Abstract
A number of studies of Saccharomyces cerevisiae have revealed RAD51-independent recombination events. These include spontaneous and double-strand break-induced recombination between repeated sequences, and capture of a chromosome arm by break-induced replication. Although recombination between inverted repeats is considered to be a conservative intramolecular event, the lack of requirement for RAD51 suggests that repair can also occur by a nonconservative mechanism. We propose a model for RAD51-independent recombination by one-ended strand invasion coupled to DNA synthesis, followed by single-strand annealing. The Rad1/Rad10 endonuclease is required to trim intermediates formed during single-strand annealing and thus was expected to be required for RAD51-independent events by this model. Double-strand break repair between plasmid-borne inverted repeats was less efficient in rad1 rad51 double mutants than in rad1 and rad51 strains. In addition, repair events were delayed and frequently associated with plasmid loss. Furthermore, the repair products recovered from the rad1 rad51 strain were primarily in the crossover configuration, inconsistent with conservative models for mitotic double-strand break repair.
DNA double-strand breaks (DSBs) are potentially lethal lesions that occur spontaneously during normal cellular processes, such as replication, or by treatment of cells with DNA-damaging agents. DSBs serve to initiate a variety of recombination events, such as yeast mating-type switching (44), meiotic recombination (7, 49), and rearrangement of the T-cell receptor and immunoglobulin loci (38). Repair of DSBs can occur by either homologous recombination or end joining (19). Repair by homologous recombination requires a homologous donor duplex and is considered to be a high-fidelity process, whereas the homology independent end-joining pathway is potentially mutagenic. Although all eukaryotes studied have the capacity for both pathways, the choice between pathways is likely to be determined by the nature of the DNA ends, availability of a sequence homologue, and phase of the cell cycle (25).
The yeast mating-type-switching system provides a useful tool to study double-strand break repair (DSBR) in mitotic cells. The HO endonuclease can be regulated by expression from an inducible promoter and the site for cleavage inserted into different loci (16). Repair of the induced break can be monitored by cell survival or by physical or genetic methods (31, 55). In the natural context, mating-type switching is dependent on homologous recombination (1, 17, 27, 47). However, under certain circumstances, repair of an HO-induced DSB is independent of several homologous recombination genes, including RAD51, RAD54, RAD55, and RAD57. This was unexpected because RAD51 encodes a homologue of the bacterial RecA protein (42) and appears to be the only functional strand exchange protein in mitotic cells. Rad54, Rad55, and Rad57 are accessory proteins that stimulate the strand exchange activity of Rad51 in vitro (34, 51) and by genetic analysis function in the same recombination pathway (36). When the HO cut site is placed between directly repeated sequences, repair occurs predominantly by resection of the ends to produce 3′ single-stranded tails followed by annealing of complementary sequences, a process known as single-strand annealing (SSA). This reaction is dependent on RAD52 (45), partially dependent on RAD59 (3, 46), and independent of RAD51, RAD54, RAD55, and RAD57 (15). The nucleotide excision repair proteins, Rad1 and Rad10, function in SSA by removing the 3′ nonhomologous tails that are formed by strand annealing (14). In addition, the Rad1/10 nuclease is required during gene conversion to remove regions of heterology at the break site (12). There are no apparent defects in gene conversion in rad1 mutants when the recombining sequences are homologous. The requirement for Rad1/10 in SSA is less stringent if the HO cut site is made within repeats that are completely homologous (12). Similarly, the efficiency of gene conversion when a large heterology is present at only one side of the break is high in rad1 mutants (8). It has been suggested that another nuclease similar in specificity to Rad1/10 is functional when only one 3′ nonhomologous tail must be removed. The mismatch repair proteins, Msh2 and Msh3, are also required for SSA when the repeats are short and are thought to stabilize the branched structure for cleavage by Rad1/10 (48).
Evidence for aberrant repair in the absence of RAD51 has come from studies of DSBR in diploid cells. Repair of an HO-induced DSB was found to occur primarily by gene conversion in wild-type cells, but in rad51 mutants repair occurred by one-ended strand invasion to prime DNA synthesis to the end of the chromosome (26). This type of repair, known as break-induced replication (BIR), is a nonreciprocal recombination event that is RAD52 dependent and is likely to be important for telomere maintenance (24). The implication of these findings is that homology-dependent strand invasion can occur in the absence of RAD51. This is surprising because similar events occur in bacteria to restore collapsed replication forks and depend on RecA (21). The requirement for RAD52 in these reactions suggests that the annealing function of this protein is critical in formation of the strand invasion intermediate, which is presumably stabilized when DNA synthesis initiates from the invading 3′ end (28).
Inverted repeats have been used extensively to study the genetic control of recombination because sequences in inverted orientation cannot recombine by a simple SSA mechanism (9, 37). It has been assumed that inverted repeat recombination occurs by a conservative mechanism, such as predicted by the model of Szostak et al. (52) or the synthesis-dependent strand annealing (SDSA)/migrating D-loop models (11, 29). However, the lack of a requirement for critical recombination genes that are essential for recombination between single-copy sequences has raised the issue of whether inverted repeats also recombine by a nonconservative mode. Studies using Escherichia coli have shown that DSBR between inverted repeats can occur by a recA-independent mechanism that requires the recE and recT genes (53). Surprisingly, most of the recombinant products recovered were in the crossover configuration, and both the frequency and types of products were unaffected by mutation of the Holliday junction resolvase gene, ruvC (23). In yeast, recombination between inverted repeats is dependent on RAD52, but the requirement for other genes in the RAD52 epistasis group is less stringent. The rate of spontaneous mitotic recombination between chromosomal inverted repeats was reduced more than 100-fold in rad52 strains (2, 37) and reduced more than 50-fold when the inverted repeats were on a plasmid (9, 35). In contrast, mutation of RAD51 reduced spontaneous recombination of chromosomal inverted repeats only 5- to 10-fold (2, 37). HO-induced recombination between plasmid-borne inverted repeats was dependent on RAD52 but independent of RAD51 except when the donor sequences were transcriptionally silenced (47). Although HO-induced recombination of inverted repeat plasmids occurs efficiently in rad51 mutants, intermolecular plasmid gap repair is reduced more than 100-fold in the absence of RAD51 (4). The inconsistency between these assays led us to consider alternate models for DSB-induced recombination between inverted repeats.
MATERIALS AND METHODS
Yeast strains and plasmids.
Saccharomyces cerevisiae JKM146 (Δho Δhml::ADE1 MATa-inc Δhmr::ADE1 ade1 leu2-3,112 lys5 trp1::hisG ura3-52) contains deletions of the HMR and HML loci, a noncleavable MATa-inc allele, and a GAL::HO fusion inserted at the chromosomal ADE3 locus (32). JKM146 and the rad1 derivative of JKM146, YFP103 (32), were kindly provided by J. Haber. A rad51 derivative of JKM146, LSY901, was generated by microhomologous one-step replacement with a PCR fragment designed to replace the RAD51 open reading frame (ORF) with the TRP1 ORF (5). Trp+ transformants were tested for sensitivity to γ irradiation and the disruption of RAD51 confirmed by PCR analysis. A rad1 derivative of LSY901, LSY902, was generated by one-step replacement with a DNA fragment derived from plasmid pL962 (gift of R. Keil) (39). Plasmid pFP122 contains inverted repeats of the E. coli lacZ gene, the URA3 gene for selection in yeast, and CEN4 and ARS1 elements for stable maintenance as an episome (32). One copy of lacZ contains an insertion of a 36-bp synthetic HO cut site; the other copy of lacZ contains an insertion of 36 bp, which differs from the HO cut site by one substitution and cannot be cleaved by HO (inc HO). Plasmid pJFL33 contains CEN and ARS elements, the selectable markers LEU2 and URA3, and two copies of the lacZ gene in direct repeat (12). One copy of the lacZ gene contains a 117-bp insertion from the MAT locus with a functional HO cut site; the other copy of lacZ contains a 117-bp insertion with the inc allele that is resistant to cleavage by HO. Yeast transformation was performed by the lithium acetate method (41).
