Abstract
To investigate the effect of nucleosomes on nucleotide excision repair in humans, we prepared a mononucleosome containing a (6-4) photoproduct in the nucleosome core and examined its repair with the reconstituted human excision nuclease system and with cell extracts. Nucleosomal DNA is repaired at a rate of about 10% of that for naked DNA in both systems. These results are in agreement with in vivo data showing a considerably slower rate of repair of overall genomic DNA relative to that for transcriptionally active DNA. Furthermore, our results indicate that the first-order packing of DNA in nucleosomes is a primary determinant of slow repair of DNA in chromatin.
Nucleotide excision repair is a general repair system for removing virtually all types of lesions from DNA and is the sole repair mechanism for eliminating bulky base adducts (54). The repair reaction is initiated by dual incisions bracketing the lesion which release damage in the form of 24- to 32-nucleotide-long oligomers in humans (20) and in Saccharomyces cerevisiae (16). In a biochemically defined human system, 15 polypeptides in six repair factors, XPA, RPA, XPC, TFIIH, XPG, and XPF-ERCC1, are necessary and sufficient to excise the damage from naked DNA (43, 44). However, the physiological substrate for nucleotide excision is chromatin, and hence it is conceivable that in addition to the six general repair factors other enzymes which make lesions in chromatin accessible to the excision nuclease proper play an important role in genomic DNA repair.
In vivo studies with both yeast and mammalian cells have revealed that the organization of DNA within chromatin has a strong negative effect on its repairability by the nucleotide excision repair system (37, 68). Similarly, in vivo studies have shown that transcribed DNA is repaired preferentially (4), and since transcription is invariably associated with significant chromatin remodeling (70, 81), it has been inferred that the various activators, coactivators, and remodeling and accessibility factors which play essential roles in transcription may play equally prominent roles in excision repair (37). The availability of a defined human excision nuclease system has made it possible to investigate the effect of chromatin structure on DNA repair. To do this, we used a mononucleosome as the substrate for human excision nuclease. We find that the nucleosome severely inhibits damage recognition and excision by both the purified human excision nuclease and mammalian cell extracts (CEs).
MATERIALS AND METHODS
DNA substrate.
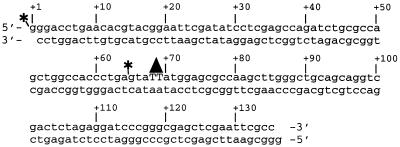
The structure of the 136-bp DNA substrate containing a unique T(6-4)T photoproduct is schematically shown in Fig. 1. The substrate DNA was prepared as described previously (45, 61). For footprinting experiments and to detect 5′ incision, the DNA was terminally radiolabeled with 32P at the 5′ end of the damage-containing strand. To detect excision (dual incision) and for electrophoretic mobility shift experiments, the substrate was internally radiolabeled with 32P at the fourth phosphodiester bond 5′ to the T(6-4)T photoproduct on the same strand.
FIG. 1.
Substrate for excision repair. The substrate was constructed by ligating six oligonucleotides. The resulting 136-bp duplex contains (6-4) photoproducts at positions +68 and +69 (triangle). The substrate was radiolabeled at either one of two sites (asterisks) by phosphorylating one of the six oligonucleotides with 32P. Internally labeled substrate was used for excision assays, and terminally labeled substrate was used to detect 5′ incision.
Proteins.
Histones from HeLa S3 cells were prepared using hydroxylapatite chromatography and a salt gradient according to published methods (32). Briefly, chromatin was prepared from Triton X-100-treated nuclei by sonication and adsorbed onto hydroxylapatite in buffer A (10 mM Tris-HCl [pH 8.0], 1 mM EDTA) containing 25 mM NaCl. Histones were eluted with 0.65, 0.93, and 2.0 M NaCl in buffer A. Fractions containing histones H2A, H2B, H3, and H4 were identified by electrophoresis on a 15% sodium dodecyl sulfate-polyacrylamide gel and used for nucleosome reconstitution.
Cell extracts (CEs) were prepared from HeLa S3 cells or AA8 Chinese hamster ovary (CHO) cells as described previously (34).
Human repair proteins (His)6-XPA, RPA, XPC-HHR23B, XPG, XPF-ERCC1, and TFIIH were prepared as described previously (3, 36, 43, 44, 53). All the repair factors except TFIIH were purified as recombinant proteins.
Nucleosome reconstitution.
Nucleosome reconstitution onto the 136-bp DNA substrate containing a unique T(6-4)T photoproduct with histone proteins H2A, H2B, H3, and H4 was carried out as described previously (31). Briefly, 1 pmol of DNA substrate was mixed with 20 μg of histone proteins in 50 μl of reconstitution buffer (10 mM Tris-HCl [pH 7.4] 1 mM EDTA, 0.2 mM phenylmethylsulfonyl fluoride) containing 1 M NaCl and incubated at 25°C for 30 min, followed by incubation for another 30 min at 4°C. The mixtures were then dialyzed against 0.6 M NaCl in reconstitution buffer for 12 h at 4°C. Finally, reaction mixtures were dialyzed against 0.05 M NaCl in reconstitution buffer for 4 h at 4°C. After reconstitution, the mononucleosome was purified away from unassembled free DNA by centrifugation through an 11-ml, 5 to 25% sucrose gradient in 10 mM HEPES-KOH (pH 7.9)–1 mM EDTA–0.1% NP-40 using an SW41 rotor (25,000 rpm, 18 h, 4°C) according to published methods (15). Reconstitution products and sucrose gradient fractions were analyzed by nondenaturing polyacrylamide gel electrophoresis (6% polyacrylamide; 1× Tris-borate-EDTA [TBE]) as described previously (31). Fractions containing mononucleosomes were pooled, dialyzed against 10 mM Tris-HCl (pH 7.4)–1 mM EDTA–50 mM NaCl, and used for excision assays and electrophoretic mobility shift experiments.
Hydroxyl radical footprinting.
Hydroxyl radical footprinting assays were carried out according to published methods (18, 78) with reconstituted mononucleosome substrate without the sucrose gradient purification step. Approximately 10 fmol of terminally radiolabeled mononucleosomes or naked DNA was treated with H2O2-iron(II)-EDTA (31). Reactions were quenched by adding 5% glycerol, and reaction mixtures were immediately applied to nondenaturing gel (6% polyacrylamide; 1× TBE) to separate the nucleosome and free-DNA substrate. The radiolabeled substrate DNAs in the nucleosome and naked-DNA bands were purified separately from the gel and analyzed by denaturing gel electrophoresis (6% polyacrylamide; 2× TBE).
Excision repair assays.
CEs from HeLa S3 cells and CHO AA8 cells or a human reconstituted system were used to measure excision or 5′ incision with the 136-bp DNA substrates in the form of a mononucleosome or free DNA as described previously (46, 52). For repair assays with CEs, 1.5 fmol of substrate DNA was incubated with 50 μg of CE at 30°C in 25 μl of excision repair buffer (32 mM HEPES-KOH [pH 7.9], 64 mM KCl, 6.4 mM MgCl2, 0.24 mM EDTA, 0.8 mM dithiothreitol, 2 mM ATP, 0.2 mg of bovine serum albumin/ml, 5.5% glycerol).
For the repair assays with the human reconstituted system, purified repair proteins, 50 ng of (His)6-XPA, 300 ng of RPA, 10 ng of XPC-HHR23B, 150 ng of TFIIH, 10 ng of XPG, and 20 ng of XPF-ERCC1, were used in a 25-μl excision repair buffer. The reaction products were purified by phenol-chloroform extraction and analyzed on a denaturing gel (8% polyacrylamide; 2× TBE). The efficiencies of excision were determined by measuring the levels of radioactivity in the bands of excised products and unexcised substrate with PhosphorImager and the ImageQuant system (Molecular Dynamics) and plotted as percentages of excision. For the repair assays, including both terminally and internally radiolabeled substrates, 0.75 fmol of each substrate was added to the reaction mixtures.
Eleotrophoretic mobility shift assays.
