Abstract
The latent membrane protein (LMP) 2A of Epstein-Barr virus (EBV) is implicated in the maintenance of viral latency and appears to function in part by inhibiting B-cell receptor (BCR) signaling. The N-terminal cytoplasmic region of LMP2A has multiple tyrosine residues that upon phosphorylation bind the SH2 domains of the Syk tyrosine kinase and the Src family kinase Lyn. The LMP2A N-terminal region also has two conserved PPPPY motifs. Here we show that the PPPPY motifs of LMP2A bind multiple WW domains of E3 protein-ubiquitin ligases of the Nedd4 family, including AIP4 and KIAA0439, and demonstrate that AIP4 and KIAA0439 form physiological complexes with LMP2A in EBV-positive B cells. In addition to a C2 domain and four WW domains, these proteins have a C-terminal Hect catalytic domain implicated in the ubiquitination of target proteins. LMP2A enhances Lyn and Syk ubiquitination in vivo in a fashion that depends on the activity of Nedd4 family members and correlates with destabilization of the Lyn tyrosine kinase. These results suggest that LMP2A serves as a molecular scaffold to recruit both B-cell tyrosine kinases and C2/WW/Hect domain E3 protein-ubiquitin ligases. This may promote Lyn and Syk ubiquitination in a fashion that contributes to a block in B-cell signaling. LMP2A may potentiate a normal mechanism by which Nedd4 family E3 enzymes regulate B-cell signaling.
In the course of infection, replication or persistence, viral gene products frequently interact with proteins that regulate signaling pathways in the host cell. This capacity to modify host cell signal transduction is typified by Epstein-Barr virus (EBV), a human herpesvirus that infects lymphoid and epithelial cells and causes infectious mononucleosis (32). EBV is also associated with a variety of human proliferative disorders, including Burkitt's lymphoma (40), undifferentiated nasopharyngeal carcinoma (24), and X-linked lymphoproliferative syndrome (10).
Following B-cell infection, EBV establishes a lifelong latent state in which the viral episome is maintained in the absence of replicative gene expression. Three types of latency have been defined, based on the expression of subsets of viral genes that appear important for maintaining the virus in a latent form (44, 45). The latent membrane proteins (LMP) 1, 2A, and 2B are membrane-spanning polypeptides that are commonly expressed in latency, in concert with EBNA1, and intersect with both CD40/tumor necrosis factor-receptor signaling pathways (LMP1) and protein-tyrosine kinases (LMP1 and LMP2A) (19, 20, 27, 30, 34).
LMP2A has an N-terminal cytoplasmic region of ∼119 residues, which is predicted to be followed by 12 membrane-spanning regions and a short C-terminal cytoplasmic tail. A variety of data suggest that LMP2A can modify signaling from the B-cell antigen receptor (BCR) through the ability of its N-terminal cytoplasmic region to bind the SH2 domains of B-cell tyrosine kinases (30). The N terminus of LMP2A becomes phosphorylated on Tyr residues and contains multiple Tyr-based motifs that can serve as docking sites for specific SH2-containing proteins (4, 14–16). In particular, Tyr residues 74 and 85 lie in a consensus ITAM motif [YXXL/I (X6–8) YXXL/I] characteristic of the signaling subunits of antigen and Fc receptors, including the immunoglobulin alpha (Ig α) and β nonpolymorphic chains of the BCR (42). Phosphorylated ITAMs bind selectively to the tandem SH2 domains of the Syk or ZAP-70 tyrosine kinases (8, 12, 18). Indeed, Syk, which binds the phospho-ITAMs of the Ig α/β BCR subunits and is required for B-cell development (9, 11, 56), becomes physically associated with LMP2A in EBV-positive lymphoblastoid cell lines (LCLs) (15) and with chimeric proteins containing the LMP2A N-terminal cytoplasmic region (1, 2). In addition, Tyr112 of LMP2A is located within a YEEA motif that, in its phosphorylated form, binds the SH2 domain of Src family tyrosine kinases, notably Lyn in B cells (16, 34).
Normal BCR signaling is initiated by a Src family kinase (SFK) which phosphorylates the Ig α/β ITAM motifs, leading to the recruitment and activation of Syk. In turn, Syk stimulates cytoplasmic enzymes, such as phospholipase C-γ2, and scaffolding proteins, such as Blnk (13, 17, 26). Since LMP2A binds both Lyn and Syk, it might be expected to perturb BCR signaling. Indeed, LMP2A impairs the ability of the cross-linked BCR to stimulate Syk and Lyn tyrosine kinase activity, induce tyrosine phosphorylation of substrates, such as phospholipase C-γ2, and mobilize calcium in B lymphocytes (32, 34). Furthermore, the inhibitory effect of LMP2A appears to require Tyr residues 74 and 85 in the ITAM motif and Tyr 112, which binds the Lyn SH2 domain (15, 16). These data suggest a model in which Lyn phosphorylates LMP2A on Tyr112, to which it binds through its SH2 domain, and then phosphorylates the ITAM motif, which subsequently engages Syk. By sequestering these tyrosine kinases away from the BCR, LMP2A may antagonize the normal process of B-lymphocyte activation and thereby represses the expression of immediate early viral genes required for replication.
However, the physiological situation may be more complex. Since LMP2A can engage SFKs and Syk in a manner very similar to that of the BCR itself, it is possible that LMP2A might, under some circumstances, function as an activator of B-cell signaling, rather than as a repressor. Indeed, a chimeric protein in which the N-terminal cytoplasmic region of LMP2A is fused to the CD8 ectodomain is able to activate B-cell signaling and induce calcium mobilization (1, 2), and transgenic expression of LMP2A induces the aberrant expression of Ig-negative cells in mice, apparently by supplanting pre-BCR signals (5). These results suggest that additional cellular factors may modulate the effects of LMP2A on lymphoid function. Indeed, LMP2A can also associate with the mitogen-activated protein kinase (36) and with the Csk tyrosine kinase in epithelial cells (49). Examination of the N-terminal cytoplasmic region of LMP2A has revealed two highly conserved PPPPY motifs, which characteristically bind a subset of WW domains found in proteins such as YAP65 and Nedd4, an E3 protein-ubiquitin ligase with a Hect catalytic domain (7, 43, 53, 55). We have therefore probed for novel LMP2A-binding partners, which recognize the LMP2A PPPPY motifs and might modify B-lymphocyte signaling proteins. We find that the LMP2A PPPPY motifs bind selectively to a subset of WW domain proteins with E3 protein-ubiquitin ligase activity, which are implicated as negative regulators of lymphoid signaling. Furthermore, we find that these proteins are able to directly ubiquitinate Lyn and Syk. Our results suggest that LMP2A serves as a molecular scaffold to recruit both tyrosine kinases and E3 protein-ubiquitin ligases, leading to ubiquitination and potential degradation of the B-cell tyrosine kinases.
MATERIALS AND METHODS
Cell lines, expression constructs, and antibodies.
The EBV-negative B-cell lines DG75 and BJAB and EBV-positive LCLs CBM-RAL-STO and IB4 were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS) and 2 mM glutamine. The adherent cell lines Cos-1 and HEK 293 were grown in Dulbecco's modified Eagle medium with high glucose content and 10% FBS. HEK 293-derived cell lines that stably express full-length LMP2A or a chimeric protein containing the N-terminal 119 amino acids of LMP2A fused to CD38 were generated by retrovirus infection and puromycin selection. A cDNA clone, pSP64-23TP, carrying the LMP2A gene from the B95-8 strain of EBV, was provided by P. J. Farrell (29). A retroviral vector, pLXPOP, was used to express LMP2A in HEK 293 cells. This was constructed from the pLNPOX vector (33) through insertion of a puromycin resistance gene into the HindIII site adjacent to the poliovirus 5′ untranslated region. In addition, a BamHI site preceding the 5′ long terminal repeat was removed, and the Tn 5 aminoglycoside 3′ phosphotransferase (Neo) gene was replaced with a multilinker of sequence 5′-GAATTCACCGGTCGACGTACGGATCCTTAATTAAGCTTATTTAAATTCGAAAGATCTGTTTAAACTCGAG-3′ (G. Winberg and R. Reynolds, unpublished data). An N-terminal FLAG epitope (DYKDDDDK)-tagged LMP2A expression construct was prepared by subcloning the EBV terminal gene (LMP2A) cDNA into pFLAG-CMV-2 (Eastman Kodak, New Haven, Conn.). A glutathione S-transferase (GST) fusion protein containing amino acid residues 1 to 122 from EBV LMP2A was generated by PCR amplification of the fragment from LMP2A cDNA and cloning into the vector pGEX-KT. LMP2A derivatives containing mutations in either or both PPPPY motifs were generated by PCR mutagenesis and authenticated by DNA sequencing. Constructs allowing for expression of individual AIP4 WW domains as GST fusion proteins in bacteria were prepared by cloning into the vector pGEX-KT synthetic oligonucleotides corresponding to the following amino acid residues: WW1, 286 to 317; WW2, 318 to 349; WW3, 398 to 429; and WW4, 438 to 469. A MYC epitope (EQKLISEEDL)-tagged KIAA0439 expression construct was prepared by subcloning cDNA provided by the Kazusa DNA Research Institute (Chiba, Japan) into pRK5-myc; this vector, which contains a CMV promoter and N-terminal MYC tag, was a gift of Jonathan Wood (Johns Hopkins University School of Medicine, Baltimore, Md.). Mutation of the KIAA0439 Hect domain catalytic Cys to Ser (C962S) was accomplished by using PCR mutagenesis and was confirmed by DNA sequencing. A MYC-tagged AIP4 expression construct was generated in the pRK5-myc vector by subcloning fragment AIP4.1, which contains the WW domain region described by Wood et al. (57), and ATCC 1011774, which encodes the C-terminal HECT domain and additional regions that overlap with AIP4.1. The AIP4 N-terminal C2 domain, amino acid residues 1 to 113, was introduced by chemical synthesis using oligonucleotides that encode the same amino acid sequence found in the mouse ortholog Itchy (37). A derivative of AIP4 (C830A) containing an inactive Hect domain was prepared by PCR mutagenesis and authenticated by DNA sequencing. A hemagglutinin (HA) epitope (YPYDVPDY)-tagged ubiquitin, T7 epitope-tagged Nedd4, and Nedd4 (C853S) expression vectors have been described previously (53). Andre Veillette (McGill University, Montreal, Canada) provided vectors that allow expression of Lyn and Syk and antibodies to these proteins. Rat anti-LMP2A monoclonal antibody (MAb) 4E11 (14) was purchased from ITN GmbH (Neuherberg, Germany). The antibodies, anti-v-H-ras rat MAb IgG1, anti-Lyn rabbit polyclonal antibody IgG, and anti-MYC mouse MAb IgG1, were purchased from Santa Cruz Biotechnology (Santa Cruz, Calif.). Mouse anti-FLAG and anti-T7 MAbs were purchased from Sigma-Aldrich Canada Ltd., (Oakville, Ontario, Canada) and Novagen (Madison, Wis.), respectively. Anti-phosphotyrosine MAb IgG2bκ clone 4G10 was purchased from Upstate Biotechnology (Lake Placid, N.Y.).
