Summary
Wheat fixes CO2 by photosynthesis into kernels to nourish humankind. Improving the photosynthesis rate is a major driving force in assimilating atmospheric CO2 and guaranteeing food supply for human beings. Strategies for achieving the above goal need to be improved. Here, we report the cloning and mechanism of CO 2 ASSIMILATION RATE AND KERNEL‐ENHANCED 1 (CAKE1) from durum wheat (Triticum turgidum L. var. durum). The cake1 mutant displayed a lower photosynthesis rate with smaller grains. Genetic studies identified CAKE1 as HSP90.2‐B, encoding cytosolic molecular chaperone folding nascent preproteins. The disturbance of HSP90.2 decreased leaf photosynthesis rate, kernel weight (KW) and yield. Nevertheless, HSP90.2 over‐expression increased KW. HSP90.2 recruited and was essential for the chloroplast localization of nuclear‐encoded photosynthesis units, for example PsbO. Actin microfilaments docked on the chloroplast surface interacted with HSP90.2 as a subcellular track towards chloroplasts. A natural variation in the hexaploid wheat HSP90.2‐B promoter increased its transcription activity, enhanced photosynthesis rate and improved KW and yield. Our study illustrated an HSP90.2–Actin complex sorting client preproteins towards chloroplasts to promote CO2 assimilation and crop production. The beneficial haplotype of Hsp90.2 is rare in modern varieties and could be an excellent molecular switch promoting photosynthesis rate to increase yield in future elite wheat varieties.
Keywords: wheat, CO2 assimilation rate, Heat Shock Protein 90, actin, kernel weight
HSP90.2 mediates the targeted sorting of nucleus‐encoded photosystem preproteins along actin microfilaments to chloroplasts to promote the CO2 assimilation rate and yield in wheat.
Introduction
Plants capture solar energy and convert atmospheric CO2 into carbohydrates to nurture the world. The source for grain filling is a limiting factor for kernels in bread wheat recombinant inbred lines (Alonso et al., 2018). Kernel number (KN) and KW determine wheat yield potential. KW is genetically stable and the primary driving force in the yield increase in the past century of wheat‐breeding practices (Li and Yang, 2017). Multiple signalling pathways control KW, including the ubiquitin‐proteasome pathway (Song et al., 2007) and G‐protein signalling (Mao et al., 2010). Any increase in photosynthetic CO2 assimilation rate increases crop production (Faralli and Lawson, 2020). In unicellular photosynthetic organisms, the artifactual fusion of photosynthetic photosystem I with hydrogenases could efficiently convert solar energy into hydrogen (Redding et al., 2022). But the manipulation of photosynthesis to improve food crops remains unclear.
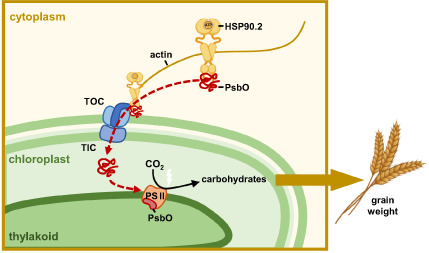
Chloroplasts are the dominant organelle to capture solar energy and produce carbohydrates and many metabolites critical for plants. In Arabidopsis, the chloroplast genome has only 88 protein‐encoding genes, while approximately 3000 nucleus‐encoded proteins depend on the host cell for allocation into chloroplasts (van Wijk, 2004). Three stromal chaperones, HSP93, cpHSP70 and HSP90C, play vital roles in translocating proteins across the chloroplast envelope and function as part of an import motor (Li et al., 2020). Two multi‐subunit translocons of the outer (TOC) and inner (TIC) membranes of the chloroplast envelope are responsible for importing preproteins into the chloroplast (Schnell, 2019). The role of molecular chaperones (HSP90) in protein accumulation in chloroplast remains elusive. Addressing the above question could be the basis for manipulating the photosystem (PS) complex and improving plant yield.
Here, we report the contribution and sorting mechanism of HSP90.2‐B to support photoassimilate and KW. We found that HSP90.2 recognized a series of nucleus‐encoded photosynthesis complex units, for example PsbO, and promoted its location in the chloroplast. Moreover, actin microfilaments bound HSP90.2 for sorting its clients to the chloroplast. A natural variation in the hexaploid wheat HSP90.2‐B promoter enhanced its transcription and protein contents and promoted photosynthesis and KW. This work illustrated the HSP90.2–Actin module mediating the targeted delivery of nucleus‐encoded chloroplast preproteins and highlighted their potential enhancing crop photosynthesis rate and yield‐related traits.
Results
Molecular cloning of CO2 ASSIMILATION KERNEL ENHANCED 1 (CAKE1)
To explore genetic elements underpinning photosynthetic CO2 assimilation rate in crops and their potential to promote grain yield, we isolated CO 2 assimilation kernel enhanced 1 (cake1) mutant with smaller grains reduced in width and length (Figure 1a). There were around 20% decreases in thousand kernel weight (TKW) in multiple environments (Figure 1b). We conducted an exome capture experiment with bulked segreant analysis (BSA) of the BC2F3 segregated samples. We detected differential SNPs enriched in chromosome 7B (Figure 1c). Next, we performed a bulked segreant analysis of RNA‐seq data (BSR) using the segregating population (Chen et al., 2020). The above data locked one candidate gene, TRITD7Bv1G071080, homeolog of TraesCS7B02G149200, in the Chinese Spring genome (Figure 1d). TRITD7Bv1G071080 encoded a Heat Shock Protein 90.2 (HSP90.2) molecular chaperone expressed in most tissues (Figure S1). The SNP (tgG to tgA) created a premature stop codon in the mutant (hsp90.2‐B1) and deleted the middle‐ and C‐terminal domains critical for the client‐ and cochaperone‐binding, respectively (Figure 1e).
Figure 1.
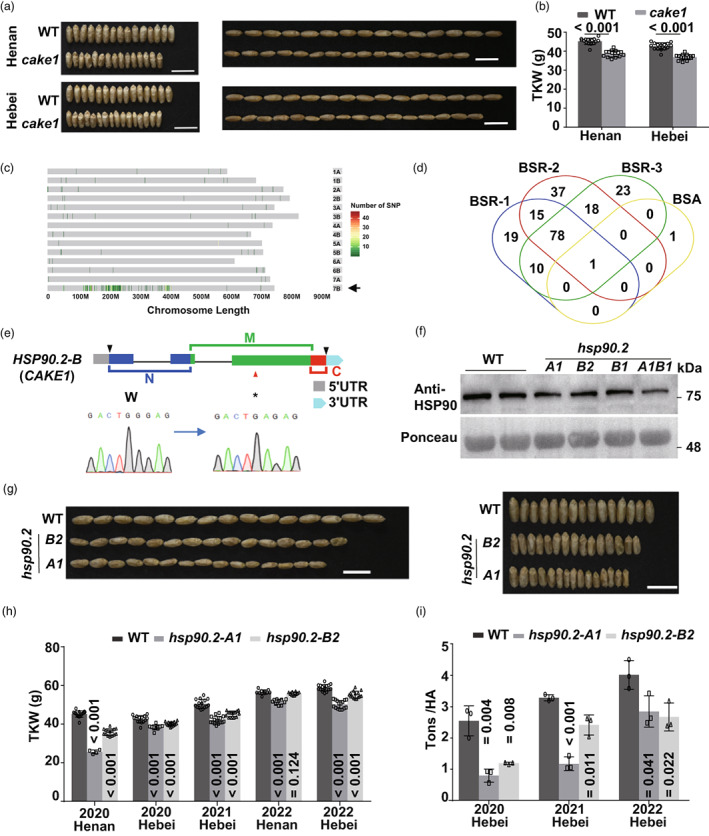
Molecular cloning of CO 2 ASSIMILATION AND KERNEL ENHANCED 1 (CAKE1). (a, b) The kernel phenotype (a) and 1000‐kernel weight (TKW, b, n ≥ 10.) of the cake1 mutant. Bars = 1 cm (hereafter). (c, d) The distribution of SNPs in the segregating population of cake1 BC2F3 lines. (e) Schematic presentation of the mutation (hsp90.2‐B1) site in the HSP90.2‐B gene. (f) Comparison of the endogenous cytosolic HSP90 protein levels in the hsp90.2‐A1, B1 and B2 single mutants and the hsp90.2 knockout mutant. (g) The grain morphology of WT, hp90.2‐A1 and hsp90.2‐B2 mutants from field trials. Bars equal to 1 cm. (h–i) The TKW (h, n ≥ 10) and yield (i, n = 3) of WT, hsp90.2‐A1 and hsp90.2‐B2 mutants in field trials. Data in this photograph represent means ± SD. A two‐tailed unpaired Students' t test P‐value showed the significance (hereafter).
