Abstract
Retinoblastoma (RB) is a childhood cancer that forms in the developing retina of young children; this tumor cannot be biopsied due to the risk of provoking extraocular tumor spread, which dramatically alters the treatment and survival of the patient. Recently, aqueous humor (AH), the clear fluid in the anterior chamber of the eye, has been developed as an organ-specific liquid biopsy for investigation of in vivo tumor-derived information found in the cell-free DNA (cfDNA) of the biofluid. However, identifying somatic genomic alterations, including both somatic copy number alterations (SCNAs) and single nucleotide variations (SNVs) of the RB1 gene, typically requires either: (1) two distinct experimental protocols—low-pass whole genome sequencing for SCNAs and targeted sequencing for SNVs—or (2) expensive deep whole genome or exome sequencing. To save time and cost, we applied a one-step targeted sequencing method to identify both SCNAs and RB1 SNVs in children with RB. High concordance (median = 96.2%) was observed in comparing SCNA calls derived from targeted sequencing to the traditional low-pass whole genome sequencing method. We further applied this method to investigate the degree of concordance of genomic alterations between paired tumor and AH samples from 11 RB eyes. We found 11/11 AH samples (100%) had SCNAs, and 10 of them (90.1%) with recurrent RB-SCNAs, while only nine out of 11 tumor samples (81.8%) had positive RB-SCNA signatures in both low-pass and targeted methods. Eight out of the nine (88.9%) detected SNVs were shared between AH and tumor samples. Ultimately, 11/11 cases have somatic alterations identified, including nine RB1 SNVs and 10 recurrent RB-SCNAs with four focal RB1 deletions and one MYCN gain. The results presented show the feasibility of utilizing one sequencing approach to obtain SCNA and targeted SNV data to capture a broad genomic scope of RB disease, which may ultimately expedite clinical intervention and be less expensive than other methods.
Keywords: retinoblastoma, aqueous humor, RB1 gene, liquid biopsy, somatic copy number alterations, variant calling, targeted sequencing
1. Introduction
While RB is considered a canonical cancer, having the first molecularly described tumor suppressor gene (RB1), surprisingly, very little is known about the molecular basis underlying the intraocular behavior of this cancer and the varying mechanisms of treatment resistance. This is mainly due to a strict contraindication against invasive biopsy for RB tumors that would risk extraocular cancer spread. Extensive molecular analysis of tumor tissue from advanced enucleated eyes has improved our understanding of RB tumorigenesis. This cancer is predominately driven by biallelic inactivation of the RB1 gene [1] through either a copy number alteration (CNA) and/or single nucleotide variants (SNVs) that negate RB1 protein function. RB1 function is crucial to RB pathology, and over 1000 RB1 SNVs have been identified that contribute to disease pathogenicity [2]. In almost half the cases, the causal event is inherited, greatly increasing the risk of tumor development in both eyes (bilateral disease) [3]. Further, in approximately 2% of patients, RB develops without any RB1 alterations and is instead driven by oncogenic amplification of MYCN on chromosome 2, which is also associated with aggressive tumor behavior [4].
Apart from loss of function alterations to RB1, deleterious variants of BCOR and CREBBP are the two most common co-occurring events at the SNV level [5,6,7]. BCOR mutations have been correlated with metastatic potential in RB patients, indicating a more aggressive disease [6], and CREBBP plays a role in regulating cell cycle and differentiation [8]. Although more work is needed to understand the impact of these genes in RB pathogenesis, both BCOR and CREBBP are thought to be tumor suppressors; thus, their inactivation may contribute to disease severity [9].
Despite this body of knowledge of RB1 mutational contributions to RB, none of it is used to improve the care we provide to these young cancer patients. While genomic analyses at the SNV and CNA levels are routinely used to profile the genetic drivers of other tumors, this is not the current clinical practice for children with RB. In the absence of tumor tissue from a biopsy, identifying these alterations for children with RB was previously not possible. This paradigm changed in 2017 when we, and then others, demonstrated that the aqueous humor (AH), the clear intraocular fluid in front of the eye, is an enriched source of tumor-derived cell-free DNA (cfDNA) in RB eyes that can serve as a liquid biopsy [10,11,12,13,14,15]. We have shown highly concordant somatic CNA (SCNA) profiles with 100 ul AH and matched tumor tissue, and with access to genomic tumor information in eyes undergoing treatment, identified a specific highly recurrent SCNA, chromosome 6p (chr6p) gain, as associated with a 10-fold increased risk of treatment failure requiring surgical removal of the eye [11,16]. This finding underlies the importance of identifying molecular drivers for intraocular disease and highlights the utility of AH in the study of RB disease without enucleation. However, to assay both SCNAs and SNVs in one clinical sample, two separate experimental protocols are required [16], which increases the overall cost and hinders the utility of expedited AH analysis for clinical use.
Methods for systematically profiling genetic disease variants have vastly improved in the next-generation sequencing era. Targeted sequencing has been applied to both AH and tumor samples to profile gene regions of interest (primarily the RB1 gene) for SNV variants, along with small insertions and deletions that negate RB1 protein function [5,16,17]. With similar intent, SCNA profiling via low-pass whole genome sequencing (WGS) has been utilized to investigate RB1 loss on chromosome 13, along with additional RB-SCNA signatures (i.e., chr6p gains), in both AH and tumor samples. However, utilizing two distinct methods to inform on SNVs and SCNAs is time-consuming and expensive, which may ultimately delay, or hasten, life-changing treatment decisions for the patient (i.e., enucleation when it is not needed). We sought to simultaneously profile genomic SNVs and SCNAs through a single targeted method to give clinicians actionable information in less time and with less incurred costs.
Here, we present a cohort of 11 patients diagnosed with RB for whom we matched tumor and AH samples. By applying a hybrid capture targeted sequencing approach, we not only systematically profile SNVs along the RB1 gene, but we also observe high intra-patient concordance of SCNAs between respective AH and tumor samples (Figure 1) and confirm all germline RB1 variants in each bioanalyte. The targeted technique also enabled the interrogation of BCOR, CREBBP, and MYCN, from which we reported deleterious variants of BCOR and CREBBP and a focal MYCN amplification. Altogether, we show how the combination of targeted SNV and SCNA detection in a single method can be utilized to profile RB and how this genomic information can be used together to understand the disease.
