Abstract
Vitiligo is an autoimmune complex pigmentation disease characterized by non-pigmented patches on the surface of the skin that affect approximately 0.5–2% population worldwide. The exact etiology is still unknown; however, vitiligo is hypothesized to be a multifactorial and genetically heterogeneous condition. Therefore, the current study is designed to investigate the anthropometric presentation and genetic spectrum of vitiligo in fifteen consanguineous Pakistani families. The clinical evaluation of participating individuals revealed varying degrees of disease severity, with 23 years as the average age of disease onset. The majority of the affected individuals had non-segmental vitiligo (NSV). Whole exome sequencing analysis revealed clustering of rare variants of known vitiligo-associated genes. For instance, in the affected individuals of family VF-12, we identified three novel rare variants of PTPN22 (c.1108C>A), NRROS (c.197C>T) and HERC2 (c.10969G>A) genes. All three variants replaced evolutionarily conserved amino acid residues in encoded proteins, which are predicted to impact the ionic interactions in the secondary structure. Although various in silico algorithms predicted low effect sizes for these variants individually, the clustering of them in affected individuals increases the polygenic burden of risk alleles. To our knowledge, this is the first study that highlights the complex etiology of vitiligo and genetic heterogeneity in multiplex consanguineous Pakistani families.
Keywords: vitiligo, non-segmental vitiligo, polygenic burden, genetic heterogeneity, allele clustering, PTPN22, HERC2, NRROS
1. Introduction
Vitiligo is a common skin pigmentation disorder with remarkable genetic and clinical heterogeneity. Vitiligo is characterized by the appearance of colorless patches on the epidermal surface of the skin. Discrepancies in the production and distribution of melanin in affected areas result in the onset of this socially stigmatizing and psychologically devastating disease. Vitiligo may affect one or both halves of the body. The vitiligo European task force assessment (VETFa) has broadly classified vitiligo into two main types, i.e., non-segmental vitiligo (NSV) or generalized vitiligo (GV) and Segmental Vitiligo (SV). Vitiligo is known to be progressive in most patients, which eventually results in complete loss of skin coloration, referred to as vitiligo Universalis (VU) in elderly patients. Various studies have reported that the worldwide incidence of vitiligo ranges between 0.5–2% [1,2]. However, there is no prior comprehensive study documenting the prevalence of vitiligo in the Pakistani population.
Vitiligo is a multifactorial, polygenic disorder that follows a non-Mendelian inheritance pattern with considerable incomplete penetrance due to complex genetic and environmental etiology [3]. A number of theories have been proposed to explain vitiligo pathogenesis that include genetics, autoimmune, auto-cytotoxic, and gene-environment interactions, which indicates that gene-environment interactions are predominantly involved in disease pathogenesis [4]. Multiplex vitiligo families partially support genetic hypotheses [5]. A report on the Korean population suggests a polygenic etiology of the disease [6]. The possible involvement of different sets of genes in melanin production, epistatic interactions, multistep regulation of the autoimmune system, and control of auto-toxicity has been implicated in disease pathogenesis [4]. The presence of certain environmental triggers, such as oxidative stress, ultraviolet radiation, and exposure to phenolic derivative compounds, may initiate vitiligo macule formation [7,8,9].
Advancement in genetic screening technologies has tremendously helped in the identification of rare variants associated with vitiligo. In the current study, we identified fifteen clinically diagnosed multiplex vitiligo families from the Punjab province of Pakistan. Whole exome sequencing was performed on seven of these families, which identified rare variants in vitiligo-associated genes. Our study also points towards the clinical and genetic complexity and locus heterogeneity of vitiligo in the inbred Pakistani population.
2. Materials and Methods
2.1. Family Enrollment and Clinical Classification
The proceedings of the current study were approved by committees of the Institutional Review Board (IRB) at the University of Maryland, School of Medicine, Baltimore, MD, USA (HP-00061036) and Pir Mher Ali Shah Arid Agriculture University Rawalpindi, Pakistan. The tenets of the Declaration of Helsinki were followed for the use of human subjects in the current study. Informed consent in writing was obtained from all participants. Fifteen vitiligo families were recruited from different regions of the Punjab province of Pakistan in this study.
Detailed family histories were obtained to document disease onset, affected area of skin, disease progression status, smoking/non-smoking status, chemical exposure, general lifestyle, eating habits, treatment etc. Family pedigrees were drawn based on the information received and verified by multiple participating family members.
2.2. Sample Collection and Isolation of DNA
From all participating individuals, 3 mL of venous blood samples were collected in an EDTA Orsin vacutainer (formerly imuMed®) and stored at 4 °C until further use. The genomic DNA was isolated from the blood samples of participants using the Thermo Scientific™ GeneJET genomic DNA purification kit (Catalog #: K0722). The concentration of DNA was quantified by NanoDrop® ND-1000 UV-Vis Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA), and DNA quality was assessed by 260/280 and 260/230 ratios.
2.3. Whole Exome Sequencing
Whole exome sequencing was performed on the probands of families VF-01, VF-02-, VF-03, VF-09, VF-10, VF-12 and VF-15. Genomic libraries were recovered for exome enrichment by using the Agilent SureSelect Human Expanded All Exon V5 kit and sequenced on an Illumina HiSeq4000 (Illumina, San Diego, CA, USA) with an average of 100× coverage [10]. Broad Institute’s Toolkit for Genome Analysis was used to analyze the sequencing data, as described previously [11,12], with the following modifications: (a) allele frequencies cut off was set to ≤0.003; (b) potential pathogenic prediction was achieved by at least 2 in silico algorithms; and (c) CADD score ≥ 6.0. Primers were designed using the online resource Primer3 (http://bioinfo.ut.ee/primer3-0.4.0/, accessed on 23 October 2018) for the variants that passed our filtration criteria; targeted DNA amplification by conventional PCR followed by Sanger sequencing was carried out for segregation studies [13].
