Abstract
Idiopathic scoliosis (IS) is a three-dimensional rotation of the spine >10 degrees with an unknown etiology. Our laboratory established a late-onset IS model in zebrafish (Danio rerio) containing a deletion in kif7. A total of 25% of kif7co63/co63 zebrafish develop spinal curvatures and are otherwise developmentally normal, although the molecular mechanisms underlying the scoliosis are unknown. To define transcripts associated with scoliosis in this model, we performed bulk mRNA sequencing on 6 weeks past fertilization (wpf) kif7co63/co63 zebrafish with and without scoliosis. Additionally, we sequenced kif7co63/co63, kif7co63/+, and AB zebrafish (n = 3 per genotype). Sequencing reads were aligned to the GRCz11 genome and FPKM values were calculated. Differences between groups were calculated for each transcript by the t-test. Principal component analysis showed that transcriptomes clustered by sample age and genotype. kif7 mRNA was mildly reduced in both homozygous and heterozygous zebrafish compared to AB. Sonic hedgehog target genes were upregulated in kif7co63/co63 zebrafish over AB, but no difference was detected between scoliotic and non-scoliotic mutants. The top upregulated genes in scoliotic zebrafish were cytoskeletal keratins. Pankeratin staining of 6 wpf scoliotic and non-scoliotic kif7co63/co63 zebrafish showed increased keratin levels within the zebrafish musculature and intervertebral disc (IVD). Keratins are major components of the embryonic notochord, and aberrant keratin expression has been associated with intervertebral disc degeneration (IVDD) in both zebrafish and humans. The role of increased keratin accumulation as a molecular mechanism associated with the onset of scoliosis warrants further study.
Keywords: idiopathic scoliosis, zebrafish, RNA sequencing, intervertebral disc degeneration, transcriptomics, histology, spine, genetics, KIF7, keratin
1. Introduction
Idiopathic scoliosis (IS) is a structural lateral curvature of the spine ≥10° with a rotatory component [1]. Although 2–3% of adolescents across populations are affected with IS [2,3], and the condition has been researched for decades, the etiology of IS initiation as well as the pathology of severe, progressive IS are essentially unknown. Research progress has been stymied by significant challenges, including (1) a known causal tissue, as IS may affect the skeleton [4,5,6,7], musculature [8,9,10], the proprioceptive system [11,12,13], the nervous system [14,15], joints and/or other musculoskeletal tissues [16]; (2) significant phenotypic and genetic heterogeneity across affected individuals and families [17,18,19,20]; and (3) the lack of a developmentally appropriate animal model.
Over the last decade, D. rerio (zebrafish) has emerged as a popular model for human scoliosis based on similar vertebral anatomy, ease of breeding, and genetic manipulation [21]. To date, multiple zebrafish genetic mutants (e.g., ptk7 [22,23], POC5 [24], kif6 [25]) have linked genes involved in cilia and ciliary motility to a progressive scoliosis, giving potential insight into molecular mechanisms that may regulate scoliosis development. These mutants exhibit ciliary motility defects and often develop hydrocephalus and/or a disruption of the Reissner fiber, a fiber aggregation along the central canal that is essential to zebrafish axial development [26,27,28]. Mechanisms related to cerebrospinal fluid (CSF) flow [23,29], specialized contacting neurons within the axial sensory system (CSF-cNs) [30], inflammation [31], and urotensin peptides [32] have been hypothesized as related to these phenotypic observations; however, a definitive mechanism related specifically to scoliosis development has not been characterized.
Our zebrafish model for scoliosis, kif7co63/co63, develops juvenile-onset spinal curvatures without obvious vertebral malformations, hydrocephalus, or malformations in the Reissner fiber [33]. As patients with IS are not yet known to have gross morphological changes to these structures and develop scoliosis during the juvenile-adolescent period, kif7co63/co63 zebrafish may uniquely mirror the human condition. KIF7 encodes a broadly conserved kinesin ciliary protein that localizes to the axonemal tip of primary cilia and binds to the plus-ends of microtubules [34]. The protein functions as a scaffold protein for ciliary function and acts as both a negative and positive regulator of the hedgehog (Hh) signaling pathway, an evolutionary conserved molecular pathway central to embryonic development, limb patterning and musculoskeletal maintenance [34], including formation and maintenance of the intervertebral disc [35]. Kif7 primarily acts by suppressing the Gli1 transcription factor. In zebrafish, Kif7 accumulates at the ciliary tip, as observed in mammals, as well as within cytoplasmic puncta, which sequester Gli1 and Gli2 and disperse in response to Hh pathway activation [36]. In humans, loss of function mutations in KIF7 have been linked to both Joubert and the rare acrocallosal syndromes (OMIM #611254), two ciliopathies with overlapping, system-wide defects including developmental disability, skeletal abnormalities and kidney disease. Scoliosis has been observed in 5–33% of Joubert syndrome cases [37,38,39], potentially related to early hypotonia, whereas the scoliosis prevalence in acrocallosal syndrome has not yet been described.
Approximately 25% of kif7co63/co63 zebrafish develop spinal curvatures as juveniles with no evidence of abnormalities in brain morphology or hydrocephaly, and no morphological changes to the central canal cilia or the Reissner fiber. Our hypothesis is that study of the kif7co63/co63 zebrafish, both with and without the scoliosis phenotype, will provide insight into potential molecular mechanisms underlying scoliosis development. To this end we performed bulk transcriptome mRNA sequencing analyses of our mutant kif7co63/co63 embryonic (4 days post fertilization) and young adult (6 weeks past fertilization [wpf]) zebrafish with and without scoliosis, as well as age-matched heterozygous and wild-type controls. Specific findings are validated via qRT-PCR and histology.
2. Materials and Methods
Kif7co63 generation: Kif7co63 zebrafish were generated by CRISPR-Cas9 as described previously [33]. Briefly, gDNA guides were generated using the CRISPOR tool [40] and targeted to exon 3 of kif7 (NM_001014816.1). Zebrafish embryos at the single-cell stage were injected with an injection mix created from 18.2 ng sgRNA, 50 ng Cas9 protein, 285 mM potassium chloride, 1 µL phenol red, and sterile water during the single cell phase (<45 min post fertilization). Embryos were analyzed for mutations the day after injection by lysing, PCR and fragment analysis (FA). We generated F4 homozygous kif7co63/co63 first by outcrossing CRISPR-Cas9 injected F0 to AB wild-type fish (ZIRC) to generate F1 zebrafish. F1 mutations were characterized by Sanger sequencing (see below). F1 zebrafish were crossed to AB (wild-type) zebrafish to generate F2 kif7co63/+. We then in crossed F2 kif7co63/+ to generate F3 kif7co63/co63. We then in-crossed F3 kif7co63/co63 to generate a pool of F4 kif7co63/co63. Two zebrafish indel lines were generated from mutations resulting in (1.) a 5 base pair (bp) deletion/14 bp insertion at exon 3 (RefSeq gene NM_001014816.1), Chr7:14433928 (kif7co63), generating a 3 amino acid insertion and (2.) a 4 bp deletion in exon 3 (RefSeq gene NM_001014816.1), Chr7:14433924 (kif7d4), resulting in a frameshift mutation. Mutants were outcrossed to AB wild-type fish every third generation to avoid genetic drift. Mutant kif7co63 was used for all experiments due to consistent percentage of scoliosis phenotype in the population [33]. A complementation test was performed using both lines to confirm the phenotype is associated with the mutation of kif7 gene [33] (Supplemental Figure S6).
2.1. Zebrafish Husbandry and Crossing
Zebrafish were housed at the University of Colorado Anschutz Medical Campus aquatic facility. Husbandry and experimental protocols were approved under the Institutional Animal Care and Use Committee Protocol #00370. Animals were maintained in a 14 h light/10 h dark cycle at 28.5 °C. Animals under anesthesia in 168 mg/L Tricaine were fin clipped for genotyping and RNA extraction. Animals were euthanized in 400 mg/L Tricaine, followed by immersion in ice water.
Zebrafish were bred in a sloped 1.7 L static tank. Males and females were placed in the tank separated by a divider. Each tank was set with a maximum of 8 total fish in an either 1:1 or 2:1 female to male ratio. Fish were placed in static tank in the afternoon after their last daily feeding. The following morning, water in the static tanks was changed and the dividers were pulled. Fish were monitored and embryos were harvested by siphoning after two hours.
