Abstract
The retinoblastoma protein (RB) represses RNA polymerase III transcription effectively both in vivo and in vitro. Here we demonstrate that the general transcription factors snRNA-activating protein complex (SNAPc) and TATA binding protein (TBP) are important for RB repression of human U6 snRNA gene transcription by RNA polymerase III. RB is associated with SNAPc as detected by both coimmunoprecipitation of endogenous RB with SNAPc and cofractionation of RB and SNAPc during chromatographic purification. RB also interacts with two SNAPc subunits, SNAP43 and SNAP50. TBP or a combination of TBP and SNAPc restores efficient U6 transcription from RB-treated extracts, indicating that TBP is also involved in RB regulation. In contrast, the TBP-containing complex TFIIIB restores adenovirus VAI but not human U6 transcription in RB-treated extracts, suggesting that TFIIIB is important for RB regulation of tRNA-like genes. These results suggest that different classes of RNA polymerase III-transcribed genes have distinct general transcription factor requirements for repression by RB.
Retinoblastoma protein (RB) is a tumor suppressor that controls cell growth by influencing cell cycle progression (8, 12, 44), differentiation (5, 15, 44), and apoptosis (23, 68). Mutations in the gene encoding RB are associated with diverse human cancers (21, 22, 27, 45). RB function is also compromised in other human malignancies through disruption of upstream control pathways or downstream targets of RB (reviewed in reference 58). The function of RB as a tumor suppressor is linked to its ability to regulate gene expression. Therefore, to fully understand the contribution of RB to cellular proliferation observed during carcinogenesis, it is important to determine the mechanisms that RB uses to regulate gene activity.
An understanding of RB function in gene regulation was revealed through its role as a modulator of E2F transcription factor activity (16, 24, 25, 59). However, RB controls additional cellular functions beyond regulating E2F activity. The intracellular concentration of RB exceeds the concentration of E2F (58), and interactions between RB and other transcription factors have been described previously (10, 34, 51). Thus, further activities performed by RB involve regulation of other genes besides E2F-responsive genes. Interestingly, RB is not limited to regulating mRNA production by RNA polymerase II but also inhibits the synthesis of rRNAs by RNA polymerase I (4) and of 5S rRNA, tRNA, and U6 snRNA by RNA polymerase III (63). It was proposed that loss of control of these genes is an important step in tumor progression because the products of genes transcribed by RNA polymerases I and III are important determinants of biosynthetic capacity (reviewed in reference 61). Repressed synthesis of nontranslated RNAs is expected to inhibit cell proliferation, presenting a significant hurdle to unregulated cell growth. Therefore, control of RNA polymerase I and III transcriptional activity may represent an essential component of growth regulation by RB.
How RB regulates RNA polymerase III activity in the cell is not clear. RNA polymerase III transcriptional activity is under cell cycle control, with higher levels observed in the late G1, S, and G2 phases of the cell cycle than in G0 and early G1 (62). The increase in RNA polymerase III activity correlates with an increase in phosphorylated RB during the G1 phase of the cell cycle. This increased activity is important because the function of RB is controlled by phosphorylation (6, 38). Hypophosphorylated RB can interact with potential target proteins to regulate their activities, whereas hyperphosphorylated RB cannot interact and, therefore, is inactive (58). RNA polymerase III activity is maximal during the cell cycle when RB is inactive. This implies that hypophosphorylated RB may target factors that function in RNA polymerase III transcription. The correlation between RB levels and RNA polymerase III activity has been further demonstrated in vivo by transient-transfection assays of adenovirus (Ad) VAI gene transcription. Transcription of this gene by RNA polymerase III is elevated in a human osteosarcoma cell line (SAOS2) that is RB deficient compared to the level of transcription in an osteosarcoma cell line (U2OS) that contains functional RB. Overexpression of RB in SAOS2 cells represses RNA polymerase III transcription, whereas RNA polymerase II transcription from the human immunodeficiency virus long terminal repeat is unaffected. Furthermore, in nuclear runoff assays, RNA polymerase III-specific transcription is diminished in nuclei isolated from wild-type mouse embryonic fibroblasts compared to that in nuclei isolated from mouse RB−/− embryonic fibroblasts, whereas wholesale RNA polymerase II activity is unchanged in RB+/+ and RB−/− embryonic fibroblasts (63). These experiments suggest that RNA polymerase III activity in vivo is regulated by RB.
We have focused on understanding the contribution of RB to repression of RNA polymerase III activity. Genes transcribed by RNA polymerase III can be subdivided into four classes. Class 1 and class 2 genes contain gene-internal promoter elements exemplified by the 5S rRNA and tRNA genes, respectively. Class 3 genes contain gene-external promoter elements exemplified by the human U6 snRNA genes. A fourth class exemplified by the Vault RNA genes contain both external and internal promoter elements (54). RNA polymerase III-transcribed genes also have distinct general transcription factor requirements, consistent with their different promoter architectures. 5S rRNA genes require TFIIIA, TFIIIB, and TFIIIC, whereas tRNA gene transcription requires only a subset of these factors, TFIIIB and TFIIIC, for full activity (50, 52, 64). In contrast, human U6 gene transcription requires the snRNA-activating protein complex (SNAPc) (48), which is also known as PSE transcription factor (PTF) (42). While TFIIIC is not required, the requirement for TFIIIB is controversial (39, 57). In addition to these general transcription factors, other transcription activator proteins, including Oct-1 (3), Sp1 (30), and Staf (43, 49), positively regulate U6 snRNA gene transcription.
The mechanism that RB utilizes to repress RNA polymerase III transcription is not known. However, potential targets for regulating RNA polymerase III activity include TFIIIB and the TFIIIC2 form of TFIIIC (7, 29). Human TFIIIB consists of the TATA box binding protein (TBP) and a tightly associated factor called TFIIB-related factor or BRF (39, 57). By analogy with Saccharomyces cerevisiae TFIIIB (26), a loosely associated factor referred to as B" may also be a component of human TFIIIB. BRF and TBP associate with RB during chromatographic fractionation of cellular extracts and during coimmunoprecipitation experiments (29). TFIIIC2 is a multiprotein complex containing five proteins (67). RB can also interact with TFIIIC2 in glutathione S-transferase (GST)-pulldown experiments from HeLa nuclear extracts (7). Together, these data indicate that RB can interact with the RNA polymerase III general transcription machinery, and this interaction may be important for RB repression of RNA polymerase III-specific gene transcription.
It is not known whether RB targets similar factors to regulate all classes of RNA polymerase III-transcribed genes. In contrast to the clear requirement of TFIIIB and TFIIIC2 for RNA polymerase III transcription of genes containing gene-internal promoter elements, neither TFIIIC (55) nor a TFIIIB complex of BRF and TBP is required for human U6 snRNA gene transcription in vitro (17, 39). Potentially, a different form of TFIIIB may function both for U6 gene transcription and regulation by RB. Other alternative spliced forms of BRF have been identified, and one form referred to as human BRF2 functions for human U6 transcription in vitro (37). It is also possible that RB targets other factors to regulate human U6 snRNA genes. One potential target is SNAPc. SNAPc is a multiprotein complex composed of at least five proteins: SNAP19 (19), SNAP43 (also called PTFγ) (20, 66), SNAP45 (also called PTFδ) (47, 66), SNAP50 (also called PTFβ) (1, 18), and SNAP190 (also called PTFα) (65). In addition, SNAPc associates with TBP (20). SNAPc binds to the proximal sequence element (PSE) contained in the core promoter regions of human U6 snRNA genes and interacts with TBP bound to the TATA box. SNAPc and TBP act cooperatively to facilitate transcription by RNA polymerase III (40). The binding of these factors to the promoter is a crucial early step in pre-initiation complex assembly at these genes, and therefore SNAPc and TBP are attractive targets for regulating human snRNA gene transcription.
Our results suggest that RB regulates different RNA polymerase III-transcribed genes by targeting different components of the general transcription machinery. The general transcription factor TFIIIB, composed of BRF and TBP, functionally restores Ad VAI gene transcription but not human U6 snRNA gene transcription in RB-treated extracts. In contrast, the general transcription factors SNAPc and TBP act cooperatively to reconstitute U6 snRNA gene transcription after RB treatment, indicating that these factors are also important for RB regulation of RNA polymerase III activity. Depleting extracts with GST bound to amino acids 379 to 928 of RB [GST-RB (379–928)] resulted in a reduction in SNAP43 levels, consistent with the idea that RB targets SNAPc. In HeLa cell nuclear extracts, a subpopulation of RB is associated with SNAPc and this association may be direct because RB interacts effectively with two components of SNAPc. These data indicate that the general transcription factors SNAPc and TFIIIB provide an important targeting mechanism governing RB function for different classes of RNA polymerase III-transcribed genes.
MATERIALS AND METHODS
Expression and purification of recombinant proteins.
