Abstract
The Hoxb1 autoregulatory element comprises three HOX-PBX binding sites. Despite the presence of HOXB1 and PBX1, this enhancer fails to activate reporter gene expression in retinoic acid-treated P19 cell monolayers. Activation requires cell aggregation in addition to RA. This suggests that HOX-PBX complexes may repress transcription under some conditions. Consistent with this, multimerized HOX-PBX binding sites repress reporter gene expression in HEK293 cells. We provide a mechanistic basis for repressor function by demonstrating that a corepressor complex, including histone deacetylases (HDACs) 1 and 3, mSIN3B, and N-CoR/SMRT, interacts with PBX1A. We map a site of interaction with HDAC1 to the PBX1 N terminus and show that the PBX partner is required for repression by the HOX-PBX complex. Treatment with the deacetylase inhibitor trichostatin A not only relieves repression but also converts the HOX-PBX complex to a net activator of transcription. We show that this activation function is mediated by the recruitment of the coactivator CREB-binding protein by the HOX partner. Interestingly, HOX-PBX complexes are switched from transcriptional repressors to activators in response to protein kinase A signaling or cell aggregation. Together, our results suggest a model whereby the HOX-PBX complex can act as a repressor or activator of transcription via association with corepressors and coactivators. The model implies that cell signaling is a direct determinant of HOX-PBX function in the patterning of the animal embryo.
HOX proteins are sequence-specific DNA-binding transcription factors that play a crucial role in the specification of anteroposterior identity in the animal embryo (20, 54). Conservation within the DNA-binding homeodomains results in different HOX proteins recognizing similar regulatory elements with only modest preferences (reviewed in reference 27). High-affinity DNA binding is achieved when HOX proteins are heterodimerized with partners of the PBC family (mammalian PBX, Drosophila Extradenticle [EXD], and Caenorhabditis elegans CEH-20) (55). Mammalian MEIS1 has been shown to independently dimerize with HOX proteins and with PBX (11, 57, 78). Recently, trimeric complexes encompassing all three homeoproteins, HOX-PBX-MEIS, have also been characterized (77, 79). The MEIS-related protein PREP1, also known as PKNOX1, can additionally form a dimer with PBX, as well as a trimeric complex with HOX and PBX partners (6, 7, 15, 34). While the majority of HOX monomers recognize a DNA core motif of TAAT (23), HOX-PBX, HOX-MEIS, and PBX-MEIS heterodimers recognize larger motifs resulting in a higher affinity and specificity of DNA binding by these homeoproteins (49).
A conserved motif with the consensus YPWM is found N terminal to the homeodomain of HOX proteins from paralogous groups 1 to 8. The YPWM motif contacts the PBX homeodomain and is strictly required for cooperative DNA binding by PBX and HOX partners (49, 50). A conserved W in HOX proteins from groups 9 and 10 performs a similar function (12).
The downstream targets of mammalian HOX proteins have been poorly characterized. The best-characterized targets are some Hox genes known to be positively autoregulated by their own products or cross-regulated by the products of other Hox genes (26, 68, 69). In these instances, HOX-PBX complexes act as activators of transcription. For example, the Hoxb1 autoregulatory element (ARE) contains three binding sites for HOX-PBX complexes. These sites are required to direct expression of a Hoxb1 transgene in rhombomere 4 (r4) of the developing hindbrain (68).
Genetic and molecular studies have provided evidence supporting a negative regulatory role for HOX proteins (43). In the case of decapentaplegic (dpp) regulation in Drosophila, repression by HOX proteins dominates over activation (9). This implies active transcriptional repression by HOX proteins (9, 25, 46). In addition, in vitro mapping studies have characterized repression domains in different HOX proteins, as well as in the PBX partner (13, 45, 75). Therefore, HOX proteins may be activators or repressors in a context-dependent manner.
By analogy to nuclear receptors, HOX-PBX complexes are likely to achieve transcriptional repression or activation through differential association with coactivators and corepressors (81). One class of coregulators are the histone acetyltransferases (HATs) and the histone deacetylases (HDACs), which modify chromatin as well as nonhistone proteins. The HATs include GCN5, PCAF, CREB-binding protein (CBP)/p300, the steroid receptor coactivator class, and the MYST family (80). On the other hand, the known HDACs include HDAC1 through -8, with class I HDACs consisting of HDAC1, HDAC2, HDAC3, and HDAC8 (homologues of the yeast RPD3 protein) and class II HDACs consisting of HDAC4, HDAC5, HDAC6, and HDAC7 (homologues of the yeast HDA1 protein) (for a review, see reference 37). HDAC1 and HDAC2 form the catalytic subunits of two characterized multiprotein complexes, the mSIN3A and Mi2 complexes (35). Additionally, HDAC3 has been shown to interact with the corepressor SMRT (28). Recent genetic evidence in C. elegans shows EGL-27, a homologue of MTA1 (a component of the Mi2-HDAC1 complex), in the same pathway as MAb-5 (86, 94), further implying that HOX proteins may interact with HDACs and other histone-modifying enzymes to accomplish their developmental program.
In this report, we present evidence for an interaction between HOX-PBX complexes and histone-modifying enzymes and show that the activity of the HOX-PBX heterodimer is determined by a regulated balance between a corepressor complex consisting of class I HDACs, mSIN3B, and N-CoR/SMRT and a coactivator complex containing CBP. We show, moreover, that activation of the protein kinase A (PKA) signaling pathway significantly potentiates the CBP-mediated transactivation by HOX-PBX complexes. We propose a model in which PKA acts as a signaling switch that converts HOX-PBX complexes from transcriptional repressors to activators.
MATERIALS AND METHODS
Cell culture and transfections.
P19 mouse embryonal carcinoma (EC) cells and human embryonic kidney HEK293 cells were cultured in alpha minimal essential medium supplemented with 10% fetal calf serum. Some experiments employed 293 T cells constitutively expressing the simian virus 40 large T antigen. Transient transfections were performed using the calcium phosphate precipitation method as described earlier (72). A lacZ reporter driven by the cytomegalovirus (CMV) enhancer was used to control for transfection efficiency in some experiments. Because the activity of the CMV enhancer appeared to change in response to PKA, a lacZ reporter driven by the Rous sarcoma virus long terminal repeat was used in transfections involving PKA. For stable transfections of P19, the cells were seeded at a density of 105 cells/10-cm plate and transfected with a total of 15 μg of DNA consisting of 9 μg of the transgene of interest (p1230 or b1-ARE-lacZ), 1 μg of PGK-puromycin, and 5 μg of pCAB-B17 as the carrier DNA (53). At 40 h posttransfection, cells were selected with 2 μg of puromycin per ml for at least 10 days. Cells were kept in monolayer or aggregated in bacterial petri dishes for 24 h in the presence or absence of treatment and then reattached in tissue culture plates overnight (73). The treatment consisted of either retinoic acid (RA) (3 × 10−7 M) or trichostatin A (TSA; concentrations ranging from 20 nM to 2 μM) or a combination of both RA and TSA. Significant cell death sometimes occurred in response to TSA; however, this was variable and dependent on drug concentration and cell context. HEK293 cells were more sensitive than P19 to TSA-induced cell death. Cells were treated with the estrogen antagonist α-hydroxytamoxifen (TOT) overnight at 10−7 M.
Antibodies.
Rabbit polyclonal antibodies raised against PBX1, mSIN3A, or mSIN3B were purchased from Santa Cruz. Rabbit polyclonal antibodies against human HDAC1 and HDAC3 were from Upstate Biotechnology. Rabbit polyclonal antibodies against HOXB1 were generously supplied by C. Largman. Mouse monoclonal antibodies against the GAL4 DNA-binding domain (DBD) (RK5C1), the hemagglutinin epitope (HA-11), and the flag epitope (M2) were purchased from Santa Cruz, Babco, and Sigma, respectively. Mouse monoclonal antibodies were recognized with horseradish peroxidase (HRP)-conjugated goat anti-mouse (κ light chain) secondary antibodies from PharMingen, and rabbit polyclonal antibodies were recognized by HRP-conjugated protein A-Sepharose (Amersham).
Plasmids.
p1230 (generous gift of R. Krumlauf) is a lacZ reporter under the control of the minimal promoter of the β globin gene. b1-ARE-lacZ consists of the ARE of the Hoxb1 gene (68) cloned by PCR amplification into the HindIII-XhoI sites of p1230. pML, pML(5xHOX-PBX), pML5xHOX, and pML5xUAS are luciferase reporters containing the adenovirus major late promoter alone, driven by 5xHOX-PBX binding sites (TGATTGAT), 5xHOX binding sites (TAAT), or 5xGAL4 binding sites, respectively (67, 72, 77). Expression plasmids for HOXA1, HOXD4, PBX1A, and PBX1A deletion mutants have been previously described (66, 77). The HOXB1 expression vector is driven by the beta-actin promoter. 89-172-HA was constructed by PCR amplification of the region encoding residues 89 to 172, followed by cloning of the product in frame with three copies of the HA epitope in the plasmid pRC/CMV (Invitrogen). Flag-HDAC1, flag-HDAC3 and E1A are described elsewhere (87, 88, 91) and were generously provided by A. Lai (McGill University). Flag-HDAC4 and flag-PCAF are described elsewhere (84, 89). Flag-N-CoR, flag-SMRT, HA-CBP, and the CBP domains were generously provided by V. Giguère and A. Tremblay (McGill University, Université de Montréal). GAL4-HOXD4N fuses the first 141 residues of HOXD4 to the GAL4 DBD and was described previously (72). HOXD4 residues 139 to 250 were fused to the GAL4 DBD to generate GAL4-HOXD4C. An expression vector for the human estrogen receptor alpha driven by the CMV enhancer was generously provided by Vincent Giguère (McGill University).
β-Galactosidase and luciferase assays.
