Abstract
We have previously shown that Sox18 is expressed in developing vascular endothelium and hair follicles during mouse embryogenesis and that point mutations in Sox18 are the underlying cause of cardiovascular and hair follicle defects in ragged (Ra) mice. Here we describe the analysis of Sox18−/− mice produced by gene targeting. Despite the profound defects seen in Ra mice, Sox18−/− mice have no obvious cardiovascular defects and only a mild coat defect with a reduced proportion of zigzag hairs. A reduction in the amount of pheomelanin pigmentation in hair shafts was also observed; later-forming hair follicles showed a reduced subapical pheomelanin band, giving Sox18−/− mice a slightly darker appearance than Sox18+/+ and Sox18+/− siblings. Sox18−/− mice are viable and fertile and show no difference in the ability to thrive relative to littermates. Because of the mild effect of the mutation on the phenotype of Sox18−/− mice, we conclude that the semidominant nature of the Ra mutations is due to a trans-dominant negative effect mediated by the mutant SOX18 proteins rather than haploinsufficiency as has been observed for other SOX genes. Due to the similarity of SOX18 to other subgroup F SOX proteins, SOX7 and −17, and the overlap in expression of these genes, functional redundancy amongst these SOX proteins could also account for the mild phenotype of Sox18−/− mice.
Members of the SOX (Sry-type HMG box) gene family encode transcription factors that have a wide range of roles in development (reviewed by Wegner [39]). SOX proteins bind DNA in a sequence-specific manner, and a heptameric SOX consensus binding motif, 5′-(A/T)(A/T)CAA(A/T)G-3′, has been identified (12). Most tissues and cell types express at least one SOX gene at one stage or another of their development (39). Moreover, many cell types or tissues express more than one SOX gene at certain times (6, 20, 22, 36, 37).
Gene targeting experiments with the mouse have assigned vital roles in development to numerous SOX genes: Sox1 in lens formation (28), Sox4 in cardiac tract outflow formation and B-lymphocyte development (32), and Sox9 in chondrogenesis (3). This has been reinforced by mutations in human SOX genes: SRY mutations in sex reversal and gonadal dysgenesis (2, 11, 13, 15), SOX9 mutations in the bone dysmorphogenesis and sex reversal syndrome campomelic dysplasia (9, 38), and SOX10 mutations in various neurocristopathies such as Waardenberg-Shah syndrome 4 (30) and the Yemenite deaf-blind hypopigmentation syndrome (4). Further, such mutations have revealed an importance of dosage for some SOX genes, with deletion or mutation of one allele of SOX9 or SOX10 resulting in a disease phenotype (9, 30, 38).
We have previously shown that point mutations in Sox18 are the underlying cause of profound cardiovascular and hair follicle defects in ragged (Ra) mice (29). Ra heterozygotes have thin, ragged coats comprised of guard hairs but lacking the later-forming auchenes and zigzags (5). Ra homozygotes, however, almost completely lack vibrissae and coat hairs, display generalized edema and cyanosis, and rarely survive past weaning (5). In a more severe Ra mutation, RaOp, heterozygotes typically show a phenotype similar to that of Ra homozygotes, whereas RaOp homozygotes die by 11.5 days postcoitus (dpc) (10, 24). The coat defects in Ra mice are due to a reduction in the total number of hair follicles, with the later-forming follicles being the most affected (33).
In Ra mice, the Sox18 mutations lead to an intact DNA-binding domain but a nonfunctional trans-activation domain (29). Given the semidominant nature of the Ra mutations (5, 10), it was not clear whether the phenotype of Ra mice was due to haploinsufficiency of Sox18, as has been described for other SOX genes, or whether there was a dominant-negative effect of the mutant SOX18 protein. To address this question, we produced mice null for Sox18, and we describe here their production and analysis.
MATERIALS AND METHODS
Targeting of Sox18 and production of chimeric mice.
Overlapping genomic clones hybridizing to Sox18 were obtained from a λ phage library constructed from 129/Sv genomic DNA which was partially digested with MboI and cloned into the BamHI sites of λDashII vector (Philippe Soriano, personal communication). Four overlapping clones were mapped using restriction digests and Southern hybridization. The 1.8-kb, 5′ flanking fragment was generated by high-fidelity PCR from a genomic clone to incorporate XbaI sites and facilitate subcloning. The 3′ flanking arm was an 11-kb fragment subcloned from a genomic clone. The 3′ and 5′ flanking sequences were cloned into a plasmid, pLoxPneo-1, that contains a neomycin resistance cassette (neor) driven by the PGK-1 promoter, all flanked by directional LoxP sites (A.N., unpublished data). The LoxP sites were included to allow the excision of intervening sequence in the presence of cre recombinase (1). A promoterless lacZ reporter cassette (19) was subcloned into the targeting vector so that an in-frame SOX18–β-galactosidase fusion protein would be produced to facilitate gene expression studies. A thymidine kinase cassette (25) was used in the targeting vector for counterselection in embryonic stem (ES) cells.
