Abstract
In normal cells, p53 is maintained at a low level by ubiquitin-mediated proteolysis, but after genotoxic insult this process is inhibited and p53 levels rise dramatically. Ubiquitination of p53 requires the ubiquitin-activating enzyme Ubc5 as a ubiquitin conjugation enzyme and Mdm2, which acts as a ubiquitin protein ligase. In addition to the N-terminal region, which is required for interaction with Mdm2, the C-terminal domain of p53 modulates the susceptibility of p53 to Mdm2-mediated degradation. To analyze the role of the C-terminal domain in p53 ubiquitination, we have generated p53 molecules containing single and multiple lysine-to-arginine changes between residues 370 and 386. Although wild-type (WT) and mutant molecules show similar subcellular distributions, the mutants display a higher transcriptional activity than WT p53. Simultaneous mutation of lysine residues 370, 372, 373, 381, 382, and 386 to arginine residues (6KR p53 mutant) generates a p53 molecule with potent transcriptional activity that is resistant to Mdm2-induced degradation and is refractory to Mdm2-mediated ubiquitination. In contrast to WT p53, transcriptional activity directed by the 6KR p53 mutant fails to be negatively regulated by Mdm2. Those differences are also manifest in HeLa cells which express the human papillomavirus E6 protein, suggesting that p53 C-terminal lysine residues are also implicated in E6-AP-mediated ubiquitination. These data suggest that p53 C-terminal lysine residues are the main sites of ubiquitin ligation, which target p53 for proteasome-mediated degradation.
Cell cycle arrest, DNA repair, and apoptosis are the most common responses to DNA damage in normal mammalian cells. Genetic instability and malignant transformation are consequences of improper DNA damage responses (9, 26, 32). The p53 protein plays an important role in the initiation of the DNA damage response, cell cycle arrest, and tumor suppression (25, 31). One of the best-characterized functions of p53 is its transcriptional activity (26, 32). p53 increases the transcription of genes coding for important cell growth inhibitors, such as the p21Waf1/Cip1 gene (10), and apoptotic genes, such as that coding for Bax (42). The identification of the Mdm2 gene as a p53 target gene, the product of which can negatively regulate p53 functions, revealed a feedback loop that regulates both p53 activity and expression of Mdm2 (3, 30, 43, 44, 65).
In normal cells p53 levels are maintained at low or undetectable levels by continual proteolytic degradation, but when cells are exposed to stress signals, such as hypoxia and DNA damage, degradation is inhibited and p53 levels rise (25, 35). Sustained degradation of p53 is mediated by the ubiquitin-proteasome pathway (34) and is carried out by the ubiquitin-activating enzyme (E1), a ubiquitin-conjugating enzyme (Ubc5), and a ubiquitin protein ligase (Mdm2) (13, 19). Ubiquitin molecules are attached to p53 by an isopeptide bond between the C terminus of ubiquitin and the ɛ-amino group of a lysine residue. Multiple ubiquitin molecules are required to target protein substrates for degradation (18). Since degradation is an efficient means to abrogate all p53 functions, p53-induced transcription of Mdm2 generates an efficient mechanism for controlling p53 levels (3, 65). Many tumors and tumor cell lines have low levels of Mdm2 because they express transcriptionally inactive p53, and as a consequence, p53 levels are very high (41). The importance of this regulatory mechanism is illustrated by the fact that deletion of the mdm2 gene in mice induces an early embryonic lethality which is rescued by deletion of the p53 gene (24). A wide variety of mechanisms have been proposed which can block the interaction of Mdm2 with p53 and as a consequence increase the levels of p53. Phosphorylation of p53 or Mdm2 by a DNA-dependent protein kinase reduces the interaction between the two molecules (37, 58), resulting in reduced ubiquitination of p53 by the ubiquitin ligase activity of Mdm2 (20). The tumor suppressor p19 ARF blocks the degradation of p53 by directly inhibiting the ubiquitin ligase activity of Mdm2 (20, 39) and sequestering Mdm2 in the nucleolus (64, 66).
Subcellular distribution plays an important role in the control of p53 transcriptional activity, and nuclear transport is dependent on three nuclear localization signals (NLSs) located in the C-terminal region of the protein (57). Fusion of the major NLS (NLSI, amino acids [aa] 316 to 322) to a heterologous cytoplasmic protein directs the chimeric protein to the nucleus (7), while NLSII (aa 370 to 377) and NLSIII (aa 380 to 386) appear to increase the efficiency of nuclear import (57). Moreover, it has been proposed that the interaction of lysine 305 with the cytoplasmic sequestration domain (aa 326 to 355) regulates p53 subcellular distribution (33). The cytoplasmic sequestration domain also contains the p53 oligomerization domain and a putative leucine-rich nuclear export signal (LR-NES) (aa 340 to 351) (60). Mdm2-mediated p53 degradation has been reported to be a cytoplasmic process dependent on an LR-NES present in Mdm2 (54). Consequently, p53 degradation is blocked by leptomycin B (12, 29), which is a competitive inhibitor of the LR-NES receptor CRM1 (1, 11, 46, 62).
The p53-Mdm2 interaction is mediated by the N-terminal regions of the proteins. Mdm2 can negatively regulate p53 transcriptional activity by binding to and occluding the p53 transcriptional activation domain (5, 43, 45). The C-terminal region of p53 modulates the susceptibility of p53 to Mdm2-mediated degradation, and deletion of the last 30 aa of p53 inhibits this process (28). Six of the C-terminal 30 aa of p53 are lysine residues which are potential sites of ubiquitination. To investigate the role of the C-terminal lysine residues in the degradation of p53, we have generated single and multiple K-to-R changes in this region of p53. The altered proteins display an increased ability to activate a p53-dependent transcriptional reporter, and this ability is maximal with the p53 mutant in which all six lysine residues have been changed to arginine residues. In vivo ubiquitination and degradation assays and in vitro ubiquitination assays suggest that p53 C-terminal lysine residues are involved in ubiquitin-mediated degradation of p53. Our results also support the notion that Mdm2-mediated ubiquitination is an important mechanism for controlling p53 activity.
MATERIALS AND METHODS
Proteins and antibodies.
Ubiquitin was purchased from Sigma. Human E1 ubiquitin-activating enzyme was purified from recombinant baculovirus-infected cells as described previously (53). Human Mdm2 (residues 6 to 491) was expressed in bacteria and purified as previously described for p53 (40). Human Ubc5 was expressed in bacteria and purified as previously described (49). Glutathione S-transferase (GST)–Mdm2 fusion protein was purified as reported previously (4). Monoclonal antibody 4B2 (5) recognizes Mdm2. Monoclonal antibody DO.1 (63) and polyclonal antibody CM1 recognize p53. Green fluorescent protein (GFP) was detected with a mixture of two mouse monoclonal antibodies (anti-GFP clones 7.1 and 13.1) purchased from Boehringer Mannheim.
Plasmid construction.
The previously described wild-type (WT) p53 BamHI-EcoRI cassette (49) was replaced to generate multiple K-to-R mutants by site-directed mutagenesis using a PCR strategy. DNA sequences were determined by the University of St. Andrews DNA sequencing facility (with an ABI 377 sequencer). The pcDNA3-Mdm2 plasmid was described previously (39). The pEGFP-C2 plasmid was purchased from Clontech.
Cell culture and transfections.
