Abstract
Esophageal squamous cell carcinoma (ESCC) is one of the most aggressive forms of human malignancy, often displaying limited therapeutic response. Here, we examine the non-steroidal anti-inflammatory drug diclofenac (DCF) as a novel therapeutic agent in ESCC using complementary in vitro and in vivo models. DCF selectively reduced viability of human ESCC cell lines TE11, KYSE150, and KYSE410 as compared with normal primary or immortalized esophageal keratinocytes. Apoptosis and altered cell cycle profiles were documented in DCF-treated TE11 and KYSE 150. In DCF-treated TE11, RNA-Sequencing identified differentially expressed genes and Ingenuity Pathway Analysis predicted alterations in pathways associated with cellular metabolism and p53 signaling. Downregulation of proteins associated with glycolysis was documented in DCF-treated TE11 and KYSE150. In response to DCF, TE11 cells further displayed reduced levels of ATP, pyruvate, and lactate. Evidence of mitochondrial depolarization and superoxide production was induced by DCF in TE11 and KYSE150. In DCF-treated TE11, the superoxide scavenger MitoTempo improved viability, supporting a role for mitochondrial reactive oxygen species in DCF-mediated toxicity. DCF treatment resulted in increased expression of p53 in TE11 and KYSE150. p53 was further identified as a mediator of DCF-mediated toxicity in TE11 as genetic depletion of p53 partially limited apoptosis in response to DCF. Consistent with the anticancer activity of DCF in vitro, the drug significantly decreased tumor burdene in syngeneic ESCC xenograft tumors and 4-nitroquinoline 1-oxide-mediated ESCC lesions in vivo. These preclinical findings identify DCF as an experimental therapeutic that should be explored further in ESCC.
Here, we identify the non-steroidal anti-inflammatory drug diclofenac as an effective experimental therapeutic for ESCC tumors in vivo. We further document diclofenac-mediated cytotoxicity in ESCC cells in vitro that is associated with cell metabolism and p53.
Introduction
Esophageal cancer is the tenth most prevalent cancer type and the sixth-leading cause of cancer-associated mortality worldwide (1). Across the globe, esophageal squamous cell carcinoma (ESCC) is the predominate histological subtype of esophageal cancer, accounting for ~80% of all esophageal cancers (2,3). ESCC most often occurs in the upper half of the esophagus, arising via malignant transformation of esophageal keratinocytes, initiating with basal cell hyperplasia then progressing to intraepithelial neoplasia, and eventually invasive carcinoma (4). The primary risk factors for ESCC are alcohol consumption and tobacco use (5,6) and common genetic events in ESCC include alterations in p53 (TP53), phosphatase and tensin homolog (PTEN), phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha (PIK3CA), cyclin D1 (CCND1), Notch receptor 1 (NOTCH1), nuclear factor, erythroid 2 like 2 (NFE2L2) and SRY-box transcription factor 2 (SOX2) (7–9). Despite recent advances in our understanding of the biology underlying esophageal carcinogenesis and improvements in clinical management of the disease, esophageal cancer remains one of the most aggressive forms of human malignancy with a 5-year survival rate of <20% and a significant portion of patients with advanced disease failing to respond to current standard of care (10). Thus, there presently exists an urgent need to identify novel approaches to treat ESCC.
Non-steroidal anti-inflammatory drugs (NSAIDs) have been of great interest for their anticancer properties as epidemiological evidence indicates that these commonly used agents limit development and progression of various cancer types, including colorectal, breast, lung, bladder and prostate cancers (11). NSAIDs act primarily on cyclooxygenase (COX) enzymes COX-1 and COX-2, the rate limiting enzymes in prostaglandin synthesis, to suppress inflammation. While COX-1 is constitutively expressed in most tissues, COX-2 expression is associated with inflammation. Increased COX-2 expression has been found in primary and metastatic ESCC lesions as compared with normal esophageal epithelium and correlates with decreased patient survival (12–14). In ESCC cells in vitro, inhibition of COX-2 by genetic depletion or NSAID treatment reduces proliferation and induces apoptotic cell death (15). Diclofenac (DCF), an NSAID that robustly inhibits COX-2 (16), has shown therapeutic promise in experimental models of various cancers (17–19), but has yet to be explored in the context of ESCC. In the current preclinical study, we employ complementary in vitro and in vivo models of ESCC to investigate therapeutic potential of DCF in this deadly disease.
Materials and methods
Cell culture
Human ESCC cell line TE11 and murine ESCC cell line AKR were obtained from Anil K. Rustgi, MD and Hiroshi Nakagawa, MD, PhD (Columbia University) in July 2017 with short tandem repeat (STR) analysis confirming their identity. TE11 and AKR were cultured in Dulbecco’s modified Eagle’s medium (10-013-CV, Corning) supplemented with 10% fetal bovine serum (FBS; PS-NB1, PEAK Serum) and 1% penicillin–streptomycin (pen-strep; 15140-122, Gibco). KYSE150 (CSC-C0423) and KYSE410 (CSC-C6791J) were purchased from Creative Bioarray in August 2021 with STR profiling confirming the identity of the cell lines. KYSE150 and KYSE410 were cultured in Roswell Park Memorial Institute (RPMI)-1640 (SH30096.01, Cytiva) supplemented with 10% FBS and 1% pen-step. Primary esophageal keratinocytes (EPC203) were generated as described previously (20) and received from Amanda Muir, MD (Children’s Hospital of Philadelphia) in June 2018. EPC2-hTERT were obtained from Anil K. Rustgi, MD and Hiroshi Nakagawa, MD, PhD (Columbia University) in July 2017 with STR analysis confirming their identity. Primary esophageal keratinocytes and EPC2-hTERT cells were cultured in keratinocyte-serum-free medium (10724-011, Gibco) and supplemented with epidermal growth factor (EGF; 10450-013, Gibco) and bovine pituitary extract (BPE; 13028-014, Gibco) as described previously (20,21). All the cells were maintained in a humidified incubator at 37°C with 5% CO2. Laxco LMI6000 inverted microscope was used to image cells. Trypsin-EDTA (25-053-CI, Corning) was used for cell detachment. Countess™ 3 Automated Cell Counter (Thermo Fisher Scientific) was used to count cells with 0.2% Trypan Blue dye to exclude dead cells. All cell lines were subjected to re-authentication by STR in July 2022.
Cell viability assay
The antiproliferative activity of DCF (D6899, Sigma) was assessed using CellTiter 96® Non-Radioactive Cell Proliferation Assay (MTT; G4000, Promega), according to the manufacturer’s instructions. Cells (TE11, KYSE150, KYSE410, EPC2T-hTERT, Primary, AKR) were plated at a density of 5 × 103 cells/well/100 µl media in 96-well plate and treated with vehicle [0.07% dimethyl sulfoxide (DMSO)] or DCF (6.25, 12.5, 25, 50, 100 and 200 µM) for 72 h. 15 µl MTT reagent was added to each well and incubated for 4 h. After addition of MTT solubilizing agent, absorbance was measured at 560 nm using Modulus microplate reader (Promega). Cell viability was determined as follows: . IC50 concentration was calculated using GraphPad Prism version 9.0.
Flow cytometry
TE11 or KYSE150 cells were plated at a density of 3–5 × 105 cells/well/3 ml media in 6-well plate and treated with vehicle (0.07% DMSO) or DCF for 48 h. TE11 cells were treated with 200 µM DCF. KYSE150 were treated with 400 µM. Apoptosis was examined with FITC Annexin V (Catalog no. 556419, BD Biosciences) and propidium iodide (PI) assay kit (Catalog no. 556463, BD Biosciences). Cell cycle distribution of DNA content was assessed with PI flow cytometry assay kit (Catalog no. ab139418, Abcam). Mitochondrial membrane potential was determined by staining cells with MitoTracker Green (Catalog no. M7514, Thermo Fisher Scientific) at a final concentration of 25 nM and MitoTracker Deep Red (Catalog no. M22426, Thermo Fisher Scientific) at a final concentration of 100 nM. Mitochondrial superoxide production was measured by staining cells with MitoSOX Red (Catalog no. M36008, Thermo Fisher Scientific) at a final concentration of 5 µM. Cells were washed with cold phosphate-buffered saline (PBS; 10010-023, Gibco) and resuspended in Annexin V binding buffer (Catalog no. 556454, BD Biosciences). Flow cytometry was performed using LSR-II flow cytometer (BD Biosciences) and data were analyzed with FlowJo version 10.0. Unstained controls for TE11 and KYSE are shown in Supplementary Figure 1, available at Carcinogenesis Online.
RNA extraction
For RNA extraction, TE11 or KYSE cells were plated at a density of 5 × 105 cells/well/3 ml media in 6-well plate and treated with vehicle (0.07% DMSO) or DCF for 48 h. TE11 cells were treated with 200 µM DCF. KYSE150 were treated with 400 µM. Total RNA was extracted and purified using RNA mini kit (Catalog no. 74104, Qiagen), according to the manufacturer’s instructions. RNA quantity and purity were assessed using Nanodrop™ 2000 (Thermo Fisher Scientific, Waltham, MA), by measuring absorbance ratios at 260/280 and 260/230. RNA integrity was also verified using RNA 6000 nano kit (Agilent) and Agilent bioanalyzer 2100. Only RNA of good quality with an absorbance ratio of at least 2.0 and RIN value of at least 9 was used for RNA-Sequencing and quantitative real-time PCR (qRT-PCR) assays.