Substrate DNA (0.75 fmol) was incubated with (His)6-XPA, RPA, or XPC-HHR23B in 12.5 μl of excision repair buffer at 30°C for 15 min. Reaction mixtures were loaded directly onto nondenaturing gels (5% polyacrylamide; 0.5× TBE). Levels of radioactivity of the bands were measured with PhosphorImager and the ImageQuant system, and the reduction of the radioactivity of the unbound-DNA band relative to that of the control (no protein) reactions was plotted as the percentage of bound DNA. For supershift assays with an antibody, a mouse anti-histone monoclonal antibody (Chemicon International) was used. Substrates were first incubated with XPC, and the antibody was then added. Reaction mixtures were analyzed on a nondenaturing 4% polyacrylamide gel.
RESULTS
Preparation of substrate.
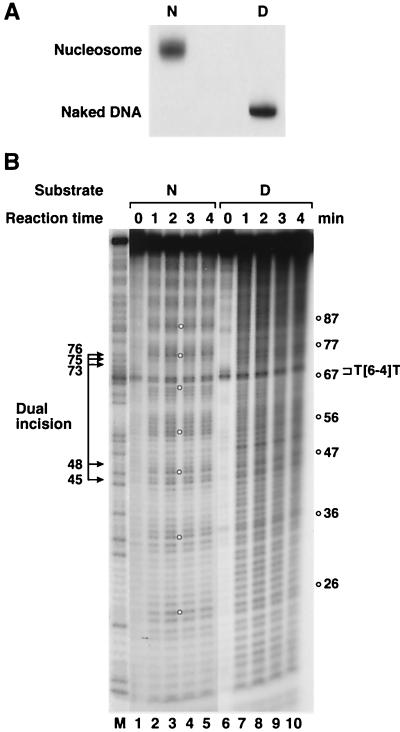
The substrate was prepared by mixing a synthetic 136-bp duplex with core histones isolated from HeLa cells. The DNA duplex was assembled by ligation of six partly overlapping oligomers and contained a T(6-4)T photoproduct in the middle of one strand and a 32P radiolabel either at the 5′ terminus of the same strand or at the fourth phosphodiester bond 5′ to the photoproduct. The sequence of the DNA substrate is shown in Fig. 1. The duplex was mixed with core histones under standard conditions for forming nucleosomes (31). When the mixture was analyzed on a nondenaturing polyacrylamide gel, about 80% of the DNA was found to be in nucleosomes. To obtain nucleosomes free of naked DNA, the nucleosomes were further purified by sucrose gradient velocity sedimentation (Fig. 2A).
FIG. 2.
Hydroxyl radical footprinting of a (6-4) photoproduct-containing mononucleosome. (A) Analysis of purified mononucleosomes on a nondenaturing 6% polyacrylamide gel. The nucleosomes were reconstituted onto a 136-bp (6-4) photoproduct-containing substrate and purified by 5 to 25% sucrose gradient sedimentation. N, nucleosome; D, naked DNA. (B) Hydroxyl radical footprint. Cleavage patterns on a 6% denaturing polyacrylamide gel are shown for nucleosomes (N; lanes 1 to 5) and naked DNA (D; lanes 6 to 10). Nucleosome or naked-DNA substrate terminally labeled at the 5′ end of the damage-containing strand was treated with hydroxyl radicals for the indicated times. The numbers to the right indicate the positions of maximum cleavage. The position of the (6-4) photoproduct and the major sites of dual incisions are also shown. The naked-DNA lanes were underexposed to the X-ray film so as to obtain intensity comparable to that of nucleosomal DNA. Lane M, Maxam-Gilbert G ladder of the DNA fragment.
To ascertain that the DNA-protein complex we obtained with core histones and the 136-bp duplex was indeed a mononucleosome, we performed hydroxyl radical footprinting on the complex. The cleavage pattern of DNA associated with histones exhibited a ca. 10-bp periodicity, consistent with a bona fide nucleosome complex, whereas the naked DNA was cleaved essentially evenly (Fig. 2B). The footprint also showed that the minor groove at the (6-4) photoproduct is positioned away from the histone surface, as evidenced by the high level of radical cleavage at and around the (6-4) photoproduct.
Effect of the nucleosome on the excision nuclease.
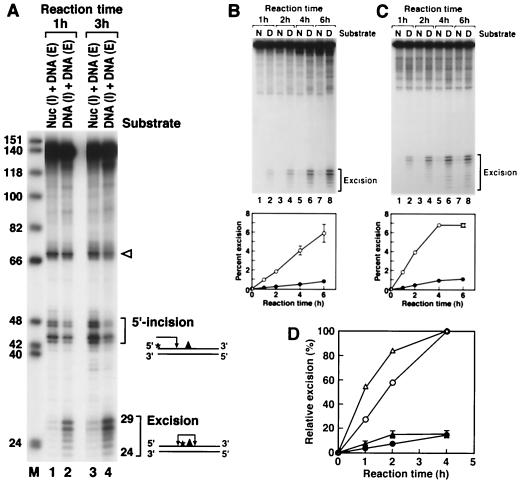
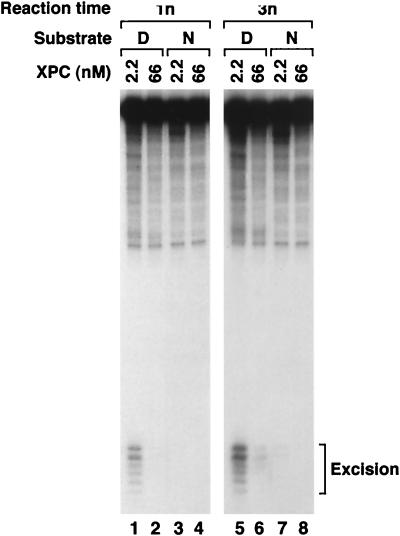
When the nucleosome containing the single (6-4) photoproduct was used as a substrate for the reconstituted human excision nuclease, a drastic inhibition of excision relative to that for the naked DNA substrate was observed (Fig. 3). We were concerned that this inhibition might have been caused by unknown contaminants present in the histone preparation which inhibited the excision nuclease nonspecifically. To address this possibility, nucleosomes containing internally labeled DNA and naked DNA with a terminal label were mixed and treated with the reconstituted excision nuclease. The labeling scheme makes it possible to detect the reaction products arising from both substrates simultaneously in a single reaction and a single lane of a polyacrylamide gel. As is apparent in Fig. 3A, even in a mixture of naked DNA and nucleosomes the excision of damage from nucleosomal DNA is specifically and severely depressed. Hence it appears that DNA in nucleosomes is a poor substrate for the human excision nuclease. Interestingly, however, the nucleosome does not change the sites of incision because the excision products exhibit the same pattern whether the substrate is naked DNA or a nucleosome (Fig. 3A).
FIG. 3.