2.2. Genotyping
At 24 h past fertilization (hpf), 8 embryos were euthanized by ice water immersion. Embryos were placed in 50 µL of lysis solution (500 µL 1 M Tris pH 8.3, 2.5 mL 1 M KCL, 1.5 mL 10% Tween, 1.5 mL 10% NP40, 44 mL ddH20). The 6 wpf zebrafish were genotyped by fin clip immediately post euthanasia. Briefly, zebrafish were immersed in 400 mg/L tricaine until gill movement stopped and tail fins were clipped with a sterile razor. Clipped fins were immersed in 50 µL lysis solution and run with the following reaction: 20 min at 95 °C, 2 min on ice, added 2.5 µL Proteinase K (Invitrogen, Waltham, MA, USA), 3 h at 55 °C, 10 min at 95 °C, and hold at 12 °C. Samples underwent the following PCR reaction using 12.5 µL 2× GOTaq Green Master Mix (Promega, Madison, WI, USA), 1 µL gDNA, 0.5 µL 10 µM F and R primers, and 10 µL sterile water (see Table 1):
Table 1.
Specifications of the PCR reaction for genotyping eight embryonic zebrafish at 24 h past fertilization (hpf).
| Initial | Denature | Anneal | Extend | Final | |
|---|---|---|---|---|---|
| Temp | 95 | 95 | 58 | 72 | 72 |
| Time | 3 min | 30 s | 30 s | 30 s | 20 min |
| 34× | |||||
Routine genotyping of zebrafish lines was performed by PCR as above, followed by gel electrophoresis using a 3% agarose gel with a 1:1 ratio of standard agarose to MetaPhor agarose (Lonza, Basel, Switzerland). The gel was run at 120 V for 1 h and 45 min followed by UV imaging (Supplemental Figure S5).
2.3. RNA Extraction and Sequencing (Novogene)
At 4 days past fertilization (dpf), 20 embryos per tube were euthanized and pooled in 1 mL of RNA lysis buffer (Zymo Research, Irvine, CA, USA). Embryos were lysed using a VWR Bead Mill Homogenizer with Lysing Matrix A beads (MP Biomedicals, Santa Ana, CA, USA). Samples were iced for 5 min, lysed for 45 s, then iced again for 5 min. This process was completed three times to ensure maximal lysis. The same protocol was used to extract RNA for 6 wpf zebrafish, but 1 fish was used per tube. RNA was extracted using the QuickRNA Miniprep (Zymo) using the on-column DNase digestion protocol.
2.4. Bioinformatic Filtering
Bioinformatic filtering was performed by the Novogene Company (Beijing, China) following an established pipeline. Briefly, Illumina image data were base called using CASAVA and stored in FASTQ format. The sequencing error rate is represented in Qphred scores. Low-quality reads (reads where uncertain nucleotides constitute >10% of the total read, or when low-quality nucleotides [base quality < 5] constituted >50% of the read) and reads containing adapters were filtered out. Sequences were mapped to the zebrafish reference genome GRCz11/danRer11 using HISAT2 v.2.0.5. Gene expression levels and differential gene expression analysis were calculated through DESeq2 [41]. Multiple testing correction was conducted using Benjamini and Hochberg’s approach for controlling the False Discovery Rate (FDR).
RT-qPCR: Complementary DNA (cDNA) was created from RNA samples using the High-Capacity cDNA Reverse Transcription Kit with RNase inhibitor (Applied Biosystems, Waltham, MA, USA) using random primers. RT-qPCR was conducted using the same RNA as that submitted for RNA sequencing (see RNA Extraction). One microgram of RNA was used per reaction. Real-time quantitative PCR primer sequences are provided in Supplemental File S1 Table S5. Primers were analyzed using IDT’s OligoAnalyzer tool (Integrated DNA Technologies, Coralville, IA, USA) to ensure length, GC content, melting temp, hairpin tm, self-dimer and heterodimer parameters are well within range. All primers also checked using BLAT (UCSC Genome Browser, University of California Santa Cruz, Santa Cruz, CA, USA) to ensure specificity. Primer efficiency was tested for all primers to ensure genes of interest and housekeeping gene is comparable. SsoAdvanced SYBR Green Supermix (Bio-Rad, Hercules, CA, USA) was used for reactions according to the manufacturer’s instructions. An amount of 2 ng of cDNA was used per reaction. Reactions were run on a Bio-Rad CFX 96 instrument at 95 °C for 10 min followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. Results were analyzed in accordance with the 2−ddCt method [42]. Embryo samples were pooled (n = 20 embryos per pool) and adult samples were run as individuals. We conducted n = 3 biological replicates per run, with all samples in triplicate. All experiments were repeated at least twice by independent technicians.
2.5. Keratin Staining and Histology
Tissue staining was performed in accordance to the protocol outlined in [43]. The 6 wpf scoliotic and non-scoliotic fish were fixed with 10% neutral buffered formalin for 3 days at 4 °C. Fish were decalcified using 20% EDTA pH 8 for 10 days at room temperature using nutator. Fish were processed and embedded in paraffin using Tissue-Tek VIP 6 AI and Tissue-Tek TEC. The 6.5 μm sections were cut and dewaxed prior to staining with Mayer’s Hematoxylin, Phloxine B, Alcian Blue, and Orange G (Electron Microscopy Sciences, Hatfield, PA, USA, 26401-04, 26401-01, 26401-02, 26401-03). Slides were visualized using Zeiss Axio Scan. Z1 at 20× using brightfield.
3. Results
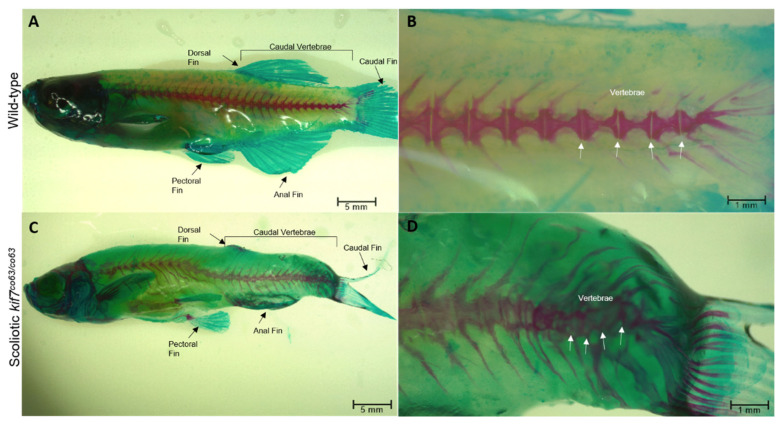
kif7co63/co63 zebrafish develop isolated spinal curvatures: Approximately 25% of kif7co63/co63 zebrafish developed isolated spinal curvatures by 6 wpf [33] (Figure 1). These zebrafish do not show obvious morphological defects within the brain, central canal, or skeleton other than the spinal curvatures [33].
Figure 1.
Approximately 25% of kif7co63/co63 zebrafish (D. rerio) show spinal curvatures, as indicated. The remaining zebrafish appear identical to wild type. Spinal curvature can be seen on the caudal part of the fish compared to wild type (A,C). Clear segmentation of vertebrae was observed in both wild-type (B) and scoliotic fish (D) as denoted by arrows.
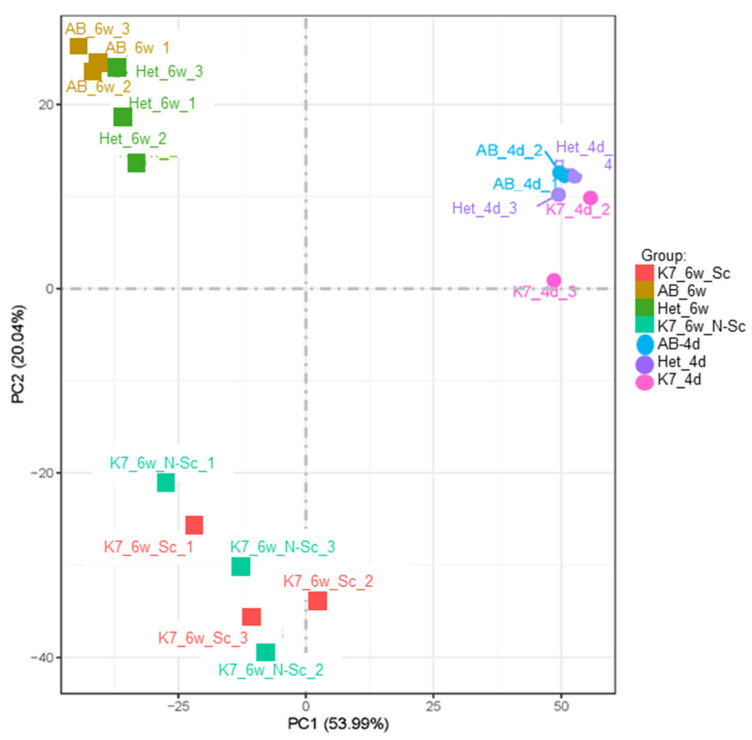
Transcriptomes cluster by sample age and genotype: To first determine overall transcriptomic changes occurring within scoliotic versus (vs.) non-scoliotic kif7co63/co63 zebrafish, we performed bulk RNA sequencing of kif7co63/co63, kif7co63/+ and wild-type (AB) zebrafish at 6 wpf, which is near the time of curvature onset (n = 3 individuals per phenotype). Additionally, we sequenced homozygous, heterozygous, and AB embryos (4 days past fertilization [dpf]; n = 3 embryo pools [20 embryos per pool] per genotype). Sequencing information is provided in Supplemental Table S1. The 6 wpf timepoint was selected for sequencing to correspond to just after the onset of spinal curvature, when scoliosis status is able to be distinguished between individuals [33]. The 4 dpf timepoint was selected due to the expression profile of kif7 (Supplemental Figure S1), which shows an increase in expression at day 5 in kif7co63/co63 embryos. At 4 dpf, embryos show a reduced expression of kif7 (0.545 fold change, 95% CI [0.450–0.660]) and are referred to as hypomorphic.
