Abstract
Wastewater-based surveillance can be a valuable tool to monitor viral circulation and serve as an early warning system. For respiratory viruses that share similar clinical symptoms, namely SARS-CoV-2, influenza, and respiratory syncytial virus (RSV), identification in wastewater may allow differentiation between seasonal outbreaks and COVID-19 peaks. In this study, to monitor these viruses as well as standard indicators of fecal contamination, a weekly sampling campaign was carried out for 15 months (from September 2021 to November 2022) in two wastewater treatment plants that serve the entire population of Barcelona (Spain). Samples were concentrated by the aluminum hydroxide adsorption-precipitation method and then analyzed by RNA extraction and RT-qPCR. All samples were positive for SARS-CoV-2, while the positivity rates for influenza virus and RSV were significantly lower (10.65 % for influenza A (IAV), 0.82 % for influenza B (IBV), 37.70 % for RSV-A and 34.43 % for RSV-B). Gene copy concentrations of SARS-CoV-2 were often approximately 1 to 2 logarithmic units higher compared to the other respiratory viruses. Clear peaks of IAV H3:N2 in February and March 2022 and RSV in winter 2021 were observed, which matched the chronological incidence of infections recorded in the Catalan Government clinical database. In conclusion, the data obtained from wastewater surveillance provided new information on the abundance of respiratory viruses in the Barcelona area and correlated favorably with clinical data.
Keywords: Influenza, Respiratory syncytial virus (RSV), SARS-CoV-2, Wastewater-based epidemiology (WBE), RT-qPCR, Surveillance
Graphical abstract
1. Introduction
The impact of the COVID-19 pandemic has highlighted the necessity for more robust estimates and models of respiratory virus circulation (World Health Organization, 2022). Outbreaks of SARS-CoV-2 now need to be monitored in addition to those of seasonal viruses, which have made a comeback after their temporary suppression by COVID-19 control measures. The most relevant are influenza viruses A and B (IAV/IBV), and respiratory syncytial virus (RSV) (García-Arroyo et al., 2022), which account for the majority of coinfections with SARS-CoV-2 (Chung et al., 2021). In temperate regions, epidemics of seasonal influenza typically occur during the winter months (Jiménez-Jorge et al., 2016); in Spain the peak incidence occurs in January–February, and that of RSV in November–December (García-Arroyo et al., 2022).
The similar symptomatology of this trio of viral threats hinders their clinical diagnosis (Chotpitayasunondh et al., 2021; Huang et al., 2020). Yet differentiation among them is crucial for monitoring outbreaks occurring outside of the usual seasonal patterns, especially in the context of a pandemic in which the community circulation of respiratory viruses has been altered by the global implementation of non-pharmaceutical interventions to mitigate the COVID-19 burden (Chow et al., 2023; Li et al., 2020). Wastewater-based surveillance (WBS), increasingly being used around the world to monitor infectious diseases (Monteiro et al., 2022; Schmitz et al., 2021; Sims and Kasprzyk-Hordern, 2020), may prove helpful in identifying the periods of highest incidence of each virus.
An efficient, highly specific, cost-effective, non-invasive, real-time surveillance technique, WBS has been widely adopted during the COVID-19 pandemic to provide comprehensive public health information and allow authorities to track the virus (Betancourt et al., 2021; Kitajima et al., 2022; Monteiro et al., 2022; Polo et al., 2020; Schmitz et al., 2021). The ability of WBS systems to detect both symptomatic and asymptomatic transmission provides a more accurate reflection of virus spread in a population (de Llanos et al., 2022; Wu et al., 2020). The detection of viral genetic material, which appears in wastewater due to fecal shedding in infected individuals (Chavarria-Miró et al., 2021; Medema et al., 2020), can alert public health agencies to the community transmission of pathogens more quickly than diagnostic tests, thus facilitating their control (Li et al., 2022; Peccia et al., 2020). WBS has already being successfully applied to monitor other viral diseases such as influenza A, RSV, poliovirus, norovirus, and hepatitis A around the world (Ahmed et al., 2023; Deshpande et al., 2003; Heijnen and Medema, 2011; Hellmér et al., 2014; Hughes et al., 2022; Koureas et al., 2023; Pintó et al., 2007). Furthermore, it has been demonstrated that SARS-CoV-2 RNA levels in wastewater correlate with incidence rates obtained from clinical epidemiological data (Peccia et al., 2020; Wolfe et al., 2021). WBS data also allow the identification of specific variants and their circulation levels (Carcereny et al., 2021; Heijnen et al., 2021). Other relevant pathogenic respiratory viruses such as influenza are detectable in human excreta, despite being primarily airborne diseases (Hirose et al., 2016; Minodier et al., 2019). Therefore, as the RNA of respiratory viruses can be detected in wastewater even when the virus is no longer infectious (Hughes et al., 2022; Wolfe et al., 2022), WBS is a valuable monitoring tool.
In response to the growing need to differentiate SARS-CoV-2 from dominant seasonal respiratory viruses, the aim of this work was to evaluate, monitor, and compare RNA levels of IAV/IBV, RSV-A/RSV-B, and SARS-CoV-2 in wastewater in the metropolitan area of Barcelona, Spain, as well as measure indicators of human fecal contamination. It was envisaged that the results would shed light on the epidemiology of influenza and RSV infection in the wake of the COVID-19 pandemic and help design tools that can discern between outbreaks of community transmitted diseases of similar symptomology such as influenza, RSV, and COVID-19.
2. Material and methods
2.1. Sample collection
The densely populated metropolitan area of Barcelona, Spain, was chosen as a model to measure the occurrence of five respiratory viruses: SARS-CoV-2, IAV/IBV, and RSV-A/RSV-B. From September 9th, 2021, to November 15th, 2022, 122 wastewater samples were collected weekly (a total of 61 weeks) on weekdays (Monday to Wednesday) between 9:00 AM-10:00 AM from two major wastewater treatment plants (WWTP1 and WWTP2) that together serve the 3.3 million inhabitants of the city of Barcelona, WWTP1 serving approximately 65 % and WWTP2 35 % of the population. Other municipalities in the province of Barcelona are also serviced by these WWTPs, as stated in official data provided by the Barcelona metropolitan area public administration webpage (AMB, P.A. of the M.A. of B, 2021a, AMB, P.A. of the M.A. of B, 2021b). Samples (200 mL) were collected in sterile polypropylene (PP) containers and kept at 4 °C until processing, which took place within 24 h after sampling.
2.2. Control viral particles
Viral particles with genetic material from each target virus, incorporated with a lentiviral vector system (Sakuma et al., 2012), were used as positive controls and to study the recovery efficiency of the method used: VIASURE Viral Influenza A, Influenza B & RSV Positive Control Kit of CerTest Biotec (Barcelona, Spain). Particles of both influenza viruses and RSVs from a stock of 109 particles/mL were diluted up to 1 particle/mL. Tested concentrations ranged from 2 particles/mL of sample up to 200 particles/mL of sample. In the case of SARS-CoV-2, as a process control, 10 μL of a stock containing 8.8 × 105 TCID50 units of the attenuated PUR46-MAD strain of the transmissible gastroenteritis enteric virus (TGEV) (Moreno et al., 2008) was added to 200 mL of each sample prior to the concentration step.
2.3. Sample concentration
Wastewater was concentrated using the adsorption-precipitation method with aluminum hydroxide. This technique has previously been implemented for the study of SARS-CoV-2 in wastewater by numerous research groups throughout Spain (Carcereny et al., 2021; Randazzo et al., 2020). It was selected for the present study over other concentration methods due to its efficiency in detecting SARS-CoV-2, while being fast, simple, and cost-effective (Pérez-Cataluña et al., 2021). All the samples were processed within 24 h after collection in a manner equivalent to that used for the analysis of SARS-CoV-2, so that the data for the different viruses could be more easily compared. Briefly, 200 mL samples were collected and their pH was adjusted to 6.0. An AlCl3 solution was added to reach a final concentration of 0.009 N and thoroughly mixed by manual shaking. The pH was adjusted again to 6.0 and the mixture was further shaken with an orbital shaker for 15 min at 150 rpm and centrifuged at 1700 ×g for 20 min. The supernatant was discarded, and the pellet resuspended in 10 mL of a 3 % beef extract solution (Becton, Dickinson and Company, Sparks, MD, US). Samples were shaken for 10 min at 200 rpm, followed by a centrifugation step at 1900 ×g for 30 min. The supernatant was discarded and the pellet resuspended in 2 mL of 1× phosphate-buffered saline solution.
