Abstract
Follicular thyroglobulin (TG) selectively suppresses the expression of thyroid-restricted transcription factors, thereby altering the expression of thyroid-specific proteins. In this study, we investigated the molecular mechanism by which TG suppresses the prototypic thyroid-restricted transcription factor, thyroid transcription factor 1 (TTF-1), in rat FRTL-5 thyrocytes. We show that the region between bp −264 and −153 on the TTF-1 promoter contains two nuclear factor I (NFI) elements whose function is involved in TG-mediated suppression. Thus, NFI binding to these elements is critical for constitutive expression of TTF-1; TG decreases NFI binding to the NFI elements in association with TG repression. NFI is a family of transcription factors that is ubiquitously expressed and contributes to constitutive and cell-specific gene expression. In contrast to the contribution of NFI proteins to constitutive gene expression in other systems, we demonstrate that follicular TG transcriptionally represses all NFI RNAs (NFI-A, -B, -C, and -X) in association with decreased NFI binding and that the RNA levels decrease as early as 4 h after TG treatment. Although TG treatment for 48 h results in a decrease in NFI protein-DNA complexes measured in DNA mobility shift assays, NFI proteins are still detectable by Western analysis. We show, however, that the binding of all NFI proteins is redox regulated. Thus, diamide treatment of nuclear extracts strongly reduces the binding of NFI proteins, and the addition of higher concentrations of dithiothreitol to nuclear extracts from TG-treated cells restores NFI-DNA binding to levels in extracts from untreated cells. We conclude that NFI binding to two NFI elements, at bp −264 to −153, positively regulates TTF-1 expression and controls constitutive TTF-1 levels. TG mediates the repression of TTF-1 gene expression by decreasing NFI RNA and protein levels, as well as by altering the binding activity of NFI, which is redox controlled.
The basic functional unit of the thyroid is the thyroid follicle; thyrotropin (TSH) is thought to be its primary regulator of function, in concert with insulin and insulin-like growth factor 1 (IGF-1). The function of each follicle is, however, heterogeneous, despite its being exposed to the same hormonal milieu in the blood. Recently it was shown that accumulated thyroglobulin (TG) in the follicular lumen is a feedback suppressor of hormonally increased thyroid function and plays a significant role in follicular heterogeneity (52–55). Thus, follicular TG selectively suppresses RNA levels of thyroid-specific proteins; the sodium iodide symporter (NIS), thyroid peroxidase (TPO), TG, and the TSH receptor (TSHR) (52–55). Suppression is transcriptional rather than being caused by changes in RNA stability, since TG decreases the promoter activity of the NIS, TG, TPO, and TSHR genes (51–55) concordantly with decreases in their RNAs. The suppressive effect of follicular TG is not duplicated by thyroid hormones or iodide nor by bovine serum albumin (BSA) or immunoglobulin (52, 54, 55). The ability of follicular TG to suppress the thyroid-specific proteins is explained by its ability to suppress the transcription activity, RNA, and protein levels of three thyroid-restricted transcription factors regulated by TSH or insulin–IGF-1, i.e., thyroid transcription factor 1 (TTF-1), Pax-8, and TTF-2, but not ubiquitous transcription factors which can also regulate the thyroid-specific genes (52–55). Thus, negative-feedback regulation by follicular TG counteracts the action of TSH or insulin–IGF-1, and TG transcriptional activity helps maintain a dynamic balance of follicular function necessary for thyroid hormone homeostasis (53, 55).
The ability of TG to suppress thyroid-restricted transcription factors is mediated by TG binding to a receptor on the apical surface of cells surrounding the follicular lumen of the thyroid follicle (51–56). This receptor is thought to be an asialoglycoprotein receptor (ASGPR), similar in structure to the liver ASGPR and previously associated with vectorial transport of TG to the follicular lumen (9, 10, 29, 37, 48, 58; F. Pacifico, N. Montuori, L. Ulianich, B. Di Jeso, L. Nitsch, L. Kohn, S. Formisano, and E. Consiglio, submitted for publication). Thus, TG binding to the ASGPR and the repressive action of TG are coordinately attenuated by neuraminidase treatment of the cells (9, 56) and coordinately effected by different macromolecular multimers of TG (9, 10, 56). Most importantly, TG suppression is abolished by an antibody against the ASGPR (56).
Specific signaling pathways activated by TG binding to the apical receptor and potentially involved in repression are under investigation (56). In this study, however, we addressed the molecular mechanism by which follicular TG modulates gene expression. We used TTF-1 as a prototypic thyroid-restricted transcription factor, since it is a major transcriptional regulator of the TG (8, 14, 50), NIS (15, 43), TPO (16, 38), and TSHR (7, 30, 49) genes. We hoped that this study would also help clarify the mechanisms which control constitutive TTF-1 gene expression and ultimately allow us to trace the TG signaling path in the reverse direction. In this report, we show that TG decreases the expression and binding of the nuclear factor I (NFI) family of proteins to the TTF-1 5′-flanking region and that NFI is a positive regulator of TTF-1 gene expression, controlling TTF-1 constitutive expression.
NFI was initially identified as a cellular factor that stimulates in vitro replication of adenovirus DNA (20, 21, 35) but was subsequently shown to play an important role in RNA transcription (5, 22, 47). The NFI family consists of four highly conserved genes (four subtypes) whose protein products are able to homodimerize and heterodimerize (18, 33). Additionally, each gene gives rise to alternatively spliced transcripts that potentially encode a number of different isoforms (17, 33, 46). The existence of a number of structurally different NFI proteins, their differential expression, and the involvement of NFI binding sites in cell-specific gene expression (5, 17, 22, 47) suggested that individual isoforms might have distinct functions. Surprisingly, and in contrast to other reports describing the contribution of these proteins to constitutive gene expression (6, 44), we show that all NFI RNAs (NFI-A, -B, -C, and -X) are transcriptionally regulated in thyrocytes and are repressible by follicular TG, which is a tissue-specific protein.
(Part of this work was presented at the 81st Annual Meeting of The Endocrine Society, abstr. OR-8-3, 1999.)
MATERIALS AND METHODS
Materials.
Bovine follicular TG was prepared by salt extraction of sliced, fresh thyroid glands, ammonium sulfate precipitation, and gel filtration chromatography on Sephacryl S-300 (Amersham Pharmacia Biotech, Uppsala, Sweden) in 0.1 M potassium phosphate (pH 7.4) (9, 10). The source of all other materials was the same as reported previously (51, 52, 54, 56).
Cells.
Buffalo rat liver 3A (BRL-3A) cells (ATCC CRL 1442) were grown in Ham's F-12 medium supplemented with 5% fetal calf serum. Nonfunctioning rat FRT thyroid cells (1) and the F1 subclone of FRTL-5 thyrocytes (Interthyr Research Foundation, Baltimore, Md.) (ATCC CRL 8305), which are diploid and have all the functional properties previously detailed (12, 24, 51–56), were grown in Coon's modified Ham's F-12 medium supplemented with 5% calf serum, 2 mM glutamine, and 1 mM nonessential amino acids. FRTL-5 cell medium also includes a six-hormone mixture (6H medium) containing bovine TSH (10−10 M), insulin (10 μg/ml), cortisol (0.4 ng/ml), transferrin (5 μg/ml), glycyl-l-histidyl-l-lysine acetate (10 ng/ml), and somatostatin (10 ng/ml).
RNA isolation and Northern analysis.
RNA was prepared using RNeasy Mini Kits (Qiagen Inc., Valencia, Calif.) and the method described by the manufacturer. For Northern blots, 20-μg portions of different RNA samples were run on denatured agarose gels, capillary blotted onto Nytran membranes (11 by 14 cm; Schleicher & Schuell), UV cross-linked, and subjected to hybridization. Probes were prepared by reverse transcription-PCR (RT-PCR) (see below). An amplified cDNA was purified from an agarose gel by using a Jetsorb kit (Genomed Inc.) and was labeled with [α-32P]dCTP by using a Ladderman labeling kit (Takara Biochemical Inc., Berkeley, Calif.). Membranes were hybridized and washed as described previously (54).
RT-PCR.
cDNA was synthesized using an Advantage RT-for-PCR Kit (Clontech Laboratories Inc., Palo Alto, Calif.). PCR was performed by “touchdown” PCR (13) using a GeneAmp 9600 PCR apparatus (Perkin-Elmer Corp., Norwalk, Conn.), Pfu DNA polymerase (Stratagene, La Jolla, Calif.), FRTL-5 cell RNA, and the following forward and reverse primer pairs (5′ → 3′, respectively): NFI-A, GGAATTCATGTATTCTCCGCTCTGTC and GGAATTCTTTTATCCCAGGTACCAGG; NFI-B, ATGGATCCCATGATGTATTCTCCC and ATGGATCCTCAGTTGCTTGTCTCCG; NFI-C, GGAATTCATGTATTCCTCCCCGCTCTG and GGAATTCGTCCTAATCCCACAAAGGG; NFI-X, GGAATTCGATGTACTCCCCGTACTGC and GGAATTCTCAGAAAGTTGCTGTCCCG; TTF-1, ACCTTACCAGGACACCATGC and TACTTCTGCTGCTTGAAGCG; and β-actin, AGCCATGTACGTAGCCATCC and TGTGGTGGTGAAGCTGTAGC. The forward and reverse primers (5′ → 3′, respectively) used to generate a specific probe for each NFI subtype were as follows: NFI-A, GGAATTCACACAGCATCACCGAC and GGAATTCCAACACTGACGAATCGG; NFI-B, GGAATTCACTTTTCCCCAGCACCAC and GGAATTCCAGTGGATGTAGTGATGG; NFI-C, GGGAATTCACACAACACCACAGGC and GGAATTCTGTCATTGCCATTGAGC; and NFI-X, GGAATTCATCAAGTGACCCTGGGAC and GGAATTCTGCTGTGGGATGTTCAG. In each case, these sequences crossed an intron-exon boundary. Each primer used for amplification of NFI cDNA contained a restriction enzyme site in its 5′ end (which is underlined), EcoRI for NFI-A, -C, and -X and BamHI for NFI-B, which was used to facilitate subcloning. All inserts were sequenced using a DNA-sequencing kit (PE Applied Biosystems, Warrington, Great Britain) and a sequencing apparatus (PE Applied Biosystems).
Plasmids.
Rat TTF-1 promoter-luciferase constructs were prepared by PCR amplification of 5′ untranslated sequences of the TTF-1 gene. Amplified genomic fragments using a rat genomic clone as a template (42) were ligated into pGL3-Basic vector (Promega, Madison, Wis.) and sequenced.
To make plasmids with one or three copies of the B element on the TTF-1 promoter, the 22-bp oligonucleotide used in DNA mobility shift assays (DMSA) was ligated to complementary 3-bp sequences, TGC and GCA (5′ → 3′ to each 5′ end), by using T4 DNA ligase and then blunt ended with Klenow fragment. The mixture was cloned in the SmaI site of pBluescript SKII (Stratagene). Inserts containing different multimers of the original oligonucleotide were purified on agarose gels, subcloned into the pGL3-Promoter vector containing a simian virus 40 (SV40) promoter and the luciferase reporter gene, and sequenced to ensure fidelity.