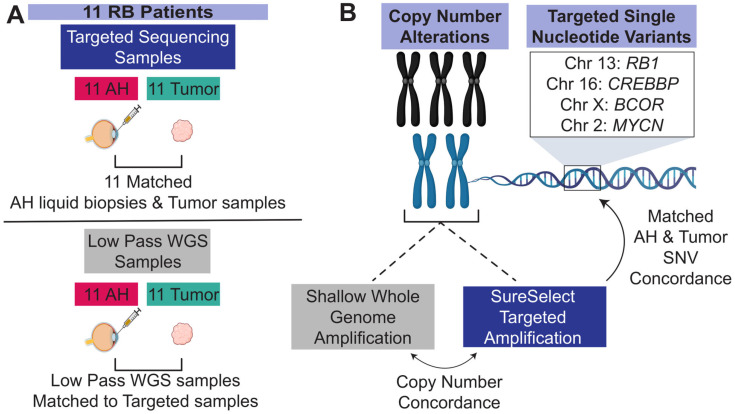
Figure 1.
Schematic of study design. (A) Eleven patients were present in the cohort. ((A) Top) Sample counts for each biospecimen collected that was processed through the dual-targeted SNV and SCNA methods are displayed. Eleven patients contained matched AH and tumor-targeted sequencing samples. ((A) Bottom) Sample count of low-pass WGS sequencing samples. (B) SCNAs from the SureSelect Targeted Sequencing approach were compared to the gold standard low-pass WGS, and concordance was calculated. The targeted reads were then used to call SNVs, and the 11 matched AH and tumor-targeted reads were directly compared for concordance. This figure was generated with BioRender.
2. Results
2.1. Patient Cohort Characteristics
This study includes 11 individuals diagnosed with RB at CHLA between 2015 and 2019 (Table 1). Cases were de-identified, and tumor stage and seeding class were reported. All patients in the cohort underwent enucleation with tumor tissue available and had matched AH sampled. Further, patient 33 has two AH samples available: one taken at diagnosis (33-dx) and one at enucleation (33-es) following therapy. Demographic and tumor information is available in Table 1. The median diagnostic age was 22 months (range, 3–35 months). The presence of a germline RB1 mutation was assayed from routine clinical testing to detect heritable mutations driving each patient’s disease. Three patients harbored germline alterations: cases 9 and 13 harbored inactivating SNVs of the RB1 gene (p.(M148Vfs*8) and p.(R320*), respectively), while case 1 had a hemizygous deletion of chromosome 13q which resulted in a loss of an RB1 allele (Table 1). All eyes were enucleated after a median of 95 days (SD = 197.5 days) following therapy.
Table 1.
Clinical characteristics of the 11 patients in the cohort.
| Case ID |
Sex | Age (Months) |
Laterality 1 | IIRC Group |
TNM Stage |
Seed Class |
Enucleation | Laser 2 Sessions | Cryo 2 Sessions |
IVM 3 | Germline RB1 Blood Test |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | M | 20 | U | E | CT3C | None | Primary | - | - | No | 13q- |
| 8 | F | 22 | U | D | CT2B | Dust | Secondary | 10 | 0 | Yes | Negative |
| 9 | F | 29 | B | D | CT2B | Dust | Secondary | 2 | 2 | No | p.(R320*) |
| 11 | F | 8 | U | D | CT2B | None | Secondary | 15 | 4 | Yes | Negative |
| 13 | M | 34 | U | D | CT2B | Sphere | Secondary | 8 | 1 | Yes | p.(M148Vfs*8) |
| 15 | M | 10 | U | D | CT2B | None | Secondary | 16 | 1 | Yes | Negative |
| 28 | F | 3 | B | D | CT2B | None | Secondary | 7 | 1 | Yes | Negative |
| 33 4 | M | 22 | U | D | CT2B | Sphere | Secondary | 3 | 1 | Yes | Negative |
| 48 | M | 18 | U | D | CT2B | All; cloud | Secondary 5 | - | - | No | Negative |
| 49 | M | 35 | U | D | CT1B | All; cloud | Primary | - | - | No | Negative |
| 50 | F | 24 | U | D | CT2B | None | Primary | - | - | No | Negative |
1 Laterality: Unilateral (U) and bilateral (B). 2 Number of laser and cryotherapy sessions received before eye enucleation. 3 IVM: Intravitreal Melphalan injections (chemotherapy) were received (yes) or not received (no). 4 Patient 33 has two AH samples: at diagnosis (33-dx) and at enucleation (33-es). 5 Patient 48 received one cycle of intra-arterial chemotherapy and then was secondarily enucleated.
2.2. Copy Number Status Is Concordant in Targeted Sequencing Samples to Low-Pass WGS Samples
From standard low-pass sequencing, which is the gold standard for SCNA determination, all 11 (100%) AH and eight out of 11 (72.7%) tumor samples were identified with one or more SCNAs (Figure S1). The AH of case 28 has a chr20q gain but with no other recurrent RB-SCNA signature (Figure S1). To have confidence in SCNA calls from targeted sequencing reads, we evaluated concordance between SCNAs from targeted reads and SCNAs from low-pass WGS reads of tumor samples (see Section 4). When comparing copy number profiles, we found that the mean values were comparable between methods, with a median concordance of 96.2% (SD = 10.7%; Figure 2A). Further, the median concordance for targeted AH to low-pass WGS AH samples was also high at 97.7% (SD = 3%; n = 8; Figure S2); however, only eight samples were analyzed due to differences in sequencing methodologies between the low-pass WGS samples (see Section 4).
