ABSTRACT
Autophagy is a catabolic cellular process that targets and eliminates superfluous cytoplasmic components via lysosomal degradation. This evolutionarily conserved process is tightly regulated at multiple levels as it is critical for the maintenance of homeostasis. Research in the past decade has established that dysregulation of autophagy plays a major role in various diseases, such as cancer and neurodegeneration. However, modulation of autophagy as a therapeutic strategy requires identification of key players that can fine tune the induction of autophagy without complete abrogation. In this Review, we summarize the recent discoveries on the mechanism of regulation of ATG (autophagy related) gene expression at the level of transcription, post transcription and translation. Furthermore, we briefly discuss the role of aberrant expression of ATG genes in the context of cancer.
Keywords: Post-translational modification, Transcription, Translation
Summary: Autophagy is an essential process that is implicated in several diseases. We discuss recent findings that uncover regulators of autophagy in mammalian cells with a focus on cancer.
Introduction
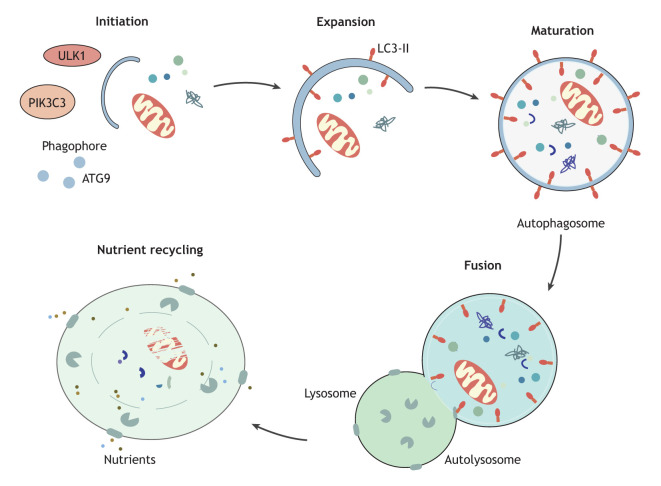
Macroautophagy (hereafter, autophagy) is an evolutionarily conserved, catabolic process by which superfluous cytoplasmic components, including protein aggregates and damaged organelles are sequestered within double-membrane structures called the autophagosome. Autophagosome biogenesis involves various steps, such as initiation, nucleation of the sequestering compartment (the phagophore), expansion, closure to form the autophagosome and fusion with a lysosome, or first with an endosome, allowing the autophagosome contents to be digested and the macromolecular products to be recycled (Dikic and Elazar, 2018). The process of autophagosome biogenesis and the subsequent fusion and delivery of superfluous cytoplasmic material to the lysosome is controlled by several ATG proteins that coordinate to initiate and complete autophagosome biogenesis (Box 1). The mechanistic roles of each of these proteins in eliciting autophagy is extensively reviewed elsewhere (Yang and Klionsky, 2010; Nakatogawa, 2020; Klionsky et al., 2021).
Box 1. Overview of autophagosome biogenesis.
Autophagosome biogenesis is characterized by five key steps – initiation, expansion, maturation, fusion and degradation-recycling (see figure). The induction of autophagosome biogenesis begins the with the phosphorylation of the unc-51 like autophagy activating kinase 1 (ULK1) complex including ULK1 (or ULK2), a serine/threonine protein kinase, RB1 inducible coiled-coil 1 (RB1CC1, also known as FIP200), autophagy related 13 (ATG13) and ATG101 (Jung et al., 2009; Hosokawa et al., 2009a; Ganley et al., 2009; Hara et al., 2008; Hosokawa et al., 2009b; Mercer et al., 2009). Following this, the ULK1 kinase phosphorylates various components of the class III phosphatidylinositol 3-kinase complex, consisting of phosphatidylinositol 3-kinase catalytic subunit type 3 (PIK3C3, also known as VPS34), beclin 1 (BECN1), phosphoinositide-3-kinase regulatory subunit 4 (PIK3R4, also known as VPS15 or p150), ATG14 and nuclear receptor binding factor 2 (NRBF2) for complex I, with UV radiation resistance associated (UVRAG) replacing ATG14 in complex II, which also includes SH3 domain containing GRB2 like, endophilin B1 (SH3GLB1) and autophagy and beclin 1 regulator 1 (AMBRA1) (Russell et al., 2013; Wold et al., 2016; Park et al., 2016; Di Bartolomeo et al., 2010). Upon activation of the lipid kinase PIK3C3, the production of phosphatidylinositol 3-phosphate, needed for the nucleation of the phagophore, increases (Nishimura et al., 2017; Cheng et al., 2014). The ATG9 trafficking system, comprised of the transmembrane protein ATG9A, ATG2 and WD repeat domain 45 (WDR45, also known as WIPI4) supplies membrane precursors to meet the demands required for autophagosome biogenesis (Itakura and Mizushima, 2010). Membrane expansion and autophagosome maturation are controlled by two ubiquitin-like conjugation systems, the ATG12–ATG5-ATG16L1 complex and the ubiquitin-like Atg8-family proteins, such as LC3, which function to conjugate the latter to the expanding phagophore (Fujita et al., 2008). Following phagophore expansion and closure, the mature autophagosome delivers the autophagic cargo along with its inner membrane to the lysosome for degradation. The contents of the autophagosome are degraded by the hydrolytic enzymes within the lysosome, and the resulting metabolites are subsequently released into the cytoplasm via transporters and are then reused by the cell (Mizushima, 2009; Broer and Gauthier-Coles, 2022).
Autophagy has roles in many diverse functions in cellular regulation, such as protein degradation, organelle homeostasis (Gatica et al., 2018), apoptosis (Marino et al., 2014), innate and adaptive immunity (Jiang et al., 2019; Metur and Klionsky, 2021), pathogen clearance (Pang et al., 2022) and maintenance of metabolic homeostasis (Lahiri et al., 2019). Accumulating evidence in the past decade has shown that dysfunctional autophagy plays a role in the pathogenesis of diseases, such as cancer (Yun and Lee, 2018; Mulcahy Levy and Thorburn, 2020; Li et al., 2020b), neurodegeneration (Nah et al., 2015) and metabolic disorders, such as diabetes, insulin resistance and obesity (Xu et al., 2021). Accordingly, autophagy is a prime target for manipulation for therapeutic purposes (Cheng et al., 2013; Galluzzi et al., 2017). However, owing to the diverse roles of autophagy, complete block or overexpression of this process can be detrimental to the cell. In line with this, modulation of induction and magnitude, concomitant with environmental cues, is required to maintain homeostasis. In the past decade, several regulators that fine tune the extent of autophagy induction by modulating the expression of ATG (autophagy related) genes have been identified. The targeted control of these regulators has tremendous therapeutic potential as they allow for more precise adjustment in manipulating autophagy for the treatment of disease.
In this Review, we summarize recent discoveries that elucidate the regulation of ATG gene and protein expression at the transcriptional, translational and post-translational levels, with a focus on cancer.
Regulation of ATG gene expression
Given the crucial and diverse functions of autophagy in cellular mechanisms, autophagy needs to be fine-tuned to avoid excessive or insufficient activity. Post-translational regulation of ATG proteins to control the extent of autophagic activity has been extensively studied. Recent investigations into the regulation of autophagy induction have identified several regulators that control ATG gene and protein expression at the transcriptional, post-transcriptional and translational levels.
Transcriptional regulation of autophagy
Regulation of ATG gene expression at the nucleus can occur through two modes – regulation by epigenetic modifications and regulation by transcription factors. In this section, we summarize studies that investigate these two modes of autophagy regulation.
Regulation by epigenetic modifications
Epigenetic modifications, including DNA and histone modifications, modulate chromatin structure, thus regulating the accessibility of transcription factors to chromatin (Lu et al., 2020).
DNA methylation refers to the methylation of cytosine on the fifth carbon atom to form 5-methylcytosine (5mC), which is catalyzed by DNA methyltransferases (DNMTs) using S-adenosylmethionine as the donor. This process usually occurs in the CpG islands within the gene promoter regions where the CpG dinucleotides are present in high frequency (Jabbari and Bernardi, 2004). DNA methylation suppresses gene transcription by blocking the binding between DNA and transcription factors. Conversely, DNA can also be demethylated either actively or passively (Moore et al., 2013). Active DNA demethylation refers to a process in which a methyl group of 5mC is removed or modified by enzymes, whereas passive DNA demethylation represents the elimination of 5mC during DNA replication.
A study focused on breast tumors revealed that the low expression of BECN1 mRNA is possibly caused by loss of heterozygosity, as well as aberrant methylation in the promoter and the intron 2 of this gene (Li et al., 2010). Consistent with this, the overexpression of tet methylcytosine dioxygenase 2 (TET2), which converts 5mC into 5-hydroxymethylcytosine (5hmC), results in decreased methylation of the BECN1 promoter, leading to an upregulation of BECN1 expression and eventually an elevated autophagy in human umbilical vascular endothelial cell (HUVEC) lines (Peng et al., 2016). Similarly, the promoter region of ULK2, an autophagy inducer gene (see Box 1), is hypermethylated in glioma cell lines, resulting in a significant downregulation of ULK2 expression and autophagy activity (Shukla et al., 2014). Furthermore, a kinase mutant of ULK2, deficient in autophagy induction, also fails to inhibit glioma cell growth, indicating that ULK2 inhibits the tumor growth in an autophagy-dependent manner. This conclusion was further validated by ectopic expression of ULK2, which induces autophagy efficiently and inhibits cell growth in wild-type cells but fails to do so in autophagy-deficient ATG5−/− cells. These data indicate that ULK2 DNA methylation impairs autophagy activity in glioma cells and is essential for gliomagenesis (Shukla et al., 2014). The methylation of the genes encoding lysosomal associated membrane protein 2 (LAMP2) and microtubule associated protein 1 light chain 3 (MAP1LC3, hereafter LC3)-family proteins are also reported to cause deficient autophagy and are associated with the pathogenesis of Danon disease and lung cancer, respectively (Chen et al., 2018; Ng et al., 2016). Additionally, the methylation of several other genes that are not directly involved in autophagy flux affect autophagy activity. For instance, the aberrant methylation in the promoter regions of nuclear receptor subfamily 4 group A member 3 (NR4A3, also known as NOR1) and SRY-box transcription factor 1 (SOX1) have been found in tumor cell lines and cause a decreased autophagy flux (Li et al., 2013; Yi et al., 2017).
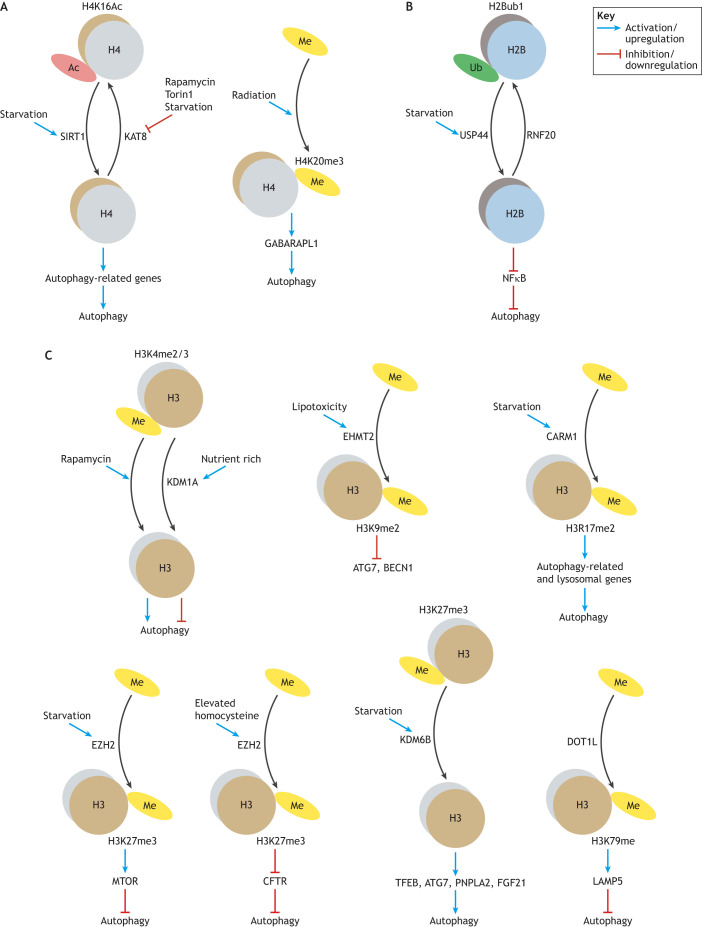
Chromatin is composed of DNA and histones. Apart from modifying DNA, histone modification also influences the overall chromatin structure and mediates the accessibility of genes, which affects their transcription. Accumulating evidence highlights the association between histone (and non-histone) modification and autophagy regulation (see Table 1 and Fig. 1).
Table 1.