The mammalian NFI expression vectors pcDNA3-NFI-A, -B, -C, and -X and prokaryotic NFI expression vectors pET41-NFI-A, -B, -C, and -X were constructed by ligating the rat NFI-A, -B, -C, and -X coding sequences with the pcDNA3 and pET41 vectors in their EcoRI site for NFI-A, -B, and -X and their BamHI site for NFI-B, respectively. All expression vectors were subjected to in vitro translation using TNT quick-coupled transcription-translation systems (Promega) and the manufacturer's protocol.
Transient-expression analysis.
A DEAE procedure was used to transfect promoter-luciferase gene constructs and expression plasmids into FRTL-5 cells, and an electroporation procedure was used to transfect FRT and BRL-3A cells. Briefly, FRTL-5 cells were grown in six-well plastic plates to about 70% confluency, washed with 2 ml of serum-free culture medium (6H0 medium), and exposed to 800 μl of a premade plasmid-DEAE mixture. The plasmid-DEAE mixture was prepared by incubating 3 μg of plasmid DNA, unless otherwise noted in individual experiments, with 40 μl of 20× DEAE (5 Prime→3 Prime, Inc., Boulder, Colo.) and 760 μl of 6H0 medium for 15 min at room temperature. FRTL-5 cells were incubated with this mixture for 2 h at 37°C in a CO2 incubator, and then 2 ml of 6H5 medium was added.
FRT and BRL-3A cells were subjected to electroporation once 80% confluency was achieved. Cells were harvested, washed, suspended at 107 cells/ml in 800 μl of Dulbecco's modified phosphate-buffered saline (PBS), and pulsed (270 V, 500-μF capacitance) using a Gene Pulser apparatus (Bio-Rad Laboratories, Richmond, Calif.). Reporter activity was measured 48 h later using a luciferase assay system (Promega) as described by the manufacturer.
Nuclear extracts.
A previously described method (24) was modified to prepare extracts from small numbers of cells. Cells were washed, scraped into 1 ml of PBS at pH 7.4, pelleted in a microcentrifuge, and resuspended in 5 volumes of buffer A (20 mM HEPES-KOH [pH 7.9], 10 mM KCl, 2 mM MgCl2, 0.1 mM EDTA) containing 0.3 M sucrose and 2% Tween 40. To release nuclei, cells were repetitively pipetted (100 to 200 times) using a micropipette with a yellow tip (200-μl capacity). Samples were overlaid on 1 ml of 1.5 M sucrose in buffer A and microcentrifuged for 10 min at 4°C. Pelleted nuclei were washed with 1 ml of buffer A, centrifuged for 30 s, and resuspended in 50 μl of buffer B (20 mM HEPES-KOH [pH 7.9], 420 mM NaCl, 2 mM MgCl2, 0.2 mM EDTA, 25% glycerol). Samples were incubated on ice for 30 min with occasional vortexing and microcentrifuged for 20 min at 4°C. Buffers A and B also contained 0.5 mM dithiothreitol (DTT), 0.5 mM phenylmethylsulfonyl fluoride, 2 ng of pepstatin A per ml, and 2 ng of leupeptin per ml. The supernatant was dialyzed for use in DMSA or DNase I protection assays and frozen in small aliquots at −70°C. Nuclear extracts for redox experiments were prepared without 0.5 mM DTT.
DNase I protection analysis.
Genomic fragments were synthesized by PCR and subcloned into pBluescript SKII. The plasmid was digested with either BglII or HindIII, end labeled with [γ-32P]ATP and T4 polynucleotide kinase, and then recut with either HindIII or BglII. The probe was purified on an 8% native polyacrylamide gel. In DNase I protection analysis, 30 μg of nuclear extract was incubated for 15 min on ice in 20 μl of binding buffer [20 mM HEPES-KOH (pH 7.9), 50 mM KCl, 2 mM MgCl2, 0.1 mM EDTA, 1 μg of poly(dI-dC)-poly(dI-dC)] and incubated for 20 min in the presence of 50,000 cpm of probe. The mixture was treated with 1 U of DNase I (Promega) for 5 min on ice before the addition of 80 μl of stopping solution (20 mM Tris-HCl [pH 8.0], 250 mM NaCl, 20 mM EDTA, 0.5% sodium dodecyl sulfate [SDS], 10 μg of proteinase K, 4 μg of sonicated calf thymus DNA). After incubation at 37°C for 15 min, samples were phenol-chloroform extracted, ethanol precipitated, and separated on an 8% sequencing gel.
DMSA.
Oligonucleotides were labeled with [γ-32P]ATP and T4 polynucleotide kinase. DMSA was performed using 3 μg of nuclear extracts or 20 ng of recombinant NFI. In some applications, a 50-fold molar excess of unlabeled oligonucleotide was added to the mixtures during a 15-min preincubation. In others, the 15-min preincubation included a polyclonal rabbit antiserum to NFI-A or its preimmune counterpart. A 32P-labeled oligonucleotide probe (50,000 cpm) was added, and the incubation was continued for 20 min at room temperature. Mixtures were analyzed on 5% native polyacrylamide gels and autoradiographed.
Recombinant NFI was produced by use of the pET system (Novagen, Madison, Wis.) as described by the manufacturer. NFI was recovered with elution buffer containing imidazole. The eluted fraction was dialyzed against 1,000 ml of buffer (20 mM HEPES-KOH [pH 7.9], 100 mM KCl, 2 mM MgCl2, 0.1 mM EDTA, 20% glycerol, 1 mM DTT, 0.1 mM phenylmethylsulfonyl fluoride, 1 ng of pepstatin A per ml) and then concentrated in a Microcon-30 concentrator (Amicon, Bedford, Mass.).
Recombinant NFI-A was used to immunize rabbits after being linked to keyhole limpet hemocyanin. The rabbit antibody produced, but not its preimmune counterpart, reacted with all recombinant NFI protein subtypes on Western blots.
Western analysis.
Nuclear extracts (15 μg) were boiled in SDS sample loading buffer (2% SDS, 10% glycerol, 100 mM dithiothreitol, 50 mM Tris-HCl [pH 6.8]) for 5 min. Samples were loaded on a 10% denaturing SDS-Tris-glycine gel (Novex, San Diego, Calif.). After electrophoresis, the proteins were transferred to a nitrocellulose membrane (Novex). The filter was blocked in PBS-T buffer (PBS with 0.6% Tween 20)–10% nonfat milk–1% BSA and then incubated with a primary antibody: an anti-NFI antibody raised against the common N terminus of the NFI isoforms (Santa Cruz Biotechnology, Inc., Santa Cruz, Calif.) diluted in PBS-T containing 1% nonfat milk. The filter was washed with PBS-T, probed with peroxidase-conjugated anti-rabbit immunoglobulin G (Santa Cruz), washed with PBS-T, and then developed using an enhanced chemiluminescence reagent (Amersham Pharmacia Biotech).
Oxidoreductive reactions.
Chemical oxidation of the thiols in NFI was performed using diamide (Sigma), an inorganic catalyst of oxidation of thiols (-SH2) to generate disulfides (-S-S-) (31). Oxidation was carried out on ice for 5 min in DMSA binding buffer without dithiothreitol, after which labeled probe was added. Chemical reduction of the disulfide to thiols was carried out using dithiothreitol at room temperature for 5 min.
Statistical significance.
All experiments were repeated at least three times with different batches of cells. Significance between experimental values was determined by two-way analysis of variance; P < 0.05 was considered significant.
RESULTS
Identification of the elements on the TTF-1 promoter that are required for TG suppression of TTF-1 gene expression.
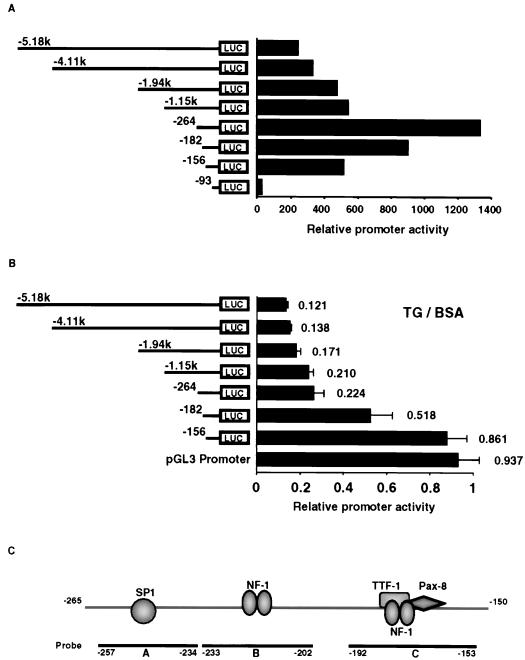
Transient transfection and reporter gene analysis were used to identify the area in the TTF-1 5′-flanking region responsible for TG-mediated repression of the rat TTF-1 gene. To do this, we measured the activity of FRTL-5 cells transfected with various TTF-1 promoter-luciferase chimeras harboring 5′ deletions of a construct containing 5.1 kb of 5′-flanking region and then exposed for 48 h to medium containing 10 mg of bovine TG or BSA per ml, as well as TSH (Fig. 1). The TG concentration used was previously determined to maximize suppression of TTF-1 RNA and protein levels and was within the physiologic range of TG present in the follicular lumen (51–56).
FIG. 1.
An element between bp −264 and −156 of the TTF-1 promoter is required for TG-mediated suppression. The ability of exogenous follicular TG to decrease TTF-1 promoter activity in functioning FRTL-5 thyroid cells was measured by transient-expression analysis. FRTL-5 cells cultured in 6H medium were transfected with 3 μg of TTF-1–luciferase chimeras containing different 5′-flanking region lengths as noted, and then exposed to medium with or without 10 mg of bovine TG per ml which was purified as described in Materials and Methods or to medium with 10 mg of crystalline BSA per ml. Promoter activity was measured 48 h later. (A) The activity of each promoter-chimera is presented relative to the pGL3 Basic plasmid with no TTF-1 promoter insert. (B) The ratio of the promoter activity treated with TG to that treated with an equivalent concentration of BSA is shown; there was no difference in promoter activity in the presence or absence of BSA. Data are the mean and standard deviation (SD) of four different experiments. (C) Schematic representation of the region between bp −264 and −156 of the TTF-1 promoter. The locations of putative NFI and TTF-1 or Pax-8 cis elements are noted. Additionally, oligonucleotides spanning the region, A, B, and C, defined by DNase I protection (Fig. 2) and used in DMSA experiments (Fig. 3), are noted.
