Significance
Tailor-made functional materials to fulfill specific requirements for different applications are of great importance in material and biomedical research. In this study, we report bola-amphiphilic dendrimers for cargo-selective delivery of DNA and small interfering RNA, both important nucleic acid therapeutics used in gene therapy. The observed distinct cargo-selective delivery performance can be ascribed to the differences in the size of nucleic acid cargos and the generation number of the dendrimer vectors. These factors dually impact the cooperative multivalent interaction in nucleic acid binding and release from the vector, thus leading to cargo-adaptive and selective delivery. This study will inspire further exploitation of tailor-made dendrimer platforms for the cargo-selective delivery of various therapeutics in precision medicine.
Keywords: dendrimer, nucleic acid delivery, nonviral vectors, gene transfection, gene silencing
Abstract
Nucleic acid therapeutics are becoming an important drug modality, offering the unique opportunity to address “undruggable” targets, respond rapidly to evolving pathogens, and treat diseases at the gene level for precision medicine. However, nucleic acid therapeutics have poor bioavailability and are chemolabile and enzymolabile, imposing the need for delivery vectors. Dendrimers, by virtue of their well-defined structure and cooperative multivalence, represent precision delivery systems. We synthesized and studied bola-amphiphilic dendrimers for cargo-selective and on-demand delivery of DNA and small interfering RNA (siRNA), both important nucleic acid therapeutics. Remarkably, superior performances were achieved for siRNA delivery with the second-generation dendrimer, yet for DNA delivery with the third generation. We systematically studied these dendrimers with regard to cargo binding, cellular uptake, endosomal release, and in vivo delivery. Differences in size both of the dendrimers and their nucleic acid cargos impacted the cooperative multivalent interactions for cargo binding and release, leading to cargo-adaptive and selective delivery. Moreover, both dendrimers harnessed the advantages of lipid and polymer vectors, while offering nanotechnology-based tumor targeting and redox-responsive cargo release. Notably, they allowed tumor- and cancer cell-specific delivery of siRNA and DNA therapeutics for effective treatment in different cancer models, including aggressive and metastatic malignancies, outperforming the currently available vectors. This study provides avenues to engineer tailor-made vectors for nucleic acid delivery and precision medicine.
Gene therapy based on nucleic acid therapeutics holds great potential to provide precision medicine and offers treatment options for diseases that are beyond the reach of traditional approaches (1). This is mainly due to the unique advantages of nucleic acid drugs including their ability to address “undruggable” targets, respond rapidly to evolving pathogens, and also treat diseases at the gene level (1–5). While DNA- or mRNA-based therapy can increase the expression of a functional protein that is defective or absent via the introduction of DNA or mRNA molecules, antisense oligonucleotide (ASO), small interfering RNA (siRNA), and microRNA (miRNA) can inhibit the expression of disease-associated genes (1–5). However, naked nucleic acids are unsuitable as pharmaceutical drugs because of their poor stability and low bioavailability. Indeed, they can be easily degraded via chemical and/or enzymatic hydrolysis. Also, they are highly hydrophilic and negatively charged, and thus cannot readily cross the cell membrane to enter cells. Furthermore, the various nucleic acid drugs such as ASO, siRNA, miRNA, mRNA, and DNA, have distinct mechanisms of action and varied physiochemical characteristics including size, sequence, molecular weight, single- or double-stranded formation, structural conformation, and flexibility, imposing the need for specific delivery systems (6–8). Thus, there is a high demand for tailor-made, safe, and robust delivery platforms that can protect, transport, and deliver various types of nucleic acid molecules into cells to reach the desired sites of action, enabling them to fulfil their therapeutic purpose.
Nucleic acid delivery can be achieved via viral and nonviral vectors (1, 6). Although viral vectors are highly efficient, increasing safety concerns surrounding their potential immunogenic, mutagenic, and inflammatory effects, coupled with their low capacity for nucleic acid cargos and expensive production costs, impel the urgent development of nonviral alternatives (1, 6). Various nonviral vectors with better safety profiles alongside simple and cost-effective production have been explored (6). Specifically, lipid and polymer vectors are the most extensively studied (9–11), with the lipid vectors being successfully implemented in the first mRNA vaccines against COVID-19 (12) and the first siRNA drug Patisiran (13). Lipid vectors mainly use a membrane-fusion mechanism to deliver nucleic acids to cells (14, 15), whereas polymers usually exploit the proton-sponge effect for endosomal release of nucleic acid cargos (11, 16, 17). Within the group of polymer vectors, dendrimers, a distinct class of polymers with radially symmetric and precisely controllable structures, have emerged as an ideal platform for engineering tailor-made vectors thanks to their well-defined architecture and cooperative multivalence (18–20).
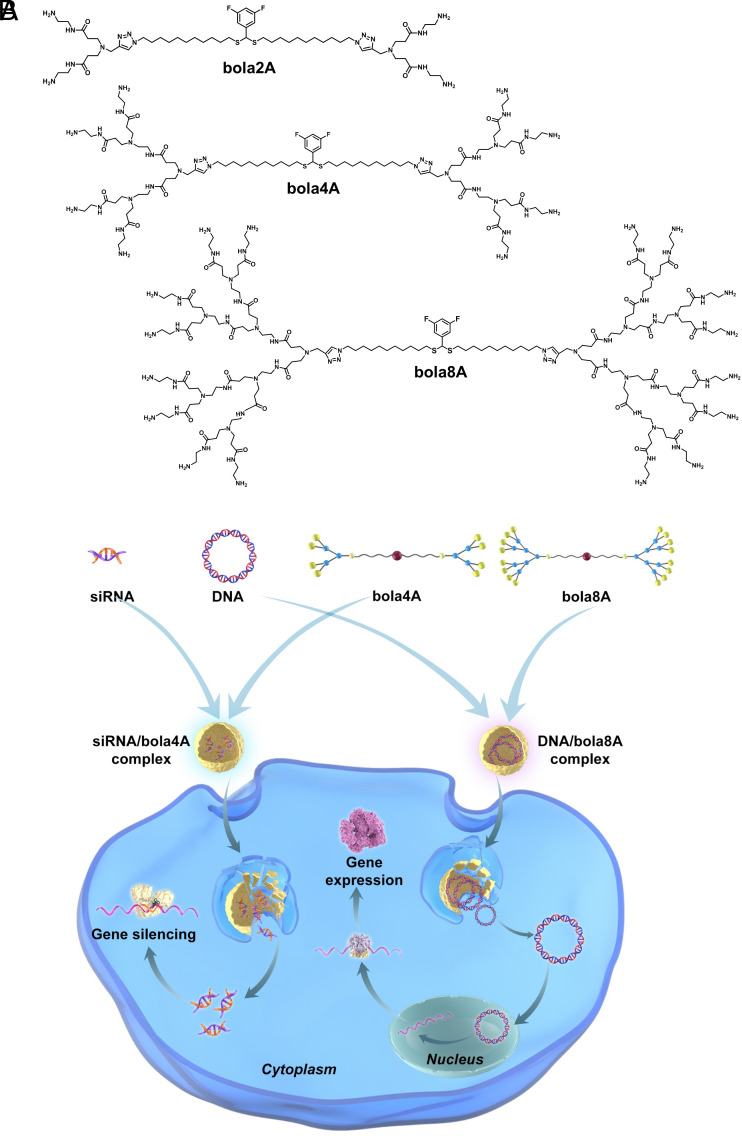
With the view to capitalizing simultaneously on the unique structural properties of dendrimers and the delivery characteristics of lipid and polymer vectors, we and others have recently developed innovative nucleic acid delivery systems based on amphiphilic dendrimers (20–29). By coupling the multivalent cooperativity of dendrimers and the self-assembling feature of lipids (30), these amphiphilic dendrimers harness the advantageous features of lipid and polymer vectors for effective delivery of nucleic acid cargos in various cell lines and animal models of different diseases (21–29, 31–34). Remarkably, these amphiphilic dendrimers self-assemble spontaneously via hydrophobic interactions of the lipid chains, while the positively charged dendron heads bind the negatively charged nucleic acids in stable complexes for delivery (20). In particular, Percec et al. have recently developed amphiphilic dendrimers for successful mRNA delivery with the single dendrimer component vectors outperforming lipid nanoparticles (LNPs) (21, 23, 24). Meanwhile, we and others elaborated bola-amphiphilic dendrimers which harbor a hydrophobic “bola-lipid” entity at the core and two hydrophilic dendrons at the two ends for siRNA delivery (27, 35). Notably, the bola-lipid scaffold was designed to mimic the strong and robust assembly properties of bola-amphiphiles observed in extremophile archae bacteria (36, 37). In particular, our bola-amphiphilic dendrimer bola4A bears a thioacetal group at the bola-lipid core (Fig. 1A), which is responsive to reactive oxygen species (ROS), enabling on-demand delivery of siRNA specifically to cancer cells in which the ROS levels are generally high (27). It is also worth noting that the presence of fluorine tags at the bola-lipid core allows tracking of the ROS-sensitive delivery using 19F-NMR (27).
Fig. 1.
Bola-amphiphilic dendrimers for cargo-specific nucleic acid delivery. (A) Chemical structures of the bola-amphiphilic dendrimers bola2A, bola4A, and bola8A studied in this work. (B) Cartoon illustration of bola-amphiphilic dendrimers bola4A and bola8A for cargo-selective and adaptive delivery of the two distinct nucleic acid types, DNA and siRNA.
