Abstract
Fragile X syndrome is the most common inherited form of mental retardation. It is caused by loss of FMR1 gene activity due to either lack of expression or expression of a mutant form of the protein. In mammals, FMR1 is a member of a small protein family that consists of FMR1, FXR1, and FXR2. All three members bind RNA and contain sequence motifs that are commonly found in RNA-binding proteins, including two KH domains and an RGG box. The FMR1/FXR proteins also contain a 60S ribosomal subunit interaction domain and a protein-protein interaction domain which mediates homomer and heteromer formation with each family member. Nevertheless, the specific molecular functions of FMR1/FXR proteins are unknown. Here we report the cloning and characterization of a Drosophila melanogaster homolog of the mammalian FMR1/FXR gene family. This first invertebrate homolog, termed dfmr1, has a high degree of amino acid sequence identity/similarity with the defined functional domains of the FMR1/FXR proteins. The dfmr1 product binds RNA and is similar in subcellular localization and embryonic expression pattern to the mammalian FMR1/FXR proteins. Overexpression of dfmr1 driven by the UAS-GAL4 system leads to apoptotic cell loss in all adult Drosophila tissues examined. This phenotype is dependent on the activity of the KH domains. The ability to induce a dominant phenotype by overexpressing dfmr1 opens the possibility of using genetic approaches in Drosophila to identify the pathways in which the FMR1/FXR proteins function.
Fragile X syndrome is the most common form of hereditary mental retardation whose effects are traced to the loss of function of a single gene, named FMR1. This syndrome affects approximately 1 in 5,000 male births and is globally distributed throughout the human population (19, 42, 61). In most cases, the disease results from the repression of FMR1 gene expression that is due to an expansion of a CGG trinucleotide repeat in the 5′ untranslated region of the gene (25, 28, 36, 45, 65, 66, 73). Subsequent methylation of this expanded repeat results in transcriptional silencing of the FMR1 gene (7, 43). A few fragile X patients with partial or complete deletions of the FMR1 gene have been identified, and these patients have phenotypes similar to those affected by the trinucleotide repeat expansion (26, 68). One patient who has a single point mutation in the FMR1 gene that replaces an isoleucine residue at amino acid 304 with asparagine (I304N) exhibits a particularly severe fragile X phenotype (17). The severity of the phenotype observed in this patient has prompted the suggestion that the I304N substitution results in a dominant-negative form of FMR1 protein (22, 53).
The FMR1 protein binds RNA in vitro and contains two types of RNA-binding motifs, KH domains and an RGG box (10, 35, 55). The RNA-binding activity of FMR1 appears to be selective. It has been estimated that FMR1 interacts with about 4% of human fetal brain mRNAs, including its own mRNA (4). The importance of the RNA-binding activity to the function of the FMR1 protein is underscored by the observation that the I304N substitution alters a highly conserved residue in the second KH domain and that this substitution impairs the ability of FMR1 to bind RNA in vitro (53) and affects its association with polyribosomes in vivo (22).
Searches for other vertebrate homologs of FMR1 and screens for proteins that interact with FMR1 led to the identification of two closely related genes, FXR1 and FXR2 (56, 74). All three proteins share extensive amino acid sequence identity or similarity over their entireties except for the carboxy-terminal end (74). They are capable of forming both heteromers and homomers via an FMR1/FXR interaction domain located near the amino termini of the proteins (57, 74) (Fig. 1). Additionally, all three proteins associate with ribosomes (22, 34, 57, 62) via a ribosome interaction domain located carboxy terminal to the second KH domain (Fig. 1). All three proteins also contain a sequence that resembles the leucine-rich nuclear export signal (NES) of the human immunodeficiency virus (HIV) Rev protein, and mutation of this putative NES in FMR1 results in mislocalization of the protein from the cytoplasm to the cell nucleus (21, 24, 58). Therefore, it has been suggested that FMR1 may shuttle between the nucleus and cytoplasm and thus may play a role in the nuclear export of as yet unidentified RNA substrates. Taken together, these results demonstrate that FMR1, FXR1, and FXR2 constitute a family of structurally related proteins that are likely to function in the transport or metabolism of specific RNA molecules. However, little is known about the potential RNA targets to which these proteins may bind and how FMR1/FXR proteins may influence the activities of such RNAs. Thus, the molecular functions of the FMR1/FXR proteins remain unknown.
FIG. 1.
Amino acid sequence alignment of hFMR1, hFXR1, hFXR2, and dFMR1 (GenBank accession no. AF305881). Identical and similar amino acid residues among all four proteins are highlighted in gray, and gaps are introduced for optimal alignment. Previously delineated functional domains of FMR1/FXR proteins are marked and include two KH domains, an FMR1/FXR interaction domain, a 60S ribosomal subunit interaction domain, and an RGG box. Highly conserved isoleucine residues that are essential for normal KH domain function are indicated with asterisks. The putative HIV Rev-like leucine-rich NES is also indicated.
Here, we report the isolation and characterization of the first invertebrate member of the FMR1/FXR gene family from Drosophila melanogaster (dfmr1). The dfmr1 gene product has considerable amino acid sequence identity/similarity with the vertebrate FMR1/FXR protein family members and, like these proteins, contains two KH domains and an RGG box, as well as a high degree of conservation in the ribosomal association and oligomerization domains. It possesses similar RNA-binding activity and displays a capacity to interact with human FMR1. Using a monoclonal antibody to dFMR1, we show that it is localized to the cytoplasm. The expression pattern of dfmr1 during Drosophila embryogenesis reflects a combination of the tissue distributions of FMR1 and the FXR proteins observed in the mouse and human embryos. We further show that overexpression of dFMR1 leads to apoptosis, indicating that the cellular dFMR1 levels must be tightly regulated. We produced dFMR1 mutants bearing point mutations analogous to the I304N substitution in each of the two KH domains and show that these mutations result in a loss of function for dFMR1 both in vitro and in vivo. The identification of FMR1 in Drosophila, along with the ability to induce a phenotype by overexpression of the protein, provides an opportunity to utilize genetic approaches in Drosophila to uncover the in vivo function(s) of the FMR1/FXR protein family.
MATERIALS AND METHODS
Isolation of cDNA clones and DNA sequencing.
Using the degenerate primers with sequences of CAG(C/T)TGGC(A/C)TC(A/C)(A/C)GATT(C/T)CA(C/T) and CTC(A/G/C/T)CCATAAAT(A/G)TG(A/G)AA(A/G/C/T)GT, PCR was performed on zebra fish genomic DNA to obtain a 180-bp DNA fragment covering the first KH domain. This fragment was then used as a probe to screen 106 plaques of a λZAP zebra fish cDNA library (generously provided by E. Weinberg), and a partial zebra fish FMR1 (zFMR1) cDNA lacking the 5′ end was isolated. The 5′ end was obtained by PCR on the zebra fish cDNA library using the T3 promoter primer which anneals to the library vector and a primer with the sequences TCCACAACCTCTTGAATCAG which anneals to the extreme 5′ end of the partial cDNA. To obtain a D. melanogaster fmr1 cDNA, the 600-bp cDNA fragment encoding amino acids 173 to 371 of zFMR1 was used as a probe to screen 106 plaques of a λZAP D. melanogaster ovarian cDNA library. The inserts of all clones were sequenced and analyzed by MacVector (Oxford Molecular Group). The full-length dFMR1 sequence was assembled from two overlapping clones.
RNA binding assay.
Binding of in vitro-translated proteins to ribohomopolymers was carried out as previously described (55) in binding buffer (10 mM Tris-HCl, 2.5 mM MgCl2, 0.5% Triton X-100, 1 μg of pepstatin A/ml, 1 μg of leupeptin/ml, 0.5% aprotinin) containing 100 or 400 mM NaCl. After binding at 4°C for 30 min, the beads bound with proteins were once washed with binding buffer containing heparin (2 mg/ml) and then four times with binding buffer. Bound proteins were eluted from the beads in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer, separated by SDS-PAGE on a 12.5% gel, and visualized by fluorography.
In vitro protein binding assays.
All the [35S]methionine-labeled proteins were produced using the TnT T7 coupled rabbit reticulocyte lysate system (Promega Biotech) in the presence of [35S]methionine (Amersham) according to the manufacturer's protocol. The dFMR1 cDNA was subcloned into the EcoRI-cleaved pGEX-5X-1 bacterial expression vector. Glutathione S-transferase (GST)– dFMR1 fusion protein was induced and purified as recommended by the manufacturer (Pharmacia). In vitro protein interaction assays were carried out as previously described (74). Briefly, GST or GST-dFMR1 (2 μg) bound to 30 μl of glutathione-Sepharose 4B resin (Pharmacia) was incubated with 5 μl of the in vitro-translated proteins in 500 μl of binding buffer (50 mM Tris-HCl [pH 7.5], 200 mM NaCl, 2 mM EDTA, 0.1% NP-40, 1 μg of leupeptin/ml, 1 μg of pepstatin A/ml, 0.5% aprotinin). Following incubation at 4°C for 1 h, the resin was washed with 1 ml of binding buffer five times. Bound proteins were eluted in SDS-PAGE sample buffer, separated by SDS-PAGE on a 12.5% gel, and visualized by fluorography.
Antibody production and immunoprecipitation.