2.4. Nucleic acid extraction
Nucleic acid was extracted within the 24 h after concentration, using a method previously described (Carcereny et al., 2021) in a class II biosafety cabinet using the Macherey-Nagel Nucleospin RNA virus kit and following the manufacturer's instructions with slight modifications to favor RNA stability and recovery (Randazzo et al., 2020). Briefly, these modifications included the initial addition of 25 μL of Plant RNA Isolation Aid reagent (Ambion™, Thermo Fisher Scientific, Vilnius, Lithuania) to 150 mL of the concentrated sample and 600 mL of lysis buffer from the NucleoSpin Virus kit, mixing by pulse-vortexing for 1 min. The homogenate was then centrifuged for 5 min at 10,000 ×g to remove the debris. This supernatant was subsequently processed according to the manufacturer's instructions, eluted in 100 mL of RNAse-free H2O and used immediately for quantification or stored at −20 °C. Quantification was performed within the 24 h after extraction.
2.5. RT-qPCR quantification
The following genes were targeted for quantification by real-time quantitative reverse transcription polymerase chain reaction (RT-qPCR): the gene encoding matrix protein M1 from influenza A and B viruses (World Health Organization, 2015)the gene encoding nucleoprotein N from RSV A and B viruses (Hu et al., 2003) and SARS-CoV-2; the IP4 fragment of the gene encoding the RNA-dependent RNA polymerase (RdRp) (Institute Pasteur, Paris) (Corman et al., 2020) that encodes viral RNA polymerase, and the gene encoding N1 (CDC 2019-Novel Coronavirus (2019-nCoV) Real-Time RT-PCR Diagnostic Panel https://www.cdc.gov/coronavirus/2019-ncov/lab/rt-pcr-panel-primer-probes.html) that encodes nucleocapsid protein. The sample was considered positive for SARS-CoV-2 when one of the two target genes was detected. The qPCR analyses of the genes encoding M1 and N (proteins of influenza viruses and RSV, respectively) were carried out using Viasure Real Time PCR Detection kits (CerTest Biotec), which contain all the necessary components to perform real-time PCR (specific primers and probes, dNTPs, buffer, polymerase, and retrotranscriptase) in a stabilized format, as well as an internal control to rule out inhibition of polymerase activity. Real-time qPCR with TaqMan hydrolysis probes was performed in class II biosafety cabinets using the StepOne Real Time PCR System (Applied Biosystems, Foster City, USA) with a 20 μL reaction mixture following the program described in Table 1 . All samples were run in duplicate (including the standards and negative controls). After the amplification reaction, virus genes were detected in the FAM channel and the internal control in the VIC/HEX/JOE channel. All viruses were evaluated at the same time. Samples were considered positive (and quantifiable) if they reached a cycle threshold (Ct) value within the values of the standard curve and if the internal control included in the reaction was amplified correctly. The standard curves were generated with serial decimal dilutions of commercial synthetic RNA. Twist Synthetic SARS-CoV-2 RNA Control 2 (Twist Biosciences, San Francisco, CA, US) was used for SARS-CoV-2 and commercial synthetic RNA included in the respective detection kits (CerTest Biotec) for RSV and influenza. The Ct values were converted into gene copy (GC)/L values using the corresponding standard curves and volumes tested. The GC number was defined as the mean of the data obtained from duplicate analysis. A sample was considered negative if it showed no amplification signal and the internal control gave a positive signal. Inhibition of the PCR reaction can be ruled out by the amplification of the internal control included in the reaction. If the negative control gave a signal or the positive control lacked a signal, the results were considered invalid, and the samples were analyzed directly and with dilutions to counteract the possible presence of reaction inhibitory compounds.
Table 1.
q RT-PCR conditions for influenza viruses, RSV and SARS-CoV-2.
| Virus | Step | Temperature | Time | Number of cycles | Limit of detection (GC/L) |
|---|---|---|---|---|---|
| RSV | Reverse transcription | 45 °C | 15 min | 1 | 7.31 × 103 |
| Initial denaturalization | 95 °C | 2 min | 1 | ||
| Denaturalization | 95 °C | 10 s | 45 | ||
| Annealing/extension | 60 °C | 50 s | |||
| Influenza | Reverse transcription | 45 °C | 15 min | 1 | 6.01 × 103 |
| Initial denaturalization | 95 °C | 2 min | 1 | ||
| Denaturalization | 95 °C | 10 s | 45 | ||
| Annealing/extension | 60 °C | 50 s | |||
| SARS-CoV-2 (N1) | Reverse transcription | 50 °C | 10 min | 1 | 1.90 × 104 |
| Initial denaturalization | 95 °C | 3 min | 1 | ||
| Denaturalization | 95 °C | 3 s | 45 | ||
| Annealing/extension | 55 °C | 30 s | |||
| SARS-CoV-2 (IP4) | Reverse transcription | 55 °C | 20 min | 1 | 2.93 × 104 |
| Initial denaturalization | 95 °C | 3 min | 11 | ||
| Denaturalization | 95 °C | 15 s | 50 | ||
| Annealing/extension | 58 °C | 30 s | |||
| Final extension | 40 °C | 30 s | 1 |
For SARS-CoV-2 determination, RT-qPCR detection of genes encoding IP4 and N1 were used, following the protocol described by the Institute Pasteur, Paris (Corman et al., 2020) for the gene IP4 and the CDC protocol (CDC 2019-Novel Coronavirus (2019-nCoV) Real-Time RT-PCR Diagnostic Panel https://www.cdc.gov/coronavirus/2019-ncov/lab/rt-pcr-panel-primer-probes.html) for the N1 gene. RT-qPCR amplification, data interpretation, and quantification of TGEV in the process control and the SARS-CoV-2 viral targets were performed as previously described (Carcereny et al., 2021; Chavarria-Miró et al., 2021).
2.6. Detection of reference fecal indicators
E. coli was chosen as a bacterial fecal indicator. Detection was achieved following the ISO protocol (ISO 9308-1) for the enumeration of E. coli and coliform bacteria based on membrane filtration and subsequent culture in a chromogenic agar medium (Anonymous, 2014). Somatic coliphages were used as a viral fecal indicator and detected following the double agar layer technique (ISO 10705: 2) (Anonymous, 2000).
2.7. Statistical analysis
Computation of data and statistical tests and charts were performed using GraphPad Prism 9 (GraphPad Software, San Diego, CA, US). Weekly influenza and RSV case numbers in the two WWTP catchments were compared with combined IAV/IBV and combined RSV A/RSV B wastewater RNA concentration (log10 GC/L) using Spearman's rank correlation (ρ). In the event where there were two wastewater RNA measurements during the period, the higher value was used. Spearman's rank correlation was also used to evaluate the correlation between microbial indicators and the respiratory viruses in wastewater. The Mann-Whitney test was used to define the differences on virus occurrence between the two WWTP and to evaluate the differences between the counts of microbial indicators in the samples; evaluations were based on significance levels of p = 0.05. As concentrations in wastewater are likely log-normally distributed, for statistical analysis data was 10-log transformed.
3. Results and discussion
3.1. Detection of SARS-CoV-2, IAV/IBV, and RSV-A/RSV-B in wastewater samples
The efficiency of viral concentration and extraction methods was tested by spiking wastewater with a stock containing a known quantity of TGEV particles (used as a process control) and particles containing influenza and RSV genetic material. Although the concentration and extraction methods were originally devised for the detection of SARS-CoV-2 nucleic acids, they were successfully applied here to measure the respiratory viruses IAV/IBV and RSV-A/B.
The recovery rates from wastewater reached 20.93 % ± 14.8 for influenza virus and 17.10 % ± 15.5 for RSV. The average recovery rate for TGEV was 43.15 % ± 25.7 confirming the viability and efficiency of the method for analysis of respiratory viruses other than SARS-CoV-2 in wastewater. No significant differences in TGEV recovery rates were observed between the two WWTPs (44.0 % ± 24.2 for WWTP1 and 41.10 % ± 26.4 for WWTP2).
