Abstract
A rhabdoid colorectal tumor (RCT) is a rare cancer with aggressive clinical behavior. Recently, it has been recognized as a distinct disease entity, characterized by genetic alterations in the SMARCB1 and Ciliary Rootlet Coiled-Coil (CROCC). We here investigate the genetic and immunophenotypic profiling of 21 RCTs using immunohistochemistry and next-generation sequencing. Mismatch repair-deficient phenotypes were identified in 60% of RCTs. Similarly, a large proportion of cancers exhibited the combined marker phenotype (CK7-/CK20-/CDX2-) not common to classical adenocarcinoma variants. More than 70% of cases displayed aberrant activation of the mitogen-activated protein kinase (MAPK) pathway with mutations prevalently in BRAF V600E. SMARCB1/INI1 expression was normal in a large majority of lesions. In contrast, ciliogenic markers including CROCC and γ-tubulin were globally altered in tumors. Notably, CROCC and γ-tubulin were observed to colocalize in large cilia found on cancer tissues but not in normal controls. Taken together, our findings indicate that primary ciliogenesis and MAPK pathway activation contribute to the aggressiveness of RCTs and, therefore, may constitute a novel therapeutic target.
Keywords: rhabdoid colorectal tumors, rare cancers, Ciliary Rootlet Coiled-Coil (CROCC), SMARCB1
1. Introduction
Patients with rare cancers account for as much as 24% of all cancer diagnoses. Commonly, patients with rare cancers have fewer treatment opportunities and are understudied at the level of genomic targets. A malignant rhabdoid tumor is a rare and extremely aggressive entity. The most frequent location of this tumor is the kidney with a prevalence in childhood. Adult forms are uncommon, and they are characterized by a poor prognosis. The extrarenal localization of these tumors has been described mainly in the central nervous system (called atypical teratoid/rhabdoid tumors), liver, soft tissues, colon, and rectum [1,2,3].
The localization in the colon and rectum is uncommon and, since 1990, fewer than 30 cases of rhabdoid colorectal tumors (RCTs) have been reported [4,5]. RCTs often arise in the right colon and in elderly patients [6,7]. Although the exact incidence and risk factors associated with rhabdoid carcinomas are unknown, Agaimy et al. reported that the most common site of this type of tumor in the gastrointestinal tract was the stomach, followed by the colon, small intestine, and esophagus [8]. The loss of the chromatin remodeling gene SMARCB1 (INI1) due to inactivating mutation and/or deletion is characteristic of malignant rhabdoid tumors arising in childhood [6,7]. In contrast, the loss of SMARCB1 is uncommon in RCTs, supporting the hypothesis that RCTs exhibit distinct molecular features from their pediatric counterpart [8,9,10]. As a consequence, the World Health Organization (WHO) classifies RCTs as carcinosarcomas, within the broader colorectal tumor family.
Over the last five years, new investigations on the molecular causes of RCTs identified key defects in the centrosome [9,10]. The centrosome is a biomolecular condensate composed of two microtubule-based, barrel-shaped centrioles, and a surrounding network of proteins named pericentriolar material (PCM) that is able to build and position the mitotic spindle and cilia in metazoans [11]. During cilia assembly, typically occurring at the quiescent G0 phase of the cell cycle (post-cytokinesis), basal bodies facilitate the formation of the ciliary rootlet, a fibrous structure composed mainly of rootletin also called CROCC (ciliary rootlet coiled-coil). Mutant CROCC has been found in a subset of RCTs [9,10,12]. In addition to its function at the base of the basal body, rootletin is also a core component of a proteinaceous centrosome linker, a poorly understood cohesion machinery that tethers the proximal ends of the two centrioles during the interphase [9,12,13].
Defects in the structure and function of primary cilia lead to a range of multifaceted disease phenotypes termed ciliopathies [14]. A number of key signaling pathways, including Hedgehog and Wnt pathways, are facilitated by primary cilia. This finding supports the hypothesis that primary cilia could have a role in cancer [15,16]. However, the way primary cilia regulate tumorigenesis seems to differ between tumor types and within tumor subtypes [15,16]. We therefore aimed to investigate the role of primary cilia in RCTs.
We here show that alterations in primary cilia and MAPK contribute to the aggressiveness of RCTs with dismal prognosis.
2. Materials and Methods
2.1. Patients
Twenty-one primary RCTs and matched normal, formalin-fixed paraffin-embedded (FFPE) samples were recruited from different medical institutions: (a) Konkuk University School of Medicine, Seoul, Korea; (b) Hospital Santariskiu Clinics, National Affiliate of Vilnius University, Vilnius, Lithuania; (c) Mater Salutis’ Hospital, ULSS9 “Scaligera” Legnago, Verona, Italy; (d) University of Genova, Italy ([9] and Ref therein).
Tumors with pleomorphic, giant, anaplastic, or undifferentiated histology were not included. The hematoxylin and eosin–stained (H&E) glass slides were independently reviewed by pathologists (A.R., F.G., L.M.) to confirm the diagnosis of RCT [9]. The twelve RCTs included in this study have been previously described [9]. The remaining cases (RCI-RCXII) have been identified and recruited for the present work. The tumors were staged using the conventional tumor, node, metastasis (TNM) staging system. No patient received chemotherapy or radiation therapy prior to surgery. Those patients who had a family history of intestinal dysfunction, CRC or who had taken non-steroidal anti-inflammatory drugs on a regular basis were not included. The diversity of the group’s ethnicity was not evaluated as more than 90% of the population was Caucasian. The overall survival (OS) time was available and defined as the time elapsed between the start of first-line chemotherapy and death. Those patients who were alive or lost in follow-up were censored at the last date they were known to be alive. Other clinical parameters, including biochemical and anthropometric variables, were not available. As treatment regimes after surgery were available for some but not all of the patients who participated, this information was not considered. The study was approved by the ethical committees according to the institutes’ ethical regulations on research conducted on human tissues. AOU Legnago-Verona: Ethics Committee Approval n. CA 2207 2016, September, 2016.