The cDNA fragment encoding the amino-terminal 580 amino acids of dFMR1 was cloned into the His tag-containing pET28(a) vector (Novagen) to create the expression plasmid pET-dFMR1(N). His-dFMR1(N) fusion protein was induced and purified using a His-binding resin column as described by the manufacturer (Novagen). The purified protein was dialyzed against phosphate-buffered saline (PBS) and used to immunize mice. Hybridoma production and screening were performed essentially as previously described (13) except that the hybridomas were screened by Western blot analysis using Drosophila Schneider 2 (S2) tissue culture cell extract. Antibody specificity was determined by immunoprecipitation of in vitro-translated dFMR1 in the presence of the detergent Empigen BB. Briefly, 5 μl of in vitro-translated, [35S]methionine-labeled proteins was mixed with 2 μl of antibodies from mouse ascites fluid and incubated with 50 μl of protein A-Sepharose beads (Pharmacia) in 450 μl of PBS containing 1% Empigen BB, 1 mM EDTA, and 0.1 mM dithiothreitol at 4°C for 1 h. Unbound proteins were removed by washing the beads in binding buffer several times. Bound proteins were eluted with SDS-PAGE sample buffer, separated by SDS-PAGE, and visualized by fluorography.
Immunofluorescence microscopy on S2 cells.
S2 cells were fixed onto polylysine-treated slides, fixed in PBS containing 2% formaldehyde for 30 min at room temperature, and permeabilized with acetone for 3 min at −20°C as previously described (13). Mouse ascites fluid of hybridoma 6A15 at 1:1,000 dilution was used, followed by fluorescein isothiocyanate-conjugated goat anti-mouse F(ab′)2.
Northern blot analysis.
Total RNA isolated from D. melanogaster ovaries and poly(A)+ RNA isolated from flies of different embryonic stages were separated by electrophoresis in a formaldehyde–1.2% agarose gel in MOPS (morpholinepropanesulfonic acid) buffer at 2 μg per lane. RNA was transferred to nitrocellulose membrane and probed with the 32P-labeled AatII/NcoI dfmr1 restriction fragment.
Western blot analysis.
S2 cell extract and protein extract from D. melanogaster embryos were prepared in SDS-PAGE sample buffer and separated by SDS-PAGE on a 12.5% gel. The amount of total protein in each lane was normalized by quantitative Coomassie blue staining. Immunoblotting and antibody probing were carried out essentially as described elsewhere (46), using 6A15 ascites fluid at a 1:1,000 dilution. Imaginal disc tissue from Drosophila larvae was lysed in a hypotonic buffer as described by Pan and Rubin (44). Proteins were separated by SDS-PAGE on a 10% gel, transferred to a nitrocellulose membrane, and detected with a 1:2,000 dilution of 6A15 ascites fluid and a 1:10,000 dilution of the mouse anti-β-tubulin antibody E7 (14).
Drosophila manipulations.
Fly stocks were maintained on cornmeal-molasses medium at 25°C. Targeted overexpression of dfmr1 was achieved by cloning the wild-type or mutant dfmr1 open reading frame into the pUAST vector (9) or downstream of sevenless promoter and enhancer regions described elsewhere (8) and generating transformed flies through germ line transformation (9, 51, 59). Transformed stocks with UAS-dfmr1 alleles were crossed to GAL4 driver stocks as described in Results, and progeny were monitored for phenotypes.
Drosophila tissue processing.
Wings were dissected from flies of interest, mounted in DPX (Fluka), and photographed using a Leica DMR microscope and a Hamamatsu C5810 charge-coupled device camera. To examine eye phenotypes by scanning electron microscopy, adult flies of appropriate genotypes went through a graded series of ethanol dehydration (70, 80, 90, 95, and 100% ethanol, at least 12 h per grade) and were dried using hexamethyldisilazane (Sigma). Flies were coated with gold-platinum and examined on a JEOL 6300 or JEOL T33A scanning electron microscope. Adult retinal morphology was examined as described by Carthew and Rubin (11). Briefly, adult eyes were dissected, fixed in glutaraldehyde and osmium tetroxide, dehydrated in ethanol, and then embedded in Durcapan resin (Fluka). Sections (1 μm) were cut on a Sorval MT-1 Porter-Blum ultramicrotome, stained with toluidine blue, and examined as described above for wing tissue. Eye-antennal imaginal discs were stained with acridine orange as described by Ye and Fortini (72). Eye-antennal discs from third-instar larvae were dissected in Ringer's solution and placed in acridine orange (0.2 mg/ml) in Ringer's solution for 4 min. Tissue was then placed into fresh Ringer's solution, immediately examined by fluorescence microscopy, and photographed.
Nucleotide sequence accession number.
The dfmr1 and zFMR1 cDNA sequences have been deposited in GenBank (accession no. AF305881 and AF305882, respectively).
RESULTS
Cloning of a Drosophila FMR1/FXR homolog.
To screen for potential Drosophila homologs of FMR1/FXR, we first isolated a lower-vertebrate homolog to use as a probe. Amino acid sequence comparison of FMR1/FXR homologs cloned from mammalian species revealed that the amino-terminal half, containing two KH domains, is highly conserved. Therefore, we designed degenerate oligonucleotide primers within the first KH domain and performed PCR on zebra fish genomic DNA. PCR using these primers yielded a product of the expected size (ca. 180 bp), and its sequences revealed that it indeed corresponded to a zebra fish homolog of mammalian FMR1 (data not shown). The entire zFMR1 coding region was obtained by hybridization screens and by assembling sequences from two overlapping clones from a cDNA library. Sequence comparison between zFMR1 and human FMR1 (hFMR1) showed a high degree of amino acid conservation, particularly in a 200-amino-acid region containing the two KH domains (not shown).
A probe comprising the two KH domains of zFMR1 was then used for hybridization screens on a Drosophila ovarian cDNA library. Six positive clones which contained identical overlapping sequences were isolated. A composite full-length cDNA sequence termed dfmr1 was assembled from the sequences of two overlapping clones. This putative homolog encodes a protein of 681 amino acids. As shown in Fig. 1, the amino acid sequence alignment of dFMR1 to hFMR1, hFXR1, and hFXR2 reveals a significant degree of conservation among all four proteins. This is particularly noticeable in the regions corresponding to previously delineated functional domains and their relative orientations within the primary structure of the proteins. Notably, dFMR1 contains all of the key structural elements of FMR1/FXR proteins that are involved in RNA binding. The two KH domains are nearly 75% identical and 85% similar between dFMR1 and hFMR1. Highly conserved isoleucine residues implicated in KH domain function (53) are present in the dFMR1 and the FMR1/FXR KH domains. The RGG box, the other type of RNA-binding motif found in hFMR1, is also found in dFMR1. A 40-amino-acid region which mediates protein-protein interactions among FMR1/FXR proteins (57) is also highly conserved, showing about 50% identity with dFMR1. Finally, a leucine-rich region, which has been shown to be involved in the binding of FMR1/FXR proteins to the 60S ribosomal subunit (57), is also highly conserved in dFMR1. This region may serve to determine the subcellular localization of FMR1, because isoforms lacking this domain are localized to the nucleus rather than the cytoplasm (58). Examination of this leucine-rich sequence in hFMR1 revealed a potential HIV Rev-protein kinase inhibitor (PKI)-type NES that consists of four critically spaced, large hydrophobic amino acids, including leucine, isoleucine, methionine, and valine (21, 24). FXR1 and FXR2 also contain this putative signal. In dFMR1, this leucine-rich region exhibits 70% overall sequence identity and 80% similarity to the human homologs. However, one of the leucine residues that is critical for NES function is changed to glutamine, suggesting that dFMR1 may lack nuclear export activity. Nonetheless, dFMR1 and its vertebrate counterparts clearly display a very high degree of conservation at the primary structure level.
dFMR1 has RNA-binding and protein-protein interaction properties similar to those of the hFMR1 and hFXR proteins.
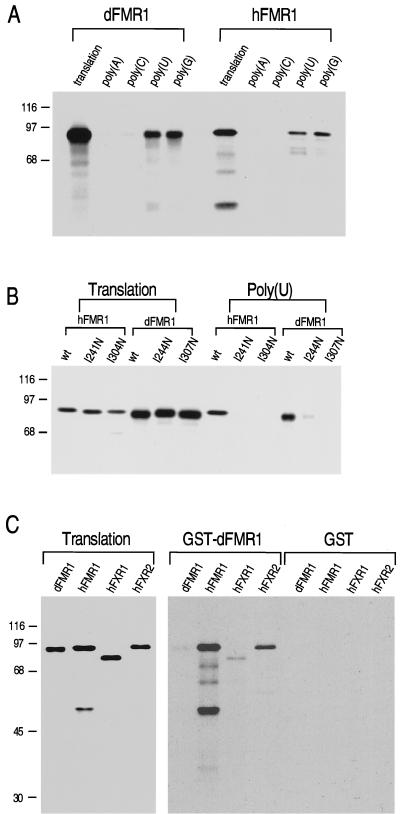
Previous studies demonstrated that FMR1 has RNA-binding activity which is conferred by the two KH domains and the RGG box (4, 53, 55). In an RNA homopolymer binding assay, in vitro-translated, 35S-labeled hFMR1 protein showed strong binding to poly(G), weaker but significant binding to poly(U), and no detectable binding to poly(C) and poly(A). Under the same binding conditions, the same RNA homopolymer binding profile was observed for dFMR1 (Fig. 2A). The conservation of the RNA-binding activity between dFMR1 and hFMR1 lends further support to the conclusion that dFMR1 is functionally related to the vertebrate FMR1/FXR proteins. To assess whether the KH domains of dFMR1 confer its RNA-binding capability, point mutations predicted to inactivate the function of either KH domain were engineered into the dfmr1 cDNA by site-directed mutagenesis. A codon for a highly conserved isoleucine residue within each of the KH domains was mutated to a codon for asparagine (I244N or I307N). These KH domain mutations are analogous to a mutation identified in the FMR1 protein of a fragile X patient (17), and such analogous mutations in human FMR1 have been shown to impair RNA binding in vitro (53). Furthermore, solution structure data of these KH domains predict that these substitutions will disrupt an alpha-helix structure within the KH domain (41). RNA homopolymer binding assays with these mutant forms of dFMR1 show that either mutation impairs the ability of dFMR1 to bind poly(U) in vitro as is observed for the analogous hFMR1 mutants (Fig. 2B).