The region of RB corresponding to amino acids 379 to 928 was amplified by PCR and cloned into a pET11c-based expression vector to generate pGST-RB (379–928). This plasmid contains an N-terminal GST tag fused in frame with RB (379–928). Both SNAP43 (1–368) and SNAP50 (1–411) were constructed in a similar manner to generate pGST-SNAP43 (1–368) and pGST-SNAP50 (1–411), respectively. GST fusion proteins were expressed in Escherichia coli BL21 DE3, and extracts were prepared by sonication. Proteins were purified by binding to glutathione agarose beads (Sigma) followed by extensive washing in HEMGT-150 buffer (20 mM HEPES [pH 7.9], 0.5 mM EDTA, 10 mM MgCl2, 10% glycerol, 0.1% Tween 20) containing protease inhibitors (0.1 mM phenylmethylsulfonyl fluoride, 1 mM sodium bis-sulfate, 1 mM benzamidine, 1 μM pepstatin A) and 1 mM dithiothreitol. Bound proteins were then used directly for GST pulldown assays. For in vitro transcription experiments, GST-RB (379–928) and GST were eluted from beads in HEMGT-150 buffer containing 50 mM glutathione for 1 h at 4°C. Eluted proteins were dialyzed against Dignam buffer D (9) and concentrated by centrifugation through YM10 centricon columns (Millipore) to give a final concentration of at least 200 ng/μl. Protein expression levels, efficiency of binding to glutathione agarose beads, and protein concentrations were monitored by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and staining with Coomassie blue.
In vitro transcription assays.
In vitro transcription of the Ad VAI and human U6 snRNA genes was performed essentially as described previously (31, 32, 48). For the repression assays whose results are depicted in Fig. 1, HeLa cell nuclear extracts were preincubated with purified recombinant proteins at 30°C for 30 min. The amounts of each protein used are indicated in the figure legend. Transcription reactions (20-μl mixtures) were initiated by the addition of 0.25 μg of DNA templates (pBSM13+VAI, pU6/Hae/RA.2), nucleoside triphosphates, and transcription buffer. Transcription reactions were performed for 1 h at 30°C. Transcripts were separated by denaturing PAGE and visualized by autoradiography or by PhosphorImager analysis (Molecular Dynamics).
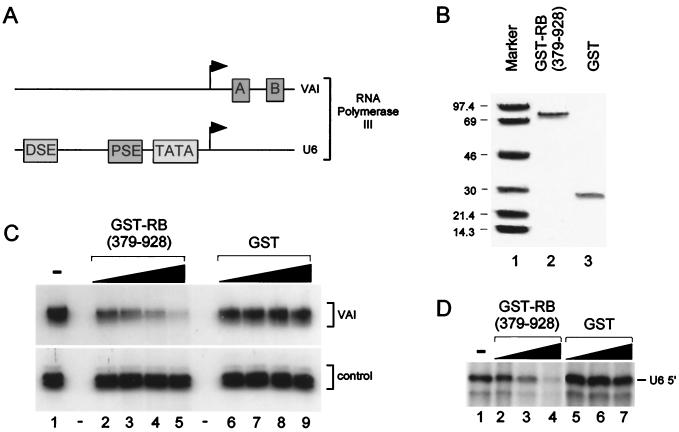
FIG. 1.
RB represses in vitro transcription by RNA polymerase III. (A) Schematic representation of the Ad VAI and human U6 snRNA promoters. (B) Analysis of GST-RB (379–928) and GST proteins used in transcription reactions. GST-RB (379–928) (lane 2) and GST (lane 3) were expressed in E. coli and purified by affinity chromatography using glutathione agarose beads and competitive elution with glutathione. After dialysis against Dignam buffer D, proteins were separated by SDS-PAGE and visualized by staining with Coomassie blue. Lane 1 contains a protein size standard. Molecular weights (in thousands) are noted at the left. (C) GST-RB (379–928) represses Ad VAI transcription by RNA polymerase III. Approximately 2 μl of HeLa cell nuclear extract (approximately 7.5 μg/μl) was incubated with 200, 400, 800, and 1,200 ng of GST-RB (379–928) (lanes 2 to 5) or GST protein (lanes 6 to 9) at 30°C for 30 min. In vitro transcription of the Ad VAI gene (top gel) was then initiated by addition of the template, cold nucleoside triphosphates, [α32P]CTP, and transcription buffer. Lane 1 shows the level of transcription with the untreated extract. Sample handling was monitored by a nonspecific RNA handling control transcript (bottom). (D) GST-RB (379–928) represses human U6 snRNA gene transcription by RNA polymerase III. Approximately 2 μl of HeLa cell nuclear extract was incubated with 100, 250, and 500 ng of GST-RB (379–928) (lanes 2 to 4) or GST protein (lanes 5 to 7). Lane 1 shows the level of transcription with the untreated extract. Correctly initiated transcripts from the U6 promoter (labeled U6 5′) were detected by RNase T1 protection essentially as described previously (31).
RB affinity depletion assays.
For human U6 snRNA gene repression assays whose results are shown in Fig. 2A, 8 μl of HeLa cell nuclear extract (7.5 μg/μl) was preincubated with 1, 2, or 3 μl of purified GST-RB (379–928) or GST (each at 1 μg/μl) for 30 min at 30°C. GST-RB (379–928) and GST were removed by affinity purification with glutathione-Sepharose beads (Pharmacia) added at a 1:1 ratio of beads to extract. Samples were then incubated at 4°C for 4 h and centrifuged to remove associated proteins. One-half of each supernatant (4.5, 5.0, and 5.5 μl of the 1-, 2-, and 3-μg treated samples, respectively) was used for in vitro transcription assays as described above. For the experiment shown in Fig. 2B, the affinity depletion reactions were scaled up to include 32 μl of nuclear extract (7.5 μg/μl) plus 10 μg of GST-RB (379–928) (200 ng/μl). For the Ad VAI gene repression assays whose results are shown in Fig. 2C, 24 μl of HeLa cell nuclear extract (7.5 μg/μl) was preincubated with approximately 14 μg of purified GST-RB (379–928) (200 ng/μl). Similar preincubation reactions were performed with either Dignam buffer D (mock depleted) or GST protein at the same amounts as were used for GST-RB (379–928). After affinity depletion, supernatants were used immediately in Ad VAI and human U6 transcription reactions as described previously. Transcription reaction mixtures were also supplemented with chromatographic fractions containing SNAPc (Mono-Q peak fraction; approximately 0.3 mg of protein per ml [20]) or TFIIIB (PII-B; approximately 0.6 mg/ml, [32]). For the experiment whose results are presented in Fig. 2B, 10 ng of recombinant human TBP (Promega) was also added as indicated. For the experiment presented in Fig. 2D, 20 μl of HeLa nuclear extract was treated with 6 μg of purified GST-RB (379–928) or GST as described above. Each supernatant (15 μl) and proteins bound to the beads after extensive washing were separated by SDS–12.5% PAGE and tested by Western blot analysis using rabbit anti-SNAP43 antiserum (CS48 [20]) or mouse monoclonal antibody (SL2 [33]).
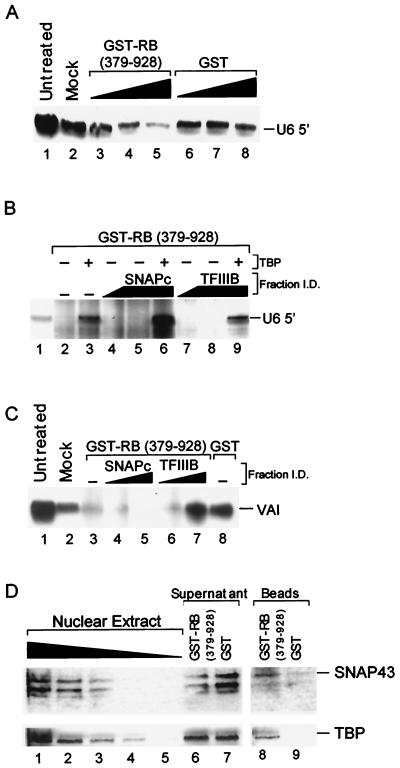
FIG. 2.
Different factors reconstitute Ad VAI and human U6 snRNA gene transcription in GST-RB (379–928)-treated extracts. (A) GST-RB (379–928) affinity depletion specifically inhibits U6 transcription. HeLa cell nuclear extracts (8 μl) were incubated with Dignam buffer D (mock lane 2) or 1, 2, or 3 μg of purified GST-RB (379–928) (lanes 3 to 5) or GST (lanes 6 to 8) for 30 min at 30°C. The recombinant proteins and associated factors were then removed by affinity purification with glutathione agarose. One-half of each treated extract was then tested for the ability to support human U6 snRNA transcription. Lane 1 shows the transcription supported by 4 μl of the untreated extract. (B) SNAPc but not TFIIIB acts cooperatively with TBP to reconstitute human U6 snRNA gene transcription. HeLa cell extracts were incubated with GST-RB (379–928) (lanes 2 to 9). After being treated with glutathione agarose beads, 5 μl of each treated extract was tested for the ability to support U6 transcription in the absence of chromatographic fractions (lane 2) or in the presence of chromatographic fractions containing SNAPc (2.7, 8, and 8 μl, lanes 4 to 6, respectively) or TFIIIB (2.7, 8, and 8 μl, lanes 7 to 9, respectively). Reactions shown in lanes 3, 6, and 9 were also complemented with 10 ng of recombinant TBP (Promega). Lane 1 shows transcription supported by 2 μl of the untreated extract. I.D., identification. (C) TFIIIB but not SNAPc reconstitutes Ad VAI gene transcription. HeLa cell nuclear extracts were incubated with Dignam buffer D (lane 2), GST-RB (379–928) (lanes 3 to 7), or GST (lane 8) for 30 min at 30°C. GST-RB (379–928) and GST were removed by incubating treated extracts with glutathione agarose beads for 4 h at 4°C. Depleted extracts (8 μl) were then tested for the ability to support VAI gene transcription in the absence of chromatographic fractions (lanes 3 and 8) or in the presence of chromatographic fractions containing SNAPc (2.7 and 8 μl, lanes 4 and 5, respectively) or TFIIIB (2.7 and 8 μl, lanes 6 and 7, respectively). Lane 1 shows transcription supported by 2 μl of the untreated extract. (D) SNAPc levels are reduced in RB-treated extracts. HeLa cell nuclear extract was treated with 6 μg of GST-RB (379–928) or GST as described above. Depleted extracts and proteins (15 μl) associated with the beads after extensive washing were separated by SDS–12.5% PAGE and tested by Western blot analysis using rabbit anti-SNAP43 antisera (top blot). The membrane was then stripped and reprobed using mouse anti-TBP antibodies (bottom panel). Lanes 1 to 5 contain 12, 6, 3, 1.5, and 0.75 μl of HeLa cell nuclear extract, respectively. The GST-RB (379–928)- and GST-treated extracts are shown in lanes 6 and 7, respectively. Lanes 8 and 9 contain proteins associated with the GST-RB (379–928) and GST agarose beads, respectively.
