Abstract
One consequence of transforming growth factor β (TGF-β) treatment is inhibition of Cdk4 synthesis, and this is dependent on p53. Here, we show that the 5′ untranslated region (UTR) of the cdk4 mRNA is both necessary and sufficient for wild-type p53-dependent TGF-β-regulated translational inhibition of cdk4. Wild-type p53 bound selectively to the 5′ UTR of the cdk4 mRNA and inhibited translation of RNAs that contain this region. RNA binding and translational control are two genetically separable functions of p53, as are specific and nonspecific RNA binding. Moreover, transactivation-defective mutants of p53 retain the ability to regulate cdk4 translation. Our findings suggest that p53 functions as a regulator of translation in response to TGF-β in vivo.
Progression through the G1 phase of the cell cycle is regulated in part by Cdk4 and its regulatory subunits, the D-type cyclins, as well as Cdk2 and cyclin E. Cdk4 and Cdk2 cooperate to functionally inactivate the retinoblastoma protein, pRb, thereby committing the cell to enter S phase. Extracellular signals influence a number of signal transduction pathways that converge on the regulation of Cdk4 activity. Transforming growth factor beta (TGF-β) is a multifunctional cytokine that has been shown to inhibit cellular proliferation by blocking progression through G1 by preventing the phosphorylation of pRb. Work in several systems indicates that Cdk4 is the primary target of TGF-β action. This cytokine has been shown to induce the Cdk4-specific inhibitors p15INK4b and p21CIP1, leading to the inactivation of D-type cyclin-dependent kinase activity (2, 8, 19). Treatment with TGF-β leading to decreased expression of the Cdc25A phosphatase and, consequently, increased tyrosine phosphorylation at inhibitory sites resulting in inactive Cdk4 has also been documented (9). In addition, it has been demonstrated that TGF-β treatment leads to the inhibition of Cdk4 synthesis (4). For a given cell system, these mechanisms are not necessarily mutually exclusive, suggesting that redundant events leading to the efficient inactivation of Cdk4 exist. The importance of inhibiting Cdk4 activity for an effective TGF-β antiproliferative response is underscored by the observation that enforced expression of Cdk4, but not Cdk2, renders cells resistant to TGF-β-induced growth arrest (4).
Previously we demonstrated that in Mv1Lu mink lung epithelial cells, which have one of the best characterized antimitogenic responses to TGF-β, downregulation of Cdk4 following TGF-β treatment occurs at the level of translation (5). The ability of the cytokine to affect cdk4 translation is dependent on functional wild-type p53. This was evidenced by the observation that expression of a dominant-negative mutant of p53 prevented inhibition of cdk4 synthesis following TGF-β treatment. A possible explanation for these phenomena was provided by the demonstration that in transiently transfected cells, wild-type, but not mutant, p53 could inhibit the translation of cdk4. Moreover, the ability of p53 to inhibit Cdk4 synthesis in this system was shown to be dependent on the 5′ untranslated region (UTR) of cdk4 (5).
The previous work left a number of important questions unanswered regarding TGF-β-mediated inhibition of Cdk4 synthesis. Here, we have addressed these issues. We have asked whether TGF-β-mediated inhibition of Cdk4 synthesis in vivo is dependent on the 5′ UTR of cdk4 and, if so, whether the presence of wild-type p53 is required for this effect. In addition, we have begun to elucidate the role played by p53 in the downregulation of cdk4 translation mediated by TGF-β. Our data indicate that this cytokine regulates cdk4 translation through the direct actions of p53 upon the cdk4 5′ UTR.
MATERIALS AND METHODS
Cell culture, transfections, and reporter assays.
Mv1Lu (ATCC CCL64) cells were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum in a 10% CO2 humidified atmosphere. Cell lines expressing a temperature-sensitive allele of p53 (ts-p53) (5) were maintained in medium supplemented with 400 μg of G418 per ml. Cell lines expressing exogenous cdk4 plasmids were maintained in medium supplemented with 0.5 of puromycin per ml. Saos-2 cells were maintained in DMEM supplemented with 10% fetal bovine serum (Fetal-Clone I; Hyclone). Mv1Lu cells and Saos-2 cells were transfected by the calcium phosphate coprecipitation method as previously described (5). Release from contact inhibition and treatment with 50 pM TGF-β (R&D Systems) were done as previously described (5).
For reporter assays, cells were washed twice with phosphate-buffered saline, scraped and pelleted, and resuspended in 100 mM KH2PO4 (pH 7.8). After three rounds of freeze-thawing, the extracts were cleared by centrifugation and used for luciferase assays (1).
Plasmids.
The mink cdk4 cDNA was amplified by PCR with or without its 5′ noncoding region and inserted into the pCMVneoBam vector to create pCMVFLminkcdk4 and pCMVΔ5′minkcdk4. The luciferase constructs contain the luciferase gene amplified by PCR and ligated into pCMVneoBam. For expression of an RNA containing the cdk4 5′ UTR fused to the luciferase coding region, the luciferase gene and the mink cdk4 5′ UTR were each amplified by PCR, the PCR products were digested with BamHI and HindIII, and a three-way ligation with BamHI-digested-pCMVneoBam was performed to generate pCMVUTRluc. The DNA sequence of the cdk4 5′ UTR was verified. To generate 5′-deleted mink Cdk4 plasmids, the cdk4 cDNA from pCMVFLminkcdk4, was isolated and subcloned into pSp72 (Promega). The resulting plasmid, pSp72FLminkcdk4, was either double digested with EcoRV and PvuII and religated (PvuII), double digested with EcoRV and BsrBI and religated (ΔBsrBI), digested with SacI and religated (ΔSacI), or double digested with EcoRV and ThaI and religated (ΔThaI). Each of these inserts was removed with BamHI and BglII and ligated back into pCMVneoBam. The p53 mutant plasmids [p53-Δ39, p53(40–359), p53-Δ96, p53(97–359), p53-(311–393), p53-Δ347, p53-Δ360, p53-Δ380, and p53-Δ390] were generated by PCR and ligated into pCMVneoBam. Mutants p53-Δ96, p53(97–359), and p53-(311–393) contain an additional ATG as a start codon. Sequencing analysis was used to verify each insert.
Metabolic labeling, immunoprecipitations, and Western blot analysis.
Metabolic labeling and immunoprecipitations were performed as previously described (5). For Western blotting, cells were lysed in RIPA-B radioimmunoprecipitation assay buffer (20 mM sodium phosphate [pH 7.4], 150 mM sodium chloride, 1% Triton X-100, 10 μg of aprotinin per ml, 10 μg of leupeptin per ml, 5 mM phenylmethylsulfonyl fluoride). Cleared extracts (30 μg) were electrophoresed on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels and transferred to Immobulon-P (Millipore) membranes. Affinity-purified anti-Cdk4 polyclonal antibody (C-22; Santa Cruz Biotechnology, Inc.) was used for immunoblotting. Polyclonal anti-Cdk4 antibody that was raised against recombinant glutathione S-transferase (GST)-Cdk4 (generous gift of C. J. Sherr) was used for immunoprecipitations.
Gel retardation assays.