Media and growth conditions.
Rich (YEPD) and synthetic complete (SC) media were made as described by Sherman et al. (41). YEP-glycerol contains 2% (wt/vol) glycerol instead of glucose as a carbon source, and YEP-galactose contains 2% galactose (wt/vol) instead of glucose as a carbon source. Cultures were grown at 30°C unless otherwise stated.
Measurement of DSBR efficiency by plasmid retention.
Each of the four strains was transformed with plasmid pFP122 and grown to saturation in SC lacking uracil (SC−Ura). This culture was used to inoculate 50 ml of YEP-glycerol to a starting concentration of about 106 cells per ml. These cultures were grown overnight to a density of 1 × 107 to 3 × 107 cells per ml. Dilutions of the cultures were then plated in parallel on YEP-glucose and YEP-galactose plates. After 3 days of growth, the colonies were replica plated to SC−Ura plates to score retention of the plasmid. For strain LSY902, replica plating was performed after 4 to 5 days of growth on YEP-galactose plates. Repair efficiency was calculated as the ratio of plasmid retention on YEP-galactose plates versus plasmid retention on YEP-glucose plates (32). The plasmid retention on YEP-glucose plates was not significantly different between strains, usually ranging between 70 and 85%. Because many of the Ura+ colonies obtained following growth on YEP-galactose contained mixed populations of parental and crossover plasmids that may be due to delayed induction of HO resulting in a round of cell division, the above procedure was changed in order to segregate plasmids. The 50-ml YEP-glycerol cultures were induced for HO expression by the addition of galactose to a final concentration of 2% and incubated for 24 h. Dilutions of cells were then plated onto YEP-glucose and incubated for 3 days prior to replica plating to SC−Ura plates.
Physical analysis of DSBR.
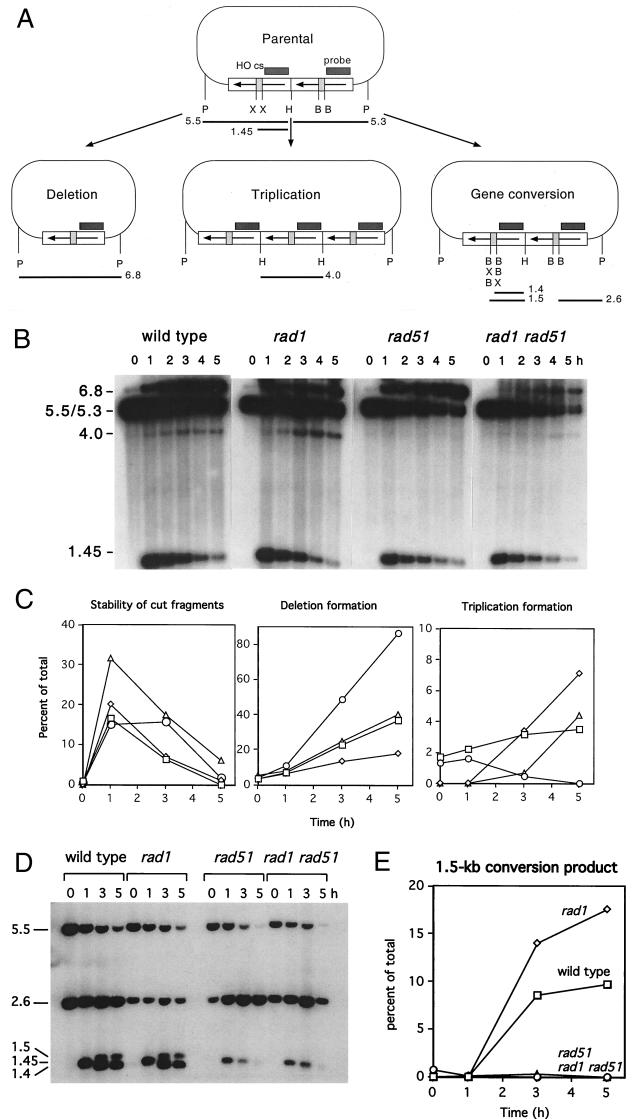
Strains transformed with plasmid pFP122 were grown as described above except that 300 ml of YEP-glycerol was used for the preinduction growth. Cultures were grown to a density of 2 × 107 to 4 × 107 per ml, and then 50-ml aliquots were removed for the 0-h time point. Galactose was added to a final concentration of 2% to induce expression of HO. Fifty-milliliter samples were removed at 1-h intervals up to 5 h following addition of galactose. DNA was extracted from these samples and digested with PstI, and fragments were separated by gel electrophoresis through 0.6% agarose. DNA fragments were transferred to nylon membranes and probed with lacZ sequences. To analyze individual repair events, DNA was extracted from 5-ml cultures derived from individual Ura+ colonies obtained after growth on YEP-galactose or after 24-h liquid induction of HO. These samples were analyzed by restriction digestion with PstI and Southern blotting to distinguish between parental (gene conversion) and crossover fragments. Repair of plasmid pJFL33 was monitored by a similar procedure except that DNA was digested with PstI and HindIII to monitor deletion and triplication products and with PstI, HindIII, and BclI to detect gene conversion products. Plasmid DNA levels were quantitated by stripping blots and reprobing with a radiolabeled MET17 DNA fragment corresponding to a unique chromosomal sequence and PhosphorImaging.
RESULTS
Experimental rationale.
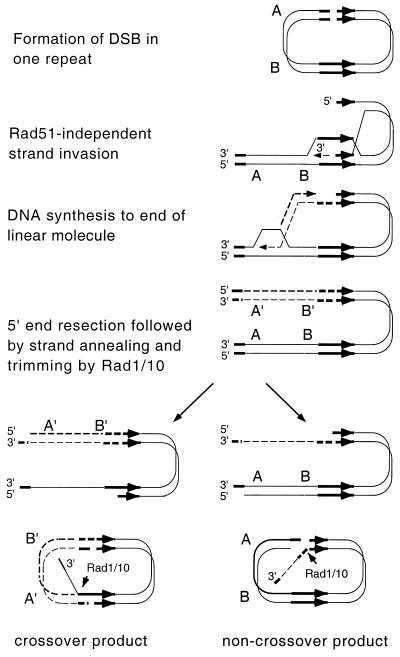
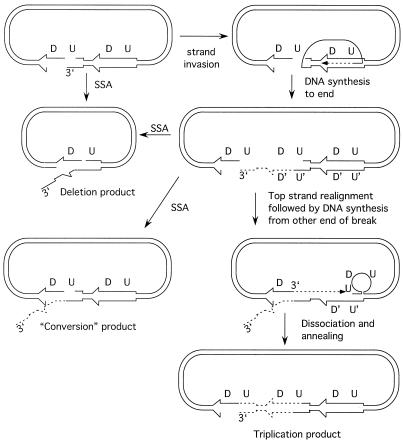
Previous studies have documented one-ended invasion to prime DNA synthesis (BIR) and also SSA in the absence of RAD51 (15, 26). Based on these observations, we considered an alternate model for the repair of DSBs within inverted repeat plasmids that might account for the high levels of recombinational repair in rad51 mutants (Fig. 1). The model proposes repair of the DSB by one-ended invasion, DNA synthesis to the other end of the plasmid, and resection of the ends to produce single-stranded tails which can then anneal through regions of homology to restore the parental configuration or form a crossover product. Although the products of the reaction have the appearance of a conservative recombination reaction, the processes used, BIR and SSA, are generally considered to be nonconservative modes of repair. It is important to note that by this model inversion of sequences between the repeats can occur without formation and resolution of a Holliday junction. Seven genes, RAD52, RAD59, RAD1, RAD10, MSH2, MSH3, and RFA1, are known to participate in strand annealing between repeated sequences. RAD52, RAD59, and RFA1 are also required for DSB-induced gene conversion (54), but RAD1 and RAD10 are not required for gene conversion if the repeats are completely homologous. However, the role of RAD1 and RAD10 in SSA is less stringent when the break is made within homologous repeats instead of between them, or when one of the repeats has heterologies flanking the HO cut site (12, 14). It would be predicted that a rad1 rad51 double mutant would be defective in DSBR of an inverted repeat plasmid if repair includes a strand annealing step to produce an intermediate with 3′ nonhomologous tails.