We are interested in further developing tailor-made amphiphilic dendrimers for precision medicine via the specific delivery of different nucleic acid therapeutics to cancer cells in tumor lesions. Particularly, we were curious as to whether the generation number of amphiphilic dendrimers would impact their delivery capacity for different nucleic acid therapeutics, such as siRNA and DNA. Therefore, in this study we specifically designed, synthesized, and characterized bola-amphiphilic polyamidoamine (PAMAM) dendrimers with different dendron generations (bola2A, bola4A, and bola8A) as illustrated in Fig. 1A, and evaluated their ability to deliver DNA and siRNA therapeutics in vitro and in vivo. Remarkably, these bola-amphiphilic dendrimers enabled cargo-specific and adaptive delivery as the second-generation dendrimer bola4A was more efficient for siRNA delivery and gene silencing, whereas the third-generation dendrimer bola8A exhibited superior performance for DNA delivery and gene transfection. We systematically studied these dendrimers in terms of their binding to nucleic acid cargos, subsequent cellular uptake, and endosomal release for siRNA-mediated gene silencing and DNA-based gene transfection. The observed differences and selectivity in the nucleic acid delivery of bola4A and bola8A could indeed be ascribed to the differences in the generation number of the dendrimer vectors and the size of the nucleic acid cargos. These factors impacted mutually and collectively on the formation and stability of the delivery complexes through varying levels of compaction and cooperative multivalent interactions for cargo binding and release, ultimately resulting in cargo-selective delivery. Most importantly, they achieved tumor- and cancer cell-specific delivery of siRNA and DNA therapeutics for potent precision cancer treatment in different cancer models including aggressive and metastatic cancers. This study provides not only evidence of effective dendrimer vector-mediated delivery of DNA and siRNA therapeutics in cancer therapy, but also inspiration for engineering tailor-made vector platforms for cargo-selective and on-demand delivery of various nucleic acid therapeutics in future precision medicine. We present below the results of this study.
Results and Discussion
Robust and Reliable Synthesis of Bola-Amphiphilic Dendrimers via Click Chemistry.
We first synthesized the bola-amphiphilic dendrimers featuring different generation numbers of the PAMAM dendron, bola2A, bola4A, and bola8A, using combined divergent and convergent approaches (SI Appendix, Fig. S1). Briefly, a hydrophobic chain bearing two azido terminals (27) was conjugated with PAMAM dendrons carrying an alkyne functionality via the Cu(I)-catalyzed azide-alkyne cycloaddition click reaction. The obtained bola-amphiphilic products were then transformed to the amine-terminated bola-amphiphilic dendrimers bola2A, bola4A, and bola8A, respectively (SI Appendix, Fig. S1). The chemical structures and integrities of all the synthesized dendrimers were analyzed and confirmed using 1H-NMR, 13C-NMR, and 19F-NMR spectroscopy, electrospray high-resolution mass spectrometry (ESI-HRMS) as well as high-performance liquid chromatography (HPLC) (SI Appendix, Fig. S2).
All the three dendrimers bola2A, bola4A, and bola8A were well soluble in water at concentrations up to 50 mM, with bola8A showing the highest solubility. This is in sharp contrast to conventional amphiphiles, which are often insoluble in water. The water solubility of these bola-dendrimers can be mainly ascribed to the two hydrophilic dendron entities, which greatly affect the dendrimer solubility in a generation-dependent manner, i.e., the higher the generation, the higher the hydrophilicity, and the more water soluble the dendrimer. The excellent solubility of these dendrimers in water is certainly advantageous for their use in biological and biomedical applications.
Neither Bola4A Nor Bola8A Shows Any Notable Toxicity, While Bola2A Is Highly Cytotoxic.
As safety is a major concern in the development of delivery vectors for nucleic acid therapeutics, we then examined the cytotoxicity of all three dendrimers (bola2A, bola4A, and bola8A) using PrestoBlue test and lactate dehydrogenase (LDH) assay. The PrestoBlue test examines metabolic toxicity by assessing the cell viability, and the LDH assay evaluates toxicity associated with cell membrane integrity by measuring the release of LDH. Results obtained from PrestoBlue and LDH assays revealed that neither bola4A nor bola8A had any notable cytotoxicity, but bola2A showed strong metabolic toxicity in all the tested cell lines, including human embryonic kidney cells (HEK293 cells), mouse fibroblast cells (L929 cells), and Madin–Darby canine kidney cells (MDCK cells) (SI Appendix, Fig. S3 A–F). The high toxicity of bola2A may be ascribed to its detergent-like characteristics as bola2A has a similar chemical structure and composition to cationic detergents. Further hemolysis assay also confirmed the high toxicity of bola2A (SI Appendix, Fig. S3G). We therefore focused all further nucleic acid delivery studies on the nontoxic bola4A and bola8A.
Bola8A Is Superior for DNA Delivery, Whereas Bola4A Excels at siRNA Delivery.
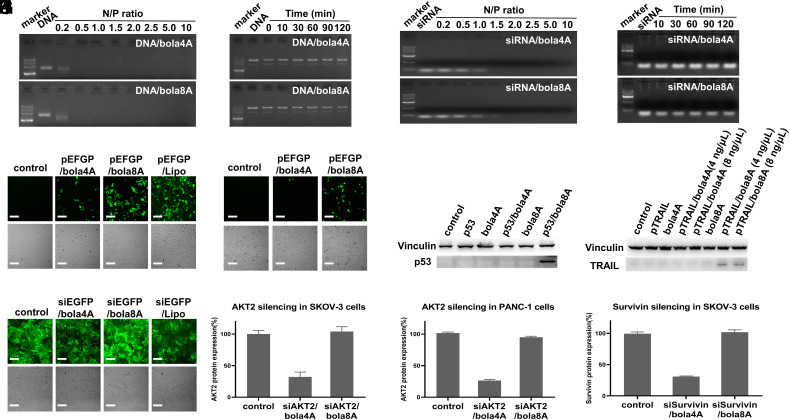
A good vector for nucleic acid delivery should be able to form stable complexes with nucleic acid molecules and protect them against degradation. We therefore inspected the complex formation between the bola-amphiphilic dendrimers (bola4A and bola8A) and different nucleic acid cargos (DNA and siRNA). The formation of dendrimer–nucleic acid complexes depends critically on the dendrimer-to-nucleic acid charge ratio (or N/P ratio), that is the total number of terminal amino groups in the cationic dendrimer divided by the total number of phosphates in the nucleic acid. Remarkably, both bola4A and bola8A formed stable complexes with plasmid DNA (Fig. 2A), even at very low N/P ratios (<0.5), and successfully protected DNA from enzymatic digestion by DNase I (Fig. 2B) as revealed by the results obtained using gel electrophoresis. However, only at N/P ratios >1.5 did bola4A and bola8A generate stable complexes with siRNA (Fig. 2C), sheltering siRNA from RNase-catalyzed degradation (Fig. 2D). The observed differences in the N/P ratios for forming stable DNA/dendrimer and siRNA/dendrimer complexes can be reasonably explained by the different sizes and lengths of DNA and siRNA molecules: The DNA cargo is a plasmid of ~4,500 base pairs and, being larger and longer, has much more negative charges to allow stronger cooperative interaction with dendrimers; accordingly, low N/P ratios (i.e., less dendrimers) are required to generate robust DNA complexes. In contrast, the shorter and smaller siRNA (21 base pairs) provides insufficient cooperative interaction and hence entails more dendrimers (or high N/P ratios) to produce stable and reliable siRNA complexes.
Fig. 2.
Bola-amphiphilic dendrimers bola4A and bola8A bind both DNA and siRNA, but exert cargo-selective delivery. Agarose gel retardation assays of (A) DNA when binding with bola4A and bola8A at N/P ratios ranging from 0.2 to 10 (DNA 200 ng per well) and (B) DNA stability against DNase digestion in the presence of dendrimer at different time points (DNA 200 ng per well, N/P ratio of 1.0). Agarose gel retardation assays of (C) siRNA when binding with bola4A and bola8A at N/P ratios ranging from 0.2 to 10 (siRNA 200 ng per well) and (D) siRNA stability against RNase in the presence of dendrimer at different time points (siRNA 200 ng per well, N/P ratio of 10). DNA and siRNA delivery mediated by the bola-amphiphilic dendrimers bola4A and bola8A, respectively. GFP protein expression in (E) SKOV-3 cells and (F) HeLa cells following treatment with the DNA/dendrimer complexes using the pEGFP (12 and 4.0 ng/μL DNA at N/P ratio of 1.0). Upper panel: fluorescent images; Lower panel: field images. Western blotting analysis of protein expression in HeLa cells upon treatment with the DNA/dendrimer complexes using (G) the plasmid DNA encoding the tumor suppressor protein p53 (p53) (12 ng/μL DNA, N/P ratio of 1.0) and (H) the plasmid DNA encoding pTRAIL (4.0 and 8.0 ng/μL DNA, N/P ratio of 1.0). (I) GFP expression after treatment of HeLa-GFP cells with siRNA/dendrimer complexes using the siRNA targeting GFP (siEGFP) (50 nM siRNA, N/P ratio of 10). Upper panel: fluorescent images; Lower panel: field images. Protein expression of protein kinase B (AKT2) in (J) SKOV-3 and (K) PANC-1 cells, and (L) Survivin in SKOV-3 cells after treatment with the siRNA/dendrimer complexes using the siRNAs targeting AKT2 (siAKT2) and Survivin (siSurvivin) respectively (50 nM siRNA, N/P ratio of 10). The commercial transfection reagent Lipo2000 (Lipo) was used as a control. pEGFP: plasmid DNA expressing GFP; p53: plasmid DNA expressing tumor suppressor protein p53; pTRAIL: plasmid DNA expressing TRAIL; siEGFP: siRNA targeting GFP: siAKT2: siRNA targeting AKT2; siSurvivin: siRNA targeting Survivin. ***P < 0.001 (mean ± SD, n = 3).
