Abstract
The Saccharomyces cerevisiae inositol polyphosphate 5-phosphatases (Inp51p, Inp52p, and Inp53p) each contain an N-terminal Sac1 domain, followed by a 5-phosphatase domain and a C-terminal proline-rich domain. Disruption of any two of these 5-phosphatases results in abnormal vacuolar and plasma membrane morphology. We have cloned and characterized the Sac1-containing 5-phosphatases Inp52p and Inp53p. Purified recombinant Inp52p lacking the Sac1 domain hydrolyzed phosphatidylinositol 4,5-bisphosphate [PtdIns(4,5)P2] and PtdIns(3,5)P2. Inp52p and Inp53p were expressed in yeast as N-terminal fusion proteins with green fluorescent protein (GFP). In resting cells recombinant GFP-tagged 5-phosphatases were expressed diffusely throughout the cell but were excluded from the nucleus. Following hyperosmotic stress the GFP-tagged 5-phosphatases rapidly and transiently associated with actin patches, independent of actin, in both the mother and daughter cells of budding yeast as demonstrated by colocalization with rhodamine phalloidin. Both the Sac1 domain and proline-rich domains were able to independently mediate translocation of Inp52p to actin patches, following hyperosmotic stress, while the Inp53p proline-rich domain alone was sufficient for stress-mediated localization. Overexpression of Inp52p or Inp53p, but not catalytically inactive Inp52p, which lacked PtdIns(4,5)P2 5-phosphatase activity, resulted in a dramatic reduction in the repolarization time of actin patches following hyperosmotic stress. We propose that the osmotic-stress-induced translocation of Inp52p and Inp53p results in the localized regulation of PtdIns(3,5)P2 and PtdIns(4,5)P2 at actin patches and associated plasma membrane invaginations. This may provide a mechanism for regulating actin polymerization and cell growth as an acute adaptive response to hyperosmotic stress.
The actin cytoskeleton plays a fundamental role in regulating cytokinesis and organelle transport. In the budding yeast Saccharomyces cerevisiae genetic and morphological evidence indicates that actin regulates cell growth. In yeast filamentous actin is found in two morphologically identified forms, cables and patches (1, 24). Actin cables are found mainly in the mother cell and extend along the axis of growth, which is asymmetrical to the emerging daughter cell. Cables are involved in regulating organelle inheritance and vesicle targeting. Actin patches are associated with plasma membrane invaginations and are motile structures that move along the plasma membrane in response to osmotic stress (6, 33, 47). It has been speculated that actin patches may be necessary machinery for maintaining secretion or endocytosis. Actin cables and patches may form part of an integrated system, as their distributions often change simultaneously (23). However, the signaling mechanisms regulating the assembly and movement of actin cables and patches in response to osmotic and other stimuli are not well understood.
The phosphoinositides are ubiquitous components of eukaryotic membranes and are critical regulators of the actin cytoskeleton and membrane trafficking (reviewed in references 11, 12, and 45). Phosphatidylinositol 4,5 bisphosphate [PtdIns(4,5)P2] serves as a precursor to a variety of second-messenger molecules and, via interactions with actin binding proteins, plays a critical role in regulating actin cytoskeletal rearrangement. The synthesis and degradation of PtdIns(4,5)P2 are mediated by a series of lipid phosphorylation and dephosphorylation reactions, governed by specific lipid kinases and phosphatases.
The enzyme family of inositol polyphosphate 5-phosphatases (5-phosphatases) regulate cellular PtdIns(4,5)P2 concentrations by dephosphorylating the position-5 phosphate from the inositol ring, forming PtdIns(4)P (28, 30). In addition, mammalian 5-phosphatases dephosphorylate other position-5 phosphate phosphoinositides and inositol phosphates in a series of signal-terminating reactions that control intracellular calcium oscillations, apoptosis, synaptic vesicle recycling, and actin cytoskeletal rearrangement (28, 30). In S. cerevisiae, four genes encoding enzymes with amino acid sequence homology to the mammalian 5-phosphatases have been identified. Three of these enzymes exhibit a structure similar to that of the mammalian 5-phosphatase, synaptojanin, and contain an N-terminal Sac1 domain, a central 5-phosphatase domain, and a C-terminal proline-rich domain. The coding sequences for these loci are designated SJL1, SJL2, and SJL3, respectively, for “synaptojanin-like” or INP51, INP52, and INP53, respectively, for inositol polyphosphate 5-phosphatases 1 to 3 (42, 43). A fourth 5-phosphatase, encoded by INP54, contains significant homology to mammalian 5-phosphatases but has no Sac1 or C-terminal proline-rich domain and specifically hydrolyzes only PtdIns(4,5)P2 (39).
Several studies of S. cerevisiae have investigated the phenotype associated with deletion of genes encoding Sac1-containing 5-phosphatases (42, 43). Single-gene disruption of any 5-phosphatase produces little change in the phenotype, suggesting the functional redundancy of these enzymes. Disruption of any two genes results in a phenotype comprising vacuolar fragmentation, abnormal plasma membrane morphology with massive plasma membrane invaginations, disorganization of polymerized actin, and cell wall thickening. Recent studies have also noted receptor-mediated and fluid-phase endocytosis abnormalities, which correlate with the severity of actin and polarity defects (41). Disruption of all three Sac1 domain-containing 5-phosphatases is lethal.
The biochemical mechanisms mediating the observed phenotype in the 5-phosphatase double mutants are currently being delineated. Disruption of the Sac1-containing 5-phosphatases, individually or in pairs, results in decreased total cellular PtdIns(4,5)P2 5-phosphatase activity and variable increases in [3H]PtdIns(4,5)P2 levels (42, 43). The cellular requirement for four yeast 5-phosphatases with overlapping phosphoinositide substrate specificities may be to localize each isoform to discrete intracellular compartments; however this has yet to be shown. In this study we investigated the intracellular location of the Sac1-containing 5-phosphatases Inp52p and Inp53p. We present evidence that in the resting cell these enzymes localize to the Triton X-100-insoluble fraction of the cell, consistent with a cytoskeletal location. Following hyperosmotic stress, Inp52p and Inp53p enzymes translocate rapidly and transiently to actin patches in the mother and daughter cells. We propose that 5-phosphatase localization at actin patches facilitates the localized hydrolysis of PtdIns(4,5)P2 and thereby actin rearrangement, which may in turn transiently regulate cell growth during hyperosmotic stress.
MATERIALS AND METHODS
Materials.
DNA-modifying and restriction enzymes were from New England Biolabs. [ortho-32P]phosphoric acid, [γ-32P]ATP, [3H]PtdIns(4)P, and [3H]PtdIns(4,5)P2 were from NEN Life Science Products. Oligonucleotides were obtained from Bresatec (Adelaide, Australia) and the Department of Microbiology, Monash University, Clayton, Australia. The pMALC2T vector was a gift from Jim Goding, Monash University; plasmid pJJ242 was a gift from Doris Germain, Peter MacCallum Cancer Institute; the pRS416 vector was from Mark Prescott, Monash University; the pPS1303 green fluorescent protein (GFP) expression vector was from Pam Silver, Dana Farber Cancer Institute, Harvard University. All other reagents were from Sigma Chemical Company (St. Louis, Mo.) unless otherwise stated. Yeast strains used in the study are listed in Table 1. Yeast strains were propagated at 30°C in standard yeast extract-peptone-dextrose (YPD) media or complete minimal media lacking nutrients to maintain selection of markers where appropriate.
TABLE 1.
Yeast strains used in this studya
| Strain | Genotype |
|---|---|
| W303 | MATa/MATα ade2-1/ade2-1 trp1-1/trp1-1, leu2-3,112/leu2-3,112 his3-11,15/his3-11,15 ura3-1/ura3-1 |
| W303α | MATα ade2-1 trp1-1 leu2-3,112 his3-11,15 ura3-1 |
| inp51 mutant | MATα ade2-1 trp1-1 leu2-3,112 his3-11,15 ura3-1 inp51::URA3 |
| inp52 mutant | MATα ade2-1 trp1-1 leu2-3,112 his3-11,15 ura3-1 inp52::HIS3 |
| inp53 mutant | MATα ade2-1 trp1-1 leu2-3,112 his3-11,15 ura3-1 inp53::TRP1 |
| inp51 inp52 mutant | MATα ade2-1 trp1-1 leu2-3,112 his3-11,15 ura3-1 inp51::URA3 inp52::HIS3 |
| inp51 inp53 mutant | MATα ade2-1 trp1-1 leu2-3,112 his3-11,15 ura3-1 inp51::URA3 inp53::TRP1 |
| inp52 inp53 mutant | MATα ade2-1 trp1-1 leu2-3,112 his3-11,15 ura3-1 inp52::HIS3 inp53::TRP1 |
All strains were from this study except for W303 and W303α, which were kind gifts from D. Germain.
Disruption of INP51, INP52, and INP53 in S. cerevisiae.
An internal fragment of INP51 corresponding to nucleotides (nt) 472 to 1947 (where the +1 nt corresponds to the first base for the initiating methionine) was amplified by PCR (incorporating an XbaI site at the 5′ end and a HindIII site at the 3′ end) and cloned into the XbaI/HindIII site of Bluescript. The construct was digested with EcoRV and EcoRI to release a 231-bp fragment (nt 1218 to 1449), which was replaced with a URA3 expression cassette obtained by digesting plasmid pJJ242 with EcoRI and PvuII (22). The URA3 gene flanked by the INP51 sequence was excised from the plasmid with NotI and XhoI, the fragment was used to transform W303 diploid cells by electroporation as described previously (3), and transformants were selected on complete minimal media lacking uracil and supplemented with 1 M sorbitol. Transformants were screened for homologous integration of the targeting construct by PCR.
The open reading frame of INP52 from nt 1747 to 2721 was replaced with a HIS3 gene. HIS3 was amplified from plasmid pRS303 (40) by PCR using sense primer GATCCAATCTGCGAATACGTCAATGAAAGGCTGTTAGAGTCAGAGTTGTACTGAGAGTGCACCAT and antisense primer TGCATGAGAA AGTTCGTCAGGACCTGTAGTTAGTGCTTCGGGATGGGTATTTCAC ACCGCATA, which each incorporate 45 bp of the INP52 sequence (underlined) flanking the HIS3 gene. W303 diploid cells were transformed with the PCR product by electroporation as described previously (3), and transformants were selected on complete minimal media lacking histidine and supplemented with 1 M sorbitol. Transformants were screened for homologous integration of the HIS3 marker by PCR.
