Summary:
The gut microbiota has been shown to be a critical regulator of anti-tumor immunity during immune checkpoint inhibitor therapies. Several bacteria that promote an anti-tumor response to immune checkpoint inhibitors have been identified in mice(1-6), and transplantation of fecal specimens from responders has been shown to improve the efficacy of anti-PD-1 therapy in melanoma patients(7, 8). However, the increased efficacy from fecal transplants is variable and how gut bacteria promote anti-tumor immunity remains unclear. Here, we show that the gut microbiome downregulates expression of PD-L2 and its binding partner Repulsive Guidance Molecule b (RGMb) to promote anti-tumor immunity and identify bacterial species that mediate this effect. PD-L1 and PD-L2 share PD-1 as a binding partner, but PD-L2 can also bind RGMb. We demonstrate that blockade of PD-L2/RGMb interactions can overcome microbiome-dependent resistance to PD-1 pathway inhibitors: antibody-mediated blockade of the PD-L2/RGMb pathway or conditional deletion of RGMb in T cells combined with anti-PD-1 or anti-PD-L1 promotes anti-tumor responses in multiple mouse tumor models that do not respond to anti-PD-1 or anti-PD-L1 alone (Germ Free mice, antibiotic treated mice, and even mice colonized with non-responder patient stool). These studies identify downregulation of the PD-L2/RGMB pathway as a specific mechanism by which the gut microbiota can promote responses to PD-1 checkpoint blockade and define a potentially effective immunological strategy for treating patients who do not respond to PD-1 cancer immunotherapy.
Antibodies blocking programmed death 1 (PD-1) or programmed death ligand 1 (PD-L1) have been approved for over 25 different tumors. However, the response rates for PD-1/PD-L1 blockade in approved indications range from 13% to 69% depending on tumor type(9, 10). There is great interest in understanding factors regulating responsiveness to PD-1 inhibitors to develop strategies to benefit more patients. Studies have shown that the gut microbiota can modulate the efficacy of PD-1 pathway inhibitors in cancer (7, 8). These findings have stimulated investigation of probiotic bacteria and fecal transplants to promote anti-tumor responses to PD-1 immunotherapy with inconsistent results, highlighting the need to understand mechanisms by which the gut microbiota can promote anti-tumor immunity. Our goal was to identify targetable immunologic mechanisms by which the gut microbiome regulates anti-tumor effects of cancer immunotherapy.
Gut microbiota enhances immunotherapy
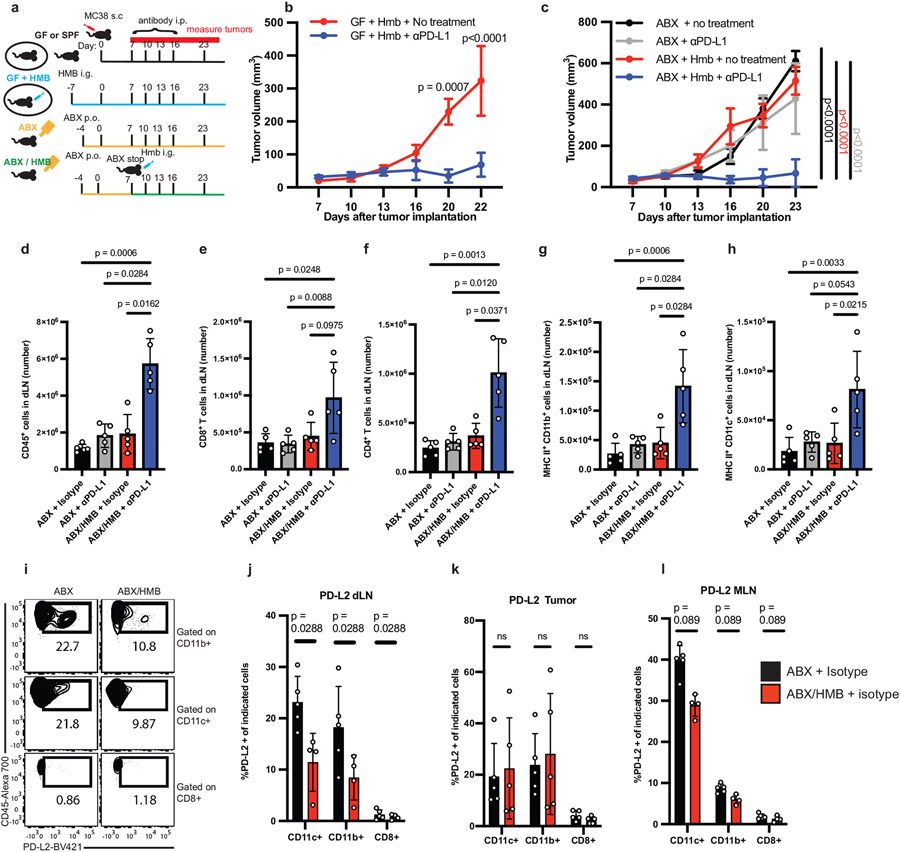
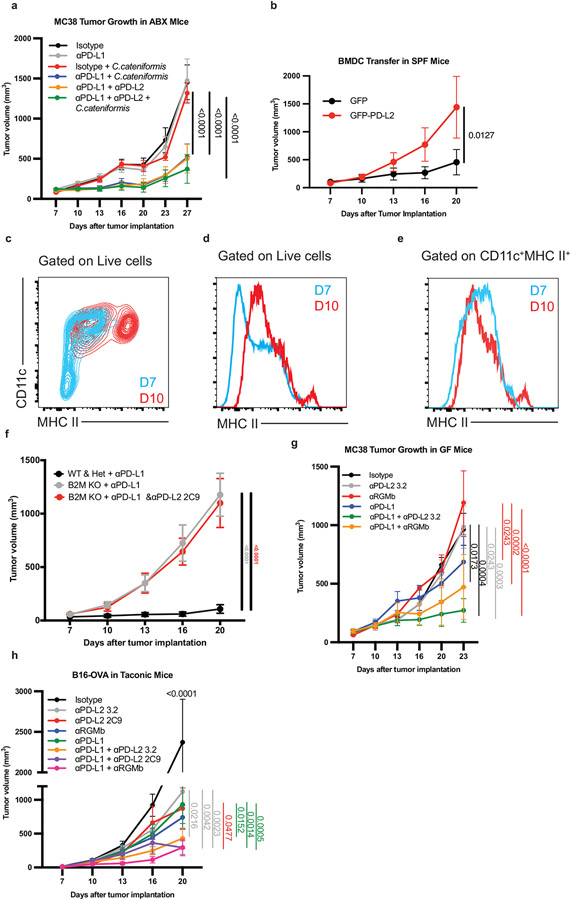
We established mouse tumor models in which the anti-tumor response depended on gut bacteria (Fig. 1a). When MC38 colon carcinoma cells were implanted subcutaneously, conventional specific pathogen-free (SPF) mice containing the vendor’s (Taconic) gut microbiome responded to anti-PD-L1 therapy (Extended Data Fig. 1a), while germ-free (GF) mice did not respond to anti-PD-L1 or anti-PD-1 (Extended Data Fig. 1b,c). Reconstitution of GF mice with healthy human microbiota (HMB) (Fig. 1b) or healthy mouse microbiota (MMB) (Extended Data Fig. 1d) (11) promoted a robust anti-tumor response to anti-PD-L1. To further investigate the immunoregulatory influence of human bacteria, we used HMB as our responder microbiota. Similar to GF mice (Extended data Fig. 1b,c), depletion of gut commensals with a cocktail of 4 oral antibiotics (ABX) (Fig. 1c, Extended Data Fig. 1e), prevented anti-PD-L1 or anti-PD-1-mediated control of MC38 tumors. In contrast, mice given ABX until day 7 after tumor implantation (pi) and then orally gavaged with HMB (termed ABX/HMB) reduced MC38 tumors in response to anti-PD-L1 or anti-PD-1 (Fig. 1c, Extended Data Fig. 1e) (11, 12). Analysis of tumors at day 24 in ABX + anti-PD-L1 mice compared to ABX/HMB + anti-PD-L1 mice revealed the ABX/HMB + anti-PD-L1 mice had a significant increase in the CD8+: regulatory T (Treg) cell ratio with increased frequency of CXCR5+TIM-3−CD8+ progenitor T cells, decreased frequency of the terminally exhausted PD-1+TIM-3+CD8+ T cells, and enhanced CD8+ T cell effector functions including increased frequencies of Granzyme B+ CD8+ T cells, TNF-α+ CD8+ T cells, , IFNα+ CD4+ and CD8+ T cells, and TNF-α+ IFNγ+ CD8+ T cells. These results indicate that HMB promotes CD8+ and CD4+ T cell function in the responding tumors (Extended Data Fig. 2a-h). The enhanced response of ABX/HMB versus ABX mice to anti-PD-L1 or anti-PD-1 allowed examination of mechanisms by which the gut microbiota regulates antitumor responses during anti-PD-L1 or anti-PD-1 treatment (Fig. 1c, Extended Data Fig. 1e).
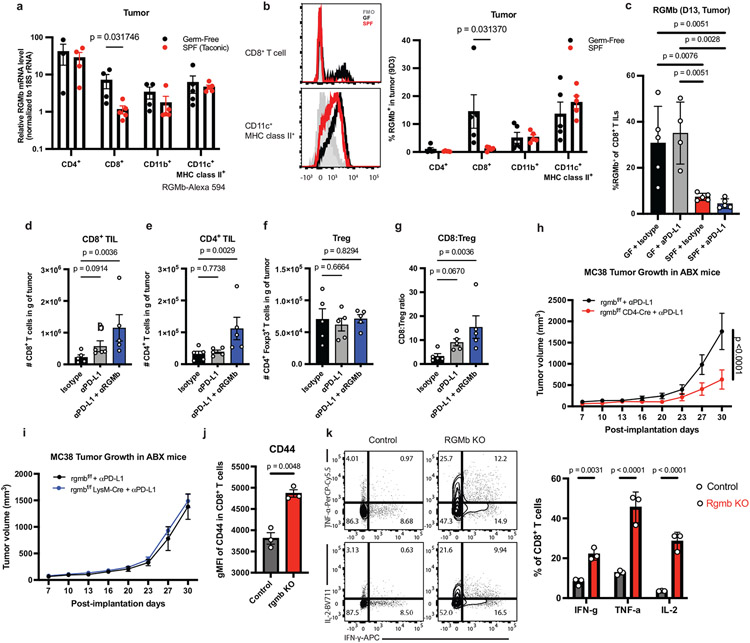
Figure 1. Microbiota promotes an effective anti-tumor response to PD-L1 blockade and suppresses PD-L2 expression.
(a) Schematic of the experimental design. s.c. subcutaneously, i.p. intraperitoneally, p.o. orally, i.g. intragastric (oral gavage). MC38 tumor growth (b) with or without anti-PD-L1 in GF + HMB mice, n = 10 mice per group, representative experiment of 4 individual experiments, significance, shown on graph, measured by two-way ANOVA and Sidak’s multiple comparisons test and (c) in ABX and ABX/HMB mice, n = 4 mice for ABX groups and n= 5 mice for ABX/HMB groups, representative experiment of 5 individual experiments, significance at day 23 is shown and was measured by two-way ANOVA with Tukey’s multiple comparisons test. For (a-b) error bars show mean and standard error of the mean (s.e.m.). (d-k) ABX and ABX/HMB mice were treated as in (a) and sacrificed on day 13 after tumor implantation. Numbers of (d) CD45+ cells, (e) CD8+ T cells, (f) CD4+ T cells, (g) MHCII+ CD11b+ cells and (h) MHCII+ CD11c+ cells in tumor draining lymph nodes (dLNs). (d-k) n = 5 mice per group, representative experiment of two individual experiments, p values indicated on graph and measured by one-way ANOVA with Dunn’s multiple comparisons test, error bars show mean and standard deviation (s.d.). (i) contour plots of representative examples of PD-L2 expression gating in dLNs. Percent of PD-L2 expression on MHCII+ CD11c+, MHCII+ CD11b+, and CD8+ T cells in (j) dLNs, n = 5 mice for ABX, n= 4 mice for ABX/HMB, (k) tumors, n = 5 mice per group, and (l) MLNs, n = 5 mice for ABX and n = 4 mice for ABX/HMB. For (j-l), significance determined by unpaired, two-tailed Mann-Whitney test and significant p values indicated on graph, error bars show s.d., representative experiment of two different experiments.
Gut microbiotas modulate PD-L2 expression
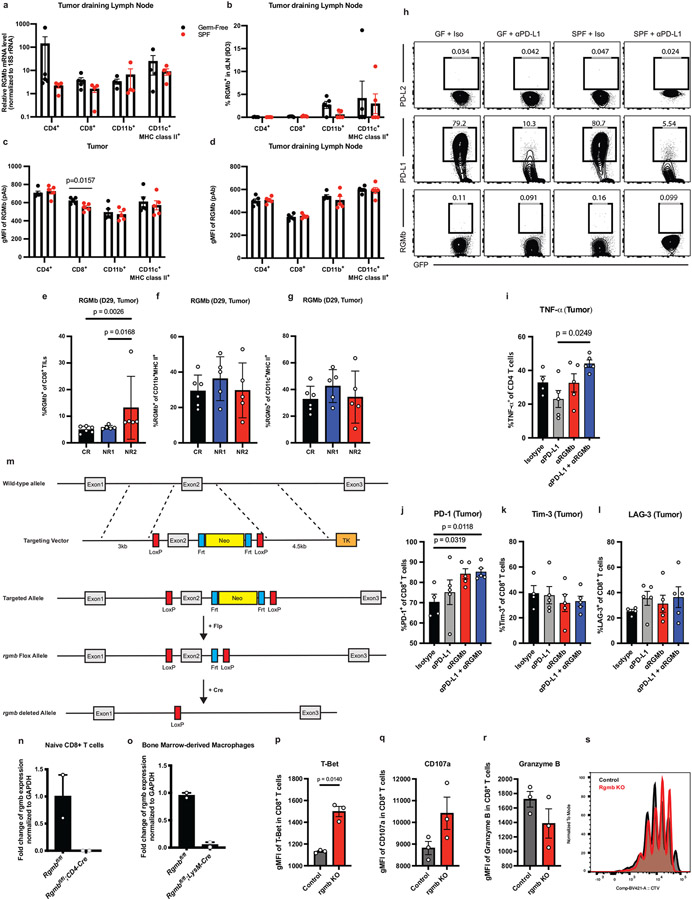
Because gut microbiotas impact the efficacy of PD-1/PD-L1 blockade, we hypothesized that gut bacteria could modulate expression of checkpoint molecules and thereby impact antitumor immune responses. Therefore, we compared expression of several immune checkpoint molecules on immune cells from tumors, tumor draining lymph nodes (dLNs), and mesenteric lymph nodes (MLNs) in ABX vs. ABX/HMB mice. To distinguish between checkpoint molecules that play a causative role in an anti-tumor response rather than are a consequence of the response, we analyzed immune cells at days 10-13 following implantation, before tumor sizes diverge (Fig. 1c). Frequencies of PD-1+, PD-1+ TIM-3+, CD44+ PD-1+, or IFNγ+ CD8+ T cells, previously shown to increase at later timepoints in the microbiota-mediated response to PD-1 blockade (3, 4), did not significantly differ at the day 13 timepoint in tumors of ABX vs. ABX/HMB mice treated with aPD-L1 (Extended Data Fig. 2i-l), indicating that this timepoint was before PD-1 blockade significantly enhanced CD8+ T cell anti-tumor function in responder (ABX/HMB + αPD-L1) mice. However, at day 13 pi, the numbers of CD45+ immune cells, CD8+ T cells, CD4+ T cells, and MHCII+ CD11b+ cells, but not MHCII+ CD11c+ cells in the dLNs were coordinately increased in ABX/HMB + anti-PD-L1 mice, compared to ABX or isotype control groups (Fig. 1d-h). We found differences in expression of immune checkpoint molecules in dendritic and myeloid cells but not T cells; PD-L2 expression was significantly decreased while PD-L1 expression was increased on CD11b+MHC class II+ and CD11c+MHC class II+ cells in the dLNs of ABX/HMB compared to ABX mice. CD80, CD86, and ICOSL showed no significant expression differences (Fig. 1i,j, Extended Data Fig. 2m-p). Thus, PD-L2 expression was downregulated while PD-L1 was increased on these populations in dLNs of responder mice. These data suggest that the gut microbiota regulates signaling pathways involved in expression of coinhibitory molecules.
Reduced PD-L2 expression in responder mice led us to hypothesize that high expression of this coinhibitory molecule in non-responder mice inhibited anti-tumor immunity. PD-L2 expression also was reduced at both day 10 and 13 on CD11c+ and CD11b+ cells in the dLNs and MLNs, but not in tumors in ABX/HMB compared to ABX mice (Fig. 1i-l; Extended Data Fig. 2q) nor at day 10 in dLNs of SPF compared to GF mice (Extended Data Fig. 2r). PD-L2 expression was still reduced at day 24 pi on CD11c+ cells in the dLNs and MLNs but not in tumors or spleens of ABX/HMB mice (Extended Data Fig. 2s-v), suggesting HMB colonization results in reduced PD-L2 expression on CD11c+ DCs in the gut and tumor dLN, but does not cause universally reduced PD-L2 expression. Anti-tumor effects of PD-1 pathway blockade derive, at least in part, from T cell responses in the dLN (13), and there is reduced PD-L2 expression in dLNs of the responder HMB mice. Therefore, we hypothesized that high PD-L2 expression on antigen presenting cells in dLNs of non-responder ABX mice is important for inhibiting anti-tumor immunity. Because anti-PD-1 was also not effective in GF or ABX mice, even though it blocks interaction with both PD-L1 and PD-L2, we considered whether PD-L2 was working through a receptor other than PD-1.