2.2. FFPE DNA Extraction
The rhabdoid component was identified by H&E staining. Manual macrodissection was used to isolate the rhabdoid component from FFPE blocks. DNA was extracted using Qiagen’s QIAamp DNA FFPE Tissue Kit according to the manufacturer’s instructions as reported [9,17].
2.3. Multipanel NGS Target Sequencing
A multigene panel was developed using AmpliSeq designer software v2.1. The panel included the following target genes: CROCC, PTEN, FBXW7, MLH1, SMARCB1, KIT, MET, PDGFRA, PLCG1, PTPN11, ARAF, ATR, and DNMT3A. PCR-amplified fragments from 20 ng of DNA were used to build an adequate library for deep sequencing in all cases. Emulsion PCR was performed with the Ion OneTouch OT2 System (Life Technologies, Carlsbad, CA, USA). A minimum coverage of 20× was obtained in all cases. The mean read length was 112 base pairs and a mean coverage of 6290× was achieved, with 94.1% target bases covered more than 100×. The quality of the obtained libraries was evaluated by the Agilent 2100 Bioanalyzer on-chip electrophoresis (Agilent, Santa Clara, CA, USA). Sequencing was run on the Ion Personal Genome Machine (Life Technologies) loaded with Ion 318 Chip v2. Data analysis, including alignment to the hg19 human reference genome and variant calling, which was performed using the Torrent Suite Software v.5.0 (Thermo fisher scientific, Whaltam, MA, USA). Filtered variants were annotated using a custom pipeline based on vcflib (https://github.com/ekg/vcflib, accessed on 17 February 2023), SnpSift, the Variant Effect Predictor (VEP) software, and NCBI RefSeq database. Additionally, alignments were visually verified with the Integrative Genomics Viewer (IGV) v2.3 to further confirm the presence of mutations identified by targeted sequencing.
2.4. Immunohistochemical Analysis
Immunohistochemical (IHC) analysis was performed on tumors and matched normal tissue sections as reported [9]. Four µm thick sections were deparaffined with BOND DEWAX Solution (Leica biosystems, Newcastle, UK), placed in graded alcohol solutions, washed, and pre-treated with the Epitope Retrieval Solution 2 (EDTA buffer pH 8.8) at 98 °C for 20 min. After the washing steps, peroxidase blocking was carried out for 5 min using the Bond Polymer Refine Detection Kit DS9800 (Leica biosystems, Newcastle, UK). Then, the sections were incubated with a rabbit primary anti-human CROCC rabbit polyclonal antibody (clone NBP1-80820, dilution 1:200, (ABCAM, Cambridge, UK)), or an anti-human γ-tubulin mouse monoclonal antibody (clone TU-30, dilution 1:100 (ABCAM, Cambridge, UK)) for 15 min, and a secondary antibody for 8 min, respectively. Subsequently, the sections were incubated with polymer (8 min), revealed with diaminobenzidene-chromogen (10 min), and stained with hematoxylin (10 min). In normal, non-pathological tissues, CROCC staining consists of up to two dot-like signals [9]. Consistently, we considered one or two signals per cell as quantitatively physiological. In contrast, abnormal staining was recorded as follows: (a) loss, less than 1 signal per cell; (b) amplified, more than 2 signals per cell. Two hundred cells in triplicate sections per sample were blindly scored by three authors (A.R., E.M., M.P.). Samples were also immunostained with the following markers: cytokeratin (CK)7, CDX2, CK20, SMARCB1/INI1, and mismatch repair (MMR) proteins (MLH1, PMS2), and scored as previously reported [9].
2.5. Dual Immunofluorescence Staining
A dual immunofluorescence method was employed to detect cilia at the level of single cells in tissue samples, as reported [9]. FFPE sections of 5 µm thickness were prepared and antigen retrieval was performed using the Universal HIER antigen retrieval reagent, 10× diluted 1:10 (ab208572, abcam), and heated at 97 °C using a decloaking chamber for 20 min. Slides were washed in washing buffer (PBS and 0.1% Tween 20 at 25 °C) followed by blocking buffer (5% normal goat serum, 0.1 M Tris-HCl, and 0.15 M NaCl, pH 7.6 at 25 °C) for 30 min at room temperature. Primary antibodies were diluted in blocking buffer and incubated at 4 °C overnight in a humidified chamber with anti-CROCC or anti-human γ-tubulin diluted 1:100. Slides were washed three times in washing buffer and incubated with secondary antibody and Dapi Staining Solution for DNA, diluted 1:10,000, for 30 min at room temperature. Secondary antibodies were: Goat Anti-Mouse IgG H&L (Alexa Fluor® 647, red) (abcam, ab150115) and Goat Anti-Rabbit IgG H&L (Alexa Fluor® 488, green) (ab150077), diluted 1:500. Slides were washed three times in washing buffer, mounted using Anti-Fade Fluorescence Mounting Medium—Aqueous, Fluoroshield (ab104135, abcam), and stored at −20 °C. Specimens were imaged using a Confocal Microscope (Nikon AXR system, 60× objective) equipped with a Ti2-E Inverted Microscope 25 mm FOV high-speed Resonant scanner and high-resolution Galvano scanner. High resolution images were acquired with NISElements C Software (N). The staining of cilia was considered abnormal if it showed a diameter greater than the diameter of signals present in normal epithelium within the same section.