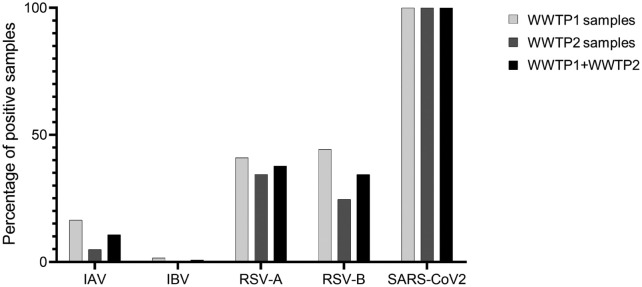
The abundance of SARS-CoV-2 differed significantly from that of the other target viruses (Fig. 1 ). While SARS-CoV-2 was detected in all samples collected from both WWTPs (100 % detection rate for the SARS-CoV-2 N1 gene), influenza virus and RSV were present in <50 % of the total samples analyzed. Specifically, IAV was observed in 10.65 % of the total samples, with a higher frequency in WWTP1 (16.39 %) than in WWTP2 (4.92 %), whereas IBV was only detected in one of the WWTP1 samples. RSV-A and RSV-B were found in 37.7 % and 34.43 % of the total samples, respectively, and more frequently in WWTP1 than WWTP2, particularly RSV-B (Fig. 1).
Fig. 1.
Positivity rate of all respiratory viruses. Percentage of positive samples for the respiratory viruses analyzed in wastewater of both WWTPs during the sampling camping.
3.2. Levels of indicators of fecal contamination in wastewater in Barcelona
E. coli and somatic coliphages in wastewater samples, analyzed following standardized protocols (Anonymous, 2000; International Organization for Standardization, 2014), showed comparable average concentrations in both WWTPs (Table 2 ). No significant differences (p > 0.05) were observed among the analyzed samples and between the WWTPs, the levels being within those reported in previous studies for wastewater sampled from the same WWTPs (Lucena et al., 2003). The microbial indicators analyzed did not correlate with the respiratory viruses. Influenza virus did not correlate with E. coli (p = 0.832) or somatic coliphages (p = 0.072), and considering that many values were below the LOD, the real correlation should be lower. Similarly, RSV did not correlate with E. coli (p = 0.334 for RSV-A and p = 0.199 for RSV- B) or with somatic coliphages (p = 0.102 (RSV-A) and 0.959 (RSV-B)). Finally, SARS-CoV-2 also did not show correlation with E. coli (p = 0.245) or somatic coliphages (p = 0.668).
Table 2.
Average concentrations of E. coli and somatic coliphages during the sampling campaign for both WWTP. No significant differences were observed among the samples (p > 0.05).
| WWTP | n | E. coli log10(CFU/mL) | Somatic coliphages log10(PFU/mL) | |
|---|---|---|---|---|
| WWTP1 | 61 | Average | 4.57 | 4.41 |
| SD | 0.17 | 0.17 | ||
| WWTP2 | 61 | Average | 4.41 | 4.32 |
| SD | 0.20 | 0.17 |
When comparing the results of molecular and culture detection methods, microorganism abundance is frequently higher according to the GC number compared to culturable indicators, as molecular analysis implies a lower inactivation of DNA and does not discriminate between live or dead, active or inactive, or infectious or non-infectious organisms (Field and Samadpour, 2007; Khan et al., 2007). However, in our samples, the fecal indicator concentrations were similar to or higher than the GC/L concentrations of the target viruses. Moreover, the indicator values remained very stable throughout the sampling campaign (p > 0.05) (Table 2), which precludes any correlation between them and the detected GC numbers of respiratory viruses. Although we observed, and some studies have reported, a correlation between fecal indicators and incidence of COVID-19 or abundance of SARS-CoV-2 (Nagarkar et al., 2022; Zhan et al., 2022), our analyses and those previously reported were carried out at the height of the pandemic, when SARS-CoV-2 levels were consistently high. Respiratory viruses are not original from the intestinal tract, and since they are pathogens, their occurrence is dependent on the presence of sick people among the population. Therefore, in a non-pandemic situation, and outside of the epidemic season, respiratory viruses will not be circulating in the population or observed in wastewater, and consequently cannot be correlated with indicators of fecal pollution.
3.3. Respiratory virus levels throughout the sampling campaign
Although the SARS-CoV-2 positivity rate in our samples was 100 %, a slight difference in frequency was observed between the two target SARS-CoV-2 genes. N1 was found in all samples, whereas IP4 was detected in 98.36 % (Supplementary Fig. 1). Although a sample was considered positive if either of the two genes were detected, N1 was used to monitor SARS-CoV-2 abundance because of its more consistent detection and higher concentration in both WWTPs.
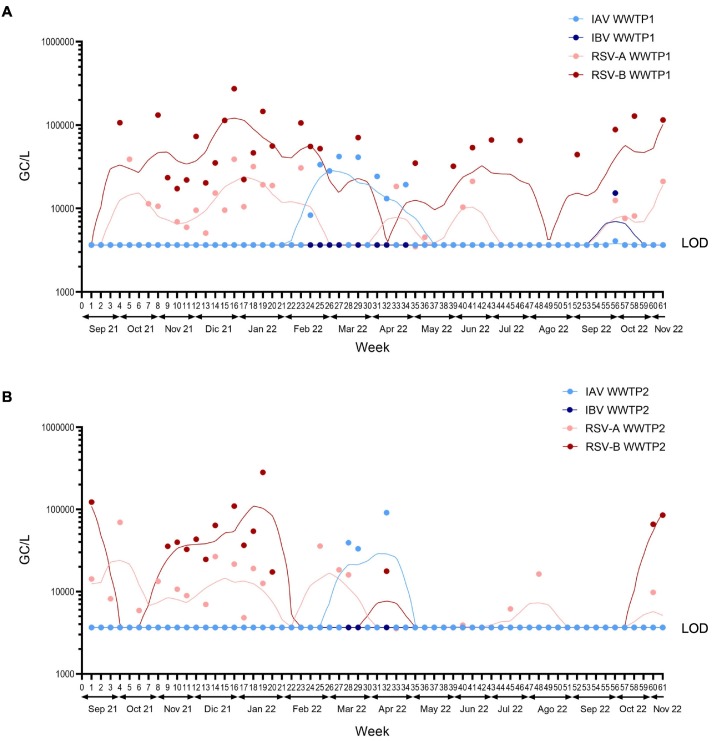
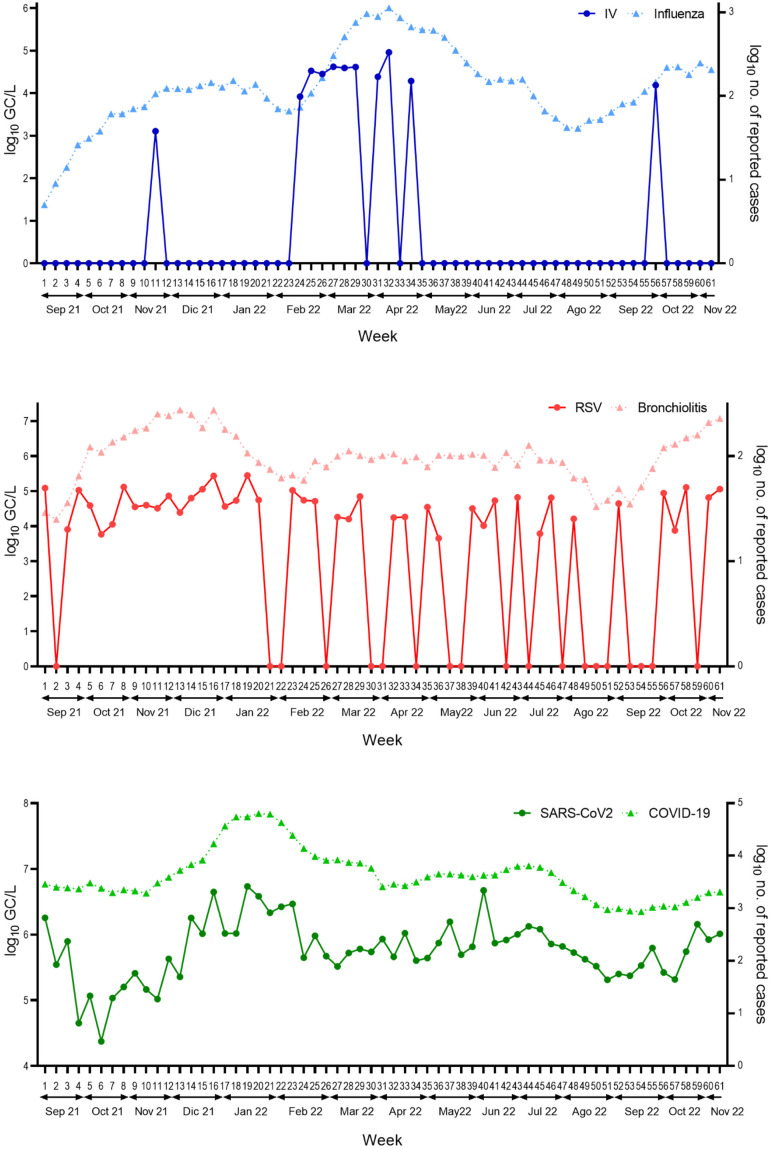
The analysis of influenza virus revealed that IAV always predominated over IBV in wastewater sampled from both WWTPs, reaching a prominent peak in late February/early March, while being practically absent the rest of the year (Fig. 2 ). This peak was more acute and longer lasting in WWTP1samples (weeks 24 to 34) (Fig. 2A). In contrast, IBV was only detected once in week 55 in wastewater from WWTP1 (Fig. 2A).