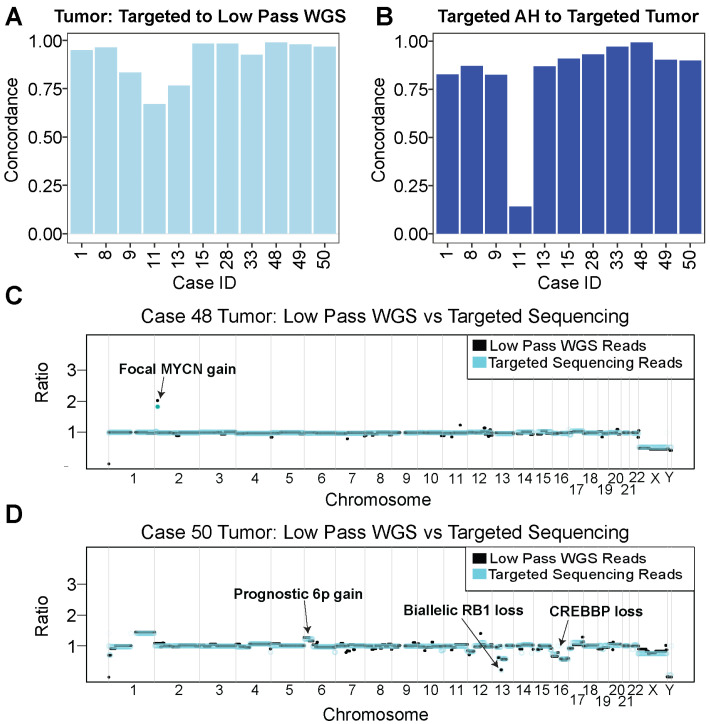
Figure 2.
Copy number alteration analysis shows high concordance between targeted AH and tumor samples. (A) Concordance of derived tumor SCNAs between targeted sequencing reads and their matched low-pass WGS reads. (B) Concordance of SCNAs between targeted AH sequencing samples and their respective targeted tumor sequencing samples. (C) Copy number representation for case 48’s tumor sample overlaying targeted sequencing reads (teal) to low-pass WGS reads (black). A focal MYCN gain was detected on chromosome 2 with both methods. (D) Copy number plot for case 50’s tumor sample overlaying targeted sequencing reads (teal) to low-pass WGS reads (black) showing focal RB1 gene deletion on chromosome 13 and CREBBP loss on chromosome 16.
Since AH and tumor concordance in low-pass WGS samples have been well documented in the literature [11], we sought to determine the concordance between AH tumor samples using our targeted method. For all sample pairs, we found a median concordance of 89.9% (SD = 23.4%; Figure 2B). Notably, case 11 had low-quality targeted tumor sequencing reads due to limited biological complexity, which led to low AH/tumor concordance. After removing case 11, the median concordance between targeted AH and targeted tumor reads raised to 90.1% (SD = 5.5%).
2.3. SCNA Profiles from Targeted Samples Recapitulate Focal Gains and Losses
Targeted sequencing has the potential to miss focal chromosomal alterations due to reduced coverage of the genome. However, we verified the sensitivity of our targeted approach by comparing our SCNA results to those of matched samples generated with low-pass WGS. Using our panel, we were able to capture relevant RB SCNAs in our samples that may drive disease and serve as prognostic indicators. MYCN is the primary driver of RB in a small fraction of cases without loss of functional RB protein; however, MYCN gains can be both with wildtype RB1 (RB1+/+/MYCNA) and as a secondary driver with a loss of the functional RB protein (RB−/−). As with other cancers, a MYCN gain serves as a poor prognostic indicator in both settings. Case 48 harbored a focal MYCN gain, which was detectable with both low-pass WGS and targeted tumor analyses (Figure 2C). Further, case 50 harbored a focal biallelic loss of the RB1 gene on chromosome 13 that was detected in both low-pass WGS and our targeted approach (Figure 2D).
2.4. Inactivating SNVs to the RB1 Gene
While patients harbored many shared passenger mutations between their respective AH and tumor samples, we were interested in detecting disease-driving variants for RB. We found that six out of the 11 cases (54.5%) harbored at least one inactivating RB1 mutation (Figure 3). In total, we detected nine pathogenic variants, of which eight were shared between the AH and tumor (88.9%; Figure 3). One deleterious variant was detected in the tumor from case 28 p.(R255*) but not in the corresponding AH. Visualization of sequencing coverage via IGV confirmed the SNV is present in the tumor but not the AH (Figure S3).
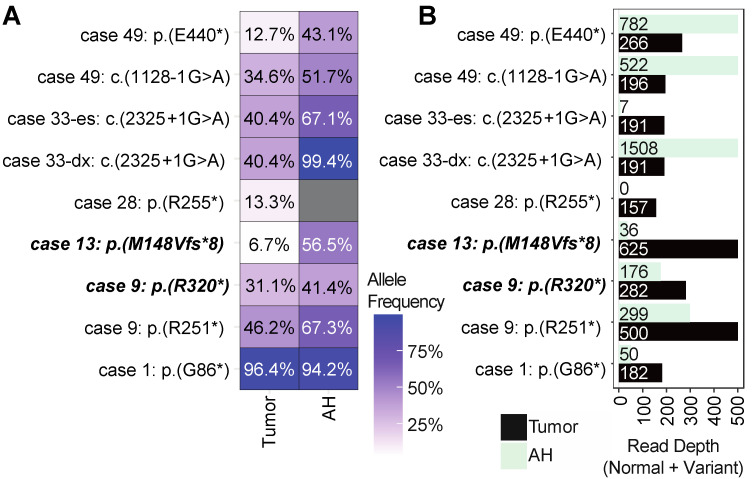
Figure 3.
SNV landscape of RB patients: (A) Allele frequency plot for deleterious SNVs detected to the RB1 gene. Case 33 had two AH samples (pt33-dx and pt33-es) but had a single tumor sample that was taken at enucleation. Cases that are bold and italicized are germline variants. Cases with asterisk (*) indicate nonsense variants that result in truncated protein. Dark grey boxes indicate the variant was not found in that sample. (B) Read depth for each genomic site in (A). Depth accounts for total reads at each specified site, including both variant reads and normal allele reads. X-axis for depth was cut off at 500 bases for legibility.