Peptide synthesis.
Synthetic peptides were prepared on an Applied Biosystems 431 peptide synthesizer using 9-fluorenylmethoxy carbonyl solid-phase chemistry. Biotin was incorporated into each peptide as the N-terminal residue during the synthesis. To allow for efficient coupling to streptavidin, an ɛ-aminocaproic acid (Aca) linker was added between the biotin moiety and the desired peptide sequence. When required, phosphotyrosine was directly incorporated into the peptide using the N-fluorenylmethoxy carbonyl-O-phosphono-l-tyrosine derivative. The peptides were isolated through a 90-min treatment of the peptide resin at room temperature in a trifluoroacetic acid solution containing a scavenger mixture of thioanisol, 1,2-ethanedithiol, and water (1.0:0.1:2.0% vol/vol). The product, cleaved from the resin and deprotected, was precipitated with cold t-butylethyl ether, collected by centrifugation, and purified using reverse-phase high-pressure liquid chromatography. The authenticity of the peptides was confirmed by amino acid analysis and mass spectrometry.
Immunoprecipitation and immunoblotting. (i) Gene pulse electroporation of vectors into B cells.
For each electroporation, approximately 107 DG75 cells were resuspended in 0.4 ml of RPMI 1640, supplemented with 10% (vol/vol) FBS and glutamine that contained 20 μg of each vector, and transferred to a 0.4-cm Gene Pulser cuvette (Bio-Rad Laboratories, Hercules, Calif.). The cells were pulsed once with 210 V at 960 μF by using a Gene Pulser equipped with a capacitance extender (Bio-Rad Laboratories). Following the pulse, the cells were diluted in 10 ml of fresh RPMI 1640 medium containing 10% (vol/vol) FBS and glutamine and were incubated at 37°C for 40 h prior to analysis. The cells were then collected by low-speed centrifugation and lysed in 1 ml of 50 mM Tris-HCl, 150 mM NaCl, 2 mM EDTA, 1% (vol/vol) NP-40, 10-μg/ml aprotinin, 10-μg/ml leupeptin, 1 mM phenylmethylsulfonyl fluoride, and 10-μg/ml pepstatin A (NP-40 buffer) on ice. Cell lysate was obtained following a 15-min centrifugation at 11,000 × g, and protein content was quantified using a bicinchoninic acid protein reagent assay (Pierce, Rockford, Ill.). Immunoprecipitations were carried out by incubating (for 18 h at 5°C) cell lysate containing approximately 1 mg of total protein, with 1 μg of primary antibody in the presence of immobilized secondary antibody. Immunoprecipitated proteins were washed three times with NP-40 buffer (1 ml each time) and resuspended in sodium dodecyl sulfate (SDS) sample buffer. Proteins were resolved using SDS–8% polyacrylamide gel electrophoresis (PAGE) and transferred to an Immobilon-P filter (Millipore Corp., Bedford, Mass.) using a semidry Western transfer apparatus (Bio-Rad Laboratories). Western blotting was carried out by treating the protein filter with a 5% skim milk powder solution in 20 mM Tris-HCl (pH 7.5), 150 mM NaCl, and 0.05% (vol/vol) Tween 20 (TBST) containing the required antibody at a concentration of 3 μg/ml. Following a 60-min incubation at room temperature, each protein filter was washed three times with excess TBST over a 15-min period and then treated for 45 min with the appropriate horseradish peroxidase-conjugated secondary antibody diluted in TBST. After being washed with TBST, the blots were developed by using chemiluminescent detection (Supersignal; Pierce) with image detection on dental film (XDBF-1 film; Eastman Kodak, Rochester, N.Y.).
(ii) Protein expression in adherent cells.
For transfection of DNA into Cos-1 cells, Lipofectin reagent (GIBCO BRL Life Technologies) was employed following the manufacturer's protocols. To express proteins in HEK 293 cells and cell lines derived from HEK 293, the calcium phosphate precipitation method described by Tong et al. (55a) was performed. In each transfection, approximately 10 μg of each expression vector was used. Following transfection of either Cos-1 or HEK 293 cells, the cells were grown for approximately 48 h and then washed in cold phosphate-buffered saline (PBS). Cell lysates were prepared in NP-40 buffer. Immunoprecipitation and Western blotting were performed as described above. To inhibit protein synthesis, a dimethyl sulfoxide solution of cycloheximide was added to a final concentration of 35 μM (10 μg/ml).
(iii) GST fusion protein binding assays.
Immobilized GST-LMP2A fusion proteins were prepared from BL21 bacterial cells that express each protein. Cell lysates were prepared in 50 mM sodium phosphate (pH 7.5), 150 mM NaCl, 2 mM EDTA, 1% (vol/vol) Triton X-100, and 2 mM benzamidine (PBS-Triton) and treated with GST-agarose beads (Sigma) for 20 min at 5°C. Immobilized proteins were isolated by collection of the beads by brief centrifugation, washed with excess PBS-Triton buffer, and analyzed for the level of protein expression by using SDS–9% PAGE with Coomassie blue staining. Equivalent amounts (approximately 10 μg) of immobilized fusion protein were incubated for 60 min at 5°C with Cos-1 cell lysates containing T7 epitope-tagged Nedd4 that were prepared as described above. The beads were then washed three times with NP-40 buffer (1 ml each time), and associated Nedd4 protein was identified by SDS–8% PAGE, followed by Western blotting using anti-T7 antibody. Similar procedures were used to monitor the interaction of LMP2A with GST-AIP4 WW domain fusion proteins. In these experiments, the presence of LMP2A was detected by Western blotting using rat anti-LMP2A MAb 4E11.
COLT protein association screen.
The synthetic peptide Biotin.Aca.S.N.E.E.P.P.P.P.pY.E.D.P.Y.W.G.N.G. was used in an expression screen of a mouse 10-day embryo cDNA library (Novagen) following the protocol for cloning of ligand targets (COLT) described by Sparks et al. (52).
Affinity chromatography and protein sequencing by quadrupole–time-of-flight mass spectrometry.
To probe for proteins that might associate with LMP2A, the synthetic peptides Biotin.Aca.S.N.E.E.P.P.P.P.pY.E.D.P.Y.W.G.N.G, Biotin.Aca.S.N.E.E.P.P.P.P.Y.E.D.P.Y.W.G.N.G, and Biotin.Aca.G.D.R.H.S.D.pY.Q.P.L.G.T.Q.S.L.pY.L.G.L.Q.H.D.G were employed as affinity reagents to probe the EBV-positive CBM-RAL-STO cell lysate. For each affinity reagent, a 100-μl-bed volume of streptavidin-agarose resin (Sigma) capable of binding 10 nmol of biotin was washed with 20 mM sodium phosphate buffer (pH 7.0) containing 150 mM NaCl (PBS) and then resuspended in a twofold-molar excess of biotinylated peptide. Following a 15-min incubation and mixing at room temperature, the resin was washed three times with excess PBS. CBM-RAL-STO cell lysate was prepared in NP-40 buffer and subjected to a 100,000 × g ultracentrifugation. For each affinity isolation, clarified cell lysate from 108 cells was treated with 1.5 nmol (15 μl) of immobilized peptide resin for 90 min at 5°C. The resin was washed three times with NP-40 buffer, and the isolated proteins were resolved using SDS–8% PAGE. Proteins were visualized by silver staining, and bands selected for mass spectrometric analysis were excised from the gel and digested with trypsin as described by Shevchenko et al. (50). Mass spectrometric measurements were carried out on a Q-Star quadrupole–time-of-flight apparatus (MDS-Sciex, Concord, Ontario, Canada).
RESULTS
LMP2A PPPPY motif binds proteins of the Nedd4 family.