HeLa, p53 null (Saos-2 and H1299), and mouse p53−/− Mdm2−/− cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum. Cells were transfected by electroporation (950 mF, 240 V; Equibio Easyject Plus) as previously described (49) or with Lipofectamine (GIBCO/BRL). pG13-luciferase (pG13-Luc) reporter assays were performed with 2 μg of reporter plasmid and 5 ng of plasmid encoding WT p53 or one of the K-to-R p53 mutants per 106 Saos-2 cells. Ten nanograms of p53-encoding and 30 ng of Mdm2-encoding plasmids were used for cotransfection into p53−/− Mdm2−/− cells. Reporter assays in HeLa cells were performed with 5 ng of WT p53 or the 6KR mutant. Empty pcDNA3 vector was used to maintain a constant level of plasmid DNA. After electroporation, cells were grown in six-well plates for 10 to 12 h and processed for luciferase activity as previously described (51). For immunofluorescence, 105 cells were cotransfected with 375 ng of plasmid encoding WT p53 or a K-to-R mutant and 2.125 μg of Bluescript plasmid DNA. Mdm2-directed degradation in vivo was performed with 200 ng of WT p53 or K-to-R mutants, 1.2 μg of Mdm2, and 600 ng of GFP-encoding plasmids per 106 cells. Empty pcDNA3 vector was used to maintain a constant level of plasmid DNA. Transfected cells were divided into two groups, and after 16 h in culture one of the groups was harvested. The second group of Mdm2-transfected cells was treated for 4 h prior to harvesting with 10 μM MG132. p53 ubiquitination in vivo was detected in 1.5 × 105 p53−/− Mdm2−/− cells transfected with 200 ng of GFP, 200 ng of WT p53 or K-to-R p53 mutants, and 600 ng of pcDNA3 or an Mdm2-encoding plasmid. After 16 h in culture, Mdm2-transfected cells were treated with 10 μM MG132 for 4 h prior to harvesting. A total of 106 HeLa cells were cotransfected with 4 μg of an SV5-tagged version of β-galactosidase (27) and 4 μg of WT p53- and 6KR-encoding plasmids. After 16 h, cells were lysed and analyzed by Western blotting.
Immunofluorescence.
After transfection, H1299 or Saos-2 cells were cultured on glass or Permanox chamber slides (Nunc). Twenty-four hours after transfection, slides were washed in phosphate-buffered saline (PBS)–1 mM MgCl2, fixed for 8 min in cold methanol-acetone (1:1), rinsed in PBS–1 mM MgCl2–0.1% Tween 20, and incubated for 1 h at room temperature with DO.1 mouse monoclonal antibody diluted in DMEM–10% fetal calf serum. Slides were washed two times in PBS–1 mM MgCl2, rinsed in PBS–1 mM MgCl2–0.1% Tween 20, and incubated with a mixture of fluorescein isothiocyanate-conjugated donkey anti-mouse immunoglobulin G in DMEM–10% fetal calf serum at the dilution specified by the manufacturer (Jackson ImmunoResearch Laboratories) for 30 min at room temperature. After two washes in PBS–1 mM MgCl2 and rinsing in PBS–1 mM MgCl2–0.1% Tween 20, the slides were mounted with Mowiol-Dabco, dried, and analyzed using a Bio-Rad MRC 600 LSM confocal microscope and custom Bio-Rad software.
Western blotting.
Whole-cell extracts were lysed in sodium dodecyl sulfate (SDS) sample buffer as described previously (8). Lysates were boiled for 10 min prior to fractionation by electrophoresis in 10% polyacrylamide gels containing SDS. Proteins were transferred to a polyvinylidene difluoride membrane (Sigma) by electroblotting. p53 (DO.1), Mdm2 (4B2), and GFP monoclonal antibodies were used as primary antibodies. Blots were developed with an enhanced chemiluminescence detection system.
Interaction of Mdm2 and p53 in vitro.
[35S]methionine-labeled in vitro-transcribed and -translated p53 proteins (10 μl) were incubated for 2 h at 4°C with 1 μg of GST or GST-Mdm2 prebound to glutathione-agarose beads in 200 μl of previously described binding buffer (21). For immunoprecipitation, [35S]methionine-labeled in vitro-cotranscribed and -cotranslated Mdm2 and p53 proteins (20 μl) were incubated for 2 h at 4°C with an equal mixture of protein A- and protein G-agarose beads (Sigma) and p53-specific polyclonal antibody CM1 in 200 μl of previously described binding buffer (21). Beads were washed as described previously (21), and bound protein was fractionated in a 10% polyacrylamide gel containing SDS. The dried gel was analyzed by phosphorimaging.
In vitro ubiquitination assays.
[35S]methionine-labeled in vitro-transcribed and -translated p53 proteins were used in ubiquitination assays as previously reported (49). Reaction products were fractionated in a 10% polyacrylamide gel containing SDS, and dried gels were analyzed by phosphorimaging (Fijix BAS 1500, MacBAS software). Ubiquitinated p53 was quantified by determining the proportion of the total radioactive p53 that migrated more slowly than unmodified p53.
RESULTS
K-to-R p53 mutants show higher transcriptional activity than does WT p53.
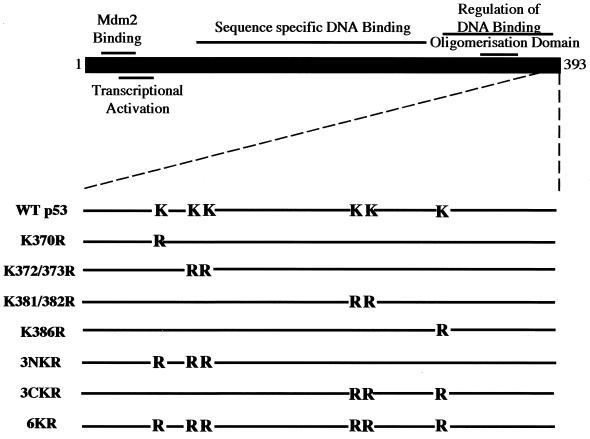
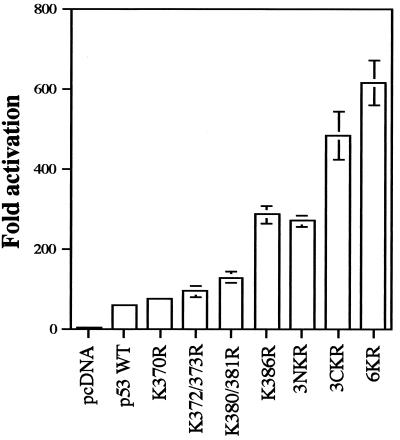
Reporter assays have been used previously to identify lysine residues which are targets for ubiquitination of transcriptionally active proteins or of transcriptional regulators (2, 51). The C-terminal 30 aa of p53 are required for Mdm2-mediated degradation (28) and contain six lysine residues which are potential targets for ubiquitination. To investigate the role of these residues on ubiquitin-proteasome-dependent degradation of p53, single and multiple K-to-R changes were introduced into the C-terminal region of p53 (Fig. 1). It was previously reported that K386R p53 has an inherently higher transcriptional activity than does WT p53 (49). To test if other K-to-R mutations have similar consequences on p53 transcriptional activity, Saos-2 cells, which lack endogenous p53, were cotransfected with the p53-responsive reporter plasmid pG13-Luc and expression plasmids for WT p53 and K-to-R p53 mutants. WT p53 activated transcription 57-fold, while all K-to-R p53 mutants showed higher transcriptional rates (Fig. 2A). The previously reported K386R mutant was the single-point mutant with the highest transcriptional activity (284-fold activation). The triple mutant 3CKR (483-fold activation) showed a higher transcriptional activity than did 3NKR (268-fold activation), suggesting that lysine residues 381, 382, and 386 may be the preferred targets for ubiquitination. However, 6KR p53, in which all six lysine residues are changed to arginine, displayed the highest transcriptional activity (613-fold activation). To confirm these results, the same p53 expression plasmids were cotransfected with the Mdm2-Luc reporter plasmid into Saos-2 cells or with the pG13-Luc reporter plasmid into H1299 p53 null cells, with similar results in each case (data not shown). This effect is likely to be mediated by Mdm-2, as p53−/− Mdm2−/− cells cotransfected with pG13-Luc and plasmids encoding p53 mutants did not show significant differences in their abilities to activate the reporter (see Fig. 8). This indicates that WT p53 and various K-to-R mutants have inherently similar transcriptional activation potential and that the observed differences in reporter activity are a reflection of the susceptibility of these proteins to ubiquitin-mediated proteolysis.
