Abstract
The cell cycle, oncogenic signaling, and topoisomerase (topo) IIα levels all influence sensitivity to anti-topo II drugs. Because the cell cycle and oncogenic signaling influence each other as well as topo IIα levels, it is difficult to assess the importance of any one of these factors independently of the others during drug treatment. Such information, however, is vital to an understanding of the cellular basis of drug toxicity. We, therefore, developed a series of analytical procedures to individually assess the role of each of these factors during treatment with the anti-topo II drug etoposide. All studies were performed with asynchronously proliferating cultures by the use of time-lapse and quantitative fluorescence staining procedures. To our surprise, we found that neither oncogene action nor the cell cycle altered topo IIα protein levels in actively cycling cells. Only a minor population of slowly cycling cells within these cultures responded to constitutively active oncogenes by elevating topo IIα production. Thus, it was possible to study the effects of the cell cycle and oncogene action on drug-treated cells while topo IIα levels remained constant. Toxicity analyses were performed with two consecutive time-lapse observations separated by a brief drug treatment. The cell cycle phase was determined from the first observation, and cell fate was determined from the second. Cells were most sensitive to drug treatment from mid-S phase through G2 phase, with G1 phase cells nearly threefold less sensitive. In addition, the presence of an oncogenic src gene or microinjected Ras protein increased drug toxicity by approximately threefold in actively cycling cells and by at least this level in the small population of slowly cycling cells. We conclude that both cell cycle phase and oncogenic signaling influence drug toxicity independently of alterations in topo IIα levels.
Topoisomerase (topo) II enzymes function to unknot and decatenate covalently closed circles of DNA. In mammalian cells a requirement for type II topo has been suggested in many aspects of DNA metabolism including replication and recombination (41). There are two isozymes of topo II with molecular masses of 170 and 180 kDa, termed topo IIα and topo IIβ (7). While the biochemical activities of the two proteins are closely related, their cellular distribution and expression characteristics differ greatly. topo IIα is expressed at low levels in quiescent cells and is induced when cells are stimulated to enter the growth phase (32). As cells become contact inhibited, the levels of topo IIα are dramatically reduced (2, 22, 24). On the other hand, topo IIβ expression does not correlate with proliferative status, remaining at constant levels in quiescent and proliferating cells (32, 39).
topo IIα is known to be expressed at high levels in tumors at both the protein and mRNA levels (19). This elevation is identified in a wide variety of tumors and is due in part to the increased growth fraction (3, 11, 17, 26). The expression of topo IIα is also increased as cells pass through the cell cycle (15), although reports vary as to the extent. In NIH 3T3 cells transformed by oncogenic ras (42) topo IIα levels increase, suggesting that oncogenic signaling might directly stimulate the topo IIα promoter, leading to increased protein levels in transformed cells. In support of this possibility, we found that oncogenic Ras is able to increase the expression of a reporter plasmid containing the basal topo IIα promoter driving a luciferase reporter gene (6). This increase, which requires ERK and JNK activities, was independent of cell cycle position (6).
Anti-topo II drugs are used clinically against a wide range of tumors (18) and target both isotypes of topo II, although a broad range of evidence indicates that topo IIα is the primary target (4). A number of studies have identified a close relationship between topo IIα levels and drug sensitivity (9, 12, 14, 17, 30, 31). Similarly, drug resistance is commonly correlated with decreased topo IIα levels (10, 28). On the other hand, factors other than topo IIα levels have been found to influence toxicity, including drug uptake, topo IIα phosphorylation (13, 16), and cellular factors involved in activation of cell death pathways (25, 28).
In order to better understand the interplay of cellular factors involved in controlling cellular response to drug treatment, we designed a series of experiments to analyze the roles of the cell cycle, oncogene action, and topo IIα levels independently of one another during treatment with an anti-topo II drug. While the level of topo IIα within the cell is believed to play a central role in determining drug toxicity (10, 36, 38, 40, 44), the considerations discussed above suggest that these levels might be altered by cell cycle position and oncogene activity. At the same time, these two physiological factors might directly influence drug toxicity independently of alterations in topo IIα levels (1, 23, 43). The challenge, therefore, is to separate the influence of the cell cycle and oncogene action on drug toxicity directly from their ability to alter topo IIα levels, and thereby to influence drug toxicity indirectly.
To address this question, we have developed a means to study the cell cycle expression characteristics of topo IIα in the presence and absence of oncogenic signaling. In addition, we have developed a means to determine cell cycle-related drug toxicity. The technical approach used relied on studies of continuously cycling cells. No attempt was made to synchronize cells in the cell cycle; rather, the cell cycle position of each individual cell was determined using either quantitative fluorescence analysis of DNA or time-lapse observations to determine the timing of mitoses (37). This technical approach has proven effective in previous studies of cyclin D1, where the expression characteristics of this protein were accurately determined in each cell cycle phase, together with the dependence of this expression on cellular Ras activity (20, 21). In the present study the same approach was utilized to determine the effect of the cell cycle and oncogene activity on the expression levels of topo IIα. We were somewhat surprised to learn that neither of these factors altered topo IIα protein expression levels in actively cycling cells. It was, therefore, possible to accurately determine the influence of cell cycle position and oncogene activity on the outcome of drug treatment without concern for topo IIα protein expression levels.
MATERIALS AND METHODS
Cell culture.
All cells were cultured in Dulbecco modified Eagle medium supplemented with 10% calf serum or fetal calf serum and antibiotics. Cells were routinely cultured in a CO2 incubator until the time of video analysis, at which time they were transferred to a moist chamber with a constantly refreshed, moistened 5% CO2 atmosphere at 37°C. The cells proliferated as well in the environmental chamber used for video analysis as in the incubator for up to 8 days (at which time the cells had become completely confluent in each environment). Cells (5 × 104 to 20 × 104) were plated on a coverslip in a 35-mm-diameter dish in preparation for video analysis. For staining purposes cells were approximately 30% confluent, while for drug toxicity studies cells were approximately 10% confluent.
Autoradiography emulsion type NTB2 was obtained from Eastman Kodak Company (New York, N.Y.). DAPI (4′,6′-diamidino-2-phenylindole; dilactate) was a product of Molecular Probes (Eugene, Oreg.). A Leica fluorescence microscope (DM 900) was used to quantitate fluorescence intensity. Leica filter cubes A, L4, and N2.1 and cube 41008 Cy5 from Chroma Technology were used to detect signals from DAPI, Cy2, Cy3, and Cy5, respectively. With those cubes, no significant crossover signal was detected except the faint crossover of the Cy5 signal to the Cy3 cube (see below). We used a cooled charge-coupled device (CCD) camera (Roper Scientific) for quantitation and Metamorph (Universal Imaging) software for image analysis as previously described (21).
Time-lapse analysis.
Time-lapse analysis was performed with a CCD camera attached to a frame capture board controlled by the National Institutes of Health Image program as previously described (37). The area of analysis was marked with two contiguous circles of different sizes using a diamond object marker (Leitz) prior to the beginning of the analysis (20). This allowed the viewing of the same area of the coverslip before and after addition of thymidine and allowed realignment of the area of analysis and identification of individual cells following staining and autoradiography. Oncogenic Ras (Leu61; 1 mg/ml) was microinjected into all the cells and only the cells within the designated circular area. There was no evidence of adverse effects of the injection or introduced protein in these analyses for more than 1 h, as judged by either morphological changes or alterations in the rate of cell division. As a control, nonspecific rat immunoglobulin (10 mg/ml) was injected into cells exactly as described above. The average cell cycle time of 16 to 18 h was increased by approximately 2 h during the period in which the cells were removed from the environmental chamber and microinjected. This lengthening of the cell cycle was due to a combination of the adverse effects of the microinjection and the reduction in the temperature of the culture during microinjection (which was performed at room temperature). In the first full cell cycle following microinjection the cells returned to the typical cell cycle time of unperturbed cultures.
Pulse-labeling with thymidine was performed in the last hour of the video analysis. Cells were first viewed for 19 to 24 h, and the stack of individual frames was saved. The position of the coverslip was noted in a final frame. The coverslip was then removed from the video apparatus, and thymidine was added (2 to 5 μCi of tritiated thymidine in 2 to 5 μl of an aqueous solution containing 2% ethanol [Amersham]). Alternately, bromodeoxyuridine (BrdU) (1×; Boehringer) was added in place of thymidine and labeled cells were detected with a fluorescent-antibody detection kit (Boehringer). The medium containing thymidine was thoroughly mixed and returned to the video apparatus. Care was taken to ensure that the same area and orientation of the coverslip were viewed after thymidine addition, and frames were collected for 60 additional minutes at the same rate as for the first stack. Finally, the frames collected after the addition of thymidine were added to the stack used in the analysis prior to thymidine addition. The end of the analysis was therefore considered the end of the thymidine labeling period. Cells were fixed immediately upon termination of the video observation to ensure that the cell positions would not change in the video analysis compared to those for the fixed slide.