Gene targeting was performed in the R1 ES cell line (27) using standard protocols (16), with the exception that ES cells were grown in the absence of feeder fibroblasts cells on gelatinized plastic tissue culture dishes in media supplemented with leukemia inhibitory factor, and 40 μg of linearized targeting vector was used in each electroporation cuvette. G418-resistant ES cell clones were screened for homologous recombination using genomic Southern blots after HindIII digestion. An external probe was used to detect a native 12-kb band or a band of 8.5 kb in targeted alleles. Two independent ES cell clones were identified as homologous recombinants. Chimeric embryos were produced from them by using CD1 donor embryos and the technique of morula aggregation (41) and transferred to pseudopregant recipients as described by Hogan et al. (14). Germ line transmission of the targeted Sox18 allele was achieved with chimeras produced from the two independently targeted ES cell lines. Analysis of the targeted mutation was conducted on mice and embryos of a 129/Sv-CD1 mixed genetic background.
Genotyping of mice and embryos.
Mice and embryos used in this study were genotyped by genomic DNA Southern hybridization (as described above for screening ES cell clones) or by PCR on genomic DNA prepared from ear punches or tail clips as described by Joyner (16). To detect the targeted allele, the neor-specific primers neoR (5′-CAA GCT CTT CAG CAA TAT CAC G-3′) and neoF (5′-ATC TCC TGT CAT CTC ACC TTG C-3′) were used. To detect the wild-type Sox18 allele, the primers Sox18-Box A (5′-CCA ACG TCT CGC CCA CCT CG-3′) and Sox18-Box B (5′-GCC GCT TCT CCG CCG TGT TC-3′) were used. Mutant embryos were always genotyped by PCR amplification from a portion of the yolk sac or allantois that had been well rinsed in a large volume of phosphate-buffered saline.
RT-PCR analysis.
cDNA was produced in a reaction mixture containing 1 μg of RNA, 1× “first-strand” buffer (Gibco BRL; 50 mM Tris-HCl [pH 8.3], 75 mM KCl, 3 mM MgCl2), 375 μM deoxynucleoside triphosphates, 100 mM dithiothreitol, 500 ng of pd(N)6 random primers (Pharmacia), 200 U of Moloney murine leukemia virus reverse transcriptase (RT) (Gibco BRL), and RNase-free MilliQ water in a total volume of 30 μl. The reaction mixture was incubated at 42°C for 1 h, and 5 μl was used in a 25-μl PCR. The primer pairs used were as follows: for detection of neor transcripts, neoR (5′-CAA GCT CTT CAG CAA TAT CAC G-3′) and neoF (5′-ATC TCC TGT CAT CTC ACC TTG C-3′); for lacZ transcripts, LacZ A (5′-CAG CAC ATC CCC CTT TCG CC-3′) and LacZ B (5′-CCA ACG CAG CAC CAT CAC CG-3′); for the 3′ portion of Sox18 downstream of the lacZ reporter and neor (encoding the trans-activation domain), Sox18-3′ A (5′-GGC TTT CCG GGC ACC CTA TG-3′) and Sox18-3′ B (5′-AAG CGG TGG AGG GCA AGG AC-3′); for the region of Sox18 encoding the HMG box, Sox18-Box A (5′-CCA ACG TCT CGC CCA CCT CG-3′) and Sox18-Box B (5′-GCC GCT TCT CCG CCG TGT TC-3′); for the 5′ region of Sox18 upstream of the lacZ reporter and neor, Sox18-5′ A (5′-TGA GAC AGT GGG AGC AGA TGG-3′) and Sox18-5′ B (5′-GCA AAG CCA AGT ACG GAG GTC 3′); and for GAPDH transcripts, GAPDH F (5′-TCG GTG TGA ACG GAT TTG-3′) and GAPDH R (5′-ATT CTC GGC CTT GAC TGT-3′).
Expression studies.
Whole-mount immunohistochemistry was done using an anti-PECAM-1 (CD31) antibody (Pharmingen, San Diego, Calif.) at a dilution of 1/250, using standard techniques described elsewhere (40).
Analysis of the coats of Sox18−/− mice.
The coat qualities of adult Sox18−/− and control mice were analyzed by visual inspection, and mice were photographed with standard color-reversal film. To survey the relative abundance of the various hair types among the coats of Sox18−/− and control mice, hairs were plucked in bunches from the middorsa of adult mice, typically 3 months of age. A broad pair of forceps (Millipore) was used to obtain three samples from each test subject. A dissecting microscope was used to count the hairs of each type according to the mouse hair classification of Dry (7). The total number of hairs from each subject varied between 160 and 380 in order to give a statistically valid sampling of hair types. Photography of mouse hairs was performed on a dissecting stereomicroscope (Leica MZ8) with bright-field illumination after hairs were dry mounted on a microscope slide with a coverslip and nail polish.
RESULTS
Targeting of the Sox18 gene.
