Abstract
This article provides a comprehensive and up-to-date overview of the repositories that contain color fundus images. We analyzed them regarding availability and legality, presented the datasets’ characteristics, and identified labeled and unlabeled image sets. This study aimed to complete all publicly available color fundus image datasets to create a central catalog of available color fundus image datasets.
Keywords: color retinal fundus images, health data, image datasets, ophthalmic diseases
1. Introduction
Research and healthcare delivery are changing in the digital age. Digital health research and deep-learning-based applications are promising to transform some of the ways we care for our patients and expand access to healthcare in both developed and underprivileged regions of the world [1,2,3]. Automated screening for diabetic retinopathy (DR) is one facet of this transformation, with deep-learning algorithms already supplementing clinical practice in different parts of the world [4,5].
As the barrier to entry for creating deep-learning-based applications significantly diminished over the last few years, many smaller companies and institutions now attempt to create their algorithms for healthcare, particularly for image-based analysis [6]. Radiology and ophthalmology are medical specialties for which deep learning is most applicable due to their reliance on images and visual analysis [7,8,9]. Color fundus photos, retinal and anterior chamber optical coherence tomography (OCT) scans, and visual field analyzer reports can all lend themselves to automatic analysis for various possible pathologies [9,10].
Creating these algorithms requires sizable numbers of initial images, both with and without pathology. Data are needed in every step of developing a deep-learning application. In the modern world, with electronic healthcare records, centralized imaging storage, and the pervasiveness of digital solutions and storage, such data are generated worldwide in massive quantities due to access, cost, and healthcare issues. Privacy and data governing laws, lack of centralized databases, heterogeneity within particular datasets, lack of or insufficient labeling, or the sheer volume of images required to access such data are often challenging. This article provides an overview of the repositories containing color fundus photos. We analyze them in terms of availability and legality of use. We present the characteristics of datasets and identify labeled and not labeled sets of images. We also analyze the origin of the datasets. This review aims to complement all publicly available color fundus image datasets to create a central catalog of what is currently available. We list the source of each dataset, their availability, and a summary of the populations represented.
Khan and colleagues have previously identified, described, listed, and jointly reviewed 94 ophthalmological imaging datasets [11]. However, this review is outdated now. We aim to provide an update on those datasets’ current state and accessibility.
Khan et al.’s [11] work includes 54 repositories of color fundus photos. At the time of writing this article, only 47 were available. In this work, we have added 73 repositories of color fundus photos, samples of the content of each of them (https://shorturl.at/hmyz3; accessed on 16 May 2023), a tool for automatic content inspection of current or future repositories, accurate access type information (form/registration/e-mail to the authors/no difficulties), degree of difficulty of access to the repository, total file size, image sizes, photo descriptions, information about additional artifacts and legality of use, along with any required papers to be cited.
In addition, we classified the datasets into the following five categories:
The availability of the datasets;
A breakdown of the legality of using the datasets;
A classification of the image descriptions;
Geographical distribution of the datasets that are available by continent and country.
2. Methods
We used the information presented in the review of publicly available ophthalmological imaging datasets [11]. Each dataset includes details about its accessibility, data access, file types, countries of origin, number of patients undergoing examination, number of all images taken, ocular diseases, types of eye examinations performed, and the device used. We have extended the information about all the datasets marked as available in the review mentioned above. We found 47 such color fundus image repositories.
We then used well-known tools to find other repositories not described in the mentioned papers. Searching for color fundus image repositories consisted of typing different types of terms into three types of search engines, including “fundus”, “retina” and “retinal image” along with the words “dataset”, “database” and “repositories”. The exact search was done in the Google search engine and the Google Dataset search engine, designed to search online datasets. Google Dataset Search is designed for online repository discovery and supports searching for tabular, graphical, and text datasets. Indexing is available for publishing their dataset with a metadata reference schema. All results from the search describe the dataset’s contents, direct links, and file format. Google searches also included terms related to images of the retina and terms related to datasets. For both searches, we considered the first ten pages of results. We found 17 unique repositories: 5 using the Google search engine and 12 using the Google Dataset search engine.
The third search engine we chose was Kaggle. Kaggle is a data science and artificial intelligence platform on which users can share their datasets and examine the datasets shared by others. Kaggle datasets are open-sourced, but to determine for what purposes these datasets can be used, we need to check the datasets’ licenses. The vast majority of Kaggle datasets are reliable. We can judge a dataset’s reliability by looking at its upvotes or reviewing the notebooks shared using the dataset. We used the same types of terms as with the Google search engine and Google Dataset search engine. We found 61 unique repositories. We investigated the actual condition of files with their total size, image sizes, information about image description, additional artifacts found in images, issues of legality of data used in scientific applications, and visualizations of sample data. We did not exclude any color fundus image datasets based on the age, sex, or ethnicity of the patients from whom data was collected. We also included datasets of all languages and geographic origins.
2.1. Dataset Checking Strategy
We noticed that the levels of access to the datasets varied: from fully accessible to available on request after sending a request to the authors. Some datasets were also unavailable. In this article, we have defined access levels as follows:
-
(1)
Fully open;
-
(2)
Available after completing a form;
-
(3)
Available after account registration (and possible approval by the authors);
-
(4)
Available after sending an email to authors and approval from them;
-
(5)
Not available.
After accessing, we manually checked each dataset described in 1 by downloading them to extract information about file status, sizes, and additional artifacts found in the images. Most available datasets were available as compressed files (ZIP/RAR/7Z), but some were available as separate files. We determined the sizes of the datasets in which the files were delivered separately by downloading all files and summing their sizes. We have prepared a tool to automatically generate the discussed information on repository contents and image samples—Ophthalmic Repository Sample Generator (Section 4.1). We also manually checked the content of each randomly selected image to see if there were any additional artifacts.
2.2. Image Descriptions and Legality of Use
All the datasets we reviewed were described on dedicated web pages or in scientific publications. Based on these sources, we have determined methods of describing images included in these datasets.
We noticed the following types of image descriptions:
-
(1)
Manually assigned labels corresponding to diagnosed ocular diseases, image quality, or described areas of interest;
-
(2)
Manual annotations on images indicating areas of interest;
-
(3)
No descriptions.
We have also extracted information on the legality of data use from websites dedicated to the datasets. We noticed the following approaches to defining the legality of the use of data contained in the datasets:
-
(1)
Notifying the authors of the datasets of the results and awaiting permission to publish the results;
-
(2)
References to the indicated articles or the dataset in the case of publication of the results;
-
(3)
No restrictions.
3. Data Availability
In the analysis, we used 121 publicly available datasets containing color fundus images [12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89,90,91,92,93,94,95,96,97,98,99,100,101,102,103,104,105,106,107,108,109,110,111,112,113,114,115,116,117,118,119,120,121,122,123,124,125,126,127].
4. Code Availability
4.1. Ophthalmic Repository Sample Generator
We developed a generator of pseudo-random samples from publicly available repositories containing color fundus photos. The generator is written in Python 3 programming language and is available on GitHub (https://github.com/betacord/OphthalmicRepositorySampleGenerator; accessed on 16 May 2023).
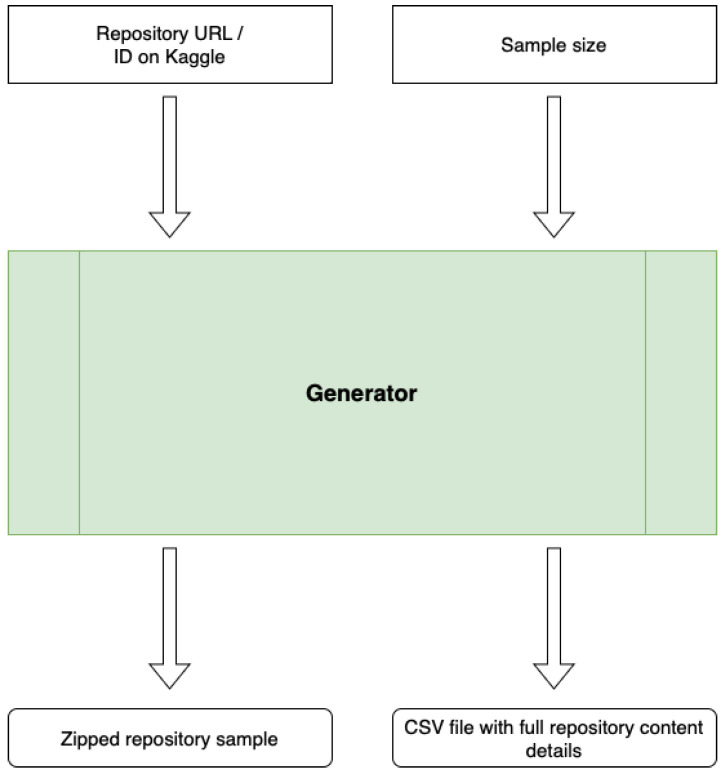
The prepared tool facilitates the manual inspection of the contents of repositories. The program obtains the URL of a given repository (or ID on the Kaggle platform) and the sample size (n). The operation result will be a pseudo-random selection of n color fundus photos from the repository and a CSV file containing extracted attributes representing the entire repository. The tool can be easily run on a local computer or in a cloud environment. The general scheme of the generator is shown in Figure 1.
Figure 1.
General scheme of the Ophthalmic Repository Sample Generator.
As an input, the generator takes the sample size, the repository URL, the data output file path, the temporary full data path, the repository sample output path, the repository type, and the output CSV file path.
The sample size is an integer number representing the size of the random output sample of photos from the repository.
A repository URL is a string representing a direct URL of the image file; e.g., for the Kaggle dataset, the schema is [username]/[dataset_id]. In the case of a Kaggle competition, it is ID.
The data output file path is a string representing the output file with the downloaded repository content.
The temporary full data path is a string representing the temporary path to which the repository will be extracted.
The repository sample output path is a string representing the path where a randomly selected repository sample will be placed.
The repository type is an integer representing the type of the repository source: 0 for classic URL, 1 for Kaggle competition, and 2 for Kaggle dataset.
The output CSV file path is a string representing the path where the CSV file will be saved (separated by;) containing information about the repository.
Therefore, external parameters characterizing the size of the generated sample of images, data source, temporary paths, source type, and paths to the output files should also be included in the tool’s run.
5. Results
In Table 1, we included the results of our review. In total, we checked 127 repositories containing color fundus images, of which 120 were currently available, and seven were unavailable due to a non-existent URL. Downloading one dataset was prevented due to a critical server error, and one dataset was delivered as a corrupt zipped file. We have described the characteristics only for the available datasets. We also generated a sample of their content and placed it in the cloud (https://shorturl.at/hmyz3; accessed on 16 May 2023).
Table 1.