Effect of the nucleosome on nucleotide excision repair. (A) Inhibition of dual incision by the nucleosome tested by the incision assay and excision assay. Mixtures of end-labeled (E) or internally labeled (I) naked DNA (DNA) and internally labeled nucleosomal DNA (Nuc) were digested with the human excision nuclease, and the products were separated on an 8% denaturing polyacrylamide gel. The reaction mixtures contained 0.75 fmol each of end-labeled and internally labeled substrates. The same internally labeled DNA preparation was used either as naked DNA or in the form of nucleosome DNA in a mixture with end-labeled naked DNA. The sources of the reaction products are shown schematically at the right. Arrowhead, cleavage at the site of the (6-4) photoproduct resulting from excessive handling of the end-labeled DNA during substrate purification. The percentages of incision and excision in the various reactions were as follows. Lane 1, 0.9 (excision) and 5.4% (incision); lane 2, 5.5 (excision) and 4.0% (incision); lane 3, 2.3 (excision) and 11.6% (incision); lane 4, 12.8 (excision) and 6.2% (incision). (B) Kinetics of excision of (6-4) photoproducts from naked DNA and the nucleosome by reconstituted human excision repair nuclease. (Top) Reaction kinetics autoradiogram. Internally labeled (6-4) substrates in the form of nucleosomes (N) or naked DNA (D) were incubated with human excision nuclease for the indicated times, and the reaction products were analyzed on an 8% denaturing polyacrylamide gel. (Bottom) Kinetic plot of averages of three experiments including the one shown at the top. The percentage of the input substrate that was excised is plotted. Bars, standard deviations (those less than 0.07% are not shown). Open circles, naked DNA; solid circles, nucleosome. (C) Kinetics of excision of (6-4) photoproducts from naked DNA and the nucleosome by HeLa CE. (Top) Autoradiogram of a kinetics experiment. Internally labeled (6-4) substrates in the form of nucleosomes or naked DNA were incubated with HeLa CE for the indicated times, and reaction products were analyzed on an 8% denaturing polyacrylamide gel. (Bottom) Kinetics plot of averages of three experiments including the one at the top. The percentage of the input substrate that was excised is plotted. Standard deviations for all data points were less than 0.2%. Open circles, naked DNA; solid circles, nucleosome. (D) Kinetics of inhibition of (6-4) photoproduct excision by nucleosomes in HeLa (DDB+) and CHO (DDB−) CEs. Internally labeled (6-4) substrates in the form of nucleosomes or naked DNA were incubated in either HeLa CE or CHO AA8 CE, and the percent excision was determined as for Fig. 5. The values were expressed relative to the percent excision with naked DNA at the 4-h time point achieved by each CE and the averages of three experiments were plotted as relative excision. Circles, HeLa CE; triangles, CHO CE; open and solid symbols, naked DNA and nucleosomes, respectively. Bars, standard deviations. For HeLa CE, the data set shown in Fig. 3C was used.
Inhibition of excision in the reconstituted system and in CE.
The severe inhibition of excision by the defined excision nuclease system raised the possibility that the core excision nuclease lacked one or more components necessary for accessing damage in nucleosomes. Hence, we carried out excision reactions with reconstituted excision nuclease and CEs in parallel using naked DNA and nucleosomes as substrates. The results shown in Fig. 3B and C indicate that the excision of damage from nucleosomes is essentially equally inhibited in the two systems. It thus appears that the excision repair factors for repairing naked DNA are also necessary and sufficient for damage recognition and repair (albeit inefficiently) in nucleosomes and that CE does not contain additional factors which increase the rate of repair.
Effect of DDB on excision repair of nucleosomes.
In addition to the six repair factors necessary for dual incision, the XPE gene product is thought to play a role in nucleotide excision repair. Some of the xeroderma pigmentosum group E (XP-E) cell lines are defective in a protein called damaged-DNA binding protein (DDB) (8, 26, 28) which binds with high specificity to (6-4) photoproducts (51). It has been found that DDB has no effect on the rate of excision by the core excision nuclease (27), and it was suggested that it may play a role in damage recognition in chromatin rather than naked DNA (50). Indeed, it was discovered that CHO cells, which are known to be deficient in global genomic repair, lack DDB activity because of gene silencing (21) and that the repair defect can be ameliorated by transfecting the cells with the gene encoding the p48 subunit (48) of the DDB heterodimer (67). Thus, it was of interest to examine the repair of nucleosomal DNA in the presence and absence of DDB. For this purpose we carried out excision reactions with nucleosome substrate and CEs from either HeLa (DDB+) cells or a CHO (DDB−) cell line. The excision reaction is inhibited to the same extent in both extracts by nucleosomes (Fig. 3D). These results suggest that DDB plays no role in damage recognition either at the level of naked DNA or nucleosomes but do not eliminate the possibility that DDB participates in damage accessibility at a higher level of chromatin organization.
Effect of nucleosomes on damage recognition.
The three basic steps of human excision nuclease are damage recognition, unwinding of the duplex, and dual incision and excision (54). We wished to know at what step the nucleosome interfered with the excision reaction. Although damage recognition by human excision nuclease is a multistep process of increasing specificity and avidity (6, 46, 74), it is generally accepted that XPA, RPA, and XPC are involved in the early steps of recognition and assembly (5, 25, 64, 74, 75). Hence, we investigated the effect of nucleosomes on the binding of these proteins to damage in nucleosomal DNA.
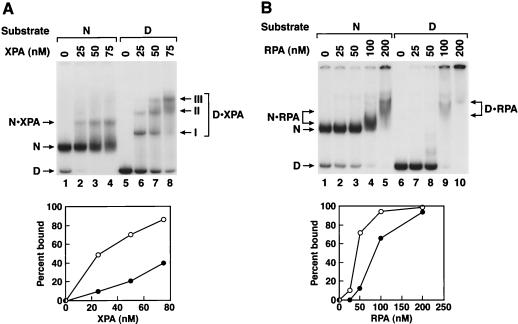
Figure 4A shows the binding of XPA to nucleosomes and naked DNA analyzed by electrophoretic mobility shift experiments. Two points of interest emerge from this figure. First, XPA binds to nucleosomal DNA with about fivefold-lower affinity than to naked DNA. Second, the protein-DNA complexes containing XPA+ naked DNA and XPA+ nucleosomes exhibit different mobilities, which indicates that the complex containing XPA+ nucleosome contains both XPA and the nucleosome core. Similar results were obtained with RPA (Fig. 4B). However, at high concentrations of RPA, complexes formed with both naked DNA and with nucleosomes did not migrate far into the gel, making detailed quantitative analysis rather difficult. Despite this shortcoming the data indicate that under appropriate experimental conditions a ternary complex of RPA-DNA-core histone does form.
FIG. 4.
Binding of damage recognition proteins to the nucleosome. XPA and RPA were incubated with naked DNA (D) or nucleosomes (N) and the DNA-protein complexes were separated on a 5% nondenaturing polyacrylamide gel. (Top) Autoradiograms; (Bottom) quantitative analysis of the binding data. Open circles, naked DNA; solid circles, nucleosome substrate. (A) Binding of XPA to the nucleosome. The single nucleosome-XPA band and the three XPA-DNA bands (I, II, III) arising from the binding of multiple XPA molecules to a single duplex are indicated. (B) Binding of RPA to the nucleosome. The RPA-nucleosome and the RPA-DNA bands are indicated. Presumably because of the high “off” rate of RPA, the RPA-nucleosome complex produces a rather “smeared” band. Similarly, with naked DNA at high concentrations of RPA multiple protein bindings retain the DNA in the origin.
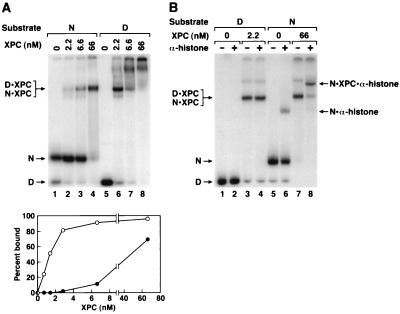
As for XPA and RPA, XPC has lower affinity for nucleosomal DNA than for naked DNA (Fig. 5A). However, in contrast to what was found for XPA and RPA, the DNA-protein complexes containing XPC and naked DNA and XPC and nucleosome have the same migration on nondenaturing gels. To determine if the complexes containing XPC and nucleosomes contained naked DNA alone (stripped-off histones) or represented XPC-nucleosome complexes, we carried out “supershift” experiments with antihistone antibodies. As seen in Fig. 5B the XPC-nucleosome complex but not the XPC-naked DNA complex was supershifted. Therefore, XPC, like XPA and RPA, can bind to nucleosomes without dissociating the DNA-histone complex.
FIG. 5.