2.4. Molecular Modeling
The Phyre2 (http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index, accessed on 15 March 2019) and protein variation predictor resource HOPE (https://www3.cmbi.umcn.nl/hope/, accessed on 15 March 2019) were applied for 3D modeling of protein structures. Visualization of the mutation impact on the 3D structure and change of ionic interaction between the residues of interest was visualized by the UCSF CHIMERA tool (https://www.cgl.ucsf.edu/chimera/, accessed on 19 March 2019). MUpro (http://mupro.proteomics.ics.uci.edu/, accessed on 27 March 2023) and I-Mutant 2.0 (http://folding.biofold.org/i-mutant/i-mutant2.0.html, accessed on 27 March 2023) were used to predict the effects of these mutations on protein structure stability on the bases of changes in Gibbs free energy (G). The evolutionary conservation status of multiple amino acids across different species was performed by Clustal Omega multiple sequence alignment (https://www.ebi.ac.uk/Tools/msa/clustalo/, accessed on 10 March 2019).
3. Results
3.1. Clinical Evaluation of Vitiligo in Multiplex Pakistani Families
After IRB approval, a total of 15 multiplex consanguineous families segregating vitiligo (Figure 1) were enrolled in this study from five different cities (Rawalpindi, Chakwal, Mianwali, Abbottabad, Islamabad) in Pakistan (Table 1). A total of 130 individuals from these families participated in our study, including 72 males (55%) and 58 females (45%). Of these enrolled subjects, 53 (33 males and 20 females) exhibited the vitiligo phenotype, while 77 individuals had no skin problem and, thus, were used as controls in our downstream genetic studies. The age range for onset vitiligo macules was between 2 to 68 years, with an average of 23 years. Among the enrolled females, non-segmental vitiligo (NSV) was the most noticeable clinical phenotype noted in forty-eight females (91%), three females (6%) had vitiligo universalis (VU), while two females (3%) reported complete recovery from their vitiligo phenotype. Almost all affected individuals reported having no other autoimmune diseases or any other comorbidity besides vitiligo, except two affected individuals that had type 1 diabetes mellitus (Table 1).
Figure 1.
Pedigrees of vitiligo families. Black-filled and empty symbols represent affected and unaffected individuals, respectively. The double line shows only consanguineous marriages. The arrow sign indicate probands that were used for whole exome sequencing. Clinical manifestations of the vitiligo patches for representative affected members are also shown below each pedigree.
Table 1.
Clinical phenotype of Vitiligo families.
| Family ID | Family Ethnicity | Individual ID | Age (Year) | Age of Onset | Gender | Phenotype | Doctoral Description of the Disease | Affected Area of Skin (%) | Comorbidity |
|---|---|---|---|---|---|---|---|---|---|
| VF-01 | Rajput | VF01_01 | 28 | 18 | Male | Affected | NSV, Generalized vitiligo | 20–30% | Nil |
| VF01_02 | 26 | 21 | Female | Affected | NSV, Generalized Vitiligo | 20–30% | Nil | ||
| VF01_04 | 17 | 14 | Female | Affected | NSV, Generalized vitiligo | 10–20% | Nil | ||
| VF-02 | Mughal | VF02_03 | 28 | 20 | Female | Affected | NSV, Generalized vitiligo | 10–20% | Nil |
| VF02_06 | 18 | 18 | Female | Affected | NSV, Generalized Vitiligo | Less than 5% | Nil | ||
| VF02_07 | 42 | 39 | Male | Recovered | Generalized Vitiligo (NSV*) | N/A | Nil | ||
| VF02_11 | 32 | 17 | Male | Affected | NSV, Generalized vitiligo | 30–40% | Nil | ||
| VF02_15 | 80 | 14 | Female | Affected | Vitiligo Universalis | 90–100% | Diabetes | ||
| VF02_16 | 79 | 19 | Male | Affected | NSV, Generalized vitiligo | 90–100% | Nil | ||
| VF02_17 | 56 | 18 | Female | Affected | NSV, Generalized vitiligo | 90–100% | Nil | ||
| VF02_18 | 23 | 11 | Female | Affected | NSV, Generalized vitiligo | 10–20% | Nil | ||
| VF02_19 | 20 | 10 | Female | Recovered | Vitiligo recovered (NSV*) | N/A | Nil | ||
| VF-03 | Awan | VF03_01 | 46 | 24 | Male | Affected | NSV, Generalized vitiligo | 30–40% | Nil |
| VF03_07 | 7 | 04 | Male | Affected | NSV, Acrofacial vitiligo | 5–10% | Nil | ||
| VF03_08 | 30 | 27 | Female | Affected | NSV, Acrofacial vitiligo | Less than 5% | Nil | ||
| VF-04 | Sardar | VF04_01 | 30 | 24 | Male | Affected | NSV, Generalized vitiligo | 20–30% | Nil |
| VF04_02 | 38 | 31 | Male | Affected | NSV, Generalized vitiligo | Less than 5% | Nil | ||
| VF04_03 | 13 | 7 | Female | Affected | NSV, Generalized vitiligo | Less than 5% | Nil | ||
| VF-05 | Mughal | VF05_01 | 47 | 33 | Male | Affected | NSV, Generalized vitiligo | 20–30% | Nil |
| VF05_04 | 22 | 21 | Male | Affected | Vitiligo started in neck region | Less than 5% | Nil | ||
| VF-06 | Rajput | VF06_01 | 31 | 11 | Male | Affected | NSV, Generalized vitiligo | 70–80% | Nil |
| VF06_02 | 25 | 13 | Male | Affected | NSV, Generalized vitiligo | 40–50% | Nil | ||
| VF-07 | Niazi | VF07_01 | 55 | 23 | Male | Affected | NSV, Generalized vitiligo | 40–50% | Nil |
| VF07_03 | 21 | 19 | Male | Affected | NSV, Generalized vitiligo | 5–10% | Nil | ||