Combined therapy in GF, ABX and SPF mice
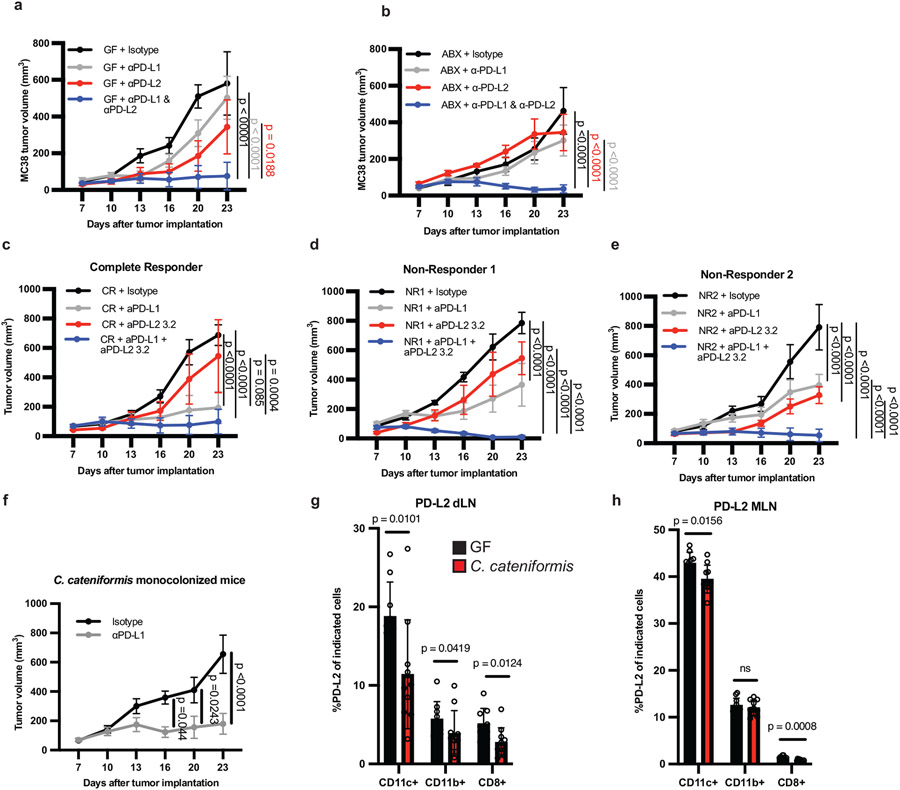
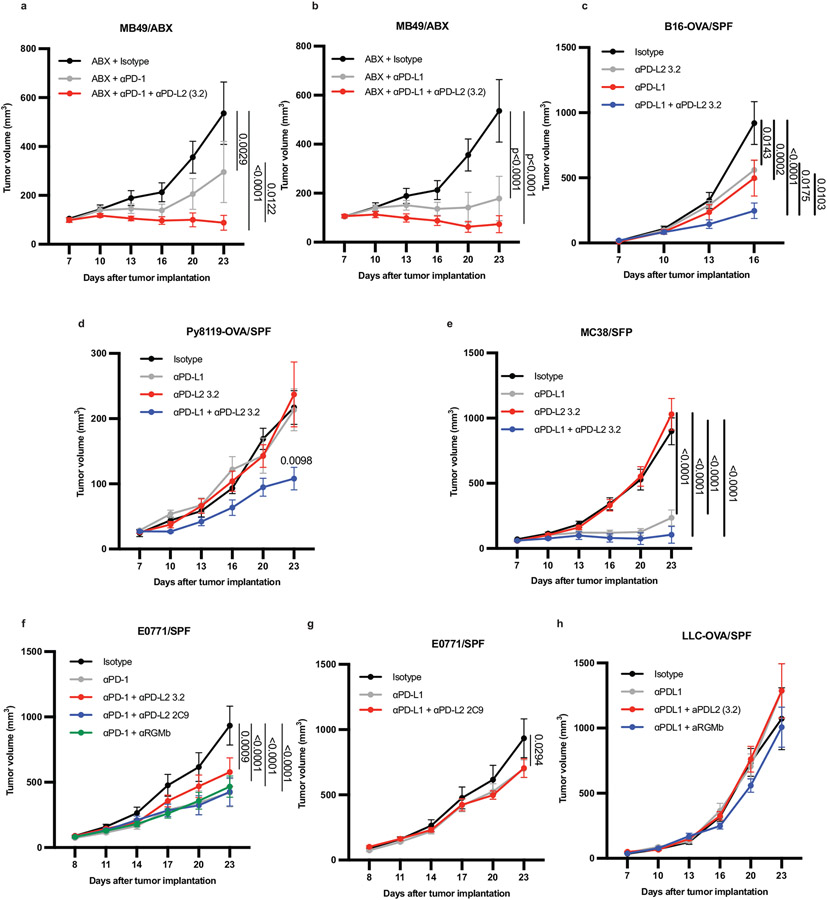
To directly investigate the contribution of upregulated PD-L2 expression to impaired anti-tumor responses in GF and ABX mice that do not respond to PD-L1 or PD-1 alone, we treated GF and ABX mice with a combination of anti-PD-L2 and anti-PD-L1 mAbs. Combined anti-PD-L1 and anti-PD-L2 (3.2 clone) treatment reduced MC38 tumors in GF and ABX mice, whereas treatment with either antibody alone failed to inhibit tumor growth (Fig. 2a, b). Combined anti-PD-L2 with anti-PD-1 or anti-PD-L1 also reduced growth in MB49 bladder tumors in ABX mice (Extended Data Fig. 3a,b). Moreover, combined anti-PD-L2 and anti-PD-L1 treatment reduced growth of B16 melanoma tumors expressing ovalbumin (B16-OVA) and Py8119 mammary tumors expressing ovalbumin (Py8119-OVA) in SPF mice, significantly better than treatment with either antibody alone (Extended Data Fig. 3c,d), suggesting that this combination therapy can improve responses in the context of an endogenous microbiota. Anti-PD-L1 + anti-PD-L2 combination therapy promoted an anti-tumor response in SPF mice with MC38 tumors, but this combination was not significantly better than anti-PD-L1 alone, which already promoted a robust response (Extended Data Fig 3e). Anti-PD-L2 treatment did not enhance the efficacy of anti-PD-L1 or PD-1 in E0771 breast cancer tumors in SPF mice or of anti-PD-L1 in Lewis Lung Carcinoma expressing ovalbumin (LLC-OVA) (Extended Data Fig. 3f-h). Therefore, combined blockade of anti-PD-L2 and anti-PD-1 or PD-L1 may be beneficial for treating multiple, but not all, cancers.
Figure 2. Anti-PD-L2 antibody blockade or colonization with C. cateniformis in combination with PD-L1 blockade promotes anti-tumor responses in non-responder GF or ABX mice.
MC38 tumor growth in (a) GF, and (b) ABX mice. (a-b) n = 4 mice for isotype, and n = 5 mice for anti-PD-L1, anti-PD-L2, and anti-PD-L1 + anti-PD-L2, data are representative of 3 experiments. (c-e) MC38 tumor growth in GF mice colonized with stool from melanoma patients treated with anti-PD-1 therapy (c) complete responder (CR) n = 6 mice for isotype, anti-PD-L1, and anti-PD-L1 + anti-PD-L2 groups, n = 5 mice for anti-PD-L2 group. (d) Nonresponder (NR1), n = 5 mice for isotype, n = 6 mice for anti-PD-L1 and anti-PD-L1 + anti-PD-L2, n = 3 for anti-PD-L2. (e) Non-responder (NR2), n = 5 mice for isotype, anti-PD-L1, anti-PD-L2 groups, n = 6 for anti-PD-L1 + anti-PD-L2 groups. (a-e) Significance measured by two-way ANOVA and Tukey’s multiple comparisons test and significant p values for day 23 are shown, error bars show mean and s.e.m. (f) MC38 tumor growth in GF mice colonized with C. cateniformis, n = 10 mice per group, significance determined by two-way ANOVA with Sidak’s multiple comparisons test and significant p values indicated on graph, error bars represent s.e.m. Percent of PD-L2 expression on indicated cells from GF mice or C. cateniformis colonized mice at day10 pi in, (g) dLN, n = 9 mice for GF and n = 10 mice for C. cateniformis (h) MLN, n = 10 mice per group. (g-h), Significance determined by unpaired, two-tailed, Mann-Whitney test and indicated on graph, error bars show mean and s.d.
Combined therapy with patient microbiota
To test the translational potential of anti-PD-L1 + anti-PD-L2 combination therapy in humans, we colonized groups of GF mice (in a blinded fashion) with feces either from one melanoma patient who had a complete response to anti-PD-1 therapy (CR) or from two melanoma patients who did not respond to anti-PD-1 therapy (non-responders, NR1 and NR2, Extended Data Fig. 4a). 16S rRNA gene sequencing of stool samples from mice colonized with patient stool for one week before tumor implantation revealed significant differences in each cohort of mice colonized with stool from a different patient (Extended Data Fig. 4a-h).
Consistent with the clinical responses of the patients to anti-PD-1, mice colonized with CR feces had significantly smaller MC38 tumors when treated with anti-PD-L1 monotherapy than mice colonized with anti-PD-L1 non-responder fecal samples from patients NR1 or NR2 (Extended Data Fig 4i) (14). Given that the response to anti-PD-1 is already good, combination therapy did not significantly increase the anti-tumor response compared to monotherapy in mice colonized with CR feces. Strikingly, combined anti-PD-L1 + anti-PD-L2 therapy significantly improved the anti-tumor response when compared to monotherapy in mice colonized with either of the NR feces and significantly increased overall survival (Fig. 2c-e, Extended Data Fig. 4j).
A gut commensal can suppress PD-L2
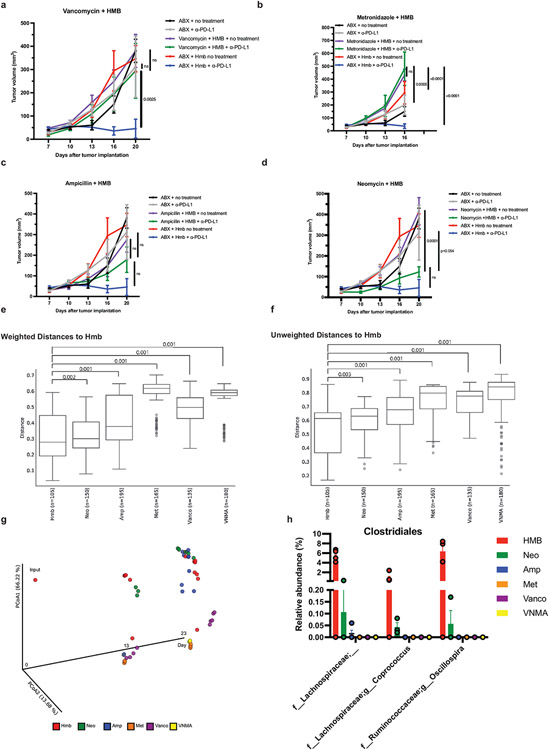
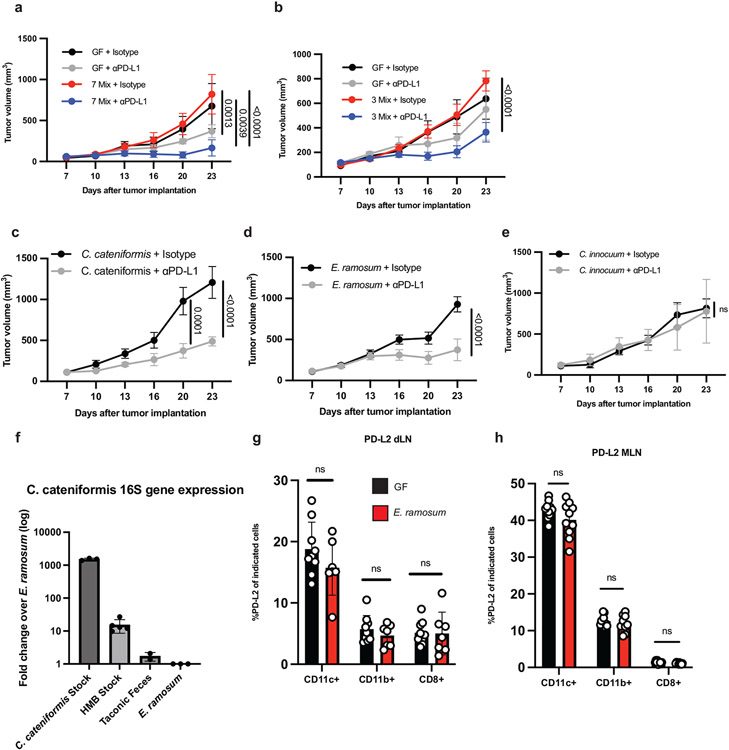
Next, we tested whether specific bacteria in HMB could induce PD-L2 downregulation on dendritic cells (DC) in the dLN and enhance anti-PD-L1-mediated anti-tumor responses. To determine which bacteria from HMB are associated with anti-PD-L1-mediated anti-tumor responses, we gavaged ABX mice with HMB at day 7 pi and maintained them on a single antibiotic for the duration of the experiment to select for different populations of gut bacteria. Vancomycin, which mainly targets Gram-positive bacteria, and metronidazole, which mainly targets anaerobic bacteria, were individually sufficient to abrogate anti-PD-L1 anti-tumor responses. Ampicillin, which can target some Gram-positive and some Gram-negative bacteria, dampened anti-PD-L1 anti-tumor responses, but tumor volumes did not statistically differ from HMB + anti-PD-L1 mice. Neomycin, which mainly targets aerobic and facultative Gramnegative bacteria, did not significantly change anti-PD-L1 anti-tumor responses compared to HMB + anti-PD-L1 (Extended Data Fig. 5a-d). These data suggest that Gram-positive anaerobes in HMB are disproportionately responsible for promoting anti-PD-L1 anti-tumor responses. 16S rRNA sequencing of stool samples at days 13 and 23 p.i. revealed significant differences in microbes between groups treated with the 4 antibiotics (Extended Data Fig. 5e-g). By comparing anti-PD-L1 responder (HMB, Neomycin) versus non-responder (VNMA, ampicillin, vancomycin, metronidazole) stool, we identified three genera from the Order Clostridiales (Gram-positive anaerobes, Extended Data Fig. 5h) and two Gram-negative bacteria (f_Porphyromonadaceae;g_Parabacteroides and f_Rikenellaceae;g_) associated with response to anti-PD-L1. Although 16S rRNA sequencing revealed that multiple genera from HMB have the potential to promote anti-tumor immunity, because our antibiotic data suggested Gram-positive anaerobes from HMB were associated with an anti-tumor response, we focused on identifying Gram-positive anaerobes from our HMB stock, potentially from the Order Clostridiales, that could promote an anti-PD-L1-mediated anti-tumor response (15, 16). Using a combination of antibiotic selection, selective media and sequencing, we identified 2 species (Coprobacillus cateniformis and Erysipelatoclostridium ramosum) that when monocolonized into mice, were sufficient to enhance anti-PD-L1-mediated anti-tumor responses (Fig. 2f, Extended Data Fig. 6a-e). Bulk 16S rRNA gene sequencing of HMB did not provide enough sequence specificity to identify C. cateniformis. However, when primers specific to C. cateniformis were used(17), C. cateniformis could be detected in HMB stock, but not in Taconic stool (Extended Data Fig. 6f). The lack of strain-specific resolution for C. cateniformis with 16S rRNA gene sequencing of stool could explain why few studies identify C. cateniformis from bulk 16S rRNA gene sequencing. Similar to other studies(17, 18), it was only when individual colonies were isolated and the full-length 16S gene was sequenced, C. cateniformis was identified. Therefore, isolation of individual strains and full-length sequencing of individual colonies may be needed to fully understand which bacterial strains may be best for effective live microorganism-enhanced immunotherapy.