The open reading frame of INP53 was disrupted by the replacement of nt 1913 to 2821 with a TRP1 gene. TRP1 was amplified by PCR from plasmid pRS304 (40) using primers GCTCTTTCTGGGAAAACTTAGTGGGTGATTGCTTAAACCAGTATGTTGTACTGAGAGTGCACCAT (sense) and TACGCGACTG GCTACTGTTTTGAGTTCTGACAGGTTGTAGTGAAGGGTATTTCACA CCGCATA (antisense), which each include 45 bp of the INP53 sequence (underlined). W303 cells were transformed as described above, and recombinants were selected on tryptophan-deficient media and screened by PCR.
Heterozygous diploid colonies were sporulated to obtain haploid homozygous deletion mutants. Double-deletion mutant strains were constructed by mating haploid strains together and selecting for diploids in appropriate selective media. Diploids were sporulated and microdissected to obtain the haploid double-deletion mutants.
Cloning of INP52.
INP52 was amplified from SEY6210 genomic DNA by PCR using synthetic oligonucleotides S-5 (CGCTCTAGAATGTACATCAACAATTTTG) and S-3 (CGTCTAGATTAATGATGATGATGATGATGATTGGGGTCGCAAGGCTTCAA). S-5 and S-3 amplified INP52 nt 1609 to 3552, encoding the 5-phosphatase and proline-rich regions, and incorporated XbaI restriction enzyme sites for cloning (boldface) and a sequence encoding six-His tag for purification (underlined). The PCR product was blunt end ligated into the vector pCRBlunt (Invitrogen), and the identity of the PCR product was confirmed as INP52 by dideoxy sequence analysis. The INP52 insert was released from the vector via an XbaI restriction digest and subcloned into the XbaI sites of vector pMALC2T to give the construct INP52-Sac1−-pMALC2T. A catalytically inactive 5-phosphatase Inp52p mutant was generated by site-directed mutagenesis of INP52-Sac1−-pCRBlunt using the primers listed in Table 2, which substitutes an alanine residue for histidine 730 in the 5-phosphatase domain. The mutant insert was cloned into the XbaI site of pMALC2T.
TABLE 2.
Constructs used in this studya
| Construct | Features | Primers (5′-) |
|---|---|---|
| pPS1303 | GFP | |
| Inp52-pPS1303 | Full-length Inp52p-GFP | GCGGATCCAAACAATGAAAATACTCCTATCG |
| GCGGATCCCATTGGGGTCGCAAGACTT | ||
| Inp5274-1183-pPS1303 | Inp52p N terminus deleted-GFP | GCGGATCCAAACAATGTTCATAGCAACCATAACAGGT |
| GCGGATCCCATTGGGGTCGCAAGACTT | ||
| Inp52537-1183-pPS1303 | Inp52p Sac1 domain deleted-GFP | GCGGATCCAAACAATGTACATCAACAATTTTGTA |
| GCGGATCCCATTGGGGTCGCAAGACTT | ||
| Inp521-1179-pPS1303 | Inp52p CAAX motif deleted-GFP | GCGGATCCAAACAATGAAAATACTCCTA |
| GCGGATCCCAGGCTTCAATGGATGAACGCT | ||
| Inp521-593-pPS1303 | Inp52p N-terminus + Sac1 domain-GFP | GCGGATCCAAACAATGAAAATACTCCTATCG |
| GCGGATCCCCAAATTTATGTTAGAATGAGT | ||
| Inp52884-1154-pPS1303 | Inp52p proline-rich domain-GFP | CGGGATCCAAACAATGAATGTGAAATTTGTTGAC |
| CGGGATCCCGCAAGGGAGTTCCTCTTC | ||
| Inp521-158-pPS1303 | Inp52p N terminus-GFP | GCGGATCCAAACAATGAAAATACTCCTATCG |
| CGGGATCCCTTGTTCTTGAGCATTCAA | ||
| Inp52H730A-pPS1303 | Full-length Inp52p 5-phosphatase dead-GFP | TCGTTAATACAGCTTTATCAGCAGGTG |
| CTGCTGATAAAGCTGTATTAACGAAACAG | ||
| Inp52-pRS416-GFP | Inp52p native promoter | GCGGATCCGTGATTCTTCAGTGAGAGCC |
| GCGGATCCCATTGGGGTCGCAAGACTT | ||
| Inp531-1107-pPS1303 | Full-length Inp53p-GFP | GCGGATCCAAACAATGATTATCTTTGTTTCAGAA |
| GCGGATCCCTTTTGGGGTCAATGGCTGCCA | ||
| Inp53512-1107-pPS1303 | Inp53p Sac1 domain deleted-GFP | CGGGATCCAAACAATGATATATATTAACAATTTC |
| GCGGATCCCTTTTGGGGTCAATGGCTGCCA | ||
| Inp531-568-pPS1303 | Inp53p N terminus + Sac1 domain-GFP | GCGGATCCAAACAATGATTATCTTTGTTTCAGAA |
| GCGGATCCCCAAGTTAATGTTGGAGGTTGA | ||
| Inp53859-1107-pPS1303 | Inp53p proline-rich domain-GFP | CGGGATCCAAACAATGAAAATAACTTTTGTTGAT |
| GCGGATCCCTTTTGGGGTCAATGGCTGCCA | ||
| Inp531-133-pPS1303 | Inp53p N terminus GFP | GCGGATCCAAACAATGATTATCTTTGTTTCAGAA |
| CGGGATCCCGGCGTCTTGATATTCAGT | ||
| Inp53-pRS416-GFP | Inp53p native promoter | GCGGATCCAAGGACATCAATAATCATGC |
| GCGGATCCCTTTTGGGGTCAATGGCTGCCA |
All constructs were developed in this study except for pPS1303, which was a kind gift from P. Silver.
Expression of recombinant Inp52p protein.
Four 100-ml cultures of cells containing INP52-Sac1−-pMALC2T or INP52-Sac1−-H730A-pMALC2T were grown at 37°C to an optical density at 600 nm of 0.5 to 0.6 in Luria broth supplemented with 2% glucose prior to induction with 0.1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 2 h at 26°C. Following induction, cells were pelleted and soluble proteins were extracted in 1/10 volume of buffer A (20 mM Tris [pH 8], 10% sucrose, 12 mM β-mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride, 2 μg of aprotinin/ml, 2 μg of leupeptin/ml, 1 mM benzamidine)–1% Triton X-100 at 4°C overnight with gentle agitation. Triton X-100 extracts were centrifuged at 15,000 × g for 15 min, and then the 40-ml supernatant was incubated with 4 ml of Talon resin (Clontech) at 4°C overnight with gentle agitation. The resin was poured into a column and washed with 20 column volumes of buffer A. Bound proteins were eluted with 4 column volumes of buffer A at pH 6.5 supplemented with 50 mM imidazole, and 1-ml fractions were collected. Fifteen-microliter aliquots of the starting material and flowthrough and eluted fractions were separated by sodium dodecyl sulfate–7.5% polyacrylamide gel electrophoresis (SDS–7.5% PAGE) (26) and either transferred to nitrocellulose membranes and immunoblotted with an affinity-purified Inp52p antibody according to standard protocols (46) or stained with Coomassie brilliant blue. The concentrations of recombinant protein in the eluted fractions were determined by comparing the results of a densitometric analysis of the Coomassie-stained gels with a standard amount of protein loaded on the gels. Immunoblots were developed using enhanced chemiluminescence (NEN Life Science Products).
Ins(1,4,5)P3, Ins(1,3,4,5)P4, PtdIns(4,5)P2, and PtdIns(3,5)P2 5-phosphatase enzyme assays.
Inositol 1,[4-32P,5-32P]-trisphosphate {Ins(1,[4-32P,5-32P])P3} was isolated from erythrocyte ghosts as previously described (14). Hydrolysis of Ins(1,[4-32P,5-32P])P3 was measured by extraction of released 32PO4 according to the method of Connolly et al. (9) using a substrate concentration of 30 μM and three linear protein concentrations in triplicate. Inositol 1,3,4,5-tetrakisphosphate [Ins(1,3,4,5)P4] 5-phosphatase assays were performed as described by Mitchell et al. (31) utilizing 5 μM [3H]Ins(1,3,4,5)P4 as the substrate. Assays were performed in triplicate using three linear protein concentrations. PtdIns(4,5)P2 5-phosphatase assays were performed as described by Matzaris et al. (29) using 5 to 250 μM [3H]PtdIns(4,5)P2 (3,500 cpm/nmol) with or without the addition of Cos-7 cell membranes prepared as described previously (29). Assays were performed for 10 min at 37°C using three linear protein concentrations in triplicate. PtdIns([3-32P],5)P2 assays were performed as described by Jackson et al. (20) except that PtdIns(5)P (Echelon Research Laboratories) was used as the substrate for PtdIns 3-kinase. Assay mixtures were incubated for 30 min at 37°C.
GFP-tagged Inp52p and Inp53p constructs.
Full-length INP52 and INP53 and deletion constructs were amplified by PCR using the primers listed in Table 2. The primers incorporated a BamHI site for cloning and were in-frame with the coding sequence for the C-terminal GFP in expression vector pPS1303. PCR products were blunt end ligated into the vector pCRBlunt (Invitrogen), and then BamHI-digested INP52 and INP53 inserts were cloned into the BglII site of pPS1303 and sequenced by dideoxy sequencing to confirm the identity of the products and ensure they were in-frame with the GFP coding sequence. A full-length catalytically inactive 5-phosphatase Inp52p mutant was constructed by replacing an SspI/ScaI fragment (nt 1650 to 3246) of full-length INP52-pCRBlunt with the corresponding fragment from INP52-Sac1−-H730A-pCRBlunt (described above). The mutant INP52 was excised from pCRBlunt with BamHI and cloned into the BglII site of pPS1303. Constructs were transformed into the corresponding null mutant cells using an S. cerevisiae EasyComp transformation kit (Invitrogen), and transformants were selected on complete media lacking uracil.
Expression of GFP-tagged 5-phosphatases.