FIG. 2.
dFMR1 and hFMR1 have similar biochemical properties. (A) Binding of dFMR1 and hFMR1 to ribonucleotide homopolymers at 100 mM NaCl. dFMR1 and hFMR1 were in vitro translated and labeled with [35S]methionine; 5 μl of each was loaded onto 30 μl of poly(A), poly(C), poly(U), or poly(G) beads for binding. After washing, retained proteins were boiled in SDS-PAGE sample buffer and analyzed by SDS-PAGE followed by fluorography. Translation lanes show 20% of the proteins used in each binding. (B) Binding profile of wild-type (wt) and mutant forms of dFMR1 (I244N and I307N) and hFMR1 (I241N and I304N) to poly(U) at 400 mM NaCl. Translation lanes show 10% of the proteins used in each binding. (C) In vitro interaction between dFMR1 and human FMR1/FXR proteins. The indicated in vitro [35S]methionine-labeled proteins were incubated with GST-dFMR1 or GST (2 μg) alone. The bound proteins were analyzed by SDS-PAGE followed by fluorography. Translation lanes show 20% of each protein used in the binding assay. Sizes are indicated in kilodaltons.
Another biochemical feature of hFMR1 and hFXR proteins is their capacity to form heteromers with other FMR1/FXR proteins (57, 74). To determine if dFMR1 has a similar capacity, we performed in vitro binding experiments to test the interaction of purified recombinant GST-dFMR1 with dFMR1 and with hFMR1 and hFXR produced by in vitro transcription and translation. As shown in Fig. 2C, GST-dFMR1 binds with the highest avidity to hFMR1, less well to hFXR2, and very weakly to hFXR1 and to itself. In reciprocal experiments in which hFMR1, hFXR1, and hFXR2 were immobilized as GST fusion proteins, the same profiles of relative binding avidity were observed (data not shown). The order of preference of binding of dFMR1 suggests that each of the FMR1/FXR proteins has a characteristic selectivity of protein-protein, as noted previously (74).
Production of monoclonal antibodies to dFMR1.
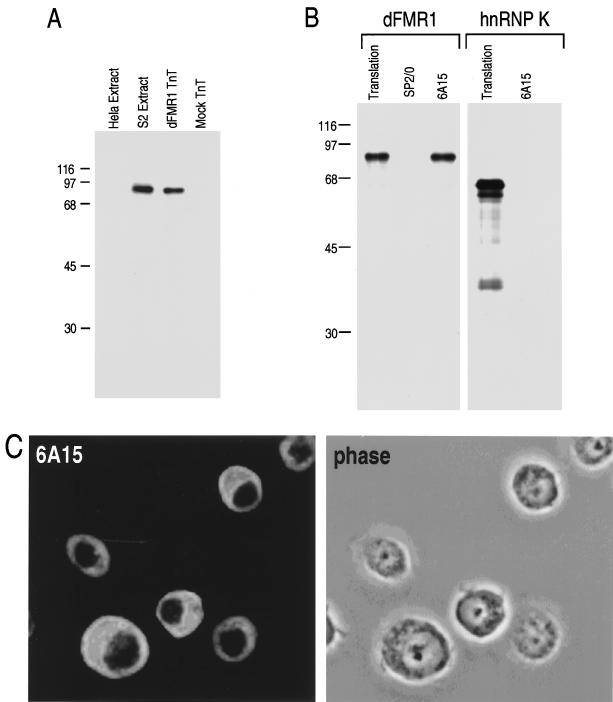
To facilitate characterization of the dFMR1 protein, we produced a monoclonal antibody to it, designated 6A15. By Western blotting, 6A15 recognizes a single major protein of approximately 85 kDa in extracts of Drosophila S2 tissue culture cells, which comigrates with the translation product of the dfmr1 cDNA. 6A15 does not cross-react with human FMR1 or FXR proteins, because no bands were detected from HeLa cell extract (Fig. 3A). The specificity of this monoclonal antibody was further demonstrated by immunoprecipitation of dFMR1 produced by in vitro transcription and translation (Fig. 3B). hnRNP K, another KH domain-containing protein (54), was not immunoprecipitated by 6A15. Since 6A15 specifically recognized dFMR1, we used this antibody further to determine the subcellular distribution of the protein in S2 cells. Like its human homologs, dFMR1 is localized predominantly to the cytoplasm at steady state (Fig. 3C).
FIG. 3.
Characterization of dFMR1 monoclonal antibody 6A15. (A) On a Western blot, 6A15 specifically recognizes dFMR1. The indicated cell extracts and in vitro-translated proteins were immunoblotted with 6A15. (B) 6A15 specifically immunoprecipitates dFMR1. Each antibody (2 μl of ascites fluid) was immobilized on protein A-Sepharose beads and incubated with in vitro-produced, [35S]methionine-labeled dFMR1 or hnRNP K. Translation lanes show 20% of each protein used in the immunoprecipitation experiment. Sizes are indicated in kilodaltons. (C) Cytoplasmic localization of dFMR1 in S2 cells by immunofluorescence microscopy using 6A15. Left, detection by 6A15 of dFMR1 largely in the cytoplasm of S2 cells; right, phase image of the same field of cells.
dFMR1 is ubiquitously expressed throughout Drosophila embryogenesis.
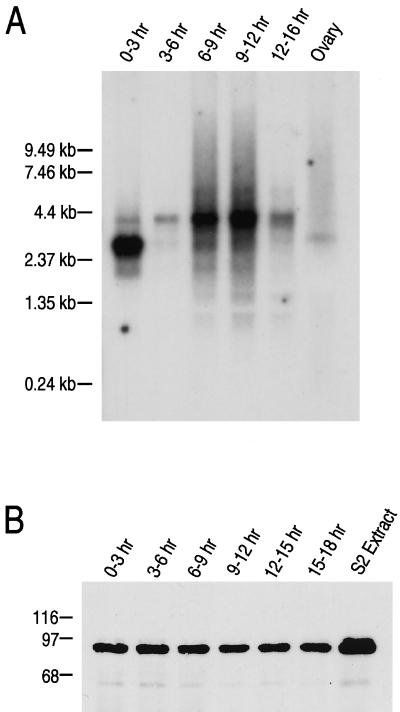
To determine the timing of expression and transcript complexity of dfmr1, we performed developmental Northern analysis using a probe derived from the dfmr1 cDNA (see Materials and Methods). A prominent 2.8-kb transcript was detected in total RNAs prepared from ovaries and in poly(A)+ RNAs prepared from 0- to 3-h embryos. At later times, from 3 to 6 h and beyond, the major transcript detected was about 4.0 kb, and its level peaked between 9 to 12 h (Fig. 4A). Although two different-sized dfmr1 transcripts are produced during development, they appear to encode proteins of the same size because a developmental Western blot probed with 6A15 detected a single protein of the expected size of 85 kDa, whose expression level remains unchanged throughout development (Fig. 4B). This suggests that the difference in the transcript sizes is most likely due to variation in the untranslated regions of the mRNAs. Indeed, fragments of the same size were amplified from RNAs from all different stages of embryogenesis by reverse transcription-PCR using three sets of primers which span the entire dfmr1 coding region (data not shown).
FIG. 4.
Expression of dfmr1 RNA and protein during embryogenesis. (A) Northern blot analysis of dfmr1 transcripts from D. melanogaster ovaries and 0- to 16-h embryos. Total RNA (2 μg) from ovaries and poly(A)+ RNA (2 μg) from embryos were resolved by electrophoresis on a formaldehyde-agarose gel, transferred to a nitrocellulose membrane, and hybridized with a 32P-labeled fragment of dfmr1 cDNA. (B) Western blot analysis of proteins expressed throughout Drosophila embryonic development from 0 to 18 h. Extracts from S2 cells and total cellular proteins from different-stage embryos were normalized by quantitative Coomassie blue staining. Equal amounts of protein were loaded in all lanes, separated by SDS-PAGE, transferred to a nitrocellulose membrane, and probed with 6A15 at a 1:1,000 dilution. Sizes are indicated in kilodaltons.