The activity of each TTF-1 promoter-luciferase chimera is presented relative to pGL3 Basic with no insert, in order to better evaluate the effect of TG (Fig. 1A). The activity of the longest construct, pTTF-1 5′-5.18k, was decreased 10-fold by TG relative to BSA or no treatment. The activity of pTTF-1 5′-264 was still repressed almost fivefold by TG treatment, even though its basal activity was sevenfold higher than that of the longest construct, pTTF-1 5′-5.18k (Fig. 1B). This 5- to 10-fold repression of TTF-1 promoter activity is consistent with the decrease in TTF-1 RNA levels observed by Northern analysis (51–56). Deletions from 5′ −264 to 5′ −156 resulted in an almost complete loss of the repression by TG (Fig. 1B), indicating that one or more elements between bp −264 and −156 of the rat TTF-1 promoter are important for TG repression. The same results were obtained if cells were exposed to TG in the absence of TSH in the medium.
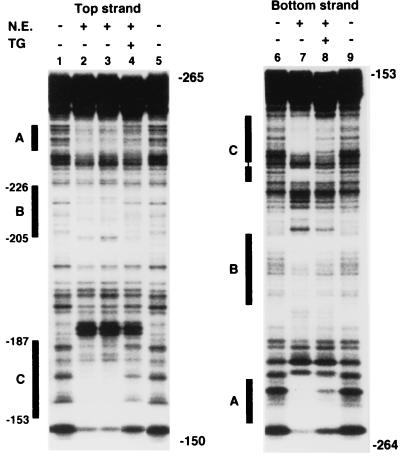
To further define the cis-acting element important for TG-mediated transcriptional repression, we performed a DNase I protection assay with a 32P-end-labeled DNA fragment from bp −264 to −153 of the TTF-1 promoter as a probe. Nuclear extracts were from FRTL-5 cells treated with TSH and with or without bovine TG. The nuclear extracts from TSH-treated cells had two distinct protected areas best seen on the bottom strand (A and C) (Fig. 2, lane 7) and one less dramatically protected area (B) (lanes 2, 3, and 7). The extract in lane 2 was from cells with no TSH in the medium, and the extract in lane 3 was from TSH-treated cells; therefore TSH did not significantly alter the protected areas. Protection is reasonably explained by the binding of one or more transcription factors; TG treatment significantly decreased the protection of the B and C regions and the binding of these factors (lanes 4 and 8).
FIG. 2.
TG treatment decreases the protection of two areas identified by DNase I protection assay in the proximal TTF-1 promoter. The coding (top) and noncoding (bottom) strands of the DNA fragment from bp −264 to −153 of the TTF-1 promoter were end labeled with [γ-32P]ATP. Footprinting analysis was performed as described in Materials and Methods in the absence (lanes 1, 5, 6, and 9) or presence (lanes 4 and 8) of 30 μg of nuclear extract (N.E.) from FRTL-5 cells treated with 10 mg of TG per ml for 48 h (lanes 4 and 8) or left untreated (lanes 2, 3, and 7). Three protected areas were identified and defined as A, B, and C sites (denoted by black bars). TG treatment results in decreased protection of the B and C sites (compare lanes 2 and 3 with lane 4 and compare lane 7 with lane 8). The extract in lane 2 was from cells with no TSH in the medium; that in lane 3 was from TSH-treated cells. TSH did not significantly alter the protected areas.
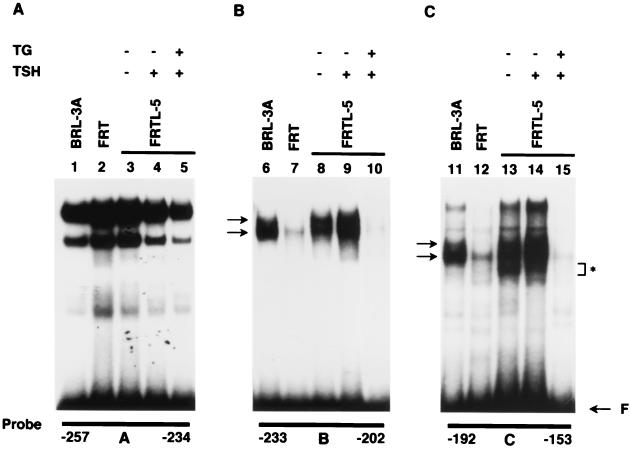
To confirm the TG-induced changes in transcription factor binding to these elements, we performed DMSA using oligonucleotides corresponding to the A, B, and C footprinted areas, bp −257 to −234, −233 to −202, and −192 to −153, respectively (Fig. 1B and 2). DMSA showed that each probe formed several complexes (Fig. 3) with extracts from FRTL-5 cells (Fig. 3, lanes 3 to 5, 8 to 10, and 13 to 15) and that TG induced significant decreases in the complexes formed by oligonucleotides B and C (compare lane 9 with lane 10 and lane 14 with lane 15). These results confirmed that a decrease in binding of one or more transcription factors to elements within the B and C oligonucleotides was associated with TG repression of TTF-1 gene expression. This was consistent with a TG-induced loss of protection in these regions in DNase I protection assays (Fig. 2).
FIG. 3.
Decreased binding of one or more transcription factors to elements within the B and C sites is closely related to TG repression of TTF-1 gene expression. The 32P-labeled oligonucleotides corresponding to the three protected areas (A, B, and C) detected in footprinting assays were incubated with 3 μg of nuclear extracts from rat liver BRL-3A cells (lanes 1, 6, and 11), nonfunctioning FRT thyrocytes (lanes 2, 7, and 12), and functioning FRTL-5 thyrocytes (lanes 3 to 5, 8 to 10, and 13 to 15). The complexes formed with B and C sites and nuclear extracts from FRTL-5 cells were markedly decreased by TG treatment (compare lane 9 with lane 10 and lane 14 with lane 15). Note that these complexes were not abundant in FRT thyrocytes (lanes 7 and 12). Extracts in lanes 4, 9, and 14 were from FRTL-5 cells exposed to TSH, and those in lanes 3, 8, and 13 were from cells treated with 10−10 M TSH. The medium of cells treated with TG also contained 10−10 M TSH.
The proteins forming these complexes appeared to be ubiquitous, since complexes were present in BRL cells (Fig. 3, lanes 6 and 11). Interestingly, nonfunctioning FRT thyrocytes (lanes 7 and 12) had low levels of these complexes compared to either FRTL-5 or BRL cells, suggesting that the levels of the factors binding to the B and C sites were low. Additionally, the presence or absence of TSH in the medium did not cause a major change in the nature or multiplicity of the complexes (Fig. 3, compare lane 8 with lane 9 and lane 13 with lane 14), consistent with the DNase I protection data (Fig. 2).
We concluded that TG decreased the binding of one or more trans factors to two cis elements within bp −264 to −153 of the 5′-flanking region. Binding of these factors was associated with constitutive TTF-1 expression in the presence or absence of TSH. Decreased binding of these factors, induced by TG, was associated with TG-induced repression of TTF-1 gene expression.
NFI proteins bind to specific cis elements identified in the B and C oligonucleotides.
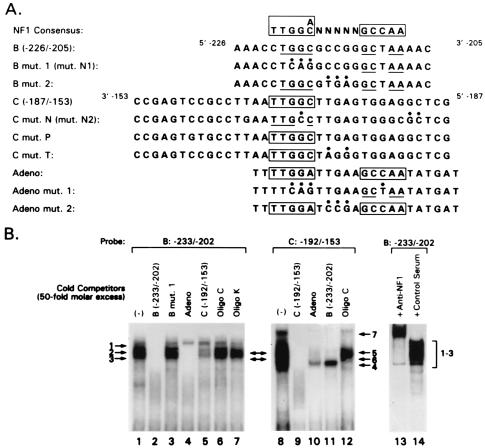
A computer-based search revealed that both the B and C sites contain a putative NFI binding site (Fig. 1B). This was evidenced when the B and C sites were compared to a consensus NFI element (19, 41) (Fig. 4A). Site B, bp −226 to −205, has an incomplete palindromic sequence of NFI consensus site (underlined), and site C, bp −187 to −153, has a complete half-site in a reverse orientation (Fig. 4A, boxed). It has been reported that NFI can bind to a single TGGC sequence (18, 44). Using the B and C site oligonucleotides as probes, we performed competition experiments using nuclear extracts from FRTL-5 cells and nonlabeled oligonucleotides with intact or mutated NFI elements to see if the complexes were with functional NFI elements (Fig. 4A).
FIG. 4.
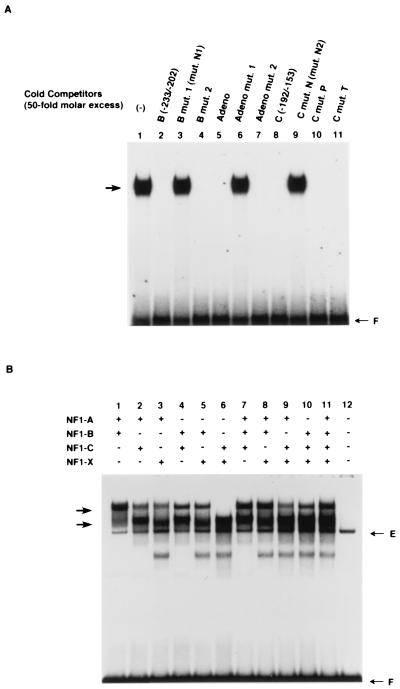
Schematic representation of the NFI consensus sequence, the B and C oligonucleotides identified in DNase I protection assays or DMSA, and mutant or related oligonucleotides used in this study, and identification of NFI binding sites on the B and C oligonucleotides of the TTF-1 proximal promoter. (A) The NFI consensus sequences are those reported previously (18, 19, 41, 44). The TTF-1 B and C region oligonucleotides and their various mutants, which we used in subsequent experiments, are compared to the NFI consensus sites. An oligonucleotide containing NFI binding sequence found on adenovirus DNA, defined as Adeno, is presented, along with its mutations; these were used as specific competitors of NFI binding. (B) DMSA reveals that oligonucleotides with the sequence of sites B and C of the proximal TTF-1 promoter contain NFI binding sites. DMSA was performed using oligonucleotides corresponding to the TTF-1 promoter B and C oligonucleotides as probes and nuclear extracts from FRTL-5 cells. Specific or nonspecific unlabeled competitors listed in panel A were used to confirm the specificity of binding and identify NFI binding sites. Two complexes formed with the B oligonucleotide (arrows 2 and 3) and with the C oligonucleotide (arrows 5 and 6) were identified as NFI proteins at this point, as discussed in the text, based on specific competition or by inhibition and supershift of the complexes with anti-NFI-A. The anti-NFI-A was not specific for this NFI subtype but reacted with all four NFI subtypes in the FRTL-5 cells, as evidenced by Western blotting of the recombinant NFI proteins isolated from the cells.