Single colonies were inoculated into 5 ml of complete media lacking uracil supplemented with 2% glucose and incubated overnight at 30°C. Cultures were diluted 1/150 into 5 ml of complete media lacking uracil supplemented with 2% raffinose and incubated at 30°C until they reached mid-log phase, and then the cells were resuspended in complete media lacking uracil supplemented with 2% galactose and induced for 4 h at 30°C. For NaCl stress of yeast cells, 0.9 M NaCl was added to the cultures for the appropriate time at the end of the induction. For latrunculin-A induced disassembly of the actin cytoskeleton, 100 μM (final concentration) latrunculin A from a 10 mM stock of latrunculin A (Molecular Probes) was added to cultures for 15 min at 30°C. Cells were fixed with 3.7% formaldehyde for 30 min at room temperature, pelleted, and incubated for 2 h in 1/10 volume of phosphate-buffered formaldehyde (35 mM K2HPO4-KH2PO4 [pH 6.5], 5 mM MgCl2, 3.7% formaldehyde). The fixed cells were washed twice with phosphate-buffered saline (PBS) and resuspended in 1 ml of PBS. One hundred-microliter aliquots of cells were treated with 1.2 μM rhodamine-phalloidin (Sigma) for 90 min or 8 U of Texas red-phalloidin (Molecular Probes)/ml for 1 h, washed three times with PBS, and then allowed to settle for 10 min on poly-l-lysine-coated slides (1 mg/ml). Excess cells were removed by washing with PBS, and then coverslips were mounted with SlowFade (Molecular Probes). Cells were analyzed with a Leica TCS-NT confocal microscope with Ar-Kr triple-line laser, with green fluorescence collected in channel 1 (488-nm excitation, 530 ± 30-nm emission) and red fluorescence collected in channel 2 (568-nm excitation; LP, 590 nm).
Expression of GFP-tagged Inp52p and Inp53p under the control of their native promoters.
The pRS416-GFP vector was constructed by cloning a BamHI/BglII GFP gene fragment from vector pFA6a-GFP(S65T)-HIS3MX6 (27) into the BamHI site of pRS416. Full-length INP52 and INP53 were amplified with 521 and 574 bp, respectively, of sequence upstream of the initiating methionine codon using primers shown in Table 2. PCR products were cloned into the BamHI site of pRS416-GFP in-frame with the C-terminal GFP gene. Constructs were transformed into the corresponding null mutant cells using an S. cerevisiae EasyComp transformation kit (Invitrogen), and transformants were selected on complete media lacking uracil. Single colonies were inoculated into complete media lacking uracil and incubated at 30°C until the cultures reached mid-log phase. Cells were hyperosmotically stressed with 0.9 M NaCl for 10 min, fixed, colocalized with phalloidin as described above, and visualized by confocal microscopy.
Preparation of yeast extracts.
Following induction of GFP-tagged 5-phosphatases, cells were resuspended in 1 ml PBSM (1 × PBS, 5 mM MgCl2, 0.2 μg of aprotinin/ml, 0.2 μg of leupeptin/ml, 1 mM benzamidine, 1 mM phenylmethylsulfonyl fluoride, 3 μg of pepstatin A/ml). An equal volume of glass beads was added, and the cells were lysed by vortexing for 12 cycles of 30 s. Lysates were centrifuged at 15,000 × g for 15 min to obtain the cytosolic fraction (supernatant). Pellets were resuspended in 1 ml of PBSM with 1% Triton X-100 added, incubated overnight at 4°C with gentle agitation, and then centrifuged at 15,000 × g for 15 min to obtain the Triton X-100-soluble (supernatant) and -insoluble (pellet) fractions. One-hundred-microliter samples were separated by SDS–7.5% PAGE, transferred to nitrocellulose, and immunoblotted with an antibody specific for GFP (P. Silver) according to standard protocols (46).
Osmohomeostasis assays.
YPD plates were supplemented with the various salts to the final specified concentrations shown in Table 3 by adding salts or sorbitol to the autoclaved media immediately prior to pouring the plates. Yeast strains were grown overnight and diluted to an optical density at 600 nm of 0.7, and 2.5-μl serial dilutions (10−1 to 10−6) of cultures were plated in duplicate and incubated overnight at 30°C. Plates were scored for growth of each mutant versus that of the wild-type strain at each dilution.
TABLE 3.
Growth of wild-type and single and double 5-phosphatase null mutants on YPD and hyperosmotic media
| Strain | Medium | Growtha at a dilution of:
|
||||||
|---|---|---|---|---|---|---|---|---|
| Neat | 10−1 | 10−2 | 10−3 | 10−4 | 10−5 | 10−6 | ||
| W303 | YPD | + | + | + | + | + | + | + |
| YPD + 0.9 M NaCl | + | + | + | + | + | + | + | |
| YPD + 1 M sorbitol | + | + | + | + | + | + | + | |
| YPD + 1.4 M sorbitol | + | + | + | + | + | + | + | |
| inp51 mutant | YPD | + | + | + | + | + | + | + |
| YPD + 0.9 M NaCl | + | + | + | + | + | + | 0 | |
| YPD + 1 M sorbitol | + | + | + | + | + | + | 0 | |
| YPD + 1.4 M sorbitol | + | + | + | + | + | + | + | |
| inp52 mutant | YPD | + | + | + | + | + | + | + |
| YPD + 0.9 M NaCl | + | + | + | + | + | + | 0 | |
| YPD + 1 M sorbitol | + | + | + | + | + | + | + | |
| YPD + 1.4 M sorbitol | + | + | + | + | + | + | + | |
| inp53 mutant | YPD | + | + | + | + | + | + | + |
| YPD + 0.9 M NaCl | + | + | + | + | + | + | 0 | |
| YPD + 1 M sorbitol | + | + | + | + | + | + | 0 | |
| YPD + 1.4 M sorbitol | + | + | + | + | + | + | + | |
| inp51 inp52 mutant | YPD | + | + | + | + | + | + | + |
| YPD + 0.9 M NaCl | + | + | + | 0 | 0 | 0 | 0 | |
| YPD + 1 M sorbitol | + | + | + | + | + | + | 0 | |
| YPD + 1.4 M sorbitol | + | + | + | + | 0 | 0 | 0 | |
| inp51 inp53 mutant | YPD | + | + | + | + | + | + | + |
| YPD + 0.9 M NaCl | + | + | + | + | + | 0 | 0 | |
| YPD + 1 M sorbitol | + | + | + | + | + | + | + | |
| YPD + 1.4 M sorbitol | + | + | + | + | + | 0 | 0 | |
| inp52 inp53 mutant | YPD | + | + | + | + | + | + | + |
| YPD + 0.9 M NaCl | + | + | + | + | 0 | 0 | 0 | |
| YPD + 1 M sorbitol | + | + | + | + | + | + | + | |
| YPD + 1.4 M sorbitol | + | + | + | + | 0 | 0 | 0 | |
+, growth; 0, no growth.
Time course of hyperosmotic stress response.
Single colonies of wild-type and null mutant strains were inoculated in 5 ml of YPD and incubated at 30°C until the cultures reached mid-log phase. Strains overexpressing GFP-tagged constructs under the control of the GAL promoter were induced as described above. Cells were hyperosmotically shocked with NaCl at a final concentration of 0.9 M for 0 to 120 min and then fixed, counterstained with Texas red-phalloidin, and analyzed by confocal microscopy as described above. Budding cells with buds less than half the size of the mother cell were scored for the presence of depolarized actin patches. For cells overexpressing GFP-tagged constructs, only cells that demonstrated expression of the recombinant protein were scored for depolarized actin patches. For each yeast strain at least 40 to 50 cells were counted in each of two separate experiments.
RESULTS
Characterization of Inp52p phosphoinositide substrate specificity.
Biochemical characterization of Inp51p, Inp52p, and Inp53p phosphoinositide and inositol phosphate substrate specificity has been reported (16, 44). Each of these enzymes specifically hydrolyzes PtdIns(4,5)P2 forming PtdIns(4)P. Inp54p has recently been shown to be a specific PtdIns(4,5)P2 5-phosphatase (39). In addition, recent studies have shown the Sac1p-like domains of Inp52p, Inp53p, and mammalian synaptojanin have intrinsic polyphosphoinositide phosphatase activity, converting PtdIns(3)P, PtdIns(4)P, and PtdIns(3,5)P2 to PtdIns (16). To determine the kinetics of Inp52p-mediated hydrolysis of PtdIns(4,5)P2, the 5-phosphatase and proline-rich region coding sequences of INP52 were cloned into expression vector pMALC2T. Following induction, the maltose binding protein–5-phosphatase recombinant fusion protein was purified from Triton X-100-extracted Escherichia coli lysates by affinity chromatography. Proteins in the starting material and flowthrough and eluted fractions were separated by SDS–7.5% PAGE and visualized by Coomassie brilliant blue staining. A single polypeptide corresponding to the predicted size of 115 kDa was present in purified fractions 4 to 12. Western blot analysis using affinity-purified antipeptide antibodies to Inp52p demonstrated the elution of a single polypeptide species of 115 kDa (data not shown).
Purified recombinant Inp52p was assayed for Ins(1,4,5)P3, Ins(1,3,4,5)P4, and PtdIns(4,5)P2 5-phosphatase activity as described in Materials and Methods. Partially purified 5-phosphatases I and II were used as a positive control for the reactions. No hydrolysis of Ins(1,4,5)P3 or Ins(1,3,4,5)P4 was observed regardless of the amount of recombinant protein present in the assay. However, significant PtdIns(4,5)P2 5-phosphatase activity was demonstrated; this activity correlated with increasing amounts of purified recombinant Inp52p (results not shown). The kinetics of PtdIns(4,5)P2 hydrolysis by Inp52p were determined using a constant amount of recombinant Inp52p and increasing concentrations of [3H]PtdIns(4,5)P2 (5 to 250 μM; 3,500 cpm/nmol) as described previously (29). The Vmax of the reaction was 125 nmol of PtdIns(4,5)P2 hydrolyzed/min/mg of recombinant Inp52p. The Km of the reaction, calculated from a Lineweaver-Burk plot, was 60 μM (results not shown). This is comparable to that for platelet cytosol 5-phosphatase II, which has an affinity constant of 45 μM for PtdIns(4,5)P2 (29). A mutation of His730 to Ala in the 5-phosphatase domain resulted in complete loss of PtdIns(4,5)P2 5-phosphatase activity, consistent with the proposed catalytic mechanism of action of the 5-phosphatases (48) (data not shown).
