Abstract
Idiopathic pulmonary fibrosis is a progressive fibrotic disease characterized by excessive deposition of (myo)fibroblast produced collagen fibrils in alveolar areas of the lung. Lysyl oxidases (LOXs) have been proposed to be the central enzymes that catalyze the cross-linking of collagen fibers. Here, we report that, while its expression is increased in fibrotic lungs, genetic ablation of LOXL2 only leads to a modest reduction of pathological collagen cross-linking but not fibrosis in the lung. On the other hand, loss of another LOX family member, LOXL4, markedly disrupts pathological collagen cross-linking and fibrosis in the lung. Furthermore, knockout of both Loxl2 and Loxl4 does not offer any additive antifibrotic effects when compared to Loxl4 deletion only, as LOXL4 deficiency decreases the expression of other LOX family members including Loxl2. On the basis of these results, we propose that LOXL4 is the main LOX activity underlying pathological collagen cross-linking and lung fibrosis.
LOXL4 is the main driver of pathological collagen crosslinking in the lung and represents a compelling drug target for IPF.
INTRODUCTION
Idiopathic pulmonary fibrosis (IPF) is a progressive fibrotic disease that is associated with high morbidity and mortality (1). With limited treatment options and a median survival of less than 3 years from diagnosis and rising prevalence due to coronavirus disease 2019 pandemic (2–5), IPF represents a huge unmet medical need for the global public health system. Triggered by microinjuries to lung airway and alveolar epithelium, IPF is proposed to be the pathological manifestation of the aberrant accumulation of myofibroblasts and the deposition of collagen fibrils in the interstitial regions (6, 7). Persistent and excessive collagen fibril formation and deposition eventually destroy normal alveolar architecture and lead to loss of lung function and, ultimately, death (8, 9).
As the common and final step of most fibrotic diseases, collagen fibril formation is a complex process involving multiple posttranslational modifications (10, 11). Once synthesized in the endoplasmic reticulum, fibrillar collagen molecules are first produced as monomeric procollagen chains with hydroxylation of a variety of prolines and lysines. These monomeric molecules are then packaged and secreted in a format of triple-helix procollagen into extracellular space where both ends of these triple-helix molecules are cleaved to enable collagen maturation. Subsequently, oxidative deamination of lysine or hydroxylysine within these mature collagen molecules initiates intra- and intermolecular cross-linking to form supramolecular collagen fibril structures (12–15), a key biochemical reaction to trigger collagen fibrillogenesis (16–19).
The main enzymes that catalyze collagen cross-linking are believed to be the lysyl oxidases (LOXs), a family of five extracellular copper-dependent amine oxidases [LOX and LOX-like 1 (LOXL1) to LOXL4] (15, 20). The importance of LOXs in homeostatic collagen cross-linking was exemplified by several genetic studies. For example, when Lox was deleted, the mice die perinatally with profound connective tissue defects and decreased elastin and collagen cross-linking (21–23). On the other hand, Loxl1 knockout mice are viable, and loss of this gene leads to marked reduction of elastin cross-linking and abnormal elastic fiber homeostasis (24–29). However, its contribution to collagen cross-linking appears to be context dependent (24–29). More recently, Loxl3 deletion in mice has only led to a minor reduction of collagen cross-linking (30, 31). The status of collagen cross-links was not reported in Loxl2 or Loxl4 knockout mice (32, 33). Overall, homeostatic collagen cross-links appear to be still largely formed in these LOX-deficient animals, indicating some functional compensatory mechanisms between LOX family members.
Given their central role in collagen cross-linking, LOXs have been perceived as potential therapeutic targets for fibrosis and cancer (34). A LOX family member, LOXL2, has attracted a great deal of attention and enthusiasm within the biomedical community on its role as the main driver of pathological collagen cross-linking in preclinical models due to the marked antifibrotic effects of antibody-based inhibition of its LOX activity (35). However, simtuzumab, a humanized LOXL2-specific blocking monoclonal antibody, has not shown any clinical benefits in several fibrotic indications including IPF (36–41). While the futility of these clinical trials has been largely attributed to the imperfect pharmacological profile of simtuzumab (20, 42), the functional role and importance of LOXs to pathological collagen cross-linking remain unresolved, and the genetic evidence to establish the causal relationship among LOXs, pathological collagen cross-linking, and lung fibrosis is lacking. To address these questions and reconcile the conflicting findings regarding the biology of collagen cross-linking, we therefore developed conditional knockout (cKO) mouse models, where Lox genes can be deleted in adulthood, to dissect the role of LOXL2 and another LOX family member, LOXL4, in pathological collagen cross-linking and lung fibrosis.
RESULTS
Expression of LOX family members in human IPF lungs
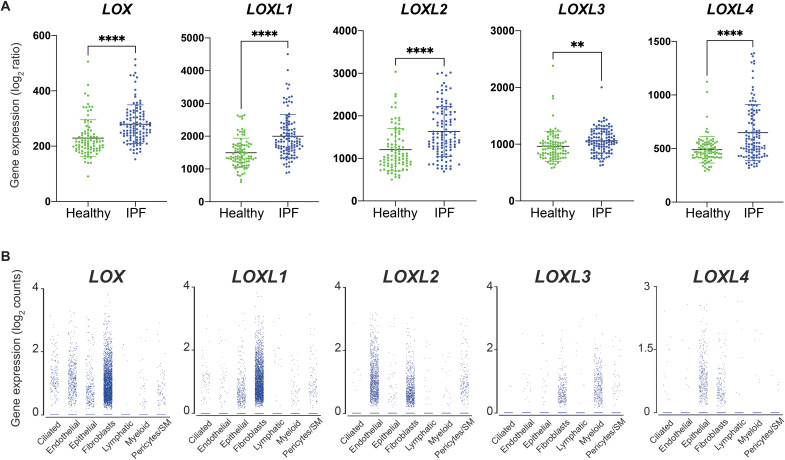
We first evaluated the expression of all five LOX family members in human IPF lungs by querying their expression in a previously reported IPF lung transcriptome dataset from human patients (GSE31962) (43). The result shows that the expression of LOX, LOXL1, LOXL2, and LOXL4 is increased in IPF lungs (Fig. 1A). LOXL3 expression appears to be only slightly increased while the trend is statistically significant (Fig. 1A). In addition, we determined the cellular source of LOXs in IPF lungs by analyzing their expression in a published single-cell transcriptome dataset (44). The result showed that the transcripts of all five LOX family members can be found in multiple cell types (Fig. 1B), suggesting that a ubiquitous promoter activity such as Rosa26 may be needed to genetically dissect the role of LOXL2, as well as other LOXs, in pathological collagen cross-linking and lung fibrosis in a mouse model.
Fig. 1. Expression of LOX family members in human IPF lungs.
(A) Expression of LOX, LOXL1, LOXL2, LOXL3, and LOXL4 in healthy and IPF human lungs. (B) Expression of LOX, LOXL1, LOXL2, LOXL3, and LOXL4 in individual cell type in human IPF lungs. SM, smooth muscle cells. Data represent means ± SD. **P < 0.01 and ****P < 0.0001. P value is calculated using one-way ANOVA.
Characterization of collagen cross-linking in the bleomycin model of lung fibrosis
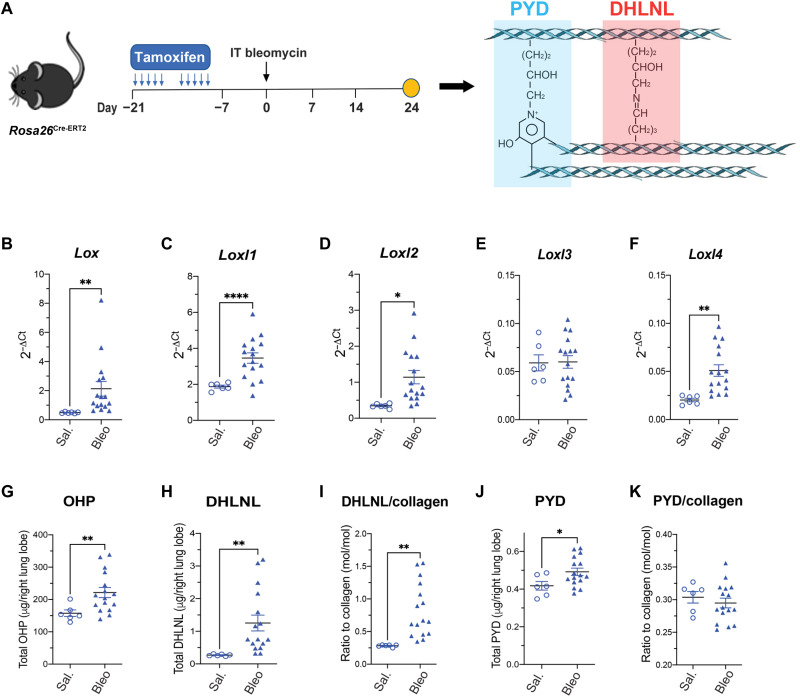
A prerequisite of genetic dissection of the role of LOXs in pathological collagen cross-linking and lung fibrosis is a preclinical animal model that can recapitulate the increased LOX expression and collagen cross-links observed in human patients with IPF (Fig. 1A) (42). In this regard, we chose to characterize the bleomycin model as it is the most widely used mouse model for lung fibrosis (45). Specifically, we challenged the mice hemizygous for a Cre-ERT2 fusion transgene driven by the Rosa26 promoter (Rosa26Cre-ERT2) with the treatment of tamoxifen and intratracheally administered bleomycin in a sequential order (Fig. 2A) because this strain will be used to generate Loxl cKO mice. In agreement with our findings in human IPF lungs (Fig. 1A), we observed that the expression of Lox, Loxl1, Loxl2, and Loxl4, but not Loxl3, was elevated in the bleomycin-challenged mouse lungs (Fig. 2, B to F). Furthermore, we quantified collagen deposition and cross-links in the Rosa26Cre-ERT2 mice treated with tamoxifen and saline or bleomycin (Fig. 2A). As expected, we found that total lung collagen content, measured by hydroxyproline (OHP), was increased remarkably in the bleomycin group (Fig. 2G). In addition, we found that total and normalized (ratio of total cross-links to total collagen) divalent [dihydroxylysinonorleucine (DHLNL)] collagen cross-links were increased in mouse lungs challenged by bleomycin compared to saline group (Fig. 2, H and I). Notably, these observations are consistent with a previous report of collagen cross-linking in human fibrotic lungs (42). On the other hand, total and normalized trivalent [pyridionoline (PYD)] collagen cross-links appeared to be either slightly increased (total) (Fig. 2J) or unchanged (normalized) (Fig. 2K), which is a previously documented phenomenon in the bleomycin model (46). Nevertheless, these results support the bleomycin model as an appropriate model to investigate the role of LOXL2, as well as other LOXs, in pathological collagen cross-linking and lung fibrosis.