RNA-Sequencing and pathway analysis
cDNA library was constructed with NEBNext® Ultra™ II Directional RNA Library Prep Kit for Illumina® (New England BioLabs). Next, pair-end 75 bp sequencing was conducted using NextSeq 500 (Illumina, San Diego, CA). Approximately, 26.5 million reads were generated per sample. Illumina reads were mapped to the reference mouse genome (GRCm38) using Rsubread 2.09 and differentially expressed genes were subsequently calculated via the DESeq2 1.34.0 packages on R. QIAGEN Ingenuity Pathway Analysis was performed on normalized count matrices and pathway plots were generated with ggplot2 on R.
Measurement of cellular ATP
Cellular ATP was detected using CellTiter-Glo® 2.0 ATP assay kit (G9241, Promega), according to the manufacturer’s instructions. TE11 cells were plated at a density of 5 × 103 cells/well/100 µl media in 96-well plate and treated with vehicle (0.07% DMSO) or 200 µM DCF for 48 h. 100 µl of CellTiter-Glo® 2.0 reagent was added to each well and mixed for 2 min on a shaker to induce cell lysis. Luminescence reading was recorded after 10 min of incubation at room temperature using Modulus microplate reader (Promega).
Measurement of oxygen consumption rate
Oxygen consumption rate (OCR) was determined using oxygen consumption rate assay kit (Catalog no. 600800, Cayman Chemical) according to the manufacturer’s instructions. TE11 cells were plated at a density of 2 × 104 cells/well/100 µl media in 96-well plate and treated with vehicle (0.07% DMSO) or 200 µM DCF for 48 h. Culture media was removed and replaced with 160 µl fresh media containing vehicle (0.07% DMSO) or 200 µM DCF. 10 µl of phosphorescent oxygen probe solution was added to each well and 100 µl of HS mineral oil was gently overlayed. Fluorescence reading was recorded kinetically at excitation and emission wavelengths of 380 and 650 nm, respectively, for ~2 h and 10 min using SpectraMax® i3x microplate reader (Molecular Devices).
Measurement of pyruvate level
Pyruvate level was measured using pyruvate assay kit (Catalog no. 700470, Cayman Chemical) according to the manufacturer’s instructions. TE11 cells were plated at a density of 3 × 105 cells/well/3 ml media in 6-well plate and treated with vehicle (0.07% DMSO) or 200 µM DCF for 48 h. 500 µl of supernatant vortexed with 500 µl of 0.5 M MPA and placed on ice for 5 min. Then the mixture was centrifuged at 10, 000g for 5 min at 4°C. The supernatant was collected and diluted at 1:2 ratio. 50 µl of assay buffer, 50 µl of cofactor mixture, 10 µl of fluorometric detector, 20 µl of supernatant and 20 µl of enzyme mixture were added to the 96-well plate. After 20-min incubation at room temperature, fluorescence reading was recorded at excitation and emission wavelengths of 530 and 585 nm, respectively, using SpectraMax® i3x microplate reader (Molecular Devices).
Measurement of lactate level
Lactate level was measured using glycolysis assay kit (Catalog no. 600450, Cayman Chemical) according to the manufacturer’s instructions. TE11 cells were plated at a density of 5 × 103 cells/well/100 µl media in 96-well plate and treated with vehicle (0.07% DMSO) or 200 µM DCF for 48 h. The 96-well plate was centrifuged at 1000 rpm for 5 min and supernatant was collected. 90 µl of assay buffer was added to a new plate and 10 µl of supernatant was transferred. 100 µl of reaction solution was added to each well and the plate was incubated at room temperature on a shaker for 30 min. The absorbance was recorded using Modulus microplate reader (Promega).
Mitochondrial reactive oxygen species inhibition
Mitochondrial reactive oxygen species (ROS) was inhibited by mitochondrial ROS scavenger MitoTempo (SML0737, Sigma). TE11 cells were plated at a density of 5 × 103 cells/well/100 µl media in 96-well plate and pretreated with 100 and 200 µM of MitoTempo for 24 h. Cells were treated with vehicle (0.07% DMSO) or 200 µM of DCF for 48 h. Cell viability was measured using CellTiter 96® Non-Radioactive Cell Proliferation Assay (MTT; Promega) as described above.
Quantitative real-time PCR
1 µg total RNA was cDNA using Maxima first strand cDNA synthesis kit (Catalog no. K1642, Thermo Fisher Scientific). Resulting cDNA was amplified with using primers listed in Supplementary Table 1, available at Carcinogenesis Online and PowerTrack™ SYBR™ green master mix (A46109, Thermo Fisher Scientific). Thermocycling condition for qRT-PCR was 95°C for 2 min, followed by 40 cycles of 95°C for 5 s and 60°C for 30 s. GAPDH was used as an internal control gene. Relative quantification of the target gene was calculated by comparative 2−∆∆CT method.
Immunoblotting
TE11 or KYSE150 cells were plated at a density of 3 × 105 cells/well/3 ml media in 6-well plate and treated with vehicle (0.07% DMSO) or DCF for 48 h. TE11 cells were treated with 200 µM DCF. KYSE150 were treated with 400 µM. Cells were lysed in cell lysis buffer (Cat# 9830 S, Cell Signaling Technology) containing protease/phosphatase inhibitor cocktail (Cat# 5872 S, Cell Signaling Technology). Protein concentration was determined by Qubit™ protein assay kit (Cat# Q33211, Invitrogen). Protein samples were solubilized in NuPAGE™ LDS Sample Buffer (Cat# NP0007, Invitrogen) and denatured with NuPAGE™ sample reducing agent (Cat# NP0009, Invitrogen) containing 50 mM dithiothreitol. 30 µg of denatured protein was fractionated on NuPAGE™ Bis-Tris 4–12% gel (Cat# NP0335BOX, Invitrogen). Following electrotransfer, Immobilon-P PVDF membranes (Cat# IPVH00010, Millipore Sigma) were blocked in blocking buffer containing 5% non-fat milk (Cat# LP0031B, Thermo Fisher Scientific) in PBST (PBS and 0.1% Tween 20) for 1 h at room temperature. Membranes were then incubated overnight with primary antibodies diluted in blocking buffer and then with the appropriate HRP-conjugated secondary antibody for 1 h at room temperature. β-Actin served as a loading control. A list of antibodies with dilutions used is provided in Supplementary Table 2, available at Carcinogenesis Online.
RNA interference and transfection
siRNA sequences directed against TP53 (J-003329-14, J-003329-15, J-00329-16, J-003329-17; Dharmacon) or a non-targeted (NT) control sequence (D-001810-01-05; Dharmacon) were transfected with Lipofectamine® RNAiMAX reagent (13778-150, Thermo Fisher Scientific), as described previously (22). Briefly, 2.5 µl of TP53 siRNA or NT siRNA and 5 µl of Lipofectamine® RNAiMAX reagent were mixed with 492.5 µl of OPTI-MEM® I (31985-062, Thermo Fisher Scientific), added in 6-well plate and incubated for 20 min at room temperature. TE11 cells were plated at a density of 2.5 × 105 cells/well/2 ml media without pen-strep and incubated for 72 h. TP53 knockdown was verified by immunoblotting method described above. TP53 depleted or NT control TE11 cells were treated with 200 µM DCF for 72 h and cell viability was determined using Countess™ 3 Automated Cell Counter (Thermo Fisher Scientific) by staining cell with 0.2% Trypan Blue dye. In addition, TP53 depleted or NT control TE11 cells were treated with 200 µM DCF for 48 h. Annexin V/PI flow cytometry, RNA extraction, and qPCR was performed as described above.
Murine models of ESCC
Syngenetic transplantation of murine ESCC cells and 4-nitroquinoline 1-oxide (4-NQO)-mediated ESCC experiments were carried out under a protocol approved by the Temple University Institutional Animal Care and Use Committee. Both male and female mice (in equal numbers) were used for 4-NQO treatment and as recipients for tumor cell transplantation studies. For transplantation studies, the murine ESCC cell line AKR (23), a generous gift from Anil K. Rustgi, MD and Hiroshi Nakagawa, MD, PhD (Columbia University), was used. Sample sizes for groups were projected based upon previous xenograft studies then adjusted following acquisition of data in initial experiment. Animals were randomized by cages upon arrival. Investigators were informed of groups during the treatment phase of experiments. AKR cells were suspended in media and implanted subcutaneously into the dorsal flanks of 10- to 12-week-old C57BL6 mice. When tumor volume reached ~125 mm3, mice were subjected to intraperitoneal (i.p.) injection of DCF (15 mg/kg), or vehicle (7% DMSO) according to a 3 days on/2 days off regimen for 11 days. Tumor volume was monitored using digital calipers and volume was calculated using the formula: . Tumor weight was measured following tumor dissection. Sample sizes for groups were projected based upon previous xenograft studies (24). For 4-NQO studies, C57BL6 mice were randomized by cage and administered 4-NQO (100 µg/ml) for 16 weeks via drinking water. 4-NQO was then withdrawn for a period of 6 weeks. Mice were subjected to i.p. injection of DCF (15 mg/kg), or vehicle (7% DMSO) according to a 3 days on/2 days off regimen for 2 weeks. Esophagi were dissected and imaged using the MVX10 microscope (Olympus) then processed for histology. Tumor area was measured by ImageJ (NIH). For all murine studies, sample sizes for groups were projected based upon previous studies (24). Investigators were informed of groups during the treatment phase of experiments. Upon processing, mouse-derived materials were given a unique identifier to blind investigators during analyses and outcome assessments.