Principal component analysis (PCA) of the bulk RNA sequencing results revealed overall clustering of transcriptomes based on zebrafish age and genotype (Figure 2). Scoliotic and non-scoliotic homozygous kif7co63/co63 also clustered together on the PCA plot, suggesting that transcriptomes were globally similar when compared to other samples.
Figure 2.
Principal Component Analysis (PCA) plot of all sequenced samples. 6 wpf (6w) kif7co63/co63 scoliotic (K7_6w_S), non-scoliotic (K7_6w_N-Sc), heterozygous (Het_6w) and AB (AB_6w) zebrafish (D. rerio) are shown, along with AB, heterozygous, and kif7co63/co63 embryonic zebrafish (4 dpf [4d]).
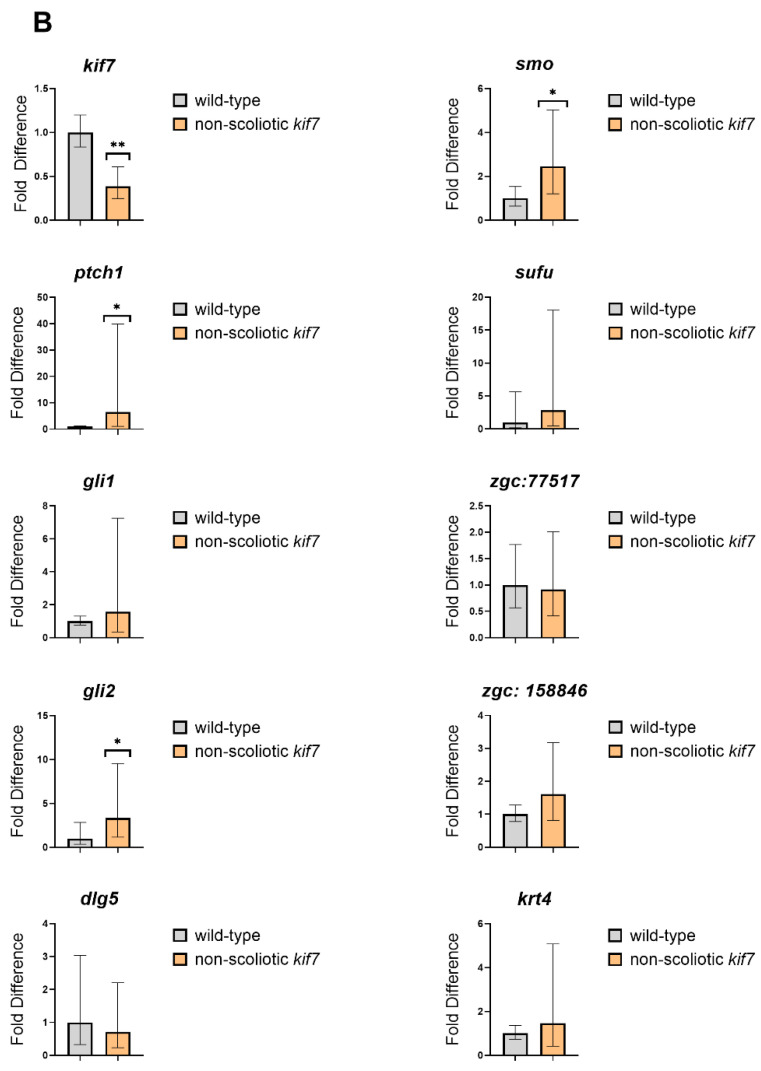
kif7co63/co63 zebrafish have reduced kif7 levels: As expected, kif7 mRNA was reduced in 6 wpf scoliotic kif7co63/co63 zebrafish compared to AB (0.5796 fold change, p = 5.76 × 10−11) (Supplemental Table S3). RT-qPCR confirmed this reduction in both 6 wpf scoliotic (0.392 fold change, 95% CI [0.314, 0.489]) and non-scoliotic zebrafish as compared to AB (0.387 fold change, 95% CI [0.247, 0.606] (Supplemental File S1, Table S7). No difference in kif7 was seen between scoliotic and non-scoliotic mutant zebrafish (1.102 fold change) by RNA sequencing or RT-qPCR. Kif7 mRNA was not detectable within 4 dpf embryos in bulk RNA sequencing; however, RT-qPCR results indicated variable kif7 expression in homozygous kif7co63/co63 embryos compared to AB embryos (0.542 fold change, 95% CI [0.142, 2.07]) (Supplemental File S1, Table S7).
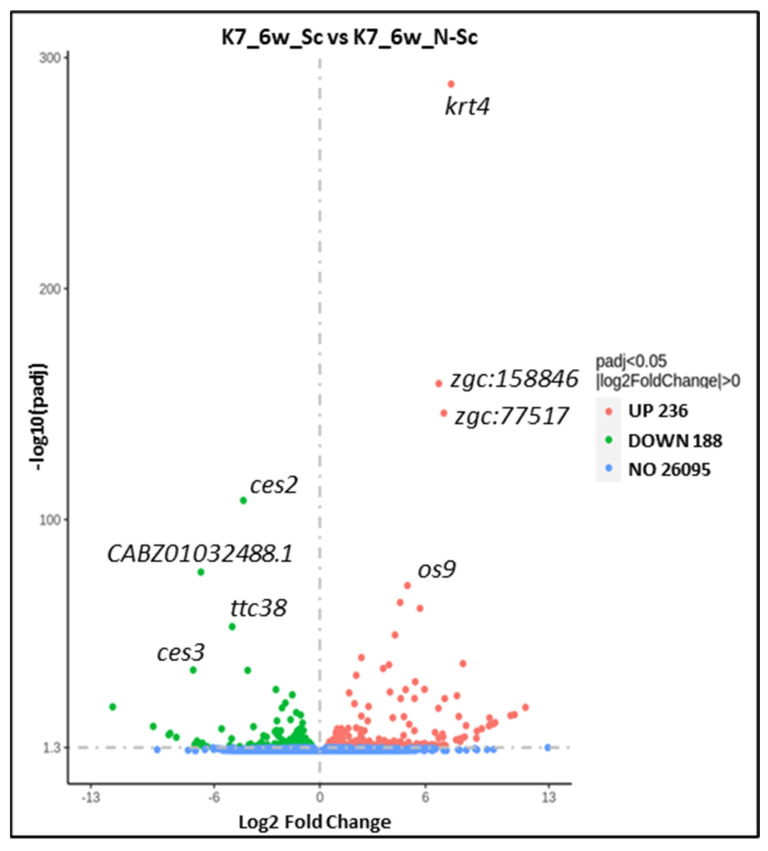
Cytoskeletal keratins are upregulated in scoliotic kif7co63/co63 zebrafish: Differential gene expression analysis showed that, between scoliotic and non-scoliotic kif7co63/co63 zebrafish, 188 genes were downregulated, and 236 transcripts were upregulated (padj < 0.05) (Supplemental Table S3). The top upregulated genes in scoliotic vs. non-scoliotic 6 wpf zebrafish were cytoskeletal keratins, including krt4 (174.85 fold change, p = 2.79 × 10−289) and zgc:158846, which is predicted to be orthologous to human KRT8 (Table 2, Figure 3); however, this upregulation of krt4 and zgc:158846 was not reflected in RT-qPCR data (Figure 4). Ces2 was the most significantly downregulated transcript (0.0494 fold change, p = 8.55 × 10−114). Gene expression analysis of upregulated transcripts showed an enrichment in 23 Gene Ontology (GO) and 16 InterPro terms, including “keratinization” (GO:0031424, p = 0.038) and “type II keratin” (IPR003054, 0.035). The most enriched terms among upregulated transcripts were “endoplasmic reticulum lumen” (GO:005788, p = 0.0018) and “somatomedin B domain” (IPR001212, p = 0.0039). Among downregulated transcripts, 28 GO and 17 InterPro terms were enriched, including the immune-specific GO terms “immunoglobulin complex, circulating”, “immunoglobulin receptor binding”, “positive regulation of B cell activation”, “complement activation, classical pathway”, “antigen binding”, and InterProt terms “Immunoglobulin-like fold”, “Immunoglobulin subtype”, “Immunoglobulin C1-set”, “Immunoglobulin V-set”, “Immunoglobulin-like domain”, and “Immunoglobulin I-set”.