Radiolabeled RNA probes (0.2 to 0.5 ng) were generated by in vitro transcription with T7 polymerase, heated at 70°C, and quenched on ice prior to incubation with 0.3 ng of baculovirus-purified p53 (80% pure; generous gift of C. Prives) or Rev (generous gift of M. L. Zapp) in binding buffer (40 mM Tris-HCl [pH 7.9], 50 mM KCl, 1 μg of bovine serum albumin [BSA] per μl, 1 mM dithiothreitol [DTT]) with 75 ng of tRNA, and 1 μl of RNasin (Promega) for 20 min. RNA-protein complexes were resolved on 4% acrylamide gels and detected by autoradiography. Immunoprecipitation-deoxycholate (IP-DOC) gel shifts were performed essentially as described previously (22). Specifically, immune complexes were washed two times with 100 mM NETN (100 mM KCl, 40 mM Tris-HCl [pH 7.9], 1 mM DTT, 0.1% NP-40) then twice with binding buffer. The pellet was incubated with 0.8% DOC in binding buffer for 10 min on ice, NP-40 was added to 1.2%, and the sample was incubated with radiolabeled probe, 75 ng of tRNA, and 1 μl of RNasin as described above.
Northern blot analyses.
Northern blotting was performed as previously described (5). To detect exogenous cdk4 RNA, the blots were probed under high-stringency conditions with an oligonucleotide that recognizes the sequence encoding the hemagglutinin (HA) tag. To detect combined endogenous and exogenous cdk4 levels, the blots were probed with a mink cdk4 cDNA. To detect luciferase and cdk4 UTR-luciferase RNAs, a luciferase cDNA was used. Northern blots to measure mink p15INK4b RNA were performed with RNA enriched for poly(A)+. The probe was a fragment of the mink p15 cDNA (19) (generous gift of J. Massagué) labeled to high specific activity by PCR with a 5′ primer corresponding to nucleotides 4 to 24 and a 3′ primer corresponding to nucleotides 115 to 136 in the human p15 sequence. To control for loading, the blots were stripped and probed with the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNA.
In vitro translation assays.
RNAs (10 to 20 ng) were heated at 70°C and quenched on ice before addition of binding buffer (40 mM Tris-HCl [pH 8.0], 1 mM DTT, 50 mM NaCl), 5 to 8 ng of protein (either p53 or BSA), 75 ng of tRNA, and 1 μl of RNasin (Promega). The binding reaction mixtures were incubated at 37°C for 20 min, followed by the addition of 20 μCi of [35S]methionine and standard components of a rabbit reticulocyte lysate system (Promega). Translation was allowed to proceed for 1 h at 30°C. Newly synthesized proteins were subjected to SDS-PAGE and autoradiography.
RESULTS
The 5′ UTR is required for Cdk4 downregulation.
In Mv1Lu cells expressing an exogenous, murine cdk4 mRNA lacking most of its 5′ UTR and all of its 3′ UTR, translation of this RNA fails to be repressed in response to TGF-β (4, 5). This suggested the possibility that TGF-β might regulate cdk4 translation through one or both of its UTRs. To test this hypothesis, we assessed whether the translational regulation of cdk4 by TGF-β requires the 5′ UTR of cdk4. Stable clones of Mv1Lu cells that express an exogenous mink cdk4 mRNA either containing the coding region with the entire 5′ UTR (henceforth referred to as the FL [full-length] cdk4 RNA) or containing the coding region and only 21 bp of the 5′ UTR (henceforth referred to as Δ5′ cdk4 RNA) were generated. In clones expressing FL cdk4 RNA, the synthesis of exogenous Cdk4 was inhibited to a similar degree as that of the endogenous Cdk4 (62% and 67% repression, respectively) following treatment with TGF-β (Fig. 1A). In contrast, in clones where the 5′ UTR of cdk4 was lacking, synthesis of this exogenous Cdk4 failed to be inhibited by TGF-β, while endogenous Cdk4 was still efficiently downregulated. The greater degree of synthesis of Cdk4 from the Δ5′ cdk4 RNA compared with the FL cdk4 RNA is likely due to the absence of the lengthy encumbered cdk4 5′ UTR. The exogenous cdk4 RNA was expressed at similar levels in clones expressing either the FL or Δ5′ cdk4 RNA or at increased levels in the FL clone relative to the Δ5′ clone (Fig. 1B). Thus, the presence of increased RNA levels is not an explanation for Δ5′ cdk4 failing to be downregulated by TGF-β. The cdk4 mRNA levels were not affected by TGF-β treatment, as determined by Northern blotting, consistent with regulation occurring at the translational level (Fig. 1B). Together, these observations suggest that TGF-β inhibits cdk4 translation only when the cdk4 mRNA contains its 5′ leader sequence.
FIG. 1.
5′ UTR-deleted cdk4 is resistant to downregulation by TGF-β. (A) Parental Mv1Lu cells and subclones stably expressing HA-tagged versions of the cdk4 RNA either containing its entire 5′ UTR (FL) or lacking its 5′ UTR (Δ5′) were treated with 50 pM TGF-β for 20 h. During the last 3 h of treatment, the cells were metabolically labeled with [35S]methionine, and specific immunoprecipitations for Cdk4 were then performed. Protein samples were electrophoresed in a 10% SDS–polyacrylamide gel. (B) RNA was harvested from parental and Cdk4 clones untreated or treated for 17 h with TGF-β, Northern blotted, and probed sequentially with mink cdk4, an oligonucleotide encoding the HA tag, and GAPDH, as indicated.
TGF-β-induced Cdk4 downregulation is mediated via its 5′ UTR.
To determine if the 5′ UTR of cdk4 is sufficient for sensitivity to TGF-β-induced translational downregulation, Mv1Lu cell clones expressing an mRNA that contains the 5′ UTR of mink cdk4 fused to the coding region of the luciferase reporter were generated. Following treatment with TGF-β, luciferase activity was markedly downregulated in cells expressing this mRNA (Fig. 2A, upper panel). In contrast, in control clones expressing luciferase RNA lacking the cdk4 5′ UTR, luciferase activity was unaffected by TGF-β treatment.
FIG. 2.
TGF-β inhibits expression of a cdk4 5′ UTR-containing RNA. (A) (Top) Luciferase activity was measured in an asynchronous population of Mv1Lu cells stably expressing either luciferase mRNA without a 5′ leader sequence (luc) or luciferase mRNA fused to the cdk4 5′ UTR (UTR-luc) that had been incubated in the absence or presence of 50 pM TGF-β for the indicated lengths of time. Values are the percentage of luciferase activity in treated populations relative to that in untreated populations harvested at identical times (set to 100%). Experiments were done in triplicate. Error bars indicate the standard deviation from the mean. (Middle) Cell cycle distribution in parallel cultures was determined by flow cytometry. Results from a representative experiment are shown. (Bottom) RNA was collected at the indicated times and subjected to Northern blotting with probes for luciferase (luc) or GAPDH. (B) Assays for luciferase activity, cell cycle distribution, and Northern blotting were performed as for panel A, except that the cultures were contact inhibited for at least 3 days, trypsinized, and then replated into growth medium in the absence or presence of TGF-β. An arrow indicates the time when untreated cells begin to enter S phase.