Fig. 2.
Viral titers of RSV and influenza virus. Values are expressed as gene copies/L (GC/L) of influenza virus and RSV during the sampling campaign in wastewater from A) WWTP1 and B) WWTP2. Dots show the results of each sampling point for each virus and lines show the spline smoothers for each virus. Bottom line indicates the limit of detection (LOD) of Influenza virus (blue) and RSV (red). Non-detects have been replaced with LOD values/2.
Regarding RSV, a peak of detection of both subtypes was apparent during the winter months (weeks 8 to 20 and 55 to 61) in samples from both WWTPs (Fig. 2). Sporadic occurrences of RSV-A/B in WWTP1 (Fig. 2A) and mainly RSV-A in WWTP2 (Fig. 2B) were also observed during the year without a clearly defined peak of detection.
Overall, a higher load of influenza virus and RSV was found in wastewater sampled from WWTP1 than WWTP2. According to the environmental data provided by the Barcelona Metropolitan Government (AMB, P.A. of the M.A. of B, 2021a, AMB, P.A. of the M.A. of B, 2021b), WWTP1 has a treatment capacity equivalent to 3 million inhabitants, 50 % higher than WWTP2, which would explain its higher positivity rate. However, the differences for each virus between both WWTP were not significant (IV p = 0.08; RSV p = 0.312 and SARS-CoV-2 p = 0.175).
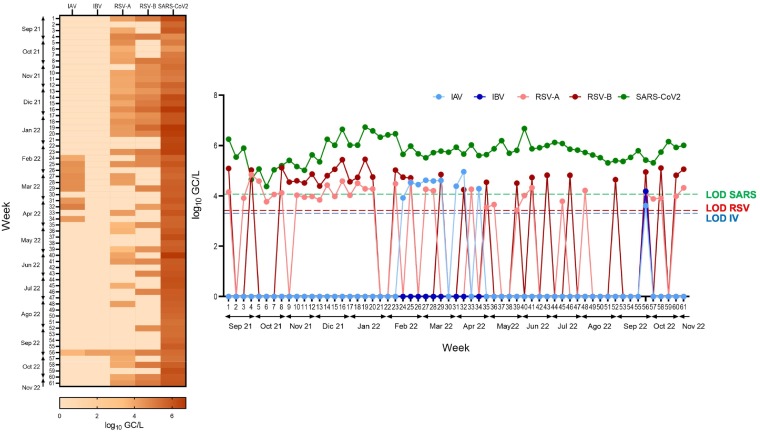
Considering the results of both WWTPs together, selecting the highest values of each virus obtained each week, during the 2021/2022 epidemic season, the SARS-CoV-2 N1 gene was detected at an average of 105 GC/L (Fig. 3), with a higher concentration than the IP4 gene throughout the sampling campaign (Fig. S1). This finding is consistent with data reported worldwide and may be explained by the higher sensitivity of the N1 versus the IP4 assay (Etievant et al., 2020). SARS-CoV-2 concentration increased slightly during the winter season (Week 13 to 23), but always remained between 1 and 2 logarithms higher than the other analyzed viruses throughout the period (Fig. 3).
Fig. 3.
Comparison of RSV, influenza virus and SARS-CoV-2. (A) Comparison of SARS-CoV-2, influenza A/B, and RSV-A/B concentrations (log10 GC/L) during the study, considering the highest value obtained in both WWTPs on each sampling day. (B) Heatmap showing the abundance of each virus along the study. The legend shows the highest value corresponding to the most intense color expressed in log10 units. Dotted lines indicated the LOD of each virus.
When detected, the concentrations of the genes M1 (influenza) and N (RSV) were in the range of 104–105 GC/L, being consistently lower than those of the SARS-CoV-2 genes in the corresponding samples (Fig. 3). RSV was detected in a higher number of samples than influenza virus throughout the epidemic season, although the concentrations were similar. These findings were in line with our expectations, as the detection of influenza and RSV has declined globally since the onset of the COVID-19 pandemic (Adlhoch et al., 2021; Groves et al., 2021; Olsen et al., 2021), likely due to the use of non-pharmacological interventions to control the spread of SARS-CoV-2, such as social distancing, mask-wearing, and implementation of lockdown measures, which suppressed the circulation of all respiratory viruses (Huang et al., 2020; Sanz-Muñoz et al., 2021).
3.4. Comparison of WBS data with clinical records from public health databases
Epidemiological data on the incidence of respiratory diseases were collected from public databases in the Catalonia Autonomous Community (Departament de Salut, Generalitat de Catalunya (Health Department Govern of Catalonia), 2022). We searched for cases of COVID-19 (indicative of SARS-CoV-2), influenza (IAV/IBV), and bronchiolitis (RSVA/B) diagnosed in hospitals and primary care centers during the weeks of the sampling campaign in the area serviced by the two WWTPs. Bronchiolitis was chosen as RSV is its leading etiological agent, more than other respiratory viruses. Unfortunately, the infectious agent causing the bronchiolitis cases was not indicated in the dataset. Althoughpneumonia can also be developed in more severe cases of RSV infection, it is not so common and may also be caused by several other infectious agents. It cannot be discarded that bronchiolitis is caused by other infectious agents, but even so, RSV is the most commonly associated agent (Leung et al., 2005; Pickles and DeVincenzo, 2015; Piedimonte and Perez, 2014).
The data provided by the SIVIC (Departament de Salut, Generalitat de Catalunya (Health Department Government of Catalonia), 2022) indicate a total of 413,663 diagnosed cases of COVID-19 in comparison with 50,494 and 20,779 diagnosed cases of influenza and bronchiolitis, respectively, within the region covered by the WWTPs during the sampling campaign.
When comparing the viral load in the wastewater samples with the clinical data, similar tendencies were found in the patterns of viral concentrations and incidence of associated diseases/symptoms (Fig. 4 ), particularly for influenza and COVID-19 cases that show an increase in the detection of IAV and SARS-CoV-2 respectively in some period just prior to the beginning of the increase in the numbers of both diseases. Accordingly, there is a significant correlation between the detection of influenza virus and the number of cases (p = 0.03) and between SARS-CoV-2 detection and the number of COVID-19 cases (p = 0.0001). In contrast, no correlation was found between bronchiolitis and RSV (p = 0.39), perhaps because the clinical disease used as marker is less specific of RSV infections. Therefore, the results evaluating the potential of WBS for early warning of different infectious agents may vary depending on the use of the appropriate clinical data for each disease. Moreover, the number of reported clinical cases is likely to be an underestimation, as individuals with mild or asymptomatic infections usually do not visit health care centers. This was apparent in COVID-19, whose incidence according to the clinical records declined more significantly than the SARS-CoV-2 concentration in wastewater (Fig. 4). The difference in detection levels was even more striking for RSV, which is significantly underreported in individuals over 14 years of age due to mild or inexistent symptoms (Heppe-Montero et al., 2022; Koureas et al., 2023). According to the clinical data, cases of influenza outnumbered those of RSV, whereas WBS data indicated that RSV was always predominant, with detection of influenza virus largely restricted to the epidemic season, unlike RSV. Possible explanations include different rates of viral shedding in feces, differences in viral stability in wastewater or, as mentioned, a higher number of asymptomatic RSV cases in the population.
Fig. 4.