In general, when the patient harbored a shared AH and tumor mutation, the variant allele frequency (VAF) for the mutation was higher in the AH (Figure 3A). Interestingly, we were able to detect a germline frameshift mutation for case 13 p.(M148Vfs*8) in both the AH and tumor, but the VAF for the tumor was 6.7%, which was significantly lower than 56.5% observed in AH, suggesting a higher normal (i.e., non-RB) cell fraction existing in the tumor sample (Figure 3A). For case 9, we detected a germline mutation p.(R320*) and a second deleterious mutant p.(R251*); the VAF was higher in the AH for both mutants.
Case 33 was a unique patient because we had an AH sample from diagnosis (case 33-dx) and one from secondary enucleation included in the analysis (case 33-es). A splicing variant (c.2325 + 1G>A) was identified in all samples, with high read depths found in the tumor sample and the AH sample at diagnosis (33-dx; Figure 3A,B). Although the targeted sample for case 33-es failed QC (7 total reads were mapped to the genomic location, which was below our cut-off of 10; Figure 3B), we retained this variant since it was present at diagnosis (case 33-dx) and in the enucleated tumor tissue with high confidence. Similarly, we detected a deleterious variant p.(R455*) on the RB1 gene for case 15 that was only present in the AH but not the tumor (VAF = 90%) that also failed QC (nine reads mapped). Since this variant had low read depth and was only present in the AH and not the tumor, it was discarded; further analysis for case 15 may prove whether this mutation is legitimate.
2.5. RB-SCNA Signatures Observed in the Cohort
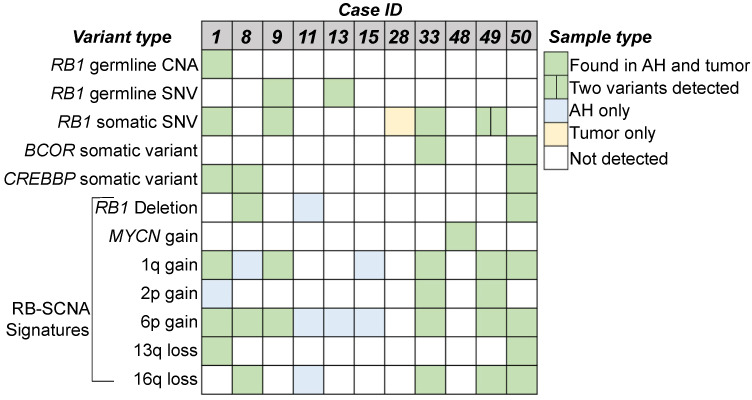
Genomic and genetic alterations for these RB cases are summarized in Figure 4, showing RB signatures, including RB-SCNAs and/or RB1 SNVs that were identified in 10 (90.1%) AH samples and nine tumor samples (81.8%). While our methods detected a MYCN gain for case 48, immunohistochemical staining of the tumor tissue for RB protein showed a lack of nuclear staining. Thus, while an SNV was not detected, there was a loss of functional RB protein suggesting the RB1 gene may have been epigenetically silenced and not that this is a tumor driven primarily by MYCN [18]. Finally, although the AH of case 28 shows neither RB-SCNA nor RB1 SNVs, it does have a significant gain on chromosome 20, indicating the existence of somatic alteration (Figure S1).
Figure 4.
Summary of variants observed in the Cohort. Germline and somatic RB1 SCNAs and SNVs, and other RB-signature genetic variants are displayed. Germline variants detected in blood (separate clinical diagnostic test), AH, and tumor for cases 1, 9, and 13. Cases 33-dx and 33-es were merged; they have the same RB1 somatic SNV detected at diagnosis and enucleation. Variants depicted are found in both targeted and tumor samples.
2.6. Deleterious Variants Detected That Impact BCOR and CREBBP
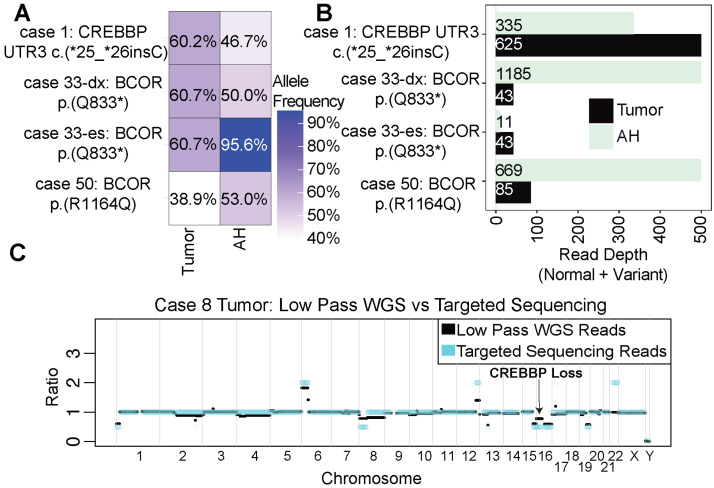
In addition to RB1 variants, we detected deleterious SNVs (n = 3) and SCNAs (n = 2) to the BCOR or CREBBP genes in 5 cases (45.4%). All SNV variants were shared between AH and tumor samples (Figure 5A,B). We also detected a chromosome 16p loss in case 50 (Figure 2D) and case 8 (Figure 5C) that resulted in the loss of a CREBBP allele. No activating MYCN SNVs were detected in the cohort.
Figure 5.
Mutational landscape to CREBBP and BCOR: (A) Allele frequency plot for deleterious mutations to BCOR and CREBBP detected in the cohort. Cases with asterisk (*) indicate nonsense variants that result in truncated protein. (B) Read depth for each genomic site in (A). Depth accounts for total reads at each specified site, including both variant reads and normal allele reads. X-axis for depth was cut off at 500 bases for legibility. (C) SCNA plot for case 8 displaying a loss to chromosome 16 impacting CREBBP.