Histological analysis
Whole esophagi were dissected and fixed with 4% paraformaldehyde for 12 h at 4°C. Tissues were washed with PBS then stored in 70% ethanol at 4°C prior to paraffin embedding. Slides were subjected to hematoxylin and eosin (H&E) staining and immunohistochemistry (IHC) for p53 (CM-5; P53-CM5P-L; Leica) at 1:200 dilution and EDTA antigen retrieval for 64 min using Ventana Autostainer (Roche). A pathologist (A.J.K.-S.) blinded to treatment parameters scored p53 labeling index. Slides were imaged using Leica DM 1000 LED microscope.
Statistical analyses
All data were analyzed as mean ± SD except in vivo data which were analyzed as mean ± SEM. Differences between groups were determined by t-test or ANOVA with indicated post hoc test using GraphPad Prism 9.0 software. P< 0.05 was considered significant.
Results
DCF inhibits cell viability, induces apoptosis, and alters cell cycle in ESCC cells in vitro
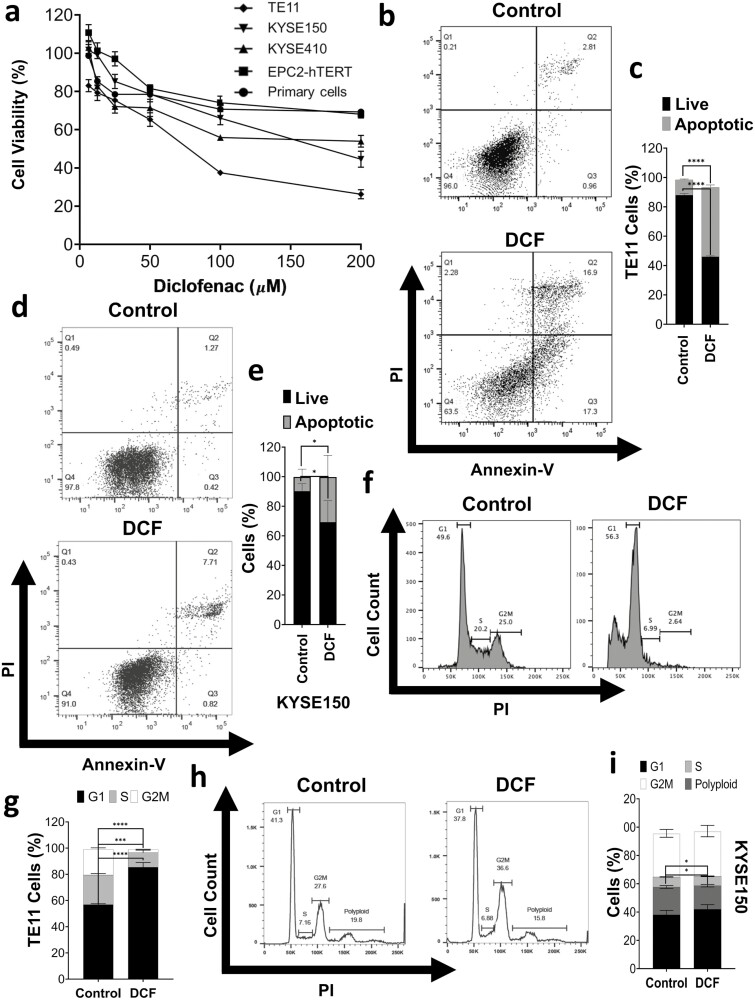
We first examined the effect of DCF on viability in ESCC cell lines as well as normal control cells using MTT assays. DCF markedly inhibited the viability of TE11, KYSE150 and KYSE410 ESCC cells in a dose-dependent manner (Figure 1a). The drug’s effect was most potent in TE11 with a reduction of cell viability to ~25% and an IC50 concentration of 70.47 µM. IC50 concentrations for KYSE150 and KYSE410 were 167.3 and 187.9 µM, respectively. In contrast, respective IC50 concentrations of 354.6 and 874.7 µM were observed in immortalized normal EPC2-hTERT cells and primary normal esophageal keratinocytes. In TE11 and KYSE 150, diminished viability in response to DCF was associated with induction of apoptosis (Figure 1b–e; Supplementary Figure 2, available at Carcinogenesis Online). DCF treatment also impacted cell cycle in both TE11 and KYSE150 (Figure 1f–i). In TE11, a robust increase in the proportion of cells in the G1 phase of the cell cycle along with significant reduction of S and G2M phase populations was observed (Figure 1f and g). In KYSE150, a portion of which are polyploid, a significant reduction of both the S phase and polyploid fractions was observed (Figure 1h and i).
Figure 1.
DCF inhibition of cell viability is associated with apoptosis and cycle alterations. (a) Cell viability was measured by MTT assay in ESCC cell lines (TE11, KYSE150, and KYSE410), normal immortalized esophageal keratinocyte cell line EPC2-hTERT, and primary esophageal keratinocytes treated with DCF at indicated doses for 72 h. Dose–response curves are indicated for respective cells. (b–i) TE11 or KYSE150 cells were treated for 48 h with 200 or 400 µM DCF, respectively. Apoptosis was measured by Annexin-V/PI flow cytometry with representative dot plot in (b, d) and bar chart summarizes date from three independent experiments in (c, e). (f–i) Cell cycle was analyzed by PI flow cytometry with representative dot plot in (f, h) and bar chart summarizes date from three independent experiments in (g, i). Data in (c, e, g, i) shown as mean ± SD; *P < 0.05; ***P < 0.001; ****P < 0.0001 by t-test; n = 3.
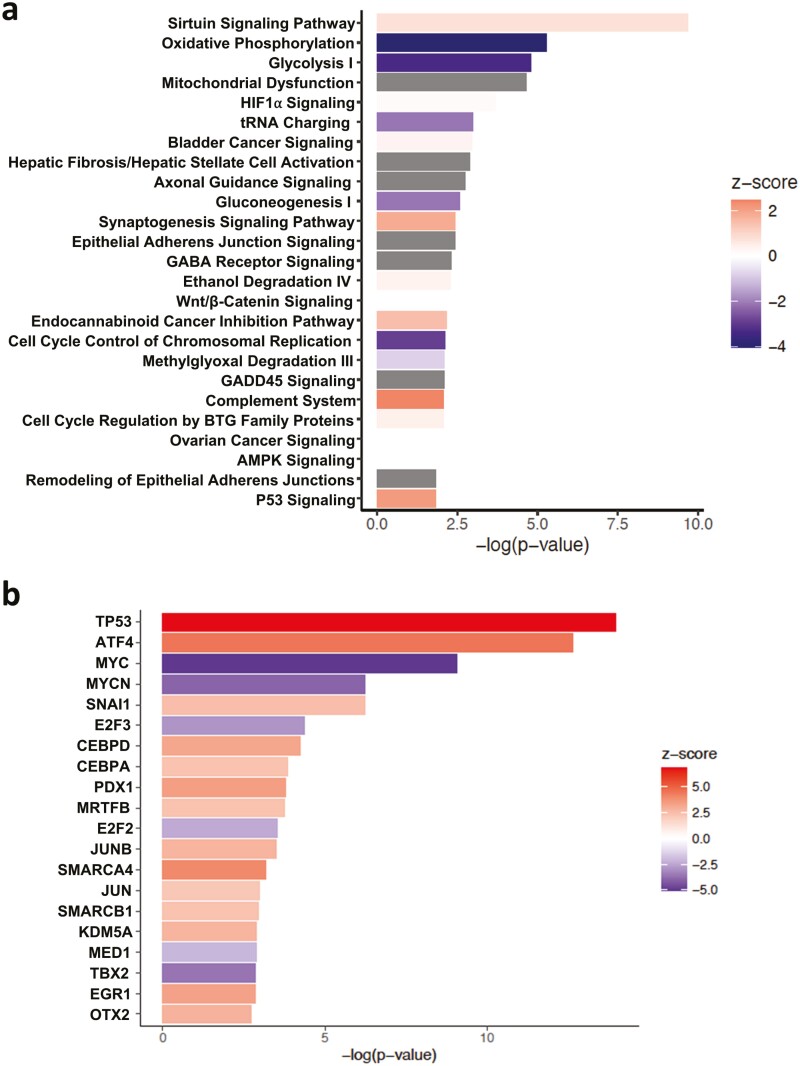
In order to examine the molecular mechanisms underlying DCF-mediated impairment of viability in ESCC cells, we performed transcriptomic profiling in TE11 cells treated with DCF or vehicle. RNA-Sequencing revealed that DCF significantly altered the expression of 2786 genes in TE11 at a cutoff fold change (FC) value of 1.50 (P < 0.05) (Supplementary File S1). Ingenuity Pathway Analysis (IPA) ranked the canonical pathways based on −log(P-value) and predicted their activation z-score (Supplementary File S1). DCF significantly modulated 63 canonical pathways in TE11 (P < 0.05). Top 20 pathways with −log(P-value) and activation z-score are shown in Figure 2a. IPA predicted DCF-mediated activation of p53 signaling as well as inhibition of metabolic pathways oxidative phosphorylation, glycolysis I, tRNA charging and gluconeogenesis. Although IPA could not predict the activation state of mitochondrial dysfunction, this was also among the most significantly altered canonical pathways in DCF-treated TE11 cells. We further performed IPA upstream regulator analysis to identify transcription regulators and kinases predicted to be activated or inhibited in TE11 cells treated with DCF. IPA ranked tumor suppressor TP53 and oncogene MYC as the top transcription factors predicted to be, respectively, activated or inhibited in DCF-treated TE11 cells (Figure 2b). IPA further ranked MAP4K4 and STK11 (LKB1) as the top kinases predicted to be, respectively, activated or inhibited in DCF-treated TE11 cells (Supplementary Figure 3, available at Carcinogenesis Online).
