-ammonium molybdate (MG-AG) solution was added and vortex immediately to inactivate the enzyme. After 2 min, 10 μL 34%(w/v) sodium citrate was added and vortex immediately. Absorbance at 650 nm was measured after 5 min. The amount of inorganic phosphate (Pi) released was calculated using a standard curve generated by a known amount of Pi using a K2HPO4 solution. Data were fitted to the Michaelis–Menten equation using Microsoft Excel and GraphPad Prism 9.
Isolation of PG.
PG was isolated essentially as described (33, 49). Briefly, 400 mL strain HMS0002 (50) was grown in brain heart infusion broth at 37 °C in 5% CO2 until OD600 reaches ~0.4. Cells were collected by centrifugation at 5,000 × g for 10 min at RT. Pellets were resuspended in 15 mL 1× PBS and immediately transferred to 10 mL boiling 10% SDS. The suspension was boiled for 30 min and centrifuged at 10,000 × g for 10 min at RT. The crude PG pellets were washed four times with 0.1× Phosphate-Buffered Saline (PBS) and once with 2 mL water, then resuspended in 1 mL reaction buffer (50 mM Tris-HCl pH 7.4, 1mM MgCl2). Next, 2 µL RNase I (10 mg/mL, EN0531) and 2 µL DNase I (M0303S) were added and the mixture was incubated at 37 °C for 1 h. Digested PG was collected by centrifugation at 20,000 × g for 5 min at RT, resuspended in 1 mL of 1× PBS, and digested by adding 200 µg trypsin (Sigma) and incubated overnight at 37 °C. After collecting the PG by centrifugation, the pellets were washed three times with 1 mL 1% SDS, once with 1 mL 8M LiCl with 15 min incubation at 37 °C, once with 1 mL 0.1M EDTA with 15 min incubation at 37 °C, and finally three times with 1 mL water. The purified PG was resuspended in 1 mL water and stored at 4 °C.
Dye Release Assay.
PG was labeled with Ramazol brilliant blue as described (33). Briefly, 478 µL PG suspension was mixed with 56.3 µL 0.2M Ramazol Brilliant Blue R (Sigma R8001 in water), 28.2 µL 5M NaOH, and 456.3 µL water. The mixture was incubated at 37 °C for 18 h. PG was collected by centrifugation at 20,000 × g for 5 min at RT, and washed extensively with water until the supernatant fraction became colorless. Labeled PG was resuspended in ddH2O and stored at 4 °C.
To remove the trace amount of unbound dye, ramazol brilliant blue (RBB)-labeled PG was washed once with water before use. To digest the PG, 20 μL RBB-labelled PG was mixed with the indicated proteins dissolved in 80 μL reaction buffer (25 mM Tris-HCl pH 7.5, 150 mM NaCl, 2 mM ATP-Mg2+) and incubated at 37 °C for 2 h. The final protein concentration is approximately 2 μM. To stop the reaction, the mixture was placed at 95 °C for 5 min and centrifuged at 20,000 × g for 5 min at RT. Then, 80 µL of the supernatant fraction was transferred carefully to a 96-well transparent plate and their absorbance at 595 nm was measured.
Electron Microscopy Sample Preparation and Data Acquisition.
For sample preparation, 3.5 μL of the protein sample at a concentration of 3 mg/mL was applied to glow-discharged Quantifoil holey carbon grids (1.2/1.3, 400 mesh). To obtain the ATP-bound complex, 2 mM ATP-Mg2+ was added to the sample and incubated at 37 °C for 8 min before freezing. Grids were blotted for 3 to 4.5 s with 100% relative humidity and plunge-frozen in liquid ethane cooled by liquid nitrogen using a Vitrobot System (Gatan). Cryo-EM data were collected at liquid nitrogen temperature on a Titan Krios electron microscope (Thermo Fisher Scientific), equipped with a K3 Summit direct electron detector (Gatan) and GIF Quantum energy filter. All cryo-EM movies were recorded in counting mode with SerialEM4 (51) with a slit width of 20 eV from the energy filter. Movies were acquired at nominal magnifications of 81 k or 105 k, corresponding to a calibrated pixel size of 1.105 or 0.858 Å on the specimen level. The total exposure time of each movie was 6 s, resulting in a total dose range between 40 and 55 electrons per Å2, fractionated into 48 frames. More details of electron microscopy data collection parameters are listed in SI Appendix, Table S1.
Electron Microscope Image Processing.
