Abstract
Protein kinase A-dependent derepression of the human prodynorphin gene is regulated by the differential occupancy of the Dyn downstream regulatory element (DRE) site. Here, we show that a direct protein-protein interaction between DREAM and the CREM repressor isoform, αCREM, prevents binding of DREAM to the DRE and suggests a mechanism for cyclic AMP-dependent derepression of the prodynorphin gene in human neuroblastoma cells. Phosphorylation in the kinase-inducible domain of αCREM is not required for the interaction, but phospho-αCREM shows higher affinity for DREAM. The interaction with αCREM is independent of the Ca2+-binding properties of DREAM and is governed by leucine-charged residue-rich domains located in both αCREM and DREAM. Thus, our results propose a new mechanism for DREAM-mediated derepression that can operate independently of changes in nuclear Ca2+.
Transcriptional derepression is an important mechanism for the accurate control of gene expression. Transcriptional repressors can bind directly to DNA or act indirectly by interacting with other DNA-associated proteins (23, 32). DREAM, a calcium-binding protein, represses basal expression of target genes through specific interaction with downstream regulatory elements (DREs) in the DNA (5, 6). Release of binding of DREAM from the DRE results in derepression, a process that is regulated by Ca2+ and protein kinase A (PKA) activation (5, 6). Other central players in the nuclear response to cyclic AMP (cAMP) and Ca2+ are activator and repressor basic region-leucine zipper (LZ) transcription factors that bind to cAMP-responsive promoter elements (CREs) (10, 15, 25). They include proteins encoded by the CREB and CREM genes whose function is tightly regulated via phosphorylation by several kinases, including PKA and Ca2+-calmodulin-dependent kinases (8, 12, 13). As such, they represent the convergence point for various signaling cascades.
The transcriptional repressor DREAM contains four EF hands, of which three (II, III, and IV) are responsible for the binding of calcium ions. In the absence of stimulated levels of nuclear calcium, DREAM binds with high affinity to the DRE sequence as a tetramer. Upon stimulation and increase in intracellular calcium, DREAM detaches from DNA without disruption of the tetrameric structure (6). The regulation by intracellular Ca2+ of DREAM binding to DRE sites is a general mechanism that depends primarily on the EF-hand domains of DREAM. Mutation of two key amino acids within any of the functional EF hands results in mutated DREAM forms that stay bound to DNA also after calcium stimulation. Since DREAM binds to DRE sites as a tetramer, DREAM mutants insensitive to Ca2+ behave as dominant negative mutants in a background of wild-type DREAM (unpublished observation). Similarly, PKA activation also results in loss of DREAM binding to the DRE and derepression of the target gene prodynorphin in human neuroblastoma cells (5). The molecular mechanism or the domains in DREAM that mediate this derepression by cAMP are unknown, and consensus domains for PKA phosphorylation have not been identified in DREAM (6). Moreover, derepression of DRE-dependent transcription is cell specific, further supporting the idea that the mechanism in this case is not intrinsic to the DREAM molecule but involves a more elaborated process perhaps implicating other proteins or cell-specific mechanisms.
Recently, three proteins related to DREAM, named KchIP1 to -3, have been found in a two-hybrid screening to interact with the amino-terminal domain of Kv4.2 potassium channels (2). One of them, KchIP-3, is identical to DREAM, and the interaction with the Kv4 potassium channels modulates A-type potassium currents in a Ca2+-dependent manner (2). The interaction with the potassium channel occurs whether calcium is present or not. However, the change in KchIP-3/DREAM conformation that follows binding to calcium profoundly affects channel properties (2). Interestingly, A-type potassium currents are also modulated by PKA, although the mechanism remains elusive (18). Furthermore, also using a yeast two-hybrid screening, another protein identical to DREAM, named calsenilin, was found to interact with the carboxy-terminal region of presenilin-2 (4). In this work, mutants of calsenilin were not described, and a possible regulation of the interaction by calcium or PKA activation remains to be determined. Taken together, these results indicate that DREAM, KchIP-3, or calsenilin might have pleiotropic functions through the interaction with specific DNA sequences and/or with proteins in different cell compartments (2, 4, 6).
In this study, we aimed to determine whether CREM or CREB proteins functionally interact with DREAM and are involved in the derepression at DRE sites observed after forskolin treatment in NB69 and SK-N-MC human neuroblastoma cells (5). Results from transient transfection experiments as well as in vitro interactions using recombinant proteins demonstrate that a calcium-independent interaction between αCREM and DREAM mediates unbinding of DREAM from DRE sites and derepression of the prodynorphin gene after forskolin treatment in human neuroblastoma cells.
MATERIALS AND METHODS
Cell culture, transfection, and CAT analysis.
Cells were grown in Dulbecco modified Eagle medium (DMEM) (HEK293) or DMEM-F12 (NB69 and SK-NMC) supplemented with 10% fetal calf serum, 2 mM Glutamax-I, and 50 μg of gentamicin/ml. Transfections by calcium phosphate precipitation and chloramphenicol acetyltransferase (CAT) activity assays were performed as described elsewhere (5). A total amount of 4.5 μg of plasmid DNA was used per 35-mm-diameter dish. The reporter plasmid pHD3CAT and the expression vectors pDREAM and pEFmutDREAM have been previously described (5, 6). Expression vectors for CREB and CREM proteins have been described elsewhere (9, 10, 15). In cotransfection experiments with two expression vectors, the DNA ratio was always 1:1. Mutations in DREAM and CREM were introduced by site-directed mutagenesis using the Quick-Change method (Stratagene).
EMSA.