Table 2.
Table of top 10 downregulated and top 10 upregulated transcripts (by p value) for genetically identical scoliotic vs. non-scoliotic 6 wpf kif7co63/co63 zebrafish (D. rerio). Corresponding zebrafish genes and predicted human gene orthologs are provided (via ZFIN). p adjusted indicates values after multiple testing correction.
| Fold Change | Log2 Fold Change | p Value | p Adjusted | Gene Name | Predicted Human Ortholog |
|---|---|---|---|---|---|
| 0.0494 | −4.34 | 8.55 × 10−113 | 5.20 × 10−109 | ces2 | CES2 |
| 0.0093 | −6.75 | 1.12 × 10−81 | 5.44 × 10−78 | CABZ01032488.1 | Unknown |
| 0.0317 | −4.98 | 8.66 × 10−58 | 2.34 × 10−54 | ttc38 | TTC38 |
| 0.0068 | −7.19 | 8.69 × 10−39 | 1.41 × 10−35 | ces3 | CES3 |
| 0.0583 | −4.10 | 1.23 × 10−38 | 1.88 × 10−35 | si:dkey-167k11.5 | CCDC134 |
| 0.1780 | −2.49 | 3.19 × 10−30 | 3.89 × 10−27 | pip4p1a | PIP4P1 |
| 0.3392 | −1.56 | 7.42 × 10−28 | 7.53 × 10−25 | ctsd | CTSD |
| 0.2588 | −1.95 | 2.89 × 10−24 | 2.43 × 10−21 | si:ch211-195h23.3 | CARD8 |
| 0.0003 | −11.76 | 1.63 × 10−22 | 1.24 × 10−19 | si:dkeyp-73d8.9 | Unknown |
| 0.2253 | −2.15 | 4.62 × 10−22 | 3.31 × 10−19 | lig1 | LIG1 |
| 174.85 | +7.45 | 1.14 × 10−293 | 2.79 × 10−289 | krt4 | KRT8 |
| 107.63 | +6.75 | 1.13 × 10−163 | 1.37 × 10−159 | zgc:158846 (krtt2c6) | KRT7,8,9 |
| 131.60 | +7.04 | 1.00 × 10−150 | 8.13 × 10−147 | zgc:77517 (krtt1c6) | KRT18 |
| 31.34 | +4.97 | 9.44 × 10−76 | 3.83 × 10−72 | os9 | OS9 |
| 23.59 | +4.56 | 2.27 × 10−68 | 7.90 × 10−65 | si:ch73-308m11.1 | GIMAP8 |
| 51.27 | +5.68 | 9.23 × 10−66 | 2.81 × 10−62 | si:ch211-11p18.6 | Unknown |
| 19.29 | +4.27 | 3.63 × 10−54 | 8.84 × 10−51 | CR774195.1 | Unknown |
| 5.13 | +2.36 | 2.49 × 10−44 | 5.52 × 10−41 | tmem97 | TMEM97 |
| 280.14 | +8.13 | 1.12 × 10−41 | 2.27 × 10−38 | myl6 | MYL6 |
| 15.14 | +3.92 | 4.14 × 10−41 | 7.77 × 10−38 | BX322618.1 | Unknown |
Figure 3.
Volcano plot of 6 wpf kif7co63/co63 scoliotic (S) vs. non-scoliotic (N-Sc) zebrafish (D. rerio). Each point represents a single transcript, and gene names are provided for the top four up- and downregulated transcripts.
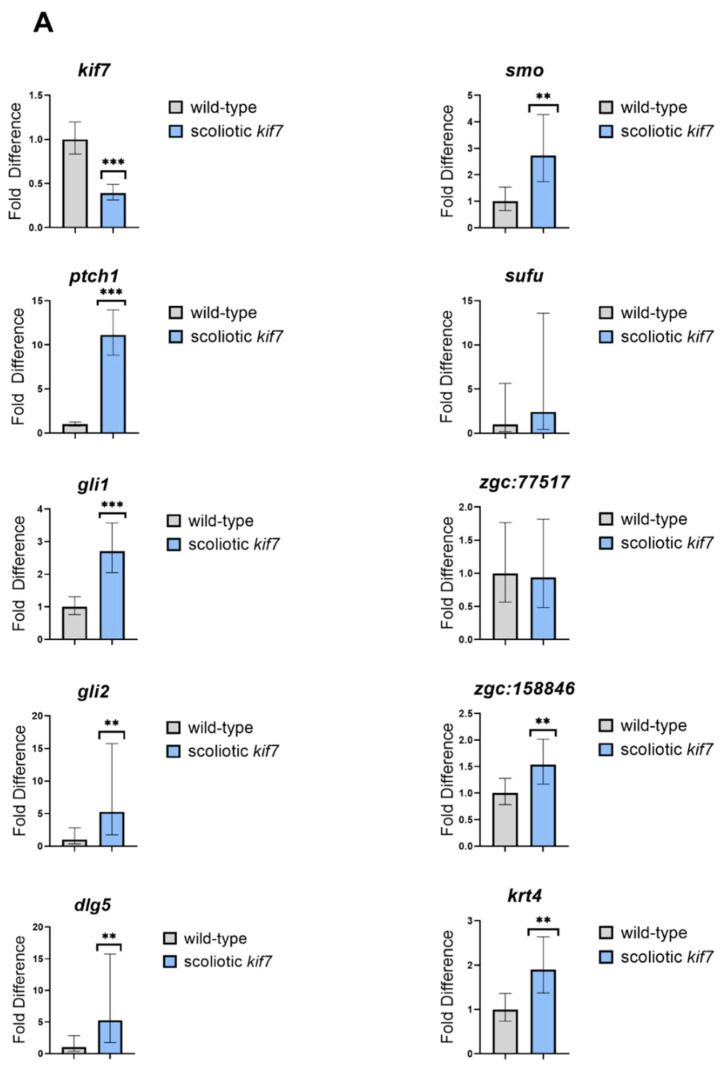
Figure 4.
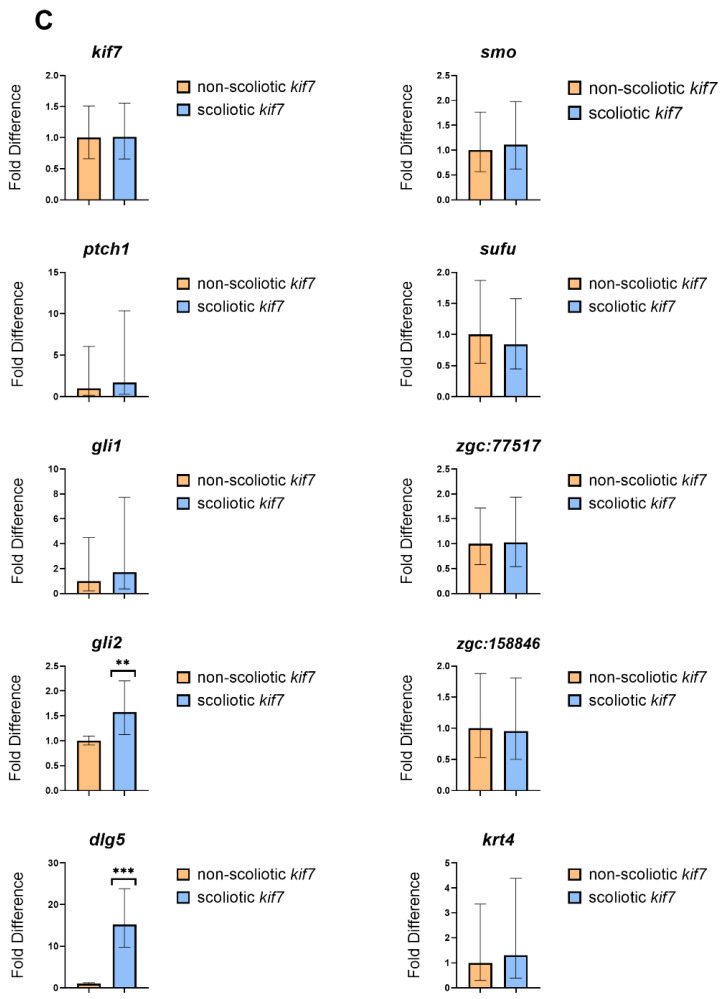
RT-qPCR of 6wpf zebrafish (D. rerio). Comparisons between wild-type and scoliotic kif7co63/co63 (A), wild-type and non-scoliotic kif7co63/co63 (B), and scoliotic kif7co63/co63 and non-scoliotic kif7co63/co63 (C) was performed for genes in the shh signaling pathway and the keratin genes observed in the RNA sequencing data. Error bars represent 95% confidence intervals. * indicates p < 0.05, ** indicates p < 0.01, and *** indicates p < 0.001.