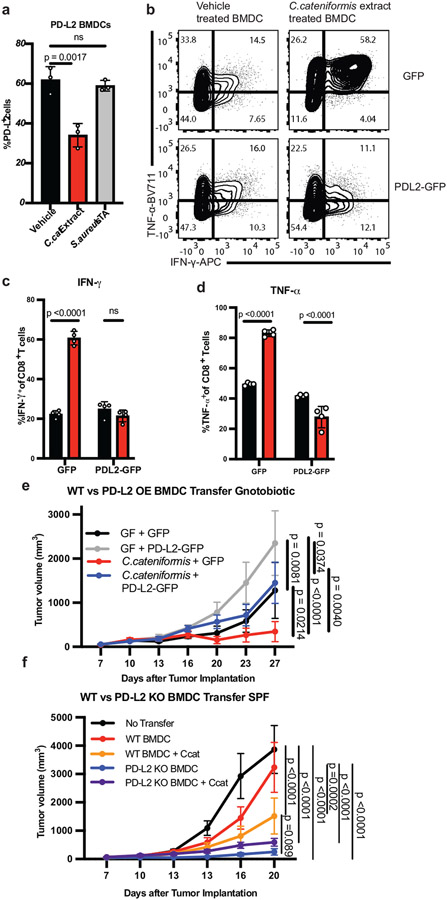
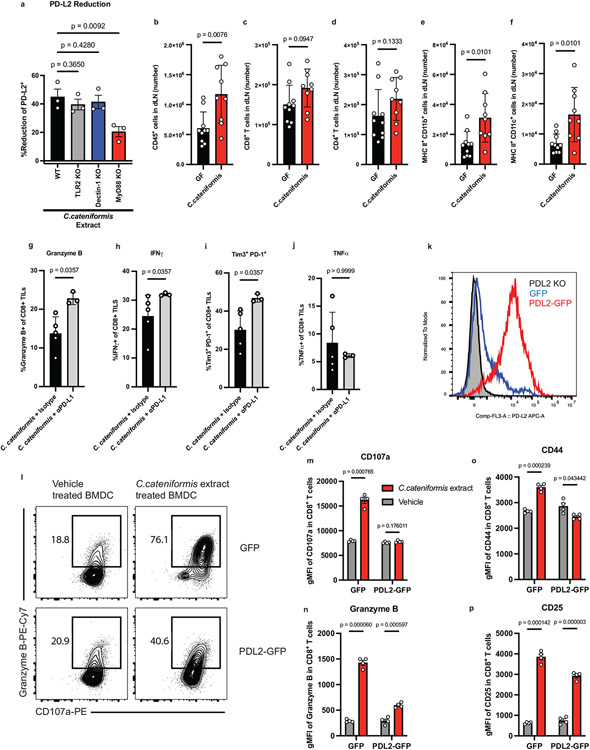
Colonization of GF mice with C. cateniformis, but not E. ramosum significantly reduced PD-L2 expression in CD11b+ and CD11c+ cells in the tumor dLNs and reduced PD-L2 expression in CD11c+ cells in the MLNs (Fig. 2g,h, Extended Data Fig. 6g-h). Additionally, treatment of bone marrow-derived dendritic cells (BMDCs) with soluble surface extracts of C. cateniformis downregulated PD-L2 expression (Fig. 3a) and this suppression was dependent on MYD88 (Extended Data Fig 7a). These data demonstrate that C. cateniformis can downregulate PD-L2 expression on DCs in vivo and in vitro.
Figure 3. C. cateniformis promotes anti-tumor immunity via PD-L2 downregulation.
(a) % PD-L2 expressing BMDCs treated with vehicle, 10 μg/ml surface extracts from C. cateniformis (C. cateniformis extract), or 10 μg/ml lipoteichoic acid (LTA) from Staphylococcus aureus, as a control for Gram-positive surface molecules, n = 3 wells per group, representative experiment of 2 experiments, significance measured by one-way ANOVA with Tukey’s multiple comparisons test, error bars show mean and s.d. (b-d) BMDCs transduced with GFP lentivirus (GFP) or lentivirus expressing GFP and PD-L2 (PD-L2-GFP) were treated with C. cateniformis extract 24 hours before co-culture with CD8+ T cells. Expression measured by flow cytometry. (b) Example of flow cytometry plots of IFNγ and TNFα expression by CD8+ T cells. Quantification of % of (c) IFNγ+ and (d) TNFα+ CD8+ T cells displayed (b). (c-d) Significance measured by two-way ANOVA with Sidak’s multiple comparisons test, significant p values shown, n = 4 wells per group, error bars show mean and s.d. (e) B16-OVA tumor growth in GF and C. cateniformis monocolonized mice injected with GFP or GFP-PD-L2 (PD-L2 overexpression) BMDCs at tumor site 3 days after tumor implantation. N = 9 mice for GF + GFP and C. cateniformis + GFP-PD-L2, n = 8 mice for GF + GFP-PD-L2, n = 10 mice for C. cateniformis + GFP (f) B16-OVA tumor growth in SPF mice injected at tumor site with WT or PD-L2 KO BMDCs treated with or without C. cateniformis extract. n=10 mice per group. (e-f) Significance measured by Tukey’s multiple comparisons test, significant p values indicated, error bars show mean and s.e.m.
As observed with ABX/HMB compared to ABX mice, C. cateniformis monocolonized mice versus GF mice also had increased frequencies of dLN CD45+ cells with significant increases specifically in MHCII+ CD11b+ and MHCII+ CD11c+ cells 13 days pi (Extended Data Fig 7b-f). At day 18 pi, C. cateniformis + anti-PD-L1 mice had increased percentages of tumor infiltrating CD8+ T cells expressing Granzyme B, IFNγ, and TIM-3+ PD-1+ (like ABX/HMB mice) but not TNFα (Extended Data Fig. 7g-j) indicating C. cateniformis recapitulates many of the beneficial immunologic effects of HMB but that other bacteria in HMB have additional immunologic effects.
PD-L2 suppression on dendritic cells
To directly investigate whether C. cateniformis impacts immune function by downregulating PD-L2 expression or whether PD-L2 expression is a marker for other immunologic activities, we transduced BMDCs with lentivirus expressing either GFP (GFP) or GFP and PD-L2 (PD-L2-GFP) (Extended Data Fig. 7k). GFP BMDCs pre-treated with soluble C. cateniformis surface extracts in vitro stimulated OT-I CD8+ T cells to express increased IFNγ, TNFα, CD107a, Granzyme B, CD44, and CD25 compared to vehicle control DC, indicating that C. cateniformis treatment of BMDCs promotes an increased proinflammatory CD8+ T cell response in vitro (Fig. 3b-d, Extended Data Fig. 7l-p). Conversely, pretreatment of PD-L2-GFP BMDCs with C.cateniformis extract did not have these effects. IFNγ, TNFα, CD107a, and CD44 expression were not increased, Granzyme B was modestly increased, and CD25 expression was similarly increased (Fig. 3b-d Extended Data Fig. 7l-p). These data show that many of the immunomodulatory effects of C. cateniformis can be negated by forced PD-L2 expression consistent with the idea that C. cateniformis downregulation of PD-L2 expression is the mechanism by which this microbe enhances anti-PD-L1 therapy.
When C. cateniformis was orally gavaged at the time anti-PD-L1 treatment was given, the resulting anti-tumor response was as robust as anti-PD-L1 + anti-PD-L2 treatment in ABX mice. The combination of C. cateniformis + anti-PD-L1 + anti-PD-L2 did not further enhance the anti-tumor response, suggesting that C. cateniformis and anti-PD-L2 have overlapping mechanisms of action (Extended Data Fig. 8a).
To determine if C.cateniformis’ anti-tumor properties depend on suppression of PD-L2 expression on DCs, we transferred GFP or PD-L2 GFP BMDCs into B16-OVA tumors in GF and C. cateniformis monocolonized mice three days after tumor implantation. GFP BMDCs promoted significantly more robust anti-tumor response in C. cateniformis monocolonized mice compared to GF mice, supporting our hypothesis that C. cateniformis enhances DC mediated anti-tumor immunity (Fig. 3e). Strikingly, transfer of PD-L2 overexpressing DCs resulted in significantly larger tumor volumes compared to transfer of GFP DCs in C. cateniformis, GF mice (Fig. 3e), and SPF mice (Extended Data Fig. 8b) indicating that high levels of PD-L2 expression on DCs inhibit anti-tumor immunity. C. cateniformis’ anti-tumor properties are dependent on PD-L2 downregulation, as C. cateniformis cannot promote robust anti-tumor immunity in the presence of high PD-L2. Thus, our in vitro and in vivo data demonstrate that C. cateniformis’ anti-tumor effects depend on downregulation of PD-L2 expression on DCs.
To directly show that C. cateniformis promotes anti-tumor immunity via downregulation of PD-L2 on DCs, we implanted WT or PD-L2 KO DCs with or without C. cateniformis treatment into tumor sites in SPF mice with B16-OVA tumors. Because our data showed that transfer of GFP BMDCs promoted robust anti-tumor immunity in SPF mice (Extended Data Fig. 8b) we sought to find a BMDC transfer model in which WT BMDC transfer was less efficacious and thus could be enhanced by PD-L2 suppression. We hypothesized that less mature BMDCs that had been cultured for 6-7 days, and expressed lower levels of MHCII compared to more mature BMDCs that had been cultured for 10 days (Extended Data Fig. 8c-e) and used in Figure 3e, would be less immunostimulatory. Though transfer of day 6 cultured WT BMDCs slightly delayed B16-OVA tumor growth, by day 20 there was no significant difference between no transfer and WT BMDC transferred tumors (Fig 3f). Injection of WT BMDC tumors pre-treated with C. cateniformis, however, was sufficient to promote a robust anti-tumor response, indicating C. cateniformis’ immunomodulatory impact on DCs is sufficient to promote anti-tumor immunity. Importantly, transfer of PD-L2 KO DCs promoted a robust anti-tumor response that could not be further enhanced by pretreatment with C.cateniformis (Fig. 3f). These data indicate that inhibition of PD-L2 expression on DCs plays a key role in promoting anti-tumor immunity and C. cateniformis treatment cannot enhance anti-tumor immunity independent of PD-L2 downregulation.
Targeting PD-L2/RGMb interactions
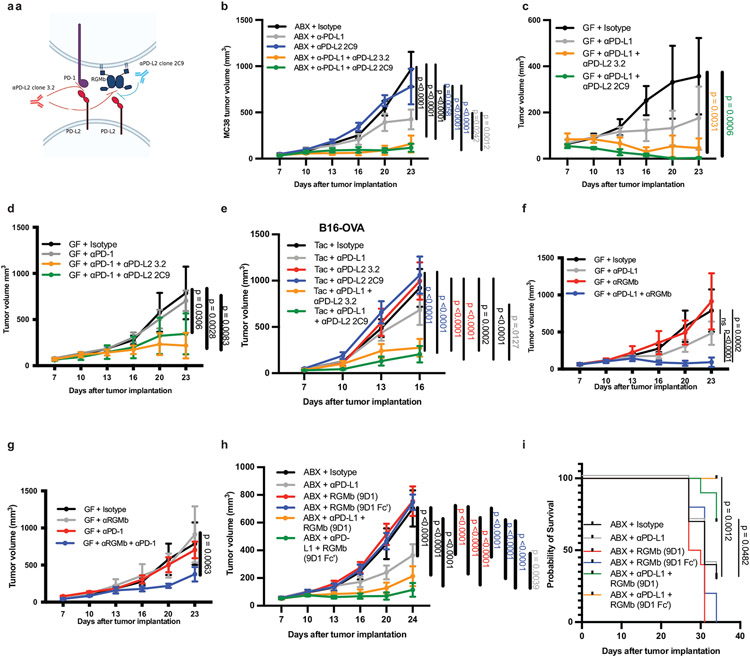
Based on our observation that combined blockade of PD-1 and PD-L2 gave better antitumor responses than PD-1 blockade alone (Extended Data Fig. 3a), we hypothesized the PD-L2 effect is mediated by a receptor other than PD-1. Consequently, we investigated whether the interaction of PD-L2 with RGMb rather than PD-1 led to microbiome-dependent resistance to anti-PD-L1 or anti-PD-1 cancer therapy. We tested whether blocking the PD-L2/PD-1 interaction or the PD-L2/RGMb interaction was necessary to promote an anti-tumor response to PD-L1 blockade by comparing the effects of two functionally distinct PD-L2 blocking antibodies in combination with anti-PD-L1 in GF or ABX mice. The 2C9 anti-PD-L2 clone selectively blocks the RGMb/PD-L2 interaction, whereas the 3.2 anti-PD-L2 clone blocks both RGMb/PD-L2 and PD-1/PD-L2 interactions (Fig. 4a)(19, 20). While monotherapy with anti-PD-L1 or either anti-PD-L2 clone did not suppress MC38 tumor growth in ABX or GF mice, administration of either the 3.2 clone or the 2C9 clone together with anti-PD-L1(Fig. 4b,c) or anti-PD-1(Fig. 4d) elicited a potent anti-tumor response in ABX and GF mice, showing that blockade of PD-L2 interaction with RGMb was sufficient. Similarly, anti-PD-L1 plus either anti-PD-L2 antibody reduced growth of the less immunogenic B16 OVA tumor (Fig. 4e). β2m−/− mice lacking CD8+ T cells and implanted with MC38 tumor cells failed to respond to combined treatment with anti-PD-L1 and the 2C9 anti-PD-L2 clone, suggesting that CD8+ T cells are needed for the anti-tumor response to this combination treatment (Extended Data Fig. 8f).
Figure 4. PD-L2/RGMb blockade is sufficient to promote anti-tumor responses in mice that do not respond to anti-PD-1 or anti-PD-L1 alone.
(a) Diagram of PD-L2 binding to PD-1 or RGMb, and blocking specificity of anti-PD-L2 antibodies. Anti-PD-L2 clone 3.2 blocks both PD-L2/PD-1 and PD-L2/RGMb interactions. Anti-PD-L2 clone 2C9 blocks only the PD-L2/RGMb interaction(19). MC38 tumor growth in (b) ABX mice, n = 5 mice per group for all except n= 4 mice for anti-PD-L2 2C9 group and (c) GF mice, n = 5 mice per group. Data are representative of 2-3 different experiments. (d) MC38 tumor growth in GF mice given isotype (n= 4 mice), anti-PD-1 (n= 5 mice), or anti-PD-1 combined with either anti-PD-L2 clone (n= 5 mice per group). (e) B16-OVA tumor growth in Taconic SPF mice given isotype (n = 10 mice), anti-PD-L1 (n = 10 mice), anti-PD-L2 3.2 (n= 10 mice), anti-PD-L2 2C9 (n = 10 mice), or anti-PD-L1 with either anti-PD-L2 clone (n = 9 mice each), representative experiment of 3 experiments. Tumor growth curves in GF mice with MC38 tumor cells and (f) treated with isotype (n = 4 mice), anti-RGMb (n = 4 mice), anti-PD-L1 (n = 5 mice) or anti-PD-L1 + anti-RGMb (n = 5 mice), representative experiment of 2 experiments or (g) treated with isotype (n = 4 mice), anti-RGMb (n = 4 mice), anti-PD-1 (n = 5 mice) or anti-PD-1 + aRGMb (n = 5 mice), representative of 2 different experiments. (h) MC38 tumor growth in ABX mice with MC38 tumors and treated with isotype, anti-RGMb (mIgG2a), anti-RGMb (mIgG2a-LALA-PG, termed Fc’), anti-PD-L1, anti-PD-L1 + anti-RGMb (mIgG2a), or anti-PD-L1 + anti-RGMb (Fc’), n = 10 mice per group and (i) corresponding survival curve, significance between anti-PD-L1 versus anti-PD-L1 + either anti-RGMb antibody determined by Log-rank (Mantel-Cox) test. (b-h) Significance determined by two-way ANOVA with Tukey’s multiple comparisons test, error bars show mean and s.e.m.
Next, we investigated the effects of combined administration of an anti-RGMb mAb and anti-PD-L1 or anti-PD-1 in the MC38 tumor model in GF mice. Anti-RGMb (9D1 clone (19)) attenuated tumor growth in GF mice when given with either anti-PD-L1 or anti-PD-1 (Fig. 4f, g), in contrast to the poor response to anti-RGMb, anti-PD-L1 or anti-PD-1 alone. Anti-PD-L1 combined with either anti-PD-L2 antibody or anti-RGMb similarly promoted a robust anti-tumor response in GF mice with MC38 tumors (Extended Data Fig. 8g) as well as in Taconic SPF mice with B16-OVA tumors (Extended Data Fig. 8h). To determine whether the anti-RGMb antibody exerts its anti-tumor effects by blocking pathway activity or by depleting cells, we compared recombinant anti-RGMb 9D1 antibody with a wild-type (mouse IgG2a) or effectorless Fc (mouse IgG2a-LALA-PG) in which the Fc portion is not able to bind to the Fc receptor, and therefore cannot function to deplete RGMb positive cells. Both RGMb antibodies attenuated tumor growth in ABX + anti-PD-L1 treated mice, and significantly increased survival compared to anti-PD-L1 therapy alone, indicating that RGMb blockade is sufficient and depletion of RGMb positive cells is not part of the mechanism (Fig 4h,i). Given that anti-PD-1 therapy benefits from combination with anti-PD-L2 and that PD-1/PD-L1 blockade combined with anti-PD-L2 2C9 clone or anti-RGMb promotes as robust an anti-tumor effect as when combined with the anti-PD-L2 3.2 clone, these findings demonstrate that the RGMb/PD-L2 pathway plays an important role in limiting the response to anti-PD-1 or anti-PD-L1 immunotherapy in a GF or colonized host.
