Abstract
The RAG-2 gene encodes a component of the V(D)J recombinase which is essential for the assembly of antigen receptor genes in B and T lymphocytes. Previously, we reported that the transcription factor BSAP (PAX-5) regulates the murine RAG-2 promoter in B-cell lines. A partially overlapping but distinct region of the proximal RAG-2 promoter was also identified as an important element for promoter activity in T cells; however, the responsible factor was unknown. In this report, we present data demonstrating that c-Myb binds to a Myb consensus site within the proximal promoter and is critical for its activity in T-lineage cells. We show that c-Myb can transactivate a RAG-2 promoter-reporter construct in cotransfection assays and that this transactivation depends on the proximal promoter Myb consensus site. By using a chromatin immunoprecipitation (ChIP) strategy, fractionation of chromatin with anti-c-Myb antibody specifically enriched endogenous RAG-2 promoter DNA sequences. DNase I genomic footprinting revealed that the c-Myb site is occupied in a tissue-specific fashion in vivo. Furthermore, an integrated RAG-2 promoter construct with mutations at the c-Myb site was not enriched in the ChIP assay, while a wild-type integrated promoter construct was enriched. Finally, this lack of binding of c-Myb to a chromosomally integrated mutant RAG-2 promoter construct in vivo was associated with a striking decrease in promoter activity. We conclude that c-Myb regulates the RAG-2 promoter in T cells by binding to this consensus c-Myb binding site.
Antigen receptor genes are assembled during B- and T-cell development by a series of site-specific DNA recombination reactions known as V(D)J recombination (58). The lymphocyte-specific gene products RAG-1 and RAG-2 are essential components of the V(D)J recombinase complex (38, 44, 51). Together, they recognize recombination signal sequences which flank rearranging gene segments and introduce double-stranded DNA breaks between these signals and gene-containing DNA segments (16). A null mutation in either gene prevents V(D)J recombination and completely blocks lymphocyte development at the early progenitor stage (38, 51).
The RAG-1 and RAG-2 genes are physically linked in the genomes of all chordates in which they have been studied; they are convergently transcribed and separated by approximately 8 kb of DNA (50). Coupled with the recent observation that the RAG proteins have DNA transposase activity in vitro, these unusual structural features of the RAG locus have led to the suggestion that RAG-1 and RAG-2 were once part of a transposable-element system (1, 26, 56).
Transcription of the RAG-1 and RAG-2 genes is limited to specific stages of B- and T-cell development. Transcription can be detected in the earliest T- and B-cell progenitors and remains high until the complete assembly of the T-cell receptor (TCR) β chain gene or the immunoglobulin (Ig) heavy-chain gene (19, 60, 63). During the midstages of lymphoid development, RAG transcription diminishes significantly, coincident with several rounds of cell division. Pre-T and pre-B cells then exit the cell cycle and increase RAG transcription, leading to rearrangement of the TCR α or Ig light-chain loci (19, 63). RAG transcription continues in T cells until positive selection occurs, at which time expression is extinguished via a TCR-dependent signal (60). The situation in B cells is more complex. RAG transcription generally stops when a complete Ig molecule is expressed on the cell surface (19). However, if that Ig recognizes self-antigen, RAG expression is stimulated, and recombination continues in a process known as receptor editing (15, 35, 57). Thus, regulated RAG expression contributes to self-tolerance of the B-cell repertoire. Finally, the RAG genes are not expressed in mature peripheral T and B cells; however, there are some data which suggest that transcription [as well as V(D)J recombination] can be reactivated in B cells during an antigen-specific immune response (20, 21, 24, 25, 46). Given the complex regulation and critical involvement of the RAG genes in lymphocyte development, efforts have been made to decipher the molecular basis of their transcriptional regulation.
Previous studies performed in our own laboratory and in others have described the general structure of the murine and human RAG-1 and RAG-2 promoters (13, 31, 32, 65). We reported recently that unlike the RAG-1 promoter, the murine RAG-2 promoter displays cell-type specificity in transient-transfection assays (32). A RAG-2 promoter-reporter construct containing the transcription start site and 279 bp of 5′ flanking DNA was fully active in both T- and B-cell lines. Surprisingly, we found that a 5′ promoter deletion mutant extending to nucleotide −71 with respect to the start site retained full activity in B cells but lost approximately 70% of its activity in T cells. Further deletion of the promoter to position −45 eliminated nearly all activity in both B and T cells (32). The DNA sequences of the murine and human RAG-2 promoters are identical between nucleotides −70 and −50. Within this region, we identified a binding site for the B-cell-specific transcription factor BSAP (Pax-5) and showed that BSAP binds this sequence both in vitro and in vivo. Furthermore, mutations which disrupt BSAP binding greatly diminish B-cell-specific promoter activity. T cells, however, do not express BSAP but still require sequences in the conserved −70 to −50 promoter region for RAG-2 promoter activity (32). Recent reports suggested that c-Myb, a hematopoietic lineage-restricted transcription factor, is preferentially expressed in developing T but not B cells (2, 11) (see below). In the experiments described below, we have gone on to determine that c-Myb binds to the RAG-2 promoter and is critical for RAG-2 promoter activity in T cells.
MATERIALS AND METHODS
Cells and cell culture.
Thymocytes used in the chromatin immunoprecipitation (ChIP) assay were isolated from 3-week-old C57BL/6J mice (Jackson Laboratory). Jurkat, a mature human T-cell lymphoma line, and 2017 (54), an Abelson virus-transformed immature murine T-cell line, were grown at 37°C and 5% CO2 in RPMI 1640 (Mediatech) supplemented with 10% fetal calf serum, penicillin-streptomycin, l-glutamine, and 50 μM β-mercaptoethanol. 293T, a human embryonic kidney cell line expressing simian virus 40 T antigen, and HeLa (human cervical carcinoma) cells were grown in Dulbecco's modified Eagle's medium (Mediatech) with 1 g of glucose per liter, supplemented with 10% fetal calf serum, l-glutamine, penicillin-streptomycin, and 50 μM β-mercaptoethanol.
Plasmid constructs.
The RAG-2 promoter constructs used in the various cotransfection assays have been described previously (32). The eukaryotic c-Myb expression construct was created by cloning the murine c-Myb cDNA (a gift from Chi Dang) into the pEFBneo vector (a modified version of pEF-BOS [37]). The basic stable-transfection reporter construct consisted of the PGK-neo drug resistance cassette and the green fluorescent protein (GFP) cDNA, intron, and polyadenylation signal from pGreenLantern (Gibco-BRL) separated by the pBSK polylinker. Tandem copies of the 1.2-kb chicken β-globin insulator were excised from pJC13 (10) and cloned into the XbaI site of the polylinker. The RAG-2 promoter from −279 to +123 was cloned in the proper orientation upstream of the GFP cDNA. A 4-kb HindIII fragment containing the Eβ enhancer core was cloned into polylinker sites upstream of the promoter and downstream from the insulator sequences.
Transient-transfection reporter assay.
For cotransfection experiments, 10 μg of the −279 to +123 reporter, 2 μg of either empty pEFBneo vector or pEFBneoMyb, and 100 ng of p-CMV-β-gal were used. 293T cells were transfected with GenePorter reagent (Gene Therapy Systems) and 2017 cells were transfected with Superfect reagent (Qiagen) according to the manufacturers' instructions. The cells were harvested as described previously (32). Luciferase activity was measured with a luminometer (Analytical Luminescence Laboratories). β-Galactosidase assays were performed with the Galacto-Light Plus Kit (Tropix) according to the manufacturer's instructions. Luciferase activity for each sample was normalized to the β-galactosidase assay control.