Electrophoretic mobility shift assays (EMSA) using recombinant proteins were performed as described elsewhere (5, 6) with 50 ng of each interacting protein in a ratio of 1:1, unless otherwise indicated. Recombinant CREM and CREB proteins were prepared as described elsewhere (17). Recombinant DREAM was purified by phenyl-Sepharose (Pharmacia) chromatography as described previously (34). Amounts of recombinant proteins were measured by the Bradford method, and batch-to-batch variability was analyzed by silver staining or by immunoblotting after polyacrylamide gel electrophoresis. In vitro phosphorylation by PKA (26) was performed with 50 ng of the purified catalytic subunit of PKA (Sigma) and incubation for 1 h at 30°C. Where indicated, calcium was added to the incubation reaction at a final concentration of 10 μM.
Pull-down experiments.
Recombinant αCREM or αCREMS68A proteins (0.5 μg) were incubated with an excess of His-tagged DREAM or DREAM protein for 1 h at 37°C in a final volume of 100 μl of binding buffer (10 mM HEPES [pH 7.9], 100 mM NaCl, 100 mM KCl, 8 mM MgCl2, 20% glycerol, 1 mM β-mercaptoethanol, 0.5 mM phenylmethylsulfonyl fluoride). When indicated, 200 ng of PKA and/or 50 μM CaCl2 were added to the reaction. After 1 h, 10 μl of Ni-nitrilotriacetic acid agarose (Quiagen) was added, and the mixture was incubated for an additional 30 min at room temperature and constant shaking. Complexes were then centrifuged, washed three times in 1 ml of binding buffer, and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. For the Western blot analysis, antibodies for CREB (NEN) or CREM (Santa Cruz Biotechnology) were used as recommended by the supplier.
RESULTS
Overexpression of αCREM derepresses DRE-dependent transcription.
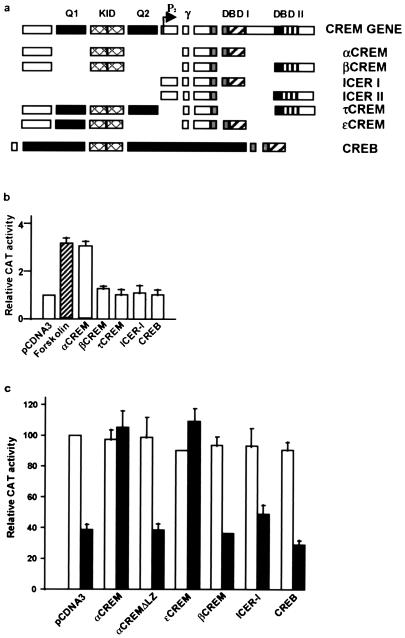
To determine whether CREM or CREB proteins functionally interact with DREAM and are involved in the derepression at DRE sites after forskolin treatment (5), we performed cotransfection experiments in NB69 cells with pHD3CAT, a reporter that contains a DRE site but no CRE site (5), together with expression vectors for CREB or the different CREM and ICER isoforms (Fig. 1a). Interestingly, we found that overexpression of αCREM mimicked the effect of forskolin and induced expression from the pHD3CAT reporter (Fig. 1b). The effect was specific since none of the other CREM isoforms tested or CREB were able to derepress pHD3CAT (Fig. 1b). Similar results were obtained after transient transfections in SK-NMC cells (data not shown). To exclude a direct effect of αCREM binding to the pHD3CAT reporter, we performed similar experiments with HEK293 cells, a cell line that does not express DREAM (6) and does not have detectable levels of CREM proteins (unpublished results). Overexpression of DREAM in HEK293 cells repressed basal expression of the pHD3CAT reporter as previously described (6), while overexpression of CREM isoforms or CREB failed to alter the basal expression of pHD3CAT (Fig. 1c). However, when cotransfected with DREAM, αCREM completely abolished the repression by DREAM (Fig. 1c). ɛCREM, a weak repressor isoform similar to αCREM but containing a glutamine-rich Q1 domain (3), also blocked the effect of DREAM (Fig. 1a and c). Importantly, deletion of DNA-binding/dimerization LZ domain I at the C terminus of αCREM (αCREMΔLZ) or substitution with LZ domain II (βCREM) did not block DREAM-mediated repression (Fig. 1a and c). Furthermore, we did not observe derepression of the pHD3CAT reporter after coexpression of DREAM with an ICER isoform (I, Iγ, II, or IIγ) (Fig. 1c and data not shown). ICER proteins are generated from the alternative P2 promoter within the 3′ end of the CREM gene and therefore lack the N-terminal half of CREM containing the kinase-inducible domain (KID) (9, 10, 21) (Fig. 1a). These data are in agreement with the results for NB69 and SK-NMC cells and point toward a functional interaction between αCREM and DREAM, which displaces DREAM from the DRE sites.
FIG. 1.
Effect of αCREM on DRE-dependent transcription. (a) Scheme showing the modular structure of the CREM and CREB genes and the different isoforms used. DBD, DNA-binding domain. (b) Transactivation by αCREM of the DRE-containing reporter pHD3CAT after transient transfection in NB69 cells. For comparison, the effect of forskolin treatment is shown (hatched bar), as well as the lack of effect of other CREM isoforms, ICER-I, or CREB. (c) Repression by DREAM (black bars) of the pHD3CAT reporter in HEK293 cells is relieved after cotransfection with αCREM or ɛCREM. For comparison, the lack of effect of αCREMΔLZ, βCREM, ICER-I, or CREB is shown. White bars represent the corresponding control transfections in the absence of DREAM.
Recombinant αCREM blocks binding of DREAM to DRE sites.