We further evaluated both bola4A and bola8A for their efficacy in DNA delivery using a DNA plasmid encoding enhanced green fluorescent protein (pEGFP) as a model. Surprisingly, although both dendrimers bound with plasmid DNA at very low N/P ratios (Fig. 2A), little or no GFP expression was observed after treatment with pEGFP/bola4A, whereas transfection with pEGFP/bola8A produced considerable GFP expression in all the tested cell lines, including human ovarian cancer SKOV-3 cells, human cervical cancer HeLa cells, and human prostate cancer PC-3 cells (Fig. 2 E and F and SI Appendix, Fig. S4). Compared with the commercial transfection reagent Lipo2000, which caused severe cell damage and considerable toxicity (Fig. 2E), no adverse effects on cell growth were observed with either bola4A or bola8A, alone or in complex with DNA, confirming the good safety profile of these two dendrimers. Further evaluation of DNA transfection using a plasmid encoding the tumor suppressor protein 53 (p53) (38) and the tumor necrosis factor (TNF)-related apoptosis-inducing ligand (pTRAIL) (39) delivered by bola4A and bola8A also confirmed that effective DNA transfection had occurred only with bola8A, whereas no expression of p53 or TRAIL was observed with bola4A as the delivery vector (Fig. 2 G and H).
We next examined the performance of bola4A and bola8A for siRNA delivery using siRNA targeting GFP (siEGFP) in HeLa-GFP cells expressing GFP. Interestingly, while both dendrimers formed stable complexes with siRNA at N/P ratios >1.5 (Fig. 2C), the fluorescence intensity of GFP in HeLa-GFP cells was notably reduced following treatment with siEGFP/bola4A, whereas no marked change was observed with siEGFP/bola8A (Fig. 2I and SI Appendix, Fig. S5). Importantly, the use of bola4A generated a level of silencing comparable to that observed with the commercial transfection reagent Lipo2000 while being devoid of any noticeable toxicity. To further investigate the siRNA delivery capacity of bola4A, we evaluated the gene silencing in different cancer cell lines using siRNAs targeting protein kinase B (AKT2), Survivin and heat shock protein 27 (HSP27), respectively. AKT2, Survivin and HSP27 are all involved in cancer development and drug resistance, and are considered as promising targets in cancer treatment (40–43). As shown in Fig. 2 J–L and SI Appendix, Fig. S6, all the siRNAs delivered by bola4A led to effective down-regulation of the corresponding protein expression, whereas no prominent gene silencing effect was observed with bola8A as the delivery vector in all the tested cancer cell lines (SKOV-3, PANC-1, and PC-3).
Moreover, unlike the commercial transfection reagent Lipo2000 that exhibited striking cytotoxicity toward all the tested cell types, neither of the two dendrimers, alone or in complex with either scramble siRNA or DNA, showed any marked toxicity (metabolic, hemolytic or cell membrane damage) (SI Appendix, Fig. S7), further confirming the excellent safety/activity profiles of bola4A and bola8A for nucleic acid delivery.
Underlying Rationale for the Cargo-Selective Nucleic Acid Delivery.
To understand the distinct cargo-selective delivery performance of the two dendrimers bola4A and bola8A, we studied the nucleic acid/dendrimer complexes and their cellular uptake, endosome escape, and cargo release, all of which are prerequisites for nucleic acid delivery.
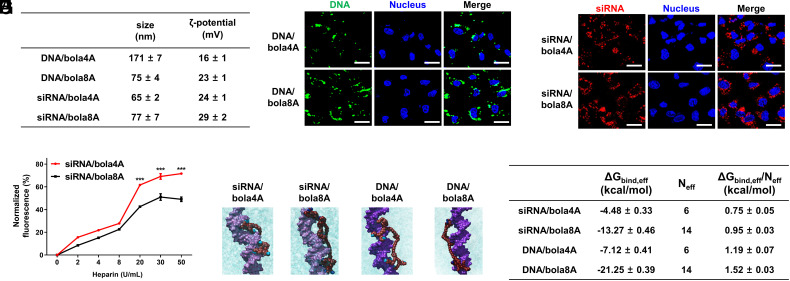
For effective delivery, the dendrimer/cargo complex should be not only stable but also small in size with a positive surface charge. We therefore examined the size and surface potential of the nucleic acid/dendrimer complexes using dynamic light scattering (DLS). For the DNA complexes, the average sizes of the DNA/bola4A and DNA/bola8A complexes were 171 and 75 nm, with ζ-potentials of +16 and +23 mV, respectively. The smaller size of the DNA/bola8A complexes (Fig. 3A) indicated that a higher compaction of DNA was achieved using bola8A compared with bola4A. This can be ascribed to the higher dendron generation number of bola8A, which can provide more and stronger cooperative and multivalent interactions with the DNA molecules, hence compacting them into smaller nanoparticles. Interestingly, both bola4A and bola8A formed siRNA complexes of similar size of ~70 nm (Fig. 3A). The ζ-potentials of the siRNA/bola4A and siRNA/bola8A complexes were also similar, with values of +23 and +26 mV, respectively. The similar size and ζ-potentials of the siRNA/dendrimer complexes can be explained by the small size and short sequence of the siRNA molecule, which appear to have little impact on the size and surface charge of the complexes formed with bola4A and bola8A.
Fig. 3.
Physicochemical characterization of and rationale behind the cargo-selective delivery performance of bola4A and bola8A. (A) The sizes and the ζ-potentials of the DNA/dendrimer complex and the siRNA/dendrimer complex obtained with DNA (24 ng/μL) at an N/P ratio of 2 and siRNA (1.0 μM) at an N/P ratio of 10. (B and C) Cellular uptake and intracellular trafficking of DNA and siRNA delivered by the bola-amphiphilic dendrimers bola4A and bola8A. Confocal imaging of the cellular uptake of (B) the DNA/dendrimer complexes (12 ng/μL YOYO-1-labeled DNA, N/P ratio of 1.0) and (C) the siRNA/dendrimer complexes (50 nM Cy5-labeled siRNA, N/P ratio of 10) in SKOV-3 cells, evaluated using confocal microscopy. The green channel image shows the YOYO-1-labeled DNA (green), the red channel image shows the Cy5-labeled siRNA (red), and the blue channel image shows the nuclei of the SKOV-3 cells stained by Hoechst33342 (blue). (D) The siRNA release from the siRNA/dendrimer complexes assessed using heparin-coupled ethidium bromide (EB) fluorescence assays. ***P < 0.001 versus siRNA/bola4A or siRNA/bola8A, and significance was determined using two-way ANOVA (mean ± SD, n = 3). Atomistic MD simulations of bola4A and bola8A in the presence of siRNA (E and F) and DNA (G and H), respectively. Bola4A atoms are shown as “firebrick spheres,” with the terminal charged amine groups highlighted in deep sky-blue, while bola8A atoms are depicted as dark red spheres with the terminal charged amines colored in navy blue. The siRNA (“orchid” light purple) and the DNA (dark purple) are portrayed as their van der Waals surface and the oxygen atoms in water are shown as cyan transparent spheres. Hydrogen atoms, ions, and counterions (Na+ and Cl−) are omitted for clarity. (I) Binding data of bola4A and bola8A with siRNA and DNA as derived from atomistic MD simulations: free energy of effective binding (ΔGbind,eff), number of effective charges (Neff), and effective-charge-normalized free energy of binding (ΔGbind,eff/Neff) for the nucleic acid/dendrimer complexes are listed.
For DNA delivery, we next used DNA labeled with the green fluorescent dye YOYO-1 to evaluate the cellular uptake of the DNA/dendrimer complexes in SKOV-3 cells. Both confocal microscopy and flow cytometry revealed a considerably higher intensity of YOYO-1 green fluorescence signals in cells treated with DNA/bola8A compared with DNA/bola4A (Fig. 3B and SI Appendix, Fig. S8A), highlighting the more efficient cellular uptake of DNA/bola8A compared with DNA/bola4A. The higher uptake of DNA/bola8A can be ascribed to the larger dendrons within bola8A, which can compact DNA to a smaller size with a higher positive surface charge (Fig. 3A), allowing for more effective cell internalization compared with bola4A. This enhanced cellular uptake explains and corroborates the superior DNA delivery performance of bola8A compared with bola4A.
For siRNA delivery, we used red Cy5-labeled siRNA and green fluorescent lysotracker to track the cellular uptake and intracellular trafficking of the siRNA/dendrimer complexes. Remarkably, both Cy5-siRNA/bola4A and Cy5-siRNA/bola8A showed similar cellular uptake kinetics (Fig. 3C and SI Appendix, Fig. S8B) and endosome release (SI Appendix, Fig. S8C). We further assessed the siRNA release from the siRNA/dendrimer complexes using a heparin displacement method. The siRNA release was greater from siRNA/bola4A than from siRNA/bola8A (Fig. 3D). The difference in the efficiency of the siRNA release can explain the superior siRNA delivery performance of bola4A compared with bola8A. Bearing two larger dendrons, bola8A likely imposes more cooperative multivalency and therefore has stronger interactions with the siRNA compared with bola4A; thus, more stable siRNA/bola8A complexes are formed making siRNA release difficult and allowing little or no gene silencing. In contrast, bola4A has two smaller dendrons and hence, forms less cooperative multivalent interactions with siRNA, making siRNA release easier for more efficient gene silencing. These results corroborated the observed superior gene silencing mediated by siRNA/bola4A, also highlighting the importance of the appropriately balanced cooperative multivalent interactions in nucleic acid binding and release for successful delivery.