Stable-transfection reporter assay.
Twenty micrograms of each linearized construct in 40 μl of sterile phosphate-buffered saline was mixed with 107 cells resuspended in 800 μl of culture medium. The cells were in the logarithmic phase of growth. The mixture of cells and DNA was electroporated at 250 V and 960 μF in a 0.4-cm-gap electroporation cuvette (Bio-Rad). The cells were grown in 50 ml of medium for 48 h without G418, and then 3 × 104 cells were seeded into each well of two 24-well plates in the presence of 2 mg of G418 (Life Technologies)/ml. After 12 to 14 days of selection, all wells were positive, and they were pooled. This ensured an adequate number of founder cells, each of which represents an independent integration event.
G418-resistant cells were subjected to flow cytometry analysis and ChIP assay. FACStar PLUS and CELLQuest software (Becton Dickinson) were used for fluorescence-activated cell sorter analysis.
ChIP.
The formaldehyde cross-linking and immunoprecipitation experiments were done as reported by Boyd and colleagues (9). In brief, 37% formaldehyde solution (Fisher Scientific) was added directly to cell culture medium at a final concentration of 1%. In the case of the mouse thymocytes, the solution was added to a single-cell suspension of thymocytes in culture medium. Cross-linking was allowed to occur at room temperature for 10 min. The cells were lysed, and the nuclei were collected and resuspended in sonication buffer. Chromatin was sonicated to an average length of 400 to 800 bp, and the suspension was precleared with blocked Staphylococcus A cells. Chromatin from 4.5 × 107 cells was then incubated with 4 μg of c-Myb-specific rabbit polyclonal antibody (for Jurkat cells, anti-human c-Myb; for mouse thymocytes, anti-mouse c-Myb [Santa Cruz]), affinity-purified rabbit IgG (Jackson Immunoresearch), or no antibody and rotated overnight at 4°C. The immune complexes were precipitated, washed, and eluted. After precipitation, supernatant from the “no-antibody” sample was processed to the cross-link reversal step and analyzed as unfractionated input chromatin. Cross-links were reversed. After proteinase K digestion, samples were phenol extracted, and DNA was ethanol precipitated and resuspended in 30 μl of H2O. Two microliters of immunoprecipitate or 50 ng of total input DNA was used for 28 cycles of PCR amplification. PCR products were analyzed by electrophoresis on a 2% agarose gel and visualized by ethidium bromide staining or denatured and transferred onto a Hybond-N membrane (Amersham) for Southern blot analysis. The following primers were used in the ChIP assays: human RAG-2 promoter, hR2U (5′GTGAATTGTGTTGCCATTGTTGC) and humR2P-2 (5′TGTGTGCCTACAGATGTTC); human RAG-2 coding exon, hRAG-2exon2/F (5′GTTCTTCTGCTGAAAGTTC) and HRAG-2exon2/B (5′GGTGATGGAAACAACAAAAG); human β-actin, hBA1 (5′CATGTGCAAGGCCGGCTTCG) and hBA2 (5′GAAGGTGTGGTGCCAGATTT); mouse RAG-2 promoter, R2F1 (5′CAACCATCACAGGGGTGCAG) and R2R2 (5′GCCTACAGATGTTCCAGTGAG); and mouse Ig kappa locus, Jk1F (5′ACCAGATTCTGGCACTCTCC) and JkREV (5′GAGTAAGATTTTATACATCATTTTTAGACA).
DNase I genomic footprinting.
DNase I genomic footprinting was performed as described by Weinmann et al. (62). In brief, Jurkat or HeLa cells were harvested and resuspended in cold NP-40 lysis buffer. Samples were incubated on ice for 5 min, and intact nuclei were pelleted and resuspended in DNase I digestion buffer. Genomic DNA was purified from Jurkat cells by proteinase K treatment, phenol extraction, and ethanol precipitation. DNase I (Boehringer Mannheim Biochemicals) was added to nuclei (from 3 × 106 cells) or pure genomic DNA (from 8 × 106 cells) to various final concentrations (nuclei, 2 to 10 U/sample; genomic DNA, 0.2 to 0.8 U/sample) and incubated on ice for 5 min. Reactions were stopped and processed for DNA purification. DNA was resuspended in H2O, and its concentration was determined by spectrophotometry. One microgram of DNA was subjected to ligation-mediated PCR. The PCR cycles were as follows: for primer extension, 95°C for 5 min, 59°C for 30 min, and 76°C for 10 min; for amplification of ligated products, 95°C for 4 min, 60°C for 2 min, and 76°C for 3 min for one cycle, 20 cycles of 95°C for 1 min, 60°C for 2 min, and 76°C for 3 min 15 s/cycle, and final extension for 10 min at 76°C; for labeling reactions, 95°C for 4 min, 61°C for 2 min, and 76°C for 10 min for one cycle and two cycles of 95°C for 1 min, 61°C for 2 min, and 76°C for 10 min/cycle. The following locus-specific primers were used: primer extension, 5′ TGCCGCTAAACCAGGTATTAA; amplification, 5′ TTAATTGTCAGCACTTGGGGA; end labeling, 5′ATTGTCAGCACTTGGGGAAGA; and DNA sequencing, 5′ ATCTTTGCCGCTAAACCAGGT. Samples were heated to 90°C for 5 min and analyzed by electrophoresis on a 6% polyacrylamide–7 M urea denaturing gel alongside a DNA sequencing ladder. The gels were dried and exposed to PhosphorImager screens.
RESULTS
c-Myb activates the RAG-2 promoter in cotransfected cell lines.
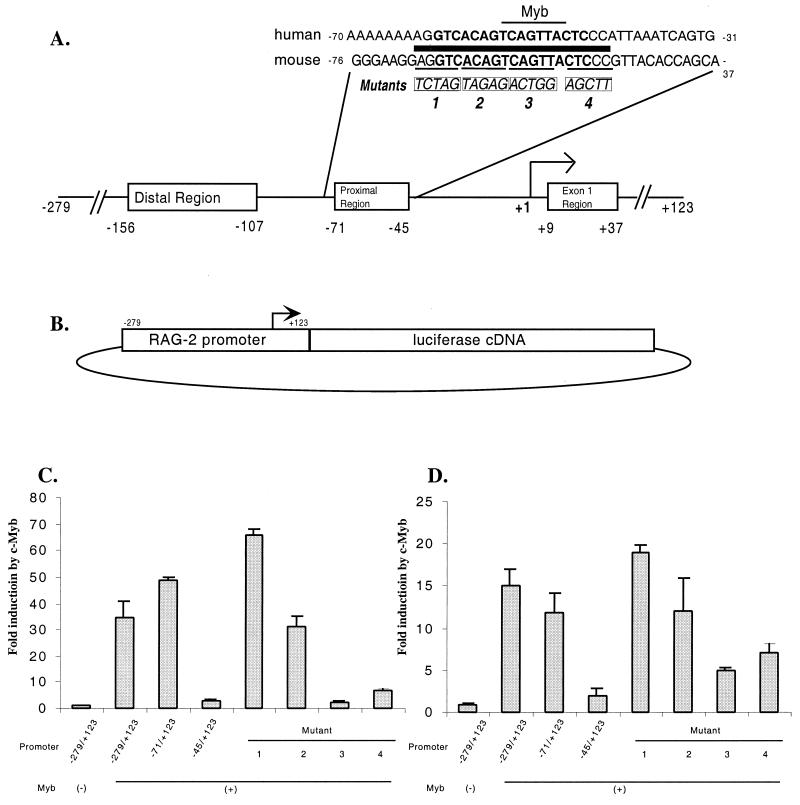
During characterization of the BSAP binding site in the −70 to −50 region of the murine RAG-2 promoter, we generated a series of 5-bp substitution mutations across this interval (Fig. 1A). An analysis of these promoter mutations suggested that distinct DNA sequences within this small region were necessary for promoter activity in B- and T-lineage cell lines (32).
