Abstract
The Saccharomyces cerevisiae mRNA capping enzyme consists of two subunits: an RNA 5′-triphosphatase (Cet1) and an mRNA guanylyltransferase (Ceg1). In yeast, the capping enzyme is recruited to the RNA polymerase II (Pol II) transcription complex via an interaction between Ceg1 and the phosphorylated carboxy-terminal domain of the Pol II largest subunit. Previous in vitro experiments showed that the Cet1 carboxy-terminal region (amino acids 265 to 549) carries RNA triphosphatase activity, while the region containing amino acids 205 to 265 of Cet1 has two functions: it mediates dimerization with Ceg1, but it also allosterically activates Ceg1 guanylyltransferase activity in the context of Pol II binding. Here we characterize several Cet1 mutants in vivo. Mutations or deletions of Cet1 that disrupt interaction with Ceg1 are lethal, showing that this interaction is essential for proper capping enzyme function in vivo. Remarkably, the interaction region of Ceg1 becomes completely dispensable when Ceg1 is substituted by the mouse guanylyltransferase, which does not require allosteric activation by Cet1. Although no interaction between Cet1 and mouse guanylyltransferase is detectable, both proteins are present at yeast promoters in vivo. These results strongly suggest that the primary physiological role of the Ceg1-Cet1 interaction is to allosterically activate Ceg1, rather than to recruit Cet1 to the Pol II complex.
Eukaryotic and viral mRNAs are modified at their 5′ end by a cap structure which consists of a 7-methylguanosine moiety attached to the 5′ terminus via a 5′-5′ linkage. Cellular mRNA capping enzyme is a bifunctional enzyme: RNA 5′-triphosphatase removes the γ-phosphate from the 5′ end of the RNA substrate to leave a diphosphate end, and GTP::mRNA guanylyltransferase subsequently transfers GMP from GTP to the 5′-diphosphate RNA end. A separate enzyme, RNA (guanine-7-)-methyltransferase, adds a methyl group to the N-7 position of the guanine cap to leave m7GpppN1- (35).
Capping enzyme from Saccharomyces cerevisiae is a heterodimer of RNA triphosphatase and guanylyltransferase subunits (20) encoded by the CET1 and CEG1 genes, respectively. Both genes are essential for cell viability (34, 39). CET1 and CEG1 homologs from Schizosaccharomyces pombe and Candida albicans functionally replace the S. cerevisiae genes (36, 42, 44). The fungal guanylyltransferase subunits have amino acid similarity to viral and metazoan guanylyltransferases, indicating a common reaction mechanism (11, 41). In contrast to the two subunit yeast enzymes, capping enzyme from higher eukaryotes are a single polypeptide consisting of an amino-terminal RNA triphosphatase domain and a carboxy-terminal guanylyltransferase domain. Both mouse (MCE or MCE1) and human (HCE or HCE1) enzymes can replace CEG1 and CET1 in vivo (16, 17, 24, 43, 47). Interestingly, the higher eukaryotic RNA triphosphatase domains belong to the PTP (protein tyrosine phosphatase) superfamily (27, 38, 47) and do not resemble the fungal phosphatases (39, 44).
Cellular capping enzymes are recruited to the phosphorylated carboxy-terminal domain (CTD-P) of the RNA polymerase II (Pol II) largest subunit (2, 27, 47). In vitro studies showed that Ceg1 directly binds to CTD-P (3, 27), while Cet1 does not (3). Surprisingly, covalent enzyme-GMP complex formation by Ceg1 is inhibited by binding to CTD-P, and Cet1 is required to reactivate Ceg1 (3). In the mammalian system, the guanylyltransferase domain interacts with CTD-P (17, 47), whereas the RNA triphosphatase domain does not (17).
The RNA 5′-triphosphatase activity of Cet1 is carried in its C-terminal region (amino acids 265 to 549) (16, 30, 39). Previously, our in vitro results showed that residues 205 to 265 of Cet1 are necessary and sufficient for both the interaction with Ceg1 and the allosteric activation of Ceg1 on CTD-P (3). Here we further analyze this interaction in vivo. We find that the region of Cet1 that interacts with Ceg1 is normally essential but becomes dispensable if Ceg1 is replaced by the mouse guanylyltransferase. This strongly suggests that the Ceg1-Cet1 interaction is essential for the positive allosteric activation of Ceg1 but not for delivering Cet1 to the transcription complex.
MATERIALS AND METHODS
DNA cloning methods.
Supplementary tables describing the construction of plasmids and oligonucleotides used in this study are available from the Buratowski Lab website (http://tfiib.med.harvard.edu/pubs/Takase.html). Standard methods were used. Yeast plasmids were based on the pRS series (37). PCRs for the construction of plasmids and site-directed mutagenesis were carried out with Vent DNA polymerase (New England BioLabs).
Genetic manipulations of S. cerevisiae.
Table 1 lists the S. cerevisiae strains used in this study. Plasmids were introduced into yeast using a modified lithium acetate transformation protocol (8). Medium preparation, plasmid shuffling, and other yeast manipulations were performed by standard methods (1, 10).
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| YSB455 | MATa/MATα ura3-52/ura3-52 leu2Δ1/leu2Δ1 trp1Δ63/trp1Δ63 his3Δ200/his3Δ200 lys2Δ202/lys2Δ202 | 30 |
| YSB532 | MATα ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2Δ202 cet1Δ1::TRP1 (pRS316-CET1) | This study |
| YSB533 | MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2Δ202 cet1Δ1::TRP1 (pRS316-CET1) | This study |
| YSB540 | MATα ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2Δ202 cet1Δ1::TRP1 (pRS313-CET1) | This study |
| YSB709 | MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2Δ202 cet1Δ1::TRP1 (pRS315-CET1) | This study |
| YSB710 | MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2Δ202 cet1Δ1::TRP1 [pRS315-CET1(pro+205–549)] | This study |
| YSB711 | MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2Δ202 cet1Δ1::TRP1 [pRS315-CET1(pro+220–549)] | This study |
| YSB712 | MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2Δ202 cet1Δ1::TRP1 [pRS315-CET1(pro+235–549)] | This study |
| YSB713 | MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2Δ202 cet1Δ1::TRP1 [pRS425-CET1(pro+235–549)] | This study |
| YSB715 | MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2Δ202 cet1Δ1::TRP1 (pRS313-cet1-401) | This study |
| YSB717 | MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2Δ202 cet1Δ1::TRP1 (pRS313-cet1-438) | This study |
| YSB718 | MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2Δ202 cet1Δ1::TRP1 (pRS313-cet1-448) | This study |
| YSB719 | MATα ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2Δ202 cet1Δ1::TRP1 ceg1Δ3::LYS2 (pRS316-CEG1-CET1) | This study |
| YSB230 | MATα ura3-52 leu2-3,112 his3Δ200 ceg1Δ1::HIS3 (pRS315-ceg1-63) | 7 |
| HF7c | MATa ura3-52 leu2-3,112 trp1-901 his3Δ200 ade2-101 lys2-801 gal4-542 gal80-538 LYS2::GAL1UAS-GAL1TATA-HIS3 URA3::GAL417(mer × 3)-CYC1TATA-LacZ | 5 |
To construct the strain containing the CET1 disruption, the diploid strain YSB455 was transformed with a 2.5-kb NotI/EcoRI fragment of pRS316-cet1Δ1 (CEN/ARS, URA3) in which the portion encoding N-terminal region (amino acids 1 to 265) of CET1 open reading frame was replaced with TRP1 gene. In sporulating and dissecting tetrads of Trp+ transformants, viability segregated 2:2, and all viable spores were Trp−. The diploid carrying the cet1Δ::TRP1 locus was transformed with pRS316-CET1 and sporulated to create haploid strains YSB532 and YSB533.