The N-terminal cytoplasmic region of EBV LMP2A has two PPPPY motifs (Fig. 1), which might bind protein modules, such as WW or SH3 domains. To assess the ability of the LMP2A PPPPY motifs to bind cellular proteins, a COLT (52) screen of a 10-day mouse embryo cDNA expression library was performed using a biotinylated synthetic peptide based on the more N-terminal proline-rich motif Biotin.Aca.S.N.E.E.P.P.P.P.Y.E.D.P.Y.W.G.N.G (PPPPY). This procedure identified cDNA encoding fragments of Nedd4 (accession no. 2088623), a Nedd4-related molecule (accession no. KIAA0439), and the SH2/SH3 adapter Grb2, indicating that these proteins are capable of binding in vitro to the proline-rich LMP2A peptide.
FIG. 1.
Amino acid sequence of the cytoplasmic N-terminal region of LMP2A. The two PPPPY motifs are underlined in bold type. Also indicated in bold type are the ITAM and YEEA motifs, which, when phosphorylated, become ligands for the B-cell tyrosine kinases Syk and Lyn, respectively.
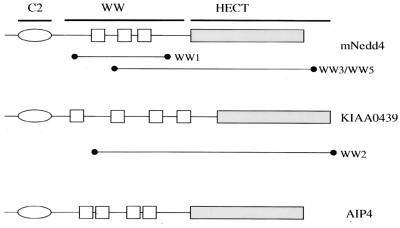
Full-length rat Nedd4 has an N-terminal C2 domain involved in membrane targeting (39), followed by three WW domains and a C-terminal Hect domain that mediates the transfer of ubiquitin to target proteins. Important for catalyzing the transfer of ubiquitin is a Cys residue within the Hect domain, which undergoes a transient covalent attachment to ubiquitin via a thioester linkage (46, 47, 54) (Fig. 2). Nedd4 is the prototype for a family of related proteins conserved from yeast to vertebrates (23) that includes KIAA0439. Of the individual Nedd4 and KIAA0439 cDNA clones isolated (Fig. 2), each encoded two or more complete WW domains, consistent with the possibility that the LMP2A proline-rich motifs might serve as ligands for specific interaction domains of host cell proteins. To investigate further the ability of this peptide to interact with Nedd4-like proteins, rabbit polyclonal antibodies to KIAA0439 were prepared and used to detect proteins precipitated from lysates of Cos-1 cells with the PPPPY peptide immobilized on streptavidin-agarose beads. We also tested a phosphotyrosine-containing (pY) peptide derivative, Biotin.Aca.S.N.E.E.P.P.P.P.pY.E.D.P.Y.W.G.N.G (PPPPpY), which was immobilized in a similar fashion. Specific binding of KIAA0439 was detected to the nonphosphorylated peptide, but no association was observed with the corresponding phosphotyrosine-containing peptide, indicating that phosphorylation of the PPPPY motif inhibits KIAA0439 binding (data not shown). Only a single Grb2 cDNA, which encoded the SH2 domain and C-terminal SH3 domain, was isolated. Its identification in the screen was most likely due to recognition of the PPPPY peptide by the C-terminal Grb2 SH3 domain. Attempts to confirm the Grb2 association through probing of Cos-1 lysates with the immobilized PPPPY and PPPPpY peptides were not successful, suggesting that the binding of the Grb2 to the PPPPY motif is likely to be of low affinity and of questionable physiological significance.
FIG. 2.
Domain organization of mouse Nedd4, KIAA0439, and AIP4. Also illustrated are the protein fragments of Nedd4 (WW1, WW3, WW5) and KIAA0439 (WW2) that were isolated from a COLT screen of a 10-day mouse embryo expression library probed with the LMP2A-derived synthetic peptide Biotin.Aca.S.N.E.E.P.P.P.P.Y.E.D.P.Y.W.G.N.G.
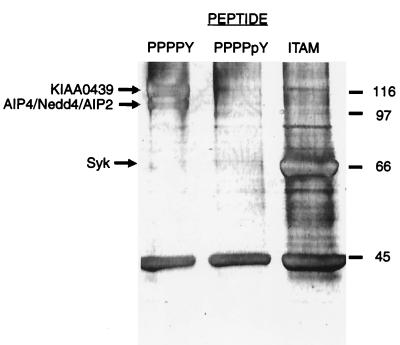
These results suggest that the PPPPY motifs of LMP2A potentially bind host proteins with WW domains. To extend these observations to a more relevant cell type, we investigated which proteins from EBV-positive human B-cell lines might associate with the LMP2A proline-rich sequence by using affinity chromatography and quadrupole–time-of-flight mass spectrometry protein identification. Figure 3 shows a silver-stained gel of proteins from the EBV-positive CBM-RAL-STO cell line that precipitate with the immobilized peptides PPPPY, PPPPpY (included as a control which should not bind WW domain proteins), and a diphosphotyrosine peptide corresponding to the ITAM sequence found in LMP2A, Biotin .Aca . G . D . R . H . S . D . pY . Q . P . L . G . T . Q . S . L . pY . L . G . L . Q . H . D . G (ITAM). Only the PPPPY and ITAM affinity matrices precipitated specific proteins in high abundance. The PPPPY peptide selectively bound protein species of 97 and 115 kDa, whereas the ITAM phosphopeptide bound primarily to a protein of approximately 70 kDa. These polypeptides were digested with trypsin, and the resulting peptides were analyzed by mass spectrometry. This unambiguously identified the human B-cell proteins that bind the EBV LMP2A PPPPY peptide as members of the Nedd4 family of proteins, namely, KIAA0439, AIP4 (37, 57), AIP2/WWP2 (38), and Nedd4 (23, 28) (Table 1). In contrast, the polypeptide that specifically associated with the ITAM motif was identified as the Syk tyrosine kinase.
FIG. 3.
Specific B-cell proteins bind to affinity reagents derived from ligand motifs found in LMP2A. Immobilized LMP2A-derived synthetic peptides based on PPPPY, PPPPpY, and ITAM motifs were treated with clarified lysates from a CBM-RAL-STO B-cell line. Associated proteins were resolved by SDS–9% PAGE, visualized by silver staining, and identified by quadrupole–time-of-flight (mass spectrometry) (Table 1). Numbers are for molecular mass markers (kilodaltons).
TABLE 1.
Proteins identified by quadrupole–time-of-flight mass spectrometry
| Protein | Accession no. | (M + 2H)2+ peaka | Peptide identified (peptide position, amino acid residue) |
|---|---|---|---|
| KIAA0439 | O43165 | 535.3 | EWFFLLSK (686–693) |
| 561.3 | LLQFVTGTSR (918–927) | ||
| 796.9 | LLMAVENAQGFEGVD (981–995) | ||
| 829.5 | SLSSPTVTLSAPLEGAK (466–482) | ||
| AIP4 | AAC04845 | 423.2 | LDLPPYK (711–717) |
| 483.7 | TTTYIDPR (339–346) | ||
| 522.8 | ITQWEDPR (299–306) | ||
| 568.3 | FIAMALFHGK (480–489) | ||
| 702.4 | SALDNGPQIAYVR (350–362) | ||
| 717.4 | FIDTGFSLPFYK (490–501) | ||
| Nedd4 | P46934 | 535.3 | EWFFLISK (618–625) |
| 561.3 | LLQFVTGTSR (850–859) | ||
| 573.3 | AVLMMDSEKR (838–847) | ||
| 590.8 | LWIEFDGEK (600–608) | ||
| 641.3 | LLFEVFDENR (114–123) | ||
| WWP2 | ACC51325 | 466.3 | AGVALPFEK (9–17) |
| Syk | P43405 | 416.2 | LLTLEDK (369–375) |
| 433.7 | ISDFGLSK (510–517) | ||
| 540.8 | GSEVTAMLEK (578–587) | ||
| 554.8 | ELGSGNFGTVK (376–386) | ||
| 647.3 | YLEESNFVHR (484–493) |
(M + 2H)2+, observed mass of the doubly charged parent ion.
LMP2A shows selective association with the AIP4 and KIAA0439 C2/WW/HECT domain proteins.
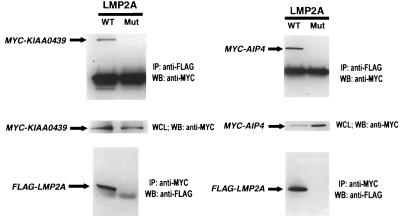
To investigate whether the identified C2/WW/Hect domain proteins associate with the full-length LMP2A protein, we electroporated a mammalian protein expression vector encoding LMP2A into a human B-cell line (DG75), with a FLAG-tagged epitope at its N terminus, along with either Myc-tagged AIP4 or KIAA0439 expression vectors. In preliminary experiments that used wild-type AIP4 or KIAA0439, protein expression levels were below our detection limits. However, variant forms of these proteins, each containing an Ala or Ser substitution of the Cys residue critical for Hect domain activity, were readily detected and coimmunoprecipitated with LMP2A (Fig. 4), regardless of which partner was subjected to immunoprecipitation.
FIG. 4.
The E3 ubiquitin-ligase proteins KIAA0439 and AIP4 associate with LMP2A when overexpressed in the B-cell line DG75. A FLAG epitope-tagged LMP2A mammalian expression vector was gene pulsed into DG75 cells with either a MYC epitope-tagged KIAA0439 C962S or AIP4 C830A expression vector. Control experiments using a derivative of LMP2A unable to bind WW domain proteins, due to mutation of both PPPPY motifs to PPPPA, are also shown. Following the gene pulse, the cells were grown for 40 h and then lysed in NP-40 buffer. Lysates were split equally and immunoprecipitated with either anti-MYC or anti-FLAG for 18 h at 5°C. Proteins were resolved by SDS–8% PAGE and subjected to Western blotting for detection of the associated FLAG or MYC epitope-tagged protein. WT, wild type; Mut, mutant; IP, immunoprecipitation; WB, Western blotting; WCL, whole-cell lysak.