Fig. 2. Collagen cross-linking and Lox/Loxl expression in the bleomycin model.
(A) Schematic regime of Rosa26Cre-ERT2 mice under intratracheal (IT) bleomycin challenge. Lungs were harvested for terminal analyses on day 24 after bleomycin challenge. (B to F) mRNA expression of Lox family members from lung tissue. (G) Lung total OHP. (H to K) Quantification of lung collagen cross-linking by (H) total DHLNL, (I) DHLNL normalized by collagen, (J) total PYD, and (K) PYD normalized by collagen. Saline, n = 6; bleomycin, n = 16. Data represent means ± SD. *P < 0.05, **P < 0.01, and ****P < 0.0001. P value is calculated using unpaired, two-tailed t test (B) to (K).
Modest contribution of LOXL2 to pathological collagen cross-linking in the lung
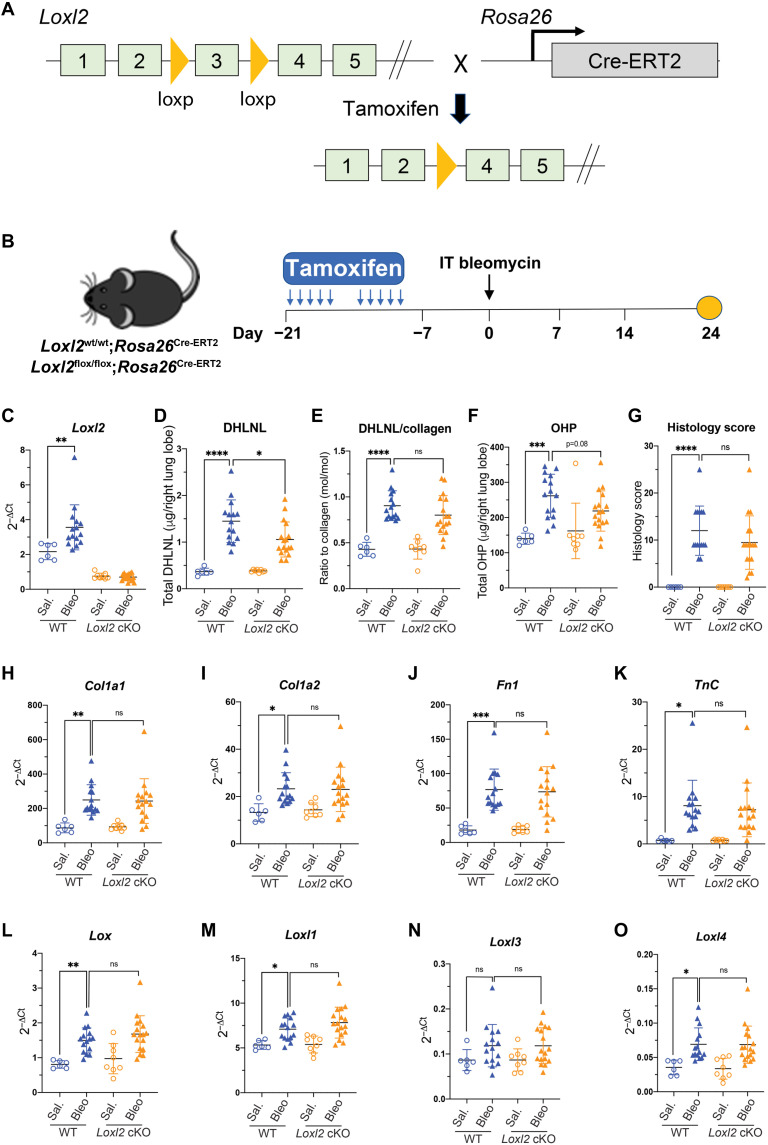
To bypass postnatal lethality reported in Loxl2-deficient mice (32), we generated mice homozygous for the floxed (cKO) or wild-type (WT) Loxl2 alleles and hemizygous for a Cre-ERT2 fusion transgene driven by the Rosa26 promoter. In these cKO mice, both Loxl2 alleles can be deleted when tamoxifen is administered (Fig. 3A). Thus, we investigated whether inducible deletion of Loxl2 affects pathological collagen cross-linking and lung fibrosis in the bleomycin model (Fig. 3B).
Fig. 3. LOXL2 modestly contributes to pathological collagen cross-linking but not fibrosis in the bleomycin model.
(A) Schematic conditional Loxl2 targeting. The exon 3 of Loxl2 gene has been flanked by loxP site. The Loxl2flox/flox mice breed with Rosa26Cre-ERT2 mice to generate Loxl2 cKO mice (Loxl2flox/flox; Rosa26Cre-ERT2) where Cre promoted recombination can be induced by tamoxifen. (B) Schematic regime of tamoxifen-induced Loxl2 deletion followed by intratracheal bleomycin challenge. Lungs were harvested for terminal analyses on day 24 after bleomycin challenge. (C) mRNA expression shows the efficient deletion of Loxl2 in the lungs from cKO mice. (D and E) Quantification of lung collagen cross-linking by (D) total DHLNL and (E) DHLNL normalized by collagen. (F and G) Quantification of lung fibrosis by (F) total OHP and (G) histology score. (H to K) mRNA expression of selective profibrotic genes from lung tissue. (L to O) mRNA expression of Lox family members from lung tissue. WT: saline, n = 6; bleomycin, n = 15; Loxl2 cKO: saline, n = 8; bleomycin, n = 17. Data represent means ± SD. *P < 0.05, **P < 0.01, and ****P < 0.0001. P value is calculated using one-way analysis of variance (ANOVA). ns, not significant.
We first confirmed that tamoxifen induced substantial loss of Loxl2 transcript in the lungs in the presence or absence of bleomycin when compared to WT controls (Fig. 3C). Next, we measured collagen cross-links and total OHP and found that Loxl2 deletion only results in a modest but statistically significant reduction of total DHLNL in the bleomycin-challenged mouse lungs (Fig. 3D). The other readouts including normalized DHLNL and total OHP levels are numerically lower in the bleomycin-treated Loxl2 cKO lungs, while this trend is not statistically significant (Fig. 3, E and F). In addition, we observed that total and normalized PYD level, histology score, and the expression of profibrotic genes such as Col1a1, Col1a2, Fn1, and Tnc were not affected by Loxl2 deletion (Fig. 3, G to K, and fig. S1). We also found that Loxl2 deletion does not affect the expression of other Lox family members in the lungs (Fig. 3, L to O), ruling out the possibility that the expression of other Lox family members may be increased to compensate for the loss of LOXL2. Overall, these data strongly argue against the proposed important role of LOXL2 in lung fibrosis (35) and suggest that other Lox family members should be explored for their contribution to this pathological process.
LOXL4 is the main LOX underlying pathological collagen cross-linking and lung fibrosis
To identify the main LOX underlying pathological collagen cross-linking and lung fibrosis, we switched our focus to LOXL4 due to the following two reasons: (i) LOXL4 expression is highly increased in both human IPF lungs and the bleomycin-challenged fibrotic mouse lungs (Figs. 1A and 2F); and (ii) unlike LOX and LOXL1, LOXL4 appears not to contribute to normal homeostasis of connective tissues (33). We first investigated upstream regulatory mechanisms underlying LOXL4 induction, as this may be relevant to its potential role in lung fibrosis. Through a screen of a variety of profibrotic and proinflammatory cytokines and stimuli, we found that LOXL4 expression can be induced in lung epithelial cells by transforming growth factor β (TGFβ) ligands such as TGFβ1, TGFβ2, and TGFβ3, and in lung fibroblasts by TGFβ1/2/3 and interleukin-1β (IL-1β) (fig. S2), which is consistent with a previous report (47) and suggests that LOXL4 expression could be induced by both proinflammatory and profibrotic signals. Next, we explored whether LOXL4 affects several profibrotic processes including fibroblast activation, proliferation, collagen production, and epithelial-mesenchymal transition (EMT) in vitro. We did not observe that either treatment of recombinant LOXL4 protein or overexpression of LOXL4 affects these processes in vitro (figs. S3 and S4). Complementarily, we did not find that genetic or pharmacological inhibition of LOXL2 or LOXL4 affects EMT and lung fibroblast activation in vitro (fig. S5). Collectively, these results argue against a direct role of LOXL4 in these profibrotic processes, which have been suggested for other LOX family members (32, 48, 49).