EM data were processed by CryoSPARC (52). Dose-fractionated movies collected using K3 Summit direct electron detector were subjected to motion correction using the program MotionCor2 (53). A sum of all frames of each movie was calculated following a dose-weighting scheme and used for all image-processing steps except defocus determination. CTFFIND4 (54) was used to calculate defocus values of the summed images from all movie frames without dose weighting. Particle picking was performed using the blob picker followed by the template picker. 2D and 3D classification and 3D refinement were carried out using “2D classification,” “ab-initio reconstruction” and “heterogenous refinement.” Refinements were done using “homogenous refinement” and “nonuniform refinement.” The overall resolutions were estimated based on the gold-standard criterion of Fourier shell correlation = 0.143. Local resolution was estimated by “local resolution estimation.”
Model Building and Refinement for FtsEX and its Complexes with EnvC.
The initial model of FtsE and FtsX was generated by AlphaFold2 (34). The initial model of EnvC was generated by SWISS-MODEL server based on the E. coli FtsX PLD–EnvC complex (PDB code: 6TPI). These models were rigid-body fitted to our cryo-EM maps in the University of California, San Francisco (UCSF) Chimera (55), manually rebuilt in Coot (56), and refined using real space refinement in Phenix (57). Restraints for ATP were generated with phenix_elbow program using its isomeric SMILES string files obtained from the PDB chemical component dictionary through Ligand Expo. Ligands were manually docked into the cryo-EM maps in Coot, followed by iterative real-space refinements in Phenix. Final models were validated with statistics from Ramachandran plots, MolProbity scores, and clash scores with the program in Phoenix (see SI Appendix, Table S1 for details). Figures were generated using UCSF Chimera.
Model Building for the Complex Structure of FtsEX/EnvC/AmiB.
To model the FtsEX/EnvC/AmiB supercomplex in the presence of ATP, we first built the FtsEX complex with the adjacent CCD domain of EnvC using the improved EM density map obtained from focused refinement at 4.5 Å resolution. This model was then docked into the 5.8 Å EM map of the supercomplex. Although the periplasmic region, including the membrane-distal region of the CCD domain of EnvC, the dLytM domain of EnvC, and AmiB, were of low resolution, we were able to manually build an initial model with the help of a homologous model of AmiB generated using SwissModel. We also modeled the interaction between dLytM and the catalytic domain of AmiB as their size is large with clear features. At this point, our modeling suggested that the RH2 region of EnvC (AA271-298) is in proximity to the AMIN domain. While we were only left with one small lobe of density for this region, we observed that multiple orientations could achieve good fitting. Furthermore, the exact interaction at the interface remained elusive due to the low resolution of the map.
To address this issue, we used AlphaFold to predict the detailed interaction models between dLytMEnvC and the catalytic domain of AmiB, the RH2 region with the AMINAmiB, and the region of EnvC (AA20-270), including the coiled-coil domain and the RH1 (SI Appendix, Fig. S15). The predicted models revealed an extensive interface between the RH-binding cleft of dLytMEnvC and α6 of the AmiB catalytic domain; On contrast, an autoinhibited state was predicted for AimB alone. We replaced the original models of EnvC and AmiB with these AlphaFold predicted models to create a detailed interaction at the interface. After this, we were able to settle the orientation of AmiNAmiB and the interacting RH2EnvC using the predicted structure on the membrane-distal side of the CCD domain. After further rough refinement against the 5.8 Å EM map for the supercomplex, we achieved a decent fitting with the coiled-coil domain of EnvC largely extended by RH1 and AmiB with the catalytic site exposed for PG hydrolysis. However, the 40-residue linker region of AmiB (AA136-175) connecting the AMIN and catalytic domain was not built in the final model.
Supplementary Material
Appendix 01 (PDF)
ATP binding leading to PLD restraining.
EnvC activation caused by the restraining of PLD upon ATP binding.
Acknowledgments
We are grateful to David Roper at the University of Warwick and Malcolm Winkler at Indiana University Bloomington, for helpful discussion and comment on the project. We thank Zongli Li at Harvard Medical School and Yumei Wang at Institute of Physics at China, for their help in initial sample screening. We also thank all Luo lab members for their constructive comments on this study. This work is supported by grants from the National University of Singapore Start-up grant (the Ministry of Education Academic Research (MOE) Tier 1 Grants 21-0053-A0001, 22-3449-A0001, and 22-3448-A0001 to M.L., NUHSRO/2017/070/SU/01 to L.-T.S.), and the National Research Foundation Fellowship (NRF) (NRFF11-2019-0005 to L.-T.S.), the MOE Tier 2 Fund (MOE-T2EP30222-0015 to M.L., and MOE-T2EP30220-0012 to L.-T.S.). In Spain, work was supported by grants PID2020-115331GB-100 funded by MCIN/AEI/10.13039/501100011033 and CRSII5_198737/1 by the Swiss NSF to J.A.H. In US work was supported by the Howard Hughes Medical Institute and the NIH R01AI083365 to T.G.B.