For dual movies before and after drug treatment, the first movie comprised the 20 to 24 h prior to drug treatment, with the 80 min during which the cells were exposed to the drug as described above for thymidine included in the first movie. Following an extensive washing of the plate the second movie of the same area of cells was initiated and continued uninterrupted for 50 to 75 h. Each cell in the last frame following drug treatment was numbered and monitored backwards in the first movie to determine the time of mitosis prior to drug treatment and therefore the age of the treated cell. The cell was then monitored through the second movie to determine its fate. Once a cell divided only one of its daughters was monitored. Upon the second division the cell was determined to be normal and was not monitored further. If at any time the original cell or its daughter passed through abortive mitosis (a mitotic event including condensation of the chromosomes which resulted in only one cell) or cell death (characterized by cell fragmentation), it was given this classification. Cells which divided only once following treatment were carefully noted, and their numbers were analyzed. They behaved essentially the same as cells which divided twice but were not included in any of the results presented.
Microinjection and antibody staining.
The procedure for microinjection was identical to that previously reported (21). Cells to be injected were plated on a 22-mm-square coverslip and cultured in a 35-mm-diameter plate. The cells were inverted in culture medium during injection and placed on the stage of an upright microscope. The micropipette (outside diameter of less than 1 μm) was positioned within the culture medium just below the coverslip, and injections were performed by bringing the micropipette up into the cell while forcing the outflow of sample. Prior to injection the circles described above were scribed on the back side of the coverslip with a Leitz object marker, and the injections into all the cells enclosed by this circle were performed. Immediately following injection the coverslip was placed in the environmental chamber and time-lapse analysis was initiated. Injected cells were also identified by staining with an antibody against nonspecific rat immunoglobulin (10 mg/ml) included as a marker in Ras injections. The topo IIα levels were determined with a monoclonal antibody specific to the p170 alpha subunit of human topo II (catalog no. 2010-1; Topogen, Columbus, Ohio). The cells were counterstained with a Cy3-linked secondary antibody. Nuclei were isolated from NIH 3T3 cells (or these cells transformed by src, ras, or raf) following swelling in hypotonic buffer (107 cells/ml; 5 mM KH2PO4 [pH 7.2], 2 mM MgCl2, 10 mM β-mercaptoethanol, 0.1 mM Na2EDTA, protease inhibitors). The cells were broken with 20 strokes with a Dounce homogenizer on ice and collected by centrifugation at 1,000 × g for 6 min.
Analysis of data.
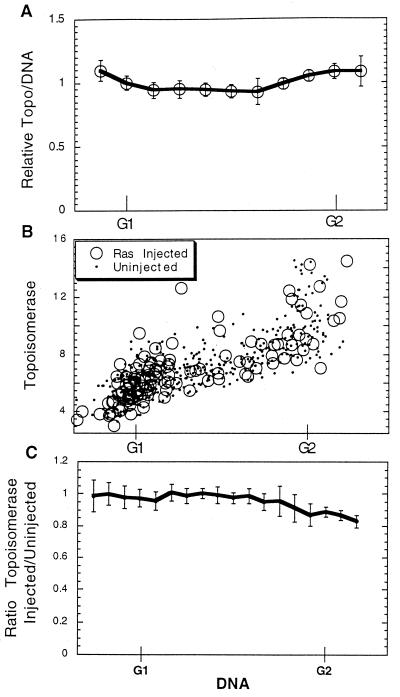
Two forms of data presentation are utilized. The first consists of raw data in which the results for each cell are presented. To assess the results of numerous determinations, average values are also presented. To obtain average numbers, all the cells within a single analysis were considered to determine the G1 and G2 phase DNA readings. Each cell was then given a DNA reading based on this range for the specific experiment. Then, the topo IIα/DNA ratio for each cell was determined, and the average of all cells within the experiment was computed. The topo IIα/DNA ratio for each cell was then divided by the average for all cells in a given experiment. In this way, the DNA and topo IIα/DNA ratio readings for each cell were normalized for a given experiment. This made it possible to compare the results for different experiments even though the staining intensity in different experiments might vary slightly. For average determinations, the average for each experiment in a given DNA range was determined and then the values from all experiments were compared to determine the mean topo IIα/DNA ratio ± the standard error (SE) for all the experiments in a given DNA range. This calculation was performed for cells in all DNA ranges, and these numbers were then plotted versus the DNA level (Fig. 3A). Similar calculations were performed to compare the ratios of topo IIα levels in injected cells to those in uninjected cells (Fig. 3C) and the average DNA or topo IIα levels in time-lapse analyses (Fig. 4C). For the comparison of Western blotting and fluorescence readings the procedure was as previously described (20), except that the Western analysis was performed by blotting with an anti-topo IIα antibody followed by a horseradish peroxidase-coupled antimouse antibody coupled to ECL (Amersham). The luciferase levels were recorded on film and quantitated using the National Institutes of Health Image program. For comparison, average fluorescence readings were determined at each time period by staining with the fluorescent anti-topo IIα stain or by staining DNA with DAPI and quantitating the levels of fluorescence in the nuclear regions of over 250 cells. These readings were then averaged to obtain the values reported in Fig. 1.
FIG. 3.
Topo IIα levels through the cell cycle. (A) The topo IIα and DNA fluorescence levels of individual cells were determined in nine separate experiments similar to that shown in Fig. 2B. The ratio of topo IIα level to DNA level for each cell was determined, and the ratio of this value to the average value for all the cells in a given experiment is reported. This number, therefore, indicates the relative topo IIα/DNA ratio for each cell compared to those for all the cells of a given experiment. A cell with a value of 1.0 would have an average topo IIα/DNA ratio in that experiment. For comparison, cells of each experiment were divided into groups according to their DNA content, and the average topo IIα/DNA ratio for each group was determined. Groups with the same DNA content from all nine experiments were then compared to determine the mean ratio of topo IIα level to DNA level (± SE) for this DNA range. This value was then plotted versus the DNA level. (B) Oncogenic Ras (Leu61; 1 mg/ml) was microinjected into cells within an asynchronous NIH 3T3 culture which was incubated for 24 h, fixed, and stained for topo IIα and DNA. DNA-associated fluorescence is plotted versus topo IIα-associated fluorescence. (C) Results from six separate experiments like those shown in panel B were compared. Cells in each experiment were divided into groups according to their DNA content, and the average topo IIα level for injected cells in a group was divided by the average level for uninjected cells. The ratios obtained in this way for all experiments were then combined, and the mean ratio of the topo IIα level in injected cells to that in uninjected cells (± SE) for each DNA range was determined. This value was plotted versus the DNA content for each group. Because there was little difference between cells injected with oncogenic Ras and uninjected cells, data concerning control injections are not shown.
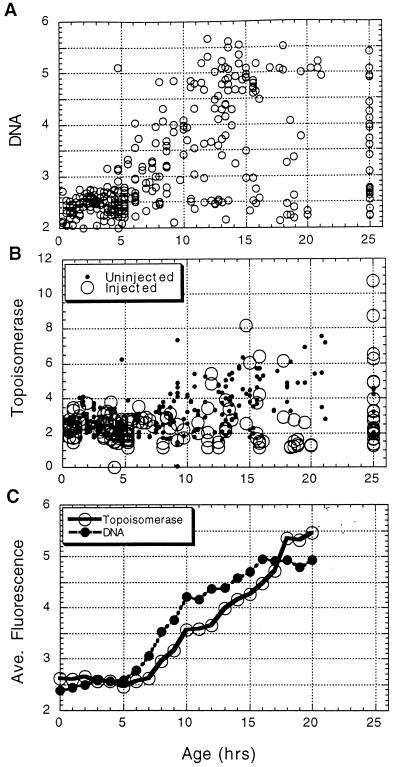
FIG. 4.
Time-lapse studies of topo IIα through the cell cycle. Cells were monitored in time-lapse for 25 h, fixed, and stained for topo IIα and DNA. These levels for each individual cell were then determined by photography and image analysis as described previously. The age of each cell was determined by analysis of the time-lapse movie. (A) The age of each cell plotted versus its DNA content. (B) The topo IIα content of each cell plotted versus its age. (C) Cells were grouped according to age, and the average DNA and topo IIα contents were determined and plotted versus age. It is clear that while most cells cycle as expected, a small population progress through the cell cycle much more slowly. The cells which failed to divide during the entire time-lapse analysis are displayed as having an age of 25 h.