Luciferase assays and liquid β-galactosidase assays were performed as described previously (67). β-Galactosidase plate assays were performed after fixation of the cells with a solution of 2% formaldehyde–0.2% glutaraldehyde in phosphate-buffered saline (PBS) for 5 min at 4°C. The cells were washed with PBS for three times and then stained at 37°C with a solution composed of 5 mM ferrocyanide, 5 mM ferricyanide, 1 mg of X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside)/ml, and 2 mM MgCl2 in PBS.
Immunoprecipitation assays.
At 40 h posttransfection, the cells were harvested and lysed on ice for 30 min with 500 μl of a low-stringency buffer containing 150 mM KCl. Whole-cell extracts were precleared with protein A- or protein G-Sepharose (depending on the source of the primary antibody used) for 30 min. Precleared lysates were incubated with 0.5 to 2 μg of primary antibody for 2 h, followed by the addition of 20 μl of a 50% slurry of protein A- or protein G-Sepharose for 2 to 18 h. Precipitates were washed six times with the lysis buffer and eluted by boiling in 2× sample buffer for 15 min. Eluted proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and analyzed after Western blotting to polyvinylidene difluoride membranes (Millipore). Secondary antibodies used in Western analysis were HRP conjugated and were detected by enhanced chemiluminescence (NEN Life Sciences). To immunoprecipitate flag-epitope-tagged proteins, a similar protocol was used except that M2 beads (Sigma) were used instead of protein G-Sepharose, and flag peptides (Sigma) were used to elute the precipitated proteins prior to boiling.
RESULTS
TSA relieves the transcriptional repression of HOX-PBX-responsive enhancers.
The induction of Hoxb1 upon RA treatment of mouse embryos is mediated directly by a 3′ RA response element (RARE) (51) and indirectly by an ARE (68). The Hoxb1 ARE consists of three cooperative binding sites for HOX-PBX heterodimers (Fig. 1A, top panel). Two paralog group 1 HOX proteins, HOXB1 (18) and HOXA1 (M. Phelan and M. S. Featherstone, unpublished observations), can activate transcription through the Hoxb1 ARE. Both gain- and loss-of-function experiments show that HOXA1 and HOXB1 regulate Hoxb1 expression in the embryonic hindbrain (5, 68, 82, 93). These effects are very likely to be mediated by the Hoxb1 ARE, as has been demonstrated in one case (68). In addition to HOXB1 and PBX, coexpression of PREP1 stimulates reporter gene expression through the Hoxb1 ARE in transfected cells (6). Together, these results suggest that the presence of first group HOX proteins, PBX, and members of the MEIS/PREP family would be sufficient to activate transcription through the Hoxb1 ARE.
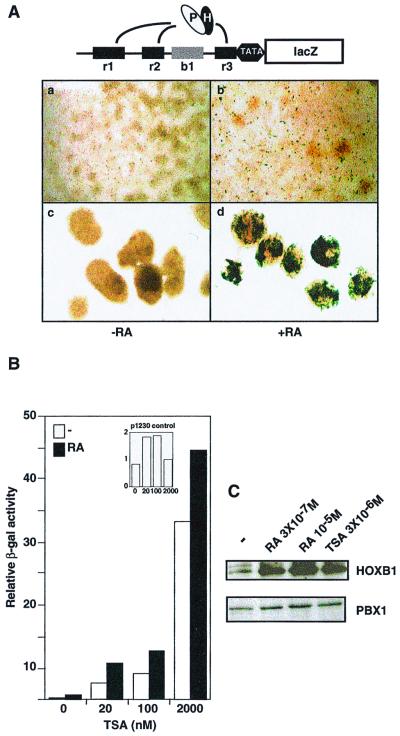
FIG. 1.
TSA relieves the transcriptional repression of HOX-PBX-responsive enhancers. (A) The upper panel schematically represents the b1-ARE-lacZ reporter used to stably transfect P19 cells. The black boxes r1, r2, and r3 represent three previously characterized HOX-PBX binding sites (71). The gray box b1 denotes block 1, a region of homology conserved across species. Ovals labeled “P” and “H” denote the PBX-HOX complex. In the lower panel, a stably transfected transgene containing the Hoxb1 ARE (b1-ARE-lacZ) was active in RA (3 × 10−7 M)-treated P19 cells only if the cells were aggregated during RA exposure for 24 h (panel d) but not if the cells were kept cultured in monolayers (panel b). P19 cell monolayers are shown in panels a and b, while cell aggregates are shown in panels c and d. The cells in panels b and d were treated with RA at 3 × 10−7 M for 24 h. (B) TSA induces the activity of the b1-ARE-lacZ in monolayers in the presence or absence of RA. Liquid β-galactosidase assays were carried out on P19 cells stably transfected with the b1-ARE-lacZ and cultured in monolayer. Monolayers were treated with either RA (3 × 10−7 M), TSA (20 nM to 2 μM), or a combination of both for 24 h. In the inset, similar assays were performed using a control transgene lacking the Hoxb1 ARE (p1230). (C) HOXB1 and PBX1 are induced in P19 cell monolayers in response to RA or TSA. Western analysis was performed using whole-cell extracts from P19 cells cultured in monolayers in the absence or presence of treatment. RA was used at 3 × 10−7 M or 10−5 M, and TSA was at 3 μM.
P19 EC cells differentiate along the neural pathway when aggregated in the presence of RA (73). While RA-treated P19 cell monolayers fail to form neurons and glia, the products of the Hoxb1, Hoxa1, Pbx, Meis, and Prep genes are induced (21, 36, 40, 62). We therefore expected that a stable integrated transgene carrying the Hoxb1 ARE driving lacZ (b1-ARE-lacZ) would be active in RA-treated P19 cell monolayers. Surprisingly, b1-ARE-lacZ was poorly active in P19 EC cells when cultured in monolayers in the presence of RA (Fig. 1Ab). The transgene was efficiently activated only when RA-treated cells were also aggregated (Fig. 1Ad), suggesting that cell aggregation provides a signal required for HOXB1-PBX complexes to activate transcription.
An alternative explanation for these results is that the site of integration imposed constraints on the activity of the Hoxb1 ARE. However, these experiments were done on populations of multiple clones representing many different sites of integration. Another possibility is that HOXB1, PBX, and MEIS/PREP proteins unexpectedly failed to accumulate upon RA treatment. This was not the case, as revealed by Western blot analysis (Fig. 1C). HOXB1 and PBX1 were both detected in P19 cell monolayers treated with RA at either of two concentrations. HOXB1 showed the most dramatic induction, while PBX1 was already present in untreated cells and was modestly induced upon RA treatment. MEIS1 was also present before and after RA treatment (data not shown).
We hypothesized that in the absence of cell aggregation, HOXB1-PBX complexes could recruit HDACs to the Hoxb1 ARE, thereby establishing a transcriptionally inactive condensed chromatin. To test this hypothesis, we treated the cells in monolayer with TSA, an HDAC inhibitor, and measured the reporter activity (Fig. 1B). As little as 20 nM TSA induced lacZ expression directed by the Hoxb1 ARE, thereby circumventing the need for cell aggregation. In fact, TSA efficiently induced reporter gene expression in the absence of both RA and aggregation.
To investigate the effect of TSA on endogenous gene expression, we performed Western blot analysis on TSA-treated cultures. Figure 1C shows that TSA efficiently induced the expression of the endogenous Hoxb1 gene, while PBX1 (Fig. 1C) and MEIS1 (data not shown) showed a moderate increase over preexisting levels. Thus, TSA-treated cultures express all three homeoprotein families implicated in activation through the Hoxb1 ARE. In contrast, TSA had no effect on a stably integrated control transgene (p1230) that lacks the Hoxb1 ARE, establishing the specificity of this effect (Fig. 1B, inset). Together, these results suggest that HOXB1-PBX complexes recruit HDACs in vivo to repress transcription directed by the Hoxb1 ARE. TSA treatment inhibits HDAC activity, thereby inducing both the endogenous Hoxb1 gene and the b1-ARE-lacZ reporter.
The Hoxb1 ARE used above is 150 bp long and may contain binding sites for TSA-responsive transcription factors other than PBX or HOX proteins. To specifically test the response of HOX-PBX complexes to TSA, we transfected HEK293 cells with an artificial luciferase reporter, pML(5xHOX-PBX), driven solely by five HOX-PBX binding sites in front of a minimal promoter. pML(5xHOX-PBX) was repressed fivefold relative to the parental vector pML lacking HOX-PBX binding sites (Fig. 2), again implicating HOX-PBX complexes in transcriptional repression. While pML was induced <2-fold by TSA, pML(5xHOX-PBX) was activated 12-fold (Fig. 2), further supporting a role for HDACs in repression mediated by HOX-PBX complexes.
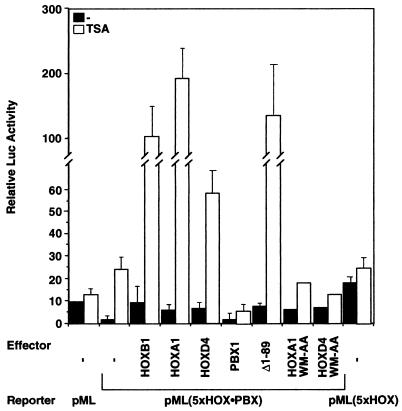
FIG. 2.
PBX is required for the HOX-PBX response to TSA. pML(5xHOX-PBX), a reporter driven by five HOX-PBX binding sites, is repressed in transiently transfected HEK293 cells compared to pML, which lacks HOX-PBX binding sites. pML(5xHOX-PBX) is significantly activated by TSA (2 μM, 24 h) both in the absence or presence of overexpressed HOX and PBX1A proteins. Removal of residues 1 to 89 of PBX1A (Δ1–89) greatly increases reporter activation by TSA. pML(5xHOX), containing five sites for monomeric HOX binding, is not repressed in 293 cells and is not further activated by TSA treatment. Overexpression of HOXA1 or HOXD4, but not of A1 WM-AA or D4 WM-AA, transactivates transcription in the presence of TSA. All transfections were repeated at least three times in duplicate except for the A1 WM-AA and D4 WM-AA experiments, which were done once in duplicate.