To further support this rationale, we performed a quantitative characterization of bola4A and bola8A when interacting, respectively, with siRNA and DNA using atomistic molecular dynamics (MD) simulations (34). Specifically, in line with the MM/PBSA theory (44), we assessed the effective free energy of binding ΔGbind,eff, that is the contribution to binding provided by the dendrimer-positive charges in persistent, efficient and effective interaction with the nucleic acid molecule. To estimate ΔGbind,eff for each nucleic acid/dendrimer complex, the number of effective charged terminal branches of bola4A and bola8A engaged in nucleic acid binding (Neff) was accurately identified, and their individual contribution toward the overall binding energy calculated through a per-residue free energy decomposition technique (45).
For the siRNA/bola4A complex, 6/8 of the dendrimer’s positive charges (75%) were effectively involved in complexing siRNA (Fig. 3E), resulting in a charge-normalized effective free energy of binding (ΔGbind,eff/Neff) value of −0.75 kcal/mol (Fig. 3I). On the other hand, bola8A was not only able to exploit 14/16 (88%) positive charges to persistently bind siRNA (Fig. 3F), but these charges also acted more effectively, as demonstrated by their more favorable ΔGbind,eff/Neff value of −0.95 kcal/mol (Fig. 3I). This higher negative value of ΔGbind,eff/Neff clearly allows bola8A to form a stronger complex with siRNA compared to that formed with bola4A; concomitantly, however, this robust binding is somewhat detrimental to an efficient and optimized release of the biological cargo, as highlighted by the experimental data in Fig. 3D.
Interestingly, these dendrimers exploited the same number of positive charges in their interactions with DNA. Indeed, 6/8 for bola4A (Fig. 3G) and 14/16 for bola8A (Fig. 3H) were detected as being efficiently involved in the binding with the DNA molecule. On the other hand, binding capacity for both dendrimers with DNA was greater compared to siRNA, and the stabilization provided by the electrostatic interaction was more efficient; in fact, more favorable normalized effective free energy of binding values were calculated for both systems: ΔGbind,eff/Neff = −1.19 kcal/mol for DNA/bola4A and ΔGbind,eff/Neff = −1.52 kcal/mol for DNA/bola8A (Fig. 3I). These results agree with the experimental data from the agarose gel retardation assays (Fig. 2 A and C) in which lower N/P ratios were required for the dendrimers to generate more stable complexes with DNA than with siRNA. Moreover, our simulation results further support the hypothesis that a higher density of positive charges of bola8A leads to a more cooperative interaction with the larger DNA and, eventually, to a greater compaction of the DNA/dendrimer complex.
Integrated Delivery Advantages of Both Lipid and Polymer Vectors.
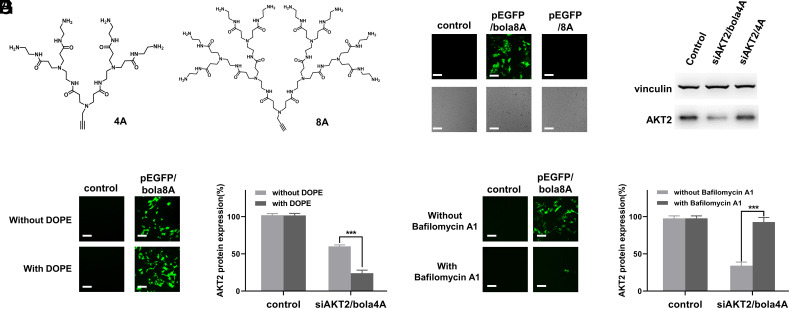
The bola-amphiphilic dendrimers were conceived to combine the delivery mechanisms of lipid and polymeric dendrimer vectors. To confirm the need for this combination for the desired delivery effect, we assessed the nucleic acid delivery performance of the PAMAM dendrons 4A and 8A (Fig. 4A) alone. Compared with the bola-amphiphilic dendrimers bola4A and bola8A, neither 4A nor 8A alone exhibited any delivery activity for siRNA and DNA (Fig. 4 B and C). This finding supports the potential benefits of integrating the delivery mechanisms of both lipids and polymers, such as offered by bola4A and bola8A.
Fig. 4.
Both Bola4A and Bola8A benefit from the combined delivery advantages of lipid and dendrimer vectors for effective nucleic acid delivery. (A) Chemical structure of the hydrophilic PAMAM dendrons 4A and 8A. (B) DNA delivery mediated by 8A for gene transfection compared with that mediated by bola8A. Upper panel: fluorescent images; Lower panel: field images. (C) siRNA delivery mediated by 4A for gene silencing compared with that mediated by bola4A. (D) EGFP protein expression upon treatment with the pEGFP/bola8A complex (12 ng/μL DNA, N/P ratio of 1.0) and (E) AKT2 protein downregulation upon treatment with the siAKT2/bola4A complex (50 nM siRNA, N/P ratio of 5.0) in the presence and absence of the fusogenic helper lipid DOPE. (F) GFP protein expression upon treatment with the pEGFP/bola8A complex (12 ng/μL DNA, N/P ratio of 1.0), and (G) AKT2 protein downregulation upon treatment with the siAKT2/bola4A complex (50 nM siRNA, N/P ratio of 10) in the presence and absence of the proton pump inhibitor, bafilomycin A1. All experiments were performed in SKOV-3 cells. (Scale bar, 200 μm). ***P < 0.001, and significance was determined using two-way ANOVA (mean ± SD, n = 3).
As lipid vectors mainly use the membrane-fusion mechanism for nucleic acid delivery, the fusogenic phospholipid 1,2-dioleoyl-sn-glycerol-3-phosphoethanolamine (DOPE), which promotes membrane fusion, is often used as a helper lipid and is included in LNP formulations to enhance the delivery efficiency (46, 47). To verify whether the bola-amphiphilic dendrimers exploit the fusion mechanism of delivery used by lipid vectors, we assessed the nucleic acid delivery performance of bola4A and bola8A in the presence of DOPE. Indeed, bola8A-mediated DNA delivery and gene transfection was considerably enhanced in the presence of DOPE (Fig. 4D), as was bola4A-mediated siRNA delivery and gene silencing (Fig. 4E). These results highlight that both bola4A and bola8A harbor lipid-like vector delivery characteristics.
Polymer or dendrimer vectors often make use of the proton-sponge effect to promote endosome release of nucleic acid cargo (48). Bafilomycin A1, a proton pump inhibitor, can inhibit endosome acidification and hence, prevent activation of the proton-sponge effect. To examine whether the bola-dendrimers exploit the proton-sponge effect used by PAMAM dendrimer vectors to promote delivery via endosome release, we evaluated the delivery performance of bola4A and bola8A in the presence of bafilomycin A1. Remarkably, both bola8A-mediated DNA delivery for gene transfection (Fig. 4F) and bola4A-mediated siRNA delivery for gene silencing (Fig. 4G) were markedly abated after incubation with bafilomycin A1. These results demonstrate that the delivery performance of both bola4A and bola8A is associated with endosome acidification and does indeed exploit the proton-sponge effect enabled by the PAMAM dendron components. Collectively, these results confirm that both bola-amphiphilic dendrimers bola4A and bola8A indeed combine the beneficial delivery characteristics of lipids and polymers for effective nucleic acid delivery.
Cancer Cell-Specific and Tumor-Targeted Delivery for Precision Cancer Treatment.
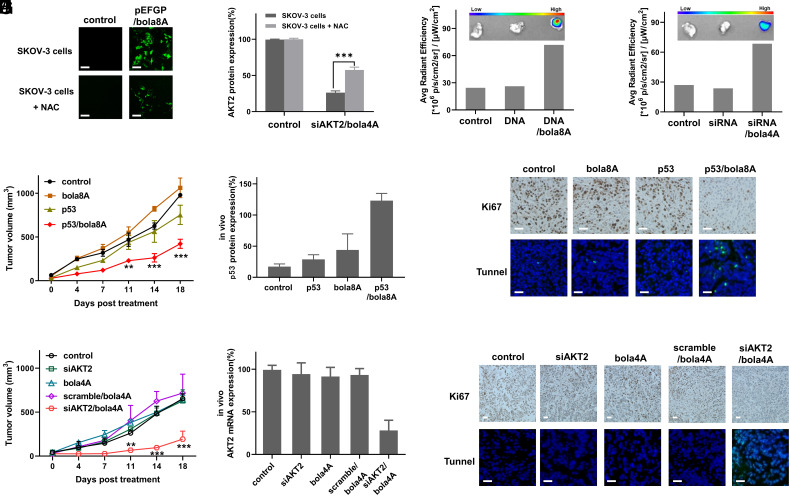
Selective cell- and tissue-specific delivery of nucleic acid drugs presents a formidable challenge. The bola-amphiphilic dendrimers bola4A and bola8A, by virtue of a ROS-responsive thioacetal group contained within, are expected to achieve on-demand specific delivery to cancer cells, which are often characterized by the presence of high ROS levels (27). The bola8A-mediated delivery of the plasmid DNA pEGFP was indeed specific to cancer cells having a high ROS content, as confirmed by a drastically decreased gene transfection observed following treatment with the antioxidant N-Acetyl-L-cysteine (NAC), which reduced the ROS levels in SKOV-3 cells (Fig. 5A and SI Appendix, Fig. S9). Similarly, the bola4A-mediated siRNA delivery was also specific to ROS-rich cancer cells, with NAC-treated cells showing significantly reduced gene silencing (Fig. 5B). Therefore, both bola4A and bola8A are able to mediate effective and specific delivery to cancer cells with high ROS content.