3. Discussion
We report the multi-sample genomic analysis of RB-related disease drivers at SCNA and SNV levels by applying a single targeted sequencing method and provide further evidence that the AH can be used as a liquid biopsy approach for the non-invasive evaluation and monitoring of RB disease. This study is the first to examine the SCNA status of both AH and tumor samples while simultaneously analyzing RB1, BCOR, CREBBP, and MYCN for SNV disease drivers. We were able to confidently call RB-signature variants for 10/11 patients in the cohort, including inactivating RB1 gene variants in nine cases and a focal MYCN gain for case 48. Case 15 had no detectable RB1 variants. There are multiple reasons why pathogenic variants are not detected in all cases, including known epigenetic-related silencing events that inactivate RB1 protein function [18].
SCNA profiling of both AH and blood cfDNA has been previously established as an effective liquid biopsy approach by our group and others. In the present study, we found that our target capture approach was highly concordant with the current gold standard of low-pass WGS for calling SCNAs in AH and tumor samples from RB patients (Figure 2A). We were also able to confirm all germline RB1 CNAs that were detected by the clinical blood test for both the AH and tumor in our assay (Table 1, Figure 3 and Figure 4). Finally, the targeted approach was able to detect a focal MYCN gain in case 48 (Figure 2C), highlighting the sensitivity of the assay.
SNV profiling showed strong concordance in the detection of pathogenic RB1 variants between the AH and tumor samples (Figure 3). Taken together, we found eight out of nine (88.9%) disease-driving SNVs in the AH (88.9%), thereby further supporting AH as an alternative liquid biopsy approach for RB. Case 28 was the one outlier where we found a deleterious variant p.(R255*) that was detected in the tumor with a low fraction but not AH (Figure 3 and Figure S3). However, AH SCNA, in this case, harbors clear evidence of the somatic alteration of the disease.
Overall, the VAFs in AH samples were found to be higher than those in matched tumor samples. This observation could be attributed to either the presence of normal cells in the tumor biopsy, which could lower the VAFs, or poor sequencing quality leading to noisy data. For instance, in case 49, the copy number profiles show that the ratio of the median of the segmented values in the tumor is much less than that of the AH sample (Figure S1: Case 49 tumor versus AH). The tumor profile appears to be condensed compared to the AH profile. At the SNV level, the tumor VAF for the p.(E440*) mutation in case 49 was only 12.7%, whereas the AH VAF was much higher at 43.1% (Figure 3). These findings further support the possibility of non-RB cells, such as associated retinal cells contaminating the tumor sample during preparation; this will be especially likely in secondarily enucleated eyes wherein the majority of the tumor has undergone necrosis, and the specimen is highly calcified. We also identified a germline SNV in case 13 (p.(M148Vfs*8)) that was confirmed by our analysis (Table 1). However, the VAF of this mutation was found to be significantly lower in the tumor sample (6.7%) than in the AH sample (56.5%; Figure 3). We speculate that this discrepancy could be attributed to the presence of more non-cancerous cells in the tumor sample, as evidenced by the respective copy number profiles of the tumor and AH samples (Figure S1: Case 13 tumor versus Case 13 AH). Specifically, the tumor samples appeared nearly diploid, while the AH samples displayed clear RB-SCNA signatures.
While the blood has been utilized to profile RB1 somatic SNVs [19], the data presented here suggests the AH is superior due to its high tumor fraction. Even with the high-quality data observed in the AH, a substantial amount of signal from the disease-driving splicing variant from case 33-es (c.2325 + 1G>A) was lost following treatment, which underlines the challenge and needs for high DNA concentrations for mutational analysis [20]. Though direct AH-blood SCNA comparisons have been done previously [21], further direct comparisons between the blood, AH, and tumor in SNV analysis for RB are needed.
While this method provided promising results, improvements can be made that can increase its accuracy. For example, probe optimization would benefit future studies. An alternative probe design, which encompasses boundaries slightly outside the genes of interest, could increase efficiency. Extending probe coverage would also allow for the profiling of potential regulatory sequences in the intronic regions of the genes. Lastly, deep sequencing in an unbiased approach, such as whole exome or whole genome sequencing, would be more expensive but may uncover disease-driving mutants outside the scope of the four genes we profiled in this study.
Retinoblastoma tumor tissue cannot be biopsied due to the risk of metastatic spread, which means that obtaining tumor tissue for molecular analysis requires surgical removal of the eye. However, the primary objective of therapy is to treat cancer while preserving the eye, which precludes access to tumor tissue. Our analysis further supports the realization that AH samples collected in vivo can serve as a liquid biopsy for RB, and we have demonstrated the feasibility and accuracy of a combined SCNA-SNV analysis from a single sample and single targeted assay in a time-efficient and cost-effective manner [22,23]. These findings represent a significant clinical advance in the diagnosis and treatment of RB, with the potential to improve outcomes and quality of life for patients and their families.
4. Materials and Methods
Statement of research ethics: The established biorepository and collection of coded clinical data was approved by the Children’s Hospital Los Angeles Institutional Review Board. This study adhered to the tenets of the Declaration of Helsinki and was in accordance with the Health Insurance Portability and Accountability Act. All patients provided written informed consent via a legal guardian for all procedures performed.
Sample collection and processing: Biospecimens were collected as previously reported [16]. Following specimen extraction, samples were stored at −80 °C until processing. All samples underwent cfDNA isolation and sequencing within 1 month of extraction. cfDNA extraction and processing was previously described [16].
Probe design and efficiency: Targeted SNV probes were designed by Agilent SureSelect DNA (Agilent, Santa Clara, CA, USA) [24] and covered portions of the RB1, MYCN, BCOR, and CREBBP genes in accordance with the human genome reference version hg19. Probes summed to cover 55,097 bp in total (Supplementary Materials File S1).