Overexpression of HOXB1, HOXA1, or HOXD4 enhanced the activation of pML(5xHOX-PBX) by TSA (Fig. 2), confirming the involvement of HOX proteins in this effect. In contrast, the TSA response was dampened by the overexpression of PBX1A (Fig. 2). Interestingly, deletion of the first 89 residues of PBX1A rendered the derivative protein highly TSA sensitive, resulting in an almost 100-fold activation of pML(5xHOX-PBX). We suggest explanations for this effect in Discussion.
PBX is required for repression by HOX-PBX and for the response to TSA.
The above results implicate HOX proteins in transcriptional activation through HOX-PBX binding sites, whereas PBX had a repressive effect. To assess the importance of PBX for repression and the TSA response, we examined an independent reporter, pML(5xHOX), driven by monomeric HOX binding sites. In contrast to pML(5xHOX-PBX), pML(5xHOX) was not repressed in 293 cells and was not activated by TSA (Fig. 2). This result argues that PBX is required for the repression observed on pML(5xHOX-PBX) and for activation by TSA on this reporter. Reciprocally, HOX proteins cannot activate transcription efficiently in the absence of a PBX partner.
In a complementary test, we used derivatives of HOXA1 and HOXD4 harboring mutations in the conserved YPWM motif (A1 WM-AA and D4 WM-AA, respectively). This mutation has been previously shown not to affect the stability of HOXD4 (72) and to abolish interaction between HOX and PBX proteins (66, 67, 76, 77). As shown in Fig. 2, while overexpression of HOXA1 or HOXD4 greatly enhanced the TSA effect on pML(5xHOX-PBX), this was abolished with A1 WM-AA and D4 WM-AA. These findings demonstrate that interaction of HOX with PBX is required for the TSA response of pML(5xHOX-PBX). To explain these results, we propose a model whereby physical interaction between HOX and PBX is required for association with coactivators and corepressors, respectively (see Discussion).
PBX1 interacts with class I HDACs.
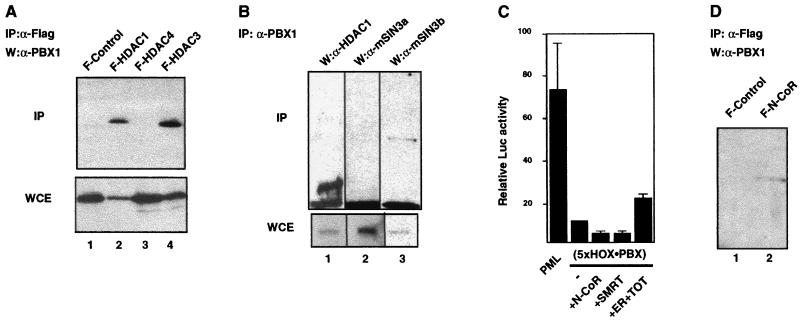
As shown above, PBX is required for TSA-sensitive repression mediated by HOX-PBX binding sites. The simplest explanation for this finding is that PBX directly interacts with one or more HDACs. To test this, we performed immunoprecipitation experiments using whole-cell extracts from transfected 293 T cells. Flag-epitope-tagged HDAC1 and HDAC3, but not HDAC4, resulted in coprecipitation of PBX1 (Fig. 3A). This interaction is specific and shows a preference for the class I HDACs by HOX-PBX complexes. More stringently, rabbit polyclonal antibodies that specifically recognize PBX1 coprecipitated the endogenous HDAC1 and mSIN3B (Fig. 3B, lanes 1 and 3). Interestingly, no interaction was observed with mSIN3A (Fig. 3B, lane 2) or with Mi2α or -β (data not shown). However, as shown in Fig. 3D (lane 2), N-CoR, known to repress transcription in an mSIN3A complex (58), coprecipitated with PBX1 in vivo. Thus, N-CoR/SMRT may associate with mSIN3B in the absence of mSIN3A.
FIG. 3.
The HOX-PBX complex associates with class I HDACs in vivo and represses transcription in a mSIN3B/N-CoR/SMRT-dependent manner. (A) PBX1 coprecipitates with class I HDACs (HDAC1 and HDAC3, lane 2 and 4) but not with HDAC4 (lane 3) or from cells transfected with the empty flag vector (F-control) (lane 1). Immunoprecipitations were done with lysates from 293 T cells cotransfected with plasmids expressing PBX1A and flag-tagged HDAC1 (F-HDAC1), F-HDAC3, F-HDAC4, or F-control. Flag-tagged proteins were immunoprecipitated with M2 beads (Sigma), and the precipitates were eluted with flag peptides (Sigma) and analyzed by Western blotting using rabbit polyclonal antibodies against PBX1 (Santa Cruz). “IP” and “WCE” denote immunoprecipitates and whole-cell extracts used in Western blot analysis. “W” denotes the antibody used in Western analysis. (B) Coprecipitation of endogenous HDAC1 and mSIN3B (but not mSIN3A) with rabbit polyclonal antibodies against PBX1. 293 T cells were transfected with a plasmid expressing PBX1A but not with plasmids expressing HDAC1, mSIN3B, or mSIN3A. Immunoprecipitates with anti-PBX1 antibodies (IP: α-PBX1) were analyzed in Western blots with antibodies against HDAC1 (W:α-HDAC1), mSIN3a (W:α-mSIN3a), and mSIN3b (W:α-mSIN3b). (C) The repression of pML(5xHOX-PBX) in 293 T cells is exerted by N-CoR/SMRT-corepressor complexes. Overexpression of either N-CoR or SMRT further repressed pML(5xHOX-PBX). This repression can be partially relieved by sequestering the endogenous N-CoR/SMRT with overexpressed estrogen receptor (ER) bound to the estrogen antagonist TOT (see Materials and Methods). (D) Immunoprecipitation of PBX1 from cells expressing flag-tagged N-CoR (F-N-CoR, lane 2) but not from cells transfected with the empty flag vector (F-control, lane 1).
To functionally characterize these interactions, we examined the effects of N-CoR and SMRT on pML(5xHOX-PBX). As shown in Fig. 3C, overexpression of either N-CoR or SMRT potentiated the repression observed with pML(5xHOX-PBX) in 293 T cells. Overexpression of an antagonist-bound estrogen receptor, in an attempt to titrate the endogenous levels of N-CoR/SMRT (41), resulted in a partial relief of repression of pML(5xHOX-PBX). These data suggest that N-CoR/SMRT complexes are recruited by HOX-PBX within the cell to exert significant repression effects on downstream targets.
Region from residues 89 to 172 in the PBX1 N terminus interacts with HDAC1.
In PBX1, three N-terminal repression domains (corresponding to regions B, C, and D in Fig. 4A) have been previously mapped (45). To directly characterize whether one of these repression domains recruits the HDAC complex, we generated multiple in-frame deletions in PBX1A (Fig. 4A) and examined the in vivo association with HDAC1. Immunoprecipitation studies were carried out with extracts from 293 T cells cotransfected with plasmids expressing flag-tagged HDAC1 along with PBX1A deletion derivatives. Following immunoprecipitation with anti-flag antibodies, the precipitates were analyzed by Western analysis using polyclonal antibodies against PBX1 or anti-HA antibodies in the cases of Δ89-HA and 89–172-HA. Fig. 4B shows that the PBX1 N terminus (ΔC232) is sufficient for HDAC1 binding.
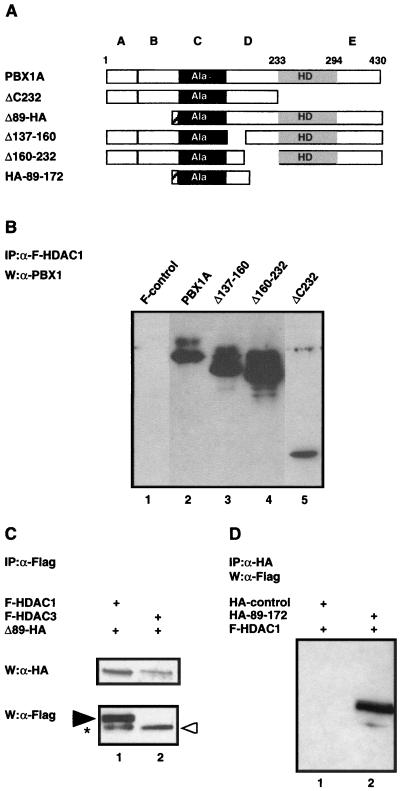
FIG. 4.
Region C of PBX1A is responsible for the interaction with HDAC1. (A) Schematic representations of wild-type PBX1A and PBX1A deletion mutants. The subdivision of the PBX1A N terminus into four domains labeled A, B, C, and D is as previously described (45). The striped rectangle indicates the position of the HA tag in Δ1–89 and in HA-89–172. (B) The PBX1A N terminus interacts with HDAC1. Binding studies similar to those described in the legend to Fig. 3A were carried out for PBX1A and PBX1A mutants with flag-tagged HDAC1 immunoprecipitated on M2 beads and eluted with flag peptide. Anti-PBX1 antibodies were used for the Western analysis. (C) Regions A and B of PBX1A are dispensable for interaction with HDAC1 and -3. The experiment was done as described in the legend to panel B except that the Δ1–89 mutant was tagged with the HA epitope and was recognized in Western analysis by anti-HA antibodies (Babco). The black arrowhead indicates HDAC1, the white arrowhead indicates HDAC3, and the asterisk indicates an HDAC1 degradation product. (D) HDAC1 coprecipitates with the region from residues 89 to 172 of PBX1A. Cells were transfected with a vector expressing flag-tagged HDAC1 and either an empty HA vector (HA-control) or one expressing the HA-tagged region from residues 89 to 172 of PBX1. Immunoprecipitation (IP) experiments were carried out with anti-HA antibodies, and anti-flag antibodies were used in the Western analysis.