2.6. High Resolution Melting (HRM) Analysis
Loss of heterozygosity (LOH) analysis was performed using fluorescent-labeled forward primers, followed by fragmental analysis detection on Rotor-gene Q 5plex HRM (Qiagen, Hilden, DE, USA), as reported [9]. Tumoral and matched normal DNA samples were analyzed for CROCC LOH using three markers: D1S3391, D1S1443 and D1S3669. LOH in SMARCB1 was investigated with the markers D22S301 and D22S345, as reported [9]. PCR conditions for the multiplex PCR consisted of: initial denaturation at 95 °C for 15 min, followed by 19 cycles of 95 °C 30 s, 55 °C 90 s, 72 °C 1 min, and a final extension of 72 °C 30 min. PCR reactions were performed in a 10 μL reaction mixture by using 50 ng template DNA, 0.2 mM dNTPs (Roche, Penzberg, DE, USA), 0.4 U FastStart Taq DNA Polymerase (Roche, Penzberg, DE, USA), 1× fluorescent dye LCGreen Plus (Idaho Technology, Salt Lake City, UT, USA), 2 mM MgCl2, and forward and reverse primers (0.5 mM each) for each gene segment. PCR conditions were optimized to temperatures between 52 °C and 64 °C for each segment. After 30 cycles of amplification, PCR products underwent an additional 1 min at 98 °C and then 5 min at 40 °C to promote heteroduplex formation. Each capillary was then transferred to the High-Resolution Melter instrument (HR-1) (Idaho Technology, Salt Lake City, UT, USA) for high-resolution melting and curve analysis. Samples were melted at 0.2 °C/s ramp rate. Melting profiles were analyzed with HR-1 software using fluorescence normalization, temperature shift and conversion to difference and derivative plots. To confirm the reliability of the HRM assay, selected paired non-neoplastic tissue samples showing differences or showing no difference in melting profiles were purified and then sequenced in both directions using the Big Dye Terminator 1.1 Cycle Sequencing kit (Applied Biosystems, Foster City, CA, USA). The sequencing reaction was performed on an automatic sequencer ABI PRISM 310 Genetic Analyzer and sequences were analyzed using BioEdit program.
2.7. Statistical Analysis
Patients were compared for survival outcomes, using both Kaplan-Meier survival curves. Differences in Kaplan-Meier curves were tested using the log-rank test. Genetic changes among subgroups were tested for statistical significance using the t-test (2-tailed). Statistical analyses were conducted by GeneSpring R/bioconductor v.12.5 and R based package and GraphPad Prism 5.
3. Results
3.1. Clinical Information and Follow-Up
A total of 21 primary RCTs diagnosed between 2007 and 2015 were included in this study.
The large majority of tumors (20/21; 95%) resulted as composites and only one (1/21; 5%) was diagnosed as a pure RCT, in which rhabdoid morphology is observed in virtually all tumor cells.
At diagnosis, the age of the patients ranged between 49 and 87 years (mean 71 years). The male-to-female ratio was 8:13 (38% versus 62%), respectively. The neoplasms were predominantly located in the right side of the colon (13/21; 62%), followed by left side (6/21; 28%) and rectum (2/21; 10%). The median tumor size was 6 cm (with a range of 3.5–10 cm). The most common tumor stage was pT3 (13/21; 62%%), followed by pT4 (8/21, 38%). Loco-regional lymph nodes were positive (N1, N2) in the majority of patients (14/21, 67%) and negative (N0) in a third of patients (7/21, 33%). Distant metastases (M1) were identified in 4 cases and localized to the liver. One case (RCIV) presented multiple liver and lung metastases. The available clinicopathological characteristics are reported in Table 1. The median overall survival of patients was 13.1 months (with a range of 1–43 months) (Table 1). Of the clinicopathological variables, only patients with lymph node involvement (N1–N2) had shorter overall survival than those without lymph node metastasis (Table 1).
Table 1.
Clinicopathological features of RCTs enrolled in this study. n.a. = not available, OS = Overall Survival.
| n° Case | Years | Center | Age (ys) | Gender | Site | Size (Max) | T | N | M | Type | OS (Months) | Status |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RCT I | 2009 | Legnago | 73 | Female | Right colon | 10 | 4 | 1b | none | composite | 6 | dead |
| RCT II | 2009 | Benevento | 71 | Male | Right colon | 10 | 4 | 1a | none | pure | 8 | dead |
| RCT III | 2009 | Seoul | 62 | Male | Sigma | 4.5 | 3 | 1a | none | composite | 36 | alive |
| RCT IV | 2010 | Seoul | 83 | Female | Rectum | 6.5 | 4 | 2b | liver, lung | composite | 1 | dead |
| RCT V | 2011 | Vilnius | 49 | Male | Sigma | 7 | n.a. | 2b | none | composite | 7 | dead |
| RCT VI | 2007 | Legnago | 63 | Male | Left colon | 6 | 3 | 2a | none | composite | 1 | dead |
| RCT VII | 2014 | Legnago | 71 | Female | Right colon | 8 | 4 | 2a | none | composite | 8 | dead |
| RCT VIII | 2015 | Legnago | 66 | Male | Sigma | 8 | 4 | 0 | n.a. | composite | n.a. | n.a. |
| RCT IX | 2015 | Legnago | 85 | Male | Anastomosis post-Bilroth | n.v. | 4 | 0 | n.a. | composite | 1 | dead |
| RCT X | 2015 | Genova | 71 | Female | Rectum | 5 | 3 | 2b | none | composite | 1 | dead |
| RCT XI | 2014 | Genova | 40 | Female | Right colon | 4 | 4a | 2b | none | composite | 18 | dead |
| RCT XII | 2014 | Genova | 72 | Female | Right colon | 5.2 | 3 | 0 | none | composite | 33 | alive |
| RCT XIII | 2015 | Genova | 87 | Female | Right colon | 7 | 3 | 0 | none | composite | 18 | alive |
| RCT XIV | 2015 | Genova | 78 | Female | Trasversum colon | 3.5 | 3 | 2b | liver | composite | 13 | dead |
| RCT XV | 2013 | Genova | 65 | Female | Right colon | 3.5 | 3 | 0 | none | composite | 36 | alive |
| RCT XVI | 2014 | Genova | 81 | Male | Right colon | 7 | 3 | 1a | none | composite | 7 | dead |
| RCT XVII | 2013 | Genova | 76 | Female | Right colon | 8.5 | 3 | 0 | none | composite | 43 | alive |
| RCT XVIII | 2014 | Genova | 73 | Male | Right colon | 5 | 3 | 2b | liver | composite | 7 | dead |
| RCT XIX | 2015 | Genova | 81 | Female | Right colon | 7 | 3 | 1a | liver | composite | 5 | dead |
| RCT XX | 2015 | Genova | 86 | Female | Right colon | 4 | 4b | 0 | none | composite | 1 | dead |
| RCT XXI | 2015 | Alaska | 74 | Female | Right colon | 4.5 | 3 | positive | none | composite | n.a. | dead |
3.2. Immunophenotypic Profiling of RCTs
Of the 21 tumors tested by IHC, 13 (62%) exhibited loss of nuclear expression of MMR proteins (MLH1/PMS2) and they were classified as deficient (MMR-d). The remaining 8 tumors (38%) exhibited intact expression of all mismatch repair proteins and they were classified as MMR proficient (MMR-p).