| VF-08 | Shiekh | VF08_01 | 69 | 19 | Female | Affected | NSV, Generalized vitiligo | 20–30% | Nil |
| VF08_04 | 21 | 17 | Male | Affected | NSV, Generalized vitiligo | 5–10% | Nil | ||
| VF-09 | Awan | VF09_01 | 64 | 59 | Female | Affected | NSV, Generalized vitiligo | 20–30% | Nil |
| VF09_05 | 17 | 15 | Male | Affected | NSV, Generalized vitiligo | Less than 5% | Nil | ||
| VF09_07 | 19 | 15 | Female | Affected | NSV, Generalized vitiligo | Less than 5% | Nil | ||
| VF09_10 | 9 | 2 | Female | Affected | NSV, Generalized vitiligo | 20–30% | Nil | ||
| VF-10 | Malik | VF10_01 | 79 | 33 | Male | Affected | NSV, Generalized vitiligo | 60–70% | Nil |
| VF10_04 | 83 | 42 | Male | Affected | NSV, Generalized vitiligo | 70–80% | Nil | ||
| VF10_07 | 80 | 40 | Female | Affected | NSV, Generalized vitiligo | 40–50% | Nil | ||
| VF10_10 | 35 | 27 | Male | Affected | NSV, Generalized vitiligo | 40–50% | Nil | ||
| VF10_11 | 51 | 33 | Male | Affected | NSV, Generalized vitiligo | 50–60% | Nil | ||
| VF10_14 | 12 | 7 | Male | Affected | NSV, Generalized vitiligo | 5–10% | Nil | ||
| VF10_16 | 28 | 17 | Male | Affected | NSV, Generalized vitiligo | 40–50% | Nil | ||
| VF-11 | Qazi | VF11_01 | 56 | 11 | Female | Affected | Vitiligo Universalis | 90–100% | Nil |
| VF11_02 | 58 | 53 | Male | Affected | Vitiligo just started at the skull | Less than 1% | Nil | ||
| VF11_03 | 19 | 11 | Male | Affected | NSV, Generalized vitiligo | 5–10% | Nil | ||
| VF-12 | Malik | VF12_01 | 57 | 43 | Male | Affected | NSV, Generalized vitiligo | 30–40% | Diabetes |
| VF12_05 | 26 | 15 | Male | Affected | NSV, Generalized vitiligo | 30–40% | Nil | ||
| VF12_06 | 23 | 22 | Male | Affected | NSV, Generalized vitiligo | Less than 5% | Nil | ||
| VF12_07 | 35 | 24 | Male | Affected | NSV, Generalized vitiligo | 20–30% | Nil | ||
| VF12_09 | 79 | 68 | Female | Affected | NSV, Generalized vitiligo | 5–10% | Nil | ||
| VF-13 | Rajput | VF13_01 | 74 | 66 | Male | Affected | NSV, Generalized vitiligo | 30–40% | Nil |
| VF13_04 | 13 | 9 | Male | Affected | NSV, Generalized vitiligo | 5–10% | Nil | ||
| VF-14 | Chaudary | VF14_01 | 67 | 31 | Female | Affected | NSV, Generalized vitiligo | 15–20% | Nil |
| VF14_03 | 47 | 38 | Male | Affected | NSV, Generalized vitiligo | 30–40% | Nil | ||
| VF14_04 | 15 | 9 | Female | Affected | NSV, Generalized vitiligo | 10–20% | Nil | ||
| VF-15 | Rajput | VF15_01 | 77 | 19 | Male | Affected | Vitiligo Universalis | 90–100% | Nil |
| VF15_04 | 21 | 14 | Male | Affected | NSV, Generalized vitiligo | 20–30% | Nil | ||
| VF15_07 | 21 | 13 | Male | Affected | NSV, Generalized vitiligo | 30–40% | Nil |
NSV: Non segmental vitiligo; NSV*: Non-segmental vitiligo (Recovered); N/A: Not applicable.
Inter- and intra-familial variability in the areas of skin discoloration among the affected individuals was identified (Table 1). For instance, family VF-02 is an extended vitiligo family having nine affected individuals (Figure 1). Three of the affected individuals (VF02_15, _16, _17) had 90–100% of their body with vitiligo patches and thus present VU (Table 1), while the other affected individuals had much fewer patches (Table 1). Intriguingly, according to family history interviews, one affected female (VH02_07) recovered from vitiligo without any medication. In contrast, the three affected individuals of family VF-04 had relatively less progressive disease severity (Table 1).
3.2. Genetic Analysis of Families with Vitiligo
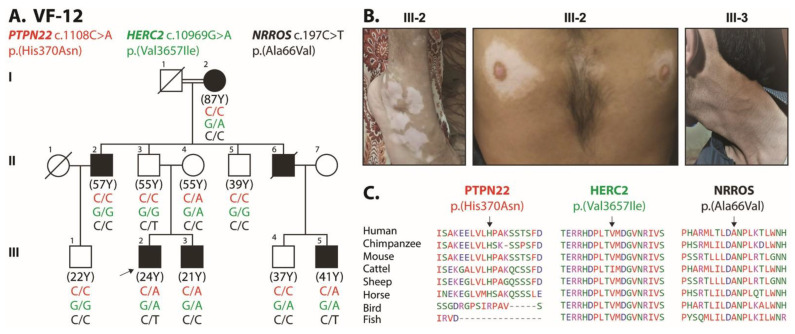
To decipher the genetic risk factors contributing to the vitiligo phenotype in the enrolled families, we performed whole exome sequencing on index patients from seven families along with ethnically matched normal control samples. After quality control, we used in silico panel to filter variants in the known vitiligo-associated genes [14]. Next, we selected variants of the genes that had (a) an allele frequency of ≤0.001; (b) a CADD score of ≥7.0; and (c) predicted damage by at least one in silico algorithm. Table 2 lists all variants of known vitiligo genes that passed these criteria. Although various in silico algorithms predicted low effect size for these variants individually, the clustering of these variants in affected individuals increases the polygenic burden of risk alleles (Table 2); we found potentially pathogenic variants of three known vitiligo-associated genes, PTPN22, HERC2 and NRROS, in a varying combination of zygosity among affected individuals of family VF-12 (Figure 2). Proband III-2, having the most severe phenotype, was found heterozygous for the rare missense variants (Figure 2) of PTPN22 (c.1108C>A; p.(His370Asn)), HERC2 (c.10969G>A; p. (Val3657Ile)), and NRROS (c.197C>T; p.(Ala66Val)). Different family members inherited different combinations of these potentially pathogenic variants, and hence disease severity and allelic burden vary among these individuals (Figure 2 and Table 1). Variants found in the PTPN22, HERC2 and NRROS genes have been previously known for their association with NSV in different ethnicities around the world [15,16,17,18].