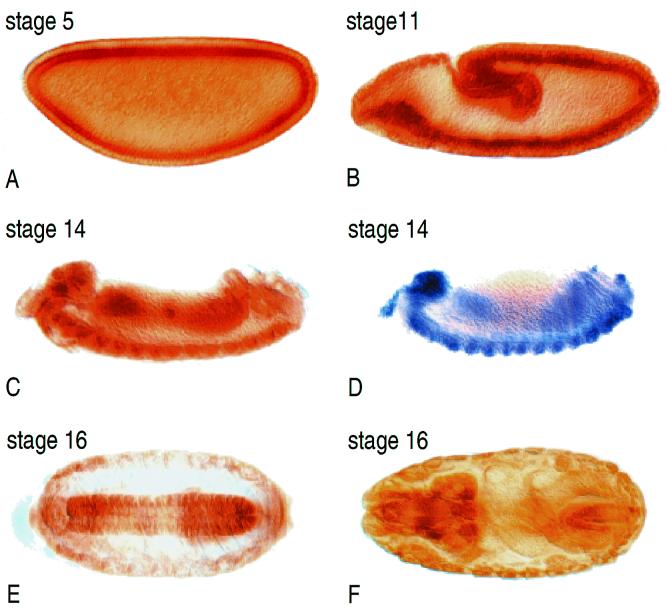
To examine the tissue distribution of dFMR1 during embryogenesis, we performed whole-mount in situ staining using the full-length dfmr1 cDNA as a hybridization probe and whole-mount immunostaining using 6A15. Both approaches revealed the same tissue distribution pattern (compare Fig. 5C and D). From the time that the egg is laid to early gastrulation, dFMR1 protein is uniformly distributed in the embryo (Fig. 5A and data not shown). At midgastrulation (stage 11), the protein is expressed everywhere but there is a discernible concentration in the mesoderm (Fig. 5B). After gastrulation (stage 14), dFMR1 is uniformly distributed, with significantly elevated levels in the mesoderm, ventral nerve cord, and brain (Fig. 5C and D). At stage 16, expression in the ventral nerve cord and brain is more pronounced and elevated staining in the muscle is also detected (Fig. 5E and F). Overall, dFMR1 expression is widespread, with more pronounced expression in the central nervous system and in muscles. In situ hybridization and immunostaining studies of mammalian FMR1 and FXR in mouse and human embryos revealed expression in all tissues, with FMR1 and FXR2 displaying the highest levels in the central nervous system and testis (1, 3, 5, 20, 29, 30, 63) and FXR1 being more prominent in muscles (16, 33). Thus, the dFMR1 expression profile resembles the combined expression pattern of its mammalian homologs.
FIG. 5.
In vivo expression of dfmr1. The dfmr1 transcript and its protein product are widely distributed throughout the developing Drosophila embryo. (A) Blastoderm (stage 5) embryo with ubiquitous distribution of dFMR1 protein. (B) Stage 11 embryo with elevated concentrations of dFMR1 in the mesoderm. (C and D) Protein (C) and transcript (D) localization in stage 14 embryos. Increased levels of expression in both RNA and protein are seen in the brain and ventral nerve cord. (E and F) At stage 16, dFMR1 expression is still high in the brain, and expression in muscles can be detected. All embryos are oriented so that anterior is left. Panels A to D are lateral views; panels E and F are ventral views.
Overexpression of dFMR1 leads to apoptosis.
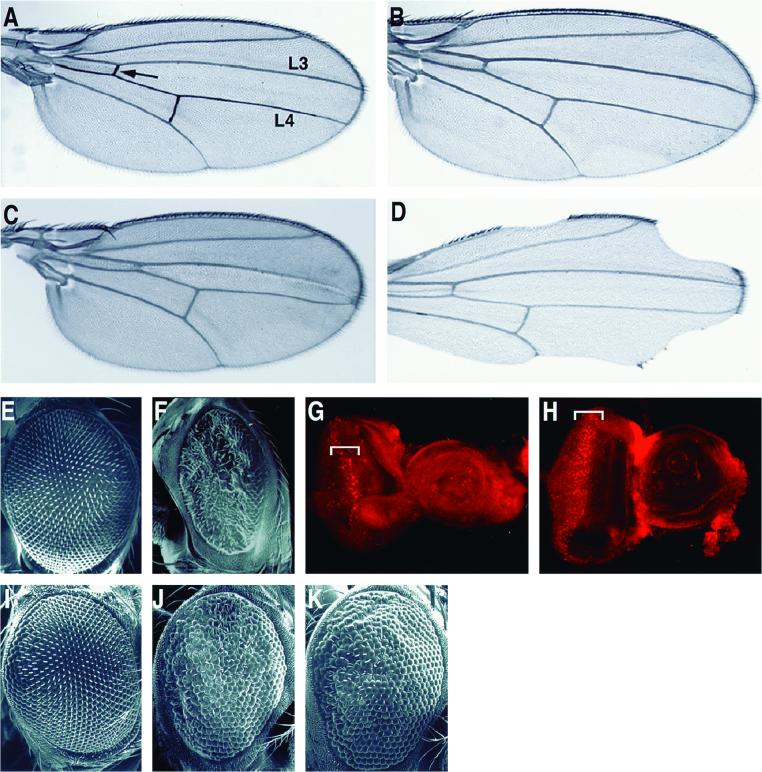
Given that there are no known loss-of-function alleles of dfmr1 and that a majority of genes in Drosophila do not mutate to a readily detectable phenotype (40), we tested the effect of overexpressing dFMR1 protein. Overexpression of a gene can lead to phenotypes that are often relevant to the function of the expressed gene product (48–50). To overexpress dFMR1 in developing tissues, we used the GAL4-UAS system (9) or sevenless promoter/enhancer sequences (8) that direct expression of dFMR1 in a subset of cell types in the larval eye-antennal imaginal disc. Transgenic flies containing a copy of UAS-dfmr1 (see Materials and Methods) were crossed to a variety of promoters that direct GAL4 expression in easily visualized tissues. Overexpression of dfmr1 under the control of vestigial (vg) or decapentaplegic (dpp) promoters, which direct expression in developing wing tissue, caused transgenic wings to suffer a loss of cells in the region of the wing where the promoters for these genes function. Expression of UAS-dfmr1 through dpp-GAL4, which is strongly expressed at the anterior/posterior margin of the developing wing blade, led to a decrease in cells between longitudinal veins 3 and 4, as well as loss of the anterior crossvein (Fig. 6C). A loss of cells at the margin of the wings was observed with 100% penetrance when UAS-dfmr1 was expressed under the control of vg-GAL4 (Fig. 6D). Finally, UAS-dfmr1 expression was directed by sevenless-GAL4 (sev-GAL4), which drives expression in a subset of the photoreceptor cells, the mystery cells, and the cone cells behind the morphogenetic furrow during eye development (6, 8, 64). Overexpression of dFMR1 in the eye leads to a severe rough eye phenotype (compare Fig. 6F and E).
FIG. 6.
Overexpression of dFMR1 induces apoptosis. (A to D) Overexpression of dfmr1 in wing tissue leads to cell loss. (A and B) UAS-dfmr1 or vg-GAL4 wings by themselves have no obvious defects. No other GAL4 driver stocks used in this study have any visible phenotypes. The arrow in panel A denotes the anterior crossvein; L3 and L4 refer to longitudinal veins 3 and 4. (C) Expression of UAS-dfmr1 under dpp-GAL4 control leads to wings with missing anterior crossveins and with fewer cells between the L3 and L4 veins. (D) vg-GAL4-driven expression of UAS-dfmr1 leads to a loss of cells at wing margins. (E and F) Overexpression of dfmr1 in the eye leads to a severe rough eye phenotype. Scanning electron micrographs show adult eyes from control sevenless-GAL4/+ (sev-GAL4) (E) and sev-GAL4/UAS-dfmr1 (F). (G and H) Eye-antennal imaginal discs from sev-GAL4/+ (G) and sev-GAL4/UAS-dfmr1 (H) larvae stained with acridine orange, a marker for apoptotic activity. Relatively few acridine orange-stained cells are seen in a sev-GAL4/+ disc, while significantly more cells stained with acridine orange are present (outlined by bracket) when UAS-dfmr1 is expressed by sev-GAL4. (I to K) The rough eye phenotype induced by sevenless overexpression of dfmr1 is partially suppressed by coexpression of DIAP1/THREAD an inhibitor of apoptotic activity. (I) Eye from a GMR-diap1/+ fly, which is wild type in appearance. (J) Eye from a sev-dfmr1 fly, showing disorganized ommatidia over the eye surface. (K) Eye from a GMR-diap/sev-dfmr1 fly shows a partial rescue over the phenotype seen in panel J, with more organized ommatidia present.
The phenotypes caused by overexpression of dFMR1 could result from cells failing to adopt an appropriate fate, defects in cell proliferation, or cell death by apoptosis or necrosis. The staining of cells with the dye acridine orange is a reliable marker for apoptosis and does not mark cells that are dying through necrosis (2, 72). Eye-antennal imaginal discs from third-instar larvae of control (sev-GAL4/+) flies or those expressing dFMR1 under sev-GAL4 control were dissected and stained with acridine orange to qualitatively assess levels of cell death. In control eye-antennal discs, relatively little apoptosis occurs (Fig. 6G). In contrast, eye-antennal discs overexpressing dFMR1 showed a large number of cells behind the morphogenetic furrow that have taken up the dye (Fig. 6H). Furthermore, we found that the rough eye phenotype was suppressed when the apoptotic inhibitor DIAP1/THREAD (27) was cooverexpressed in the eye-antennal disc (compare Fig. 6K and J). These results indicate that the overexpression of dFMR1 leads to cell death by apoptosis and that any deficiencies in proliferation or cell fate decisions that may contribute to the observed phenotypes may very well be a secondary consequence of the apoptotic events.
The effects of overexpressing dFMR1 are dependent on the function of the KH domains.