(A) Binding of XPC to nucleosomal DNA. XPC was incubated with naked DNA (D) or nucleosomes (N) and DNA-protein complexes were separated on a 5% nondenaturing gel. (Top) Autoradiogram; (Bottom) quantitative analysis of the binding data. The data points for lower concentrations of XPC were obtained from a separate experiment. The main retarded bands with either naked DNA or nucleosomes comigrate. With naked DNA high XPC concentrations led to multiple protein binding and a smear extending all the way to the origin. (B) Characterization of XPC-nucleosome complexes with an antihistone antibody. To the XPC-DNA and XPC-nucleosome reaction mixtures antihistone monoclonal antibodies were added where indicated, and the DNA-protein complexes were separated on a 4% nondenaturing polyacrylamide gel. The nucleosome-XPC (N · XPC), nucleosome-XPC-antihistone antibody (N · XPC · α-histone), and DNA-XPC (D · XPC) bands are indicated. Note that at a high antibody concentration there was nonspecific binding of the antibody to DNA and hence in the supershift experiments less-than-saturating amounts of antibody were used, resulting in supershift of only a fraction of the histone-containing complexes (lanes 6 and 8).
It should be noted that, because of the relatively low selectivity (affinity for damaged nucleotides/affinity for undamaged nucleotides) of XPA, RPA, and XPC proteins (75), even though with shorter oligomers preferential binding to damaged DNA can be detected (74, 75), with 136-bp duplexes no difference between the binding to the undamaged control and to the (6-4) substrate could be discerned by gel mobility shift assay with either naked DNA or nucleosomes (data not shown). Hence, all the nucleosome binding experiments were carried out with damaged DNA only.
Is XPC a DNA accessibility factor?
XPC is required for global (nontranscribed) genomic repair and is dispensable in transcription-coupled repair (72). These findings have raised the distinct possibility that XPC may play a role in making DNA in chromatin accessible to human excision nuclease (2). Our finding that XPC can bind to nucleosomal DNA and convert the mononucleosome completely to an XPC-nucleosome complex at physiologically relevant XPC concentrations is consistent with such a role. To test this model, we carried out excision reactions with two concentrations of XPC: 2.2 nM, which we have found to be optimal for excision with naked DNA in our assay system, and 66 nM, which converts >70% of nucleosomal DNA into an XPC-nucleosome complex (Fig. 5A, lane 4). Figure 6 shows that the higher concentration of XPC inhibits excision from both naked DNA and nucleosomes; intermediate concentrations inhibited excision in proportion with the degree of binding to nucleosomes (data not shown). Thus, our data do not support a model for XPC as an accessibility factor and are in agreement with previous findings that the preformed XPC-DNA complex reduces the rate of excision by the reconstituted human excision nuclease (75).
FIG. 6.
Inhibition of excision by high XPC concentrations. Naked DNA (D) or nucleosomes (N) were incubated with human excision nuclease reconstituted with the indicated concentrations of XPC, and the reaction products were analyzed on an 8% denaturing polyacrylamide gel. The excision levels as percentages of input substrate were as follows: lane 1, 1.3%; lane 2, 0.1%; lane 3, not detectable; lane 4, not detectable; lane 5, 3.3%; lane 6, 0.7%; lane 7, 0.3%; lane 8, not detectable. Note that with 66 nM XPC most of the nucleosome DNA is in the form of XPC-nucleosome complexes (cf. Fig. 5A, lane 4).
DISCUSSION
Effect of nucleosome on damage formation and repair.
In eukaryotes, the chromatin structure profoundly affects replication, transcription, and repair by interfering with the accessibility of DNA to enzymes which carry out these processes (68, 70). Factors which affect the accessibility of chromosomal DNA to replication and transcription enzymes have been identified and investigated in in vitro systems (33, 73). Less is known about the modulation of DNA excision repair in chromatin. Pioneering in vivo work by Meijer and Smerdon (37) and Thoma (68) has provided the conceptual framework for investigating the effect of chromatin structure on repair in vitro. In vivo data have shown that the chromatin structure has significant effects both on DNA damage formation and repair (37, 49). Thus, of the two major UV photoproducts, cyclobutane pyrimidine dimers (Pyr⋄Pyr) were found to form more or less randomly throughout the chromatin whereas (6-4) photoproducts were formed at about twofold higher frequency in the linker region than in the nucleosome core (41, 65).
Regarding the effect of nucleosomes on repair, the first evidence suggesting an inhibitory effect was the finding that 24 and 48 h following UV irradiation of human fibroblasts there were more cyclobutane dimers in the nuclease-resistant fraction of chromatin than in the nuclease-accessible fraction (77). This was interpreted to mean that nucleosome-free DNA was repaired at a faster rate than nucleosomal DNA. Analysis of damage distribution at the nucleosomal level revealed that Pyr⋄Pyr dimers were produced with a 10.3-bp periodicity in the core nucleosome and that this periodicity was maintained during the repair period, indicating that there was no preferential repair of Pyr⋄Pyr along the nucleosome (23). More-detailed studies of the effect of chromatin structure on repair have been carried out in yeast using a minichromosome with well-defined nucleosome phasing and transcriptionally active and inactive regions. These studies (60, 66) conclusively showed that both Pyr⋄Pyr and (6-4) photoproducts were repaired at faster rates in nucleosome-free regions and in the linker DNA than were photoproducts in the nucleosome core.
DDB and chromatin repair.
Despite the commonly held belief that there are chromatin-remodeling and accessibility factors necessary for excision repair there is scarce in vivo data for the presence of such factors. The only known candidate for such a function is DDB. This protein is a heterodimer of 125- and 48-kDa subunits (29), and it binds to DNA containing (6-4) photoproducts with high specificity and avidity (51) and to DNA containing other lesions such as pyrimidine dimers with moderate to poor specificity. The DDB activity is missing in about 30% of XP-E cell lines (8, 26, 28) because of mutations in the small subunit (48), and XP-E cell lines are defective in global genomic repair (22). In addition, it has been found that the commonly used Chinese hamster cell lines lack DDB activity because of gene silencing of DDB2 encoding the p48 subunit (21) and are also defective in global genomic repair (22). Expression of p48 in these cell lines by transfection restores the DDB activity in CE and global genome repair activity in vivo (67). Thus, it was proposed that DDB functions as an accessibility factor for lesions in nontranscribed chromatin (21, 67).
We have found that nucleosomal DNA is repaired at about 10% the rate of naked DNA by the reconstituted excision nuclease and by CEs which contain or lack DDB. Thus, our results suggest that DDB does not function as an accessibility or remodeling factor at the nucleosome level. It is conceivable, however, that it may function as an accessibility factor at higher levels (30-nm fiber of packed nucleosomes or even higher-order structures) of chromatin organization. The role of DDB in repair is complex, however, on the basis of recent findings that DDB interacts specifically with transcription factor E2F1 and stimulates its activity (19, 47, 58). It is possible that DDB functions as an activator of transcription of excision repair genes. Clearly, more work is needed to understand the effect of DDB on excision repair; our study simply indicates that DDB does not stimulate the repair of either naked DNA or DNA at the nucleosome level of organization.
In vitro systems.
In this study, using a nucleosomal substrate with a lesion at a defined position and the six-factor human excision nuclease or mammalian CEs we investigated the effect of compaction of DNA in the nucleosome on nucleotide excision repair. We find that damage within the nucleosome core is excised at about 10% the rate of damage in naked DNA by both the reconstituted excision nuclease and the whole-cell extract. These findings suggest that the nucleosome structure is a serious impediment for human excision nuclease but that, in addition to the six general repair factors, there are no nucleosome accessibility factors specific for nucleotide excision repair.
Although we investigated the repair of a (6-4) photoproduct in a single location in the nucleosome and only in one rotational setting, our results may be applicable to lesions anywhere in the nucleosome core and in any rotational setting because in vivo data indicate that these two factors are not important for the relative rates of repair of UV lesions in mammalian cells (23). Our conclusion is also in agreement with in vivo data showing that photoproducts in linker DNA are repaired more rapidly than the nucleosomal photoproducts in human cells (65) and in nucleosome-free regions of a yeast minichromosome with a well-characterized nucleosome organization (60). If there were a nucleosome accessibility factor specific for excision repair, one would expect that in vivo the rates of damage removal from nucleosomal and nucleosome-free DNA would be comparable. It is reasonable to suggest, then, that one or more of the three damage recognition factors themselves function as accessibility factors of limited capacity for overcoming the inhibitory effect of nucleosomes partially, so as to carry out repair at a rate that is fast enough to be of significance in survival and in mutation avoidance.