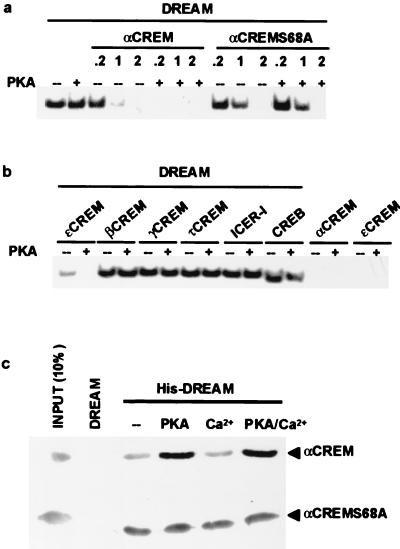
To confirm a specific αCREM-DREAM interaction, recombinant proteins were analyzed by gel mobility shift using the DRE as a probe. The results showed that αCREM does not bind to the DRE probe, but that the formation of αCREM-DREAM heteromers displaced DREAM from the DRE site in a protein ratio-dependent manner (Fig. 2a and c). Ratios of αCREM to DREAM in the order of 0.2:1 or lower did not modify the DREAM-DRE retarded band, but ratios higher than 1:1 eliminated the DRE band (Fig. 2a). Since phosphorylation of Ser68 in the N-terminal part of αCREM is an important determinant of its activity (13, 21), we checked whether this affects the interaction with DREAM. In vitro phosphorylation of αCREM using purified PKA (26) increased its ability to block the DREAM-DRE retarded band (Fig. 2a). The effect of PKA was specific since it was not observed when we used the phosphorylation mutant αCREMS68A (Fig. 2a) and was blocked by coincubation with H89, a selective inhibitor of PKA (data not shown). Consistent with the absence of PKA phosphorylation sites in DREAM (6), incubation of DREAM with PKA did not modify its binding to the DRE (Fig. 2a). Furthermore, in agreement with the transfection results (Fig. 1b and c), ɛCREM also displaced the DREAM-DRE retarded band, while other CREM isoforms or CREB failed to affect the DRE band whether phosphorylated or not (Fig. 2b). Similar results were obtained using nuclear extracts from NB69 cells as a source of endogenous DREAM and nuclear extracts from stably transfected COS cell lines overexpressing the different CREM isoforms (data not shown). Moreover, since Ca2+ is important for the binding of DREAM to the DRE, we checked whether Ca2+ affects the interaction with αCREM. Pull-down experiments using His-tagged recombinant DREAM protein showed that the interaction with αCREM was not modified by the presence of Ca2+ even at concentrations that preclude binding of DREAM to the DRE (Fig. 2c). Interestingly, the potentiating effect of PKA on the DREAM-αCREM interaction was also observed in pull-down experiments with αCREM and confirmed by its absence when we used the phosphorylation mutant αCREMS68A (Fig. 2c). Taken together, these results support a specific protein-protein interaction between αCREM and DREAM resulting in loss of binding to DRE sites. Since Ca2+ does not affect the interaction, unbinding of DREAM from DRE sites by Ca2+ and by αCREM are independent mechanisms that could operate synergistically.
FIG. 2.
In vitro analysis of the DREAM-αCREM interaction. (a) EMSA with a DRE probe showing the interaction between recombinant DREAM and αCREM proteins at different ratios and its modulation by PKA phosphorylation. (b) EMSA with a DRE probe showing the interaction between DREAM and αCREM and the lack of effect of other CREM isoforms, ICER, and CREB. A DREAM/CRE-binding protein ratio of 1:1 was used. For comparison, the interaction between αCREM and DREAM is shown. The lack of binding of αCREM and ɛCREM to the DRE probe is shown. (c) Pull-down experiments showing that the DREAM-αCREM interaction is increased after PKA phosphorylation and not affected by calcium. The lack of effect of PKA on the phosphorylation mutant αCREMS68A is shown.
Two LCD domains in αCREM mediate the interaction with DREAM.
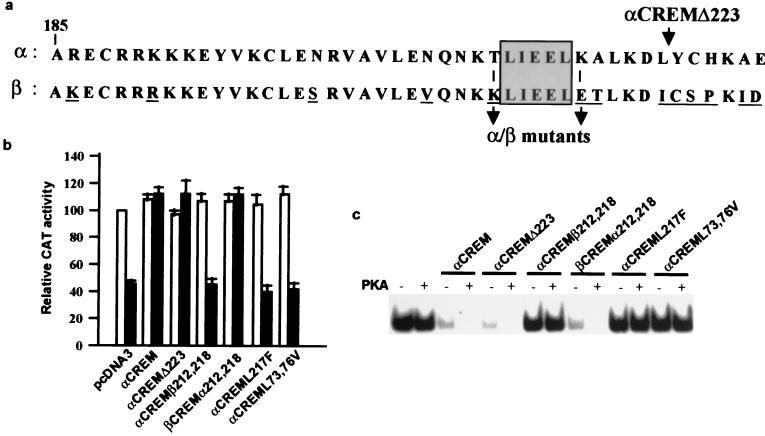
We next wanted to investigate the domains in αCREM responsible for the interaction with DREAM. Based on the results with the different CREM isoforms, we could hypothesize that at least two domains in αCREM are involved, one located within the LZ domain I and a second in the N-terminal half of α/ɛCREM including the KID but not the Q1 domain (Fig. 1a). Direct comparison of LZ domain I (αCREM) and domain II (βCREM) revealed differences in 13 amino acids, 6 of which are at the C-terminal end (Fig. 3a). Premature termination of αCREM by insertion of a TAA stop codon at position 224 resulted in a truncated αCREM protein, αCREMΔ223, that retained its capacity to interact with DREAM in transfection experiments (Fig. 3B) and in band shift assays (Fig. 3c). This result indicates that the difference in these six residues between αCREM and βCREM is not responsible for their difference in interaction with DREAM. Of the other different amino acids, two nonconservative substitutions at positions 212 and 218 were notable since they flank the sequence LIEEL, which could correspond to a leucine-charged residue-rich domain (LCD) (14, 22, 31). Substitution of the flanking amino acids T212 and K218 in αCREM by the corresponding amino acids in βCREM, K, and E, respectively, originated an αCREM mutant (αCREMβ212,218) which failed to block DREAM repression in transfection experiments (Fig. 