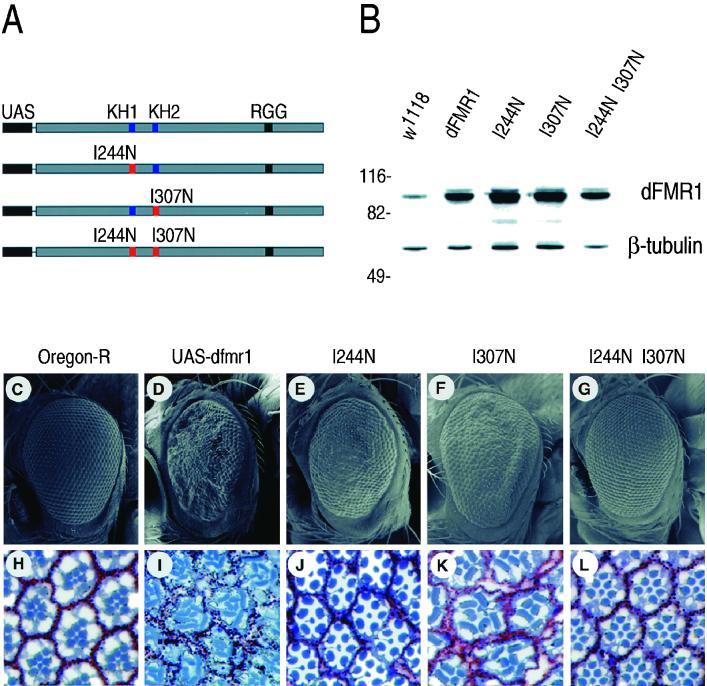
One potential problem with an analysis dependent on overexpression is nonspecific effects caused by the overabundance of a protein. To assess whether the overexpression phenotype is specific to a normal function of the dFMR1 protein, dfmr1 alleles containing isoleucine-to-asparagine changes (I244N and I307N) in either or both KH domains were expressed behind UAS (Fig. 7A). Using sev-GAL4 as a driver, transgenic stocks were selected for lines that exogenously expressed wild-type and mutant forms of dFMR1 at approximately equal levels to allow for quantitative comparisons of the phenotypic effect (Fig. 7B).
FIG. 7.
The phenotype elicited by overexpressing dFMR1 is dependent on the functions of its KH domains. (A) Schematic of UAS-dfmr1 alleles used for this study. Highly conserved isoleucine residues known to be essential for RNA-binding function were mutated to asparagine to inactivate either one or both of the KH domains. (B) Western blot analysis of extracts prepared from eye-antennal imaginal discs of w1118 or flies expressing the transgenes described in panel A under sev-GAL4 control. An antibody that recognizes β-tubulin was used as a loading control. (C to G) Scanning electron micrographs of adult eyes expressing dfmr1. (C) Oregon-R; (D) sev-GAL4 expression of UAS-dfmr1; (E) sev-GAL4 expression of UAS-dfmr1I244N allele; (F) sev-GAL4 expression of UAS-dfmr1I307N allele; (G) sev-GAL4 expression of UAS-dfmr1I244N, I307N double mutant. (H to L) Cross sections of ommatidia from eyes of the above-mentioned genotypes. Sections (1 μm) from fixed eyes were stained with toluidine blue and examined by microscopy. (H) Oregon-R; (I) sev-GAL4 expression of UAS-dfmr1; (J) sev-GAL4 expression of UAS-dfmr1I244N allele; (K) sev-GAL4 expression of UAS-dfmr1I307N allele; (L) sev-GAL4 expression of UAS-dfmr1I244N, I307N double mutant.
Adult eyes from the transgenic stocks were examined by scanning electron microscopy. Mutations in either KH domain significantly ameliorated the degree of eye roughness compared to flies overexpressing a wild-type copy of dfmr1 (compare Fig. 7D to 7E and F). These results indicate that the observed overexpression phenotypes are due to increased dFMR1 activity. However, that a milder rough eye phenotype was observed with the overexpression of either of the KH domain mutations indicates that these mutations do not remove all of the activity of dFMR1 as observed by this assay. sev-GAL4-driven expression of a UAS-dfmr1 allele where both KH domains were mutated has little or no discernible phenotype (Fig. 7G), suggesting that most or all activity of the sev-GAL4-expressed dFMR1 requires functional KH domains. Examination of ommatidial cross sections taken through the photoreceptor cells from eyes of transgenic flies showed several phenotypes (Fig. 7H to L). Ommatidia overexpressing wild-type dFMR1 had occasional missing photoreceptor cells, and pigment cells were often missing (Fig. 7I). These phenotypes can be explained from the apoptotic events occurring in the developing eye imaginal disc. Ommatidial assembly is an ordered process of cell recruitment with photoreceptor cells forming a cluster, subsequently joined by cells destined to function as cone cells, followed by pigment cells and bristle cells (69, 70). The apoptotic events likely reduce the number of cells available to form an ommatidial cluster. In addition, rhabdomere structure is altered in photoreceptor cells overexpressing dFMR1. An interesting observation in this regard is that overexpression of the wild type or I307N mutant, both of which contained an intact first KH domain, altered the shape of the rhabdomeres within photoreceptor cells, whereas overexpressing the I244N mutant or the double mutant did not (compare Fig. 7I and K to H, J, and L). Whether the two KH domains function cooperatively in binding all RNA substrates or whether they can have independent functions cannot be determined at this time.
Dominant-negative functions for the I244N and I307N substitutions in dFMR1 protein?
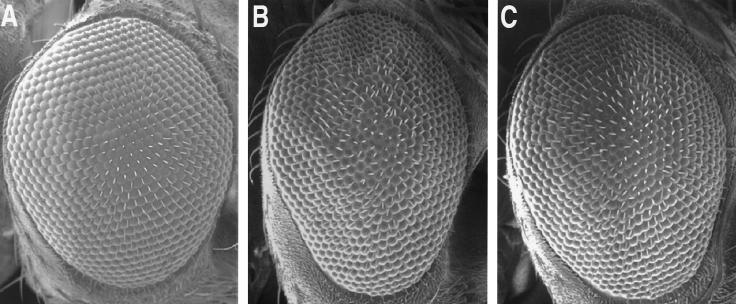
A severe case of fragile X has been described in which there is a single missense mutation in the second KH domain of the FMR1 gene that substitutes an asparagine residue for the normal isoleucine residue (I304N). The extreme phenotype of this patient, as well as observations that the I304N mutation causes abnormally sized RNP particles to form (22) and that mutations mapping to the KH domains of some genes elicit stronger phenotypes than null alleles of the same gene (31, 38), has suggested that the effects of the I304N mutation in hFMR1 may be due to a dominant-negative rather than a loss-of-function effect. To determine whether the phenotype elicited by the dFMR1 I307N substitution was due to a dominant-negative or a loss-of-function effect, we generated Drosophila stocks that in addition to overexpressing dFMR1 I307N had a deficiency in a wild-type copy of dfmr1. The deficiency Df(3R)by62 (85D11-85F16) removes one copy of dfmr1 as assessed by quantitative Southern hybridization (data not shown). If the I307N mutation of dFMR1 acts as a dominant negative, removal of one wild-type copy of the gene would be predicted to increase the severity of the rough eye phenotype. In contrast, if the I307N mutation represented a partial loss of function, removal of one wild-type copy of dfmr1 would be expected to have no effect on the eye or perhaps even lessen the rough phenotype. The comparison of UASdfmr1I307N/+; sev-GAL4/+, and UASdfmr1I307N/+; sev-GAL4/Df(3R)by62 flies indicates no obvious enhancement of eye roughness (compare Fig. 8B and C) and perhaps shows a lessening of the defect in flies with the dfmr1 deficiency. Identical results were observed for the mutation in the first KH domain of dFMR1 (I244N [data not shown]). These results indicate that the isoleucine-to-asparagine changes in the KH domains of dFMR1 act as loss-of-function mutations in the genetic background used for these experiments.
FIG. 8.
The I307N mutation of dFMR1 acts as a loss-of-function mutation. Scanning electron micrographs show adult eyes from Oregon-R (A), UAS-dfmr1I307N/+; sev-GAL4/+ (B), and UAS-dfmr1I307N/+; sev-GAL4/Df(3R)by62 (C) flies.
DISCUSSION
An invertebrate homolog of the FMR1/FXR gene family.
dFMR1 represents the first invertebrate homolog of the FMR1/FXR gene family. Like the vertebrate members of the FMR1/FXR gene family, dFMR1 contains two KH domains that are 85% similar in amino acid sequence to those of hFMR1, as well as an RGG box. Indeed, dFMR1 binds homopolymeric RNAs with a profile similar to that of hFMR1. Previously defined domains that mediate the interaction of FMR1/FXR proteins with the 60S ribosomal subunit and mediate protein-protein interaction among FMR1/FXR family members are also highly conserved in dFMR1. Also like its vertebrate counterparts, dFMR1 is mostly localized to the cytoplasm. We have also shown that the two KH domains in dFMR1 have activity in vivo. The high degree of conservation of the functional domains between dFMR1 and vertebrate FMR1/FXR proteins strongly suggests that these proteins all have similar biological functions.
The characterization of several cDNAs identified by the hybridization screening for the Drosophila FMR1 homologs, as well yeast two-hybrid screens with dFMR1 as bait, failed to identify any additional family members in Drosophila (unpublished results). Moreover, search of the recently completed Drosophila genome sequence indicates that dFMR1 is a unique, single gene without related genes in this organism. Notably, no apparent FMR1/FXR homologs are found in Caenorhabditis elegans and Saccharomyces cerevisiae, two other lower eukaryotes for which the entire genome has been sequenced. It thus appears likely that dfmr1 is a prototype of the FMR1/FXR gene family and that it evolved to give rise to a family of three related FMR1/FXR genes in mammals.