Histone modifications involved in autophagy regulation
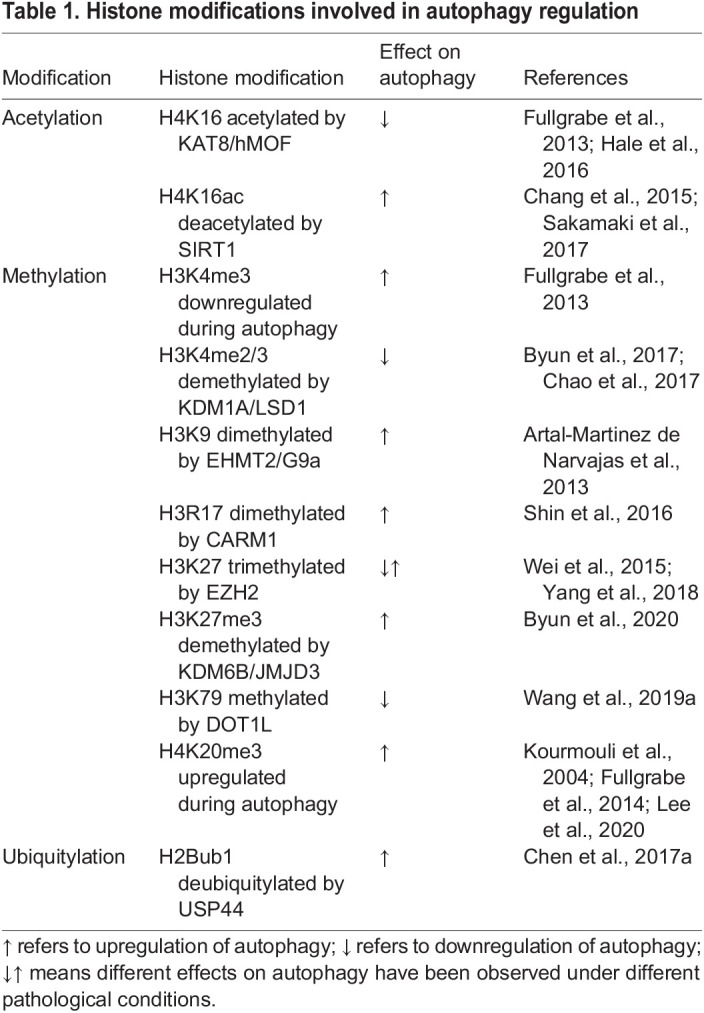
Fig. 1.
Histone modifications and autophagy regulation. Histone modifications regulate autophagy by modulating chromatin structure and gene expression. Here, we show the histone modifications, such as acetylation (Ac), methylation (Me) and ubiquitylation (Ub), implicated in autophagy regulation for the indicated histone proteins. (A) Histone modifications on histone 4. (B) Histone modification on histone 2B. (C) Histone modification on histone 3. Of note, different effects for the same histone modification on autophagy regulation have been observed for several histone modifications. The specific modifiers are included in the figure for those histone modifications with known modifiers. See the text for reference to specific model systems and for further details.
Histone acetylation is catalyzed by histone acetylases (HATs) and is generally considered to activate or increase gene expression by relaxing the chromatin to create an open, accessible conformation for the binding of transcription factors (Bannister and Kouzarides, 2011). Several studies have demonstrated that histone acetylation is implicated in autophagy regulation. The reduction of H4K16ac resulting from decreased lysine acetyltransferase 8 (KAT8, also known as hMOF or MYST1) activity induces autophagy in mammalian cells (Fullgrabe et al., 2013; Hale et al., 2016). In line with this, the deacetylation of H4K16ac by the deacetylase sirtuin 1 (SIRT1) also plays a role in autophagy stimulation (Sakamaki et al., 2017; Chang et al., 2015) (Fig. 1A).
Histone methylation is also involved in autophagy regulation, and it can either activate or inhibit autophagy-related gene expression, depending on the specific site and the degree of methylation. Downregulation of H3K4me3 has been observed in both yeast and mammalian cells during autophagy induced by rapamycin, although the causal link between reduced H3K4me3 and elevated autophagy has not been explored (Fullgrabe et al., 2013). In contrast, demethylation of H3K4me2/3 by nuclear receptor subfamily 0 group B member 2 (NR0B2, or SHP), which recruits the lysine demethylase 1A (KDM1A, or LSD1) suppresses autophagy activity in mice (Byun et al., 2017), and the activation of autophagy by LSD1 inhibition is also observed in cancer cells (Chao et al., 2017) (Fig. 1C). Similarly, opposing effects of H3K27 trimethylation by enhancer of zeste 2 polycomb repressive complex 2 subunit (EZH2) on autophagy regulation have also been reported (Wei et al., 2015; Yang et al., 2018) (Fig. 1C). Moreover, the inhibition of autophagy by the methyltransferase euchromatic histone lysine methyltransferase 2 (EHMT2, or G9a), which methylates H3K9me2 on the promoters of LC3B, WD repeat domain, phosphoinositide interacting 1 (WIPI1) and tumor protein p53 inducible nuclear protein 2 (TP53INP2, or DOR), is observed in human, mouse and Drosophila melanogaster (Artal-Martinez de Narvajas et al., 2013). H3K27me3 is also associated with autophagy regulation. Fibroblast growth factor 21 (FGF21), which is induced upon fasting, activates hepatic autophagy via demethylation of histone H3K27me3 to epigenetically induce transcription of autophagy-related genes, including ATG7, ULK1, patatin like phospholipase domain containing 2 (PNPLA2, or ATGL), TFEB and PPARG coactivator 1α (PPARGC1A, or PGC-1α) (Byun et al., 2020). The pivotal role of H3R17 dimethylation by coactivator associated arginine methyltransferase 1 (CARM1) in autophagy induction has been demonstrated in mammals (Shin et al., 2016) (Fig. 1C). Enhanced levels of lysosomal associated membrane protein family member 5 (LAMP5) owing to the action of the H3K79 histone methyltransferase DOT1-like histone lysine methyltransferase (DOT1L) interact with ATG5 to impair autophagy flux (Wang et al., 2019a) (Fig. 1C). In addition, a link between histone H4K20 trimethylation and upregulation of autophagy has been reported in several studies (Kourmouli et al., 2004; Fullgrabe et al., 2014; Lee et al., 2020).
Finally, although histone ubiquitylation is a less well-studied modification in terms of autophagy regulation, it has also been reported to be an important epigenetic switch for autophagy regulation (Chen et al., 2017a). That study demonstrated that H2B (H2B clustered histone) monoubiquitylation is downregulated by the deubiquitylase ubiquitin specific peptidase 44 (UPS44) in response to starvation in human cell lines, which eventually led to autophagy activation (Chen et al., 2017a) (Fig. 1B).
Overall, the regulation of autophagy by histone modification is a complex and multifaceted process and further research is needed to better understand the underlying mechanisms.
Regulation by transcription factors
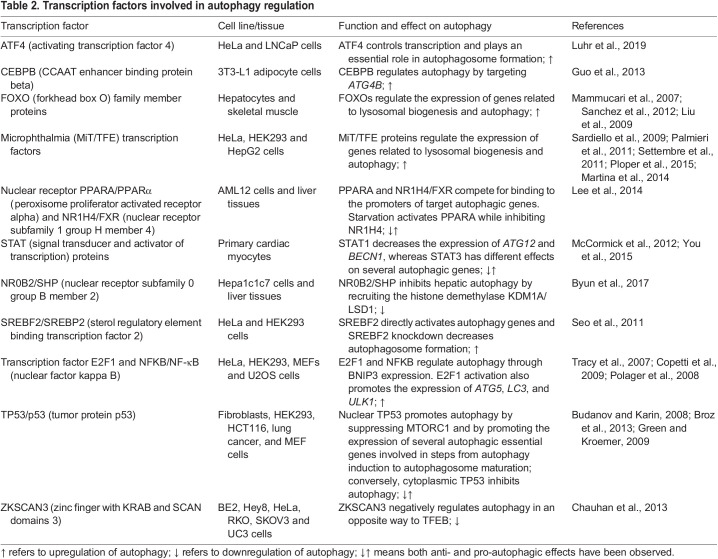
The induction of autophagy is accompanied by an increase of ATG gene expression in the nucleus, which is orchestrated by various transcription factors. Studies in the past decade have identified several transcription factors that regulate autophagy by either promoting or inhibiting the expression of autophagy-related and lysosomal genes at the level of RNA abundance. Here, we present a brief overview of transcription factors that are involved in the regulation of autophagy (Li et al., 2017b; Shin et al., 2016) (see also Table 2).
Table 2.
Transcription factors involved in autophagy regulation
Transcription factor EB (TFEB) is one of the key transcription factors involved in autophagy regulation. TFEB is a member of the microphthalmia/transcription factor E (MiT/TFE) family, along with melanocyte inducing transcription factor (MITF), TFEB, TFEC and transcription factor binding to IGHM enhancer 3 (TFE3) (Hemesath et al., 1994). Pattern discovery analysis of the promoter regions of many lysosomal genes showed that these genes have one or more so-called coordinated lysosomal expression and regulation (CLEAR) motifs, which can be preferentially recognized by MiT/TFE members (Sardiello et al., 2009). TFEB has emerged as a key regulator of autophagy as it mediates the expression of genes involved in lysosomal biogenesis, as well as many other genes involved in various autophagy steps, from genes encoding proteins critical for autophagy initiation (ATG9B, BECN1, NRBF2 and WIPI1; see Box 1) to genes for proteins that play important roles in the fusion of autophagosomes with lysosomes, for example, RAB7A and UV radiation resistance-associated (UVRAG) (Palmieri et al., 2011; Settembre et al., 2011). Furthermore, MITF and TFE3 also regulate autophagy flux in a similar manner to TFEB, modulating the expression of genes involved in lysosomal biogenesis as well as genes involved in different steps of autophagy (Martina et al., 2014; Ploper et al., 2015).
The forkhead box O (FOXO) family of transcription factors also regulates autophagy at the transcriptional level. Among the members of the FOXO family, FOXO3 was the first member reported to positively regulate the expression of several autophagy genes, including BCL2-interacting protein 3 (Bnip3), Becn1, Lc3b and GABA type A receptor associated protein like 1 (Gabarap1) in skeletal muscle (Mammucari et al., 2007; Sanchez et al., 2012). During osteoblast differentiation, elevated reactive oxygen species (ROS) levels resulting from an increase in mitochondrial respiration are mitigated by the induction of ATG gene transcription by FOXO3 (Gomez-Puerto et al., 2016). Furthermore, FOXO1 was also identified as a regulator for different autophagy genes at the transcriptional level including Pik3c3, Atg12 and Gabarapl1 (Liu et al., 2009). FOXO1 can also induce autophagy in human cancer cell lines under oxidative stress or serum-starvation conditions; however, this process is dependent on FOXO1 acetylation and its direct binding with ATG7 instead of its transcriptional activity (Zhao et al., 2010). Although a role for FOXO4 in autophagy regulation has been discovered subsequently (Matsuzaki et al., 2018), FOXO6 has not yet been found to be linked to autophagy regulation. Although the above studies suggest that FOXO transcription factors are positive regulators of autophagy, depletion of FOXO1, FOXO3, and FOXO4 results in an increase in autophagy during neuronal development, suggesting an underlying negative regulation of autophagy by the FOXO family (Schaffner et al., 2018). This study highlights the context-dependent regulation of autophagy; this is especially important to consider in cancer, as tumors originating from different tissues might have different modes of regulated autophagy.
Post-transcriptional and translational regulation of autophagy
Although context-specific transcription of ATG genes confers regulation of autophagy induction, numerous studies have identified essential players that are involved in the regulation of ATG gene expression at the post-transcriptional and translational levels, such as non-coding RNAs, mRNA modification and RNA-binding proteins (RBPs). In some cases, these regulators might affect the stability of the mRNA or translational efficiency by association with actively translating polysomes via RBPs, thereby modulating the abundance of ATG proteins and subsequently autophagy levels as discussed below.
Regulation by microRNAs
MicroRNAs (miRNAs) are a class of non-coding RNA of an average 22 nucleotides in length. In most cases, miRNAs regulate gene expression through binding to the 3′-untranslated region (3′ UTR) of target mRNA, leading to their degradation or translation inhibition, although interactions between miRNAs and other regions of mRNA are also reported (O'Brien et al., 2018; Xu et al., 2014). In addition, miRNAs can both directly bind and regulate ATG genes and control ATG gene expression indirectly, which makes ATG gene regulation by miRNAs complicated.
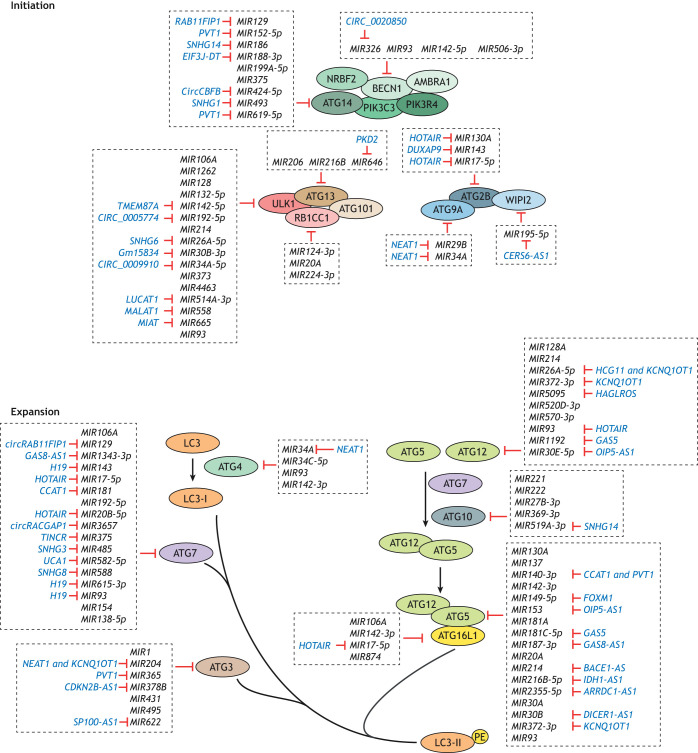
MIR30A, which targets BECN1, was the first identified autophagic miRNA (Zhu et al., 2009). Subsequently, a wide range of miRNAs have been found to affect autophagy activity by regulating ATG gene expression, with many of them associated with certain diseases, including cancers, neurodegenerative diseases and cardiac pathologies, which have been reviewed elsewhere (Akkoc and Gozuacik, 2020; Gozuacik et al., 2017; Xu et al., 2012). In the past several years, the list of autophagy-regulating miRNAs has grown considerably. Here, we summarize the miRNAs recently identified that directly bind to ATG mRNAs (Table S1; Fig. 2).