FIG. 1.
Schematic representation of p53 mutants with C-terminal K-to-R changes.
FIG. 2.
p53 mutants containing C-terminal K-to-R substitutions show higher transcriptional activity than does WT p53. Saos-2 cells were cotransfected by electroporation with expression plasmids for WT p53 or K-to-R mutants and the pG13-Luc reporter plasmid. Each point is the mean of four independent transfections, with error bars representing 1 standard deviation.
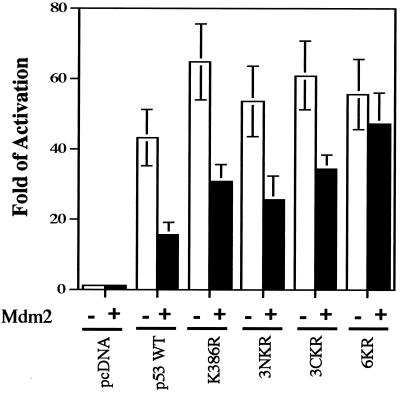
FIG. 8.
Negative regulation of p53 transcriptional activity by Mdm2 requires ubiquitin-mediated degradation. p53−/− Mdm2−/− cells were cotransfected with the pG13-Luc reporter plasmid and expression plasmids for WT p53 or the 6KR mutant, together with an empty vector or an Mdm2 expression plasmid. Twelve hours after transfection, reporter activity was determined. Each point is the mean of five independent transfections, with error bars representing 1 standard deviation.
Subcellular localization of K-to-R p53 mutants is similar to that of WT p53.
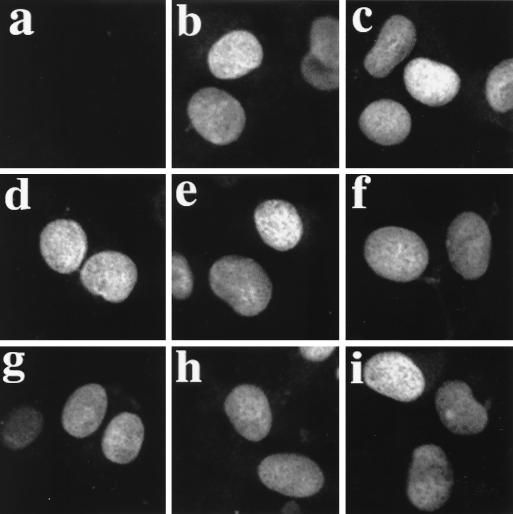
p53 lysine residues 370, 372, 373, 381, 382, and 386 represent the basic amino acid core of both p53 NLSII and NLSIII (57). Although NLSI (aa 316 to 322) is the predominant nuclear import signal in p53 (7, 57) and conservative K-to-R changes were introduced to create the p53 mutants, it was important to eliminate the possibility that the differences in transcriptional activity were a consequence of altered nuclear import. To establish this point, H1299 cells were transfected with WT p53 or K-to-R p53 mutants, and the subcellular localization of p53 was determined by indirect immunofluorescence with a p53-specific monoclonal antibody. WT p53 and K-to-R p53 mutants showed similar subcellular distributions. Ninety percent of transfected cells showed exclusive nuclear localization of immunoreactive p53 (Fig. 3). In the remaining 10% of transfected cells, p53 was found in both the nucleus and the cytoplasm. Similar results were obtained in Saos-2 cells (data not shown). These results suggest that the differences in transcriptional activity observed between WT p53 and K-to-R mutants cannot be explained by differences in subcellular localization.
FIG. 3.
Subcellular localization of WT p53 and mutants with C-terminal K-to-R changes. H1299 cells were transfected with empty pcDNA3 vector (a) and plasmids expressing WT p53 (b), 6KR (c), 3NKR (d), 3CKR (e), K372/373R (f), K381/382R (g), K370R (h), and K386R (i). Indirect immunofluorescence with antibody against p53 DO.1 is shown.
C-terminal lysine residues of p53 control Mdm2-directed degradation of p53.
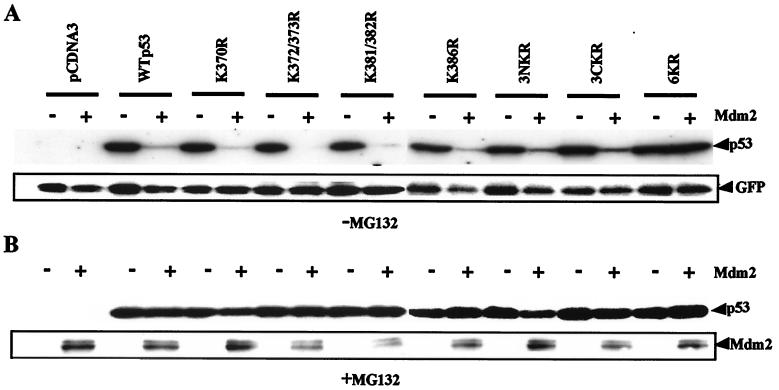
To assess the role of p53 C-terminal lysine residues in Mdm2-directed degradation of p53, K-to-R p53 mutants were either expressed alone or coexpressed with Mdm2 in Saos-2 cells. p53 and Mdm2 expression were analyzed by Western blotting, and p53 molecules containing single or double K-to-R changes all appeared to be susceptible to Mdm2-induced degradation (Fig. 4A). In contrast, p53 mutant 6KR was resistant to Mdm2-mediated degradation. Under these conditions, Mdm2 was detected only when the film was overexposed but was at a relatively constant level in each case (data not shown). GFP expressed from a cotransfected plasmid was used as an internal control and indicated that the observed differences were not due to variations in gel loading or transfection efficiency. The remaining 50% of transfected cells were pretreated with proteasome inhibitor MG132 for 4 h prior to harvest and Western blot analysis, as described above. As expected, MG132 blocked Mdm2-directed degradation of p53 proteins (Fig. 4B). Concomitant accumulation of p53 and Mdm2 after MG132 treatment confirmed that degradation of both proteins is proteasome mediated. Although 6KR was resistant to Mdm2-mediated degradation, limited degradation could be observed when Mdm2 was expressed at high levels, suggesting that alternative lysine residues can be used to mediate p53 degradation (data not shown). Thus, lysine residues 370, 372, 373, 381, 382, and 386 have an important role in the Mdm2-mediated degradation of p53.
FIG. 4.
Mdm2-mediated degradation is dependent on lysine residues in the C-terminal region of p53. Saos-2 cells were electroporated with plasmids encoding GFP, WT p53, or K-to-R p53 mutants, together with empty pcDNA3 vector or Mdm2 vector as indicated. (A) Twenty-four hours after transfection, whole-cell extracts were prepared and analyzed by Western blotting with anti-p53, anti-Mdm2, and anti-GFP monoclonal antibodies. (B) Transfected cells were treated with 10 μM MG132 for 4 h prior to harvest. Western blotting against p53 and Mdm2 was performed as described above.