Previously, by using minichromosomes, attempts to investigate the effect of chromatin structure on human nucleotide excision repair in vitro have been made (63, 76). In those studies randomly damaged minichromosomes were used as the substrate, whole-cell extract was used as the source of human excision nuclease, and incorporation of radiolabeled nucleotides into DNA (repair synthesis) was used to measure repair. The two studies arrived at different conclusions. In one study (76), it was found that assembly of a damaged plasmid into a minichromosome suppressed the repair synthesis that was observed with naked DNA, while the second study (63) reported that with naked DNA there was high background repair synthesis into undamaged DNA in naked plasmid control reactions, which was eliminated to yield true damage-dependent repair synthesis in damage-containing minichromosomes.
In a more recent study, with CE or reconstituted excision nuclease as the enzyme source, randomly damaged naked plasmid DNA or minichromosomes as the substrate, and the repair synthesis assay as the probe it was reported that there was no difference in the initial rates (up to 2 h) of repair of naked DNA and minichromosome DNA (1). Even after 2 h, nucleosomal DNA was repaired at about 80% of the rate of naked DNA. These results, which at face value appear to be contradictory to the findings reported in this paper, can be reconciled with our results as follows. The (6-4) photoproduct is repaired at a 5- to 10-fold-faster rate than Pyr⋄Pyr by human excision nuclease both in vivo (40, 65) and in vitro (44, 46), and thus most of the repair synthesis observed with human cell-free systems and UV-irradiated DNA is due to the removal of the (6-4) photoproducts (59, 79). Since (6-4) photoproducts form preferentially in the linker region of chromatin (41, 65), the repair synthesis observed in vitro with UV-irradiated minichromosomes is most likely due to the excision of (6-4) photoproducts from the linker region (1). In contrast, in our study we used a defined substrate which contained the (6-4) photoproduct in the nucleosome core to specifically address the question of nucleosome structure on excision, and we found that the nucleosome is a potent inhibitor of excision. Since the same level of inhibition was observed whether purified proteins or whole-cell extract was used for repair, our data also indicate that there is no cellular factor specific for repair to increase the accessibility of damage in the nucleosome core to the excision nuclease system. We discovered that the nucleosome reduces the DNA affinity of the three factors, XPA, RPA, and XPC, known to be involved in the early steps of damage recognition (64, 74, 75) by a factor of 5 to 10, which is roughly equivalent to the inhibition factor of excision by nucleosomes. Thus, it is likely that the nucleosome inhibits excision repair mainly by interfering with the earliest steps of the rather elaborate nucleotide excision repair system.
Transcription and repair.
There are several chromatin-remodeling/nucleosome accessibility factors for transcription in eukaryotes (30, 73) which are essential for cell survival. Considering the importance of nucleotide excision repair for maintaining cellular and organismal integrity it may seem surprising that there is no direct evidence for the existence of such factors specific for repair. However, looked at from a different perspective, the transcription accessibility factors may legitimately be considered repair accessibility factors as well because of the coupling of repair to transcription (17). Sequences transcribed by RNA polymerase II are repaired at a 5- to 10-fold-faster rate than nontranscribed sequences (4) and, importantly, this rate enhancement is due exclusively to the enhanced rate of repair of the template strand; the coding strand is repaired at the rate of general genomic repair (38).
Apparently, RNA polymerase stalled at a lesion constitutes a signal for the assembly of the excision nuclease at the transcriptional block site and hence functions as a high-specificity damage recognition factor (13, 17). Transcription-coupled repair occurs in Escherichia coli as well (39), and it involves active recruiting of repair factors to the site of transcriptional block by a transcription-repair coupling factor (55, 56). No such details are available at present for eukaryotic transcription-repair coupling because of the lack of an in vitro system. Nevertheless, the phenomenology of the process allows us to make some general statements regarding excision repair and chromatin remodeling/accessibility factors. First, because these factors are necessary for, or aid in, transcription initiation and elongation and since transcription stimulates repair, these factors are both transcription (directly) and repair (indirectly) accessibility factors. Second, since the nontranscribed strand (coding strand) is repaired at the general genomic repair rate (38, 72), it appears that the transient unfolding of chromatin, which must occur during transcription, is not sufficient to accelerate the repair rate because lesions in the coding strand do not slow the rate of RNA polymerase progression (10, 57). It is unclear at present whether transcription-repair coupling in eukaryotes occurs by an active mechanism as it does in prokaryotes (active recruiting of repair factors to the site of transcriptional block by a transcription-repair coupling factor) or is the consequence of having a long-lived RNA polymerase-RNA-DNA ternary complex and the accompanying open chromatin conformation and nucleosome mobility at the site of occlusion (57, 69, 71). Regardless of the mechanism, clearly transcription-coupled repair is a form of repair aided by chromatin-remodeling/nucleosomal DNA accessibility factors.
Finally, the requirement for repair-specific accessibility factors deserves some comment. While lack of transcription because of the absence of a remodeling/accessibility factor might prove lethal to the cell, lack of rapid repair because of a missing accessibility factor is mostly harmless unless the lesion is within an essential gene or within an active replicon. For lesions within transcribed sequences the problem has been solved by transcription-coupled repair, and, for replication, it has been solved by the presence of DNA polymerases capable of error-prone or error-free DNA synthesis (9, 24, 35). Hence, lesions in nontranscribed DNA can be repaired at the slow rate imposed by the packing of DNA into chromatin without seriously endangering the well-being of the cell. In this regard, it is noteworthy that nucleosome folding of damaged DNA inhibited the activity of the prokaryotic repair enzymes E. coli photolyase and T4 endonuclease V (which do not use a nucleosome substrate in nature) drastically (11, 31), whereas the much more complex human excision nuclease was inhibited by only a factor of 10, consistent with the notion that the human excision nuclease has evolved to work on nucleosomal DNA, albeit less efficiently than on naked DNA. However, it is also conceivable that the repair accessibility factor(s) is damage inducible and as such would have not been detected in our in vitro system. Indeed, damage-inducible protein GADD45 was reported to bind to UV-irradiated mononucleosomes (7). However, this binding resulted in inhibition rather than stimulation of T4 endonuclease V, and hence its relevance to chromatin repair is uncertain.
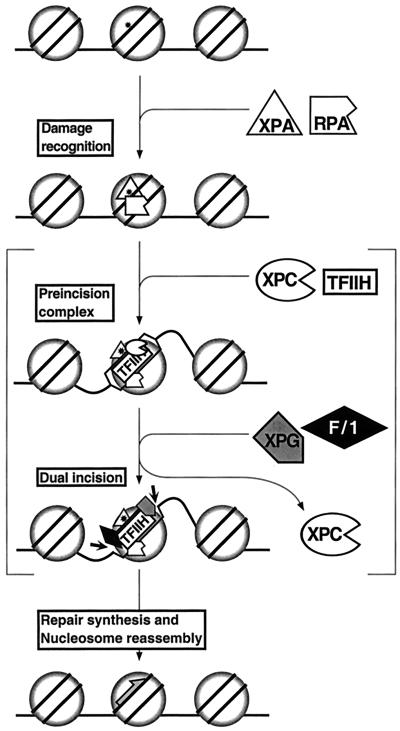
Figure 7 is a model for repairing DNA damage in nucleosomes by human excision nuclease. The model incorporates the findings reported in this paper as well as other existing data on this subject. The initial damage recognition by XPA and RPA occurs without disrupting the nucleosome. Subsequent assembly of TFIIH-XPC may disrupt the nucleosome and forms a preincision complex in which the DNA around the damage is unwound by about 20 bp (12, 46, 74). Then, XPG and XPF-ERCC1 nucleases are recruited concomitant with displacement of XPC, which functions as a molecular matchmaker (74, 75). Following the dual incision the excision nuclease complex disassembles in a manner coupled with repair synthesis, which in turn is coupled with nucleosome reassembly. Our data simply show that assembly and excision can occur on nucleosomal DNA; it does not give any information on the fate of the nucleosome during and after excision. The multiple DNA-protein complexes which exist in the postexcision reaction mixture with naked DNA (43) make such an analysis impractical. However, an in vivo study has shown that nascent repair patches are preferentially in the nuclease-sensitive fraction of the chromatin (2), consistent with movement or disassembly of nucleosomes during excision or repair synthesis. Similarly, in an in vitro study with randomly damaged plasmid DNA and Xenopus oocyte CE it was found that repair synthesis was accompanied by nucleosome assembly in a CAF1 (chromatin assembly factor 1)-dependent reaction (14, 42), as occurs during replicative DNA synthesis (62). Additional experiments of higher resolution are needed to test the specific steps of this model.