Copy number alteration analysis: Copy number variation was performed on the targeted reads with the R package CopywriteR (version 3.16)—which takes advantage of off-target reads in targeted sequencing data—using the suggested parameters [25]. Samples were aligned with BWA-MEM (version 0.7.17). Bins were set to 500 kb for the hg19 genome, and a female gDNA control with no copy number alterations was used as a baseline for all samples (Figure S1).
WGS reads were analyzed as previously described [26] with some modifications. Briefly, samples were 150 base-pairs paired-end sequenced at a depth of 1–2 million reads on Illumina HiSeq 4000. Sequencing reads were aligned via BWA-MEM to the hg19 reference. Reads were mapped to 5000 bins spanning the human genome and were then normalized for GC content. Count data were segmented via the R package DNAcopy (version 1.70.0) [27].
SCNA concordance determination: We measured concordance between copy number profiles as previously described [11]. For comparing the targeted versus low-pass sequencing data, we divided the segmented reads from the targeted sample by the WGS sample. Analogously, for comparing tumors to AH samples, segmented means were divided for the tumor by the segmented means for the respective AH. Samples in question were considered concordant if their ratio was between 0.8–1.2 within each bin. Concordant bins were then set to 1; bins whose ratios were either below 0.8 or above 1.2 were considered discordant and set to 0. Each bin was then normalized for size and then summed together to give a final concordance value between 0–1.
RB-SCNA signatures—defined here as 1q gain, 2p gain, 6p gain, 13q loss, and/or 16q loss—were manually inspected for concordance between tumor and AH samples in both the shallow and targeted sequencing approaches. Other RB-SCNA (i.e., 17q gain and 7q gain) signatures may exist [15], but we did not evaluate the signature concordance between samples for those alterations.
Concordance was not calculated for three pairs of AH-targeted and low-pass WGS samples from cases 11, 49, and 50. These samples were single-end 50 bp sequenced; therefore, concordance metrics would not reflect the rest of the cohort that was paired-end 150 bp sequenced (Figure S1).
Single nucleotide variant calling: Targeted samples were paired-end sequenced at 150 bp on an Illumina HiSeq 4000 (Fulgent, Inc., Temple City, CA, USA). FastQ (version 2.0.1) files were aligned to hg19 with BWA-MEM (version 0.7.17). Reads falling within the probed regions (RB1, BCOR, CREBBP, and MYCN) were considered. SAMtools (version 1.15-1) was used to sort by coordinates [28]. Picard (version 2.27.4) was utilized to mark duplicates. The bam files were then indexed via SAMtools and utilized as the input for the Mutect2 (version 4-4.3) pipeline. For Mutect2 analysis, both the germline resource and panel of normals provided by GATK were utilized [29]. The read orientation (via the learn orientation model), pile-up summaries, and contamination artifacts were all calculated and utilized for the filtering of Mutect2 variant calls. The variant effect was determined through the Annovar pipeline (version 2020-06-08) [30], and specifically, SIFT, Polyphen2, and gnomAD scores were utilized to classify mutation severity (i.e., deleterious or tolerable) [31,32,33]. Unless otherwise specified, mutations of interest were then filtered to require a read depth greater than 10. Final mutation calls were then viewed through the Integrated Genome Viewer to confirm the calls [34]. For further validation, all Mutect2 calls were cross-checked with an internal bioinformatic pipeline at the CHLA Center for Personalized Medicine [35].
Acknowledgments
We would like to thank the retinoblastoma cancer patients and their families for their participation and for providing specimens for the advancement of cancer research. We are grateful for the research support provided by CHLA Team RAPIDO: Brianne Brown, Mark Reid, Dilshad Contractor & Armine Begijanmasihi, and Shreya Sirivolu. We are also grateful for the research support provided at CSI-Cancer by Nikki Higa.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms24108606/s1.
Author Contributions
Conceptualization, M.J.S., L.X. and J.L.B.; methodology, L.X, J.L.B., M.J.S. and R.K.P.; validation, L.X., J.L.B., V.Y. and M.J.S.; formal analysis, L.X. and M.J.S.; investigation, L.X. and J.L.B.; resources, L.X., J.L.B., V.Y., C.-C.P., J.H. and P.K.; data curation, L.X. and M.J.S.; writing—original draft preparation, M.J.S.; writing—review and editing, M.J.S., R.K.P., S.P., J.H., P.K., L.X. and J.L.B.; visualization, M.J.S.; supervision, J.H., L.X. and J.L.B.; project administration, L.X. and J.L.B.; funding acquisition, P.K., L.X. and J.L.B. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
This study was conducted under the Institutional Review Board approval CHLA-17-0028 at Children’s Hospital Los Angeles and conformed to the requirements of the United States Insurance and Privacy Act and to the tenants of the Declaration of Helsinki. Approval data was 23 June 2017 and has been renewed annually. All patients included in the analysis provided written informed consent for an Institutional Review Board-approved biorepository at Children’s Hospital Los Angeles via a legal guardian.
Informed Consent Statement
Written informed consent was obtained from patients included in this study via a legal guardian.
Data Availability Statement
The data presented in this text are available in the text and Supplementary Materials. Additional data and analysis pipelines and scripts utilized in this manuscript are available upon request.
Conflicts of Interest
The following authors have no conflict of interest to disclosure: M.J.S., R.K.P., S.P., V.Y., C.P. and P.K.; J.L.B. has financial support from the National Cancer Institute of the National Institute of Health Award Number K08CA232344, The Wright Foundation, Children’s Oncology Group/St. Baldrick’s Foundation, The Knights Templar Eye Foundation, Hyundai Hope on Wheels, Childhood Eye Cancer Trust, Children’s Cancer Research Fund, The Berle & Lucy Adams Chair in Cancer Research, The Larry and Celia Moh Foundation, The Institute for Families, Inc., Children’s Hospital Los Angeles, and an unrestricted departmental grant from Research to Prevent Blindness. L.X has financial support from The Knights Templar Eye Foundation and Children’s Hospital Los Angeles Saban Research Institute, Research Career Development Award J.L.B and L.X. have filed a provisional patent application entitled Aqueous Humor Cell-Free DNA for Diagnostic and Prognostic Evaluation of Ophthalmic Disease 62/654,160 (Berry, Xu, Hicks).