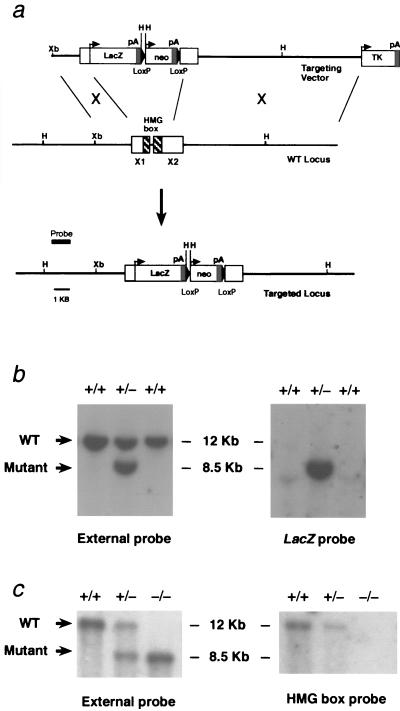
The gene targeting strategy was designed to produce mice null for Sox18 by removing the region encoding the HMG box (DNA-binding domain) and replacing it with a lacZ reporter cassette and a neor cassette, both of which contain transcription termination sequences. Southern blot analysis of genomic DNA from Sox18−/− and Sox18+/− mice confirmed that the Sox18 locus had been targeted as expected, with the region encoding the HMG box being removed and replaced by the lacZ and neor cassettes (Fig. 1). However, analysis of Sox18−/− and Sox18+/− embryos at 9.5 dpc, a stage when Sox18 is expressed strongly in the developing vascular system (29), failed to show β-galactosidase staining (data not shown). Sequencing of the targeting vector confirmed that an in-frame fusion protein comprising the N-terminal 89 amino acids of SOX18 and β-galactosidase (data not shown) should have been produced. It is unclear why the lacZ reporter cassette is nonfunctional in Sox18−/− and Sox18+/− embryos, though one possibility is that the N-terminal 89 amino acids of SOX18 interfered with the enzymatic activity of β-galactosidase.
FIG. 1.
Disruption of Sox18 by gene targeting. (a) Structure of Sox18 and gene targeting by homologous recombination. The targeting vector was designed to replace the regions of exon 1 and 2 (X1 and X2) encoding the HMG box domain (hatched boxes) with a lacZ reporter cassette (LacZ) and a neor selection cassette (neo). The probe was designed for screening for homologous recombinants in ES cells by HindIII digestion of genomic DNA. pA, polyadenylation site; TK, thymidine kinase cassette; Xb, XbaI restriction site; H, HindIII restriction site. (b) Detection of homologous recombinants in ES cells. (Left) Southern blotting of G418-resistant genomic DNA digested with HindIII (using the external probe) revealed a wild-type band of approximately 12 kb and a targeted band of 8.5 kb as expected. (Right) Probing the same Southern blot with an internal probe (lacZ fragment) revealed a targeted band of 8.5 kb, as expected, indicating that there was a single homologous recombination event. (c) Genotyping of Sox18−/− mice by Southern blotting. (Left) The same strategy was used as for screening ES cells, revealing the genotype of wild-type, heterozygous, and homozygous knockout mice. (Right) Probing the same Southern blot with a probe specific to the HMG box-encoding region confirmed that this region was removed in the targeted loci.
RT-PCR analysis of the mutant transcript.
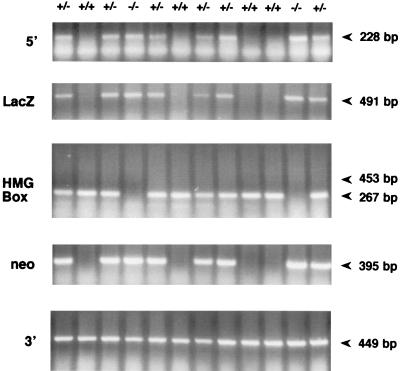
To analyse which transcripts were produced from the targeted Sox18 locus, RT-PCR was conducted on RNA from mutant and wild-type embryos. RT-PCR analysis confirmed that there were no transcripts from the HMG box domain-encoding region from the targeted allele (Fig. 2). As can be seen in Fig. 2, the pattern of neor and lacZ expression was also as expected, being present only in embryos carrying a targeted locus. Analysis of the region encoding the trans-activation domain (downstream of the lacZ and neor cassettes), however, revealed that this portion of the gene was transcribed from both wild-type and targeted loci. In addition, a portion of the coding region 5′ to the lacZ and neor cassettes, and common to both wild-type and targeted loci, appeared to be transcribed only from the targeted locus. It is known that this region is transcribed under normal circumstances and that mRNA secondary structure does not interfere with reverse transcription in this region, since this sequence is present in cDNA clones derived by reverse transcription (8). We are therefore unable to explain the lack of expression of the 5′ region of Sox18 observed in these experiments.
FIG. 2.
RT-PCR analysis of transcripts from the Sox18−/− and wild-type loci. At 9.5 dpc, littermates were used to provide RNA samples. Shown are results of RT-PCR of a region 5′ to the HMG box domain expected to be in both the wild-type and mutant transcripts (labeled 5′), RT-PCR of a portion of the lacZ reporter, expected to be present only in mutant transcripts (labeled LacZ), and RT-PCR spanning the HMG box region and 186-bp intron (labeled HMG Box). This also served as a control for genomic DNA contamination. Genomic bands would be 453 bp in this reaction. Also shown are results of RT-PCR of a portion of the neor marker, expected to be present only in mutant transcripts (labeled neo) and RT-PCR of a region of the trans-activation domain (labeled 3′).
Sox18−/− mice are viable and fertile.
Germ line transmission of the targeted Sox18 allele was achieved by breeding male chimeras derived from the two independently targeted ES cell lines. Genotyping of litters from intercrosses of F1 Sox18+/− heterozygotes revealed that Sox18−/− mice appeared in Mendelian ratios. Breeding studies also indicated that the Sox18−/− mice could interbreed, proving that they were fertile (data not shown).
Anatomical analysis of Sox18−/− embryos.