Fig. 2.
The regulation of ATG gene expression by non-coding RNAs. The expression of ATG genes can be regulated by miRNAs. In addition, ncRNAs and circular RNAs can function as competitive endogenous RNAs (ceRNAs) to control the expression of ATG genes through binding to miRNAs. It is of note that these types of regulation are usually observed under different cell contexts and can have different implications for human diseases. For further details see Table S1. PE, phosphatidylethanolamine.
Genes encoding ATG proteins that function in almost all stages of autophagy are found to be regulated by miRNAs, with ULK1, ATG5, ATG7 and ATG12 being especially targeted (Table S1; Fig. 2). One ATG gene can be targeted by several miRNAs, but it is of note that these studies were mostly performed under different backgrounds, including using different cell lines and autophagy stimuli, such as drug treatment and hypoxia conditions (Table S1). Taking ULK1 as an example (see Box 1), more than 15 miRNAs were discovered that directly bind ULK1 mRNA and eventually affect ULK1 expression and autophagy activity. These miRNAs were discovered and explored in different cell types, such as Mir214 in hepatocytes and rat renal proximal tubule cells (Lee et al., 2021; Ma et al., 2020b), MIR514A-3p in non-small cell lung cancer cells (Shen et al., 2020) and MIR192-5p in acute myeloid leukemia cells (Li et al., 2021a). In addition, the role of MIR93 and MIR4463 in regulating ULK1 expression under hypoxia conditions has been explored (Li et al., 2017a; He et al., 2022). Similarly, several miRNAs target ATG5. For instance, MIR142, MIR93 and MIR137 have been studied in hepatocellular carcinoma cells, glioma stem-like cells and pancreatic cancer cells, respectively, with regard to their function in chemotherapy-induced autophagy (Zhang et al., 2018; Huang et al., 2019; Wang et al., 2019b). Because miRNA expression is highly sensitive and specific to genetic background and cellular contexts (Peng and Croce, 2016), the targeting of one ATG by multiple miRNAs allows the strict control of autophagy under different conditions. In some cases, a specific miRNA targets not only a single but several ATG genes. For example, MIR106A binds to ULK1, ATG7 and ATG16L1 mRNAs in macrophages (Liu et al., 2020b), and MIR93 targets BECN1, ATG4B (autophagy related 4B cysteine peptidase) and ATG5 in glioma stem-like cells (Huang et al., 2019). Some miRNAs also target multiple ATG mRNAs in different contexts. For instance, MIR142-3p targets ATG5 and ATG16L1 in hepatocellular carcinoma cells (Zhang et al., 2018), but can also bind ATG4C in macrophages (Qu et al., 2021). MIR130A is another example; it regulates ATG2B expression in vascular smooth muscle cells (Zheng et al., 2021) and gastrointestinal stromal tumor cells (Zhang et al., 2021a) and targets ATG5 in hepatic cells (Duan et al., 2019).
As mentioned previously, miRNAs also regulate ATG gene expression indirectly. For instance overexpressing MIR21 upregulates ULK1 expression and promotes autophagy in non-small cell lung cancer (NSCLC) cells; however, ULK1 is not a predicted direct target of MIR21, indicating that there could be some intermediates that are targeted by this miRNA (Li et al., 2018). In addition, cisplatin-resistant ovarian cancer cells have lower MIR29C-3p expression and higher autophagy activity. Further exploration found that MIR29C-3p inhibits ATG14 expression through binding and downregulating its transcription factor, FOXP1, in ovarian cancer cells; accordingly, overexpressing MIR29C-3p inhibits cisplatin resistance partially through downregulation of FOXP1, ATG14 and eventually autophagy (Hu et al., 2020).
Regulation by long non-coding RNA and circular RNA
Long non-coding RNA (lncRNA) comprise another class of non-coding RNAs; they are longer than 200 nucleotides (Statello et al., 2021) and emerged as regulators participating in a collection of physiological processes, including autophagy (Yang et al., 2017). lncRNAs often regulate ATG genes as competing endogenous RNAs (ceRNAs) to modulate autophagic miRNAs (Table S1). For instance, in recently published studies, the lncRNA PVT1 has been found as the ceRNA of several autophagic miRNAs, including MIR365 (Yang et al., 2019), MIR140-3p (Wang et al., 2022), MIR152 (Yu et al., 2020) and MIR619-5p (Zhou et al., 2020), through which PVT1 (Pvt1 oncogene) induces ATG gene expression and thus promotes autophagy. In addition, HOX transcript antisense RNA (HOTAIR), H19 imprinted maternally expressed transcript (H19) and nuclear paraspeckle assembly transcript 1 (NEAT1), whose roles in cancer progression have been reported previously (Hajjari and Salavaty, 2015; Yang et al., 2021; Dong et al., 2018), are also found as the ceRNA of multiple autophagic miRNAs and, hence, affect ATG gene expression and autophagy (Li et al., 2020c; Liu et al., 2020a; Pan and Zhou, 2020; Wu et al., 2019, 2020; Zhang et al., 2021a).
Apart from functioning as a ceRNA, lncRNAs also regulate ATG gene expression through stabilizing their mRNAs. ZNF649-AS1, which has a higher expression in trastuzumab-resistant breast cancer cells, binds and recruits pyrimidine tract-binding protein 1 (PTBP1) to ATG5 mRNA, which helps in the stabilization of ATG5 mRNA and its expression. Similarly, IDH1-AS1 can stabilize ATG5 mRNA through binding to PTBP3 and enhancing its interaction with ATG5 mRNA in prostate cancer cells (Zhang et al., 2019b). Another lncRNA, EIF3J-DT, is reported to upregulate ATG14 expression level through both competitively binding (sponging) ATG14-targeting MIR188-3p and directly interacting with and stabilizing ATG14 mRNA (Luo et al., 2021). Additionally, overexpressing HULC downregulates ATG7 and LC3 mRNA levels in epithelial ovarian carcinoma (Chen et al., 2017b), and overexpression of ZNNT1 upregulates ATG12 expression in uveal melanoma (Li et al., 2020a), but the mechanisms are not clear.
Circular RNAs (circRNAs) are another class of noncoding RNAs discovered in recent years (Meng et al., 2017), and a role in regulating ATG genes and autophagy as ceRNAs is emerging (Table S1). For instance, CIRC_0005774 and CIRC_0009910 function as ceRNAs of ULK1 targeting MIR192-5p and MIR34A-5p, respectively (Li et al., 2021a; Cao et al., 2020), and circRAB11FIP1 and circRACGAP1 can upregulate ATG7 expression through sponging MIR129 and MIR3657, respectively (Zhang et al., 2021b; Ma et al., 2020a). It is of note that most of the studies focusing on circRNA and ATG gene expression are conducted in cancer cells, and the altered expression of these circRNAs is usually correlated with cancer phenotypes (Table S1), which points to circRNAs as new potential biomarkers for cancer (Meng et al., 2017).
Regulation by RNA modification
N6-methyladenosine (m6A) is the most abundant post-transcriptional RNA modification in eukaryotes and is recognized by regulators, such as methyltransferase (‘writers’), demethylases (‘erasers’) and RBPs (‘readers’). m6A contributes to diverse aspects of RNA function, such as pre-mRNA splicing, nuclear transport, translation and stability. Thus, abnormal m6A modifications can have detrimental effects and have been shown to result in the development of human diseases (Chen et al., 2021).
Several studies have revealed the link between m6A modification and autophagy. Notably, m6A modification decreases the abundance of the ATG transcripts. Regulators that ‘write’ m6A are negative regulators of autophagy owing to the decrease in ATG transcript abundance, which results in autophagy inhibition. Conversely, m6A ‘erasers’ positively regulate ATG transcript abundance and thus autophagy. Additionally, RBPs that are ‘readers’ can specifically bind to the m6A methylation site, and exert their influence on RNA function. A screen using small interfering RNAs (siRNAs) specifically targeting genes that encode the regulators of m6A modification with the goal of identifying regulators of autophagy revealed the FTO α-ketoglutarate dependent dioxygenase as a positive regulator through its post-transcriptional regulation of ULK1 (Jin et al., 2018). FTO is an m6A ‘eraser’, and therefore, to investigate the presence of m6A methylation on ULK1 transcripts, the authors performed m6A-Seq, an immunocapturing approach using m6A-specific antibodies to precipitate RNA to identify transcriptome-wide localization of this modification. Analysis of the m6A modification sites on ULK1 transcripts using m6A-Seq showed that ULK1 is modified in the 3′ UTR region. The m6A-modified ULK1 transcripts could then be targeted for degradation by YTH N6-methyladenosine RNA-binding protein F2 (YTHDF2), unless reversed by FTO, which, as an ‘eraser’, demethylates the transcript to positively regulate its abundance (Jin et al., 2018). In adipogenesis, deletion of FTO decreases the expression of ATG5 and ATG7, abrogating autophagosome formation and, thus, inhibiting autophagy in a manner similar to post-transcriptional regulation of ULK1 (Wang et al., 2020). Furthermore, in hypoxia-reoxygenation treated neonatal mouse ventricular cardiomyocytes, aberrant m6A methylation of TFEB by methyltransferase 3, N6-adenosine-methyltransferase complex catalytic subunit (METTL3), a methyltransferase or ‘writer’, inhibits autophagy flux by reducing the expression of Tfeb mRNA (Song et al., 2019). These studies have broadened our understanding of the post-transcriptional regulation of autophagy by exploring the effect of m6A modification on autophagy. This interplay extends to critical roles in the regulation of diseases such as obesity, heart disease and cancer (Chen et al., 2021).
Regulation by RBPs
RBPs elicit regulatory roles in gene expression by modulating the functional state of RNA commensurate with the cellular context. By regulating all steps of the RNA lifecycle, such as transcription, splicing, modification, translation and decay, RBPs control regulatory networks that are critical to maintaining cellular homeostasis. Several recent studies have revealed the intricate interplay between RNA metabolism and RBPs that modulate autophagy activity via post-transcriptional and translational regulatory modalities.
In searching for RBPs that regulate autophagy, a high-throughput siRNA screen targeting 1530 RBPs in MCF-7 cells determined that ablation of the eukaryotic translation initiation factors 5A (EIF5A) and 4A3 (EIF4A3) causes a decrease in GFP–LC3B puncta in autophagy-inducing conditions (Lubas et al., 2018; Sakellariou et al., 2021). Association of EIF5A with ribosomes increases during starvation conditions, suggesting that recruitment of EIF5A to ribosomes can reflect the repertoire of the actively translated pool of transcripts that promote autophagy. Analysis of newly synthesized proteins by liquid chromatography mass spectrometry (LC-MS) determined that ATG3 protein levels are decreased upon EIF5A ablation and revealed that EIF5A assists the translation of ATG3 through a hard-to-translate motif called the DDG motif. This facilitates the lipidation of LC3B and thus the promotion of autophagosome biogenesis (Lubas et al., 2018). Interestingly, in B cells, the polyamine spermidine post-translationally modifies EIF5A by adding the unusual amino acid hypusine (Zhang et al., 2019a). Hypusinated EIF5A is required for the translation of hard-to-read motifs and is also important for the translation of TFEB, a key autophagy regulator (Zhang et al., 2019a).
The human embryonic lethal abnormal vision (ELAV) family is a highly conserved family of RBPs that consists of four members, ELAV like RNA binding protein 1 (ELAVL1, also known as HuA or HuR), ELAVL2 (HuB or Hel-N1), ELAVL3 (HuC) and ELAVL4 (HuD). These RBPs are characterized by the presence of three RNA recognition motifs (RRMs), with RRM2 and RRM3 connected by a flexible linker region (Maris et al., 2005). The ELAV family RBPs stabilize mRNAs and subsequently activate their translation. Analyses of their targets have revealed these RBPs regulate autophagy by stabilization and translational upregulation of ATG transcripts. For example, in pancreatic β-cells, ELAVL4 associates with ATG5 mRNA (Kim et al., 2014; Lee et al., 2012). Here, ELAVL4 binds to the 3′ UTR of ATG5 mRNA. Ablation or overexpression of ELAVL4 does not affect endogenous ATG5 mRNA levels; however, the protein levels of ATG5 are significantly reduced following silencing of ELAVL4 (Kim et al., 2014). Furthermore, analysis of translation of ATG5 mRNA by polysome fractionation confirms that ELAVL4 enhances ATG5 translation by increasing its association with actively translating polysome fractions. ELAVL4 promotes autophagosome biogenesis by increasing ATG5 abundance, therefore acting as a positive regulator of autophagy (Kim et al., 2014). Furthermore, ELAVL1 was found to bind to ATG5, ATG12 and ATG16L1 mRNAs at their 3′ UTR regions, and it regulates autophagy by enhancing translation of these transcripts (Ji et al., 2019). Studies focusing on elucidating the role of ELAVL1 in hypoxia-induced autophagy demonstrate that ELAVL1 binds to ATG7 and ATG16L1 in the coding and 3′ UTR regions, respectively, which results in the upregulation of the protein levels and enhanced autophagosome formation (Palanisamy et al., 2019). Another study determined that ELAVL1 activates autophagy by stabilizing ATG7 transcripts to suppress senescence in diabetic nucleus pulposus (NP) cells (Shao et al., 2021). Together, these studies show that ELAV proteins, such as ELAVL4 and ELAVL1, are important regulators of ATG mRNA stability and promote autophagy by enhancing ATG mRNA translation through association with their 3′ UTRs. It is worth noting that ATG mRNAs can also be negatively regulated by RBPs. For instance, ZFP36 ring finger protein (ZFP36, also known as TTP) binds to the 3′ UTR of ATG16L1 mRNA to recruit deadenylation and degradation factors during ferroptosis, thus acting as a negative regulator of autophagy (Zhang et al., 2020).