Characteristics of the open access datasets.
| ID | Name | Access | Technical Details | Epidemiological Details | Legality | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Access Type | Ease of Access | Number of Photos | Photo Sizes | Photo Descriptions | Size of the Repository | Additional Artifacts in the Photos | Repository URL | Number of Patients | Eye Diseases | Country of Origin | |||
| 1 | 1000FIWC | OAAR | 2 | 1000 | Different: 3000 × 3152 px; 1728 × 2592 px; … | Labels | 777 MB | Absence | [12] | NR | DR, G | China | Citation: [128] |
| 2 | ACDLTFARIFP | OA | 1 | 2206 | 350 × 346 × 3 px | Absence | 294 MB | Absence | [13] | NR | NR | NR | Citation: [129] |
| 3 | ACRFAODAWDCMFERLRLASMAGD Version 1 | OA | 1 | 2560 | Different: 456 × 951 px; 576 × 760 px; … | Labels | 302 MB | Absence | [14] | NR | DME, CSR, AMD, G | UAE | Citation: [130] |
| 4 | ACRFAODAWDCMFERLRLASMAGD Version 2 | OA | 1 | 2988 | Different: 456 × 951 px; 576 × 760 px; … | Labels | 1597 MB | Absence | [15] | NR | G | UAE | Citation: [131] |
| 5 | ACRFAODAWDCMFERLRLASMAGD Version 3 | OA | 1 | 3100 | Different: 1934 × 2032 px; 576 × 760 px; … | Labels | 2969 MB | Absence | [16] | NR | DME, CSR, AMD, G | UAE | Citation: [132] |
| 6 | ACRFAODAWDCMFERLRLASMAGD Version 4 | OA | 1 | 2560 | Different: 1934 × 2032 px; 576 × 760 px; … | Labels | 1433.6 MB | Absence | [17] | NR | DME, CSR, AMD, G | UAE | Citation: [133,134] |
| 7 | ACRIMA | OA | 3 | 705 | Different: 349 × 349 px; 871 × 871 px; … | Labels | 23 MB | Absence | [18] | NR | G | Spain | Citation: [135] |
| 8 | APTOS | OAAL | 2 | 5590 | Different: 640 × 480 px; 2416 × 1736; … | Labels | 9748 MB | Absence | [19] | NR | DR | India | NR |
| 9 | Arteriovenous Nicking | OA | 3 | 90 | 401 × 401 px | Labels | 29 MB | Absence | [20] | NR | NR | NR | NR |
| 10 | CHHSIE | OA | 3 | 28 | 999 × 960 px | Annotations | 3 MB | Absence | [21] | 14 | HE | UK | NR |
| 11 | CTDRD | OAAR | 2 | 35,136 | Different: 4712 × 3163 px; 2213 × 2205 px; … | Absence | 40,755 MB | Absence | [22] | NR | DR | NR | NR |
| 12 | Calibration level 1 | OAAR | 2 | 3200 | Different: 1424 × 2144 px; 1536 × 2048 px; | Labels | 8181 MB | Absence | [23] | NR | DR | India | Citation: [136] |
| 13 | Cardiacemboli | OAAR | 2 | 943 | Different: 3264 × 4928 px; 2048 × 3072 px; … | Labels | 767 MB | Absence | [24] | NR | CE | NR | NR |
| 14 | DDR | OAAR | 2 | 12524 | Different: 512 × 512 px; 1536 × 2048 px; … | Labels | 3348 MB | Absence | [25] | NR | DR | China | Citation: [137] |
| 15 | DEFRIOVAA | OA | 3 | 50 | Different: 798 × 658 px; 886 × 719 px; … | Absence | 43.2 MB | Absence | [26] | 50 | DED | Netherlands | NR |
| 16 | DFFIFTSODR Version 0.1 | OA | 1 | 396 | 2124 × 2056 px | Labels | 238 MB | Absence | [27] | NR | DR | Paraguay | Citation: [138] |
| 17 | DFFIFTSODR Version 0.2 | OA | 1 | 757 | Different: 1444 × 1444 px; 2056 × 2124 px; … | Labels | 375 MB | Absence | [28] | NR | DR | Paraguay | Citation: [138] |
| 18 | DFFIFTSODR Version 0.3 | OA | 1 | 1437 | Different: 1028 × 1052 px; 1029 × 1062 px | Labels | 1536 MB | Absence | [29] | NR | DR | Paraguay | Citation: [138] |
| 19 | DHRFID | OAAR | 2 | 6542 | Different: 3456 × 5184 px; 1904 × 2460 px; … | Labels | 857 MB | Absence | [30] | NR | DR, G, O | NR | NR |
| 20 | DOFIFVSDOHRDRAP | OA | 1 | 100 | 1504 × 1000 px | Absence | 192 MB | Presence | [31] | NR | HR, DR, P | Pakistan | Citation: [139] |
| 21 | DOOAFI Version 1 | OA | 1 | 50 | 2032 × 1934 px | Absence | 33 MB | Absence | [32] | NR | G | NR | Citation: [140] |
| 22 | DOOAFI Version 2 | OA | 1 | 50 | 2032 × 1 934 px | Absence | 33 MB | Absence | [33] | NR | G | NR | Citation: [140] |
| 23 | DR (resized) | OAAR | 2 | 70,234 | Different: 278 × 278 px; 774 × 1024 px; … | Labels | 8038 MB | Absence | [34] | NR | DR | China | NR |
| 24 | DR 1 | OA | 3 | 1077 | 640 × 480 px | Labels | 200 MB | Absence | [35] | NR | DED | Brazil | Citation: [141] |
| 25 | DR 2 | OA | 3 | 520 | Different: 873 × 500 px; 877 × 582 px; 876 × 581 px; … | Labels | 200 MB | Absence | [36] | NR | DED | Brazil | Citation: [141] |
| 26 | DR 3 | OAAR | 2 | 13251 | Different: 1184 × 1792 px; 2048, 3072 px; … | Labels | 14,848 MB | Absence | [38] | NR | DR | NR | NR |
| 27 | DR 4 | OAAR | 2 | 105375 | 512 × 786 px | Labels | 12,697 MB | Absence | [37] | NR | DR | NR | NR |
| 28 | DR 5 | OAAR | 2 | 3662 | Different: 358 × 474 px; 1226 × 1844 px; … | Labels | 8908 MB | Absence | [39] | NR | DR | NR | NR |
| 29 | DR 6 | OAAR | 2 | 301 | 2056 × 2124 px | Labels | 128 MB | Absence | [40] | NR | DR | NR | NR |
| 30 | DR 7 | OAAR | 2 | 103 | Different: 3264 × 4928 px; 2000 × 3008 px; … | Absence | 101 MB | Absence | [41] | NR | NR | NR | NR |
| 31 | DR2015DCR | OAAR | 2 | 35,126 | 224 × 224 px | Labels | 1014 MB | Absence | [42] | NR | DR | NR | NR |
| 32 | DR224 × 2242019 | OAAR | 2 | 3662 | 224 × 224 px | Labels | 238 MB | Absence | [43] | NR | DR | India | NR |
| 33 | DR224 × 224GF | OAAR | 2 | 3662 | 224 × 224 px | Labels | 426 MB | Absence | [44] | NR | DR | India | NR |
| 34 | DR224 × 224GI | OAAR | 2 | 3662 | 224 × 224 px | Labels | 157 MB | Absence | [45] | NR | DR | India | NR |
| 35 | DRA | OAAR | 2 | 35,126 | Different: 1024 × 1024 px; 779 × 1024 px; … | Labels | 1024 MB | Absence | [46] | NR | DR | NR | NR |
| 36 | DRB | OAAR | 2 | 49,703 | 512 × 512 px | Labels | 2048 MB | Absence | [47] | NR | DR | NR | NR |
| 37 | DRBDC | OAAR | 2 | 88,700 | Different: 4752 × 3168 px; 2560 × 1920 px; … | Labels | 98,017 MB | Absence | [48] | NR | DR | NR | NR |
| 38 | DRC | OAAR | 2 | 70,234 | Different: 278 × 278 px; 727 × 1024 px; … | Labels | 8192 MB | Absence | [49] | NR | DR | NR | NR |
| 39 | DRC #2 | OAAR | 2 | 2608 | 224 × 224 px | Absence | 33 MB | Absence | [50] | NR | NR | NR | NR |
| 40 | DRC 3 | OAAR | 2 | 2608 | 224 × 224 px | Absence | 33 MB | Absence | [51] | NR | NR | NR | NR |
| 41 | DRD | OAAR | 2 | 2750 | 256 × 256 px | Labels | 350 MB | Absence | [52] | NR | DR | NR | NR |
| 42 | DRD 2 | OAAR | 2 | 2111 | Different: 554 × 512 px; 424 × 512 px; … | Labels | 269 MB | Absence | [53] | NR | DR | NR | NR |
| 43 | DRD 3 | OAAR | 2 | 3662 | 224 × 224 px | Labels | 349 MB | Absence | [54] | NR | DR | NR | NR |
| 44 | DRD 4 | OAAR | 2 | 12,844 | 224 × 224 px | Labels | 725 MB | Absence | [55] | NR | DR | NR | NR |
| 45 | DRHARMDAGI | OA | 3 | 39 | Different: 3216 × 2136 px; 2816 × 1880 px; … | Labels | 10 MB | Presence | [56] | 38 | DED, HR, G, AMD | UK | Citation: [142] |
| 46 | DRIFONS | OA | 3 | 110 | 600 × 400 px | Annotations | 2.5 MB | Presence | [57] | 55 | HR | Spain | Citation: [143] |
| 47 | DRIFVS | OAAR | 2 | 40 | 565 × 584 px | Annotations | 30 MB | Absence | [58] | 400 | DED | Netherlands | NR |
| 48 | DRO | OAAR | 2 | 35,128 | Different: 278 × 278 px; 738 × 1024 px; … | Labels | 6656 MB | Absence | [59] | NR | DR | NR | NR |
| 49 | DRPD | OAAR | 2 | 13,970 | 128 × 128 px | Labels | 46 MB | Absence | [60] | NR | DR | NR | NR |
| 50 | DRR300 × 300C | OAAR | 2 | 16,798 | Different: 300 × 300 px; 271 × 273 px; … | Labels | 123 MB | Absence | [61] | NR | DR | NR | NR |
| 51 | DRS | OAAR | 2 | 10024 | Different: 315 × 400 px; 1957 × 2196 px; … | Absence | 10,240 MB | Absence | [62] | NR | NR | NR | NR |
| 52 | DRTW | OAAR | 2 | 37254 | Different: 278 × 278 px; 899 × 1024 px; … | Labels | 4915 MB | Absence | [63] | NR | DR | NR | NR |
| 53 | DRU | OAAR | 2 | 34882 | Different: 3456 × 5184 px; 1957 × 2196 px; … | Labels | 32,768 MB | Absence | [64] | NR | DR | China | NR |
| 54 | DR_2000 | OAAR | 2 | 2000 | Different: 3456 × 5184; 2056 × 3088 px; … | Labels | 2048 MB | Absence | [65] | NR | DR | China | NR |