FIG. 1.
c-Myb transactivates the RAG-2 promoter through a proximal-region binding site. (A) Critical sequences in the murine RAG-2 promoter. The major transcription start site is numbered +1. The boxed regions represent sequences important for promoter activity in transiently transfected Jurkat T cells (32). The human and murine proximal-region DNA sequences are shown, with the BSAP binding site in boldface and a potential c-Myb site indicated by the thin horizontal bar above the sequence. The thick bar denotes a 20-bp region of complete identity between murine and human sequences. The sites and DNA sequences of four substitution mutations (labeled 1, 2, 3, and 4) are shown in boxes below the sequence. (B) Diagram of the transient-transfection assay vector for RAG-2 promoter activity. Each substitution mutation was analyzed in the context of the full −279 to +123 promoter. The arrow indicates the transcription start site. (C) Luciferase assay analysis of transient transfections into 2017 murine pre-T cells. The cells were cotransfected with the RAG-2 promoter-reporter construct and either empty expression vector (−) or c-Myb expression vector (+). Two 5′ promoter deletion mutants (−71/+123 and −45/+123) and four 5-nucleotide substitution mutations (labeled 1 to 4) were also tested. The results are shown as the fold increase in luciferase activity with c-Myb cotransfection compared to the activity with empty vector (the actual luminometer unit readings were 2,759 ± 434 for the wild-type promoter cotransfected with an empty expression vector and 194,886 ± 228 for the wild-type promoter cotransfected with the c-Myb expression vector). The data shown are the average (+ standard deviation) of two experiments, each performed in duplicate and adjusted for transfection efficiency using β-galactosidase. (D) Luciferase assay analysis of transient transfections into 293T human embryonic kidney cells as described for panel C. The actual luminometer unit readings were 1,723 ± 96 for the wild-type promoter cotransfected with an empty expression vector and 16,573 ± 67 for the wild-type promoter cotransfected with the c-Myb expression vector.
A transcription factor database analysis of the proximal regulatory region of the murine RAG-2 promoter revealed a perfect match to the consensus binding site for the hematopoietic lineage transcription factor, c-Myb (PyAACG/TG [8]) (Fig. 1A). The murine and human RAG-2 promoter sequences are identical from positions −70 to −50 and contain this consensus c-Myb binding site. Two other potential transcription factor binding sites in the proximal promoter region (a second c-Myb site and an Ikaros site) were not located in this region of identity, and we did not pursue them further.
To test whether c-Myb can transactivate the murine RAG-2 promoter, we cotransfected cell lines with variants of a RAG-2 promoter–luciferase reporter construct (Fig. 1B) and either an empty expression vector or a murine c-Myb expression vector. Each transfection was done in duplicate, and each experiment was performed at least twice. The reporter construct itself had very little activity in 2017, a mouse pre-T-cell line which expresses very low levels of endogenous RAG-2 and c-Myb mRNAs (Fig. 1C) (reference 54 and data not shown). Cotransfection of the c-Myb expression vector resulted in an almost 40-fold increase in luciferase activity (Fig. 1C). To localize the DNA sequences responsible for this effect, different truncations of the promoter were tested in cotransfection experiments. A 5′ deletion extending to nucleotide −71 retained the same sensitivity to c-Myb coexpression as the wild-type promoter, while further truncation deleting the proximal region almost abolished the stimulatory effect of c-Myb on the RAG-2 promoter. We further analyzed our set of 5-bp substitution mutations across this proximal region in the context of the full −279 to +123 promoter (Fig. 1A). In the presence of c-Myb, mutants 1 and 2 still gave an approximately 35- to 65-fold increase in promoter activity, while mutants 3 and 4, which disrupt the Myb site and a sequence immediately adjacent to that site, respectively, resulted in a significant decrease in promoter activity. When 293T, a human embryonic kidney cell line, was tested in the cotransfection assay, the same pattern of promoter activity was observed (Fig. 1D). Overall, c-Myb stimulated the RAG-2 promoter to a greater extent in 2017 cells than in 293T cells. This may be due to the fact that 2017 is an immature T-cell line, which may provide lymphoid-specific factors to cooperate with c-Myb, resulting in higher levels of reporter gene expression.
Similar cotransfection studies failed to reveal any stimulatory effect of c-Myb on RAG-2 promoter activity in Jurkat cells (a human T-cell lymphoma line which expresses RAG-2 [data not shown] [see below]). Our previous studies showed that the cloned RAG-2 promoter is very active in these cells (32). Western blot analysis also showed that Jurkat cells express a high level of c-Myb protein (data not shown). Reporter gene expression may already be saturated in this system without introducing any exogenous activators.
c-Myb binds to the RAG-2 promoter in vivo.
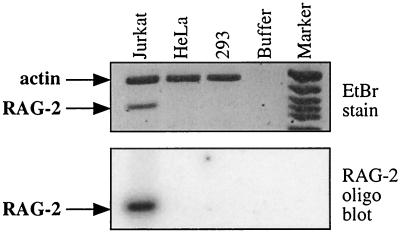
Although Jurkat cells were reported to lack V(D)J recombinase activity (33, 49), we could easily detect endogenous RAG-2 mRNA in this cell line by reverse transcription-PCR analysis (Fig. 2) (14). Since both the endogenous RAG-2 gene and the RAG-2 promoter-reporter construct were expressed in Jurkat cells, we proceeded to use this cell line to explore the role of c-Myb in regulating the RAG-2 promoter in vivo.
FIG. 2.
Jurkat T cells express a readily detectable amount of RAG-2 mRNA. Total RNA was purified from Jurkat, HeLa, and 293 cells and converted to cDNA by random-primed reverse transcription. The cDNAs were analyzed by multiplex PCR for β-actin and RAG-2 transcripts. On top is a digital image of an ethidium bromide (EtBr)-stained gel, and on the bottom is a phosphorimage of the same gel blotted to a membrane and probed with an internal RAG-2-specific radiolabeled oligonucleotide.
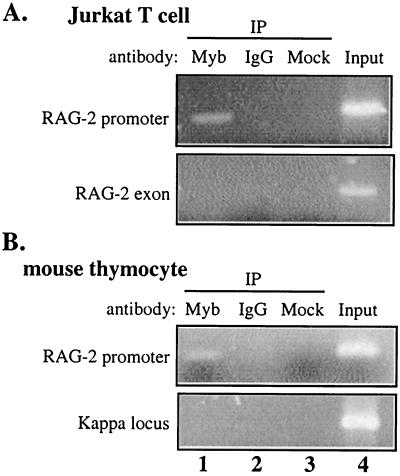
Boyd and colleagues recently adapted a chromatin cross-linking and immunoprecipitation (ChIP) protocol to characterize the binding of mammalian transcription factors to a promoter region in intact living cells (9). Using this ChIP technique, we investigated whether c-Myb binds to the RAG-2 promoter in vivo. Jurkat cells were treated with formaldehyde. After being sheared to an average length of 600 nucleotides, cross-linked chromatin purified from these cells was subjected to immunoprecipitation using either c-Myb-specific antibody or control rabbit IgG. After reversal of the cross-linking, immunoprecipitated DNA was purified and subjected to PCR analysis using primers specific for the human RAG-2 promoter and for other control DNA sequences. As shown in Fig. 3A, fractionation of chromatin with anti-c-Myb antibody, but not the control IgG, specifically enriched for endogenous RAG-2 promoter DNA (the amplified region is from −102 to +9). DNA from the β-actin locus (data not shown) or the major RAG-2 exon, which is approximately 10 kb away from the promoter, was not enriched with either antibody (Fig. 3A).