Author contributions
J.A.H., T.G.B., L.-T.S., and M.L. designed research; X.X. and J.L. performed research; W.-Z.C., M.A.P., and J.S. contributed new reagents/analytic tools; X.X., J.L., J.A.H., T.G.B., L.-T.S., and M.L. analyzed data and wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Contributor Information
Juan A. Hermoso, Email: xjuan@iqfr.csic.es.
Thomas Bernhardt, Email: thomas_bernhardt@hms.harvard.edu.
Lok-To Sham, Email: lsham@nus.edu.sg.
Min Luo, Email: dbslmin@nus.edu.sg.
Data, Materials, and Software Availability
Five 3D cryo-EM density maps of FtsEX and its complexes in the presence and absence of bound ATP have been deposited in the Electron Microscopy Data Bank under accession codes: EMDB-35203 (ATP-free FtsEX); EMDB-35204 (ATP-bound FtsEE163QX); EMDB-35201 (ATP-free FtsEX complexed with EnvC); EMDB-35213 (ATP-bound FtsEE163QX complexed with both EnvC and AmiB) and EMDB-35205 (ATP-bound FtsEE163QX complexed with EnvC, obtained by focused refinement of EMDB-35213 data with a mask on FtsEX region). Four atomic models have been deposited in the Protein Data Bank under accession codes 8I6Q (ATP-free FtsEX); 8I6R (ATP-bound FtsEE163QX); 8I6O (ATP-free FtsEX complexed with EnvC); 8I6S (ATP-bound FtsEE163QX complexed with EnvC). All data presented are available in the main text or the SI Appendix.
Supporting Information
References
- 1.Typas A., Banzhaf M., Gross C. A., Vollmer W., From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat. Rev. Microbiol. 10, 123–136 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Singh S. K., SaiSree L., Amrutha R. N., Reddy M., Three redundant murein endopeptidases catalyse an essential cleavage step in peptidoglycan synthesis of Escherichia coli K12. Mol. Microbiol. 86, 1036–1051 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Uehara T., Bernhardt T. G., More than just lysins: Peptidoglycan hydrolases tailor the cell wall. Curr. Opin. Microbiol. 14, 698–703 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McQuillen R., Xiao J., Insights into the structure, function, and dynamics of the bacterial cytokinetic FtsZ-ring. Annu. Rev. Biophys. 49, 309–341 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Du S., Lutkenhaus J., At the heart of bacterial cytokinesis: The Z ring. Trends Microbiol. 27, 781–791 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsang M. J., Bernhardt T. G., Guiding divisome assembly and controlling its activity. Curr. Opin. Microbiol. 24, 60–65 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mahone C. R., Goley E. D., Bacterial cell division at a glance. J Cell Sci 133 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heidrich C., Ursinus A., Berger J., Schwarz H., Holtje J. V., Effects of multiple deletions of murein hydrolases on viability, septum cleavage, and sensitivity to large toxic molecules in Escherichia coli. J. Bacteriol. 184, 6093–6099 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heidrich C., et al. , Involvement of N-acetylmuramyl-L-alanine amidases in cell separation and antibiotic-induced autolysis of Escherichia coli. Mol. Microbiol. 41, 167–178 (2001). [DOI] [PubMed] [Google Scholar]
- 10.Yang D. C., Tan K., Joachimiak A., Bernhardt T. G., A conformational switch controls cell wall-remodelling enzymes required for bacterial cell division. Mol. Microbiol. 85, 768–781 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gerding M. A., et al. , Self-enhanced accumulation of FtsN at division sites and roles for other proteins with a SPOR domain (DamX, DedD, and RlpA) in Escherichia coli cell constriction. J. Bacteriol. 191, 7383–7401 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alcorlo M., et al. , Structural basis of denuded glycan recognition by SPOR domains in bacterial cell division. Nat. Commun. 10, 5567 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cook J., et al. , Insights into bacterial cell division from a structure of EnvC bound to the FtsX periplasmic domain. Proc. Natl. Acad. Sci. U.S.A. 117, 28355–28365 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pichoff S., Du S., Lutkenhaus J., Roles of FtsEX in cell division. Res. Microbiol. 170, 374–380 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alcorlo M., Martinez-Caballero S., Molina R., Hermoso J. A., Regulation of lytic machineries by the FtsEX complex in the bacterial divisome. Subcell Biochem. 99, 285–315 (2022). [DOI] [PubMed] [Google Scholar]