| 55 | DR_201010 | OAAR | 2 | 35,136 | Different: 2560 × 1920 px; 2592 × 1944 px; … | Labels | 38,809 MB | Absence | [66] | NR | DR | NR | NR |
| 56 | DeepDRiD | OA | 1 | 320 | Different: 1725 × 2230 px; 3072 × 3900; … | Absence | 1464 MB | Absence | [67] | NR | NR | NR | Citation: [138] |
| 57 | DiaRetDB1 V2.1 | OAAR | 2 | 89 | 1152 × 1500 px | Annotations | 137 MB | Absence | [68] | NR | DR | Finland | Citation: [144] |
| 58 | Diabetic | OAAR | 2 | 2769 | Different: 342 × 512 px; 434 × 512 px; … | Labels | 353 MB | Absence | [69] | NR | DR | NR | NR |
| 59 | Diabetic Retinopathy Detection Processed | OAAR | 2 | 51,500 | Different: 224 × 224 px; 400 × 400 px; … | Labels | 2764 MB | Absence | [70] | NR | DR | NR | NR |
| 60 | Drishti-GS | OAAR | 2 | 101 | Different: 1845 × 2050 px; 1763 × 2047 px; … | Annotations | 341 MB | Absence | [71] | NR | NR | NR | NR |
| 61 | Drishti-GS1 | OAAR | 2 | 101 | Different: 2048 × 1760 px; 2049 × 1749; … | Annotations, labels | 350 MB | Absence | [72] | NR | G | India | Citation: [145,146] |
| 62 | EOptha Diabetic Retinopathy | OAAR | 2 | 46 | Different: 1696 × 2544 px; 1000 × 1504 px; … | Annotations | 21 MB | Absence | [73] | NR | DR | NR | NR |
| 63 | EPACS | OA | 2 | 88702 | Different: 4752 × 3168 px; 4928 × 3264 px; … | Labels | 84,203 MB | Absence | [74] | NR | DED | USA | NR |
| 64 | Eye Dataset Workshop | OAAR | 2 | 12734 | Different: 3264 × 3264 px; 2592 × 2592 px; … | Labels | 672 MB | Absence | [75] | NR | DR | NR | NR |
| 65 | FID | OAAR | 2 | 32 | 605 × 700 px | Absence | 1 MB | Absence | [76] | NR | HE | NR | NR |
| 66 | FIRD | OA | 3 | 268 | 2912 × 2912 px | Annotations | 264 MB | Absence | [77] | 39 | NR | Greece | Citation: [147] |
| 67 | Fundus | OAAR | 2 | 1600 | Different: 1725 × 2230 px; 1727 × 2232 px; … | Labels | 890 MB | Absence | [78] | NR | DR | NR | NR |
| 68 | Fundus Images | OAAR | 2 | 650 | Different: 2048 × 3072 px; 2048 × 3085 | Labels | 206 MB | Absence | [79] | NR | G | NR | NR |
| 69 | Fundus_DR | OAAR | 2 | 61830 | 256 × 256 px | Labels | 1710 MB | Absence | [80] | NR | DR | NR | NR |
| 70 | Fundusvessels | OAAL | 1 | 3909 | 565 × 584 × 3 px | Absence | 100 MB | Absence | [98] | NR | NR | China | Citation: [148] |
| 71 | GFDR 2 | OAAR | 2 | 3662 | 224 × 224 px | Labels | 349 MB | Absence | [81] | NR | DR | NR | NR |
| 72 | Glaucoma Fundus | OA | 3 | 1542 | 240 × 240 px | Labels | 118.2 MB | Absence | [82] | 1542 | G | Republic of Korea | Citation |
| 73 | HEIME | OA | 3 | 169 | 2196 × 1958 px | Annotations | 300 MB | Absence | [83] | 910 | DED | USA | Citation: [149] |
| 74 | HRFQA | OA | 3 | 45 | 3888 × 2592 px | Labels | 68.1 MB | Absence | [84] | 45 | DED | Germany and Czech Republic | Citation: [150] |
| 75 | HRFQS | OA | 3 | 36 | 3504 × 2336 px | Annotations, labels | 73 MB | Absence | [84] | 18 | NR | Germany and Czech Republic | Citation: [151] |
| 76 | IARMD | OAAR | 2 | 1200 | 2124 × 2056 px | Labels | 606 MB | Absence | [85] | NR | AMD | China | NR |
| 77 | IDRID | OAAR | 2 | 516 | 4288 × 2848 px | Labels | 970 MB | Absence | [86] | NR | DED | India | Citation: [152] |
| 78 | INSFPIOTRAR | OAAF | 2 | 40 | 2392 × 2048 px | Annotations | 80 MB | Absence | [87] | NR | G | USA | Citation: [153] |
| 79 | INSFPIOTRS | OAAF | 2 | 30 | 768 × 1019 px | Annotations, labels | 80 MB | Presence | [87] | 15 | G | USA | Citation: [154] |
| 80 | IPM | OAAR | 2 | 1200 | 2124 × 2056 px | Labels | 608 MB | Absence | [88] | NR | M | China | Citation: [155] |
| 81 | IRC | OAAR | 2 | 1032 | Different: 2848 × 3408 px; 2848 × 3712 px; … | Labels | 486 MB | Absence | [89] | NR | DR, DME | India | NR |
| 82 | ISBI_2021_Retina_23 | OAAR | 2 | 2560 | Different: 1424 × 2144 px; 1536 × 2048 px; … | Labels | 6400 MB | Absence | [90] | NR | O | NR | NR |
| 83 | ISBI_RETINA_TEST | OAAR | 2 | 640 | Different: 1424 × 2144 px; 1536 × 2048 px; … | Labels | 1597 MB | Absence | [91] | NR | O | NR | NR |
| 84 | JSIEC | OA | 3 | 1000 | 3046 × 2572 px | Labels | 778 MB | Absence | [92] | NR | DED | China | Citation: [129] |
| 85 | Jichi DR | OA | 3 | 9939 | 1272 × 1272 px | Labels | 845 MB | Absence | [93] | 2740 | DED | Japan | NR |
| 86 | LSABG | OAAE | 1 | 4854 | 500 × 500 px | Labels | 300 MB | Absence | [94] | NR | G | China | Citation: [156] |
| 87 | Messidor-2 | OAAF | 2 | 1748 | 2240 × 1488 px | Absence | 2355 MB | Absence | [95] | 874 | DED | France | Citation: [157] |
| 88 | ODIR | OAAR | 1 | 8000 | 2048 × 1536 | Labels | 1228 MB | Absence | [96] | 5000 | DED, HR, G, AMD, C, M, O | China | NR |
| 89 | ODR | OAAR | 2 | 14,392 | Different: 188 × 250 px; 1607 × 2139 px; … | Labels | 2078 MB | Absence | [97] | 5000 | D, G | China | NR |
| 90 | OFOAOS | OAAR | 1 | 3909 | 740 × 740 × 3 px | Annotations | 100 MB | Absence | [98] | NR | NR | China | NR |
| 91 | Papila Version 1 | OA | 1 | 490 | 2576 × 1934 px | Annotations, labels | 563 MB | Absence | [99] | NR | M, H, A | Paraguay | Citation: [158] |
| 92 | Papila Version 2 | OA | 1 | 488 | 2576 × 1934 px | Annotations, labels | 563 MB | Absence | [100] | NR | M, H, A | Spain | Citation: [158] |
| 93 | R20152019BDI | OAAR | 2 | 94,292 | Different: 149 × 1024 px; 825 × 1024 px; … | Labels | 18,432 MB | Absence | [101] | NR | DR | Indie | NR |
| 94 | REFUGE Challenge 2020 | OAAR | 1 | 1200 | Different: 2124 × 2056 px; 1634 × 1634 px | Annotations, labels | NR | Absence | [102] | NR | G | China | Citation: [159] |
| 95 | RFGC | OAAR | 2 | 1200 | 1634 × 1634 px | Labels | 1433 MB | Absence | [103] | NR | G | China | NR |
| 96 | RFI | OAAR | 2 | 21,746 | Different: 314 × 336 px; 753 × 1024 px; … | Labels | 2048 MB | Absence | [104] | NR | DR, M, C | NR | NR |
| 97 | RFIFGA | OA | 3 | 750 | 2376 × 1584 px | Annotations | 13,209 MB | Absence | [105] | NR | G | Saudi Arabia, France | Citation: [160] |
| 98 | RFIR | OAAR | 2 | 270 | Different: 2912 × 2912 px; 2912 × 2912 px; … | Annotations | 456 MB | Presence | [106] | NR | NR | NR | Citation: [147] |
| 99 | RIM-ONE Version 2 | OA | 3 | 455 | Different: 398 × 401 px; 517 × 494 px; … | Annotations, labels | 12 MB | Absence | [107] | NR | G | Spain | NR |
| 100 | RIM-ONE Version 3 | OA | 3 | 159 | 2144 × 1424 px | Annotations, labels | 227 MB | Absence | [107] | NR | G | Spain | NR |
| 101 | RITE | OAAR | 2 | 100 | 512 × 512 px | Annotations | 33 MB | Absence | [108] | NR | NR | USA | Citation: [161] |
| 102 | RODRD | OAAF | 3 | 1120 | 2000 × 1312 px | Labels | 4710 MB | Absence | [109] | 70 | DED | Netherlands | Citation: [162] |
| 103 | RVCSF | OAAR | 2 | 226 | Different: 224 × 224 px; 605 × 700 px | Annotations | 6 MB | Absence | [110] | NR | NR | NR | NR |
| 104 | Retina | OAAL | 2 | 601 | Different: 1848 × 1224 px; 2592 × 1728; … | Labels | 3072 MB | Absence | [111] | NR | G, C, RD | NR | NR |
| 105 | Retina Online Challenge | OAAF | 2 | 100 | 768 × 576 px | Annotations | 26 MB | Absence | [112] | NR | DED | Netherlands | NR |
| 106 | Retina_Quality | OAAR | 2 | 26,052 | Different: 3246 × 3245 px; 2258 × 2257 px; … | Labels | 3788 MB | Absence | [113] | NR | DR | NR | NR |
| 107 | Retinagen | OAAR | 2 | 500 | Different: 605 × 700 px; 1106 × 1280 px; … | Absence | 23 MB | Absence | [114] | NR | NR | NR | NR |
| 108 | Retinal Vessel Tortuosity | OAAF | 2 | 60 | 1200 × 900 px | Annotations | 16 MB | Markers | [115] | 34 | HR | Italy | Citation, inform: [163] |
| 109 | Retinal_tiny | OAAR | 2 | 2062 | 512 × 512 | Labels | 104 MB | Absence | [116] | NR | DR | NR | NR |
| 110 | SAOTR | OA | 3 | 397 | 700 × 605 px | Annotations, labels | 361 MB | Absence | [117] | NR | DED | USA | NR |
| 111 | SDRD | OAAR | 2 | 1939 | 224 × 224 px | Labels | 188 MB | Absence | [118] | NR | DR | NR | NR |
