Abstract
Transcription factors of the CREB family control the expression of a large number of genes in response to various signaling pathways. Regulation mediated by members of the CREB family has been linked to various physiological functions. Classically, activation by CREB is known to occur upon phosphorylation at an essential regulatory site (Ser133 in CREB) and the subsequent interaction with the ubiquitous coactivator CREB-binding protein (CBP). However, the mechanism by which selectivity is achieved in the identification of target genes, as well as the routes adopted to ensure tissue-specific activation, remains unrecognized. We have recently described the first tissue-specific coactivator of CREB family transcription factors, ACT (activator of CREM in testis). ACT is a LIM-only protein which associates with CREM in male germ cells and provides an activation function which is independent of phosphorylation and CBP. Here we characterize a family of LIM-only proteins which share common structural organization with ACT. These are referred to as four-and-a-half-LIM-domain (FHL) proteins and display tissue-specific and developmentally regulated expression. FHL proteins display different degrees of intrinsic activation potential. They provide powerful activation function to both CREB and CREM when coexpressed either in yeast or in mammalian cells, specific combinations eliciting selective activation. Deletion analysis of the ACT protein shows that the activation function depends on specific arrangements of the LIM domains, which are essential for both transactivation and interaction properties. This study uncovers the existence of a family of tissue-specific coactivators that operate through novel, CBP-independent routes to elicit transcriptional activation by CREB and CREM. The future identification of additional partners of FHL proteins is likely to reveal unappreciated aspects of tissue-specific transcriptional regulation.
Transcription factors of the CREB family are involved in the regulation of gene expression in response to a number of signaling pathways (15). Proteins issued from CREB and CREM genes play central roles in many physiological processes, including memory and long-term potentiation, circadian rhythms, pituitary function, and spermatogenesis (17, 54).
CREB and CREM belong to the basic domain-leucine zipper (bZip) class of proteins. These factors bind, as homo- or heterodimers, to a DNA sequence known as the cyclic AMP-responsive element, which is present in the regulatory region of various target genes (40, 54). The N-terminal half of CREB and CREM contains a modular activation domain (AD) that is divided into two independent regions (28, 34, 49). The first region comprises two glutamine-rich domains, Q1 and Q2. These flank a second region, called the phosphorylation box (17), also known as the kinase-inducible domain (28), which contains a cluster of sites phosphorylated by various kinases that regulate the transactivation potential of these proteins (15).
Various proteins are known to physically associate with the CREB and CREM AD. The Q2 domain constitutively interacts with the TATA-binding protein-associated factor TAF130, a subunit of the TFIID complex (21). The phosphorylation box is required for binding to the large proteins CREB-binding protein (CBP) and p300 (1, 4, 11, 32, 37). CBP and p300 are ubiquitously expressed coactivators that function by interacting with basal transcription factors, such as TFIIB (32), TATA-binding protein (60), and RNA helicase A (44), and/or by modifying the chromatin state through their histone acetyltransferase activity (5, 47). Interaction with CBP and/or p300 requires the phosphorylation of a specific serine residue (Ser133 in CREB and Ser117 in CREM) (48, 51), which can be triggered by a variety of kinases, such as cyclic AMP-dependent kinase A (29), mitogen-activated p90rsk (16, 66), stress-regulated mitogen-activated protein kinase-activated protein kinase 2 (61), and mitogen- and stress-activated kinases (14). Thus, proteins of the CREB family operate as nuclear targets of a number of converging transduction pathways and are implicated in multiple cellular responses.
Although modulation of CREB activity by specific transduction pathways has been extensively studied, little is known about the selectivity code by which proteins of the CREB family regulate the expression of different sets of genes in response to specific external stimuli. One intriguing possibility is that the interaction with specific cofactors may lead to the formation of different transcriptional complexes with diverse promoter specificities. Thus, the use of different coactivators could lead to tissue-specific CREB- and CREM-mediated transcription.
Recently, we have reported that CREM transcriptional activity can be stimulated by interaction with a tissue-specific coactivator, activator of CREM in testis (ACT) (22, 23). ACT is a factor belonging to the class of LIM-only (LMO) proteins with a characteristic organization of four and a half LIM domains (FHL). These are structural motifs composed of two adjacent zinc fingers that are known to be involved in protein-protein interaction (56). ACT expression is testis specific and temporally coordinated with CREM during germ cell differentiation. Upon binding to the CREM AD, ACT powerfully stimulates CREM transcriptional activity in a phosphorylation- and CBP-independent manner (23).
In this study, we show that ACT shares a high degree of similarity with a group of proteins which constitute a novel family of transcriptional coactivators. These are members of the FHL protein family, which are defined by their characteristic secondary arrangement of LIM domains. Two family members, FHL1 (SLIM1) and FHL3 (SLIM2), were initially identified by their expression in skeletal muscle (36, 41). FHL2 (DRAL, SLIM3) was isolated as a gene whose expression is down-regulated in rhabdomyosarcoma cells (9, 25). Another member of the family, FHL4, which is expressed only in the testis (42), has been described more recently.
Here we present a comparative analysis of the various FHL proteins with respect to their expression profile, association potential, and functional properties. These ACT-like proteins differ from each other both in terms of transactivation capability and specificity of interaction with members of the CREB family. We also show that the members of the FHL family are able to form homo- and heterocomplexes but that a specific combination code exists. Furthermore, we provide evidence that both the activation and protein-protein interaction properties of these factors depend on specific arrangements of the individual LIM domains.
MATERIALS AND METHODS
Yeast plasmid constructions.
FHL1 and FHL4 cDNAs, obtained by PCR from a mouse embryo cDNA library, and FHL2 and FHL3 cDNAs, obtained by PCR from a human heart cDNA library, were cloned into pGBT9 and pGAD424 vectors. pGBT-CREM, pGBT-ACT, and pGAD-ACT have been described (23). The CREB AD was subcloned from the plasmid pG4CREBΔLZ (43) into the pGBT9 vector. The Sp1 AD (amino acids [aa] 132 to 485) was obtained by PCR from the plasmid pSP1-778C (12) and cloned into the pGBT9 plasmid. LMO-2, muscle LIM protein (MLP), and LIM domain binding factor (LDB) open reading frames (ORFs) were obtained by PCR from a mouse embryo cDNA library and cloned into the pGAD424 vector (LMO-2 and MLP) or a pGBT9 plasmid (LDB). ACT mutants were obtained by PCR using the Quikchange site-directed mutagenesis kit (Stratagene). ACT ΔLIM1/2 mutant lacks aa 2 to 36; ΔLIM1, aa 37 to 97; ΔLIM2, aa 98 to 158; ΔLIM3, aa 159 to 217; ΔLIM4, aa 218 to 284; ΔLIM3,4, aa 159 to 284; ΔLIM2,3,4, aa 98 to 284; and ΔLIM1/2-1, aa 2 to 97. All constructs were verified by sequencing.
Yeast analysis.
Yeast transformation and the β-galactosidase assay were performed in Y190 yeast strain, as described in the Clontech Matchmaker two-hybrid system protocol. β-Galactosidase activity was calculated in Miller units, and results are means of three to four independent experiments. For Western analysis, 8 ml of mid-log-phase cultures was harvested by centrifugation, resuspended in 200 μl of Laemmli buffer, and boiled for 10 min. Glass beads were added to the lysates, vortexed for 5 min, and centrifuged to remove insoluble material. Western blot analysis was performed as previously described (19). Gal4 fusion proteins were detected with anti-Gal4 DNA-binding domain (Gal4DBD) and anti-Gal4AD monoclonal antibodies (Santa Cruz Biotechnology).