Funding Statement
This study is supported by The Miriam and Sheldon G. Adelson Medical Research Foundation (M.J.S.; R.K.P.). J.L.B has grant support from the following: National Cancer Institute of the National Institute of Health (K08CA232344), The Wright Foundation, Children’s Oncology Group/St. Baldrick’s Foundation, The Knights Templar Eye Foundation, Hyundai Hope on Wheels, Danhaki Family Foundation, Childhood Eye Cancer Trust, and Children’s Cancer Research Fund. Other research support comes from The Berle & Lucy Adams Chair in Cancer Research, The Larry and Celia Moh Foundation, A. Linn Murphree, MD, Chair in Ocular Oncology, The Institute for Families, Inc., Children’s Hospital Los Angeles, and an unrestricted departmental grant from Research to Prevent Blindness.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Rao R., Honavar S.G. Retinoblastoma. Indian J. Pediatr. 2017;84:937–944. doi: 10.1007/s12098-017-2395-0. [DOI] [PubMed] [Google Scholar]
- 2.Lohmann D.R., Gallie B.L. Retinoblastoma: Revisiting the Model Prototype of Inherited Cancer. Am. J. Med. Genet. C Semin. Med. Genet. 2004;129C:23–28. doi: 10.1002/ajmg.c.30024. [DOI] [PubMed] [Google Scholar]
- 3.Dimaras H., Kimani K., Dimba E.A.O., Gronsdahl P., White A., Chan H.S.L., Gallie B.L. Retinoblastoma. Lancet. 2012;379:1436–1446. doi: 10.1016/S0140-6736(11)61137-9. [DOI] [PubMed] [Google Scholar]
- 4.Dimaras H., Corson T.W. Retinoblastoma, the Visible CNS Tumor: A Review. J. Neurosci. Res. 2019;97:29–44. doi: 10.1002/jnr.24213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Afshar A.R., Pekmezci M., Bloomer M.M., Cadenas N.J., Stevers M., Banerjee A., Roy R., Olshen A.B., Van Ziffle J., Onodera C., et al. Next-Generation Sequencing of Retinoblastoma Identifies Pathogenic Alterations beyond RB1 Inactivation That Correlate with Aggressive Histopathologic Features. Ophthalmology. 2020;127:804–813. doi: 10.1016/j.ophtha.2019.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Francis J.H., Richards A.L., Mandelker D.L., Berger M.F., Walsh M.F., Dunkel I.J., Donoghue M.T.A., Abramson D.H. Molecular Changes in Retinoblastoma beyond RB1: Findings from Next-Generation Sequencing. Cancers. 2021;13:149. doi: 10.3390/cancers13010149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McEvoy J.D., Dyer M.A. Genetic and Epigenetic Discoveries in Human Retinoblastoma. Crit. Rev. Oncog. 2015;20:217–225. doi: 10.1615/CritRevOncog.2015013711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wydner K.L., Bhattacharya S., Eckner R., Lawrence J.B., Livingston D.M. Localization of Human CREB-Binding Protein Gene (CREBBP) to 16p13.2-p13.3 by Fluorescence in Situ Hybridization. Genomics. 1995;30:395–396. [PubMed] [Google Scholar]
- 9.Kooi I.E., Mol B.M., Massink M.P.G., Ameziane N., Meijers-Heijboer H., Dommering C.J., van Mil S.E., de Vries Y., van der Hout A.H., Kaspers G.J.L., et al. Somatic Genomic Alterations in Retinoblastoma beyond RB1 Are Rare and Limited to Copy Number Changes. Sci. Rep. 2016;6:25264. doi: 10.1038/srep25264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berry J.L., Xu L., Murphree A.L., Krishnan S., Stachelek K., Zolfaghari E., McGovern K., Lee T.C., Carlsson A., Kuhn P., et al. Potential of Aqueous Humor as a Surrogate Tumor Biopsy for Retinoblastoma. JAMA Ophthalmol. 2017;135:1221–1230. doi: 10.1001/jamaophthalmol.2017.4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berry J.L., Xu L., Kooi I., Murphree A.L., Prabakar R.K., Reid M., Stachelek K., Le B.H.A., Welter L., Reiser B.J., et al. Genomic cfDNA Analysis of Aqueous Humor in Retinoblastoma Predicts Eye Salvage: The Surrogate Tumor Biopsy for Retinoblastoma. Mol. Cancer Res. 2018;16:1701–1712. doi: 10.1158/1541-7786.MCR-18-0369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerrish A., Stone E., Clokie S., Ainsworth J.R., Jenkinson H., McCalla M., Hitchcott C., Colmenero I., Allen S., Parulekar M., et al. Non-Invasive Diagnosis of Retinoblastoma Using Cell-Free DNA from Aqueous Humour. Br. J. Ophthalmol. 2019;103:721–724. doi: 10.1136/bjophthalmol-2018-313005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Le Gall J., Dehainault C., Benoist C., Matet A., Lumbroso-Le Rouic L., Aerts I., Jiménez I., Schleiermacher G., Houdayer C., Radvanyi F., et al. Highly Sensitive Detection Method of Retinoblastoma Genetic Predisposition and Biomarkers. J. Mol. Diagn. 2021;23:1714–1721. doi: 10.1016/j.jmoldx.2021.08.014. [DOI] [PubMed] [Google Scholar]
- 14.Raval V., Racher H., Wrenn J., Singh A.D. Aqueous Humor as a Surrogate Biomarker for Retinoblastoma Tumor Tissue. J. AAPOS. 2022;26:137.e1–137.e5. doi: 10.1016/j.jaapos.2022.03.005. [DOI] [PubMed] [Google Scholar]