A CET1 CEG1 double disruption strain was created as follows: a 4.8-kb EcoRI/SalI fragment from pBS-LYS2 containing the LYS2 gene was subcloned into the EcoRI and XhoI sites of pBSKS(+)(ΔRI-X)-CEG1 (6). The resulting plasmid [pBSKS(+)(ΔRI-X)-ceg1Δ3::LYS2] releases a 5.5-kb fragment of ceg1Δ3::LYS2 with NotI/PstI digestion. YSB532 was transformed with pRS313-CET1, and transformants were selected which were His+ and resistant to 5-fluoro-orotic acid (5-FOA). This strain (YSB540) was then transformed with pRS316-CEG1 (6) and the 5.5-kb fragment from pBSKS(+)(ΔRI-X)-ceg1Δ3::LYS2, and transformants were selected which were Trp+, Ura+, and Lys+ and 5-FOA sensitive. Then, pRS315-CEG1 (6) was introduced, and pRS316-CEG1 was shuffled out with 5-FOA. Finally, pRS316-CEG1-CET1 was used for transformation, and Trp+ Lys+ Ura+ His− Leu− cells were selected to generate YSB719.
Yeast two-hybrid assays were carried out with strain HF7c using the GAL4-dependent HIS3 reporter gene (5). pAS1 plasmid was used for expression of Gal4 DNA binding domain fusions (4). Either pGAD-C1 (21) or pY2 (31) was used for Gal4 activation domain fusions. Leu+ Trp+ transformants were selected and tested for their viability in the absence of histidine on plates containing 1 mM 3-aminotriazole.
Site-directed mutagenesis.
Site-directed mutagenesis was performed using a PCR-mediated method (14) with three mutagenic primers: CET1P238/Y241mut, CET1P245/W247mut, and CET1W251/P253mut.
To introduce the PCR product into yeast, a single-step method based on gap repair was used (13, 28). The mutagenized DNA was transformed into YSB533 together with a 7.6-kb BglII/BglII fragment of pRS315-CET1 which removes much of the coding region but carries overlap with each end of the PCR product to allow recombination. After transformation, the wild-type CET1/URA3 plasmid was shuffled out on medium containing 5-FOA. Plasmid DNA was isolated from 5-FOA resistant cells and sequenced to confirm the mutations.
Isolation of cet1 conditional alleles by random mutagenesis.
The CET1 gene was randomly mutagenized by a PCR-based misincorporation method (28). Reactions contained 1 U of Taq DNA polymerase, 0.3 μM CET1-A and CET1-B as primers, 0.25 mM MnCl2, and biased deoxynucleoside triphosphate concentrations (0.4 mM dGTP, dCTP, and dTTP and 0.1 mM dATP). The reaction was cycled 30 times for 1 min at 94°C, 1 min at 57°C, and 2 min at 72°C.
The mutagenized DNA was transformed into YSB533 together with a 7.6-kb BglII/BglII fragment of pRS315-CET1 as described above. After shuffling out the wild-type CET1 plasmid, FOA-resistant cells were replica plated at 16, 30, and 37°C to screen for heat sensitivity and cold sensitivity. Plasmid DNA was isolated from heat- and cold-sensitive mutants and retransformed to YSB533 to confirm plasmid linkage. Several tight alleles were selected for further analysis and sequenced.
Preparation of antibodies.
Anti-Cet1 polyclonal antiserum was raised in a rabbit by immunizing with a bacterially produced carboxy-terminal region of Cet1 (amino acids 265 to 549) fused with polyhistidine tag and 13 extra residues at the amino terminus [His7-Cet1(265–549) (3)]. The protein was purified by chromatography over Ni2+-nitrilotriacetic acid (NTA) agarose (Qiagen) and S-Sepharose FF (Pharmacia).
Anti-Abd1 polyclonal antiserum was raised in a rabbit by immunizing with a bacterially produced polyhistidine-tagged Abd1 protein. His7-Abd1 was purified from the soluble fraction by Ni2+-NTA agarose chromatography (26) and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (12).
For immunoaffinity purification of anti-Cet1 antibody, Cet1(265–549) fused to glutathione S-transferase was expressed in BL21(DE3) cells using pSBET-GEXCT-CET1(265–549), a derivative of pSBET and pETGEXCT (32, 33). Lysates were prepared by sonication in buffer B (Tris-HCl, pH 8.0; 300 mM KCl; 1 mM EDTA; 1 mM dithiothreitol [DTT]; 1 mM phenylmethylsulfonyl fluoride [PMSF]; 0.5% [vol/vol] NP-40). Purification and cleavage of the fusion protein was carried out as described earlier (9), with the following modifications. Soluble extract (100,000 × g supernatant fraction) were incubated in batch with glutathione agarose (Sigma) for 2 h at 4°C on a rotator. The resin was successively washed with about 5 bed volumes of PBST and 1 bed volume of buffer C (50 mM Tris-HCl, pH 8.0; 150 mM KCl; 2.5 mM CaCl2; 1 mM DTT; 1 mM PMSF). Beads were then resuspended in buffer C (50% [vol/vol]), and 5 μg of thrombin (Calbiochem) was added. The cleavage reaction was for 40 h at 4°C on a rotator. The resin was then poured into a column and eluted with 2 bed volumes of buffer C. Cleaved Cet1(265–549) was dialyzed against buffer D (20 mM Tris-HCl, pH 7.5; 20 mM KCl; 1 mM EDTA; 1 mM DTT; 1 mM PMSF). The protein was crosslinked to CNBr-activated Sepharose 4B (Pharmacia), and antibody was immunopurified as described elsewhere (12).
Yeast whole-cell extract preparation and protein analysis.
Whole-cell extracts from S. cerevisiae were prepared by glass bead disruption of cells in lysis buffer (20 mM Tris-HCl, pH 7.9; 1 mM EDTA; 200 mM KCl; 10% [vol/vol] glycerol; 1 mM DTT; 1 mM PMSF). Equivalent amounts of protein from each sample were then subjected to either immunoblot analysis with enhanced chemiluminescence detection or enzyme-GMP formation assay with 3 μM [α-32P]GTP (30,000 to 60,000 cpm/pmol; NEN) (6).
Immunoprecipitation was carried out as described previously (2) with minor modifications. Forty micrograms of yeast whole-cell extract protein was incubated for 1 h at room temperature with antibody in binding buffer (20 mM Tris-HCl, pH 7.5; 1 mM EDTA; 50 mM KCl; 20% [vol/vol] glycerol; 1 mM DTT; 1 mM PMSF; 0.05% [vol/vol] NP-40; 500 ng of bovine serum albumin per μl) with a 10-μl bed volume of protein A-Sepharose CL-4B (Pharmacia). The precipitate was washed three times (1 ml each) with washing buffer (20 mM Tris-HCl, pH 7.5; 50 mM KCl; 0.1% [vol/vol] NP-40) and once with 1 ml of reaction buffer (20 mM Tris-HCl, pH 7.5; 5 mM MgCl2). The beads were then resuspended in 20 μl of enzyme-GMP complex assay buffer (20 mM Tris-HCl, pH 7.5; 5 mM MgCl2; 5 mM DTT; 3 μM [α-32P]GTP), and the reaction mixture was incubated for 15 min at 30°C. The labeled guanylyltransferase-GMP complex was resolved by SDS-PAGE and visualized by using a phosphorimager.
Chromatin immunoprecipitation.
Preparation of chromatin from S. cerevisiae, immunoprecipitation, and quantitative analysis of precipitated DNA were carried out as described previously (22, 23).
RESULTS
Deletion analysis of Cet1 in vivo.