Two-hybrid interaction and in vitro protein binding experiments suggest that FMR1/FXR proteins have higher affinity toward heteromers than homomers. The capacity of these proteins to associate with each other indicates that regulation of the relative concentration of various FMR1/FXR homo- or heterocomplexes is an important determinant in influencing the function of FMR1 (74). dFMR1 can bind the human FMR1/FXR proteins in vitro with relative avidity in the order hFMR1 > hFXR2 > hFXR1 > dFMR1. However, the significance of the conservation of protein-protein interaction activity in dFMR1 is unclear, as no other FMR1/FXR family members have been identified in Drosophila. It is possible that the conserved domain that mediates heteromerization in the vertebrate family members serves as a protein-protein interaction domain that enables dFMR1 to associate with other, yet to be identified proteins.
The discovery of a leucine-rich sequence with the capacity to serve as an NES in hFMR1 and hFXR led to the proposal that these proteins may shuttle between the nucleus and the cytoplasm and possibly bind to as yet unidentified cellular RNAs and mediate their export to the cytoplasm (21, 24). However, the putative NES of the mammalian counterparts is not well conserved in dFMR1. Despite the overall similarity of this region between dFMR1 and its human homologs, one critical leucine residue in hFMR1 is a glutamine (Gln-371) in dFMR1 instead. Thus, it is not likely that dFMR1 has nuclear export activity. Indeed, when injected into the nuclei of Xenopus oocytes, dFMR1 remains in the nucleus, whereas the majority of hFMR1 is efficiently exported to the cytoplasm in the same system (U. Fischer, L. Wan, and G. Dreyfuss, unpublished data). This suggests that dFMR1 lacks efficient export activity compared to hFMR1, which raises a question of the biological significance of the putative NESs in FMR1/FXR proteins.
The KH domains of dFMR1 are essential for its function.
Our results from analyzing mutations in the KH domains of dFMR1 indicate that each KH domain is required for in vivo function, as judged by the rough eye phenotypes elicited upon overexpression of the mutant dFMR1 bearing a single point mutation in either of the two KH domains. The importance of RNA-binding activity to dFMR1 function is underscored by the observation that mutation of either KH domain of dFMR1 affects its homopolymeric RNA-binding activity and that overexpression of a dfmr1 allele where both KH domains have been inactivated leads to no obvious phenotype.
The I244N and I307N substitutions of dFMR1 appear to act as loss-of-function mutations based on their reduced in vivo activity and on genetic criteria, where removal of one wild-type copy of dfmr1 is shown to have no enhancing effect on a rough eye phenotype induced by overexpression of dfmr1 alleles with single point mutations in either KH domain. Whether the reduced activity of dFMR1 with an inactivated KH domain comes from a necessity for both KH domains to function together to bind RNA or whether each KH domain may have some individual functions is not clear. The tandem RNA-binding domains of hnRNP A1 have been shown to have both shared and distinct functions (39). Identification of in vivo substrates for dFMR1 will be needed to further address this issue.
The phenotypes caused by overexpression of dFMR1 are due at least in large part to apoptosis, as judged by positive acridine orange staining and genetic suppression of apoptotic effects by an inhibitor of apoptosis, DIAP1/THREAD, which may function to inhibit the activity of caspases (18). That DIAP1/THREAD can suppress the effects of overexpressing dFMR1 places the activity of dFMR1 upstream or in parallel to the activity of caspases. Determination of whether the effect of dFMR1 on activation of the apoptotic pathway is direct or indirect will require studies of loss-of-function alleles of dfmr1. Toxicity of FMR1/FXR proteins through overexpression is probably not unique to dFMR1 in Drosophila since a recent study by Ceman et al. noted an inability to express FLAG-hFMR1 in cell lines that had a relatively high endogenous level of hFMR1 (12).
Drosophila as a model system to study the in vivo functions of FMR1/FXR proteins.
The RNA-binding activity of the FMR1/FXR proteins is one biochemical property of these proteins that has been experimentally demonstrated. Human FMR1 has been reported to bind approximately 4% of human fetal brain mRNAs, thus displaying a degree of selectivity for RNA substrates (4), but other than the FMR1 mRNA, RNA substrates to which the FMR1/FXR proteins may bind have not been identified. Loss of function of FMR1 in mice leads to abnormalities in dendritic spine processing and maturation, and FMR1 mRNA is rapidly translated in response to neurotransmitter at synapses (15, 67). Thus, it has been proposed that vertebrate FMR1 protein is essential for some aspects of neural development. Specific functions for FXR1 and FXR2 are unknown. Given the considerable overlap in the expression patterns between the three vertebrate members of the FMR1/FXR family and that all three proteins can be isolated as RNPs, it has been speculated that these proteins may be functionally redundant to some degree (3). This makes it more difficult to genetically dissect the function of these proteins in mammals.
The study of Drosophila homologs of various human genes involved in human genetic disorders, such as Alzheimer's disease and Huntington's disease, has provided new insight into fundamental aspects of protein function (23, 37, 60, 71). Function-based genetic screens have been extremely useful as a way to characterize molecular pathways. The FMR1/FXR genes were first identified in vertebrate organisms; however, such organisms are not readily amenable to large-scale function-based screens. The identification of a single highly conserved homolog of the FMR1/FXR family in Drosophila and the ability to elicit a dominant phenotype through overexpression of the protein should facilitate the identification of RNAs and proteins that function in the dfmr1 pathway through the use of genetic modifier screens. Similar screens have been invaluable for studies of the other pathways in Drosophila, such as the ras pathway, where previously unknown genes have been discovered by such approaches (32, 47, 52). Identification of modifying loci whose transcripts are bound by the dFMR1 protein should help to uncover the function of FMR1/FXR proteins and may provide insights into the molecular basis and the pathogenesis of fragile X syndrome.
ACKNOWLEDGMENTS
We are grateful to Qing Liu, Chun-Pyn Shen, Haruhiko Siomi, Mikiko Siomi, Fan Wang, and Yan Zhang for reagents, technical assistance, and helpful discussions. Special thanks go to Gigi Gray-Board, Gerald Harrison, and Doug Yates for advice and assistance with scanning electron microscopy. We thank members of our laboratories, especially Naoyuki Kataoka, Sara Nakielny, Bernard Charroux, and Westley Friesen, for critical reading and suggestions on the manuscript.
This work was supported by grants from the National Institutes of Health to T.A.J. and to G.D. G.D. is an Investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Abitbol M, Menini C, Delezoide A L, Rhyner T, Vekemans M, Mallet J. Nucleus basalis magnocellularis and hippocampus are the major sites of FMR-1 expression in the human fetal brain. Nat Genet. 1993;4:147–153. doi: 10.1038/ng0693-147. [DOI] [PubMed] [Google Scholar]
- 2.Abrams J M, White K, Fessler L I, Steller H. Programmed cell death during Drosophila embryogenesis. Development. 1993;117:29–43. doi: 10.1242/dev.117.1.29. [DOI] [PubMed] [Google Scholar]
- 3.Agulhon C, Blanchet P, Kobetz A, Marchant D, Faucon N, Sarda P, Moraine C, Sittler A, Biancalana V, Malafosse A, Abitbol M. Expression of FMR1, FXR1, and FXR2 genes in human prenatal tissues. J Neuropathol Exp Neurol. 1999;58:867–880. doi: 10.1097/00005072-199908000-00009. [DOI] [PubMed] [Google Scholar]
- 4.Ashley C T, Jr, Wilkinson K D, Reines D, Warren S T. FMR1 protein: conserved RNP family domains and selective RNA binding. Science. 1993;262:563–566. doi: 10.1126/science.7692601. [DOI] [PubMed] [Google Scholar]
- 5.Bachner D, Steinbach P, Wohrle D, Just W, Vogel W, Hameister H, Manca A, Poustka A. Enhanced Fmr-1 expression in testis. Nat Genet. 1993;4:115–116. doi: 10.1038/ng0693-115. [DOI] [PubMed] [Google Scholar]