Fig. 5.
Both bola4A and bola8A mediated cancer cell- and tumor-specific delivery with high efficacy. (A) Imaging of GFP protein expression in SKOV-3 cells and NAC-treated SKOV-3 cells following incubation with the plasmid DNA pEGFP complexed with bola8A (12 ng/μL DNA, N/P ratio of 1.0). (Scale bar, 200 μm). (B) Western blotting analysis of AKT2 protein expression in SKOV-3 cells and NAC-treated SKOV-3 cells upon treatment with siAKT2 delivered by bola4A (50 nM siRNA, N/P ratio of 10). ***P < 0.001 and significance was determined using two-way ANOVA (mean ± SD, n = 3). Accumulation of Cy5-labeled (C) DNA/bola8A complex (0.25 mg/kg Cy5-labeled DNA, 0.20 mg/kg bola8A, N/P ratio of 1.0) and (D) siRNA/bola4A complex (1.0 mg/kg Cy5-labeled siRNA, 3.9 mg/kg bola4A, N/P ratio of 5.0) in SKOV-3 xenograft mice after intravenous administration. (E–G): HeLa xenograft mice received intravenous injections of PBS buffer (control), p53 alone, bola8A alone, or p53/bola8A complex (1.0 mg/kg DNA, 0.70 mg/kg bola8A, N/P ratio of 1.0): (E) Tumor growth, (F) p53 protein expression, (G) Tumor cell proliferation revealed by immunohistochemistry staining for Ki67 (Upper), and tumor cell apoptosis detected by the TUNEL assay (Lower) after treatment. **P < 0.01, ***P < 0.001 versus control or p53/bola8A, and significance was determined using two-way ANOVA (mean ± SD, n = 3). (Scale bar, 200 μm). (H–J) SKOV-3 xenograft mice received intravenous injections of PBS buffer (control), siAKT2 alone, bola4A alone, scramble/bola4A complex, or siAKT2/bola4A complex (1.0 mg/kg siRNA, 3.9 mg/kg bola4A, N/P ratio of 5.0): (H) Tumor growth, (I) AKT2 protein expression, (J) Tumor cell proliferation revealed by immunohistochemistry staining for Ki67 (Upper), and cell apoptosis detected by the TUNEL assay (Lower) after treatment. **P < 0.01, ***P < 0.001 versus control or siAKT2/bola4A, and significance was determined using two-way ANOVA (mean ± SD, n = 3). (Scale bar, 200 μm). pEGFP: plasmid DNA expressing GFP protein, p53: plasmid DNA expressing tumor suppressor protein p53, siAKT2: siRNA targeting AKT2, scramble: scramble siRNA.
Considering the ability of both bola4A and bola8A to form small and stable nanoparticles with the nucleic acid molecules, we expected they could thereby effectively enter into and accumulate within tumor tissues via the enhanced permeability and retention (EPR) effect. The leaky vasculature and dysfunctional lymphatic drainage responsible for this phenomenon in the tumor microenvironment (49), enables tumor-targeted delivery. We assessed the tumor-homing of the Cy5-labeled siRNA and DNA, respectively, delivered by bola4A and bola8A in tumor-xenograft mice. As shown in Fig. 5C and SI Appendix, Fig. S10 A and B, Cy5-DNA delivered by bola8A accumulated efficiently in the tumor via the EPR effect, whereas no fluorescent signal was detected in tumors from mice treated with Cy5-DNA alone or with PBS buffer. A similar enriched accumulation in tumor lesions was observed for Cy5-siRNA delivered by bola4A (Fig. 5D and SI Appendix, Fig. S10 C and D). These findings demonstrate the capacity of both dendrimers to provide preferential homing in on tumors for nucleic acid delivery thanks to their ability to form nanoparticles with siRNA and DNA, thereby enabling them to exploit the EPR effect.
Based on the favorable tumor-homing feature, we wanted to assess the performance of bola4A and bola8A to deliver nucleic acid therapeutics for precision cancer treatment. To this end, we chose siRNA targeting AKT2 (siAKT2) and p53 plasmid DNA as the anticancer nucleic acid therapeutics. p53 is an important tumor suppressor protein in many cancers (38), and activating p53 using plasmid DNA encoding p53 represents a promising therapeutic approach for cancer treatment (50). It is to note that the first approved gene therapy product made use of p53 DNA delivered by an adenovirus in treating head and neck cancer (51). Also, AKT2 is a key multifunctional protein associated with tumor growth, proliferation, invasion, metastasis, and drug resistance, and it is therefore considered as a potential target for precision cancer treatment using siRNA therapeutics.
We used tumor-xenograft mouse models issued from the aggressive SKOV-3 ovarian cancer and HeLa cervical cancer cell lines to evaluate the anticancer potency of p53 DNA and siAKT2 delivered by bola8A and bola4A, respectively. Notably, mice treated with p53/bola8A showed significantly reduced tumor growth and tumor weight when compared with the untreated mice or those treated with p53 alone or bola8A alone (Fig. 5E and SI Appendix, Fig. S11A). This can be ascribed to the successful delivery of p53 DNA mediated by bola8A, which produced significantly enhanced p53 expression (Fig. 5F and SI Appendix, Fig. S11B) for effective tumor suppression. Further immunohistochemical analysis confirmed that cancer cell proliferation was indeed considerably inhibited in tumor lesions after treatment with p53/bola8A (Fig. 5G). In addition, TUNNEL-positive cells were significantly increased in the tumor tissues (Fig. 5G and SI Appendix, Fig. S11C), highlighting the induction of apoptosis following treatment with p53/bola8A.
Likewise, over 75% reduction of tumor growth and tumor weight was observed for mice treated with the siAKT2 delivered by bola4A, but not in those treated with siAKT2 alone, bola4A alone, or the scramble siRNA/bola4A complex (Fig. 5H and SI Appendix, Fig. S12A). Examination of AKT2 expression in tumor tissues revealed effective silencing of AKT2 at both mRNA and protein levels in the group treated with siAKT2/bola4A (Fig. 5I and SI Appendix, Fig. S12B). In addition, immunochemistry analysis showed considerably fewer Ki-67-positive cells (Fig. 5J) but much more TUNNEL-positive cells in the tumor tissues from mice treated with siAKT2/bola4A (Fig. 5J and SI Appendix, Fig. S12C), indicating apoptosis induction as the principal mechanism behind the observed anticancer effect.
Importantly, throughout the treatment period, all mice had a normal life showing no unusual behavior or noticeable weight loss (SI Appendix, Figs. S11D and S12D). Moreover, we observed no pathological changes in any organs (SI Appendix, Figs. S11E and S12E) or any major serum biomarker (SI Appendix, Figs. S11F and S12F), confirming that our dendrimer-based nucleic acid delivery systems are indeed safe and devoid of notable adverse effects.
Excellent Performance against Cancer Metastasis.
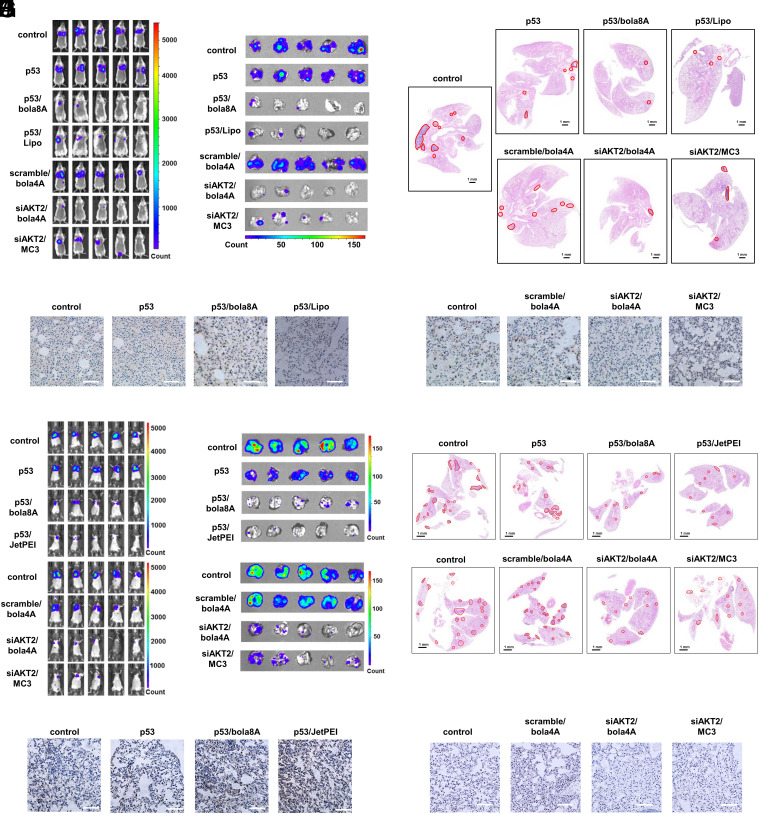
Encouraged by the tumor tissue- and cancer cell-specific delivery mediated by bola8A and bola4A alongside their excellent safety profiles, we wished to challenge the vectors further by assessing their capacity to deliver nucleic acid therapeutics to and effectively treat metastatic cancers, as metastasis is the key cause of failure of cancer therapy and mortality. To this end, we used the lung metastasis cancer models induced by either the murine triple-negative breast cancer 4T1 or the mouse melanoma cancer B16-F10. Specifically, luciferase-tagged 4T1 (4T1-luc) and B16-F10 (B16-F10-luc) cells were injected into the tail vein of BALB/c and C57BL6 female mice, respectively, and tumor metastases were detected in mice using bioluminescence imaging (Fig. 6). The commercial transfection reagent Lipofectamine 2000, JetPEI, and MC3 (MC3: DLin-MC3-DMA, which is an FDA-approved LNP for siRNA delivery) (2, 15) were used as positive controls.