K-to-R mutations in the C terminus of p53 do not affect its interaction with Mdm2.
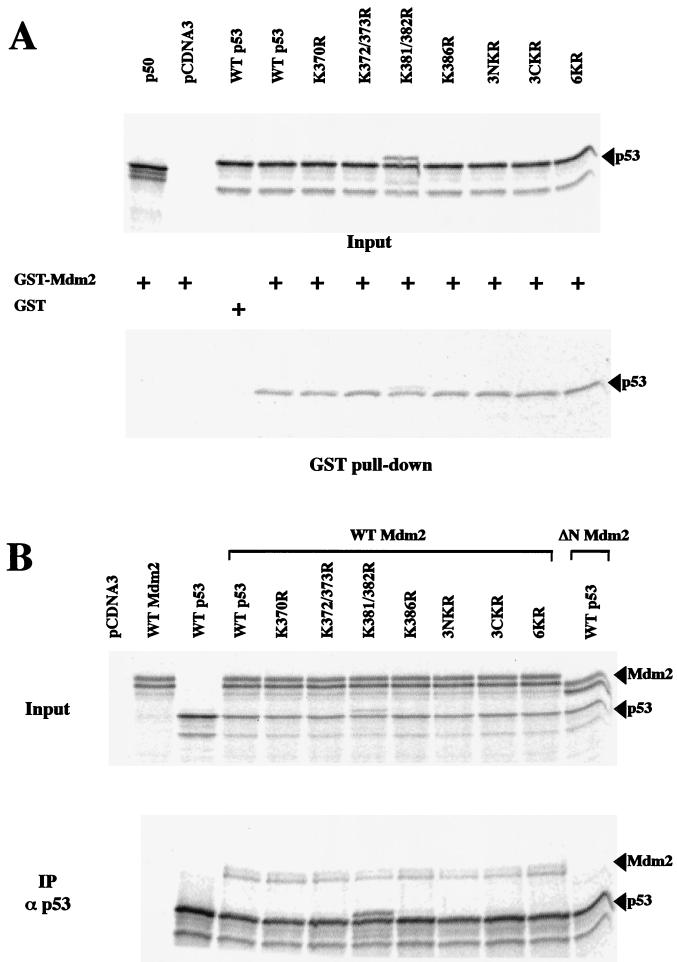
In addition to the N-terminal region, which is required for interaction with Mdm2, it has been suggested that the conformation of the C-terminal region of p53 may be important for targeting Mdm2-dependent p53 ubiquitination (20). To exclude the possibility that the observed resistance of 6KR to Mdm2-mediated degradation results from a failure to bind Mdm2, this interaction was analyzed in vitro. In vitro-translated and 35S-labeled WT p53 and K-to-R mutants were allowed to interact with GST-Mdm2, and the bound proteins were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) and phosphorimaging. No differences were observed between the binding abilities of WT and mutant p53 molecules (Fig. 5A). The observed interactions were specific, since GST alone did not interact with p53 and GST-Mdm2 failed to interact with 35S-labeled NF-κB p50 (Fig. 5A). Similar results were obtained by coimmunoprecipitation of Mdm2 and p53 molecules (which were generated by cotranslation in vitro) using anti-p53 antibody (Fig. 5B). Thus, C-terminal K-to-R point mutations do not affect the ability of p53 to be recognized by Mdm2.
FIG. 5.
K-to-R changes in the p53 C-terminal region do not affect p53-Mdm2 interaction. (A) 35S-labeled NF-κB (p50), WT p53, and K-to-R p53 mutants generated by in vitro transcription and translation were incubated with GST-Mdm2 or GST as indicated. GST pull-down assays were performed at 4°C. Mdm2-associated material was fractionated by SDS-PAGE, and the dried gel was analyzed by phosphorimaging (lower panel). (B) For coimmunoprecipitation, 35S-labeled Mdm2, ΔN Mdm2, WT p53, and K-to-R p53 mutants generated by in vitro transcription and translation were incubated with protein A- and protein G-agarose beads and p53-specific polyclonal antibody CM1, as indicated (IPα p53). p53-associated material was fractionated by SDS-PAGE, and the dried gel was analyzed by phosphorimaging (lower panel).
p53 C-terminal lysine residues are targets for Mdm2-dependent ubiquitination of p53.
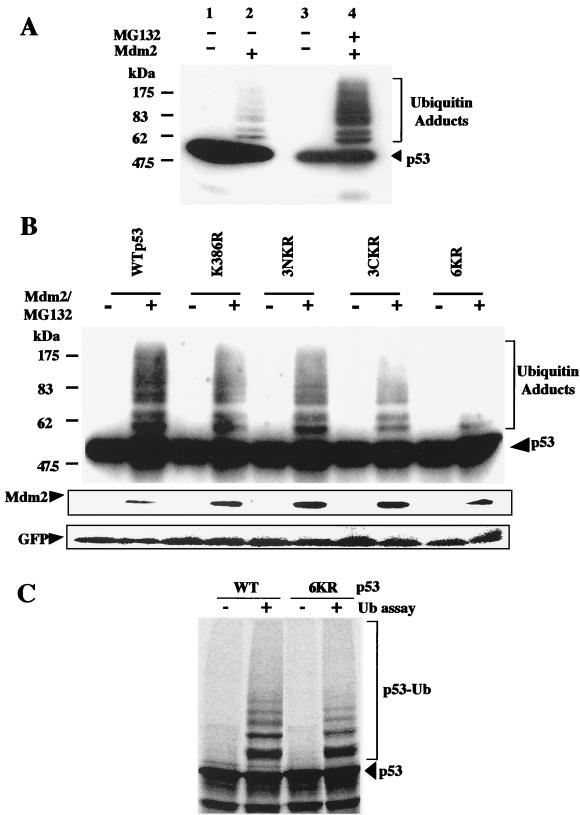
Since lysine residues are targets for ubiquitination, it is likely that the inability of the 6KR mutant to undergo Mdm2-dependent degradation is due to the fact that it cannot be ubiquitinated. To test this hypothesis, WT p53 and K-to-R mutants were expressed alone or together with Mdm2 in p53−/− Mdm2−/− cells. Twelve hours after transfection, Mdm2-transfected cells were exposed to 10 μM MG132 for 4 h. Whole-cell extracts were analyzed by Western blotting with the DO.1 monoclonal antibody. Slowly migrating forms of p53, consistent with ubiquitination, were observed with WT p53 and the K386R and 3NKR mutants (Fig. 6A and B). In the absence of proteasome inhibitor, p53 ubiquitinated forms were observed only when film was overexposed (Fig. 6A, lanes 1 and 2). The appearance of slowly migrating forms was reduced for the 3CKR mutant and virtually eliminated for the 6KR mutant. GFP expressed from a cotransfected plasmid was also analyzed by Western blotting, and the results indicated that the observed differences were not due to variations in gel loading or transfection efficiency. The relatively small proportion of p53 that accumulated as ubiquitinated products was a consequence of the intracellular removal of ubiquitin by multiple ubiquitin C-terminal hydrolases. Similar results were obtained for the H1299 and Saos-2 cell lines (data not shown). Ubiquitination of WT p53 and the 6KR mutant was also compared in an in vitro system containing 35S-labeled substrate, ubiquitin, and purified recombinant E1, Ubc5, and Mdm2 (49). Phosphorimager quantification of ubiquitin adducts indicated that ubiquitination of the 6KR mutant was reduced by 40% compared to that of WT p53 (Fig. 6C). We conclude that C-terminal lysine residues are involved in the Mdm2-mediated ubiquitination of p53.