Sonic hedgehog signaling (Shh) genes are largely unchanged in kif7co63/co63 zebrafish: Kif7 is known to function both within sonic hedgehog signaling (Shh) pathway and as an organizer of the axonemal tip of the primary cilia compartment [34]. Common transcriptional targets of hedgehog signaling include ptch1 and gli1 [44,45], which can serve as indicators of Shh pathway activation. We observed that ptch1 transcripts were elevated in both 4 dpf embryos (9.816 fold change, p = 1.16 × 10−5) and 6 wpf scoliotic zebrafish (50.96 fold change, p = 4.02 × 10−35) compared to AB. Gli1 was slightly upregulated in 6 wpf scoliotic kif7co63/co63 zebrafish compared to AB (1.461 fold change, p =1.05 × 10−5). Additional RT-qPCR data for Shh pathway genes are provided in Supplemental Table S4. Cilia genes as listed in the SYSCILIA database [46] appear to be largely unchanged in kif7co63/co63 zebrafish as compared to AB (Supplemental Table S4. Although we did not obtain usable data for Shh genes in scoliotic vs. non-scoliotic kif7co63/co63 zebrafish through RNA sequencing, RT-qPCR results suggested that there was little difference in these genes (Figure 4, Supplemental Table S6), with the exception of dlg5a (15.225 fold change, 95% CI [9.727, 23.830]) and gli2 (1.574 fold change, 95% CI [1.123–2.206]).
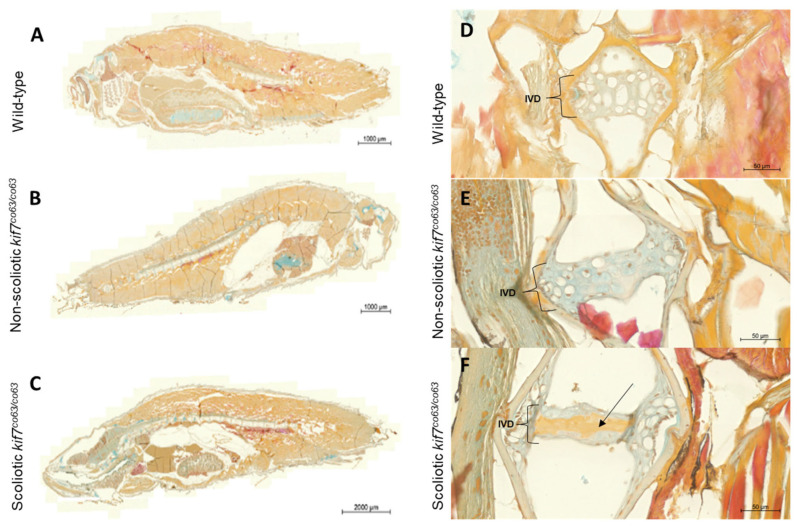
Pankeratin staining confirms an upregulation of keratins in scoliotic kif7co63/co63 zebrafish: To confirm an enrichment of keratins in scoliotic kif7co63/co63 zebrafish compared to genotype-matched kif7co63/co63 non-scoliotic zebrafish, we performed pankeratin staining of 6 wpf kif7co63/co63 zebrafish. Scoliotic zebrafish displayed increased levels of keratins and prekeratins across muscle tissues in whole mount sections (Figure 5). Additionally, scoliotic zebrafish displayed increased keratins within the intervertebral disc (IVD).
Figure 5.
Keratin staining of wild-type (A,D), non-scoliotic (B,E), and scoliotic kif7co63/co63 (C,F) 6 wpf zebrafish (D. rerio). Keratin staining of whole cross section fish (A–C) and selective caudal vertebrae (D–F) depicting the intervertebral disk (IVD) are shown. The IVD is distinguished by vacuolated cells clearly in the wild-type and non-scoliotic kif7co63/co63 fish, but vacuolated cells are less visible in scoliotic kif7co63/co63. Blue: glycosaminoglycans; orange to red: prekeratin to keratin; brown: nuclei. Increased keratin/prekeratin can be seen in the IVD of scoliotic fish compared to non-scoliotic fish and wild type, as denoted by the black arrow. N = 3 zebrafish were analyzed for each group (wild type, scoliotic kif7co63/co63, and non-scoliotic kif7co63/co63).
4. Discussion
In this work, we build on previous findings in which human data supported by a unique zebrafish animal model suggest that mutations in KIF7 may contribute to the IS phenotype [33]. Exploration of our unique zebrafish model, kif7co63/co63, which develops spinal curvatures during the juvenile period, supports the role of distinct molecular mechanisms which underlie the pathology of the scoliosis phenotype. Our results continue to support the role of hypomorphic mutations in KIF7 as contributory to IS pathogenesis.
As expected, kif7 mRNA was reduced in both scoliotic and non-scoliotic kif7co63 zebrafish. Overall, zebrafish transcriptomes clustered primarily by sample age and genotype. Regardless of scoliotic phenotype, homozygous kif7co63/co63 zebrafish clustered together on the PCA plot (Figure 1), indicating that the transcriptomes of these zebrafish were overall similar.
Kif7 as a ciliary kinesin has dual roles within the cell; one, in the sonic hedgehog (Shh) signaling pathway and, second, as a ciliary regulator [34]. Kif7 is broadly expressed in human [47] and zebrafish [48]. In mouse, Kif7 functions as both a negative and positive regulator of the Shh pathway within the primary cilium downstream of Smoothened (Smo) and upstream of Gli transcription factors [49]. Ptch1, which encodes Patched, the primary receptor for Shh, is noted to be upregulated in 4 day (9.816 fold change, p = 1.16 × 10−05) and 6 wpf scoliotic kif7co63/co63 zebrafish (50.96 fold change, p = 4.02 × 10−35) as compared to age-matched AB zebrafish. Although we did not obtain usable bulk RNAseq data for Shh genes in scoliotic vs. non-scoliotic kif7co63/co63 zebrafish, RT-qPCR data showed little difference in Shh genes in 6 wpf scoliotic vs. non-scoliotic kif7co63/co63 zebrafish, with the exception of dlg5a (15.225-fold change, 95% CI [9.727, 23.830]). Dlg5a binds directly to Kif7 [50], and was observed as upregulated in kif7co63/co63 embryos over controls within our previous study [33]. Dlg5 is also a regulator of Gli1 protein ubiquitination and degradation and, most recently, is recognized as a major factor of Shh signaling, particularly in relation to glioblastoma tumors, reflecting its known role in critical processes involving cell–cell adhesion during neural development [51,52]. Although we cannot rule out the possibility that Shh signaling plays a tissue-specific role in spine morphogenesis in kif7co63/co63 zebrafish, as Shh target genes ptch1 and gli1-2 were largely unchanged between scoliotic and non-scoliotic mutants, the upregulation of dlg5 could represent alternative functional mechanisms of cellular interactions.
We observed striking differences in several other transcripts between scoliotic and non-scoliotic kif7co63/co63 zebrafish, notably a large upregulation in the zebrafish cytoskeletal keratins krt4 and zgc:158846 (krtt2c6), which are orthologous to human KRT8 (fold change= 174.85, p = 2.79 × 10−289) and KRT7, −8 and −9 (fold change = 107.63, p = 1.37 × 10−159). This upregulation of zebrafish keratin krt4 was too variable between specimens to draw a definitive conclusion by RT-qPCR, which may have been due to a difference in the methodologies used [53]. However, we also observed increased levels of keratins and prekeratin proteins in the dermis, musculature, and intervertebral disc (IVD) of scoliotic kif7co63/co63 zebrafish as compared to non-scoliotic genotype- and age-matched mutant zebrafish. These key transcriptomic differences may arise spontaneously and contribute to the development of spinal curvatures in scoliotic mutants, or they may be a secondary effect that develops after the initiation of spinal curvatures.
