Abstract
Surgical site infections represent a significant clinical problem. Herein, this study aimed to develop a nanofiber-based dressing capable of local sustained delivery of immunomodulating compounds including 1α,25-dihydroxyvitamin D3 (1,25(OH)2D3) and VID400, a CYP24A1 inhibitor, for inducing expression of the endogenous cathelicidin antimicrobial peptide (CAMP) gene which encodes the hCAP18 protein that is processed into the LL-37 peptide. Nanofiber wound dressings with co-encapsulation of 1,25(OH)2D3 and VID400 were prepared by electrospinning. Both 1,25(OH)2D3 and VID400 were successfully loaded into nanofibers with encapsulation efficiencies larger than 90% and exhibited a sustained release from nanofibers over 4 weeks. Treatment with 1,25(OH)2D3/VID400-co-loaded poly(ϵ-caprolactone) nanofibers significantly induced hCAP18/LL-37 expression in monocytes, neutrophils, and keratinocytes in vitro. In addition, administration of 1,25(OH)2D3/VID400 nanofiber membranes dramatically increased expression of hCAP18/LL-37 in skin wounds of human CAMP transgenic mice and artificial wounds of human skin explants. The 1,25(OH)2D3 and VID400 containing nanofiber dressings enhanced innate immunity by inducing antimicrobial peptide production more efficiently than free drug alone or 1,25(OH)2D3 loaded nanofibers. Together, 1,25(OH)2D3/VID400 embedded nanofiber dressings presented in this study show potential in preventing surgical site infections.
Keywords: 1α,25-dihydroxyvitamin D3; CYP24A1 inhibitor; co-delivery; nanofiber dressings; antimicrobial peptide LL-37
Graphical Abstract
INTRODUCTION
Surgical site infections (SSIs) have been historically associated with increased morbidity and mortality but remain a clinical issue in modern healthcare.1 Regardless of today's surgical protocols, between 2% and 5% of all surgical interventions result in a SSI.2 As of 2017, the Centers for Disease Control and Prevention (CDC) estimated that SSIs occurred in at least 1.9% of all surgical patients; however, this number is most likely not representative of the total number of SSIs cases since about 50% of SSIs occur after hospital discharge.3 SSIs also incur a considerable economic burden. In the United States alone, the management of SSIs costs over $3 billion USD per year.4
The most common method to treat SSIs is antibiotic therapy;5 however, due to the abuse of various antibiotics in recent decades, antibiotic resistance has become a serious problem in the field of biomedicine.6 The World Health Organization (WHO) warns by 2050, the global death toll from antibiotic-resistant strains of pathogens may affect 10 million people per year.7 There is an urgent need for the development of new antimicrobial and anti-infection methods in the post-antibiotic era. Among various approaches, enhancing innate immunity is a promising direction. Innate immunity constitutes the first line of defense and includes specific cells of hematopoietic origin that produce various effector molecules to activate mechanisms for eliminating pathogens.8 These include non-professional (NK cells) and professional phagocytes (monocytes/macrophages, dendritic cells and granulocytes).9 Furthermore, epithelial cells are fundamentally important for forming a continuous defense barrier.10
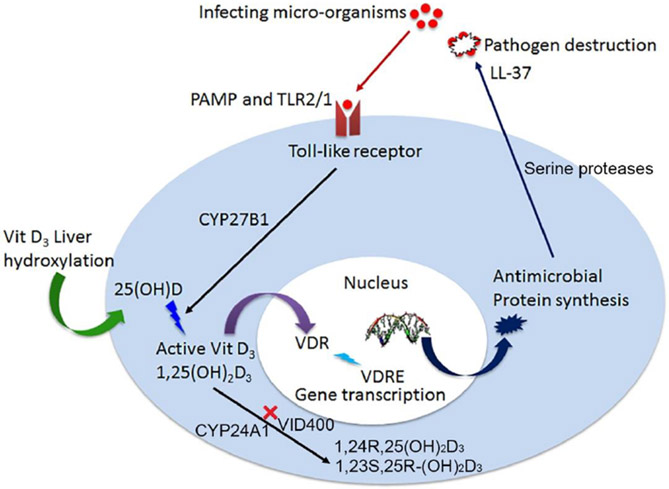
The bioactive form of vitamin D3, 1,25-dihydroxyvitamin D3 (1,25(OH)2D3) strongly induces CAMP gene expression in monocytes, macrophages, dendritic and epithelial barrier cells through the vitamin D receptor (VDR).11-14 In our previous work, we reported nanofibers loaded with 25(OH)vitamin D3 (25D3) or 1,25(OH)2D3 can induce higher CAMP gene expression in HaCat and U937 cells than the free drug at an equivalent dose.15, 16 Another target gene of VDR is CYP24A1, which encodes the 24-hydroxylase, a protein that limits the amount of 1,25(OH)2D3 in the body.17 Inhibitors of CYP24A1 are expected to extend the half-life of 1,25(OH)2D3 and increase its endogenous levels.18 The inhibitor, N-(2-(1H-Imidazol-1-yl)-2-phenylethyl)-4'-chloro-[1,1'-biphenyl]-4-carboxamide (VID400), directly binds to the heme iron of the CYPs via an azole nitrogen and to other parts of the substrate site which has potential to selectively inhibit CYP24A1.19 Therefore, a promising strategy to prevent the catabolism of 1,25(OH)2D3 and possibly potentiate the action on CAMP induction could involve inhibiting CYP24A1 activity. As shown in Figure 1, specific pathogen associated molecular patterns (PAMP) are recognized by Toll-like receptors initiating the local intra-cellular conversion of active vitamin D. Active vitamin D3 is bound by the vitamin D receptor (VDR) initiating gene transcription at specific DNA sequences, vitamin D response elements (VDRE).20 Following protein synthesis of antimicrobial peptides, like cathelicidin (LL-37), pathogen destruction ensues. This intracrine system of active vitamin D production and immunity is dependent on the concentration of 25(OH)D or 1,25(OH)2D3.21 Inactivation of 1,25(OH)2D3 occurs via C-23 and C-34 oxidation pathways, catalyzed by CYP24A1. CYP24A1 inhibitor VID400 can inhibit the activity of CYP24A1 by occupying its substrate site and reduce the inactivation of 1,25(OH)2D3, which may result in promotion of cathelicidin/LL-37 production.
Figure 1.
Schematic showing antimicrobial peptide production is vitamin D dependent. CYP24A1 inhibitor can reduce the conversion of 1,25(OH)2D3 to 1,24R,25(OH)2D3 and 1,23S,25R-(OH)2D3.
In this work, we report the co-delivery of 1,25(OH)2D3 and CYP24A1 inhibitor VID400 using electrospun nanofibers in an attempt to develop a nanofiber dressing containing 1,25(OH)2D3 and a CYP24A1-specific inhibitor to potentiate the induction of CAMP expression. Briefly, we generated biodegradable electrospun nanofiber dressings to up-regulate CAMP gene expression and induce endogenous antimicrobial peptide hCAP18/LL-37 expression in keratinocytes, monocytes, neutrophils, skin wounds of humanized transgenic mice and artificial wounds of human skin explants. We found after treating with 1,25(OH)2D3/VID400-co-loaded poly(ϵ-caprolactone) (PCL) nanofibers, the hCAP18/LL-37 expression in monocytes, neutrophils, and keratinocytes, skin wounds of human CAMP transgenic mice and artificial wounds of human skin explants was significantly induced. Our findings supported the use of 1,25(OH)2D3/VID400-co-loaded nanofibers as wound dressings for potentially preventing SSIs.