Microbiota impacts RGMb expression
To further understand how the gut microbiota regulate the RGMb/PD-L2 interaction, we compared RGMb expression in MC38 tumor-bearing SPF and GF mice. RGMb is expressed in the nervous system, epithelial cells, and immune system, where it is most highly expressed by macrophages(19, 21, 22). Surprisingly, the transcript levels of RGMb in CD8+ tumor infiltrating T cells were 6.1-fold higher in GF mice compared to SPF mice (Fig. 5a). Similarly, RGMb protein expression, measured by a monoclonal antibody (9D3 clone) or a polyclonal antibody (Fig. 5b, Extended Data Fig. 9a-d), was significantly higher in CD8+ tumor infiltrating T cells from GF mice compared to SPF mice. Differences in RGMb expression in other immune cell subsets were not significant (Fig. 5b), suggesting that RGMb expression on T cells plays an important role in CD8+ T cell mediated anti-tumor immunity. Analysis of patient stool-colonized mice at day 29 pi revealed that NR2 tumors had significantly higher expression of RGMb on CD8+ T cells, but not on CD11b+ or CD11c+ DCs, suggesting that certain non-responder patients might have a microbiota resulting in increased RGMb expression on T cells (Extended Data Fig. 9e-g). We also found significantly fewer RGMb-expressing intratumoral CD8+ T cells in SPF + anti-PD-L1 or isotype treated mice compared to GF + anti-PD-L1 or isotype treated mice (Fig 5c). MC38 tumor cells expressing GFP showed high expression of PD-L1 as reported (23) but undetectable RGMb or PD-L2 expression when implanted in SPF or GF mice, suggesting RGMb or PD-L2 expression on immune cells and not the tumor is responsible for influencing anti-tumor immunity (Extended Data Fig. 9h). Taken together, these findings demonstrate that blockade of PD-L2/RGMb interactions can overcome microbiome-dependent resistance to anti-PD-1 or anti-PD-L1.
Figure 5. RGMb on T cells regulates anti-tumor immunity in GF mice.
(a) Levels of RGMb RNA expression in tumor-infiltrating CD4+ T cells (n = 3 tumors for GF and n = 4 for SPF), CD8+ T cells (n = 4 GF, n = 5 SPF), CD11c+MHC class II+ cells (n = 5 for GF and SPF) and CD11b+ cells (n = 4 for GF, and n= 5) sorted from MC38 tumors in GF and SPF mice on day 11 after implantation, and quantified by qPCR. (b) Surface expression of RGMb protein on tumor-infiltrating leukocytes isolated from MC38 tumors on day 13 after implantation, measured by flow cytometry using monoclonal antibody (clone 9D3) against RGMb. Representative histograms of expression of RGMb on CD8+ T cells (upper left) and CD11c+MHC class II+ cells (lower left) in GF (black) and SPF (red) mice are shown and frequencies of RGMb+ cells within indicated cell populations quantified (right). N = 5 mice per group. (a-b) Significance measured by unpaired, two-tailed, Mann-Whitney test. Data are representative from 2 different experiments, error bars show mean and s.d. (c) RGMb expression on CD8+ T cells in MC38 tumors day 11 pi from GF and SPF mice treated with isotype and anti-PD-L1, n = 5 mice for GF + Isotype and SPF groups, N= 4 mice for GF + anti-PD-L1, significance determined by one-way ANOVA and Sidak’s multiple comparisons test, significant p values indicated on graph, error bars show mean and s.d. (d-g) MC38 tumors were harvested at day 18 pi from GF mice treated with indicated antibodies and total numbers of (d) CD8+ T cells, (e) CD4+ T cells and (f) Treg cells in tumors were measured. (g) The ratio of CD8+ to Treg was calculated. Significance measured by non-parametric one-way ANOVA and Dunn’s multiple comparisons test, n = 5 mice per group, error bars show mean and s.d. MC38 tumor growth in (h) rgmbf/f (n = 8 mice) vs rgmbf/f CD4-Cre (n = 10 mice) or (i) rgmbf/f (n = 7 mice) vs rgmbf/f LysM-Cre (n = 12 mice) mice treated with ABX and given anti-PD-L1. Significance at day 30 is shown and was measured by two-way ANOVA and Sidak’s multiple comparisons test, error bars show mean and s.e.m. Data are representative from 2 different experiments. WT BMDCs were co-cultured with WT or RGMb KO T-cells and CD8+ T-cells were analyzed by flow cytometry for (j) Mean Fluorescence Intensity (MFI) of CD44 and (k) frequency of cells expressing IFNγ, TNFα, and IL-2. Significance measured by unpaired, two-tailed, Mann-Whitney test, error bars show mean and s.d.
RGMb functions on T cells
To determine how anti-RGMb treatment affected immune responses in GF mice, we analyzed immune cell subsets at day 18 after MC38 tumor implantation. Combined anti-PD-L1 and anti-RGMb therapy in GF mice led to a significant increase in the numbers of intratumoral CD8+ and CD4+ T cells, compared to isotype controls (Fig. 5d,e). Anti-PD-L1 treatment alone did not significantly change the numbers of intratumoral CD8+ and CD4+ T cells in GF mice consistent with other findings (15). The numbers of intratumoral Treg cells were similar in GF mice given anti-PD-L1, anti-PD-L1 plus anti-RGMb or isotype control antibody, consequently, the CD8+ to Treg cell ratio was increased in the tumors of GF mice treated with anti-PD-L1 and anti-RGMb (Fig. 5f,g). Even as early as day 11 pi, anti-RGMb combined with anti-PD-L1 treatment significantly increased Tumor Necrosis Factor α (TNF-α) production by CD4+ T cells (Extended Data Fig. 9i), suggesting a potential role for RGMb in regulating pro-inflammatory cytokine expression by T cells. PD-1 expression on CD8+ tumor infiltrating T cells was increased in anti-RGMb treated groups without upregulation of the co-inhibitory receptors TIM-3 and LAG-3 (Extended Data Fig. 9j-l). These data suggest that combined anti-PD-L1 and anti-RGMb treatment acts on the T cells to enhance anti-tumor immunity.
To directly investigate the function of RGMb on specific cell types in regulating antitumor immunity, we developed RGMb conditional knockout mice (RGMbfl/fl) and crossed them with CD4-Cre to delete RGMb in T cells or with LysM-Cre mice to delete RGMb in macrophages and granulocytes (Extended Data Fig. 9m-o). Deletion of RGMb in T cells, but not macrophages and granulocytes, improved anti-tumor responses in ABX mice given anti-PD-L1 (Fig. 5h, i). Furthermore, in co-cultures of WT BMDCs with WT or RGMb KO CD8+ T cells, the RGMb KO T-cells displayed a more activated phenotype with increased mean fluorescence intensity of CD44 (Fig 5j), T-bet, and a slight but not significant increase of CD107a (Extended Data Fig. 9p,q) and significantly increased percentages of cells expressing IFNγ, IL-2, and TNFα (Fig 5k). Both sets of T-cells had high levels of proliferation and Granzyme B MFI, but were not significantly different (Extended Data Fig. 9r-s). Together, these findings reveal a novel, inhibitory function for RGMb on T cells and provide a deeper mechanistic understanding of the role of RGMb in anti-tumor immunity.
Discussion
Current understanding of how the gut microbiota can enhance cancer immunotherapy is still at an early stage. FMTs have been shown to increase the efficacy of anti-PD-1 therapy in patients by 30%(7, 8) but probiotics did not enhance patient survival. In fact, an inverse correlation between use of probiotics and anti-PD-L1 response has been reported(24). There are multiple ways gut bacteria can promote anti-tumor immunity. For example, different bacteria can secrete inosine (5), a hydrolase that generates muropeptides (6), or STING agonists that enhance the response to immunotherapy(14). Further work is needed to define mechanisms by which the gut microbiota enables the adaptive immune system to overcome the immunosuppressive tumor microenvironment.
By investigating immune responses in microbiome-dependent mouse cancer models, we discovered a new mechanism by which the microbiota can promote response to anti-PD-1 and anti-PD-L1: by downregulating PD-L2 and RGMb expression on immune cells in responder mice. Upregulated expression of PD-L2 and RGMb in non-responder GF or ABX mice is functionally significant because blockade of the PD-L2/RGMb pathway (using either anti-PD-L2 or anti-RGMb antibodies) combined with anti-PD-1 or anti-PD-L1 promotes anti-tumor responses in GF or ABX mice that do not respond to anti-PD-1 or anti-PD-L1 alone. The requirement for RGMb or PD-L2 blockade to potentiate the effect of PD-1 blockade in GF mice suggests that PD-L2 inhibitory effect may be largely via RGMb/PD-L2 interactions and not PD-1/PD-L2 interactions. Combined anti-PD-L1 and anti-PD-L2 therapy enhanced tumor clearance and survival compared to monotherapy in mice colonized with stool from NR patients, suggesting that this combination therapy may have translational relevance, particularly in patients with a non-responder microbiota.
Monocolonization with C. cateniformis is sufficient to promote response to anti-PD-L1 therapy and suppress PD-L2 expression on DCs in MLNs and tumor dLNs. Our data suggest this signal could originate in the MLNs and travel to tumor dLNs. Notably, incubation of BMDCs with C. cateniformis surface extracts suppressed PD-L2 expression and injection of C. cateniformis-treated WT BMDCs or PD-L2 KO BMDCs promoted a robust anti-tumor response compared to controls. Conversely, transfer of PD-L2 overexpressing BMDCs resulted in significantly larger tumors than WT BMDCs transfer in C. cateniformis mice. Together, these studies demonstrate that PD-L2 expression on DCs plays a key role in limiting anti-tumor immunity and that C.cateniformis promotes anti-tumor immunity by downregulating PD-L2 expression on DCs. Our finding that PD-L2 KO DCs potently promote an anti-tumor response suggests a novel strategy for cellular therapy. As previous studies have shown that bacteria from different phyla can have overlapping immunoregulatory consequences and strains of the same species can produce different immunoregulatory outcomes, C. cateniformis is likely not the only strain in human microbiota that can mediate this effect (25). We isolated a second organism that also promotes anti-tumor immunity to anti-PD-L1 but does not significantly suppress PD-L2. Therefore, while PD-L2 suppression by some bacteria can promote anti-tumor immunity, this is not the only mechanism by which gut microbes can impact immune function during immunotherapy.
RGMb is expressed by several cell types including neuronal cells, epithelial cells, and immune cells(21, 22, 26). In the immune system, RGMb has been implicated in mucosal tolerance and asthma(19, 27). RGMb plays an important role in neuronal development, kidney function, and most recently, global conditional RGMb knockout has been shown to alter the gut microbiome and increase susceptibility to Dextran Sulfate Sodium (DSS)-induced colitis(28). Thus, the PD-L2/RGMb axis may be a key mediator of tolerogenic interactions between the microbiota and the host immune system. Our work using RGMb conditional knockout mice lacking RGMb only on T cells reveals a novel and unanticipated role for RGMb on T cells in anti-tumor immunity, as previous studies have focused on RGMb function on DCs and myeloid cells(19, 29).
In conclusion, by investigating how the gut microbiome modulates anti-tumor immunity, we identified attenuation of PD-L2 and RGMB expression as an immunoregulatory mechanism that informs a possible therapeutic strategy to overcome resistance to PD-1 blockade. We propose a two-step model in which certain gut bacteria suppress PD-L2 expression on DCs in lymph nodes and PD-L2lo DCs play a critical role in activating CD8+ T cells to promote antitumor immunity, whereas PD-L2high DCs interact with RGMb on CD8+ T cells to inhibit CD8+ T cell responses. RGMb expression is maintained at a low level in the tumor microenvironment of a host with a favorable gut microbiota, whereas unfavorable gut microbiota may induce upregulation on CD8+ TILs. Blockade of PD-L2/RGMb interactions combined with anti-PD-1 or anti-PD-L1 can overcome microbiome-dependent resistance to monotherapies with PD-1 pathway inhibitors and provides a novel strategy for treating patients who do not respond to PD-1 cancer immunotherapy. Our study provides an innovative approach to identify new cancer immunotherapy targets using the gut microbiome as a discovery platform.
Materials and Methods
Mice
6-week old C57BL/6 female mice were purchased from Taconic Biosciences. βm-deficient mice were obtained from The Jackson Laboratory. Germ-free mice were maintained in the germ-free facility at Harvard Medical School. All experimental mice were housed in specific pathogen-free conditions or germ-free isolators. RGMb conditional knockout mice were generated by homologous integration of a construct, which contains LoxP sites flanking exon 2 of the rgmb gene and Neomycin cassette (Extended Data Fig. 9m). Neo flanked by Frt sites was removed by breeding mice with germ-line transmission of the homologously integrated construct with a Flp deleter strain. RGMb conditional knockout mice were further crossed with LysM-Cre (Jackson Laboratory, #004781) or CD4-Cre mice (Jackson Laboratory, #022071), to generate Macrophage-specific or T-cell specific conditional knockout mice, respectively. All mice were used in accordance with animal care guidelines from Harvard Medical School Standing Committee on Animals and the National Institutes of Health. Mice were housed by the facilities operated by Harvard Center for Comparative Medicine under the following housing conditions: 12:12 hr light-dark cycle, main room temperature 68-79°F and humidity 40-65%. All mice were used in accordance with animal care guidelines from Harvard Medical School Standing Committee on Animals and the National Institutes of Health.
Tumor models and antibody treatment
Mice were anesthetized with 2.5% 2,2,2,-Tribromoethanol (Avertin, Sigma Aldrich Cat# T48402-25G) diluted in DPBS and injected subcutaneously in the abdominal flank with 2.5 x 105 MC38 tumor cells, B16-OVA tumor cells, LLC-OVA, MB49 or Py8119-OVA cells. 2.5 x 105 E0771 cells were injected into the mammary fat pad. Whenever GF or ABX mice were directly compared to SPF, HMB, or ABX/HMB, the same tumor cell harvest was implanted into all groups of mice on the same day. Mice were given the following antibodies intraperitoneally: 100μg of anti-PD-L1 antibody (clone 10F.9G2), anti-PD-1 antibody (clone RMP1-14), anti-PD-L2 antibody (clone 3.2), anti-PD-L2 antibody (clone GF17.2C9), anti-RGMb antibody (clone 307.9D1), recombinant anti-RGMb antibody (clone 307.9D1 Fv with either wild-type mouse IgG2a Fc or L234A/L235A/P329G (LALA-PG) triple mutant Fc effector silent antibody (30), rat IgG2b isotype control (LTF-2, BioXCell Cat# BE0090) or rat IgG2a isotype control (2A3 BioXCell Cat#BE0089), individually or in combination as indicated, on days 7, 10, 13, 16 post tumor implantation. Tumor volume was determined by the volume formula for an ellipsoid: 0.5 x D x d2, where D is the longer diameter and d is the shorter diameter. Mice were humanely euthanized when tumors were greater than 30% ulcerated for SPF or greater than 50% ulcerated for gnotobiotic or reached the volume of 2000 mm3 or greater, in accordance with our IACUC protocol.
Antibiotic treatment and fecal transfer
Antibiotics were given via drinking water. For broad-spectrum antibiotic treatment (ABX), 0.5mg/mL of Vancomycin, 1mg/mL of Neomycin, 1mg/mL of Metronidazole and 1mg/mL of Ampicillin were added to drinking water containing Splenda (4g/L). Antibiotic water was refreshed every three days. To confirm antibiotic activity, fecal matter was resuspended in PBS (1 ml/pellet) and cultured on Trypticase soy agar (TSA) and Brucella blood agar plates in aerobic and anaerobic conditions, respectively. For human microbiota (HMB) or mouse microbiota (MMB) treatments, colon contents from 10 C57BL/6 HMB mice or MMB mice (11, 12) were squeezed into a pre-weighed 50 ml tube and resuspended in Brain Heart Infusion media at 100 mg/ml in the anaerobic chamber. Aliquots were kept at −80 °C. GF mice were orally gavaged with 200 μl of HMB or MMB stock one week before tumor implantation. For ABX/HMB mice, antibiotics were stopped at day 7 p.i. and mice were orally gavaged with 200 μl of HMB stock (11, 12). For individual antibiotic experiments, mice were treated with broad spectrum antibiotics until day 7 pi at which point they were orally gavaged with HMB and continued with only a single antibiotic in the drinking water until the end of the experiment.
Patient stool samples
For in vivo studies involving human samples, stool samples were obtained from melanoma patients treated with immune checkpoint blockade (ICB; anti-PD-1). Fresh frozen fecal samples were used, including one responder (CR, Sample ID 274(14)) and two non-responders (NR1, sample ID 376 (unpublished) and NR2 Sample ID 426(14))). Patients were selected based on their tumor progression and overall survival after surgery and subsequent treatment with PD-1 blockade (pembrolizumab or nivolumab). Response to treatment was assessed with radiographic imaging and according to Response Evaluation Criteria in Solid Tumors (RECIST 1.1)(31); patients with complete or partial response or stable disease ≥6 months were classified as responders and those with stable disease < 6 months or progressive disease were classified as non-responders. The mean patient age was 56.3 years (50-62), and mean BMI was 29.9 kg/m2 (20.4-42.31). The complete responder sample was taken 2 years post-treatment. Complete response was identified 3 years prior to fecal sample collection. The non-responder samples were taken during treatment, 3 months to 1 year after the start of ICB. Samples were collected under approved IRB protocols 2012-0846 and PA15-0232. Fresh stool samples were collected in stool collection cups, shipped overnight on ice to the Melanoma Clinical Database, Tissue Resource and Translational Pathology Core (MELCORE) at the University of Texas MD Anderson Cancer Center. Samples were then aliquoted and stored at −80 C.
FMT Tumor Studies
For FMT studies, stool samples were prepared in an anaerobic chamber. One gram of stool was diluted in 10 mL PBS + 10% Glycerol. Diluted stool samples were filtered using a 100 μm cell strainer, aliquoted, and stored at −80 °C. 200 μl of diluted patient stool was administered to mice via oral gavage 8, 6, and 4 days prior to tumor implantation.