To determine if the time course of translational inhibition coincided with the time course of TGF-β-induced cell cycle arrest, the clones were analyzed at successive times following TGF-β treatment. Within 7 h of TGF-β treatment of actively proliferating cells, luciferase activity was downregulated by 30 to 40% in clones expressing the cdk4 5′ UTR-luciferase mRNA compared to that in an untreated population (Fig. 2A). This was also roughly the same time at which the cells began to accumulate in the G1 phase (Fig. 2A, middle panel). In clones expressing the luciferase mRNA lacking the cdk4 5′ UTR, TGF-β was without effect on luciferase activity, despite the ability of these cells to undergo a normal TGF-β-induced arrest. Luciferase RNA levels were also unchanged by the cytokine, suggesting that regulation is occurring at the level of translation (Fig. 2A, lower panel). Together, these observations suggest that the cdk4 5′ UTR is capable of conferring responsiveness to TGF-β-induced translational downregulation upon a heterologous RNA and that translational repression of a cdk4 5′ UTR-containing RNA occurs with a time course similar to Cdk4 downregulation and coincides with or precedes a TGF-β-induced growth arrest.
Because cells are only sensitive to the antiproliferative and Cdk4 inhibitory effects of TGF-β when exposed to the cytokine in early to mid-G1 phase (4, 13), we repeated the above experiment with cells that were treated with TGF-β immediately following release from a contact-inhibited, quiescent state. TGF-β treatment of cells expressing the cdk4 5′ UTR-luciferase mRNA following release from quiescence resulted in a rapid reduction in luciferase activity compared with untreated cells also released from quiescence (Fig. 2B). Luciferase activity was repressed by approximately 20% within 9 h of TGF-β treatment following release from contact inhibition, which was prior to the time when untreated cells began to enter S phase. Similar results are also observed for inhibition of Cdk4 by TGF-β. Luciferase activity in control cells that express a luciferase RNA without the cdk4 5′ UTR was again unchanged by TGF-β treatment under these conditions. The luciferase RNA levels in TGF-β-treated cells were also unchanged by the cytokine, supporting our hypothesis that reduction in luciferase activity following TGF-β treatment is regulated at the translational level (Fig. 2B). These results, like those from experiments with proliferating cells, demonstrate a strong correlation between the time during which downregulation of Cdk4 occurring through its 5′ UTR takes place and the time during which TGF-β induces cell cycle arrest.
Wild-type p53 is required for TGF-β-induced, 5′ UTR-dependent downregulation of Cdk4.
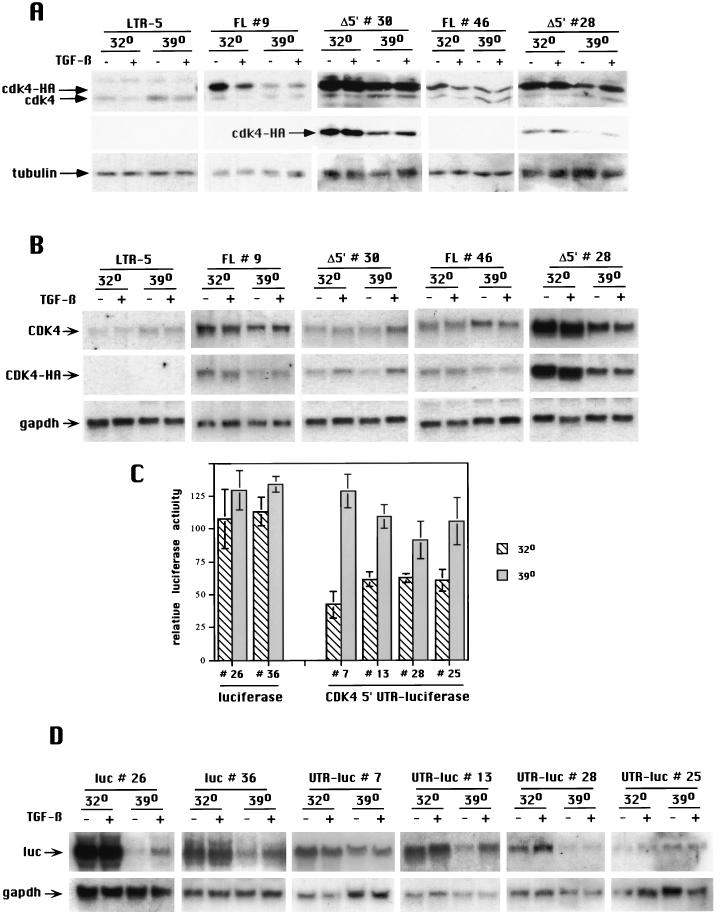
Previously we demonstrated that in cells expressing mutant p53, Cdk4 synthesis fails to be inhibited by TGF-β (5). In these studies, Mv1Lu cell clones that expressed ts-p53 (Val 135) were used. When the cells were cultured at the nonpermissive temperature (39°C, mutant p53), they failed to arrest and to inhibit Cdk4 synthesis in response to TGF-β. Parental Mv1Lu cells express wild-type p53, and in the ts-p53 clones grown at the nonpermissive temperature, the mutant form of p53 acts in a dominant-negative manner over endogenous p53 (5). We asked if the p53-dependent Cdk4 downregulation by TGF-β also requires the 5′ UTR of cdk4 by generating ts-p53 clones that express either of the two mink cdk4-HA mRNAs (FL and Δ5′) described above. When ts-p53 cells expressing an exogenous FL cdk4 RNA containing its entire 5′ UTR were cultured at 32°C (wild-type p53), synthesis of both exogenous and endogenous Cdk4 was inhibited in response to TGF-β (Fig. 3A). In contrast, when the cells were cultured at the nonpermissive temperature, neither endogenous nor exogenous Cdk4 was downregulated in response to TGF-β. Importantly, in ts-p53 cells expressing exogenous Δ5′ cdk4 RNA that lacks a 5′ UTR, the exogenous Cdk4 failed to be downregulated by TGF-β treatment at either 32 or 39°C, supporting the notion that inhibition of Cdk4 synthesis is mediated via the 5′ UTR. Although the levels of exogenous Cdk4-HA protein were unexpectedly lower after culture at 39°C compared to those at 32°C, this was a consequence of changes in RNA abundance (Fig. 3B) and thus was likely due to changes in transcriptional activity of the cytomegalovirus (CMV) promoter used to drive expression of exogenous Cdk4-HA. Northern blot analysis also confirmed that although Cdk4 protein levels were repressed by TGF-β under conditions in which p53 is present in its wild-type form, the mRNA levels were unaltered by TGF-β treatment (Fig. 3B). These observations are consistent with the requirement for wild-type p53 in TGF-β-mediated Cdk4 downregulation (5) and indicate that the 5′ UTR of cdk4 is essential in this setting.
FIG. 3.