FIG. 6.
p53 C-terminal lysine residues are targets for Mdm2-mediated ubiquitination. (A) Ubiquitination of p53 in vivo. p53−/− Mdm2−/− cells were cotransfected with WT p53 and empty pcDNA3 vector or an Mdm2 expression plasmid and incubated with proteasome inhibitor MG132, as indicated. Whole-cell extracts were analyzed by Western blotting with DO.1 anti-p53 monoclonal antibody. (B) p53−/− Mdm2−/− cells were cotransfected with a GFP-encoding plasmid and WT p53 or K-to-R mutants, together with empty pcDNA3 vector or an Mdm2 expression plasmid. Cells were treated and analyzed as described above. After being stripped, the membrane was incubated with Mdm2 and GFP antibodies. (C) In vitro ubiquitination (Ub) assays were performed with 35S-labeled WT p53 and the 6KR mutant. Reaction products were fractionated by SDS-PAGE, and the dried gel was analyzed by phosphorimaging. p53-Ub, ubiquitin-conjugated forms of p53.
Role of p53 C-terminal lysine residues in E6- and E6-AP-mediated p53 degradation.
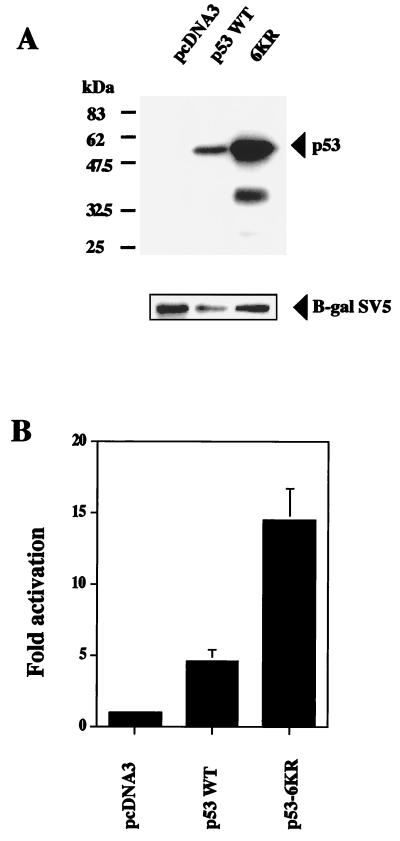
To investigate the role of p53 C-terminal lysine residues in E6-AP-mediated degradation, HeLa cells which express the E6 protein of human papillomavirus (HPV) were cotransfected with WT p53 or the 6KR mutant, together with SV5-tagged β-galactosidase expression plasmids. Twenty-four hours after transfection, whole-cell extracts were analyzed by Western blotting with a monoclonal antibody recognizing p53. Higher levels of the 6KR mutant than of WT p53 were observed (Fig. 7A). Similar results were obtained with a polyclonal antibody to p53 (data not shown). The increased accumulation of 6KR compared to WT p53 was not due to variations in transfection efficiency, because these differences were not reflected in the levels of the cotransfected SV5-tagged β-galactosidase used as an internal control. To confirm these results, HeLa cells were cotransfected with the pG13-Luc reporter plasmid and expression plasmids for WT p53 or the 6KR mutant. The 6KR mutant showed three times more transcriptional activity than did WT p53 (Fig. 7B). Thus, it is likely that p53 C-terminal lysine residues are also targets for E6-AP-mediated regulation.
FIG. 7.
p53 C-terminal lysine residues are targets for E6-AP-mediated degradation. (A) HeLa cells which express HPV E6 protein were cotransfected with WT p53 or the 6KR mutant and SV5-tagged β-galactosidase expression plasmids. Twenty-four hours after transfection, whole-cell extracts were analyzed by Western blotting with a monoclonal antibody to p53 (DO.1). After being stripped, the membrane was blotted with SV5 antibody (β-gal SV5). (B) HeLa cells were cotransfected with the pG13-Luc reporter plasmid and expression plasmids for WT p53 or the 6KR mutant. Twelve hours after transfection, reporter activity was determined. Each point is the mean of four independent transfections, with error bars representing 1 standard deviation.
The susceptibility of p53 to ubiquitination correlates with the ability of Mdm2 to negatively regulate p53-dependent transcription.
To evaluate the contribution of p53 degradation to the negative regulation of p53 functions by Mdm2, we cotransfected p53−/− Mdm2−/− cells with WT p53 and the K386R, 3NKR, 3CKR, and 6KR mutants and the p53 reporter plasmid pG13-Luc in the absence or presence of an Mdm2 expression plasmid. It was reasoned that K-to-R mutants resistant to Mdm2-mediated ubiquitination and degradation may show some resistance to the Mdm2-mediated negative regulation of p53-dependent transcription. Mdm2 expression reduced WT p53-dependent transcription by 60 to 70% (Fig. 8). Reductions of 40 to 60% of transcriptional activity were observed when the K386R, 3NKR, and 3CKR mutants were cotransfected with Mdm2 (Fig. 8). In contrast, 6KR transcriptional activity was not significantly reduced by Mdm2 (Fig. 8). Our results suggest that ubiquitin-proteasome-mediated degradation of p53 is a major function of Mdm2 that contributes to the negative regulation of p53-dependent transcriptional activity.
DISCUSSION
Degradation is the only mechanism which abrogates all functions of p53, and in normal cells this is accomplished by the ubiquitin-proteasome system (34). To identify lysine residues which are the sites of modification by ubiquitin, a series of p53 molecules were created that contain lysine-to-arginine changes in the C-terminal region of the protein. Using p53-dependent reporter assays, we demonstrated that the p53 molecules containing K-to-R changes display a higher transcriptional activity, which is a reflection of their increased stability and resistance to ubiquitin modification (Fig. 2). In addition to acting as a ubiquitin protein ligase (19), it has been reported that Mdm2 negatively regulates p53 transcriptional activity by binding to the transactivation domain of p53 (45, 65). Since p53 mutants which accumulate may also induce more efficient synthesis of Mdm2, differences in transcription are more obvious at early stages after transfection. The observed transcriptional differences between K-to-R p53 mutants cannot be explained as a consequence of conformational changes induced by the mutations, since reporter assays performed in p53−/− Mdm2−/− cells showed no significant differences in the transactivation potential of WT and mutant versions of p53 in the absence of Mdm2 (Fig. 8). This result also suggests that Mdm2 is the key regulatory molecule for the control of p53 stability. In HPV-transformed cells p53 is regulated by the viral E6 protein, which, in conjunction with E6-AP, targets p53 for ubiquitin-mediated proteolysis. The 6KR mutant also appears to be more resistant to degradation than WT p53 when introduced into HeLa cells which constitutively express HPV E6. Because complex formation between E6 and p53 is not mediated through the C terminus of p53 (6, 36), and since WT p53 and 6KR showed similar subcellular distributions in HeLa cells (data not shown), we conclude that E6-directed ubiquitination may also target the C-terminal lysines of p53. However, it has been reported that p53 mutants lacking residues 370 to 393 or containing lysine-to-isoleucine changes at positions 381, 382, and 386 are efficiently degraded after cotransfection with HPV E6 into p53−/− mouse fibroblasts (6). Differences in the levels of E6 between transfected and transformed cells and the extent to which Mdm2 is activated by p53 under these different conditions may go some way to explaining the variance of these results. In an in vitro degradation system it was noted that E6-mediated degradation of p53 containing K-to-I changes at positions 381, 382, and 386 was less efficient than that of WT p53 (6).