Recent studies have demonstrated that a novel PtdIns designated PtdIns(3,5)P2 is rapidly but transiently formed in yeast in response to osmotic stress (13). Purified recombinant Inp52p dephosphorylated PtdIns([3-32P],5)P2 forming PtdIns([3-32P])P, with little hydrolysis observed in the absence of Inp52p (data not shown). High-pressure liquid chromatography analysis of the reaction products confirmed that the 5-phosphatase domain of Inp52p hydrolyzes PtdIns(3,5)P2 to form PtdIns(3)P. Other potential substrates such as PtdIns(4)P, which is hydrolyzed by synaptojanin (7), and PtdIns(3)P were not metabolized by recombinant Inp52p, which lacks the Sac1 domain (data not shown). Collectively, these studies demonstrate that the 5-phosphatase domain of Inp52p demonstrates a unique substrate specificity for a member of this enzyme family, as it hydrolyzes the position-5 phosphates from PtdIns(4,5)P2 and PtdIns(3,5)P2. The kinetics of PtdIns(4,5)P2 hydrolysis are comparable to those of mammalian homologues.
Intracellular localization of Inp52p and Inp53p.
The characterization of the intracellular location of the three Sac1-containing 5-phosphatases may help delineate the cellular mechanisms mediating the phenotype associated with the double-null mutant 5-phosphatase strains, i.e., thick cell walls and massive plasma membrane invaginations.
To define the intracellular localization of the three Sac1 domain-containing 5-phosphatases, Inp51p, Inp52p, and Inp53p were expressed as N-terminal fusion proteins with GFP, under the control of a galactose-inducible promoter, in the corresponding 5-phosphatase null mutant strains. Following induction of fusion protein expression, fractionated yeast cell lysates were analyzed by Western blotting using antibodies to the GFP tag to confirm that the recombinant proteins were intact. Inp52p-GFP and Inp53p-GFP were expressed as 159- and 151-kDa polypeptides respectively, consistent with their predicted molecular masses (data not shown). Recombinant Inp51p-GFP was extensively proteolyzed and therefore was not further characterized (results not shown). Other studies have also reported difficulties in expressing a full-length intact recombinant Inp51p (44), suggesting that this protein is extremely sensitive to degradation.
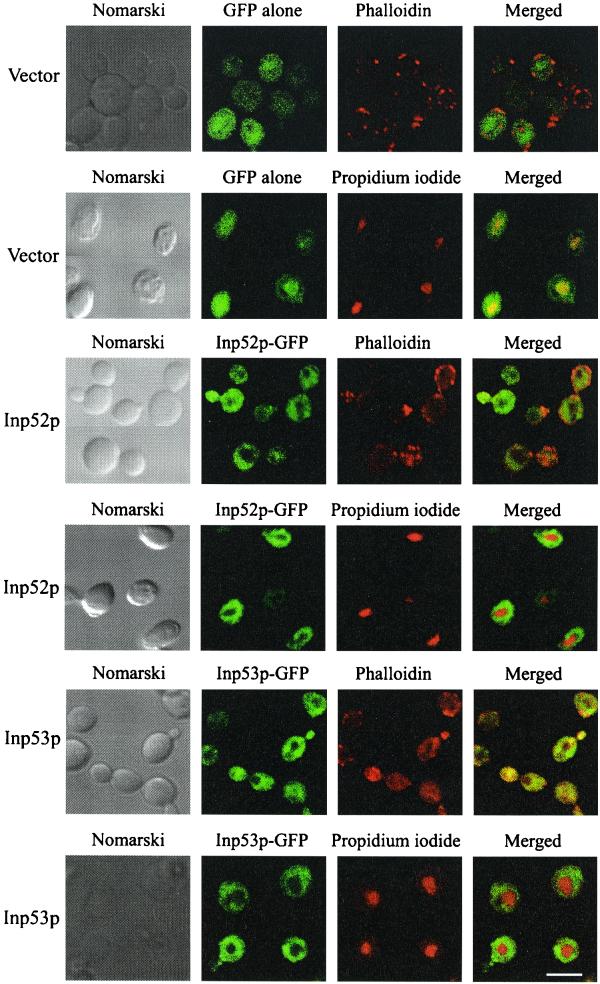
Characterization of the intracellular location of Inp52p and Inp53p was undertaken by confocal microscopy of yeast expressing recombinant GFP-tagged 5-phosphatases. In the presence of 2% raffinose, which does not induce the GAL promoter, Inp52p-GFP and Inp53p-GFP were not expressed. Following incubation in the presence of galactose for 4 h, Inp52p-GFP and Inp53p-GFP were both expressed diffusely through the cell but were excluded from the nucleus as determined by colocalization with propidium iodide (Fig. 1). Neither recombinant 5-phosphatase clearly colocalized with actin patches, which were visualized by staining the cells with phalloidin (Fig. 1) (47). In control studies GFP was expressed diffusely throughout the cell including the nucleus, when expressed in either inp52 (Fig. 1) or inp53 null mutant cells (data not shown).
FIG. 1.
Intracellular localization of Inp52p and Inp53p in unstressed cells. Inp52p and Inp53p were cloned in-frame with GFP under the control of a galactose-inducible promoter and induced in the corresponding null mutant 5-phosphatase yeast strains. Expression of GFP, Inp52p-GFP, and Inp53p-GFP recombinant proteins was induced by growing the cells in galactose for 4 h. Nuclei were visualized by counterstaining with propidium iodide. Actin patches were stained with phalloidin. Bar, 5 μm.
We have demonstrated that recombinant Inp52p-GFP and Inp53p-GFP are functional 5-phosphatases, as expression of these recombinant proteins rescued the delocalized actin patch phenotype of inp52 inp53 double-null mutant 5-phosphatase strains. In addition, expression of either of these two recombinant proteins was able to rescue the lethal phenotype of the inp51 inp52 inp53 triple-null mutant strain (results not shown).
Localization of Inp52p and Inp53p in response to hyperosmotic stress.
S. cerevisiae demonstrates stereotypic patterns of budding, with haploid cells showing an axial budding pattern, while diploids display a bipolar pattern from either pole (5). Yeast cells respond to osmotic stress by rearrangement of actin in an acute adaptive response. A rapid change in the actin cytoskeleton, comprising collapse of actin cables and delocalization of cortical actin patches from sites of active growth in the bud to the mother cell, is observed (6). As PtdIns(4,5)P2 regulates actin polymerization and Inp52p and Inp53p have been shown to hydrolyze this phosphoinositide, we investigated the effects of osmotic stress on the intracellular location of these 5-phosphatases.
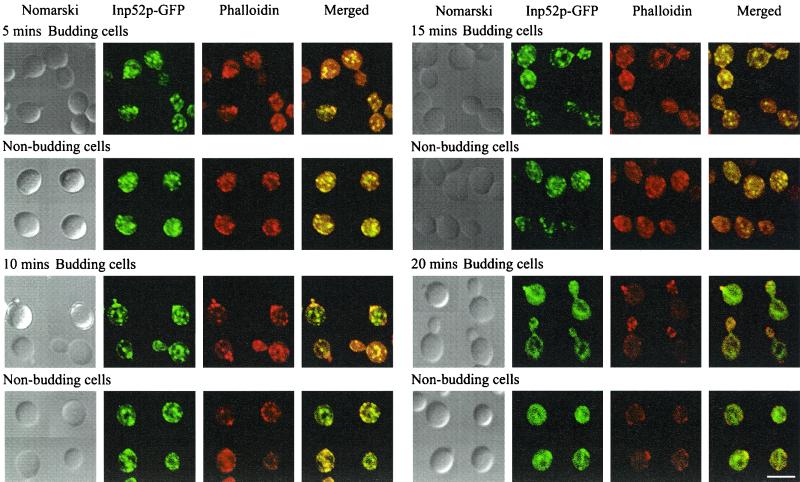
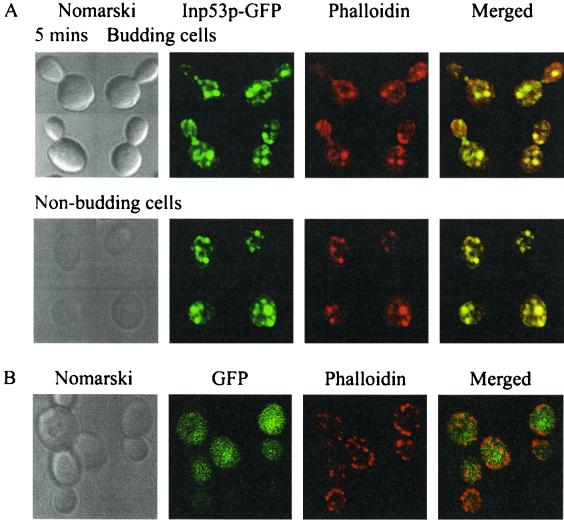
Expression of Inp52p-GFP and Inp53p-GFP was induced by galactose treatment for 4 h, followed by hyperosmotic NaCl treatment to a final concentration of 0.9 M for 5, 10, 15, 20, or 30 min. Cells were fixed by the addition of formaldehyde directly to the culture. Cortical actin patches were visualized by counterstaining with phalloidin. In the nonstressed cell, actin patches were concentrated at the growing bud. In budding cells, hyperosmotic stress treatment resulted in actin patch translocation from sites of active cell growth in the bud to the mother cell (Fig. 2 and 3A) (6). Nonbudding cells showed little change in actin patch localization. The intracellular location of both Inp52p-GFP and Inp53p-GFP changed significantly within 5 min of hyperosmotic stress in both budding and nonbudding cells. Both recombinant enzymes rapidly translocated to an intracellular location that colocalized with cortical actin patches in both the mother and daughter cells (Fig. 2 and 3A). In addition, Inp52p-GFP and Inp53p-GFP colocalized with cortical actin patches in nonbudding cells. Cells expressing Inp53p-GFP demonstrated fewer, larger actin patches following stress than Inp52p-GFP-expressing cells (see below). The association of the 5-phosphatases with actin patches was transient, and within 20 min of treatment both Inp52p-GFP (Fig. 2) and Inp53p-GFP (data not shown) relocated back to a diffuse cellular distribution in more than 50% of cells.
FIG. 2.
Inp52p-GFP colocalizes with actin patches following hyperosmotic stress. Cells expressing Inp52p-GFP were treated with 0.9 M NaCl for the indicated times, fixed with formaldehyde, and stained with rhodamine-phalloidin. Cells were analyzed by confocal microscopy. Regions of colocalization between Inp52p-GFP and rhodamine-phalloidin appear yellow in the merged images. Bar, 5 μm.
FIG. 3.