MATERIALS AND METHODS
Materials.
N-(2-(1H-Imidazol-1-yl)-2-phenylethyl)-4'-chloro-[1,1'-biphenyl]-4-carboxamide (VID400) was purchased from Aobious (Gloucester, MA, USA). 1α,25(OH)2D3 was purchased from Santa Cruz Biotechnology, Inc. (Dallas, TX, USA). Poly(ϵ-caprolactone) (PCL; molecular weight (Mw) = 70–90 kDa) and Pluronic® F-127 were obtained from Sigma-Aldrich (St. Louis, MO, USA).
Fabrication and Characterization of 1,25(OH)2D3- and 1,25(OH)2D3/VID400-loaded Nanofibers.
1,25(OH)2D3 or 1,25(OH)2D3/VID400 was encapsulated in PCL nanofibers using electrospinning as described in our previous studies.16 Briefly, PCL was dissolved in a solvent mixture consisting of dichloromethane (DCM, Thermo Fisher Scientific, MA, USA) and dimethylformamide (DMF, Thermo Fisher Scientific, MA, USA) with a ratio of 4:1 (v/v) at a concentration of 9% (w/v). To enhance the hydrophilicity of fibers, 1% (w/v) pluronic F-127 was added to the solution. The stock solutions of 1,25(OH)2D3 or 1,25(OH)2D3/VID400 prepared by dissolving 1 mg 1,25(OH)2D3 or 1 mg 1,25(OH)2D3 and 10 mg VID400 in 1 mL DMSO (Thermo Fisher Scientific, MA, USA) were added to the polymer (PCL) solution with initial drug loadings ranging from 250 μg/g to 2500 μg/g. The polymer solution was pumped at a flow rate of 0.6 ml/h using a syringe pump, while an electrical potential of 15 kV was applied between the spinneret (a 22-gage needle) and a grounded collector located 15 cm from the spinneret. A rotating drum collected membranes composed of random fibers with a rotating speed less than 100 rpm. The morphology and diameter of the nanofiber samples were characterized by a scanning electron microscope (SEM, FEI, Quanta 200, OR, USA) following our previous studies.
In Vitro Release of 1,25(OH)2D3 and VID400 from Nanofibers.
In vitro release of 1,25(OH)2D3 and VID400 from the nanofibers was evaluated by immersing 10 mg fiber samples in 10 ml phosphate-buffered saline (PBS) at 37 °C. The supernatants were collected at each time point and replaced with fresh PBS. Drug loading and encapsulation efficiency were determined following our previous studies. 1,25(OH)2D3/VID400-loaded nanofiber samples were first dissolved in glacial acetic acid at the concentration of 10 mg/ml, and then the solutions were diluted 100-fold with glacial acetic acid, and further diluted 100-fold with PBS. The 1,25(OH)2D3 concentrations of collected samples were determined using a 1,25(OH)2D3 ELISA kit according to the manufacturer’s instructions (Cayman Chemical, CA, USA). The VID400 concentrations were determined by high-performance liquid chromatography (HLPC, Agilent Technologies). Briefly, drug release solution was prepared by using PBS and 1% bovine serum albumin. The 1,25(OH)2D3/VID400-loaded nanofiber samples were suspended at a concentration of 20 mg/mL of drug release solution and incubated at 37°C with constant stirring. At different time points, suspensions were centrifuged and 1 mL drug release solution was taken and replaced with fresh drug release solution. A mixture of 0.5% ammonium acetate solution/0.2% diisopropylamine solution in methanol was applied as the mobile phase at the flow rate of 1.0 mL/ min. Samples were analyzed at 240 nm.
Cell Culture and Treatments.
The human keratinocytes HaCaT cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM, Gibco, MA, USA) with 10% fetal bovine serum (FBS, Gibco, MA, USA). Human leukemia HL-60 cells and monocytes U937 cells were cultured in RPMI-1640 media with 10% FBS. The cultures were maintained at 37°C with 5% CO2. 5.0 × 105, 2.0 × 105 and 1.0 × 105 cells were seeded in 6-cm dishes for 1, 3 and 5 days respectively. To evaluate the effect of 1,25(OH)2D3 and VID400 on these three cell lines. The media was then replaced by DMEM containing 0.52% DMSO (control) or 200 nM 1,25(OH)2D3 and 0 to 2000 nM VID400. U937 and HL-60 cells were pelleted by 300 × g centrifugation for 5 min and then resuspended in fresh complete media. Cells were seeded in 6-cm culture dishes and incubated with RPMI-1640 containing 0.52% DMSO (control) or 200 nM 1,25(OH)2D3 and 0 to 2000 nM VID400. All the nanofiber samples were sterilized by γ-radiation at a dose of 15 kGy prior to use for both in vitro and in vivo tests. HaCaT cells were incubated for 1, 3 and 5 days with DMEM containing 1 mg/ml pristine PCL nanofibers, 1,25(OH)2D3-loaded PCL nanofibers, or 1,25(OH)2D3/VID400-loaded PCL nanofibers. Similarly, HL60 and U937 cells were incubated with RPMI-1640 containing 1 mg/ml pristine PCL nanofibers, 1,25(OH)2D3-loaded PCL nanofibers, or 1,25(OH)2D3/VID400-loaded PCL nanofibers for 1, 3 and 5 days. Meanwhile, the in vitro cell toxicity was evaluated using Alamar Blue™ Cell Viability Reagent (Thermo Fisher Scientific, MA, USA). U937, HL60, HaCaT and HDF-α were incubated with different nanofiber samples at the concentration of 1 mg/ml.
RNA Isolation and qPCR Analysis.
Total RNA was isolated using a RNeasy Mini Kit according to the manufacturer’s protocol (Qiagen, Hilden, Germany). RNA concentration and purity were determined using a NanoDrop™ One Microvolume UV-Vis Spectrophotometer (Thermo Fisher Scientific). Then, RNA was converted to cDNA using a qScript® cDNA Synthesis Kit (Quantabio, Beverly, MA, USA) as instructed by the manufacturer using a PCR machine (Bio-Rad Laboratories, CA, USA). Real-time polymerase-chain-reaction (qPCR) reactions were performed using a StepOnePlus Real-time PCR System (Applied Biosystem, Thermo fisher Scientific) with SsoAdvanced Universal SYBR Green Supermix as instructed by the manufacturer (Bio-Rad Laboratories). The primers for the human CAMP gene used for qPCR were as follows.
CAMP primer Forward: 5’-AGGGGCTCCTTTGACATCAG-3’
Reverse: 5’-GGGTAGGGCACACACTAGGA-3’
qPCR was performed for 45 cycles (15 s at 95 °C and 1 min at 60 °C). The relative mRNA expression in each sample was calculated based on its Ct value comparison to the Ct of a housekeeping gene GADPH. The data were presented as 2−ΔΔCt, an arbitrary unit. All amplified products showed single peak in the melting curve analysis. qPCR was performed in triplicate for each sample. Experiments were performed at least three independent times.
In Vitro Induction of hCAP18/LL-37.