- 16.Yang D. C., et al. , An ATP-binding cassette transporter-like complex governs cell-wall hydrolysis at the bacterial cytokinetic ring. Proc. Natl. Acad. Sci. U.S.A. 108, E1052–1060 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reddy M., Role of FtsEX in cell division of Escherichia coli: Viability of ftsEX mutants is dependent on functional SufI or high osmotic strength. J. Bacteriol. 189, 98–108 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmidt K. L., et al. , A predicted ABC transporter, FtsEX, is needed for cell division in Escherichia coli. J. Bacteriol. 186, 785–793 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arends S. J., Kustusch R. J., Weiss D. S., ATP-binding site lesions in FtsE impair cell division. J. Bacteriol. 191, 3772–3784 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pichoff S., Du S., Lutkenhaus J., Disruption of divisome assembly rescued by FtsN-FtsA interaction in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 115, E6855–E6862 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Du S., Henke W., Pichoff S., Lutkenhaus J., How FtsEX localizes to the Z ring and interacts with FtsA to regulate cell division. Mol. Microbiol. 112, 881–895 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Du S., Pichoff S., Lutkenhaus J., Roles of ATP hydrolysis by FtsEX and Interaction with FtsA in regulation of septal peptidoglycan synthesis and hydrolysis. mBio 11, e01247-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meisner J., et al. , FtsEX is required for CwlO peptidoglycan hydrolase activity during cell wall elongation in Bacillus subtilis. Mol. Microbiol. 89, 1069–1083 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sham L. T., Barendt S. M., Kopecky K. E., Winkler M. E., Essential PcsB putative peptidoglycan hydrolase interacts with the essential FtsXSpn cell division protein in Streptococcus pneumoniae D39. Proc. Natl. Acad. Sci. U.S.A. 108, E1061–1069 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gaday Q., et al. , FtsEX-independent control of RipA-mediated cell separation in Corynebacteriales. Proc. Natl. Acad. Sci. U.S.A. 119, e2214599119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mavrici D., et al. , Mycobacterium tuberculosis FtsX extracellular domain activates the peptidoglycan hydrolase, RipC. Proc. Natl. Acad. Sci. U.S.A. 111, 8037–8042 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alcorlo M., Straume D., Lutkenhaus J., Havarstein L. S., Hermoso J. A., Structural Characterization of the essential cell division protein ftse and its interaction with FtsX in Streptococcus pneumoniae. mBio 11 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rued B. E., et al. , Structure of the large extracellular loop of FtsX and its interaction with the essential peptidoglycan hydrolase PcsB in Streptococcus pneumoniae. mBio 10, e02622-18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fu Y., Bruce K. E., Rued B., Winkler M. E., Giedroc D. P., 1H, 13C, 15N resonance assignments of the extracellular loop 1 domain (ECL1) of Streptococcus pneumoniae D39 FtsX, an essential cell division protein. Biomol. NMR Assign 10, 89–92 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crow A., Greene N. P., Kaplan E., Koronakis V., Structure and mechanotransmission mechanism of the MacB ABC transporter superfamily. Proc. Natl. Acad. Sci. U.S.A. 114, 12572–12577 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sharma S., et al. , Mechanism of LolCDE as a molecular extruder of bacterial triacylated lipoproteins. Nat. Commun. 12, 4687 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tang X., et al. , Structural basis for bacterial lipoprotein relocation by the transporter LolCDE. Nat. Struct. Mol. Biol. 28, 347–355 (2021). [DOI] [PubMed] [Google Scholar]
- 33.Uehara T., Parzych K. R., Dinh T., Bernhardt T. G., Daughter cell separation is controlled by cytokinetic ring-activated cell wall hydrolysis. EMBO J. 29, 1412–1422 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jumper J., et al. , Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rocaboy M., et al. , The crystal structure of the cell division amidase AmiC reveals the fold of the AMIN domain, a new peptidoglycan binding domain. Mol. Microbiol. 90, 267–277 (2013). [DOI] [PubMed] [Google Scholar]
