Significance
Gastric cancer (GC) is one of the leading causes of cancer-related deaths with high degree of intratumoral heterogeneity. Herein, we propose a treatment strategy for chemoresistant GC patients via deciphering metabolic plasticity of Stem-like/Epithelial-to-mesenchymal transition/Mesenchymal-type GC cells. This study provides detailed molecular mechanisms behind the metabolic plasticity of stemness-high GC cells, from chromatin architecture to transcriptional drivers ATF4/CEBPB, and the importance of reactive oxygen species scavenging activity mediated by mitochondrial 1C metabolism. Further understanding of 1C metabolism may lead to development of therapeutic strategies for chemoresistant GC patients, as verified in the patient-derived cancer organoids.
Keywords: gastric cancer, metabolic plasticity, glutaminase, 3 phosphoglycerate dehydrogenase
Abstract
Molecular classification of gastric cancer (GC) identified a subgroup of patients showing chemoresistance and poor prognosis, termed SEM (Stem-like/Epithelial-to-mesenchymal transition/Mesenchymal) type in this study. Here, we show that SEM-type GC exhibits a distinct metabolic profile characterized by high glutaminase (GLS) levels. Unexpectedly, SEM-type GC cells are resistant to glutaminolysis inhibition. We show that under glutamine starvation, SEM-type GC cells up-regulate the 3 phosphoglycerate dehydrogenase (PHGDH)-mediated mitochondrial folate cycle pathway to produce NADPH as a reactive oxygen species scavenger for survival. This metabolic plasticity is associated with globally open chromatin structure in SEM-type GC cells, with ATF4/CEBPB identified as transcriptional drivers of the PHGDH-driven salvage pathway. Single-nucleus transcriptome analysis of patient-derived SEM-type GC organoids revealed intratumoral heterogeneity, with stemness-high subpopulations displaying high GLS expression, a resistance to GLS inhibition, and ATF4/CEBPB activation. Notably, coinhibition of GLS and PHGDH successfully eliminated stemness-high cancer cells. Together, these results provide insight into the metabolic plasticity of aggressive GC cells and suggest a treatment strategy for chemoresistant GC patients.
Gastric cancer (GC) is a highly heterogeneous adenocarcinoma. The Cancer Genome Atlas (TCGA) project and Asian Cancer Research Group (ACRG) have defined clinically relevant molecular subtypes of GC (1, 2). The genetically stable subtype from the TCGA project and epithelial-to-mesenchymal transition (EMT) subtype from the ACRG study, although not identical, have a high percentage of diffuse histological types, and present a worse prognosis than intestinal histological types (3). We have subtyped GC patients in the Yonsei cohort into five groups—gastric, inflammatory, intestinal, mixed, and stem-like subtypes—with the stem-like subtype displaying the most aggressive phenotype and resistance to chemotherapy (4). However, there are currently no targeted treatments available for this aggressive subtype, which we refer to as SEM (stem-like/ EMT/ mesenchymal) in this study. We believe a thorough understanding of the molecular characteristics in aggressive SEM-type GC is warranted.
Glutamine is a ready source of carbon and nitrogen for numerous biosynthetic pathways (5). Glutamine first undergoes glutaminolysis to become glutamate via the action of glutaminase (GLS) (6). While GLS2 (liver-type) is expressed primarily in the liver, brain, and pancreas, GLS (kidney-type) is broadly expressed in various tissues including cancers (6). With the known importance of GLS in tumorigenesis and cancer cell proliferation (7), GLS has been widely investigated as a therapeutic target for cancer cells, particularly those in a high mesenchymal state (8). Indeed, selective inhibitors of GLS are currently being tested in clinical trials in refractory solid tumors (9, 10). As we previously reported, SEM-type GC is chemoresistant while maintaining a high mesenchymal state (4), raising the potential for GLS to be an effective therapeutic target.
However, cancer cells can adapt their metabolism to survive through activating compensatory metabolic pathways. Furthermore, intratumoral metabolic heterogeneity leads to the cell-to-cell difference in sensitivity to the anticancer drugs within a tumor, resulting in incomplete treatment response and acquired therapeutic resistance (11). Particularly, cancer stem cells often exhibit chemoresistance due to their quiescent state and slow proliferation rate. As cancer stem cells are known to be critical in the formation and maintenance of liver metastases in aggressive GC (12), failure in the elimination of cancer stem cells can result in reemergence of the disease. Therefore, there is an urgent, unmet need for a therapeutic regimen that enables simultaneous elimination of each cancer cell subpopulation including cancer stem cells, which display a high degree of plasticity (13).
Here, we demonstrate that while SEM-type GC is GLS-positive, it exhibits resistance to GLS inhibition. SEM-type GC cells have a globally open chromatin structure that is accessible to binding with ATF4 and CEBPB, contributing to the activation of genes involved in the PHGDH-driven salvage pathway that controls mitochondrial reactive oxygen species (ROS) in response to GLS inhibition. Single-nucleus transcriptome analysis revealed that GLS-positive clusters in organoids derived from SEM-type GC patients express high levels of cancer stem cell marker CD44 (14) and WNT signaling (12). The combinatorial inhibition of GLS and PHGDH reduces the survival of stem-like GC. Thus, we demonstrate the effectiveness of combination therapy targeting chemoresistant stemness-high GC.
Results
SEM-Type GC Has Distinct Metabolic Features Compared to Intestinal-Type GC.
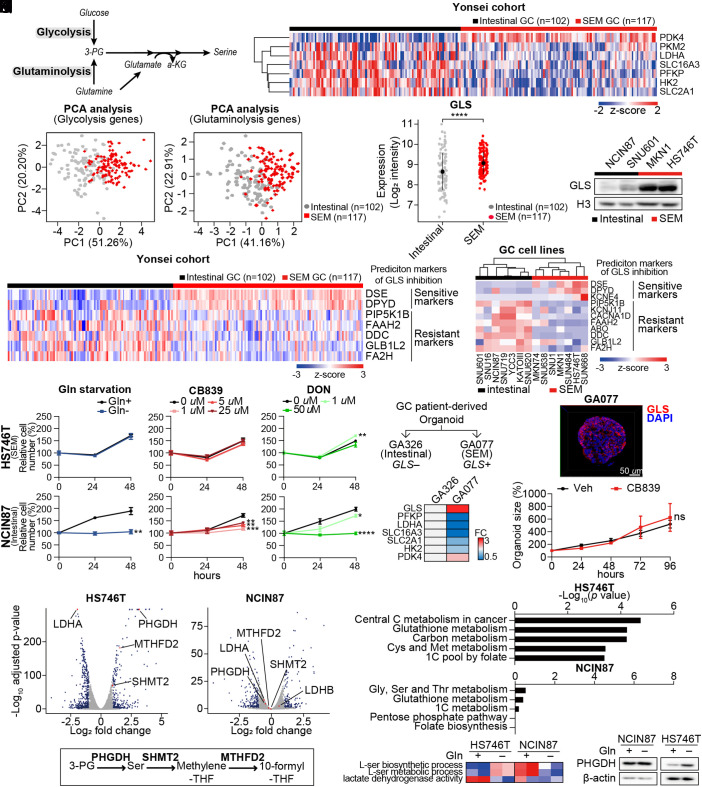
Glycolysis and glutaminolysis provide 3-phosphoglycerate (3-PG) for serine biosynthesis in various cancer cells (Fig. 1A) (15). In the Yonsei cohort, compared with the intestinal subtype GC tumors that maintain high epithelial integrity (4), we observed that SEM-type GC tumors which have up-regulated EMT gene signatures and the worst prognoses (4) exhibited downregulation of glycolysis-related genes including pyruvate kinase M2 (16), lactate dehydrogenase A (LDHA), solute carrier family 16 member 3 (SLC16A3), phosphofructokinase-platelet (PFKP) (17), hexokinase 2 (HK2), and solute carrier family two member one (SLC2A1) (Fig. 1B). In contrast, expression of the pyruvate kinase 4 (PDK4) gene in SEM-type GC cells was largely up-regulated, preventing pyruvate from entering the TCA cycle, implying that SEM-type GC cells are less glycolytic and that the metabolized glucose does not undergo mitochondrial oxidation (Fig. 1B). Principal component analysis (PCA) with seven glycolysis genes consistently highlighted subtype-specific differences in the glycolytic pathway between intestinal and SEM-type GC tumors (Fig. 1C). PCA with glutaminolysis-related genes also exhibited distinct clustering between SEM-type GC and intestinal subtype GC tumors (Fig. 1D and SI Appendix, Fig. S1A). Indeed, although genes related to glutaminolysis and regulation of protein metabolic process are mostly down-regulated in SEM-type GC patients (SI Appendix, Fig. S1 B and C), the gene expression of GLS, which is responsible for the first step of glutaminolysis, was up-regulated in SEM-type GC tumors (Fig. 1E). Consistent with this, GLS protein levels were also elevated in the SEM-type GC cell lines (Fig. 1F). Previously, a pan-cancer metabolic signature was reported to predict responsiveness to GLS inhibition (8). Based on the pan-cancer metabolic signature, SEM-type GC tumors and cells exhibited up-regulated gene expression of GLS inhibition-sensitive markers, implying that SEM-type GC would have high GLS dependency (Fig. 1G). Likewise, SEM-type GC cell lines were also predicted to be codependent on GLS and glutathione synthesis (Fig. 1H and SI Appendix, Fig. S1 D and E).