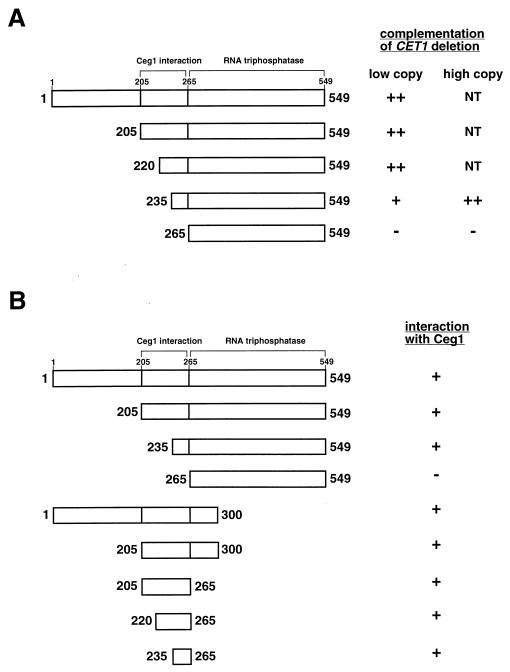
We constructed a truncation series in Cet1 and tested them for function in vivo by plasmid shuffling (Fig. 1A). Each mutant was expressed under the control of the native CET1 promoter on a centromeric plasmid. CET1(205–549) supported cell viability with no apparent difference in growth from wild-type cells (colonies formed after 2 days). Therefore, the N-terminal 205 amino acids of Cet1 are dispensable in vivo, in agreement with the findings of Ho et al. (16). In contrast, CET1(265–549) cells were unable to grow even though this region is sufficient for its enzymatic activity (30). Amino acids 205 to 265 are required for interaction with Ceg1 in vitro (3), so it appears this interaction is essential for cell viability.
FIG. 1.
Deletion analysis of Cet1. (A) Complementation by plasmid shuffling. CET1 shuffling strain YSB533 was transformed with low-copy-number (CEN/ARS) or high-copy-number (2μm) plasmids carrying full-length or truncated versions of Cet1 as indicated. Transformants were tested for complementation on plates containing 5-FOA incubated at 30°C. Scoring: ++, grew like wild-type (colonies formed after 2 days); +, formed colonies after 4 days; −, formed no colonies after 7 days; NT, not tested. (B) Interaction with Ceg1 as tested by yeast two-hybrid assay. The reporter strain HF7c bearing a Gal4-binding domain-Ceg1 fusion (pG4BD-CEG1orf) was transformed with plasmids encoding the indicated fragments of Cet1 fused to the Gal4 transactivation domain. Transformants were tested for activation of the Gal4 responsive-His3 fusion reporter by the ability to grow on plates lacking histidine and containing 1 mM 3-aminotriazole. The plates were scored after 2 days at 30°C.
To further define the minimal region of the interaction site, we tested additional deletion mutants. CET1(220–549) cells grow indistinguishably from wild-type CET1 cells. CET1(235–549) cells grew slowly, forming colonies after 4 days. However, when this mutant was overexpressed from a high-copy-number plasmid, colonies formed after 2 days. CET1(265–549) did not complement growth even when overexpressed. Therefore, the minimal region of Cet1 necessary for the interaction with Ceg1 appears to be amino acids 235 to 265. To verify this conclusion, we used a yeast two-hybrid assay. Ceg1 protein fused to the DNA binding domain of Gal4 protein was coexpressed with Cet1 fragments fused to the activation domain of Gal4 and tested for the ability to activate a Gal4-dependent reporter gene (Fig. 1B). Cet1(265–549) did not interact with Ceg1, confirming our in vitro results (3). However, a fusion protein containing residues 235 to 265 was able to interact with Ceg1.
Mutational analysis of the Ceg1 interaction region of Cet1.
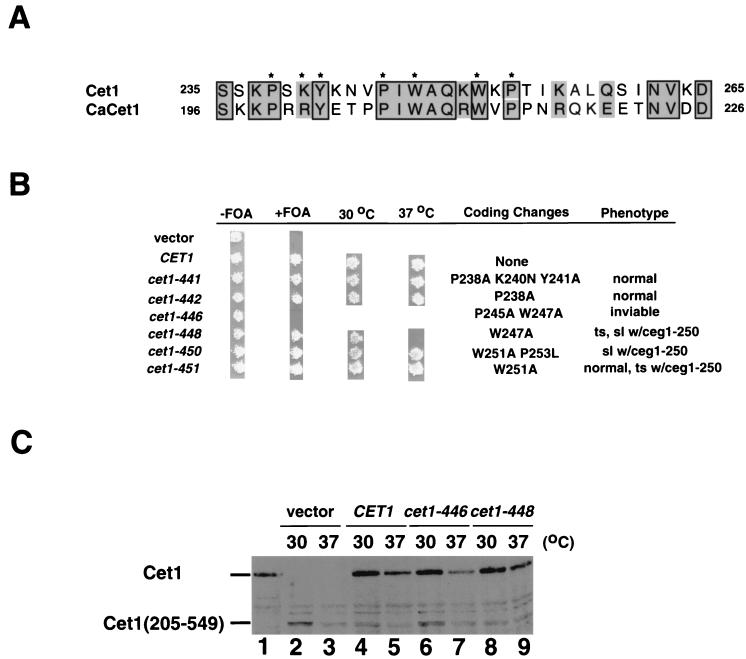
Our results and those of others (19, 24) indicated that one or more residues between amino acids 235 and 265 must be critical for binding to Ceg1. On the basis of the alignment between Cet1 and its C. albicans homologue (CaCet1 [44]) (Fig. 2A), we mutagenized six conserved residues to generate the following alleles: cet1-441 (P238A, K240N, and Y241A), cet1-442 (P238A), cet1-446 (P245A, W247A), cet1-448 (W247A), cet1-450 (W251A and P253L), and cet1-451 (W251A). We expressed these mutants in the Δcet1 strain on a centromeric plasmid and tested them for complementation by plasmid shuffling. Cells containing cet1-441, cet1-442, cet1-450, and cet1-451 were viable without any abnormal phenotype. In contrast, cet1-446 was lethal, and cells with cet1-448 grew normally at 30°C but not at 37°C. Therefore, it is likely that both Trp-247 and Pro-245 of Cet1 are important for interaction with Ceg1. Yeast strains containing the cet1-446 and cet1-448 alleles were tested by immunoblotting, and it was found that these mutant proteins were stably expressed at both permissive and nonpermissive temperatures (Fig. 2C).
FIG. 2.
Analysis of the interaction region of Cet1. (A) Amino acid sequence alignment between Cet1 (GenBank accession no. AB008799, residues 235 to 265 [39]) and CaCET1 (AB016242, residues 196 to 226 [44]). Asterisks indicate the residues of Cet1 mutated in this study. (B) Phenotypes of site-directed mutants. Wild-type CET1 and six mutant alleles were transformed into YSB533 (−FOA) and tested for complementation by plasmid shuffling (+FOA). Alleles that supported viability were further tested for growth at 30 and 37°C. The coding changes and phenotypes of the alleles are listed to the right. ts, temperature sensitivity; sl, synthetic lethality. ceg1–250 is a temperature-sensitive allele of CEG1 (7). (C) Immunoblot analysis of mutant proteins. YSB710 containing the Cet1(205–549) allele was transformed with pRS313 (lanes 2 and 3), pRS313-CET1 (lanes 4 and 5), pRS313-cet1-446 (lanes 6 and 7), and pRS313-cet1-448 (lanes 8 and 9). Lane 1 had the extract from wild-type yeast. Cultures of 100 ml of the indicated strains were grown at 30°C to an optical density at 600 nm of 0.4. Cultures were split, resuspended in 50 ml of medium prewarmed to the indicated temperature, and further cultured for 90 min at that temperature. Twenty micrograms of whole-cell extract from each culture was analyzed by immunoblotting with anti-Cet1 antibody.