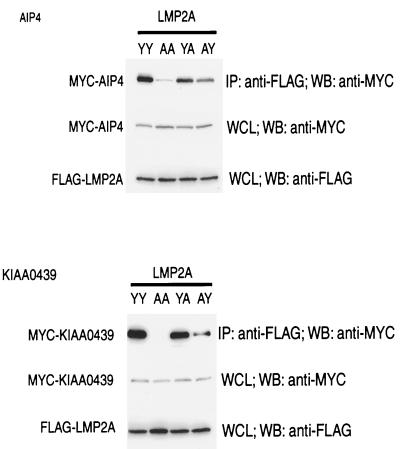
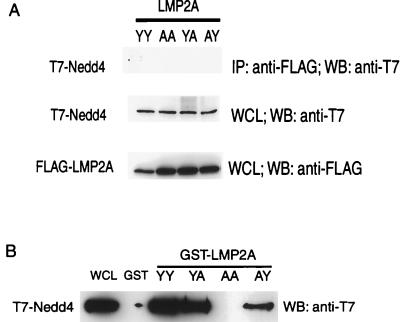
To further probe the interaction between C2/WW/Hect domain proteins and LMP2A, we cotransfected Cos-1 cells with FLAG-tagged LMP2A and either Myc-AIP4, Myc-KIAA0439, or T7-tagged Nedd4. As observed in the DG75 B cells, the level of C2, WW, and Hect domain protein expression was much higher for Hect mutants than for the wild-type proteins. Using the Hect Cys-to-Ala mutant proteins, LMP2A was found to coimmunoprecipitate with AIP4 or KIAA0439 (Fig. 5) but did not detectably interact with Nedd4 (Fig. 6A). In this assay we investigated the specificity of the interaction of LMP2A with AIP4 or KIAA0439 and analyzed the role of the PPPPY motifs in the formation of these complexes by expressing engineered LMP2A derivatives that contained tyrosine-to-alanine substitutions in either or both PPPPY motifs. These substitutions were based on previous biochemical and structural data demonstrating that the corresponding tyrosine is critical for the recognition of PPXY sites by a subclass of WW domains (7). The LMP2A mutant proteins containing PPPPA substitutions were stably expressed in Cos-1 cells. However, no interaction between LMP2A and the Nedd4 family proteins was observed in transfected cells when both PPPPY sites were changed to PPPPA (Y60A/Y101A). The single substitution of Tyr60 in the more N-terminal PPPPY site (Y60A) strongly reduced the extent of complex formation but did not entirely abrogate binding of LMP2A to AIP4 or KIAA0439. Substitution of Tyr101 alone in the C-terminal PPPPY motif (Y101A) resulted in a more modest decrease in LMP2A binding to Nedd4 family members. These results suggest that both PPPPY motifs can contribute to the association of LMP2A with AIP4 and KIAA0439 in vivo and that together they are essential for complex formation. Furthermore, the more N-terminal PPPPY motif appears to be more efficient in binding to AIP4 and KIAA0439 in cells than is the more C-terminal PPPPY site.
FIG. 5.
The LMP2A PPPPY motifs are essential for binding E3 ubiquitin-ligase proteins KIAA0439 and AIP4. Wild-type and PPPPY motif mutants (YY, wild type; AA, Y60A/Y101A; YA, Y101A; AY, Y60A) of a FLAG epitope-tagged LMP2A mammalian expression vector were transfected into Cos-1 cells with either MYC epitope-tagged KIAA0439 or AIP4 expression vectors. Lysates were then subjected to anti-FLAG immunoprecipitations (IP), and associated proteins were identified by SDS–8% PAGE with Western blotting for MYC epitope-tagged protein. WB, Western blotting; WCL, whole-cell lysate.
FIG. 6.
Nedd4 can be precipitated with N-terminal LMP2A GST fusion protein but does not associate with full-length LMP2A when both proteins are overexpressed in Cos-1 cells. (A) Wild type (YY) and PPPPY motif (AA, Y60A/Y101A; YA, Y101A; AY, Y60A) mutants of a FLAG epitope-tagged LMP2A mammalian expression vector were transfected into Cos-1 cells with the T7 epitope-tagged Nedd4 expression vector. Lysates were then subjected to anti-FLAG immunoprecipitations (IP), and associated protein was identified by SDS–8% PAGE with Western blotting (WB) for T7 epitope-tagged protein. (B) Cos-1 cell lysates containing T7 epitope-tagged Nedd4 protein were treated with immobilized GST or GST fusion proteins containing the N-terminal 122 amino acid residues of wild-type LMP2A or PPPPY mutants. Associated Nedd4 was identified by SDS–9% PAGE with Western blotting (WB) for the T7 epitope-tagged protein. WCL, whole-cell lysate.
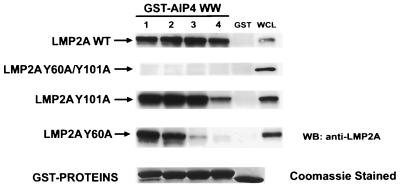
To investigate which of the four WW domains of AIP4 has the potential to interact with LMP2A, GST fusion proteins containing individual WW domains were tested for their ability to associate with wild-type or mutant forms of LMP2A in cell lysates (Fig. 7). Each of the four AIP4 WW domains bound to wild-type LMP2A but not to the Y60A/Y101A mutant lacking both Tyr60 and Tyr101 in the two PPPPY motifs. WW domains 1 to 3 also bound to the LMP2A Y101A mutant, but binding of this mutant to WW domain 4 was markedly reduced. WW domains 1 and 2 still bound to the Y60A LMP2A mutant but less efficiently than to the wild-type protein or to the Y101A mutant, consistent with the in vivo data that the N-terminal PPPPY motif is more effective in binding AIP4 than is the C-terminal motif. Furthermore, WW domain 3 bound poorly to the Y60A mutant, and binding by WW domain 4 was barely detectable. These results indicate that all four of the AIP4 WW domains have the capacity to interact with wild-type LMP2A and that WW domains 1 and 2 can recognize both PPPPY motifs. WW domain 3 bound preferentially to the N-terminal PPPPY motif, and WW domain 4 showed only weak binding to the individual sites. Thus, it is likely that a single AIP4 molecule can simultaneously interact with both PPPPY motifs on an individual LMP2A chain through two distinct WW domains. We cannot exclude the possibility that the two PPPPY motifs within a single chain of LMP2A could bind independently to two separate AIP4 molecules, but it seems probable that a bidentate interaction would be favored. In either case, the remaining WW domains might engage additional LMP2A molecules or contact other ligands, potentially leading to the formation of a larger complex. It is interesting to note that AIP4 WW domain 4 lacks the more C-terminal of two conserved Trp residues, which is replaced by a Tyr in AIP4 WW domain 4. Since this Trp forms part of the ligand-binding surface of other WW domains, its absence in WW domain 4 might influence the efficiency or specificity of ligand recognition. Regardless, our data indicate that there are some differences in the ligand-binding properties of the four AIP4 WW domains.
FIG. 7.
Individual AIP4 WW domains can precipitate LMP2A. Immobilized GST fusion proteins of individual AIP4 WW domains were tested for their ability to precipitate from lysates of wild-type LMP2A (LMP2A WT) or Y60A/Y101A, Y101A, or Y60A LMP2A mutant proteins expressed transiently in DG75 B cells. Western blotting (WB) with anti-LMP2A antibody identified precipitated LMP2A. The numerical designation of each AIP4 WW domain corresponds to its position from the N terminus of the protein. WCL, whole-cell lysate.
Although we were unable to detect a complex between Nedd4 and LMP2A in a cotransfection assay, a GST fusion protein which contained the N-terminal 122 amino acids of LMP2A was able to precipitate Nedd4 from a lysate of transfected Cos-1 cells (Fig. 6B). Using this approach, we investigated the specificity of Nedd4 towards individual LMP2A PPPPY sites and found a selectivity for intact LMP2A similar to that observed with AIP4 and KIAA0439. However, the failure to identify Nedd4 in association with LMP2A in transfected cells suggests that the WW domains of Nedd4 are likely to have a lower affinity for the LMP2A PPPPY motifs than are the WW domains of AIP4 or KIAA0439. Alternatively, it is possible that Nedd4 is less accessible to LMP2A in vivo, for example through localization to a distinct subcellular compartment. These data provide evidence for selective binding of specific C2/WW/Hect domain proteins to LMP2A.
An endogenous complex between LMP2A and C2/WW/Hect domain proteins.