To test if HSP90.2 is CAKE1, we first acquired a pre‐mature stop mutant line of the HSP90.2‐B gene (hsp90.2‐B2) and one pre‐mature stop mutant line in the A genome homeolog gene (hsp90.2‐A1) (Figure S2) from an open Kronos EMS‐mutant database (Krasileva et al., 2017). We backcrossed the above mutants twice to remove off‐target EMS mutations and crossed them to generate an hsp90.2 double mutant to knock out the HSP90.2 gene in tetraploid wheat. The endogenous HSP90 protein level was lower in the single‐ and double‐mutant lines (Figure 1f). There was only a faint HSP90 band in the hsp90.2 knockout mutant. We predicted that the residual signal could come from other cytosolic HSP90 homologs (Figure S3).
In the above single and double mutants, the dysfunction of HSP90.2 curbed grain development and reduced grain sizes (Figure 1g) and TKW (Figure 1h) in multiple field trials at different locations/years. The average yield was reduced by 30%–60% in field trials consecutively over 3 years (Figure 1i). The above hsp90.2 mutants produced smaller grains, with significant reductions in the length, width and thickness (Figure S4). In consistence, the mutant grains accumulated less starch but more protein (Table 1). The above loss‐of‐functional data demonstrated that HSP90.2 played a remarkable role in determining wheat kernel weight, an essential component of final yield.
Table 1.
Agricultural traits of WT, hsp90.2‐A1 and related mutants in field trials
| WT | hsp90.2‐A1 | hsp90.2‐B1 | hsp90.2‐B2 | hsp90.2 | |
|---|---|---|---|---|---|
| SPN |
14.0 ± 1.43 n = 31 |
14.0 ± 1.28 P = 0.4966, n = 36 |
14.5 ± 1.82 P = 0.0796, n = 37 |
13.32 ± 1.38 P = 0.0274, n = 43 |
13.46 ± 1.48 P = 0.0915, n = 30 |
| BF (N) |
10.95 ± 2.07 n = 26 |
9.79 ± 2.73 P = 0.05, n = 25 |
9.67 ± 2.9 P = 0.034, n = 33 |
9.95 ± 2.09 P = 0.032, n = 42 |
9.48 ± 3.25 P = 0.033, n = 24 |
| SPW (g) |
2.72 ± 0.39 n = 26 |
2.53 ± 0.43 P = 0.032, n = 36 |
2.54 ± 0.48 P = 0.05, n = 37 |
2.25 ± 0.30 P < 0.0001, n = 42 |
2.40 ± 0.38 P = 0.001, n = 30 |
| SPL (cm) |
5.83 ± 0.43 n = 31 |
5.63 ± 0.45 P = 0.0031, n = 36 |
6.03 ± 0.84 P = 0.0132, n = 37 |
5.65 ± 0.67 P = 0.0845, n = 43 |
5.73 ± 0.60 P = 0.2016, n = 30 |
| GNS |
36.19 ± 4.66 n = 31 |
39.19 ± 7.76 P = 0.0323, n = 36 |
35.68 ± 7.10 P = 0.1856, n = 37 |
24.93 ± 4.95 P < 0.0001, n = 43 |
27.97 ± 3.30 P < 0.0001, n = 30 |
| GWS (g) |
2.11 ± 0.34 n = 31 |
1.94 ± 0.36 P = 0.0287, n = 36 |
1.92 ± 0.42 P = 0.0314, n = 37 |
1.35 ± 0.27 P < 0.0001, n = 43 |
1.58 ± 0.20 P < 0.0001, n = 30 |
| Pro (%) |
10.68 ± 1.36 n = 55 |
12.28 ± 1.89 P < 0.0001, n = 62 |
12.01 ± 1.15 P < 0.0001, n = 39 |
14.97 ± 1.87 P < 0.0001, n = 60 |
12.22 ± 1.87 P < 0.0001, n = 56 |
| Star (%) |
78.46 ± 1.00 n = 55 |
76.21 ± 1.91 P < 0.0001, n = 62 |
77.09 ± 1.44 P < 0.0001, n = 39 |
72.76 ± 1.83 P < 0.0001, n = 60 |
77.00 ± 1.58 P < 0.0001, n = 56 |
Note: Data are represented as mean ± SD, and a one‐tailed unpaired t test indicates P‐values.
Abbreviations: BF, stem‐breaking force; GNS, Grain number per spike; GWS, Grain weight per spike; SPW, spike weight; SPL, spike length; SPN, Spikelet number; SPL, Spike length (cm), Pro, protein content; Star, starch content.
HSP90.2 positively regulates the CO2 assimilation rate and kernel weight
To further exclude the effects of potential off‐target EMS mutations tightly linked with cake1, we suppressed the endogenous HSP90.2 by RNAi using the N‐ or C‐terminus of HSP90.2. The HSP90.2 RNAi line accumulated less HSP90 protein (Figure 2a), which mimicked the hsp90.2 mutants (Figure 1f). In a controlled condition, the leaves of the HSP90.2 RNAi lines showed early senescence that WT (Figure 2b). There were significant reductions in the chlorophyll contents (Figure 2c) and CO2 assimilation rates (Figure 2d) in the HSP90.2 RNAi lines.
Figure 2.