3b). Moreover, αCREMβ212,218 did not interact with recombinant DREAM in retardation assays (Fig. 3c). Conversely, substitution of the flanking amino acids K212 and E 218 in βCREM by T and K, respectively, created the mutant βCREMα212,218, which was able to interact with DREAM and block DREAM repression and the DRE retarded band (Fig. 3b and c). Furthermore, mutation of the putative LCD in αCREML217F prevented its interaction with DREAM (Fig. 3b and c). These results indicate that the LCD sequence TLIEELK in the LZ of αCREM is necessary for the interaction with DREAM that prevents binding to the DRE. However, the absence of interaction between ICER-I, which contains the LCD motif at the LZ and DREAM, indicates that this motif is necessary but not sufficient. To investigate residues in the N-terminal half of αCREM, absent in ICER-I, that are necessary for the interaction with DREAM, we focused on the region containing the KID domain since the interaction is affected by phosphorylation at serine 68 in αCREM. Again we observed the presence of a putative LCD motif, ILNEL, located at position 72 in αCREM. Mutation of the two L residues at positions 73 and 76 to V resulted in mutant αCREML73,76V, which did not block DREAM-mediated repression and did not show any interaction with DREAM in vitro (Fig. 3b and c). These results identified the sequence ILNEL located within the KID region as part of the second region necessary for the interaction with DREAM. Interestingly, both LCD sequences in αCREM necessary for the interaction with DREAM are conserved in τCREM-I and CREB (10, 15), but neither of these proteins blocked binding of DREAM to the DRE (Fig. 1 and 2 and data not shown). This suggests that the spacing between the two LCD sequences in αCREM is crucial and that the insertion of the glutamine-rich Q2 domain in CREB or τCREM-I prevents the interaction. The results obtained with ɛCREM support this assumption. Furthermore, this opens the possibility that CREB isoforms lacking the Q2 domain (16) could also interact with DREAM and mediate DRE-dependent derepression. Taken together, these results reveal the presence in CRE-binding proteins of a new type of LCD (L/IL/IxxL) that in the case of αCREM is responsible for its two-site interaction with DREAM.
FIG. 3.
Two LCDs in αCREM are necessary for the interaction with DREAM. (a) Alignment of LZ domains I and II from αCREM and βCREM, respectively, showing the 13-amino-acid difference and the positions of the different mutations and the truncated form αCREMΔ223. The putative LCD is boxed. Transient transfections in HEK293 cells (b) and EMSA with recombinant proteins (c) show the lack of interaction of αCREMβ212,218, αCREML217F, and αCREML73,76V with DREAM. The gain of function in mutant βCREMα212,218 is also shown. Values of CAT activity after transfection with wild-type or mutant αCREM, in the absence (open bars) or presence (black bars) of DREAM, are relative to basal acetylation of reporter pHD3CAT in cotransfection with empty reporter vector. Equal amounts of DREAM and αCREM proteins were used in the EMSA.
Two LCDs in DREAM mediate the interaction with αCREM.
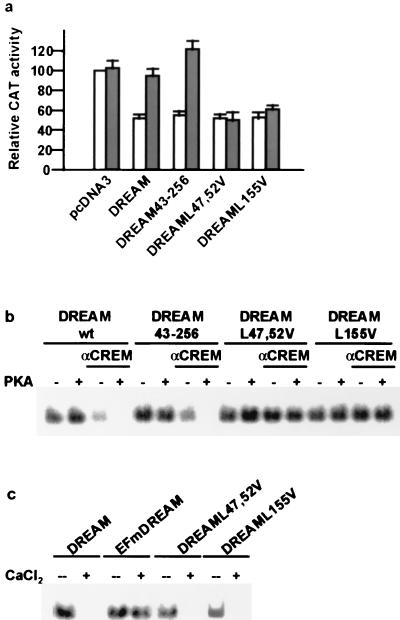
To investigate the domains in DREAM responsible for the interaction with αCREM, we searched for LCDs within DREAM since it has been shown that the mutual interaction between CREB-binding protein (CBP) and p/CIP depends on LCDs located in both proteins (31). Interestingly, we identified three putative LCDs within the DREAM sequence, at positions 15, 47, and 155. Absence of LCD-15 in a truncated DREAM construct that starts at M43 (DREAM43-256) did not modify DREAM repressor activity or its interaction with αCREM in transfection experiments (Fig. 4a) or in vitro (Fig. 4b). A double mutation within LCD-47, CLVKWIL, a sequence that resembles more an LCD of the CoRNR box type (19), yielded a mutant, DREAML47,52V, that still repressed transcription from DRE reporters and did not interact with αCREM in transfection experiments (Fig. 4a) or in vitro (Fig. 4b). Finally, a single mutation within LCD-155, LSILL, created the mutant DREAML155V, whose binding to DRE sites was not affected by the presence of αCREM in transfection experiments (Fig. 4a) or in vitro (Fig. 4b). Importantly, DREAM mutants unable to interact with αCREM because of the mutation at the LCDs still bound to DRE in a calcium-dependent manner (Fig. 4c) as previously reported for wild-type DREAM (6). These results identified two LCD sequences in DREAM that are necessary for the interaction with αCREM, reinforcing the specificity of the interaction. However, these results do not imply a direct interaction between LCDs in DREAM and αCREM, although a mutual interaction between LCDs cannot be excluded.
FIG. 4.