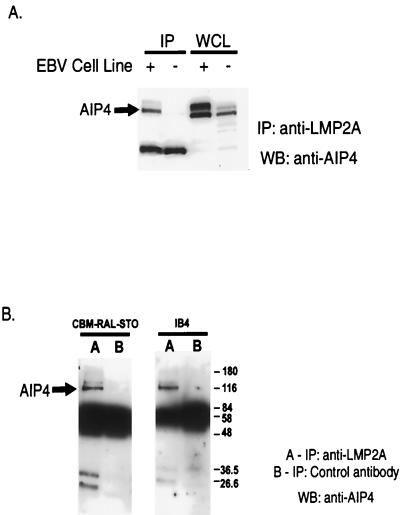
To investigate whether an endogenous complex can form between EBV LMP2A and the Nedd4 family of proteins in human B cells, we raised a polyclonal rabbit antiserum to the AIP4 protein, which cross-reacts with the closely related KIAA0439 polypeptide (data not shown). We then investigated whether these proteins could coimmunoprecipitate with LMP2A, using either the EBV-negative BJAB B-cell line as a control or the EBV-positive CBM-RAL-STO line. Cell lysates were immunoprecipitated using a rat monoclonal antibody directed against LMP2A, and the precipitated proteins were separated and blotted with anti-AIP4 antiserum. This assay identified complexes between endogenous LMP2A and both AIP4 and KIAA0439, which were specifically detected in the EBV-positive CBM-RAL-STO B-cell line (Fig. 8A). As shown in Fig. 8B, whereas LMP2A antibody was able to coprecipitate AIP4 from EBV-positive CBM-RAL-STO and IB4 cell lines, a control rat monoclonal antibody directed against v-Ras was not. This evidence lends further support to the hypothesis that a constitutive complex between LMP2A and C2/WW/Hect domain proteins exists in EBV-infected B cells.
FIG. 8.
LMP2A associates with endogenous AIP4 in EBV-positive B-cell lines. (A) LMP2A immunoprecipitations, using rat anti-LMP2A MAb 4E11 from cell lysates of either an EBV-negative (−) cell line, BJAB, or an EBV-positive (+) B-cell line, CBM-RAL-STO, were probed for the presence of AIP4 by Western blotting using anti-AIP4 antibodies. (B) LMP2A immunoprecipitations and control immunoprecipitations using an irrelevant anti-rat MAb IgG1 from cell lysates of two EBV-positive B-cell lines, CBM-RAL-STO and IB4, were probed for AIP4. IP, immunoprecipitation; WB, Western blotting, WCL, whole-cell lysate. Numbers are molecular mass in kilodaltons.
The N-terminal cytoplasmic region of LMP2A is sufficient to bind AIP4 in cells.
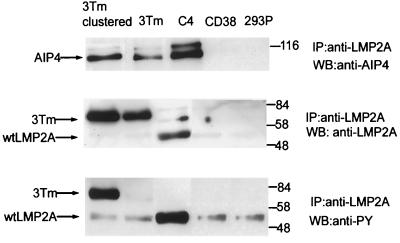
The data presented above suggest that AIP4 and related E3 protein-ubiquitin ligases form an endogenous complex with LMP2A in a fashion that is mediated by the AIP4 WW domains and the PPPPY motifs of LMP2A. To investigate whether the N-terminal region of LMP2A containing the PPPPY sequences is sufficient to bind AIP4 in cells, we made use of a chimeric protein in which the extracellular and transmembrane regions of the type II transmembrane protein CD38 are fused to the N-terminal 177 amino acids of LMP2A. In this chimeric protein, the N-terminal region of LMP2A should adopt its normal orientation with respect to the membrane. Although the CD38-LMP2A chimera is expressed at the cell surface of HEK 293 cells (L. Matskova and G. Winberg, unpublished results), it is not significantly tyrosine phosphorylated until it is cross-linked with anti-CD38 antibodies (Fig. 9). Nonetheless, the CD38-LMP2A protein coprecipitated with AIP4 from a lysate of HEK 293 cells even prior to cross-linking. These data indicate firstly that the N-terminal region of LMP2A is both necessary and sufficient for binding to AIP4 and secondly that this interaction is not dependent on tyrosine phosphorylation.
FIG. 9.
The N-terminal cytoplasmic region of LMP2A is sufficient to bind AIP4. Four HEK 293 cell lines that stably express full-length LMP2A (C4), a CD38-LMP2A chimeric molecule (3Tm), a control vector expressing the CD38 portion of the chimeric molecule (CD38), or vector alone (293P) were tested for their ability to form LMP2A-AIP4 complexes with endogenous AIP4. Clustering CD38-LMP2A, through treatment of the cells with anti-CD38 antibody, shows little effect on association with AIP4. However, clustering dramatically increases the level of tyrosine phosphorylation, as determined by immunoblotting with anti-phosphotyrosine (anti-PY) 4G10 antibody. Full-length LMP2A is also tyrosine phosphorylated when expressed in HEK 293 cells. IP, immunoprecipitation; WB, Western blotting; wt, wild type. Numbers are molecular mass in kilodaltons.
Ubiquitination of Lyn and Syk in LMP2A-containing cells.
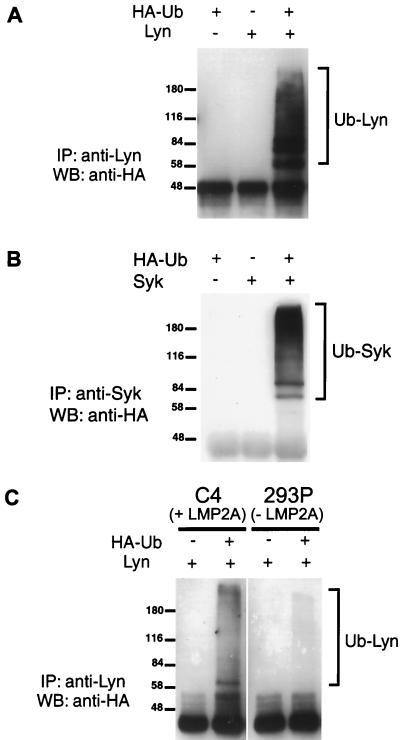
These results suggest that LMP2A serves as a scaffold to recruit not only cytoplasmic tyrosine kinases through SH2-mediated interactions but also E3 protein-ubiquitin ligases through the binding of PPPPY motifs to specific WW domain proteins. Thus, LMP2A may function not only to physically recruit signaling proteins involved in BCR signaling but may potentially target such proteins for ubiquitination and degradation. In an effort to establish a functional role for the formation of a complex between LMP2A and the Nedd4 class of E3 protein-ubiquitin ligases, we investigated the possibility that the other proteins known to associate with LMP2A might undergo ubiquitination. We focused our attention on the Src family tyrosine kinase Lyn, since previous work has indicated that the level of Lyn is reduced in LMP2A-positive B-cell lines (34). Furthermore, recent data have suggested that SFKs are prone to ubiquitin-mediated degradation in the activated state (21, 22, 35). In addition, we tested the B-cell tyrosine kinase Syk for ubiquitination. The assay for ubiquitination involved cotransfection of HEK 293 cells with the relevant kinase and a vector encoding hemagglutinin (HA)-tagged ubiquitin and monitoring for ubiquitinated species by blotting of either anti-Lyn or Syk immunoprecipitates with anti-HA antibody. As shown in Fig. 10A and B, both Lyn and Syk are polyubiquitinated in this assay. These higher-molecular-weight species were not observed in the absence of transfected HA-tagged ubiquitin or substrate kinase.
FIG. 10.
Lyn and Syk undergo ubiquitination; Lyn ubiquitination is augmented in an LMP2A-expressing cell line. (A) HEK 293 cells were cotransfected with Lyn and HA-ubiquitin expression vectors along with control transfections using each vector separately. Ubiquitinated Lyn species were isolated by immunoprecipitation with anti-Lyn antibody, resolved by SDS–8.3% PAGE, and detected by Western blotting using anti-HA antibody. (B) Ubiquitination of Syk in HEK 293 cells was monitored as described for panel A, with transfection of the Syk expression vector and immunoprecipitation using anti-Syk antibody. (C) Lyn was transfected into either a HEK 293 cell line stably expressing LMP2A (C4) or a control cell line (293P) in the presence or absence of an HA-ubiquitin expression vector. Ubiquitinated Lyn species were isolated by immunoprecipitation with anti-Lyn antibody and detected by Western blotting using anti-HA antibody. Numbers are molecular mass in kilodaltons. IP, immunoprecipitation; WB, Western blotting; Ub, ubiquitin; +, DNA vector added; −, DNA vector not added.
To test whether LMP2A might play a role in catalyzing Lyn ubiquitination, we expressed Lyn either in a HEK 293 line that stably expresses EBV LMP2A (C4) or in a control cell line (293P). The results are shown in Fig. 10C. In the control 293P cell line, Lyn was converted to higher-molecular-weight species recognized by anti-HA antibodies, indicative of ubiquitination. These higher-molecular-weight species were not observed in the absence of HA-tagged ubiquitin. The abundance of ubiquitinated Lyn species identified by this assay was substantially increased in the C4 cell line, which expresses LMP2A. These results suggest that the Lyn tyrosine kinase has a propensity to undergo ubiquitination in 293 cells, which is enhanced by the coexpression of LMP2A.
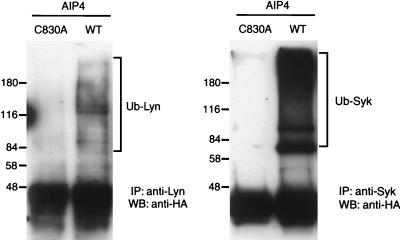
As shown in Fig. 9, the HEK 293-derived cell line stably expressing LMP2A (C4) contains endogenous E3 protein-ubiquitin ligases, including AIP4 and KIAA0439, which are potentially responsible for Lyn ubiquitination. To test this possibility, we expressed either wild-type AIP4 or the C830A AIP4 mutant (which removes a Cys essential for HECT domain catalytic activity) in LMP2A-expressing C4 cells also transfected with Lyn and HA-tagged ubiquitin (Fig. 11). Since the C830A protein can readily bind to LMP2A through its WW domains, it would be anticipated to have a dominant-negative function. Although wild-type AIP4 did not significantly increase Lyn ubiquitination, indicating that E3 activity is not limiting in this assay, the C830A dominant-negative mutant effectively blocked the appearance of ubiquitinated Lyn species. These results are consistent with the notion that Nedd4 family members are responsible for LMP2A-mediated Lyn ubiquitination in 293 cells.