When the B oligonucleotide was the radiolabeled probe, at least three complexes were formed (Fig. 4B, lane 1, arrows 1 to 3). All complexes were competed by the same unlabeled oligonucleotide (lane 2) but not by an oligonucleotide which has a 3-bp substitution mutation in the NFI binding site, mut.N1 (Fig. 4A and B, lane 3). These results indicate that the three complexes are specific and that a mutation in the NFI cis element can abolish the binding of all complexes. The two higher-mobility complexes (Fig. 4B, arrows 2 and 3) were also competed completely by oligonucleotide Adeno, which has an NFI consensus sequence (Fig. 4A and B, lane 4) but no other sequence common to oligonucleotide B. They were partially competed by the C oligonucleotide with the separate putative NFI half-element (Fig. 4B, lane 5) but not by an oligonucleotide from the TG promoter, termed oligo C (8, 16, 49, 50), which has a TTF-1 and Pax-8 binding site but no NFI binding site (lane 6), or an oligonucleotide containing the TTF-2 site of the TG promoter, termed oligo K (lane 7), also with no NFI binding site.
When the C oligonucleotide was used as probe, at least four complexes were formed (Fig. 4B, lane 8), which are identified as complexes 4 to 7. Complexes 5 and 6 with the TTF-1 C oligonucleotide had the same mobility as complexes 2 and 3 with the TTF-1 B oligonucleotide (Fig. 4B). All complexes were specific, as evidenced by self-competition (Fig. 4B, compare lanes 8 and 9). Two of these complexes (arrows 5 and 6) were competed by the Adeno oligonucleotide (lane 10) and by an oligonucleotide corresponding to the B site oligonucleotide in the minimal TTF-1 promoter (lane 11). There was no competition of complexes 5 and 6 by oligo C of the TG promoter, which contains TTF-1 and Pax-8 sites but no NFI sites (lane 12). Finally, a polyclonal rabbit antibody to recombinant NFI-A, produced using NFI-A cloned from FRTL-5 cells (see below), inhibited complex formation and caused a supershift of the complexes, unlike its control preimmune counterpart (compare lanes 13 and 14). Although made against NFI-A, the antibody reacted with all NFI proteins in FRTL-5 cell extracts, as evidenced by Western blotting (data not shown). From these data, we concluded that oligonucleotides B and C from the minimal TTF-1 promoter each contain a cis element whose sequence is related to consensus NFI elements and is a binding site for NFI or NFI-related proteins.
Interestingly, the highest-mobility complex (Fig. 4B, arrow 4) formed with the TTF-1 C site oligonucleotide was competed by oligo C of the rat TG promoter (lane 12), suggesting that TTF-1 or Pax-8 could bind to this oligonucleotide. This is consistent with the existence of a putative TTF-1 or Pax-8 site in this region, as noted in Fig. 1B; however, the TTF-1 or Pax-8 site was clearly functionally distinct from the NFI site. A separate report will detail the role of these putative TTF-1 and Pax-8 binding sites and any functional relationship to the NFI sites or the converse.
We next asked whether NFI proteins, rather than NFI-like proteins, were bound to the NFI elements within oligonucleotides B and C from the minimal TTF-1 promoter, since NFI-like proteins can bind to almost the same sequences as the NFI consensus sequence and have been described (32, 34, 45). We cloned all of the NFI family proteins (NFI-A, -B, -C, and -X), which are the most abundant NFI isoforms expressed in FRTL-5 cells. We used RT-PCR and mRNA purified from FRTL-5 cells (Fig. 5). Primers were designed so that all amplified fragments contained both translation initiation and termination codons of the longest cDNAs of the different NFI proteins known so far in the sequence data banks. We sequenced all of the clones (Fig. 5A), subcloned them into pcDNA3 mammalian expression vectors, and confirmed their ability to generate an appropriately sized protein by using in vitro transcription-translation (Fig. 5B). Cloning of the NFI cDNAs enabled us to use both prokaryotic and eukaryotic NFI proteins, expressed in Escherichia coli or in mammalian cells, to measure binding.
FIG. 5.
NFI proteins in FRTL-5 cells; sequence and expression by in vitro translation. NFI subtypes were cloned by PCR using FRTL-5 cell RNA (see Materials and Methods). Each was sequenced (A) and subjected to in vitro translation (B) as described in Materials and Methods. In vitro translation was performed using 1 μg of expression vector carrying full cDNA sequences of each NFI subtype and [35S]methionine as described in Materials and Methods. All proteins had the molecular masses expected from the sizes of the open reading frames.
Using bacterium- or mammal-expressed NFI proteins, we showed that the NFI elements in B and C site oligonucleotides formed a distinct complex with recombinant NFI proteins. This is illustrated for bacterially expressed NFI-A protein and the radiolabeled B site oligonucleotide in Fig. 6A, lane 1. The specificity of this complex was confirmed using multiple oligonucleotides as cold competitors (Fig. 6A). Thus, NFI binding to radiolabeled B oligonucleotide was completely eliminated by unlabeled B oligonucleotide (Fig. 6A, compare lanes 1 and 2) and by B mut.2, which does not have an NFI site mutation (Fig. 4A and 6A, lane 4), but not by B mut.1, which has a mutation in the NFI site (Fig. 4A and 6A, lane 3). NFI binding to radiolabeled oligonucleotide B was also inhibited by the Adeno oligonucleotide, which has an NFI site (Fig. 4A and 6A, lane 5), and by the Adeno mut.2 oligonucleotide, which does not have an NFI site mutation (Fig. 4A and 6A, lane 7), but not by the Adeno mut.1 oligonucleotide with mutations in the NFI sites (Fig. 4A and 6A, lane 6). Similar competition results were obtained with the C region oligonucleotide of the TTF-1 promoter without mutations in the NFI site (Fig. 4A and 6A, lanes 8, 10, and 11) compared to the C region oligonucleotide with an NFI mutation, C mut.N2 (Fig. 4A and 6A, lane 9). Identical results were found with radiolabeled oligonucleotide B of the TTF-1 promoter using recombinant bacterial or mammalian NFI-B, -C, and -X proteins (data not shown). Results were also the same using radiolabeled oligonucleotide C of the TTF-1 minimal promoter as probe (data not shown).
FIG. 6.
The NFI elements identified in the B and C regions of the TTF-1 minimal promoter bind recombinant NFI proteins, and all NFI subtypes can bind to the B or C sites and generate a specific set of complexes involving combinations of homo- and heterodimers. (A) To confirm that NFI proteins, not NFI-like proteins, bind to both sites B and C in the minimal TTF-1 promoter, DMSA was performed using bacterially expressed NFI proteins and radiolabeled oligonucleotides with the sequence of each site. The representative result shown in this panel uses recombinant NFI-A protein and radiolabeled B site oligonucleotide as a probe (lane 1). Binding is inhibited by an unlabeled self-oligonucleotide or by unlabeled C oligonucleotide with intact NFI sites (lanes 2, 3, 8, 10, and 11) but not if these unlabeled competitors had an NFI site mutation (lane 3 and 9). An unrelated oligonucleotide containing no sequence similarity other than the NFI site, Adeno, also was inhibitory (lane 5) except if there was an NFI mutation (lane 6). See Fig. 4A for sequences of these oligonucleotides and their mutations. Identical binding results were found using recombinant NFI-B, -C, and -X proteins and with all recombinant NFI proteins and the radiolabeled C site oligonucleotide as probe. Incubations and DMSA were performed as in Fig. 3. (B) The labeled B site oligonucleotide was incubated with nuclear extracts from COS-7 cells transfected with various combinations of expression plasmids for each NFI subtype and subjected to DMSA. The E arrow shows the complex generated with endogenous NFI proteins from COS-7 cells. Note that even by adding nuclear extracts transfected with all four NFI subtypes in the binding solution, the major complexes appeared to be two bands, just as seen in DMSA using nuclear extracts from FRTL-5 cells. Incubations and DMSA were performed as in Fig. 3. The results in panels A and B are representative of three separate experiments with different batches of cells and extracts.
Using NFI proteins exogenously expressed in mammalian COS-7 cells, we also could show that all NFI subtypes bound to the B or C site oligonucleotides and generated a specific set of complexes as a result of combinations of their homo- and heterodimers. This is illustrated in Fig. 6B with the radiolabeled B site oligonucleotide, but the results with the C site oligonucleotide were identical (data not shown). It was notable that a particular preference for the binding by each NFI subtype combination was not detected. Moreover, even by adding nuclear extracts from cells transfected with all four NFI expression plasmids in the binding solution, the major complexes appeared to be two bands with the approximate mobilities of the complexes seen in DMSA using nuclear extracts from FRTL-5 cells (compare Fig. 6B, lane 11, arrows, with Fig. 3, arrows). As expected from the data in Fig. 6A, unlabeled Adeno oligonucleotide completely inhibited the formation of these complexes but unlabeled Adeno with an NFI mutation did not (data not shown). The complex, designated E, is formed by nuclear extracts from COS-7 cells which were transfected with vector alone.
We conclude from the sum of these data that the B and C sites in the minimal TTF-1 promoter, whose protection is reversed by TG treatment of FRTL-5 cells, are NFI cis elements that interact with NFI-A, -B, -C, and -X proteins which are present in FRTL-5 cells. TG treatment of cells decreases the NFI protein interaction with these elements.
Functional roles of the two NFI binding elements on the minimal TTF-1 promoter.
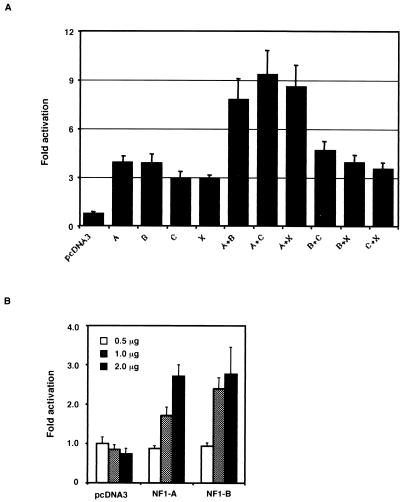
To characterize the roles of the NFI binding elements, we constructed heterologous promoter-luciferase chimeric plasmids by inserting one or three copies of the B or C site oligonucleotides into a plasmid with an SV40 promoter (Fig. 7A, top). FRTL-5, FRT, and BRL-3A cells were transfected with each chimera, and the luciferase activity was measured 48 h after transfection. In FRTL-5 cells, the constructs with the B site oligonucleotide caused higher activity as a function of copy number (Fig. 7A). The increase in promoter activity caused by inserting this element 5′ to the SV40 promoter was weaker in BRL-3A cells and minimal in FRT cells (Fig. 7A). The increase in promoter activity in FRTL-5 cells was abolished if the B or C site oligonucleotides had an NFI site mutation the same as that eliminating their ability to bind NFI proteins (see below). These results indicate that the B site works as an enhancer in FRTL-5 cells and in BRL-3A cells but has minimal activity in FRT cells. Taken together with the results of DMSA showing a very low level of specific NFI complexes in FRT cells (Fig. 3, lanes 7 and 12), this enhancer activity seems to be related to the abundance of the NFI complexes formed with this fragment. The same results were obtained using constructs containing one or three copies of the C oligonucleotide (data not shown).
FIG. 7.