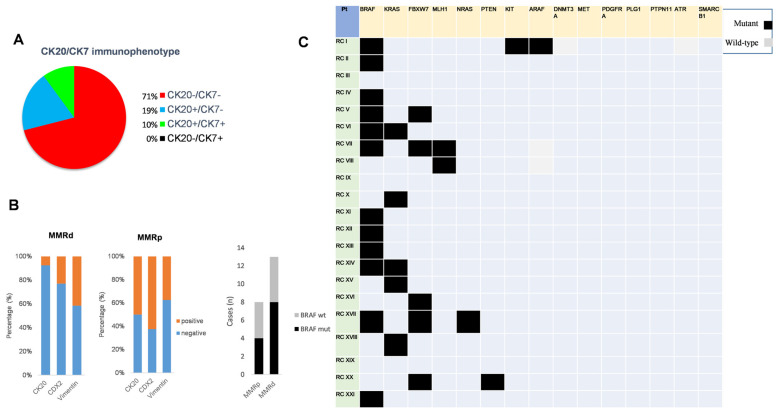
Notably, CK20, a specific marker for classical colorectal carcinoma, was expressed in 5/21 (24%) cases and the remaining 16 tumors (76%) were negative. CK7 resulted as absent in the large majority of cases 18/21 (85.7%) and just one case was positive. Therefore, the combined evaluation of both markers revealed that the CK20−/CK7− pattern included the greatest proportion of cases 15/21 (71%), followed by CK20+/CK7− in 4/21 (19%), CK20+/CK7+ in 2/21 (10%), and CK20−/CK7+ in 0/21 (0%) (Figure 1A). CDX2 resulted as positive in 8/21 (38%) and negative in 13/21 (62%) tumors, respectively.
Figure 1.
Immunophenotypic and genetic features of RCTs. (A) The combined CK20/CK7 immunostaining profiles in RCTs is shown by a pie chart; (B) Left: CK20, CDX2 and vimentin expression in mismatch repair-deficient (MMR-d) and mismatch repair-proficient (MMR-p) subgroups is shown by graphs. Right: BRAF mutants or wild-type tumors are related to MMR-d and MMR-p subgroups; (C) The recurrence of mutant (black squares) and wild-type (grey squares) in the indicated genes for each patient is shown by a panel.
We then evaluated the expression of vimentin, a marker for the epithelial to mesenchymal transition (EMT). Vimentin-positive staining was found in 8/21 (38%) of RCTs, while the majority of cases were vimentin negative (13/21 (62%)) (Table 2).
Table 2.
Immunohistochemistry Profile of the RCTs.
| n° Case | MMR | CK7 | CK20 | CDX-2 | Vimentin |
|---|---|---|---|---|---|
| RCT I | MMRd (mlh1/PMS2) | neg. | neg | neg. | ++ |
| RCT II | MMRd (mlh1/PMS2) | neg. | neg | neg. | ++ |
| RCT III | MMRp | n.a. | ++ | ++ | neg. |
| RCT IV | MMRp | n.a. | neg | ++ | ++ |
| RCT V | MMRp | neg. | neg | neg. | ++ |
| RCT VI | MMRp | neg. | neg | neg. | ++ |
| RCT VII | MMRd (mlh1/PMS2) | neg. | neg | neg. | ++ |
| RCT VIII | MMRd (mlh1/PMS2) | neg. | neg | ++ | neg. |
| RCT IX | MMRd (mlh1/PMS2) | neg. | Neg. (10%+) | ++ | Focally + |
| RCT X | MMRp | neg. | ++ | ++ | neg |
| RCT XI | MMRp | ++ | ++ | ++ | neg |
| RCT XII | MMRd (mlh1/PMS2) | neg. | neg | neg | ++ |
| RCT XIII | MMRd (mlh1/PMS2) | neg. | neg | neg | neg |
| RCT XIV | MMRd (mlh1/PMS2) | neg. | ++ | ++ | neg |
| RCT XV | MMRp | neg. | ++ | ++ | neg |
| RCT XVI | MMRd (mlh1/PMS2) | neg. | neg | neg | neg |
| RCT XVII | MMRd (mlh1/PMS2) | neg. | neg | neg | neg |
| RCT XVIII | MMRp | neg. | neg | neg | neg |
| RCT XIX | MMRd (mlh1/PMS2) | neg. | neg | neg | neg |
| RCT XX | MMRd (mlh1/PMS2) | neg. | neg | neg. | neg |
| RCT XXI | MMRd (mlh1/PMS2) | neg. | neg | neg. | n.a. |
Notably, negative expression of the epithelial markers CDX2 and CK20 was identified in more than 80% of MMR-d cancers compared to less than 50% of MMR-p cancers (Figure 1B). It is also of note that the IHC profile of vimentin was independent of MMR status (Figure 1B). The immunohistochemistry profiles of MMR, epithelial, and mesenchymal markers are shown in (Table 2).