FIG. 3.
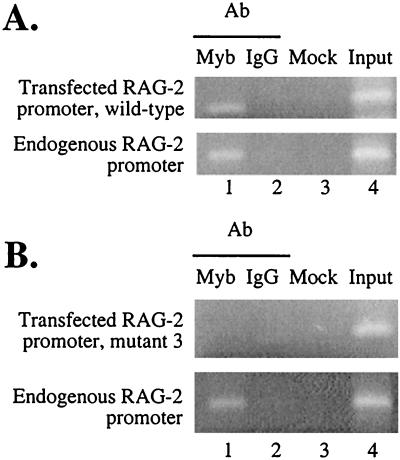
Anti-c-myb antibody specifically enriches RAG-2 promoter DNA sequences in a (ChIP) assay. Chromatin proteins were cross-linked to DNA in intact Jurkat T cells (A) and in freshly purified mouse thymocytes (B) by formaldehyde treatment, and purified nucleoprotein complexes were fractionated using either anti-Myb antibody (lanes 1), nonspecific IgG (lanes 2), or no antibody (lanes 3). The precipitated DNA fractions were analyzed by PCR for the presence of the proximal RAG-2 promoter region, RAG-2 major exon DNA, or a region of the Ig kappa locus. In each case, the input DNA was used as a positive control (lanes 4). Amplification products were analyzed on a 2% agarose gel and visualized by ethidium bromide staining. IP, immunoprecipitate.
We further investigated whether c-Myb binds to the endogenous RAG-2 promoter in a primary tissue. A single-cell suspension was made from whole thymus from 3-week-old mice and subjected to the ChIP assay as described above. Immunoprecipitated DNA was analyzed by PCR using primers specific for the mouse RAG-2 promoter (Fig. 1A; the amplified region is from −163 to −6) and for control DNA sequences. Genomic sequences from the murine RAG-2 promoter, but not the Ig kappa locus, were specifically enriched by the c-Myb antibody. We could not detect precipitation of Ig kappa locus DNA by either anti-c-Myb antibody or rabbit IgG (Fig. 3B).
The PCR products were also examined by Southern blotting and quantified by PhosphorImager (data not shown). No signals were detected from mock precipitation lanes. The signal from anti-c-Myb antibody-precipitated DNA was compared to that from control antibody-precipitated DNA. The differences ranged from 70- to 100-fold for the RAG-2 promoter region and from 0.5- to 7-fold for control regions. These ChIP experiments provided strong evidence that c-Myb was bound in the vicinity of the proximal region of the RAG-2 promoter.
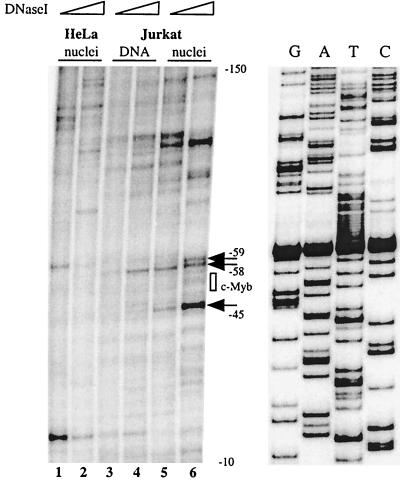
In order to determine with higher resolution the site of factor binding, we performed in vivo DNase I genomic footprinting on Jurkat T cells and nonlymphoid HeLa cells. Purified intact nuclei or genomic DNA was treated with increasing concentrations of DNase I, and cleavages were mapped by ligation-mediated PCR. To test the specificity of the assay, an RsaI restriction enzyme-treated sample of Jurkat genomic DNA was first examined. The unique cleavage introduced by the enzyme in the promoter-proximal region was visualized as a single intense band migrating at the expected position in a denaturing gel (data not shown). As shown in Fig. 4, a DNase-hypersensitive site was detected at position −45 of the human promoter (which corresponds to position −51 in the mouse promoter), just on the 3′ side of the putative c-Myb binding site, in Jurkat nuclei but not in the purified Jurkat DNA or HeLa nucleus samples. Two additional DNase I-sensitive bands were present on the 5′ side of the Myb site at positions −59 and −58. This was distinct from the pattern observed in either pure DNA or HeLa nucleus samples. These data suggested that the c-Myb site may be occupied in vivo in a lymphoid-cell-specific fashion. When taken together with the ChIP data, these experiments led us to the preliminary conclusion that c-Myb binds to the Myb site at positions −54 to −49 of the human RAG-2 promoter in vivo (corresponding to positions −60 to −55 in the mouse promoter). Other differences between the various samples in this DNase footprint analysis suggest that additional factors also bind specifically to the RAG-2 promoter in vivo (e.g., a hypersensitive site at position −100).
FIG. 4.
The RAG-2 promoter proximal region is occupied in the vicinity of the c-Myb site in vivo. Purified nuclei from Jurkat (lanes 5 and 6) and HeLa (lanes 1 and 2) cells or purified genomic DNA from Jurkat cells (lanes 3 and 4) were treated with increasing concentrations of DNase I. DNase-treated DNAs were then repurified and subjected to linker ligation and PCR amplification. Radiolabeled extension products were electrophoresed on a denaturing polyacrylamide gel alongside a DNA sequencing ladder, and the dried gel was analyzed by phosphorimaging. This ligation-mediated PCR analysis reveals DNase-sensitive sites on the top strand of the promoter through the use of a radiolabeled bottom-strand primer. The numbers indicate positions with respect to the transcription start site. The arrows point to in vivo DNase I-hypersensitive sites in Jurkat cells, and the position of the c-Myb binding site is indicated (box). The DNA sequencing ladder shows the bottom-strand sequence.
The Myb site is critical for promoter activity and required for recruitment of c-Myb to the RAG-2 promoter in vivo.
To probe the functional consequences of the interaction between c-Myb and its binding site in the proximal RAG-2 promoter in a chromosomal context, we analyzed a series of Jurkat cell lines stably transfected with reporter constructs containing the murine RAG-2 promoter.
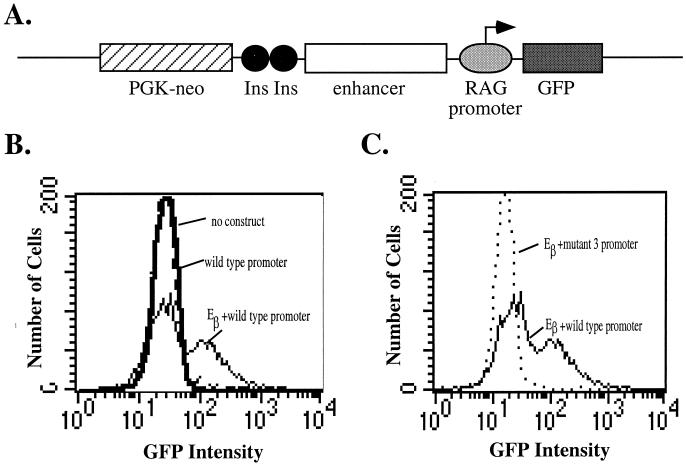
The design of the reporter construct is shown in Fig. 5A. It consists of a GFP cDNA under the control of the −279 to +123 region of the murine RAG-2 promoter. The vector contains a neomycin resistance cassette expressed from the PGK promoter and separated from the reporter region by two copies of the chicken β-globin insulator (10). The reporter was stably transfected into Jurkat T cells, and the G418-resistant transfectant pool was assayed for GFP expression by flow cytometry. Initial experiments showed that the RAG-2 promoter-containing construct, in the absence of an enhancer, gave a fluorescence pattern indistinguishable from that of untransfected cells (Fig. 5B). We interpreted this negative result to indicate that either the promoter itself cannot drive sufficient GFP expression to be detectable above the background or it is unable to overcome the repressive effects of chromatin structure. When the TCR β locus enhancer Eβ was inserted upstream of the RAG-2 promoter, 30 to 50% of transfectant cells expressed GFP (Fig. 5B and C). When mutation 3 was introduced into the promoter, less than 0.5% of transfected cells expressed GFP in the presence of Eβ (Fig. 5C), leading us to conclude that the Myb binding site was essential for promoter activity within a chromatin context.