Fig. 1.
GLS-high SEM-type GC cells are resistant to GLS inhibition via upregulation of 1C metabolic pathway. (A) Schematic of glycolysis and glutaminolysis. (B) Analyses of transcriptomic profiles of tumors of GC patients in the Yonsei cohort. The expression levels of glycolysis-related genes were compared according to the subtypes: SEM (n = 117) and intestinal (n = 102). (C) PCA of GC patients in the Yonsei cohort with glycolysis-related genes introduced in B. (D) PCA of GC patients in the Yonsei cohort with glutaminolysis-related genes introduced in 1B. (E) Expression levels of GLS compared between SEM-type (n = 117) and intestinal subtype GC patients (n = 102) in the Yonsei cohort. (F) Immunoblotting of GLS in gastric cell lines. Histone H3 was used as the loading control. (G and H) Heatmap displaying the expression levels of sensitive markers (DSE, DPYD, and KCNE4) and resistance markers (PIP5K1B, KCNJ11, CACNA1D, FAAH2, ABO, DDC, GLB1L2, and FA2H) for GLS inhibition in the Yonsei cohort and GC cell lines (SRP078289). (I and J) Proliferation assays were performed for 48 h in HS746T and NCIN87 cells under glutamine-deficient and drug-treated conditions. CB839, GLS inhibitor; DON, glutamine antagonist. (K and L) Diagram describes selection of tumors for generation of patient-derived cancer organoids. Heatmap shows fold change in GLS and glycolysis-associated genes. (M) Immunocytochemistry image of GLS in GA077 stained with DAPI. (Scale bar: 50 μm.) (N) Sizes of patient-derived cancer organoid GA077 with or without 5 μM CB839 were measured. Organoid size measured by averaging short and long diameters (Right). n ≥ 5 (O) Transcriptomic profiles of glutamine starvation in HS746T and NCIN87 were analyzed via RNA sequencing analysis. Genes with fold change greater than 1.5 or less than −1.5 with adjusted P-values smaller than 0.05 are marked in blue. PHGDH, MTHFD2, SHMT2, and LDHA genes are marked in red. (P) Schematic of 1C metabolic pathway. (Q) KEGG analysis of differentially expressed genes. The P-values are less than 0.05. (R) GSVA was performed with pathways from MsigDB v7.4. Gene set enrichment score represented as a heatmap. (S) Immunoblot showing the protein levels of PHGDH upon glutamine starvation in HS746T and NCIN87. β-actin was used as a loading control. ns, no significance, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, Wilcox test for 1E and two-tailed Student’s t test for I, J, and N.
GLS-High SEM-Type GC Cells Are Resistant to GLS Inhibition or Glutamine Starvation via Upregulation of 1C Metabolic Pathway.
Treatment with the GLS inhibitor, CB839, blocked glutaminolysis as shown by the decrease in glutamate levels and increase in glutamine levels measured via liquid chromatography-mass spectrometry analysis (SI Appendix, Fig. S2A) and induced profound changes in the transcriptomic profile including upregulation of pathways related to purine metabolism (SI Appendix, Fig. S2 B and C). Surprisingly, treatment with GLS inhibitors (CB839 and BPTES), glutamine antagonist (DON), or glutamine starvation did not affect the viability of the SEM-type GC cell line, HS746T (Fig. 1I and SI Appendix, Fig. S2D); in contrast, the intestinal-type GC cell line, NCIN87 exhibited a greater decrease in cell number (Fig. 1J and SI Appendix, Fig. S2D).
To test the effect of GLS inhibition in a more physiological setting, we generated patient-derived cancer organoids. GA077 organoids were derived from a SEM-type GC patient, and GA326 organoids were derived from an intestinal subtype GC patient (Fig. 1K). In accordance with the transcriptomic profiles of patients and cell lines, GA077 organoids exhibited elevated mRNA levels of GLS and PDK4, whereas glycolysis-related gene expression (PFKP, LDHA, SLC16A3, SLC2A1, and HK2) was down-regulated compared with GA326 (Fig. 1L). Although immunocytochemical analyses confirmed abundant protein levels of GLS (Fig. 1M), the GA077 organoids were not affected by CB839 treatment in terms of proliferation (Fig. 1N).
To identify the underlying mechanisms promoting the resistance to GLS inhibition or glutamine starvation, we performed RNA sequencing analysis on HS746T and NCIN87 cells with or without glutamine. Interestingly, there was a significant upregulation of PHGDH expression only in SEM-type GC cell lines under glutamine starvation (Fig. 1O and SI Appendix, Fig. S2 E and F). Moreover, the most down-regulated gene with the highest P-value was LDHA, indicating that glutamine starvation redirects glucose flux from glycolysis to serine synthesis and subsequent folate cycle-related pathways (Fig. 1P). Pathway analysis also revealed a change in the 1C pool by folate and methionine metabolism and elevation of the serine metabolism pathway in a HS746T-specific manner (Fig. 1 Q and R) (18). The elevated protein levels of PHGDH were detectable only in HS746T cells upon glutamine starvation (Fig. 1S).
The folate cycle using THF as a carrier of 1C unit is coupled to the methionine cycle using SAM as a carrier. (19) With serine transferring its 1C unit to THF by being converted into glycine via the action of serine hydroxymethyltransferase (SHMT), the folate cycle participates in the oligonucleotide synthesis pathway (SI Appendix, Fig. S3A). Consistently, metabolite analysis of pan-cancer cell lines from Cancer Cell Line Encyclopedia showed a decrease in adenosine monophosphate and guanosine monophosphate (GMP) in SEM-type GC cell lines (SI Appendix, Fig. S3B). Moreover, the level of methionine was higher in SEM-type GC cell lines while s-adenosyl homocysteine, a product of methionine after 1C unit is transferred out, was less abundant (SI Appendix, Fig. S3B). In accordance with the findings in the metabolome, SEM-type GC cells showed a lower level of SHMT2, the major mitochondrial isoform of SHMT, along with higher 5-methyl-THF-homocysteine methyltransferase (MTR) levels (SI Appendix, Fig. S3C). MTR is an enzyme which transfers 1C unit to homocysteine, resulting in the production of methionine. Thus, SEM-type GC cells are mainly replenishing the 1C unit in the methionine cycle, rather than the folate cycle (SI Appendix, Fig. S3D). As a result, the level of SAM was highly elevated in SEM-type GC cells measured via SAM assay (SI Appendix, Fig. S3E). As in the cell lines, GC patients of EMT type in the ACRG cohort had up-regulated MTR and down-regulated SHMT2 (SI Appendix, Fig. S3F). Also, MTR and SHMT2 were inversely correlated in TCGA STAD (SI Appendix, Fig. S3G). In the Yonsei cohort, MTR was distinctively elevated in stem-like subtype patients compared to the downregulation of folate cycle-related genes (SHMT1, SHMT2, MTHFD1, MTHFD1L, and MTHFD2) (SI Appendix, Fig. S3H). These results imply that glutamine starvation activates mitochondrial 1C metabolic pathway in SEM-type GC cells, which put emphasis on methionine cycle, instead of folate cycle, at endogenous state.