FIG. 7.
Model for nucleotide excision repair with nucleosomal substrate. Damage in the nucleosome is recognized by XPA and RPA. The binding of XPC and TFIIH leads to open-complex formation. Brackets indicate that there might be nucleosome disassembly or movement at this stage although direct experimental evidence for these events is lacking. XPG and XPF-ERCC1 are recruited to the complex, and XPC leaves the complex. The damage-containing oligonucleotide is excised by dual incision mediated by XPG and XPF-ERCC1. Repair synthesis, ligation, and nucleosome reassembly occur in a coupled series of reactions (2, 42). Circle, histone; asterisk, DNA damage; gray half-arrow, repair synthesis.
ACKNOWLEDGMENTS
We thank T. Bessho, L. Lindsey-Boltz, J. Reardon, and C. Selby for useful discussions and J. Reardon and C. Selby for critical reading of the manuscript.
This work was supported by National Institutes of Health grant GM 32833.
REFERENCES
- 1.Araki M, Masutani C, Maekawa T, Watanabe Y, Yamada A, Kusumoto R, Sakai D, Sugasawa K, Ohkuma Y, Hanaoka F. Reconstitution of damage DNA excision reaction from SV40 minichromosomes with purified nucleotide excision repair proteins. Mutat Res. 2000;459:147–160. doi: 10.1016/s0921-8777(99)00067-1. [DOI] [PubMed] [Google Scholar]
- 2.Baxter B K, Smerdon M J. Nucleosome unfolding during DNA repair in normal and xeroderma pigmentosum (group C) human cells. J Biol Chem. 1998;273:17517–17524. doi: 10.1074/jbc.273.28.17517. [DOI] [PubMed] [Google Scholar]
- 3.Bessho T, Sancar A, Thompson L H, Thelen M P. Reconstitution of human excision nuclease with recombinant XPF-ERCC1 complex. J Biol Chem. 1997;272:3833–3837. doi: 10.1074/jbc.272.6.3833. [DOI] [PubMed] [Google Scholar]
- 4.Bohr V A, Smith C A, Okumoto D S, Hanawalt P C. DNA repair in an active gene: removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell. 1985;40:359–369. doi: 10.1016/0092-8674(85)90150-3. [DOI] [PubMed] [Google Scholar]
- 5.Burns J L, Guzder S N, Sung P, Prakash S, Prakash L. An affinity of human replication protein A for ultraviolet-damaged DNA: implications for damage recognition in nucleotide excision repair. J Biol Chem. 1996;271:11607–11610. doi: 10.1074/jbc.271.20.11607. [DOI] [PubMed] [Google Scholar]
- 6.Buschta-Hedayat N, Buterin T, Hess M T, Missura M, Naegeli H. Recognition of nonhybridizing base pairs during nucleotide excision repair of DNA. Proc Natl Acad Sci USA. 1999;96:6090–6095. doi: 10.1073/pnas.96.11.6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carrier F, Georgel P T, Pourquier P, Blake M, Kontny H U, Antinore M J, Gariboldi M, Myers T G, Weinsterin J N, Pommier Y, Fornace A J., Jr Gadd 45, a p53-responsive stress protein, modifies DNA accessibility on damaged chromatin. Mol Cell Biol. 1999;19:1673–1685. doi: 10.1128/mcb.19.3.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chu G, Chang E. Xeroderma pigmentosum group E cells lack a nuclear factor that binds to damaged DNA. Science. 1988;242:564–567. doi: 10.1126/science.3175673. [DOI] [PubMed] [Google Scholar]
- 9.Cordonnier A M, Fuchs R P P. Replication of damaged DNA: molecular defect in xeroderma pigmentosum variant cells. Mutat Res. 1999;435:111–119. doi: 10.1016/s0921-8777(99)00047-6. [DOI] [PubMed] [Google Scholar]
- 10.Donahue B A, Yin S, Taylor J S, Reines D, Hanawalt P C. Transcript cleavage by RNA polymerase II arrested by a cyclobutane pyrimidine dimer in the DNA template. Proc Natl Acad Sci USA. 1994;91:8502–8506. doi: 10.1073/pnas.91.18.8502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Evans D H, Linn S. Excision repair of pyrimidine dimers from simian virus 40 minichromosome in vitro. J Biol Chem. 1984;259:10252–10259. [PubMed] [Google Scholar]
- 12.Evans E, Moggs J G, Hwang J R, Egly J M, Wood R D. Mechanism of open complex and dual incision formation by human nucleotide excision repair factors. EMBO J. 1997;16:6559–6573. doi: 10.1093/emboj/16.21.6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friedberg E C. Relationships between DNA repair and transcription. Annu Rev Biochem. 1996;65:15–42. doi: 10.1146/annurev.bi.65.070196.000311. [DOI] [PubMed] [Google Scholar]
- 14.Gaillard P H L, Martini E M D, Kaufman P D, Stillman B, Moustacchi E, Almouzni G. Chromatin assembly coupled to DNA repair: a new role of chromatin assembly factor I. Cell. 1996;86:887–896. doi: 10.1016/s0092-8674(00)80164-6. [DOI] [PubMed] [Google Scholar]
- 15.Golding A, Chandler S, Ballestar E, Wolffe A P, Schlissel M S. Nucleosome structure completely inhibits in vitro cleavage by the V(D)J recombinase. EMBO J. 1999;18:3712–3723. doi: 10.1093/emboj/18.13.3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guzder S N, Habraken Y, Sung P, Prakash L, Prakash S. Reconstitution of yeast nucleotide excision repair with purified Rad proteins, replication protein A, and transcription factor TFIIH. J Biol Chem. 1995;270:12973–12976. doi: 10.1074/jbc.270.22.12973. [DOI] [PubMed] [Google Scholar]
- 17.Hanawalt P C. Transcription-coupled repair and human disease. Science. 1994;266:1957–1958. doi: 10.1126/science.7801121. [DOI] [PubMed] [Google Scholar]
- 18.Hayes J J, Clark D J, Wolffe A P. Histone contributions to the structure of DNA in the nucleosome. Proc Natl Acad Sci USA. 1991;88:6829–6833. doi: 10.1073/pnas.88.15.6829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hayes S A, Shiyanov P, Chen X, Raychaudhuri P. DDB, a putative DNA repair protein, can function as a transcriptional partner of E2F1. Mol Cell Biol. 1998;18:240–249. doi: 10.1128/mcb.18.1.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang J C, Svoboda D L, Reardon J T, Sancar A. Human nucleotide excision nuclease removes thymine dimers from DNA by incising the 22nd phosphodiester bond 5′ and the 6th phosphodiester bond 3′ to the photodimer. Proc Natl Acad Sci USA. 1992;89:3664–3668. doi: 10.1073/pnas.89.8.3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hwang B J, Toering S, Francke U, Chu G. p48 activates a UV-damaged DNA binding factor and is defective in xeroderma pigmentosum group E cells that lack the binding activity. Mol Cell Biol. 1998;18:4391–4399. doi: 10.1128/mcb.18.7.4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hwang B J, Ford J M, Hanawalt P C, Chu G. Expression of the p48 xeroderma pigmentosum gene is p53-dependent and is involved in global genomic repair. Proc Natl Acad Sci USA. 1999;96:424–428. doi: 10.1073/pnas.96.2.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jensen K A, Smerdon M J. DNA repair within nucleosome cores of UV-irradiated human cells. Biochemistry. 1990;29:4773–4782. doi: 10.1021/bi00472a005. [DOI] [PubMed] [Google Scholar]