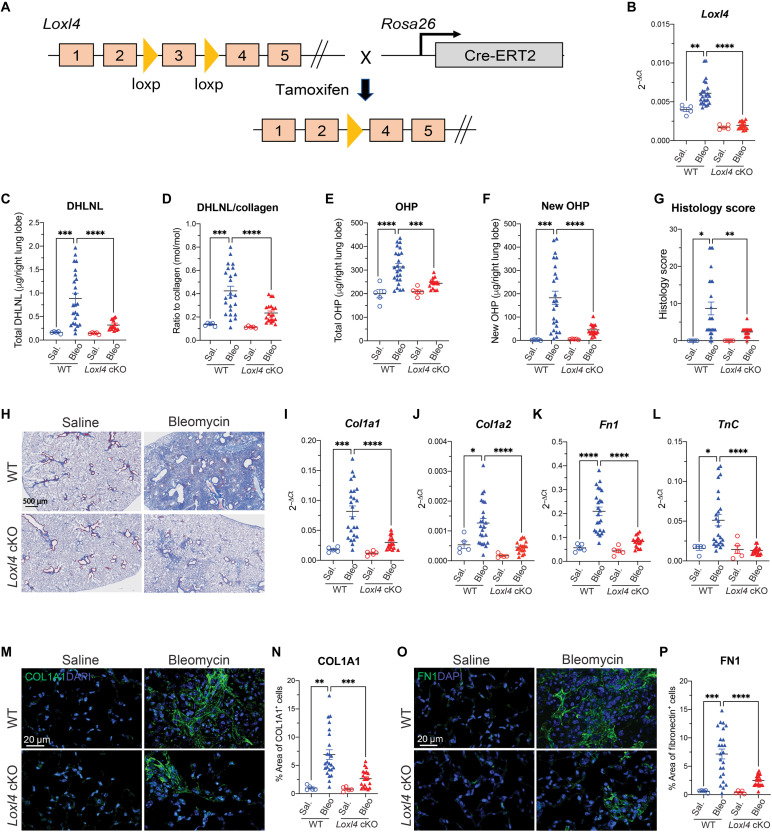
To assess its in vivo role in lung fibrosis, we therefore generated Loxl4 cKO mice (Loxl4flox/flox; Rosa26Cre-ERT2) as depicted (Fig. 4A) and investigated whether inducible deletion of Loxl4 affects pathological collagen cross-linking and lung fibrosis in the bleomycin model (Fig. 4A and fig. S6A). The data showed a markedly reduction of total and normalized DHLNL by more than 70% (Fig. 4, C and D) in bleomycin-challenged lungs from Loxl4 cKO mice. Notably, Loxl4 deletion appears to decrease total and normalized PYD in bleomycin-treated fibrotic lungs as well (fig. S6, B and C). Lung fibrosis quantified by total OHP, newly synthesized OHP, and histology score was also greatly decreased in Loxl4 cKO mice challenged with bleomycin (Fig. 4, E to H). We also observed that the bleomycin-induced expression of profibrotic genes was almost normalized in the lungs from Loxl4 cKO mice (Fig. 4, I to L). In addition, we confirmed the decrease in type I collagen and fibronectin protein levels in Loxl4 cKO mouse fibrotic lungs through immunofluorescent analysis (Fig. 4, M to P). These results, therefore, not only suggest a requirement of collagen cross-linking for lung fibrosis but also support LOXL4 as the main LOX-mediating pathological collagen cross-linking and fibrosis in the lung.
Fig. 4. Loxl4 cKO mice are protective against bleomycin-induced lung fibrosis.
(A) Schematic conditional Loxl4 targeting. The exon 3 of Loxl4 gene has been flanked by loxP site. The Loxl4flox/flox mice breed with Rosa26Cre-ERT2 mice to generate Loxl4 cKO mice (Loxl4flox/flox; Rosa26Cre-ERT2) where Cre promoted recombination can be induced by tamoxifen. (B) mRNA expression shows the efficient deletion of Loxl4 in the lungs from cKO mice. (C and D) Quantification of lung collagen cross-linking by (C) total DHLNL and (D) DHLNL normalized by collagen. (E and F) Quantification of lung fibrosis by (E) total OHP and (F) newly synthesized OHP. (G) Histology score of lung fibrosis lesion. (H) Representative images of trichrome staining of lung fibrosis. (I to L) mRNA expression of selective profibrotic genes from lung tissue. (M and N) (M) Representative immunofluorescent images of COL1A1 in lung tissue with quantification of (N) percentage of COL1A1+ area. (O and P) (O) Representative immunofluorescent images of FN1 in lung tissue with quantification of (P) percentage of FN1+ area. WT: saline, n = 5; bleomycin, n = 24; Loxl4 cKO: saline, n = 5; bleomycin, n = 17. Data represent means ± SD. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. P value is calculated using one-way ANOVA. DAPI, 4′,6-diamidino-2-phenylindole.
LOXL4 deficiency disrupts profibrotic YAP/TAZ and TGFβ signaling in myofibroblasts during lung fibrosis
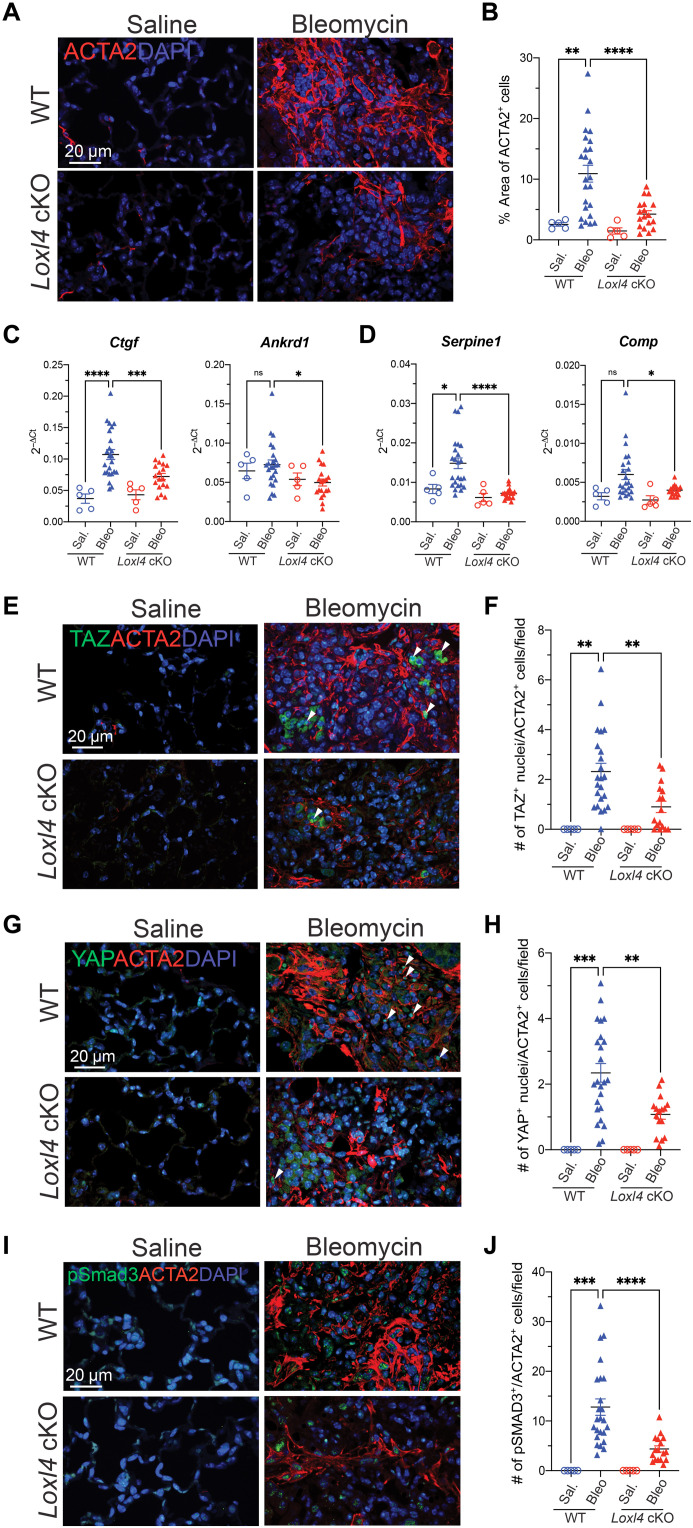
The notable antifibrotic phenotype of Loxl4 cKO mice prompts us to investigate the mechanism underlying LOXL4’s central role in lung fibrosis. In this regard, we noticed that α–smooth muscle actin (ACTA2)–positive myofibroblasts were markedly reduced in the fibrotic lungs from bleomycin-challenged Loxl4 cKO mice (Fig. 5, A and B), and this phenomenon cannot be attributed to decreased proliferation or increased apoptosis of lung myofibroblasts in the absence of LOXL4 (fig. S7). As LOXL4 appears not to contribute to lung fibroblast activation directly (figs. S3 to S5), we speculate that LOXL4 and its dependent pathological collagen cross-linking may promote myofibroblast activation via indirect stimulation of profibrotic signaling pathways such as yes-associated protein (YAP)/transcriptional coactivator with PDZ-binding motif (TAZ) and TGFβ signaling in vivo (50–55).
Fig. 5. LOXL4 deficiency disrupts TGFβ signaling and inactivates myofibroblasts in the bleomycin injured lungs.
(A and B) (A) Representative immunofluorescent images of ACTA2 in lung tissue with quantification of (B) percentage of ACTA2+ area. (C and D) mRNA expression of selective (C) YAP/TAZ target genes and (D) TGFβ target genes from lung tissue. (E and F) (E) Representative immunofluorescent images of TAZ and ACTA2 in lung tissue with quantification of (F) number of TAZ+ nuclei/ACTA2+ cells per field. (G and H) (G) Representative immunofluorescent images of YAP and ACTA2 in lung tissue with quantification of (H) number of YAP+ nuclei/ACTA2+ cells per field. Arrowheads present (E) TAZ+ nuclei/ACTA2+ cells and (G) YAP+ nuclei/ACTA2+ cells. (I and J) (I) Representative immunofluorescent images of pSMAD3 and ACTA2 in lung tissue with quantification of (J) number of pSMAD3+/ACTA2+ cells per field. Data in (B), (F), (H), and (J) represent mean value of five random fields per image. Data represent means ± SD. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. P value is calculated using one-way ANOVA.