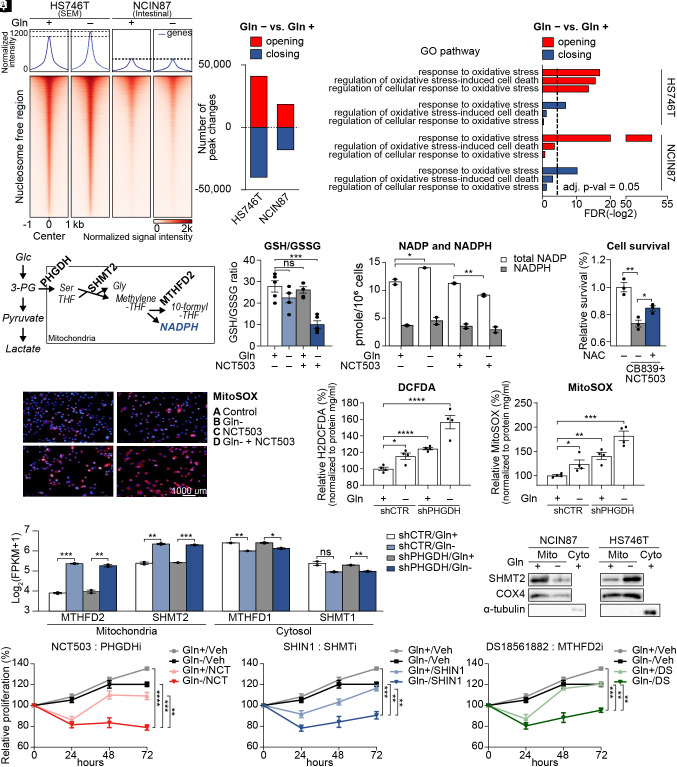
PHGDH-Driven Salvage Pathway Inhibits Accumulation of Mitochondrial ROS.
Stem cells maintain globally open chromatin to facilitate accessibility of target motifs by transcription factors (20–22). To determine SEM-type-specific responses and metabolic robustness during glutamine starvation, we conducted an assay for transposase-accessible chromatin using sequencing (ATAC-seq). Differences in chromatin accessibility between the cell lines were apparent in nucleosome-free regions. However, only HS746T exhibited a net increase in accessibility upon glutamine starvation (Fig. 2A). In addition, the number of opening and closing peaks upon glutamine starvation was greater in HS746T than in NCIN87 cells (Fig. 2B). Regions with increased chromatin accessibility following glutamine starvation were enriched in pathways related to the oxidative stress responses in both cell lines (Fig. 2C). However, pathways that protect against oxidative stress, such as regulation of oxidative stress-induced cell death and regulation of cellular response to oxidative stress, were only enriched in the newly opened regions of HS746T cells (Fig. 2C).
Fig. 2.
PHGDH-driven salvage pathway inhibits mitochondrial ROS production. (A) Chromatin accessibility heatmap of HS746T and NCIN87 with or without glutamine. Color intensity represents chromatin accessibility. (B) Numbers of peak changes after glutamine starvation were compared (adjusted P < 0.05). (C) Pathway analysis of the most variable peaks introduced in B associated with glutamine starvation. Dotted line indicates adjusted P-value = 0.05. (D) Diagram explaining the mitochondrial folate cycle using PHGDH, SHMT2, and MTHFD2 as a mediating enzymes. (E) GSH/GSSG ratio was measured with glutamine starvation and NCT503 treatment. (F) Total intracellular NADPH and NADP levels in glutamine starvation or NCT503-treated HS746T, normalized to cell number. (G) Cell survival with CB839 and NCT503 treatments in combination with N-acetyl-L-cysteine(NAC) (H) Mitochondrial superoxide visualized with MitoSOX with or without glutamine starvation and NCT503 treatment in HS746T cells. (I) Cellular ROS was measured via fluorescence assay using H2DCFDA in HS746T stable cell lines with or without glutamine. (J) Mitochondrial ROS was measured via fluorescence assay using MitoSOX in HS746T stable cell lines with or without glutamine. (K) RNA sequencing was performed on shControl or shPHGDH-stable HS746T cells with or without glutamine. Expression levels of mitochondrial folate cycle-related genes (MTHFD2 and SHMT2) and cytosolic folate cycle-related genes (MTHFD1and SHMT1) with or without glutamine were marked as log2(FPKM+1). (L) Immunoblot showing the mitochondrial protein level of SHMT2 upon glutamine starvation in HS746T and NCIN87 cells after mitochondrial isolation. (M–O) Proliferation assays for 72 h in HS746T cells. NCT503, PHGDH inhibitor; SHIN1, SHMT1 and SHMT2 inhibitor; DS18561882, MTHFD2 inhibitor. ns, no significance, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, Benjamini–Hochberg adjusted P-value for 2K and two-tailed Student’s t test for E–G, I, J, and M–O.
Metabolic stress, including glutamine starvation, produces ROS (23), and consecutive activation of PHGDH, SHMT2, and MTHFD2 leads to the production of NADPH to minimize ROS-induced cellular stress (Fig. 2D). NADPH is an important reducing agent that eliminates cellular ROS and maintains redox homeostasis (24). As PHGDH was the most up-regulated gene with the lowest adjusted P-value (Fig. 1O), we investigated the combinatorial effect of glutamine starvation and PHGDH inhibitor NCT503 in SEM-type GC cells (25). Indeed, the ratio of reduced glutathione (GSH) to oxidized glutathione (GSSG) decreased when HS746T cells were cultured without glutamine along with the pharmaceutical inhibition of PHGDH (Fig. 2E). The amount of NADP, the oxidized form of NADPH, was also lowest in PHGDH-inhibited cells with glutamine deficiency (Fig. 2F). Indeed, cell death induced by cotreatment with CB839 and NCT503 was partially rescued by the addition of the antioxidant N-acetylcysteine (NAC) (Fig. 2G). Mitochondrial ROS estimated by MitoSOX was also the highest with NCT503 treatment and glutamine starvation (Fig. 2H).
Next, we generated an HS746T shPHGDH stable cell line to stably knockdown gene expression of PHGDH and measured oxidative stress in the absence of glutamine. Both oxidative stress and mitochondrial ROS were the highest in glutamine-deprived HS746T shPHGDH cells (Fig. 2 I and J). To examine the changes in the transcriptomic landscape including ROS-related genes, we performed RNA sequencing analysis on the shPHGDH stable cells (SI Appendix, Fig. S4 A and B). Interestingly, the mitochondrial forms of SHMT and MTHFD were significantly up-regulated (Fig. 2K). Given that folate metabolism is crucial for the cellular NADPH supply (26), only the mitochondrial isoforms of folate cycle-related genes were up-regulated (SI Appendix, Fig. S4 C and D), highlighting the importance of scavenging mitochondrial ROS upon glutamine starvation. The upregulation of SHMT2 was apparent in HS746T upon glutamine starvation, whereas NCIN87 cells exhibited a reduction in SHMT2 protein levels (Fig. 2L and SI Appendix, Fig. S4E). We next tested the effect of simultaneous inhibition of the salvage pathway along with glutamine starvation. We treated glutamine-starved HS746T cells with PHGDH inhibitor (NCT503), SHMT1 and two inhibitor (SHIN1), and MTHFD2 inhibitor (DS18561882). Pharmacological inhibition of the salvage pathway successfully inhibited cell proliferation in combination with glutamine starvation in chemoresistant SEM-type GC cells (Fig. 2 M–O), implying that the salvage pathway is crucial for the survival of chemoresistant SEM-type GC cells undergoing glutamine starvation.