Δ89 is highly responsive to TSA (Fig. 2), suggesting that the HDAC interaction region in PBX1A is C terminal to residue 89. As shown in Fig. 4C, Δ89-HA associated with HDAC1 and HDAC3 in whole-cell extracts, mapping the region of interaction with HDAC1 to PBX1A region C or D. Two deletions in region D were therefore tested and found to be dispensable for HDAC1 binding (Δ137–160 and Δ160–232, Fig. 4A and B). These data imply that region C is important for the recruitment of the HDAC complex by PBX1.
A deletion mutant of region C was not stable in mammalian cells. To address whether region C is sufficient for interaction with HDAC1, we used anti-HA antibodies to immunoprecipitate a fusion protein containing the HA epitope fused in frame to residues 89 to 172 spanning region C of PBX1A. As seen in Fig. 4D, HDAC1 coprecipitated with HA–89-172 (lane 2) but not with an HA control (lane 1). The above data indicate that, while the region B repression mechanism is TSA insensitive, region C recruits HDACs to repress transcription.
The HOXD4 activation domain binds the HAT-C/H3 domain of CBP.
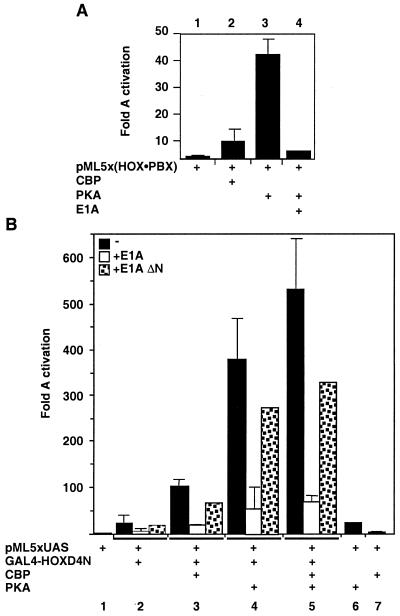
Treatment with TSA led to large increases in transcription from natural and artificial enhancers bearing HOX-PBX binding sites (Fig. 1 and 2). Activation of pML(5xHOX-PBX) exceeded a simple loss of repression relative to pML (Fig. 2A). These results show that TSA reveals a transcriptional activation function of the HOX-PBX heterodimer. Transcriptional activation is achieved through recruitment of coactivators by enhancer-bound proteins. One such coactivator is CBP. To assess its involvement in transcriptional activation by HOX-PBX complexes, we overexpressed CBP in 293 T cells. CBP stimulated expression from pML(5xHOX-PBX) 10- to 12-fold, similar to the activation obtained by TSA treatment (Fig. 5A, lane 2). This result suggested that PBX, HOX, or both recruited CBP to target promoters.
FIG. 5.
CBP enhances the transactivation potential of HOX-PBX complexes and is required to transduce PKA signaling. (A) pML(5xHOX-PBX) is activated by overexpression of CBP in 293 T cells and is superactivated by the catalytic domain of PKA. Activation by PKA is inhibited by overexpression of E1A. (B) A fusion of the N terminus of HOXD4 to the GAL4 DNA-binding domain (GAL4-HOXD4N) is able to transactivate transcription from a heterologous promoter driven by 5xGAL4 binding sites [pML(5xUAS)] (lanes 1 and 2, black bars). CBP potentiates the transactivation function of HOXD4N on this reporter (lane 3, black bar) in a manner sensitive to E1A (white bar) but not to E1A ΔN (dotted bar), a mutant deficient in CBP binding. PKA stimulates HOXD4N transactivation in a CBP-dependent manner (lanes 4 and 5).
We have previously characterized an activation domain in the proline-rich N-terminal half of HOXD4 (73). We therefore tested whether the HOXD4 activation domain (HOXD4N, residues 3 to 141) could recruit CBP to a target promoter. Figure 5B (lanes 1, 2, and 3, black bars) shows that overexpression of CBP potentiates transactivation by a GAL4-HOXD4N fusion protein on the GAL4-responsive reporter pML(5xUAS). In contrast, depletion of endogenous CBP by overexpression of the oncoprotein E1A neutralizes the coactivation effect seen with overexpressed CBP. E1A also inhibits the initial activation observed with HOXD4N (Fig. 5B, compare white bars in lanes 2 and 3 to black bars in lanes 1, 2, and 3). A deletion mutant of E1A that cannot bind CBP is unable to affect transcription significantly (dotted bars in Fig. 5B). These results show that the transactivation function of HOXD4N is mediated by endogenous CBP. We also note that E1A interacts with the coactivator p300 through this same domain. None of our data excludes an interaction between HOX proteins and p300, in addition to CBP. Likewise, PCAF is expected to bind CBP in association with HOX (89).
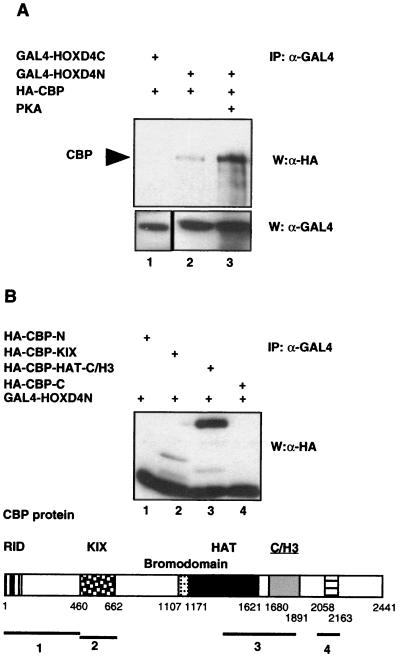
In vivo mapping studies were carried out to determine the respective domains of interactions between HOXD4 and CBP. A fusion of GAL4 to the HOXD4 N terminus (GAL4-HOXD4N) but not to the C terminus (GAL4-HOXD4C) coprecipitated with CBP, consistent with the N-terminal transactivation function of HOXD4 (Fig. 6A, lanes 1 and 2). To map the domains in CBP required for HOX binding, immunoprecipitation experiments were carried out with extracts from 293 T cells cotransfected with plasmids expressing GAL4-HOXD4N and one of four HA-tagged CBP domains: CBP-N, CBP-KIX, CBP-HAT-C/H3, or CBP-C (Fig. 6B). Analysis of the precipitates was carried out by Western blot analysis with anti-HA antibodies. The four CBP domains used in this experiment were expressed at equivalent amounts in 293 T cells (data not shown). Figure 6B shows that the HAT-C/H3 domains of CBP constitute the region of interaction with HOXD4N.
FIG. 6.
(A) Interactions between the HOXD4 N terminus and CBP. GAL4-HOXD4N or GAL4-HOXD4C were immunoprecipitated with antibodies against the GAL4 DBD. Interaction with HA-tagged CBP (HA-CBP) in the presence or in the absence of overexpressed PKA was assessed by Western analysis using anti-HA antibodies. (B) The HOXD4 N terminus coprecipitates with the CBP HAT-C/H3 domains. Immunoprecipitation studies were performed on whole-cell extracts from 293 T cells cotransfected with GAL4-HOXD4N along with four HA-tagged domains of CBP: HA-CBP-N (amino acids 1 to 460), HA-CBP-KIX (amino acids 460 to 662), HA-CBP-HAT-C/H3 (amino acids 1450 to 1903), or HA-CBP-C (amino acids 2040 to 2170). Immunoprecipitation (IP) was performed with antibodies against the GAL4 DBD, and the CBP domains were detected by Western analysis using anti-HA antibodies. The schematic representation of the CBP protein is as described by Chariot et al. (14).
PKA signaling stimulates HOX-PBX promoters.
The above results show that PBX and HOX proteins directly contact transcriptional corepressors and coactivators, respectively. What determines whether the HOX-PBX complex will have a net activating or repressive effect on gene expression? Our studies in P19 EC cells show that aggregation provides a signal that converts HOX-PBX complexes from repressors to activators. This conversion is dependent on cell aggregation. Among other possibilities, aggregation may increase the concentration of secreted growth factors or allow presentation of surface-bound ligands to receptors on adjacent cells. Signaling via cyclic AMP (cAMP) second messenger is mediated by PKA. PKA has been implicated in the activation function of a number of transcription factors, including the homeoprotein PIT1. Given the known role of CBP in mediating the effects of PKA on transcriptional activation (2, 24), we tested the ability of PKA to convert HOX-PBX complexes from transcriptional repressors to activators.
Overexpression of the catalytic domain of PKA significantly stimulated pML(5xHOX-PBX) in 293 T cells (Fig. 5A). This effect was mediated through HOX-PBX binding sites since PKA had a minimal effect (2.6-fold) on pML lacking the HOX-PBX binding sites. This result suggests a link between the activation of the intracellular cAMP signal transduction pathway and the activity of HOX-PBX complexes.
We examined the impact of PKA signaling on transactivation of the GAL4-responsive reporter pML(5xUAS) by the GAL4-HOXD4N fusion protein. Figure 5B (lane 4) shows that PKA stimulated this reporter 500-fold in a HOXD4N-dependent manner. The PKA stimulation requires CBP since depletion of endogenous CBP by overexpression of E1A inhibited this effect (lanes 4 and 5, white bars). Overexpression of PKA along with GAL4-HOXD4N and CBP-HA resulted in increased amounts of CBP coprecipitates with equivalent amounts of HOXD4N (Fig. 6A, lane 3). These data suggest that the recruitment of CBP by the activation domain of HOXD4 is facilitated in the presence of PKA. This further suggests a mechanism by which DNA-bound HOX-PBX complexes could be switched from repressors to activators through enhanced association with CBP.