Figure 2.
IPA of transcriptomic changes in DCF-treated TE11 cells. TE11 cells were treated with 200 µm DCF for 48 h then subjected to transcriptomic profiling by RNA-Sequencing. IPA predicted changes in canonical pathways in (a) and transcriptional regulators in (b). IPA ranking of top canonical pathways and transcription factors is based on −log(P-value). IPA predicted activation state is based on z-score with positive value indicating activation and negative value indicating inhibition of respective pathways. No activation state is available for the canonical pathways with gray colored bar.
Alterations in metabolic pathways contribute to DCF-mediated cytotoxic activity in TE11 and KYSE150 ESCC cells
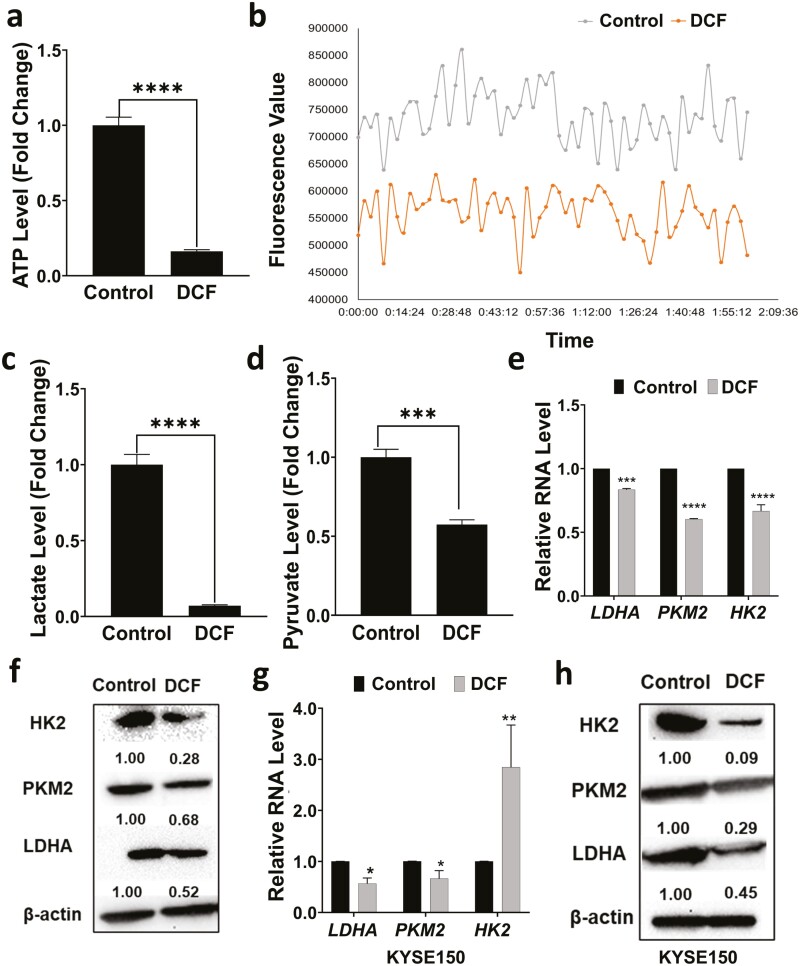
As three of the top 5 pathways predicted by IPA to impacted by DCF treatment in TE11 cells were related to metabolism (i.e. oxidative phosphorylation, glycolysis and mitochondria dysfunction) (Figure 2a), we continued to explore the effect of DCF on metabolism in ESCC cells. DCF-treated TE11 cells displayed a dramatic decrease in ATP level as compared with vehicle-treated controls (Figure 3a). Consistent with diminished energy production in response to DCF, we identified reduction in OCR (Figure 3b) as well as decreased lactate and pyruvate levels in DCF-treated TE11 cells (Figure 3c and d). We also evaluated expression of glycolysis-associated genes and proteins in TE11 and KYSE150 cells. DCF significantly reduced the expression of the glycolysis-associated enzymes, pyruvate kinase M (PKM2) and lactate dehydrogenase A (LDHA) in both cell lines (Figure 3e–h). Expression of hexokinase (HK)2, which catalyzes the first step in glucose metabolism, was diminished at the protein level in both TE11 and KYSE150 (Figure 3f and h); however, decreased HK2 RNA in response to DCF was only found in TE11 (Figure 3e). In KYSE150, a significant induction of HK2 RNA was detected (Figure 3g).
Figure 3.
DCF suppresses energy metabolism in ESCC cells. TE11 or KYSE150 cells were treated for 48 h with 200 or 400 µM DCF, respectively. TE11 were assessed for ATP level in (a), OCR in (b), lactate level in (c) and pyruvate level in (d). (e–h) In TE11 and KYSE150 cells, RNA level of glycolysis-associated genes was evaluated by qPCR in (e, g), and protein level of indicated glycolysis enzymes by immunoblotting in (f, h). In (f, h) densitometry values determined the level of indicated proteins relative to β-actin. Data in (a, c–e, g) shown as mean ± SD relative to vehicle-treated cells; *P < 0.05; **P < 0.01; ***P < 0.005; ****P < 0.001 by t-test; n = 3.
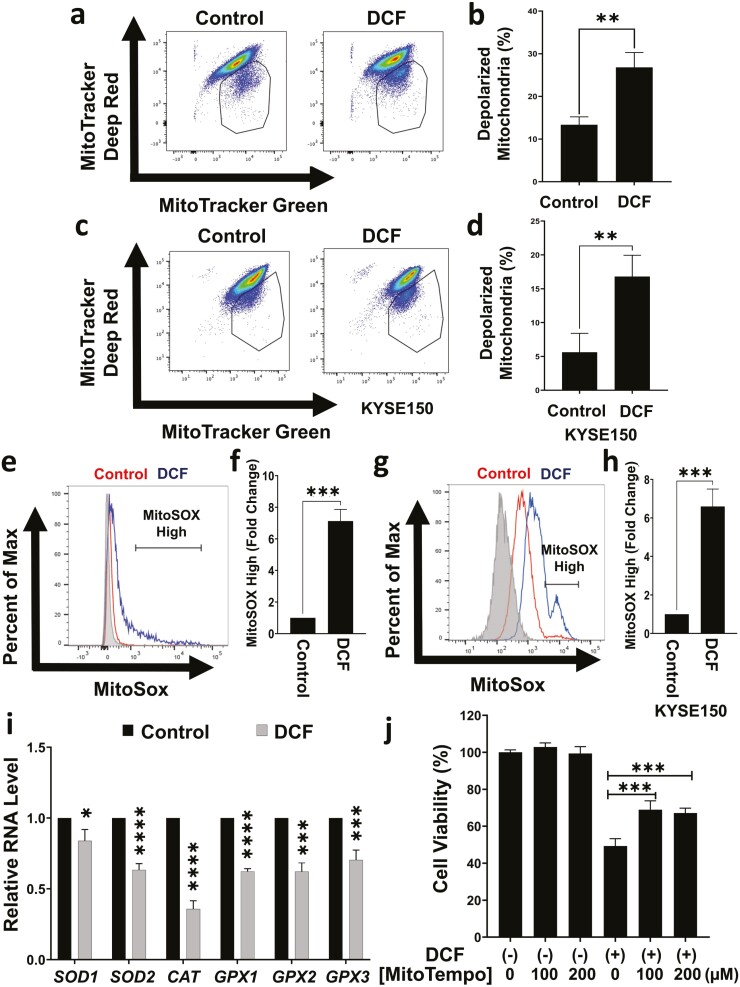
We continued to evaluate effects of DCF on mitochondria in ESCC cells. DCF promoted membrane depolarization in TE11 and KYSE150 cells (Figure 4a–d). As IPA predicted increased synthesis, production, and metabolism of ROS (Supplementary Figure 4, available at Carcinogenesis Online), we next measured mitochondrial superoxide (O2−) radical levels in DCF-treated ESCC cells. DCF elevated mitochondrial O2− radical production in TE11 and KYSE150 as evidenced by a significant increase the percentage of cells with high fluorescence for the O2− indicating dye MitoSOX (Figure 4e–h). Moreover, expression of antioxidant genes SOD1/2, CAT and GPX1/2/3 was decreased in DCF-treated TE11 cells (Figure 4i). We continued to pretreat TE11 cells with the mitochondrial antioxidant MitoTempo to determine if mitochondrial dysfunction contributes to the anticancer activity of DCF. Cell viability was significantly increased in DCF-treated TE11 cells after MitoTempo pretreatment (Figure 4j). Taken together, these findings identify metabolic dysfunction as a mechanism contributing to ESCC cell death in response to DCF.