FIG. 11.
An inactive HECT domain form of AIP4 reduces the level of Lyn and Syk ubiquitination in LMP2A-expressing HEK 293 cells. A HEK 293-derived cell line that stably expresses LMP2A (C4) was used for transfection of HA-ubiquitin with either Lyn or Syk expression vectors. Also transfected was wild-type AIP4 (WT) or an inactive form of AIP4 (C830A). The presence of ubiquitinated Lyn (Ub-Lyn) or Syk (Ub-Syk) was detected by immunoprecipitation (IP) with anti-Lyn or anti-Syk antibody, resolution of associated proteins by SDS–8% PAGE, and Western blotting (WB) using anti-HA antibody. Numbers are molecular mass in kilodaltons.
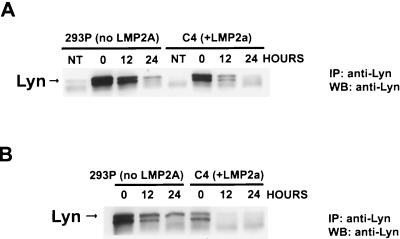
To investigate whether LMP2A influences Lyn abundance, we measured Lyn levels in lysates from C4 cells, which express LMP2A, or control 293P cells transfected with Lyn cDNA (Fig. 12). When cycloheximide was used to block protein synthesis, Lyn levels in the control 293P cells were decreased by 10% following 12 h of cycloheximide treatment; however, in the C4 LMP2A-expressing cells, Lyn levels were decreased by 60%. Similar results regarding Lyn stability were obtained when both Lyn and Syk cDNAs were transfected. These results are consistent with LMP2A acting to destabilize Lyn protein.
FIG. 12.
Lyn protein levels are reduced in the presence of LMP2A. (A) Lyn cDNA was transfected into a HEK 293 cell line stably expressing LMP2A (C4) or a control cell line (293P). Prior to isolation of lysate, cells were treated with 35 μM cycloheximide for the indicated period. The abundance of Lyn in lysates of equal protein concentration was determined by immunoprecipitation (IP) with anti-Lyn antibody followed by SDS–8.3% PAGE and detection by Western blotting (WB) using anti-Lyn antibody. (B) Both Lyn and Syk cDNA were transfected, and the experiment was performed as described above.
Previous work has suggested that the stability of Syk is not affected by LMP2A expression in LCLs. Surprisingly, however, we have found that Syk is also ubiquitinated in C4 cells cotransfected with Syk and HA-ubiquitin expression vectors. In addition, Syk ubiquitination is blocked in C4 cells expressing the C830A AIP4 mutant (Fig. 11). Thus far, we have not observed any effect of LMP2A on Syk stability, although it is possible that ubiquitination has a subtle effect on a subset of activated Syk molecules bound to LMP2A. Alternatively, Syk ubiquitination may not lead to its destruction but could potentially result in its functional inactivation.
DISCUSSION
The PPPPY motifs of LMP2A are highly conserved among different EBV-related viruses, such as the rhesus monkey herpesvirus (cercopithecine herpesvirus 15), suggesting that they are functionally important. Both by probing cDNA expression libraries with biotinylated peptides corresponding to the LMP2A PPPPY motifs and by using such peptides to isolate LMP2A-binding proteins from B-cell lysates, we have identified a family of WW domain proteins that selectively bind the LMP2A PPPPY motifs.
WW domains are small protein modules of ∼30 amino acids that form a three-stranded antiparallel β-sheet capable of binding proline-rich motifs that adopt a polyproline type II helix (31, 41, 55). The prototypic family of WW domains, which includes the Nedd4 E3 protein-ubiquitin ligase, recognizes motifs with the consensus PPXY (6, 7, 53), although additional WW domain families bind distinct proline-rich motifs, such as PPPPLP in the case of formin-binding proteins (3). Nedd4 is the founding member of a group of E3 enzymes with an N-terminal C2 domain, involved in membrane localization, multiple WW domains, and a C-terminal Hect domain that serves to transfer ubiquitin from an E2 enzyme to a target protein bound to the WW domains (23). The WW domains of Nedd4, for example, bind PY motifs in the β and γ subunits of the epithelial Na+ channel (EnaC), leading to targeted proteolysis of the channel. Mutations in the EnaC β and γ subunits which abrogate this interaction cause a hypertensive disorder, Liddle's syndrome (48, 51).
Although the WW domains of Nedd4 bind LMP2A in vitro, we have been unable to detect an association between these two proteins in cells. Rather, LMP2A appears to preferentially bind the WW domains of related Hect domain proteins, notably AIP4 and KIAA0439. We have investigated the basis for this interaction in a number of different ways. First, AIP4 and KIAA0439 were initially isolated through their ability to bind a PPPPY peptide derived from the LMP2A N-terminal region and were identified by mass spectrometry. Using epitope-tagged forms of AIP4 and KIAA0439 and LMP2A, we found that the WW domains of these proteins bound to LMP2A both in GST pull-down and coimmunoprecipitation experiments with transfected Cos-1 cells. The ability of LMP2A to recognize AIP4 or KIAA0439 was dependent on the LMP2A PPPPY motifs. Substitution of the Tyr residue in each PPPPY motif with Ala caused a decrease in binding, whereas substitution of both PPPPY Tyr residues entirely abrogated binding of LMP2A to AIP4 and KIAA0439. Since LMP2A binds stably to AIP4 and KIAA0439 but not to the closely related Nedd4, these data indicate that the PPPPY motifs of LMP2A may contain residues that confer specificity for the AIP4 and KIAA0439 WW domains. Furthermore, AIP4 can potentially bind to LMP2A through multiple WW domains, raising the possibility that these act cooperatively to bind LMP2A. Consistent with the notion that the N-terminal cytoplasmic region of LMP2A is both necessary and sufficient for binding to AIP4, a CD38-LMP2A chimera, containing only the N-terminal 177 residues of LMP2A, associated with AIP4 in 293 cells. This interaction was independent of clustering with anti-CD38 antibodies, whereas tyrosine phosphorylation of CD38-LMP2A required cross-linking. Thus, the binding of C2/WW/Hect domain proteins to LMP2A can be dissociated from recognition by tyrosine kinases.
The association of AIP4 and KIAA0439 with LMP2A is apparently of physiological significance, since coimmunoprecipitation experiments using LMP2A-positive B cells have identified endogenous complexes of AIP4 and KIAA0439 with EBV LMP2A. Furthermore, transfection of tagged LMP2A and AIP4 into the EBV-negative B-cell line DG75 was sufficient to elicit a stable complex between AIP4 and LMP2A.
These data strongly suggest that LMP2A recruits specific C2/WW/Hect domain E3 protein-ubiquitin ligases, which may then selectively target B-cell signaling proteins for ubiquitination and degradation. Of interest, recent results indicate that SFKs are sensitive to ubiquitination by Hect domain E3 enzymes. Indeed, SFKs physically interact with E6AP, the prototypic Hect domain protein (35), and activated forms of SFKs are preferential substrates for ubiquitin-mediated degradation (21, 22). Consistent with these observations, we find that Lyn becomes ubiquitinated in HEK 293 cells in a fashion that is markedly enhanced by the expression of LMP2A. Furthermore, Lyn ubiquitination is suppressed by a mutant form of AIP4 with a substitution of a conserved Cys in the Hect domain, which forms a thioester linkage with ubiquitin. The C830A AIP4 mutant likely acts as a dominant negative to poison the protein complexes required for Lyn ubiquitination. Consistent with a role for LMP2A in promoting Lyn degradation, Lyn was destabilized in HEK 293 cells transfected with LMP2A, compared with wild-type cells.
Surprisingly, we also found that Syk could undergo ubiquitination in C4 293 cells, in a fashion that was blocked by expression of the C830A AIP4 mutant. Although Syk levels, unlike Lyn, are not reported to decrease as a consequence of LMP2A expression in LCLs, it is nonetheless possible that LMP2A acts to recruit a small subpopulation of activated Syk which is targeted for destruction. Syk ubiquitination might also directly impair Syk kinase activity.
While this study was in preparation, Ikeda et al. (25) also reported the identification of AIP4 and WWP2/AIP2 in association with LMP2A. Our results are generally consistent with this observation. However, we extend their observations by demonstrating that LMP2A binding partners include KIAA0439. Furthermore, we show that AIP4 can potentially interact with LMP2A through multiple WW domains. Importantly, we show that LMP2A is able to promote the ubiquitination of B-cell tyrosine kinases in a fashion that appears dependent on the activity of Nedd4 family E3 protein-ubiquitin ligases and correlates with destabilization of the Lyn tyrosine kinase. Furthermore, we set these observations in a physiological context by establishing that endogenous AIP4 and LMP2A form a complex in EBV-positive B cells.
Taken together, these data suggest a modified view of LMP2A function in regulating B-cell signaling. SFKs evidently phosphorylate LMP2A at Tyr 112 and bind to this site through their SH2 domain. In this form, the SFK must be in an enzymatically active state, since its SH2 domain is no longer available to make an intramolecular interaction with the inhibitory pTyr site in its own C-terminal tail. The activated SFK then phosphorylates the ITAM motif, resulting in Syk recruitment. However, the LMP2A-bound SFK is still active and might stimulate rather than antagonize BCR signaling. The recruitment of C2/WW/Hect domain proteins to LMP2A may therefore serve to limit the pool of activated SFKs by promoting their ubiquitination and degradation. It will be of interest in the future to explore the consequences of Syk ubiquitination. Clearly, there may be other, as yet unidentified, targets for ubiquitination by AIP4 and related E3 enzymes that contribute to the effects of EBV LMP2A.