To quantify the induction of hCAP18/LL-37, HaCaT, HL60 and U937 cells were seeded in 6-cm culture dishes at 5.0 × 105, 2.0 × 105 and 1.0 × 105 cells per dish, and incubated for 1, 3 and 5 days and treated with different formulations as described above. Subconfluent HaCaT cells and U937 and HL60 cell suspensions were washed with PBS twice, pelleted and resuspended in 300 μl of the M-PER mammalian protein extraction reagent (Thermo Fisher Scientific) containing 0.1% protease inhibitor cocktail (Sigma-Aldrich). The total protein concentration was quantified using a MicroBCA assay kit (Thermo Fisher Scientific). The amount of hCAP18/LL-37 in each cell lysate was determined using an ELISA assay kit as instructed by the manufacturer (Hycult Biotech, PA, USA).
Ex Vivo Induction of CAMP Gene and Antimicrobial Peptide in Human Skin Tissue.
The human skin tissues were collected from patients who underwent plastic surgery, and the IRB protocol was approved by the University of Nebraska Medical Center (Protocol #152-14-EP). Human skin tissues were incubated in DMEM within 2 h after collection from patients who underwent plastic surgery. The skin tissue was cut into 2 cm × 2 cm pieces. PCL was formed in a sheet with a size of 2 cm × 2 cm × 0.2 cm using a customized mold. The tissue was fixed on the PCL sheet by three or four staple clips at the corners of each tissue and then placed in a 6-cm diameter culture dish. Approximately, 7 ml DMEM medium containing 10% FBS was added to each dish to maintain the dermal layer in contact with the medium and the epidermal layer exposed to the air. After incubation for 1 day, a 1 mm deep wound was generated in the center of each skin fragment using an 8-mm diameter punch. Nanofiber discs were cut using an 8-mm diameter punch and inserted into each wound (each group = 6 samples). Both the wound tissue and 2-mm border from around the wound were harvested using a 10-mm punch at various time points and homogenized in 0.5-ml tissue lysis buffer at 4 °C. Total RNA was isolated using an RNeasy Mini Kit and CAMP gene expression was evaluated by qPCR as described above. The amount of hCAP18/LL-37 in 100 μL of supernatant was determined by ELISA as described above.
In Vivo Antimicrobial Peptide Induction.
In vivo LL-37 expression of 1,25(OH)2D3/VID400-loaded PCL nanofibers induction was evaluated in the CAMPTg/Tg:KO/KO transgenic mouse wound model. This animal study was conducted following approval by the Oregon State University’s IACUC in accordance with animal protocol # IACUC-2021-019. Briefly, protein samples were extracted from day 3 skin wounds of CAMPTg/Tg:KO/KO mice treated with pristine PCL nanofibers, 1,25(OH)2D3-loaded PCL nanofibers, or 1,25(OH)2D3/VID400-loaded PCL nanofibers (each group = 6 samples). Western blots were performed using specific anti-hCAP18 antibody, and β-actin was used as a loading control. The induction of hCAP18 expression in day 3 skin wounds post treatment with different nanofibers was quantified. Immunofluorescent staining for hCAP18/LL-37 protein and the macrophage marker F4/80 was performed on day 3 samples post wounding in the presence of different nanofibers as described before.16
Statistical Analysis.
The data were presented as the mean ± standard deviation, and statistical analysis was performed using SPSS 13.0 and GraphPad 8.0 software. T-test and one-way analysis of variance with Tukey’s multiple comparison post-test were used to determine significance. The values of p < 0.05 were considered statistically significant.
RESULTS
Fabrication and Characterization of Nanofiber Formulations.
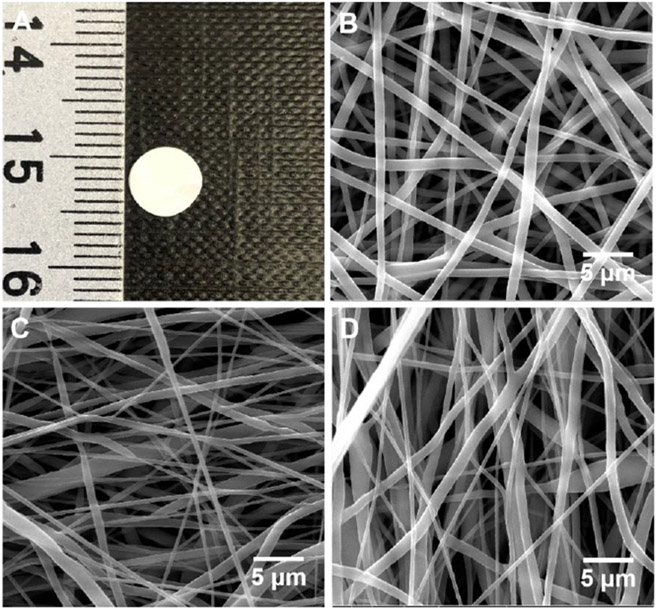
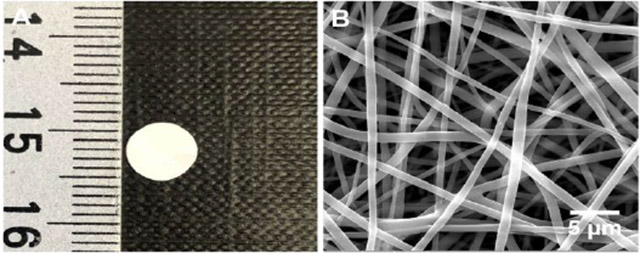
In this study, we co-encapsulated 1,25(OH)2D3 and VID400 in electrospun nanofibers to ensure their sustained delivery at wound or surgical sites. We selected PCL as a carrier material because it is a biocompatible and biodegradable polymer used in FDA-approved medical devices for certain clinical applications22,, and we applied the additive pluronic F127 to increase the hydrophilicity of the nanofibers.23 In this study, the PCL nanofibers primarily served as dressings for releasing 1,25(OH)2D3 and VID400 molecules in a sustained manner instead of serving as scaffolds for cell infiltration and tissue regeneration; therefore, the degradation of PCL nanofibers was not considered. Figure 2A shows a photograph of 6-mm nanofiber disc. Figure 2 B-D show SEM images of pristine PCL/pluronic F-127 nanofibers, 1,25(OH)2D3-loaded PCL/pluronic F127 nanofibers, and 1,25(OH)2D3/VID400-loaded PCL/pluronic F127 nanofibers. All the nanofibers possessed a cylindrical shape with a smooth surface and diameters ranging from about 300 nm to 1 μm.
Figure 2.
Morphology characterization. (A) Photograph shows a 1,25(OH)2D3/VID400-loaded PCL nanofiber membrane with a diameter of 6 mm. (B-D) SEM images of (B) PCL/pluronic F127 nanofibers, (C) 1,25(OH)2D3-loaded PCL/pluronic F127 nanofibers, and (D) 1,25(OH)2D3/VID400-loaded PCL/pluronic F127 nanofibers.