3.3. Identification of Actionable Mutations in RCTs
The next-generation sequencing (NGS) analysis revealed the mutation V600E in BRAF as the most frequent with 12/21 (57%) (Figure 1C). Mutations in BRAF occurred regardless of MMR status (Figure 1B). Mutations in KRAS were identified in 5/21 (24%) cases, 2 of which co-occurred with BRAF mutations in the same lesion. The mutation profiles clearly indicated prevalent activating mutations in the RAS-RAF-MAPK pathway (15/21 (71.4%)) (Figure 1C). Recurrent mutations were also identified in the gene encoding for F-box and WD repeat domain containing 7 (FBXW7), with 5 of 21 (23.8%) cases (Figure 1C).
3.4. Defects in Primary Cilia and Rhabdoid Dedifferentiation
Although SMARCB1/INI1 is widely used as a marker of rhabdoid tumors, its relevance has not yet been comprehensively investigated in RCTs.
We thus analyzed SMARCB1/INI1 expression by IHC. Surprisingly, we found that only 3/21 cases (14%) exhibited loss of INI1 expression. In contrast, the large majority of RCTs (18/21 (86%)) exhibited an intense expression of SMARCB1/INI1 (Table 3).
Table 3.
The DNA mutational profile and Loss of heterozigosity (LOH) at SMARC1/INI1 and CROCC loci is correlated with immunohistochemistry (IHC). Results: n.a. = not available, neg. = negative.
| n° Case | CROCC Gene | CROCC Loh | CROCC Ihc | SMARCB1/INI1 Gene | smarcb1/ini1 loh | INI1 ihc |
|---|---|---|---|---|---|---|
| RCT I | c481G>T; c.3705-2A>G | present | Loss (<1 signal/cell) | wild type | present | neg. |
| RCT II | c5654T>C | present | Loss (<1 signal/cell) | wild type | present | ++ |
| RCT III | wildtype | present | Loss (<1 signal/cell) | wild type | present | ++ |
| RCT IV | wildtype | present | Loss (<1 signal/cell) | wild type | present | ++ |
| RCT V | wildtype | present | Loss (<1 signal/cell) | wild type | present | ++ |
| RCT VI | wildtype | present | Loss (<1 signal/cell) | wild type | present | neg. |
| RCT VII | wildtype | present | Loss (<1 signal/cell) | wild type | intact | ++ |
| RCT VIII | wildtype | intact | Loss (<1 signal/cell) | wild type | intact | ++ |
| RCT IX | wildtype | intact | Loss (<1 signal/cell) | wild type | present | ++ |
| RCT X | wildtype | present | Loss (<1 signal/cell) | wild type | intact | ++ |
| RCT XI | Leu1159Phe | present | Loss (<1 signal/cell) | wild type | present | ++ |
| RCT XII | wildtype | intact | Loss (<1 signal/cell) | wild type | present | ++ |
| RCT XIII | wildtype | intact | Loss (<1 signal/cell) | wild type | intact | ++ |
| RCT XIV | wildtype | intact | Loss (<1 signal/cell) | wild type | present | ++ |
| RCT XV | wildtype | present | Loss (<1 signal/cell) | wild type | present | ++ |
| RCT XVI | Ala1510Thr | intact | Loss (<1 signal/cell) | wild type | present | ++ |
| RCT XVII | Ser1320Ile + Arg1659His | intact | Loss (<1 signal/cell) | wild type | present | ++ |
| RCT XVIII | wildtype | present | Loss (<1 signal/cell) | wild type | present | ++ |
| RCT XIX | wildtype | intact | Loss (<1 signal/cell) | wild type | present | ++ |
| RCT XX | wildtype | present | Loss (<1 signal/cell) | wild type | present | ++ |
| RCT XXI | wildtype | n.a. | Loss (<1 signal/cell) | wild type | n.a. | neg. |
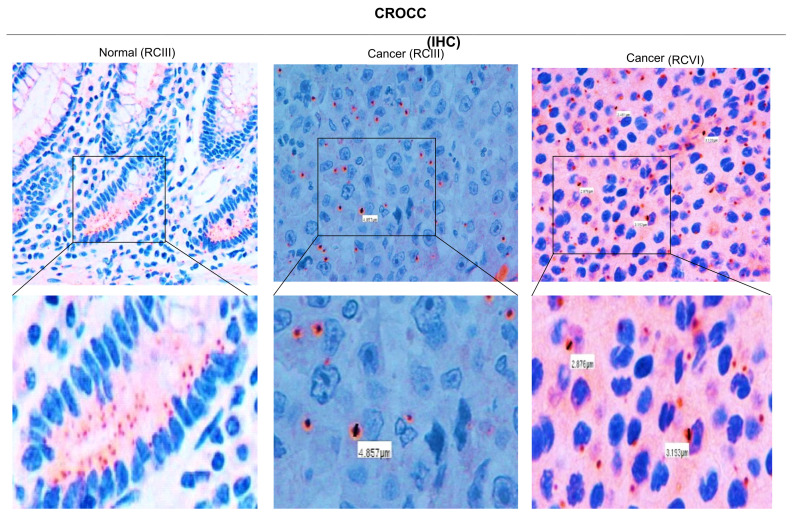
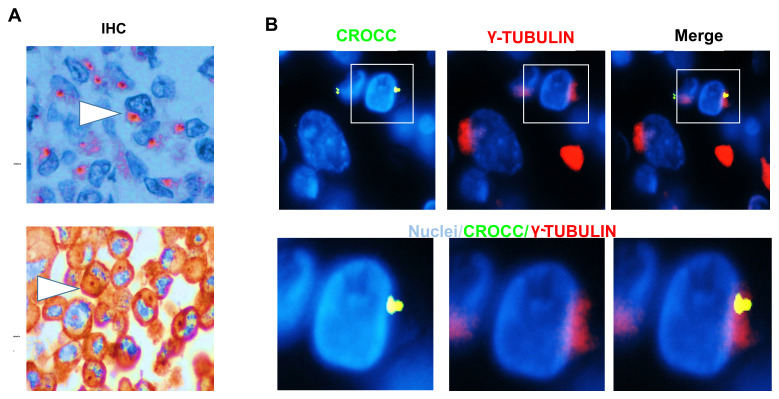
As mutations in the ciliogenic gene CROCC have been reported in RCTs, we tested its expression by IHC. We found that the large majority of RCTs (19/21; 90%) exhibited no CROCC staining (Table 3). Notably, CROCC-negative coexisted with CROCC-positive cells, having large and apparently abnormal cilia compared to those detected in the matched normal mucosa (Figure 2). Single and abnormal large cilia were confirmed using IHC for γ-tubulin (Figure 3A). To verify the presence of abnormal cilia within tissues, we used immunofluorescence to investigate the colocalization of CROCC and γ-tubulin at cilia. We found that CROCC and γ-tubulin colocalized and appeared as large foci at ciliary basal bodies (Figure 3B).