Fig. 6.
Effective inhibition of tumor metastasis using DNA and siRNA therapeutics delivered by bola8A and bola4A, respectively, in lung metastatic cancer model. (A–E) 4T1-luc metastatic tumor-bearing mice received intravenous injections of PBS buffer (control), p53 alone, p53/bola8A complex, p53/Lipo complex (2.0 mg/kg DNA, 1.5 mg/kg bola8A, N/P ratio of 1.0), siAKT2 alone, siAKT2/bola4A complex, or siAKT2/MC3 complex (1.0 mg/kg siRNA, 3.9 mg/kg bola4A, N/P ratio of 5.0) (n = 5): (A) In vivo bioluminescence imaging of 4T1-luc tumor metastases in the mice. (B) Ex vivo bioluminescence imaging of 4T1-luc tumor metastases in the lung at the experimental end point post treatment. (C) Histological analysis of lung tissues from 4T1-luc metastatic tumor-bearing mice at the experimental end point post treatment. The metastatic lesions (red solid outlines) were identified as cell clusters with darkly stained nuclei. (Scale bars, 1 μm). (D) p53 and (E) AKT2 protein expression revealed by immunohistochemistry staining after treatments. (Scale bar, 200 μm). (F–J) B16-F10-luc metastatic tumor-bearing mice received intravenous injections of PBS buffer (control), p53 alone, p53/bola8A complex, p53/JetPEI complex (2.0 mg/kg DNA, 1.5 mg/kg bola8A, N/P ratio of 1.0), siAKT2 alone, siAKT2/bola4A complex, or siAKT2/MC3 complex (1.0 mg/kg siRNA, 3.9 mg/kg bola4A, N/P ratio of 5.0) (n = 5): (F) in vivo bioluminescence imaging of B16-F10-luc tumor metastases in the mice. (G) Ex vivo bioluminescence imaging of B16-F10-luc tumor metastases in the lung tissue or images of excised lung tissues at the experimental end point post treatment. (H) Histological analysis of lung tissues from B16-F10-luc metastatic tumor-bearing mice at the experimental end point post treatment. The metastatic lesions (red solid outline) were identified as cell clusters with darkly stained nuclei. (Scale bars, 1.0 μm). (I) p53 and (J) AKT2 protein expression revealed by immunohistochemistry staining after treatments. (Scale bar, 200 μm). p53: plasmid DNA expressing tumor suppressor protein p53, siAKT2: siRNA targeting AKT2.
Remarkably, treatment with either p53/bola8A or siAKT2/bola4A led to a dramatic reduction, and in some cases, complete suppression of tumor metastases in the lung as revealed by the decrease in bioluminescent signals detected both in vivo and ex vivo in 4T1 and B16-F10 lung metastasis models (Fig. 6 A, B, F, and G). Quantitative analysis of the bioluminescent images confirmed significant inhibition of metastases in the treatment groups in comparison with the nontreatment group and control groups (SI Appendix, Fig. S13 A–F). Further histological analysis showed that much fewer and smaller tumor nodules were detected in the lung after treatment with either p53/bola8A or siAKT2/bola4A in both 4T1 and B16-F10 lung metastasis models (Fig. 6 C and H and SI Appendix, Fig. S13 G–I). Such potent inhibition of breast cancer and melanoma metastasis was indeed achieved by both the effective activation of p53 or the silencing of AKT2 following the successful delivery of the corresponding nucleic acid therapeutics using bola4A and bola8A, respectively (Fig. 6 D, E, I, and J). Importantly, both p53/bola8A and siAKT2/bola4A exhibited stronger or comparable inhibition of tumor metastasis compared to that allowed using Lipofectamine 2000, JetPEI, and MC3-based delivery (Fig. 6 and SI Appendix, Figs. S13 and S14). Altogether, these data demonstrate that the systemic delivery of siRNA and DNA therapeutics using the corresponding bola-amphiphilic dendrimers bola4A and bola8A significantly prevented the progression of metastatic cancer.
Further in vivo safety assessments in healthy mice demonstrated that neither dendrimer, whether alone or in complex with their corresponding nucleic acid cargo, induced any adverse effects including inflammatory responses and organ dysfunction or damage, as demonstrated by cytokine assay (SI Appendix, Fig. S15 A–C), blood biochemistry analysis (SI Appendix, Fig. S15 D–F), and organ histochemical analysis (SI Appendix, Fig. S15G). The excellent in vivo safety profiles of both bola4A and bola8A are certainly promising with regard to their potential for the in vivo delivery of nucleic acid therapeutics.
Collectively, our results demonstrate the capacity and excellent performance of the bola-amphiphilic dendrimers bola4A and bola8A to safely deliver nucleic acid therapeutics in the treatment of various aggressive and metastatic cancers, and highlight their great promise for further implementation in precision cancer treatment.
Conclusion
In this study, we studied the bola-amphiphilic dendrimers bola4A and bola8A as tailor-made vector platforms for cargo-selective delivery of DNA and siRNA therapeutics in precision cancer treatment. They outperformed the current gold-standard vectors and showed cargo-selective delivery. Specifically, the third-generation dendrimer bola8A exhibited superior performance for DNA delivery, compared with the second-generation dendrimer bola4A, whereas bola4A was more efficient for siRNA delivery than bola8A. The observed difference and selectivity in nucleic acid delivery performance of bola4A and bola8A can be ascribed to the difference both in the size of the nucleic acid cargos and the generation number of the bola-amphiphilic dendrimers.
Compared to siRNA, DNA has a longer sequence and is larger in size, thus requiring the higher generation dendrimer bola8A for delivery. The increased number and strength of the cooperative and multivalent interactions offered by bola8A enabled the efficient compaction of the large-cargo DNA into smaller nanoparticles thus favoring their cellular uptake and hence their delivery and gene transfection. Therefore, the higher generation dendrimer bola8A outperformed the lower generation dendrimer bola4A for DNA delivery. In contrast, the small siRNA cargo can be encapsulated readily and similarly by both bola4A and bola8A, resulting in siRNA delivery complexes of similar size and surface potential, which enabled comparable cellular uptake and endosome escape. However, the siRNA release was more efficient from the siRNA/bola4A complex because the smaller dendrons in bola4A provided more appropriately balanced cooperative multivalent interactions for siRNA binding and release, compared with bola8A. Consequently, bola4A was more effective than bola8A for siRNA delivery.
Remarkably, bola4A and bola8A not only exploit the delivery advantages of both lipid and polymer vectors, they also harbor nanotechnology-based tumor targeting and ROS-responsive cargo release in cancer cells. The result is excellent performance in tumor- and cancer cell-specific delivery of, respectively, siRNA and DNA in different cancer models, both in vitro and in vivo. Using AKT2 siRNA and p53 plasmid DNA as the nucleic acid therapeutics delivered by bola4A and bola8A, respectively, effective anticancer activity and therapeutic effect was achieved in both SKOV-3 ovarian cancer and HeLa cervical cancer xenograft mouse models, as well as lung metastasis models induced by the murine triple-negative 4T1 breast cancer and the mouse B16-F10 melanoma. These results provide experimental evidence in support of the implementation of tailored dendrimer vectors for tissue- and cell-specific delivery of nucleic acid therapeutics in precision cancer treatment.
It should be mentioned that the generation number of bola-amphiphilic dendrimers had a considerable impact, not only on the cargo selectivity for nucleic acid delivery, but also on the toxicity profiles. Both bola4A and bola8A had no noticeable toxicity, while the first-generation dendrimer bola2A showed considerable toxicity, likely because of its detergent-like structure and characteristics. These findings should inspire the further design and development of safe and effective vectors for nucleic acid delivery using dendritic motifs. It is to mention that high-generation dendrimers are often more toxic, more difficult to prepare, and also associated with more structural defects, compared with low-generation dendrimers. Both bola4A and bola8A are amphiphilic dendrimers of low generation and can be readily synthesized in a defect-free state, further highlighting the beneficial characteristics of these amphiphilic dendrimers for nucleic acid delivery.
Noteworthy is also the high water-solubility of both bola4A and bola8A and the ease with which these dendrimers form complexes with their corresponding nucleic acid cargo by simply mixing the dendrimer and nucleic acid solutions. This is in sharp contrast with many lipid and polymer vectors as well as conventional amphiphilic vectors, often insoluble in water and requiring organic solvents and multiple recipients in complex formulations (11) which can lead to instability and even toxicity. For example, the current clinically used LNP delivery systems are composed of four different lipid components and require stringent formulation recipes and procedures, with often laborious preparation and unstable formulations leading to complicated storage (9, 15). The single recipient formulations and good water-solubility of both bola4A and bola8A certainly represent extraordinary advantages for their easy and convenient use, encouraging their future translation into real biomedical applications.