Regulation of autophagy in cancer
It is well accepted that autophagy plays dual roles in cancer. In the early stages of tumorigenesis, autophagy acts as a tumor suppressor by preventing the accumulation of intracellular waste. In established tumors, autophagy exerts a pro-survival role by providing the cancer cells with energy and nutrients during periods of stress, such as nutrient deprivation and hypoxia (Ariosa et al., 2021). Even though ATG genes are not highly targeted by single-nucleotide mutations (Lebovitz et al., 2015), aberrant ATG gene expression has been found in a wide range of cancers. Reduced expression of BECN1, the first ATG protein linked with cancer (Liang et al., 1999), has been shown in ovarian carcinomas (Lin et al., 2013), NSLSC (Zhou et al., 2013) and gastric cancer (Cao et al., 2016). Importantly, these studies also demonstrate the association between low BECN1 expression and poor prognosis. ATG5 and ATG7 are the other two ATG genes that have received great attention due to their essential roles in the autophagy process. The mode of dysregulation of ATG5 and ATG7 is cancer dependent. Some cancers harbor elevated levels of ATG5 and ATG7, whereas they are downregulated in others. In chemoresistant gastric cancer cells, ATG5 expression is upregulated, and inhibition of ATG5 sensitizes these cells to the treatment (Ge et al., 2014). In addition, a recent study indicates high ATG5 expression pan-cancer, especially in solid tumors, which is significantly associated with poor patient prognosis in most cases (Xu et al., 2021). Elevated ATG7 expression is found in some bladder cancer and lung cancer patients (Zhu et al., 2017, 2019; Sun et al., 2016). Furthermore, a higher ATG7 expression level is associated with lower overall survival rate in breast cancer patients, indicating a prognostic value of ATG7 (Desai et al., 2013). In contrast, significantly reduced ATG5 expression is detected in cancer tissue from colorectal cancer patients compared with their adjacent normal mucosa (Cho et al., 2012). A change in ULK1 expression has also been demonstrated in a recent pan-cancer study; lower ULK1 expression compared with the normal tissue is found in brain, gynecological and esophageal cancer types, but higher ULK1 expression has been discovered in lung squamous cell carcinoma (Kumar and Papaleo, 2020). The fact that ATG gene expression can be altered to opposite directions in cancer cells might be explained by the difference in cancer type, the stage of the tumor and drug resistance, which further indicates the complex association between cancer and autophagy.
The change of ATG gene expression in cancer cells could result from dysregulation at different levels. In cancer, the DNA methylation status of some ATG genes is altered compared with that in normal cells or tissues. For instance, in early-stage melanoma, the ATG5 promoter region is hypermethylated, which results in lower ATG5 expression; here, reduced colony formation after overexpressing ATG5 indicates that lower expression of ATG5 contributes to early tumorigenesis (Liu et al., 2013). In addition, hypermethylation in the promoters of ATG2B, ATG4D, ATG9A and ATG9B is seen in specimens of invasive ductal carcinoma, which results in a lower expression of these genes (Zhang et al., 2016). In contrast, lung adenocarcinoma cells that are resistant to erlotinib have higher levels of demethylated MAP1LC3A, which leads to increased expression of this gene and induction of autophagy (Nihira et al., 2014). Transcription of ATG genes in cancer cells can also be different from normal cells due to mutations in autophagy transcription factors that affect their function, expression level and cellular localization. Mutations in tumor protein p53 (TP53) are the most common genetic alternation in human cancers (Parrales and Iwakuma, 2015) and cancer-associated mutations, such as R273H and R175H, reduce the transcription of ATG12 and BECN1 (Cordani et al., 2016). In addition, higher TFEB expression and nuclear localization are reported in multiple cancers (Blessing et al., 2017; Fang et al., 2017; Giatromanolaki et al., 2015), which might lead to increased ATG gene expression and autophagy activity.
At the post-transcription level, the ncRNAs with a role in regulating ATG gene expression can be differently expressed in cancer cells compared to normal tissue. For instance, ATG3-targeting MIR1, ATG5-targeting MIR153-3p and ATG7-targeting MIR138-5p are downregulated in drug-resistant NSCLC cells or tissues (Hua et al., 2018; Pan et al., 2019; Zhang et al., 2019c), and accordingly, higher expression of ATG3 and ATG5 are reported in the first two studies. lncRNAs are usually upregulated in cancer cells and, in most cases, their levels are correlated with tumor proliferation and drug resistance (Zhang and Lu, 2020). The lncRNA HOTAIR is upregulated in colorectal cancer cells and tissues, especially after radiotherapy, which leads to higher ATG12 expression through sponging of the ATG12-targeting MIR93. The fact that knocking down HOTAIR and ATG12 can both inhibit autophagy and potentiate radiosensitivity indicates that HOTAIR mediates radioresistance through upregulating autophagy (Liu et al., 2020c). A similar phenotype is seen in gastrointestinal stromal tumors, where HOTAIR expression is upregulated in recurrent tumors and after imatinib treatment. Downregulation of HOTAIR and ATG2B, an ATG gene regulated by HOTAIR through MIR130A, can both induce imatinib sensitivity (Zhang et al., 2021a). Similarly, the lncRNA KCNQ1 opposite strand/antisense transcript 1 (KCNQ1OT1) is upregulated in radiotherapy-resistant lung adenocarcinoma cells, which increases ATG5 and ATG12 expression through sponging MIR372, resulting in higher autophagy activity. The fact that knocking down KCNQ1OT1 or overexpressing MIR372 promotes radiosensitivity, and further overexpressing ATG5 and ATG12 suppresses this effect, connects increased autophagy with the resistance to radiotherapy (He et al., 2020). Another lncRNA, PVT1, is upregulated and promotes the expression of ATG genes and autophagy in hepatocellular carcinoma (Yang et al., 2019) and lung cancer cells (Wang et al., 2022), and its role in promoting cancer cell proliferation or chemoresistance is also demonstrated in these studies. Further experiments assessing whether downregulating ATG genes regulated by PVT1 can reach a similar effect to that seen upon knocking down PVT1 might help to elucidate how autophagy participates in the PVT1-mediated cancer cell proliferation and chemoresistance.
M6A methylation, another post-transcriptional regulatory mechanism, influences tumor development and drug resistance by modifying ATG transcripts. In human hepatocellular carcinoma (HCC), significant overexpression of the m6A reader YTH N6-methyladenosine RNA-binding protein F1 (YTHDF1) is associated with poor prognosis (Zhao et al., 2018). Upon further investigation, it was revealed that YTHDF1 binds to and promotes translation of m6A methylated ATG2A and ATG14 (Li et al., 2021b). Deficiency of YTHDF1 does not affect the stability of ATG2A and ATG14; however, there is a moderate shift of the transcripts to non-polysome fractions. Conversely, these two ATG transcripts are shifted to the highly translating polysome fractions upon overexpression of YTHDF1, suggesting that YTHDF1 promotes autophagy in hypoxic conditions via translation of ATG2A and ATG14 (Li et al., 2021b). Patients suffering from advanced HCC are frequently treated with sorafenib, but they are susceptible to developing a resistance to the therapy (Ben Mousa, 2008). Analysis of patient tumors with acquired sorafenib resistance showed that METTL3 is significantly downregulated (Lin et al., 2020). Further investigation utilizing RNA m6A-Seq identified FOXO3 as being methylated at the 3′ UTR by METTL3 (Lin et al., 2020). Depletion of METTL3 in sorafenib-resistant cells reduces the stability of FOXO3. In this study, the authors found that FOXO3 directly negatively affects the expression of ATG3, ATG5, ATG7, ATG12, ATG16L1 and LC3B. Overexpression of METTL3 or FOXO3 sensitizes the cells to sorafenib treatment by downregulating autophagy. Therefore, the METTL3-FOXO3-autophagy axis is a key regulator of sorafenib resistance in HCC (Lin et al., 2020).
These recently published reports underscore the close correlation between ATG gene dysregulation and cancer, and suggest the potential of ATG genes and proteins as therapeutic targets or biomarkers for diagnosis and prognosis.
Conclusions
As a critical cellular pathway with diverse roles in maintaining organismal homeostasis, autophagy needs to be tightly controlled. Tremendous progress has been made in the past decade in broadening our understanding of the molecular mechanisms and regulatory networks that fine-tune the induction and extent of autophagy by controlling ATG gene expression. We have highlighted here several proteins that exert regulatory control at the transcriptional, post-translational and translational levels. Furthermore, we have provided a brief overview of dysregulation of autophagy at these levels caused by either mutations or differential expression of regulators. Although autophagy is routinely implicated in cancer, and ATG genes are favorable prognostic markers due to their differential expression, not all ATG subtypes are similarly changed in all types of cancer. The autophagy-independent roles of these ATG genes adds to the complexity of this pathway and therefore yield different prognostic value as cancer markers. Furthermore, the correlation between the regulators of ATG gene expression and the type of cancer also remains obscure as the function of these genes is again dependent on the tissue type. This lack of clarity might be due in part to the dual roles of regulators involved at the levels of transcriptional and post-transcriptional regulation. However, advances in genome sequencing and precision medicine provide an opportunity to target these regulators to elicit the desired autophagic response. Therefore, studies uncovering regulators of autophagy will continue to be important and useful in designing treatments for cancer.
Supplementary Material
Acknowledgements
We apologize to authors whose work could not be cited due to space limitations.
Footnotes
Funding
Our work in this area is supported by the National Institute of General Medical Sciences (GM131919). Deposited in PMC for release after 12 months.