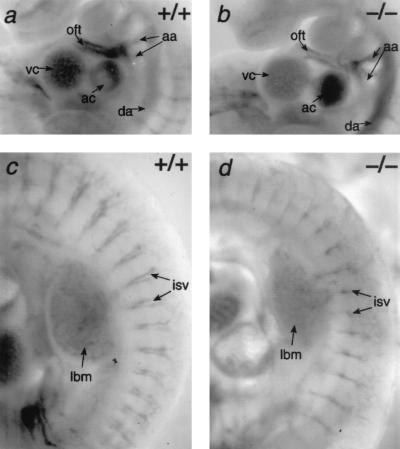
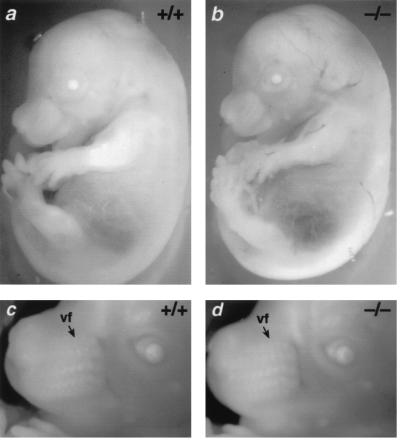
As Sox18−/− mice appeared to have no reduced viability or fertility, there were likely to be no major cardiovascular defects associated with the targeted mutation. Immunohistochemistry using an antibody to the vascular endothelium marker PECAM-1 (CD31) revealed that there were no major defects in the gross morphology of the hearts of Sox18−/− embryos (Fig. 3). In addition, the dorsal aortae, intersomitic vessels, and vessels of the limb bud mesenchyme appeared grossly normal (Fig. 3). There did not appear to be any hemorrhage or edema in these embryos at 9.5 dpc. Likewise, at 14.5 dpc, no gross abnormalities, hemorrhage, or edema was detected in Sox18−/− embryos (Fig. 4). Furthermore, vibrissa follicles in Sox18−/− embryos had developed in numbers comparable to those in littermates at the appropriate stage (Fig. 4).
FIG. 3.
Analysis of vascular development in Sox18−/− embryos at 9.5 dpc. (a and b) PECAM-1 (CD31) immunostaining of Sox18+/+ and Sox18−/− embryos, respectively, showing staining of the aortic arches (aa), atrial chamber (ac), dorsal aorta (da), outflow tract (oft), and ventricular chamber (vc). (c and d) Embryos shown in panels a and b, respectively, showing staining of the intersomitic vessels (isv) and the vasculature of the limb bud mesenchyme (lbm).
FIG. 4.
Gross morphology and vibrissa formation in Sox18−/− embryos. (a and b) Gross morphology of Sox18+/+ and Sox18−/− embryos, respectively, at 14.5 dpc. (c and d) Close-up view of the embryos in panels a and b, respectively, showing the grossly visible vibrissa follicle (vf) formation.
Growth rate of Sox18−/− mice.
As a general index of the well-being of Sox18−/− mice, and to try to detect any subtle difference in the physiologies of these mice, their growth rate relative to that of littermate controls was measured in terms of total weight. This analysis was performed using male littermates from both targeted lines. Over a 2-month period, no statistically significant difference was observed between Sox18−/− mice and Sox18+/− and Sox18+/+ littermate controls at any stage (data not shown).
Coat color of Sox18−/− mice.
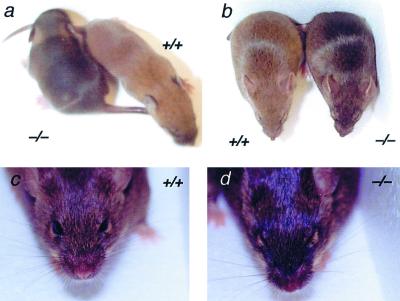
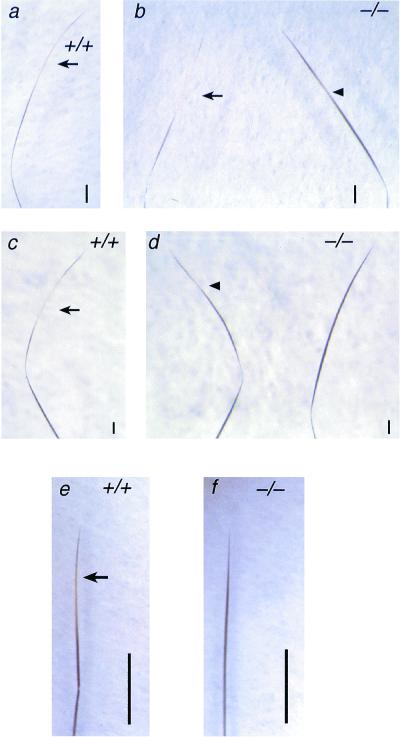
Segregation of alleles of coat color genes among the stock of Sox18−/− mice was observed due to the mixed genetic background of CD1 and 129/Sv. This facilitated the analysis of coat formation, with numerous pigmentation phenotypes, including albino, chinchilla, and agouti, appearing in the stock of Sox18−/− mice. Sox18−/− mice had a slightly darker appearance than littermate controls on agouti (Fig. 5) and chinchilla (data not shown) backgrounds. Otherwise, no visible difference was detected in the coats of Sox18−/− mice. Microscopic examination of the various hair types was conducted to further analyze the basis of the different coat pigmentation of Sox18−/− mice. Again, this was done using mice with an agouti phenotype to facilitate examination of the subapical pheomelanin band typical of many agouti hairs. Examples of the four main types of pelage hairs—the guard hairs, awls, auchenes, and zigzags—appeared morphologically normal, though there was a difference in the pigmentation of many hair types between Sox18−/− mice and littermate controls (Fig. 6). Guard hairs, which never have a subapical pheomelanin band, appeared the same in Sox18−/− mice and controls upon microscopic examination (Fig. 6). Some awls from wild-type mice showed a subapical pheomelanin band, though most, like all those from Sox18−/− mice, did not. Many auchenes and zigzags from Sox18−/− mice showed either a reduced or completely absent pheomelanin band.