DISCUSSION
Two observations suggested to us that HOX-PBX complexes may recruit transcriptional corepressors to target promoters. First, the Hoxb1 ARE is inactive in RA-treated P19 cell monolayers despite the presence of HOXB1 and PBX1 but is activated in response to the HDAC inhibitor TSA (Fig. 1). Second, repression by multimerized HOX-PBX binding sites is likewise alleviated by TSA treatment (Fig. 2). Transcriptional activation through the Hoxb1 ARE or multimerized HOX-PBX binding sites further suggested that HOX-PBX complexes recruit transcriptional coactivators. In support of this suggestion, a repression domain in the PBX1 N terminus binds a corepressor complex containing class I HDACs in association with N-CoR/SMRT and mSIN3B (Fig. 3 and 4). Conversely, the proline-rich activation domain of HOXD4 binds the CBP coactivator. We provided additional evidence that the HOX-PBX complex can be switched from a repressor to an activator of transcription through the action of signaling cascades (Fig. 5, 6, and 7). Specifically, the HOX-PBX complex becomes a CBP-dependent transcriptional activator in response to PKA. Thus, the transcriptional activity of the HOX complex in a specific tissue at a given developmental stage may come under the control of signaling cues such as intracellular cAMP.
FIG. 7.
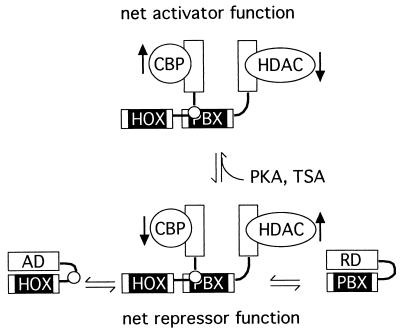
A model for activation and repression by HOX-PBX complexes. The N-terminal activation and repression domains of HOX and PBX proteins are believed to make intramolecular contact with their respective homeodomains (8, 42, 43, 59, 74). Heterodimerization on cooperative sites on DNA, and perhaps additional interactions with members of the MEIS/PREP family, exposes HOX and PBX N termini, thereby freeing them for interaction with coactivators and corepressors such as CBP and HDAC1 and -3. Under some cellular contexts, the net activity of bound corepressors exceeds that of the activators (bottom, “net repressor function”). However, in response to enhanced PKA signaling or P19 cell aggregation, increased coactivator and/or decreased corepressor function shifts the balance toward net activation (top). This could be accomplished by an increase in the amount of coactivator or by increased affinity for the HOX N terminus. In parallel, decreases in the amount or affinity of corepressor for PBX could contribute to the switch. Treatment with TSA would exert the same overall effect by inhibiting bound HDACs. The model is simplified and does not exclude other possible interactions. The black vertical arrows denote increases or decreases in HAT or HDAC activity. AD, HOX activation domain; RD, PBX repression domain C; black box, homeodomain; small white circle, HOX YPWM motif.
Repression of HOX-PBX targets is mediated by PBX-corepressor interactions.
PBX1 has been previously shown to possess three repression domains in its N terminus (45). Our results indicate that PBX1 represses transcription through both TSA-sensitive and -insensitive mechanisms. We found that the first N-terminal repression domain of PBX1 (domain B) represses transcription in a TSA-resistant fashion. By contrast, the second N-terminal repression domain (within region C) associates with class I HDACs. Recently, others have shown that PBX1A binds N-CoR and SMRT through its C terminus (4). The set of PBX1A derivatives employed here does not refute this finding. Rather, the cumulative data suggest that PBX1A contains more than one docking site for corepressor complexes.
The corepressors N-CoR and SMRT are known to repress transcription in an mSIN3A complex (58). In addition, SMRT has been shown to function in an HDAC3 complex (28). The presence of mSIN3B and not mSIN3A in the corepressor complex recruited by PBX1 is a novel indication of an interaction between N-CoR/SMRT and mSIN3B.
Overexpression of wild-type PBX1A inhibits TSA-mediated activation of a reporter bearing multiple HOX-PBX binding sites (Fig. 2, lane 5). By contrast, removal of the first 89 residues of PBX1A, or overexpression of HOX proteins, confers a strong TSA response. Two nonexclusive explanations are possible. First, residues 1 to 89 of PBX1A may harbor a TSA-insensitive repression domain. This could be mediated by direct contact to a repressor, or indirectly though members of the MEIS/PREP family which bind PBX proteins through this N-terminal domain (11). This could explain the enhanced TSA response with Δ1–89 but would not explain the dampened response with wild-type PBX1A. Another explanation is that increased levels of PBX1A promote the formation of PBX-PBX homodimers at the target promoter. Such homodimers have been described in the literature (8, 59) and would be expected to form on the multimerized binding sites in pML(5xHOX-PBX). In theory, the PBX homodimer would compete with HOX-PBX heterodimers for DNA binding, recruiting only corepressors to the target promoter and thereby dampening the response to TSA. Deletion of the first 89 residues from PBX1A severely impairs homodimerization (K. Shanmugam and M. S. Featherstone, unpublished observations) without affecting heterodimerization with at least some HOX partners (77). Thus, Δ1–89 would promote binding by HOX-PBX heterodimers at the expense of PBX homodimers, resulting in more efficient recruitment of coactivators.
Residues 1 to 89 of PBX1 are deleted in the oncoprotein E2A-PBX (32). Thus, the increased transcriptional activation function and the concomitant oncogenicity of E2A-PBX may be due to both the loss of a repression domain, as well as to the recruitment of HATs by the E2A activation domain (16, 33, 52). The HDAC1 binding domain in PBX1 (domain C) is retained in E2A-PBX. Consistent with this, TSA potentiates the activation observed with E2A-PBX (unpublished observations). Thus, treatment with TSA may potentiate B-cell transformation.
Domain C of PBX1 spans a short stretch of nine alanine residues and impinges on the conserved PBC-A and -B domains. The PBC domains are highly conserved across species. In contrast, the alanine stretch is conserved in mammals and flies but is absent in the C. elegans CEH-20 protein. Monotonic alanine regions have been implicated in repressor function (29, 44, 47); however, at this time the highly conserved portions of PBC-A and -B are equally plausible candidates for direct interaction with repressor complexes.
CBP modifies HOXD4 function and transduces PKA stimulation of HOX-PBX promoters.
We have shown that the proline-rich activation domain of HOXD4 physically interacts with the HAT-C/H3 domain of the CBP coactivator. Interestingly, the interaction between HOXD4 and CBP seems to be conserved through evolution, since Deformed, the Drosophila orthologue of Hoxd4, has been shown genetically to interact with Nejire, encoding a transcriptional adapter belonging to the CBP/p300 family (22). A previous study has shown physical interaction between CBP and the N terminus of HOXB7 (14). Using truncated versions of each protein in vitro and in transfections, their sites of interaction were mapped to the HOXB7 N terminus and two regions in CBP, including the C/H3 domain and the extreme C terminus. Together with another study showing interaction between the N terminus of the HOX-like protein PDX and the CBP C/H3 domain (4), these findings suggest a common mechanism used by homeoproteins to activate transcription.
To date, four Hox genes, namely Hoxb1, Hoxa4, Hoxb4, and Hoxd4, have been shown to contain RAREs and AREs in their flanking regions (26, 30, 39, 51, 56, 63, 69, 70, 83, 92). The HOX interaction region in CBP centering on the C/H3 domain is different from the nuclear receptor interaction region (RID) (10, 31). This suggests that one CBP molecule could simultaneously bind both retinoid receptor and HOX family members. This may result in synergistic recruitment of CBP to Hox gene promoters, thereby integrating the activities of retinoid receptors and HOX proteins.
Interactions between HOX and CBP can explain some of the phenotypes resulting from Cbp loss-of-function mutations. In humans, the Rubinstein-Taybi syndrome is caused by point mutations in the Cbp gene and is characterized by craniofacial deformations, broad thumbs, broad big toes, severe mental retardation, and increased tumor incidence (65). In the mouse, targeted disruptions of Cbp and p300 have revealed the importance of these cofactors in embryonic development (90). In Drosophila, mutations in Cbp cause embryonic lethality as well as pattern defects (1). Some of these defects are reminiscent of those caused by mutations in Hox genes (38) and can be partly explained by the finding that CBP modifies HOX transcriptional activities.
Genetic and molecular studies in Drosophila have led to a model whereby the N-terminal activation domain of HOX proteins is masked due to direct or indirect contact with the HOX homeodomain (42, 43). The model further suggests that this inhibition is relieved upon a conformational change provoked by cooperative DNA binding of HOX with PBX. In this model, DNA-bound HOX monomers are repressors, while HOX-EXD (or HOX-PBX) heterodimers are activators. Our data are consistent with aspects of this model. First, TSA is able to activate a promoter driven by HOX-PBX dimer binding sites but not one driven by HOX monomer binding sites. Second, mutations in the HOX YPWM motif that abrogate interaction with PBX also abolish the TSA response, even on HOX-PBX cooperative binding sites. Both of these observations would be expected if PBX is required to unmask the HOX activation domain, thereby permitting interaction with CBP. However, the very fact that the HOX-PBX complex is responsive to TSA suggests a repressor function mediated by interaction with HDACs, consistent with data reported here and elsewhere that PBX functions as a repressor and binds corepressors (4, 45).
In addition, we did not observe transcriptional repression by HOX monomers under our conditions. HOX monomer binding sites do not repress basal transcription [Fig. 2, compare pML to pML(5xHOX)], and HOX mutants that are incapable of interacting with PBX partners do not behave as transcriptional repressors (Fig. 2) (72). Rather, our data suggest that the HOX-PBX complex can act as both a transcriptional repressor and activator, depending on the cellular context (Fig. 7). We argue that this context can be influenced by cell-cell signaling, since aggregation is required to activate the Hoxb1 ARE in RA-treated P19 cells. Monolayers of P19 cells can be induced down the neural pathway by combined treatment with forskolin, an activator of PKA signaling, and a factor secreted by cells resembling primitive streak mesoderm (71). This is consistent with a role for PKA in the activation of the Hoxb1 ARE. We also note that aggregation of P19 cells has been proposed to influence the activity of the MYOD muscle-specific transcription factor through effects on chromatin (3).