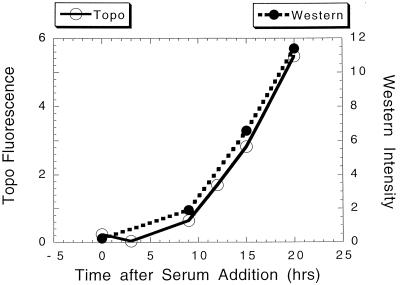
FIG. 1.
Comparison of topo IIα protein levels and fluorescence intensity. NIH 3T3 cells were rendered quiescent by culture in low serum (0.5% calf serum) for 48 h. Serum was then added to separate cultures, and at the times indicated lysates were prepared for quantitative Western analysis. For comparison, a parallel plate was fixed and stained for topo IIα with a specific, indirect fluorescent stain. Images of the stained cultures were collected with a CCD camera (together with images of the DNA stained with DAPI). The total amount of topo IIα-associated fluorescence was determined for each nucleus. The average intensity of 200 to 400 individual nuclei is plotted versus the relative amount of topo IIα protein determined by quantitating the topo IIα protein band in Western analysis.
Mitotic shakeoff was performed with 10 large NIH 3T3 plates at near confluence as previously described (20). These were rapidly replated in prewarmed medium in several small cultures. These individual cultures were treated with drug separately at the times indicated or were mock treated. After drug treatment and replacement with conditioned medium the plates were cultured for 3 days and the cells were counted.
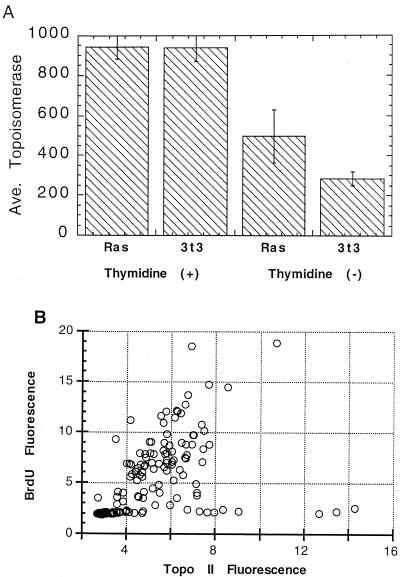
For the determination of topo IIα levels in slowly cycling cells, NIH 3T3 cells, transformed by oncogenic ras or not transformed, were cultured with tritiated thymidine, washed, fixed, and stained for topo IIα. The culture was then autoradiographed, and the cells without any labeling were separately photographed and analyzed to determine the topo IIα fluorescence. For comparison, a group of labeled cells were also analyzed for topo IIα levels to determine the staining pattern for random cells of the culture. Most cells not labeled with thymidine failed to express high levels of topo IIα, but approximately 20% of these cells in the transformed culture had topo IIα levels dramatically elevated above those of normal G1 phase cells. The results displayed (Fig. 5A) reflect the average values obtained.
FIG. 5.
Ras stimulation of topo IIα in slowly cycling NIH 3T3 cells. (A) NIH 3T3 cells and a clone of these transformed by oncogenic ras (Val12) were labeled with [3H]thymidine for 20 h, fixed, stained for topo IIα, and autoradiographed. The topo IIα levels of individual slowly cycling cells, which failed to incorporate thymidine, in both ras-transformed and untransformed cultures were determined, and the means (± SE) are presented. For comparison, the average topo IIα level of 200 cycling, thymidine-labeled cells in each culture was also determined. (B) src-transformed NIH 3T3 cells were labeled with BrdU for 24 h, fixed, and stained for BrdU and topo IIα. The topo IIα-associated fluorescence is plotted versus the BrdU-associated fluorescence for all cells in the culture. Slowly cycling cells form a line along the bottom of the profile with low BrdU staining.
RESULTS
Our goal is to determine the effects of the cell cycle, oncogenic activity, and topo IIα protein levels on the outcome of treatment with the anti-topo II drug etoposide. It was first necessary to determine exactly how topo IIα protein levels are affected by the cell cycle and oncogene action. This required a detailed cell cycle analysis of topo IIα protein levels. Our approach was to determine the cell cycle positions of individual cells in an asynchronous culture. This approach not only allowed analysis of cells in all cell cycle phases simultaneously but also eliminated any potential complication associated with methods of cell cycle synchronization. The cell cycle phase was determined in two separate ways. In the first, cells were fixed and DNA was stained with DAPI. Previous studies have conclusively demonstrated that the fluorescence intensity of the cell stained in this way is proportional to DNA content. Moreover, the analysis of monolayer cells was found to be extremely accurate due to the high-resolution optics involved, the ability to analyze fluorescence specifically within the nuclear region, and the low level of cytoplasmic interference within the nuclear region in flattened monolayer cells (20). In the second technique, cell cycle position was assessed by monitoring cells in time-lapse for 24 h and then determining the age of each cell (or the time since passing through mitosis) at the time of fixation (21).
To determine the level of topo IIα protein in each cell, the cells were stained with an antibody specific to topo IIα. It was first necessary to demonstrate that this staining procedure resulted in fluorescence levels proportional to those of topo IIα protein within the cell. To accomplish this, topo IIα protein levels and fluorescence were compared in cells synchronized by serum deprivation and restimulation. At various times following serum addition to quiescent NIH 3T3 cultures, lysates were collected for Western analysis, while cells in parallel cultures were fixed and stained for topo IIα and DNA. The amount of topo IIα protein at each time point was determined by quantitating the intensity of the Western band. This was then compared to fluorescence intensity by measuring the fluorescence of 200 to 400 cells in a parallel culture and determining the average fluorescence (21). The results of a typical analysis (Fig. 1) indicate a close correlation between the average fluorescence intensity of the topo IIα stain and the absolute amount of topo IIα protein within the culture. In separate analyses, it was found that only full-length topo IIα was present in isolated nuclei, thus reducing the possibility that the results obtained reflected partially degraded protein (data not shown). It is, therefore, possible to measure fluorescence intensity and determine the relative amounts of topo IIα in individual cells.
Increase of topo IIα protein through the cell cycle.
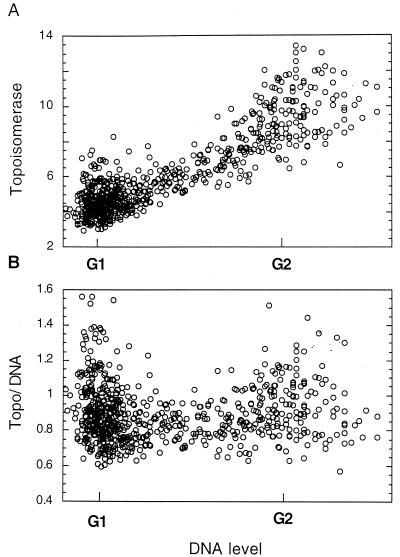
The levels of topo IIα through the cell cycle of actively cycling cells were next determined by fixing and staining an asynchronous culture of NIH 3T3 cells for topo IIα and DNA as described above. Separate images of each fluorochrome were collected from the same cells. The nuclear region of each cell was identified from the DNA stain, after which the total amounts of DNA- and topo IIα-associated fluorescence for each nucleus were determined. When the DNA fluorescence levels for individual cells are plotted versus topo IIα-associated fluorescence, it can be seen that the amounts of topo IIα continuously increased as the cells progress from G1 to G2 phase (Fig. 2A). While the amounts of topo IIα protein increase during passage through the cell cycle, it was somewhat surprising, based on previously reported data, that the increase was not larger than the one observed. Most cell constituents passively increase in amount during the cell cycle due merely to the increase in cell size and mass. It is critical to determine if this might be the explanation for the slight increase in topo IIα protein observed above.
FIG. 2.
Topo IIα expression through the cell cycle. (A) An asynchronous culture of NIH 3T3 cells was fixed and stained for DNA and topo IIα. The levels of each fluorochrome were determined by image analysis, and the DNA levels for each cell were plotted versus the topo IIα levels, with the G1 and G2 levels of DNA indicated. (B) To determine relative topo IIα levels, the topo IIα-associated fluorescence value was divided by the DNA-associated fluorescence and this ratio was plotted versus the DNA level (and therefore cell cycle position).
To test this possibility, the total amount of topo IIα fluorescence in each cell was divided by the DNA-associated fluorescence for that cell. This normalized topo IIα value was then plotted versus that for DNA. When the amount of topo IIα protein relative to DNA content was displayed in this way, it was clear that the increase in topo IIα protein through the cell cycle was roughly equal to the increase in DNA content within the cell (Fig. 2B). The increase in topo IIα levels was, therefore, reflective only of the overall increase in cell size as cells progress through the cell cycle. (A very few cells with high topo IIα levels during G1 and G2 phases can largely be accounted for by the presence in these populations of rounded cells in mitosis, which give artificially high fluorescence values [Fig. 2].) This type of analysis was extended to a number of other cell types, including tumor cells, with identical results.