Results of respiratory virus detection in wastewater and of number of cases reported in public health databases. A) Detection of SARS-CoV-2, influenza virus, and RSV during the sampling campaign in both WWTPs (left Y-axis log10GC/L). B) Primary care and hospital diagnostics in the area covered by the two WWTPs (right Y-axis log10 number of reported cases).
The higher number of reported influenza cases than of RSV was surprising, as the seasonal epidemic activity of RSV in Catalonia is longer than that of influenza virus, as reflected in epidemiological data from previous years in the SIVIC public database (Departament de Salut, Generalitat de Catalunya (Health Department Govern of Catalonia), 2022). A study carried out with data obtained before the pandemic found that RSV cases were most frequently detected early in the season, in late autumn and winter (November to January), whereas the incidence of IAV reached a clear peak in the later winter months (February to March)(García-Arroyo et al., 2022). In the present study, IBV was detected only once in wastewater samples throughout the sampling campaign and sporadic evidence of both RSV subtypes was observed along the year.
Although RSV infection is diagnosed less frequently than other respiratory viruses and is generally associated with children, its occurrence in adults is of concern, since asymptomatic individuals can infect susceptible individuals, including children under 14 years of age, who can develop harsher symptoms (Mitchell et al., 2017). In the aftermath of the COVID-19 pandemic, this issue is of even greater importance, as the development of herd immunity was hindered by the suppression of most respiratory viruses other than SARS-CoV-2 (Ando et al., 2023; Di Mattia et al., 2021).
As RSV-infected individuals seeking hospital treatment are generally only those with severe symptoms, clinical data do not accurately reflect the real infection dynamics in the population. In contrast, undiagnosed or asymptomatic infections can be detected in wastewater (Heppe-Montero et al., 2022). This hypothesis is supported by the data collected from wastewater in the metropolitan region of Barcelona, where RSV was found to be more common and longer-lasting than suggested by clinical records, being detected throughout the year and not only in seasonal epidemics.
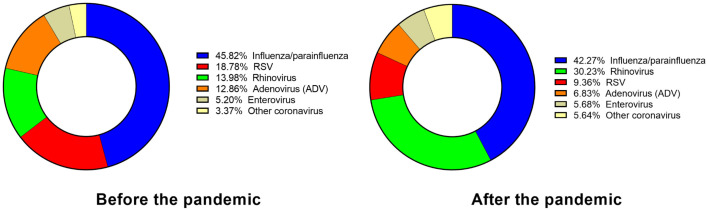
Another important finding from the analysis of clinical data is the resurgence of several respiratory viruses, some being more reported than before the COVID-19 pandemic and others less so (Fig. 5 ). Out of all the respiratory viruses isolated in hospitals in Catalonia during the study period, 47 % corresponded to SARS-CoV-2. The next most common were influenza and parainfluenza viruses (42.3 %), 95 % of the former being identified as IAV, which also predominated in wastewater samples. Next in frequency were rhinoviruses (30.2 %), that were reported twice as much as before the pandemic (14 %) (Fig. 5). The detection of RSV infection was lower compared to influenza and rhinovirus, representing 9.4 % of the total, and lower than before the pandemic (Fig. 5). Other respiratory viruses were detected in minor percentages (Fig. 5), with adenoviruses showing a decrease in comparison with pre-pandemic data.
Fig. 5.
Changes in the proportion of respiratory viruses identified in hospitals before and after the pandemic. Proportion (%) of respiratory viruses (non-SARS-CoV-2) identified in clinical laboratories in Catalan hospitals during this study compared with the data obtained before the SARS-CoV-2 pandemics.
The alteration of viral community dynamics by the drastic reduction in seasonal respiratory virus transmission during the COVID-19 pandemic (Ando et al., 2023; Chow et al., 2023) has stimulated interest in monitoring the evolution of virus concentrations in wastewater. Consequently, wastewater-based epidemiology has been implemented as a robust tool to predict outbreaks and changes in the occurrence of respiratory viruses (Chavarria-Miró et al., 2021; Randazzo et al., 2020; Viveros et al., 2022). The WBS data of SARS-CoV-2 and IAV obtained in the present study correlate with clinical data. IAV showed clearly defined peaks of incidence coinciding with an increase in the number of cases. However IAV was only detected in some time points in wastewater while there were influenza cases along the whole period. Weak or moderate correlation of IAV with influenza disease has previously been reported (Ahmed et al., 2023; Wolfe et al., 2022). The lack of detection of influenza virus at certain times in our study even though there was detection of clinical cases could be attributed to different factors; among others the proportion of influenza-infected individuals that shed the virus fecally (Chan et al., 2011; Hirose et al., 2016), the viral fecal load and the duration of the viral shedding after the disease (Chan et al., 2011) or the higher levels of IAV found in solid fractions as opposed to liquid fractions of wastewater (Wolfe et al., 2022).
Correlation was also found for SARS-CoV-2 concentrations in wastewater and COVID-19. Some correspondence could also be observed between RSV and the number of cases, although without statistical correlation. This is probably because RSV causes a higher number of asymptomatic cases and/or milder symptomatic infections that are not reported and because bronchiolitis is a less clear clinical marker. These results support the value of WBS as a diagnostics tool in clinical settings, when the reflecting clinical cases serves as an epidemiological signal to confirm outbreaks of a specific virus, as it was previously shown in other studies (Ahmed et al., 2023; Hughes et al., 2022). In terms of anticipating the peaks of infection, WBS is less useful, as the outbreaks can only be predicted a few days beforehand, unless the report of the clinical cases is also delayed, in which case the WBS could provide the peak information significantly before the clinical report. The potential role of WBS systems in preventive health measures should be assessed in studies of other respiratory viruses such as parainfluenza or rhinovirus, whose circulation after the COVID-19 pandemic has increased in the population worldwide in an unpredictable way (Chow et al., 2023). This resurgence has been observed all around the globe, and predictions of the behavior of respiratory viruses after pandemic measures have not been entirely accurate (Agca et al., 2021; Groves et al., 2021; Hodjat et al., 2021).
4. Conclusions
This study has provided significant data on the presence and evolution of respiratory viruses in the city of Barcelona, revealing different epidemiological patterns to those before the COVID-19 pandemic. Some respiratory viruses, especially RSV, were detected at a higher rate in wastewater compared to the number of reported clinical cases, which could be attributed to non-attendance at health care centers of asymptomatic individuals or those with mild symptoms. As these undetected cases can result in a spread of infection and cause outbreaks, they should be regulated. This function could feasibly be performed by WBS, a reliable and cost-effective tool that can provide relevant information about viral dynamics in a population independently of clinical records. The epidemiological study of wastewater needs further research to realize its potential as a system that can confirm and anticipate outbreaks of infectious diseases.
The following is the supplementary data related to this article.
Comparison between log10GC/L concentrations of SARS-CoV-2 IP4 and N1 genes during the sampling campaign in both WWTPs.
CRediT authorship contribution statement
D. Toribio-Avedillo: Investigation (analysis of RSV and Influenza virus); Data search; Writing- Original draft preparation.
C. Gómez-Gómez: Investigation (analysis of RSV and Influenza virus); Validation.
L. Sala-Comorera: Data search, Formal analysis (statistics), Visualization (chart and figures design).
L. Rodriguez-Rubio Reviewing and Editing.
A. Carcereny: Investigation (analysis of SARS-CoV-2); Validation.
D. Garcia-Pedemonte: Investigation (analysis of SARS-CoV-2); Validation.
R.M. Pintó: Conceptualization, Methodology (Viral concentration).
S. Guix. Conceptualization, Methodology (Viral concentration).
B. Galofre: Resources (Sampling and Contacts with the managers of WWTP).
A. Bosch: Conceptualization, Supervision, Writing, Reviewing and Editing.
S. Merino: Supervision and Methodology (viral DNA extraction), Validation; Data curation.
M. Muniesa Conceptualization, Funding acquisition, Project management, Writing, Reviewing and Editing.
All authors: Reviewing and Editing.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:
Maite Muniesa reports financial support was provided by Ministerio de Innovación y Ciencia. Maite Muniesa reports financial support was provided by Government of Catalonia.