Our finding that CBP-HOX activation of downstream targets is significantly enhanced by PKA suggests a mechanism for conversion of HOX-PBX complexes from transcriptional repressors to activators. PKA was previously shown to be important for the transactivation of bovine CYP17 by PBX, as well as the oncoprotein E2A-PBX, via a cAMP response sequence (CRS) (61). The CRS in the promoter of CYP17 is very closely linked to a PBX response sequence that should accommodate cooperative binding by HOX-PBX in vitro. This suggests that the CRS response to PKA could be mediated by a HOX partner via CBP.
CBP contains a defined PKA phosphorylation site at serine 1772 shown to be important for mediating PKA-stimulated activation by the homeoprotein PIT1 (85). Our results likewise suggest that CBP phosphorylation by PKA is the signal transduction step required for HOXD4 to activate transcription in response to increased intracellular cAMP. We demonstrated increased association of the HOXD4 activation domain with CBP upon increased PKA signaling (Fig. 5A). How is this achieved? The levels of CBP are greatly increased in 293 cells expressing the catalytic subunit of PKA (unpublished observations). This increase may be sufficient to account for the greater association between HOXD4 and CBP upon PKA stimulation.
A role for PKA in HOX function in the embryo has not been clearly demonstrated. However, patterning by the hedgehog signaling pathway in flies and mice involves antagonizing the PKA pathway (19, 60). Our results suggest that PKA may also impinge on patterning mediated by the HOX family. Hox genes are known to determine the morphogenetic outcome of cell signaling in fly imaginal discs (64). In C. elegans, genetic studies have shown that a HOX protein determines the developmental consequences of RAS signaling (48). On theoretical grounds, HOX proteins were predicted to interpret cell signaling events in vertebrates as well (17). Our results support this suggestion.
In summary, we have demonstrated that HOX-PBX can function as an activator or a repressor through differential interactions with coregulators. Moreover, we have shown that PKA serves as a signaling switch that converts HOX-PBX from repressor to activator, implying that cell signaling is an important determinant of HOX-PBX function in the patterning of the animal embryo.
ACKNOWLEDGMENTS
We thank C. Largman for a gift of anti-HOXB1 antibody and R. Krumlauf for the p1230 lacZ reporter. We are grateful to A. Lai and P. Branton for their advice and gifts of vectors for F-HDAC1, F-HDAC3, E1A, and E1A ΔN. We thank A. Tremblay and V. Giguère for gifts of vectors for N-CoR, SMRT, ER, and the CBP domains; Y. Zhang for the Mi2 antibodies; and members of the Featherstone lab for helpful discussions.
M.S. is the recipient of a Medical Research Council of Canada Studentship. X.-J.Y. is a scholar of the Medical Research Council of Canada. M.S.F. is a Chercheur-Boursier of the Fonds de la Recherche en Santé du Québec. This work was funded by grants to X.-J.Y. and M.S.F. from the Medical Research Council of Canada.
REFERENCES
- 1.Akimaru H, Chen Y, Dai P, Hou D X, Nonaka M, Smolik S M, Armstrong S, Goodman R H, Ishii S. Drosophila CBP is a co-activator of cubitus interruptus in hedgehog signalling. Nature. 1997;386:735–738. doi: 10.1038/386735a0. [DOI] [PubMed] [Google Scholar]
- 2.Arias J, Alberts A S, Brindle P, Claret F X, Smeal T, Karin M, Feramisco J, Montminy M. Activation of cAMP and mitogen responsive genes relies on a common nuclear factor. Nature. 1994;370:226–229. doi: 10.1038/370226a0. [DOI] [PubMed] [Google Scholar]
- 3.Armour C, Garson K, McBurney M W. Cell-cell interaction modulates myoD-induced skeletal myogenesis of pluripotent P19 cells in vitro. Exp Cell Res. 1999;251:79–91. doi: 10.1006/excr.1999.4567. [DOI] [PubMed] [Google Scholar]
- 4.Asahara H, Dutta S, Kao H Y, Evans R, Montminy M. Pbx-Hox heterodimers recruit coactivator-corepressor complexes in an isoform-specific manner. Mol Cell Biol. 1999;19:8219–8225. doi: 10.1128/mcb.19.12.8219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barrow J R, Stadler H S, Capecchi M R. Roles of Hoxa1 and Hoxa2 in patterning the early hindbrain of the mouse. Development. 2000;127:933–944. doi: 10.1242/dev.127.5.933. [DOI] [PubMed] [Google Scholar]
- 6.Berthelsen J, Zappavigna V, Ferretti E, Mavilio F, Blasi F. The novel homeoprotein Prep1 modulates Pbx-Hox protein cooperativity. EMBO J. 1998;17:1434–1445. doi: 10.1093/emboj/17.5.1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berthelsen J, Zappavigna V, Mavilio F, Blasi F. Prep1, a novel functional partner of Pbx proteins. EMBO J. 1998;17:1423–1433. doi: 10.1093/emboj/17.5.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calvo K R, Knoepfler P, McGrath S, Kamps M P. An inhibitory switch depressed by pbx, hox, and Meis/Prep1 partners regulates DNA-binding by pbx1 and E2a-pbx1 and is dispensable for myeloid immortalization by E2a-pbx1. Oncogene. 1999;18:8033–8043. doi: 10.1038/sj.onc.1203377. [DOI] [PubMed] [Google Scholar]
- 9.Capovilla M, Botas J. Functional dominance among Hox genes: repression dominates activation in the regulation of dpp. Development. 1998;125:4949–4957. doi: 10.1242/dev.125.24.4949. [DOI] [PubMed] [Google Scholar]
- 10.Chakravarti D, LaMorte V J, Nelson M C, Nakajima T, Schulman I G, Juguilon H, Montminy M, Evans R M. Role of CBP/p300 in nuclear receptor signalling. Nature. 1996;383:99–103. doi: 10.1038/383099a0. [DOI] [PubMed] [Google Scholar]
- 11.Chang C P, Jacobs Y, Nakamura T, Jenkins N A, Copeland N G, Cleary M L. Meis proteins are major in vivo DNA binding partners for wild-type but not chimeric Pbx proteins. Mol Cell Biol. 1997;17:5679–5687. doi: 10.1128/mcb.17.10.5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang C P, Shen W F, Rozenfeld S, Lawrence H J, Largman C, Cleary M L. Pbx proteins display hexapeptide-dependent cooperative DNA binding with a subset of Hox proteins. Genes Dev. 1995;9:663–674. doi: 10.1101/gad.9.6.663. [DOI] [PubMed] [Google Scholar]
- 13.Chariot A, Castronovo V, Le P, Gillet C, Sobel M E, Gielen J. Cloning and expression of a new HOXC6 transcript encoding a repressing protein. Biochem J. 1996;319:91–97. doi: 10.1042/bj3190091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chariot A, van Lint C, Chapelier M, Gielen J, Merville M P, Bours V. CBP and histone deacetylase inhibition enhance the transactivation potential of the HOXB7 homeodomain-containing protein. Oncogene. 1999;18:4007–4014. doi: 10.1038/sj.onc.1202776. [DOI] [PubMed] [Google Scholar]
- 15.Chen H, Rossier C, Nakamura Y, Lynn A, Chakravarti A, Antonarakis S E. Cloning of a novel homeobox-containing gene, PKNOX1, and mapping to human chromosome 21q22.3. Genomics. 1997;41:193–200. doi: 10.1006/geno.1997.4632. [DOI] [PubMed] [Google Scholar]
- 16.Cleary M L. Oncogenic conversion of transcription factors by chromosomal translocations. Cell. 1991;66:619–622. doi: 10.1016/0092-8674(91)90105-8. [DOI] [PubMed] [Google Scholar]
- 17.Davidson E H. Spatial mechanisms of gene regulation in metazoan embryos. Development. 1991;113:1–26. doi: 10.1242/dev.113.1.1. [DOI] [PubMed] [Google Scholar]
- 18.Di Rocco G, Mavilio F, Zappavigna V. Functional dissection of a transcriptionally active, target-specific Hox-Pbx complex. EMBO J. 1997;16:3644–3654. doi: 10.1093/emboj/16.12.3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Epstein D J, Marti E, Scott M P, McMahon A P. Antagonizing cAMP-dependent protein kinase A in the dorsal CNS activates a conserved sonic hedgehog signaling pathway. Development. 1996;122:2885–2894. doi: 10.1242/dev.122.9.2885. [DOI] [PubMed] [Google Scholar]
- 20.Favier B, Dollé P. Development functions of mammalian Hox genes. Mol Hum Reprod. 1997;3:115–131. doi: 10.1093/molehr/3.2.115. [DOI] [PubMed] [Google Scholar]
- 21.Ferretti E, Marshall H, Popperl H, Maconochie M, Krumlauf R, Blasi F. Segmental expression of Hoxb2 in r4 requires two separate sites that integrate cooperative interactions between Prep1, Pbx and Hox proteins. Development. 2000;127:155–166. doi: 10.1242/dev.127.1.155. [DOI] [PubMed] [Google Scholar]
- 22.Florence B, McGinnis W. A genetic screen of the Drosophila X chromosome for mutations that modify Deformed function. Genetics. 1998;150:1497–1511. doi: 10.1093/genetics/150.4.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gehring W J, Qian Y Q, Billeter M, Furukubo-Tokunaga K, Schier A F, Resendez-Perez D, Würthrich K. Homeodomain-DNA recognition. Cell. 1994;78:211–223. doi: 10.1016/0092-8674(94)90292-5. [DOI] [PubMed] [Google Scholar]