It is unknown specifically how kif7 mutations may lead to increased expression of keratins, a known product of keratinocytes, in hypomorphic kif7co63/co63 zebrafish. Due to the role of Shh signaling in the pathogenesis of basal cell carcinoma of the skin, the role of Kif7 has been studied in keratinocytes. This work has been primarily performed in the mouse as the model system; however, studies within the zebrafish have shown similarities of epidermal ontogeny [54,55]. In keratinocytes, Kif7 has been shown to have a crucial role as a positive regulator of Shh signaling through its regulatory function of the Gli transcription factors [56]. In embryonic keratinocytes, inactivation of both Kif7 and Sufu, an additional regulator of Gli transcription factors, leads to a loss of epidermal differentiation and follicular fate, while in the adult this loss leads to the induction of basal cell carcinoma [57]. As seen in histological sectioning (Figure 4), our hypomorphic kif7co63/co63 zebrafish with scoliosis exhibited significant upregulation of keratin expression overall. The identification of the specific mechanism as to what drives this strong expression of keratin within the muscle and epithelium of the hypomorphic scoliotic kif7co63/co63 zebrafish remains to be determined.
As stated, the specific human orthologues of the top differentially expressed genes within our expression data are KRT7 and KRT8. KRT7 and KRT8 encode keratin-7 and keratin-8, respectively, and are members of the type II keratin family that are broadly expressed and form intermediate filaments in the cytoplasm of epithelial cells. KRT7 expression has a role in epithelial–mesenchymal transition and its dysregulation has been highly associated with various types of cancers and tumor progression [58,59,60]. Cytokeratins of the intermediate filament subgroup, which includes KRT8, have been observed in adult mouse skeletal muscle [61]. Keratin-8, in addition to keratin-19, links the contractile apparatus of striated muscle to dystrophin [62]. More recently, KRT8, KRT18 and KRT19, all recognized markers of notochordal cells, have been found to be expressed in a subpopulation of adult nucleus pulposus (NP) cells. NP cell degeneration is associated with intervertebral disc degeneration (IVDD), a prominent health problem worldwide [63,64]. The finding of these cellular markers within the NP has allowed for study of the intervertebral disc (IVD) and its degeneration in both zebrafish and humans [65]. The role of keratins within the IVD as a potential molecular mechanism associated with the onset of scoliosis warrants further study. Future explorations will focus on isolating the tissue or cell type driving this expression pattern.
Ces2 and TTC38 were significantly downregulated within the scoliotic homozygous kif7co63/co63 zebrafish when compared to non-scoliotic homozygous kif7co63/co63 fish. Both genes are related to metabolic abnormalities through enzymatic reactions of various drugs and endogenous compounds (via NCBI RefSeq [66]), to which there is no known relationship to axial skeletal development.
Collectively, we present transcriptomic data that associate an upregulation of cytokeratins with the phenotype of scoliosis within a kif7co63/co63 zebrafish animal model. This comparative transcriptomics analysis has significance in the determination of unique differentially expressed genes as related to the scoliosis phenotype. This initial step, when followed with gene-set-enrichment analyses and orthology mapping, has the potential to reveal significant associations with biological relevance to the observed phenotype of idiopathic scoliosis.
Acknowledgments
The authors thank Ryan Gray (University of Texas at Austin) for his helpful discussions and advice, Christine Archer for her helpful zebrafish husbandry support, Terrin Geohring (Tissue and Molecular Imaging Core, Colorado Program for Musculoskeletal Research, University of Colorado Anschutz) for assistance with zebrafish staining and imaging, and Bruce Appel for his helpful discussions regarding zebrafish histology and results.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/genes14051058/s1, File S1: Supplementary genotyping and RNA sequencing data; Table S1 and Table S3: Differentially Expressed Genes; Table S4: SHH and Cilia Genes.
Author Contributions
Conceptualization, N.H.M. and E.T.; methodology, M.C., C.W., A.M., R.N., M.J. and D.G.; validation, M.C. and C.W.; formal analysis, E.T. and M.C.; original draft preparation, E.T. and M.C.; writing—review and editing, E.T., M.C. and N.H.M.; supervision, N.H.M.; project administration, A.M.; resources, N.H.M., funding acquisition, N.H.M. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
The animal study protocol was approved by the Institutional Animal Care and Use Committee Review Board of the University of Colorado Anschutz (protocol #00370).
Informed Consent Statement
Not applicable.
Data Availability Statement
The data necessary for confirming the conclusions of this article are represented fully within the article and its supplementary files, including the complete lists of differentially expressed genes for all RNA sequencing samples (Supplemental Table S3). Raw data files are available through NCBI Gene Expression Omnibus (GEO), accession number: GSE231606.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research was generously funded by the Scoliosis Research Society SRS-Cotrel Basic Science Research Grant, the National Institutes of Health R01-AR068292, Children’s Hospital Colorado and the University of Colorado Anschutz Department of Orthopedics.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Miller N.H., Schwab D.L., Sponseller P.D., Manolio T.A., Pugh E.W., Wilson A.P. Characterization of Idiopathic Scoliosis in a Clinically Well-Defined Population. Clin. Orthop. Relat. Res. 2001;392:349–357. doi: 10.1097/00003086-200111000-00045. [DOI] [PubMed] [Google Scholar]
- 2.Roach J.W. Adolescent idiopathic scoliosis. Orthop. Clin. N. Am. 1999;30:353–365. doi: 10.1016/S0030-5898(05)70092-4. [DOI] [PubMed] [Google Scholar]
- 3.Asher M.A., Burton D.C. Adolescent idiopathic scoliosis: Natural history and long term treatment effects. Scoliosis. 2006;1:2. doi: 10.1186/1748-7161-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng J.C.Y., Qin L., Cheung C.S.K., Sher A.H.L., Lee K.M., Ng S.W.E., Guo X. Generalized Low Areal and Volumetric Bone Mineral Density in Adolescent Idiopathic Scoliosis. J. Bone Miner. Res. 2000;15:1587–1595. doi: 10.1359/jbmr.2000.15.8.1587. [DOI] [PubMed] [Google Scholar]
- 5.Cheung C.S.K., Lee W.T.K., Tse Y.K., Lee K.M., Guo X., Qin L., Cheng J.C.Y. Generalized osteopenia in adolescent idiopathic scoliosis—Association with abnormal pubertal growth, bone turnover, and calcium intake? Spine. 2006;31:330–338. doi: 10.1097/01.brs.0000197410.92525.10. [DOI] [PubMed] [Google Scholar]
- 6.Guo X., Chau W.W., Chan Y.L., Cheng J.Y. Relative anterior spinal overgrowth in adolescent idiopathic scoliosis: Results of disproportionate endochondral-membranous bone growth. J. Bone Jt. Surg. Br. 2003;85:1026–1031. doi: 10.1302/0301-620X.85B7.14046. [DOI] [PubMed] [Google Scholar]
- 7.Man G.C.-W., Wong J.H., Wang W.W.-J., Sun G.-Q., Yeung B.H.-Y., Ng T.-B., Lee S.K.-M., Ng B.K.-W., Qiu Y., Cheng J.C.-Y. Abnormal melatonin receptor 1B expression in osteoblasts from girls with adolescent idiopathic scoliosis. J. Pineal Res. 2011;50:395–402. doi: 10.1111/j.1600-079X.2011.00857.x. [DOI] [PubMed] [Google Scholar]
- 8.Blanco G., Coulton G.R., Biggin A., Grainge C., Moss J., Barrett M., Berquin A., Maréchal G., Skynner M., Van Mier P., et al. The kyphoscoliosis (ky) mouse is deficient in hypertrophic responses and is caused by a mutation in a novel muscle-specific protein. Hum. Mol. Genet. 2001;10:9–16. doi: 10.1093/hmg/10.1.9. [DOI] [PubMed] [Google Scholar]
- 9.Latalski M., Danielewicz-Bromberek A., Fatyga M., Latalska M., Kröber M., Zwolak P. Current insights into the aetiology of adolescent idiopathic scoliosis. Arch. Orthop. Trauma Surg. 2017;137:1327–1333. doi: 10.1007/s00402-017-2756-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yagci G., Ayhan C., Yakut Y. Effectiveness of basic body awareness therapy in adolescents with idiopathic scoliosis: A randomized controlled study1. J. Back Musculoskelet. Rehabil. 2018;31:693–701. doi: 10.3233/BMR-170868. [DOI] [PubMed] [Google Scholar]
- 11.Blecher R., Krief S., Galili T., Biton I.E., Stern T., Assaraf E., Levanon D., Appel E., Anekstein Y., Agar G., et al. The Proprioceptive System Masterminds Spinal Alignment: Insight into the Mechanism of Scoliosis. Dev. Cell. 2017;42:388–399.e3. doi: 10.1016/j.devcel.2017.07.022. [DOI] [PubMed] [Google Scholar]
- 12.Haumont T., Gauchard G.C., Lascombes P., Perrin P.P. Postural Instability in Early-Stage Idiopathic Scoliosis in Adolescent Girls. Spine. 2011;36:E847–E854. doi: 10.1097/BRS.0b013e3181ff5837. [DOI] [PubMed] [Google Scholar]