- 24.Johnson R E, Kondratick C M, Prakash S, Prakash L. hRAD30 mutations in the variant form of xeroderma pigmentosum. Science. 1999;285:263–265. doi: 10.1126/science.285.5425.263. [DOI] [PubMed] [Google Scholar]
- 25.Jones C J, Wood R D. Preferential binding of the xeroderma-pigmentosum group A complementing protein to damaged DNA. Biochemistry. 1993;32:12096–12104. doi: 10.1021/bi00096a021. [DOI] [PubMed] [Google Scholar]
- 26.Kataoka H, Fujiwara Y. UV damage-specific DNA-binding protein in xeroderma pigmentosum complementation group E. Biochem Biophys Res Commun. 1991;175:1139–1143. doi: 10.1016/0006-291x(91)91684-5. [DOI] [PubMed] [Google Scholar]
- 27.Kazanstev A, Mu D, Nichols A F, Zhao X, Linn S, Sancar A. Functional complementation of xeroderma pigmentosum complementation group E by replication protein A in an in vitro system. Proc Natl Acad Sci USA. 1996;93:5014–5018. doi: 10.1073/pnas.93.10.5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keeney S, Wein H, Linn S. Biochemical heterogeneity in xeroderma pigmentosum complementation group E. Mutat Res. 1992;273:49–56. doi: 10.1016/0921-8777(92)90049-9. [DOI] [PubMed] [Google Scholar]
- 29.Keeney S, Chang G J, Linn S. Characterization of a human DNA damage binding protein implicated in xeroderma pigmentosum E. J Biol Chem. 1993;268:21293–21300. [PubMed] [Google Scholar]
- 30.Kingston R E, Narlikar G J. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev. 1999;13:2339–2352. doi: 10.1101/gad.13.18.2339. [DOI] [PubMed] [Google Scholar]
- 31.Kosmoski J V, Smerdon M J. Synthesis and nucleosome structure of DNA containing a UV photoproduct at a specific site. Biochemistry. 1999;38:9485–9494. doi: 10.1021/bi990297h. [DOI] [PubMed] [Google Scholar]
- 32.Lee D Y, Hayes J J, Pruss D, Wolffe A P. A positive role for histone acetylation in transcription factor access to nucleosomal DNA. Cell. 1993;72:73–84. doi: 10.1016/0092-8674(93)90051-q. [DOI] [PubMed] [Google Scholar]
- 33.LeRoy G, Orphanides G, Lane W S, Reinberg D. Requirement of RSF and FACT for transcription of chromatin templates in vitro. Science. 1998;282:1900–1904. doi: 10.1126/science.282.5395.1900. [DOI] [PubMed] [Google Scholar]
- 34.Manley J L, Fire A, Cano A, Sharp P A, Gefter M L. DNA-dependent transcription of adenovirus genes in a soluble whole-cell extract. Proc Natl Acad Sci USA. 1980;77:3855–3859. doi: 10.1073/pnas.77.7.3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Masutani C, Kusumoto R, Yamada A, Dohmae N, Yokoi M, Yuasa M, Araki M, Iwai S, Takio K, Hanaoka F. The XPV (xeroderma pigmentation variant) gene encodes DNA polymerase η. Nature. 1999;399:700–704. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- 36.Matsunaga T, Park C H, Bessho T, Mu D, Sancar A. Replication protein A confers structure-specific endonuclease activities to the XPF-ERCC1 and XPG subunits of human DNA repair excision nuclease. J Biol Chem. 1996;271:11047–11050. doi: 10.1074/jbc.271.19.11047. [DOI] [PubMed] [Google Scholar]
- 37.Meijer M, Smerdon M J. Accessing DNA damage in chromatin: insights from transcription. Bioessays. 1999;21:596–603. doi: 10.1002/(SICI)1521-1878(199907)21:7<596::AID-BIES8>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 38.Mellon I, Spivak G, Hanawalt P C. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell. 1987;51:241–249. doi: 10.1016/0092-8674(87)90151-6. [DOI] [PubMed] [Google Scholar]
- 39.Mellon I, Hanawalt P C. Induction of the Escherichia coli lactose operon selectively increases repair of its transcribed strand. Nature. 1989;342:95–98. doi: 10.1038/342095a0. [DOI] [PubMed] [Google Scholar]
- 40.Mitchell D L, Haipek C A, Clarkson J M. (6-4) photoproducts are removed from the DNA of UV-irradiated mammalian cells more efficiently than cyclobutane dimers. Mutat Res. 1985;143:109–112. doi: 10.1016/s0165-7992(85)80018-x. [DOI] [PubMed] [Google Scholar]
- 41.Mitchell D L, Nguyen T D, Cleaver J E. Nonrandom induction of pyrimidine-pyrimidone (6-4) photoproducts in ultraviolet irradiated human chromatin. J Biol Chem. 1990;265:5353–5356. [PubMed] [Google Scholar]
- 42.Moggs J G, Almouzni G. Chromatin rearrangements during nucleotide excision repair. Biochimie. 1999;81:45–52. doi: 10.1016/s0300-9084(99)80037-6. [DOI] [PubMed] [Google Scholar]
- 43.Mu D, Park C H, Matsunaga T, Hsu D S, Reardon J T, Sancar A. Reconstitution of human DNA repair excision nuclease in a highly defined system. J Biol Chem. 1995;270:2415–2418. doi: 10.1074/jbc.270.6.2415. [DOI] [PubMed] [Google Scholar]
- 44.Mu D, Hsu D S, Sancar A. Reaction mechanism of human DNA repair excision nuclease. J Biol Chem. 1996;271:8285–8294. doi: 10.1074/jbc.271.14.8285. [DOI] [PubMed] [Google Scholar]
- 45.Mu D, Tursun M, Duckett D R, Drummond J T, Modrich P, Sancar A. Recognition and repair of compound DNA lesions (base damage and mismatch) by human mismatch repair and excision repair systems. Mol Cell Biol. 1997;17:760–769. doi: 10.1128/mcb.17.2.760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mu D, Wakasugi M, Hsu D S, Sancar A. Characterization of reaction intermediates of human excision repair nuclease. J Biol Chem. 1997;272:28971–28979. doi: 10.1074/jbc.272.46.28971. [DOI] [PubMed] [Google Scholar]
- 47.Nichols A F, Itoh T, Graham J A, Liu W, Yamaizumi M, Linn S. Human damage-specific DNA-binding protein p48. Characterization of XPE mutations and regulation following UV irradiation. J Biol Chem. 2000;275:21422–21428. doi: 10.1074/jbc.M000960200. [DOI] [PubMed] [Google Scholar]
- 48.Nichols A F, Ong P, Linn S. Mutations specific to the xeroderma pigmentosum group E Ddb− phenotype. J Biol Chem. 1996;271:24317–24320. doi: 10.1074/jbc.271.40.24317. [DOI] [PubMed] [Google Scholar]
- 49.Pfeifer G P. Formation and processing of UV photoproducts: effects of DNA sequence and chromatin environment. Photochem Photobiol. 1997;65:270–283. doi: 10.1111/j.1751-1097.1997.tb08560.x. [DOI] [PubMed] [Google Scholar]
- 50.Rapić Otrin V, Kuraoka I, Nordo T, McLenigan M, Eker A P M, Stefanini M, Levine A S, Wood R D. Relationship of the xeroderma pigmentosum group E DNA repair defect to the chromatin and DNA binding proteins UV-DDB and replication protein A. Mol Cell Biol. 1998;18:3182–3190. doi: 10.1128/mcb.18.6.3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reardon J T, Nichols A F, Keeney S, Smith C A, Taylor J S, Linn S, Sancar A. Comparative analysis of binding of human damaged DNA-binding protein (XPE) and Escherichia coli damage recognition protein (UvrA) to the major ultraviolet photoproducts: T[c,s]T, T[t,s]T, T[6-4]T, and T[Dewar]T. J Biol Chem. 1993;268:21301–21308. [PubMed] [Google Scholar]
- 52.Reardon J T, Thompson L H, Sancar A. Excision repair in man and the molecular basis of xeroderma pigmentosum. Cold Spring Harbor Symp Quant Biol. 1993;58:605–617. doi: 10.1101/sqb.1993.058.01.067. [DOI] [PubMed] [Google Scholar]