| 112 | SUSTech-SYSU | OAAR | 2 | 1151 | 2136 × 2880 px | Labels | 400 MB | Absence | [119] | NR | DR | China | Citation: [164] |
| 113 | Sydney Innovation Challenge 2019 | OAAR | 2 | 14,145 | 2448 × 3264 px | Labels | 18,534 MB | Absence | [120] | NR | DR | Australia | NR |
| 114 | VACRDD | OAAR | 2 | 3785 | 512 × 512 px | Labels | 61 MB | Absence | [121] | NR | DR, G, O | NR | NR |
| 115 | VIFTCOTAVR | OAAE | 1 | 56 | 768 × 576 px | Annotations | 12 MB | Absence | [122] | NR | NR | Spain | Citation: [165] |
| 116 | WIDE | OA | 3 | 30 | Different: 731 × 1300 px; 977 × 1516 px; 854 × 1393 px… | Annotations | 63 MB | Absence | [123] | 30 | AMD | USA | Citation: [166] |
| 117 | William Hoyt | OA | 3 | 850 | Different: 601 × 600 px; 596 × 600 px; … | Labels | 170 MB | Presence | [124] | NR | P | NR | NR |
| 118 | Yangxi | OA | 3 | 18,394 | 297 × 297px | Labels | 5529 MB | Absence | [125] | 5825 | AMD | China | NR |
| 119 | dr15_test | OAAR | 2 | 34,043 | 512 × 512 px | Labels | 10,240 MB | Absence | [126] | NR | DR | NR | NR |
| 120 | merged_retina_datasets | OAAR | 2 | 2451 | Different: 2847 × 3925 px; 2847 × 3414 px; … | Labels | 5816 MB | Absence | [127] | NR | DR, O | NR | NR |
Dataset acronyms: APTOS = Asia Pacific Tele-Ophthalmology Society, CHHSIE = Child Heart Health Study in England, DEFRIOVAA = Digital Extraction from Retinal Images of Veins and Arteries, DRHARMDAGI = Diabetic Retinopathy Hypertension, Age-Related Macular Degeneration and Glaucoma Images, DRIFONS = Digital Retinal Images for Optic Nerve Segmentation, DRIFVS = Digital Retinal Images for Vessel Segmentation, EPACS = Eye Picture Archive Communication System, FIRD = Fundus Image Registration Dataset, HEIME = Hamilton Eye Institute Macular Edema, HRFQS = High-Resolution Fundus Quality Segmentation, HRFQA = High-Resolution Fundus Quality Assessment, IARMD = iChallenge Age-Related Macular Degeneration, IPM = iChallenge Pathological Myopia, IDRID = Indian Diabetic Retinopathy Image Dataset, INSFPIOTRAR = Iowa Normative Set for Processing Images of the Retina—Arteriovenous Ratio, INSFPIOTRS = Iowa Normative Set for Processing Images of the Retina—Stereo, JSIEC = Joint Shantou International Eye Center, LSABG = Large-Scale Attention-based Glaucoma, RODRD = Rotterdam Ophthalmic Data Repository DR, ODIR = Ocular Disease Intelligent Recognition, RFGC = Retina Fundus Glaucoma Challenge, RFIFGA = Retinal Fundus Images for Glaucoma Analysis, SAOTR = Structured Analysis of the Retina, VIFTCOTAVR = VARPA Images for the Computation of the Arterio/Venular Ratio, DOFIFVSDOHRDRAP = Data on Fundus Images for Vessel Segmentation, Detection of Hypertensive Retinopathy, Diabetic Retinopathy and Papilledema, DFFIFTSODR Version 0.1 = Dataset from Fundus Images for the Study of Diabetic Retinopathy Version 0.1, DFFIFTSODR Version 0.2 = Dataset From Fundus Images for the Study of Diabetic Retinopathy Version 0.2, DFFIFTSODR Version 0.3 = Dataset from Fundus Images for the Study of Diabetic Retinopathy Version 0.3, ACRFAODAWDCMFERLRLASMAGD Version 1 = A Composite Retinal Fundus and OCT Dataset along with Detailed Clinical Markings for Extracting Retinal Layers, Retinal Lesions and Screening Macular and Glaucomatous Disorders Version 1, ACRFAODAWDCMFERLRLASMAGD Version 2 = A Composite Retinal Fundus and OCT Dataset along with Detailed Clinical Markings for Extracting Retinal Layers, Retinal Lesions and Screening Macular and Glaucomatous Disorders Version 2, ACRFAODAWDCMFERLRLASMAGD Version 3 = A Composite Retinal Fundus and OCT Dataset along with Detailed Clinical Markings for Extracting Retinal Layers, Retinal Lesions and Screening Macular and Glaucomatous Disorders Version 3, ACRFAODAWDCMFERLRLASMAGD Version 4 = A Composite Retinal Fundus and OCT Dataset along with Detailed Clinical Markings for Extracting Retinal Layers, Retinal Lesions and Screening Macular and Glaucomatous Disorders Version 4, ACDLTFARIFP = A CycleGAN Deep Learning Technique for Artifact Reduction in Fundus Photography, OFOAOS = ORIGA for OD and OC Segmentation, DeepDRiD = Deep-Diabetic- Retinopathy- Image-Dataset- (DeepDRiD), DOOAFI Version 1 = Data on OCT and Fundus Images Version 1, DOOAFI Version 2 = Data on OCT and Fundus Images Version 2, DR224 × 224GF = Diabetic Retinopathy 224 × 224 Gaussian Filtered, DR2015DCR = Diabetic Retinopathy 2015 Data Colored Resized, DRA = Diabetic Retinopathy Arranged, RFIR = Retina Fundus Image Registration, DR224 × 2,242,019 = Diabetic Retinopathy 224 × 224 (2019 Data), DR224 × 224GI = Diabetic Retinopathy 224 × 224 Grayscale Images, Drishti-GS = Drishti-GS—RETINA DATASET FOR ONH SEGMENTATIO N, DRU = Diabetic Retinopathy Unziped, RVCSF = Retinal Vessel Combine Same Format, DRR300 × 300C = Diabetic-Retinopathy-Resized-300 × 300-Cropped, VACRDD = VietAI Advance Course—Retinal Disease Detection, 1000FIWC = 1000 Fundus Images with 39 Categories, DR (resized) = Diabetic Retinopathy (resized), DHRFID = Derbi_Hackathon _Retinal_Fundus _Image_Dataset, SDRD = Small_Diabetic_ Retinopathy_Dataset, DRD = Diabetic Retinopathy Dataset, R20152019BDI = Resized 2015 & 2019 Blindness Detection Images, DRPD = Diabetic Retinopathy Preprocessed Dataset, DRC = Diabetic Retinopathy Classified, DRD 2 = Diabetic Retinopathy Detection, DRB = Diabetic_Retinopathy_Balanced, DRD 3 = Diabetic Retinopathy Detection, DRBDC = Diabetic Retinopathy Blindness Detection c_data, DRO = Diabetic Retinopathy Organized, Diabetic Retinopathy Detection Processed = Diabetic Retinopathy Detection Processed, CTDRD = Cropped-Train-Diabetic-Retinopathy-Detection, DRTW = Diabetic-Retinopathy-Train- Validation, GFDR 2 = Gaussian_Filtered _Diabetic_Retinopathy, DRC #2 = Diabetic Retinopathy Classification #2, DRC 3 = Diabetic Retinopathy Classification, Sydney Innovation Challenge 2019 = Sydney Innovation Challenge 2019. Access type: OA = Open access, OAAR = Open access after registration, OAAE = open access after sending email, OAAR = open access after login, OAJC = open access after joining the contest and acceptance of the registration, NR= not reported. Diseases: G = glaucoma, HE = healthy eyes, DR = diabetic retinopathy, C = cataracts, RD = retinal diseases, DED = diabetic eye disease, HR = hypertensive retinopathy, AMD = age-related macular degeneration, M = myopia, O = other diseases, P = papilledema, H = hyperopia, A = astigmatism, CSR = central serous retinopathy, DME = diabetic macular edema, CE = cardiac embolism, D = diabetes, RVO = retinal vascular occlusion.
5.1. Data Access
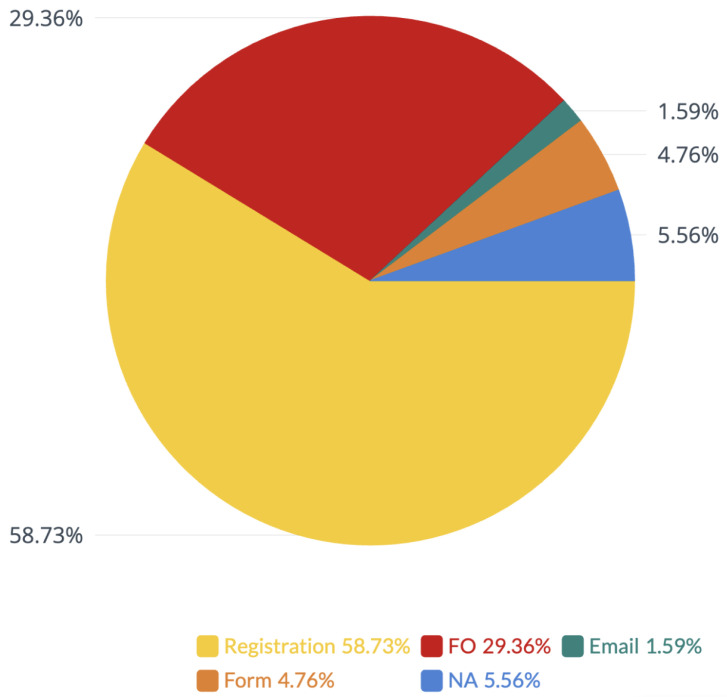
Out of the 127 available datasets, we marked 37 as fully open, 6 as available after completing a form, 75 as available after account registration, 2 as available after sending an email to authors and approval from them, and 7 as not available, as can be seen in Figure 2.
Figure 2.
Availability of datasets. FO = fully open, FORM = available after completing a form, Registration = available after account registration (and possible approval by the authors), Email = available after sending an email to authors and approval from them, NA = not available.
5.2. Characteristics of Datasets
Almost all (122 out of 124) of the datasets could be downloaded as zipped files, and only 2 could be downloaded separately. There was a problem with the extension on three of the zipped files that contained datasets. In 59 datasets, all images had the exact dimensions (in pixels), but there were 68 unique ones. Images from nine different datasets contained additional artifacts such as dates, numbers, digits, color scales, markers, icons, arteries, vessels, veins, and key points marked on the photographs.