FIG. 1.
A model for RAD51-independent recombination between inverted repeats. Following introduction of a DSB in one of the repeats, one end is resected to produce a 3′ single-stranded tail, which invades the other repeat. DNA synthesis is primed from the invading end and proceeds to the end of the plasmid, coupled with lagging-strand synthesis. The sequences at the end of the linear intermediate have homology with the internal repeats. Resection and strand annealing can produce parental or inversion products. The inverted repeats are shown by thick arrows, newly synthesized DNA is indicated by dashed lines, and sequences between the repeats are designated A and B.
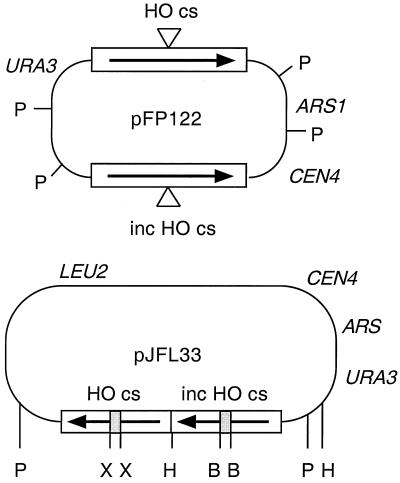
To test this hypothesis, we constructed a set of isogenic strains that contain no chromosomal HO cut sites, an integrated copy of a GAL-HO fusion gene, and null alleles of RAD1 and/or RAD51. Plasmid pFP122, which contains inverted repeats of lacZ, one disrupted by an HO cut site and the other containing an almost identical insertion except for a nucleotide substitution that prevents cleavage by HO, was used as a substrate (32) (Fig. 2). As the cut lacZ recipient has complete homology to the donor except at one nucleotide, repair by gene conversion should be independent of RAD1 function (12).
FIG. 2.
Plasmid substrates. Plasmid pFP122 contains two copies of the lacZ gene oriented as inverted repeats. Both copies are interrupted by an insertion of 36 bp containing either the HO cut site (cs) or a point mutation to prevent HO cleavage, inc HO cut site. The plasmid contains the URA3 gene for selection in yeast and ARS and CEN elements for stable replication as an episome. After induction of HO, repair can occur by gene conversion to form noncrossover or crossover products, which can be distinguished by restriction digestion with PstI (P). Failure to repair results in plasmid loss and a Ura− phenotype. Plasmid pJFL33 contains direct repeats of lacZ interrupted by an insertion of 117 bp with the HO or inc HO cut site, URA3 and LEU2 genes, and ARS and CEN elements for stable maintenance. Restriction sites for BclI (B), HindIII (H), PstI (P), and XhoI (X) are shown.
Reduced efficiency of repair in rad51 and rad1 rad51 strains.
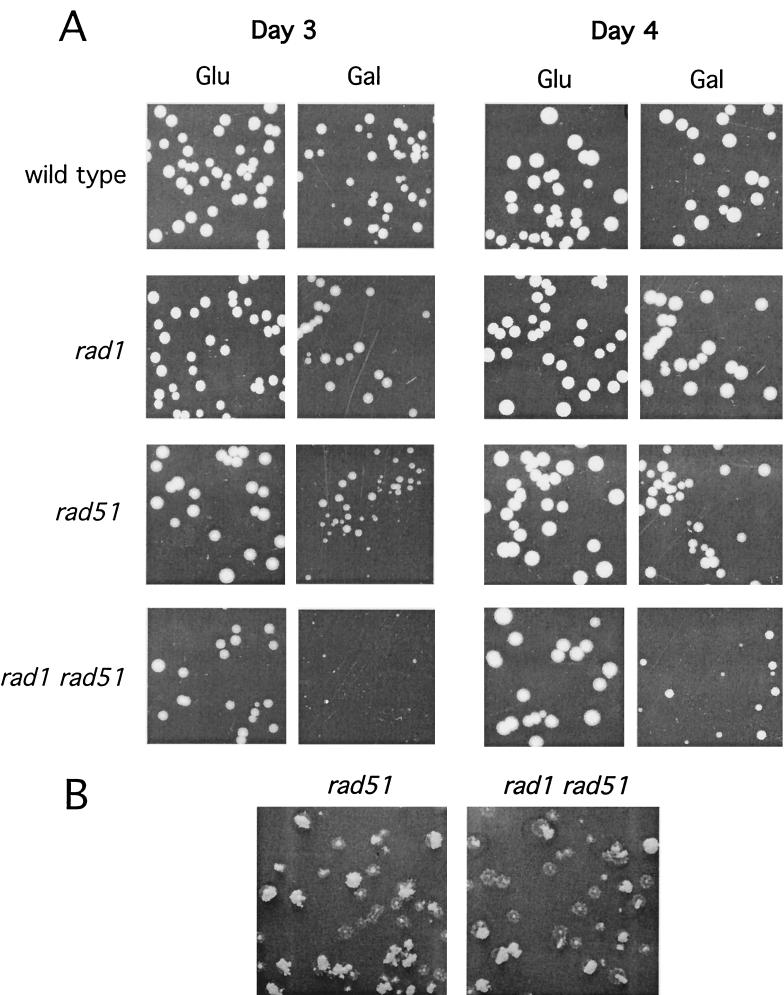
To measure the efficiency of repair, cultures of each strain containing pFP122 were plated on medium containing glucose to repress transcription of HO or on YEP-galactose medium to induce expression of HO. After 3 days of growth, the colonies were replica plated to SC−Ura to determine the percentage that retained the plasmid. Retention of the plasmid after growth on YEP-galactose is indicative of repair; Ura− colonies derive from cells that lacked that plasmid prior to plating or fail to repair the plasmid. Plasmid loss prior to plating is controlled by parallel plating on YEP-glucose. Thus, the ratio of plasmid retention after growth on YEP-galactose to plasmid retention after growth on YEP-glucose provides a measure of repair efficiency. In wild-type and rad1 strains, repair efficiency was about 71 to 74% (Table 1). The colonies were 2 to 3 mm in diameter after growth for 3 days on YEP-galactose (Fig. 3); when colonies were replica plated to SC−Ura, very few sectored colonies were found. In rad51 strains, repair efficiency was reduced to 50% and more than half of the Ura+ colonies were sectored, with half or more of the colony containing Ura− cells (Fig. 3). Repair efficiency was further reduced in the rad1 rad51 double mutant. Most of the colonies from the rad1 rad51 strain grew slowly on YEP-galactose (Fig. 3). The untransformed rad1 rad51 strain grew normally on YEP-galactose, indicating that the slow-growth defect was a consequence of the DSB present on pFP122 (data not shown). Only 21% of the colonies from the rad1 rad51 strain grown on YEP-galactose were Ura+ and most were sectored, often with only one-eighth of the colony growing on SC−Ura medium (Fig. 3). These results indicate a reduced efficiency of repair in rad51 and rad1 rad51 strains. The slow growth suggests delayed repair of the DSB on pFP122, which signals cell cycle arrest. RAD9-dependent cell cycle arrest has previously been shown for plasmid and chromosomal HO-induced DSBs (6, 40). The growth defect conferred by growth on galactose-containing medium reflects delayed repair; repaired Ura+ products obtained after growth on galactose showed normal growth. Sectored colonies could arise by perpetuation of the broken plasmid through one or more cell cycles or could reflect a nonreciprocal mode of repair in which more than one parental plasmid is used to generate a single repaired product. Nonreciprocal repair could occur if the cells have more than one copy of the CEN plasmid, such as in the G2 phase of the cell cycle, or if the plasmid copy number is higher than one in G1.