Table 2.
Genetic analysis of index patients for known vitiligo gene variants.
| Family | Chr | Gene | Ref | Alt | c.DNA Change | Protein Change | SNP ID | rs Number | ExAC | CADD | SIFT | Polyphen | Mutation Taster |
Mutation Assessor |
Genotyping of Proband |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| VF-01 | 2 | IFIH1 | T | C | c.A1046G | p.K349R | NM_022168 | rs72650664 | 0.0035 | 14.59 | T | B | D | N | Het |
| 11 | GSTP1 | A | C | c.A601C | p.N201H | NM_000852 | rs145957405 | - | 12.59 | D | P | D | M | Het | |
| 11 | GSTP1 | A | C | c.A602C | p.N201T | NM_000852 | rs376074280 | 0.00001677 | 7.554 | T | B | D | M | Het | |
| 12 | ATXN2 | TGC | - | c.534_536del | p.178_179del | NM_002973 | rs193922927 | 0 | - | - | - | - | - | Het | |
| 15 | HERC2 | CT | - | c.5780_5781de | p.K1927fs | NM_004667 | - | - | - | - | - | - | - | Het | |
| 16 | RPGRIP1L | T | A | c.A278T | p.E93V | NM_015272 | rs765876839 | 0.00000825 | 18.56 | T | P | D | L | Het | |
| X | IL1RAPL1 | TG | - | c.63_64del | p.V21fs | NM_014271 | rs147274241 | . | . | . | . | . | . | Het | |
| VF-02 | 19 | BCL2L12 | G | A | c.G542A | p.R181Q | NM_138639 | rs762286281 | 0.0003 | 19.56 | T | D | D | L | Het |
| VF-03 | 8 | TG | C | T | c.C3902T | p.P1301L | NM_003235 | rs549184203 | 0.0002 | 21.1 | D | D | D | M | Het |
| 11 | GSTP1 | A | G | c.A1G | p.M1V | NM_000852 | rs1448870282 | - | 10.05 | D | B | - | - | Het | |
| VF-09 | 3 | GPX1 | C | A | c.G442T | p.A148S | NM_001329455 | rs6446261 | - | 6.871 | T | B | D | L | Het |
| 3 | GPX1 | GCC | - | c.33_35del | p.11_12del | NM_001329455 | rs17838762 | - | - | - | - | - | - | Het | |
| 9 | TLR4 | A | G | c.A1084G | p.K362E | NM_003266 | rs539153708 | 0.0002 | 12.84 | T | P | N | N | Het | |
| 10 | CASP7 | C | T | c.C481T | p.R161W | NM_033340 | rs185649982 | 0 | 0 | T | B | N | - | Het | |
| 21 | UBASH3A | G | A | c.G1666A | p.E556K | NM_018961 | rs116752327 | 0.0001 | 15.6 | D | D | D | M | Het | |
| VF-10 | 1 | PTPRC | T | A | c.T566A | p.I189N | NM_002838 | rs201715157 | 0.0013 | 7.444 | D | B | N | L | Het |
| 2 | FARP2 | A | G | c.A134G | p.H45R | NM_014808 | rs150312458 | 0.0034 | 8.045 | T | B | N | N | Het | |
| 8 | TG | G | A | c.G2963A | p.R988H | NM_003235 | rs16893332 | 0.0062 | 11.02 | T | B | P | N | Het | |
| 14 | STRN3 | G | T | c.C53A | p.P18H | NM_001083893 | rs1036923688 | . | 15.33 | D | B | D | N | Het | |
| 16 | CDH1 | G | A | c.G188A | p.R63Q | NM_004360 | rs587780117 | 0.00005765 | 12.03 | T | B | N | M | Het | |
| 17 | NOS2 | G | A | c.C2084T | p.P695L | NM_000625 | rs767807366 | 0.00004954 | 19.85 | T | P | D | M | Het | |
| VF-12 | 1 | PTPN22 | G | T | c.C1108A | p.H370N | NM_015967 | rs72650671 | 0.0023 | 6.752 | T | P | N | M | Het |
| 10 | ACTA2 | C | T | - | Splicing | - | rs112687898 | - | 26 | - | - | D | - | Het | |
| 15 | HERC2 | C | T | c.G10969A | p.V3657I | NM_004667 | rs139953376 | 0.0025 | 3.685 | T | B | N | N | Het | |
| 20 | RALY | G | A | c.G694A | p.G232S | NM_016732 | rs779745009 | 0.0085 | 7.476 | T | B | N | N | Het | |
| VF-15 | 1 | PTPRC | A | G | c.A1808G | p.D603G | NM_002838 | rs754699279 | 0.00004967 | 12.77 | T | P | D | L | Het |
| 3 | TGFBR2 | G | A | c.G1283A | p.R428H | NM_001024847 | rs143095746 | 0.0004 | 17.14 | D | B | D | L | Het | |
| 3 | GPX1 | GCC | - | c.33_35del | p.11_12del | NM_001329455 | rs17838762 | - | - | - | - | - | - | Het |
CADD: Combined Annotation Dependent Depletion, https://cadd.gs.washington.edu/, accessed on 4 February 2019; ExAC: Exome Aggregation Consortium, http://exac.broadinstitute.org/, accessed on 4 February 2019; SIFT: Sorting intolerant from tolerant, https://sift.bii.a-star.edu.sg/, accessed on 4 February 2019; Mutation Taster: Mutation impact testing, http://www.mutationtaster.org/, accessed on 4 February 2019.