16S sequencing of mouse stool
DNA from frozen stool samples was extracted using QIAamp Fast DNA Stool Mini Kit (Qiagen Cat#51604) for Extended Data Figure 11 or by Phenol Chloroform Extraction as in (32) for Extended Data Figure 9. Purified DNA was quantified by Qubit dsDNA HS Assay (Thermo Fisher Cat#Q32854) and normalized. The V4 region of the 16S rRNA gene was amplified with primers 515F and 806R as previously described(33), and ~390 bp amplicons were purified and quantified by Qubit dsDNA HS Assay and combined with equal mass to make a pooled library. The pooled library was multiplexed and sequenced (Illumina MiSeq, 251 nt x 2 pair-end reads with 12 nt index reads) at the Harvard Biopolymers Facility. Raw sequencing data were processed with QIIME2 pipelines(34). In brief, raw sequencing data were imported to QIIME2 and demultiplexed, then DADA2 were used for sequence, quality control, and feature table construction. The feature table was further used for beta diversity analysis, taxonomic analysis, and differential abundance testing using QIIME2. We used QIIME2_DADA2 to convert raw sequencing data into Amplicon Sequencing Variant ASV-level feature identification, and then performed differential abundance testing on all different taxonomy levels. We then focused on the genus level because of the resolution limitation of the 16S V4 region. We identified in total 5 genera significantly associated with response to anti-PD-L1 groups with an abundance ratio (R/NR) over 5. Beta group significance was determined by permutational analysis of variance (PERMANOVA). Identification of taxa associated with different groups was determined using Analysis of Composition of Microbiomes (ANCOM).
Isolation of Gram-positive strains from HMB
Isolation of Gram-positive strains from HMB was performed in an anaerobic chamber. Feces from HMB mice were collected and resuspended in PBS at 100 mg/ml. Serial dilutions of HMB stock were plated on Brain Heart Infusion plates made according to the manufacturer’s instructions (BD Cat# 241830) and stored in the anaerobic chamber for three days. Individual isolates were selected from these plates and cultured in Reinforced Clostridial Medium (BD Cat#218081) overnight in the anaerobic chamber. Stocks were made from overnight cultures, diluted so that they were 20% glycerol, and stored at −80 °C. Stocks were plated on BD BBL Brucella plates (BD Cat#8807311) in the anaerobic chamber to confirm viability and colonies were sequenced to identify strains. Individual colonies were resuspended in 50 μl of DMSO and heated at 95 °C for 15 minutes and quickly spun down to access bacterial DNA. For PCR of full length 16S bacteria sequence, 2 μl of bacterial DNA was mixed with 10 μl 5x GC Buffer (New England BioLabs Cat#B0519S) +1μ1 of 1 μM dNTPs + 2.5 μl each of 10 μM forward (27F AGA GTT TGA TCM TGG CTC AG) and reverse primers (1492R CGG TTA CCT TGT TAC GAC TT) + 1 μl PhusionDNA Polymerase (New England BioLabs Cat#M0530) + 31 μl water. PCR was performed according to manufacturer’s instructions. PCR reactions were purified using QIAquick PCR purification kit (Qiagen Cat#28104) and eluted with 30° C of water. 16S rDNA PCR product was cloned using ZeroBlunt TOPO PCR Cloning Kit (Thermo Fisher Scientific #K280020) according to manufacturer’s instructions and Sanger sequenced at the Harvard Biopolymers Facility using the (M13 Forward 5′-GTAAAACGACGGCCAG-3)or (M13 Reverse 5′-CAGGAAACAGCTATGAC-3′) primers. Individual sequences were identified using NCBI BlastN. Species were identified by having >99% identity to published sequences.
On the first dilution plate in which individual colonies could be observed, we isolated and sequenced 24 different colonies and identified 7 different species of bacteria, five Gram-positive anaerobes (Blautia hydrogenotrophica, Clostridium orbiscindens, Clostridium innocuum, Coprobacillus cateniformis and Ersyipelatoclostridium ramosum), one facultative Gram-positive (Longicatena caecimuris), and one Gram-negative anaerobe (Phocaeicola dorei). The combination of these 7 different species promoted an anti-tumor response to anti-PD-L1 therapy in GF mice (Extended Data Fig. 6a). From these 7, we next tested the combination of the 3 Gram-positive anaerobes that have been reported as human commensal bacteria (C. innocuum, C. cateniformis, E. ramosum), and found this combination was also sufficient to promote an antitumor response in GF mice (Extended Data Fig. 6b). We further monocolonized mice with each of these bacteria and found colonization with either C.cateniformis or E. ramosum was sufficient to promote anti-tumor immunity to anti-PD-L1(Fig 2f, Extended Data Fig. 6c,d), whereas colonization with C. innocuum did not (Extended Fig 6e).
Bacterial DNA isolation for qPCR
C.cateniformis, E. ramosum, and HMB stocks were pelleted, and pellets and Taconic stool samples were extracted by Phenol Chloroform Extraction as in (32). Reactions comprised of 50 ng of extracted DNA as template, 10 μl SSoAdvanced Universal SYBRGreen Super Mix (BioRad1725270), 10 μl PCR-grade water, and 0.25 μl of forward and reverse primers at 10μM and analyzed on a QuantStudio5. (Universal forward: 5’-CTCCTACGGGAGGCAGCAG-3’, Universal reverse: 5’-TTACCGCGG CTGCTGGCAC-3’ C. cateniformis F: 5’-ACCGCATAGGTGAAGGGGTC-3’, C. cateniformis-R: GAATCATTTCCTATTCATA(17)). Cycle conditions were 95°C for 3 min, followed by 39 cycles (95°C for 10 sec, 55°C for 30 sec, 95°C for 10 sec, 65°C for 5 sec, 95°C for 5 sec). For each sample, C. cateniformis Ct values were normalized to universal 16S Ct values.
Preparation of Bacteria
Coprobacillis cateniformis, Erysipelatoclostrium ramosum, and Clostridium innocuum stocks were plated on Brucella plates and stored in the anaerobic chamber for 3 days. Individual colonies were inoculated in chopped meat medium (Anaerobe Systems Cat#AS-811). OD600 was measured and 200 μl of overnight culture was orally gavaged into GF mice in isolators one week before tumor implantation.
For C. cateniformis bacterial extracts, individual colonies were inoculated in Reinforced Clostridia media overnight in the anaerobic chamber. One ml of overnight culture was used to inoculate 10 L of Reinforced Clostridia Media and culture was left to grow for 2-3 days. Surface extracts were isolated as in (35).
Cell lines
MC38 and B16-OVA(36) cells were cultured in DMEM supplemented with 10% FBS and 1% penicillin/streptomycin at 37°C in the presence of 5% CO2. B16-OVA cells were selected for 2 days with 2μg/ml puromycin after thawing a new batch of cells. E0771 (ATCC Cat#CRL3461), LLC (ATCC Cat#CRL1642) and MB49 (Sigma Cat#SCC148) were cultured in DMEM supplemented with 10% FBS and 1% penicillin/streptomycin at 37°C in the presence of 5% CO2. PY8119 (ATCC Cat#CRL3278) was cultured in F12 media supplemented with 5% FBS and 1% penicillin/streptomycin at 37°C in the presence of 5% CO2. To create OVA-expressing cell lines of LLC and PY8119, lentivirus was produced by transfecting 293x cells with the packaging plasmids pMD2.G (Addgene ID: 12259) and psPAX2 (Addgene ID: 12260), and the OVA expression plasmid (pLX305-BlastR-OVA). LLC and PY8119 cells were transduced with lentiviral supernatants and polybrene as a transduction enhancer. Transduced cells were then selected with Blasticidin and confirmed to express the SIINFEKL peptide by flow cytometry following stimulation with IFNγ. All cell lines were confirmed mycoplasma negative.
Bone marrow-derived dendritic cell (BMDC) generation
Bone marrow cells were isolated from mouse femurs and cultured in RPMI 1640 media supplemented with 10% FBS, 1% penicillin/streptomycin, 10mM HEPES, 1mM Sodium Pyruvate, 55μM β-mercaptoethanol and 20 ng/ml of GM-CSF (R&D Systems, Cat#315-03) and 20 ng/ml of IL-4 (BioLegend, Cat#574302) for BMDC at 37°C in the presence of 5% CO2. GM-CSF and IL-4 were refreshed every two days.
In vitro CD8+ T cell differentiation and proliferation assay
Naïve CD8+ T cells were purified from splenocytes of OT-I (C57BL/6-Tg(TcraTcrb)1100Mjb/J) RGMb flox/flox and OT-I RGMb flox/flox CD4-Cre+ mice using the Naïve CD8+ T cell isolation kit (Miltenyi Biotec, Cat#130-093-543). The purified naïve CD8+ T cells were labeled with 5μM Cell Trace Violet Proliferation dye (Thermo Fisher Scientific Cat#C34557), and co-cultured with BMDCs at the ratio of 5:1 in the presence of 100U/ml of IL-2 (PeproTech Cat#200-02) and OVA257-264 peptide (Anaspec Cat#AS-60193-1) for 72hrs. For measurement of cytokine production, the cells were restimulated with Golgi inhibitors and PMA/Ionomycin for 5 hrs, followed by intracellular cytokine staining.
Tumor-Infiltrating Leukocyte Isolation
Tumors were harvested on post-implantation days 10-16, mechanically dissociated, and incubated in DPBS containing calcium, magnesium and 250 units/mL of Type 1 Collagenase (Worthington Biochemical Corporation) for 20 minutes at 37°C with gentle rocking. After filtration, tumor-infiltrating lymphocytes were isolated by Percoll density gradient (40%/70%) centrifugation at 800 x g for 20 minutes without brake. The interface of the Percoll layers were recovered for further analyses.
Flow cytometry and cell sorting
Primary mouse cells were isolated from draining lymph node, mesenteric lymph nodes and tumors. Single cell suspensions were prepared with a 70μm cell strainer and incubated with TruStain FcX (BioLegend Cat#101319) to block Fc receptors prior to staining. Cells were stained with LIVE/DEAD™ Fixable Near-IR Dead Cell Stain Kit (Thermo Fisher Scientific Cat#L34975) according to the manufacturer’s instructions. Surface antigen staining was performed in DPBS containing 1% FBS and 2 mM EDTA, followed by intracellular staining using the eBioscience™ FoxP3/Transcription Factor Staining Buffer Set (Thermo Fisher Scientific Cat#00-5523-00). For intracellular cytokine staining, isolated cells were stimulated with 50 ng/mL of Phorbol 12-myristate 13-acetate and 500 ng/mL of ionomycin for 5 hours in the presence of 1X GolgiPlug protein transport inhibitor (BD Biosciences Cat#555029) and 1X GolgiStop protein transport inhibitor (BD Biosciences Cat#554724) prior to intracellular staining. The following antibodies were used for staining procedures: BUV395 anti-mouse CD8β clone H35-17.2 (BD Biosciences Cat#740278 1:200), Brilliant Violet 421 anti-mouse CD8β clone YTS156.7.7 (BioLegend Cat#126629 1:200), PerCP-Cy5.5 anti-mouse CD8β clone YTS156.7.7 (BioLegend Cat#126610 1:200), Alexa Flour 700 anti-mouse CD8α clone 53-6.7 (BioLegend Cat#100730 1:200), BUV395 anti-mouse I-A/I-E clone 2G9 (BD Biosciences Cat#743876 1:500), Pacific Blue anti-mouse I-A/I-E clone M5/114.15.2 (BioLegend Cat#107620 1:500), BUV737 anti-mouse CD45.2 clone 104 (BD Biosciences Cat#564880 1:200), Brilliant Violet 711 anti-mouse CD44 clone IM7 (BioLegend Cat#103057 1:200), Brilliant Violet 605 anti-mouse CD44 clone IM7 (BioLegend Cat#103047 1:200), Alexa Fluor 700 anti-mouse CD45 clone 30-F11 (BioLegend Cat#103128 1:200), Brilliant Violet 510 antimouse CD45 clone 30-F11 (BioLegend Cat#103138 1:200), PE-Texas Red anti-mouse CD4 clone RM4-5 (Thermos Fisher Scientific Cat#MCD0417 1:200), BUV564 anti-mouse CD4 clone GK1.5 (BD Biosciences Cat#612923 1:200), PE anti-mouse CD4 clone GK1.5 (BioLegend Cat#100408 1:200), PE-Cy7 anti-mouse IL-17A clone TC11-18H10.1 (BioLegend Cat#506922 1:200), Brilliant Violet 510 anti-mouse CD366 clone 5D12/TIM-3 (BD Biosciences Cat#747625 1:200), Brilliant Violet 711 anti-mouse CD366 clone 7D3 (BD Biosciences Cat#565566 1:200), PE-CF594 anti-mouse CD11b clone M1/70 (BD Biosciences Cat#562317 1:200), BUV805 antimouse CD11b clone M1/70 (BD Biosciences Cat#741934 1:200), Brilliant Violet 605 antimouse CD11b clone M1/70 (BD Biosciences Cat#563015 1:200), BUV496 anti-mouse CD3ε clone 145-2C11 (BD Biosciences Cat#564661 1:200), Brilliant Violet 570 anti-mouse CD3ε clone 17A2 (BioLegend Cat#100225 1:200), PE-Cy7 anti-mouse CD3ε clone 17A2 (BioLegend Cat#100220 1:200), Brilliant Violet 570 anti-mouse TCRβ clone H57-597 (BioLegend Cat#109231 1:200), BUV615 anti-mouse TCRβ clone H57-597 (BD Biosciences Cat#751212 1:200), APC anti-mouse IFNγ clone XMG1.2 (BioLegend Cat#505810 1:200), PerCP-eFluor 710 anti-mouse CD274 clone MIH5 (Thermo Fisher Scientific Cat#46-5982-82 1:200), BUV737 anti-mouse CD80 clone 16-10A1 (BD Biosciences Cat#564670 1:200), Pe-Cy7 anti-mouse CD11c clone HL3 (BD Biosciences, Cat#558079 1:200), Brilliant Violet 510 anti-mouse CD11c clone HL3 (BD Biosciences Cat#562949 1:200), FITC anti-mouse CD11c clone N418 (BioLegend Cat#117306 1:200), PE anti-mouse CD11c clone N418 (BioLegend Cat#117307 1:200), PerCP-Cy5.5 anti-mouse TNF-a clone MP6-XT22 (BioLegend Cat#506322 1:200), Brilliant Violet 711 anti-mouse TNF-a clone MP6-XT22 (BioLegend Cat#506349 1:200), FITC anti-human/mouse Granzyme B clone GB11 (BioLegend Cat#515403 1:200), PE-Cy7 anti-human/mouse Granzyme B clone QA16A02 (BioLegend Cat#372214 1:200), Brilliant Violet 605 anti-mouse CD279 clone 29F.1A12 (BioLegend Cat#135220 1:200), PE-Cy7 anti-mouse Galectin-9 clone RG9-35 (BioLegend Cat#136113 1:200), Brilliant Violet 510 anti-mouse CD40 clone 3/23 (BD Biosciences Cat#745041 1:200), PerCP-Cy5.5 anti-human/mouse/rat CD278 clone C398.4A (BioLegend Cat#313518 1:200), PE-Cy7 anti-mouse CD252 clone RM134L (BioLegend Cat#108813 1:200), Biotinylated anti-mouse RGMb polyclonal antibody (R&D Systems Cat#BAF3597 1:200), APC streptavidin (BioLegend Cat#405243 1:200), Brilliant Violet 605 anti-mouse CD86 clone GL1 (BD Biosciences Cat#563055 1:200), PE anti-mouse CD275 (BioLegend Cat#107405 1:200), Brilliant Violet 421 anti-mouse CD273 clone TY25 (BioLegend Cat#107219 1:200), APC anti-mouse CD273 clone TY25 (BioLegend Cat#107210 1:200), PE anti-mouse CD223 clone eBioC9B7W (Thermo Fisher Scientific Cat#12-2231-82 1:200), PE-Cy7 T-bet clone 4B10 (BioLegend Cat#644823 1:200), PE anti-mouse CD107a clone 1D4B (BioLegend Cat#121612 1:200), BV711 anti-mouse IL-2 clone JES6-5H4 (BioLegend Cat#503837 1:200). Alexa Fluor 594 anti-RGMb clone 9D3 antibody (1:100) was generated in Gordon Freeman’s laboratory as previously described(29). Flow cytometry analyses were performed on a BD™ LSR II or BD FACSymphony™. For cell sorting, a BD FACSAria™ II was used. Data were collected using BD DIVA 9 software analyzed using FlowJo (version 10.8.1) software. Gating strategies displayed in Extended Data Fig. 10.
Quantitative real-time PCR analysis
Cells were sorted by FACS BD Aria II and lysed with RLT lysis buffer. RNA was extracted using an Qiagen RNeasy Plus Mini kit (Qiagen Cat#74236). cDNA was generated using a SuperScript™ VILO™ cDNA synthesis kit (Thermo Fisher Scientific Cat#11754250), and analyzed on a LightCycler 96 machine and LightCycler 96 1.1 software using Applied Biosystems™ TaqMan™ Fast Advanced Master Mix (Fisher scientific Cat#44-445-57) and Taqman probes (ThermoFisher Scientific Cat#4331182, Cat#4319413E).
Lentiviral production
Lentiviruses were generated using LV-MAX™ lentiviral production system (Thermo Fisher Cat# A35684). In brief, HEK293F cells were transfected with a mixture of pLV-C-GFPSpark lentiviral vector (Sino biological) or pLV-mPD-L2-GFPSpark lentiviral vector and LV-MAX™ lentiviral packaging mix (Thermo Fisher Cat# A43237) using the LV-MAX™ transfection reagent kit (Thermo Fisher Cat# 35348), following the manufacturer’s instruction. The supernatants from the cell culture were obtained and concentrated using Lenti-X concentrator (Takara Cat# 631232).