An NFI element works as an enhancer in FRTL-5 cells; both NFI elements are important for maximal expression of the TTF-1 gene as well as TG suppression. (A) We constructed heterologous promoter-luciferase chimeric plasmids by inserting one or three copies of the B elements 5′ to the SV40 promoter (top). A 1-μg portion of each chimera or the vector control was transfected into FRTL-5, FRT, and BRL-3A cells, and the luciferase activity was measured 48 h later. Data are the mean and SD of three different experiments. The B element works as an enhancer in FRTL-5 cells and in BRL-3A cells but has minimal activity in FRT cells. (B) The 224-bp TTF-1 promoter-luciferase chimeric plasmid containing two NFI binding sites was used to construct mutant plasmids containing 3-bp substitution mutations of either NFI element (mut.N1 and mut.N2 in Fig. 4A) or both (mut.N1+N2) (top). A 3-μg portion of each chimera was transfected into FRTL-5 cells, and the promoter activity was measured 48 h later in cells exposed or not exposed to 10 mg of purified 19S bovine TG per ml. Data are the mean and SD of three different experiments. Data are expressed as relative light units (R.L.U.), which are arbitrarily defined in each experiment.
To confirm the enhancer activity of this element in the context of the native TTF-1 promoter, we introduced a 3-bp substitution mutation of either or both of the NFI binding sites into a TTF-1 promoter-luciferase chimeric plasmid; these are designated mut.N1, mut.N2, and mut.N1+N2, respectively (Fig. 7B, top). We compared the promoter activity of these mutants with that of the wild-type counterpart, pTTF-1 5′−224 (Fig. 7B). Promoter activity was decreased to one-fourth of control levels by mut.N1 and to one-fifth by mut.N2 or mut.N1+N2. Moreover, the ability of TG to suppress TTF-1 activity was lost in all of the NFI mutations, mut.N1, mut.N2, and mut.N1+N2. The N1 and N2 mutations eliminate the ability of NFI proteins to bind to the NFI element in oligonucleotide B or C, as evidenced in Fig. 6A, lanes 3 and 9. mut.N2 does not abolish the binding of TTF-1 or Pax-8 to oligonucleotide C (data not shown).
These results indicate that both NFI binding elements are required for maximal expression of the TTF-1 gene and TG suppression. Since a mutation in the NFI element in oligonucleotide C of the TTF-1 minimal promoter, mut.N2, can abolish the TTF-1 promoter activity, NFI binding to this NFI element may be essential for transcription initiation; i.e., it may be involved in the assembly of the basal transcription machinery. This possibility is raised because the major and minor transcription start sites of the TTF-1 proximal promoter gene exist between bp −198 and −125 with respect to the translation initiation codon (23, 36) and because this NFI element is within the two start sites, i.e., at bp −173 to −169.
Summing all data presented thus far, the results support the conclusion that TG-mediated negative regulation is caused mainly by decreased binding of NFI proteins that work as activators when binding to NFI elements within the B and C sites on the TTF-1 promoter. NFI binding sites behave as enhancer elements and control constitutive TTF-1 gene expression even in the presence of TSH-cyclic AMP (cAMP), which can decrease TTF-1 RNA levels (24, 30, 49).
All major NFI isoforms enhance TTF-1 promoter activity.
Using FRT cells, we cotransfected the bp −224 TTF-1 promoter-luciferase chimera (Fig. 7B) with expression plasmids containing the full-length cDNAs encoding the major NFI proteins (NFI-A, -B, -C, and -X) that we had cloned from FRTL-5 cells (Fig. 5). As noted above, FRT cells had only low levels of NFI-A, -B, -C, and -X, as evidenced by the faint complexes seen with the B and C site oligonucleotides in DMSA (Fig. 3, lanes 7 and 12). All the major NFI isoforms increased TTF-1 promoter activity individually or in combination (Fig. 8A). Significantly, the combinations containing NFI-A had a greater transactivation activity, suggesting that heterodimers containing NFI-A play an important role in maximal TTF-1 gene expression (Fig. 8A). Cotransfection of the NFI vectors with the TTF-1 promoter-reporter plasmid increased promoter activity in a concentration-dependent manner, as illustrated for NFI-A and NFI-B (Fig. 8B). In each case, the same experiment performed side by side with either mut.N1, mut.N2, or mut.N1+N2 had no significant activity, as separately illustrated in Fig. 7B (data not shown). Thus, in the experiment in Fig. 7A, activity with each mutant was not significantly different from that of the pcDNA3 control and in the experiment in Fig. 7B, 2.0 μg of NFI-A or NFI-B had no effect on activity, compared to pcDNA3, using any of the −224 NFI site mutant promoter-luciferase chimeras.
FIG. 8.
Cotransfection of all NFI expression vectors with the 224-bp TTF-1 promoter-luciferase chimeric plasmid can increase its promoter activity in a concentration-dependent manner. (A) The 224-bp TTF-1 promoter-luciferase chimeric plasmid was cotransfected with 0.5 μg of the expression vectors for NFI-A, -B, -C, -X, or combinations thereof, into FRT cells which form only faint NFI complexes in DMSA (Fig. 3). The promoter activity was measured 48 h later and expressed as fold activation over the control, whose activity is determined by cotransfection with 0.5 μg of vacant vector. Data are the mean and SD of three different experiments. Note that the combinations containing NFI-A had a greater activity of transactivation. (B) FRT cells were cotransfected with the 224-bp TTF-1 promoter-luciferase chimeric plasmid and different amounts of the expression vectors for NFI-A or NFI-B. Activity was measured as in panel A. Activity in panel A was not significantly different from that of the pcDNA3 control when using the bp −224 TTF-1 promoter-luciferase chimera with 3-bp substitution mutations of either NFI element (mut.N1 and mut.N2 in Fig. 4A and 7B) or both (mut.N1+N2). Similarly, in panel B, 2.0 μg of NFI-A or NFI-B had no effect on activity, compared to pcDNA3, using any of the bp −224 NFI site mutant promoter-luciferase chimeras, i.e., mut.N1, mut.N2, or mut.N1+N2.
TG modulates NFI expression in FRTL-5 cells.
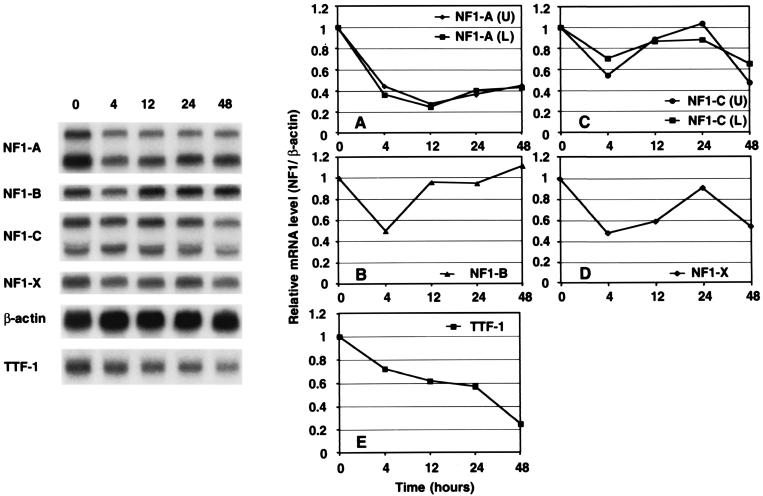
To investigate whether all or some of the subtypes of NFI proteins are regulated by TG, we performed Northern analysis using total RNA from FRTL-5 cells maintained in complete medium containing all six hormones including TSH. We used specific probes recognizing the different NFI subtypes and measured their RNA levels as a function of time after treatment of cells with 10 mg of bovine TG per ml (Fig. 9). As early as 4 h after addition of TG to the culture medium, expression of the different NFI subtypes was repressed 50% or more by comparison to control values, except for the larger RNA of NFI-C, which was repressed almost 30% (Fig. 9A to D). At 4 h after TG treatment, the degree of decrease of the levels of the different NFI RNAs was greater than that of TTF-1 RNA, whose levels were 70% of control values (Fig. 9E).
FIG. 9.
TG can decrease NFI RNA levels in FRTL-5 cells. FRTL-5 cells cultured in complete 6H medium with TSH were washed, and the incubation was continued in 6H medium containing 10 mg of TG per ml. Before (0 h) or after (4, 12, 24, and 48 h) TG treatment, total RNA was prepared and 20 μg was subjected to Northern analysis. Blots were sequentially hybridized with probes for NFI-A, -B, -C, and -X, TTF-1, and β-actin. A representative blot is presented. After quantitative analysis, the ratio of TTF-1 to β-actin (E) or of each NFI subtype to β-actin (A to D) was calculated. The values are expressed relative to the respective control values (0 h). Data are the mean of three independent experiments.
Between 4 and 24 h after TG addition, there were transient increases in NFI-B, -C, and -X RNA levels (Fig. 9B to D), but the levels of RNA were still lower than that of the control. At 24 h, the RNA levels of all NFI subtypes except NFI-B were decreased by comparison to the control (Fig. 9).
Of interest, however, RNA levels of NFI-A were continuously lower than that of the control through 48 h (Fig. 9A). Taken together with the result of cotransfection experiments (Fig. 8), the decreased NFI-A RNA level seems to correlate best with the decrease in TTF-1 RNA. The repression of TTF-1 RNA by TG did not require some newly synthesized factors, since it was not abolished by cycloheximide (data not shown).
We determined whether changes in RNA levels reflected changes in NFI proteins. We performed Western analysis using an NFI-specific antibody which recognizes a common amino acid sequence of all NFI subtypes and nuclear extracts from FRTL-5 cells treated with TG for 48 h or left untreated (Fig. 10). Western analysis showed that TG treatment changed the spectrum of dominant NFI subtypes (isoforms) (Fig. 10A) and, when quantified with an image analyzer, decreased total NFI protein levels to 60% of control values (Fig. 10B). It is important to note that changes in the dominant NFI subtypes as determined by Western analysis, based on their estimated molecular weights, correlated well with the concomitant changes in RNA levels of NFI subtypes (compare Fig. 10A with Fig. 9).
FIG. 10.
TG can decrease NFI protein levels in FRTL-5 cells. (A) Western analysis was performed using an NFI-specific antibody that recognizes common amino acid sequences of all NFI subtypes, an antibody to actin, and 15 μg of nuclear extracts treated with TG for 48 h or left untreated, as described in Materials and Methods and in parallel to the studies in Fig. 9. A representative blot from one experiment is presented. (B) After quantitative analysis, the ratio of total NFI protein to actin was calculated. The values were compared and expressed relative to the values from extracts from cells which were not exposed to TG. Data are the mean and SD from three independent experiments.
These results suggest that the decrease in the level of TTF-1 RNA induced by TG is in part due to the ability of TG to decrease NFI RNA and protein levels of all the major NFI subtypes in FRTL-5 cells, in particular NFI-A, which has the greatest ability to increase TTF-1 promoter activity in combination with other subtypes (Fig. 8).