Acknowledgements
This work was supported by the Generalitat de Catalunya's program “Replegar-se per créixer: l'impacte de les pandèmies en un món sense fronteres visibles” (PANDÈMIES 2020) (2020PANDE00055) and partially supported by the Spanish Ministerio de Innovación y Ciencia (PID2020-113355GB-I00), the Agencia Estatal de Investigación (AEI) and the European regional fund (ERF). Authors are members of consolidated research groups of the Generalitat of Catalunya: Microbiologia d'aigües relacionada amb la salut (MARS) (2017SGR0170) and Virus Entèrics group (2021SGR01089), the Institut de Recerca de l'Aigua/University of Barcelona, and the Institut de Recerca en Nutrició i Seguretat Alimentària (INSA-UB), University of Barcelona, recognized as a Maria de Maeztu Unit of Excellence by grant CEX2021-001234-M (funded by MCIN/AEI/10.13039/501100011033). Authors want to thank the CerTest company for their contribution providing some of the kits for this study, the staff from the WWTPs for helping in the collection of the samples, and to M.T. Polo for their technical assistance. Authors declare no conflict of interest. C. G-G. has a grant from the university of Barcelona and L. R-R. is lecturer of the Serra-Hunter program, Generalitat de Catalunya.
Editor: Warish Ahmed
Data availability
Data will be made available on request.
References
- Adlhoch C., Mook P., Lamb F., Ferland L., Melidou A., Amato-Gauci A.J., Pebody R., Abovyan R., Sargsyan S., Redlberger-Fritz M., Karaban I., Shmialiova N., Bossuyt N., Thomas I., Vukmir N.R., Dedeic-Ljubovic A., Petrovic G., Tabain I., Pieridou D., Karagiannis C., Jirincova H., Kyncl J., Vestergaard L.S., Trebbien R., Sadikova O., Dotsenko L., Ikonen N., Lyytikäinen O., Enouf V., Jung Y.J., Buda S., Dürrwald R., Gioula G., Kossyvakis T., Molnár Z., Rózsa M., Aspelund G., Löve A., Domegan L., Dunford L., Kaufman Z., Mandelboim M., Bella A., Puzelli S., Nazym T., Aidar U., Nikiforova R., Pakarna G., Gargasiene G., Muralyte S., Mossong J., Abdelrahman T., Melillo J.M., Melillo T., Druc A., Apostol M., Meijer A., Fouchier R.A.M., Golubinka B., Kochinski D., Paulsen T.H., Hungnes O., Guiomar R., Rodrigues A.P., Popescu R., Popovici O., Sominina A., Danilenko D., Dimitrijevic D., Socan M., Prosenc K., Sanchez F.P., Delgado-Sanz C., Byström E., Carnahan A.S., Gonçalves A.R., Born R., Emine A.V.C.I., Ayse Basak A.L.T.A.S., Demchyshyna I., Dykhanovska T., Sinnathamby M., Bernal J.L. Very little influenza in the WHO European Region during the 2020/21 season, weeks40 2020 to 8 2021. Eurosurveillance. 2021;26:1–8. doi: 10.2807/1560-7917.ES.2021.26.11.2100221. [DOI] [Google Scholar]
- Agca H., Akalin H., Saglik I., Hacimustafaoglu M., Celebi S., Ener B. Changing epidemiology of influenza and other respiratory viruses in the first year of COVID-19 pandemic. J. Infect. Public Health. 2021;14:1186–1190. doi: 10.1016/j.jiph.2021.08.004. [DOI] [PubMed] [Google Scholar]
- Ahmed W., Bivins A., Stephens M., Metcalfe S., Smith W.J.M., Sirikanchana K., Kitajima M., Simpson S.L. Occurrence of multiple respiratory viruses in wastewater in Queensland, Australia: potential for community disease surveillance. Sci. Total Environ. 2023;864 doi: 10.1016/J.SCITOTENV.2022.161023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AMB, P.A. of the M.A. of B . AMB; 2021. Besòs WWTP Technical data [WWW Document]https://www.amb.cat/en/web/ecologia/aigua/instalacions-i-equipaments/detall/-/equipament/edar-del-besos/275728/11818 URL. [Google Scholar]
- AMB, P.A. of the M.A. of B . AMB; 2021. Prat de Llobregat WWTP Technical data [WWW Document]https://www.amb.cat/es/web/ecologia/aigua/instalacions-i-equipaments/detall/-/equipament/edar-del-prat-de-llobregat/276285/11818 URL. [Google Scholar]
- Ando H., Ahmed W., Iwamoto R., Ando Y., Okabe S., Kitajima M. Impact of the COVID-19 pandemic on the prevalence of influenza A and respiratory syncytial viruses elucidated by wastewater-based epidemiology. Sci. Total Environ. 2023;880 doi: 10.1016/J.SCITOTENV.2023.162694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anonymous . 2000. ISO 10705-2: Water Quality. Detection and Enumeration of Bacteriophages -Part 2: Enumeration of Somatic Coliphages. [Google Scholar]
- Anonymous . 2014. ISO 9308-1:2014 Water Quality — Enumeration of Escherichia coli and Coliform Bacteria — Part 1: Membrane Filtration Method for Waters With Low Bacterial Background Flora. [Google Scholar]
- Betancourt W.Q.Q., Schmitz B.W.W., Innes G.K.K., Prasek S.M.M., Pogreba Brown K.M.M., Stark E.R.R., Foster A.R.R., Sprissler R.S.S., Harris D.T.T., Sherchan S.P.P., Gerba C.P.P., Pepper I.L.L. COVID-19 containment on a college campus via wastewater-based epidemiology, targeted clinical testing and an intervention. Sci. Total Environ. 2021;779 doi: 10.1016/j.scitotenv.2021.146408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carcereny A., Martínez-Velázquez A., Bosch A., Allende A., Truchado P., Cascales J., Romalde J.L., Lois M., Polo D., Sánchez G., Pérez-Cataluña A., Díaz-Reolid A., Antón A., Gregori J., Garcia-Cehic D., Quer J., Palau M., Ruano C.G., Pintó R.M., Guix S. Monitoring emergence of the SARS-CoV-2 B.1.1.7 variant through the Spanish National SARS-CoV-2 Wastewater Surveillance System (VATar COVID-19) Environ. Sci. Technol. 2021;55:11756–11766. doi: 10.1021/ACS.EST.1C03589/SUPPL_FILE/ES1C03589_SI_001.PDF. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan M.C.W., Lee N., Chan P.K.S., To K.F., Wong R.Y.K., Ho W.S., Ngai K.L.K., Sung J.J.Y. Seasonal influenza A virus in feces of hospitalized adults. Emerg. Infect. Dis. 2011;17:2038. doi: 10.3201/EID1711.110205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavarria-Miró G., Anfruns-Estrada E., Martínez-Velázquez A., Vázquez-Portero M., Guix S., Paraira M., Galofré B., Sánchez G., Pintó R.M., Bosch A. Time evolution of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in wastewater during the first pandemic wave of COVID-19 in the metropolitan area of Barcelona, Spain. Appl. Environ. Microbiol. 2021;87:1–9. doi: 10.1128/AEM.02750-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chotpitayasunondh T., Fischer T.K.K., Heraud J.M.M., Hurt A.C.C., Monto A.S.S., Osterhaus A., Shu Y., Tam J.S.S. Influenza and COVID-19: what does co-existence mean? Influenza Other Respir. Viruses. 2021;15:407–412. doi: 10.1111/irv.12824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow E.J., Uyeki T.M., Chu H.Y. The effects of the COVID-19 pandemic on community respiratory virus activity. Nat. Rev. Microbiol. 2023;21:195. doi: 10.1038/S41579-022-00807-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung H.Y., Jian M.J., Chang C.K., Lin J.C., Yeh K.M., Chen C.W., Chiu S.K., Wang Y.H., Liao S.J., Li S.Y., Hsieh S.S., Tsai S.H., Perng C.L., Yang J.R., Liu M.T., Chang F.Y., Shang H.S. Novel dual multiplex real-time RT-PCR assays for the rapid detection of SARS-CoV-2, influenza A/B, and respiratory syncytial virus using the BD MAX open system. Emerg. Microbes Infect. 2021;10:161–166. doi: 10.1080/22221751.2021.1873073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K.W., Bleicker T., Brünink S., Schneider J., Schmidt M.L., Mulders D.G.J.C., Haagmans B.L., Van Der Veer B., Van Den Brink S., Wijsman L., Goderski G., Romette J.L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P.G., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Llanos R., Cejudo-Marín R., Barneo M., Pérez-Cataluña A., Barberá-Riera M., Rebagliato M., Bellido-Blasco J., Sánchez G., Hernández F., Bijlsma L. Monitoring the evolution of SARS-CoV-2 on a Spanish university campus through wastewater analysis: a pilot project for the reopening strategy. Sci. Total Environ. 2022;845 doi: 10.1016/j.scitotenv.2022.157370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Departament de Salut, Generalitat de Catalunya (Health Department Govern of Catalonia), Agencia de Salut Pública (Public Health Agency) Sistema d'informació per a la Vigilancia d'infeccions a Catalunya (Information System for Surveillance of Infections in Catalonia) [WWW Document] 2022. https://sivic.salut.gencat.cat/ URL.