To determine if there might be a slight cell cycle-related alteration in topo IIα protein, results from nine separate experiments were compared. For this comparison, cells within each experiment were grouped according to DNA content and the average topo IIα/DNA ratio for each of these groups was determined. Data from all nine separate experiments were then compared to determine the mean topo IIα/DNA ratio (± SE) for each DNA range. When this mean value was plotted versus the DNA level, the possibility that a slight reduction in the topo IIα/DNA ratio might exist during S phase became apparent (Fig. 3A). While the difference was only approximately 10%, it might indicate a larger difference in the size of a particular cellular pool of topo IIα.
Time-lapse analysis of topo IIα levels.
To confirm the above results, topo IIα expression through the cell cycle was analyzed using time-lapse to determine cell cycle position. Cells of an asynchronous culture were monitored for 25 h prior to fixation and staining for topo IIα. The level of topo IIα-associated fluorescence was determined for each cell as described above. Individual cells were then monitored in the time-lapse movie to determine their ages, or how long prior to fixation they had passed through mitosis (29). In previous studies we have shown that most NIH 3T3 cells less than 5 h old are in G1 phase, that cells 5 to 12 h old are in S phase, that cells older than 12 h are most likely in G2 phase, and that the average generation time was 16 to 17 h (20, 37). As an indication of the cell cycle characteristics of these cultures, the ages of individual cells in a typical analysis are plotted versus their DNA contents (Fig. 4A). Although most cells in the culture behave as described above, a small number display slower transit through the cell cycle. The topo IIα-associated fluorescence also increased with age (Fig. 4B). To relate topo IIα levels to DNA levels as these cells progressed through the cell cycle, cells were grouped according to age and the average topo IIα fluorescence levels for each group were determined, as was the average DNA fluorescence level. When the average topo IIα and the average DNA levels were plotted versus age, it was clear that topo IIα levels and DNA content increased together through the cell cycle in this time-lapse analysis (Fig. 4C). Based on all the above data, therefore, we conclude that the topo IIα content of cycling cells increases roughly in proportion to the increase in DNA content through the cell cycle in all cell types analyzed.
Ras injection and topo IIα levels.
We next analyzed the effects of constitutive oncogenic signaling on topo IIα levels. Oncogenic Ras (L61; 1 mg/ml) was microinjected into cells of an asynchronous culture, and the topo IIα and DNA levels were determined 24 h later by fluorescence photography and image analysis. No increase in the topo IIα levels was observed following Ras injection during any cell cycle phase (Fig. 3B). As above, for a critical comparison the results of several separate experiments were combined. Cells in individual experiments were divided into groups according to DNA content. For each group the topo IIα levels for all Ras-injected cells were compared to topo IIα levels for uninjected, neighboring cells. Results from all six experiments were combined to determine the mean ratio of the topo IIα level in injected cells to that in uninjected cells (± SE) for each DNA group (Fig. 3C). The topo IIα levels for injected and uninjected cells were remarkably similar, particularly during the first half of the cell cycle, although a slight reduction might have been induced by Ras during G2 phase. Similar results were obtained when Ras-injected cells were compared to cells injected with rat immunoglobulin as a control.
Ras stimulation of topo IIα in slowly cycling cells.
To confirm the above results, oncogenic Ras was injected into cells, which were then monitored for 25 h in time-lapse prior to fixation and analysis of topo IIα levels. As above, no overall stimulation of topo IIα levels by Ras could be observed (Fig. 4B). However, this analytical approach allowed the identification of cells with lengthened cell cycle phases. Cells which failed to divide in the 25 h of the time-lapse observation are represented as having an age of 25 h. The topo IIα levels in these cells were low as expected, except in the Ras-injected cells. In some of these slowly cycling, Ras-injected cells the topo IIα levels increased dramatically (Fig. 4B). Such cells would not have been apparent without time-lapse analysis. These cells are considered potentially interesting because they might serve as a culture model for tumor cells. Like most cells of a tumor, they retain proliferative capacity but cycle slowly. The fact that these cells responded to Ras injection by increasing topo IIα levels, therefore, is an observation which was carefully verified.
A second analysis of slowly cycling cells was performed by comparing ras-transformed and untransformed NIH 3T3 cells in asynchronous culture. Each cell type was cultured with [3H]thymidine for 20 h prior to staining for topo IIα. Cells which cycle slowly enough to avoid passing through any part of S phase during this time would remain unlabeled. In the ras-transformed culture these unlabeled cells contained a significantly higher topo IIα level than the unlabeled, untransformed cells (Fig. 5A). This result confirms that the oncogenic signaling resulted in topo IIα elevation in the slowly cycling cells. As above, the difference between transformed and untransformed cells was due to a few noncycling ras-transformed cells which expressed extremely high topo IIα levels.
As a final confirmation that oncogenic signaling can lead to high topo IIα levels in slowly cycling cells, an analysis was performed on NIH 3T3 cells transformed by a separate oncogene, src. This activated tyrosine kinase functions upstream of cellular Ras and was expected to stimulate many of the same targets as Ras. The transformed cells were labeled with BrdU for 24 h and then stained for BrdU and topo IIα. For each cell the BrdU fluorescence was plotted versus the topo IIα fluorescence. Slowly cycling cells in this analysis would fail to incorporate BrdU. As above, a proportion of these slowly cycling cells displayed high topo IIα levels (Fig. 5B). While only a fraction of these cells expressed high topo IIα, the highest topo IIα content in the culture was found in these non-BrdU-labeled, slowly cycling cells. These combined results indicate that, while the topo IIα levels of actively cycling cells appear to be held in check by cell size and DNA levels, when a cell pauses in its proliferation for a period of time, Ras activity is able to stimulate the production of high levels of topo IIα protein.
Drug toxicity and the cell cycle.
Once the effect of the cell cycle and oncogenic activity on topo IIα levels was determined, it was possible to determine the influence of each of these factors on drug toxicity. This comparison was simplified by the fact that neither the cell cycle nor oncogenic activity altered topo IIα levels, so that the effect of each could be determined without concern that secondary effects on topo IIα levels were involved. Drug toxicity studies were performed in asynchronous cultures with two consecutive time-lapse movies of the same cells separated by a brief (80-min) treatment with etoposide (2 to 10 μM). From the first movie (24 h) the cell cycle position of each individual cell at the time of drug treatment was determined, while from the second movie (50 to 75 h) the fate of each treated cell was analyzed (Fig. 6). This allowed analysis of the effects of the cell cycle on drug toxicity without treatments required to induce cell cycle synchrony. Cells were poisoned with 5-azacytidine (100 to 500 μM) or tritiated thymidine (2 mCi/ml) (8, 34) as controls in separate analyses.
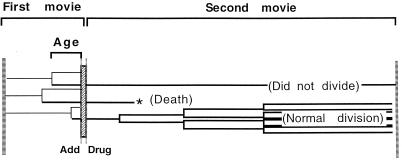
FIG. 6.
Experimental procedure for cell cycle toxicity analysis. In order to determine the effects of the cell cycle on drug toxicity, NIH 3T3 cells were monitored in time-lapse for 25 h, the final 80 min of which was in the presence of the drug. The drug was removed from the culture, and a second movie of 50 to 70 h was immediately initiated, making certain that the same cells were monitored throughout. Each cell is illustrated by a single horizontal line, with the branch point representing mitosis. Age is the time between the initiation of drug treatment and the preceding mitosis. The fates of treated cells are determined during the second movie, with the most common fates illustrated.
Drug-treated cells displayed a variety of fates in the 2 to 3 days following treatment. Cells were considered normal if they divided at least twice during this time. They were considered to have been poisoned if they failed to divide, passed through an abortive mitosis, or underwent a rapid cell death. The typical appearance of rapid cell death and abortive mitosis is depicted in Fig. 7 (see the figure legend for a web site leading to video clips of these events). For abnormal cells, the failure to divide was most common within the first 3 days following drug treatment. In prolonged analyses, however, those cells which failed to divide in the first 3 days generally exhibited either abortive mitosis or rapid cell death within 6 days following treatment (data not shown). Many cells divided only once in the second time-lapse analysis. These cells were carefully monitored throughout and were found to behave similarly to normal cells but are not included in any of the calculations discussed below.