RNA analysis.
Total RNA was extracted from mouse tissues as previously described (10) and analyzed by RNase protection (24). To score for the expression of different FHL genes, internal fragments from murine FHL cDNAs (FHL1, from +27 to +384; FHL2, from +564 to +839; FHL3, from +615 to +835; FHL4, from +277 to +719) were subcloned into pBluescript SK(−). RNA probes were prepared using an in vitro transcription kit (Promega). ACT and CREM riboprobes were already described (23, 39). In all RNase protection analyses, transfer RNA was used as a control for nonspecific protection. A mouse β-actin riboprobe was used as an internal control to monitor the loading of equal amounts of RNA (fragment from +193 to +331 of the mouse coding sequence).
In situ hybridization analysis was performed as described (38). ACT and FHL4 antisense riboprobes were prepared as described for RNase protection. For control of nonspecific hybridization, consecutive sections were hybridized with ACT and the FHL4 sense RNA probe (not shown).
Transfections and reporter gene assay.
COS cells, maintained in Dulbecco modified Eagle medium with 5% fetal calf serum, were transfected by the calcium phosphate coprecipitation technique (53). In each sample, the total amount of expression vector DNA was kept constant by the addition of pJΩ7. Chloramphenicol acetyltransferase (CAT) and luciferase activity was assayed as described (24); 0.1 μg of a CMVβ-gal plasmid was included in each transfection to monitor for transfection efficiency. The c-fos-CAT (FC8-CAT [55]), pCycA-Luc (19), pG4CREB, and pG4CREB-Ala133 (18) plasmids have been described. pJΩ7-FHL expression plasmids were obtained by subcloning FHL ORFs into the pJΩ7 plasmid. pGal4Luc contains five Gal4 binding sites upstream from the herpes thymidine kinase promoter region (−109 to +52), cloned in the pGL2 basic vector (Promega).
Coimmunoprecipitation assays.
The FHL expression vector pCS2Myc-FHLs was constructed by inserting FHL ORFs into the pCS2Myc plasmid (52). COS cells were transfected with 10 μg of each plasmid and harvested after 48 h in 1 ml of EBC (50 mM Tris-HCl [pH 8.0], 170 mM NaCl, 0.5% NP-40, 50 mM NaF) containing 1 mM phenylmethylsulfonyl fluoride and 10 μg of aprotinin and leupeptin per ml each. Lysates were incubated at 4°C for 3 h with 10 μl of anti-GAL4DBD monoclonal antibody. The beads were washed four times in NETN (10 mM Tris-HCl [pH 8.0], 250 mM NaCl, 5 mM EDTA, 0.5% NP-40). Western blot analysis of immunocomplexes was performed by using anti-myc (9E10) and anti-CREB (New England Biolabs) antibodies.
RESULTS
FHL: a family of LIM-only proteins.
We have recently reported the isolation of ACT, an LMO that is expressed exclusively in spermatids (23). Databank searches for sequence similarity revealed that ACT has a high degree of homology with a previously identified group of proteins (Fig. 1A), which includes FHL1 (also known as SLIM1), FHL2 (DRAL/SLIM3), FHL3 (SLIM2), and FHL4 (9, 25, 36, 41, 42). Members of this family are characterized by a specific arrangement of LIM motifs, being composed of four and a half LIM domains, with the half-domain always located in the N terminal. Among these proteins, FHL2 is the most similar to ACT, showing 60% identity and 80% similarity at the amino acid level, followed by FHL3 with 68% similarity and FHL1 and FHL4 with 62%. The homology between these proteins and ACT is distributed throughout the whole sequence (Fig. 1A), although selected stretches show a higher degree of identity (indicated by orange, green, grey, and black boxes) (Fig. 1A legend). Apart from the cysteine and histidine residues within the zinc fingers of the LIM domains required for the interaction with the Zn2+ ions, other amino acids are highly conserved (black and grey boxes), which may also be involved in determining the structural conformation of repeated LIM domains. Alignment of the different LIM domain sequences within any given FHL gene reveals no strong sequence conservation (Fig. 1B). However, comparison of individual LIM domains at equivalent positions among the different FHL proteins demonstrates significant homology within each group (Fig. 1B). These results are consistent with the hypothesis that the FHL gene family is derived by duplication from a single ancestral gene during evolution. From this analysis, it is also interesting to note that the first half-domain and the third LIM domain are significantly less conserved in ACT with respect to FHL2 and FHL3, which may suggest a possible specialized function for these domains. The presence of FHL2 homologues in Amphioxus (57) and in Caenorhabditis elegans underscores the evolutionary conservation of this family.
FIG. 1.
Sequence comparison of FHL proteins. (A) Full-length protein sequences (except for ceF25h5.1A) of FHL proteins from different species were aligned using the CLUSTAL X program (63). The amino acid one-letter code is used. The sequences have been divided into three groups. The character background of the amino acid one-letter code is red (FHL1 and FHL4), blue (FHL3), or yellow (FHL2 and ACT), respectively, to highlight sequence conservation (80% identity) within a group. Conservation between any two groups is shown by color combination coding: combination of groups 1 and 2, violet (blue and red); groups 1 and 3, orange (yellow and red); and groups 2 and 3, green (blue and yellow). Identical residues in the alignment are shown as white characters with a black background. Similarly, conserved residues (80% identity) in the alignment are shown with a grey background. In addition, for overall 80% residue conservation, if amino acids within a group show conservation higher than 80%, the amino acid one-letter code is given the group's specific color. Abbreviations: m, mouse; r, rat; h, human; a, Amphioxus; ce, C. elegans. (B) Unrooted tree deduced from an alignment of the different LIM domain sequences within any given murine FHL gene. The unrooted tree was created using the neighbor-joining method. The numbers in the interior branches are bootstrap values for 1,000 resamplings.
A hallmark of FHL proteins: tissue-specific expression.
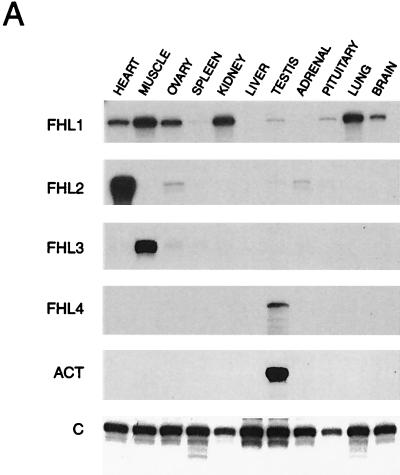
On the basis of previous reports suggesting that FHL genes show tissue-specific expression patterns (25, 36, 42), we decided to compare quantitatively their expression levels in a variety of mouse tissues. Given the high sequence similarity shared by FHL genes, we measured their expression by RNase protection analysis due to the high sensitivity and specificity of this assay. All FHL genes were expressed in a tissue-specific manner, although with distinct patterns. FHL2 and FHL3 were dominantly expressed in the heart and in skeletal muscle, respectively (Fig. 2A). Low but detectable levels of FHL2 transcript were also detected in the ovary, testis, and adrenal and pituitary glands, while FHL3 was also expressed in the ovary, spleen, and adrenal glands. FHL4 was exclusively expressed in the testis. FHL1 was the only member of the family displaying a wider range of expression, its transcript being present at high levels in the heart, muscle, ovary, kidney, lung, and brain.