FIG. 5.
Coat color of Sox18−/− mice. The mice shown in panels a and b are the same two male agouti littermates. (a) Sox18+/+ and Sox18−/− littermates at 8 days after birth. Note the darker color of the Sox18−/− sibling. (b) Sox18+/+ and Sox18−/− littermates at 2 months after birth; the color difference is less pronounced than at 8 days. (c and d) Vibrissae of the Sox18+/+ and Sox18−/− littermates, respectively, at 3 months after birth, showing no obvious differences.
FIG. 6.
Pigmentation of hair types in Sox18−/− mice. The hair samples shown are from agouti littermates. (a and b) Subapical pheomelanin banding in auchenes from Sox18+/+ and Sox18−/− mice, respectively. Arrows indicate pheomelanin bands. The arrowhead indicates a greatly reduced pheomelanin band. Bars, 200 μm. (c and d) Subapical pheomelanin banding in zigzags from Sox18+/+ and Sox18−/− mice, respectively. The arrow indicates a pheomelanin band. The arrowhead indicates a greatly reduced pheomelanin band. Note the absence of any pheomelanin in the zigzag on the right in panel d. Bars, 100 μm. (e and f) Awls from Sox18+/+ and Sox18−/− mice, respectively. Note the presence of a subapical pheomelanin band in an awl from a Sox18+/+ mouse (indicated by an arrow) but not in an awl from a Sox18−/− mouse. Bars, 1 mm.
Hair formation in Sox18−/− mice.
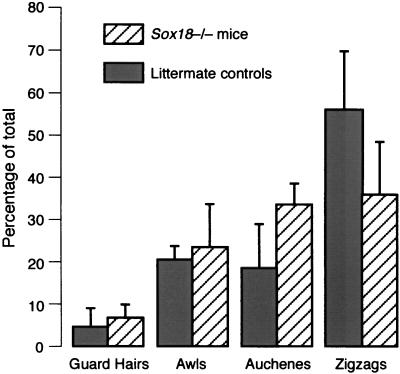
As some hair types are malformed and underrepresented in Ra mice, namely the later-forming auchenes and zigzag hairs, we conducted a survey of the proportion of pelage hair types amongst Sox18−/− mice. From Fig. 7 it can be inferred that the proportion of guard hairs and awls was not significantly different between Sox18−/− and littermate controls. There was a marked difference, however, in the proportion of zigzag hairs, which represented 56% of total hairs in wild-type mice and 36% in Sox18−/− mice. It is worth noting that vibrissae and vibrissa follicles seemed normal at all stages in Sox18−/− embryos and mice (Fig. 4 and 5).
FIG. 7.
Summary of the prevalence of hair types in Sox18−/− mice. Note the reduced proportion of zigzag hairs among the coats of Sox18−/− mice. Error bars represent standard deviations.
DISCUSSION
The inactivation of Sox18 by gene targeting has demonstrated that this gene is not vital for embryogenesis. The mild phenotype observed in Sox18−/− mice is in stark contrast to that resulting from the Ra mutations. Given the surprisingly mild effect of the mutation on the phenotype of the Sox18−/− mice, it was important to show that the targeted mutation was indeed a null mutation by genomic Southern blot. This analysis confirmed that the targeting event had removed the portion of the gene encoding the HMG box domain and replaced it with a lacZ reporter and a neor cassette. It remains formally possible that the residual, expressed, C-terminal region of SOX18, containing the trans-activation domain, may retain some function, perhaps by interaction with another protein in a manner that does not depend on DNA binding. However, given the known importance of HMG domain-mediated DNA binding for the function of other SOX transcription factors, such as SRY and SOX9, it is reasonable to assume that removal of the HMG box destroys the function of the SOX18 protein.
Despite the expression pattern of Sox18 during embryogenesis and the finding that Sox18 mutations in Ra mice have a profound effect on cardiovascular and hair follicle development (29), Sox18 does not appear to be essential for development of these tissues. That the weight of Sox18−/− mice is similar to that of littermates is also in contrast to the situation with Ra mice. Shortly after birth, Ra mice are heavier than normal because of a buildup of fluid due to edema and hemorrhage, which soon dissipates. More significantly, however, they grow more slowly than their littermates (5). The normal growth rate of Sox18−/− mice indicates that the physiology of these mice under normal conditions is not unduly affected by the targeted mutation.
In the light of the contrasting phenotypes of Ra and Sox18−/− mice, it appears that the mutant SOX18 proteins in Ra mice are acting in a trans-dominant negative manner. It is tempting to speculate that the mutant SOX18 in Ra mice interferes with another SOX protein(s), which in the presence of the targeted null mutation manages to almost fully compensate for the absence of a fully functional SOX18. One can readily envisage a situation whereby the mutant SOX18 protein from Ra mice, which have an intact HMG box but have lost the ability to trans-activate, competes with wild-type SOX18 (and any other SOX protein present with a similar target binding site) and prevents the activation of transcription of the target gene. This hypothesis is supported by the evidence that SOX proteins display affinity for similar binding sites (39) and that, at least in some tissue types, Sox18 is coexpressed with Sox7 and Sox17 (see below). Conversely, it is possible that the mutant SOX18 proteins from Ra mice have gain-of-function mutations and that they interfere with some other aspects of the cell's transcription machinery. Analysis of the phenotype of compound heterozygotes from a cross of Ra/+ and Sox18−/− mice would be informative with regard to the mode of action of these mutant proteins and might shed some light on the nature of SOX proteins. These experiments are in progress.