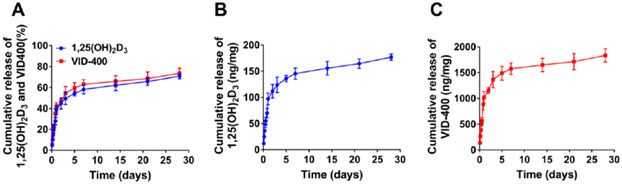
To determine the release kinetics of 1,25(OH)2D3 and VID400, we incubated the nanofibers in PBS and quantified the amount of 1,25(OH)2D3 and VID400 released into the solution. The release profiles exhibited an initial burst followed by a sustained release over 28 days (Figure 3A). It is observed that 178 ±5.9 ng 1,25(OH)2D3 (Figure 3B) and 1812 ±128.5 ng of VID400 (Figure 3C) were released from 1 mg of 1,25(OH)2D3/VID400-loaded PCL/pluronic F127 nanofiber mats, respectively, after incubation for 28 days. The 1,25(OH)2D3 loading for 1,25(OH)2D3-loaded PCL/pluronic F127 nanofibers was 241 ± 9 ng/mg, the corresponding encapsulation efficiency was 96.4 ± 3.6%. The 1,25(OH)2D3 and VID400 loadings for 1,25(OH)2D3/VID400-loaded PCL/pluronic F127 nannofibers were 240 ± 8 and 2388 ± 145 ng/mg, respectively. The corresponding encapsulation efficiencies were 96.0 ± 3.2% and 95.52 ± 3.8%, respectively.
Figure 3.
In vitro release profiles of 1,25(OH)2D3 and VID400 from nanofibers over 28 days (A) and the daily release of 1,25(OH)2D3 (B) and VID400 (C).
Induction of CAMP Gene Expression In Vitro.
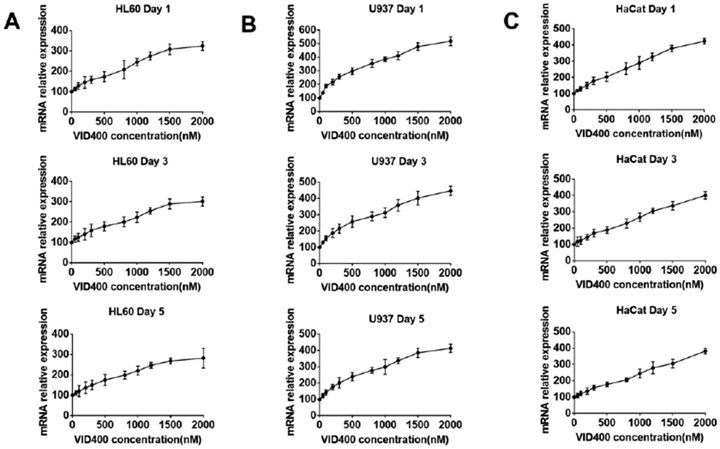
To assess the effect of 1,25(OH)2D3 and VID400 on the expression of CAMP, we exposed the human keratinocyte cell line HaCaT, promyelocytic cell line HL60 and pro-monocytic cell line U937 to 200 nM 1,25(OH)2D3 and concentrations of VID400 ranging from 0 to 2000 nM for 1, 3, 5 days, respectively. The levels of CAMP mRNA were significantly up-regulated after 1, 3, and 5 days of exposure, with a maximum induction of 5-fold at 2000 nM VID400 when incubated with 200 nM 1,25(OH)2D3 (Figure 4). We chose this concentration range for VID400 as we tested the in vitro cell toxicity of different concentrations of VID400 in combination with 200 nM 1,25(OH)2D3, and we found a VID400 concentration higher than 2500 nM caused significant cell toxicity to these three types of cells at day 3 (Figure S1).
Figure 4.
CAMP gene relative expression of (A) HL60, (B) U937, (C) HaCaT cells after treatment with 200 nM 1,25(OH)2D3 and 0-2000 nM VID400 for 1, 3, and 5 days.
Quantification of Induced Antimicrobial Peptide In Vitro.
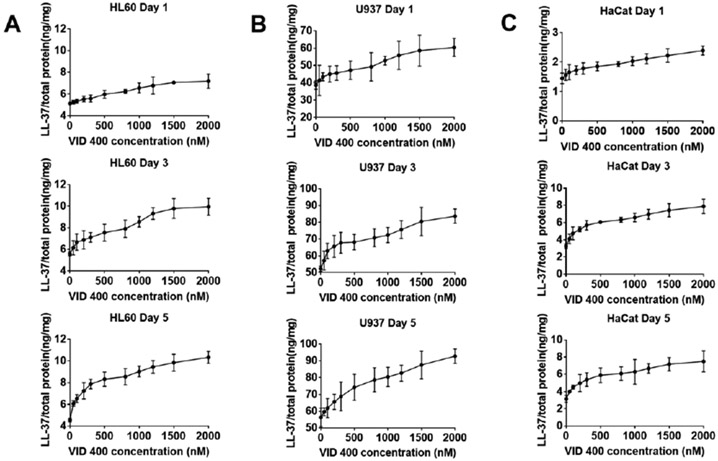
To determine if the induction of cathelicidin mRNA levels correlated to an increase in protein expression, we quantified hCAP18/LL-37 levels using an ELISA assay. The three cell lines were treated with 200 nM 1,25(OH)2D3 and 0-2000 nM VID400 as free drug, and total cell lysates were collected for analysis. After treatment for 1, 3 and 5 days, increased levels of hCAP18/LL37 were observed. As shown in Figure 5, administration of 200 nM 1,25(OH)2D3 and 2000 nM VID400 for 5 days induced the highest amount of hCAP18/LL-37 in HaCaT cells among the treatment groups.
Figure 5.
LL-37 expression level of (A) HL60, (B) U937, (C) HaCaT cells after treatment with 200 nM 1,25(OH)2D3 and 0-2000 nM VID400 for 1, 3, and 5 days.
In Vitro Cell Toxicity Assay.
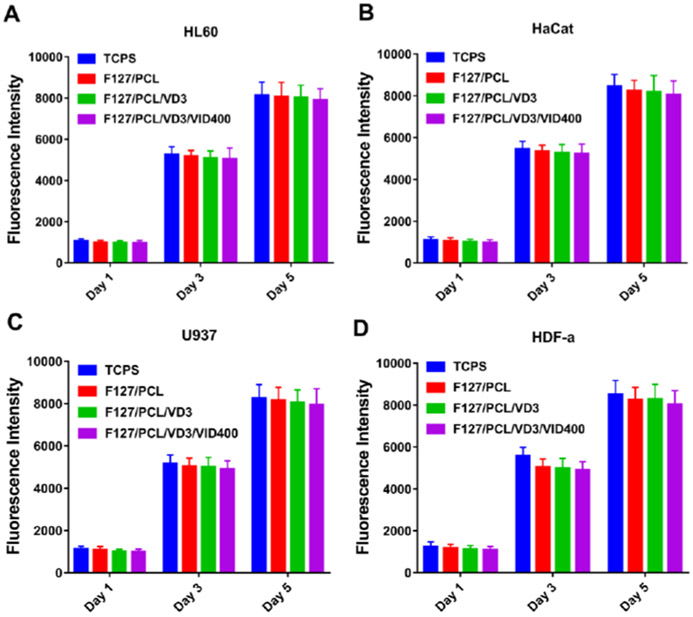
To test the cytotoxicity of the nanofiber dressings, we tested the effect of nanofiber membranes at the concentration of 1 mg/ml on the proliferation of HaCaT, HL60 and U937 cells. As shown in Figure 6, drug-loaded nanofiber membranes had no significant influences on the proliferation of HaCaT, HL60 and U937 cells compared with control groups tissue culture polystyrene (TCPS) and PCL/Pluronic F127 nanofibers. Overall, the cell viability results revealed no significant cytotoxicity with 1,25(OH)2D3-loaded PCL/pluronic F127 nanofibers and 1,25(OH)2D3/VID400-co-loaded PCL/pluronic F127 nanofibers in direct contact with cells. Therefore, the 1,25(OH)2D3/VID400-co-loaded PCL/pluronic F127 nanofibers have excellent cytocompatibility, supporting their application as wound dressings.