Figure 2.
Pedigree of vitiligo family and identified causative mutation. (A) Polygenic pattern of disease-causing alleles segregating in multiplex Pakistani vitiligo family. Black-filled and empty symbols represent affected and unaffected individuals, respectively. Symbols filled in gray represent individuals that recovered (<5%) from vitiligo. Double line shows only consanguineous marriages. The genotypes (wild type, heterozygous) of the identified mutant alleles are also shown for each of the participating family members. Arrow sign represents the proband used for whole exome sequencing. (B) Vitiligo patches were observed in the affected individuals of VF-12 family. (C) Clustal-W multiple amino acid sequence alignments of orthologous proteins show evolutionarily conserved mutated residues across different species.
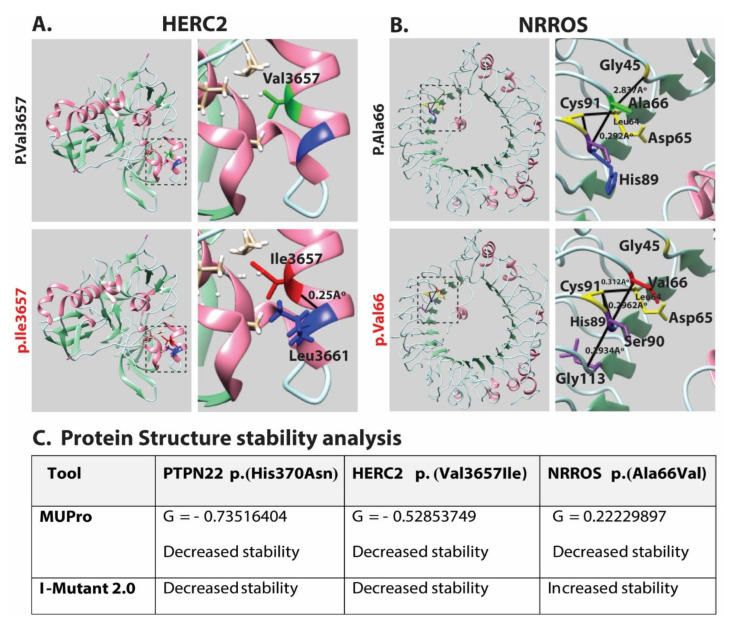
Amino acid conservation studies have shown that the variants identified in PTPN22, HERC2 and NRROS genes are present in the evolutionarily conserved regions of the encoded proteins (Figure 2C). Next, we performed molecular modeling analysis using the Phyre2 online resource for retrieving structural information based on protein homology present in the protein database. Using this approach, we were able to model the two missense variants found in the HERC2 and NROSS proteins (Figure 3). The lack of available crystalline structural information for PTPN22 restrained further analysis from predicting the effect of the mutation on protein structure and function. A different pattern of interresidual bonding interaction between wild type and mutant protein was observed for the p.Val 3657Ile variant of HERC2 (Figure 3A). The modeling data predicted that p.Val3657Ile substitution might impact protein folding due to the formation of an additional H-bond between p.Ile3657 and p.Leu3661 (Figure 3A). Similarly, the p.Ala66Val variant of NRROS is predicted to greatly hamper the microscopic interaction in the surrounding area of 10 Å by differential H-bonding patterns (Figure 3B). Surrounding the p.Ala66 residue in WT NRROS, there are four prominent hydrogen bonds: between p.Gly45 and p.Asp65 (2.837 Å); p.Cys91 and Leu64 (0.3129 Å); p.Cys91 and p.His89 (0.374 Å); and p.Ala66 and p.His89 (2.921 Å; Figure 3B). In contrast, the p.Ala66Val substitution exhibits a different set of H-bonding capabilities, resulting in the loss of H-bonds between p.Gly45 and p.Asp65 and the formation of two new H-bond bonds between p.Val66 and p.His89 (2.962 Å), and between p.Gly113 and p.Ser90 (2.293 Å; Figure 3B). This new pattern of interaction may induce torsion, leading to improper protein folding and properties, which may have an effect on function. A noticeable decrease in Gibbs free energy was recorded in PTPN22 (−0.73516404), HERC2 (−0.52853749) and NRROS (−0.22229897) by MUpro, indicating a decrease in protein 3D structure stability, as corroborated by the I-Mutant prediction program. Overall, the three identified variants have been anticipated to have deleterious effects on protein structures, rendering the proteins unstable (Figure 3C).
Figure 3.
Three-dimensional protein structural modeling encoded from variants identified in the current study. Structures of both wild type (shown in green) and mutated residues (shown in red) are highlighted. Solid black lines represent the interactions with neighboring amino acids. Stick representation in green shows wild type residue, while red color shows mutated amino acid. Helices are shown in magenta, sheets in light green and loops in cyan color. (A) Missense variant in HERC2 p.(Val3657Ile), wild type residue is relatively smaller in size than the mutant. Mutant residue p.(Ile 3657), shown in red, forms H-bonds with neighboring p.(Leu 3661), shown in blue, and bond length is 0.2561 Å, which may have an impact on protein folding and functionality. (B) Similarly, for missense variant of NRROS p.(Ala66Val), wild type residue p.(Ala66), shown in green, forms H-bonds with p.(His89), shown in blue. Neighboring residue, shown in yellow with solid black line, represents H-bonds in the wild type. Mutant model represents different sets of interactions in the adjacent region of mutation. p.(Val66), shown in red, forms H-bonds with p.(His89), shown in blue, with different bond lengths causing loss of H-bonding between p.(Gly45) and p.(Asp65), shown in yellow, in wild type. Formation of new intra residual H-bonding between p.(Ser90) and p.(Gly113) is shown in violet color. (C) The underlying mutations negatively impacted protein structures, and the encoded proteins were predicted to become unstable by MUpro and I-mutant.