Inp53p-GFP localizes to actin patches after hyperosmotic stress. (A) Cells expressing Inp53p-GFP were treated with 0.9 M NaCl for 5 min, formaldehyde fixed, stained with rhodamine-phalloidin, and then analyzed by confocal microscopy. Regions of colocalization between Inp53p-GFP and rhodamine-phalloidin appear yellow in the merged images. (B) inp52 null mutant cells expressing GFP alone were treated with 0.9 M NaCl for 10 min, fixed, and stained with phalloidin.
In control studies no change in the cytosolic location of recombinant GFP was observed in response to hyperosmotic stress when GFP was expressed in either inp52 (Fig. 3B) or inp53 (data not shown) null mutant cells, indicating that the translocation of GFP–5-phosphatase fusion proteins to actin patches was not due to the GFP moiety. In addition, recombinant GFP-tagged Inp54p, which does not contain an N-terminal Sac1 domain or C-terminal proline-rich domain, did not relocate to actin patches following stress treatment (results not shown). To further establish that GFP does not mediate 5-phosphatase translocation to actin patches, cells expressing N-terminal or C-terminal hemagglutinin (HA)-tagged Inp52p were treated with 0.9 M NaCl for 10 min and analyzed by confocal microscopy. The HA-tagged constructs displayed similar hyperosmotic stress responses, with the recombinant 5-phosphatase colocalizing with actin patches (results not shown).
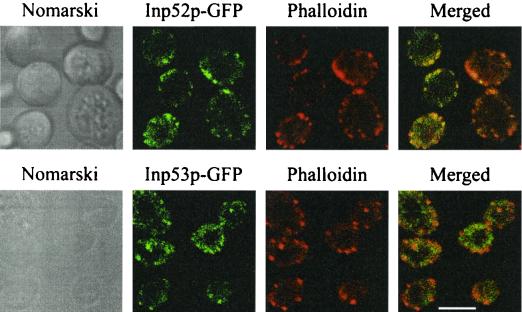
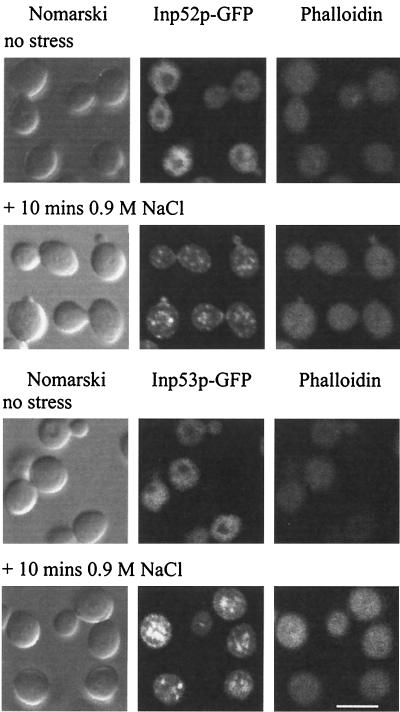
To ensure that overexpression of the GFP-tagged 5-phosphatases did not result in aberrant translocation to actin patches in response to stress, both 5-phosphatases were expressed as GFP fusion proteins under the control of their native promoters and subjected to 10 min of 0.9 M NaCl stress. The 5-phosphatase–GFP fluorescence was significantly less intense than when the 5-phosphatases were overexpressed under the control of the GAL promoter; however both 5-phosphatases still formed patches that colocalized with actin patches, as demonstrated by counterstaining with phalloidin following hyperosmotic stress (Fig. 4). It is also noteworthy that decreased expression of Inp53p resulted in actin patches that resembled those of wild-type and Inp52p-expressing cells, suggesting that overexpression of Inp53p affects the number and size of delocalized actin patches.
FIG. 4.
Inp52p and Inp53p expressed under the control of their native promoters localize to actin patches following hyperosmotic stress. Cells expressing Inp52p-GFP or Inp53p-GFP under the control of the respective native 5-phosphatase promoter were treated with 0.9 M NaCl for 10 min, formaldehyde fixed, stained with phalloidin, and then analyzed by confocal microscopy. Bar, 5 μm.
Cytoskeletal rearrangement occurs in response to other solutes in addition to NaCl (6). The ability of Inp52p-GFP to translocate to actin patches following different hyperosmotic stimuli was examined at 5 and 10 min after addition of a number of solutes. Glucose (0.6 M) or glycerol (0.76 M) had no effect on the localization of the Inp52p-GFP protein or on the normal distribution of actin patches within yeast. KCl (0.6 M) treatment caused partial delocalization of actin patches to the mother cell in budding yeast but no change in the distribution of Inp52p-GFP. Treatment of cells with 1 M sorbitol had no effect on the polarized localization of the actin patches or on the location of Inp52p. However, hyperosmotic shock using 1.4 M sorbitol or 0.5 M CaCl2 resulted in delocalization of actin patches to the mother cell in budding yeast and translocation of Inp52p-GFP to actin patches (data not shown).
Translocation of Inp52p and Inp53p to actin patches is independent of actin.
Cortical actin patch assembly is a hierarchical process in which assembly factors initially associate with the plasma membrane, followed by recruitment of actin-nucleating factors and incorporation of actin filaments and finally actin-dependent proteins (38). In order to determine whether the translocation of Inp52p and Inp53p to patches following hyperosmotic stress is dependent on an intact actin cytoskeleton, cells expressing the GFP-tagged 5-phosphatases were treated with latrunculin-A. In previous studies latrunculin-A has been shown to completely disrupt the yeast actin cytoskeleton within minutes (4). In nonstressed cells exposed to latrunculin-A for 15 min, Inp52p-GFP and Inp53p-GFP displayed an apparent cytosolic localization similar to that of nontreated cells (Fig. 5). Cortical actin patches were not detected by phalloidin staining. Following 10 min of hyperosmotic stress, both 5-phosphatases localized to patch-like structures identical to those observed in nontreated cells (Fig. 5), implying that the translocation of Inp52p and Inp53p is actin independent. Several other proteins such as Sla1 and Sla2 form patches in the absence of an intact actin cytoskeleton following latrunculin-A treatment, suggesting that these proteins localize to patches upstream of actin in patch assembly (4).
FIG. 5.
Inp52p-GFP and Inp53p-GFP translocate to patches independent of actin. Cells expressing Inp52p-GFP or Inp53p-GFP were treated with 100 μM latrunculin-A for 15 min at 30°C and then either left untreated or treated with 0.9 M NaCl for 10 min, formaldehyde fixed, stained with phalloidin, and analyzed by confocal microscopy. Bar, 5 μm.
Analysis of motifs within Inp52p and Inp53p regulating enzyme localization in response to hyperosmotic stress.
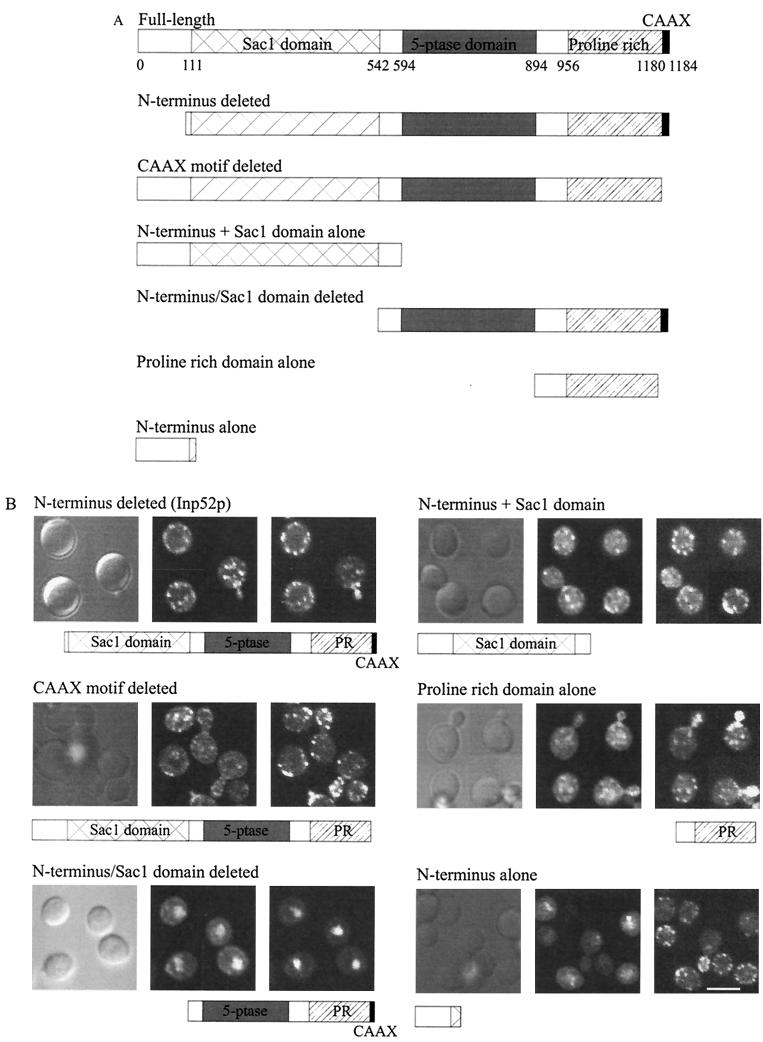
In order to determine which domains within Inp52p and Inp53p are responsible for the intracellular location of these enzymes, in particular, the hyperosmotic stress-induced translocation to actin patches, various regions of the INP52 gene (Fig. 6A) or INP53 gene (Fig. 7A) were cloned upstream of the GFP gene in vector pPS1303 and the constructs were transformed into the corresponding null mutant cells. Expression of the mutant recombinant proteins was induced with galactose for 4 h, and recombinant proteins were analyzed by confocal microscopy.
FIG. 6.
Analysis of Inp52p domains which mediate the intracellular localization in hyperosmotically stressed cells. Inp52p-GFP deletion mutant constructs were generated as described in Materials and Methods. (A) Schematic representation of wild-type and mutant Inp52p-GFP fusion proteins. 5-ptase, 5-phosphatase. (B) Confocal microscopy of yeast expressing mutant Inp52p-GFP fusion proteins (middle panels) as shown in Fig. 7A. Cells were treated for 10 min with 0.9 M NaCl prior to fixation. All cells were stained with phalloidin (right panels) except cells expressing Inp52p-GFP with the N terminus and Sac1 domain deleted, which were stained with propidium iodide to visualize the nucleus. Bar, 5 μm.
FIG. 7.