Conservative changes of K to R do not have consequences for other p53 properties. Similar subcellular distributions were observed for K-to-R p53 mutants and WT p53 when transiently expressed in p53 null cells. Conservative changes in NLSII and NLSIII do not affect the NLS function, and it has been shown that NLSI is the predominant NLS of p53 (7, 57). Likewise, K-to-R p53 mutants show binding to Mdm2 similar to that of WT p53. Thus, the transcriptional activity of K-to-R p53 mutants cannot be explained by differences in nuclear localization or Mdm2 binding.
Multiple sites for ubiquitin ligation are often required to target a substrate for degradation (2, 51, 56, 61). Only the 6KR mutant was resistant to Mdm2-mediated ubiquitination and degradation under the experimental conditions used. Although a dramatic reduction in the detection of ubiquitinated forms of the 6KR mutant was observed in vivo, it should be noted that when the Western blot was exposed for a long time, some ubiquitinated forms could still be detected (data not shown), suggesting that neighboring lysine residues, such as K351 and/or K357, may also be used as alternative ubiquitination sites. This seems to be the case in vitro, where only a 40% reduction in the amount of ubiquitinated forms of 6KR was observed compared to that of WT p53. The remaining 60% of ubiquitination in the 6KR mutant could also be explained by the excess of E1, E2, and E3 used in our in vitro assay or the absence of other cofactors regulating p53 ubiquitination. For instance, the interaction of p19 ARF with Mdm2 inhibits the ubiquitin ligase activity of Mdm2 (20, 39), and retinoblastoma protein has been proposed to block Mdm2-mediated degradation of p53 (21). Together, our results suggest that the previously reported resistance to Mdm2-mediated degradation observed in a p53 mutant lacking the extreme C-terminal 30 aa (28) can be explained by the failure of this mutant to be efficiently ubiquitinated (20).
Reporter assays performed in p53−/− Mdm2−/− cells show that exogenous Mdm2 is incapable of inhibiting the transcriptional activity of the 6KR mutant (Fig. 8). Therefore, the predominant function of Mdm2 is to act as a ubiquitin protein ligase rather than as a direct transcriptional inhibitor (13, 19, 20, 39). This is consistent with other published work which indicates that under special circumstances, when p53 and Mdm2 are present in the same cellular compartment, no inhibition of p53-dependent transcription is observed. Thus, when p53 and Mdm2 are retained in the nucleolus by treatment with leptomycin B, p53 is transcriptionally active (29). Likewise, c-Abl nonreceptor tyrosine kinase appears to stabilize p53 by inhibiting Mdm2-mediated degradation. In this situation, p53 retains its transcriptional activity even though the amount of p53 bound to Mdm2 does not change (59). It is now widely accepted that several regions in p53 are required for Mdm2-mediated degradation. It has been recently reported that a region from aa 92 to 112 of p53 functions as a degradation signal for Mdm2 (15). Interestingly, this region is part of an extensive N-terminal PEST region in p53 (52) that is presumably exposed on the surface of the protein, since it is susceptible to proteolysis in vitro (47). Gu et al. (15) also recognize that in addition to the N-terminal region of p53, the C-terminal region of p53 also contributes to Mdm2-mediated degradation of p53. Thus, it appears that Mdm2 binds to the N terminus of p53 but directs ubiquitin ligation at the C terminus of p53.
A complex requirement for degradation is not confined to p53, and striking similarities can be recognized between the degradation of the NF-κB inhibitor protein IκBα and that of p53. Degradation of both molecules seems to require both C- and N-terminal regions, one of which contains a PEST region, while the other contains lysine residues which are the targets for ubiquitination (27, 48, 50–52). Moreover, the IκBα and p53 lysine residue implicated in SUMO-1 conjugation is also involved in ubiquitination (8, 49). In fact, p53 mutant K386R, which cannot conjugate SUMO-1 (49), is also the single-point mutation with the highest transcriptional rate, suggesting that lysine residue 386 may be a major ubiquitination site. Since ubiquitin conjugation of the K386R mutant is possible, we can conclude that SUMO-1 and ubiquitin conjugation of p53 do not compete for the same lysine residue. However, since the C-terminal region of p53 is recognized by both ubiquitin and SUMO-1 enzymes, an interference between the two conjugation systems cannot be excluded. The balance between SUMO-1 and ubiquitin conjugation may be critical for controlling p53 activity, because SUMO-1 conjugation activates p53 transcriptional activity (14, 49), while ubiquitination reduces p53 levels and as a consequence abrogates these functions.
Other posttranslational modifications in the C terminus of p53 have also been reported. Phosphorylation of serine residues 376, 378, and 392 (17, 22, 23, 38) and acetylation of lysine residues 320 and 382 (16, 55) of p53 enhance sequence-specific DNA binding in vitro. Interestingly, lysine residue 382 becomes acetylated and lysine residue 386 is conjugated to SUMO-1 after cells are exposed to UV light (49, 55). In addition to their consequences for the transcriptional activity of p53, those modifications may subtly influence p53 ubiquitination by altering the selection of lysine residues that are available for ubiquitination. Thus, phosphorylation, acetylation, and SUMO-1 conjugation may contribute to the induction of p53 responses after DNA damage, whereas the role of ubiquitination is to maintain a low steady-state level of p53 in normal cells. This fine balance allows the p53 system to quickly respond to signals from the extracellular environment by dramatically increasing the levels and activity of p53.
ACKNOWLEDGMENTS
We thank Alex Houston, University of St. Andrews, for DNA sequencing and Mark Rolfe, Mitotix, for provision of the E1 baculovirus.
This work was supported by the Medical Research Council, the Biotechnology and Biological Research Council, and the Cancer Research Campaign. D.P.L. is a Gibb fellow of the Cancer Research Campaign.