Figure 6.
In vitro cell toxicity of different nanofiber formulations for 1, 3, and 5 days. (A) HL60, (B) U937, (C) HaCaT, and (D) HDF-α. TCPS: tissue culture polystyrene. F127/PCL: PCL/pluronic F-127 blend nanofibers. F127/PCL/VD3: 1,25(OH)2D3 loaded PCL/pluronic F-127 blend nanofibers. F127/PCL/VD3/VID400: 1,25(OH)2D3 and VID400 coloaded PCL/pluronic F-127 blend nanofibers.
1,25(OH)2D3/VID400-loaded Nanofiber Formulations Induced CAMP Gene Expression In Vitro.
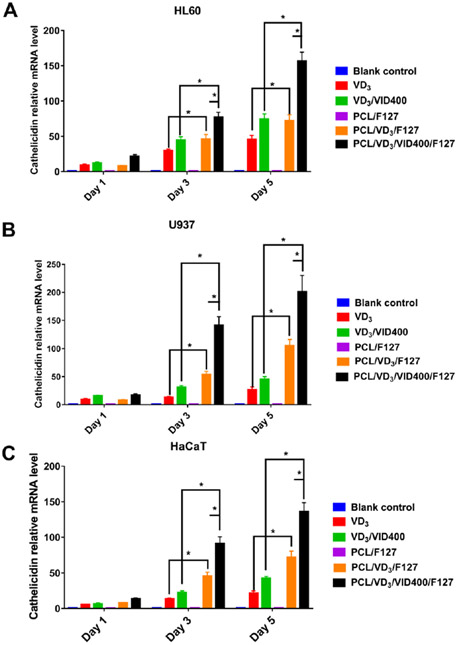
To examine the capability in inducing CAMP gene expression, the U937, HL60 and HaCaT cells were treated with 1,25(OH)2D3 and 1,25(OH)2D3/VID400-loaded nanofibers for 1, 3 and 5 days (Figure 7 A-C). As determined by qPCR, both 1,25(OH)2D3 and 1,25(OH)2D3/VID400-loaded nanofiber significantly induced CAMP expression in U937, HL60 and HaCaT cells, but was strongest in the U937 cells (Figure 7A). CAMP induction continued throughout the 5 days of treatment and was significantly higher by the drug-loaded nanofibers than by either free 1,25(OH)2D3 or 1,25(OH)2D3/VID400. Meanwhile, the 1,25(OH)2D3/VID400 loaded nanofibers induced higher levels of CAMP than the 1,25(OH)2D3 loaded nanofibers, revealing that the CYP24A1 inhibitor VID400 enhanced CAMP expression.
Figure 7.
CAMP gene relative expression of (A) HL60, (B) U937, (C) HaCaT cells after treatment with different nanofiber formulations for 1, 3, and 5 days. Blank control: no treatment. VD3: free 1,25(OH)2D3. VD3/VID400: free 1,25(OH)2D3 and VID400. F127/PCL: PCL/pluronic F-127 blend nanofibers. F127/PCL/VD3: 1,25(OH)2D3 loaded PCL/pluronic F-127 blend nanofibers. F127/PCL/VD3/VID400: 1,25(OH)2D3 and VID400 co-loaded PCL/pluronic F-127 blend nanofibers. (*p<0.05.)
1,25(OH)2D3/VID400-loaded Nanofiber Formulations Induced Antimicrobial Peptide Production In Vitro.
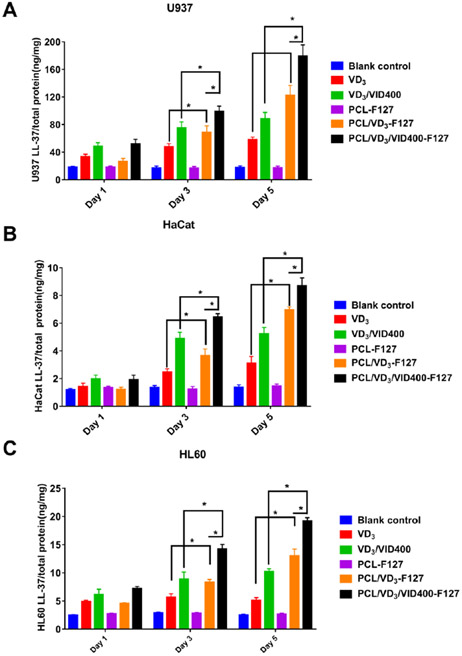
The induction of hCAP18/LL-37 in HL60, U937 and HaCaT cells was significantly higher when incubated with 1 mg/ml 1,25(OH)2D3-loaded and 1,25(OH)2D3/VID400 loaded PCL nanofibers for 3 and 5 days than the control and free drugs in HL60 (Figure 8 A), U937 (Figure 8 B) and HaCaT (Figure 8 C), respectively. Similarly, administration of 1 mg/ml 1,25(OH)2D3/VID400-loaded PCL nanofibers induced the highest amount of hCAP18/LL-37 in HL60, U937 and HaCaT cells among the treatment groups after incubation for 3 and 5 days. In addition, U937 cells produced higher amounts of hCAP18/LL-37 than HaCaT and HL60 cells under the same conditions.
Figure 8.
LL-37 expression of (A) HL60, (B) U937, (C) HaCaT cells after treatment with different nanofiber formulations for 1, 3, and 5 days. Blank control: no treatment. VD3: free 1,25(OH)2D3. VD3/VID400: free 1,25(OH)2D3 and VID400. F127/PCL: PCL/pluronic F-127 blend nanofibers. F127/PCL/VD3: 1,25(OH)2D3 loaded PCL/pluronic F-127 blend nanofibers. F127/PCL/VD3/VID400: 1,25(OH)2D3 and VID400 coloaded PCL/pluronic F-127 blend nanofibers. (*p<0.05)
CAMP Gene Expression and Antimicrobial Peptide Production Ex Vivo.
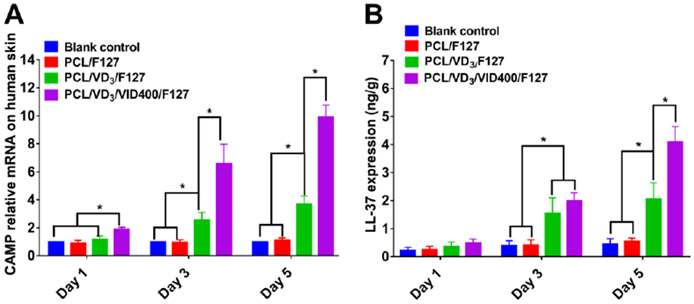
We further examined the ability of nanofiber formulations to induce hCAP18/LL-37 expression in ex vivo human skin explants. A 1,25(OH)2D3-loaded or 1,25(OH)2D3/VID400-loaded PCL nanofiber membrane was placed in a 1-mm deep wound in human skin explants and cultured for 1, 3 and 5 days. As shown in Figure 9A, the 1,25(OH)2D3/VID400-loaded PCL nanofibers induced CAMP in the wound to significantly higher levels at days 3 and 5 as compared with the vehicle control group. We quantified the expression of hCAP18/LL-37 by ELISA and the results showed a similar trend as the CAMP mRNA (Figure 9B). The 1,25(OH)2D3/VID400 nanofiber dressings induced the highest level of hCAP18/LL-37 at the wounds.