FIG. 5.
The c-Myb binding site is essential for RAG-2 promoter activity in an integrated reporter construct in Jurkat T cells. (A) Reporter construct used in the stable-transfection assay. Linearized constructs contain a GFP cDNA and the wild-type or mutant 3 RAG-2 promoter (−279 to +123) in the presence or absence of the TCR β locus enhancer (Eβ). Ins, the 1.2-kb chicken β-globin insulator sequence (10), included to prevent the neomycin resistance cassette (PGK-neo) from influencing GFP activity. (B and C) Reporter constructs containing the wild-type or mutant 3 RAG-2 promoter in the presence or absence of Eβ were transfected into Jurkat cells by electroporation. Transfected cells were selected as a pool in G418 and then analyzed by flow cytometry for GFP expression. The fluorescence-activated cell sorter histograms of untransfected (dashed line) and wild-type promoter-transfected cells (bold solid line) are coincident. The thin solid line represents transfectants containing the wild-type promoter and Eβ, and the dotted line represents the mutant 3 promoter and Eβ.
Jurkat cells transfected with either the wild-type murine RAG-2 promoter plus Eβ or the mutation 3-containing promoter plus Eβ were also analyzed by the ChIP assay. Immunoprecipitates were analyzed for the endogenous human RAG-2 promoter and the transfected murine RAG-2 promoter. As shown in Fig. 6, both the transfected wild-type murine and endogenous human RAG-2 promoter regions, but not human RAG-2-coding exon DNA (data not shown), were specifically enriched by c-Myb antibody (Fig. 6A). Furthermore, the integrated mutant RAG-2 promoter was not enriched in this assay (Fig. 6B), suggesting that this sequence is critical for recruitment of c-Myb to the promoter region in vivo, and this recruitment is well correlated with promoter activity.
FIG. 6.
The c-Myb site in the proximal region is essential for in vivo recruitment of c-Myb to the RAG-2 promoter. Pools of G418-resistant cells containing the wild-type (A) or mutant 3 (B) RAG-2 promoter analyzed in Fig. 5 were subjected to the ChIP assay using anti-c-Myb antibody (Ab) (lanes 1) or nonspecific IgG (lanes 2). Mock (lanes 3) indicates samples processed in the absence of antibody, and input (lanes 4) is DNA prior to immunoprecipitation. Primers specific for either the transfected or endogenous RAG-2 promoter were used in PCR analysis of the various precipitated chromatin fractions. Images of ethidium bromide-stained gels are shown.
DISCUSSION
Regulated expression of RAG-1 and RAG-2 is essential for lymphocyte development. Recent transgenic studies have revealed distinct DNA sequence requirements for RAG expression in B and T cells. Reporter constructs containing as little as 2 kb of genomic DNA sequence 5′ of the RAG-2 promoter are active in B but not T cells, whereas similar constructs containing 9 kb of upstream sequence are active in both lineages (39). In a separate study, DNA sequences 5′ of the RAG-2 promoter were found to be involved in the coordinate expression of RAG-1 and RAG-2 in T and B cells while sequences on the RAG-1 side of the locus were only important for expression of RAG-1. Perhaps surprisingly, no regulatory elements were identified in the intergenic region (64). Our own previous studies revealed distinct requirements for RAG-2 promoter activity in B and T cells (32). In B-lineage cells, BSAP bound to a promoter-proximal DNA sequence and was critical for promoter activity. This factor is not expressed in the T lineage, however. In the present study, we found that the hematopoietic lineage transcription factor c-Myb binds to the RAG-2 promoter in T cells, where it is important for RAG-2 transcription.
We showed that a c-Myb expression vector can transactivate a RAG-2 promoter-reporter construct in a transient-transfection assay (Fig. 1C and D). When serial deletions and substitution mutations of this promoter were tested, we found that c-Myb activates luciferase reporter gene expression through a c-Myb consensus binding site completely conserved between the mouse and human promoters and located in a region critical for promoter activity in the transient-transfection assay. Mutation of a sequence adjacent to the conserved c-Myb site also interferes with RAG-2 promoter activity and its induction by c-Myb. This may indicate that c-Myb binds the RAG-2 promoter cooperatively with a second, adjacently bound factor. We went on to show that c-Myb was bound to this consensus site in vivo in Jurkat T cells by using a ChIP assay combined with in vivo DNase I footprinting. Enrichment of RAG-2 promoter sequences by c-Myb-specific antibody in the ChIP assay provides strong evidence that c-Myb is binding in the vicinity of the promoter (Fig. 3A). We obtained the same result using mouse thymocytes (Fig. 3B), which actively express RAG genes. A chromosomally integrated RAG-2 promoter construct with a mutant c-Myb site was not specifically immunoprecipitated (Fig. 6B). In a complementary set of experiments, in vivo DNase footprinting revealed hypersensitive cleavage adjacent to the c-Myb site in Jurkat cell nuclei but not in non-lymphoid-cell nuclei or in purified genomic DNA (Fig. 4). This is consistent with the c-Myb expression pattern as well as the tissue specificity of the murine RAG-2 promoter.
Using an electrophoretic mobility shift assay and Jurkat cell nuclear extract, we were unable to detect c-Myb binding to a 40-bp oligonucleotide probe spanning the RAG-2 proximal-promoter c-Myb binding site (data not shown). This may be due to the inadequacy of our in vitro binding and analysis conditions. Alternatively, it may indicate that c-Myb must cooperate with various other factors to efficiently bind its site in the RAG-2 promoter. First, as we demonstrated previously, a DNA sequence located at a distal position (between positions −156 and −107 [Fig. 1A]) is required for RAG-2 promoter activity in T-lineage cells (32). Whichever transcription factor binds to this region of the promoter may influence the binding of c-Myb to its site in the proximal region. For example, c-Myb collaborates with NF-M to induce mim-1 gene expression, which is restricted to differentiating granulocytes (43). In cooperation with core binding factor, c-Myb regulates the TCR δ enhancer (22, 23). A recent report provided in vitro evidence that GATA-3 may regulate the RAG-2 promoter as a T-cell-specific factor (30). Further studies will be required to identify factors which collaborate with c-Myb in the regulation of the RAG-2 promoter in T cells.
The Myb transcription factor family.