REFERENCES
- 1.Askjaer P, Jensen T H, Nilsson J, Englmeier L, Kjems J. The specificity of the CRM1-Rev nuclear export signal interaction is mediated by RanGTP. J Biol Chem. 1998;273:33414–33422. doi: 10.1074/jbc.273.50.33414. [DOI] [PubMed] [Google Scholar]
- 2.Baldi L, Brown K, Franzoso G, Siebenlist U. Critical role for lysine-21 and lysine-22 signal-induced, ubiquitin-mediated proteolysis of IκBα. J Biol Chem. 1996;271:376–379. doi: 10.1074/jbc.271.1.376. [DOI] [PubMed] [Google Scholar]
- 3.Barak Y, Juven T, Haffner R, Oren M. mdm2 expression is induced by wild type p53 activity. EMBO J. 1993;12:461–468. doi: 10.1002/j.1460-2075.1993.tb05678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bottger V, Bottger A, Howard S F, Picksley S M, Chene P, Garcia-Echeverria C, Hochkeppel H K, Lane D P. Identification of novel mdm2 binding peptides by phage display. Oncogene. 1996;13:2141–2147. [PubMed] [Google Scholar]
- 5.Chen J, Marechal V, Levine A J. Mapping of the p53 and mdm-2 interaction domains. Mol Cell Biol. 1993;13:4107–4114. doi: 10.1128/mcb.13.7.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crook T, Ludwig R L, Marston N J, Willkomm D, Vousden K H. Sensitivity of p53 lysine mutants to ubiquitin-directed degradation targeted by human papillomavirus E6. Virology. 1996;217:285–292. doi: 10.1006/viro.1996.0115. [DOI] [PubMed] [Google Scholar]
- 7.Dang C V, Lee W M. Nuclear and nucleolar targeting sequences of c-erb-A, c-myb, N-myc, p53, HSP70, and HIV tat proteins. J Biol Chem. 1989;264:18019–18023. [PubMed] [Google Scholar]
- 8.Desterro J M P, Rodriguez M S, Hay R T. SUMO-1 modification of IκBα inhibits NF-κB activation. Mol Cell. 1998;2:233–239. doi: 10.1016/s1097-2765(00)80133-1. [DOI] [PubMed] [Google Scholar]
- 9.Eizenberg O, Faber-Elman A, Gottlieb E, Oren M, Rotter V, Schwartz M. Direct involvement of p53 in programmed cell death of oligodendrocytes. EMBO J. 1995;14:1136–1144. doi: 10.1002/j.1460-2075.1995.tb07097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.el-Deiry W S, Tokino T, Velculescu V E, Levy D B, Parsons R, Trent J M, Lin D, Mercer W E, Kinzler K W, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 11.Fornerod M, Ohno M, Yoshida M, Mattaj I W. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 12.Freedman D A, Levine A J. Nuclear export is required for degradation of endogenous p53 by MDM2 and human papillomavirus E6. Mol Cell Biol. 1998;18:7288–7293. doi: 10.1128/mcb.18.12.7288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuchs S Y, Adler V, Buschmann T, Wu X, Ronai Z. Mdm2 association with p53 targets its ubiquitination. Oncogene. 1998;17:2543–2547. doi: 10.1038/sj.onc.1202200. [DOI] [PubMed] [Google Scholar]
- 14.Gostissa M, Hengstermann A, Fogal V, Sandy P, Schwarz S E, Scheffner M, Del Sal G. Activation of p53 by conjugation to the ubiquitin-like protein SUMO-1. EMBO J. 1999;18:6462–6471. doi: 10.1093/emboj/18.22.6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gu J, Chen D, Rosenblum J, Rubin R M, Yuan Z-M. Identification of a sequence element from p53 that signals for Mdm2-targeted degradation. Mol Cell Biol. 2000;20:1243–1253. doi: 10.1128/mcb.20.4.1243-1253.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gu W, Roeder R T. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 17.Herrmann C P, Kraiss S, Montenarh M. Association of casein kinase II with immunopurified p53. Oncogene. 1991;6:877–884. [PubMed] [Google Scholar]
- 18.Hershko A, Ciechanover A. The ubiquitin system for protein degradation. Annu Rev Biochem. 1992;61:761–807. doi: 10.1146/annurev.bi.61.070192.003553. [DOI] [PubMed] [Google Scholar]
- 19.Honda R, Tanaka H, Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 1997;420:25–27. doi: 10.1016/s0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- 20.Honda R, Yasuda H. Association of p19(ARF) with Mdm2 inhibits ubiquitin ligase activity of Mdm2 for tumor suppressor p53. EMBO J. 1999;18:22–27. doi: 10.1093/emboj/18.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hsieh J K, Chan F S, O'Connor D J, Mittnacht S, Zhong S, Lu X. RB regulates the stability and the apoptotic function of p53 via MDM2. Mol Cell. 1999;3:181–193. doi: 10.1016/s1097-2765(00)80309-3. [DOI] [PubMed] [Google Scholar]
- 22.Hupp T R, Lane D P. Allosteric activation of latent p53 tetramers. Curr Biol. 1994;4:865–875. doi: 10.1016/s0960-9822(00)00195-0. [DOI] [PubMed] [Google Scholar]
- 23.Hupp T R, Meek D W, Midgley C A, Lane D P. Regulation of the specific DNA binding function of p53. Cell. 1992;71:875–886. doi: 10.1016/0092-8674(92)90562-q. [DOI] [PubMed] [Google Scholar]
- 24.Jones S N, Roe A E, Donehower L A, Bradley A. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature. 1995;378:206–208. doi: 10.1038/378206a0. [DOI] [PubMed] [Google Scholar]
- 25.Kastan M B, Onyekwere O, Sidransky D, Vogelstein B, Craig R W. Participation of p53 protein in the cellular response to DNA damage. Cancer Res. 1991;51:6304–6311. [PubMed] [Google Scholar]
- 26.Ko L J, Prives C. p53: puzzle and paradigm. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 27.Kroll M, Conconi M, Desterro M J, Marin A, Thomas D, Friguet B, Hay R T, Virelizier J L, Arenzana-Seisdedos F, Rodriguez M S. The carboxy-terminus of IκBα determines susceptibility to degradation by the catalytic core of the proteasome. Oncogene. 1997;15:1841–1850. doi: 10.1038/sj.onc.1201560. [DOI] [PubMed] [Google Scholar]
- 28.Kubbutat M H G, Ludwig R L, Ashcroft M, Vousden K H. Regulation of Mdm2-directed degradation by the C terminus of p53. Mol Cell Biol. 1998;18:5690–5698. doi: 10.1128/mcb.18.10.5690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lain S, Midgley C, Sparks A, Lane E B, Lane D P. An inhibitor of nuclear export activates the p53 response and induces the localization of HDM2 and p53 to U1A-positive nuclear bodies associated with the PODs. Exp Cell Res. 1999;248:457–472. doi: 10.1006/excr.1999.4433. [DOI] [PubMed] [Google Scholar]
- 30.Lane D P. Awakening angels. Nature. 1998;394:616–617. doi: 10.1038/29166. [DOI] [PubMed] [Google Scholar]
- 31.Lane D P. Cancer. p53, guardian of the genome. Nature. 1992;358:15–16. doi: 10.1038/358015a0. [DOI] [PubMed] [Google Scholar]
- 32.Levine A J. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 33.Liang S H, Hong D, Clarke M F. Cooperation of a single lysine mutation and a C-terminal domain in the cytoplasmic sequestration of the p53 protein. J Biol Chem. 1998;273:19817–19821. doi: 10.1074/jbc.273.31.19817. [DOI] [PubMed] [Google Scholar]
- 34.Maki C G, Huibregtse J M, Howley P M. In-vivo ubiquitination and proteasome-mediated degradation of p53. Cancer Res. 1996;56:2649–2654. [PubMed] [Google Scholar]
- 35.Maltzman W, Czyzyk L. UV irradiation stimulates levels of p53 cellular tumor antigen in nontransformed mouse cells. Mol Cell Biol. 1984;4:1689–1694. doi: 10.1128/mcb.4.9.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mansur C P, Marcus B, Dalal S, Androphy E J. The domain of p53 required for binding HPV 16 E6 is separable from the degradation domain. Oncogene. 1995;10:457–465. [PubMed] [Google Scholar]