Figure 9.
Cathelicidin gene relative expression (A) and LL-37 expression (B) via treating fresh ex vivo human skin tissue with different nanofibers in Day 1, 3 and 5. Blank control: no treatment. F127/PCL: PCL/pluronic F-127 blend nanofibers. F127/PCL/VD3: 1,25(OH)2D3 loaded PCL/pluronic F-127 blend nanofibers. F127/PCL/VD3/VID400: 1,25(OH)2D3 and VID400 coloaded PCL/pluronic F-127 blend nanofibers. (*p<0.05)
Induction Cathelicidin Protein/LL-37 In Vivo.
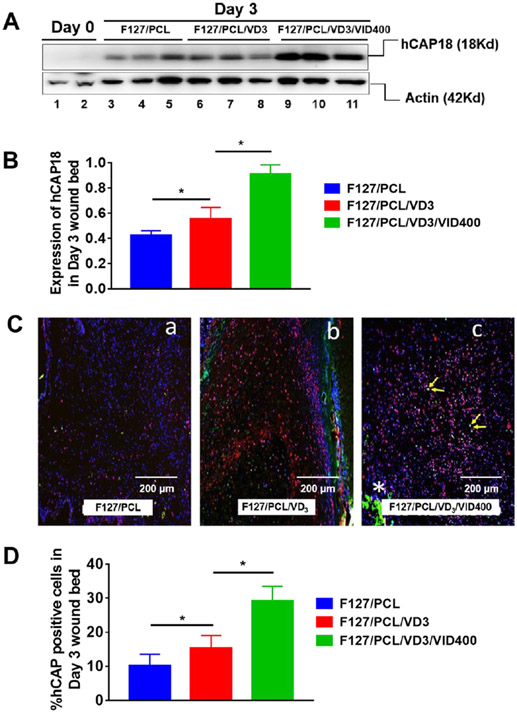
We further tested 1,25(OH)2D3 and VID400 co-loaded nanofiber membranes in excisional skin wounds created in human CAMP transgenic mice (Figure 10). After day 3 of implantation, the results showed co-delivery of 1,25(OH)2D3 and VID400 greatly enhanced hCAP18/LL-37 expression/production compared to PCL pristine nanofibers and PCL nanofibers loaded with 1,25(OH)2D3 alone (Figure 10A and B). We detected increased numbers of hCAP18+ cells in the wound bed of 1,25(OH)2D3-loaded PCL nanofibers treated skin wounds, which was further increased in the skin wounds treated with 1,25(OH)2D3 and VID400 co-loaded nanofibers (Figure 10C). We observed hCAP18+F4/80+ macrophages in the wound bed (Figure 10C, panel c, yellow arrows).
Figure 10.
1,25(OH)2D3 and 1,25(OH)2D3/VID400 nanofiber membranes promote LL-37 expression in the CAMPTg/Tg:KO/KO transgenic mouse wound model. (A) Induction of hCAP18 expression in Day 3 skin wounds post treatment with nanofibers containing 1,25(OH)2D3 and 1,25(OH)2D3/VID400. Protein samples were extracted from day 3 skin wounds of CAMPTg/Tg:KO/KO mice treated with F127/PCL only, F127/PCL/VD3 and F127/PCL/VD3/VID400. Western blots were performed using specific anti-hCAP18 antibody. Actin was used as a loading control. (B) Induction of hCAP18 expression in day 3 skin wounds post treatment with nanofibers containing 1,25(OH)2D3 and 1,25(OH)2D3/VID400. Significant induction of hCAP18 was observed in 1,25(OH)2D3 and 1,25(OH)2D3/VID400 loaded nanofibers compared to the PCL control. (C) Immunofluorescence staining of hCAP18/LL-37 protein (in red) and F4/80 (green) on day 3 samples post wounding in the presence of F127/PCL fibers alone and with 1,25(OH)2D3, as well as 1,25(OH)2D3/VID400. Nuclei were counterstained with DAPI (in blue). (a) F127/PCL; (b) F127/PCL/VD3 and (c) F127/PCL/VD3/VID400. Increased number of hCAP18+ cells were detected in the wound bed of F127/PCL/VD3 treated skin wounds, which was further increased in the skin wounds with F127/PCL/VD3/VID400 nanofibers. (D) Quantitative analysis of the number of hCAP18+ cells detected in the wound bed in day 3 for skin wounds with post-treatment of nanofibers containing 1,25(OH)2D3 and 1,25(OH)2D3/VID400. F127/PCL: PCL/pluronic F-127 blend nanofibers. F127/PCL/VD3: 1,25(OH)2D3 loaded PCL/pluronic F-127 blend nanofibers. F127/PCL/VD3/VID400: 1,25(OH)2D3 and VID400 coloaded PCL/pluronic F-127 blend nanofibers. (*p<0.05)
DISCUSSION
CYP24A1 plays the key role in tuning levels and function of active vitamin D.24 Therefore, inhibition of CYP24A1 opens up a very wide field of possible applications ranging from basic research to the prevention and treatment of diseases, including SSIs.25 There is general agreement that unbalanced high and/or long-lasting expression of CYP24A1 can contribute to the pathology of diseases that otherwise would respond to endogenous or supplemented vitamin D in a favorable way like e.g. chronic kidney disease, bone disease, cancers, and psoriasis.26 In these cases, inhibition of CYP24A1 could be an appropriate strategy to increase the lifetime and thereby the function of active vitamin D.27
The first identified inhibitors for CYP24A1 gene were antifungal imidazole derivatives, such as ketoconazole and liarozole. However, they lack specificity because they inhibit steroidogenesis by interfering broadly with cytochrome P450 enzyme systems.28 Shuster et al. first reported VID400 as a CYP24A1 inhibitor.19 VID400 showed the desired qualities as a strong, selective CYP24A1 inhibitor (IC50: 15.2±3.5 nM) that exhibited only moderate inhibition of CYP27B1 (IC50: 616.17±113.2 nM) and was selected as a candidate for development.24 In addition, VID400 suppressed the degradation of endogenous 1,25(OH)2D3 by blocking CYP24A1 activity.29 Thus, administration of vitamin D compounds with CYP24A1 inhibitory property may enhance and prolong the activity of 1, 25(OH)2D3 and other VDR agonists in target cells.