Analysis of the domains mediating translocation of Inp53p to actin patches following hyperosmotic stress. Inp53p-GFP deletion mutant constructs were prepared as described in Materials and Methods. (A) Schematic diagram of the Inp53p mutant constructs. 5-ptase, 5-phosphatase; proline, proline-rich region. (B) Cells expressing Inp53p-GFP mutants (middle panels) were analyzed by confocal microscopy following 10 min of 0.9 M NaCl stress, fixed, and stained with phalloidin (right panels). Bar, 5 μm.
A mutant Inp52p lacking the N-terminal 111 amino acids colocalized with actin patches following hyperosmotic stress, indicating that this region is not required for actin patch association (Fig. 6B). Deletion of the putative CAAX motif from Inp52p did not alter the localization of recombinant Inp52p in hyperosmotically stressed yeast (Fig. 6B). The CAAX motif therefore is not predicted to play a role in the translocation of Inp52p to actin patches. These results are consistent with our observation that the proposed Inp52p CAAX motif does not conform to the consensus sequence for a CAAX box, as it lacks the aliphatic residues in the middle of the motif.
To determine whether the Sac1 domain of Inp52p plays a role in the localization of the protein, a construct lacking the N terminus and Sac1 domain, containing the 5-phosphatase and proline-rich domains alone, and a construct consisting of the N terminus and Sac1 domain were expressed in inp52 null mutant cells. Removal of both the N-terminal 111 amino acids plus the Sac1 domain resulted in expression of the mutant recombinant Inp52p in the nuclei of stressed cells and diffusely throughout the cytosol, as determined by colocalization with propidium iodide (Fig. 6B). As this recombinant protein at 83 kDa is too large to enter the nucleus by passive diffusion, a putative nuclear localization signal (KKKSKPK, amino acids 1155 to 1161) may mediate nuclear relocation following removal of the Sac1 domain. The protein containing only the N terminus and Sac1 domain translocated to actin patches following 10 min of hyperosmotic stress (Fig. 6B). However, some recombinant protein remained in an apparently cytosolic localization. Deletion of the proline-rich domain of Inp52p resulted in the formation of large protein aggregates in both nonstressed and hyperosmotically stressed yeast cells probably due to protein misfolding (data not shown).
The proline-rich domain alone translocated to actin patches following hyperosmotic stress; however this recombinant protein did not localize to patches as efficiently as the full-length protein (Fig. 6B). These results suggest that both (i) the N terminus and Sac1 domain and (ii) the proline-rich domain of Inp52p play a role in mediating the translocation of the protein to actin patches following hyperosmotic stress.
To determine whether similar domains also mediate translocation of Inp53p to actin patches, a number of deletion constructs were generated (Fig. 7A) and expressed in inp53 null mutant cells. The Inp53p N terminus and Sac1 domain did not translocate to actin patches following hyperosmotic stress (Fig. 7B). A longer deletion mutant, which included the 5-phosphatase domain (proline-rich domain deleted) was also constructed to ensure that the lack of translocation did not result from the misfolding of the truncated protein. However, this mutant lacking the proline-rich domain also failed to localize to actin patches under hyperosmotic stress conditions (Fig. 7B), indicating that the N terminus and Sac1 domain of Inp53p do not play a role in mediating 5-phosphatase translocation to actin patches.
To determine whether the proline-rich domain of Inp53p is responsible for mediating actin patch localization, two constructs, one with the N terminus and Sac1 domain deleted and one consisting of the proline-rich domain alone, were generated and expressed in inp53 null mutant cells. Both these mutant recombinant proteins translocated to actin patches following stress (Fig. 7B), suggesting that the Inp53p proline-rich domain localizes the enzyme to actin patches.
As the N terminus and Sac1 domain of Inp52p, but not of Inp53p, contributed to actin patch localization, we analyzed the amino acid sequences of both Inp52p and Inp53p for variations that may explain these differences. A serine-rich sequence in the Sac1 domain of Inp52p between amino acids 132 and 150, which is not present in Inp53p, was identified. In order to determine whether this motif mediates translocation to actin patches, constructs comprising amino acids 1 to 158 of Inp52p and the corresponding region of Inp53p, (amino acids 1 to 133), were expressed as GFP fusion proteins. Neither construct translocated to actin patches upon hyperosmotic stress (Fig. 6B and 7B), suggesting that other sequences within Inp52p are required for relocation of the 5-phosphatase.
Collectively, these results indicate that Inp52p and Inp53p translocate to actin patches following hyperosmotic shock. Sequences within the N terminus and Sac1 domain of Inp52p and the proline-rich domains of both Inp52p and Inp53p play a critical role in mediating the translocation of Inp52p and Inp53p to actin patches following hyperosmotic stress.
Growth of single and double 5-phosphatase null mutants on hyperosmotic media.
We investigated the ability of 5-phosphatase null mutant strains to grow under hyperosmotic conditions. Serial dilutions of mid-log-phase cultures of wild-type and single and double 5-phosphatase null mutants were plated on YPD, YPD plus 0.9 M NaCl, YPD plus 1 M sorbitol, or YPD plus 1.4 M sorbitol. Wild-type cells grew equally well on all media to a dilution of 10−6 (Table 3). Cells containing a null mutation of any one Sac1 domain containing 5-phosphatase displayed growth comparable to that of wild-type yeast in YPD and hyperosmotic media, with colonies detected in 10−5 to 10−6 dilutions. Although all double knockouts grew on YPD and YPD plus 1 M sorbitol to dilutions of 10−5 to 10−6, these mutant yeasts demonstrated a significantly reduced ability to grow on YPD plus 0.9 M NaCl and YPD plus 1.4 M sorbitol. The inp51 inp52 null mutant was the most severely affected (10−2 dilution for YPD plus 0.9 M NaCl; 10−3 dilution for 1.4 M sorbitol), followed by the inp52 inp53 mutant (10−3 dilution) with the inp51 inp53 mutant (10−4 dilution) moderately impaired. We noted a consistent correlation between the concentrations of hyperosmotic media (0.9 M NaCl or 1.4 M sorbitol) required to induce the translocation of the 5-phosphatases Inp52p and Inp53p to actin patches and the osmosensitive phenotype of null mutant stains. For example lower concentrations of sorbitol (1 M), which did not induce relocation of 5-phosphatases to actin patches, did not induce osmosensitivity in 5-phosphatase double-null mutants. Srinivasan et al. (42) previously reported only the inp51 inp52 double mutant was inhibited by NaCl; however this study did not quantitate the growth of yeast strains using serial dilutions and may therefore have missed the impaired growth of the other 5-phosphatase mutants on hyperosmotic media.
Overexpression of Inp52p and Inp53p reduces the duration of the osmotic stress response.
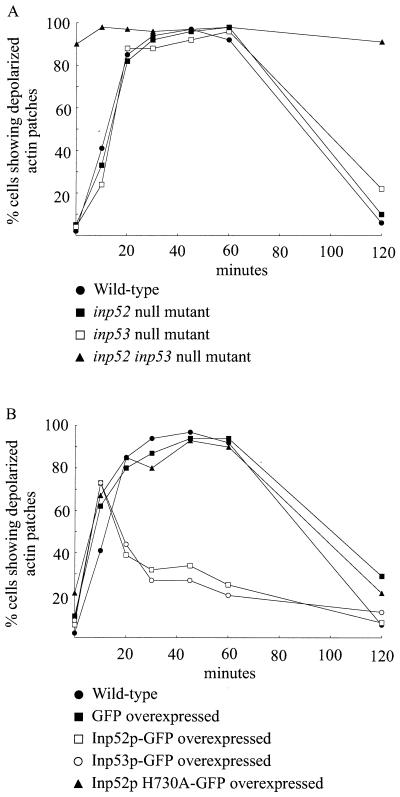
Following hyperosmotic shock, actin patches depolarize from sites of growth in the bud to the mother cell. Over time this phenomenon is reversed as the osmotic gradient is restored and actin patches repolarize to the bud. As Inp52p and Inp53p localize to cortical actin patches following stress, we investigated the temporal correlation of stress-induced depolarization and repolarization in wild-type cells versus that in 5-phosphatase null mutants and cells overexpressing Inp52p or Inp53p or mutant Inp52p (H730A), which lacks PtdIns(4,5)P2 5-phosphatase activity. As shown by previous studies (6), the majority of osmotically stressed yeast cells showed evidence of depolarized actin patches after 20 min of treatment, and these patches remained depolarized until 60 min poststress and then returned to a polarized state over the next 60 min (Fig. 8A). Cells containing a single inp52 or inp53 null mutation or overexpressing GFP alone exhibited a response similar to that of wild-type cells (Fig. 8). Cells lacking both INP52 and INP53, however, did not demonstrate normal polarization of actin patches at any point during osmotic stress (Fig. 8A), suggesting that the 5-phosphatases play a role in cytoskeletal polarity.
FIG. 8.
Time course of actin patch depolarization and repolarization in response to hyperosmotic stress. Cells were treated with 0.9 M NaCl for 0 to 120 min, fixed, stained with phalloidin, and analyzed by confocal microscopy. The percentages of budding cells with buds less than half the size of the mother cell exhibiting depolarized actin patches were determined as described in Materials and Methods. For each yeast strain 40 to 50 cells were counted. Results shown are from one representative experiment of two. In overexpression studies only cells that demonstrated expression of the GFP-tagged recombinant protein were scored for depolarized actin patches.
Overexpression of either Inp52p or Inp53p resulted in a dramatic decrease in the time required for the actin patches to repolarize to the bud (Fig. 8B). At 10 min poststress ∼70% of the cells expressing Inp52p or Inp53p exhibited depolarized actin patches. Within 30 min of initiation of osmotic stress, a large proportion of the cells exhibited repolarized actin patches, in contrast to wild-type cells, almost all of which maintained depolarized patches. This suggests that Inp52p and Inp53p play a significant role in the repolarization of actin patches. In order to determine whether the enzyme activity encoded by the 5-phosphatase domain was essential for the reduction in the duration of the stress response, a single point mutation was made in the 5-phosphatase domain of Inp52p. This mutation, which substitutes an alanine residue for histidine 730 in the conserved 5-phosphatase catalytic domain, results in complete loss of PtdIns(4,5)P2 hydrolyzing activity. Overexpression of this catalytically inactive mutant resulted in a response similar to that observed in wild-type cells (Fig. 8B), indicating that PtdIns(4,5)P2 5-phosphatase activity plays a critical role in the repolarization of actin patches within the cell, independent of the Sac1 domain.