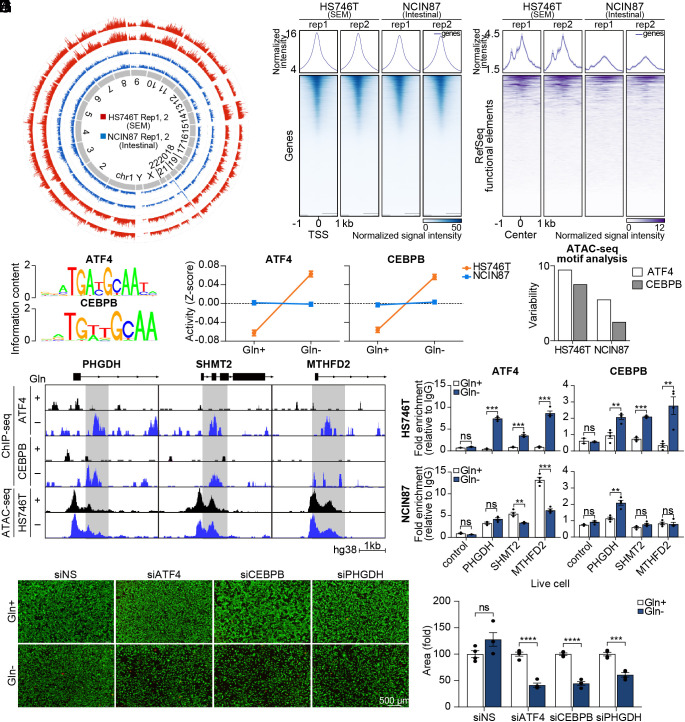
The Landscape of Chromatin Accessibility Shows ATF4 and CEBPB as Transcriptional Drivers of the PHGDH-Driven Salvage Pathway.
As seen in stem cells, which have a globally open chromatin (20–22), the chromatin of HS746T was less condensed than that of NCIN87 (Fig. 3A). Although the difference in accessibility was minimal in promoter regions, HS746T chromatin exhibited greater accessibility in the functional element regions (Fig. 3 B and C). To explore the mechanisms of activation of the PHGDH-driven salvage pathway, we integrated RNA-seq and ATAC-seq data of HS746T with or without glutamine. Strikingly, only 0.9% of differentially expressed genes in the RNA-seq were found to have differentially accessible peaks at the corresponding promoter regions (SI Appendix, Fig. S5 A and B), which indicates that upregulation of PHGDH-mediated mitochondrial folate cycle-related genes is not due to the increased accessibility of the promoters. Motif analysis was performed with RNA-seq data and ATAC-seq data. We first calculated the activity score of DNA motifs using the Integrated System for Motif Activity Response Analysis (ISMARA) algorithm to predict regulatory motifs and relevant target genes (27). Interestingly, the activity of ATF4 and CEBPB was specifically increased in HS746T upon glutamine starvation (Fig. 3 D and E). In addition, ATAC-seq motif analysis via chromVAR (28) showed a higher degree of predicted variability of ATF4 and CEBPB motifs in HS746T (Fig. 3F). While non-SEM GC cell lines showed various responses to glutamine starvation in terms of ATF4/CEBPB activation and endoplasmic reticulum (ER) stress signaling, SEM-type GC cell lines consistently showed upregulation of ATF4/CEBPB with active ER stress signaling pathway (Fig. 3G and SI Appendix, Fig. S5C). To identify whether ATF4 and CEBPB bind directly to the promoter region of stress-response genes, we performed chromatin immunoprecipitation–sequencing (ChIP-seq) in HS746T. As expected, the recruitment of ATF4 and CEBPB to the promoter regions of PHGDH, SHMT2, and MTHFD2 was significantly increased upon glutamine starvation (Fig. 3G). Concerning that the accessibility of the promoter regions of PHGDH, SHMT2, and MTHFD2 is not increased upon glutamine starvation in ATAC-seq data (Fig. 3G), ATF4 and CEBPB are the major contributing factors in the increase in the expression of PHGDH, SHMT2, and MTHFD2. These findings were confirmed with conventional ChIP-qPCR, while NCIN87 did not show increased binding of ATF4 and CEBPB to the promoter regions of PHGDH, SHMT2, and MTHFD2 upon glutamine starvation (Fig. 3H). Knockdown of ATF4 and CEBPB with siRNA successfully inhibited mRNA expression of PHGDH, SHMT2, and MTHFD2 (SI Appendix, Fig. S5 D and E). Also, ATF4, CEBPB, and PHGDH knockdown decreased cell viability with metabolic stress, such as glutamine starvation, implying that PHGDH induced by ATF4/CEBPB is crucial for cell viability (Fig. 3 I and J).
Fig. 3.
The landscape of chromatin accessibility shows ATF4 and CEBPB as transcriptional drivers of PHGDH-driven salvage pathway. (A) Circular plot showing genome-wide chromatin accessibility. (B) Heatmap of ATAC-Seq peaks in promoter regions. Color intensity represents chromatin accessibility. The peaks are aligned with the transcription start site (TSS) as the center. (C) Heatmap of ATAC-Seq peaks in RefSeq functional element regions. (D) Oligonucleotide sequence of ATF4- and CEBPB-binding site. (E) ISMARA with RNA sequencing data was performed. Motif activity is shown as a z-score. (F) Variability of motif activity upon glutamine starvation was computed with chromVAR. (G) ATF4 and CEBPB ChIP sequencing analysis were performed. The genome browser shows ChIP-seq profiles within the PHGDH, SHMT2, and MTHFD2 loci. (H) ChIP assay of the MTHFD2, PHGDH, and SHMT2 promoters using ATF4 and CEBPB antibodies in glutamine-deprived HS746T and NCIN87 cells. Data are represented as fold-change. (n = 4) (I) Microscopy images of HS746T cells with or without glutamine along with ATF4, CEBPB, or PHGDH knockdown. Live cells are shown with green color and dead cells are shown with red color. (Scale bar; 500 µm.) (J) The number of live cells was counted. (n = 4) ns, no significance, **P < 0.01, ***P < 0.001, ****P < 0.0001, two-tailed Student’s t test for H and J.
Single-Nucleus Transcriptome Analysis Reveals Intratumoral Heterogeneity along with High ATF4/ CEBPB Regulon Activity in Cell Subpopulations with High EMT Signature.