RB-SNAPc coimmunoprecipitation.
Approximately 300 μl of HeLa cell nuclear extract was incubated with 2 μg of mouse anti-RB (clone G3-245; Pharmingen) or anti-hemagglutinin (HA) (12CA5) antibody overnight at 4°C. Samples were diluted with 1 ml of HEMGT-150 containing protease inhibitors, and 20 μl of protein G agarose beads (Gibco-BRL) was added to each reaction mixture. Samples were further incubated at 4°C for 4 h. Antibody beads were washed three times in HEMGT-150 containing protease inhibitors (1 ml), and bound proteins were eluted by boiling in 1× Laemmli buffer prior to size fractionation by SDS–12.5% PAGE. SNAP43 was detected by Western blot analysis using an antibody specific to SNAP43 (CS48 [20]). To perform the reciprocal immunoprecipitations, approximately 300 μl of HeLa cell nuclear extract was incubated with 50 μl of protein A agarose beads (Boehringer Mannheim) precoupled with either rabbit anti-SNAP43 antibody or preimmune-phase antibodies. Reaction mixtures were incubated for 2 h at 4°C with mixing. Beads were washed extensively in Dignam buffer D (100 mM KCl) containing protease inhibitors, and bound proteins were then competitively eluted in 200 μl of Dignam buffer D containing either a specific peptide (CSH375 [20]) or a nonspecific peptide, each at 1 mg/ml. Eluted samples were precipitated with trichloroacetic acid (TCA). Precipitates were redissolved in 1× Laemmli buffer and size fractionated by SDS–12.5% PAGE. Full-length RB was detected by Western blot analysis using mouse monoclonal antibodies directed against an epitope contained within amino acids 300 to 380 (G3-245; Pharmingen).
Protein chromatography and EMSA analysis.
Nuclear extracts were prepared from HeLa cells by the method of Dignam et al. (9). SNAPc- and TFIIIB-containing fractions were generated essentially as described previously (20, 32, 48). The SNAPc fractions used were from the Mono-Q step of purification. The TFIIIB fractions used were from the P11-B step of purification. PSE-specific DNA binding by SNAPc was assayed by electrophoretic mobility shift assay (EMSA) as described previously (48).
GST pulldown assays.
Individual SNAPc subunits and full-length RB were individually expressed in vitro using rabbit reticulocyte lysates (TNT; Promega), and proteins were labeled with [35S]methionine. GST pulldown reactions were performed using 20 μl of glutathione agarose beads containing approximately 1 μg of GST-RB (379–928), GST-SNAP50 (1–411), GST-SNAP43 (1–368), or GST or beads alone. These were individually incubated with 10 μl of 35S-labeled proteins for 2 h at 4°C in 1 ml of HEMGT-150 containing protease inhibitors and 1 mM dithiothreitol. The specific combinations of proteins used are indicated in the legend of Fig. 5. Beads were washed extensively in HEMGT-150, and bound proteins were separated by SDS–17% PAGE. Proteins were stained with Coomassie blue to ensure equivalent loadings of GST-tagged proteins in the samples. Associated radioactive proteins were detected by autoradiography.
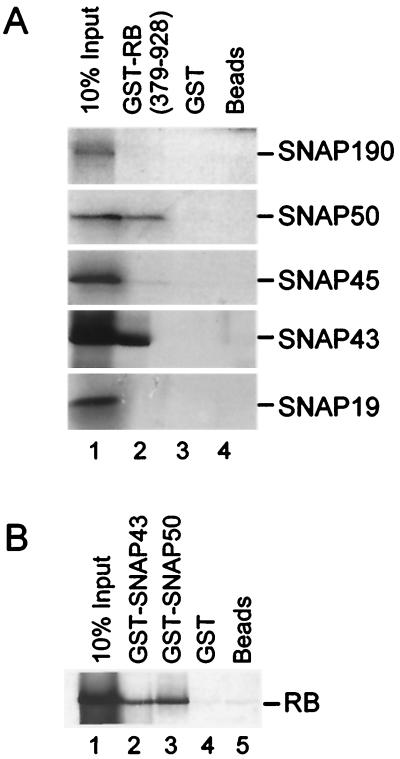
FIG. 5.
RB interacts with two components of SNAPc. (A) GST pulldown experiment performed to test interactions between individual SNAPc subunits and GST-RB (379–928). Each SNAPc subunit was expressed in vitro using rabbit reticulocyte lysates, and proteins were labeled with [35S]methionine. Lane 1 contains 10% of the 35S-labeled proteins used as inputs. These were tested for interactions with GST-RB (379–928) (lane 2), GST (lane 3), or glutathione agarose beads alone (lane 4). Proteins were size fractionated by SDS–12.5% PAGE and visualized by autoradiography. The identity of each SNAPc subunit is indicated. (B) Full-length RB was expressed in vitro and labeled with [35S]methionine. Lane 1 contains 10% of 35S-labeled RB used as input, which was tested for interaction with GST-SNAP43 (lane 2), GST-SNAP50 (lane 3), GST (lane 4), and beads alone (lane 5) as described above.
RESULTS
RB represses RNA polymerase III transcription.
RB is an important regulator of cellular growth, and its ability to perform this function can partially be attributed to regulation of RNA polymerase III activity. RB contains 928 amino acids and can be divided into at least three regions: the N-terminal region from amino acids 1 to 378, the A/B region from amino acids 393 to 772, and the C region from amino acids 768 to 869. Most functions ascribed to RB, including tumor suppressor activity and interactions with regulatory target proteins, require the A/B and/or C regions (56, 60).
To determine the function of RB in regulating RNA polymerase III activity, recombinant RB containing the A/B and C regions was tested for its ability to repress in vitro transcription by RNA polymerase III. Specifically, in vitro transcription assays of the Ad VAI gene and a human U6 snRNA gene were performed to compare levels of regulation of RNA polymerase III transcription by RB for genes containing gene-internal (class 2) and gene-external (class 3) promoter elements. Schematic representation of the core promoters of the genes used for this study are shown in Fig. 1A. The Ad VAI gene contains gene-internal A and B box control elements that are also characteristic of human tRNA genes. The core promoter regions of human U6 snRNA genes contain a PSE and a TATA box. In addition, the U6 gene contains a distal sequence element (DSE) that recruits Oct-1 to activate U6 transcription. The GST-RB (379–928) and GST proteins typically used for these experiments are shown in Fig. 1B. GST-RB (379–928) and GST were each expressed in E. coli and were purified to homogeneity by affinity purification using glutathione agarose beads. In each case, the full-length protein is the most prevalent species observed. To determine the effect of these proteins on RNA polymerase III transcription, increasing amounts of purified GST-RB (379–928) and GST were added to HeLa cell nuclear extracts and these extracts were tested for the ability to support Ad VAI transcription. As shown in Fig. 1C, GST-RB (379–928) inhibited transcription of the Ad VAI gene (top gel, lanes 2 to 5) compared to levels observed for the untreated extract (lane 1). This repression appears to be specific because addition of equivalent amounts of the GST control protein had no significant effect on Ad VAI transcription (lanes 6 to 9). The repression observed for RB is not limited to classical RNA polymerase III-transcribed genes containing gene-internal promoter elements. As shown in Fig. 1D, increasing amounts of GST-RB (379–928) significantly reduced RNA polymerase III transcription, which was correctly initiated from the human U6 promoter (lanes 2 to 4). Again, comparable levels of the GST control protein had no significant effect in these assays (lanes 5 to 7). Therefore, RB effectively represses in vitro transcription by RNA polymerase III.
The human U6 snRNA and Ad VAI promoters have distinct factor requirements for transcriptional repression by RB.
In order to repress gene transcription by RNA polymerase III, RB must target specific factors required for transcription of these genes. To identify these factors, chromatographic fractions previously demonstrated to reconstitute human U6 and Ad VAI transcription were tested for their ability to restore transcription in extracts that were treated with GST-RB (379–928). Potentially, these chromatographic fractions also contain the factor(s) that is targeted by RB, and these factors may act as a dominant inhibitor of RB function. Both Ad VAI and human U6 transcription can be reconstituted by using a combination of fractions obtained from the purification of HeLa cell extracts over a phosphocellulose P-11 column. These fractions include the P11-B fraction (containing RNA polymerase III as well as 0.38 M TFIIIB [32]) and the P11-C fraction (containing RNA polymerase III, TFIIIC, and SNAPc). For human U6 transcription, the P11-C fraction can be replaced with a chromatographic fraction further enriched for SNAPc (Mono-Q) and additional recombinant TBP (48). When recombinant TBP and chromatographic fractions containing SNAPc were initially tested for their ability to directly restore human U6 transcription in RB-treated extracts, none were able to counter RB repression (data not shown), perhaps because these reaction mixtures contain an excess of RB (379–928). Therefore, whether chromatographic fractions can restore transcription in RB-repressed extracts that have had GST-RB (379–928) removed by affinity purification was tested. First, the amounts of GST-RB (379–928) required for specific repression under these conditions were established. To perform these experiments, GST-RB (379–928) or GST was preincubated with HeLa cell nuclear extracts. Subsequently, GST-RB (379–928) was removed by affinity purification with glutathione agarose beads and extracts were then tested for the ability to support U6 transcription. As shown in Fig. 2A, diminished U6 transcription was observed with extracts affinity depleted with increasing amounts of GST-RB (379–928) (lanes 3 to 5) compared to that observed with extracts treated with similar amounts of GST protein (lanes 6 to 8). Therefore, GST-RB (379–928) can specifically repress human U6 transcription under these conditions and transcription levels remain low after removal of GST-RB (379–928) by affinity depletion.