This hypothesis was initially supported by gene expression profiling, as we found the YAP/TAZ and TGFβ target gene expression sharply declined in the fibrotic lungs from Loxl4 cKO mice (Fig. 5, C and D). Immunofluorescent analysis further confirmed that nuclear signal of YAP and TAZ, a hallmark of YAP/TAZ activation, decreased in the myofibroblasts of fibrotic lungs from Loxl4 cKO mice (Fig. 5, E to H). In addition, we examined the abundance of phosphorylated SMAD3 (pSMAD3), a hallmark of TGFβ activation, in the lungs from WT or Loxl4 cKO mice challenged with bleomycin and found lower pSMAD3 signal in the myofibroblasts of fibrotic lungs from Loxl4 cKO mice (Fig. 5, I and J). We also observed decreased expression of TGFβ ligands such as Tgfb2 and Tgfb3 but not Tgfb1 in the lungs from bleomycin-challenged Loxl4 cKO mice (fig. S8). Hence, our results support an indirect role of LOXL4 in YAP/TAZ and TGFβ activation during lung fibrosis and implicate that pathological collagen cross-linking may be important in amplification of these key profibrotic signaling pathways, myofibroblast activation, and ultimate formation of a profibrotic microenvironment in the lung.
LOXL2 deficiency does not further decrease pathological collagen cross-linking and lung fibrosis in the absence of LOXL4
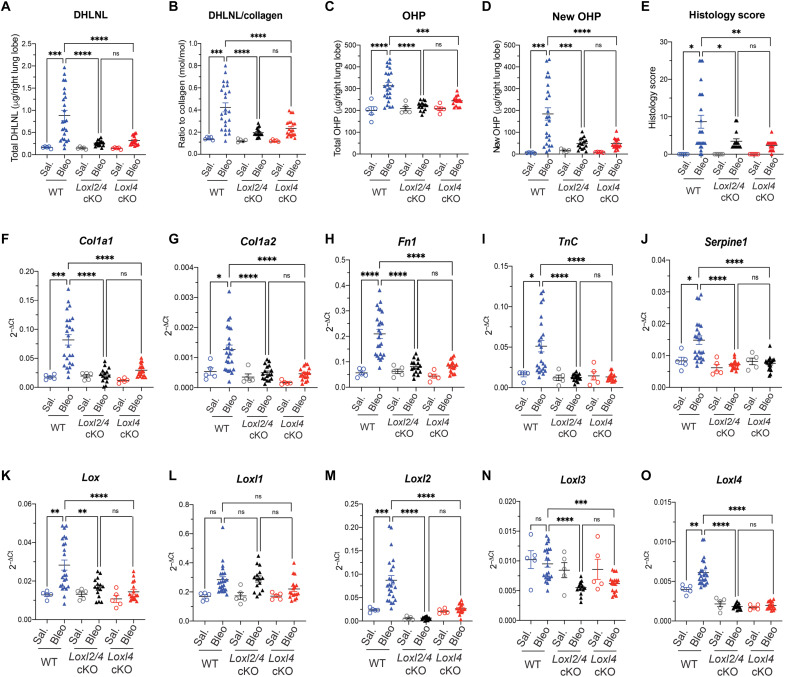
As both LOXL2 and LOXL4 contribute to pathological collagen cross-linking, we asked whether LOXL2 and LOXL4 may synergistically mediate lung fibrosis. To address this question, we crossed Loxl2 cKO and Loxl4 cKO mice to generate Loxl2/4 cKO(Loxl2flox/flox; Loxl4flox/flox; Rosa26Cre-ERT2) mice, which were then tested along with Loxl4 cKO mice in the same bleomycin-challenged experiment (fig. S6A). We found that simultaneous deletion of Loxl2 and Loxl4 did not further reduce pathological collagen cross-linking (DHLNL and PYD) when compared to single deletion of Loxl4 (Fig. 6, A and B, and fig. S6, D and E). Similarly, lung fibrosis measured by total OHP, newly synthesized OHP, and histology score was comparable between Loxl2/4 cKO and Loxl4 cKO mice (Fig. 6, C to E). We observed little difference in the expression of profibrotic genes including Col1a1, Col1a2, Fn1, TnC, and Serpine1 either (Fig. 6, F to J). In this regard, we noticed that Loxl4 deletion decreases the expression of all the other LOX family members including Lox, Loxl2, and Loxl3 but not Loxl1 in the bleomycin-challenged fibrotic mouse lungs (Fig. 6, K to O). This finding may help explain little add-on or synergistic effect of LOXL2 deficiency on top of LOXL4 deficiency with respect to pathological collagen cross-linking and lung fibrosis and further support a critical role of LOXL4 in perpetuating a profibrotic microenvironment.
Fig. 6. LOXL2 deficiency has no extra protective effects against bleomycin-induced pathological collagen cross-linking and lung fibrosis in the absence of LOXL4.
(A and B) Quantification of lung collagen cross-linking by (A) total DHLNL and (B) DHLNL normalized by collagen. (C to E) Quantification of lung fibrosis by (C) total OHP, (D) newly synthesized OHP, and (E) histology score of fibrosis lesion. (F to J) mRNA expression of selective profibrotic genes from lung tissue. (K to O) mRNA expression of Lox family members from lung tissue. WT: saline, n = 5; bleomycin, n = 24; Loxl2/4 cKO: saline, n = 5; bleomycin, n = 15; Loxl4 cKO: saline, n = 5; bleomycin, n = 17. Data represent means ± SD. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. P value is calculated using one-way ANOVA.
DISCUSSION
Since its mouse surrogate exhibited impressive antifibrotic effects in preclinical models (35), simtuzumab had garnered considerable enthusiasm from the entire biomedical community as it was regarded as a breakthrough to directly target collagen fibrillogenesis. However, the initial excitement of this molecule abated with a series of setback in clinical trials. Several reasons have been proposed to explain the futility of simtuzumab in human patients who include, but not limited to, potential intracellular activity of LOXL2, the poor potency of simtuzumab, and the insufficient capability of antibody-based therapeutics to penetrate fibrotic milieu. Our data presented in this study, however, argue that LOXL2 may not be a suitable target for fibrotic diseases such as IPF at all as the LOXL2 contribution to pathological collagen cross-linking and lung fibrosis in the bleomycin model appears to be either marginal or null, which is conceivably not translatable to human patients. Although the limitations of simtuzumab’s molecular profile may still account for its poor performance in human trials, our study exemplifies the utmost necessity of genetic validation of drug targets when possible.
So, which LOX could be the culprit of pathological collagen cross-linking and lung fibrosis if there was any? Genetic dissection helped us find LOXL4 as a previously underappreciated LOX family member that functions as a master regulator of pathological collagen cross-linking underlying lung fibrogenesis. While LOXL4 deficiency leads to a marked decrease in pathological collagen cross-link underlying lung fibrosis, we did not observe any disturbance of homeostatic collagen cross-linking and deposition in the lung in the absence of LOXL4. Furthermore, we did not notice any overtly adverse phenotypes in Loxl4 cKO mice either. Consistent to these observations, Loxl4 knockout mice appear to be largely normal as well (33). These findings, thus, position LOXL4 in a “sweet” spot for pharmacological manipulation of pathological collagen cross-linking driving lung fibrosis progression without perturbing normal homeostasis of connective tissues, which is a serious safety concern of targeting this biology.
Another surprising but not unexpected finding from our study is that LOXL4-mediated pathological collagen cross-linking appears to be the main driver of profibrotic microenvironment in the lung. On the basis of a series of elegant in vitro studies (51, 53, 56–60), it has long been proposed that fibrosis and fibrosis-triggered tissue stiffening can induce a self-sustaining fibroblast activation to nucleate a highly fibrotic microenvironment that forms a vicious feedback loop to perpetuate the disease progression. So, to the best of our knowledge, Loxl4 cKO mouse model provides the first evidence to establish a direct causal relationship between fibrosis and fibroblast activation by genetic manipulation of collagen matrix. Furthermore, the indispensability of LOXL4 in pathological collagen cross-linking and lung fibrosis supports the concept that the matrix-fibroblast cross-talk driving the progression of lung fibrosis and suggests that this mechanism may be druggable by LOXL4 inhibition.
Last, we propose that the Loxl4 cKO mouse model may represent a unique opportunity to delineate the reciprocal relationship between fibrosis and other pathophysiological processes. Fibrosis has long been viewed as the chronic consequence or outcome of other pathophysiological processes such as inflammation, autoimmunity, metabolic imbalance, and cancer, in that these abnormalities usually precede fibrosis and selective modulation of these processes can prevent fibrosis (6, 8, 9). However, whether fibrosis can perpetuate the progression of these pathological processes remains largely unknown probably because of the lack of the genetic means to manipulate fibrosis in a specific manner. Hence, the Loxl4 cKO mouse model may be an invaluable genetic tool to overcome this technical barrier by providing an effective means to manipulate pathological collagen cross-linking and fibrosis directly. Additional studies are therefore warranted to test this mouse model in a wide spectrum of disease settings where pathological collagen cross-linking occurs.
In sum, our data provide the genetic evidence that LOXL4, but not LOXL2, is required for pathological collagen cross-linking and lung fibrosis. This discovery not only establishes the causal relationship between pathological collagen cross-linking and lung fibrosis but also supports LOXL4 as a previously unrecognized therapeutic target for fibrotic lung diseases such as IPF.