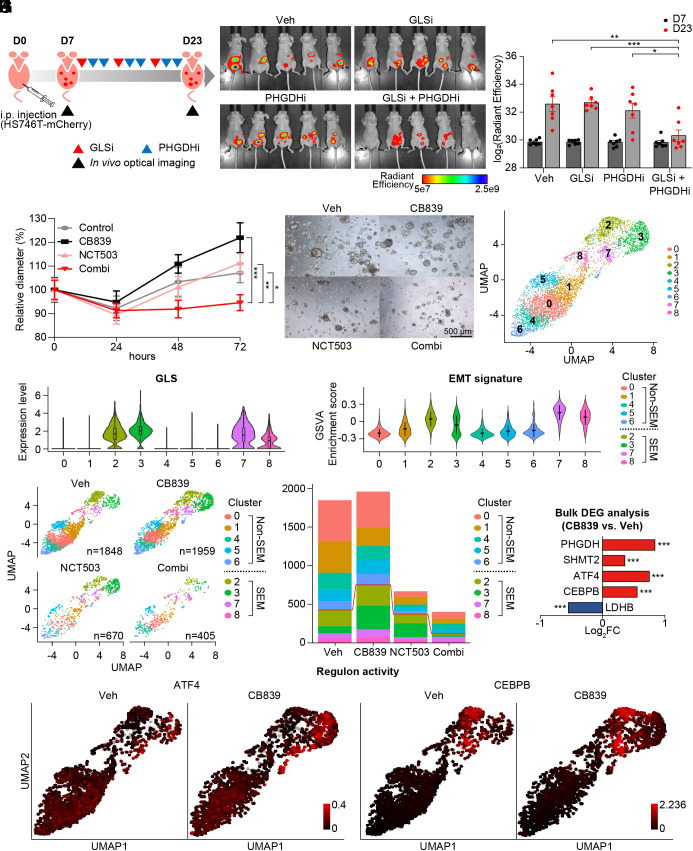
Next, the efficacy of combinational therapy was evaluated in a more physiological setting. As metastasis is the leading cause of the mortality of GC patients and SEM-type GC cells are more inclined to invade surrounding tissue and metastasize, we generated the GC peritoneal dissemination model (29) which reflects the nature of SEM-type GC via intraperitoneal injection of HS746T stable cell line with mCherry expression. Peritoneum is a common site of GC metastasis and the GC peritoneal dissemination mouse model successfully mimics metastatic and dissemination tendencies of SEM-type GC (29). This peritoneal dissemination mouse model was employed to evaluate the actions of GLS inhibitor (30) and PHGDH inhibitor (25). In accordance with the cytotoxic effect of GLS/PHGDH inhibition in GC cell lines, combinational therapy successfully inhibited tumor growth in the mouse model without a change in mean body weight (Fig. 4 A–C and SI Appendix, Fig. S6A). Also, in SEM-type patient-derived GC organoids, coinhibition of GLS and PHGDH resulted in a statistically significant decrease in proliferation compared to single treatments in SEM-type GC organoids (Fig. 4 D and E and SI Appendix, Fig. S6B).
Fig. 4.
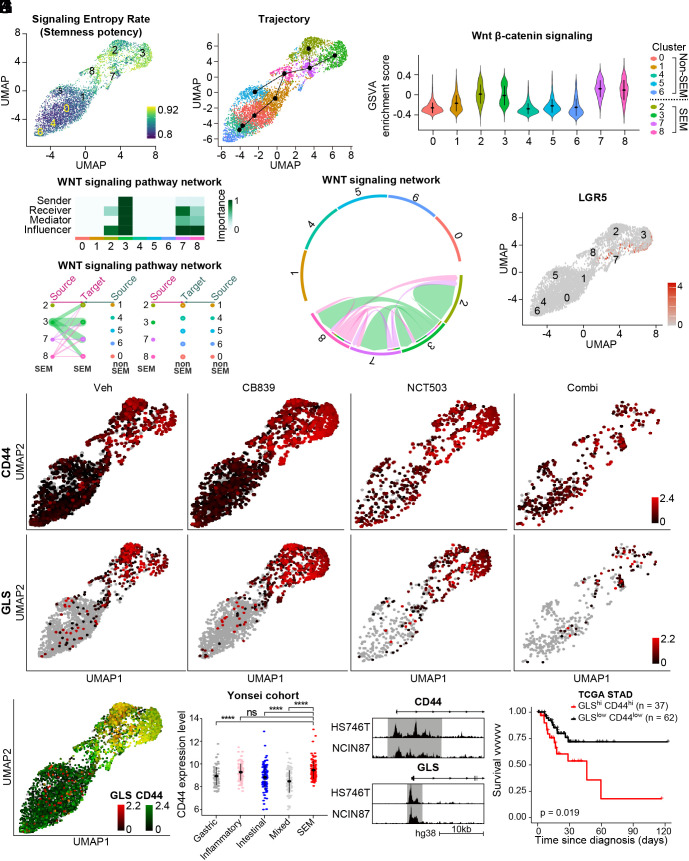
GLS inhibition activates ATF4/CEBPB-mediated transcriptional network in EMT signature-enriched clusters in patient-derived cancer organoids. (A) Diagram to explain the process in vivo experiment with vehicle, BPTES (12.5 mg/kg), NCT503 (40 mg/kg), or combination of BPTES and NCT503. (B) Representative picture of mice (n = 5) with tumor volume measured via in vivo optical imaging system. Total radiant efficiency (p/sec/cm2/sr/μW/cm2) was measured in peritoneal area. (C) Total radiant efficiency was compared in every group (n =7) before (D7) and after (D23) three cycles of BPTES/NCT503 injection. (D) Patient-derived cancer organoids GA077 were treated with 5 μM CB839, 50 μM NCT503, or both. The size was measured as the average of the short and long diameters (n ≥ 18). (E) Representative microscopy images of the organoid GA077 after different drug treatments. (F) Single-nucleus RNA sequencing was performed on GA077 with vehicle, CB839 (5 μM), NCT503 (50 μM) or both. Uniform manifold approximation projection (UMAP) visualization of cells is shown after integration. (G) The expression level of GLS is shown as a violin plot in the identified cell clusters. (H) GSVA was performed to calculate enrichment score with GO pathway “HALLMARK_ EPITHELIAL_MESENCHYMAL_TRANSITION” from MSigDB v7.4. (I) UMAP visualization of cells under different conditions: vehicle, CB839 (5 μM), NCT503 (50 μM) or both. (J) Fraction of cells from each sample for each cluster. (K) Gene expression level was compared as a bulk tumor. (L) Regulon activity (ATF4 and CEBPB) is marked as a color on the corresponding UMAP. *P < 0.05, **P < 0.01, ***P < 0.001, two-tailed Student’s t test for C and D and Benjamini–Hochberg adjusted P-value for K.
Next, we performed single-nucleus RNA sequencing analysis on GC organoids with different drug treatments. Single-nucleus sequencing of the four conditions resulted in transcriptomic information for a total of 4,882 cells (Fig. 4F). Clustering revealed nine cell subtypes that were conserved across the different drug treatments (Fig. 4F). Among the nine clusters, clusters 2, 3, 7, and 8 showed high expression of GLS as seen in SEM-type tumors and cell lines (Fig. 4G). Intratumoral heterogeneity in terms of GLS expression level was detected also in other SEM-type GC organoids (SI Appendix, Fig. S7A). Consistently, clusters 2, 3, 7, and 8 exhibited high EMT signatures and low ROS pathway enrichment scores (Fig. 4H and SI Appendix, Fig. S7B).
We next investigated the drug response of each cluster (Fig. 4I). In terms of cell number, GLS inhibition did not affect the population of each cluster (Fig. 4J). Analysis of patient-derived cancer organoids as a bulk tumor confirmed the upregulation of PHGDH, SHMT2, ATF4, and CEBPB with GLS inhibition (Fig. 4K). As GLS inhibition induces activation of the salvage pathway led by ATF4 and CEBPB, CB839 treatment up-regulated the expression levels of ATF4, and CEBPB, specifically in EMT-type clusters (SI Appendix, Fig. S7C). Reconstruction of the gene regulatory network using SCENIC determined that the activity of the ATF4 and CEBPB regulon, which denotes a gene regulatory network with significant motif enrichment, increased upon GLS inhibition in only EMT-type cell clusters (Fig. 4L). The increase in expression levels of PHGDH and SHMT2, as well as GLS and MTHFD2, was also apparent in EMT-type clusters (SI Appendix, Fig. S7D).
Combinatorial Inhibition of GLS and PHGDH Simultaneously Targets Cell Subpopulations Including Stemness-High Cancer Cells.
Although PHGDH is essential for cell proliferation (31), each cancer cell has a different growth rate (32). Indeed, the expression of cyclin-dependent kinase 4 (CDK4) in patient-derived cancer organoids has been detected in non-EMT type clusters (SI Appendix, Fig. S7E). Along with a wide range of vulnerabilities to PHGDH inhibition, EMT-type cell clusters show resistance to NCT503 (SI Appendix, Fig. S7F). In particular, Cluster 3 was not affected by PHGDH inhibition while up-regulating its EMT signature (SI Appendix, Fig. S7G). Thus, intratumoral heterogeneity was observed in patient-derived cancer organoids with diverse cellular metabolism and drug sensitivity, and the inhibition of GLS appeared to activate the ATF4/CEBPB-mediated transcriptional network in EMT-type cell clusters.