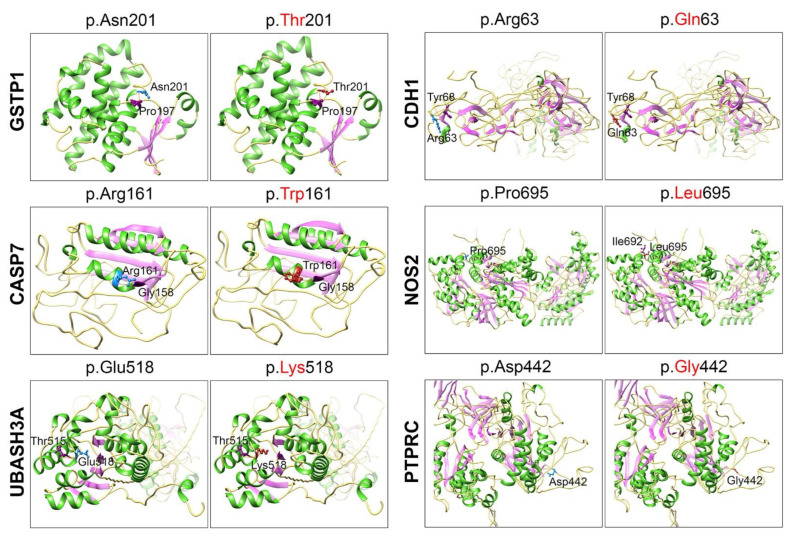
Using this in silico approach, we also modeled six missense variants found in the affected individuals of our vitiligo families (Table 2) and predicted their potential pathogenic impact on the encoded proteins (Figure 4). These include p.Asn201Thr (GSTP1), p.Arg63Gln (CDH1), p.Arg161Trp (CASP7), p.Pro695Leu (NOS2), p.Glu518Lys (UBASH3A), and p.Asp442Gly (PTPRC) variants (Figure 4). The p.Asn201Thr variant of GSTP1 is located within the GST C-terminal domain, and due to different sizes and ionic properties, it might impact the folding of the domain (Figure 4). A similar impact was predicted for the p.Arg63Gln variant of CDH1, p.Arg161Trp of CASP7, and p.Asp442Gly of PTPRC (Figure 4). The p.Pro695Leu of NOS2 is predicted to form a new H-bond with p.Ile692 (Figure 4). Moreover, the WT Proline residue is known to be very rigid and therefore induce a special backbone conformation; therefore, substitution with Leucine would affect this conformation. Finally, the p.Glu518Lys variant of UBASH3A is predicted to alter the salt bridge with Arginine at position 406 and Arginine at position 485 (Figure 4), and thus it ultimately affects protein folding and function. Overall, the above missense variants are predicted to have an impact on protein folding and secondary structure, which might lead to impaired function.
Figure 4.
WT residue is shown in dodger blue color, mutant in firebrick (red) and hydrogen-bond (orange color line) associated amino acids in dark magenta. Amino acid atoms are shown in ball and stick forms. Secondary structure: Helix: Green strands: pink-purple and coils: golden-yellow in color.
4. Discussion
Vitiligo is a disease of complex etiology, whereby the involvement of genetic contributors in disease pathogenesis has been emphasized greatly since the advent of modern genetic technologies. Previously, several genetic association studies have identified risk alleles responsible for various types of vitiligo in different ethnicities [17,19,20,21,22,23,24]. In the present study, we reported clinical and genetic findings in fifteen highly inbred consanguineous Pakistani families. Our study highlights the genetic heterogeneity of the disease in terms of risk allele clustering as well as associated risk allele burden in affected individuals. We report the identification of several rare variants of known vitiligo-associated genes, including three novel missense variants in PTPN22 (c.1108C>A; p.(His370Asn)), NRROS (c.197C>T; p.(Ala66Val)) and HERC2 (c.10969G>A; p. (Val3657Ile)) in family VF-12. In silico analysis of the risk variants for pathogenicity determination predicted a low to moderate pathogenic impact of all the identified variants; however, clustering of mutated alleles in an individual collectively contributed to the onset of vitiligo.
Autoimmune-mediated melanocyte destruction is a common phenomenon in vitiligo pathogenesis that involves intrinsic defects within melanocytes and an autoimmune system that targets these cells [25]. PTPN22, which encrypts genetic information for lymphoid-specific intracellular phosphatase, is the main inhibitor of T-cell activation [19]. Many autoimmune diseases, including type 1 diabetes, arthritis, systemic lupus and vitiligo, have also been previously associated with PTPN22 variants [4]. NSV was found to be strongly associated with PTPN22 (c.1858C>T; p. (Arg620Trp)) in the cohorts from the United Kingdom, Gujarat, India and Romania in three different case-control studies [15,26]. Similarly, HERC2, which encodes for the E3 ubiquitin-protein, provides protection against ionizing radiation and DNA damage related to radiation. Previous studies have shown that an elevated risk of vitiligo is associated with HERC2 variations, which have a significant impact on determining skin and iris color besides the OCA2. Previous studies on Caucasian and Chinese Han cohorts also reported a significant association between the T allele of rs6583331 SNP of NRROS and vitiligo [17]. However, in our cohort, a novel vitiligo-associated variant of NRROS was identified. Our outcomes are in agreement with previous findings, thus indicating an increased risk for intra-familial NSV development linked to NRROS.