To determine the identities of factors targeted by GST-RB (379–928), the ability of chromatographic fractions to reconstitute human U6 gene transcription in GST-RB (379–928) affinity-depleted extracts was then assessed. In the experiment whose results are shown in Fig. 2B, the GST-RB (379–928) depletion conditions were similar to those shown in lane 5 of Fig. 2A; however, less extract was tested for transcription and thus the starting level of transcription was reduced. Under these conditions, U6 transcription was abolished by GST-RB (379–928) affinity depletion (lane 2) compared to that in the untreated extract (lane 1). Addition of recombinant TBP alone restored significant activity to depleted extracts (lane 3). Therefore, TBP or a TBP-containing complex required for U6 snRNA gene transcription is functionally limiting in these RB-treated extracts. Neither fractions containing SNAPc (lanes 4 and 5) nor TFIIIB (lanes 7 and 8) restored transcription when added alone, even though these fractions contained significant levels of TBP. However, when chromatographic fractions containing SNAPc were complemented with recombinant TBP, enhanced activity was now observed (lane 6). This level is approximately threefold greater than that observed for TBP alone (lane 3) and significantly greater than that observed for SNAPc alone (lane 5). The cooperative effect observed for TBP with SNAPc is specific because this effect was not observed with TBP complemented with chromatographic fractions containing TFIIIB (lane 9). Therefore, alone, SNAPc cannot restore transcription, but together, SNAPc and recombinant TBP act cooperatively to restore U6 snRNA transcriptional activity to fractions treated with GST-RB (379–928).
The above results suggest that RB may target TBP or alternatively target SNAPc to control TBP activity but that TFIIIB may not be involved in RB repression of these genes. However, TFIIIB was previously described as a target for RB repression of RNA polymerase III activity (7, 29). To determine whether RNA polymerase III-transcribed genes with intragenic promoter elements have similar factor requirements for relief from RB repression, the ability of chromatographic fractions to restore Ad VAI transcription was also tested. As shown in Fig. 2C, the GST-RB (379–928)-treated extract was compromised for Ad VAI transcriptional activity (lane 3) compared to that of either mock-treated (lane 2) or untreated (lane 1) HeLa cell nuclear extracts. Transcriptional activity was not restored by addition of chromatographic fractions containing SNAPc (lanes 4 and 5). This result was expected because SNAPc is not required for transcription of these genes. However, Ad VAI transcriptional activity is effectively reconstituted by addition of increasing amounts of chromatographic fractions containing TFIIIB (lanes 6 and 7). Transcription in these samples is comparable to levels obtained with either the mock-treated (lane 2) or GST-treated (lane 8) extracts. Therefore, fractions containing TFIIIB effectively reconstitute Ad VAI transcription, and this reconstitution is specific because restoration is not observed with SNAPc-containing fractions. These data are consistent with those previously described (7, 29) and support the hypothesis that TFIIIB is one target for gene regulation by RB.
One explanation for the reduced U6 transcription following affinity depletion with GST-RB (379–928) is that TBP or higher-order complexes containing TBP and SNAPc are removed. Thus, affinity depletion of nuclear extracts using GST-RB (379–928) should also result in a measurable reduction in the levels of these factors. Therefore, a Western blot analysis was performed using anti-SNAP43 antibody (CS48 [20]) and anti-TBP antibody (SL2 [33]) to determine whether treatment of nuclear extracts with GST-RB (379–928) alters the level of SNAPc or TBP. As shown in Fig. 2D (top blots), depleting extracts with GST-RB (379–928) resulted in a significant decrease in SNAP43 levels (lane 6) compared to amounts present in extracts depleted with the GST control protein (lane 7). Comparison of SNAP43 in the GST-RB (379–928)-treated sample and in decreasing amounts of HeLa cell nuclear extract (lanes 1 to 5) indicated that at least 50% of endogenous SNAP43 was removed by affinity depletion with GST-RB (379–928). This same membrane was then reprobed using antibodies directed against TBP, and the results are shown in Fig. 2D (lower blots). In this case, no significant difference in TBP levels was observed for the GST-RB (379–928)- and GST-treated samples, suggesting that TBP is not effectively depleted in these assays. This may mean that RB does not target TBP. Alternatively, since TBP is present in numerous TBP-containing complexes and RB may target only some of these complexes, the removal of a minor proportion of the total TBP may not be detectable in these assays. Indeed, TBP was associated with the GST-RB (379–928) agarose beads (lane 8) but not with the GST-agarose beads (lane 9), suggesting that a minor proportion of the total TBP can associate specifically with GST-RB (379–928). Our results are also consistent with the notion that GST-RB (379–928) targets SNAPc in a stable fashion.
Endogenous RB associates with SNAPc.
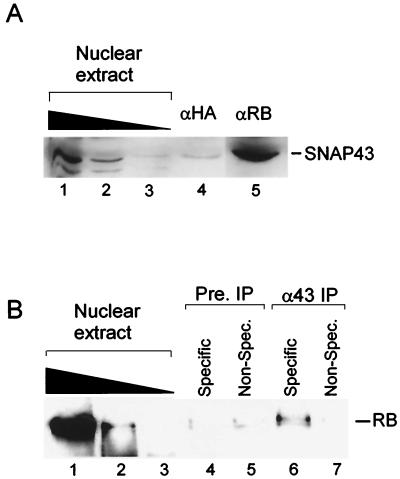
The above-described experiments suggest that high levels of GST-RB (379–928) can target SNAPc to potentially control human U6 gene transcription. However, it is important to determine whether an association between endogenous RB and SNAPc is possible. To determine whether endogenous RB and SNAPc can interact, coimmunoprecipitation experiments were performed by incubating HeLa cell nuclear extracts with either anti-HA or anti-RB antibody. Proteins bound by these antibodies were eluted by boiling in Laemmli buffer and separated by SDS-PAGE for analysis by anti-SNAP43 antibody Western blotting. As shown in Fig. 3A, significant levels of SNAP43 were detected in the samples immunoprecipitated with the anti-RB (lane 5) but not with the anti-HA (lane 4) antibody. To confirm these results, the reciprocal experiment was performed by testing the coimmunoprecipitation of RB through immunoprecipitation of the SNAP43 subunit of SNAPc (Fig. 3B). In these experiments the immunoprecipitation methodology was modified to reduce the high nonspecific background that was observed due to cross-reaction of the secondary antibody with the heavy chain from the anti-SNAP43 antibody. HeLa cell nuclear extracts were incubated with agarose beads covalently cross-linked with either rabbit anti-SNAP43 (CS48 [20]) or preimmune-phase antibody. Each sample was then divided in half. Bound proteins were competitively eluted with either the specific peptide used to generate the anti-SNAP43 antibody (1 mg/ml, peptide CSH375 [20]) or a nonspecific peptide. These elution conditions were chosen to minimize the disruption of protein-protein interactions and reduce contamination of the eluted proteins with the heavy chain from the SNAP43 antibodies. Eluted proteins were then concentrated by precipitation with TCA, size fractionated by SDS–12.5% PAGE, and analyzed by Western blot analysis using antibodies directed against RB. As shown in Fig. 3, a significant amount of endogenous RB is coimmunoprecipitated using anti-SNAP43 antibodies and eluted with the specific peptide (lane 6). The coimmunoprecipitation of RB with SNAPc is specific because it is not nonspecifically eluted from the anti-SNAP43 antibodies (lane 7) and it also is not observed under any immunoprecipitation conditions using preimmune-phase antibodies (lanes 4 and 5). Therefore, a subpopulation of endogenous RB associates with endogenous SNAPc.
FIG. 3.
Endogenous RB is associated with SNAPc. (A) Endogenous SNAP43 is coimmunoprecipitated with RB. Approximately 300 μl of HeLa cell nuclear extract was incubated with mouse anti-RB (G3-245; Pharmingen) (lane 5) or anti-HA (12CA5) (lane 4) antibodies (αRB and αHA, respectively) overnight. Protein-antibody complexes were removed by affinity purification using protein G agarose beads (Gibco-BRL). The beads were washed extensively, and bound proteins were resolved by SDS–12.5% PAGE. SNAP43 association was detected by Western blot analysis using antibodies specific to SNAP43 (CS48 [20]). Lanes 1 to 3 show the amount of SNAP43 present in 10, 3, and 1 μl of nuclear extract. (B) Endogenous RB coimmunoprecipitated with SNAPc. HeLa cell nuclear extracts were incubated with antibodies directed against the SNAP43 subunit of SNAPc (lanes 6 and 7) or rabbit preimmune-phase antibodies (lanes 4 and 5) covalently coupled to protein A agarose beads (Boehringer Mannheim). After protein binding and extensive washing, each reaction mixture was divided in half. Bound proteins were competitively eluted in buffer containing either a specific peptide (CSH375 [20]) (lanes 4 and 6) or an irrelevant peptide (CSH374) (lanes 5 and 7). Eluted proteins were precipitated with TCA and size fractionated by SDS–12.5% PAGE, and the levels of RB were detected by Western blot analysis. Lanes 1 to 3 show the levels of RB detected in 10, 3, and 1 μl of HeLa cell nuclear extract after precipitation with TCA. Pre. IP, preimmune-phase antibody immunoprecipitation; α43 IP, anti-SNAP43 antibody immunoprecipitation; Non-Spec., nonspecific.