FIG. 2.
Expression of FHL genes. (A) Ten micrograms of total RNA from the indicated mouse tissues was analyzed by RNase protection assay by using specific riboprobes as described in Materials and Methods. (B) ACT and FHL4 mRNA expression in adult mouse testis sections by in situ hybridization. (C) Expression of ACT, FHL4, and CREM during testis development. RNAs were extracted from testes of mice at different ages (as indicated), and 10 μg was analyzed by RNase protection assay, using ACT-, FHL4-, and CREM-specific riboprobes. A β-actin riboprobe was used as an internal control.
The highly similar expression profiles of ACT and FHL4, as well as the crucial effect exerted by ACT on CREM activation function (23), prompted us to analyze in further detail FHL4 expression during spermatogenesis. In situ hybridization analysis of testis sections revealed that FHL4 expression was specific to the seminiferous tubules, while no signal was detected in the interstitial spaces (Fig. 2B). However, in contrast to ACT, whose expression is restricted to certain tubule sections corresponding to specific differentiation stages, FHL4 expression appeared to be more generalized and present in almost all the tubule sections analyzed. Thus, while ACT expression parallels the well-characterized pattern of CREM (23), FHL4 expression appears to be less specific to stages of differentiation. The different temporal appearance of FHL4 and ACT transcripts was also observed during germ cell differentiation. During the first wave of spermatogenesis, germ cells are synchronized in their development and the timing of appearance of the different cell types is well characterized (46). RNase protection analysis of RNA prepared from 1- to 4-week-old mouse testis demonstrates that FHL4 expression was already detectable at the end of the 3rd week (Fig. 2C). This period corresponds during testis development to the accumulation of spermatocyte cells (46). In contrast, ACT expression was detectable only at the end of the 4th week, when spermatid cells accumulate in the tubule cell population. These results demonstrate that FHL4 is expressed earlier than ACT during male germ cell development. In conclusion, the expression of FHL proteins is regulated in a tissue- and development-specific manner.
FHL proteins have different intrinsic activation potentials.
Unlike previously described LMO proteins, ACT was found to contain a powerful AD that stimulates transcription in both yeast and mammalian cells (23). To elucidate whether this property is shared by the other members of the FHL family, we fused FHL ORFs to Gal4DBD to generate chimeric FHL proteins provided with an autonomous DNA-binding capability. These constructs were used to test directly the transactivation potential of FHL proteins upon expression in yeast. As shown in Fig. 3A, FHL3 strongly stimulated transcription of a β-galactosidase reporter gene, being even more powerful than ACT; this indicates the presence of an autonomous AD within this protein. A significantly less pronounced, but reproducible, stimulation of transcription was observed with FHL2 and FHL4. However, no induction of β-galactosidase activity was detected with FHL1. Western blot analysis of protein extracts prepared from the transformed yeast cells showed that the differences in the transactivation capability were not due to different stability or expression levels of the proteins, as shown in Fig. 3A (top). These results are of interest, as they reveal that the only FHL protein with activation capacity comparable to that of ACT is FHL3, although sequence similarity between the two proteins is not particularly high (Fig. 1B). Instead, FHL2, which displays the highest degree of homology with ACT, has a weaker activation potential. These results suggest that a major determinant for activation potential may reside in the relative organization of the LIM domains.
FIG. 3.
Transactivating properties of FHL proteins. (A) Y190 yeast cells, bearing the Gal1 upstream activation sequence-lacZ reporter gene, were transformed with Gal4DBD-FHL expression vectors and analyzed for β-galactosidase activity. Fold activation was calculated with respect to the β-galactosidase activity obtained when yeast cells were cotransformed with the pGBT9 empty vector (bottom). Expression levels of FHL proteins were determined by Western blot analysis using an anti-Gal4DBD antibody (top). (B) Analysis of the transactivating properties of ACT mutant proteins. (Upper left) Schematic representation of the ACT deletion mutants used in the transactivation assay. LIM domains are indicated by black boxes, and numbers refer to their relative position within the protein. All ACT mutants are fused to the Gal4DBD. (Lower left) Western blot analysis of extracts prepared from the transformed yeast using an anti-Gal4DBD antibody. (Right) β-Galactosidase activity of yeast transformed with the indicated constructs. The fold activation value obtained with wild-type ACT was defined as 100. (C) Analysis of the transactivating property of a chimeric ACT-FHL2 protein. ACT protein, in which the sequence aa 195 to 200 was replaced with the corresponding amino acids of FHL2, was fused to the Gal4DBD (top) and analyzed by transactivation assay in yeast (bottom). β-Galactosidase activity was calculated in Miller units. C, control.
In order to further characterize the transcriptional activity of the various FHL proteins, we decided to assess which LIM domain was responsible for the transactivation property. With this aim, we generated a number of mutant ACT proteins and analyzed their activity in Gal4DBD fusions (Fig. 3B). The results of this analysis indicate that various LIM domains contribute differentially to the ACT transactivation function. The N-terminal half-domain and the first and third LIM domains appear to be important for the activation function, as deletion of each of these domains strongly reduced the activity of the protein. In contrast, a minor role seems to be played by the second and fourth LIM domains, since mutants lacking each of these domains were only slightly impaired in their ability to stimulate transcription. However, none of these deletions leads to a complete inactivation of the protein, indicating that full activity requires contribution of all domains. Analysis of mutants with deletion of multiple LIM domains confirmed this conclusion. Indeed, deletion of the two C-terminal LIM domains severely impaired but did not completely abolish the capacity of ACT to stimulate transcription, while mutants lacking either the first domain and the N-terminal LIM half-domain or the last three LIM motifs were totally inactive. Thus, a specific combination of LIM domains appears to be responsible for full transcriptional activity.
Although ACT and FHL2 present the highest level of homology within the whole family (Fig. 1A), they display a striking difference in activation potential (Fig. 3A). This functional diversity prompted us to map the critical residues in these proteins responsible for the observed differences. We tested a chimeric ACT-FHL2 protein designed on the basis of the alignment analysis (Fig. 1). We replaced the aa 195-to-200 sequence within the third LIM domain of the ACT protein with the corresponding region of FHL2. This sequence was chosen because it appeared to be the most divergent stretch between the two proteins. Indeed, this region is negatively charged in ACT, while the corresponding domain in FHL2 is rich in basic residues. Moreover, this region maps within the third LIM domain of ACT, which appears important for its transactivating function. The chimeric protein (ACT>QLSGQR) (Fig. 3C) was fused to the Gal4DBD and analyzed in a transactivation assay in yeast. Surprisingly, the substitution did not alter the activation capability of ACT (Fig. 3C), suggesting that this region is not responsible for the different transactivation potentials of ACT and FHL2. This result confirms the conclusions of the deletion analysis (Fig. 3B), indicating that activation potential requires a specific combination of LIM domains.
Differential and selective interaction of FHL proteins with CREB and CREM.