The 5′ UTR of cdk4 mediates p53 dependence in TGF-β-induced translational inhibition of cdk4. (A) Mv1Lu cells expressing a temperature-sensitive allele of p53 (LTR5) (5) and derivative cell lines expressing exogenous cdk4 either containing its entire 5′ UTR (FL) or lacking its 5′ UTR (Δ5′) were cultured at the permissive temperature (32°C) or the nonpermissive temperature (39°C) in the presence or absence of TGF-β for 27 h (32°C) or 24 h (39°C). Whole-cell lysates were prepared and analyzed by Western blotting with specific antiserum to Cdk4. A shorter exposure time for the Δ5′ clones is shown to emphasize the resistance to TGF-β-induced downregulation for the exogenous, HA-tagged Cdk4. The membranes were stripped and blotted with antitubulin antibody as a loading control. (B) Northern blot analysis of RNA harvested from the Mv1Lu derivative lines in panel A after culture at the indicated temperatures in the presence or absence of TGF-β. After probing with a Cdk4 cDNA, the membranes were sequentially stripped and reprobed with an oligonucleotide encoding the HA tag and then a GAPDH cDNA. (C) Luciferase activity was measured in Mv1Lu cells expressing both a temperature-sensitive allele of p53 and either luciferase mRNA without a 5′ leader sequence (luciferase) or luciferase mRNA fused to the cdk4 5′ UTR (CDK4 5′ UTR-luciferase) that had been cultured as described for panel A. The bar graph shows luciferase activity after TGF-β treatment relative to untreated cell populations cultured under the same conditions (set at 100). Experiments were done in triplicate. Error bars indicate the standard deviation from the mean. (D) Northern blot analysis of RNA harvested from the Mv1Lu derivative lines shown in panel C. Membranes were probed with a luciferase cDNA (luc), stripped, and reprobed with a GAPDH cDNA.
We next asked whether the 5′ UTR of cdk4 was sufficient for TGF-β-induced translational inhibition in a p53-dependent manner. Mv1Lu ts-p53 clones that expressed either the cdk4 5′ UTR-luciferase mRNA or the luciferase RNA lacking a 5′ UTR (described above) were generated. When cells expressing the cdk4 5′ UTR-luciferase mRNA were cultured at 32°C, luciferase activity was markedly reduced in TGF-β-treated cells compared to untreated populations (Fig. 3C). In cells expressing the mRNA without the cdk4 5′ UTR, luciferase activity was unaffected by TGF-β treatment. When the cells were cultured at 39°C, luciferase activity was not affected by TGF-β in either clones expressing luciferase mRNA or in clones expressing the cdk4 5′ UTR-luciferase mRNA, consistent with the notion that the 5′ UTR of cdk4 is sufficient to mediate p53-dependent translational inhibition induced by TGF-β. Analysis of luciferase RNA levels by Northern blotting supported the idea that regulation of luciferase activity by TGF-β was occurring at the translational level because the RNA levels were unchanged under the same conditions in which repression of luciferase activity is observed (Fig. 3D).
Wild-type p53 can bind the Cdk4 mRNA.
Given our observations that translational inhibition of Cdk4 (by TGF-β) required p53 and the cdk4 5′ UTR, along with reports that p53 has a high affinity for single-stranded RNA and possesses nucleic acid reannealing (antihelicase) activity, we hypothesized that p53 was directly involved in regulating Cdk4 translation and might bind to the cdk4 RNA (5′ UTR) as part of the mechanism by which it inhibits Cdk4 translation. We assessed whether p53 could interact with the cdk4 RNA by gel shift analysis using baculovirus-purified recombinant p53 and a radiolabeled cdk4 5′ UTR RNA. Using this assay, we detected a specific RNA-protein complex that contained p53. The presence of p53 was demonstrated by the ability of p53-specific antibodies to shift the mobility of the complex (Fig. 4A). To test the specificity of this interaction, a nonspecific probe, the Rev responsive element (RRE) (27) in an RNA of a similar size and G-C content was used. p53 bound selectively to the cdk4 RNA, because only weak binding to the nonspecific RNA was detected (Fig. 4B). In contrast, the HIV-1 Rev protein was capable of interacting specifically with the RRE, its RNA target, but not with the cdk4 RNA (Fig. 4B). The weak interaction observed between p53 and the RRE probe is likely due to p53's ability to bind nonspecifically to RNA (10). The specificity of the complex was confirmed by competition experiments. An excess of unlabeled cdk4 5′ UTR RNA successfully competed for the interaction between p53 and the radiolabeled cdk4 RNA, whereas the RRE RNA and the reverse complement of the CDK4 5′ UTR were much less effective at competing for binding to p53 (Fig. 4C). Again, p53's weak nonspecific RNA binding activity is the likely reason that the RRE and the antisense cdk4 5′ UTR RNAs were able to partly compete for p53 binding.
FIG. 4.
Wild-type p53 can bind selectively to the cdk4 5′ UTR RNA. (A) Baculovirus-purified p53 was analyzed for RNA binding activity in a gel shift assay with 32P-labeled cdk4 5′ UTR (230 nucleotides) as a probe. Addition of monoclonal anti-p53 antibodies (Bp53-12, DO-I) induced a “supershift” of the RNA-protein complex. Migration of the probe in the absence of added protein is shown in the first lane (−). (B) The specificity of p53's interaction with RNA was analyzed by gel shift with either the RRE or the cdk4 5′ UTR as a probe. Migrations of free, unbound RRE, and cdk4 5′ UTR probes are shown in the first and fourth lanes, respectively (−). Binding of baculovirus-purified Rev protein to its target sequence, the RRE, is indicated by the large arrow. Binding of p53 to the RRE or the cdk4 5′ UTR is indicated by the small arrow. The autoradiograph in panel B was exposed for approximately twice as long as those in panel A, C, or D to show the weak binding of p53 to the RRE. (C) Increasing amounts of unlabeled RNA were included in the gel shift reactions to compete for binding to p53. In the left panel, 13- to 40-fold molar excess of competitor RNA compared to labeled RNA was included. In the right panel, 100- to 400-fold molar excess of competitor RNA was included. specific, cdk4 5′ UTR; antisense, the reverse complement of cdk4 5′ UTR. (D) Saos-2 cells were transfected with either wild-type p53, a mutant form of p53 (143A), or vector alone. After transfection, p53 was immunoprecipitated (IP) with the indicated antibodies, released by DOC treatment, and used in gel shift analysis. (Bottom panel) immunoprecipitates prepared in parallel were resolved on SDS-PAGE and blotted with anti-p53 antibody. Bac-p53, p53 purified from recombinant baculovirus.
As an alternative approach to assess p53's ability to bind the cdk4 5′ UTR, we used p53 isolated in vivo from mammalian cells. Following transfection of p53-expressing plasmids into Saos-2 cells (which lack endogenous p53), p53 was immunoprecipitated and the immune complexes were treated with sodium deoxycholate (DOC) to release p53, which was then used in RNA gel shift analysis. Wild-type p53 isolated in this way bound to the cdk4 5′ UTR RNA as well as the baculovirus-purified form (Fig. 4D). In contrast, immunoprecipitation of a mutant form of p53 (143A) failed to generate an RNA-protein complex. A control monoclonal antibody (MAb 240) that does not recognize wild-type p53 was unable to immunoprecipitate proteins that bound to the cdk4 5′ UTR. Similarly, if the cells were transfected with vector alone, an RNA-protein complex was not detected. Together, these observations suggest that wild-type p53, but not a mutant form of p53, can interact specifically with the 5′ UTR of the cdk4 RNA.