Figure 2.
Immunohistochemistry of CROCC in RCT tissues. Left: CROCC IHC in colon normal tissues. Down panel: magnifications show that CROCC is expressed in normal mucosa. Right: CROCC immunohistochemistry in tumor tissues (RCIII and RCVI). Down panels: magnifications show that CROCC is absent in about 50% of tumor cells. Immunosignals foci near the nuclei appear as single and large foci. The size of CROCC-positive foci is shown by white rectangles.
Figure 3.
CROCC and γ-tubulin alterations in RCT tissues. (A) CROCC and γ-tubulin immunohistochemistry in RCT. Positive foci are shown by white arrows. (B) RCT tissue stained with γ-tubulin (red) and CROCC (green). Nuclei in blue. The magnification shows the colocalization of CROCC/γ-tubulin in abnormal cilia. Bar 10 μm.
We next investigated the DNA mutational profile and LOH at SMARC1/INI1 and CROCC loci. The mutations and LOH in CROCC were identified in 5/21 (23.8%) and 12/21 (57%) cases, respectively. Of these, three cases exhibited the co-occurrence of missense mutation and LOH. In contrast, no mutations were identified in SMARCB1/INI1, while LOH was evidenced in 16/21 (76%) cases (Table 3). Notably, three cases with LOH in SMARCB1/INI1 exhibited negative INI1 staining by IHC.
4. Discussion
RCTs represent a distinct entity that occurs in adult-elderly patients (31–87 years), mostly in the right colon without predisposition towards any one sex [9,18,19].
The diagnostic hallmark of an RCT is the rhabdoid morphology characterized by the presence of “large round neoplastic cells with glassy eosinophilic cytoplasm containing hyaline-like inclusion bodies, eccentric nuclei and macronucleoli” [8,9,20].
In this study, we analyzed a large series of 21 RCTs, 12 of which have already been described in the literature. The median overall survival time of patients was extremely short at “13.1 months” [8,9].
We included in the study RCTs with two pathological features: (a) mononuclear round cells with conspicuous eccentric macronucleoli and (b) binucleated large–medium sized cells with prominent nucleoli. Tumors with pleomorphic, giant, anaplastic, or undifferentiated colorectal carcinoma were excluded.
The lack of or negative expression of SMARCB1/INI1 was considered the predominant event in the development of pediatric rhabdoid tumors. However, the exact occurrence of SMARCB1/INI1 loss in RCTs remains unknown. We found that SMARCB1/INI1 expression was positive in the large majority of RCTs (85%), supporting the notion that RCTs are not genetically related to their pediatric counterparts [20].
A recent study on 3051 cases of classical CRC showed that SMARCB1/INI1 is deleted in 14 cases (0.46%), confirming the previous data obtained by our group [7,8,9,21]. Therefore, the role of SMARCB1/INI1 in the pathogenesis of RCTs may not be as relevant as initially thought.
The malfunctioning of the MMR repair pathway in RCTs is unknown. Unexpectedly, we found that more than half of RCTs are MMR-d cancers. In addition, the loss of expression of CDX2/CK20 was often associated with the MMR-d (MLH1/PMS2) subgroup [22]. The low prevalence of vimentin expression suggests that it is not a diagnostic marker of rhabdoid differentiation. Further studies are needed to understand whether the EMT process is involved in the rhabdoid pathogenesis.
The BRAF V600E mutation was identified in about 60% of RCTs. Interestingly, more than 70% of lesions displayed activating mutations in the MAPK pathway. These findings support the idea that RCTs prevalently arise and progress through serrated precursor lesions. However, mutations in BRAF were not correlated with MMR status. Thus, BRAF mutations and MMR, rather than causative events, could shape a permissive microenvironment for the progression to rhabdoid carcinomas. However, the precise molecular events need to be further investigated in future studies.