5.3. The Legality of Use and Image Descriptions
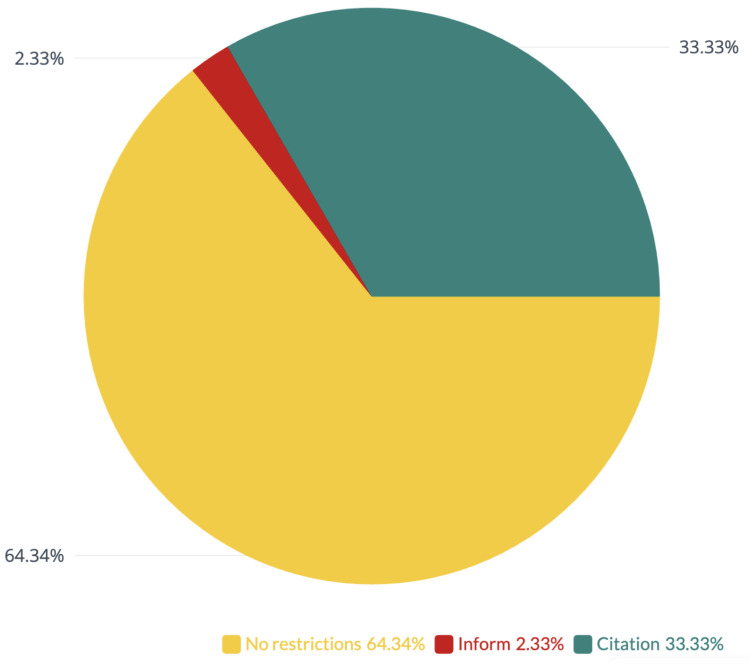
Out of the 127 datasets, the authors of 3 of them provided a note about the need to inform them about the obtained results. Authors of 44 datasets provided information about the need to cite the indicated works using the provided data and publishing the results. Over two-thirds, or 80 datasets, had no restrictions on use. Figure 3 shows the full breakdown of the legality of using data contained in the datasets.
Figure 3.
Breakdown of the legality of using data. Inform = notifying the authors of the datasets of the results and awaiting permission to publish the results, Citation = references to the indicated articles in the case of publication of the results.
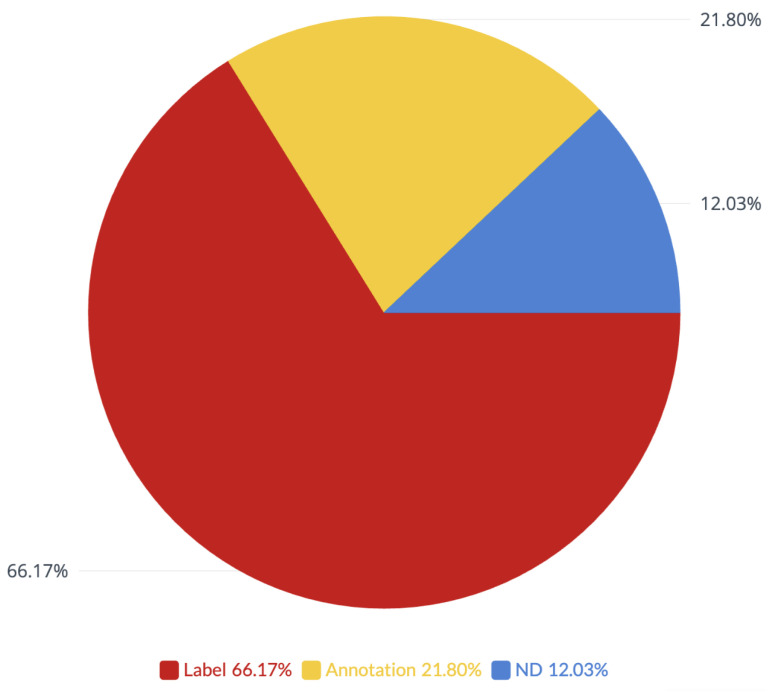
Eighty-nine datasets were labeled with the images assigned to them. Thirty datasets had areas of interest labeled on the images. Sixteen datasets did not have any descriptions. A full breakdown of the image descriptions is shown in Figure 4.
Figure 4.
Breakdown of the image descriptions. Label = manually assigned labels corresponding to diagnosed ocular diseases, image quality or described areas of interest, Annotation = manual annotations on images indicating areas of interest, ND = no descriptions.
6. Discussion
Publicly available datasets remain important in digital health research and innovation in ophthalmology. Although, on the whole, the number of publicly available color fundus image datasets is growing, it is an ongoing process with new datasets arriving and older datasets becoming inaccessible. A central repository or listing for ophthalmic datasets, coupled with the low discoverability of many of the datasets, constitutes a significant barrier to access to high-quality representative data suitable to a given purpose. However, this article provided an up-to-date review and discussion of available color fundus image datasets.
Health data poverty, in this case, scarcity of color fundus image datasets originating from underprivileged regions, particularly Africa, is cause for concern. Although the relationship between patients’ ethnicity, background, and other attributes and fundus features is not clearly documented, the lack of representative datasets might lead to ethnic or geographical bias and poor generalizability in deep-learning applications. The recent relative lack of datasets might mean underrepresented regions miss out on future data-driven screening and healthcare solutions benefits.
The study published by Khan and colleagues was the first comprehensive and systematic listing of public ophthalmological imaging databases. In their work, out of 121 datasets identified through various searching strategies, only 94 were deemed truly available, with 27 databases being inaccessible even after multiple attempts spaced weeks apart [11]. Therefore roughly one-fourth of datasets were inaccessible at the time of their review in work mentioned above. It is in line with our findings in writing this update. Out of 94 datasets marked available by the authors, only 74 were available at the time of preparing this article, just over 1 year from the initial paper publication of Khan and colleagues and just 15 months after the first online publishing. Therefore, access to over one-fifth (21%) of datasets was lost in fewer than 2 years, similar to the 22% found inaccessible in the original review. Almost all of the datasets that became unavailable since the publishing of Khan’s study were offline—the dedicated websites are unreachable, with two unavailable due to errors.
Of the unavailable datasets, 7 became unavailable during the identification, verification, and review of the newly discovered datasets out of 127 color fundus image repositories initially identified for this analysis. Although the initial period between publishing the individual datasets and becoming inaccessible has yet to be discovered, it is clear that datasets going offline or otherwise becoming unreachable is an ongoing process, and information on availability can quickly become obsolete. It is important to note that while Khan et al. published a list of datasets containing OCT and other imaging modalities, this review focuses specifically on datasets of color fundus images [11].
Given adequate citation of sources, all but three of the datasets allowed unrestricted access and publishing of results for scientific, non-commercial purposes. The two exceptions required approval from dataset authors before publishing any results, which may limit their usability in scientific regard, leaving potential publication opportunities to the whim of original dataset authors. More than half of datasets do not impose any restrictions and do not explicitly require citations, though quoting sources is one of the fundamental ethical principles of scientific use.
Most datasets contain additional information about individual images. More than half of the datasets (66%) contained manually assigned text-based labels corresponding to diagnosed ocular diseases, image quality, or described areas of interest. One-fifth (22%) of datasets contained annotations indicating areas of interest or pathology. Only 12% of datasets contained raw images without metadata for individual images.
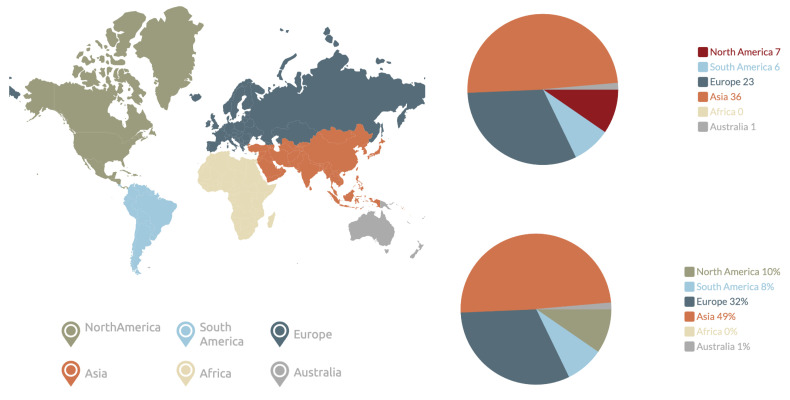
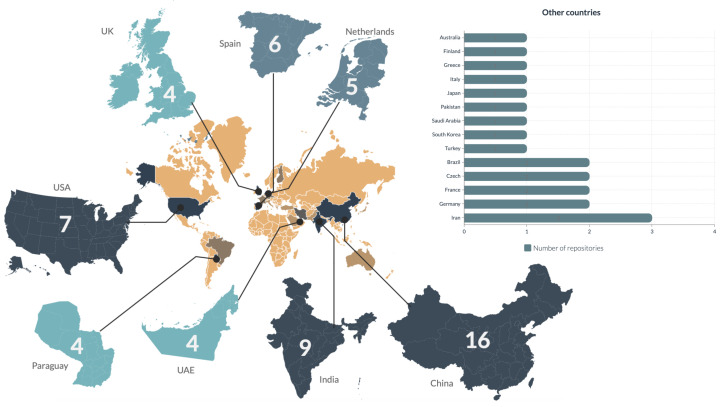
Figure 5 and Figure 6 explore the geographical origins of the datasets.
Figure 5.
Geographical distribution of the available datasets by continent (where origins of the dataset could be determined).
Figure 6.
Number of datasets originating from individual countries (where origins of the dataset could be determined).
Almost half of the datasets for which a region of origin could be established originated from Asia, with Europe making up another one-third. Overall, out of 73 datasets, 24 originated from outside of Europe or Asia, with none of the datasets originating from Africa and a nearly equal split between North and South America. The distribution of datasets available from individual countries is shown in Figure 6. Although dataset origin relates to the location of the person or organization sharing the dataset and does not necessarily represent the origin of patients’ images, the complete lack of images from Africa is concerning.
Africa, particularly sub-Saharan Africa, is a vastly underserved region with one of the lowest numbers of ophthalmologists in the population globally [167]. There are, on average, three ophthalmologists per million populations in sub-Saharan Africa, compared to about 80 in developed countries [167]. Although this is likely one of the reasons for the lack of available datasets from the region, it is also the rationale for the need for datasets from this region. Digital healthcare solutions, including deep-learning software, may help alleviate some of the healthcare disparities in the region. However, these require development or at least validation on the target validation to avoid any potential for racial or other population-specific trait bias [168]. It remains steadfast in the case of color fundus images where other than background fundus pigmentation, the influence of patients’ attributes such as age, sex, or race on fundus features and their variations are not well known. It is also currently being tackled using deep-learning methods [169]. The suspicion of poor generalizability in populations outside of the ethnic or geographical scope of the initial training image data and, subsequently, the need for the development and validation of multi-ethnic populations is not a new concept in the automated analysis of color fundus images [11,149,170,171]. Serener et al. have shown that the performance of deep-learning algorithms for detecting diabetic retinopathy in color fundus images varies based on geographical or ethnic traits of the training and validation populations [171].
7. Conclusions
Open datasets are still crucial for digital health research and innovation in ophthalmology. Even though the public has access to more datasets with color fundus images, new datasets are always being added, making older datasets inaccessible. There are only a few places to store or list ophthalmic datasets, and many of them are hard to find, making it hard to obtain high-quality, representative data useful for a given purpose. This paper discussed the many color fundus image datasets that are now available and gave an up-to-date review.
Acknowledgments
The authors thank Siamak Yousefi and his team from the University of Tennessee, Memphis, USA, and Paweł Borkowski from the Rzeszow University of Technology, Poland, for their help in identifying new repositories.