The mutant CET1 alleles were also tested in combination with the ceg1-250 allele (7) with the thought that this strain might be more sensitized to perturbation of Cet1. We previously found that the temperature-sensitive phenotype of ceg1-250 can be suppressed by overexpression of CET1 (3). In this assay, cet1-448 and cet1-450 exhibited synthetic lethality with ceg1-250, while cet1-451 becomes temperature sensitive in the presence of ceg1-250 (data not shown). These additional phenotypes suggest that Trp-251 and Pro-253 may also contribute to the Cet1-Ceg1 interface.
Mutational analysis of the CET1 catalytic region.
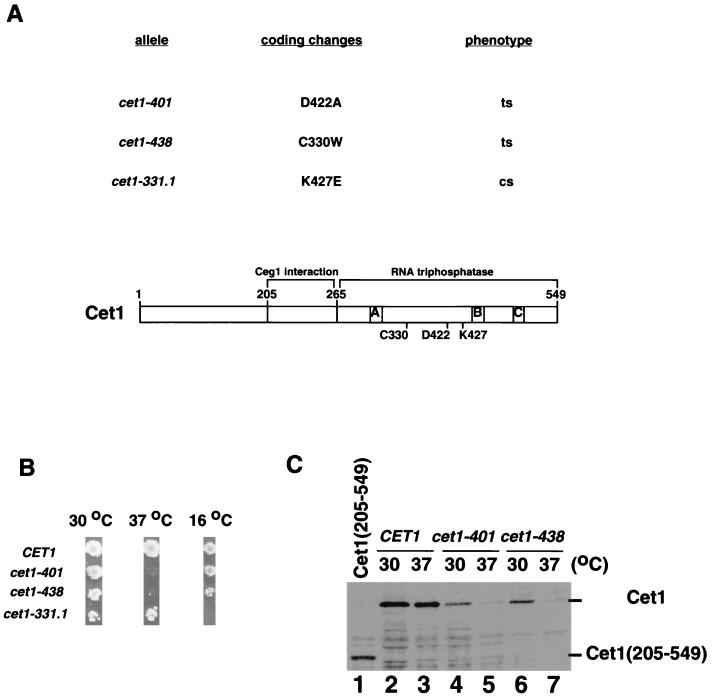
Several temperature-sensitive alleles of CET1 were isolated using random mutagenesis by PCR-based misincorporation. All mutants isolated in this screen were in the catalytic region (amino acids 265 to 549). Some mutants had multiple changes, so single point mutant alleles were constructed and tested to isolate the changes relevant to their temperature sensitivities. cet1-401 (D422A) and cet1-438 (C330W) showed a severe growth defect at 37°C, while cet1-331.1 (K427E) grew normally at 30 and 37°C but not at 16°C (Fig. 3A and B).
FIG. 3.
Conditional alleles of CET1 mutated in the catalytic region. (A) cet1 point mutants. The upper table lists the coding changes and the phenotypes of three mutant alleles. ts, temperature sensitive, unable to grow at 37°C; cs, cold sensitive, unable to grow at 16°C. The lower schematic shows the location of these coding changes relative to three motifs (motifs A, B, and C [15]) conserved among Cet1, C. albicans Cet1 (44), and Ctl1, a second RNA triphosphatase from S. cerevisiae (29, 30). (B) Conditional phenotypes. Mutants were introduced into cells by plasmid shuffling and spotted on new plates for 2 days at 30 and 37°C and for 6 days at 16°C. (C) Temperature-sensitive mutants in the catalytic region are unstable at the nonpermissive temperature. Extracts were prepared from cells grown at the temperatures indicated as described in the legend to Fig. 2 and tested for Cet1 protein by immunoblotting. Lanes: 2 and 3, wild-type CET1; 4 and 5, cet1-401; 6 and 7, cet1-438. Lane 1 is extract from YSB710 containing only Cet1(205–549).
The recent crystal structure of Cet1(241–539) shows that the triphosphatase is a barrel of multiple β sheets (25). D422 is located in a turn between two antiparallel β sheets (β7 and β8) and is partially solvent accessible. It is likely that this residue contributes to overall folding of the enzyme. C330 is in the β3 sheet and appears to contribute to the Cet1-Cet1 dimer interface within the crystal. Interestingly, K427 is in the β8 sheet at the edge of the Cet1 active-site tunnel, suggesting that the cold sensitivity of cet1-331.1 might be due to defective interactions with the substrate RNA. Sequence alignment of Cet1 with CaCet1 and Ctl1-Cth1 (another S. cerevisiae RNA phosphatase not associated with capping enzyme [29, 30]) shows that K427 is conserved in Ctl1 and an arginine in CaCet1, that C330 is only conserved in CaCet1, and that D422 is not conserved in the other two proteins. Immunoblotting showed that both cet1-401 (D422A) and cet1-438 (C330W) were degraded at the nonpermissive temperature (Fig. 3C). The cold-sensitive allele cet1-331.1 produced a protein that was stable at 16°C (data not shown).
Suppression of cet1 conditional phenotypes by overexpression of guanylyltransferase.
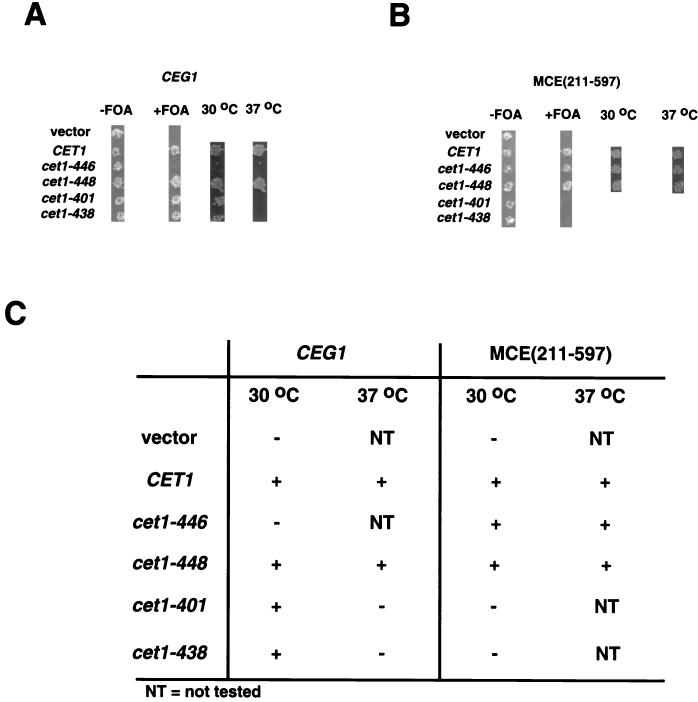
Overexpression of Cet1 suppresses some ceg1 temperature-sensitive alleles, presumably by stabilizing the mutant Ceg1 protein (3, 16). We tested the converse, whether overexpression of Ceg1 could suppress cet1 conditional phenotypes. Either Ceg1 or the mouse guanylyltransferase domain [Mce(211–597)] were expressed from high-copy plasmids in a Δceg1 Δcet1 strain. A URA3-marked plasmid carrying wild-type CET1 was replaced with the indicated CET1 alleles by plasmid shuffling, and the resulting strains were tested for viability at 30 and 37°C (Fig. 4A and B).
FIG. 4.