MATERIALS AND METHODS
Generation of inducible Loxl2 and Loxl4 knockout mouse
Homologous recombination and mouse embryonic stem (ES) cell technology (61–63) was used to generate a genetically modified mouse strain with Loxl2 and Loxl4 cKO alleles. To generate Loxl2-flox/flox alleles, a gene-targeting vector constructed with a 2020–base pair (bp) arm of 5′ homology corresponds to GRCm38/mm10 chr14: 69,642,553 to 69,644,572 and a 2006-bp arm of 3′ homology corresponds to chr14: 69,644,987 to 69,646,992. The 414-bp region flanked by loxP sites (exon 3) corresponds to chr14: 69,644,573 to 69,644,986. To generate Loxl4-flox/flox alleles, a gene-targeting vector constructed with a 1354-bp arm of 5′ homology corresponds to GRCm38/mm10 chr19: 42,607,859 to 42,609,212 and a 2154-bp arm of 3′ homology corresponds to chr19: 42,605,266 to 42,607,422. The 436-bp region flanked by loxP sites (exon 3) corresponds to chr19: 42,607,423 to 42,607,858.
The final vector was confirmed by DNA sequencing, linearized, and used to target C57BL/6J ES cells by standard methods (G418-positive and ganciclovir-negative selection) (64). C57BL/6J ES cells (65) were electroporated with 20 μg of linearized targeting vector DNA and cultured under drug selection. Positive clones were identified by long-range polymerase chain reaction (PCR), followed by sequence confirmation. Correctly targeted ES cells were subjected to karyotyping. Euploid gene-targeted ES cell clones were treated with adeno-FLP to remove glycolytic enzyme phosphoglycerate kinase (PGK) neomycin, ES cell clones were tested to identify clones with no copies of the PGK neomycin cassette, and the correct sequence of the targeted allele was verified. The presence of the Y chromosome was verified before microinjection into albino C57BL/6J embryos. Germline transmission was obtained after crossing resulting chimeras with C57BL/6J females. Genomic DNA from pups born was screened by long-range PCR to verify the desired gene-targeted structure before mouse colony expansion. Genotyping primers used to identify Loxl2 germline transmission were the following: primer 1, ATCCCTTTGTAAACCGTCAG; primer 2, AGTAGATATTGTCCAACCAGATG; primer 3, ATCCTTCAACTTGCCTGTG; expected WT 382 bp, conditional 416 bp, and knockout 359 bp. Genotyping primers used to identify Loxl4 germline transmission were the following: primer 1, GCAGACACTCAGAAGATGTT; primer 2, CTTAGGAAGTAACTGATGGACA; primer 3, TGCTGTTGTTTGGCTTGA; expected WT 363 bp, conditional 449 bp, and knockout 298 bp.
Induced excision of Loxl2 and Loxl4 in adult mice
All animal studies were conducted in accordance with the Guide for the Care and Use of Laboratory Animals, published by the National Institutes of Health (NIH) (NIH Publication 8523, revised 1985). The Institutional Animal Care and Use Committee at Genentech reviewed and approved all animal protocols. To generate Loxl2 and Loxl4 cKO mice, Loxl2-flox/flox and Loxl4-flox/flox mice were bred to the Rosa26-Cre-ERT2 mouse strain (66) to produce Loxl2-flox/flox;Rosa26-Cre-ERT2 (Loxl2 cKO), Loxl4-flox/flox;Rosa26-Cre-ERT2(Loxl4 cKO), Loxl2-flox/flox; Loxl4-flox/flox;Rosa26-Cre-ERT2 (Loxl2/4 cKO) mice. Loxl2-WT/WT; Loxl4-WT/WT; Rosa26-Cre-ERT2 (WT) mice were used as controls in this study. Mice from all groups were injected intraperitoneally daily for five consecutive days with tamoxifen (T5648, Sigma-Aldrich) in sunflower oil (S5007-1L, Sigma-Aldrich) at 80 mg/kg. Animals then rest for 2 days and received second dose of tamoxifen again for five consecutive days. Subsequent studies were initiated at least 1 week after the completion of tamoxifen treatment.
Bleomycin model of lung fibrosis
The study was carried out in compliance with the Animal Research: Reporting of in vivo Experiments (ARRIVE) guidelines. Adult mice (>12 weeks) were randomized on the basis of prestudy weights to minimize variance between experimental and control groups. For intratracheal dosing, all mice were lightly anesthetized with isoflurane in an induction chamber. Once anesthetized, the animals were removed from the chamber and manually restrained, the mouth of the animal was opened, and the tongue was set aside. A 1-ml syringe with 50 μl of sterile injectable isotonic saline or bleomycin [0.72 U/kg (0703-3155-01, Teva) in 50 μl sterile isotonic saline] was connected to a 24-gauge gavage needle. The gavage needle was inserted into the trachea, and a dose of either vehicle or bleomycin was delivered intratracheally. After delivery, animals were monitored continuously until fully awake and ambulatory. Mouse whole lungs were harvested at day 24 after bleomycin administration. In saline group, n = 5 to 6 in each genotyping. In bleomycin group, n = 16 in Loxl2 cKO, n = 17 in Loxl4 cKO, n = 15 in Loxl2/4 cKO, and n = 24 in WT control littermates.
Histology analysis
Formalin-fixed samples of mouse lungs were embedded as a whole, processed to one slide per animal, and stained with Masson’s trichrome. The extent of pulmonary fibrosis was scored according to the following criteria:
1) Interstitial fibrosis pattern—number of foci: 0, none detected; 1, ≤10; 2, ≤15; 3, >15/all sections, but distinct; 4, multifocally coalescent or locally extensive; 5, diffuse;
2) Interstitial fibrosis—size of foci: 0, none detected; 1, largest focus ≤ area of ~2 alveolar spaces; 2, largest focus ≤ area of ~4 alveolar spaces; 3, coalescent (>4 patent alveolar spaces); 4, locally extensive (60 to 90% of an entire lobe); 5, diffuse (>90% of an entire lobe);
3) Total scores: number of foci × size of foci.
Immunofluorescence staining
Four-micrometer sections of formalin-fixed and paraffin-embedded specimens were deparaffinated, followed by antigen retrieval using Target Retrieval (S1700, Dako). The sections were subsequently blocked with phosphate-buffered saline plus 10% donkey serum and 3% bovine serum albumin and stained with anti–ACTA2-Cy3(1:400; C6198, Sigma-Aldrich), anti-collagen 1 alpha 1 (COL1A1) (1:200; ab279711, Abcam), anti-fibronectin 1 (FN1) (1:200; SAB5700724, Sigma-Aldrich), anti-pSMAD3 (0.5 μg/ml; LS-B64-50, LifeSpan BioSciences), anti-TAZ (1:400; ZRB1260, Sigma-Aldrich), anti-YAP (1:200; 4912, Cell Signaling Technology), anti-Ki67 (1:200; 14-5698-82, eBioscience), or anti-γH2AX (1:200; MAB15111, Abnova).
Collagen cross-linking and OHP measurement
The right lung lobes were weighted after harvested and snap-frozen in liquid nitrogen. Lung lobes were then reduced using 0.4 M sodium borohydride (MP Biomedicals, CA) in 10 μM NaOH (Sigma-Aldrich, MO) and washed by 0.2 M acetic acid (Thermo Fisher Scientific, IL) and high-performance liquid chromatography (HPLC) grade water (Thermo Fisher Scientific, MA) until the pH value was neutral. Lung lobes were freeze-dried and hydrolyzed in 6 N HCl (Thermo Fisher Scientific, IL) at 110°C overnight. After cooled to room temperature, the hydrolysates were then dried using vacuum concentrator.
For analysis of collagen cross-links, the dried hydrolysates were reconstituted with 7% heptafluorobutyric acid (HFBA) (Thermo Fisher Scientific, IL) and centrifuge. Clean supernatants were transferred to HPLC vial for LC–tandem mass spectrometry analysis. Chromatographic separation of OHP, PYD, and DHLNL were performed on a reverse-phase column (Zorbax SB-Aq 5 μm, 3.0 mm × 150 mm) using Sciex ExionLC system (Sciex, CA). The temperatures of the column oven and autosampler were set at 40° and 15°C, respectively. The LC flow rate was 0.2 ml/min. Mobile phase A is 0.1% HFBA in water, and mobile phase B is 0.1% HFBA in acetonitrile. For OHP analysis, the extracts were diluted with internal standard D3-OHP (1 μg/ml) (Quidel, CA). The HPLC was coupled to a 6500+ QTRAP mass spectrometer (Sciex, CA) operated under positive ionization mode with the following source settings: turbo ion spray source at 350°C under N2 nebulization at 20 psi, N2 heater gas at 10 psi, curtain gas at 30 psi, collision-activated dissociation gas pressure was held at medium, turbo ion spray voltage at 5500 V, declustering potential (DP) at 20 V, entrance potential at 10 V. Collision energy (CE) at 25 V for OHP, 40 V for PYD, 28 V for DHLNL, and collision cell exit potential at 10 V. Sample analysis was performed in multiple reaction monitoring mode. The transitions for OHP, PYD, and DHLNL were 132 > 68, 429 > 267, and 308 > 128, respectively. All cross-links were quantified using linear calibration curve of standards [OHP (Sigma-Aldrich, MO), DHLNL (Santa Cruz Biotechnology, CA), PYD (Quidel, CA)]. For collagen cross-link normalization, total DHLNL and PYD levels were divided by the amount of collagen in lung tissue. The collagen amount is calculated from OHP, assuming that it comprises ~14% of the total amino and imino acid content of fibrillar collagen (67).