Altogether, this study has provided insight into how the modulation of bola-amphiphilic dendrimers can affect solubility, toxicity, cargo-selectivity, and adaptivity for nucleic acid delivery. Considering that nucleic acid therapeutics encompasses various single- and double-stranded nucleic acid molecules, including ASO, aptamer, siRNA, miRNA, mRNA, and DNA, all of which have different physiochemical properties, including length, size, molecular weight, structural conformation, and flexibility, as well as distinct mechanisms of action, we foresee a bright future for amphiphilic dendrimer platforms in the cargo-specific and on-demand delivery of nucleic acid therapeutics in precision medicine.
Materials and Methods
Synthesis and Characterization of Bola4A and Bola8A.
The synthetic protocols of bola4A and bola8A are detailed in SI Appendix. bola4A: 1H NMR (500 MHz, CDCl3/CD3OD) δ 7.92 (s, 2H, CH), 7.11 (dt, J = 6.4, 3.2 Hz, 2H, ArH), 6.86 (tt, J = 8.9, 2.3 Hz, 1H, ArH), 4.98 (s, 1H, CH), 4.46 (t, J = 7.2 Hz, 4H, CH2), 3.90 (s, 4H, CH2), 3.34 (t, J = 6.1 Hz, 24H, CH2), 2.93 to 2.76 (m, 40H, CH2), 2.74 to 2.57 (m, 12H, CH2), 2.57 to 2.36 (m, 24H, CH2), 2.07 to 1.92 (m, 4H, CH2), 1.73 – 1.57 (m, 4H, CH2), 1.50 to 1.27 (m, 28H, CH2). 13C NMR (125 MHz, CDCl3/CD3OD) δ 173.79, 173.22, 164.56, 164.16, 162.11, 161.89, 145.23, 143.31, 123.66, 110.65, 110.45, 102.77, 52.16, 50.20, 49.90, 49.02, 48.57, 48.40, 48.23, 48.06, 47.89, 47.72, 47.55, 47.17, 41.66, 40.76, 37.30, 33.58, 33.32, 32.13, 30.08, 29.22, 28.88, 28.82, 28.52, 26.29. 19F NMR (471 MHz, CDCl3/CD3OD) δ −110.49. ESI-HRMS: calcd. for C95H178N32S2O12F2 [M+3H]3+ 516.6007, found 516.6005. HPLC (RT = 19.6 min). bola8A: 1H NMR (300 MHz, CDCl3/CD3OD) δ 7.83 (s, 2H, CH), 7.02 (d, J = 6.3 Hz, 2H, ArH), 6.77 (tt, J = 9.1, 2.4 Hz, 1H, ArH), 4.89 (s, 1H, CH), 4.36 (t, J = 7.1 Hz, 4H, CH2), 3.80 (s, 4H, CH2), 3.24 (t, J = 6.0 Hz, 56H, CH2), 2.90 to 2.66 (m, 88H, CH2), 2.66 to 2.48 (m, 28H, CH2), 2.48 to 2.26 (m, 56H, CH2), 1.96 to 1.83 (m, 4H, CH2), 1.61 to 1.45 (m, 4H, CH2), 1.44 to 1.13 (m, 28H, CH2). 13C NMR (125 MHz, CDCl3/CD3OD) δ 173.86, 173.37, 173.19, 161.95, 161.85, 145.89, 143.39, 123.78, 110.61, 110.41, 102.78, 102.58, 52.15, 50.05, 49.80, 49.07, 48.27, 48.10, 47.93, 47.76, 47.59, 47.42, 47.25, 47.09, 41.17, 40.53, 37.27, 33.80, 33.46, 33.34, 32.03, 30.02, 29.19, 28.84, 28.77, 28.44, 26.22. 19F NMR (471 MHz, CDCl3/CD3OD) δ −111.02. ESI-HRMS: calcd. for C175H338F2N64O28S26+, [M+6H]6+ 649.1148, found 649.1147. HPLC (RT = 15.3 min).
In Vitro Transfection of Nucleic Acid/Dendrimer Complexes.
DNA delivery.
SKOV-3, HeLa, and PC-3 cells were, respectively, seeded in 24-well plates in the presence of medium supplemented with 10% fetal bovine serum (FBS) 1 d prior to transfection. The DNA/bola–dendrimer complex solution was prepared at the N/P ratio of 1.0 in the absence of 10% FBS before transfection. Then, the cells were transfected with the DNA/bola–dendrimer complexes for 8 h at 37 °C. After incubation, the DNA/bola–dendrimer complex solution was replaced with the culture medium supplemented with 10% FBS. Then the cells were maintained under normal growth conditions for 48 h. The commercial transfection reagent Lipofectamine 2000 (Lipo2000) was used as a positive control. The protein expression of the target gene was detected as described in SI Appendix, Materials and Methods.
siRNA delivery.
One day before transfection, HeLa-GFP cells were seeded in 24-well plates in a medium containing 10% FBS; PC-3, SKOV-3, and PANC-1 cells were, respectively, seeded in 6-well plates in a medium containing 10% FBS. A solution of the siRNA/bola–dendrimer complexes was prepared at the N/P ratio of 10 in the absence of 10% FBS before transfection. Then, the cells were transfected with the siRNA/bola–dendrimer complexes for 8 h at 37 °C. After incubation, the siRNA/bola–dendrimer complex solution was replaced with the culture medium supplemented with 10% FBS. Then the cells were maintained under normal growth conditions for 72 h. The commercial transfection reagent Lipofectamine 2000 was used as a positive control. The silencing effect of the target gene was measured as described in SI Appendix, Materials and Methods.
In Vivo DNA and siRNA Delivery in Animal Models.
Animals involved in this work were maintained in China Pharmaceutical University Laboratory Animal Center. All procedures for animal experiments were approved by the Institutional Animal Care and Use Committee of China Pharmaceutical University (approval numbers of “2021-10-012” and “2022-07-006”) and performed in accordance with the guidelines and policies. Mice bearing either xenograft tumors or metastasis tumors were used in this study. For DNA delivery, mice were intravenously administrated with DNA/bola8A, using the commercial gene transfection reagent lipofectamine 2000 (Lipo) or in vivo-JetPEI as the positive control. For siRNA delivery, mice were intravenously injected with siRNA/bola4A, using Dlin-MC3-DMA (MC3), an FDA-approved LNP for siRNA delivery, as the positive control.
A full description of the Materials and Methods is provided in SI Appendix, Materials and Methods.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
This work was supported by National Key Research & Development Program of China for International S&T Cooperation Projects (2018YFE0117800) (X.L. and Y.W.), the Ligue Nationale Contre le Cancer (L.P.), the National Natural Science Foundation of China (No. 81701815) (X.L.), the State Key Laboratory of Natural Medicines at China Pharmaceutical University (No. SKLNMZZ202007) (X.L.), Bourse Eiffel (J.C.), the CAI YUAN PEI program (P.C.), China Scholarship Council (J.C., L.D., W. Lan, and P.C.), the French National Research Agency under the frame of the Era-Net EURONANOMED European Research projects “Target4Cancer,” “NANOGLIO,” “iNanoGUN,” and H2020 NMBP “SAFE-N-MEDTECH” (L.P., X.L., and Y.W.; under grant agreement No. 814607). This publication is based upon work from European Cooperation in Science and Technology (COST) Action CA 17140 “Cancer Nanomedicine from the Bench to the Bedside” supported by COST. We thank Jie Zhao (Pharmaceutical Animal Experimental Center of China Pharmaceutical University) for her kind help with in vivo experimental operation guidance.
Author contributions
X.L. and L.P. designed research; J.C., D.Z., B.L., K.S., P.C., Ying Li, W. Lin, L.D., E.L., W. Lan, and S.P. performed research; Q.L., C.J., C.Z., and J.I. contributed new reagents/analytic tools; J.C., D.Z., P.C., Ying Li, W. Lin, L.D., Y.W., E.L., W. Lan, Yun Li, A.T., S.P., X.L., and L.P. analyzed data; and J.C., D.Z., Ying Li, W. Lin, L.D., E.L., S.P., X.L., and L.P. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Contributor Information
Xiaoxuan Liu, Email: xiaoxuanliu@cpu.edu.cn.