Two LCDs in DREAM are necessary for the interaction with αCREM. Transient transfection in HEK293 cells (a) and EMSA with recombinant proteins (b) show that DREAM LCD mutants DREAML47,52V and DREAML155V are no longer able to interact with αCREM. On the other hand, truncated DREAM43-256 missing a putative LCD at position 15 still interacts with αCREM. Values of CAT activity after transfection with wild-type or mutant DREAM, with (gray bars) or without (open bars) αCREM, are relative to basal acetylation of reporter pHD3CAT in cotransfection with empty reporter vector. Equal amounts of αCREM and DREAM proteins were used for the EMSA. (c) EMSA with a DRE probe showing the unaffected sensitivity to Ca2+ of the DREAM LCD mutants. For comparison, the lack of Ca2+ sensitivity of the dominant negative mutant EFmDREAM is shown.
Derepression of the prodynorphin gene is mediated by the αCREM-DREAM interaction.
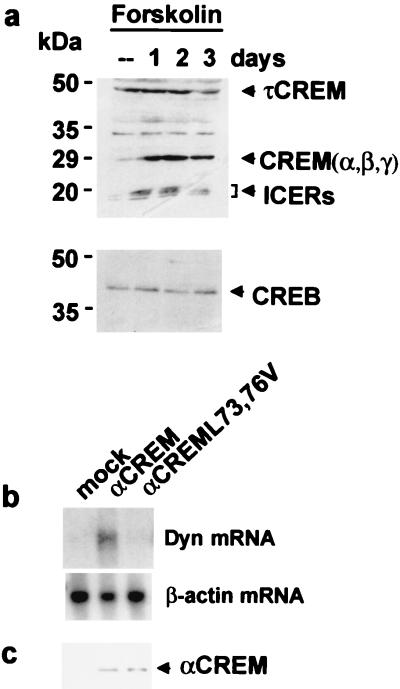
The results described above support a mechanism of derepression at DRE sites based on a cell-specific increase in αCREM and/or its phosphorylation after PKA activation and the interaction with DREAM. To investigate whether this mechanism of derepression indeed mediates the in vivo derepression of the prodynorphin gene in human neuroblastoma cells, we used Western blot analysis to monitor changes in CREM proteins after forskolin administration. Previously, we had reported that treatment of SK-NMC cells with forskolin resulted in a robust and sustained loss of the DREAM-DRE interaction and a parallel accumulation of prodynorphin mRNA (5). The effect was maximal 2 to 3 days after treatment (5). Western blot analysis with an antibody able to recognize all CREM and ICER isoforms showed an increase of CREM repressor isoforms in the 30-kDa range (including αCREM) as well as in ICER isoforms (Fig. 5a). Interestingly, the accumulation of CREM repressor proteins was also maximal 2 to 3 days after treatment. Moreover, the effect of forskolin on the accumulation of CREM repressors was specific, since levels of τCREM or CREB proteins were not modified at any time after forskolin treatment (Fig. 5a). Conversely, in NB69 cells a rapid and transient increase in CREM repressor proteins (data not shown) correlates with a brief increase in prodynorphin mRNA (5). Thus, a temporal correlation could be observed between the unbinding of DREAM from the DRE, the induction of the target gene prodynorphin throughout derepression (5), and the sustained (SK-NMC) or transient (NB69) cell-specific accumulation of CREM repressor proteins in human neuroblastoma cells after forskolin exposure. These data strongly suggest that a specific interaction between αCREM and DREAM in vivo derepresses expression of the prodynorphin gene. To further substantiate this correlation, we analyzed prodynorphin mRNA in SK-NMC cells after overexpression of αCREM or a mutant (αCREML73,76V) unable to interact with DREAM. Confirming our model of derepression, αCREM overexpression resulted in a robust increase in prodynorphin mRNA, while the mutant did not modify basal levels of the transcript (Fig. 5b). Western blot analysis after CREM overexpression showed similar levels of wild-type and mutant αCREM proteins in respective cell extracts (Fig. 5c).
FIG. 5.
The αCREM-DREAM interaction directs prodynorphin gene expression in human neuroblastoma cells after forskolin treatment. (a) Western blot analysis of the accumulation of CREM repressor proteins in SK-NMC cells at different times following forskolin treatment. The lack of effect on the levels of τCREM and CREB proteins is also shown. (b) Northern blot showing the induction of prodynorphin mRNA in SK-NMC cells after αCREM overexpression and the lack of effect of mutant αCREML73,76V. Levels of β-actin are shown as a control of the loading of each lane. (C) Western blot analysis showing similar levels of overexpressed wild-type or mutant αCREM in SK-NMC cells.
DISCUSSION
Here we have identified a novel mechanism of DRE-dependent transcriptional derepression triggered by PKA activation. It involves a specific protein-protein interaction between DREAM and αCREM that is mediated by LCD motifs present in both proteins and results in loss of binding of the transcriptional repressor DREAM to DRE sites. Furthermore, we have demonstrated that the expression of a bona fide target gene of DREAM repression/derepression, the prodynorphin gene, is increased in human neuroblastoma cells as a consequence of the αCREM-DREAM interaction.
Expression of the CREM gene is controlled by an upstream regulatory region (P1) that gives rise to repressors and activators of transcription and an intragenic regulatory region (P2) that produces repressor ICER proteins (9, 25). The CREM gene contains several exons, and the different CREM or ICER isoforms are generated by differential splicing of the primary transcripts (Fig. 1a) (21). The interaction with DREAM is specific for repressor CREM isoforms α and ɛ, while other repressor isoforms (β and γ) and ICER or activator τCREM isoforms (I and II) or CREB do not block binding of DREAM to DRE sites. These results reveal for the first time a functional difference among CREM repressor isoforms in the ability to uncouple DREAM binding to DRE sites. Since differences in nuclear distribution or function for the various repressor CREM isoforms have not been shown, the functional meaning of their differential interaction with DREAM is presently not understood. On the other hand, since the KID and LZ domains are essential for CREM dimerization, binding to CRE sites, and repressor function, further studies will address the possibility that binding of DREAM to these domains can specifically influence CRE-dependent repression through its interaction with αCREM or ɛCREM.