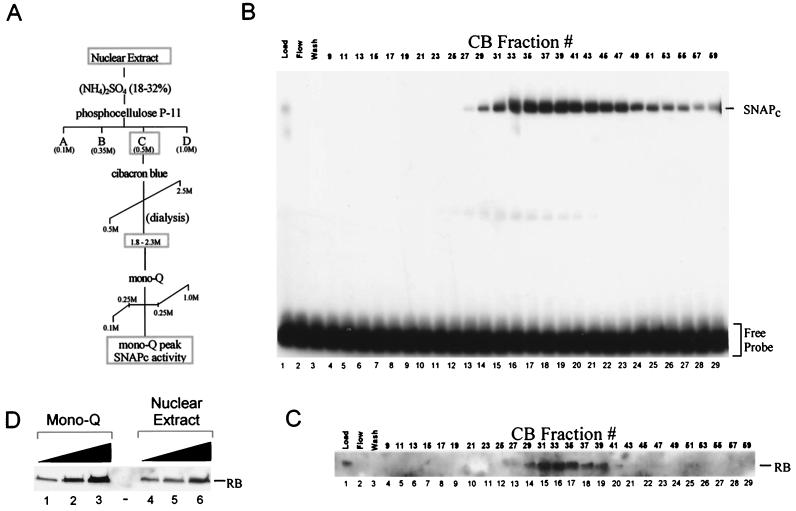
If the association between RB and SNAPc is stable, then it is possible that a significant amount of endogenous RB will cofractionate with SNAPc during extensive chromatographic purification of SNAPc. Therefore, to further test the association between endogenous RB and SNAPc, SNAPc was purified from HeLa cell extracts and the copurification of RB was monitored by Western blot analysis. The purification scheme typically used to fractionate SNAPc is shown in Fig. 4A. This scheme applies to the purification of SNAPc from both HeLa cell nuclear and S-100 extracts. Briefly, HeLa cell extracts were selectively precipitated by ammonium sulfate prior to fractionation using a phosphocellulose P-11 column. Proteins bound to this column were step eluted in buffers containing increasing concentrations of KCl to generate the P11-A, -B, -C, and -D fractions. The majority of SNAPc is present in the P11-C fraction. The P11-C fraction was then directly passed over a Cibacron blue (CB) affigel column, and bound proteins were eluted with a linear gradient from 500 mM KCl (0% ethylene glycol) to 2.5 M KCl (25% ethylene glycol). Fractions generated from this column were dialyzed against Q100 buffer and then tested for DNA binding activity as previously described (48). EMSA of the CB fractions revealed that the majority of PSE binding activity is present in CB fractions 29 to 59 (Fig. 4B). This peak corresponds to fractions eluted at KCl concentrations between 1.8 and 2.5 M KCl and is indicative of SNAPc activity. These fractions were then tested for the presence of RB by Western blot analysis, and the results are shown in Fig. 4C. A significant level of RB is present in the CB fractions, with a peak observed in fractions 29 to 39, indicating that RB cofractionates with SNAPc. However, RB is present only in a subset of the fractions containing significant levels of SNAPc activity. For example, CB fractions 39, 41, and 43 contain similar levels of DNA binding activity and yet contain distinctly different levels of RB. This finding suggests that RB is associated with a subpopulation of SNAPc.
FIG. 4.
Endogenous RB cofractionates with SNAPc during chromatographic purification. (A) Schematic representation of the chromatographic purification used to purify SNAPc. (B) Characterization of chromatographic fractions for PSE binding activity. Approximately 10-μl aliquots of the P11-C fraction (load, ca. 0.5 mg of protein per ml) and fractions obtained from the CB step of purification were tested for DNA binding activity by EMSA using radioactive probes containing a high-affinity mouse U6 PSE and the TATA box. The positions of the unbound probe (free probe) and SNAPc bound to DNA (SNAPc) are labeled. The peak of SNAPc is contained in fractions 27 to 59 and corresponds to fractions eluted in buffer containing between 1.8 and 2.5 M KCl. (C) Anti-RB antibody Western blot analysis of SNAPc-containing fractions shown in panel B. Approximately 10-μl aliquots of each fraction were tested for the presence of RB using mouse anti-RB monoclonal antibodies that recognize an epitope between amino acids 300 and 380 of human RB (antibody G3-245; Pharmingen). Significant levels of RB are detected in CB fractions 29 to 39. (D) Anti-RB antibody Western blot analysis of highly purified SNAPc fractions. The peak fractions of SNAPc activity from the CB column (CB fractions 29 to 59) were pooled and further purified by anion-exchange column chromatography using a Mono-Q HR 5/5 column (Pharmacia). The peak of SNAPc elutes from this column as a single peak in buffer containing 250 mM KCl. Increasing amounts of the Mono-Q peak fractions (lanes 1 to 3, approximately 0.8, 2.4, and 7.5 μg of total protein, respectively) and nuclear extract starting material (lanes 4 to 6, approximately 10, 30, and 100 μg of total protein, respectively) were analyzed by SDS–12.5% PAGE and Western blotting using antibodies directed against RB.
To further characterize the association of RB with SNAPc, the peak of DNA binding activity from the CB column was purified by anion-exchange chromatography using a Mono-Q column (Pharmacia). The vast majority of SNAPc is eluted from this column isocratically in buffer containing 250 mM KCl. By this stage of purification, SNAPc has been purified approximately 2,000-fold (data not shown). To test for the presence of RB in these fractions, increasing amounts of the Mono-Q peak (3, 10, and 30 μl, ca. 0.25 mg of protein per ml) were analyzed for the presence of RB, and the results are shown in Fig. 4D. Again, significant levels of RB are present in these fractions containing highly purified SNAPc (lanes 1 to 3). More RB is detected in these Mono-Q fractions than in HeLa cell nuclear extracts (1, 3, and 10 μl, ca. 10 mg of protein per ml) shown in lanes 4 to 6. These data indicate that RB is approximately one- to threefold more concentrated in the Mono-Q fractions than in HeLa cell nuclear extracts. Correspondingly, this represents an estimated 30- to 100-fold purification of RB during the purification of SNAPc. Therefore, a subpopulation of endogenous RB is associated with a subpopulation of endogenous SNAPc.
RB can interact with SNAPc.
RB associates with SNAPc as detected both by coimmunoprecipitation from nuclear extracts and by cofractionation during SNAPc purification. However, it is not known whether this association is mediated by additional factors or whether RB directly targets SNAPc. Therefore, to determine whether direct interactions between RB and SNAPc are possible, GST pulldown assays were performed with GST-RB (379–928) and individual subunits of SNAPc. Each SNAPc subunit was expressed separately in rabbit reticulocyte lysates, and proteins were labeled with [35S]methionine. These labeled proteins were then mixed with GST-RB (379–928) that had been prebound to glutathione agarose beads. Proteins were also tested for interactions with GST protein or beads alone as negative controls. The results of these GST pulldown assays are shown in Fig. 5A. Significant interactions were observed between GST-RB (379–928) and two SNAPc subunits: SNAP43 and SNAP50. These interactions are specific, as no interaction was detected between these proteins and either the GST samples or the control samples with beads alone. In contrast, no significant interactions were observed between GST-RB (379–928) and any other SNAPc subunits. As an additional control, the reciprocal experiment was performed (Fig. 5B). Full-length RB (1–928) was expressed in vitro and labeled with [35S]methionine and was then tested for interactions with GST-SNAP43 and GST-SNAP50. Again, full-length RB interacted with both SNAP43 and SNAP50, and this interaction was specific because little cross-reaction with the samples with GST alone or beads alone was observed. Therefore, the association of RB with SNAPc previously observed may involve direct protein-protein interactions between RB and SNAPc.
DISCUSSION
RB is an important tumor suppressor protein that acts to regulate cell growth in part by controlling progression through the cell cycle. Typically, RB is thought to act by regulating expression of genes that are important for executing specific cell cycle functions. The discovery that RB represses transcription by RNA polymerases I (4) and III (63) reveals that RB also acts to regulate expression of highly transcribed genes encoding nontranslated RNAs.
The ability of RB to regulate gene expression is determined by targeting RB to specific gene promoters. Control of RNA polymerase II transcription typically involves recruiting RB to gene promoters by direct protein-protein interactions with the transcriptional regulatory protein E2F (11). Interactions between RB and other regulatory proteins are also important for RB function. Nonetheless, RB does not directly regulate transcription of most genes by RNA polymerase II. In contrast, RB appears to generally repress RNA polymerase III activity (63). This suggests that RB interacts with the general transcriptional machinery required for RNA polymerase III transcription to effect repression. Indeed, the general transcription factor TFIIIB, composed of TBP and BRF, is important for expression of 5S rRNA and tRNA genes and also appears important for regulation of these genes by RB. In our experiments, addition of chromatographic fractions containing TFIIIB restored Ad VAI transcription to extracts that were affinity depleted with GST-RB (379–928). This result suggests that TFIIIB is limiting for Ad VAI transcription in extracts affinity depleted with GST-RB (379–928) and is consistent with the previously suggested hypothesis that RB targets TFIIIB for repression of these genes (7, 29).