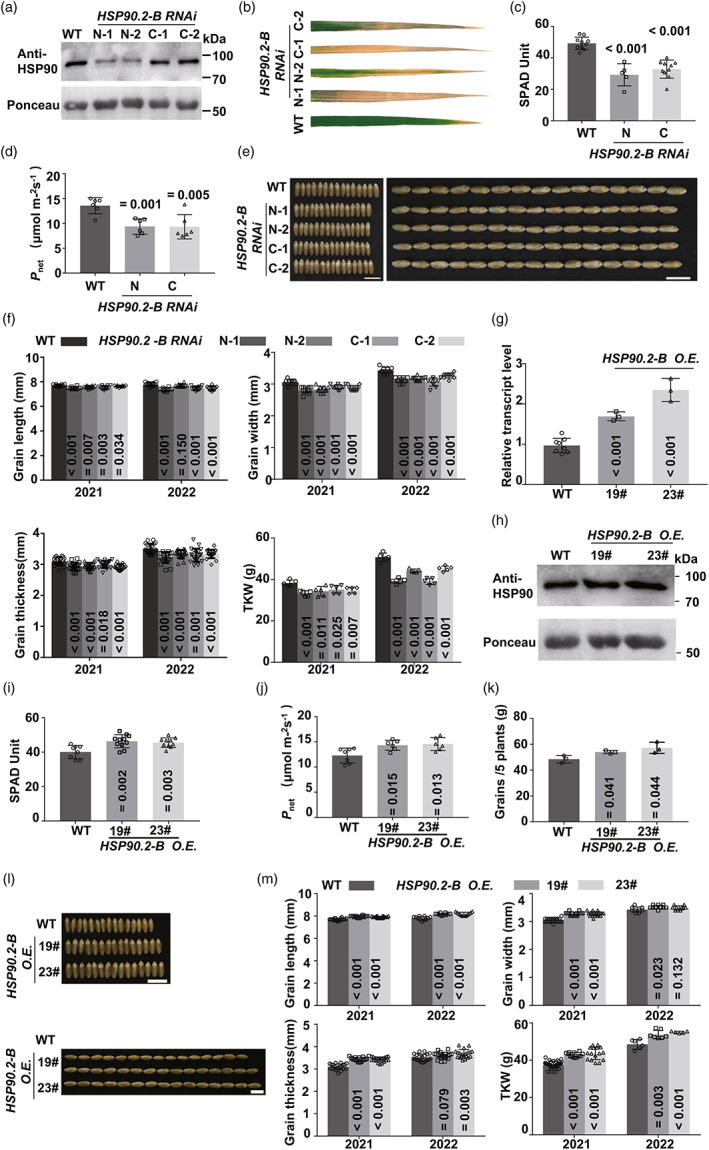
Genetic validation of HSP90.2‐B as CAKE1. (a) The endogenous HSP90 protein levels in WT and HSP90.2 RNAi transgenic lines. (b–d) Comparison of the leaves phenotype (b), chlorophyll contents (c, n ≥ 8) and photosynthesis rates (d, n = 6) between the HSP90.2 RNAi transgenic lines and WT. (e) Comparison of the grain shapes from WT and HSP90.2 RNAi lines. Bars = 1 cm. (f) Phenotypic data of grains from WT, and HSP90.2 RNAi lines, including grain length, width, thickness and TKW (n ≥ 10). (g, h) The gene expression level (g, n ≥ 4) and endogenous HSP90 protein contents (h) in WT and HSP90.2 over‐expression (HSP90.2 O.E.) lines. (i, j) Comparison of the chlorophyll contents (i, n = 6) and photosynthesis rates (j, n = 6) between the HSP90.2 O.E. transgenic lines and WT. (k) The effect of HSP90.B O.E. transgenic on wheat yield (five plants per row). (l, m) Comparison of the grain shapes (l) and phenotypic statistics (m, n ≥ 10) from WT and HSP90.2 O.E. lines.
The HSP90.2 RNAi lines produced smaller grains in a controlled environment (Figure S5) and the field condition (Figure 2e). The suppression of HSP90.2 significantly decreased grain length, width, thickness and KW in a field trial (Figure 2f) and the growth chamber (Figure S5). These results could exclude potential roles of off‐target EMS mutations in the mutant phenotypes and further confirmed that HSP90.2 was necessary to maintain regular CO2 assimilation rate and kernel weight.
To test whether HSP90.2 is CAKE1, we created the HSP90.2 over‐expression (HSP90.2 O.E.) lines in the Kronos genetic background. The over‐expression of HSP90.2 boosted the total transcript level of HSP90.2 (Figure 2g). The endogenous HSP90 protein accumulated in the HSP90.2 O.E. lines (Figure 2h). In a growth chamber, the leaves of the HSP90.2 O.E. lines contained more chlorophyll (Figure 2i). The CO2 assimilation rates were significantly higher in HSP90.2 O.E. lines than in WT (Figure 2j).
In a field trial, the yield of HSP90.2 O.E. lines was higher than WT (Figure 2k). The transgenic lines produced bigger grains in the field (Figure 2l) and the growth chambers (Figure S6). The grains of HSP90.2 O.E. lines showed significant increases in the length, width, thickness and KW compared with WT in field trials (Figure 2m) and the controlled environment (Figure S6). Therefore, HSP90.2 was necessary and sufficient to enhance photo‐assimilation and increase kernel size/weight.
HSP90.2 recognizes nucleus‐encoded photosynthesis complex proteins and is critical for PsbO accumulation in chloroplasts
To explore the mechanism underlying the regulation of HSP90.2 on photo assimilation, we examined the photosynthesis‐related traits of hsp90.2 mutants. In the field, leaves of the hsp90.2 mutants contained lower chlorophylls than WT (Figure 3a). The photosynthesis rate declined in the hsp90.2‐A1, ‐B1 and hsp90.2 knockout‐mutant lines (Figure 3b). We then analysed the proteins interacting with HSP90.2. We identified multiple photosynthesis complexes (PS) members, including an extrinsic member PsbO (Table S1) (Ferreira et al., 2004). PsbO is the extrinsic member to stabilize and protects the core complex (Wei et al., 2016). Therefore, we used PsbO as a model to study the protection of HSP90.2‐B on photosystem preproteins.
Figure 3.
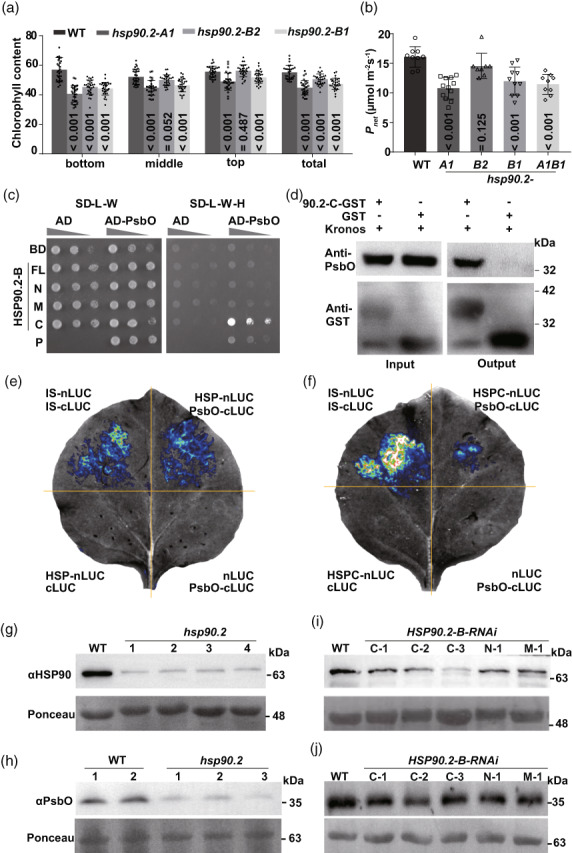
HSP90.2 positively regulates the photosynthesis rate and directly targets PsbO for its accumulation in the chloroplast. (a, b) The chlorophyll contents (a, n ≥ 20) and photosynthesis rates (b, n ≥ 10) in WT and hsp90.2 mutants. (c) The in vitro protein–protein interaction between PsbO and different domains of HSP90.2. (d) The binding between recombinant HSP90.2 C‐terminus and PsbO proteins. (e, f) The in vivo protein–protein interaction between full‐length HSP90.2 (e) and HSP90.2 C‐terminus (f) with PsbO. The WKS1.1 Interlinker START (IS) domain formed a homodimer and worked as the positive control. (g) The endogenous content of cytosolic HSP90 protein in the hsp90.2 knockout mutant. (h) The accumulation of PsbO in the chloroplasts of the hsp90.2 knockout mutant. (i) The endogenous content of cytosolic HSP90 protein in the HSP90.2 RNAi transgenic lines. (j) The accumulation of PsbO in the chloroplasts of the HSP90.2 RNAi transgenic lines.