- 6.Banerjee U, Renfranz P J, Pollock J A, Benzer S. Molecular characterization and expression of sevenless, a gene involved in neuronal pattern formation in the Drosophila eye. Cell. 1987;49:281–291. doi: 10.1016/0092-8674(87)90569-1. [DOI] [PubMed] [Google Scholar]
- 7.Bell M V, Hirst M C, Nakahori Y, MacKinnon R N, Roche A, Flint T J, Jacobs P A, Tommerup N, Tranebjaerg L, Froster-Iskenius U, et al. Physical mapping across the fragile X: hypermethylation and clinical expression of the fragile X syndrome. Cell. 1991;64:861–866. doi: 10.1016/0092-8674(91)90514-y. [DOI] [PubMed] [Google Scholar]
- 8.Bowtell D D, Kimmel B E, Simon M A, Rubin G M. Regulation of the complex pattern of sevenless expression in the developing Drosophila eye. Proc Natl Acad Sci USA. 1989;86:6245–6249. doi: 10.1073/pnas.86.16.6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brand A H, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 10.Burd C G, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 11.Carthew R W, Rubin G M. seven in absentia, a gene required for specification of R7 cell fate in the Drosophila eye. Cell. 1990;63:561–577. doi: 10.1016/0092-8674(90)90452-k. [DOI] [PubMed] [Google Scholar]
- 12.Ceman S, Brown V, Warren S T. Isolation of an FMRP-associated messenger ribonucleoprotein particle and identification of nucleolin and the fragile X-related proteins as components of the complex. Mol Cell Biol. 1999;19:7925–7932. doi: 10.1128/mcb.19.12.7925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choi Y D, Dreyfuss G. Monoclonal antibody characterization of the C proteins of heterogeneous nuclear ribonucleoprotein complexes in vertebrate cells. J Cell Biol. 1984;99:1997–2004. doi: 10.1083/jcb.99.6.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chu D T, Klymkowsky M W. The appearance of acetylated alpha-tubulin during early development and cellular differentiation in Xenopus. Dev Biol. 1989;136:104–117. doi: 10.1016/0012-1606(89)90134-6. [DOI] [PubMed] [Google Scholar]
- 15.Comery T A, Harris J B, Willems P J, Oostra B A, Irwin S A, Weiler I J, Greenough W T. Abnormal dendritic spines in fragile X knockout mice: maturation and pruning deficits. Proc Natl Acad Sci USA. 1997;94:5401–5404. doi: 10.1073/pnas.94.10.5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coy J F, Sedlacek Z, Bachner D, Hameister H, Joos S, Lichter P, Delius H, Poustka A. Highly conserved 3′ UTR and expression pattern of FXR1 points to a divergent gene regulation of FXR1 and FMR1. Hum Mol Genet. 1995;4:2209–2218. doi: 10.1093/hmg/4.12.2209. [DOI] [PubMed] [Google Scholar]
- 17.De Boulle K, Verkerk A J, Reyniers E, Vits L, Hendrickx J, Van Roy B, Van den Bos F, de Graaff E, Oostra B A, Willems P J. A point mutation in the FMR-1 gene associated with fragile X mental retardation. Nat Genet. 1993;3:31–35. doi: 10.1038/ng0193-31. [DOI] [PubMed] [Google Scholar]
- 18.Deveraux Q L, Reed J C. IAP family proteins—suppressors of apoptosis. Genes Dev. 1999;13:239–252. doi: 10.1101/gad.13.3.239. [DOI] [PubMed] [Google Scholar]
- 19.de Vries B B, Halley D J, Oostra B A, Niermeijer M F. The fragile X syndrome. J Med Genet. 1998;35:579–589. doi: 10.1136/jmg.35.7.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Devys D, Lutz Y, Rouyer N, Bellocq J P, Mandel J L. The FMR-1 protein is cytoplasmic, most abundant in neurons and appears normal in carriers of a fragile X premutation. Nat Genet. 1993;4:335–340. doi: 10.1038/ng0893-335. [DOI] [PubMed] [Google Scholar]
- 21.Eberhart D E, Malter H E, Feng Y, Warren S T. The fragile X mental retardation protein is a ribonucleoprotein containing both nuclear localization and nuclear export signals. Hum Mol Genet. 1996;5:1083–1091. doi: 10.1093/hmg/5.8.1083. [DOI] [PubMed] [Google Scholar]
- 22.Feng Y, Absher D, Eberhart D E, Brown V, Malter H E, Warren S T. FMRP associates with polyribosomes as an mRNP, and the I304N mutation of severe fragile X syndrome abolishes this association. Mol Cell. 1997;1:109–118. doi: 10.1016/s1097-2765(00)80012-x. [DOI] [PubMed] [Google Scholar]
- 23.Fortini M E, Bonini N M. Modeling human neurodegenerative diseases in Drosophila: on a wing and a prayer. Trends Genet. 2000;16:161–167. doi: 10.1016/s0168-9525(99)01939-3. [DOI] [PubMed] [Google Scholar]
- 24.Fridell R A, Benson R E, Hua J, Bogerd H P, Cullen B R. A nuclear role for the fragile X mental retardation protein. EMBO J. 1996;15:5408–5414. [PMC free article] [PubMed] [Google Scholar]
- 25.Fu Y H, Kuhl D P, Pizzuti A, Pieretti M, Sutcliffe J S, Richards S, Verkerk A J, Holden J J, Fenwick R G, Jr, Warren S T, et al. Variation of the CGG repeat at the fragile X site results in genetic instability: resolution of the Sherman paradox. Cell. 1991;67:1047–1058. doi: 10.1016/0092-8674(91)90283-5. [DOI] [PubMed] [Google Scholar]
- 26.Gedeon A K, Baker E, Robinson H, Partington M W, Gross B, Manca A, Korn B, Poustka A, Yu S, Sutherland G R, et al. Fragile X syndrome without CCG amplification has an FMR1 deletion. Nat Genet. 1992;1:341–344. doi: 10.1038/ng0892-341. [DOI] [PubMed] [Google Scholar]
- 27.Hay B A, Wassarman D A, Rubin G M. Drosophila homologs of baculovirus inhibitor of apoptosis proteins function to block cell death. Cell. 1995;83:1253–1262. doi: 10.1016/0092-8674(95)90150-7. [DOI] [PubMed] [Google Scholar]
- 28.Heitz D, Rousseau F, Devys D, Saccone S, Abderrahim H, Le Paslier D, Cohen D, Vincent A, Toniolo D, Della Valle G, et al. Isolation of sequences that span the fragile X and identification of a fragile X-related CpG island. Science. 1991;251:1236–1239. doi: 10.1126/science.2006411. [DOI] [PubMed] [Google Scholar]
- 29.Hergersberg M, Matsuo K, Gassmann M, Schaffner W, Luscher B, Rulicke T, Aguzzi A. Tissue-specific expression of a FMR1/beta-galactosidase fusion gene in transgenic mice. Hum Mol Genet. 1995;4:359–366. doi: 10.1093/hmg/4.3.359. [DOI] [PubMed] [Google Scholar]
- 30.Hinds H L, Ashley C T, Sutcliffe J S, Nelson D L, Warren S T, Housman D E, Schalling M. Tissue specific expression of FMR-1 provides evidence for a functional role in fragile X syndrome. Nat Genet. 1993;3:36–43. doi: 10.1038/ng0193-36. [DOI] [PubMed] [Google Scholar]
- 31.Jones A R, Schedl T. Mutations in gld-1, a female germ cell-specific tumor suppressor gene in Caenorhabditis elegans, affect a conserved domain also found in Src-associated protein Sam68. Genes Dev. 1995;9:1491–1504. doi: 10.1101/gad.9.12.1491. [DOI] [PubMed] [Google Scholar]
- 32.Karim F D, Chang H C, Therrien M, Wassarman D A, Laverty T, Rubin G M. A screen for genes that function downstream of Ras1 during Drosophila eye development. Genetics. 1996;143:315–329. doi: 10.1093/genetics/143.1.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khandjian E W, Bardoni B, Corbin F, Sittler A, Giroux S, Heitz D, Tremblay S, Pinset C, Montarras D, Rousseau F, Mandel J. Novel isoforms of the fragile X related protein FXR1P are expressed during myogenesis. Hum Mol Genet. 1998;7:2121–2128. doi: 10.1093/hmg/7.13.2121. [DOI] [PubMed] [Google Scholar]
- 34.Khandjian E W, Corbin F, Woerly S, Rousseau F. The fragile X mental retardation protein is associated with ribosomes. Nat Genet. 1996;12:91–93. doi: 10.1038/ng0196-91. [DOI] [PubMed] [Google Scholar]
- 35.Kiledjian M, Dreyfuss G. Primary structure and binding activity of the hnRNP U protein: binding RNA through RGG box. EMBO J. 1992;11:2655–2664. doi: 10.1002/j.1460-2075.1992.tb05331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kremer E J, Pritchard M, Lynch M, Yu S, Holman K, Baker E, Warren S T, Schlessinger D, Sutherland G R, Richards R I. Mapping of DNA instability at the fragile X to a trinucleotide repeat sequence p(CCG)n. Science. 1991;252:1711–1714. doi: 10.1126/science.1675488. [DOI] [PubMed] [Google Scholar]
- 37.Li Z, Karlovich C A, Fish M P, Scott M P, Myers R M. A putative Drosophila homolog of the Huntington's disease gene. Hum Mol Genet. 1999;8:1807–1815. doi: 10.1093/hmg/8.9.1807. [DOI] [PubMed] [Google Scholar]
- 38.Mahone M, Saffman E E, Lasko P F. Localized Bicaudal-C RNA encodes a protein containing a KH domain, the RNA binding motif of FMR1. EMBO J. 1995;14:2043–2055. doi: 10.1002/j.1460-2075.1995.tb07196.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mayeda A, Munroe S H, Xu R M, Krainer A R. Distinct functions of the closely related tandem RNA-recognition motifs of hnRNP A1. RNA. 1998;4:1111–1123. doi: 10.1017/s135583829898089x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miklos G L, Rubin G M. The role of the genome project in determining gene function: insights from model organisms. Cell. 1996;86:521–529. doi: 10.1016/s0092-8674(00)80126-9. [DOI] [PubMed] [Google Scholar]