TABLE 1.
Repair efficiency and distribution of Ura+ products
| Relevant genotype | Repair efficiencya | No. of Ura+ coloniesb
|
|||||
|---|---|---|---|---|---|---|---|
| Total analyzed | Noncrossover | Crossover | Mixedc | Aberrantd | % Crossoversb | ||
| 3-day plate induction | |||||||
| RAD1 RAD51 | 77.3 ± 9.3 | 50 (58) | 36 (44) | 1 (9) | 8 | 5 | 2 (16) |
| rad1 RAD51 | 74.2 ± 8.1 | 50 (67) | 25 (42) | 3 (20) | 17 | 4 | 6 (30) |
| RAD1 rad51 | 50.4 ± 8.3 | 34 (37) | 19 (22) | 9 (12) | 3 | 3 | 26 (32) |
| rad1 rad51 | 21.4 ± 7.3 | 52 (60) | 14 (22) | 29 (37) | 8 | 1 | 55 (62) |
| 24-h liquid induction | |||||||
| RAD1 RAD51 | 54.3 | 18 | 17 | 1 | 0 | 0 | 6 |
| rad1 RAD51 | 54.5 | 18 (19) | 15 (16) | 1 (2) | 1 | 1 | 6 (11) |
| RAD1 rad51 | 25.8 | 18 | 12 | 6 | 0 | 0 | 33 |
| rad1 rad51 | 10.8 | 18 (19) | 6 | 9 (10) | 1 | 2 | 50 (53) |
Percentage of Ura+ colonies after HO induction (see Materials and Methods).
Numbers in parentheses count the mixed populations as two one-crossover and one noncrossover product.
Mixed population of crossover and noncrossover products.
Plasmids with restriction fragments inconsistent with either crossover or noncrossover products.
FIG. 3.
Delayed plasmid repair and plasmid loss in rad51 mutants. (A) Strains containing the inverted-repeat plasmid were plated onto solid medium containing either glucose or galactose, and colony size was analyzed after 3 or 4 days of growth. (B) Colonies were replica plated to SC−Ura after 4 to 5 days of growth on YEP-galactose plates to measure plasmid retention and colony sectoring.
Aberrant mode of repair in the rad1 rad51 strain.
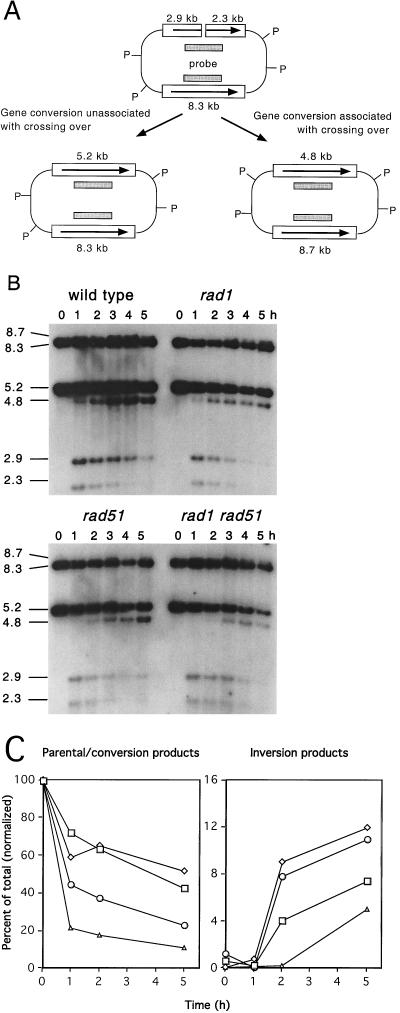
DNA was isolated from independent Ura+ colonies from each of the strains after growth on YEP-galactose plates and digested with PstI to distinguish between parental and crossover configurations of the recombination products (Table 1). Seventy-two percent of the products obtained from the wild-type strain had the parental configuration of PstI fragments, and 16% contained mixtures of parental and crossover bands. Because gene conversion unassociated with crossing over generates PstI fragments of the same size as the unrecombined plasmid, the high number of noncrossover plasmids could represent plasmids that had escaped HO cleavage. Four independent Ura+ colonies containing apparent gene conversion products were induced with galactose in liquid medium; samples of cells before and after a 1-h induction were taken; DNA was isolated and analyzed by Southern blotting. All four were resistant to digestion with HO, indicating that the HO site had been converted during the initial induction on YEP-galactose plates. Thus, plasmids obtained after growth on YEP-galactose undergo efficient induction of HO and are repaired primarily to form noncrossover products in the wild-type strain. The rad1 strain also showed a high percentage of mixed products and fewer noncrossover products than the wild-type strain. Fewer mixed products were recovered from the rad51 and rad1 rad51 double mutant, and a greater proportion of crossover products were obtained, particularly from the double mutant. The observation of more than 50% crossing over in the double mutant suggests that the residual repair does not occur by the normal mechanisms.
The mixed products might be due (i) to cells that were induced in the S or G2 phase of the cell cycle and reflect independent repair of two individual plasmids or (ii) to inefficient induction of HO endonuclease so that a round of cell division occurred on the YEP-galactose plates prior to HO induction. To segregate recombinant plasmids, the procedure was modified to include a 24-h liquid induction with galactose prior to plating on YEP-glucose and subsequent replica plating to SC−Ura (see Materials and Methods). Fewer mixed products were obtained using this protocol, and Ura+ plasmids recovered from the wild-type strain showed an even lower association of crossing over. However, the trend of lower plasmid survival and increased levels of inversion was maintained in the rad51 strains. In all of the strains we recovered some products with restriction fragments inconsistent with either crossover or noncrossover recombinants; these were classified as aberrant (8).
Kinetics of repair of the inverted-repeat plasmid.
To determine whether there was an alteration in the kinetics of repair in the mutant strains, HO was induced in liquid cultures, and samples of cells were removed at various times after HO induction for Southern blot analysis (Fig. 4). Because the PstI digestion products indicative of gene conversion unassociated with crossing over are the same size as the parental fragments, only crossover products can be identified by this analysis. HO was efficiently induced in all of the strains. The kinetics of repair were similar in the rad1 and wild-type strains, with crossover products detectable 1 h after induction of HO and reaching a value of 7 to 12% (normalized for plasmid loss) at 5 h. The PstI fragments corresponding to the parental or conversion products were of similar intensity through the time course. Repair was delayed in the rad51 and rad1 rad51 strains, with crossover products appearing 2 to 3 h after HO induction. There was a decrease in the intensity of the PstI fragments corresponding to the parental/conversion bands in both rad51 strains, which indicates reduced levels of gene conversion and/or increased levels of plasmid loss (Fig. 4). Quantitation of the plasmid sequences relative to chromosomal sequences confirmed loss of the plasmid in the rad1 rad51 strain following HO induction, and values of products were normalized to account for plasmid loss. Consistent with the analysis of Ura+ recombinants, there was a bias in recovery of crossover fragments relative to the conversion/parental band in the rad51 and rad1 rad51 strains. Although the 8.7- and 4.8-kb PstI fragments are assumed to arise by a completed crossover event, fragments of the same size would be generated by strand invasion and DNA synthesis to the end of the linearized plasmid. Consistent with the delay in repair, HO cut fragments accumulated in the rad51 strain and to even higher levels in rad1 rad51 double mutant. Five hours after HO induction, cut fragments represented 10% of the total plasmid DNA in the rad1 rad51 strain, compared with 2.2% in wild-type, 3.3% in rad1, and 3.8% in rad51 strains.