Different phenotypes of cet1 in the presence of overexpressed yeast and mouse guanylyltransferases. The CET1/CEG1 double shuffling strain YSB719 was transformed with: pRS425-CEG1 (2μm, LEU2) (A) or pAD5-MCE(211–597) (2μm, LEU2, which expresses mouse guanylyltransferase domain tagged with the epitope of influenza virus hemagglutinin (HA) under the control of ADH1 promoter) (B) Leu+ isolates were subsequently transformed with pRS313 (vector; carries HIS3 and CEN/ARS) or derivatives thereof carrying wild-type or mutant CET1 alleles. Leu+ His+ transformants were grown in the presence of 5-FOA to shuffle out the wild-type genes carried on pRS316-CEG1-CET1. After 2 days, FOA-resistant cells were spotted on new plates and further incubated for 2 days at 30 or 37°C as indicated. (C) Summary of the results shown in panels A and B.
Overexpression of Ceg1 clearly suppressed the temperature sensitivity of cet1-448, which is mutated in the subunit interaction region (Fig. 4A). In contrast, the temperature-sensitive phenotypes of the catalytic domain mutants cet1-401 and cet1-438 were not suppressed. This allele-specific suppression indicates that increased levels of Ceg1 can overcome a weakened subunit interaction with Cet1 but cannot bypass defects in Cet1 catalytic activity. Interestingly, cet1-446 was still lethal in the presence of increased Ceg1, suggesting that its subunit interaction defect is too severe to be suppressed.
When the mouse guanylyltransferase domain [Mce(211–597)] was substituted for Ceg1, the results were somewhat different (Fig. 4B). Not only were the catalytic domain mutants cet1-401 and cet1-438 not suppressed, they could not even support viability in the presence of Mce(211–597). Like Ceg1, Mce(211–597) also suppressed the cet1-448 conditional phenotype. Remarkably, subunit interaction mutant cet1-446, which cannot support viability in the presence of CEG1, supported growth even at 37°C in the presence of Mce(211–597).
Expression of mouse capping enzyme guanylyltransferase bypasses the requirement for the interaction domain of Cet1.
The physiological role of Cet1 amino acids 205 to 265 could be to recruit RNA triphosphatase activity (residues 265 to 549 of Cet1) to the Pol II initiation complex via binding to Ceg1 and/or to activate Ceg1 guanylyltransferase bound to the phosphorylated CTD (3). The mammalian RNA triphosphatase domain is structurally and mechanistically unrelated to Cet1 (27, 47). In the mammalian system, the phosphorylated CTD interacts with the guanylyltransferase domain and not the RNA triphosphatase domain (17, 47). In contrast to the fungal guanylyltransferase inhibition (3), the ability of the mouse guanylyltransferase (both the full-length enzyme and the isolated guanylyltransferase domain) to form the enzyme-GMP complex is stimulated by binding to phosphorylated CTD (18). Therefore, mammalian guanylyltransferase expressed in yeast presumably does not need to be allosterically activated by Cet1. On the other hand, Cet1 still must get to the transcription complex to carry out the first step of mRNA capping. Therefore, we expected that the mouse guanylyltransferase domain would allow us to assess the two functions of Cet1(205–265) in vivo.
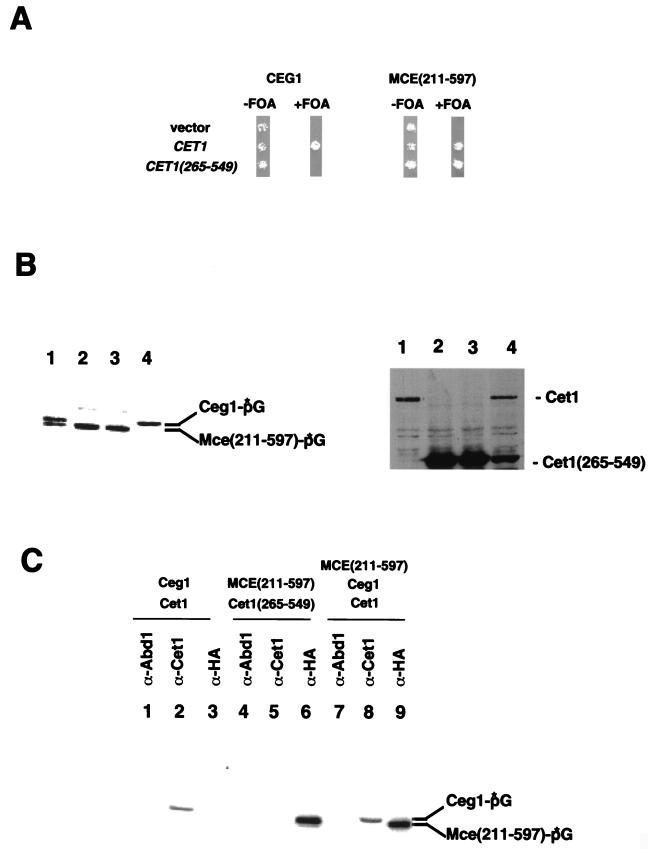
Mammalian full-length enzymes complement null and conditional mutants of CEG1 and/or CET1 if expressed under a strong constitutive promoter and/or from a high copy plasmid (16, 24, 43, 47). Δceg1 Δcet1 cells with MCE under the control of the native CET1 promoter on a centromeric plasmid grew very slowly (Y. Takase, unpublished observations). Cells with MCE(211–597) and CET1(265–549) grew poorly if the latter was supplied by a centromeric plasmid (Takase, unpublished). Therefore, we coexpressed Cet1(265–549) and Mce(211–597) from high-copy vectors and tested their ability to replace the wild-type Ceg1 and Cet1 by plasmid shuffling in Δceg1 Δcet1 cells (Fig. 5A). As previously observed, Cet1(265–549) did not support cell growth when guanylyltransferase activity was supplied by Ceg1. However, in cells expressing the mouse guanylyltransferase domain, Cet1(265–549) supported growth almost as well as full-length Cet1. Using enzyme-GMP complex formation (Fig. 5B, left panel) and immunoblotting (Fig. 5B, right panel), neither full-length Cet1 or Ceg1 protein was detected (lane 3), confirming that the wild-type copies of the corresponding genes were shuffled out of these cells. These results clearly demonstrate that amino acids 205 to 265 of Cet1 are not absolutely required under all conditions.
FIG. 5.
Coexpression of Cet1(265–549) and Mce(211–597) in Δceg1 Δcet1 cells supports viability in the absence of any subunit interaction. (A) Plasmid shuffling. YSB719 cells transformed with either pRS425-CEG1 (left panel) or pAD5-MCE(211–597) (right panel) were subsequently transformed with: pRS313 (vector); pRS313-CET1 (CET1); pRS423-CET1(265–549) [CET1(265–549), a 2μ plasmid that expresses the truncated protein from the native CET1 promoter]. Leu+ His+ isolates were plated on medium containing 5-FOA to shuffle out the pRS316-CEG1-CET1. The plates were then incubated at 30°C for 2 days. (B) Protein analysis. Whole-cell extracts were prepared from YSB719 transformed with the following: lane 1, pAD5-MCE(211–597); lanes 2 and 3, pAD5-MCE(211–597) and pRS423-CET1(265–549); and lane 4, pRS425-CEG1 and pRS423-CET1(265–549). Cells were grown in liquid media selective for the transformed plasmids. In lane 3, cells had been further selected for loss of pRS316-CEG1-CET1 by growth on medium containing FOA. The left panel shows guanylyltransferase-GMP complex formation. A total of 10 μg of protein of whole-cell extract was incubated with [α-32P]GTP and subjected to SDS-PAGE. Complexes were detected by using a phosphorimager. The right panel shows immunoblotting analysis. Forty micrograms of protein was assayed with anti-Cet1 antibody. Interestingly, the wild-type CEG1/CET1 plasmid was lost even without selection on FOA (lane 2), indicating that the combination of mouse guanylyltransferase and the truncated Cet1 supported viability as well or better than the wild-type yeast capping enzyme. (C) Immunoprecipitation. A total of 40 μg of extract protein from YSB719 transformants containing the indicated capping enzyme components was immunoprecipitated with the indicated antibodies. Pellets were assayed for guanylyltransferase-GMP intermediate formation. Antibodies used: lanes 1, 4, and 7, anti-Abd1 [S. cerevisiae (guanine-7-)-methyltransferase]; lanes 2, 5, and 8, anti-Cet1; lanes 3, 6, and 9, anti-HA (recognizing the mouse guanylyltransferase domain). Lanes 1 to 3, 4 to 6, and 7 to 9 correspond to the strains in lanes 4, 3, and 1, respectively, of Panel B.