- 36.Peters N. T., et al. , Structure-function analysis of the LytM domain of EnvC, an activator of cell wall remodelling at the Escherichia coli division site. Mol. Microbiol. 89, 690–701 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Navarro P. P., et al. , Cell wall synthesis and remodelling dynamics determine division site architecture and cell shape in Escherichia coli. Nat. Microbiol. 7, 1621–1634 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Erickson H. P., How bacterial cell division might cheat turgor pressure - a unified mechanism of septal division in Gram-positive and Gram-negative bacteria. Bioessays 39 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brunet Y. R., Wang X., Rudner D. Z., SweC and SweD are essential co-factors of the FtsEX-CwlO cell wall hydrolase complex in Bacillus subtilis. PLoS Genet 15, e1008296 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lim H. C., et al. , Identification of new components of the RipC-FtsEX cell separation pathway of Corynebacterineae. PLoS Genet. 15, e1008284 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marmont L. S., Bernhardt T. G., A conserved subcomplex within the bacterial cytokinetic ring activates cell wall synthesis by the FtsW-FtsI synthase. Proc. Natl. Acad. Sci. U.S.A. 117, 23879–23885 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meier E. L., et al. , FtsEX-mediated regulation of the final stages of cell division reveals morphogenetic plasticity in Caulobacter crescentus. PLoS Genet. 13, e1006999 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou X., et al. , Sequential assembly of the septal cell envelope prior to V snapping in Corynebacterium glutamicum. Nat. Chem. Biol. 15, 221–231 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Crawford M. A., et al. , Identification of the bacterial protein FtsX as a unique target of chemokine-mediated antimicrobial activity against Bacillus anthracis. Proc. Natl. Acad. Sci. U.S.A. 108, 17159–17164 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Margulieux K. R., Fox J. W., Nakamoto R. K., Hughes M. A., CXCL10 acts as a bifunctional antimicrobial molecule against Bacillus anthracis. mBio 7, e00334-16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bruce K. E., Rued B. E., Tsui H. T., Winkler M. E., The opp (AmiACDEF) oligopeptide transporter mediates resistance of serotype 2 Streptococcus pneumoniae D39 to killing by chemokine CXCL10 and other antimicrobial peptides. J. Bacteriol. 200, e00745-17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carlson M. L., et al. , The Peptidisc, a simple method for stabilizing membrane proteins in detergent-free solution. Elife 7, e34085 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lanzetta P. A., Alvarez L. J., Reinach P. S., Candia O. A., An improved assay for nanomole amounts of inorganic phosphate. Anal. Biochem. 100, 95–97 (1979). [DOI] [PubMed] [Google Scholar]
- 49.Barendt S. M., et al. , Influences of capsule on cell shape and chain formation of wild-type and pcsB mutants of serotype 2 Streptococcus pneumoniae. J. Bacteriol. 191, 3024–3040 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chua W. Z., et al. , High-throughput mutagenesis and cross-complementation experiments reveal substrate preference and critical residues of the capsule transporters in Streptococcus pneumoniae. mBio 12, e0261521 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mastronarde D. N., Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005). [DOI] [PubMed] [Google Scholar]
- 52.Punjani A., Rubinstein J. L., Fleet D. J., Brubaker M. A., cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 53.Zheng S. Q., et al. , MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rohou A., Grigorieff N., CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pettersen E. F., et al. , UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 56.Emsley P., Cowtan K., Coot: Model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 57.Adams P. D., et al. , PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
ATP binding leading to PLD restraining.
EnvC activation caused by the restraining of PLD upon ATP binding.
Data Availability Statement
Five 3D cryo-EM density maps of FtsEX and its complexes in the presence and absence of bound ATP have been deposited in the Electron Microscopy Data Bank under accession codes: EMDB-35203 (ATP-free FtsEX); EMDB-35204 (ATP-bound FtsEE163QX); EMDB-35201 (ATP-free FtsEX complexed with EnvC); EMDB-35213 (ATP-bound FtsEE163QX complexed with both EnvC and AmiB) and EMDB-35205 (ATP-bound FtsEE163QX complexed with EnvC, obtained by focused refinement of EMDB-35213 data with a mask on FtsEX region). Four atomic models have been deposited in the Protein Data Bank under accession codes 8I6Q (ATP-free FtsEX); 8I6R (ATP-bound FtsEE163QX); 8I6O (ATP-free FtsEX complexed with EnvC); 8I6S (ATP-bound FtsEE163QX complexed with EnvC). All data presented are available in the main text or the SI Appendix.