Author Contributions
Conceptualization: A.E.G. and T.K.; methodology: T.K.; data collection: T.K.; project administration: A.E.G.; supervision: A.E.G.; software: T.K. and A.M.Z.; formal analysis and visualizations: A.M.Z.; writing original draft: A.E.G., P.B., A.M.Z. and T.K; writing-review and editing, A.M.Z. and T.K. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data sharing not applicable No new data were created or analyzed in this study. Data sharing is not applicable to this article.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research received no external funding.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Ibrahim H., Liu X., Zariffa N., Morris A.D., Denniston A.K. Health data poverty: An assailable barrier to equitable digital health care. Lancet Digit. Health. 2021;3:e260–e265. doi: 10.1016/S2589-7500(20)30317-4. [DOI] [PubMed] [Google Scholar]
- 2.Esteva A., Robicquet A., Ramsundar B., Kuleshov V., DePristo M., Chou K., Cui C., Corrado G., Thrun S., Dean J. A guide to deep learning in healthcare. Nat. Med. 2019;25:24–29. doi: 10.1038/s41591-018-0316-z. [DOI] [PubMed] [Google Scholar]
- 3.Norgeot B., Glicksberg B.S., Butte A.J. A call for deep-learning healthcare. Nat. Med. 2019;25:14–15. doi: 10.1038/s41591-018-0320-3. [DOI] [PubMed] [Google Scholar]
- 4.Grzybowski A., Brona P., Lim G., Ruamviboonsuk P., Tan G.S.W., Abramoff M., Ting D.S.W. Artificial intelligence for diabetic retinopathy screening: A review. Eye. 2019;34:451–460. doi: 10.1038/s41433-019-0566-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grzybowski A., Brona P. A pilot study of autonomous artificial intelligence-based diabetic retinopathy screening in Poland. Acta Ophthalmol. 2019;97:e1149–e1150. doi: 10.1111/aos.14132. [DOI] [PubMed] [Google Scholar]
- 6.Miotto R., Wang F., Wang S., Jiang X., Dudley J.T. Deep learning for healthcare: Review, opportunities and challenges. Briefings Bioinform. 2017;19:1236–1246. doi: 10.1093/bib/bbx044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Litjens G., Kooi T., Bejnordi B.E., Setio A.A.A., Ciompi F., Ghafoorian M., van der Laak J.A., van Ginneken B., Sánchez C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017;42:60–88. doi: 10.1016/j.media.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 8.Bluemke D.A., Moy L., Bredella M.A., Ertl-Wagner B.B., Fowler K.J., Goh V.J., Halpern E.F., Hess C.P., Schiebler M.L., Weiss C.R. Assessing Radiology Research on Artificial Intelligence: A Brief Guide for Authors, Reviewers, and Readers from the Editorial Board. Radiology. 2020;294:487–489. doi: 10.1148/radiol.2019192515. [DOI] [PubMed] [Google Scholar]
- 9.Aggarwal R., Sounderajah V., Martin G., Ting D.S.W., Karthikesalingam A., King D., Ashrafian H., Darzi A. Diagnostic accuracy of deep learning in medical imaging: A systematic review and meta-analysis. NPJ Digit. Med. 2021;4:65. doi: 10.1038/s41746-021-00438-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ting D.S.W., Pasquale L.R., Peng L., Campbell J.P., Lee A.Y., Raman R., Tan G.S.W., Schmetterer L., Keane P.A., Wong T.Y. Artificial intelligence and deep learning in ophthalmology. Br. J. Ophthalmol. 2018;103:167–175. doi: 10.1136/bjophthalmol-2018-313173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khan S.M., Liu X., Nath S., Korot E., Faes L., Wagner S.K., Keane P.A., Sebire N.J., Burton M.J., Denniston A.K. A global review of publicly available datasets for ophthalmological imaging: Barriers to access, usability, and generalisability. Lancet Digit. Health. 2021;3:e51–e66. doi: 10.1016/S2589-7500(20)30240-5. [DOI] [PubMed] [Google Scholar]
- 12. [(accessed on 16 May 2023)]. Available online: https://www.kaggle.com/datasets/linchundan/fundusimage1000.
- 13. [(accessed on 16 May 2023)]. Available online: https://data.mendeley.com/datasets/dh2x8v6nf8/1.
- 14. [(accessed on 16 May 2023)]. Available online: https://data.mendeley.com/datasets/trghs22fpg/1.
- 15. [(accessed on 16 May 2023)]. Available online: https://data.mendeley.com/datasets/trghs22fpg/2.
- 16. [(accessed on 16 May 2023)]. Available online: https://data.mendeley.com/datasets/trghs22fpg/3.
- 17. [(accessed on 16 May 2023)]. Available online: https://data.mendeley.com/datasets/trghs22fpg/4.
- 18. [(accessed on 16 May 2023)]. Available online: https://figshare.com/s/c2d31f850af14c5b5232.
- 19. [(accessed on 16 May 2023)]. Available online: https://www.kaggle.com/c/aptos2019-blindness-detection/data.
- 20. [(accessed on 16 May 2023)]. Available online: https://people.eng.unimelb.edu.au/thivun/projects/AV_nicking_quantification/
- 21. [(accessed on 16 May 2023)]. Available online: https://blogs.kingston.ac.uk/retinal/chasedb1/
- 22. [(accessed on 16 May 2023)]. Available online: https://www.kaggle.com/datasets/gilescodes/cropped-train-diabetic-retinopathy-detection.
- 23. [(accessed on 16 May 2023)]. Available online: https://www.kaggle.com/datasets/andrewmvd/retinal-disease-classification.
- 24. [(accessed on 16 May 2023)]. Available online: https://www.kaggle.com/datasets/fareesamasroor/cardiacemboli.
- 25. [(accessed on 16 May 2023)]. Available online: https://www.kaggle.com/datasets/mariaherrerot/ddrdataset.
- 26. [(accessed on 16 May 2023)]. Available online: https://medicine.uiowa.edu/eye/abramoff/
- 27. [(accessed on 16 May 2023)]. Available online: https://zenodo.org/record/4532361#.Yrr5sOzP1W4.
- 28. [(accessed on 16 May 2023)]. Available online: https://zenodo.org/record/4647952#.YtRttC-plQI.
- 29. [(accessed on 16 May 2023)]. Available online: https://zenodo.org/record/4891308#.YtRwDS-plQI.
- 30. [(accessed on 16 May 2023)]. Available online: https://www.kaggle.com/datasets/nikkich9/derbi-hackathon-retinal-fundus-image-dataset.
- 31. [(accessed on 16 May 2023)]. Available online: https://data.mendeley.com/datasets/3csr652p9y/1.
- 32. [(accessed on 16 May 2023)]. Available online: https://data.mendeley.com/datasets/2rnnz5nz74/1.
- 33. [(accessed on 16 May 2023)]. Available online: https://data.mendeley.com/datasets/2rnnz5nz74/2.
- 34. [(accessed on 16 May 2023)]. Available online: https://www.kaggle.com/datasets/tanlikesmath/diabetic-retinopathy-resized.
- 35. [(accessed on 16 May 2023)]. Available online: https://figshare.com/articles/dataset/Advancing_Bag_of_Visual_Words_Representations_for_Lesion_Classification_in_Retinal_Images/953671/2.
- 36. [(accessed on 16 May 2023)]. Available online: https://figshare.com/articles/dataset/Advancing_Bag_of_Visual_Words_Representations_for_Lesion_Classification_in_Retinal_Images/953671/3.
- 37. [(accessed on 16 May 2023)]. Available online: https://www.kaggle.com/datasets/mustaqimabrar/diabeticretinopathy.
- 38. [(accessed on 16 May 2023)]. Available online: https://www.kaggle.com/datasets/rutujachaudhari/diabetic-retinopathy.
- 39. [(accessed on 16 May 2023)]. Available online: https://www.kaggle.com/datasets/himanshuagarwal1998/diabetic-retinopathy.
- 40. [(accessed on 16 May 2023)]. Available online: https://www.kaggle.com/datasets/analaura000/diabetic-retinopathy.
- 41. [(accessed on 16 May 2023)]. Available online: https://www.kaggle.com/datasets/kameshwarandhayalan/diabetic-retinopathy.
- 42. [(accessed on 16 May 2023)]. Available online: https://www.kaggle.com/datasets/sovitrath/diabetic-retinopathy-2015-data-colored-resized.
- 43. [(accessed on 16 May 2023)]. Available online: https://www.kaggle.com/datasets/sovitrath/diabetic-retinopathy-224x224-2019-data.
- 44. [(accessed on 16 May 2023)]. Available online: https://www.kaggle.com/datasets/sovitrath/diabetic-retinopathy-224x224-gaussian-filtered.
- 45. [(accessed on 16 May 2023)]. Available online: https://www.kaggle.com/datasets/sovitrath/diabetic-retinopathy-224x224-grayscale-images.
- 46. [(accessed on 16 May 2023)]. Available online: https://www.kaggle.com/datasets/amanneo/diabetic-retinopathy-resized-arranged.
- 47. [(accessed on 16 May 2023)]. Available online: https://www.kaggle.com/datasets/kushagratandon12/diabetic-retinopathy-balanced.
- 48. [(accessed on 16 May 2023)]. Available online: https://www.kaggle.com/datasets/makrovh/diabetic-retinopathy-blindness-detection-c-data.
- 49. [(accessed on 16 May 2023)]. Available online: https://www.kaggle.com/datasets/dola1507108/diabetic-retinopathy-classified.
- 50. [(accessed on 16 May 2023)]. Available online: https://www.kaggle.com/competitions/diabetic-retinopathy-classification-2/data.
- 51. [(accessed on 16 May 2023)]. Available online: https://www.kaggle.com/competitions/retinopathy-classification-sai/data.
- 52. [(accessed on 16 May 2023)]. Available online: https://www.kaggle.com/datasets/sachinkumar413/diabetic-retinopathy-dataset.
- 53. [(accessed on 16 May 2023)]. Available online: https://www.kaggle.com/datasets/ahmedghazal54/diabetic-retinopathy-detection.
- 54. [(accessed on 16 May 2023)]. Available online: https://www.kaggle.com/datasets/poojita2305/diabetic-retinopathy-detection.
- 55. [(accessed on 16 May 2023)]. Available online: https://www.kaggle.com/datasets/mostafaeltalawy/diabetic-retinopathy-dataset.
- 56. [(accessed on 16 May 2023)]. Available online: https://personalpages.manchester.ac.uk/staff/niall.p.mcloughlin/
- 57. [(accessed on 16 May 2023)]. Available online: http://www.ia.uned.es/~ejcarmona/DRIONS-DB.html.
- 58. [(accessed on 16 May 2023)]. Available online: https://drive.grand-challenge.org.
- 59. [(accessed on 16 May 2023)]. Available online: https://www.kaggle.com/datasets/dola1507108/diabetic-retinopathy-organized.
- 60. [(accessed on 16 May 2023)]. Available online: https://www.kaggle.com/datasets/sachinkumar413/diabetic-retinopathy-preprocessed-dataset.
- 61. [(accessed on 16 May 2023)]. Available online: https://www.kaggle.com/datasets/shuvokkr/diabeticresized300.