The human U6 gene family has distinctly different promoter elements contained entirely in the 5′ regions flanking these genes. In vitro, RB also effectively represses transcription of these genes and in our assays repression of U6 gene transcription is observed at moderately lower concentrations of RB than that required for repression of Ad VAI gene transcription (Fig. 1 and data not shown). Many explanations might explain the differential regulation of these two genes by RB, but one explanation that we favor is that RB specifically interacts with different factors specialized for transcription of these genes and that these interactions are important for repression. Transcription of human U6 snRNA genes by RNA polymerase III requires the general transcription factors SNAPc, BRF2, and TBP (37, 48), whereas TFIIIB, consisting of BRF and TBP, appears not to be required (39). Fractions containing TFIIIB were unable to restore U6 snRNA gene transcription in extracts that were affinity depleted with GST-RB (379–928). Therefore, TFIIIB appears to be important for RB repression of Ad VAI but not human U6 snRNA gene transcription.
In order to test whether the general transcription factor SNAPc has a role in mediating RB regulation, chromatographic fractions containing SNAPc were added to GST-RB (379–928)-treated extracts. In these experiments, SNAPc fractions did not restore U6 snRNA gene transcription, which suggests that SNAPc is not involved in RB repression. One possibility is that RB, also present in these fractions, represses SNAPc activity. We consider this possibility unlikely because SNAPc fractions obtained by biochemical fractionation efficiently restore U6 transcription in extracts immunodepleted of endogenous SNAPc (data not shown), suggesting that the levels of RB present are not inhibitory for SNAPc function. A second possibility is that other factors not contained in the SNAPc fractions are targeted and removed from extracts by affinity depletion with GST-RB (379–928). Indeed, recombinant TBP alone restored transcription from the U6 promoter when it was added to GST-RB (379–928)-affinity-depleted extracts. This result suggests that TBP or higher-order complexes containing TBP are important for RB regulation of human U6 snRNA genes.
Although recombinant TBP alone restored U6 transcription, the levels of transcription observed were increased by the addition of fractions containing SNAPc, but not TFIIIB, to reaction mixtures containing recombinant TBP. This result suggests that SNAPc is also involved in the pathway for RB regulation of human U6 genes. Consistent with these observations, the levels of endogenous SNAPc were reduced in nuclear extracts affinity depleted with an excess of GST-RB (379–928). One function of SNAPc is to recruit TBP to the TATA box contained in human U6 snRNA gene promoters (40). However, high levels of recombinant TBP can overcome the requirement for SNAPc for U6 transcription (data not shown). The ability of TBP to function alone in these assays, therefore, may be because high levels of TBP can bypass the requirement for SNAPc. Thus, RB may control SNAPc to affect TBP activity at these promoters. If RB targets SNAPc to repress human U6 transcription, then it was expected that there should be a measurable association between endogenous RB and SNAPc. Indeed, a fraction of the total endogenous RB associates with SNAPc. First, a modest level of RB was observed to coimmunoprecipitate with SNAPc. Furthermore, copurification of RB with SNAPc was observed during the biochemical fractionation of SNAPc. In support of these results, RB can interact with the SNAP43 and SNAP50 subunits of SNAPc, which suggests that direct interactions between RB and SNAPc may be important for regulating U6 gene expression. These results, however, do not rule out the possibility that additional factors may also contribute to the function of RB at these promoters. It remains to be determined whether the recently described alternatively spliced forms of human BRF (37) also participate in RB repression of human U6 gene transcription.
For RNA polymerase II transcription, two models have been proposed to explain the mechanism for gene repression by RB. In one model, RB binds E2F at the promoter to abrogate the potential of E2F to activate transcription. E2F recruits the general transcription factor TFIID to the promoter in a TFIIA-dependent manner, and this promoter is sensitive to the presence of RB. After TFIID-TFIIA complex formation, gene expression becomes resistant to repression by RB (46). In a second model, RB represses transcription of some cell cycle-responsive genes that contain E2F binding sites via recruitment of a histone deactylase (HDAC) (2, 35, 36). Thus, transcriptional repression by RB may involve modification of chromatin structure. HDACs remove acetyl groups from histones and consequently reconfigure the chromatin structure to a nonpermissive state for transcription. The recruitment of HDACs by RB may be direct (13, 36) or require a tethering protein (28). However, not all promoters are sensitive to recruitment of HDACs, and thus it appears that these two models for RB repression are promoter selective (35).
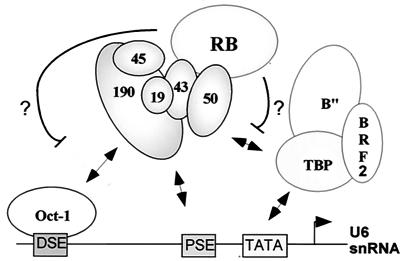
How does RB regulate U6 snRNA gene transcription by RNA polymerase III? Potentially, histone deacetylation is an attractive model to explain regulation of U6 snRNA gene transcription in vivo. In chromatin reconstitution experiments, the human U6 snRNA gene contains a positioned nucleosome located between the DSE and the PSE (53). Transcription of the U6 wild-type gene is enhanced after chromatin assembly, and thus, modification of histones may potentially repress gene activity. However, in our in vitro system we observed repression of RNA polymerase III transcription using naked DNA templates. Thus, it appears that chromatin modification is not essential for repression of RNA polymerase III in vitro. In an alternative model, RB acts to inhibit preinitiation complex assembly at human U6 promoters. For example, RB could disrupt the interaction between the transcriptional activator Oct-1 and SNAPc (Fig. 6). Direct contacts between the Oct-1 POU domain and the SNAP190 subunit of SNAPc facilitate SNAPc binding to the PSE (14, 41). Interactions between RB and SNAPc may prevent interactions between Oct-1 and SNAPc and therefore prevent recruitment of SNAPc to human U6 gene promoters. This model is reminiscent of the role of RB in repressing RNA polymerase II transcription by modulating E2F-mediated pre-initiation complex assembly with TFIID and TFIIA (46). RB may also interfere with pre-initiation complex assembly by preventing DNA binding by SNAPc or TBP. By binding to SNAPc, RB may directly prevent this core promoter complex from binding to the PSE in the promoter of human U6 snRNA genes. Another interesting possibility is that RB disrupts communication between SNAPc and TBP. The binding of SNAPc to the PSE and of recombinant TBP to the TATA box is cooperative, and this is important for transcription of the U6 snRNA gene (40). Therefore, RB may disrupt TBP recruitment by interfering with the function of SNAPc for TBP recruitment. Interestingly, SNAPc interacts well with TBP, and this interaction may involve the SNAP43 subunit (20), which also binds RB in our assays. Thus, potential interactions between SNAP43 and RB may modulate simultaneous interactions between SNAP43 and TBP.
FIG. 6.
Model for RB repression of human U6 snRNA gene transcription. There are several mechanisms by which RB may repress U6 snRNA gene expression by RNA polymerase III. RB may interact with SNAPc and prevent DNA binding by SNAPc. Alternatively, RB may disrupt protein-protein communications that are important for U6 transcription. Targeting interactions between SNAPc and either the transcriptional activator protein Oct-1 or TBP are predicted to repress human U6 snRNA gene transcription. Finally, RB may also interact with both SNAPc and TBP-BRF2-B" simultaneously to block further preinitiation complex assembly.
RB plays an important role in coordinating RNA polymerase III activity, and our data indicate that RB does this by targeting TFIIIB and SNAPc or TBP. Clearly, regulating TBP is important and RB appears to target core-promoter complexes that are important for TBP function. TFIIIB and SNAPc play crucial early roles in pre-initiation complex assembly at RNA polymerase III gene promoters, and therefore, these are attractive targets for regulating RNA polymerase III activity. By understanding the mechanisms by which expression of nontranslated RNAs is controlled, we can define the contribution of these RNAs to the regulation of normal cell growth. Importantly, the availability of essential nontranslated RNAs may act to limit cell growth and RB repression of genes encoding these RNAs likely has to be overcome prior to tumor progression.
ACKNOWLEDGMENTS
We are indebted to Nouria Hernandez for her generous support, helpful advice, and contribution of numerous reagents. We also gratefully thank Beicong Ma for constructing the GST-RB (379–928) expression plasmid and David Arnosti, Zachary Burton, and Craig Hinkley for critical reading of the manuscript.
This work was supported by grants from the American Cancer Society (RPG-00-263-01-GMC) and the Michigan State University Intramural Research Grant Program. Generous support was also provided by the Michigan State University College of Human Medicine and College of Natural Science.