Reverse transcription quantitative PCR
Total RNA was purified using RNeasy kit (74004, QIAGEN) and treated with deoxyribonuclease I (18047019, Life Technologies). Complementary DNA synthesis was carried out with iScript RT Supermix (1708841, Bio-Rad). Quantitative PCR was performed in technical triplicates using TaqMan assays (4304437, Thermo Fisher Scientific) for examining human gene expression and SYBR Green assays (1725124, Bio-Rad) for examining mouse gene expression. The relative standard curve method was used for quantitation, and expression levels were calculated by normalization to Hprt. Sequences of primers were provided in table S1.
Western blot
Western blot was carried out in total protein extracts as previously described (68). Equal amounts of protein lysates were separated by SDS–polyacrylamide gel electrophoresis, transferred to a nitrocellulose membrane, and subjected to immunoblotting analysis using following primary antibodies: anti-FLAG (1:1000; F3165, Sigma-Aldrich) and anti–glyceraldehyde-3-phosphate dehydrogenase (1:1000; 5174, Cell Signaling Technology).
Cell-based assays
Human lung epithelial cell line A549 (CCL-185, American Type Culture Collection) was cultured in RPMI 1640 medium supplemented with l-glutamine and 10% fetal bovine serum (FBS) (SH30070.01, Cytiva). Primary human lung fibroblasts (HLFs) were isolated from crude whole lung single-cell suspension and cultured in Dulbecco’s modified Eagle’s medium supplemented with l-glutamine and 10% FBS.
For upstream inducer screen, cells (A549 or HLF) were grown in desired culture medium until reaching 70 to 80% confluency and starved in serum-free medium overnight. Cells were then stimulated with tumor necrosis factor–α (10 ng/ml; 210-TA-020/CF, R&D Systems), IL-1β (10 ng/ml; 201-LB-005/CF, R&D Systems), IL-6 (10 ng/ml; 7270-IL-010/CF, R&D Systems), IL-13 (10 ng/ml; 213-ILB-025/CF, R&D Systems), TGFβ1 (10 ng/ml; 240-B-002/CF, R&D Systems), TGFβ2 (10 ng/ml; 302-B2-002/CF, R&D Systems), TGFβ3 (10 ng/ml; 243-B3-002/CF, R&D Systems), Wnt family member 3a (WNT3A) (500 ng/ml; 5036-WN-010/CF, R&D Systems), lipopolysaccharide (100 ng/ml; 82857-67-8, Sigma-Aldrich), or 20% FBS for 24 hours, followed by gene expression analysis using reverse transcription quantitative PCR (RT-qPCR).
For EMT and fibroblast activation assays, recombinant LOXL4 protein and FLAG-tagged LOXL4 expression plasmid were synthesized internally. Cells (A549 or HLFs) were treated with recombinant LOXL4 protein (5 μg/ml) or transfected with plasmids expressing FLAG-tagged LOXL4 by Lipofectamine 3000 (A549) (L3000001, Invitrogen) or electroporation (HLFs) using P3 primary cell 4D-Nucleofector (V4XP-3024, Lonza). Treated or transfected cells were cultured without perturbation for 48 hours, followed by cell viability analysis using CellTiter-Glo assay (G7570, Promega) and gene expression analysis using RT-qPCR.
For LOXL2/4 loss-of-function assays, small interfering RNA (siRNAs) were purchased from PerkinElmer (Dharmacon), and β-aminopropionitrile (BAPN) was synthesized internally. Transfection of siRNA as indicated was carried out at a concentration of 20 nM using RNAiMAX transfection reagent (13778075, Invitrogen). Cells (A549 or HLFs) were seeded and grown in desired culture medium for 24 hours before transfection. Transfected cells were then cultured with or without TGFβ1 (10 ng/ml) for 24 hours before 72-hour termination point. Pan-LOX activity inhibition was performed using BAPN at concentrations of 100 and 5 μM, for A549 and HLFs, respectively, with or without TGFβ1 (10 ng/ml). Treated cells were cultured without perturbation till the end of termination point that was 96 hours for A549 and 16 hours for HLFs. Harvested cells were further analyzed for gene expression using RT-qPCR.
Quantification and statistical analysis
ImageJ was used for immunofluorescent quantification of five random fields per image. GraphPad Prism was used for statistical analysis. Statistical details of experiments can be found in figure legends, including the statistical tests used and value and definition of n. Differences are considered to be statistically significant when P < 0.05.
Acknowledgments
We thank the Genentech microinjection, animal production, genetic analysis, histology and fluorescence-activated cell sorting laboratories for technical assistance and the Genentech Center for Advanced Light Microscopy for imaging. We also thank R. Pappu for critical review of this work and M. Lamoureux and B. Grellman for animal husbandry.
Funding: The funding for this project was provided by Genentech/Roche.
Author contributions: H.-Y.M. designed the study, performed the experiments, analyzed the data, and wrote the manuscript. E.-N.N. designed the study, performed the experiments, and analyzed the data. Q.L., W.R.W., and W.S. performed analysis of OHP and collagen cross-links and contributed to the data interpretation. P.C. and H.Be. contributed to the histological analysis of fibrotic lung tissues. Z.H., A.A., S.J., A.W., and H.Br. performed the bleomycin experiments. H.Br. and C.E. helped design and supervised the bleomycin studies. M.R.-G. designed the targeting vector and supervised the generation of the Loxl2 and Loxl4 cKO mouse line. L.T. and R.N. contributed to generation of Loxl2 and Loxl4 cKO mouse line. N.D. designed the study, performed the experiments, analyzed the data, wrote the manuscript with H.-Y.M., and supervised the study.
Competing interests: All authors are or were employees of Genentech/Roche. The authors declare that they have no other competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Loxl2/4 cKO mouse strains generated in this study can be provided by Genentech pending scientific review and a completed material transfer agreement. Requests for these mouse strains should be submitted to N.D. (ding.ning@gene.com). Investigators may also request these mouse strains from Genentech by submitting a request form at www.gene.com/gene/reagents-program/request.do.
Supplementary Materials
This PDF file includes:
Figs. S1 to S8
Table S1
REFERENCES AND NOTES
- 1.D. S. Kim, H. R. Collard, T. E. King Jr., Classification and natural history of the idiopathic interstitial pneumonias. Proc. Am. Thorac. Soc. 3, 285–292 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.A. Nalbandian, K. Sehgal, A. Gupta, M. V. Madhavan, C. McGroder, J. S. Stevens, J. R. Cook, A. S. Nordvig, D. Shalev, T. S. Sehrawat, N. Ahluwalia, B. Bikdeli, D. Dietz, C. der-Nigoghossian, N. Liyanage-Don, G. F. Rosner, E. J. Bernstein, S. Mohan, A. A. Beckley, D. S. Seres, T. K. Choueiri, N. Uriel, J. C. Ausiello, D. Accili, D. E. Freedberg, M. Baldwin, A. Schwartz, D. Brodie, C. K. Garcia, M. S. V. Elkind, J. M. Connors, J. P. Bilezikian, D. W. Landry, E. Y. Wan, Post-acute COVID-19 syndrome. Nat. Med. 27, 601–615 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.P. M. George, A. U. Wells, R. G. Jenkins, Pulmonary fibrosis and COVID-19: The potential role for antifibrotic therapy. Lancet Respir. Med. 8, 807–815 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.L. Richeldi, H. R. Collard, M. G. Jones, Idiopathic pulmonary fibrosis. Lancet 389, 1941–1952 (2017). [DOI] [PubMed] [Google Scholar]
- 5.G. Raghu, B. Rochwerg, Y. Zhang, C. A. Garcia, A. Azuma, J. Behr, J. L. Brozek, H. R. Collard, W. Cunningham, S. Homma, T. Johkoh, F. J. Martinez, J. Myers, S. L. Protzko, L. Richeldi, D. Rind, M. Selman, A. Theodore, A. U. Wells, H. Hoogsteden, H. J. Schünemann; American Thoracic Society; European Respiratory society; Japanese Respiratory Society; Latin American Thoracic Association , An official ATS/ERS/JRS/ALAT clinical practice guideline: Treatment of idiopathic pulmonary fibrosis. An update of the 2011 clinical practice guideline. Am. J. Respir. Crit. Care Med. 192, e3–e19 (2015). [DOI] [PubMed] [Google Scholar]
- 6.T. A. Wynn, Integrating mechanisms of pulmonary fibrosis. J. Exp. Med. 208, 1339–1350 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.W. R. Coward, G. Saini, G. Jenkins, The pathogenesis of idiopathic pulmonary fibrosis. Ther. Adv. Respir. Dis. 4, 367–388 (2010). [DOI] [PubMed] [Google Scholar]
- 8.N. C. Henderson, F. Rieder, T. A. Wynn, Fibrosis: From mechanisms to medicines. Nature 587, 555–566 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.S. L. Friedman, D. Sheppard, J. S. Duffield, S. Violette, Therapy for fibrotic diseases: Nearing the starting line. Sci. Transl. Med. 5, 167sr161 (2013). [DOI] [PubMed] [Google Scholar]