References
- Akkoc, Y. and Gozuacik, D. (2020). MicroRNAs as major regulators of the autophagy pathway. Biochim. Biophys. Acta Mol. Cell Res. 1867, 118662. 10.1016/j.bbamcr.2020.118662 [DOI] [PubMed] [Google Scholar]
- Ariosa, A. R., Lahiri, V., Lei, Y., Yang, Y., Yin, Z., Zhang, Z. and Klionsky, D. J. (2021). A perspective on the role of autophagy in cancer. Biochim. Biophys. Acta. Mol. Basis. Dis. 1867, 166262. 10.1016/j.bbadis.2021.166262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artal-Martinez De Narvajas, A., Gomez, T. S., Zhang, J. S., Mann, A. O., Taoda, Y., Gorman, J. A., Herreros-Villanueva, M., Gress, T. M., Ellenrieder, V., Bujanda, L.et al. (2013). Epigenetic regulation of autophagy by the methyltransferase G9a. Mol. Cell Biol. 33, 3983-3993. 10.1128/MCB.00813-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister, A. J. and Kouzarides, T. (2011). Regulation of chromatin by histone modifications. Cell Res. 21, 381-395. 10.1038/cr.2011.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Mousa, A. (2008). Sorafenib in the treatment of advanced hepatocellular carcinoma. Saudi. J. Gastroenterol. 14, 40-42. 10.4103/1319-3767.37808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blessing, A. M., Rajapakshe, K., Reddy Bollu, L., Shi, Y., White, M. A., Pham, A. H., Lin, C., Jonsson, P., Cortes, C. J., Cheung, E.et al. (2017). Transcriptional regulation of core autophagy and lysosomal genes by the androgen receptor promotes prostate cancer progression. Autophagy 13, 506-521. 10.1080/15548627.2016.1268300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broer, S. and Gauthier-Coles, G. (2022). Amino acid homeostasis in mammalian cells with a focus on amino acid transport. J. Nutr. 152, 16-28. 10.1093/jn/nxab342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broz, D. K., Mello, S. S., Bieging, K. T., Jiang, D. D., Dusek, R. L., Brady, C. A., Sidow, A. and Attardi, L. D. (2013). Global genomic profiling reveals an extensive p53-regulated autophagy program contributing to key p53 responses. Genes Dev. 27, 1016-1031. 10.1101/gad.212282.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budanov, A. V. and Karin, M. (2008). p53 target genes Sestrin1 and Sestrin2 connect genotoxic stress and mTOR signaling. Cell 134, 451-460. 10.1016/j.cell.2008.06.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byun, S., Kim, Y. C., Zhang, Y., Kong, B., Guo, G., Sadoshima, J., Ma, J., Kemper, B. and Kemper, J. K. (2017). A postprandial FGF19-SHP-LSD1 regulatory axis mediates epigenetic repression of hepatic autophagy. Embo J. 36, 1755-1769. 10.15252/embj.201695500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byun, S., Seok, S., Kim, Y. C., Zhang, Y., Yau, P., Iwamori, N., Xu, H. E., Ma, J., Kemper, B. and Kemper, J. K. (2020). Fasting-induced FGF21 signaling activates hepatic autophagy and lipid degradation via JMJD3 histone demethylase. Nat. Commun. 11, 807. 10.1038/s41467-020-14384-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, Q. H., Liu, F., Yang, Z. L., Fu, X. H., Yang, Z. H., Liu, Q., Wang, L., Wan, X. B. and Fan, X. J. (2016). Prognostic value of autophagy related proteins ULK1, Beclin 1, ATG3, ATG5, ATG7, ATG9, ATG10, ATG12, LC3B and p62/SQSTM1 in gastric cancer. Am. J. Transl. Res. 8, 3831-3847. [PMC free article] [PubMed] [Google Scholar]
- Cao, H. X., Miao, C. F., Sang, L. N., Huang, Y. M., Zhang, R., Sun, L. and Jiang, Z. X. (2020). Circ_0009910 promotes imatinib resistance through ULK1-induced autophagy by sponging miR-34a-5p in chronic myeloid leukemia. Life Sci. 243, 117255. 10.1016/j.lfs.2020.117255 [DOI] [PubMed] [Google Scholar]
- Chang, C., Su, H., Zhang, D., Wang, Y., Shen, Q., Liu, B., Huang, R., Zhou, T., Peng, C., Wong, C. C.et al. (2015). AMPK-dependent phosphorylation of GAPDH triggers Sirt1 activation and is necessary for autophagy upon glucose starvation. Mol. Cell 60, 930-940. 10.1016/j.molcel.2015.10.037 [DOI] [PubMed] [Google Scholar]
- Chao, A., Lin, C. Y., Chao, A. N., Tsai, C. L., Chen, M. Y., Lee, L. Y., Chang, T. C., Wang, T. H., Lai, C. H. and Wang, H. S. (2017). Lysine-specific demethylase 1 (LSD1) destabilizes p62 and inhibits autophagy in gynecologic malignancies. Oncotarget 8, 74434-74450. 10.18632/oncotarget.20158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan, S., Goodwin, J. G., Chauhan, S., Manyam, G., Wang, J., Kamat, A. M. and Boyd, D. D. (2013). ZKSCAN3 is a master transcriptional repressor of autophagy. Mol. Cell 50, 16-28. 10.1016/j.molcel.2013.01.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, S., Jing, Y., Kang, X., Yang, L., Wang, D. L., Zhang, W., Zhang, L., Chen, P., Chang, J. F., Yang, X. M.et al. (2017a). Histone H2B monoubiquitination is a critical epigenetic switch for the regulation of autophagy. Nucleic Acids Res. 45, 1144-1158. 10.1093/nar/gkw1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, S., Wu, D. D., Sang, X. B., Wang, L. L., Zong, Z. H., Sun, K. X., Liu, B. L. and Zhao, Y. (2017b). The lncRNA HULC functions as an oncogene by targeting ATG7 and ITGB1 in epithelial ovarian carcinoma. Cell Death Dis. 8, e3118. 10.1038/cddis.2017.486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y., Schnitzler, K. L., Ma, Y., Nenkov, M., Theis, B. and Petersen, I. (2018). The clinical influence of autophagy-associated proteins on human lung cancer. Dis. Markers 2018, 8314963. 10.1155/2018/8314963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X., Wang, J., Tahir, M., Zhang, F., Ran, Y., Liu, Z. and Wang, J. (2021). Current insights into the implications of m6A RNA methylation and autophagy interaction in human diseases. Cell Biosci. 11, 147. 10.1186/s13578-021-00661-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, Y., Ren, X., Hait, W. N. and Yang, J. M. (2013). Therapeutic targeting of autophagy in disease: biology and pharmacology. Pharmacol. Rev. 65, 1162-1197. 10.1124/pr.112.007120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, J., Fujita, A., Yamamoto, H., Tatematsu, T., Kakuta, S., Obara, K., Ohsumi, Y. and Fujimoto, T. (2014). Yeast and mammalian autophagosomes exhibit distinct phosphatidylinositol 3-phosphate asymmetries. Nat. Commun. 5, 3207. 10.1038/ncomms4207 [DOI] [PubMed] [Google Scholar]
- Cho, D. H., Jo, Y. K., Kim, S. C., Park, I. J. and Kim, J. C. (2012). Down-regulated expression of ATG5 in colorectal cancer. Anticancer Res. 32, 4091-4096. [PubMed] [Google Scholar]
- Copetti, T., Bertoli, C., Dalla, E., Demarchi, F. and Schneider, C. (2009). p65/RelA modulates BECN1 transcription and autophagy. Mol. Cell. Biol. 29, 2594-2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordani, M., Oppici, E., Dando, I., Butturini, E., Dalla Pozza, E., Nadal-Serrano, M., Oliver, J., Roca, P., Mariotto, S., Cellini, B.et al. (2016). Mutant p53 proteins counteract autophagic mechanism sensitizing cancer cells to mTOR inhibition. Mol. Oncol. 10, 1008-1029. 10.1016/j.molonc.2016.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai, S., Liu, Z., Yao, J., Patel, N., Chen, J., Wu, Y., Ahn, E. E., Fodstad, O. and Tan, M. (2013). Heat shock factor 1 (HSF1) controls chemoresistance and autophagy through transcriptional regulation of autophagy-related protein 7 (ATG7). J. Biol. Chem. 288, 9165-9176. 10.1074/jbc.M112.422071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Bartolomeo, S., Corazzari, M., Nazio, F., Oliverio, S., Lisi, G., Antonioli, M., Pagliarini, V., Matteoni, S., Fuoco, C., Giunta, L.et al. (2010). The dynamic interaction of AMBRA1 with the dynein motor complex regulates mammalian autophagy. J. Cell Biol. 191, 155-168. 10.1083/jcb.201002100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dikic, I. and Elazar, Z. (2018). Mechanism and medical implications of mammalian autophagy. Nat. Rev. Mol. Cell Biol. 19, 349-364. 10.1038/s41580-018-0003-4 [DOI] [PubMed] [Google Scholar]
- Dong, P., Xiong, Y., Yue, J., Hanley, S. J. B., Kobayashi, N., Todo, Y. and Watari, H. (2018). Long non-coding RNA NEAT1: a novel target for diagnosis and therapy in human tumors. Front. Genet. 9, 471. 10.3389/fgene.2018.00471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan, X., Liu, X., Li, W., Holmes, J. A., Kruger, A. J., Yang, C., Li, Y., Xu, M., Ye, H., Li, S.et al. (2019). Microrna-130a downregulates HCV replication through an atg5-dependent autophagy pathway. Cells 8, 338. 10.3390/cells8040338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang, L. M., Li, B., Guan, J. J., Xu, H. D., Shen, G. H., Gao, Q. G. and Qin, Z. H. (2017). Transcription factor EB is involved in autophagy-mediated chemoresistance to doxorubicin in human cancer cells. Acta Pharmacol. Sin. 38, 1305-1316. 10.1038/aps.2017.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita, N., Itoh, T., Omori, H., Fukuda, M., Noda, T. and Yoshimori, T. (2008). The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol. Biol. Cell. 19, 2092-2100. 10.1091/mbc.e07-12-1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullgrabe, J., Lynch-Day, M. A., Heldring, N., Li, W., Struijk, R. B., Ma, Q., Hermanson, O., Rosenfeld, M. G., Klionsky, D. J. and Joseph, B. (2013). The histone H4 lysine 16 acetyltransferase hMOF regulates the outcome of autophagy. Nature 500, 468-471. 10.1038/nature12313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullgrabe, J., Klionsky, D. J. and Joseph, B. (2014). The return of the nucleus: transcriptional and epigenetic control of autophagy. Nat. Rev. Mol. Cell Biol. 15, 65-74. 10.1038/nrm3716 [DOI] [PubMed] [Google Scholar]
- Galluzzi, L., Bravo-San Pedro, J. M., Levine, B., Green, D. R. and Kroemer, G. (2017). Pharmacological modulation of autophagy: therapeutic potential and persisting obstacles. Nat. Rev. Drug Discov. 16, 487-511. 10.1038/nrd.2017.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganley, I. G., Lam Du, H., Wang, J., Ding, X., Chen, S. and Jiang, X. (2009). ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J. Biol. Chem. 284, 12297-12305. 10.1074/jbc.M900573200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatica, D., Lahiri, V. and Klionsky, D. J. (2018). Cargo recognition and degradation by selective autophagy. Nat. Cell Biol. 20, 233-242. 10.1038/s41556-018-0037-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge, J., Chen, Z., Huang, J., Chen, J., Yuan, W., Deng, Z. and Chen, Z. (2014). Upregulation of autophagy-related gene-5 (ATG-5) is associated with chemoresistance in human gastric cancer. PLoS One 9, e110293. 10.1371/journal.pone.0110293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giatromanolaki, A., Kalamida, D., Sivridis, E., Karagounis, I. V., Gatter, K. C., Harris, A. L. and Koukourakis, M. I. (2015). Increased expression of transcription factor EB (TFEB) is associated with autophagy, migratory phenotype and poor prognosis in non-small cell lung cancer. Lung Cancer 90, 98-105. 10.1016/j.lungcan.2015.07.008 [DOI] [PubMed] [Google Scholar]
- Gomez-Puerto, M. C., Verhagen, L. P., Braat, A. K., Lam, E. W., Coffer, P. J. and Lorenowicz, M. J. (2016). Activation of autophagy by FOXO3 regulates redox homeostasis during osteogenic differentiation. Autophagy 12, 1804-1816. 10.1080/15548627.2016.1203484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gozuacik, D., Akkoc, Y., Ozturk, D. G. and Kocak, M. (2017). Autophagy-regulating microRNAs and cancer. Front. Oncol. 7, 65. 10.3389/fonc.2017.00065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green, D. R. and Kroemer, G. (2009). Cytoplasmic functions of the tumour suppressor p53. Nature, 458, 1127-1130. 10.1038/nature07986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, L., Huang, J. X., Liu, Y., Li, X., Zhou, S. R., Qian, S. W., Liu, Y., Zhu, H., Huang, H. Y., Dang, Y. J.et al. (2013). Transactivation of Atg4b by C/EBP beta promotes autophagy to facilitate adipogenesis. Mol. Cell. Biol. 33, 3180-3190. 10.1128/Mcb.00193-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajjari, M. and Salavaty, A. (2015). HOTAIR: an oncogenic long non-coding RNA in different cancers. Cancer Biol. Med. 12, 1-9. 10.7497/j.issn.2095-3941.2015.0006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale, C. M., Cheng, Q., Ortuno, D., Huang, M., Nojima, D., Kassner, P. D., Wang, S., Ollmann, M. M. and Carlisle, H. J. (2016). Identification of modulators of autophagic flux in an image-based high content siRNA screen. Autophagy 12, 713-726. 10.1080/15548627.2016.1147669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara, T., Takamura, A., Kishi, C., Iemura, S., Natsume, T., Guan, J. L. and Mizushima, N. (2008). FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol, 181, 497-510. 10.1083/jcb.200712064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, H., Song, X., Yang, Z., Mao, Y., Zhang, K., Wang, Y., Su, B., Li, Q., Chen, H. and Li, Y. (2020). Upregulation of KCNQ1OT1 promotes resistance to stereotactic body radiotherapy in lung adenocarcinoma by inducing ATG5/ATG12-mediated autophagy via miR-372-3p. Cell Death Dis. 11, 883. 10.1038/s41419-020-03083-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, X., Wu, F., Zhou, Y., Wu, J., Long, Y. and Zhou, X. (2022). miR-4463 regulates hypoxia-induced autophagy and apoptosis by targeting ULK1 in endothelial cells. Front. Biosci. (Landmark Ed), 27, 175. 10.31083/j.fbl2706175 [DOI] [PubMed] [Google Scholar]