REFERENCES
- 1.Bai L, Wang Z, Yoon J B, Roeder R G. Cloning and characterization of the beta subunit of human proximal sequence element-binding transcription factor and its involvement in transcription of small nuclear RNA genes by RNA polymerases II and III. Mol Cell Biol. 1996;16:5419–5426. doi: 10.1128/mcb.16.10.5419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brehm A, Miska E A, McCance D J, Reid J L, Bannister A J, Kouzarides T. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature. 1998;391:597–601. doi: 10.1038/35404. [DOI] [PubMed] [Google Scholar]
- 3.Carbon P, Murgo S, Ebel J P, Krol A, Tebb G, Mattaj L W. A common octamer motif binding protein is involved in the transcription of U6 snRNA by RNA polymerase III and U2 snRNA by RNA polymerase II. Cell. 1987;51:71–79. doi: 10.1016/0092-8674(87)90011-0. [DOI] [PubMed] [Google Scholar]
- 4.Cavanaugh A H, Hempel W M, Taylor L J, Rogalsky V, Todorov G, Rothblum L I. Activity of RNA polymerase I transcription factor UBF blocked by Rb gene product. Nature. 1995;374:177–180. doi: 10.1038/374177a0. [DOI] [PubMed] [Google Scholar]
- 5.Chen P L, Riley D J, Chen Y, Lee W H. Retinoblastoma protein positively regulates terminal adipocyte differentiation through direct interaction with C/EBPs. Genes Dev. 1996;10:2794–2804. doi: 10.1101/gad.10.21.2794. [DOI] [PubMed] [Google Scholar]
- 6.Chen P L, Scully P, Shew J Y, Wang J Y, Lee W H. Phosphorylation of the retinoblastoma gene product is modulated during the cell cycle and cellular differentiation. Cell. 1989;58:1193–1198. doi: 10.1016/0092-8674(89)90517-5. [DOI] [PubMed] [Google Scholar]
- 7.Chu W M, Wang Z, Roeder R G, Schmid C W. RNA polymerase III transcription repressed by Rb through its interactions with TFIIIB and TFIIIC2. J Biol Chem. 1997;272:14755–14761. doi: 10.1074/jbc.272.23.14755. [DOI] [PubMed] [Google Scholar]
- 8.Cobrinik D, Dowdy S F, Hinds P W, Mittnacht S, Weinberg R A. The retinoblastoma protein and the regulation of cell cycling. Trends Biochem Sci. 1992;17:312–315. doi: 10.1016/0968-0004(92)90443-d. [DOI] [PubMed] [Google Scholar]
- 9.Dignam J D, Martin P L, Shastry B S, Roeder R G. Eukaryotic gene transcription with purified components. Methods Enzymol. 1983;101:582–598. doi: 10.1016/0076-6879(83)01039-3. [DOI] [PubMed] [Google Scholar]
- 10.Dunaief J L, Strober B E, Guha S, Khavari P A, Alin K, Luban J, Begemann M, Crabtree G R, Goff S P. The retinoblastoma protein and BRG1 form a complex and cooperate to induce cell cycle arrest. Cell. 1994;79:119–130. doi: 10.1016/0092-8674(94)90405-7. [DOI] [PubMed] [Google Scholar]
- 11.Dyson N. The regulation of E2F by pRB-family proteins. Genes Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- 12.Ewen M E, Sluss H K, Sherr C J, Matsushime H, Kato J, Livingston D M. Functional interactions of the retinoblastoma protein with mammalian D-type cyclins. Cell. 1993;73:487–497. doi: 10.1016/0092-8674(93)90136-e. [DOI] [PubMed] [Google Scholar]
- 13.Ferreira R, Magnaghi-Jaulin L, Robin P, Harel-Bellan A, Trouche D. The three members of the pocket proteins family share the ability to repress E2F activity through recruitment of a histone deacetylase. Proc Natl Acad Sci USA. 1998;95:10493–10498. doi: 10.1073/pnas.95.18.10493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ford E, Strubin M, Hernandez N. The Oct-1 POU domain activates snRNA gene transcription by contacting a region in the SNAPc largest subunit that bears sequence similarities to the Oct-1 coactivator OBF-1. Genes Dev. 1998;12:3528–3540. doi: 10.1101/gad.12.22.3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gu W, Schneider J W, Condorelli G, Kaushal S, Mahdavi V, Nadal-Ginard B. Interaction of myogenic factors and the retinoblastoma protein mediates muscle cell commitment and differentiation. Cell. 1993;72:309–324. doi: 10.1016/0092-8674(93)90110-c. [DOI] [PubMed] [Google Scholar]
- 16.Helin K, Lees J A, Vidal M, Dyson N, Harlow E, Fattaey A. A cDNA encoding a pRB-binding protein with properties of the transcription factor E2F. Cell. 1992;70:337–350. doi: 10.1016/0092-8674(92)90107-n. [DOI] [PubMed] [Google Scholar]
- 17.Henry R W, Ford E, Mital R, Mittal V, Hernandez N. Crossing the line between RNA polymerases: transcription of human snRNA genes by RNA polymerases II and III. Cold Spring Harbor Symp Quant Biol. 1998;63:111–120. doi: 10.1101/sqb.1998.63.111. [DOI] [PubMed] [Google Scholar]
- 18.Henry R W, Ma B, Sadowski C L, Kobayashi R, Hernandez N. Cloning and characterization of SNAP50, a subunit of the snRNA-activating protein complex SNAPc. EMBO J. 1996;15:7129–7136. [PMC free article] [PubMed] [Google Scholar]
- 19.Henry R W, Mittal V, Ma B, Kobayashi R, Hernandez N. SNAP19 mediates the assembly of a functional core promoter complex (SNAPc) shared by RNA polymerases II and III. Genes Dev. 1998;12:2664–2672. doi: 10.1101/gad.12.17.2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henry R W, Sadowski C L, Kobayashi R, Hernandez N. A TBP-TAF complex required for transcription of human snRNA genes by RNA polymerase II and III. Nature. 1995;374:653–656. doi: 10.1038/374653a0. [DOI] [PubMed] [Google Scholar]
- 21.Horowitz J M, Park S H, Bogenmann E, Cheng J C, Yandell D W, Kaye F J, Minna J D, Dryja T P, Weinberg R A. Frequent inactivation of the retinoblastoma anti-oncogene is restricted to a subset of human tumor cells. Proc Natl Acad Sci USA. 1990;87:2775–2779. doi: 10.1073/pnas.87.7.2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horowitz J M, Yandell D W, Park S H, Canning S, Whyte P, Buchkovich K, Harlow E, Weinberg R A, Dryja T P. Point mutational inactivation of the retinoblastoma antioncogene. Science. 1989;243:937–940. doi: 10.1126/science.2521957. [DOI] [PubMed] [Google Scholar]
- 23.Hsieh J K, Chan F S, O'Connor D J, Mittnacht S, Zhong S, Lu X. RB regulates the stability and the apoptotic function of p53 via MDM2. Mol Cell. 1999;3:181–193. doi: 10.1016/s1097-2765(00)80309-3. [DOI] [PubMed] [Google Scholar]
- 24.Johnson D G, Schwarz J K, Cress W D, Nevins J R. Expression of transcription factor E2F1 induces quiescent cells to enter S phase. Nature. 1993;365:349–352. doi: 10.1038/365349a0. [DOI] [PubMed] [Google Scholar]
- 25.Kaelin W G, Jr, Krek W, Sellers W R, DeCaprio J A, Ajchenbaum F, Fuchs C S, Chittenden T, Li Y, Farnham P J, Blanar M A, et al. Expression cloning of a cDNA encoding a retinoblastoma-binding protein with E2F-like properties. Cell. 1992;70:351–364. doi: 10.1016/0092-8674(92)90108-o. [DOI] [PubMed] [Google Scholar]
- 26.Kassavetis G A, Bartholomew B, Blanco J A, Johnson T E, Geiduschek E P. Two essential components of the Saccharomyces cerevisiae transcription factor TFIIIB: transcription and DNA-binding properties. Proc Natl Acad Sci USA. 1991;88:7308–7312. doi: 10.1073/pnas.88.16.7308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaye F J, Kratzke R A, Gerster J L, Horowitz J M. A single amino acid substitution results in a retinoblastoma protein defective in phosphorylation and oncoprotein binding. Proc Natl Acad Sci USA. 1990;87:6922–6926. doi: 10.1073/pnas.87.17.6922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lai A, Lee J M, Yang W M, DeCaprio J A, Kaelin W G, Jr, Seto E, Branton P E. RBP1 recruits both histone deacetylase-dependent and -independent repression activities to retinoblastoma family proteins. Mol Cell Biol. 1999;19:6632–6641. doi: 10.1128/mcb.19.10.6632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Larminie C G, Cairns C A, Mital R, Martin K, Kouzarides T, Jackson S P, White R J. Mechanistic analysis of RNA polymerase III regulation by the retinoblastoma protein. EMBO J. 1997;16:2061–2071. doi: 10.1093/emboj/16.8.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lescure A, Tebb G, Mattaj I W, Krol A, Carbon P. A factor with Sp1 DNA-binding specificity stimulates Xenopus U6 snRNA in vivo transcription by RNA polymerase III. J Mol Biol. 1992;228:387–394. doi: 10.1016/0022-2836(92)90828-8. [DOI] [PubMed] [Google Scholar]
- 31.Lobo S M, Hernandez N. A 7 bp mutation converts a human RNA polymerase II snRNA promoter into an RNA polymerase III promoter. Cell. 1989;58:55–67. doi: 10.1016/0092-8674(89)90402-9. [DOI] [PubMed] [Google Scholar]
- 32.Lobo S M, Tanaka M, Sullivan M L, Hernandez N. A TBP complex essential for transcription from TATA-less but not TATA-containing RNA polymerase III promoters is part of the TFIIIB fraction. Cell. 1992;71:1029–1040. doi: 10.1016/0092-8674(92)90397-u. [DOI] [PubMed] [Google Scholar]
- 33.Lobo-Ruppert S M, McCulloch V, Meyer M, Bautista C, Falkowski M, Stunnenberg H G, Hernandez N. Monoclonal antibodies directed against the amino-terminal domain of human TBP cross-react with TBP from other species. Hyrbridoma. 1996;15:55–68. doi: 10.1089/hyb.1996.15.55. [DOI] [PubMed] [Google Scholar]