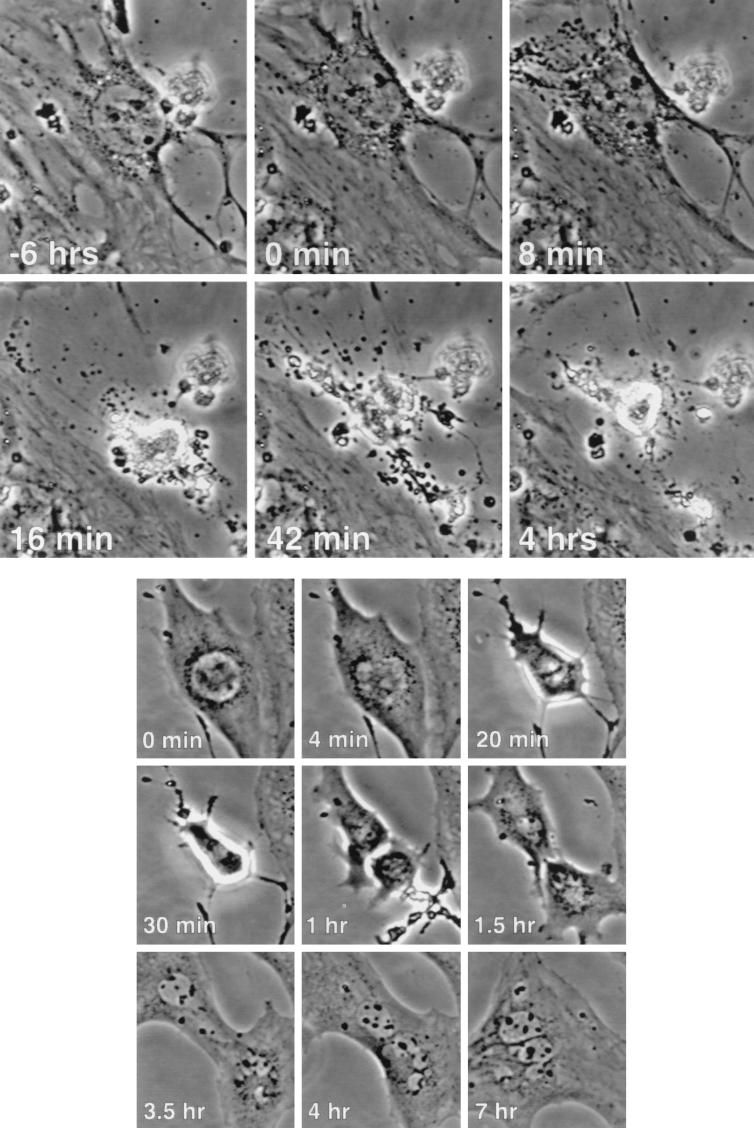
FIG. 7.
Rapid cell death and abortive mitosis. (Top) A small proportion of NIH 3T3 cells treated with etoposide experienced a sudden, dramatic cell death. A typical event is pictured, with the relative times indicated. Note that the most dramatic effects took place between frames 8 min apart. (Bottom) A similar presentation of a cell passing through an abortive mitosis following etoposide treatment is made, with the relative times indicated. (A video presentation of each of these events is presented at http://www.lerner.ccf.org/labs/stacey/dir.cgi, labeled RD2 and AM1).
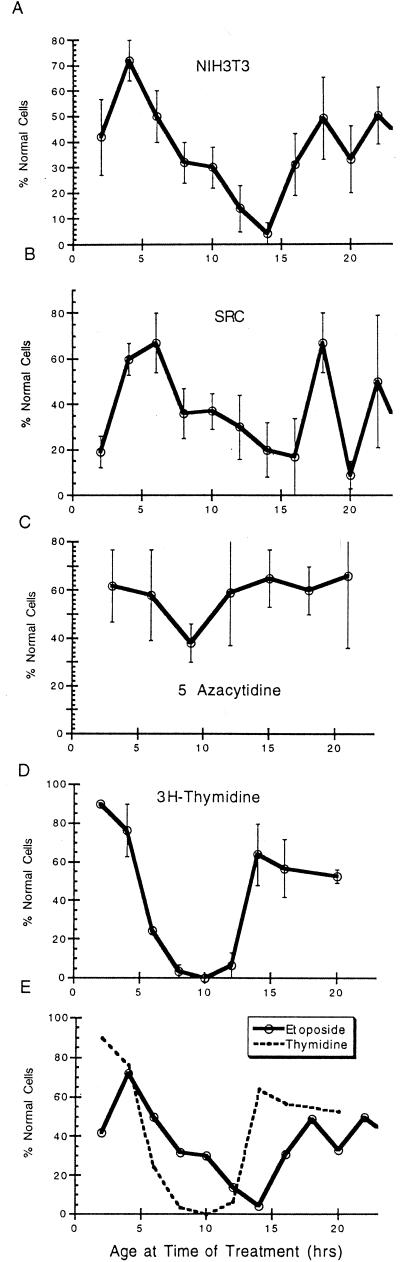
Because the number of cells analyzed in any given experiment was limited, each toxicity experiment was repeated a number of times and the highly consistent data from each analysis were combined to yield a cumulative profile (Fig. 8). Etoposide treatment of NIH 3T3 cells (7 μM) was most toxic when administered between 8 and 16 h following mitosis, when the cells were from mid-S phase to early G2 phase. Cells treated during G1 phase were much more likely to remain normal than cells treated during this sensitive phase. Interestingly, the toxicity of etoposide was reduced as cells progressed late into G2 phase (Fig. 8A). The validity of these results is emphasized by the fact that this experiment was performed independently at least six times with highly similar results. As a control cells were treated in the same way with another toxic agent, 5-azacytidine. In this case there was little cell cycle-specific killing, but the small effect seen was opposite to that obtained with etoposide, with the greatest survival of cells during S and G2 phases (Fig. 8C).
FIG. 8.
Cell cycle and drug toxicity. (A) Six individual experiments were performed as described for Fig. 6. In these experiments NIH 3T3 cells were treated with etoposide (7 μM) to determine cell cycle-specific toxicity. The age and fate of all treated cells were determined, the cells were separated according to age, and the proportion of normal cells for each age period was calculated. The results for all six separate experiments for each age range were then compared to yield the mean percentage of normal cells (± SE) for cells of a given age. This number is plotted versus the age at the time of treatment. (B) A similar analysis was tabulated from five separate experiments using src-transformed NIH 3T3 cells treated with 2 μM etoposide. (C) NIH 3T3 cells were treated with 5-azacytidine (150 μM) in three separate experiments. (D) Two separate experiments in which NIH 3T3 cells were treated for 80 min with [3H]thymidine (2.5 μCi/ml; 79 Ci/mM) were performed. (E) The results from panels A and D are plotted together for comparison.
To more specifically relate the results with etoposide to specific cell cycle stages, and as another control, cells were poisoned with tritiated thymidine, which was found to be specific for S phase toxicity as expected. The experimental strategy was exactly as described above. The results with labeled thymidine were dramatic, with an almost quantitative killing of cells in S phase and little toxicity in other cell cycle stages (Fig. 8D). Not only does this result emphasize the validity of our cell cycle analytical approach, it clearly identifies the position of S phase in cells from 5 to 13 h old (Fig. 8D). It is therefore apparent that the maximal toxicity of etoposide is seen not in a given cell cycle stage but rather is localized from mid-S phase to early G2 phase (Fig. 8E).
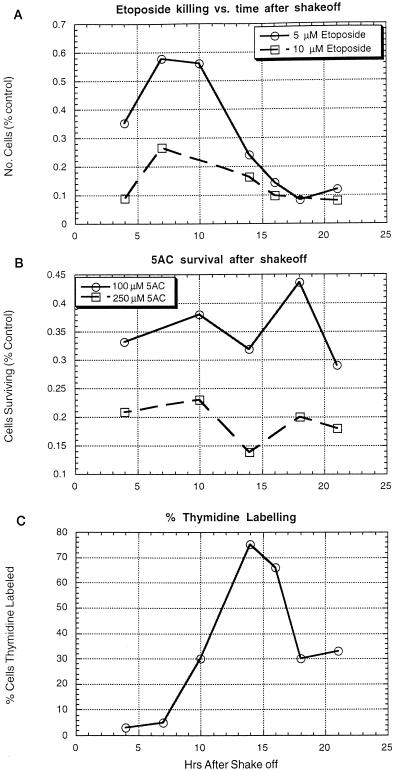
The cell cycle toxicity experiments described above were next confirmed by treatment of cells synchronized by mitotic shakeoff. Mitotic NIH 3T3 cells were collected, replated, and treated with etoposide or 5-azacytidine for 80 min at various times after replating. The effects of the drug treatment were then determined by counting the cells after 3 days. To determine the timing of S phase entry following mitotic detachment, a set of parallel cultures was labeled with tritiated thymidine at various times following replating (Fig. 9). While the delay between mitosis and S phase was much greater following mitotic detachment than in actively cycling cells, the cell cycle-related toxicity profiles were similar. As with the asynchronous cells, etoposide-induced toxicity in the shakeoff cells increased beginning in approximately mid-S phase until it reached a maximum late in the cell cycle. With 5-azacytidine, the toxicity during S phase might have been somewhat reduced compared to that during other cell cycle phases (Fig. 9). From these studies it can be concluded that the cell cycle has an important effect on etoposide toxicity, with greatest sensitivity during late S and G2 phases.
