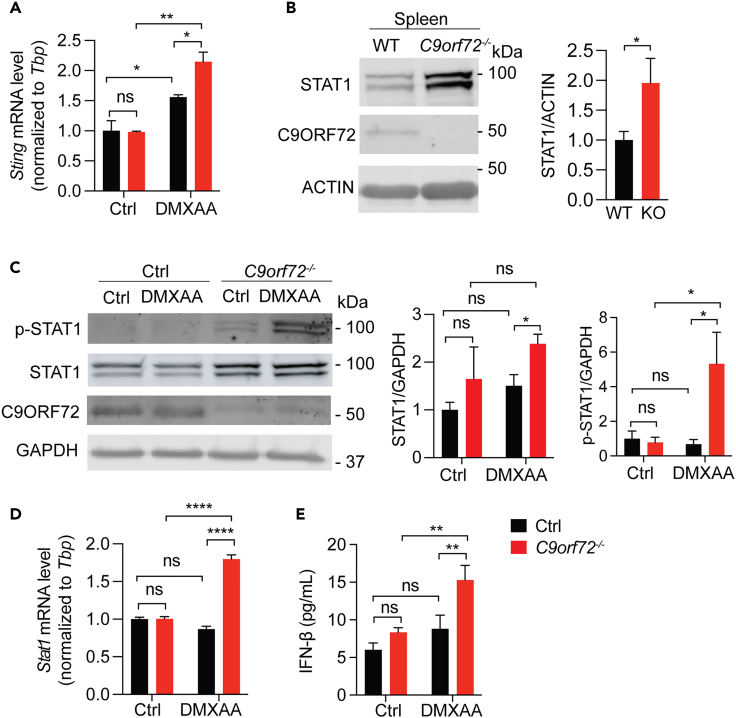
Figure 3.
The loss of C9ORF72 leads to JAK-STAT activation
(A) RT-qPCR analysis of Sting levels in control and C9orf72−/− RAW264.7 cells untreated or treated with DMXAA for 16h. Data represent the mean ± SEM. Statistical significance was analyzed by two-way ANOVA (n = 3), ns = not significant, ∗p < 0.05, ∗∗p < 0.01.
(B) Protein levels of STAT1 in spleen lysates from 6-month-old WT and C9orf72−/− mice were analyzed using western blot. Mixed male and female mice were used. Data represent the mean ± SEM. Statistical significance was analyzed by unpaired one-tail Student’s t test (n = 4), ns = not significant, ∗p < 0.05.
(C) Protein levels of STAT1 and p-STAT1 in control or C9orf72−/− RAW264.7 cells untreated or treated with DMXAA for 2h were analyzed using western blot and normalized to GAPDH. Data represent the mean ± SEM. Statistical significance was analyzed by unpaired two-tail Student’s t test (n = 3), ns = not significant, ∗p < 0.05.
(D) RT-qPCR analysis of Stat1 levels in control and C9orf72−/− RAW264.7 cells untreated or treated with DMXAA for 16h. Data represent the mean ± SEM. Statistical significance was analyzed by two-way ANOVA (n = 3), ns = not significant, ∗p < 0.05, ∗∗∗∗p < 0.0001.
(E) IFN-β levels in control and C9orf72−/− RAW264.7 cells untreated or treated with DMXAA for 16h were measured using ELISA. Data represent the mean ± SEM. Statistical significance was analyzed by two-way ANOVA (n = 3), ns = not significant, ∗∗p < 0.01.