- 41.Musco G, Stier G, Joseph C, Castiglione Morelli M A, Nilges M, Gibson T J, Pastore A. Three-dimensional structure and stability of the KH domain: molecular insights into the fragile X syndrome. Cell. 1996;85:237–245. doi: 10.1016/s0092-8674(00)81100-9. [DOI] [PubMed] [Google Scholar]
- 42.Nussbaum R L, Ledbetter D H. Fragile X syndrome: a unique mutation in man. Annu Rev Genet. 1986;20:109–145. doi: 10.1146/annurev.ge.20.120186.000545. [DOI] [PubMed] [Google Scholar]
- 43.Oberle I, Rousseau F, Heitz D, Kretz C, Devys D, Hanauer A, Boue J, Bertheas M F, Mandel J L. Instability of a 550-base pair DNA segment and abnormal methylation in fragile X syndrome. Science. 1991;252:1097–1102. doi: 10.1126/science.252.5009.1097. [DOI] [PubMed] [Google Scholar]
- 44.Pan D, Rubin G M. Kuzbanian controls proteolytic processing of Notch and mediates lateral inhibition during Drosophila and vertebrate neurogenesis. Cell. 1997;90:271–280. doi: 10.1016/s0092-8674(00)80335-9. [DOI] [PubMed] [Google Scholar]
- 45.Pieretti M, Zhang F P, Fu Y H, Warren S T, Oostra B A, Caskey C T, Nelson D L. Absence of expression of the FMR-1 gene in fragile X syndrome. Cell. 1991;66:817–822. doi: 10.1016/0092-8674(91)90125-i. [DOI] [PubMed] [Google Scholar]
- 46.Pinol-Roma S, Choi Y D, Dreyfuss G. Immunological methods for purification and characterization of heterogeneous nuclear ribonucleoprotein particles. Methods Enzymol. 1990;181:317–325. doi: 10.1016/0076-6879(90)81132-e. [DOI] [PubMed] [Google Scholar]
- 47.Rebay I, Chen F, Hsiao F, Kolodziej P A, Kuang B H, Laverty T, Suh C, Voas M, Williams A, Rubin G M. A genetic screen for novel components of the Ras/mitogen-activated protein kinase signaling pathway that interact with the van gene of Drosophila identifies split ends, a new RNA recognition motif-containing protein. Genetics. 2000;154:695–712. doi: 10.1093/genetics/154.2.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rine J. Gene overexpression in studies of Saccharomyces cerevisiae. Methods Enzymol. 1991;194:239–251. doi: 10.1016/0076-6879(91)94019-9. [DOI] [PubMed] [Google Scholar]
- 49.Rorth P. A modular misexpression screen in Drosophila detecting tissue-specific phenotypes. Proc Natl Acad Sci USA. 1996;93:12418–12422. doi: 10.1073/pnas.93.22.12418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rorth P, Szabo K, Bailey A, Laverty T, Rehm J, Rubin G M, Weigmann K, Milan M, Benes V, Ansorge W, Cohen S M. Systematic gain-of-function genetics in Drosophila. Development. 1998;125:1049–1057. doi: 10.1242/dev.125.6.1049. [DOI] [PubMed] [Google Scholar]
- 51.Rubin G M, Spradling A C. Genetic transformation of Drosophila with transposable element vectors. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- 52.Simon M A, Bowtell D D, Dodson G S, Laverty T R, Rubin G M. Ras1 and a putative guanine nucleotide exchange factor perform crucial steps in signaling by the sevenless protein tyrosine kinase. Cell. 1991;67:701–716. doi: 10.1016/0092-8674(91)90065-7. [DOI] [PubMed] [Google Scholar]
- 53.Siomi H, Choi M, Siomi M C, Nussbaum R L, Dreyfuss G. Essential role for KH domains in RNA binding: impaired RNA binding by a mutation in the KH domain of FMR1 that causes fragile X syndrome. Cell. 1994;77:33–39. doi: 10.1016/0092-8674(94)90232-1. [DOI] [PubMed] [Google Scholar]
- 54.Siomi H, Matunis M J, Michael W M, Dreyfuss G. The pre-mRNA binding K protein contains a novel evolutionarily conserved motif. Nucleic Acids Res. 1993;21:1193–1198. doi: 10.1093/nar/21.5.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Siomi H, Siomi M C, Nussbaum R L, Dreyfuss G. The protein product of the fragile X gene, FMR1, has characteristics of an RNA-binding protein. Cell. 1993;74:291–298. doi: 10.1016/0092-8674(93)90420-u. [DOI] [PubMed] [Google Scholar]
- 56.Siomi M C, Siomi H, Sauer W H, Srinivasan S, Nussbaum R L, Dreyfuss G. FXR1, an autosomal homolog of the fragile X mental retardation gene. EMBO J. 1995;14:2401–2408. doi: 10.1002/j.1460-2075.1995.tb07237.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Siomi M C, Zhang Y, Siomi H, Dreyfuss G. Specific sequences in the fragile X syndrome protein FMR1 and the FXR proteins mediate their binding to 60S ribosomal subunits and the interactions among them. Mol Cell Biol. 1996;16:3825–3832. doi: 10.1128/mcb.16.7.3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sittler A, Devys D, Weber C, Mandel J L. Alternative splicing of exon 14 determines nuclear or cytoplasmic localisation of fmr1 protein isoforms. Hum Mol Genet. 1996;5:95–102. doi: 10.1093/hmg/5.1.95. [DOI] [PubMed] [Google Scholar]
- 59.Spradling A C, Rubin G M. Transposition of cloned P elements into Drosophila germ line chromosomes. Science. 1982;218:341–347. doi: 10.1126/science.6289435. [DOI] [PubMed] [Google Scholar]
- 60.Struhl G, Greenwald I. Presenilin is required for activity and nuclear access of Notch in Drosophila. Nature. 1999;398:522–525. doi: 10.1038/19091. [DOI] [PubMed] [Google Scholar]
- 61.Sutherland G R, Hecht F, Mulley J C, Glover T W, Hecht B S. Fragile sites on human chromosomes. New York, N.Y: Oxford University Press; 1986. [Google Scholar]
- 62.Tamanini F, Meijer N, Verheij C, Willems P J, Galjaard H, Oostra B A, Hoogeveen A T. FMRP is associated to the ribosomes via RNA. Hum Mol Genet. 1996;5:809–813. doi: 10.1093/hmg/5.6.809. [DOI] [PubMed] [Google Scholar]
- 63.Tamanini F, Willemsen R, van Unen L, Bontekoe C, Galjaard H, Oostra B A, Hoogeveen A T. Differential expression of FMR1, FXR1 and FXR2 proteins in human brain and testis. Hum Mol Genet. 1997;6:1315–1322. doi: 10.1093/hmg/6.8.1315. [DOI] [PubMed] [Google Scholar]
- 64.Tomlinson A, Bowtell D D, Hafen E, Rubin G M. Localization of the sevenless protein, a putative receptor for positional information, in the eye imaginal disc of Drosophila. Cell. 1987;51:143–150. doi: 10.1016/0092-8674(87)90019-5. [DOI] [PubMed] [Google Scholar]
- 65.Verkerk A J, Pieretti M, Sutcliffe J S, Fu Y H, Kuhl D P, Pizzuti A, Reiner O, Richards S, Victoria M F, Zhang F P, et al. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991;65:905–914. doi: 10.1016/0092-8674(91)90397-h. [DOI] [PubMed] [Google Scholar]
- 66.Vincent A, Heitz D, Petit C, Kretz C, Oberle I, Mandel J L. Abnormal pattern detected in fragile-X patients by pulsed-field gel electrophoresis. Nature. 1991;349:624–626. doi: 10.1038/349624a0. [DOI] [PubMed] [Google Scholar]
- 67.Weiler I J, Irwin S A, Klintsova A Y, Spencer C M, Brazelton A D, Miyashiro K, Comery T A, Patel B, Eberwine J, Greenough W T. Fragile X mental retardation protein is translated near synapses in response to neurotransmitter activation. Proc Natl Acad Sci USA. 1997;94:5395–5400. doi: 10.1073/pnas.94.10.5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wohrle D, Kotzot D, Hirst M C, Manca A, Korn B, Schmidt A, Barbi G, Rott H D, Poustka A, Davies K E, et al. A microdeletion of less than 250 kb, including the proximal part of the FMR-1 gene and the fragile-X site, in a male with the clinical phenotype of fragile-X syndrome. Am J Hum Genet. 1992;51:299–306. [PMC free article] [PubMed] [Google Scholar]
- 69.Wolff T, Ready D F. Pattern formation in the Drosophila retina. In: Bate M, Arias A M, editors. The development of Drosophila melanogaster. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1993. pp. 1277–1325. [Google Scholar]
- 70.Yamamoto D. Molecular dynamics in the developing Drosophila eye. R. G. Austin, Tex: Landes Company; 1996. [Google Scholar]
- 71.Ye Y, Lukinova N, Fortini M E. Neurogenic phenotypes and altered Notch processing in Drosophila Presenilin mutants. Nature. 1999;398:525–529. doi: 10.1038/19096. [DOI] [PubMed] [Google Scholar]
- 72.Ye Y, Fortini M E. Apoptotic activities of wild-type and Alzheimer's disease-related mutant presenilins in Drosophila melanogaster. J Cell Biol. 1999;146:1351–1364. doi: 10.1083/jcb.146.6.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yu S, Pritchard M, Kremer E, Lynch M, Nancarrow J, Baker E, Holman K, Mulley J C, Warren S T, Schlessinger D, et al. Fragile X genotype characterized by an unstable region of DNA. Science. 1991;252:1179–1181. doi: 10.1126/science.252.5009.1179. [DOI] [PubMed] [Google Scholar]
- 74.Zhang Y, O'Connor J P, Siomi M C, Srinivasan S, Dutra A, Nussbaum R L, Dreyfuss G. The fragile X mental retardation syndrome protein interacts with novel homologs FXR1 and FXR2. EMBO J. 1995;14:5358–5366. doi: 10.1002/j.1460-2075.1995.tb00220.x. [DOI] [PMC free article] [PubMed] [Google Scholar]