- 24.Goldman P S, Tran V K, Goodman R H. The multifunctional role of the co-activator CBP in transcriptional regulation. Rec Prog Horm Res. 1997;52:103–119. [PubMed] [Google Scholar]
- 25.González-Reyes A, Macías A, Morata G. Autocatalysis and phenotypic expression of Drosophila homeotic gene Deformed: its dependence on polarity and homeotic gene function. Development. 1992;116:1059–1068. doi: 10.1242/dev.116.4.1059. [DOI] [PubMed] [Google Scholar]
- 26.Gould A, Morrison A, Sproat G, White R A H, Krumlauf R. Positive cross-regulation and enhancer sharing—two mechanisms for specifying overlapping Hox expression patterns. Genes Dev. 1997;11:900–913. doi: 10.1101/gad.11.7.900. [DOI] [PubMed] [Google Scholar]
- 27.Graba Y, Aragnol D, Pradel J. Drosophila Hox complex downstream targets and the function of homeotic genes. Bioessays. 1997;19:379–388. doi: 10.1002/bies.950190505. [DOI] [PubMed] [Google Scholar]
- 28.Guenther M G, Lane W S, Fischle W, Verdin E, Lazar M A, Shiekhattar R. A core SMRT corepressor complex containing HDAC3 and TBL1, a WD40-repeat protein linked to deafness. Genes Dev. 2000;14:1048–1057. [PMC free article] [PubMed] [Google Scholar]
- 29.Han K, Manley J L. Functional domains of the Drosophila Engrailed protein. EMBO J. 1993;12:2723–2733. doi: 10.1002/j.1460-2075.1993.tb05934.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang D, Chen S W, Langston A W, Gudas L J. A conserved retinoic acid responsive element in the murine Hoxb-1 gene is required for expression in the developing gut. Development. 1998;125:3235–3246. doi: 10.1242/dev.125.16.3235. [DOI] [PubMed] [Google Scholar]
- 31.Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin S C, Heyman R A, Rose D W, Glass C K, Rosenfeld M G. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- 32.Kamps M P, Look A T, Baltimore D. The human t(1;19) translocation in pre-B ALL produces multiple nuclear E2A-Pbx1 fusion proteins with differing transforming potentials. Genes Dev. 1991;5:358–368. doi: 10.1101/gad.5.3.358. [DOI] [PubMed] [Google Scholar]
- 33.Kamps M P, Murre C C, Sun X-H, Baltimore D. A new homeobox gene contributes the DNA binding domain of the t(1;19) translocation protein in pre-B ALL. Cell. 1990;60:547–555. doi: 10.1016/0092-8674(90)90658-2. [DOI] [PubMed] [Google Scholar]
- 34.Knoepfler P S, Calvo K R, Chen H, Antonarakis S E, Kamps M P. Meis1 and pKnox1 bind DNA cooperatively with Pbx1 utilizing an interaction surface disrupted in oncoprotein E2a-Pbx1. Proc Natl Acad Sci USA. 1997;94:14553–14558. doi: 10.1073/pnas.94.26.14553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Knoepfler P S, Eisenman R N. Sin meets NuRD and other tails of repression. Cell. 1999;99:447–450. doi: 10.1016/s0092-8674(00)81531-7. [DOI] [PubMed] [Google Scholar]
- 36.Knoepfler P S, Kamps M P. The Pbx family of proteins is strongly upregulated by a post-transcriptional mechanism during retinoic acid-induced differentiation of P19 embryonal carcinoma cells. Mech Dev. 1997;63:5–14. doi: 10.1016/s0925-4773(97)00669-2. [DOI] [PubMed] [Google Scholar]
- 37.Kouzarides T. Histone acetylases and deacetylases in cell proliferation. Curr Opin Genet Dev. 1999;9:40–48. doi: 10.1016/s0959-437x(99)80006-9. [DOI] [PubMed] [Google Scholar]
- 38.Krumlauf R. Hox genes in vertebrate development. Cell. 1994;78:191–201. doi: 10.1016/0092-8674(94)90290-9. [DOI] [PubMed] [Google Scholar]
- 39.Langston A W, Gudas L J. Identification of a retinoic acid responsive enhancer 3′ of the murine homeobox gene Hox-1.6. Mech Dev. 1992;38:217–227. doi: 10.1016/0925-4773(92)90055-o. [DOI] [PubMed] [Google Scholar]
- 40.LaRosa G J, Gudas L J. Early retinoic acid-induced F9 teratocarcinoma stem cell gene ERA-1: alternate splicing creates transcripts for a homeobox-containing protein and one lacking the homeobox. Mol Cell Biol. 1988;8:3906–3917. doi: 10.1128/mcb.8.9.3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lavinsky R M, Jepsen K, Heinzel T, Torchia J, Mullen T M, Schiff R, Del-Rio A L, Ricote M, Ngo S, Gemsch J, Hilsenbeck S G, Osborne C K, Glass C K, Rosenfeld M G, Rose D W. Diverse signaling pathways modulate nuclear receptor recruitment of N-CoR and SMRT complexes. Proc Natl Acad Sci USA. 1998;95:2920–2925. doi: 10.1073/pnas.95.6.2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li X, McGinnis W. Activity regulation of Hox proteins, a mechanism for altering functional specificity in development and evolution. Proc Natl Acad Sci USA. 1999;96:6802–6807. doi: 10.1073/pnas.96.12.6802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li X, Murre C, McGinnis W. Activity regulation of a Hox protein and a role for the homeodomain in inhibiting transcriptional activation. EMBO J. 1999;18:198–211. doi: 10.1093/emboj/18.1.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Licht J D, Hanna-Rose W, Reddy J C, English M A, Ro M, Grossel M, Shaknovich R, Hansen U. Mapping and mutagenesis of the amino-terminal transcriptional repression domain of the Drosophila Kruppel protein. Mol Cell Biol. 1994;14:4057–4066. doi: 10.1128/mcb.14.6.4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu Q, Kamps M P. Selective repression of transcriptional activators by Pbx1 does not require the homeodomain. Proc Natl Acad Sci USA. 1996;93:470–474. doi: 10.1073/pnas.93.1.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lufkin T, Dierich A, LeMeur M, Mark M, Chambon P. Disruption of the Hox-1.6 homeobox gene results in defects in a region corresponding to its rostral domain of expression. Cell. 1991;66:1105–1119. doi: 10.1016/0092-8674(91)90034-v. [DOI] [PubMed] [Google Scholar]
- 47.Mailly F, Berube G, Harada R, Mao P L, Phillips S, Nepveu A. The human cut homeodomain protein can repress gene expression by two distinct mechanisms: active repression and competition for binding site occupancy. Mol Cell Biol. 1996;16:5346–5357. doi: 10.1128/mcb.16.10.5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maloof J N, Kenyon C. The Hox gene lin-39 is required during C. elegans vulval induction to select the outcome of Ras signaling. Development. 1998;125:181–190. doi: 10.1242/dev.125.2.181. [DOI] [PubMed] [Google Scholar]
- 49.Mann R S, Affolter M. Hox proteins meet more partners. Curr Opin Genet Dev. 1998;8:423–429. doi: 10.1016/s0959-437x(98)80113-5. [DOI] [PubMed] [Google Scholar]
- 50.Mann R S, Chan S-K. Extra specificity from extradenticle: the partnership between HOX and PBX/EXD homeodomain proteins. Trends Genet. 1996;12:258–262. doi: 10.1016/0168-9525(96)10026-3. [DOI] [PubMed] [Google Scholar]
- 51.Marshall H, Studer M, Pöpperl H, Aparicio S, Kuriowa A, Brenner S, Krumlauf R. A conserved retinoic acid response element required for early expression of the homeobox gene Hoxb-1. Nature. 1994;370:567–571. doi: 10.1038/370567a0. [DOI] [PubMed] [Google Scholar]
- 52.Massari M, Grant P, Pray-Grant M G, Berger S L, Workman J L, Murre C. A conserved motif present in a class of helix-loop-helix proteins activates transcription by direct recruitment of the SAGA complex. Mol Cell. 1999;4:63–73. doi: 10.1016/s1097-2765(00)80188-4. [DOI] [PubMed] [Google Scholar]
- 53.McBurney M W, Yang X, Jardine K, Cormier M. A role for RNA processing in regulating expression from transfected genes. Somat Cell Mol Genet. 1998;24:203–215. doi: 10.1023/b:scam.0000007123.70630.40. [DOI] [PubMed] [Google Scholar]
- 54.McGinnis W, Krumlauf R. Homeobox genes and axial patterning. Cell. 1992;68:283–302. doi: 10.1016/0092-8674(92)90471-n. [DOI] [PubMed] [Google Scholar]
- 55.Monica K, Galili N, Nourse J, Saltman D, Cleary M L. PBX2 and PBX3, new homeobox genes with extensive homology to the human proto-oncogene PBX1. Mol Cell Biol. 1993;11:6149–6157. doi: 10.1128/mcb.11.12.6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Morrison A, Moroni M C, Ariza-McNaughton L, Krumlauf R, Mavilio F. In vitro and transgenic analysis of a human hoxd4 retinoid-responsive enhancer. Development. 1996;122:1895–1907. doi: 10.1242/dev.122.6.1895. [DOI] [PubMed] [Google Scholar]
- 57.Moskow J J, Bullrich F, Huebner K, Daar I O, Buchberg A M. Meis1, a PBX1-related homeobox gene involved in myeloid leukemia in BXH-2 mice. Mol Cell Biol. 1995;15:5434–5443. doi: 10.1128/mcb.15.10.5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nagy L, Kao H Y, Chakravarti D, Lin R J, Hassig C A, Ayer D E, Schreiber S L, Evans R M. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell. 1997;89:373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- 59.Neuteboom S T C, Murre C. Pbx raises the DNA binding specificity but not the selectivity of Antennapedia Hox proteins. Mol Cell Biol. 1997;17:4696–4706. doi: 10.1128/mcb.17.8.4696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Noveen A, Jiang T X, Chuong C M. cAMP, an activator of protein kinase A, suppresses the expression of sonic hedgehog. Biochem Biophys Res Commun. 1996;219:180–185. doi: 10.1006/bbrc.1996.0202. [DOI] [PubMed] [Google Scholar]