We have previously shown that ACT is able to bind to CREM and CREB and modulate their transcriptional activity (23). The high sequence similarity shared by all FHL proteins (Fig. 1) prompted us to investigate their interaction with the ADs of CREB and CREM. FHL proteins, fused to Gal4AD, were analyzed, in a yeast two-hybrid assay, for their ability to interact with the ADs of CREM and CREB fused to the Gal4DBD (Fig. 4A). In the same assay we also tested two other LMO proteins, LMO-2 and MLP, which show only a limited homology with the FHL group of proteins but are nevertheless known to interact with other transcription factors, such as GATA1, TAL1, and MyoD (31, 64). The specific association and coactivation function exerted by ACT are confirmed in the present assay (Fig. 4B). Indeed, ACT was the only protein which bound efficiently to the CREM AD. A weaker but significant interaction was observed between FHL2 and CREM. No detectable interaction was observed with the other LMO factors. Strikingly, different results are obtained with CREB. In this case, FHL2 and FHL3 strongly interacted with CREB, with an affinity higher than that of ACT (Fig. 4C). This result is particularly remarkable, considering the very high homology known to exist between the ADs of CREB and CREM (15). Noteworthy is also the case of FHL2, which displays the highest homology with ACT within the FHL family (Fig. 1) yet functions as a powerful coactivator of CREB but not of CREM.
FIG. 4.
FHL proteins bind differentially to CREB, CREM, and Sp1. (A) Protein extracts prepared from transformed yeast were analyzed by Western blotting using an anti-GAL4AD antibody. Gal4AD-FHL fusion proteins were expressed in yeast (Y190 strain) together with the Gal4DBD-CREM (B), Gal4DBD-CREB (C), and Gal4DBD-Sp1 (D) fusion proteins. β-Galactosidase activity is reported as a ratio of the values obtained when CREM, CREB, and Sp1 were coexpressed with the various FHL proteins to the values obtained when CREB, CREM, and Sp1 were expressed alone (fold induction). In the absence of Gal4DBD-CREM, -CREB, and -Sp1, Gal4AD-FHL fusion proteins show no significant induction of β-galactosidase activity (data not shown).
In order to further establish the specificity of the interactions observed, we tested the ability of ACT, FHL2, and FHL3 to bind to Sp1, a transcription factor which, like CREB and CREM, bears a glutamine-rich AD (12), although this shows no significant homology with CREB and CREM ADs. No binding between any of the FHL proteins and Sp1 was detected (Fig. 4D), demonstrating the selectivity of the interaction between FHL and the distinct members of the CREB family.
FHL proteins form homo- and heterocomplexes.
Many LMO proteins have recently been observed to act as dimers (13). Given the coexpression of certain FHL proteins, we wondered whether they were able to interact with each other. The dimerization between LMO proteins is believed to occur either by direct interaction between specific LIM domains or indirectly by association with the LDB/NLI protein (13). In the latter case, LDB is able to recruit two LMO proteins to the same complex through the formation of homodimers. To elucidate whether association can occur also among members of the FHL family, we first tested their ability to form homocomplexes and to bind to LDB in a two-hybrid assay. We found that ACT, FHL2, and FHL3 were able to interact with themselves, although to different degrees, while FHL1 and FHL4 apparently lack the ability for homologous interaction (Fig. 5A). Importantly, no interaction was detected between any of the FHL proteins and LDB (data not shown). The ability to form heterocomplexes was then tested by analyzing different combinations of FHL proteins. Table 1 shows that a specificity code in the intermolecular interactions exists between different members of the FHL family. For example, FHL2 associated efficiently only with FHL3 and ACT, while FHL4 was able to interact only with ACT. As already observed for the homoassociation analysis, FHL1 showed no apparent interaction property.
FIG. 5.
Association properties of FHL proteins. (A) Y190 yeast cells were transformed with Gal4DBD-FHL expression vectors in the presence or absence of the corresponding Gal4AD-FHL expression vectors and analyzed for β-galactosidase activity. (B) ACT mutants described for Fig. 3B were expressed in Y190 yeast cells in the presence or absence of the Gal4AD-ACT expression plasmid. Fold activation is reported as a ratio of the value obtained with the Gal4DBD constructs in the presence of the Gal4AD-ACT fusion to the value measured with DBD fusions alone. Note that the apparently higher values obtained for some mutants with respect to ACT reflect the lower transactivation levels of the mutants when expressed alone, as compared to expression with ACT (Fig. 3B).
TABLE 1.
Heteroassociation properties of FHL proteinsa
| Gal4AD vector | Induction values obtained with Gal4DBD constructs:
|
||||
|---|---|---|---|---|---|
| ACT | FHL1 | FHL2 | FHL3 | FHL4 | |
| ACT | +++ | ||||
| FHL1 | − | − | |||
| FHL2 | ++ | − | ++ | ||
| FHL3 | + | − | ++ | + | |
| FHL4 | + | − | − | − | + |
Y190 yeast cells were transformed with the Gal4DBD-FHL expression vectors in the presence or absence of the Gal4AD-FHL expression vectors and analyzed for β-galactosidase activity. Fold activation is calculated as a ratio of the value obtained with the Gal4DBD constructs in the presence of the Gal4AD fusions to the value measured with DBD fusions alone. Abbreviations: +, induction values between 2 and 3; ++, values between 5 and 10; +++, values between 20 and 40; −, no induction. Data are means from three independent transfections, with variations not exceeding 10%.
To gain further insights into the mechanism by which FHL proteins interact with themselves, we decided to analyze the role of the different LIM domains in mediating intermolecular interactions. To this aim, we assessed the ability of various ACT mutants carrying deletions of single LIM domains to interact with the wild-type ACT protein. Strikingly, most mutations had very little effect on interaction with ACT. Deletion of the second LIM domain was the only mutation that drastically reduced the ability of ACT to interact with itself (Fig. 5B), indicating that this specific LIM domain is necessary for the association function.
FHL proteins differentially modulate CRE-dependent transcription in mammalian cells.
The interaction between CREB and the different FHL proteins was further investigated by coimmunoprecipitation experiments after ectopic expression in mammalian cells. COS cells were cotransfected with a Gal4-CREB expression vector in combination with different Myc-tagged FHL expression vectors. Protein extracts were subjected to immunoprecipitation using an anti-Gal4 antibody. Western blot analysis using the anti-Myc antibody revealed that FHL2, FHL3, and ACT efficiently associate with CREB in the immunoprecipitated complexes from cotransfected cells (Fig. 6A). No FHL1 and FHL4 proteins were coimmunoprecipitated by the anti-Gal4 antibody, thus paralleling the situation observed in yeast cells.
FIG. 6.
FHL2 and FHL3 function as CREB coactivators in mammalian cells. (A) FHL2 and FHL3 coimmunoprecipitate with CREB in vivo. COS cells were cotransfected with pSVGal4DBD-CREB (Gal4-CREB) and pCS2Myc-tagged FHL expression vectors. Cells were harvested 48 h after transfection, and lysates were subjected to immunoprecipitation with an anti-Gal4DBD antibody. Whole extracts (left) and immunocomplexes (right) were analyzed by Western blotting using both anti-CREB and anti-Myc antibodies as indicated. (B) FHL2 and FHL3 stimulate CREB-mediated transcription. COS cells were transfected with 1 μg of a pGal4-luciferase reporter plasmid and 0.5 μg of pSVGal4DBD-CREB (Gal4-CREB) or pSVGal4DBD-CREB133Ala (Gal4-CREB133) expression vectors in the presence or absence of 5 μg of the pCMV-FHL expression vectors. Luciferase activity was determined 24 h after transfection, and the value obtained in the absence of FHL expression vectors was defined as 1. No significant variation of luciferase activity was observed in the absence of Gal4-CREB (not shown). (C and D) Cyclin A and c-fos promoters are activated by FHL2, FHL3, and ACT. The cyclin A promoter (−1050 to +241) fused to the luciferase gene (C) and the c-fos promoter (also known as FC8; from −220 to +42) fused to the CAT gene (D) were transfected in the presence or absence of 5 μg of the pCMV-FHL expression vectors. Fold activations were calculated as shown in panel B.