FIG. 4.
Repair is delayed in rad51 mutants. (A) Substrate and products detected by PstI (P) digestion. (B) Kinetic analysis of repair in each strain. Samples were removed prior to HO induction (0 h) and at hourly intervals after HO induction. DNA samples were digested with PstI and fragments were separated on 0.6% agarose gels prior to transfer. Sizes are indicated in kilobases. (C) Hybridized filters were analyzed using a Molecular Dynamics PhosphorImager, and the recombination products were normalized to unique chromosomal sequences to account for plasmid loss. □, wild type; ◊, rad1 mutant; ○, rad51 mutant; ▵, rad1 rad51 double mutant.
RAD51 is required for gene conversion.
To further test the requirement for RAD51 in strand invasion and gene conversion, we used a plasmid similar to the one described above except containing direct repeats of the lacZ gene (pJFL33). Previous results from the Haber lab have shown that RAD1 is not required for DSB-induced conversion of this plasmid (12). An aberrant product, containing three copies of the lacZ gene, was recovered from repair of pJFL33 in the rad1 strain and thought to arise by one-ended strand invasion, replication to the end of the cut plasmid, and then strand annealing, a model similar to the one we propose for repair of inverted-repeat plasmids (Fig. 1). Ivanov et al. (15) used a similar direct-repeat plasmid, but without the HO-inc insertion within the donor allele, to show that RAD51 is not required for SSA. In their experiments, gene conversion was reduced 20-fold in the rad51 strain, which they attributed to competition between the inefficient conversion pathway and the more efficient SSA pathway. To circumvent problems in interpretation arising from highly efficient SSA, we monitored repair of pJFL33 in rad1 rad51 strains to determine whether gene conversion events could occur in the plasmid context.
Kinetic analysis of DSBR was performed as described above for the inverted-repeat plasmid. DNA samples prior to and after HO induction were digested with HindIII and PstI to detect deletion products formed by SSA and with BclI, HindIII, and PstI to detect gene conversion products. SSA to generate the 6.8-kb deletion product occurred with high efficiency in the wild-type strain and the rad51 mutant (Fig. 5). The deletion product resulting from SSA was reduced twofold in the rad1 strain but was present at the wild-type level in the rad1 rad51 double mutant (40% of the plasmid DNA). A DNA fragment of about 4 kb was produced in the wild-type strain and most likely corresponds to the triplication product observed by Fishman-Lobell and Haber (12) (Fig. 6). This novel species was even more abundant in the rad1 mutant, was barely detectable in the rad51 mutant, but was present in the rad1 rad51 double mutant. Conversion of the restriction site polymorphisms that flank the HO cut site during repair results in the formation of two novel species of 1.4 and 1.5 kb (Fig. 5). The 1.5-kb conversion product was clearly apparent in the rad1 and wild-type strains but was reduced at least 10-fold in both rad51 and rad1 rad51 strains. The DNA fragment produced by HO cleavage is 1.45 kb, and processing of this fragment by nucleases changes the mobility so that it migrates at a similar position as the 1.4-kb conversion product. Quantitation was performed only on the 1.5-kb product and therefore is likely to underestimate the conversion defect in the rad51 strains. By both plating assays (data not shown) and quantitation of Southern blots, there was no defect in plasmid retention following HO induction in the wild-type, rad1, and rad51 strains but a threefold loss in the rad1 rad51 double mutant.
FIG. 5.
Kinetic analysis of repair of plasmid pJFL33. (A) pJFL33 contains direct repeats of lacZ interrupted by a 117-bp insertion of the HO cut site (cs) or inc HO cut site. Repair of the HO-induced DSB can occur by SSA to delete one copy of lacZ, by gene conversion unassociated with crossing over, or by formation of a triplication of lacZ. Each of these products can be distinguished by restriction digestion with PstI (P), HindIII (H), and BclI (B). HO cleavage produces a fragment of 1.45 kb, the SSA product produces a 6.8-kb PstI fragment, and the triplication product generates a 4-kb HindIII fragment. The conversion product produces PstI/HindIII fragments of the same size as the parental plasmid but can be monitored by the appearance of 1.4- and 1.5-kb BclI/HindIII fragments generated by conversion of restriction site polymorphisms flanking the HO cut site. X, XhoI. (B) DNA samples were digested with PstI and HindIII to detect the deletion and triplication products. Sizes are indicated in kilobases. (C) The percentages of total plasmid DNA represented by the HO cut fragments, deletion products, and triplication products were quantitated by phosphorimaging and shown graphically. (D) DNA samples were digested with PstI, HindIII, and BclI to detect the conversion products. Sizes are indicated in kilobases. (E) Quantitation of the 1.5-kb conversion product shown in panel D. □, wild type; ◊, rad1 mutant; ○, rad51 mutant; ▵, rad1 rad51 double mutant.
FIG. 6.
Model for BIR/SSA within the direct-repeat plasmid (12). One-ended invasion of the unbroken lacZ gene to prime DNA synthesis to the end of the linear molecule results in duplication of the lacZ gene. Depending on how the complementary sequences pair, a deletion or apparent conversion can result. If the upstream (U) sequence of the broken repeat pairs with the upstream (U′) sequence of the unbroken repeat, and the newly synthesized downstream (D′) sequences pair with the broken repeat (D), then DNA synthesis could initiate from the other side of the break to create an additional repeat. Dissociation and realignment are required to generate the triplication product. The deletion and conversion products require clipping of a 3′ heterologous tail, whereas the triplication product has no tails to be removed and thus is favored in rad1 strains.
DISCUSSION
Conservative and nonconservative repair pathways.
The repair of DSBs by homologous recombination is generally considered to be a high-fidelity event in which two parental duplexes generate two recombinant duplexes (conservative recombination). Conservative recombination is predicted by both the DSBR and SDSA models (11, 29, 52). Both models predict strand invasion by the 3′ single-stranded tail formed on one side of the broken duplex to prime DNA synthesis from the donor duplex. After limited DNA synthesis, the invading strand is extruded or the branched structure is cleaved by a junction resolvase. Nonconservative repair events are characterized by a net loss of DNA. For example, during SSA, two DNA duplexes are used to generate a single recombinant duplex. Here, we present evidence for a RAD51-independent pathway of repair that generates products similar to those predicted from a conservative reaction but incorporates features of nonconservative recombination.
Inverted-repeat recombination.