Figure 4.
DCF induces mitochondrial membrane depolarization and mitochondrial ROS production in TE11 and KYSE150 cells. TE11 or KYSE150 cells were treated with for 48 h with 200 or 400 µM DCF, respectively. (a–d) Flow cytometry assessed mitochondrial membrane potential using MitoTracker Deep Red (membrane potential sensitive dye) and MitoTracker Green (membrane potential insensitive dye). Representative dot plot with gate indicating depolarized mitochondria in (a, c) and bar diagram summarizing data from three independent experiments in (b, d). (e–h) Flow cytometry assessed mitochondrial superoxide using MitoSOX. Representative dot plot with gate indicating MitoSOX-high fraction in (e, g) and bar diagram summarizing data from three independent experiments in (f, h). (i) RNA level of indicated antioxidant genes was evaluated in TE11 cells by qPCR relative to vehicle-treated control cells. upon treatment with DCF. (j) Prior to addition of DCF, TE11 cells were pretreated the ROS scavenger MitoTempo for 24 h cells. Data in (b, d, f, h, i) shown as mean ± SD relative to vehicle-treated cells; *P < 0.05; **P < 0.01; *** P < 0.005; ****P< 0.001 by t-test; n = 3. Data in (j) shown as mean ± SD; ***P < 0.005 by ANOVA with Tukey’s post hoc test; n = 3.
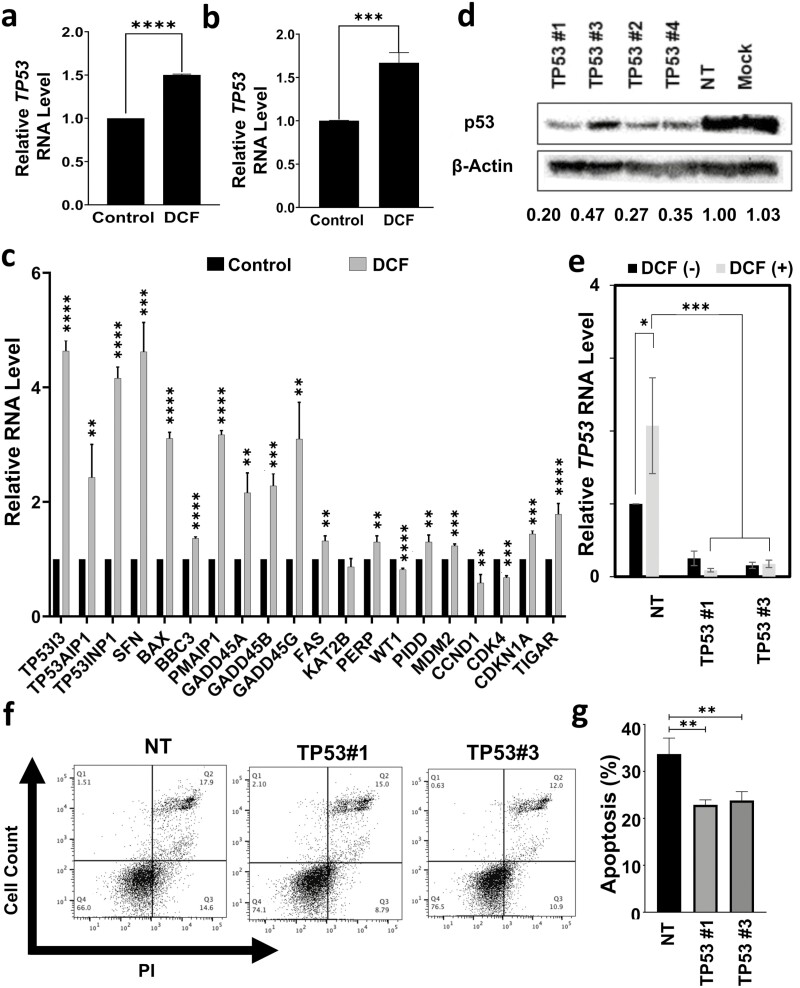
p53 plays a role in DCF-mediated apoptosis of TE11 cells
As IPA predicted activation of p53 signaling in TE11 cells treated with DCF (Figure 2a and b) and p53 is a critical player in ESCC biology, we continued by exploring the role of p53 in DCF-mediated cytotoxicity. p53 induction at the level of RNA was detected in response to DCF in both TE11 and KYSE150 cells (Figure 5a and b). Additionally, increased expression of various p53 target genes was identified in TE11 (Figure 5c). Among p53 target genes, DCF upregulated those associated with cell cycle arrest (e.g. CDKN1A, GADD45A/B/G, SFN), and apoptosis (e.g. TP53, TP53INP1, TP53AIP1, TP53I3, BAX, BBC3, PMAIP1, FAS, PERP, PIDD) while downregulating those associated with cell cycle progression (e.g. CCND1, CDK4) (Figure 5c). To functionally investigate the role of p53 signaling in the TE11 response to DCF, we knocked down p53 expression (Figure 5d and e). With p53 knockdown, we observed increased viability of DCF-treated TE11 cells (Supplementary Figure 5, available at Carcinogenesis Online) coupled with reduced apoptotic cell death (Figure 5f and g). Notably, p53 depletion did not fully suppress DCF-mediated cell death (Supplementary Figure 6, available at Carcinogenesis Online), indicating that other pathways are involved in the cytotoxicity of DCF. These findings suggest that DCF activates p53 signaling to inhibit proliferation and induce apoptosis of TE11 cells.
Figure 5.
p53 contributes to DCF-mediated cytotoxicity in TE11 cells. TE11 or KYSE150 cells were treated for 48 h with 200 or 400 µM DCF, respectively. RNA level of p53 was evaluated by qPCR relative to vehicle-treated control cells in TE11 in (a) or KYSE150 in (b). (c) In TE11 cells, RNA level of indicated p53 target genes was evaluated by qPCR relative to vehicle-treated control cells. (d) Protein level of p53 in TE11 cells was determined by immunoblotting 3 days following transfection of siRNA oligos targeting p53 or non-targeting siRNA oligos. β-Actin was used as a loading control. TE11 cells transfected with indicated siRNA oligos were treated with 200 µM DCF for 72 (e) or 48 (f, g) h. Cells were assessed for RNA level of TP53 by qPCR in (e), and apoptosis by Annexin V/PI flow cytometry with representative FACS dot plots shown in (f) with dot plots summarizing data from three independent experiments in (g). Data in (a, b, d, c, e, g) shown as mean ± SEM; *P < 0.05; **P < 0.01; ***P < 0.005; ****P < 0.001 by t-test; n = 3.
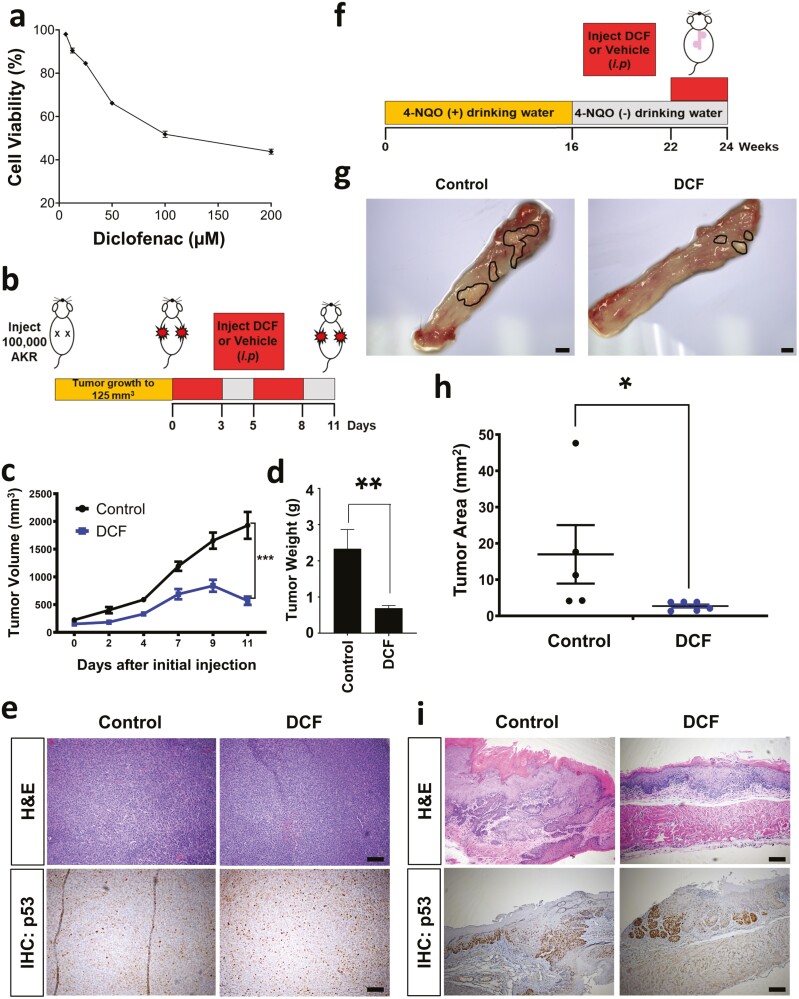
DCF inhibits ESCC cell growth in vivo
Given the apparent antitumor activity of DCF in ESCC cells in vitro, we finally aimed to determine how this drug impacts ESCC in vivo. To investigate the antitumor activity of DCF, a mouse syngeneic ESCC tumor model was employed. DCF reduced the viability of the murine ESCC cell line AKR cells in a dose-dependent manner in vitro with an IC50 concentration of 126.0 µM (Figure 6a). We then examined the effect of DCF on established AKR cells in vivo using a syngeneic xenotransplantation model (Figure 6b). DCF significantly reduced tumor growth and weight in this model system (Figure 6c and d). IHC for p53 failed to reveal a significant difference between AKR xenograft tumors in mice treated with vehicle or DCF (Figure 6e). Finally, we evaluated the impact of DCF on tumorigenesis in the 4-NQO carcinogen-driven ESCC model (Figure 6f) which mimics the effects of tobacco. Mice were treated with 4-NQO for 16 weeks followed by an 8-week wash out period. At week 22 of this protocol, mice were treated with DCF or vehicle control. After 2 weeks of treatment, DCF-treated mice displayed a marked decrease in tumor burden as compared with their vehicle-treated counterparts (Figure 6g and h). p53 staining was also not significantly altered in 4-NQO-treated mice upon DCF treatment (Figure 6i; Supplementary Figure 7, available at Carcinogenesis Online). These studies confirmed potent in vivo anticancer activity of DCF as a single agent using two independent murine ESCC models. In contrast to studies with ESCC cell lines in vitro, DCF failed to impact p53 in these two in vivo ESCC models.