Wild-type p53 inhibits Cdk4 translation in vitro.
Interaction with the cdk4 RNA could provide a means for p53 to regulate cdk4 translation. Prior to initiation of translation, the ribosome is thought to progressively scan the 5′ UTR for potential start codons (11, 12). With p53 bound to the cdk4 5′ UTR, ribosomal scanning could be sterically hindered, resulting in 40S subunit stalling and the failure of translation to initiate. To determine if p53 can directly inhibit cdk4 translation, we translated cdk4 in vitro, using rabbit reticulocyte lysates, in the presence or absence of p53 or in the presence of another protein, BSA. Translation of either human or mink cdk4 RNA containing their respective 5′ UTR was markedly inhibited by the presence of p53. In contrast, translation of RNAs lacking a 5′ UTR was completely unaffected by the presence of p53 (Fig. 5A and B). Given that the cdk4 5′ UTR contains features that are known to markedly inhibit translation, we were not surprised to observe that the cdk4 RNA lacking its 5′ UTR was consistently translated more efficiently than the cdk4 RNA containing its cognate 5′ UTR. A mutant form of p53, 143A, shown to be incapable of binding RNA (Fig. 4D) (17), was defective for cdk4 translational repression (Fig. 5B).
FIG. 5.
Wild-type p53 can repress cdk4 translation in a cell-free assay. (A, top panel) In vitro translation of human cdk4 from either a cdk4 RNA containing its entire 5′ UTR (full-length CDK4) or a cdk4 RNA lacking a 5′ UTR (Δ5′ CDK4) in the presence of BSA (−) or baculovirus-purified p53 protein (+). Samples of the in vitro translation mixtures were analyzed on a 10% SDS–polyacrylamide gel. (A, bottom panel) Parallel assays were conducted in which the RNA was collected both preceding and following incubation in the cell-free translation assay and subsequently analyzed by Northern blotting with Cdk4 cDNA as a probe. retic, reticulocyte. (B) In vitro translation of mink cdk4 in the presence of baculovirus-purified wild-type (wt) or mutant (143A) p53. Samples were analyzed as in panel A. (C) [top panel]) In vitro translation of luciferase from either an RNA containing the cdk4 5′ UTR fused to the luciferase coding region (CDK4 5′ UTR-luciferase) or from an RNA lacking a 5′ UTR (Δ5′ luciferase) in the presence of BSA (−) or baculovirus-purified p53 protein (+). Samples were analyzed as in panel A. (C [bottom panel]) Same as panel A, bottom panel, except a luciferase cDNA was used as a probe for the Northern blot.
To ascertain whether the 5′ UTR of cdk4 alone could mediate p53-induced translational inhibition, luciferase RNAs either lacking a 5′ noncoding sequence or containing the cdk4 5′ UTR were translated in the absence or presence of p53. p53 inhibited translation from the cdk4 5′ UTR-containing transcript, but had no effect on the luciferase RNA that lacked a 5′ noncoding region (Fig. 5C). Because the abundance of the UTR-containing transcripts was as constant as that of the transcripts lacking a 5′ UTR following incubation with p53 and reticulocyte lysates (Fig. 5, lower panels), these results cannot be explained by differential stabilities of the transcripts. Our observations that wild-type p53 can inhibit translation of an RNA containing the cdk4 5′ UTR, together with the previous demonstration that p53 can bind to the cdk4 5′ UTR (Fig. 4), suggest that translational inhibition may occur as a result of a p53-RNA interaction.
Selective RNA binding and regulation by p53 require the region from −100 to −64 in the cdk4 5′ UTR.
To localize the region of the cdk4 RNA required for p53 interaction, we tested a series of cdk4 5′ UTR deletion mutants for their ability to bind p53 (Fig. 6A). cdk4 cDNAs with deletion of up to the first 127 nucleotides remained capable of binding to p53 by gel shift analysis (Fig. 6B). However, deletion of an additional 36 nucleotides from the cdk4 RNA rendered the RNA incapable of interaction with p53. To exclude the possibility that an RNA be of a minimal length for p53 binding, we tested a second set of deletion constructs that were each lengthened by the addition of 53 nucleotides of the cdk4 RNA coding region. We observed an identical pattern of binding by gel shift analysis when comparing the longer RNA probes with the shorter matched RNAs (not shown). This observation suggests that p53 interaction is determined by a specific region of the RNA and not its length. In either situation, deletion to position −64 (relative to the initiating AUG) in cdk4 created an RNA molecule that no longer interacted with p53.
FIG. 6.
Localization of the RNA region in ∼cdk4 that is essential for p53 interaction and p53-induced repression. (A) Schematic representation of the Cdk4 plasmids used that express RNAs containing 5′ UTRs of various lengths. (B) 32P-labeled cdk4 5′ UTR RNAs were synthesized in vitro from the plasmids noted in panel A, incubated with baculovirus-purified p53 protein at either 25°C (lanes 2, 5, 8, and 11) or 37°C (lanes 3, 6, 9, and 12), and subjected to gel shift analysis. (C) Saos-2 cells were transfected with various derivatives of CMV driven mink cdk4 with or without wild-type human p53 expression plasmid. After transfection, cells were metabolically labeled, and specific immunoprecipitations for Cdk4 were performed. Samples were analyzed on a 10% SDS–polyacrylamide gel. (D) Saos-2 cells were transfected as described above. After transfection, total RNA was harvested and used for Northern analysis. The blot was probed with a Cdk4 cDNA and reprobed with GAPDH.
We next asked whether the ability of p53 to bind to the cdk4 5′ UTR correlated with its ability to repress cdk4 translation. To address this question, expression vectors for Cdk4 and wild-type p53 were cotransfected into the p53-null Saos-2 cells, and synthesis of Cdk4 protein was monitored by metabolic labeling and immunoprecipitation. When cdk4 cDNAs with deletion of up to the first 127 nucleotides were cotransfected with p53, p53 inhibited their translation to an extent similar to that of the FL RNA (Fig. 6C). However, similar to the RNA binding results, deletion of an additional 36 nucleotides from the RNA rendered cdk4 nearly insensitive to p53 regulation. Repression of Cdk4 synthesis by p53 was not a consequence of changes in cdk4 RNA levels, as demonstrated by Northern blotting, consistent with our hypothesis that p53 regulates Cdk4 at the translational level. Taken together with the gel shift results, these findings suggest that the region in the cdk4 RNA from −100 to −64, relative to the initiating AUG, is essential for both p53 interaction and p53-mediated translational regulation.
Analysis of p53 mutants for abilities to bind cdk4 RNA and repress its translation.
To determine what aspects of the p53 protein are involved in translational regulation and RNA binding, we utilized a panel of p53 mutants in the cotransfection and gel shift assays. The p53 protein can be divided into distinct domains defined in terms of separable activities. These include a transcriptional activation region at the N terminus (residues 1 to 43), a sequence-specific DNA-binding domain (residues 100 to 300), an oligomerization domain (residues 320 to 360), and a carboxy-terminal domain containing activities for nonspecific nucleic acid binding and nucleic acid reannealing (antihelicase) (residues 330 to 393) (10).