- Deshpande J.M., Shetty S.J., Siddiqui Z.A. Environmental surveillance system to track wild poliovirus transmission. Appl. Environ. Microbiol. 2003;69:2919–2927. doi: 10.1128/AEM.69.5.2919-2927.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Mattia G., Nenna R., Mancino E., Rizzo V., Pierangeli A., Villani A., Midulla F. During the COVID-19 pandemic where has respiratory syncytial virus gone? Pediatr. Pulmonol. 2021;56:3106–3109. doi: 10.1002/ppul.25582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etievant S., Bal A., Escuret V., Brengel-Pesce K., Bouscambert M., Cheynet V., Generenaz L., Oriol G., Destras G., Billaud G., Josset L., Frobert E., Morfin F., Gaymard A. Performance assessment of SARS-CoV-2 PCR assays developed by who referral laboratories. J. Clin. Med. 2020;9:1–10. doi: 10.3390/jcm9061871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field K.G., Samadpour M. Fecal source tracking, the indicator paradigm, and managing water quality. Water Res. 2007;41:3517–3538. doi: 10.1016/j.watres.2007.06.056. [DOI] [PubMed] [Google Scholar]
- García-Arroyo L., Prim N., del Cuerpo M., Marín P., Roig M.C., Esteban M., Labeaga R., Martí N., Berengua C., Gich I., Navarro F., Rabella N. Prevalence and seasonality of viral respiratory infections in a temperate climate region: a 24-year study (1997–2020) Influenza Other Respir. Viruses. 2022;16:756. doi: 10.1111/IRV.12972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groves H.E., Piché-Renaud P.P., Peci A., Farrar D.S., Buckrell S., Bancej C., Sevenhuysen C., Campigotto A., Gubbay J.B., Morris S.K. The impact of the COVID-19 pandemic on influenza, respiratory syncytial virus, and other seasonal respiratory virus circulation in Canada: a population-based study. Lancet Reg. Health Am. 2021;1 doi: 10.1016/j.lana.2021.100015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heijnen L., Medema G. Surveillance of influenza A and the pandemic influenza a (H1N1) 2009 in sewage and surface water in the Netherlands. J. Water Health. 2011;9:434–442. doi: 10.2166/wh.2011.019. [DOI] [PubMed] [Google Scholar]
- Heijnen L., Elsinga G., de Graaf M., Molenkamp R., Koopmans M.P.G.P.G., Medema G. Droplet digital RT-PCR to detect SARS-CoV-2 signature mutations of variants of concern in wastewater. Sci. Total Environ. 2021;799 doi: 10.1016/j.scitotenv.2021.149456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmér M., Paxéus N., Magnius L., Enache L., Arnholm B., Johansson A., Bergström T., Norder H. Detection of pathogenic viruses in sewage provided early warnings of hepatitis A virus and norovirus outbreaks. Appl. Environ. Microbiol. 2014;80:6771–6781. doi: 10.1128/AEM.01981-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heppe-Montero M., Gil-Prieto R., del Diego Salas J., Hernández-Barrera V., Gil-de-Miguel Á. Impact of respiratory syncytial virus and influenza virus infection in the adult population in Spain between 2012 and 2020. Int. J. Environ. Res. Public Health. 2022;19:14680. doi: 10.3390/IJERPH192214680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose R., Daidoji T., Naito Y., Watanabe Y., Arai Y., Oda T., Konishi H., Yamawaki M., Itoh Y., Nakaya T. Long-term detection of seasonal influenza RNA in faeces and intestine. Clin. Microbiol. Infect. 2016;22:813.e1–813.e7. doi: 10.1016/J.CMI.2016.06.015. [DOI] [PubMed] [Google Scholar]
- Hodjat P., Christensen P.A.A., Subedi S., Bernard D.W.W., Olsen R.J.J., Long S.W.W. The reemergence of seasonal respiratory viruses in Houston, Texas, after relaxing COVID-19 restrictions. Microbiol. Spectr. 2021;9:2020–2022. doi: 10.1128/spectrum.00430-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu A., Colella M., Tam J.S., Rappaport R., Cheng S.M. Simultaneous detection, subgrouping, and quantitation of respiratory syncytial virus A and B by real-time PCR. J. Clin. Microbiol. 2003;41:149–154. doi: 10.1128/JCM.41.1.149-154.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes B., Duong D., White B.J., Wigginton K.R., Chan E.M.G., Wolfe M.K., Boehm A.B. Respiratory syncytial virus (RSV) RNA in wastewater settled solids reflects RSV clinical positivity rates. Environ. Sci. Technol. Lett. 2022;9:173–178. doi: 10.1021/acs.estlett.1c00963. [DOI] [Google Scholar]
- International Organization for Standardization . 2014. ISO 9308-1:2014 - Water Quality -- Enumeration of Escherichia coli and Coliform Bacteria -- Part 1: Membrane Filtration Method for Waters With Low Bacterial Background Flora. [Google Scholar]
- Jiménez-Jorge S., Delgado-Sanz C., De Mateo S., Pozo F., Casas I., Larrauri A. Vigilancia del virus respiratorio sincitial en el marco del Sistema de Vigilancia de la Gripe en España, 2006-2014. Enferm. Infecc. Microbiol. Clin. 2016;34:117–120. doi: 10.1016/j.eimc.2014.12.012. [DOI] [PubMed] [Google Scholar]
- Khan I.U.H., Gannon V., Kent R., Koning W., Lapen D.R., Miller J., Neumann N., Phillips R., Robertson W., Topp E., van Bochove E., Edge T.A. Development of a rapid quantitative PCR assay for direct detection and quantification of culturable and non-culturable Escherichia coli from agriculture watersheds. J. Microbiol. Methods. 2007;69:480–488. doi: 10.1016/j.mimet.2007.02.016. [DOI] [PubMed] [Google Scholar]
- Kitajima M., Murakami M., Iwamoto R., Katayama H., Imoto S. COVID-19 wastewater surveillance implemented in the Tokyo 2020 Olympic and Paralympic Village. J. Travel Med. 2022;29 doi: 10.1093/JTM/TAAC004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koureas M., Mellou K., Vontas A., Kyritsi M., Panagoulias I., Koutsolioutsou A., Mouchtouri V.A., Speletas M., Paraskevis D., Hadjichristodoulou C. Wastewater levels of respiratory syncytial virus associated with influenza-like illness rates in children-a case study in Larissa, Greece (October 2022-January 2023) Int. J. Environ. Res. Public Health. 2023;20:5219. doi: 10.3390/IJERPH20065219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung A.K.C., Kellner J.D., Davies H.D. Respiratory syncytial virus bronchiolitis. J. Natl. Med. Assoc. 2005;97:1708–1713. doi: 10.2165/00151829-200605060-00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Pei S., Chen B., Song Y., Zhang T., Yang W., Shaman J. Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS-CoV-2) Science. 2020;368:489–493. doi: 10.1126/SCIENCE.ABB3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Mazurowski L., Dewan A., Carine M., Haak L., Guarin T.C., Dastjerdi N.G., Gerrity D., Mentzer C., Pagilla K.R. Longitudinal monitoring of SARS-CoV-2 in wastewater using viral genetic markers and the estimation of unconfirmed COVID-19 cases. Sci. Total Environ. 2022;817 doi: 10.1016/j.scitotenv.2022.152958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucena F., Méndez X., Morón A., Calderón E., Campos C., Guerrero A., Cárdenas M., Gantzer C., Shwartzbrood L., Skraber S., Jofre J. Occurrence and densities of bacteriophages proposed as indicators and bacterial indicators in river waters from Europe and South America. J. Appl. Microbiol. 2003;94:808–815. doi: 10.1046/j.1365-2672.2003.01812.x. [DOI] [PubMed] [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environ. Sci. Technol. Lett. 2020;7:511–516. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- Minodier L., Masse S., Capai L., Blanchon T., Ceccaldi P.E., van der Werf S., Hanslik T., Charrel R., Falchi A. Risk factors for seasonal influenza virus detection in stools of patients consulting in general practice for acute respiratory infections in France, 2014-2016. Influenza Other Respir. Viruses. 