Figure 6.
DCF inhibits ESCC in vivo. (a) Cell viability was measured by MTT assay in murine ESCC cell line AKR treated with DCF at indicated doses for 72 h. Dose–response curve is shown ±SD; n = 3. (b) Schematic overview of experimental design for syngenetic transplantation. (c–e) AKR cells were transplanted into C57Black6 mice. Once tumors reached ~125 mm3 mice were administered DCF (15 mg/kg) or vehicle (7% DMSO) by i.p. injection according to a 3 days on/2 days off regimen for 11 days. Tumor volume is shown in (c) and tumor weight is shown in (d). Data shown in (c, d) as mean ± SEM; *P < 0.05; **P < 0.01 by t-test; n = 5 tumors from DCF-treated mice; n = 5 tumors from vehicle-treated mice. (e) Representative brightfield images of H&E or p53 IHC staining in histological tissue sections. Scale bars, 100 µm. (f) Schematic overview of experimental design for 4-NQO-mediated ESCC model. (g–i) C57Black6 mice were administered 4-NQO (100 µg/ml) for 16 weeks via drinking water. 4-NQO was then withdrawn for a period of 6 weeks. Starting at week 22, mice were subjected to i.p. injection of DCF (15 mg/kg) or vehicle (7% DMSO) according to a 3 days on/2 days off regimen for 2 weeks. (g) Representative brightfield images of macroscopic mouse esophagus. Tumors are outlined in black. Scale bars, 1 mm. (h) Quantification of tumor burden was measured using images in (g). Data shown as mean ± SEM; *P < 0.05 by t-test; n = 6 DCF-treated mice; n = 5 vehicle-treated mice. (f) Representative brightfield images of H&E or p53 IHC staining in histological tissue sections. Scale bars, 100 µm.
Discussion
In the current study, we demonstrate the anticancer activity of DCF in ESCC using complementary in vitro and in vivo model systems. Our in vitro studies indicated that DCF more efficiently impairs viability of human and murine ESCC cell lines as compared with normal human primary or immortalized esophageal keratinocytes. As current standard of care in advanced ESCC includes neoadjuvant chemo- and/or radiotherapy (25), both of which have off-target effects, the observed selectivity of DCF for ESCC cells makes this drug an attractive candidate for further investigation. Our in vivo studies revealed that DCF effectively reduces tumor growth in syngeneic ESCC xenograft tumors and in the 4-NQO-induced murine model of ESCC. Several studies have evaluated the relationship between the use of aspirin, which inhibits both COX-1 and COX-2, and ESCC incidence as well as progression in human patients with both positive and negative associations reported (26–29). However, effects of DCF use in ESCC patients have yet to be explored to the best of our knowledge. It must be noted that the selective COX-2 inhibitor Celecoxib (200 mg, twice daily) failed to show chemopreventative activity in individuals with mild or moderate esophageal dysplasia in a randomized placebo-controlled study (30). The authors of this study note that dosage and timing of Celecoxib treatment may account for a lack of chemoprevention in their study as 400 mg twice daily Celecoxib was demonstrated to be required to reduce colonic neoplasia (31) and COX-2 expression increases with ESCC progression (32). The DCF dosing regimen that we employed was based upon published studies exploring the drug’s effects in mouse models of cancer (33); however, DCF dosage should be carefully considered in future studies. In humans, daily dose of NSAIDs routinely used in clinical practice is 1.0–2.5 mg/kg which is significantly lower than the 15 mg/kg DCF dose employed in our studies. Anecdotally, we did not observe gross signs of toxicity associated with DCF treatment in these model systems; however, future studies will carefully evaluate effects of DCF on the intestine, kidneys, and cardiovascular system as toxicity at these sites is associated with NSAIDs (11) and may complicate translation of our findings into ESCC patients. Should DCF treatment move into clinical trials, the use of a slurry may provide a potential means to increase exposure of the drug to esophageal mucosa. Furthermore, while the current study provides evidence that DCF has antitumor activity in the context of established ESCC lesions and cell lines, it is possible that DCF may also influence ESCC development, progression, and/or response to therapy.
While DCF has been shown to have antitumor properties in experimental systems of several cancer types (17–19), the mechanisms underlying these effects have yet to be fully elucidated and are likely to vary by tissue and cell type. In the current study, RNA-Sequencing in DCF-treated TE11 cells identified alterations in cellular metabolism as a potential contributor to the drug’s toxicity. In response to DCF, TE11 cells displayed reduction in ATP, lactate, and pyruvate levels. Additionally, both TE11 and KYSE150 cells demonstrated downregulation of several glycolytic enzymes in response to DCF treatment. These findings suggest that impaired glycolysis accompanies DCF-mediated cytotoxicity in ESCC cells which is consistent with evidence of enhanced glycolysis, potentially contributing to the Warburg effect, in ESCC. Gene expression profiling in Asian ESCC patients displayed enrichment of the glycolysis pathway (34). Upregulation of HK2, PKM2, and LDHA has been demonstrated in ESCC tumors with HK2 and PKM2 expression negatively correlating with disease progression, invasion, and survival (35,36). Additionally, LDHA depletion in ESCC cells suppressed growth and migration in vitro as well as xenograft tumor growth in vivo (36). DCF-treated TE11 and KYSE150 cells also exhibited evidence of mitochondrial dysfunction. A functional role for mitochondrial superoxide in DCF-mediated cytotoxicity was indicated in TE11 as the superoxide scavenger MitoTempo rescued cell viability in response to DCF. DCF-mediated induction of cell death and cell cycle arrest has also been linked to mitochondrial dysfunction in colon and liver cancer cells (18,19). Our own work has demonstrated that autophagy serves as a critical mechanism to limit mitochondrial dysfunction and oxidative stress during epithelial–mesenchymal transition (EMT)-mediated generation of ESCC cancer stem cells (CSCs) defined by high expression of CD44 (CD44H cells) (21). As DCF has been shown to limit expression of CD44 in colorectal cancer cells and to impair autophagy flux in hepatocytes (37,38), it is tempting to speculate that DCF-mediated suppression of ESCC may involve suppression of autophagy and subsequent depletion of CD44H CSCs.
Our RNA-Sequencing data also predicted activation of p53 signaling in TE11 cells treated with DCF. Gene expression analysis confirmed that DCF-induced expression of TP53 in TE11 and KYSE150 cells. Expression of p53 target genes, including TP53INP1, TP53I3, TP53AIP1, GADD45A/B/G and BBC3, was further documented in TE11 cells. These genes have been associated with reduced cell proliferation and induction of cell death in cancer cells (39–42). Genetic depletion of p53 in TE11 cells limited cell death in response to DCF, supporting a role for p53 in DCF-mediated cytotoxicity. TE11 cells have a missense mutation in exon 4 of TP53 (43). TP53 is mutated in 83% of ESCC patients (44) and mutant p53 been shown to promote malignant features of ESCC, including invasion and metastasis (24,45–47). DCF-mediated toxicity does not appear to be dependent upon the presence of mutant p53, however, as the drug limited viability in KYSE150 and KYSE410 ESCC cells both of which have wild-type p53 (43). Notably, TE11 cells were more sensitive to DCF than either KYSE150 or KYSE410, raising the possibility that mutant p53 may augment response to the drug. It will also be of interest to determine if p53 plays a role in the suppression of glycolysis and induction of mitochondrial dysfunction that we identified in TE11 cells. However, as DCF-associated cytotoxicity in TE11 cells is not fully abrogated with p53 knockdown, metabolic alterations may contribute to cell death independent of p53. In the oral cavity/tongue, 4-NQO induces p53 even in the absence of histologic changes and p53 mutation is detectable in hyperplastic and dysplastic lesions (48). Interestingly, DCF failed to impact p53 level in either of two murine immunocompetent ESCC models employed in the current study, perhaps as p53 activation was already present at the time of DCF treatment. It is possible DCF impacts the spectrum of p53 targets rather than expression; however, further experiments are necessary to investigate this. Alternatively, p53 may not be required for ESCC response to DCF in vivo. DCF-mediated inhibition of COX-2 is likely to have effects on the inflammatory tumor microenvironment. Moving forward, the described animal models will facilitate delineation of immune-mediated mechanisms as well as the functional role of p53 (both mutant and wild-type) in the antitumor effects of DCF. Xenografts and 3D organoids derived from ESCC patients will also be employed to define the impact of DCF on human esophageal cells, both malignant and non-malignant (49,50).