In the yeast two‐hybrid (Y2H) system, PsbO interacted with the C‐terminus of HSP90.2, suggesting their direct binding in vitro (Figure 3c). Moreover, the recombinant HSP90.2 C‐terminus pulled PsbO from the wheat leaf total protein extract, indicating their potential interaction in vivo (Figure 3d). Furthermore, the full‐length HSP90.2 bound PsbO, producing a positive signal in a split‐luciferase experiment (Figure 3e). The C‐terminus of HSP90.2 was sufficient for the binding with PsbO in the split‐luciferase experiment (Figure 3f). The above experimental results supported the PPI between HSP90.2 and PsbO. But how could HSP90.2 contribute to the biological function of its clients?
To investigate the effect of HSP90.2 on its clients, we tested the distribution of PsbO in hsp90.2 knockout mutant and dysfunction lines. The tetraploid wheat hsp90.2 mutation knocked out the endogenous HSP90.2 and showed an apparent reduction in the total content of cytosolic HSP90 (Figure 3g). In the hsp90.2 knockout mutant, the PsbO declined in the chloroplast (Figure 3h).
We next confirmed the role of HSP90.2 in PsbO accumulation inside chloroplast using the HSP90.2 RNAi transgenic lines. In different transgenic lines suppressing HSP90.2 by its fragments, the endogenous content of HSP90 declined by differential ratios (Figure 3i). The content of PsbO in chloroplast was significantly lower in the transgenic lines than in WT (Figure 3j). The above data confirmed that HSP90.2 played a critical role in the accumulation of PsbO in chloroplasts.
Actin microfilaments mediate the sorting of HSP90.2/clients to chloroplasts
To address the effect of HSP90.2 on clients' translocation, we analysed the interacting members of HSP90.2 and noticed multiple actin members (Table S2). HSP90.2 interacted with actin (TraesCS1A02G020500.1) through different functional domains in a Y2H experiment (Figure 4a). The recombinant actin‐His fusion protein was detected in the pull‐down sample of HSP90.2‐GST, not in GST control, confirming the direct PPI between actin and HSP90.2 in vitro (Figure 4b).
Figure 4.
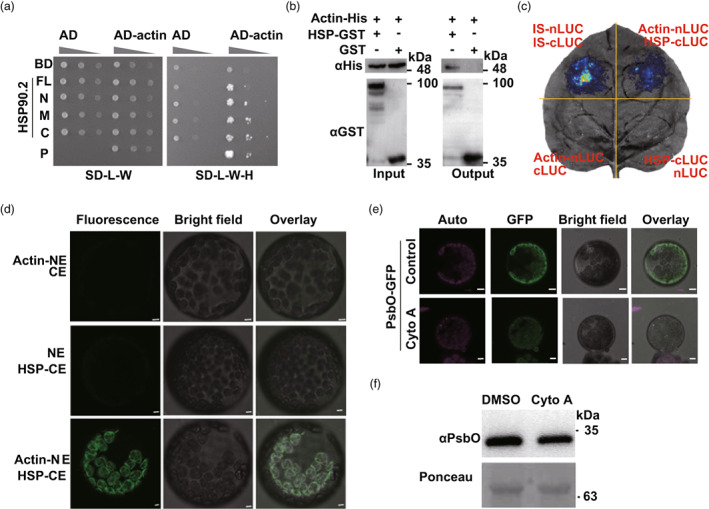
Actin microfilaments mediate the sorting of HSP90.2/clients towards chloroplasts. (a) The in vitro protein–protein interaction between HSP90.2 and Actin. (b) The binding between recombinant the full‐length HSP90.2 and Actin proteins. (c) The in vivo protein–protein interaction between HSP90.2 and Actin. (d) The subcellular binding sites of HSP90.2 and Actin. (e, f) The effect of an inhibitor against Actin polymerization on the accumulation of PsbO‐GFP (e) of endogenous PsbO (f) inside the chloroplast. Bar = 10 μm.
Next, we examined the in vivo PPI between HSP90.2 and actin. The HSP90.2 interacted with actin, which released a luciferase signal in a split‐luciferase system (Figure 4c). The PPI between HSP90.2 and actin occurred at the peridermal region of chloroplasts (Figure 4d). Actin filaments coprecipitate with chloroplasts in an intact out membrane‐depended manner (Kumatani et al., 2006). Exogenous inhibitors against actin polymerization (cytochalasins A, Cyto A) (Himes and Houston, 1976) remarkably reduced the GFP fluorescence of PsbO‐GFP in the chloroplast (Figure 4e). The endogenous PsbO protein accumulated at lower levels in the Cyto A‐treated chloroplasts (Figure 4f). The above data confirmed that HSP90.2 and actin microfilaments were crucial for accumulating preproteins into the chloroplast.
A natural HSP90.2 beneficial allele enhances the CO2 assimilation rate, kernel weight and yield
We analysed the relationships with genome resequencing results to investigate the relationship of the HSP90.2‐Actin module with grain weight in natural populations (Wang et al., 2020). We detected hexaploid wheat cultivars possessing HSP90.2‐B haplotype II (Hsp90.2‐B HapII) with tandem SNPs in the promoter region (Figure 5a). We crossed two elite varieties containing different HSP90.2‐B haplotypes and developed RIL lines. The above SNPs promoted the expression level of HSP90.2‐B in the offspring with the Hsp90.2‐B‐HapII (Figure 5b). The endogenous HSP90 protein was more abundant in lines with Hsp90.2‐B‐HapII than Hsp90.2‐B‐HapI (Figures 5c,d).
Figure 5.
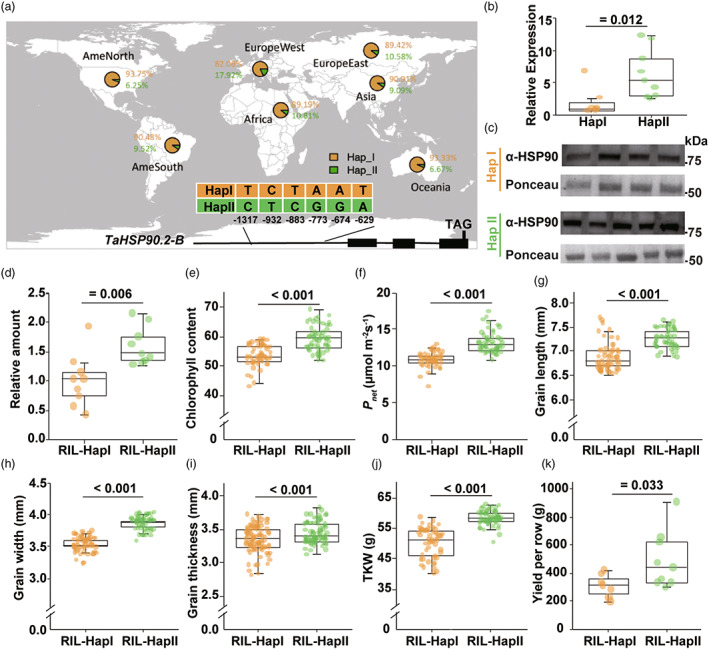
A natural variation in the HSP90.2 promoter enhanced its function and increased wheat yield. (a) The schematic presentation and distribution of tandem single nucleotide polymorphisms (SNPs) in the promoter region of the HSP90.2 gene. (b) The accumulation of cytosolic HSP90 protein in RIL lines with Hsp90.2‐B‐HapI or Hsp90.2‐B ‐HapII. (c, d) Comparison of transcriptional level (c) or protein contents (d) in Hsp90.2‐B‐HapI or Hsp90.2‐B‐HapII RIL lines. (e–f) The net photosynthesis rates and chlorophyll contents in the flag leaf at the grain filling stage in Hsp90.2‐B‐HapI or Hsp90.2‐B‐HapII RIL lines. (g–i) The kernel length (g), width (h) and thickness (i) of RIL lines with Hsp90.2‐B‐HapI or Hsp90.2‐B‐HapII. n ≥ 10. (j, k) The kernel weight (j, n ≥ 10) and single‐row yields (k, n ≥ 5) of RIL lines with Hsp90.2‐B‐HapI or Hsp90.2‐B‐HapII.