Next, we wished to test whether the distinct ability of the FHL proteins to bind CREB might result in differential modulation of its transcriptional activity. Using a Gal4-based reporter, CREB-mediated activation was strongly enhanced by ACT and FHL3 and to a lesser extent by FHL2 (Fig. 6B). These results are consistent with the ability to bind to CREB displayed by the various FHL proteins. Importantly, FHL2 and FHL3 do not require phosphorylation of CREB at Ser133 to exert their effect, as already demonstrated for ACT (23). Indeed, stimulation of CREB activity was also observed in the presence of a CREB mutant bearing a Ser→Ala substitution at position 133, which prevents phosphorylation at this site (29) and association with CBP (11).
CREB is known to regulate the expression of a large number of genes in a variety of physiological contexts. Two important targets of CREB-mediated regulation are the c-fos and the cyclin A genes. In the c-fos promoter, a CRE sequence located at position −60 is important for directing expression of the c-fos gene in response to a variety of extracellular stimuli (26, 30, 55, 58). In the cyclin A promoter, a CRE site at position −80 is involved in inducing expression of the gene during the G1-to-S transition of the cell cycle (19, 33, 65, 67). We wished to assess whether the FHL proteins may modulate the transcriptional activity of these CRE-containing promoters by interacting with CREB. We observed that FHL3, ACT, and FHL2 readily enhanced the transcription of reporter genes driven by the c-fos and cyclin A promoters (Fig. 6C and D). Taken together, these results indicate that two other members of the FHL family, with expression patterns different from those of ACT, may also function as coactivators of CRE-mediated transcription in mammalian cells.
DISCUSSION
An increasing body of evidence points to the involvement of some LMO proteins in transcription regulation (13). One interesting example is LMO-2, an LMO protein required for erythropoietic differentiation, which binds to and regulates the activity of the GATA-1 and TAL-1 transcription factors (50). Another example is the LMO MLP, an important regulator of muscle differentiation, which is able to interact with and modulate the function of MyoD (2, 3, 31). Recently, we have shown that the transcriptional activity of CREM is stimulated by interaction with ACT, an LMO expressed exclusively in male germ cells (23). ACT bears some unique features: it contains an intrinsic AD and stimulates CREM activity in a phosphorylation-independent manner, thus providing an alternative to the classical scenario. Here, we provide evidence that CREB transcriptional activity can also be modulated by the interaction with different LMO proteins expressed in a tissue-specific manner.
ACT shares the same structural organization and a high degree of sequence homology with a group of proteins constituted by FHL1, FHL2, FHL3, and FHL4. The hallmark of the FHL proteins is the presence of four LIM domains and a LIM half-motif located at the amino terminus of the protein. Our comparative analysis shows that the similarity among the FHL proteins is distributed throughout the sequence and suggests that these genes originated from a common ancestor by gene duplication. This is also supported by the fact that the order of the different homologous LIM domains is conserved.
The different FHL genes show distinct patterns of expression. FHL2 and FHL3 are expressed at high levels in the heart and muscle, respectively. Similar to that for ACT, FHL4 expression seems to be exclusively restricted to the testis, even though the FHL4 transcript appears earlier than ACT during germ cell differentiation. FHL1 is expressed in a broader range of tissues, such as the muscle, heart, kidney, lung, brain, and ovary. However, it has recently been shown that diverse isoforms of the FHL1 protein, containing different numbers of LIM domains, can be generated by alternative splicing (8, 35, 62). This raises the possibility that different isoforms of FHL1 might have a more restricted expression pattern.
ACT represents the first example of a tissue-specific coactivator involved in the regulation of CRE-dependent transcription. Here, we show that two other FHL proteins, FHL2 and FHL3, share this property with ACT. However, remarkable differences exist among these factors with respect to their relative affinity for CREB and CREM. Interestingly, this different specificity seems to correlate with their respective expression patterns. Thus, ACT is able to bind to both CREM and CREB ADs but shows a higher affinity for CREM, which is coexpressed at high levels in spermatid cells. FHL2 and FHL3, which interact specifically with CREB, are expressed mainly in the heart and skeletal muscle, respectively, where CREB is more abundant than the activator CREM (data not shown). Another level of complexity is provided by the fact that CREB and CREM can form heterodimers with each other (54). Thus, it remains to be verified whether FHL proteins might have a different affinity for the heterodimers with respect to CREB and CREM homodimers.
Recently, it has been reported that FHL2 binds to the androgen receptor and modulates its transcriptional activity (43). These findings strongly support the hypothesis that FHL proteins may constitute a family of coregulators involved in the modulation of tissue-specific gene expression by integrating the activity of different transcription factors.
FHL1 and FHL4 do not show a strong interaction with members of the CREB family, indicating that this property is not a common feature of FHL proteins. Our observations suggest that, despite their structural similarity, different members of the FHL family have evolved specialized functions in vivo. Therefore, it is possible that FHL1 and FHL4 might regulate the activity of other transcription factors. Indeed, it has been recently demonstrated that a splicing isoform of FHL1, containing only the first two and a half LIM domains, can interact and negatively regulate the activity of RBP-J, a transcription factor involved in the Notch signaling pathway (62). Taken together, these results indicate that the various FHL proteins may be involved in the formation of different specific transcriptional complexes.
Besides the ability to interact with members of the CREB family, we have found that some FHL proteins are able to form homo- or heterocomplexes. For example, ACT and FHL2 strongly homoassociate and interact with each other. Moreover, ACT binds to FHL3 and FHL4, though to a lesser extent than does FHL2. Given the observed coexpression of some of the FHL proteins, these results raise the possibility that other FHL proteins may be recruited by indirect interaction to form complexes with CREB or CREM at the promoter level.
Additional evidence that different members of FHL have evolved specialized functions emerges from analysis of their transcriptional activation properties. ACT and FHL3 have a strong AD and FHL2 and FHL4 stimulate transcription at intermediate levels, while FHL1 apparently lacks any transactivation property. Given the high similarity of FHL2 and ACT (Fig. 1), it is noteworthy to underscore the observed difference in their transactivation capability. Indeed, the results obtained with the FHL2-ACT chimeric protein suggest that the activation capability depends on the specific assembly of a number of structural determinants. In addition, we have also found that the chimeric protein is able to interact with CREM with an affinity similar to that of ACT (data not shown). Therefore, a fine mutational analysis will be required to identify the structural motifs that confer activation specificity and protein-protein interaction selectivity to ACT and FHL2. Although our results suggest that only certain FHL proteins are involved in stimulating transcription, it remains to be verified whether the transcriptional properties of other proteins may only be apparent in a more physiological context. Possibly, some FHL proteins may need to interact with other specific cofactors in order to exert transcriptional activity.