- 10.K. E. Kadler, Fell Muir Lecture: Collagen fibril formation in vitro and in vivo. Int. J. Exp. Pathol. 98, 4–16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.J. K. Mouw, G. Ou, V. M. Weaver, Extracellular matrix assembly: A multiscale deconstruction. Nat. Rev. Mol. Cell Biol. 15, 771–785 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.D. R. Eyre, M. A. Weis, J. J. Wu, Advances in collagen cross-link analysis. Methods 45, 65–74 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.H. A. Lucero, H. M. Kagan, Lysyl oxidase: An oxidative enzyme and effector of cell function. Cell. Mol. Life Sci. 63, 2304–2316 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.D. R. Eyre, M. J. Glimcher, Collagen cross-linking. Isolation of cross-linked peptides from collagen of chicken bone. Biochem. J. 135, 393–403 (1973). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.M. Yamauchi, M. Sricholpech, Lysine post-translational modifications of collagen. Essays Biochem. 52, 113–133 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.K. E. Kadler, Y. Hojima, D. J. Prockop, Assembly of collagen fibrils de novo by cleavage of the type I pC-collagen with procollagen C-proteinase. Assay of critical concentration demonstrates that collagen self-assembly is a classical example of an entropy-driven process. J. Biol. Chem. 262, 15696–15701 (1987). [PubMed] [Google Scholar]
- 17.E. G. Canty, K. E. Kadler, Procollagen trafficking, processing and fibrillogenesis. J. Cell Sci. 118, 1341–1353 (2005). [DOI] [PubMed] [Google Scholar]
- 18.D. J. Hulmes, Building collagen molecules, fibrils, and suprafibrillar structures. J. Struct. Biol. 137, 2–10 (2002). [DOI] [PubMed] [Google Scholar]
- 19.K. E. Kadler, D. F. Holmes, J. A. Trotter, J. A. Chapman, Collagen fibril formation. Biochem. J. 316, 1–11 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.W. Chen, A. Yang, J. Jia, Y. V. Popov, D. Schuppan, H. You, Lysyl oxidase (LOX) family members: Rationale and their potential as therapeutic targets for liver fibrosis. Hepatology 72, 729–741 (2020). [DOI] [PubMed] [Google Scholar]
- 21.J. M. Mäki, J. Räsänen, H. Tikkanen, R. Sormunen, K. Mäkikallio, K. I. Kivirikko, R. Soininen, Inactivation of the lysyl oxidase gene Lox leads to aortic aneurysms, cardiovascular dysfunction, and perinatal death in mice. Circulation 106, 2503–2509 (2002). [DOI] [PubMed] [Google Scholar]
- 22.I. K. Hornstra, S. Birge, B. Starcher, A. J. Bailey, R. P. Mecham, S. D. Shapiro, Lysyl oxidase is required for vascular and diaphragmatic development in mice. J. Biol. Chem. 278, 14387–14393 (2003). [DOI] [PubMed] [Google Scholar]
- 23.J. M. Mäki, R. Sormunen, S. Lippo, R. Kaarteenaho-Wiik, R. Soininen, J. Myllyharju, Lysyl oxidase is essential for normal development and function of the respiratory system and for the integrity of elastic and collagen fibers in various tissues. Am. J. Pathol. 167, 927–936 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.X. Liu, Y. Zhao, J. Gao, B. Pawlyk, B. Starcher, J. A. Spencer, H. Yanagisawa, J. Zuo, T. Li, Elastic fiber homeostasis requires lysyl oxidase-like 1 protein. Nat. Genet. 36, 178–182 (2004). [DOI] [PubMed] [Google Scholar]
- 25.A. Yang, X. Yan, H. Xu, X. Fan, M. Zhang, T. Huang, W. Li, W. Chen, J. Jia, H. You, Selective depletion of hepatic stellate cells-specific LOXL1 alleviates liver fibrosis. FASEB J. 35, e21918 (2021). [DOI] [PubMed] [Google Scholar]
- 26.A. Yang, X. Yan, X. Fan, Y. Shi, T. Huang, W. Li, W. Chen, J. Jia, H. You, Hepatic stellate cells-specific LOXL1 deficiency abrogates hepatic inflammation, fibrosis, and corrects lipid metabolic abnormalities in non-obese NASH mice. Hepatol. Int. 15, 1122–1135 (2021). [DOI] [PubMed] [Google Scholar]
- 27.W. Zhao, A. Yang, W. Chen, P. Wang, T. Liu, M. Cong, A. Xu, X. Yan, J. Jia, H. You, Inhibition of lysyl oxidase-like 1 (LOXL1) expression arrests liver fibrosis progression in cirrhosis by reducing elastin crosslinking. Biochim. Biophys. Acta Mol. Basis Dis. 1864, 1129–1137 (2018). [DOI] [PubMed] [Google Scholar]
- 28.P. S. Bellay, C. Shimbori, C. Upagupta, S. Sato, W. Shi, J. Gauldie, K. Ask, M. Kolb, Lysyl oxidase-like 1 protein deficiency protects mice from adenoviral transforming growth factor-β1–induced pulmonary fibrosis. Am. J. Respir. Cell Mol. Biol. 58, 461–470 (2018). [DOI] [PubMed] [Google Scholar]
- 29.L. Alsofi, E. Daley, I. Hornstra, E. F. Morgan, Z. D. Mason, J. F. Acevedo, R. A. Word, L. C. Gerstenfeld, P. C. Trackman, Sex-linked skeletal phenotype of lysyl oxidase like-1 mutant mice. Calcif. Tissue Int. 98, 172–185 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.J. Zhang, Z. Liu, T. Zhang, Z. Lin, Z. Li, A. Zhang, X. Sun, J. Gao, Loss of lysyl oxidase-like 3 attenuates embryonic lung development in mice. Sci. Rep. 6, 33856 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.J. Zhang, R. Yang, Z. Liu, C. Hou, W. Zong, A. Zhang, X. Sun, J. Gao, Loss of lysyl oxidase-like 3 causes cleft palate and spinal deformity in mice. Hum. Mol. Genet. 24, 6174–6185 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.A. Martin, F. Salvador, G. Moreno-Bueno, A. Floristán, C. Ruiz-Herguido, E. P. Cuevas, S. Morales, V. Santos, K. Csiszar, P. Dubus, J. J. Haigh, A. Bigas, F. Portillo, A. Cano, Lysyl oxidase-like 2 represses Notch1 expression in the skin to promote squamous cell carcinoma progression. EMBO J. 34, 1090–1109 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.H. Li, J. Guo, Y. Jia, W. Kong, W. Li, LOXL4 abrogation does not exaggerate angiotensin II-induced thoracic or abdominal aortic aneurysm in mice. Genes (Basel) 12, 513 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.P. C. Trackman, Lysyl oxidase isoforms and potential therapeutic opportunities for fibrosis and cancer. Expert Opin. Ther. Targets 20, 935–945 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.V. Barry-Hamilton, R. Spangler, D. Marshall, S. McCauley, H. M. Rodriguez, M. Oyasu, A. Mikels, M. Vaysberg, H. Ghermazien, C. Wai, C. A. Garcia, A. C. Velayo, B. Jorgensen, D. Biermann, D. Tsai, J. Green, S. Zaffryar-Eilot, A. Holzer, S. Ogg, D. Thai, G. Neufeld, P. van Vlasselaer, V. Smith, Allosteric inhibition of lysyl oxidase-like-2 impedes the development of a pathologic microenvironment. Nat. Med. 16, 1009–1017 (2010). [DOI] [PubMed] [Google Scholar]
- 36.A. J. Muir, C. Levy, H. L. A. Janssen, A. J. Montano-Loza, M. L. Shiffman, S. Caldwell, V. Luketic, D. Ding, C. Jia, B. J. McColgan, J. G. McHutchison, G. Mani Subramanian, R. P. Myers, M. Manns, R. Chapman, N. H. Afdhal, Z. Goodman, B. Eksteen, C. L. Bowlus; for the GS‐US‐321‐0102 Investigators , Simtuzumab for primary sclerosing cholangitis: Phase 2 study results with insights on the natural history of the disease. Hepatology 69, 684–698 (2019). [DOI] [PubMed] [Google Scholar]
- 37.S. A. Harrison, M. F. Abdelmalek, S. Caldwell, M. L. Shiffman, A. M. Diehl, R. Ghalib, E. J. Lawitz, D. C. Rockey, R. A. Schall, C. Jia, B. McColgan, J. McHutchison, G. M. Subramanian, R. P. Myers, Z. Younossi, V. Ratziu, A. J. Muir, N. H. Afdhal, Z. Goodman, J. Bosch, A. J. Sanyal; GS-US-321-0105 and GS-US-321-0106 Investigators , Simtuzumab is ineffective for patients with bridging fibrosis or compensated cirrhosis caused by nonalcoholic steatohepatitis. Gastroenterology 155, 1140–1153 (2018). [DOI] [PubMed] [Google Scholar]
- 38.S. Verstovsek, M. R. Savona, R. A. Mesa, H. Dong, J. D. Maltzman, S. Sharma, J. Silverman, S. T. Oh, J. Gotlib, A phase 2 study of simtuzumab in patients with primary, post-polycythaemia vera or post-essential thrombocythaemia myelofibrosis. Br. J. Haematol. 176, 939–949 (2017). [DOI] [PubMed] [Google Scholar]
- 39.G. Raghu, K. K. Brown, H. R. Collard, V. Cottin, K. F. Gibson, R. J. Kaner, D. J. Lederer, F. J. Martinez, P. W. Noble, J. W. Song, A. U. Wells, T. P. M. Whelan, W. Wuyts, E. Moreau, S. D. Patterson, V. Smith, S. Bayly, J. W. Chien, Q. Gong, J. J. Zhang, T. G. O'Riordan, Efficacy of simtuzumab versus placebo in patients with idiopathic pulmonary fibrosis: A randomised, double-blind, controlled, phase 2 trial. Lancet Respir. Med. 5, 22–32 (2017). [DOI] [PubMed] [Google Scholar]