Like Ra mice, Sox18−/− mice are most affected in the later-forming hair types, with a reduction in the number of zigzags. Also like Ra mice, Sox18−/− mice are darker than their littermates, reflecting a reduction in pheomelanin pigment. Interestingly, another Sox gene, Sox10, has been implicated in pigmentation. Sox10 is expressed in neural crest cells as they migrate to numerous sites in the developing embryo (21), and mutations in Sox10 result in, apart from other things, hypopigmentation (4, 30, 35). One population of neural crest cells is the melanoblasts, which migrate through the mesenchymal layer beneath the ectoderm before populating the dermis and differentiating to melanocytes. Melanocytes that populate the hair bulb, adjacent to the dermal papilla, donate melanosomes to the matrix keratinocytes, giving hair its visible pigmentation.
There is no evidence that migrating neural crest cells, which express Sox10, also express Sox18. Cells that will give rise to the dermal papilla, adjacent to the hair bulb, do, however, express Sox18, at least in a transient manner (29). Perturbation of Sox18 function in Ra and Sox18−/− mice does not result in the hypopigmentation of hairs, though it does affect the production of pheomelanin in hairs. This may be due to the effect Sox18 mutations have on the agouti signaling peptide: cells that express the agouti gene reside in the dermal papillae (26). These are the same cell types as, or a population of cells in close proximity to, those that express Sox18 transiently during hair follicle maturation. It is a distinct possibility that normal Sox18 function is needed for agouti-expressing cells to fulfill their role completely. It seems unlikely that the receptor antagonized by the agouti signaling peptide is adversely affected by Sox18 mutations, since in Ra and Sox18−/− mice eumelanin is produced by hair follicles, indicating that melanocyte-stimulating hormone α and its receptor are functioning normally.
Binding site selection, analyses of chimeric proteins, and phenotype rescue experiments indicate that there is scope for redundancy amongst SOX proteins (18, 39). Other evidence for functional redundancy includes the rescue of phenotypes by related SOX proteins: mouse SOX2 can rescue the defects in the differentiation of the midline glia caused by a mutation in the Drosophila gene Dichaete (34). In addition, domains of different SOX proteins may be interchanged with little change in function of the chimeric protein; chimeric SOX1-SOX9 proteins displayed the ability to activate transcription after binding the appropriate target sequences (δ-crystallin enhancer, DC5, for SOX1 and Col2a1, enhancer, COL2C2, for SOX9), demonstrating that the HMG box domains are interchangeable (17). Further, functional redundancy has been documented amongst other developmentally expressed gene families, such as the MyoD family (31) and Hox family (23). There is some overlap in the expression of Sox7-, -17, and -18 in the developing vascular endothelium (Jane Olsson, personal communication), and it therefore remains a distinct possibility given the mild phenotype of Sox18−/− mice that there is functional redundancy amongst SOX proteins. To address the question of redundancy amongst these proteins, particularly subgroup F SOX proteins, the relevant knockout mice, once available, can be used to generate double and triple knockouts to test the hypothesis of functional redundancy.
Gene targeting in the mouse and identification of human mutations have revealed vital roles for SOX genes, with, in the cases of SOX9 and SOX10, both copies of the gene needed for normal development. This study provides the first evidence of a SOX gene that is largely dispensable for embryonic development. This study should contribute towards the understanding of possible interactions of SOX proteins in various embryonic tissues and the identification of downstream target genes.
ACKNOWLEDGMENTS
This work was funded by the National Heart Foundation (Australia), the Queensland Cancer Fund, and the National Health and Medical Research Council (Australia). P.K. is an Australian Research Council Senior Research Fellow.
We thank Anne Hardacre for invaluable help in the maintenance of the animal breeding colony and Philippe Soriano for the gift of the 129/Sv genomic library used for the production of the Sox18 targeting vector.