Adoptive transfer of BMDC
WT or PD-L2 KO bone marrow cells were cultured in the presence of 20 ng/ml of GM-CSF for 6 days to generate BMDCs, followed by pulsing with 100 μg/ml of Ovalbumin protein (Endofit, Cat#vac-pova) for 24 hrs in the presence of 10 μg/ml of C.cateniformis extract or vehicle control. On post-implantation day 3, 5 x 105 of OVA-pulsed BMDCs were injected subcutaneously into B16-OVA tumor bearing mice, at the tumor site (37). For PD-L2 overexpressing BMDCs, the cells were infected with lentivirus at MOI of 30 by spin-inoculation for 90 min at 25 °C in the presence of 8 μg/ml of polybrene (Millipore Sigma, Cat# TR-1003G) on BMDC culture day 3 and day 4. On day 7, GFP expressing BMDCs were sorted by FACS, followed by subsequent culture for 2 days in the presence of 20 ng/ml of GM-CSF. GFP expressing BMDCs were pulsed with OVA protein for 24 hrs, and 2 x 105 cells were injected subcutaneously into the B16-OVA tumor site on the post-implantation day 3.
Statistical analysis
Statistical analyses were performed using GraphPad Prism 9 software. Statistical significance was determined by p-value < 0.05 by using an unpaired Student’s t-test for comparing two groups or non-parametric one-way ANOVA for comparisons with more than two groups. Two-way ANOVA was used for tumor growth curves with multiple groups, followed by Bonferroni’s, Tukey’s, or Sidak’s multiple comparison test as indicated. All the data are represented as mean ± s.e.m. or s.d. P-values were denoted in figures, not significant (n.s.).
Extended Data
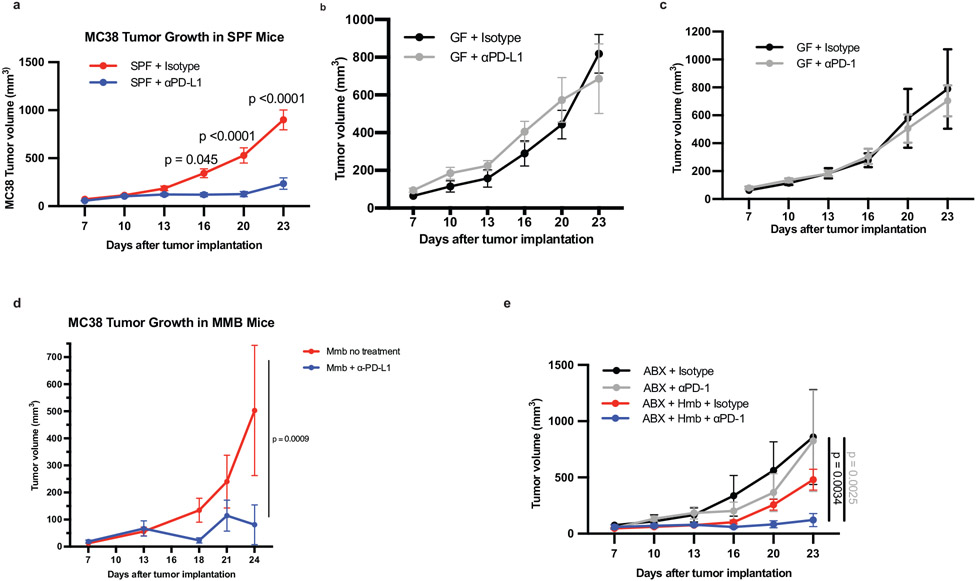
Extended Data Fig. 1. SPF, MMB, and HMB promote anti-tumor responses to PD-1/PD-L1 blockade.
MC38 tumor growth with or without anti-PD-L1 in (a) Taconic SPF n= 10 mice per group. Significance indicated on graph and measured by Two Way ANOVA and Sidak’s multiple comparisons test or (b) GF n = 5 mice per group, performed at the same time as (a), representative experiment of 14 individual experiments. MC38 tumor cells were implanted subcutaneously in (c) GF mice n=4 mice for isotype group and n=5 mice for anti-PD-1 group. (d) GF mice were orally gavaged with MMB one week before implantation of MC38 tumor cells. N=3 mice for no treatment group and n=6 mice for anti-PD-L1 group. Significance indicated on graph and measured by Two Way ANOVA and Sidak’s multiple comparisons test. (e) MC38 tumor cells were implanted subcutaneously in ABX or ABX/HMB mice and treated with anti-PD-1 according to Figure 1a, and monitored for tumor growth. N=3 mice for ABX + isotype group and n= 4 mice for ABX + anti-PD-1, ABX/HMB + Isotype, and ABX/HMB + anti-PD-1 groups. Significance indicated on graph and measured by Two-Way ANOVA and Tukey’s multiple comparisons test. (a-e) Error bars show mean and s.e.m.
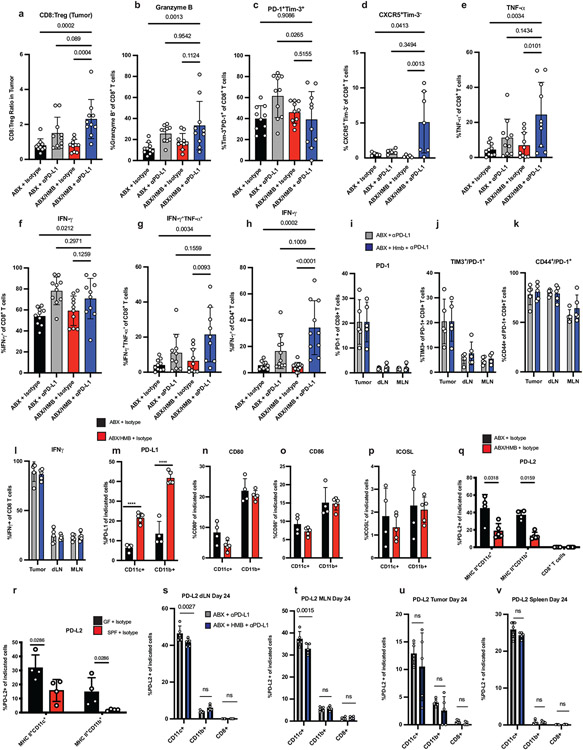
Extended Data Fig. 2. Immune cell responses in ABX, ABX/HMB, GF, and SPF mice.
MC38 tumor cells were implanted subcutaneously in ABX and ABX/HMB mice. Mice were treated with isotype or anti-PD-L1 on days 7, 10, 13, and 16, followed by sacrificed on day 24 after tumor implantation. For detection of cytokines, tumor-infiltrating lymphocytes were stimulated with PMA/Ionomycin for 5 hours in the presence of Golgi inhibitors. (a) Ratio of CD8+ T cells to Treg cells n=10 mice per group (b) Percent of Granzyme B+ CD8+ T cells n= 10 mice per group (c) Frequency of PD-1+TIM-3+ n= 10 mice per group or (d) CXCR5+ TIM-3− among CD8+ T cells in tumors n=5 mice per group for ABX groups and ABX/HMB+ Isotype group and n=7 for ABX/HMB + anti-PD-L1 group. Percent of (e)TNF-α+ n= 10 mice per group (f) IFNγ+n=10 mice per group, and (g) TNF-α+IFNγ+ CD8+ T cells n = 10 mice per group for ABX groups and ABX/HMB + isotype and n= 9 mice for ABX/HMB + anti-PD-L1 (h) Percent of IFNγ+ CD4+ T cells n = 10 mice per group for ABX groups and ABX/HMB + anti-PD-L1 and n=12 mice per group for ABX/HMB + isotype. Mice were analyzed on day 24 after tumor implantation, representative of two independent experiments. Significance measured by non-parametric one-way ANOVA and Dunn’s multiple comparisons and P values are indicated on graphs, error bars show mean and s.d. (i-l) MC38 tumor cells were implanted subcutaneously in ABX and ABX/HMB mice. Mice were treated with isotype or anti-PD-L1 on days 7 and 10 and sacrificed on day 13 after tumor implantation. (i) Percent of PD-1+ CD8+ T cells in tumors, dLNs, and mesenteric lymph nodes (MLNs) n= 5 mice per group (j) Percent TIM-3+ among PD-1+ CD8+ T cells in Tumors, dLNs, and MLNs n= 5 mice per group for all except MLN ABX/HMB + anti-PD-L1 n= 4 mice per group (k) Percent CD44+ expression on PD-1+ CD8+ T cells in tumors, dLNs, and MLNs n = 4 mice per group (l) Percent IFNγ+ CD8+ T cells in tumors, dLNs, and MLNs n= 5 mice per group. (i-l) Significance determined by non-parametric one-way ANOVA with Dunn’s multiple comparisons test, error bars show mean and s.d. Expression of (m) PD-L1, (n) CD80, (o) CD86, and (p) ICOSL on CD11c+ MHCII+ and CD11b+ MHCII+ cells in draining lymph nodes of ABX and ABX/HMB mice implanted with MC38 tumor cells subcutaneously and treated with isotype control mAb as in Figure 1a. Mice were analyzed on day 13 after tumor implantation. N=4 mice in ABX group and n=5 mice in ABX/HMB group. Significance measured by unpaired two-tailed, Mann-Whitney test. Expression of PD-L2 on CD11c+ MHCII+ , CD11b+ MHCII+ cells and CD8+ T cells in draining lymph nodes of (q) ABX vs. ABX/HMB mice n n= 4 mice per group for ABX and n=5 mice per group for ABX/HMB or (r) GF vs. Taconic SPF mice n = 4 mice per group, implanted with MC38 tumor cells subcutaneously and treated with isotype control mAb as in Figure 1a. Mice were analyzed on day 10 after tumor implantation. Significance measured by unpaired, two-tailed Mann-Whitney test, and significant P values indicated on graphs. ABX and ABX/HMB mice were sacrificed 24 days after implantation with MC38 tumor cells. PD-L2 was measured on MHCII+ CD11c+, MHCII+ CD11b+, and CD8+ T-cells in (s) draining lymph nodes n = 5 mice per group, (t) MLN n = 5 mice for ABX and n=4 mice for ABX/HMB, (u) tumors n = 5 mice per group and (v) spleen n= 5 mice per group. Significance measured by Two-Way ANOVA and Sidak’s multiple comparison’s test. Significant P values indicated on graph. Representative experiment of two different experiments. Error bars show mean and s.d.
Extended Data Fig. 3. Combination therapy with anti-PD-1 or anti-PD-L1 plus anti-PD-L2 or anti-RGMb promotes an anti-tumor response in multiple, but not all, tumor types in SPF mice.
MC38, B16-OVA, MB49, Py8119-OVA, LLC-OVA cells were implanted subcutaneously, and E0771 cells were injected into mammary fat pad of C57BL/6 mice with the indicated microbiota. The mice were treated with four doses of anti-PD-L1 or anti-PD-1 with/without anti-PD-L2 3.2 or anti-PD-L2 2C9 or anti-RGMb 9D1 one week after tumor implantation as indicated. (a) Growth of MB49 tumors in ABX mice given isotype, anti-PD-1 alone or combined with anti-PD-L2 3.2 n =10 mice per group for Isotype and anti-PD-1 + anti PD-L2 3.2 groups, n = 9 mice for anti-PD-1 group (b) Growth of MB49 tumors in ABX mice given isotype, anti-PD-L1 alone or combined anti-PD-L2 3.2 n = 10 mice per group (c) Growth of B16-OVA tumors in Taconic SPF mice given isotype, anti-PD-L1, anti-PD-L2 3.2 or anti-PD-L1 + anti-PD-L2. n= 5 mice per group for isotype and anti-PD-L2 3.2 groups and n= 10 mice per group for anti-PD-L1 and anti-PD-L1 + anti-PD-L2 3.2 groups. (d) Growth of Py8119-OVA in Taconic SPF mice given isotype or anti-PD-L1 and/or anti-PD-L2 3.2. n= 5 mice per group for all except n=4 mice for anti-PD-L2 3.2 group (e) Growth of MC38 tumors in Taconic SPF mice given isotype, anti-PD-L1, anti-PD-L2 3.2, or anti-PD-L1 + anti-PD-L2 n = 10 mice per group for all except n= 9 mice for anti-PD-L2 3.2 group. (f-g) Growth of E0771 tumors in Taconic SPF mice (f) given isotype, or anti-PD-1 combined with anti-PD-L2 3.2, anti-PD-L2 2C9 or anti-RGMb 9D1 n = 10 mice per group or (g) given isotype n=10 mice, anti-PD-L1 alone n= 9 mice or combined with anti-PD-L2 n=5 mice. (h) Growth of LLC-OVA tumors in Taconic SPF mice given isotype, anti-PD-L1, or anti-PD-L1 and either anti-PD-L2 3.2 or anti-RGMb 9D1. n=10 mice per group. Significance measured by two-way ANOVA and Tukey’s multiple comparisons test. Significant P vales are designated on graphs. Error bars show mean and s.e.m.
Extended Data Fig. 4. 16S sequencing of fecal samples, tumor growth, and survival of mice colonized with melanoma patient stool.
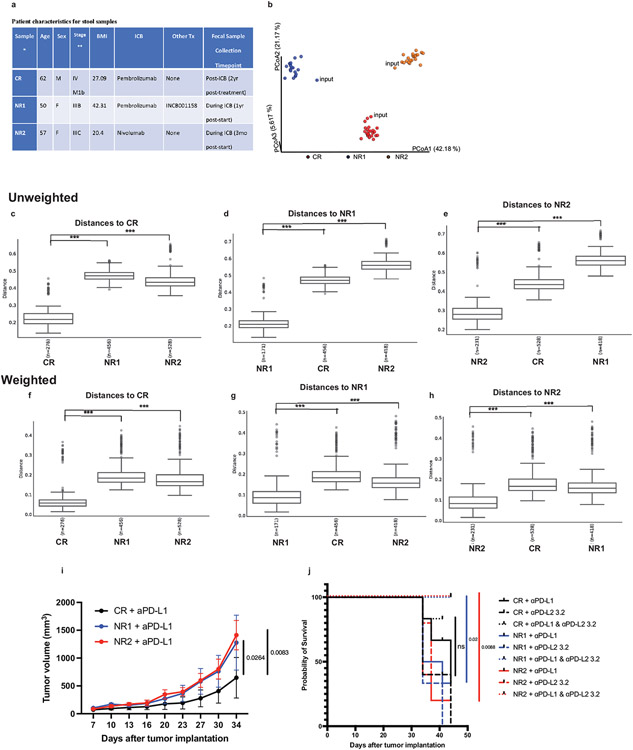
(a) Patient characteristics for stool samples. *Samples were characterized as complete responder (CR) or non-responder (NR) based on RECIST 1.1 response criteria. Mean age is 56.3 years, mean BMI is 29.9 kg/m2. **Stage at start of ICB treatment. (b-h) Bacterial community configuration and distances in fecal samples collected from three different sets of mice colonized with melanoma patient stool: complete responder (CR), Non-responder 1 (NR1) and Non-responder 2 (NR2) as well as input. (b) Principal coordinates analysis (PCoA) of unweighted UniFrac distance measurements based on the 16S sequencing analysis of the composition of bacterial communities at 7 day after gavage. Each colored circle represented a fecal community sampled from a mouse belonging to the indicated donor group or input gavage sample. Pairwise unweighted UniFrac distances of the composition of bacterial communities in fecal samples within (c) CR and across NR1 and NR2 groups compared to CR (d) NR1 and across CR and NR2 groups compared to NR1 and (e) NR2 and across CR and NR1 groups compared to NR2. Pairwise weighted UniFrac distances of the composition of bacterial communities in fecal samples within (f) CR and across NR1 and NR2 groups compared to CR (g) NR1 and across CR and NR2 groups compared to NR1 and (h) NR2 and across CR and NR1 groups compared to NR2. n= number of pairwise distances calculated between all samples in each group compared to the comparator group. *** q-values=0.001, PERMANOVA. Minima, maxima, center, bounds, and percentiles of boxplots shown in source data. GF mice were colonized with stool from Responder (R) or Non-Responder (NR) melanoma patients treated with anti-PD-1, implanted with MC38 tumors, and given anti-PD-L1, anti-PD-L2 or anti-PD-L1 plus anti-PD-L2 mAbs. (i) Tumor growth in mice treated with anti-PD-L1 monotherapy. N=6 mice for CR and NR1 groups and n= 5 mice for NR2 group. Significance determined by 2 way ANOVA and Tukey’s multiple comparisons test and significant p values are indicated on graph. Error bars show standard error of the mean. (j) Survival of mice given anti-PD-L1, anti-PD-L2 or anti-PD-L1 plus anti-PD-L2 mAbs. Survival defined as number of live mice with tumors <2 cm3 or <50% ulcerated. N= 6 for CR + anti-PD-L1, CR+ anti-PD-L1 + anti-PD-L2, NR1 + anti-PD-L1, NR1 + anti-PD-L1 and anti-PD-L2, NR2 + anti-PD-L1 and anti-PD-L2, n=5 for CR + anti-PD-L2, NR2 anti-PD-L1, NR2 anti-PD-L2, n=3 for NR1 + anti-PD-L2. Significance of anti-PD-L1 monotherapy versus combination therapy for each group of mice shown and significance indicated on graph.