- Hemesath, T. J., Steingrimsson, E., Mcgill, G., Hansen, M. J., Vaught, J., Hodgkinson, C. A., Arnheiter, H., Copeland, N. G., Jenkins, N. A. and Fisher, D. E. (1994). Microphthalmia, a critical factor in melanocyte development, defines a discrete transcription factor family. Genes Dev. 8, 2770-2780. 10.1101/gad.8.22.2770 [DOI] [PubMed] [Google Scholar]
- Hosokawa, N., Hara, T., Kaizuka, T., Kishi, C., Takamura, A., Miura, Y., Iemura, S., Natsume, T., Takehana, K., Yamada, N.et al. (2009a). Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol. Biol. Cell 20, 1981-1991. 10.1091/mbc.e08-12-1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa, N., Sasaki, T., Iemura, S., Natsume, T., Hara, T. and Mizushima, N. (2009b). Atg101, a novel mammalian autophagy protein interacting with Atg13. Autophagy 5, 973-979. 10.4161/auto.5.7.9296 [DOI] [PubMed] [Google Scholar]
- Hu, Z., Cai, M., Zhang, Y., Tao, L. and Guo, R. (2020). miR-29c-3p inhibits autophagy and cisplatin resistance in ovarian cancer by regulating FOXP1/ATG14 pathway. Cell Cycle, 19, 193-206. 10.1080/15384101.2019.1704537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua, L., Zhu, G. and Wei, J. (2018). MicroRNA-1 overexpression increases chemosensitivity of non-small cell lung cancer cells by inhibiting autophagy related 3-mediated autophagy. Cell Biol. Int. 42, 1240-1249. 10.1002/cbin.10995 [DOI] [PubMed] [Google Scholar]
- Huang, T., Wan, X., Alvarez, A. A., James, C. D., Song, X., Yang, Y., Sastry, N., Nakano, I., Sulman, E. P., Hu, B.et al. (2019). MIR93 (microRNA -93) regulates tumorigenicity and therapy response of glioblastoma by targeting autophagy. Autophagy 15, 1100-1111. 10.1080/15548627.2019.1569947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura, E. and Mizushima, N. (2010). Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy 6, 764-776. 10.4161/auto.6.6.12709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabbari, K. and Bernardi, G. (2004). Cytosine methylation and CpG, TpG (CpA) and TpA frequencies. Gene, 333, 143-149. 10.1016/j.gene.2004.02.043 [DOI] [PubMed] [Google Scholar]
- Ji, E., Kim, C., Kang, H., Ahn, S., Jung, M., Hong, Y., Tak, H., Lee, S., Kim, W. and Lee, E. K. (2019). RNA Binding protein HuR promotes autophagosome formation by regulating expression of autophagy-related proteins 5, 12, and 16 in human hepatocellular carcinoma cells. Mol. Cell. Biol. 39, e00508-18. 10.1128/MCB.00508-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, G. M., Tan, Y., Wang, H., Peng, L., Chen, H. T., Meng, X. J., Li, L. L., Liu, Y., Li, W. F. and Shan, H. (2019). The relationship between autophagy and the immune system and its applications for tumor immunotherapy. Mol. Cancer 18, 17. 10.1186/s12943-019-0944-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, S., Zhang, X., Miao, Y., Liang, P., Zhu, K., She, Y., Wu, Y., Liu, D. A., Huang, J., Ren, J.et al. (2018). m(6)A RNA modification controls autophagy through upregulating ULK1 protein abundance. Cell Res. 28, 955-957. 10.1038/s41422-018-0069-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung, C. H., Jun, C. B., Ro, S. H., Kim, Y. M., Otto, N. M., Cao, J., Kundu, M. and Kim, D. H. (2009). ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol. Biol. Cell 20, 1992-2003. 10.1091/mbc.e08-12-1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, C., Kim, W., Lee, H., Ji, E., Choe, Y. J., Martindale, J. L., Akamatsu, W., Okano, H., Kim, H. S., Nam, S. W.et al. (2014). The RNA-binding protein HuD regulates autophagosome formation in pancreatic beta cells by promoting autophagy-related gene 5 expression. J. Biol. Chem. 289, 112-121. 10.1074/jbc.M113.474700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky, D. J., Petroni, G., Amaravadi, R. K., Baehrecke, E. H., Ballabio, A., Boya, P., Bravo-San Pedro, J. M., Cadwell, K., Cecconi, F., Choi, A. M. K.et al. (2021). Autophagy in major human diseases. EMBO J. 40, e108863. 10.15252/embj.2021108863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourmouli, N., Jeppesen, P., Mahadevhaiah, S., Burgoyne, P., Wu, R., Gilbert, D. M., Bongiorni, S., Prantera, G., Fanti, L., Pimpinelli, S.et al. (2004). Heterochromatin and tri-methylated lysine 20 of histone H4 in animals. J. Cell Sci. 117, 2491-2501. 10.1242/jcs.01238 [DOI] [PubMed] [Google Scholar]
- Kumar, M. and Papaleo, E. (2020). A pan-cancer assessment of alterations of the kinase domain of ULK1, an upstream regulator of autophagy. Sci. Rep. 10, 14874. 10.1038/s41598-020-71527-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahiri, V., Hawkins, W. D. and Klionsky, D. J. (2019). Watch what you (Self-) eat: autophagic mechanisms that modulate metabolism. Cell Metab. 29, 803-826. 10.1016/j.cmet.2019.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebovitz, C. B., Robertson, A. G., Goya, R., Jones, S. J., Morin, R. D., Marra, M. A. and Gorski, S. M. (2015). Cross-cancer profiling of molecular alterations within the human autophagy interaction network. Autophagy 11, 1668-1687. 10.1080/15548627.2015.1067362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, E. K., Kim, W., Tominaga, K., Martindale, J. L., Yang, X., Subaran, S. S., Carlson, O. D., Mercken, E. M., Kulkarni, R. N., Akamatsu, W.et al. (2012). RNA-binding protein HuD controls insulin translation. Mol. Cell 45, 826-835. 10.1016/j.molcel.2012.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J. M., Wagner, M., Xiao, R., Kim, K. H., Feng, D., Lazar, M. A. and Moore, D. D. (2014). Nutrient-sensing nuclear receptors coordinate autophagy. Nature 516, U112-U291. 10.1038/nature13961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, T. G., Kim, S. Y., Kim, H. R., Kim, H. and Kim, C. H. (2020). Radiation induces autophagy via histone H4 lysine 20 trimethylation in non-small cell lung cancer cells. Anticancer Res. 40, 2537-2548. 10.21873/anticanres.14224 [DOI] [PubMed] [Google Scholar]
- Lee, D. H., Park, S. H., Ahn, J., Hong, S. P., Lee, E., Jang, Y. J., Ha, T. Y., Huh, Y. H., Ha, S. Y., Jeon, T. I.et al. (2021). Mir214-3p and Hnf4a/Hnf4alpha reciprocally regulate Ulk1 expression and autophagy in nonalcoholic hepatic steatosis. Autophagy 17, 2415-2431. 10.1080/15548627.2020.1827779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, N., Li, X., Li, S., Zhou, S. and Zhou, Q. (2013). Cisplatin-induced downregulation of SOX1 increases drug resistance by activating autophagy in non-small cell lung cancer cell. Biochem. Biophys. Res. Commun. 439, 187-190. 10.1016/j.bbrc.2013.08.065 [DOI] [PubMed] [Google Scholar]
- Li, W., Yang, Y., Ba, Z., Li, S., Chen, H., Hou, X., Ma, L., He, P., Jiang, L., Li, L.et al. (2017a). MicroRNA-93 Regulates Hypoxia-Induced Autophagy by Targeting ULK1. Oxid. Med. Cell Longev. 2017, 2709053. 10.1155/2017/2709053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X., Yu, W., Qian, X., Xia, Y., Zheng, Y., Lee, J. H., Li, W., Lyu, J., Rao, G., Zhang, X.et al. (2017b). Nucleus-translocated ACSS2 promotes gene transcription for lysosomal biogenesis and autophagy. Mol. Cell 66, 684-697.e9. 10.1016/j.molcel.2017.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S., Zeng, X., Ma, R. and Wang, L. (2018). MicroRNA-21 promotes the proliferation, migration and invasion of non-small cell lung cancer A549 cells by regulating autophagy activity via AMPK/ULK1 signaling pathway. Exp. Ther. Med. 16, 2038-2045. 10.3892/etm.2018.6370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, P., He, J., Yang, Z., Ge, S., Zhang, H., Zhong, Q. and Fan, X. (2020a). ZNNT1 long noncoding RNA induces autophagy to inhibit tumorigenesis of uveal melanoma by regulating key autophagy gene expression. Autophagy 16, 1186-1199. 10.1080/15548627.2019.1659614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X., He, S. and Ma, B. (2020b). Autophagy and autophagy-related proteins in cancer. Mol. Cancer 19, 12. 10.1186/s12943-020-1138-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X., Zhou, Y., Yang, L., Ma, Y., Peng, X., Yang, S., Li, H. and Liu, J. (2020c). LncRNA NEAT1 promotes autophagy via regulating miR-204/ATG3 and enhanced cell resistance to sorafenib in hepatocellular carcinoma. J. Cell Physiol. 235, 3402-3413. 10.1002/jcp.29230 [DOI] [PubMed] [Google Scholar]
- Li, Q., Luan, Q., Zhu, H., Zhao, Y., Ji, J., Wu, F. and Yan, J. (2021a). Circular RNA circ_0005774 contributes to proliferation and suppresses apoptosis of acute myeloid leukemia cells via circ_0005774/miR-192-5p/ULK1 ceRNA pathway. Biochem. Biophys. Res. Commun. 551, 78-85. 10.1016/j.bbrc.2021.02.058 [DOI] [PubMed] [Google Scholar]
- Li, Q., Ni, Y., Zhang, L., Jiang, R., Xu, J., Yang, H., Hu, Y., Qiu, J., Pu, L., Tang, J.et al. (2021b). HIF-1alpha-induced expression of m6A reader YTHDF1 drives hypoxia-induced autophagy and malignancy of hepatocellular carcinoma by promoting ATG2A and ATG14 translation. Signal Transduct. Target Ther. 6, 76. 10.1038/s41392-020-00453-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Z. D., Chen, B., Wu, Y. Q., Jin, F., Xia, Y. J. and Liu, X. J. (2010). Genetic and epigenetic silencing of the beclin 1 gene in sporadic breast tumors. Bmc Cancer 10, 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, X. H., Jackson, S., Seaman, M., Brown, K., Kempkes, B., Hibshoosh, H. and Levine, B. (1999). Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 402, 672-676. 10.1038/45257 [DOI] [PubMed] [Google Scholar]
- Lin, H. X., Qiu, H. J., Zeng, F., Rao, H. L., Yang, G. F., Kung, H. F., Zhu, X. F., Zeng, Y. X., Cai, M. Y. and Xie, D. (2013). Decreased expression of Beclin 1 correlates closely with Bcl-xL expression and poor prognosis of ovarian carcinoma. PLoS One 8, e60516. 10.1371/journal.pone.0060516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, Z., Niu, Y., Wan, A., Chen, D., Liang, H., Chen, X., Sun, L., Zhan, S., Chen, L., Cheng, C.et al. (2020). RNA m6A methylation regulates sorafenib resistance in liver cancer through FOXO3-mediated autophagy. EMBO J. 39, e103181. 10.15252/embj.2019103181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H. Y., Han, J. M., Cao, S. Y., Hong, T., Zhuo, D. G., Shi, J. B., Liu, Z. Q. and Cao, W. H. (2009). Hepatic autophagy is suppressed in the presence of insulin resistance and hyperinsulinemia inhibition of foxo1-dependent expression of key autophagy genes by insulin. J. Biol. Chem. 284, 31484-31492. 10.1074/jbc.M109.033936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H., He, Z., Von Rutte, T., Yousefi, S., Hunger, R. E. and Simon, H. U. (2013). Down-regulation of autophagy-related protein 5 (ATG5) contributes to the pathogenesis of early-stage cutaneous melanoma. Sci. Transl. Med. 5, 202ra123. 10.1126/scitranslmed.3005864 [DOI] [PubMed] [Google Scholar]