The increases in the expression level and protein content promoted the biological function of Hsp90.2‐B. In field conditions, the lines with Hsp90.2‐B‐HapII accumulated more chlorophyll than those with HapI at the grain filling stage (Figure 5e). The net CO2 assimilation rates were markedly higher in RIL lines with Hsp90.2‐B‐HapII than HapI (Figure 5f). As a result, the grain length, width and thickness of Hsp90.2‐B‐HapII lines were significantly higher than the HapI lines (Figure 5g–i). The Hsp90.2‐B‐HapII plants showed significant increases in TKW (Figure 5j) and yield (Figure 5k). Therefore, the Hsp90.2‐B‐HapII beneficial allele propelled photosynthesis rate and yield potentials, similar to over‐expression lines.
Discussion
This work illustrated the contribution of HSP90.2 to wheat CO2 assimilation rate and kernel weight. Actin microfilaments bound HSP90.2 to increase the chance of HSP90.2/clients reaching chloroplasts. The HSP90.2–Actin network is critical in allocating chloroplast preproteins, promoting CO2 assimilation and crop yield.
The discovery of the HSP90.2–Actin interaction provided information for the previous model about sorting preproteins into chloroplasts. From pea chloroplast envelope membranes, Toc159 co‐immunoprecipitated with actin (Jouhet and Gray, 2009a), which raised the question of whether an actin‐TOC–TIC–VIPP1 complex could mediate the chloroplast importation of photosynthesis proteins (Jouhet and Gray, 2009b). However, there needed to be follow‐up reports. Based on the subcellular localization patterns of HSP90.2‐B–actin interaction (Figure 4d), we hypothesized that the floating actin microfilaments extended from the chloroplast could increase the chance for HSP90.2B/clients to reach the chloroplast. Disturbations of HSP90.2B or actin polymerization reduced the accumulation of PsbO in chloroplasts (Figures 3h and 4f) and photosynthesis. By folding its clients, for example PsbO, HSP90.2 positively regulated the CO2 assimilation rate as evidenced by the agricultural traits of cake1 mutant and the RNAi transgenic lines. The photosynthetic members could be regulated at the transcription level, post‐translational modification and protein homeostasis (Gao et al., 2022; Jonwal et al., 2022). This work illustrated a positive, not linear correlation between the HSP90.2 function and the CO2 assimilation rate or yield. Based on field tests, HSP90.2 accounted for 30–60% of wheat yield (Figure 1). Hence, the HSP90.2–Actin complex contributed remarkably to wheat production.
The Hsp90.2‐B–HapII discovered in hexaploid wheat cultivars could increase the resilience of wheat to changing environmental conditions by promoting the expression level of HSP90.2. Environmental temperature displays an apparent effect on wheat photosynthesis and growth. The optimal temperature for photosynthesis ranges from 20 to 26 °C, while photosynthesis is significantly reduced in temperature lower than 13 °C (Jolliffe and Tregunna, 1968). The biomass production for wheat maximizes at 25/19 °C but decreases at 19/16 °C and further declines at 13/10 °C, suggesting the inhibition of low temperature on wheat growth (Nagai and Makino, 2009). HSP90.2, as a heat shock protein, is suppressed by low temperatures, which is unfavourable for the accumulation of photosystem elements in chloroplasts to confer photosynthesis (Figure 3). Nevertheless, the Hsp90.2‐B–HapII variation could increase the expression of HSP90.2 in the RIL population (Figure 5b). It is worth testing if this beneficial variation could enhance HSP90.2 in different environmental conditions to increase photosynthesis and yield.
The HSP90–SGT1–RAR1 complex protects a wide range of disease‐resistance proteins (Takahashi et al., 2003). Arabidopsis HSP90 binds RPP4 (RECOGNITION OF PERONOSPORA PARASITICA 4) and is essential for RPP4‐mediated temperature‐dependent cell death and resistance against pathogens (Bao et al., 2014). HSP90.2‐B–HapII mainly resided in varieties from the Yangtze River wheat‐growing region, where wheat faced high fungal disease pressures, including powdery mildew and head blast. Modern Chinese cultivars harbouring the HSP90.2‐HapII, EEN1, Yangmai158 and Yangmai12 have excellent resilience to fungal stress. Our preliminary work demonstrated that HSP90.2 positively regulates wheat resistance against powdery mildew and stripe rust. Therefore, HSP90.2 has the potential to balance disease resistance and yield in wheat.
Despite the application in past breeding programs, the beneficial HSP90.2‐B–HapII only accounts for 10% of the world's natural wheat populations. The highest ratio (18%) of HSP90.2‐B–HapII existed in European varieties (Figure 5a), where the average wheat yield was 7.15 MT/HA in the last two decades (based on https://ourworldindata.org/grapher/wheat‐yields). In developing countries, for example Pakistan, the ratio is as low as 2% (based on the data of Awais Rasheed), and the average wheat yield was only 2,23 MT/HA in the last two decades. As HSP90.2‐B‐HapII positively regulated wheat yield, we predicted that its ratio could partially contribute to the above difference. Therefore, around 90% of current wheat varieties possess the potential to incorporate the HSP90.2‐B–HapII allele to increase their photosynthesis and yield prospects.
Materials and methods
Plant materials and growth conditions
The EMS mutants and transgenic lines used in this study were in Triticum turgidum (tetraploid, AABB genome), Kronos ecotype background. Wild‐type (WT) seeds were treated with 0.7% EMS overnight at 25 °C with slow rotation (60 rpm) to create the EMS‐mutant library (Uauy et al., 2009). We isolated EMS mutants with advantages in grain weight at M5 generation. We used BC2F3 generation plants for the segregation test and selected homozygous plants for subsequent analyses.
For HSP90.2‐B haplotype analysis, we searched Wheat Union Database in the Genomic variation data sets of wheat (http://wheat.cau.edu.cn/Wheat_SnpHub_Portal/) and identified cultivars with polymorphism in HSP90.2‐B (Wang et al., 2020). We acquired the seeds for corresponding varieties (Hao et al., 2020; Zhou et al., 2020), confirmed the genotype, and increased them with single seeds in a growth chamber. Zhou11550 was crossed with Zhongmai895 to create a double‐parental segregating population. Twenty‐five seeds of the above varieties were sown in 1‐mitre rows in October on a farm in Yuanyang, Henan Province (113.97′E, 35.05′N), China, with a completely random distribution design. The mutants were grown in Wuyi, Hebei Province (115.89′ E, 37.80′N), China, with a Latin Square design. The transgenic seeds of HSP90.2‐B RNAi or HSP90.2‐B O.E. were sown on a local farm in December at Fudan University, Shanghai, China (121.52′E, 31.27′N).