- 53.Reardon J T, Mu D, Sancar A. Overproduction, purification, and characterization of the XPC subunit of the human DNA repair excision nuclease. J Biol Chem. 1996;271:19451–19456. doi: 10.1074/jbc.271.32.19451. [DOI] [PubMed] [Google Scholar]
- 54.Sancar A. DNA excision repair. Annu Rev Biochem. 1996;65:43–81. doi: 10.1146/annurev.bi.65.070196.000355. [DOI] [PubMed] [Google Scholar]
- 55.Selby C P, Sancar A. Molecular mechanism of transcription-repair coupling. Science. 1993;260:53–58. doi: 10.1126/science.8465200. [DOI] [PubMed] [Google Scholar]
- 56.Selby C P, Sancar A. Mechanisms of transcription-repair coupling and mutation frequency decline. Microbiol Rev. 1994;58:317–329. doi: 10.1128/mr.58.3.317-329.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Selby C P, Drapkin R, Reinberg D, Sancar A. RNA polymerase II stalled at a thymine dimer: footprint and effect on excision repair. Nucleic Acids Res. 1997;25:787–793. doi: 10.1093/nar/25.4.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shiyanov P, Hayes S A, Donepudi M, Nichols A F, Linn S, Slagle B L, Raychaudhuri P. The naturally occurring mutants of DDB are impaired in stimulating nuclear transport of the p125 subunit and E2F1-activated transcription. Mol Cell Biol. 1999;19:4935–4943. doi: 10.1128/mcb.19.7.4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sibghat-Ullah A, Sancar Substrate overlap and functional competition between human nucleotide excision repair and E. coli photolyase and (A)BC excision nuclease. Biochemistry. 1990;29:5711–5718. doi: 10.1021/bi00476a011. [DOI] [PubMed] [Google Scholar]
- 60.Smerdon M J, Thoma F. Site-specific DNA repair at the nucleosome level in a yeast minichromosome. Cell. 1990;61:675–684. doi: 10.1016/0092-8674(90)90479-x. [DOI] [PubMed] [Google Scholar]
- 61.Smith C A, Taylor J S. Preparation and characterization of a set of deoxyoligonucleotide 49-mers containing site-specific cis-syn trans-syn I, (6-4) and Dewar pyrimidone photoproducts of thymidyl-(3′→5′) thymidine. J Biol Chem. 1993;268:11143–11151. [PubMed] [Google Scholar]
- 62.Smith S, Stillman B. Purification and characterization of CAF-I, a human cell factor required for chromatin assembly during DNA replication in vitro. Cell. 1989;58:15–25. doi: 10.1016/0092-8674(89)90398-x. [DOI] [PubMed] [Google Scholar]
- 63.Sugasawa K, Masutani C, Hanaoka F. Cell-free repair of UV-damaged simian virus 40 chromosome in human cell extracts. I. Development of a cell-free system detecting excision repair of UV-irradiated SV40 chromosomes. J Biol Chem. 1993;268:9098–9104. [PubMed] [Google Scholar]
- 64.Sugasawa K, Ng J M Y, Masutani C, Iwai S, van der Spek P J, Eker A P M, Hanaoka F, Bootsma D, Hoeijmakers J H J. Xeroderma pigmentosum group C protein complex is the initiator of global genome nucleotide excision repair. Mol Cell. 1998;2:223–232. doi: 10.1016/s1097-2765(00)80132-x. [DOI] [PubMed] [Google Scholar]
- 65.Suquet C, Mitchell D L, Smerdon M J. Repair of UV-induced (6-4) photoproducts in nucleosome core DNA. J Biol Chem. 1995;270:16507–16509. doi: 10.1074/jbc.270.28.16507. [DOI] [PubMed] [Google Scholar]
- 66.Tanaka S, Livingstone-Zatchej M, Thoma F. Chromatin structure of the yeast URA3 gene at high resolution provides insight into structure and positioning of nucleosomes in the chromosomal context. J Biol Chem. 1996;257:919–934. doi: 10.1006/jmbi.1996.0212. [DOI] [PubMed] [Google Scholar]
- 67.Tang J Y, Hwang B J, Ford J M, Hanawalt P C, Chu G. Xeroderma pigmentosum p48 gene enhances global genomic repair and suppresses UV-induced mutagenesis. Mol Cell. 2000;5:737–744. doi: 10.1016/s1097-2765(00)80252-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Thoma F. Light and dark in chromatin repair: repair of UV-induced DNA lesions by photolyase and nucleotide excision repair. EMBO J. 1999;18:6585–6598. doi: 10.1093/emboj/18.23.6585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tijsterman M, De Pril R, Tasseron-DeJong J G, Brouwer J. RNA polymerase II transcription suppresses nucleosomal modulation of UV-induced (6-4) photoproducts and cyclobutane pyrimidine dimer repair in yeast. Mol Cell Biol. 1999;19:934–940. doi: 10.1128/mcb.19.1.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tyler J K, Kadonaga J T. The “dark side” of chromatin remodeling: repressive effects on transcription. Cell. 1999;99:443–446. doi: 10.1016/s0092-8674(00)81530-5. [DOI] [PubMed] [Google Scholar]
- 71.Venema J, Mullenders L H, Natarajan A T, van Zeeland A A, Mayne L V. The genetic defect in cockayne syndrome is associated with a defect in repair of UV-induced DNA damage in transcriptionally active DNA. Proc Natl Acad Sci USA. 1990;87:4707–4711. doi: 10.1073/pnas.87.12.4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Venema J, van Hoffen A, Karcagi V, Natarajan A T, van Zeeland A A, Mullenders L H F. Xeroderma pigmentosum complementation group C cells remove pyrimidine dimers selectively from the transcribed strand of active genes. Mol Cell Biol. 1991;11:4128–4134. doi: 10.1128/mcb.11.8.4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vignali M, Hassan A H, Neely K E, Workman J L. ATP-dependent chromatin-remodeling complexes. Mol Cell Biol. 2000;20:1899–1910. doi: 10.1128/mcb.20.6.1899-1910.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wakasugi M, Sancar A. Assembly, subunit composition, and footprint of human DNA repair excision nuclease. Proc Natl Acad Sci USA. 1998;95:6669–6674. doi: 10.1073/pnas.95.12.6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wakasugi M, Sancar A. Order of assembly of human DNA excision repair nuclease. J Biol Chem. 1999;274:18759–18768. doi: 10.1074/jbc.274.26.18759. [DOI] [PubMed] [Google Scholar]
- 76.Wang Z, Wu X, Friedberg E C. Nucleotide excision repair by human cell extracts is suppressed in reconstituted nucleosomes. J Biol Chem. 1991;266:22472–22478. [PubMed] [Google Scholar]
- 77.Wilkins R J, Hart R W. Preferential DNA repair in human cells. Nature. 1974;247:35–36. doi: 10.1038/247035a0. [DOI] [PubMed] [Google Scholar]
- 78.Wolffe A P, Hayes J J. Transcription factor interactions with model nucleosomal templates. Methods Mol Genet. 1993;2:314–330. [Google Scholar]
- 79.Wood R D. Repair of pyrimidine dimer ultraviolet light photoproducts by human cell-extracts. Biochemistry. 1990;28:8287–8292. doi: 10.1021/bi00447a005. [DOI] [PubMed] [Google Scholar]
- 80.Wood R D. DNA repair in eukaryotes. Annu Rev Biochem. 1996;65:135–167. doi: 10.1146/annurev.bi.65.070196.001031. [DOI] [PubMed] [Google Scholar]
- 81.Workman J L, Kingston R E. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu Rev Biochem. 1998;67:545–579. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]