Extended Data Fig. 5. Individual antibiotic treatments and 16S sequencing show Grampositive species are associated with an anti-tumor response.
All mice were treated with Vancomycin, Neomycin, Metronidazole, and Ampicillin (VNMA) in the drinking water 4 days before tumor implantation until day 7 pi. VNMA mice continued with the antibiotic cocktail for the duration of the experiment. VNMA + HMB mice stopped VNMA at day 7 and were orally gavaged with HMB stock. Mice given individual antibiotics stopped VNMA at day 7 pi and water was replaced with an individual antibiotic and mice were orally gavaged with HMB stock. MC38 tumor growth curves in mice receiving HMB + (a) Vancomycin (Vanco) n=5 mice per group (b) Metronidazole (Met) n= 5 mice per group (c) Ampicillin (AMP) n = 5 mice per group and (d) Neomycin (Neo) n = 5 mice for no treatment and n=3 mice for anti-PD-L1 group with or without anti-PD-L1. N =4 for VNMA groups and n=5 for HMB groups. Significance determined by two-way ANOVA and Tukey’s multiple comparisons test and significant p values between individual antibiotic treatments versus HMB or VNMA are shown. Error bars show mean and s.e.m. (e-f) Pairwise weighted and unweighted UniFrac distances of the composition of bacterial communities in fecal samples within HMB and across treatment groups compared to HMB. Each dot represented the weighted or unweighted UniFrac distance between the configuration of bacterial populations as judged from the relative abundances of its members, determined by 16S sequencing, in fecal samples collected from members of the indicated treatment group. q-values were indicated for each group comparisons as determined by permutational analysis of variance (PERMANOVA) for beta diversity group significance. Minima, maxima, center, bounds, and percentiles of boxplots shown in source data. (g) Principal coordinates analysis (PCoA) of weighted UniFrac distance measurements based on the 16S sequencing data of the composition of bacterial communities in the fecal samples of mice with different treatment at day 13 and 23 pi. Each colored circle represented a fecal community sampled from a mouse belonging to the indicated antibiotic treatment group or input gavage sample. (h) The relative abundance of the taxa in the order of Clostridiales in each treatment group at day 23 pi after anti-PD-L1 treatment. These identified taxa were significantly (W-statistic=39, 35 and 34 respectively, p-value<0.05) associated with response to anti-PD-L1 by differential abundance testing of taxa between the responder group (HMB n= 5, Neomycin n = 3) to the non-responder group (ABX n = 3, Ampicillin n = 4, Vancomycin n =3, and Metronidazole n = 2) using ANCOM (Analysis of Composition of Microbiomes).
Extended Data Fig. 6. Narrowing down bacteria isolated from HMB stocks that promote anti-tumor immunity and suppress PD-L1.
MC38 tumor growth in gnotobiotic mice colonized one week prior to tumor implantation with (a) Mix of overnight cultures of Longicatena caecimuris, Blautia hydrogenotrophica, Clostridium orbiscindens, Clostridium innocuum, Phocaeicola dorei, Coprobacillus cateniformis, and Ersyipelatoclostridium ramosum n= 4 mice for GF + Isotype, n= 5 mice per group for GF + anti-PD-L1, 7 mix + isotype, and 7 mix + anti-PD-L1 (b) mix of overnight cultures of C. innocuum, C. cateniformis, E. ramosum n= 4 mice for GF + isotype, n= 5 mice for GF + anti-PD-L1, n = 10 mice for 3 mix + Isotype, n= 9 mice for 3mix + anti-PD-L1 (c) C. cateniformis n= 5 mice per group (d) E. ramosum n= 10 mice per group (e) C. innocuum n= 5 mice for isotype and n= 4 mice for anti-PD-L1. Significance at day 23 is shown and was determined by two-way ANOVA with Tukey’s multiple comparisons test for a and b. Significance is shown and was measured by two-way ANOVA with Sidak’s multiple comparisons test for all days was shown for c-e. Error bars for all tumor graphs show mean and s.e.m. (f) C.cateniformis-specific primers for the 16S gene were used to detect C. cateniformis in C. cateniformis stock, HMB stocks, Taconic feces, and E. ramosum stocks. C. cateniformis gene expression was normalized using the 16S universal primers in each group. N= 3 technical replicates for C. cateniformis and E. ramosum pure stocks, n=5 biological replicates from 5 HMB stocks, n= 2 biological replicates of stool samples from two different Taconic mice. To show sequence specificity, data are shown as log fold change over the normalized C. cateniformis-specific 16S levels in E .ramosum stock. Error bars represent standard deviation. GF mice or GF mice monocolonized with E. ramosum one week prior to tumor implantation were sacrificed at day 10 p.i. PD-L2 was measured on MHCII+ CD11c+, MHCII+ CD11b+, and CD8+ T cells in (g) draining lymph nodes, n= 9 mice for GF and 7 mice for E. ramosum (h) MLN, n = 10 mice per group. Significance measured by Mann-Whitney test and error bars show mean and s.d.
Extended Data Figure 7. C. cateniformis and its surface extracts impact immune function.
(a) BMDCs from WT, TLR2 KO, Dectin-1 KO, and MyD88 KO mice were treated with surface extracts isolated from C. cateniformis pellets. PD-L2 expression was measured by flow cytometry. Percent reduction (compared to vehicle treatment) of PD-L2 expressing dendritic cells is shown. Significance measured by Kruskal-Wallis test and P values compared to WT are indicated on graph. Error bars show mean and s.d., n = 3 wells of BMDCs per group. (b-f) Isotype treated-GF versus GF mice monocolonized with C. cateniformis one week prior to tumor implantation were sacrificed at day 13 p.i. and dLNs were analyzed by flow cytometry. Frequencies of (b) CD45+ cells, (c) CD8+ T cells (d) CD4+ T cells (e) MHCII+ CD11b+ cells and (f) MHCII+CD11c+ cells. (b-f) n= 10 mice for GF and n= 9 mice for C. cateniformis, significance measured by unpaired, two-tailed Mann-Whitney test and P values are indicated on graphs, error bars show mean and s.d. (g-j) GF mice were monocolonized one week prior to tumor implantation and treated with either isotype or anti-PD-L1. Mice were sacrificed at day 18 p.i. and tumors were harvested for analysis. Frequencies of CD8+ T cells expressing (g) Granzyme B (h) IFNγ (i) TIM-3 and PD-1 and (j) TNFα. For (g-j) n= 5 mice for isotype and n= 3 mice for anti-PD-L1, significance determined by unpaired, two-tailed Mann-Whitney tests and P values are indicated on graphs, error bars show mean and s.d. (k) Histograms of PD-L2 expression on BMDCs measured by flow cytometry. Red= BMDCs transduced with PD-L2 GPF lentivirus, Blue = BMDCs transduced with control GFP lentivirus, Gray = BMDCs from PD-L2 KO mice. (l-p) BMDCs transduced with GFP lentivirus (GFP) or lentivirus expressing GFP and PD-L2 (PD-L2-GFP) were treated with C. cateniformis extract 24 hours before co-culture with CD8+ T cells. Expression measured by flow cytometry. (l) Example of flow cytometry plots of Granzyme B and CD107a expression on CD8+ T cells. Quantification of mean fluorescence intensity of (m) CD107a and (n) Granzyme B expression on CD8+ T cells displayed in (l). Quantification of mean fluorescence intensity of (o) CD44 and (p) CD25 on CD8+ T cells. (m-p) Significance determined by unpaired, two-tailed t tests, with Welch correction and P values indicated on graph, n = 4 wells of BMDC-CD8+ T cell co-culture per group, error bars show mean and s.d.
Extended Data Fig. 8. C. cateniformis treatment and PD-L2 blockade impacts anti-tumor immunity.
(a) Taconic mice were treated with broad spectrum antibiotics one week prior to tumor implantation. On days 7, 10, 13, 16 C. cateniformis or PBS was orally gavaged and antibodies were administered i.p. Significance between anti-PD-L1 + C. cateniformis treatment and the other groups at day 27 is shown. Significance measured by Two way ANOVA and Tukey’s multiple comparisons test, n = 10 mice per group, error bars show mean and s.e.m. (b) B16-OVA tumor growth in SPF mice that received GFP or PD-L2-GFP BMDCs at tumor site 3 days after tumor implantation. Significance indicated on graph and measured by 2way ANOVA and Sidak’s multiple comparisons test, n= 10 mice per group, error bars show mean and s.e.m. (c-e) BMDCs were cultured for 7 (blue) or 10 (red) days and MHCII expression was measured by flow cytometry. (c) Expression of CD11c and MHCII gated on live cells from BMDC culture. (d) Histogram of MHCII expression gate on live cells from BMDC culture. (e) Histogram of MHCII expression gated on CD11c+ MHCII+ cells. (f) Growth of MC38 tumor cells implanted subcutaneously in β2m−/− mice (B2M KO), β2m+/− (Het), and WT littermate controls. n= 5 mice per group for WT/het + anti-PD-L1 and B2M KO + anti-PD-L1 and n= 3 mice for B2M KO + anti-PD-L1 and anti-PD-L2. Significance indicated on graph measured by one-way ANOVA and Tukey’s multiple comparisons, error bars show mean and s.e.m. Tumor growth of (g) MC38 tumor cells in GF mice treated with Isotype (n= 5 mice), anti-PD-L1 (n= 5 mice), anti-PD-L2 clone 3.2 (n= 5 mice), anti-RGMb (n= 4 mice), anti-PD-L1 + anti-PD-L2 3.2 (n= 4 mice), or anti-PD-L1 + anti-RGMb (n= 5 mice) and (h) B16-OVA tumor cells in Taconic mice treated with Isotype (n= 5 mice), anti-PD-L1 (n= 10 mice), anti-PD-L2 clone 3.2 (n= 5 mice), anti-PD-L2 clone 2C9 (n= 5 mice), anti-RGMb (n= 5 mice), anti-PD-L1 + anti-PD-L2 3.2 (n= 10 mice), anti-PD-L1 + anti-PD-L2 2C9 (n= 10 mice), or anti-PD-L1 + anti-RGMb (n= 10 mice). (g,h) Significance measured by Two-way ANOVA and Tukey’s multiple comparisons, P values are shown on graph, error bars show mean and s.e.m.
Extended Data Fig. 9. RGMb expression is modulated by the gut microbiota.
Tumor draining lymph nodes from GF and SPF mice implanted with MC38 tumor cells subcutaneously, as in Figure 1a, were analyzed on day 11 after implantation. (a) Relative mRNA expression of RGMb in tumor draining lymph nodes of indicated cells. The levels of rgmb transcripts were normalized to expression of an internal control gene 18S rRNA. N = 5 mice for GF and n= 4 mice for SPF and (b-d) cell surface expression of RGMb protein in CD4+ T cells, CD8+ T cells, CD11c+ MHCII+ and CD11b+ cells. (b) Frequencies of RGMb -expressing CD4+ T cells, CD8+ T cells, CD11c+ MHCII+ and CD11b+ cells were measured using 9D3 clone mAb in dLN, n = 5 mice per group. (c-d) Geometric Mean Fluorescent Intensity (gMFI) of RGMb was assessed in indicated populations from (c) tumor and (d) tumor draining lymph nodes using aRGMb polyclonal antibody, n = 5 mice per group. Significance measured by unpaired, two tailed Mann-Whitney test and significant p values indicated on graph, error bars show mean and s.d. (e-g) GF mice were colonized with stools from three patients who received anti-PD-1 therapy and responded or did not respond - Complete Responder (CR, n = 6) or Non-Responder 1(NR1, n = 5) or Non-Responder 2 (NR2, n = 5). The mice were injected subcutaneously with MC38 tumor cells and treated with rat IgG2b isotype control. Frequencies of RGMb-expressing tumor-infiltrating (e) CD8+ T cells, (f) CD11b+MHC II+ cells, and (g) CD11c+MHC II+ cells were examined on post-implantation day 29. Significance measured by non-parametric one-way ANOVA with Dunn’s multiple comparisons test and significant p values indicated on graph. Error bars show mean and s.d. (h) MC38 tumor cells expressing GFP were implanted in GF or SPF mice. The mice were treated with two doses of isotype control or anti-PD-L1 one week after tumor implantation, and tumors were harvested 13 days after tumor implantation. MC38-GFP cells were isolated and examined to measure PD-L2 (Upper), PD-L1 (Middle), and RGMb (Lower) expression by flow cytometry. Representative of 5 mice per group. (i) GF mice were implanted with MC38 tumor cells subcutaneously and treated with indicated antibodies as in Figure 1a. Tumor-infiltrating CD4+ T cells were isolated on day 11 after tumor implantation and stimulated with PMA/Ionomycin for 5 hours. Frequencies of TNF-α producing cells among CD4+ T cell population were measured by intracellular staining and flow cytometry. N= 4 mice for isotype and n = 5 mice for anti-PD-L1, anti-RGMb, and anti-PD-L1 + anti-RGMb groups. Significance measured by non-parametric one-way ANOVA with Dunn’s multiple comparisons and indicated on graph. Error bars show mean and s.d. (j-l) GF mice were implanted with MC38 tumor cells subcutaneously and treated with indicated antibodies as in Figure 1a. Cells in tumor and tumor draining lymph node were analyzed on day 11 after tumor implantation. Frequencies of tumor infiltrating CD8+ T cells expressing (j) PD-1 (k) TIM-3 and (l) LAG-3. N= 4 mice for isotype and n = 5 mice for anti-PD-L1, anti-RGMb, anti-PD-L1 + anti-RGMb groups.
Significance measured by non-parametric one-way ANOVA with Dunn’s multiple comparisons and significant p values indicated on graphs. Error bars show mean and s.d.
(m) Strategy to generate RGMb conditional knockout mice (n) Validation of CD4-Cre mediated deletion of RGMb in peripheral naïve CD8 T cells by qPCR (o) Validation of LysM-Cre mediated deletion of RGMb in bone marrow derived macrophages by qPCR, n = 2 mice per group. Error bars show mean and s.d. (p-s) WT or RGMb KO CD8+ T-cells were co-cultured with WT BMDCs. CD8+ T cells were analyzed by flow cytometry for (p) Mean Fluorescence Intensity (MFI) of T-Bet (q) MFI of CD107a (r) MFI of Granzyme B and (s) proliferation measured by Cell Trace Violet. N=3 per group. Representative experiment of 3 experiments. Significance determined by unpaired Mann-Whitney test and significant p values indicated on graphs. Error bars show mean and s.d.
Extended Data Fig 10. Gating strategies for flow cytometric analysis.
The following gating schemes were used to define CD11c+ MHC II+, CD11b+ MHC II+ cells, CD8+ T cells, CD4+ T cells and Treg cells in (a) draining lymph nodes and mesenteric lymph nodes and (b) tumor-infiltrating immune cells. (c) Gating strategy to examine OT-I cells stimulated by OVA-loaded BMDCs. (d) Gating strategy used to analyze tumor cells from the mice implanted with MC38 tumor cells expressing GFP. Gating strategies used to sort CD4+ T cells, CD8+ T cells, CD11c+MHC II+ cells and CD11b+ cells from (e) tumor-infiltrating leukocytes and (f) tumor draining lymph nodes.
Acknowledgments:
We would like to thank Tsering Yanortsang and Jaime Ramos for their assistance with Germ Free mice. We also thank the members of Sharpe lab and Kasper lab for insightful discussions.
Funding:
Quark Ventures A31696 (JSP, FSG, DLK, AHS)
National Institutes of Health National Institute of Childhood Health and Human Disease T32: 5T32HD55148-10 (FSG)
National Institute of Health / National Cancer Institute 5F32CA247072-02 (JSP)
P50CA206963 and P50CA101942 (GJF) and P01 AI56299 (AHS and GJF)
CPRIT Training Award RP210028 (EMP)
National Institute of Health / National Cancer Institute 1F32CA260769-01 (GM)
Footnotes
Competing interests:
GJF has patents/pending royalties on the PD-1/PD-L1 pathway from Roche, Merck MSD, Bristol-Myers-Squibb, Merck KGA, Boehringer-Ingelheim, AstraZeneca, Dako, Leica, Mayo Clinic, and Novartis. GJF has a patent on RGMb in cancer immunotherapy. GJF has served on advisory boards for Roche, Bristol-Myers-Squibb, Xios, Origimed, Triursus, iTeos, NextPoint, IgM, Jubilant, Trillium, GV20, IOME, and Geode. GJF has equity in Nextpoint, Triursus, Xios, iTeos, IgM, Trillium, Invaria, GV20, and Geode.
AHS has patents/pending royalties on the PD-1 pathway from Roche and Novartis. AHS is on advisory boards for Surface Oncology, SQZ Biotechnologies, Elpiscience, Selecta, Bicara and Monopteros, Bicara, Fibrogen, and Alixia. She also is on scientific advisory boards for the Massachusetts General Cancer Center, Program in Cellular and Molecular Medicine at Boston Children's Hospital, the Human Oncology and Pathogenesis Program at Memorial Sloan Kettering Cancer Center, Glaxo Smith Kline and Janssen. She is an academic editor for the Journal of Experimental Medicine. AHS has received research funding from Novartis, Roche, UCB, Ipsen, Merck, AbbVie, Moderna, Vertex and Erasca unrelated to this project.