- 13.Missaoui B., Portero P., Bendaya S., Hanktie O., Thoumie P. Posture and equilibrium in orthopedic and rheumatologic diseases. Neurophysiol. Clin. 2008;38:447–457. doi: 10.1016/j.neucli.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 14.Burwell R.G., Aujla R.K., Grevitt M.P., Dangerfield P.H., Moulton A., Randell T.L., Anderson S.I. Pathogenesis of adolescent idiopathic scoliosis in girls—A double neuro-osseous theory involving disharmony between two nervous systems, somatic and autonomic expressed in the spine and trunk: Possible dependency on sympathetic nervous system and hormones with implications for medical therapy. Scoliosis. 2009;4:24. doi: 10.1186/1748-7161-4-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herman R., MacEwen G.D. Idiopathic scoliosis; A visio-vestibular disorder of the central nervous system. In: Zorab P.A., Siegler D., Cardiothoracic Institute (London, England), editors. Scoliosis, 1979. Volume xvi. Academic Press; London, UK: New York, NY, USA: 1980. 289p [Google Scholar]
- 16.Hadley-Miller N., Mims B., Milewicz D.M. The potential role of the elastic fiber system in adolescent idiopathic scoliosis. J. Bone Jt. Surg. 1994;76:1193–1206. doi: 10.2106/00004623-199408000-00010. [DOI] [PubMed] [Google Scholar]
- 17.Clough M., Justice C.M., Marosy B., Miller N.H. Males with familial idiopathic scoliosis: A distinct phenotypic subgroup. Spine. 2010;35:162–168. doi: 10.1097/BRS.0b013e3181b7f1a7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dunwoodie S.L., Kusumi K. The Genetics and Development of Scoliosis. Volume XII. Springer International Publishing; Cham, Switzerland: 2018. 199p [Google Scholar]
- 19.Edery P., Margaritte-Jeannin P., Biot B., Labalme A., Bernard J.-C., Chastang J., Kassai B., Plais M.-H., Moldovan F., Clerget-Darpoux F. New disease gene location and high genetic heterogeneity in idiopathic scoliosis. Eur. J. Hum. Genet. 2011;19:865–869. doi: 10.1038/ejhg.2011.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Justice C.M., Bishop K., Carrington B., Mullikin J.C., Swindle K., Marosy B., Sood R., Miller N.H., Wilson A.F. Evaluation of IRX Genes and Conserved Noncoding Elements in a Region on 5p13.3 Linked to Families with Familial Idiopathic Scoliosis and Kyphosis. G3. 2016;6:1707–1712. doi: 10.1534/g3.116.029975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boswell C.W., Ciruna B. Understanding Idiopathic Scoliosis: A New Zebrafish School of Thought. Trends Genet. 2017;33:183–196. doi: 10.1016/j.tig.2017.01.001. [DOI] [PubMed] [Google Scholar]
- 22.Hayes M., Gao X., Yu L.X., Paria N., Henkelman R.M., Wise C.A., Ciruna B. ptk7 mutant zebrafish models of congenital and idiopathic scoliosis implicate dysregulated Wnt signalling in disease. Nat. Commun. 2014;5:4777. doi: 10.1038/ncomms5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grimes D.T., Boswell C.W., Morante N.F.C., Henkelman R.M., Burdine R.D., Ciruna B. Zebrafish models of idiopathic scoliosis link cerebrospinal fluid flow defects to spine curvature. Science. 2016;352:1341–1344. doi: 10.1126/science.aaf6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patten S., Margaritte-Jeannin P., Bernard J.-C., Alix E., Labalme A., Besson A., Girard S., Fendri K., Fraisse N., Biot B., et al. Functional variants of POC5 identified in patients with idiopathic scoliosis. J. Clin. Investig. 2015;125:1124–1128. doi: 10.1172/JCI77262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buchan J.G., Gray R.S., Gansner J.M., Alvarado D.M., Burgert L., Gitlin J.D., Gurnett C.A., Goldsmith M.I. Kinesin family member 6 (kif6) is necessary for spine development in zebrafish. Dev. Dyn. 2014;243:1646–1657. doi: 10.1002/dvdy.24208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cantaut-Belarif Y., Sternberg J.R., Thouvenin O., Wyart C., Bardet P.-L. The Reissner Fiber in the Cerebrospinal Fluid Controls Morphogenesis of the Body Axis. Curr. Biol. 2018;28:2479–2486.e4. doi: 10.1016/j.cub.2018.05.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Troutwine B.R., Gontarz P., Konjikusic M.J., Minowa R., Monstad-Rios A., Sepich D.S., Kwon R.Y., Solnica-Krezel L., Gray R.S. The Reissner Fiber Is Highly Dynamic In Vivo and Controls Morphogenesis of the Spine. Curr. Biol. 2020;30:2353–2362.e3. doi: 10.1016/j.cub.2020.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rose C.D., Pompili D., Henke K., Van Gennip J.L., Meyer-Miner A., Rana R., Gobron S., Harris M.P., Nitz M., Ciruna B. SCO-Spondin Defects and Neuroinflammation Are Conserved Mechanisms Driving Spinal Deformity across Genetic Models of Idiopathic Scoliosis. Curr. Biol. 2020;30:2363–2373.e6. doi: 10.1016/j.cub.2020.04.020. [DOI] [PubMed] [Google Scholar]
- 29.Catanzariti J.F., D’hulster-Hocquet L., Le Berre M., Delaplace C., Boukelifa M., Thevenon A. Adolescent idiopathic scoliosis (AIS), cerebrospinal fluid (CSF) flow and ciliopathy. Ann. Phys. Rehabil. Med. 2017;60:e80–e81. doi: 10.1016/j.rehab.2017.07.221. [DOI] [Google Scholar]
- 30.Sternberg J.R., Prendergast A.E., Brosse L., Cantaut-Belarif Y., Thouvenin O., Orts-Del’immagine A., Castillo L., Djenoune L., Kurisu S., McDearmid J.R., et al. Pkd2l1 is required for mechanoception in cerebrospinal fluid-contacting neurons and maintenance of spine curvature. Nat. Commun. 2018;9:3804. doi: 10.1038/s41467-018-06225-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Gennip J.L.M., Boswell C.W., Ciruna B. Neuroinflammatory signals drive spinal curve formation in zebrafish models of idiopathic scoliosis. Sci. Adv. 2018;4:eaav1781. doi: 10.1126/sciadv.aav1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang X., Jia S., Chen Z., Chong Y.L., Xie H., Feng D., Wu X., Song D.Z., Roy S., Zhao C. Cilia-driven cerebrospinal fluid flow directs expression of urotensin neuropeptides to straighten the vertebrate body axis. Nat. Genet. 2018;50:1666–1673. doi: 10.1038/s41588-018-0260-3. [DOI] [PubMed] [Google Scholar]
- 33.Terhune E.A., Cuevas M.T., Monley A.M., Wethey C.I., Chen X., Cattell M.V., Bayrak M.N., Bland M.R., Sutphin B., Trahan G.D., et al. Mutations in KIF7 implicated in idiopathic scoliosis in humans and axial curvatures in zebrafish. Hum. Mutat. 2020;42:392–407. doi: 10.1002/humu.24162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He M., Subramanian R., Bangs F., Omelchenko T., Liem K.F., Jr., Kapoor T.M., Anderson K.V. The kinesin-4 protein Kif7 regulates mammalian Hedgehog signalling by organizing the cilium tip compartment. Nat. Cell Biol. 2014;16:663–672. doi: 10.1038/ncb2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rajesh D., Dahia C.L. Role of Sonic Hedgehog Signaling Pathway in Intervertebral Disc Formation and Maintenance. Curr. Mol. Biol. Rep. 2018;4:173–179. doi: 10.1007/s40610-018-0107-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maurya A.K., Ben J., Zhao Z., Lee R.T.H., Niah W., Ng A.S.M., Iyu A., Yu W., Elworthy S., van Eeden F.J.M., et al. Positive and Negative Regulation of Gli Activity by Kif7 in the Zebrafish Embryo. PLoS Genet. 2013;9:e1003955. doi: 10.1371/journal.pgen.1003955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brancati F., Dallapiccola B., Valente E.M. Joubert Syndrome and related disorders. Orphanet J. Rare Dis. 2010;5:20. doi: 10.1186/1750-1172-5-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bachmann-Gagescu R., Dempsey J.C., Phelps I.G., O’Roak B.J., Knutzen D.M., Rue T.C., Ishak G.E., Isabella C.R., Gorden N., Adkins J., et al. Joubert syndrome: A model for untangling recessive disorders with extreme genetic heterogeneity. J. Med. Genet. 2015;52:514–522. doi: 10.1136/jmedgenet-2015-103087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Elhassanien A.F., Alghaiaty H.A.-A. Joubert syndrome: Clinical and radiological characteristics of nine patients. Ann. Indian Acad. Neurol. 2013;16:239–244. doi: 10.4103/0972-2327.112480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haeussler M., Schönig K., Eckert H., Eschstruth A., Mianné J., Renaud J.-B., Schneider-Maunoury S., Shkumatava A., Teboul L., Kent J., et al. Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 2016;17:148. doi: 10.1186/s13059-016-1012-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmittgen T.D., Livak K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 43.Dane E.T., Herman D.L. Haematoxylin-Phloxine-Alcian Blue-Orange G Differential Staining of Prekeratin, Keratin and Mucin. Stain. Technol. 1963;38:97–101. doi: 10.3109/10520296309061171. [DOI] [PubMed] [Google Scholar]
- 44.Cohen M., Kicheva A., Ribeiro A., Blassberg R., Page K.M., Barnes C.P., Briscoe J. Ptch1 and Gli regulate Shh signalling dynamics via multiple mechanisms. Nat. Commun. 2015;6:6709. doi: 10.1038/ncomms7709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sasai N., Toriyama M., Kondo T. Hedgehog Signal and Genetic Disorders. Front. Genet. 2019;10:1103. doi: 10.3389/fgene.2019.01103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Van Dam T.J., Wheway G., Slaats G.G., Huynen M.A., Giles R.H. The SYSCILIA gold standard (SCGSv1) of known ciliary components and its applications within a systems biology consortium. Cilia. 2013;2:7. doi: 10.1186/2046-2530-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.The Genotype-Tissue Expression (GTEx) Project KIF7. [(accessed on 21 April 2023)]. Available online: https://www.gtexportal.org/home/gene/KIF7.