First we determined whether p53-regulated translational control of cdk4 can occur through a p53 transcriptional transactivation-independent pathway by using a transcriptionally inert mutant of p53 lacking the N-terminal 39 amino acids. Coexpression of Cdk4 with such an N-terminally-deleted mutant of p53 in Saos-2 cells caused inhibition of Cdk4 expression when the FL cdk4 cDNA was used, but not when the Δ5′ cdk4 cDNA was used, similar to the actions of wild-type p53 (Fig. 7B). In contrast, mutants of p53 missing larger regions of the N terminus (p53-Δ79 and p53-Δ96) were incapable of regulating Cdk4 synthesis (Fig. 7B and Table 1). When tested for binding to the cdk4 RNA, the shorter N-terminally-deleted p53 (Δ39) bound as well as wild-type p53, and p53s harboring larger deletions of the N terminus (Δ79 and Δ96), although defective for translational control, could still bind reasonably well to the cdk4 RNA (Fig. 7C and Table 1). Together, these data suggest that the transcriptional transactivation domain of p53 is not required, but the region between amino acids 43 and 80 is essential for p53 to regulate translation of cdk4. Moreover, these results suggest that functions of p53 in addition to its ability to bind RNA are required for translational inhibition.
FIG. 7.
Analysis of p53 mutants for RNA binding and translational regulation capabilities. (A) Expression of wild-type and mutant forms of p53 following transfection into Saos-2 cells was assayed by immunoblotting with a mixture of the monoclonal antibodies PAb240 and PAb421. (B) Saos-2 cells were transfected with either human CDK4 containing its 5′ UTR or lacking its 5′ UTR and with or without wild-type p53 or mutant forms of p53. Following transfection, cells were metabolically labeled, and specific immunoprecipitations for Cdk4 were performed. (C) Saos-2 cells were transfected with wild-type and mutant forms of p53. Following transfection, p53 was immunoprecipitated with anti-p53 antibodies, released by DOC treatment, and analyzed by gel shift with a 32P-labeled cdk4 5′ UTR riboprobe, except in those lanes (labeled RRE) where the RRE was used as the riboprobe. The p53(311–393)-Cdk4 protein-RNA complex is indicated by the large arrow, and the p53(311–393)-RRE protein-RNA complex is indicated by the small arrow. FREE, riboprobe without addition of immunoprecipitated protein.
TABLE 1.
Summary of mutant p53 analysis
| Mutanta | RNA bindingb | Translational inhibitionc |
|---|---|---|
| N terminal | ||
| Δ39 | + | +/− |
| Δ79 | +/− | − |
| Core domain | ||
| 97–359 | − | − |
| 40–359 | +/− | +/− |
| C terminal | ||
| Δ291 | − | − |
| Δ343 | − | − |
| Δ347 | − | − |
| Δ360 | + | + |
| Δ370 | + | + |
| Δ380 | + | + |
| Δ390 | + | + |
| 392A | + | + |
| 311–393 | + | +/− |
Proteins were detected by immunoblotting.
RNA binding activity was determined by gel shift analysis.
Translational inhibition was assayed by cotransfection with Cdk4.
To determine whether the ability of p53 to regulate translation and bind to the cdk4 RNA might be dependent upon an intact C-terminal region, mutants of p53 truncated at the carboxy terminus were tested. p53 mutants that were lacking up to 33 residues from the C terminus, p53(1–360), p53(1–370), p53(1–380), and p53(1–390), were still capable of inhibiting Cdk4 synthesis in cotransfection assays. These mutants also bound to the cdk4 RNA in a manner similar to wild-type p53. On the other hand, mutants of p53 that were lacking larger portions of the C terminus [p53(1–291), p53(1–343), and p53(1–347)] were incapable of interacting with the CDK4 5′ UTR and failed to regulate cdk4 translation (Fig. 7B and Table 1). To exclude the possibility that p53 mutants lacking larger regions of the C terminus were defective for translational regulation and specific RNA binding because they had lost their ability to oligomerize, we tested a p53 molecule that contains p53(1–343) fused to the GCN4 dimerization domain (p53-GCN4) (17). Reconstitution of dimerization function did not rescue the ability of p53(1–343) to either inhibit Cdk4 synthesis or bind to the cdk4 RNA, suggesting that the inability to dimerize does not explain why these mutants fail to inhibit Cdk4 expression (Table 1). Thus, our observations suggest that in the context of the full-length protein, the extreme C-terminal region of p53 (amino acids 360 to 393) is dispensable for translational regulation and specific RNA binding, while adjacent regions (N terminal to residue 360) are essential. In addition, we found that p53 lacking both N- and C-terminal domains [p53(40–359)] still inhibited Cdk4 synthesis and could also bind effectively to the cdk4 RNA. However, elimination of amino acids 40 to 96 from this mutant, resulting in p53(97–359), completely abolished these activities, illustrating the necessity for these residues (Fig. 7B and Table 1). This result is consistent with our findings that p53-Δ96 fails to regulate Cdk4 translation.
When an isolated carboxy domain of p53 (residues 311 to 393) was tested for its ability to repress Cdk4 synthesis, we observed some inhibition of Cdk4 synthesis, albeit not to the same extent as with the wild-type protein. Thus, the p53 C-terminal fragment alone (amino acids 311 to 360) may have some intrinsic ability to regulate cdk4 translation in this system. This fragment of p53 was also competent for binding to the cdk4 RNA, yet it bound to the nonspecific RNA, the RRE, as well as it did to the cdk4 RNA (Fig. 7). Given that a nonspecific nucleic acid binding region has been mapped to residues 330 to 393, these data suggest that interactions through the C terminus may be entirely nonspecific, while other regions of the p53 molecule could provide specificity. Because p53(311–393) has lost its ability to selectively recognize the cdk4 RNA, these data also suggest that nonspecific RNA binding and specific RNA binding can be genetically separated.
DISCUSSION
Treatment with TGF-β potently inhibits Cdk4 synthesis and causes a cell cycle arrest in G1 phase. Here, we employed Mv1Lu cells to test in vivo the necessity for the cdk4 5′ UTR in Cdk4 downregulation in response to TGF-β. We show, for the first time, that the 5′ UTR of the cdk4 mRNA is essential for TGF-β-mediated cdk4 translational inhibition and, moreover, that this region of the cdk4 RNA can confer TGF-β-induced translational regulation upon a heterologous RNA. TGF-β-mediated translational regulation through the cdk4 5′ UTR occurs with similar kinetics as does endogenous Cdk4 regulation in response to TGF-β, consistent with the possibility that TGF-β-mediated Cdk4 regulation occurs entirely through the 5′ UTR. In addition, the 5′ UTR of cdk4 is both necessary and sufficient for p53-dependent regulation of cdk4 in response to TGF-β. p53 is not only capable of binding to a specific region of the cdk4 5′ UTR, but also can directly repress Cdk4 translation in vitro. Together, our data support a model in which TGF-β inhibits cdk4 translation via p53 interaction with the cdk4 5′ UTR. Given the importance of downregulating Cdk4 in the TGF-β growth arrest pathway, consideration of these results is crucial to understanding the mechanisms of TGF-β action.