- Liu, F., Ai, F. Y., Zhang, D. C., Tian, L., Yang, Z. Y. and Liu, S. J. (2020a). LncRNA NEAT1 knockdown attenuates autophagy to elevate 5-FU sensitivity in colorectal cancer via targeting miR-34a. Cancer Med. 9, 1079-1091. 10.1002/cam4.2746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, K., Hong, D., Zhang, F., Li, X., He, M., Han, X., Zhang, G., Xu, G., Stonehouse, N. J., Jiang, Z.et al. (2020b). MicroRNA-106a inhibits autophagy process and antimicrobial responses by targeting ULK1, ATG7, and ATG16L1 during mycobacterial infection. Front. Immunol. 11, 610021. 10.3389/fimmu.2020.610021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y., Chen, X., Chen, X., Liu, J., Gu, H., Fan, R. and Ge, H. (2020c). Long non-coding RNA HOTAIR knockdown enhances radiosensitivity through regulating microRNA-93/ATG12 axis in colorectal cancer. Cell Death Dis. 11, 175. 10.1038/s41419-020-2268-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, Y., Chan, Y. T., Tan, H. Y., Li, S., Wang, N. and Feng, Y. (2020). Epigenetic regulation in human cancer: the potential role of epi-drug in cancer therapy. Mol. Cancer 19, 79. 10.1186/s12943-020-01197-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubas, M., Harder, L. M., Kumsta, C., Tiessen, I., Hansen, M., Andersen, J. S., Lund, A. H. and Frankel, L. B. (2018). eIF5A is required for autophagy by mediating ATG3 translation. EMBO Rep. 19, e46072. 10.15252/embr.201846072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luhr, M., Torgersen, M. L., Szalai, P., Hashim, A., Brech, A., Staerk, J. and Engedal, N. (2019). The kinase PERK and the transcription factor ATF4 play distinct and essential roles in autophagy resulting from tunicamycin-induced ER stress. J. Biol. Chem. 294, 8197-8217. 10.1074/jbc.RA118.002829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, Y., Zheng, S., Wu, Q., Wu, J., Zhou, R., Wang, C., Wu, Z., Rong, X., Huang, N., Sun, L.et al. (2021). Long noncoding RNA (lncRNA) EIF3J-DT induces chemoresistance of gastric cancer via autophagy activation. Autophagy 17, 4083-4101. 10.1080/15548627.2021.1901204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, L., Wang, Z., Xie, M., Quan, Y., Zhu, W., Yang, F., Zhao, C., Fan, Y., Fang, N., Jiang, H.et al. (2020a). Silencing of circRACGAP1 sensitizes gastric cancer cells to apatinib via modulating autophagy by targeting miR-3657 and ATG7. Cell Death Dis., 11, 169. 10.1038/s41419-020-2352-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, Z., Li, L., Livingston, M. J., Zhang, D., Mi, Q., Zhang, M., Ding, H. F., Huo, Y., Mei, C. and Dong, Z. (2020b). p53/microRNA-214/ULK1 axis impairs renal tubular autophagy in diabetic kidney disease. J. Clin. Invest. 130, 5011-5026. 10.1172/JCI135536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mammucari, C., Milan, G., Romanello, V., Masiero, E., Rudolf, R., Del Piccolo, P., Burden, S. J., Di Lisi, R., Sandri, C., Zhao, J.et al. (2007). FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metabolism. 6, 458-471. 10.1016/j.cmet.2007.11.001 [DOI] [PubMed] [Google Scholar]
- Marino, G., Niso-Santano, M., Baehrecke, E. H. and Kroemer, G. (2014). Self-consumption: the interplay of autophagy and apoptosis. Nat. Rev. Mol. Cell Biol. 15, 81-94. 10.1038/nrm3735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maris, C., Dominguez, C. and Allain, F. H. (2005). The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 272, 2118-2131. 10.1111/j.1742-4658.2005.04653.x [DOI] [PubMed] [Google Scholar]
- Martina, J. A., Diab, H. I., Lishu, L., Jeong, A. L., Patange, S., Raben, N. and Puertollano, R. (2014). The nutrient-responsive transcription factor TFE3 promotes autophagy, lysosomal biogenesis, and clearance of cellular debris. Sci. Signal 7, ra9. 10.1126/scisignal.2004754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki, T., Alvarez-Garcia, O., Mokuda, S., Nagira, K., Olmer, M., Gamini, R., Miyata, K., Akasaki, Y., Su, A. I., Asahara, H.et al. (2018). FoxO transcription factors modulate autophagy and proteoglycan 4 in cartilage homeostasis and osteoarthritis. Sci. Transl. Med. 10, eaan0746. 10.1126/scitranslmed.aan0746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mccormick, J., Suleman, N., Scarabelli, T. M., Knight, R. A., Latchman, D. S. and Stephanou, A. (2012). STAT1 deficiency in the heart protects against myocardial infarction by enhancing autophagy. J. Cell. Mol. Med. 16, 386-393. 10.1111/j.1582-4934.2011.01323.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng, S., Zhou, H., Feng, Z., Xu, Z., Tang, Y., Li, P. and Wu, M. (2017). CircRNA: functions and properties of a novel potential biomarker for cancer. Mol. Cancer 16, 94. 10.1186/s12943-017-0663-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer, C. A., Kaliappan, A. and Dennis, P. B. (2009). A novel, human Atg13 binding protein, Atg101, interacts with ULK1 and is essential for macroautophagy. Autophagy 5, 649-662. 10.4161/auto.5.5.8249 [DOI] [PubMed] [Google Scholar]
- Metur, S. P. and Klionsky, D. J. (2021). Adaptive immunity at the crossroads of autophagy and metabolism. Cell Mol. Immunol. 18, 1096-1105. 10.1038/s41423-021-00662-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima, N. (2009). Physiological functions of autophagy. Curr. Top Microbiol. Immunol. 335, 71-84. 10.1007/978-3-642-00302-8_3 [DOI] [PubMed] [Google Scholar]
- Moore, L. D., Le, T. and Fan, G. (2013). DNA methylation and its basic function. Neuropsychopharmacology 38, 23-38. 10.1038/npp.2012.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulcahy Levy, J. M. and Thorburn, A. (2020). Autophagy in cancer: moving from understanding mechanism to improving therapy responses in patients. Cell Death Differ 27, 843-857. 10.1038/s41418-019-0474-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nah, J., Yuan, J. and Jung, Y. K. (2015). Autophagy in neurodegenerative diseases: from mechanism to therapeutic approach. Mol. Cells 38, 381-389. 10.14348/molcells.2015.0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatogawa, H. (2020). Mechanisms governing autophagosome biogenesis. Nat. Rev. Mol. Cell Biol. 21, 439-458. 10.1038/s41580-020-0241-0 [DOI] [PubMed] [Google Scholar]
- Ng, K. M., Mok, P. Y., Butler, A. W., Ho, J. C., Choi, S. W., Lee, Y. K., Lai, W. H., Au, K. W., Lau, Y. M., Wong, L. Y.et al. (2016). Amelioration of X-Linked Related Autophagy Failure in Danon Disease With DNA Methylation Inhibitor. Circulation 134, 1373-1389. 10.1161/CIRCULATIONAHA.115.019847 [DOI] [PubMed] [Google Scholar]
- Nihira, K., Miki, Y., Iida, S., Narumi, S., Ono, K., Iwabuchi, E., Ise, K., Mori, K., Saito, M., Ebina, M.et al. (2014). An activation of LC3A-mediated autophagy contributes to de novo and acquired resistance to EGFR tyrosine kinase inhibitors in lung adenocarcinoma. J. Pathol. 234, 277-288. 10.1002/path.4354 [DOI] [PubMed] [Google Scholar]
- Nishimura, T., Tamura, N., Kono, N., Shimanaka, Y., Arai, H., Yamamoto, H. and Mizushima, N. (2017). Autophagosome formation is initiated at phosphatidylinositol synthase-enriched ER subdomains. EMBO J. 36, 1719-1735. 10.15252/embj.201695189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'brien, J., Hayder, H., Zayed, Y. and Peng, C. (2018). Overview of MicroRNA Biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. (Lausanne), 9, 402. 10.3389/fendo.2018.00402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palanisamy, K., Tsai, T. H., Yu, T. M., Sun, K. T., Yu, S. H., Lin, F. Y., Wang, I. K. and Li, C. Y. (2019). RNA-binding protein, human antigen R regulates hypoxia-induced autophagy by targeting ATG7/ATG16L1 expressions and autophagosome formation. J. Cell Physiol. 234, 7448-7458. 10.1002/jcp.27502 [DOI] [PubMed] [Google Scholar]
- Palmieri, M., Impey, S., Kang, H. J., Di Ronza, A., Pelz, C., Sardiello, M. and Ballabio, A. (2011). Characterization of the CLEAR network reveals an integrated control of cellular clearance pathways. Hum. Mol. Genet. 20, 3852-3866. 10.1093/hmg/ddr306 [DOI] [PubMed] [Google Scholar]
- Pan, R. and Zhou, H. (2020). Exosomal transfer of lncRNA H19 promotes erlotinib resistance in non-small cell lung cancer via miR-615-3p/ATG7 Axis. Cancer Manag. Res. 12, 4283-4297. 10.2147/cmar.s241095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, X., Chen, Y., Shen, Y. and Tantai, J. (2019). Knockdown of TRIM65 inhibits autophagy and cisplatin resistance in A549/DDP cells by regulating miR-138-5p/ATG7. Cell Death Dis. 10, 429. 10.1038/s41419-019-1660-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang, Y., Wu, L., Tang, C., Wang, H. and Wei, Y. (2022). Autophagy-Inflammation Interplay During Infection: Balancing Pathogen Clearance and Host Inflammation. Front Pharmacol, 13, 832750. 10.3389/fphar.2022.832750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, J. M., Jung, C. H., Seo, M., Otto, N. M., Grunwald, D., Kim, K. H., Moriarity, B., Kim, Y. M., Starker, C., Nho, R. S.et al. (2016). The ULK1 complex mediates MTORC1 signaling to the autophagy initiation machinery via binding and phosphorylating ATG14. Autophagy 12, 547-564. 10.1080/15548627.2016.1140293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrales, A. and Iwakuma, T. (2015). Targeting oncogenic mutant p53 for cancer therapy. Front. Oncol. 5, 288. 10.3389/fonc.2015.00288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, Y. and Croce, C. M. (2016). The role of MicroRNAs in human cancer. Signal Transduct. Target Ther. 1, 15004. 10.1038/sigtrans.2015.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, J., Yang, Q., Li, A. F., Li, R. Q., Wang, Z., Liu, L. S., Ren, Z., Zheng, X. L., Tang, X. Q., Li, G. H.et al. (2016). Tet methylcytosine dioxygenase 2 inhibits atherosclerosis via upregulation of autophagy in ApoE-/- mice. Oncotarget 7, 76423-76436. 10.18632/oncotarget.13121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploper, D., Taelman, V. F., Robert, L., Perez, B. S., Titz, B., Chen, H. W., Graeberd, T. G., Von Euw, E., Ribas, A. and De Robertis, E. M. (2015). MITF drives endolysosomal biogenesis and potentiates Wnt signaling in melanoma cells. Proc. Natl. Acad. Sci. USA 112, E420-E429. 10.1073/pnas.1424576112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polager, S., Ofir, M. and Ginsberg, D. (2008). E2F1 regulates autophagy and the transcription of autophagy genes. Oncogene 27, 4860-4864. 10.1038/onc.2008.117 [DOI] [PubMed] [Google Scholar]
- Qu, Y., Gao, Q., Wu, S., Xu, T., Jiang, D. and Xu, G. (2021). MicroRNA-142-3p inhibits autophagy and promotes intracellular survival of Mycobacterium tuberculosis by targeting ATG16L1 and ATG4c. Int. Immunopharmacol. 101, 108202. 10.1016/j.intimp.2021.108202 [DOI] [PubMed] [Google Scholar]
- Russell, R. C., Tian, Y., Yuan, H., Park, H. W., Chang, Y. Y., Kim, J., Kim, H., Neufeld, T. P., Dillin, A. and Guan, K. L. (2013). ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat. Cell Biol. 15, 741-750. 10.1038/ncb2757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamaki, J. I., Wilkinson, S., Hahn, M., Tasdemir, N., O'prey, J., Clark, W., Hedley, A., Nixon, C., Long, J. S., New, M.et al. (2017). Bromodomain protein BRD4 is a transcriptional repressor of autophagy and lysosomal function. Mol. Cell 66, 517-532.e9. 10.1016/j.molcel.2017.04.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakellariou, D., Tiberti, M., Kleiber, T. H., Blazquez, L., Lopez, A. R., Abildgaard, M. H., Lubas, M., Bartek, J., Papaleo, E. and Frankel, L. B. (2021). eIF4A3 regulates the TFEB-mediated transcriptional response via GSK3B to control autophagy. Cell Death Differ 28, 3344-3356. 10.1038/s41418-021-00822-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez, A. M. J., Csibi, A., Raibon, A., Cornille, K., Gay, S., Bernardi, H. and Candau, R. (2012). AMPK promotes skeletal muscle autophagy through activation of forkhead FoxO3a and interaction with Ulk1. J. Cell. Biochem. 113, 695-710. 10.1002/jcb.23399 [DOI] [PubMed] [Google Scholar]
- Sardiello, M., Palmieri, M., Di Ronza, A., Medina, D. L., Valenza, M., Gennarino, V. A., Di Malta, C., Donaudy, F., Embrione, V., Polishchuk, R. S.et al. (2009). A gene network regulating lysosomal biogenesis and function. Science 325, 473-477. 10.1126/science.1174447 [DOI] [PubMed] [Google Scholar]