REFERENCES
- 1.Abremski K, Hoess R. Bacteriophage P1 site-specific recombination. Purification and properties of the Cre recombinase protein. J Biol Chem. 1984;259:1509–1514. [PubMed] [Google Scholar]
- 2.Berta P, Hawkins J R, Sinclair A H, Taylor A, Griffiths B L, Goodfellow P N, Fellous M. Genetic evidence equating SRY and the male sex determining gene. Nature. 1990;348:448–450. doi: 10.1038/348448A0. [DOI] [PubMed] [Google Scholar]
- 3.Bi W, Deng J M, Zhang Z, Behringer R R, de Crombrugghe B. Sox9 is required for cartilage formation. Nat Genet. 1999;22:85–89. doi: 10.1038/8792. [DOI] [PubMed] [Google Scholar]
- 4.Bondurand N, Kuhlbrodt K, Pingault V, Enderich J, Sajus M, Tommerup N, Warburg M, Hennekam R C, Read A P, Wegner M, Goossens M. A molecular analysis of the Yemenite deaf-blind hypopigmentation syndrome: SOX10 dysfunction causes different neurocristopathies. Hum Mol Genet. 1999;8:1785–1789. doi: 10.1093/hmg/8.9.1785. [DOI] [PubMed] [Google Scholar]
- 5.Carter T C, Phillips R J S. Ragged, a semidominant coat texture mutant in the house mouse. J Hered. 1954;45:151–154. [Google Scholar]
- 6.Collignon J, Sockanathan S, Hacker A, Cohen-Tannoudji M, Norris D, Rastan S, Episkopou V, Stevanovic M, Goodfellow P N, Lovell-Badge R. A comparison of the properties of Sox-3 with Sry and two related genes, Sox-1 and Sox-2. Development. 1996;122:509–520. doi: 10.1242/dev.122.2.509. [DOI] [PubMed] [Google Scholar]
- 7.Dry F W. The coat of the mouse (Mus musculus) J Genet. 1926;16:287–340. [Google Scholar]
- 8.Dunn T L, Mynett-Johnson L, Hosking B M, Koopman P A, Muscat G E O. Sequence and expression of Sox-18, a new HMG-box transcription factor. Gene. 1995;161:223–225. doi: 10.1016/0378-1119(95)00280-j. [DOI] [PubMed] [Google Scholar]
- 9.Foster J W, Dominguez-Steglich M A, Guioli S, Kwok C, Weller P A, Weissenbach J, Mansour S, Young I D, Goodfellow P N, Brook J D, Schafer A J. Campomelic dysplasia and autosomal sex reversal caused by mutations in an SRY-related gene. Nature. 1994;372:525–530. doi: 10.1038/372525a0. [DOI] [PubMed] [Google Scholar]
- 10.Green E L, Mann S J. Opossum, a semi-dominant lethal mutation affecting hair and other characteristics of mice. J Heredity. 1961;52:223–227. [Google Scholar]
- 11.Harley V R, Jackson D I, Hextall P J, Hawkins J R, Berkovitz G D, Sockanathan S, Lovell-Badge R, Goodfellow P N. DNA binding activity of recombinant SRY from normal males and XY females. Science. 1992;255:453–456. doi: 10.1126/science.1734522. [DOI] [PubMed] [Google Scholar]
- 12.Harley V R, Lovell-Badge R, Goodfellow P N. Definition of a consensus DNA binding site for SRY. Nucleic Acids Res. 1994;22:1500–1501. doi: 10.1093/nar/22.8.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hawkins J R, Taylor A, Berta P, Levilliers J, Van der Auwera B, Goodfellow P N. Mutational analysis of SRY: nonsense and missense mutations in XY sex reversal. Hum Genet. 1992;88:471–474. doi: 10.1007/BF00215684. [DOI] [PubMed] [Google Scholar]
- 14.Hogan B, Beddington R, Costantini F, Lacy E. Manipulating the mouse embryo: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- 15.Jäger R J, Anvret M, Hall K, Scherer G. A human XY female with a frame shift mutation in the candidate testis-determining gene SRY. Nature. 1990;348:452–454. doi: 10.1038/348452a0. [DOI] [PubMed] [Google Scholar]
- 16.Joyner A L. Gene targeting: a practical approach. Oxford, United Kingdom: Oxford University Press; 1993. [Google Scholar]
- 17.Kamachi Y, Cheah K S, Kondoh H. Mechanism of regulatory target selection by the SOX high-mobility-group domain proteins as revealed by comparison of SOX1/2/3 and SOX9. Mol Cell Biol. 1999;19:107–120. doi: 10.1128/mcb.19.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kamachi Y, Uchikawa M, Kondoh H. Pairing SOX off: with partners in the regulation of embryonic development. Trends Genet. 2000;16:182–187. doi: 10.1016/s0168-9525(99)01955-1. [DOI] [PubMed] [Google Scholar]
- 19.Korn R, Schoor M, Neuhaus H, Henseling U, Soininen R, Zachgo J, Gossler A. Enhancer trap integrations in mouse embryonic stem cells give rise to staining patterns in chimaeric embryos with a high frequency and detect endogenous genes. Mech Dev. 1992;39:95–109. doi: 10.1016/0925-4773(92)90029-j. [DOI] [PubMed] [Google Scholar]
- 20.Kuhlbrodt K, Herbarth B, Sock E, Enderich J, Hermans-Borgmeyer I, Wegner M. Cooperative function of POU proteins and SOX proteins in glial cells. J Biol Chem. 1998;273:16050–16057. doi: 10.1074/jbc.273.26.16050. [DOI] [PubMed] [Google Scholar]