Decreased binding of NFI proteins by nuclear extracts from cells treated with TG is restored by higher concentrations of DTT in the binding assay mixture.
Although DMSA revealed that TG treatment for 48 h caused a major decrease in the formation of NFI complexes with the NFI elements in oligonucleotides B and C of the minimal TTF-1 promoter (Fig. 3, lanes 10 and 11), NFI proteins were still detectable in FRTL-5 cell nuclear extracts by Western analysis, using NFI-specific antibody (Fig. 10). To investigate additional mechanisms that might be involved in the decreased binding of NFI proteins, we first examined the possibility of regulation by phosphorylation. Treatment of nuclear extracts with potato acid phosphatase or calf intestinal phosphatase and with various phosphatase inhibitors had no effect in DMSA (data not shown).
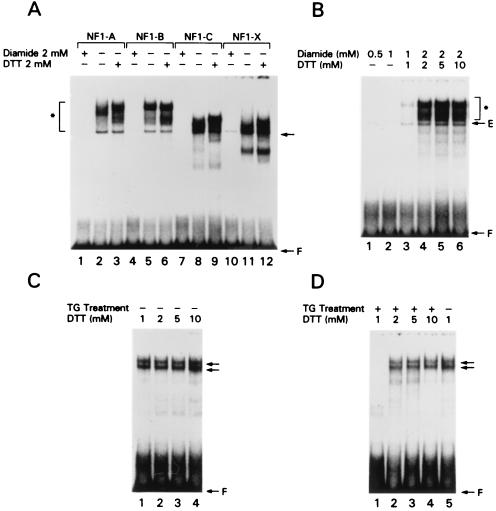
We next examined the possibility of redox regulation of the binding. Using the B site oligonucleotide of the TTF-1 promoter as the radiolabeled probe, we confirmed that it bound all NFI subtypes exogenously expressed in COS-7 cells under our standard conditions (Fig. 11A, lanes 2, 5, 8, and 11). Consistent with previous reports concerning the redox sensitivity of NFI proteins (3, 4, 40), we showed that treatment of nuclear extracts with the oxidizing reagent diamide markedly decreased the binding activity of all NFI subtypes exogenously expressed in COS-7 cells to the B site oligonucleotide (lanes 1, 4, 7, and 10). In contrast, all NFI subtypes exhibited a greater ability to bind the B site oligonucleotide if nuclear extracts contained higher concentrations of the reducing reagent DTT (lanes 3, 6, 9, and 12). Moreover, the DNA binding ability of FRTL-5 cell nuclear extracts preoxidized by diamide was fully restored by subsequent incubation with DTT (Fig. 11B, compare lanes 1 to 3 with lanes 4 to 6). Thus, oxidizing-reagent inactivation of the binding of NFI proteins to the NFI element in the B site oligonucleotide was fully reversible.
FIG. 11.
The binding of all NFI subtypes is regulated by their redox state; TG regulates the redox state. (A) DMSA was performed using the radiolabeled B site oligonucleotide and nuclear extracts from COS-7 cells transfected with the expression plasmids of each NFI subtype. An arrow shows the complex generated with endogenous NFI proteins from COS-7 cells. Nuclear extracts were treated with 2 mM diamide in lanes 1, 4, 7, and 10, with 2 mM DTT in lanes 3, 6, 9, and 12, and with neither diamide nor DTT in lanes 2, 5, 8, and 11. The DNA binding activity of all NFI subtypes was markedly attenuated by oxidizing reagent and considerably increased by reducing reagent. (B) Nuclear extracts from exogenously expressed NFI-A protein from COS-7 cells were incubated with 0.5 mM (lane 1), 1.0 mM (lane 2), and 2.0 mM (lanes 3 to 6) diamide, incubated with increasing amounts of DTT for 48 h (lanes 3 to 6), and subjected to DMSA. An arrow shows the complex generated with endogenous NFI proteins from COS-7 cells. To a large extent, high concentrations of DTT restored the DNA binding of NFI proteins from FRTL-5 cells treated with TG (compare lanes 4 and 5). (C and D) Nuclear extracts from FRTL-5 cells which were not treated with TG were incubated with increasing amounts of DTT (C, lanes 1 to 4) and compared to extracts from cells treated with follicular TG for 48 h and also incubated with increasing amounts of DTT (D, lanes 1 to 4).
To determine if the decreased binding of NFI proteins that is exhibited by nuclear extracts from FRTL-5 cells treated with TG is due in part to a redox change in NFI, we tested the DNA binding activity of extracts from cells which were treated (Fig. 11D) or not treated (Fig. 11C) with TG and which contained different DTT concentrations in the extract. Nuclear extracts from FRTL-5 cells not treated with TG showed almost full binding activity in the usual buffer containing 1 mM DTT (Fig. 11C, lane 1). In contrast, those treated with TG did not show any significant binding using the same concentration of DTT (Fig. 11D, lane 1). In the presence of higher concentrations of DTT (Fig. 11D, lanes 2 to 4), the DNA binding ability of TG-treated nuclear extracts was restored to a level similar to that exhibited by extracts from cells not exposed to TG. In contrast (Fig. 11C), the same concentrations of DTT had no effect on the NFI complexes formed by extracts from cells not exposed to TG. These results suggest that TG decreases the binding of NFI proteins not only by decreasing their amounts but also by causing their oxidation.
Of interest, the complex with the C site oligonucleotide, which appears to result from an interaction with Pax-8 or TTF-1 (Fig. 3, arrow 7), is also abolished by TG treatment (Fig. 3, lane 15). Since TTF-1 and Pax-8 binding can be attenuated by oxidative inactivation (2, 25), TG not only decreases Pax-8 and TTF-1 RNA levels (51–56) but also decreases their redox state, as is the case for NFI.
DISCUSSION
The present report addresses two issues. First, it addresses the mechanism by which follicular TG suppresses expression of TTF-1, a critical thyroid-restricted transcription factor. Second, it presents evidence for a novel, non-thyroid-specific regulatory mechanism which controls constitutive expression of the tissue-restricted trans factor TTF-1. Although the first issue was our initial and primary question at the start of these experiments, the second became an unexpected integral part of understanding the TG results. We therefore discuss our findings on constitutive TTF-1 gene expression first.
Previous studies demonstrated that TTF-1 plays a critical role in thyroid morphogenesis (28) and in the regulation of a number of genes critically involved in thyroid function. Thus, TTF-1 regulates the gene expression of all known thyroid-specific or -restricted proteins: NIS, TPO, TG, and TSHR (7, 8, 15, 16, 30, 36, 38, 49, 52–55). TSH-cAMP and follicular TG can each down regulate TTF-1 gene expression (24, 30, 49, 51–56); however, little is known about what regulates TTF-1 constitutive expression.
TTF-1 contains major transcription start sites on the proximal 5′-flanking region (23, 36), but alternative transcription start sites located far upstream of the proximal promoter have also been reported (39). In the course of our studies to define elements on the TTF-1 5′-flanking region that are involved in the ability of follicular TG to suppress TTF-1 expression (see below), we noted that TG suppression involved the proximal 5′-flanking region, bp −257 to −156. We then showed that this region has two NFI sites within nucleotides −233 to −202 and −192 to −153 and that their deletion eliminates constitutive TTF-1 expression. The NFI elements within these regions serve as enhancers of TTF-1 gene expression and function by binding all four major NFI proteins present in functioning thyrocytes: NFI-A, NFI-B, NFI-C, and NFI-X. Consistent with this, we show that overexpression of each up regulates TTF-1 promoter activity.
The TTF-1-proximal promoter does not have a functioning TATA box. In genes without a TATA box, initiation of RNA polymerase II-directed transcription is mediated by DNA sequence-specific activator proteins that can interact with components of the transcription initiation complex. NFI is a family of constitutive and sequence-specific binding proteins which stimulate transcription in many promoters (5, 17, 22, 47) and interact with TFIIB (27), one of the important components of the transcription machinery. Although we show that both NFI binding sites are important for maximal expression of TTF-1 gene, they are not equivalent. The most proximal site lies between the major and minor transcription start sites of the proximal promoter of the TTF-1 gene, bp −198 and −125 with respect to the translation initiation codon (23, 36), and its mutation (mut.N2) can alone abolish TTF-1 promoter activity and the binding of NFI proteins to the NF-1 element. For these reasons, we suggest the NFI sites may be essential for transcription initiation of the TTF-1 gene and constitutive expression. NFI-A data are consistent with this suggestion.
Thus, it has been reported that a motif with the amino acid sequence SPTSPSY, existing in a core domain of NFI-CTF1, is important for transcriptional activation and is strongly related to the heptapeptide repeat, YSPTSPS, present in the carboxy-terminal domain of RNA polymerase II (26, 27, 57). When we cloned the NFI subtypes expressed in FRTL-5 cells, only NFI-A had a similar sequence motif, SPTSPTY. The data in this report, which show that NFI-A significantly increases NFI transactivation activity when present along with other NFI subtypes, would be consistent with a role in transcription initiation. This is under investigation.
Since NFI proteins are ubiquitous, they may regulate TTF-1 gene expression not only in thyrocytes but also in other cells where TTF-1 is expressed. Two points are notable in this respect. First, nonfunctioning FRT thyrocytes have low levels of NFI able to activate the activity of a 224-bp TTF-1 promoter construct containing two NFI binding sites. The low levels of NFI in these cells, along with low levels of TTF-1 expression, raise the possibility that negative regulation of NFI expression or a failure in NFI expression may be an important factor in determining a thyroid-specific, or perhaps even a tissue-specific, phenotype. Second, although TSH can decrease TTF-1 gene expression (24, 30, 49), the effect of TSH does not seem to be mediated by NFI proteins or elements, given their identical protection and binding by nuclear extracts from cells exposed or not exposed to TSH. The role of the TTF-1 or Pax-8 site and the SP1 site in the region between bp −257 and −153 and their role in TSH-cAMP suppression of TTF-1 are not known and are the subject of work in progress.
As pointed out above, we started this work to examine the molecular mechanism by which TG suppresses thyroid-restricted transcription factor gene expression. We used TTF-1 as a prototype thyroid-restricted transcription factor because it regulated NIS, TSHR, TG, and TPO gene expression. Suppression of thyroid-restricted or -specific transcription factors by follicular TG is thought to be an important means by which thyroid homeostasis is maintained and by which the thyroid is able to secrete thyroid hormones in a regulated manner. Follicular TG is the feedback regulator counteracting the transcriptional actions of TSH (51–55).
In this report we demonstrate that TG decreases the expression of all NFI subtypes in FRTL-5 thyroid cells. Thus, the RNAs of all NFI subtypes are decreased after 4 h of TG treatment as an early response to TG and, with one exception, the RNA levels are lower than control values 48 h after TG treatment. We show that there is an associated decrease in NFI protein levels and correlate these decreases with the decrease in TTF-1 RNA levels. Thus, 48 h after TG treatment, the molecular weights of the dominant NFI subtypes changed concomitant with changes in their RNA levels and the total NFI protein levels decreased to almost 60% of control values. We show that NFI-A has the strongest transactivation activity when present with other NFI subtypes, and we currently think that regulation of NFI-A is at the core of the mechanism by which TG suppresses TTF-1 gene expression levels. We also believe that the change in the amount of the dominant NFI subtypes, rather than a change in the NFI isoforms, is the major action of TG, since we did not detect changes in the sizes of cDNA fragments generated by RT-PCR using mRNAs treated with TG (data not shown).