In parallel with the comparative analysis of the various FHL proteins, deletion studies of the ACT protein were performed in order to identify functional domains. ACT mutants carrying deletions of individual LIM domains were tested in transactivation and interaction assays. The results from this analysis suggest that the transactivation property of ACT resides in a combination of different LIM domains, the half-domain and first and third domains being required for an efficient transcriptional activation. In contrast, ACT's ability to interact with itself depends mostly on the presence of the second LIM domain. These results, in combination with the reported characterization of ACT-CREM interaction, clearly indicate that different LIM domains are responsible for specific functions of the protein. According to the evidence that LIM domains mediate protein-protein interactions, our results suggest that the different LIM domains within the FHL proteins represent specific surfaces for different functional interactions. Crystallography studies will help to elucidate how the organization of the LIM domains within the three-dimensional structure of the protein allows multiple interactions to take place.
CREB and CREM have important roles in the regulation of proliferation and differentiation in different cellular systems. For example, the expression of a number of early-response or cell cycle genes is regulated by CRE sequences present in their promoter regions (40, 54). CREB and CREM are involved in the regulatory processes linked to the differentiation of various cell types, as shown by gene targeting or expression of dominant-negative mutants in transgenic mice (6, 7, 45, 59). Here we show that FHL2, FHL3, and ACT could be directly involved in these differentiation programs, as they are able to stimulate the activity of the CRE-containing c-fos and cyclin A promoters upon interaction with CREB and CREM. Little is known about the role of CREB or CREM in the development and physiology of skeletal muscle or the heart, the tissues where FHL2 and FHL3 are more abundantly expressed. It has been shown that CREB phosphorylation is induced in cardiac cells after beta-adrenergic stimulation (27). Interestingly, transgenic mice bearing a dominant negative isoform of CREB develop dilated cardiomyopathy, implicating a role for CREB in the regulation of cardiac myocyte function (20).
We propose that in specific cellular systems, CREB or CREM activity might be modulated by the interaction with tissue-specific cofactors of the FHL family. The presence of these coactivators provides a further level of regulation of CREM and CREB activity, in addition to phosphorylation and association with CBP. We anticipate that the activity of several other transcription factors will be found to be modulated by FHL proteins. An important future goal will be to elucidate whether the presence of distinct FHL proteins, in a specific tissue, is required for the activation of selected subsets of genes.
ACKNOWLEDGMENTS
We thank J.M. Wurtz for advice with computer alignment programs; E. Heitz and M. Rastegar for technical help; and N. Foulkes, A. Morlon, B. Macho-Mellitzer, and all members of the Sassone-Corsi laboratory for discussions and help.
G.M.F. is supported by a postdoctoral fellowship from the Schering Foundation and D.D.C. by a fellowship of the Fondation de la Recherche Médicale. This work was supported by grants from Centre National de la Recherche Scientifique, Institut National de la Santé et de la Recherche Médicale, Hôpital Universitaire de Strasbourg, Fondation de la Recherche Médicale, Université Louis Pasteur, and Association pour la Recherche sur le Cancer.
G.M.F. and D.D.C. contributed equally to this work.
REFERENCES
- 1.Arany Z, Newsome D, Oldread E, Livingston D M, Eckner R. A family of transcriptional adaptor proteins targeted by the E1A oncoprotein. Nature. 1995;374:81–84. doi: 10.1038/374081a0. [DOI] [PubMed] [Google Scholar]
- 2.Arber S, Halder G, Caroni P. Muscle LIM protein, a novel essential regulator of myogenesis, promotes myogenic differentiation. Cell. 1994;79:221–231. doi: 10.1016/0092-8674(94)90192-9. [DOI] [PubMed] [Google Scholar]
- 3.Arber S, Hunter J J, Ross J J, Hongo M, Sansig G, Borg J, Perriard J C, Chien K R, Caroni P. MLP-deficient mice exhibit a disruption of cardiac cytoarchitectural organization, dilated cardiomyopathy, and heart failure. Cell. 1997;88:393–403. doi: 10.1016/s0092-8674(00)81878-4. [DOI] [PubMed] [Google Scholar]
- 4.Arias J, Alberts A S, Brindle P, Claret F X, Smeal T, Karin M, Feramisco J, Montminy M. Activation of cAMP and mitogen responsive genes relies on a common nuclear factor. Nature. 1994;370:226–229. doi: 10.1038/370226a0. [DOI] [PubMed] [Google Scholar]
- 5.Bannister A J, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 6.Barton K, Muthusamy N, Chanyangam M, Fischer C, Clendenin C, Leiden J M. Defective thymocyte proliferation and IL-2 production in transgenic mice expressing a dominant-negative form of CREB. Nature. 1996;379:81–85. doi: 10.1038/379081a0. [DOI] [PubMed] [Google Scholar]
- 7.Blendy J A, Kaestner K H, Weinbauer G F, Nieschlag E, Schutz G. Severe impairment of spermatogenesis in mice lacking the CREM gene. Nature. 1996;380:162–165. doi: 10.1038/380162a0. [DOI] [PubMed] [Google Scholar]
- 8.Brown S, McGrath M J, Ooms L M, Gurung R, Maimone M M, Mitchell C A. Characterization of two isoforms of the skeletal muscle LIM protein 1, SLIM1. J Biol Chem. 1999;274:27083–27091. doi: 10.1074/jbc.274.38.27083. [DOI] [PubMed] [Google Scholar]
- 9.Chan K K, Tsui S K, Lee S M, Luk S C, Liew C C, Fung K P, Waye M M, Lee C Y. Molecular cloning and characterization of FHL2, a novel LIM domain protein preferentially expressed in human heart. Gene. 1998;210:345–350. doi: 10.1016/s0378-1119(97)00644-6. [DOI] [PubMed] [Google Scholar]
- 10.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 11.Chrivia J C, Kwok R P, Lamb N, Hagiwara M, Montminy M R, Goodman R H. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- 12.Courey A J, Tjian R. Analysis of Sp1 in vivo reveals multiple transcriptional domains, including a novel glutamine-rich activation motif. Cell. 1988;55:887–898. doi: 10.1016/0092-8674(88)90144-4. [DOI] [PubMed] [Google Scholar]
- 13.Dawid I B, Breen J J, Toyama R. LIM domains: multiple roles as adapters and functional modifiers in protein interactions. Trends Genet. 1998;14:156–162. doi: 10.1016/s0168-9525(98)01424-3. [DOI] [PubMed] [Google Scholar]