Domain analysis and site-directed mutagenesis of the CREM proteins have identified two LCD motifs located within the KID and the LZ domains of αCREM that are responsible for the interaction with DREAM. Absence of the N-terminal LCD in ICER isoforms or deletion of the C-terminal LCD in αCREMΔLZ completely prevents the interaction. Moreover, the spacing of the two LCDs seems to be critical, since activator τCREM-I and CREB, which contain the Q2 transactivation domain between the LCDs, do not block binding of DREAM to DRE sites. Removal of the Q2 domain in ɛCREM restores the interaction with DREAM. The LCD motif was first found in nuclear coactivators (NCoA-1 and p/CIP) or corepressors (N-CoR and SMRT) and has been implicated in protein-protein interactions with nuclear hormone receptors or CBP (14, 19, 22, 31). Moreover, LCDs in the N- and C-terminal ends of CBP mediate the interaction with nuclear receptors and p/CIP, respectively (31). To date, two classes of LCDs have been defined: the NR (nuclear receptor) box and the CoRNR box, whose consensus sequences are LxxLL and L/IxxV/II, respectively, where x denotes any amino acid (19). Interestingly, both LCDs in αCREM (ILNEL and LIEEL) have an antiparallel orientation compared to the NR consensus box, which could define a third type of LCD. However, the functional meaning, if any, of the antiparallel orientation is not known. It has been shown that the α-helical structure of the LCD binds to a hydrophobic groove located in the ligand-binding domain of the target nuclear receptor (29), and the interaction is often regulated by the amino acids flanking the LCD (14, 22, 29, 32). This is particularly significant in the case of the C-terminal LCD in αCREM, where the flanking residues, different from those in a similar C-terminal LCD in βCREM, confer the differential ability to interact with DREAM. Likewise, two LCD motifs in DREAM are necessary for the interaction with αCREM. Noteworthy, the two LCDs in DREAM conform to the CoRNR and NR boxes, respectively, and have the consensus parallel orientation. Whether the two LCDs in CREM interact directly with the two LCDs in DREAM is not known. Future studies using nuclear magnetic spectroscopy to resolve the solution structure of DREAM bound and not bound to the αCREM LCDs will clarify this point. More importantly, to our knowledge this is the first description of functional LCDs in proteins other than nuclear receptors and their interacting coactivators and corepressors. Thus, our results increase the functional importance of LCD motifs in the context of gene expression as well as our understanding about how protein interactions are orchestrated in the nucleus.
The interaction between CREM and DREAM does not require PKA-dependent phosphorylation of αCREM; however, the interaction is strengthened after CREM phosphorylation. Thus, a rapid phosphorylation of preexisting low levels of αCREM or the accumulation of αCREM protein could initiate the derepression of prodynorphin after forskolin treatment in human neuroblastoma cells. The rapid time course of DREAM unbinding in NB69 cells after forskolin treatment (5) could suggest a rapid and reversible posttranslational modification, i.e., phosphorylation of αCREM. On the other hand, the slow time course and the stability of prodynorphin induction in SK-NMC cells, where a significant increase in prodynorphin mRNA becomes noticeable at 12 h and is maximum 2 days after forskolin treatment (5), points to an increase in CREM protein levels. PKA-dependent transcriptional activation of the CREM gene is restricted to the intragenic P2 promoter, while P1-dependent transcription of CREM does not respond to cAMP stimulation (10, 25). Thus, an increased accumulation of P1-derived transcripts via increased transcription is unlikely to occur. However, a specific effect at the posttranscriptional level has been proposed to occur in testes during development (11) and in supraoptic neurons following osmotic stimulation (24), resulting in the specific accumulation of activator or repressor CREM isoforms, respectively. The mechanism for this could involve a selective alteration of the differential splicing process or a differential change in the stability of P1-derived transcripts. The selective accumulation of CREM repressor proteins in SK-NMC and NB69 cells after forskolin treatment, with no change in the levels of CREM activator τ isoforms, supports a selective change at the posttranscriptional level after PKA activation. However, the molecular mechanisms that control the differential processing of CREM transcripts and its regulation following PKA activation are not known.
The interaction between CREM and DREAM prevents binding to the DRE. The mechanism by which αCREM disrupts DREAM binding to DRE is not known. Apparently, the mechanism does not involve a direct competition for the binding to DNA, since αCREM does not show any affinity for the DRE and DREAM mutants unable to interact with CREM still bind to the DRE. Alternatively, CREM may affect the stability of DREAM tetramers, which are required for efficient interaction with the DRE site (6). Experiments using a reverse two-hybrid protocol may help to identify the domain responsible for DREAM oligomerization and whether αCREM affects this process.
Our results showing that DREAM interacts with α/ɛCREM further indicate the pleiotropic functionality of the DREAM/KchIP-3/calsenilin protein able to participate in different cellular functions through the interaction with DNA or with various proteins, including α/ɛCREM, Kv4 potassium channels, and presenilin-2 (2, 4). Studies using DREAM as bait in a yeast two-hybrid screening try to identify new targets for DREAM interaction are under way. The multifunctionality of DREAM as well as its nuclear and cytosolic locations (6), with specific functions in each compartment, has previously been described for calmodulin, another Ca2+-binding protein. It has been shown that calmodulin interacts with slow-desensitizing voltage-dependent potassium channels in the cell membrane and modulates their permeability (27), controls the activity of many cytosolic enzymes (reviewed in references 1 and 20), and regulates transcription upon binding to helix-loop-helix nucleoproteins (7) and CaM-dependent kinases in the nucleus (8, 28). However, unlike DREAM, calmodulin does not directly regulate transcription since calmodulin binding to DNA has not been demonstrated.