We further evaluated key factors related to the intratumoral heterogeneity, attempting to find an effective treatment strategy that would target every cell cluster. EMT-type cancer cells demonstrate high plasticity, which is known to be associated with the acquisition of stem-like traits (33). As expected, the signaling entropy rate which estimates differentiation potency of individual cells was remarkably high in EMT-type clusters 2, 3, 7, and 8 (Fig. 5A). Analysis of developmental lineages using RNA velocity (34) and trajectory inference with Slingshot (35) also indicate high lineage potency in clusters 2, 3, 7, and 8 (Fig. 5B and SI Appendix, Fig. S8A).
Fig. 5.
Coinhibition of GLS and PHGDH shows high efficacy in targeting stemness-high cell clusters. (A) Single-cell entropy (SCENT) analysis. Signaling entropy at the single-cell level was estimated across a population of cells and colored corresponding to the score in the UMAP plot. (B) Inferred cell lineage with Slingshot (C) GSVA was performed with WNT β-catenin signaling pathway from MsigDB v7.4. (D) A heatmap showing the relative importance of each cell cluster based on the computed four network centrality measures of the WNT signaling network. (E) Hierarchical plot showing the inferred intracellular network for the WNT signaling pathway. The color of the circles differs among clusters, and the edge width indicates the communication probability. (F) Inferred WNT signaling network with edge width representing the communication probability. (G) The expression level of LGR5 was plotted on the corresponding UMAP. (H) The expression levels of CD44 and GLS were plotted on the corresponding UMAP. (I) CD44 and GLS were coplotted on the corresponding UMAP with color intensity corresponding to the expression level. (J) The expression level of CD44 is compared according to the subtype in the Yonsei cohort: mixed (n = 99), gastric (n = 89), SEM (n = 117), intestinal (n = 102), and inflammatory (n = 90). (K) The genome browser shows ATAC-seq profiles in the promoter region of CD44 and GLS in HS746T and NCIN87, respectively. (L) Kaplan–Meier plot for CD44high/GLShigh (n = 37) and CD44low/GLSlow group (n = 62) in TCGA stomach adenocarcinoma cohort. One hundred and fifty tumor samples with highest or lowest gene expression were marked as high or low, resulting in the CD44high/GLShigh and CD44low/GLSlow group. Logrank P = 0.019. ns, no significance, ****P < 0.0001, Wilcox test for J.
The Wingless-INT (WNT) signaling pathway promotes self-renewal activity in various types of cells including tumor cells (36), while also being involved in inducing EMT in tumors (37). In addition, WNT signaling is a critical pathway for retaining the stemness of the organoid (38). We found that the GSVA enrichment score for the WNT/β-catenin signaling pathway was markedly high in EMT-type clusters 2, 3, 7, and 8 (Fig. 5C). Analysis of cell-to-cell communication via CellChat revealed active signaling communication among EMT-type cell clusters via the inferred WNT signaling network (Fig. 5 D–F and SI Appendix, Fig. S8B). Cluster 3 showed the highest communication probability (Fig. 5F). In contrast, non-EMT type cell clusters showed active communication through the macrophage migration inhibitory factor signaling pathway, which enhances tumor growth (39), and protease-activated receptors’ (PARs) signaling pathway (SI Appendix, Fig. S8C). Gene markers of cluster 3 include a well-known intestinal stem cell marker, LGR5, which is a WNT signaling pathway signature gene (12, 40) (Fig. 5G). CD44 is a prominent WNT signaling target as well as a cancer stem cell marker (14). We found that CD44-positive cells were present even after the treatment only with a GLS inhibitor or PHGDH inhibitor (Fig. 5H). However, coinhibition of GLS and PHGDH successfully removed CD44-positive cells and decreased the number of cells across all cell clusters, compared to PHGDH inhibition alone, which failed to eradicate stemness-high cancer cells (Fig. 5H). Coinhibition of GLS and PHGDH was most successful in targeting the GLS-positive subpopulation of cells (Fig. 5H). GLS was also coexpressed with CD44 within the cancer organoid (Fig. 5I). Consistent with the patient-derived organoid data, SEM-type GC patients showed the highest CD44 expression (Fig. 5J). ATAC-sequencing results also demonstrated a higher degree of chromatin openness in HS746T in the promoter regions of CD44 and GLS (Fig. 5K). In TCGA stomach adenocarcinoma data, we found that patients with elevated levels of CD44 and GLS had a significantly worse outcome in terms of disease-specific survival, validating our findings in a separate cohort (Fig. 5L).
Discussion
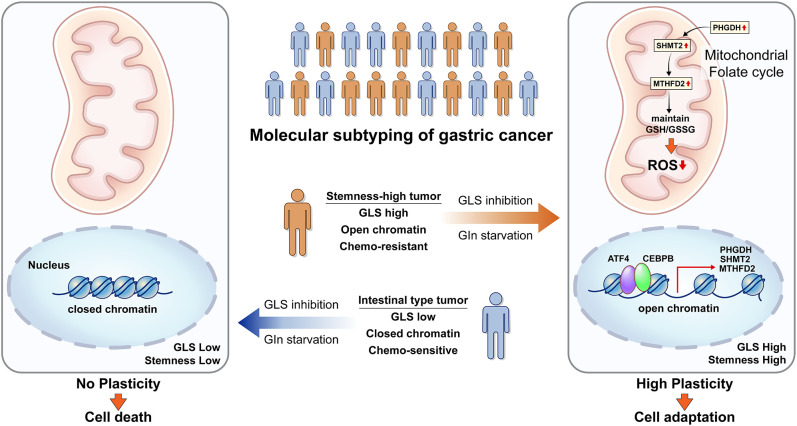
On the basis of the difference in gene expression involved in glycolysis and glutaminolysis between intestinal and SEM-type GC, we examined the subtype-specific metabolic pathways in GC under nutrient stress, such as glutamine starvation. Although SEM-type GC has been predicted to be vulnerable to GLS inhibition or glutamine starvation, we reveal that ATF4/CEBPB activates the PHGDH-driven salvage pathway to protect stemness-high cancer cells against mitochondrial ROS in metabolic stress such as glutamine starvation or GLS inhibition (Fig. 6).
Fig. 6.
The proposed model of how SEM-type GC cells survive in glutamine starvation or GLS inhibition.
An emerging view is that the development of anticancer drugs targeting metabolic vulnerabilities of cancer would be a therapeutic strategy for cancer treatment. Numerous drugs targeting diverse aspects of nutrient transport and utilization are currently going through clinical trials for various cancers (41). Furthermore, metabolic alterations have been shown to play pivotal roles in the sensitivity of cancer cells to first-line chemotherapy. Cisplatin-resistant cancer cells have been reported to exhibit dependency on glutamine for nucleotide biosynthesis (42). Also, cancer cells meet metabolically harsh environment during the course of tumor metastasis. SEM-type GC cells, which are experts in invasion, are expected to have enhanced level of metabolic reprogramming for their survival under the course of metastasis. These findings highlight the potential of therapeutic strategies targeting the metabolic vulnerabilities of chemoresistant cancer cells by identification of key metabolic gene signatures.