Ling Peng, Email: ling.peng@univ-amu.fr.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Kulkarni J. A., et al. , The current landscape of nucleic acid therapeutics. Nat. Nanotechnol. 16, 630–643 (2021). [DOI] [PubMed] [Google Scholar]
- 2.Setten R. L., Rossi J. J., Han S. P., The current state and future directions of RNAi-based therapeutics. Nat. Rev. Drug Discov. 18, 421–446 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Dunbar C. E., et al. , Gene therapy comes of age. Science 359, eaan4672 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Pardi N., Hogan M. J., Porter F. W., Weissman D., mRNA vaccines–a new era in vaccinology. Nat. Rev. Drug Discov. 17, 261–279 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sahin U., Kariko K., Tureci O., mRNA-based therapeutics–developing a new class of drugs. Nat. Rev. Drug Discov. 13, 759–780 (2014). [DOI] [PubMed] [Google Scholar]
- 6.Gupta A., Andresen J. L., Manan R. S., Langer R., Nucleic acid delivery for therapeutic applications. Adv. Drug. Deliv. Rev. 178, 113834 (2021). [DOI] [PubMed] [Google Scholar]
- 7.Paunovska K., Loughrey D., Dahlman J. E., Drug delivery systems for RNA therapeutics. Nat. Rev. Genet. 23, 265–280 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roberts T. C., Langer R., Wood M. J. A., Advances in oligonucleotide drug delivery. Nat. Rev. Drug Discov. 19, 673–694 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Samaridou E., Heyes J., Lutwyche P., Lipid nanoparticles for nucleic acid delivery: Current perspectives. Adv. Drug Deliv. Rev. 154–155, 37–63 (2020). [DOI] [PubMed] [Google Scholar]
- 10.Piotrowski-Daspit A. S., Kauffman A. C., Bracaglia L. G., Saltzman W. M., Polymeric vehicles for nucleic acid delivery. Adv. Drug. Deliv. Rev. 156, 119–132 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kumar R., et al. , Polymeric delivery of therapeutic nucleic acids. Chem. Rev. 121, 11527–11652 (2021). [DOI] [PubMed] [Google Scholar]
- 12.Mullard A., Pfizer’s COVID-19 vaccine secures first full FDA approval. Nat. Rev. Drug Discov. 20, 728 (2021). [DOI] [PubMed] [Google Scholar]
- 13.Ledford H., Gene-silencing technology gets first drug approval after 20-year wait. Nature 560, 291–292 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Xu Y., Szoka F. C. Jr., Mechanism of DNA release from cationic liposome/DNA complexes used in cell transfection. Biochemistry 35, 5616–5623 (1996). [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y., Sun C., Wang C., Jankovic K. E., Dong Y., Lipids and lipid derivatives for RNA delivery. Chem. Rev. 121, 12181–12277 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boussif O., et al. , A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: Polyethylenimine. Proc. Natl. Acad. Sci. U.S.A. 92, 7297–7301 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sonawane N. D., Szoka F. C. Jr., Verkman A. S., Chloride accumulation and swelling in endosomes enhances DNA transfer by polyamine-DNA polyplexes. J. Biol. Chem. 278, 44826–44831 (2003). [DOI] [PubMed] [Google Scholar]
- 18.Dufes C., Uchegbu I. F., Schatzlein A. G., Dendrimers in gene delivery. Adv. Drug. Deliv. Rev. 57, 2177–2202 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Yang J., Zhang Q., Chang H., Cheng Y., Surface-engineered dendrimers in gene delivery. Chem. Rev. 115, 5274–5300 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Chen J., Zhu D., Liu X., Peng L., Amphiphilic dendrimer vectors for RNA delivery: State-of-the-art and future perspective. Acc. Mater. Res. 3, 484–497 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang D., et al. , The unexpected importance of the primary structure of the hydrophobic part of one-component ionizable amphiphilic Janus dendrimers in targeted mRNA delivery activity. J. Am. Chem. Soc. 144, 4746–4753 (2022). [DOI] [PubMed] [Google Scholar]
- 22.Chen J., et al. , Synthesis and use of an amphiphilic dendrimer for siRNA delivery into primary immune cells. Nat. Protoc. 16, 327–351 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang D., et al. , One-component multifunctional sequence-defined ionizable amphiphilic Janus dendrimer delivery systems for mRNA. J. Am. Chem. Soc. 143, 12315–12327 (2021). [DOI] [PubMed] [Google Scholar]
- 24.Zhang D., et al. , Targeted delivery of mRNA with one-component ionizable amphiphilic Janus dendrimers. J. Am. Chem. Soc. 143, 17975–17982 (2021). [DOI] [PubMed] [Google Scholar]
- 25.Dhumal D., et al. , An ionizable supramolecular dendrimer nanosystem for effective siRNA delivery with a favorable safety profile. Nano Res. 14, 2247–2254 (2021). [Google Scholar]
- 26.Cheng Q., et al. , Dendrimer-based lipid nanoparticles deliver therapeutic FAH mRNA to normalize liver function and extend survival in a mouse model of hepatorenal tyrosinemia type I. Adv. Mater. 30, e1805308 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Liu X., et al. , A fluorinated bola-amphiphilic dendrimer for on-demand delivery of siRNA, via specific response to reactive oxygen species. Adv. Funct. Mater. 26, 8594–8603 (2016). [Google Scholar]
- 28.Liu X., et al. , Adaptive amphiphilic dendrimer-based nanoassemblies as robust and versatile siRNA delivery systems. Angew. Chem. Int. Ed. 53, 11822–11827 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu T., et al. , An amphiphilic dendrimer for effective delivery of small interfering RNA and gene silencing in vitro and in vivo. Angew. Chem. Int. Ed. 51, 8478–8484 (2012). [DOI] [PubMed] [Google Scholar]
- 30.Percec V., et al. , Self-assembly of Janus dendrimers into uniform dendrimersomes and other complex architectures. Science 328, 1009–1014 (2010). [DOI] [PubMed] [Google Scholar]
- 31.Garofalo S., et al. , Natural killer cells modulate motor neuron-immune cell cross talk in models of amyotrophic lateral sclerosis. Nat. Commun. 11, 1773 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dong Y., et al. , A dual targeting dendrimer-mediated siRNA delivery system for effective gene silencing in cancer therapy. J. Am. Chem. Soc. 140, 16264–16274 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Chahal J. S., et al. , Dendrimer-RNA nanoparticles generate protective immunity against lethal Ebola, H1N1 influenza, and Toxoplasma gondii challenges with a single dose. Proc. Natl. Acad. Sci. U.S.A. 113, E4133–E4142 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen C., et al. , Mastering dendrimer self-assembly for efficient siRNA delivery: From conceptual design to in vivo efficient gene silencing. Small 12, 3667–3676 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zeng H., Johnson M. E., Oldenhuis N. J., Tiambeng T. N., Guan Z., Structure-based design of dendritic peptide bolaamphiphiles for siRNA delivery. ACS Cent. Sci. 1, 303–312 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fuhrhop J. H., Wang T., Bolaamphiphiles. Chem. Rev. 104, 2901–2937 (2004). [DOI] [PubMed] [Google Scholar]
- 37.Fariya M., Jain A., Dhawan V., Shah S., Nagarsenker M. S., Bolaamphiphiles: A pharmaceutical review. Adv. Pharm. Bull. 4, 483–491 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Levine A. J., p53: 800 million years of evolution and 40 years of discovery. Nat. Rev. Cancer 20, 471–480 (2020). [DOI] [PubMed] [Google Scholar]
- 39.von Karstedt S., Montinaro A., Walczak H., Exploring the TRAILs less travelled: TRAIL in cancer biology and therapy. Nat. Rev. Cancer 17, 352–366 (2017). [DOI] [PubMed] [Google Scholar]
- 40.Manning B. D., Toker A., AKT/PKB signaling: Navigating the network. Cell 169, 381–405 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu J., et al. , Heat shock proteins and cancer. Trends Pharmacol. Sci. 38, 226–256 (2017). [DOI] [PubMed] [Google Scholar]
- 42.Rocchi P., et al. , Heat shock protein 27 increases after androgen ablation and plays a cytoprotective role in hormone-refractory prostate cancer. Cancer Res. 64, 6595–6602 (2004). [DOI] [PubMed] [Google Scholar]
- 43.Altieri D. C., Survivin, cancer networks and pathway-directed drug discovery. Nat. Rev. Cancer 8, 61–70 (2008). [DOI] [PubMed] [Google Scholar]
- 44.Massova I., Kollman P. A., Combined molecular mechanical and continuum solvent approach (MM-PBSA/GBSA) to predict ligand binding. Perspect. Drug Discov. Des. 18, 113–135 (2000). [Google Scholar]
- 45.Tsui V., Case D. A., Theory and applications of the generalized Born solvation model in macromolecular Simulations. Biopolymers 56, 275–291 (2001). [DOI] [PubMed] [Google Scholar]
- 46.Cheng X., Lee R. J., The role of helper lipids in lipid nanoparticles (LNPs) designed for oligonucleotide delivery. Adv. Drug. Deliv. Rev. 99, 129–137 (2016). [DOI] [PubMed] [Google Scholar]
- 47.Hattori Y., Suzuki S., Kawakami S., Yamashita F., Hashida M., The role of dioleoylphosphatidylethanolamine (DOPE) in targeted gene delivery with mannosylated cationic liposomes via intravenous route. J. Control. Release 108, 484–495 (2005). [DOI] [PubMed] [Google Scholar]
- 48.Xu E., Saltzman W. M., Piotrowski-Daspit A. S., Escaping the endosome: Assessing cellular trafficking mechanisms of non-viral vehicles. J. Control. Release 335, 465–480 (2021). [DOI] [PubMed] [Google Scholar]
- 49.Fang J., Nakamura H., Maeda H., The EPR effect: Unique features of tumor blood vessels for drug delivery, factors involved, and limitations and augmentation of the effect. Adv. Drug. Deliv. Rev. 63, 136–151 (2011). [DOI] [PubMed] [Google Scholar]
- 50.Huang J., Current developments of targeting the p53 signaling pathway for cancer treatment. Pharmacol. Ther. 220, 107720 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang W. W., et al. , The first approved gene therapy product for cancer ad-p53 (Gendicine): 12 years in the clinic. Hum. Gene. Ther. 29, 160–179 (2018). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
All study data are included in the article and/or SI Appendix.