This scheme suggests that the functional output of LMP2A depends on the coordinate binding of multiple modular proteins. LMP2A might therefore have different biological activities in distinct cell types or during different activation states of the B cell, depending on the relative expression of its SH2 and WW domain ligands.
The ability of LMP2A to promote AIP4-mediated ubiquitination of Lyn and Syk raises the question whether the viral protein is simply potentiating an existing mechanism through which antigen receptor signaling is negatively regulated. That this might be the case is suggested by the phenotype of mice with a loss-of-function mutation in the AIP4/Itch gene (37). On a C57BL/6J background, such mice develop hyperplasia of lymphoid, hematopoietic, and epithelial cells, leading to an inflammatory syndrome which is ultimately lethal. This suggests that AIP4 plays a physiological role in restraining signaling in lymphoid cells, potentially through the targeted proteolysis of SFKs. In this vein, it will be of interest to identify physiological binding partners for the AIP4 WW domains.
ACKNOWLEDGMENTS
This work was supported by grants from the Medical Research Council of Canada and the National Cancer Institute of Canada to T.P. and from the Swedish Cancer Society to I.E. and grant no. K1999-06X-012622-02B from the Swedish Medical Research Council to G.W. and L.M. R.I. is a Research Fellow of the National Cancer Institute of Canada supported with funds provided by the Terry Fox Run.
REFERENCES
- 1.Alber G, Kim K-M, Weiser P, Riesterer C, Carsetti R, Reth M. Molecular mimicry of the antigen receptor signalling motif by transmembrane proteins of the Epstein-Barr virus and the bovine leukaemia virus. Curr Biol. 1993;3:333–339. doi: 10.1016/0960-9822(93)90196-u. [DOI] [PubMed] [Google Scholar]
- 2.Beaufils P, Choquest D, Mamoun R Z, Malissen B. The (YXXL/I)2 signalling motif found in the cytoplasmic segments of the bovine leukaemia virus envelope protein and Epstein-Barr virus latent membrane protein 2A can elicit early and late lymphocyte activation events. EMBO J. 1993;12:5105–5112. doi: 10.1002/j.1460-2075.1993.tb06205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bedford M T, Chan D C, Leder P. FBP WW domains and the Abl SH3 domain bind to a specific class of proline-rich ligands. EMBO J. 1997;16:2376–2383. doi: 10.1093/emboj/16.9.2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burkhardt A L, Bolen J B, Kieff E, Longnecker R. An Epstein-Barr virus transformation-associated membrane protein interacts with src family tyrosine-kinases. J Virol. 1992;66:5161–5167. doi: 10.1128/jvi.66.8.5161-5167.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caldwell R G, Brown R C, Longnecker R. Epstein-Barr virus LMP2A-induced B-cell survival in two unique classes of EmuLMP2A transgenic mice. J Virol. 2000;74:1101–1113. doi: 10.1128/jvi.74.3.1101-1113.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen H I, Einbond A, Kwak S J, Linn H, Koepf E, Peterson S, Kelly J W, Sudol M. Characterization of the WW domain of human yes-associated protein and its polyproline-containing ligands. J Biol Chem. 1997;272:17070–17077. doi: 10.1074/jbc.272.27.17070. [DOI] [PubMed] [Google Scholar]
- 7.Chen H I, Sudol M. The WW domain of Yes-associated protein binds a proline-rich ligand and differs from consensus established for Src homology 3-binding modules. Proc Natl Acad Sci USA. 1995;92:7819–7823. doi: 10.1073/pnas.92.17.7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng A M, Chan A C. Protein tyrosine kinases in thymocyte development. Curr Opin Immunol. 1997;9:528–533. doi: 10.1016/s0952-7915(97)80106-9. [DOI] [PubMed] [Google Scholar]
- 9.Cheng A M, Rowley B, Pao W, Hayday A, Bolen J B, Pawson T. Syk tyrosine kinase required for mouse viability and B-cell development. Nature. 1995;378:303–306. doi: 10.1038/378303a0. [DOI] [PubMed] [Google Scholar]
- 10.Coffey A J, Brooksbank R A, Brandau O, Oohashi T, Howell G R, Bye J M, Cahn A P, Durham J, Heath P, Wray P, Pavitt R, Wilkinson J, Leversha M, Huckle E, Shaw-Smith C J, Dunham A, Rhodes S, Schuster V, Porta G, Yin L, Serafini P, Sylla B, Zollo M, Franco B, Bentley D R. Host response to EBV infection in X-linked lymphoproliferative disease results from mutations in an SH2-domain encoding gene. Nat Genet. 1998;20:129–135. doi: 10.1038/2424. [DOI] [PubMed] [Google Scholar]
- 11.Cornall R J, Cheng A M, Pawson T, Goodnow C C. Role of Syk in B-cell development and antigen-receptor signaling. Proc Natl Acad Sci USA. 2000;97:1713–1718. doi: 10.1073/pnas.97.4.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeFranco A L. Transmembrane signaling by antigen receptors of B and T lymphocytes. Curr Opin Cell Biol. 1995;7:163–175. doi: 10.1016/0955-0674(95)80024-7. [DOI] [PubMed] [Google Scholar]
- 13.DeFranco A L. The complexity of signaling pathways activated by the BCR. Curr Opin Immunol. 1997;9:296–308. doi: 10.1016/s0952-7915(97)80074-x. [DOI] [PubMed] [Google Scholar]
- 14.Fruehling S, Lee S K, Herrold R, Frech B, Laux G, Kremmer E, Grasser F A, Longnecker R. Identification of latent membrane protein 2A (LMP2A) domains essential for the LMP2A dominant-negative effect on B-lymphocyte surface immunoglobulin signal transduction. J Virol. 1996;70:6216–6226. doi: 10.1128/jvi.70.9.6216-6226.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fruehling S, Longnecker R. The immunoreceptor tyrosine-based activation motif of Epstein-Barr virus LMP2A is essential for blocking BCR-mediated signal transduction. Virology. 1997;235:241–251. doi: 10.1006/viro.1997.8690. [DOI] [PubMed] [Google Scholar]
- 16.Fruehling S, Swart R, Dolwick K M, Kremmer E, Longnecker R. Tyrosine 112 of latent membrane protein 2A is essential for protein tyrosine kinase loading and regulation of Epstein-Barr virus latency. J Virol. 1998;72:7796–7806. doi: 10.1128/jvi.72.10.7796-7806.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fu C, Turck C W, Kurosaki T, Chan A C. BLNK: a central linker protein in B cell activation. Immunity. 1998;9:93–103. doi: 10.1016/s1074-7613(00)80591-9. [DOI] [PubMed] [Google Scholar]
- 18.Futterer K, Wong J, Grucza R A, Chan A C, Waksman G. Structural basis for Syk tyrosine kinase ubiquity in signal transduction pathways revealed by the crystal structure of its regulatory SH2 domains bound to a dually phosphorylated ITAM peptide. J Mol Biol. 1998;281:523–537. doi: 10.1006/jmbi.1998.1964. [DOI] [PubMed] [Google Scholar]
- 19.Gires O, Kohlhuber F, Kilger E, Baumann M, Kieser A, Kaiser C, Zeidler R, Scheffer B, Ueffing M, Hammerschmidt W. Latent membrane protein 1 of Epstein-Barr virus interacts with JAK3 and activates STAT proteins. EMBO J. 1999;18:3064–3073. doi: 10.1093/emboj/18.11.3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gires O, Zimber-Strobl U, Gonnella R, Ueffing M, Marschall G, Zeidler R, Pich D, Hammerschmidt W. Latent membrane protein 1 of Epstein-Barr virus mimics a constitutively active receptor molecule. EMBO J. 1997;16:6131–6140. doi: 10.1093/emboj/16.20.6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hakak Y, Martin G S. Ubiquitin-dependent degradation of active Src. Curr Biol. 1999;9:1039–1042. doi: 10.1016/s0960-9822(99)80453-9. [DOI] [PubMed] [Google Scholar]
- 22.Harris K F, Shoji I, Cooper E M, Kumar S, Oda H, Howley P M. Ubiquitin-mediated degradation of active Src tyrosine kinase. Proc Natl Acad Sci USA. 1999;96:13738–13743. doi: 10.1073/pnas.96.24.13738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harvey K F, Kumar S. Nedd4-like proteins: an emerging family of ubiquitin-protein ligases implicated in diverse cellular functions. Trends Cell Biol. 1999;9:166–169. doi: 10.1016/s0962-8924(99)01541-x. [DOI] [PubMed] [Google Scholar]
- 24.Hausen A, Brink A A, Craanen M E, Middeldorp J M, Meijer C J, van der Brule A J. Unique transcription pattern of Epstein-Barr virus (EBV) in EBV-carrying gastric adenocarcinomas: expression of the transforming BARF1 gene. Cancer Res. 2000;60:2745–2748. [PubMed] [Google Scholar]