FIG. 9.
Drug toxicity after mitotic selection. At 90-min intervals mitotic NIH 3T3 cells were detached mechanically from asynchronous cultures and replated in several parallel plates. At the indicated times following replating each culture was treated with the indicated concentration of etoposide (A) or 5-azacytidine (AC) (B) for 80 min. The cultures were washed and cultured for an additional 3 days before trypsinization and counting. The numbers reported indicate the numbers of cells compared to those for mock-treated cultures. (C) To determine cell cycle characteristics, cells were labeled with tritiated thymidine at various times following replating, immediately fixed, and autoradiographed to determine the proportion of cells in S phase.
Oncogenic signaling and toxicity.
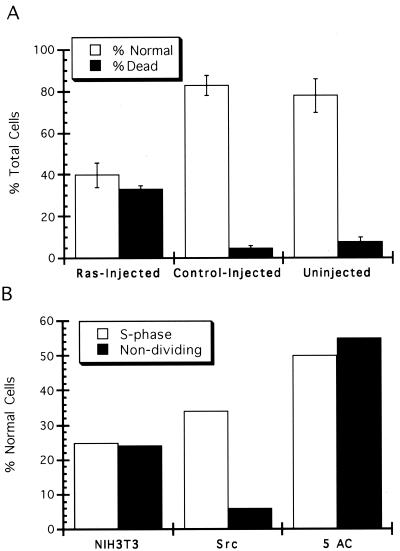
Experiments were last performed to determine the effect of oncogenic activity on etoposide toxicity. Experiments similar to those described above were repeated with NIH 3T3 cells transformed by the src oncogene. As previously reported (5), these cells were much more sensitive to etoposide toxicity than untransformed cells. Experiments were performed with 2 μM etoposide so that the proportion of cells killed would be approximately equal to that observed when untransformed cells were treated with 7 μM etoposide. While the sensitivity of src-transformed cells increased, the overall cell cycle characteristics of killing were similar to those observed with untransformed cells, with the possible exception that the overall cell cycle effects of toxicity might have been reduced in the transformed cells (Fig. 8B). To extend this observation, NIH 3T3 cells were injected with oncogenic Ras protein in a defined area of the culture and the toxicity of etoposide (2 μM) was determined with the time-lapse analysis. In this case the overall proportions of normal cells for the injected and uninjected neighboring cells were determined. As a control, similar experiments were performed by microinjecting rat immunoglobulin as a negative control. In repeated experiments, a dramatically increased sensitivity of Ras-containing cells was observed. The proportion of poisoned cells increased over threefold following Ras injection (Fig. 10A). Unfortunately, the overall numbers of these cells were too small to make definite conclusions regarding the cell cycle profile of toxicity following Ras injection. Since we know that the overall levels of topo IIα do not increase following Ras injection, this clearly indicates that oncogenic activity sensitizes the cells to etoposide independently of alterations in topo IIα levels.
FIG. 10.
Oncogenic transformation and etoposide toxicity. (A) The time-lapse analysis of toxicity (Fig. 6) was applied to cells injected with oncogenic Ras (Leu61; 1 mg/ml). The proportion of the cells which divided normally or which were killed was determined in three separate experiments, and the combined results are presented. For comparison, the results with uninjected, neighboring cells are also presented, together with the results following microinjection of a control protein (10 mg of rat immunoglobulin/ml). (B) Survival of slowly cycling cells following drug treatment. Cells which failed to divide in the movie prior to drug treatment from all experiments presented in Fig. 8A to C are considered. The proportion of these cells which divided normally is presented, together with the proportion of cells in S phase at the time of drug treatment which remained normal following treatment. Results for NIH 3T3 cells treated with 7 μM etoposide, src-transformed NIH 3T3 cells treated with 2 μM etoposide, and NIH 3T3 cells treated with 150 μM 5-azacyticine are presented. These results are based on a total of 67 nondividing cells in all experiments.
A final consideration relates to those few cells which failed to pass through mitosis prior to treatment in the above time-lapse toxicity studies (Fig. 8). These cells were considered interesting because they responded to Ras injection by increasing topo IIα to high levels in some cases and because they are considered to be a potential culture model for cells of a tumor, most of which also cycle slowly. While the numbers of these cells were small, when the total number from all the numerous experiments described above were considered, it appeared that src transformation greatly increased the etoposide toxicity in these slowly cycling cells (Fig. 10B). This is interesting, since it is known that the topo IIα levels increase in only a small proportion of slowly cycling src-transformed cells, yet a high proportion of these cells displayed increased toxicity to etoposide treatment. This again emphasizes the role of oncogenic transformation in potentiating drug toxicity independently of topo IIα levels, although final conclusions from the slowly cycling cells await further work to identify increased numbers of such cells.
DISCUSSION
A number of technical approaches were developed to separately assess the roles of the cell cycle and oncogenic signaling in the sensitivity to the anti-topo II drug etoposide, independently of changes in topo IIα levels. It was first necessary to determine the influence of the cell cycle and oncogene activity on topo IIα expression. In these studies we avoided working with synchronized cell populations to avoid potential complications resulting from treatments required to induce cell cycle synchrony. Instead, an analytical approach was utilized to accurately assess cell cycle-specific protein expression levels in asynchronous cultures by determining the cell cycle positions of individual cells using quantitative fluorescence staining of DNA or time-lapse analysis. This approach has proven useful in analyzing the cell cycle expression levels of cyclin D1 protein and in determining the signaling requirements for this cell cycle-regulated expression pattern (21). In the present study, it was first necessary to directly demonstrate that the fluorescence intensity of cells stained with a specific antibody stain against topo IIα accurately reflected the amount of protein within the cell. Once this relationship was established, it was possible to determine topo IIα expression characteristics of individual cells. We were surprised to find that there was no specific increase in topo IIα protein as cells progress through the cell cycle. This was the case for NIH 3T3 cells as well as for all other cell lines analyzed (including HeLa cells, T24 [bladder tumor] cells, MCF-7 and 468 [breast tumor lines] cells, and MRC-5 diploid human fibroblasts). It is not clear if topo IIα levels are linked directly to DNA levels within the cell or if another related cellular factor is involved. Moreover, contrary to our initial beliefs, there was absolutely no induction of topo IIα levels in oncogene-containing cells. These observations conflict with some published reports most probably because we have studied only actively cycling cells. In studies with serum-deprived cells, the addition of serum induces not only a change in the cell cycle but also, more importantly, a total change in growth phase. Topo IIα is known to be tremendously sensitive to changes in growth state, with high levels of expression in proliferating cells (2, 43).
The only exception to this pattern was observed in cells within the asynchronous culture which exhibited an extended cell cycle time. In perhaps 20% of these cells oncogenic Ras and src were able to induce high levels of topo IIα protein expression. To explain these results, we predict that topo IIα levels are strictly tied to the level of a cellular factor which increases proportionally through the cell cycle. Our studies have conclusively shown, however, that a portion of the topo IIα promoter is responsive to Ras activity (6). In cells which remain viable but which are temporarily removed from active cycling this portion of the topo IIα promoter apparently becomes dominant and the cell responds by producing high levels of topo IIα protein. This fact might be responsible for the high levels of topo IIα protein within some tumors (10) which, like the slowly cycling NIH 3T3 cells, retain proliferative capacity but which are temporarily not passing actively through the cell cycle. Indeed, the slowly cycling cells in this study, like many tumor cells, were able to reenter the pool of actively cycling cells at any time, as indicated by the fact that (except in transformed cells) they were as likely as the general cellular population to divide normally following drug treatment.