SEM-type GC remains one of the most difficult GCs due to drug resistance to standard treatment guidelines. Exploiting metabolic vulnerability would be a promising therapeutic strategy for the treatment of SEM-type GC. Although its gene signature predicted sensitivity to GLS inhibition in SEM-type GC, the SEM-type GC cells showed strong resistance to GLS inhibition or glutamate starvation due to upregulation of the PHGDH-driven salvage pathway. In accordance with our findings, a recent study has also shown that single-drug treatment targeting GLS with CB-839 only exhibited a very limited antitumor effect in glutamine-dependent liver cancer cells (43). Impressively, CB-839 treatment has been shown to increase metabolic vulnerability in glutamine-dependent liver cancer with antimetabolic drugs, including OXPHOS inhibitors, PPP inhibitors, and PHGDH inhibitors (43). In our study, combinatorial inhibition of GLS and PHGDH was able to decrease the tumor burden significantly in in vivo model with peritoneal metastasis, which is a debilitating form of metastasis in advanced GC. These results clearly indicate that concomitant inhibition of GLS and PHGDH represents a therapeutic strategy for the treatment of refractory tumors like SEM-type GC.
Most cancer cells proliferate rapidly using glycolysis. Since serine replenishes the 1C unit used in the folate cycle, PHGDH diverts part of the glycolytic flux into the de novo serine synthesis pathway to facilitate the folate cycle and support oligonucleotide synthesis (15, 44). Accordingly, PHGDH which catalyzes the rate-limiting step of de novo serine synthesis has been widely studied due to its protumorigenic effects (31, 45). However, little is known about the role of PHGDH in slow-growing cancer cells, such as dormant cancer cells, which do not rely much on glycolysis. In fact, PHGDH is turned “off” in SEM-type GC cells in stress-free conditions. This study reveals that cancer cells with mesenchymal features can turn PHGDH, SHMT2, and MTHFD2 “on” under metabolic stress to control ROS. It has been reported that GLS inhibition in hepatocellular carcinoma attenuated stemness by increasing cellular ROS, which subsequently suppresses the WNT/beta-catenin pathway (46). However, SEM-type GC cells were able to maintain ROS homeostasis and stemness via activation of a PHGDH-driven mitochondrial folate cycle.
During cancer progression, cancer cells face a metabolically harsh environment. Due to limited nutrient supplies, intermediate metabolites are alternatively consumed for ATP production, putting an emphasis on ROS production rather than ROS elimination (47). Our results point to the role of the PHGDH-driven salvage pathway as the central antioxidant system of stemness-high GC cells under glutamine starvation. To explore the reasons for the rapid response detected only in SEM-type GC cell lines upon the same metabolic stress, we profiled the chromatin accessibility of two different subtypes of GC cell lines. As stem cells have an open chromatin devoid of heterochromatin as a hallmark (48), the SEM-type GC cell line showed a high degree of chromatin accessibility and further increase in openness upon glutamine starvation. This key feature in chromatin architecture activates the ATF4/CEBPB-driven gene regulatory network and subsequent gene expression regulatory program (49). As chromatin architecture has been known to be associated with chemotherapy-induced dormancy and drug resistance (50), targeting chromatin structure could represent a method to eliminate remnant cancer cells after chemotherapy or to resensitize cancer cells to anticancer drugs. Their high chromatin accessibility allows SEM-type GC cells to activate the salvage pathway, enabling successful defense against oxidative stress and survive under metabolically stressful condition.
In summary, we have deciphered in detail the molecular characteristics of SEM-type GC cells including their chromatin structure, gene signatures, and defense mechanism to environmental stress. We propose the necessity of combination therapy that simultaneously targets GLS and PHGDH for the treatment of chemoresistant SEM-type GC.
Materials and Methods
Clinical Cohort.
Fresh frozen tumor tissues and matched clinical data were obtained from patients with GC who had undergone curative intent gastrectomy at the Yonsei Cancer Center (Seoul, Republic of Korea). This study was approved by the institutional review board of Severance. All samples were collected after obtaining written informed consent from the patients. With collected samples, we generated two batches of cohort data set (n = 497; GSE13861 and GSE84437; Illumina HUmanHT-12 v3.0 Expression BeadChip array).
Transcriptome Analysis and Patient Annotation.
Analyses were conducted in the R language environment. Datasets were merged using the “Combatting Batch Effects When Combining Batches of Gene Expression Microarray Data (ComBat)” analysis method. Unsupervised clustering was conducted with the R package “Algorithms and Framework for Nonnegative Matrix Factorization”. For annotation of subtypes, the gene set enrichment analysis was performed using the R “GSEABase” package and predefined gene sets from the Molecular Signature Database (MSigDB; www.broadinstitute.org/msigdb).
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
This work was supported by the National Research Foundation (NRF-2021R1A2C2009749) and Ministry of Health and Welfare (HR18C0012) to S.F.; National Research Foundation (NRF-2018R1A5A2025079) to J.-w.K.; Korea Health Industry Development Institute funded by the Ministry of Health & Welfare (H14C1324) to J-H.C.; National Research Foundation (NRF- 2020R1C1C1010489) to H.K. (Hyunkyung Kim). B.K.Y. is supported by the Seok-San Biomedical Science Scholarship, Yonsei University College of Medicine. R.M.E. holds the March of Dimes Chair in Molecular and Developmental Biology at the Salk Institute and is supported by grants from the NIH (P30 CA0141945 and R01 CA220468), The Lustgarten Foundation (122215393), the Samuel Waxman Cancer Research Foundation (SWCRF Investigator Award), The NOMIS Foundation (Science of Health), the Don and Lorraine Freeberg Foundation and the David C. Copley Foundation. We thank MID (Medical Illustration & Design) for providing excellent support with medical illustration.
Author contributions
B.K.Y., Hyeonhui Kim, J.-w.K., Hyunkyung Kim, R.M.E., J.-H.C., and S.F. designed research; B.K.Y., Hyeonhui Kim, S.K.O., S. Jo, M.K., K.-H.C., N.H., S.L., and S. Jin performed research; B.K.Y., Hyeonhui Kim, T.G.O., S.K.O., S. Jo, M.K., K.-H.C., N.H., S.L., S. Jin, A.R.A., R.T.Y., M.D., J.-w.K., Hyunkyung Kim, R.M.E., J.-H.C., and S.F. analyzed data; B.K.Y., Hyeonhui Kim, S. Jo, and N.H. generated tumor organoids; T.G.O. performed single cell analysis and provided technical support for the project; A.R.A., M.D., and R.M.E. interpreted results; J.-H.C. collected clinical information of the patients; and B.K.Y., Hyeonhui Kim, A.R.A., R.T.Y., M.D., J.-w.K., Hyunkyung Kim, R.M.E., J.-H.C., and S.F. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Contributor Information
Jae-woo Kim, Email: japol13@yuhs.ac.
Hyunkyung Kim, Email: hyunkkim@korea.ac.kr.
Ronald M. Evans, Email: evans@salk.edu.
Jae-Ho Cheong, Email: jhcheong@yuhs.ac.
Sungsoon Fang, Email: sfang@yuhs.ac.
Data, Materials, and Software Availability
The data relevant to the manuscript are uploaded for public access. Single-cell nuclei RNA sequencing analysis of the patient-derived GC organoid is deposited in Gene Expression Omnibus with the accession number GSE196836 (51), and ATAC sequencing, ChIP sequencing, and RNA sequencing data are deposited in NCBI Sequence Read Archive with the BioProject ID PRJNA808205.