The TG-induced decreases in NFI RNA and protein levels account in part for decreases in the levels of NFI complexes with NFI cis elements that we show exist on the proximal promoter and that we associate with decreased TTF-1 gene expression. This results in decreased TTF-1 gene expression because NFI elements and their binding of NFI controls constitutive expression and normally enhances TTF-1 gene expression. In short, TG acts by down regulating constitutive TTF-1 expression by its action on NFI proteins.
Although decreases in NFI complex levels were detected in DMSA using nuclear extracts from FRTL-5 cells treated with TG for 48 h, NFI proteins were still detectable in Western blots, suggesting that TG caused a potential posttranslational modification of the NFI subtypes, which contributed to decreased binding. It had been reported that the DNA binding activity of NFI is regulated by redox (3, 4, 40) and phosphorylation (11) reactions. We tested both possibilities and showed that treatment of nuclear extracts with oxidizing reagent can almost abolish the binding ability of all NFI subtypes; this decrease is reversed by DTT. While the mechanism of oxidative inactivation of DNA binding by NFI has been studied extensively in vitro (3, 4, 40), the physiological relevance of the reversible oxidation of NFI proteins is still not clear. Nevertheless, in this report, we demonstrate that excess reducing agent can restore the decreased NFI binding ability of nuclear extracts from FRTL-5 cells treated with TG toward the normal value. This suggests that the change of the redox state of NFI proteins may be another factor in TG action; i.e., TG-decreased NFI binding and TG-decreased TTF-1 gene expression might be partially caused by the ability of TG to increase the oxidized state of NFI proteins. In a separate report we show that this effect is specific for TG and is not mimicked by transforming growth factor β, which can down regulate TTF-1 expression by a different effect on NFI proteins (unpublished data).
It remains to be determined how TG changes the intracellular redox status and whether this phenomenon is relevant to in vivo regulation of thyrocyte function by follicular TG. Given that TG is a substrate for oxidation in the generation of thyroid hormone, TG could be functioning to stimulate an oxidative “burst” that generates reactive oxygen species that could oxidize NFI and other transcription factors. While this mechanism is speculative, it would be a relatively simple mechanism of coupling NFI activity levels to the level of TG. Of interest, we have observed that TG-decreased binding of NFI protein is more apparent when using nuclear extracts from aged FRTL-5 cells, i.e., cells passaged more than 20 times (data not shown). The suggestion that aged FRTL-5 cells are more sensitive to oxidative stress and oxidative changes in NFI proteins may be physiologically relevant in the context of aged cells in general.
In sum, this report describes the possible mechanism to explain how follicular TG suppresses the gene expression of TTF-1, which is one of the important thyroid-restricted transcription factors controlling thyroid-specific expression of critical genes controlling thyroid function. We show that NFI sites and NFI binding control constitutive TTF-1 gene expression even in the presence of TSH-cAMP, which can independently decrease TTF-1 gene expression. We further show that the ability of TG to decrease this binding is associated with suppression of TTF-1 gene expression. We describe two mechanisms for decreased NFI binding, a decrease in the profile of dominant NFI subtypes, particularly a decrease in the NFI-A subtype, and an increase in their oxidative state. We do not know if this mechanism will be a general phenomenon for all thyroid-restricted transcription factors, nor do we know if it will be generalized to all cells that can interact with TG and have TTF-1 as an important regulatory factor. Nevertheless, we anticipate that further pursuit of these findings, i.e., understanding how TG decreases NFI gene expression transcriptionally, will generate a more complete and perhaps a general mechanism for controlling TTF-1 constitutive expression as well as regulation.
REFERENCES
- 1.Ambesi-Impiombato F S, Coon H G. Thyroid cells in culture. Int Rev Cytol Suppl. 1979;10:163–172. doi: 10.1016/s0074-7696(08)60619-1. [DOI] [PubMed] [Google Scholar]
- 2.Arnone M I, Zannini M, Di Lauro R. The DNA binding activity and the dimerization ability of the thyroid transcription factor I are redox regulated. J Biol Chem. 1995;270:12048–12055. doi: 10.1074/jbc.270.20.12048. [DOI] [PubMed] [Google Scholar]
- 3.Bandyopadhyay S, Gronostajski R M. Identification of a conserved oxidation-sensitive cysteine residue in the NFI family of DNA-binding proteins. J Biol Chem. 1994;269:29949–29955. [PubMed] [Google Scholar]
- 4.Bandyopadhyay S, Starke D W, Mieyal J J, Gronostajski R M. Thioltransferase (glutaredoxin) reactivates the DNA-binding activity of oxidation-inactivated nuclear factor I. J Biol Chem. 1998;273:392–397. doi: 10.1074/jbc.273.1.392. [DOI] [PubMed] [Google Scholar]
- 5.Bienz M. A CCAAT box confers cell-type-specific regulation on the Xenopus hsp70 gene in oocytes. Cell. 1986;46:1037–1042. doi: 10.1016/0092-8674(86)90703-8. [DOI] [PubMed] [Google Scholar]
- 6.Chaudhry A Z, Lyons G E, Gronostajski R M. Expression patterns of the four nuclear factor I genes during mouse embryogenesis indicate a potential role in development. Dev Dyn. 1997;208:313–325. doi: 10.1002/(SICI)1097-0177(199703)208:3<313::AID-AJA3>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 7.Civitareale D, Castelli M P, Falasca P, Saiardi A. Thyroid transcription factor 1 activates the promoter of the thyrotropin receptor gene. Mol Endocrinol. 1993;7:1589–1595. doi: 10.1210/mend.7.12.8145764. [DOI] [PubMed] [Google Scholar]
- 8.Civitareale D, Lonigro R, Sinclair A J, Di Lauro R. A thyroid-specific nuclear protein essential for tissue-specific expression of the thyroglobulin promoter. EMBO J. 1989;8:2537–2542. doi: 10.1002/j.1460-2075.1989.tb08391.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Consiglio E, Salvatore G, Rall J E, Kohn L D. Thyroglobulin interactions with thyroid plasma membranes. The existence of specific receptors and their potential role. J Biol Chem. 1979;254:5065–5076. [PubMed] [Google Scholar]
- 10.Consiglio E, Shifrin S, Yavin Z, Ambesi-Impiombato F S, Rall J E, Salvatore G, Kohn L D. Thyroglobulin interactions with thyroid membranes. Relationship between receptor recognition of N-acetylglucosamine residues and the iodine content of thyroglobulin preparations. J Biol Chem. 1981;256:10592–10599. [PubMed] [Google Scholar]
- 11.Cooke D W, Lane M D. The transcription factor nuclear factor I mediates repression of the GLUT4 promoter by insulin. J Biol Chem. 1999;274:12917–12924. doi: 10.1074/jbc.274.18.12917. [DOI] [PubMed] [Google Scholar]
- 12.Corda D, Marcocci C, Kohn L D, Axelrod J, Luini A. Association of the changes in cytosolic Ca2+ and iodide efflux induced by thyrotropin and by the stimulation of alpha 1-adrenergic receptors in cultured rat thyroid cells. J Biol Chem. 1985;260:9230–9236. [PubMed] [Google Scholar]
- 13.Don R H, Cox P T, Wainwright B J, Baker K, Mattick J S. ‘Touchdown’ PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 1991;19:4008. doi: 10.1093/nar/19.14.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Donda A, Vassart G, Christophe D. Isolation and characterization of the canine thyroglobulin gene promoter region. Biochim Biophys Acta. 1991;1090:235–237. doi: 10.1016/0167-4781(91)90107-w. [DOI] [PubMed] [Google Scholar]
- 15.Endo T, Kaneshige M, Nakazato M, Ohmori M, Harii N, Onaya T. Thyroid transcription factor-1 activates the promoter activity of rat thyroid Na+/I− symporter gene. Mol Endocrinol. 1997;11:1747–1755. doi: 10.1210/mend.11.11.0012. [DOI] [PubMed] [Google Scholar]
- 16.Francis-Lang H, Price M, Polycarpou-Schwarz M, Di Lauro R. Cell-type-specific expression of the rat thyroperoxidase promoter indicates common mechanisms for thyroid-specific gene expression. Mol Cell Biol. 1992;12:576–588. doi: 10.1128/mcb.12.2.576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gil G, Smith J R, Goldstein J L, Slaughter C A, Orth K, Brown M S, Osborne T F. Multiple genes encode nuclear factor 1-like proteins that bind to the promoter for 3-hydroxy-3-methylglutaryl-coenzyme A reductase. Proc Natl Acad Sci USA. 1988;85:8963–8967. doi: 10.1073/pnas.85.23.8963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gounari F, De Francesco R, Schmitt J, van der Vliet P, Cortese R, Stunnenberg H. Amino-terminal domain of NFI binds to DNA as a dimer and activates adenovirus DNA replication. EMBO J. 1990;9:559–566. doi: 10.1002/j.1460-2075.1990.tb08143.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gronostajski R M. Analysis of nuclear factor I binding to DNA using degenerate oligonucleotides. Nucleic Acids Res. 1986;14:9117–9132. doi: 10.1093/nar/14.22.9117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gronostajski R M, Nagata K, Hurwitz J. Isolation of human DNA sequences that bind to nuclear factor I, a host protein involved in adenovirus DNA replication. Proc Natl Acad Sci USA. 1984;81:4013–4017. doi: 10.1073/pnas.81.13.4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hay R T. Origin of adenovirus DNA replication. Role of the nuclear factor I binding site in vivo. J Mol Biol. 1985;186:129–136. doi: 10.1016/0022-2836(85)90263-3. [DOI] [PubMed] [Google Scholar]
- 22.Hennighausen L, Fleckenstein B. Nuclear factor 1 interacts with five DNA elements in the promoter region of the human cytomegalovirus major immediate early gene. EMBO J. 1986;5:1367–1371. doi: 10.1002/j.1460-2075.1986.tb04368.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ikeda K, Clark J C, Shaw-White J R, Stahlman M T, Boutell C J, Whitsett J A. Gene structure and expression of human thyroid transcription factor-1 in respiratory epithelial cells. J Biol Chem. 1995;270:8108–8114. doi: 10.1074/jbc.270.14.8108. [DOI] [PubMed] [Google Scholar]