DISCUSSION
We have shown that the yeast 5-phosphatases Inp52p and Inp53p translocate transiently to actin patches in both budding and nonbudding yeast during hyperosmotic stress. Overexpression of these 5-phosphatases results in a rapid repolarization of actin patches which is dependent on PtdIns(4,5)P2 5-phosphatase enzyme activity. The proline-rich domains of these proteins and the Sac1 domain of Inp52p appear to be important for localization to actin patches. Recombinant Inp52p hydrolyzes PtdIns(4,5)P2 with kinetics similar to those of mammalian 5-phosphatases and also hydrolyzes the recently identified phosphoinositide PtdIns(3,5)P2. It is noteworthy that PtdIns(3,5)P2 is generated transiently in yeast following hyperosmotic stress. Previous studies have shown that Inp53p also hydrolyzes PtdIns(4,5)P2 (16). Collectively, these studies suggest that the relocation of Inp52p and Inp53p to actin patches during osmotic stress may result in the localized hydrolysis of PtdIns(4,5)P2, thereby regulating cytoskeletal organization.
Regulation of PtdIns(3,5)P2 by Inp52p and Inp53p.
PtdIns(3,5)P2 is a recently identified phosphoinositide present in both yeast and higher eukaryotic cells. The lipid is synthesised in yeast by the actions of Fab1p, a specific PtdIns(3)P 5-kinase, from a preexisting pool of PtdIns(3)P (10). A role for PtdIns(3,5)P2 in regulating the actin cytoskeleton has not been shown; rather the phenotype associated with loss of function of Fab1p is a dramatic enlargement of the vacuole (50), indicating that PtdIns(3,5)P2 plays a role in regulating vacuole membrane homeostasis. We have been unable to show localization of Inp52p or Inp53p with vacuole membranes using a variety of conditions, in particular, hyperosmotic stress, despite the demonstration that the Sac1 domains of both enzymes hydrolyze this phosphoinositide. In resting yeast we have demonstrated a diffuse expression of these GFP-tagged recombinant 5-phosphatases throughout the cytoplasm.
The osmotic stress-induced localization of these 5-phosphatases to the actin cytoskeleton, and in particular actin patches, does not preclude an association with the vacuole membrane. In budding yeast a small proportion of the vacuole is polarized along actin cables to the emerging bud site, consistent with actin playing a primary role in vacuole movement (18). Electron microscopy studies have also revealed that cytoskeletal fibers extend into the vacuolar space (37). In addition, in nonbudding yeast colocalization studies have revealed that the vacuoles associate with many of the actin cables and cortical actin patches at the site of bud emergence (18). Therefore the relocation of Inp52p and Inp53p to actin patches following hyperosmotic stress should allow access of these 5-phosphatases to PtdIns(3,5)P2 as it is rapidly synthesized in the vacuole membrane. It is tempting to speculate that the abnormal actin polymerization associated with 5-phosphatase null mutant yeast may lead to fragmentation of the vacuoles as they move along actin cables during yeast budding. This hypothesis may explain the fragmented vacuoles observed in 5-phosphatase disruptants.
Role of the Sac1 and proline-rich domains in 5-phosphatase intracellular location.
We have shown that the Sac1 domain of Inp52p and the proline-rich regions of both Inp52p and Inp53p mediate the association of the proteins with actin patches during periods of osmotic stress. The Sac1 domain represents a 300-amino-acid conserved protein module that is present in the product of the SAC1 gene, three of the four yeast 5-phosphatases, and the mammalian homologue synaptojanin. Recent studies have indicated the Sac1 domains of Inp52p, Inp53p, and synaptojanin contain phosphoinositide phosphatase activity. In addition, loss of function of Sac1p results in a dramatic accumulation of PtdIns(4)P, PtdIns(3)P, and PtdIns(3,5)P2 (16). It is noteworthy that although the yeast Sac1 protein has been localized to the endoplasmic reticulum and Golgi (49), it plays a role in the regulation of the actin cytoskeleton. Temperature-sensitive sac1 mutants grown at the nonpermissive temperature demonstrate loss of visible cables and delocalized actin patches randomly distributed between the mother cell and the bud (8, 35). Similarly, various spliced variants of synaptojanin are localized via the distinct C-terminal proline-rich regions to various compartments including the mitochondria; however the translocation of either of these Sac1-containing proteins has not been determined under stress conditions (34). The proline-rich domains of Inp52p and Inp53p contain multiple proline-rich motifs which could potentially allow interactions of the 5-phosphatases with a number of different proteins, in particular, with SH3 domains. We propose that under conditions of hyperosmotic stress, Inp52p and Inp53p translocate to actin patches, where they interact with patch proteins and regulate phosphoinositides and thereby the actin cytoskeleton.
Colocalization of actin patches and finger-like invaginations.
We investigated the intracellular location of Inp52p and Inp53p in an attempt to further define the cellular mechanisms mediating the thickened cell wall and massive plasma membrane invaginations observed in 5-phosphatase null mutant yeast. Mulholland et al. (33), using immunoelectron microscopy of ultrathin sections, demonstrated a relationship between plasma membrane invaginations and the accumulation of cortical actin observed in actin patches. In nonstressed cells, actin patches display a polarized localization, being present at sites of active cell growth (1, 24). These patches surround plasma membrane invaginations and appear to be attached to cables, which extend into the cell and which are postulated to transport cell wall components and possibly endocytic vesicles (21, 25, 33). In addition, it has been suggested that the plasma membrane invaginations are sites of cell wall synthesis (15) and are surrounded by cortical actin patches, which play an important role in the construction of the cell wall (25, 33). Patch components have also been postulated to be present in low concentrations at sites on the plasma membrane other than cortical actin patches, where they may contribute to receptor internalization (32, 38).
In yeast, PtdIns(4,5)P2 is localized predominantly in the plasma membrane (36) and binds to actin-regulatory proteins such as profilin, cofilin, and capping protein to inhibit their functions (2, 17, 19). Hydrolysis of PtdIns(4,5)P2 releases actin-regulatory proteins resulting in actin reorganization. There is a striking similarity between the phenotype of cortical actin patch component null mutants and that of double 5-phosphatase null mutants, both of which comprise cell wall thickening, depolarized actin patches under nonstressed conditions, osmosensitivity, and defects in endocytosis (reviewed in reference 38). We propose that the yeast 5-phosphatases Inp52p and Inp53p regulate PtdIns(4,5)P2 at actin patches at the plasma membrane to regulate cytoskeletal polarization, endocytosis, and localized cell wall generation.
Role of the 5-phosphatases in the yeast hyperosmotic stress response.
Yeasts respond to osmotic pressure with changes in the actin cytoskeleton resulting in collapse of the osmotic gradient and disorganization of the actin cytoskeleton (6, 21). Cortical actin patches depolarize away from sites of active cell growth and actin cables collapse, resulting in a temporary pause in cell growth. The translocation of increased concentrations of 5-phosphatases to actin patches in response to hyperosmotic stress may mediate the localized hydrolysis of PtdIns(4,5)P2 and thereby provide a mechanism allowing rapid repolarization of the actin cytoskeleton following hyperosmotic stress. Loss of function of both Inp52p and Inp53p is associated with depolarization of the actin cytoskeleton, while overexpression of 5-phosphatases results in a dramatic repolarization of actin patches in response to osmotic stress, which is dependent on PtdIns(4,5)P2 5-phosphatase activity. Collectively, these studies provide evidence for the mechanisms by which inositol polyphosphate 5-phosphatases may serve to regulate actin cytoskeletal reorganization following hyperosmotic stress.
ACKNOWLEDGMENTS
This research was funded by a grant (9606077) from the Australian Research Council.
Confocal images were obtained at the Biomedical Confocal Imaging Facility of Monash University.