- 62. [(accessed on 16 May 2023)]. Available online: https://www.kaggle.com/datasets/makjn10/diabetic-retinopathy-small.
- 63. [(accessed on 16 May 2023)]. Available online: https://www.kaggle.com/datasets/dantealonso/diabeticretinopathytrainvalidation.
- 64. [(accessed on 16 May 2023)]. Available online: https://www.kaggle.com/datasets/saipavansaketh/diabetic-retinopathy-unziped.
- 65. [(accessed on 16 May 2023)]. Available online: https://www.kaggle.com/datasets/zhizhid/dr-2000.
- 66. [(accessed on 16 May 2023)]. Available online: https://www.kaggle.com/datasets/aviagarwal123/dr-201010.
- 67. [(accessed on 16 May 2023)]. Available online: https://github.com/deepdrdoc/DeepDRiD/blob/master/README.md.
- 68. [(accessed on 16 May 2023)]. Available online: https://www.kaggle.com/datasets/nguyenhung1903/diaretdb1-v21.
- 69. [(accessed on 16 May 2023)]. Available online: https://www.kaggle.com/datasets/muhamedahmed/diabetic.
- 70. [(accessed on 16 May 2023)]. Available online: https://www.kaggle.com/datasets/alisalen/diabetic-retinopathy-detection-processed.
- 71. [(accessed on 16 May 2023)]. Available online: https://www.kaggle.com/datasets/lokeshsaipureddi/drishtigs-retina-dataset-for-onh-segmentation.
- 72. [(accessed on 16 May 2023)]. Available online: http://cvit.iiit.ac.in/projects/mip/drishti-gs/mip-dataset2/Home.php.
- 73. [(accessed on 16 May 2023)]. Available online: https://www.kaggle.com/datasets/diveshthakker/eoptha-diabetic-retinopathy.
- 74. [(accessed on 16 May 2023)]. Available online: https://www.kaggle.com/c/diabetic-retinopathy-detection.
- 75. [(accessed on 16 May 2023)]. Available online: https://www.kaggle.com/datasets/bishalbanerjee/eye-dataset.
- 76. [(accessed on 16 May 2023)]. Available online: https://www.kaggle.com/datasets/iamachal/fundus-image-dataset.
- 77. [(accessed on 16 May 2023)]. Available online: https://projects.ics.forth.gr/cvrl/fire/
- 78. [(accessed on 16 May 2023)]. Available online: https://www.kaggle.com/datasets/izander/fundus.
- 79. [(accessed on 16 May 2023)]. Available online: https://www.kaggle.com/datasets/klmsathishkumar/fundus-images.
- 80. [(accessed on 16 May 2023)]. Available online: https://www.kaggle.com/datasets/spikeetech/fundus-dr.
- 81. [(accessed on 16 May 2023)]. Available online: https://www.kaggle.com/datasets/balnyaupane/gaussian-filtered-diabetic-retinopathy.
- 82. [(accessed on 16 May 2023)]. Available online: https://dataverse.harvard.edu/dataset.xhtml?persistentId=doi:10.7910/DVN/1YRRAC.
- 83. [(accessed on 16 May 2023)]. Available online: https://github.com/lgiancaUTH/HEI-MED.
- 84. [(accessed on 16 May 2023)]. Available online: https://www5.cs.fau.de/research/data/fundus-images/
- 85. [(accessed on 16 May 2023)]. Available online: http://ai.baidu.com/broad/introduction.
- 86. [(accessed on 16 May 2023)]. Available online: https://idrid.grand-challenge.org/Rules/
- 87. [(accessed on 16 May 2023)]. Available online: https://medicine.uiowa.edu/eye/inspire-datasets.
- 88. [(accessed on 16 May 2023)]. Available online: http://ai.baidu.com/broad/subordinate?dataset=pm.
- 89. [(accessed on 16 May 2023)]. Available online: https://www.kaggle.com/datasets/dineswarreddy/indian-retina-classification.
- 90. [(accessed on 16 May 2023)]. Available online: https://www.kaggle.com/datasets/bachaboos/isbi-2021-retina-23.
- 91. [(accessed on 16 May 2023)]. Available online: https://www.kaggle.com/datasets/bachaboos/isbi-retina-test.
- 92. [(accessed on 16 May 2023)]. Available online: https://www.kaggle.com/linchundan/fundusimage1000.
- 93. [(accessed on 16 May 2023)]. Available online: https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0179790#sec006.
- 94. [(accessed on 16 May 2023)]. Available online: https://github.com/smilell/AG-CNN.
- 95. [(accessed on 16 May 2023)]. Available online: https://www.adcis.net/en/third-party/messidor2/
- 96. [(accessed on 16 May 2023)]. Available online: https://odir2019.grand-challenge.org/Download/
- 97. [(accessed on 16 May 2023)]. Available online: https://www.kaggle.com/datasets/andrewmvd/ocular-disease-recognition-odir5k.
- 98. [(accessed on 16 May 2023)]. Available online: https://aistudio.baidu.com/aistudio/datasetdetail/122940.
- 99. [(accessed on 16 May 2023)]. Available online: https://figshare.com/articles/dataset/PAPILA/14798004/1.
- 100. [(accessed on 16 May 2023)]. Available online: https://figshare.com/articles/dataset/PAPILA/14798004.
- 101. [(accessed on 16 May 2023)]. Available online: https://www.kaggle.com/datasets/benjaminwarner/resized-2015-2019-blindness-detection-images.
- 102. [(accessed on 16 May 2023)]. Available online: https://refuge.grand-challenge.org/
- 103. [(accessed on 16 May 2023)]. Available online: https://ai.baidu.com/broad/download?dataset=gon.
- 104. [(accessed on 16 May 2023)]. Available online: https://www.kaggle.com/datasets/kssanjaynithish03/retinal-fundus-images.
- 105. [(accessed on 16 May 2023)]. Available online: https://deepblue.lib.umich.edu/data/concern/data_sets/3b591905z?locale=en.
- 106. [(accessed on 16 May 2023)]. Available online: https://www.kaggle.com/datasets/andrewmvd/fundus-image-registration.
- 107. [(accessed on 16 May 2023)]. Available online: http://medimrg.webs.ull.es/research/downloads/
- 108. [(accessed on 16 May 2023)]. Available online: https://www.kaggle.com/datasets/priyanagda/ritedataset.
- 109. [(accessed on 16 May 2023)]. Available online: http://www.rodrep.com/longitudinal-diabetic-retinopathy-screening—description.html.
- 110. [(accessed on 16 May 2023)]. Available online: https://www.kaggle.com/datasets/aifahim/retinal-vassel-combine-same-format.
- 111. [(accessed on 16 May 2023)]. Available online: https://www.kaggle.com/jr2ngb/cataractdataset.
- 112. [(accessed on 16 May 2023)]. Available online: http://webeye.ophth.uiowa.edu/ROC/
- 113. [(accessed on 16 May 2023)]. Available online: https://www.kaggle.com/datasets/beatrizsimoes/retina-quality.
- 114. [(accessed on 16 May 2023)]. Available online: https://www.kaggle.com/datasets/hebamohamed/retinagen.
- 115. [(accessed on 16 May 2023)]. Available online: http://bioimlab.dei.unipd.it/Retinal%20Vessel%20Tortuosity.htm.
- 116. [(accessed on 16 May 2023)]. Available online: https://www.kaggle.com/datasets/ustinianw/retinal-tiny.
- 117. [(accessed on 16 May 2023)]. Available online: http://cecas.clemson.edu/~ahoover/stare/
- 118. [(accessed on 16 May 2023)]. Available online: https://www.kaggle.com/datasets/balnyaupane/small-diabetic-retinopathy-dataset.
- 119. [(accessed on 16 May 2023)]. Available online: https://www.kaggle.com/datasets/mariaherrerot/the-sustechsysu-dataset.
- 120. [(accessed on 16 May 2023)]. Available online: https://www.kaggle.com/competitions/innovation-challenge-2019/data.
- 121. [(accessed on 16 May 2023)]. Available online: https://www.kaggle.com/competitions/vietai-advance-retinal-disease-detection-2020/data.
- 122. [(accessed on 16 May 2023)]. Available online: http://www.varpa.es/research/ophtalmology.html#vicavr.
- 123. [(accessed on 16 May 2023)]. Available online: http://people.duke.edu/~sf59/Estrada_TMI_2015_dataset.htm.
- 124. [(accessed on 16 May 2023)]. Available online: https://novel.utah.edu/Hoyt/
- 125. [(accessed on 16 May 2023)]. Available online: https://zenodo.org/record/3393265#.XazZaOgzbIV.
- 126. [(accessed on 16 May 2023)]. Available online: https://www.kaggle.com/datasets/nawa393/dr15_test.
- 127. [(accessed on 16 May 2023)]. Available online: https://www.kaggle.com/datasets/makorromanuel/merged-retina-datasets.