To test for an interaction between Mce(211–597) and Cet1(265–549), we immunoprecipitated whole-cell extracts from cells expressing these two proteins (Fig. 5C). As expected, antibody against the (guanine-7-)-methyltransferase Abd1 did not pull down either Ceg1 or Mce1(211–597) (lanes 1, 4, and 7). Therefore, there is no stable interaction between Abd1 and Ceg1 or Cet1, as also suggested by yeast two-hybrid assay (45; Takase, unpublished). The interaction between full-length Cet1 and Ceg1 was clearly detected (lane 2) even in the presence of excess mouse guanylyltransferase (lane 8). Under the same conditions, no Mce(211–597) was pulled down with either full-length Cet1 (lane 8) or Cet1(265–549) (lane 5). Therefore, Cet1(265–549) must be recruited to the Pol II transcription complex by a mechanism other than binding to Mce(211–597).
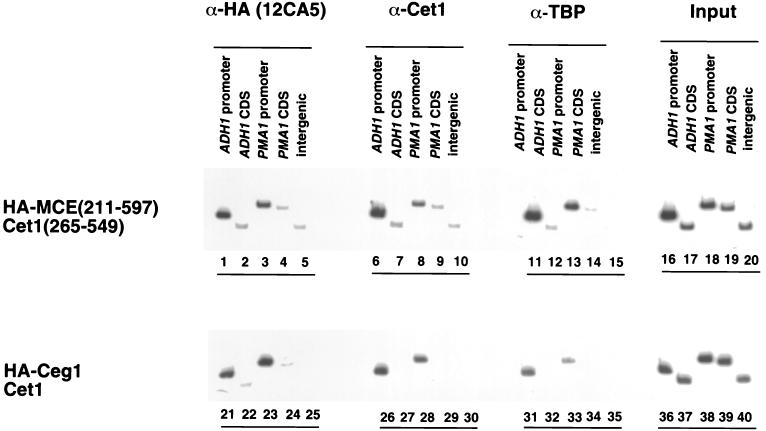
To address whether Cet1(265–549) is still recruited to promoters in the absence of Ceg1 interaction, chromatin immunoprecipitation was performed (22, 23). We recently found that capping enzyme subunits are normally associated with promoter but not coding regions, while the Abd1 mRNA methyltransferase associates with both (22). Formaldehyde cross-linked, sheared chromatin was immunoprecipitated with anti-Cet1(265–549) serum, a monoclonal antibody against the epitope-tagged guanylyltransferases [Mce(211–597) or Ceg1], or anti-TBP control antibodies. PCR was used to quantitate precipitated DNA containing promoter or coding sequences of the ADH1 and PMA1 genes (Fig. 6). Both Mce(211–597) and Cet1(265–549) are found at promoter regions, which is similar to the pattern seen with full-length Ceg1 and Cet1. This indicates either that Cet1(265–549) can be targeted to promoters independently of any guanylyltransferase or that Cet1(265–549) is associated with the mouse guanylyltransferase domain despite our best efforts to detect such an interaction.
FIG. 6.
Cet1(265–549) is localized to promoters in the absence of an interaction with guanylyltransferase. Chromatin immunoprecipitation was performed as previously described (22, 23) to localize capping enzyme components. Chromatin was prepared from strains containing Cet1(265–549) and HA-tagged Mce(211–597) (lanes 1 to 20) or wild-type capping enzyme (HA-tagged Ceg1+Cet1, lanes 21 to 40) after cross-linking with formaldehyde. Chromatin was immunoprecipitated with anti-HA monoclonal antibody 12CA5 (lanes 1 to 5 and 21 to 25) to monitor guanylyltransferases, anti-Cet1 serum (lanes 6 to 10 and 26 to 30), and anti-TBP (lanes 11 to 15 and 31 to 35) serum. DNA coprecipitated with each protein was de-cross-linked and quantitated by PCR using primers specific for the promoter or coding sequences (CDS) of the ADH1 and PMA1 genes. In addition, an Intergenic primer pair was used to measure background cross-linking to nontranscribed regions of DNA. A total of 1/20,000 of the de-cross-linked input chromatin was used as a PCR control for different primer pair efficiencies. PCR products were separated on an 8% acrylamide gel and visualized by using a phosphorimager.
Based on the results presented above, it seems exceedingly unlikely that the primary role of Cet1 amino acids 205 to 265 is to recruit Cet1 to the promoter via Ceg1 interaction. Because the requirement for this region is dependent upon the source of guanylyltransferase activity, we suggest that the most important function of the Cet1 interaction region is to allosterically activate Ceg1 bound to the phosphorylated CTD of Pol II.
DISCUSSION
The Cet1 mRNA triphosphatase plays two crucial roles in capping. Its carboxy-terminal region (amino acids 265 to 549) catalyzes the first step of cap synthesis while a second region (amino acids 205 to 265) binds to Ceg1 and allosterically activates the guanylyltransferase when it is bound to the phosphorylated CTD of Pol II (3). In this report, we study the interaction between the yeast capping enzyme subunits and come to the surprising conclusion that its primary role is not to recruit Cet1 to the promoter. Because the requirement for Cet1 amino acids 205 to 265 is observed with guanylyltransferase from S. cerevisiae but not from mammals, it is likely that the major purpose of the Ceg1-Cet1 interaction is the allosteric activation previously documented in vitro.
Both plasmid shuffling and two-hybrid analysis (Fig. 1) indicate that amino acids 235 to 265 of Cet1 are necessary and sufficient for interaction with Ceg1. We found that at least four residues in this region (P238, W247, W251, and P253) are important for this function (Fig. 2). Recently, an alanine scanning study of amino acids 247 to 251 found that a W247A-Q249A mutant is nonviable and a K250A-W251A mutant causes conditional lethality, but a P245A-I246A mutant did not affect cell viability (19, 24). Overall, our results are in good agreement with those studies. The crystal structure study of Cet1(241–539) shows that this region is exposed on the surface, with the side chains of both W247 and W251 accessible for binding to Ceg1 (25). The interaction studies are also supported by allele specific suppression of cet1 mutants by CEG1 overexpression (Fig. 4). The temperature-sensitive cet1-448 allele is mutated in the subunit interaction domain and makes a stable protein. Increased levels of Ceg1 suppress the conditional phenotype of cet1-448 by driving the Ceg1-Cet1 interaction. In contrast, catalytic domain mutants cet1-401 and cet1-438 are unstable and are not significantly suppressed by Ceg1 overexpression.
Since the mouse guanylyltransferase domain supports viability in a Δceg1 strain (17; T. Takagi, unpublished observations), it might be predicted that Mce(211–597) would bind to full-length Cet1 and guide it to the phosphorylated CTD. Peptide-affinity chromatography showed that Mce(211–597) can bind weakly to residues 232 to 265 of Cet1 in vitro (19). However, glycerol gradient sedimentation (16), yeast two-hybrid assay (Takagi, unpublished), and immunoprecipitation of yeast extracts (Fig. 5C) argue against any physiological interaction between mouse guanylyltransferase and the yeast triphosphatase. Interestingly, Mce(211–597) suppresses the lethality of the interaction domain mutant cet1-446, whereas Ceg1 overexpression does not (Fig. 4). This suggests that the manner of suppression of cet1-448 by Mce(211–597) is different from that of Ceg1. Also, cet1-401 and cet1-438 cannot support viability if Ceg1 is replaced with mouse guanylyltransferase (Fig. 4C). We speculate that binding to Ceg1 helps stabilize these triphosphatase mutants and that Mce(211–597) cannot carry out this function because it does not bind to Cet1.