Supporting Information
References
- 1.Cancer Genome Atlas Research, Comprehensive molecular characterization of gastric adenocarcinoma. Nature 513, 202–209 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cristescu R., et al. , Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat. Med. 21, 449–456 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Sohn B. H., et al. , Clinical significance of four molecular subtypes of gastric cancer identified by the cancer genome atlas project. Clin. Cancer Res. 23, 4441–4449 (2017), 10.1158/1078-0432.CCR-16-2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheong J. H., et al. , Predictive test for chemotherapy response in resectable gastric cancer: A multi-cohort, retrospective analysis. Lancet Oncol. 19, 629–638 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Wise D. R., Thompson C. B., Glutamine addiction: A new therapeutic target in cancer. Trends Biochem. Sci. 35, 427–433 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Z., et al. , Targeting glutaminolysis: New perspectives to understand cancer development and novel strategies for potential target therapies. Front. Oncol. 10, 589508 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang J. B., et al. , Targeting mitochondrial glutaminase activity inhibits oncogenic transformation. Cancer Cell 18, 207–219 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daemen A., et al. , Pan-cancer metabolic signature predicts co-dependency on glutaminase and de novo glutathione synthesis linked to a high-mesenchymal cell state. Cell Metab. 28, 383–399.e389 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Harding J. J., et al. , A phase I dose-escalation and expansion study of telaglenastat in patients with advanced or metastatic solid tumors. Clin. Cancer Res. 27, 4994–5003 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gross M. I., et al. , Antitumor activity of the glutaminase inhibitor CB-839 in triple-negative breast cancer. Mol. Cancer Ther. 13, 890–901 (2014). [DOI] [PubMed] [Google Scholar]
- 11.Pribluda A., de la Cruz C. C., Jackson E. L., Intratumoral heterogeneity: From diversity comes resistance. Clin. Cancer Res. 21, 2916–2923 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Fatehullah A., et al. , A tumour-resident Lgr5(+) stem-cell-like pool drives the establishment and progression of advanced gastric cancers. Nat. Cell Biol. 23, 1299–1313 (2021). [DOI] [PubMed] [Google Scholar]
- 13.Shiozawa Y., Nie B., Pienta K. J., Morgan T. M., Taichman R. S., Cancer stem cells and their role in metastasis. Pharmacol. Ther. 138, 285–293 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zeilstra J., et al. , Deletion of the WNT target and cancer stem cell marker CD44 in Apc(Min/+) mice attenuates intestinal tumorigenesis. Cancer Res. 68, 3655–3661 (2008). [DOI] [PubMed] [Google Scholar]
- 15.Locasale J. W., et al. , Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat. Genet. 43, 869–874 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou K., et al. , VDAC2 interacts with PFKP to regulate glucose metabolism and phenotypic reprogramming of glioma stem cells. Cell Death Dis. 9, 988 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Webb B. A., et al. , Structures of human phosphofructokinase-1 and atomic basis of cancer-associated mutations. Nature 523, 111–114 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanzelmann S., Castelo R., Guinney J., GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 14, 7 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Locasale J. W., Serine, glycine and one-carbon units: Cancer metabolism in full circle. Nat. Rev. Cancer 13, 572–583 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meshorer E., et al. , Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev. Cell 10, 105–116 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Efroni S., et al. , Global transcription in pluripotent embryonic stem cells. Cell Stem Cell 2, 437–447 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gaspar-Maia A., Alajem A., Meshorer E., Ramalho-Santos M., Open chromatin in pluripotency and reprogramming. Nat. Rev. Mol. Cell Biol. 12, 36–47 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reid M. A., et al. , The B55alpha subunit of PP2A drives a p53-dependent metabolic adaptation to glutamine deprivation. Mol. Cell 50, 200–211 (2013). [DOI] [PubMed] [Google Scholar]
- 24.Xiao W., Wang R. S., Handy D. E., Loscalzo J., NAD(H) and NADP(H) redox couples and cellular energy metabolism. Antioxid. Redox Signal. 28, 251–272 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pacold M. E., et al. , A PHGDH inhibitor reveals coordination of serine synthesis and one-carbon unit fate. Nat. Chem. Biol. 12, 452–458 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fan J., et al. , Quantitative flux analysis reveals folate-dependent NADPH production. Nature 510, 298–302 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Balwierz P. J., et al. , ISMARA: Automated modeling of genomic signals as a democracy of regulatory motifs. Genome Res. 24, 869–884 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schep A. N., Wu B., Buenrostro J. D., Greenleaf W. J., chromVAR: Inferring transcription-factor-associated accessibility from single-cell epigenomic data. Nat. Methods 14, 975–978 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang C., Awasthi N., Schwarz M. A., Schwarz R. E., Establishing a peritoneal dissemination xenograft mouse model for survival outcome assessment of experimental gastric cancer. J. Surg. Res. 182, 227–234 (2013). [DOI] [PubMed] [Google Scholar]
- 30.Xiang Y., et al. , Targeted inhibition of tumor-specific glutaminase diminishes cell-autonomous tumorigenesis. J. Clin. Invest. 125, 2293–2306 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Possemato R., et al. , Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature 476, 346–350 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weedon-Fekjaer H., Lindqvist B. H., Vatten L. J., Aalen O. O., Tretli S., Breast cancer tumor growth estimated through mammography screening data. Breast Cancer Res. 10, R41 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mani S. A., et al. , The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 133, 704–715 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.La Manno G., et al. , RNA velocity of single cells. Nature 560, 494–498 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Street K., et al. , Slingshot: Cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics 19, 477 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reya T., Morrison S. J., Clarke M. F., Weissman I. L., Stem cells, cancer, and cancer stem cells. Nature 414, 105–111 (2001). [DOI] [PubMed] [Google Scholar]
- 37.Schmalhofer O., Brabletz S., Brabletz T., E-cadherin, beta-catenin, and ZEB1 in malignant progression of cancer. Cancer Metastasis Rev. 28, 151–166 (2009). [DOI] [PubMed] [Google Scholar]
- 38.Nanki K., et al. , Divergent routes toward Wnt and R-spondin niche independency during human gastric carcinogenesis. Cell 174, 856–869.e817 (2018). [DOI] [PubMed] [Google Scholar]
- 39.Guda M. R., et al. , Pleiotropic role of macrophage migration inhibitory factor in cancer. Am. J. Cancer Res. 9, 2760–2773 (2019). [PMC free article] [PubMed] [Google Scholar]
- 40.Yan H. H. N., et al. , A comprehensive human gastric cancer organoid biobank captures tumor subtype heterogeneity and enables therapeutic screening. Cell Stem Cell 23, 882–897.e811 (2018). [DOI] [PubMed] [Google Scholar]
- 41.Lemberg K. M., Gori S. S., Tsukamoto T., Rais R., Slusher B. S., Clinical development of metabolic inhibitors for oncology. J. Clin. Invest. 132, e148550 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Obrist F., et al. , Metabolic vulnerability of cisplatin-resistant cancers. EMBO J 37, e98597 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jin H., et al. , A powerful drug combination strategy targeting glutamine addiction for the treatment of human liver cancer. Elife 9, e56749 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vander Heiden M. G., Cantley L. C., Thompson C. B., Understanding the warburg effect the metabolic requirements of cell proliferation. Science 324, 1029–1033 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li X., et al. , ATF3 promotes the serine synthesis pathway and tumor growth under dietary serine restriction. Cell Rep. 36, 109706 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li B., et al. , Targeting glutaminase 1 attenuates stemness properties in hepatocellular carcinoma by increasing reactive oxygen species and suppressing Wnt/beta-catenin pathway. EBioMedicine 39, 239–254 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao Y., et al. , ROS signaling under metabolic stress: Cross-talk between AMPK and AKT pathway. Mol. Cancer 16, 79 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Melendez-Ramirez C., et al. , Dynamic landscape of chromatin accessibility and transcriptomic changes during differentiation of human embryonic stem cells into dopaminergic neurons. Sci. Rep. 11, 16977 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Feng Y., Liu X., Pauklin S., 3D chromatin architecture and epigenetic regulation in cancer stem cells. Protein Cell 12, 440–454 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang L., et al. , Chromatin accessibility regulates chemotherapy-induced dormancy and reactivation. Mol. Ther. Nucleic Acids 26, 269–279 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yoon B. K., Mitochondrial folate italic confers metabolic plasticity to stemness-high gastric cancer in response to glutaminase inhibition. NCBI GEO. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE196836. Deposited 16 February 2022. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
The data relevant to the manuscript are uploaded for public access. Single-cell nuclei RNA sequencing analysis of the patient-derived GC organoid is deposited in Gene Expression Omnibus with the accession number GSE196836 (51), and ATAC sequencing, ChIP sequencing, and RNA sequencing data are deposited in NCBI Sequence Read Archive with the BioProject ID PRJNA808205.