- Schaffner, I., Minakaki, G., Khan, M. A., Balta, E. A., Schlotzer-Schrehardt, U., Schwarz, T. J., Beckervordersandforth, R., Winner, B., Webb, A. E., Depinho, R. A.et al. (2018). FoxO function is essential for maintenance of autophagic flux and neuronal morphogenesis in adult neurogenesis. Neuron 99, 1188-1203.e6. 10.1016/j.neuron.2018.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo, Y. K., Jeon, T. I., Chong, H. K., Biesinger, J., Xie, X. H. and Osborne, T. F. (2011). Genome-wide localization of SREBP-2 in hepatic chromatin predicts a role in autophagy. Cell Metabolism 13, 367-375. 10.1016/j.cmet.2011.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre, C., Di Malta, C., Polito, V. A., Garcia-Arencibia, M., Vetrini, F., Erdin, S., Erdin, S. U., Huynh, T., Medina, D., Colella, P.et al. (2011). TFEB links autophagy to lysosomal biogenesis. Science 332, 1429-1433. 10.1126/science.1204592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao, Z., Ni, L., Hu, S., Xu, T., Meftah, Z., Yu, Z., Tian, N., Wu, Y., Sun, L., Wu, A.et al. (2021). RNA-binding protein HuR suppresses senescence through Atg7 mediated autophagy activation in diabetic intervertebral disc degeneration. Cell Prolif 54, e12975. 10.1111/cpr.12975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, Q., Xu, Z. and Xu, S. (2020). Long noncoding RNA LUCAT1 contributes to cisplatin resistance by regulating the miR514a3p/ULK1 axis in human nonsmall cell lung cancer. Int. J. Oncol. 57, 967-979. 10.3892/ijo.2020.5106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin, H. J., Kim, H., Oh, S., Lee, J. G., Kee, M., Ko, H. J., Kweon, M. N., Won, K. J. and Baek, S. H. (2016). AMPK-SKP2-CARM1 signalling cascade in transcriptional regulation of autophagy. Nature 534, 553-557. 10.1038/nature18014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla, S., Patric, I. R., Patil, V., Shwetha, S. D., Hegde, A. S., Chandramouli, B. A., Arivazhagan, A., Santosh, V. and Somasundaram, K. (2014). Methylation silencing of ULK2, an autophagy gene, is essential for astrocyte transformation and tumor growth. J. Biol. Chem. 289, 22306-22318. 10.1074/jbc.M114.567032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, H., Feng, X., Zhang, H., Luo, Y., Huang, J., Lin, M., Jin, J., Ding, X., Wu, S., Huang, H.et al. (2019). METTL3 and ALKBH5 oppositely regulate m(6)A modification of TFEB mRNA, which dictates the fate of hypoxia/reoxygenation-treated cardiomyocytes. Autophagy 15, 1419-1437. 10.1080/15548627.2019.1586246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Statello, L., Guo, C. J., Chen, L. L. and Huarte, M. (2021). Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 22, 96-118. 10.1038/s41580-020-00315-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, S., Wang, Z., Tang, F., Hu, P., Yang, Z., Xue, C., Gong, J., Shi, L. and Xie, C. (2016). ATG7 promotes the tumorigenesis of lung cancer but might be dispensable for prognosis predication: a clinicopathologic study. Onco Targets Ther, 9, 4975-4981. 10.2147/OTT.S107876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tracy, K., Dibling, B. C., Spike, B. T., Knabb, J. R., Schumacker, P. and Macleod, K. F. (2007). BNIP3 is an RB/E2F target gene required for hypoxia-induced autophagy. Mol. Cell. Biol. 27, 6229-6242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, W. T., Han, C., Sun, Y. M., Chen, Z. H., Fang, K., Huang, W., Sun, L. Y., Zeng, Z. C., Luo, X. Q. and Chen, Y. Q. (2019a). Activation of the lysosome-associated membrane protein LAMP5 by DOT1L serves as a bodyguard for MLL fusion oncoproteins to evade degradation in leukemia. Clin. Cancer Res. 25, 2795-2808. 10.1158/1078-0432.CCR-18-1474 [DOI] [PubMed] [Google Scholar]
- Wang, Z. C., Huang, F. Z., Xu, H. B., Sun, J. C. and Wang, C. F. (2019b). MicroRNA-137 inhibits autophagy and chemosensitizes pancreatic cancer cells by targeting ATG5. Int. J. Biochem. Cell Biol. 111, 63-71. 10.1016/j.biocel.2019.01.020 [DOI] [PubMed] [Google Scholar]
- Wang, X., Wu, R., Liu, Y., Zhao, Y., Bi, Z., Yao, Y., Liu, Q., Shi, H., Wang, F. and Wang, Y. (2020). m(6)A mRNA methylation controls autophagy and adipogenesis by targeting Atg5 and Atg7. Autophagy 16, 1221-1235. 10.1080/15548627.2019.1659617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J., Dong, Z., Sheng, Z. and Cai, Y. (2022). Hypoxia-induced PVT1 promotes lung cancer chemoresistance to cisplatin by autophagy via PVT1/miR-140-3p/ATG5 axis. Cell Death Discov. 8, 104. 10.1038/s41420-022-00886-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, F. Z., Cao, Z., Wang, X., Wang, H., Cai, M. Y., Li, T., Hattori, N., Wang, D., Du, Y., Song, B.et al. (2015). Epigenetic regulation of autophagy by the methyltransferase EZH2 through an MTOR-dependent pathway. Autophagy 11, 2309-2322. 10.1080/15548627.2015.1117734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wold, M. S., Lim, J., Lachance, V., Deng, Z. and Yue, Z. (2016). ULK1-mediated phosphorylation of ATG14 promotes autophagy and is impaired in Huntington's disease models. Mol. Neurodegener 11, 76. 10.1186/s13024-016-0141-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Z., Cai, L., Lu, J., Wang, C., Guan, J., Chen, X., Wu, J., Zheng, W., Li, Q. and Su, Z. (2019). MicroRNA-93 mediates cabergoline-resistance by targeting ATG7 in prolactinoma. J. Endocrinol. 240, 1-13. 10.1530/JOE-18-0203 [DOI] [PubMed] [Google Scholar]
- Wu, Z., Zheng, Y., Xie, W., Li, Q., Zhang, Y., Ren, B., Cai, L., Cheng, Y., Tang, H., Su, Z.et al. (2020). The long noncoding RNA-H19/miRNA-93a/ATG7 axis regulates the sensitivity of pituitary adenomas to dopamine agonists. Mol. Cell Endocrinol 518, 111033. 10.1016/j.mce.2020.111033 [DOI] [PubMed] [Google Scholar]
- Xu, J., Wang, Y., Tan, X. and Jing, H. (2012). MicroRNAs in autophagy and their emerging roles in crosstalk with apoptosis. Autophagy 8, 873-882. 10.4161/auto.19629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, W., San Lucas, A., Wang, Z. and Liu, Y. (2014). Identifying microRNA targets in different gene regions. BMC Bioinformatics 15 Suppl. 7, S4. 10.1186/1471-2105-15-s7-s4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, J., Kitada, M., Ogura, Y. and Koya, D. (2021). Relationship between autophagy and metabolic syndrome characteristics in the pathogenesis of atherosclerosis. Front. Cell Dev. Biol. 9, 641852. 10.3389/fcell.2021.641852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Z. and Klionsky, D. J. (2010). Mammalian autophagy: core molecular machinery and signaling regulation. Curr. Opin. Cell Biol. 22, 124-131. 10.1016/j.ceb.2009.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, L., Wang, H., Shen, Q., Feng, L. and Jin, H. (2017). Long non-coding RNAs involved in autophagy regulation. Cell Death Dis. 8, e3073. 10.1038/cddis.2017.464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, A., Jiao, Y., Yang, S., Deng, M., Yang, X., Mao, C., Sun, Y., Ding, N., Li, N., Zhang, M.et al. (2018). Homocysteine activates autophagy by inhibition of CFTR expression via interaction between DNA methylation and H3K27me3 in mouse liver. Cell Death Dis. 9, 169. 10.1038/s41419-017-0216-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, L., Peng, X., Jin, H. and Liu, J. (2019). Long non-coding RNA PVT1 promotes autophagy as ceRNA to target ATG3 by sponging microRNA-365 in hepatocellular carcinoma. Gene 697, 94-102. 10.1016/j.gene.2019.02.036 [DOI] [PubMed] [Google Scholar]
- Yang, J., Qi, M., Fei, X., Wang, X. and Wang, K. (2021). LncRNA H19: a novel oncogene in multiple cancers. Int. J. Biol. Sci. 17, 3188-3208. 10.7150/ijbs.62573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi, M., Yang, J. B., Li, W. J., Li, X. L., Xiong, W., Mccarthy, J. B., Li, G. Y. and Xiang, B. (2017). The NOR1/OSCP1 proteins in cancer: from epigenetic silencing to functional characterization of a novel tumor suppressor. J. Cancer 8, 626-635. 10.7150/jca.17579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- You, L. K., Wang, Z. G., Li, H. S., Shou, J. W., Jing, Z., Xie, J. S., Sui, X. B., Pan, H. M. and Han, W. D. (2015). The role of STAT3 in autophagy. Autophagy 11, 729-739. 10.1080/15548627.2015.1017192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, F., Dong, B., Dong, P., He, Y., Zheng, J. and Xu, P. (2020). Hypoxia induces the activation of hepatic stellate cells through the PVT1-miR-152-ATG14 signaling pathway. Mol. Cell Biochem. 465, 115-123. 10.1007/s11010-019-03672-y [DOI] [PubMed] [Google Scholar]
- Yun, C. W. and Lee, S. H. (2018). The roles of autophagy in cancer. Int. J. Mol. Sci. 19, 3466. 10.3390/ijms19113466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H. and Lu, B. (2020). The roles of ceRNAs-mediated autophagy in cancer chemoresistance and metastasis. Cancers (Basel) 12, 2926. 10.3390/cancers12102926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X., Li, C., Wang, D., Chen, Q., Li, C. L. and Li, H. J. (2016). Aberrant methylation of ATG2B, ATG4D, ATG9A and ATG9B CpG island promoter is associated with decreased mRNA expression in sporadic breast carcinoma. Gene 590, 285-292. 10.1016/j.gene.2016.05.036 [DOI] [PubMed] [Google Scholar]
- Zhang, K., Chen, J., Zhou, H., Chen, Y., Zhi, Y., Zhang, B., Chen, L., Chu, X., Wang, R. and Zhang, C. (2018). PU.1/microRNA-142-3p targets ATG5/ATG16L1 to inactivate autophagy and sensitize hepatocellular carcinoma cells to sorafenib. Cell Death Dis. 9, 312. 10.1038/s41419-018-0344-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H., Alsaleh, G., Feltham, J., Sun, Y., Napolitano, G., Riffelmacher, T., Charles, P., Frau, L., Hublitz, P., Yu, Z.et al. (2019a). Polyamines control eIF5A hypusination, TFEB translation, and autophagy to reverse B cell senescence. Mol. Cell 76, 110-125.e9. 10.1016/j.molcel.2019.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, N., Li, Z., Bai, F. and Zhang, S. (2019b). PAX5-induced upregulation of IDH1-AS1 promotes tumor growth in prostate cancer by regulating ATG5-mediated autophagy. Cell Death Dis. 10, 734. 10.1038/s41419-019-1932-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, W., Dong, Y. Z., Du, X., Peng, X. N. and Shen, Q. M. (2019c). MiRNA-153-3p promotes gefitinib-sensitivity in non-small cell lung cancer by inhibiting ATG5 expression and autophagy. Eur. Rev. Med. Pharmacol. Sci. 23, 2444-2452. 10.26355/eurrev_201903_17391 [DOI] [PubMed] [Google Scholar]
- Zhang, Z., Guo, M., Li, Y., Shen, M., Kong, D., Shao, J., Ding, H., Tan, S., Chen, A., Zhang, F.et al. (2020). RNA-binding protein ZFP36/TTP protects against ferroptosis by regulating autophagy signaling pathway in hepatic stellate cells. Autophagy 16, 1482-1505. 10.1080/15548627.2019.1687985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J., Chen, K., Tang, Y., Luan, X., Zheng, X., Lu, X., Mao, J., Hu, L., Zhang, S., Zhang, X.et al. (2021a). LncRNA-HOTAIR activates autophagy and promotes the imatinib resistance of gastrointestinal stromal tumor cells through a mechanism involving the miR-130a/ATG2B pathway. Cell Death Dis. 12, 367. 10.1038/s41419-021-03650-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z., Zhu, H. and Hu, J. (2021b). CircRAB11FIP1 promoted autophagy flux of ovarian cancer through DSC1 and miR-129. Cell Death Dis. 12, 219. 10.1038/s41419-021-03486-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, X., Chen, Y., Mao, Q., Jiang, X., Jiang, W., Chen, J., Xu, W., Zhong, L. and Sun, X. (2018). Overexpression of YTHDF1 is associated with poor prognosis in patients with hepatocellular carcinoma. Cancer Biomark 21, 859-868. 10.3233/CBM-170791 [DOI] [PubMed] [Google Scholar]
- Zhao, Y., Yang, J., Liao, W. J., Liu, X. Y., Zhang, H., Wang, S., Wang, D. L., Feng, J. N., Yu, L. and Zhu, W. G. (2010). Cytosolic FoxO1 is essential for the induction of autophagy and tumour suppressor activity. Nat. Cell Biol. 12, U665-U88. 10.1038/ncb2069 [DOI] [PubMed] [Google Scholar]
- Zheng, L., Wang, Z., Li, Z., Wang, M., Wang, W. and Chang, G. (2021). MicroRNA-130a inhibits proliferation of vascular smooth muscle cells by suppressing autophagy via ATG2B. J. Cell Mol. Med. 25, 3829-3839. 10.1111/jcmm.16305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, W., Yue, C., Deng, J., Hu, R., Xu, J., Feng, L., Lan, Q., Zhang, W., Ji, D., Wu, J.et al. (2013). Autophagic protein Beclin 1 serves as an independent positive prognostic biomarker for non-small cell lung cancer. PLoS ONE 8, e80338. 10.1371/journal.pone.0080338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, C., Yi, C., Yi, Y., Qin, W., Yan, Y., Dong, X., Zhang, X., Huang, Y., Zhang, R., Wei, J.et al. (2020). LncRNA PVT1 promotes gemcitabine resistance of pancreatic cancer via activating Wnt/beta-catenin and autophagy pathway through modulating the miR-619-5p/Pygo2 and miR-619-5p/ATG14 axes. Mol. Cancer 19, 118. 10.1186/s12943-020-01237-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, H., Wu, H., Liu, X., Li, B., Chen, Y., Ren, X., Liu, C. G. and Yang, J. M. (2009). Regulation of autophagy by a beclin 1-targeted microRNA, miR-30a, in cancer cells. Autophagy 5, 816-823. 10.4161/auto.9064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, J., Li, Y., Tian, Z., Hua, X., Gu, J., Li, J., Liu, C., Jin, H., Wang, Y., Jiang, G.et al. (2017). ATG7 overexpression is crucial for tumorigenic growth of bladder cancer In Vitro and in vivo by targeting the ETS2/miRNA196b/FOXO1/p27 Axis. Mol. Ther. Nucleic Acids 7, 299-313. 10.1016/j.omtn.2017.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, J., Tian, Z., Li, Y., Hua, X., Zhang, D., Li, J., Jin, H., Xu, J., Chen, W., Niu, B.et al. (2019). ATG7 Promotes Bladder Cancer invasion via autophagy-mediated increased ARHGDIB mRNA stability. Adv. Sci. (Weinh), 6, 1801927. 10.1002/advs.201801927 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.