The Myb family of transcription factors include three members, A-Myb, B-Myb, and c-Myb (45). Although all three proteins share a highly conserved DNA binding domain and specifically recognize the same hexanucleotide consensus sequence, subtle differences exist in the binding preferences of different members (27, 36). Moreover, studies suggest that each member interacts with distinct collections of cellular factors and performs distinct biological functions (42, 45). A-Myb expression is restricted to male germ cells, ovaries, and germinal-center B lymphocytes and is not found at significant levels in immature hematopoietic lineages, including bone marrow cells and T cells (18). A-Myb homozygous null mutant mice show defects in spermatogenesis and female breast development, as well as growth abnormalities (59). These data lead us to believe that A-Myb is not involved in RAG expression. B-Myb is expressed in dividing cells of a wide variety of tissues, and it is a key regulator of the cell cycle (48, 52). B-Myb has been suggested to play a role in cellular differentiation (45), but its expression closely parallels cell proliferation rather than differentiation status and particular cell type (29, 47, 48). For this reason, B-Myb is less likely to be an activator of RAG expression, since RAG transcription is diminished in proliferating cells. An antagonistic relationship between c-Myb and B-Myb has been noted (12, 61). This raises the possibility that B-Myb plays a role in inhibiting RAG expression, perhaps during active periods of cell division in early lymphoid cell development. Since Jurkat cells do express RAG mRNA, a different model system would be needed to test this hypothesis. c-Myb is prevalently expressed in immature hematopoietic cells, where genetic analyses have shown that it plays an essential role in lymphocyte development (see below). In this regard, it is worth noting that the anti-c-Myb antiserum used in the ChIP experiments reported here (Fig. 3 and 6) lacks cross-reactivity with B-Myb. Thus, we conclude that c-Myb, rather than another Myb family member, is involved in regulating RAG-2 expression.
The Role of c-Myb in T-cell development.
The c-Myb gene is primarily expressed in immature lymphoid, erythroid, and myeloid cells (45). This transcription factor is also present in developing airway epithelium, hair follicles, and gastrointestinal crypt epithelial cells, as well as tooth buds and the thyroid primordium during development (11). Within the lymphoid lineage, mature T and B cells can reactivate c-Myb expression following in vitro mitogenic stimulation (17). Many tumor cell lines of T- and B-lymphocyte origin also express high levels of c-Myb (4, 6, 7). Recent studies of normal developing tissue, however, suggested that c-Myb is preferentially expressed in the T, but not B, lineage (2, 11). Primary cells at various stages of development were examined by reverse transcription PCR for expression of different transcription factors (2). While pro- and pre-T cells showed similar significant amounts of c-Myb mRNA, pro- and pre-B cells totally lacked c-Myb mRNA expression. The same expression pattern was shared by GATA-3, a T-cell-specific transcription factor, but not by other factors examined, such as GATA-1 and -2, PU-1, and c/EBPα. Other workers have characterized c-Myb expression using in situ hybridization (11). In the developing thymus, high levels of c-Myb RNA were observed in the cortex, the area populated with immature thymocytes expressing RAG-1 and RAG-2 and undergoing V(D)J recombination at TCR loci. When adult bone marrow was examined, a strong c-Myb signal was noted in only a small percentage of blastoid-appearing cells, which are likely precursors of erythroid and myeloid lineages.
Mice which are homozygous for a null mutation in the c-Myb gene exhibit a specific failure of fetal liver hematopoiesis and die before the onset of definitive lymphopoiesis (40). The vital role of c-Myb in T-lymphocyte differentiation was confirmed in a recently reported study which used homozygous null c-Myb ES cells to generate chimeric mice using the RAG-deficient blastocyst complementation assay (3). The chimeric mice were generated by injecting c-Myb−/− RAG-1+/+ ES cells into blastocysts generated from RAG-1−/− mice. Since RAG-deficient mice do not generate mature T and B cells, the authors were able to examine the ability of c-Myb-deficient progenitors to contribute to T-cell development in this chimera. They showed that T-cell development was blocked before the onset of gene rearrangement at the CD44lo CD25−stage, the beginning of definitive T-cell differentiation. Thus, in the absence of c-Myb, the V(D)J recombinase is not expressed in developing T cells. Taken together with our finding that c-Myb regulates the RAG-2 promoter, these data strongly suggest that the critical role of c-Myb in early T-cell development may be, at least in part, due to its ability to regulate expression of the RAG-2 gene.
In addition to its critical role in the commitment of precursors to the T-cell lineage, c-Myb also seems to be involved in later stages of T-cell development. The CD4, TCR γ, and TCR δ genes, each of which encodes a protein required for the selection of various T-cell subsets, have been identified as c-Myb targets (23, 28, 41, 53). A dominant-interfering c-Myb mutant severely affected thymocyte maturation in transgenic mice, apparently by interfering with the expression of antiapoptotic proteins (5, 55), suggesting a role for c-Myb in late T-cell development. A recent report has suggested that like B cells, mature T cells might reactivate RAG expression under very unusual circumstances (34). Regulated c-Myb expression might also contribute to this phenomenon.
ACKNOWLEDGMENTS
We thank Peggy J. Farnham (University of Wisconsin) for sharing the chromatin immunoprecipitation protocol and Kathryn E. Boyd (Yale) for generous technical assistance with this assay. We also thank Stephen T. Smale (UCLA) for his DNase I genomic footprinting protocol. We are grateful to the late Eugenia Spanopoulou for providing us with 293T cells and the pEF-BOS expression vector and Chi Dang (Hopkins) for sharing with us the murine c-Myb cDNA. We also thank Astar Winoto, Alan Friedman, Shau-Ku Huang, Laurent Bentolila, Hans Brightbill, Jamie Geier, and various members of Schlissel laboratory for critical reading of the manuscript.
This research was funded in part by NIH grants RO1 AI40227 and HL48702 to M.S.S., who also acknowledges the support of a Leukemia Society Scholarship.
REFERENCES
- 1.Agrawal A, Eastman Q, Schatz D G. Transposition mediated by RAG-1 and RAG-2 and its implications for the evolution of the immune system. Nature. 1998;394:744–751. doi: 10.1038/29457. [DOI] [PubMed] [Google Scholar]
- 2.Akashi K, Traver D, Miyamoto T, Weissman I L. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;404:193–197. doi: 10.1038/35004599. [DOI] [PubMed] [Google Scholar]