The final and strongest piece of evidence that mouse guanylyltransferase does not function by binding to the Ceg1 interaction domain of Cet1 is that this region (amino acids 205 to 265 of Cet1) becomes completely dispensable in the presence of the MCE(211–597). Guanylyltransferase from some species can modify a triphosphate end of RNA in vitro to form an unusual tetraphosphate cap structure, GppppN1- (40, 46). However, Δceg1 Δcet1 cells with Mce(211–597) still require the RNA triphosphatase. Therefore, the mouse guanylyltransferase bypasses the requirement for the interaction domain of Cet1 but not the requirement for the catalytic domain. We tested whether the two functions of Cet1 could be supplied on different proteins. Coexpression of Cet1(1–265) and Cet1(265–549) in the presence of Ceg1 did not support viability (Takase, unpublished), but this may be due to instability or inability of Cet1(1–265) protein to localize in the nucleus.
How does Cet1(265–549) get to the nascent mRNA when not chaperoned by guanylyltransferase? In vitro experiments suggest that Cet1 itself does not bind to the phosphorylated CTD (3). We did not detect any interaction of Cet1 with Mce(211–597) by immunoprecipitation (Fig. 5C) or yeast two-hybrid assay. Nevertheless, in vivo cross-linking of Cet1 in the presence of the mouse guanylyltransferase shows that it is still present at the promoter. Therefore, there must be some guanylyltransferase-independent pathway for the recruitment of Cet1 to the transcription complex, perhaps via interactions with the RNA, with another part of the polymerase, or with some other promoter-localized factor.
In summary, we find that the Ceg1-interacting domain of the mRNA triphosphatase Cet1 is essential for viability but not for delivering Cet1 to the promoter. The requirement for this domain is alleviated when the yeast guanylyltransferase Ceg1 is substituted with the mammalian guanylyltransferase. Since Ceg1, but not the mouse guanylyltransferase, is allosterically activated by Cet1, we propose that this is the essential function of the interaction between yeast capping enzyme subunits. So far, all yeast capping enzymes studied consist of two subunits. If the allosteric interaction turns out to be a general property of fungal capping enzymes, one can envision targeting this feature to design antifungal drugs that would not affect the mammalian host enzyme.
ACKNOWLEDGMENTS
We thank A. Sharrocks (University of Newcastle) for pETGEXCT, A. Shatkin and R. Pilluta (Center for Advanced Biotechnology and Medicine, Piscataway, N.J.) for pG-MCE, and F. Winston (Harvard Medical School) for pAD5 and FB235. We are grateful to members of the Buratowski lab, particularly L. Fresco-Cohen for Abd1 antiserum, C. Rodriguez for contributions in the initial phase of this project, V. Polodny for help sequencing, and R. Buratowski for help in making tables.
This work was supported by NIH grant GM56663 to S.B. T.T. was a Senior Postdoctoral Fellow of the American Cancer Society, Massachusetts Division, Inc. S.B. is a Scholar of the Leukemia and Lymphoma Society.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1991. [Google Scholar]
- 2.Cho E-J, Takagi T, Moore C R, Buratowski S. mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev. 1997;11:3319–3326. doi: 10.1101/gad.11.24.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cho E-J, Rodriguez C R, Takagi T, Buratowski S. Allosteric interaction between capping enzyme subunits and the RNA polymerase II carboxy-terminal domain. Genes Dev. 1998;12:3482–3487. doi: 10.1101/gad.12.22.3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Durfee T, Becherer K, Chen P-L, Yeh S-H, Yang Y, Kilburn A E, Lee W-H, Elledge S J. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- 5.Feilotter H E, Hannon G J, Ruddell C J, Beach D. Construction of an improved host strain for two hybrid screening. Nucleic Acids Res. 1994;22:1502–1503. doi: 10.1093/nar/22.8.1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fresco L D, Buratowski S. Active site of the mRNA-capping enzyme guanylyltransferase from Saccharomyces cerevisiae: similarity to the nucleotidyl attachment motif of DNA and RNA ligases. Proc Natl Acad Sci USA. 1994;91:6624–6628. doi: 10.1073/pnas.91.14.6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fresco L D, Buratowski S. Conditional mutants of the yeast mRNA capping enzyme show that the cap structure enhances, but is not required for, mRNA splicing. RNA. 1996;2:584–595. [PMC free article] [PubMed] [Google Scholar]
- 8.Gietz D, St. Jean A, Woods R A, Schiestl R. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guan K, Dixon J E. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal Biochem. 1991;192:262–267. doi: 10.1016/0003-2697(91)90534-z. [DOI] [PubMed] [Google Scholar]
- 10.Guthrie C, Fink G R, editors. Methods in enzymology. 194. Guide to yeast genetics and molecular biology. San Diego, Calif: Academic Press, Inc.; 1991. [PubMed] [Google Scholar]
- 11.Hakansson K, Doherty A J, Shuman S, Wigley D B. X-ray crystallography reveals a large conformational change during guanylyl transfer by mRNA capping enzyme. Cell. 1997;89:545–553. doi: 10.1016/s0092-8674(00)80236-6. [DOI] [PubMed] [Google Scholar]
- 12.Harlow E, Lane D, editors. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- 13.Hirschhorn J N, Bortvin D B, Ricupero-Hovasse S L, Winston F. A new class of histone H2A mutations in Saccharomyces cerevisiae causes specific transcription defects in vivo. Mol Cell Biol. 1995;15:1999–2009. doi: 10.1128/mcb.15.4.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 15.Ho C K, Pei Y, Shuman S. Yeast and viral RNA 5′ triphosphatases comprise a new nucleoside triphosphatase family. J Biol Chem. 1998;273:34151–34156. doi: 10.1074/jbc.273.51.34151. [DOI] [PubMed] [Google Scholar]
- 16.Ho C K, Schwer B, Shuman S. Genetic, physical, and functional interactions between the triphosphatase and guanylyltransferase components of the yeast mRNA capping apparatus. Mol Cell Biol. 1998;18:5189–5198. doi: 10.1128/mcb.18.9.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ho C K, Sriskanda V, McCracken S, Bentley D, Schwer B, Shuman S. The guanylyltransferase domain of mammalian mRNA capping enzyme binds to the phosphorylated carboxyl-terminal domain of RNA polymerase II. J Biol Chem. 1998;273:9577–9585. doi: 10.1074/jbc.273.16.9577. [DOI] [PubMed] [Google Scholar]
- 18.Ho C K, Shuman S. Distinct roles for CTD Ser-2 and Ser-5 phosphorylation in the recruitment and allosteric activation of mammalian mRNA capping enzyme. Mol Cell. 1999;3:405–411. doi: 10.1016/s1097-2765(00)80468-2. [DOI] [PubMed] [Google Scholar]