2019;13:398–406. doi: 10.1111/irv.12523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell I., Defoy I., Grubb E. Burden of respiratory syncytial virus hospitalizations in Canada. Can. Respir. J. 2017;2017:1–9. doi: 10.1155/2017/4521302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro S., Rente D., Cunha M.V., Gomes M.C., Marques T.A., Lourenço A.B., Cardoso E., Álvaro P., Silva M., Coelho N., Vilaça J., Meireles F., Brôco N., Carvalho M., Santos R. A wastewater-based epidemiology tool for COVID-19 surveillance in Portugal. Sci. Total Environ. 2022;804 doi: 10.1016/J.SCITOTENV.2021.150264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno J.L., Zúñiga S., Enjuanes L., Sola I. Identification of a coronavirus transcription enhancer. J. Virol. 2008;82:3882–3893. doi: 10.1128/JVI.02622-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarkar M., Keely S.P., Jahne M., Wheaton E., Hart C., Smith B., Garland J., Varughese E.A., Braam A., Wiechman B., Morris B., Brinkman N.E. SARS-CoV-2 monitoring at three sewersheds of different scales and complexity demonstrates distinctive relationships between wastewater measurements and COVID-19 case data. Sci. Total Environ. 2022;816 doi: 10.1016/J.SCITOTENV.2021.151534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen S.J., Winn A.K., Budd A.P., Prill M.M., Steel J., Midgley C.M., Kniss K., Burns E., Rowe T., Foust A., Jasso G., Merced-Morales A., Davis C.T., Jang Y., Jones J., Daly P., Gubareva L., Barnes J., Kondor R., Sessions W., Smith C., Wentworth D.E., Garg S., Havers F.P., Fry A.M., Hall A.J., Brammer L., Silk B.J. Changes in influenza and other respiratory virus activity during the COVID-19 pandemic — United States, 2020–2021. MMWR Morb. Mortal. Wkly Rep. 2021;70:1013–1019. doi: 10.15585/mmwr.mm7029a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peccia J., Zulli A., Brackney D.E.E., Grubaugh N.D.D., Kaplan E.H.H., Casanovas-Massana A., Ko A.I.I., Malik A.A.A., Wang D., Wang M., Warren J.L.L., Weinberger D.M.M., Arnold W., Omer S.B.B. Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. Nat. Biotechnol. 2020;38:1164–1167. doi: 10.1038/s41587-020-0684-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Cataluña A., Cuevas-Ferrando E., Randazzo W., Falcó I., Allende A., Sánchez G. Comparing analytical methods to detect SARS-CoV-2 in wastewater. Sci. Total Environ. 2021;758 doi: 10.1016/J.SCITOTENV.2020.143870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickles R.J., DeVincenzo J.P. Respiratory syncytial virus (RSV) and its propensity for causing bronchiolitis. J. Pathol. 2015;235:266–276. doi: 10.1002/path.4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piedimonte G., Perez M.K. Respiratory syncytial virus infection and bronchiolitis. Pediatr. Rev. 2014;35:519. doi: 10.1542/PIR.35-12-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pintó R.M., Alegre D., Domíngueza A., El-Senousy W.M., Sánchez G., Villena C., Costafreda M.I., Aragonès L., Bosch A. Hepatitis A virus in urban sewage from two Mediterranean countries. Epidemiol. Infect. 2007;135:270. doi: 10.1017/S0950268806006753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo D., Quintela-Baluja M., Corbishley A., Jones D.L., Singer A.C., Graham D.W., Romalde J.L. Making waves: wastewater-based epidemiology for COVID-19 - approaches and challenges for surveillance and prediction. Water Res. 2020;186 doi: 10.1016/J.WATRES.2020.116404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo W., Cuevas-Ferrando E., Sanjuán R., Domingo-Calap P., Sánchez G. Metropolitan wastewater analysis for COVID-19 epidemiological surveillance. Int. J. Hyg. Environ. Health. 2020;230 doi: 10.1016/J.IJHEH.2020.113621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma T., Barry M.A., Ikeda Y. Lentiviral vectors: basic to translational. Biochem. J. 2012;443:603–618. doi: 10.1042/BJ20120146. [DOI] [PubMed] [Google Scholar]
- Sanz-Muñoz I., Tamames-Gómez S., Castrodeza-Sanz J., Eiros-Bouza J.M., de Lejarazu-Leonardo R.O. Social distancing, lockdown and the wide use of mask; a magic solution or a double-edged sword for respiratory viruses epidemiology? Vaccines (Basel) 2021;9:595. doi: 10.3390/VACCINES9060595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz B.W., Innes G.K., Prasek S.M., Betancourt W.Q., Stark E.R., Foster A.R., Abraham A.G., Gerba C.P., Pepper I.L. Enumerating asymptomatic COVID-19 cases and estimating SARS-CoV-2 fecal shedding rates via wastewater-based epidemiology. Sci. Total Environ. 2021;801 doi: 10.1016/J.SCITOTENV.2021.149794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims N., Kasprzyk-Hordern B. Future perspectives of wastewater-based epidemiology: monitoring infectious disease spread and resistance to the community level. Environ. Int. 2020;139 doi: 10.1016/j.envint.2020.105689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viveros M.L., Azimi S., Pichon E., Roose-Amsaleg C., Bize A., Durandet F., Rocher V. Wild type and variants of SARS-COV-2 in Parisian sewage: presence in raw water and through processes in wastewater treatment plants. Environ. Sci. Pollut. Res. 2022;29:67442–67449. doi: 10.1007/s11356-022-22665-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe M.K., Topol A., Knudson A., Simpson A., White B., Vugia D.J., Yu A.T., Li L., Balliet M., Stoddard P., Han G.S., Wigginton K.R., Boehm A.B. High-frequency, high-throughput quantification of SARS-CoV-2 RNA in wastewater settled solids at eight publicly owned treatment works in Northern California shows strong association with COVID-19 incidence. mSystems. 2021;6 doi: 10.1128/MSYSTEMS.00829-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe M.K., Duong D., Bakker K.M., Ammerman M., Mortenson L., Hughes B., Arts P., Lauring A.S., Fitzsimmons W.J., Bendall E., Hwang C.E., Martin E.T., White B.J., Boehm A.B., Wigginton K.R. Wastewater-based detection of two influenza outbreaks. Environ. Sci. Technol. Lett. 2022;9:687–692. doi: 10.1021/ACS.ESTLETT.2C00350. [DOI] [Google Scholar]
- World Health Organization . WHO; 2015. A Manual for Estimating Disease Burden Associated With Seasonal Influenza; pp. 1–124. [Google Scholar]
- World Health Organization (WHO) 2022. Environmental Surveillance for SARS-COV-2 to Complement Public Health Surveillance - Interim Guidance. [Google Scholar]
- Wu F., Zhang J., Xiao A., Gu X., Lee L., Armas F., Kauffman K. SARS-CoV-2 titers in wastewater are higher than expected. mSystems. 2020;5:1–9. doi: 10.1128/mSystems.00614-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan Q., Babler K.M., Sharkey M.E., Amirali A., Beaver C.C., Boone M.M., Comerford S., Cooper D., Cortizas E.M., Currall B.B., Foox J., Grills G.S., Kobetz E., Kumar N., Laine J., Lamar W.E., Mantero A.M.A., Mason C.E., Reding B.D., Robertson M., Roca M.A., Ryon K., Schürer S.C., Shukla B.S., Solle N.S., Stevenson M., Tallon J.J., Thomas C., Thomas T., Vidović D., Williams S.L., Yin X., Solo-Gabriele H.M. Relationships between SARS-CoV-2 in wastewater and COVID-19 clinical cases and hospitalizations, with and without normalization against indicators of human waste. ACS ES&T Water. 2022;2:1992–2003. doi: 10.1021/ACSESTWATER.2C00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparison between log10GC/L concentrations of SARS-CoV-2 IP4 and N1 genes during the sampling campaign in both WWTPs.
Data Availability Statement
Data will be made available on request.