- 14.Deak M, Clifton A D, Lucocq L M, Alessi D R. Mitogen- and stress-activated protein kinase-1 (MSK1) is directly activated by MAPK and SAPK2/p38, and may mediate activation of CREB. EMBO J. 1998;17:4426–4441. doi: 10.1093/emboj/17.15.4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Cesare D, Fimia G M, Sassone-Corsi P. Signaling routes to CREM and CREB: plasticity in transcriptional activation. Trends Biochem Sci. 1999;24:281–285. doi: 10.1016/s0968-0004(99)01414-0. [DOI] [PubMed] [Google Scholar]
- 16.De Cesare D, Jacquot S, Hanauer A, Sassone-Corsi P. Rsk-2 activity is necessary for epidermal growth factor-induced phosphorylation of CREB protein and transcription of c-fos gene. Proc Natl Acad Sci USA. 1998;95:12202–12207. doi: 10.1073/pnas.95.21.12202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Cesare D, Sassone-Corsi P. Transcriptional regulation by cyclic AMP-responsive factors. Prog Nucleic Acid Res Mol Biol. 2000;64:343–369. doi: 10.1016/s0079-6603(00)64009-6. [DOI] [PubMed] [Google Scholar]
- 18.de Groot R P, den Hertog J, Vandenheede J R, Goris J, Sassone-Corsi P. Multiple and cooperative phosphorylation events regulate the CREM activator function. EMBO J. 1993;12:3903–3911. doi: 10.1002/j.1460-2075.1993.tb06068.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Desdouets C, Matesic G, Molina C A, Foulkes N S, Sassone-Corsi P, Brechot C, Sobczak-Thepot J. Cell cycle regulation of cyclin A gene expression by the cyclic AMP-responsive transcription factors CREB and CREM. Mol Cell Biol. 1995;15:3301–3309. doi: 10.1128/mcb.15.6.3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fentzke R C, Korcarz C E, Lang R M, Lin H, Leiden J M. Dilated cardiomyopathy in transgenic mice expressing a dominant-negative CREB transcription factor in the heart. J Clin Investig. 1998;101:2415–2426. doi: 10.1172/JCI2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferreri K, Gill G, Montminy M. The cAMP-regulated transcription factor CREB interacts with a component of the TFIID complex. Proc Natl Acad Sci USA. 1994;91:1210–1213. doi: 10.1073/pnas.91.4.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fimia G M, De Cesare D, Sassone-Corsi P. Mechanisms of activation by CREB and CREM: phosphorylation, CBP, and a novel coactivator, ACT. Cold Spring Harbor Symp Quant Biol. 1998;63:631–642. doi: 10.1101/sqb.1998.63.631. [DOI] [PubMed] [Google Scholar]
- 23.Fimia G M, De Cesare D, Sassone-Corsi P. CBP-independent activation of CREM and CREB by the LIM-only protein ACT. Nature. 1999;398:165–169. doi: 10.1038/18237. [DOI] [PubMed] [Google Scholar]
- 24.Foulkes N S, Borrelli E, Sassone-Corsi P. CREM gene: use of alternative DNA-binding domains generates multiple antagonists of cAMP-induced transcription. Cell. 1991;64:739–749. doi: 10.1016/0092-8674(91)90503-q. [DOI] [PubMed] [Google Scholar]
- 25.Genini M, Schwalbe P, Scholl F A, Remppis A, Mattei M G, Schafer B W. Subtractive cloning and characterization of DRAL, a novel LIM-domain protein down-regulated in rhabdomyosarcoma. DNA Cell Biol. 1997;16:433–442. doi: 10.1089/dna.1997.16.433. [DOI] [PubMed] [Google Scholar]
- 26.Ginty D D, Bonni A, Greenberg M E. Nerve growth factor activates a Ras-dependent protein kinase that stimulates c-fos transcription via phosphorylation of CREB. Cell. 1994;77:713–725. doi: 10.1016/0092-8674(94)90055-8. [DOI] [PubMed] [Google Scholar]
- 27.Goldspink P H, Russell B. Physiological role of phosphorylation of the cyclic AMP response element binding protein in rat cardiac nuclei. Cell Tissue Res. 1996;285:379–385. doi: 10.1007/s004410050653. [DOI] [PubMed] [Google Scholar]
- 28.Gonzalez G A, Menzel P, Leonard J, Fischer W H, Montminy M R. Characterization of motifs which are critical for activity of the cyclic AMP-responsive transcription factor CREB. Mol Cell Biol. 1991;11:1306–1312. doi: 10.1128/mcb.11.3.1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gonzalez G A, Montminy M R. Cyclic AMP stimulates somatostatin gene transcription by phosphorylation of CREB at serine 133. Cell. 1989;59:675–680. doi: 10.1016/0092-8674(89)90013-5. [DOI] [PubMed] [Google Scholar]
- 30.Iordanov M, Bender K, Ade T, Schmid W, Sachsenmaier C, Engel K, Gaestel M, Rahmsdorf H J, Herrlich P. CREB is activated by UVC through a p38/HOG-1-dependent protein kinase. EMBO J. 1997;16:1009–1022. doi: 10.1093/emboj/16.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kong Y, Flick M J, Kudla A J, Konieczny S F. Muscle LIM protein promotes myogenesis by enhancing the activity of MyoD. Mol Cell Biol. 1997;17:4750–4760. doi: 10.1128/mcb.17.8.4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kwok R P, Lundblad J R, Chrivia J C, Richards J P, Bachinger H P, Brennan R G, Roberts S G, Green M R, Goodman R H. Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature. 1994;370:223–226. doi: 10.1038/370223a0. [DOI] [PubMed] [Google Scholar]
- 33.Lamas M, Molina C, Foulkes N S, Jansen E, Sassone-Corsi P. Ectopic ICER expression in pituitary corticotroph AtT20 cells: effects on morphology, cell cycle, and hormonal production. Mol Endocrinol. 1997;11:1425–1434. doi: 10.1210/mend.11.10.9987. [DOI] [PubMed] [Google Scholar]
- 34.Laoide B M, Foulkes N S, Schlotter F, Sassone-Corsi P. The functional versatility of CREM is determined by its modular structure. EMBO J. 1993;12:1179–1191. doi: 10.1002/j.1460-2075.1993.tb05759.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee S M, Li H Y, Ng E K, Or S M, Chan K K, Kotaka M, Chim S S, Tsui S K, Waye M M, Fung K P, Lee C Y. Characterization of a brain-specific nuclear LIM domain protein (FHL1B) which is an alternatively spliced variant of FHL1. Gene. 1999;237:253–263. doi: 10.1016/s0378-1119(99)00251-6. [DOI] [PubMed] [Google Scholar]
- 36.Lee S M Y, Tsui S K W, Chan K K, Garcia-Barcelo M, Waye M M Y, Fung K P, Liew C C, Lee C Y. Chromosomal mapping, tissue distribution and cDNA sequence of four-and-a-half LIM domain protein 1 (FHL1) Gene. 1998;216:163–170. doi: 10.1016/s0378-1119(98)00302-3. [DOI] [PubMed] [Google Scholar]
- 37.Lundblad J R, Kwok R P S, Laurence M E, Harter M L, Goodman R H. Adenoviral E1A-associated protein p300 as a functional homologue of the transcriptional co-activator CBP. Nature. 1995;374:85–88. doi: 10.1038/374085a0. [DOI] [PubMed] [Google Scholar]
- 38.Mellstrom B, Naranjo J R, Foulkes N S, Lafarga M, Sassone-Corsi P. Transcriptional response to cAMP in brain: specific distribution and induction of CREM antagonists. Neuron. 1993;10:655–665. doi: 10.1016/0896-6273(93)90167-p. [DOI] [PubMed] [Google Scholar]