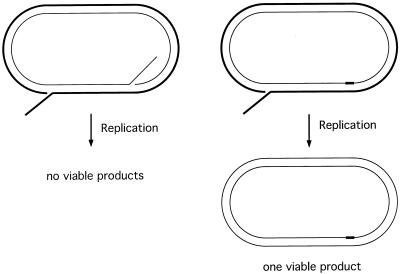
Recombination between sequences oriented as inverted repeats is considered to be a conservative event because simple strand annealing is not envisioned to give rise to a viable product. However, spontaneous recombination between chromosomal inverted repeats is reduced only 4- to 10-fold, and DSB-induced recombination between plasmid-borne repeats is reduced only 2-fold, by mutation of RAD51. Because intermolecular recombination is strongly dependent on RAD51 (1, 4, 10), we propose that recombination events between repeated sequences must occur by an unusual mechanism that does not normally operate within unique sequences. A model that could account for some of the recombination events between inverted repeats in rad51 mutants is presented (4) (Fig. 1). This model predicts strand invasion as envisioned by the DSBR and SDSA models, but DNA synthesis primed from the invading strand proceeds to the end of the DNA molecule (the other side of the break). The linear intermediate, which has short regions of shared homology between the terminii and internal sequences, can then repair by SSA to produce either crossover or noncrossover products. We found an increase in the number of crossover recombinants in rad51 strains, consistent with the model. These events are unlikely to occur in RAD51 cells because a rad1 mutation does not reduce the efficiency of inverted-repeat recombination. In contrast, a rad1 mutation decreased inverted-repeat recombination in rad51 strains and also increased the number of crossover products recovered. Although the overall efficiency of repair in the rad51 strain was reduced only two- to threefold by the rad1 mutation, the kinetics of repair were delayed. We assume the delay is caused by a limiting step, such as removal of the 3′ nonhomologous tail predicted to form during strand annealing. As shown in Fig. 5, the efficiency of SSA between direct repeats is reduced only twofold in the rad1 strain, consistent with the defect observed using the inverted-repeat plasmid. When there is homology between donor and recipient repeats, the annealed intermediate will have only one 3′ heterologous tail to be removed (Fig. 7). In contrast, the intermediate formed when the donor and recipient repeats lack homology flanking the cut site will have 3′ heterologous tails on both sides of the annealed intermediate. The requirement for RAD1 in SSA and gene conversion appears to be far less stringent when only one heterologous tail has to be removed (8, 12). This could be explained if ligation of the nicked strands on one side of the annealed intermediate occurs and the plasmid then undergoes replication (Fig. 7). The discontinuous strand containing the 3′ flap might not be recovered, but the continuous strand should be completely replicated, yielding a viable product. Such a mechanism might contribute to the plasmid loss observed in the rad1 rad51 double mutant.
FIG. 7.
Plasmid recovery through replication bypass of the Rad1 trimming step. The annealed intermediate formed by SSA when the HO cut site is within heterologous sequences is predicted to have two 3′ heterologous tails flanking the annealed region. This molecule is predicted to form inviable products if replicated. When SSA occurs between repeats that are completely homologous, only one 3′ heterologous tail is predicted to form. If the other strand is ligated and the plasmid undergoes replication, one viable daughter should be generated.
In RAD51 cells, we imagine the extent of DNA synthesis is limited to the region of shared homology between the repeats, and RAD51 may be important for this step as well as the initial homology search and strand invasion. A true strand exchange might limit DNA synthesis by coupling synthesis to strand exchange via branch migration or by coupling the second strand invasion to the first one. The model described (Fig. 1) could also apply to chromosomal repeats, but invasion from only one of the two ends would result in a successful recombination event. The synergistic decrease in the rate of spontaneous recombination between chromosomal inverted repeats in a rad1 rad51 mutant is consistent with the proposed model (36).
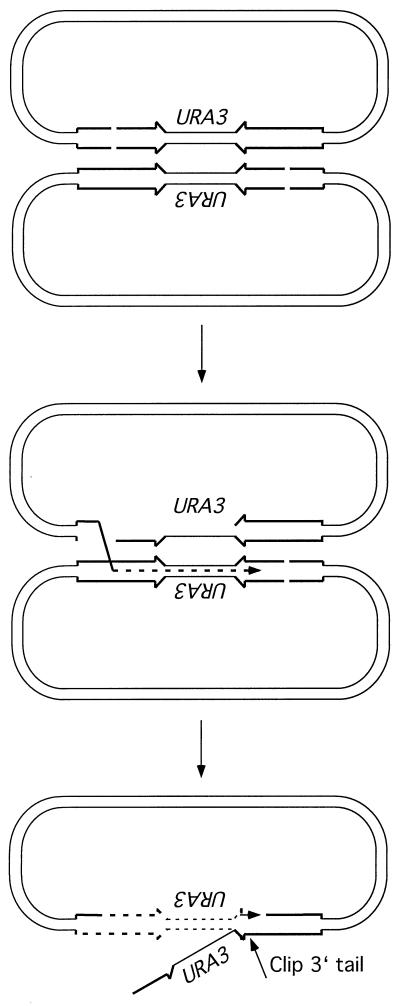
The observation of 62% crossover products recovered from the rad1 rad51 strain is inconsistent with the DSBR and SDSA models but is more consistent with BIR, which predicts 100% crossing over (Table 1). One mechanism that could account for the high levels of crossover events is intermolecular recombination between misaligned sister chromatids. If two plasmids paired with the lacZ repeats in an antiparallel configuration, then BIR initiated from one repeat could replicate through the intervening DNA, forming an apparent inversion (Fig. 8). After displacement of the invading strand, pairing could occur between the newly synthesized strand and the other lacZ repeat. In this way, the integrity of one of the cut plasmids would be restored and an apparent crossover would result. Although this model predicts a RAD1-dependent trimming step to remove the 3′ nonhomologous tail, it is clear from the analysis of direct-repeat recombination that the efficiency of SSA is reduced only twofold in the rad1 mutant (Fig. 5) (12).
FIG. 8.
Intermolecular recombination to produce an inversion product. If two plasmids pair in an antiparallel configuration, BIR initiated from one cut repeat, coupled with lagging-strand synthesis, will duplicate the intervening sequences to form an inversion. DNA synthesis will be blocked at the break in the second plasmid, leading to strand displacement, annealing, and removal of the 3′ heterologous tail, resulting in formation of an inversion product.
Direct-repeat recombination by BIR and SSA?
Kinetic analysis of DSBR of the direct-repeat plasmid indicated low levels of conversion in both rad51 and rad1 rad51 strains. However, of the Ura+ products recovered from the rad1 rad51 strain, 17% were apparent conversion products (data not shown). Because no conversion products were detected 5 h after HO induction, we assume this pathway, or mismatch repair, is highly inefficient in the rad1 rad51 double mutant. Although it is possible that conversions arise by the same pathway as operates in wild-type cells, another possibility is BIR coupled to SSA as shown in Fig. 6. The linear intermediate generated by BIR from the direct-repeat plasmid could undergo annealing in three ways to generate all three classes of products. Although we consider this to be an aberrant pathway that might occur only in rad51 mutants, there is some evidence for unusual events in mammalian cells that are best explained by BIR extending beyond homology followed by end joining (18).
The role of RAD1 in SSA.
The substrates used in this study contain an inc HO cut site within the donor locus and so have only a single base pair mismatch between donor and recipient repeats. SSA of the direct-repeat plasmid, pJFL33, containing the inc mutation is reduced only two- to three-fold in rad1 strains (Fig. 5) (12). Thus, plasmid survival in rad1 rad51 strains could be due to the residual level of SSA that still occurs in the absence of RAD1. When the donor cassette lacks homology to the recipient (by insertion of an HO cut site within the recipient), or if a break is made within unique sequences between direct repeats, then SSA is reduced more than 15-fold in rad1 strains. The putative annealed intermediate formed in these two cases is different. When there is homology between donor and recipient repeats, the annealed intermediate will have only one 3′ heterologous tail to be removed, whereas the intermediate formed when the break is between the repeats, or between breaks that lack homology with the donor, will have 3′ heterologous tails on both sides of the annealed intermediate. As described above, the requirement for RAD1 appears to be far less stringent when only one heterologous tail has to be removed.
Role of RAD52 in RAD51-independent recombination.
RAD52 is required for virtually every mitotic recombination event in S. cerevisiae. All of the RAD51-independent events require RAD52 and show a partial requirement for RAD59, which encodes a homologue of Rad52. The model in Fig. 1 predicts two steps at which Rad52 could act. Rad52 could be involved in the initial strand invasion step and/or to promote strand annealing of the resected intermediate. Although we cannot easily distinguish between these steps, we would argue that Rad52 is involved in strand invasion because BIR requires Rad52 but does not involve SSA (26).