- 24.Ikuyama S, Shimura H, Hoeffler J P, Kohn L D. Role of the cyclic adenosine 3′,5′-monophosphate response element in efficient expression of the rat thyrotropin receptor promoter. Mol Endocrinol. 1992;6:1701–1715. doi: 10.1210/mend.6.10.1333054. [DOI] [PubMed] [Google Scholar]
- 25.Kambe F, Nomura Y, Okamoto T, Seo H. Redox regulation of thyroid-transcription factors, Pax-8 and TTF-1, is involved in their increased DNA-binding activities by thyrotropin in rat thyroid FRTL-5 cells. Mol Endocrinol. 1996;10:801–812. doi: 10.1210/mend.10.7.8813721. [DOI] [PubMed] [Google Scholar]
- 26.Kim T K, Roeder R G. CTD-like sequences are important for transcriptional activation by the proline-rich activation domain of CTF1. Nucleic Acids Res. 1994;22:251. doi: 10.1093/nar/22.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim T K, Roeder R G. Proline-rich activator CTF1 targets the TFIIB assembly step during transcriptional activation. Proc Natl Acad Sci USA. 1994;91:4170–4174. doi: 10.1073/pnas.91.10.4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kimura S, Hara Y, Pineau T, Fernandez-Salguero P, Fox C H, Ward J M, Gonzalez F J. The T/ebp null mouse: thyroid-specific enhancer-binding protein is essential for the organogenesis of the thyroid, lung, ventral forebrain, and pituitary. Genes Dev. 1996;10:60–69. doi: 10.1101/gad.10.1.60. [DOI] [PubMed] [Google Scholar]
- 29.Kohn L. Thyroglobulin interactions with thyroid membranes: implications for regulation of thyroid hormone formation in thyroglobulin. Prog Endocr Res Ther. 1985;2:171–190. [Google Scholar]
- 30.Kohn L D, Shimura H, Shimura Y, Hidaka A, Giuliani C, Napolitano G, Ohmori M, Laglia G, Saji M. The thyrotropin receptor. Vitam Horm. 1995;50:287–384. doi: 10.1016/s0083-6729(08)60658-5. [DOI] [PubMed] [Google Scholar]
- 31.Kosower N S, Kosower E M. Formation of disulfides with diamide. Methods Enzymol. 1987;143:264–270. doi: 10.1016/0076-6879(87)43050-4. [DOI] [PubMed] [Google Scholar]
- 32.Krohn K, Rozovsky I, Wals P, Teter B, Anderson C P, Finch C E. Glial fibrillary acidic protein transcription responses to transforming growth factor-beta1 and interleukin-1 beta are mediated by a nuclear factor-1-like site in the near-upstream promoter. J Neurochem. 1999;72:1353–1361. doi: 10.1046/j.1471-4159.1999.721353.x. [DOI] [PubMed] [Google Scholar]
- 33.Kruse U, Sippel A E. Transcription factor nuclear factor I proteins form stable homo- and heterodimers. FEBS Lett. 1994;348:46–50. doi: 10.1016/0014-5793(94)00585-0. [DOI] [PubMed] [Google Scholar]
- 34.Lee M, Song H, Park S, Park J. Transcription of the rat p53 gene is mediated by factor binding to two recognition motifs of NFI-like protein. Biol Chem. 1998;379:1333–1340. doi: 10.1515/bchm.1998.379.11.1333. [DOI] [PubMed] [Google Scholar]
- 35.Leegwater P A, van Driel W, van der Vliet P C. Recognition site of nuclear factor I, a sequence-specific DNA-binding protein from HeLa cells that stimulates adenovirus DNA replication. EMBO J. 1985;4:1515–1521. doi: 10.1002/j.1460-2075.1985.tb03811.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lonigro R, De Felice M, Biffali E, Macchia P E, Damante G, Asteria C, Di Lauro R. Expression of thyroid transcription factor 1 gene can be regulated at the transcriptional and posttranscriptional levels. Cell Growth Differ. 1996;7:251–261. [PubMed] [Google Scholar]
- 37.Miquelis R, Alquier C, Monsigny M. The N-acetylglucosamine-specific receptor of the thyroid. Binding characteristics, partial characterization, and potential role. J Biol Chem. 1987;262:15291–15298. [PubMed] [Google Scholar]
- 38.Mizuno K, Gonzalez F J, Kimura S. Thyroid-specific enhancer-binding protein (T/EBP): cDNA cloning, functional characterization, and structural identity with thyroid transcription factor TTF-1. Mol Cell Biol. 1991;11:4927–4933. doi: 10.1128/mcb.11.10.4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakazato M, Endo T, Saito T, Harii N, Onaya T. Transcription of the thyroid transcription factor-1 (TTF-1) gene from a newly defined start site: positive regulation by TTF-1 in the thyroid. Biochem Biophys Res Commun. 1997;238:748–752. doi: 10.1006/bbrc.1997.7383. [DOI] [PubMed] [Google Scholar]
- 40.Novak A, Goyal N, Gronostajski R M. Four conserved cysteine residues are required for the DNA binding activity of nuclear factor I. J Biol Chem. 1992;267:12986–12990. . (Erratum, 268:26032, 1993.) [PubMed] [Google Scholar]
- 41.Nowock J, Borgmeyer U, Puschel A W, Rupp R A, Sippel A E. The TGGCA protein binds to the MMTV-LTR, the adenovirus origin of replication, and the BK virus enhancer. Nucleic Acids Res. 1985;13:2045–2061. doi: 10.1093/nar/13.6.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oguchi H, Kimura S. Multiple transcripts encoded by the thyroid-specific enhancer-binding protein (T/EBP)/thyroid-specific transcription factor-1 (TTF-1) gene: evidence of autoregulation. Endocrinology. 1998;139:1999–2006. doi: 10.1210/endo.139.4.5933. [DOI] [PubMed] [Google Scholar]
- 43.Ohmori M, Endo T, Harii N, Onaya T. A novel thyroid transcription factor is essential for thyrotropin-induced up-regulation of Na+/I− symporter gene expression. Mol Endocrinol. 1998;12:727–736. doi: 10.1210/mend.12.5.0101. [DOI] [PubMed] [Google Scholar]
- 44.Paonessa G, Gounari F, Frank R, Cortese R. Purification of a NFI-like DNA-binding protein from rat liver and cloning of the corresponding cDNA. EMBO J. 1988;7:3115–3123. doi: 10.1002/j.1460-2075.1988.tb03178.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reifel-Miller A E, Calnek D S, Grinnell B W. Tyrosine phosphorylation regulates the DNA binding activity of a nuclear factor 1-like repressor protein. J Biol Chem. 1994;269:23861–23864. [PubMed] [Google Scholar]
- 46.Santoro C, Mermod N, Andrews P C, Tjian R. A family of human CCAAT-box-binding proteins active in transcription and DNA replication: cloning and expression of multiple cDNAs. Nature. 1988;334:218–224. doi: 10.1038/334218a0. [DOI] [PubMed] [Google Scholar]
- 47.Shaul Y, Ben-Levy R, De-Medina T. High affinity binding site for nuclear factor I next to the hepatitis B virus S gene promoter. EMBO J. 1986;5:1967–1971. doi: 10.1002/j.1460-2075.1986.tb04451.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shifrin S, Kohn L D. Binding of thyroglobulin to bovine thyroid membranes. Role of specific amino acids in receptor recognition. J Biol Chem. 1981;256:10600–10605. [PubMed] [Google Scholar]
- 49.Shimura H, Okajima F, Ikuyama S, Shimura Y, Kimura S, Saji M, Kohn L D. Thyroid-specific expression and cyclic adenosine 3′,5′-monophosphate autoregulation of the thyrotropin receptor gene involves thyroid transcription factor-1. Mol Endocrinol. 1994;8:1049–1069. doi: 10.1210/mend.8.8.7997232. [DOI] [PubMed] [Google Scholar]
- 50.Sinclair A J, Lonigro R, Civitareale D, Ghibelli L, Di Lauro R. The tissue-specific expression of the thyroglobulin gene requires interaction between thyroid-specific and ubiquitous factors. Eur J Biochem. 1990;193:311–318. doi: 10.1111/j.1432-1033.1990.tb19339.x. [DOI] [PubMed] [Google Scholar]
- 51.Suzuki K, Mori A, Saito J, Moriyama E, Ullianich L, Kohn L D. Follicular thyroglobulin suppresses iodide uptake by suppressing expression of the sodium/iodide symporter gene. Endocrinology. 1999;140:5422–5430. doi: 10.1210/endo.140.11.7124. [DOI] [PubMed] [Google Scholar]
- 52.Suzuki K, Lavaroni S, Mori A, Ohta M, Saito J, Pietrarelli M, Singer D S, Kimura S, Katoh R, Kawaoi A, Kohn L D. Autoregulation of thyroid-specific gene transcription by thyroglobulin. Proc Natl Acad Sci USA. 1998;95:8251–8256. doi: 10.1073/pnas.95.14.8251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Suzuki K, Lavaroni S, Mori A, Yamamoto K, Yi X, Miyagi E, Katoh R, Kohn L, Kawaoi A. Thyroglobulin: a master regulator of follicular function via transcriptional suppression of thyroid specific genes. Acta Histochem Cytochem. 1999;32:111–119. [Google Scholar]
- 54.Suzuki K, Mori A, Lavaroni S, Miyagi E, Ulianich L, Katoh R, Kawaoi A, Kohn L D. In vivo expression of thyroid transcription factor-1 RNA and its relation to thyroid function and follicular heterogeneity: identification of follicular thyroglobulin as a feedback suppressor of thyroid transcription factor-1 RNA levels and thyroglobulin synthesis. Thyroid. 1999;9:319–331. doi: 10.1089/thy.1999.9.319. [DOI] [PubMed] [Google Scholar]
- 55.Suzuki K, Mori A, Lavaroni S, Ulianich L, Miyagi E, Saito J, Nakazato M, Pietrarelli M, Shafran N, Grassadonia A, Kim W B, Consiglio E, Formisano S, Kohn L D. Thyroglobulin regulates follicular function and heterogeneity by suppressing thyroid-specific gene expression. Biochimie. 1999;81:329–340. doi: 10.1016/s0300-9084(99)80078-9. [DOI] [PubMed] [Google Scholar]
- 56.Ulianich L, Suzuki K, Mori A, Nakazato M, Pietrarelli M, Goldsmith P, Pacifico F, Consiglio E, Formisano S, Kohn L D. Follicular thyroglobulin (TG) suppression of thyroid-restricted genes involves the apical membrane asialoglycoprotein receptor and TG phosphorylation. J Biol Chem. 1999;274:25099–25107. doi: 10.1074/jbc.274.35.25099. [DOI] [PubMed] [Google Scholar]
- 57.Wendler W, Altmann H, Ludwig-Winnacker E. Transcriptional activation of NFI/CTF1 depends on a sequence motif strongly related to the carboxyterminal domain of RNA polymerase II. Nucleic Acids Res. 1994;22:2601–2603. doi: 10.1093/nar/22.13.2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zheng G, Marino M, Zhao J, McCluskey R T. Megalin (gp330): a putative endocytic receptor for thyroglobulin (Tg) Endocrinology. 1998;139:1462–1465. doi: 10.1210/endo.139.3.5978. [DOI] [PubMed] [Google Scholar]