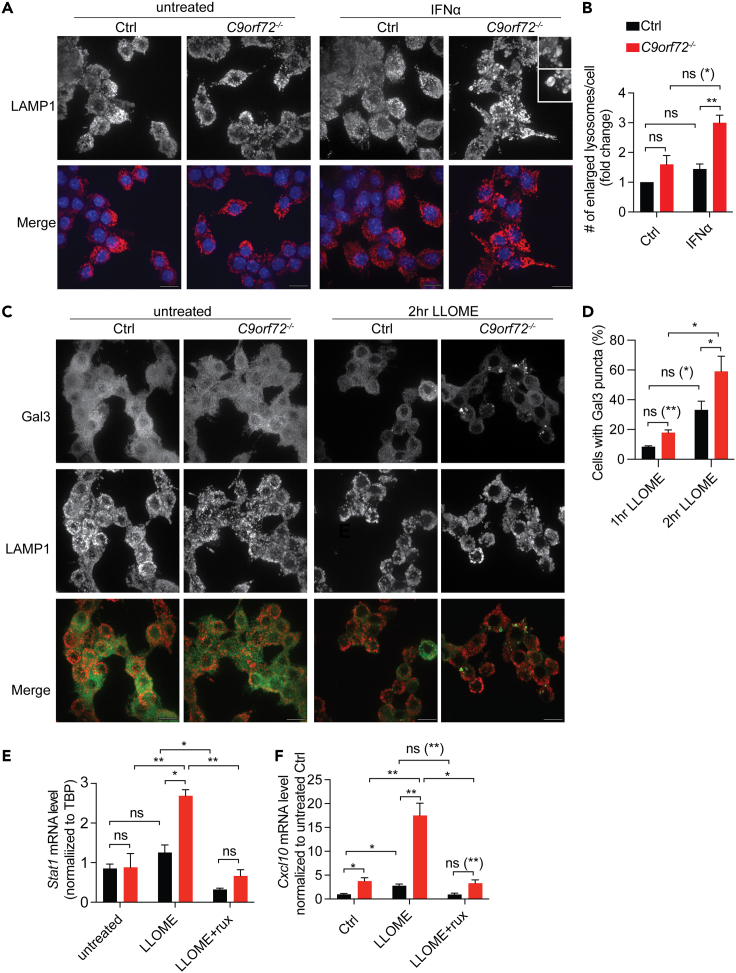
Figure 6.
Impairment of lysosome integrity due to C9ORF72 loss results in increased JAK-STAT-mediated inflammation
(A and B) Immunostaining of lysosome marker LAMP1 in control or C9orf72−/− RAW264.7 cells without and with 16h of IFNα treatment (scale bar = 10 μm). The number of lysosomes with a diameter larger than 0.75 μm in each cell was counted and normalized to untreated control RAW264.7 cells. 106–142 cells from three independent experiments (142 (Ctrl); 178 (Ctrl IFNa); 106 (C9orf72−/− Ctrl); 117 (C9orf72−/− IFNα)) were quantified for the experiment in (A). Data represent the mean ± SEM. Statistical significance was analyzed by two-way ANOVA. Groups that exhibit non-significant differences with two-way ANOVA were re-analyzed using unpaired two-tailed Student’s t test, and the results are indicated in parentheses. ns = not significant, ∗∗p < 0.01.
(C and D) Immunostaining of Galectin 3 and LAMP1 in control or C9orf72−/− RAW264.7 cells without and with 2h LLOME (1 μM) treatment (scale bar = 10 μm). The number of cells with Galectin-3 puncta was quantified and normalized to the total number of cells in each condition. Total number of cells from four independent experiments analyzed: Ctrl 1h LLOME n = 525, C9orf72−/− 1h LLOME n = 492, Ctrl 2h LLOME n = 319, C9orf72−/− 2h LLOME n = 294. Data represent the mean ± SEM. Statistical significance was analyzed by two-way ANOVA. Groups that exhibit non-significant differences with two-way ANOVA were re-analyzed using unpaired two-tailed Student’s t test, and the results are indicated in parentheses. ns = not significant, ∗p < 0.05.
(E and F) Control or C9orf72−/− RAW264.7 cells are either untreated (Ctrl), or treated with 4hrs of LLOME then recover for 12hrs in normal cell medium (LLOME), or treated with ruxolitinib for 16hrs before 4 h of LLOME treatment and then recover for 12hrs in normal cell medium with ruxolitinib (LLOME+rux). The mRNA level of Stat1 and Cxcl10 was analyzed by RT-qPCR. Data represent the mean ± SEM. Statistical significance was analyzed by two-way ANOVA (n = 3). Groups that exhibit non-significant differences with two-way ANOVA were re-analyzed using unpaired two-tailed Student’s t test, and the results are indicated in parentheses. ns = not significant, ∗p < 0.05, ∗∗p < 0.01.