- 3.Allen R D, III, Bender T P, Siu G. c-Myb is essential for early T cell development. Genes Dev. 1999;13:1073–1078. doi: 10.1101/gad.13.9.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Asahina M, Ishiguro N, Wu D, Goryo M, Davis W C, Okada K. The proto-oncogene c-myb is expressed in sporadic bovine lymphoma, but not in enzootic bovine leukosis. J Vet Med Sci. 1996;58:1169–1174. doi: 10.1292/jvms.58.12_1169. [DOI] [PubMed] [Google Scholar]
- 5.Badiani P, Corbella P, Kioussis D, Marvel J, Weston K. Dominant interfering alleles define a role for c-Myb in T-cell development. Genes Dev. 1994;8:770–782. doi: 10.1101/gad.8.7.770. [DOI] [PubMed] [Google Scholar]
- 6.Bender T P, Kuehl W M. Differential expression of the c-myb proto-oncogene marks the pre-B cell/B cell junction in murine B lymphoid tumors. J Immunol. 1987;139:3822–3827. [PubMed] [Google Scholar]
- 7.Bender T P, Thompson C B, Kuehl W M. Differential expression of c-myb mRNA in murine B lymphomas by a block to transcription elongation. Science. 1987;237:1473–1476. doi: 10.1126/science.3498214. [DOI] [PubMed] [Google Scholar]
- 8.Biedenkapp H, Borgmeyer U, Sippel A E, Klempnauer K H. Viral myb oncogene encodes a sequence-specific DNA-binding activity. Nature. 1988;335:835–837. doi: 10.1038/335835a0. [DOI] [PubMed] [Google Scholar]
- 9.Boyd K E, Wells J, Gutman J, Bartley S M, Farnham P J. c-Myc target gene specificity is determined by a post-DNA binding mechanism. Proc Natl Acad Sci USA. 1998;95:13887–13892. doi: 10.1073/pnas.95.23.13887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chung J H, Whiteley M, Felsenfeld G. A 5′ element of the chicken beta-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell. 1993;74:505–514. doi: 10.1016/0092-8674(93)80052-g. [DOI] [PubMed] [Google Scholar]
- 11.Ess K C, Witte D P, Bascomb C P, Aronow B J. Diverse developing mouse lineages exhibit high-level c-Myb expression in immature cells and loss of expression upon differentiation. Oncogene. 1999;18:1103–1111. doi: 10.1038/sj.onc.1202387. [DOI] [PubMed] [Google Scholar]
- 12.Foos G, Grimm S, Klempnauer K H. Functional antagonism between members of the myb family: B-myb inhibits v-myb-induced gene activation. EMBO J. 1992;11:4619–4629. doi: 10.1002/j.1460-2075.1992.tb05564.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuller K, Storb U. Identification and characterization of the murine Rag 1 promoter. Mol Immunol. 1997;34:939–954. doi: 10.1016/s0161-5890(97)00000-x. [DOI] [PubMed] [Google Scholar]
- 14.Gaffney P M, Lund J, Miller J S. FLT-3 ligand and bone marrow stroma-derived factors promote CD3 gamma, CD3 delta, CD3 zeta, and RAG-2 gene expression in primary human CD34+LIN−DR− marrow progenitors. Blood. 1998;91:1662–1670. [PubMed] [Google Scholar]
- 15.Gay D, Saunders T, Camper S, Weigert M. Receptor editing: an approach by autoreactive B cells to escape tolerance. J Exp Med. 1993;177:999–1008. doi: 10.1084/jem.177.4.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gellert M. Recent advances in understanding V(D)J recombination. Adv Immunol. 1997;64:39–64. doi: 10.1016/s0065-2776(08)60886-x. [DOI] [PubMed] [Google Scholar]
- 17.Golay J, Capucci A, Arsura M, Castellano M, Rizzo V, Introna M. Expression of c-myb and B-myb, but not A-myb, correlates with proliferation in human hematopoietic cells. Blood. 1991;77:149–158. [PubMed] [Google Scholar]
- 18.Golay J, Facchinetti V, Ying G, Introna M. The A-myb transcription factor in neoplastic and normal B cells. Leuk Lymphoma. 1997;26:271–279. doi: 10.3109/10428199709051776. [DOI] [PubMed] [Google Scholar]
- 19.Grawunder U, Leu T M J, Schatz D G, Werner A, Rolink A G, Melchers F, Winkler T H. Down-regulation of RAG-1 and RAG-2 gene expression in pre-B cells after functional immunoglobulin heavy chain rearrangement. Immunity. 1995;3:601–608. doi: 10.1016/1074-7613(95)90131-0. [DOI] [PubMed] [Google Scholar]
- 20.Han S, Dillon S, Zheng B, Shimoda M, Schlissel M, Kelsoe G. V(D)J recombinase activity in a subset of germinal center B lymphocytes. Science. 1997;278:301–305. doi: 10.1126/science.278.5336.301. [DOI] [PubMed] [Google Scholar]
- 21.Han S, Zheng B, Schatz D, Spanopoulou E, Kelsoe G. Neoteny in lymphocytes: RAG-1 and Rag 2 expression in germinal center B cells. Science. 1996;274:2094–2097. doi: 10.1126/science.274.5295.2094. [DOI] [PubMed] [Google Scholar]
- 22.Hernandez-Munain C, Krangel M S. c-Myb and core-binding factor/PEBP2 display functional synergy but bind independently to adjacent sites in the T-cell receptor delta enhancer. Mol Cell Biol. 1995;15:3090–3099. doi: 10.1128/mcb.15.6.3090. . (Erratum, 15:4659.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hernandez-Munain C, Krangel M S. Regulation of the T-cell receptor delta enhancer by functional cooperation between c-Myb and core-binding factors. Mol Cell Biol. 1994;14:473–483. doi: 10.1128/mcb.14.1.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hikida M, Mori M, Takai T, Tomochika K, Hamatani K, Ohmori H. Reexpression of RAG-1 and RAG-2 genes in activated mature mouse B cells. Science. 1996;274:2092–2094. doi: 10.1126/science.274.5295.2092. [DOI] [PubMed] [Google Scholar]
- 25.Hikida M, Ohmori H. Rearrangement of λ light chains in mature B cells in vitro and in vivo. Function of reexpressed recombination-activating gene (RAG) products. J Exp Med. 1998;187:795–799. doi: 10.1084/jem.187.5.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hiom K, Melek M, Gellert M. DNA transposition by the RAG-1 and RAG-2 proteins: a possible source of oncogenic translocations. Cell. 1998;94:463–470. doi: 10.1016/s0092-8674(00)81587-1. [DOI] [PubMed] [Google Scholar]
- 27.Howe K M, Watson R J. Nucleotide preferences in sequence-specific recognition of DNA by c-myb protein. Nucleic Acids Res. 1991;19:3913–3919. doi: 10.1093/nar/19.14.3913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hsiang Y H, Goldman J P, Raulet D H. The role of c-Myb or a related factor in regulating the T cell receptor gamma gene enhancer. J Immunol. 1995;154:5195–5204. [PubMed] [Google Scholar]
- 29.Kamano H, Burk B, Noben-Trauth K, Klempnauer K H. Differential splicing of the mouse B-myb gene. Oncogene. 1995;11:2575–2582. [PubMed] [Google Scholar]
- 30.Kishi H, Wei X C, Jin Z X, Fujishiro Y, Nagata T, Matsuda T, Muraguchi A. Lineage-specific regulation of the murine RAG-2 promoter: GATA-3 in T cells and pax-5 in B cells. Blood. 2000;95:3845–3852. [PubMed] [Google Scholar]
- 31.Kurioka H, Kishi H, Isshiki H, Tagoh H, Mori K, Kitagawa T, Nagata T, Dohi K, Muragachi A. Isolation and characterization of a TATA-less promoter for the human RAG-1 gene. Mol Immunol. 1996;33:1059–1066. doi: 10.1016/s0161-5890(96)00062-4. [DOI] [PubMed] [Google Scholar]
- 32.Lauring J, Schlissel M S. Distinct factors regulate the murine RAG-2 promoter in B- and T-cell lines. Mol Cell Biol. 1999;19:2601–2612. doi: 10.1128/mcb.19.4.2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lieber M R, Hesse J E, Mizuuchi K, Gellert M. Developmental stage specificity of the lymphoid V(D)J recombination activity. Genes Dev. 1987;1:751–761. doi: 10.1101/gad.1.8.751. [DOI] [PubMed] [Google Scholar]