- 61.Ogo A, Waterman M R, Kamps M P, Kagawa N. Protein kinase A-dependent transactivation by the e2a-pbx1 fusion protein. J Biol Chem. 1995;270:25340–25343. doi: 10.1074/jbc.270.43.25340. [DOI] [PubMed] [Google Scholar]
- 62.Oulad-Abdelghani M, Chazaud C, Bouillet P, Sapin V, Chambon P, Dollé P. Meis2, a novel mouse Pbx-related homeobox gene induced by retinoic acid during differentiation of P19 embryonal carcinoma cells. Dev Dynam. 1997;210:173–183. doi: 10.1002/(SICI)1097-0177(199710)210:2<173::AID-AJA9>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 63.Packer A I, Crotty D A, Elwell V A, Wolgemuth D J. Expression of the murine Hoxa4 gene requires both autoregulation and a conserved retinoic acid response element. Development. 1998;125:1991–1998. doi: 10.1242/dev.125.11.1991. [DOI] [PubMed] [Google Scholar]
- 64.Percival-Smith A, Weber J, Gilfoyle E, Wilson P. Genetic characterization of the role of the two HOX proteins, Proboscipedia and Sex Combs Reduced, in determination of adult antennal, tarsal, maxillary palp and proboscis identities in Drosophila melanogaster. Development. 1997;124:5049–5062. doi: 10.1242/dev.124.24.5049. [DOI] [PubMed] [Google Scholar]
- 65.Petrij F, Giles R H, Dauwerse H G, Saris J J, Hennekam R C, Masuno M, Tommerup N, van Ommen G J, Goodman R H, Peters D J, et al. Rubinstein-Taybi syndrome caused by mutations in the transcriptional co-activator CPB. Nature. 1995;376:348–351. doi: 10.1038/376348a0. [DOI] [PubMed] [Google Scholar]
- 66.Phelan M, Featherstone M. Distinct HOX N-terminal arm residues are responsible for specificity of DNA recognition by HOX monomers and HOX-PBX heterodimers. J Biol Chem. 1997;272:8635–8643. doi: 10.1074/jbc.272.13.8635. [DOI] [PubMed] [Google Scholar]
- 67.Phelan M L, Rambaldi I, Featherstone M S. Cooperative interactions between HOX and PBX proteins mediated by a conserved peptide motif. Mol Cell Biol. 1995;15:3989–3997. doi: 10.1128/mcb.15.8.3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pöpperl H, Bienz M, Studer M, Chan S-K, Aparacio S, Brenner S, Mann R, Krumlauf R. Segmental expression of Hoxb-1 is controlled by a highly conserved autoregulatory loop dependent on exd/pbx. Cell. 1995;81:1031–1042. doi: 10.1016/s0092-8674(05)80008-x. [DOI] [PubMed] [Google Scholar]
- 69.Pöpperl H, Featherstone M S. An autoregulatory element of the murine Hox-4.2 gene. EMBO J. 1992;11:3673–3680. doi: 10.1002/j.1460-2075.1992.tb05452.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pöpperl H, Featherstone M S. Identification of a retinoic acid response element upstream of the murine Hox-4.2 gene. Mol Cell Biol. 1993;13:257–265. doi: 10.1128/mcb.13.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pruitt S C. Discrete endogenous signals mediate neural competence and induction in P19 embryonal carcinoma stem cells. Development. 1994;120:3301–3312. doi: 10.1242/dev.120.11.3301. [DOI] [PubMed] [Google Scholar]
- 72.Rambaldi I, Nagy Kovàcs E, Featherstone M S. A proline-rich transcriptional activation domain in murine HOXD-4 (HOX-4.2) Nucleic Acids Res. 1994;22:376–382. doi: 10.1093/nar/22.3.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rudnicki M A, McBurney M W. Cell culture methods and induction of differentiation of embryonal carcinoma cell lines. Oxford, England: IRL Press; 1987. [Google Scholar]
- 74.Saleh, M., H. Huang, N. C. Green, and M. S. Featherstone. A conformational change in PBX1A is necessary for its nuclear localization. Exp. Cell Res., in press. [DOI] [PubMed]
- 75.Schnabel C A, Abate-Shen C. Repression by HoxA7 is mediated by the homeodomain and the modulatory action of its N-terminal-arm residues. Mol Cell Biol. 1996;16:2678–2688. doi: 10.1128/mcb.16.6.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shanmugam K, Featherstone M S, Saragovi H U. Residues flanking the HOX YPWM motif contribute to cooperative interactions with PBX. J Biol Chem. 1997;272:19081–19087. doi: 10.1074/jbc.272.30.19081. [DOI] [PubMed] [Google Scholar]
- 77.Shanmugam K, Green N C, Rambaldi I, Saragovi H U, Featherstone M S. PBX and MEIS as non-DNA-binding proteins in trimeric complexes with HOX proteins. Mol Cell Biol. 1999;19:7577–7588. doi: 10.1128/mcb.19.11.7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shen W-F, Rozenfeld S, Lawrence H J, Largman C. The Abd-B-like Hox homeodomain proteins can be subdivided by the ability to form complexes with Pbx1a on a novel DNA target. J Biol Chem. 1997;272:8198–8206. doi: 10.1074/jbc.272.13.8198. [DOI] [PubMed] [Google Scholar]
- 79.Shen W F, Rozenfeld S, Kwong A, Köm Ves L G, Lawrence H J, Largman C. HOXA9 forms triple complexes with PBX2 and MEIS1 in myeloid cells. Mol Cell Biol. 1999;19:3051–3061. doi: 10.1128/mcb.19.4.3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sterner D E, Berger S L. Acetylation of histones and transcription-related factors. Microbiol Mol Biol Rev. 2000;64:435–459. doi: 10.1128/mmbr.64.2.435-459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 82.Studer M, Gavalas A, Marshall H, Ariza-McNaughton L, Rijli F M, Chambon P, Krumlauf R. Genetic interactions between Hoxa1 and Hoxb1 reveal new roles in regulation of early hindbrain patterning. Development. 1998;125:1025–1036. doi: 10.1242/dev.125.6.1025. [DOI] [PubMed] [Google Scholar]
- 83.Studer M, Pöpperl H, Marshall H, Kuroiwa A, Krumlauf R. Role of a conserved retinoic acid response element in rhombomere restriction of Hoxb-1. Science. 1994;265:1728–1732. doi: 10.1126/science.7916164. [DOI] [PubMed] [Google Scholar]
- 84.Wang A H, Bertos N R, Vezmar M, Pelletier N, Crosato M, Heng H H, Th'ng T, Han J, Yang X J. HDAC4, a human histone deacetylase related to yeast HDA1, is a transcriptional corepressor. Mol Cell Biol. 1999;19:7816–7827. doi: 10.1128/mcb.19.11.7816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xu L, Lavinsky R M, Dasen J S, Flynn S E, McInerney E M, Mullen T M, Heinzel T, Szeto D, Korzus E, Kurokawa R, Aggarwal A K, Rose D W, Glass C K, Rosenfeld M G. Signal-specific co-activator domain requirements for Pit-1 activation. Nature. 1998;395:301–306. doi: 10.1038/26270. [DOI] [PubMed] [Google Scholar]
- 86.Xue Y, Wong J, Moreno G T, Young M K, Cote J, Wang W. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol Cell. 1998;2:851–861. doi: 10.1016/s1097-2765(00)80299-3. [DOI] [PubMed] [Google Scholar]
- 87.Yang W M, Inouye C, Zeng Y, Bearss D, Seto E. Transcriptional repression by YY1 is mediated by interaction with a mammalian homolog of the yeast global regulator RPD3. Proc Natl Acad Sci USA. 1996;93:12845–12850. doi: 10.1073/pnas.93.23.12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yang W M, Yao Y L, Sun J M, Davie J R, Seto E. Isolation and characterization of cDNAs corresponding to an additional member of the human histone deacetylase gene family. J Biol Chem. 1997;272:28001–28007. doi: 10.1074/jbc.272.44.28001. [DOI] [PubMed] [Google Scholar]
- 89.Yang X J, Ogryzko V V, Nishikawa J, Howard B H, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 90.Yao T P, Oh S P, Fuchs M, Zhou N D, Ch'ng L E, Newsome D, Bronson R T, Li E, Livingston D M, Eckner R. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell. 1998;93:361–372. doi: 10.1016/s0092-8674(00)81165-4. [DOI] [PubMed] [Google Scholar]
- 91.Yee S P, Rowe D T, Tremblay M L, McDermott M, Branton P E. Identification of human adenovirus early region 1 products by using antisera against synthetic peptides corresponding to the predicted carboxy termini. J Virol. 1983;46:1003–1013. doi: 10.1128/jvi.46.3.1003-1013.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhang F, Nagy Kovacs E, Featherstone M S. Murine Hoxd4 expression in the CNS requires multiple elements including a retinoic acid response element. Mech Dev. 2000;96:79–89. doi: 10.1016/s0925-4773(00)00377-4. [DOI] [PubMed] [Google Scholar]
- 93.Zhang M, Kim H-J, Marshall H, Gendron-Maguire M, Lucas A D, Baron A, Gudas L J, Gridley T, Krumlauf R, Grippo J F. Ectopic Hoxa-1 induces rhombomere transformation in mouse hindbrain. Development. 1994;120:2431–2442. doi: 10.1242/dev.120.9.2431. [DOI] [PubMed] [Google Scholar]
- 94.Zhang Y, LeRoy G, Seelig H P, Lane W S, Reinberg D. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell. 1998;95:279–289. doi: 10.1016/s0092-8674(00)81758-4. [DOI] [PubMed] [Google Scholar]