The fact that neither the cell cycle nor oncogene action influenced topo IIα levels in cycling NIH 3T3 made it possible to study the effects of these two factors on drug toxicity, without concern for alterations in topo IIα levels. Previous work with synchronized cells pointed to a close correlation between S phase and drug toxicity (30). In these studies, drug treatment between two time-lapse observations allowed determination of the cell cycle phase at the time of drug treatment and the effects of this treatment on individual cells. The results of numerous studies established that etoposide was approximately threefold more toxic when added to cells in late S and G2 phases than when added in G1 phase. (Similar results were obtained with cells collected by mitotic detachment.) The exact cell cycle timing of this sensitive period was determined and compared to that for treatment with labeled thymidine. The highly selective killing of cells only in S phase following tritiated thymidine treatment was remarkable and emphasizes the validity of our procedure for determining cell cycle-related toxicity. Finally, the cell cycle toxicity pattern was specific to etoposide, as it was not seen with another toxic agent, 5-azacytidine. While a 10% increase in the levels of topo IIα through the cell cycle exactly correlated with the period of increased drug sensitivity (Fig. 3A), we are skeptical that this small difference could account for the dramatic differences in toxicity observed (Fig. 8A). Moreover, the alterations in drug sensitivity described below with oncogene-containing cells could not be accounted for by even slight alterations in topo IIα levels. We have not considered the potential role of topo IIβ in these studies, because the expression of this protein is not increased as cells enter a growing phase (35), even though there is evidence that it can serve as a target of anti-topo II drugs (10, 33).
While further study is required, we consider it significant that the topo IIα expression characteristics of slowly cycling cells were so different from those seen in actively proliferating cells. An effective antitumor treatment must target cells which are not actively cycling, since these make up the majority of all tumors. This study demonstrates that such cells, which are neither actively cycling nor quiescent, are likely to have unique biological characteristics. A study of such cells might be highly informative relative to the behavior of cells within a tumor.
When src-transformed cells were analyzed, it was clear, as previously reported, that oncogenic transformation increased the sensitivity to etoposide. Approximately equal killing was obtained in transformed cells with etoposide at 30% of the level used with untransformed cells. Interestingly, while the cell cycle effects of toxicity might have been diminished somewhat in transformed cells, transformed cells showed the same general pattern of increased toxicity late in the cell cycle as untransformed cells. Moreover, increased toxicity was observed in the slowly cycling cells transformed by oncogenic src, despite the fact that only a small number of these cells had increased topo IIα protein levels. To eliminate the possibility that clonal effects might have been responsible for the increased sensitivity with src-transformed NIH 3T3 cells, microinjection of oncogenic Ras was also shown to dramatically increase toxicity to etoposide. It is, therefore, concluded that the action of oncogenes from two separate classes is able to dramatically sensitize NIH 3T3 cells to etoposide toxicity, even though no changes in topo IIα levels result. This result is similar to that of a recent study of tumor cells where the presence of an oncogenic ras mutation correlated to sensitivity to anti-topo II drugs (27).
We conclude, therefore, that both the cell cycle and oncogene action play significant roles in determination of the effect of treatment with etoposide, without altering topo IIα expression levels. The explanation for this observation is not presently known. It is possible that these two physiological factors might influence the phosphorylation of topo IIα or its intermolecular associations. On the other hand, cell cycle and oncogene action might affect a molecule totally distinct from topo IIα, which plays a significant role in controlling drug toxicity. Further study will be required to answer these questions.
ACKNOWLEDGMENTS
We thank R. Ganopathi and G. Sa for helpful discussions and S. Kaufmann for a critical review of the manuscript.
This work was supported by PHS service grant GM52271.
REFERENCES
- 1.Adachi N, Kobayashi M, Koyama H. Cell cycle-dependent regulation of the mouse DNA topoisomerase IIalpha gene promoter. Biochem Biophys Res Commun. 1997;230:105–109. doi: 10.1006/bbrc.1996.5893. [DOI] [PubMed] [Google Scholar]
- 2.Boege F, Andersen A, Jensen S, Zeidler R, Kreipe H. Proliferation-associated nuclear antigen Ki-S1 is identical with topoisomerase II alpha. Delineation of a carboxy-terminal epitope with peptide antibodies. Am J Pathol. 1995;146:1302–1308. [PMC free article] [PubMed] [Google Scholar]
- 3.Brown M S, Holden J A, Rahn M P, Perkins S L. Immunohistochemical staining for DNA topoisomerase IIa in Hodgkin's disease. Am J Clin Pathol. 1998;109:39–44. doi: 10.1093/ajcp/109.1.39. [DOI] [PubMed] [Google Scholar]
- 4.Burden D A, Osheroff N. Mechanism of action of eukaryotic topoisomerase II and drugs targeted to the enzyme. Biochim Biophys Acta. 1998;1400:139–154. doi: 10.1016/s0167-4781(98)00132-8. [DOI] [PubMed] [Google Scholar]
- 5.Chen G, Shu J, Stacey D W. Oncogenic transformation potentiates apoptosis, S-phase arrest and stress-kinase activation by etoposide. Oncogene. 1997;15:1643–1651. doi: 10.1038/sj.onc.1201347. [DOI] [PubMed] [Google Scholar]
- 6.Chen G, Templeton D, Suttle D P, Stacey D W. Ras stimulates DNA topoisomerase IIa through MEK: a link between oncogenic signaling and a therapeutic target. Oncogene. 1999;18:7149–7160. doi: 10.1038/sj.onc.1203149. [DOI] [PubMed] [Google Scholar]
- 7.Chung T D, Drake F H, Tan K B, Per S R, Crooke S T, Mirabelli C K. Characterization and immunological identification of cDNA clones encoding two human DNA topoisomerase II isozymes. Proc Natl Acad Sci USA. 1989;86:9431–9435. doi: 10.1073/pnas.86.23.9431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clements J M, Craig S, Gearing A J, Hunter M G, Heyworth C M, Dexter T M, Lord B I. Biological and structural properties of MIP-1 alpha expressed in yeast. Cytokine. 1992;4:76–82. doi: 10.1016/1043-4666(92)90040-x. [DOI] [PubMed] [Google Scholar]
- 9.Davies S M, Robson C N, Davies S L, Hickson I D. Nuclear topoisomerase II levels correlate with the sensitivity of mammalian cells to intercalating agents and epipodophyllotoxins. J Biol Chem. 1988;263:17724–17729. [PubMed] [Google Scholar]
- 10.Dingemans A M, Pinedo H M, Giaccone G. Clinical resistance to topoisomerase-targeted drugs. Biochim Biophys Acta. 1998;1400:275–288. doi: 10.1016/s0167-4781(98)00141-9. [DOI] [PubMed] [Google Scholar]
- 11.Fogt F, Nikulasson S T, Holden J A, Alder S A, Hallgrimsson J, Jessup M J, O'Brien M J, Lavin P T, Goldman H. Topoisomerase II alpha expression in normal, inflammatory, and neoplastic conditions of the gastric and colonic mucosa. Mod Pathol. 1997;10:296–302. [PubMed] [Google Scholar]
- 12.Fry A M, Chresta C M, Davies S M, Walker M C, Harris A L, Hartley J A, Masters J R, Hickson I D. Relationship between topoisomerase II level and chemosensitivity in human tumor cell lines. Cancer Res. 1991;51:6592–6595. [PubMed] [Google Scholar]
- 13.Ganapathi R, Constantinou A, Kamath N, Dubyak G, Grabowski D, Krivacic K. Resistance to etoposide in human leukemia HL-60 cells: reduction in drug-induced DNA cleavage associated with hypophosphorylation of topoisomerase II phosphopeptides. Mol Pharmacol. 1996;50:243–248. [PubMed] [Google Scholar]
- 14.Giaccone G, Gazdar A F, Beck H, Zunino F, Capranico G. Multidrug sensitivity phenotype of human lung cancer cells associated with topoisomerase II expression. Cancer Res. 1992;52:1666–1674. [PubMed] [Google Scholar]