CO2 assimilation rates of the flag leaves at the grain filling stage were analysed with an Li‐COR 6400XT using a standard leaf room (LI‐COR Corporate, Lincoln, NE). The fan speed was 500 rpm, and the mixer setting was ‘off’. The similar position on the same leaf of 10 individual plants was analysed to collect data for each sample. ΔCO2 was quantified and compared between the samples in the morning. We used a DA 7200 NIR Diode Array Based Feed Analyser (PerkinElmer Inc., Shanghai, China) according to the instruction to quantify the starch and protein contents of the grain in the field trail (Hebei, 2022). The third nodes were collected and used to analyse the stem‐breaking force with an automatic Push‐pull gauge (ZHIQU Precision Instruments, Guangzhou, Guangdong, China).
Bulked segregant analysis to clone CAKE1
We extracted genomic DNA from flag leave of BC2F3 generation plants with the cake1 phenotype. The DNA was digested and subjected to an exome capture analysis targeted to 260 M/300 M sequences covering the 150 000 genes annotated based on the IWGSC and AK58 genome sequences according to the user manual (Chengdu Tiancheng Future Technology, Sichuan, China). According to the user manual, we further extracted RNA from mature leaves of 1‐month BC2F3 generation plants using TRIzol Reagent (Merck Sigma‐Aldrich, Chaoyang District, Beijing, China).
We mixed RNA samples from 15 individual homozygous plants together equally into each group. We collected three groups from WT or homozygous mutants to construct PE300 libraries for sequencing in the Illumina Hiseq4000 (Meiji Bio‐Tech, Shanghai, China). We got over 6 Giga of high‐quality data for each group and analysed them to filter homozygous SNPs with reads higher than 2. Deferentially expressed genes were screened by ¦fold change¦ > 2, FDR (q value) < 0.001, and TPM above 20 in any sample using the MA‐plot‐based method with the Random sampling model (MARS). Gene assembly and annotation were conducted according to wheat assembly (TGACv1.1).
Gene cloning, recombinant protein purification, and pull‐down analysis
HSP90.2‐B cDNA was cloned from the cDNA of WT plants (Table S2), integrated into pDNOR207 by BP reaction (Invitrogen, Life Technologies, Carlsbad, CA) and confirmed by Sanger sequencing. The N‐, middle‐ and C‐terminals of HSP90.2‐B were amplified from the above construct and incorporated into pDNOR207 by BP reaction. The coding sequence of actin was cloned from the cDNA of WT plants and integrated into pDNOR207 by BP reaction.
The actin (TraesCS1A02G020500.1) gene was incorporated into a gateway‐compatible destination expression pHIS9 (PT7:His9‐AttR: TT7) LR reaction. The full‐length cDNA and the C‐terminal fragments of HSP90.2‐B were inserted into pET 41b in the frame with GST (PT7:HSP90.2‐GST:TT7, PT7:HSP90.2C‐GST:TT7). The above plasmids were transformed into E. Coli, Rosetta, by a heat shock method. Recombinant proteins were purified from Rosetta (Gou et al., 2009). For protein induction, 0.5 mm IPTG was added into cell cultures at OD600 around 0.8 in a shaker set at 18 °C. According to the user manual, cells were collected from overnight cultures, sonicated to lysis and purified by Ni‐NTA His•Bind® Resin or Glutathione−Agarose (MilliporeSigma, Merck Limited, Shanghai, China).
For the pull‐down experiment, the recombinant proteins were mixed with equal volume and mixed in an eight‐fold GST binding buffer with a rotating mixer at 4 °C for 2 h. Then, 5 μL of prewashed Glutathione−Agarose was added and mixed for two more hours. To remove the unbound proteins, we pelleted the agarose and washed the samples with 10 folds of GST washing buffer four times. We eluted the proteins with GST elution buffer and treated the samples with a 2× SDS loading buffer. For the semi‐pull‐down experiment, we extracted total protein from 2‐week seedlings with 2× Plant active protein extraction buffer (pH 7.5 Tris–HCl 100 mm, EDTA 2 mm, NaCl 300 mm, Triton X‐100 1%, glycerin 10%). We performed the pull‐down experiment as described aforementioned.
We analysed the contents of PsbO with a monoclonal anti‐PsbO antibody (Agrisera, Vännäs, Sweden). We quantified the content of His9‐actin with an HRP‐conjugated monoclonal anti‐His antibody (Abmart, Xuhui District, Shanghai, China). We developed the above samples with a basic ECL kit (AB Clonal) in the Tanon 5200 Multi imaging system (Tanon Science & Technology Co., Ltd., Shanghai, China).
Subcellular localization and transgenic plants
For subcellular localization analysis, HSP90.2‐B was incorporated into pMDC83 (Pro35S: HSP90.2‐B‐GFP) by LR reaction, transformed into Agrobacterium GV3101 and infiltrated into tobacco leaves. At 2 DPI, we cut the leaves into 0.5–1 mm strips with a new sharp blade and then transferred them into the prepared enzyme solution (1.25% Cellulase R10, 0.3% Macerozyme R10, 0.4 m Mannitol, 20 mm KCl, 20 mm MES‐KOH pH 5.7, 10 mm CaCl2 and 0.1% BSA). We performed the enzymolysis in the dark with slight oscillation (60–80 rpm) for 4–6 h. We resuspended the samples with equal volume W5 solution (154 mm NaCl, 125 mm CaCl2, 5 mm KCl and 2 mm MES‐KOH pH 5.7) and gently shook the samples for 15 s to fully release the protoplast. We filtered the protoplasts through a filter cloth into a 50 mL round bottom centrifuge tube and collected them by horizontal centrifugation at 100 g for 10 min. We discarded the supernatant and resuspended the protoplast in an appropriate MMG solution (0.6 m D‐Mannitol, 15 mm MgCl2, 2 mm MES‐KOH pH 5.7). We captured the images using 488‐nm laser excitation and 500‐ to 550‐nm long‐pass emission filters under a Leica TCS SP8 (Leica Microsystems, Mannheim, Germany). Chloroplast autofluorescence was imaged at 600 nm over a long‐pass emission filter (Curtis and Grossniklaus, 2003).
To over‐express HSP90.2‐B in wheat, we incorporated the HSP90.2‐B full‐length cDNA into a gateway transgenic vector (P Ubi :AttR‐Flag:T NOS ) under the drive of maize ubiquitin (Proubi:HSP90.2‐Flag) by LR reaction. We incorporated the N‐terminal or C‐terminal fragments of HSP90.2‐B into the gateway‐compactable pANDA vector (Wang et al., 2019). According to earlier reports, we transformed the above constructs into WT immature embryos with Agrobacterium (Gou et al., 2015). For transgenic wheat analyses, the transgene was first verified by PCR primers on different exons of HSP90.2‐B with varying sizes from cDNA and genomic DNA. We then extracted total proteins from transgenic lines or WT leaves and analysed them in a Western Blot experiment with a polyclonal anti‐HSP90 antibody generated with the full‐length HSP90.2 protein (Abmart, Xuhui District, Shanghai, China).