- 40.A. B. Benson III, Z. A. Wainberg, J. R. Hecht, D. Vyushkov, H. Dong, J. Bendell, F. Kudrik, A phase II randomized, double-blind, placebo-controlled study of simtuzumab or placebo in combination with gemcitabine for the first-line treatment of pancreatic adenocarcinoma. Oncologist 22, 241–e15 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.E. G. Meissner, M. McLaughlin, L. Matthews, A. M. Gharib, B. J. Wood, E. Levy, R. Sinkus, K. Virtaneva, D. Sturdevant, C. Martens, S. F. Porcella, Z. D. Goodman, B. Kanwar, R. P. Myers, M. Subramanian, C. Hadigan, H. Masur, D. E. Kleiner, T. Heller, S. Kottilil, J. A. Kovacs, C. G. Morse, Simtuzumab treatment of advanced liver fibrosis in HIV and HCV-infected adults: Results of a 6-month open-label safety trial. Liver Int. 36, 1783–1792 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.M. G. Jones, O. G. Andriotis, J. J. W. Roberts, K. Lunn, V. J. Tear, L. Cao, K. Ask, D. E. Smart, A. Bonfanti, P. Johnson, A. Alzetani, F. Conforti, R. Doherty, C. Y. Lai, B. Johnson, K. N. Bourdakos, S. V. Fletcher, B. G. Marshall, S. Jogai, C. J. Brereton, S. J. Chee, C. H. Ottensmeier, P. Sime, J. Gauldie, M. Kolb, S. Mahajan, A. Fabre, A. Bhaskar, W. Jarolimek, L. Richeldi, K. M. A. O'Reilly, P. D. Monk, P. J. Thurner, D. E. Davies, Nanoscale dysregulation of collagen structure-function disrupts mechano-homeostasis and mediates pulmonary fibrosis. eLife 7, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.I. V. Yang, C. D. Coldren, S. M. Leach, M. A. Seibold, E. Murphy, J. Lin, R. Rosen, A. J. Neidermyer, D. F. McKean, S. D. Groshong, C. Cool, G. P. Cosgrove, D. A. Lynch, K. K. Brown, M. I. Schwarz, T. E. Fingerlin, D. A. Schwartz, Expression of cilium-associated genes defines novel molecular subtypes of idiopathic pulmonary fibrosis. Thorax 68, 1114–1121 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.D. J. DePianto, J. A. V. Heiden, K. B. Morshead, K. H. Sun, Z. Modrusan, G. Teng, P. J. Wolters, J. R. Arron, Molecular mapping of interstitial lung disease reveals a phenotypically distinct senescent basal epithelial cell population. JCI Insight 6, e143626 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.A. Moeller, K. Ask, D. Warburton, J. Gauldie, M. Kolb, The bleomycin animal model: A useful tool to investigate treatment options for idiopathic pulmonary fibrosis? Int. J. Biochem. Cell Biol. 40, 362–382 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.M. L. Decaris, M. Gatmaitan, S. FlorCruz, F. Luo, K. Li, W. E. Holmes, M. K. Hellerstein, S. M. Turner, C. L. Emson, Proteomic analysis of altered extracellular matrix turnover in bleomycin-induced pulmonary fibrosis. Mol. Cell. Proteomics 13, 1741–1752 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.O. Busnadiego, J. González-Santamaría, D. Lagares, J. Guinea-Viniegra, C. Pichol-Thievend, L. Muller, F. Rodríguez-Pascual, LOXL4 is induced by transforming growth factor β1 through Smad and JunB/Fra2 and contributes to vascular matrix remodeling. Mol. Cell. Biol. 33, 2388–2401 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.H. Peinado, M. del Carmen Iglesias-de la Cruz, D. Olmeda, K. Csiszar, K. S. K. Fong, S. Vega, M. A. Nieto, A. Cano, F. Portillo, A molecular role for lysyl oxidase-like 2 enzyme in snail regulation and tumor progression. EMBO J. 24, 3446–3458 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.L. Ma, C. Huang, X. J. Wang, D. E. Xin, L. S. Wang, Q. C. Zou, Y. N. S. Zhang, M. D. Tan, Y. M. Wang, T. C. Zhao, D. Chatterjee, R. A. Altura, C. Wang, Y. S. Xu, J. H. Yang, Y. S. Fan, B. H. Han, J. Si, X. Zhang, J. Cheng, Z. Chang, Y. E. Chin, Lysyl oxidase 3 is a dual-specificity enzyme involved in STAT3 deacetylation and deacetylimination modulation. Mol. Cell 65, 296–309 (2017). [DOI] [PubMed] [Google Scholar]
- 50.J. J. Tomasek, G. Gabbiani, B. Hinz, C. Chaponnier, R. A. Brown, Myofibroblasts and mechano-regulation of connective tissue remodelling. Nat. Rev. Mol. Cell Biol. 3, 349–363 (2002). [DOI] [PubMed] [Google Scholar]
- 51.P. J. Wipff, D. B. Rifkin, J. J. Meister, B. Hinz, Myofibroblast contraction activates latent TGF-β1 from the extracellular matrix. J. Cell Biol. 179, 1311–1323 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.D. J. Tschumperlin, G. Ligresti, M. B. Hilscher, V. H. Shah, Mechanosensing and fibrosis. J. Clin. Invest. 128, 74–84 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.M. Shi, J. Zhu, R. Wang, X. Chen, L. Mi, T. Walz, T. A. Springer, Latent TGF-β structure and activation. Nature 474, 343–349 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.X. He, M. F. Tolosa, T. Zhang, S. K. Goru, L. Ulloa Severino, P. S. Misra, C. M. McEvoy, L. Caldwell, S. G. Szeto, F. Gao, X. Chen, C. Atin, V. Ki, N. Vukosa, C. Hu, J. Zhang, C. Yip, A. Krizova, J. L. Wrana, D. A. Yuen, Myofibroblast YAP/TAZ activation is a key step in organ fibrogenesis. JCI Insight 7, e146243 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.M. M. Mia, M. K. Singh, New insights into Hippo/YAP signaling in fibrotic diseases. Cell 11, (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.F. Liu, J. D. Mih, B. S. Shea, A. T. Kho, A. S. Sharif, A. M. Tager, D. J. Tschumperlin, Feedback amplification of fibrosis through matrix stiffening and COX-2 suppression. J. Cell Biol. 190, 693–706 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.F. Liu, D. Lagares, K. M. Choi, L. Stopfer, A. Marinković, V. Vrbanac, C. K. Probst, S. E. Hiemer, T. H. Sisson, J. C. Horowitz, I. O. Rosas, L. E. Fredenburgh, C. Feghali-Bostwick, X. Varelas, A. M. Tager, D. J. Tschumperlin, Mechanosignaling through YAP and TAZ drives fibroblast activation and fibrosis. Am. J. Physiol. Lung Cell. Mol. Physiol. 308, L344–L357 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Y. Zhou, X. Huang, L. Hecker, D. Kurundkar, A. Kurundkar, H. Liu, T. H. Jin, L. Desai, K. Bernard, V. J. Thannickal, Inhibition of mechanosensitive signaling in myofibroblasts ameliorates experimental pulmonary fibrosis. J. Clin. Invest. 123, 1096–1108 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.A. J. Booth, R. Hadley, A. M. Cornett, A. A. Dreffs, S. A. Matthes, J. L. Tsui, K. Weiss, J. C. Horowitz, V. F. Fiore, T. H. Barker, B. B. Moore, F. J. Martinez, L. E. Niklason, E. S. White, Acellular normal and fibrotic human lung matrices as a culture system for in vitro investigation. Am. J. Respir. Crit. Care Med. 186, 866–876 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.M. W. Parker, D. Rossi, M. Peterson, K. Smith, K. Sikström, E. S. White, J. E. Connett, C. A. Henke, O. Larsson, P. B. Bitterman, Fibrotic extracellular matrix activates a profibrotic positive feedback loop. J. Clin. Invest. 124, 1622–1635 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.M. R. Kuehn, A. Bradley, E. J. Robertson, M. J. Evans, A potential animal-model for Lesch-Nyhan syndrome through introduction of Hprt mutations into mice. Nature 326, 295–298 (1987). [DOI] [PubMed] [Google Scholar]
- 62.O. Smithies, R. G. Gregg, S. S. Boggs, M. A. Koralewski, R. S. Kucherlapati, Insertion of DNA sequences into the human chromosomal β-globin locus by homologous recombination. Nature 317, 230–234 (1985). [DOI] [PubMed] [Google Scholar]
- 63.K. R. Thomas, K. R. Folger, M. R. Capecchi, High frequency targeting of genes to specific sites in the mammalian genome. Cell 44, 419–428 (1986). [DOI] [PubMed] [Google Scholar]
- 64.R. J. Newman, M. Roose-Girma, S. Warming, Efficient conditional knockout targeting vector construction using co-selection BAC recombineering (CoSBR). Nucleic Acids Res. 43, e124 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.M. Gertsenstein, L. M. J. Nutter, T. Reid, M. Pereira, W. L. Stanford, J. Rossant, A. Nagy, Efficient generation of germ line transmitting chimeras from C57BL/6N ES cells by aggregation with outbred host embryos. PLOS ONE 5, e11260 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.J. Seibler, B. Zevnik, B. Küter-Luks, S. Andreas, H. Kern, T. Hennek, A. Rode, C. Heimann, N. Faust, G. Kauselmann, M. Schoor, R. Jaenisch, K. Rajewsky, R. Kühn, F. Schwenk, Rapid generation of inducible mouse mutants. Nucleic Acids Res. 31, e12 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.W. J. Marshall, M. Lapsley, A. P. Day, R. M. Ayling, Clinical Biochemistry: Metabolic and Clinical Aspects, Third Edition (2014).
- 68.N. Ding, H. Zhou, P. O. Esteve, H. G. Chin, S. Kim, X. Xu, S. M. Joseph, M. J. Friez, C. E. Schwartz, S. Pradhan, T. G. Boyer, Mediator links epigenetic silencing of neuronal gene expression with x-linked mental retardation. Mol. Cell 31, 347–359 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S8
Table S1