- 19.Ho C K, Lehman K, Shuman S. An essential surface motif (WAQKW) of yeast RNA triphosphatase mediates formation of the mRNA capping enzyme complex with RNA guanylyltransferase. Nucleic Acids Res. 1999;27:4671–4678. doi: 10.1093/nar/27.24.4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Itoh N, Yamada H, Kaziro Y, Mizumoto K. Messenger RNA guanylyltransferase from Saccharomyces cerevisiae: large scale purification, subunit functions, and subcellular localization. J Biol Chem. 1987;262:1989–1995. [PubMed] [Google Scholar]
- 21.James P, Halladay J, Craig E A. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Komarnitsky P B, Cho E-J, Buratowski S. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 2000;14:2452–2460. doi: 10.1101/gad.824700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuras L, Struhl K. Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature. 1999;399:609–613. doi: 10.1038/21239. [DOI] [PubMed] [Google Scholar]
- 24.Lehman K, Schwer B, Ho C K, Rouzankina I, Shuman S. A conserved domain of yeast RNA triphosphatase flanking the catalytic core regulates self-association and interaction with the guanylyltransferase component of the mRNA capping apparatus. J Biol Chem. 1999;274:22668–22678. doi: 10.1074/jbc.274.32.22668. [DOI] [PubMed] [Google Scholar]
- 25.Lima C D, Wang L K, Shuman S. Structure and mechanism of yeast RNA triphosphatase: an essential component of the mRNA capping apparatus. Cell. 1999;99:533–543. doi: 10.1016/s0092-8674(00)81541-x. [DOI] [PubMed] [Google Scholar]
- 26.Mao X, Schwer B, Shuman S. Yeast mRNA cap methyltransferase is a 50-kilodalton protein encoded by an essential gene. Mol Cell Biol. 1995;15:4167–4174. doi: 10.1128/mcb.15.8.4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCracken S, Fong N, Rosonina E, Yankulov K, Brothers G, Siderovski D, Hessel A, Foster S, S, Shuman S, Bentley D L Amgen EST Program. 5′-Capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes Dev. 1997;11:3306–3318. doi: 10.1101/gad.11.24.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mulhard D, Hunter R, Parker R. A rapid method for localized mutagenesis of yeast genes. Yeast. 1992;8:79–82. doi: 10.1002/yea.320080202. [DOI] [PubMed] [Google Scholar]
- 29.Pei Y, Ho C K, Schwer B, Shuman S. Mutational analyses of yeast RNA triphosphatases highlight a common mechanism of metal-dependent NTP hydrolysis and a means of targeting enzymes to pre-mRNAs in vivo by fusion to the guanylyltransferase component of the capping apparatus. J Biol Chem. 1999;274:28865–28874. doi: 10.1074/jbc.274.41.28865. [DOI] [PubMed] [Google Scholar]
- 30.Rodriguez C R, Takagi T, Cho E-J, Buratowski S. A Saccharomyces cerevisiae RNA 5′-triphosphatase related to mRNA capping enzyme. Nucleic Acids Res. 1999;27:2181–2188. doi: 10.1093/nar/27.10.2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sadowski I, Bell B, Broad P, Hollis M. GAL4 fusion vectors for expression in yeast or mammalian cells. Gene. 1992;118:137–141. doi: 10.1016/0378-1119(92)90261-m. [DOI] [PubMed] [Google Scholar]
- 32.Schenk P M, Baumann S, Mattes R, Steinbiss H-H. Improved high-level expression system for eukaryotic genes in Escherichia coli using T7 RNA polymerase and rare argtRNAs. BioTechniques. 1995;19:196–200. [PubMed] [Google Scholar]
- 33.Sharrocks A D. A T7 expression vector for producing N- and C-terminal fusion proteins with glutathione S-transferase. Gene. 1994;138:105–108. doi: 10.1016/0378-1119(94)90789-7. [DOI] [PubMed] [Google Scholar]
- 34.Shibagaki Y, Itoh N, Yamada H, Nagata S, Mizumoto K. mRNA capping enzyme: isolation and characterization of the gene encoding mRNA guanylyltransferase subunit from Saccharomyces cerevisiae. J Biol Chem. 1992;267:9521–9528. [PubMed] [Google Scholar]
- 35.Shuman S. Capping enzyme in eukaryotic mRNA synthesis. Prog Nucleic Acids Res Mol Biol. 1995;50:101–129. doi: 10.1016/s0079-6603(08)60812-0. [DOI] [PubMed] [Google Scholar]
- 36.Shuman S, Liu Y, Schwer B. Covalent catalysis in nucleotidyl transfer reactions: essential motifs in Saccharomyces cerevisiae RNA capping enzyme are conserved in Schizosaccharomyces pombe and viral capping enzymes and among polynucleotide ligases. Proc Natl Acad Sci USA. 1994;91:12046–12050. doi: 10.1073/pnas.91.25.12046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takagi T, Moore C R, Diehn F, Buratowski S. An RNA 5′-triphosphatase related to the protein tyrosine phosphatases. Cell. 1997;89:867–873. doi: 10.1016/s0092-8674(00)80272-x. [DOI] [PubMed] [Google Scholar]
- 39.Tsukamoto T, Shibagaki Y, Imajoh-Ohmi S, Murakoshi T, Suzuki M, Nakamura A, Gotoh H, Mizumoto K. Isolation and characterization of the yeast mRNA capping enzyme β subunit gene encoding RNA 5′-triphosphatase, which is essential for cell viability. Biochem Biophys Res Commun. 1997;239:116–122. doi: 10.1006/bbrc.1997.7439. [DOI] [PubMed] [Google Scholar]
- 40.Wang D, Shatkin A J. Synthesis of Gp4N and Gp3N compounds by guanylyltransferase purified from yeast. Nucleic Acids Res. 1984;12:2303–2315. doi: 10.1093/nar/12.5.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang S P, Deng L, Ho C K, Shuman S. Phylogeny of mRNA capping enzyme. Proc Natl Acad Sci USA. 1997;94:9573–9578. doi: 10.1073/pnas.94.18.9573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamada-Okabe T, Shimmi O, Doi R, Mizumoto K, Arisawa M, Yamada-Okabe H. Isolation of the mRNA-capping enzyme and ferric-reductase-related genes from Candida albicans. Microbiology. 1996;142:2515–2523. doi: 10.1099/00221287-142-9-2515. [DOI] [PubMed] [Google Scholar]
- 43.Yamada-Okabe T, Doi R, Shimmi O, Arisawa M, Yamada-Okabe H. Isolation and characterization of a human cDNA for mRNA 5′-capping enzyme. Nucleic Acids Res. 1998;26:1700–1706. doi: 10.1093/nar/26.7.1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamada-Okabe T, Mio T, Matsui M, Kashima Y, Arisawa M, Yamada-Okabe H. Isolation and characterization of the Candida albicans gene for mRNA 5′-triphosphatase: association of mRNA 5′-triphosphatase and mRNA 5′-guanylyltransferase activities is essential for the function of mRNA 5′-capping enzyme in vivo. FEBS Lett. 1998;435:49–54. doi: 10.1016/s0014-5793(98)01037-0. [DOI] [PubMed] [Google Scholar]
- 45.Yamada-Okabe T, Mio T, Kashima Y, Matsui M, Arisawa M, Yamada-Okabe H. The Candida albicans gene for mRNA 5′-cap methyltransferase: Identification of additional residues essential for catalysis. Microbiology. 1999;145:3023–3033. doi: 10.1099/00221287-145-11-3023. [DOI] [PubMed] [Google Scholar]
- 46.Yu L, Martins A, Deng L, Shuman S. Structure-function analysis of the triphosphatase component of vaccinia virus mRNA capping enzyme. J Virol. 1997;71:9837–9843. doi: 10.1128/jvi.71.12.9837-9843.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yue Z, Maldonado E, Pillutla R, Cho H, Reinberg D, Shatkin A J. Mammalian capping enzyme complements mutant Saccharomyces cerevisiae lacking mRNA guanylyltransferase and selectively binds to the elongation form of RNA polymerase II. Proc Natl Acad Sci USA. 1997;94:12898–12903. doi: 10.1073/pnas.94.24.12898. [DOI] [PMC free article] [PubMed] [Google Scholar]