In vitro studies have shown that Rad52 catalyzes the annealing of complementary single-stranded DNAs and stimulates strand exchange by the Rad51 protein (28, 30, 43, 50). The annealing function of Rad52 is likely to be biologically relevant, as Rad52 is required for SSA in vivo (45). Human Rad52 is reported to promote D-loop formation in vitro, but the reaction is very inefficient (22). It is possible that accessory proteins are required for strand invasion by Rad52, or in vivo strand invasion might occur by Rad52-catalyzed strand annealing between the 3′ single-stranded tail and transiently unwound donor duplex DNA formed during transcription or replication. We predict that recombination between transcriptionally silent regions will show a stronger requirement for Rad51 because the sequences are inaccessible to Rad52-promoted annealing. This is consistent with the requirement for RAD51 in mating-type switching and DSB-induced recombination between HMR and MAT oriented as inverted repeats on plasmids (47).
Similarities between the Rad52 and RecET/lambda Red pathways.
Inverted-repeat plasmids have been extensively used to study the genetic control of recombination in E. coli (20). DSB-induced recombination is recA dependent but does not require recA in a recBC sbcA strain background (53). The sbcA mutation activates the recET operon of a cryptic prophage present in some E. coli strains. recT encodes an annealing protein with functional similarity to Redβ (33) and Rad52 that can also catalyze strand exchange in vitro (13). We suggest that recA-independent recombination of inverted-repeat plasmids in E. coli could occur by a similar mechanism to the one we propose for RAD51-independent recombination and require an annealing protein, such as RecT or lambda β. Surprisingly, plasmid inverted-repeat recombination mediated by the RecET pathway in E. coli is independent of ruvC, suggesting that Holliday junction processing is not required; this observation is consistent with the model presented.
In summary, we present evidence in support of BIR coupled with SSA to repair DSBs within repeated sequences. These events may not occur in RAD51 strains but occur with low efficiency in rad51 mutants and can account for the recombination observed in these strains.
ACKNOWLEDGMENTS
We thank James Haber for providing the plasmids and strains used for this study and for helpful discussions. We thank Elizabeth Morgan and Sylvie Moreau for help in scanning yeast colonies and phosphorimaging, and we thank members of the Symington laboratory and W. K. Holloman for discussions and critical reading of the manuscript.
This work was supported by Public Health Service grant NIH GM54099 from the National Institutes of Health.
REFERENCES
- 1.Aboussekhra A, Chanet R, Adjiri A, Fabre F. Semidominant suppressors of Srs2 helicase mutations of Saccharomyces cerevisiae map in the RAD51 gene, whose sequence predicts a protein with similarities to procaryotic RecA proteins. Mol Cell Biol. 1992;12:3224–3234. doi: 10.1128/mcb.12.7.3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aguilera A. Genetic evidence for different RAD52-dependent intrachromosomal recombination pathways in Saccharomyces cerevisiae. Curr Genet. 1995;27:298–305. doi: 10.1007/BF00352096. [DOI] [PubMed] [Google Scholar]
- 3.Bai Y, Davis A P, Symington L S. A novel allele of RAD52 that causes severe DNA repair and recombination deficiencies only in the absence of RAD51 or RAD59. Genetics. 1999;153:1117–1130. doi: 10.1093/genetics/153.3.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartsch S, Kang L E, Symington L S. RAD51 is required for the repair of plasmid double-stranded DNA gaps from either plasmid or chromosomal templates. Mol Cell Biol. 2000;20:1194–1205. doi: 10.1128/mcb.20.4.1194-1205.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bennett C B, Lewis A L, Baldwin K K, Resnick M A. Lethality induced by a single site-specific double-strand break in a dispensable yeast plasmid. Proc Natl Acad Sci USA. 1993;90:5613–5617. doi: 10.1073/pnas.90.12.5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao L, Alani E, Kleckner N. A pathway for generation and processing of double-strand breaks during meiotic recombination in S. cerevisiae. Cell. 1990;61:1089–1101. doi: 10.1016/0092-8674(90)90072-m. [DOI] [PubMed] [Google Scholar]
- 8.Colaiacovo M P, Paques F, Haber J E. Removal of one nonhomologous DNA end during gene conversion by a RAD1- and MSH2-independent pathway. Genetics. 1999;151:1409–1423. doi: 10.1093/genetics/151.4.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dornfeld K J, Livingston D M. Plasmid recombination in a rad52 mutant of Saccharomyces cerevisiae. Genetics. 1992;131:261–276. doi: 10.1093/genetics/131.2.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elias-Arnanz M, Firmenich A A, Berg P. Saccharomyces cerevisiae mutants defective in plasmid-chromosome recombination. Mol Gen Genet. 1996;252:530–538. doi: 10.1007/BF02172399. [DOI] [PubMed] [Google Scholar]
- 11.Ferguson D O, Holloman W K. Recombinational repair of gaps in DNA is asymmetric in Ustilago maydis and can be explained by a migrating D-loop model. Proc Natl Acad Sci USA. 1996;93:5419–5424. doi: 10.1073/pnas.93.11.5419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fishman-Lobell J, Haber J E. Removal of nonhomologous DNA ends in double-strand break recombination: the role of the yeast ultraviolet repair gene RAD1. Science. 1992;258:480–484. doi: 10.1126/science.1411547. [DOI] [PubMed] [Google Scholar]
- 13.Hall S D, Kolodner R D. Homologous pairing and strand exchange promoted by the Escherichia coli RecT protein. Proc Natl Acad Sci USA. 1994;91:3205–3209. doi: 10.1073/pnas.91.8.3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ivanov E L, Haber J E. RAD1 and RAD10, but not other excision repair genes, are required for double-strand break-induced recombination in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:2245–2251. doi: 10.1128/mcb.15.4.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ivanov E L, Sugawara N, Fishman-Lobell J, Haber J E. Genetic requirements for the single-strand annealing pathway of double-strand break repair in Saccharomyces cerevisiae. Genetics. 1996;142:693–704. doi: 10.1093/genetics/142.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jensen R E, Herskowitz I. Directionality and regulation of cassette substitution in yeast. Cold Spring Harbor Symp Quant Biol. 1984;49:97–104. doi: 10.1101/sqb.1984.049.01.013. [DOI] [PubMed] [Google Scholar]
- 17.Johnson R, Symington L S. Functional differences and interactions among the yeast RecA homologues Rad51, Rad55, and Rad57. Mol Cell Biol. 1995;15:4843–4850. doi: 10.1128/mcb.15.9.4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson R D, Jasin M. Sister chromatid gene conversion is a prominent double-strand break repair pathway in mammalian cells. EMBO J. 2000;19:3398–3407. doi: 10.1093/emboj/19.13.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanaar R, Hoeijmakers J H, van Gent D C. Molecular mechanisms of DNA double strand break repair. Trends Cell Biol. 1998;8:483–489. doi: 10.1016/s0962-8924(98)01383-x. [DOI] [PubMed] [Google Scholar]
- 20.Kobayashi I, Takahashi N. Double-stranded gap repair of DNA by gene conversion in Escherichia coli. Genetics. 1988;119:751–757. doi: 10.1093/genetics/119.4.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kogoma T. Recombination by replication. Cell. 1996;85:625–627. doi: 10.1016/s0092-8674(00)81229-5. [DOI] [PubMed] [Google Scholar]
- 22.Kurumizaka H, Aihara H, Kagawa W, Shibata T, Yokoyama S. Human Rad51 amino acid residues required for Rad52 binding. J Mol Biol. 1999;291:537–548. doi: 10.1006/jmbi.1999.2950. [DOI] [PubMed] [Google Scholar]