Beyond effects on metabolism and p53, our RNA-Sequencing data are hypothesis-generating, identifying canonical pathways, transcriptional regulators and kinases that may contribute to DCF-mediated cytotoxicity in ESCC. Sirtuin signaling pathway and hypoxia-inducible factor (HIF)-1α signaling are among the top 5 canonical pathways predicted to be associated with response to DCF in TE11 ESCC cells (Figure 2a). Members of the Sirtuin family have been linked to ESCC with cancer-promoting roles suggested for SIRT1, 2, 3 and 6 (51). HIF-1α has also been shown to promote ESCC via mechanisms that include induction of EMT and activation of Wnt/β-catenin signaling (52–54). SIRT1-mediated deacetylation of p53 inhibits its transcriptional activity (55), providing a potential mechanism through which SIRT1 may limit p53-mediated cytotoxicity in DCF-treated ESCC cells. Moreover, HIF-1α signaling has been linked to activation of the COX-2–prostaglandin E synthase–prostaglandin–E2 axis in esophageal keratinocytes upon exposure to hypoxia (56). Thus, it is possible that activation of Sirtuins and HIF-1α signaling may represent mechanisms of resistance to DCF-mediated ESCC cell death. Should this be the case, inhibition of these pathways may augment DCF-mediated antitumor activity. In addition to p53, IPA predicted upregulation of transcriptional factor ATF4 (Figure 2b), which is a critical mediator of endoplasmic reticulum (ER) stress pathway along with PERK (EIF2AK3), the latter of which is a kinase predicted to be upregulated in DCF-treated TE11 cells. As such, it will be of interest to determine if ER stress contributes to DCF-mediated toxicity in ESCC.
Taken together, the current study presents DCF as a promising experimental therapeutic agent that effectively targets ESCC in vitro and in vivo. Mechanistically, the anticancer activity of DCF in ESCC cells in vitro is associated with activation of p53 signaling and suppression of energy metabolism pathways. Furthermore, DCF exhibits potent antitumor effects in vivo that is not associated with increased p53 expression. In sum, these preclinical findings provide a foundation to support further exploration of the therapeutic potential of DCF in ESCC.
Supplementary Material
Acknowledgements
We thank Amir Yarmahmoodi, manager of the Lewis Katz School of Medicine Flow Cytometry Core, and members of the Fox Chase Cancer Center Histopathology Facility and Genomics Facility for technical support.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- COX
cyclooxygenase
- DCF
diclofenac
- DMSO
dimethyl sulfoxide
- ESCC
esophageal squamous cell carcinoma
- FBS
fetal bovine serum
- HIF
hypoxia-inducible factor
- IHC
immunohistochemistry
- NSAID
non-steroidal anti-inflammatory drug
- OCR
oxygen consumption rate
- PBS
phosphate-buffered saline
- PI
propidium iodide
- qRT-PCR
quantitative real-time PCR
- ROS
reactive oxygen species
- STR
short tandem repeat
Contributor Information
Mohammad Faujul Kabir, Fels Cancer Institute for Personalized Medicine, Temple University Lewis Katz School of Medicine, Philadelphia, PA, USA.
Jazmyne L Jackson, Fels Cancer Institute for Personalized Medicine, Temple University Lewis Katz School of Medicine, Philadelphia, PA, USA.
Annie D Fuller, Fels Cancer Institute for Personalized Medicine, Temple University Lewis Katz School of Medicine, Philadelphia, PA, USA.
Leonny Gathuka, Fels Cancer Institute for Personalized Medicine, Temple University Lewis Katz School of Medicine, Philadelphia, PA, USA.
Adam L Karami, Fels Cancer Institute for Personalized Medicine, Temple University Lewis Katz School of Medicine, Philadelphia, PA, USA.
Don-Gerard Conde, Fels Cancer Institute for Personalized Medicine, Temple University Lewis Katz School of Medicine, Philadelphia, PA, USA.
Alena Klochkova, Fels Cancer Institute for Personalized Medicine, Temple University Lewis Katz School of Medicine, Philadelphia, PA, USA.
Anbin Mu, Fels Cancer Institute for Personalized Medicine, Temple University Lewis Katz School of Medicine, Philadelphia, PA, USA.
Kathy Q Cai, Histopathology Facility, Fox Chase Cancer Center, Philadelphia, PA, USA.
Andres J Klein-Szanto, Histopathology Facility, Fox Chase Cancer Center, Philadelphia, PA, USA.
Amanda B Muir, Department of Pediatrics, Division of Gastroenterology, Hepatology, and Nutrition, Children’s Hospital of Philadelphia, Philadelphia, PA, USA.
Kelly A Whelan, Fels Cancer Institute for Personalized Medicine, Temple University Lewis Katz School of Medicine, Philadelphia, PA, USA; Department of Cancer & Cellular Biology, Temple University Lewis Katz School of Medicine, Philadelphia, PA, USA.
Funding
This work was supported by the following grants: NIH R21CA256465 (K.A.W.); Willian J. Avery Fellowship (M.F.K.); NIH R01DK121159 (K.A.W.); NIH R01DK121159-S1 (J.L.J.); NIH P30CA006927 (K.Q.C., A.J.K.-S.; PI: Jonathan Chernoff, Fox Chase Cancer Center); NIH T32GM142606 (A.D.F.; PIs: Xavier Graña, Jonathan Soboloff, Temple University).
Authors’ contributions
Data collection and analysis: M.F.K., J.L.J., A.D.F., L.G., A.L.K., D.-G.C., A.K., A.B.M., K.Q.C., A.J.K.-S. and K.A.W. Study design and interpretation: M.F.K., J.L.J., A.D.F., A.B.M., A.J.K.-S. and K.A.W. Manuscript writing and editing: M.F.K., J.L.J., A.D.F., A.B.M. and K.A.W.
Data availability
All data supporting findings of this study are available within the article and its supplementary information files. Sequencing data files generated in this study along with their associated metadata are deposited into GEO database (accession code GSE227127).