- 34.McMahan C J, Fink P J. RAG reexpression and DNA recombination at T cell receptor loci in peripheral CD4+ T cells. Immunity. 1998;9:637–647. doi: 10.1016/s1074-7613(00)80661-5. [DOI] [PubMed] [Google Scholar]
- 35.Melamed D, Benschop R, Cambier J, Nemazee D. Developmental regulation of B lymphocyte immune tolerance compartmentalizes clonal selection from receptor selection. Cell. 1998;92:173–182. doi: 10.1016/s0092-8674(00)80912-5. [DOI] [PubMed] [Google Scholar]
- 36.Mizuguchi G, Nakagoshi H, Nagase T, Nomura N, Date T, Ueno Y, Ishii S. DNA binding activity and transcriptional activator function of the human B-myb protein compared with c-MYB. J Biol Chem. 1990;265:9280–9284. [PubMed] [Google Scholar]
- 37.Mizushima S, Nagata S. pEF-BOS, a powerful mammalian expression vector. Nucleic Acids Res. 1990;18:5322. doi: 10.1093/nar/18.17.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mombaerts P, Iacomini J, Johnson R S, Herrup K, Tonegawa S, Papaioannou V E. RAG-1 deficient mice have no mature B and T lymphocytes. Cell. 1992;68:869–877. doi: 10.1016/0092-8674(92)90030-g. [DOI] [PubMed] [Google Scholar]
- 39.Monroe R J, Chen F, Ferrini R, Davidson L, Alt F W. RAG-2 is regulated differentially in B and T cells by elements 5′ of the promoter. Proc Natl Acad Sci USA. 1999;96:12713–12718. doi: 10.1073/pnas.96.22.12713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mucenski M L, McLain K, Kier A B, Swerdlow S H, Schreiner C M, Miller T A, Pietryga D W, Scott W J, Potter S S. A functional c-myb gene is required for normal murine fetal hepatic hematopoiesis. Cell. 1991;65:677–689. doi: 10.1016/0092-8674(91)90099-k. [DOI] [PubMed] [Google Scholar]
- 41.Nakayama K, Yamamoto R, Ishii S, Nakauchi H. Binding of c-Myb to the core sequence of the CD4 promoter. Int Immunol. 1993;5:817–824. doi: 10.1093/intimm/5.8.817. [DOI] [PubMed] [Google Scholar]
- 42.Ness S A. The Myb oncoprotein: regulating a regulator. Biochim Biophys Acta. 1996;1288:F123–F139. doi: 10.1016/s0304-419x(96)00027-3. [DOI] [PubMed] [Google Scholar]
- 43.Ness S A, Kowenz-Leutz E, Casini T, Graf T, Leutz A. Myb and NF-M: combinatorial activators of myeloid genes in heterologous cell types. Genes Dev. 1993;7:749–759. doi: 10.1101/gad.7.5.749. [DOI] [PubMed] [Google Scholar]
- 44.Oettinger M A, Schatz D G, Gorka C, Baltimore D. RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science. 1990;248:1517–1523. doi: 10.1126/science.2360047. [DOI] [PubMed] [Google Scholar]
- 45.Oh I H, Reddy E P. The myb gene family in cell growth, differentiation and apoptosis. Oncogene. 1999;18:3017–3033. doi: 10.1038/sj.onc.1202839. [DOI] [PubMed] [Google Scholar]
- 46.Papavasiliou F, Casellas R, Suh H, Qin X-F, Besmer E, Pelanda R, Nemazee D, Rajewsky K, Nussenzweig M. V(D)J recombination in mature B cells: a mechanism for altering antibody responses. Science. 1997;278:298–301. doi: 10.1126/science.278.5336.298. [DOI] [PubMed] [Google Scholar]
- 47.Sala A, Watson R. B-Myb protein in cellular proliferation, transcription control, and cancer: latest developments. J Cell Physiol. 1999;179:245–250. doi: 10.1002/(SICI)1097-4652(199906)179:3<245::AID-JCP1>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 48.Saville M K, Watson R J. B-Myb: a key regulator of the cell cycle. Adv Cancer Res. 1998;72:109–140. doi: 10.1016/s0065-230x(08)60701-0. [DOI] [PubMed] [Google Scholar]
- 49.Schatz D G, Oettinger M A, Baltimore D. The V(D)J recombination activating gene, RAG-1. Cell. 1989;59:1035–1048. doi: 10.1016/0092-8674(89)90760-5. [DOI] [PubMed] [Google Scholar]
- 50.Schatz D G, Oettinger M A, Schlissel M S. V(D)J recombination: molecular biology and regulation. Annu Rev Immunol. 1992;10:359–383. doi: 10.1146/annurev.iy.10.040192.002043. [DOI] [PubMed] [Google Scholar]
- 51.Shinkai Y, Rathbun G, Lam K, Oltz E, Stewart V, Mendelsohn M, Charron J, Datta M, Young F, Stall A M, Alt F W. RAG-2 deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell. 1992;68:855–867. doi: 10.1016/0092-8674(92)90029-c. [DOI] [PubMed] [Google Scholar]
- 52.Sitzmann J, Noben-Trauth K, Kamano H, Klempnauer K H. Expression of B-Myb during mouse embryogenesis. Oncogene. 1996;12:1889–1894. [PubMed] [Google Scholar]
- 53.Siu G, Wurster A L, Lipsick J S, Hedrick S M. Expression of the CD4 gene requires a Myb transcription factor. Mol Cell Biol. 1992;12:1592–1604. doi: 10.1128/mcb.12.4.1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Spolski R, Miescher G, Erard R, Risser R, MacDonald H R, Mak T W. Regulation of expression of T cell gamma chain, L3T4 and Ly-2 messages in Abelson/Moloney virus-transformed T cell lines. Eur J Immunol. 1988;18:295–300. doi: 10.1002/eji.1830180218. [DOI] [PubMed] [Google Scholar]
- 55.Taylor D, Badiani P, Weston K. A dominant interfering Myb mutant causes apoptosis in T cells. Genes Dev. 1996;10:2732–2744. doi: 10.1101/gad.10.21.2732. [DOI] [PubMed] [Google Scholar]
- 56.Thompson C. New insights into V(D)J recombination and its role in the evolution of the immune system. Immunity. 1995;3:531–539. doi: 10.1016/1074-7613(95)90124-8. [DOI] [PubMed] [Google Scholar]
- 57.Tiegs S, Russell D, Nemazee D. Receptor editing in self-reactive bone marrow B cells. J Exp Med. 1993;177:1009–1020. doi: 10.1084/jem.177.4.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tonegawa S. Somatic generation of antibody diversity. Nature. 1983;302:575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- 59.Toscani A, Mettus R V, Coupland R, Simpkins H, Litvin J, Orth J, Hatton K S, Reddy E P. Arrest of spermatogenesis and defective breast development in mice lacking A-myb. Nature. 1997;386:713–717. doi: 10.1038/386713a0. [DOI] [PubMed] [Google Scholar]
- 60.Turka L A, Schatz D G, Oettinger M A, Chun J J M, Gorka C, Lee K, McCormack W T, Thompson C B. Thymocyte expression of RAG-1 and RAG-2: termination by T cell receptor cross-linking. Science. 1991;253:778–781. doi: 10.1126/science.1831564. [DOI] [PubMed] [Google Scholar]
- 61.Watson R J, Robinson C, Lam E W. Transcription regulation by murine B-myb is distinct from that by c-myb. Nucleic Acids Res. 1993;21:267–272. doi: 10.1093/nar/21.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Weinmann A S, Plevy S E, Smale S T. Rapid and selective remodeling of a positioned nucleosome during the induction of IL-12 p40 transcription. Immunity. 1999;11:665–675. doi: 10.1016/s1074-7613(00)80141-7. [DOI] [PubMed] [Google Scholar]
- 63.Wilson A, Held W, MacDonald H R. Two waves of recombinase gene expression in developing thymocytes. J Exp Med. 1994;179:1355–1360. doi: 10.1084/jem.179.4.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yu W, Misulovin Z, Suh H, Hardy R R, Jankovic M, Yannoutsos N, Nussenzweig M C. Coordinate regulation of RAG-1 and RAG-2 by cell type-specific DNA elements 5′ of RAG-2. Science. 1999;285:1080–1084. doi: 10.1126/science.285.5430.1080. [DOI] [PubMed] [Google Scholar]
- 65.Zarrin A, Fong I, Malkin L, Marsden P, Berinstein N. Cloning and characterization of the human recombination activating gene 1 (RAG-1) and RAG-2 promoter regions. J Immunol. 1997;159:4382–4394. [PubMed] [Google Scholar]