We recently found that the depletion of CROCC in BRAF-mutant colorectal cancer cells promotes the acquisition of rhabdoid features [8]. Primary cilia play an important role in the regulation of cell signaling pathways but have seldom been studied in rare cancer variants. Our study confirms the prominent role of centrosome/cilia in the development of rhabdoid features, expanding the spectrum of lesions at the CROCC locus that characterize such rare lesions.
Rhabdoid cells often exhibited single and aberrantly larger cilia as compared to normal tissues, suggesting an elevated level of genetic heterogeneity and chromosomal instability typical of rare cancer variants. By contrast, CROCC has been reported as significantly increased in classical CRC [9,23]. Therefore, our study suggests that RCTs and classical CRCs exhibit different abnormalities in primary cilia. Interestingly, our data indicate that the loss of primary cilia could potentiate BRAF/MAPK pathway activation, thereby explaining the rapid malignant progression and resistance to therapies of these neoplasms. Recent studies suggest that the primary cilium is a critical determinant in controlling tumor pathway switching. For example, in basal cell carcinoma, the primary cilium acts as an important lineage gatekeeper, preventing Hedgehog to Ras/MAPK pathway switching [24]. We propose that the duplication or division of the primary cilia could not occur under normal circumstances in RCTs, leading to marked defects in cellular signaling and microtubules dynamics. Thus, the close inverse association between mutations in the MAPK pathway and loss of primary cilia could be clinically exploitable in the development of selective therapies for patients with RCTs.
5. Conclusions
Taken together, our findings indicate that primary ciliogenesis and MAPK pathway activation contribute to the aggressiveness of RCTs and, therefore, may constitute a novel therapeutic target.
Author Contributions
A.R. Conceptualization, methodology, investigation, data curation, writing-original draft prepararion, supervision, project administration, F.G. methodology, validation, writing-original draft prepararion, writing—review and editing, L.M. methodology, validation, writing-original draft prepararion, writing—review and editing, M.S. methodology, software, M.F. data curation, supervision, M.P.C. methodology, validation, G.M. methodology, A.S. (Antonio Sassano). methodology, P.P. methodology, resources, data curation, A.V. resources, data curation, G.S. data curation, G.G. investigation, data curation, E.D.U. investigation, data curation, L.C. software, A.S. (Aldo Scarpa). supervision, writing—original draft preparation, project administration, funding acquisition F.F. resources, visualization, M.P. Conceptualization, methodology, investigation, writing-original draft prepararion, supervision, project administration, writing—review and editing. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Institutional Review Board approval no. 997CESC from the Ethics Committee (Comitato Etico di Verona e Rovigo dell’Azienda Ospedaliera Universitaria Integrata) on 7 September 2016, documented by the CESC prot. 42160 on 9 September 2016, and formalized by the General Manager with deliberation no. 458 of 16 September 2016, communicated with protocol 51319 on 23 September 2016.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research received no external funding.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Frierson H.F., Mills S.E., Innes D.J. Malignant rhabdoid tumor of the pelvis. Cancer. 1985;55:1963–1967. doi: 10.1002/1097-0142(19850501)55:9<1963::AID-CNCR2820550922>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 2.Kodet R., Newton W.A., Sachs N., Hamoudi A.B., Raney R.B., Asmar L., Gehan E.A. Rhabdoid tumors of soft tissues: A clinic-pathologic study of 26 cases enrolled on the Intergroup Rhabdomyosarcoma Study. Hum. Pathol. 1991;22:674–684. doi: 10.1016/0046-8177(91)90289-2. [DOI] [PubMed] [Google Scholar]
- 3.Miyagi J., Tsuhako K., Kinjo T., Iwamasa T., Hashimoto H., Ishikawa S. Rhabdoid tumour of the lung is a dedifferentiated phenotype of pulmonary adenocarcinoma. Histopathology. 2000;37:37–44. doi: 10.1046/j.1365-2559.2000.00906.x. [DOI] [PubMed] [Google Scholar]
- 4.Kojima M., Miyake T., Ueki T., Ohta H., Kushima R., Shiohara M., Mizuta H., Iida H., Yamaguchi T., Kaida S., et al. INI1-negative colorectal undifferentiated carcinoma with rhabdoid features and postoperative rapidly growing liver metastases: A case report and review of the literature. Surg. Case Rep. 2021;7:104. doi: 10.1186/s40792-021-01189-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Remo A., Cecchini M.P., Benati D., Bernardi P., Manfrin E., Giordano G., Bonomi F., Parcesepe P., Fassan M., Colombari R., et al. CROCC-mutated rhabdoid colorectal carcinoma showing in intercellular spaces lamellipodia and cellular projections revealed by electron microscopy. Virchows Arch. 2019;475:245–249. doi: 10.1007/s00428-019-02554-4. [DOI] [PubMed] [Google Scholar]
- 6.Biegel J.A., Zhou J.Y., Rorke L.B., Stenstrom C., Wainwright L.M., Fogelgren B. Germ-line and acquired mutations of INI1 in atypical teratoid and rhabdoidtumors. Cancer Res. 1999;59:74–79. [PubMed] [Google Scholar]