- 23.Kusano K, Sunohara Y, Takahashi N, Yoshikura H, Kobayashi I. DNA double-strand break repair: genetic determinants of flanking crossing-over. Proc Natl Acad Sci USA. 1994;91:1173–1177. doi: 10.1073/pnas.91.3.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Le S, Moore J K, Haber J E, Greider C W. RAD50 and RAD51 define two pathways that collaborate to maintain telomeres in the absence of telomerase. Genetics. 1999;152:143–152. doi: 10.1093/genetics/152.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lewis L K, Kirchner J M, Resnick M A. Requirement for end-joining and checkpoint functions, but not RAD52-mediated recombination, after EcoRI endonuclease cleavage of Saccharomyces cerevisiae DNA. Mol Cell Biol. 1998;18:1891–1902. doi: 10.1128/mcb.18.4.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Malkova A, Ivanov E L, Haber J E. Double-strand break repair in the absence of RAD51 in yeast: a possible role for break-induced DNA replication. Proc Natl Acad Sci USA. 1996;93:7131–7136. doi: 10.1073/pnas.93.14.7131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malone R E, Esposito R E. The RAD52 gene is required for homothallic interconversion of mating type and spontaneous mitotic recombination in yeast. Proc Natl Acad Sci USA. 1980;77:503–507. doi: 10.1073/pnas.77.1.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mortensen U H, Bendixen C, Sunjevaric I, Rothstein R. DNA strand annealing is promoted by the yeast Rad52 protein. Proc Natl Acad Sci USA. 1996;93:10729–10734. doi: 10.1073/pnas.93.20.10729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nassif N, Penney J, Pal S, Engels W R, Gloor G B. Efficient copying of nonhomologous sequences from ectopic sites via P-element-induced gap repair. Mol Cell Biol. 1994;14:1613–1625. doi: 10.1128/mcb.14.3.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.New J H, Sugiyama T, Zaitseva E, Kowalczykowski S C. Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein A. Nature. 1998;391:407–410. doi: 10.1038/34950. [DOI] [PubMed] [Google Scholar]
- 31.Nickoloff J A, Singer J D, Hoekstra M F, Heffron F. Double-strand breaks stimulate alternative mechanisms of recombination repair. J Mol Biol. 1989;207:527–541. doi: 10.1016/0022-2836(89)90462-2. [DOI] [PubMed] [Google Scholar]
- 32.Paques F, Haber J E. Two pathways for removal of nonhomologous DNA ends during double-strand break repair in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:6765–6771. doi: 10.1128/mcb.17.11.6765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Passy S I, Yu X, Li Z, Radding C M, Egelman E H. Rings and filaments of beta protein from bacteriophage lambda suggest a superfamily of recombination proteins. Proc Natl Acad Sci USA. 1999;96:4279–4284. doi: 10.1073/pnas.96.8.4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Petukhova G, Stratton S, Sung P. Catalysis of homologous DNA pairing by yeast Rad51 and Rad54 proteins. Nature. 1998;393:91–94. doi: 10.1038/30037. [DOI] [PubMed] [Google Scholar]
- 35.Prado F, Aguilera A. Role of reciprocal exchange, one-ended invasion crossover and single-strand annealing on inverted and direct repeat recombination in yeast: different requirements for the RAD1, RAD10, and RAD52 genes. Genetics. 1995;139:109–123. doi: 10.1093/genetics/139.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rattray A J, Symington L S. Multiple pathways for homologous recombination in Saccharomyces cerevisiae. Genetics. 1995;139:45–56. doi: 10.1093/genetics/139.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rattray A J, Symington L S. Use of a chromosomal inverted repeat to demonstrate that the RAD51 and RAD52 genes of Saccharomyces cerevisiae have different roles in mitotic recombination. Genetics. 1994;138:587–595. doi: 10.1093/genetics/138.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roth D B, Nakajima P B, Menetski J P, Bosma M J, Gellert M. V(D)J recombination in mouse thymocytes: double-strand breaks near T cell receptor delta rearrangement signals. Cell. 1992;69:41–53. doi: 10.1016/0092-8674(92)90117-u. [DOI] [PubMed] [Google Scholar]
- 39.Rothstein R J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- 40.Sandell L L, Zakian V A. Loss of a yeast telomere: arrest, recovery, and chromosome loss. Cell. 1993;75:729–739. doi: 10.1016/0092-8674(93)90493-a. [DOI] [PubMed] [Google Scholar]
- 41.Sherman F, Fink G, Hicks J. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1986. [Google Scholar]
- 42.Shinohara A, Ogawa H, Ogawa T. Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell. 1992;69:457–470. doi: 10.1016/0092-8674(92)90447-k. [DOI] [PubMed] [Google Scholar]
- 43.Shinohara A, Ogawa T. Stimulation by Rad52 of yeast Rad51-mediated recombination. Nature. 1998;391:404–407. doi: 10.1038/34943. [DOI] [PubMed] [Google Scholar]
- 44.Strathern J N, Klar A J, Hicks J B, Abraham J A, Ivy J M, Nasmyth K A, McGill C. Homothallic switching of yeast mating type cassettes is initiated by a double-stranded cut in the MAT locus. Cell. 1982;31:183–192. doi: 10.1016/0092-8674(82)90418-4. [DOI] [PubMed] [Google Scholar]
- 45.Sugawara N, Haber J E. Characterization of double-strand break-induced recombination: homology requirements and single-stranded DNA formation. Mol Cell Biol. 1992;12:563–575. doi: 10.1128/mcb.12.2.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sugawara N, Ira G, Haber J E. DNA length dependence of the single-strand annealing pathway and the role of Saccharomyces cerevisiae RAD59 in double-strand break repair. Mol Cell Biol. 2000;20:5300–5309. doi: 10.1128/mcb.20.14.5300-5309.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sugawara N, Ivanov E L, Fishman-Lobell J, Ray B L, Wu X, Haber J E. DNA structure-dependent requirements for yeast RAD genes in gene conversion. Nature. 1995;373:84–86. doi: 10.1038/373084a0. [DOI] [PubMed] [Google Scholar]
- 48.Sugawara N, Paques F, Colaiacovo M, Haber J E. Role of Saccharomyces cerevisiae Msh2 and Msh3 repair proteins in double-strand break-induced recombination. Proc Natl Acad Sci USA. 1997;94:9214–9219. doi: 10.1073/pnas.94.17.9214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun H, Treco D, Schultes N P, Szostak J W. Double-strand breaks at an initiation site for meiotic gene conversion. Nature. 1989;338:87–90. doi: 10.1038/338087a0. [DOI] [PubMed] [Google Scholar]
- 50.Sung P. Function of yeast Rad52 protein as a mediator between replication protein A and the Rad51 recombinase. J Biol Chem. 1997;272:28194–28197. doi: 10.1074/jbc.272.45.28194. [DOI] [PubMed] [Google Scholar]
- 51.Sung P. Yeast Rad55 and Rad57 proteins form a heterodimer that functions with replication protein A to promote DNA strand exchange by Rad51 recombinase. Genes Dev. 1997;11:1111–1121. doi: 10.1101/gad.11.9.1111. [DOI] [PubMed] [Google Scholar]
- 52.Szostak J W, Orr-Weaver T L, Rothstein R J, Stahl F W. The double-strand-break repair model for recombination. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- 53.Takahashi N K, Kusano K, Yokochi T, Kitamura Y, Yoshikura H, Kobayashi I. Genetic analysis of double-strand break repair in Escherichia coli. J Bacteriol. 1993;175:5176–5185. doi: 10.1128/jb.175.16.5176-5185.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Umezu K, Sugawara N, Chen C, Haber J E, Kolodner R D. Genetic analysis of yeast RPA1 reveals its multiple functions in DNA metabolism. Genetics. 1998;148:989–1005. doi: 10.1093/genetics/148.3.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.White C I, Haber J E. Intermediates of recombination during mating type switching in Saccharomyces cerevisiae. EMBO J. 1990;9:663–673. doi: 10.1002/j.1460-2075.1990.tb08158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]