- 15.Luo Y., Xu M., Yang L., Yao Y., Berry J.L., Xu L., Wen X., He X., Han M., Fan X., et al. Correlating Somatic Copy Number Alteration in Aqueous Humour cfDNA with Chemotherapy History, Eye Salvage and Pathological Features in Retinoblastoma. Br. J. Ophthalmol. 2023 doi: 10.1136/bjo-2022-322866. in press . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu L., Shen L., Polski A., Prabakar R.K., Shah R., Jubran R., Kim J.W., Biegel J., Kuhn P., Cobrinik D., et al. Simultaneous Identification of Clinically Relevant RB1 Mutations and Copy Number Alterations in Aqueous Humor of Retinoblastoma Eyes. Ophthalmic Genet. 2020;41:526–532. doi: 10.1080/13816810.2020.1799417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Devarajan B., Prakash L., Kannan T.R., Abraham A.A., Kim U., Muthukkaruppan V., Vanniarajan A. Targeted next Generation Sequencing of RB1 Gene for the Molecular Diagnosis of Retinoblastoma. BMC Cancer. 2015;15:320. doi: 10.1186/s12885-015-1340-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li H.-T., Xu L., Weisenberger D.J., Li M., Zhou W., Peng C.-C., Stachelek K., Cobrinik D., Liang G., Berry J.L. Characterizing DNA Methylation Signatures of Retinoblastoma Using Aqueous Humor Liquid Biopsy. Nat. Commun. 2022;13:5523. doi: 10.1038/s41467-022-33248-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kothari P., Marass F., Yang J.L., Stewart C.M., Stephens D., Patel J., Hasan M., Jing X., Meng F., Enriquez J., et al. Cell-Free DNA Profiling in Retinoblastoma Patients with Advanced Intraocular Disease: An MSKCC Experience. Cancer Med. 2020;9:6093–6101. doi: 10.1002/cam4.3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Im D.H., Pike S., Reid M.W., Peng C.-C., Sirivolu S., Grossniklaus H.E., Hubbard G.B., Skalet A.H., Bellsmith K.N., Shields C.L., et al. A Multicenter Analysis of Nucleic Acid Quantification Using Aqueous Humor Liquid Biopsy in Retinoblastoma: Implications for Clinical Testing. Ophthalmol. Sci. 2023;3:100289. doi: 10.1016/j.xops.2023.100289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berry J.L., Xu L., Polski A., Jubran R., Kuhn P., Kim J.W., Hicks J. Aqueous Humor Is Superior to Blood as a Liquid Biopsy for Retinoblastoma. Ophthalmology. 2020;127:552–554. doi: 10.1016/j.ophtha.2019.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Christodoulou E., Yellapantula V., O’Halloran K., Xu L., Berry J.L., Cotter J.A., Zdanowicz A., Mascarenhas L., Amatruda J.F., Ostrow D., et al. Combined Low-Pass Whole Genome and Targeted Sequencing in Liquid Biopsies for Pediatric Solid Tumors. NPJ Precis. Oncol. 2023;7:21. doi: 10.1038/s41698-023-00357-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dunn P., Albury C.L., Maksemous N., Benton M.C., Sutherland H.G., Smith R.A., Haupt L.M., Griffiths L.R. Next Generation Sequencing Methods for Diagnosis of Epilepsy Syndromes. Front. Genet. 2018;9:20. doi: 10.3389/fgene.2018.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giuffre A., Joshi S., Ravi H., Pabon-Pena C., Novak B., Visitacion M., Hamady M., Useche F., Arezi B., Buehler B., et al. Overview of the Agilent Technologies SureSelectTM Target Enrichment System. J. Biomol. Tech. 2011;22:S30. [Google Scholar]
- 25.Kuilman T., Velds A., Kemper K., Ranzani M., Bombardelli L., Hoogstraat M., Nevedomskaya E., Xu G., de Ruiter J., Lolkema M.P., et al. CopywriteR: DNA Copy Number Detection from off-Target Sequence Data. Genome Biol. 2015;16:49. doi: 10.1186/s13059-015-0617-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baslan T., Kendall J., Ward B., Cox H., Leotta A., Rodgers L., Riggs M., D’Italia S., Sun G., Yong M., et al. Optimizing Sparse Sequencing of Single Cells for Highly Multiplex Copy Number Profiling. Genome Res. 2015;25:714–724. doi: 10.1101/gr.188060.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seshan V.E., Olshen A. DNAcopy: DNA Copy Number Data Analysis. R Package Version. 2023. [(accessed on 1 December 2022)]. Available online: https://bioconductor.org/packages/release/bioc/html/DNAcopy.html.
- 28.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map Format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Benjamin D., Sato T., Cibulskis K., Getz G., Stewart C., Lichtenstein L. Calling Somatic SNVs and Indels with Mutect2. bioRxiv. 2019 doi: 10.1101/861054. [DOI] [Google Scholar]
- 30.Yang H., Wang K. Genomic Variant Annotation and Prioritization with ANNOVAR and wANNOVAR. Nat. Protoc. 2015;10:1556–1566. doi: 10.1038/nprot.2015.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumar P., Henikoff S., Ng P.C. Predicting the Effects of Coding Non-Synonymous Variants on Protein Function Using the SIFT Algorithm. Nat. Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 32.Koch L. Exploring Human Genomic Diversity with gnomAD. Nat. Rev. Genet. 2020;21:448. doi: 10.1038/s41576-020-0255-7. [DOI] [PubMed] [Google Scholar]
- 33.Adzhubei I., Jordan D.M., Sunyaev S.R. Predicting Functional Effect of Human Missense Mutations Using PolyPhen-2. Curr. Protoc. Hum. Genet. 2013;76:7–20. doi: 10.1002/0471142905.hg0720s76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M. Integrative Genomics Viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chapman B., Kirchner R., Pantano L., DeSmet M., Beltrame L., Khtiaisteva T., Namenko S., Savliev V., Guimera R.V., Sytchey I., et al. Bcbio-Nextgen. 2023. [(accessed on 1 February 2023)]. Available online: https://zenodo.org/record/5781867#.ZFo3ZM5BxPY.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this text are available in the text and Supplementary Materials. Additional data and analysis pipelines and scripts utilized in this manuscript are available upon request.