Finally, as in the case of the interaction between KchIP-3/DREAM and Kv4 potassium channels (2) or the interaction between calmodulin and neuromodulin, inducible nitric oxide synthase, or myosin I (30, 33), the physical interaction between DREAM and αCREM is also observed in the absence of calcium binding to DREAM. This indicates that transcriptional derepression at DRE sites can be independently achieved through at least two distinct signaling pathways, calcium and PKA activation. Since cellular stimulation by hormones or activation of membrane receptors is often followed by concomitant elevations in intracellular calcium and cAMP, both mechanisms can cooperatively derepress DRE-dependent transcription. The fact that binding of DREAM to DRE sites is controlled by two major signaling pathways may suggest that DRE-dependent derepression is a step necessary for the transcriptional activation of many genes. Identification of DRE sites in a number of genes activated by calcium and/or PKA activation (5, 6; unpublished results) supports this proposal. Future studies using transgenic mice overexpressing dominant negative mutants of DREAM, unable to respond to calcium and/or unable to interact with αCREM, will help to elucidate the physiological importance of the transcriptional repressor DREAM and the functional meaning of the αCREM-DREAM interaction.
ACKNOWLEDGMENTS
Work in this laboratory is supported by grants from DGICYT, CAM, Europharma SA, and Janssen-Cilag SA to J.R.N.
F.L. and A.M.C contributed equally to this work.
We thank N. S. Foulkes, M. Lamas, D. Martin-Zanca, and P. Sassone-Corsi for discussions, A. Aranda, M. Montminy, R. Perona, and P. Sassone-Corsi for plasmids, and D. Campos for technical assistance.
REFERENCES
- 1.Agell N, Aligue R, Alemany V, Castro A, Jaime M, Pujol M J, Rius E, Serratosa J, Taules M, Bachs O. New nuclear functions for calmodulin. Cell Calcium. 1998;23:115–121. doi: 10.1016/s0143-4160(98)90109-9. [DOI] [PubMed] [Google Scholar]
- 2.An W F, Bowlby M R, Betty M, Cao J, Ling H-P, Mendoza G, Hinson J W, Mattson K I, Strassle B W, Trimmer J S, Rhodes K J. Modulation of A-type potassium channels by a family of calcium sensors. Nature. 2000;403:553–556. doi: 10.1038/35000592. [DOI] [PubMed] [Google Scholar]
- 3.Brindle P, Linke S, Montminy M R. Protein-kinase A-dependent activator in transcription factor CREB reveals new roles for CREM repressors. Nature. 1993;364:821–824. doi: 10.1038/364821a0. [DOI] [PubMed] [Google Scholar]
- 4.Buxbaum J D, Choi E-K, Luo Y, Lilliemook C, Crowley A C, Merriam D E, Wasco W. Calsenilin: a calcium-binding protein that interacts with the presenilins and regulates the levels of a presenilin fragment. Nat Med. 1999;4:1177–1181. doi: 10.1038/2673. [DOI] [PubMed] [Google Scholar]
- 5.Carrion A M, Mellström B, Naranjo J R. Protein kinase A-dependent derepression of the human prodynorphin gene via differential binding to an intragenic silencer element. Mol Cell Biol. 1998;18:6921–6929. doi: 10.1128/mcb.18.12.6921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carrion A M, Link W A, Ledo F, Mellström B, Naranjo J R. DREAM is a Ca2+-regulated transcriptional repressor. Nature. 1999;398:80–84. doi: 10.1038/18044. [DOI] [PubMed] [Google Scholar]
- 7.Corneliussen B, Holm M, Waltersson Y, Onions J, Hallberg B, Thornell A, Grundström T. Calcium/calmodulin inhibition of basic-helix-loop-helix transcription factor domains. Nature. 1994;368:760–764. doi: 10.1038/368760a0. [DOI] [PubMed] [Google Scholar]
- 8.Deisseroth K, Heist E K, Tsien R W. Translocation of calmodulin to the nucleus supports CREB phosphorylation in hippocampal neurons. Nature. 1998;392:198–202. doi: 10.1038/32448. [DOI] [PubMed] [Google Scholar]
- 9.Delmas V, Laoide B, Masquillier D, deGroot R P, Foulkes N S, Sassone-Corsi P. Alternative usage of initiation codons in mRNA encoding cAMP-responsive-element modulator generates regulators with opposite functions. Proc Natl Acad Sci USA. 1992;89:4226–4230. doi: 10.1073/pnas.89.10.4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Foulkes N S, Borrelli E, Sassone-Corsi P. CREM gene: use of alternative DNA-binding domains generate multiple antagonists of cAMP-induced transcription. Cell. 1991;64:739–749. doi: 10.1016/0092-8674(91)90503-q. [DOI] [PubMed] [Google Scholar]
- 11.Foulkes N S, Schlotter F, Pevet P, Sassone-Corsi P. Pituitary hormone FSH directs the CREM functional switch during spermatogenesis. Neuron. 1993;10:655–665. doi: 10.1038/362264a0. [DOI] [PubMed] [Google Scholar]
- 12.Gonzalez G A, Yamamoto K K, Fischer W H, Karr D, Menzel P, Biggs III W H, Vale W W, Montminy M R. A cluster of phosphorylation sites on cyclic AMP-regulated nuclear factor CREB predicted by its sequence. Nature. 1989;337:749–752. doi: 10.1038/337749a0. [DOI] [PubMed] [Google Scholar]