- 25.Ikeda M, Ikeda A, Longan L C, Longnecker R. The Epstein-Barr virus latent membrane protein 2A PY motif recruits WW domain-containing ubiquitin-protein ligases. Virology. 2000;268:178–191. doi: 10.1006/viro.1999.0166. [DOI] [PubMed] [Google Scholar]
- 26.Ishiai M, Kurosaki M, Pappu R, Okawa K, Ronko I, Fu C, Shibata M, Iwamatsu A, Chan A C, Kurosaki T. BLNK required for coupling Syk to PLC gamma 2 and Rac1-JNK in B cells. Immunity. 1999;10:117–125. doi: 10.1016/s1074-7613(00)80012-6. [DOI] [PubMed] [Google Scholar]
- 27.Kieff E. Epstein-Barr virus and its replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2343–2396. [Google Scholar]
- 28.Kumar S, Tomooka Y, Noda M. Identification of a set of genes with developmentally down-regulated expression in the mouse brain. Biochem Biophys Res Commun. 1992;185:1155–1161. doi: 10.1016/0006-291x(92)91747-e. [DOI] [PubMed] [Google Scholar]
- 29.Laux G, Perricaudet M, Farrell P J. A spliced Epstein-Barr virus gene expressed in immortalized lymphocytes is created by circularization of the linear viral genome. EMBO J. 1988;7:769–774. doi: 10.1002/j.1460-2075.1988.tb02874.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Longnecker R, Miller C L. Regulation of Epstein-Barr virus latency by latent membrane protein 2. Trends Microbiol. 1996;4:38–42. doi: 10.1016/0966-842x(96)81504-6. [DOI] [PubMed] [Google Scholar]
- 31.Macias M J, Hyvonen M, Baraldi E, Schultz J, Sudol M, Saraste M, Oschkinat H. Structure of the WW domain of a kinase-associated protein complexed with a proline-rich peptide. Nature. 1996;382:646–649. doi: 10.1038/382646a0. [DOI] [PubMed] [Google Scholar]
- 32.Masucci M G, Ernberg I. Epstein-Barr virus: adaptation to a life within the immune system. Trends Microbiol. 1994;2:125–130. doi: 10.1016/0966-842x(94)90599-1. [DOI] [PubMed] [Google Scholar]
- 33.Miller A D, Rosman G J. Improved retroviral vectors for gene transfer and expression. BioTechniques. 1989;7:980–982. , 984–986, 989–990. [PMC free article] [PubMed] [Google Scholar]
- 34.Miller C L, Burkhardt A L, Lee J H, Stealey B, Longnecker R, Bolen J B, Kieff E. Integral membrane protein 2 of Epstein-Barr virus regulates reactivation from latency through dominant negative effects on protein-tyrosine kinases. Immunity. 1995;2:155–166. doi: 10.1016/s1074-7613(95)80040-9. [DOI] [PubMed] [Google Scholar]
- 35.Oda H, Kumar S, Howley P M. Regulation of the Src family tyrosine kinase Blk through E6AP mediated ubiquitination. Proc Natl Acad Sci USA. 1999;96:9957–9562. doi: 10.1073/pnas.96.17.9557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Panousis C G, Rowe D T. Epstein-Barr virus latent membrane protein 2 associates with and is a substrate for mitogen-activated protein kinase. J Virol. 1997;71:4752–4760. doi: 10.1128/jvi.71.6.4752-4760.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perry W L, Hustad C M, Swing D A, O'Sullivan T N, Jenkins N A, Copeland N G. The itchy locus encodes a novel ubiquitin protein ligase that is disrupted in a 18H mice. Nat Genet. 1998;18:143–146. doi: 10.1038/ng0298-143. [DOI] [PubMed] [Google Scholar]
- 38.Pirozzi G, McConnell S J, Uveges A J, Carter J M, Sparks A B, Kay B K, Fowlkes D M. Identification of novel human WW domain-containing proteins by cloning of ligand targets. J Biol Chem. 1997;272:14611–14616. doi: 10.1074/jbc.272.23.14611. [DOI] [PubMed] [Google Scholar]
- 39.Plant P J, Yeger H, Staub O, Howard P, Rotin D. The C2 domain of the ubiquitin protein ligase Nedd4 mediates Ca2+-dependent plasma membrane localization. J Biol Chem. 1997;272:32329–32336. doi: 10.1074/jbc.272.51.32329. [DOI] [PubMed] [Google Scholar]
- 40.Raab-Traub N, Flynn K, Klein C, Pizza G, De Vinci C, Occhiuzzi L, Farneti G, Caliceti U, Pirodda E. EBV associated malignancies. J Exp Pathol. 1987;3:449–456. [PubMed] [Google Scholar]
- 41.Ranganathan R, Lu K P, Hunter T, Noel J P. Structural and functional analysis of the mitotic rotamase Pin1 suggests substrate recognition is phosphorylation dependent. Cell. 1997;89:875–886. doi: 10.1016/s0092-8674(00)80273-1. [DOI] [PubMed] [Google Scholar]
- 42.Reth M. Antigen receptor tail clue. Nature. 1989;338:383–384. [PubMed] [Google Scholar]
- 43.Rotin D. WW (WWP) domains: from structure to function. Curr Top Microbiol Immunol. 1998;228:115–133. doi: 10.1007/978-3-642-80481-6_5. [DOI] [PubMed] [Google Scholar]
- 44.Rowe M, Lear A L, Croom-Carter D, Davies A H, Rickinson A B. Three pathways of Epstein-Barr virus gene activation from EBNA1-positive latency in B lymphocytes. J Virol. 1992;66:122–131. doi: 10.1128/jvi.66.1.122-131.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rowe M, Rowe D T, Gregory C D, Young L S, Farrell P J, Rupani H, Rickinson A B. Differences in B cell growth phenotype reflect novel patterns of Epstein-Barr virus latent gene expression in Burkitt's lymphoma cells. EMBO J. 1987;6:2743–2751. doi: 10.1002/j.1460-2075.1987.tb02568.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scheffner M, Huibregtse J M, Vierstra R D, Howley P M. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell. 1993;75:495–505. doi: 10.1016/0092-8674(93)90384-3. [DOI] [PubMed] [Google Scholar]
- 47.Scheffner M, Nuber U, Huibregtse J M. Protein ubiquitination involving an E1–E2-E3 enzyme ubiquitin thioester cascade. Nature. 1995;373:81–83. doi: 10.1038/373081a0. [DOI] [PubMed] [Google Scholar]
- 48.Schild L, Lu Y, Gautschi I, Schneeberger E, Lifton R P, Rossier B C. Identification of a PY motif in the epithelial Na channel subunits as a target sequence for mutations causing channel activation found in Liddle syndrome. EMBO J. 1996;15:2381–2387. [PMC free article] [PubMed] [Google Scholar]
- 49.Scholle F, Longnecker R, Raab-Traub N. Epithelial cell adhesion to extracellular matrix proteins induces tyrosine phosphorylation of the Epstein-Barr virus latent membrane protein 2: a role for C-terminal Src kinase. J Virol. 1999;73:4767–4775. doi: 10.1128/jvi.73.6.4767-4775.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 51.Shimkets R A, Warnock D G, Bositis C M, Nelson-Williams C, Hansson J H, Schambelan M, Gill J R, Jr, Ulick S, Milora R V, Findling J W. Liddle's syndrome: heritable human hypertension caused by mutations in the beta subunit of the epithelial sodium channel. Cell. 1994;79:407–414. doi: 10.1016/0092-8674(94)90250-x. [DOI] [PubMed] [Google Scholar]
- 52.Sparks A B, Hoffman N G, McConnell S J, Fowlkes D M, Kay B K. Cloning of ligand targets: systematic isolation of SH3 domain-containing proteins. Nat Biotech. 1996;14:741–744. doi: 10.1038/nbt0696-741. [DOI] [PubMed] [Google Scholar]
- 53.Staub O, Dho S, Henry P, Correa J, Ishikawa T, McGlade J, Rotin D. WW domains of Nedd4 bind to the proline-rich PY motifs in the epithelial Na+ channel deleted in Liddle's syndrome. EMBO J. 1996;15:2371–2380. [PMC free article] [PubMed] [Google Scholar]
- 54.Staub O, Gautschi I, Ishikawa T, Breitschopf K, Ciechanover A, Schild L, Rotin D. Regulation of stability and function of the epithelial Na+ channel (ENaC) by ubiquitination. EMBO J. 1997;16:6325–6336. doi: 10.1093/emboj/16.21.6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sudol M. Structure and function of the WW domain. Prog Biophys Mol Biol. 1996;65:113–132. doi: 10.1016/s0079-6107(96)00008-9. [DOI] [PubMed] [Google Scholar]
- 55a.Tong J, Oyamada H, Demaurex N, Grinstein S, McCarthy T V, MacLennan D H. Caffeine and halothane sensitivity of intracellular Ca2+release is altered by 15 calcium release channel (ryanodine receptor) mutations associated with malignant hyperthermia and/or central core disease. J Biol Chen. 1997;272:26332–26339. doi: 10.1074/jbc.272.42.26332. [DOI] [PubMed] [Google Scholar]
- 56.Turner M, Mee P J, Costello P S, Williams O, Price A A, Duddy L P, Furlong M T, Geahlen R I, Tybulewicz V L. Perinatal lethality and blocked B-cell development in mice lacking the tyrosine kinase Syk. Nature. 1995;378:298–302. doi: 10.1038/378298a0. [DOI] [PubMed] [Google Scholar]
- 57.Wood J D, Yuan J, Margolis R L, Colomer V, Duan K, Kishi J, Kaminsky Z, Kleiderlein J J, Jr, Sharp A H, Ross C A. Atrophin-1, the DRPLA gene product, interacts with two families of WW domain-containing proteins. Mol Cell Neurosci. 1998;11:149–160. doi: 10.1006/mcne.1998.0677. [DOI] [PubMed] [Google Scholar]