- 7.Wang J., Andrici J., Sioson L., Clarkson A., Sheen A., Farzin M., Toon C.W., Turchini J., Gill A.J. Loss of INI1 expression in colorectal carcinoma is associated with high tumor grade, poor survival, BRAFV600E mutation, and mismatch repair deficiency. Hum. Pathol. 2016;55:83–90. doi: 10.1016/j.humpath.2016.04.018. [DOI] [PubMed] [Google Scholar]
- 8.Agaimy A., Rau T.T., Hartmann A., Stoehr R. SMARCB1 (INI1)-negative rhabdoid carcinomas of the gastrointestinal tract: Clinicopathologic and molecular study of a highly aggressive variant with literature review. Am. J. Surg. Pathol. 2014;38:910–920. doi: 10.1097/PAS.0000000000000173. [DOI] [PubMed] [Google Scholar]
- 9.Remo A., Manfrin E., Parcesepe P., Ferrarini A., Han H.S., Mickys U., Laudanna C., Simbolo M., Malanga D., Oliveira D.M., et al. Centrosome linker-induced tetraploid segregation errors link rhabdoid phenotypes and lethal colorectal cancers. Mol. Cancer Res. 2018;16:1385–1395. doi: 10.1158/1541-7786.MCR-18-0062. [DOI] [PubMed] [Google Scholar]
- 10.Remo A., Li X., Schiebel E., Pancione M. The Centrosome Linker and Its Role in Cancer and Genetic Disorders. Trends Mol. Med. 2020;26:380–393. doi: 10.1016/j.molmed.2020.01.011. [DOI] [PubMed] [Google Scholar]
- 11.Woodruff J.B., Ferreira Gomes B., Widlund P.O., Mahamid J., Honigmann A., Hyman A.A. The Centrosome Is a Selective Condensate that Nucleates Microtubules by Concentrating Tubulin. AA. Cell. 2017;169:1066–1077.e10. doi: 10.1016/j.cell.2017.05.028. [DOI] [PubMed] [Google Scholar]
- 12.Vlijm R., Li X., Panic M., Rüthnick D., Hata S., Herrmannsdörfer F., Kuner T., Heilemann M., Engelhardt J., Hell S.W., et al. STED nanoscopy of the centrosome linker reveals a CEP68-organized, periodic rootletin network anchored to a C-Nap1 ring at centrioles. Proc. Natl. Acad. Sci. USA. 2018;115:E2246–E2253. doi: 10.1073/pnas.1716840115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dang H., Schiebel E. Emerging roles of centrosome cohesion. Open Biol. 2022;12:220229. doi: 10.1098/rsob.220229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang B., Liang Z., Liu P. Functional aspects of primary cilium in signaling, assembly and microenvironment in cancer. J Cell. Physiol. 2021;236:3207–3219. doi: 10.1002/jcp.30117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Michaud E.J., Yoder B.K. The primary cilium in cell signaling and cancer. Cancer Res. 2006;66:6463–6467. doi: 10.1158/0008-5472.CAN-06-0462. [DOI] [PubMed] [Google Scholar]
- 16.Corbit K.C., Aanstad P., Singla V., Norman A.R., Stainier D.Y., Reiter J.F. Vertebrate Smoothened functions at the primary cilium. Nature. 2005;437:1018–1021. doi: 10.1038/nature04117. [DOI] [PubMed] [Google Scholar]
- 17.Simbolo M., Gottardi M., Corbo V., Fassan M., Mafficini A., Malpeli G., Lawlor R.T., Scarpa A. DNA qualification workflow for next generation sequencing of histopathological samples. PLoS ONE. 2013;8:e62692. doi: 10.1371/journal.pone.0062692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bando H., Ohtsu A., Yoshino T. Therapeutic landscape and future direction of metastatic colorectal cancer. Nat. Rev. Gastroenterol. Hepatol. 2023;20:306–322. doi: 10.1038/s41575-022-00736-1. [DOI] [PubMed] [Google Scholar]
- 19.Remo A., Fassan M., Vanoli A., Bonetti L.R., Barresi V., Tatangelo F., Gafà R., Giordano G., Pancione M., Grillo F., et al. Morphology and Molecular Features of Rare Colorectal Carcinoma Histotypes. Cancers. 2019;11:1036. doi: 10.3390/cancers11071036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glückstein M.-I., Dintner S., Arndt T.T., Vlasenko D., Schenkirsch G., Agaimy A., Müller G., Märkl B., Grosser B. Comprehensive Immunohistochemical Study of the SWI/SNF Complex Expression Status in Gastric Cancer Reveals an Adverse Prognosis of SWI/SNF Deficiency in Genomically Stable Gastric Carcinomas. Cancers. 2021;13:3894. doi: 10.3390/cancers13153894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pancione M., Remo A., Zanella C., Sabatino L., Blasi A.D., Laudanna C., Astati L., Rocco M., Bifano D., Piacentini P., et al. The chromatin remodelling component SMARCB1/INI1 influences the metastatic behavior of colorectal cancer through a gene signature mapping to chromosome 22. J. Transl. Med. 2013;11:297. doi: 10.1186/1479-5876-11-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lugli A., Tzankov A., Zlobec I., Terracciano L.M. Differential diagnostic and functional role of the multi-marker phenotype CDX2/CK20/CK7 in colorectal cancer stratified by mismatch repair status. Mod. Pathol. 2008;21:1403–1412. doi: 10.1038/modpathol.2008.117. [DOI] [PubMed] [Google Scholar]
- 23.Yasar B., Linton K., Slater C., Byers R. Primary cilia are increased in number and demonstrate structural abnormalities in human cancer. J. Clin. Pathol. 2017;70:571–574. doi: 10.1136/jclinpath-2016-204103. [DOI] [PubMed] [Google Scholar]
- 24.Kuonen F., Huskey N.E., Shankar G., Jaju P., Whitson R.J., Rieger K.E., Atwood S.X., Sarin K.Y., Oro A.E. Loss of Primary Cilia Drives Switching from Hedgehog to Ras/MAPK Pathway in Resistant Basal Cell Carcinoma. J. Investig. Dermatol. 2019;139:1439–1448. doi: 10.1016/j.jid.2018.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]