- 128.Cen L.P., Ji J., Lin J.W., Ju S.T., Lin H.J., Li T.P., Wang Y., Yang J.F., Liu Y.F., Tan S., et al. Automatic detection of 39 fundus diseases and conditions in retinal photographs using deep neural networks. Nat. Commun. 2021;12:4828. doi: 10.1038/s41467-021-25138-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yoo T.K. A CycleGAN Deep Learning Technique for Artifact Reduction in Fundus Photography. Graefe’s Arch. Clin. Exp. Ophthalmol. 2020;258:1631–1637. doi: 10.1007/s00417-020-04709-5. [DOI] [PubMed] [Google Scholar]
- 130.Hassan T. A Composite Retinal Fundus and OCT Dataset along with Detailed Clinical Markings for Extracting Retinal Layers, Retinal Lesions and Screening Macular and Glaucomatous Disorders (dataset 1) 2021. [DOI]
- 131.Hassan T. A Composite Retinal Fundus and OCT Dataset along with Detailed Clinical Markings for Extracting Retinal Layers, Retinal Lesions and Screening Macular and Glaucomatous Disorders (dataset 2) 2021. [DOI]
- 132.Hassan T. A Composite Retinal Fundus and OCT Dataset along with Detailed Clinical Markings for Extracting Retinal Layers, Retinal Lesions and Screening Macular and Glaucomatous Disorders (dataset 3) 2021. [DOI]
- 133.Hassan T., Akram M.U., Masood M.F., Yasin U. Deep structure tensor graph search framework for automated extraction and characterization of retinal layers and fluid pathology in retinal SD-OCT scans. Comput. Biol. Med. 2019;105:112–124. doi: 10.1016/j.compbiomed.2018.12.015. [DOI] [PubMed] [Google Scholar]
- 134.Hassan T., Akram M.U., Werghi N., Nazir M.N. RAG-FW: A Hybrid Convolutional Framework for the Automated Extraction of Retinal Lesions and Lesion-Influenced Grading of Human Retinal Pathology. IEEE J. Biomed. Health Inform. 2021;25:108–120. doi: 10.1109/JBHI.2020.2982914. [DOI] [PubMed] [Google Scholar]
- 135.Diaz-Pinto A., Morales S., Naranjo V., Köhler T., Mossi J.M., Navea A. CNNs for automatic glaucoma assessment using fundus images: An extensive validation. Biomed. Eng. Online. 2019;18:29. doi: 10.1186/s12938-019-0649-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Pachade S., Porwal P., Thulkar D., Kokare M., Deshmukh G., Sahasrabuddhe V., Giancardo L., Quellec G., Mériaudeau F. Retinal Fundus Multi-Disease Image Dataset (RFMiD): A Dataset for Multi-Disease Detection Research. Data. 2021;6:14. doi: 10.3390/data6020014. [DOI] [Google Scholar]
- 137.Li T., Gao Y., Wang K., Guo S., Liu H., Kang H. Diagnostic assessment of deep learning algorithms for diabetic retinopathy screening. Inf. Sci. 2019;501:511–522. doi: 10.1016/j.ins.2019.06.011. [DOI] [Google Scholar]
- 138.Benítez V.E.C., Matto I.C., Román J.C.M., Noguera J.L.V., García-Torres M., Ayala J., Pinto-Roa D.P., Gardel-Sotomayor P.E., Facon J., Grillo S.A. Dataset from fundus images for the study of diabetic retinopathy. Data Brief. 2021;36:107068. doi: 10.1016/j.dib.2021.107068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Akbar S., Sharif M., Akram M.U., Saba T., Mahmood T., Kolivand M. Automated techniques for blood vessels segmentation through fundus retinal images: A review. Microsc. Res. Tech. 2019;82:153–170. doi: 10.1002/jemt.23172. [DOI] [PubMed] [Google Scholar]
- 140.Raja H. Data on OCT and Fundus Images. 2020. [DOI] [PMC free article] [PubMed]
- 141.Pires R., Jelinek H.F., Wainer J., Valle E., Rocha A. Advancing Bag-of-Visual-Words Representations for Lesion Classification in Retinal Images. PLoS ONE. 2014;9:e96814. doi: 10.1371/journal.pone.0096814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Holm S., Russell G., Nourrit V., McLoughlin N. DR HAGIS—a fundus image database for the automatic extraction of retinal surface vessels from diabetic patients. J. Med. Imaging. 2017;4:014503. doi: 10.1117/1.JMI.4.1.014503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Carmona E.J., Rincón M., García-Feijoó J., de-la Casa J.M.M. Identification of the optic nerve head with genetic algorithms. Artif. Intell. Med. 2008;43:243–259. doi: 10.1016/j.artmed.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 144.Kauppi T., Kalesnykiene V., Kamarainen J.K., Lensu L., Sorri I., Raninen A., Voutilainen R., Uusitalo H., Kalviainen H., Pietila J. The DIARETDB1 diabetic retinopathy database and evaluation protocol; Proceedings of the British Machine Vision Conference 2007; Warwick, UK. 10–13 September 2007; [DOI] [Google Scholar]
- 145.Sivaswamy J., Chakravarty A., Joshi G.D., Syed T.A. A Comprehensive Retinal Image Dataset for the Assessment of Glaucoma from the Optic Nerve Head Analysis. JSM Biomed. Imaging Data Pap. 2015;2:1004. [Google Scholar]
- 146.Sivaswamy J., Krishnadas S.R., Datt Joshi G., Jain M., Syed Tabish A.U. Drishti-GS: Retinal image dataset for optic nerve head(ONH) segmentation; Proceedings of the 2014 IEEE 11th International Symposium on Biomedical Imaging (ISBI); Beijing, China. 29 April–2 May 2014; pp. 53–56. [DOI] [Google Scholar]
- 147.Hernandez-Matas C., Zabulis X., Triantafyllou A., Anyfanti P., Douma S., Argyros A.A. FIRE: Fundus Image Registration dataset. Model. Artif. Intell. Ophthalmol. 2017;1:16–28. doi: 10.35119/maio.v1i4.42. [DOI] [Google Scholar]
- 148.Akbar S., Hassan T., Akram M.U., Yasin U., Basit I. AVRDB: Annotated Dataset for Vessel Segmentation and Calculation of Arteriovenous Ratio. 2017.
- 149.Giancardo L., Meriaudeau F., Karnowski T.P., Li Y., Garg S., Tobin K.W., Chaum E. Exudate-based diabetic macular edema detection in fundus images using publicly available datasets. Med. Image Anal. 2012;16:216–226. doi: 10.1016/j.media.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kohler T., Budai A., Kraus M.F., Odstrcilik J., Michelson G., Hornegger J. Automatic no-reference quality assessment for retinal fundus images using vessel segmentation; Proceedings of the 26th IEEE International Symposium on Computer-Based Medical Systems; Porto, Portugal. 20–22 June 2013; [DOI] [Google Scholar]
- 151.Budai A., Bock R., Maier A., Hornegger J., Michelson G. Robust Vessel Segmentation in Fundus Images. Int. J. Biomed. Imaging. 2013;2013:1–11. doi: 10.1155/2013/154860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Prasanna Porwal S.P. Indian Diabetic Retinopathy Image Dataset (IDRiD) 2018. [DOI]
- 153.Niemeijer M., Xu X., Dumitrescu A.V., Gupta P., van Ginneken B., Folk J.C., Abramoff M.D. Automated Measurement of the Arteriolar-to-Venular Width Ratio in Digital Color Fundus Photographs. IEEE Trans. Med. Imaging. 2011;30:1941–1950. doi: 10.1109/TMI.2011.2159619. [DOI] [PubMed] [Google Scholar]
- 154.Tang L., Garvin M.K., Lee K., Alward W.L.W., Kwon Y.H., Abramoff M.D. Robust Multiscale Stereo Matching from Fundus Images with Radiometric Differences. IEEE Trans. Pattern Anal. Mach. Intell. 2011;33:2245–2258. doi: 10.1109/TPAMI.2011.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Fu H., Li F., Orlando J.I., Bogunović H., Sun X., Liao J., Xu Y., Zhang S., Zhang X. ADAM: Automatic Detection Challenge on Age-Related Macular Degeneration. 2020. [DOI]
- 156.Li L., Xu M., Wang X., Jiang L., Liu H. Attention Based Glaucoma Detection: A Large-scale Database and CNN Model. arXiv. 2019 doi: 10.1109/TMI.2019.2927226.1903.10831 [DOI] [PubMed] [Google Scholar]
- 157.Decenciere E., Zhang X., Cazuguel G., Lay B., Cochener B., Trone C., Gain P., Ordonez R., Massin P., Erginay A., et al. Feedback on a publicly distributed image database: The messidor database. Image Anal. Stereol. 2014;33:231. doi: 10.5566/ias.1155. [DOI] [Google Scholar]
- 158.Kovalyk O., Morales-Sánchez J., Verdú-Monedero R., Sellés-Navarro I., Palazón-Cabanes A., Sancho-Gómez J.L. PAPILA. 2022. [DOI] [PMC free article] [PubMed]
- 159.Orlando J.I., Fu H., Breda J.B., van Keer K., Bathula D.R., Diaz-Pinto A., Fang R., Heng P.A., Kim J., Lee J., et al. REFUGE Challenge: A unified framework for evaluating automated methods for glaucoma assessment from fundus photographs. Med. Image Anal. 2020;59:101570. doi: 10.1016/j.media.2019.101570. [DOI] [PubMed] [Google Scholar]
- 160.Almazroa A. Retinal Fundus Images for Glaucoma Analysis: The Riga Dataset. 2018. [DOI]
- 161.Hu Q., Abràmoff M.D., Garvin M.K. Advanced Information Systems Engineering. Springer; Berlin/Heidelberg, Germany: 2013. Automated Separation of Binary Overlapping Trees in Low-Contrast Color Retinal Images; pp. 436–443. [DOI] [PubMed] [Google Scholar]
- 162.Adal K.M., van Etten P.G., Martinez J.P., van Vliet L.J., Vermeer K.A. Accuracy Assessment of Intra- and Intervisit Fundus Image Registration for Diabetic Retinopathy Screening. Investig. Ophthalmol. Vis. Sci. 2015;56:1805–1812. doi: 10.1167/iovs.14-15949. [DOI] [PubMed] [Google Scholar]
- 163.Grisan E., Foracchia M., Ruggeri A. A Novel Method for the Automatic Grading of Retinal Vessel Tortuosity. IEEE Trans. Med. Imaging. 2008;27:310–319. doi: 10.1109/TMI.2007.904657. [DOI] [PubMed] [Google Scholar]
- 164.Lin L., Li M., Huang Y., Cheng P., Xia H., Wang K., Yuan J., Tang X. The SUSTech-SYSU dataset for automated exudate detection and diabetic retinopathy grading. Sci. Data. 2020;7:409. doi: 10.1038/s41597-020-00755-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Vázquez S.G., Cancela B., Barreira N., Penedo M.G., Rodríguez-Blanco M., Seijo M.P., de Tuero G.C., Barceló M.A., Saez M. Improving retinal artery and vein classification by means of a minimal path approach. Mach. Vis. Appl. 2012;24:919–930. doi: 10.1007/s00138-012-0442-4. [DOI] [Google Scholar]
- 166.Estrada R., Allingham M.J., Mettu P.S., Cousins S.W., Tomasi C., Farsiu S. Retinal Artery-Vein Classification via Topology Estimation. IEEE Trans. Med. Imaging. 2015;34:2518–2534. doi: 10.1109/TMI.2015.2443117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Resnikoff S., Felch W., Gauthier T.M., Spivey B. The number of ophthalmologists in practice and training worldwide: A growing gap despite more than 200,000 practitioners. Br. J. Ophthalmol. 2012;96:783–787. doi: 10.1136/bjophthalmol-2011-301378. [DOI] [PubMed] [Google Scholar]
- 168.Holst C., Sukums F., Radovanovic D., Ngowi B., Noll J., Winkler A.S. Sub-Saharan Africa—the new breeding ground for global digital health. Lancet Digit. Health. 2020;2:e160–e162. doi: 10.1016/S2589-7500(20)30027-3. [DOI] [PubMed] [Google Scholar]
- 169.Müller S., Koch L.M., Lensch H., Berens P. A Generative Model Reveals the Influence of Patient Attributes on Fundus Images; Proceedings of the Medical Imaging with Deep Learning; Zurich, Switzerland. 6–8 July 2022. [Google Scholar]
- 170.Ting D.S.W., Cheung C.Y.L., Lim G., Tan G.S.W., Quang N.D., Gan A., Hamzah H., Garcia-Franco R., Yeo I.Y.S., Lee S.Y., et al. Development and Validation of a Deep Learning System for Diabetic Retinopathy and Related Eye Diseases Using Retinal Images From Multiethnic Populations With Diabetes. JAMA. 2017;318:2211. doi: 10.1001/jama.2017.18152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Serener A., Serte S. Geographic variation and ethnicity in diabetic retinopathy detection via deeplearning. Turk. J. Electr. Eng. Comput. Sci. 2020;28:664–678. doi: 10.3906/elk-1902-131. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing not applicable No new data were created or analyzed in this study. Data sharing is not applicable to this article.