Besides the above-mentioned genes, as shown in Table 2, the majority of the variants identified in our cohort are present in proteins that function in innate immune responses, inflammation reaction, control of cell apoptosis, anti-oxidative damage, and have a role in detoxification reactions. For instance, IFIH1 is an interferon-induced helicase C domain-containing protein, an innate immune receptor that acts as a cytoplasmic sensor of viral nucleic acids and plays a major role in sensing viral infection and in the activation of a cascade of antiviral responses, including the induction of type I interferons and pro-inflammatory cytokines [25]. The IL1RAPL1-Interleukin-1 receptor accessory protein-like 1 regulates secretion and presynaptic differentiation through inhibition of the activity of the N-type voltage-gated calcium channel and is also involved in the activation of the MAP kinase JNK. TLR4 is a transmembrane protein, a member of the Toll-like receptor family, which belongs to the pattern recognition receptor (PRR) family. TLR4 participates in intracellular signaling NF-κB and inflammatory cytokine production, which is responsible for activating the innate immune system. CASP7 encodes a member of the Cysteine-Aspartic acid protease (caspase) family and plays a central role in the execution phase of cell apoptosis and survival of melanocytes [4]. Similarly, the BCL2L12 protein family and their interactors include inhibitors and inducers of cell death, regulate and mediate the process by which mitochondria contribute to cell death known as the intrinsic apoptosis pathway, and thus regulate the survival of pigmented cells.
In conclusion, the convergence theory of vitiligo development bridges all previously known etiologies and describes vitiligo as a multivariate, complex, and multifactorial disorder [9]. Vitiligo is not caused merely by the presence of an underlying genetic variation or only by exposure to some specific external factor such as stimuli or defective melanocytes and an internal autoimmune condition but is a result of an intricate combination of all these etiological contributors. Therefore, it is important to acknowledge that the underlying molecular pathways affected in one patient do not always lead to the same level of pathogenicity in other individuals. Such attributes of vitiligo define that identification of individual-specific pathways is significant for the targeted development of more effective therapeutics [26]. Polygenic Risk Score (PRS) estimation is a trending approach for the genetic description of complex disorders, including vitiligo. However, there always remains quite a fair chance for the involvement of additional genes or novel variants in the known vitiligo-associated genes. In this regard, it has been recently described that the phenomenon of clustering of risk alleles in an inbred family increases polygenic risk burden [5], and our study further supports this notion.
Acknowledgments
We are thankful to all the participants of the study and health care professionals for their cooperation in clinical investigations. We are very thankful to Muhammad Naeem, Fahmida (Quaid-e-Azam University, Islamabad, Pakistan) and Ehsanullah (National Institute of eye, NIH, USA) for their technical support.
Author Contributions
Conceptualization, P.A.S. and Z.M.A.; methodology, P.A.S.; software, R.I. and U.H.; validation, U.H., M.I. and R.I.; formal analysis, R.I.; investigation, R.I. and M.N.u.A.; resources, Z.M.A., S.S. and G.K.R.; data curation, R.I. and U.H.; writing—original draft preparation, R.I. and Z.M.A.; writing—review and editing, R.I. and Z.M.A.; visualization, R.I.; supervision, P.A.S. and Z.M.A.; project administration, P.A.S.; funding acquisition, P.A.S. and Z.M.A. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
This study was conducted in accordance with the Declaration of Helsinki, and approved by the Institutional Review Board/Ethics committee of Pir Mehr Ali Shah Arid Agriculture University, Rawalpindi (No. PMAS-AAUR/IEC/15 dated 11 October 2017).
Informed Consent Statement
Informed consent was obtained from all subjects included in the study.
Data Availability Statement
The data presented in this study are available on request from Corresponding authors. The data is not publically available due to confidentiality of patient’s information.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This study was partially supported by International Research Support Initiative Program, Higher Education Commission of Pakistan and by National Institute of Arthritis and Musculoskeletal and Skin Disease, National Institutes of Health, Bethesda, MD, USA (R01AR077563 to ZMA).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Berqvist C., Ezzedine K. Vitiligo: A Review. Dermatology. 2020;236:571–592. doi: 10.1159/000506103. [DOI] [PubMed] [Google Scholar]
- 2.Garcia-Melendez M.E., Salinas-Santander M., Sanchez-Dominguez C., Gonzalez-Cardenas H., Cerda-Flores R.M., Ocampo-Candiani J., Ortiz-López R. Protein tyrosine phosphatase PTPN22 +1858C/T polymorphism is associated with active vitiligo. Exp. Med. 2014;8:1433–1437. doi: 10.3892/etm.2014.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spritz R.A. Recent progress in the genetics of generalized vitiligo. J. Genet. Genom. 2011;38:271–278. doi: 10.1016/j.jgg.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Czajkowski R., Mecinska-Jundzill K. Current aspects of vitiligo genetics. Postep. Derm. Alergol. 2014;31:247–255. doi: 10.5114/pdia.2014.43497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roberts G.H.L., Paul S., Yorgov D., Santorico S.A., Spritz R.A. Family Clustering of Autoimmune Vitiligo Results Principally from Polygenic Inheritance of Common Risk Alleles. Am. J. Hum. Genet. 2019;105:364–372. doi: 10.1016/j.ajhg.2019.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim S.M., Chung H.S., Hann S.-K. The genetics of vitiligo in Korean patients. Int. J. Dermatol. 1998;37:908–910. doi: 10.1046/j.1365-4362.1998.00549.x. [DOI] [PubMed] [Google Scholar]