- 13.Groot R P, Hertog J, Vandenheede J R, Goris J, Sassone-Corsi P. Multiple and cooperative phosphorylation events regulate the CREM activator function. EMBO J. 1993;12:3903–3911. doi: 10.1002/j.1460-2075.1993.tb06068.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heery D M, Kalkhoven E, Hoare S, Parker M G. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 15.Hoeffler J P, Meyer T E, Yun Y, Jameson J L, Habener J F. Cyclic AMP-responsive DNA-binding protein: structure based on a cloned placental cDNA. Science. 1988;242:1430–1433. doi: 10.1126/science.2974179. [DOI] [PubMed] [Google Scholar]
- 16.Hoeffler J P, Meyer T E, Waeber G, Habener J F. Multiple adenosine 3′,5′-monophosphate response element DNA-binding proteins generated by gene diversification and alternative exon splicing. Mol Endocrinol. 1990;4:920–930. doi: 10.1210/mend-4-6-920. [DOI] [PubMed] [Google Scholar]
- 17.Hoeffler J P, Lustbader J W, Chen C-Y. Identification of multiple nuclear factors that interact with cyclic adenosine 3′,5′-monophosphate response element-binding protein and activating transcription factor-2 by protein-protein interactions. Mol Endocrinol. 1991;5:256–266. doi: 10.1210/mend-5-2-256. [DOI] [PubMed] [Google Scholar]
- 18.Hoffman D A, Johnston D. Downregulation of transient K+ channels in dendrites of hippocampal CA1 pyramidal neurons by activation of PKA and PKC. J Neurosci. 1998;18:3521–3528. doi: 10.1523/JNEUROSCI.18-10-03521.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu X, Lazar M A. The CoRNR motif controls the recruitment of corepressors by nuclear receptors. Nature. 1999;402:93–96. doi: 10.1038/47069. [DOI] [PubMed] [Google Scholar]
- 20.Ikura M. Calcium binding and conformational response in EF-hand proteins. Trends Biochem Sci. 1996;21:14–17. [PubMed] [Google Scholar]
- 21.Laoide B M, Foulkes N S, Schlotter F, Sassone-Corsi P. The functional versatility of CREM is determined by its modular structure. EMBO J. 1993;12:1179–1191. doi: 10.1002/j.1460-2075.1993.tb05759.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.LeDouarin B, Nielsen A L, Garnier J M, Ichinose H, Jeanmougin F, Losson R, Chambon P. A possible involvement of TIF1 alpha and TIF1 beta in the epigenetic control of transcription by nuclear receptors. EMBO J. 1996;15:6701–6715. [PMC free article] [PubMed] [Google Scholar]
- 23.Maldonado E, Hampsey M, Reinberg D. Repression: targeting the heart of the matter. Cell. 1999;99:455–458. doi: 10.1016/s0092-8674(00)81533-0. [DOI] [PubMed] [Google Scholar]
- 24.Mellström B, Naranjo J R, Foulkes N S, Lafarga M, Sassone-Corsi P. Transcriptional response to cAMP in brain: basal and induced expression of CREM antagonists. Neuron. 1993;10:655–665. doi: 10.1016/0896-6273(93)90167-p. [DOI] [PubMed] [Google Scholar]
- 25.Molina C, Foulkes N S, Lalli E, Sassone-Corsi P. Inducibility and negative autoregulation of CREM: an alternative promoter directs the expression of ICER, an early responsive represor. Cell. 1993;75:875–886. doi: 10.1016/0092-8674(93)90532-u. [DOI] [PubMed] [Google Scholar]
- 26.Nichols M, Weih F, Schmid W, DeVack C, Kowenz-Leutz E, Luckow B, Boshart M, Schültz G. Phosphorylation of CREB affects its binding to high and low affinity sites: implications for cAMP-induced gene transcription. EMBO J. 1992;11:3337–3346. doi: 10.1002/j.1460-2075.1992.tb05412.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sah P. Ca2+-activated K+ currents in neurons: types, physiological roles and modulation. Trends Neurochem Sci. 1996;19:150–154. doi: 10.1016/s0166-2236(96)80026-9. [DOI] [PubMed] [Google Scholar]
- 28.Santella L, Carafoli E. Calcium signaling in the cell nucleus. FASEB J. 1997;11:1091–1109. [PubMed] [Google Scholar]
- 29.Shiau A K, Barstad D, Loria P M, Cheng L, Kushner P J, Agard D A, Greene G L. The structural basis of estrogen receptor/coactivator recognition and the antagonism by tamoxifen. Cell. 1998;95:927–937. doi: 10.1016/s0092-8674(00)81717-1. [DOI] [PubMed] [Google Scholar]
- 30.Swanljung-Collins H, Collins J H. Brush border myosin I has a calmodulin/phosphatidylserine switch and tail actin-binding. Adv Exp Med Biol. 1994;358:205–213. doi: 10.1007/978-1-4615-2578-3_19. [DOI] [PubMed] [Google Scholar]
- 31.Torchia J, Rose D W, Inostroza J, Kamei Y, Westin S, Glass C K, Rosenfeld M G. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature. 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- 32.Torchia J, Glass C K, Rosenfeld M G. Co-activators and co-repressors in the integration of transcriptional responses. Curr Opin Cell Biol. 1998;10:373–383. doi: 10.1016/s0955-0674(98)80014-8. [DOI] [PubMed] [Google Scholar]
- 33.Yuan T, Vogel H J, Sutherland C, Walsh M P. Characterization of the Ca2+-dependent and -independent interactions between calmodulin and its binding domain of inducible nitric oxide synthase. FEBS Lett. 1998;431:210–214. doi: 10.1016/s0014-5793(98)00750-9. [DOI] [PubMed] [Google Scholar]
- 34.Zozulya S, Stryer L. Calcium-myristoyl protein switch. Proc Natl Acad Sci USA. 1992;89:11569–11573. doi: 10.1073/pnas.89.23.11569. [DOI] [PMC free article] [PubMed] [Google Scholar]