Our hypothesis that p53 may have a direct role in regulating cdk4 translation in response to TGF-β in vivo is consistent with several lines of evidence pointing to a role for p53 in protein translation. Cytoplasmic p53 has been found in association with ribosomes (7), and p53 has been reported to regulate its own translation via its 5′ leader sequence (16). p53 has also been detected in complexes containing the L5 ribosomal protein (15), has been found to be covalently bound to the 5.8S ribosomal RNA (6, 21), and has a higher binding affinity for RNA than for DNA (17). While we have shown that p53 can interact with the cdk4 RNA and can inhibit translation of RNAs containing the 5′ UTR of cdk4, our results do not distinguish between p53 directly contacting the cdk4 RNA and p53 interacting with the RNA through an associated molecule. Thus, although we propose that p53 directly regulates cdk4 translation in vivo, we cannot exclude other indirect roles for p53. Importantly, we found that RNA binding by p53 is necessary but insufficient for optimal translational inhibition. Thus, the mechanism by which p53 regulates translation is most likely due to a dynamic activity (e.g., catalytic) of the protein rather than a passive result of p53 binding stoichiometrically to the RNA, consistent with the findings of others (16).
The abilities to bind nonspecifically to nucleic acids, to reanneal nucleic acids, and to inhibit helicases are all activities of p53 that have been mapped to the C-terminal region (residues 311 to 393) of the protein (10). Our results suggest that in the context of the full-length protein, the carboxy-terminal 33 amino acids of p53 are dispensable for translational regulation and specific RNA binding, but an adjacent region (residues 347 to 360) is essential for these activities. The antihelicase and reannealing activities attributed to p53 might allow it to inhibit the translation of cdk4 by stabilizing hairpin structures in the 5′ UTR by promoting pairing of matched strands of RNA and/or by inhibiting unwinding. Our results suggest that reannealing is not equivalent to specific RNA binding. Although a truncated p53 protein encompassing amino acids 311 to 393 is sufficient to promote nucleic acid reannealing (26) we found that this domain alone, in contrast to the full-length protein, lacks the ability to recognize specific RNA fragments. Moreover, the p53-Δ360 mutant retains specific RNA binding, but the alternative spliced form of murine p53 (which lacks amino acids 364 to 390 and contains 17 new residues) cannot reanneal nucleic acids (26). Our observation that this C-terminal fragment (amino acids 311 to 393) of p53 is partially defective for translational regulation also suggests that the reannealing activity of p53 is insufficient to modulate translation. Moreover, the ability to recognize specific RNA as opposed to nonspecific binding is a requirement for optimal translational regulation. Finally, these results suggest that specific and nonspecific RNA binding by p53 can be genetically separated. p53 mutants that retain an intact C-terminal domain but lack other regions of the protein (Δ79 and Δ96) are completely defective for translational regulation, but bind to RNA. This suggests an interplay between protein domains, possibly involving negative regulation. Altogether, the C-terminal functions of p53 most likely depend upon communication with the rest of the protein, similar to the manner in which the C-terminal region of p53 regulates the sequence-specific DNA binding activity of its core domain (14). Finally, since p53 is a predominantly nuclear protein, it may bind to the target RNA in the nucleus and alter its capacity to be translated prior to its export into the cytoplasm.
The results described here show striking parallels to another p53-dependent growth arrest pathway. Growth arrest by the Gas1 membrane-associated protein has been demonstrated to require p53 (3). Moreover, the N-terminal transcriptional transactivation-containing domain of p53 is dispensable for this arrest (3). Thus, the requirement for p53 in the Gas1 growth arrest pathway requires a function separate from its transcriptional transactivation. This result is analogous to our findings that p53 translational inhibition of cdk4 does not require the N terminus of p53. Moreover, both p53-dependent growth arrest by Gas1 and p53-dependent growth arrest by TGF-β share a requirement for the proline-rich region of p53 extending from amino acid 61 to amino acid 94. Our observations that the p53-Δ39 mutant retains the ability to regulate Cdk4 synthesis and that p53-Δ96 and p53-Δ79 mutants were defective for translational regulation of cdk4 suggest that residues 43 to 80 of p53 are important for this function. Growth arrest by Gas1 was shown to require residues 63 to 85 of p53, and mutation of four prolines in this region eliminated p53's ability to cooperate with Gas1 in promoting a G1 phase arrest (20). In a third setting, H1299 cells, a p53 mutant lacking this proline-rich region is competent to activate transcription, but cannot inhibit proliferation (24). Proline-rich domains are thought to provide an interaction surface for SH3 domains. A role for SH3 interactions involving RNA-binding proteins is not unprecedented (23, 25). Since many SH3-containing molecules are involved in signaling from the cell surface, the proline-rich region of p53 could act as a receptor for signals transduced from the TGF-β receptor. Together these observations suggest that the p53 function required in the Gas1 pathway, like the TGF-β pathway, might be translational regulation.
We found that a specific RNA region in the cdk4 5′ UTR could mediate both RNA binding by p53 and translational inhibition by p53. This region (from nucleotides −100 to −64 relative to the initiating AUG), is predicted to be part of a stable hairpin structure in the context of the entire 5′ UTR. Interestingly, disruption at nucleotide position −64 severs one of the most conserved regions in the cdk4 5′ UTR. From position −72 to position −53, the sequence is 84% identical across four species: human, mink, pig, and rat. Thus, this sequence or the structure it acquires is likely to be critical to cdk4 regulation. Given that deletion of small segments can dramatically change the predicted secondary structure of an RNA, further studies involving mutagenesis of single nucleotides that subtly disrupt the RNA structure will be most informative in defining whether p53 is recognizing a specific RNA secondary structure.
TGF-β growth inhibition might occur via multiple mechanisms dependent on several factors and/or environmental conditions. Our results suggest that in mink lung epithelial cells, wild-type p53 is essential to TGF-β-mediated cell cycle arrest. p53 is necessary for translational regulation of cdk4 through its 5′ UTR in these cells, and a link between the effects of p53 on cdk4 translation and TGF-β-mediated growth arrest is indicated.
ACKNOWLEDGMENTS
We thank Carol Prives for supplying purified p53 protein; Maria Zapp for providing purified Rev protein and for help with the RNA gel shift assay; Peter Howley, Arnold Levine, Jennifer Pietenpol, and Karen Vousden for p53 mutant expression plasmids; Joan Massagué for the mink p15 cDNA; Charles Sherr for GST-Cdk4; and Bert Vogelstein for the pCMVneoBam vector. We also thank Peter Adams, Justin Lamb, Christine McMahon, Elizabeth Neuman, and Monique Yoakim for critical reading of the manuscript; Xiaoping He and Teena Kohli for excellent technical assistance; and our colleagues for valuable discussions.