We treated the protoplasts expressing PsbO‐GFP with 4 μg/mL cytochalasin A (Cyto A, Shyuanye, Shanghai, China) or DMSO as the negative control for 12 h. We then observed the samples with a Leica TCS SP8 (Leica Microsystems, Mannheim, Germany) confocal laser scanning microscope. We used the same settings as formerly mentioned to compare the fluorescence strength.
Protein–protein interaction assays
Following a previous report, we cloned different fragments of HSP90.2‐B into the Gateway‐compatible BD vector pLAW11 (Gou et al., 2015). PsbO full‐length cDNA in pDONR207 was incorporated into the Gateway‐compatible AD vector pLAW10 by LR clonase II (11 791 100; Invitrogen, ThermoFisher, Shanghai, China). We then transformed target plasmids into Y187 and Y2H Gold yeast cells using a LiAc‐mediated transformation procedure. The cells were confirmed by PCR and then mated following the Clontech user manual.
HSP90.2‐B, HSP90.2‐B C‐terminal, PsbO, and actin were incorporated into a Gateway‐compatible split‐luciferase system. We transformed the above plasmids into Agrobacterium GV3101, infiltrated the cells into tobacco leaves and kept the plants in the dark for 48 h. We added 1 mL of the 1 μg/mL firefly luciferase substrate (CAT: 40901ES01; Yeasen Biotechnology (Shanghai) Co., Ltd., Pudong District, Shanghai, China) and then monitored the interaction with Tanon 5200 Multi imaging system (Tanon Science & Technology Co., Ltd., Shanghai, China).
Actin and HSP90.2 fragments were incorporated into a Gateway‐compatible bimolecular fluorescence complementation (BiFC) system (Bracha‐Drori et al., 2004), which was then infiltrated into tobacco leaves and kept the plants in the dark for 48 h, and then observed under a Leica TCS SP8 (Leica, Microsystems, Mannheim, Germany).
Chloroplast isolation and protein analysis
We isolated the chloroplasts from wheat grown in the field (WT and hsp90.2) at the bolting stage with chloroplast extract buffer solution (20 mm tricine‐NaOH pH 8.4, 300 mm sorbitol, 10 mm KCl, 10 mm Na‐EDTA, 0.25% (w/v) BSA, 4.5 mm sodium ascorbate, 5 mm L‐cysteine) as described before. Subsequently, we collected the total extracts and separated the intact chloroplast as described before (Wang et al., 2019). We separated the crude chloroplast samples through a 30% and 80% Percoll Cushion buffer (20 mm tricine‐NaOH pH 8.4, 300 mm sorbitol, 10 mm KCl, 10 mm Na‐EDTA, 0.25% (w/v) BSA, 4.5 mm sodium ascorbate, 5 mm L‐cysteine).
We quantified the protein contents in the above samples, separated them in an SDS‐PAGE and examined them with anti‐PsbO monoclonal antibodies (Agrisera, Vännäs, Sweden) at 1:1000 dilution in a Western blot experiment. We used goat anti‐mouse IgG HRP (CAT: M21001S; Abmart, Xuhui District, Shanghai, China) as the secondary antibody at 1:5000 dilution and developed it according to the manufacturer's user manual.
Accession numbers
The RNA‐seq raw data in this work have been deposited into the National Center for Biotechnology Information center Gene Expression Omnibus (GEO) Database (https://www.ncbi.nlm.nih.gov/geo/) under accession number PRJNA862299. The Y2H data was under accession number PRJNA827186.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Author contributions
Y.Y. performed the experiments, analysed the data and drafted the manuscript. N.Y.G. and C.D. X.L. helped with the field test. C.S., Z.D., D.C. and F.C. created the RIL line and analysed the haplotype distribution. A.W. analysed the haplotype in Pakistan. C.H. and X.Z. contributed wheat varieties. M.W. and G.G. helped quantify starch and protein contents. Z.N. and Q.S. discussed the results and revised the manuscript. J.G. conceived and advised on all the experiments and wrote the manuscript.
Supporting information
Figure S1 The expression profiles of the HSP90 family genes in wheat. Note that we normalized and extracted the data from GSE12508 in NCBI.
Figure S2 The genotype of yield profiles of the hsp90.2‐A1 and hsp90.2‐B2 mutants. Data in this photograph represent means ± SD. A two‐tailed unpaired Students' t test P value showed the significance (hereafter) (n ≥ 10).
Figure S3 The phylogenetic analysis of the HSP90 family genes in wheat.
Figure S4 The agricultural traits of hsp90.2 knockout mutant in field trials (n ≥ 10).
Figure S5 The phenotype and agricultural traits of HSP90.2 RNAi transgenic lines in a controlled environment (n ≥ 10).
Figure S6 The phenotype and agricultural traits of HSP90.2 over‐expression transgenic lines in a controlled environment (n ≥ 10).
Table S1 The wheat photosynthesis members in the HSP90.2 interactome.
Table S2 The wheat actin members in the HSP90.2 interactome.
Acknowledgements
We thank Dr. Yan Yan at Fudan University for preparing the transgenic lines. We thank Prof. Jorge Dubcvosky for his helpful advice. The Research described here was supported by the National Key Research and Development Program of China (2022YFF1002902) and the National Natural Science Foundation of China (31972350 and 32072057).
Contributor Information
Feng Chen, Email: fengchen@henau.edu.cn.
Jin‐Ying Gou(缑金营), Email: jygou@cau.edu.cn.
Data availability statement
The corresponding author's data supporting this study's findings are available upon reasonable request.