- 21.Kuhlbrodt K, Herbarth B, Sock E, Hermans-Borgmeyer I, Wegner M. Sox10, a novel transcriptional modulator in glial cells. J Neurosci. 1998;18:237–250. doi: 10.1523/JNEUROSCI.18-01-00237.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lefebvre V, Li P, de Crombrugghe B. A new long form of Sox5 (L-Sox5), Sox6 and Sox9 are coexpressed in chondrogenesis and cooperatively activate the type II collagen gene. EMBO J. 1998;17:5718–5733. doi: 10.1093/emboj/17.19.5718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maconochie M, Nonchev S, Morrison A, Krumlauf R. Paralogous Hox genes: function and regulation. Annu Rev Genet. 1996;30:529–556. doi: 10.1146/annurev.genet.30.1.529. [DOI] [PubMed] [Google Scholar]
- 24.Mann S J. The phenogenetics of hair mutants in the house mouse: Opossum and Ragged. Genet Res. 1963;4:1–11. [Google Scholar]
- 25.Mansour S L, Thomas K R, Capecchi M R. Disruption of the proto-oncogene int-2 in mouse embryo-derived stem cells: a general strategy for targeting mutations to non-selectable genes. Nature. 1988;336:348–352. doi: 10.1038/336348a0. [DOI] [PubMed] [Google Scholar]
- 26.Millar S E, Miller M W, Stevens M E, Barsh G S. Expression and transgenic studies of the mouse agouti gene provide insight into the mechanisms by which mammalian coat color patterns are generated. Development. 1995;121:3223–3232. doi: 10.1242/dev.121.10.3223. [DOI] [PubMed] [Google Scholar]
- 27.Nagy A, Rossant J, Nagy R, Abramow-Newerly W, Roder J C. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc Natl Acad Sci USA. 1993;90:8424–8428. doi: 10.1073/pnas.90.18.8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nishiguchi S, Wood H, Kondoh H, Lovell-Badge R, Episkopou V. Sox1 directly regulates the gamma-crystallin genes and is essential for lens development in mice. Genes Dev. 1998;12:776–781. doi: 10.1101/gad.12.6.776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pennisi D, Gardner J, Chambers D, Hosking B, Peters J, Muscat G, Abbott C, Koopman P. Mutations in Sox18 underlie cardiovascular and hair follicle defects in ragged mice. Nat Genet. 2000;24:434–437. doi: 10.1038/74301. [DOI] [PubMed] [Google Scholar]
- 30.Pingault V, Bondurand N, Kuhlbrodt K, Goerich D E, Prehu M O, Puliti A, Herbarth B, Hermans-Borgmeyer I, Legius E, Matthijs G, Amiel J, Lyonnet S, Ceccherini I, Romeo G, Smith J C, Read A P, Wegner M, Goossens M. SOX10 mutations in patients with Waardenburg-Hirschsprung disease. Nat Genet. 1998;18:171–173. doi: 10.1038/ng0298-171. [DOI] [PubMed] [Google Scholar]
- 31.Rudnicki M A, Jaenisch R. The MyoD family of transcription factors and skeletal myogenesis. Bioessays. 1995;17:203–209. doi: 10.1002/bies.950170306. [DOI] [PubMed] [Google Scholar]
- 32.Schilham M W, Oosterwegel M A, Moerer P, Ya J, de Boer P A J, van de Wetering M, Verbeek S, Lamers W H, Kruisbeek A M, Cumano A, Clevers H. Defects in cardiac outflow tract formation and pro-B-lymphocyte expansion in mice lacking Sox-4. Nature. 1996;380:711–714. doi: 10.1038/380711a0. [DOI] [PubMed] [Google Scholar]
- 33.Slee J. The morphology and development of ragged—a mutant affecting the skin and hair of the house mouse. I. Adult morphology. J Genet. 1957;55:100–121. [Google Scholar]
- 34.Soriano N S, Russell S. The Drosophila SOX-domain protein Dichaete is required for the development of the central nervous system midline. Development. 1998;125:3989–3996. doi: 10.1242/dev.125.20.3989. [DOI] [PubMed] [Google Scholar]
- 35.Southard-Smith E M, Kos L, Pavan W J. Sox10 mutation disrupts neural crest development in Dom Hirschsprung mouse model. Nat Genet. 1998;18:60–64. doi: 10.1038/ng0198-60. [DOI] [PubMed] [Google Scholar]
- 36.Stock D W, Buchanan A V, Zhao Z, Weiss K M. Numerous members of the Sox family of HMG box-containing genes are expressed in developing mouse teeth. Genomics. 1996;37:234–237. doi: 10.1006/geno.1996.0548. [DOI] [PubMed] [Google Scholar]
- 37.Uwanogho D, Rex M, Cartwright E J, Pearl G, Healy C, Scotting P, Sharpe P T. Embryonic expression of the chicken Sox2, Sox3 and Sox11 genes suggests an interactive role in neural development. Mech Dev. 1994;49:23–36. doi: 10.1016/0925-4773(94)00299-3. [DOI] [PubMed] [Google Scholar]
- 38.Wagner T, Wirth J, Meyer J, Zabel B, Held M, Zimmer J, Pasantes J, Bricarelli F D, Keutel J, Hustert E, Wolf U, Tommerup N, Schempp W, Scherer G. Autosomal sex reversal and campomelic dysplasia are caused by mutations in and around the SRY-related gene SOX9. Cell. 1994;79:1111–1120. doi: 10.1016/0092-8674(94)90041-8. [DOI] [PubMed] [Google Scholar]
- 39.Wegner M. From head to toes: the multiple facets of Sox proteins. Nucleic Acids Res. 1999;27:1409–1420. doi: 10.1093/nar/27.6.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wheatley S C, Isacke C M, Crossley P H. Restricted expression of the hyaluronan receptor, CD44, during postimplantation mouse embryogenesis suggests key roles in tissue formation and patterning. Development. 1993;119:295–306. doi: 10.1242/dev.119.2.295. [DOI] [PubMed] [Google Scholar]
- 41.Wood S A, Allen N D, Rossant J, Auerbach A, Nagy A. Non-injection methods for the production of embryonic stem cell-embryo chimaeras. Nature. 1993;365:87–89. doi: 10.1038/365087a0. [DOI] [PubMed] [Google Scholar]