- 39.Molina C A, Foulkes N S, Lalli E, Sassone-Corsi P. Inducibility and negative autoregulation of CREM: an alternative promoter directs the expression of ICER, an early response repressor. Cell. 1993;75:875–886. doi: 10.1016/0092-8674(93)90532-u. [DOI] [PubMed] [Google Scholar]
- 40.Montminy M. Transcriptional regulation by cyclic AMP. Annu Rev Biochem. 1997;66:807–822. doi: 10.1146/annurev.biochem.66.1.807. [DOI] [PubMed] [Google Scholar]
- 41.Morgan M J, Madgwick A J A. Slim defines a novel family of LIM-proteins expressed in skeletal muscle. Biochem Biophys Res Commun. 1996;225:632–638. doi: 10.1006/bbrc.1996.1222. [DOI] [PubMed] [Google Scholar]
- 42.Morgan M J, Madgwick A J A. The fourth member of the FHL family of LIM proteins is expressed exclusively in the testis. Biochem Biophys Res Commun. 1999;255:251–255. doi: 10.1006/bbrc.1999.0180. [DOI] [PubMed] [Google Scholar]
- 43.Müller J M, Isele U, Metzger E, Rempel A, Moser M, Pscherer A, Breyer T, Holubarsch C, Buettner R, Schüle R. FHL2, a novel tissue-specific coactivator of the androgen receptor. EMBO J. 2000;19:359–369. doi: 10.1093/emboj/19.3.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nakajima T, Uchida C, Anderson S F, Lee C G, Hurwitz J, Parvin J D, Montminy M. RNA helicase A mediates association of CBP with RNA polymerase II. Cell. 1997;90:1107–1112. doi: 10.1016/s0092-8674(00)80376-1. [DOI] [PubMed] [Google Scholar]
- 45.Nantel F, Monaco L, Foulkes N S, Masquilier D, Le Meur M, Henriksen K, Dierich A, Parvinen M, Sassone-Corsi P. Spermiogenesis deficiency and germ-cell apoptosis in CREM-mutant mice. Nature. 1996;380:159–162. doi: 10.1038/380159a0. [DOI] [PubMed] [Google Scholar]
- 46.Nebel B R, Amarose A P, Hackett E M. Calendar of gametogenesis development in the prepubertal male mouse. Science. 1961;134:832–833. doi: 10.1126/science.134.3482.832. [DOI] [PubMed] [Google Scholar]
- 47.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 48.Parker D, Ferreri K, Nakajima T, La M V, Evans R, Koerber S C, Hoeger C, Montminy M R. Phosphorylation of CREB at Ser-133 induces complex formation with CREB-binding protein via a direct mechanism. Mol Cell Biol. 1996;16:694–703. doi: 10.1128/mcb.16.2.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Quinn P G. Distinct activation domains within cAMP response element-binding protein (CREB) mediate basal and cAMP-stimulated transcription. J Biol Chem. 1993;268:16999–17009. [PubMed] [Google Scholar]
- 50.Rabbitts T H. LMO T-cell translocation oncogenes typify genes activated by chromosomal translocations that alter transcription and developmental processes. Genes Dev. 1998;12:2651–2657. doi: 10.1101/gad.12.17.2651. [DOI] [PubMed] [Google Scholar]
- 51.Radhakrishnan I, Perez-Alvarado G C, Parker D, Dyson H J, Montminy M R, Wright P E. Solution structure of the KIX domain of CBP bound to the transactivation domain of CREB: a model for activator:coactivator interactions. Cell. 1997;91:741–752. doi: 10.1016/s0092-8674(00)80463-8. [DOI] [PubMed] [Google Scholar]
- 52.Rupp R A W, Snider L, Weintraub H. Xenopus embryos regulate the nuclear localization of XMyoD. Genes Dev. 1994;8:1311–1323. doi: 10.1101/gad.8.11.1311. [DOI] [PubMed] [Google Scholar]
- 53.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 54.Sassone-Corsi P. Transcription factors responsive to cAMP. Annu Rev Cell Dev Biol. 1995;11:355–377. doi: 10.1146/annurev.cb.11.110195.002035. [DOI] [PubMed] [Google Scholar]
- 55.Sassone-Corsi P, Visvader J, Ferland L, Mellon P L, Verma I M. Induction of proto-oncogene fos transcription through the adenylate cyclase pathway: characterization of a cAMP-responsive element. Genes Dev. 1988;2:1529–1538. doi: 10.1101/gad.2.12a.1529. [DOI] [PubMed] [Google Scholar]
- 56.Schmeichel K L, Beckerle M C. The LIM domain is a modular protein-binding interface. Cell. 1994;79:211–219. doi: 10.1016/0092-8674(94)90191-0. [DOI] [PubMed] [Google Scholar]
- 57.Schubert M, Holland N D, Holland L Z. Amphioxus AmphiDRAL encoding a LIM-domain protein: expression in the epidermis but not in the presumptive neuroectoderm. Mech Dev. 1998;76:203–205. doi: 10.1016/s0925-4773(98)00120-8. [DOI] [PubMed] [Google Scholar]
- 58.Sheng M, Thompson M A, Greenberg M E. CREB: a Ca2+-regulated transcription factor phosphorylated by calmodulin-dependent kinases. Science. 1991;252:1427–1430. doi: 10.1126/science.1646483. [DOI] [PubMed] [Google Scholar]
- 59.Struthers R S, Vale W W, Arias C, Sawchenko P E, Montminy M R. Somatotroph hypoplasia and dwarfism in transgenic mice expressing a non-phosphorylatable CREB mutant. Nature. 1991;350:622–624. doi: 10.1038/350622a0. [DOI] [PubMed] [Google Scholar]
- 60.Swope D L, Mueller C L, Chrivia J C. CREB-binding protein activates transcription through multiple domains. J Biol Chem. 1996;271:28138–28145. doi: 10.1074/jbc.271.45.28138. [DOI] [PubMed] [Google Scholar]
- 61.Tan Y, Rouse J, Zhang A, Cariati S, Cohen P, Comb M J. FGF and stress regulate CREB and ATF-1 via a pathway involving p38 MAP kinase and MAPKAP kinase-2. EMBO J. 1996;15:4629–4642. [PMC free article] [PubMed] [Google Scholar]
- 62.Taniguchi Y, Furukawa T, Tun T, Han H, Honjo T. LIM protein KyoT2 negatively regulates transcription by association with the RBP-J DNA-binding protein. Mol Cell Biol. 1998;18:644–654. doi: 10.1128/mcb.18.1.644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thompson J D, Gibson T J, Plewniak F, Jeanmougin F, Higgins D G. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wadman I A, Osada H, Grutz G G, Agulnick A D, Westphal H, Forster A, Rabbitts T H. The LIM-only protein Lmo2 is a bridging molecule assembling an erythroid, DNA-binding complex which includes the TAL1, E47, GATA-1 and Ldb1/NLI proteins. EMBO J. 1997;16:3145–3157. doi: 10.1093/emboj/16.11.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang E H, Zou S, Tjian R. TAFII250-dependent transcription of cyclin A is directed by ATF activator proteins. Genes Dev. 1997;11:2658–2669. doi: 10.1101/gad.11.20.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xing J, Ginty D D, Greenberg M E. Coupling of the RAS-MAPK pathway to gene activation by RSK2, a growth factor-regulated CREB kinase. Science. 1996;273:959–963. doi: 10.1126/science.273.5277.959. [DOI] [PubMed] [Google Scholar]
- 67.Yoshizumi M, Hsieh C-M, Zhou F, Tsai J-C, Patterson C, Perrella M A, Lee M-E. The ATF site mediates downregulation of the cyclin A gene during contact inhibition in vascular endothelial cells. Mol Cell Biol. 1995;15:3266–3272. doi: 10.1128/mcb.15.6.3266. [DOI] [PMC free article] [PubMed] [Google Scholar]