References
- Alonso, M.P. , Abbate, P.E. , Mirabella, N.E. , Aramburu Merlos, F. , Panelo, J.S. and Pontaroli, A.C. (2018) Analysis of sink/source relations in bread wheat recombinant inbred lines and commercial cultivars under a high yield potential environment. Eur. J. Agron. 93, 82–87. [Google Scholar]
- Bao, F. , Huang, X. , Zhu, C. , Zhang, X. , Li, X. and Yang, S. (2014) Arabidopsis HSP90 protein modulates RPP4‐mediated temperature‐dependent cell death and defense responses. New Phytol. 202, 1320–1334. [DOI] [PubMed] [Google Scholar]
- Bracha‐Drori, K. , Shichrur, K. , Katz, A. , Oliva, M. , Angelovici, R. , Yalovsky, S. and Ohad, N. (2004) Detection of protein–protein interactions in plants using bimolecular fluorescence complementation. Plant J. 40, 419–427. [DOI] [PubMed] [Google Scholar]
- Chen, Y. , Yan, Y. , Wu, T.T. , Zhang, G.L. , Yin, H. , Chen, W. , Wang, S. et al. (2020) Cloning of wheat keto‐acyl thiolase 2B reveals a role of jasmonic acid in grain weight determination. Nat. Commun. 11, 6266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis, M.D. and Grossniklaus, U. (2003) A gateway cloning vector set for high‐throughput functional analysis of genes in planta. Plant Physiol. 133, 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faralli, M. and Lawson, T. (2020) Natural genetic variation in photosynthesis: an untapped resource to increase crop yield potential? Plant J. 101, 518–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira, K.N. , Iverson, T.M. , Maghlaoui, K. , Barber, J. and Iwata, S. (2004) Architecture of the photosynthetic oxygen‐evolving center. Science 303, 1831–1838. [DOI] [PubMed] [Google Scholar]
- Gao, Y. , Thiele, W. , Saleh, O. , Scossa, F. , Arabi, F. , Zhang, H. , Sampathkumar, A. et al. (2022) Chloroplast translational regulation uncovers nonessential photosynthesis genes as key players in plant cold acclimation. Plant Cell 34, 2056–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou, J.Y. , Yu, X.H. and Liu, C.J. (2009) A hydroxycinnamoyltransferase responsible for synthesizing suberin aromatics in Arabidopsis . Proc. Natl. Acad. Sci. U. S. A. 106, 18855–18860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou, J.Y. , Li, K. , Wu, K. , Wang, X. , Lin, H. , Cantu, D. , Uauy, C. et al. (2015) Wheat stripe rust resistance protein WKS1 reduces the ability of the thylakoid‐associated ascorbate peroxidase to detoxify reactive oxygen species. Plant Cell 27, 1755–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao, C. , Jiao, C. , Hou, J. , Li, T. , Liu, H. , Wang, Y. , Zheng, J. et al. (2020) Resequencing of 145 landmark cultivars reveals asymmetric sub‐genome selection and strong founder genotype effects on wheat breeding in China. Mol. Plant 13, 1733–1751. [DOI] [PubMed] [Google Scholar]
- Himes, R.H. and Houston, L.L. (1976) The action of cytochalasin A on the in vitro polymerization of brain tubulin and muscle G‐Actin. J. Supramol. Struct. 5, 81–90. [DOI] [PubMed] [Google Scholar]
- Jolliffe, P.A. and Tregunna, E.B. (1968) Effect of temperature, CO2 concentration, and light intensity on oxygen inhibition of photosynthesis in wheat leaves. Plant Physiol. 43, 902–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonwal, S. , Verma, N. and Sinha, A.K. (2022) Regulation of photosynthetic light reaction proteins via reversible phosphorylation. Plant Sci. 321, 111312. [DOI] [PubMed] [Google Scholar]
- Jouhet, J. and Gray, J.C. (2009a) Interaction of Actin and the chloroplast protein import apparatus. J. Biol. Chem. 284, 19132–19141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouhet, J. and Gray, J.C. (2009b) Is chloroplast import of photosynthesis proteins facilitated by an Actin‐TOC‐TIC‐VIPP1 complex? Plant Signal. Behav. 4, 986–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasileva, K.V. , Vasquez‐Gross, H.A. , Howell, T. , Bailey, P. , Paraiso, F. , Clissold, L. , Simmonds, J. et al. (2017) Uncovering hidden variation in polyploid wheat. Proc. Natl. Acad. Sci. USA 114, E913–E921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumatani, T. , Sakurai‐Ozato, N. , Miyawaki, N. , Yokota, E. , Shimmen, T. , Terashima, I. and Takagi, S. (2006) Possible association of Actin filaments with chloroplasts of spinach mesophyll cells in vivo and in vitro . Protoplasma 229, 45–52. [DOI] [PubMed] [Google Scholar]
- Li, W. and Yang, B. (2017) Translational genomics of grain size regulation in wheat. Theor. Appl. Genet. 130, 1765–1771. [DOI] [PubMed] [Google Scholar]
- Li, H.M. , Schnell, D. and Theg, S.M. (2020) Protein import motors in chloroplasts: on the role of chaperones. Plant Cell 32, 536–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao, H. , Sun, S. , Yao, J. , Wang, C. , Yu, S. , Xu, C. , et al. (2010) Linking differential domain functions of the GS3 protein to natural variation of grain size in rice. Proc. Indian National Sci. Acad. 107, 19579–19584. 10.1073/pnas.1014419107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai, T. and Makino, A. (2009) Differences between rice and wheat in temperature responses of photosynthesis and plant growth. Plant Cell Physiol. 50, 744–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redding, K.E. , Appel, J. , Boehm, M. , Schuhmann, W. , Nowaczyk, M.M. , Yacoby, I. and Gutekunst, K. (2022) Advances and challenges in photosynthetic hydrogen production. Trends Biotechnol. 40, 1313–1325. [DOI] [PubMed] [Google Scholar]
- Schnell, D.J. (2019) The TOC GTPase receptors: regulators of the fidelity, specificity and substrate profiles of the general protein import machinery of chloroplasts. Protein J. 38, 343–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, X.-J. , Huang, W. , Shi, M. , Zhu, M.-Z. and Lin, H.-X. (2007) A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat. Genet. 39, 623–630. 10.1038/ng2014 [DOI] [PubMed] [Google Scholar]
- Takahashi, A. , Casais, C. , Ichimura, K. and Shirasu, K. (2003) HSP90 interacts with RAR1 and SGT1 and is essential for RPS2‐mediated disease resistance in Arabidopsis. Proc. Natl. Acad. Sci. USA 100, 11777–11782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uauy, C. , Paraiso, F. , Colasuonno, P. , Tran, R.K. , Tsai, H. , Berardi, S. , Comai, L. et al. (2009) A modified TILLING approach to detect induced mutations in tetraploid and hexaploid wheat. BMC Plant Biol. 9, 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S. , Li, Q.P. , Wang, J. , Yan, Y. , Zhang, G.L. , Yan, Y. , Zhang, H. et al. (2019) YR36/WKS1‐mediated phosphorylation of PsbO, an extrinsic member of photosystem II, inhibits photosynthesis and confers stripe rust resistance in wheat. Mol. Plant 12, 1639–1650. [DOI] [PubMed] [Google Scholar]
- Wang, W. , Wang, Z. , Li, X. , Ni, Z. , Hu, Z. , Xin, M. , Peng, H. et al. (2020) SnpHub: an easy‐to‐set‐up web server framework for exploring large‐scale genomic variation data in the post‐genomic era with applications in wheat. Gigascience 9, giaa060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, X. , Su, X. , Cao, P. , Liu, X. , Chang, W. , Li, M. , Zhang, X. et al. (2016) Structure of spinach photosystem II‐LHCII supercomplex at 3.2 A resolution. Nature 534, 69–74. [DOI] [PubMed] [Google Scholar]
- van Wijk, K.J. (2004) Plastid proteomics. Plant Physiol. Biochem. 42, 963–977. [DOI] [PubMed] [Google Scholar]
- Zhou, Y. , Zhao, X. , Li, Y. , Xu, J. , Bi, A. , Kang, L. , Xu, D. et al. (2020) Triticum population sequencing provides insights into wheat adaptation. Nat. Genet. 52, 1412–1422. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 The expression profiles of the HSP90 family genes in wheat. Note that we normalized and extracted the data from GSE12508 in NCBI.
Figure S2 The genotype of yield profiles of the hsp90.2‐A1 and hsp90.2‐B2 mutants. Data in this photograph represent means ± SD. A two‐tailed unpaired Students' t test P value showed the significance (hereafter) (n ≥ 10).
Figure S3 The phylogenetic analysis of the HSP90 family genes in wheat.
Figure S4 The agricultural traits of hsp90.2 knockout mutant in field trials (n ≥ 10).
Figure S5 The phenotype and agricultural traits of HSP90.2 RNAi transgenic lines in a controlled environment (n ≥ 10).
Figure S6 The phenotype and agricultural traits of HSP90.2 over‐expression transgenic lines in a controlled environment (n ≥ 10).
Table S1 The wheat photosynthesis members in the HSP90.2 interactome.
Table S2 The wheat actin members in the HSP90.2 interactome.
Data Availability Statement
The corresponding author's data supporting this study's findings are available upon reasonable request.