- 34.Lu J, Danielsen M. Differential regulation of androgen and glucocorticoid receptors by retinoblastoma protein. J Biol Chem. 1998;273:31528–31533. doi: 10.1074/jbc.273.47.31528. [DOI] [PubMed] [Google Scholar]
- 35.Luo R X, Postigo A A, Dean D C. Rb interacts with histone deacetylase to repress transcription. Cell. 1998;92:463–473. doi: 10.1016/s0092-8674(00)80940-x. [DOI] [PubMed] [Google Scholar]
- 36.Magnaghi-Jaulin L, Groisman R, Naguibneva I, Robin P, Lorain S, Le Villain J P, Troalen F, Trouche D, Harel-Bellan A. Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature. 1998;391:601–605. doi: 10.1038/35410. [DOI] [PubMed] [Google Scholar]
- 37.McCulloch V, Hardin P, Peng W, Ruppert J M, Lobo-Ruppert S M. Alternatively spliced hBRF variants function at different RNA polymerase III promoters. EMBO J. 2000;19:4134–4143. doi: 10.1093/emboj/19.15.4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mihara K, Cao X R, Yen A, Chandler S, Driscoll B, Murphree A L, T'Ang A, Fung Y K. Cell cycle-dependent regulation of phosphorylation of the human retinoblastoma gene product. Science. 1989;246:1300–1303. doi: 10.1126/science.2588006. [DOI] [PubMed] [Google Scholar]
- 39.Mital R, Kobayashi R, Hernandez N. RNA polymerase III transcription from the human U6 and adenovirus type 2 VAI promoters has different requirements for human BRF, a subunit of human TFIIIB. Mol Cell Biol. 1996;16:7031–7042. doi: 10.1128/mcb.16.12.7031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mittal V, Hernandez N. Role for the amino-terminal region of human TBP in U6 snRNA transcription. Science. 1997;275:1136–1140. doi: 10.1126/science.275.5303.1136. [DOI] [PubMed] [Google Scholar]
- 41.Mittal V, Ma B, Hernandez N. SNAP(c): a core promoter factor with a built-in DNA-binding damper that is deactivated by the Oct-1 POU domain. Genes Dev. 1999;13:1807–1821. doi: 10.1101/gad.13.14.1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Murphy S, Yoon J B, Gerster T, Roeder R G. Oct-1 and Oct-2 potentiate functional interactions of a transcription factor with the proximal sequence element of small nuclear RNA genes. Mol Cell Biol. 1992;12:3247–3261. doi: 10.1128/mcb.12.7.3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Myslinski E, Krol A, Carbon P. ZNF76 and ZNF143 are two human homologs of the transcriptional activator staf. J Biol Chem. 1998;273:21998–22006. doi: 10.1074/jbc.273.34.21998. [DOI] [PubMed] [Google Scholar]
- 44.Novitch B G, Mulligan G J, Jacks T, Lassar A B. Skeletal muscle cells lacking the retinoblastoma protein display defects in muscle gene expression and accumulate in S and G2 phases of the cell cycle. J Cell Biol. 1996;135:441–456. doi: 10.1083/jcb.135.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Onadim Z, Hogg A, Baird P N, Cowell J K. Oncogenic point mutations in exon 20 of the RB1 gene in families showing incomplete penetrance and mild expression of the retinoblastoma phenotype. Proc Natl Acad Sci USA. 1992;89:6177–6181. doi: 10.1073/pnas.89.13.6177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ross J F, Liu X, Dynlacht B D. Mechanism of transcriptional repression of E2F by the retinoblastoma tumor suppressor protein. Mol Cell. 1999;3:195–205. doi: 10.1016/s1097-2765(00)80310-x. [DOI] [PubMed] [Google Scholar]
- 47.Sadowski C L, Henry R W, Kobayashi R, Hernandez N. The SNAP45 subunit of the small nuclear RNA (snRNA) activating protein complex is required for RNA polymerase II and III snRNA gene transcription and interacts with the TATA box binding protein. Proc Natl Acad Sci USA. 1996;93:4289–4293. doi: 10.1073/pnas.93.9.4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sadowski C L, Henry R W, Lobo S M, Hernandez N. Targeting TBP to a non-TATA box cis-regulatory element: a TBP-containing complex activates transcription from snRNA promoters through the PSE. Genes Dev. 1993;7:1535–1548. doi: 10.1101/gad.7.8.1535. [DOI] [PubMed] [Google Scholar]
- 49.Schaub M, Myslinski E, Schuster C, Krol A, Carbon P. Staf, a promiscuous activator for enhanced transcription by RNA polymerases II and III. EMBO J. 1997;16:173–181. doi: 10.1093/emboj/16.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Segall J, Matsui T, Roeder R G. Multiple factors are required for the accurate transcription of purified genes by RNA polymerase III. J Biol Chem. 1980;255:11986–11991. [PubMed] [Google Scholar]
- 51.Sellers W R, Novitch B G, Miyake S, Heith A, Otterson G A, Kaye F J, Lassar A B, Kaelin W G., Jr Stable binding to E2F is not required for the retinoblastoma protein to activate transcription, promote differentiation, and suppress tumor cell growth. Genes Dev. 1998;12:95–106. doi: 10.1101/gad.12.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shastry B S, Ng S Y, Roeder R G. Multiple factors involved in the transcription of class III genes in Xenopus laevis. J Biol Chem. 1982;257:12979–12986. [PubMed] [Google Scholar]
- 53.Stunkel W, Kober I, Seifart K H. A nucleosome positioned in the distal promoter region activates transcription of the human U6 gene. Mol Cell Biol. 1997;17:4397–4405. doi: 10.1128/mcb.17.8.4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vilalta A, Kickhoefer V A, Rome L H, Johnson D L. The rat vault RNA gene contains a unique RNA polymerase III promoter composed of both external and internal elements that function synergistically. J Biol Chem. 1994;269:29752–29759. [PubMed] [Google Scholar]
- 55.Waldschmidt R, Wanandi I, Seifart K H. Identification of transcription factors required for the expression of mammalian U6 genes in vitro. EMBO J. 1991;10:2595–2603. doi: 10.1002/j.1460-2075.1991.tb07801.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang J Y, Knudsen E S, Welch P J. The retinoblastoma tumor suppressor protein. Adv Cancer Res. 1994;64:25–85. doi: 10.1016/s0065-230x(08)60834-9. [DOI] [PubMed] [Google Scholar]
- 57.Wang Z, Roeder R G. Structure and function of a human transcription factor TFIIIB subunit that is evolutionarily conserved and contains both TFIIB- and high-mobility-group protein 2-related domains. Proc Natl Acad Sci USA. 1995;92:7026–7030. doi: 10.1073/pnas.92.15.7026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weinberg R A. The retinoblastoma protein and cell cycle control. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 59.Weintraub S J, Prater C A, Dean D C. Retinoblastoma protein switches the E2F site from positive to negative element. Nature. 1992;358:259–261. doi: 10.1038/358259a0. [DOI] [PubMed] [Google Scholar]
- 60.Welch P J, Wang J Y. Disruption of retinoblastoma protein function by coexpression of its C pocket fragment. Genes Dev. 1995;9:31–46. doi: 10.1101/gad.9.1.31. [DOI] [PubMed] [Google Scholar]
- 61.White R J. Regulation of RNA polymerases I and III by the retinoblastoma protein: a mechanism for growth control? Trends Biochem Sci. 1997;22:77–80. doi: 10.1016/s0968-0004(96)10067-0. [DOI] [PubMed] [Google Scholar]
- 62.White R J, Gottlieb T M, Downes C S, Jackson S P. Cell cycle regulation of RNA polymerase III transcription. Mol Cell Biol. 1995;15:6653–6662. doi: 10.1128/mcb.15.12.6653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.White R J, Trouche D, Martin K, Jackson S P, Kouzarides T. Repression of RNA polymerase III transcription by the retinoblastoma protein. Nature. 1996;382:88–90. doi: 10.1038/382088a0. [DOI] [PubMed] [Google Scholar]
- 64.Willis I M. RNA polymerase III. Genes, factors and transcriptional specificity. Eur J Biochem. 1993;212:1–11. doi: 10.1111/j.1432-1033.1993.tb17626.x. [DOI] [PubMed] [Google Scholar]
- 65.Wong M W, Henry R W, Ma B, Kobayashi R, Klages N, Matthias P, Strubin M, Hernandez N. The large subunit of basal transcription factor SNAPc is a Myb domain protein that interacts with Oct-1. Mol Cell Biol. 1998;18:368–377. doi: 10.1128/mcb.18.1.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yoon J B, Roeder R G. Cloning of two proximal sequence element-binding transcription factor subunits (gamma and delta) that are required for transcription of small nuclear RNA genes by RNA polymerases II and III and interact with the TATA-binding protein. Mol Cell Biol. 1996;16:1–9. doi: 10.1128/mcb.16.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yoshinaga S K, L'Etoile N D, Berk A J. Purification and characterization of transcription factor IIIC2. J Biol Chem. 1989;264:10726–10731. [PubMed] [Google Scholar]
- 68.Zacksenhaus E, Jiang Z, Chung D, Marth J D, Phillips R A, Gallie B L. pRb controls proliferation, differentiation, and death of skeletal muscle cells and other lineages during embryogenesis. Genes Dev. 1996;10:3051–3064. doi: 10.1101/gad.10.23.3051. [DOI] [PubMed] [Google Scholar]