- 48.Tay S.Y., Ingham P.W., Roy S. A homologue of the Drosophila kinesin-like protein Costal2 regulates Hedgehog signal transduction in the vertebrate embryo. Development. 2005;132:625–634. doi: 10.1242/dev.01606. [DOI] [PubMed] [Google Scholar]
- 49.Liem K.F., Jr., He M., Ocbina P.J.R., Anderson K.V. Mouse Kif7/Costal2 is a cilia-associated protein that regulates Sonic hedgehog signaling. Proc. Natl. Acad. Sci. USA. 2009;106:13377–13382. doi: 10.1073/pnas.0906944106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chong Y.C., Mann R.K., Zhao C., Kato M., Beachy P.A. Bifurcating action of Smoothened in Hedgehog signaling is mediated by Dlg5. Genes Dev. 2015;29:262–276. doi: 10.1101/gad.252676.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang S.-H.J., Celic I., Choi S.-Y., Riccomagno M., Wang Q., Sun L.O., Mitchell S.P., Vasioukhin V., Huganir R.L., Kolodkin A.L. Dlg5 regulates dendritic spine formation and synaptogenesis by controlling subcellular N-cadherin localization. J. Neurosci. 2014;34:12745–12761. doi: 10.1523/JNEUROSCI.1280-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kundu S., Nandhu M.S., Longo S.L., Longo J.A., Rai S., Chin L.S., E Richardson T., Viapiano M.S. The scaffolding protein DLG5 promotes glioblastoma growth by controlling Sonic Hedgehog signaling in tumor stem cells. Neuro-Oncol. 2022;24:1230–1242. doi: 10.1093/neuonc/noac001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Everaert C., Luypaert M., Maag J.L., Cheng Q.X., Dinger M.E., Hellemans J., Mestdagh P. Benchmarking of RNA-sequencing analysis workflows using whole-transcriptome RT-qPCR expression data. Sci. Rep. 2017;7:1559. doi: 10.1038/s41598-017-01617-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chan K.Y., Yan C.-C.S., Roan H.-Y., Hsu S.-C., Tseng T.-L., Hsiao C.-D., Hsu C.-P., Chen C.-H. Skin cells undergo asynthetic fission to expand body surfaces in zebrafish. Nature. 2022;605:119–125. doi: 10.1038/s41586-022-04641-0. [DOI] [PubMed] [Google Scholar]
- 55.Hou R., Kim J.E., Plikus M.V. Keratinocytes cut corners on the cell cycle for the sake of skin barrier integrity. Dev. Cell. 2022;57:1437–1438. doi: 10.1016/j.devcel.2022.06.001. [DOI] [PubMed] [Google Scholar]
- 56.Lee R.T.H., Asharani P.V., Carney T.J. Basal Keratinocytes Contribute to All Strata of the Adult Zebrafish Epidermis. PLoS ONE. 2014;9:e84858. doi: 10.1371/journal.pone.0084858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li Z.J., Nieuwenhuis E., Nien W., Zhang X., Zhang J., Puviindran V., Wainwright B., Kim P.C.W., Hui C.-C. Kif7 regulates Gli2 through Sufu-dependent and -independent functions during skin development and tumorigenesis. Development. 2012;139:4152–4161. doi: 10.1242/dev.081190. [DOI] [PubMed] [Google Scholar]
- 58.Chen S., Su T., Zhang Y., Lee A., He J., Ge Q., Wang L., Si J., Zhuo W., Wang L. Fusobacterium nucleatum promotes colorectal cancer metastasis by modulating KRT7-AS/KRT7. Gut Microbes. 2020;11:511–525. doi: 10.1080/19490976.2019.1695494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.An Q., Liu T., Wang M.Y., Yang Y.J., Zhang Z.D., Liu Z.J., Yang B. KRT7 promotes epithelial-mesenchymal transition in ovarian cancer via the TGF-beta/Smad2/3 signaling pathway. Oncol. Rep. 2021;45:481–492. doi: 10.3892/or.2020.7886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen F., Chen Z., Guan T., Zhou Y., Ge L., Zhang H., Wu Y., Jiang G.-M., He W., Li J., et al. N6-Methyladenosine Regulates mRNA Stability and Translation Efficiency of KRT7 to Promote Breast Cancer Lung Metastasis. Cancer Res. 2021;81:2847–2860. doi: 10.1158/0008-5472.CAN-20-3779. [DOI] [PubMed] [Google Scholar]
- 61.O’Neill A., Williams M.W., Resneck W.G., Milner D.J., Capetanaki Y., Bloch R.J. Sarcolemmal Organization in Skeletal Muscle Lacking Desmin: Evidence for Cytokeratins Associated with the Membrane Skeleton at Costameres. Mol. Biol. Cell. 2002;13:2347–2359. doi: 10.1091/mbc.01-12-0576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stone M.R., O’Neill A., Catino D., Bloch R.J. Specific Interaction of the Actin-binding Domain of Dystrophin with Intermediate Filaments Containing Keratin 19. Mol. Biol. Cell. 2005;16:4280–4293. doi: 10.1091/mbc.e05-02-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Richardson S.M., Ludwinski F.E., Gnanalingham K.K., Atkinson R.A., Freemont A.J., Hoyland J.A. Notochordal and nucleus pulposus marker expression is maintained by sub-populations of adult human nucleus pulposus cells through aging and degeneration. Sci. Rep. 2017;7:1501. doi: 10.1038/s41598-017-01567-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mohanty S., Pinelli R., Dahia C.L. Characterization of Krt19 (CreERT) allele for targeting the nucleus pulposus cells in the postnatal mouse intervertebral disc. J. Cell. Physiol. 2020;235:128–140. doi: 10.1002/jcp.28952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kague E., Turci F., Newman E., Yang Y., Brown K.R., Aglan M.S., Otaify G.A., Temtamy S.A., Ruiz-Perez V.L., Cross S., et al. 3D assessment of intervertebral disc degeneration in zebrafish identifies changes in bone density that prime disc disease. Bone Res. 2021;9:39. doi: 10.1038/s41413-021-00156-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.O’Leary N.A., Wright M.W., Brister J.R., Ciufo S., Haddad D., McVeigh R., Rajput B., Robbertse B., Smith-White B., Ako-Adjei D., et al. Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44:D733–D745. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data necessary for confirming the conclusions of this article are represented fully within the article and its supplementary files, including the complete lists of differentially expressed genes for all RNA sequencing samples (Supplemental Table S3). Raw data files are available through NCBI Gene Expression Omnibus (GEO), accession number: GSE231606.