References
- 1. Sung, H., et al. (2021) Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin., 71, 209–249. [DOI] [PubMed] [Google Scholar]
- 2. Rustgi, A.K., et al. (2014) Esophageal carcinoma. N. Engl. J. Med., 371, 2499–2509. [DOI] [PubMed] [Google Scholar]
- 3. Arnold, M., et al. (2015) Global incidence of oesophageal cancer by histological subtype in 2012. Gut, 64, 381–387. [DOI] [PubMed] [Google Scholar]
- 4. Ohashi, S., et al. (2015) Recent advances from basic and clinical studies of esophageal squamous cell carcinoma. Gastroenterology, 149, 1700–1715. [DOI] [PubMed] [Google Scholar]
- 5. Freedman, N.D., et al. (2007) A prospective study of tobacco, alcohol, and the risk of esophageal and gastric cancer subtypes. Am. J. Epidemiol., 165, 1424–1433. [DOI] [PubMed] [Google Scholar]
- 6. Pandeya, N., et al. (2013) Sex differences in the proportion of esophageal squamous cell carcinoma cases attributable to tobacco smoking and alcohol consumption. Cancer Epidemiol, 37, 579–584. [DOI] [PubMed] [Google Scholar]
- 7. Network, C.G.A.R., et al. (2017). Integrated genomic characterization of oesophageal carcinoma. Nature, 541(7636), 169–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lin, D.C., et al. (2018) Genomic and epigenomic aberrations in esophageal squamous cell carcinoma and implications for patients. Gastroenterology, 154, 374–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang, K., et al. (2015) Comprehensive genomic profiling of advanced esophageal squamous cell carcinomas and esophageal adenocarcinomas reveals similarities and differences. Oncologist, 20, 1132–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jemal, A., et al. (2011) Global cancer statistics. CA Cancer J. Clin., 61, 69–90. [DOI] [PubMed] [Google Scholar]
- 11. Gurpinar, E., et al. (2014) NSAIDs inhibit tumorigenesis, but how? Clin. Cancer Res., 20, 1104–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu, J.F., et al. (2006) Cyclooxygenase-2 expression in squamous cell carcinoma of the esophagus. Dis. Esophagus, 19, 350–354. [DOI] [PubMed] [Google Scholar]
- 13. Zhi, H., et al. (2006) Significance of COX-2 expression in human esophageal squamous cell carcinoma. Carcinogenesis, 27, 1214–1221. [DOI] [PubMed] [Google Scholar]
- 14. Kase, S., et al. (2004) Expression of cyclooxygenase-1 and cyclooxygenase-2 in human esophageal mucosa, dysplasia and carcinoma. Pathobiology, 71, 84–92. [DOI] [PubMed] [Google Scholar]
- 15. Zhang, L., et al. (2010) Effects of the inhibition of cyclooxygenase-2 on human esophageal cancer cells: inhibition of cell proliferation and induction of apoptosis. Pathol. Oncol. Res., 16, 39–45. [DOI] [PubMed] [Google Scholar]
- 16. Sciulli, M.G., et al. (2005) The future of traditional nonsteroidal antiinflammatory drugs and cyclooxygenase-2 inhibitors in the treatment of inflammation and pain. Pharmacol. Rep., 57 (suppl), 66–85. [PubMed] [Google Scholar]
- 17. Galisteo, A., et al. (2021) Diclofenac N-derivatives as therapeutic agents with anti-inflammatory and anti-cancer effect. Int. J. Mol. Sci., 22, 5067–5090. doi: 10.3390/ijms22105067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Choi, S., et al. (2022) Diclofenac: a nonsteroidal anti-inflammatory drug inducing cancer cell death by inhibiting microtubule polymerization and autophagy flux. Antioxidants (Basel), 11, 1009–1029. doi: 10.3390/antiox11051009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Arisan, E.D., et al. (2018) Diclofenac induced apoptosis via altering PI3K/Akt/MAPK signaling axis in HCT 116 more efficiently compared to SW480 colon cancer cells. Mol. Biol. Rep., 45, 2175–2184. [DOI] [PubMed] [Google Scholar]
- 20. Chandramouleeswaran, P.M., et al. (2016) Preferential secretion of thymic stromal lymphopoietin (TSLP) by terminally differentiated esophageal epithelial cells: relevance to eosinophilic esophagitis (EoE). PLoS One, 11, e0150968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Whelan, K.A., et al. (2017) Autophagy supports generation of cells with high CD44 expression via modulation of oxidative stress and Parkin-mediated mitochondrial clearance. Oncogene, 36, 4843–4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kinugasa, H., et al. (2015) Mitochondrial SOD2 regulates epithelial-mesenchymal transition and cell populations defined by differential CD44 expression. Oncogene, 34, 5229–5239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Predina, J.D., et al. (2011) Neoadjuvant in situ gene-mediated cytotoxic immunotherapy improves postoperative outcomes in novel syngeneic esophageal carcinoma models. Cancer Gene Ther., 18, 871–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Natsuizaka, M., et al. (2017) Interplay between Notch1 and Notch3 promotes EMT and tumor initiation in squamous cell carcinoma. Nat. Commun., 8, 1758–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Reichenbach, Z.W., et al. (2019) Clinical and translational advances in esophageal squamous cell carcinoma. Adv. Cancer Res., 144, 95–135. [DOI] [PubMed] [Google Scholar]
- 26. Jiang, J., et al. (2022) Effect of taking aspirin before diagnosis on the prognosis of esophageal squamous cell carcinoma and analysis of prognostic factors. J. Int. Med. Res., 50, 3000605221089799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Araujo, J.L., et al. (2016) Prediagnosis aspirin use and outcomes in a prospective cohort of esophageal cancer patients. Therap. Adv. Gastroenterol., 9, 806–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. van Staalduinen, J., et al. (2016) The effect of aspirin and nonsteroidal anti-inflammatory drug use after diagnosis on survival of oesophageal cancer patients. Br. J. Cancer, 114, 1053–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Arai, J., et al. (2022) Chemoprevention of oesophageal squamous-cell carcinoma and adenocarcinoma: a multicentre retrospective cohort study. Digestion, 103, 192–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Limburg, P.J., et al. (2005) Randomized, placebo-controlled, esophageal squamous cell cancer chemoprevention trial of selenomethionine and celecoxib. Gastroenterology, 129, 863–873. [DOI] [PubMed] [Google Scholar]
- 31. Steinbach, G., et al. (2000) The effect of celecoxib, a cyclooxygenase-2 inhibitor, in familial adenomatous polyposis. N. Engl. J. Med., 342, 1946–1952. [DOI] [PubMed] [Google Scholar]
- 32. Shamma, A., et al. (2000) Up-regulation of cyclooxygenase-2 in squamous carcinogenesis of the esophagus. Clin. Cancer Res., 6, 1229–1238. [PubMed] [Google Scholar]
- 33. Pantziarka, P., et al. (2016) Repurposing Drugs in Oncology (ReDO)—diclofenac as an anti-cancer agent. Ecancermedicalscience, 10, 610–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang, F., et al. (2019) Gene expression profiling reveals distinct molecular subtypes of esophageal squamous cell carcinoma in Asian populations. Neoplasia, 21, 571–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li, W., et al. (2014) Expression patterns of three regulation enzymes in glycolysis in esophageal squamous cell carcinoma: association with survival. Med. Oncol., 31, 118–126. [DOI] [PubMed] [Google Scholar]
- 36. Yao, F., et al. (2013) LDHA is necessary for the tumorigenicity of esophageal squamous cell carcinoma. Tumour Biol., 34, 25–31. [DOI] [PubMed] [Google Scholar]
- 37. Yilmaz, C., et al. (2021) Diclofenac down-regulates COX-2 induced expression of CD44 and ICAM-1 in human HT29 colorectal cancer cells. Naunyn Schmiedebergs Arch. Pharmacol., 394, 2259–2272. [DOI] [PubMed] [Google Scholar]
- 38. Jung, S.H., et al. (2020) Diclofenac impairs autophagic flux via oxidative stress and lysosomal dysfunction: implications for hepatotoxicity. Redox Biol., 37, 101751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nakamura, Y. (2004) Isolation of p53-target genes and their functional analysis. Cancer Sci., 95, 7–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wei, Q., et al. (2012) MiR-17-5p. targets TP53INP1 and regulates cell proliferation and apoptosis of cervical cancer cells. IUBMB Life, 64, 697–704. [DOI] [PubMed] [Google Scholar]
- 41. Chu, Y., et al. (2018) PIG3 suppresses gastric cancer proliferation by regulating p53-mediated apoptosis. J. Biol. Regul. Homeost. Agents, 32, 1185–1189. [PubMed] [Google Scholar]
- 42. Patel, K., et al. (2022) Roles for GADD45 in development and cancer. Adv. Exp. Med. Biol., 1360, 23–39. [DOI] [PubMed] [Google Scholar]
- 43. Furukawa, H., et al. (2018) PRIMA-1 induces p53-mediated apoptosis by upregulating Noxa in esophageal squamous cell carcinoma with TP53 missense mutation. Cancer Sci., 109, 412–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Song, Y., et al. (2014) Identification of genomic alterations in oesophageal squamous cell cancer. Nature, 509, 91–95. [DOI] [PubMed] [Google Scholar]
- 45. Wong, G.S., et al. (2013) Periostin cooperates with mutant p53 to mediate invasion through the induction of STAT1 signaling in the esophageal tumor microenvironment. Oncogenesis, 2, e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Grugan, K.D., et al. (2013) A common p53 mutation (R175H) activates c-Met receptor tyrosine kinase to enhance tumor cell invasion. Cancer Biol. Ther., 14, 853–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tang, Q., et al. (2021). Mutant p53 regulates Survivin to foster lung metastasis. Genes Dev, 35, 528–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Osugi, Y. (1996) p53 expression in various stages of 4-nitroquinoline 1-oxide induced carcinoma in the rat tongue. J. Osaka Dent. Univ., 30, 29–35. [PubMed] [Google Scholar]
- 49. Nakagawa, H., et al. (2020) Modeling epithelial homeostasis and reactive epithelial changes in human and murine three-dimensional esophageal organoids. Curr. Protoc. Stem Cell Biol., 52, e106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kijima, T., et al. (2019) Three-dimensional organoids reveal therapy resistance of esophageal and oropharyngeal squamous cell carcinoma cells. Cell. Mol. Gastroenterol. Hepatol., 7, 73–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Otsuka, R., et al. (2022) Role of sirtuins in esophageal cancer: current status and future prospects. World J. Gastrointest. Oncol., 14, 794–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Cui, Y., et al. (2016) STAT3 regulates hypoxia-induced epithelial mesenchymal transition in oesophageal squamous cell cancer. Oncol. Rep., 36, 108–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lv, Z., et al. (2019). HIF-1α promotes the stemness of oesophageal squamous cell carcinoma by activating the Wnt/β-catenin pathway. Oncol. Rep., 42, 726–734. [DOI] [PubMed] [Google Scholar]
- 54. Tang, K., et al. (2022) HIF-1α stimulates the progression of oesophageal squamous cell carcinoma by activating the Wnt/β-catenin signalling pathway. Br. J. Cancer, 127, 474–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Vaziri, H., et al. (2001). hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell, 107, 149–159. [DOI] [PubMed] [Google Scholar]
- 56. Lee, J.J., et al. (2010) Hypoxia activates the cyclooxygenase-2-prostaglandin E synthase axis. Carcinogenesis, 31, 427–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting findings of this study are available within the article and its supplementary information files. Sequencing data files generated in this study along with their associated metadata are deposited into GEO database (accession code GSE227127).