- 7.Laddha N.C., Dwivedi M., Mansuri M.S., Gani A.R., Ansarullah M., Ramachandran A.V., Dalai S., Begum R. Vitiligo: Interplay between oxidative stress and immune system. Exp. Derm. 2013;22:245–250. doi: 10.1111/exd.12103. [DOI] [PubMed] [Google Scholar]
- 8.Yu S., Doycheva D.M., Gamdzyk M., Yang Y., Lenahan C., Li G., Li D., Lian L., Tang J., Lu J., et al. Activation of MC1R with BMS-470539 attenuates neuroinflammation via cAMP/PKA/Nurr1 pathway after neonatal hypoxic-ischemic brain injury in rats. J. Neuroinflamm. 2021;18:26. doi: 10.1186/s12974-021-02078-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alikhan A., Felsten L.M., Daly M., Petronic-Rosic V. Vitiligo: A comprehensive overview Part I. Introduction, epidemiology, quality of life, diagnosis, differential diagnosis, associations, histopathology, etiology, and work-up. J. Am. Acad. Derm. 2011;65:473–491. doi: 10.1016/j.jaad.2010.11.061. [DOI] [PubMed] [Google Scholar]
- 10.Noman M., Ishaq R., Bukhari S.A., Ahmed Z.M., Riazuddin S. Delineation of Homozygous Variants Associated with Prelingual Sensorineural Hearing Loss in Pakistani Families. Genes. 2019;10:1031. doi: 10.3390/genes10121031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Richard E.M., Santos-Cortez R.L.P., Faridi R., Rehman A.U., Lee K., Shahzad M., Acharya A., Khan A.A., Imtiaz A., Chakchouk I., et al. Global genetic insight contributed by consanguineous Pakistani families segregating hearing loss. Hum. Mutat. 2019;40:53–72. doi: 10.1002/humu.23666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Riazuddin S., Hussain M., Razzaq A., Iqbal Z., Shahzad M., Polla D.L., Song Y., van Beusekom E., Khan A.A., Tomas-Roca L., et al. Exome sequencing of Pakistani consanguineous families identifies 30 novel candidate genes for recessive intellectual disability. Mol. Psychiatry. 2017;22:1604–1614. doi: 10.1038/mp.2016.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jaworek T.J., Kausar T., Bell S.M., Tariq N., Maqsood M.I., Sohail A., Ali M., Iqbal F., Rasool S., Riazuddin S., et al. Molecular genetic studies and delineation of the oculocutaneous albinism phenotype in the Pakistani population. Orphanet. J. Rare Dis. 2012;7:44. doi: 10.1186/1750-1172-7-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Debasis d., Vivek, Manchanda, Singh P., Ankita, Gupta I. A Compendium of Genes and Variations Associated with Vitiligo. 2023. [(accessed on 12 March 2023)]. Available online: https://vitivar.igib.res.in/genes.php.
- 15.Canton I., Akhtar S., Gavalas N.G., Gawkrodger D.J., Blomhoff A., Watson P.F., Weetman A.P., Kemp E.H. A single-nucleotide polymorphism in the gene encoding lymphoid protein tyrosine phosphatase (PTPN22) confers susceptibility to generalised vitiligo. Genes Immun. 2005;6:584–587. doi: 10.1038/sj.gene.6364243. [DOI] [PubMed] [Google Scholar]
- 16.Passeron T., Ortonne J.P. Activation of the unfolded protein response in vitiligo: The missing link? J. Investig. Derm. 2012;132:2502–2504. doi: 10.1038/jid.2012.328. [DOI] [PubMed] [Google Scholar]
- 17.Tang X., Fang F., Yang J., Zheng X., Fan M., Wang L., Zhang A. Association Study Reveals One Susceptibility Locus with Vitiligo in the Chinese Han Population. Genet. Test. Mol. Biomark. 2019;23:791–796. doi: 10.1089/gtmb.2019.0129. [DOI] [PubMed] [Google Scholar]
- 18.Liu J., Zhang Z., Chai L., Che Y., Min S., Yang R. Identification and characterization of a unique leucine-rich repeat protein (LRRC33) that inhibits Toll-like receptor-mediated NF-κB activation. Biochem. Biophys. Res. Commun. 2013;434:28–34. doi: 10.1016/j.bbrc.2013.03.071. [DOI] [PubMed] [Google Scholar]
- 19.Rajendiran K.S., Rajappa M., Chandrashekar L., Thappa D.M. Association of PTPN22 gene polymorphism with non-segmental vitiligo in South Indian Tamils. Postep. Derm. Alergol. 2018;35:280–285. doi: 10.5114/ada.2018.76225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheong K.A., Kim N.H., Noh M., Lee A.Y. Three new single nucleotide polymorphisms identified by a genome-wide association study in Korean patients with vitiligo. J. Korean Med. Sci. 2013;28:775–779. doi: 10.3346/jkms.2013.28.5.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rodrigues M., Ezzedine K., Hamzavi I., Pandya A.G., Harris J.E., Vitiligo Working Group New discoveries in the pathogenesis and classification of vitiligo. J. Am. Acad. Dermatol. 2017;77:1–13. doi: 10.1016/j.jaad.2016.10.048. [DOI] [PubMed] [Google Scholar]
- 22.Laddha N.C., Dwivedi M., Shajil E.M., Prajapati H., Marfatia Y.S., Begum R. Association of PTPN22 1858C/T polymorphism with vitiligo susceptibility in Gujarat population. J. Derm. Sci. 2008;49:260–262. doi: 10.1016/j.jdermsci.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 23.Bekker-Jensen S., Rendtlew Danielsen J., Fugger K., Gromova I., Nerstedt A., Lukas C., Bartek J., Lukas J., Mailand N. HERC2 coordinates ubiquitin-dependent assembly of DNA repair factors on damaged chromosomes. Nat. Cell. Biol. 2010;12:80–86. doi: 10.1038/ncb2008. [DOI] [PubMed] [Google Scholar]
- 24.Jin Y., Birlea S.A., Fain P.R., Ferrara T.M., Ben S., Riccardi S.L., Cole J.B., Gowan K., Holland P.J., Bennett D.C., et al. Genome-wide association analyses identify 13 new susceptibility loci for generalized vitiligo. Nat. Genet. 2012;44:676–680. doi: 10.1038/ng.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jin Y., Andersen G.H.L., Santorico S.A., Spritz R.A. Multiple Functional Variants of IFIH1, a Gene Involved in Triggering Innate Immune Responses, Protect against Vitiligo. J. Invest. Derm. 2017;137:522–524. doi: 10.1016/j.jid.2016.09.021. [DOI] [PubMed] [Google Scholar]
- 26.Kundu R.V., Mhlaba J.M., Rangel S.M., Le Poole I.C. The convergence theory for vitiligo: A reappraisal. Exp. Dermatol. 2019;28:647–655. doi: 10.1111/exd.13677. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data presented in this study are available on request from Corresponding authors. The data is not publically available due to confidentiality of patient’s information.