- 37.Mayo L D, Turchi J J, Berberich S J. Mdm-2 phosphorylation by DNA-dependent protein kinase prevents interaction with p53. Cancer Res. 1997;57:5013–5016. [PubMed] [Google Scholar]
- 38.Meek D W, Simon S, Kikkawa U, Eckhart W. The p53 tumour suppressor protein is phosphorylated at serine 389 by casein kinase II. EMBO J. 1990;9:3253–3260. doi: 10.1002/j.1460-2075.1990.tb07524.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Midgley C A, Desterro J M, Saville M K, Howard S, Sparks A, Hay R T, Lane D P. An N-terminal p14ARF peptide blocks Mdm2-dependent ubiquitination in vitro and can activate p53 in vivo. Oncogene. 2000;19:2312–2323. doi: 10.1038/sj.onc.1203593. [DOI] [PubMed] [Google Scholar]
- 40.Midgley C A, Fisher C J, Bartek J, Vojtesek B, Lane D, Barnes D M. Analysis of p53 expression in human tumours: an antibody raised against human p53 expressed in Escherichia coli. J Cell Sci. 1992;101:183–189. doi: 10.1242/jcs.101.1.183. [DOI] [PubMed] [Google Scholar]
- 41.Midgley C A, Lane D P. p53 protein stability in tumour cells is not determined by mutation but is dependent on Mdm2 binding. Oncogene. 1997;15:1179–1189. doi: 10.1038/sj.onc.1201459. [DOI] [PubMed] [Google Scholar]
- 42.Miyashita T, Reed J C. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell. 1995;80:293–299. doi: 10.1016/0092-8674(95)90412-3. [DOI] [PubMed] [Google Scholar]
- 43.Momand J, Zambetti G P, Olson D C, George D, Levine A J. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell. 1992;69:1237–1245. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- 44.Oliner J D, Kinzler K W, Meltzer P S, George D L, Vogelstein B. Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature. 1992;358:80–83. doi: 10.1038/358080a0. [DOI] [PubMed] [Google Scholar]
- 45.Oliner J D, Pietenpol J A, Thiagalingam S, Gyuris J, Kinzler K W, Vogelstein B. Oncoprotein MDM2 conceals the activation domain of tumour suppressor p53. Nature. 1993;362:857–860. doi: 10.1038/362857a0. [DOI] [PubMed] [Google Scholar]
- 46.Ossareh-Nazari B, Bachelerie F, Dargemont C. Evidence for a role of CRM1 in signal-mediated nuclear protein export. Science. 1997;278:141–144. doi: 10.1126/science.278.5335.141. [DOI] [PubMed] [Google Scholar]
- 47.Pavletich N P, Chambers K A, Pabo C O. The DNA-binding domain of p53 contains the four conserved regions and the major mutation hot spots. Genes Dev. 1993;7:2556–2564. doi: 10.1101/gad.7.12b.2556. [DOI] [PubMed] [Google Scholar]
- 48.Rechsteiner M, Rogers S W. PEST sequences and regulation by proteolysis. Trends Biochem Sci. 1996;21:267–271. [PubMed] [Google Scholar]
- 49.Rodriguez M S, Desterro J M, Lain S, Midgley C A, Lane D P, Hay R T. SUMO-1 modification activates the transcriptional response of p53. EMBO J. 1999;18:6455–6461. doi: 10.1093/emboj/18.22.6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rodriguez M S, Michalopoulos I, Arenzana-Seisdedos F, Hay R T. Inducible degradation of IκBα in vitro and in vivo requires the acidic C-terminal domain of the protein. Mol Cell Biol. 1995;15:2413–2419. doi: 10.1128/mcb.15.5.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rodriguez M S, Wright J, Thompson J, Thomas D, Baleux F, Virelizier J L, Hay R T, Arenzana-Seisdedos F. Identification of lysine residues required for signal-induced ubiquitination and degradation of IκBα in vivo. Oncogene. 1996;12:2425–2435. [PubMed] [Google Scholar]
- 52.Rogers S, Wells R, Rechsteiner M. Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science. 1986;234:364–368. doi: 10.1126/science.2876518. [DOI] [PubMed] [Google Scholar]
- 53.Rolfe M, Beerromero P, Glass S, Eckstein J, Berdo I, Theodoras A, Pagano M, Draetta G. Reconstitution of p53-ubiquitinylation reactions from purified components—the role of human ubiquitin-conjugating enzyme Ubc4 and E6-associated protein (E6AP) Proc Natl Acad Sci USA. 1995;92:3264–3268. doi: 10.1073/pnas.92.8.3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roth J, Dobbelstein M, Freedman D A, Shenk T, Levine A J. Nucleo-cytoplasmic shuttling of the hdm2 oncoprotein regulates the levels of the p53 protein via a pathway used by the human immunodeficiency virus rev protein. EMBO J. 1998;17:554–564. doi: 10.1093/emboj/17.2.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sakaguchi K, Herrera J E, Saito S, Miki T, Bustin M, Vassilev A, Anderson C W, Appella E. DNA damage activates p53 through a phosphorylation-acetylation cascade. Genes Dev. 1998;12:2831–2841. doi: 10.1101/gad.12.18.2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Scherer D C, Brockman J A, Chen Z, Maniatis T, Ballard D W. Signal-induced degradation of IκBα requires site-specific ubiquitination. Proc Natl Acad Sci USA. 1995;92:11259–11263. doi: 10.1073/pnas.92.24.11259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shaulsky G, Goldfinger N, Ben-Ze'ev A, Rotter V. Nuclear accumulation of p53 protein is mediated by several nuclear localization signals and plays a role in tumorigenesis. Mol Cell Biol. 1990;10:6565–6577. doi: 10.1128/mcb.10.12.6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shieh S Y, Ikeda M, Taya Y, Prives C. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell. 1997;91:325–334. doi: 10.1016/s0092-8674(00)80416-x. [DOI] [PubMed] [Google Scholar]
- 59.Sionov R V, Moallem E, Berger M, Kazaz A, Gerlitz O, Ben-Neriah Y, Oren M, Haupt Y. c-Abl neutralizes the inhibitory effect of Mdm2 on p53. J Biol Chem. 1999;274:8371–8374. doi: 10.1074/jbc.274.13.8371. [DOI] [PubMed] [Google Scholar]
- 60.Stommel J M, Marchenko N D, Jimenez G S, Moll U M, Hope T J, Wahl G M. A leucine-rich nuclear export signal in the p53 tetramerization domain: regulation of subcellular localization and p53 activity by NES masking. EMBO J. 1999;18:1660–1672. doi: 10.1093/emboj/18.6.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Treier M, Staszewski L M, Bohmann D. Ubiquitin-dependent c-jun degradation in-vivo is mediated by the delta-domain. Cell. 1994;78:787–798. doi: 10.1016/s0092-8674(94)90502-9. [DOI] [PubMed] [Google Scholar]
- 62.Ullman K S, Powers M A, Forbes D J. Nuclear export receptors: from importin to exportin. Cell. 1997;90:967–970. doi: 10.1016/s0092-8674(00)80361-x. [DOI] [PubMed] [Google Scholar]
- 63.Vojtesek B, Dolezalova H, Lauerova L, Svitakova M, Havlis P, Kovarik J, Midgley C A, Lane D P. Conformational changes in p53 analysed using new antibodies to the core DNA binding domain of the protein. Oncogene. 1995;10:389–393. [PubMed] [Google Scholar]
- 64.Weber J D, Taylor L J, Roussel M F, Sherr C J, Bar-Sagi D. Nucleolar Arf sequesters Mdm2 and activates p53. Nat Cell Biol. 1999;1:20–26. doi: 10.1038/8991. [DOI] [PubMed] [Google Scholar]
- 65.Wu X, Bayle J H, Olson D, Levine A J. The p53-mdm-2 autoregulatory feedback loop. Genes Dev. 1993;7:1126–1132. doi: 10.1101/gad.7.7a.1126. [DOI] [PubMed] [Google Scholar]
- 66.Zhang Y, Xiong Y. Mutations in human ARF exon 2 disrupt its nucleolar localization and impair its ability to block nuclear export of MDM2 and p53. Mol Cell. 1999;3:579–591. doi: 10.1016/s1097-2765(00)80351-2. [DOI] [PubMed] [Google Scholar]