REFERENCES
- 1.Adams A E, Pringle J R. Relationship of actin and tubulin distribution to bud growth in wild-type and morphogenetic-mutant Saccharomyces cerevisiae. J Cell Biol. 1984;98:934–945. doi: 10.1083/jcb.98.3.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amatruda J F, Cooper J A. Purification, characterization, and immunofluorescence localization of Saccharomyces cerevisiae capping protein. J Cell Biol. 1992;117:1067–1076. doi: 10.1083/jcb.117.5.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1991. [Google Scholar]
- 4.Ayscough K R, Stryker J, Pokala N, Sanders M, Crews P, Drubin D G. High rates of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed using the actin inhibitor latrunculin-A. J Cell Biol. 1997;137:399–416. doi: 10.1083/jcb.137.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chant J, Pringle J R. Patterns of bud-site selection in the yeast Saccharomyces cerevisiae. J Cell Biol. 1995;129:751–765. doi: 10.1083/jcb.129.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chowdhury S, Smith K W, Gustin M C. Osmotic stress and the yeast cytoskeleton: phenotype-specific suppression of an actin mutation. J Cell Biol. 1992;118:561–571. doi: 10.1083/jcb.118.3.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chung J K, Sekiya F, Kang H S, Lee C, Han J S, Kim S R, Bae Y S, Morris A J, Rhee S G. Synaptojanin inhibition of phospholipase D activity by hydrolysis of phosphatidylinositol 4,5-bisphosphate. J Biol Chem. 1997;272:15980–15985. doi: 10.1074/jbc.272.25.15980. [DOI] [PubMed] [Google Scholar]
- 8.Cleves A E, Novick P J, Bankaitis V A. Mutations in the SAC1 gene suppress defects in yeast Golgi and yeast actin function. J Cell Biol. 1989;109:2939–2950. doi: 10.1083/jcb.109.6.2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Connolly T M, Bross T E, Majerus P W. Isolation of a phosphomonoesterase from human platelets that specifically hydrolyzes the 5-phosphate of inositol 1,4,5-trisphosphate. J Biol Chem. 1985;260:7868–7874. [PubMed] [Google Scholar]
- 10.Cooke F T, Dove S K, McEwen R K, Painter G, Holmes A B, Hall M N, Michell R H, Parker P J. The stress-activated phosphatidylinositol 3-phosphate 5-kinase Fab1p is essential for vacuole function in S. cerevisiae. Curr Biol. 1998;8:1219–1222. doi: 10.1016/s0960-9822(07)00513-1. [DOI] [PubMed] [Google Scholar]
- 11.Corvera S, D'Arrigo A, Stenmark H. Phosphoinositides in membrane traffic. Curr Opin Cell Biol. 1999;11:460–465. doi: 10.1016/S0955-0674(99)80066-0. [DOI] [PubMed] [Google Scholar]
- 12.De Camilli P, Emr S D, McPherson P S, Novick P. Phosphoinositides as regulators in membrane traffic. Science. 1996;271:1533–1539. doi: 10.1126/science.271.5255.1533. [DOI] [PubMed] [Google Scholar]
- 13.Dove S K, Cooke F T, Douglas M R, Sayers L G, Parker P J, Michell R H. Osmotic stress activates phosphatidylinositol-3,5-bisphosphate synthesis. Nature. 1997;390:187–192. doi: 10.1038/36613. [DOI] [PubMed] [Google Scholar]
- 14.Downes C P, Mussat M C, Michell R H. The inositol trisphosphate phosphomonoesterase of the human erythrocyte membrane. Biochem J. 1982;203:169–177. doi: 10.1042/bj2030169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gabriel M, Kopecka M. Disruption of the actin cytoskeleton in budding yeast results in formation of an aberrant cell wall. Microbiology. 1995;141:891–899. doi: 10.1099/13500872-141-4-891. [DOI] [PubMed] [Google Scholar]
- 16.Guo S, Stolz L E, Lemrow S M, York J D. SAC1-like domains of yeast SAC1, INP52, and INP53 and of human synaptojanin encode polyphosphoinositide phosphatases. J Biol Chem. 1999;274:12990–12995. doi: 10.1074/jbc.274.19.12990. [DOI] [PubMed] [Google Scholar]
- 17.Haarer B K, Lillie S H, Adams A E, Magdolen V, Bandlow W, Brown S S. Purification of profilin from Saccharomyces cerevisiae and analysis of profilin-deficient cells. J Cell Biol. 1990;110:105–114. doi: 10.1083/jcb.110.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hill K L, Catlett N L, Weisman L S. Actin and myosin function in directed vacuole movement during cell division in Saccharomyces cerevisiae. J Cell Biol. 1996;135:1535–1549. doi: 10.1083/jcb.135.6.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iida K, Moriyama K, Matsumoto S, Kawasaki H, Nishida E, Yahara I. Isolation of a yeast essential gene, COF1, that encodes a homologue of mammalian cofilin, a low-M(r) actin-binding and depolymerizing protein. Gene. 1993;124:115–120. doi: 10.1016/0378-1119(93)90770-4. [DOI] [PubMed] [Google Scholar]
- 20.Jackson S P, Schoenwaelder S M, Matzaris M, Brown S, Mitchell C A. Phosphatidylinositol 3,4,5-trisphosphate is a substrate for the 75 kDa inositol polyphosphate 5-phosphatase and a novel 5-phosphatase which forms a complex with the p85/p110 form of phosphoinositide 3-kinase. EMBO J. 1995;14:4490–4500. doi: 10.1002/j.1460-2075.1995.tb00128.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnston G C, Prendergast J A, Singer R A. The Saccharomyces cerevisiae MYO2 gene encodes an essential myosin for vectorial transport of vesicles. J Cell Biol. 1991;113:539–551. doi: 10.1083/jcb.113.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones J S, Prakash L. Yeast Saccharomyces cerevisiae selectable markers in pUC18 polylinkers. Yeast. 1990;6:363–336. doi: 10.1002/yea.320060502. [DOI] [PubMed] [Google Scholar]
- 23.Karpova T S, McNally J G, Moltz S L, Cooper J A. Assembly and function of the actin cytoskeleton of yeast: relationships between cables and patches. J Cell Biol. 1998;142:1501–1517. doi: 10.1083/jcb.142.6.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kilmartin J V, Adams A E. Structural rearrangements of tubulin and actin during the cell cycle of the yeast Saccharomyces. J Cell Biol. 1984;98:922–933. doi: 10.1083/jcb.98.3.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kopecka M, Gabriel M. Actin cortical cytoskeleton and cell wall synthesis in regenerating protoplasts of the Saccharomyces cerevisiae actin mutant DBY 1693. Microbiology. 1995;141:1289–1299. doi: 10.1099/13500872-141-6-1289. [DOI] [PubMed] [Google Scholar]
- 26.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 27.Longtine M S, McKenzie III A, Demarini D J, Shah N G, Wach A, Brachat A, Philippsen P, Pringle J R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 28.Majerus P W. Inositols do it all. Genes Dev. 1996;10:1051–1053. doi: 10.1101/gad.10.9.1051. [DOI] [PubMed] [Google Scholar]
- 29.Matzaris M, Jackson S P, Laxminarayan K M, Speed C J, Mitchell C A. Identification and characterization of the phosphatidylinositol-(4, 5)-bisphosphate 5-phosphatase in human platelets. J Biol Chem. 1994;269:3397–3402. [PubMed] [Google Scholar]
- 30.Mitchell C A, Brown S, Campbell J K, Munday A D, Speed C J. Regulation of second messengers by the inositol polyphosphate 5-phosphatases. Biochem Soc Trans. 1996;24:994–1000. doi: 10.1042/bst0240994. [DOI] [PubMed] [Google Scholar]
- 31.Mitchell C A, Connolly T M, Majerus P W. Identification and isolation of a 75-kDa inositol polyphosphate-5-phosphatase from human platelets. J Biol Chem. 1989;264:8873–8877. [PubMed] [Google Scholar]
- 32.Mulholland J, Konopka J, Singer-Kruger B, Zerial M, Botstein D. Visualization of receptor-mediated endocytosis in yeast. Mol Biol Cell. 1999;10:799–817. doi: 10.1091/mbc.10.3.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mulholland J, Preuss D, Moon A, Wong A, Drubin D, Botstein D. Ultrastructure of the yeast actin cytoskeleton and its association with the plasma membrane. J Cell Biol. 1994;125:381–391. doi: 10.1083/jcb.125.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nemoto Y, De Camilli P. Recruitment of an alternatively spliced form of synaptojanin 2 to mitochondria by the interaction with the PDZ domain of a mitochondrial outer membrane protein. EMBO J. 1999;18:2991–3006. doi: 10.1093/emboj/18.11.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Novick P, Osmond B C, Botstein D. Suppressors of yeast actin mutations. Genetics. 1989;121:659–674. doi: 10.1093/genetics/121.4.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patton J L, Lester R L. Phosphatidylinositol phosphate, phosphatidylinositol bisphosphate, and the phosphoinositol sphingolipids are found in the plasma membrane and stimulate the plasma membrane H(+)-ATPase of Saccharomyces cerevisiae. Arch Biochem Biophys. 1992;292:70–76. doi: 10.1016/0003-9861(92)90052-x. [DOI] [PubMed] [Google Scholar]
- 37.Penman J, Penman S. Resinless section electron microscopy reveals the yeast cytoskeleton. Proc Natl Acad Sci USA. 1997;94:3732–3735. doi: 10.1073/pnas.94.8.3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pruyne D, Bretscher A. Polarization of cell growth in yeast. J Cell Sci. 2000;113:571–585. doi: 10.1242/jcs.113.4.571. [DOI] [PubMed] [Google Scholar]
- 39.Raucher D, Stauffer T, Chen W, Shen K, Guo S, York J D, Sheetz M P, Meyer T. Phosphatidylinositol 4,5-bisphosphate functions as a second messenger that regulates cytoskeleton-plasma membrane adhesion. Cell. 2000;100:221–228. doi: 10.1016/s0092-8674(00)81560-3. [DOI] [PubMed] [Google Scholar]
- 40.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Singer-Kruger B, Nemoto Y, Daniell L, Ferro-Novick S, De Camilli P. Synaptojanin family members are implicated in endocytic membrane traffic in yeast. J Cell Sci. 1998;111:3347–3356. doi: 10.1242/jcs.111.22.3347. [DOI] [PubMed] [Google Scholar]
- 42.Srinivasan S, Seaman M, Nemoto Y, Daniell L, Suchy S F, Emr S, De Camilli P, Nussbaum R. Disruption of three phosphatidylinositol-polyphosphate 5-phosphatase genes from Saccharomyces cerevisiae results in pleiotropic abnormalities of vacuole morphology, cell shape, and osmohomeostasis. Eur J Cell Biol. 1997;74:350–360. [PubMed] [Google Scholar]
- 43.Stolz L E, Huynh C V, Thorner J, York J D. Identification and characterization of an essential family of inositol polyphosphate 5-phosphatases (INP51, INP52 and INP53 gene products) in the yeast Saccharomyces cerevisiae. Genetics. 1998;148:1715–1729. doi: 10.1093/genetics/148.4.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stolz L E, Kuo W J, Longchamps J, Sekhon M K, York J D. INP51, a yeast inositol polyphosphate 5-phosphatase required for phosphatidylinositol 4,5-bisphosphate homeostasis and whose absence confers a cold-resistant phenotype. J Biol Chem. 1998;273:11852–11861. doi: 10.1074/jbc.273.19.11852. [DOI] [PubMed] [Google Scholar]
- 45.Toker A. The synthesis and cellular roles of phosphatidylinositol 4,5-bisphosphate. Curr Opin Cell Biol. 1998;10:254–261. doi: 10.1016/s0955-0674(98)80148-8. [DOI] [PubMed] [Google Scholar]
- 46.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Waddle J A, Karpova T S, Waterston R H, Cooper J A. Movement of cortical actin patches in yeast. J Cell Biol. 1996;132:861–870. doi: 10.1083/jcb.132.5.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Whisstock, J. C., S. Romero, R. Gurung, H. Nandurkar, L. Ooms, S. P. Bottomley, and C. A. Mitchell. The inositol polyphosphate 5-phosphatases and the apurinic/apyrimidinic base excision repair endonucleases share a common mechanism for catalysis. J. Biol. Chem., in press. [DOI] [PubMed]
- 49.Whitters E A, Cleves A E, McGee T P, Skinner H B, Bankaitis V A. SAC1p is an integral membrane protein that influences the cellular requirement for phospholipid transfer protein function and inositol in yeast. J Cell Biol. 1993;122:79–94. doi: 10.1083/jcb.122.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamamoto A, DeWald D B, Boronenkov I V, Anderson R A, Emr S D, Koshland D. Novel PI(4)P 5-kinase homologue, Fab1p, essential for normal vacuole function and morphology in yeast. Mol Biol Cell. 1995;6:525–539. doi: 10.1091/mbc.6.5.525. [DOI] [PMC free article] [PubMed] [Google Scholar]