S.J.M. was supported by a fellowship from the American Cancer Society. T.S. was supported by a fellowship from the National Institutes of Health. G.P.Z. is supported by grant CA63230 from the National Cancer Institute, by Cancer Center Core grant 5 P30 CA21765, and by the American Lebanese Syrian Associated Charities (ALSAC) of St. Jude Children's Research Hospital. This work was supported by grants from the National Cancer Institute (CA65842) and the Dana-Farber/Sandoz Drug Discovery Program to M.E.E. M.E.E. is a Scholar for the Leukemia Society of America.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: Greene and Wiley Interscience; 1996. [Google Scholar]
- 2.Datto M B, Li Y, Panus J F, Howe D J, Xiong Y, Wang X F. Transforming growth factor beta induces the cyclin-dependent kinase inhibitor p21 through a p53-independent mechanism. Proc Natl Acad Sci USA. 1995;92:5545–5549. doi: 10.1073/pnas.92.12.5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Del Sal G, Ruaro E M, Utrera R, Cole C N, Levine A J, Schneider C. Gas1-induced growth suppression requires a transactivation-independent p53 function. Mol Cell Biol. 1995;15:7152–7160. doi: 10.1128/mcb.15.12.7152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ewen M E, Sluss H K, Whitehouse L L, Livingston D M. TGF-β inhibition of cdk4 synthesis is linked to cell cycle arrest. Cell. 1993;74:1009–1020. doi: 10.1016/0092-8674(93)90723-4. [DOI] [PubMed] [Google Scholar]
- 5.Ewen M E, Oliver C J, Sluss H K, Miller S J, Peeper D S. p53-dependent repression of CDK4 translation in TGF-β-induced G1 cell-cycle arrest. Genes Dev. 1995;9:204–217. doi: 10.1101/gad.9.2.204. [DOI] [PubMed] [Google Scholar]
- 6.Fontoura B M A, Sorokina E A, David E, Carroll R B. p53 is covalently linked to 5.8S rRNA. Mol Cell Biol. 1992;12:5145–5151. doi: 10.1128/mcb.12.11.5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fontoura B M A, Atienza C A, Sorokina E A, Morimoto T, Carroll R B. Cytoplasmic p53 polypeptide is associated with ribosomes. Mol Cell Biol. 1997;17:3146–3154. doi: 10.1128/mcb.17.6.3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hannon G J, Beach D. p15INK4B is a potential effector of TGF-β-induced cell cycle arrest. Nature. 1994;371:257–261. doi: 10.1038/371257a0. [DOI] [PubMed] [Google Scholar]
- 9.Iavarone A, Massague J. Repression of the CDK activator Cdc25A and cell-cycle arrest by cytokine TGF-β in cells lacking the CDK inhibitor p15. Nature. 1997;387:417–422. doi: 10.1038/387417a0. [DOI] [PubMed] [Google Scholar]
- 10.Ko L J, Prives C. p53: puzzle and paradigm. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 11.Kozak M. An analysis of vertebrate mRNA sequences: intimations of translational control. J Cell Biol. 1991;115:887–903. doi: 10.1083/jcb.115.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kozak M. Initiation of translation in prokaryotes and eukaryotes. Gene. 1999;234:187–208. doi: 10.1016/s0378-1119(99)00210-3. [DOI] [PubMed] [Google Scholar]
- 13.Laiho M, DeCaprio J A, Ludlow J W, Livingston D M, Massagué J. Growth inhibition of TGF-β linked to suppression of retinoblastoma protein phosphorylation. Cell. 1990;62:175–185. doi: 10.1016/0092-8674(90)90251-9. [DOI] [PubMed] [Google Scholar]
- 14.Levine A J. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 15.Marechal V, Elenbaas B, Piette J, Nicolas J-C, Levine A J. The ribosomal L5 protein is associated with mdm-2 and mdm-2–p53 complexes. Mol Cell Biol. 1994;14:7414–7420. doi: 10.1128/mcb.14.11.7414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mosner J, Mummenbrauer T, Bauer C, Sczakiel G, Grosse F, Deppert W. Negative feedback regulation of wild-type p53 biosynthesis. EMBO J. 1995;14:4442–4449. doi: 10.1002/j.1460-2075.1995.tb00123.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oberosler P, Hloch P, Ramsperger U, Stahl H. p53-catalyzed annealing of complementary single-stranded nucleic acid. EMBO J. 1993;12:2389–2396. doi: 10.1002/j.1460-2075.1993.tb05893.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pientenpol J A, Tokino T, Thiagalingam S, El-Deiry W S, Kinzler K W, Vogelstein B. Sequence-specific transcriptional activation is essential for growth suppression by p53. Proc Natl Acad Sci USA. 1994;91:1998–2002. doi: 10.1073/pnas.91.6.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reynisdottir I, Polyak K, Iavarone A, Massague J. Kip/cip and ink4 cdk inhibitors cooperate to induce cell cycle arrest in response to TGF-β. Genes Dev. 1995;9:1831–1845. doi: 10.1101/gad.9.15.1831. [DOI] [PubMed] [Google Scholar]
- 20.Ruaro E M, Collavin L, Del Sal G, Haffner R, Oren M, Levine A J, Schneider C. A proline-rich motif in p53 is required for transactivation independent growth arrest as induced by Gas1. Proc Natl Acad Sci USA. 1997;94:4675–4680. doi: 10.1073/pnas.94.9.4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Samad A, Carroll R B. The tumor suppressor p53 is bound to RNA by a stable covalent linkage. Mol Cell Biol. 1991;11:1598–1606. doi: 10.1128/mcb.11.3.1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shirodkar S, Ewen M, DeCaprio J A, Morgan J, Livingston D M, Chittenden T. The transcription factor E2F interacts with the retinoblastoma product and a p107-cyclin A complex in a cell cycle-regulated manner. Cell. 1992;68:157–166. doi: 10.1016/0092-8674(92)90214-w. [DOI] [PubMed] [Google Scholar]
- 23.Taylor S J, Shalloway D. An RNA-binding protein associated with src through its SH2 and SH3 domains in mitosis. Nature. 1994;368:867–871. doi: 10.1038/368867a0. [DOI] [PubMed] [Google Scholar]
- 24.Walker K K, Levine A J. Identification of a novel p53 functional domain that is necessary for efficient growth suppression. Proc Natl Acad Sci USA. 1996;93:15335–15340. doi: 10.1073/pnas.93.26.15335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weng Z, Thomas S M, Rickles R J, Taylor J A, Brauer A W, Seidel-Dugan C, Michael W M, Dreyfuss G, Brugge J S. Identification of Src, Fyn, and Lyn SH3-binding proteins: implications for a function of SH3 domains. Mol Cell Biol. 1994;14:4509–4521. doi: 10.1128/mcb.14.7.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu L, Bayle J H, Elenbaas B, Pavletich N P, Levine A J. Alternatively spliced forms in the carboxy-terminal domain of the p53 protein regulate its ability to promote annealing of complementary single strands of nucleic acids. Mol Cell Biol. 1995;15:497–504. doi: 10.1128/mcb.15.1.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zapp M L, Stern S, Green M R. Small molecules that selectively block RNA binding of HIV-1 rev protein inhibit rev function and viral production. Cell. 1993;74:969–978. doi: 10.1016/0092-8674(93)90720-b. [DOI] [PubMed] [Google Scholar]