- 15.Goswami P C, Roti R J, Hunt C R. The cell cycle-coupled expression of topoisomerase IIα during S phase is regulated by mRNA stability and is disrupted by heat shock or ionizing radiation. Mol Cell Biol. 1996;16:1500–1508. doi: 10.1128/mcb.16.4.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grabowski D R, Dubyak G R, Rybicki L, Hidaka H, Ganapathi R. Tumor cell resistance to topoisomerase II poisons: role for intracellular free calcium in the sensitization by inhibitors or calcium-calmodulin-dependent enzymes. Biochem Pharmacol. 1998;56:345–349. doi: 10.1016/s0006-2952(98)00159-2. [DOI] [PubMed] [Google Scholar]
- 17.Guinee D J, Holden J A, Benfield J R, Woodward M L, Przygodzki R M, Fishback N F, Koss M N, Travis W D. Comparison of DNA topoisomerase II alpha expression in small cell and nonsmall cell carcinoma of the lung. In search of a mechanism of chemotherapeutic response. Cancer. 1996;78:729–735. doi: 10.1002/(SICI)1097-0142(19960815)78:4<729::AID-CNCR6>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 18.Hande K R. Clinical applications of anticancer drugs targeted to topoisomerase II. Biochim Biophys Acta. 1998;1400:173–184. doi: 10.1016/s0167-4781(98)00134-1. [DOI] [PubMed] [Google Scholar]
- 19.Hasegawa T, Isobe K, Nakashima I, Shimokata K. Higher expression of topoisomerase II in lung cancers than normal lung tissues: different expression pattern from topoisomerase I. Biochem Biophys Res Commun. 1993;195:409–414. doi: 10.1006/bbrc.1993.2058. [DOI] [PubMed] [Google Scholar]
- 20.Hitomi M, Stacey D W. Cellular ras and cyclin D1 are required during different cell cycle periods in cycling NIH 3T3 cells. Mol Cell Biol. 1999;19:4623–4632. doi: 10.1128/mcb.19.7.4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hitomi M, Stacey D W. Cyclin D1 production in cycling cells depends on ras in a cell-cycle-specific manner. Curr Biol. 1999;9:1075–1084. doi: 10.1016/s0960-9822(99)80476-x. [DOI] [PubMed] [Google Scholar]
- 22.Hochhauser D, Stanway C A, Harris A L, Hickson I D. Cloning and characterization of the 5′-flanking region of the human topoisomerase II alpha gene. J Biol Chem. 1992;267:18961–18965. [PubMed] [Google Scholar]
- 23.Isaacs R J, Davies S L, Sandri M I, Redwood C, Wells N J, Hickson I D. Physiological regulation of eukaryotic topoisomerase II. Biochim Biophys Acta. 1998;1400:121–137. doi: 10.1016/s0167-4781(98)00131-6. [DOI] [PubMed] [Google Scholar]
- 24.Isaacs R J, Harris A L, Hickson I D. Regulation of the human topoisomerase IIalpha gene promoter in confluence-arrested cells. J Biol Chem. 1996;271:16741–16747. doi: 10.1074/jbc.271.28.16741. [DOI] [PubMed] [Google Scholar]
- 25.Kaufmann S H. Cell death induced by topoisomerase-targeted drugs: more questions than answers. Biochim Biophys Acta. 1998;1400:195–211. doi: 10.1016/s0167-4781(98)00136-5. [DOI] [PubMed] [Google Scholar]
- 26.Kawanami K, Nakamura T, Ono M, Kusano T, Okada K, Kikuchi A, Adachi N, Kohno K, Higashi K, Kuwano M. Decreased DNA topoisomerase II alpha expression and cold-sensitive growth in a mouse mammary cancer cell line resistant to etoposide and doxorubicin. Oncol Res. 1996;8:197–206. [PubMed] [Google Scholar]
- 27.Koo H M, Gray-Goodrich M, Kohlhagen G, McWilliams M J, Jeffers M, Vaigro-Wolff A, Alvord W G, Monks A, Paull K D, Pommier Y, Vande Woude G F. The ras oncogene-mediated sensitization of human cells to topoisomerase II inhibitor-induced apoptosis. J Nat Cancer Inst. 1999;91:236–244. doi: 10.1093/jnci/91.3.236. [DOI] [PubMed] [Google Scholar]
- 28.Larsen A K, Skladanowski A. Cellular resistance to topoisomerase-targeted drugs: from drug uptake to cell death. Biochim Biophys Acta. 1998;1400:257–274. doi: 10.1016/s0167-4781(98)00140-7. [DOI] [PubMed] [Google Scholar]
- 29.Larsson O, Zetterberg A. Existence of a commitment program for mitosis in early G1 in tumour cells. Cell Prolif. 1995;28:33–43. doi: 10.1111/j.1365-2184.1995.tb00037.x. [DOI] [PubMed] [Google Scholar]
- 30.Markovits J, Pommier Y, Kerrigan D, Covey J M, Tilchen E J, Kohn K W. Topoisomerase II-mediated DNA breaks and cytotoxicity in relation to cell proliferation and the cell cycle in NIH 3T3 fibroblasts and L1210 leukemia cells. Cancer Res. 1987;47:2050–2055. [PubMed] [Google Scholar]
- 31.Mo Y Y, Ameiss K A, Beck W T. Overexpression of human DNA topoisomerase II alpha by fusion to enhanced green fluorescent protein. BioTechniques. 1998;25:1052–1057. doi: 10.2144/98256cr04. [DOI] [PubMed] [Google Scholar]
- 32.Negri C, Chiesa R, Cerino A, Bestagno M, Sala C, Zini N, Maraldi N M, Astaldi R G. Monoclonal antibodies to human DNA topoisomerase I and the two isoforms of DNA topoisomerase II: 170- and 180-kDa isozymes. Exp Cell Res. 1992;200:452–459. doi: 10.1016/0014-4827(92)90195-e. [DOI] [PubMed] [Google Scholar]
- 33.Perrin D, van Hille B, Hill B T. Differential sensitivities of recombinant human topoisomerase IIalpha and beta to various classes of topoisomerase II-interacting agents. Biochem Pharmacol. 1998;56:503–507. doi: 10.1016/s0006-2952(98)00082-3. [DOI] [PubMed] [Google Scholar]
- 34.Ponchio L, Conneally E, Eaves C. Quantitation of the quiescent fraction of long-term culture-initiating cells in normal human blood and marrow and the kinetics of their growth factor-stimulated entry into S-phase in vitro. Blood. 1995;86:3314–3321. [PubMed] [Google Scholar]
- 35.Prosperi E, Sala E, Negri C, Oliani C, Supino R, Astraldi R G, Bottiroli G. Topoisomerase II alpha and beta in human tumor cells grown in vitro and in vivo. Anticancer Res. 1992;12:2093–2099. [PubMed] [Google Scholar]
- 36.Rudolph P, MacGrogan G, Bonichon F, Frahm S O, de Mascarel I, Trojani M, Durand M, Avril A, Coindre J M, Parwaresch R. Prognostic significance of Ki-67 and topoisomerase IIalpha expression in infiltrating ductal carcinoma of the breast. A multivariate analysis of 863 cases. Breast Cancer Res Treat. 1999;55:61–71. doi: 10.1023/a:1006159016703. [DOI] [PubMed] [Google Scholar]
- 37.Stacey D W, Hitomi M, Kanovsky M, Gan L, Johnson E M. Cell cycle arrest and morphological alterations following microinjection of NIH3T3 cells with Pur alpha. Oncogene. 1999;18:4254–4261. doi: 10.1038/sj.onc.1202795. [DOI] [PubMed] [Google Scholar]
- 38.Towatari M, Adachi K, Marunouchi T, Saito H. Evidence for a critical role of DNA topoisomerase IIalpha in drug sensitivity revealed by inducible antisense RNA in a human leukaemia cell line. Br J Haematol. 1998;101:548–551. doi: 10.1046/j.1365-2141.1998.00713.x. [DOI] [PubMed] [Google Scholar]
- 39.Turley H, Comley M, Houlbrook S, Nozaki N, Kikuchi A, Hickson I D, Gatter K, Harris A L. The distribution and expression of the two isoforms of DNA topoisomerase II in normal and neoplastic human tissues. Br J Cancer. 1997;75:1340–1346. doi: 10.1038/bjc.1997.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vassetzky Y S, Alghisi G C, Gasser S M. DNA topoisomerase II mutations and resistance to anti-tumor drugs. Bioessays. 1995;17:767–774. doi: 10.1002/bies.950170906. [DOI] [PubMed] [Google Scholar]
- 41.Wang J C. DNA topoisomerases. Annu Rev Biochem. 1985;54:665–697. doi: 10.1146/annurev.bi.54.070185.003313. [DOI] [PubMed] [Google Scholar]
- 42.Woessner R D, Chung T D, Hofmann G A, Mattern M R, Mirabelli C K, Drake F H, Johnson R K. Differences between normal and ras-transformed NIH-3T3 cells in expression of the 170kD and 180kD forms of topoisomerase II. Cancer Res. 1990;50:2901–2908. [PubMed] [Google Scholar]
- 43.Woessner R D, Mattern M R, Mirabelli C K, Johnson R K, Drake F H. Proliferation- and cell cycle-dependent differences in expression of the 170 kilodalton and 180 kilodalton forms of topoisomerase II in NIH-3T3 cells. Cell Growth Differ. 1991;2:209–214. [PubMed] [Google Scholar]
- 44.Zhou Z, Zwelling L A, Kawakami Y, An T, Kobayashi K, Herzog C, Kleinerman E S. Adenovirus-mediated human topoisomerase II alpha gene transfer increases the sensitivity of etoposide-resistant human breast cancer cells. Cancer Res. 1999;59:4618–4624. [PubMed] [Google Scholar]