In this study, we aimed to prepare 1,25(OH)2D3/VID400-co-loaded PCL nanofibers serving as wound dressings which may enhance innate immunity by significantly inducing the endogenous production of hCAP/LL-37. Herein, we mainly consider the application for external wounds. However, if applying this material to internal injuries, we could switch to other polymers that can degrade faster. Electrospun nanofibers are an ideal topical delivery system for the co-delivery of multiple agents because of their proven capacity to encapsulate and deliver physicochemically diverse drugs and ability to modulate drug release kinetics over both short and long time.30, 31 Electrospun nanofiber mats have demonstrated outstanding properties such as high porosity, superior mechanical performance, flexible surface, high surface area and length/diameter ratio.32, 33 Because degradation is slow, the drug release from PCL nanofibers is primarily diffusion of molecules through and desorption from nanofibers. Thus, we could alter the release profiles of PCL by reducing/increasing the nanofiber diameters by varying processing parameters including the solution concentration, solution flow rate, and strength of the electrical field during electrospinning. In addition, altering the crystallinity provides another approach to adjust release profiles through annealing. We could also control the porosity of the fibers and add water-soluble additives to the fibers to modulate release profiles based on the needs (e.g., suppressing the infection for a short or long period). In this study, we demonstrated that administration of encapsulated 1,25(OH)2D3 and VID400 using nanofiber formulations was markedly more efficient for inducing hCAP/LL-37 than equivalent amounts of corresponding free drugs in vitro and in vivo. We showed that electrospun nanofibers provide a sustained release strategy to enhance the efficacy of 1,25(OH)2D3 and VID400.
It is known that injury, infection or inflammation upregulate cathelicidin expression.34, 35 To more closely simulate the clinical situation, we utilized an ex vivo human partial-thickness wound model and an established full-thickness wound mouse model for vitamin D-induced human CAMP transgene expression in mice lacking the murine Camp gene (CAMPTg/Tg:KO/KO) to examine how the 1,25(OH)2D3/VID400-loaded PCL nanofibers performed.36 We applied our nanofibers to wounds in both systems and observed that 1,25(OH)2D3/VID400-loaded PCL electrospun nanofibers enhanced hCAP18/LL37 expression to higher levels than with 1,25(OH)2D3 alone. These findings support our hypothesis that using CYP24A1 inhibitors to prevent the local catabolism of 1,25(OH)2D3 enhances CAMP expression. Therefore, our 1,25(OH)2D3/VID400-loaded PCL electrospun nanofibers provide a potential wound dressing to enhance antimicrobial peptide expression, thus supporting our efforts to generate wound dressings that modulate the immune response to prevent SSIs.
CONCLUSIONS
We demonstrated the local delivery of 1,25(OH)2D3 and VID400 using electrospun nanofibers as a carrier. VID400 could significantly up-regulate the cathelicidin gene relative expression in HL60, U937 and HaCaT cells in combination with 1,25(OH)2D3. Co-incubation with 1,25(OH)2D3/VID400-loaded PCL nanofibers induced significantly higher cathelicidin gene and protein expression in HL60, U937 and HaCaT cells at day 5 compared with free drugs in vitro. Additionally, 1,25(OH)2D3/VID400-loaded nanofibers induced higher hCAP18/LL-37 protein expression in human skin explants. Finally, 1,25(OH)2D3/VID400-loaded nanofibers induced higher hCAP18/LL-37 protein expression when treating skin wounds in human CAMP transgenic mice. Our findings suggest locally sustained delivery of 1,25(OH)2D3/VID400 from nanofiber dressings could significantly enhance innate immunity by inducing antimicrobial peptide production for preventing SSIs.
Supplementary Material
ACKNOWLEDGMENTS
This work was partially supported by startup funds from the University of Nebraska Medical Center (UNMC), National Institute of General Medical Science (NIGMS) of the National Institutes of Health under Award Numbers R01GM123081 and R01GM138552, UNMC Regenerative Medicine Program pilot grant, Nebraska Research Initiative grant, and NE LB606. I.E.L. was supported by the 2019 Audrey and George Varsevelt LPI Graduate Fellowship, the 2019–20 Mark Spoonenburgh LPI Graduate Fellowship, and the 2020–21 P.F. and Nellie Buck Yerex Graduate Scholarship from OSU.
Contributor Information
Yajuan Su, Department of Surgery-Transplant and Mary & Dick Holland Regenerative Medicine Program, University of Nebraska Medical Center, Omaha, Nebraska 68198, United States.
Gitali Ganguli-Indra, Department of Pharmaceutical Sciences, College of Pharmacy, Oregon State University, Corvallis, Oregon, 97331, United States; Knight Cancer Institute, Oregon Health & Science University, Portland, Oregon, 97239, United States.
Nilika Bhattacharya, Department of Pharmaceutical Sciences, College of Pharmacy, Oregon State University, Corvallis, Oregon, 97331, United States.
Isabelle E. Logan, Linus Pauling Institute, Department of Biochemistry and Biophysics, Oregon State University, Corvallis, Oregon, 97331, United States
Arup K. Indra, Department of Pharmaceutical Sciences, College of Pharmacy, Oregon State University, Corvallis, Oregon, 97331, United States; Knight Cancer Institute, Oregon Health & Science University, Portland, Oregon, 97239, United States; Department of Dermatology, Oregon Health & Science University, Portland, Oregon, 97239, United States; Linus Pauling Institute, Department of Biochemistry and Biophysics, Oregon State University, Corvallis, Oregon, 97331, United States
Adrian F. Gombart, Linus Pauling Institute, Department of Biochemistry and Biophysics, Oregon State University, Corvallis, Oregon, 97331, United States
Shannon L. Wong, Department of Surgery-Plastic Surgery, University of Nebraska Medical Center, Omaha, Nebraska 68198, United States
Jingwei Xie, Department of Surgery-Transplant and Mary & Dick Holland Regenerative Medicine Program, University of Nebraska Medical Center, Omaha, Nebraska 68198, United States; Department of Mechanical and Materials Engineering, College of Engineering, University of Nebraska-Lincoln, Lincoln, Nebraska 68588, United States.