SSW is on the advisory board for Asylia Therapeutics and reports compensation from Ridgeline Therapeutics. JAW reports compensation for speaker’s bureau and honoraria from Imedex, Dava Oncology, Omniprex, Illumina, Gilead, PeerView, Physician Education Resource, MedImmune and Bristol-Myers Squibb and serves as a consultant / advisory board member for Roche/Genentech, Novartis, AstraZeneca, GlaxoSmithKline, Bristol-Myers Squibb, Merck, Micronoma, and Biothera Pharmaceuticals, with stock options for Micronoma.
Data and materials availability: 16S sequencing data has been deposited to NIH. Bio Project number: PRJNA936792.
References
- 1.Vétizou M, Pitt JM, Daillère R, Lepage P, Waldschmitt N, Flament C, Rusakiewicz S, Routy B, Roberti MP, Duong CPM, Poirier-Colame V, Roux A, Becharef S, Formenti S, Golden E, Cording S, Eberl G, Schlitzer A, Ginhoux F, Mani S, Yamazaki T, Jacquelot N, Enot DP, Bérard M, Nigou J, Opolon P, Eggermont A, Woerther P-L, Chachaty E, Chaput N, Robert C, Mateus C, Kroemer G, Raoult D, Boneca IG, Carbonnel F, Chamaillard M, Zitvogel L, Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science. 350, 1079–1084 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Routy B, Chatelier EL, Derosa L, Duong CPM, Alou MT, Daillère R, Fluckiger A, Messaoudene M, Rauber C, Roberti MP, Fidelle M, Flament C, Poirier-Colame V, Opolon P, Klein C, Iribarren K, Mondragón L, Jacquelot N, Qu B, Ferrere G, Clémenson C, Mezquita L, Masip JR, Naltet C, Brosseau S, Kaderbhai C, Richard C, Rizvi H, Levenez F, Galleron N, Quinquis B, Pons N, Ryffel B, Minard-Colin V, Gonin P, Soria J-C, Deutsch E, Loriot Y, Ghiringhelli F, Zalcman G, Goldwasser F, Escudier B, Hellmann MD, Eggermont A, Raoult D, Albiges L, Kroemer G, Zitvogel L, Gut microbiome influences efficacy of PD-1–based immunotherapy against epithelial tumors. Science. 359, 91–97 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM, Benyamin FW, Lei YM, Jabri B, Alegre M-L, Chang EB, Gajewski TF, Commensal Bifidobacterium promotes antitumor immunity and facilitates anti–PD-L1 efficacy. Science. 350, 1084–1089 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tanoue T, Morita S, Plichta DR, Skelly AN, Suda W, Sugiura Y, Narushima S, Vlamakis H, Motoo I, Sugita K, Shiota A, Takeshita K, Yasuma-Mitobe K, Riethmacher D, Kaisho T, Norman JM, Mucida D, Suematsu M, Yaguchi T, Bucci V, Inoue T, Kawakami Y, Olle B, Roberts B, Hattori M, Xavier RJ, Atarashi K, Honda K, A defined commensal consortium elicits CD8 T cells and anti-cancer immunity. Nature. 565, 600–605 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Mager LF, Burkhard R, Pett N, Cooke NCA, Brown K, Ramay H, Paik S, Stagg J, Groves RA, Gallo M, Lewis IA, Geuking MB, McCoy KD, Microbiome-derived inosine modulates response to checkpoint inhibitor immunotherapy. Science (2020), doi: 10.1126/science.abc3421. [DOI] [PubMed] [Google Scholar]
- 6.Griffin ME, Espinosa J, Becker JL, Luo J-D, Carroll TS, Jha JK, Fanger GR, Hang HC, Enterococcus peptidoglycan remodeling promotes checkpoint inhibitor cancer immunotherapy, 8 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baruch EN, Youngster I, Ben-Betzalel G, Ortenberg R, Lahat A, Katz L, Adler K, Dick-Necula D, Raskin S, Bloch N, Rotin D, Anafi L, Avivi C, Melnichenko J, Steinberg-Silman Y, Mamtani R, Harati H, Asher N, Shapira-Frommer R, Brosh-Nissimov T, Eshet Y, Ben-Simon S, Ziv O, Khan MAW, Amit M, Ajami NJ, Barshack I, Schachter J, Wargo JA, Koren O, Markel G, Boursi B, Fecal microbiota transplant promotes response in immunotherapy-refractory melanoma patients. Science. 371, 602–609 (2021). [DOI] [PubMed] [Google Scholar]
- 8.Davar D, Dzutsev AK, McCulloch JA, Rodrigues RR, Chauvin J-M, Morrison RM, Deblasio RN, Menna C, Ding Q, Pagliano O, Zidi B, Zhang S, Badger JH, Vetizou M, Cole AM, Fernandes MR, Prescott S, Costa RGF, Balaji AK, Morgun A, Vujkovic-Cvijin I, Wang H, Borhani AA, Schwartz MB, Dubner HM, Ernst SJ, Rose A, Najjar YG, Belkaid Y, Kirkwood JM, Trinchieri G, Zarour HM, Fecal microbiota transplant overcomes resistance to anti-PD-1 therapy in melanoma patients. Science. 371, 595–602 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ribas A, Wolchok JD, Cancer immunotherapy using checkpoint blockade. Science. 359, 1350–1355 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao B, Zhao H, Zhao J, Efficacy of PD-1/PD-L1 blockade monotherapy in clinical trials. Ther Adv Med Oncol. 12 (2020), doi: 10.1177/1758835920937612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chung H, Pamp SJ, Hill JA, Surana NK, Edelman SM, Troy EB, Reading NC, Villablanca EJ, Wang S, Mora JR, Umesaki Y, Mathis D, Benoist C, Relman DA, Kasper DL, Gut Immune Maturation Depends on Colonization with a Host-Specific Microbiota. Cell. 149, 1578–1593 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Surana NK, Kasper DL, Moving beyond microbiome-wide associations to causal microbe identification. Nature. 552, 244–247 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fransen MF, Schoonderwoerd M, Knopf P, Camps MGM, Hawinkels LJAC, Kneilling M, van Hall T, Ossendorp F, Tumor-draining lymph nodes are pivotal in PD-1/PD-L1 checkpoint therapy. JCI Insight. 3, doi: 10.1172/jci.insight.124507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lam KC, Araya RE, Huang A, Chen Q, Di Modica M, Rodrigues RR, Lopes A, Johnson SB, Schwarz B, Bohrnsen E, Cogdill AP, Bosio CM, Wargo JA, Lee MP, Goldszmid RS, Microbiota triggers STING-type I IFN-dependent monocyte reprogramming of the tumor microenvironment, Cell 184, 5338–5356 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, Prieto PA, Vicente D, Hoffman K, Wei SC, Cogdill AP, Zhao L, Hudgens CW, Hutchinson DS, Manzo T, de Macedo MP, Cotechini T, Kumar T, Chen WS, Reddy SM, Sloane RS, Galloway-Pena J, Jiang H, Chen PL, Shpall EJ, Rezvani K, Alousi AM, Chemaly RF, Shelburne S, Vence LM, Okhuysen PC, Jensen VB, Swennes AG, McAllister F, Sanchez EMR, Zhang Y, Chatelier EL, Zitvogel L, Pons N, Austin-Breneman JL, Haydu LE, Burton EM, Gardner JM, Sirmans E, Hu J, Lazar AJ, Tsujikawa T, Diab A, Tawbi H, Glitza IC, Hwu WJ, Patel SP, Woodman SE, Amaria RN, Davies MA, Gershenwald JE, Hwu P, Lee JE, Zhang J, Coussens LM, Cooper ZA, Futreal PA, Daniel CR, Ajami NJ, Petrosino JF, Tetzlaff MT, Sharma P, Allison JP, Jenq RR, Wargo JA, Gut microbiome modulates response to anti–PD-1 immunotherapy in melanoma patients. Science. 359, 97–103 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matson V, Fessler J, Bao R, Chongsuwat T, Zha Y, Alegre M-L, Luke JJ, Gajewski TF, The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science. 359, 104–108 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kageyama A, Benno Y, Coprobacillus catenaformis Gen. Nov., Sp. Nov., a New Genus and Species Isolated from Human Feces. Microbiology and Immunology. 44, 23–28 (2000). [DOI] [PubMed] [Google Scholar]
- 18.Silveira-Nunes G, Durso DF, de O. LRA Jr., Cunha EHM, Maioli TU, Vieira AT, Speziali E, Corrêa-Oliveira R, Martins-Filho OA, Teixeira-Carvalho A, Franceschi C, Rampelli S, Turroni S, Brigidi P, Faria AMC, Hypertension Is Associated With Intestinal Microbiota Dysbiosis and Inflammation in a Brazilian Population. Frontiers in Pharmacology. 11 (2020) (available at https://www.frontiersin.org/article/10.3389/fphar.2020.00258). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiao Y, Yu S, Zhu B, Bedoret D, Bu X, Francisco LM, Hua P, Duke-Cohan JS, Umetsu DT, Sharpe AH, DeKruyff RH, Freeman GJ, RGMb is a novel binding partner for PD-L2 and its engagement with PD-L2 promotes respiratory tolerance. J Exp Med. 211, 943–959 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pauken KE, Torchia JA, Chaudhri A, Sharpe AH, Freeman GJ, Emerging concepts in PD-1 checkpoint biology. Seminars in Immunology. 52, 101480 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Severyn CJ, Shinde U, Rotwein P, Molecular biology, genetics and biochemistry of the repulsive guidance molecule family. Biochem J. 422, 393–403 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xia Y, Cortez-Retamozo V, Niederkofler V, Salie R, Chen S, Samad TA, Hong CC, Arber S, Vyas JM, Weissleder R, Pittet MJ, Lin HY, Dragon (Repulsive Guidance Molecule b) Inhibits IL-6 Expression in Macrophages. The Journal of Immunology. 186, 1369–1376 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Juneja VR, McGuire KA, Manguso RT, LaFleur MW, Collins N, Haining WN, Freeman GJ, Sharpe AH, PD-L1 on tumor cells is sufficient for immune evasion in immunogenic tumors and inhibits CD8 T cell cytotoxicity. J Exp Med. 214, 895–904 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spencer CN, McQuade JL, Gopalakrishnan V, McCulloch JA, Vetizou M, Cogdill AP, Khan MAW, Zhang X, White MG, Peterson CB, Wong MC, Morad G, Rodgers T, Badger JH, Helmink BA, Andrews MC, Rodrigues RR, Morgun A, Kim YS, Roszik J, Hoffman KL, Zheng J, Zhou Y, Medik YB, Kahn LM, Johnson S, Hudgens CW, Wani K, Gaudreau P-O, Harris AL, Jamal MA, Baruch EN, Perez-Guijarro E, Day C-P, Merlino G, Pazdrak B, Lochmann BS, Szczepaniak-Sloane RA, Arora R, Anderson J, Zobniw CM, Posada E, Sirmans E, Simon J, Haydu LE, Burton EM, Wang L, Dang M, Clise-Dwyer K, Schneider S, Chapman T, Anang N-AAS, Duncan S, Toker J, Malke JC, Glitza IC, Amaria RN, Tawbi HA, Diab A, Wong MK, Patel SP, Woodman SE, Davies MA, Ross MI, Gershenwald JE, Lee JE, Hwu P, Jensen V, Samuels Y, Straussman R, Ajami NJ, Nelson KC, Nezi L, Petrosino JF, Futreal PA, Lazar AJ, Hu J, Jenq RR, Tetzlaff MT, Yan Y, Garrett WS, Huttenhower C, Sharma P, Watowich SS, Allison JP, Cohen L, Trinchieri G, Daniel CR, Wargo JA, Dietary fiber and probiotics influence the gut microbiome and melanoma immunotherapy response. Science (2021), doi: 10.1126/science.aaz7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Geva-Zatorsky N, Sefik E, Kua L, Pasman L, Tan TG, Ortiz-Lopez A, Yanortsang TB, Yang L, Jupp R, Mathis D, Benoist C, Kasper DL, Mining the Human Gut Microbiota for Immunomodulatory Organisms. Cell. 168, 928–943.e11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xia Y, Babitt JL, Bouley R, Zhang Y, Da Silva N, Chen S, Zhuang Z, Samad TA, Brenner GJ, Anderson JL, Hong CC, Schneyer AL, Brown D, Lin HY, Dragon Enhances BMP Signaling and Increases Transepithelial Resistance in Kidney Epithelial Cells. J Am Soc Nephrol. 21, 666–677 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu S, Leung KM, Kim H-Y, Umetsu SE, Xiao Y, Albacker LA, Lee H-J, Umetsu DT, Freeman GJ, DeKruyff RH, Blockade of RGMb inhibits allergen-induced airways disease. Journal of Allergy and Clinical Immunology. 144, 94–108.e11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi Y, Zhong L, Li Y, Chen Y, Feng S, Wang M, Xia Y, Tang S, Repulsive Guidance Molecule b Deficiency Induces Gut Microbiota Dysbiosis and Increases the Susceptibility to Intestinal Inflammation in Mice. Frontiers in Microbiology. 12 (2021) (available at https://www.frontiersin.org/article/10.3389/fmicb.2021.648915). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tan CL, Kuchroo JR, Sage PT, Liang D, Francisco LM, Buck J, Thaker YR, Zhang Q, McArdel SL, Juneja VR, Lee SJ, Lovitch SB, Lian C, Murphy GF, Blazar BR, Vignali DAA, Freeman GJ, Sharpe AH, PD-1 restraint of regulatory T cell suppressive activity is critical for immune tolerance. Journal of Experimental Medicine. 218 (2020), doi: 10.1084/jem.20182232. [DOI] [PMC free article] [PubMed] [Google Scholar]
Extended References
- 30.Lo M, Kim HS, Tong RK, Bainbridge TW, Vernes J-M, Zhang Y, Lin YL, Chung S, Dennis MS, Zuchero YJY, Watts RJ, Couch JA, Meng YG, Atwal JK, Brezski RJ, Spiess C, Ernst JA, Effector-attenuating Substitutions That Maintain Antibody Stability and Reduce Toxicity in Mice*. Journal of Biological Chemistry. 292, 3900–3908 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S, Mooney M, Rubinstein L, Shankar L, Dodd L, Kaplan R, Lacombe D, Verweij J, New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1). European Journal of Cancer. 45, 228–247 (2009). [DOI] [PubMed] [Google Scholar]
- 32.Wu M, McNulty NP, Rodionov DA, Khoroshkin MS, Griffin NW, Cheng J, Latreille P, Kerstetter RA, Terrapon N, Henrissat B, Osterman AL, Gordon JI, Genetic determinants of in vivo fitness and diet responsiveness in multiple human gut Bacteroides. Science. 350, aac5992 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, Knight R, Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. The ISME Journal. 6, 1621–1624 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, Bai Y, Bisanz JE, Bittinger K, Brejnrod A, Brislawn CJ, Brown CT, Callahan BJ, Caraballo-Rodríguez AM, Chase J, Cope EK, Da Silva R, Diener C, Dorrestein PC, Douglas GM, Durall DM, Duvallet C, Edwardson CF, Ernst M, Estaki M, Fouquier J, Gauglitz JM, Gibbons SM, Gibson DL, Gonzalez A, Gorlick K, Guo J, Hillmann B, Holmes S, Holste H, Huttenhower C, Huttley GA, Janssen S, Jarmusch AK, Jiang L, Kaehler BD, Kang KB, Keefe CR, Keim P, Kelley ST, Knights D, Koester I, Kosciolek T, Kreps J, Langille MGI, Lee J, Ley R, Liu Y-X, Loftfield E, Lozupone C, Maher M, Marotz C, Martin BD, McDonald D, McIver LJ, Melnik AV, Metcalf JL, Morgan SC, Morton JT, Naimey AT, Navas-Molina JA, Nothias LF, Orchanian SB, Pearson T, Peoples SL, Petras D, Preuss ML, Pruesse E, Rasmussen LB, Rivers A, Robeson MS, Rosenthal P, Segata N, Shaffer M, Shiffer A, Sinha R, Song SJ, Spear JR, Swafford AD, Thompson LR, Torres PJ, Trinh P, Tripathi A, Turnbaugh PJ, Ul-Hasan S, van der Hooft JJJ, Vargas F, Vázquez-Baeza Y, Vogtmann E, von Hippel M, Walters W, Wan Y, Wang M, Warren J, Weber KC, Williamson CHD, Willis AD, Xu ZZ, Zaneveld JR, Zhang Y, Zhu Q, Knight R, Caporaso JG, Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nature Biotechnology. 37, 852–857 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deng L, Kasper DL, Krick TP, Wessels MR, Characterization of the Linkage between the Type III Capsular Polysaccharide and the Bacterial Cell Wall of Group BStreptococcus *. Journal of Biological Chemistry. 275, 7497–7504 (2000). [DOI] [PubMed] [Google Scholar]
- 36.LaFleur MW, Nguyen TH, Coxe MA, Miller BC, Yates KB, Gillis JE, Sen DR, Gaudiano EF, Abosy RA, Freeman GJ, Haining WN, Sharpe AH, PTPN2 regulates the generation of exhausted CD8+ T cell subpopulations and restrains tumor immunity. Nat Immunol. 20, 1335–1347 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dixon KO, Tabaka M, Schramm MA, Xiao S, Tang R, Dionne D, Anderson Ana. C., Rozenblatt-Rosen O, Regev A, Kuchroo VK, TIM-3 restrains anti-tumour immunity by regulating inflammasome activation. Nature. 595, 101–106 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]