REFERENCES
- (1).Wenzel RP Minimizing surgical-site infections. N. Engl. J. Med 2010, 362, 75. [DOI] [PubMed] [Google Scholar]
- (2).Gottrup F. Prevention of surgical-wound infections, N. Engl. J. Med 2000, 342, 202–204. [DOI] [PubMed] [Google Scholar]
- (3).Berríos-Torres SI; Umscheid CA; Bratzler DW; Leas B; Stone EC; Kelz RR; Reinke CE; Morgan S; Solomkin JS; Mazuski JE; Dellinger EP; Itani KMF; Berbari EF; Segreti J; Parvizi J; Blanchard J; Allen G; Kluytmans JAJW; Donlan R; Schecter WP; Healthcare Infection Control Practice Advisory Committee. Centers for disease control and prevention guideline for the prevention of surgical site infection, JAMA Surg. 2017, 152, 784–791. [DOI] [PubMed] [Google Scholar]
- (4).Shiroky J; Lillie E; Muaddi H; Sevigny M; Choi WJ; Karanicolas PJ The impact of negative pressure wound therapy for closed surgical incisions on surgical site infection: a systematic review and meta-analysis. Surgery 2020, 167, 1001–1009. [DOI] [PubMed] [Google Scholar]
- (5).Eagye KJ; Kim A; Laohavaleeson S; Kuti JL; Nicolau DP Surgical site infection: does inadequate antibiotic therapy affect patient outcomes?. Surg. Infect. (Larchmt) 2009, 10, 323–331. [DOI] [PubMed] [Google Scholar]
- (6).Spellberg B. The future of antibiotics. Crit. Care 2014, 18, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Morehead MS; Scarbrough C Emergence of global antibiotic resistance. Prim. Care 2018, 45, 467–484. [DOI] [PubMed] [Google Scholar]
- (8).Bergman P; Raqib R; Rekha RS; Agerberth B; Gudmundsson GH Hosted directed therapy against infection by boosting innate immunity. Front. Immunol 2020, 11, 1209–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Silva MT; Correia-Neves M Neutrophils and macrophages: the main partners of phagocyte cell systems. Front. Immunol 2012, 3, 174–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Peterson LW; Artis D Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat. Rev. Immunol 2014, 14, 141–153. [DOI] [PubMed] [Google Scholar]
- (11).Gombart AF; Luong QT; Koeffler HP Vitamin D compounds: activity against microbes and cancer. Anticancer Res. 2006, 26, 2531–2542. [PubMed] [Google Scholar]
- (12).Lowry MB; Guo C; Borregaard N; Gombart AF Regulation of the human cathelicidin antimicrobial peptide gene by 1α,25-dihydroxyvitamin D3 in primary immune cells. J. Steroid Biochem. Mol. Biol 2014, 143, 183–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Schauber J; Dorschner RA; Coda AB; Büchau AS; Liu PT; Kiken D; Helfrich YR; Kang S; Elalieh HZ; Steinmeyer A; Zügel U; Bikle DD; Modlin RL; Gallo RL Injury enhances TLR2 function and antimicrobial peptide expression through a vitamin D-dependent mechanism. J. Clin. Invest 2007, 117, 803–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Liu PT; Stenger S; Li H; Wenzel L; Tan BH; Krutzik SR; Ochoa MT; Schauber J; Wu K; Meinken C; Kamen DL; Wagner M; Bals R; Steinmeyer A; ZüGel U; Gallo RL; Eisenberg D; Hewison M; Hollis BWJ; Adams S; Bloom BR; Modlin RL Toll-like receptor triggering of a vitamin D-mediated human antimicrobial response. Science, 2006, 311, 1770–1773. [DOI] [PubMed] [Google Scholar]
- (15).Jiang J; Chen G; Shuler FD; Wang CH; Xie J Local sustained delivery of 25-hydroxyvitamin D3 for production of antimicrobial peptide. Pharm. Res 2015, 32, 2851–2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Jiang J; Zhang Y; Indra AK; Ganguli-Indra G; Le MN; Wang H; Hollins RR; Reilly DA; Carlson MA; Gallo RL 1α,25-dihydroxyvitamin D3-eluting nanofibrous dressings induce endogenous antimicrobial peptide expression. Nanomedicine 2018, 13, 1417–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Deeb KK; Trump DL; Johnson CS Vitamin D signaling pathways in cancer: potential for anticancer therapeutics. Nat. Rev. Cancer 2007, 7, 684–700. [DOI] [PubMed] [Google Scholar]
- (18).Jenkinson C. The vitamin D metabolome: an update on analysis. Cell Biochem. Funct 2019, 37, 408–423. [DOI] [PubMed] [Google Scholar]
- (19).Schuster I; Egger H; Nussbaumer P; Kroemer RT Inhibitors of vitamin D hydroxylases: structure-activity relationships. J. Cell. Biochem 2003, 88, 372–380. [DOI] [PubMed] [Google Scholar]
- (20).Pike JW; Meyer MB The vitamin D receptor: new paradigms for the regulation of gene expression by 1,25-dihydroxyvitamin D-3. Rheum. Dis. Clin. North Am 2012, 38, 13–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Hewison M. Vitamin D and the immune system: new perspectives on an old theme. Rheum. Dis. Clin 2012, 38, 125–139. [DOI] [PubMed] [Google Scholar]
- (22).Puhl S; Li L; Meinel L; Germershaus O Controlled protein delivery from electrospun non-wovens: novel combination of protein crystals and a biodegradable release matrix. Mol. Pharm 2014, 11, 2372–2380. [DOI] [PubMed] [Google Scholar]
- (23).Su Y; Wang H; Mishra B; Narayana JL; Jiang J; Reilly DA; Hollins RR; Carlson MA; Wang G; Xie J Nanofiber dressings topically delivering molecularly engineered human cathelicidin peptides for the treatment of biofilms in chronic wounds. Mol. Pharm 2019, 16, 2011–2020. [DOI] [PubMed] [Google Scholar]
- (24).Schuster I. Cytochromes P450 are essential players in the vitamin D signaling system. Biochim. Biophys. Acta 2011, 1814, 186–199. [DOI] [PubMed] [Google Scholar]
- (25).Schuster I; Bernhardt R Inhibition of cytochromes P450: existing and new promising therapeutic targets. Drug Metab. Rev 2007, 39, 481–499. [DOI] [PubMed] [Google Scholar]
- (26).Bai X; Miao D; Xiao S; Qiu D; St-Arnaud R; Petkovich M; Gupta A; Goltzman D; Karaplis AC CYP24 inhibition as a therapeutic target in FGF23-mediated renal phosphate wasting disorders. J. Clin. Invest 2016, 126, 667–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Chiellini G; Rapposelli S; Zhu J; Massarelli I; Saraceno M; Bianucci AM; Plum LA; Clagett-Dame M; DeLuca HF Synthesis and biological activities of vitamin D-like inhibitors of CYP24 hydroxylase. Steroids 2012, 77, 212–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).St-Arnaud R; Jones G Chapter 6-CYP24A1: structure, function, and physiological role. Vitamin D (Fourth Edition) 2018, 1, 81–95. [Google Scholar]
- (29).King AN; Beer DG; Christensen PJ; Simpson RU; Ramnath N The vitamin D/CYP24A1 story in cancer. Anti-Cancer Agents Med. Chem 2010, 10, 213–224. [DOI] [PubMed] [Google Scholar]
- (30).Ulubayram K; Calamak S; Shahbazi R; Eroglu I Nanofibers based antibacterial drug design, delivery and applications. Curr. Pharm. Des 2015, 21, 1930–1943. [DOI] [PubMed] [Google Scholar]
- (31).Liu W; Thomopoulos S; Xia Y Electrospun nanofibers for regenerative medicine. Adv. Healthcare Mater 2012, 1, 10–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Xue J; Xie J; Liu W; Xia Y Electrospun nanofibers: new concepts, materials, and applications. Acc. Chem. Res 2017, 50, 1976–1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Chen M; Li YF; Besenbacher F Electrospun nanofibers-mediated on-demand drug release. Adv. Healthcare Mater 2014, 3, 1721–1732. [DOI] [PubMed] [Google Scholar]
- (34).Dürr UH; Sudheendra U; Ramamoorthy A LL-37, the only human member of the cathelicidin family of antimicrobial peptides. Biochim. Biophys. Acta Rev. Biomembr 2006, 1758, 1408–1425. [DOI] [PubMed] [Google Scholar]
- (35).Yu JR; Navarro J; Coburn JC; Mahadik B; Molnar J; Holmes IV JH; Nam AJ; Fisher JP Current and future perspectives on skin tissue engineering: key features of biomedical research, translational assessment, and clinical application. Adv. Healthcare Mater 2019, 8, 1801471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Lowry MB; Guo C; Zhang Y; Fantacone ML; Logan IE; Campbell Y; Zhang W; Le M; Indra AK; Ganguli-Indra G; Xie J; Gallo RL; Koeffler H,P; Gombart AF J. Steroid Biochem. Mol. Biol 2020, 198, 105552. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.